Significance
The dynamic voltage-sensing domain–pore domain (VSD-PD) coupling process during the gating of voltage-gated ion channels remains largely unknown despite decades of intensive study. The voltage-gated potassium channel KCNQ1, which plays a key role in controlling the heart rhythm, provides a good model to dissect the dynamic VSD-PD coupling due to its unique two-open-state gating mechanism. In this study, we determined high-resolution cryo-electron microscopy structures of full-length human KCNQ1 with two VSD-PD coupling enhancers — the membrane lipid PIP2 (phosphatidylinositol 4,5-bisphosphate) and a small-molecule ML277. These structures uncover the molecular mechanisms for these VSD-PD coupling enhancers to activate the KCNQ1 channel and pave the way to develop new therapeutics for anti-arrhythmia.
Keywords: KCNQ1, PIP2, ML277, E-M coupling
Abstract
The cardiac KCNQ1 potassium channel carries the important IKs current and controls the heart rhythm. Hundreds of mutations in KCNQ1 can cause life-threatening cardiac arrhythmia. Although KCNQ1 structures have been recently resolved, the structural basis for the dynamic electro-mechanical coupling, also known as the voltage sensor domain–pore domain (VSD-PD) coupling, remains largely unknown. In this study, utilizing two VSD-PD coupling enhancers, namely, the membrane lipid phosphatidylinositol 4,5-bisphosphate (PIP2) and a small-molecule ML277, we determined 2.5–3.5 Å resolution cryo-electron microscopy structures of full-length human KCNQ1-calmodulin (CaM) complex in the apo closed, ML277-bound open, and ML277-PIP2-bound open states. ML277 binds at the “elbow” pocket above the S4-S5 linker and directly induces an upward movement of the S4-S5 linker and the opening of the activation gate without affecting the C-terminal domain (CTD) of KCNQ1. PIP2 binds at the cleft between the VSD and the PD and brings a large structural rearrangement of the CTD together with the CaM to activate the PD. These findings not only elucidate the structural basis for the dynamic VSD-PD coupling process during KCNQ1 gating but also pave the way to develop new therapeutics for anti-arrhythmia.
The KCNQ1 potassium channel, together with its auxiliary KCNE1 subunit, carries the slow delayed-rectifier (IKs) current that is important for repolarizing the cardiac action potential (1–3). Loss-of-function mutations in the KCNQ1 channel have been found to induce functional defects to the IKs current, leading to long QT syndrome type 1 (LQT1) that predisposes patients to life-threatening cardiac arrhythmia (1, 4). KCNQ1 thus is also known as the KvLQT1 channel. LQT1 is the most common subtype of long QT syndromes, responsible for 30–35% of all cases (5). Clinical studies have identified hundreds of variants in KCNQ1 that can cause LQT1 (1, 4, 6), reinforcing the importance of the KCNQ1 channel in maintaining heart rhythm.
KCNQ1 belongs to the voltage-activated potassium channel KV7 subfamily. It adopts the canonical structural organization of the homo-tetrameric KV superfamily with domain-swapped architecture (7, 8). Each subunit contains six transmembrane segments (S1-S6); the S1-S4 form the voltage-sensing domain (VSD), and the S5-S6 constitute the pore domain (PD). The C-terminal domain (CTD) interacts with the calcium-modulated protein calmodulin (CaM) (9, 10). Upon membrane depolarization, the VSD activates and induces conformational changes to open the PD via interactions between the VSD and the PD (VSD-PD coupling, or electro-mechanical coupling). We recently elucidated the two-open-state gating mechanism of the KCNQ1 channel (6, 11–14). Similar to other KV channels such as the Shaker channel, the VSD of KCNQ1 activates in two steps: from the resting state via an experimentally resolvable intermediate state to the fully activated state (6, 11–20). However, unlike the Shaker channel that opens only when the VSD moves to the fully activated state, KCNQ1 opens when the VSD occupies both the intermediate and fully activated conformations, leading to intermediate-open (IO) and activated-open (AO) states (Fig. 1A). The underlying VSD-PD coupling process was proposed as a two-stage “hand-and-elbow” VSD-PD coupling mechanism, where intrasubunit interactions at the “hand” (between the C terminus of the S4-S5 linker and the S6) are responsible for the IO state, and intersubunit interactions at the “elbow” (between the corner of S4 and S4-S5 linker and the neighboring S5′ and S6′) are in charge of the AO state (13). Of note, a membrane lipid phosphatidylinositol 4,5-bisphosphate (PIP2) is required for the VSD-PD coupling of both IO and AO states, because depleting PIP2 disrupts the overall VSD-PD coupling and results in a closed pore with normal VSD activation (21).
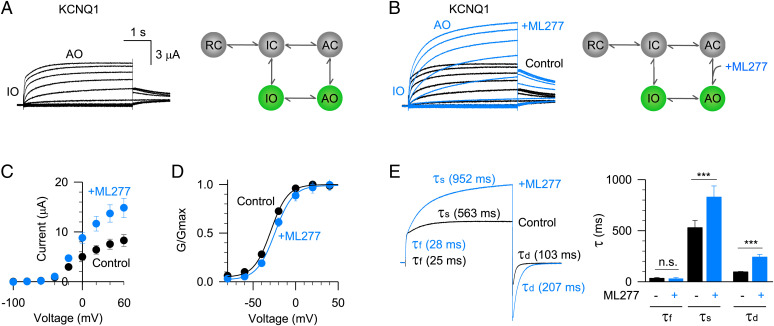
Fig. 1.
ML277 activates the KCNQ1 channel. (A) Left, the KCNQ1 activation currents exhibit two phases: a fast phase approximates the intermediate-open (IO) state, and the slow phase approximates the activated-open (AO) state. The depolarization voltages were from −120 to +60 mV in 20 mV increments for 4 s and then stepped back to −40 mV to record the tail currents. Right, a five-state kinetic model recapitulates the unique gating process of the KCNQ1 channel (RC: resting closed, IC: intermediate closed, AC: activated closed). (B) Left, 1 µM ML277 activates the KCNQ1 currents. Current recorded before (black) and after (blue) adding ML277 were superimposed to show the ML277 effects. Right, ML277 enhances the VSD-PD coupling transition of the AO state. (C) Averaged current–voltage (I–V) relations of KCNQ1 channel before (black, n = 6) and after (blue, n = 6) adding ML277. (D) Normalized G–V relations of KCNQ1 channel before (black, n = 6) and after (blue, n = 6) adding ML277. Data points were fitted with a Boltzmann equation. (E) Left, activation and deactivation time constants (τ) of KCNQ1 currents before (black) and after (blue) adding ML277. The voltage was +60 mV for 4 s to activate KCNQ1 and then stepped to –120 mV for 2 s to deactivate KCNQ1. The activation time constants of the fast phase (τf) and slow phase (τs) were 25 and 563 ms for control, and 28 and 952 ms after adding ML277. The deactivation time constants (τd) were 103 ms for control and 207 ms after adding ML277. Right, averaged results of time constants (τf, τs, and τd) of KCNQ1 currents before (black) and after (blue) adding ML277 (n ≥ 8). n.s.: nonsignificant. The P values are 0.69 for τf, 0.00073 for τs, and 0.00011 for τd.
KCNQ1 associates with the auxiliary KCNE1 subunit in the heart (1–3). The association of KCNE1 profoundly changes the KCNQ1 channel function. For example, it slows down the activation and deactivation rate, removes the inactivation, right-shifts the G–V relation, and changes the ion permeation and drug sensitivity (2, 3). These function changes may occur because the KCNE1 suppresses the IO state but enhances the AO state, rendering the IKs channel with only AO state openings (6, 11, 12). Therefore, the AO state is physiologically more important in the heart. Enhancing the AO state to rescue the defective IKs currents of LQT1 mutations represents a potential strategy for anti-arrythmia therapy. Tremendous efforts have been made to identify KCNQ1 and KCNQ1+KCNE1 (IKs) activators (22–35). However, the lack of structural mechanism for any of these activators that bind and modulate KCNQ1 largely impedes the development of new therapeutics. Among these activators, a small-molecule ML277 exhibits several unique properties: 1) it effectively activates the KCNQ1 channel with an EC50 of 260 nM (30, 36), more potent than other known KCNQ1/IKs activators; 2) it exclusively activates the KCNQ1 channel without altering other key cardiac ion channels (such as hERG, CaV1.2, and NaV1.5) or neuronal KCNQ (KCNQ2-KCNQ5) channels (30, 36); 3) it shortens the action potential duration in human-induced pluripotent stem cell-derived cardiomyocytes (including LQT1 conditions) and guinea pig cardiomyocytes (36–39); and 4) we recently found that ML277 can specifically activate the AO state current by enhancing the VSD-PD coupling (6, 13). All these properties enable ML277 as a promising candidate for treating cardiac arrythmia.
