Abstract
Coccidioidomycosis is a fungal infection that is endemic in the southwestern United States. Infection is more severe in blacks and Filipinos, which suggests that there is a genetic basis for susceptibility to this infection in humans. We found that there is also a difference in resistance to Coccidioides immitis infection among inbred mouse strains: B6 mice are susceptible, while DBA/2 mice are resistant (T. N. Kirkland and J. Fierer, Infect. Immun. 40:912–916, 1983). In this paper we report the results of our efforts to map the genes responsible for resistance to this infection in mice. Mice were infected by intraperitoneal inoculation, and 15 days later the numbers of viable fungi in their lungs and spleens were enumerated. We also determined the amounts of interleukin-10 mRNA made in the infected lungs. These three phenotypes were mapped as quantitative traits by using the 26 available lines of recombinant inbred mice derived from a cross between B6 and DBA/2 mice. The best associations were those between the regions near the Lv locus on chromosome 4 and the Tnfr1 locus on chromosome 6. We then infected backcross mice [(B6 × DBA/2) × B6] and confirmed these associations; 14 of 16 (87%) mice that were heterozygous at both Lv and Tnfr1 were resistant to infection, whereas only 4 of 16 (25%) mice that were homozygous B6 at both loci were resistant. These are the first genetic loci to be associated with susceptibility to C. immitis, but there may be additional genes involved in murine resistance to this infection.
Coccidioidomycosis is one of the mycoses endemic in the United States. The fungus Coccidioides immitis grows as a mold in the desert soil in the southwestern states, and infection is acquired by inhalation of arthroconidia (8). In the lung the fungus converts to the spherule form, which is a pathognomonic structure (4, 25). The majority of infected people develop delayed hypersensitivity manifested by a positive skin test, and they recover spontaneously (24). However, 5 to 10% of infections are not self-limited, and in many of those patients, infections disseminate to extrapulmonary sites (4). Patients with disseminated infections do not develop a positive skin test but do make high titers of antibody (16, 23). Thus, it is likely that patients who recover spontaneously mount Th1 immune responses, whereas the others mount Th2 responses.
Although the risk of progressive (disseminated) infection is relatively small, it is not equal in all populations. Patients with T-lymphocyte deficiencies are more susceptible (1, 9). Among previously healthy people, Filipinos and blacks have 5- to 10-fold higher rates of disseminated coccidioidomycosis than do Caucasians (3, 6, 15, 22). Even in military populations, where the risk of exposure and access to medical care are similar for all groups, blacks and Filipinos have higher rates of disseminated coccidioidomycosis (6, 15, 17). This suggests that there is a genetic basis for susceptibility to this infection in humans.
In order to try to find an animal model that could be used to study the genetics of resistance to C. immitis, we infected several inbred strains of mice (11). We found that DBA/2 mice are more than 1,000-fold more resistant to intraperitoneal (i.p.) infection with C. immitis than B6 and BALB/c mice, and that resistance is the dominant phenotype. These results were confirmed by Cox et al., who infected mice by the respiratory route (2). We recently showed that susceptible strains of mice make higher levels of interleukin-10 (IL-10) and that IL-10-deficient mice are resistant to infection (5). IL-4-deficient mice are also more resistant, but less so than the IL-10-deficient mice. Magee and Cox also demonstrated that IL-4 impairs the ability of BALB/c mice to resist infection (13).
In this paper we have started to map the genes in mice that determine resistance to i.p. infection with C. immitis and influence the IL-10 response to infection. We have used a set of recombinant inbred (RI) mice that were derived from a cross between B6 (susceptible) and DBA/2 (resistant) mice (B×D). Each line of RI mice is completely inbred and is a result of recombination between the two progenitor strains at the F2 generation, with 18 generations of subsequent inbreeding fixing each locus. Thus, each RI line is a patchwork of the two progenitors, and at every genetic locus an RI line has either the B6 or the DBA/2 allele. More than 1,000 loci have been mapped in each RI line. However, since the progenitor strains are genetically related to varying degrees, not all alleles will be polymorphic in any given RI set. The expression of a phenotype (such as resistance to an infection) can be determined in the different RI lines and then compared to the strain distribution pattern of all known genetic loci in order to look for evidence of linkage.
