Abstract
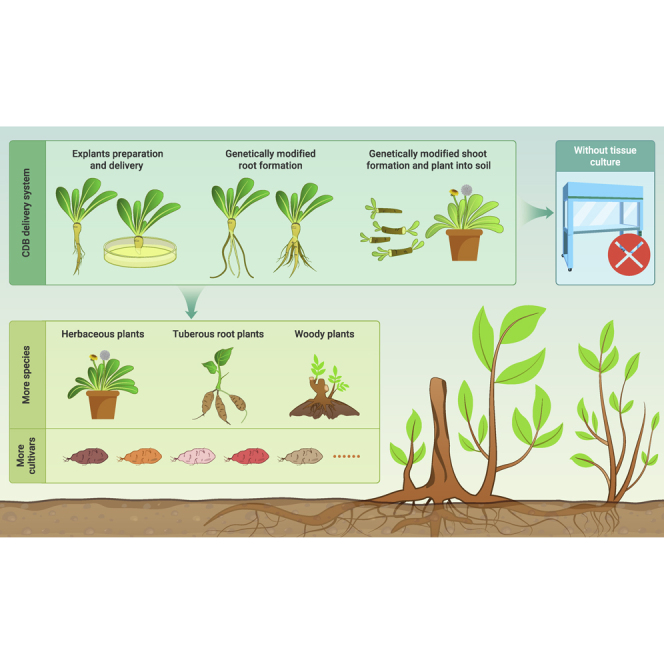
Of the more than 370 000 species of higher plants in nature, fewer than 0.1% can be genetically modified due to limitations of the current gene delivery systems. Even for those that can be genetically modified, the modification involves a tedious and costly tissue culture process. Here, we describe an extremely simple cut–dip–budding (CDB) delivery system, which uses Agrobacterium rhizogene to inoculate explants, generating transformed roots that produce transformed buds due to root suckering. We have successfully used CDB to achieve the heritable transformation of plant species in multiple plant families, including two herbaceous plants (Taraxacum kok-saghyz and Coronilla varia), a tuberous root plant (sweet potato), and three woody plant species (Ailanthus altissima, Aralia elata, and Clerodendrum chinense). These plants have previously been difficult or impossible to transform, but the CDB method enabled efficient transformation or gene editing in them using a very simple explant dipping protocol, under non-sterile conditions and without the need for tissue culture. Our work suggests that large numbers of plants could be amenable to genetic modifications using the CDB method.
Graphical abstract
Public summary
-
•
Gene delivery is the biggest hurdle in plant genetics research and breeding
-
•
Current delivery methods rely on tedious and costly tissue culture process
-
•
Current delivery methods can be applied to only a very small number of plants
-
•
The cut-dip-budding (CDB) delivery method is tissue-culture free and does not need sterile condition
-
•
The CDB method is extremely simple and potentially applicable to many plants
Introduction
Genome-editing technologies have revolutionized plant research and have the potential to significantly improve crops. The major obstacle to fully realizing this potential is the delivery of gene-editing tools into plant cells.1 Gene-editing tools are often delivered into plant cells by one of two means: Agrobacterium tumefaciens-mediated transformation or particle bombardment. These deliveries, however, are often inefficient. Besides requiring the tedious process of tissue culture, the transformation efficiencies of these delivery methods are plant species and genotype dependent.2 Moreover, most plant species are recalcitrant to genetic transformation. Even for the major crop species for which transformation protocols have been developed and perfected, the process is far from routine.1 This represents serious challenges to the practical application of gene-editing technology.
An improvement to the traditional methods of transformation is the use of developmental regulators during the process. In maize and sorghum, co-delivering transgenes with Wus2 and Bbm, which function as developmental regulators, expedites the production of transgenic plants.3,4 Similarly, overexpression of the wheat gene TaWOX5 from the WUSCHEL family increases the transformation efficiency in wheat with less genotype dependency than other methods.5 The chimeric growth factor GRF4–GIF1 can increase the efficiency and speed of regeneration in wheat, triticale, and rice and increases the number of transformable wheat genotypes.6 Unfortunately, the effects of developmental genes on transformation are less significant for dicot species, even though positive effects on transformation efficiency have been reported for canola, soybean, and sugarbeet when the Arabidopsis GRF5 gene was ectopically expressed in the explant tissues.7 Even for monocot species, the effects of developmental genes have been inconsistent.
