Abstract
Many mucosal pathogens, including Neisseria gonorrhoeae, produce proteases that cleave immunoglobulin A (IgA), the predominant immunoglobulin class produced at mucosal surfaces. While considerable circumstantial evidence suggests that IgA1 protease contributes to gonococcal virulence, there is no direct evidence that N. gonorrhoeae requires IgA1 protease activity to infect a human host. We constructed a N. gonorrhoeae iga mutant without introducing new antibiotic resistance markers into the final mutant strain and used human experimental infection to test the ability of the mutant to colonize the male urethra and to cause gonococcal urethritis. Four of the five male volunteers inoculated with the Iga− mutant became infected. In every respect—clinical signs and symptoms, incubation period between inoculation and infection, and the proportion of volunteers infected—the outcome of human experimental infection with FA1090iga was indistinguishable from that previously reported for a variant of parent strain FA1090 matching the mutant in expression of Opa proteins, lipooligosaccharide, and pilin. These results indicate that N. gonorrhoeae does not require IgA1 protease production to cause experimental urethritis in males.
Immunoglobulin A (IgA) is the predominant Ig produced at mucosal surfaces. While the extent of its contribution to host defenses is unclear, IgA appears to perform a number of potentially beneficial functions, including inhibition of bacterial adherence (3). Many pathogens that colonize mucosal surfaces produce IgA1 proteases, proteins that cleave within the heavy (α)-chain hinge of the IgA isotype IgA1 to produce intact Fab and Fc fragments (reviewed in references 25 and 26). IgA1 proteases are generally made by pathogenic members of the genera Neisseria and Haemophilus but not by nonpathogenic members. The mucosal secretions of patients colonized with certain IgA1 protease-producing bacteria possess IgA1 protease activity and/or contain IgA1 fragments, and it has often been theorized that IgA1 protease may facilitate colonization by cleaving secretory IgA1 (2, 16).
Almost all known strains of Neisseria gonorrhoeae, the causative agent of the sexually transmitted disease gonorrhea, produce IgA1 protease (18, 24, 27, 28). Each strain produces one of two similar types of the enzyme (type 1 or type 2), which cleave different bonds (Pro-Ser and Pro-Thr, respectively) in the 18-amino-acid hinge region of human IgA1. Production of gonococcal IgA1 protease involves self-directed secretion and autocatalytic processing of a larger precursor protein encoded by the iga gene (reviewed in reference 17). Processing leads to release of the mature protease plus two smaller fragments, the α and γ peptides. Recently, alternative roles in gonococcal pathogenesis other than cleavage of IgA1 at mucosal surfaces have been proposed for IgA1 protease and for the α peptide. IgA1 protease cleaves synaptobrevin II in vitro and, when introduced into the cytosol of bovine chromaffin cells, blocks exocytosis (1). The type 2 IgA1 protease cleaves LAMP1 (lysosome/late endosome-associated membrane protein 1), and intracellular N. gonorrhoeae localizes to LAMP1-positive compartments in both epithelial cells and phagocytes (10). Lin et al. (20) also demonstrated cleavage of LAMP1 by neisserial type 2 IgA1 protease and found that LAMP1 was degraded at a higher rate in an epithelial cell line infected with wild-type N. gonorrhoeae than in uninfected cells or in cells infected with an iga mutant. The iga mutant grew poorly relative to the wild-type strain in these epithelial cells (20). Pohlner et al. (29) recently showed that the α peptide targets eukaryotic nuclei and proposed that it may influence host cell gene expression. These findings suggest that IgA1 protease and other iga gene products may contribute to neisserial pathogenesis at intracellular stages of the infection process. However, a gonococcal iga mutant was not impaired in its ability to invade human fallopian tube organ cultures (5).
The role played by IgA1 protease in infecting a host with an intact immune system has never been addressed. For N. gonorrhoeae, infection occurs only in humans, and no convenient animal models of mucosal infection are available. However, human challenge studies can be used to assess the role of specific virulence factors in gonococcal pathogenesis. A human model of infection is particularly well suited for assessment of gonococcal IgA1 protease function, because neisserial IgA1 proteases cleave only the IgA of humans and closely related primates (reviewed in reference 21). Experimental infection of male volunteers with N. gonorrhoeae is safe and successfully reproduces the signs and symptoms of natural infection (4, 14, 23, 32, 35, 36). In this study, we tested the ability of an N. gonorrhoeae FA1090 iga mutant (designated FA1090iga) to infect male subjects in the human challenge model of urethral infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
N. gonorrhoeae FA1090 was originally a cervical isolate from a patient with disseminated gonococcal infection; it is streptomycin resistant (Smr) and is sensitive to both ceftriaxone and ciprofloxacin. FA1090 variants A21 and A22 have been characterized and used previously in human experimental infections (4, 12, 33). The A22 stock was created by a single passage of the original A21 variant and was given a new designation; the two variants are identical in all tested phenotypes (4, 9, 12). Gonococci were grown as previously described (4). For IgA1 protease activity assays, gonococci were grown in gonococcal base (GCB) broth plus Kellogg’s supplements and 5 mM sodium bicarbonate at 37°C with shaking (14). Where appropriate, the medium was supplemented with erythromycin (5 μg/ml) or streptomycin (10.5 mg/ml). Escherichia coli DH5αMCR (Life Technologies, Gaithersburg, Md.) and SURE (Stratagene, La Jolla, Calif.) were grown at 37°C in Luria-Bertani broth or on Luria-Bertani agar supplemented with 100 μg of ampicillin per ml and 250 μg of erythromycin per ml where appropriate. For blue/white β-galactosidase activity detection, the plates were spread with 40 μl of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (20 mg/ml) and 4 μl of isopropyl-β-d-thiogalactopyranoside (IPTG) (200 mg/ml) before the bacteria were plated.