MATERIALS AND METHODS
Mice.
Mice were purchased from Jackson Laboratories, Bar Harbor, Maine, and housed four to a cage with free access to food and water. The Animal Research Committee at the Veterans Administration (VA) approved the experiments. After the mice were infected, they were housed in an isolator with HEPA-filtered air. Female mice 7 to 12 weeks of age were used in all experiments.
Infection.
Mice were infected i.p. with 500 to 800 CFU of the RS strain of C. immitis as described previously (5, 27). Mice were sacrificed 15 days later (unless otherwise stated), and their spleens and left lungs were removed for quantitative culture on Mycosel agar. The right lungs were used to extract RNA for cytokine analysis, as previously described. There were 10 RI mice in each group, and at least 4 B6 and 4 DBA/2 mice were used as controls for each experiment. The experiment was repeated if the infection was not sufficiently severe in the B6 mice (one or more deaths). The DBA/2 mice were used as the standard for resistance to infection.
Genetic analysis. (i) B×D.
The median number of CFU per lung was calculated for each B×D line by performing quantitative cultures of the homogenized lungs. This value was used to determine linkages with marker loci in the B×D RI lines by using Map Manager QT (14). We defined resistance to infection as a function of the number of CFU per lung and performed quantitative trait locus (QTL) mapping, using Map Manager QTb23. This program does both simple regression, to find marker loci which are significantly associated with the quantitative trait, and interval mapping, to identify locations among mapped marker loci which are candidates for the position of a QTL. The program calculates a likelihood ratio statistic for the interval mapping, producing a chi-square statistic at each genomic position. True genomewide P values were also computed based on 1,000 random permutations of the phenotypes, with the maximum logarithm of the odds favoring linkage (LOD) recorded for each permutation.
(ii) Backcross.
(B6 × DBA/2)F1 mice were bred from parents obtained from Jackson Laboratories, and at the age of 8 weeks they were mated with B6 mice. Female backcross offspring were infected at the age of 6 to 8 weeks. Each experiment included 10 to 15 backcross mice and 6 females of the F1 and B6 parental strains as controls. The B6 mice were used to ensure that the inoculum was adequate in each experiment. Because of experiment-to-experiment variation in the inoculum, the definition of resistance was normalized to the F1 controls. Backcross mice were classified as resistant if the number of CFU per lung was no greater than the highest value found in an F1 (resistant) control in that experiment. DNA was prepared from the tail of each animal and used for genotyping. We purchased PCR primers from Genetics Research (Huntsville, Ala.) to amplify D4Mit142, a polymorphic SSLP that maps to within 1 centimorgan (cM) of the Lv gene. We synthesized PCR primers according to published sequences to amplify the polymorphic region of the Tnfr1 gene (26). In both cases, the PCR products were run on 4% agarose gels, and the two alleles were distinguished by size. Each mouse was classified as homozygous B6 or heterozygous at each locus. The distribution of genotypes was compared by the Fisher exact test, and we ran a test for a trend in the proportion susceptible to infection as a function of the number of B6 alleles (7).
