Abstract
Actinobacillus actinomycetemcomitans, a gram-negative bacterium isolated from the human mouth, has been implicated in the pathogenesis of early-onset periodontitis. Primary isolates cultured from subgingival plaque exhibit an adherent, rough colony phenotype which spontaneously converts to a nonadherent, smooth phenotype upon in vitro subculture. The rough colony variant produces abundant fimbriae and autoaggregates, while the smooth colony variant is planktonic and produces scant fimbriae. To begin to understand the significance of colony variation in biofilm formation by A. actinomycetemcomitans, outer membrane protein profiles of four isogenic rough and smooth colony variants were compared by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Two proteins with relative molecular masses of 43 and 20 kDa were expressed by the rough colony variants exclusively. Expression of these proteins was not found to be dependent on growth phase, oxygen tension, or type of complex medium. N-terminal amino acid sequences of these proteins obtained by Edman degradation were compared with sequences from the University of Oklahoma A. actinomycetemcomitans genome database. Two contiguous open reading frames (ORFs) encoding proteins having sequence homology with these proteins were identified. The 43-kDa protein (RcpA [rough colony protein A]) was similar to precursor protein D of the general secretion pathway of gram-negative bacilli, while the 20-kDa protein (RcpB [rough colony protein B]) appeared to be unique. The genes encoding these proteins have been cloned from A. actinomycetemcomitans 283 and sequenced. A BLASTX (gapped BLAST) search of the surrounding ORFs revealed homology with other fimbria-related proteins. These data suggest that the genes encoding the 43-kDa (rcpA) and 20-kDa (rcpB) proteins may be functionally related to each other and to genes that may encode fimbria-associated proteins.
Actinobacillus actinomycetemcomitans, a gram-negative, capnophilic coccobacillus, is a resident of the indigenous flora of the oral cavity, where it can be recovered from saliva, the tongue, and the buccal mucosa (38, 58, 70). However, its primary ecological niche is the microbial biofilm, subgingival dental plaque (51, 58). A. actinomycetemcomitans has been associated with the pathogenesis of localized juvenile periodontitis, which is a form of early-onset periodontitis, and some forms of refractory adult periodontitis (56–58, 63, 64, 70). A. actinomycetemcomitans has also been associated with various extraoral infections, including endocarditis, pericarditis, brain abscess, meningitis, osteomyelitis, thyroid abscess, and urinary tract infection (7, 30, 37, 40, 42, 54).
An interesting property of A. actinomycetemcomitans is its ability to produce at least two distinct colonial morphologies on solid media in vitro. Typically, when A. actinomycetemcomitans is first isolated from the gingival sulcus, it expresses a rough colony phenotype characterized by a translucent, dull, circular colony with irregular borders that pits the agar; upon further incubation, the pit deepens and the colony embeds into the agar, leaving a central star-shaped or cross-cigar-shaped pattern after the colony is scraped away (24, 43, 46, 52, 59). When rough colonies are inoculated into broth medium, growth appears along the vessel walls as granular, autoaggregating adherent cells, leaving a clear broth. Within a few in vitro subcultures on solid media, however, the colonies convert to a smooth phenotype, characterized by a more opaque, glistening, circular colony with a regular border which does not pit or embed into the agar (24). In broth, the smooth cells grow as a turbid, homogeneous suspension with no adherent cells (24). An intermediate colony variant characterized by a smooth, adherent, nonembedding colony on agar may also be identified. When grown in broth, cells of this phenotype autoaggregate and adhere to the walls of the tube (24).
Multiple colony morphologies are seen in other pathogenic organisms and may be considered a form of phase variation (29). The expression of surface proteins of many gram-negative bacteria, including the opacity protein (OapA) of Haemophilus influenzae, the opacity and fimbrial proteins of Neisseria gonorrhoeae and Neisseria meningitidis, and the type 1 fimbriae of Escherichia coli and other enteric species, are known to phase vary (13, 36, 50, 60, 62, 66). However, unlike the rapid reversal of phenotype typical in other organisms, the colonial morphology shift in A. actinomycetemcomitans has rarely been observed in vitro to change from the smooth to the rough colony phenotype (6, 24, 35, 39, 46, 66). To date, the environmental factors influencing colony phase variation in A. actinomycetemcomitans have not been determined.
Colonial morphology reflects the differential expression of components on surfaces of bacterial cells within the colony (5). Some of these surface components are likely adhesins involved in colonization of the host. A direct correlation has been noted between the rough colony (star-positive) morphology and the presence of fimbriae (24, 46). The ultrastructure of the rough colony variants of A. actinomycetemcomitans demonstrates abundant peritrichous, bundle-forming fimbriae, compared to the smooth colony variants, which show little or no fimbriae but numerous blebs or vesicles (24, 46, 51, 52). Rough colony variants expressing fimbriae adhere to both hydroxyapatite and saliva-coated hydroxyapatite better than smooth colony variants (46). Thus, adhesion of A. actinomycetemcomitans is thought to be mediated, at least in part, by fimbriae. However, there is evidence for other nonfimbrial adhesins contained in the microvesicles and extracellular amorphous material produced by this organism (24, 26, 34, 35, 47). The available evidence suggests that the smooth colony variant is able to invade epithelial cells in vitro better than the rough colony variant (35). Nevertheless, the actual role of each of the variants and surface proteins in the pathogenesis of localized juvenile periodontitis is unknown.
To date, no proteins other than fimbriae have been associated with the rough colony phenotype. We report here the identification of two proteins, of 43 and 20 kDa, from outer membrane preparations unique to the rough colony variants of several strains of A. actinomycetemcomitans. Our goals were to describe the association of the rough colony phenotype and the expression of these proteins, to identify the genes that encode these proteins, and to characterize their relationship to each other.
MATERIALS AND METHODS
Bacterial strains.
Four isogenic rough/smooth pairs of A. actinomycetemcomitans (strains 29, 283, and 361, obtained from C. K. Chen, University of Southern California; strain A26, obtained from J. J. Zambon, State University of New York at Buffalo [SUNYaB]) were studied. Initial isolates of the rough phenotype were obtained from subgingival plaques of periodontal patients cultured in the clinical microbiology laboratories of the University of Southern California or SUNYaB. Two additional established laboratory strains exhibiting only the smooth colony phenotype were also studied: strain SUNYaB 75 (serotype a) and type strain NCTC 9710 (also designated ATCC 33384; serotype c) (71). A. actinomycetemcomitans strains were routinely cultivated from frozen stocks either on tryptic soy blood agar supplemented with 0.5% (wt/vol) yeast extract (Difco), hemin (5 μg/ml), and menadione (0.2 μg/ml) or in Trypticase soy broth (BBL) supplemented with 0.6% (wt/vol) yeast extract (Difco) and 0.04% (wt/vol) NaHCO3 (Baker) (pH 7.2) (TSBY broth) at 37°C in 5% CO2 in air or anaerobically (5% CO2, 10% H2, 85% N2) for 24 to 48 h. For comparison, Todd-Hewitt broth (Difco) with 1.0% (wt/vol) yeast extract and brain heart infusion broth (Difco) with 0.6% (wt/vol) yeast extract were also used. All broth cultures were grown statically.
E. coli TOP10F′ (Invitrogen) or DH5α was cultivated in Luria-Bertani (LB) broth (pH 7.2) with shaking for 16 to 18 h at 37°C with appropriate antibiotic selection. Recombinant clones were identified by blue-white colony selection on LB agar plates containing ampicillin (100 μg/ml; Sigma), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 μg/ml; Sigma), and isopropyl-β-d-thiogalactopyranoside (IPTG; 22 μg/ml; Sigma).
Generation and isolation of isogenic rough/smooth pairs of A. actinomycetemcomitans.