Recombinant DNA techniques and PCR.
Restriction enzymes, DNA-modifying enzymes, and buffers were obtained from New England Biolabs (Beverly, Mass.) and Life Technologies. PCR reagents were obtained from Boehringer Mannheim (Indianapolis, Ind.), and Life Technologies. Oligonucleotides were synthesized by the University of North Carolina Nucleic Acids Core Facility or by Life Technologies. Unless otherwise noted, PCR mixtures contained 50 pmol of each primer, 250 mM each deoxynucleoside triphosphate, 2.5 to 5.0 U of Taq polymerase, and manufacturer’s recommended buffer plus MgCl2. DNA was sequenced at the University of North Carolina at Chapel Hill. Automated DNA Sequencing Facility on a model 373A DNA Sequencer (Applied Biosystems, Foster City, Calif.) with the Taq DyeDeoxy Terminator cycle-sequencing kit (Applied Biosystems). Probes for colony blots were prepared by enhanced chemiluminescence direct-labeling or 3′-oligolabeling methods (Amersham, Arlington Heights, Ill.). When 3′-oligolabeling was used, the hybridization temperatures ranged from 13 to 17°C below the oligonucleotide melting temperature. Southern blots were performed by standard methods with hybridization temperatures 11°C below the oligonucleotide melting temperature (31).
Construction of the N. gonorrhoeae FA1090 iga mutant.
We constructed a derivative of the vector pUC19 containing the gonococcal uptake sequence that is required for transformation of N. gonorrhoeae (7). Complementary oligonucleotides Upt1 (5′-GGGCAAGCTTGCCGTCTGAAAAGCTTGGGC-3′) and Upt2 (5′-GCCCAAGCTTTTCAGACGGCAAGCTTGCCC-3′) (100 pmol each) were boiled for 5 min and then annealed by slow cooling to room temperature. The annealed oligonucleotides, containing one copy of the gonococcal uptake sequence flanked by HindIII sites (underlined), were digested with HindIII and cloned into the HindIII site of plasmid pUC19 to create plasmid pUPT1.
A 1,350-bp fragment of the N. gonorrhoeae FA1090 iga gene was amplified by PCR with primers Iga1 and Iga2, which were derived from the sequence of the iga gene of strain MS11 (28). The amplified fragment was part of the segment of the iga gene encoding the mature IgA1 protease product (Fig. 1). Primers Iga1 and Iga2 corresponded to regions that are conserved in neisserial iga genes from different strains (8, 22, 28). Iga1 (5′-GGGAATTCGATAACGCCGGCACCGTCAAAGG-3′; nucleotides 1082 to 1104 of MS11 iga) introduced an EcoRI site (underlined), and Iga2 (5′-CCGGATCCCCGGTATAGTCCGAGCGCACGC-3′; nucleotides 2451 to 2430) introduced a BamHI site (underlined). N. gonorrhoeae FA1090 chromosomal DNA served as template. The PCR consisted of 30 cycles of 2 min at 96°C, 2 min at 55°C, and 3.5 min at 72°C. Following BamHI and EcoRI digestion of the PCR product and of pUPT1, the Iga1-Iga2 PCR product was cloned into the pUPT1 multiple-cloning site, creating plasmid pUI. The partial sequence (>450 bp) of the pUI insert confirmed the identity of the cloned fragment. After we cloned the iga fragment, the sequence of the FA1090 iga gene became available from the gonococcal genome-sequencing project; the sequence of the clone was 99% identical to the preliminary sequence from the FA1090 genome project (30).
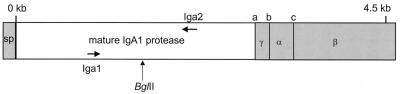
FIG. 1.
Illustration of the gonococcal iga gene, based on the sequence published by Pohlner et al. (28). The portion of the iga gene encoding the mature IgA1 protease is unshaded. Primers Iga1 and Iga2 were used in PCR to amplify the iga gene fragment for cloning. The BglII site shown was used subsequently for insertional inactivation of the gene, as described in the text. Shaded areas encode portions of the protein that are not part of the mature protease. The signal peptide (SP) and β domain (β) remain in the inner and outer membranes, respectively, during export, and the α peptide (α) and γ peptide (γ) are cleaved off following export to produce the mature IgA1 protease. This export model is reviewed by Klauser et al. (17).
