Abstract
Abstract
Caseous lymphadenitis (CLA) is a disease that affects small ruminants, and the best way to prevent its spread on a herd is through immunoprophylaxis. Thus, we aimed to evaluate the MBP:PLD:CP40 fusion protein as a new CLA immunogen. The fusion protein was constructed by combining Corynebacterium pseudotuberculosis PLD and CP40 proteins with maltose-binding protein (MBP) as an intrinsic adjuvant. The antigenicity, allergenic potential, prediction of B epitopes, binding to MHC receptors, and docking on the Toll-Like 2 receptor were evaluated in silico. MBP:PLD:CP40 was expressed and purified. 40 BALB/c were divided into four groups (G1 – control, G2 – Saponin, G3 – MBP:PLD:CP40, and G4 – rPLD + rCP40). Total IgG, IgG1, and IgG2a were quantified, and the expressions of cytokines after splenocyte in vitro stimulation were assessed. Mice were challenged 42 days after the first immunization. The in silico analysis showed that MBP:PLD:CP40 has immunogenic potential, does not have allergic properties, and can dock on the TRL2 receptor. MBP:PLD:CP40 stimulated the production of IgG1 antibodies in a fivefold proportion to IgG2a, and TNF and IL-17 were significantly expressed in response to the antigenic stimuli. When rPLD and rCP40 were used together for immunization, they could induce IFN-γ and IL-12, but with no detectable antibody production. The G3 and G4 groups presented a survival of 57.14% and 42.86%, respectively, while the G1 and G2 mice were all dead 15 days after the challenge. MBP:PLD:CP40 partially protected the mice against C. pseudotuberculosis infection and can be considered a potential new CLA immunogen.
Key points
• The fusion protein induced more IgG1 than IgG2a antibodies;
• The fusion protein also induced the expression of the TNF and IL-17 cytokines;
• Mice inoculated with MBP:PLD:CP40 presented a 57.14% survival.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00253-022-12279-1.
Keywords: Caseous lymphadenitis, Immunoinformatics, Vaccine development, Recombinant proteins
Introduction
Corynebacterium pseudotuberculosis is the etiologic agent of caseous lymphadenitis (CLA), a disease that affects small ruminants and causes economic losses worldwide (Dorella et al. 2006). Antibiotics cannot penetrate the capsule formed around the CLA’s granulomatous lesions, and in this way, the treatment of the disease is difficult, and vaccination can be considered the most effective preventive measure (Dorella et al. 2009).
Commercial vaccines against CLA are currently available; however, the efficacy of these vaccines can vary among sheep and goats, and abscess formation is still observed in vaccinated animals (Leal et al. 2018). Thus, experimental immunogens are being tested to improve safety and protection rates (Pinho et al. 2021a).
C. pseudotuberculosis phospholipase D (PLD) and CP40 proteins have already been used as CLA vaccine targets, with the first inducing significant antibody production and the second activating cellular immunity (Selim et al. 2010; Silva et al. 2014). Thus, it would be interesting to test PLD combined with CP40, since it would include effective activators of both cellular and humoral immune responses (Leal et al. 2018). Maltose Binding Protein (MBP) has already been described as an intrinsic adjuvant for fusion proteins (Gong et al. 2015), since it can bind to Toll-Like 2 receptors (Liu et al. 2017; Wang et al. 2015). In this way, the objective of this study was to develop a MBP:PLD:CP40 fusion protein and then characterize its immunogenicity and immunoprophylaxis properties in silico and in vivo.
Materials and methods
In silico analyses
Physicochemical analysis, 3D structure prediction, antigenicity, and allergenic potential
The physicochemical properties of the MBP:PLD:CP40 fusion protein were analyzed using the constructed amino acid sequence through the ProtParam software (http://web.expasy.org/protparam). The analyzed parameters were (1) molecular weight; (2) theoretical isoelectric point (pI); (3) instability index; (4) in vitro and in vivo estimated half-life; (4) aliphatic index; (5) the grand average of hydropathicity (GRAVY); and (6) number of amino acids (Gasteiger et al. 2005).
The RaptorX tool was used to predict and select the best tertiary structure (PMID: 22814390). The SAVE PROCHECK server (https://saves.mbi.ucla.edu) (Laskowski et al. 1993) was used to generate a Ramachandran graph. The structural representation was made using the Chimera software (UCSF, San Francisco, CA).
The antigenicity analysis was performed through ANTIGENpro (http://scratch.proteomics.ics.uci.edu/) (Cheng et al. 2005). The assessment of the allergenic potential was performed using the software AlgPred (http://www.imtech.res.in/raghava/algpred/index.html) (Saha and Raghava 2006).
Prediction of B epitopes and binding to class I and II MHC molecules
For the prediction of B lymphocyte epitopes, the Immune Epitope Database and Analysis Resource (IEDB) server (http://www.iedb.org/) were used. Additionally, analyses based on the hydrophilicity scale method (Parker et al. 1986), solvent accessibility scale method (Emini et al. 1985), and β-turn prediction method (Chou and Fasman 1978) were also performed. The predictions of the possible binding to the major histocompatibility complex (MHC) class I and II molecules were performed at the IEDB server using the NetMHCpan EL 4.1 (Reynisson et al. 2020) and 2.22 (Wang et al. 2010) methods.
Prediction of docking on the Toll-like 2 receptor
The docking analysis between the MBP:PLD:CP40 fusion protein and the mouse-specific Toll-like 2 (TLR2) receptor (PDB 5d3i, with modifications) was performed using the SwarmDock software (https://bmm.crick.ac.uk/~svc-bmm-swarmdock) (Torchala et al. 2013). The 3D structures of the fusion protein and the receptor were made using the Chimera software (UCSF), and the representation of the binding was performed with the LIGPLOT v.2.2 software (Wallace et al. 1995).
Fusion protein construction and expression
Nucleotide sequence and vector for the expression of the MBP:PLD:CP40 fusion protein
The fusion protein sequence was constructed based on the gene sequences that encode the C. pseudotuberculosis PLD (WP_013240889.1) and CP40 (WP_138130220.1) proteins. The genes were merged entirely, with substitutions of the active site amino acids (PLD H19A-H53A and CP40 D150A-D153A-E157A). The inclusion of a rigid linker (EAAAKEAAAKEAAAKE) between PLD and CP40 proteins was performed (Lu and Feng 2008). Additionally, the sequence GGENLYF was added between MBP and PLD as a TEV (Tobacco Etch Virus) protease cleavage site. The sequence was expressed and cloned into a pMAL-c2X (Plasmid #75286) vector, containing the maltose-binding protein (MBP) sequence.
Cloning and expression
rPLD and rCP40 recombinant proteins, used in a separately way for immunization, ELISA and cellular immunity assays, were expressed as previously described. Briefly, for rPLD and rCP40, the plasmids previously constructed pAE/pld (Silva et al. 2018) and pAE/cp40 (Droppa-Almeida et al. 2016) were introduced by heat shock into the expression strain E. coli BL21 (DE3) Star. Expression of recombinant proteins was induced by the addition of 1 mM of isopropyl-α-D-thiogalactoside (IPTG) to the culture medium supplemented with ampicillin (100 µg/mL), followed by incubation in an orbital shaker (200 rpm) at 37 °C for 3 h. After, the confirmation of expression of rPLD and rCP40 were performed by western blotting using a horseradish peroxidase (HRP)-conjugated anti-6 × His tag monoclonal antibody (Sigma–Aldrich, Saint Louis, MO). Then, the cells were lysate by sonication in buffer containing 8 M urea for solubilization of proteins and the purification was performed by affinity chromatography on a HisTrap™ Sepharose nickel column (GE Healthcare Life Sciences, Marlborough, MA). After, a dialysis with PBS buffer was performed to refold the proteins.
The MBP:PLD:CP40 fusion protein was expressed as follows: 80 ng of the pMAL-c2X recombinant vector was used to transform thermocompetent E. coli (Origami) cells. Afterward, the transformation product was stirred at 250 rpm for 1 h at 37 °C in SOC liquid medium. Then, the bacteria were selected in LB selective medium (solid) containing ampicillin (100 µg/mL) and subsequently inoculated into individual tubes containing 10 mL of LB selective medium with ampicillin (100 µg/mL) under constant agitation and aeration for 16 h at 37 °C. These cultures were then diluted 100 times in fresh LB medium containing antibiotics and incubated at 37 °C until reaching a 600 nm optical density between 0.4 and 0.6. Expression induction was performed at 20 °C with 0.4 mM IPTG. After expression, the culture was centrifuged at 8000 × g, the supernatant was discarded, and the precipitate was resuspended in a cell lysis buffer, sonicated (4 pulses of 40 s) and centrifuged at 15,000 × g for 30 min at 4 °C. Additionally, the MBP:PLD:CP40 was treated with TEV-protease for 16 h to show the possibility of cleavage of MBP, obtaining the PLD:CP40 protein (not used in this particular study). Afterwards, the samples were then applied to a 15% SDS-PAGE and electrotransferred to a nitrocellulose membrane. The membrane was incubated with an anti-polyhistidine monoclonal antibody (Sigma-Aldrich) diluted 1:10,000 in PBS Tween-20 0.05% (PBST) and subsequently with an alkaline phosphatase-conjugated anti-mouse IgG antibody (Sigma-Aldrich) diluted 1:30,000 in PBST. The reaction was detected with a solution containing H2O2 and BCIP/NBT (5-bromo-4chloro-3-indolylphosphate/nitro-bluetetrazolium).
