Abstract
We present a case of unexplained familial breast cancer (BC) from six family members, including four affected and two unaffected women, for whom clinical genetic testing panels were inconclusive. Exome sequencing data revealed heterozygous and rare germline variants to be inherited in an autosomal dominant manner in the family, in addition to several unclassified mutations in DNA repair and cell cycle-regulating genes that were not included in the family’s clinical genetic testing. A rare MYC-N11S germline mutation with conflicting interpretations of pathogenicity in the literature, and predicted to be deleterious, was present in all affected individuals. Whole exome sequencing provided a more comprehensive picture of inherited BC in this family that was missed by cancer gene panels alone.
Keywords: Breast cancer, Cancer intervention, Genetics, Genetic screening / counselling
Background
Clinical cancer panel testing often fails to capture the full complexities of genetic aberrations that may contribute to tumorigenesis. This case report underscores the complexity of genetic testing as it relates to inherited breast cancer (BC) and the importance and limitations of whole exome sequencing (WES) for discovery of germline mutations in familial BC. It is important to assess the family pedigree when a patient presents with BC as the risk of developing BC is correlated with how many first-degree, second-degree and third-degree relatives are affected. As many as 30% of BC cases are familial; however, only 5–10% of BC cases have clear genetic component such as high penetrance BRCA1 or BRCA2 mutations.1
Case presentation
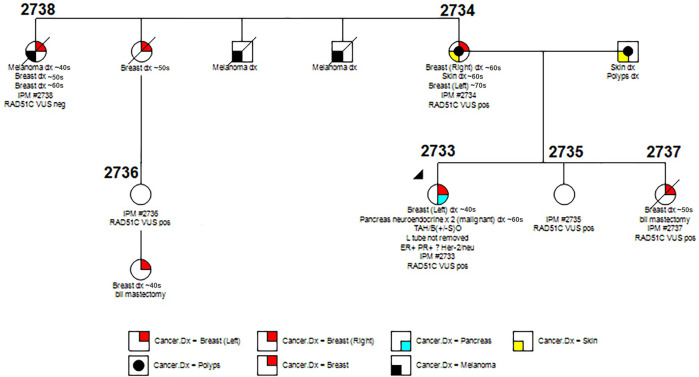
A Caucasian woman (2733, proband) presenting with BC in her forties underwent multicancer panel. Results were negative for all genes screened except for RAD51C V169A, a variant of unknown significance (VUS), which several other labs classify as likely benign in ClinVar. She also presented with a malignant pancreatic neuroendocrine tumour several years later. A multigenerational history of breast, sarcoma, melanoma, pancreatic and colon cancer, etc was also present in the family (figure 1). A first-degree relative (2734) was diagnosed with bilateral BC in both the right and left breasts a few years apart. She was screened using a comprehensive cancer panel consisting of 29 genes that was negative except for the same RAD51C-V169A variant as the proband, classified by the lab as likely benign. Clinical gene panel information was not available for two other affected family members, a first-degree and second-degree relative. One other first-degree relative and one third-degree relative were unaffected but participated in our study to aid in modelling heritability as recommended by the American College of Medical Genetics (ACMG).2 Unlike WES, the multicancer panels used for these individuals (table 1) lack coverage for many informative genes such as MYC and, as such, are not always able to provide causative variants.
Figure 1.
Pedigree for multigenerational case of breast cancer.
Table 1.