In this study, we combine cryo-electron microscopy (cryo-EM) and electrophysiology techniques to reveal the structural mechanisms of the modulation of KCNQ1 by VSD-PD coupling enhancers PIP2 and ML277. We present 2.5–3.5 Å resolution cryo-EM structures of human KCNQ1-CaM complex in the apo closed, ML277-bound open, and two ML277-PIP2-bound open states. While PIP2 binds at the cleft formed by VSD and PD induces a large structural rearrangement of the CTD of KCNQ1 and its surrounding CaM, ML277 binds at the “elbow” pocket formed by the S4-S5 linker, S5, and the neighboring S5′ and S6′, and directly activates KCNQ1 without affecting the CTD. Our study not only elucidates the molecular basis for the activation of the KCNQ1 by enhancing VSD-PD coupling with two different small molecules, but also paves the way to develop new therapeutics for treating cardiac arrhythmia.
Results
ML277 and PIP2 Enhance the VSD-PD Coupling of KCNQ1.
The KCNQ1 channel has two different open states, IO and AO (6, 11–14). The activation currents exhibit double-exponential kinetics –– the fast component approximates the IO state current and the slow component approximates the AO state current (11). This unique gating process can be illustrated in a five-state scheme as shown in Fig. 1A. We have systematically investigated the functional effects of ML277 on the KCNQ1 channel expressed in Xenopus oocytes and found that ML277 specifically enhances the AO state VSD-PD coupling of the KCNQ1 channel (6) (Fig. 1B). For example, 1 µM ML277 almost doubled the current amplitude at +60 mV (Fig. 1C). The superimposed currents recorded before and after adding ML277 showed that ML277 only increased the slow component AO state current, leaving the fast component IO state current unaltered (Fig. 1B). Consistent with this result, ML277 specifically changed the activation time constant of the AO state. For example, at +60 mV depolarization, the activation time constant of the slow component (τs) was changed from 563 to 952 ms, while the activation time constant of the fast component (τf) changed little (from 25 to 28 ms, Fig. 1E). The deactivation time constant (τd) also changed from 103 to 207 ms (Fig. 1E). These observations agree with our previous simulation results of kinetic modeling. ML277 can slow down the exit transitions from the AO to stabilize the conformational energy of the AO state (6).
We then also analyzed ML277-induced changes in the voltage-dependent activation of the KCNQ1 channel. The conductance–voltage (G–V) relation of the KCNQ1 channel contains the G–V relations of both the IO and AO states, and the G–V relation of the AO state is more positive than that of the IO state (11, 12). Unlike normal activators that potentiate ion channel currents and left-shift the G–V relation, ML277 slightly shifted the G–V relation to a more positive voltage, with ΔV50 of ∼6 mV (Fig. 1D). This intriguing result suggests that ML277 enhances the occupancy of the AO state and thereby right-shifts the G–V relation. Besides current size, kinetics, and the voltage-dependent activation, ML277 also changed the ion permeation property closer to that of the AO state without modifying the VSD activation (6). All the evidence consistently supports the mechanism that ML277 specifically enhances the AO state VSD-PD coupling of the KCNQ1 channel.
The membrane lipid PIP2, on the other hand, mediates the VSD-PD coupling processes of both IO and AO states. Depletion of PIP2 disrupts the VSD-PD coupling and results in a channel pore that can no longer open upon VSD activation (21). Taken together, these results indicate that the VSD-PD coupling of the KCNQ1 channel can be effectively modulated by both the endogenous lipid molecule PIP2 and the exogenous small-molecule ML277.
Structural Determination of KCNQ1-CaM with Different VSD-PD Coupling Enhancers.
To reveal activation mechanisms of human KCNQ1 by these VSD-PD coupling enhancers, we determined cryo-EM structures of KCNQ1-CaM using protein samples in the absence and presence of ML277 and/or PIP2 (Fig. 2 and SI Appendix, Figs. S1–S4 and Table S1). Different from the previously reported structures of the human KCNQ1 complexes (apo KCNQ1-CaM complex, PDB 6UZZ, KCNQ1-CaMapo-6UZZ; apo KCNQ1-CaM-KCNE3 complex, PDB 6V00, KCNQ1-CaM-KCNE3apo-6V00; PIP2-bound KCNQ1-CaM-KCNE3 complex, PDB 6V01, KCNQ1-CaM-KCNE3PIP2-6V01) that were obtained from a truncated KCNQ1 sample (residues 76–620) (8), we determined cryo-EM structures of the full-length human KCNQ1 channel in three different conditions: 1) the apo-state structure of the KCNQ1-CaM complex (KCNQ1-CaMapo) at 3.5 Å resolution (Fig. 2A and SI Appendix, Fig. S1); 2) the ML277-bound KCNQ1-CaM structure (KCNQ1-CaMML277) at 2.6 Å resolution (Fig. 2A and SI Appendix, Fig. S2); and 3) the PIP2- and ML277-bound KCNQ1-CaM structures in two different conformations, which were designated as KCNQ1-CaMML277-PIP2-A at 3.1 Å resolution and KCNQ1-CaMML277-PIP2-B at 2.5 Å resolution (Fig. 2A and SI Appendix, Fig. S3). We also endeavored to capture the structure of KCNQ1-CaM in the PIP2-only condition. However, the protein particles in the presence of PIP2 alone seemed heterogeneous and dynamic, resulting in two 3D reconstructions at 6–7 Å resolutions, which will be analyzed in Discussion (SI Appendix, Fig. S4).
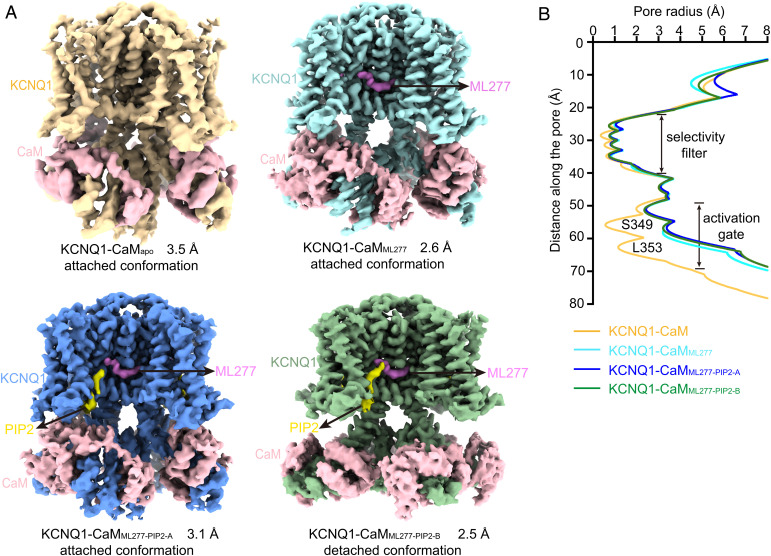
Fig. 2.
Structures of KCNQ1-CaM in different ligand-bound states. (A) The cryo-EM maps of KCNQ1-CaMapo (Upper Left), KCNQ1-CaMML277 (Upper Right), KCNQ1-CaMML277-PIP2-A (Lower Left), and KCNQ1-CaMML277-PIP2-B (Lower Right). KCNQ1, ML227, PIP2, and CaM are labeled. (B) Pore radii along the ion permeation pathway in different KCNQ1-CaM structures. Key residues forming the two constriction layers of the activation gate in the closed KCNQ1 are labeled.
In all the three ligand-bound structures, i.e., KCNQ1-CaMML277, KCNQ1-CaMML277-PIP2-A, and KCNQ1-CaMML277-PIP2-B, the density for the ML277 molecule was unambiguously resolved in the EM map, and therefore its binding configuration was confidently assigned (Fig. 2A and SI Appendix, Figs. S2 and S3). In the structures of KCNQ1-CaMML277-PIP2-A and KCNQ1-CaMML277-PIP2-B, the density for the inositol 1,4,5-trisphosphate head group of PIP2 were well defined in the EM map, whereas the density for the flexible fatty acid chains of PIP2 was poorly resolved, likely due to their relatively weak interactions with the protein (Fig. 2A and SI Appendix, Fig. S3).