Protein purification
The purification was performed through a Ni-Sepharose affinity chromatography (GE Healthcare Life Sciences). Briefly, the resin was washed and eluted by gradually increasing imidazole concentrations. The fractions were analyzed on a 15% SDS-PAGE and concentrated in 10 kDa centrifuge devices (Amicon, Miami, FL). Subsequently, another purification step was performed by size exclusion chromatography on a Superdex G75 10/300 column (GE Healthcare Life Sciences) using the AKTA-pure system (Cytiva, Marlborough, MA). The samples were evaluated by Dynamic Light Scattering (DLS) in a Zetasizer Nano series S90 device (Malvern Instruments, Malvern, UK), and data acquisition was performed from 10 cycles of the 30 s at a constant temperature of 25 °C. The purified protein was also tested in an immunoenzymatic assay, using serum samples from goats and sheep naturally infected or not by C. pseudotuberculosis in different dilutions, and the MBP:PLD:CP40 purified protein as antigen and in different concentrations, with the objective to verify the recognition of the fusion protein by antibodies of animals with caseous lymphadenitis.
Circular dichroism spectroscopy
Circular dichroism spectroscopy was performed on a Jasco J-815 spectropolarimeter (Jasco, Portland, OR) with a Peltier-type temperature control system. The far UV–CD spectrum of MBP:PLD:CP40 fusion protein was collected from 260 to 197 nm at 25 °C in a 0.05 cm quartz cuvette. A scan speed of 50 nm/min was used with a response time of 1.0 s, spectral bandwidth of 1.0 nm and spectral resolution of 0.1 nm. The signal was averaged over 10 scans. Each spectrum was acquired independently twice. The protein was diluted to 5 μM in 20 Mm Tris and 100 mM NaCl, pH 7.4. The contribution of the buffer was subtracted from the protein spectra. Percentages of the secondary structure in solution were calculated with the CONTINLL software of the CDPro package, using the reference set of proteins SP43.
In vivo analyses
Animal model and ethical aspects
Forty BALB/c mice (males, 8 weeks old—from Instituto Gonçalo Moniz—Fiocruz-BA, Salvador, Brazil) were used in this study. This project was approved by the Ethics Committee of the School of Veterinary Medicine and Zootechnics of the Federal University of Bahia (permission number 19/2021).
Experimental groups, sampling, immunization, and challenge
The animals were divided into four groups of 10 animals each, and immunizations were performed subcutaneously at 0 and 28 days. Groups were immunized as follows: G1—200 µL of a sterile saline solution; G2—200 µL of saline solution containing 7.5 µg of saponin; G3—200 µl of sterile saline containing 50 µg of the MBP:PLD:CP40 fusion protein and 7.5 µg of saponin; G4—200 µL of sterile saline containing 25 µg of rPLD, 25 µg of rCP40, and 7.5 µg of saponin. Blood was sampled by retro-orbital sinus puncture at 0, 14, 28, and 42 days after the first immunization. All blood samplings were performed under anesthesia with 10% ketamine (100 mg/kg) and 2% xylazine (10 mg/kg). The blood samples were centrifuged at 1500 × g for 15 min and stored at -20 °C. The challenge was performed on seven animals from each group two weeks after the last immunization by intraperitoneally injecting 200 µL of a solution containing 104 CFU of the virulent C. pseudotuberculosis MIC-6 strain (Ref. Seq. NZ_CP019769.1), and the animals were observed for 30 days. All animals were necropsied after death by veterinarian pathologists, and lesions were macroscopically evaluated.
Humoral immune response evaluation
The production of total IgG, IgG1, and IgG2a was evaluated as previously described (Bezerra et al. 2021), with minor modifications. Briefly, ELISA plates were coated with 100 µl of rPLD and/or rCP40 at a concentration of 2.5 µg/ml, at 4 °C for 16 h. Serum samples diluted 1:100 in PBST-1% skimmed milk was then applied at 100 µl/well, and incubated at 37 °C for 1 h. After six washes with PBST, the anti-mouse total IgG (Sigma-Aldrich), IgG1 (Invitrogen, Waltham, MA), and IgG2a (Invitrogen) conjugated with horseradish peroxidase diluted 1:5000 in PBST-1% skimmed milk were applied to the plates, which were incubated at 37 °C for 1 h. After additional six washes with PBST, the reaction was developed with a solution of OPD (ortho phenylenediamine) and hydrogen peroxide for 10 min, stopped with 2 N sulfuric acid (H2SO4), and then read at 492 nm.
Cellular immune response evaluation
Forty-two days after the first immunization, three mice from each experimental group were euthanized by cervical dislocation under anesthesia and had their spleens aseptically collected. The spleens were macerated through a cell strainer in a petri dish containing 5 mL of HBBS-HANK’S buffer (Sigma-Aldrich), transferred into tubes, and centrifuged at 500 × g for 7 min at 4 °C; the pellet was resuspended in ACK lysing buffer (Thermo Fisher Scientific, Waltham, MA) and left at room temperature for 5 min; the lysis reaction was stopped with 10 mL of PBS and centrifuged at 500 × g for 7 min at 4 °C. The pellet was then resuspended in DMEM medium (Thermo Fisher Scientific) containing 1% antibiotic/antimycotic solution (Sigma-Aldrich) and 10% inactivated fetal bovine serum (Sigma-Aldrich). The cells were counted and had the concentration adjusted to 1.5 × 107 cells/mL, and then 1 mL of this solution was placed in each well of sterile 24-well culture plates.
Cells were stimulated with rPLD and rCP40 proteins (10 µg/mL each), concanavalin A (CoA) (10 µg/mL) as a positive control, and culture medium as a negative control. The plates were incubated for 48 h, and each well content was centrifuged at 500 × g at 4 °C for 10 min. The pellets were resuspended in TRIzol (Invitrogen), and the transformation of RNA into cDNA was made using the High-capacity Reverse Transcriptase kit (Applied Biosystems, Waltham, MA). Finally, the IL-4, IL-12, IL-17, IFN-γ and TNF cDNAs were quantified using the Stratagene Mx3005P Real-Time PCR System (Agilent Technologies, Santa Clara, CA) and specific primers; the results were normalized using the glycerol-3-phosphate dehydrogenase (GPDH) gene (Dummer et al. 2014; Overbergh et al. 1999).
Statistical analyses
Statistical analyses were performed using the software GraphPad v. 8 (GraphPad Software, California, USA). The humoral and cellular immune response quantification followed a normal distribution based on the Kolmogorov–Smirnov test. Therefore, one-way ANOVA and Holm-Šídák’s multiple comparisons test compared differences among the groups. The percent of survival was calculated dividing the number of dead animals by the total number of animals of each group. The Log-rank and Fisher’s tests were performed to verify the differences between the survival curves (kinetics of mortality during 30 days).
Results
MBP:PLD:CP40 sequence and 3D structure
The proposed 3D model can be visualized in Fig. S1. The fusion protein contains the amino acid residues encoding for maltose binding protein (386 AA), PLD (282 AA), CP40 (348 AA), His-tag, TevSite (7 AA), and the rigid linker (16 AA), as shown in Table S1. The analysis of the secondary structure of the protein showed that it has 33% alpha-Helix, 16% beta-sheets, and 50% coil structure (Fig. S2). The Ramachandran graph shows that the MBP:PLD:CP40 protein 3D structure presented 89.5% of the amino acids in favorable regions (Fig. S3).
Physicochemical analysis, antigenicity, and allergenic potential
The MBP:PLD:CP40 fusion protein presented antigenicity in the in silico analysis and was non-allergenic, showing no IgE epitopes and no similarity with known allergens. Additionally, through the ProtParam server, it is speculated that the protein presents stability even at high temperatures and is hydrophilic due to the low instability and high aliphatic indices. In addition, it has a significant half-life. These data are presented in Table 1.
Table 1.
