Abstract
Nearly one third of Saccharomyces cerevisiae protein coding sequences correspond to duplicate genes, equally split between small-scale duplicates (SSD) and whole-genome duplicates (WGD). While duplicate genes have distinct properties compared to singletons, to date, there has been no systematic analysis of their positional preferences. In this work, we show that SSD and WGD genes are organized in distinct gene clusters that occupy different genomic regions, with SSD being more peripheral and WGD more centrally positioned close to centromeric chromatin. Duplicate gene clusters differ from the rest of the genome in terms of gene size and spacing, gene expression variability and regulatory complexity, properties that are also shared by singleton genes residing within them. Singletons within duplicate gene clusters have longer promoters, more complex structure and a higher number of protein–protein interactions. Particular chromatin architectures appear to be important for gene evolution, as we find SSD gene-pair co-expression to be strongly associated with the similarity of nucleosome positioning patterns. We propose that specific regions of the yeast genome provide a favourable environment for the generation and maintenance of small-scale gene duplicates, segregating them from WGD-enriched genomic domains. Our findings provide a valuable framework linking genomic innovation with positional genomic preferences.
INTRODUCTION
Gene duplication is widely accepted to be a major source of genomic innovation, occurring with relatively high frequency, through various mechanisms and affecting large proportions of chromosomes or even entire genomes (1,2). Duplicated genes may be the result of either localized, small-scale duplication (SSD), through replication slippage or ectopic recombination (3), or of cataclysmic events of whole-genome duplication (WGD) leading to temporary polyploidy (4,5). Both phenomena are more common than one might expect (6,7) but genomes are not replete with multiple copies of genes. This is because once a gene is duplicated, it becomes subject to strong evolutionary constraints that vary in both type and extent. Functional redundancy and the energy cost of maintaining two identical genes leads to the most likely outcome, which is the subsequent erosion and eventual loss of one of the two copies, through the accumulation of deleterious mutations (8–10). In some cases, however, duplicated genes may diverge to the point of acquiring diversified functions, which leads to the fixation of both copies. This process also varies, depending on whether the two copies both diverge to assume subsets of the original functions of the ancestral gene (sub-functionalization) or if one adopts a novel role, while the other preserves the original (neo-functionalization) (11,12).
The way genomic and other constraints drive the fate of gene duplicates has been investigated in great depth in the case of the budding yeast Saccharomyces cerevisiae. Nearly one third of S. cerevisiae genes are the result of gene duplication, roughly half of which originate from a massive whole-genome duplication event that occurred ∼100 million years ago (13–15), while the rest are the result of continuously occurring, small-scale duplications. A number of properties appear to be particular to yeast duplicated genes in general. Since relatively early, they were shown to be over-represented among highly expressed genes (16), to reside in early replicating chromatin (17), to have more complex promoters (18,19) and, overall, fewer genetic interactions than non-duplicated, singleton genes (20).
The gene duplication mechanism may also be associated with different properties between ‘ohnologues’, that is whole-genome (WGD) and paralogs, which are small-scale duplicate (SSD) (21). Knock-out studies showed WGD genes to be less essential and genetic interaction profiling suggested WGD pairs to be functionally more similar than SSD ones (22). WGD are also s more likely to form part of protein complexes (23). SSD genes, on the other hand, were found to have greater expression divergence (19), to be more prone to accumulate non-synonymous substitutions (24), as well as new protein–protein interactions (25). Together these observations draw associations with the evolutionary fate of gene duplicates, with WGD being more likely to be maintained due to subfunctionalization and the maintenance of dosage balance, while SSD through the adoption of divergent expression patterns and the acquisition of novel functions (26).
Until now, most of the analyses have treated the two daughter genes as linked or entangled from their birth through an ‘invisible thread’, regardless of their genomic context. Nevertheless the genomic environment, in which a gene is embedded, is an important determinant of its function and evolution, shaped by epigenetic factors and access by transcriptional regulators in the 3D genomic landscape (27,28). Beyond sequence and functional conservation, the spatial organization of yeast genes has been the subject of extensive research. The conservation of pre- and post-WGD gene arrangement has been thoroughly documented in (29). The order of genes in linear chromosomes has been associated with the length of their intergenic spacers (30), as has the directionality of their transcription (31) and their co-expression in conserved clusters (32), while it has been suggested that large intergenic spacers may decouple the transcriptional interference between adjacent genes and thus increase their expression divergence (33). In the context of gene duplication evolution, reports have shown a strong tendency for WGD (but not SSD) to be located in genomic areas with increased non-synonymous substitution rate (mutational hotspots) (34). WGD genes are also documented to very rarely undergo small-scale duplication (35). Experimental evidence for the evolutionary fate of gene duplication being context-dependent came from an innovative approach (36), in which a duplicated copy of the IFA38 gene was found to be more likely to escape deletion when positioned in tandem with the ancestral gene.
It is, thus, quite plausible that the position in which a gene is found and the overall genomic ‘context’ may shape not only its regulatory and expression patterns, but also its evolutionary fate. Even more so, one may hypothesize, that particular areas of the genome may represent more or less ‘permissive’ environments for the occurrence and maintenance of gene duplicates. Having previously identified positional preferences for genes associated with sequence composition (37) gene regulation (28,38) and chromatin structure (39), in this work we focus on such context-dependent properties for gene duplicates. Starting from the observation of extensive gene spatial clustering for both WGD and SSD genes, we go on to define genomic domains of WGD/SSD enrichment and to examine how the properties of these regions may be affecting not only the duplicate but also the singleton genes they harbour. We find SSD and WGD being largely segregated in distinct parts of the yeast genome. We further uncover a number of structural, regulatory and functional properties of gene duplicates that are domain-specific and which are, in addition, partially shared by neighbouring singleton genes. Our findings may support a model for the evolution of gene duplication events, according to which, the yeast genome may be divided in areas with differential capacity for genetic innovation, driven primarily by the divergence of the genes’ regulatory sequences. We find that SSD duplication preferentially occurs in confined areas of the yeast genome that constitute genomic ‘niches’ favourable for faster divergence, thus enabling the emergence of novel functions.
MATERIALS AND METHODS
Genome segmentation into duplicate gene-enriched clusters
We obtained SSD and WGD lists and genomic coordinates, as compiled by Fares et al. (25). Chromosomal coordinates of yeast genes were obtained from UCSC using the sacCer2 assembly (June 2008) to achieve maximum compatibility with all the datasets used. Out of these we were able to compile a large number of structural, transcriptional and regulatory properties for a subset of 5799 genes that precluded non-protein coding genes and most of S. cerevisiae's dubious ORFs.
We then proceeded to partition the yeast genome in three major compartments consisting of: (a) SSD-enriched clusters, (b) WGD-enriched clusters and (c) the remaining part of the genome. SSD and WGD gene clusters were created by extending the coordinates of each duplicate gene for 10 kb in both directions. These segments were merged with adjacent segments containing the same type of genes to form duplicate-enriched clusters. Mixed genomic regions, in which SSD- and WGD-clusters overlapped, were then assigned to the status of the most prevalent gene type and consequently merged to the adjacent cluster of the same type. That is, for a region lying between clusters of different type, we counted the number of SSD and WGD genes and converted the segment to the type having the greatest number of genes. Duplicate genes that were farther than 10kb from a gene of the same type (SSD or WGD) were assigned as solitary-gene clusters. These constituted less than 10% of the total duplicate genes (94/1032 for SSD and 83/1090 for WGD). Gene clusters were thus defined on the basis of self-containment prerequisite and each cluster extended in order to contain the minimal amount of contiguous genes, that allowed it to include all duplicate genes within a certain distance threshold. Regions of the genome that were not associated with either SSD or WGD genes, were assigned as ‘Complement’ clusters, exclusively comprising singleton genes.
