Significance
Understanding how plant cells sense and transduce heat signals has critical implications for improving plant heat tolerance. Here, we demonstrate that under heat stress, the phosphatase PP2AB′β dephosphorylates and stabilizes the histone deacetylase HDA9 in the cytoplasm. The nucleoporin HOS1 then translocates HDA9 into the nucleus, where the zinc-finger transcription factor YY1 helps recruit it to target genes to regulate gene expression via histone deacetylation. The cytoplasm-to-nucleus translocation of HDA9 in response to heat is conserved in wheat and rice. Thus, we identified a mechanism by which plant cells transduce heat signals from the cytoplasm to the nucleus and regulate gene expression, which could be used for crop improvement.
Keywords: HDA9 nuclear relocation, heat signal transduction, dephosphorylation, HOS1, YY1
Abstract
Heat stress limits plant growth, development, and crop yield, but how plant cells precisely sense and transduce heat stress signals remains elusive. Here, we identified a conserved heat stress response mechanism to elucidate how heat stress signal is transmitted from the cytoplasm into the nucleus for epigenetic modifiers. We demonstrate that HISTONE DEACETYLASE 9 (HDA9) transduces heat signals from the cytoplasm to the nucleus to play a positive regulatory role in heat responses in Arabidopsis. Heat specifically induces HDA9 accumulation in the nucleus. Under heat stress, the phosphatase PP2AB′β directly interacts with and dephosphorylates HDA9 to protect HDA9 from 26S proteasome-mediated degradation, leading to the translocation of nonphosphorylated HDA9 to the nucleus. This heat-induced enrichment of HDA9 in the nucleus depends on the nucleoporin HOS1. In the nucleus, HDA9 binds and deacetylates the target genes related to signaling transduction and plant development to repress gene expression in a transcription factor YIN YANG 1–dependent and –independent manner, resulting in rebalance of plant development and heat response. Therefore, we uncover an HDA9-mediated positive regulatory module in the heat shock signal transduction pathway. More important, this cytoplasm-to-nucleus translocation of HDA9 in response to heat stress is conserved in wheat and rice, which confers the mechanism significant implication potential for crop breeding to cope with global climate warming.
Heat stress (HS) adversely affects plant survival, growth, and development. Rises in global temperature have severe deleterious consequences for plant growth and development and substantially reduce crop yields (1, 2). During evolution, plants have acquired sophisticated heat signal transduction mechanisms to respond to daily and seasonal variation in temperature (3, 4). The mechanisms underlying plant thermotolerance have been dissected in detail and rely on heat shock factors (HSFs), heat shock proteins (HSPs), reactive oxygen species, phospholipids, and calcium signaling pathways, as well as phytohormone networks (5). Phytochrome B (phyB) and other photosensory receptors, plasma membrane–localized CYCLIC NUCLEOTIDE GATED CALCIUM CHANNELs (CNGCs) and EARLY FLOWERING 3 (ELF3) in Arabidopsis as well as Thermo-tolerance 3.1 (TT3.1) in rice, may function as thermosensors (6–9). Although great progress has been achieved in elucidating the molecular mechanisms of plant heat sensing, how plant cells specifically relay heat signals remains elusive.
Plant growth and development are plastic processes under genetic and epigenetic regulation. Epigenetic regulation is essential for plant adaptation to surrounding environments; epigenetic regulators modulate chromatin conformation and composition, thus affecting gene expression (10). The histone acetylation code, which is critical to transcriptional regulation and genome integrity, is written by histone acetyltransferases (HATs) and erased by histone deacetylases (HDACs). Increased histone acetylation (hyperacetylation) is typically associated with enhanced gene expression, while hypoacetylation represses transcription (11, 12). HATs and HDACs thus influence gene expression and are involved in numerous plant development and stress response pathways. HDACs are highly conserved among eukaryotes. In Arabidopsis, 18 HDAC isoforms have been identified and divided into three different subfamilies: the REDUCED POTASSIUM DEPENDENCY PROTEIN 3 (RPD3/HDA1) subfamily, the HISTONE DEACETYLASE 2 (HD2) subfamily, and the SILENT INFORMATION REGULATOR PROTEIN 2 (SIR2) or sirtuin subfamily (13).
The RPD3-like subfamily member HISTONE DEACETYLASE 9 (HDA9) is integral to plant biological processes including seed germination, flowering, senescence, leaf development, and resistance to stress through its regulation of histone deacetylation and thus gene expression (14–22). HDA9 interacts with the transcription factor WRKY53 to negatively regulate plant sensitivity to salt and drought stress (23, 24). Moreover, POWERDRESS (PWR), which acts in the same complex as HDA9, is essential for thermomorphogenesis in Arabidopsis (17, 18, 25). HDA9 is stabilized in response to high ambient temperature (27 °C) and mediates thermomorphogenesis through deacetylation at YUCCA8, eviction of H2A.Z, and binding of PHYTOCHROME INTERACTING FACTOR 4 (PIF4) (25–27). Together, these reports showed that HDA9 is part of a thermomorphogenesis pathway (28). However, the exact function of HDA9 in plant heat shock signal transduction and responses is unclear. Moreover, a positive role for HDA9 in stress responses has not been reported to date.
Protein phosphorylation is a major posttranslational modification that is integral to many signaling cascades, including environmental stress responses. Protein phosphorylation and dephosphorylation rely on kinases and phosphatases, respectively (29), which are crucial for the regulation of protein activity and stability. Plant PROTEIN PHOSPHATASE 2A (PP2A) complexes function in cell division, phytohormone signaling, development, and biotic stress resistance by dephosphorylating target proteins (30–32). Emerging evidence also indicates that phosphorylation/dephosphorylation modulates nucleocytoplasmic distribution of proteins during signal transduction (33–35). Nuclear pore complexes (NPCs) are the main channels controlling nucleocytoplasmic transport and comprise ∼30 nucleoporins (NUPs) (36). HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 (HOS1), one of the outer-ring components of NPCs (36, 37), is integral to biological processes in plants including flowering, cold, and high-temperature responses (37–42). Whether heat induces changes in the phosphorylation status of HS signaling transduction components and how the components transduce heat signals from the cytoplasm to the nucleus to regulate the genomic chromatin modification landscape are not well-understood.
In this study, we demonstrated that HDA9 relays heat signals to regulate plant heat responses by gradually accumulating in the nucleus in its nonphosphorylated form upon exposure to heat, a process that is fine-tuned by PP2AB′β, a B′ regulatory subunit of PP2A. PP2AB′β directly dephosphorylates HDA9 at high temperatures to protect HDA9 from degradation by the 26S proteasome, leading to the translocation of nonphosphorylated HDA9 to the nucleus. Heat-induced enrichment of HDA9 in the nucleus was dependent on the nucleoporin HOS1. In the nucleus, HDA9 binds and deacetylates the target genes to repress gene expression in a C2H2-type zinc-finger transcription factor YIN YANG 1 (YY1)–dependent and –independent manner. This distinct switch of HDA9 from the cytoplasm to the nucleus in response to HS is conserved in the crop plants wheat and rice. Thus, our findings uncover an elaborate, conserved heat signal transduction mechanism, elucidating how HS signal is transmitted from the cytoplasm into the nucleus for epigenetic modifiers, which has significant implications for crop breeding to cope with global climate warming.
Results
HDA9 Responds Specifically to Heat in Arabidopsis.
To investigate whether HDACs participate in heat responses, we treated wild-type (WT, Col) Arabidopsis seedlings with trichostatin A (TSA), a specific HDAC inhibitor (24, 43). Seedlings grew normally on Murashige and Skoog (MS) medium containing 3 μM TSA or dimethyl sulfoxide (DMSO) (as negative control) under normal growth conditions at 22 °C. However, under a previously established HS treatment protocol (SI Appendix, Fig. S1A) (44), TSA-treated seedlings exhibited increased sensitivity to HS, as evidenced by their lower survival rate compared with untreated seedlings (SI Appendix, Fig. S1 B–D). This result is consistent with a previous report that inhibitors of HDACs diminish temperature-induced hypocotyl elongation (25). This suggests that HDACs are required in the HS response in Arabidopsis.
To determine which HDACs respond to HS, we obtained mutants in class I HDACs of the RPD3-like superfamily for HS phenotypic analysis. Similar to Col, all hdac mutants grew normally under normal conditions. However, hda9 plants displayed significant defects in their thermotolerance in response to HS (Fig. 1 A and B), while mutants of HDA6, HDA7, and HDA19, which are from the same subfamily and have the highest sequence identities with HDA9, showed normal responses to HS treatment (SI Appendix, Fig. S1 E–G). This result indicates that HDA9 specifically functions in HS responses. Moreover, a genomic copy of HDA9 tagged with a FLAG tag fully rescued the compromised thermotolerance and survival rate of the hda9 mutant (HDA9pro:HDA9-FLAG/hda9) (18) after HS treatment (Fig. 1 A and B), as did a genomic copy of HDA9 tagged with green fluorescent protein (GFP) (HDA9pro:HDA9-GFP/hda9) (SI Appendix, Fig. S1 H and I). HDA9 therefore plays a positive regulatory role in heat responses.
