Abstract
Lung cancer is the leading cause of death and morbidity from malignant neoplasms worldwide, and its poor prognosis places a heavy burden on patients. A large percentage of lung cancer cases are associated with smoking. A significant number of non-smokers also develop the disease, suggesting an epigenetic and genetic mechanism for the development of lung cancer. The current situation with the diagnosis and treatment of lung cancer remains grim, and effective therapeutic targets and molecular markers are urgently needed. Circular RNAs (circRNAs) are covalently closed non-coding RNAs that have received much attention due to their biological properties such as conservatism, stability, and tissue specificity. Many studies have shown that circRNAs are involved in the regulation of lung cancer through various mechanisms, such as microRNA adsorption, and play an important role in the early diagnosis, treatment, and prognosis of lung cancer. In recent years, it has become increasingly clear that circRNAs are involved in the proliferation, migration, and invasion of lung cancer cells. Differentially expressed circRNAs can be used as non-invasive diagnostic and prognostic markers of lung cancer. This article summarizes the current advances of circRNAs in the diagnosis, treatment and prognosis of lung cancer.
Keywords: Noncoding RNAs, Circular RNAs, Lung cancer, Biology function, Biomarker
1. Introduction
Lung cancer is the most common malignant tumor disease in the world, posing a serious threat to human life and health. According to statistics, in 2018, there were about 2.1 million new cases of lung cancer and 1.8 million deaths from lung cancer worldwide, with the morbidity and mortality ranking first among all cancer types [1]. According to histological types, lung cancer can be divided into small cell lung cancer and non-small cell lung cancer, among which small cell lung cancer and non-small cell lung cancer account for about 15% of the total lung cancers, respectively and 85% [2]. Although clinical diagnosis and treatment methods have improved, the 5-year survival rate of lung cancer is still not optimistic due to untimely diagnosis, limited beneficiary population, and drug resistance of patients. In addition, the lack of relatively specific tumor markers adds challenges to the diagnosis, treatment and prognosis of lung cancer. Therefore, it is necessary to deeply study the molecular mechanism of lung cancer to explore potential biomarkers and therapeutic targets for lung cancer.
Circular RNA (circRNA) is a special kind of endogenous non-coding RNA. As early as the 1970s, circRNA was found to exist in RNA viruses [3,4]. However, due to the limitations of the technology at the time, circRNAs were considered to be by-products of the splicing process, so they did not receive widespread attention [5]. In recent years, with the development of high-throughput sequencing technology and bioinformatics, circRNAs have been discovered in large numbers and gradually become a research hotspot in the field of RNA. At present, many studies have confirmed that circRNA can participate in the regulation of the occurrence and development of lung cancer, and is expected to provide new ideas for the diagnosis, treatment and prognosis of lung cancer [[6], [7], [8], [9]].
2. Biological functions of circular RNAs
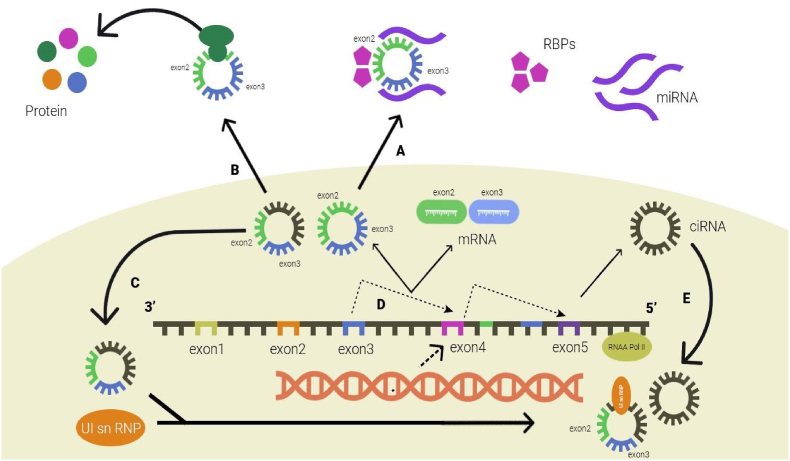
CircRNAs are covalently closed noncoding RNA molecules that are ubiquitous in eukaryotic transcriptomes. CircRNAs are usually divided into exonic circRNAs (ecRNAs), intronic circRNAs (ciRNAs), and exon-intron circRNAs (EIciRNAs) according to their sources [10]. Among them, exonic circRNAs are the most common. Unlike linear RNA, circRNA does not have a cap structure at the 5′ end and a polyadenylation tail at the 3′ end, which can resist the degradation of exonuclease RNase R, so circRNA is more stable and has a longer half-life than linear RNA [11]. Research also found that circRNAs show good species conservation [12]. In addition, the expression of circRNAs is tissue specific and developmental stage specific, suggesting that circRNAs may be involved in the regulation of various pathophysiological processes in the body [13]. The functional studies of circRNAs mostly focus on the following aspects: 1) Adsorb miRNAs as molecular sponges. Competing endogenous RNA (ceRNA) mechanism points out that RNAs with the same miRNA response elements (MREs) can competitively bind miRNAs, thereby regulating each other's expression (Fig. 1) [14].
Fig. 1.