The resulting segmentation partitions the genome in three types of domains with comparable extent, with 29% of the genome belonging to SSD Clusters, 32% to WGD Clusters and the remaining 39% to Complement Clusters (the percentages of contained genes were almost identical). The spatial distribution, gene content and location in the genome of these clusters is highly non-random and may be summarized in Figures 1D–F.
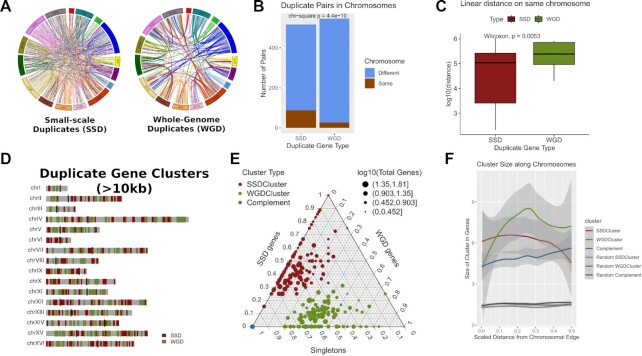
Figure 1.
Duplicate genes segregate into different regions of the yeast genome. (A) Circos Plots joining gene duplicate pairs in the yeast genome. Left: Whole Genome Duplicates (WGD), Right: Small-Scale Duplicates (SSD). Light blue lines join duplicate pairs on different chromosomes, while brown lines join pairs located on the same chromosome. (B) Number of gene duplicate pairs found on the same or on different chromosomes. (C) Distribution of linear distances (log10-transformed) between gene duplicate pairs that are located on the same chromosomes. (D) Genome domainogram showing the locations of gene duplicate clusters along the sixteen chromosomes of the yeast genome. For reasons of clarity, only clusters with length greater or equal to 10kb are shown. (E) Ternary plot showing the gene content of duplicate and complement clusters. The scale on each side of the triangle is relative and corresponds to the gene content in one of SSD, WGD and singleton gene categories. The colour of the dots corresponds to the cluster type. As all Complement Clusters contain exclusively singleton genes they all converge to one point at the lower left vertex of the plot. Size of the points corresponds to the size of the cluster in total number of genes. Notice how SSD and WGD clusters are occupying different parts of the plot, suggestive of their relative purity in duplicate genes of a specific type. (F) Composite plot of actual and random duplicated gene clusters showing the average cluster size (in number of genes) along a scaled distance from the chromosomal edge. Shaded bands correspond to standard error of the mean.
Genomic coordinates in one and three dimensions
Distances to the centromeres and the chromosomal edges were calculated as ratios over the entire chromosomal arm length as described in (28). Coding density was calculated as the percentage of coding sequences spanning a region of eleven genes, symmetrically flanking the gene in question.
For the three dimensional coordinates we used the published conformational model of the yeast genome (40) which has been resampled at gene resolution. We obtained gene positions by linearly interpolating the model's control points to approximate the center base pair of each gene, which resulted in each gene being represented as a set of three coordinates in arbitrary space. Assuming the mean coordinates of all genes to correspond to the center of the genome in 3D space, we calculated its euclidean distance from each gene and then took quantiles of these distances to assign genes into three sections: Central, bottom quartile (lowest 25% of the distances), Intermediate, middle half (>25% and < 75% of the distances) and Peripheral, top quartile (highest 25% of the distances).
Enrichment analysis
Enrichments of genome coordinates and set overlaps were calculated as described in (41). The positional overlaps between gene coordinates were calculated with BedTools (42) and the enrichment was defined as the observed-over-expected ratio of the coordinates’ overlap. The expected value was calculated as the product of the two independent proportions of gene coordinates over the total genome size. Significance was assessed with a permutation test, by shuffling the smaller of the two coordinate sets, while keeping the same number and size distribution of its segments. In all cases, one thousand such shuffles were performed. The reported P-values corresponded to the proportion of times a random shuffle yielded a value more extreme than the one observed.
Gene age
The phylogenetic age of S. cerevisiae genes was determined by phylostratigraphy. We performined BLASTP (43) searches on all available Fungi proteomes in GenBank (794 unique species excluding S. cerevisiae, 1266 total proteomes, downloaded December 2019) with an E-value cutoff of 1E–3. Age was defined as the most recent common ancestor of species that shared a homologue. The NCBI Taxonomy common tree was used, resulting in genes classified in the following phylogenetic ages: species-specific, genus (Saccharomyces), family (saccharomycetaceae), order (saccharomycetales), division (ascomycota) or kingdom (fungi).
Conservation, sequence divergence and structural constraint data
Sequence conservation was measured using phastCons (44) precalculated scores for an alignment of seven Saccharomyces species. Divergence between duplicate gene pairs was obtained from (25) and was calculated at the level of amino acid sequence, taking into account the age of the duplicate (for SSD genes).
Structural constraints were assessed with the use of two models: the deep learning model of Routhier and colleagues (45) which attempts to capture the effect of nucleotide substitutions on nucleosome positioning and our ownab initio model based on the variability of nucleosome positioning predictive scores (46).
Aggregation of phastCons and structural constraint values was performed through the calculation of a mean score over the segment under question, thus taking the average of all single-nucleotide-resolution values over the complete length of each gene. Average gene profiles were created as vectors of binned averaged values for 1000-nucleotide regions symmetrically flanking each gene's transcription start site, in bins of 10 nt.
Nucleosome positioning and gene regulation
Nucleosome positioning data were obtained from a genome wide MNase profiling (47). Average gene profiles were created as described above for conservation scores and for the same regions and bin size. Nucleosome positioning similarities for duplicate gene pairs were calculated as Pearson correlation coefficients of the nucleosome positioning 10nt-bin profiles.
We used a dataset of highly reliable conserved transcription factor binding sites (48) to assign a set of transcription factors (TF), to each gene in our duplicate dataset. Transcriptional regulation similarity between gene duplicate pairs was calculated as the Jaccard index of their corresponding TFs. The Jaccard similarity between two sets, is defined as the ratio of the size of their intersection over that of their union.
Co-expression scores and transcriptional variability
We obtained normalized expression data from a compendium of ∼2400 experimental conditions from the SPELL database (49). We used SPELL’s pre-calculated Adjusted Co-expression Score (ACS) as a measure of gene co-expression. In order to assess expression variability, we calculated the standard deviation of gene expression levels for each gene across all conditions and then normalized it across genes with the use of a z-score.
Protein complexity, protein–protein interactions and functional enrichment
We used the PFAM database (50) API to assign the predicted functional protein domains to the protein sequences of the complete set of genes. For each protein we then calculated the proportion of its sequence being attributed to a PFAM domain as a proxy for protein complexity. Protein-protein interactions were obtained from the STRING database (51). Functional entanglement was assessed as the number of GO terms associated with each gene. Functional enrichments were assessed with the use of gProfiler (52) and suitably adopted custom R functions.
Association rules
Association rules were extracted from the sets of genes against transcriptional regulators using the data by MacIsacc and colleagues (48) (see above). We employed the arules R package (53,54) through the Apriori algorithm and extracted the top 10% most significant associations in terms of lift values. Lift corresponds to the strength of the association between regulator and gene target, controlling for the prevalence of the regulator in the dataset. The results were presented in the form of association networks between regulators.
Additional methods
Compiled datasets and code used in this study, as well as a description of the methodology may be found at the accompanying github site: https://github.com/christoforos-nikolaou/YeastSegmentationDuplicates.
RESULTS
Duplicate genes are segregated in the yeast genome
Because of their mode of duplication, which predominantly takes place in tandem, small-scale duplicate (henceforth SSD) gene pairs tend to be found in close proximity to each other and are also more often located on the same chromosome (Figure 1A, B). As expected, SSD genes, when found on the same chromosome, are very often juxtaposed to each other (Figure 1C). On the other hand, whole-genome duplicate (from now on WGD) genes are preferentially localized in regions that maintain synteny and which conserve the ancestral juxtaposition (Figure 1A). Both types of genes are therefore likely to cluster in linear genomic space. Our starting hypothesis was that clusters of SSD and WGD genes occupy distinct areas of the yeast genome.