Fig. 1.
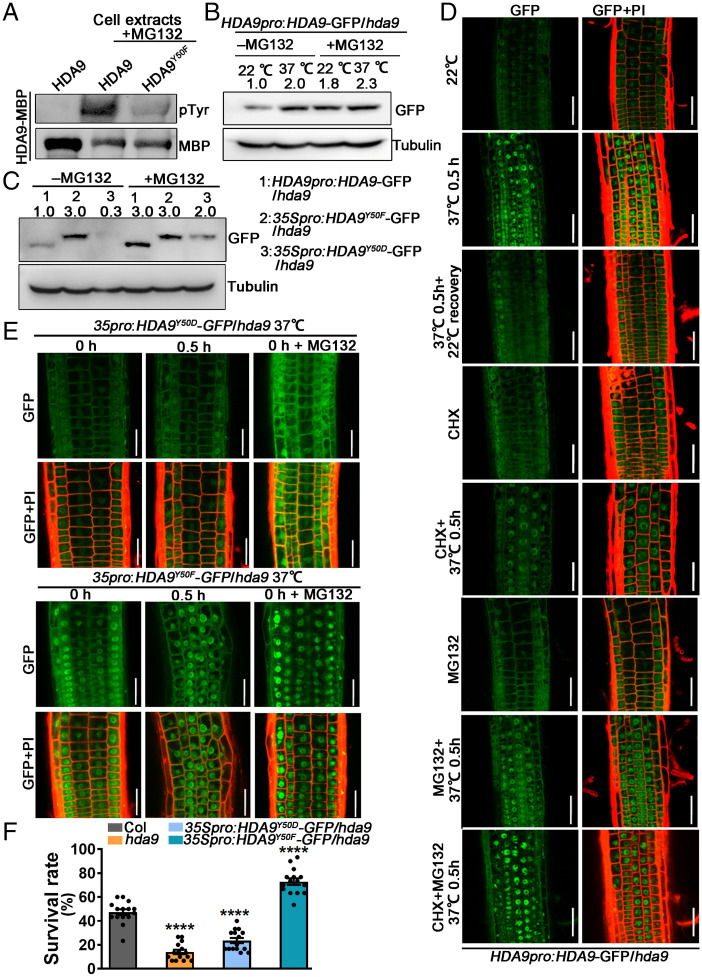
HDA9 abundance specifically responds to heat in Arabidopsis. (A) Phenotypic analysis of Col, hda9, and the complementation line HDA9pro:HDA9-FLAG/hda9 grown at 22 °C or exposed to HS treatment. (B) Survival rates of seedlings shown in A. Each point represents an individual data point. Data are shown as mean ± SE from all seedlings. At least three independent biological replicates were performed (****P < 0.0001; ns, no significant difference; as determined by two-tailed Student’s t test). (C) Immunoblot analysis of HDA9-GFP abundance in crude cell extracts (total; Left) and cytosolic (cytoplasm; Middle) and nuclear (nucleus; Right) fractions from the HDA9pro:HDA9-GFP/hda9 complemented line under control (22 °C), 37 °C (0.5 h), 150 mM NaCl, or 50 μM ABA treatment and then subjected to immunoblot analysis with an anti-GFP antibody. Anti-tubulin and anti-histone H3 antibodies served as loading controls and fraction markers. The titration of control samples (loading 4×, 2×, 1×, and 0.5×) is shown in SI Appendix, Fig. S19A. The control samples in C are equivalent to 0.5× (total), 2× (cytoplasm), and 1× (nucleus) of the titration shown in SI Appendix, Fig. S19A. Three independent biological replicates are shown in Dataset S9. The signal can be compared with the experimental samples. At least three independent biological replicates were performed. (D) HDA9-GFP fluorescence from HDA9pro:HDA9-GFP/hda9 roots subjected to 37 °C HS treatment for the indicated time. Confocal images of GFP fluorescence (green; Upper) and propidium iodide (PI; red) merged with GFP fluorescence (Lower) are shown. (Scale bars, 50 μm.) (E) Relative fluorescence intensity of HDA9-GFP (background was subtracted), as analyzed by ImageJ. Thirty nuclei in the same field of view were observed and quantified. Data are shown as mean ± SE.
To understand how HDA9 contributes to the heat response, we examined HDA9 accumulation before and after exposure to HS (0.5 h at 37 °C). Both immunoblot and fluorescence observations of HDA9-GFP indicated that heat promotes the accumulation of HDA9 (Fig. 1C and SI Appendix, Fig. S2A). Given that HDA9 was previously shown to be involved in salt and abscisic acid (ABA) signaling (13, 23), we examined HDA9 accumulation upon treatment with 150 mM NaCl or 50 µM ABA. We observed no HDA9 enrichment following either treatment but instead a decrease in HDA9-GFP abundance relative to control conditions (Fig. 1C and SI Appendix, Fig. S2A), indicating that HDA9 may be degraded in response to NaCl and ABA stress, in agreement with the proposed negative regulatory role for HDA9 in these stress responses (13, 23).
We detected equal HDA9-GFP fluorescence in the cytosol and the nucleus under normal conditions (SI Appendix, Fig. S2A); however, HDA9-GFP abundance dramatically increased in the nucleus in response to HS but not NaCl or ABA treatments (SI Appendix, Fig. S2A). We confirmed the subcellular distribution of HDA9 after HS treatment using a nuclear–cytoplasmic fractionation assay, which revealed HDA9 in both compartments in the absence of HS (Fig. 1C), in line with previous findings (15, 18). Notably, after HS treatment, HDA9 accumulation increased in total protein extracts and in the nuclear fraction but decreased in the cytosolic fraction (Fig. 1C), indicating that heat either induces HDA9 translocation from the cytoplasm to the nucleus or promotes HDA9 stability in the nucleus.
To examine the dynamics of HDA9 accumulation, we treated HDA9pro:HDA9-GFP/hda9 seedlings with HS. We observed an increase in HDA9 accumulation in the nucleus starting after 5 min of HS treatment (Fig. 1 D and E). This suggests that HDA9 responds quickly to heat signals. Furthermore, when HDA9pro:HDA9-GFP/hda9 seedlings were treated at different temperatures ranging from 22 to 37 °C, we observed a gradual enrichment in HDA9 in the nucleus with increasing temperature. HDA9 abundance returned to its 22 °C levels when seedlings were allowed to recover from HS treatment (SI Appendix, Fig. S2 B and C), indicative of a precise mechanism regulating the enrichment and turnover of nuclear HDA9 in response to temperature changes.
To further explore the function of HDA9, we generated and selected 35Spro:HDA9-GUS (β-GLUCURONIDASE)–overexpressing lines (OE1 to OE3) harboring a single insertion and with clear overexpression of HDA9 (SI Appendix, Fig. S3). However, HDA9-overexpressing lines exhibited a comparable heat resistance to Col seedlings (SI Appendix, Fig. S3), indicating that the increased HDA9 expression did not enhance heat resistance or HDA9 was fine-tuned at posttranscriptional levels.
Heat Promotes HDA9 Dephosphorylation and Translocation to the Nucleus.
Phosphorylation and dephosphorylation can modulate the nucleocytoplasmic partitioning and stability of proteins (45). We thus analyzed the phosphorylation status of HDA9 under normal and HS conditions. Accordingly, we immunoprecipitated HDA9-FLAG with an anti-FLAG antibody and analyzed the immunoprecipitates by Phos-tag polyacrylamide-gel electrophoresis (PAGE), in which phosphorylated proteins migrate in the gel more slowly than nonphosphorylated proteins (34). We detected slower-migrating bands representing putative phosphorylated HDA9 forms in the total and cytoplasmic fractions, which were nearly abolished by the addition of calf intestinal alkaline phosphatase (CIP) (SI Appendix, Fig. S4A). However, HDA9 immunoprecipitated from the nucleus showed no evidence of phosphorylation, either before or after HS treatment (SI Appendix, Fig. S4A), indicating that only nonphosphorylated HDA9 accumulates in the nucleus. Notably, HS treatment decreased the levels of phosphorylated HDA9 in total and cytoplasmic fractions (SI Appendix, Fig. S4A). These results indicate that heat induces HDA9 dephosphorylation and promotes its accumulation in the nucleus.