The KCNQ1-CaMapo structure is in the closed state, as indicated by the <1 Å pore radius at the activation gate of the channel calculated by the program HOLE (Fig. 2B) (40). By contrast, all three ligand-bound structures of KCNQ1-CaM complex are in an open state, with the pore radii expending to ∼2.5 Å at the activation gate (Fig. 2B), similar to that in the previously reported PIP2-bound open-state structure KCNQ1-CaM-KCNE3PIP2-6V01 (8). In addition, we observe two conformations of KCNQ1-CaM based on different interactions between CaM and KCNQ1. In the structures of KCNQ1-CaMapo, KCNQ1-CaMML277, and KCNQ1-CaMML277-PIP2-A, CaM forms direct contact with VSD; therefore, we call this an “attached conformation” (Fig. 2A). In the structure of KCNQ1-CaMML277-PIP2-B, however, CaM is detached from VSD of KCNQ1; we designate KCNQ1-CaMML277-PIP2-B a “detached conformation” (Fig. 2A).
The KCNQ1-CaMapo Structure Shows Activated VSDs but Closed Pore.
The overall structure of KCNQ1-CaMapo is similar to the previously reported KCNQ1-CaMapo-6UZZ structure (Fig. 3A and SI Appendix, Fig. S5) (8). In the CTD of KCNQ1, three α-helices, namely, HA, HB, and HC, were resolved. Four HC helices from four subunits assemble into a coiled-coil that runs perpendicular to the membrane along the channel central axis below the activation gate (Fig. 3A). Within one subunit, the HA and HB are wrapped by the CaM (Fig. 3 A and B). The linker between the fifth and sixth helix of CaM (H5-H6 linker) forms direct interactions with S0 and the S2-S3 linker by hydrogen bonds (Fig. 3B).
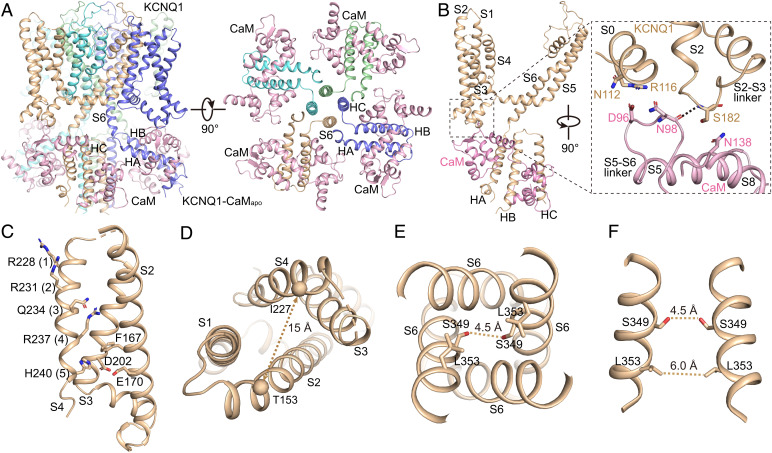
Fig. 3.
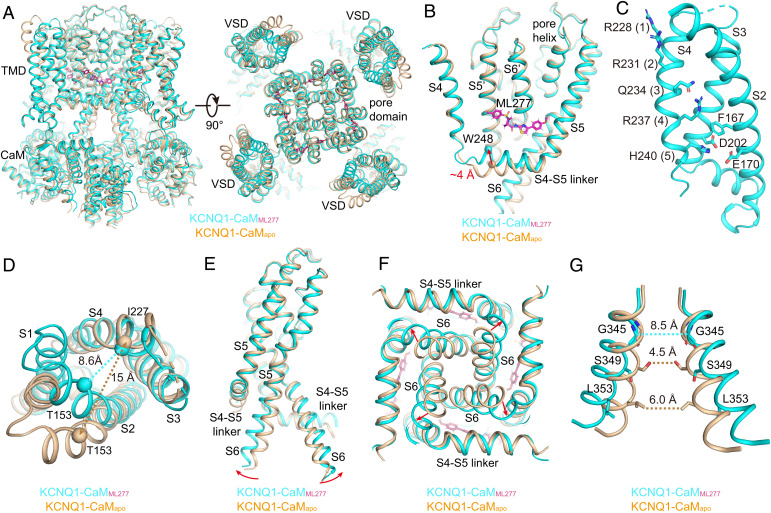
The structure of KCNQ1-CaMapo. (A) The cartoon model of KCNQ1-CaMapo in the side view (Left) and top view (Right). Each subunit of KCNQ1 is colored individually, and the CaM is in pink. The TMD of KCNQ1 is not shown in the top view. (B) Interactions between the VSD of KCNQ1 and CaM. The dashed line shows the hydrogen bond between KCNQ1 and CaM. (C) The VSD structure of KCNQ1-CaMapo with S1 omitted for clarity. Side chains of gating charge residues and residues forming the CTC are shown as sticks. (D) A large solvent-accessible cavity formed in the extracellular side of VSD. The dashed line shows the distance between Cα atoms of Thr153 in S2 and Ile227 in S4 (in Å). (E) The pore domain of KCNQ1-CaMapo. The dashed line shows diagonal atom-to-atom distances between two Ser residues. (F) Ser349 and Leu353 form two constriction layers in the closed activation gate of KCNQ1-CaMapo. The dashed lines show diagonal atom-to-atom distances between two Ser and Leu residues (in Å).
In the transmembrane domain (TMD), KCNQ1 adopts a canonical fold of voltage-gated ion channels in a domain-swapped arrangement (Fig. 3A). Upon membrane depolarization, the VSD of KCNQ1 undergoes a stepwise activation process: from the resting state via the intermediate state and finally to the fully activated state (6, 11–14). In the intermediate state, the third gating charge Gln234 (Q3) is in the gating charge transfer center (CTC) (19) (or the hydrophobic plug) (18), which is formed by residues Phe167, Glu170, and Asp202; while in the fully activated state, the fifth gating charge His240 (H5) is interacting with the CTC (8, 14). Our KCNQ1-CaM structures were determined under 0 mV, a voltage that allows the VSD to activate in both the intermediate and the fully activated states (11, 13). Consistent with this, the S4 in KCNQ1-CaMapo seems dynamic and is resolved at a relatively lower resolution than PD (SI Appendix, Fig. S1). While we tentatively modeled S4 in the fully activated state with H5 in the CTC (Fig. 3C), the local density suggests that in some particles, S4 may be in a partially activated state (SI Appendix, Fig. S1), similar to the intermediate-state structure of the isolated VSD of KCNQ1 determined by NMR (14). Due to the dynamics of S4, the interactions between S4 and S1-S2 at the outer leaflet of the membrane are weak, with a large solvent-accessible cavity formed in the extracellular side of VSD (Fig. 3D). The activation gate of KCNQ1 is closed by two constriction layers formed by Ser349 and Leu353 (Fig. 3 E and F). Thus, KCNQ1-CaMapo adopts a decoupled conformation, with activated VSDs and a closed gate.
Similarly, the previously reported KCNQ1-CaMapo-6UZZ structure is also in a decoupled conformation with activated VSDs and a closed gate (8). Structural alignment of KCNQ1-CaMapo-6UZZ and our KCNQ1-CaMapo reveals almost-identical structures except for a major difference in the VSDs at the outer leaflet of the membrane (SI Appendix, Fig. S5). Different from the VSD in KCNQ1-CaMapo that forms a large extracellular-facing cavity, the VSD in KCNQ1-CaMapo-6UZZ tightly packs, with the S4 clearly resolved in the fully activated state (8). The different conformations of the VSDs may be caused by different sample preparations (the detergent GDN used for KCNQ1-CaMapo vs. Digitonin for KCNQ1-CaMapo-6UZZ) and (or) different constructs of KCNQ1 (the full-length KCNQ1 for KCNQ1-CaMapo vs. the truncated KCNQ1 with residues 76–620 for KCNQ1-CaMapo-6UZZ).
ML277 Enhances the VSD-PD Coupling and Induces the Pore Opening.
Then we sought to resolve KCNQ1-CaM structures with ML277 and PIP2 to rescue the VSD-PD coupling. In the structure of KCNQ1-CaMML277, ML277 binds in the hydrophobic pocket formed by the S4-S5 linker and S5 from one subunit, and S5′ and S6′ from the adjacent subunit (Fig. 4 A–C). It mainly interacts with the nearby hydrophobic residues, including Trp248, Leu251, and Val255 in the S4-S5 linker, Leu262 and Leu266 in S5, Leu271 in S5′, and Phe335 and Phe339 in S6′ (Fig. 4B). In addition, the carbonyl of Phe335 in S6′ forms a hydrogen bond with the amide group in ML277 (Fig. 4B). ML277 does not directly interact with the pore helix, and its binding site is below the classical drug binding site in the fenestration in voltage-gated calcium channels (41, 42). The ML277 binding site also differs from those of the anti-epileptic drug retigabine in KCNQ2 and the activator ML213 in KCNQ4 (43–45), although they have some overlap near the conserved Pro in S6 (Pro343 in KCNQ1) (SI Appendix, Fig. S6).
