Abstract
The CD14 molecule expressed on monocytes and macrophages is a high-affinity receptor for bacterial lipopolysaccharide (LPS) and hence an important component of the innate immune system. LPS binding protein (LBP) is required to facilitate the binding of LPS to CD14 in vitro and is necessary for the induction of an inflammatory response to LPS in vivo. Here we show that CD14 and LBP can also bind to lipoteichoic acid from the gram-positive bacterium Bacillus subtilis. Although CD14 does not interact with intact B. subtilis organisms, a brief exposure of the bacteria to serum converts them into a form which can bind to CD14 in an LBP-dependent reaction. When serum-pretreated B. subtilis organisms are incubated with the myelomonocytic cell line U937, which expresses CD14, the bacteria are rapidly phagocytosed. The phagocytosis is strictly dependent both on LBP and on CD14. These in vitro results suggest that LBP plays a role in the innate response not only to gram-negative but also to gram-positive infections.
Receptors of the innate immune system have been selected for their ability to recognize molecules which are present on microorganisms but not on cells of the host (8). These target molecules are typically broadly distributed among major groups of microorganisms and are required for their survival. One well-characterized example of such a target molecule is lipopolysaccharide (LPS), the principal endotoxin of gram-negative bacteria (13). The CD14 molecule expressed on monocytes and macrophages serves as a high-affinity receptor for LPS (19), and the binding of LPS results in cellular activation and the induction of an inflammatory response. However, the interaction of LPS with CD14 is slow unless it is catalyzed by the serum protein LPS binding protein (LBP) (14), which is therefore essential for the cellular response to LPS in vivo (6). Monocytes expressing CD14 are able to bind not only free LPS but also intact gram-negative bacteria, and this binding, which likewise requires catalysis by LBP (7), is followed by phagocytosis (4). These interactions involving LBP and CD14 are believed to be pivotal to the innate response to a gram-negative bacterial infection. Recent in vitro experiments have shown that CD14 can bind not only LPS but also components of the gram-positive bacterial cell wall (5, 9, 10, 12, 15, 17, 18). Of particular interest is the observation that CD14 can bind fragments of gram-positive cell walls generated by boiling the bacteria in sodium dodecyl sulfate solution, followed by mechanical disruption and acetone extraction (12). Nevertheless, in these experiments it is unclear whether the cell wall components recognized by CD14 are likely to be exposed on the bacterial surface in vivo. The role of LBP in innate recognition of gram-positive bacteria has not yet been established.
Here we show that both LBP and CD14 can bind to lipoteichoic acid (LTA) derived from the gram-positive bacterium Bacillus subtilis. In addition, whole bacteria are recognized by CD14 in an LBP-dependent reaction but only after preincubation with serum. Electron microscopy suggests that the serum pretreatment results in the opening up of the bacterial cell wall. When myelomonocytic cells expressing CD14 bind serum-pretreated bacteria, the bacteria may be phagocytosed. The CD14-LBP system may thus play a role in countering gram-positive bacterial infections.
MATERIALS AND METHODS
Reagents.
