Abstract
Background
Diagnosis and management of latent tuberculosis (TB) infections are one of the challenges of eradicating pulmonary TB. A critical aspect of controlling pulmonary TB spread is early diagnosis. One TB biological marker type under evaluation is microRNAs (miRNAs). Mycobacterium tuberculosis infection causes epigenetic changes. The upregulation of miRNA-29a-3p suppresses the immune response by post-transcriptionally inhibiting interferon (INF)-γ expression in T cells, increasing susceptibility to pulmonary TB. This study aimed to assess miRNA-29a-3p expression as a biomarker of active and latent pulmonary TB infection.
Methods
This case-control study included 50 individuals with active TB, 33 household contacts with a positive IFN-γ release assay (IGRA), and 30 healthy controls. An enzyme-linked immunosorbent assay-based IGRA was used to determine latent pulmonary TB infection in household contacts. Quantitative real-time PCR was used to quantify miRNA-29a-3p expression. Data analysis used analyses of variance and receiver operating characteristic (ROC) curves.
Results
miRNA-29a-3p expression differed significantly between active TB, latent TB, and healthy participants (controls; p = <0.001. ROC curve analysis showed that miRNA-29a-3p expression had 86% sensitivity and 73% specificity with an area under the ROC curve (AUC) of 0.763 (95% confidence interval [CI]: 0.668–0.858). The miRNA-29a-3p ROC curve had 84.8% sensitivity and 70% specificity with an AUC of 0.808 (95% CI: 0.698–0.919) for latent TB. Additionally, miRNA-29a-3p expression was significantly correlated with active (p < 0.0001) and latent (p < 0.0001) pulmonary TB. However, miRNA-29a-3p expression was not significantly correlated with INF-γ levels in patients with active (R = 0.005; p = 0.62) and latent (R = 0.010; p = 0.38) pulmonary TB or healthy controls (R = 0.060; p = 0.19).
Conclusion
miRNA-29a-3p expression was increased in patients with active and latent pulmonary TB. Therefore, miRNA-29a-3p represents a potential biomarker for latent and active pulmonary TB. However, IFN-γ levels were not correlated with miR-29a-3p expression.
Keywords: miRNA-29a-3p, IFN-γ, Active pulmonary tuberculosis, Latent pulmonary tuberculosis
Highlights
-
•
One biological marker being evaluated for tuberculosis is microRNA (miRNA).
-
•
Mycobacterium tuberculosis infection causes epigenetic changes.
-
•
miRNA-29a-3p upregulation suppresses the immune response by inhibiting INF-γ expression in T cells.
-
•
miRNA-29a-3p expression was elevated in active and latent pulmonary tuberculosis.
-
•
miRNA-29a-3p is a potential biomarker for latent and active pulmonary tuberculosis.
1. Introduction
Tuberculosis (TB) diagnosis remains a global problem, with misdiagnoses increasing morbidity and mortality. Therefore, TB infection biomarkers are needed to diagnose active and latent pulmonary TB. One TB biological marker type being evaluated is microRNA (miRNA). In several circumstances, including infectious diseases, diabetes, heart disease, cancer, psoriasis, and pregnancy, miRNA has been proposed as a new bio-diagnostic. Mycobacterium tuberculosis is one of many microbes whose infection causes alterations in host epigenetic patterns. Epigenetic processes change gene expression without altering its DNA sequence. They include DNA methylation, histone modification, and miRNAs [1,2]. As a biomarker of disease diagnosis, treatment outcome, and prognosis, miRNAs afford the opportunity as molecular diagnostic markers can apply to various infectious diseases.
Additionally, miRNAs have been associated with various illnesses, such as immunological, cardiac, infectious diseases, and cancer [3]. When the disease develops, the affected organ can release specific miRNAs into the blood. Blood miRNA levels differ appreciably in individuals with several disorders compared to healthy individuals. To date, miRNAs have been investigated and used as molecular diagnostic markers for cancer, diabetes, psychiatric disorders, heart disease, and infectious diseases [4,5].
Several previous studies have used microarrays to identify miRNAs in macrophages and peripheral blood mononuclear cells in the serum, blood, and sputum of TB patients as candidate TB biomarkers [[2], [3], [4], [5], [6], [7], [8], [9]]. A previous study on circulating miRNA in patients with active pulmonary TB identified 59 upregulated and 33 downregulated miRNAs compared to healthy controls [10]. One upregulated miRNA was miRNA-29a-3p, which suppresses the immune response by post-transcriptionally inhibiting interferon (INF)-γ expression in T cells, increasing TB susceptibility [[11], [12], [13], [14]].
In this study, we assessed miRNA-29a-3p as a potential biomarker for early diagnosis of both active and latent TB, which could help prevent the spread of pulmonary TB [4].