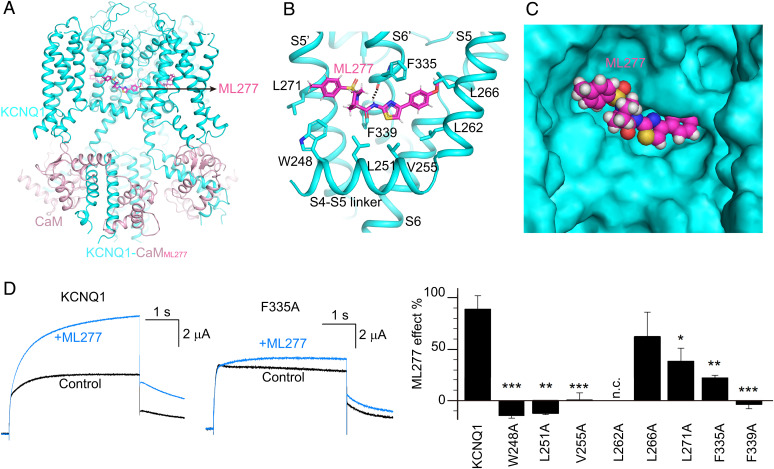
Fig. 4.
The ML277 binding mode in KCNQ1. (A) The overall structure of KCNQ1-CaMML277. KCNQ1, CaM, and ML277 are colored in cyan, pink, and magenta, respectively. (B) The binding site of ML277 in KCNQ1. Side chains of residues in KCNQ1 involved in the hydrophobic interactions with ML277 are shown as sticks. The dashed line indicates the hydrogen bond between ML277 and Phe335. (C) The binding pocket of ML277 in KCNQ1. KCNQ1 is rendered as a surface model, and ML277 is shown as spheres. (D) Left, representative currents of WT KCNQ1 and the F335A mutant before (black) and after (blue) adding 1 µM ML277. Currents were recorded at +40 mV for 4 s and then back to −40 mV to induce the tail currents. Right, averaged results of 1 µM ML277-induced current increase on WT KCNQ1 and its mutations. L262A showed no current (n.c.). n ≥ 4. The P values are 0.00017 for W248A, 0.0011 for L251A, 0.00013 for V255A, 0.30 for L266A, 0.048 for L271A, 0.0016 for F335A, and 0.00091 for F339A.
Using ML277 as a pharmacological tool, we previously have identified residues that are critical to the VSD-PD coupling (6). These residues form a hydrophobic pocket that matches well with the ML277 binding site identified in this study. To further validate the ML277 binding site, we performed electrophysiology assays on the KCNQ1 with mutations at the ML277-interacting residues. Among the eight residues we tested, the mutant L266A showed similar ML277 effect as the wild type (WT), whereas L271A and F335A displayed a significantly reduced ML277 effect (Fig. 4D). For example, at +40 mV, 1 µM ML277 increased the current of WT KCNQ1 by 89%, whereas it only increased the current of F335A by 22% (Fig. 4D). Furthermore, mutations W248A, L251A, V255A, and F339A almost abolished the ML277 effect (Fig. 4D). Of note, the mutant L262A had no detectable current, limiting further testing of the ML277 effect on it (Fig. 4D). Taken together, these functional studies support the ML277 binding site observed in the structure of KCNQ1-CaMML277.
In comparison with those in KCNQ1-CaMapo, the CTD and CaM in KCNQ1-CaMML277 remain almost unchanged, and KCNQ1-CaMML277 is still in the attached conformation (Fig. 5A). Of note, the binding of ML277 induces several major conformational changes to the TMD of KCNQ1 (SI Appendix, Movie S1). First, induced by hydrophobic interactions between ML277 and the S4-S5 linker, the N-terminal end of S4-S5 linker moves up by ∼4 Å (Fig. 5B and SI Appendix, Movie S2). Second, along with the upward movement of the S4-S5 linker, S4 becomes less dynamic and is resolved unambiguously at the fully activated state with H5 interacting with the CTC (Fig. 5C and SI Appendix, Fig. S2). Third, the N-terminal half of S4 now forms strong interactions with the S1-S2 linker, as well as its connecting S1 C-terminal half and S2 N terminus, with Thr153 in S2 moving closer to Ile227 in S4 by ∼7 Å (Fig. 5D). Fourth, due to the hydrophobic packing between the S4-S5 linker and S6 of the same subunit, the upward movement of the S4-S5 linker further causes the bending of the S6 C-terminal half at the conserved “PAG” segment away from the central axis of the channel pore (Fig. 5 E and F and SI Appendix, Movie S2) (46, 47). Consequently, the minimum atom-to-atom cross distance at the activation gate expands from 4.5 to 8.5 Å (Fig. 5G and SI Appendix, Movie S3), which likely allows the passage of the hydrated potassium ions. Thus, ML277 itself is sufficient to rescue the defective VSD-PD coupling from the KCNQ1-CaMapo and induce the pore opening. Since the CTD and CaM remain almost unchanged (attached), this ML277-induced opening of the activation gate in KCNQ1-CaM seems different from the PIP2-induced one in KCNQ1-CaM-KCNE3, which involves a large conformational change in the CTD of KCNQ1 and CaM (detached) (8). The observation that the ML277-induced opening of KCNQ1 is independent of PIP2 is supported by a previous electrophysiology study, which showed that ML277 can activate KCNQ1 with the PIP2 depleted by the Ciona intestinalis voltage sensor-containing phosphatase upon membrane depolarization (37).
Fig. 5.
ML277-induced conformational changes of KCNQ1. (A) Structural comparison of KCNQ1-CaMapo (wheat) and KCNQ1-CaMML277 (cyan) in the side view (Left) and top view (Right). (B) ML277 induces an upward movement of the S4-S5 linker. Arrow indicates the distance between the N-terminal residue Trp248 in the S4-S5 linkers in two structures. (C) The VSD in KCNQ1-CaMML277 with S1 omitted for clarity. Side chains of gating charge residues and residues forming the CTC are shown as sticks. (D) Structural differences of VSDs in KCNQ1-CaMapo (wheat) and KCNQ1-CaMML277 (cyan) in the top view. The dashed lines show the distances between Cα atoms of Thr153 in S2 and Ile227 in S4 in two structures. (E) Structural differences of PDs in KCNQ1-CaMapo and KCNQ1-CaMML277 in the side view. For clarity, only two opposing subunits are shown. Red arrows indicate the outward bending of S6 C-terminal halves upon ML277 binding. (F) Structural differences of PDs in KCNQ1-CaMapo and KCNQ1-CaMML277 in the bottom view. (G) Structural differences of the activation gates in KCNQ1-CaMapo and KCNQ1-CaMML277 in the side view. The dashed lines show diagonal atom-to-atom distances between constriction-forming residues (in Å). The gate in KCNQ1-CaMML277 opens up.
PIP2 Induces a Large Structural Rearrangement of the CTD and CaM.
