Highlights
-
•
TMEM100 expression was shown to decrease in PCa patients, and low TMEM100 expression was associated with tumor stage and metastasis.
-
•
Overexpression of TMEM100 suppressed PCa progression by inhibiting the FAK/PI3K/AKT signaling pathway.
-
•
TMEM100 could regulate SCNN1D by inhibiting FAK/PI3K/AKT signaling in PCa cell lines.
Keywords: Transmembrane protein 100, Prostate cancer, Progression, FAK/PI3K/AKT pathway
Abbreviations: DAB, 3,3′-diaminobenzidine; EMT, Epithelial-mesenchymal transition; FAK, Focal adhesion kinase; GEPIA, Gene Expression Profiling Interactive Analysis; HRP, Horseradish peroxidase; IHC, Immunohistochemistry; OS, Overall survival; P13K, Phosphatidylinositol-3-kinase; PFS, Progression-free survival; Pca, Prostate cancer; STRING, Search tool for the retrieval of interacting genes/proteins; SCNN1D, Sodium Channel Epithelial 1 Subunit Delta; TMEM, Transmembrane
Abstract
The effects of transmembrane (TMEM) proteins in the progression of prostate cancer (PCa) remain unknown. This study aims to explore the functions of TMEM100 in PCa. To explore the expression, regulation, and effects of TMEM100 in PCa, two PCa cell lines and 30 PCa tissue samples with adjacent control tissues were examined. Online databases, immunohistochemistry, immunofluorescence, western blot, flow cytometry, colony formation, wound healing, transwell assays, and xenograft mouse models were used to explore effects of TMEM100 relevant to PCa. TMEM100 expression was shown to decrease in PCa patients, and low TMEM100 expression was associated with tumor stage and metastasis. Overexpression of TMEM100 suppressed PCa progression by inhibiting the FAK/PI3K/AKT signaling pathway. Tumor size was smaller in TMEM100 overexpressing PCa cells in xenograft mice than in control mice. We also found that TMEM100 could regulate SCNN1D by inhibiting FAK/PI3K/AKT signaling in PCa cell lines. Taken together, our findings indicate that TMEM100 is a tumor suppressor that plays a vital role in preventing PCa proliferation, migration, and invasion through inhibition of FAK/PI3K/AKT signaling. These studies suggest that TMEM100 can be used as a predictive biomarker and therapeutic target.
Introduction
Prostate cancer (PCa) is the most commonly diagnosed cancer affecting men's health, with significant increase in its incidence worldwide [1]. In 2020, 1414,259 patients were diagnosed with PCa, and 375,304 deaths were reported globally [2]. Early stage diagnosis of PCa is difficult and approximately 5% of newly diagnosed patients showed evidence of distant metastasis [3]. Exploring the underlying mechanisms of PCa is critical to expand PCa therapy options.
Focal adhesion kinase (FAK) is a non-receptor tyrosine kinase that classically transduces cell adhesion signaling to regulate various biological processes, including cell growth, embryonic development, and tumor invasion [4]. It has been found that FAK knockdown can antagonize depression-induced PCa invasion and metastasis, indicating that FAK is a potential new target for PCa treatment [5]. Previous studies have proven that FAK can promote p53 degradation through ubiquitination, leading to PCa cell proliferation and metastasis, and can increase phosphorylation levels of phosphatidylinositol-3-kinase (PI3K) and protein kinase B (AKT) to promote cell proliferation and epithelial-mesenchymal transition (EMT) of PCa cells [6, 7]. A series of components contribute to regulation of FAK/PI3K/AKT signaling, including receptor tyrosine kinases, integrins, and cytokine receptors [8]. Therefore, the identification of key regulators of FAK/PI3K/AKT signaling in PCa is especially relevant in exploring the best strategies to treat PCa.
Tumor progression is a complex biological behavior, with various genes and pathways reported to be involved in cell invasion processes [9]. The transmembrane (TMEM) protein family comprises components of various cell membranes, including endoplasmic reticulum, plasma membrane, and cytoskeleton membranes [10]. To date, various TMEM proteins have been discovered to participate in cellular processes including autophagy, protein glycosylation, and epidermal keratinization [11]. Recent studies have shown that many TMEM proteins serve as critical regulators of tumor progression. For example, TMEM48 and TMEM97 are potential biomarkers of lung cancer [12]. In addition, TMEM17 silencing inhibits invasion, migration, and EMT through AKT/GSK3β signaling pathway in breast cancer cells [13].
TMEM100, a two-pass transmembrane protein, has been confirmed to have prognostic value in various tumors, including renal cell carcinoma, gastric cancer, and non-small cell lung cancer [14, 15]. By regulating the activity of hypoxia-inducible factor-1, TMEM100 inhibited gastric cancer cell proliferation and metastasis in vivo and in vitro [16]. The Sodium Channel Epithelial 1 Subunit Delta (SCNN1D) is involved in the transfer of cellular components [17]. Analysis of data obtained from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) and Gene Expression Profiling Interactive Analysis (GEPIA) databases indicates that TMEM100 may interact with SCNN1D. However, the cytological effect(s) of TMEM100 and SCNN1D interaction in PCa remains unknown.
We investigated the functions and mechanisms of TMEM100 activity in PCa. Overexpression of TMEM100 inhibited PCa progression via FAK/PI3K/AKT signaling pathway, and SCNN1D may play a vital role in this process. Therefore, TMEM100 has the potential to serve as a biomarker for PCa diagnosis and treatment.
Materials and methods
Clinical specimens
A total of 30 PCa and adjacent noncancerous prostate tissues were acquired from the Renmin Hospital of Wuhan University (Wuhan, China) between June 2020 and January 2022. All patients were diagnosed by two independent pathologists, and the collected tissue samples were used for immunohistochemistry (IHC) and western blot analysis. This study was approved by the Ethics Committee of Renmin Hospital (approval number: WDRM2019-Y068) and we collected written consent from each patient. Tumors and adjacent noncancerous tissues were collected. Tissues were fixed with 4% paraformaldehyde at 26 °C for 48 h, embedded in paraffin, and sectioned to 5-μm thick sections for immunohistochemistry staining. Alternatively, tissues were immediately preserved at -80 °C for protein extraction.
Cell culture and transfection
Two PCa cell lines, PC3 and DU145, were obtained from Procell (Wuhan, China) and cultured in RPMI-1640 media supplemented with 10% fetal bovine serum (Gibco, CA, United States), 1% penicillin (Sigma, MO, USA) and 1% streptomycin (Sigma, MO, USA). The cell lines were cultured in a humidified incubator at 37 °C containing 5% CO2.
For TMEM100 overexpression experiments, PC3 and DU145 cells were transfected with lentiviruses Lv-TMEM100 and Lv-Vector (OBiO Biotechnology, Shanghai, China); PCa cells were cultured in 6-well plates approximately 8–10 h, then inoculated with the corresponding lentiviruses in the presence of polybrene (8 μg/ml). After incubation with virus for 24 h, the inoculating medium was replaced with complete medium supplemented with puromycin (3 μg/ml) to kill noninfected cells. Transfection efficiency was confirmed by western blot.
