Abstract
Antitumor immune polarization is a key predictor of clinical outcomes to cancer therapy. An emerging concept influencing clinical outcome involves the spatial location of CD8+ T cells, within the tumor. Our earlier work demonstrated immunosuppressive effects of NOS2 and COX2 tumor expression. Here, we show that NOS2/COX2 levels influence both the polarization and spatial location of lymphoid cells including CD8+ T cells. Importantly, elevated tumor NOS2/COX2 correlated with exclusion of CD8+ T cells from the tumor epithelium. In contrast, tumors expressing low NOS2/COX2 had increased CD8+ T cell penetration into the tumor epithelium. Consistent with a causative relationship between these observations, pharmacological inhibition of COX2 with indomethacin dramatically reduced tumor growth of the 4T1 model of TNBC in both WT and Nos2- mice. This regimen led to complete tumor regression in ∼20–25% of tumor-bearing Nos2- mice, and these animals were resistant to tumor rechallenge. Th1 cytokines were elevated in the blood of treated mice and intratumoral CD4+ and CD8+ T cells were higher in mice that received indomethacin when compared to control untreated mice. Multiplex immunofluorescence imaging confirmed our phenotyping results and demonstrated that targeted Nos2/Cox2 blockade improved CD8+ T cell penetration into the 4T1 tumor core. These findings are consistent with our observations in low NOS2/COX2 expressing breast tumors proving that COX2 activity is responsible for limiting the spatial distribution of effector T cells in TNBC. Together these results suggest that clinically available NSAID’s may provide a cost-effective, novel immunotherapeutic approach for treatment of aggressive tumors including triple negative breast cancer.
Abbreviations: ER, Estrogen Receptor; NOS2, Nitric Oxide Synthase 2; COX2, Cyclooxygenase 2; TNBC, Triple Negative Breast Cancer; NSAID, Non Steroidal Anti-inflammatory Drug; SER, Substance Enhancement Ratio; CODEX, Co-detection by indexing
1. Introduction
Breast cancer is the most common tumor globally among women and the leading cause of cancer-related deaths among women in North America [1,2]. It is a heterogeneous disease that can be classified as estrogen receptor positive (ER+) or the more aggressive ER negative (ER-) and triple negative (ER-, PR-, HER2-) subtypes. The upregulation of proinflammatory signaling pathways has been reported in breast cancer, suggesting a regulatory impact on the immune system [3]. In ER- and triple negative breast cancer (TNBC), nitric oxide synthase-2 (NOS2) and cyclooxygenase-2 (COX2) have been described as independent predictors of disease outcome [[4], [5], [6]]. Recently, elevated tumor coexpression of NOS2/COX2 demonstrated strong predictive power associated with poor survival among ER-patients as defined by a hazard ratio (HR) of 21 [7]. In addition, NOS2-derived NO and COX2-derived prostaglandin E2 (PGE2) were shown to promote feed-forward NOS2/COX2 crosstalk where NO induced COX2 and PGE2 induced NOS2, as well as other downstream signaling targets [7]. While NOS2-derived NO is a driver of oncogenesis, chemoresistance, metastasis, and immunosuppression, COX2 supports both NOS2 expression and mediates immune suppression both systemically and within the tumor microenvironment (TME) [[7], [8], [9], [10], [11]]. Studies have shown that NOS inhibition improved treatment efficacy by down regulating IL-10 within the TME [12,13]. Importantly, a recent phase 1/2 clinical trial has demonstrated improved clinical outcome defined by an overall response rate of 45.8% in patients with chemorefractory, locally advanced breast cancer (LABC) and metastatic TNBC that received the pan-NOS inhibitor L-NMMA and aspirin combined with taxane [14]. Importantly, this study also revealed that 27.3% of treated LABC patients achieved pathological complete response at surgery where remodeling of the tumor immune environment was observed in patients responding to this therapy [14]. Given that COX2 promotes immune suppression within the TME, targeting both NOS2/COX2 may further improve outcome and thus provide a novel option in breast cancer treatment by augmented antitumor immune response [9,10].
Recent studies have shown that the localization of CD8+ T cells provides important insight into tumor response and patient survival [15]. Tumors with enhanced CD8+ T cell density and penetration into the tumor epithelium have improved outcomes compared to tumors where CD8+ T cells are absent, sparse, or spatially restricted to the tumor margin or stroma [15]. Tumor expression of NOS2 and COX2 is predictive of ineffective immune response and poor outcomes, however, how their expression impacts the spatial distribution of immune cells remains unclear [4,5,7,8].
Multiplex spatial imaging provides a powerful tool to improve our understanding of how the spatial localization of tumor cellular neighborhoods including the tumor immune microenvironment impacts clinical outcome. Here, we spatially examined TNBC tumors and found that those high in both NOS2 and COX2 showed an immunosuppressed phenotype with reduced infiltrating CD8+ T cells that were restricted to the tumor margin or stroma. Modeling of TNBC in mice showed that the clinically available NSAID indomethacin substantially reduced tumor growth, restored T cell numbers and resulted in their improved spatial distribution within the tumor. This work proves that COX2 activity is responsible for limiting the spatial distribution of effector T cells in TNBC and suggests that NSAIDs may provide a cost-effective, novel immunotherapeutic approach for treatment of aggressive tumors including TNBC.
2. Results
2.1. NOS2Lo/COX2Lo tumors exhibit enhanced T cell infiltration
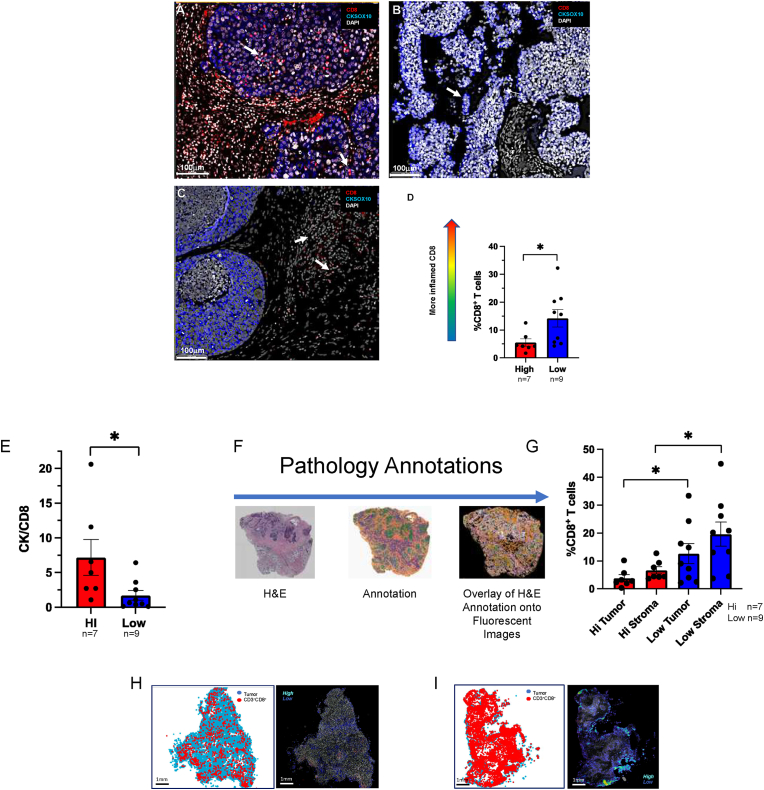
Tumor NOS2/COX2 co-expression predicts poor clinical outcome via promotion of breast cancer disease progression by several mechanisms including altered immune signaling [[7], [8], [9], [10], [11], [12],14]. Herein, we explored the impact of NOS2/COX2 signaling on 1) immune polarization, 2) the spatial location of immune mediators, and 3) tumor response and outcome. Immune status was evaluated by multiplex imaging in 26 ER-breast tumors previously examined for NOS2/COX2 expression [4,5,7]. From these tumors, 16 were selected for spatial analyses based upon TNBC phenotype and NOS2Lo/COX2Lo vs NOS2Hi/COX2Hi tumor expression. Multiplex imaging was performed to evaluate CD8+ T cell infiltration and spatial location associated with NOS2Lo/COX2Lo vs NOS2Hi/COX2Hi tumor expression. Fluorescent antibodies targeting CD3, CD4, CD8, CD68, FOXP3, PDL1, PD1, together with KRT1 (CK-1) and SOX10 (CKSOX10) as tumor markers, were used to spatially define the immune microenvironment in 16 TNBC tumors. Fig. 1A demonstrates enhanced infiltration of CD8+ T cells (red) into the tumor core (CKSOX10, blue) of a NOS2Lo/COX2Lo expressing tumor. In contrast, NOS2Hi/COX2Hi expressing tumors exhibited immune deserts lacking CD8+ T cells (Fig. 1B) or markedly reduced CD8+ T cell infiltration, with stromal restriction as shown in Fig. 1C, which has been associated with poor clinical outcome [15]. Fig. 1B also shows dispersed clusters of tumor cells in a NOS2Hi/COX2Hi tumor (Fig. 1B white arrow), which may be indicative of more invasive tumor phenotypes that support poor clinical outcome as previously described [7]. When quantified, NOS2Hi/COX2Hi tumors had significantly fewer infiltrating CD8+ T cells when compared to NOS2Lo/COX2Lo tumors (Fig. 1D). To further examine the influence of NOS2/COX2 tumor expression on CD8+ T cell infiltration, the ratio of the tumor marker CKSOX10 to the CD8+ T cell marker, which is a more direct regional tumor-to-CD8 comparison, was determined. The CKSOX10/CD8+ T cell ratio provides a metric of both CD8+ T cell infiltration and density where higher numbers represent tumors with reduced CD8+ T cell infiltration associated with immune deserts. As seen in Fig. 1E, NOS2Hi/COX2Hi tumors had significantly higher tumor CKSOX10/CD8+ T cell ratios when compared to NOS2Lo/COX2Lo tumors. These observations support the role of elevated tumor NOS2/COX2 in the regulation of CD8+ T cell density and penetration into the tumor core. To further examine the CD8+ T cell regional distribution, tumor and stroma were spatially annotated on serial H&E sections as summarized in Fig. 1F, and these annotations were overlaid onto fluorescent images to determine the T cell spatial distribution. Using this approach, Fig. 1G shows a marked increase in CD8+ T cell distribution in both the tumor and stroma regions of NOS2Lo/COX2Lo tumors when compared to NOS2Hi/COX2Hi tumors. Fig. 1H shows sparse CD8+ T cells with less aggregation in a NOS2Hi/COX2Hi tumor. In contrast, Fig. 1I shows dramatically increased CD8+ T cell density and aggregation deep in the tumor core in a NOS2Lo/COX2Lo tumor. These results further implicate a regulatory role of elevated NOS2/COX2 tumor expression in abated CD8+ T cell penetration into the tumor core.