Primers ER1 and ER2 were used to amplify the 2.3-kb ermC′-rpsL cassette from plasmid pFLOB4250 (13). The PCR conditions were the same as those described for amplification of iga. ER1 (5′-GCGGCAGATCTTGCCCGTTACAGAAAGTGG-3′) and ER2 (5′-GGCTCTGAGATCTTAAACGGTTGACAAGCCG-3′) introduced BglII sites (underlined) flanking the ermC′-rpsL cassette in the resultant PCR product. The ER1-ER2 PCR product was digested with BglII and cloned into the unique BglII site within the iga gene in pUI, resulting in plasmid pUIE.
Complementary oligonucleotides IL1 (5′-GGAGATCTAGTTAATTAACGTCAGATCTGG-3′) and IL2 (5′-CCAGATCTGACGTTAATTAACTAGATCTCC-3′) (100 pmol each) were annealed and purified as described above. The annealed oligonucleotides, containing a central PacI site (doubly underlined) flanked by BglII sites (underlined), were cloned into the BglII site within the iga gene in plasmid pUI. The resulting plasmid, containing the IL1-IL2 linker (designated IgaStop) within the iga gene, was designated pUIL. IgaStop created a frameshift and encoded translational stops in all three frames.
Gonococcal transformations were performed by a plate-spot transformation method. Piliated gonococci were streaked on GCB agar, and approximately 1 μg of DNA in 1× SSC (0.15 M NaCl, 0.015 sodium citrate) was spotted on the freshly streaked bacteria. After overnight incubation, colonies from the areas in which DNA was spotted were streaked onto selective media and incubated overnight; the resulting colonies were single-colony purified and rescreened for appropriate antibiotic resistance and sensitivity. Plasmid pUIE was transformed into N. gonorrhoeae FA1090 A22, selecting for erythromycin resistance and screening for streptomycin sensitivity. An erythromycin-resistant, streptomycin-sensitive transformant was transformed with plasmid pUIL, selecting for streptomycin resistance and screening for erythromycin sensitivity. A transformant that was Iga− by both Southern blot analysis and IgA1 protease activity assay was designated FA1090iga.
IgA1 protease activity assay.
IgA1 protease activity assays were performed as described by St. Geme et al. (34) with minor modifications. Gonococci were grown in GCB broth to a turbidity of 150 Klett units as measured with a Klett-Summerson colorimeter (approximately 4 × 108 CFU per ml). Aliquots of the cultures were centrifuged for 2 min at 15,000 × g in a microcentrifuge. Culture supernatant (10 μl) was added to 5 μl of human IgA1 (0.5 mg/ml; Calbiochem, La Jolla, Calif.), and chloramphenicol was added to a final concentration of 2 μg/ml. Samples were incubated overnight at 37°C, subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% polyacrylamide gel with Laemmli buffers, and transferred to 0.45-μm-pore-size Optitran supported nitrocellulose (Schleicher & Schuell, Keene, N.H.) (19). The filters were probed with anti-human IgA (α-chain specific)-peroxidase conjugate (1:5,000) (Sigma, St. Louis, Mo.) followed by the enhanced chemiluminescence substrate (Amersham, Arlington Heights, Ill.).
Preparation of FA1090iga inoculum.
We isolated a variant of strain FA1090iga that was matched in expression of variable surface components to FA1090 A21 and A22, the variants we have used in the majority of the human challenge experiments with this strain. FA1090 A21 and A22 are Opa negative and piliated and express three lipooligosaccharide (LOS) species, including one that binds monoclonal antibody (MAb) 3F11. Pilin expressed by these variants is derived entirely from the pilS6c1 storage copy; the variants produce full-length pilin plus some S-pilin (reference 9 and data not shown). The LOS phenotype was characterized by SDS-PAGE of proteinase K-treated lysates as previously described (12). Expression of Opa proteins was assessed by colony immunoblotting with MAbs specific for the Opa proteins of strain FA1090 and by observation of the colony opacity phenotype under a dissecting microscope, as previously described (12). Piliation was assessed by (i) determination of colony morphology, (ii) Western blot analysis of pilin expression with a polyclonal anti-pilin antiserum, (iii) competence for genetic transformation by a quantitative assay measuring the transformation frequency for the chromosomal rifampin resistance marker, and (iv) determination of the DNA sequence of the pilE expression locus. The pilE locus was amplified with primers pilstart (5′-GAGATAAACGCATAAAATTTCACC-3′) and sp3a (5′-CCGGAACGGACGACCCCG-3′), using FA1090iga freezer stock diluted in water and boiled as the template (33). The PCR product was sequenced directly with sequencing primer pilAW (5′-CCTACCAAGACTACACCGCCC-3′).
After isolation of an appropriate variant, designated FA1090iga 15b, the variant was expanded and a suspension of cells was divided into multiple aliquots, which were stored at −70°C in freezer storage medium (FSM) (12). One vial of this group was thawed and cultured, and all of the above phenotypes were reconfirmed. In addition, we determined that (i) the mutant strain grew on GCB agar containing VCN selective supplement (Oxoid, Basingstoke, United Kingdom) (used for culture of specimens from experimentally infected subjects), (ii) the mutant strain survived in urine as well as the wild-type parent did (a property that might affect the recovery of viable organisms from urine specimens), and (iii) the relationship between the turbidity of a suspension measured with a Klett-Summerson colorimeter and CFU per milliliter was the same as that for the parent strain (the inoculation procedure involves suspending the organisms in phosphate-buffered saline to a Klett reading that was previously determined to correspond to the desired number of CFU). The sensitivity of the mutant strain to ceftriaxone and ciprofloxacin was confirmed by the Clinical Microbiology Laboratory of University of North Carolina Hospitals.