In order to quantify this observation, we created clusters of SSD and WGD genes by merging genomic regions containing genes of the same type within a distance of 10kb (see Methods). One first indication for the strong clustering of duplicate genes was that the size distribution of the created, extended regions was larger than the corresponding ones for random selections of equal numbers of genes (P ≤ 0.001 for 1000 random permutations). The created SSD/WGD-clusters clearly occupy distinct genomic spaces in the yeast genome (Figure 1D).
Even if this segregation is, to some extent, expected by the way these clusters are constructed, one can see that the clusters’ both relative and absolute positions on the genome, as well as their gene content, strongly deviate from randomness. The average number of SSD and WGD genes within their clusters is 3.1 and 3.4, respectively, which is more than 3-fold increased compared to the expected values of 0.86 and 0.91, if one assumes a uniform distribution of genes across the genome (P = 0.0017). Figure 1E shows a ternary plot with the gene content for the three types of genes (SSD, WGD and singletons) in each cluster type (Complement clusters exclusively contain singletons and thus all coincide in the low-left vertex of the plot). SSD and WGD clusters are distributed along their corresponding axes, which suggests increased ‘purity’ for genes of the same type (P = 0.003 and P = 0.0012 compared to random clusters for SSD and WGD respectively, Supplementary Figure S1). Their sizes, in terms of gene number, are also significantly larger from randomly created clusters, assuming the same number of duplicates and total number of clusters (Supplementary Figure S1).
The localization of SSD and WGD in different genomic areas has already been documented for vertebrates through the study of copy number variation (CNV), which may be largely responsible for the creation of SSD but to which WGD are largely refractory (35,55). In Figure 1F, we have plotted the average cluster size against its scaled distance from the corresponding chromosomal edge (see Methods) for both real and random gene clusters. It is clear that SSD clusters tend to occupy the edges of chromosomes, while the opposite is true for WGD ones, which appear to be enriched towards their centers. Complement clusters show only a small preference for the overall gene-denser chromosomal centers. These preferences are not an artefact of random gene content fluctuations as may be seen in a direct comparison with random clusters, which are also considerably smaller in size and are uniformly distributed along the chromosomal arms.
Duplicate gene clusters occupy regions with different architectural preferences
The segregation of SSD and WGD, shown in Figures 1D and F, effectively divides the genome in three domains, one for each type of gene duplicate and the remaining complementary part of the genome, exclusively containing singleton, non-duplicated, genes. We went on to analyze particular spatial tendencies for these three compartments of the yeast genome. The enrichment of SSD clusters towards the chromosomal edges, when compared to WGD-clusters and singletons is quantitatively supported by a comparison of the distributions of scaled distances from the chromosomal edges for genes residing in the three genomic domains (Supplementary Figure S2). Interestingly this tendency is also visible at the 3D level, with SSD clusters being preferentially positioned in the most peripheral regions of the genome according to the conformational model of (40) as opposed to more central positions for WGD ones (Supplementary Figure S3).
Duplicate genes are also found to have particular preferences in terms of size in both genic (the part of the gene between transcription start and end site) and their associated non-genic sequences. Gene length is directly associated with functional complexity as longer genes may accommodate a larger number of functional domains (56). In a similar way, the surrounding non-coding space may be associated with regulatory complexity, as longer promoters and gene upstream regions may provide the platform for a greater number of transcription factor binding sites (57). A simple comparison of gene lengths shows duplicate genes to be significantly longer than singletons (Figure 2A). This is partly explained by their enrichment in genes of greater age which are generally longer (Supplementary Figure S4) but also on the basis of the more complex evolutionary patterns of duplicate genes, which are more prone to maintain a large number of functions and thus greater length (20,58,59). Singleton genes have, on average, shorter lengths, independently of their position in the genome.
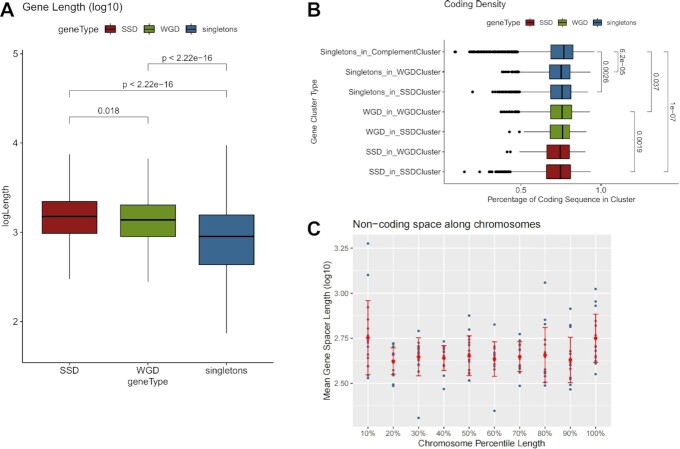
Figure 2.
Gene duplicates are localized in areas with more extended non-coding space. (A) Distribution of gene lengths (log10-transformed) for SSD, WGD and singleton genes. Significance is denoted with adjusted p-values of a Mann–Whitney test. (B) Distribution of coding density for regions around genes from different gene clusters (see text for details). Significant differences are denoted with adjusted P-values of a Mann–Whitney test. (C) Mean gene spacer length (log10-transformed) along 10 quantiles in each of the sixteen yeast chromosomes. Dots correspond to mean values for the given percentile for each chromosome. Red bars correspond to standard deviation.
From an evolutionary viewpoint, the size of the proximal non-coding sequences may be equally important for functional divergence through the establishment of novel regulatory relationships. Adjacent genes that involve SSD duplicate pairs tend to have wider spacing (33) while duplicate genes with small distances are more likely to be deleted (9). Both SSD and WGD are generally flanked by longer gene spacers (Supplementary Figure S5), but this is a feature that extends in duplicate gene clusters also affecting singletons. We compared the coding sequence density (see Materials and Methods) for the clusters of SSD, WGD and singleton genes (Figure 2B) and found SSD clusters to be located in regions of the genome with the largest proportion of non-coding sequences. Moreover, both WGD and singleton genes were found to be embedded in areas of smaller coding density when found in the SSD-type gene clusters.
An explanation for this observation is the proximity of SSD gene clusters to the chromosomal edges. In the past, we have reported coding density to drop near the ends of chromosomes (28) and indeed there is trend for longer gene spacers to be enriched towards the chromosomal edges (Figure 2C). These observations are suggestive of a general tendency of the genomic environment that does not only affect genes of one particular category. In all, we find that the lengths of both coding and non-coding sequences, associated with gene duplicates, are dependent on the broader genomic area, in which these are embedded.
SSD genes diverge at the regulatory level in a spatially-dependent manner
The increased size of non-coding spacers in gene duplicates and in particular SSD, prompted us to look closer into their gene upstream regions for possible sequence, structural and functional constraints. In order to assess sequence constraint, we created aggregate phastCons plots along the length of a region spanning 500bp either side of the transcription start site of each gene. In all eukaryotes, sequence conservation is, in general, lower in the gene upstream region because of relaxed sequence constraints in non-coding DNA. In genomes with small non-coding spacers between genes, such as yeast, a drop in sequence conservation is relatively sharp but no differences in this pattern are expected between genes. We were thus surprised to find that, the drop in conservation in the promoter region is sharper for both SSD and WGD genes compared to singletons, especially in the region immediately upstream of the gene's transcription start site (Figure 3A). This reduction in promoter sequence conservation, somewhat more pronounced in SSD compared to WGD, may be associated to more relaxed constraints in terms of gene regulation for duplicate genes against singletons, which is compatible with the general view of duplicate genes being more prone to diverge into new functions. What is more interesting, while coding sequence conservation appears to be independent from the region in which the gene is located, this is not the case for the reduced promoter conservation, which follows a strong spatial pattern along the genome and is associated with the surrounding duplicate gene environment (Figure 3B).