For the construction of Si-SCNN1D cells, a small interfering RNA targeting a sequence in SCNN1D was synthesized (RiboBio, China). PCa cells were plated in 6-well plates at 2 × 105 cells/plate and cultured for 24 h. According to the manufacturer's instructions, Si-SCNN1D and scrambled control siRNA were transfected into PCa cell lines using lipofectamine 2000 (Invitrogen, MA, USA). Targeting sequences for SNCC1D siRNA were: 5′-AGCCAGUACUUUGGGAGCATT-3′.
TMEM100 siRNA was used to reduce the TMEM100 expression. PCa cell lines in the exponential phase of growth were plated in six-well plates at 2 × 105 cells/plate and cultured for 24 h. After that, transfection of scramble control and TMEM100 siRNA (all from GenePharma, Shanghai, China) in PCa cell lines were carried out by using lipofectamine 2000 (Invitrogen, MA, USA) in accordance with the manufacturer's instructions. TMEM100 siRNA Sequences were: 5′-CAGACUUUAUGUUCAUAGUUCUUCCUUC-3′.
Cell viability assay
Cell viability was quantified using a cell counting kit-8 (CCK8, Beyotime) assay. Briefly, PC3 and DU145 cells were seeded into 96-well culture plates at a concentration of 2 × 103 cells/well in 100 μl of medium at 37 °C for 24 h. Media was aspirated and cells were rinsed with phosphate-buffered saline, then treated with 10 μl/well CCK-8 at 37 °C for 2 h. OD values under 450 nm were determined by a microplate reader (Bio-Rad Laboratories, Benicia, CA, USA).
Colony formation assay
PC3 and DU145 cells were seeded into each well of a 6-well plate at a density of 1000 cells/well and incubated for two weeks. Resultant colonies were fixed with 10% methanol for 15 min and stained with 1% crystal violet at 37 °C for 20 min. Visible colonies were counted under a microscope.
Wound healing assay
PC3 and DU145 cells were cultured in 6-well plates for 24 h to achieve appropriate density. The medium was replaced with serum-free medium, and a straight scratch was made using a 200 μl pipette tip. After scratching, images were captured at 0 h and 48 h and analyzed using Image J software.
Transwell invasion assays
Transwell invasion assays were carried out using Biocoat Matrigel Invasion Chambers (Corning, NY). PCa cells (3 × 104) were placed in the upper chamber in 200 μl medium with 2% FBS, and 700 μl of medium containing 10% FBS was added to the lower chamber. Chambers were washed with PBS after incubation at 37 °C for 48 h, fixed with 4% paraformaldehyde, and stained with 0.1% crystal violet (G1014, Servicebio, China) for 3 min. Cells in the inner layer were then carefully removed using cotton swabs and images were obtained with an Olympus microscope.
Apoptosis and cell cycle assays
An Annexin V-PE/7-ADD detection kit (BD Bioscience, USA) was used to detect cell apoptosis. A cell suspension with a concentration of 106 cells/ml was plated in a 6-well plate and cultured overnight. The collected cells were washed with PBS three times and 500 μl Binding Buffer was added to resuspend the pellet. Annexin V-PE and 7-ADD were added, and cells were incubated in the dark for 10 min. Finally, flow cytometry (Beckman Coulter, USA) was used to detect the apoptosis rate of the cells. For cell cycle analysis, cells were fixed with 70% ethanol overnight at 4 °C, and stained with a working solution containing 450 μl propidium iodide and 50 μl RNase A for 30 min. Stained cells were detected by flow cytometry.
Immunohistochemistry (IHC) staining
TMEM100, vimentin, and Ki67 proteins were detected by IHC as previously described [18]. Briefly, 4-µm paraffin-embedded sections were deparaffinized, subjected to antigen retrieval using citrate buffer (pH 6.0) at 95 °C for 10 min, and treated with 0.3% H2O2 at room temperature for 10 min to block endogenous peroxidase activity. Next, the sections were incubated with blocking reagent QuickBlock™ (cat. no. P0260; Beyotime Institute of Biotechnology), then incubated with primary antibody: anti-TMEM100 (cat.no.PA5–48260; 1:100; ThermoFisher Scientific), anti-Vimentin (cat.no.ab92547; 1:200; Abcam), or anti-Ki67 (cat.no.D3B5; 1:50; CST) at 4 °C for 12 h. After washing, incubation with secondary antibodies (cat.no.TA130017–1; 1:5000;biotinylated goat anti-rabbit; OriGene Technologies) was performed for 1 h at 37 °C. Peroxidase activity was visualized using 3,3′-diaminobenzidine at room temperature for 5 min. Positively stained areas for KIM-1, based on unit area, were calculated as the percentage of all examined areas using ImageJ software (version 1.8.0; National Institutes of Health).
Tunel detection
A Tunel assay (Roche Applied Science) was performed in accordance with the manufacturer's instructions. Tissue samples were prepared, and apoptotic cells were labeled by terminal transferase-mediated dUTP nick-end labeling. Finally, apoptotic cells were stained brown and quantified under a microscope.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and permeabilized with 0.3% Triton X-100 solution. Then, cells were blocked with 5% BSA and incubated at 4 °C overnight with primary antibodies against Ki67 (12075; 1:50; CST), Vimentin (10366–1-AP; 1:100; Proteintech), or N-cadherin (13116, 1:200; CST). After washing with PBS, cells were incubated with CY3-conjugated secondary antibodies (1:1000; ThermoFisher Scientific, MA, USA) and the fluorescent nuclear stain DAPI. Images were captured using a fluorescence microscope (Nikon, Tokyo, Japan).
Western blot analysis
RIPA buffer was used to extract proteins from cells or tissue samples. Total protein was measured via a bicinchoninic acid (BCA) Protein Kit (Beyotime, China). Proteins were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes. The membranes were blocked with 5% skim milk for 1 h at 25 °C and incubated for 12 h at 4 °C with primary antibodies: anti-TMEM100 (cat.no.A16653; 1:1000; ABclonal), anti-E-cadherin (cat.no.20874–1-AP; 1:5000; Proteintech), anti-N-cadherin (cat.no.ab76011; 1:10000; Abcam), anti-MMP-9 (cat.no.ab76003; 1:2000; Abcam), anti-vimentin (cat.no.10366–1-AP; 1:2000; Proteintech), anti-Bax (cat.no.50599–2; 1:5000; Proteintech), anti-Bcl-2 (cat.no.265931; 1:1000; Proteintech), anti-Caspase-3 (cat.no.19677–1; 1:100; Proteintech), anti-CyclinD1 (cat.no.60186–1; 1:5000; Proteintech), anti-p-FAK (cat.no.8556; 1:1000; CST), anti-FAK (cat.no.71433; 1:1000; CST), anti-p-PI3K (cat.no.17366; 1:1000; CST), anti-PI3K (cat.no.T40064; 1:1000; Abmart), anti-p-AKT (cat.no.4060; 1:1000; CST), anti-AKT (cat.no.60203; 1:5000; Proteintech), anti-Beta actin (cat.no.20536; 1:1000; Proteintech). Blots were incubated with HRP-conjugated secondary antibodies (cat. no. SA00001–2; 1:1000; ProteinTech) for 2 h at 27 °C. Target bands were visualized with an ECL system (cat.no.BL520A; Biosharp Life Sciences) and protein band images were quantified by densitometry using Image J software (version 1.8.0; National Institutes of Health).