Two days prior to inoculation, FA1090iga 15b was streaked onto GCB agar, using a freezer stock that had never been thawed. The following day (approximately 20 h before inoculation of volunteers), colonies were observed under a dissecting microscope. Two separate suspensions of colonies were made by picking approximately 30 colonies with appropriate transparent, piliated morphology into a small amount of GCB broth. These suspensions were streaked onto fresh GCB agar plates. The Opa-negative phenotype of the two suspensions was confirmed by immunoblotting with anti-FA1090 Opa MAbs. On the day of the trial, the colonies cultured from the two suspensions were examined under a dissecting microscope to confirm that the colonies were predominantly (>90%) piliated and transparent. The inoculum was prepared with some colonies derived from each of the two suspensions. The bacterial inoculum was suspended in phosphate-buffered saline, passed through a 1.2-μm-pore-size filter, and quantitated as previously described (4, 12).
Experimental infection of human subjects.
Male subjects were recruited and screened as previously described (4), and procedures for experimental infection of subjects were as previously described (4). The subjects gave informed consent and were compensated for their participation. They were examined by a physician at least twice daily for signs and symptoms of infection, and urethral swab specimens were collected, Gram stained, and cultured as soon as a subject developed a urethral exudate. Urine specimens from all subjects were cultured quantitatively each morning, and the total number of CFU washed from the urethra in each urine specimen was calculated. The volumes of the urine specimens and thus the concentrations of organisms in those specimens were widely variable; the culture results were therefore expressed as total CFU/urine specimen. The subjects were treated with ceftriaxone or ciprofloxacin immediately upon the appearance of signs or symptoms of infection. Subjects who did not become infected were treated at the end of the 5-day trial. Urethral swab specimens and colonies cultured from infected subjects were stored in FSM at −70°C. The protocols were reviewed and approved by the Institutional Review Board, the Institutional Biosafety Committee, and the General Clinical Research Center Advisory Board of the University of North Carolina at Chapel Hill.
Genotypic analysis of trial reisolates.
Colonies cultured from experimentally infected subjects were picked individually from primary isolation plates into 25 μl of FSM in microtiter wells and stored at −70°C. To confirm the inactivation of the iga gene, a portion of the gene was amplified by PCR with oligonucleotides Iga1 and Iga2 as described above, using Platinum Taq polymerase (Life Technologies). For the template in the PCRs, 2 μl of the FSM stock of one colony was added to the complete PCR mixture, which was incubated for 10 min at 94°C before the cycling program was started. The resulting PCR products were digested with PacI and analyzed by gel electrophoresis.
RESULTS
Construction of an Iga− mutant of N. gonorrhoeae FA1090.
To study the role played by gonococcal IgA1 protease in the early stages of urethral infection, we constructed an Iga− mutant of strain FA1090 (FA1090iga) for use in experimental human infection. The FA1090 genome (30) contains a single iga gene with features characteristic of one encoding a type 2 IgA1 protease (28). To minimize any potential risk to trial subjects, we used a two-step transformation procedure for constructing gonococcal mutants containing no new antibiotic resistance markers (13). Strain FA1090 is Smr and erythromycin sensitive (Erms). In the first step of the strain construction (described in detail in Materials and Methods), the FA1090 chromosomal iga gene was insertionally inactivated by introducing an ermC′-rpsL cassette into it by allelic exchange. Transformants were selected by the ermC′-encoded Ermr. The introduction of rpsL, which encodes E. coli ribosomal protein S12, conferred Sms on the transformants, due to the dominance of Sms over Smr in an Sms/Smr merodiploid. In the second step, the ermC′-rpsL cassette in one of the Ermr Sms first-step transformants was replaced by allelic exchange with an iga allele containing the IgaStop linker, which insertionally inactivated the gene by both changing its reading frame and introducing translational termination codons in all three frames. Transformants were selected for Smr and screened for Erms. The second step thus restored the Smr Erms phenotype of the parent strain. One of the Smr Erms transformants (shown below to have the correct genotype and IgA1 protease phenotype) was designated FA1090iga. We isolated a variant of FA1090iga (variant 15b) that was identical to the parent (FA1090 variant A22) with regard to variable attributes that might influence infectivity: LOS type, Opa protein expression (>90% Opa negative), and piliation (P+, with >99% identical pilE DNA sequences).
