Abstract

The branched-chain amino acid transaminases (BCATs) are enzymes that catalyze the first reaction of catabolism of the essential branched-chain amino acids to branched-chain keto acids to form glutamate. They are known to play a key role in different cancer types. Here, we report a new structural class of BCAT1/2 inhibitors, (trifluoromethyl)pyrimidinediones, identified by a high-throughput screening campaign and subsequent optimization guided by a series of X-ray crystal structures. Our potent dual BCAT1/2 inhibitor BAY-069 displays high cellular activity and very good selectivity. Along with a negative control (BAY-771), BAY-069 was donated as a chemical probe to the Structural Genomics Consortium.
Introduction
The branched-chain amino acid transaminases (BCATs) catalyze the catabolism of the essential branched-chain amino acids (BCAAs) valine, leucine, and isoleucine to branched-chain keto acids by using α-ketoglutarate (α-KG) as the recipient of the α-amino group to form glutamate.1 The two known isoenzymes are the cytosolic BCAT1 and the mitochondrial BCAT2. Whereas the expression of mitochondrial BCAT2 is rather ubiquitous, the expression of BCAT1 is restricted to specific organs and tissues. This includes the brain and neurons of the peripheral nervous system2 where the activity of BCAT1 serves as a major nitrogen source for the synthesis of the neurotransmitter glutamate.3 BCAT1 is overexpressed in many tumor types, including primary glioblastoma,4 breast cancer,5 AML,6,7 NSCLC,8 and prostate cancer,9 and plays a role in tumorigenesis and metabolism. Furthermore, BCAT1 might be involved in chemotherapy resistance10 and could be part of a local immunoevasion by glioblastoma.11 Thus, BCAT1 is an interesting target for the development of tumor therapies. The principal targetability of the BCAT enzymes has already been shown as several BCAT inhibitors have been reported.12−17 Among them is a pyrazolopyrimidinone-derived compound A developed by GSK/University of Strathclyde. A has suitable pharmacokinetic features that allowed for in vivo proof of concept in mice.16 Its published in vitro BCAT1/2 profile is presented in Figure 1. Also depicted in Figure 1 are a benzimidazole compound B identified by GSK/Northeastern University13 and a BCAT1 inhibitor C reported by Pfizer in 2002 for the treatment of neurodegenerative diseases.14,15 We resynthesized both A and C using published procedures and obtained IC50 values of 81 nM on BCAT1 and 21 nM on BCAT2 for A, largely in line with the published data in Figure 1. For C, we determined the IC50 values to be 1500 nM on BCAT1 and 8300 nM on BCAT2. We also determined the pKa values for these compounds and found a pKa value of 6.3 for A and 8.3 for C. In line with the anionic nature of its natural substrates, the amino acids leucine, valine, and isoleucine, BCAT1 shows a tendency to bind negatively charged compounds.14−17 Known BCAT binders with acidic properties include A, as well as several carboxylic acid fragments GSK identified in a fragment screen.17
Figure 1.
Previously reported BCAT inhibitors.17 The activities listed are taken from the literature.13,15,16
As part of our research activities in the area of tumor metabolism, our team set out to identify a BCAT lead-like inhibitor with primary activity on BCAT1 as a tool for in vitro, or ideally in vivo, proof of concept.4 Since more recent literature also suggested a role for BCAT2 in cancer,8 we considered some cross-reactivity with BCAT2 acceptable and thought it could even lead to additional benefit.
Results and Discussion
Screening and Hit Identification
As our primary aim was to identify a potent BCAT1 inhibitor with cellular activity, we developed a biochemical assay for the detection of BCAT1 activity. This assay utilizes leucine dehydrogenase (LeuDH) which catalyzes the nicotinamide adenine dinucleotide (NADH)-dependent reduction of the BCAA α-ketoisocaproate (α-KIC) to leucine. This gives a coupled readout, where NADH consumption was measured by fluorescent and luminescent readout (Figure 2). An in-house library containing 788,762 small-molecule compounds was screened using dimethyl sulfoxide as a negative control. To distinguish BCAT1 inhibitors from inhibitors of the cascade enzymes, in a second measurement, α-KIC was added to the reaction. The inhibitors of the cascade enzymes show an effect in this setup and were consequently eliminated. Primary hits thus obtained were tested in the dose response for confirmation of their BCAT1 target activity, and their IC50 values were determined. Based on the hypothesis that additional BCAT2 activity may contribute to the desired antiproliferative activity, we also profiled BCAT1-confirmed hits against BCAT2 in dose response. This data was supposed to further guide the selection of clusters if BCAT1/BCAT2 selectivity ratios spread widely. Our criterion for the final hit list was an IC50 value of lower than 20 μM for BCAT1 enzyme. Applying this criterion, we identified 399 compounds with the potential for inhibiting BCAT1 protein (Figure 2).
Figure 2.
Biochemical BCAT1 HTS assay and hit selection.
Selectivity profiling of the hitlist revealed a high correlation between BCAT1 and BCAT2 IC50 values, providing evidence that the development of a BCAT1-focused dual inhibitor, as was our goal, appeared practically feasible from the obtained chemical matter. At the same time, development of a BCAT1-selective tool compound, which may additionally be of scientific value to clarify the phenotypical impact of the individual BCAT enzymes, appeared a lot more difficult. Here, 59% of all hits and 71% of all hits with submicromolar BCAT1 activity were acidic, confirming the preference of BCAT1 for negatively charged compounds. Since highly charged compounds tend to have more difficulties penetrating membranes, we anticipated that target potency and cell permeabilities may become conflicting optimization parameters, difficult to improve both at the same time. To allow for balancing charge-assisted interactions relevant for target affinity with potential pharmacokinetic liabilities, our cluster prioritization process favored compounds containing structural motifs whose acidity could be readily modulated by chemical variations. A series of aryl-substituted pyrimidinediones (Figure 3) appeared as attractive starting points, combining reasonably good BEI and LLE values18−20 (17.87 and 4.47, respectively, for compound 1) and log D = 1.9, with low micromolar activity in the cellular mechanistic assay (data not shown). 1 showed fair Caco2 intestinal permeability in the apical to basolateral direction (30.0 nm/s), combined with an efflux ratio of 6.2.
Figure 3.
Co-crystal structures of human BCAT1 in complex with 1, 2, and substrate-mimic 3PP. (A) HTS hits 1 (left) and 2 (right), with IC50 values from the biochemical assays. (B) Superimposition of the crystal structures of BCAT1 in complex with 1 (PDB 7NWB chain A, ligand carbon atoms in green, protein carbon atoms in gray, and cofactor PLP in magenta) and in complex with 3PP (PDB 7NTR, for clarity, only the ligand is shown, with carbon atoms in yellow). (C) Same superimposition as B, now rotated and with the molecular surface to show the view from the solvent into the deep active site cavity. (D) Same view as B into the active site of BCAT1 in complex with 2 (chain A, ligand carbon atoms in cyan, protein carbon atoms in gray, and cofactor PLP in magenta (PDB 7NWC). (E) Superimposition of the co-crystal structures of BCAT1 with 1 and 2 (surface only shown for 1).
Chemistry efforts to build up early SAR focused on analogues around two compounds. The first was compound 2, which represents an efficient binder given its low molecular weight, and the second was the aryl ether variant compound 1, which so far had the best potency, albeit at a slightly reduced BEI. Some of our initial SAR findings around the pyrimidinedione core hinted at a nonadditive SAR (data not shown), but the SAR trends could be rationalized better when the X-ray structures of two cluster representatives in complex with hBCAT1 were solved. We were able to co-crystallize BCAT1 with compound 1, compound 2, and 3-phenylpropionic acid (3PP), a product mimic which has been reported as an additive that facilitates crystallization of BCAT1.21 BCAT1 forms a homodimer in solution and also in all published crystal structures,15,21 with every homodimer containing two identical active sites. While each chain forms the major part of one binding site, respectively, the second chain contributes one additional loop (see the Supporting Information, Figure S1). The active site is located in a deep cavity where the cofactor pyridoxal phosphate (PLP) is covalently bound to a lysine residue (Lys222) at the back wall of the deep pocket (Figure 3B,C). The crystal structure with 3PP (which can be viewed as deaminated phenylalanine) revealed the binding pocket for small BCAAs (Supporting Information, Figure S1). The crystal structures of BCAT1 in complex with compounds 1 and 2 showed that both inhibitors bind within this active site and directly in front of the PLP cofactor (Figure 3). Although chemically closely related, compounds 1 and 2 adopt very different binding modes. Compound 1 places its methylphenyl moiety into the hydrophobic section of the binding pocket where it superimposes well with the phenyl ring of 3PP. In contrast, compound 2 binds in an inverted fashion and places its trifluoromethyl moiety into this hydrophobic pocket. As a consequence, the respective pyrimidinedione cores do not overlap. Rather, the hydrogen bond formed between one of the carbonyl oxygen atoms and the backbone amide nitrogen of Val175 represents the only feature the binding modes have in common, and the exit vectors of the N-substituents point in almost opposing directions.
Altogether, the binding mode of the aryl ether subseries and the exit vectors it provided appeared to offer more options for the improvement of potency. This was in contrast to 2, where we observed no or little gain in potency for the first analogues (compounds and data not shown). We therefore focused additional SAR exploration on the aryl ether variations of the pyrimidinedione core and built SAR around compound 1.
Structure–Activity Relationship (SAR) Learning Cycles
Aryl Substitutions
As position 2′ of the derived benzonitrile-substituted pyrimidinediones is known to undergo nucleophilic aromatic substitution (SNAr) reactions,22,23 we started to explore the northern part (see Supporting Information Scheme S4), which was enhanced with libraries generated using combinatorial chemistry.24,25 Selected examples are represented in Table 1.
Table 1. Exploration of Position 2′ of Benzonitrile-Substituted Pyrimidinediones for BCAT1 and BCAT2 Activities (Selected Examples)a.
IC50 values given are arithmetic mean values over at least two independent IC50 determinations.
Compounds 1 and 3–8 are assumed to be racemates; for further information, see the Supporting Information.
Modification at position 2′ of the benzonitrile moiety afforded additional compounds in the three-digit nanomolar range regarding the BCAT1 activity. Ortho mono-substitution was important, in accordance with the X-ray co-crystal structure of compound 1, which shows the o-methyl group nicely filling the space between the Phe49 side chain and the Leu173 backbone of BCAT1. Docking26−28 with a core constraint on the pyrimidinedione suggested that ortho, ortho di-substitution with small substituents should be possible. This was confirmed by the enzymatic data (compounds 3–6). While the improvement in IC50 values compared to compound 1 was small, the o-o-dimethyl (compound 3) and o-chloro (compound 8) were the best substituents identified, the latter reflecting that the proximity of Tyr90.OH and the Arg163.Nz allow for nonaliphatic substituents to be introduced at this position.
We further investigated position 5′ of the benzonitrile moiety (Table 2), keeping the o-methyl-substituted phenyl ether as in compound 1. Analysis of the X-ray co-crystal structure of 1 suggested that sterically bulkier substituents could be added to position 5′ to fill the corresponding part of the binding pocket. In line with docking calculations keeping the pyrimidinedione core fixed, many groups were tolerated. We observed a clear preference, though, for sterically less-demanding groups, for example, methyl (10), methoxy (12), or halogen (11, 15, 17), which showed a 2-fold improved activity for BCAT1 relative to 1 or 9. The co-crystal structures of 10 and 12 confirmed the binding mode observed with compound 1 (see the Supporting Information, Figure S2). We assume that the generated compounds with substitution on the 5′-position should induce axial chirality (see the Atropisomerism section).
Table 2. Exploration of Position 5′ of Benzonitrile-Substituted Pyrimidinediones for BCAT1 and BCAT2 Activitiesa.
IC50 values given are arithmetic mean values over at least two independent IC50 determinations.
Compounds 1 and 10–17 are assumed to be racemates; for further information, see the Supporting Information.
Besides showing similar target potencies, the most potent compounds 12 and 15 also displayed similar pharmacokinetic in vitro profiles (the Fmax values in rat microsomes were 95% for 12 and 88% for 15). We elected to continue with the methoxy series, for which we had higher amounts of starting material available.
Core
Pyrimidinediones are well-known heterocycles in the literature.29 Given the influence the acidity of the 1-NH atom of the pyrimidinedione core30 has on potency, as well as on permeability, we specifically searched for chemical handles to modulate the pKa value of the pyrimidine 1-NH. In line with the estimations obtained using SimulationsPlus’ pKa predictor,31−33 substitution of the 6-position of the pyrimidinedione core proved a strong modulator of pKa values in this series.
The SAR data shown in Table 3 summarize the effects that different electron-withdrawing (12, 18–24, 27, and 28) or electron-donating (25 and 26) groups have on potency, on Caco-2 cell permeability, and on the measured pKa values of 1-NH. Altogether, no simple correlation between these endpoints can be observed.
Table 3. Exploration of Position 6 of Benzonitrile-Substituted Pyrimidinediones for BCAT1 Activity and Intestinal Caco-2 Permeability and Measured pKa Valuesa.
IC50 values given are arithmetic mean values over at least two independent IC50 determinations.
Compounds 12 and 18–28 are assumed to be racemates; for further information, see the Supporting Information. n.a.: data not available.
While calculated log D values largely capture the observed trends in passive permeability (data not shown), efflux ratios show no correlation with any simpler endpoint. Compared to compound 12, 6-substituents increasing the TPSA34 values led to an increased efflux out of the cell. Amide 28, which is only mildly acidic, illustrates that this effect appears unrelated to the pKa value of the compound.
Combining target potency with good cell permeation appears a challenge, as anticipated. Within the series, the most potent compound 24 (BCAT1 IC50 = 60 nM, BEI 14.75), carrying a phenylsulfonyl substituent, showed only low permeation in the apical to basolateral direction (Papp A–B = 0.5 nm/s) and the highest efflux ratio of 34. Compound 26, while showing no efflux, was much less potent on BCAT1. The only substituent with an attractive efflux ratio (<1) identified further to trifluoromethyl (Table 3, compound 12), which also showed similar BCAT1 activity, was α,α-difluorobenzyl (compound 21). With a pKa value of 7.0, this compound was significantly less acidic than 12. The most acidic compound, methyl sulfone 23, showed no improvement in potency.
Interestingly, structural variations at these positions also have an effect on target affinity that is only partly related to the change in pKa values. Variation of the 6-substituent of the pyrimidinedione core surprisingly revealed a broader SAR than had been anticipated based on the X-ray co-crystal structure of compound 1, in which the trifluoromethyl group is accommodated in a rather small subpocket. To better understand the unexpected target activity that substituents as large as phenylsulfonyl (compound 24) or α,α-difluorobenzyl (compound 21) could mediate, further protein–ligand complex structures were solved by X-ray crystallography. The binding modes of the 5′-fluoro variant35 of 24 and the 5′-fluoro variant of 21 resembled one another closely but featured yet another, and so far unprecedented, orientation of the pyrimidinedione core in the BCAT1 pocket (Figure 4).
Figure 4.
Co-crystal structures of BCAT1 in complex with the 5′-fluoro analogue of 21 (carbon atoms in green, PDB accession code 7NXN) and in complex with the 5′-fluoro analogue of 24 (carbon atoms of protein in gray and of inhibitor in orange, PDB accession code 7NXO), shown from two different orientations (A,B). The carbon atoms of the cofactor PLP are depicted in magenta. Shown are the active sites in the B chains. For clarity, the molecular surface is shown only in chain A and only for 24.
We had initially anticipated that the rigid pyrimidinedione core, featuring three polar groups, would form specific interactions in the binding pocket, which would be conserved over multiple analogues. However, the co-crystal structures we obtained surprisingly exhibit multiple binding modes. While the intrinsic acidity of the pyrimidinedione core is a relevant driver for target binding in the series, flexibility in the pocket allowed for multiple orientations of the core depending on the peripheral substituents.
The binding mode of compound 21 (Figure 4) appeared less attractive as a basis for further compound optimization than the shared binding mode of compounds 1 and 12 for two reasons. First, the X-ray structure of 5′-F analogue of 21 (PDB 7NXN) shows the whole aryl-ether moiety in a fairly solvent-exposed position, not engaging in tight interactions with the target protein. This is reflected in a BEI of 15.27 for 21, showing that the added molecular weight compared to 12 (BEI 18.07) does not translate into an improved IC50. Second, the binding mode of 21 provides no simple exit vector to grow into the subpocket flanked by Phe49. In the co-crystal structure of 12 (PDB 7NWM), this subpocket is nicely filled by the central phenyl ring. As a result, the possibilities to efficiently fill the remaining unoccupied space in the pocket of 21 appear limited. Given the higher attractiveness for further chemical modifications, we therefore continued to optimize the series based on the shared binding mode of compounds 1 and 12.
Role of the Cyano Group
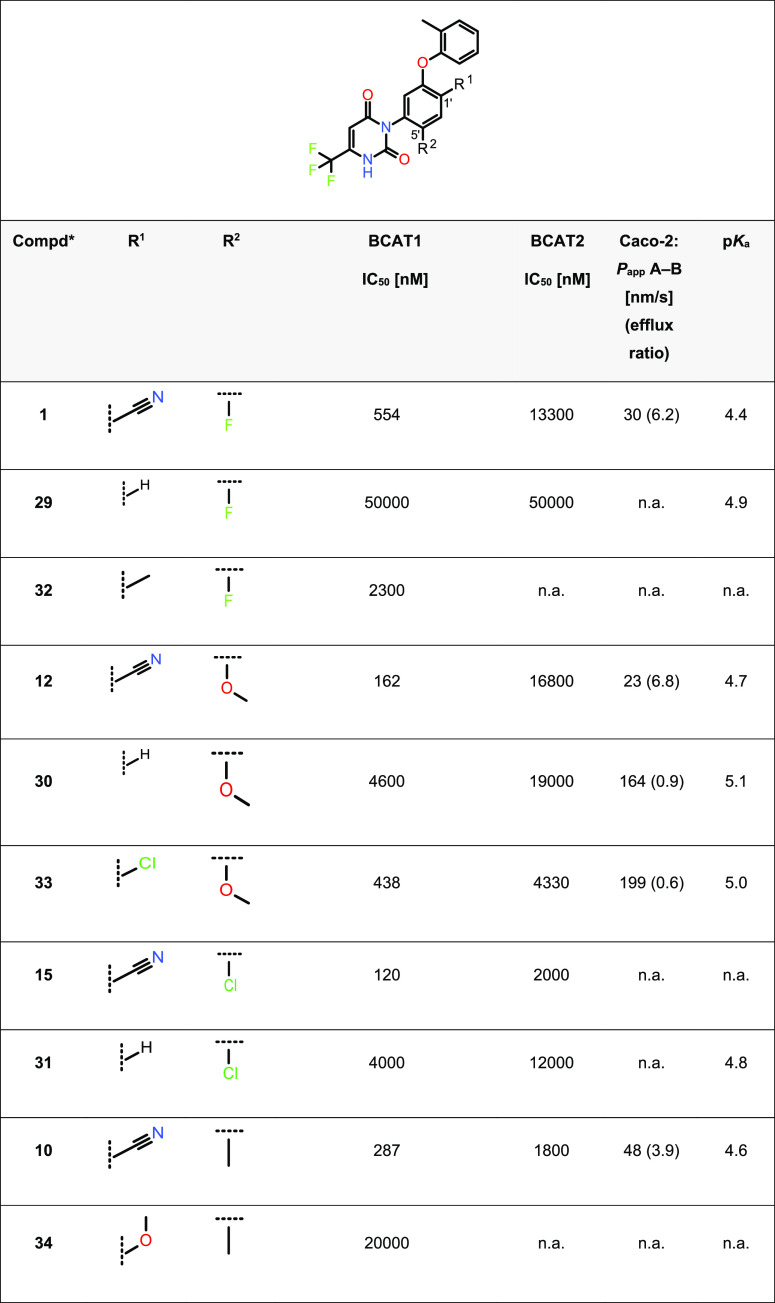
Replacement of the 1′-cyano group led to pronounced structure activity and structure property relationships, and its removal even features an affinity cliff in our pyrimidinedione series. Table 4 depicts data from some matched pairs or series36−38 (compounds 1, 29, and 32; compounds 12, 30, and 33; and compounds 15 and 31 as well as 10 and 34, respectively, share the same 5′ substituent while varying the 1′ position). In the X-ray co-crystal structure of compound 1, this cyano group did not form any specific polar interactions with BCAT1 but nicely filled a small pocket between the side chains of Lys99, Phe49, and Ala334. Generally, the 1′-position of the central phenyl ring also provides a relevant handle for modulating acidity. Electron-withdrawing groups at this position decrease the pKa value recognizably (see, e.g., the series 12, 30, and 33 in Table 4). Complete removal of the cyano substituent results in significantly higher pKa values, which is also reflected in the Caco-2 permeability (see 30). From a structural point of view, it would leave an unoccupied subpocket underneath the Ala334 side chain. In line with this, the fragment molecular orbital (FMO) group contribution calculations (see the Supporting Information) showed a loss of binding energy of 7.4 kcal/mol upon removal of the cyano group. Not surprisingly, the 1′H-carrying compounds 29, 30, and 31 showed only very weak to no activity in the enzymatic BCAT1 assay. The 1′-methyl analogue 32 showed a BCAT1 activity of 2.3 μM (IC50 value). With an experimental pKa value of 5.0, the chloro analogue 33 showed a more acidic characteristic compared to 32 while being less acidic than 15. Combining a three-digit nanomolar BCAT1 activity with good apical to basolateral permeability (no hint of efflux), 33 eventually provided the balance between target activity and cell permeability we had been looking for. The 1′-chloro substituent was thus identified as a good replacement for the cyano group. Replacement of the cyano by a methoxy group, as in compound 34 (Table 4), was not tolerated.
