Abstract
The plateau zokor (Myospalax baileyi) is a small subterranean rodent endemic to China that lives alone in sealed underground burrows at altitudes ranging from 2000 to 4200 m above sea level on the Tibetan Plateau. Due to the unique environmental factors in the Tibetan Plateau, intestinal parasites in the local population may be more likely to develop host-adapted genotypes. We therefore conducted an epidemiological survey of common intestinal parasites in plateau zokors on the Tibetan plateau to estimate their actual gastrointestinal parasite status.
Two areas with high populations of plateau zokor in Xunhua County, Qinghai Province were selected as sampling sites, and a total of 98 zokors were trapped. Four parasites, Cryptosporidium spp., Enterocytozoon bieneusi, Giardia lamblia and Blastocystis hominis, were tested in the faecal samples. The results showed that a new genotype of Cryptosporidium sp. was identified by amplification and sequencing of a portion of the small subunit ribosomal RNA (SSU rRNA) gene with an infection rate of 1.0% (1/98), and new genotypes of E. bieneusi were identified by amplification and sequencing of a portion of the internal transcribed spacer (ITS) region of the ribosomal RNA gene sequences with an infection rate of 4.1% (4/98). Neither of the two intestinal parasites, G. lamblia and B. hominis, was detected.
Keywords: Plateau zokor, Cryptosporidium sp., Enterocytozoon bieneusi, Novel genotypes, Host-adapted
Graphical abstract
Highlights
-
•
First survey of plateau zokors for four protozoa species.
-
•
New genotypes of two protozoa identified.
-
•
Host-adapted genotypes of the parasite in plateau zokors.
1. Introduction
The plateau zokor (Myospalax baileyi) is a small subterranean rodent endemic to China that lives alone year-round in sealed burrows underground at altitudes of 2000–4200 m on the Tibetan Plateau (Kang et al., 2020a; Pu et al., 2019). Faced with a harsh environment of high humidity, limited oxygen, high CO2 concentration, low temperature, and food scarcity, the body structure and physiological functions of Plateau zokor have evolved to adapt to the subterranean tunnel system(Shao et al., 2015). It is now become a good model for studying differentiation, selection, and species formation(Kang et al., 2022).
Cryptosporidium spp. and E. bieneusi have been shown to have multiple host-adapted genotypes (Song et al., 2021; Xiao et al., 2002)Cryptosporidium sp. is a widespread zoonotic parasite, the main symptom of which is diarrhoea, capable of infecting 240 species of animals, including humans, and can cause serious public health emergencies in outbreaks (Ryan et al., 2018; Zahedi and Ryan, 2020). Enterocytozoon bieneusi are a group of intracellular, specific parasitic eukaryotes that can infect almost all vertebrates, including humans, are often associated with HIV or AIDS patients, making them an important opportunistic pathogen in humans (Li et al., 2019; Stentiford et al., 2016). Giardia lamblia is a genus of intestinal flagellates that infect a wide range of vertebrate hosts, and are found in a variety of mammals, including humans, pets and livestock (Feng and Xiao, 2011). Blastocystis hominis is a globally distributed intestinal protozoan that infects humans and many species of animals(Wang et al., 2013).
Compared to some terrestrial rodents, plateau zokors are less active, and their unique geographic location and living habits make them often an ideal model for studying phenotypic and ecological adaptations (Kang et al., 2020b). This also leads to a low probability of exogenous anthropogenic influences and pathogen invasion in plateau zokors, and intra-population intestinal parasites may be more likely to occur to produce host-adapted genotypes (Cao et al., 2014; Zhao et al., 2014). Therefore, we investigated the infection status of plateau zokors with Cryptosporidium spp., E. bieneusi, G. lamblia and B. hominis on the Tibetan plateau.
2. Materials and methods
Ethical statement
This study was conducted in accordance with the Guidelines for the Care and Use of Animals in Research published by the Institute of Zoology, Chinese Academy of Sciences. This study was reviewed and approved by the Animal Ethics Committee of the Institute of Zoology, Chinese Academy of Sciences (2019FY100300-03).
2.1. Sample capture of plateau zokor
The Qinghai subspecies of Plateau zokor (Myospalax fontanierii baileyi) was selected as the host for the study, and two regions with high populations of plateau zokor in Xunhua County, Qinghai Province were selected as the sampling sites. In September 2021, we went to each of these two sites and carried out rodent trapping using the rope trap method. A total of 30 zokors were captured at site 1 (35°38′46.3 "N 102°15′02.0 "E altitude: 3553.28m) and 68 zokors were captured at site 2 (3211.91m 35°41′48.7 "N 102°14′29.1 "E). Captured plateau zokors were euthanised with isoflurane and then taken back to the local laboratory for risk assessment. After confirming that there was no potential biosafety risk, the autopsy was performed, and gender and location were recorded, as shown in Table 1.