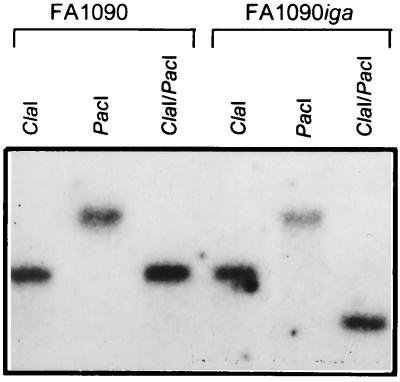
We used PCR analysis and Southern blotting to confirm that the iga gene in FA1090iga 15b was insertionally inactivated and that no duplication or rearrangement of the gene had occurred. Iga1-Iga2 PCR products from the mutant were cleaved by PacI (data not shown). A Southern blot of chromosomal DNA from the mutant digested with ClaI and PacI and probed with the Iga2 oligonucleotide showed that the probe-hybridizing fragment in FA1090iga 15b was shifted relative to that of FA1090, revealing the extra PacI site within the IgaStop linker in the mutant (Fig. 2).
FIG. 2.
Southern blot analysis of chromosomal DNA from strains FA1090 A22 and FA1090iga 15b digested with ClaI, PacI, or ClaI-PacI and probed with oligonucleotide Iga2, specific for the iga gene. The blot demonstrates the presence of the PacI site in the linker insertion used to inactivate the iga gene in the mutant strain.
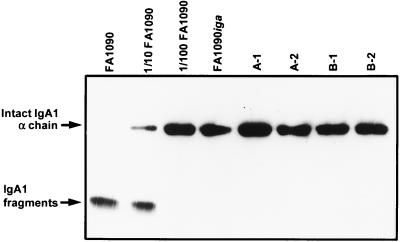
The iga mutant lacked detectable IgA1 protease function. Supernatant from a broth culture of FA1090iga 15b did not cleave human IgA1, whereas undiluted and 1:10 dilutions of culture supernatant from FA1090 A22 were able to do so (Fig. 3).
FIG. 3.
IgA1 protease activity of strain FA1090iga 15b and colonies cultured from human subjects infected with the mutant strain. Culture supernatants or dilutions of culture supernatants were incubated with human IgA1 overnight, subjected to SDS-PAGE (12% polyacrylamide), transferred to nitrocellulose, and probed with antibody to human IgA. The first three lanes contain IgA1 that was incubated with supernatant from wild-type strain FA1090 A22 cultures, undiluted, diluted 1:10, and diluted 1:100. The fourth lane contains IgA1 that was incubated with undiluted supernatant from a FA1090iga 15b culture. In the remaining lanes, IgA1 was incubated with undiluted supernatant from cultures of colonies isolated from FA1090iga 15b-infected subjects. A and B are two different subjects; 1 and 2 indicate two different colonies cultured from each subject.
Experimental human infection with FA1090iga 15b.
Five volunteers were inoculated with 106 CFU of FA1090iga 15b; four of the five became infected during the 5-day trial. Three subjects developed a urethral exudate containing gram-negative diplococci within 2 days of inoculation. The fourth infected subject developed a urethral discharge on the 4th day after inoculation. Urethral swab specimens and urine specimens from all four subjects were positive for gonococci. The fifth subject showed no signs or symptoms of infection, and no gonococci were cultured from urethral swab or urine specimens. These results are similar to those we obtained in previous human challenge experiments with wild-type FA1090 variants A21 and A22, in which 89% of subjects inoculated with 106 CFU were infected (n = 28). Subjects infected with either of these Opa-negative, P+ variants of the wild-type strain develop clinical signs of infection 1 to 4 days postinoculation (references 4, 6, 9, and 12 and our unpublished observations). The total number of CFU cultured from urine specimens obtained when subjects infected with the iga mutant developed a urethral discharge ranged from 3.2 × 104 to 1.3 × 105 total CFU/urine specimen, which is within the range normally occurring in FA1090-infected subjects (1 × 102 to 5 × 105 total CFU/urine specimen) (references 4, 9, and 12 and our unpublished observations). Similarly, leukocyte counts for subjects infected with the mutant strain, which ranged from 5 to 38 leukocytes per mm3 of urine sediment, were within the range for FA1090-infected subjects (our unpublished observations), although the values were highly variable in both cases.
Genotypic and phenotypic characterization of gonococci isolated from infected subjects.
Using PCR analysis and IgA1 protease activity assays, we confirmed that the subjects who became infected when inoculated with FA1090iga 15b were indeed infected with bacteria that had an insertionally inactivated iga gene and that lacked IgA1 protease activity. Primers Iga1 and Iga2 were used to amplify a portion of the iga gene from 24 individual colonies cultured from each of two of the infected volunteers. In all cases, the resulting PCR products were digested with PacI into two smaller fragments, showing that the iga genes of the reisolates contained the PacI site that is present in the IgaStop insertion (data not shown). To confirm that the presence of the IgaStop insertion was still associated with lack of IgA1 protease activity, we assayed supernatants from cultures grown from two IgaStop-containing colonies isolated from each of two infected subjects; all were unable to cleave human IgA1 (Fig. 3).