2. Materials and methods
2.1. Study design and subjects
This observational case-control study is reported in accordance with the “Strengthening the Reporting of Cohort Studies in Surgery” (STROCSS) guidelines [15] and has been registered with the research registry (no. 8089). Its study population was all patients diagnosed with pulmonary TB who were examined at the Makassar Community Lung Health Center, a referral center for TB cases from primary health services in Makassar, Indonesia, and those with household contact with them.
2.2. Sample collection
Research samples were obtained from pulmonary TB patients who were examined at our center and their household contacts who met the selection criteria and were included in the study. The samples required were sputum and blood for pulmonary TB patients and blood for household contacts.
This study was performed at the Hasanuddin University Medical Research Center Laboratory, Hasanuddin University Hospital (Makassar, Indonesia). The Center for Community Lung Health sampled patients with active and latent pulmonary TB. The Hasanuddin University's Health Research Ethics Committee (Makassar, Indonesia) approved this study (243/UN4.6.4.5.31/PP36/2021). This study included 113 samples consisting of 50 samples of active TB patients, 33 samples of latent TB patients and 30 samples of healthy controls.
The active pulmonary TB group comprised 50 samples from patients with positive acid-fast Bacillus smear results and a positive Mycobacteria growth indicator tube (MGIT) confirmation test. The latent pulmonary TB group comprised 63 samples from 33 household contacts with a positive enzyme-linked immunosorbent assay (ELISA)-based IFN-γ release assay (IGRA) result. The healthy control group comprised 30 individuals with negative IGRA results.
2.3. Sample processing and assessments
2.3.1. Sputum collection
Research subjects were interviewed using standardized questionnaires. The study participants’ sputum was collected for Ziehl–Nielsen staining and confirmed using liquid medium culture and an MGIT (BACTEC MGIT 960; BD, New Jersey, USA).
2.3.2. Blood collection
Briefly, 5–10 mL venous blood samples were obtained from patients with active pulmonary TB in a blood tube and household contacts in a QuantiFERON-TB Gold plus ELISA (QFT-Plus) tube (Qiagen GmbH; Qiagen, Hilden, Germany). Blood samples were centrifuged at 3000 rpm for total RNA extraction.
2.3.3. Total RNA extraction
Total RNA was extracted and purified from blood samples using the QIAamp RNA Blood Mini Kit (Qiagen; Massachusetts, USA), according to the manufacturer's instructions.
2.3.4. Complementary DNA (cDNA) synthesis
The synthesis of cDNA from the extracted RNA was performed by the reverse transcription polymerase chain reaction (PCR) method. This process was performed using the iScript cDNA Synthesis Kit (Bio-Rad; Hercules, CA, USA), according to the manufacturer's instructions. The cDNA was quantified using real-time PCR (qPCR).
2.3.5. Quantification of miRNA-29a-3p with real-time quantitative PCR (qPCR)
We quantified miRNA-29a-3p expression using qPCR with specific primers. The basic principle of this method is that only the specific miRNA-29a-3p sequence in the cDNA will be amplified by the specific primer pair. Amplification was performed with a commercial kit (miRCURY LNA miRNA PCR assay; GeneGlobe ID: YP00204698), according to the manufacturer's instructions. The specific primer sequence used for miRNA-29a was 5′-UAGCACCAUCUGAAAUCGGUUA-3’. The amplification results were detected in real-time and are reported as the cycle threshold, representing the number of amplification cycles before the amplicon surpassed the detection threshold.
2.4. Statistical analysis
Data were analyzed using SPSS Statistics v.23.0 (IBM Co.; Armonk, NY, USA). miRNA-29a-3p expression was compared between individuals in the active pulmonary TB, latent pulmonary TB, and healthy control groups using an analysis of variance (ANOVA). The ROC curve analysis was also used in this study to describe diagnostic accuracy. All results with p < 0.05 were considered statistically significant [9].
3. Results
3.1. Patient characteristics
Patient characteristics included age, sex, relationship to active TB patients, and smoking history. As shown in Table 1, of the 50 active pulmonary TB patients, 34 were male and 16 were female. Among household contacts with positive IGRA results in the latent pulmonary TB group, nine were male and 24 were female. Among household contacts with negative IGRA results or healthy participants in the healthy control group, seven were male and 23 were female. The IGRA results indicated a strong association between sex and the incidence of active TB, latent TB, and healthy controls (p ≤ 0.001). Samples were homogenous with respect to age (p = 0.562).
Table 1.