Table 1.
Sampling location and sample data information.
| Factors | Category | Sampling Location 1 | Sampling Location 2 | Total |
|---|---|---|---|---|
| Location | longitude and latitude | 35°38′46.3"N 102°15′02.0"E | 35°41′48.7"N 102°14′29.1"E | |
| Altitude(meter) | 3553.28m | 3211.91m | ||
| Gender | Female | 14 | 47 | 61 |
| Male | 16 | 21 | 37 | |
| Total | 30 | 68 | 98 |
2.2. Microscopic examination
Parasite isolation and purification was an improvement on the method described by Zajac et al. (2021). In short, the collected feces were added to a sucrose solution and then floated and shaken and mixed, and then centrifuged to take the liquid film on the surface and place it for observation under an optical microscope.
2.3. Stool sample collection and DNA extraction
Rectal contents were collected into 2 mL sterilized centrifuge tubes, tubes were labeled, placed in liquid nitrogen for quick freezing, and then transported to the laboratory in Beijing using dry ice and stored in a −80 °C refrigerator. Total genomic DNA was extracted from 200 mg Stool according to the instruction of E.Z.N.A. ® Stool DNA Kit Stool Whole Genome Extraction Kit (OMEGA Biotek INCD4015-01, USA). The extracted Stool DNA was stored in a refrigerator at −20 °C for further PCR assays.
The small subunit ribosomal RNA (SSU) gene of Cryptosporidium spp. (Xiao et al., 2001), the internal transcribed spacer (ITS) of the ribosomal RNA gene of E. bieneusi (Buckholt Michael et al., 2002), the β-giardin gene of G. lamblia (Lalle et al., 2005) and the SSU rRNA gene of B. hominis (Wang et al., 2013) were examined under nested PCR according to previous studies, respectively. Primer information and PCR procedures are detailed in Table 2. Positive control and negative control were added in each amplification. Amplified DNA was added to 1.5% agarose containing Gel-Red for electrophoresis. Finally, the results of electrophoresis were observed and analyzed by gel imaging system and photographed.
Table 2.
Forward and reverse primers used for the detection of Cryptosporidium sp., E. bieneusi, G. lamblia and B. hominis.
| Gene | Primers | Amplication protocol | Size | Ref | ||
|---|---|---|---|---|---|---|
|
Cryptosporidium sp. SSU gene |
1st | Forward | 5′-TTCTAGAGCTAATACATGCG-3′ | 94 °C: 3 min, 35 cycles: (94 °C: 45 s, 55 °C: 45 s, 72 °C:1 min) 72 °C: 7min | 1325 bp | Xiao et al., 2001 |
| PCR | Reverse | 5′-CCCATTTCCTTCGAAACAGGA-3′ | ||||
| 2nd | Forward | 5′-GGAAGGGTTGTATTTATTAGATAAAG-3′ | 94 °C: 3 min, 35 cycles: (94 °C: 45 s, 55 °C: 45 s, 72 °C:1 min) 72 °C: 7min | 864 bp | ||
| PCR | Reverse | 5′-AAGGAGTAAGGAACAACCTCCA-3′ | ||||
|
E. bieneusi ITS gene |
1st | Forward | 5′-GGTCATAGGGATGAAGAG-3′ | 94 °C: 5 min, 35 cycles: (94 °C: 30 s, 57 °C: 30 s, 72 °C:40 s) 72 °C: 7min | Buckholt et al., 2002 | |
| PCR | Reverse | 5′-TTCGAGTTCTTTCGCGCTC-3′ | 435 bp | |||
| 2nd | Forward | 5′-GCTCTGAATATCTATGGCT-3′ | 94 °C: 5 min, 35 cycles: (94 °C: 30 s, 55 °C: 30 s, 72 °C:40 s) 72 °C: 7min | |||
| PCR | Reverse | 5′-ATCGCCGACGGATCCAAGTG-3′ | 390 bp | |||
|
G. lamblia β-giardin gene |
1st | Forward | 5′-AAGCCCGACGACCTCACCCGCAGTGC-3′ | 94 °C: 5 min, 35 cycles: (94 °C: 30 s, 65 °C: 30 s, 72 °C:40 s) 72 °C: 7min | 753 bp | Lalle et al., 2005 |
| PCR | Reverse | 5′-GAGGCCGCCCTGGATCTTCGAGACGAC-3′ | ||||
| 2nd | Forward | 5′-GAACGAACGAGATCGAGGTCCG-3′ | 94 °C: 5 min, 35 cycles: (94 °C: 30 s, 55 °C: 30 s, 72 °C:40 s) 72 °C: 7min | 511 bp | ||
| PCR | Reverse | 5′-CTCGACGAGCTTCGTGTT-3′ | ||||