To identify the HDA9 residues modified by phosphorylation, we predicted all potential phosphorylation sites (SI Appendix, Fig. S5) in HDA9 and compared their positions and residues with those in HDA9 homologous proteins. We selected two putative phosphorylation sites in HDA9 located within the HDAC domain (SI Appendix, Fig. S5). Given the specific response of HDA9 to HS treatment, we focused on a single HDA9-specific tyrosine residue at position 50 (Y-50), which is unique to HDA9 from other homologous genes, for further examination (SI Appendix, Fig. S5). We generated point mutants with Y-50 replaced by phenylalanine (Y50F) to mimic a nonphosphorylated residue or by aspartic acid (Y50D) to mimic a phosphorylated residue and introduced the resulting constructs into the hda9 mutant to obtain 35Spro:HDA9Y50F-GFP/hda9 and 35Spro:HDA9Y50D-GFP/hda9 transgenic lines. Then, we immunoprecipitated HDA9-GFP, HDA9Y50F-GFP, and HDA9Y50D-GFP from the corresponding lines with an anti-GFP antibody and performed Phos-tag PAGE and sodium dodecyl sulfate (SDS)–PAGE assays. We observed a small but reproducible reduction in the phosphorylation of HDA9Y50F-GFP and HDA9Y50D-GFP lines (SI Appendix, Fig. S4B). Treating the seedlings with MG132, a specific inhibitor of the 26S proteasome (34), increased the phosphorylation signal detected in HDA9Y50D-GFP seedlings (SI Appendix, Fig. S4B). To validate these results, we tested bulk kinase and phosphatase activity directed against recombinant maltose-binding protein (MBP-HDA9) in protein extracts in a cell-free system with MG132. Notably, Tyr phosphorylation levels decreased in MBP-HDA9Y50F relative to MBP-HDA9 (Fig. 2A). Collectively, these results show that the Y-50 residue of HDA9 is phosphorylated and influences HDA9 protein abundance.
Fig. 2.
Dephosphorylation enhances the nuclear accumulation and stability of HDA9 under HS. (A) Examination of phosphorylation modifications in a cell-free system. Recombinant MBP-HDA9 was added to cell extracts from Col seedlings along with MG132. Anti-MBP and anti-pTyr antibodies were used to detect all MBP-HDA9 and phosphorylated MBP-HDA9, respectively. (B) MG132 inhibits the degradation of HDA9. HDA9pro:HDA9-GFP/hda9 seedlings grown in the absence or presence of 50 μM MG132 were treated with HS at 37 °C for 0.5 h. The titration of control samples (loading 4×, 2×, 1×, and 0.5×) is shown in SI Appendix, Fig. S19A. The control samples in B are equivalent to 1× (total) of the titration shown in SI Appendix, Fig. S19A. Three independent biological replicates are shown in Dataset S9. (C) The nonphosphorylated form of HDA9 is relatively stable. Immunoblot detection of HDA9Y50D-GFP and HDA9Y50F-GFP in seedlings grown in the absence or presence of 50 μM MG132. The titration of control samples (loading 4×, 2×, 1×, and 0.5×) are shown in SI Appendix, Fig. S19 A–C. The control samples in C are equivalent to 0.5× (total), 2×, and 0.5× of the titration shown in SI Appendix, Fig. S19 A–C, respectively. Three independent biological replicates are shown in Dataset S9. At least three independent biological replicates were performed for A–C. (D) HDA9-GFP fluorescence from HDA9pro:HDA9-GFP/hda9 seedlings subjected to the indicated treatments; 100 μM MG132 and 100 μM CHX were used. (Scale bars, 50 μm.) (E) HDA9-GFP fluorescence from 35Spro:HDA9Y50D-GFP/hda9 and 35Spro:HDA9Y50F-GFP/hda9 seedlings subjected to the indicated treatments. Confocal images of GFP fluorescence (green; Upper) and PI (red) merged with GFP fluorescence (Lower) are shown. (Scale bars, 50 μm.) (F) The survival rates of Col, hda9, and 35Spro:HDA9Y50D/Y50F-GFP/hda9 seedlings after HS treatment, as illustrated in SI Appendix, Fig. S1A. Each point represents an individual data point. Data are shown as mean ± SE of all plants from three independent biological replicates (****P < 0.0001, as determined by Student’s t test).
To determine if phosphorylation affects HDA9 stability, we treated HDA9pro:HDA9-GFP/hda9 seedlings with MG132 under normal and HS conditions and then performed immunoblotting of plant total protein extracts with an anti-GFP antibody. This revealed a marked increase in HDA9-GFP abundance in the presence of MG132 that became more pronounced under HS treatment, indicating that HDA9 is degraded by the 26S proteasome in normal conditions but this process is blocked by heat (Fig. 2B). We next treated 35Spro:HDA9Y50F-GFP/hda9 and 35Spro:HDA9Y50D-GFP/hda9 as well as HDA9-GFP seedlings with MG132 under normal conditions to examine HDA9 stability. While HDA9 levels increased by twofold in the presence of MG132, HDA9Y50F-GFP levels remained constant (Fig. 2C). In contrast, HDA9Y50D-GFP abundance dramatically increased, by over fivefold, upon MG132 treatment (Fig. 2C), revealing that phosphorylated HDA9 is specifically degraded by the 26S proteasome. Thus, the low levels of phosphorylated HDA9 in bulk HDA9 were likely responsible for the slight but reproducible rise in HDA9 abundance in the presence of MG132 (SI Appendix, Fig. S4B).
Consistent with these observations, confocal imaging showed that HS treatment increased both the abundance and relative accumulation of HDA9-GFP in the nucleus in HDA9pro:HDA9-GFP/hda9 seedlings (Fig. 2D). To distinguish whether the nuclear accumulation of HDA9 is due to increased cytoplasmic translocation or to increased protein stabilization in the nucleus, we treated the HDA9pro:HDA9-GFP/hda9 seedlings with the protein synthesis inhibitor cycloheximide (CHX) (46) and MG132 and then observed the subcellular localization of HDA9-GFP. Even after CHX application, the HDA9pro:HDA9-GFP/hda9 seedlings still showed significant nuclear enrichment of HDA9 after HS treatment (Fig. 2D). Meanwhile, the HDA9-GFP fluorescence from 35Spro:HDA9-GFP/Col seedlings was also stronger in the nucleus after HS treatment (SI Appendix, Fig. S6A). These results indicate that the increased level of HDA9 in the nucleus under HS treatment is not due to higher expression of the HDA9 gene. Adding MG132 increased the protein stability of HDA9 under both standard and HS conditions (Fig. 2 B–D), but it increased nuclear enrichment of HDA9 only under HS treatment (Fig. 2D). When CHX and MG132 were added simultaneously, there was still a significant accumulation of HDA9-GFP signals in the nucleus after HS treatment (Fig. 2D). We obtained similar results using 35Spro:HDA9-GFP/Col seedlings treated with MG132 (SI Appendix, Fig. S6A). These results show that the accumulation of HDA9 in the nucleus is most likely due to cytoplasmic translocation rather than to promotion of protein stability in the nucleus.
Importantly, HDA9Y50D-GFP from 35Spro:HDA9Y50D-GFP/hda9 seedlings displayed a weak signal that was insensitive to HS treatment but increased in the presence of MG132, indicating that HDA9 phosphorylation is critical for HDA9 stability (Fig. 2E). HDA9Y50D-GFP was concentrated in the cytoplasm and not in the nucleus, in line with the exclusive nuclear localization of nonphosphorylated HDA9 (Fig. 2E). In contrast, 35Spro:HDA9Y50F-GFP/hda9 showed stronger GFP signals than HDA9pro:HDA9-GFP/hda9 seedlings in both the cytoplasm and the nucleus. In addition, HDA9Y50F-GFP was concentrated in the nucleus under HS treatment, and its abundance was unaffected by MG132 treatment (Fig. 2 C and E). These results indicate that nonphosphorylated HDA9 is stable and that exposure to high temperature promotes the relocation of nonphosphorylated HDA9 into the nucleus, not the stabilization of HDA9 in the nucleus. In agreement with this, 35Spro:HDA9Y50F-GFP/hda9 seedlings exhibited gain-of-function tolerance for HS more than Col seedlings, whereas 35Spro:HDA9Y50D-GFP/hda9 seedlings still showed compromised thermotolerance close to that of hda9 seedlings (Fig. 2F and SI Appendix, Fig. S6B). These results indicate that translocation of nonphosphorylated HDA9 into the nucleus is required for HDA9-mediated heat responses.
PP2AB′β Interacts with and Dephosphorylates HDA9 to Stabilize HDA9 in Response to Heat.