Fig. 1.
Multiplex Fluorescence Imaging evaluating altered CD8+ T cell infiltration in TNBC NOS2Lo/COX2Lo vs NOS2Hi/COX2Hi expressing tumors (n = 16). Spatial distribution at 100 μm magnification showing CD8+ T cells (red) relative to the CKSOX10 (blue) tumor marker in A) fully inflamed NOS2Lo/COX2Lo tumor with increased CD8+ T cell penetration (white arrows) into the tumor core. B) immune desert region lacking CD8+ T cells in NOS2Hi/COX2Hi tumor. Dispersed clusters of tumor cells are noted (white arrow), which may be indicative of invasive tumor cells. C) stroma restricted CD8+ T cells (white arrows) in NOS2Hi/COX2Hi tumor. D) Percent CD8+ T cells are significantly elevated in NOS2Lo/COX2Lo (n = 9, blue) vs NOS2Hi/COX2Hi (n = 7, red) expressing tumors. E) Elevated CKSOX10/CD8+ T cell ratio in NOS2Hi/COX2Hi vs NOS2Lo/COX2Lo tumors suggesting reduced CD8+ T cell infiltration into the tumor core of NOS2Hi/COX2Hi when compared to NOS2Lo/COX2Lo tumors. F) Pathology annotations for spatial distribution analysis of CD8+ T cells in NOS2Lo/COX2Lo vs NOS2Hi/COX2Hi tumors. H&E-stained serial sections were annotated for viable tumor (green), necrosis (purple), and stroma (orange) regions; these annotations were overlaid onto fluorescent images to determine T cell spatial distribution. G) Quantification of %CD8+ T cells distributed in tumor vs stroma of NOS2Lo/COX2Lo vs NOS2Hi/COX2Hi expressing tumors. CD8+ T cell spatial distributions (Tumor: blue, CD3+CD8+ cells: red) and density heat maps reflecting T cell aggregation (color coded High (cyan) Low (blue)) in whole tumor images at 1 mm magnification showing H) fewer CD8+ T cells with less aggregation in a NOS2Hi/COX2Hi tumor. In contrast, I) shows densely infiltrated CD8+ T cells with higher aggregation in a fully Inflamed NOS2Lo/COX2Lo tumor. Significance determined by Mann Whitney two-tail test; NOS2Lo/COX2Lo (n = 9), NOS2Hi/COX2Hi (n = 7); *p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.2. Indomethacin engages the immune system for long term tumor-free survival in Nos2- mice
Our earlier work has shown that murine 4T1 tumor cells can express high Nos2 and Cox2 levels and that Nos2/Cox2 blockade limited tumor growth [16,17]. Thus, 4T1 tumor bearing mice are representative of NOS2Hi/COX2Hi breast tumors. Next, the influence of tumor Nos2/Cox2 expression on tumor immune status was explored in immunocompetent 4T1 tumor-bearing BALB/c mice. We utilized indomethacin (INDO), a clinically available NSAID that has been shown to efficiently target tumors expressing COX2 due to the slow rate of release of INDO from the COX2 enzyme when compared to other NSAIDs such as aspirin [18,19]. In addition, INDO also increased expression of the PGE2 consumptive enzyme 15-PGDH (Supplemental Fig. 1), unlike other agents such as celecoxib. These key features make INDO an ideal candidate for studying the impact of COX2/PGE2 on the disease progression of cancer. To avoid potential toxicity associated with the dual NOS inhibitor/NSAID combination, a Nos2- mouse was utilized [20,21].
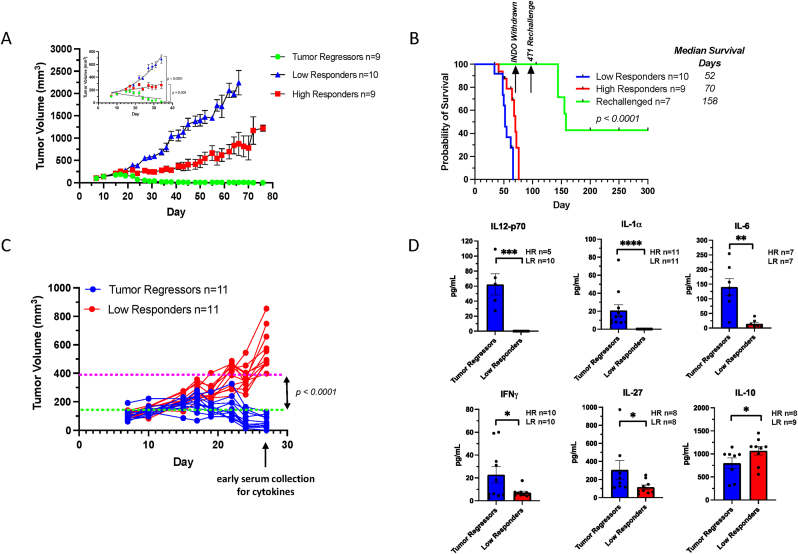
When compared to untreated mice, dramatically reduced 4T1 tumor growth in WT and Nos2- mice treated with INDO was previously observed [17]. Herein, 4T1 tumor-bearing Nos2- mice exhibited a moderate reduction in tumor growth as described by substance enhancement ratio (SER: ratio of slopes of treated mice to untreated mice) of 1.14 in mice reaching tumor volumes of 500 mm3, while INDO treatment of WT mice was more effective at limiting tumor burden (SER 1.86). However, Cox2 inhibition in INDO-treated Nos2- mice showed markedly reduced tumor growth with SER 5.59, where only 30% of mice were able to reach a tumor volume of 1000 mm3 (Supplemental Table 1). The remaining mice continued to show partial as well as complete responses as demonstrated by the survival curve shown in Supplemental Fig. 2 where 2 of 10 Nos2- mice resisted 4T1 rechallenge following INDO withdrawal and thus appeared to be cured. Further exploration of the observed cure revealed that Nos2/Cox2 blockade in INDO-treated Nos2- mice could be categorized into groups of low and high responders to INDO treatment, as well as mice with regressing tumors (Fig. 2A). The animals with regressing tumors were taken off INDO treatment for 30 days and then the mice were rechallenged at 14 weeks with 4T1 tumor cells in the ipsilateral mammary fat pad. The 4T1 rechallenge tumors grew more slowly and three of seven mice remained tumor-free and stayed in remission for five months or longer (Fig. 2B), which supported our earlier work [17]. These results suggest that resistance to tumor rechallenge was mediated by a robust immune response associated with Nos2/Cox2 blockade.
Fig. 2.
Orthotopic 4T1 tumor-bearing Nos2- mice treated with INDO. Tumor growth curves for INDO-treated Nos2- mice showing A) mice with Tumor Regression (n = 9 green), High Responders (n = 9 red), and Low Responders (n = 10 blue). Panel A inset shows non-linear regression analysis demonstrating significant growth differences between Tumor Regressors and Low Responders (p < 0.0001) as well as High and Low Responders (p = 0.005). B) Survival analysis of Rechallenged (n = 7 green), High Responders (n = 9 red), and Low Responders (n = 10 blue). INDO was withdrawn after 10 weeks of treatment, and the mice with regressing tumors were rechallenged with 4T1 tumor cells at 14 weeks. Three mice resisted tumor rechallenge and remained tumor-free for five months or longer before being euthanized. C) For cytokine analysis, blood serum was collected from Tumor Regressor and Low Responder mice on day 27, which demonstrated clear separation in tumor growth. D) Serum cytokine analyses show significantly elevated IL-12p70, IL-1α, IL-6, and IFNγ reflective of proinflammatory, antitumor immune responses in the Tumor Regressors, while immunosuppressive IL-10 is significantly elevated in Low Responders. *p ≤ 0.05, **p ≤ 0.001, ***p = 0.0003, ****p ≤ 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To explore altered immune response in low vs high responders to Nos2/Cox2 blockade, serum cytokine analysis was performed on serum previously collected at day 27, consistent with an approximate time point when separation in tumor volume between low responding and high responding mice began as shown in Fig. 2C. Serum cytokine analysis revealed considerably higher levels of proinflammatory antitumor Th1 cytokines including IL-12, IL-1α, IL-6, and IFNγ implicating a more robust systemic immune response and restored immune surveillance in high responders to INDO treatment (Fig. 2D). Moreover, responding mice demonstrated increased IL-27 (Fig. 2D), which suppresses FOXP3+ Treg populations, thus providing additional supportive evidence that the high responders exhibit a more proinflammatory, antitumor immune microenvironment by limiting immunosuppressive Th2 immune polarization. In contrast, low responding mice produced low levels of IL-27 and proinflammatory cytokines, as well as higher levels of circulating immune suppressive Th2 cytokine IL10 (Fig. 2D). These results support a role of enhanced proinflammatory antitumor immune response associated with improved survival in Tumor Regressors when compared to Low Responders (Fig. 2B).