RNA extraction and quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
TRIzol reagent (Invitrogen; Thermo Fisher Scientific) was used to extract RNA. Reverse-transcription was performed using a Takara reagent kit (Takara Biotechnology, Otsu, Japan) to obtain cDNA, followed by qPCR amplification using SYBR Green Master Mix (Yeasen, Shanghai, China) according to the manufacturer's protocol. The primers used in this work are as follows: TMEM100, forward primer sequence: 5′-GGTTCTTGTGGAGCCATAGG - 3′ and reverse primer sequence: 5′-CTCTCCTTCTCTTGGTCTCTCT - 3′; GAPDH, forward primer sequence: 5′-GCATCGAGATCCCTCCAAAAT - 3′ and reverse primer sequence: 5′ -GATGTTTGTCATACTTCTCATGG - 3′.
Xenografts in mice
This study was approved by the Animal Experiment Ethics Committee of Wuhan University. Ten male BALA/C nude mice (4 weeks old) were obtained from the Hubei Provincial Center for Disease Control and Prevention (Wuhan, China). All mice were kept under specific pathogen free conditions and were allowed access to food and water ad libitum. The mice were randomly distributed into an Lv-vector group and an Lv-TMEM100 group after 7 d of adaptation to the laboratory animal facility of Wuhan University. Lv-vector and Lv-TMEM100 mice were inoculated subcutaneously in the back with DU145 cells (5 × 106) resuspended in 100μl of PBS. The tumor formation status of mice was monitored and recorded every 5 d by measuring tumor width (W) and length (L) with calipers. Four weeks post-injection, mice were euthanized, and tumors were harvested for IHC and Tunel staining. Tumor volume (V) was calculated using the formula V = (W2 × L)/2.
Databases used and Kegg pathway analysis
Several online databases or tools were used to obtain and further analyze data in our current study, including the TCGA database (https://portal.gdc.cancer.gov), Gene Expression Profiling Interactive Analysis (http://gepia.cancer-pku.cn/) GEO database (http://www.ncbi.nlm.nih.gov/geo), cBioPortal database (http://www.cbioportal.org/) and Xiantao academic website (https://www.xiantao.love/).
Statistical analyses
Experimental results are presented as means ± SD. GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA) was used to conduct the Statistical analysis. Student's t-test and one-way analysis of variance (ANOVA) was performed for Statistical analysis. Log-rank test was used for overall survival and progression-free survival. P < 0.05 was considered to have statistical significance.
Results
TMEM100 exhibits lower expression level in cancer tissues compared to control tissues
As seen in Fig. 1A, the TMEM100 gene is one member of the transmembrane protein family, and maps to chromosome 17. The TCGA databases were utilized to further investigate TMEM100 expression levels in PCa. As shown in Fig. 1B, TMEM100 expression level was downregulated in various cancer types relative to corresponding healthy controls, including bladder carcinoma, breast cancer, esophageal squamous cancer, and prostate cancer. Furthermore, TMEM100 expression level decreased significantly in PCa tissues compared with their adjacent healthy prostate tissues (Fig. 1C and D). A total of 30 paired PCa tissues and adjacent healthy tissues were obtained for further experiments. We carried out IHC using these samples, and the results showed strong positive expression of TMEM100 in the healthy tissues (Fig. 1M). Western blot analysis revealed that TMEM100 protein significantly decreased in PCa specimens relative to that in paired control prostate tissues (Fig. 1L). Overall, TMEM100 exhibits lower expression level in PCa compared with healthy tissues, suggesting that it may be playing a tumor suppression role in PCa.
Fig. 1.
The chromosomal location, expression characteristics of TMEM100 gene, and its correlation with clinicopathological features in PCa (A) The TMEM gene on Chr17. (B) TMEM100 expression profile across all tumor samples and normal tissues. (C) The difference expression of TMEM100 in PCa tissues and adjacent normal tissues. (D) The difference expression of TMEM100 in PCa tissues and paired normal tissues. (E-G) Relative expression levels of TMEM100 in TCGA database with T stage (E), N stage (F), M stage (G). (H-I) Overall survival (H) and progress free interval (I) curve of PCa patients with low and high TMEM100 expression in TCGA database. (J) progress free interval in GEO database (GSE16560). (K) ROC curve showed the efficiency of TMEM100 expression level to distinguishing PCa tissue from non-tumor tissue (L) Western blot of TMEM100 protein expression in adjacant tissues and PCa tissues. (M) Representative IHC staining of TMEM100 in adjacant tissues, low-stage PCa, moderate-stage PCa and high-stage PCa and the quantitative evaluation of TMEM100 staining intensity. Scale bars = 200 μm or 50 μm, respectively. *<0.05, **<0.01, ***<0.001.
TMEM100 expression is correlated with PCa progression
According to TCGA database analysis results, TMEM100 expression decreased with tumor progression (Fig. 1E, F and G). Similarly, IHC results showed that TMEM100 expression decreased with increases in tumor grade in PCa tissues (Fig. 1M). Furthermore, KM plotter analysis showed a significant correlation between high TMEM100 expression level and overall survival (OS) and progression-free survival (PFS) in PCa patients (Fig. 1H, I, and J). In addition, the results of ROC analysis showed the power of TMEM100 in predicting outcomes of healthy prostate tissue and cancer tissue (Fig. 1K). These results indicate that TMEM100 expression correlates positively with PCa patient prognosis.
TMEM100 inhibits PCa cell proliferation
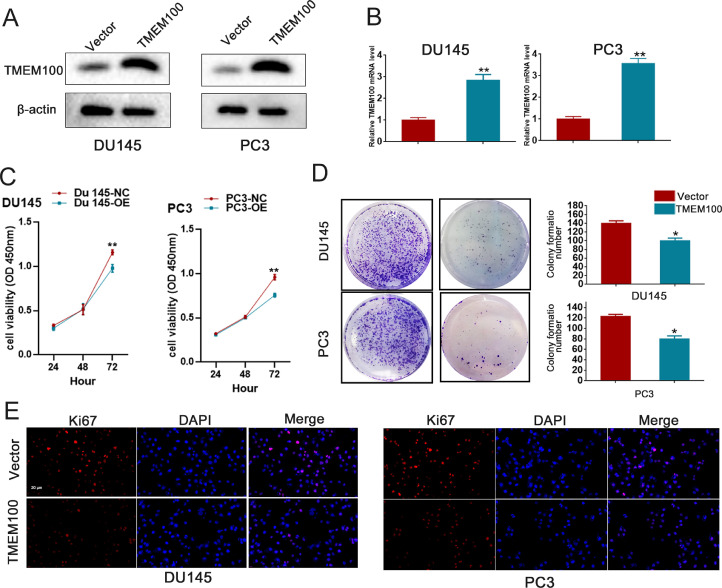
To explore the effect of TMEM100 on PCa cell proliferation, stable TMEM100-overexpressing cells were constructed in PC3 and DU145 cell lines, and their expression levels were verified by western blot and qRT-PCR (Fig. 2A and B). CCK8 assays indicated that overexpression of TMEM100 significantly inhibited PC3 and DU145 cell proliferation (Fig. 2C). Likewise, colony numbers were significantly decreased when TMEM100 was overexpressed. In addition, the results of Ki67 immunofluorescence were consistent with the results of CCK8 and colony formation assays (Fig. 2D and E). In addition, we also designed siRNA against TMEM100 to explore the role of endogenous TMEM100. qRT-PCR showed the that si-TMEM100 decreased TMEM100 expression as is shown in the supply Fig. 1A-B. Meanwhile, CCK-8 assay indicated that inhibition of TMEM100 enhanced proliferation of PC3 and DU145 cells. In summary, these results showed that overexpression of TMEM100 decreased the proliferation of PCa cells, while silencing TMEM100 enhanced the proliferation of PCa cells.