Rough phenotype isolates from frozen stocks were cultured on tryptic soy blood agar plates under anaerobic conditions at 37°C for 24 to 48 h. A single colony exhibiting the typical rough phenotype was subcultured in TSBY broth at 37°C under anaerobic conditions for 24 h. Adherent cells were scraped from the sides of the tube, and 1 ml of culture was transferred to 4 ml of fresh TSBY broth. Several daily subcultures in TSBY broth were performed to achieve conversion from the rough phenotype to a homogeneous, turbid smooth phenotype. Colony morphology of the final broth culture was assessed by inoculating a blood agar plate. Subcultures of both the initial rough colony isolate and its smooth colony isogenic variant were frozen in TSBY broth with 15% (vol/vol) glycerol at −70°C. For each experiment, fresh subcultures were started from these stock cultures to ensure a uniform colony phenotype.
Preparation of outer membrane-enriched fractions.
Outer membrane-enriched fractions were prepared from all four isogenic rough/smooth pairs of A. actinomycetemcomitans by a modification of the method of Barenkamp et al. (2). Briefly, each phenotype was subcultured from frozen stocks onto blood agar and incubated at 37°C anaerobically for 48 h. A single colony was transferred to 5 ml of TSBY broth and incubated at 37°C anaerobically for 24 to 48 h. The 5 ml of the starter culture was then diluted in 45 ml of TSBY broth and incubated at 37°C anaerobically for 24 h, at which time the cells were harvested by centrifugation and processed immediately or frozen at −70°C. Cells were suspended in 10 mM HEPES (pH 7.4) containing protease inhibitor cocktail (Sigma) at a concentration of 25 μl/100 mg (wet weight) of cells. The suspension was sonicated in an ice bath, using a microtip on a Branson 450 Sonifier for 10 cycles of 30-s duration. Cell debris was removed by centrifugation for 10 min at 2,000 × g. The total membrane pellet obtained by ultracentrifugation of the supernatant at 105,000 × g for 1 h (Beckman Ti50 rotor) was then suspended in 1% sodium lauroylsarcosine (Sarkosyl; Sigma) in 10 mM HEPES (pH 7.4) and rocked for 1 h at room temperature. The Sarkosyl-insoluble, outer membrane-enriched fraction was pelleted by ultracentrifugation at 105,000 × g for 1 h to separate it from the Sarkosyl-soluble, inner membrane-enriched fraction. The outer membrane-enriched pellet was suspended in 500 μl of distilled water with an additional 5 μl of protease inhibitor cocktail, quantitated for protein, aliquoted, and stored at −70°C. The inner membrane-enriched fraction was also aliquoted and stored at −70°C. For each experiment, a fresh aliquot was used and not subjected to additional freeze/thaw cycles.
Expression of the 43- and 20-kDa proteins under various conditions.
To determine if anaerobiosis affected the expression of these proteins, the rough colony variant of A. actinomycetemcomitans 283 (283R) was cultured from frozen stock on blood agar at 37°C under anaerobic conditions or in 5% CO2 in air and designated the initial culture. After 48 h, a single colony of the characteristic rough phenotype was subcultured to 5 ml of TSBY broth and incubated for 24 h in the designated atmosphere. A final subculture was made by diluting the 5-ml culture 1:10 in TSBY broth and incubating it under the same atmosphere. The following cultures were compared: initial and final cultures grown exclusively under anaerobic conditions; initial and final cultures grown exclusively in 5% CO2 in air; and initial cultures grown anaerobically and subcultures grown aerobically. Cells from the 50-ml broth cultures were harvested in late log to early stationary phase and stored at −70°C until analyzed.
The effect of culture medium on the expression of these proteins was examined by comparing the outer membrane preparations from cultures grown in TSBY broth, brain heart infusion broth, and Todd-Hewitt broth. A. actinomycetemcomitans 283R was inoculated onto blood agar and incubated at 37°C under anaerobic conditions for 48 h. A single colony of the characteristic rough phenotype was subcultured into 5 ml of each type of broth medium and incubated at 37°C under anaerobic conditions for 24 h. The starter culture was diluted 1:10 in the same medium and incubated under the same conditions. Cells were harvested in late log to early stationary phase and stored at −70°C until use.
To determine if the 43- and 20-kDa proteins were expressed during different phases of growth, frozen stocks of A. actinomycetemcomitans 283R and 283S (smooth colony variant) were inoculated onto blood agar at 37°C and incubated under anaerobic conditions. A single colony representative of each phenotype was inoculated into 5 ml of TSBY broth and incubated overnight. The starter culture was diluted 1:10 in 50 ml of TSBY broth. After an overnight incubation under the same conditions, the cultures were diluted to an optical density at 495 of approximately 0.050 in fresh medium and incubated at 37°C anaerobically. A 50-ml aliquot was removed at 5 h (lag phase), 12 h (log phase), and 24 h (early stationary phase). Cells were harvested at each time point and frozen at −70°C. Outer membrane-enriched fractions were prepared from the cell pellets, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and stained with Coomassie blue or silver.
SDS-PAGE, Western blotting, and N-terminal amino acid sequencing.
Optimal separation of outer membrane preparations was achieved on 12.5% Laemmli gels run under denaturing conditions in a Tall Mighty Small gel apparatus (Hoefer) (27). Preparations were evaluated by using equal protein concentrations estimated by A280, using bovine serum albumin as a standard. Samples were prepared in an equal volume of sample buffer (0.125 M Tris-HCl [pH 6.8], 4% [wt/vol] SDS, 20% [vol/vol] glycerol, 10% [vol/vol] 2-mercaptoethanol) and heated for 5 min at 100°C unless otherwise indicated. The apparent molecular masses of the proteins of interest were determined by comparison with bands of known protein standards. Gels were stained with 0.025% Coomassie brilliant blue R-250 (Bio-Rad), silver (33), or periodic acid-Schiff (PAS) (11). Western blot analysis was performed using alkaline phosphatase for detection as previously described (45).
For N-terminal amino acid sequencing, outer membrane preparations were applied to 9 or 16% denaturing Laemmli gels and transblotted onto Immobilon-PSQ (Millipore) to recover the 43- and 20-kDa proteins, respectively. The blots were stained with fresh 0.1% (wt/vol) Coomassie blue according to the Applied Biosystems protocol. When the blot was dried, the desired bands were excised and sequenced by Edman degradation using an Applied Biosystem peptide sequencer. The N-terminal amino acid sequences obtained were submitted to the University of Oklahoma A. actinomycetemcomitans genome database (63a) for comparison with deduced amino acid sequences of A. actinomycetemcomitans HK1651.
The sensitivity of the outer membrane proteins to proteolytic degradation was determined by diluting the outer membrane preparations with an equal volume of 20 mM Tris-HCl (pH 7.8) containing 1% (wt/vol) SDS and 10 mM EDTA to which proteinase K (Sigma) was added to a concentration of 60 μg/ml in a total reaction volume of 100 μl. Samples were incubated at 37°C for 0 and 2 h and at 65°C for 1 h. Enzymatic activity was terminated by boiling the sample at 100°C for 5 min. Proteins were analyzed by SDS-PAGE as described above.
Polyclonal antibody preparation.
Monospecific polyclonal antibodies to the 43- and 20-kDa proteins of A. actinomycetemcomitans were prepared by injecting 8- to 10-kg New Zealand White rabbits with gel-purified antigen from A. actinomycetemcomitans 283R. To prepare antigen, outer membrane preparations (1 mg/gel) were separated on 9 or 16% Laemmli nondenaturing SDS-polyacrylamide preparative gels to optimally separate the 43- or 20-kDa bands, respectively. Four gels were used for each antigen. After staining with Coomassie brilliant blue, the appropriate band was excised from the gels, macerated in a tissue homogenizer, mixed with an equal volume of TiterMax (CytRx) adjuvant, and sonicated in a 3-ml syringe according to the manufacturer’s instructions to obtain the proper emulsion. Each rabbit was injected with approximately 100 μg of protein in four sites and boosted with the same material 2 weeks later. After 2 additional weeks, the animals were bled for assessment of antibody titer. Sufficient antibody concentration (titer of at least 1:1,000) was obtained after one boost for the 20-kDa antigen. However, due to a low titer of antibody specific for the 43-kDa protein, a third boost 3 weeks later with total outer membrane preparation from A. actinomycetemcomitans 283R was required. The inoculum was prepared by first dialyzing the outer membrane preparation exhaustively against water to reduce the detergent concentration before mixing with TiterMax. Sufficient antibody to the 43-kDa antigen was obtained after this boost.