Biological functions of circular RNAs. (A) circRNAs act as miRNA sponges. (B) circRNAs bind to proteins, such as RBP and MBL. (C) circRNAs act as translation templates. (D) circRNAs regulate transcription. (E) circRNAs regulate protein expression.
At present, most circRNAs studies focus on the mechanism of miRNA molecular sponge; 2) regulate the transcription of parental genes by binding to RNA polymerase II [15]; 3) interact with RNA-binding proteins to play biological roles (Table 1) [12]; 4) translate proteins. For example, Yang et al. found that Circ-FBXW7 encodes a protein that inhibits the occurrence of gliomas [26].
Table 1.
Functions of circRNAs.
| Function | Example | Ref. |
|---|---|---|
| miRNA sponge | circ-HIPK3 circ-PRKCI |
[16,17] |
| Histone methylation | Circ-ANRIL | [18] |
| Protein sponge | circ-Foxo3 | [19] |
| RNA maturation | circ-ANRIL | [20] |
| RNAP II elongation | circ-EIF3J circ-PAIP2 |
[21] |
| Translation regulator | circ-PABPN1 | [22] |
| Alternative splicing | circ-Mbl | [23] |
| Protein translation (including m6A-driven) | circ-ZNF609 | [24,25] |
3. circRNAs and the diagnosis of lung cancer
Early and accurate diagnosis is critical to the treatment of lung cancer. Although a variety of diagnostic methods have been used in clinical practice, the current methods still have room for improvement due to reasons such as cost, accuracy, and patient acceptance. Therefore, it is still necessary to explore the diagnostic markers of lung cancer. CircRNAs have the advantages of conservation, stability, and specificity, so they have the potential to become emerging markers of lung cancer [27]. A meta-analysis of the Chinese lung cancer population pooled 8 studies on the diagnostic efficacy of circRNAs in lung cancer tissue and blood. The area under curve (AUC) of characteristic curve (ROC) was 0.78, suggesting that circRNAs have diagnostic potential in the Chinese lung cancer population [28].
3.1. The diagnostic value of blood circRNAs
Compared with traditional biopsy, liquid biopsy has the advantages of simple operation, less invasiveness, and low cost, so the research prospect is broad. At present, some literatures have preliminarily confirmed that plasma circRNAs have good diagnostic ability, such as circRNA-002178, circMAN1A2 and so on [29,30]. Chen et al. used high-throughput sequencing technology to identify differentially expressed circRNAs in plasma exosomes from lung adenocarcinoma (LUAD) patients [31]. Compared with the control group, the expression of 105 circRNAs was increased, and the expression of 78 circRNAs was decreased. Further research found that the expressions of hsa_circ_0001492 and hsa_circ_0001346 were significantly up-regulated in the early stage of LUAD, but were almost undetectable in the plasma of the control group, suggesting that hsa_circ_0001492 and hsa_circ_0001346 may be candidate markers for early LUAD diagnosis. Liu et al. detected and analyzed the differential expression of hsa_circ_0005962 and hsa_circ_0086414 in the plasma of LUAD patients [32]. The combined diagnosis AUC of the two reached 0.81, suggesting that dual circRNAs may be used as non-invasive biomarkers for the diagnosis of LUAD. In addition, blood circRNA may be related to tumor progression, and the expression of hsa_circ_0005962 in LUAD patients was significantly decreased after surgery compared with preoperative ones. The expression level of hsa_circ_0086414 was correlated with epidermal growth factor receptor (EGFR) mutation. Compared with wild-type patients, hsa_circ_0086414 was highly expressed in EFGR mutant patients. This study demonstrates the multi-faceted application value of blood circRNAs. Of course, in order to realize the clinical translation of blood circRNA lung cancer diagnosis, a larger sample size and more in-depth mechanism exploration are still needed.
3.2. Diagnostic value of circRNAs in lung cancer tissues
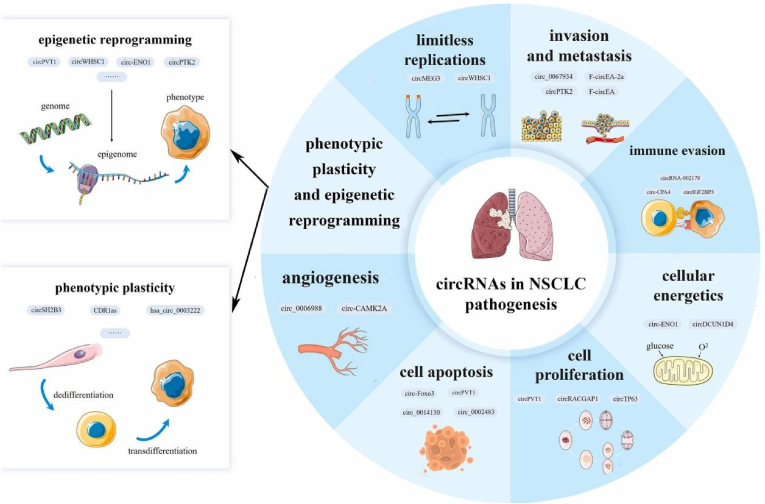
Wang et al. found that in distinguishing non-small cell lung cancer from normal tissues, the AUCs of hsa_circ_0077837 and hsa_circ_0001821 were 0.921 and 0.863, respectively, showing the diagnostic value of these two circRNAs for lung cancer [33]. Liu et al. confirmed that the expression of hsa_circ_11780 was significantly decreased in non-small cell lung cancer tissues and cell lines, and patients with low expression of hsa_circ_11780 had a greater risk of developing larger tumors (>3 cm), distant metastasis and poor survival prognosis [34]. Zhao et al. analyzed 61 pairs of paired lung cancer and paracancerous tissues and found that hsa_circ_0037515 and hsa_circ_0037516 were lowly expressed in non-small cell lung cancer, and their AUCs were 0.81 and 0.82, respectively, which also showed good diagnostic ability (Fig. 2) [35]. The combined AUC of hsa_circ_0037515 and hsa_circ_0037516 increased to 0.90, indicating the importance of circRNA joint diagnosis in lung cancer tissue.
