Abstract
Background
Esophageal cancer (EC) is one of the most lethal cancers. Esophageal squamous cell carcinoma (ESCC) is the most common histological subtype in Asian people. Diverse microRNAs, such as miR-375, have been confirmed to be involved in the process of tumorigenesis and metastasis. However, the underlying mechanism through which miR-375 acts in ESCC patients remains unknown.
Methods
We used The Cancer Genome Atlas (TCGA) database to analyze the association between miR-375 and the survival rate in patients with esophageal squamous cell carcinoma. Real Time quantitative PCR (RT-qPCR) analysis was performed to evaluate the level of miR-375 in EC tissues and cells. A luciferase reporter assay was used to confirm the target gene of miR-375. A colony formation assay as well as flow cytometric and transwell invasion experiments were employed to examine the effects of miR-375 and peroxiredoxin 1 (PRDX1) on ESCC cells. A tumor xenograft mouse model was then used to investigate the role of miR-375 on tumor growth in vivo. Moreover, we performed rescue experiments to evaluate the effect of PRDX1 on ESCC progression.
Results
miR-375 expression was significantly downregulated in both ESCC clinical tissues and serum, and the reduction of miR-375 was remarkably linked to a poor prognosis in ESCC. Further investigation illustrated that aberrant expression of miR-375 dampened the growth and infiltration of ESCC cells both in vitro and in vivo. Bioinformatics and luciferase reporter analysis verified that the transcript of PRDX1 is a direct target of miR-375 and its expression in ESCC cells was found to be inversely modulated by miR-375. Moreover, the tumor formation experiment in nude mice confirmed that miR-375 can effectively dampen tumor growth in xenograft tumor mice models. Notably, over-expression of PRDX1 effectively counteracted the tumor-suppressing capabilities of miR-375.
Conclusions
We demonstrated the antitumor effect of miR-375 on ESCC by targeting PRDX1 both in vitro and in vivo.
Keywords: miR-375, esophageal squamous cell carcinoma (ESCC), peroxiredoxin 1 (PRDX1), tumor suppressor
Introduction
Esophageal cancer (EC) is the seventh most prevalent malignancy and the sixth leading cause of cancer mortality globally (1). Esophageal squamous cell carcinoma (ESCC) is the most common pathogenic type of EC, particularly in the high-incidence countries of East Asia, including China, where it accounts for almost 90% of all EC cases (2). At present, despite significant breakthroughs in diagnostic and treatment technology, the prognosis and outcome in ESCC remain dismal, with a 5-year overall survival rate of less than 30% for advanced stages (3). Hence, elucidating the molecular basis that accounts for the progress and prognosis of ESCC is pivotal for developing novel treatment approaches to enhance survival rates.
MicroRNAs (miRNAs) are a family of non-coding RNAs that are approximately 22 nucleotides in length and play critical roles in gene regulation at the post-transcriptional level in both plants and animals (4,5). Translational repression or message RNA (mRNA) degradation can occur as a result of the docking of the RNA-induced silencing complex (RISC) to complementary sites in the 3' untranslated region of target mRNA guided by miRNAs (6). miRNAs are involved in cell proliferation, apoptosis, differentiation, maturation, metabolism and other processes, and therefore play an important role in tumor pathology (7-9). Among them, multiple miRNAs are dysregulated and can serve as either tumor repressors or oncogenes in almost all types of cancers (10). These elevated or decreased miRNAs might be utilized as signatures for tumor diagnosis and prognosis, and new cancer therapy can be developed by manipulating miRNAs (11).
Recently, numerous miRNAs have been documented to participate in the pathological processes of ESCC and can be used as potential diagnostic and prognosis biomarkers (12-14). Among them, miR-375 has been confirmed to be remarkably downregulated and acts as a tumor repressor targeting numerous oncogenic genes in several cancer types, including gastric cancer (15), colorectal cancer (16), oral squamous cell carcinoma (17), head and neck squamous cell carcinoma (18), hepatocellular carcinoma (19), and ESCC (20-22). Nonetheless, the precise mechanism by which miR-375 acts in ESCC remains unclear.
This study is the first to establish that miR-375 functions as a tumor suppressor by directly regulating the peroxiredoxin 1 (PRDX1) gene, which is an antioxidant enzyme (23) and a pivotal intracellular intermediate balancing cell survival with apoptosis in ESCC (24,25). Real-time quantitative polymerase chain reaction (RT-qPCR) assay highlighted the significant downregulation of miR-375 in human ESCC specimens, serum, and ESCC cell lines. Also, the data from the Kaplan-Meier Plotter website illustrated that miR-375 content in ESCC patients is positively linked to their rate of survival. Moreover, aberrant miR-375 expression was found to dampen proliferation, metastasis, and infiltration, but promoted the apoptosis of ESCC cells, as demonstrated by functional studies. Bioinformatics and luciferase reporter analyses showed that individuals with ESCC have a gene called peroxiredoxin 1 (PRDX1) that is a direct target of miR-375. Overall, miR-375 was found to exert a critical modulatory function in the pathophysiological process of ESCC by targeting PRDX1. We present the following article in accordance with the ARRIVE reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-929/rc).
Methods
Patients and clinical samples
In total, 27 ESCC specimens along with matched neighboring non-malignant ESCC tissues from patients who underwent surgeries between August 2017 and April 2020 were received from Nanjing Lishui People’s Hospital (Nanjing, China). The clinical specimens were kept at −80 ℃ for further study. We also acquired 2 mL of peripheral blood from each of the aforementioned ESCC patients and 25 healthy individuals. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Nanjing Lishui People’s Hospital and informed consent was taken from all the patients.
Cell culture
The China Cell Culture Center (Shanghai, China) provided TE-1, TE-10, and ECA-109 ESCC cells coupled with healthy Het-1A esophageal epithelial cells. All cells were inoculated at 5% carbon dioxide (CO2) and 37 ℃ in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, United States) enriched with 10% (v/v) Fetal Bovine Serum (FBS) (Gibco, CA), as well as 1% penicillin (v/v) and streptomycin (Gibco, United States).
Transfection of cells with the miRNA mimic and plasmid
The synthetic miR-375 mimics and scramble mimics (mock) were provided by Genepharma Company (Shanghai, China). The sequences of the miR-375 mimic are as follows: (5'-3'): UUUGUUCGUUCGGCUCGCGUGA. The PRDX1 coding sequence was cloned and propagated into a pcDNA3.1 plasmid. We planted the cells in six-well plates and then transfected them using the Lipofectamine 2000 system (Invitrogen, USA) according to the manufacturer’s instructions.