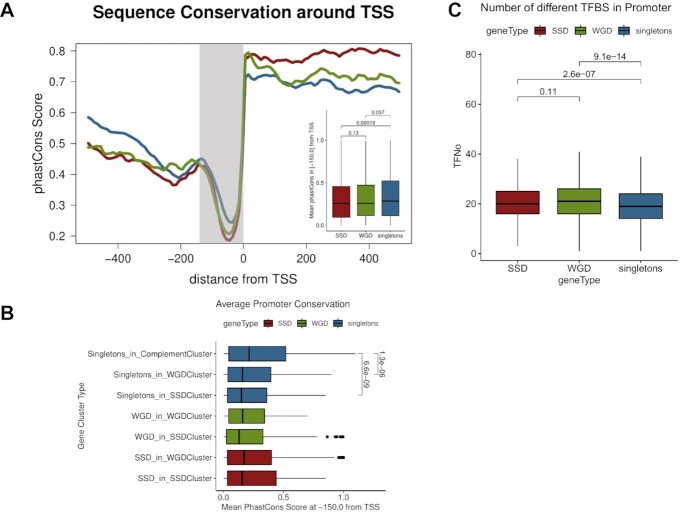
Figure 3.
Sequence constraints are more relaxed in the promoters of duplicate genes. (A) Aggregate mean conservation (phastCons) along a region spanning 500 bp either side of TSS for singleton, SSD and WGD genes. Embedded boxplot represents mean phastCons scores for the highlighted, proximal promoter region (150 bp upstream to TSS). Values in brackets correspond to adjusted P-values of a Mann–Whitney test. (B) Distribution of mean conservation in the proximal promoter region (150 bp upstream to TSS) measured as mean phastCons scores for different gene clusters. Values next to brackets denote adjusted P-values of a Mann–Whitney test. (C) Distributions of numbers of predicted transcription factor binding sites (TFBS) in the gene upstream regions (300 bp upstream to TSS) for SSD, WGD and singleton genes. TFBS obtained from a set of predictions based on comparative sequence analysis (48).
Singleton genes found in WGD clusters have less conserved promoters and this is further reduced in SSD clusters. It should be noted here that the estimated divergence between the duplicate gene pairs (25) is smaller in SSD genes compared to WGD ones, regardless of their position in the genome. Thus the apparent, more relaxed constraints in the promoter region are more likely to be associated with gene regulation and not overall higher divergence rates. We found support for this in the increased number of transcription factor binding sites (TFBS) identified at the promoters of different gene types. Both SSD and WGD have significantly greater numbers of conserved TFBS in an equally-sized gene upstream region (up to 300 bp upstream of the TSS) when compared to singleton genes (Figure 3C).
Together, these observations suggest that, the combination of more extensive non-coding space in SSD clusters and relaxed sequence constraints in the promoters of the contained genes, may be the primary driving force for their functional divergence, occurring primarily at the regulatory level. Furthermore, there are strong indications of a spatially dependent relaxation of promoter constraints that occurs in areas enriched for SSD genes and which affects both singletons and WGD genes as well.
Increased chromatin structural complexity for gene duplicates extends in broad genomic regions
We have recently described the organization of the yeast genome in extended regions with similar nucleosomal patterns which are, in addition, associated with a number of functional and regulatory characteristics (39). We used a public dataset (47) to analyze the nucleosome occupancy patterns around the transcription start sites of SSD, WGD and singleton genes. The results showed marked differences in the gene upstream regions with both types of duplicate genes having more ‘shallow’ nucleosome free regions (NFR), compared to a clear and deep NFR for singleton genes (Figure 4A). Notably, this is not affected by the sample size, as the mean nucleosome occupancy at the promoter remains significantly lower even for a random selection of 1000 singleton genes (Supplementary Figure S6). Moreover, it appears to be position-independent as singleton genes have strong NFRs, regardless of the cluster they are found in.
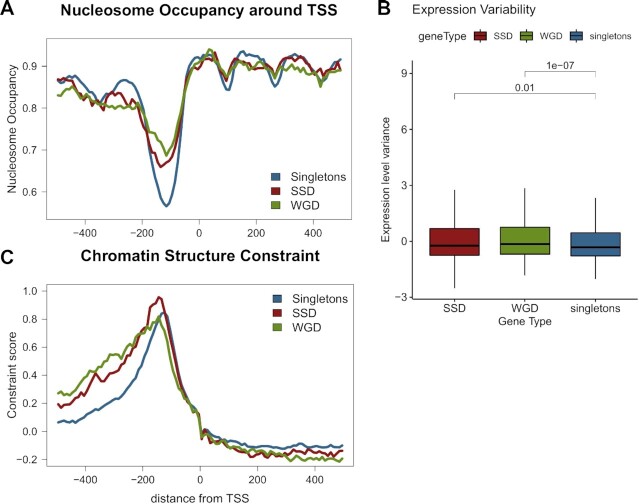
Figure 4.
Nucleosome positioning patterns in gene duplicates suggest more complex promoter structure. (A) Mean nucleosome occupancy along a region spanning 500 bp either side of TSS for singleton, SSD and WGD genes. Nucleosome occupancy was calculated in one hunded 10 bp bins, as the fraction of the region overlapping a positioned nucleosome. Positions obtained from (47). (B) Distribution of expression variability for SSD, WGD and singleton genes. Expression variability was assessed as the z-score of the variance of gene expression values from the SPELL database. Values over brackets denote adjusted P-values of a Mann–Whitney test. (C) Chromatin structure constraint score as measured with the mutation score (45) along a region spanning 500 bp either side of TSS for singleton, SSD and WGD genes.
Genes with strong NFR are generally subject to less complex regulation, as they do not require chromatin remodelling to allow for transcriptional activation by regulators (60,61). They are thus enriched among constitutively expressed genes with more stable expression levels. Indeed, this is supported by our assessment of transcriptional variability on the basis of data collected by the SPELL database (49), which show significantly smaller variability in mRNA levels for singleton genes compared to both SSD and WGD ones (Figure 4B). As the variability in mRNA levels is here measured between a broad spectrum of different conditions it may be seen as a proxy for expression plasticity, which is found to be increased in the case of duplicate genes. This comes in agreement with the observation of lower TFBS number in singletons.
The way sequence constraints may be affecting the positioning of nucleosomes has been the focal point of both experimental (60,62) as well as computational works (61,63). In the past, we have suggested an ab initio method to assess structural constraints on the primary DNA sequence (46) based on our own model for nucleosome positioning (64). A more recent work, using a deep learning model, has presented evidence of strong sequence constraints affecting nucleosome positioning in gene upstream regions (45). We used both this deep learning model of mutation impact on nucleosome positioning, as well as our own model of structural robustness (46) (see Materials and Methods) to assess chromatin-related constraints around the TSS of duplicate genes and singletons. Both analyses produced similar patterns that show more elevated constraints in the gene upstream regions of duplicate genes compared to singletons. (Figure 4C, Supplementary Figure S7).
In the case of duplicated genes the constraints were also more extended upstream, which may be related to the overall larger size of their gene upstream regions (see above). The increased structural constraints of gene duplicates are also spatially associated, in a way similar to sequence constraints. What should be noted is that most of the constraint observed for singletons is due to singleton genes found in SSD and WGD clusters (Supplementary Figure S8), as the mean structural conservation of singletons in SSD/WGD clusters is significantly higher than the one of singletons in the rest of the genome (Supplementary Figure S9). Together, these observations point towards strong structural constraints in the promoters of genes that are found in the areas of the genome, where duplicate genes are preferentially positioned.