Anti-CD14 monoclonal antibody (MAb) My4 (immunoglobulin G2b) was purchased from Coulter Electronics (Krefeld, Germany). Anti-CD14 MAb biG 14 (immunoglobulin G2a) was obtained from Biometec GmbH (Greifswald, Germany), and Fab fragments were prepared from it as previously described (4). Isotypically identical control antibodies (anti-keyhole limpet hemocyanin) were from Becton Dickinson (Heidelburg, Germany). Histidine-tagged recombinant murine LBP and recombinant human soluble CD14 were purified from supernatants of transfected CHO cells as previously described (4, 11). Normal mouse serum (NMS) and LPS from Salmonella minnesota Re 595 were purchased from Sigma (Munich, Germany). For the experiment shown in Fig. 2B mouse serum and plasma were obtained from BALB/c mice. Plasma was prepared from blood anticoagulated with 0.011 M sodium citrate. For all experiments serum and plasma were heat inactivated at 56°C for 1 h. LTA derived from B. subtilis was purchased from Sigma. The purity of the commercially available LTA has not been established. We therefore prepared LTA from B. subtilis DSMZ 1087 according to a method of Fischer et al. (2, 3). Briefly, a hot-phenol–water extraction of disrupted bacteria was followed by dialysis of the aqueous phase and subsequent hydrophobic interaction chromatography (fast protein liquid chromatography with octyl-Sepharose CL-4B; Pharmacia, Uppsala, Sweden). Glass materials, tubing, and the column material were rendered pyrogen free with 0.1 M NaOH and washed with pyrogen-free water. The phosphorus contents of the different fractions were determined by a phosphomolybdene blue assay. LTA is eluted in the late fractions which contain the highest amount of phosphate. The LTA was lyophylized until use in the respective experiments. This LTA preparation was negative in the Limulus amebocyte lysate assay, i.e., it was contaminated with less than 100 pg of LPS per mg of LTA. The LTA-containing fractions were capable of inducing release of tumor necrosis factor alpha in human whole blood. When the LTA preparation was subjected to an additional anion-exchange chromatography experiment (fast protein liquid chromatography with DEAE-Sepharose Fast Flow; Pharmacia), the material eluted as a single symmetric peak. LTA and cytokine-inducing activity still coeluted, indicating that LTA represented the cytokine-inducing activity.
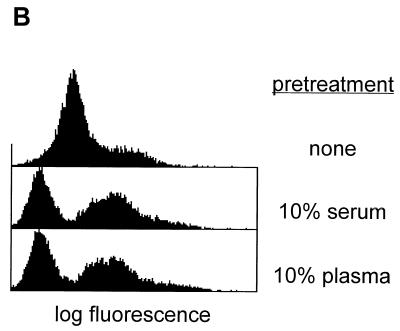
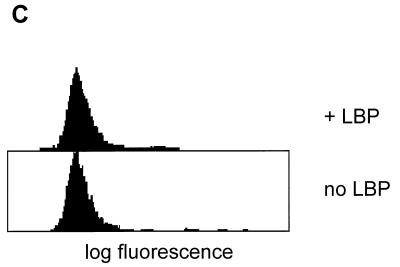
FIG. 2.
(A) Binding of serum-pretreated FITC-labeled B. subtilis to human CD14 expressed on CHO cells. Cells were incubated with labeled bacteria in the presence or absence of 1 μg of recombinant LBP per ml. Binding in the presence of LBP is abolished by inclusion of (i) a 20-fold excess of serum-pretreated unlabeled B. subtilis cells, (ii) Salmonella minnesota Re 595 LPS at a final concentration of 20 μg/ml, or (iii) 10 μg of anti CD14 MAb My4 per ml (but not by the same amount of an isotype-matched control). The ratio of cells to bacteria was 1:20. (B) Binding of plasma-pretreated FITC-labeled B. subtilis to human CD14 expressed on CHO cells. B. subtilis organisms were not pretreated (upper), pretreated with serum (middle), or pretreated with plasma (lower). Binding to CHO cells expressing human CD14 was determined in the presence of 1 μg of recombinant LBP per ml. (C) B. subtilis does not bind to the mutant CD14(39–41, 43–44A), which lacks the binding sites for LPS and E. coli.
Bacteria.
B. subtilis 168 was grown in Luria-Bertani broth at 37°C to mid-log phase, and cells were collected by centrifugation. The number of bacteria present was determined by plating serial dilutions. Bacteria were heat inactivated by treatment at 60°C for 1 h, washed twice in sterile phosphate-buffered saline (PBS) (without Ca2+ and Mg2+) and resuspended in PBS to achieve an optical density a 550 nm of 2.0.
FITC labeling of B. subtilis and analysis of bacterial binding to CHO cells expressing human CD14.