Considerable background antibodies to A. actinomycetemcomitans outer membrane components were evident in prebleed samples, possibly as a result of natural colonization by Pasteurella species, which are closely related to A. actinomycetemcomitans. Rabbits were chosen for inoculation based on an absence of reactivity to the 43- and 20-kDa components in the prebleed samples. Antisera from final bleeds were extensively adsorbed to remove background antibodies to obtain monospecific antisera. Undiluted rabbit antiserum was sequentially adsorbed with whole cell pellets of the smooth phenotype from A. actinomycetemcomitans 9719 and 283; two different strains were used to better adsorb both strain-specific and cross-reactive antibodies. Final adsorption was done on nitrocellulose discs, previously adsorbed with A. actinomycetemcomitans 283S outer membrane preparation and blocked with 3% skim milk.
PCR, gene cloning, and sequencing of the 43- and 20-kDa protein genes.
There were two overlapping open reading frames (ORFs) in the contig from the A. actinomycetemcomitans database strain HK1651 that could encode the N-terminal amino acid sequence from the 43-kDa protein. One ORF encoded a 50.1-kDa (hereafter designated 50-kDa) protein, while the other encoded a 47.8-kDa protein. Therefore, two forward PCR primers were constructed for the N terminus of the putative gene encoding the 43-kDa protein. The 50-kDa forward primer was 5′-CACGCGGTGTATTCATCA-3′, and the 47.8-kDa forward primer was 5′-TGCTGTGCAGTTCTCGGA-3′. One reverse PCR primer, 5′-ATGCAATGGAGGCCGTAA-3′, was constructed since both overlapping reading frames terminated at the same position (see Fig. 3A and B). The PCR program was as follows: 94°C for 45 s, 65°C for 30 s, and 72°C for 1 min 30 s for 35 cycles. The PCR primer pair 50-forward and 50-reverse was used to amplify the gene from genomic DNA of strain 283R. Similarly, PCR primer pair 47.8-forward and 50-reverse was used to amplify its gene from the same genomic DNA. Forward and reverse PCR primers were also constructed for the 20-kDa protein putative gene sequence based on the nucleotide sequence containing the N-terminal peptides in the same contig of A. actinomycetemcomitans HK1651. The ORF containing the 20-kDa nucleotide sequence was predicted to encode an 18.9-kDa protein, hereafter designated the 20-kDa protein. The 20-forward primer was 5′-ATTCTTCCATACGACTCC-3′, and the 20-reverse primer was 5′-TAGTACGTGCACTGTCTG-3′ (see Fig. 3B and C). The PCR program was as follows: 94°C for 45 s, 54°C for 30 s, and 72°C for 1 min 30 s for 35 cycles. The PCR products were subsequently cloned into the TA cloning vector pCR2.1 (Invitrogen) according to the manufacturer’s protocol and transformed into E. coli host strain TOP10F′ (Invitrogen). Plasmids, prepared either by the alkaline lysis method (4) or with a Wizard SV Plus miniprep kit (Promega), were submitted to the CAMBI Nucleic Acid Sequencing Facility at SUNYaB for sequencing. Nucleotide sequencing was performed for each gene from both strands by the dideoxynucleotide chain termination method (49) using the DyeDeoxy terminator cycle sequencing protocol (Applied Biosystems). Sequences were analyzed with HIBIO DNASIS (Hitachi Software Engineering) and PSORT (43a) software. GenBank homology searches were performed by using BLASTX (gapped BLAST) (1).
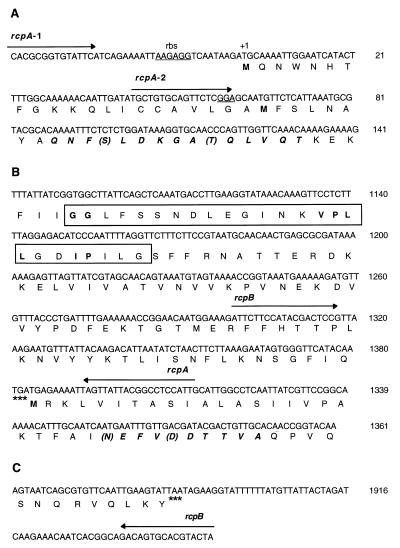
FIG. 3.
Partial nucleotide sequences of the rcpA and rcpB genes from A. actinomycetemcomitans HK1651. (A) Upstream sequence and 5′ terminus of the rcpA gene encoding the 43-kDa protein; (B) 3′ terminus of rcpA and the 5′ terminus of the rcpB gene encoding the 20-kDa protein; (C) 5′ terminus of rcpB and downstream sequence. PCR primers are indicated by arrows according to direction and labeled with the corresponding gene. Potential ribosome-binding sites (rbs; Shine-Dalgarno sequences) are underlined. PCR primer rcpA-1 is the 50-forward primer, and rcpA-2 is the 47.8-forward primer. Amino acid residues corresponding to the N-terminal amino acid sequences (bold italics) and mismatches (parentheses) from the deduced amino acid sequence of the protein purified from A. actinomycetemcomitans 283R are noted. Stop codons are denoted by ∗∗∗; the secretion protein motif, GGX12VP(L/F)LXXIPXIGXL(F/L), is enclosed by a box, with the consensus bases in bold.
PCR, gene cloning, and sequencing of a gene fragment spanning from orfA to orf1.
PCR was used to confirm that the previously identified flp (fimbrial precursor protein) gene and immediately downstream orfA to orfC (23) were contiguous to orf1 to orf5 identified in this study (see Fig. 4). A forward primer was made to the 3′ end of orfA based on the nucleotide sequence from A. actinomycetemcomitans 304-a (5′-GGCGCCATAGTTAGCA-3′), and a reverse primer was made to the 5′ end of orf1 just after the overlap region with orfC (5′-GTCGTAGCCTCTCTTAGG-3′), based on the nucleotide sequence from the database strain HK1651 (see Fig. 4). Genomic DNA from A. actinomycetemcomitans 283R was used as the template. The touchdown PCR program was as follows: 94°C for 1 min, 58°C for 1 min, and 72°C for 1.5 min for four cycles. The annealing temperature was sequentially reduced by two degrees to 48°C; four cycles of amplification were performed at each annealing temperature. The amplified PCR product was cloned into the TA cloning vector pGEM-T (Promega), transformed into E. coli DH5α and sequenced as described above.
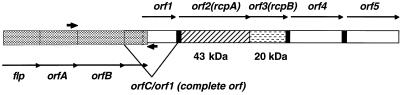
FIG. 4.
Putative fimbrial operon of A. actinomycetemcomitans. The gene fragment containing the flp gene and downstream regions described by Inoue et al. (23) are indicated by 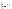 . Immediately downstream is the gene fragment described in this study (orf1 to orf5) containing the rcpA gene encoding the 43-kDa protein and the rcpB gene encoding the 20-kDa protein. Together, orfC and orf1 form a complete ORF with start and stop codons. The vertical broken line demarcates the overlap region between orf1 and orfC. Thin arrows indicate the direction of transcription of all ORFs. Heavy arrows depict the locations of PCR primers used to confirm the continuity of these gene fragments.