Structural constraints at the promoter are independent of sequence conservation but may be shaping the gene expression of gene duplicates
The existence of structural constraints in duplicate gene clusters appears to contrast the relaxed sequence conservation in their promoters. We went on to examine the association between the two and were not surprised to observe a significant negative correlation (overall Spearman's rho = −0.229, P ≤ 10−57). This inverse relationship between sequence conservation and structural constraint at the gene promoters appears to be a general characteristic of all genes, even though it is stronger for singletons and SSD compared to WGD (Figure 5A). Thus, it appears that a relaxation at the sequence level may be counteracted at the level of chromatin structure.
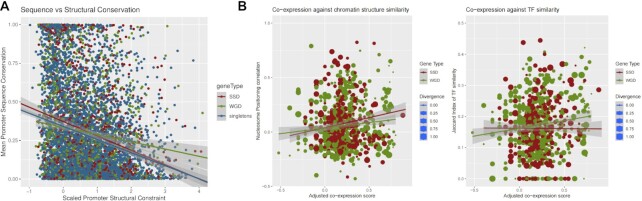
Figure 5.
Increased structural constraints are associated with gene duplication status. (A) Two-dimensional density plot of sequence constraint (as mean conservation score) versus structural constraint (as mean mutation score by (45) for the same regions of 200 bp to 50bp upstream of the TSS for all yeast genes (overall Spearman's rho = −0.229, P ≤ 10−57). Each line corresponds to a linear regression fit. Shaded band corresponds to standard error. (B) Left: two-dimensional scatterplots of gene pair co-expression, measured as scaled Adjusted co-expression Score (obtained from SPELL) against nucleosome positioning correlation, measured as the Pearson correlation coefficient of nucleosome occupancy profiles between gene pairs. Right: Gene pair co-expression against TF similarity measured as the Jaccard index of TFBS found in the promoters of each gene pair. Red: SSD gene pairs, Green: WGD gene pairs. Size of points proportional to the estimated diversity between the genes in each pair.
What is more, the inverse relationship between sequence conservation and structural constraint in the gene upstream regions, denotes two distinct promoter architectures. On one hand there are more conserved, small promoters with clear nucleosome-free regions pertaining to singleton genes, which overall have less complex regulation and smaller expression variability. On the other, broader promoters with stronger structural constraints and more complex nucleosomal patterns are representative of duplicate genes, which, in turn, have more complex regulatory patterns and greater transcriptional plasticity.
The co-regulation of gene duplicates has been shown to be dependent on both their linear (65) and 3D proximity (66) but the role of chromatin structure in the modulation of gene regulation has not been investigated. We analyzed the effect of nucleosome positioning on gene duplicates by comparing the structural similarity of the sequence around the TSS with an adjusted co-expression score for each duplicate gene pair. Structural similarity was calculated as the Pearson correlation coefficient of nucleosome positioning patterns for an area of 1000 bp symmetrically flanking the TSS (see Materials and Methods). The adjusted co-expression score (ACS) was obtained from the SPELL database (49) as a weighted correlation of gene expression levels estimated for >2400 different experimental conditions. We found a small, yet significant correlation for both types of gene duplicates (all duplicates Spearman's rho = 0.089, P = 0.0035), which was even stronger for SSD (SSD Spearman's rho = 0.109, P = 0.015) (Figure 5B).
This correlation between structural similarity and gene co-expression was independent of the sequence divergence between the gene pair. A number of studies have suggested that divergence is more pronounced at the level of regulation than at that of gene sequence or function (18,33). In order to assess this association, we performed the same analysis comparing the adjusted co-expression score with the regulatory similarity, as assessed with the Jaccard similarity of common TFs having a conserved binding site at the genes’ promoters (see Materials and Methods). The effect of regulatory similarity on co-expression is comparable (all duplicates Spearman's rho = 0.096, P = 0.0017) but in this case it is the WGD genes that show the stronger association (WGD Spearman's rho = 0.119, P = 0.0053) (Figure 5B). Together, these results suggest that the local chromatin environment may shape the expression patterns of duplicate genes in a way that is as strong as the one exhibited by transcription factor binding. Moreover, they paint a complex picture of the modulation of functional divergence. SSD genes appear to be more dependent on structural properties than TF binding, while the opposite is the case for WGD ones (Figure 5B).
Singleton gene functional and regulatory properties are dependent on their location in the genome
Combined, our observations suggest that a set of spatial, structural and regulatory properties define different genomic ‘niches’ that are occupied preferentially by SSD and WGD duplicates. Given that a number of these properties affect the broader environment and are shared by proximal singleton genes, one hypothesis is that singletons found in duplicate gene clusters may partly share the evolutionary history of nearby duplicate genes. This may be particularly interesting for the case of singletons in SSD gene clusters, as SSD genes occupy genomic regions which appear to be more prone to complex regulation (longer non-coding regions, stronger chromatin constraints and more relaxed sequence promoter conservation). We wanted to examine the possibility that a set of singleton genes, sharing SSD-like properties, may constitute remnants of a duplication event and which may be, in part, maintaining some characteristics that distinguish them from genes that have not recently undergone gene duplication.
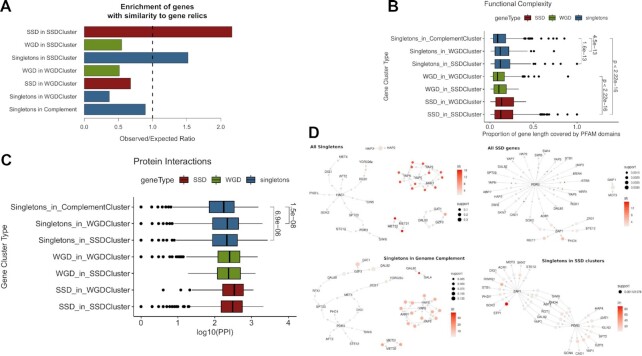
One way to examine this is by analyzing the similarity of genomic sequences that contain some residual similarity with existing genes and may thus represent gene ‘relics’, products of a duplication event, which have acquired a sufficient amount of substitutions to render them indistinguishable from intergenic DNA. We used a dataset of 124 such relics from the yeast genome, bearing similarity with 149 distinct genes, identified through a stringent sequence similarity analysis (67). Genes with similarity to gene relics were preferentially positioned in SSD gene clusters. In addition, they were enriched in both SSD and singletons, but not WGD genes residing in these clusters (Figure 6A). This is suggestive of more frequent duplication events in the areas of the genome lying towards the chromosomal edges, with longer intergenic spacers. It also indicates that a significant proportion of singleton genes found in these regions constitute remnants of small-scale duplications. This hypothesis is further supported by the fact that the relics themselves are preferentially found in SSD gene clusters, with an observed/expected ratio, o/e = 2.08 (P = 0.003), compared to a clear depletion in WGD clusters (o/e = 0.51, P = 0.021).
Figure 6.
Functional and regulatory complexity is increased in duplicate-enriched yeast genomic territories. (A) Relative enrichment of genes with similarity to gene relics in the different cluster categories. Values correspond to observed over expected ratios, with expected values calculated from 1000 random permutations of the gene relic similar genes. All ratios were significant with at least P ≤ 0.05 for the permutation tests. (B) Distribution of functional complexity measured as the proportion of gene length attributed to a known PFAM domain for genes from different gene clusters. Values next to brackets denote adjusted P-values of a Mann–Whitney test. (C) Distribution of the number of protein–protein interactions (log10-transformed) for genes from different gene clusters. Values next to brackets denote adjusted p-values of a Mann–Whitney test. (D) Association rule networks created for distinct sets of genes against transcriptional regulators whose binding sites were found in the genes’ promoters. Networks describe the top 10% most significant associations between regulators that share gene targets, omitting these targets for simplicity.