Cancer panel genes tested in 2733 (proband) and 2734 (first-degree relative)
| 79 gene cancer panel screened for in 2733 | (Sequence changes and exonic deletions/duplications), ALK, APC, ATM, AXIN2, BAP1, BARD1, BLM, BMPR1A, BRCA1, BRCA2, BRIP1, CASR, CDC73, CDH1, CDK4, CDKN1B, CDKN1C, CDKN2A, CEBPA, CHEK2, CTNNA1, DICER1, DIS3L2, EGFR, EPCAM, FH, FLCN, GATA2, GPC3, GREM1, HOXB13, (Promoter region deletion/duplication testing only), HRAS, KIT, MAX, MEN1, MET, MLH1, MSH2, MSH6, MUTYH, NBN, NF1, NF2, PALB2, PDGFRA, PHOX2B, PMS2, POLD1, POLE, PRKAR1A, PTCH1, PTEN, RAD50, RAD51C, RAD51D, RB1, RECQL4, RET, RUNX1, SDHA, SDHAF2, SDHB, SDHC, SDHD, SMAD4, SMARCA4, SMARCB1, SMARCE1, STK11, SUFU, TERC, TERT, TMEM127, TP53, TSC1, TSC2, VHL, WRN, WT1. Sequence changes only: EGFR (c.2369C>T, pThr790Met variants), HOXB13 (c.251G>A, p.Gly84Glu variant), MITF (c.952G>A, p.Glu318Lys variant), SDHA. |
| 29 gene cancer panel screened for in 2734 | APC, ATM, AXIN2, BARD1, BMPR1A, BRCA1, BRCA2, BPRIP1, CDH1, CDK4, CDKN2A, CHEK2, EPCAM, FANCC, MLH1, MSH2, MSH6, MUTYH, NBN, PALB2, PMS2, PTEN, RAD51C, RAD51D, SMAD4, STK11, TP53, VHL, XRCC2 |
Investigations
To evaluate the possibility that rare variants with low penetrance could be contributing to susceptibility to BC in this family, we performed WES and modelled heritability on a 34 megabase region of saliva DNA from six family members, including four affected and two unaffected women. We were interested in rare germline mutations which followed an autosomal dominant pattern of inheritance. We used Sanger sequencing and confirmed the germline RAD51C-V169A VUS in three affected individuals, 2733, 2734, and 2737 and two unaffected individuals 2735 and 2736. One affected member, 2738, did not carry the RAD51C variant. These findings suggest that this VUS may be a passenger rather than a causative variant. WES data revealed an additional 92 germline mutations that had a low alternative allele frequency in the general population (≤5%) and were inherited in an autosomal dominant manner. These are listed in online supplemental table 1. Genomewide association studies show that common genetic variants only explain a subset of inherited risk for BC.3 Rare variants may account for a significant portion of unexplained inherited cancers and those that follow an autosomal dominant inheritance are more likely to be causative of disease and be relevant diagnostic targets. One potentially causative mutation following these criteria was a heterozygous A→G change in MYC (rs4645959) that caused an N11S amino acid change. Following ACMG guidelines, the predicted impact of this mutation from SIFT, PolyPhen and PathoMan was probably damaging/deleterious/pathogenic as shown in table 2. We also observed this mutation to occur in a promoter and enhancer region identified by ChIP-seq data in breast epithelial tissue from ENCODE, the encyclopedia of DNA elements. Conflicting interpretations of pathogenicity or association with BC highlight the need to study this variant in more detail.4 5 We were also able to identify several deletions, insertions, duplications and breakend pairs corresponding to intra and interchromosomal translocations or complex structural variants (SVs) in all six family members. While there were no SVs that were inherited in an autosomal dominant manner in the family, one simple inversion was detected in 3 of the 4 affected individuals. (2733, 2737, 2738) with a breakpoint at chr4:86 063 and chr4:367 272 involving the genes ZNF595, ZNF718, ZNF876P, ZNF732, ZNF141. This inversion was not detected in one affected first-degree relative or unaffected individuals. A limitation of WES is that it may fail to capture many SVs that would otherwise be detected through whole genome sequencing and, thus, the possibility of inherited SVs contributing to heritability cannot be ruled out. Many rare mutations of interest that might explain the inheritance of BC in this family were missed by clinical cancer panel testing but were revealed through WES. This suggests a need to significantly expand targeted cancer gene panels to be more comprehensive of cancer-associated genes as many common variants in cancer-associated genes do not fully explain the patterns of inherited BC.
Table 2.