RESULTS
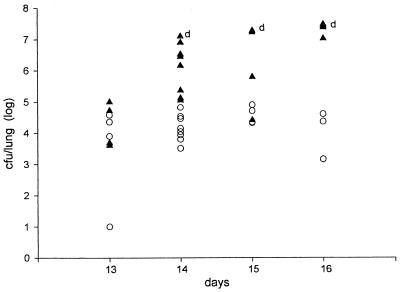
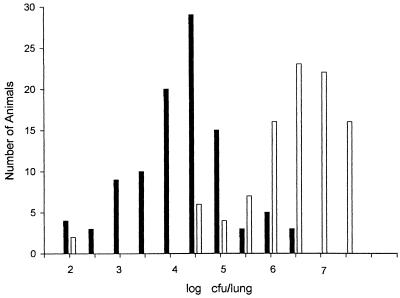
We first compared the time courses of infection in B6 and DBA/2 mice that were infected i.p. with ∼500 CFU of arthroconidia (Fig. 1). There was no evidence of infection in the lungs of either strain from days 10 to 12 after infection, and then the disease progressed rapidly in B6 mice. B6 mice began to die on day 14 after infection, and only one survived to day 16, whereas all DBA/2 mice survived to day 16. Therefore, for subsequent experiments we chose day 15 as the time of sacrifice, assuming that this would be the point of maximum infection in surviving B6 mice, when we would find the largest difference in numbers of CFU per lung between susceptible and resistant mice. On day 15 after infection the median number of CFU per lung was >100 times higher in B6 mice than in DBA/2 mice. However, there was some overlap between the two groups, and the spread of values within each group was >2 log units (Fig. 2). This variability was seen both within experiments and between experiments.
FIG. 1.
Comparison of the time courses of C. immitis infection in resistant DBA/2 mice (open circles) and susceptible B6 mice (solid triangles), as measured by quantitative colony counts in their lungs. Each point represents a single mouse. Dead mice were assigned 107 CFU/lung, the number recovered from the most heavily infected live mouse. The dead mice are indicated with a “d.” The one remaining B6 mouse died on day 16 and was necropsied immediately.
FIG. 2.
Distribution of values for CFU per lung on day 15 after infection with ∼500 arthroconidia injected i.p. Open bars, values for DBA/2 mice; solid bars, values for B6 mice. For simplicity, mice with colony counts between the 0.5 log markers were grouped together into the next category.
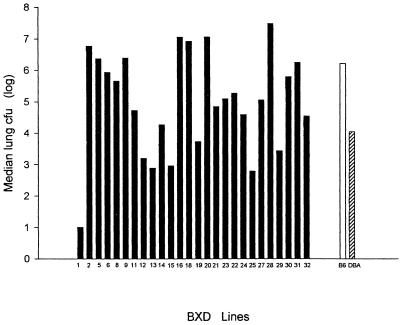
In Fig. 3 we show the median numbers of CFU per lung for the 26 RI lines and the two progenitor strains. Four B×D lines (lines 16, 18, 19, and 28) had median numbers of CFU per lung that were >10 times the number of CFU found in the susceptible B6 parent, and these strains also had higher mortality rates (data not shown). In five B×D lines the median numbers of CFU per lung were <10% of the median number in the resistant DBA/2 strain. Thus, ∼25% of the B×D lines were more resistant and 25% were more susceptible than either parental strain, which suggests that resistance to C. immitis is a polygenic trait and may be due to two unlinked loci, and that the B6 strain carries one or more genes that contribute to resistance. The same distribution was seen among B×D strains when we analyzed the numbers of CFU per spleen (data not shown). Therefore, we analyzed these phenotypes as quantitative or continuous traits. We used Map Manager QTb23 to find genetic loci that were associated with these phenotypes in B×D lines. We identified the two loci that had the highest associations with both phenotypes (CFU per lung and CFU per spleen): Tnfr1 (p55) on chromosome 6 and Lv (aminolevulinate dehydratase) (10) on chromosome 4 (Table 1). Using the geometric mean rather than the median values for CFU per lung or per spleen to define the QTL phenotypes did not appreciably change the results. We previously used the designation Cms for the gene that is responsible for resistance to C. immitis (12), so we called the resistance loci on chromosomes 4 and 6 Cms1 and Cms2, respectively. The permutation-based P values for these associations are not as striking because they are true genomewide values.
FIG. 3.