|
B. hominis SSU rRNA gene |
1st | Forward | 5′-GGGATCCTGATCCTTCCGCAGGTTCACCTAC-3′ | 94 °C: 5 min, 35 cycles: (94 °C: 30 s, 65 °C: 45 s, 72 °C:40 s) 72 °C: 7min | 1800 bp | Wang et al., 2013 |
| PCR | Reverse | 5′-GGAAGCTTATCTGGTTGATCCTGCCAGTA-3′ | ||||
| 2nd | Forward | 5′-GGAGGTAGTGGACAATAAATC-3′ | 94 °C: 5 min, 35 cycles: (94 °C: 30 s, 54 °C: 45 s, 72 °C:40 s) 72 °C: 7min | 1100 bp | ||
| PCR | Reverse | 5′-CGTTCATGATGAACAATTAC-3′ | ||||
2.4. Gene sequencing and phylogenetic analysis
The secondary PCR positive products were sequenced and spliced in both directions by Shenzhen Huada Gene Co Ltd (Beijing, China). After sequencing results were compared, DNAMAN software was used to correct splicing and assembly. We selected the host genotypes and Cryptosporidium sp. Environment sequences including chipmunk, muskrat, Microtus arvalis, muskrat genotype II, Brandt's vole genotype V, C. hominis, C. ubiquitum and others as reference genes. Due to the significant sequence differences among the copies of Eimeria SSU rRNA gene, the sequences of the three Eimeria species were selected for rooting of SSU rRNA phylogenetic trees. Similarly, samples including those covering Ebpc WbEb1 B13, B18, MUL5, D, CDZ211, JLNB-3, JLNB-6, horse 1, 5, ST32, Peru 16, BEB4, WbEb4, YZZ157, MS2, BEB6, BEB7, C4 and other taxa were selected as reference genes and Enterocytozoon epatopenaei was used as an outgroup control in the construction of the tree and was used for rooting of the ITS gene phylogenetic tree. Cryptosporidium and Enterocytozoon species/genotypes were determined by aligning with reference sequences available in GenBank database with the ClustalX 2.1 software package. The phylogenetic relationships of Cryptosporidium sp. and E. bieneusi were constructed using the maximum-likelihood (ML) under MEGA 7.0 (Mello, 2018) to construct phylogenetic evolutionary trees, while the reliability was checked using Bootstrap with 500 replicates.
3. Results
3.1. Cryptosporidium spp., E. bieneusi, G. lamblia and B. hominis infection rates in plateau zokors
The microscopic examination of the collected fecal samples showed that cysts and oocysts of Cryptosporidium spp., E. bieneusi, G. lamblia and B. hominis were not detected. Then PCR was used to detect Cryptosporidium sp. SSU gene, E. bieneusi ITS gene, G. lamblia β-giardin gene and B. hominis SSU rRNA gene by amplification in nested PCR, and only the samples after successful sequence alignment in the sequencing results were considered as positive samples. Of 98 samples, a total of 4 samples gave bands at the right place in the PCR for the Cryptosporidium spp. PCR, but only one could be successfully sequenced. And a total of 6 samples gave bands at the right place in the PCR for the E. bieneusi PCR, 4 samples could be successfully sequenced. Cryptosporidium spp. was not detected at site 1 (0/30), 1.5% (1/68) at site 2 and 1.0% (1/98) overall. E. bieneusi were detected at site 1 in 13.3% (4/30), not found positive at location 2 (0/68) and total infection was 4.1% (4/98).
The sequences of the 18S rRNA locus of Cryptosporidium sp. QH219-58 were sequenced to 853bp and submitted to GenBank to obtain the sequence accession number: OM403638. The ITS sequences of E. bieneusi QH219-5, QH219-9, QH219-13, QH219-21 were 397bp, 393bp, 392bp and 392bp, respectively. The sequences were submitted to GenBank and the sequence accession numbers were obtained: OM406189, OM406190, OM406191 and OM406192.