Table 4. Exploration of Positions 1′ and 5′ of Benzonitrile-Substituted Pyrimidinediones for BCAT1 and BCAT2 Activities and Intestinal Caco-2 Permeabilitya.
IC50 values given are arithmetic mean values over at least two independent IC50 determinations.
Compounds 1, 12, 15, and 29–34 are assumed to be racemates; for further information, see the Supporting Information. n.a.: data not available.
Despite improved Caco-2 permeability and target IC50 value, 33 (BEI 17.23) showed no cellular mechanistic activity (assays see below). We thus resorted to biochemical activity as an optimization parameter to be further improved. Guided by the available X-ray structural data, we broadened the SAR exploration around the central phenyl ring to include the 6-position of the benzonitrile.
Annulated Structures
In a series of virtual analogues modifying the central phenyl ring, naphthalene analogues appeared particularly attractive, as docking experiments showed that the annulated ring could engage in a π–π-interaction with the Phe49 side chain, thus filling unoccupied space in the binding pocket. Since free energy perturbation39−41 calculations did not reproduce experimentally available SAR data well, we turned to FMO calculations42 to estimate the potency gain associated with the annulation of a phenyl ring. Using the method described in the Supporting Information, the binding energy of 35 was calculated to be 0.9 kcal/mol stronger than that of compound 12 (ΔE = −0.9 kcal/mol). When reducing the in silico design into practice, synthesis and testing of the annulated compounds very pleasingly led to a boost in BCAT1 and BCAT2 activities, as summarized in Table 5. Indeed, cyano-substituted annulated compound 35 attained a two-digit nanomolar activity in the BCAT1 assay. Furthermore, its corresponding atropisomers 35a and 35b could be isolated. However, an efflux ratio of 6.2, along with a pKa value of 4.8, was suboptimal for further optimization of 35. In line with the SAR data displayed in Table 4, chloro-substituted annulated compound 36 was less active; nevertheless, its atropisomer 36a displayed a BCAT1 IC50 activity of 31 nM, translating into a BEI of 19.28. At the same time, it showed no efflux and, compared to 35a, a considerably less acidic pKa value of 5.7 (for the matched pair 12 and 33, the shift in pKa going from a nitrile to a chloro substituent was only 0.3, while it is 0.9 for the pair 35a and 36a). For SAR comparison, the corresponding α,α-difluorobenzyl-substituted annulated compound 37 was synthesized as well, and its atropisomers separated. In the absence of a co-crystal structure of 37, the molecular basis for the similar target activities of the separated atropisomers remains unknown, and so do the absolute stereochemistries of 37a and 37b.
Table 5. BCAT1 and BCAT2 Activities of Annulated Compounds and Intestinal Caco-2 Permeabilitiesa.
IC50 values given are arithmetic mean values over at least two independent IC50 determinations.
Compounds 35, 36, and 37 are racemates.
A second approach toward annulated structures was inspired by data published on GSK’s benzimidazole-based BCAT2 inhibitor series.13 An overlay of the respective X-ray structure (PDB accession code 5HNE) with our X-ray structure of 12 revealed remarkable similarities in the placement of aromatic moieties in the substrate pocket in front of the PLP cofactor as well as in the orientation of the exit vectors initiating from the central cores (along the 4-C–N central bond in 12) (see Figure 5B). A hybrid structure of our trifluoromethyl-substituted pyrimidinedione core and GSK’s compound B13 was designed. To enable attachment of the pyrimidinedione via one of its nitrogen atoms, the benzimidazole of GSK’s compound B was dropped in favor of an imidazopyridine system (compound 38, Figure 5A). When tested in our biochemical BCAT1 assay, the potency of this hybrid was disappointing, with an IC50 value of 850 nM.
Figure 5.
Hybrid design of compound 38. (A) Structure of compound 38, combining substructural elements of compound B13 with the trifluoromethyl-pyrimidinedione core, exemplified by compound 12. (B) Superposition of two X-ray co-crystal structures that inspired the design: Co-crystal structure of BCAT2 with the compound B13 (PDB accession code 5HNE, carbon atoms in wheat) and BCAT1 structure in complex with 12 (chain A, ligand carbon atoms in cyan, PDB 7NWM). (C) Co-crystal structure of BCAT1 with hybrid compound 38 (PDB accession code 7NY9), protein carbon atoms in gray and inhibitor carbon atoms in yellow).
Nonetheless, compound 38 allowed for an X-ray structure in complex with BCAT1 to be solved (Figure 5C, PDB 7NY9). The elucidated binding mode of 38 provided an explanation for the mediocre potency. Once more, the binding mode of the pyrimidinedione had flipped (cf. Figure 3): rather than forming a hydrogen bond to Lys79, the methyl amide group is placed between the side chain of Gln234 and the backbone segment Gln243 to Val245, with a hydrogen bond being formed between Val245NH and the carbonyl oxygen of the methyl amide. Unlike compound 38, the naphthalene analogues 35 and 36 showed the aspired improvement in potency compared to the monocyclic series and, when characterized by X-ray crystallography in complex with the target (Figure 6), showed the conserved binding mode. In both co-crystal structures, the naphthalene moiety fills a hydrophobic surface groove and inserts between the side chains of Phe49 and Tyr193. The general binding mode of the compounds in the BCAT1 pocket is unaffected by the nitrile to chloro exchange, although the former fits the subpocket between the side chains of Lys99, Phe49, and Ala334 somewhat more snugly. While 35 and 36 represent mixtures of atropisomers (see below), the electron densities show a single isomer being bound in the pocket, thus illustrating the specificity of the recognition properties of the optimized core, featuring an elevated rotation barrier around the single bond between the naphthyl and the pyrimidinedione core (see discussion below). As a consequence, the more active atropisomers 35a and 36a (Table 5) were assigned M-helical chirality. The X-ray data also nicely illustrate why the P-helical enantiomers 35b and 36b show much weaker target activity: in the corresponding binding mode (keeping the position of the naphthalene-phenyl-ether fixed), they could not engage in the same charge-assisted hydrogen bond to Gln244.
Figure 6.
(A): X-ray co-crystal structures of BCAT1 in complex with 35a (PDB accession code 7NY2, inhibitor carbon atoms in blue). (B) X-ray co-crystal structure of 36a (PDB accession code 7NYA, inhibitor carbon atoms in green).
We also determined the co-crystal structure of BCAT1 in complex with compound A and confirmed the binding mode of this series in BCAT2 as previously published for a related compound by GSK16 (PDB accession code 5BWX; see the Supporting Information, Figure S3).
Atropisomerism
The importance of atropisomerism in drug discovery is highly recognized.44−46 Atropisomers are stereoisomers which arise due to hindered rotation around a single bond. Within our chemical probe program,47 we have previously disclosed benzoisoquinolinediones presenting this interesting feature.48 A similar situation is reflected here with the pyrimidinediones: the more sterically demanding the group at position 5′, the less the free rotation around the single bond connecting the two aromatic systems. To systematically explore this effect, we undertook quantum mechanical calculations (see the Supporting Information).49 The calculated rotational energy barrier was investigated for some examples (compounds 1, 10, 13, and 16) in comparison to the 5′-unsubstituted derivative 9 (see Table 2), and the data is presented in Figure S5. Compound 1 appears to represent a class I atropisomer according to the categorization proposed by LaPlante et al.,50 whereas compounds 10, 13, and 16 are class II atropisomers with rotational energy barrier values of 22.3–24.6 kcal/mol. In contrast to the small R groups in Table 2, an annulated ring had a marked effect on the rotational energy barrier between the central aromatic system and the pyrimidinedione core. Experimentally, we found that separation of the atropisomers was not possible for the 5′-fluoro derivatives. The atropisomers of all other compounds could be easily separated using chiral HPLC. Comparing these findings to the cutoffs suggested by LaPlante et al.,50 interconversion of our atropisomers surprisingly occurs at somewhat higher calculated rotational energy barrier.
The separated atropisomers 36a and 36b of compound 36 were individually submitted to our enzymatic assay, which revealed that the more active atropisomer 36a, characterized to be M-helical by means of its X-ray co-crystal structure with BCAT1, has an IC50 of 31 nM (BCAT1), while the other atropisomer 36b has only micromolar activity (see Table 5).
Cellular Mechanistic Activity and Proliferation
To study the translation of target potency into cellular effects, we developed a cellular mechanistic assay which measures the levels of leucine in the medium of different tumor cell lines (U-87 MG and MDA-MB-231). As the enzyme BCAT catalyzes the consumption of leucine, a decrease in BCAAs reflects the cellular BCAT activity. In general, the test compounds showed substantially weaker cellular activity than biochemical activity (Table 6). As shown in Figure S19 (see the Supporting Information), a decent correlation was seen between biochemical BCAT1 activity and cellular activity in U-87 MG cells (which show high expression of BCAT1); however, some compounds with potent biochemical activity only displayed weak activity in cells. This might be explained by low permeability into cells. 36a likely owes its remarkably small shift in IC50, when comparing biochemical and cellular data, to its good cell permeability properties.
Table 6. BCAT1 and BCAT2 Activities and BCAA Measurement in U-87 MG and MDA-MB-231 Cell Lines for Inhibitors 35 and 36 and Their Respective Atropisomers a and ba.
| BCAA IC50 |
||||
|---|---|---|---|---|
| compd | BCAT1 IC50 | BCAT2 IC50 | U-87 MG cell line | MDA-MB-231 cell line |
| 35 | 70 nM | 270 nM | 5 μM | 5 μM |
| 35a | 90 nM | 290 nM | 2.7 μM | 780 nM |
| 35b | 3.5 μM | 5.5 μM | n.a. | 4.7 μM |
| 36(43) | 186 nM | 1.2 μM | 1.8 μM | 1 μM |
| 36a | 31 nM | 153 nM | 358 nM | 874 nM |
| 36b | 2.3 μM | 2.7 μM | 2 μM | n.a. |
Strong loss of compound 36a potency was observed in a biochemical BCAT1 assay in the presence of bovine serum albumin or human serum albumin. The IC50 shifted to the micromolar range in the presence of 2% serum albumin (see Supporting Information, Figure S17). High plasma protein binding might thus limit the unbound compound concentrations and partially contribute to the lack of efficacy of 36a in in vitro assays with serum albumin (e.g., proliferation assays).
As compound 36a (now termed BAY-069) presented the best cellular activities, it was selected as an in vitro probe candidate.
To further address the potential of BCAT1/2 as therapeutic targets in oncology, we tested the impact of compound 36a (BAY-069) and compound A (see Figure 1) on the proliferation of U-87 MG (high BCAT1 expression) and MDA-MB-231 (high BCAT2 expression) cells (in cell culture medium with 10% FCS). Importantly, despite their ability to increase the BCAA levels in these cell lines, both BAY-069 and compound A failed to inhibit cell proliferation (see the Supporting Information, Table S5). Furthermore, a consistent lack of an antiproliferative effect for both compounds was observed in an extended panel of cell lines with high BCAT1 and/or BCAT2 expression, including SEM, CAL-51, HCC-33, and NCI-H2110 cells (see the Supporting Information, Table S5 and Figure S18). Additionally, compound A was similarly inactive in 3D proliferation experiments performed in U-87 MG cells (in cell culture medium containing 1 or 10% FCS) (data not shown). However, several studies have demonstrated a rather weak effect of BCAT1 knockdown in in vitro assays like proliferation,4,5,8 on glutamate levels,51 and on colony numbers52 compared to the achieved antitumor effects in the subsequent in vivo studies. Presumably, this highlights the dependence of the effects from the tumor environment and other host factors.
In Vitro and In Vivo Pharmacokinetic Profile
The in vitro metabolic stability of compound 36a (BAY-069) was high after incubation with human liver microsomes and moderate after incubation with rat hepatocytes, resulting in a high and moderate predicted maximal oral bioavailability (Fmax), respectively (Table 7). Permeability through Caco-2 cell monolayers was high with no hint of efflux, suggesting good oral absorption characteristics. Studies of the in vivo pharmacokinetics (PK) of BAY-069 in rats (Table 7) revealed a favorable pharmacokinetic profile after iv dosing with low blood clearance (CLblood), moderate volume of distribution at steady state (Vss), and intermediate terminal half-life (t1/2). The observed oral availability was high and in accordance with the predicted bioavailability based on iv clearance, indicating unmitigated absorption. Plasma protein binding in the tested species was high with a fraction unbound (fu) of 0.14% in the mouse and 0.055% in the rat.
Table 7. Pharmacokinetic Properties of Compound 36a (BAY-069)a.
| BAY-069 (36a) | |||
|---|---|---|---|
| metabolic stability | CLblood[L/h/kg] | Fmax [%] | |
| human liver microsomes | 0.11 | 92 | |
| rat hepatocytes | 1.8 | 56 | |
| Caco-2 permeability | PappA–B[nm/s] | PappB–A [nm/s] | efflux ratio |
| 252 | 122 | 0.48 | |
| species | CLblood[L/h/kg] | Vss[L/kg] | t1/2 [h] iv | AUCnorm[kg·h/L] iv | AUCnorm[kg h/L] po | F [%] po |
|---|---|---|---|---|---|---|
| rat | 0.64 | 0.25 | 1.6 | 2.9 | 2.5 | 89 |
Doses applied to rats were 0.3 mg/kg i.v. and 0.6 mg/kg for p.o. administration.
Further studies of the in vivo PK with orally administered single doses of compound 36a (BAY-069) at 25, 50, and 100 mg/kg in female NMRI nude mice revealed an overproportional increase in the plasma levels of BAY-069 in the animals (see the Supporting Information). However, at the highest dose used, the unbound plasma concentrations of BAY-069 reached, but did not exceed, the estimated half-maximal inhibitory concentration (IC50) determined with the cellular BCAA assay. The unbound brain-to-plasma partition ratio (Kpuu, brain) was 0.22.
Synthesis
Compound 36a (BAY-069) was synthesized using a five-step procedure starting from commercially available 2-bromo-4-nitronaphthalen-1-amine (Scheme 1) which was submitted to a copper(I)-catalyzed Ullmann-type reaction to provide the methylaryl ether 39. The aniline moiety of 39 was then transformed into the corresponding aryl chloride 40 in 45% yield using a Sandmeyer-type reaction in the presence of tert-butyl nitrite as a diazotization reagent. Thereafter, the nitro group was reduced smoothly using iron powder under acidic conditions. The resulting amine 41 was converted into methyl carbamate 42 using standard conditions. Finally, pyrimidinedione 36 was produced in 38% yield in a one-pot two-step protocol with sodium hydride and ethyl 3-amino-4,4,4-trifluorobut-2-enoate. Separation of the atropisomers by chiral HPLC provided compound 36a (BAY-069). All additional pyrimidinedione derivatives described in this manuscript (1–35 and 38) were synthesized in a similar fashion (see the Supporting Information).
Scheme 1. Synthetic Route for Compound 36.

Reagents and conditions: (a) o-cresol, CuCl, Cs2CO3, 2,2,6,6-tetramethylheptane-3,5-dione, 1-methyl-2-pyrrolidinone (NMP), 120 °C, overnight, and 23%; (b) CuCl2, tBuONO, MeCN, 70 °C, 2 h, and 45%; (c) Fe, AcOH, rt, 2 h, and 74%; (d) methyl chloroformate, pyridine, rt, 4 h, and 56%; (e) (i) NaH, ethyl 3-amino-4,4,4-trifluorobut-2-enoate DMF, 0 °C, and 10 min; and (ii) addition of 42, 100 °C, 24 h, and 38%.
Selectivity of Compound 36a (BAY-069)
BAY-069 was evaluated in a biochemical assay for aspartate transaminases (GOT1 and GOT2, glutamate-oxaloacetate transaminase) and showed no activity (IC50 > 50 μM). We also tested BAY-069 in an in-house protease panel (30 proteases) and kinase panel (30 kinases). No activity (IC50 > 10 μM) was detected except for one protease with an IC50 = 6 μM and one kinase hit with an IC50 = 2 μM. Furthermore, BAY-069 was additionally evaluated in the Eurofins LeadProfilingScreen panel, which contains 77 potential targets, and no inhibition or stimulation exceeding 45% was noted in the presence of 10 μM compound [residual activities for thromboxane synthase (45%) and GABA transporter (46%); see the Supporting Information, Table S6]. To identify potential additional targets of the (trifluoromethyl)pyrimidinedione series outside the protein family, the co-crystal structure of compound 35 was used to query the PDB for similar binding pockets independent of sequence similarities using CavBase.53 No significant hits were identified, providing further support that we have not overlooked any potential off-targets and that the series appears selective for BCATs.
Negative Control
To successfully probe the effect of a BCAT1 inhibitor in vitro, a negative control with the same chemotype and only minimal structural changes compared to the active probe is usually seen as highly desirable to best retain the off-target activity profile of 36a (BAY-069). In a recent publication, Lee and Schapira54 evaluated and nicely demonstrated the possible risk associated with the use of negative controls. For future considerations, the development of two chemically unrelated probes would be justified.