Variation in severity of C. immitis infection among 26 B×D RI lines. Each point is the median value for CFU per lung for each RI line. There were at least 10 mice in each group, except for B×D group 13, which had only 4 mice because of limited availability of this line. Values for the control B6 and DBA/2 mice are shown for comparison.
TABLE 1.
Linkage between resistance phenotypes and genetic loci in B×D micea
| Locus | CFU/lungb
|
CFU/spleenb
|
IL-10 mRNAc
|
|||
|---|---|---|---|---|---|---|
| LOD | P valued | LOD | P valued | LOD | P valued | |
| Tnfr1 | 3.06 | 0.00018 (0.13) | 2.8 | 0.00033 (0.62) | 2.41 | 0.00087 (0.45) |
| Lv | 2.6 | 0.00050 (0.15) | 2.0 | 0.00289 (0.80) | 2.06 | 0.00204 (0.37) |
Determined as described in Materials and Methods.
Median value on day 15 after infection.
Molecules per microgram of RNA in the lung.
The permutation-based P value is given in parentheses.
The associations of these two loci with the susceptibility phenotype were not strong enough to exclude the possibility of a false-positive association (type 1 error). However, the number of CFU per lung, the number of CFU per spleen, and the IL-10 mRNA level all mapped to the same regions on chromosomes 4 and 6. This encouraged us to conduct further experiments to try to confirm these linkages.
Because resistance to C. immitis is a dominant phenotype (11), we backcrossed (B6 × DBA/2)F1 × B6 mice and tested all the progeny for resistance to C. immitis. As shown in Table 2, 14 of 16 mice that were heterozygous at both loci were resistant to infection, while only 4 of 16 that were homozygous B6 at both loci were resistant (P = 0.0012 by the Fisher exact test). Note that about 50% of the mice that were heterozygous at either locus were resistant. A chi-square test for a linear trend in the table (with singly heterozygous mice combined) was highly significant (Χ2 = 12.52; P = 0.0004) (7).
TABLE 2.
Relationship between resistance to infection and the genotype of Tnfr1 and Lv in (B6 × DBA/2) × B6 backcross mice
| Phenotypea | No. of mice with the following genotypesb (Tnfr1/Lv):
|
|||
|---|---|---|---|---|
| BB/BB | BB/BD | BD/BB | BD/BD | |
| Susceptible | 12 | 9 | 13 | 2 |
| Resistant | 4 | 4 | 17 | 14 |
Individual backcross mice were classified as resistant if the number of CFU/lung was no higher than the highest value for the resistant (DBA/2 × B6)F1 controls in each individual experiment.
B, B6; D, DBA/2.
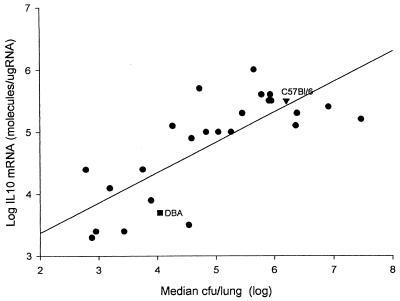
Because we know that susceptible strains of inbred mice make high levels of IL-10 after infection and that IL-10 knockout (KO) mice are resistant to C. immitis (5), we reasoned that the genetic control over infection was at least in part due to genetic controls on the IL-10 response to coccidioidomycosis. As shown in Fig. 4, there was a strong correlation (r = 0.77) between the median number of C. immitis CFU per lung and the amount of IL-10 mRNA in the lung. Therefore, it was not unexpected that when we mapped the IL-10 response as a QTL, we found linkages with the same two loci near Tnfr1 and Lv (Table 1).
FIG. 4.
Correlation between median values for CFU and IL-10 mRNA levels in 26 B×D RI lines. IL-10 mRNA was measured by a quantitative reverse transcription-PCR assay, pooling equal amounts of RNA from four mice in each group. Each point is a single RI line (r = 0.77).