Physicochemical parameters, antigenicity, and allergenic potential of the MBP:PLD:CP40 fusion protein. The physicochemical parameters were analyzed using the ProtParam server. Antigenicity was analyzed on the ANTIGENpro server. The allergenic potential was analyzed using the AlgPred server
| Parameter | Result |
|---|---|
| Allergenic potential | None |
| Antigenicity | ANTIGENpro = 0.846011 |
| Number of amino acids | 1039 |
| Molecular weight | 115.11 kDa |
| Theoretical isoelectric point | 6.02 |
| Instability index | 22.80 |
| Aliphatic index | 74.28 |
| GRAVY | − 0.525 |
| Estimated half-life | 30 h (mammalian reticulocytes, in vitro) |
| > 20 h (yeast, in vivo) | |
| > 10 h (E. coli, in vivo) |
B lymphocyte epitopes and binding analysis to MHC I and II
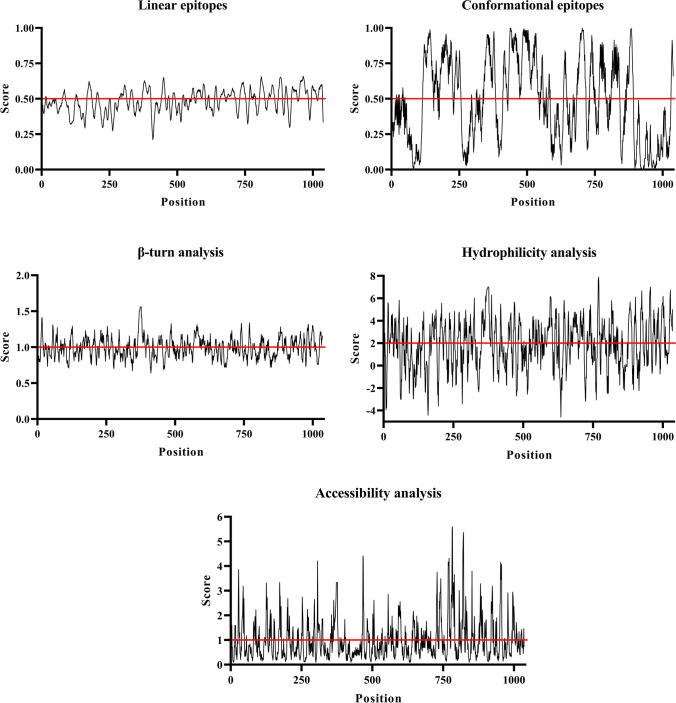
The fusion protein shows a range of possible regions that can be recognized as linear or conformational epitopes (Fig. 1). Additionally, the analyses of β-turn, epitope accessibility, and hydrophilicity show possible epitopes that can be recognized by B lymphocytes (Fig. 1). Table 2 shows the specific sequences most likely to bind to class I and II MHC molecules from mice, chosen for binding to all tested alleles, and overlap sequences that can bind to both molecules.
Fig. 1.
Prediction of MBP:PLD:CP40 fusion protein linear and conformational epitopes likely to be recognized by B cells, and epitope analyses through amino acid characterization. The parameters evaluated were β-turn, accessibility, and hydrophilicity of conformational epitopes likely to be recognized by B cells. The cut-off point was established as recommended by the IEDB server and is represented by a red line within the graph
Table 2.
Fusion protein aminoacid sequences most likely to bind to class I and II MHC from mice (Mus musculus). The underlined sequences represent the overlapping regions for MHC I and II
| Protein | Start | End | Sequence |
|---|---|---|---|
| PLD | 394 | 408 | QSMASPASTANRPVY |
| 420 | 434 | VDDAVAIGANALEID | |
| 449 | 463 | IPTSAGATAEEIFKH | |
| 506 | 520 | YLEPAGVRVLYGFYK | |
| 592 | 606 | LRKSSEARDQGKLGK | |
| 626 | 640 | KANVDGLIFGFKITH | |
| 653 | 667 | AIKRWVDKHSATHHL | |
| CP40 | 697 | 711 | QAPLKASPGHADKVG |
| 794 | 808 | VVRTVGAQLLLNKIK | |
| 859 | 873 | QWQLRKIMGAFSELM | |
| 917 | 931 | LAQTYDKGTKESIDQ | |
| 953 | 967 | EENDTNRFLTAVGEV | |
| 1002 | 1016 | GDDFTTLKPTDFAFT |
Docking results for the binding with TLR2
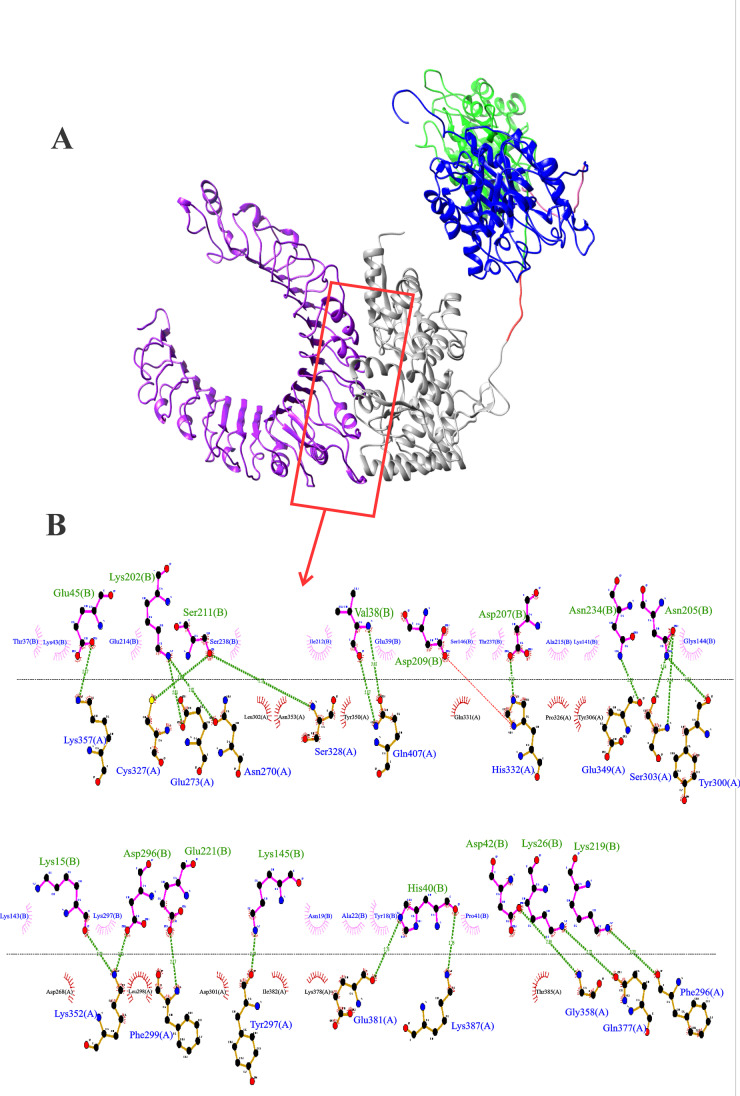
Figure 2 shows the representation of the docking between the fusion protein and TLR2. It was possible to confirm that the MBP:PLD:CP40 fusion protein has several docking regions to the TLR2 receptor. The interaction in which there was a greater variation in the Gibbs free energy and the possible bonds between the MBP and the receptor are shown in Fig. 2. It also can be seen the residue interactions in Fig. 2B, presenting 21 hydrogen bonds and 29 hydrophobic interactions, denoting a stable ligation between the MBP and the TLR2. Specifically, Glu45, Lys 202, Ser211, Val38, Asp207, Asn234, Asn205, Lys16, Asp296, Glu221, Lys145, His40, Asp 42, Lys2,6 and Lys219 amino acid residues of the MBP protein are able to form hydrogen bonds with the Lys357. Cys327, Glu273, Asn270, Ser328, Gln407, His332, Glu349, Ser303, Tyr 320, Lys 352, Phe 299, Tyr 297, Glu381, Lys 387, Gly 328, Gln377, and Phe 296 amino acid residues of the TLR2. Regarding the hydrophobic interactions, the amino acid residues of the MPB Thr37, Lys43, Glu214, Ser238, Ile212, Glu39, Ser146, Thr237, Ala215, Lys 141, Gly144, Lys143, Lys297, Asn19, Ala22, Tyr18 and Pro41, and the Leu302, Asn353, Tyr350, Gln331, Pro326, Tyr306, Asp268, Leu298, Asp301, Ile382, Lys378, and Thr385 amino acid residues of the TLR are envolved.
Fig. 2.
Anchoring of the MBP:PLD:CP40 fusion protein to the TLR2 receptor. A Structure generated by the Chimera software; TLR2 (receptor) in purple and MBP:PLD:CP40 protein (ligand) with MBP (grey), TEV protease site (red), PLD (green) rigid linker (pink), and CP40 (blue). B Residue interactions between proteins provided by the LIGPLOT v.2.2 software. Green lines represent hydrogen bonds, semicircles denote hydrophobic interactions. It can be seen 21 hydrogen bonds and 29 hydrophobic interactions between MBP and the TLR2. The MBP aminoacid envolved in the hydrogen bonds are written in green and followed by the letter (B), and the TLR2 residues envolved in these bonds are written in blue and followed by the letter (A). The MBP and TLR2 amino acid residues that participate in the hydrophobic interactions are represented inside pink and red semicircles, respectively
Expression and purification of MBP:PLD:CP40
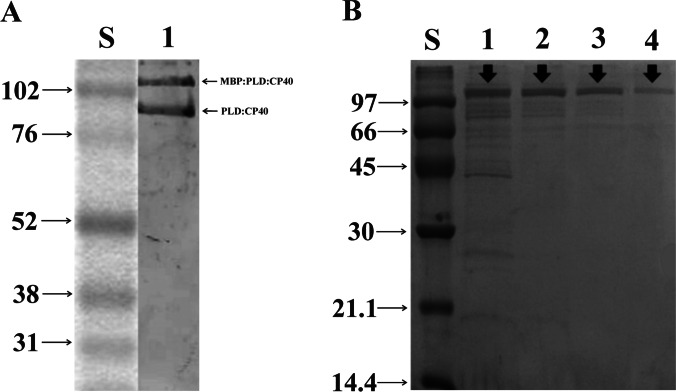
The MBP:PLD:CP40 protein was expressed the expected molecular weight of 115 kDa and that the MBP part can be cleaved and the PLD:CP40 protein can be obtained (Fig. 3A). The purity of the samples after four eluating conditions (imidazole 40 mM, 60 mM, 80 mM, and 400 mM) (Fig. 3B). After obtaining a significant degree of purity, the DLS analysis showed that the protein is monodispersed and free of protein aggregates and secondary structures (Fig. S4). Additionally, the expressed protein could be recognized by antibodies from serum samples of sheep and goats naturally infected by C. pseudotuberculosis, as seen by the differences of the OD results in C. pseudotuberculosis-negative and positive animals (Table S2).