While an inactive enantiomer can often provide a suitable negative probe, in our case, 36b still showed an unsatisfyingly high cellular activity. As an alternative approach, through our SAR insights shown in Table 4, we expected that removal of the chlorine atom should lead to a major drop in affinity (see above, role of the Cyano Group). We identified a structurally close pyrimidinedione compound 43 (BAY-771) with good lead-like properties and high permeability in Caco-2 cells (no hint of efflux), as shown in Table 8.55 Compared to the matched pair 30, 33 (ΔpKa = 0.1) in Table 4, the shift in pKa replacing a chlorine by a hydrogen atom is increased in the annulated series (matched pair 36a, 43, ΔpKa = 0.91).
Table 8. Overall Profile of Inactive Control Compound 43 (BAY-771).
The annulated pyrimidinedione 43 shows very weak inhibitory activity in the BCAT1 biochemical assay and no activity in BCAT2. The BCAA activity of compound 43 in the U-87 MG cell line is surprisingly close to the enzymatic activity, resulting in an only 17-fold selectivity relative to the active probe 36a (BAY-069) in this assay.
Conclusions
(Trifluoromethyl)pyrimidinediones56 were identified as inhibitors of BCAT1/2 using an HTS campaign. SAR analysis, together with X-ray crystallographic studies, revealed that the binding of the pyrimidinedione core is largely driven by its acidity, while the interaction partners in the pocket and thus the resulting binding modes within the active site of BCAT1 vary considerably depending on the substitution pattern of the inhibitor. Based on the experimental binding mode of compound 1, and guided by protein–structure-based design, optimization of the series led to annulated compounds that showed atropisomerism and eventually combined high affinity with good cell permeability. The optimized BCAT1/2 inhibitor, BAY-069 (36a), displays high cellular activity. Its overall in vivo pharmacokinetic profile and its selectivity in various in vitro panels suggest that BAY-069 is suitable for in vivo experiments.
BAY-069 was accepted by the Structural Genomics Consortium (SGC)57 as a donated chemical probe47,58,59 to support open science. BAY-069 and the corresponding negative control BAY-771 (43) are available free of charge through the SGC Donated Chemical Probes program.57 We are convinced that BAY-069 due to its unique selectivity profile will be a valuable tool to better understand the biology and pharmacology of BCAT inhibition in the field of tumor metabolism in the future, as an addition to the previously reported compounds A, B, and C.
Experimental Section
Chemistry
General Comments
Chemical names were generated using ACD/Name Batch or AutoNom 2000, following IUPAC nomenclature.
Stereochemistry
In certain cases, atropisomerism could be induced, and the compounds are assumed to be obtained as racemic mixtures of atropisomers. In the cases where the atropisomeric ratio was investigated, the ratio and the retention times of the corresponding atropisomers are given in the analytical data part of the corresponding racemates. In some cases, the atropisomers were separated using chiral HPLC. The HPLC methods used for separation are described in the individual cases, along with the corresponding analytical data. For the atropisomeric pairs, the atropisomer with the shorter retention time (LC–MS) was defined as atropisomer 1 (atrop1) and that with the longer retention time as atropisomer 2 (atrop2). The absolute configuration of the obtained atropisomers was not determined.
Analytical Methods
All NMR spectroscopy results were recorded on Bruker AVANCE III HD spectrometers. 1H NMR spectra were obtained at 400 MHz and referenced to the residual solvent signal (2.50 ppm for [D]6DMSO). All spectra were obtained at ambient temperature (22 ± 1 °C). Peak forms and multiplicities are specified as apparent in the spectra; potential higher order effects have not been considered. Chemical shifts (δ) are reported in parts per million (ppm) and coupling constants (J) in hertz (Hz). Spin multiplicities are reported as s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, bs = broad singlet, and mc = centered multiplet. Optical rotations were measured on a P2000 JASCO polarimeter using a 1 mL microcell (10 cm thickness and 3 mm diameter) and the α-d-line of sodium at 20 °C.
LC–MS Methods (Analytic)
Method 1: instrument: Shimadzu LCMS, UFLC 20-AD, LCMS-2020 MS detector; column: Ascentis Express C18 2.7 μM, 50 × 3.0 mm; eluent A: water + 0.05 vol % TFA, eluent B: acetonitrile + 0.05 vol % TFA; gradient: assigned for each compound; flow: 1.2 mL/min; temperature: 40 °C; PDA scan: 190–400 nm.
Method 2: instrument: Shimadzu LCMS, UFLC 20-AD, LCMS-2020 MS detector; column: CORTECS C18 2.7 μM, 50 × 2.1 mm; eluent A: water + 0.1 vol % formic acid, eluent B: acetonitrile + 0.10 vol % formic acid; gradient: assigned for each compound; flow: 1.2 mL/min; temperature: 40 °C; PDA scan: 190–400 nm.
Method 3: instrument: Waters Acquity UPLC–MS Single Quad; column: Acquity UPLC BEH C18 1.7 μM, 50 × 2.1 mm; eluent A: water + 0.1 vol % formic acid (99%), eluent B: acetonitrile; gradient: 0–1.6 min 1–99% B, 1.6–2.0 min 99% B; flow: 0.8 mL/min; temperature: 60 °C; DAD scan: 210–400 nm.
Method 4: instrument: Waters AutoPurification MS Single Quad; column: Waters XBridge C18 5 μM, 100 × 30 mm; eluent A: water + 0.1 vol % formic acid (99%), eluent B: acetonitrile; gradient: 0–5.5 min 5–100% B; flow: 70 mL/min; temperature: 25 °C; DAD scan: 210–400 nm.
Method 5: instrument: Agilent 1290 UPLC–MS 6200 TOF; column: BEH C18 1.7 μm, 50 × 2.1 mm; eluent A: water + 0.05% formic acid (99%); eluent B: acetonitrile + 0.05% formic acid (99%); gradient: 0–1.7, 2–90% B, 1.7–2.0, 90% B; flow 1.2 mL/min; temperature: 60 °C; DAD scan: 190–400 nm.
Reagents
All reagents for which the synthesis is not described below are either commercially available or were synthesized according to literature procedures. All final products were at least 95% pure, as determined by analytical HPLC, apart from intermediates and where otherwise indicated.
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-5-fluoro-2-(2-methylphenoxy)benzonitrile (1)
Commercially available 4-[2,6-dioxo-4′-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2,5-difluorobenzo-nitrile (CAS: 162926-25-2;23 500 mg, 1.58 mmol), potassium carbonate (545 mg, 3.94 mmol), and 2-methylphenol (160 μL, 1.6 mmol) were suspended in DMSO (5.5 mL), and the reaction was heated to 110 °C for 21 h. Upon completion of the reaction, the mixture was filtered and concentrated under vacuum. The crude material was purified by HPLC-HT, affording the desired product (409 mg, 61%) as a colorless solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.15 (s, 3H, Me), 6.35 (br s, 1H, 3′-H), 7.05 (d, J = 5.7 Hz, 1H), 7.11 (dd, J = 1.3, 7.8 Hz, 1H), 7.23 (td, J = 7.4, 1H), 7.31 (t, J = 7.4 Hz, 1H), 7.39 (dd, J = 0.8, 7.4 Hz, 1H), 8.17 (d, J = 9.1 Hz, 1H), 12.71 (br s, 1H, NH); LCMS (ESI+) m/z: 406 [M + H]+.
2-(2,6-Dimethylphenoxy)-4-[2,6-dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-5-fluorobenzonitrile (3)
Commercially available 4-[2,6-dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2,5-difluorobenzonitrile (CAS: 162926-25-2,23 100 mg, 315 μmol), potassium carbonate (109 mg, 788 μmol), and 2,6-dimethylphenol (38.5 mg, 315 μmol) were suspended in DMSO (2 mL), and the reaction was heated to 110 °C for 21 h. Upon completion of the reaction, the mixture was filtered and concentrated under vacuum. The crude material was purified by HPLC-HT, affording the title compound (29 mg, 21%) as an ochre solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.09 (s, 6H), 6.34 (s, 1H), 6.85 (d, J = 5.8 Hz, 1H), 7.14–7.23 (m, 3H), 8.16 (d, J = 8.9 Hz, 1H), 12.66 (br s, NH) ppm; LCMS (ESI+) m/z: 418 [M – H]−.
2-(2,6-Difluorophenoxy)-4-[2,6-dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-5-fluorobenzonitrile (4)
Commercially available 4-[2,6-dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2,5-difluorobenzonitrile (CAS: 162926-25-2,23 100 mg, 315 μmol), potassium carbonate (109 mg, 788 μmol), and 2,6-difluorophenol (41.0 mg, 315 μmol) were suspended in DMSO (2 mL), and the reaction was heated to 110 °C for 21 h. Upon completion of the reaction, the mixture was filtered and concentrated under vacuum. The crude material was purified by HPLC-HT, affording the title compound (12 mg, 8%) as an ochre solid. 1H NMR (400 MHz, [D]6 DMSO): δ 6.39 (s, 1H), 7.34–7.50 (4H), 8.23 (d, J = 9.1 Hz, 1H), 12.78 (br s, NH) ppm LCMS (ESI−) m/z: 427 [M + H]−.
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2-(2-ethyl-6-methylphenoxy)-5-fluorobenzonitrile (5)
Commercially available 4-[2,6-dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2,5-difluorobenzonitrile (CAS: 162926-25-2,23 100 mg, 315 μmol), potassium carbonate (109 mg, 788 μmol), and 2-ethyl-6-methylphenol (42.9 mg, 315 μmol) were suspended in DMSO (2 mL), and the reaction was heated to 110 °C for 21 h. Upon completion of the reaction, the mixture was filtered and concentrated under vacuum. The crude material was purified by HPLC-HT, affording the title compound (23 mg, 16%) as an ochre solid. 1H NMR (400 MHz, [D]6 DMSO): δ 1.10 (t, J = 7.6 Hz, 3H), 2.07 (s, 3H), 2.42–2.50 (m, 2H)*, 6.33 (s, 1 H), 6.84 (d, J = 6.1 Hz, 1H), 7.18–7.25 (m, 3H), 8.16 (d, J = 9.1 Hz, 1H), 12.64 (br s, NH) ppm; *overlay with DMSO peak LCMS (ESI+) m/z: 432 [M – H]−.
2-(2-Chloro-6-methylphenoxy)-4-[2,6-dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-5-fluorobenzonitrile (6)
Commercially available 4-[2,6-dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2,5-difluorobenzonitrile (CAS: 162926-25-2,23 100 mg, 315 μmol), potassium carbonate (109 mg, 788 μmol), and 2-chloro-6-methylphenol (74.2 mg, 520 μmol) were suspended in DMSO (1.6 mL), and the reaction was heated to 110 °C for 17 h. Upon completion of the reaction, the mixture was filtered and concentrated under vacuum. The crude material was purified by HPLC-HT, affording the title compound (37.8 mg, 17%) as a beige solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.16 (s, 3H), 6.34 (s, 1H), 6.94 (d, J = 5.6 Hz, 1H), 7.30 (t, J = 7.8 Hz, 1H), 7.39, 7.50 (2 m, 1H each), 8.19 (d, J = 9.1 Hz, 1H), 12.68 (br s, NH) ppm; LCMS (ESI+) m/z: 440 [M + H]+.
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-5-fluoro-2-phenoxybenzonitrile (7)
Commercially available 4-[2,6-dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2,5-difluorobenzonitrile (CAS: 162926-25-2,23 100 mg, 315 μmol), potassium carbonate (109 mg, 788 μmol), and phenol (29.7 mg, 315 μmol) were suspended in DMSO (1.6 mL), and the reaction was heated to 110 °C for 21 h. Upon completion of the reaction, the mixture was filtered and concentrated under vacuum. The crude material was purified by HPLC-HT, affording the title compound (63 mg, 49% yield) as a colorless solid. 1H NMR (400 MHz, [D]6 DMSO): δ 6.38 (s, 1H), 7.18, 7.29, 7.49 (3 m, 2H each), 8.18 (d, J = 9.1 Hz, 1H), 12.75 (br s, NH) ppm; LCMS (ESI+) m/z: 390 [M – H]−.
Compound 8 was synthesized using a different synthetic route:
2-(2-Chlorophenoxy)-4-[2,6-dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-5-fluorobenzonitrile (8)
To a cooled suspension of commercially available NaH (CAS: 7440-23-5, 60% in mineral oil, 34.6 mg, 865 μmol) in DMF (1.5 mL), ethyl-3-amino-4,4,4-trifluorobut-2-enoate (120 μL, 810 μmol, dissolved in 0.3 mL DMF) was added dropwise. The reaction mixture was stirred at RT for around 40 min (until no formation of gas was observed anymore). Then, carbamate (65, 227 mg, 85% purity, 576 μmol, dissolved in 1.2 mL DMF) was added, and the reaction solution was heated to 90 °C for 18 h. Upon completion of the reaction, the mixture was poured into water, the resulting precipitate was filtered off, and the aqueous solution was acidified until pH = 3 with an aqueous solution of 2 M HCl (1.3 mL). The solution was then diluted with DCM. The organic phase was extracted twice. The combined organic layers were washed with brine and dried with sodium sulfate. After filtration, the solvent was removed under vacuum. The resulted residue was purified using HPLC-HT (basic), affording the desired title product in two fractions (72 mg with 90% purity; 47.6 mg with 95% purity; 45% yield) as both colorless solids. Analytics of the 95% pure fraction: 1H NMR (400 MHz, [D]6 DMSO): δ 6.35 (s, 1H), 7.14 (d, J = 5.8 Hz, 1H), 7.37, 7.48 (2 m, 2H, 1H), 7.68 (dd, J = 7.9, 1.4 Hz, 1H) 8.20 (d, J = 8.9 Hz, 1H), 12.73 (br s, NH) ppm; LCMS (ESI+) m/z: 426 [M + H]+.
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2-(2-methylphenoxy)benzonitrile (9)
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2-fluorobenzonitrile (79, 70.0 mg, 234 μmol), potassium carbonate (80.8 mg, 585 μmol), and 2-methylphenol (24 μL, 230 μmol) were suspended in DMSO (2.1 mL), and the reaction was heated to 110 °C for 20 h. Upon completion of the reaction, the mixture was filtered and concentrated under vacuum. The crude material was purified by HPLC-HT, affording the title compound (50 mg, 42%) as a colorless solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.14 (s, 3H), 6.25 (s, 1H), 6.82 (m, 1H), 7.11 (d, J = 7.9 Hz, 1H), 7.19–7.25 (m, 2H), 7.31 (m, 1H), 7.38 (d, J = 7.4 Hz, 1H), 8.00 (d, J = 8.1 Hz, 1H), 12.45 (br s, NH) ppm; LCMS (ESI+) m/z: 388 [M + H]+.
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-5-methyl-2-(2-methylphenoxy)benzonitrile (10)
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2-fluoro-5-methylbenzonitrile (80, 100 mg, 319 μmol), potassium carbonate (110 mg, 798 μmol), and 2-methylphenol (33 μL, 320 μmol) were suspended in DMSO (2 mL), and the reaction was heated to 110 °C for 21 h. Upon completion of the reaction, the mixture was filtered and concentrated under vacuum. The crude material was purified by HPLC-HT, affording the title compound (18 mg, 13%) as a light brown solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.04, 2.13 (2 s, 3H each), 6.30, 6.88 (2 s, 1H each), 7.06 (dd, J = 7.9, 1.1 Hz, 1H), 7.21, 7.29, 7.36 (3 m, 1H each), 7.92 (s, 1H), 12.53 (br s, NH) ppm; LCMS (ESI+) m/z: 401 [M + H]+.
5-Bromo-4-[2,6-dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2-(2-methylphenoxy)benzonitrile (11)
To a suspension of sodium hydride (337 mg, 8.4 mmol, 60% in mineral oil) in N,N-dimethylformamide (50 mL) was added ethyl 3-amino-4,4,4-trifluorobut-2-enoate (1.5 g, 8.4 mmol) at 0 °C. After stirring at this temperature for 30 min, methyl-carbamate (70, 1.90 g, 4.20 mmol, 80% purity) was added to the above slurry solution. The resulting mixture was stirred at 100 °C overnight under a nitrogen atmosphere. After cooling to room temperature (rt), saturated ammonium chloride solution was added at 0 °C, and the resulting mixture was extracted with ethyl acetate. The combined organic layer was dried over anhydrous sodium sulfate and concentrated in vacuo. The residue was purified by silica gel column chromatography (petroleum ether: ethyl acetate = 1: 1) to afford 1.1 g (50% yield) of the product as a white solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.13 (s, 3H), 6.21 (br, 1H), 6.96 (s, 1H), 7.15 (d, 1H), 7.21–7.26 (m, 1H), 7.30–7.34 (m, 1H), 7.39 (d, 1H), 8.42 (s, 1H), 12.68 (br, 1H) ppm; LCMS (ESI−) m/z: 464 [M – H]−.
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-5-methoxy-2-(2-methylphenoxy)benzonitrile (rac) (12)
2-Bromo-4-[2,6-dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-5-methoxybenzonitrile (91, 600 mg, 1.54 mmol), 2-methylphenol (220 μL, 2.20 mmol), N,N-dimethylglycine (42.0 mg, 408 μmol), copper(I) iodide (39.5 mg, 208 μmol), and cesium carbonate (1.20 g, 3.69 mmol) were heated in DMF (30 mL) for 18 h at 140 °C. Upon completion of the reaction, work-up, and purification, the title compound was obtained (35 mg, 6%) as an ochre solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.18 (s, 3 H), 3.79 (s, 3H), 6.26 (br s, 1 H), 6.97 (m, 2H), 7.17, 7.27, 7.35 (3 m, 1H each), 7.73 (s, 1H), 12.56 (br s, 1H); LCMS (ESI−) m/z: 416 [M – H]−. Atropisomeric ratio: atrop 1/atrop 2 = 50:34 (16% impurities); tR (atrop1) = 3.20 min; tR (atrop2) = 4.99 min. The atropisomeric ratio was determined using the following chiral HPLC method: instrument: Agilent HPLC 1260; column: Chiralpak ID 3 μm 100 × 4.6 mm; eluent A: hexane + 0.1 vol % diethylamine (99%); eluent B: 2-propanol; gradient: 20–50% B in 7 min; flow: 1.4 mL/min; temperature: 25 °C; DAD 254 nm.
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-5-hydroxy-2-(2-methylphenoxy)benzonitrile (13)
To a solution of 5-bromo-4-[2,6-dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2-(2-methylphenoxy)benzonitrile (11, 1.00 g, 1.90 mmol) in dry 1,4-dioxane (30 mL) were added bis(pinacolato)diboron (1.00 g, 3.90 mmol), potassium acetate (570 mg, 5.8 mmol), and [1,1′-bis(diphenylphosphino)ferrocene]dichloropalladium(II) (141 mg, 0.2 mmol). The resulting mixture was stirred at 90 °C overnight under a nitrogen atmosphere. After cooling to rt, hydrogen peroxide (0.6 mL, 5.8 mmol, 30% aqueous solution) was added, and the resulting mixture was stirred at rt for another 1 h. Upon completion of the reaction, the solvent was removed in vacuo, and the residue was diluted with water. The resulting mixture was extracted with ethyl acetate, and the combined organic layers were dried over anhydrous sodium sulfate. The solvent was removed in vacuo, and the residue was purified by Prep-HPLC [eluent A: water, eluent B: acetonitrile; gradient 30% B to 60% B in 15 min] to afford the title compound (250 mg, 26% yield) as a white solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.08 (s, 3H), 6.29 (s, 1H), 6.90–6.93 (m, 2H), 7.13 (t, 1H), 7.24 (t, 1H), 7.32–7.35 (m, 2H), 10.28 (s, 1H), 12.67 (br, 1H); LCMS (ESI+) m/z: 404 [M + H]+.