Heat-inactivated B. subtilis organisms (2.3 × 109) in 10 ml of PBS were labeled with fluorescein isothiocyanate (FITC) by incubation with 5(6)-carboxyfluorescein-N-hydroxysuccinimide ester (Fluos; Boehringer, Mannheim, Germany). A solution containing 0.9 ml of 2-mg/ml Fluos in dimethyl sulfoxide was added to the bacteria, and the mixture was shaken in the dark for 2 h. After labeling, the bacteria were pelleted at 5,000 × g for 6 min, washed twice with PBS, and resuspended in 10 ml of PBS. The FITC-labeled bacteria were stored at 4°C and used within 2 weeks. With the exception of the experiments shown in Fig. 1 and 2B, the labeled bacteria were pretreated by incubation in PBS containing 10% NMS at 37°C for 2 h. Serum-pretreated bacteria were washed twice and taken up in the original volume of PBS.
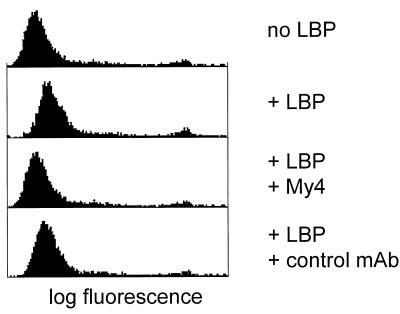
FIG. 1.
Binding of FITC-labeled B. subtilis to human CD14 expressed on CHO cells. Cells were incubated with labeled bacteria in the presence or absence of 1 μg of recombinant LBP per ml. The marginal binding in the presence of LBP is abolished by inclusion of 10 μg of anti CD14 MAb My4 per ml in the incubation mixture but not by the same amount of an isotype-matched control. The ratio of cells to bacteria was 1:20.
The binding of FITC-labeled bacteria to CHO cells expressing the human CD14 gene was carried out as previously described (7). Cells were briefly trypsinized, collected by centrifugation, washed twice in PBS, and resuspended in ice-cold PBS containing 0.1% sodium azide and 1% bovine serum albumin. Cells at a final concentration of 106/ml were incubated with FITC-labeled B. subtilis at 4°C for 20 min at a cell-to-bacterium ratio of 1:20. In some experiments a 10-fold excess of unlabeled bacteria was added to the binding reaction mixture as a competitor. After cells were washed twice, fluorescence intensities were measured in a FACScan.
Gel shift assay.
Binding of LTA to CD14 and LBP was assayed by a gel shift method (18). Briefly, the amounts of soluble CD14 or LBP indicated in Fig. 4 were mixed with different amounts of LTA and incubated overnight at 37°C in a final volume of 10 μl of PBS. Mixtures were loaded onto Tris-glycine gels (pH 8.8; Bio-Rad Laboratories, Munich, Germany) and separated by native polyacrylamide gel electrophoresis. CD14 and LBP were detected by Western blotting with a rabbit anti-human CD14 antiserum (1) or a rabbit anti-mouse LBP antiserum (Biometec GmbH).
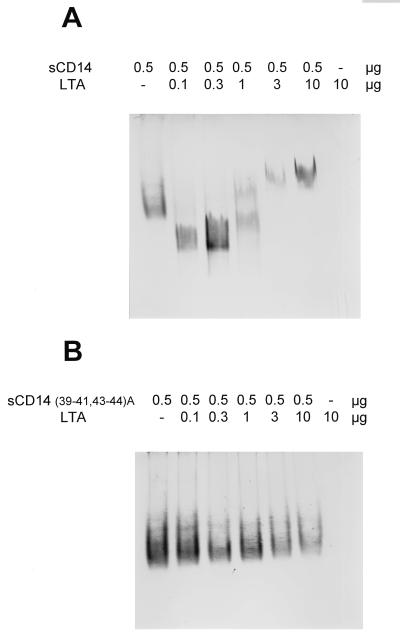
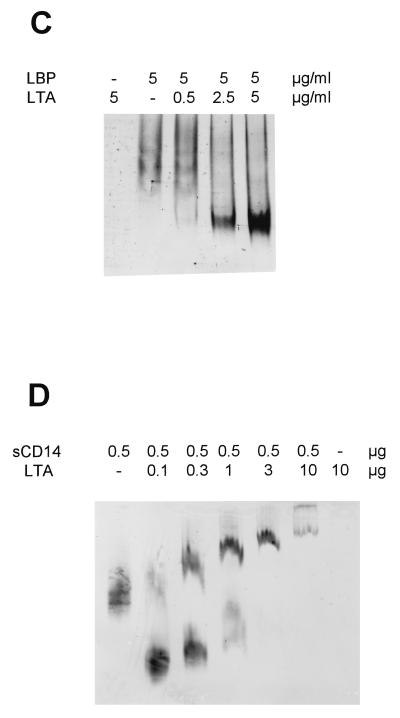
FIG. 4.