Fig. 3.
Expression and purification evaluation of the MBP:PLD:CP40 fusion protein. A Western blot using anti-polyhistidine antibody for the detection of MBP:PLD:CP40 and PLD:CP40. S—Molecular weight in kDa; 1—MBP:PLD:CP40 and PLD:CP40. B 15% SDS-PAGE evidencing the results of the fusion protein purification. S—Molecular weight in kDa. Washes were performed with different concentrations of imidazole: 1 – 40 mM; 2 – 60 mM; 3 – 80 mM, and 4—elution with 400 mM. Black arrows indicate the presence of the respective proteins
Circular dichroism spectroscopy
The UV-CD spectrum characteristic of MBP:PLD:CP40 is shown in Fig. S5; additionally, the percentages of the secondary structure in solution are shown in Table S3. These results indicate that the MBP:PLD:CP40 fusion protein is correctly folded, and suitable for performing biological assays.
Humoral immune response
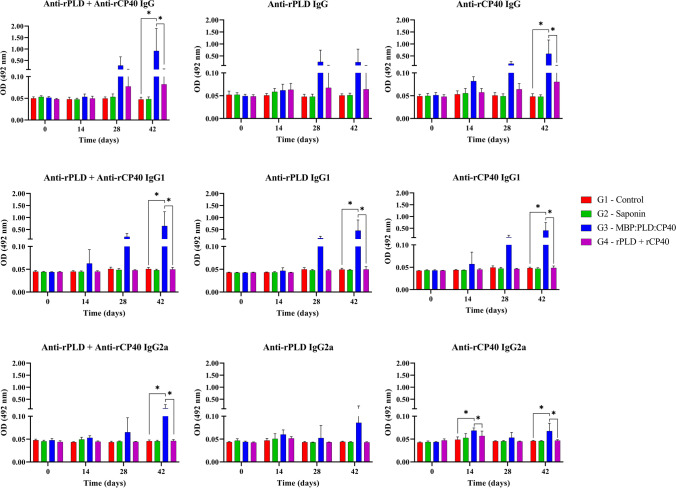
rPLD- and rCP40-specific antibodies were produced after immunization with the fusion protein. Total IgG antibodies were significantly (p < 0.05) produced in the G3 group 42 days after first immunization; however, an increase in antibody titers can be seen at 28 days after immunization (Fig. 4). Anti-rCP40 total IgG was more produced than anti-rPLD in the G3 group, as shown in Fig. 4 (p < 0.05).
Fig. 4.
Total IgG, IgG1, and IgG2a quantification during 42 days after the first immunization. Anti-rPLD, anti-rCP40 and anti-rPLD + anti-rCP40 total IgG, IgG1, and IgG2a were evaluated for all experimental groups: G1 – Control; G2 – Saponin; G3 – MBP:PLD:CP40 fusion protein; and G4 – rPLD + rCP40. Results are expressed as means of optical density (OD 492 nm) and standard errors; statistical differences (p < 0.05) among the groups are expressed as an “*” within each graph
The production of anti-rPLD and anti-rCP40 IgG1 antibodies (p < 0.05) was only significant in the G3 group after 42 days of immunization (p < 0.05), and statistical differences between G3 and G4 groups (Fig. 4). Regarding the IgG2a subtype, it was noticed a less pronounced production of this antibody when compared to IgG1 (Table 3), with no significant production of anti-rPLD (p < 0.05) in the G3 group (Fig. 4). Anti-rCP40 IgG2a antibodies were significantly produced at 14 and 42 days after the first immunization in the G3 group, with no significant production at 28 days, and presenting higher titles than the G4 group. No significant production was found in the control groups (G1 and G2) for any of the antibodies assessed (Fig. 4).
Table 3.
Ratio between the production of IgG subtypes IgG1 and IgG2a in the G3 group. The ratio was based on the mean value for each analysis at 42 days after the first immunization of the G3 group (MBP:PLD:CP40 fusion protein)
| Specific antibodies | IgG1/IgG2a |
|---|---|
| anti-rPLD | 4.83 |
| anti-rCP40 | 5.24 |
| anti-rPLD + rCP40 | 5.14 |
Cellular immune response
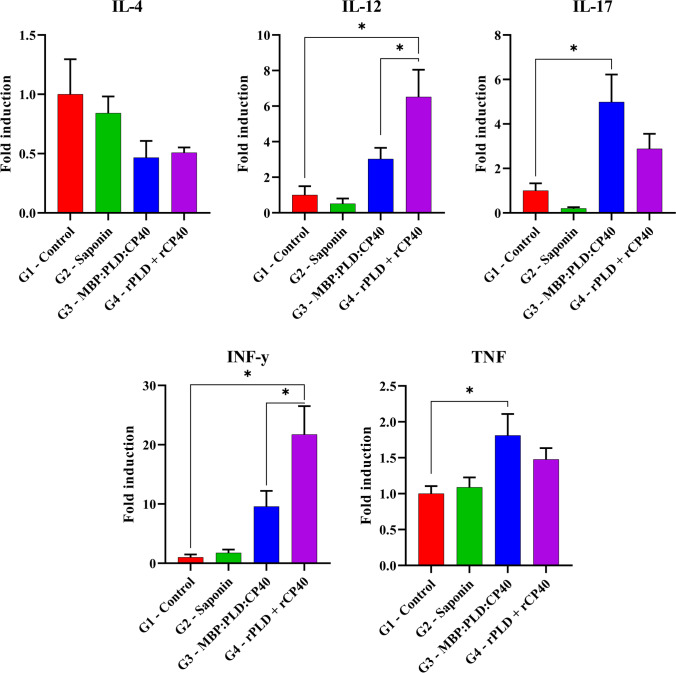
The IL-4 mRNA quantification showed no significant differences among the groups (p < 0.05). The G4 group significantly expressed more IL-12 and IFN-γ than the G3 group (p < 0.05). Regarding IL-17 and TNF, only the G3 group presented a significant expression when compared to the control group (p < 0.05). Higher IL-12 and IFN-γ levels were found in the G4 group (Fig. 5). G1 and G2 groups did not present significant levels of any of the cytokines evaluated herein.
Fig. 5.
Cytokine production profile after in vitro stimulation. G1 – Control; G2 – Saponin; G3 – MBP:PLD:CP40 fusion protein; and G4 – rPLD + rCP40. Forty-two days after the first immunization, mice were euthanized and the splenocytes were obtained, and stimulated. The quantification of IL-4, IL-12, IL-17, IFN-γ, and TNF expression was made through real-time PCR. Results are presented as the means of fold induction and standard errors. The symbol “*” indicate significant statistical differences between groups (p < 0.05), as defined by the one way ANOVA and Holm-Šídák’s multiple comparisons tests
Survival analysis
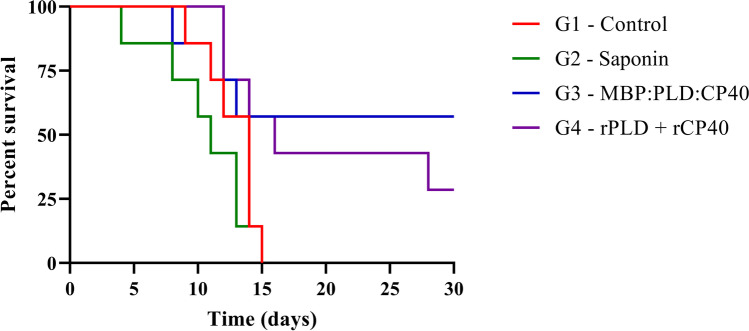
After the virulent C. pseudotuberculosis MIC-6 strain challenge, a survival curve was constructed based on a 30-day monitoring (Fig. 6). All animals from G1 and G2 groups were dead after 15 days of challenge, and the immunized G3 and G4 groups had a final percent survival of 57.14% (4/7) and 42.86% (3/7), respectively after the 30 days of observation. Through the Log-rank test, all curves were significantly different (p < 0.05), and the final percent survival of the G3 group was significantly higher than the G4 group (p < 0.05). Additionally, lesions were detected on the kidneys, liver, lungs, spleen, and heart. The total number of lesions was lower in the fusion protein group than the other groups – 05 lesions in the mice inoculated with the fusion protein, 13 in the control group, 16 in the saponin group, and 13 in the group inoculated with rPLD and rCP40 (Table 4).
Fig. 6.
Survival curve after experimental challenge with the virulent MIC-6 strain of C. pseudotuberculosis. A 30-days follow-up of seven animals from each group: G1 – Control; G2 – Saponin; G3 – MBP:PLD:CP40 fusion protein; and G4 – rPLD + rCP40. Results are expressed as percent of survival. The G3 and G4 curves were statistically different (p < 0.05) from the G1 and G2 curves, as defined by the Log-rank and Fisher’s tests
Table 4.