. Immediately downstream is the gene fragment described in this study (orf1 to orf5) containing the rcpA gene encoding the 43-kDa protein and the rcpB gene encoding the 20-kDa protein. Together, orfC and orf1 form a complete ORF with start and stop codons. The vertical broken line demarcates the overlap region between orf1 and orfC. Thin arrows indicate the direction of transcription of all ORFs. Heavy arrows depict the locations of PCR primers used to confirm the continuity of these gene fragments.
Southern blotting.
Genomic DNA was prepared by using a Puregene gram-positive bacterial and yeast DNA isolation kit (Gentra Systems). For Southern blots, genomic DNA was digested to completion overnight with BamHI and SalI and electrophoresed on 0.7% (wt/vol) agarose (Life Technologies) gels. Restricted DNA was transferred to Hybond-N membranes (Amersham) by capillary action and cross-linked to the membranes by UV light fixation. PCR-generated DNA probes were labeled with biotin by using the BioPrime system (Life Technologies). Hybridization was done under stringent conditions; for detection of DNA, the Photogene nucleic acid detection system (version 2.0; Life Technologies) was used according to the manufacturer’s protocol.
Nucleotide sequence accession numbers.
The nucleotide sequences of the rcpA and rcpB genes have been deposited in GenBank under accession no. AF139249 and AF139250, respectively.
RESULTS
Variation in colony morphology of A. actinomycetemcomitans.
Four isogenic rough/smooth pairs of A. actinomycetemcomitans (strains 29, 283, A26, and 361) were studied. For each pair, the smooth colony variant was obtained by successive subculture of the rough colony variant in broth medium, although the morphology shift also occurred on solid media after additional subcultures. When adherent cells from TSBY broth were scraped from the sides of the tube and diluted 1:5 in fresh TSBY broth, the broth phenotype gradually changed with each subsequent daily transfer from small autoaggregating clumps of totally adherent cells in a clear broth to large clumps of semiadherent cells in a clear broth, to large clumps of nonadherent cells in a clear broth, to smaller nonadherent clumps in a turbid broth, and finally to no adherent cells in a turbid broth. Subculture of the broth at day 5 to blood agar yielded colonies of both rough and smooth colony morphologies. After an additional five subcultures to TSBY broth, there was no further alteration of the phenotype of the turbid broth, which when plated on agar yielded exclusively large, nonadherent, nonpitting, smooth colonies. No spontaneous conversion of the smooth to the rough colony variant was observed for any of the smooth colony variants studied.
Comparison of outer membrane protein profiles from rough and smooth colony variants.
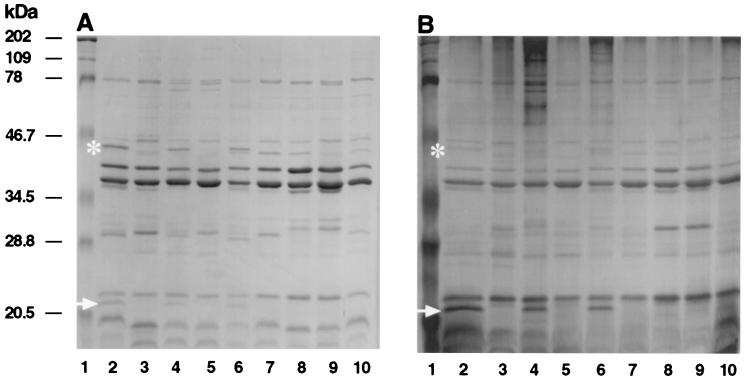
Colonies of each of the paired strains were scraped from TSBY agar plates, suspended in phosphate-buffered saline, and centrifuged to recover cell pellets and supernatants. Cells were also harvested from TSBY broth, suspended in 10 mM HEPES (pH 7.4), sonicated, and centrifuged briefly to remove particulates. The cell pellets, supernatants (both undiluted or concentrated), and cell lysates were examined by SDS-PAGE and stained with Coomassie blue and/or silver. A large number of closely spaced bands were visualized. No definitive differences between phenotypic variants were noted (data not shown). However, when inner and outer membrane-enriched preparations were examined, several consistent differences were noted between the rough and smooth variants of each strain. Components of 43 and 20 kDa present in the rough colony variants were absent from the smooth colony variants on Coomassie blue-stained gels (Fig. 1A). There also appeared to be an increase in the amount of a 30-kDa component in the smooth colony variants. Silver staining enhanced the intensity of the 20-kDa component but decreased the intensity of both the 43-kDa and 30-kDa components (Fig. 1B). PAS staining did not stain any of these components (data not shown). However, some PAS staining was evident below 20 kDa in each of the strains, presumably due to the binding to residual lipopolysaccharide in the preparations; in some rough/smooth pairs, PAS also stained components of between 100 and 200 kDa which may be surface polysaccharides.
FIG. 1.
Comparison of outer membrane protein profiles from rough colonies of A. actinomycetemcomitans 29, 361, and 283 (lanes 2, 4, and 6) and smooth isogenic variants (lanes 3, 5, and 7) and smooth colony variants NCTC 9710, ATCC 33384, and SUNYaB 75 (lanes 8 through 10) separated by SDS-PAGE, and stained with Coomassie blue (A) and silver (B). Asterisks and arrows indicate the 43- and 20-kDa proteins, respectively. Molecular mass markers are in lanes 1.
Initial characterization of the 43- and 20-kDa proteins.
The 43- and 20-kDa components disappeared after treatment with proteinase K (data not shown), suggesting that these components were proteins.
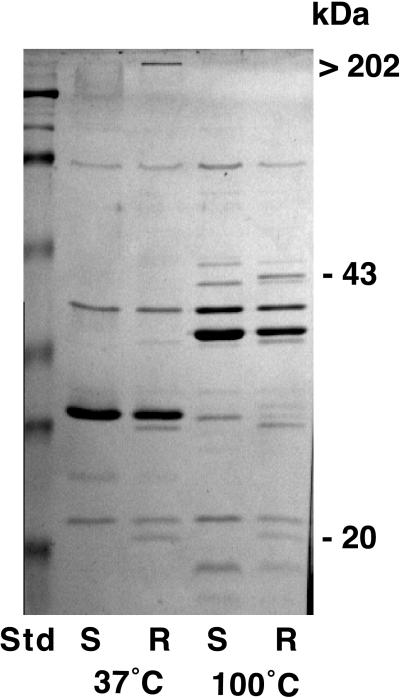
To determine heat modifiability and subunit formation of the proteins, separate samples of outer membrane preparation containing 2-mercaptoethanol in the SDS-PAGE sample buffer were heated for 5 min at 37, 60, 70, 80, and 100°C. The 43-kDa protein did not appear until 70°C, while the 20-kDa band was present at all temperatures tested (data not shown). Comparison of outer membrane preparation samples from rough and smooth colony variants of strain 283 heated to 37 and 100°C showed the presence of a high-molecular-weight band on a Coomassie brilliant blue-stained gel that barely entered the separating gel, only in the rough colony variant and only at 37°C. This band disappeared, and the 43-kDa band appeared after heating at 100°C (Fig. 2). The same banding pattern was noted with samples suspended in buffer without 2-mercaptoethanol heated to 37 and 100°C.
FIG. 2.
Effect of heat on mobility of the 43- and 20-kDa proteins separated by SDS-PAGE and stained with Coomassie blue. A. actinomycetemcomitans 283 smooth (S) and rough (R) colony variants in sample buffer were heated to 37°C and 100°C, as indicated. Molecular mass markers (Std) are as indicated in Fig. 1.
Immunoblots of these samples probed with the anti-43-kDa antibody showed reactivity with the 43-kDa band only in rough colony variants heated to 100°C and no reactivity with the 20-kDa protein (data not shown). Several attempts were made to transfer the high-molecular-weight component to either nylon or nitrocellulose membrane, using various transfer times, amperage, and buffer conditions. When transblots were stained with 0.1% Coomassie blue to verify protein transfer, the high-molecular-weight component was not visible even though the 200-kDa standard was clearly evident. Also, the high-molecular-weight component was still present in the stained gel after transblotting. Therefore, no conclusive statement can be made regarding the reactivity of the anti-43-kDa protein antibody with the high-molecular-weight band. Similarly, immunoblots of the same samples probed with the anti-20-kDa protein antibody showed reactivity only with the 20-kDa component of the rough colony variants under all sample conditions tested (data not shown).
