Abstract
Duchenne muscular dystrophy (DMD) is a myopathy characterized by progressive muscle weakness caused by a mutation in the dystrophin gene on the X chromosome. We recently showed that a medium-chain triglyceride-containing ketogenic diet (MCTKD) improves skeletal muscle myopathy in a CRISPR/Cas9 gene-edited rat model of DMD. We examined the effects of the MCTKD on transcription profiles in skeletal muscles of the model rats to assess the underlying mechanism of the MCTKD-induced improvement in DMD. DMD rats were fed MCTKD or normal diet (ND) from weaning to 9 months, and wild-type rats were fed with the ND, then tibialis anterior muscles were sampled for mRNA-seq analysis. Pearson correlation heatmaps revealed a one-node transition in the expression profile between DMD and wild-type rats. A total of 10,440, 11,555 and 11,348 genes were expressed in the skeletal muscles of wild-type and ND-fed DMD rats the MCTKD-fed DMD rats, respectively. The MCTKD reduced the number of DMD-specific mRNAs from 1624 to 1350 and increased the number of mRNAs in common with wild-type rats from 9931 to 9998. Among 2660 genes were differentially expressed in response to MCTKD intake, the mRNA expression of 1411 and 1249 of them was respectively increased and decreased. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses suggested that the MCTKD significantly suppressed the mRNA expression of genes associated with extracellular matrix organization and inflammation. This suggestion was consistent with our previous findings that the MCTKD significantly suppressed fibrosis and inflammation in DMD rats. In contrast, the MCTKD significantly increased the mRNA expression of genes associated with oxidative phosphorylation and ATP production pathways, suggesting altered energy metabolism. The decreased and increased mRNA expression of Sln and Atp2a1 respectively suggested that Sarco/endoplasmic reticulum Ca2+-ATPase activation is involved in the MCTKD-induced improvement of skeletal muscle myopathy in DMD rats. This is the first report to examine transcription profiles in the skeletal muscle of CRISPR/Cas9 gene-edited DMD model rats and the effect of MCTKD feeding on it.
Keywords: Duchenne muscular dystrophy, Ketogenic diet, Transcriptome, Ketone bodies, Skeletal muscle
Abbreviations: DAPC, dystrophin-associated glycoprotein complex; DEG, differentially expressed gene; DMD, Duchenne muscular dystrophy; FPKM, fragments per kilobase of exons per million mapped reads; GO, gene ontology; KD, ketogenic diet; KR, ketogenic ratio; MCT, medium-chain triglycerides; MCTKD, medium-chain triglycerides containing ketogenic diet; ND, normal diet; PC, principal component; SERCA, sarco/endoplasmic reticulum (SR) Ca2+ ATPase; TA, tibialis anterior; WT, wild type
Highlights
-
•
We evaluated the effects of an MCTKD on the global transcriptome of DMD rats.
-
•
DMD rats are suitable models of human DMD for assessing transcriptome changes.
-
•
MCTKD suppressed fibrosis and inflammatory pathways at the transcriptional level.
-
•
MCTKD upregulated oxidative phosphorylation and ATP production pathways.
-
•
MCTKD might activate SERCA at the transcriptional level.
1. Introduction
Duchenne muscular dystrophy (DMD) is an X-linked myopathy caused by a deficiency of dystrophin protein in muscle. Dystrophin is a component of the dystrophin-glycoprotein complex that connects the cytoskeleton to the basement membrane [1]. Defects in dystrophin cause persistent muscle degeneration, necrosis, fibrosis, and adipose tissue replacements [2]. Despite therapeutic advances, irreversible skeletal muscle weakness with consequent loss of mobility, inevitable progressive cardiomyopathy, and respiratory failure, eventually lead to the death of affected individuals [3]. Therefore, new therapeutic methods that could complement existing strategies and enhance treatment effectiveness for this lethal disease need to be identified.
Several animal models of DMD have been created to develop effective therapeutic options. The most popular animal model is the mdx mouse, but the lifespans of mdx and WT mice are similar, and the pathology is milder than that of human DMD. We generated rat models of DMD using CRISPR/Cas9 genome editing and found that skeletal muscle injury and the cardiomyopathy phenotype was more severe in the DMD rats than the mdx mice [[4], [5], [6]]. A change in the DMD transcriptome in skeletal muscles has been identified in humans and mice. The skeletal muscle transcriptome of DMD in boys is characterized by an increased inflammatory response, extracellular matrix remodeling, muscle regeneration pathways, and the reduced transcription of genes involved in energy metabolism [7,8]. These trends are similar in mouse models [9,10], but some genes clearly differ between mdx and human DMD. Currently, there are no data on the DMD rat transcriptome, and whether the phenotype of DMD rats is more similar to humans than to mdx mice even at the transcriptional level remains unclear.
Ketogenic diets (KD) are high-fat, low-carbohydrate diets that promote the production of ketone bodies in the liver, and they have been applied to patients with intractable epilepsy since the 1920s. A ketogenic diet containing medium chain triglycerides (MCTKD) was developed in the 1970s as an alternative to the classical ketogenic diet. Medium chain triglycerides are absorbed and transported more efficiently than other types of fat and will yield more ketones per unit of dietary energy [11]. Therefore, less total fat is needed on an MCT diet, which can include more protein and carbohydrate food sources. We recently found that dietary intervention with an MCTKD improves skeletal muscle function and pathology in DMD rats [12], suggesting that it could be a novel dietary treatment option for patients with DMD. Furthermore, MCTKD significantly inhibited necrosis, inflammation, and fibrosis in the skeletal muscles of DMD rats and promoted regeneration by stimulating satellite cell proliferation.