Macroscopy evaluation of lesions found on mice experimentally infect by C. pseudotuberculosis. Organs were obtained after necropsy and were then analyzed by veterinarian pathologists
| Organs | Lesions | |||
|---|---|---|---|---|
| G1 | G2 | G3 | G4 | |
| Kidneys | Multifocal granulomatous nephritis (n = 4) | Multifocal granulomatous nephritis (n = 5) | Multifocal granulomatous nephritis (n = 2) | Multifocal granulomatous nephritis (n = 5) |
| Liver | Mild degeneration and multifocal hepatitis/necrosis; focal granulomatous hepatitis (n = 5) | Mild degeneration and multifocal hepatitis/necrosis; focal granulomatous hepatitis (n = 5) | Mild degeneration and multifocal hepatitis/necrosis; focal granulomatous hepatitis (n = 2) | Mild degeneration and multifocal hepatitis/necrosis; focal granulomatous hepatitis (n = 5) |
| Lungs | Focal granulomatous pneumonia (n = 1) | – | – | – |
| Spleen | – | Focal granulomatous splenitis (n = 2) | – | – |
| Heart | – | – | Focal granulomatous myocarditis (n = 1) | Focal granulomatous myocarditis (n = 1) |
| Systemic | Congestion and hemorrhagic lesions (n = 3) | Congestion and hemorrhagic lesions (n = 4) | – | Congestion and hemorrhagic lesions (n = 2) |
| Total of lesions | 13 | 16 | 5 | 13 |
Discussion
Nowadays, one of the most used tools for vaccine development is reverse vaccinology, which uses immunoinformatics to evaluate new targets and predict epitopes capable of being recognized and presented by the immune system (Rappuoli 2001). Thus, we used immunoinformatics approaches to develop the MBP:PLD:CP40 fusion protein, and tested its immunoprophylaxis potential in vivo using a murine model.
The novelty regarding C. pseudotuberculosis immunogens that were employed in the MBP:PLD:CP40 design is the presence of the maltose-binding protein (MBP), which has already been described in the literature as an intrinsic adjuvant in vaccine models based on fusion proteins, inducuing the immune system activation through binding to Toll-Like 2 and 4 receptors (Gong et al. 2015; Liu et al. 2017; Wang et al. 2015), which can be found on antigen-presenting cells (APCs). Nevertheless, MBP lacks other important adjuvant functions and cannot be the only adjuvant molecule used in a vaccine construct, and saponins are largely used for C. pseudotuberculosis vaccine trials for preventing the protein denaturation and aiding the protein to be released in a slower manner, as seen in recent studies (Bezerra et al. 2021; Pinho et al. 2021b). It was seen that the fusion protein binds to the TLR2 receptor through the MBP, with a total of 21 hydrogen bonds and 29 hydrophobic interactions, and can assist in the recognition of the antigen and its presentation to T cells through the activation of antigen presenting cells (Kumar et al. 2019). The role of TLR2 functions in C. pseudotuberculosis infection is not yet described; however, it plays an important role in infections by mycobacteria through the recognition of several mycobacterial components and inflammatory responses (Hu and Spaink 2022). Additionally, it was already been described that mycobacteria may use TLR2 inhibition as an invading mechanism, through the PPE51 protein which inhibits autophagy by suppressing this receptor (Strong et al. 2022).
Although immunoinformatics analyses are of great help in terms of time/cost parameters, predictions are not formal evidence, and they do not entirely replace traditional vaccinal research methods (Oli et al. 2020), and the confirmation of the results obtained in silico is required. The expressed MBP:PLD:CP40 fusion protein, presented an approximate molecular weight of 115 kDa, as previously predicted, and despite the high molecular weight and the presence of three whole proteins, it was stable as previously in silico predicted and did not present any interfering secondary structures that could hampered its immunological functions, as seen by the DLS and circular dichroism results.
The MBP:PLD:CP40 protein presented several epitopes for B lymphocytes, both linear and conformational, bearing several regions with accessible and hydrophilic β-turn conformations, capable of being recognized and interact with antibodies (Novotný et al. 1986). Due to its large size and presence of several B and T epitopes, this fusion protein may modulate both humoral and cellular immune responses in a more diverse way than quimeric antigens that present a limited number of epitopes (Baranov et al. 2021). It can also be cited that quimeric and synthetic peptide vaccines can present low solubility and stability (Patarroyo et al. 2002), and in this way they may remain in the host organism for a short time, significantly interfering in its capacity to interact with immune cells. The in vivo analysis confirmed the immunoinformatic results by showing a significant production of IgG antibodies after immunization with the fusion protein, mainly IgG1 subclass, associated with a lower presence of IgG2a. A higher production of IgG1 in mice, when compared to IgG2a, is a consequence of a tendency to develop a Th2 response (Mills et al. 2000). As Th1 and Th2 cells secrete different cytokines, humoral immune responses develop through the production of distinct antibody isotype patterns, with Th1-type immunity associated with the IgG2a subclass and Th2-type with antibodies of the IgG1 subclass in mice (Germann et al. 1995).
It was already described that high IgG2a levels, in the absence of detectable IgG1, were able to protect mice immunized with rCP40 (Droppa-Almeida et al. 2016). Recent studies aiming to develop a new CLA vaccine have shown a predominance of IgG2a production over the IgG1 subclass (Bezerra et al. 2021; Rezende et al. 2020; Silva et al. 2020). Differently, Silva and collaborators (Silva et al. 2014) have also employed the rCP40 alone and in combination with a mutant C. pseudotuberculosis strain (CP09) as an immunogen and observed a more pronounced Th2 response, which is characterized by the induction of the production of neutralizing antibodies, leading to a partial protection against C. pseudotuberculosis infection through the neutralization of exotoxins, such as PLD. This fact was also seen herein, with the MBP:PLD:CP40 leading to a five-fold higher production of IgG1 when compared to IgG2a, suggesting that the fusion protein elicited a response characterized by a Th2 profile rather than a Th1 response. It must be also considered that Th2 antagonizes Th1 responses (Abdelaziz et al. 2021) and, as the development of granulomes is a result of exacerbated cellular immune responses based on excessive chemotaxis (Guler et al. 2021), the Th2 response can also regulate this process and inhibit the formation of caseous lymphadenitis lesions.
The pathways that make Th cells specialize into Th1 or Th2 profiles are regulated by different stimuli, such as the presence of specific cytokines and immunologically active hormones, the dose and route of antigen administration, and the type of APCs stimulating the T cells, and the lack of Th1 profile stimuli could lead to the polarization to Th2 response triggered by MBP:PLD:CP40 (Abebe and Bjune 2009). Considering this situation, it is possible that the protein dose used in the present study drove the cellular response to a Th2 profile, and its partial protection can be related to the neutralization of the exotoxins produced by C. pseudotuberculosis, or even to the inhibition of cytokines that drive the formation of caseous granulomes, since IFN-γ, IL-12 and TNF are crucial for this particular process and are significantly produced when a Th1 profile is induced, and this induction can be inhibited by a Th2 profile (Munk and Emoto 1995; Ferluga et al. 2020).
Another fact that must be considered in the Th2 polarization induced by the vaccinal formulations is the adjuvant that was used herein. Saponins are traditionally used in veterinary vaccines with significant result results (Fernandes et al. 2008); however, it can induce, in some cases, a more pronounced humoral response than a Th1-mediated one (Wang 2021). Saponin was used in this experiment since it was already employed as an adjuvant in other caseous lymphadenitis vaccinal trials (Bezerra et al. 2021; Droppa-Almeida et al. 2021), and is being now used even in commercial veterinary vaccines (Fernandes et al. 2008); however, the use of an adjuvant that can drive to a higher production of IFN-γ, with a consequent bacterial clearance through activated phagocytosis, can be included in further trials using the fusion protein.
The fusion protein stimulated the production of both anti-PLD and anti-CP40 specific antibodies, despite what was previously reported in the literature about C. pseudotuberculosis PLD and CP40 proteins, where the first is an inducer of substantial antibody production and the second stimulate cellular immunity (Selim et al. 2010; Silva et al. 2014). Although specific antibody production was nearly equal between the two proteins, anti-CP40 was more produced than anti-PLD, especially considering that the total IgG and IgG2a subclass showed no statistical difference in anti-PLD production. Interestingly, no antibody production was observed when rPLD and rCP40 were combined to immunize mice. This fact could have happened due to the amount of protein herein used (25 µg each) since it has already been described a significant antibody production when rPLD or rCP40 alone were used in a higher quantity (50 µg) (Leal et al. 2018; Pinho et al. 2021b; Silva et al. 2014). Class II MHC genes regulate the humoral antibody response to protein antigens (Egea et al. 1991) in mice and other mammalian species. Furthermore, TLRs are necessary for further antibody production, and TLR signaling is required for DC maturation and T and B cell activation (Delgado et al. 2009); therefore, a low amount of rPLD and rCP40 could hamper antibody production due to the lack of B cell stimulation.