LTA derived from the cell wall of B. subtilis forms complexes with CD14 (A) and LBP (C) but not with CD14(39–41, 43–44A) (B). Soluble CD14 and LBP were incubated overnight with LTA in the amounts indicated and separated by native polyacrylamide gel electrophoresis. CD14 and LBP were detected by Western blotting. CD14 binding at limiting ligand concentrations results in complexes which migrate faster than the native protein, while CD14 binding at excess ligand concentrations results in complexes that migrate more slowly. (D) The binding to CD14 was repeated with a second highly purified preparation of LTA. s, soluble.
Scanning electron microscopy.
Bacteria were fixed with glutaraldehyde and applied to poly-l-lysine-coated glass plates. After treatment with osmium tetroxide, the samples were processed through critical-point drying and then examined in a Zeiss DSM 904A scanning electron microscope.
Analysis of phagocytosis of FITC-labeled B. subtilis by U937 cells.
CD14 expression was induced on U937 cells with vitamin D3 as previously described (7). Cells were collected, washed three times with ice-cold PBS, and resuspended in PBS containing 1% bovine serum albumin. Induced or noninduced U937 cells (1.5 × 105) were incubated at 37°C in a shaking water bath with 20 μl of serum-pretreated FITC-labeled B. subtilis organisms (cell-to-bacterium ratio, 1:50) in the presence or absence of 5 μg of LBP per ml in a final volume of 100 μl. After 10 min, 100 μl of Phagotest trypan blue quenching solution (Orpegen Pharma, Heidelberg, Germany) was added. The cells were washed twice with 3 ml of ice-cold PBS containing 0.1% NaN3 and analyzed in a FACScan.
RESULTS AND DISCUSSION
LBP mediates binding of serum-pretreated B. subtilis to membrane-bound CD14.
FITC-labeled B. subtilis organisms do not detectably bind to CHO cells expressing human CD14 in the absence of LBP. In the presence of LBP there is marginal binding which appears to be dependent on CD14 since it can be inhibited by the anti-CD14 MAb My4 but not by the isotype-matched control antibody (Fig. 1). However, when the labeled B. subtilis cells were first pretreated with 10% NMS at 37°C for 2 h, significant binding was seen in the presence but not in the absence of LBP (Fig. 2A). This binding is not an artifact induced by the labeling procedure since it can be inhibited by a 20-fold excess of unlabeled serum-pretreated bacteria. The interaction is CD14 dependent since it is specifically blocked by the anti CD14 MAb My4 and by an excess of LPS. The binding of B. subtilis is potentiated in a similar way by preincubation with plasma (Fig. 2B). This fact demonstrates that the potentiation is not dependent on the presence of activated components of the clotting cascade. Finally, CHO cells expressing a mutant CD14, CD14(39–41, 43–44A), which is incapable of recognizing LPS and Escherichia coli (16), fail to bind the serum-pretreated bacteria (Fig. 2C). These results show that both CD14 and LBP are directly involved in the binding of serum-pretreated B. subtilis and that the LPS binding site on CD14 is required for the interaction.