Here, we analyzed the global transcriptome of skeletal muscles from CRISPR/Cas9 gene-edited DMD model rats. We also examined the effects of an MCTKD on transcriptional changes in DMD rats to understand the cellular and molecular processes involved in the beneficial effects of MCTKD on skeletal muscle myopathy in DMD.
2. Materials and methods
2.1. Ethics approval
All animal experiments and handling procedures complied with the guidelines of the National Institute of Advanced Industrial Science and Technology (AIST), The University of Tokyo, and relevant international guidelines for the proper care and use of laboratory animals. The Institutional Animal Care and Use Committees at AIST and The University of Tokyo approved the experimental protocol (Permission Nos: 2020-358 and P20-012, respectively).
2.2. Rat model of DMD
We previously established a Wistar-Imamichi rat model with an out-of-frame mutation in the DMD gene using CRISPR/Cas9 gene-editing. The DMD rats have deletion in exons 3 and insertion and conversion in exon 16 in the DMD gene. The DMD rats were perpetuated by crossing WT male rats with females harboring a heterozygous mutation in the Dmd gene. All rats were maintained in groups (2–4 per cage) at 23 °C under a 12-h light/dark cycle (lights on at 8 a.m.) and food and water were supplied ad libitum.
2.3. Dietary intervention
Male DMD rats were randomly assigned to groups after weaning at 3 weeks of age (n = 6 each) and fed with the AIN93 M normal diet (ND) or the MCTKD. The controls were WT littermates fed with the ND (n = 4). The dietary intervention continued from the age of 3 weeks to 9 months. Two types of MCTKD were used to rapidly increase blood ketone level after the dietary intervention started. The rats were fed for the first 10 days with an MCTKD containing a ketogenic ratio (KR) of fat weight to carbohydrate + protein weight of 2.0, followed thereafter by an MCTKD with a KR of 1.4.
2.4. Extraction of RNA and mRNA-seq analysis
Total RNA was extracted from frozen tibialis anterior (TA) muscles disrupted in RNAiso Plus (Takara Bio Inc. Kusatsu, Japan) using a Micro Smash MS-100R (Tommy Seiko Co., Ltd., Tokyo, Japan). Messenger RNA was purified from total RNA using poly-T oligo-attached magnetic beads. After fragmentation, the first strand cDNA was synthesized using random hexamer primers, followed by second strand cDNA synthesis using dTTP. After end repair, a poly(A) tail was added to double-stranded cDNA, then ligated with an RNA adaptor, followed by size selection, amplification, and purification. The library was checked with Qubit, quantified using real-time RT-PCR and size distribution was determined using a bioanalyzer. The libraries were sequenced on an Illumina NovaSeq 6000 sequencing platform (Illumina In., San Diego, CA, USA), and 150 bp paired-end reads were generated.
2.5. Sequence data analysis
Clean reads were obtained from the raw data in FASTQ format by removing low quality reads and reads with poly(N) sequences. The clean paired-end reads were mapped to a reference genome using Hisat2 v2.0.5. Uniquely mapped reads accounted for ∼93% in all samples, and only uniquely mapped reads were subsequently analyzed. The read numbers mapped to each gene were counted using featureCounts v1.5.0-p3. Genes with fragments per kilobase of exons per million mapped reads (FPKM) > 1 were considered as being expressed. Differential expression was analyzed using the DESeq2R package (1.20.0). Genes with a P < 0.05 identified by DESeq2 were considered as being differentially expressed.
2.6. Gene function analysis
We analyzed the pathways enriched by differentially expressed genes (DEGs) using Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) and the clusterProfiler R package, in which gene length bias was corrected. The GO and KEGG categories with corrected P < 0.05 were considered as being significantly enriched.
2.7. Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from rat frozen TA muscles using RNAiso Plus (Takara Bio Inc., Otsu, Japan). Complementary DNA was then synthesized using PrimeScript™ RT reagent kits with gDNA Eraser (Takara Bio). Quantitative RT-PCR (qRT-PCR) proceeded on a LightCycler™ (Roche Diagnostics, Mannheim, Germany), using SYBR® Premix Ex Taq™ II (Takara Bio Inc) under the following amplification conditions: 45 cycles of 95 °C for 5 s, 57 °C for 10 s, and 72 °C for 10 s. Supplementary Table 1 shows the qRT-PCR primer sets, and the amount of target mRNA was normalized to the amount of Cmas.
2.7. Statistical analysis
The significance of differences (p) in RNA-seq data between two groups was assessed using negative binomial distribution tests. Expression determined from qRT-PCR results was compared using one-way ANOVA followed by Tukey-Kramer multiple comparison tests.