Due to the presence of epitopes capable of being recognized by several alleles of mouse MHC I and II, it was predicted that the fusion protein would activate the cellular immune system when tested in vivo. IL-4, like many cytokines, can affect a wide range of target cells in a variety of ways, and it plays a distinct and essential role in regulating antibody production, inflammation, and the development of effector T cell responses (Brown and Hural 2017). Nevertheless, when production of IL-4 was assessed, no significant mRNA expression was observed in mice immunized with the MBP:PLD:CP40 fusion protein, nor when rPLD and rCP40 were used herein. Only a subset of activated T cells, mast cells, and basophils express detectable amounts of IL-4, and stimulation of IL-4 mRNA expression is transient: peak levels are only detected six hours postinduction and then fall precipitously (Brown and Hural 2017).
IL-12 has been recognized as a cytokine that connects innate and adaptive immunity because it primarily induces IFN-γ production from Th and NK cells in the early stages of the immune response and induces the differentiation of T cells in Th1 effectors, appropriate for protection against bacterial infection (Méndez-Samperio 2010). Interestingly, the immunization with MBP:PLD:CP40 did not induce a Th2 profile cytokine (IL-4) nor Th1 profile’s IFN-γ and IL-12 cytokines. However, when rPLD and rCP40 were applied as an immunogen in mice, we observed a significantly higher production of IFN-γ and IL-12. IFN‑γ acts mainly through phagosome maturation, programmed death activation of host cells, and ROS generation (Ghanavi et al. 2021), acting as an essential molecule in intracellular pathogens’ elimination. The fact that MBP:PLD:CP40 did not significantly induce such cytokines could be related to the recognition of the MBP by the TLR2, since the engagement of these ligands during bacterial phagocytosis can lead, in particular cases, to the inhibition of cytokine synthesis and decreased cytokine responsiveness (Huynh et al. 2011); this fact must be considered since the TLR2 recognizes the fusion protein through the MBP and this protein is not present when rPLD and rCP40 were separately used to immunize mice. It can also be speculated that the formation of secondary structures that is usually seen in the expression of large proteins such as the fusion protein use in this study, can hamper the correct immune activation by the fusion antigen. This was not the case of our fusion protein, since the DLS and the circular dichroism spectroscopy assays showed not only a correct folding, but also the absence of such structures; however, secondary structures can be formed in in vivo scenarios (Tretyachenko et al. 2017).
While MBP:PLD:CP40 failed to induce IL-4, IL-12, and IFN-γ, there was a significant production of TNF after antigenic stimulation in vitro. TNF is primarily produced by macrophages early after a challenge and is involved in the innate phase of the immune response, playing a critical role in the defense against intracellular pathogens, acting as a pro-inflammatory factor, and is costimulatory to B cells (Körner et al. 2010). Although a high recognition of MBP:PLD:CP40 by TLR2 could suppress cytokine signaling (Huynh et al. 2011), it is likewise plausible that this recognition profile has induced the production of TNF observed herein for the fusion protein. Similarly, the lack of TNF stimulation in the group immunized with rPLD and rCP40 can be due to a failure to significant stimulate the innate immune system. It is noteworthy to say that the significant production of TNF seen in the fusion protein group could be led to the partial protection seen after challenge since it has already been described that a more efficient and rapid innate immune response can lead to a better prognosis of caseous lymphadenitis (Bastos et al. 2013).
Interestingly, the fusion protein induced a significant production of IL-17 whereas rPLD and rCP40 did not, and although the role of IL-17 in C. pseudotuberculosis infection is yet unclear, Th17 cells are widely considered to guide cellular traffic towards infected lesions (Orme and Ordway 2016). IL-17 is a significant pro-inflammatory cytokine with immunomodulatory functions, one of which is to coordinate tissue inflammation by promoting the expression of pro-inflammatory cytokines such as TNF (Abebe and Bjune 2009), which could also explain the significant levels of TNF observed in the MBP:PLD:CP40 group. Unfortunately, there are no records on the importance of IL-17 in the C. pseudotuberculosis infection; however, IL-17 is induced during both the innate and adaptive immune responses to Mycobacterium bovis (a microorganism phylogenetically related to C. pseudotuberculosis) infection, and is needed for granuloma development (Curtis and Way 2009), being plausible to infer that this may also occur in C. pseudotuberculosis infection. Recent studies on vaccine development for CLA have shown a specific production of IL-17 after in vitro solenocyte stimulation (Bezerra et al. 2021; Silva et al. 2020), as seen herein. Although the immunomodulation of Th17 by vaccines is a very recent and unexploited issue, it was already described that different antigenic emulsions, even those containing saponins (Khademi et al. 2019; Phelps et al. 2020), were able to induce immunoprotective responses associated with IL-17 production (Doligalska et al. 2013).
Considering both humoral and cellular immune responses elicited by immunization of mice with MBP:PLD:CP40, it was observed a more pronounced production of IgG1 rather than IgG2a, and significant expression of TNF and IL-17 cytokine mRNAs, and the fusion protein conferred a 57.14% protection after challenge with a virulent strain of C. pseudotuberculosis. A 42.86% protection was observed after mice immunized herein with rPLD and rCP40 were experimentally challenged; however, this group presented a significant production of IFN-γ and IL-12 but no significant antibody production. It is noteworthy to say that low antigen doses induce, in some cases, an exclusively cell-mediated reaction. Medium doses produce a cell-mediated response that changes into a humoral one over time; and large doses produce faster responses, sometimes with a scarcely detectable cell-mediated response (Bretscher 2014), which can be seen when lower doses of antigens are used. In this study, it was used 25 µg of rPLD and rCP40 each, since it was a control for the 50ug fusion protein dose, and in this way this lower dose could be responsible for this partial protection and immune induction.
An antigenic formulation containing an E. coli bacterin expressing the PLD protein was already tested in a murine model, which conferred a 40% protection; in this same study, when a vaccine composed only of rPLD was used, it could be observed a 30% protection (Pinho et al. 2021b). The use of a genetically inactivated rPLD as a vaccinal antigen, in the same way that was made in our study but conducted in sheep, was able to protect 44% of the vaccinated sheep against a virulent C. pseudotuberculosis challenge (Hodgson et al. 1999). Regarding the CP40 protein, when peptides derived from this protein sequence were tested in a murine model, they conferred protection levels that varied from 10 to 40% (Droppa-Almeida et al. 2021). The 57.14% percent survival that was reached in our study when the fusion protein was used suggest this vaccinal formulation can represent a higher number of epitopes that are able to elicit both the humoral and cellular response, acting on a complementary way to induce a more protective response, along with the adjuvant effect of the MBP. In a similar way, Barral et al. (2019) was able to develop an immunoenzymatic assay combining rPLD and rCP40 that presented higher levels of sensitivity and specificity than other immunodiagnosis assays that were developed to detect anti-C. pseudotuberculosis antibodies in goats and sheep (Menzies et al. 2004; Rebouças et al. 2013; Rezende et al. 2016).
In conclusion, the immunoinformatics analyses predicted that the MBP:PLD:CP40 fusion protein is antigenic, does not have allergenic potential, and contains multiple epitopes that can be recognized by B lymphocytes and presented to T lymphocytes via MHC I and II molecules. The interaction with innate immune receptors can also be seen in the prediction of protein binding to the Toll-Like 2 receptor, demonstrating its intrinsic adjuvant role. In vivo analyses revealed that the fusion protein conferred a 57.14% percent survival following a virulent challenge, higher than the 42.86% reported when rPLD and rCP40 were used, with a considerable production of IgG1 antibody, as well as TNF and IL-17 cytokines. Hence, the MBP:PLD:CP40 fusion protein is a potential antigen for immunoprophylaxis of caseous lymphadenitis; however, more studies involving dosage adjustments in a small ruminant model are needed to understand better its protective efficacy in the primary hosts of C. pseudotuberculosis.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are grateful to Francisca Soares and Luiz Eduardo Lacerda (LABIMUNO-UFBA) for technical assistance.
Author contribution
TDB and RWP conceived and designed the study. All authors performed experimental procedures and analyzed the data. TDB drafted the manuscript. TDB and RWP proofread the manuscript, which all authors revised and approved for submission.
Funding
This study was funded by the Fundação de Apoio à Pesquisa e Extensão (FAPEX). TDB and MAK are PhD fellows from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). VA is a Research Fellow from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). RWP is a Technical Development fellow from CNPQ (Proc. 313350/2019–1).
Data availability
All data generated or analyzed during this study are included in this article (and its supplementary information files).
Declarations
Ethics approval
This project was approved by the Ethics Committee of the School of Veterinary Medicine and Zootechnics of the Federal University of Bahia (permission number 19/2021).