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2-(2-methylphenoxy)-5-propoxybenzonitrile (14)
To a suspension of sodium hydride (38 mg, 0.9 mmol, 60% in mineral oil) in N,N-dimethylformamide (5 mL) was added ethyl 3-amino-4,4,4-trifluorobut-2-enoate (173 mg, 0.9 mmol) at 0 °C. After stirring at this temperature for 30 min, methyl 4-cyano-2-propoxy-5-(o-tolyloxy)phenylcarbamate (71, 200 mg, 0.5 mmol) was added to the above mixture, and the resulting solution was stirred at 100 °C overnight under a nitrogen atmosphere. After cooling to rt, ammonium chloride solution was added at 0 °C, and the solvent was removed in vacuo. The residue was purified by Prep-HPLC [mobile phase A: water (0.1% NH4HCO3), mobile phase B: acetonitrile; gradient: 25% B to 50% B in 8 min] to afford 65.6 mg (31% yield) of the product as a white solid. 1H NMR (400 MHz, [D]6 DMSO): δ 0.84 (t, 3H), 1.56–1.64 (m, 2H), 2.19 (s, 3H), 3.99 (t, 2H), 6.27 (s, 1H), 6.94 (s, 1H), 6.98 (d, 1H), 7.17 (t, 1H), 7.26 (t, 1H), 7.35 (d, 1H), 7.72 (s, 1H), 12.54 (br, 1H) ppm; LCMS (ESI+) m/z: 446 [M + H]+.
5-Chloro-4-[2,6-dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2-(2-methylphenoxy)benzonitrile (15)
To a slurry of sodium hydride (152 mg, 3.8 mmol, 60% in mineral oil) in N,N-dimethylformamide (50 mL) were added ethyl-3-amino-4,4,4-trifluorobut-2-enoate (690 mg, 3.8 mmol) and methyl [2-chloro-4-cyano-5-(2-methylphenoxy)phenyl]carbamate (72, 1.0 g, 2.5 mmol), at 0 °C. The resulting mixture was stirred at 100 °C overnight under a nitrogen atmosphere. After cooling to rt, ammonium chloride solution was added, and the resulting mixture was extracted with ethyl acetate. The combined organic layer was concentrated in vacuo, and the residue was purified by Prep-HPLC [mobile phase A: water (0.1% FA), mobile phase B: acetonitrile; gradient: 40% B to 60% B in 8 min] to afford 480 mg (44%) of the product as a light yellow solid. It was separated by Chiral-HPLC [Column: CHIRAL ART Cellulose-SB, 2 × 25 cm, 5 μm; mobile phase A: n-hexane (0.1% FA), mobile phase B: IPA; gradient: 20% B to 20% B in 16 min; RT1 = 7.55 min, RT2 = 9.20 min] to afford 161.3 mg (33%) of the first eluting isomer as a light yellow solid and 142 mg (29% yield) of second eluting isomer as a light yellow solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.13 (s, 3H), 6.37 (s, 1H), 7.10 (s, 1H), 7.16 (d, 1H), 7.26 (t, 1H), 7.32 (t, 1H), 7.39 (d, 1H), 8.35 (s, 1H), 12.68 (br, 1H) ppm; LCMS (ESI−) m/z: 420 [M – H]−.
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2-(2-methylphenoxy)-5-(trifluoromethoxy)benzonitrile (16)
To a solution of 4-(2,6-dioxo-4-(trifluoromethyl)-2,3-dihydropyrimidin-1(6H)-yl)-5-hydroxy-2-(o-tolyloxy)benzonitrile (13, 200 mg, 0.4 mmol, 82% purity) in (trifluoromethyl)benzene/toluene (12 mL, v/v = 2: 1) were added AgOTf (522 mg, 2.0 mmol), selectfluor (288 mg, 0.8 mmol), N-fluorobenzenesulfonimide (256 mg, 0.8 mmol), CsF (371 mg, 2.4 mmol), 2-fluoropyridine (189 mg, 2.0 mmol), and TMSCF3 (289 mg, 2.0 mmol). The resulting mixture was stirred at rt overnight under a nitrogen atmosphere. Upon completion of the reaction, water was added, and the resulting mixture was extracted with ethyl acetate. The combined organic layer was dried over anhydrous sodium sulfate and concentrated in vacuo. The residue was purified by Prep-HPLC [Column: Xselect C18 5 μm 19 × 150 mm, mobile phase A: water (0.1% NH4HCO3), mobile phase B: acetonitrile; gradient: 25% B to 55% B in 8 min] to afford 5.2 mg (3% yield) of the product as a white solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.15 (s, 3H), 6.21 (br, 1H), 7.01 (s, 1H), 7.17 (d, 1H), 7.25 (t, 1H), 7.35 (t, 1H), 7.40 (d, 1H), 8.29 (s, 1H), 12.68 (br, 1H) ppm; LCMS (ESI−) m/z: 470 [M – H]−.
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-5-iodo-2-(2-methylphenoxy)benzonitrile (17)
To a slurry of sodium hydride (47.0 mg, 1.20 mmol, 60% in mineral oil) in N,N-dimethylformamide (5 mL) were added ethyl 3-amino-4,4,4-trifluorobut-2-enoate (215 mg, 1.20 mmol) and methyl 4-cyano-2-iodo-5-(o-tolyloxy)phenylcarbamate (73, 300 mg, 0.6 mmol) at 0 °C. The resulting mixture was stirred at 100 °C overnight under a nitrogen atmosphere. Upon completion of the reaction, ammonium chloride solution was added at 0 °C, and the resulting mixture was extracted with ethyl acetate. The combined organic layer was dried over anhydrous sodium sulfate and concentrated in vacuo. The residue was purified by Prep-HPLC [Column: mobile phase A: water (0.1% FA), mobile phase B: acetonitrile; gradient: 40% B to 60% B in 8 min] to afford 113.2 mg (37% yield) of the product as a white solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.12 (s, 3H), 6.33 (s, 1H), 6.96 (d, 1H), 7.14 (d, 1H), 7.21–7.26 (m, 1H), 7.29–7.34 (m, 1H), 7.38 (d, 1H), 8.50 (s, 1H), 12.66 (br, 1H) ppm; LCMS (ESI−) m/z: 512 [M – H]−.
4-[4-(Difluoromethyl)-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl]-5-methoxy-2-(2-methylphenoxy)benzonitrile (18)
4-[4-(Difluoromethyl)-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl]-2-fluoro-5-methoxybenzonitrile [83 (20.0 mg, 64.3 μmol)], commercially available 2-methylphenol (8.0 μL, 77 μmol), and potassium carbonate (22.2 mg, 161 μmol) were stirred in DMSO (850 μL) for 3 days at 110 °C. Upon completion of the reaction, the mixture was filtered and purified by HT HPLC, affording the title compound (1 mg; 3%) as an ochre solid. 1H NMR (400 MHz, MeOD): δ 2.24, 3.84 (2s, 3H each), 5.89 (s, 1H), 6.58 (t, J = 6.7 Hz, 1H), 6.77 (s, 1H), 6.95 (d, J = 8.11 Hz, 1H), 7.12, 7.20, 7.28 (3 m, 1H each), 7.53 (s, 1H) ppm; LCMS (ESI+) m/z: 399 [M + H]+.
4-{4-[Chloro(difluoro)methyl]-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}-5-methoxy-2-(2-methylphenoxy)benzonitrile (rac) (19)
4-{4-[Chloro(difluoro)methyl]-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}15-2-fluoro-5-methoxybenzonitrile (81, 120 mg, 70% purity, 243 μmol), 2-methylphenol (31.5 mg, 292 μmol), and potassium carbonate (84.0 mg, 608 μmol) were stirred in DMSO (3.2 mL) for 21 h at 110 °C. An additional equivalent of 2-methylphenol was added, and the mixture was stirred for additional 18 h. Upon completion of the reaction, the mixture was filtered and purified by HT HPLC, affording the title compound (5 mg, 4%) as an ochre solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.18, 3.80 (2 s, 3H each), 6.19 (s, 1H), 6.97 (d, J = 7.4 Hz, 1H), 7.01 (s, 1H), 7.17, 7.26, 7.35 (3 m, 1H each), 7.74 (s, 1H), 12.50 (br s, NH) ppm; LCMS (ESI+) m/z: 434 [M + H]+.
4-[2,6-Dioxo-4-(pentafluoroethyl)-3,6-dihydropyrimidin-1(2H)-yl]-5-methoxy-2-(2-methylphenoxy)benzonitrile (20)
2-Bromo-4-[2,6-dioxo-4-(pentafluoroethyl)-3,6-dihydropyrimidin-1(2H)-yl]-5-methoxybenzonitrile (84, 90.0 mg, 204 μmol, 2-methylphenol (31.4 mg, 290 μmol), N,N-dimethylglycine (5.59 mg, 54.2 μmol), copper(I) iodide (5.26 mg, 27.6 μmol), and cesium carbonate (160 mg, 491 μmol) were heated in DMF (2 mL) for 18 h at 140 °C. Upon completion of the reaction, work-up, and purification, the title compound was obtained (10 mg, 9%) as an ochre solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.18 (s, 3 H), 3.80 (s, 3 H), 6.99, 7.17, 7.27, 7.36 (4 m, 2H, 1H, 1H, 1H), 7.74/s, 1 H), 12.45 (br s, NH) ppm; LCMS (ESI+) m/z: 467 [M + H]+.
4-{4-[Difluoro(phenyl)methyl]-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}-5-methoxy-2-(2-methylphenoxy)benzonitrile (21)
2-Bromo-4-{4-[difluoro(phenyl)methyl]-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}-5-methoxybenzonitrile (85, 60.0 mg, 134 μmol), commercially available 2-methylphenol (CAS: 95-48-7, 17 μL, 160 μmol), N,N-dimethylglycine (3.66 mg, 35.5 μmol), copper(I) iodide (3.44 mg, 18.1 μmol), and cesium carbonate (105 mg, 321 μmol) were heated in DMF for 18 h at 140 °C. Upon completion of the reaction, work-up, and purification, the title compound was obtained as a brown solid (6 mg, 8% yield). 1H NMR (400 MHz, [D]6 DMSO): δ 2.18, 3.78 (2 s, 3H each), 5.98 (s, 1H), 6.94 dd, J = 7.9, 1.0 Hz, 1H), 7.01 (br s, 1H), 7.15 (td, J = 7.4, 1.1 Hz, 1H), 7.25 (td, J = 7.7, 1.3 Hz, 1H), 7.34 (d, J = 6.8 Hz, 1H), 7.54–7.60, 7.69–7.71 (2 m, 3H each), 12.02 (br s, NH) ppm; LCMS (ESI+) m/z: 475 [M + H]+.
4-(4-Bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl)-5-methoxy-2-(2-methylphenoxy)benzonitrile (22)
To a stirred solution of 5-methoxy-2-(o-tolyloxy)-4-(2,4,6-trioxotetrahydropyrimidin-1(2H)-yl)benzonitrile (64, 6.5 g, 0.017 mol) in acetonitrile (65 mL) was added phosphoryl bromide (POBr3, 12.8 g, 0.045 mol). The resulting mixture was stirred at 110 °C under microwave irradiation for 30 min. After completion of the reaction, the solvent was removed under reduced pressure. The crude residue was treated with saturated aqueous solution of sodium bicarbonate (500 mL), and the mixture was then extracted twice with ethyl acetate (250 mL each). The combined organic layers were concentrated under vacuum. The residue was purified using a RP C18 80 g column, eluting 40% acetonitrile in water to afford the title compound (3.02 g, 39%) as an off-white solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.18, 3.76 (2 s, 3H each), 5.68, 6.72 (2 bs, 1H each), 6.92 (d, J = 7.8 Hz, 1H), 7.15 (td, J = 7.4, 1.1 Hz, 1H), 7.25 (td, J = 7.6, 1.5 Hz, 1H), 7.34 (d, J = 7.8 Hz, 1H), 7.64 (s, 1H), 12.40 (br s, NH, 1H) ppm; LCMS (ESI+) m/z: 428 [M + H]+.
4-[4-(Methanesulfonyl)-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl]-5-methoxy-2-(2-methylphenoxy)benzonitrile (23)
In a pressure vessel, aforementioned 4-[4-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl]-5-methoxy-2-(2-methylphenoxy)benzonitrile (22, 100 mg, 234 μmol) was dissolved in DMSO (1.9 mL) under an argon atmosphere. The commercially available copper catalyst copper(I) trifluoromethanesulfonate benzene complex (CAS: 42152-46-5, 13.1 mg, 23.4 μmol), commercially available sodium methanesulfinate (CAS: 20277-69-4, 36.0 mg, 353 μmol), and commercially available ligand (±)-trans-1,2-diaminocyclohexane (CAS: 1121-22-8, 11 μL, 96 μmol) were added, and the reaction mixture was heated to 110 °C for 20 h. After cooling, the reaction mixture was filtered and purified by HPLC, affording the title compound as a light yellow solid (42 mg, 38% yield). 1H NMR (400 MHz, [D]6 DMSO): δ 2.17, 3.41, 3.80 (3 s, 3H each), 6.16 (br s, 1H), 6.93 (s, 1H), 6.98 (d, J = 8.1 Hz, 1H), 7.17 (t, J = 7.4 Hz, 1H), 7.27 (t, J = 7.7 Hz, 1H), 7.35 (d, J = 7.1 Hz, 1H), 7.74 (s, 1H), 12.42 (br s, NH) ppm; LCMS (ESI+) m/z: 428 [M + H]+. Atropisomeric ratio: atrop 1/atrop 2 = 50:49 (1% impurities); Rt (atrop1) = 4.99 min; Rt (atrop2) = 6.25 min. The atropisomeric ratio was determined using the following chiral HPLC method: instrument: Waters Alliance 2695 Agilent HPLC 1260; column: Chiralpak IE 3 μm 100 × 4.6 mm; eluent A: hexane + 0.1 vol % TFA (99%); eluent B: 2-propanol; isocratic: 20% B; flow: 1.4 mL/min; T: 25 °C; DAD 254 nm.
4-[4-(Benzenesulfonyl)-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl]-5-methoxy-2-(2-methylphenoxy)benzonitrile (24)
In a pressure vessel, aforementioned 4-[4-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl]-5-methoxy-2-(2-methylphenoxy)benzonitrile (22, 500 mg, 1.17 mmol) was dissolved in DMSO (9.6 mL) under an argon atmosphere. The commercially available copper catalyst copper(I) trifluoromethanesulfonate benzene complex (CAS: 42152-46-5, 65.3 mg, 117 μmol), commercially available sodium benzenesulfinate (CAS: 873-55-2, 289 mg, 1.76 mmol), and commercially available ligand (±)-trans-1,2-diaminocyclohexane (CAS: 1121-22-8, 57 μL, 480 μmol) were added, and the reaction mixture was heated to 110 °C for 20 h. After cooling, the reaction mixture was filtered and purified by HPLC, affording the title compound as a dark yellow solid (113 mg, 19%). 1H NMR (400 MHz, [D]6 DMSO): δ 2.15, 3.75 (2s, 3H each), 6.35 (s, 1H), 6.93–6.96 (m, 2H), 7.14 (t, J = 7.4 Hz, 1H), 7.23 (t, J = 7.3 Hz, 1H), 7.32 (d, J = 7.1 Hz, 1H), 7.68–7.76, 7.84–7.68, 8.09–8.13 (3 m, 3H, 1H, 2H), 12.47 (br s, NH) ppm; LCMS (ESI+) m/z: 489 [M + H]+. Atropisomeric ratio: atrop 1/atrop 2 = 44:44 (12% impurities); Rt (atrop1) = 2.39 min; Rt (atrop2) = 2.85 min. The atropisomeric ratio was determined using the following chiral HPLC method: instrument: Waters Alliance 2695; column: Amylose SA 3 μm 100 × 4.6 mm; eluent A: 2-methoxy-2-methylpropane + 0.1 vol % diethylamine (99%); eluent B: methanol; isocratic: 50% A + 50% B; flow: 1.4 mL/min; T: 25 °C; DAD: 254 nm.
4-(4-Cyclopropyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl)-5-methoxy-2-(2-methylphenoxy)benzonitrile (25)
In a pressure vessel, aforementioned 4-[4-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl]-5-methoxy-2-(2-methylphenoxy)benzonitrile (22, 250 mg, 584 μmol), commercially available catalyst palladium(II) acetate (CAS: 3375-31-3, 6.92 mg, 11.7 μmol), sodium carbonate (161 mg, 1.17 mmol), commercially available potassium cyclopropyl(trifluoro)borate (CAS: 1065010-87-8, 95.0 mg, 642 μmol), and commercially available tricyclohexylphosphine (CAS: 2622-14-2, 6.55 mg, 23.4 μmol) were suspended in toluene (4.9 mL) under an argon atmosphere. The reaction mixture was heated to 90 °C for 18 h. After cooling, the mixture was filtered and washed with ethyl acetate. The residue was purified by HPLC, affording the target compound as a light yellow solid (5 mg, 2%, 85% purity). 1H NMR (400 MHz, MeOD): δ 0.91, 1.09, 1.69 (3 m, 2H, 2H, 1H), 2.24, 3.83 (2s, 3H each), 5.35 (s, 1H), 6.69 (s, 1H), 6.95 (d, J = 8.1 Hz, 1H), 7.12 (m, 1H), 7.19 (dd, J = 7.9, 1.5 Hz, 1H), 7.29 (d, J = 7.1 Hz, 1H), 7.51 (s, 1H) ppm; LCMS (ESI+) m/z: 389 [M + H]+.