Isolation of total and serum RNA and RT-qPCR
TRIzol reagent (Invitrogen) or mirVana PARIS System (Ambion, Texas, USA) were respectively employed to isolate total or serum RNA. Subsequently, RNA quantitation and quality determination were performed on the Nanodrop 2000 spectrophotometer platform (Thermo Fisher Scientific, United States). Thereafter, TaqMan miRNA (miR-375 and U6 snRNA) probes (Applied Biosystems, USA) were utilized to perform RT-qPCR according to the manufacturer’s protocol (26) for mature microRNAs or mRNA using the SYBR Green (Applied Biosystems) method. RT-qPCR was carried out on the 7900 detection system (Applied Biosystems), with U6 small nuclear RNA (snRNA) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) serving as normalization standards for miRNA and mRNA, respectively. A comparative threshold cycle method was used and values were computed using the 2−ΔΔCt equation. Each reaction in this experiment was run three separate times. The qRT-PCR primer sequences are listed in Table 1.
Table 1. Primers used for RT-qPCR.
| Gene name | Forward (5'-3') | Reverse (5'-3') |
|---|---|---|
| GAPDH | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG |
| PRDX1 | CCACGGAGATCATTGCTTTCA | AGGTGTATTGACCCATGCTAGAT |
| Cyclin D1 | GCTGCGAAGTGGAAACCATC | CCTCCTTCTGCACACATTTGAA |
| Bcl-2 | GGTGGGGTCATGTGTGTGG | CGGTTCAGGTACTCAGTCATCC |
| E-Cadherin | CGAGAGCTACACGTTCACGG | GGGTGTCGAGGGAAAAATAGG |
| N-Cadherin | TCAGGCGTCTGTAGAGGCTT | ATGCACATCCTTCGATAAGACTG |
| Bax | CCCGAGAGGTCTTTTTCCGAG | CCAGCCCATGATGGTTCTGAT |
| PCNA | CCTGCTGGGATATTAGCTCCA | CAGCGGTAGGTGTCGAAGC |
RT-qPCR, real-time quantitative polymerase chain reaction.
CCK-8 assay
A Cell Counting Kit-8 (CCK-8) assay kit (Dojindo, Kumamoto, Japan) was used to determine cell proliferation. 1.0×103 cells were seeded into 96-well plates and incubated for 48 h. After 12 hours of post-transfection with the microRNA mimic, mock, or plasmid, the cells were inoculated with a 20 µL CCK-8 solution and the absorbance at 450 nm was measured using a microplate reader (BioTek, USA) after 2 h of incubation at 37 ℃. It was necessary to assay each sample in triplicate.
Clonogenic assay of cells in vitro
A cell colony formation assay was performed as described previously (27). Briefly, the cells were treated with a microRNA mimic, mock, or plasmid for 48 h. Next, we harvested the cells and re-suspended them in a 1 mL culture medium into every well of a 24-well plate. The cells were incubated to grow for 2 weeks at 37 ℃. We then removed all of the media and fixed the colonies in 4% Paraformaldehyde Solution (PFA) before staining the cells using crystal violet dye. Under the microscope, the formation of colonies in three different regions of each experiment was counted.
Transwell assay
Transwell chambers (Corning, United States) were used to determine cell infiltration and migration. The Transwell compartment was pre-smeared with 50 µL of Matrigel for detection of cell infiltration but was utilized without the Matrigel coating when detecting cell migration. After being inoculated into a 24-well plate in a growth medium enhanced with 2% FBS at 37 ℃, the treated cells were transferred to the lower compartment of the plate and placed in 600 µL of culture medium enriched with 10% FBS. Following 48 h of growth, the cells were fixed for 30 min. After removing the cells that had not migrated with cotton swabs, the migrating cells were stained with 0.2% crystal violet and incubated at 37 ℃ for 10 minutes. Five random fields of cells were counted under microscope.
Plasmid construction and luciferase reporter assay
The PRDX1 3'-UTR partial sequence, which contains the binding site or mutant binding site, was generated and inserted into a dual-luciferase pmirGLO plasmid (Promega, United States) by GenScript Corporation (Nanjing, China). For the luciferase enzyme reporter experiment, the cells were inoculated into 24-well plates and co-transfected with 0.5 µg firefly luciferase enzyme reporter plasmid and an equivalent amount of scrambled mock or miR-375 mimic using the Lipofectamine 2000 system (Invitrogen, USA) for 48 h. The activity of the luciferase enzyme was determined using a Dual-Luciferase Enzyme Reporter Assay Kit (Promega) according to the manufacturer's instructions, with the Renilla luciferase signal acting as a control.
Animal studies
We construct the tumor graft models to investigate the antitumor effect of miR-375 in vivo. Experiments were performed under a project license (No. 20180815003) granted by the Animal Care & Welfare Committee of Southeast University, in compliance with Chinese National Standard (GBT35823-2018) guidelines for the care and use of animals. A protocol was prepared before the study without registration. The Weitong Lihua Company (Beijing, China) provided us with 6-week-old male BALB/c nudes. The nude mice were housed in a specific pathogen-free (SPF) environment (50%±10% relative humidity, 25±1 ℃), and 5 mice per cage. A total of 10 nude mice were prepared for the experiments, 5 each in the experimental group or the control group. To construct the tumor graft models, TE-1 cells were inoculated with either a lentiviral vector over-expressing miR-375 (Lenti-miR-375) or a lentiviral-mock vector (Lenti-mock vector). 2×106 cells per mouse (n=5) were inoculated subcutaneously into the flanks of the mice after treatment. The length (L), width (W) and height (H) of tumor was measured every 4 days and the tumor volume (V) was calculated by the formula V =π/6×L×W×H. The nude mice were put to death by cervical dislocation after 24 days. The tumors were subsequently removed and weighed. Some tissues were used to extract mRNA and RT-qPCR was used to detect oncogenes such as Cyclin D1, PCNA, N-cadherin, B-cell lymphoma-2 (Bcl-2) and tumor suppressor genes such as E-cadherin and Bax. Each procedure was performed by the same person to minimize errors, and the mice were processed in the same order. The mice grouping was known to the designers and data analysts, but not to the breeders and operators.
Western blotting
Cell and tissue lysis was performed using a lysis buffer enriched with inhibitor cocktail (Sigma, USA). The Bicinchoninic Acid (BCA) assay (Pierce, United States) was used to determine the amount of protein in the sample. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was used to fractionate 50 µL samples, which were then transferred to a Polyvinylidene Fluoride (PVDF) membrane (Roche, USA) for analysis. Next, the membranes were blocked with 5% non-fat milk in a Tris-Buffered Saline and Tween 20 TBST buffer for 1 h at room temperature, and overnight inoculation with primary antibodies against PRDX1 (Cell Signaling Technology, United States) and GAPDH (Abcam, Cambridge, United States) was carried out at 4 ℃.