Fig. 2.
The role of circRNAs in the pathogenesis of non-small cell lung cancer.
4. circRNAs and the treatment of lung cancer
Previous studies have found that circRNA can act as a regulatory molecule to promote or inhibit the occurrence and development of lung cancer, so regulating the expression level of circRNA is of great significance to the malignant biological behavior of lung cancer. At present, many studies have explored the mechanism of malignant biological behavior of lung cancer based on the ceRNA mechanism of circRNAs (Table 2).
Table 2.
Summary of circRNA acting on malignant biological behaviors of lung cancer through ceRNA mechanism.
| circRNA | Dysregulation | Cell lines | Function | Sponge target | Ref. |
|---|---|---|---|---|---|
| circ_11780 | Down | A549, H226 | Proliferation (−), migration (−), invasion (−) | miR-544a | [34] |
| circGFRA1 | Up | A549, H838 | Proliferation (+) | miR-188–3p | [36] |
| circ_0012673 | Up | A549, H23 | Proliferation (+), apoptosis (−), migration (+), EMT (+) | miR-320a | [38] |
| circ-0000211 | Up | A549, H1299, H1650 | Migration (+), invasion (+) | miR-622 | [39] |
| circ-ABCB10 | Up | A549, H292 | Proliferation (+), migration (+) | miR-556–3p | [40] |
| circ_0000326 | Up | A549, H1299 | Proliferation (+), apoptosis (−), migration (+) | miR-338–3p | [41] |
| circ_0014130 | Up | PC-9, A549 | Proliferation (+), apoptosis (−), invasion (+) | miR-136–5p | [42] |
| circ-SOX4 | Up | A549, SPC-A1 | Proliferation (+), migration (+), invasion (+) | miR-1270 | [43] |
| circCCDC66 | Up | A549, H1299 | Proliferation (+), apoptosis (−), migration (+), invasion (+) | miR-33a-5p | [44] |
| circCDR1as | Up | A549, Calu-3 | Proliferation (+), apoptosis (−), migration (+), invasion (+) | miR-219a-5p | [45] |
| circ_0058124 | Up | A549, H1975 | Proliferation (+), apoptosis (−), migration (+), invasion (+) | miR-1297 | [46] |
| circ-MTO1 | Down | A549, SPC-A1 | Proliferation (−) | miR-17 | [47] |
| cMras | Down | A549, H1299 | Proliferation (−), migration (−) | miR-567 | [48] |
| circ-IGF1R | Down | PC9, A549 | Migration (−), invasion (−) | miR-1270 | [49] |
| circCRIM1 | Down | A549, H1299, SPC-A1 | Migration (−), invasion (−) | miR-93, miR-182 | [50] |
| circ_0007059 | Down | A549, H1975 | Proliferation (−), EMT (−) | miR-378 | [51] |
| circ_0006427 | Down | SPC-A1, Calu-3 | Proliferation (−), migration (−), invasion (−) | miR-6783–3p | [52] |
| circPTPRA | Down | H23, H1755, H522 | Migration (−), invasion (−), EMT (−) | miR-96–5p | [53] |
| circSMARCA5 | Down | A549 | Proliferation (−), migration (−), invasion (−) | miR-19b-3p | [54] |
| circ_0002483 | Down | A549, H1299 | Proliferation (−), migration (−), invasion (−) | miR-182–5p | [55] |
| circ_0078767 | Down | A549, H23 | Proliferation (−), apoptosis (+), invasion (−) | miR-330–3p | [56] |
For example, Yao et al. found that circGFRA1 was up-regulated in lung cancer cells and promoted the malignant proliferation of lung cancer through the circGFRA1/miR-188–3p/PI3K/AKT pathway [36]. As a serine-threonine protein kinase, LIMK1 participates in epithelial-mesenchymal transition (EMT) by affecting the actin cytoskeleton and regulates tumor progression [37]. Qin et al. found that circ_0012673 was highly expressed in lung cancer tissues and cell lines [38]. The adsorption of miR-320a by circ_0012673 sponge resulted in increased expression of the downstream target protein LIMK1, thereby inhibiting lung cancer cell apoptosis and promoting its proliferation, migration and EMT process.