DISCUSSION
The well-documented association of IgA1 protease production with pathogens that colonize mucosal surfaces has suggested that this proteinase may contribute to the virulence of such bacteria as N. gonorrhoeae, N. meningitidis, and Haemophilus influenzae. Because the role played by IgA in host mucosal defenses is not fully understood, it is difficult to predict how IgA1 cleavage would benefit an IgA1 protease-producing organism. However, recent findings suggest that IgA1 protease and other iga gene products may influence the intracellular fate of gonococci (1, 10, 20, 29). Using the human challenge model, we assessed the role played by IgA1 protease in the early stages of gonococcal infection. The Iga− mutant of N. gonorrhoeae FA1090 caused gonococcal urethritis in male volunteers. In all respects, including the proportion of inoculated volunteers infected, the signs and symptoms of infection, the period of incubation between inoculation and infection, and the numbers of CFU cultured from urine specimens, the outcome of infection with FA1090iga 15b was indistinguishable from the results we previously described for infection of subjects with wild-type strain FA1090 variants A21 or A22 (references 4, 6, 9, and 12 and our unpublished observations). These results indicate that N. gonorrhoeae does not require production of IgA1 protease to successfully colonize the male urethra and cause urethritis. Because the sites of infection and types of infection caused by different IgA1 protease-producing pathogens vary considerably, it is not appropriate to extrapolate from this result with the gonococcus and attempt to draw conclusions about the contribution that IgA1 protease makes to the virulence of organisms other than gonococci.
Practical limitations preclude the study of large numbers of subjects in this model of experimental gonococcal infection. While we can reach broad conclusions about the ability of a particular mutant to cause infection and can to some extent characterize the severity of that infection, we cannot detect slight differences in infectivity that may exist between mutant and wild-type strains. The fact that the gonococcal Iga− mutant can cause urethritis in this model of infection does not mean that the mutant is completely unimpaired in its ability to initiate an infection. However, it is possible to render a gonococcal strain noninfectious in the human challenge model by mutational inactivation of a single gene encoding a critical virulence factor, as Cornelissen et al. (6) recently demonstrated with a transferrin receptor mutant. Additionally, we know that the experimental-infection model is sufficiently sensitive to detect an attenuated phenotype with an intermediate level of infection in which inoculated volunteers have positive urine cultures but do not develop the normal clinical symptoms of gonococcal infection (9). Any impairment of the infectivity of the iga mutant, if such impairment exists, must be relatively subtle.
For ethical reasons, we believe that treatment of human subjects in these experimental trials must be initiated at the onset of signs or symptoms of infection. Our assessment of the events that occur in volunteers is limited to the early stages of the infection process, including colonization and the subsequent inflammatory response, and excludes later times, when an acquired immune response would come into play. It is possible that Iga− bacteria would not exhibit decreased virulence until after the host initiated a specific antibody response to infection. Perhaps IgA1 protease function does not contribute to uncomplicated infection but rather aids in establishing invasive complications such as pelvic inflammatory disease. Alternatively, IgA1 protease may play little or no role in a naive host but may contribute to reinfection of a previously exposed host by cleaving antibodies raised during an earlier infection.
Given the potential for serious complications of gonococcal infection in women, only male subjects can be studied by experimental infection. We can draw no conclusions from our data about the role that IgA1 protease may play in female gonococcal infection. The tissues of the female reproductive system differ significantly from those of the male urogenital tract, and within the female reproductive tract itself there are several different anatomical sites at which the level and types of Ig present can vary. To further complicate matters, both IgA and IgG levels in the fallopian tubes, uterus, and cervix fluctuate over the course of the menstrual cycle (15, 38). Clearly, IgA1 protease may play a role in pathogenesis in females that differs significantly from its role in males. However, in a recent study, Hedges et al. (11) found no evidence for IgA1 protease activity in cervical mucus or vaginal wash samples from women with uncomplicated gonorrhea. As these authors pointed out, their results do not exclude the possibility that IgA1 protease is a more potent virulence factor in female infection when gonococci are located in the subepithelium rather than in the lumen or on the mucosal surface.
For several reasons, IgA1 protease has been viewed as an appealing vaccine candidate for both N. gonorrhoeae and N. meningitidis. It is produced by virtually all strains of gonococci and meningococci, and neutralizing antibodies recognize epitopes shared by the proteases of both species (21, 22). In a recent study of vaccinated Gambian children, Thiesen et al. (37) found that N. meningitidis IgA1 protease was highly immunogenic and that anti-IgA1 protease antibodies in serum persisted or increased over a 5-year period. In the event that mucosal responses to IgA1 protease are also strong and sustained, IgA1 protease may prove to be a useful vaccine component for some types of infection or reinfection. However, our results suggest that targeting IgA1 protease alone would not be sufficient to protect males from uncomplicated infection with N. gonorrhoeae, since urethral infection occurred in the absence of IgA1 protease activity.
ACKNOWLEDGMENTS
We thank Jo Ann Dempsey, Irving Hoffman, Andrea Reed, Luigi Troiani, and the staff of the Verne S. Caviness General Clinical Research Center for assistance with the human challenge experiments; Gloria Thomson for technical assistance; Mike Koomey for generously providing anti-pilin antisera; and Nan Guyer and Jo Ann Dempsey for helpful comments on the manuscript.