Subsequently, the membranes were rinsed in TBST buffer, and the blots were inoculated for 1 hour with the secondary antibody labeled with Horseradish Peroxidase (HRP) and then detected with enhanced chemiluminescent (ECL) reagents (Pierce, Rockford, USA). The autoradiographic intensity of each bar graph was scanned and calculated by using Image J software (National Institutes of Health, USA).
Flow cytometry assay
The rate of cell apoptosis was investigated using flow cytometry and an Annexin V-FITC/PI apoptosis detection system (BD Biosciences, New Jersey, USA) as per the manufacturer’s protocol. Briefly, the treated cells were inoculated and collected via spinning at 400 × g for 5 minutes. The cells were re-suspended in 300 µL docking buffer and stained in the dark using Annexin V-FITC and propidium iodide at room temperature for 15 min. Cell apoptosis was detected using a flow cytometry platform (BD Biosciences, New Jersey, USA).
Statistical analysis
The data were expressed as means ± standard deviation (SD) for a minimum of three separate experiments. SPSS 22.0 (SPSS, Chicago, IL, USA) software was used to conduct statistical analysis. Statistical comparisons between the two groups were performed by Student's t-test, with P<0.05 signifying statistical significance.
Results
miR-375 levels are lower in ESCC tissues, serum, and ESCC cell lines
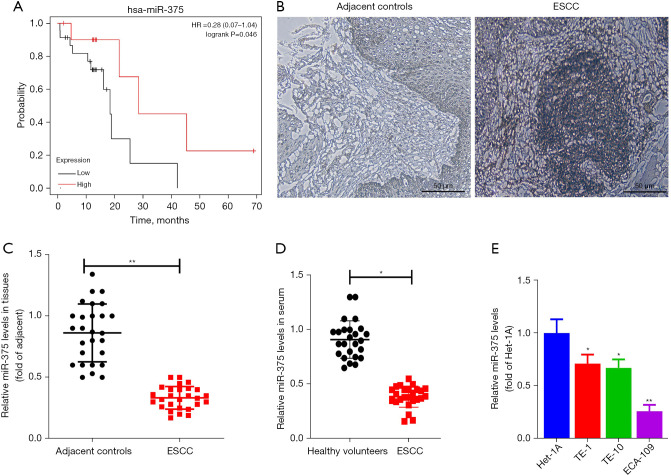
To elucidate the association between prognosis and miR-375 in ESCC patients, we assessed the miR-375 levels and survival rates of ESCC patients using The Cancer Genome Atlas (TCGA) database and Kaplan-Meier Plotter website using the TCGA assembler (28). As illustrated in Figure 1A, the high level of miR-375 in patients corresponds to a better prognosis and greater survival rate. This data indicated that miR-375 is associated with a protective phenotype in ESCC progression.
Figure 1.
miR-375 levels were lower in serum, ESCC tissues, and ESCC cell lines. (A) Survival analysis of miR-375 in ESCC patients from TCGA databases. (B) ESCC tissues and adjacent tissues under the microscope (H&E Staining). (C) TaqMan probe-based RT-qPCR assessment of miR-375 levels in ESCC tissues and corresponding adjacent noncancerous clinical samples. (D) TaqMan probe-based RT-qPCR assessment of miR-375 levels in serum with ESCC and healthy volunteers. (E) The relative levels of miR-375 in TE-1, ECA-109, and TE-10 ESCC cells, and the non-malignant cancer cell line, Het-1A, were examined by RT-qPCR. Data are presented as the mean ± SD of three independent experiments. *P<0.05. **P<0.01. ESCC, esophageal squamous cell carcinoma; RT-qPCR, real-time quantitative polymerase chain reaction.
To further understand the function of miR-375 in ESCC, we first determined the miR-375 level in ESCC tissues and serum. A total of 27 pairs of ESCC tissues and non-malignant neighboring tissues were obtained from Nanjing Lishui People’s Hospital. The clinicopathological properties of individuals with ESCC are shown in Table 2. ESCC tissues and adjacent tissues (hematoxylin-eosin staining) were observed under microscope to confirm the pathological features (Figure 1B).
Table 2. Clinicopathological characteristics of patients with ESCC.
| Variables | Total |
|---|---|
| Gender | |
| Male | 17 |
| Female | 10 |
| Age, years | |
| ≤60 | 20 |
| >60 | 7 |
| T stage | |
| T1 + T2 | 13 |
| T3 + T4 | 14 |
| N stage | |
| N0 | 10 |
| N1-3 | 8 |
| Nx | 9 |
| M stage | |
| M0 | 14 |
| M1 | 13 |
| Stage | |
| Unknown | 3 |
| I + II | 14 |
| III + IV | 10 |
ESCC, esophageal squamous cell carcinoma.
We also isolated the total RNA from these paired samples and serum. The miR-375 level was verified using the TaqMan probe-based RT-qPCR method (26). We assessed the miR-375 level in the 27 pairs of ESCC and the corresponding adjacent noncancerous clinical tissues. As illustrated in Figure 1C, the miR-375 level was markedly reduced in ESCC tissues in contrast to the controls. Additionally, the miR-375 content in serum with ESCC was significantly decreased in contrast to the healthy volunteers (Figure 1D), which is congruous with previous reports (29,30). Consistently, miR-375 expression was also remarkably decreased in three types of ESCC cells in contrast to the Het-1A non-tumorous esophageal cell line (Figure 1E). These findings suggest that miR-375 exerts an anti-tumor effect in ESCC.
miR-375 dampens invasion and proliferation but triggers apoptosis in ESCC cells in vitro
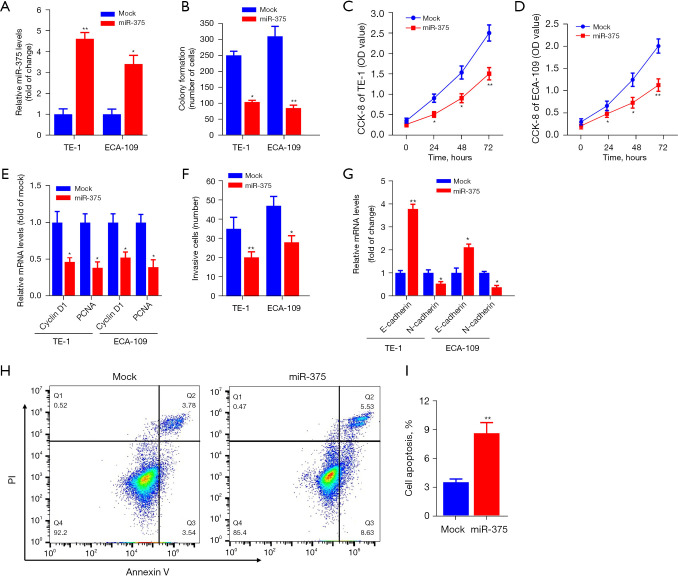
To determine the precise biological function of miR-375 in ESCC, we performed gain-of-function experiments by transfecting ESCC cell lines with a miR-375 mimic or scramble mimic control (mock). RT-qPCR analysis indicated that enhanced miR-375 expression significantly boosted miR-375 content in TE-1 and ECA-109 cells (Figure 2A).