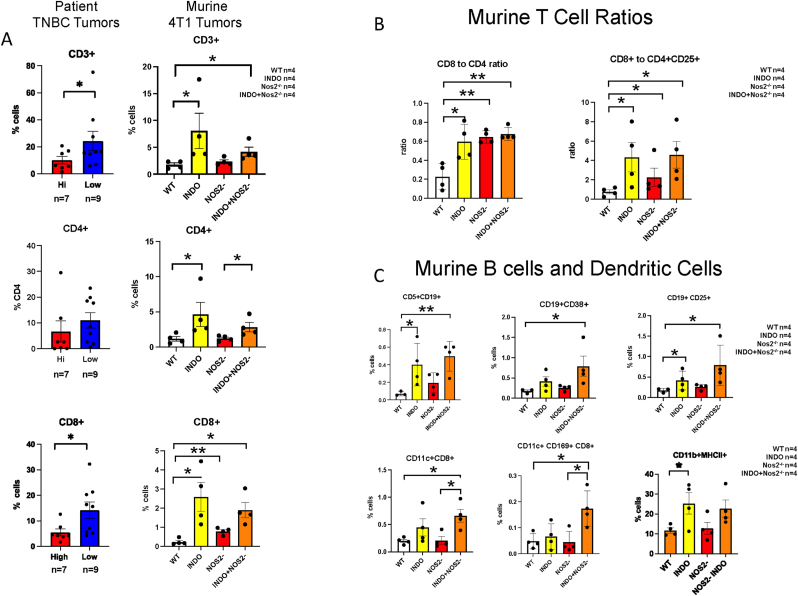
Nos2/Cox2 Blockade Alters Immune Polarization Within the TME. To further explore how Nos2/Cox2 inhibition alters the tumor immune microenvironment, flow cytometry (FACS), RNAseq gene expression, and multiplex imaging (CODEX) analyses were performed on 4T1 tumors. FACS and gene expression analyses revealed significant increases in CD45+ immune cells and CD11b+ and F4/80+ myeloid cells in the INDO-treated tumors (Supplemental Fig. 3). While CD4+ T cells trended higher, multiplex imaging demonstrated significantly increased CD3+ and CD8+ T cell expression in NOS2Lo/COX2Lo when compared to NOS2Hi/COX2Hi TNBC tumors (Fig. 3A, Fig. 1). In support of these observations, CODEX multiplex imaging analyses also revealed elevated CD3+, CD4+, and CD8+ T cells in WT+INDO and Nos2- + INDO treated mice (Fig. 3A). In addition, CD8+ T cells were also elevated in Nos2-mice when compared to WT controls (Fig. 3A). Significantly elevated CD8+ to CD4+ T cell and CD8+ T cell to CD4+CD25+ Treg ratios were observed, implicating increased proinflammatory phenotypes (Fig. 3B). Also, enhanced CD11c+CD8+ and CD11c+CD8+CD169+ phenotypes implicate increased cross-priming and expansion of cytolytic T cells while elevated CD11b+MHCII+ phenotypes implicated increased antigen presenting cells (Fig. 3C). Together, these results further suggest an augmented proinflammatory immune landscape in response to INDO treatment of WT and Nos2-mice.
Fig. 3.
Multiplex imaging analyses showing altered lymphoid populations in patient TNBC and murine 4T1 tumors. A) Ultivue (patient TNBC tumors) and CODEX (murine 4T1 tumors) multiplex image analyses. Increased CD3+ and CD8+ T cells were observed in NOS2Lo/COX2Lo (n = 9) vs NOS2Hi/COX2Hi (n = 7) TNBC tumors, while CD4+ T cells trended higher in NOS2Lo/COX2Lo tumors. Increased CD3+, CD4+, and CD8+ T cells were observed in INDO-treated WT and Nos2- 4T1 tumors (n = 4 per group). Elevated CD8+ T cells were also observed in Nos2- when compared to WT tumors. B) comparison of CD8+ to CD4+ and CD8+ to CD4+CD25+ cell ratios in 4T1 tumors from WT and Nos2- mice treated ± INDO indicating augmented antitumor and reduced immunosuppressive T cell phenotypes. C) Increased CD19+ B cell, CD11c+ dendritic cell, and CD11b+ MHCII+ proinflammatory macrophage activated phenotypes were observed in 4T1 tumors from INDO-treated mice (*p < 0.05, **p < 0.01).
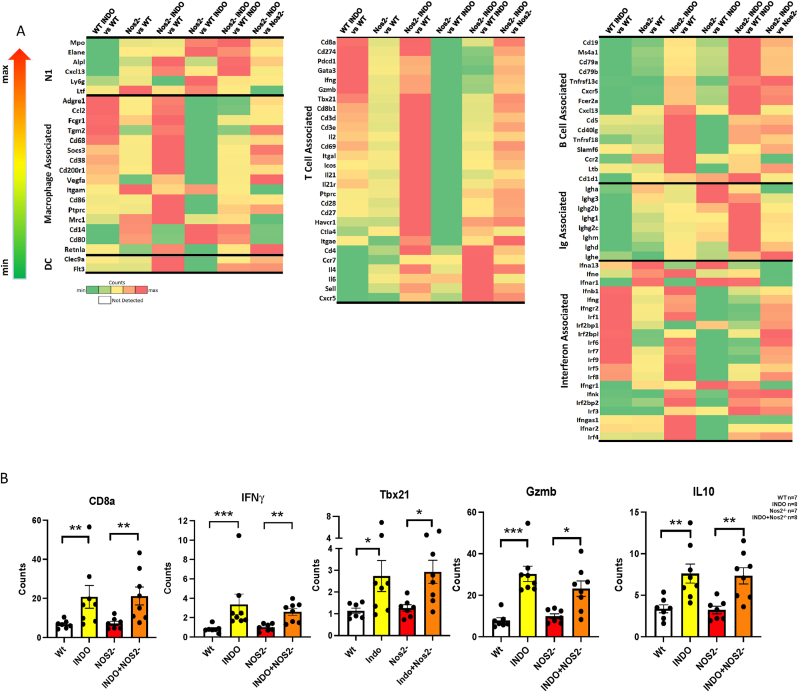
Additional supportive evidence of a proinflammatory, antitumor immune landscape is provided by RNAseq gene expression showing elevated Th1 genes in INDO-treated mice (Fig. 4A), including increased IFNγ, Gzmb, and Tbx21 as well as reduced IL10 expression, implicating augmented cytolytic CD8+ T cell function (Fig. 4AB). However, higher CD19+ and CD20+ (Ms4a1) B cell and Ig-associated gene expressions indicative of B cell activation (i.e., CD79a, CD79b, Tnfrsf13c, IgM, IgD) was observed in Nos2- mice (Fig. 4A). B cell gene expression profiles and CODEX analysis showed increased CD79 B-cell receptor as well as increased Ig-associated gene expression and elevated CD19+CD38+ and CD19+CD25+ cells implicating mature, antibody producing B cell phenotypes (Fig. 3, Fig. 4A), while the Bregs marker CD1d1 was generally reduced (Fig. 4A) [22,23]. Nos2/Cox2 blockade also promoted increased proinflammatory N1 neutrophil biomarkers including myeloperoxidase (Mpo), neutrophil elastase (Elane) and lactoferrin (Ltf) expression (Fig. 4A). Interestingly, when compared to WT, WT + INDO suppresses B cell and N1 neutrophil markers (Fig. 4A). Together, these results show that Nos2/Cox2 blockade promotes a proinflammatory, antitumor immune microenvironment involving increased N1/M1/Th1 antitumor immune phenotypes.
Fig. 4.
Gene expression heatmaps show altered innate and adaptive biomarker expression in 4T1 tumors from Control and INDO-treated WT and Nos2- mice. A) 4T1 tumors from WT and Nos2-mice treated with INDO showed increased DC, T cell, Ifn, and macrophage-associated gene signatures. Tumors from Nos2-mice showed increased N1 neutrophils, B cell activation biomarkers, and Ig-associated gene signatures. B) Gene expression quantification showing significantly increased CD8a, Ifnγ, Tbx21, Gzmb, and reduced IL10 in INDO-treated mice, which implicate increased cytolytic CD8+ T cell function. Heatmaps and their color scale were generated per gene across different comparison pairs using the fold change values obtained from the differential gene detection algorithm, gene-specific analysis (GSA) in Partek Flow with default settings. Max and Min on the Counts color scale bar are used to indicate the direction of fold change. WT (n = 7, gray), INDO (n = 8, yellow), Nos2- (n = 7, red), INDO + Nos2- (n = 8, orange).
*p ≤ 0.01, **p ≤ 0.001, ***p ≤ 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Identification of Spatially Distinct Immune Phenotypes. Next, CODEX images were analyzed to assess the effects of Nos2/Cox2 blockade on the spatial distributions of immune cells in the 4T1 TNBC model. When compared to the untreated WT control (Fig. 5A), CODEX imaging revealed increased density and penetration of CD8+ T cell populations into the tumor core in the Nos2- and INDO-treated tumors (Fig. 5B,C,D), which was consistent with the observations in NOS2Lo/COX2Lo vs NOS2Hi/COX2Hi TNBC tumors shown in Fig. 1. When compared to the untreated WT and Nos2- tumors (Fig. 5 E,G), increased CD8+ T cell aggregation was observed in the INDO-treated WT and Nos2- tumors, which is shown in the spatial heatmaps in Fig. 5F,H (arrows). Cytolytic T cells secrete IFNγ and granzyme B to facilitate tumor killing. To further characterize the CD8+ T cell phenotype, RNAscope analyses was performed. When compared to untreated WT Control (Fig. 5I,M) enhanced Ifnγ and Gzmb was observed in Nos2- Control and INDO-treated tumors, while immunosuppressive IL10 expression did not change (Fig. 5 panels J,K,L,N,O,P, and Q). Moreover, the elevated Ifnγ and Gzmb was spatially oriented in the tumor core where the CD8+ T cells were localized (Fig. 5J,K,L,N,O,P). Also, spatial localization in the whole tumor revealed an interesting feature where IL10 was mainly expressed on the tumor margins in all samples (Fig. 5I and J,K,L). In addition to increased CD8+ T cells, elevated immune populations including CD4+ T cells, CD19+ B cells and antigen presenting CD11c+ DC were also observed in the Nos2- and INDO-treated tumors as shown in Supplemental Fig. 4. When compared to CD8+ T cell spatial orientation, CD4+ T cells and CD19+ B cells were observed at or near the tumor margins. The spatial orientation of CD8+ T cells along with Ifnγ, Gzmb, and IL10 expression provides additional evidence that Nos2/Cox2 inhibition leads to enhanced cytolytic T cell function and DC occurring in the tumor core that were surrounded by CD4+ T cells and B cells, which may be more conducive for antigen presentation and the development of adaptive and humoral immunity (Fig. 5, Supplemental Fig. 4). Taken together these results show that Nos2/Cox2 blockade promotes a robust immune response in 4T1 tumors that is similar to the immune microenvironment observed in NOS2Lo/COX2Lo TNBC tumors (Fig. 1, Fig. 2).