This work was supported by Public Health Service grant U01 AI 31496 (J.G.C. and M.S.C.) and training grant T32 AI 07001 (D.M.J.) from the National Institute of Allergy and Infectious Diseases. D.B.J. was supported by an Office of Naval Research predoctoral fellowship.
REFERENCES
- 1.Binscheck T, Bartels F, Bergel H, Bigalke H, Yamasaki S, Hayashi T, Niemann H, Pohlner J. IgA protease from Neisseria gonorrhoeae inhibits exocytosis in bovine chromaffin cells like tetanus toxin. J Biol Chem. 1995;270:1770–1774. doi: 10.1074/jbc.270.4.1770. [DOI] [PubMed] [Google Scholar]
- 2.Blake M, Holmes K K, Swanson J. Studies on gonococcus infection. XVII. IgA1-cleaving protease in vaginal washings from women with gonorrhoea. J Infect Dis. 1979;139:89–92. doi: 10.1093/infdis/139.1.89. [DOI] [PubMed] [Google Scholar]
- 3.Brown T A. Immunity at mucosal surfaces. Adv Dent Res. 1996;10:62–65. doi: 10.1177/08959374960100011201. [DOI] [PubMed] [Google Scholar]
- 4.Cohen M S, Cannon J G, Jerse A E, Charniga L M, Isbey S F, Whicker L G. Human experimentation with Neisseria gonorrhoeae: rationale, methods, and implications for the biology of infection and vaccine development. J Infect Dis. 1994;169:532–537. doi: 10.1093/infdis/169.3.532. [DOI] [PubMed] [Google Scholar]
- 5.Cooper M D, McGee Z A, Mulks M H, Koomey J M, Hindman T L. Attachment to and invasion of human fallopian tube mucosa by an IgA1 protease-deficient mutant of Neisseria gonorrhoeae and its wild-type parent. J Infect Dis. 1984;150:737–744. doi: 10.1093/infdis/150.5.737. [DOI] [PubMed] [Google Scholar]
- 6.Cornelissen C N, Kelley M, Hobbs M M, Anderson J E, Cannon J G, Cohen M S, Sparling P F. The transferrin receptor expressed by gonococcal strain FA1090 is required for experimental infection of human male volunteers. Mol Microbiol. 1998;27:611–616. doi: 10.1046/j.1365-2958.1998.00710.x. [DOI] [PubMed] [Google Scholar]
- 7.Elkins C, Thomas C E, Seifert H S, Sparling P F. Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J Bacteriol. 1991;173:3911–3913. doi: 10.1128/jb.173.12.3911-3913.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halter R, Pohlner J, Meyer T F. Mosaic-like organization of IgA protease genes in Neisseria gonorrhoeae generated by horizontal genetic exchange in vivo. EMBO J. 1989;8:2737–2744. doi: 10.1002/j.1460-2075.1989.tb08415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamrick, T. et al. Unpublished data.
- 10.Hauck C R, Meyer T F. The lysosomal/phagosomal membrane protein h-lamp-1 is a target of the IgA1 protease of Neisseria gonorrhoeae. FEBS Lett. 1997;405:86–90. doi: 10.1016/s0014-5793(97)00163-4. [DOI] [PubMed] [Google Scholar]
- 11.Hedges S R, Mayo M S, Kallman L, Mestecky J, Hook III E W, Russell M W. Evaluation of immunoglobulin A1 (IgA1) protease and IgA1 protease inhibitory activity in human female genital infection with Neisseria gonorrhoeae. Infect Immun. 1998;66:5826–5832. doi: 10.1128/iai.66.12.5826-5832.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jerse A E, Cohen M S, Drown P M, Whicker L G, Isbey S F, Seifert H S, Cannon J G. Multiple gonococcal opacity proteins are expressed during experimental urethral infection in the male. J Exp Med. 1994;179:911–920. doi: 10.1084/jem.179.3.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston, D. M., and J. G. Cannon. Construction of mutant strains of Neisseria gonorrhoeae lacking new antibiotic resistance markers using a two gene cassette with positive and negative selection. Gene, in press. [DOI] [PubMed]
- 14.Kellogg D S, Peacock W L, Deacon W E, Brown L, Pirkle C I. Neisseria gonorrhoeae. 1. Virulence genetically linked to clonal variation. J Bacteriol. 1963;85:1274–1279. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelsall B L, Strober W. Host defenses at mucosal surfaces. In: Rich R R, editor. Clinical immunology principles and practice. St. Louis, Mo: Mosby-Year Book, Inc.; 1996. pp. 299–332. [Google Scholar]
- 16.Kilian M, Reinholdt J, Lomholt H, Poulsen K, Frandsen E V G. Biological significance of IgA1 proteases in bacterial colonization and pathogenesis: critical evaluation of experimental evidence. APMIS. 1996;104:321–338. doi: 10.1111/j.1699-0463.1996.tb00724.x. [DOI] [PubMed] [Google Scholar]
- 17.Klauser T, Pohlner J, Meyer T F. The secretion pathway of IgA protease-type proteins in Gram-negative bacteria. Bioessays. 1993;15:799–805. doi: 10.1002/bies.950151205. [DOI] [PubMed] [Google Scholar]