4-(2,6-Dioxo-4-phenyl-3,6-dihydropyrimidin-1(2H)-yl)-5-methoxy-2-(2-methylphenoxy)benzonitrile (26)
A mixture of aforementioned 4-[4-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl]-5-methoxy-2-(2-methylphenoxy)benzonitrile (22, 250 mg, 584 μmol), commercially available phenylboronic acid (CAS: 95-48-7, 71.2 mg, 584 μmol), and commercially available potassium fluoride (CAS: 7789-23-3, 102 mg, 1.75 mmol in dioxane (2.4 mL) was stirred under an argon atmosphere at rt. Then, commercially available tris(dibenzylideneacetone)dipalladium(0) (CAS: 51364-51-3, 26.7 mg, 29.2 μmol, 0.05 equiv) and commercially available tri-tert-butylphosphoniumtetrafluoroboronate (CAS: 131274-22-1, 16.9 mg, 58.4 μmol) were added to the brown reaction mixture, which was then heated to 110 °C for 18 h. Upon completion of the reaction, the reaction mixture was concentrated and then purified using HPLC-HT, affording the title compound as a yellow glittery solid (30 mg, 11% yield). 1H NMR (400 MHz, [D]6 DMSO): δ 2.21, 3.61 (2 s, 3H each), 5.99 (br s, 1H), 6.96 (dd, J = 8.2, 0.9 Hz, 1H)*, 6.97 (s, 1H), 7.16 (td, J = 7.4, 7.4, 1.1 Hz, 1H), 7.26 (td, J = 7.7, 1.3 Hz, 1H), 7.35 (m, 1H), 7.49–7.56, 7.73–7.76 (2 m, 3H each), 11.54 (br s, NH) ppm; LCMS (ESI+) m/z: 426 [M + H]+. Atropisomeric ratio: atrop 1/atrop 2 = 50:47 (3% impurities); Rt (atrop1) = 2.73 min; Rt (atrop2) = 3.38 min. The atropisomeric ratio was determined using the following chiral HPLC method: Instrument: Agilent HPLC 1260; column: cellulose SC 3 μm 100 × 4.6 mm; eluent A: hexane + 0.1 vol % TFA; eluent B: 2-propanol; gradient: 20–50% B in 7 min; flow 1.4 mL/min; T: 25 °C; DAD 254 nm. The corresponding atropisomers were separated using preparative chiral HPLC: Instrument: Labomatic HD5000, Labocord-5000; Gilson GX-241, Labcol Vario 4000; column: cellulose SC 5 μm 250 × 30 mm; eluent A: hexane + 0.1 vol % TFA; eluent B: 2-propanol; gradient: 20–50% B in 7 min; flow 40.0 mL/min; UV 254 nm.
4-{2,6-Dioxo-4-[4-(trifluoromethyl)phenyl]-3,6-dihydropyrimidin-1(2H)-yl}-5-methoxy-2-(2-methylphenoxy)benzonitrile (27)
A mixture of aforementioned 4-(4-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl)-5-methoxy-2-(2 methylphenoxy)benzonitrile (22, 100 mg, 234 μmol), commercially available [4-(trifluoromethyl)phenyl]boronic acid (CAS: 128796-39-4, 44.4 mg, 234 μmol), and commercially available potassium fluoride (CAS: 7789-23-3, 40.7 mg, 701 μmol) in dioxane (970 μL) was stirred under an argon atmosphere at rt. Then, commercially available tris(dibenzylideneacetone)dipalladium(0) (CAS: 51364-51-3, 10.7 mg, 11.7 μmol, 0.05 equiv) and commercially available tri-tert-butylphosphoniumtetrafluoroboronate (CAS: 131274-22-1, 6.77 mg, 23.4 μmol) were added to the reaction mixture, which was heated to 110 °C for 18 h. Upon completion of the reaction, the reaction mixture was purified using HPLC-HT to afford the title compound as a light yellow solid (38 mg, 30%). NMR (400 MHz, [D]6 DMSO): δ 2.20, 3.82 (2s, 3H each), 6.10 (d, J = 1.5 Hz, 1H), 6.95–6.98 (m, 2H), 7.17 (t, J = 7.4 Hz, 1H), 7.25 (t, J = 7.7 Hz, 1H), 7.35 (d, J = 7.4 Hz, 1H), 7.75 (s, 1H), 7.88, 7.96 (2 d, J = 8.4 Hz, 1H each), 11.71 (br s, NH) ppm; LCMS (ESI+) m/z: 494 [M + H]+.
1-[4-Cyano-2-methoxy-5-(2-methylphenoxy)phenyl]-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxamide (28)
Aforementioned 4-(4-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl)-5-methoxy-2-(2 methylphenoxy)benzonitrile (22, 70.0 mg, 163 μmol), commercially available catalyst palladium(π-cinnamyl) chloride dimer (CAS: 12131-44-1, 4.23 mg, 8.2 μmol), commercially available 1,1′-bis(diphenylphosphino)ferrocene (CAS: 12150-46-8, 4.53 mg, 8.17 μmol), and commercially available zinc cyanide (CAS: 557-21-1, 19.2 mg, 163 μmol) were placed in a 5 mL crimp sealable reaction vessel, sealed, and flushed with argon. N,N-diisopropylethylamine (57 μL, 330 μmol) and degassed N,N-dimethylacetamide (1 mL) were added, and the mixture was stirred for 14 h at 120 °C. Saturated aqueous sodium bicarbonate solution was added, and the mixture was extracted three times with dichloromethane. The combined organic layers were filtered using a water-repellent filter and concentrated in vacuo. The residue was purified by reversed-phase HPLC, yielding the title compound (32.8 mg, 51% yield), which turned out to be a carboxamide instead of the initially expected nitrile. 1H NMR (400 MHz, [D]6 DMSO): δ 2.19, 3.79 (2 s, 3H each), 6.21 (s, 1H), 6.93 (d, J = 7.7 Hz, 1H), 6.95 (s, 1H), 7.15, 7.25 (2 m, 1H each), 7.34 (d, J = 7.3 Hz, 1H), 7.72 (s, 1H), 8.05, 8.29 (2 bs, 1H each), 11.10 (br s, NH) ppm; LCMS (ESI+) m/z: 393 [M + H]+.
3-[2-Fluoro-5-(2-methylphenoxy)phenyl]-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (29)
3-(5-Bromo-2-fluorophenyl)-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (86, 50.0 mg, 142 μmol), 2-methylphenol (21.7 mg, 201 μmol), N,N-dimethylglycine (3.87 mg, 37.5 μmol), copper(I) iodide (3.64 mg, 19.1 μmol), and cesium carbonate (111 mg, 340 μmol) were heated in DMF (2.8 mL) for 18 h at 140 °C. Upon completion of the reaction, work-up, and purification, the title compound was obtained as a brown solid (3 mg, 5% yield, 90% purity). 1H NMR (400 MHz, [D]6 DMSO): δ 2.18 (s, 3H), 6.35 (br s, 1H), 6.94 (dd, J = 8.11, 1.0 Hz, 1H), 7.04, 7.13 (2 m, 1H each), 7.13 (td, J = 7.7, 7.7, 1.3 Hz, 1H), 7.32–7.40 (m, 2H), 12.69 (br s, NH) ppm; LCMS (ESI+) m/z: 381 [M + H]+.
3-[2-Methoxy-5-(2-methylphenoxy)phenyl]-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (30)
3-(5-bromo-2-methoxyphenyl)-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (87, 37.0 mg, 101 μmol), 2-methylphenol (15 μL, 140 μmol), N,N-dimethylglycine (2.77 mg, 26.9 μmol), copper(I) iodide (2.61 mg, 13.7 μmol), and cesium carbonate (79.2 mg, 243 μmol) were heated in DMF (2 mL) for 18 h at 140 °C. Upon completion of the reaction, work-up, and purification, the title compound was obtained (9 mg, 19%) as an ochre solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.21, 3.72 (2 s, 3H each), 6.31 (br s, 1H), 6.84 (d, J = 7.7 Hz, 1H), 6.94 (m, 1H), 7.00 (dd, J = 7.0, 7.0 Hz, 1H), 7.06 (m, 1H), 7.14 (d, J = 9.1 Hz, 1H), 7.19, 7.30 (2 m, 1H each), 12.52 (br s, NH) ppm; LCMS (ESI+) m/z: 392 [M + H]+.
3-[2-Chloro-5-(2-methylphenoxy)phenyl]-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (31)
3-(5-Bromo-2-chlorophenyl)-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (82, 150 mg, 406 μmol), cesium carbonate (317 mg, 974 μmol), 2-methylphenol (62.3 mg, 576 μmol), N,N-dimethylglycine (11.1 mg, 108 μmol), and copper iodide (10.4 mg, 54.8 μmol) were suspended in DMF (8 mL), and the reaction was heated to 140 °C for 18 h. Upon completion of the reaction, the mixture diluted with ammoniac and stirred for further minutes at rt. Ethyl acetate and water were added. The phases were separated, and the organic phase was extracted twice. The combined organic layers were washed with brine and dried with sodium sulfate. After filtration, the solvent was removed under vacuum. The crude material was purified by HPLC-HT, affording the title compound (50 mg, 42% yield) as a colorless solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.15 (s, 3H), 6.39 (s, 1H), 7.0 (m, 2H), 7.10 (d, J = 3.0 Hz, 1H), 7.17, 7.27, 7.35 (3 m, 1H each), 7.59 (d, J = 8.9 Hz, 1H), 12.67 (br s, NH) ppm; LCMS (ESI+) m/z: 397 [M + H]+.
3-[2-Fluoro-4-methyl-5-(2-methylphenoxy)phenyl]-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (32)
3-(5-Bromo-2-fluoro-4-methylphenyl)-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (88, 115 mg, 313 μmol), cesium carbonate (245 mg, 752 μmol), 2-methylphenol (48.1 mg, 445 μmol), N,N-dimethylglycine (8.56 mg, 83.0 μmol), and copper(I) iodide (8.05 mg, 42.3 μmol) were suspended in DMF (6.2 mL), and the reaction was heated to 140 °C for 18 h. Upon completion of the reaction, the mixture was diluted with ammoniac and stirred for further minutes at rt. Ethyl acetate and water were added. The phases were separated, and the organic phase was extracted twice. The combined organic layers were washed with brine and dried with sodium sulfate. After filtration, the solvent was removed under vacuum. The crude material was purified by HPLC-HT, affording the title compound (10.0 mg, 8% yield) as a colorless solid. 1H NMR (400 MHz, MeOD): δ 2.24, 2.31 (2 s, 3H each), 6.20 (s, 1H), 6.68 (d, J = 6.6 Hz, 1H), 6.76 (d, J = 7.9 Hz, 1H), 7.00 (td, J = 7.4, 7.4 Hz, 1H), 7.11 (t, J = 7.8 Hz, 1H), 7.20–7.24 (m, 2H) ppm; LCMS (ESI+) m/z: 395 [M + H]+.
3-[4-Chloro-2-methoxy-5-(2-methylphenoxy)phenyl]-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (33)
Similar to previously described procedures, 3-(5-bromo-4-chloro-2-methoxyphenyl)-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (89, 160 mg, 400 μmol), commercially available 2-methylphenol (CAS: 95-48-7, 59 μL, 570 μmol), N,N-dimethylglycine (10.9 mg, 106 μmol), copper(I) iodide (10.3 mg, 54.1 μmol), and cesium carbonate (313 mg, 961 μmol) were heated in DMF (7.9 mL) for 18 h at 140 °C. Upon completion of the reaction, work-up, and purification, the title compound was obtained as a dark brown solid (16 mg, 8% yield). 1H NMR (400 MHz, [D]6 DMSO): δ 2.24, 3.77 (2 s, 3H each), 6.28 (br s, 1H), 6.68 (dd, J = 8.1, 0.8 Hz, 1H), 7.04 (m, 1H), 7.17 (td, J = 7.7, 7.7, 1.3 Hz, 1H), 7.29 (d, J = 7.4 Hz, 1H), 7.42 (br s, 1H), 12.53 (br s, MH) ppm; LCMS (ESI+) m/z: 427 [M + H]+.
3-[4-Methoxy-2-methyl-5-(2-methylphenoxy)phenyl]-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (34)
Similar to previously described procedures, 3-(5-bromo-4-methoxy-2-methylphenyl)-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (90, 100 mg, 264 μmol), commercially available 2-methylphenol (CAS: 95-48-7, 27 μL, 260 μmol), 2,2,6,6-tetramethylheptane-3,5-dione (55 μL, 260 μmol), copper(I) iodide (25.1 mg, 132 μmol), and cesium carbonate (172 mg, 528 μmol) were heated in N-methylpyrrolidone (500 μL) for 18 h at 140 °C. Upon completion of the reaction, work-up, and purification, the title compound was obtained as a brown solid (8 mg, 85% purity, 6% yield). 1H NMR (400 MHz, [D]6 DMSO): δ 2.02, 2.22, 3.80 (3 s, 3H each), 6.28 (br s, 1H), 6.61 (d, J = 7.8 Hz, 1H), 6.81 (s, 1H), 6.97 (td, J = 7.4, 7.4, 0.9 Hz, 1H), 7.11 (m, 2H), 7.23 (d, J = 7.1 Hz, 1H), 12.45 (br s, NH) ppm; LCMS (ESI+) m/z: 407 [M + H]+.
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2-(2-methylphenoxy)naphthalene-1-carbonitrile (35)
According to GP2, to a suspension of sodium hydride (60% in mineral oil) (1.17 g, 29.3 mmol, in 40 mL DMF) was added a solution of commercially available ethyl (2Z)-3-amino-4,4,4-trifluorobut-2-enoate (CAS: 372-29-2, 5.36 g, 29.3 mmol, in 10 mL DMF) at 0 °C, and the mixture was stirred for 10 min. Then, the mixture was slowly added to a solution of methyl [4-cyano-3-(2-methylphenoxy)naphthalen-1-yl]carbamate (78, 5.0 g, 14.7 mmol, in 10 mL DMF). The resulting mixture was allowed to warm up to rt and was then heated at 100 °C for 24 h under a nitrogen atmosphere. Upon completion of the reaction, the reaction mixture was cooled to 0 °C, and then saturated aqueous ammonium chloride solution was added at 0 °C. The solvent was removed in vacuo, the residue was diluted with water, the mixture was extracted with ethyl acetate, the combined organic layers were dried over anhydrous sodium sulfate and filtered, the solvent was removed in vacuo, and the residue was purified by Prep-HPLC (column: Sunfire C18, 5 μm, (19*150 mm); eluent A: 0.1% TFA in water, eluent B: acetonitrile, flow: 20.0 mL/min; gradient: time/% B: 0/5, 1/5, 5/20, 5.1/100, 8/100, 8.1/5, 10/5) to afford the title compound (2.50 g, 38%) as an off-white solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.18 (s, 3H), 6.33 (s, 1 H), 7.16 (d, 1H), 7.24–7.35 (m, 3H), 7.41 (d, 1H), 7.61 (t, 1H), 7.86 (t, 1H), 7.93 (d, 1 H), 8.15 (d, 1H), 12.55 (br, 1 H) ppm; LC–MS (method C, 0–2.00 min 10–95% B): Rt = 1.50 min; MS (ESI+) m/z: 438 [M + H]+.
The corresponding atropisomers were separated using preparative chiral HPLC: column: CHIRALPAK IA, 2 × 25 cm, 5 μm; mobile phase A: hexane (0.1% TFA)-HPLC, mobile phase B: ethanol-HPLC; gradient: 10% B to 10% B in 20 min to afford 27.2 mg (37% yield) of 35a as a white solid and 26.8 mg (36% yield) of 35b (second eluting isomer) as a white solid.
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2-(2-methylphenoxy)naphthalene-1-carbonitrile (35a)
tr = 10.365. 1H NMR (400 MHz, [D]6 DMSO): δ 2.07 (s, 3H), 6.36 (s, 1H), 7.15 (d, 1H), 7.26 (t, 1H), 7.31–7.35 (m, 2H), 7.41 (d, 1H), 7.60 (t, 1H), 7.86 (t, 1H), 7.95 (d, 1H), 8.15 (d, 1H), 12.57 (br, 1H) ppm; LC–MS (method C, 0–2.00 min 10–95% B): Rt = 1.42 min; MS (ESI+) m/z: 438 [M + H]+.
4-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-2-(2-methylphenoxy)naphthalene-1-carbonitrile (35b)
tr = 14.427. 1H NMR (400 MHz, [D]6 DMSO): δ 2.18 (s, 3H), 6.36 (s, 1H), 7.15 (d, 1H), 7.25 (t, 1H), 7.31–7.35 (m, 2H), 7.41 (d, 1H), 7.60 (t, 1H), 7.84 (t, 1H), 7.95 (d, 1H), 8.15 (d, 1H), 12.57 (br, 1H) ppm; LC–MS (method C, 0–2.00 min 10–95% B): Rt = 1.43 min; MS (ESI+) m/z: 438 [M + H]+.
3-[4-Chloro-3-(2-methylphenoxy)naphthalen-1-yl]-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (36)
To a suspension of sodium hydride (60% in mineral oil; 1.17 g, 29.3 mmol) in DMF (40 mL) was added a solution of ethyl 3-amino-4,4,4-trifluorobut-2-enoate (5.36 g, 29.3 mmol) in DMF (10 mL) at 0 °C, and the mixture was stirred for 10 min. Then, a solution of methyl [4-chloro-3-(2-methylphenoxy)naphthalen-1-yl]carbamate (42; 5.0 g, 14.7 mmol) in DMF (10 mL) was slowly added. The resulting mixture was allowed to warm to rt and was then heated at 100 °C for 24 h under a nitrogen atmosphere. The progress of the reaction was monitored by TLC. Upon completion of the reaction, the mixture was cooled to 0 °C, and then saturated aqueous ammonium chloride solution was added. The solvent was removed under reduced pressure, the residue was diluted with water, and the mixture was extracted with ethyl acetate. The combined organic layers were dried over anhydrous sodium sulfate and filtered, and the solvent was removed under reduced pressure. The residue was purified by preparative HPLC [mobile phase A: water (10 mmol/L NH4HCO3), mobile phase B: acetonitrile; gradient: 25% B to 65% B in 8 min] to afford 36 (2.50 g, 38% yield) as an off-white solid. 1H NMR (400 MHz, [D]6DMSO): δ 2.08 (s, 3H), 6.36 (s, 1H), 6.85 (d, 1H), 7.13 (t, 1H), 7.23 (t, 1H), 7.36 (d, 1H), 7.38 (s, 1H), 7.57 (t, 1H), 7.78 (t, 1H), 7.86 (d, 1H), 8.31 (d, 1H), 12.55 (s, 1H) ppm. LC–MS (method 2): tR = 1.63 min; MS (ESI+) m/z: 447 [M + H]+. Atropisomeric ratio: atrop1 (BAY-069, 36a)/atrop2 (36b) = 49:51 (1% impurities); tR (atrop1, BAY-069) = 4.33 min; tR (atrop2) = 5.27 min. The atropisomeric ratio was determined using the following chiral HPLC method: instrument: Agilent HPLC 1260; column: ReproSil Chiral-NR 5 μm, 100 × 4.6 mm; eluent A: hexane + 0.1 vol % TFA, eluent B: 2-propanol; isocratic: 65% A + 35% B; flow: 1.4 mL/min; temperature: 25 °C; DAD 254 nm. The corresponding atropisomers were separated using preparative chiral HPLC: instrument: Labomatic HD-5000, Labocord-5000; Gilson GX-241, Labcol Vario 4000; column: ReproSil Chiral-NR 5 μm, 250 × 30 mm; eluent A: hexane + 0.1 vol % TFA, eluent B: 2-propanol; isocratic: 65% A + 35% B; flow: 50.0 mL/min; UV 254 nm. Optical rotation: atrop1: + 5.7, ±0.66° (off-white solid) and atrop2: −4.3°, ±0.47° (off-white solid).