Fig. 5.
Nos2/Cox2 blockade alters lymphoid spatial localization and gene expression. CD8+ T cell (red) spatial orientation in A) 4T1 tumors from untreated WT control mice showing fewer CD8+ T cells. In contrast, panels B) WT + INDO, C) Nos2-, and D) Nos2- + INDO demonstrated markedly increased CD8+ T cell density and penetration into the 4T1 tumor core. E-H) Spatial Heatmap analyses demonstrate enhanced CD8+ T cell aggregation (white arrows) in 4T1 tumors from INDO-treated mice. RNAscope analyses across serial tissue sections shown in panels I–L (1 mm magnification) and M − P (50 μm magnification) show increased IFNγ (yellow) and Gzmb (magenta) expression in Nos2-and INDO-treated mice indicating augmented cytolytic T cells function. In contrast, IL10 (green) expression associated with immunosuppression did not change. Q) Quantification showing significantly elevated IFNγ and Gzmb in INDO-treated WT and Nos2- tumors, with no change in IL10 expression. **p ≤ 0.001, ***p ≤ 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
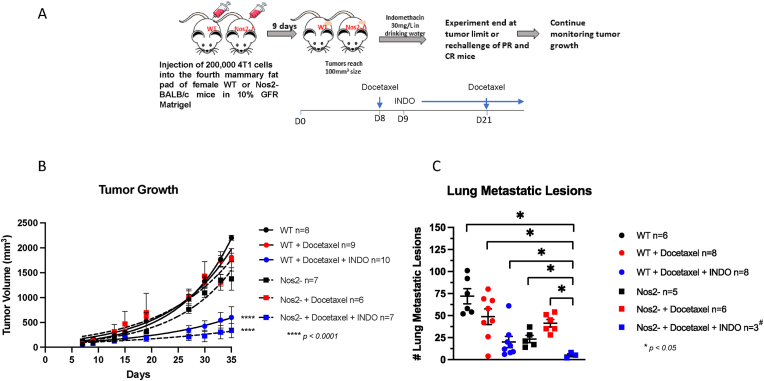
Effect of NOS2/COX2 Inhibition on Taxol Therapeutic Efficacy. Given that NOS2/COX2 inhibition promotes a robust and spatially distinct antitumor immune response, its impact on therapeutic efficacy was examined. Docetaxel is a commonly used chemotherapeutic that often results in the development of drug-resistance [24]. An earlier report demonstrated that neoadjuvant administration of a pan-NOS inhibitor (L-NMMA) with docetaxel improved therapeutic response in patients who were previously non-responsive to the taxane treatment [25]. To better understand the effects of dual inhibition in combination with docetaxel, an in vivo study was performed in the 4T1 model as summarized in Fig. 6A. While docetaxel administered in two doses provided little effect on primary 4T1 tumor growth in WT mice, treatment with docetaxel + INDO significantly improved the response (Fig. 6B). Similarly, the same dosage administered to 4T1 tumor-bearing Nos2- mice demonstrated significantly improved response by docetaxel + INDO treatment (Fig. 6B). Importantly, docetaxel + INDO treated Nos2- mice exhibited significantly reduced lung metastatic burden (Fig. 6C) as well as reductions in severe side effects and co-morbidities observed with docetaxel + INDO treatment in WT mice. Moreover, while two mice succumbed to disease and lung tissues could not be harvested for metastatic analysis, two of the seven mice (28.5%) lived tumor-free and healthy for nearly one year (343 days) after initiation of treatment. These results further demonstrate the importance of Nos2/Cox2 blockade for improved response to docetaxel (Fig. 6), which is consistent with a recently reported clinical study [14].
Fig. 6.
Nos2/Cox2 blockade improves docetaxel therapeutic efficacy. Treatment with docetaxel study design is summarized in panel A. Briefly, 200,000 4T1 tumor cells were injected and allowed to grow. Docetaxel was given in food on days 8 and 21, and INDO administration in drinking was initiated on day 9 and continued for the duration of the experiment. Panels B and C show reduced tumor growth and reduced lung metastatic burden, respectively in WT and Nos2-treated with docetaxel and INDO. #While seven Nos2-mice were treated with INDO as shown in panel B, two died, and tissues could not be harvested. Three of the remaining mice were euthanized and lungs harvested, and the other two mice were tumor-free and taken off INDO. These mice remained healthy and tumor-free for over 5 months. *p < 0.05, ****p < 0.0001.
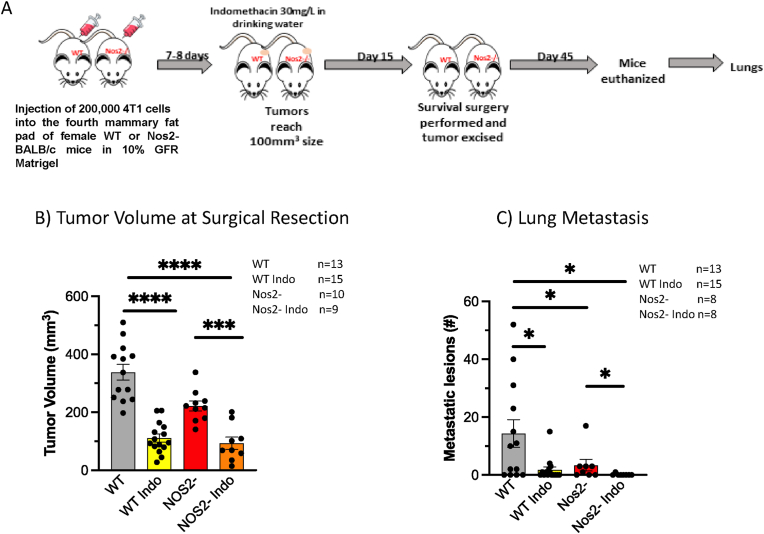
Metastasis is a key determinant of clinical outcome in cancer. Our earlier work showed that indomethacin played an important role in controlling growth of the primary tumor but had only a modest effect on lung metastatic burden. In contrast, dual Nos2/Cox2 blockade was able to significantly reduce metastatic lesions in 4T1 tumor-bearing mice [17]. To explore the role of Nos2/Cox2 in metastatic development, INDO administration was initiated when 4T1 tumors reached a size of 100 mm3 and then tumor resection/survival surgery was performed 15 days later. The mice were maintained for a total of 45 days and then euthanized, and lungs were harvested for evaluation of metastatic burden as described in the experimental design summarized in Fig. 7A. While tumor volumes at harvest were significantly decreased in treated mice (Fig. 7B), untreated WT mice exhibited significantly greater metastatic lesions when compared to INDO-treated and Nos2- mice (Fig. 7C, Table 1). Importantly, only one detectable metastatic lesion was observed among eight INDO-treated Nos2- mice, which was less than 1% of the lesions observed in untreated WT mice (Fig. 7C, Table 1). These results suggest that neoadjuvant Nos2/Cox2 blockade may provide a beneficial option for TNBC patients undergoing surgical resection.
Fig. 7.
Nos2/Cox2 blockade improves surgical therapeutic efficacy. A schematic display of the experimental design of survival surgery summarized in panel A. Briefly, 200,000 4T1 tumor cells were injected and allowed to grow to 100 mm3 tumor volumes when INDO administration in drinking water began. Fifteen days later tumor resection/survival surgery was performed, and the mice were maintained on INDO-drinking water for 45 days. The mice were then euthanized, and lungs removed and inflated in Bouin’s solution. B) Nos2/Cox2 inhibition reduces tumor volume at surgical resection. C) Nos2/Cox2 inhibition reduces lung metastatic burden following surgical resection. *p < 0.05, ***p = 0.0001, ****p < 0.0001.
Table 1.
Summary of lung metastatic lesions in all mice after survival surgery. The total number of metastatic lesions in all mice per group, as well as the number of mice per group, and the metastatic index is shown. The Metastatic Index is defined as #Mets/#Mice. * p values were determined using Welch’s correction test.
| Total #Mets | #Mice | Metastatic Index | P value$ | |
|---|---|---|---|---|
| WT | 186 | 13 | 14.31 | |
| Nos2- | 27 | 8 | 3.38 | 0.026* |
| WT INDO | 27 | 15 | 1.80 | 0.024* |
| Nos2- INDO | 1 | 8 | 0.13 | 0.012* |
$Welch’s Correction Test.
*Compared to WT.
3. Discussion
The coinhibitory immune ligand programmed cell death ligand 1 (PD-L1; CD274) is elevated in TNBC. However, when compared to other cancers, these advanced tumors are less responsive to checkpoint inhibitors that target these ligands [26]. In addition, advanced TNBC patients present with increased drug resistance due in part to the presence of immune suppressive cell types, making this disease more difficult to treat [27]. Indeed, poor clinical outcome is, in part, due to elevated immunosuppressive tumor microenvironments. Herein, we show that elevated tumor Nos2/Cox2 expression creates an immunosuppressive tumor microenvironment, as blockade of these pathways promotes proinflammatory immune polarization. Tumor NOS2 and/or COX2 overexpression occurs in numerous aggressive tumors including, lung, glioma, liver, pancreatic, cervical, gastric, esophageal, and ER-breast cancer. In ER-breast cancer patients, elevated NOS2/COX2 tumor expression is a strong predictor of poor survival [4,5,7]. Interestingly, p53 negatively regulates NOS2 and COX2 expression. Mutations in p53 are often associated with poor clinical outcome of aggressive tumors, which may in part lead to the elevated and chronic expression of NOS2 and COX2 [28,29]. Moreover, tumor NOS2 expression correlates with p53 mutation and predicts poor survival in ER-breast tumors [4]. Herein, we show that elevated NOS2/COX2 signaling directly impacts both the immune response and spatial orientation of CD8+ T cells in TNBC. Importantly, Nos2/Cox2 blockade markedly improved the immune profile and tumor infiltration of cytolytic CD8+ T cells, resulting in reduced tumor burden in 4T1 tumor-bearing mice. These results suggest that NOS2/COX2 overexpression promotes an immunosuppressive tumor immune microenvironment.