Impact classifications of the rs4645959 mutation in MYC N11S
| MYC N11S(rs4645959) | SIFT | PolyPhen | PathoMan | InterVar | ClinVar |
| Deleterious | Probably damaging | Likely pathogenic | N/A | N/A |
bcr-2022-251336supp001.pdf (90.4KB, pdf)
Differential diagnosis
At the time of diagnosis, only one RAD51C-V169A VUS was found in the proband and a first-degree relative using comprehensive cancer panels on a clinical basis. A previous study performed a mitomycin C assay, which did not show a significant phenotype of the RAD51C-V169A variant compared with wild-type RAD51C in DT40 cells.6 Variant prediction algorithms SIFT and PolyPhen did not predict this variant to be deleterious, which suggested that other mutations might be contributing to the familial BC phenotype in these individuals that were not included in their cancer panels. We were able to identify 92 mutations that followed an autosomal dominant pattern of inheritance in the family and were also present in less than 5% in a general population, including a predicted deleterious variant, MYC-N11S with conflicting pathogenicity interpretations in the literature.4 5
Discussion
Low penetrance variants in genes not included in standard genetic testing panels could contribute to susceptibility to BC as well as other cancers observed in this family. These results may explain the contribution of germline mutations to increased susceptibility to inherited BC. Other case reports have highlighted the importance of exome sequencing in uncovering potentially pathogenic variants in genes not typically included in custom panels such as SLC18A3, CACNA1H and TSC2.7–9 In our case, the proto-oncogene MYC, which is a transcription factor involved in cell growth, proliferation and metabolism, had an N11S variant with conflicting interpretations of pathogenicity in the literature.4 5 This mutation was also found to overlap with promoter and enhancer regions identified by ChIP-Seq data in breast epithelial tissue from ENCODE, suggesting that this variant may affect gene expression. In one cohort of over 700 individuals, a study found 54% increased risk for familial BC in BRCA-negative individuals carrying the heterozygous MYC-N11S mutation. The mutation was detected in 55/650 cases (7.8%) and 58/1037 controls (5.3%).4 However, in a Canadian population with ovarian, breast or a combination of cancers, the MYC-N11S mutation was not found to be associated with overall BC risk.5 Furthermore, immunohistochemical (IHC) detection of MYC has been shown to be blocked by the MYC-N11S variant, which highlights the need for genetic sequencing to detect tumours with shared biology that IHC fails to capture.10 The conflicting interpretations of this variant warrant its further investigation and screening in inherited BC cases.
While it is true that, in comparison with standard multigene panels, WES enables analysis of all coding gene regions, it also presents uncertainty in risk assessment in the form of increased complexity and volume of data, and a need for cancer genetic expert interpretations to safely communicate information to the patient. WES identifies numerous variants, many of which are labelled as a VUS and cannot presently contribute to risk assessment. ACMG guidelines advise against the use of VUS in clinical decision-making, which limits the usefulness of WES as a clinical tool for risk assessment. However, follow-up testing is encouraged by ACMG, which may create further evidence that could allow a variant to be reclassified as either benign or pathogenic.11 In instances where variants are rare, additional reports from WES to publicly accessible databases, such as dbSNP, would be beneficial, especially in instances of cosegregation of a variant with disease in multiple generations. In our study, individuals in the family without a variant in question should not be considered to have average BC risk because other rare low penetrance variants that remain incompletely classified may contribute to BC risk. Furthermore, WES still only covers around 2% of the human genome leaving many regulatory regions, SVs and copy-number variations of interest out of the picture.12 Age of onset of disease in affected family members must also be considered when calculating risk in unaffected family members without the suspected causative variant as younger individuals may develop BC later in life making that individual a false negative in familial studies. The use of WES in clinic presents its own challenges but may be a beneficial tool in instances where standard multigene panel testing remains inconclusive and expanded reporting of many VUS may lead to more inclusive gene panels in future BC testing.
Patient’s perspective.
This long unknown journey of genetic testing has been a time of hoping for answers. Our family has had so many losses from cancer and no explanation why. Having had cancer two times myself, I know the scare of the diagnosis, and the joy of the final treatment. I know the horror of the doctor telling you, you are never really cancer free. I pray the findings of this gene testing will help my family and all future generations to maybe someday be spared the cancer diagnosis or worse the heart-breaking loss of the relatives we love so much. Thank you so much for the hard work and research!