3. Results
3.1. Correlation and principal component analysis of mRNA-seq data
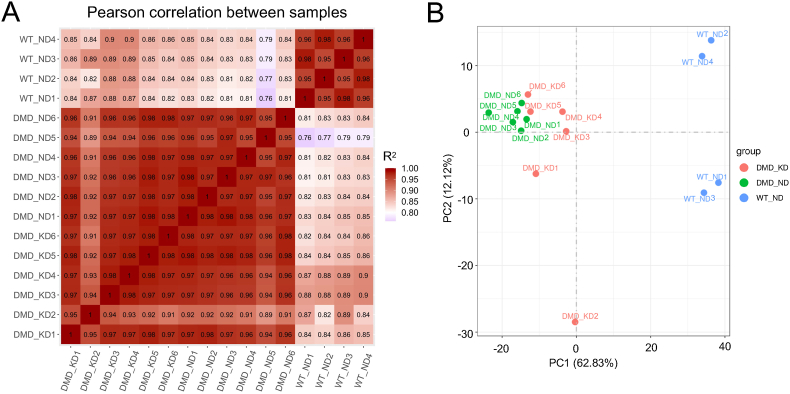
Samples of RNA obtained from TA muscles were analyzed using mRNA-seq, and >93% of reads were obtained as unique maps. Messenger RNA expression was quantified then normalized for each individual. Pearson correlation analysis of mRNA-seq data showed that all samples closely correlated within groups (Pearson correlation coefficient, R2 = ∼0.91; Fig. 1A). A single node transition was identified between the WT and DMD groups, and the R2 between tended to be larger between the WT rats and the MCTKD-fed DMD rats than the ND-fed DMD rats. The first and second principal components (PCs) explained 62.83% and 12.12% of the total variation in mRNA expression, respectively (Fig. 1B). The divergence of the first principal component (PC1) generally reflected differences between groups, with a particularly clear divergence between DMD and WT rats. Similar to the Pearson correlation results, the MCTKD-fed DMD rats were located between, but were much closer to those given the ND-fed DMD rats than WT rats on the PC1 axis.
Fig. 1.
Gene expression correlations among ND-fed DMD rats, MCTKD-fed DMD rats and WT rats.
(A) Pearson correlation heatmaps and (B) principal component analysis of gene expression (FPKM) among ND-fed DMD rats (n = 6), MCTKD-fed DMD rats (n = 6) and WT rats (n = 4). Ratios of variance are shown as PCs.
3.2. Analysis of DEGs
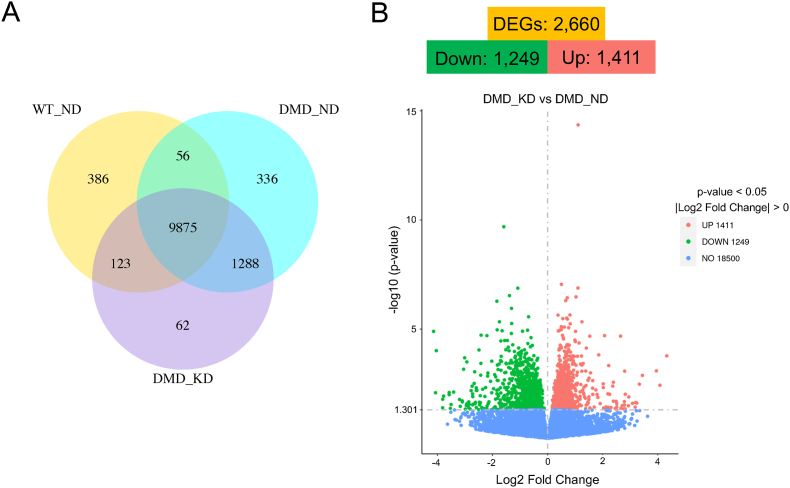
Genes with an average of >1.0 FPKM of in each group were defined as expressed, and the number of genes that were independently expressed and co-expressed in each group were compared (Fig. 2A). Overall 10,440, 11,555 and 11,348 genes were expressed in the TA muscles of WT rats, and the DMD rats fed with the ND and the MCTKD, respectively. The MCTKD reduced the number of genes expressed only in DMD rats from 1624 to 1350 and increased the number of mRNAs in common with WT rats from 9931 to 9998. The mRNA expression of 6130 and 5142 genes was significantly increased, and that of 4992 and 4159 was decreased, respectively, in TA muscles of ND-fed DMD rats and the MCTKD-fed DMD rats compared with WT rats (Supplementary Fig. 1). A comparison between MCTKD-fed DMD rats and ND-fed DMD rats revealed that the expression of 1411 and 1249 of 2660 DEGs was respectively increased and decreased in response to the MCTKD.
Fig. 2.
Expression of DEGs.
(A) Venn diagram of co-expression among ND-fed DMD rats, MCTKD-fed DMD rats and WT rats. Genes with fragments per kilobase of exons per million mapped reads (FPKM) > 1 were considered as being expressed. (B) Volcano plots of DEGs between ND-fed DMD rats and MCTKD-fed DMD rats.
3.3. Enrichment analysis of DEGs downregulated by MCTKD
We used GO and KEGG to identify biological processes and signaling pathways, respectively, which were associated with downregulated DEGs. The 4992 downregulated DEGs in ND-fed DMD rats compared with WT rats, were enriched in the biological processes of generation of precursor metabolites and energy, mitochondrion organization, mitochondrial respiratory chain complex assembly, energy derivation by oxidation of organic compounds, and oxidative phosphorylation (Supplementary Fig. 2A). The signaling pathways enriched in downregulated DEGs in ND-fed DMD rats compared with WT rats were thermogenesis, oxidative phosphorylation, and pathways of neurodegeneration - multiple diseases, Huntington disease, and Parkinson disease (Supplementary Fig. 2B).