Within the TME, NOS2-derived NO and COX2-derived PGE2 can impact different aspects of the local immune response by tuning the area to favor an immunosuppressive microenvironment [10,30,31]. Pro-oncogenic NO levels (100–300 nM) influence T cell function through the induction of apoptosis of CD4+/CD8+ Th1 phenotypes while favoring CD4+/CD8+ Tregs that produce the immunosuppressive cytokine IL-10 [12,[32], [33], [34]]. In addition, these levels of NO activate TGFβ as well as IL-10, which prevents M1 macrophage development [35]. Also, NO can inhibit immune infiltration of neutrophils, monocytes, and lymphoid cells through regulation of vascular adhesion molecules, such as ICAM, VCAM and MCP-1 thereby creating leukocyte deserts [36]. Herein, the development of the immune architecture shows several particularly strong changes in the immune profile in response to Nos2/Cox2 blockade including increased T cells, B cells, N1 neutrophils and dendritic cells (Fig. 4). COX2 inhibition in WT mice clearly increased T cells populations with increased Ifng, Ifngr, Irf1, and Irf7 expression regardless of NOS2 status (Fig. 4). However, INDO-treatment of WT mice suppressed N1 neutrophils and B cells (Fig. 4A). In contrast, INDO-treated Nos2- mice led to increased N1 neutrophils and B cell infiltration (Fig. 4A), which suggests that Nos2/Cox2 blockade offers complementary antitumor immune regulatory components. According to the canonical pathway analysis summarized in Supplemental Fig. 5, Cox2 inhibition reprogrammed the downstream signaling cascade, which involved augmented IFNγ signaling and the activation of several T cell-related pathways including T Cell Receptor Signaling, Natural Killer Cell Signaling, ICOS-ICOSL Signaling in T Helper Cells, and Crosstalk between Dendritic Cells and Natural Killer Cells. In contrast, INDO treatment of Nos2- tumor-bearing mice increased B cells and N1 neutrophils (Fig. 4). Together, Nos2/Cox2 blockade augmented T and B cells as well as N1 neutrophils, which restored immune surveillance and dramatically improved the antitumor response.
When expressed by the tumor, PGE2 has a multidimensional impact on the overall tone of the immune microenvironment by favoring immunosuppression and anti-inflammatory mechanisms [10]. Herein, we show that COX inhibition by indomethacin promotes proinflammatory/antitumor Th1 phenotypes of CD4+ and CD8+ T cells, which reduced tumor growth and metastatic burden and improved survival of 4T1 tumor-bearing mice. Like NO, PGE2 increases IL10 and TGFβ Th2 cytokines, which block DC maturation and antigen presentation [9,10]. In general, PGE2 suppresses Th1, cytolytic T cells and NK cells through cAMP-dependent mechanisms while attracting Treg and MDSC immunosuppressive phenotypes [9,10]. Indeed, a key mechanism of immune suppression by PGE2 is its cAMP-dependent effect on T cells [10]. Early studies have shown that PGE2 promotes T cell anergy and limited T cell responsiveness [37]. Importantly, feedforward tumor NOS2/COX2 regulation augments NO/PGE2 levels in a synergistic manner leading to a chronically immunosuppressed tumor immune microenvironment [7].
Mature neutrophils play a key role in tumor eradication through increased MPO and reactive oxygen species (ROS) generation. Herein, we show that 4T1 tumors grown on a Nos2- background in the presence of INDO increased the proportion of mature neutrophils in the tumor. In contrast, the higher proportion of immature neutrophils in the WT control, due to excessive GMC-SF production by the tumor promotes immunosuppression as well as setting the metastatic niche in the lung [38]. While NO will drive many of the oncogenic pathways for EMT and migration, reduced pulmonary Ly6G was identified, which is important in seeding the metastatic niches in the lung [39]. Moreover, primary tumors from Nos2- + INDO-treated mice exhibited elevated expression of neutrophil activation markers implicating limited immunosuppression and the promotion of antitumor neutrophil phenotypes by the dual treatment (Fig. 5). This suggests a role for elevated ROS, which is known to be involved in tumor eradication [40]. In contrast, Nos2-derived NO scavenges ROS directly, thus serving as an antioxidant, which protects the tumor [41]. Importantly, elevated neutrophil infiltration identified in patients who responded to NOS inhibition therapy suggests increased proportions of mature neutrophils is a key antitumor mechanism associated with NOS2 blockade in both mice and humans [14].
Another important feature of NOS2- + INDO combination therapy is increased B cell infiltration. The RNAseq analysis identified the modulation of several B cell-associated genes. The RNAseq observations were further supported by CODEX analyses, which demonstrated an increase in B cell and APC density (Fig. 4, Supplemental Fig. 4) as well as reduced tumor growth and metastasis indicating a potentiated antitumor response. Importantly, these results are consistent with a recent clinical study, where chemo-resistant metastatic breast cancer patients who responded to docetaxel and adjuvant NOS inhibition exhibited markedly increased B cell penetration into the tumor [14]. Taken together, NOS2/COX2 blockade polarizes the tumor immune microenvironment favoring a potent proinflammatory, antitumor immune response.
The spatial localization of proinflammatory immune phenotypes is also influenced by NOS2/COX2 blockade, which led to dramatically increased cytolytic CD8+ T cell penetration deep into the tumor core. A layered effect was observed, reminiscent of an “Inside-out lymph node” consisting of cytolytic CD8+ T cells surrounded by separate layers of CD4+ T cells and B cells (Fig. 5, Supplemental Fig. 4). Recently, tertiary lymphoid structures (TLS) have been detected in tumors, which correlate with improved survival [42,43]. This configuration suggests an orthogonality in the lymphoid population that is like lymph node structures. Furthermore, the spatial orientation of CD25+ T cells are on the margins and not in the cytolytic core, suggesting that NOS2/COX2 inhibition not only changes the immune tone but also the spatial arrangement of immune mediators. Importantly, this novel spatial orientation provides an opportunity for the T cells, CD11c+ DC, and B cells to cross-train each other leading to a more positive therapeutic response [44]. A study describing an in situ vaccine utilizing an IL12 agent combined with radiation demonstrated that T cell-based antigen recognition along with humoral response through cross education was important for a stable resistance [45]. In addition, elevated neutrophil density was observed in the necrotic cores of treated tumors. The spatial localization of antitumor neutrophils and cytolytic CD8+ T cells in the tumor core suggests a cooperative mechanism of these immune mediators [46]. Given that neutrophils home to and lead T cells into necrotic areas, these results suggest a layering sequence of immune processing and activation [47]. Together, the observed lymphoid layering may provide a unique and important spatial orientation associated with NOS2/COX2 targeted therapies that maximizes antitumor crosstalk between immune cells.
3.1. Limitations of the study
The mouse model used in this study is Nos2 deficient, but the tumor is not. The Nos2-mouse represents systemic/host effects of abated Nos2-derived NO signaling while tumor Nos2 expression remains intact. Nonetheless, the current study clearly shows that targeting systemic Nos2 is an important component in the success of potential clinical outcomes, which is consistent with results of a recent clinical trial where remodeling of the tumor immune microenvironment was observed in 27.3% of locally advanced BC patients treated with the pan-NOS inhibitor L-NMMA, aspirin, and taxane who achieved pathological complete response at surgery [14]. Our previous work has demonstrated that NOS2-derived NO promotes tumor invasion and metastasis [4] as well as the perpetuation of NOS2/COX2 expression by a NOS2 and COX2 feedforward loop, where NOS2-derived NO induced COX2 and COX2-derived PGE2 induced NOS2 [7]. Further exploration of these mechanisms will require new tools, which we are currently developing (i.e., 4T1 and MB231 NOS2 knockouts) to tease out the impact of host versus tumor interactions with respect to different tumor niches in the primary lesion and metastatic tissue beds. A future goal is to explore different clinically available drug combinations.
In addition, while INDO clearly augments a proinflammatory tumor immune microenvironment, it may also impart COX independent effects. COX-independent mechanisms of NSAIDs including indomethacin have been reported [48]. To explore COX-independent mechanisms our RNAseq gene expression data was examined for NSAID-induced proteins associated with COX-independent effects where we found modest alterations in PDE5a, PPARd, RXRa, and SP1 (Supplemental Table III).
Also, the Nos2 deficient Cox inhibited model is not investigated in a model of NOS2Lo/COX2Lo tumors, which is relevant from a clinical perspective. Nonetheless, indomethacin is a powerful inhibitor of PGE2 generation while increasing expression of the PGE2 consumptive enzyme 15-PGDH (Supplemental Fig. 1). Together, these characteristics would be consistent with a COX2Lo tumor that would generate reduced levels of PGE2 and should also be considered therapeutically. This is consistent with Supplemental Fig. 1 demonstrating significant elevations in 15-PDGH in Nos2- + INDO-treated mice.
4. Conclusions
In summary, the therapeutic importance of a proinflammatory tumor microenvironment indicates a need for therapeutic interventions that modulate tumor inflammation. The current study implicates tumor NOS2/COX2 expression as key immunosuppressive effectors that could limit the efficacy of immune therapies. Importantly, the clinical availability of NOS2/COX2 targeting agents may provide a novel inexpensive opportunity for therapeutically unresponsive tumors [14].