DISCUSSION
We have begun mapping the genes responsible for genetic resistance to C. immitis in mice. When RI lines were first conceived of, it was with the idea of using them to analyze complex phenotypes (19). Initially we used the B×D RI set for mapping, which has both advantages and disadvantages. The advantages are that 26 B×D lines are available from Jackson Laboratories, many genetic markers have already been mapped in this set of mice, and the data set can be analyzed with available statistical programs. The limitation of the B×D set is that it contains only 26 RI lines, which means that one is testing only 26 individual crosses. As a result, it is difficult to map a multigenic trait with a high degree of assurance. The more genes that contribute to a phenotype, the more subjects are needed to accurately map the QTL (19).
Another advantage of RI lines is that the mice are inbred, and so multiple genetically identical subjects can be tested. This allowed us to assign a phenotype to the RI lines (median CFU per lung or per spleen 15 days after i.p. infection) more precisely than would have been possible for individual mice. This was particularly important for analyzing resistance to this infection, because, as shown in Fig. 2, there was even variability in the number of CFU per lung in the two parental strains. When there is overlap in the distribution of the phenotype (number of CFU per lung in this case), it becomes more difficult to analyze QTL. Some of the variability in colony counts could have been due to the biology of replication of the fungus. In tissue, C. immitis grows as spherules that mature to contain thousands of endospores. A single intact spherule will give rise to one colony when cultured in vitro, as will each endospore. Therefore, the rupture of a spherule to release endospores can raise colony counts substantially. This probably also accounts for the rapidity with which the infection progresses in susceptible B6 mice (Fig. 1). Because the growth of the fungus is so dynamic, it is possible that small variations in the inoculum could result in large variations in colony counts 15 days after infection, which could also account for some of the variability. We also recognize that we are mapping the genes that determine the numbers of fungi in the lungs and spleens after i.p. infection, which may not always be the same as the susceptibility to infection measured in other tissues, or susceptibility to respiratory infection.
Despite these difficulties with mapping in the B×D lines, we identified a region on chromosome 6 near Tnfr1 and another on chromosome 4 near Lv that were linked in B×D mice to resistance to infection and to IL-10 response to infection with C. immitis. The LOD scores for these linkages were not high enough to exclude the possibility that these were chance associations, but the same two loci were identified for each of the three phenotypes. We then confirmed those linkages by infecting (B6 × DBA/2)F1 × B6 backcross mice. Eighty-seven percent (14 of 16) of the mice that were heterozygous at both Tnfr1r and Lv were resistant to infection, whereas only 25% (4 of 16) of the mice that were B6 at both loci were resistant. Mice that were heterozygous at only one of the two loci showed intermediate resistance, a result that is consistent with the additive effect of the two loci. Taken together, we believe we have good evidence that there are genes on chromosomes 4 and 6 that influence resistance to coccidioidomycosis.
We have recently reported that DBA/2 mice make less IL-10 in response to infection with C. immitis than do three susceptible inbred strains (B6, BALB/c, and CAST/Ei) (5). We also showed that IL-10 KO mice are resistant to C. immitis. In this study we showed that in the 26 B×D lines there was a strong correlation between the amount of IL-10 mRNA made in the lungs and the number of fungi growing in the lungs on day 15 after infection (Fig. 4). These results with the RI lines give more weight to the association between IL-10 production and susceptibility to infection.
It has long been recognized that there is great variability in the severity of coccidioidomycosis, ranging from asymptomatic infection to overwhelming disease (4, 8). Such variation could be due to differences in the dose or the virulence of a pathogen, prior immunity, or intercurrent illnesses that affect resistance to infection, but in many cases there is no apparent explanation for the variability in the severity of the disease. It is often assumed that some of the variability is due to genetic differences in the hosts, but few genes, that make humans or animals susceptible to specific pathogens have been identified (20).