To investigate the biochemical mechanism by which HDA9 mediates the heat signal, we isolated the HDA9 protein complex by immunoaffinity purification followed by mass spectrometry (IP-MS) using HDA9pro:HDA9-FLAG/hda9 seedlings. We identified 25 unique peptides matching HDA9, as well as 9 unique peptides corresponding to the known HDA9-interacting protein PWR (18), verifying the high reliability of our IP-MS assay. Besides HDA9, we obtained the greatest number of unique enriched peptides (15) for the largely cytosolic protein PP2AB′β (SI Appendix, Fig. S7), which is the regulatory subunit of PP2A and contributes to abiotic and biotic stress responses by assisting the PP2A enzyme complex in the dephosphorylation of target proteins (47, 48). To validate the interaction between HDA9 and PP2AB′β, we performed a yeast two-hybrid assay and verified that HDA9 directly interacts with PP2AB′β in yeast (SI Appendix, Fig. S8A), which was confirmed in vivo with firefly luciferase complementation imaging (LCI) assays in Nicotiana benthamiana leaves (SI Appendix, Fig. S8 B and C). Notably, the HDA9–PP2AB′β interaction was enhanced by HS treatment (SI Appendix, Fig. S8 B and C). As HDA9 and PP2AB′β expression was driven by a constitutive promoter in the firefly luciferase system, we deduced that heat enhances the interaction of HDA9 and PP2AB′β rather than their respective transcript levels. Notably, compared with intact HDA9, the phosphorylation-mimic form of HDA9 (HDA9Y50D) showed a stronger affinity for PP2AB′β while its nonphosphorylatable form (HDA9Y50F) exhibited normal binding toward PP2AB′β that did not increase upon HS treatment (SI Appendix, Fig. S8 A–C), indicating that PP2AB′β preferentially interacts with phosphorylated HDA9. To test whether the interaction between HDA9Y50D and PP2AB′β occurred in the cytosol, we performed bimolecular fluorescence complementation (BiFC) assays in tobacco epidermal cells and found yellow fluorescent protein (YFP) fluorescence signal in the cytosol (SI Appendix, Fig. S8D), implying a direct interaction between HDA9Y50D and PP2AB′β in the cytosol in planta. We further confirmed the heat-induced promotion of the HDA9–PP2AB′β interaction in vivo by coimmunoprecipitation experiments using 35Spro:PP2AB′β-GFP transgenic lines. For equal amounts of immunoprecipitated PP2AB′β-GFP, we detected twofold as much coprecipitated HDA9 from HS-treated seedlings relative to control conditions (Fig. 3A). Collectively, these results indicated that HDA9 is a substrate of dephosphorylation by PP2AB′β and that heat promotes the interaction between the two proteins.
Fig. 3.
PP2AB′β plays a positive regulatory role in response to HS by dephosphorylating HDA9. (A) Coimmunoprecipitation (Co-IP) assays. Anti-GFP antibody was used for IP and anti-HDA9 antibody was used to detect HDA9 in Col and a PP2AB′β-overexpressing line (35Spro:PP2AB′β-GFP). The titration of control samples (loading 4×, 2×, 1×, and 0.5×) is shown in SI Appendix, Fig. S19 D and E. The control samples in A are equivalent to 1× of the titration shown in SI Appendix, Fig. S19 D and E. Three independent biological replicates are shown in Dataset S9. (B) Immunoblot analysis of HDA9 abundance in Col and the pp2ab′β mutant before (0 h) and after HS treatment (37 °C for 0.5 h). (C) The loss of PP2AB′β in the pp2ab′β mutant increases the phosphorylation level of HDA9. Equal amounts of immunoprecipitated HDA9 protein were separated by Phos-tag SDS-PAGE and then subjected to immunoblot analysis with an anti-HDA9 antibody. The slower mobility seen for HDA9 in the Phos-tag gel represents the phosphorylated form of HDA9 (pHDA9). As loading control (Bottom), equal amounts of the immunoprecipitated HDA9 protein were separated by regular SDS-PAGE and then subjected to immunoblot analysis. The titration of control samples (loading 4×, 2×, 1×, and 0.5×) is shown in SI Appendix, Fig. S19H. The control samples in C are equivalent to 2× of the titration shown in SI Appendix, Fig. S19H. At least three independent biological replicates were performed for A–C (Dataset S9). (D) The pp2ab′β mutation greatly reduces HDA9-GFP accumulation in the nucleus after HS treatment. Confocal images of HDA9-GFP in the roots of HDA9pro:HDA9-GFP/hda9, HDA9pro:HDA9-GFP/pp2ab′β, 35Spro:HDA9Y50D-GFP/pp2ab′β, and 35Spro:HDA9Y50F-GFP/pp2ab′β transgenic lines grown on MS medium, before (0 h) or after HS treatment (37 °C for 0.5 h). Confocal images of GFP fluorescence (green; Left) and PI (red) merged with GFP fluorescence (Right) are shown. (Scale bars, 50 μm.) (E) Survival rates of Col, pp2ab′β, 35Spro:HDA9Y50D-GFP/pp2ab′β, and 35Spro:HDA9Y50F-GFP/pp2ab′β seedlings after HS, as illustrated in SI Appendix, Fig. S1A. For E, each point represents an individual data point. Data are shown as mean ± SE of all plants from three independent biological replicates (****P < 0.0001, **P < 0.01, *P < 0.05; ns, no significant difference; as determined by Student’s t test).
We further investigated the role of PP2AB′β in regulating HDA9 stability. We determined that HDA9 abundance decreased in pp2ab′β seedlings exposed to HS treatment (Fig. 3B), indicating that PP2AB′β stabilizes HDA9. Next, we tested whether PP2AB′β affected the phosphorylation status of HDA9 in vivo through Phos-tag PAGE analysis of protein extracts from Col and pp2ab′β seedlings grown under normal and HS conditions. HDA9 phosphorylation levels increased in pp2ab′β relative to Col in both conditions (Fig. 3C), indicating that the loss of PP2AB′β prevents the dephosphorylation of HDA9 and destabilizes the protein. We next characterized the subcellular localization of HDA9-GFP derived from the HDA9pro:HDA9-GFP construct in the hda9 and pp2ab′β mutant backgrounds. Under the same laser energy settings for confocal imaging, HDA9-GFP fluorescence, particularly in the nucleus, was much weaker in pp2ab′β even in normal growth conditions and did not increase upon HS treatment. The 35Spro:HDA9Y50F-GFP/pp2ab′β seedlings produced stronger GFP signals in both the cytoplasm and the nucleus compared with pHDA9:gHDA9-GFP/pp2ab′β and 35Spro:HDA9Y50D-GFP/pp2ab′β seedlings (Fig. 3D). Collectively, these results suggest that heat promotes the interaction between PP2AB′β and phosphorylated HDA9 to dephosphorylate and stabilize HDA9. Based on the observation that only nonphosphorylated HDA9 accumulates in the nucleus, combined with the higher abundance of nonphosphorylatable HDA9Y50F-GFP in the nucleus in normal conditions (Fig. 2E), we propose that reversible dephosphorylation of HDA9 by PP2A fine-tunes the ratio between phosphorylated and nonphosphorylated HDA9 to determine how much nuclear HDA9 accumulates in response to temperature changes.
To test this hypothesis, we assessed the thermotolerance of Col, pp2ab′β, and transgenic lines overexpressing HDA9Y50F (35Spro:HDA9Y50F-GFP/pp2ab′β) or HDA9Y50D (35Spro:HDA9Y50D-GFP/pp2ab′β) in response to HS treatment. The pp2ab′β mutant was more sensitive than Col to HS treatment and was similar to the hda9 mutant in this regard (Fig. 1A). This HS-sensitive phenotype was rescued by the 35Spro:HDA9Y50F-GFP transgene but not the 35Spro:HDA9Y50D-GFP transgene (Fig. 3E and SI Appendix, Fig. S8E), indicating that hda9 is epistatic to pp2ab′β and that HDA9 functions downstream of PP2AB′β in response to heat.
HDA9 Nuclear Accumulation after HS Is Dependent on HOS1.
To investigate the molecular mechanisms by which HDA9 enters the nucleus after HS, we analyzed the proteins that interacted with HDA9 in the IP-MS results. We found that several nucleoporins were specifically coprecipitated with HDA9-GFP from plant protein extracts (SI Appendix, Fig. S7), indicating a strong association of HDA9 with components of the NPC. One of these interacting proteins was a temperature-sensitive NPC component, HOS1 (36). We first examined the HDA9–HOS1 interaction using LCI assays in N. benthamiana leaves. Notably, like intact HDA9, HDA9Y50D and HDA9Y50F bound HOS1 (SI Appendix, Fig. S9A). Then, we confirmed the HDA9–HOS1 interaction in vivo by coimmunoprecipitation experiments using 35Spro:HOS1-MYC transgenic lines. For equal amounts of immunoprecipitated HOS1-MYC, we detected a twofold increase of coprecipitated HDA9 from seedlings under HS relative to seedlings under control conditions (Fig. 4A), suggesting that heat promoted the interaction between HDA9 and HOS1.
Fig. 4.