By eliminating the need for tissue culture to obtain transformed plants, the floral dipping transformation protocol for Arabidopsis thaliana was an important development.8 The ideal solution for obtaining gene-edited plants would be the development of simplified protocols that, like the floral dipping method, do not require tissue culture. Many attempts have been made to engineer a plant virus-based system to deliver gene-editing tools into plants without the need for tissue culture. Even though delivery of single guide RNA (sgRNA) or gene-editing tools was successful,9,10,11 tissue culture was still needed to obtain heritable gene editing in all plant virus-based delivery systems reported to date.
Here, we describe a system for delivering genetic-modification tools to plants; we call this the cut–dip–budding (CDB) delivery system. Using the CDB delivery system in the current study, we successfully transformed six plant species in several families. With this system, genetically modified plants can be obtained without tissue culture, enabling stable genetic modification and genome editing of previously non-transformable or difficult-to-transform plant species.
Results
Transformation of Taraxacum kok-saghyz Rodin with the CDB delivery system
“Root suckering” refers to the developing of shoots from adventitious shoot primordia on roots. Many plant species in nature have developed this clonal propagation feature, enabling them to regenerate complete plants from roots (Figure S1).12 We hypothesized that the root-suckering ability of plants could be used to deliver genetic tools without the need for tissue culture. We tested our hypothesis first in the Russian dandelion T. kok-saghyz Rodin (TKS). We chose TKS not only because it has a strong root-suckering ability in the absence of any hormone treatment (Figure S1) but also because it has substantial economic potential, ie, TKS can produce high-quality rubber.13 Further improvement in its yield and other agronomic traits via gene editing could help TKS become a commercially viable and competitive rubber crop.14
TKS seedlings that were 3–4 weeks old were cut near the shoot–root junction in a non-sterile environment; the upper parts were then inoculated with Agrobacterium rhizogenes K599 at the cut site and cultured in vermiculite in an incubator (Figures 1A–1D). Because A. rhizogenes contains a root-inducing plasmid, hairy roots were formed about 2 weeks later. Some of these hairy roots were transgenic and GFP positive (Figures 1E and 1F). Their development was similar to that of hairy roots that were transgene negative. Explants with GFP-positive roots were transferred to nutrient-rich soil for further cultivation. After about 8–10 weeks, the GFP-positive roots had continued to grow and looked like the secondary roots of wild-type (WT) TKS plants (Figures 1G and 1H). Because root segments of WT TKS can regenerate buds without hormone treatment or tissue culture (Figure S1), we reasoned that transgene-positive roots could do the same. We detached GFP-positive roots, cut them into 2- to 3-cm-long root segments (Figures 1I and 1J), and placed the segments on the surface of moist, non-sterile soil for budding. After about 1 week, these GFP-positive root segments began to produce green buds that developed into normal-appearing shoots (Figure 1K). These shoots were GFP positive as indicated by the weak but clear GFP fluorescence (Figure 1L). We confirmed that these shoots were transgenic by PCR analysis of GFP (Figure S2A) and by TAIL-PCR detection of the transfer DNA insertion site (Figure S2B). These shoots grew normally into the reproductive stage and set seeds (Figure 1M).
Figure 1.
The CDB delivery system and genetic transformation of TKS
(A) CDB delivery system workflow. Seedlings that were 3–4 weeks old were cut and used as explants. The reporter gene or gene-editing constructs were delivered to plant cells near the cut site via A. rhizogenes. Hairy roots were formed after several weeks. The GFP-positive roots were segmented and cultured to give rise to transgene-positive or gene-edited shoots.
(B) A 3- to 4-week-old seedling.
(C) Explant. Gray arrow indicates the site of infection.
(D) Explants inoculated with Agrobacterium were cultured in vermiculite.
(E and F) Formation of GFP-positive hairy root of TKS.