The two loci that we have identified are almost certainly not the only genes involved in determining resistance to C. immitis. BALB/c and B6 mice are both highly susceptible to C. immitis, and the F1 generation from these two strains is also susceptible, so they do not complement each other’s defects (11). However, in separate experiments we found that (CAST/Ei × B6)F1 mice were more resistant to C. immitis than either parent strain (they complement each other), evidence that B6 mice have at least one beneficial allele (5a). This is reminiscent of the genetics of resistance to salmonella infections in mice, in which the Nramp1 gene plays a dominant role (21). However, Sebastiani et al. recently reported that B6 mice, which are susceptible because they carry a mutant Nramp1, have at least two Salmonella resistance genes that were detected because B6 mice complemented another susceptible strain (18). A similar mapping strategy may reveal additional genes in B6 mice for resistance to C. immitis.
ACKNOWLEDGMENTS
This work was supported in part by the VA Research Service and NIH PO1AI37232, and by NIH P30CA16508 and RO1gm58934 (F. Wright).
REFERENCES
- 1.Ampel N M, Dols C L, Galgiani J N. Coccidioidomycosis during human immunodeficiency virus infection: results of a prospective study in a coccidioidal endemic area. Am J Med. 1993;94:235–240. doi: 10.1016/0002-9343(93)90054-s. [DOI] [PubMed] [Google Scholar]
- 2.Cox R A, Kennell W, Boncyk L, Murphy J W. Induction and expression of cell-mediated immune responses in inbred mice infected with Coccidioides immitis. Infect Immun. 1988;56:13–17. doi: 10.1128/iai.56.1.13-17.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox R A, Vivas J R, Gross A, Lecara G, Miller E, Brummer E. In vivo and in vitro cell-mediated responses in coccidioidomycosis. I. Immunologic responses of persons with primary, asymptomatic infections. Am Rev Respir Dis. 1976;114:937–943. doi: 10.1164/arrd.1976.114.5.937. [DOI] [PubMed] [Google Scholar]
- 4.Drutz D J, Catanzaro A. Coccidioidomycosis. Part I. Am Rev Respir Dis. 1978;117:559–583. doi: 10.1164/arrd.1978.117.3.559. [DOI] [PubMed] [Google Scholar]
- 5.Fierer J, Walls L, Eckmann L, Yamamoto T, Kirkland T N. Importance of interleukin-10 in genetic susceptibility of mice to Coccidioides immitis. Infect Immun. 1998;66:4397–4402. doi: 10.1128/iai.66.9.4397-4402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Fierer, J. Unpublished data.
- 6.Fischl M A, Dickinson G M, Sinave C, Pitchenik A E, Cleary T J. Salmonella bacteremia as manifestation of acquired immunodeficiency syndrome. Arch Intern Med. 1986;146:113–115. [PubMed] [Google Scholar]
- 7.Fleiss J L. Statistical methods for rates and proportions. New York, N.Y: Wiley; 1981. [Google Scholar]
- 8.Galgiani J N. Coccidioidomycosis. West J Med. 1993;159:153–171. [PMC free article] [PubMed] [Google Scholar]
- 9.Hall K A, Sethi G K, Rosado L J, Martinez J D, Huston C L, Copeland J G. Coccidioidomycosis and heart transplantation. J Heart Lung Transplant. 1993;12:525–526. [PubMed] [Google Scholar]
- 10.Hutton J J, Coleman D L. Linkage analyses using biochemical variants in mice. II. Levulinate dehydratase and autosomal glucose 6-phosphate dehydrogenase. Biochem Genet. 1969;3:517–523. doi: 10.1007/BF00485612. [DOI] [PubMed] [Google Scholar]