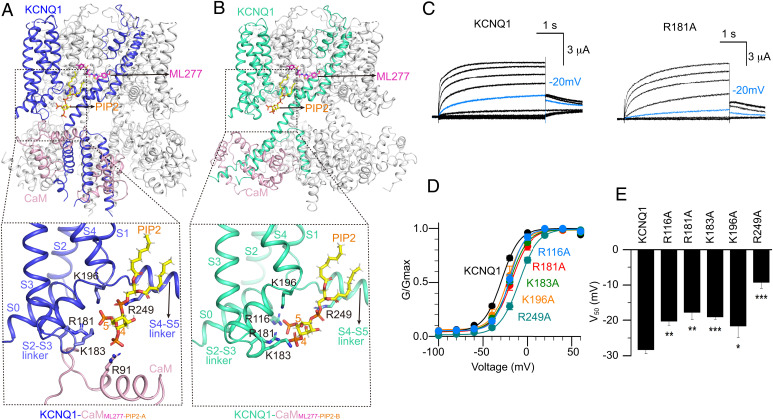
In physiological condition, the endogenous lipid molecule PIP2 is required for the VSD-PD coupling of the KCNQ1 channel (21). To further investigate the key role of PIP2 in KCNQ1 channel gating, we resolved KCNQ1-CaM structures in the presence of both PIP2 and ML277. PIP2-ML277-bound KCNQ1-CaM structures exhibited clearly two different conformations, KCNQ1-CaMML277-PIP2-A and KCNQ1-CaMML277-PIP2-B (Fig. 2A). In both structures, PIP2 binds at the cleft formed by the S2-S3 linker, S3, S4, and S4-S5 linker (Fig. 6 A and B). The inositol 1,4,5-trisphosphate head group of PIP2 interacts with several positive-charged residues, including Arg181, Lys183, and Lys196 in the S2-S3 linker, and Arg249 in the S4-S5 linker (Fig. 6 A and B). In addition, PIP2 in KCNQ1-CaMML277-PIP2-A also forms a salt bridge with Arg91 in CaM, whereas in KCNQ1-CaMML277-PIP2-B, it loses the interaction with the CaM and forms a new interaction with another residue Arg116 in S0 (Fig. 6 A and B). This PIP2 binding site is similar to the observation in the previously reported structure KCNQ1-CaM-KCNE3PIP2-6V01 (8). To further confirm this PIP2 binding site, we performed electrophysiology experiments on the KCNQ1 with mutations on the PIP2-interacting residues. All the five mutants, R116A, R181A, K183A, K196A, and R249A, showed mild and consistent right-shifted G–V relations in comparison with the WT KCNQ1, suggesting the important roles of these residues for the activation of KCNQ1 (Fig. 6 C–E). Similar results were also observed in the PIP2-binding residue mutants of neuronal KCNQ4 (44).
Fig. 6.
The binding modes of ML277 and PIP2 in KCNQ1. (A) The overall structure of KCNQ1-CaMML277-PIP2-A and a zoomed-in view of the PIP2 binding site. KCNQ1, CaM, ML277, and PIP2 are colored in blue, pink, magenta, and yellow, respectively. Side chains of positively charged residues involved in the PIP2 recognition are shown as sticks. (B) The overall structure of KCNQ1-CaMML277-PIP2-B and a zoomed-in view of the PIP2 binding site. KCNQ1, CaM, ML277, and PIP2 are colored in green, pink, magenta, and yellow, respectively. Side chains of positively charged residues involved in the PIP2 recognition are shown as sticks. (C) Activation currents of WT KCNQ1 and the R181A mutant. Currents were recorded from –120 mV to +60 mV in 20 mV increments for 4 s and then back to −40 mV to induce the tail currents. Currents recorded at –20 mV were shown in blue to indicate right-shifted voltage-dependent activation of R181A. (D) Normalized G–V relations of WT KCNQ1 channel (V50 = −28.4 ± 1.1 mV, n = 10), R116A (V50 = −20.3 ± 1.2 mV, n = 3), R181A (V50 = −17.8 ± 2.0 mV, n = 3), K183A (V50 = −19.1 ± 0.7 mV, n = 5), K196A (V50 = −21.7 ± 3.2 mV, n = 3), R249A (V50 = −9.3 ± 1.6 mV, n = 5). Data points were fitted with a Boltzmann equation. (E) Averaged results of V50 for WT KCNQ1 and its mutants. The P values are 0.0028 for R116A, 0.0018 for R181A, 0.000071 for K183A, 0.037 for K196A, and 0.0000094 for R249A.
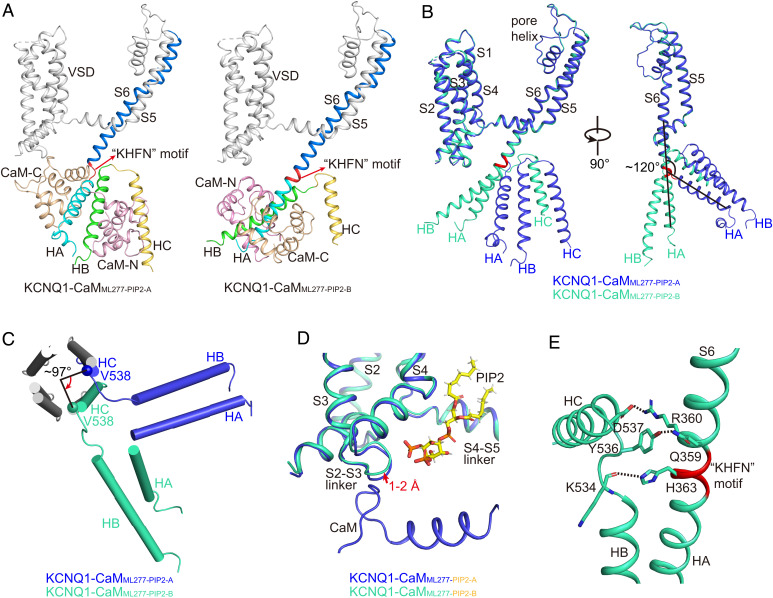
KCNQ1-CaMML277-PIP2-A is virtually the same as the ML277-bound structure KCNQ1-CaMML277. KCNQ1-CaMML277-PIP2-B, by contrast, displays several structural differences in the CTD and CaM, although in the TMD, they all maintain the same activated VSD and opened pore (Fig. 2B and SI Appendix, Fig. S7). In the CTD, S6 and the HA helix, which is connected by a loop linker of residues KHFN with an ∼120° angle in KCNQ1-CaMML277-PIP2-A, becomes a joint continuous helix in KCNQ1-CaMML277-PIP2-B (Fig. 7 A and B). This structure remodeling induces an almost 180° rotation of HA, HB, and the surrounding CaM. HB, which sits at the right side of HA in KCNQ1-CaMML277-PIP2-A, moves to the left side of HA in KCNQ1-CaMML277-PIP2-B (Fig. 7 A and B and SI Appendix, Movies S4 and S5). Consequently, the coiled-coil-forming helix HC, which is directly connected to HB, rotates by ∼97° (Fig. 7C and SI Appendix, Movie S6). In summary, like KCNQ1-CaMML277, KCNQ1-CaMML277-PIP2-A is still in the attached conformation (Fig. 6A and SI Appendix, Fig. S7), whereas KCNQ1-CaMML277-PIP2-B is in the detached conformation due to the dissociation of CaM and HA/HB from VSD and their following rotation (Fig. 6B).
Fig. 7.
Structural comparisons of KCNQ1-CaMML277-PIP2-A and KCNQ1-CaMML277-PIP2-B. (A) Different structure arrangements of CTD and CaM in KCNQ1-CaMML277-PIP2-A and KCNQ1-CaMML277-PIP2-B. S6, S6-HA linker, HA, HB, and HC helices of KCNQ1 are colored in marine, red, cyan, green, and yellow, respectively. The N-lobe and C-lobe of CaM are shown in wheat and pink, respectively. (B) Structural comparison of one KCNQ1 subunit in KCNQ1-CaMML277-PIP2-A (blue) and KCNQ1-CaMML277-PIP2-B (green) in two side views. The S6-HA linker ‘‘KHFN’’ motif that undergoes structural rearrangement from a loop to a helix is colored in red. (C) Structural comparison of the HA, HB, and HC helices in KCNQ1-CaMML277-PIP2-A (blue) and KCNQ1-CaMML277-PIP2-B (green) in the top view with TMD and HA/HB from the other three subunits omitted for clarity. The N-terminal residue Val538 of HC rotates by ∼97° from KCNQ1-CaMML277-PIP2-A to KCNQ1-CaMML277-PIP2-B. (D) The binding of PIP2 induces a 1–2 Å movement of the S2-S3 linker in KCNQ1-CaMML277-PIP2-B (green) in comparison with that in KCNQ1-CaMML277-PIP2-A (blue). The PIP2 in KCNQ1-CaMML277-PIP2-A is not shown for clarity. (E) Interactions between S6 and HB-HC linker. The dashed lines indicate the salt bridge Arg360-Asp537 and the hydrogen bonds Gln359-Tyr536 and His363-Lys534.