(G and H) Positive hairy roots continued to develop.
(I and J) GFP-positive root segment.
(K and L) Transgene-positive shoots formed on a GFP-positive root segment.
(M) Transformed shoots that were planted into soil grew normally to the reproductive stage.
In (E), (F), (I), (J), (L), and (I), the sample on the left is untransformed control. In (F), (H), (J), and (I), samples were illuminated with fluorescent light. Red arrow indicates GFP fluorescence in transformed tissue. Bar: 1 cm.
This CDB delivery system was tested again using another reporter gene, RUBY, that can be directly visualized with the unaided eye.15 Roots stably expressing RUBY were obtained and gave rise to transgenic shoots (Figures S3 and S4). These results indicated that the CDB delivery system could be used to reproducibly create transgenic TKS plants.
Efficient delivery of gene-editing tools by the CDB delivery system
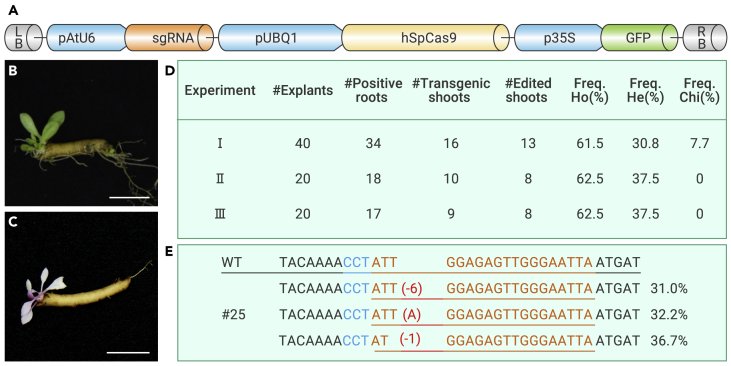
We next determined whether the CDB delivery system can be used to deliver gene-editing tools in order to create genome-edited TKS. Phytoene desaturase (PDS) was chosen as the target for easy identification of gene-edited plants. PDS is a key enzyme in the carotenogenic pathway, and its disruption results in albino plants. To knock out the TKS PDS gene, we inserted AtU6-driven sgRNA and AtUBQ1-driven Cas9 expression cassettes before the GFP cassette (Figure 2A) for simultaneous delivery of sgRNA and Cas9 as well as reporter GFP into the plant cells. Even though TKS is diploid (2n = 2X = 16),13 the TKS cultivar we used in the experiment has three copies of the PDS gene. Albino shoots appeared only when all three copies of PDS were edited (Figures 2B and 2C).
Figure 2.
Gene editing in TKS using the CDB deliver system
(A) Schematic of the TKS gene-editing vector.
(B) A shoot that was formed on a wild-type root segment.
(C) Albino phenotype of a homozygous TkPDS knockout shoot.
(D) The CDB delivery system efficiently delivered gene-editing tools and generated edited shoots in three independent experiments.
(E) Sequencing results of a representative TkPDS knockout albino seedling.
The sgRNA target sequence is shown in orange, the protospacer-adjacent motif is shown in blue, and mutations are shown in red. Bar: 1 cm.
We carried out a CDB experiment three times (experiments I–III) with the PDS gene in TKS as the target. In experiment I, which used the CDB protocol with 40 explants, 34 transgene-positive roots were obtained (Figure 2D). Among them, 16 generated transgene-positive shoots, 13 of which were successfully edited at PDS loci. Among the 13 edited plants, eight were homozygous (ie, all three copies of PDS were edited, and albino shoots were produced; Figure 2E), four were heterozygous (one or two copies of PDS were edited), and one was chimeric. Experiments II and III each used 20 explants. In both of these repeat experiments, similar percentages of homozygous and heterozygous edits were obtained (Figure 2D), suggesting that the CDB system is a highly reliable method for delivering gene-editing tools and for obtaining gene-edited plants.