4.1. circRNAs and lung cancer immunotherapy
Tumor cells are able to express a variety of mechanisms to evade the immune system and create conditions for their own growth. Programmed death protein 1 (PD-1) is a transmembrane protein, which has been found to be expressed on the surface of almost all types of tumor cells, and participates in tumor immune escape by interacting with PD-L1 mechanism [57]. In recent years, immune checkpoint inhibitors (ICIs) targeting PD-1/PD-L1 have provided a powerful weapon for lung cancer treatment Wang et al. found that circRNA-002178 was abnormally highly expressed in lung adenocarcinoma tissues, and promoted the expression of PD-L1 in lung cancer cells by adsorbing miR-34 [29]. At the same time, lung cancer cells can secrete exosomal circRNA-002178 and deliver it to T cells, which promotes the expression of PD-1 in T cells by inhibiting miR-28–5p. Literature confirmed that CXCR4 is involved in the process of cytotoxic T lymphocyte depletion and induction of anti-PD-1 drug resistance [58]. Zhang et al. found that the circFGFR1/miR-381–3p/CXCR4 pathway plays an immunosuppressive effect by promoting the resistance of lung cancer cells to anti-PD-1 drugs [59]. It is suggested that circRNAs can participate in tumor immune escape mechanism, and the combined use of related pathway inhibitors is expected to improve clinical efficacy and provide new ideas for tumor immunotherapy.
4.2. circRNAs and drug resistance in lung cancer
With the continuous advent of anti-tumor drugs, it has brought more hope to lung cancer patients, but the problem of drug resistance is still a major problem that plagues clinical treatment. Therefore, it is urgent to further explore the drug resistance mechanism of lung cancer in order to find efficient biomarkers or therapeutic targets. Studies have found that some circRNAs can participate in the drug resistance process of lung cancer (Table 3).
Table 3.
Summary of the effects of circRNA on tumor drug sensitivity.
| CircRNA | Cell lines | Drugs | Sensitivity | Ref. |
|---|---|---|---|---|
| circAKT3 | A549, H1299 | Cisplatin | Down-regulated | [6] |
| circ-ABCB10 | A549, H292 | Cisplatin | Down-regulated | [40] |
| circ_0002483 | A549, H1299 | Paclitaxel | Up-regulated | [55] |
| circZFR | A549, H522 | Cisplatin | Down-regulated | [62] |
| circ_0076305 | A549, H1650 | Cisplatin | Down-regulated | [63] |
| circ_0004015 | A549, HCC827 | Gefitinib | Down-regulated | [64] |
| circ_0003998 | A549, H1299 | Docetaxel | Down-regulated | [65] |
| circ_0001946 | A549 | Cisplatin | Up-regulated | [66] |
| circESRP1 | H69, H446 | Cisplatin, etoposide | Up-regulated | [67] |
| circ-SMARCA5 | H1299, H1437 | Cisplatin, gemcitabine | Up-regulated | [68] |
Hong et al. found that circCPA4 acts as a molecular sponge of let-7, and its down-regulation can affect programmed death-ligand 1 (PD-L1) to reduce its expression, thereby inhibiting the growth and development of non-small cell lung cancer cells migration and EMT process [60]. In addition, non-small cell lung cancer-derived PD-L1-containing exosomes can promote their stem cell properties and enhance the tolerance of non-small cell lung cancer cells to cisplatin. Li et al. reported that circ_0002483 could reduce the expression level of miR-182–5p, relieve its inhibition of target molecules GRB2, FOXO1, and FOXO3, thereby enhancing the sensitivity of non-small cell lung cancer to paclitaxel [55]. CircRNA_103762 is highly expressed in lung cancer and induces multidrug resistance in lung cancer by inhibiting the target protein CHOP [61].
5. circRNAs and prognosis of lung cancer
Prognostic monitoring of patients with lung cancer is a key link in evaluating the effect of clinical diagnosis and treatment, and is of great significance for adjusting drug regimens and improving patient survival time. Studies have confirmed that a variety of circRNAs can be used as independent prognostic indicators of lung cancer patients and are closely related to the survival of lung cancer patients, such as circSMARCA5, circ_11780, circCRIM1 [30,50,68]. Liu et al. performed RT-qPCR detection on tumor tissues of 93 non-small cell lung cancer patients and found that hsa_circ_11780 was abnormally low expressed, and patients with low expression of hsa_circ_11780 tended to have larger tumors with distant metastasis and more severe tumor according to tumor-lymph node- Metastasis (TNM) staging [34]. Survival analysis by Kaplan-Meier method showed that non-small cell lung cancer patients with low expression of hsa_circ_11780 had shorter overall survival (OS). circHIPK3 is derived from exon 2 of the oncogene HIPK3 in the chromosome 11p13 region. Chen et al. found that knockdown of circHIPK3 could inhibit the proliferation, migration, and invasion of non-small cell lung cancer cell lines A549, H838, and H1299, and induce the occurrence of autophagy, while circHIPK3 and linHIPK3 antagonized the regulation of autophagy [9]. CircHIPK3:linHIPK3 (C:L) ratio can reflect the autophagy level of tumor cells. For patients with advanced non-small cell lung cancer, high C:L ratio (>0.49) is an effective indicator of low survival rate. These results suggest that the autophagy regulator circHIPK3 has potential clinical application value as a prognostic factor. EGFR-tyrosine kinase inhibitors (EGFR-TKIs) are an important treatment option for non-small cell lung cancer patients with sensitive EGFR mutations. Liu et al. detected 1377 differentially expressed circRNAs by sequencing the plasma circRNAs of non-small cell lung cancer patients in the effective and ineffective groups after using EGFR-TKI gefitinib [69]. RT-qPCR detection confirmed that hsa_circ_0109320 and hsa_circ_0134501 were highly expressed in the gefitinib effective group. Further research found that the high expression of hsa_circ_0109320 was associated with better progression-free survival (PFS) in patients, suggesting that hsa_circ_0109320 may be a biomarker reflecting the efficacy of gefitinib. Fu et al. found that the expression of hsa_circRNA_012515 was significantly increased in non-small cell lung cancer tissues and cells, especially in gefitinib-resistant cell lines [70]. In addition, the up-regulation of hsa_circRNA_012515 was closely related to lymph node metastasis, tumor stage and prognosis of patients. Non-small cell lung cancer patients with high expression of hsa_circRNA_012515 had shorter OS and PFS. We also found that hsa_circRNA_012515 was expressed at higher levels in stage III/IV patients compared with stage I/II non-small cell lung cancer patients. Thus, hsa_circRNA_012515 has good clinical correlation and may be a biomarker for predicting poor prognosis of non-small cell lung cancer patients.