Learning points.
Comprehensive cancer panels commonly used in clinic are not entirely ‘comprehensive’ and many lack coverage over known cancer-associated genes.
Rare and low penetrance variants in genes not included in standard genetic testing panels could contribute to susceptibility to familial cancers.
The MYC-N11S mutation (rs4645959) has conflicting interpretation of pathogenicity in current literature and may be a possible cause of familial cancer development.
The American College of Medical Genetics guidelines for interpretation of variants rely, in-part, on variants reported to show cosegregation with the disease in multiple affected family members and, as such, modelling heritability in the family pedigree is beneficial for interpretation of a variant of unknown significance.
Acknowledgments
We would like to thank our funding sources. The research reported in this publication was supported by the Zimmerman Family Endowment and by the National Institutes of Health under award number 5T32LM012415-04.
Footnotes
Contributors: LB: conducting sequencing, writing and editing of manuscript. MB: genetic counselor/medical geneticist, writing and editing of manuscript. JB: writing and editing of manuscript.
Funding: This study was funded by Penn State Biomedical Big Data to Knowledge (B2D2K) Training Program (5T32LM012415-04).
Case reports provide a valuable learning resource for the scientific community and can indicate areas of interest for future research. They should not be used in isolation to guide treatment choices or public health policy.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s)
References
- 1.Collaborative Group on Hormonal Factors in Breast Cancer . Familial breast cancer: collaborative reanalysis of individual data from 52 epidemiological studies including 58,209 women with breast cancer and 101,986 women without the disease. Lancet 2001;358:1389–99. 10.1016/S0140-6736(01)06524-2 [DOI] [PubMed] [Google Scholar]
- 2.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet Med 2015;17:405–24. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H, Ahearn TU, Lecarpentier J, et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat Genet 2020;52:572–81. 10.1038/s41588-020-0609-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wirtenberger M, Hemminki K, Försti A, et al. C-MYC Asn11Ser is associated with increased risk for familial breast cancer. Int J Cancer 2005;117:638–42. 10.1002/ijc.21225 [DOI] [PubMed] [Google Scholar]
- 5.Figueiredo JC, Knight JA, Cho S, et al. Polymorphisms cMyc-N11S and p27-V109G and breast cancer risk and prognosis. BMC Cancer 2007;7:1–8. 10.1186/1471-2407-7-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 2010;42:410–4. 10.1038/ng.569 [DOI] [PubMed] [Google Scholar]
- 7.Wulczyn K, Perez-Reyes E, Nussbaum RL, et al. Primary aldosteronism associated with a germline variant in CACNA1H. BMJ Case Rep 2019;12:e229031. 10.1136/bcr-2018-229031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zureick AH, McFadden KA, Mody R, et al. Successful treatment of a TSC2-mutant glioblastoma with everolimus. BMJ Case Rep 2019;12:e227734. 10.1136/bcr-2018-227734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamond A, Buckley D, O'Dea J, et al. Variants of SLC18A3 leading to congenital myasthenic syndrome in two children with varying presentations. BMJ Case Rep 2021;14:e237799. 10.1136/bcr-2020-237799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collinge B, Ben-Neriah S, Chong L, et al. The impact of MYC and BCL2 structural variants in tumors of DLBCL morphology and mechanisms of false-negative Myc IHC. Blood 2021;137:2196–208. 10.1182/blood.2020007193 [DOI] [PubMed] [Google Scholar]
- 11.Miller DT, Lee K, Gordon AS, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2021 update: a policy statement of the American College of medical genetics and genomics (ACMG). Genet Med 2021;23:1391–8. 10.1038/s41436-021-01171-4 [DOI] [PubMed] [Google Scholar]
- 12.Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet 2011;12:745–55. 10.1038/nrg3031 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bcr-2022-251336supp001.pdf (90.4KB, pdf)