Loss of HOS1 function represses HDA9 nuclear accumulation after HS. (A) Co-IP assays. Anti-MYC antibody was used for IP and an anti-HDA9 antibody was used to detect HDA9 in Col and a HOS1-overexpressing line (35Spro:HOS1-MYC). The titration of control samples (loading 4×, 2×, 1×, and 0.5×) is shown in SI Appendix, Fig. S19F. The control samples in A are equivalent to 2× of the titration shown in SI Appendix, Fig. S19F. At least three independent biological replicates were performed (Dataset S9). (B) Colocalization of HDA9-GFP, HDA9Y50D-GFP, HDA9Y50F-GFP, and HOS1-mCherry in transgenic plant roots. The GFP, mCherry, and overlapped fluorescence are indicated by green, blue, and yellow arrows, respectively. (Scale bars, 50 μm.) (C) The hos1 mutation repressed HDA9-GFP accumulation in the nucleus after HS treatment. Confocal images of HDA9-GFP in the roots of 35Spro:HDA9Y50D-GFP/hos1 and 35Spro:HDA9Y50F-GFP/hos1 transgenic lines grown on MS medium, before (0 h) or after HS treatment (37 °C for 0.5 h). Confocal images of GFP fluorescence (green; Left) and PI (red) merged with GFP fluorescence (Right) are shown. (Scale bars, 50 μm.) (D) Survival rates of Col, hos1, 35Spro:HDA9Y50D-GFP/hos1, and 35Spro:HDA9Y50F-GFP/hos1 seedlings after HS, as illustrated in SI Appendix, Fig. S1A. Each point represents an individual data point. Data are shown as mean ± SE of all plants from three independent biological replicates (****P < 0.0001, as determined by Student’s t test). (E) Euclidean distance hierarchical clustering of gene expression profiles among Col, hda9, hos1, 35Spro:HDA9Y50F-GFP/hda9, and 35Spro:HDA9Y50F-GFP/hos1 under control and HS conditions 37 °C (0.5 h). All expressed genes, of which mean Fragments Per Kilobase of transcript per million mapped reads (FPKM) ≥1 in at least one sample, were subjected to hierarchical clustering analysis. Heatmap showing the Z scores (scaled and centered expression) transformed from FPKM values of expressed genes in the indicated samples. Columns represent sequenced samples; rows indicate individual expressed genes; and the dendrogram above the columns indicates relatedness among samples. The color range in the legend indicates the scaled expression level ranging from −2 (blue, below average expression) to +2 (red, above average expression).
To verify the in vivo association of HDA9 and HOS1, we then examined the colocalization of the two proteins. We obtained dual-expression lines by crossing transgenic gHOS1-mCherry plants (37) with HDA9-GFP, HDA9Y50D-GFP, or HDA9Y50F-GFP plants. In agreement with a previous study (37), we found that HOS1-mCherry was predominantly located at the nuclear envelope (Fig. 4B). HDA9Y50F-GFP protein showed more overlapping distribution with HOS1-mCherry than HDA9-GFP and HDA9Y50D-GFP at the nuclear envelope in dual-transgenic plants (Fig. 4B). Collectively, these results indicated that HDA9 and HOS1 coexist at the nuclear envelope and that heat promotes their interaction.
The overlap of HDA9 and HOS1 at the nuclear envelope prompted us to determine if the HDA9 nuclear accumulation could be affected by HOS1 in a temperature-dependent manner. To this end, we analyzed the subcellular localization of the HDA9Y50D-GFP and HDA9Y50F-GFP proteins in hos1 plants, the T-DNA insertion knockout mutants (Fig. 4C). HS treatment did not increase the nuclear accumulation of the HDA9Y50F-GFP fusion proteins in hos1 as it did in hda9 (Figs. 2D and 4C), which was confirmed by the nucleocytoplasmic fractionation assay using 35Spro:HDA9Y50F-GFP/hda9 and 35Spro:HDA9Y50F-GFP/hos1 complemented lines (SI Appendix, Fig. S9B). These results suggest that a lack of HOS1 represses the nuclear accumulation of HDA9 after HS. We examined the thermotolerance phenotypes of Col and hos1 seedlings alongside hos1 transgenic lines harboring the 35Spro:HDA9Y50D-GFP and 35Spro:HDA9Y50F-GFP transgenes to determine if HOS1 could affect HDA9 function. Consistently, the hos1 mutant was more sensitive than Col to HS treatment, as reported previously (41). In addition, 35Spro:HDA9Y50F-GFP/hos1 seedlings were as sensitive to HS as the hos1 mutant (Fig. 4D and SI Appendix, Fig. S9C), in contrast to the enhanced thermotolerance exhibited by 35Spro:HDA9Y50F-GFP/hda9 (Fig. 2F and SI Appendix, Fig. S6B) and 35Spro:HDA9Y50F-GFP/pp2ab′β seedlings (Fig. 3E and SI Appendix, Fig. S8E). This indicates that HOS1 is required for HDA9 nuclear accumulation and function in plant thermotolerance.
To investigate the role of HOS1 in HDA9-mediated heat responses, we performed transcriptome analysis by messenger RNA sequencing (RNA-seq) of Col, hda9, 35Spro:HDA9Y50F-GFP/hda9, hos1, and 35Spro:HDA9Y50F-GFP/hos1 plants under normal and HS conditions. Hierarchical clustering analysis revealed that the transcriptome profile of hda9, but not 35Spro:HDA9Y50F-GFP/hda9, grouped with Col under normal conditions. This indicates that the gene expression profile in hda9 is similar to that in Col, likely due to redundant genes, and that the HDA9Y50F transgene has a gain-of-function effect on gene expression, consistent with the nuclear enrichment of HDA9Y50F and increased heat tolerance of plants expressing HDA9Y50F (Fig. 2F and SI Appendix, Fig. S6B). Under HS conditions, the 35Spro:HDA9Y50F transgene rescued the gene expression defects of hda9, confirming its role in heat responses, while hda9 and hos1 grouped together, consistent with their heat-sensitive phenotype (Figs. 1A and 4D). Moreover, 35Spro:HDA9Y50F-GFP/hos1 clustered closely with hos1 under normal and HS conditions (Fig. 4E), showing that HOS1 is required for HDA9Y50F function, in line with the impaired HDA9Y50F localization in 35Spro:HDA9Y50F-GFP/hos1 plants (Fig. 4C and SI Appendix, Fig. S9B).
Heat Enhances Formation of an HDA9–YY1 Complex Involved in Thermotolerance.
Our IP-MS analysis also identified the transcription factor YY1 as a protein copurifying with HDA9, with five unique peptides (SI Appendix, Fig. S7). YY1 acts as a dual-function transcription factor with both repression and activation domains that each recruit different cofactors to target loci (49). We confirmed the HDA9–YY1 interaction using a yeast two-hybrid assay (SI Appendix, Fig. S10A). We then performed a coimmunoprecipitation assay with an anti-GFP antibody in protein extracts from 35Spro:YY1-GFP transgenic seedlings to assess the HDA9–YY1 interaction. Indeed, YY1-GFP coimmunoprecipitated with HDA9, indicating that HDA9 interacts with YY1 in vivo (Fig. 5A). To test whether the interaction between HDA9 and YY1 occurred in the nucleus, we performed BiFC assays in tobacco epidermal cells and found the YFP fluorescence signal only in the nucleus (SI Appendix, Fig. S10B), implying a direct HDA9–YY1 interaction in the nucleus of living tobacco cells. Importantly, more HDA9 was coimmunoprecipitated by YY1 upon HS treatment than under control conditions, for equal amounts of precipitated YY1 (Fig. 5A), suggesting that heat either enhances the HDA9–YY1 interaction or promotes the relocation of HDA9 to the nucleus, where it can more easily bind to YY1. We then employed the firefly LCI assay to distinguish between these possibilities. The interactions between YY1 and both HDA9 and the HDA9Y50F variant increased in response to HS treatment to the same extent, and HDA9Y50F showed a similar binding affinity for YY1 under normal conditions compared with intact HDA9 (Fig. 5B and SI Appendix, Fig. S10C). As HDA9Y50F was more abundant in the nucleus than intact HDA9, even in normal conditions (Fig. 2E), we conclude that heat may enhance the strength of the interaction between HDA9 and YY1.
Fig. 5.
Heat enhances the interaction between HDA9 and YY1. (A and B) Co-IP (A) and LCI (B) assays show that HDA9 and HDA9Y50F interact with YY1 and that HS treatment enhances these interactions. The titration of control samples (loading 4×, 2×, 1×, and 0.5×) for A is shown in SI Appendix, Fig. S19G. The control samples in A are equivalent to 0.5× of the titration shown in SI Appendix, Fig. S19G. At least three independent biological replicates were performed (Dataset S9). (C) Survival rates of Col, yy1, 35Spro:HDA9Y50F-GFP/yy1, and 35Spro:HDA9Y50F-GFP/hda9 seedlings, as illustrated in SI Appendix, Fig. S1A. Each point represents an individual data point. For C, data are shown as mean ± SE of all plants from three independent biological replicates (****P < 0.0001, *P < 0.05, as determined by Student’s t test).
As an independent genetic validation of these results, we examined the thermotolerance phenotypes of Col and yy1 seedlings alongside yy1 transgenic lines harboring the 35Spro:HDA9Y50F-GFP transgene. The yy1 mutant was more sensitive than Col to HS treatment; in addition, 35Spro:HDA9Y50F-GFP/yy1 seedlings were as sensitive to HS as the yy1 mutant (Fig. 5C and SI Appendix, Fig. S10D), in contrast to the enhanced thermotolerance exhibited by 35Spro:HDA9Y50F-GFP/hda9 seedlings (Fig. 2F), indicating that YY1 is required for HDA9-mediated plant thermotolerance.
HDA9 Transmits Heat Signals to Regulate Gene Expression in the Nucleus.