Expression of the 43- and 20-kDa proteins under various environmental conditions. (i) Oxygen tension and protein expression.
Previously published studies suggested that the number of fimbriae may be elevated when A. actinomycetemcomitans is grown under anaerobic conditions (52). If either the 43-kDa or 20-kDa protein was related to fimbriae, perhaps its expression would be increased by anaerobiosis. To determine if anaerobiosis affected the production of these proteins, outer membrane-enriched fractions from rough phenotype cells initially grown anaerobically and shifted to 5% CO2 in air were compared with those of cells grown exclusively either anaerobically or in 5% CO2 in air. Qualitatively, as observed on Coomassie blue-stained SDS-polyacrylamide gels, both 43- and 20-kDa proteins were present in outer membrane preparations under all conditions tested (data not shown). These data suggest that rough phenotype organisms are able to produce both proteins irrespective of the atmosphere used.
(ii) Culture medium.
The 43- and 20-kDa proteins were produced from the rough colony variants cultured in all of the three media tested. Growth in defined medium was not attempted, as previous reports described unsuccessful efforts to grow this organism in such medium (53, 61). Therefore, based on results with the complex media used in these experiments, the culture medium does not appear to influence the gross expression of these proteins.
(iii) Growth phase.
Both 43- and 20-kDa proteins were present at each time point selected, representing the lag, exponential, and stationary phases. Expression of these proteins did not appear to be dependent on growth.
N-terminal amino acid sequence of the 43- and 20-kDa proteins from A. actinomycetemcomitans 283R.
The N-terminal amino acid sequence of the 43-kDa protein was determined to be QNFKLDKGAIQLVQT (Fig. 3A). This sequence was compared to the sequence provided by the University of Oklahoma A. actinomycetemcomitans genome database. A significant match of the protein sequence (86% identity and 86% similarity) was localized to a single contig. Two overlapping ORFs within the contig in the −1 frame contained this sequence encoding proteins of 50.1 and 47.8 kDa (Fig. 3A). A potential ribosome-binding site (Shine-Dalgarno sequence), AAGAGG, was located 9 bases upstream from the start codon (ATG) of the ORF encoding the 50-kDa protein (55). Another potential ribosome-binding site (GGA) was located 4 bases upstream of the 47.8-kDa start codon. A gapped BLAST (BLASTX) database search of this ORF revealed 30% identity and 50% similarity to the general secretory protein D precursor (22, 44) and 50% similarity to proteins related to type 4 fimbriae of several gram-negative bacteria (20, 22, 44). The product of the ORF encoding the 50-kDa protein was predicted by the PSORT program to be an outer membrane protein with a signal sequence of 29 amino acids. The 3′ terminus of this predicted signal sequence adjoined the 5′ end of the N-terminal amino acid sequence of the gel-purified protein.
Submission of the N-terminal amino acid sequence of the 20-kDa protein, SEFVNDTTVA, to the A. actinomycetemcomitans genome database revealed a match of the protein sequence with an ORF having 80% identity and 100% similarity on the same contig as the 43-kDa protein gene sequence (Fig. 3B). The deduced amino acid sequence encoded a protein of 18.9 kDa. Interestingly, this ORF was contiguous with the 43-kDa protein putative gene sequence described above. In fact, the termination codon of the 43-kDa protein putative gene sequence contained the start codon of the 20-kDa protein putative gene sequence in the −3 frame (Fig. 3B). No discernible Shine-Dalgarno sequence was evident upstream of the 20-kDa start codon. Analysis of the ORF by the PSORT program predicted that this was either a periplasmic or outer membrane protein with a signal sequence of 23 amino acids. The N terminus of this predicted sequence was very close to the N-terminal amino acid sequence of the purified protein. A potential glycosylation site of NDTT was contained within the N-terminal amino acid sequence, although this protein did not stain with the carbohydrate stain PAS. The greatly enhanced visualization of this protein with silver stain suggests that this protein may have associated carbohydrate moieties.
PCR, cloning, and sequencing of the 43- and 20-kDa proteins.
The PCR primer pair constructed for the ORF encoding a 50-kDa protein (Fig. 3A and B) and used to amplify the gene from genomic DNA of A. actinomycetemcomitans 283R yielded a 1.4-kb product. Similarly, the PCR primer pair for the ORF encoding a 47.8-kDa protein used to amplify its gene from the same genomic DNA yielded a 1.3-kb product. Also, forward and reverse PCR primers were constructed for the 20-kDa putative gene (Fig. 3B and C). This PCR primer pair yielded a 648-bp product. These PCR products were subsequently cloned into the TA cloning vector pCR2.1, transformed into E. coli, and sequenced from the plasmid. The nucleotide sequence of the 1.4-kb insert was 97% identical, and that of the 648-bp insert was 93% identical, to the sequence from the A. actinomycetemcomitans HK1651 genome database.
PCR, cloning, and sequencing of a gene fragment spanning orfA to orf1.
A PCR primer pair was constructed to amplify the gene fragment spanning from orfA, a gene fragment previously identified by Inoue et al. (23), to orf1 identified in the present study to determine their continuity. Using a forward primer derived from orfA and a reverse primer based on orf1, a 770-bp fragment was amplified from genomic DNA of A. actinomycetemcomitans 283R (Fig. 4). This PCR product was cloned into the TA cloning vector pGEM-T, transformed into E. coli, and sequenced from the plasmid. The nucleotide sequence of the insert from strain 283R was 93% identical to the sequence from database strain HK1651.
Presence of the 43- and 20-kDa protein genes in other strains of A. actinomycetemcomitans.
Genomic DNAs from A. actinomycetemcomitans A26S, 283R, 283S, and 361S were digested overnight with BamHI, XhoI, SalI, BamHI/SalI, and XhoI/SalI. Considering the A. actinomycetemcomitans database sequence of the 43- and 20-kDa proteins, these enzymes were predicted not to cut within the gene(s) of interest; SalI cuts upstream of the gene encoding the 50-kDa protein (within orf1), whereas BamHI and XhoI cut near one another downstream of the gene encoding the 20-kDa protein (within orf4). Upon Southern blotting, the biotinylated 1.4-bp PCR product from the 50-kDa protein hybridized to a single 4.1-kb band produced by BamHI/SalI digest as expected in all rough and smooth strains tested (data not shown). However, the XhoI/SalI digest produced a single band ranging from 7.2 to 10.1 kb, depending on the strain used. The BamHI/SalI digests confirm the conserved gene size among the strains. Also, the gene apparently is present in only one copy in the genomes of all A. actinomycetemcomitans strains tested. The XhoI site present in the strain sequenced by the genome project is evidently absent from the strains in our collection, but this finding awaits confirmation.
Similarly, when the same Southern blot was probed with the 648-bp PCR product from the 20-forward/20-reverse primer pair, it hybridized to a single band which varied from 4.1 to 4.3 kb in the BamHI/SalI digest of both the rough and smooth colony variants of all the strains tested (data not shown). This finding also confirms the conserved size of this gene among different strains and that it is present in single copy in the A. actinomycetemcomitans chromosome. The A. actinomycetemcomitans database was searched with the complete deduced amino acid sequences for both the 43-kDa and 20-kDa proteins. Each complete sequence was present in only one contig, confirming that the genes are present as a single copy in the genome.