- 11.Kirkland T N, Fierer J. Inbred mouse strains differ in resistance to lethal Coccidioides immitis infection. Infect Immun. 1983;40:912–916. doi: 10.1128/iai.40.3.912-916.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirkland T N, Fierer J. Genetic control of resistance to Coccidioides immitis: a single gene that is expressed in spleen cells determines resistance. J Immunol. 1985;135:548–552. [PubMed] [Google Scholar]
- 13.Magee D M, Cox R A. Roles of gamma interferon and interleukin-4 in genetically determined resistance to Coccidioides immitis. Infect Immun. 1995;63:3514–3519. doi: 10.1128/iai.63.9.3514-3519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manley K F. A Macintosh program for storage and analysis of experimental genetic mapping data. Mamm Genome. 1993;4:303–313. doi: 10.1007/BF00357089. [DOI] [PubMed] [Google Scholar]
- 15.Pappagianis D. Epidemiology of coccidioidomycosis. In: McGinnis M R, editor. Current topics in medical mycology. New York, N.Y: Springer-Verlag; 1988. pp. 199–238. [DOI] [PubMed] [Google Scholar]
- 16.Pappagianis D, Zimmer B L. Serology of coccidioidomycosis. Clin Microbiol Rev. 1990;3:247–268. doi: 10.1128/cmr.3.3.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rush W L, Dooley D P, Blatt S P, Drehner D M. Coccidioidomycosis: a persistent threat to deployed populations. Aviat Space Environ Med. 1993;64:653–657. [PubMed] [Google Scholar]
- 18.Sebastiani G, Olien L, Gauthier S, Skamene E, Morgan K, Gros P, Malo D. Mapping of genetic modulators of natural resistance to infection with Salmonella typhimurium in wild-derived mice. Genomics. 1998;47:180–186. doi: 10.1006/geno.1997.5116. [DOI] [PubMed] [Google Scholar]
- 19.Silver L M. Classical linkage analysis and mapping panels. In: Silver L M, editor. Mouse genetics. Concepts and applications. New York, N.Y: Oxford University Press; 1995. pp. 195–263. [Google Scholar]
- 20.Skamene E. Genetic regulation of host resistance to bacterial infection. Rev Infect Dis. 1983;5(Suppl. 4):S823–S831. doi: 10.1093/clinids/5.supplement_4.s823. [DOI] [PubMed] [Google Scholar]
- 21.Skamene E, Gros P, Forget A, Kongshavn P A L, St. Charles C, Taylor B A. Genetic regulation of resistance to intracellular pathogens. Nature. 1982;297:506–510. doi: 10.1038/297506a0. [DOI] [PubMed] [Google Scholar]
- 22.Smith C E, Beard R R, Whiting E G, Rosenberger H G. Varieties of coccidioidal infection in relation to the epidemiology and control of the diseases. Am J Public Health. 1946;36:1394–1402. doi: 10.2105/ajph.36.12.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith C E, Saito M T, Simons S A. Pattern of 39,500 serologic tests in coccidioidomycosis. JAMA. 1956;160:546–552. doi: 10.1001/jama.1956.02960420026008. [DOI] [PubMed] [Google Scholar]
- 24.Stevens D A, Levine H B, TenEyck D R. Dermal sensitivity to different doses of spherulin and coccidioidin. Chest. 1974;65:530–533. doi: 10.1378/chest.65.5.530. [DOI] [PubMed] [Google Scholar]
- 25.Sun S H, Sekhon S S, Huppert M. Electron microscopic studies of saprobic and parasitic forms of Coccidioides immitis. Sabouraudia. 1979;17:265–273. doi: 10.1080/00362177985380391. [DOI] [PubMed] [Google Scholar]
- 26.Takao S, Mykytyn K, Jacob C O. Novel DNA polymorphism in the mouse tumor necrosis factor receptors type 1 and type 2. Immunogenetics. 1993;37:199–203. doi: 10.1007/BF00191885. [DOI] [PubMed] [Google Scholar]
- 27.Walch H A, Kalvoda A. Immunization of mice with induced mutants of Coccidioides immitis. I. Characterization of mutants and preliminary studies of their use as viable vaccines. Sabouraudia. 1971;9:173–184. [PubMed] [Google Scholar]