While the ML277-bound KCNQ1-CaM is stable in the attached conformation, the ML277-PIP2-bound KCNQ1-CaM seems dynamic and toggles between the attached and the detached conformations. How does PIP2 induce the structural rearrangement of the CTD and the CaM during the voltage gating? In KCNQ1-CaMML277-PIP2-A, the phosphate at position 5 of the inositol in PIP2 interacts with the S2-S3 linker and the phosphate at position 4 binds in CaM (Fig. 6A), whereas in KCNQ1-CaMML277-PIP2-B, both phosphate groups interact with the S2-S3 linker and S0 (Fig. 6B), which induces a 1–2 Å movement of the S2-S3 linker closer to the 1,4,5 trisphosphate head group of PIP2 (Fig. 7D). This small movement may disturb the interactions between the S2-S3 linker and CaM and cause the release of CaM. On the other hand, the folding of the KHFN motif between S6 and HA from a loop linker to an α helix is likely energy favorable, as the salt bridge Arg360-Asp537 and the hydrogen bonds Gln359-Tyr536 and His363-Lys534 are introduced at the interface of S6 and the HB-HC linker (Fig. 7E). Therefore, once CaM is detached from the VSD, HA/HB and the surrounding CaM may spontaneously undergo an almost 180° rotation, generating a continuous helix from S6 and HA. This structural rearrangement of CTD and CaM usually accompanies the opening of the activation gate across the KCNQ family (8, 44).
Discussion
ML277 is a Direct Activator of KCNQ1.
In this study, we report four structures of the KCNQ1-CaM complex, one in the apo closed state, one in the ML277-bound open state, and two in the ML277-PIP2-bound open states. These different conformations of the KCNQ1-CaM complex provide mechanistic insights into the ligand activation of the channel. More specifically, since both PIP2 and ML277 play key roles in enhancing the VSD-PD coupling, these findings elucidate the dynamic VSD-PD coupling process of KCNQ1.
ML277 is a long-shaped molecule. It binds in the “elbow” pocket above the S4-S5 linker and runs almost parallel to the S4-S5 linker. We previously have identified multiple pairs of interactions between two neighboring subunits at this pocket that are critical to the VSD-PD coupling (13). The hydrophobic packing between ML277 and the S4-S5 linker pulls the S4-S5 linker to move up, which may strengthen these coupling interactions and increase the open probability of the channel. This activation of KCNQ1 by ML277, which does not affect the CTD and the CaM, is not observed in other structures of the KCNQ family (8, 43, 44), supporting the unique feature of ML277 in enhancing the VSD-PD coupling (6). The open-state conformation of the KCNQ family was first observed in the PIP2-bound KCNQ1-KCNE3-CaMPIP2-6V01, which indicates that the activation of KCNQ1 involves a large structural rearrangement of the CTD and the CaM (8). Here, the ML277-bound KCNQ1-CaMML277 structure provides a second activation mode, which shows that this large structural rearrangement of the CTD and the CaM is not essential for the activation of KCNQ1.
Since ML277 exclusively changes the gating properties of the AO state (6, 13), ML277-induced conformational changes, especially the upward movement at the N-terminal of the S4-S5 linker, is more likely to correlate with the AO state gating process, and the KCNQ1-CaMML277-PIP2-B represents the AO state. These findings also provide direct evidence that supports the “hand-and-elbow” VSD-PD coupling mechanism across the KV channel superfamily (13). The Fedida Laboratory recently found that ML277 has profound effects on KCNQ1 single-channel kinetics and amplitude (48). Due to the lack of high-resolution structure for KCNQ1-CaMPIP2, the structural basis for this single-channel amplitude change requires further study.
ML277 Provides Clues for the Development of New KCNQ Activators.
Neuronal KCNQ2-5 channels play important roles in maintaining the membrane potential and are important drug targets for the treatment of epilepsy, pain, and deafness (49–52). Previously, structures of KCNQ2 and KCNQ4 in complex with different activators such as retigabine, ztz240, and ML213 were reported (43–45), yet no high-resolution structure of KCNQ in the open state with these activators alone was captured. By contrast, ML277 alone is able to activate KCNQ1 efficiently, and the open-state structure of KCNQ1-CaMML277 was readily obtained. While retigabine and ML213 mainly interact with the middle parts of S5 and S6 and likely favor the transition from closed to open states, ML277 forms extensive hydrophobic interactions with the S4-S5 linker and directly triggers its upward motion, which induces the opening of the activation gate. Thus, the binding mode and activation mechanism of ML277 are different from those of retigabine and ML213. The ML277-bound open-state structure of KCNQ1 will guide the design of new drugs targeting KCNQ channels.
The structural and sequence alignments of KCNQ also explain why ML277 displays 100-fold higher sensitivity to KCNQ1 over KCNQ2 and KCNQ4 (30). In KCNQ1, Phe335 and Leu266 are directly involved in the recognition of ML277 (Fig. 4B). In KCNQ2 and KCNQ4, these two residues are replaced by Ile/Leu and Trp residues (Ile300 and Trp236 in KCNQ2, and Leu306 and Trp242 in KCNQ4), respectively (SI Appendix, Fig. S6). The Trp residue with a large side chain that plays an essential role in the binding of retigabine would likely block the access of ML277 to the binding pocket. The Ile or Leu residue, on the other hand, has a smaller side chain and probably weakens interactions between ML277 and KCNQ2 or KCNQ4.
KCNEs May Alter Interactions between ML277 and KCNQ1.
In the presence of saturating amounts of KCNE1 (or KCNE3), ML277 no longer activates the channel complex (36). Structural comparison of KCNQ1-CaM-KCNE3apo-6V00 and KCNQ1-CaMapo-6UZZ shows that the binding of KCNE3 between S1 and S6 induces a 2∼3 Å shift of both S5 and S6 at the outer leaflet of the membrane (SI Appendix, Fig. S8A) (8). This KCNE3-induced conformational change of S5 and S6 would likely prevent the binding of ML277, as revealed by the structural alignment of KCNQ1-CaM-KCNE3apo-6V00 and KCNQ1-CaMML277, where the side chain of Phe335 in S6 in KCNQ1-CaM-KCNE3apo-6V00 would form clashes with ML277 (SI Appendix, Fig. S8B). Therefore, although KCNE3 does not occupy the ML277 binding site in KCNQ1, it induces conformational change of S5 and S6 and narrows the potential binding pocket of ML277.
Structural Comparison of ML277-Bound Human and Xenopus KCNQ1-CaM.
During the review of this paper, the Fedida Laboratory reported the structure of ML277-bound Xenopus KCNQ1-CaM (PBD 7CTI, xKCNQ1-CaMML277-7CTI) (53). Generally, structures of KCNQ1-CaMML277 and xKCNQ1-CaMML277-7CTI reveal the same binding site of ML277. Meanwhile, two differences are observed. First, the configuration of ML277 is slightly different. In the xKCNQ1-CaMML277-7CTI, the carbonyl in ML277 points to S6 and introduces electrostatic repulsion with the mainchain carbonyl of Phe335, whereas in our KCNQ1-CaMML277 structure, it points outwards (SI Appendix, Fig. S9 A and B). Second, the two structures capture different channel states. The xKCNQ1-CaMML277-7CTI structure remains in a closed state, while the human KCNQ1-CaMML277 is in an open state (SI Appendix, Fig. S9 C and D).
PIP2 Triggers the Structural Rearrangement of the CTD and CaM.
Previously, the structure of the PIP2-bound KCNQ1-KCNE3-CaMPIP2-6V01 showed how PIP2 is recognized by KCNQ1 in the detached conformation (8). Moreover, structural comparison of apo closed KCNQ1-KCNE3-CaMapo-6V00 and PIP2-bound open-state KCNQ1-KCNE3-CaMPIP2-6V01 reveals how the large structural rearrangement of KCNQ1 CTD and CaM accompanies the opening of the channel (SI Appendix, Fig. S10). However, the PIP2-bound KCNQ in the attached conformation is unknown, and how PIP2 activates KCNQs remains inconclusive. Our observations in the structures of KCNQ1-CaMML277-PIP2-A and KCNQ1-CaMML277-PIP2-B not only recapitulate the large structural rearrangement of KCNQ1 CTD and CaM (Figs. 6B and 7 A and B), but also provide insight into the PIP2-mediated gating processes. During the VSD activation, the binding mode of PIP2 may change and modulate the channel function. For example, the AO state has higher PIP2 sensitivity than the IO state (12). As the PIP2 binds close to S4, the outward movement of S4 during VSD activation may induce conformational changes to the PIP2 binding site and initiate the structural rearrangement of the CTD and CaM. The subtle change at the S2-S3 linker may disrupt the interaction between the S2-S3 linker and CaM, as well as the interaction between PIP2 and CaM, resulting in the release of the CTD and CaM from the VSD (Fig. 7D). In vivo, this release will favor the large structural rearrangement of the CTD and CaM because S6 and HA will form one stable secondary structure, and new interactions at the interface between S6 and the HB-HC linker are established (Fig. 7E). In vitro, however, this does not ensure a stable open-state structure. In our experiment, by adding PIP2 alone, we captured two low-resolution structures of KCNQ1-CaMPIP2, one in the attached conformation and the other likely in an intermediate state between attached and detached conformations (SI Appendix, Fig. S4). Similarly, the PIP2-retigabine-bound KCNQ2 structure we previously reported was also in an intermediate state (43). In the cases of KCNQ1-KCNE3-CaMPIP2-6V01 and KCNQ4-CaMML213-PIP2, the auxiliary subunit KCNE3 and the activator ML213 likely help stabilize the open state (8, 44). The ML277 in the structure of KCNQ1-CaMML277-PIP2-B also helps to stabilize the open state.