The 4159 DEGs downregulated in MCTKD-fed DMD rats compared with WT rats were enriched in the biological processes of generation of precursor metabolites and energy, cellular respiration, energy derivation by oxidation of organic compounds, oxidative phosphorylation, and mitochondrion organization (Supplementary Fig. 3A). The signaling pathways enriched in DEGs downregulated in MCTKD-fed DMD rats compared with WT rats comprised thermogenesis, oxidative phosphorylation, Parkinson disease, diabetic cardiomyopathy, and pathways of neurodegeneration - multiple diseases (Supplementary Fig. 3B).
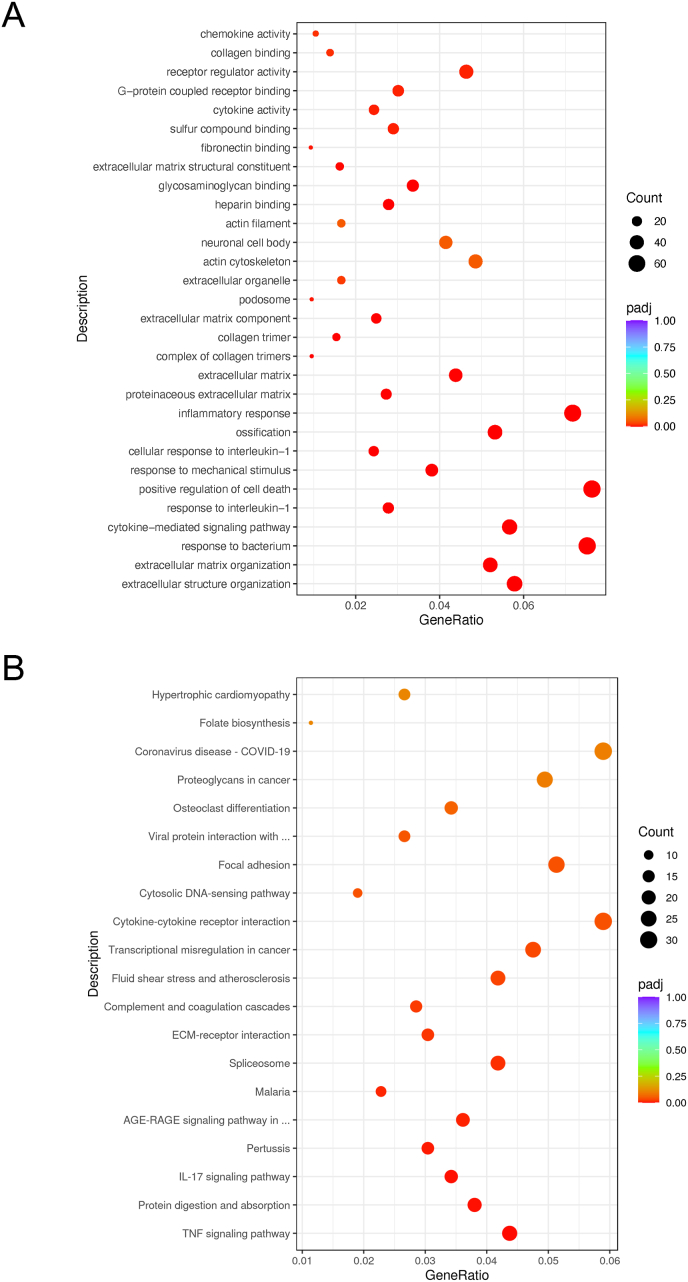
The 1249 DEGs downregulated in MCTKD-fed DMD rats compared with ND-fed DMD rats were enriched in the biological processes of extracellular structure organization, extracellular matrix organization, response to bacterium, cytokine-mediated signaling, and responses to interleukin-1 (Fig. 3A). The signaling pathways enriched in these DEGs comprised TNF signaling, protein digestion and absorption, IL-17 signaling, pertussis, and advanced glycation endproducts-receptor for advanced glycation endproducts (AGE-RAGE) in diabetic complications (Fig. 3B).
Fig. 3.
Gene ontology and KEGG pathway analyses of downregulated DEGs in ND-fed DMD rats and MCTKD-fed DMD rats.
(A) GO and (B) KEGG pathway analyses of downregulated genes in ND-fed DMD rats and MCTKD-fed DMD rats.
3.4. Enrichment of DEGs upregulated by MCTKD intake
We used GO and KEGG to identify biological processes and signaling pathways associated with upregulated DEGs.
The 6130 upregulated DEGs in ND-fed DMD rats compared with WT rats were enriched in the biological processes of leukocyte migration, inflammatory response, extracellular structure organization, extracellular matrix organization, and regulation of cell activation (Supplementary Fig. 4A). The signaling pathways enriched in these DEGs were cytokine-cytokine receptor interaction, viral protein interaction with cytokine and cytokine receptor, chemokine signaling pathway, complement and coagulation cascades, and hematopoietic cell lineage (Supplementary Fig. 4B).
The 5142 upregulated DEGs in MCTKD-fed DMD rats compared with WT rats were enriched in the biological processes of leukocyte migration, inflammatory response, chemotaxis, taxis, and extracellular matrix organization (Supplementary Fig. 5A) and the signaling pathways of cytokine-cytokine receptor interaction, viral protein interaction with cytokine and cytokine receptor, chemokine signaling pathway, complement and coagulation cascades, and hematopoietic cell lineage (Supplementary Fig. 5B).