DISCUSSION
In this study, two proteins of 43 and 20 kDa specific for the rough colony phenotype of A. actinomycetemcomitans have been identified in outer membrane preparations. The expression of these proteins, which were found to be immunogenic in rabbits, was not dependent on growth phase or type of complex culture medium. These proteins are produced under both anaerobic and aerobic (5% CO2 in air) conditions. However, Northern blot analysis of RNA will need to be done to determine if there are quantitative differences in expression. No differences in molecular mass of the 20-kDa protein were noted on silver-stained gels in samples heated to 37 or 100°C or in buffer with or without 2-mercaptoethanol. However, the 43-kDa protein did not appear in gels until samples were heated to at least 70°C. When samples from rough phenotypes were heated to lower temperatures, a high-molecular-weight component appeared to just enter the gel. Heat did not appear to alter the migration of other components with the exception of the previously described 29-kDa heat-modifiable protein (69). Immunoblots of outer membrane proteins probed with anti-43-kDa or anti-20-kDa protein polyclonal antibodies showed reactivity only with the homologous components. These results suggest that both 43- and 20-kDa proteins are monomeric. However, the possibility of structural relationship between the 43-kDa protein and the high-molecular-weight component remains undetermined since there was incomplete transfer of high-molecular-weight components from the gel to the membrane in Western blotting.
The rough colony (star-positive) phenotype has been associated with the presence of abundant fimbriae (24, 46, 52). In previous studies (23), fimbrial preparations from A. actinomycetemcomitans contained both 54- and 6.5-kDa proteins when analyzed by SDS-PAGE and Coomassie blue staining. The 54-kDa protein, a product of the fup gene, is thought to be a fimbria-associated protein (15, 24, 25). The 6.5-kDa protein, encoded by the flp gene, is proposed to be the putative fimbrial subunit (23). The first 20 amino acids of the mature Flp protein are similar to N. gonorrhoeae type 4 fimbriae (23). Downstream of the flp gene are three potential ORFs (orfA, orfB, and orfC) (GenBank accession no. AB005741) (23) (Fig. 4). orfA encodes a protein with no significant homology to fimbrial proteins; orfB encodes a protein similar to Bacillus subtilis type 4 prepilin-like protein specific leader peptidase; and orfC, while truncated at the 3′ end, encodes a protein resembling TraB, which is involved in F-pilus assembly in E. coli. All three ORFs are transcribed in the same orientation as the flp gene (23).
Our sequence data suggest that the genes encoding the 43- and 20-kDa proteins adjoin each other on the A. actinomycetemcomitans chromosome and are two of five predicted major ORFs (orf1 to orf5), all transcribed in the same orientation as orfA, orfB, and orfC (Fig. 4). In fact, there is a region of overlap between the A. actinomycetemcomitans genomic fragment sequenced by Inoue et al. (23) and the genes identified in this study. orf1 from our study (truncated at 5′ end, 232 amino acids) contains 141 bases from the truncated 3′ end of orfC described by Inoue et al. (Fig. 4) (23). The nucleotide sequence of this overlap region from the A. actinomycetemcomitans database strain HK1651 is 99% identical, and the deduced amino acid sequence is 97% identical, to that of A. actinomycetemcomitans 304-a used in the study by Inoue et al. (23). We have confirmed that flp and orfA to orfC are immediately upstream of orf1 to orf5 by PCR, cloning, and sequencing of a fragment amplified by using primers from the 3′ end of orfA and 5′ end of orf1 just after the overlap region (Fig. 4). The deduced amino acid sequence from the C terminus of Orf1 also shares 30% identity to Ag43, the bipartite “fluffing protein” of E. coli K-12 (8, 17, 41). The exact function of this protein is unknown, but it is associated with rough/smooth colony morphology and autoaggregation in E. coli (17, 18).
orf2 (460 amino acids), which we have designated here rcpA (rough colony protein A), encodes the 43-kDa protein, which has similarity (50%) to protein D of the general secretion pathway (GspD) in several gram-negative organisms (48), and to PilQ (53%), an assembly protein of type 4 fimbriae in both Pseudomonas aeruginosa and N. gonorrhoeae (9, 31). The general secretory pathway is used to transport a variety of macromolecules across the outer membrane, including type 4 fimbriae (14, 22). Bacterial homologues within the GspD protein family have been proposed to function as a gatekeepers by interacting with substrates bound for export through the outer membrane (32). The 43-kDa protein shares with these homologues the secretion protein motif (consensus sequence GGX12VP[L/F]LXXIPXIGXL[F/L]) located near the C terminus (Fig. 3B) (14, 21, 31). The homologue in N. gonorrhoeae, PilQ (renamed from Omc), is proposed to function in the terminal steps of organelle biogenesis by acting as a pilus channel or pore (9).
orf3 (167 amino acids), which we have designated here rcpB (rough colony protein B), starts within the stop codon of rcpA on a different reading frame and encodes the 20-kDa protein, which appears to be unique. orf4 (374 amino acids) also appears to encode a unique protein. orf5 (352 amino acids) encodes a protein with similarity to TrbB (47%) in E. coli and Enterobacter aerogenes (a conjugal transfer protein), protein E (45%) of the general secretion pathway of various gram-negative bacilli, PilB (50%) of P. aeruginosa and TapB (47%) of Aeromonas sp. (fimbrial assembly proteins), and PilT (42%) of N. gonorrhoeae (a twitching motility protein associated with type 4 pili). Near the C terminus of Orf5 and each of these proteins is a Walker A box motif (consensus sequence GXXGXGK[T/S]), a putative ATP-binding region found in many nucleotide-binding proteins (65). Further, Orf5 appears to lack a signal sequence and is predicted to be located in the cytoplasm, based on PSORT protein analysis.
Several pathogenic bacteria, including N. gonorrhoeae and P. aeruginosa, possess type 4 fimbriae, which are typically bundle forming and polar and are associated with twitching motility (19, 28, 67). While A. actinomycetemcomitans fimbriae frequently form bundles, they appear to be peritrichous (24, 46, 52), and twitching motility has not yet been described for this bacterium. However, the similarity of the protein sequence of the purported fimbrial subunit, Flp, and several of the adjacent ORFs with type 4 fimbria assembly proteins (3, 28, 67, 68) suggests the presence of similar fimbriae in A. actinomycetemcomitans. Thus, as proposed for other organisms, the conserved regions of these ORFs likely indicate similarities in the function of the encoded proteins, including protein secretion, DNA transfer, and assembly of fimbriae (20).
The biogenesis of fimbriae involves many genes, including those that encode the major subunit(s), minor components, proteins required for biogenesis and assembly, and regulatory proteins (22, 29). Based on its similarity to the GspD family of proteins, especially PilQ, its location in the outer membrane, and its association with the fimbriated rough colony variant of A. actinomycetemcomitans, the 43-kDa protein may function in fimbrial assembly and expression. The contiguous arrangement of rcpA and rcpB on the A. actinomycetemcomitans chromosome suggests that the transcription and function of RcpA are likely coupled to those of RcpB. Similarly, the GspD proteins often require a second protein to function (10, 12, 16). RNA analysis and mutagenesis studies of these genes are in progress. Mutants deficient in RcpA and RcpB will be tested in functional assays to help elucidate relationship to each other and their role in adhesion and colonization. These studies should lead to a better understanding of the role each colony phenotype plays in the pathogenesis of A. actinomycetemcomitans-mediated disease.
ACKNOWLEDGMENTS
We thank Howard K. Kuramitsu for critical review of the manuscript and Jeffrey D. Rogers for critical review of the manuscript and assistance in preparation of the figures.