3.2. Phylogenetic analysis of SSU rRNA gene in Cryptosporidium sp
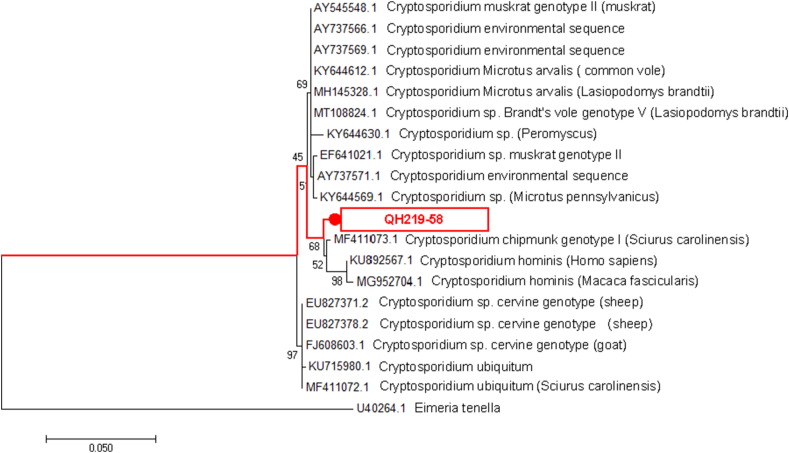
A phylogenetic tree was constructed by comparing with SSU rRNA gene sequence of reference Cryptosporidium sp. (Fig. 1). The genotypes of Cryptosporidium sp. found in plateau zokors were significantly different from other known host genotypes of Cryptosporidium, aggregating with the Cryptosporidium chipmunk genotype I.
Fig. 1.
Phylogenetic relationships of Cryptosporidium sp. genotypes identified in the present study and other known genotypes and species on GenBank was inferred by a maximum-likelihood phylogenetic analysis of SSU rRNA gene sequences using the Tamura 3-parameter model and with 500 replicates. The Eimeria (GenBank: U40264.1) were used as the outgroup. The red circles and squares indicate the novel genotypes identified in this study. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Phylogenetic analysis of ITS gene in E. bieneusi
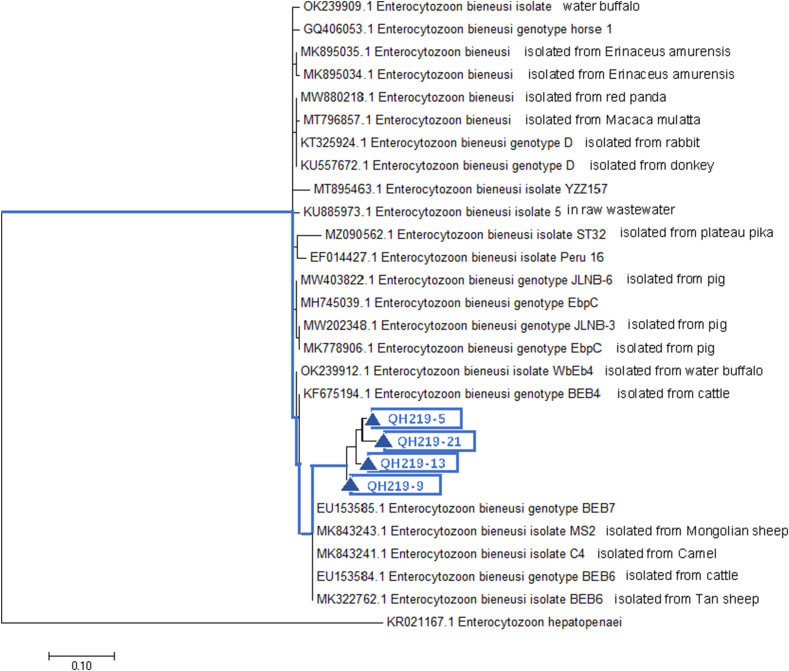
A phylogenetic tree was constructed by comparing and analyzing the ITS gene sequence with that of the reference E. bieneusi (Fig. 2). The genotypes of E. bieneusi detected in plateau zokor were significantly different from those of other known E. bieneusi genotypes, forming a separate group.
Fig. 2.