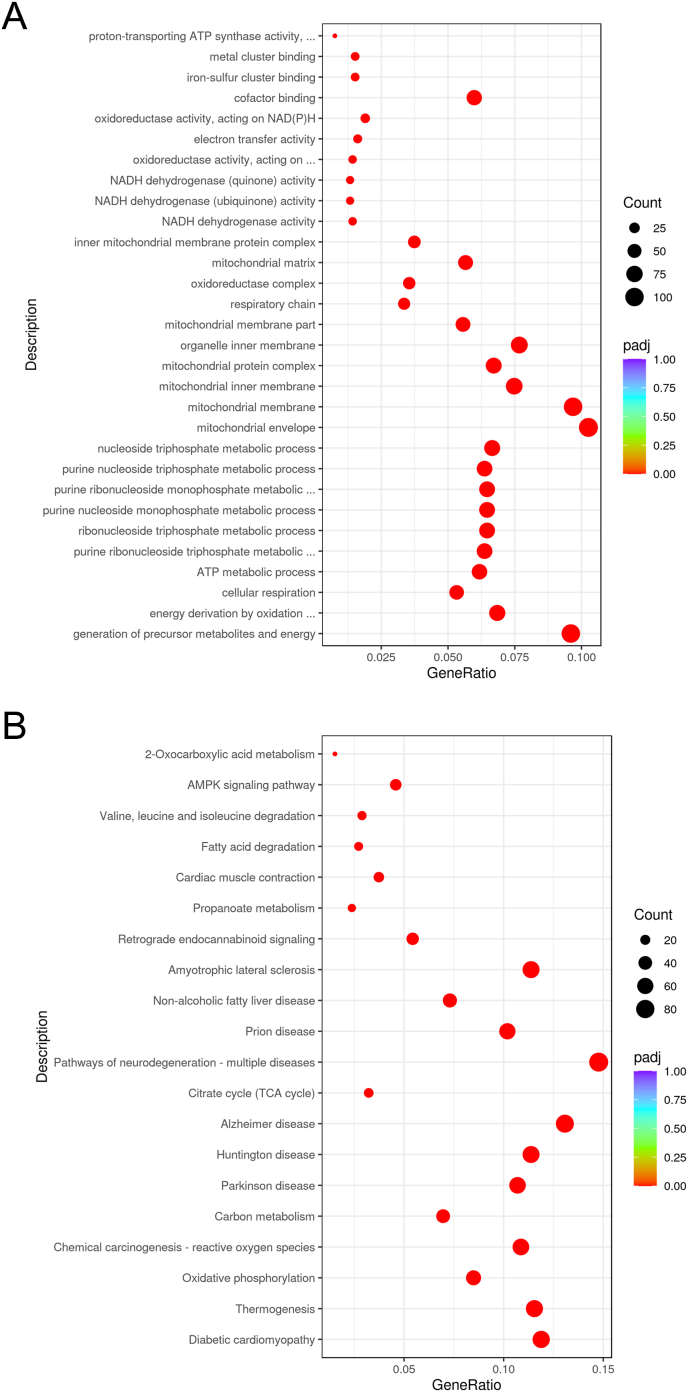
The 1411 DEGs upregulated in MCTKD-fed DMD rats compared with ND-fed DMD rats, were enriched in the biological processes of generation of precursor metabolites and energy, energy derivation by oxidation of organic compounds, cellular respiration, ATP metabolic process, and purine ribonucleoside triphosphate metabolic process (Fig. 4A). The enriched signaling pathways were diabetic cardiomyopathy, thermogenesis, oxidative phosphorylation, chemical carcinogenesis - reactive oxygen species, and carbon metabolism (Fig. 4B).
Fig. 4.
Gene ontology and KEGG pathway analysis of upregulated DEGs in ND-fed DMD rats and MCTKD-fed DMD rats.
(A) GO and (B) KEGG pathway analysis of upregulated genes in ND-fed DMD rats and MCTKD-fed DMD rats.
3.5. Expression of known DMD related genes
We compared the expression of genes involved in calcium overload and structural proteins such as dystrophin-associated glycoprotein complex (DAPC) to determine the effects of the MCTKD on the expression of genes associated with DMD progression. Supplementary Fig. 6 shows relative mRNA expression profiles of the genes associated with sarco/endoplasmic reticulum (SR) Ca2+ ATPase (SERCA) activity, Sln, Ryr1, Atp2a1 and Atp2a2. The MCTKD minimally affected the expression Atp2a2, but significantly decreased the mRNA expression of Sln, a SERCA inhibitor and significantly increased that of Ryr1 and Atp2a1 in MCTKD-fed DMD rats compared with ND-fed DMD rats. Gene expression of the structural protein DAPC was minimally affected by the MCTKD, compared with the ND-fed DMD rats (Supplementary Fig. 7). We validated the expression of genes associated with SERCA activity, DAPC, and inflammation using qRT-PCR to avoid RNA-seq bias (Supplementary Fig. 9‒11). We then assessed whether the relatively severe disease phenotype of DMD rats simulates DMD-related gene expression in patients with DMD. We compared our results from DMD rats with previous findings of mdx mice and patients with DMD (Supplementary Fig. 8). The expression of genes involved in ECM formation tended to be enhanced in DMD rats compared with mdx mice.