- 18.Koomey J M, Falkow S. Nucleotide sequence homology between the immunoglobulin A1 protease genes of Neisseria gonorrhoeae, Neisseria meningitidis, and Haemophilus influenzae. Infect Immun. 1984;43:101–107. doi: 10.1128/iai.43.1.101-107.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lin L, Ayala P, Larson J, Mulks M, Fukuda M, Carlsson S R, Enns C, So M. The Neisseria type 2 IgA1 protease cleaves LAMP1 and promotes survival of bacteria within epithelial cells. Mol Microbiol. 1997;24:1083–1094. doi: 10.1046/j.1365-2958.1997.4191776.x. [DOI] [PubMed] [Google Scholar]
- 21.Lomholt H. Molecular biology and vaccine aspects of bacterial immunoglobulin A1 proteases. APMIS Suppl. 1996;62:1–28. doi: 10.1111/j.1600-0463.1996.tb05580.x. [DOI] [PubMed] [Google Scholar]
- 22.Lomholt H, Lind I, Kilian M. Neisseria gonorrhoeae IgA1 proteases share epitopes recognized by neutralizing antibodies. Vaccine. 1995;13:1213–1219. doi: 10.1016/0264-410x(95)00057-8. [DOI] [PubMed] [Google Scholar]
- 23.Mahoney J F, Van Slyke C J, Cutler J C, Blum H L. Experimental gonococci urethritis in human volunteers. Am J Syphilis Gonorrhea Vener Dis. 1946;30:1–39. [PubMed] [Google Scholar]
- 24.Mulks M H, Knapp J S. Immunoglobulin A1 protease types of Neisseria gonorrhoeae and their relationship to auxotype and serovar. Infect Immun. 1987;55:931–936. doi: 10.1128/iai.55.4.931-936.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulks M H, Shoberg R J. Bacterial immunoglobulin A1 proteases. Methods Enzymol. 1994;235:543–554. doi: 10.1016/0076-6879(94)35169-4. [DOI] [PubMed] [Google Scholar]
- 26.Plaut A G, Bachovchin W W. IgA-specific prolyl endopeptidases: serine type. Methods Enzymol. 1994;244:137–151. doi: 10.1016/0076-6879(94)44012-3. [DOI] [PubMed] [Google Scholar]
- 27.Plaut A G, Gilbert J V, Artenstein M S, Capra J D. Neisseria gonorrhoeae and Neisseria meningitidis: extracellular enzyme cleaves human immunoglobulin A. Science. 1975;190:1103–1105. doi: 10.1126/science.810892. [DOI] [PubMed] [Google Scholar]
- 28.Pohlner J, Halter R, Beyreuther K, Meyer T F. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature (London) 1987;325:458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- 29.Pohlner J, Langenberg U, Wolk U, Beck S C, Meyer T F. Uptake and nuclear transport of Neisseria IgA1 protease-associated proteins in human cells. Mol Microbiol. 1995;17:1073–1083. doi: 10.1111/j.1365-2958.1995.mmi_17061073.x. [DOI] [PubMed] [Google Scholar]
- 30.Roe, B. A., S. Clifton, and D. W. Dyer. Gonococcal genome sequencing project. Unpublished data.
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Schneider H, Griffiss J M, Boslego J W, Hitchcock P J, Zahos K M, Apicella M A. Expression of paragloboside-like lipooligosaccharides may be a necessary component of gonococcal pathogenesis in men. J Exp Med. 1991;174:1601–1606. doi: 10.1084/jem.174.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seifert H S, Wright C J, Jerse A E, Cohen M S, Cannon J G. Multiple gonococcal pilin antigenic variants are produced during experimental human infections. J Clin Investig. 1994;93:2744–2749. doi: 10.1172/JCI117290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.St. Geme J W, III, de la Morena M L, Falkow S. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol Microbiol. 1994;14:217–233. doi: 10.1111/j.1365-2958.1994.tb01283.x. [DOI] [PubMed] [Google Scholar]
- 35.Swanson J, Robbins K, Barrera O, Corwin D, Boslego J, Ciak J, Blake M, Koomey J M. Gonococcal pilin variants in experimental gonorrhea. J Exp Med. 1987;165:1344–1357. doi: 10.1084/jem.165.5.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanson J, Barrera O, Sola J, Boslego J. Expression of outer membrane protein II by gonococci in experimental gonorrhea. J Exp Med. 1988;168:2121–2129. doi: 10.1084/jem.168.6.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiesen B, Greenwood B, Brieske N, Achtman M. Persistence of antibodies to meningococcal IgA1 protease versus decay of antibodies to group A polysaccharide and Opc protein. Vaccine. 1997;15:209–219. doi: 10.1016/s0264-410x(96)00138-7. [DOI] [PubMed] [Google Scholar]
- 38.Wira C R, Kaushic C. Mucosal immunity in the female reproductive tract: effect of sex hormones on immune recognition and responses. In: Kiyono H, Ogra P L, McGhee J R, editors. Mucosal vaccines. San Diego, Calif: Academic Press, Inc.; 1996. pp. 375–388. [Google Scholar]