Figure 2.
miR-375 dampens the proliferation and infiltration of ESCC cells but triggers apoptosis. (A) The miR-375 levels in TE-1 and ECA-109 cells after transfection with a miR-375 mimic or mimic control (mock) were monitored for 48 h using RT-qPCR. (B) Influence of miR-375 over-expression on colony formation in TE-1 and ECA-109 cells detected by colony generation assay. The proliferation ability of TE-1 (C) and ECA-109 (D) cells was determined by the CCK-8 assay in miR-375 or mock cell transfects. (E) Cells from the TE-1 and ECA-109 lines were transfected with miR-375 or a mock control, and the relative amounts of Cyclin D1 and PCNA mRNA were determined 24 h after transfection. (F) Transwell assays were used to determine the effect of miR-375 overexpression on the respective infiltration abilities of TE-1 and ECA-109 cells. (G) At 24 hours after transfection with miR-375 or a mock control, the relative levels of E-cadherin and N-cadherin mRNA in TE-1 and ECA-109 cells were determined. (H,I) Flow cytometry was used to detect cell apoptosis in TE-1 cells after transfection with miR-375 or a mock control, respectively. The data are presented as the mean ± SD of three independent experiments. *P<0.05. **P<0.01. RT-qPCR, real-time quantitative polymerase chain reaction.
The regulatory role of miR-375 on the growth, invasion along with apoptosis of TE-1, as well as ECA-109 cells was also investigated. Figure 2B shows that ectopic miR-375 expression significantly decreased the colony generation capacity of TE-1 and ECA-109 cells in contrast to the mock control. The CCK-8 data revealed that miR-375 transfection markedly repressed the growth and viability of TE-1 (Figure 2C) and ECA-109 (Figure 2D) cells, respectively.
The Cyclin D1 and proliferating cell nuclear antigen (PCNA) are considered to be oncogenes, which are upregulated in different cancer tissues and promoting tumor cell proliferation and migration (31,32). We determined whether miR-375's effect on ESCC cell development involves these two genes. As shown in Figure 2E, overexpression of miR-375 in TE-1 and ECA-109 cells significantly decreased the expression of these two genes compared to a mock control. Additionally, transwell invasion data illustrated that forced expression of miR-375 markedly restrained the invasive potential of TE-1 and ECA-109 cells (Figure 2F).
To elucidate the specific molecular mechanisms implicated in esophageal cell migration, we assessed the expression of E-cadherin and N-cadherin genes, which are involved in the migration and infiltration of ESCC cells in both TE-1 and ECA-109 cells (33). The results illustrated that miR-375 increased the E-cadherin content but reduced the content of N-cadherin (Figure 2G). Moreover, flow cytometry illustrated that the aberrant expression of miR-375 can considerably increase the ESCC cell apoptotic rate compared with the vehicle in TE-1 cells (Figure 2H,2I). These data confirmed that miR-375 dampens cell proliferation and migration but induces cell apoptosis in vitro.
miR-375 effectively dampened tumor growth in xenograft tumor mice model in vivo
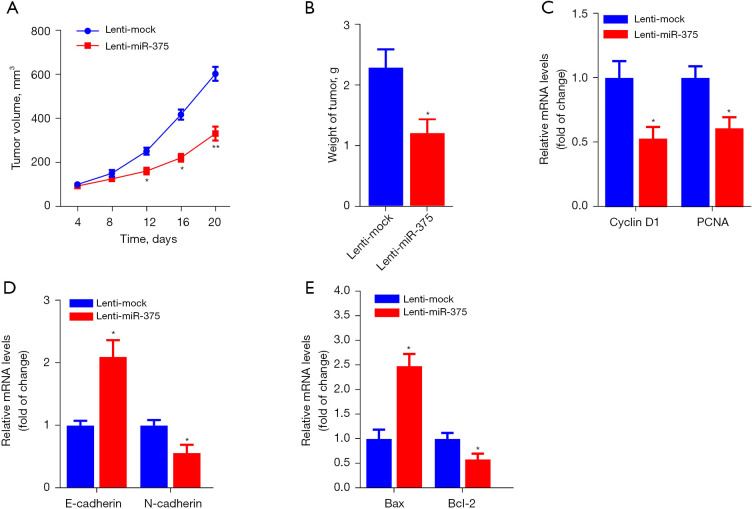
To determine the antitumor effect of miR-375 in vivo, a xenograft mouse model using ESCC cell lines was established. Firstly, TE-1 cells inoculated with lentivirus-packaged miR-375 or lenti-mock were implanted into nude mice by subcutaneous injection. The tumors were measured every 4 days and after 24 days, all the nude mice were sacrificed and tumors were removed and weighed. The xenograft experiments showed that inoculation with the miR-375-overexpression lentivirus obviously dampened the the growth of tumor volume and weight in contrast to the mock group (Figure 3A,3B).
Figure 3.
Overexpression of miR-375 can significantly suppress tumor growth of xenograft nude mice. Stable TE-1 cell transfects of Lenti-miR-375 or Lenti-mock were inoculated subcutaneously into nude mice (n=5). (A) The time course of tumor volume after injection of miR-375 or the mock lentivirus. (B) Quantitative assessment of the tumor weight. RT-qPCR assessment of Cyclin D1 and PCNA (C), E-cadherin and N-cadherin (D), and Bax and Bcl-2 (E) levels from the excised tumor RNA samples. The data are presented as the mean ± SD of three independent experiments. *P<0.05. **P<0.01. RT-qPCR, real-time quantitative polymerase chain reaction.
Moreover, the RT-qPCR assay showed that the oncogenes such as Cyclin D1, PCNA, N-cadherin, and B-cell lymphoma-2 (Bcl-2) were significantly reduced, whereas tumor suppressor genes, including E-cadherin and Bax, were markedly increased in tumor tissues by treatment of miR-375 in contrast to the mock controls (Figure 3C-3E). Collectively, these findings illustrate that miR-375 promotes apoptosis and dampens migration and invasion in esophageal carcinoma xenografts in vivo.