Fig. 2.
Overexpression of TMEM100 inhibits the proliferation of PCa cells in vitro (A-B) The overexpressing efficiency of TMEM100 was verified by Western blot and qRT-PCR in DU145 and PC3 cells. (C-E) Effects of TMEM100 overexpression on cell proliferation by CCK-8 (D), colony formation (E) and Ki67 (F) assays in DU145 and PC3 cells. Scale bars = 20 μm. *P<0.05, **P<0.01 VS control group.
TMEM100 overexpression inhibits PCa cell migration, invasion, and EMT in vitro
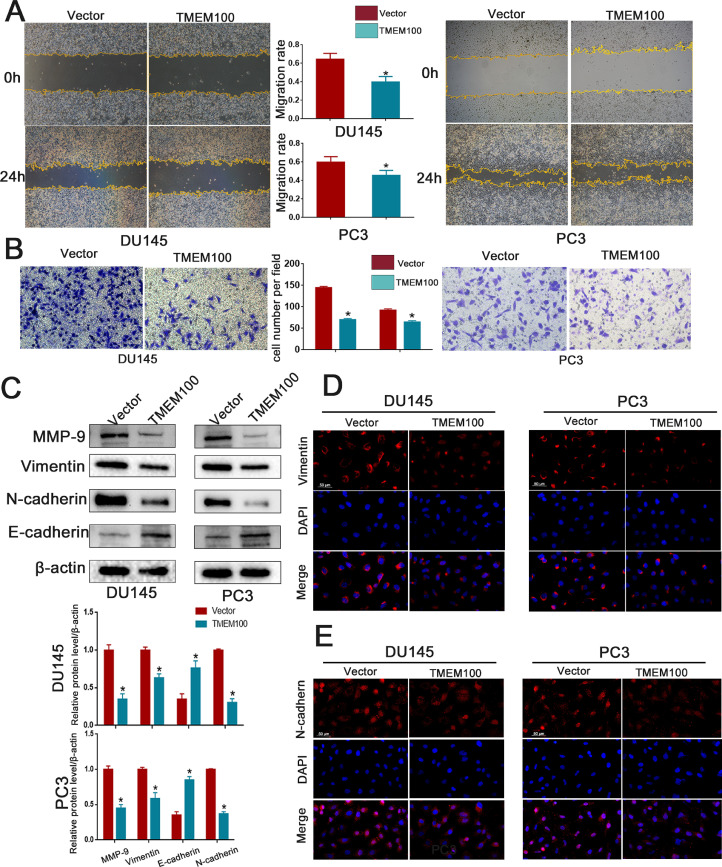
Given that EMT is the initial step in tumor cell invasion and metastasis, we investigated the action of TMEM100 in PCa cell EMT. A wound-healing assay was performed to quantify the effect of TMEM100 overexpression on PCa cell motility. TMEM100 overexpression significantly impaired PCa cell motility (Fig. 3A). Transwell assays disclosed that PC3 and DU145 cell invasiveness was remarkably reduced by TMEM100 overexpression (Fig. 3B). Western blot analysis showed that TMEM100 overexpression significantly decreased mesenchymal cell marker proteins, including Vimentin, N-cadherin, and MMP-9, and increased the epithelial cell marker E-cadherin (Fig. 3C). Immunofluorescence results are consistent with western blotting (Fig. 3D and E) Collectively, these results show that TMEM100 overexpression inhibits the migration, invasion, and EMT of PCa cells in vitro.
Fig. 3.
Overexpression of TMEM100 inhibits the migration, invasion and EMT of PCa cells in vitro (A) Effects of TMEM100 overexpression on migration by wounding healing assays in DU145 and PC3 cells. (B) Effects of TMEM100 overexpression on invasion by transwell assays in DU145 and PC3 cells. (C) Changes in the EMT-related proteins were detected by Western blot in DU145 and PC3 cells. (D-E) Representative fluorescence images of Vimentin and N-cadherin in DU145 and PC3 cells. Scale bars = 50 μm. *P<0.05 VS control group.
TMEM100 overexpression inhibits the cell cycle and promotes PCa cell apoptosis in vitro
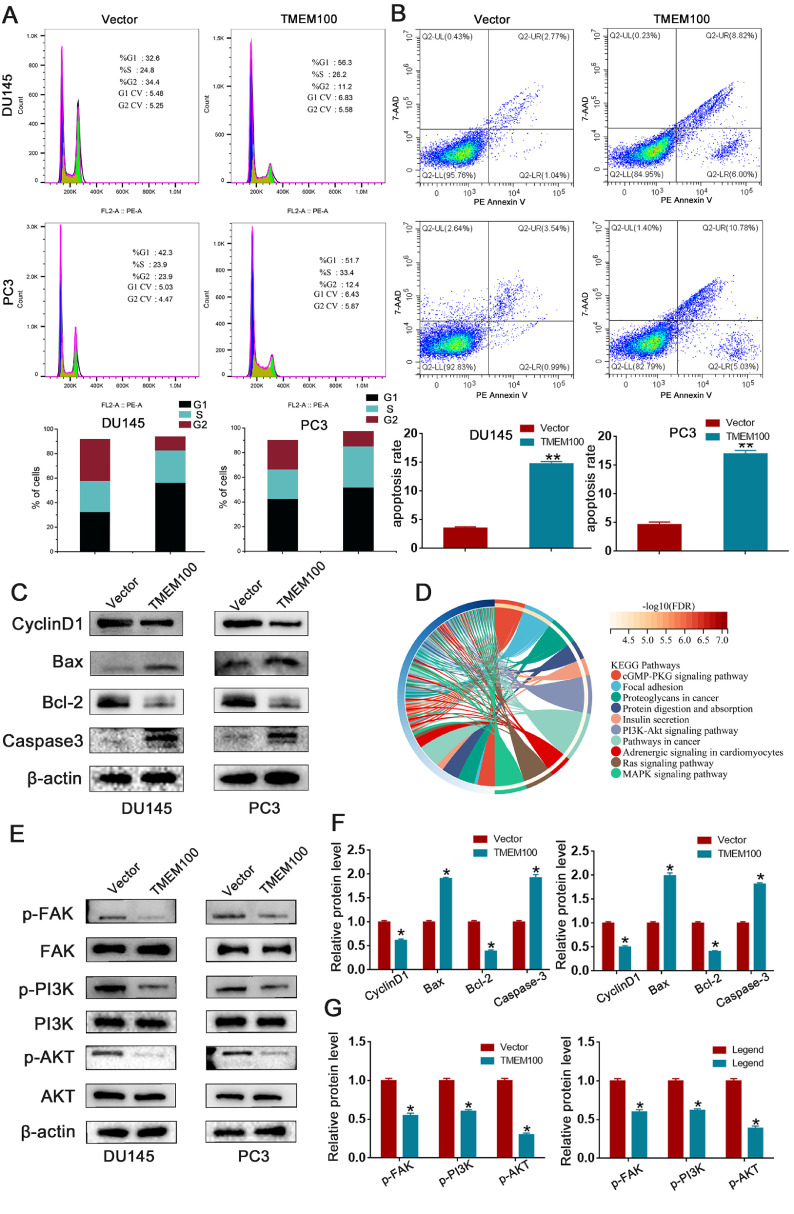
Flow cytometry was used to explore the effect of TMEM100 on the cell cycle and apoptosis. As shown in Fig. 4A, overexpression of TMEM100 in PCa cells increased the percentage of cells in G1 phase and decreased the percentage in G2 phase. Consistent with the result of flow cytometry, western blot analysis affirmed that cyclin D1 was decreased in TMEM100 overexpressing cells compared with the vector control cells (Fig. 4C and F). We also observed a marked increase in the apoptotic cell population with TMEM100 overexpression (Fig. 4B). Western blot results also indicated that TMEM100 overexpression significantly increased the expression of apoptotic proteins including Bax and caspase-3, and decreased expression of the anti-apoptotic protein Bcl-2 (Fig. 4C and F). In summary, TMEM100 overexpression inhibits the cell cycle and induces cell apoptosis in vitro.