Study participant characteristics in the active TB, latent TB, and healthy control groups.
| Characteristic | Groups |
p | ||
|---|---|---|---|---|
| Active TB n = 50 | Latent TB n = 33 | Healthy controls n = 30 | ||
| Sex | ||||
|
34 (30.1%) | 9 (8.0%) | 7 (6.2%) | |
|
16 (14.2%) | 24 (21.2%) | 23 (20.4%) | <0.001 |
| Age | 41.04 ± 12.32 | 39.03 ± 12.66 | 37.83 ± 15.74 | 0.562 |
| Relationship | ||||
|
6 (9.5%) | 3 (4.8%) | ||
|
7 (11.1%) | 7 (11.1%) | ||
|
16 (25.4%) | 12 (19.0%) | 0.266 | |
|
3 (4.8%) | 2 (3.2%) | ||
|
1 (1.6%) | 6 (9.5%) | ||
| Smoking history | ||||
|
33 (29.2%) | 9 (8.0%) | 6 (5.3%) | <0.001 |
|
17 (15.0%) | 24 (21.2%) | 24 (21.2%) | |
Considering the relationship of household contacts to active TB patients, there were no significantly more household contacts who were partners (husband or wife; 25.4%; p = 0.266) than children, parents, and other family relationships (e.g., relatives, cousins, or in-laws) or nonfamilial relationships (e.g., friends/friends or neighbors). Samples were homogeneous for smoking history (p < 0.001).
3.2. miRNA-29a-3p Expression in active TB, latent TB, and healthy participants
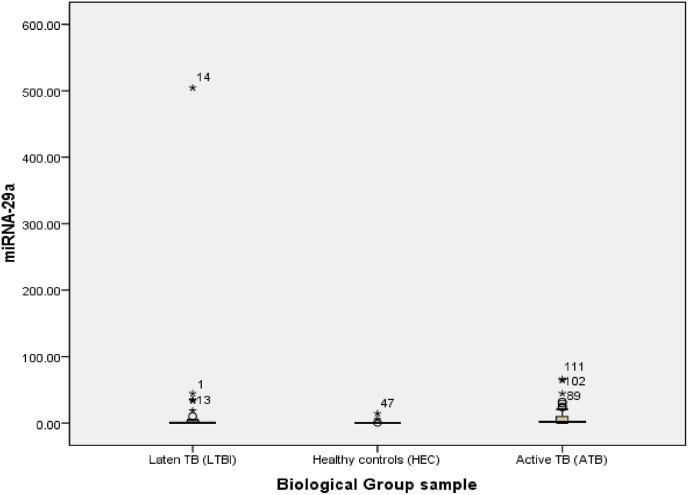
The average qPCR-based miRNA-29a-3p expression was 21.32 in the latent TB group and 8.69 in the active TB group, and 0.83 in the healthy controls (Fig. 1). A statistical analysis using the ANOVA test showed that miRNA-29a-3p expression was significantly higher in the latent TB group than in the active TB and healthy control groups (p < 0.001; Table 2). The difference in miRNA-29a-3p expression was only significant between the active TB group and the healthy control (p-value = 0.002). There was no difference in expression between the active TB group and latent TB (p = 0.803) as well as latent TB with healthy controls (p = 0.466) (p-value >0.05).
Fig. 1.
miRNA-29a-3p Expression in the Active TB, Latent TB, and Healthy Control Groups. Differences in miRNA-29a-3p expression between groups were significant based on an ANOVA test (p < 0.001).
Table 2.
The miRNA-29a-3p Expression Levels in Active TB, Latent TB, and Healthy Control Groups.
| Group | N | Mean | SD | SE | 95% CI | p | |
|---|---|---|---|---|---|---|---|
| Active TB | 50 | 8.69 | 15.18 | 2.14 | 4.37 | 13.00 | <0.001 |
| Latent TB | 33 | 21.32 | 87.62 | 15.25 | −9.74 | 52.39 | |
| Healthy control | 30 | 0.83 | 2.73 | 0.46 | −0.18 | 1.85 | |
Key: SD, standard deviation; SE, standard error of the mean.
3.3. Correlation of miRNA-29a-3p Expression with IFN-γ protein levels in active and latent pulmonary TB patients
The ELISA method was used to quantify IFN-γ levels in T cells from patients with active and latent pulmonary TB and healthy controls. IFN-γ protein levels varied significantly among the three groups based on an ANOVA test (p < 0.001; Table 3). Average IFN-γ levels were 73.92 in the active pulmonary TB group, 33.75 in the latent pulmonary TB group, and 0.42 in the healthy control group. Therefore, IFN-γ levels are more significant in active pulmonary TB patients than in latent pulmonary TB patients and healthy controls. There was a significant difference in IFN-γ between the three groups (p-value <0.001). When compared between two groups, active TB and latent TB (p-value <0.001), active and healthy TB (p-value <0.001), and latent and healthy TB (p-value <0.001).
Table 3.