3-[4-Chloro-3-(2-methylphenoxy)naphthalen-1-yl]-6-[difluoro(phenyl)methyl]pyrimidine-2,4(1H,3H)-dione (37)
According to GP2, to a suspension of sodium hydride (60% in mineral oil) (1.17 g, 29.3 mmol) in DMF (40 mL) was added a solution of ethyl 3-amino-4,4-difluoro-4-phenylbut-2-enoate (48, 4.24 g, 17.6 mmol) in DMF (10 mL) at 0 °C and stirred for 10 min. Then, it was slowly added a solution of methyl [4-chloro-3-(2-methylphenoxy)naphthalen-1-yl]carbamate (42, 5.0 g, 14.66 mmol) in DMF (10 mL). The resulting mixture was allowed to warm up to rt for 10 min and was then heated at 100 °C for 24 h under a nitrogen atmosphere. Upon completion of the reaction, the reaction mixture was cooled to 0 °C, and saturated aqueous ammonium chloride solution was added at 0 °C. The solvent was removed in vacuo, the residue was diluted with water, the mixture was extracted with ethyl acetate, the combined organic layers were dried over anhydrous sodium sulfate and filtered, and the solvent was removed in vacuo. The residue was purified by Prep-HPLC to afford the title compound (2.1 g, 14%) as an off-white solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.25 (s, 3H), 6.04 (br s, 1H), 6.80 (dd, J = 7.9, 1.0 Hz, 1H), 7.11 (td, J = 7.4, 7.4, 1.1 Hz, 1H), 7.20 (td, J = 7.7, 7.7, 1.3 Hz, 1H), 7.34 (dd, J = 7.4, 0.8 Hz, 1H), 7.34 (br s, 1H), 7.55–7.62, 7.75–7.79 (2 m, 4H each), 8.30 (d, J = 8.1 Hz, 1H), 12.09 (br s, 1H) ppm; LCMS (ESI+) m/z: 505 [M + H]+.
3-[4-Chloro-3-(2-methylphenoxy)naphthalen-1-yl]-6-[difluoro(phenyl)methyl]pyrimidine-2,4(1H,3H)-dione (37a)
Rt = 1.44 min; LCMS (ESIpos) m/z: 505 [M + H]+; optical rotation [α] −1.0, ±0.55°; honey-like sticky oil.
3-[4-Chloro-3-(2-methylphenoxy)naphthalen-1-yl]-6-[difluoro(phenyl)methyl]pyrimidine-2,4(1H,3H)-dione (37b)
Rt = 1.44 min; LCMS (ESI+) m/z: 505 [M + H]+; optical rotation [α] +4.7°, ±0.68°; off-white solid.
3-[2,6-Dioxo-4-(trifluoromethyl)-3,6-dihydropyrimidin-1(2H)-yl]-N-methyl-2-(pyridin-2-yl)imidazo[1,2-a]pyridine-7-carboxamide (38)
To a slurry of sodium hydride (35 mg, 0.88 mmol, 60% in mineral oil) in DMF (8 mL) were added ethyl-3-amino-4,4,4-trifluorobut-2-enoate (107 mg, 0.58 mmol) and methyl[7-(methylcarbamoyl)-2-(pyridin-2-yl)imidazo[1,2-a]pyridin-3-yl]carbamate (95; 200 mg, 0.58 mmol). The resulting mixture was stirred at 100 °C overnight under a nitrogen atmosphere. After cooling to 0 °C, aqueous ammonium chloride solution was added, and the resulting mixture was concentrated in vacuo. The residue was purified by Prep-HPLC [mobile phase A: water (0.1% NH4HCO3), mobile phase B: acetonitrile; gradient: 15% B to 50% B in 12 min] to afford 72.9 mg (29% yield) of the title compound as a yellow solid. 1H NMR (400 MHz, [D]6 DMSO): δ 2.84 (d, 3H), 6.43 (s, 1H), 7.31 (t, 1H), 7.42 (d, 1H), 7.91 (t, 1H), 8.20 (s, 1H), 8.22 (s, 1H), 8.46 (d 1H), 8.61 (br, 1H), 8.73 (d, 1H), 12.75 (br, 1H) ppm; LC–MS (method 4, 0–3.00 min 10–95% B): Rt = 0.67 min; MS (ESI+) m/z: 431 [M + H]+. Atropisomeric ratio: atrop 1/atrop 2 = 51:49 (impurities present); Rt (atrop1) = 5.65 min; Rt (atrop2) = 7.70 min. The atropisomeric ratio was determined using the following chiral HPLC method: Instrument: Agilent HPLC 1260; column: Cellulose SC 3 μm 100 × 4.6 mm; eluent A: hexane + 0.1 vol % diethylamine (99%); eluent B: ethanol; isocratic: 50% A + 50% B; flow: 1.4 mL/min; temperature: 25 °C; DAD 254 nm. The racemate was separated into its single atropisomers using preparative chiral HPLC: instrument: Instrument: Labomatic HD5000, Labocord-5000; Gilson GX-241, Labcol Vario 4000, column: cellulose SC 5 μm 250 × 30 mm; eluent A: hexane + 0.1 vol % diethylamine (99%); eluent B: ethanol; isocratic: 50% A + 50% B; flow 40.0 mL/min; UV 254 nm.
2-(2-Methylphenoxy)-4-nitronaphthalen-1-amine (39)
To a solution of commercially available 2-bromo-4-nitronaphthalen-1-amine (CAS: 63240-26-6; 21.0 g, 74.7 mmol) in NMP (150 mL) were added o-cresol (80.8 g, 746 mmol), copper(I) chloride (7.40 g, 74.7 mmol), cesium carbonate (48.6 g, 149 mmol), and 2,2,6,6-tetramethylheptane-3,5-dione (27.4 g, 149 mmol). The resulting mixture was stirred at 120 °C overnight under a nitrogen atmosphere and then cooled to rt. The solvent was removed under reduced pressure, and the residue was diluted with water. The resulting mixture was extracted with ethyl acetate, and the combined organic layer was dried over anhydrous sodium sulfate and filtered. The solvent was removed under reduced pressure, and the residue was purified by silica gel column chromatography (petroleum ether/ethyl acetate 2:1) to afford 39 (5.50 g, 90% purity, 23% yield) as a brown solid. LC–MS (method 1, 0–2.00 min 10–95% B): tR = 1.04 min; MS (ESI+) m/z: 295 [M + H]+.
1-Chloro-2-(2-methylphenoxy)-4-nitronaphthalene (40)
To a solution of 2-(2-methylphenoxy)-4-nitronaphthalen-1-amine (39; 550 mg, 1.70 mmol) in acetonitrile (8 mL) were added copper(II) chloride (37.7 g, 280 mmol) and tert-butyl nitrite (28.9 g, 280 mmol). The resulting mixture was stirred at 70 °C for 2 h under a nitrogen atmosphere. After completion of the reaction, the mixture was concentrated under reduced pressure, and the residue was purified by silica gel column chromatography (petroleum ether/ethyl acetate 10:1) to afford 40 (280 mg, 45% yield, 85% purity) as an orange solid. LC–MS (method 5): tR = 1.60 min; MS (ESI+) m/z: 313.9 [M + H]+.
4-Chloro-3-(2-methylphenoxy)naphthalen-1-amine (41)
To a solution of 1-chloro-2-(2-methylphenoxy)-4-nitronaphthalene (40; 280 mg, 0.80 mmol, 85% purity) in acetic acid (20 mL) was added iron powder (339 mg, 6.0 mmol). The resulting mixture was stirred at rt for 2 h. After consumption of the starting material, the reaction mixture was filtered, and the filtrate was concentrated to obtain a residue which was partitioned between ethyl acetate (25 mL) and water (25 mL). The organic phase was washed with brine solution, dried over anhydrous sodium sulfate, filtered, and concentrated under reduced pressure. The residue was purified by silica gel column chromatography (petroleum ether/ethyl acetate 2:1) to afford 41 (190 mg, 74% yield, 84% purity) as a light yellow solid. LC–MS (method 2, 0–3.00 min 5–95% B): tR = 1.59 min; MS (ESI+) m/z: 284 [M + H]+.
Methyl [4-Chloro-3-(2-methylphenoxy)naphthalen-1-yl]carbamate (42)
To an ice-cooled solution of 4-chloro-3-(2-methylphenoxy)naphthalen-1-amine (41; 190 mg, 0.60 mmol) in pyridine (20 mL), methyl chloroformate (159 mg, 1.69 mmol) was added dropwise. After complete addition, the corresponding suspension was stirred at rt for 4 h. The mixture was extracted with ethyl acetate. The organic phase was washed with water and brine and dried over anhydrous sodium sulfate. The solvent was removed under reduced pressure, and the residue was purified by silica gel column chromatography (petroleum ether/ethyl acetate 2:1) to afford 42 (150 mg, 56% yield, 72% purity) as a light yellow solid. LC–MS (method 2, 0–1.50 min 5–95% B): tR = 1.05 min; MS (ESI+) m/z: 342 [M + H]+.
3-[3-(2-Methylphenoxy)naphthalen-1-yl]-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (BAY-771, 43)
To a solution of 3-(3-bromonaphthalen-1-yl)-6-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione (45; 100 mg, 260 μmol) in DMF (5.1 mL) were added o-cresol (39.9 mg, 369 μmol), copper(I) chloride (6.68 mg, 35.1 μmol), cesium carbonate (203 mg, 623 μmol), and N,N-dimethylglycine (7.10 mg, 68.8 μmol). The resulting mixture was stirred at 140 °C overnight under a nitrogen atmosphere. Upon completion of the reaction, the mixture was cooled, then diluted with aqueous ammonia solution, and further stirred at rt for 10 min. Subsequently, ethyl acetate and water were added, and the layers were very carefully separated. The aqueous layer was extracted with ethyl acetate. The combined organic layers were washed with brine and dried with sodium sulfate. After filtration, the solvent was removed under reduced pressure. The resulting residue was purified using HT-HPLC (acid), affording 43 as a purple solid (46 mg, 41% yield). 1H NMR (400 MHz, [D]6DMSO): δ 2.22 (s, 3H), 6.41 (s, 1H), 7.05 (dd, 1H), 7.19 (td, 1H), 7.26–7.31, 7.38–7.43 (2 m, 2H, 3H), 7.51 (m, 1H), 7.71, 7.87 (2 d, 1H each), 12.63 (br s, 1H) ppm. LCMS (ESI−) m/z: 412 [M – H]−.
Measurement of Ionization Constants (pKa)
Ionization constants were determined using spectral gradient analysis.60 The test compound was dissolved in DMSO at 10 mM. Using a pion T3 instrument that performs automated acid–base titrations, the test compound was mixed with a continuously changing pH. As the pH changed, the fraction of the ionized compound changed as well. The absorption of an UV chromophore near the ionization center changed with ionization. The resulting UV spectrum changed with pH. The pKa was the inflection point of the absorption curve.
Solubility from DMSO in buffer pH 6.5: Aqueous solubility at pH 6.5 was determined by an orientating high-throughput screening method.61 Solubility was determined in PBS buffer pH 6.5 containing 1% DMSO. Test compounds were applied as 1 mm DMSO solution. After the addition of PBS buffer pH 6.5, solutions were shaken for 24 h at rt. The undissolved material was removed by filtration. The compound dissolved in the filtrate was quantified by HPLC-UV. The response was fitted to a one-point standard curve prepared in DMSO.
Biological Assays
BCAT1 and BCAT2 Biochemical Assay
BCAT catalyzes transamination of the BCAA leucine to α-KIC. The BCAT enzymatic reaction was measured in a coupled readout with LeuDH, which catalyzes the NADH-dependent reduction of α-KIC to leucine. NADH consumption was measured by fluorescent readout.
The biochemical reactions were performed at 32 °C in 384-well plates using a reaction volume of 51 μL and the following assay buffer conditions: 50 mM HEPES (2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid) pH 8.0, 50 mM aqueous ammonium sulfate, 1 mM ethylenediaminetetraacetic acid, 2 mM dithiothreitol, 0.01% BSA, 0.005% Brij 35 detergent, 0.5 μM PLP, 10 μM NADH, 50 μM α-KG, and 100 μM leucine. The BCAT enzyme was used in a final concentration of 0.75 nM and the LeuDH enzyme in a final concentration of 0.1 nM. Test compounds were used in a concentration range between 0.005 and 30 μM. All IC50 values are derived from two independent measurements for every given concentration, and we determined at least two such IC50 values for every compound in every assay. The final DMSO concentration was 2%. The reaction was initiated by the addition of substrates (leucine, α-KG, NADH). Enzyme activity was monitored kinetically over 90 min by the consumption of NADH fluorescence (extinction 340 nm/emission 480 nm). Recombinant human BCAT1 enzyme (catalogue no. ATGP1479) and recombinant human BCAT2 enzyme (catalogue no. ATGP1523) were purchased from ATGen (South Korea). Other chemicals were purchased from Sigma-Aldrich. Fluorescence measurements were obtained using a BMG CLARIOstar plate reader spectrophotometer. The decrease in fluorescence is proportional to BCAT activity. IC50 values were determined by interpolation from plots of relative fluorescence versus inhibitor concentration.
Protein Production, Crystallization, and Co-complex Structure Determination
Human full-length BCAT1 was expressed in Escherichia coli, purified via Ni affinity chromatography, tag-cleavage, anion exchange chromatography, and size exclusion chromatography and crystallized via the hanging drop method. Inhibitor co-complexes were formed via soaking. Diffraction data sets collected at synchrotron sources were processed using XDS62 and XDSapp,63 and the structures were solved via molecular replacement using software Phaser,64 rebuilt using COOT,65 and refined using Refmac.66 Ligand parameters were calculated using ProDrg.67 All details are described in the Supporting Information.
Cell Culture
Cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) or the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). They were maintained in the recommended cell culture media at 37 °C in 5% CO2.
BCAT Cellular Mechanistic Assays
Levels of leucine were measured in the medium of different tumor cell lines. The enzyme BCAT catalyzes the consumption of leucine.
Cells (U-87 MG and MDA-MB-231 from ATCC) were grown in DMEM containing 10% fetal calf serum (FCS). They were harvested with trypsin and seeded into 96-well plates with the medium devoid of FCS. The cells were incubated overnight at 37 °C in an atmosphere containing 5% CO2. The next day, test compounds were added to each cell well starting with 10 μM in a 1:3 dilution. The final concentration of DMSO was 0.1%, and DMSO controls were included. The plates were then placed in an incubator at 37 °C with 5% CO2 for 24 h. Leucine was measured in an enzymatic readout with LeuDH and diaphorase. The medium was deproteinized with perchloric acid; 30 μL of 1 M aqueous perchloric acid was added to each well, and the plates were centrifuged at 2000 rpm. Then, 5 μL of 3.2 M aqueous potassium carbonate was added to each well, and the plates were centrifuged at 2000 rpm. Aliquots (20 μL) were removed and incubated with 40 μL of buffer (50 mM TrispH 7.5, 0.5% glycerin, 0.01% Tween, 0.05% BSA with 0.125 U/mL LeuDH, 0.1 unit/mL diaphorase, 1 mM NAD+, and 2 μM resazurin). The conversion of resazurin into resorufin was detected by fluorescence spectroscopy (extinction 540 nm/emission 600 nm). Chemicals and enzymes were purchased from Sigma-Aldrich. Fluorescence measurements were obtained using a BMG CLARIOstar plate reader spectrophotometer. The increase in fluorescence is proportional to leucine concentration. IC50 values were determined by interpolation from plots of relative fluorescence versus inhibitor concentration.
Proliferation Assays
For proliferation studies, cells were plated in cell culture media at a density of 500 cells/30 μL/well in 384-well black plates (Corning no. 3571). Sister wells were plated in a separate plate for time zero determination, and all plates were incubated overnight at 37 °C. The next day, the test compounds were added in seven serial dilutions, each 1:3, starting at 50 μM down to 70 nM compound concentration using a Tecan HP D300 digital dispenser, and the plates were incubated at 37 °C for 72 h. The time zero plate was measured by adding 30 μL/well of CellTiter-Glo solution (Promega, no. G7573), followed by incubation for 2 min in an orbital shaker to induce cell lysis. After 10 min if additional incubation at rt, the luminescence was read with a PHERAstar FS microplate reader (BMG Labtech). After 72 h of incubation, the plates were measured as described for the time zero plate. Control wells, containing cells with culture medium and DMSO, were used to determine the control cell growth at 72 h compared to the initial number of cells (time zero value). To distinguish between cell growth inhibition and cell kill, the luminescence values after 72 h were corrected for the mean luminescence observed for the time zero wells at the day of test compound addition (time zero value). IC50 values, defined as the test compound concentration that corresponds to a reduction of cellular growth by 50% when compared with the values of DMSO control cells, were calculated using the dose–response curve Master Spreadsheet (Bella software).
Pharmacokinetics
In Vitro Metabolic Stability in Human Liver Microsomes
The in vitro metabolic stability of test compounds was determined by incubation at 1 μM in a suspension of liver microsomes (Sekisui XenoTech) in 100 mM phosphate buffer pH 7.4 (NaH2PO4·H2O + Na2HPO4·2H2O) and at a protein concentration of 1 mg/mL at 37 °C. The microsomes were activated by adding a cofactor mix containing 8 mM glucose-6-phosphate, 0.5 mM NADP, and 1 IU/mL glucose-6-phosphate dehydrogenase in phosphate buffer pH 7.4. The metabolic assay was started shortly afterward by adding the test compound to the incubation at a final volume of 0.55 mL (or 1.21 mL). The organic solvent in the incubations was limited to ≤1% DMSO and acetonitrile. During incubation, the microsomal suspensions were continuously shaken at 550 rpm (or 375 rpm), and aliquots were taken at 2, 8, 16, 30, 45, and 60 min, to which a double volume of cold acetonitrile was immediately added. Samples were frozen at −20 °C overnight and, after thawing, subsequently centrifuged for 15 min at 3700 rpm. The supernatant was analyzed with an Agilent 1200 HPLC system with LC–MS/MS detection. The half-life of a test compound was determined from the concentration–time plot. From the half-life, the intrinsic clearances were calculated. Together with the additional parameters, liver blood flow, specific liver weight, and microsomal protein content, the hepatic in vivo blood clearance (CLblood) and the maximal oral bioavailability (Fmax) were calculated using the “well-stirred” liver model:68 CLintrinsic′ [mL/min/kg] = kel [1/min]/((mg protein/volume of incubation [mL])·fu,inc)·(mg protein/liver weight [g])·(specific liver weight [g liver/kg body weight]); CLblood,well-stirred [L/h/kg] = (QH [L/h/kg]·fu,blood·CLintrinsic′ [L/h/kg])/(QH [L/h/kg] + fu,blood·CLintrinsic′ [L/h/kg]); Fmax = 1 – CLblood/QH; and using the following parameter values: liver blood flow, 1.32 L/h/kg, specific liver weight, 21 g/kg, microsomal protein content, 40 mg/g; fu,inc and fu,blood are taken as 1.