This study was supported in part by grant DE 09838 (F.A.S.) and postdoctoral training grant DE 07034 (E.M.H.) from the National Institute of Dental Research.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barenkamp S J, Munson R S, Jr, Granoff D M. Subtyping isolates of Haemophilus influenzae type b by outer-membrane protein profiles. J Infect Dis. 1981;143:668–676. doi: 10.1093/infdis/143.5.668. [DOI] [PubMed] [Google Scholar]
- 3.Bieber D, Ramer S W, Wu C-Y, Murray W H, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 4.Birnboim H D, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1522. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun W. Bacterial genetics. W. B. Philadelphia, Pa: Saunders Co.; 1965. [Google Scholar]
- 6.Brinton J J., Jr Non-flagellar appendages of bacteria. Nature. 1959;183:782–786. doi: 10.1038/183782a0. [DOI] [PubMed] [Google Scholar]
- 7.Burgher L W, Loomis G W, Ware F. Systemic infection due to Actinobacillus actinomycetemcomitans. Am J Clin Pathol. 1973;60:412–415. doi: 10.1093/ajcp/60.3.412. [DOI] [PubMed] [Google Scholar]
- 8.Diderichsen B. flu, a metastable gene controlling surface properties of Escherichia coli. J Bacteriol. 1980;141:858–867. doi: 10.1128/jb.141.2.858-867.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drake S L, Koomey M. The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol Microbiol. 1995;18:975–986. doi: 10.1111/j.1365-2958.1995.18050975.x. [DOI] [PubMed] [Google Scholar]
- 10.Drake S L, Sandstedt S A, Koomey M. PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass-multimer. Mol Microbiol. 1997;23:657–668. doi: 10.1046/j.1365-2958.1997.2511618.x. [DOI] [PubMed] [Google Scholar]
- 11.Fairbanks G, Steck T L, Wallach D F H. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971;10:2606. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- 12.Filloux A, Bally M, Ball G, Akrim M, Tommassen J, Lazdunski A. Protein secretion in gram-negative bacteria; transport across the outer membrane involves common mechanisms in different bacteria. EMBO J. 1990;9:4323–4329. doi: 10.1002/j.1460-2075.1990.tb07881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gally D L, Bogan J A, Eisenstein B I, Blomfield I C. Environmental regulation of the Fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J Bacteriol. 1993;175:6186–6193. doi: 10.1128/jb.175.19.6186-6193.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genin S, Boucher C A. A superfamily of proteins involved in different secretion pathways in gram-negative bacteria: modular structure and specificity of the N-terminal domain. Mol Gen Genet. 1994;243:112–118. doi: 10.1007/BF00283883. [DOI] [PubMed] [Google Scholar]
- 15.Harano K, Yamanaka A, Okuda K. An antiserum to a synthetic fimbrial peptide of Actinobacillus actinomycetemcomitans blocked adhesion of the microorganism. FEMS Microb Lett. 1995;130:279–286. doi: 10.1111/j.1574-6968.1995.tb07732.x. [DOI] [PubMed] [Google Scholar]
- 16.Hardie K R, Lory S, Pugsley A P. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 1996;15:978–988. [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson I R, Meehan M, Owen P. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol Lett. 1997;149:115–120. doi: 10.1111/j.1574-6968.1997.tb10317.x. [DOI] [PubMed] [Google Scholar]
- 18.Henderson I R, Meehan M, Owen P. A novel regulatory mechanism for a novel phase-variable outer membrane protein. In: Paul P S, Francis D H, Benfield D A, editors. Mechanisms in the pathogenesis of enteric diseases. New York, N.Y: Plenum Press; 1997. pp. 349–355. [Google Scholar]
- 19.Henrichsen J. Twitching motility. Annu Rev Microbiol. 1983;37:81–93. doi: 10.1146/annurev.mi.37.100183.000501. [DOI] [PubMed] [Google Scholar]
- 20.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 21.Huang J-C, He S Y, Bauer D W, Collmer A. The Pseudomonas syringae pr. syringae 61 hrpH product, an envelope protein required for elicitation of the hypersensitive response in plants. J Bacteriol. 1992;174:6878–6885. doi: 10.1128/jb.174.21.6878-6885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hultgren S J, Jones C H, Normark S. Bacterial adhesins and their assembly. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2730–2756. [Google Scholar]
- 23.Inoue T, Tanimoto I, Ohta H, Kato K, Murayama Y, Kazuhiro F. Molecular characterization of low-molecular-weight component protein, Flp, in Actinobacillus actinomycetemcomitans fimbriae. Microbiol Immunol. 1998;42:253–258. doi: 10.1111/j.1348-0421.1998.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 24.Inouye T, Ohta H, Kokeguchi S, Fukui K, Kato K. Colonial variation and fimbriation of Actinobacillus actinomycetemcomitans. FEMS Microbiol Lett. 1990;69:13–18. doi: 10.1016/0378-1097(90)90405-f. [DOI] [PubMed] [Google Scholar]
- 25.Ishihara K, Honma K, Miura T, Kato T, Okuda K. Cloning and sequence analysis of the fimbriae associated protein (fap) gene from Actinobacillus actinomycetemcomitans. Microb Pathog. 1997;23:63–69. doi: 10.1006/mpat.1997.0137. [DOI] [PubMed] [Google Scholar]
- 26.Kagermeier A S, London J. Actinobacillus actinomycetemcomitans strains Y4 and N27 adhere to hydroxyapatite by distinctive mechanisms. Infect Immun. 1985;47:654–658. doi: 10.1128/iai.47.3.654-658.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lauer P, Albertson N H, Koomey M. Conservation of genes encoding components of a type IV pilus assembly/two-step protein export pathway in Neisseria gonorrhoeae. Mol Microbiol. 1993;8:357–368. doi: 10.1111/j.1365-2958.1993.tb01579.x. [DOI] [PubMed] [Google Scholar]
- 29.Low D, Braaten B, van der Woude M. Fimbriae. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 146–157. [Google Scholar]
- 30.Martin B F, Derby B M, Budzilovich G N, Ransohoff J. Brain abscesses due to Actinobacillus actinomycetemcomitans. Neurology. 1967;17:833–837. doi: 10.1212/wnl.17.9.833. [DOI] [PubMed] [Google Scholar]
- 31.Martin P R, Hobbs M, Free P D, Jeske Y, Mattick J S. Characterization of pilQ, a new gene required for the biogenesis of type 4 fimbriae in Pseudomonas aeruginosa. Mol Microbiol. 1993;9:857–868. doi: 10.1111/j.1365-2958.1993.tb01744.x. [DOI] [PubMed] [Google Scholar]
- 32.Martinez A, Ostrovsky P, Nunn D N. Identification of an additional member of the secretin superfamily of proteins in Pseudomonas aeruginosa that is able to function in type II protein secretion. Mol Microbiol. 1998;28:1235–1246. doi: 10.1046/j.1365-2958.1998.00888.x. [DOI] [PubMed] [Google Scholar]
- 33.Merril C R, Goldman D, Van Keuren M L. Simplified silver protein detection and image enhancement methods in polyacrylamide gels. Electrophoresis. 1982;3:17–23. [Google Scholar]
- 34.Meyer D H, Fives-Taylor P M. Evidence that extracellular components function in adherence of Actinobacillus actinomycetemcomitans to epithelial cells. Infect Immun. 1993;61:4933–4936. doi: 10.1128/iai.61.11.4933-4936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer D H, Screenivasan P D, Fives-Taylor P M. Evidence for invasion of a human oral cell line by Actinobacillus actinomycetemcomitans. Infect Immun. 1991;59:2719–2726. doi: 10.1128/iai.59.8.2719-2726.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mims C A. The pathogenesis of infectious disease. New York, N.Y: Academic Press; 1976. [Google Scholar]
- 37.Muhle I, Rau J, Ruskin J. Vertebral osteomyelitis due to Actinobacillus actinomycetemcomitans. JAMA. 1979;241:1824–1825. [PubMed] [Google Scholar]
- 38.Muller H-P, Lange D E, Muller R F. Actinobacillus actinomycetemcomitans recovery from extracrevicular locations of the mouth. Oral Microbiol Immunol. 1993;6:344–348. doi: 10.1111/j.1399-302x.1993.tb00609.x. [DOI] [PubMed] [Google Scholar]
- 39.Old D C, Corneil I, Gibson L F, Thomson A D, Duguid J P. Fimbriation, pellicle formation and the amount of growth of salmonellas in broth. J Gen Microbiol. 1968;51:1–16. doi: 10.1099/00221287-51-1-1. [DOI] [PubMed] [Google Scholar]
- 40.Overholt B F. Actinobacillus actinomycetemcomitans endocarditis. Arch Intern Med. 1966;117:99–102. [PubMed] [Google Scholar]
- 41.Owen P, Caffrey P, Josefson L-G. Identification and partial characterization of a novel bipartite protein antigen associated with the outer membrane of Escherichia coli. J Bacteriol. 1987;169:3770–3777. doi: 10.1128/jb.169.8.3770-3777.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Page M I, King E O. Infection due to Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. N Engl J Med. 1966;275:181–188. doi: 10.1056/NEJM196607282750403. [DOI] [PubMed] [Google Scholar]
- 43.Preus H R, Namork E, Olsen I. Fimbriation of Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 1988;3:93–94. doi: 10.1111/j.1399-302x.1988.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 43a.PSORT WWW Server. 12 June 1998, posting date. PSORT software. [Online.] http://psort.nibb.ac.jp/. [14 April 1999, last date accessed.]