Reflections of the evolutionary history of such ‘vestigial’ singleton genes may be found in their functional properties. According to the definition of functional entanglement by (59), functionalities associated to specific protein domains may be structurally constrained and thus restrict the way genes evolve. Entangled, constrained functions do not allow for the evolutionary divergence and sub-functionalization and a gene duplicate pair with highly constrained functions is more likely to revert to a singleton state. We used the predicted PFAM domains as a proxy for protein functional domains and assessed the percentage of the gene covered by a known PFAM as a measure of its functional complexity. We found that singletons in WGD and (even more) in SSD gene clusters have significantly increased fractions of their length assigned to a functional domain (Figure 6B) even though they are of similar length (see Figure 2A).
Increased functional complexity for singletons from gene duplicate clusters may suggest greater overall involvement in protein–protein interactions (PPI). As already suggested in (25), gene duplicates tend to have more PPIs. This may be attributed to a number of characteristics already reported in the literature and other that we have identified above, such as increased protein sequence length, regulatory complexity, expression variability and functional entanglement. It was thus interesting to see that singleton genes residing in duplicate clusters also tend to have increased numbers of PPIs (Figure 6C). Interaction preferences are not limited to protein–protein but extend to regulatory relationships. We calculated the relative enrichments of transcriptional regulator binding sites in genes belonging to each category and found the profiles of singletons in SSD clusters bearing stronger similarities with SSD genes than with singletons overall (Supplementary Figure S10), being enriched in binding sites of XBP1, MET4, STP1 and STB4 among others.
As enrichment values can also be biased towards over-represented regulators in the dataset, we further explored regulatory interactions through an association rules analysis, that aims to capture significant associations by better controlling for very common regulators (see Materials and Methods). By this point, we were not surprised to see that the association rules learned, were also position-specific, with singleton and SSD genes exhibiting regulatory associations largely defined by the region in which they were located (Figure 6D). A number of regulators, including PDR3, ARR1 and ZAP1, were found to be strongly associated with both singletons and SSD genes when found in SSD clusters, suggesting the existence of pervasive, position-specific regulatory preferences.
DISCUSSION
Recent advances at both experimental and theoretical levels have provided evidence for the structural organization of eukaryote genomes (68–70). Even though, yeast has a rather small and dense genome in comparison to multicellular eukaryotes, a certain degree of organization exists at both one (71–73) and three dimensions (74,75). The existence of an underlying conformational genome scaffold alludes to the possibility of a concomitant functional compartmentalization, aspects of which may be seen at the localized chromatin structure of the genes’ promoters (39) as well as at more generalized ‘architectural’ properties, such as gene spacing and promoter complexity (28).
In this work, we show that a certain degree of functional segmentation in the yeast genome is strongly associated with the localized potential for genomic innovation. We find gene duplicates to be concentrated in specific areas of the genome and, moreover, strongly segregating depending on their mode of duplication. This is probably expected for Whole-Genome Duplicates as they largely maintain synteny. The fact however, that they show a tendency to be more centrally positioned is unexpected. A plausible explanation for this, is that the less dense genomic space at the chromosomal edges constitutes a more dynamic environment, where rapid turnover of gene duplication has gradually eroded the syntenic structure. In exchange, it is these areas that preferentially host genes with stress-related and condition-specific functions (28). Small-scale duplicates, which are in general more prone to diverge into acquiring such novel functions, are preferentially located towards chromosomal edges. This is consistent with the view of small-scale duplicates being intrinsically faster evolving, which has been reported for primates (76). This positional segregation offers likely explanations for the evolutionary fate of both gene duplicates as well as the singleton genes that are found in the same areas.
A sufficient amount of genome space is required for a duplicate to emerge without interrupting nearby genes and regulatory elements. In addition, it is more likely to be maintained, if it is long enough to accommodate a number of functions that will allow its divergence (59). Both of these conditions are met in the regions of the chromosomal edges which are less gene-dense, with long non-coding spacers, thus constituting a genomic ‘niche’ that is more permissive for genomic innovation. Reflections of this may be seen in the functional enrichment of genes found in SSD clusters, with a strong over-representation of transmembrane proteins and transporters. Genes found in WGD clusters are, in contrast, primarily enriched in basal metabolic pathways and functions (Supplementary Figure S11). A general association of SSD genes with more specialized functions is also supported by the transcriptional regulator enrichments (Figure 6D, Supplementary Figure S10), with the majority being related to stress response and the use of alternative energy and nutrient resources. A plausible scenario for the confinement of such genes into specific chromosomal areas is that it may be conferring an advantageous genomic ‘division of labour’, whereby novel functions may be explored in certain parts of the genome, minimizing the possibility of interference with regions hosting more stably expressed, constitutive genes. Stress response genes are known to be more likely to revert to singleton state when duplicated (18). In this sense SSD gene clusters, located at the chromosomal edges, would correspond to dynamic regions of high duplicate turnover, driving genomic innovation. An additional, strong indication for this is the enrichment of gene ‘relics’ in these areas.
The mechanism, through which the exploration of genomic innovation takes place, is far from understood, but our findings point to chromatin structure being an unexpected, yet crucial property in this respect. Both SSD and WGD genes have singular nucleosomal architectures at their promoters, that are both quantitatively and qualitatively different from the ones of singleton genes, suggesting that a structurally complex promoter is a primal property of gene duplicates. This is reflected on the increased and more extended structural constraints observed not only for gene duplicates, but also, residually, for singletons found in duplicate clusters. This finding appears at first counter-intuitive, especially when one sees its inverse correlation with gene conservation at the promoters (Figure 5A). However, we should consider that the structural constraints discussed herein are related to the maintenance of nucleosome positioning, which is only loosely associated with specific sequence signatures (46,61,77). This means that a gene promoter's structural profile may be modulated with some minimal prerequisites of sequence constraint, in a way that is permissive for the exploratory process of genomic innovation. Expression and eventually functional divergence may thus be achieved with a more complex promoter structure with minimal changes at the level of gene sequence, at least at the early stages of the divergence process. This is supported by a number of observations including the higher correlation of gene co-expression with nucleosome positioning similarity than with sequence divergence (Figure 5B). Interestingly, the correlation between duplicate pair gene co-expression and nucleosome positioning is almost 3-fold increased for duplicate genes lying close to the chromosomal edges compared to those being more central (cc = 0.198 for duplicates lying within 10% of the length of a chromosomal arm from the edge as opposed to cc = 0.068 for genes lying at the opposite 90%). Our findings, related to nucleosome positioning structure, structural constraints and their relationship with expression divergence are highly indicative of local chromatin structure being a ‘soft’ constraint, which acts permissively for the exploration of novel functionalities, without compromising the duplication and reverting into singleton. Such events are obviously the majority even in the ‘permissive’ SSD niches as indicated by the similarity of singletons in these areas with gene relics (Figure 6A).
Overall, our observations regarding the properties of singleton genes that co-localize with gene duplicates, are supportive of our main hypothesis, that specific genomic niches are more tolerant to gene duplicates. New duplication events are expected to be preferentially taking place in genomic subcompartments with specific properties. These properties are implicit in duplicate genes, regardless of their clustering, as clusters of variable gene size show only marginal differences (Supplementary Figure S12). Singletons found in these subcompartments residually carry many of the attributes of gene duplicates and could be speculated to constitute remnants of recent duplication events. In all, our findings point towards an architectural segregation of function, regulation and evolvability in the yeast genome, which, buffered by chromatin structure, creates permissive environments for both neo- and sub-functionalization and thus creates preferential ‘niches’ for gene duplicates. This apparent link of genome innovability with positional preferences may shed new light to the evolutionary dynamics of eukaryote genomes and provide a valuable framework for more nuanced approaches in the field of synthetic biology.