Phylogenetic relationships of E. bieneusi genotypes identified in the present study and other known genotypes deposited on GenBank was inferred by a maximum-likelihood phylogenetic analysis of ITS sequences using the Tamura 3-parameter model with 500 replicates. The Enterocytozoon hepatopenaei (GenBank: KR021167.1) was used as the outgroup. The blue triangle and squares indicate the novel genotypes identified in this study, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Rodents are considered to be important zoonotic hosts for Cryptosporidium spp. (Zhang et al., 2022) and E. bieneusi (Ni et al., 2021). To our knowledge, this study is the first time to investigate the infection status of Cryptosporidium sp. and E. bieneusi in plateau zokors. Many studies have shown that Cryptosporidium sp. can infect humans and animals, and many Cryptosporidium sp. species/genotypes of public health significance have been identified. However, it has been rarely reported in rodents, especially in plateau zokors. These results indicate that the prevalence of Cryptosporidium sp. infection in plateau zokors was not only lower than that of the Chinese Qinghai vole (8.9%, 8/90) and plateau pika (6.25% (4/64) in the same region (Zhang et al., 2018),but also lower than that of rodent populations in other countries (Horčičková et al., 2019; Stenger et al., 2018). The global prevalence of Cryptosporidium sp. in rodents is estimated to be about 17% (Taghipour et al., 2020), which is higher than our detection results. The E. bieneusi in plateau zokors is lower than the average infection rate of 10.0% (50/498) in rodents from five regions within Gansu Province (Xu et al., 2020).
In this study, neither G. lamblia nor B. hominis were detected positively, which may be related to the low number of parasite infections and the low intensity of infection. Based on these results we speculate that this may be related to the unique living environment and habits of plateau zokors. An increase in rodent population density can cause an increase in infectious disease infection rates (Krawczyk et al., 2020), but plateau zokors often live alone underground, reducing the probability of suffering from disease infestation. Coupled with the fact that intra-population encounters are mostly possible during spring breeding, low-frequency contact within populations reduces parasite transmission (Cai et al., 2018; Jonsson et al., 2010).
Most of the morphological differences between Cryptosporidium spp. and E. bieneusi species cannot be distinguished by the naked eye, and identification based on oocyst morphology alone has limitations, which can be compensated by molecular identification methods that can analyze genetic differences between different Cryptosporidium species at the genetic level (Ryan et al., 2014; Xiao, 2010). The 18S rRNA gene of Cryptosporidium sp. is responsible for encoding the multicopy repeat sequence of the SSU rRNA, which is approximately 1850 bp in length and is the marker gene with both informative and functional roles, making it the most widely used genotyping tool for identifying host-adapted Cryptosporidium (Xiao and Feng, 2017). Host adaptation is a general phenomenon in the genus Cryptosporidium, and specific genotypes are usually associated with specific animal groups (Xiao et al., 2002). The new genotype identified in this study is evolutionarily related to C. chipmunk genotype I and Cryptosporidium hominis. C. chipmunk genotype I, originally identified in rodents (chipmunks, squirrels, and deer mice) and watershed runoff in New York, has been identified as a novel zoonotic pathogen in humans (Guo et al., 2015; Xu et al., 2019), and the disease has had small outbreaks in Sweden and poses a significant public health threat (Bujila et al., 2021). Therefore, the Cryptosporidium sp. found in plateau zokors may not only be a new host-adapted genotype, but also a novel zoonotic pathogen.
Sequence analysis of the ITS of the ribosomal RNA gene of E. bieneusi has been widely used in E. bieneusi typing studies (Widmer and Akiyoshi, 2010). Based on the high diversity of ITS, more than 500 genotypes have been identified, some of which are host-adapted genotypes (Karim et al., 2014, 2015; Santín and Fayer, 2011; Wei et al., 2019). It has been demonstrated that environmental and host isolation characteristics make differences in the distribution of E. bieneusi genotypes (Wei et al., 2019). New genotypes may exist in different hosts, for example, ten novel genotypes (WR1-WR10) were identified in wild rodents in Poland in 2015 (Perec-Matysiak et al., 2015). A study in the USA found that the E. bieneusi prevalent in animals from Texas in 2011 may be a prairie dog-specific genotype (Roellig et al., 2015).
In the present experiment, phylogenetic analysis using the ML method showed that all ITS representative gene sequences of E. bieneusi identified in this study were evolutionarily distant from the pre-existing nucleotide sequences, forming a separate branch, indicating that it may be a plateau zokor-specific genotype, and based on the differences in its molecular characteristics and host specificity, we therefore named the newly identified genotype as zokor genotype of Enterocytozoon bieneusi.
5. Conclusions
In summary, this study enriched the host range and genotype database of Cryptosporidium spp. and E. bieneusi by molecular biological detection detection and phylogenetic analysis of host-adapted genotypes in plateau zokor populations in the Tibetan Plateau region, providing a new case for environmental isolation that may lead to special species reservoirs of more host-adapted genotypes of parasites.
Author contributions statement
Hongxuan He, Bin Hu contributed to the conception of the study; Jiamin Wang, Shuairan Zhang performed the experiment; Yanan Xing, Shuyi Han contributed significantly to analysis and manuscript preparation; Bo wang helped perform the analysis with constructive discussions; Bin Hu performed the data analyses and wrote the manuscript.