PRDX1 is a direct target of miR-375, which negatively modulates its expression in ESCC cells
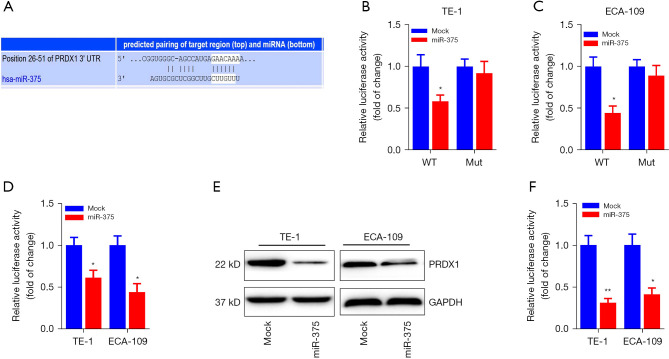
To investigate the potential mechanism by which miR-375 regulates ESCC progression, we searched the prospective miR-375 gene targets via bioinformatics analysis (34). Using the miRNA target forecasting program (TargetScan), the PRDX1 mRNA transcript was hypothesized to be a miR-375 target gene. As illustrated in Figure 4A, the 3'-untranslated region (3'-UTR) of the PRDX1 mRNA transcript contains one putative miR-375 target site. Moreover, the seed site (core sequence comprising the first 2–8 bases of the miRNA) and cognate targets were completely base-paired.
Figure 4.
PRDX1 is a direct target of miR-375. (A) Bioinformatics identification of miR-375 docking sites within the PRDX1 mRNA 3’-UTR. The paired bases are designated using a black line. (B,C) A luciferase enzyme reporter assays were conducted in TE-1 and ECA-109 transfected with miR-375 mimics or mimic controls as well as luciferase reporter plasmids. (D) The relative levels of PRDX1 mRNA in TE-1/ECA-109 cell transfects of the miR-375 mimic or mock were detected using a RT-qPCR assay. (E) Western blot analysis was used to determine the relative protein level of PRDX1 in TE-1 and ECA-109 cells transfected with a miR-375 mimic or mock. (F) The quantitative protein level of PRDX1 was assessed via western blot as designated in the grey scale. The data are presented as mean ± SD of three independent experiments. *P<0.05. **P<0.01. WT, wildtype; Mut, mutant; RT-qPCR, real-time quantitative polymerase chain reaction.
We utilized the luciferase reporter assay to assess whether miR-375 could directly attach to the PRDX1 3'-UTR and determine whether it was effective. The PRDX1 3'-UTR harboring a wild-type or mutant docking site was propagated into a dual-luciferase reporter plasmid. These data indicated that forced miR-375 expression significantly diminished the luciferase enzyme activity of wild-type PRDX1 3'-UTR luciferase vector, while overexpression of miR-375 in the TE-1 and ECA-109 cells did not have any effect on the activity of the luciferase enzyme produced by the mutant PRDX1 3'-UTR vector (Figure 4B,4C).
To further validate PRDX1 as a direct target of miR-375, we examined the modulatory effect of miR-375 on PRDX1 expression in ESCC cells. As illustrated in Figure 4D-4F, aberrant miR-375 expression markedly down-regulated the PRDX1 mRNA and protein levels in both TE-1 and ECA-109 cells compared to the mock control. Taken together, these findings suggest that PRDX1 is a direct target gene of miR-375.
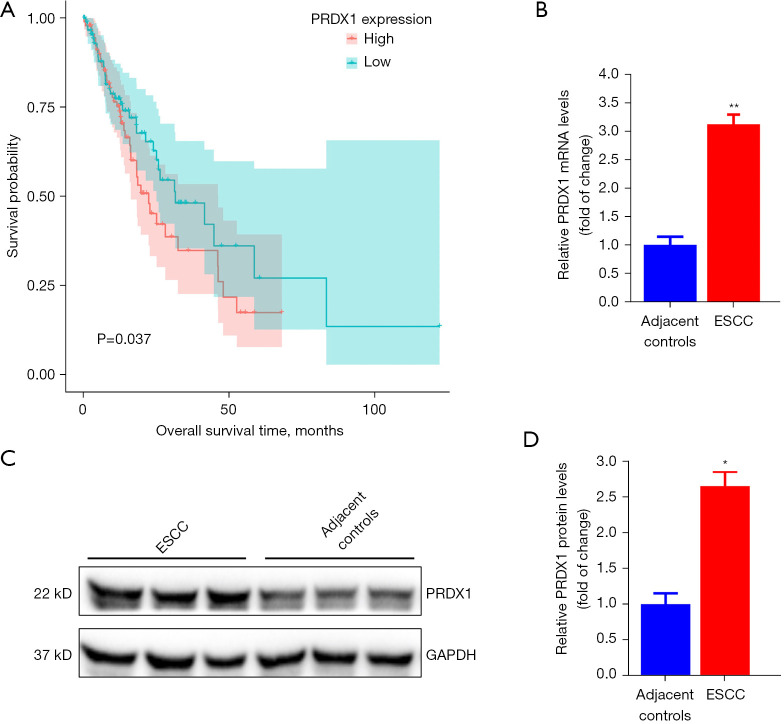
PRDX1 is up-regulated in ESCC tissues and correlates with ESCC patient prognosis
To further verify the function of PRDX1 in ESCC, we explored the association between the PRDX1 level and the survival rate of individuals with esophageal adenocarcinoma specimens in the esca-tcga-pan-can-atlas_2018. As illustrated in Figure 5A, high levels of PRDX1 in patients were associated with a worse prognosis and poor survival rate. Using the collected compared ESCC samples, we established that the PRDX1 level was markedly up-regulated in ESCC samples in contrast to the mRNA (Figure 5B) and protein (Figure 5C,5D) levels in neighboring non-malignant specimens. These data demonstrate that PRDX1 acts as an oncogene in ESCC progression.
Figure 5.
PRDX1 levels are reduced in ESCC tissues. (A) Survival analysis of PRDX1 in ESCC patients from TCGA datasets. (B) PRDX1 mRNA expression in ESCC tissues and neighboring non-malignant samples was assessed via RT-qPCR assay. (C) The relative protein levels of PRDX1 in ESCC tissues and neighboring non-malignant samples were analyzed by western blot. (D) Protein quantitation of PRDX1 was detected via western blot and evaluated in greyscale. The data are presented as mean ± SD of three independent experiments. *P<0.05. **P<0.01. ESCC, esophageal squamous cell carcinoma; RT-qPCR, real-time quantitative polymerase chain reaction.