5. Materials and methods
5.1. Tissue collection and immunohistochemical analysis of patient tumor sections: Ultivue®
Tumor specimens were obtained from breast cancer patients recruited at the University of Maryland (UMD) Medical Center, the Baltimore Veterans Affairs Medical Center, Union Memorial Hospital, Mercy Medical Center, and the Sinai Hospital in Baltimore between 1993 and 2003. Informed consent was obtained from all patients. The collection of tumor specimens, survey data, and clinical and pathological information (UMD protocol no. 0298229) was reviewed and approved by the UMD Institutional Review Board (IRB) for the participating institutions. The research was also reviewed and approved by the NIH Office of Human Subjects Research (OHSR no. 2248). Breast tumor NOS2/COX2 expression was analyzed previously by IHC using 1:250 diluted NOS2 antibody and 1:50 diluted COX2 antibody (no. 610328 and 610204, respectively, BD Biosciences, San Diego, CA) and scored by a pathologist [4,5]. For NOS2 staining, a combination score of intensity and distribution were used to categorize the immunohistochemical NOS2 stains where intensity received a score of 0–3 if the staining was negative, weak, moderate, or strong. The NOS2 distribution received scores of 0–4 for distributions <10%, 10–30%, >30–50%, >50–80% and >80% positive cells [4]. For COX2 staining, scores of negative to weak [1,2] or moderate to strong [3,4] were categorized as low or high, respectively [5]. Herein, NOS2 and COX2 expressions were also analyzed by fluorescent staining performed on the Leica Biosystems Bond RX autostainer using the Bond Polymer Refine Kit (Leica Biosystems DS9800), with omission of the PostPrimary reagent, DAB and Hematoxylin. After antigen retrieval with EDTA (Bond Epitope Retrieval 2), sections were incubated for 30 min with COX2 (Cell Signaling Technology no. 12282, 1:100), followed by the Polymer reagent and OPAL Fluorophore 520 (AKOYA). The COX2 antibody complex was stripped by heating with Bond Epitope Retrieval 2. Sections were then incubated for 30 min with NOS2 antibody (Abcam no. ab15323, 1:50), followed by the Polymer reagent and OPAL Fluorophore 690. The NOS2 antibody complex was stripped by heating with Bond Epitope Retrieval 2 and then stained with CD8 (Abcam no. 101500, 1:100. Sections were stained with DAPI and coverslipped with Prolong Gold AntiFade Reagent (Invitrogen). Images were captured using the Aperio Scanscope FL whole slide scanner. The original IHC previously reported [4,5] and fluorescent NOS2/COX2 staining results were generally consistent.
Formalin-fixed paraffin embedded (FFPE) tissue sectioned at 4 μm and mounted on SuperFrost Plus slides were stained with a FixVUE Immuno-8™ Kit (formerly referred to as UltiMapper® kits, Ultivue Inc., Cambridge, MA, USA; CD8, PD-1, PD-L1, CD68, CD3, CD8, FoxP3, and pan-CK/SOX10 cocktail) using the antibody conjugated DNA-barcoded multiplexed immunofluorescence (mIF) method [1]. These kits include the required buffers and reagents to run the assays: antibody diluent, pre-amplification mix, amplification enzyme and buffer, fluorescent probes and corresponding buffer, and nuclear counterstain reagent. Hematoxylin and Eosin (H&E) and mIF staining was performed using the Leica Biosystems BOND RX autostainer. Before performing the mIF staining, FFPE tissue sections were baked vertically at 60–65 °C for 30 min to remove excess paraffin prior to loading on the BOND RX. The BOND RX was used to stain the slides with the recommended FixVUE (UltiMapper) protocol. During assay setup, the reagents from the kit were prepared and loaded onto the autostainer in Leica Titration containers. Solutions for epitope retrieval (ER2, Leica Biosystems cat# AR9640), BOND Wash (Leica Biosystems cat# AR9590), along with all other BOND RX bulk reagents were purchased from Leica). During this assay, the sample was first incubated with a mixture of all 8 antibody conjugates, next the DNA barcodes of each target were simultaneously amplified to improve the sensitivity of the assay. Fluorescent probes conjugated with complementary DNA barcodes were then added to the sample to bind and label the first round of 4 targets; a Round 1 fluorescent image was then acquired. Next, a gentle signal removal step was used to remove the fluorescent probes of the first set of markers before adding the fluorescent probes specific for the second set of 4 markers before imaging the slide a second time to acquire the Round 2 fluorescent image. There was no need for quenching, bleaching or other means to minimize signal between rounds. Before each round of imaging, the stained slides were mounted in Prolong Gold Anti-Fade mountant (Thermo Fisher Scientific, cat# P36965 and coverslipped (Fisherbrand Cover Glass 22 × 40 mm, #1.5). Digital immunofluorescence images were scanned at 20 × magnification. Round 1 and 2 images were co-registered and stacked with Ultivue UltiStacker software. The Immuno8 images used the following marker/fluorophore combinations: FITC (CD8 Round 1 (R1), CD3 Round 2(R2)), TRITC (PD-1 R1,CD4 R2), Cy5 (PD-L1 R1, FoxP3 R2), Cy7 (CD68 R1, panCK/Sox10 R2) and the custom kit used the following combinations: FITC (Alpha SMA R1, CD68/CD163 R2), TRITC (Arg-1 R1, CD15 R2), Cy5 (iNOS R1, IDO1 R2), Cy7 (CD20 R1, CD14 R2). The digital images were then analyzed using HALO™ software [49].
5.2. In vivo studies
Animal care was provided at the NCI-Frederick Animal Facility according to procedures outlined in the Guide for Care and Use of Laboratory Animals. Our facility is accredited by the Association for Accreditation of Laboratory Animal Care International and follows the Public Health Service Policy for the Care and Use of Laboratory Animals. Female BALB/c mice obtained from the Frederick Cancer Research and Development Center Animal Production Area were used for the in vivo studies and housed five per cage. Eight to ten-week-old female WT and Nos2- BALB/c mice were shaved a day prior to tumor injection and then were injected subcutaneously into the fourth mammary fat pad with 200,000 4T1 TNBC cells. Tumor measurements began one week after tumor cell injection, using a Vernier caliper and calculated in cubic millimeter volumes according to the following equation.
5.3. [(Short diameter)2 × long diameter]/2
Upon reaching tumor size of 100 mm3, tumor-bearing mice were divided into groups and treatment with 30 mg/L indomethacin in drinking water was initiated. The water was changed every Monday Wednesday Friday and treatment continued for the duration of the experiment unless otherwise specified. For the combination treatment with docetaxel, taxane was administered at 20 mg/kg of the drug in gel-chow (Nutra gel-chow, Bio-Serv, Flemington, NJ) on day 8 and day 21 after tumor injection. For this study, Indomethacin treatment began on the following day and continued as mentioned previously. Experimental end point varied according to the study. Mice were euthanized at or before the tumors reach 2000 mm3 size. For the survival surgery, tumors were resected when WT control tumors reached 400 mm3 size. Upon ending the experiment, lungs were fixed in Bouin’s solution and metastatic lesions were counted. Survival studies associated with the effects of dual inhibition on immune response to tumor were also performed; indomethacin treatment was stopped in the mice achieving complete remission i.e., cured mice, and these mice were then injected subcutaneously in the fourth mammary fat pad on the opposite side with 200,000 4T1 cells. Tumor growth was monitored in these rechallenged mice for a period of 5 months or until the tumor in the rechallenge site reached 2000 mm3 size, whichever was earlier.
5.4. Immunophenotyping of tumors by flow cytometry
Mice were euthanized and tumors were collected and dissociated by mechanical dissociation (Miltenyi GentleMACS) in lysis buffer containing Collagenase and DNase in 5% RPMI. Red blood cells were removed by incubating in ACK lysis buffer and washing with phosphate buffered saline (PBS). Cells were counted and equal numbers of cells were stained with the Live/Dead Aqua reagent (Amcyan) (1:1000) in PBS for 30 min followed by PBS wash and 20min at 4 °C with Fc blocker (1:200) in Sorter buffer (1%FBS, 1 mM EDTA in PBS). The cells were then stained with a panel of fluorophore-tagged antibodies against various immune cell markers including CD45-FITC, CD3-BV785, CD4-PECy7, CD8a-PerCPCy5.5, CD19-BV605, Tim3-APC, CD62L-PE, CD45-BV605, CD11b-PerCPCy5.5, CD11c-APCCy7, F4/80- APC, Ly6G-BV711, Ly6C-PECy7, CD206-FITC, PDL1-PE, MHCII (MHC IA/IE)-BV421. Samples were incubated for 20min at 4 °C, washed and read on a flow cytometer. Respective unstained cells and FMO (Fluorescence Minus One) controls were used to set the positive gates during acquisition. Samples were acquired using the low/medium flow rate setting on the BD LSRII Sorp flow cytometer, normalized to tumor weight and analysis was performed using FlowJo software.
5.5. Serum cytokine analysis
Blood was collected from mice by retroorbital bleed and saved in Microtainer® Blood tubes (BD Biosciences, Cat. No. 365967). The vials were left undisturbed for 30 min at room temperature, spun down at 2000 rpm for 10 min at RT and serum was collected, aliquoted and stored at −80 °C. Samples were completely thawed and centrifuged prior to use. Multiple freeze-thaws were strictly avoided. Serum cytokine analysis was performed using LEGENDPlex Mouse Inflammation Panel from BioLegend (Cat. No. 740446) according to manufacturer’s instructions. This method employs the Sandwich ELISA principle. Briefly, 2x diluted serum samples were incubated on a shaker for 2 h at RT with mixed beads tagged with antibodies against 13 different cytokines. The beads are distinguishable by size and the level of APC fluorophore on their surfaces. The beads were then washed and incubated with biotinylated-detection antibodies on a shaker for 1 h at RT followed by a 30min incubation with Streptavidin-PE. Samples and standards were washed and read on the BD LSR Fortessa flow cytometer in the PE and APC channels and data was analyzed using the LEGENDPlex software. Cytokine values exceeding the standard curve were not obtained for some samples.
5.6. RNA sequencing of bulk tumor
In brief, 30 tissue samples (WT n = 7, WT + INDO n = 8, Nos2-n = 7, Nos2- + INDO n = 8) were harvested after eight days of treatment with indomethacin and stored at −80 °C. Two 10 μm slices were homogenized in the presence of TRIzol (ThermoFisher) and further purified with affinity column (RNeasy Mini Kit, Qiagen) following the manufacturer’s protocols. Extracted RNAs underwent RNA quality check in a bioanalyzer and only samples with a RIN (RNA Integrity Number) larger than 6 were used to make the RNAseq library prep. Sample libraries were prepped with the Illumina Stranded Total RNA Prep and paired-end sequencing performed according to the manufacturer protocol and sequenced in a NovaSeq 600 sequencing system. Reads of the samples were trimmed for adapters and low-quality bases using Cutadapt. Sequencing data were exported and then uploaded to the Partek Flow server for subsequent sample normalization and QC steps using the build-in RNAseq Data Analysis workflow. Differentially expressed gene lists were generated with the Partek GSA algorithm which applies multiple statistical models to each individual gene in order to account for each gene’s varying responses to different experimental factors, and different data distributions. A 2-fold cutoff and p-value < 0.05 filter was applied to finalize the gene lists.