DATA AVAILABILITY
Compiled datasets, code used, as well as an outline of the methodology may be found at the accompanying github site: https://github.com/christoforos-nikolaou/YeastSegmentationDuplicates.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Maria Malliarou and all previous members of the Nikolaou lab at the University of Crete for constructive comments.
Contributor Information
Athanasia Stavropoulou, Medical School, University of Crete, Heraklion 70013, Greece; Computational Genomics Group, Biomedical Sciences Research Center “Alexander Fleming”, Athens 16672, Greece.
Emilios Tassios, Medical School, University of Crete, Heraklion 70013, Greece; Computational Genomics Group, Biomedical Sciences Research Center “Alexander Fleming”, Athens 16672, Greece.
Maria Kalyva, European Bioinformatics Institute, EMBL-EBI, Wellcome Genome Campus, Hinxton, Cambridgeshire, CB10 1SD, UK.
Michalis Georgoulopoulos, Hellenic Open University, Patras 26335, Greece.
Nikolaos Vakirlis, Computational Genomics Group, Biomedical Sciences Research Center “Alexander Fleming”, Athens 16672, Greece.
Ioannis Iliopoulos, Medical School, University of Crete, Heraklion 70013, Greece.
Christoforos Nikolaou, Computational Genomics Group, Biomedical Sciences Research Center “Alexander Fleming”, Athens 16672, Greece; Hellenic Open University, Patras 26335, Greece.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
Athanasia Stavropoulou and Emilios Tassios were funded by an internal BSRC ‘Alexander Fleming’ Grant (to Christoforos Nikolaou); Nikos Vakirlis was funded by a Greek State Scholarships Foundation (IKY) post-doctoral fellowship; Greece and the European Union (European Social Fund- ESF) through the Operational Programme ‘Human Resources Development, Education and Lifelong Learning’ in the context of the project ‘Reinforcement of Postdoctoral Researchers - 2nd Cycle’ [MIS-5033021], implemented by the State Scholarships Foundation. This work was partially supported by a FONDATION SANTE2020-2021 grant to Christoforos Nikolaou.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ohno S. Evolution by gene duplication. 1970; NY: Springer-Verlag. [Google Scholar]
- 2. Acharya D., Ghosh T.C.. Global analysis of human duplicated genes reveals the relative importance of whole-genome duplicates originated in the early vertebrate evolution. BMC Genomics. 2016; 17:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang J. Evolution by gene duplication: an update. Trends Ecol. Evol. 2003; 18:292–298. [Google Scholar]
- 4. Otto S.P., Whitton J.. Polyploid incidence and evolution. Annu. Rev. Genet. 2000; 34:401–437. [DOI] [PubMed] [Google Scholar]
- 5. Sémon M., Wolfe K.H.. Consequences of genome duplication. Curr. Opin. Genet. Dev. 2007; 17:505–512. [DOI] [PubMed] [Google Scholar]
- 6. Graur D., Li W.-H.. Fundamentals of molecular evolution sinauer. 2000; [Google Scholar]
- 7. Lynch M., Conery J.S.. The origins of genome complexity. Science. 2003; 302:1401–1404. [DOI] [PubMed] [Google Scholar]
- 8. Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., De Montigny J., Marck C., Neuvéglise C., Talla E.et al.. Genome evolution in yeasts. Nature. 2004; 430:35–44. [DOI] [PubMed] [Google Scholar]
- 9. Van Hoek M.J.A., Hogeweg P.. The role of mutational dynamics in genome shrinkage. Mol. Biol. Evol. 2007; 24:2485–2494. [DOI] [PubMed] [Google Scholar]
- 10. Innan H., Kondrashov F.. The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 2010; 11:97–108. [DOI] [PubMed] [Google Scholar]
- 11. He X., Zhang J.. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005; 169:1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sandve S.R., Rohlfs R.V., Hvidsten T.R.. Subfunctionalization versus neofunctionalization after whole-genome duplication. Nat. Genet. 2018; 50:908–909. [DOI] [PubMed] [Google Scholar]
- 13. Wolfe K.H., Shields D.C.. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997; 387:708–713. [DOI] [PubMed] [Google Scholar]
- 14. Kellis M., Birren B.W., Lander E.S.. Proof and evolutionary analysis of ancient genome duplication in the yeast saccharomyces cerevisiae. Nature. 2004; 428:617–624. [DOI] [PubMed] [Google Scholar]
- 15. Wolfe K.H. Origin of the yeast whole-genome duplication. PLoS Biol. 2015; 13:e1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seoighe C., Wolfe K.H.. Yeast genome evolution in the post-genome era. Curr. Opin. Microbiol. 1999; 2:548–554. [DOI] [PubMed] [Google Scholar]
- 17. Guan Y., Dunham M.J., Troyanskaya O.G.. Functional analysis of gene duplications in saccharomyces cerevisiae. Genetics. 2007; 175:933–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wapinski I., Pfeffer A., Friedman N., Regev A.. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007; 449:54–61. [DOI] [PubMed] [Google Scholar]
- 19. Papp B., Pál C., Hurst L.D.. Evolution of cis-regulatory elements in duplicated genes of yeast. Trends Genet. 2003; 19:417–422. [DOI] [PubMed] [Google Scholar]
- 20. Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., Ding H., Koh J.L.Y., Toufighi K., Mostafavi S.et al.. The genetic landscape of a cell. Science (80-.). 2010; 327:425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baudot A., Jacq B., Brun C.. A scale of functional divergence for yeast duplicated genes revealed from analysis of the protein–protein interaction network. Genome Biol. 2004; 5:R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hakes L., Pinney J.W., Lovell S.C., Oliver S.G., Robertson D.L.. All duplicates are not equal: the difference between small-scale and genome duplication. Genome Biol. 2007; 8:R209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hakes L., Lovell S.C., Oliver S.G., Robertson D.L.. Specificity in protein interactions and its relationship with sequence diversity and coevolution. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:7999–8004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keane O.M., Toft C., Carretero-Paulet L., Jones G.W., Fares M.A.. Preservation of genetic and regulatory robustness in ancient gene duplicates of saccharomyces cerevisiae. Genome Res. 2014; 24:1830–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fares M.A., Keane O.M., Toft C., Carretero-Paulet L., Jones G.W.. The roles of whole-genome and small-scale duplications in the functional specialization of Saccharomyces cerevisiae genes. PLoS Genet. 2013; 9:e1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mattenberger F., Sabater-Muñoz B., Toft C., Fares M.A.. The phenotypic plasticity of duplicated genes in saccharomyces cerevisiae and the origin of adaptations. G3 Genes, Genomes, Genet. 2017; 7:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De S., Babu M.M.. Genomic neighbourhood and the regulation of gene expression. Curr. Opin. Cell Biol. 2010; 22:326–333. [DOI] [PubMed] [Google Scholar]
- 28. Tsochatzidou M., Malliarou M., Papanikolaou N., Roca J., Nikolaou C.. Genome urbanization: clusters of topologically co-regulated genes delineate functional compartments in the genome of Saccharomyces cerevisiae. Nucleic Acids Res. 2017; 45:5818–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Byrne K.P., Wolfe K.H.. The yeast gene order browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005; 15:1456–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Poyatos J.F., Hurst L.D.. The determinants of gene order conservation in yeasts. Genome Biol. 2007; 8:R233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sugino R.P., Innan H.. Natural selection on gene order in the genome reorganization process after whole-genome duplication of yeast. Mol. Biol. Evol. 2012; 29:71–79. [DOI] [PubMed] [Google Scholar]
- 32. Sémon M., Duret L.. Evolutionary origin and maintenance of coexpressed gene clusters in mammals. Mol. Biol. Evol. 2006; 23:1715–1723. [DOI] [PubMed] [Google Scholar]
- 33. Byrnes J.K., Morris G.P., Li W.H.. Reorganization of adjacent gene relationships in yeast genomes by whole-genome duplication and gene deletion. Mol. Biol. Evol. 2006; 23:1136–1143. [DOI] [PubMed] [Google Scholar]