HDA9 plays important roles in flowering time, aging, and other aspects of plant development (17–21). The binding of HDA9 to target loci is the first, essential step for removing histone acetylation and thus decreasing transcriptional activation after accumulation of HDA9 in the nucleus. Therefore, to investigate the role of HDA9 as a heat signal transducer from its stable function in plant development, we examined the in vivo genomic localization of HDA9 by performing chromatin immunoprecipitation sequencing (ChIP-seq) with anti-FLAG using HDA9pro:HDA9-FLAG/hda9 and Col seedlings under control and HS conditions. Based on two biological replicates of the ChIP-seq experiment (SI Appendix, Fig. S11A and Dataset S1), we in total identified 6,087 and 4,678 HDA9-binding peaks under HS and normal conditions, respectively (Dataset S2). Annotation of these ChIP-seq peaks identified 5,009 HDA9-binding genes (Dataset S3), among which 1,328 were heat-induced HDA9 targets, which we termed HS-specific genes, consistent with the nuclear accumulation of HDA9 under HS. In addition, 310 genes displayed loss of HDA9 binding under HS, and these were termed control-specific genes (Fig. 6A). Among the remaining 3,371 HDA9-binding genes shared in control and HS conditions, a set of 243 genes had much more HDA9 binding under HS compared with the control (Fig. 6A and Dataset S3). Therefore, combining the 1,328 HS-specific genes with the 243 shared genes with increased HDA9 binding, we identified 1,571 genes as bound by HDA9 after it accumulates in the nucleus under HS. HDA9 mainly occupied the gene-body regions in control-specific HDA9-binding genes, but occupied the region 1 kb upstream of transcript start sites (TSSs) in shared and HS-specific HDA9-binding genes (SI Appendix, Fig. S12A). In contrast to the control-specific genes, the shared and HS-specific HDA9-binding genes exhibited significantly higher HDA9 levels in HS compared with control conditions (SI Appendix, Fig. S12B), which suggested that HS enhanced HDA9 occupancy in these two types of genes.
Fig. 6.
Identification of HDA9-mediated direct target genes during heat signal transduction. (A) Comparison between HDA9-binding genes identified in control and HS conditions. The number in parentheses indicates the number of shared HDA9-binding genes showing significantly higher HDA9-binding levels in HS compared with the control condition, which is indicated by the arrow. (B) Enriched GO functional categories of the three types of HDA9-binding genes. The color in each cell indicates −log10-transformed FDR of the GO enrichment according to the scale shown, and cells with gray color indicate no significance. (C) The significantly enriched motifs in the 200-bp regions surrounding the summit of HDA9-binding peaks in control and HS. The number and percentage of peaks containing the motif in control and HS conditions are shown. Beneath the HS peaks, the numbers and percentages indicate the counts and fractions of the 1,239 HS peaks with a YY1 motif present in shared and HS-specific gene clusters, respectively. (D) Metaplot of average normalized HDA9-FLAG ChIP-seq signals at regions from 2K bp up- to downstream of the YY1-dependent (Left) and -independent (Right) HDA9-binding genes under HS conditions in HDA9pro:HDA9-FLAG/hda9 and HDA9pro:HDA9-FLAG/yy1. TTS, transcription termination site. −2Kb and 2Kb represent 2K bp upstream of the TSS and 2K bp downstream of the TTS. The y axis represents the log2-transformed value of HDA9-FLAG ChIP-seq reads relative to those of WT. (E) Venn diagram showing the overlap among HDA9-binding genes in HS conditions and HS-induced hypoacetylated and down-regulated genes identified in Col. (F) Integrated Genomics Viewer snapshots of normalized read counts merged from different biological replicates for HDA9-FLAG (anti-FLAG) and H3K9ac ChIP-seq (anti-H3K9ac) as well as RNA-seq at the loci of WRKY33. (G) ChIP-qPCR validation of HDA9 enrichment and H3K9ac level as well as qRT-PCR validation of the expression level of WRKY33. pTA3 and UBC30 (AT5G25760) were used as an internal control for ChIP-qPCR and qRT-PCR, respectively. Error bars denote SEM of three biological replicates. Data points are indicated in the bar charts. For anti-FLAG ChIP-qPCR, different letters indicate significant differences based on one-way ANOVA with Tukey’s Honest Significant Difference test (P < 0.05). For anti-H3K9ac ChIP-qPCR and qRT-PCR, asterisks indicate statistically significant differences between HS and control conditions (**P < 0.01, *P < 0.05, as determined by two-tailed Student’s t test).
HDA9-binding genes are implicated in signal transduction.
Gene Ontology (GO) analysis of HDA9-binding genes revealed that GO terms related to signal transduction, including “signal transduction” and “protein kinase cascade,” and biosynthesis, metabolism, and signaling pathways of various phytohormones like jasmonic acid (JA), ethylene, and ABA, were highly enriched in the HDA9-binding genes under heat treatment; these were also enriched in the shared genes (Fig. 6B). Surprisingly, GO terms related to HS such as “response to stress,” “response to heat,” and “heat acclimation” were enriched in the control-specific genes (Fig. 6B). These results indicated that HDA9, as a heat signal transducer, binds to genes involved in signal transduction and phytohormone functions to regulate plant development in response to heat. At the same time, HDA9 releases from genes related to HS to derepress these genes, ensuring an active HS response.
HDA9 binding of target loci is partially dependent on YY1.
Motif discovery analysis found that the DNA sequence NNDSCWTNN, a potential YY1-binding motif (50), showed significant enrichment and occurred in 12 and 20% of the detected peaks of control and HS conditions, respectively (Fig. 6C), indicating that the binding of the HDA9–YY1 complex to target loci was increased after HS. Out of 1,239 HS peaks with the YY1 motif, 872 (70%) are present in both the control and the HS condition and 367 (30%) are HS-specific (Fig. 6C). GO enrichment analysis revealed that no specific GO terms were significantly enriched in HDA9-binding genes with the YY1 motif under control conditions (SI Appendix, Fig. S13). However, the HDA9-binding genes with the YY1 motif under HS conditions were mainly associated with the response to stress, phytohormone-related (JA, ABA, and ethylene) responses, as well as signaling processes (SI Appendix, Fig. S13), which was similar to the HS-specific HDA9-binding genes (Fig. 6B). Considering this and the observation that YY1 can interact with HDA9 (Fig. 5), we next explored whether the loss of function of YY1 affects the genomic occupancy of HDA9. To this end, we conducted ChIP-seq using anti-FLAG and comparing HDA9pro:HDA9-FLAG/yy1 with HDA9pro:HDA9-FLAG/hda9. Using false discovery rate (FDR) <0.05 and fold change (FC) <0.75 as cutoffs, we identified 108 out of 1,571 HS-induced HDA9-binding genes, referred to as YY1-dependent HDA9-binding genes, exhibiting a significantly higher extent of decreased HDA9-binding levels in HDA9pro:HDA9-FLAG/yy1 under HS conditions than YY1-independent genes (1,463) (Fig. 6D and Dataset S3). The enriched GO terms of these 108 genes are highly similar to the ones of HS-specific HDA9-binding genes (Fig. 6B and SI Appendix, Fig. S14A). Besides, some GO terms were specifically enriched in the YY1-dependent HDA9-binding genes, such as JA metabolic/biosynthetic processes (SI Appendix, Fig. S14A). Furthermore, unlike the YY1-independent ones, the YY1-dependent HDA9-binding genes containing the YY1 motif (26 out of 108; 24%) showed more decreased levels of HDA9 binding than the ones without the YY1 motif (82 out of 108; 76%) in HDA9-FLAG/yy1 compared with HDA9-FLAG/hda9 (SI Appendix, Fig. S14B), which suggested that the YY1 motif contributes to the YY1-dependent HDA9 binding. Collectively, the results indicated that HS-induced recruitment of HDA9 to target loci is partially dependent on YY1.
HDA9 binding affects local histone acetylation.
Next, we examined whether HDA9 binding is associated with changes in histone acetylation and gene expression before and after HS treatment. As HDA9 removes H3K9 acetylation marks (H3K9ac) in vivo (17), we conducted ChIP-seq using a specific anti-H3K9ac antibody in 7-d-old Col seedlings before and after 30-min HS treatment, together with RNA-seq (Dataset S1). Reproducibility between independent biological replicates of anti-H3K9ac ChIP-seq and RNA-seq was high (SI Appendix, Fig. S11 B and C). First, we observed that the H3K9ac level of HDA9-binding loci was higher than non–HDA9-binding regions in both control and HS conditions, indicating that HDA9 preferentially binds the loci with high levels of H3K9ac (SI Appendix, Fig. S15).
Then, we detected 916 HS-induced hyperacetylated and 3,577 hypoacetylated peaks (FDR < 0.05) in Col (Dataset S4), corresponding to 869 and 3,407 genes (Dataset S5), respectively, along with 3,073 and 4,793 transcriptionally up-regulated and down-regulated genes (Dataset S6). Considering the role of HDA9 in HS signal transduction and H3K9 deacetylation, we compared the 1,571 HDA9-binding genes with 3,407 hypoacetylated and 4,793 down-regulated genes in Col under heat treatment. This identified 543 HDA9-binding genes with decreased histone acetylation, of which the 100 down-regulated genes were considered the primary HDA9 target genes (Fig. 6E and Dataset S7).