Serum treatment opens up the structure of the bacterial cell wall.
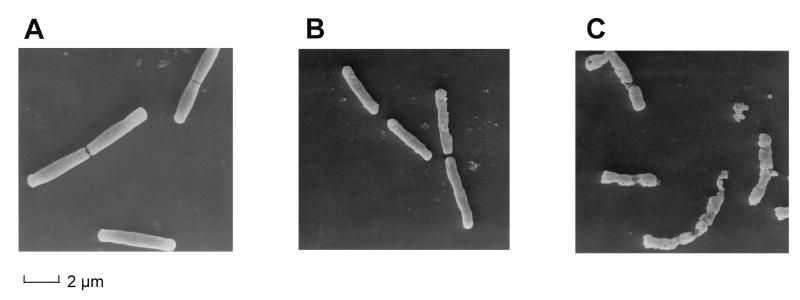
The experiments shown in Fig. 1 and 2 demonstrate that heat-treated FITC-labeled B. subtilis organisms are not effectively bound to CD14 unless the bacterial preparation is first incubated in serum. We therefore asked whether the serum pretreatment has any demonstrable effect on the gross morphology of the bacteria. As shown in the scanning electron micrographs in Fig. 3A, fresh log-phase B. subtilis organisms have a smooth surface which takes on a somewhat roughened appearance after heat treatment (Fig. 3B). However, incubation in 10% NMS (Fig. 3C) results in considerable damage and the breaking up of almost all bacteria. We are currently trying to identify the serum factor(s) involved in this process. Thus, serum treatment of the bacteria makes available structures which are essential for the LBP-mediated interaction with CD14. The low degree of binding to CD14 which is seen prior to serum treatment (Fig. 1) may be due to the presence of a small fraction of bacteria which was disrupted during heat inactivation.
FIG. 3.
Serum treatment of B. subtilis opens up the bacterial cell wall. Mid-log-phase cells were examined by transmission electron microscopy either directly (A), after heat inactivation (B), or after heat inactivation and exposure to 10% NMS for 2 h at 37°C (C).
Both LBP and CD14 bind in vitro to LTA derived from the cell wall of B. subtilis.
Because LTA is a major constituent of the outer cell wall of gram-positive organisms, we have used a gel shift assay to test whether LBP and CD14 are able to interact with it. As shown in Fig. 4, both proteins are shifted after incubation with LTA, indicating the formation of stable CD14-LTA (Fig. 4A) and LBP-LTA (Fig. 4C) complexes. Again, the formation of CD14-LTA complexes depends on the presence of an intact LPS binding site, since the soluble form of CD14(39–41, 43–44A) is incapable of binding to LTA (Fig. 4B). Since the LTA content of the Sigma preparation is unclear, we repeated the binding to wild-type CD14 using a second highly purified sample of LTA (Fig. 4D). The results of this similar gel shift demonstrated that binding is indeed to LTA rather than to some contaminant in the Sigma preparation.
These results point to LTA being a putative ligand for recognition both by LBP and by CD14. These proteins may act as pattern recognition elements (8) by detecting a three-dimensional similarity between part of the LPS molecule on gram-negative bacteria and the LTA molecule on gram-positive bacteria.
CD14- and LBP-dependent phagocytosis of serum-pretreated B. subtilis by the myelomonocytic cell line U937.
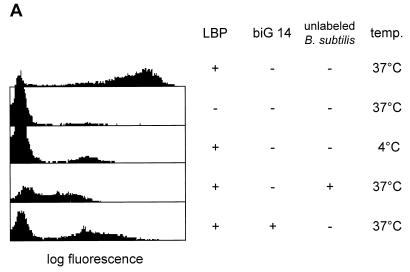
To determine whether the binding of the bacteria to CD14 might have physiologically relevant consequences, we looked for phagocytosis of B. subtilis by myeloid cells. We have previously established a flow cytometric assay to show that vitamin D3-induced U937 cells bind and internalize gram-negative bacteria in a CD14- and LBP-dependent fashion (4). In this assay a quenching reagent abolishes the fluorescence signal from labeled bacteria bound to the outside of the cell. A signal can be obtained only from those labeled bacteria which have been protected from the quenching agent by internalization. We have used this assay to study the fate of serum-pretreated B. subtilis cells after they bound to the CD14 receptor. As shown in Fig. 5, vitamin D3-induced U937 cells can internalize serum-pretreated B. subtilis and this requires the presence of both LBP and CD14. This phagocytosis process is dependent on CD14, since it can be effectively blocked by Fab fragments of the anti CD14 MAb biG 14.