Data availability statement
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
Declaration of competing interest
All the authors have no conflict of interest to declare.
Acknowledgments
This study was financially supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA19050204), National Key R&D Program of China, National Forestry and Grassland Adminstration, China, the Major Program of National Natural Science Foundation of China (No. 32090023), Beijing Innovation Consortium of Agriculture Research System (BAIC04-2021).
Contributor Information
Bin Hu, Email: hubin@ioz.ac.cn.
Jiamin Wang, Email: 627008328@qq.com.
Shuairan Zhang, Email: 897506531@qq.com.
Bo Wang, Email: wangboqueen@qq.com.
Yanan Xing, Email: 2537240310@qq.com.
shuyi Han, Email: hanshuyi13@163.com.
Hongxuan He, Email: hehx@ioz.ac.cn.
References
- Buckholt Michael A., Lee John H., Tzipori S. Prevalence of Enterocytozoon bieneusi in swine: an 18-month survey at a slaughterhouse in Massachusetts. Appl. Environ. Microbiol. 2002;68:2595–2599. doi: 10.1128/AEM.68.5.2595-2599.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujila I., Troell K., Fischerström K., Nordahl M., Killander G., Hansen A., Söderlund R., Lebbad M., Beser J. Cryptosporidium chipmunk genotype I - an emerging cause of human cryptosporidiosis in Sweden. Infect. Genet. Evol. 2021;92 doi: 10.1016/j.meegid.2021.104895. [DOI] [PubMed] [Google Scholar]
- Cai Z., Wang L., Song X., Tagore S., Li X., Wang H., Chen J., Li K., Frenkel Z., Gao D., et al. Adaptive transcriptome profiling of subterranean zokor, myospalax baileyi, to high- altitude stresses in tibet. Sci. Rep. 2018;8:4671. doi: 10.1038/s41598-018-22483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.-F., Nie X.-H., Zhang T.-Z., Du S.-Y., Duszynski D.W., Bian J.-H. Four new coccidia (Apicomplexa: Eimeriidae) from the plateau zokor, myospalax baileyi thomas (Rodentia: myospalacinae), a subterranean rodent from Haibei area, Qinghai Province, China. Syst. Parasitol. 2014;87:181–186. doi: 10.1007/s11230-013-9466-z. [DOI] [PubMed] [Google Scholar]
- Feng Y., Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Cebelinski E., Matusevich C., Alderisio K.A., Lebbad M., McEvoy J., Roellig D.M., Yang C., Feng Y., Xiao L. Subtyping novel zoonotic pathogen Cryptosporidium chipmunk genotype I. J. Clin. Microbiol. 2015;53:1648–1654. doi: 10.1128/JCM.03436-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horčičková M., Čondlová Š., Holubová N., Sak B., Květoňová D., Hlásková L., Konečný R., Sedláček F., Clark M., Giddings C., et al. Diversity of Cryptosporidium in common voles and description of Cryptosporidium alticolis sp. n. and Cryptosporidium microti sp. n. (Apicomplexa: Cryptosporidiidae) Parasitology. 2019;146:220–233. doi: 10.1017/S0031182018001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson C.B., Figueiredo L.T.M., Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin. Microbiol. Rev. 2010;23:412–441. doi: 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Liu Q., Yao B., Hou Q., Su J. Pleistocene climate-driven diversification of plateau zokor (Eospalax baileyi) in the eastern Qinghai-Tibet Plateau. Ecol. Indicat. 2022;139 [Google Scholar]
- Kang Y., Su J., Yao B., Ji W., Hegab I.M., Hanafy A.M., Zhang D. Geometric morphometric analysis of the plateau zokor (Eospalax baileyi) revealed significant effects of environmental factors on skull variations. Zoology (Jena) 2020;140 doi: 10.1016/j.zool.2020.125779. [DOI] [PubMed] [Google Scholar]
- Kang Y., Su J., Yao B., Ji W., Hegab I.M., Hanafy A.M., Zhang D. Geometric morphometric analysis of the plateau zokor (Eospalax baileyi) revealed significant effects of environmental factors on skull variations. Zoology (Jena) 2020;140 doi: 10.1016/j.zool.2020.125779. [DOI] [PubMed] [Google Scholar]