IFN-γ protein levels in the active TB, latent TB, and healthy control groups.
| Group | N | Mean | SD | SE | 95% CI | p | |
|---|---|---|---|---|---|---|---|
| Active TB | 50 | 73.92 | 48.77 | 6.89 | 60.05 | 87.78 | <0.001 |
| Latent TB | 33 | 33.75 | 39.16 | 6.18 | 19.86 | 47.64 | |
| Healthy control | 30 | 0.42 | 0.21 | 0.03 | 0.34 | 0.50 | |
Key: SD, standard deviation; SE, standard error of the mean.
3.4. The receiver operating characteristic (ROC) curve analysis
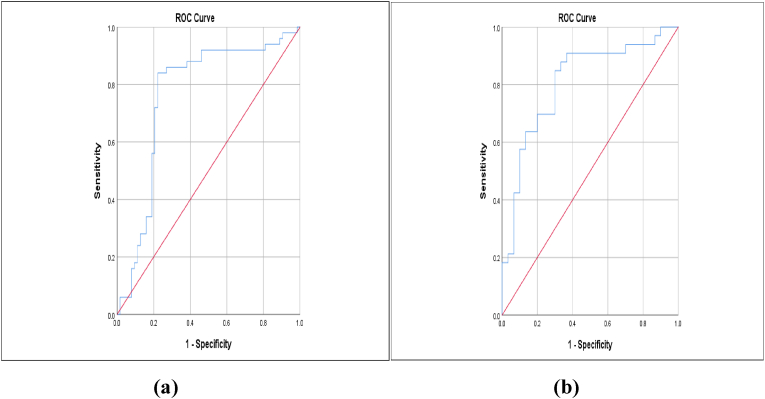
A ROC curve (Fig. 2) using a cut-off value of 0.01 demonstrates the potential of miRNA-29a-3p as a sensitive biomarker for active pulmonary TB infection, with 86.0% sensitivity, 73.0% specificity, and an area under the ROC curve (AUC) of 0.76 (95% confidence interval [CI]: 0.668–0.858). Moreover, miRNA-29a-3p expression was significantly correlated with active pulmonary TB (p < 0.001; Table 4).
Fig. 2.
miRNA-29a-3p Expression as a Potential Biomarker of active and latent TB.
a) Potential of miRNA-29a-3p expression as a biomarker of active TB infection (p ≤ 0.0001);
b) Potential of miRNA-29a-3p expression as a biomarker of latent TB infection (p ≤ 0.0001).
Table 4.
Potential of miRNA-29a-3p Expression as a Biomarker of Active and Latent TB.
| Target | Group | P | Sensitivity | Specificity |
|---|---|---|---|---|
| miRNA-29a-3p | Active TB | <0.001 | 86.0% | 73.0% |
| Latent TB | <0.001 | 84.8% | 70.0% |
Furthermore, the potential of miRNA-29a-3p expression as a sensitive biomarker of latent pulmonary TB infection is shown by a ROC curve (Fig. 2) using a cut-off of 0.01, with 84.8% sensitivity, 70.0% specificity, and an AUC of 0.808 (95% CI: 0.698–0.919). Moreover, miRNA-29a-3p expression was significantly correlated with latent pulmonary TB (p < 0.001; Table 4).
4. Discussion
Misdiagnoses that might raise morbidity and mortality rates are a persistent concern in diagnosing TB. In countries where TB is endemic, national TB programs still rely on direct smear microscopy, solid cultures, radiography, skin testing, and the tuberculin skin test (TST) [9]. Sputum production for a TB confirmatory test is not permitted in children aged under six. TST is not specific for the identification of M. tuberculosis. The gold standard for diagnosing TB, M. tuberculosis culture, is time-consuming and unsuitable for extrapulmonary TB. Due to the difficulty of sputum sampling, identifying smear-negative and extrapulmonary TB continues to pose a significant clinical challenge in pediatric TB. The newly developed FERON-TB test can aid in TB cases, but it cannot distinguish between latent and active TB or be used to manage TB patients taking anti-TB medications. Therefore, a more effective diagnostic technique is required based on alternative specimens, including blood, feces, and urine, that can be obtained from individuals of all ages. MiRNAs are one biological indicator being examined for TB. A novel bio-diagnostic called miRNA has been developed and is widely associated with conditions such as diabetes, heart disease, cancer, psoriasis, pregnancy, and infectious disorders. M. tuberculosis is one of many microbes whose infection can alter host epigenetic patterns. The epigenetic processes change gene expression without changing its DNA sequence. These include DNA methylation, histone modification, and miRNA [1,16].
The function of miRNA, a type of single-stranded RNA molecule 18–25 nucleotides long, is to limit the function of its target genes at the post-transcriptional stage of gene expression. miRNA is an RNA that does not code for a protein. Instead, its final transcript interacts with the target gene's messenger RNA (mRNA). The miRNA works in conjunction with other regulatory components, including transcription factors, to regulate mRNA translation. Most miRNAs are located in regions of the genome that were believed to be non-coding. Approximately 2%–5% of human genes throughout the genome are thought to encode miRNAs. In many cases, polycistronic transcripts encode miRNAs. It is estimated that miRNAs regulate more than one-third of human genes because one miRNA might have several target mRNAs [17,18].