In Vitro Metabolic Stability in Rat Hepatocytes
Hepatocytes from Han/Wistar rats (Charles River) were isolated via a two-step perfusion method. After perfusion, the liver was carefully removed from the rat: the liver capsule was opened, and the hepatocytes were gently shaken out into a Petri dish with ice-cold Williams’ medium E (WME). The resulting cell suspension was filtered through sterile gauze into 50 mL Falcon tubes and centrifuged at 50g for 3 min at rt. The cell pellet was resuspended in WME (30 mL) and centrifuged twice through a Percoll gradient at 100 g. The hepatocytes were washed again with WME and resuspended in medium containing 5% FCS. Cell viability was determined by Trypan blue exclusion. For the metabolic stability assay, liver cells were distributed in WME containing 5% FCS to glass vials at a density of 1.0 × 106 vital cells/mL. The test compound was added to a final concentration of 1 μM. During incubation (37 °C), the hepatocyte suspensions were continuously shaken at 580 rpm, and aliquots were taken at 2, 8, 16, 30, 45, and 90 min, to which an equal volume of cold MeOH was immediately added. The samples were frozen at −20 °C overnight and, after thawing, subsequently centrifuged for 15 min at 3000 rpm. The supernatant was analyzed with an Agilent 1200 HPLC system with LC–MS/MS detection. The half-life of a test compound was determined from the concentration–time plot. From the half-life, the intrinsic clearance was calculated. Together with the additional parameters’ liver blood flow, specific liver weight, and amount of liver cells in vivo and in vitro, the hepatic in vivo blood clearance (CLblood) and the maximal oral bioavailability (Fmax) were calculated using the “well-stirred” liver model.68 The following parameter values were used: liver blood flow, 4.2 L/h/kg; specific liver weight, 32 g/kg body weight; liver cells in vivo, 1.1 × 108 cells/g liver; and liver cells in vitro, 1.0 × 106/mL.
Caco-2 Permeability Assay
Caco-2 cells (purchased from the DSMZ) were seeded at a density of 4.5 × 105 cells/well on 24-well insert plates, 0.4 μm pore size, 0.3 cm2 (Costar), and grown for 13–15 d in DMEM supplemented with 10% FCS, 1% GlutaMAX (100×, Gibco), 100 U/mL penicillin, 100 μg/mL streptomycin (Gibco), and 1% nonessential amino acids (100×, Thermo Fisher Scientific). Cells were maintained at 37 °C in a humidified 5% CO2 atmosphere. The medium was changed every 2–3 d. The bidirectional transport assay for the evaluation of Caco-2 permeability was undertaken in 24-well insert plates using a robotic system (Tecan). Before the assay was run, the culture medium was replaced by transport medium (FCS-free HEPES carbonate transport buffer pH 7.2). For the assessment of monolayer integrity, the transepithelial electrical resistance (TEER) was measured. Only monolayers with a TEER of at least 400 Ω·cm2 were used. Test compounds were predissolved in DMSO and added either to the apical or basolateral compartment at a final concentration of 2 μM. Evaluation was done in triplicate. Before and after incubation at 37 °C for 2 h, the samples were taken from both compartments and analyzed, after precipitation with MeOH, by LC–MS/MS. The apparent permeability coefficient (Papp) was calculated both for the apical to basolateral (A → B) and the basolateral to apical (B → A) direction using the following equation: Papp = (Vr/P0) (1/S) (P2/t), where Vr is the volume of the medium in the receiver chamber, P0 is the measured peak area of the test compound in the donor chamber at t = 0, S is the surface area of the monolayer, P2 is the measured peak area of the test compound in the acceptor chamber after incubation for 2 h, and t is the incubation time. The efflux ratio (ER) basolateral (B) to apical (A) was calculated by dividing Papp B–A by Papp A–B.
Estimation of Plasma Protein Binding by Equilibrium Dialysis
Binding of test compounds to mouse and rat plasma proteins was measured by equilibrium dialysis in a 96-well format using the HT dialysis equipment made of Teflon and a semipermeable membrane (regenerated cellulose, MWCO 12–14 K). The membrane separated the plasma from the buffer side (50 mM phosphate buffer, pH 7.4), each filled with 150 μL. The test compound was added in a concentration of 3 μM to the plasma and dialyzed for 7 h at 37 °C under moderate shaking. The unbound fraction of the test compound passes the membrane and distributes on both sides until equilibrium is reached. The relative compound concentration (peak area ratios analyte/internal standard) of the plasma and the buffer side was measured by LC–MS/MS analysis. For this, both sides were matrix-matched, that is, diluted with buffer and plasma to achieve the same matrix (10% plasma) and subsequently were precipitated with methanol. From the quotient of buffer and plasma concentration, the mean free (unbound) fraction (fu) was calculated (at least N = 3 replicates, CV %). Stability and recovery controls were included. Additionally, the test compound was dialyzed in buffer against buffer to estimate nonspecific binding to the equipment and/or the membrane and to ensure equilibrium. Due to the osmotic pressure of the plasma proteins, a dilution of the plasma takes place during the incubation (volume shift). The potential imprecision was addressed by inclusion of an empirical factor in the calculation of fu. Establishment of equilibrium in buffer/buffer dialysis and stability in the plasma was at least 80%; the recovery in buffer/buffer dialysis was aimed to be >30%. A free fraction of <1% was designated as high, between 1 and 10% as moderate, and >10% as low plasma protein binding.
Estimation of Binding to Brain Tissue by Equilibrium Dialysis
Brain homogenate was prepared freshly from frozen mouse brain by dilution with phosphate buffer (usually 1:4 m w/v) using a potter homogenisator. Binding of test compounds to brain homogenate was measured by equilibrium dialysis in a 96-well format using the HT dialysis equipment made of Teflon and a semipermeable membrane (regenerated cellulose, MWCO 3.5 K). The membrane separated the homogenate from the buffer side (50 mM phosphate buffer, pH 7.4), each filled with 150 μL. The test compound was added in a concentration of 3 μM to the brain homogenate and dialyzed for 22–24 h at 37 °C under moderate shaking. The unbound fraction of the test compound passes the membrane and distributes on both sides until equilibrium is reached. The relative compound concentration (peak area ratios analyte/internal standard) of the homogenate and the buffer side was measured by LC–MS/MS analysis. For this, both sides were diluted with buffer and brain homogenate to achieve the same matrix (10% brain homogenate) and subsequently were precipitated with methanol. From the quotient of buffer and brain homogenate concentration, the mean free (unbound) fraction (fu) in diluted brain homogenate (fu′) was calculated. Fraction unbound (fu) in brain tissue is calculated using the dilution factor (a) according to the law of mass action
Stability and recovery controls were included. Additionally, the test compound was dialyzed in buffer against buffer to estimate nonspecific binding to the equipment and/or the membrane and to ensure equilibrium. Establishment of equilibrium and stability in brain homogenate was at least 80%; the recovery in brain homogenate was aimed to be >30%. A free fraction of <1% was designated as high, between 1 and 10% as moderate, and of >10% as low brain tissue binding.
In Vivo PK in Rats
All animal experiments were conducted in accordance with the German animal welfare laws and were approved by local authorities. For in vivo pharmacokinetic experiments, test compounds were administered to male Wistar rats (Charles River) intravenously at doses of 0.5–1 mg/kg and intragastrically at doses of 1–2 mg/kg formulated as solutions using solubilizers such as PEG400 in well-tolerated amounts. For PK after intravenous (iv) administration, test compounds (0.3 mg/kg) were given to male rats as iv bolus, and blood samples were taken at 2 min, 8 min, 15 min, 30 min, 45 min, 1 h, 2 h, 4 h, 7 h, and 24 h after sampling. For PK after intragastral (ig) administration, test compounds (0.6 mg/kg) were given ig to fasted rats, and blood samples were taken at 5 min, 15 min, 30 min, 45 min, 1 h, 2 h, 4 h, 7 h, and 24 h after dosing. Blood was taken from the vena jugularis, collected into lithium heparin tubes (Monovette, Sarstedt), and centrifuged for 15 min at 3000 rpm. An aliquot of 100 μL from the supernatant (plasma) was taken and precipitated by the addition of 400 μL cold acetonitrile. Samples were frozen at −20 °C overnight and subsequently thawed and centrifuged at 3000 rpm, 4 °C for 20 min. Aliquots of the supernatants were analyzed using an Agilent HPLC system with LC–MS/MS detection. PK parameters were calculated by noncompartmental analysis (Microsoft Excel-based). PK parameters derived from concentration–time profiles after iv administration: CLplasma, total plasma clearance of test compound (in L/h/kg); CLblood, total blood clearance of test compound, CLplasma·Cp/Cb (in L/h/kg) with Cp/Cb being the ratio of concentrations in the plasma and blood. PK parameters calculated from concentration–time profiles after ig administration: Cmax, maximal plasma concentration (in mg/L); Cmax, norm, Cmax divided by the administered dose (in kg/L); Tmax, time point at which Cmax was observed (in h). Parameters calculated from both iv and ig concentration–time profiles: AUCnorm, area under the concentration–time curve from t = 0 to infinity (extrapolated) divided by the administered dose (in kg·h/L); AUC(0–t last)norm, area under the concentration–time curve from t = 0 to the last time point for which plasma concentrations could be measured divided by the administered dose (in kg·h/L); t1/2, terminal half-life (in h); F, oral bioavailability, AUCnorm after ig administration divided by AUCnorm after iv administration (in %).
In Vivo Pharmacology Studies
All animal experiments were conducted in accordance with the German animal welfare laws and were approved by local authorities. Female 6–8 week-old NMRI nude mice were obtained from Charles River (Sulzfeld, Germany). To determine the blood plasma levels of compound, BAY-069 (36a) was administered orally at doses of 25, 50, and 100 mg/kg. At each time point (1, 3, 7, and 24 h), three animals were euthanized, and blood samples and brains were collected for further analysis. Brain samples were mixed with Tris-HCl buffer in a ratio of 1:5 (v/v) and homogenized using a TissueLyser. Blood samples were centrifuged for 15 min at 3000 rpm. Aliquots of the supernatant (plasma) and brain homogenates were mixed with acetonitrile in a ratio of 1:5 (v/v) and stored in a freezer at −20 °C overnight. After thawing and mixing, samples were centrifuged at 3000 rpm, 4 °C for 20 min. Aliquots of the supernatants were analyzed using an HPLC system with LC–MS/MS detection.
Acknowledgments
We are indebted to Andrea Haegebarth, Marcus Bauser, Ingo Hartung, Knut Eis, Ashley Eheim, Holger Hess-Stumpp, Jan Huebner, and Georg Kettschau for their support throughout the project. The expert technical assistance of Lisa Schaelicke, Stefanie Wolf, Qiuwen Wang, Paul Bednarski, Stefanie Klar, Kirsten Ostertag, Wolfgang Schröter, Annette Zietz, Silvia Schätzlein, Xinlong Huang, Chonh Wang, Sasha Wu, Maria Hallmann, and Tina Stromeyer is gratefully acknowledged. We thank the staff at beamline BL14.1 at Helmholtz-Zentrum Berlin, at beamline ID29 at the European Synchrotron Radiation Facility (ESRF) in Grenoble, and at beamline P11 at PETRA III at the Deutsches Elektronen-Synchrotron (a member of the Helmholtz Association) for access to synchrotron radiation and support during data collection and the staff at MOLOX GmbH for data collection services. The support of Kay Greenfield in preparing the manuscript is gratefully acknowledged. We thank Joseph Bluck, Georg Kettschau, and Clara Christ for careful proofreading, and we are grateful to our unknown reviewers for very constructive feedback and advice.
Glossary
Abbreviations
- α-KG
α-ketoglutarate
- α-KIC
α-ketoisocaproate
- atrop
atropisomer
- BEI
binding efficiency index
- BCAA
branched-chain amino acid
- BCAT
branched-chain amino acid transaminases
- CL
blood clearance
- DAD
diode array detector
- DMEM
Dulbecco’s modified Eagle’s medium
- LeuDH
leucine dehydrogenase
- LLE
lipophilic ligand efficiency
- 3PP
3-phenylpropionic acid
- PLP
pyridoxal phosphate
- NMP
N-methyl-2-pyrrolidon
- PDA
photodiode array
- TPSA
total polar surface area
- UPLC–MS
ultra-performance liquid chromatography–mass spectrometry
- WME
Williams’ medium
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.2c00441.
Accession Codes
The coordinates and structure factors for the described crystal structures have been deposited with the Protein Data Bank (PDB). The PDB accession codes are 7NTR (3PP), 7NWA (compound A), 7NWB (compound 1), 7NWC (compound 2), 7NWE (compound 10), 7NWM (compound 12), 7NXN (compound 21 5-F), 7NXO (compound 24 5-F), 7NY2 (compound 35), 7NY9 (compound 38), and 7NYA (compound 36/BAY-069). Authors will release the atomic coordinates upon article publication.
Author Present Address
# Nuvisan ICB GmbH, Müllerstrasse 178, 13353 Berlin, Germany
Author Present Address
¶ AWE Aldermaston, Reading, Berkshire, RG7 4PR, U.K.
Author Present Address
∇ F. Hoffmann-La Roche Ltd, Grenzacherstrasse 124, CH-4070 Basel, Switzerland.
Author Present Address
○ Research & Development, Pharmaceuticals, Bayer U.S. LLC, Bayer Research & Innovation Center, 238 Main Street, Cambridge, MA 02142, United States.
Author Contributions
The manuscript was written with contributions from all authors. All authors have given approval to the final version of the manuscript.
The authors declare the following competing financial interest(s): J.G., R.C.H., K.Z., S.K., C.L., D.N., C.L., R.N., U.G., M.D., H.R., and L.B. are or have been employees and stockholders of Bayer AG.
Supplementary Material
References
- Harper A. E.; Miller R. H.; Block K. P. Branched-chain amino acid metabolism. Annu. Rev. Nutr. 1984, 4, 409–454. 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- Sweatt A. J.; Wood M.; Suryawan A.; Wallin R.; Willingham M. C.; Hutson S. M. Branched-chain amino acid catabolism: unique segregation of pathway enzymes in organ systems and peripheral nerves. Am. J. Physiol.: Endocrinol. Metab. 2004, 286, E64–E76. 10.1152/ajpendo.00276.2003. [DOI] [PubMed] [Google Scholar]
- Yudkoff M. Brain metabolism of branched-chain amino acids. Glia 1997, 21, 92–98. . [DOI] [PubMed] [Google Scholar]
- Tönjes M.; Barbus S.; Park Y. J.; Wang W.; Schlotter M.; Lindroth A. M.; Pleier S. V.; Bai A. H. C.; Karra D.; Piro R. M.; Felsberg J.; Addington A.; Lemke D.; Weibrecht I.; Hovestadt V.; Rolli C. G.; Campos B.; Turcan S.; Sturm D.; Witt H.; Chan T. A.; Herold-Mende C.; Kemkemer R.; König R.; Schmidt K.; Hull W. E.; Pfister S. M.; Jugold M.; Hutson S. M.; Plass C.; Okun J. G.; Reifenberger G.; Lichter P.; Radlwimmer B. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat. Med. 2013, 19, 901–908. 10.1038/nm.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thewes V.; Simon R.; Hlevnjak M.; Schlotter M.; Schroeter P.; Schmidt K.; Wu Y.; Anzeneder T.; Wang W.; Windisch P.; Kirchgäßner M.; Melling N.; Kneisel N.; Büttner R.; Deuschle U.; Sinn H. P.; Schneeweiss A.; Heck S.; Kaulfuss S.; Hess-Stumpp H.; Okun J. G.; Sauter G.; Lykkesfeldt A. E.; Zapatka M.; Radlwimmer B.; Lichter P.; Tönjes M. The branched-chain amino acid transaminase 1 sustains growth of antiestrogen-resistant and ERα-negative breast cancer. Oncogene 2017, 36, 4124–4134. 10.1038/onc.2017.32. [DOI] [PubMed] [Google Scholar]
- Raffel S.; Falcone M.; Kneisel N.; Hansson J.; Wang W.; Lutz C.; Bullinger L.; Poschet G.; Nonnenmacher Y.; Barnert A.; Bahr C.; Zeisberger P.; Przybylla A.; Sohn M.; Tönjes M.; Erez A.; Adler L.; Jensen P.; Scholl C.; Fröhling S.; Cocciardi S.; Wuchter P.; Thiede C.; Flörcken A.; Westermann J.; Ehninger G.; Lichter P.; Hiller K.; Hell R.; Herrmann C.; Ho A. D.; Krijgsveld J.; Radlwimmer B.; Trumpp A. Author Correction: BCAT1 restricts αKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature 2018, 560, E28 10.1038/s41586-018-0403-9. [DOI] [PubMed] [Google Scholar]
- Raffel S.; Falcone M.; Kneisel N.; Hansson J.; Wang W.; Lutz C.; Bullinger L.; Poschet G.; Nonnenmacher Y.; Barnert A.; Bahr C.; Zeisberger P.; Przybylla A.; Sohn M.; Tönjes M.; Erez A.; Adler L.; Jensen P.; Scholl C.; Fröhling S.; Cocciardi S.; Wuchter P.; Thiede C.; Flörcken A.; Westermann J.; Ehninger G.; Lichter P.; Hiller K.; Hell R.; Herrmann C.; Ho A. D.; Krijgsveld J.; Radlwimmer B.; Trumpp A. BCAT1 restricts αKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature 2017, 551, 384–388. 10.1038/nature24294. [DOI] [PubMed] [Google Scholar]
- Mayers J. R.; Torrence M. E.; Danai L. V.; Papagiannakopoulos T.; Davidson S. M.; Bauer M. R.; Lau A. N.; Ji B. W.; Dixit P. D.; Hosios A. M.; Muir A.; Chin C. R.; Freinkman E.; Jacks T.; Wolpin B. M.; Vitkup D.; Vander Heiden M. G. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras -driven cancers. Science 2016, 353, 1161–1165. 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay I.; Wang J.; Qin M.; Gao L.; Holtz R.; Vessella R. L.; Leach R. W.; Gelman I. H. Src promotes castration-recurrent prostate cancer through androgen receptor-dependent canonical and non-canonical transcriptional signatures. Oncotarget 2017, 8, 10324–10347. 10.18632/oncotarget.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W.; Shao Y.; Peng Y. MicroRNA-218 inhibits tumor growth and increases chemosensitivity to CDDP treatment by targeting BCAT1 in prostate cancer. Mol. Carcinog. 2017, 56, 1570–1577. 10.1002/mc.22612. [DOI] [PubMed] [Google Scholar]
- Silva L. S.; Poschet G.; Nonnenmacher Y.; Becker H. M.; Sapcariu S.; Gaupel A. C.; Schlotter M.; Wu Y.; Kneisel N.; Seiffert M.; Hell R.; Hiller K.; Lichter P.; Radlwimmer B. Branched-chain ketoacids secreted by glioblastoma cells via MCT 1 modulate macrophage phenotype. EMBO Rep. 2017, 18, 2172–2185. 10.15252/embr.201744154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H.; Zhou J.; Sundersingh F. S.; Summerfield J.; Somers D.; Messer J. A.; Satz A. L.; Ancellin N.; Arico-Muendel C. C.; Bedard K. L.; Beljean A.; Belyanskaya S. L.; Bingham R.; Smith S. E.; Boursier E.; Carter P.; Centrella P. A.; Clark M. A.; Chung C. W.; Davie C. P.; Delorey J. L.; Ding Y.; Franklin G. J.; Grady L. C.; Herry K.; Hobbs C.; Kollmann C. S.; Morgan B. A.; Kaushansky L. J.; Zhou Q. Discovery, SAR, and X-ray Binding Mode Study of BCATm Inhibitors from a Novel DNA-Encoded Library. ACS Med. Chem. Lett. 2015, 6, 919–924. 10.1021/acsmedchemlett.5b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H.; Zhou J.; Sundersingh F.; Messer J. A.; Somers D. O.; Ajakane M.; Arico-Muendel C. C.; Beljean A.; Belyanskaya S. L.; Bingham R.; Blazensky E.; Boullay A. B.; Boursier E.; Chai J.; Carter P.; Chung C. W.; Daugan A.; Ding Y.; Herry K.; Hobbs C.; Humphries E.; Kollmann C.; Nguyen V. L.; Nicodeme E.; Smith S. E.; Dodic N.; Ancellin N. Discovery and Optimization of Potent, Selective, and in Vivo Efficacious 2-Aryl Benzimidazole BCATm Inhibitors. ACS Med. Chem. Lett. 2016, 7, 379–384. 10.1021/acsmedchemlett.5b00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bora K. M.; Hu L.-Y.; Kesten S. R.; Lei H.; Moreland D. W.; Rafferty M. F.; Ryder T. R.; Scholten J. D.; Wustrow D. J.. Branched chain amino acid-dependent aminotransferase inhibitors and their use in the treatment of neurodegenerative diseases. WO 2002024672 A3, 2002.