- Karim M.R., Dong H., Li T., Yu F., Li D., Zhang L., Li J., Wang R., Li S., Li X., et al. Predomination and new genotypes of Enterocytozoon bieneusi in captive nonhuman primates in zoos in China: high genetic diversity and zoonotic significance. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim M.R., Wang R., Dong H., Zhang L., Li J., Zhang S., Rume F.I., Qi M., Jian F., Sun M., et al. Genetic polymorphism and zoonotic potential of Enterocytozoon bieneusi from nonhuman primates in China. Appl. Environ. Microbiol. 2014;80:1893–1898. doi: 10.1128/AEM.03845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk A.I., van Duijvendijk G.L.A., Swart A., Heylen D., Jaarsma R.I., Jacobs F.H.H., Fonville M., Sprong H., Takken W. Effect of rodent density on tick and tick-borne pathogen populations: consequences for infectious disease risk. Parasites Vectors. 2020;13 doi: 10.1186/s13071-020-3902-0. 34-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalle M., Pozio E., Capelli G., Bruschi F., Crotti D., Cacciò S.M. Genetic heterogeneity at the beta-giardin locus among human and animal isolates of Giardiaduodenalis and identification of potentially zoonotic subgenotypes. Int. J. Parasitol. 2005;35:207–213. doi: 10.1016/j.ijpara.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Li W., Feng Y., Zhang L., Xiao L. Potential impacts of host specificity on zoonotic or interspecies transmission of Enterocytozoon bieneusi. Infect. Genet. Evol. 2019;75 doi: 10.1016/j.meegid.2019.104033. [DOI] [PubMed] [Google Scholar]
- Mello B. Estimating TimeTrees with MEGA and the TimeTree Resource. Mol. Biol. Evol. 2018;35:2334–2342. doi: 10.1093/molbev/msy133. [DOI] [PubMed] [Google Scholar]
- Ni H.B., Sun Y.Z., Qin S.Y., Wang Y.C., Zhao Q., Sun Z.Y., Zhang M., Yang D., Feng Z.H., Guan Z.H., et al. Molecular detection of cryptosporidium spp. and Enterocytozoon bieneusi infection in wild rodents from six provinces in China. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.783508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perec-Matysiak A., Buńkowska-Gawlik K., Kváč M., Sak B., Hildebrand J., Leśniańska K. Diversity of Enterocytozoon bieneusi genotypes among small rodents in southwestern Poland. Vet. Parasitol. 2015;214:242–246. doi: 10.1016/j.vetpar.2015.10.018. [DOI] [PubMed] [Google Scholar]
- Pu P., Lu S., Niu Z., Zhang T., Zhao Y., Yang X., Zhao Y., Tang X., Chen Q. Oxygenation properties and underlying molecular mechanisms of hemoglobins in plateau zokor %28%29. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2019;317:R696–R708. doi: 10.1152/ajpregu.00335.2018. [DOI] [PubMed] [Google Scholar]
- Roellig D.M., Salzer J.S., Carroll D.S., Ritter J.M., Drew C., Gallardo-Romero N., Keckler M.S., Langham G., Hutson C.L., Karem K.L., et al. Identification of Giardia duodenalis and Enterocytozoon bieneusi in an epizoological investigation of a laboratory colony of prairie dogs, Cynomys ludovicianus. Vet. Parasitol. 2015;210:91–97. doi: 10.1016/j.vetpar.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan U., Fayer R., Xiao L. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. 2014;141:1667–1685. doi: 10.1017/S0031182014001085. [DOI] [PubMed] [Google Scholar]
- Ryan U., Hijjawi N., Xiao L. Foodborne cryptosporidiosis. Int. J. Parasitol. 2018;48 doi: 10.1016/j.ijpara.2017.09.004. [DOI] [PubMed] [Google Scholar]
- Santín M., Fayer R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res. Vet. Sci. 2011;90:363–371. doi: 10.1016/j.rvsc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Shao Y., Li J.-X., Ge R.-L., Zhong L., Irwin D.M., Murphy R.W., Zhang Y.-P. Genetic adaptations of the plateau zokor in high-elevation burrows. Sci. Rep. 2015;5 doi: 10.1038/srep17262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.-Y., Wang K.-S., Yang J.-F., Mao H.-M., Pu L.-H., Zou Y., Ma J., Zhu X.-Q., Zou F.-C., He J.-J. Prevalence and novel genotypes identification of Enterocytozoon bieneusi in dairy Cattle in yunnan Province, China. Animals. 2021;11:3014. doi: 10.3390/ani11113014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger B.L.S., Horčičková M., Clark M.E., Kváč M., Čondlová Š., Khan E., Widmer G., Xiao L., Giddings C.W., Pennil C., et al. Cryptosporidium infecting wild cricetid rodents from the subfamilies Arvicolinae and Neotominae. Parasitology. 2018;145:326–334. doi: 10.1017/S0031182017001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stentiford G.D., Becnel J.J., Weiss L.M., Keeling P.J., Didier E.S., Williams B.A.P., Bjornson S., Kent M.L., Freeman M.A., Brown M.J.F., et al. Microsporidia-emergent pathogens in the global food Chain (trends in parasitology 32, 336-348; April 2, 2016) Trends Parasitol. 2016;32:657. doi: 10.1016/j.pt.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Taghipour A., Olfatifar M., Foroutan M., Bahadory S., Malih N., Norouzi M. Global prevalence of Cryptosporidium infection in rodents: a systematic review and meta-analysis. Prev. Vet. Med. 2020;182 doi: 10.1016/j.prevetmed.2020.105119. [DOI] [PubMed] [Google Scholar]