4. Discussion
We aimed to determine the underlying mechanism of the ameliorative effects of MCTKD on DMD skeletal muscle myopathy. We therefore analyzed the effects of MCTKD on the transcriptional profiles of TA muscles in DMD rats. Pearson correlation heatmaps and PCA revealed remarkably different gene expression profiles between DMD and WT rats. We found far more DEGs in the TA muscle of ND-fed DMD rats than the MCTKD-fed DMD rats. The results of the GO and KEGG enrichment analyses of DEGs suggested that the MCTKD significantly suppressed the mRNA expression of genes associated with extracellular matrix composition and inflammation, and significantly activated that of genes related to oxidative phosphorylation and ATP production pathways. The analysis of genes associated with DMD pathogenesis suggested that MCTKD activated SERCA. This is the first study to analyze the transcription profiles and effects of an MCTKD on the skeletal muscle of CRISPR/Cas9 gene-edited DMD model rats.
Although fibrosis is a hallmark of DMD, little or none is detectable in the fore or hind limbs of mdx mice (the most popular mouse models of DMD), until they reach an advanced age [13,14]. In contrast, fibrosis is evident 3-month-old DMD rat [4]. Consistent with these facts, we found here that the expression of genes involved in ECM formation such as COL1A1, COL1A2, LUM, BGN, and COL6A1 was significantly enhanced in DMD rats compared with mdx mice (Supplementary Fig. 8). The mRNA expression of the ADAM10, COL4A3, LAMB2, P4HA2, SERPINH1, and TGFB3, is increased in human DMD but decreased or unchanged in mdx and DMD rats, which might reflect species differences between human and rodents. Like human DMD, the mRNA expression of genes associated with inflammation was also increased in DMD rats compared with mdx mice (Supplementary Fig. 8). These findings of at least fibrosis and inflammation indicated that DMD rats are suitable models of human DMD and transcriptome changes.
Our results indicated that MCTKD exerts two major effects on the DMD rat transcriptome. One is the suppression of mRNA levels of genes involved in inflammation and fibrosis pathways. This was consistent with our previous observations of fewer CD11b-positive cells and less fibrosis in TA sections of MCTKD-fed DMD rats [12]. The MCTKD might reduce inflammation as the immune system adapts to a reduced glucose supply and energy metabolism switches to mitochondrial fatty acid oxidation, ketogenesis, and ketolysis that occur during prolonged caloric restriction and fasting [[15], [16], [17], [18], [19]]. Only corticosteroid-based anti-inflammatory therapy has been approved for DMD [20]. Since anti-inflammatory therapy suppresses fibrosis and improves DMD myopathy, MCTKD might improve DMD via the suppression of inflammatory pathways and the subsequent improvement of fibrosis with ketone bodies.
The other effect of MCTKD was to increase gene expression associated with oxidative phosphorylation and mitochondrial function. This might be explained by changes in SERCA. The sarcolemma is disrupted during muscle contraction and relaxation in DMD, causing extracellular Ca2+ to flow through the membrane cleft into the cytoplasm, resulting in increased concentrations of cytosolic Ca2+ [21]. Abundant SERCA on the sarcoplasmic reticulum actively takes up > 70% of cytosolic Ca2+ in the sarcoplasmic reticulum [22]. Sarcolipin inhibits SERCA and is abnormally elevated in the muscles of patients and animal models of DMD [23]. In our experiments, MCTKD suppressed and increased the expression of Sln and Atp2a1 respectively, suggesting SERCA activation. Lowering the gene expression of sarcolipin ameliorates dystrophic pathology in a dystrophin/utrophin double KO mutant mouse model [23], and Atp2a1 overexpression improves the mdx phenotype [24]. Sustained elevated cytosolic Ca2+ levels cause mitochondrial dysfunction due to the formation of mitochondrial permeability transition pores [25,26], and pharmacological SERCA activation reduces cytosolic Ca2+ levels, restores mitochondrial respiratory function and improves mdx pathogenesis [27]. Therefore, the MCTKD might regulate cytosolic Ca2+ concentrations and restore mitochondrial function by increasing Atp2a1 and suppressing Sln expression. This might have increased the gene expression of oxidative phosphorylation, increased ATP production, and restored muscle function.
This study has some limitations. The functional consequences of identified changes in gene expression in vivo or in vitro require investigation. We also investigated only steady-state mRNA expression that might reflect either an active transcription process or RNA stability. In addition, although RNA-seq is currently one of the best methods for quantifying gene expression, and RNA-seq data closely correlates with qRT-PCR data, it has some bias regarding gene length and genes with low expression. The present RNA-seq and qRT-PCR results revealed consistent trends for most genes, except Utrn and Dtnb genes, the expression of which was extremely low in RNA-seq. This likely resulted in the low number of RNA-seq reads and low accuracy. Another critical limitation of this study is that the gene expression of distinct cell subpopulations was simply averaged because it was bulk tissue RNA-seq, although the present study aimed to evaluate the global effect of MCTKD on TA muscles of DMD rats. Because skeletal muscle comprises several cell populations, whether the changes at the transcriptional level in the tissue bulk RNA-seq in this study are due to differences in cell populations or in levels of transcripts of individual cells is unclear. Confirmation of gene expression in individual cells will support more comprehensive information.
In summary, this study revealed the genome-wide expression profile of a CRISPR/Cas9 gene-edited rat model of DMD. The findings indicated that the MCTKD suppresses the expression of genes associated with extracellular matrix composition and inflammation in model rats and activates the expression of genes associated with oxidative phosphorylation and ATP production pathways in DMD. Our results deepen understanding of the molecular mechanisms underlying the improvement of skeletal muscle myopathy induced by MCTKD and should lead to the development promising new therapeutic strategies to combat DMD.