- 34. Fares M.A., Sabater-Muñoz B., Toft C.. Genome mutational and transcriptional hotspots are traps for duplicated genes and sources of adaptations. Genome Biol. Evol. 2017; 9:1229–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Makino T., McLysaght A.. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:9270–9274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Naseeb S., Ames R.M., Delneri D., Lovell S.C.. Rapid functional and evolutionary changes follow gene duplication in yeast. Proc. R. Soc. B Biol. Sci. 2017; 284:20171393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Papanikolaou N., Trachana K., Theodosiou T., Promponas V., Iliopoulos I.. Gene socialization: gene order, GC content and gene silencing in salmonella. BMC Genomics. 2009; 10:597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nikolaou C., Bermúdez I., Manichanh C., García-Martinez J., Guigó R., Pérez-Ortín J.E.J.E., Roca J.. Topoisomerase II regulates yeast genes with singular chromatin architectures. Nucleic Acids Res. 2013; 41:9243–9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nikolaou C. Invisible cities: segregated domains in the yeast genome with distinct structural and functional attributes. Curr. Genet. 2018; 64:247–258. [DOI] [PubMed] [Google Scholar]
- 40. Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y.J., Lee C., Shendure J., Fields S., Blau C.A., Noble W.S.. A three-dimensional model of the yeast genome. Nature. 2010; 465:363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Andreadis C., Nikolaou C., Fragiadakis G.S., Tsiliki G., Alexandraki D. Rad9 interacts with aft1 to facilitate genome surveillance in fragile genomic sites under non-DNA damage-inducing conditions in s. cerevisiae. Nucleic Acids Res. 2014; 42:12650–12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quinlan A., Hall I.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. States D.J., Gish W.. Combined use of sequence similarity and codon bias for coding region identification. J. Comput. Biol. 1994; 1:39–50. [DOI] [PubMed] [Google Scholar]
- 44. Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S.et al.. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005; 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Routhier E., Pierre E., Khodabandelou G., Mozziconacci J.. Genome-wide prediction of DNA mutation effect on nucleosome positions for yeast synthetic genomics. Genome Res. 2020; 31:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nikolaou C., Althammer S., Beato M., Guigó R.. Structural constraints revealed in consistent nucleosome positions in the genome of s. cerevisiae. Epigenetics Chromatin. 2010; 3:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jiang C., Pugh B.F.. A compiled and systematic reference map of nucleosome positions across the saccharomyces cerevisiae genome. Genome Biol. 2009; 10:R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. MacIsaac K.D., Wang T., Gordon D.B., Gifford D.K., Stormo G.D., Fraenkel E.. An improved map of conserved regulatory sites for saccharomyces cerevisiae. BMC Bioinformatics. 2006; 7:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hibbs M.A., Hess D.C., Myers C.L., Huttenhower C., Li K., Troyanskaya O.G.. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 2007; 23:2692–2699. [DOI] [PubMed] [Google Scholar]
- 50. Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J.et al.. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021; 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P.et al.. The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021; 49:D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reimand J., Kull M., Peterson H., Hansen J., Vilo J.. G:Profiler-a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007; 35:W193–W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hahsler M., Grün B., Hornik K.. Mining association rules and frequent itemsets. J. Stat. Softw. 2005; 14:1–25. [Google Scholar]
- 54. Hahsler M., Grün B., Hornik K., Buchta C.. Introduction toarules – A computational environment for mining association rules and frequent item sets. 2022; https://cran.r-project.org/web/packages/arules/vignettes/arules.pdf.
- 55. Makino T., McLysaght A., Kawata M.. Genome-wide deserts for copy number variation in vertebrates. Nat. Commun. 2013; 4:2283. [DOI] [PubMed] [Google Scholar]
- 56. Lopes I., Altab G., Raina P., de Magalhães J.P.. Gene size matters: an analysis of gene length in the human genome. Front. Genet. 2021; 12:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. David L., Huber W., Granovskaia M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M.. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:5320–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vitkup D., Kharchenko P., Wagner A.. Influence of metabolic network structure and function on enzyme evolution. Genome Biol. 2006; 7:R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kuzmin E., Vandersluis B., Ba A.N.N., Wang W., Koch E.N., Usaj M., Khmelinskii A., Usaj M.M., Leeuwen J.Van, Kraus O.et al.. Exploring whole-genome duplicate gene retention with complex genetic interaction analysis. Science. 2020; 368:eaaz5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yuan G.-C., Liu Y.-J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J.. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005; 309:626–630. [DOI] [PubMed] [Google Scholar]
- 61. Kaplan N., Moore I.K., Fondufe-Mittendorf Y., Gossett A.J., Tillo D., Field Y., LeProust E.M., Hughes T.R., Lieb J.D., Widom J.et al.. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009; 458:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Segal E., Fondufe-Mittendorf Y., Chen L., Thåström A., Field Y., Moore I.K., Wang J.P.Z., Widom J.. A genomic code for nucleosome positioning. Nature. 2006; 442:772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Babbitt G., Kim Y.. Inferring natural selection on fine-scale chromatin organization in yeast. Mol. Biol. Evol. 2008; 25:1714–1727. [DOI] [PubMed] [Google Scholar]
- 64. Tilgner H., Nikolaou C., Althammer S., Sammeth M., Beato M., Valcárcel J., Guigó R.. Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 2009; 16:996–1001. [DOI] [PubMed] [Google Scholar]
- 65. Lan X., Pritchard J.K.. Coregulation of tandem duplicate genes slows evolution of subfunctionalization in mammals. Science. 2016; 352:1009–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ibn-Salem J., Muro E.M., Andrade-Navarro M.A.. Co-regulation of paralog genes in the three-dimensional chromatin architecture. Nucleic Acids Res. 2017; 45:81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lafontaine I., Fischer G., Talla E., Dujon B.. Gene relics in the genome of the yeast Saccharomyces cerevisiae. Gene. 2004; 335:1–17. [DOI] [PubMed] [Google Scholar]
- 68. van Steensel B., Furlong E.E.M.. The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol. 2019; 20:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dekker J., Marti-Renom M.a, Mirny L.a. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 2013; 14:390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tanay A., Cavalli G.. Chromosomal domains: epigenetic contexts and functional implications of genomic compartmentalization. Curr. Opin. Genet. Dev. 2013; 23:197–203. [DOI] [PubMed] [Google Scholar]
- 71. Kouzine F., Gupta A., Baranello L., Wojtowicz D., Ben-Aissa K., Liu J., Przytycka T.M., Levens D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 2013; 20:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Henikoff J.G., Belsky J.A., Krassovsky K., MacAlpine D.M., Henikoff S.. Epigenome characterization at single base-pair resolution. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:18318–18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Janga S.C., Collado-Vides J., Babu M.M.. Transcriptional regulation constrains the organization of genes on eukaryotic chromosomes. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:15761–15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rutledge M.T., Russo M., Belton J.M., Dekker J., Broach J.R.. The yeast genome undergoes significant topological reorganization in quiescence. Nucleic Acids Res. 2015; 43:8299–8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Duan Z., Andronescu M., Schutz K., Lee C., Shendure J., Fields S., Noble W.S., Anthony Blau C.. A genome-wide 3C-method for characterizing the three-dimensional architectures of genomes. Methods. 2012; 58:277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. O’Toole Á.N., Hurst L.D., McLysaght A.. Faster evolving primate genes are more likely to duplicate. Mol. Biol. Evol. 2018; 35:107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ioshikhes I., Hosid S., Pugh B.F.. Variety of genomic DNA patterns for nucleosome positioning. Genome Res. 2011; 21:1863–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Compiled datasets, code used, as well as an outline of the methodology may be found at the accompanying github site: https://github.com/christoforos-nikolaou/YeastSegmentationDuplicates.