6. Conclusions
With the deepening of research, the connection between circRNA and lung cancer is becoming increasingly prominent. On the one hand, circRNAs act as tumor-promoting or tumor-suppressing factors to regulate the biological behaviors of lung cancer, such as proliferation, metastasis, apoptosis, and autophagy, regulate the sensitivity of chemotherapy or targeted drugs and the efficacy of immunotherapy, and provide a preliminary theoretical basis for adjuvant clinical treatment. On the other hand, the differential expression of circRNAs in tissue or blood shows a certain correlation in the early diagnosis and prognosis evaluation of lung cancer, and is expected to become a potential biomarker of lung cancer. However, the current circRNA research is still in the early stage, most researchers focus on the exploration of the adsorption function of miRNA sponges, and many mechanisms have not yet been elucidated [[71], [72], [73], [74], [75]]. Its clinical relevance research is also limited to a small number of samples, and its translational value remains to be questioned. It is believed that there will be more breakthroughs in the field of circRNA in the future, providing more ideas for the diagnosis and treatment of lung cancer.
Funding
This work was supported by the Bashkir State Medical University Strategic Academic Leadership Program (PRIORITY-2030).
Author contributions
Albert Sufianov: Conceptualization, Writing – original draft, Project administration. Sema Begliarzade: Writing – review & editing, Investigation, Project administration. Aferin Beilerli: Formal analysis, Methodology, and original draft. Yanchao Liang: Resources, Data curation. Tatiana Ilyasova: Validation, Visualization. Ozal Beylerli: Supervision, Funding acquisition. All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Contributor Information
Albert Sufianov, Email: sufianov@gmail.com.
Sema Begliarzade, Email: semanagiyeva@yandex.ru.
Aferin Beilerli, Email: abeilerli@mail.ru.
Yanchao Liang, Email: liangyanchao@hrbmu.edu.cn.
Tatiana Ilyasova, Email: Iltanya67@yandex.ru.
Ozal Beylerli, Email: obeylerli@mail.ru.
References
- 1.Bray F., Ferlay J., Soerjomataram I., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Molina J.R., Yang P., Cassivi S.D., et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008;83(5):584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanger H.L., Klotz G., Riesner D., et al. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U. S. A. 1976;73(11):3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sufianov A., Begliarzade S., Ilyasova T., Liang Y., Beylerli O. MicroRNAs as prognostic markers and therapeutic targets in gliomas. Noncoding RNA Res. 2022 Jul 6;7(3):171–177. doi: 10.1016/j.ncrna.2022.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocquerelle C., Mascrez B., Hetuin D., et al. Mis-splicing yields circular RNA molecules. Faseb. J. 1993;7(1):155–160. doi: 10.1096/fasebj.7.1.7678559. [DOI] [PubMed] [Google Scholar]
- 6.Xu Y., Jiang T., Wu C., et al. CircAKT3 inhibits glycolysis balance in lung cancer cells by regulating miR-516b-5p/STAT3 to inhibit cisplatin sensitivity. Biotechnol. Lett. 2020;42(7):1123–1135. doi: 10.1007/s10529-020-02846-9. [DOI] [PubMed] [Google Scholar]
- 7.Beylerli O., Khasanov D., Gareev I., Valitov E., Sokhatskii A., Wang C., Pavlov V., Khasanova G., Ahmad A. Differential non-coding RNAs expression profiles of invasive and non-invasive pituitary adenomas. Noncoding RNA Res. 2021 Jun 30;6(3):115–122. doi: 10.1016/j.ncrna.2021.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beilerli A., Begliarzade S., Sufianov A., Ilyasova T., Liang Y., Beylerli O. Circulating ciRS-7 as a potential non-invasive biomarker for epithelial ovarian cancer: an investigative study. Noncoding RNA Res. 2022 Jul 31;7(3):197–204. doi: 10.1016/j.ncrna.2022.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X., Mao R., Su W., et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha signaling in STK11 mutant lung cancer. Autophagy. 2020;16(4):659–671. doi: 10.1080/15548627.2019.1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang C., Tan S., Li J., Liu W.R., Peng Y., Li W. CircRNAs in lung cancer - biogenesis, function and clinical implication. Cancer Lett. 2020 Nov 1;492:106–115. doi: 10.1016/j.canlet.2020.08.013. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki H., Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int. J. Mol. Sci. 2014;15(6):9331–9342. doi: 10.3390/ijms15069331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Memczak S., Jens M., Elefsinioti A., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 13.Xia S., Feng J., Lei L., et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Briefings Bioinf. 2017;18(6):984–992. doi: 10.1093/bib/bbw081. [DOI] [PubMed] [Google Scholar]