PRDX1 is a pivotal factor in the regulation of ESCC progression by miR-375
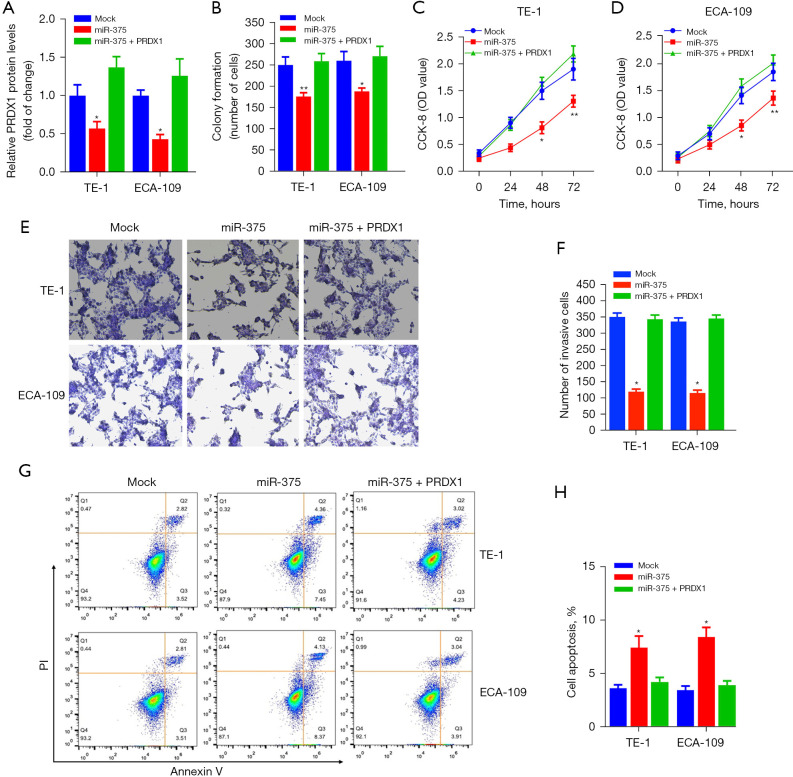
We performed rescue experiments to verify whether PRDX1 contributes to the miR-375-mediated tumor-suppressive effect in ESCC cells. We firstly transfected the PRDX1 expression vector (pcDNA-PRDX1) into miR-375-overexpression cells. As illustrated in Figure 6A, pcDNA-PRDX1 vector significantly increased the PRDX1 expression in cells, while aberrant expression of miR-375 markedly abolished the expression of PRDX1 by pcDNA-PRDX1 plasmid compared to the mock control in TE-1 and ECA-109 cells.
Figure 6.
miR-375 inhibits ESCC cell growth by directly targeting PRDX1. (A) The relative mRNA levels of PRDX1 in TE-1/ECA-109 cell co-transfects of a miR-375 mimics and pcDNA3.1-PRDX1 vector for 48 h. (B) Using a clonogenic assay, we determined the effect of PRDX1 overexpression and miR-375 on TE-1/ECA-109 cell colony development. (C,D) A CCK-8 assay was used to evaluate cell proliferation in TE-1/ECA-109 cells that had been transfected with miR-375 or a sham control, with or without PRDX1 overexpression. (E,F) A transwell assay was used to analyze cell invasion following transfection with miR-375 or a sham control in TE-1 and ECA-109 cells, with or without PRDX1 overexpression (0.2% crystal violet staining, original magnification 400×). (G,H) To detect apoptosis, a flow cytometry assessment was conducted using a mock control or miR-375 overexpression TE-1/ECA-109 cells, with/without PRDX1 over-expression. Data are presented as mean ± SD of three independent experiments. *P<0.05. **P<0.01. ESCC, esophageal squamous cell carcinoma.
In addition, Clonogenic assay and CCK-8 assay exhibited that forced expression of PRDX1 partially abolished the miR-375-mediated antitumor effect on ESCC cell proliferation and viability (Figure 6B-6D). To further assess the role of the PRDX1 on ESCC cell migration and invasion, we established that the dampening role of miR-375 on TE-1/ECA-109 cell migration was abolished via transfection of the pcDNA-PRDX1 plasmid (Figure 6E,6F). Moreover, the apoptosis assay indicated that forced expression of PRDX1 into miR-375-overexpressing TE-1/ECA-109 cells dramatically reversed the pro-apoptotic role of miR-375 (Figure 6G,6H). Taken together, these data illustrated that miR-375 modulates the proliferation, migration, infiltration, and apoptosis of ESCC cells by targeting the PRDX1 gene.
Discussion
ESCC is a highly malignant disease with a poor 5-year-survival rate. Although multiple investigations have documented that miRNAs participate in several aspects of the ESCC progression process, further investigations are pivotal to ascertaining the underlying mechanism of a miRNA-regulated network in ESCC cells. Herein, we purposed to ascertain whether miR-375 is clinically relevant to the survival rate of ESCC patients and to evaluate the expression pattern, biological role, and modulatory molecular mechanisms of miR-375. Through TCGA database and Kaplan-Meier curve analysis, we found that miR-375 confers a protective phenotype on patients exhibiting elevated miR-375 expression and predicts longer overall survival (OS). Furthermore, we discovered that miR-375 levels were decreased in ESCC samples, serum, and cell lines, showing that miR-375 exerts an anti-tumor effect. Increased miR-375 expression drastically repressed the proliferation, metastasis, and infiltration of ESCC cells but induced their apoptosis. The xenograft experiment illustrated that miR-375 dampened tumor growth in vivo. Bioinformatics analysis demonstrated that miR-375 inversely modulated the PRDX1 mRNA transcript, thus acting as a tumor suppressor during ESCC development and progression.
MiRNAs is a hot topic in the study of malignant tumors. A large number of miRNAs have been found to be significantly differentially expressed in cancer tissues or serum of ESCC patients compared with healthy tissues or serum (35). MiRNAs can act either oncogenes or tumor suppressors in ESCC by participating in cell proliferation, migration and apoptosis or other processes (36). It is worth noting that many studies have found that intervening or increasing the expression of certain miRNAs has a significant impact on the development of ESCC (37). On the other hand, miRNAs can also be used as potential markers for ESCC diagnosis and prognosis prediction (38,39).
As an evolutionarily conserved pancreatic islet-distinct miRNA, miR-375 was first identified as a regulator of insulin secretion in the pancreatic islets (40). Further research verified that miR-375, which is a multi-functional miRNA, is involved in the development of islets, blood sugar balance, cell proliferation, mucosal immunity, and more significantly, tumorigenesis (41). miR-375 is downregulated in various cancers and is implicated in the modulation of numerous pivotal oncogenes (15,16,19), implying that miR-375 may function as an antitumor miRNA in cancer cells. Diminished levels of miR-375 in the circulatory system or tissue may reflect a poor prognosis in several malignant cancers, including ESCC (42).
Peroxiredoxins (PRDXs) constitute a ubiquitous family of redox-modulating proteins with a role in modulating reactive oxygen species (ROS) (23). Although PRDX1 was initially identified as an antioxidant enzyme, its physiological role in the oxidation-reduction balance during tumorigenesis is currently unknown (25). Previous investigations have found that the PRDX1 level is elevated in ESCC tissues (25,43). Moreover, numerous studies have documented PRDX1 overexpression in ESCC cells compared to non-malignant esophageal epithelial cells (44,45). A study has also shown that the elevated expression of PRDX1 can promote the development and growth of ESCC and that down-regulated PRDX1 expression can reduce tumorigenesis (46). These findings illustrate that PRDX1 may act as an oncogene in ESCC. Nonetheless, the unambiguous role and detailed molecular mechanism of PRDX1 in ESCC remains enigmatic.