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abdelaziz MH, Ji X, Wan J, Abouelnazar FA, Abdelwahab SF, Xu H. Mycobacterium-induced Th1, helminths-induced Th2 Cells and the potential vaccine candidates for allergic asthma: imitation of natural infection. Front Immunol. 2021;12:696734. doi: 10.3389/fimmu.2021.696734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abebe F, Bjune G. The protective role of antibody responses during Mycobacterium tuberculosis infection. Clin Exp Immunol. 2009;157:235–243. doi: 10.1111/j.1365-2249.2009.03967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov MV, Kumar M, Sacanna S, Thutupalli S, van den Bogaart G. Modulation of immune responses by particle size and shape. Front Immunol. 2021;11:607945. doi: 10.3389/fimmu.2020.607945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral TD, Mariutti RB, Arni RK, Santos AJ, Loureiro D, Sokolonski AR, Azevedo V, Borsuk S, Meyer R, Portela RD (2019) A panel of recombinant proteins for the serodiagnosis of caseous lymphadenitis in goats and sheep. Microb Biotechnol 12:1313–1323. 10.1111/1751-7915.13454 [DOI] [PMC free article] [PubMed]
- Bastos BL, Loureiro D, Raynal JT, Guedes MT, Vale VL, Moura-Costa LF, Guimarães JE, Azevedo V, Portela RW, Meyer R. Association between haptoglobin and IgM levels and the clinical progression of caseous lymphadenitis in sheep. BMC Vet Res. 2013;9:254. doi: 10.1186/1746-6148-9-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra FSB, Silva MTO, Rezende AFS, Lopes AS, Pinho RB, Seixas FK, Collares TV, Portela RWD, Azevedo V, Borsuk S. Saponin-adjuvanted recombinant vaccines containing rCP00660, rCP09720 or rCP01850 proteins against Corynebacterium pseudotuberculosis infection in mice. Vaccine. 2021;18:2568–2574. doi: 10.1016/j.vaccine.2021.03.062. [DOI] [PubMed] [Google Scholar]
- Bretscher PA. On the mechanism determining the TH1/TH2 phenotype of an immune response, and its pertinence to strategies for the prevention, and treatment, of certain infectious diseases. Scand J Immunol. 2014;79:361–376. doi: 10.1111/sji.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MA, Hural J. Functions of IL-4 and control of its expression. Crit Rev Immunol. 2017;37:181–212. doi: 10.1615/CritRevImmunol.v37.i2-6.30. [DOI] [PubMed] [Google Scholar]
- Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33:W72–76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921. [DOI] [PubMed] [Google Scholar]
- Curtis MM, Way SS. Interleukin-17 in host defence against bacterial, mycobacterial and fungal pathogens. Immunology. 2009;126:177–185. doi: 10.1111/j.1365-2567.2008.03017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado MF, Coviello S, Monsalvo AC, Melendi GA, Hernandez JZ, Batalle JP, Diaz L, Trento A, Chang HY, Mitzner W, Ravetch J, Melero JA, Irusta PM, Polack FP. Lack of antibody affinity maturation due to poor Toll-like receptor stimulation leads to enhanced respiratory syncytial virus disease. Nat Med. 2009;15:34–41. doi: 10.1038/nm.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doligalska M, Joźwicka K, Laskowska M, Donskow-Łysoniewska K, Pączkowski C, Janiszowska W. Changes in Heligmosomoides polygyrus glycoprotein pattern by saponins impact the BALB/c mice immune response. Exp Parasitol. 2013;135:524–531. doi: 10.1016/j.exppara.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Dorella FA, Pacheco LG, Oliveira SC, Miyoshi A, Azevedo V. Corynebacterium pseudotuberculosis: microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Vet Res. 2006;37:201–218. doi: 10.1051/vetres:2005056. [DOI] [PubMed] [Google Scholar]
- Dorella FA, Pacheco LG, Seyffert N, Portela RW, Meyer R, Miyoshi A, Azevedo V. Antigens of Corynebacterium pseudotuberculosis and prospects for vaccine development. Expert Rev Vaccines. 2009;8:205–213. doi: 10.1586/14760584.8.2.205. [DOI] [PubMed] [Google Scholar]
- Droppa-Almeida D, Vivas WL, Silva KK, Rezende AF, Simionatto S, Meyer R, Lima-Verde IB, Delagostin O, Borsuk S, Padilha FF. Recombinant CP40 from Corynebacterium pseudotuberculosis confers protection in mice after challenge with a virulent strain. Vaccine. 2016;34:1091–1096. doi: 10.1016/j.vaccine.2015.12.064. [DOI] [PubMed] [Google Scholar]
- Droppa-Almeida D, da Silva GA, Gaspar LMDAC, Pereyra BBS, Nascimento RJM, Borsuk S, Franceschi E, Padilha FF. Peptide vaccines designed with the aid of immunoinformatic against Caseous Lymphadenitis promotes humoral and cellular response induction in mice. PLoS One. 2021;16:e0256864. doi: 10.1371/journal.pone.0256864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dummer LA, Araujo IL, Finger PF, Santos AG, Jr, Rosa MC, Conceição FR, Fischer G, van Drunen Littel-van den Hurk, Leite FP. Immune responses of mice against recombinant bovine herpesvirus 5 glycoprotein D. Vaccine. 2014;32:2413–2419. doi: 10.1016/j.vaccine.2014.03.011. [DOI] [PubMed] [Google Scholar]
- Egea E, Iglesias A, Salazar M, Morimoto C, Kruskall MS, Awdeh Z, Schlossman SF, Alper CA, Yunis EJ. The cellular basis for lack of antibody response to hepatitis B vaccine in humans. J Exp Med. 1991;173:531–538. doi: 10.1084/jem.173.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emini EA, Hughes JV, Perlow DS, Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55:836–839. doi: 10.1128/JVI.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferluga J, Yasmin H, Al-Ahdal MN, Bhakta S, Kishore U. Natural and trained innate immunity against Mycobacterium tuberculosis. Immunobiology. 2020;225:151951. doi: 10.1016/j.imbio.2020.151951. [DOI] [PubMed] [Google Scholar]
- Fernandes AP, Costa MM, Coelho EA, Michalick MS, de Freitas E, Melo MN, Luiz Tafuri W, Resende Dde M, Hermont V, Abrantes Cde F, Gazzinelli RT. Protective immunity against challenge with Leishmania (Leishmania) chagasi in beagle dogs vaccinated with recombinant A2 protein. Vaccine. 2008;26:5888–5895. doi: 10.1016/j.vaccine.2008.05.095. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The proteomics protocols handbook: springer protocols handbooks. Nova Jersey: Humana Press; 2005. pp. 571–607. [Google Scholar]
- Germann T, Bongartz M, Dlugonska H, Hess H, Schmitt E, Kolbe L, Kölsch E, Podlaski FJ, Gately MK, Rüde E. Interleukin-12 profoundly up-regulates the synthesis of antigen-specific complement-fixing IgG2a, IgG2b and IgG3 antibody subclasses in vivo. Eur J Immunol. 1995;25:823–829. doi: 10.1002/eji.1830250329. [DOI] [PubMed] [Google Scholar]
- Ghanavi J, Farnia P, Farnia P, Velayati AA. The role of interferon-gamma and interferon-gamma receptor in tuberculosis and nontuberculous mycobacterial infections. Int J Mycobacteriol. 2021;10:349–357. doi: 10.4103/ijmy.ijmy_186_21. [DOI] [PubMed] [Google Scholar]
- Gong Z, Martin-Garcia JM, Daskalova SM, Craciunescu FM, Song L, Dörner K, Hansen DT, Yang JH, LaBaer J, Hogue BG, Mor TS, Fromme P. Biophysical characterization of a vaccine candidate against hiv-1: the transmembrane and membrane proximal domains of HIV-1 gp41 as a maltose binding protein fusion. PLoS ONE. 2015;10:e0136507. doi: 10.1371/journal.pone.0136507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler R, Ozturk M, Sabeel S, Motaung B, Parihar SP, Thienemann F, Brombacher F. Targeting molecular inflammatory pathways in granuloma as host-directed therapies for tuberculosis. Front Immunol. 2021;12:733853. doi: 10.3389/fimmu.2021.733853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson AL, Carter K, Tachedjian M, Krywult J, Corner LA, McColl M, Cameron A. Efficacy of an ovine caseous lymphadenitis vaccine formulated using a genetically inactive form of the Corynebacterium pseudotuberculosis phospholipase D. Vaccine. 1999;17:802–808. doi: 10.1016/s0264-410x(98)00264-3. [DOI] [PubMed] [Google Scholar]