FIG. 5.
Serum-pretreated B. subtilis organisms are phagocytosed by vitamin D3-induced U937 cells in a CD14- and LBP-dependent fashion. (A) Phagocytosis was performed at 37 or 4°C in the presence or absence of 2.5 μg of murine recombinant LBP per ml. Internalization of the labeled bacteria is inhibitable with Fab fragments of the anti-CD14 MAb biG 14 or with a 20-fold excess of unlabeled bacteria (added where indicated). The ratio of cells to labeled bacteria was 1:50. (B) The same experiment as described for panel A was carried out with uninduced U937 cells, which do not express CD14.
These results demonstrate that serum pretreatment of this gram-positive bacterium facilitates an LBP-mediated interaction between some component of the bacterial cell wall and the pattern recognition element CD14 on the surface of a phagocyte, leading to phagocytosis. We further show that LTA—a major component of the gram-positive bacterial wall—is able to bind to CD14 in an LBP-dependent fashion. Further work will be necessary to determine whether an LTA-CD14 interaction is actually involved in the binding of serum-pretreated B. subtilis to CD14 and, if so, whether this interaction alone is sufficient to trigger phagocytosis or cellular activation. Recent reports (17, 18) indicate that CD14 can also interact with the peptidoglycan component of the gram-positive cell wall. It is therefore very possible that CD14 as a pattern recognition receptor utilizes more than one target molecule on the bacterium. Whatever the precise nature of the ligands, these interactions may be significant in the destruction and removal of invading gram-positive bacteria in vivo.
ACKNOWLEDGMENTS
We thank E. Friebe and R. Görlich for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft through grants SFB217 and DFG IIB6Ste 587/2-2.
REFERENCES
- 1.Bufler P, Stiegler G, Schuchmann M, Hess S, Krüger C, Stelter F, Schütt C, Eckershorn C, Engelmann H. Soluble LPS receptor (CD14) is released via two different mechanisms from human monocytes and CD14 transfectants. Eur J Immunol. 1995;25:604–610. doi: 10.1002/eji.1830250244. [DOI] [PubMed] [Google Scholar]
- 2.Fischer W. Molecular analysis of lipid macroamphiphiles by hydrophobic interaction chromatography, exemplified with lipoteichoic acids. Anal Biochem. 1993;208:49–56. doi: 10.1006/abio.1993.1007. [DOI] [PubMed] [Google Scholar]
- 3.Fischer W, Koch H U, Haas R. Improved preparation of lipoteichoic acids. Eur J Biochem. 1983;133:523–530. doi: 10.1111/j.1432-1033.1983.tb07495.x. [DOI] [PubMed] [Google Scholar]
- 4.Grunwald U, Fan X, Jack R S, Workalemahu G, Kallies A, Stelter F, Schütt C S. Monocytes can phagocytose Gram-negative bacteria by a CD14 dependent mechanism. J Immunol. 1996;157:4119–4125. [PubMed] [Google Scholar]
- 5.Gupta D, Kirkland T N, Viriyakosol S, Dziarski R. CD14 is a cell-activating receptor for bacterial peptidoglycan. J Biol Chem. 1996;271:23310–23316. doi: 10.1074/jbc.271.38.23310. [DOI] [PubMed] [Google Scholar]
- 6.Jack R S, Fan X, Bernheiden M, Rune G, Ehlers M, Weber A, Kirsch G, Mentel R, Fürll B, Freudenberg M, Schmitz G, Stelter F, Schütt C. Lipopolysaccharide binding protein is required to combat a murine Gram-negative infection. Nature. 1997;389:742–745. doi: 10.1038/39622. [DOI] [PubMed] [Google Scholar]