- Wang W., Cuttell L., Bielefeldt-Ohmann H., Inpankaew T., Owen H., Traub R.J. Diversity of Blastocystis subtypes in dogs in different geographical settings. Parasites Vectors. 2013;6:215. doi: 10.1186/1756-3305-6-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z., HuanHuan Z., LiuLian X., TianMing M., Gang L. Research on molecular epidemiology of Enterocytozoon bieneusi in rodents. Chin. J. Zoonoses. 2019;35:1139–1143. [Google Scholar]
- Widmer G., Akiyoshi D.E. Host-specific segregation of ribosomal nucleotide sequence diversity in the microsporidian Enterocytozoon bieneusi. Infect. Genet. Evol. : J. Mol. Epidemiol. Evol. Gene. Infect. Dis. 2010;10:122–128. doi: 10.1016/j.meegid.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 2010;124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- Xiao L., Feng Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017;8–9:14–32. doi: 10.1016/j.fawpar.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Singh A., Limor J., Graczyk T.K., Gradus S., Lal A. Molecular characterization of cryptosporidium oocysts in samples of raw surface water and wastewater. Appl. Environ. Microbiol. 2001;67:1097–1101. doi: 10.1128/AEM.67.3.1097-1101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Sulaiman I.M., Ryan U.M., Zhou L., Atwill E.R., Tischler M.L., Zhang X., Fayer R., Lal A.A. Host adaptation and host-parasite co-evolution in Cryptosporidium: implications for taxonomy and public health. Int. J. Parasitol. 2002;32:1773–1785. doi: 10.1016/s0020-7519(02)00197-2. [DOI] [PubMed] [Google Scholar]
- Xu J., Wang X., Jing H., Cao S., Zhang X., Jiang Y., Yin J., Cao J., Shen Y. Identification and genotyping of Enterocytozoon bieneusi in wild Himalayan marmots (Marmota himalayana) and Alashan ground squirrels (Spermophilus alashanicus) in the Qinghai-Tibetan Plateau area (QTPA) of Gansu Province, China. Parasites Vectors. 2020;13:367. doi: 10.1186/s13071-020-04233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Guo Y., Roellig D.M., Feng Y., Xiao L. Comparative analysis reveals conservation in genome organization among intestinal Cryptosporidium species and sequence divergence in potential secreted pathogenesis determinants among major human-infecting species. BMC Genom. 2019;20:406. doi: 10.1186/s12864-019-5788-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedi A., Ryan U. Cryptosporidium - an update with an emphasis on foodborne and waterborne transmission. Res. Vet. Sci. 2020;132:500–512. doi: 10.1016/j.rvsc.2020.08.002. [DOI] [PubMed] [Google Scholar]
- Zajac A.M., Conboy G.A., Little S.E., Reichard M.V. John Wiley & Sons; 2021. Veterinary Clinical Parasitology. [Google Scholar]
- Zhang K., Fu Y., Li J., Zhang L. Public health and ecological significance of rodents in Cryptosporidium infections. One Health. 2022;14 doi: 10.1016/j.onehlt.2021.100364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Jian Y., Li X., Ma L., Karanis G., Karanis P. The first report of Cryptosporidium spp. in Microtus fuscus (Qinghai vole) and Ochotona curzoniae (wild plateau pika) in the Qinghai-Tibetan Plateau area, China. Parasitol. Res. 2018;117:1401–1407. doi: 10.1007/s00436-018-5827-5. [DOI] [PubMed] [Google Scholar]
- Zhao F., Ma J.-Y., Cai H.-X., Su J.-P., Hou Z.-B., Zhang T.-Z., Lin G.-H. Molecular identification of Taenia mustelae cysts in subterranean rodent plateau zokors (Eospalax baileyi) Dongwuxue Yanjiu. 2014;35:313–318. doi: 10.13918/j.issn.2095-8137.2014.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.