- 14.Salmena L., Poliseno L., Tay Y., et al. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z., Huang C., Bao C., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 16.Tian F., Wang Y., Xiao Z., Zhu X. Circular RNA CircHIPK3 promotes NCIH1299 and NCI-H2170 cell proliferation through miR-379 and its target IGF1. Zhongguo Fei Ai Za Zhi. 2017;20(7):459–467. doi: 10.3779/j.issn.1009-3419.2017.07.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu M., Xia W., Chen R., Wang S., Xu Y., Ma Z., Xu W., Zhang E., Wang J., Fang T., et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78(11):2839–2851. doi: 10.1158/0008-5472.CAN-17-2808. [DOI] [PubMed] [Google Scholar]
- 18.Burd C.E., Jeck W.R., Liu Y., Sanof H.K., Wang Z., Sharpless N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001210–e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du W.W., Yang W., Chen Y., Wu Z., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2016;38(18):1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 20.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northof B.H., Nicolaou A., et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7(17429):10–1038. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22(3):256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 22.Abdelmohsen K., Panda A.C., Munk R., Grammatikakis I., Dudekula D.B., De S., Kim J., Noh J.H., Kim K.M., Martindale J.L., et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation byCircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 24.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y., et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y., Gao X., Zhang M., et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110(3):304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han B., Chao J., Yao H. Circular RNA and its mechanisms in disease: from the bench to the clinic. Pharmacol. Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Xiao Z., Chen X., Lu X., et al. Accuracy evaluation of circular RNA in diagnosing lung cancer in a Chinese population. Dis. Markers. 2019;2019 doi: 10.1155/2019/7485389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J., Zhao X., Wang Y., et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11(1):32. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan C.M., Wang J.P., Tang Y.Y., et al. circMAN1A2 could serve as a novel serum biomarker for malignant tumors. Cancer Sci. 2019;110(7):2180–2188. doi: 10.1111/cas.14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen F., Huang C., Wu Q., et al. Circular RNAs expression profiles in plasma exosomes from early-stage lung adenocarcinoma and the potential biomarkers. J. Cell. Biochem. 2020;121(3):2525–2533. doi: 10.1002/jcb.29475. [DOI] [PubMed] [Google Scholar]
- 32.Liu X.X., Yang Y.E., Liu X., et al. A two-circular RNA signature as a noninvasive diagnostic biomarker for lung adenocarcinoma. J. Transl. Med. 2019;17(1):50. doi: 10.1186/s12967-019-1800-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C., Tan S., Liu W.R., et al. RNA-Seq profiling of circular RNA in human lung adenocarcinoma and squamous cell carcinoma. Mol. Cancer. 2019;18(1):134. doi: 10.1186/s12943-019-1061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y., Yang C., Cao C., et al. Hsa_circ_RNA_0011780 represses the proliferation and metastasis of non-small cell lung cancer by decreasing FBXW7 via targeting miR-544a. OncoTargets Ther. 2020;13:745–755. doi: 10.2147/OTT.S236162. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Zhao D., Liu H., Liu H., et al. Downregulated expression of hsa_circ_0037515 and hsa_circ_0037516 as novel biomarkers for non-small cell lung cancer. Am J Transl Res. 2020;12(1):162–170. [PMC free article] [PubMed] [Google Scholar]
- 36.Yao J., Xu G., Zhu L., et al. circGFRA1 enhances NSCLC progression by sponging miR-188-3p. OncoTargets Ther. 2020;13:549–558. doi: 10.2147/ott.S230795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao Q., Li R., Zhou R., et al. LIM kinase 1 interacts with myosin-9 and alpha-actinin-4 and promotes colorectal cancer progression. Br. J. Cancer. 2017;117(4):563–571. doi: 10.1038/bjc.2017.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin H., Liu J., Du Z.H., et al. Circular RNA hsa_circ_0012673 facilitates lung cancer cell proliferation and invasion via miR-320a/LIMK18521 axis. Eur. Rev. Med. Pharmacol. Sci. 2020;24(4):1841–1852. doi: 10.26355/eurrev_202002_20362. [DOI] [PubMed] [Google Scholar]