Tumor initiation and development are determined by a vast and complex molecular network. Our study does not answer the question regarding which molecular signals affect the expression changes of miR-375 itself. In previous studies, researchers found that single nucleotide polymorphisms (SNPs) play an important role in the miRNA maturation process by affecting Drosha enzyme processing as well as Dicer enzyme processing and function (47,48). MiRNA expression is also regulated by DNA methylation (49). In recent years, many studies have found that long noncoding RNAs (lncRNAs) have the role of sponge, they can competitively adsorb miRNA, thereby inhibiting the regulation of miRNA on target genes (50,51). In future research, we will explore the causes and mechanisms of miR-375 expression changes in tumors from an epigenetic perspective and explore the feasibility of miRNA-375 as an anti-tumor drug.
Conclusions
In this study, we discovered for the first time that miR-375 acts as a tumor suppressor by directly altering the PRDX1 target gene. Our result highlights a profound function of the miR-375/PRDX1 signaling pathway in regulating ESCC cell proliferation and migration and suggests that miR-375 is expected to be a new and potentially non-invasive method for the diagnosis and treatment of ESCC.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by the Nanjing Medical Science and Technology Development Fund (Grant No. YKK18221), the Talent Scientific Research Start-up Foundation of Yijishan Hospital, Wannan Medical College (Grant No. YR201806), the Nature and Science Fund of Key Cultivate Program from Wannan Medical College, CN (Grant No. WK2018ZF06), and the Science Fund for Key Program of Three Creative Research of Yijishan Hospital of Wannan Medical College (Grant No. Z1810).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Nanjing Lishui People’s Hospital and informed consent was taken from all the patients. Animal experiments were performed under a project license (No. 20180815003) granted by the Animal Care & Welfare Committee of Southeast University, in compliance with Chinese National Standard (GBT35823-2018) guidelines for the care and use of animals.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-929/rc
Data Sharing Statement: Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-929/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-929/coif). The authors have no conflicts of interest to declare.
(English Language Editor: A. Kassem)
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Li S, Chen H, Man J, et al. Changing trends in the disease burden of esophageal cancer in China from 1990 to 2017 and its predicted level in 25 years. Cancer Med 2021;10:1889-99. 10.1002/cam4.3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xi Y, Lin Y, Guo W, et al. Multi-omic characterization of genome-wide abnormal DNA methylation reveals diagnostic and prognostic markers for esophageal squamous-cell carcinoma. Signal Transduct Target Ther 2022;7:53. 10.1038/s41392-022-00873-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5. 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 6.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci U S A 2003;100:9779-84. 10.1073/pnas.1630797100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang JK, Wang Z, Li G. MicroRNA-125 in immunity and cancer. Cancer Lett 2019;454:134-45. 10.1016/j.canlet.2019.04.015 [DOI] [PubMed] [Google Scholar]
- 8.Tapeh BEG, Mosayyebi B, Mansoori B, et al. Emerging Molecular Functions of MicroRNA-9: Cancer Pathology and Therapeutic Implications. Anticancer Agents Med Chem 2021;21:2304-14. 10.2174/1871520621666210217094839 [DOI] [PubMed] [Google Scholar]
- 9.Ali Syeda Z, Langden SSS, Munkhzul C, et al. Regulatory Mechanism of MicroRNA Expression in Cancer. Int J Mol Sci 2020;21:1723. 10.3390/ijms21051723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong FY, Bai T, Zhang DY, et al. Regulation Mechanism of MicroRNAs in Non-Small Cell Lung Cancer. Curr Pharm Des 2018;23:5973-82. 10.2174/1381612823666170714153424 [DOI] [PubMed] [Google Scholar]
- 11.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med 2014;20:460-9. 10.1016/j.molmed.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 12.Bobryshev YV, Orekhov AN, Chistiakov DA. MicroRNAs in Esophageal Adenocarcinoma: Functional Significance and Potential for the Development of New Molecular Disease Markers. Curr Pharm Des 2015;21:3402-16. 10.2174/1381612821666150311124418 [DOI] [PubMed] [Google Scholar]
- 13.Fassan M, Baffa R, Kiss A, et al. MicroRNA dysregulation in esophageal neoplasia: the biological rationale for novel therapeutic options. Curr Pharm Des 2013;19:1236-41. [DOI] [PubMed] [Google Scholar]
- 14.Gao S, Zhao ZY, Zhang ZY, et al. Prognostic Value of MicroRNAs in Esophageal Carcinoma: A Meta-Analysis. Clin Transl Gastroenterol 2018;9:203. 10.1038/s41424-018-0070-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsukamoto Y, Nakada C, Noguchi T, et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res 2010;70:2339-49. 10.1158/0008-5472.CAN-09-2777 [DOI] [PubMed] [Google Scholar]
- 16.Xu X, Chen X, Xu M, et al. miR-375-3p suppresses tumorigenesis and partially reverses chemoresistance by targeting YAP1 and SP1 in colorectal cancer cells. Aging (Albany NY) 2019;11:7357-85. 10.18632/aging.102214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Sun X, Song B, et al. MiR-375/SLC7A11 axis regulates oral squamous cell carcinoma proliferation and invasion. Cancer Med 2017;6:1686-97. 10.1002/cam4.1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nohata N, Hanazawa T, Kikkawa N, et al. Tumor suppressive microRNA-375 regulates oncogene AEG-1/MTDH in head and neck squamous cell carcinoma (HNSCC). J Hum Genet 2011;56:595-601. 10.1038/jhg.2011.66 [DOI] [PubMed] [Google Scholar]
- 19.He XX, Chang Y, Meng FY, et al. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene 2012;31:3357-69. 10.1038/onc.2011.500 [DOI] [PubMed] [Google Scholar]
- 20.Kong KL, Kwong DL, Chan TH, et al. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut 2012;61:33-42. 10.1136/gutjnl-2011-300178 [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita T, Hanazawa T, Nohata N, et al. The functional significance of microRNA-375 in human squamous cell carcinoma: aberrant expression and effects on cancer pathways. J Hum Genet 2012;57:556-63. 10.1038/jhg.2012.75 [DOI] [PubMed] [Google Scholar]