5.7. CODEX® analysis
The CODEX protocol was performed according to Akoya User Manual, revision B.0. Square (22 × 22 mm) glass coverslips (72204–10, Electron Microscopy Sciences) were pre-treated with l-Lysine (Sigma, St. Louis, MO) overnight at room temperature. Coverslips were rinsed in distilled water, dried, and stored at room temperature. Fresh frozen tissue blocks were sectioned (10 μm) on treated coverslips and stored in a coverslip storage box (Qintay, LLC) at −80 °C until further use. CODEX antibodies, reagents (including those for conjugation of additional antibodies), and instrumentation were purchased from Akoya Biosciences (Marlborough, MA). Antibodies labeled for CODEX included CD279, CD86, Ki67, E-cadherin, CD19, PIMO, CD31, CD49f, vimentin, F4-80, alphaSMA, CD44v6, Ly6C, NOS2, CD206, CD25, CD11c, CD274, CD44, CD24, MHCII, CD3, CD90, CD5, CD71, CD45, CD4, CD169, CD38, CD8a, Ly6G, CD11b. Tissue sections were stained with an antibody cocktail consisting of 0.5–1 μl of each antibody per tissue. CODEX assays were performed according to the manufacturer’s recommendations. Fluorescent oligonucleotide plates were prepared in black 96-well plates for image acquisition. Each CODEX cycle contains four fluorescent channels (three for antibody visualization and one for nuclear stain). For each cycle, up to three fluorescent oligonucleotides (5 μL each) were added to a final volume of 250 μL of plate buffer (containing Hoechst nuclear stain). For blank (empty) cycles, 5 μL of plate buffer was substituted for fluorescent oligonucleotides. Plates were sealed and kept at 4 °C until use. For imaging, the CODEX coverslip was mounted onto a custom-designed plate holder and securely tightened onto the stage of a Keyence BZ-X810 inverted fluorescence microscope. Cycles of hybridization, buffer exchange, image acquisition, and stripping were then performed using an Akoya CODEX instrument. Briefly, that instrument performs hybridization of the fluorescent oligonucleotides in a hybridization buffer, imaging of tissues in CODEX buffer, and stripping of fluorescent oligonucleotides in the stripping buffer. CODEX multicycle automated tumor imaging of was performed using a CFI Plan Apo 20x/0.75 objective (Nikon). The multipoint function of the BZ-X viewer software (BZ-X ver. 1.3.2, Keyence) was manually programmed to align with the center of each tumor and set to 10 Z stacks. Nuclear stain (DAPI, 1:600 final concentration) was imaged in each cycle at an optimized exposure time of roughly 10 ms. The respective channels were imaged in the automated run using optimized exposure times. Raw TIFF images produced during image acquisition were processed using the CODEX image processer. The processer concatenates Z-stack images, performs drift compensation based on alignment of nuclear stain across images, and removes the out-of-focus light using the Microvolution deconvolution algorithm (Microvolution). The processer also corrects for non-uniform illumination and subtracts the background and artefacts using blank imaging cycles without fluorescent oligonucleotides. The output of this image processing was tiled images corresponding to all fluorescence channels and imaging cycles that were then visualized and analyzed using HALO software (Version 3.3.2541.383, Indica Labs Inc.). Segmentation of cells was performed using the nuclear channel and the cell cytoplasm was defined as a fixed width ring around each nucleus. Nuclear segmentation settings were optimized by visual verification of segmentation performance on random subsets of cells aiming to minimize the number of over segmentations, under segmentation, detected artefacts and missed cells. Cell type Annotation and Differential Marker Analysis Cell populations were gated as follows. All nucleated cells were first identified by positive nuclear signals. Cell phenotypes were defined based upon biomarker expression as judged by expert visual inspection.
5.8. RNAscope
RNAscope (Advanced Cell Diagnostics (ACD) Bio, Newark, CA) fluorescence in situ hybridization (FISH) was performed on 5 μm cryosections of samples using the RNA-scope Multiplex Fluorescent Reagent Kit V2 (ACD) according to the manufacturer’s instructions. In brief, RNA-FISH was performed on all tumor samples by the Molecular Pathology Laboratory at the National Cancer Institute at Frederick (Frederick, MD). Serial cryosections of each sample were also processed for H&E (Hematoxylin and Eosin) and CODEX staining as well as RNAseq analyses. Histopathology was performed by a board-certified veterinary pathologist. RNAscope target probe staining was performed using a Leica Biosystems Bond Rx automated IHC/FISH slide staining system (Leica Biosystems, Wetzlar, Germany). For each set of RNA in situ hybridization probes stained, an RNAscope positive control and negative control probe were included to serve as an assay control. All target RNAscope probes were tested in a separate pilot study for the validation of probe specificity and localization. 20X digital fluorescent images were acquired on an Aperio ScanScope FL Scanner (Leica Biosystems), and object cell fluorescence intensity for each probe was quantified with HALO Imaging Software (Indica Labs, Albuquerque, NM). A classifier was built to distinguish between viable and necrotic tumor areas for each sample. Poorly stained areas were excluded and then the fluorescent channels were adjusted by eye for each probe and tissues were quantified. All data were exported to Excel (Microsoft, Redmond, WA) and Prism (GraphPad, San Diego, CA) for subsequent data and statistical analyses.
5.9. Image analysis using HALO
CODEX and InSituPlex images were analyzed using HALO V3.3 (Indica Labs) available through the NCI HALO Image Analysis Resource. Registered and stacked InSituPlex images and processed CODEX images were fused to generate afi composite images for analysis. Segmentation of cells was performed using the nuclear channel and the cell cytoplasm was defined as a fixed width ring (2 μm) around each nucleus. Nuclear segmentation settings were optimized by visual verification of segmentation performance on random subsets of cells aiming to minimize the number of over segmentations, under segmentation, detected artefacts and missed cells. Signal thresholding for each individual signal was defined based upon biomarker expression as judged by expert visual inspection independently on each image. Cellular phenotypes were set based on the combination if signals described in Supplemental Table II using HALO and/or positive and negative criteria. Regions of viable tumor, stroma, and necrosis were annotated manually on H&E-stained TNBC breast tumor slides. The H&E annotations were fused with Ultivue and NOS2/COX2 stained images to spatially localize the expression patterns of NOS2/COX2 with respect to immune phenotypes including CD8+ T cells.
5.10. Statistical analysis
Survival analysis, one-way ANOVA with Tukey's multiple comparisons test or Mann-Whitney test was employed to assess statistical significance using the GraphPad Prism software. Significance is reported as *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001.
Funding
This work was funded in whole or in part by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research NIH, under Contract HHSN261200800001E (VS, LAR, RC, DDB, THD, RK, SL, SMH, DWM, SKA, SA, DAW). This research was supported in part by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research and 5N91019D00024 (AW, ALW, DAS, WFH, EFE, DB, AW, MP, SKA, SJL), Cancer Research UK/Royal College of Surgeons of England Clinician Scientist Fellowship C39297/A22748 (RJB), NIH R01CA238727, NIH U01CA253553, and John S Dunn Research Foundation (STCW), NCI grant no. U54 CA210181, the Breast Cancer Research Foundation (BCRF), the Moran Foundation, Causes for a Cure, philanthropic support from M. Neal and R. Neal, and the Center for Drug Repositioning and Development Program (CREDO) (JCC), Science Foundation Ireland (SFI) grant number 17/CDA/4638, and a SFI and European Regional Development Fund (ERDF) grant number 13/RC/2073 (SAG).
Contributions
VS, LAR, RC, DWM, SJL, DAW conceptualized and designed the study. VS, LAR, RC, AJW, DDB, DB, AW, THD, MP performed experiments. VS, LAR, RC, NK, ALW, EFE, DAS, WH, MP analyzed data. VS, LAR, RC, ALW, EFE, WH, RJB, RK, SL, STCW, MP, SMH, DWM, SKA, JCC, SAG, SA, SJL, DAW interpreted data. VS, LAR, DAW wrote the paper. Data.
Availability
all data associated with the study are present in the paper or the Supplementary Materials.
Declaration of competing interest
The authors have no conflict of interests.
Acknowledgements
The authors wish to thank Tim Back, Tim Gower, and Adrienne Kimmel for their technical expertise. This project was funded in whole or in part with Federal funds from the National Cancer Institute, NIH, under Contract HHSN261200800001E (VS, LAR, RYSC, NK, DDB, THD, RJK, SL, SMH, DWM, SKA, SA, DAW). This research was supported in part by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research and 5N91019D00024 (AJW, ALW, EFE, DB, AW, DAS, WFH, MP, SKA, SJL), NIH R01CA238727, NIH U01CA253553, and John S Dunn Research Foundation (STCW), NCI grant no. U54 CA210181, the Breast Cancer Research Foundation (BCRF), the Moran Foundation, Causes for a Cure, philanthropic support from M. Neal and R. Neal, and the Center for Drug Repositioning and Development Program (CREDO) (JCC), Breast Cancer Now Grant 2013MayPR019 (SAG). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102529.
Contributor Information
Stephen J. Lockett, Email: locketts@mail.nih.gov.