Author contributions
Y.F. designed the study, conducted the experiments, analyzed the data, and wrote the manuscript. K.Y. designed the study, revised the manuscript, and supervised the study. H.S. designed the study and performed the experiments. M.H. provided advice concerning MCTKD composition. T.A. and S.A. revised the manuscript and supervised the study. K.O. designed the study, revised the manuscript, and supervised the study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by JSPS KAKENHI Grant Number 22K18345. We express our sincere thanks to Masanari Ikeda, Riku Yamaguchi, and Julian Larrick for assistance with tissue sampling.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2022.101378.
Contributor Information
Keitaro Yamanouchi, Email: akeita@g.ecc.u-tokyo.ac.jp.
Katsutaka Oishi, Email: k-ooishi@aist.go.jp.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
The RNA-seq data generated in this study is available in the NCBI database under the accession number GSE210799.
References
- 1.Lapidos K.A., Kakkar R., Mcnally E.M. The dystrophin glycoprotein complex. Circ. Res. 2004;94(8):1023–1031. doi: 10.1161/01.RES.0000126574.61061.25. Apr 30. [DOI] [PubMed] [Google Scholar]
- 2.Verhaart I.E.C., Aartsma-Rus A. Therapeutic developments for Duchenne muscular dystrophy. Nat. Rev. Neurol. 2019;15(7):373–386. doi: 10.1038/s41582-019-0203-3. Jul. [DOI] [PubMed] [Google Scholar]
- 3.Mercuri E., Muntoni F. Muscular dystrophies. Lancet. 2013;381(9869):845–860. doi: 10.1016/S0140-6736(12)61897-2. Mar 9. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura K., Fujii W., Tsuboi M., Tanihata J., Teramoto N., Takeuchi S., Naito K., Yamanouchi K., Nishihara M. Generation of muscular dystrophy model rats with a CRISPR/Cas system. Sci. Rep. 2014;4:5635. doi: 10.1038/srep05635. Jul 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugihara H., Teramoto N., Nakamura K., Shiga T., Shirakawa T., Matsuo M., Ogasawara M., Nishino I., Matsuwaki T., Nishihara M., Yamanouchi K. Cellular senescence-mediated exacerbation of Duchenne muscular dystrophy. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-73315-6. Oct 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugihara H., Kimura K., Yamanouchi K., Teramoto N., Okano T., Daimon M., Morita H., Takenaka K., Shiga T., Tanihata J., Aoki Y., Inoue-Nagamura T., Yotsuyanagi H., Komuro I. Age-Dependent echocardiographic and pathologic findings in a rat model with Duchenne muscular dystrophy generated by CRISPR/Cas9 genome editing. Int. Heart J. 2020;61(6):1279–1284. doi: 10.1536/ihj.20-372. Nov 28. [DOI] [PubMed] [Google Scholar]
- 7.Haslett J.N., Sanoudou D., Kho A.T., Bennett R.R., Greenberg S.A., Kohane I.S., Beggs A.H., Kunkel L.M. Gene expression comparison of biopsies from Duchenne muscular dystrophy (DMD) and normal skeletal muscle. Proc. Natl. Acad. Sci. U. S. A. 2002;99(23):15000–15005. doi: 10.1073/pnas.192571199. Nov 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pescatori M., Broccolini A., Minetti C., Bertini E., Bruno C., D'Amico A., Bernardini C., Mirabella M., Silvestri G., Giglio V., Modoni A., Pedemonte M., Tasca G., Galluzzi G., Mercuri E., Tonali P.A., Ricci E. Gene expression profiling in the early phases of DMD: a constant molecular signature characterizes DMD muscle from early postnatal life throughout disease progression. Faseb. J. 2007;21(4):1210–1226. doi: 10.1096/fj.06-7285com. Apr. [DOI] [PubMed] [Google Scholar]
- 9.Chemello F., Wang Z., Li H., McAnally J.R., Liu N., Bassel-Duby R., Olson E.N. Degenerative and regenerative pathways underlying Duchenne muscular dystrophy revealed by single-nucleus RNA sequencing. Proc. Natl. Acad. Sci. U. S. A. 2020;117(47):29691–29701. doi: 10.1073/pnas.2018391117. Nov 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Pelt D.W., Kharaz Y.A., Sarver D.C., Eckhardt L.R., Dzierzawski J.T., Disser N.P., Piacentini A.N., Comerford E., McDonagh B., Mendias C.L. Multiomics analysis of the mdx/mTR mouse model of Duchenne muscular dystrophy. Connect. Tissue Res. 2021;62(1):24–39. doi: 10.1080/03008207.2020.1791103. Jan. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y.M. Medium-chain triglyceride (MCT) ketogenic therapy. Epilepsia. 2008;49(8):33–36. doi: 10.1111/j.1528-1167.2008.01830.x. Nov. [DOI] [PubMed] [Google Scholar]
- 12.Fujikura Y., Sugihara H., Hatakeyama M., Oishi K., Yamanouchi K. Ketogenic diet with medium-chain triglycerides restores skeletal muscle function and pathology in a rat model of Duchenne muscular dystrophy. Faseb. J. 2021;35(9) doi: 10.1096/fj.202100629R. Sep. [DOI] [PubMed] [Google Scholar]