- 22.Wu W, Zhang Y, Li X, et al. miR-375 Inhibits the Proliferation, Migration and Invasion of Esophageal Squamous Cell Carcinoma by Targeting XPR1. Curr Gene Ther 2021;21:290-8. 10.2174/1566523220666201229155833 [DOI] [PubMed] [Google Scholar]
- 23.Ding C, Fan X, Wu G. Peroxiredoxin 1 - an antioxidant enzyme in cancer. J Cell Mol Med 2017;21:193-202. 10.1111/jcmm.12955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoshino I, Matsubara H, Akutsu Y, et al. Tumor suppressor Prdx1 is a prognostic factor in esophageal squamous cell carcinoma patients. Oncol Rep 2007;18:867-71. 10.3892/or.18.4.867 [DOI] [PubMed] [Google Scholar]
- 25.Ren P, Ye H, Dai L, et al. Peroxiredoxin 1 is a tumor-associated antigen in esophageal squamous cell carcinoma. Oncol Rep 2013;30:2297-303. 10.3892/or.2013.2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 2005;33:e179. 10.1093/nar/gni178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crowley LC, Christensen ME, Waterhouse NJ. Measuring Survival of Adherent Cells with the Colony-Forming Assay. Cold Spring Harb Protoc 2016. 10.1101/pdb.prot087171 [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y, Qiu P, Ji Y. TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat Methods 2014;11:599-600. 10.1038/nmeth.2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komatsu S, Ichikawa D, Takeshita H, et al. Prognostic impact of circulating miR-21 and miR-375 in plasma of patients with esophageal squamous cell carcinoma. Expert Opin Biol Ther 2012;12 Suppl 1:S53-9. 10.1517/14712598.2012.681373 [DOI] [PubMed] [Google Scholar]
- 30.Komatsu S, Ichikawa D, Takeshita H, et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer 2011;105:104-11. 10.1038/bjc.2011.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu P, Zhou K, Lu S, et al. Modulation of aryl hydrocarbon receptor inhibits esophageal squamous cell carcinoma progression by repressing COX2/PGE2/STAT3 axis. J Cell Commun Signal 2020;14:175-92. 10.1007/s12079-019-00535-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu YC, Lam KY, Law S, et al. Profiling of differentially expressed cancer-related genes in esophageal squamous cell carcinoma (ESCC) using human cancer cDNA arrays: overexpression of oncogene MET correlates with tumor differentiation in ESCC. Clin Cancer Res 2001;7:3519-25. [PubMed] [Google Scholar]
- 33.Labernadie A, Kato T, Brugués A, et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat Cell Biol 2017;19:224-37. 10.1038/ncb3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15-20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- 35.Cui D, Cheung AL. Roles of microRNAs in tumorigenesis and metastasis of esophageal squamous cell carcinoma. World J Clin Oncol 2021;12:609-22. 10.5306/wjco.v12.i8.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harada K, Baba Y, Ishimoto T, et al. The role of microRNA in esophageal squamous cell carcinoma. J Gastroenterol 2016;51:520-30. 10.1007/s00535-016-1161-9 [DOI] [PubMed] [Google Scholar]
- 37.Liu H, Ren G, Zhu L, et al. The upregulation of miRNA-146a inhibited biological behaviors of ESCC through inhibition of IRS2. Tumour Biol 2016;37:4641-7. 10.1007/s13277-015-4274-5 [DOI] [PubMed] [Google Scholar]
- 38.Wen J, Wang G, Xie X, et al. Prognostic Value of a Four-miRNA Signature in Patients With Lymph Node Positive Locoregional Esophageal Squamous Cell Carcinoma Undergoing Complete Surgical Resection. Ann Surg 2021;273:523-31. 10.1097/SLA.0000000000003369 [DOI] [PubMed] [Google Scholar]
- 39.Xue J, Jia E, Ren N, et al. Identification of prognostic miRNA biomarkers for esophageal cancer based on The Cancer Genome Atlas and Gene Expression Omnibus. Medicine (Baltimore) 2021;100:e24832. 10.1097/MD.0000000000024832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004;432:226-30. 10.1038/nature03076 [DOI] [PubMed] [Google Scholar]
- 41.Yan JW, Lin JS, He XX. The emerging role of miR-375 in cancer. Int J Cancer 2014;135:1011-8. 10.1002/ijc.28563 [DOI] [PubMed] [Google Scholar]
- 42.Luo HS, Wu DH. Identification of miR-375 as a potential prognostic biomarker for esophageal squamous cell cancer: A bioinformatics analysis based on TCGA and meta-analysis. Pathol Res Pract 2019;215:512-8. 10.1016/j.prp.2019.01.009 [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Wang K, Zhang J, et al. Using proteomic approach to identify tumor-associated proteins as biomarkers in human esophageal squamous cell carcinoma. J Proteome Res 2011;10:2863-72. 10.1021/pr200141c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gong F, Hou G, Liu H, et al. Peroxiredoxin 1 promotes tumorigenesis through regulating the activity of mTOR/p70S6K pathway in esophageal squamous cell carcinoma. Med Oncol 2015;32:455. 10.1007/s12032-014-0455-0 [DOI] [PubMed] [Google Scholar]
- 45.Gong F, Liu H, Li J, et al. Peroxiredoxin 1 is involved in disassembly of flagella and cilia. Biochem Biophys Res Commun 2014;444:420-6. 10.1016/j.bbrc.2014.01.081 [DOI] [PubMed] [Google Scholar]
- 46.Chen Q, Li J, Yang X, et al. Prdx1 promotes the loss of primary cilia in esophageal squamous cell carcinoma. BMC Cancer 2020;20:372. 10.1186/s12885-020-06898-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Ru J, Meng X, et al. Role of SNPs in the Biogenesis of Mature miRNAs. Biomed Res Int 2021;2021:2403418. 10.1155/2021/2403418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papaconstantinou I, Stamatis K, Tzathas C, et al. The role of variations within microRNA in inflammatory bowel disease. Eur J Gastroenterol Hepatol 2013;25:399-403. 10.1097/MEG.0b013e32835c34ea [DOI] [PubMed] [Google Scholar]
- 49.Fuso A, Raia T, Orticello M, et al. The complex interplay between DNA methylation and miRNAs in gene expression regulation. Biochimie 2020;173:12-6. 10.1016/j.biochi.2020.02.006 [DOI] [PubMed] [Google Scholar]
- 50.Tian W, Jiang C, Huang Z, et al. Comprehensive analysis of dysregulated lncRNAs, miRNAs and mRNAs with associated ceRNA network in esophageal squamous cell carcinoma. Gene 2019;696:206-18. 10.1016/j.gene.2019.02.051 [DOI] [PubMed] [Google Scholar]
- 51.Xue ST, Zheng B, Cao SQ, et al. Long non-coding RNA LINC00680 functions as a ceRNA to promote esophageal squamous cell carcinoma progression through the miR-423-5p/PAK6 axis. Mol Cancer 2022;21:69. 10.1186/s12943-022-01539-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as