Fig. 4.
Overexpression of TMEM100 inhibits the cell cycle and apoptosis of PCa cells and regulate FAK/PI3K/AKT signaling pathway. (A) Effects of TMEM100 overexpression on cell cycle by flow cytometry in DU145 and PC3 cells. (B) Effects of TMEM100 overexpression on cell apoptosis by flow cytometry in DU145 and PC3 cells. (C) Changes in the apoptotic and cell cycle-related proteins were detected by Western blot in DU145 and PC3 cells. (D) The analysis results of enrichment of KEEG pathway. (E) Changes in the FAK/PI3K/AKT signaling pathway proteins were detected by Western blot in DU145 and PC3 cells. (F) The qualification of apoptotic and cell cycle-related proteins by densitometry (G) The qualification of FAK/PI3K/AKT signaling pathway proteins by densitometry. *P<0.05 VS control group.
TMEM100 regulates the FAK/PI3K/AKT pathway
To further elucidate how TMEM100 affects PCa progression, we performed KEEG analysis of TMEM100 in PCa. As is shown in Fig. 4D, the top affected canonical signaling pathways were found: cGMP-PKG, Focal adhesion (FAK), Proteoglycans in cancer, and the PI3K/AKT signaling pathway. Both PI3K and AKT are downstream genes of the FAK signaling pathway. Hence, our study revealed an effect of TMEM100 on FAK/PI3K/AKT signaling. Western blot analysis showed that phosphorylation of FAK (p-FAK), PI3K (p-PI3K), and AKT (p-AKT) decreased in PC3 and DU145 cells after TMEM100 overexpression (Fig. 4E and G). These results indicate that TMEM100 is a regulator of the FAK/PI3K/AKT pathway, as predicted by our KEEG analysis.
TMEM100 overexpression inhibits PCa cell growth in vivo
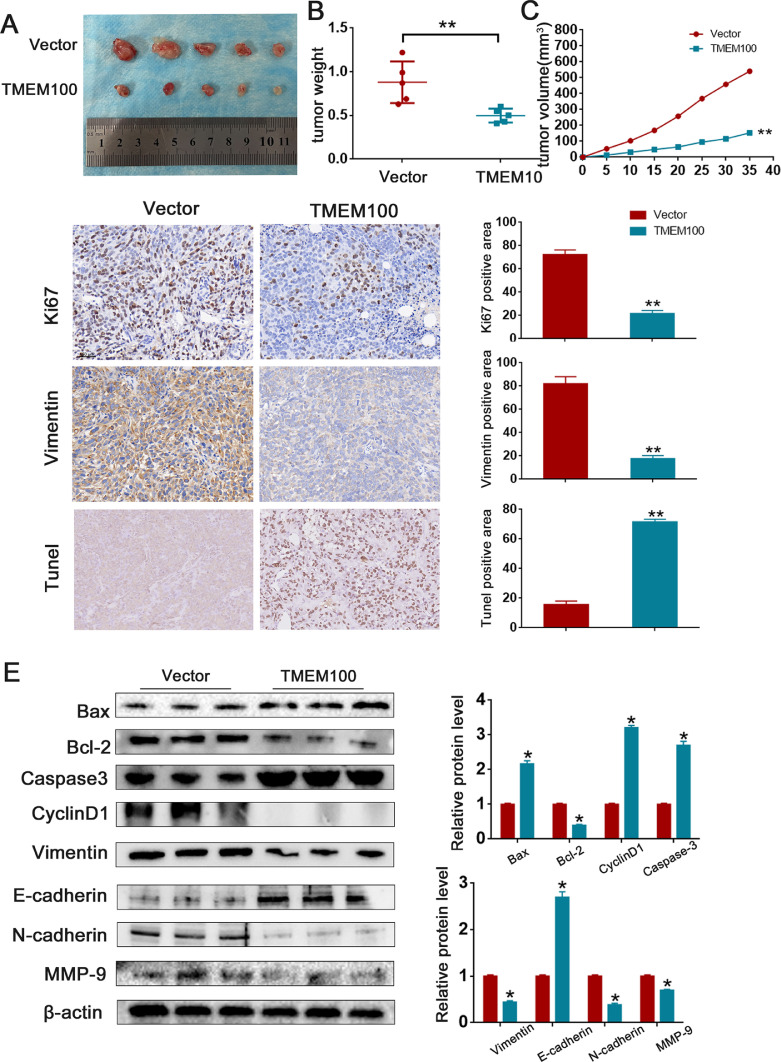
Our results indicate that TMEM100 negatively regulates cell growth in vitro. We further explored its role in the modulation of PCa progression in vivo. PC3/Lv-TMEM100 cells and PC-3/Lv-vector cells were injected subcutaneously into nude mice. As shown in Fig. 5A-C, tumors in the TMEM100 group grew slower than those in the control group. Furthermore, tumors in mice injected with Lv-TMEM100 cells showed a significant regression of tumor growth compared with the control mice. Western blot, Tunel Assay, and IHC staining showed that TMEM100 overexpression inhibits EMT, affects cell cycle distribution, and promotes cell apoptosis in vivo (Fig. 5D and E).
Fig. 5.
Overexpression of TMEM100 inhibits the growth of PCa cells in vivo.
(A) Representative images of xenograft tumors in nude mice. (B) The tumor weight of xenografts. (C) The growth curves of xenografts. (D) TUNEL staining images and IHC of Vimentin and Ki67 in control group and TMEM100 overexpression group. Scale bars = 200 μm. (E) Changes in the apoptotic, EMT and cell cycle-related proteins were detected by Western blot in vivo. *P<0.05, **P<0.01 VS control group.