Materials and Methods
Protein Expression and Structural Determination.
The human KCNQ1 gene with a C-terminal strep tag was expressed in HEK293S cells at 30 °C for 48 h using the BacMam system. Protein was extracted with 1.5% n-dodecyl-β-D-maltoside (Anatrace) and 0.3% cholesteryl hemisuccinate Tris salt (Anatrace) at 4 °C for 3 h. After purification with Strep-Tactin Sepharose resin (IBA) and size-exclusion chromatography on a Superose 6 10/300 GL column (GE Healthcare), the final sample was concentrated to 8 mg/mL for single-particle analysis. The cryo-EM grids were prepared using a Mark IV Vitrobot (FEI). Micrographs were acquired with a Titan Krios microscope (FEI) operated at 300 kV with a K2 summit direct electron detector (Gatan) via SerialEM (54) following standard procedure. The defocus range was set from –1.1 to –1.3 μm. Data were dose fractionated to 40 frames with a dose rate of 8 e−/pixel/s and a total exposure time of 8 s, corresponding to a total dose of ∼64 e−/Å2. Image processing and three-dimensional reconstruction were performed following standard procedures, and the final resolution was estimated by the gold-standard Fourier shell correlation = 0.143 criterion (SI Appendix, Figs. S1–S4 and Table S1). The structure of human KCNQ1-CaM complex (PDB: 6UZZ) (8) was used as the initial model for model building.
Electrophysiology.
KCNQ1 point mutations were made by overlap extension and high-fidelity PCR, and were verified by DNA sequencing. complementary RNAs (cRNAs) of KCNQ1 were synthesized by T7 polymerase transcription kit (Thermo Fisher Scientific). Xenopus oocytes were used for KCNQ1 channel expression. KCNQ1 cRNAs (9.2 ng) were injected into each oocyte with a microinjector (RWD R-480 model). After injection, oocytes were kept in the ND96 solution (in mM): 96 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2, 5 Hepes, 2.5 CH3COCO2Na, 1:100 Pen-Strep, pH 7.6) at 18 °C for 2–6 d to allow sufficient KCNQ1 channel expression. A two-electrode voltage clamp was used to record the whole-oocyte currents. Currents were sampled at 1 kHz and filtered at 2 kHz. Microelectrodes were made with a Sutter puller (P-1000) with resistances at ∼1 MΩ when filled with 3 M KCl. For ML277 experiments, ML277 stock (Sigma Aldrich, in dimethyl sulfoxide [DMSO]) was added to the bath and diluted to 1 µM. All recordings were performed at room temperature (21–23 °C). Data were analyzed with Clampfit (Axon Instruments, Inc.) (55), SigmaPlot (SPSS, Inc.) (56), and IGOR (Wavemetrics) (57). G–V curves were fitted a Boltzmann equation 1/(1+exp(−z*F*(V−V50)/RT)), where V is the voltage, z is the equivalent valence, V50 is the half-maximal voltage, F is the Faraday constant, R is the gas constant, and T is the absolute temperature. For activation (τf and τs) and deactivation (τd) time constants of KCNQ1 currents in Fig. 1, KCNQ1 activation currents were fitted with a double-exponential equation to get the τf and τs for control, and were fitted separately with a single exponential function to get the τf and τs for currents after adding ML277.
Supplementary Material
Acknowledgments
Single-particle cryo-electron microscopy data were collected at Center of Cryo-Electron Microscopy at Zhejiang University and the Cryo-Electron Microscopy Facility of Hubei University. We thank Dr. Huaiyu Yang for the discussion on ligand activation mechanisms of KCNQ channels. We thank Dr. Xing Zhang and Dr. Shenghai Chang for support in facility access and data acquisition. This work was supported in part by the Ministry of Science and Technology (2020YFA0908501 and 2018YFA0508100 to J.G.), National Natural Science Foundation of China (31870724 to J.G., 82030108 to W.Y., 32171221 to P.H., and 81800231 to Y.H.), Zhejiang Provincial Natural Science Foundation (LR19C050002 to J.G.), and Fundamental Research Funds for the Central Universities (2021FZZX001-28 to J.G.). This work was also funded by Dr. Neher's Biophysics Laboratory for Innovative Drug Discovery (Grant no. 001/2020/ALC, FDCT-20-051-SKL, and 0006/2021/AMJ to P.H.) as supported by the Macau Science and Technology Development Fund. W.Y. and J.G. are supported by MOE Frontier Science Center for Brain Science & Brain-Machine Integration, Zhejiang University.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2207067119/-/DCSupplemental.
Data, Materials, and Software Availability
The cryo-electron microscopy density maps and coordinates of KCNQ1-CaMapo, KCNQ1-CaMML277, KCNQ1-CaMML277-PIP2-A, and KCNQ1-CaMML277-PIP2-B have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-33316, EMD-33317, EMD-33318, and EMD-33319, respectively, and in the RCSB Protein Data Bank under accession codes 7XNI, 7XNK, 7XNL, and 7XNN, respectively (58, 59, 60, 61, 62, 63, 64, 65). All study data are included in the article and/or supporting information.
References
- 1.Wang Q., et al. , Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat. Genet. 12, 17–23 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Sanguinetti M. C., et al. , Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature 384, 80–83 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Barhanin J., et al. , K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature 384, 78–80 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Schwartz P. J., Crotti L., Insolia R., Long-QT syndrome: From genetics to management. Circ. Arrhythm. Electrophysiol. 5, 868–877 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohnen M. S., et al. , Molecular pathophysiology of congenital long QT syndrome. Physiol. Rev. 97, 89–134 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou P., Shi J., White K. M., Gao Y., Cui J., ML277 specifically enhances the fully activated open state of KCNQ1 by modulating VSD-pore coupling. eLife 8, e48576 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun J., MacKinnon R., Cryo-EM structure of a KCNQ1/CaM complex reveals insights into congenital long QT syndrome. Cell 169, 1042–1050.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun J., MacKinnon R., Structural basis of human KCNQ1 modulation and gating. Cell 180, 340–347.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shamgar L., et al. , Calmodulin is essential for cardiac IKS channel gating and assembly: Impaired function in long-QT mutations. Circ. Res. 98, 1055–1063 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Ghosh S., Nunziato D. A., Pitt G. S., KCNQ1 assembly and function is blocked by long-QT syndrome mutations that disrupt interaction with calmodulin. Circ. Res. 98, 1048–1054 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Hou P., et al. , Inactivation of KCNQ1 potassium channels reveals dynamic coupling between voltage sensing and pore opening. Nat. Commun. 8, 1730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaydman M. A., et al. , Domain-domain interactions determine the gating, permeation, pharmacology, and subunit modulation of the IKs ion channel. eLife 3, e03606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou P., et al. , Two-stage electro-mechanical coupling of a KV channel in voltage-dependent activation. Nat. Commun. 11, 676 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor K. C., et al. , Structure and physiological function of the human KCNQ1 channel voltage sensor intermediate state. eLife 9, e53901 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bezanilla F., Perozo E., Stefani E., Gating of Shaker K+ channels: II. The components of gating currents and a model of channel activation. Biophys. J. 66, 1011–1021 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho-de-Souza J. L., Bezanilla F., Nonsensing residues in S3-S4 linker’s C terminus affect the voltage sensor set point in K+ channels. J. Gen. Physiol. 150, 307–321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delemotte L., Tarek M., Klein M. L., Amaral C., Treptow W., Intermediate states of the Kv1.2 voltage sensor from atomistic molecular dynamics simulations. Proc. Natl. Acad. Sci. U.S.A. 108, 6109–6114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacroix J. J., Bezanilla F., Control of a final gating charge transition by a hydrophobic residue in the S2 segment of a K+ channel voltage sensor. Proc. Natl. Acad. Sci. U.S.A. 108, 6444–6449 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao X., Lee A., Limapichat W., Dougherty D. A., MacKinnon R., A gating charge transfer center in voltage sensors. Science 328, 67–73 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva J. R., et al. , A multiscale model linking ion-channel molecular dynamics and electrostatics to the cardiac action potential. Proc. Natl. Acad. Sci. U.S.A. 106, 11102–11106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaydman M. A., et al. , Kv7.1 ion channels require a lipid to couple voltage sensing to pore opening. Proc. Natl. Acad. Sci. U.S.A. 110, 13180–13185 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salata J. J., et al. , A novel benzodiazepine that activates cardiac slow delayed rectifier K+ currents. Mol. Pharmacol. 