This study examined 33 household contacts with positive ELISA-based IGRA results in the latent TB group and 30 individuals with negative IGRA results in the healthy control group. According to the patient characteristics, there were three patient groups with active pulmonary TB: those with positive IGRA household contacts, those with negative IGRA household contacts, and healthy controls (p < 0.001). This result is similar to World Health Organization data showing that more males than females are diagnosed with smear-positive TB [19,20].
Several studies have shown that active TB patients and latent TB and healthy control participants have different gene expression profiles in macrophages and natural killer (NK) cells [10,21]. Based on research by Fu et al. [10] and Sharbati et al. [21] that miR-29, which was originally found to be an mRNA repressor in viral infection specifically targeting the HIV-1 3′UTR region, has been reported to be downregulated in NK cells during systemic infection due to Listeria monocytogenes or Mycobacterium bovis infection, leading to depression of IFN-y as the target. The miR-29a mediated pathway increases host resistance to Listeria infection. In contrast, miR-29a was upregulated in patients with active pulmonary TB serum and sputum compared with healthy controls. Mir-29a and miR-let-7e were also induced in human macrophages in Mycobacterium avium infection, where the targets were caspases 7 and 3 in the apoptotic pathway. In this way, inhibition of apoptosis in Mycobacteria infection is controlled by miRNAs. MiRNAs control changes in cellular make-up and related gene expression in TB patients. It has been shown that several miRNAs control T cell differentiation and operation. Additionally, miRNAs have been shown to play a significant role in controlling the activity of NK cells, macrophages, innate immune cells, and dendritic cells [22].
MiRNAs significantly regulate the primary activities of dendritic cells, macrophages, and NK cells. Several studies have shown alterations in gene expression in macrophages and NK cells in cases of active and latent TB and healthy individuals. miRNAs regulate gene expression changes and T cell differentiation and function [23].
MiRNA-29a-3p is one miRNA associated with M. tuberculosis infection. M. tuberculosis infection of host cells leads to miRNA-29a-3p overexpression. miRNA-29a-3p suppresses the immune response against M. tuberculosis by decreasing IFN-γ levels. In addition to targeting the 3′ untranslated region of the IFN-γ mRNA, miRNA-29a-3p promotes IFN-γ mRNA joining with the protein Argonaute 2 to create an RNA silencing complex, reducing post-transcriptional IFN-γ expression.
In addition, several studies have shown that miRNA-29a-3p also targets the myeloid cell leukemia-1, anti-apoptotic B-cell lymphoma 22, GTP-binding cell division cycle 42, and p85 kinase genes, demonstrating its role in regulating the apoptotic pathway. In an anti-TB response, miRNA-29a-3p overexpression in TB infection prevents macrophage phagocytosis by inhibiting IFN-γ and increasing cell apoptosis [10].
This study examined miRNA-29a-3p expression in active TB, latent TB, and healthy control groups using qPCR. Its results showed that mean miRNA-29a-3p expression was 21.32 in the latent TB group, 8.69 in the active TB group, and 0.83 in the healthy control group (Fig. 1). When we compared the active TB and healthy control groups to the latent TB group, miRNA-29a-3p mRNA levels were considerably significantly in the latent TB group based on the ANOVA test. This result is consistent with the findings of Ndzi et al. [9], who showed that hsa-miR-155-5p, hsa-miR-361-5p, and hsa-miR-29a-3p, were significantly upregulated in active TB patients compared to healthy controls. miR-29a-3p showed good performance (81.37%) in differentiating active TB patients from healthy controls and a good diagnostic role (84.35%) in differentiating active TB from latent TB. miR-29a-3p is a valuable infection marker, particularly for establishing the TB diagnosis, because it can distinguish between active TB, latent TB, and healthy controls in the blood. Additionally, blood (plasma) testing for miR-29a-3p is preferable to sputum testing since blood is simpler to collect and can be used for pediatric and extrapulmonary TB diagnosis [9,11].
Additionally, this study's findings are consistent with those of Fu et al. [10], who found that active TB patients had 59 miRNAs that were upregulated and 33 miRNAs that were downregulated compared to healthy controls. One upregulated miRNA was miRNA-29a-3p, which can increase TB susceptibility by inhibiting INF-γ expression in T cells.
Similarly, Sharbati et al. [21], Wu et al. [24], Draz et al. [25], Zhou et al. [3], and Lu et al. [4] reported that blood miRNA-29a-3p was considerably overexpressed in patients with active and latent TB compared to healthy controls. Therefore, miRNA-29a-3p has the potential to act as a candidate biomarker for pulmonary TB diagnosis.