TMEM100 participates in PCa progression by regulating SCNN1D via the FAK/PI3K/AKT signaling pathway
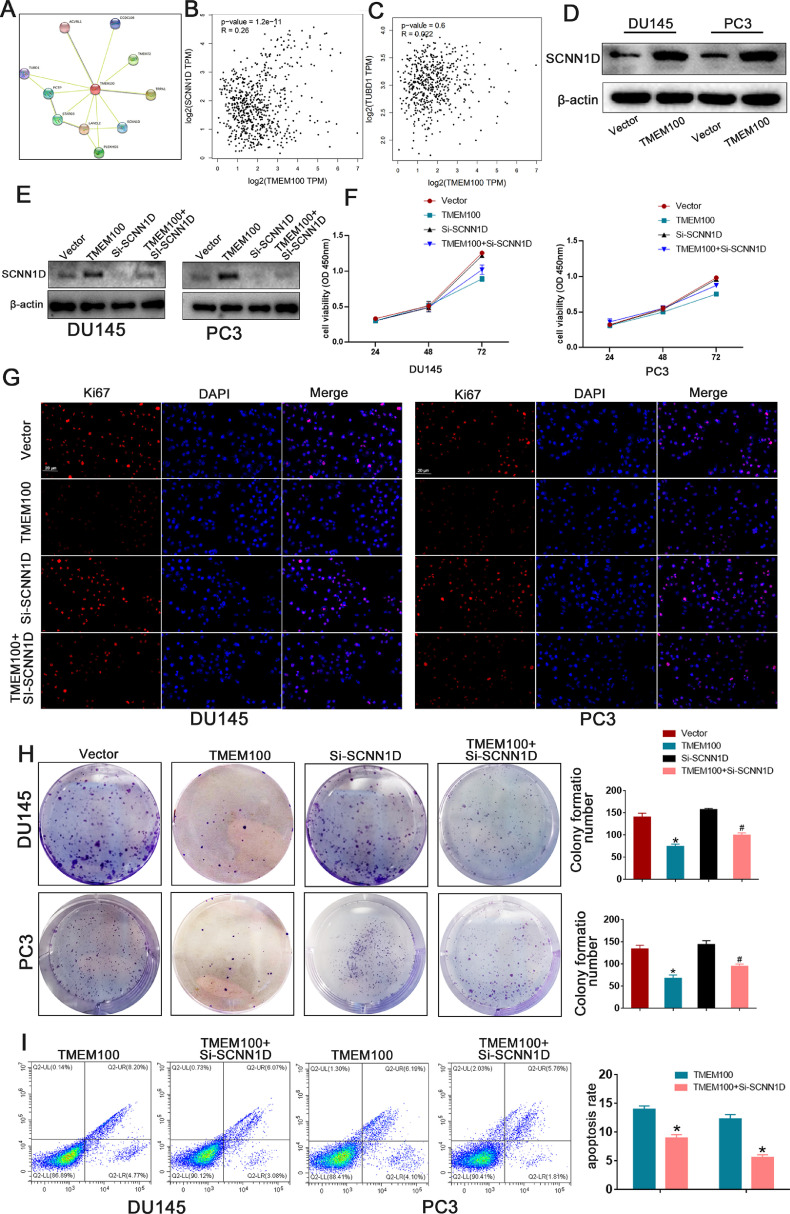
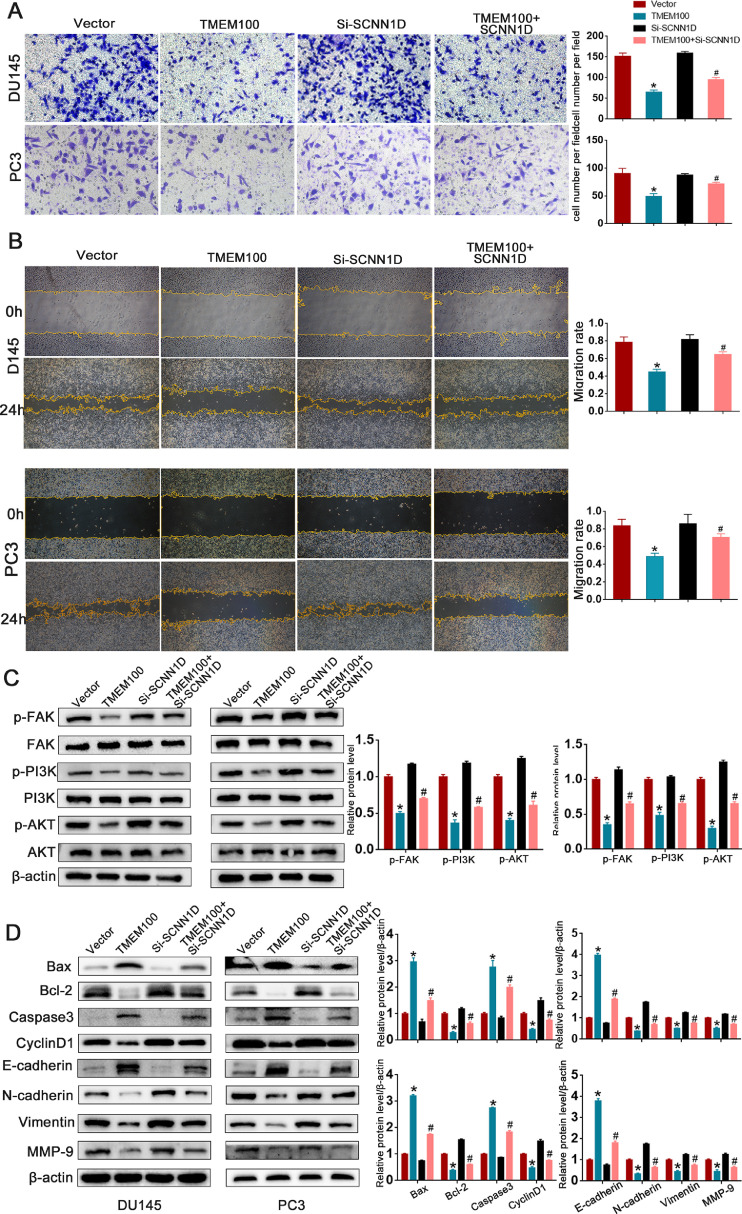
We explored the mechanism underlying the correlation between decreased TMEM100 expression and PCa progression. STRING results revealed that TMEM100 may interact with SCNN1D to regulate PCa cell growth, and a significant positive correlation between TMEM100 and SCNN1D was observed in PCa tissues (Figs. 6A and B). Correlations between TMEM100 and other genes (TUBD, TRPA1) were also explored, but we have not discovered a statistically significant correlation (Fig. 6C). Western blot analysis revealed that TMEM100 substantially increased SCNN1D expression compared to that of the controls in Fig. 6D. We also used siRNA to knockdown SCNN1D expression, and the western blot affirmed siRNA efficiency in DU145 and PC3 cells (Fig. 6E). CCK8, colony formation, Ki67, and cell apoptosis assays were implemented to explore concrete mechanisms by which the TMEM100/SCNN1D interaction influences cell behavior. SCNN1D downregulation accelerated proliferation and inhibited apoptosis of PCa cells in response to TMEM100 overexpression (Fig. 6F-I). Wound-healing and transwell assays were used to verify whether TMEM100 affected cell migration via SCNN1D. As shown in Fig. 7A and B, TMEM100 overexpression could inhibit PCa cell migration, while SCNN1D downregulation could rescue PCa cell migration ability in TMEM100 overexpressing cells. The above results were consistent with western blot analysis (Fig. 7D). As shown in Fig. 7C, western blot analysis results showed that transfection with si-SCNN1D also increases FAK/PI3K/AKT signaling in TMEM100-overexpressing PCa cells. Overall, these results indicate that TMEM100 can inhibit PCa progression via SCNN1D through the FAK/PI3K/AKT pathway.