54, 220–230 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Gao Z., Xiong Q., Sun H., Li M., Desensitization of chemical activation by auxiliary subunits: Convergence of molecular determinants critical for augmenting KCNQ1 potassium channels. J. Biol. Chem. 283, 22649–22658 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seebohm G., Pusch M., Chen J., Sanguinetti M. C., Pharmacological activation of normal and arrhythmia-associated mutant KCNQ1 potassium channels. Circ. Res. 93, 941–947 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Xiong Q., Sun H., Li M., Zinc pyrithione-mediated activation of voltage-gated KCNQ potassium channels rescues epileptogenic mutants. Nat. Chem. Biol. 3, 287–296 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Abitbol I., Peretz A., Lerche C., Busch A. E., Attali B., Stilbenes and fenamates rescue the loss of I(KS) channel function induced by an LQT5 mutation and other IsK mutants. EMBO J. 18, 4137–4148 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Silva A. M., Manville R. W., Abbott G. W., Deconstruction of an African folk medicine uncovers a novel molecular strategy for therapeutic potassium channel activation. Sci. Adv. 4, eaav0824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yazdi S., et al. , Identification of PUFA interaction sites on the cardiac potassium channel KCNQ1. J. Gen. Physiol. 153, e202012850 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liin S. I., et al. , Polyunsaturated fatty acid analogs act antiarrhythmically on the cardiac IKs channel. Proc. Natl. Acad. Sci. U.S.A. 112, 5714–5719 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattmann M. E., et al. , Identification of (R)-N-(4-(4-methoxyphenyl)thiazol-2-yl)-1-tosylpiperidine-2-carboxamide, ML277, as a novel, potent and selective K(v)7.1 (KCNQ1) potassium channel activator. Bioorg. Med. Chem. Lett. 22, 5936–5941 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redford K. E., Abbott G. W., KCNQ potassium channels as targets of botanical folk medicines. Annu. Rev. Pharmacol. Toxicol. 62, 447–464 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y., et al. , A PIP2 substitute mediates voltage sensor-pore coupling in KCNQ activation. Commun. Biol. 3, 385 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Y., et al. , Modulating the voltage sensor of a cardiac potassium channel shows antiarrhythmic effects. Proc. Natl. Acad. Sci. U.S.A. 118, e2024215118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schreiber J. A., et al. , A benzodiazepine activator locks Kv7.1 channels open by electro-mechanical uncoupling. Commun. Biol. 5, 301 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y., Eldstrom J., Fedida D., The IKs ion channel activator mefenamic acid requires KCNE1 and modulates channel gating in a subunit-dependent manner. Mol. Pharmacol. 97, 132–144 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Yu H., et al. , Dynamic subunit stoichiometry confers a progressive continuum of pharmacological sensitivity by KCNQ potassium channels. Proc. Natl. Acad. Sci. U.S.A. 110, 8732–8737 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Y., et al. , Probing binding sites and mechanisms of action of an I(Ks) activator by computations and experiments. Biophys. J. 108, 62–75 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma D., et al. , Characterization of a novel KCNQ1 mutation for type 1 long QT syndrome and assessment of the therapeutic potential of a novel IKs activator using patient-specific induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Res. Ther. 6, 39 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wuriyanghai Y., et al. , Complex aberrant splicing in the induced pluripotent stem cell-derived cardiomyocytes from a patient with long QT syndrome carrying KCNQ1-A344Aspl mutation. Heart Rhythm 15, 1566–1574 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S. P., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y., et al. , Molecular basis for ligand modulation of a mammalian voltage-gated Ca2+ channel. Cell 177, 1495–1506.e12 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Tang L., et al. , Structural basis for inhibition of a voltage-gated Ca2+ channel by Ca2+ antagonist drugs. Nature 537, 117–121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X., et al. , Molecular basis for ligand activation of the human KCNQ2 channel. Cell Res. 31, 52–61 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y., et al. , Structural insights into the lipid and ligand regulation of a human neuronal KCNQ channel. Neuron 110, 237–247.e4 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Li T., et al. , Structural basis for the modulation of human KCNQ4 by small-molecule drugs. Mol. Cell 81, 25–37.e4 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Seebohm G., et al. , Regulation of endocytic recycling of KCNQ1/KCNE1 potassium channels. Circ. Res. 100, 686–692 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Boulet I. R., Labro A. J., Raes A. L., Snyders D. J., Role of the S6 C-terminus in KCNQ1 channel gating. J. Physiol. 585, 325–337 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eldstrom J., McAfee D. A., Dou Y., Wang Y., Fedida D., ML277 regulates KCNQ1 single-channel amplitudes and kinetics, modified by voltage sensor state. J. Gen. Physiol. 153, e202112969 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown D. A., Adams P. R., Muscarinic suppression of a novel voltage-sensitive K+ current in a vertebrate neurone. Nature 283, 673–676 (1980). [DOI] [PubMed] [Google Scholar]
- 50.Jentsch T. J., Neuronal KCNQ potassium channels: Physiology and role in disease. Nat. Rev. Neurosci. 1, 21–30 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Liu Y., Bian X., Wang K., Pharmacological activation of neuronal voltage-gated Kv7/KCNQ/M-channels for potential therapy of epilepsy and pain. Handb. Exp. Pharmacol. 267, 231–251 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Jones F., Gamper N., Gao H., Kv7 channels and excitability disorders. Handb. Exp. Pharmacol. 267, 185–230 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Willegems K., et al. , Structural and electrophysiological basis for the modulation of KCNQ1 channel currents by ML277. Nat. Commun. 13, 3760 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mastronarde D. N., Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 55.S. H. Heinemann, “Guide to Data Acquisition and Analysis” in Single-Channel Recording, B. Sakmann, E. Neher, Eds. (Springer, Boston, MA, 1995). [Google Scholar]
- 56.Hilbe J. M., Review of SigmaPlot 9.0. Am. Stat. 59, 111–112 (2005). [Google Scholar]
- 57.Kline S. R., Reduction and analysis of SANS and USANS data using IGOR Pro. J. Appl. Crystallogr. 39, 895–900 (2006). [Google Scholar]
- 58.Ma D., Guo J., Human KCNQ1-CaM in apo state. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-33316. Deposited 28 April 2022.
- 59.Ma D., Guo J., Human KCNQ1-CaM in complex with ML277. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-33317. Deposited 29 April 2022.
- 60.Ma D., Guo J., Human KCNQ1-CaM-ML277-PIP2 complex in state A. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-33318. Deposited 29 April 2022.
- 61.Ma D., Guo J., Human KCNQ1-CaM-ML277-PIP2 complex in state B. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-33319. Deposited 29 April 2022.
- 62.Ma D., Guo J., Human KCNQ1-CaM in apo state. RCSB Protein Data Bank. https://www.rcsb.org/structure/7XNI. Deposited 28 April 2022.
- 63.Ma D., Guo J., Human KCNQ1-CaM in complex with ML277. RCSB Protein Data Bank. https://www.rcsb.org/structure/7XNK. Deposited 29 April 2022.
- 64.Ma D., Guo J., Human KCNQ1-CaM-ML277-PIP2 complex in state A. RCSB Protein Data Bank. https://www.rcsb.org/structure/7XNL. Deposited 29 April 2022.
- 65.Ma D., Guo J., Human KCNQ1-CaM-ML277-PIP2 complex in state B. RCSB Protein Data Bank. https://www.rcsb.org/structure/7XNN. Deposited 29 April 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-electron microscopy density maps and coordinates of KCNQ1-CaMapo, KCNQ1-CaMML277, KCNQ1-CaMML277-PIP2-A, and KCNQ1-CaMML277-PIP2-B have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-33316, EMD-33317, EMD-33318, and EMD-33319, respectively, and in the RCSB Protein Data Bank under accession codes 7XNI, 7XNK, 7XNL, and 7XNN, respectively (58, 59, 60, 61, 62, 63, 64, 65). All study data are included in the article and/or supporting information.