- 44.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers J D, Haase E M, Brown A E, Douglas C W I, Gwynn J P, Scannapieco F A. Identification and analysis of a gene (abpA) encoding a major amylase-binding protein in Streptococcus gordonii. Microbiology. 1998;144:1223–1233. doi: 10.1099/00221287-144-5-1223. [DOI] [PubMed] [Google Scholar]
- 46.Rosan B, Slots J, Lamont R J, Listgarten M A, Nelson G M. Actinobacillus actinomycetemcomitans fimbriae. Oral Microbiol Immunol. 1988;3:58–63. doi: 10.1111/j.1399-302x.1988.tb00082.x. [DOI] [PubMed] [Google Scholar]
- 47.Rosan B, Slots J, Lamont R J, Nelson G M, Listgarten M. Fimbriae of Actinobacillus actinomycetemcomitans. J Dent Res. 1988;76:179. doi: 10.1111/j.1399-302x.1988.tb00082.x. [DOI] [PubMed] [Google Scholar]
- 48.Russel M. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J Mol Biol. 1998;279:485–499. doi: 10.1006/jmbi.1998.1791. [DOI] [PubMed] [Google Scholar]
- 49.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarkarki J, Pandit N, Moxon E R, Achtman M. Variable expression of the Opc outer membrane protein in Neisseria meningitidis is caused by size variation of a promoter containing poly-cytidine. Mol Microbiol. 1994;13:207–217. doi: 10.1111/j.1365-2958.1994.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 51.Scannapieco F A, Kornman K S, Coykendall A L. Observation of fimbriae and flagella in dispersed subgingival dental plaque and fresh bacterial isolates from periodontal disease. J Periodontal Res. 1983;18:620–633. doi: 10.1111/j.1600-0765.1983.tb00399.x. [DOI] [PubMed] [Google Scholar]
- 52.Scannapieco F A, Millar S J, Reynolds H S, Zambon J J, Levine M J. Effect of anaerobiosis on the surface ultrastructure and surface proteins of Actinobacillus actinomycetemcomitans (Haemophilus actinomycetemcomitans) Infect Immun. 1987;55:2320–2323. doi: 10.1128/iai.55.9.2320-2323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Screenivasan P K, Meyer D H, Fives-Taylor P M. Factors influencing the growth and viability of Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 1993;8:361–369. doi: 10.1111/j.1399-302x.1993.tb00612.x. [DOI] [PubMed] [Google Scholar]
- 54.Serra P, Tonato M. Subacute bacterial endocarditis due to Actinobacillus actinomycetemcomitans. Am J Med. 1969;47:809–812. doi: 10.1016/0002-9343(69)90174-0. [DOI] [PubMed] [Google Scholar]
- 55.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slots J. The predominant cultivable organisms in juvenile periodontitis. Scand J Dent Res. 1976;84:1–10. doi: 10.1111/j.1600-0722.1976.tb00454.x. [DOI] [PubMed] [Google Scholar]
- 57.Slots J, Dahlen G. Subgingival microorganisms and bacterial virulence factors in periodontitis. Scand J Dent Res. 1985;93:119–127. doi: 10.1111/j.1600-0722.1985.tb01319.x. [DOI] [PubMed] [Google Scholar]
- 58.Slots J, Reynolds H S, Genco R J. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect Immun. 1980;29:1013–1020. doi: 10.1128/iai.29.3.1013-1020.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Slots J, Taichman N S. Actinobacillus actinomycetemcomitans. In: Newman M G, Nisengard R, editors. Oral microbiology and immunology. W. B. Philadelphia, Pa: Saunders Co.; 1988. pp. 251–256. [Google Scholar]
- 60.Snellings N J, Tall B D, Venkatesan M M. Characterization of Shigella type 1 fimbriae: expression, FimA sequence, and phase variation. Infect Immun. 1997;65:2462–2467. doi: 10.1128/iai.65.6.2462-2467.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spitznagel J J, Kraig E, Kolodrubetz D. The regulation of leukotoxin production in Actinobacillus actinomycetemcomitans strain JP2. Adv Dent Res. 1995;9:48–54. doi: 10.1177/08959374950090010901. [DOI] [PubMed] [Google Scholar]
- 62.Stern A, Brown M, Nickel P, Meyer T F. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell. 1986;47:61–71. doi: 10.1016/0092-8674(86)90366-1. [DOI] [PubMed] [Google Scholar]
- 63.Tanner A C R, Haffer C, Brathall G T, Visconti R A, Socransky S S. A study of the bacteria associated with advancing periodontitis in man. J Clin Periodontol. 1979;6:278–307. doi: 10.1111/j.1600-051x.1979.tb01931.x. [DOI] [PubMed] [Google Scholar]
- 63a.University of Oklahoma. 18 February, 1998, posting date. A. actinomycetemcomitans genome database. [Online.] www.genome.ou.edu/act.html. [13 april 1999, last date accessed.]
- 64.van Winkelhoff A J, Rodenburg J P, Goene R J, Abbas F, Winkel E G, de Graaff J. Metronidazole plus amoxycillin in the treatment of Actinobacillus actinomycetemcomitans associated periodontitis. J Clin Periodontol. 1989;16:128–131. doi: 10.1111/j.1600-051x.1989.tb01626.x. [DOI] [PubMed] [Google Scholar]
- 65.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;8:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Weiser J N, Chong S T H, Greenberg D, Fong W. Identification and characterization of a cell envelope protein of Haemophilus influenzae contributing to phase variation in colony opacity and nasopharyngeal colonization. Mol Microbiol. 1995;17:555–564. doi: 10.1111/j.1365-2958.1995.mmi_17030555.x. [DOI] [PubMed] [Google Scholar]
- 67.Whitchurch C B, Hobbs M, Livingston S P, Krisnapillai V, Mattick J S. Characterization of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialized protein export system widespread in eubacteria. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- 68.Whitchurch C B, Mattick J S. Escherichia coli contains a set of gene homologous to those involved in protein secretion, DNA uptake and the assembly of type-4 fimbriae in other bacteria. Gene. 1994;150:9–15. doi: 10.1016/0378-1119(94)90851-6. [DOI] [PubMed] [Google Scholar]
- 69.Wilson M E. The heat-modifiable outer membrane protein of Actinobacillus actinomycetemcomitans: relationship to OmpA proteins. Infect Immun. 1991;59:2505–2507. doi: 10.1128/iai.59.7.2505-2507.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zambon J J. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 71.Zambon J J, Slots J, Genco R J. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect Immun. 1983;41:19–27. doi: 10.1128/iai.41.1.19-27.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]