Fig. 6.
TMEM100 inhibits PCa proliferation and apoptosis by regulating SCNN1D. (A) The analysis of STRING showed that TMEM100 interacted with SCNN1D. (B) Data derived from the GEPIA database showed that there was a significantly positive correlation between TMEM100 and SNCC1D. (C) The correlation between TMEM100 and TUBD1 was also detected. There was no significantly correlation between TMEM100 and TUBD1 in PCa tissues. (D) The western blot analysis showed that enhanced TMEM100 expression increased SCNN1D expression. (E)The expression level of SCNN1D was detected by western blot. (F-H) The depletion of SCNN1D increases proliferation in TMEM100-overexpression DU145 and PC3 cells. (I) The depletion of SCNN1D inhibits apoptosis in TMEM100- overexpression DU145 and PC3 cells. Scale bars = 20 μm. *P<0.05 VS control group, #P<0.05 VS TMEM100 overexpression group.
Fig. 7.
TMEM100 inhibits migration, invasion and EMT by regulating SCNN1D through FAK/PI3K/AKT signaling pathway. (A-B) The depletion of SCNN1D increases migration and invasion ability in TMEM100-overexpression DU145 and PC3 cells. (C) The depletion of SCNN1D rescue the function that TMEM100 overexpression hindered the protein level of p-FAK, p-PI3K and p-AKT. (D) TMEM100 affected cell apoptosis, cell cycle and EMT via regulating SCNN1D by western blot analysis. *P<0.05 VS control group, #P<0.05 VS TMEM100 overexpression group.
Discussion
PCa is the most diagnosed cancer among males and imposes a heavy social and economic burden worldwide [19]. Despite advances in detection techniques and treatment strategies, the prognosis for PCa patients with metastasis remains bleak. This study identifies an underlying mechanism by which decreased TMEM100 expression affects PCa progression. Our results indicate that TMEM100 overexpression inhibits proliferation, migration, and invasion via the FAK/PI3K/AKT signaling pathway, and SCNN1D may play a vital role in this process in PCa.
Tumor invasion and metastasis, the most important features of malignant tumors, are extremely complex biological behaviors [20,21]. TMEM100 belongs to the TMEM protein family, and is widely expressed in the notochord, ventral region of the neural tube, endocardium, and the mammary glands [9,22,23]. Previous studies have shown that TMEM100 inhibits differentiation and promotes autophagy in lung cancer through PI3K/AKT signaling pathway [14]. TMEM100 participates in the formation and maintenance of blood vessels, and inhibits tumor cell metastasis in various cancers [24]. In the present study, we found that TMEM100 expression was lower in PCa samples compared to their paired prostate samples and was highly correlated with clinical outcomes in PCa patients. In addition, TMEM100 overexpression inhibited PCa cell proliferation and metastasis and exacerbated apoptosis. Finally, Lv-vector and Lv-TMEM100 cells were injected subcutaneously into nude mice to investigate the effect of TMEM100 on PCa progression in vivo. Tumor volumes in LV-TMEM100 mice were significantly smaller than those in the LV-vector group, indicating that TMEM100 regulates tumor progression in vivo and in vitro.
The human SCNN1D gene is a homolog of the degenerins in Caenorhabditis elegans, which comprise an epithelial sodium channel superfamily with the other counterparts [25]. In addition, previous studies have described SCNN1D as a vital regulator of cell migration that mediates alveolar epithelial type 2 progenitor cells [26]. SCNN1D overexpression in glioma cells can inhibit migration and proliferation, and can increase cell accumulation in G1 phase via a MAPK signaling dependent mechanism [27]. STRING and GEPIA analyses indicate that TMEM100 interacts with SCNN1D to regulate PCa cell growth, and a significant positive correlation between TMEM100 and SCNN1D was observed in PCa tissues. Western blot analysis affirmed that the SCNN1D level increases with TMEM100 overexpression. Our study showed that SCNN1D, positively regulated by TMEM100, inhibited PCa cell proliferation and metastasis. Knockdown of SCNN1D expression rescued cell viability, clone formation, migration, and invasion capability.
FAK/PI3K/AKT signaling pathway plays an important role in PCa progression, and dysregulation of this pathway affects many biological processes, including cell migration, apoptosis, and inflammatory cell infiltration [28,29]. Jing Luo et al. [8] found that 14,15-epoxyeicosatrienoic acid induces breast cancer cell EMT via FAK/PI3K/AKT signaling. KEEG results demonstrated that TMEM100 was significantly associated with FAK signaling. Moreover, TMEM100 overexpression inhibited FAK phosphorylation, thereby decreasing PI3K and AKT phosphorylation and activity. Western blot analysis indicated that the degree of FAK, PI3K, and AKT phosphorylation were significantly decreased by TMEM100 overexpression, and this effect was inhibited after SCNN1D knockdown. In summary, TMEM100 overexpression regulates PCa progression through FAK/PI3K/AKT signaling, and SCNN1D may play a vital role in this process.
One limitation of our research is that the existing experimental results do not prove that SCNN1D has a direct regulatory relationship with FAK. We will explore the relationship between SCNN1D and FAK/PI3K/AKT signaling in PCa in our future work. In conclusion, we found that TMEM100 expression was downregulated in PCa tissues and TMEM100 overexpression inhibited PCa progression by regulating FAK/PI3K/AKT signaling, and SCNN1D may play a vital role in this process (Fig. 8). Our results may provide new insights regarding the relationship between TMEM100 and FAK/PI3K/AKT signaling pathway that should help in understanding tumorigenic mechanisms in PCa, and serve as a new therapeutic target in the future.
Fig. 8.
TMEM100 inhibited prostate cancer progression by regulating the FAK/PI3K/AKT signaling pathway via SCNN1D.
Funding
This research was supported by grants from National Natural Science Foundation of China (81870471 and 81800617) and Science and Technology Major Project of Hubei Province(2019AEA170).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
this study protocol was approved by the Ethics Committee of the Renmin Hospital of Wuhan University (approval number: WDRM2019-Y068). Animal Studies: This study was permitted by the Animal Experiment Ethics Committee of Wuhan University (approval number: WDRM-20210806) and was performed in accordance with the Declaration of Helsinki.
Patient consent for publication
Not applicable.
CRediT authorship contribution statement
Zehua Ye: Conceptualization, Methodology, Software, Data curation, Writing – original draft, Validation. Yuqi Xia: Writing – review & editing, Visualization. Lei Li: Visualization, Investigation. BoJun Li: Software, Investigation. Wu Chen: Software, Investigation. Shangting Han: Data curation, Investigation. Xiangjun Zhou: Visualization, Investigation. Lijia Chen: Formal analysis, Investigation. Weimin Yu: Data curation, Investigation. Yuan Ruan: Conceptualization, Visualization, Investigation, Supervision. Fan Cheng: Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Not applicable.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101578.