- Hu L. Y.; Boxer P. A.; Kesten S. R.; Lei H. J.; Wustrow D. J.; Moreland D. W.; Zhang L.; Ahn K.; Ryder T. R.; Liu X.; Rubin J. R.; Fahnoe K.; Carroll R. T.; Dutta S.; Fahnoe D. C.; Probert A. W.; Roof R. L.; Rafferty M. F.; Kostlan C. R.; Scholten J. D.; Hood M.; Ren X. D.; Schielke G. P.; Su T. Z.; Taylor C. P.; Mistry A.; McConnell P.; Hasemann C.; Ohren J. The design and synthesis of human branched-chain amino acid aminotransferase inhibitors for treatment of neurodegenerative diseases. Bioorg. Med. Chem. Lett. 2006, 16, 2337–2340. 10.1016/j.bmcl.2005.07.058. [DOI] [PubMed] [Google Scholar]
- Bertrand S. M.; Ancellin N.; Beaufils B.; Bingham R. P.; Borthwick J. A.; Boullay A. B.; Boursier E.; Carter P. S.; Chung C. W.; Churcher I.; Dodic N.; Fouchet M. H.; Fournier C.; Francis P. L.; Gummer L. A.; Herry K.; Hobbs A.; Hobbs C. I.; Homes P.; Jamieson C.; Nicodeme E.; Pickett S. D.; Reid I. H.; Simpson G. L.; Sloan L. A.; Smith S. E.; Somers D. O.; Spitzfaden C.; Suckling C. J.; Valko K.; Washio Y.; Young R. J. The Discovery of in Vivo Active Mitochondrial Branched-Chain Aminotransferase (BCATm) Inhibitors by Hybridizing Fragment and HTS Hits. J. Med. Chem. 2015, 58, 7140–7163. 10.1021/acs.jmedchem.5b00313. [DOI] [PubMed] [Google Scholar]
- Borthwick J. A.; Ancellin N.; Bertrand S. M.; Bingham R. P.; Carter P. S.; Chung C. W.; Churcher I.; Dodic N.; Fournier C.; Francis P. L.; Hobbs A.; Jamieson C.; Pickett S. D.; Smith S. E.; Somers D. O.; Spitzfaden C.; Suckling C. J.; Young R. J. Structurally Diverse Mitochondrial Branched Chain Aminotransferase (BCATm) Leads with Varying Binding Modes Identified by Fragment Screening. J. Med. Chem. 2016, 59, 2452–2467. 10.1021/acs.jmedchem.5b01607. [DOI] [PubMed] [Google Scholar]
- Hopkins A. L.; Keserü G. M.; Leeson P. D.; Rees D. C.; Reynolds C. H. The role of ligand efficiency metrics in drug discovery. Nat. Rev. Drug Discovery 2014, 13, 105–121. 10.1038/nrd4163. [DOI] [PubMed] [Google Scholar]
- Schultes S.; de Graaf C.; Haaksma E. E. J.; de Esch I. J. P.; RobLeurs R.; Krämer O. Ligand efficiency as a guide in fragment hit selection and optimization. Drug Discov. Today Technol. 2010, 7, e157 10.1016/j.ddtec.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Shultz M. D. Setting expectations in molecular optimizations: Strengths and limitations of commonly used composite parameters. Bioorg. Med. Chem. Lett. 2013, 23, 5980–5991. 10.1016/j.bmcl.2013.08.029. [DOI] [PubMed] [Google Scholar]
- Goto M.; Miyahara I.; Hirotsu K.; Conway M.; Yennawar N.; Islam M. M.; Hutson S. M. Structural determinants for branched-chain aminotransferase isozyme-specific inhibition by the anticonvulsant drug gabapentin. J. Biol. Chem. 2005, 280, 37246–37256. 10.1074/jbc.m506486200. [DOI] [PubMed] [Google Scholar]
- Rohrbach S.; Smith A. J.; Pang J. H.; Poole D. L.; Tuttle T.; Chiba S.; Murphy J. A. Concerted Nucleophilic Aromatic Substitution Reactions. Angew. Chem., Int. Ed. Engl. 2019, 58, 16368–16388. 10.1002/anie.201902216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes M.-W.; Andree R.; Schallner O.; Santel H.-J.; Dollinger M.. Substituted diazacyclohexandi(thi)ones. WO 9531440 A1, 1994.
- Szostak J. W. Introduction: Combinatorial Chemistry. Chem. Rev. 1997, 97, 347–348. 10.1021/cr9700080. [DOI] [PubMed] [Google Scholar]
- Koppitz M.; Eis K. Automated medicinal chemistry. Drug Discov. Today 2006, 11, 561–568. 10.1016/j.drudis.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Halgren T. A.; Murphy R. B.; Friesner R. A.; Beard H. S.; Frye L. L.; Pollard W. T.; Banks J. L. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Banks J. L.; Murphy R. B.; Halgren T. A.; Klicic J. J.; Mainz D. T.; Repasky M. P.; Knoll E. H.; Shelley M.; Perry J. K.; Shaw D. E.; Francis P.; Shenkin P. S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Glide, Release 2017-2; Schrodinger, LLC: New York, 2017.
- Zhang Z.; Wallace M. B.; Feng J.; Stafford J. A.; Skene R. J.; Shi L.; Lee B.; Aertgeerts K.; Jennings A.; Xu R.; Kassel D. B.; Kaldor S. W.; Navre M.; Webb D. R.; Gwaltney S. L. Design and synthesis of pyrimidinone and pyrimidinedione inhibitors of dipeptidyl peptidase IV. J. Med. Chem. 2011, 54, 510–524. 10.1021/jm101016w. [DOI] [PubMed] [Google Scholar]
- Neiland O. Y.; Zagorskaya N. N. Electronic absorption spectra and acidity of new condensed systems of uracil-derivatives of 1,3-dithiolo[4,5-d]pyrimidine. Chem. Heterocycl. Compd. 1992, 28, 350–354. 10.1007/bf00529385. [DOI] [Google Scholar]
- ADMET Predictor, 7.1; Simulations Plus, Inc: Lancaster, CA, 2017.
- Fraczkiewicz R.In silico Prediction of Ionization. In Reference Module in Chemistry, Molecular Sciences and Chemical Engineering; Reedijk J., Ed.; Elsevier: The Netherlands, 2013; Vol. 5. [Google Scholar]
- Fraczkiewicz R.; Lobell M.; Göller A. H.; Krenz U.; Schoenneis R.; Clark R. D.; Hillisch A. Best of Both Worlds: Combining Pharma Data and State of the Art Modeling Technology To Improve in Silico pKa Prediction. J. Chem. Inf. Model. 2015, 55, 389–397. 10.1021/ci500585w. [DOI] [PubMed] [Google Scholar]
- Ertl P.; Rohde B.; Selzer P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000, 43, 3714–3717. 10.1021/jm000942e. [DOI] [PubMed] [Google Scholar]
- 5′-F variant of 21/24 was synthesized in a similar fashion to compound 21/24; for the synthesis of 21/24, see Supporting Information.
- Dossetter A. G.; Griffen E. J.; Leach A. G. Matched molecular pair analysis in drug discovery. Drug Discov. Today 2013, 18, 724–731. 10.1016/j.drudis.2013.03.003. [DOI] [PubMed] [Google Scholar]
- O’Boyle N. M.; Boström J.; Sayle R. A.; Gill A. Using matched molecular series as a predictive tool to optimize biological activity. J. Med. Chem. 2014, 57, 2704. 10.1021/jm500022q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awale M.; Riniker S.; Kramer C. Matched Molecular Series Analysis for ADME Property Prediction. J. Chem. Inf. Model. 2020, 60, 2903–2914. 10.1021/acs.jcim.0c00269. [DOI] [PubMed] [Google Scholar]
- Shivakumar D.; Harder E.; Damm W.; Friesner R. A.; Sherman W. Improving the Prediction of Absolute Solvation Free Energies Using the Next Generation OPLS Force Field. J. Chem. Theory Comput. 2012, 8, 2553–2558. 10.1021/ct300203w. [DOI] [PubMed] [Google Scholar]
- Shivakumar D.; Williams J.; Wu Y.; Damm W.; Shelley J.; Sherman W. Prediction of Absolute Solvation Free Energies using Molecular Dynamics Free Energy Perturbation and the OPLS Force Field. J. Chem. Theory Comput. 2010, 6, 1509–1519. 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- Wang L.; Berne B. J.; Friesner R. A. On achieving high accuracy and reliability in the calculation of relative protein-ligand binding affinities. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 1937–1942. 10.1073/pnas.1114017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov D. G. The fragment molecular orbital method: theoretical development, implementation in GAMESS, and applications. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2017, 7, e1322 10.1002/wcms.1322. [DOI] [Google Scholar]
- The given BCAT IC50 values for 36 may underestimate the true target activity of the racemate (as a comparison with the values for the single enantiomers also suggests). The values reported are the numerical average over all independent IC50 determinations, for two different compound preparations. For completeness, the individual averaged IC50 values were for 294 nM BCAT1/1850 nM BCAT2 for the older preparation and 42 nM BCAT1/237 nM BCAT2 for the newer preparation. Analytics showed both preparations had equally high quality.
- Nguyen T.Giving Atropisomers Another Chance; C&EN Global Enterprise, 2018; Vol. 96, p 22 [Google Scholar]
- Raffa R. B.; Pergolizzi J. V. Jr.; Taylo R. ″Atropisomeric″ Drugs: Basic Concept and Example of Application to Drug Development. Pharmacol. Pharm. 2020, 11, 1–8. 10.4236/pp.2020.111001. [DOI] [Google Scholar]
- Cardenas M. M.; Nguyen A. D.; Brown Z. E.; Heydari B. S.; Heydari B. S.; Vaidya S. D.; Gustafson J. L. Atropisomerism as inspiration for new chemistry. Arkivoc 2021, 2021, 20–47. 10.24820/ark.5550190.p011.382. [DOI] [Google Scholar]
- Müller S.; Ackloo S.; Arrowsmith C. H.; Bauser M.; Baryza J. L.; Blagg J.; Böttcher J.; Bountra C.; Brown P. J.; Bunnage M. E.; Carter A. J.; Damerell D.; Dötsch V.; Drewry D. H.; Edwards A. M.; Edwards J.; Elkins J. M.; Fischer C.; Frye S. V.; Gollner A.; Grimshaw C. E.; Hanke T.; Hartung I. V.; Hitchcock S.; Howe T.; Hughes T. V.; Laufer S.; Li V. M.; Liras S.; Marsden B. D.; Matsui H.; Mathias J.; O’Hagan R. C.; Owen D. R.; Pande V.; Rauh D.; Rosenberg S. H.; Roth B. L.; Schneider N. S.; Scholten C.; Singh Saikatendu K.; Simeonov A.; Takizawa M.; Tse C.; Thompson P. R.; Treiber D. K.; Viana A. Y.; Wells C. I.; Willson T. M.; Zuercher W. J.; Knapp S.; Mueller-Fahrnow A. Donated chemical probes for open science. eLife 2018, 7, e34311 10.7554/elife.34311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché L.; Christ C. D.; Siegel S.; Fernández-Montalván A. E.; Holton S. J.; Fedorov O.; ter Laak A.; Sugawara T.; Stöckigt D.; Tallant C.; Bennett J.; Monteiro O.; Díaz-Sáez L.; Siejka P.; Meier J.; Pütter V.; Weiske J.; Müller S.; Huber K. V. M.; Hartung I. V.; Haendler B. Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains. J. Med. Chem. 2017, 60, 4002–4022. 10.1021/acs.jmedchem.7b00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barca G. M. J.; Bertoni C.; Carrington L.; Datta D.; De Silva N.; Deustua J. E.; Fedorov D. G.; Gour J. R.; Gunina A. O.; Guidez E.; Harville T.; Irle S.; Ivanic J.; Kowalski K.; Leang S. S.; Li H.; Li W.; Lutz J. J.; Magoulas I.; Mato J.; Mironov V.; Nakata H.; Pham B. Q.; Piecuch P.; Poole D.; Pruitt S. R.; Rendell A. P.; Roskop L. B.; Ruedenberg K.; Sattasathuchana T.; Schmidt M. W.; Shen J.; Slipchenko L.; Sosonkina M.; Sundriyal V.; Tiwari A.; Galvez Vallejo J. L.; Westheimer B.; Włoch M.; Xu P.; Zahariev F.; Gordon M. S. Recent developments in the general atomic and molecular electronic structure system. J. Chem. Phys. 2020, 152, 154102. 10.1063/5.0005188. [DOI] [PubMed] [Google Scholar]
- LaPlante S. R.; Fader L. D.; Fandrick K. R.; Fandrick D. R.; Hucke O.; Kemper R.; Miller S. P. F.; Edwards P. J. Assessing atropisomer axial chirality in drug discovery and development. J. Med. Chem. 2011, 54, 7005–7022. 10.1021/jm200584g. [DOI] [PubMed] [Google Scholar]
- Cho H. R.; Hong B.; Kim H.; Park C. K.; Park S. H.; Park S.; Choi S. H. Assessment of bevacizumab resistance increased by expression of BCAT1 in IDH1 wild-type glioblastoma: application of DSC perfusion MR imaging. Oncotarget 2016, 7, 69606–69615. 10.18632/oncotarget.11901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori A.; Tsunoda M.; Konuma T.; Kobayashi M.; Nagy T.; Glushka J.; Tayyari F.; McSkimming D.; Kannan N.; Tojo A.; Edison A. S.; Ito T. Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature 2017, 545, 500–504. 10.1038/nature22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S.; Kuhn D.; Klebe G. A new method to detect related function among proteins independent of sequence and fold homology. J. Mol. Biol. 2002, 323, 387–406. 10.1016/s0022-2836(02)00811-2. [DOI] [PubMed] [Google Scholar]
- Lee J.; Schapira M.. Negative controls of chemical probes can be misleading. 2020, bioRxiv 2020.09.30.320465. [Google Scholar]
- For the synthetic route of 43, see the Supporting Information.
- Bouche L. A.; Kaulfuss S.; Zimmermann K.; Rehwinkel H.; Neuhaus R.; Hillig R.; Nguyen D.; Günther J.; Faria Alvares De Lemos C. A.. Pyrimidinedione derivatives. WO 2021063821 A1, 2021.
- SGC . For further information. please go to: https://www.sgc-ffm.uni-frankfurt.de/#!specificprobeoverview/BAY-069 (accessed May 1, 2022).
- Müller S.; Ackloo S.; Al Chawaf A.; Al-Lazikani B.; Antolin A.; Baell J. B.; Beck H.; Beedie S.; Betz U. A. K.; Bezerra G. A.; Brennan P. E.; Brown D.; Brown P. J.; Bullock A. N.; Carter A. J.; Chaikuad A.; Chaineau M.; Ciulli A.; Collins I.; Dreher J.; Drewry D.; Edfeldt K.; Edwards A. M.; Egner U.; Frye S. V.; Fuchs S. M.; Hall M. D.; Hartung I. V.; Hillisch A.; Hitchcock S. H.; Homan E.; Kannan N.; Kiefer J. R.; Knapp S.; Kostic M.; Kubicek S.; Leach A. R.; Lindemann S.; Marsden B. D.; Matsui H.; Meier J. L.; Merk D.; Michel M.; Morgan M. R.; Mueller-Fahrnow A.; Owen D. R.; Perry B. G.; Rosenberg S. H.; Saikatendu K. S.; Schapira M.; Scholten C.; Sharma S.; Simeonov A.; Sundström M.; Superti-Furga G.; Todd M. H.; Tredup C.; Vedadi M.; von Delft F.; Willson T. M.; Winter G. E.; Workman P.; Arrowsmith C. H. Target 2035 – update on the quest for a probe for every protein. RSC Med. Chem. 2022, 13, 13. 10.1039/D1MD00228G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith C. H.; Audia J. E.; Austin C.; Baell J.; Bennett J.; Blagg J.; Bountra C.; Brennan P. E.; Brown P. J.; Bunnage M. E.; Buser-Doepner C.; Campbell R. M.; Carter A. J.; Cohen P.; Copeland R. A.; Cravatt B.; Dahlin J. L.; Dhanak D.; Edwards A. M.; Frederiksen M.; Frye S. V.; Gray N.; Grimshaw C. E.; Hepworth D.; Howe T.; Huber K. V.; Jin J.; Knapp S.; Kotz J. D.; Kruger R. G.; Lowe D.; Mader M. M.; Marsden B.; Mueller-Fahrnow A.; Müller S.; O’Hagan R. C.; Overington J. P.; Owen D. R.; Rosenberg S. H.; Ross B.; Roth R.; Schapira M.; Schreiber S. L.; Shoichet B.; Sundström M.; Superti-Furga G.; Taunton J.; Toledo-Sherman L.; Walpole C.; Walters M. A.; Willson T. M.; Workman P.; Young R. N.; Zuercher W. J. The promise and peril of chemical probes. Nat. Chem. Biol. 2015, 11, 536–541. 10.1038/nchembio.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns E. H.; Di L.. pKa Methods. Drug-like Properties: Concepts, Structure Design and Methods; Academic Press: Burlington, MA, 2008; pp 271–275. [Google Scholar]
- Onofrey T.; Kazan G.; Barbagallo C.; Blodgett J.; Weiss A.. Automated Screening of Aqueous Compound Solubility in Drug Discovery in Millipore Corporation; Life Sciences Division: Danvers, MA USA, 2019. [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 133–144. 10.1107/s0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparta K. M.; Krug M.; Heinemann U.; Mueller U.; Weiss M. S. XDSAPP2.0. J. Appl. Crystallogr. 2016, 49, 1085–1092. 10.1107/s1600576716004416. [DOI] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. Phasercrystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. 10.1107/s0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 486–501. 10.1107/s0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N.; Skubák P.; Lebedev A. A.; Pannu N. S.; Steiner R. A.; Nicholls R. A.; Winn M. D.; Long F.; Vagin A. A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011, 67, 355–367. 10.1107/s0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüttelkopf A. W.; van Aalten D. M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 1355–1363. 10.1107/s0907444904011679. [DOI] [PubMed] [Google Scholar]
- Pang K. S.; Rowland M. Hepatic clearance of drugs. I. Theoretical considerations of a ″well-stirred″ model and a ″parallel tube″ model. Influence of hepatic blood flow, plasma and blood cell binding, and the hepatocellular enzymatic activity on hepatic drug clearance. J. Pharmacokinet. Biopharm. 1977, 5, 625–653. 10.1007/bf01059688. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.