Our ROC curve analysis using a cut-off of 0.01 showed that miRNA-29a-3p expression has the potential to act as a biomarker of active pulmonary TB infection, with 86% sensitivity, 73% specificity, and an AUC of 0.763 (95% CI: 0.668–0.858). Moreover, miRNA-29a-3p expression correlated significantly with active pulmonary TB (Fig. 2). In addition, miRNA-29a-3p expression showed 84.8% sensitivity, 70% specificity, and an AUC of 0.808 (95% CI: 0.698–0.919) for latent TB infection. Moreover, miRNA-29a-3p expression was significantly correlated with latent TB (Fig. 2).
IFN-γ levels in the active TB, latent TB, and healthy control groups were assessed. The ANOVA test indicated there were significant differences between groups (Table 3). However, there was no association between IFN-γ levels and miRNA-29a-3p expression in the active TB, latent TB, and healthy control groups. This result is similar to Afum-Adjei Awuah et al. [12], who investigated the dynamics of IFN-γ and miR-29a-3p expression in CD4+ T cells from patients with active TB (n = 32) and household contacts infected with M. tuberculosis (n = 19) in Ghana. There was no interdependence between miR-29a-3p and IFN-γ since they discovered no statistically significant correlation in either purified protein derivative of tuberculin-induced in active TB patients (R = 0.20; p = 0.157), and tuberculin-induced latent TB patients (R = 0.37; p = 0.118), and Staphylococcal enterotoxin B-induced in active TB patients (R = 0.20; p = 0.167), and Staphylococcal enterotoxin B-induced in latent TB patients (R = 0.22; p = 0.367). These findings are consistent with Kleinsteuber et al. [13], who found no association between miR-29a-3p expression and IFN-γ protein levels in young patients with pulmonary TB. This study was performed in Indonesia with a high TB incidence. While our findings do not rule out a role for miR-29a-3p in IFN-γ regulation, it is doubtful that miR-29a-3p has a significant inhibitory effect on CD4+ T cells during active and latent pulmonary TB infection. This finding could be due to factors contributing to IFN-γ regulation other than miR-29a-3p.
IFN-γ production in CD4 T cells has a significant impact on M. tuberculosis immunity despite not being the sole effector mechanism contributing to the CD4 T cell-mediated immune system. IFN-γ production was positively associated with miR-29a-3p expression during M. tuberculosis infection. Compared to uninfected controls, CD4+ or CD8+ T cells from Mycobacterium bovis Bacille Calmette-Guérin-infected mice showed decreased miR-29a-3p but increased IFN-γ expression. Studies have shown that miR-29a-3p targets the IFN-γ mRNA, reducing immune responses against intracellular infections. However, evidence for the connection between IFN-γ and miR-29a-3p in humans is in stark contrast. Lower expression of miR-142-3p, miR-29a-3p, miR-26a-5p, and miR-21-5p was observed in CD4+ T cells from children with active compared to latent TB. In contrast, there was no association between IFN-γ and miR-29a-3p expression in children with TB compared to healthy controls in T cells expressing M. tuberculosis-specific IFN-γ. Additionally, miR-29a-3p downregulation in T cells by antagomir did not impact IFN-γ expression after in vitro activation [26].
Our study provides an in-depth understanding of TB pathogenesis and a reasonable basis for developing reliable biomarkers for TB diagnosis. MiRNA expression in TB patients can serve as a potential biomarker for TB diagnosis. TB remains a significant infectious disease worldwide. With increasing TB infection and antibiotic resistance, rapid diagnoses are needed to achieve TB control. Moreover, most miRNAs were detected in the blood, which can be easily obtained from TB patients. Circulating miRNAs play an important role in biological pathways and can serve as biomarkers for TB detection. MiRNAs are small non-coding RNAs and negatively post-transcriptionally regulate gene expression. MiRNA panels can enable early diagnosis and expedited medical intervention decisions. Some miRNA biomarkers may also serve as viable drug candidates. MiRNA biomarkers may also be sufficient to determine the health outcome.
This study had several limitations. First, the number of latent TB patients included was small for assessing the potential of miRNA-29a-3p as a biomarker. Therefore, further research with larger sample sizes is needed to confirm our results. However, our results are consistent with previous studies regarding the use of miRNA-29a-3p as a biomarker for diagnosing active and latent TB. Second, miRNA-29a-3p expression did not affect IFN-γ expression. Therefore, further research is needed involving TB patients undergoing anti-TB therapy to clarify the role of miRNA-29a-3p in inhibiting IFN-γ in CD4 T cells during TB infection.