Appendix. Supplementary materials
References
- 1.Sandhu S., et al. Prostate cancer. Lancet North Am. Ed. 2021;398(10305):1075–1090. doi: 10.1016/S0140-6736(21)00950-8. [DOI] [PubMed] [Google Scholar]
- 2.Zhu Y., et al. Epidemiology and genomics of prostate cancer in Asian men. Nat. Rev. Urol. 2021;18(5):282–301. doi: 10.1038/s41585-021-00442-8. [DOI] [PubMed] [Google Scholar]
- 3.Chang A.J., et al. High-risk prostate cancer-classification and therapy. Nat. Rev. Clin. Oncol. 2014;11(6):308–323. doi: 10.1038/nrclinonc.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shorning B.Y., et al. The PI3K-AKT-mTOR Pathway and Prostate Cancer: at the Crossroads of AR, MAPK, and WNT Signaling. Int. J. Mol. Sci. 2020;21(12) doi: 10.3390/ijms21124507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng Y., et al. Depression promotes prostate cancer invasion and metastasis via a sympathetic-cAMP-FAK signaling pathway. Oncogene. 2018;37(22):2953–2966. doi: 10.1038/s41388-018-0177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye J., et al. Breviscapine suppresses the growth and metastasis of prostate cancer through regulating PAQR4-mediated PI3K/Akt pathway. Biomed. Pharmacother. 2020;127 doi: 10.1016/j.biopha.2020.110223. [DOI] [PubMed] [Google Scholar]
- 7.Sun W., et al. Long non-coding DANCR targets miR-185-5p to upregulate LIM and SH3 protein 1 promoting prostate cancer via the FAK/PI3K/AKT/GSK3beta/snail pathway. J. Gene Med. 2021;23(7):e3344. doi: 10.1002/jgm.3344. [DOI] [PubMed] [Google Scholar]
- 8.Luo J., et al. 14, 15-EET induces breast cancer cell EMT and cisplatin resistance by up-regulating integrin alphavbeta3 and activating FAK/PI3K/AKT signaling. J. Exp. Clin. Cancer Res. 2018;37(1):23. doi: 10.1186/s13046-018-0694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weng H.J., et al. Tmem100 Is a Regulator of TRPA1-TRPV1 Complex and Contributes to Persistent Pain. Neuron. 2015;85(4):833–846. doi: 10.1016/j.neuron.2014.12.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marx S., et al. Transmembrane (TMEM) protein family members: poorly characterized even if essential for the metastatic process. Semin. Cancer Biol. 2020;60:96–106. doi: 10.1016/j.semcancer.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 11.Hayez A., et al. High TMEM45A expression is correlated to epidermal keratinization. Exp. Dermatol. 2014;23(5):339–344. doi: 10.1111/exd.12403. [DOI] [PubMed] [Google Scholar]
- 12.Schmit K., Michiels C. TMEM Proteins in Cancer: a Review. Front. Pharmacol. 2018;9:1345. doi: 10.3389/fphar.2018.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y., et al. TMEM17 promotes malignant progression of breast cancer via AKT/GSK3beta signaling. Cancer Manag. Res. 2018;10:2419–2428. doi: 10.2147/CMAR.S168723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H., et al. TMEM100 Modulates TGF-beta Signaling Pathway to Inhibit Colorectal Cancer Progression. Gastroenterol. Res. Pract. 2021;2021 doi: 10.1155/2021/5552324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han Z., Wang T., Han S., et al. Low-expression of TMEM100 is associated with poor prognosis in non-small-cell lung cancer. Am. J. Transl. Res. 2017;9(5):2567–2578. 15. [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuang J., et al. TMEM100 expression suppresses metastasis and enhances sensitivity to chemotherapy in gastric cancer. Biol. Chem. 2020;401(2):285–296. doi: 10.1515/hsz-2019-0161. [DOI] [PubMed] [Google Scholar]
- 17.Gettings S.M., Maxeiner S., Tzika M. Two Functional Epithelial Sodium Channel Isoforms Are Present in Rodents despite Pronounced Evolutionary Pseudogenization and Exon Fusion. Mol. Biol. Evol. 2021;38(12):5704–5725. doi: 10.1093/molbev/msab271. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandhu S., Moore C.M., Chiong E., Beltran H., Bristow R.G., Williams S.G. Prostate cancer. Lancet. 2021;398(10305):1075–1090. doi: 10.1016/S0140-6736(21)00950-8. 18. [DOI] [PubMed] [Google Scholar]
- 19.Eccles S.A., Welch D.R. Metastasis: recent discoveries and novel treatment strategies. Lancet North Am. Ed. 2007;369(9574):1742–1757. doi: 10.1016/S0140-6736(07)60781-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta G.P., Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Moon E.H., et al. Generation of mice with a conditional and reporter allele for Tmem100. Genesis. 2010;48(11):673–678. doi: 10.1002/dvg.20674. [DOI] [PubMed] [Google Scholar]
- 22.Somekawa S., et al. Tmem100, an ALK1 receptor signaling-dependent gene essential for arterial endothelium differentiation and vascular morphogenesis. Proc. Natl. Acad. Sci. U S A, 2012;109(30):12064–12069. doi: 10.1073/pnas.1207210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moon E.H., Kim Y.H., Vu P.N., Yoo H., Hong K., Lee Y.J., Oh S.P. TMEM100 is a key factor for specification of lymphatic endothelial progenitors. Angiogenesis. 2020;23(3):339–355. doi: 10.1007/s10456-020-09713-1. [DOI] [PubMed] [Google Scholar]
- 24.Gettings S.M., et al. Two Functional Epithelial Sodium Channel Isoforms Are Present in Rodents despite Pronounced Evolutionary Pseudogenization and Exon Fusion. Mol. Biol. Evol. 2021;38(12):5704–5725. doi: 10.1093/molbev/msab271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao R., et al. Proliferative regulation of alveolar epithelial type 2 progenitor cells by human Scnn1d gene. Theranostics. 2019;9(26):8155–8170. doi: 10.7150/thno.37023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rooj A.K., McNicholas C.M., Bartoszewski R., et al. Glioma-specific cation conductance regulates migration and cell cycle progression. J. Biol. Chem. 2012;287(6):4053–4065. doi: 10.1074/jbc.M111.311688. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song G., et al. TIMP1 is a prognostic marker for the progression and metastasis of colon cancer through FAK-PI3K/AKT and MAPK pathway. J. Exp. Clin. Cancer Res. 2016;35(1):148. doi: 10.1186/s13046-016-0427-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fresno Vara J.A., et al. PI3K/Akt signalling pathway and cancer. Cancer Treat. Rev. 2004;30(2):193–204. doi: 10.1016/j.ctrv.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Ye Z., Xia Y., Zhou X., Li B., et al. CXCR4 inhibition attenuates calcium oxalate crystal deposition-induced renal fibrosis. Int. Immunopharmacol. 2022;107 doi: 10.1016/j.intimp.2022.108677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.