David A. Wink, Email: wink@mail.nih.gov.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Akram M., Iqbal M., Daniyal M., Khan A.U. Awareness and current knowledge of breast cancer. Biol. Res. 2017;50:33. doi: 10.1186/s40659-017-0140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan M., Chollet A. Breast cancer screening: common questions and answers. Am. Fam. Physician. 2021;103:33–41. [PubMed] [Google Scholar]
- 3.Zhang W., Borcherding N., Kolb R. IL-1 signaling in tumor microenvironment. Adv. Exp. Med. Biol. 2020;1240:1–23. doi: 10.1007/978-3-030-38315-2_1. [DOI] [PubMed] [Google Scholar]
- 4.Glynn S.A., et al. Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients. J. Clin. Invest. 2010;120:3843–3854. doi: 10.1172/JCI42059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glynn S.A., et al. COX-2 activation is associated with Akt phosphorylation and poor survival in ER-negative, HER2-positive breast cancer. BMC Cancer. 2010;10:626. doi: 10.1186/1471-2407-10-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granados-Principal S., et al. Inhibition of iNOS as a novel effective targeted therapy against triple-negative breast cancer. Breast Cancer Res. 2015;17:25. doi: 10.1186/s13058-015-0527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basudhar D., et al. Coexpression of NOS2 and COX2 accelerates tumor growth and reduces survival in estrogen receptor-negative breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2017;10 doi: 10.1073/pnas.1709119114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinecke J.L., et al. Tumor microenvironment-based feed-forward regulation of NOS2 in breast cancer progression. Proc. Natl. Acad. Sci. U. S. A. 2014 doi: 10.1073/pnas.1401799111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu B., Qu L., Yan S. Cyclooxygenase-2 promotes tumor growth and suppresses tumor immunity. Cancer Cell Int. 2015;15:106. doi: 10.1186/s12935-015-0260-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalinski P. Regulation of immune responses by prostaglandin E2. J. Immunol. 2012;188:21–28. doi: 10.4049/jimmunol.1101029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Switzer C.H., et al. S-nitrosation mediates multiple pathways that lead to tumor progression in estrogen receptor-negative breast cancer. For Immunopathol Dis Therap. 2012;3:117–124. doi: 10.1615/ForumImmunDisTher.2012006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ridnour L.A., et al. NOS inhibition modulates immune polarization and improves radiation-induced tumor growth delay. Cancer Res. 2015;75:2788–2799. doi: 10.1158/0008-5472.CAN-14-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruffell B., et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 2014;26:623–637. doi: 10.1016/j.ccell.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung A.W., et al. A phase 1/2 clinical trial of the nitric oxide synthase inhibitor L-NMMA and taxane for treating chemoresistant triple-negative breast cancer. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abj5070. [DOI] [PubMed] [Google Scholar]
- 15.Gruosso T., et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J. Clin. Invest. 2019;129:1785–1800. doi: 10.1172/JCI96313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmore A.C., et al. An in vitro tumorigenesis model based on live-cell-generated oxygen and nutrient gradients. Commun Biol. 2021;4:477. doi: 10.1038/s42003-021-01954-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Somasundaram V., et al. Inducible nitric oxide synthase-derived extracellular nitric oxide flux regulates proinflammatory responses at the single cell level. Redox Biol. 2020;28 doi: 10.1016/j.redox.2019.101354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu S., et al. Fluorescent indomethacin-dansyl conjugates utilize the membrane-binding domain of cyclooxygenase-2 to block the opening to the active site. J. Biol. Chem. 2019;294:8690–8698. doi: 10.1074/jbc.RA119.007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uddin M.J., et al. Selective visualization of cyclooxygenase-2 in inflammation and cancer by targeted fluorescent imaging agents. Cancer Res. 2010;70:3618–3627. doi: 10.1158/0008-5472.CAN-09-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agmon Y., Peleg H., Greenfeld Z., Rosen S., Brezis M. Nitric oxide and prostanoids protect the renal outer medulla from radiocontrast toxicity in the rat. J. Clin. Invest. 1994;94:1069–1075. doi: 10.1172/JCI117421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brzozowski T., Sliwowski Z., Majka J., Drozdowicz D., Konturek S.J. Urea-urease system in cytoprotection against acute mucosal damage. J. Physiol. Pharmacol. 1996;47:137–150. [PubMed] [Google Scholar]
- 22.Amu S., Gjertsson I., Brisslert M. Functional characterization of murine CD25 expressing B cells. Scand. J. Immunol. 2010;71:275–282. doi: 10.1111/j.1365-3083.2010.02380.x. [DOI] [PubMed] [Google Scholar]
- 23.Kuroda H., et al. Prognostic value of tumor-infiltrating B lymphocytes and plasma cells in triple-negative breast cancer. Breast Cancer. 2021;28:904–914. doi: 10.1007/s12282-021-01227-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cazet A.S., et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat. Commun. 2018;9:2897. doi: 10.1038/s41467-018-05220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davila-Gonzalez D., et al. Pharmacological inhibition of NOS activates ASK1/JNK pathway augmenting docetaxel-mediated apoptosis in triple-negative breast cancer. Clin. Cancer Res. 2018;24:1152–1162. doi: 10.1158/1078-0432.CCR-17-1437. [DOI] [PubMed] [Google Scholar]
- 26.Solinas C., et al. Targeting immune checkpoints in breast cancer: an update of early results. ESMO Open. 2017;2 doi: 10.1136/esmoopen-2017-000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deepak K.G.K., et al. Tumor microenvironment: challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol. Res. 2020;153 doi: 10.1016/j.phrs.2020.104683. [DOI] [PubMed] [Google Scholar]
- 28.Forrester K., et al. Nitric oxide-induced p53 accumulation and regulation of inducible nitric oxide synthase expression by wild-type p53. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2442–2447. doi: 10.1073/pnas.93.6.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Moraes E., Dar N.A., de Moura Gallo C.V., Hainaut P. Cross-talks between cyclooxygenase-2 and tumor suppressor protein p53: balancing life and death during inflammatory stress and carcinogenesis. Int. J. Cancer. 2007;121:929–937. doi: 10.1002/ijc.22899. [DOI] [PubMed] [Google Scholar]
- 30.Basudhar D., et al. Chemotherapeutic potential of diazeniumdiolate-based aspirin prodrugs in breast cancer. Free Radic. Biol. Med. 2015;83:101–114. doi: 10.1016/j.freeradbiomed.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ekmekcioglu S., Grimm E.A., Roszik J. Targeting iNOS to increase efficacy of immunotherapies. Hum. Vaccines Immunother. 2017;13:1105–1108. doi: 10.1080/21645515.2016.1276682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong K., et al. Murine myeloid dendritic cells that phagocytose apoptotic T cells inhibit the immune response via NO. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saio M., Radoja S., Marino M., Frey A.B. Tumor-infiltrating macrophages induce apoptosis in activated CD8(+) T cells by a mechanism requiring cell contact and mediated by both the cell-associated form of TNF and nitric oxide. J. Immunol. 2001;167:5583–5593. doi: 10.4049/jimmunol.167.10.5583. [DOI] [PubMed] [Google Scholar]
- 34.Niedbala W., et al. Nitric oxide induces CD4+CD25+ Foxp3 regulatory T cells from CD4+CD25 T cells via p53, IL-2, and OX40. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15478–15483. doi: 10.1073/pnas.0703725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alleva D.G., Burger C.J., Elgert K.D. Tumor-induced regulation of suppressor macrophage nitric oxide and TNF-alpha production. Role of tumor-derived IL-10, TGF-beta, and prostaglandin E2. J. Immunol. 1994;153:1674–1686. [PubMed] [Google Scholar]
- 36.Bishop-Bailey D., Burke-Gaffney A., Hellewell P.G., Pepper J.R., Mitchell J.A. Cyclo-oxygenase-2 regulates inducible ICAM-1 and VCAM-1 expression in human vascular smooth muscle cells. Biochem. Biophys. Res. Commun. 1998;249:44–47. doi: 10.1006/bbrc.1998.8966. [DOI] [PubMed] [Google Scholar]
- 37.Mannie M.D., Prevost K.D., Marinakis C.A. Prostaglandin E2 promotes the induction of anergy during T helper cell recognition of myelin basic protein. Cell. Immunol. 1995;160:132–138. doi: 10.1016/0008-8749(95)80018-e. [DOI] [PubMed] [Google Scholar]
- 38.Li W., et al. Aerobic glycolysis controls myeloid-derived suppressor cells and tumor immunity via a specific CEBPB isoform in triple-negative breast cancer. Cell Metabol. 2018;28:87–103 e106. doi: 10.1016/j.cmet.2018.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sceneay J., et al. Primary tumor hypoxia recruits CD11b+/Ly6Cmed/Ly6G+ immune suppressor cells and compromises NK cell cytotoxicity in the premetastatic niche. Cancer Res. 2012;72:3906–3911. doi: 10.1158/0008-5472.CAN-11-3873. [DOI] [PubMed] [Google Scholar]
- 40.Zhang C., et al. Artificial super neutrophils for inflammation targeting and HClO generation against tumors and infections. Adv. Mater. 2019;31 doi: 10.1002/adma.201901179. [DOI] [PubMed] [Google Scholar]
- 41.Wink D.A., et al. Mechanisms of the antioxidant effects of nitric oxide. Antioxidants Redox Signal. 2001;3:203–213. doi: 10.1089/152308601300185179. [DOI] [PubMed] [Google Scholar]
- 42.Kroeger D.R., Milne K., Nelson B.H. Tumor-infiltrating plasma cells are associated with tertiary lymphoid structures, cytolytic T-cell responses, and superior prognosis in ovarian cancer. Clin. Cancer Res. 2016;22:3005–3015. doi: 10.1158/1078-0432.CCR-15-2762. [DOI] [PubMed] [Google Scholar]
- 43.Zhang N.N., et al. Prognostic impact of tertiary lymphoid structures in breast cancer prognosis: a systematic review and meta-analysis. Cancer Cell Int. 2021;21:536. doi: 10.1186/s12935-021-02242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rastogi I., et al. Role of B cells as antigen presenting cells. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.954936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mills B.N., et al. Stereotactic body radiation and interleukin-12 combination therapy eradicates pancreatic tumors by repolarizing the immune microenvironment. Cell Rep. 2019;29:406–421 e405. doi: 10.1016/j.celrep.2019.08.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kish D.D., Gorbachev A.V., Parameswaran N., Gupta N., Fairchild R.L. Neutrophil expression of Fas ligand and perforin directs effector CD8 T cell infiltration into antigen-challenged skin. J. Immunol. 2012;189:2191–2202. doi: 10.4049/jimmunol.1102729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vono M., et al. Neutrophils acquire the capacity for antigen presentation to memory CD4(+) T cells in vitro and ex vivo. Blood. 2017;129:1991–2001. doi: 10.1182/blood-2016-10-744441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gurpinar E., Grizzle W.E., Piazza G.A. COX-independent mechanisms of cancer chemoprevention by anti-inflammatory drugs. Front. Oncol. 2013;3:181. doi: 10.3389/fonc.2013.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manesse M., Patel K.K., Bobrow M., Downing S.R. The InSituPlex((R)) staining method for multiplexed immunofluorescence cell phenotyping and spatial profiling of tumor FFPE samples. Methods Mol. Biol. 2020:585–592. doi: 10.1007/978-1-4939-9773-2_26. 2055. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.