5. Conclusion
MiRNA-29a-3p expression was increased in active pulmonary TB and latent pulmonary TB patients. In addition, miRNA-29a-3p expression was higher in latent TB than in active TB and healthy control groups. Moreover, miRNA-29a-3p is a potential biomarker to diagnose active and latent pulmonary TB infection. However, there was no significant correlation between IFN-γ protein levels and miR-29a-3p expression.
Sources of funding for your research
No funding or sponsorship.
Ethical approval
All procedure for human experiment has been approved by our institutional review board.
Consent
The research was conducted ethically in accordance with the World Medical Association Declaration of Helsinki. The patients have given their written informed consent on admission to use their prospective data base and files for research work.
Author contribution
Nirmawati Angria, Muhammad Nasrum Massi, Agussalim Bukhari, Irawaty Djaharuddin, and Oslan Jumadi initiated and designed the study. Handayani Halik performed the statistical analysis. Ahyar Ahmad, Upik Andriani Miskad, Rusdina Bte Ladju, and Arif Santoso contributed in the data processing. All authors have read and approved the final manuscript.
Registration of research studies
This study has been registered with the Research Registry no. 8089.
Guarantor
Muhammad Nassrum Massi.
Provenance and peer review
Not commissioned, externally peer-reviewed.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We express appreciation to Muhammad Faruk, M.D. for his contribution in reviewing this original research.
Contributor Information
Nirmawati Angria, Email: nirmawatiangria@gmail.com.
Muhammad Nasrum Massi, Email: nasrumm2000@yahoo.com.
Agussalim Bukhari, Email: agussalimbukhari@yahoo.com.
Irawaty Djaharuddin, Email: zalfaradhyani19@gmail.com.
Oslan Jumadi, Email: oslanj@unm.ac.id.
Ahyar Ahmad, Email: ahyarahmad@gmail.com.
Upik Andriani Miskad, Email: upik.miskad@gmail.com.
Rusdina Bte Ladju, Email: rbteladju@gmail.com.
Arif Santoso, Email: arifs777@gmail.com.
Handayani Halik, Email: handayani.halik@gmail.com.
References
- 1.Behrouzi A., Alimohammadi M., Nafari A.H., Yousefi M.H., Riazi Rad F., Vaziri F., Siadat S.D. The role of host miRNAs on Mycobacterium tuberculosis. ExRNA. 2019;1:40. doi: 10.1186/s41544-019-0040-y. [DOI] [Google Scholar]
- 2.Xu Y., Ren W., Liu Y., Zhang X., Li C., Sun Z. Tuberculosis-related miRNAs have potential as disease biomarkers. J. Tubercul. Res. 2013:17–27. doi: 10.4236/jtr.2013.12005. 01. [DOI] [Google Scholar]
- 3.Zhou M., Yu G., Yang X., Zhu C., Zhang Z., Zhan X. Circulating microRNAs as biomarkers for the early diagnosis of childhood tuberculosis infection. Mol. Med. Rep. 2016;13:4620–4626. doi: 10.3892/mmr.2016.5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu Y., Wang X., Dong H., Wang X., Yang P., Han L., Wang Y., Zheng Z., Zhang W., Zhang L. Bioinformatics analysis of microRNA expression between patients with and without latent tuberculosis infections. Exp. Ther. Med. 2019:3977–3988. doi: 10.3892/etm.2019.7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi Y., Cui L., Ge Y., Shi Z., Zhao K., Guo X., Yang D., Yu H., Cui L., Shan Y., Zhou M., Wang H., Lu Z. Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect. Dis. 2012;12:384. doi: 10.1186/1471-2334-12-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J.X., Xu J., Han Y.F., Zhu Y.B., Zhang W.J. Diagnostic values of microRNA-31 in peripheral blood mononuclear cells for pediatric pulmonary tuberculosis in Chinese patients. Genet. Mol. Res. 2015;14:17235–17243. doi: 10.4238/2015.December.16.23. [DOI] [PubMed] [Google Scholar]
- 7.Latorre I., Leidinger P., Backes C., Domínguez J., de Souza-Galvão M.L., Maldonado J., Prat C., Ruiz-Manzano J., Sánchez F., Casas I., Keller A., von Briesen H., Knobel H., Meese E., Meyerhans A. A novel whole-blood miRNA signature for a rapid diagnosis of pulmonary tuberculosis. Eur. Respir. J. 2015;45:1173–1176. doi: 10.1183/09031936.00221514. [DOI] [PubMed] [Google Scholar]