- 39.Feng D., Xu Y., Hu J., et al. A novel circular RNA, hsa-circ-0000211, promotes lung adenocarcinoma migration and invasion through sponging of hsa-miR-622 and modulating HIF1-alpha expression. Biochem. Biophys. Res. Commun. 2020;521(2):395–401. doi: 10.1016/j.bbrc.2019.10.134. [DOI] [PubMed] [Google Scholar]
- 40.Wu Z., Gong Q., Yu Y., et al. Knockdown of circ-ABCB10 promotes sensitivity of lung cancer cells to cisplatin via miR-556-3p/AK4 axis. BMC Pulm. Med. 2020;20(1):10. doi: 10.1186/s12890-019-1035-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu Y., Yu J., Huang Z., et al. Circular RNA hsa_circ_0000326 acts as a miR-338-3p sponge to facilitate lung adenocarcinoma progression. J. Exp. Clin. Cancer Res. 2020;39(1):57. doi: 10.1186/s13046-020-01556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geng Y., Bao Y., Zhang W., et al. Circular RNA hsa_circ_0014130 inhibits apoptosis in non-small cell lung cancer by sponging miR-136-5p and upregulating BCL2. Mol. Cancer Res. 2020;18(5):748–756. doi: 10.1158/1541-7786.MCR-19-0998. [DOI] [PubMed] [Google Scholar]
- 43.Gao N., Ye B. Circ-SOX4 drives the tumorigenesis and development of lung adenocarcinoma via sponging miR-1270 and modulating PLAGL2 to activate WNT signaling pathway. Cancer Cell Int. 2020;20:2. doi: 10.1186/s12935-019-1065-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y., Zhao W., Zhang S. STAT3-induced upregulation of circCCDC66 facilitates the progression of non-small cell lung cancer by targeting miR-33a-5p/KPNA4 axis. Biomed. Pharmacother. 2020;126 doi: 10.1016/j.biopha.2020.110019. [DOI] [PubMed] [Google Scholar]
- 45.Li Y., Zhang J., Pan S., et al. CircRNA CDR1as knockdown inhibits progression of non-small-cell lung cancer by regulating miR-219a-5p/SOX5 axis. Thorac Cancer. 2020;11(3):537–548. doi: 10.1111/1759-7714.13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X., Zhang Q., Yang Z. Knockdown of hsa_circ_0058124 inhibits the proliferation of human lung cancer cells by up-regulation of miR-1297. Artif. Cell Nanomed. Biotechnol. 2020;48(1):584–593. doi: 10.1080/21691401.2020.1725537. [DOI] [PubMed] [Google Scholar]
- 47.Zhang B., Chen M., Jiang N., et al. A regulatory circuit of circ-MTO1/miR-17/QKI-5 inhibits the proliferation of lung adenocarcinoma. Cancer Biol. Ther. 2019;20(8):1127–1135. doi: 10.1080/15384047.2019.1598762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Yu C., Tian F., Liu J., et al. Circular RNA cMras inhibits lung adenocarcinoma progression via modulating miR-567/PTPRG regulatory pathway. Cell Prolif. 2019;52(3) doi: 10.1111/cpr.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Z., Xiang W., Chen W., et al. Circ-IGF1R inhibits cell invasion and migration in non-small cell lung cancer. Thorac Cancer. 2020;11(4):875–887. doi: 10.1111/1759-7714.13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L., Liang Y., Mao Q., et al. Circular RNA circCRIM1 inhibits invasion and metastasis in lung adenocarcinoma through the microRNA (miR)-182/miR-93-leukemia inhibitory factor receptor pathway. Cancer Sci. 2019;110(9):2960–2972. doi: 10.1111/cas.14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao S., Yu Y., Liu L., et al. Circular RNA hsa_circ_0007059 restrains proliferation and epithelial-mesenchymal transition in lung cancer cells via inhibiting microRNA-378. Life Sci. 2019;233 doi: 10.1016/j.lfs.2019.116692. [DOI] [PubMed] [Google Scholar]
- 52.Yao Y., Hua Q., Zhou Y. CircRNA has_circ_0006427 suppresses the progression of lung adenocarcinoma by regulating miR-6783-3p/DKK1 axis and inactivating Wnt/beta-catenin signaling pathway. Biochem. Biophys. Res. Commun. 2019;508(1):37–45. doi: 10.1016/j.bbrc.2018.11.079. [DOI] [PubMed] [Google Scholar]
- 53.Wei S., Zheng Y., Jiang Y., et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193. doi: 10.1016/j.ebiom.2019.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beilerli A., Gareev I., Beylerli O., Yang G., Pavlov V., Aliev G., Ahmad A. Circular RNAs as biomarkers and therapeutic targets in cancer. Semin. Cancer Biol. 2022 Aug;83:242–252. doi: 10.1016/j.semcancer.2020.12.026. [DOI] [PubMed] [Google Scholar]