GO enrichment analysis revealed that these 100 genes were mainly associated with signaling processes and phytohormone-related responses and signaling pathways, as well as regulation of transcription (SI Appendix, Fig. S16A). Besides, we also identify 17 stringently direct HDA9-regulated target genes with an FC cutoff of 1.5× for ChIP-seq data (SI Appendix, Fig. S16B). The functions of these 17 genes (Dataset S7), consistent with the 100 genes, are mainly associated with response to JA, signaling, and response to stress (SI Appendix, Fig. S16C). Consequently, all these results indicate that the identified targets were the first set of genes to trigger the HDA9-mediated heat-signaling cascade. For example, under heat treatment, HDA9 directly binds WRKY33, which encodes a transcription factor participating in different biological processes (51–53), resulting in decreased H3K9ac and reduced expression (Fig. 6 F and G). Other than WRKY33, four other target genes also showed the same trend (SI Appendix, Fig. S17 A and B). Notably, the temporal pattern analysis revealed that WRKY33, WRKY14, TZF1, and SUVH6 with subtle H3K9ac changes after 30-min HS exhibited gradual and significant decrease of H3K9ac enrichment as well as expression levels with prolonged HS treatment compared with control conditions (SI Appendix, Fig. S17C), indicating that the binding of HDA9 to target genes, removal of H3K9ac, and alteration of gene expression are the cascaded events. Comparing our results with published data (54) showed that 479 (6%; P value = 12 × 10−23) out of the 7,866 genes with altered expression were potential WRKY33 target genes, implying that some of the primary HDA9 target genes may act upstream of the main signaling pathway and have central regulatory functions in heat signal transduction and thermotolerance.
Discussion
Typical signal transduction includes signal perception, transduction, and gene expression regulation. Although various temperature sensors have been identified, how plant cells transduce heat signals from the cytoplasm to the nucleus remained largely unknown. For example, chromatin remodeling, via evicting the histone variant H2A.Z from nucleosomes, is a critical component of the plant thermal response (10). Plasma membrane–localized CNGCs and ELF3 may also function as temperature sensors in Arabidopsis (7, 9). The plasma membrane–localized E3 ligase TT3.1 might also serve as a thermosensor (6). Here, we uncovered a conserved heat signal transduction pathway whereby plant cells transduce a heat signal from the cytoplasm to the nucleus in response to HS. The HDA9-phosphorylated Tyr residues are conserved in wheat, rice, soybean, and maize but other HDA9-related proteins in Arabidopsis are not (SI Appendix, Fig. S5). HS treatment increased the accumulation of both TaHDA9 and OsHDA9 in crop species, particularly in the nucleus (SI Appendix, Fig. S18). This heat signal transduction pathway mediated by HDA9 may be a conserved HS response among different plant species.
Different TRPV ion channels can sense different temperatures in animals (55). However, no ion channels or proteins have been found in plants that accurately reflect temperature changes. The localization of HDA9 directly reflects ambient temperature change (SI Appendix, Fig. S2B) and this response occurred within several minutes. Phosphorylation of HDA9 may act as a buffer to adjust HDA9 abundance in response to different temperatures. HDA9 physically interacts with PP2AB′β, leading to its dephosphorylation, which is enhanced by HS treatment. Heat-induced enrichment of HDA9 in the nucleus was dependent on the nucleoporin HOS1. In the nucleus, HDA9 binds and deacetylates the target genes related to signaling transduction and plant development to repress gene expression in a YY1-dependent and -independent manner (SI Appendix, Fig. S20). Whether the HDA9–PP2A complex directly senses heat or needs to interact with other effectors will require further investigation. This signal transduction and consequent changes in gene expression mainly depend on the temperature-sensitive NPC component HOS1, although we cannot rule out that other impaired genes in hos1 contribute to the heat sensitivity of hos1 mutants.
HDA9 participates in flowering time, plant aging, and multiple aspects of plant development (17–21), depending on its interacting partners. For instance, the HDA9–HOS15 complex modulates transcription to achieve a balance between growth and defense. Similarly, PWR interacts with HDA9 to facilitate thermomorphogenesis, possibly by linking histone deacetylation and H2A.Z nucleosome dynamics in plants (25, 26). In contrast with the previously reported negative regulatory role for HDA9 in salt and ABA stress (13, 23), HDA9 has a positive regulatory function during HS. This difference in function is probably due to the different transcription factors binding to HDA9. The HDA9–YY1 interaction can be enhanced by HS treatment (Fig. 5 A and B), representing another level in the hierarchy of heat sensing and signal transduction in the nucleus. Our genetics experiments (Fig. 5C) and the genomics data (Fig. 6 C and D) suggested that YY1 is partially required for the recruitment of HDA9 to target loci. HDA9 bound to the target genes via a YY1-dependent and YY1-independent manner under HS. Although our work dissected the molecular mechanism underlying HDA9-mediated HS signal transduction, there are still open questions of how HDA9 is recruited to the YY1-independent target loci and what are the functions of the YY1-dependent HDA9-binding genes in HS response.
Plant cells have complex mechanisms to respond to environmental changes. As a histone deacetylase, HDA9 acts antagonistically with histone acetylases and plays diverse roles in regulating plant development through repressing target gene expression under normal conditions (17–20). Under stress conditions such as salt treatment or in response to ABA treatment, HDA9 was degraded, ensuring the expression of genes involved in ABA and salt responses (13, 23). Here, we characterized the role of HDA9 as a heat signal transducer, which differs from its role in regular plant development and stress responses. After heat treatment, HDA9 accumulated in the nucleus. We identified 1,571 genes as HS-induced HDA9-binding genes (Fig. 6B). Interestingly, these HS-specific genes were enriched in the GO terms related to signal transduction, response to phytohormone stimulus, and biosynthesis and metabolism of phytohormones (Fig. 6C), indicating that the primary effects of HDA9 in heat signal transduction are deactivation of signaling pathways and retardation of plant development to cope with the coming HS. Moreover, we identified a set of primary HDA9 targets after 30-min HS with subtle but gradual H3K9ac changes along with the HS treatment time course (SI Appendix, Fig. S17C), which were also functionally related to signaling and plant development (SI Appendix, Fig. S16A). Given that the primary HDA9 target genes are located upstream of signaling pathways and play core regulatory functions, small gene expression changes in these genes will result in large cascading effects as the WRKY33 we tested.
On the other hand, HDA9 was removed from the gene body of genes involved in “response to heat” and “response to stress” by unknown mechanisms to derepress the gene expression, ensuring a positive response to HS. Thus, HDA9 plays a vital role in maintaining low expression and low H3K9ac levels of heat response genes (for example HSPs) under normal conditions but promoting low expression and low H3K9ac levels of genes related to signaling and development under HS to balance plant development and HS response. Consistently, the expression profiling of these genes would be impaired in hda9 (Fig. 4D and SI Appendix, Fig. S17), resulting in an HS-hypersensitive phenotype. Therefore, we discovered an elaborate HDA9-mediated heat response mechanism, in which HDA9 acts as a heat signal transducer to slow down plant development and, simultaneously, HDA9 is removed from stress response genes to positively regulate heat tolerance under HS, resulting in rebalance of plant development and heat response (SI Appendix, Fig. S20).
Climate change and the associated increased frequency of extreme temperature events seriously threaten crop yields worldwide. Given the conserved mechanism, our findings provide a potential target for breeding crops to cope with global warming.
Materials and Methods
Plant materials and growth conditions, phenotype analyses and gene mapping, plasmid construction, and generation of transgenic plants are described in SI Appendix, Materials and Methods. Detailed procedures of Western blot analysis, confocal microscopy, immunofluorescence assay, ChIP assay and library preparation, RNA isolation and RNA-seq library preparation, yeast two-hybrid assay, BiFC assay, coimmunoprecipitation assay, phylogenetic analyses, synteny mapping, and bioinformatic analyses are provided in SI Appendix, Materials and Methods. The primers and probes used in this study are listed in Dataset S8.
Supplementary Material
Acknowledgments
We thank Dr. Yongfu Fu for providing gHOS1-mCherry and 35S:HOS1-MYC transgenic seeds, and Dr. Xuehua Zhong for providing HDA9-FLAG transgenic seeds. We thank Professor Kang Chong for active discussions and valuable suggestions. We thank Dr. Lianfeng Gu and Dr. Bing Zhou for the advice on multiomics data analysis. This work was supported by the National Key Research and Development Program of China (2020YFA0509900 to X.C.), National Natural Science Foundation of China (31870243 to S.Z.), High-Level Talent Team Construction Project (225A2902D to S.Z.), Natural Science Foundation of Hebei Province (C2021205009 to S.Z.; C202105013 to Xigang Liu; C2020205028 to W.T.; C2020205006 to Xinye Liu), Advanced Talents Foundation of the Hebei Provincial Department of Science and Technology (2020HBQZYC004 to Xigang Liu), Science Foundation of Hebei Normal University (L042018B01 to J. Bai; L2019B20 to Xinye Liu), Postdoctoral Research Foundation of China (2019M651060 to J. Bai), Science and Technology Project of the Hebei Education Department (QN2020184 to Xinye Liu), and the Postgraduate Innovation Fund of Hebei Province (CXZZBS2019084 to Y.N.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2206846119/-/DCSupplemental.