- Hu W, Spaink HP. The role of TLR2 in infectious diseases caused by mycobacteria: from cell biology to therapeutic target. Biology. 2022;11:246. doi: 10.3390/biology11020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KK, Joshi SA, Brown EJ. A delicate dance: host response to mycobacteria. Curr Opin Immunol. 2011;23:464–472. doi: 10.1016/j.coi.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Khademi F, Yousefi A, Derakhshan M, Najafi A, Tafaghodi M. Enhancing immunogenicity of novel multistage subunit vaccine of Mycobacterium tuberculosis using PLGA:DDA hybrid nanoparticles and MPLA: Subcutaneous administration. Iran J Basic Med Sci. 2019;22:893–900. doi: 10.22038/ijbms.2019.33962.8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner H, McMorran B, Schlüter D, Fromm P. The role of TNF in parasitic diseases: still more questions than answers. Int J Parasitol. 2010;40:879–888. doi: 10.1016/j.ijpara.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Kumar S, Sunagar R, Gosselin E. Bacterial Protein Toll-Like-Receptor Agonists: A Novel Perspective on Vaccine Adjuvants. Front Immunol. 2019;10:1144. doi: 10.3389/fimmu.2019.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- Leal KS, Silva MTO, Rezende AFA, Bezerra FSB, Begnini K, Seixas F, Collares T, Dellagostin O, Portela RW, Azevedo V, Borsuk S. Recombinant M. bovis BCG expressing the PLD protein promotes survival in mice challenged with a C. pseudotuberculosis virulent strain. Vaccine. 2018;36:3578–3583. doi: 10.1016/j.vaccine.2018.05.049. [DOI] [PubMed] [Google Scholar]
- Liu G, Zhang Y, Zhang N, Ni W, Jie J, Jiang L, Tai G. Escherichia coli maltose-binding protein (MBP) activates mouse Th1 through TLR2-mediated MyD88-dependent pathway and TLR4-mediated TRIF-dependent pathway. Int Immunopharmacol. 2017;50:338–344. doi: 10.1016/j.intimp.2017.07.016. [DOI] [PubMed] [Google Scholar]
- Lu P, Feng MG. Bifunctional enhancement of a beta-glucanase-xylanase fusion enzyme by optimization of peptide linkers. Appl Microbiol Biotechnol. 2008;79:579–587. doi: 10.1007/s00253-008-1468-4. [DOI] [PubMed] [Google Scholar]
- Méndez-Samperio P. Role of interleukin-12 family cytokines in the cellular response to mycobacterial disease. Int J Infect Dis. 2010;14:e366–e371. doi: 10.1016/j.ijid.2009.06.022. [DOI] [PubMed] [Google Scholar]
- Menzies PI, Hwang YT, Prescott JF (2004) Comparison of an interferon-gamma to a phospholipase D enzyme-linked immunosorbent assay for diagnosis of Corynebacterium pseudotuberculosis infection in experimentally infected goats. Vet Microbiol 100:129–137. 10.1016/j.vetmic.2004.01.012 [DOI] [PubMed]
- Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol. 2000;164:6166–6173. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- Munk ME, Emoto M. Functions of T-cell subsets and cytokines in mycobacterial infections. Eur Respir J Suppl. 1995;20:668s–675s. [PubMed] [Google Scholar]
- Novotný J, Handschumacher M, Haber E, Bruccoleri RE, Carlson WB, Fanning DW, Smith JA, Rose GD. Antigenic determinants in proteins coincide with surface regions accessible to large probes (antibody domains) Proc Natl Acad Sci U S A. 1986;83:226–230. doi: 10.1073/pnas.83.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oli AN, Obialor WO, Ifeanyichukwu MO, Odimegwu DC, Okoyeh JN, Emechebe GO, Adejumo SA, Ibeanu GC. Immunoinformatics and vaccine development: an overview. Immunotargets Ther. 2020;9:13–30. doi: 10.2147/ITT.S241064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orme IM, Ordway DJ (2016) Mouse and guinea pig models of tuberculosis. Microbiol Spectr 4. 10.1128/microbiolspec.TBTB2-0002-2015 [DOI] [PubMed]
- Overbergh L, Valckx D, Waer M, Mathieu C. Quantification of murine cytokine mRNAs using real time quantitative reverse transcriptase PCR. Cytokine. 1999;11:305–312. doi: 10.1006/cyto.1998.0426. [DOI] [PubMed] [Google Scholar]
- Parker JM, Guo D, Hodges RS. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25:5425–5432. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- Patarroyo JH, Portela RW, De Castro RO, Pimentel JC, Guzman F, Patarroyo ME, Vargas MI, Prates AA, Mendes M. Immunization of cattle with synthetic peptides derived from the Boophilus microplus gut protein (Bm86) Vet Immunol Immunopathol. 2002;88:163–172. doi: 10.1016/s0165-2427(02)00154-x. [DOI] [PubMed] [Google Scholar]
- Phelps CC, Vadia S, Boyaka PN, Varikuti S, Attia Z, Dubey P, Satoskar AR, Tweten R, Seveau S. A listeriolysin O subunit vaccine is protective against Listeria monocytogenes. Vaccine. 2020;38:5803–5813. doi: 10.1016/j.vaccine.2020.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho RB, Silva MTO, Bezerra FSB, Borsuk S. Vaccines for caseous lymphadenitis: up-to-date and forward-looking strategies. Appl Microbiol Biotechnol. 2021;105:2287–2296. doi: 10.1007/s00253-021-11191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho RB, Silva MTO, Brenner G, Alves MSD, Azevedo V, Portela RD, Borsuk S. A novel approach for an immunogen against Corynebacterium pseudotuberculosis infection: an Escherichia coli bacterin expressing phospholipase D. Microb Pathog. 2021;151:104746. doi: 10.1016/j.micpath.2021.104746. [DOI] [PubMed] [Google Scholar]
- Rappuoli R. Reverse vaccinology, a genome-based approach to vaccine development. Vaccine. 2001;19:2688–2691. doi: 10.1016/s0264-410x(00)00554-5. [DOI] [PubMed] [Google Scholar]
- Rebouças MF, Loureiro D, Bastos BL, Moura-Costa LF, Hanna SA, Azevedo V, Meyer R, Portela RW. Development of an indirect ELISA to detect Corynebacterium pseudotuberculosis specific antibodies in sheep employing T1 strain culture supernatant as antigen. Pesq Vet Bras. 2013;33:1296–1302. doi: 10.1590/S0100-736X2013001100002. [DOI] [Google Scholar]
- Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezende AFS, Brum AA, Bezerra FSB, Braite DC, Sá GL, Thurow HS, Seixas FK, Azevedo V, Portela RW, Borsuk S. Assessment of the acid phosphatase CP01850 from Corynebacterium pseudotuberculosis in DNA and subunit vaccine formulations against caseous lymphadenitis. Arq Bras Med Vet Zootec. 2020;72:199–207. doi: 10.1590/1678-4162-10790. [DOI] [Google Scholar]
- Rezende AFS, Brum AA, Reis CG, Angelo HR, Leal KS, Silva MTO, Simionatto S, Azevedo V, Santos A, Portela RW, Dellagostin O, Borsuk S. In silico identification of Corynebacterium pseudotuberculosis antigenic targets and application in immunodiagnosis. J Med Microbiol. 2016;65:521–529. doi: 10.1099/jmm.0.000263. [DOI] [PubMed] [Google Scholar]
- Saha S, Raghava GP. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006;34:W202–209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selim SA, Ghoneim ME, Mohamed KF. Vaccinal efficacy of genetically inactivated phospholipase D against caseous lymphadenitis in small ruminants. Int J Microbiol Res. 2010;1:129–136. [Google Scholar]
- Silva JW, Droppa-Almeida D, Borsuk S, Azevedo V, Portela RW, Miyoshi A, Rocha FS, Dorella FA, Vivas WL, Padilha FF, Hernández-Macedo ML, Lima-Verde IB. Corynebacterium pseudotuberculosis cp09 mutant and cp40 recombinant protein partially protect mice against caseous lymphadenitis. BMC Vet Res. 2014;10:965. doi: 10.1186/s12917-014-0304-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva MTO, Bezerra FSB, de Pinho RB, Begnini KR, Seixas FK, Collares T, Portela RD, Azevedo V, Dellagostin O, Borsuk S. Association of Corynebacterium pseudotuberculosis recombinant proteins rCP09720 or rCP01850 with rPLD as immunogens in caseous lymphadenitis immunoprophylaxis. Vaccine. 2018;36:74–83. doi: 10.1016/j.vaccine.2017.11.029. [DOI] [PubMed] [Google Scholar]
- Silva M, Pinho RB, Fonseca BR, Bezerra FSB, Sousa FSS, Seixas FK, Collares T, Meyer R, Portela RW, Azevedo V, Borsuk S. NanH and PknG putative virulence factors as a recombinant subunit immunogen against Corynebacterium pseudotuberculosis infection in mice. Vaccine. 2020;38:8099–8106. doi: 10.1016/j.vaccine.2020.11.010. [DOI] [PubMed] [Google Scholar]
- Strong EJ, Wang J, Ng TW, Porcelli SA, Lee S. Mycobacterium tuberculosis PPE51 inhibits autophagy by suppressing Toll-like receptor 2-dependent signaling. mBio. 2022;13:e0297421. doi: 10.1128/mbio.02974-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchala M, Moal IH, Chaleil RA, Fernandez-Recio J, Bates PA. SwarmDock: a server for flexible protein-protein docking. Bioinformatics. 2013;29:807–809. doi: 10.1093/bioinformatics/btt038. [DOI] [PubMed] [Google Scholar]
- Tretyachenko V, Vymětal J, Bednárová L, Kopecký V, Jr, Hofbauerová K, Jindrová H, Hubálek M, Souček R, Konvalinka J, Vondrášek J, Hlouchová K. Random protein sequences can form defined secondary structures and are well-tolerated in vivo. Sci Rep. 2017;7:15449. doi: 10.1038/s41598-017-15635-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- Wang P. Natural and synthetic saponins as vaccine adjuvants. Vaccines. 2021;9:222. doi: 10.3390/vaccines9030222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yuan HY, Liu GM, Ni WH, Wang F, Tai GX. Escherichia coli maltose-binding protein induces M1 polarity of RAW264.7 Macrophage Cells via a TLR2- and TLR4-Dependent Manner. Int J Mol Sci. 2015;16:9896–9909. doi: 10.3390/ijms16059896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article (and its supplementary information files).