- 13.Gutpell K.M., Hrinivich W.T., Hoffman L.M. Skeletal muscle fibrosis in the mdx/utrn+/- mouse validates its suitability as a murine model of Duchenne muscular dystrophy. PLoS One. 2015;10(1) doi: 10.1371/journal.pone.0117306. Jan 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desguerre I., Arnold L., Vignaud A., Cuvellier S., Yacoub-Youssef H., Gherardi R.K., Chelly J., Chretien F., Mounier R., Ferry A., Chazaud B. A new model of experimental fibrosis in hindlimb skeletal muscle of adult mdx mouse mimicking muscular dystrophy. Muscle Nerve. 2012;45(6):803–814. doi: 10.1002/mus.23341. Jun. [DOI] [PubMed] [Google Scholar]
- 15.Cotter D.G., Schugar R.C., Crawford P.A. Ketone body metabolism and cardiovascular disease. Am. J. Physiol. Heart Circ. Physiol. 2013;304(8):H1060–H1076. doi: 10.1152/ajpheart.00646.2012. Apr 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shido O., Nagasaka T., Watanabe T. Blunted febrile response to intravenous endotoxin in starved rats. J. Appl. Physiol. 1985;67(3):963–969. doi: 10.1152/jappl.1989.67.3.963. (1989) Sep. [DOI] [PubMed] [Google Scholar]
- 17.Johnson J.B., Summer W., Cutler R.G., Martin B., Hyun D.H., Dixit V.D., Pearson M., Nassar M., Telljohann R., Maudsley S., Carlson O., John S., Laub D.R., Mattson M.P. Alternate day calorie restriction improves clinical findings and reduces markers of oxidative stress and inflammation in overweight adults with moderate asthma. Free Radic. Biol. Med. 2007;42(5):665–674. doi: 10.1016/j.freeradbiomed.2006.12.005. Mar 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mercken E.M., Crosby S.D., Lamming D.W., JeBailey L., Krzysik-Walker S., Villareal D.T., Capri M., Franceschi C., Zhang Y., Becker K., Sabatini D.M., de Cabo R., Fontana L. Calorie restriction in humans inhibits the PI3K/AKT pathway and induces a younger transcription profile. Aging Cell. 2013;12(4):645–651. doi: 10.1111/acel.12088. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGettrick A.F., O'Neill L.A.J. How metabolism generates signals during innate immunity and inflammation. J. Biol. Chem. 2013;288(32):22893–22898. doi: 10.1074/jbc.R113.486464. Aug 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyatake S., Shimizu-Motohashi Y., Takeda S., Aoki Y. Anti-inflammatory drugs for Duchenne muscular dystrophy: focus on skeletal muscle-releasing factors. Drug Des. Dev. Ther. 2016;10:2745–2758. doi: 10.2147/DDDT.S110163. Aug 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumura K., Campbell K.P. Dystrophin-glycoprotein complex: its role in the molecular pathogenesis of muscular dystrophies. Muscle Nerve. 1994;17(1):2–15. doi: 10.1002/mus.880170103. Jan. [DOI] [PubMed] [Google Scholar]
- 22.Periasamy M., Kalyanasundaram A. SERCA pump isoforms: their role in calcium transport and disease. Muscle Nerve. 2007;35(4):430–442. doi: 10.1002/mus.20745. Apr. [DOI] [PubMed] [Google Scholar]
- 23.Voit A., Patel V., Pachon R., Shah V., Bakhutma M., Kohlbrenner E., McArdle J.J., Dell'Italia L.J., Mendell J.R., Xie L.H., Hajjar R.J., Duan D., Fraidenraich D., Babu G.J. Reducing sarcolipin expression mitigates Duchenne muscular dystrophy and associated cardiomyopathy in mice. Nat. Commun. 2017;8(1):1068. doi: 10.1038/s41467-017-01146-7. Oct 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazala D.A., Pratt S.J.P., Chen D., Molkentin J.D., Lovering R.M., Chin E.R. SERCA1 overexpression minimizes skeletal muscle damage in dystrophic mouse models. Am. J. Physiol. Cell Physiol. 2015;308(9):C699–C709. doi: 10.1152/ajpcell.00341.2014. May 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millay D.P., Sargent M.A., Osinska H., Baines C.P., Barton E.R., Vuagniaux G., Sweeney H.L., Robbins J., Molkentin J.D. Genetic and pharmacologic inhibition of mitochondrial-dependent necrosis attenuates muscular dystrophy. Nat. Med. 2008;14(4):442–447. doi: 10.1038/nm1736. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reutenauer J., Dorchies O.M., Patthey-Vuadens O., Vuagniaux G., Ruegg U.T. Investigation of Debio 025, a cyclophilin inhibitor, in the dystrophic mdx mouse, a model for Duchenne muscular dystrophy. Br. J. Pharmacol. 2008;155(4):574–584. doi: 10.1038/bjp.2008.285. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nogami K., Maruyama Y., Sakai-Takemura F., Motohashi N., Elhussieny A., Imamura M., Miyashita S., Ogawa M., Noguchi S., Tamura Y., Kira J.I., Aoki Y., Takeda S., Miyagoe-Suzuki Y. Pharmacological activation of SERCA ameliorates dystrophic phenotypes in dystrophin-deficient mdx mice. Hum. Mol. Genet. 2021;30(11):1006–1019. doi: 10.1093/hmg/ddab100. May 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated in this study is available in the NCBI database under the accession number GSE210799.