- 7.Jack R S, Grunwald U, Stelter F, Workalemahu G, Schütt C. Both membrane bound and soluble forms of CD14 bind to Gram-negative bacteria. Eur J Immunol. 1995;25:1436–1441. doi: 10.1002/eji.1830250545. [DOI] [PubMed] [Google Scholar]
- 8.Janeway C A. The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol Today. 1992;13:11–16. doi: 10.1016/0167-5699(92)90198-G. [DOI] [PubMed] [Google Scholar]
- 9.Kusunoki T, Hailman E, Todd S C J, Lichenstein H S, Wright S D. Molecules from Staphylococcus aureus that bind CD14 and stimulate innate immune responses. J Exp Med. 1995;182:1673–1682. doi: 10.1084/jem.182.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kusunoki T, Wright S D. Chemical characteristics of Staphylococcus aureus molecules that have CD14-dependent cell-stimulating activity. J Immunol. 1996;157:5112–5117. [PubMed] [Google Scholar]
- 11.Neumeister B, Faigle M, Sommer M, Zähringer U, Stelter F, Menzel R, Schütt C, Northoff H. Low endotoxic potential of Legionella pneumophila lipopolysaccharide due to failure of interaction with the monocyte lipopolysaccharide receptor CD14. Infect Immun. 1998;66:4151–4157. doi: 10.1128/iai.66.9.4151-4157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pugin J, Heumann D, Tomasz A, Kravchenko V V, Akamatsu Y, Nishijima M, Glauser M P, Tobias P S, Ulevitch R J. CD14 is a pattern recognition receptor. Immunity. 1994;1:509–516. doi: 10.1016/1074-7613(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 13.Rietschel E T, Kirikae T, Schade F U, Mamat U, Schmidt G, Loppnow H, Ulmer A J, Zähringer U, Seydel U, Padova F D, Schreier M, Brade H. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- 14.Schumann R R, Leong S R, Flaggs G W, Gray P W, Wright S D, Mathison J C, Tobias P S, Ulevitch R J. Structure and function of lipopolysaccharide binding protein. Science. 1990;249:1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 15.Soell M, Lett E, Holveck F, Schöller M, Wachsmann D, Klein J P. Activation of human monocytes by streptococcal rhamnose glucose polymers is mediated by CD14 antigen, and mannan binding protein inhibits TNF-α release. J Immunol. 1995;154:851–860. [PubMed] [Google Scholar]
- 16.Stelter F, Bernheiden M, Menzel R, Jack R S, Witt S, Fan X, Pfister M, Schütt C. Mutation of amino acids 39–44 of human CD14 abrogates binding of lipopolysaccharide and Escherichia coli. Eur J Biochem. 1997;243:100–109. doi: 10.1111/j.1432-1033.1997.00100.x. [DOI] [PubMed] [Google Scholar]
- 17.Weidemann B, Brade H, Rietschel E T, Dziarski R, Bazil V, Kusumoto S, Flad H D, Ulmer A J. Soluble peptidoglycan-induced monokine production can be blocked by anti-CD14 monoclonal antibodies and by lipid A partial structures. Infect Immun. 1994;62:4709–4715. doi: 10.1128/iai.62.11.4709-4715.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weidemann B, Schletter J, Dziarski R, Kusumoto S, Stelter F, Rietschel E T, Flad H D, Ulmer A J. Specific binding of soluble peptidoglycan and muramyldipeptide to CD14 on human monocytes. Infect Immun. 1997;65:858–864. doi: 10.1128/iai.65.3.858-864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright S D, Ramos R A, Tobias P S, Ulevitch R J, Mathison J C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]