Data, Materials, and Software Availability
All sequencing data generated in this study have been deposited in the Sequence Read Archive at the National Center for Biotechnology Information under accession no. PRJNA743930 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA743930).
All study data are included in the article and/or supporting information. All the data will be accessible upon publication.
References
- 1.Li B., Gao K., Ren H., Tang W., Molecular mechanisms governing plant responses to high temperatures. J. Integr. Plant Biol. 60, 757–779 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Chao L. M., et al. , Arabidopsis transcription factors SPL1 and SPL12 confer plant thermotolerance at reproductive stage. Mol. Plant 10, 735–748 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Wigge P. A., Ambient temperature signalling in plants. Curr. Opin. Plant Biol. 16, 661–666 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Mittler R., Finka A., Goloubinoff P., How do plants feel the heat? Trends Biochem. Sci. 37, 118–125 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Liu J., Feng L., Li J., He Z., Genetic and epigenetic control of plant heat responses. Front. Plant Sci. 6, 267 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H., et al. , A genetic module at one locus in rice protects chloroplasts to enhance thermotolerance. Science 376, 1293–1300 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Jung J. H., et al. , A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature 585, 256–260 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Jung J. H., et al. , Phytochromes function as thermosensors in Arabidopsis. Science 354, 886–889 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Finka A., Cuendet A. F., Maathuis F. J., Saidi Y., Goloubinoff P., Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 24, 3333–3348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S. V., Wigge P. A., H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140, 136–147 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Shen Y., Wei W., Zhou D. X., Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 20, 614–621 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Liu X., et al. , Transcriptional repression by histone deacetylases in plants. Mol. Plant 7, 764–772 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Baek D., et al. , Histone deacetylase HDA9 with ABI4 contributes to abscisic acid homeostasis in drought stress response. Front. Plant Sci. 11, 143 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim W., Latrasse D., Servet C., Zhou D.-X., Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem. Biophys. Res. Commun. 432, 394–398 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Kang M.-J., Jin H.-S., Noh Y.-S., Noh B., Repression of flowering under a noninductive photoperiod by the HDA9-AGL19-FT module in Arabidopsis. New Phytol. 206, 281–294 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Zeng X., et al. , HISTONE DEACETYLASE 9 functions with Polycomb silencing to repress FLOWERING LOCUS C expression. Plant Physiol. 182, 555–565 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y. J., et al. , POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 113, 14858–14863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X., et al. , POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. eLife 5, e17214 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki M., et al. , Oligocellula1/high expression of osmotically responsive GENES15 promotes cell proliferation with HISTONE DEACETYLASE9 and POWERDRESS during leaf development in Arabidopsis thaliana. Front. Plant Sci. 9, 580 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayer K. S., et al. , HDA9-PWR-HOS15 is a core histone deacetylase complex regulating transcription and development. Plant Physiol. 180, 342–355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park H. J., et al. , HOS15 interacts with the histone deacetylase HDA9 and the evening complex to epigenetically regulate the floral activator GIGANTEA. Plant Cell 31, 37–51 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang L., et al. , HOS15 and HDA9 negatively regulate immunity through histone deacetylation of intracellular immune receptor NLR genes in Arabidopsis. New Phytol. 226, 507–522 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng Y., et al. , Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 67, 1703–1713 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y., et al. , Histone deacetylase HDA9 and WRKY53 transcription factor are mutual antagonists in regulation of plant stress response. Mol. Plant 13, 598–611 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Tasset C., et al. , POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in Arabidopsis thaliana. PLoS Genet. 14, e1007280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Woude L. C., et al. , HISTONE DEACETYLASE 9 stimulates auxin-dependent thermomorphogenesis in Arabidopsis thaliana by mediating H2A.Z depletion. Proc. Natl. Acad. Sci. U.S.A. 116, 25343–25354 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen Y., et al. , Arabidopsis histone deacetylase HDA15 directly represses plant response to elevated ambient temperature. Plant J. 100, 991–1006 (2019). [DOI] [PubMed] [Google Scholar]
- 28.de Rooij P. G. H., Perrella G., Kaiserli E., van Zanten M., The diverse and unanticipated roles of histone deacetylase 9 in coordinating plant development and environmental acclimation. J. Exp. Bot. 71, 6211–6225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai M., et al. , The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 25, 517–534 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang W., et al. , PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13, 124–131 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang R., et al. , The brassinosteroid-activated BRI1 receptor kinase is switched off by dephosphorylation mediated by cytoplasm-localized PP2A B′ subunits. Mol. Plant 9, 148–157 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y. L., et al. , Protein phosphatase 2AB′α and B′β protect centromeric cohesion during meiosis I. Plant Physiol. 179, 1556–1568 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S., et al. , MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell 28, 2866–2883 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang X., et al. , Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. eLife 7, e31636 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R., et al. , Nucleocytoplasmic trafficking and turnover mechanisms of BRASSINAZOLE RESISTANT1 in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 118, e2101838118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang A., et al. , Nuclear pore complex components have temperature-influenced roles in plant growth and immunity. Plant Cell Environ. 43, 1452–1466 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Cheng Z., et al. , Nup96 and HOS1 are mutually stabilized and gate CONSTANS protein level, conferring long-day photoperiodic flowering regulation in Arabidopsis. Plant Cell 32, 374–391 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H., et al. , The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo–cytoplasmic partitioning. Genes Dev. 15, 912–924 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung J. H., Seo P. J., Park C. M., The E3 ubiquitin ligase HOS1 regulates Arabidopsis flowering by mediating CONSTANS degradation under cold stress. J. Biol. Chem. 287, 43277–43287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J. H., Lee H. J., Jung J. H., Lee S., Park C. M., HOS1 facilitates the phytochrome B-mediated inhibition of PIF4 function during hypocotyl growth in Arabidopsis. Mol. Plant 10, 274–284 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Han S. H., Park Y. J., Park C. M., HOS1 activates DNA repair systems to enhance plant thermotolerance. Nat. Plants 6, 1439–1446 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Li C., Liu L., Teo Z. W. N., Shen L., Yu H., Nucleoporin 160 regulates flowering through anchoring HOS1 for destabilizing CO in Arabidopsis. Plant Commun. 1, 100033 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshida M., Horinouchi S., Beppu T., Trichostatin A and trapoxin: Novel chemical probes for the role of histone acetylation in chromatin structure and function. BioEssays 17, 423–430 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Zheng S. Z., et al. , Phosphoinositide-specific phospholipase C9 is involved in the thermotolerance of Arabidopsis. Plant J. 69, 689–700 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Mizoi J., et al. , Heat-induced inhibition of phosphorylation of the stress-protective transcription factor DREB2A promotes thermotolerance of Arabidopsis thaliana. J. Biol. Chem. 294, 902–917 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J. H., et al. , Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342, 628–632 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Shin J. H., Chekanova J. A., Arabidopsis RRP6L1 and RRP6L2 function in FLOWERING LOCUS C silencing via regulation of antisense RNA synthesis. PLoS Genet. 10, e1004612 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Terol J., Bargues M., Carrasco P., Pérez-Alonso M., Paricio N., Molecular characterization and evolution of the protein phosphatase 2A B′ regulatory subunit family in plants. Plant Physiol. 129, 808–822 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li T., Wu X. Y., Li H., Song J. H., Liu J. Y., A dual-function transcription factor, AtYY1, is a novel negative regulator of the Arabidopsis ABA response network. Mol. Plant 9, 650–661 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Najafabadi H. S., et al. , C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol. 33, 555–562 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Rigal A., et al. , A network of stress-related genes regulates hypocotyl elongation downstream of selective auxin perception. Plant Physiol. 187, 430–445 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu B., et al. , The ubiquitin E3 ligase SR1 modulates the submergence response by degrading phosphorylated WRKY33 in Arabidopsis. Plant Cell 33, 1771–1789 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verma V., et al. , SUMO enables substrate selectivity by mitogen-activated protein kinases to regulate immunity in plants. Proc. Natl. Acad. Sci. U.S.A. 118, e2021351118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Birkenbihl R. P., Kracher B., Roccaro M., Somssich I. E., Induced genome-wide binding of three Arabidopsis WRKY transcription factors during early MAMP-triggered immunity. Plant Cell 29, 20–38 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dhaka A., Viswanath V., Patapoutian A., Trp ion channels and temperature sensation. Annu. Rev. Neurosci. 29, 135–161 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data generated in this study have been deposited in the Sequence Read Archive at the National Center for Biotechnology Information under accession no. PRJNA743930 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA743930).
All study data are included in the article and/or supporting information. All the data will be accessible upon publication.