- 8.Meng Q.L., Liu F., Yang X.Y., Liu X.M., Zhang X., Zhang C., De Zhang Z. Identification of latent tuberculosis infection-related microRNAs in human U937 macrophages expressing Mycobacterium tuberculosis Hsp16.3. BMC Microbiol. 2014;14:37. doi: 10.1186/1471-2180-14-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ndzi E.N., Nkenfou C.N., Mekue L.M., Zentilin L., Tamgue O., Pefura E.W.Y., Kuiaté J.-R., Giacca M., Ndjolo A. MicroRNA hsa-miR-29a-3p is a plasma biomarker for the differential diagnosis and monitoring of tuberculosis. Tuberculosis. 2019;114:69–76. doi: 10.1016/j.tube.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Fu Y., Yi Z., Wu X., Li J., Xu F. Circulating MicroRNAs in patients with active pulmonary tuberculosis. J. Clin. Microbiol. 2011;49:4246–4251. doi: 10.1128/JCM.05459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kathirvel M., Saranya S., Mahadevan S. Expression levels of candidate circulating microRNAs in pediatric tuberculosis. Pathog. Glob. Health. 2020;114:262–270. doi: 10.1080/20477724.2020.1761140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Afum-Adjei Awuah A., Ueberberg B., Owusu-Dabo E., Frempong M., Jacobsen M. Dynamics of T-cell IFN- and miR-29a expression during active pulmonary tuberculosis. Int. Immunol. 2014;26:579–582. doi: 10.1093/intimm/dxu068. [DOI] [PubMed] [Google Scholar]
- 13.Kleinsteuber K., Heesch K., Schattling S., Kohns M., Sander-Jülch C., Walzl G., Hesseling A., Mayatepek E., Fleischer B., Marx F.M., Jacobsen M. Decreased expression of miR-21, miR-26a, miR-29a, and miR-142-3p in CD4+ T cells and peripheral blood from tuberculosis patients. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst J.D. Mechanisms of M. tuberculosis immune evasion as challenges to TB vaccine design. Cell Host Microbe. 2018;24:34–42. doi: 10.1016/j.chom.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathew G., Agha R., STROCSS Group STROCSS 2021: strengthening the reporting of cohort, cross-sectional and case-control studies in surgery. Int. J. Surg. 2021;96 doi: 10.1016/j.ijsu.2021.106165. [DOI] [PubMed] [Google Scholar]
- 16.Kenyorini Suradi, Surjanto E., Tuberkulin Uji. Tuberkulosis Indones. 2006;3:1–5. [Google Scholar]
- 17.Wuchty S., Arjona D., Bozdag S., Bauer P.O. Involvement of microRNA families in cancer. Nucleic Acids Res. 2012;40:8219–8226. doi: 10.1093/nar/gks627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J., Li C.X., Li Y.S., Lv J.Y., Ma Y., Shao T.T., De Xu L., Wang Y.Y., Du L., Zhang Y.P., Jiang W., Li C.Q., Xiao Y., Li X. MiRNA-miRNA synergistic network: construction via co-regulating functional modules and disease miRNA topological features. Nucleic Acids Res. 2011;39:825–836. doi: 10.1093/nar/gkq832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.UNHCR Global report 2018(有一些关于非洲的数据), World heal. Organ. 2018;63:476. https://apps.who.int/iris/handle/10665/274453 [Google Scholar]
- 20.Nurjana M.A. Faktor Risiko Terjadinya Tubercolosis Paru Usia Produktif (15-49 Tahun) di Indonesia. Media Penelit. Dan Pengemb. Kesehat. 2015;25:163–170. doi: 10.22435/mpk.v25i3.4387.163-170. [DOI] [Google Scholar]
- 21.Sharbati J., Lewin A., Kutz-Lohroff B., Kamal E., Einspanier R., Sharbati S. Integrated microrna-mrna-analysis of human monocyte derived macrophages upon mycobacterium avium subsp. hominissuis infection. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 23.Bezman N.A., Cedars E., Steiner D.F., Blelloch R., Hesslein D.G.T., Lanier L.L. Distinct requirements of MicroRNAs in NK cell activation, survival, and function. J. Immunol. 2010;185:3835–3846. doi: 10.4049/jimmunol.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L.S.H., Lee S.W., Huang K.Y., Lee T.Y., Hsu P.W.C., Weng J.T.Y. Systematic expression profiling analysis identifies specific MicroRNA-gene interactions that may differentiate between active and latent tuberculosis infection. BioMed Res. Int. 2014;2014 doi: 10.1155/2014/895179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Draz N.I., rahman el Hady S.A., Elsayed M.S., Korraa E.E.A., Abo el Magd N.M.A. Serum microRNA-29a and microRNA-361-5p as potential diagnostic biomarkers for active pulmonary tuberculosis, Egypt. J. Med. Microbiol. 2014;29:1–10. [Google Scholar]
- 26.Sinigaglia A., Peta E., Riccetti S., Venkateswaran S., Manganelli R., Barzon L. Tuberculosis-associated MicroRNAs: from pathogenesis to disease biomarkers. Cells. 2020;9:2160. doi: 10.3390/cells9102160. [DOI] [PMC free article] [PubMed] [Google Scholar]