- 55.Li X., Yang B., Ren H., et al. Hsa_circ_0002483 inhibited the progression and enhanced the Taxol sensitivity of non-small cell lung cancer by targeting miR-182-5p. Cell Death Dis. 2019;10(12):953. doi: 10.1038/s41419-019-2180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen T., Yang Z., Liu C., et al. Circ_0078767 suppresses non-small-cell lung cancer by protecting RASSF1A expression via sponging miR-330-3p. Cell Prolif. 2019;52(2) doi: 10.1111/cpr.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang X., Wang J., Deng X., et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol. Cancer. 2019;18(1):10. doi: 10.1186/s12943-018-0928-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seo Y.D., Jiang X., Sullivan K.M., et al. Mobilization of CD8(+) T cells via CXCR4 blockade facilitates PD-1 checkpoint therapy in human pancreatic cancer. Clin. Cancer Res. 2019;25(13):3934–3945. doi: 10.1158/1078-0432.Ccr-19-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang P.F., Pei X., Li K.S., et al. Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol. Cancer. 2019;18(1):179. doi: 10.1186/s12943-019-1111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hong W., Xue M., Jiang J., et al. Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC) J. Exp. Clin. Cancer Res. 2020;3(1):149. doi: 10.1186/s13046-020-01648-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Xiao G., Huang W., Zhan Y., et al. CircRNA_103762 promotes multidrug resistance in NSCLC by targeting DNA damage inducible transcript 3 (CHOP) J. Clin. Lab. Anal. 2020;34(6) doi: 10.1002/jcla.23252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H., Liu F., Qin W. Circ_0072083 interference enhances growth-inhibiting effects of cisplatin in non-small-cell lung cancer cells via miR-545-3p/CBLL1 axis. Cancer Cell Int. 2020;20:78. doi: 10.1186/s12935-020-1162-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dong Y., Xu T., Zhong S., et al. Circ_0076305 regulates cisplatin resistance of non-small cell lung cancer via positively modulating STAT3 by sponging miR-296-5p. Life Sci. 2019;239 doi: 10.1016/j.lfs.2019.116984. [DOI] [PubMed] [Google Scholar]
- 64.Zhou Y., Zheng X., Xu B., et al. Circular RNA hsa_circ_0004015 regulates the proliferation, invasion, and TKI drug resistance of non-small cell lung cancer by miR-1183/PDPK1 signaling pathway. Biochem. Biophys. Res. Commun. 2019;508(2):527–535. doi: 10.1016/j.bbrc.2018.11.157. [DOI] [PubMed] [Google Scholar]
- 65.Yu W., Peng W., Sha H., et al. Hsa_circ_0003998 promotes chemoresistance via modulation of miR-326 in lung adenocarcinoma cells. Oncol. Res. 2019;27(5):623–628. doi: 10.3727/096504018x15420734828058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang M.S., Liu J.Y., Xia X.B., et al. Hsa_circ_0001946 inhibits lung cancer progression and mediates cisplatin sensitivity in non-small cell lung cancer via the nucleotide excision repair signaling pathway. Front. Oncol. 2019;9:508. doi: 10.3389/fonc.2019.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang W., Yang Y., Wu J., et al. Circular RNA cESRP1 sensitises small cell lung cancer cells to chemotherapy by sponging miR-93-5p to inhibit TGF-beta signalling. Cell Death Differ. 2019;27(5):1709–1727. doi: 10.1038/s41418-019-0455-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tong S. Circular RNA SMARCA5 may serve as a tumor suppressor in non-small cell lung cancer. J. Clin. Lab. Anal. 2020;34(5) doi: 10.1002/jcla.23195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Y.T., Han X.H., Xing P.Y., et al. Circular RNA profiling identified as a biomarker for predicting the efficacy of Gefitinib therapy for non-small cell lung cancer. J. Thorac. Dis. 2019;11(5):1779–1787. doi: 10.21037/jtd.2019.05.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu Y., Huang L., Tang H., et al. hsa_circRNA_012515 is highly expressed in NSCLC patients and affects its prognosis. Cancer Manag. Res. 2020;12:1877–1886. doi: 10.2147/cmar.S245525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gareev I., Beylerli O., Liang Y., Xiang H., Liu C., Xu X., Yuan C., Ahmad A., Yang G. The role of MicroRNAs in therapeutic resistance of malignant primary brain tumors. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.740303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun J., Sun Z., Gareev I., Yan T., Chen X., Ahmad A., Zhang D., Zhao B., Beylerli O., Yang G., Zhao S. Exosomal miR-2276-5p in plasma is a potential diagnostic and prognostic biomarker in glioma. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.671202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beylerli O., Gareev I., Sufianov A., Ilyasova T., Zhang F. The role of microRNA in the pathogenesis of glial brain tumors. Noncoding RNA Res. 2022;7(2):71–76. doi: 10.1016/j.ncrna.2022.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gareev I., Gileva Y., Dzidzaria A., Beylerli O., Pavlov V., Agaverdiev M., Mazorov B., Biganyakov I., Vardikyan A., Jin M., Ahmad A. Long non-coding RNAs in oncourology. Noncoding RNA Res. 2021 Aug 26;6(3):139–145. doi: 10.1016/j.ncrna.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beylerli O., Gareev I., Sufianov A., Ilyasova T., Guang Y. Long noncoding RNAs as promising biomarkers in cancer. Noncoding RNA Res. 2022 Feb 25;7(2):66–70. doi: 10.1016/j.ncrna.2022.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]