CDB-mediated gene editing in sweet potato
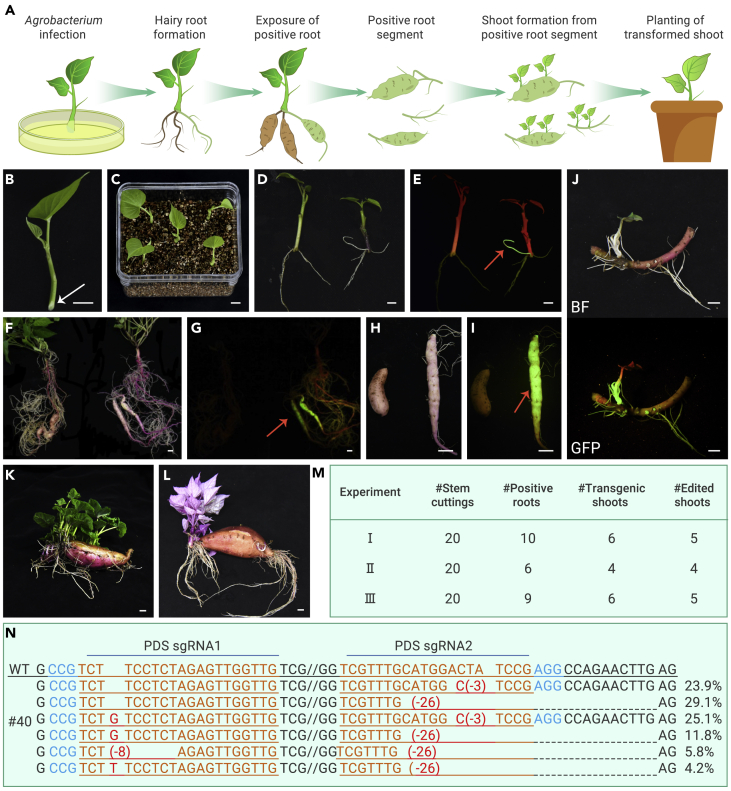
To determine whether the CDB delivery system can be applied to other species, we tried it on sweet potato (Ipomoea batatas [L.] Lam., Figure 3A). As a crop, sweet potato is important for the development of sustainable food supplies and economies in many parts of the world.16 Sweet potato is a typical tuberous root crop. Because tuberous roots can regenerate buds for vegetative propagation,17 sweet potato is an ideal crop for testing the CDB system.
Figure 3.
CDB-mediated gene editing in sweet potato
(A) Illustration of the CDB delivery process in sweet potato. Transgenes were delivered by A. rhizogene to sweet potato apical stem tissue, resulting in the induction of hairy roots under non-sterile conditions. After approximately 10 weeks, some of these hairy roots developed into tuberous roots. Transgenic (GFP-positive, indicated in brown color) roots (including tuberous roots) were harvested and allowed to bud. Transgene-positive shoots formed were transplanted into soil.
(B) Photograph of an explant used in the experiment; gray arrow points to the site of infection by A. rhizogene.
(C) Inoculated explants were inserted into vermiculite and cultured under a high-moisture environment.
(D and E) Transgenic hairy roots formed.
(F and G) Hairy roots were allowed to grow and some developed into tuberous roots.
(H and I) GFP-positive roots were cut into segments.
(J) Transgene-positive shoots formed on a root fragment.
(K) Shoots formed on a wild-type root segment.
(L) Albino shoots of a homozygous PDS knockout mutant.
(M) CDB delivery system efficiently delivered gene-editing tools into sweet potato and generated edited shoots in three independent experiments.
(N) Sequencing of the target region in PDS in an albino plant.
The sgRNA target sequence is shown in orange, the protospacer-adjacent motif is shown in blue, and mutations are shown in red. Red arrows in (E), (G), and (I) point to a GFP-positive signal. Bar: 1cm.
Apical stem cuttings of sweet potato (Figure 3B) were inoculated with Agrobacterium K599 and cultured in vermiculite in an incubator (Figure 3C). After about 3 weeks, GFP-positive transgenic hairy roots were formed (Figures 3D and 3E). These explants with induced hairy roots were transferred to nutrient-rich soil, and after about 10–12 weeks, GFP-positive hairy roots were thickened—some uniformly, and some into tuberous roots (Figures 3F and 3G). We cut the GFP-positive roots, including both the tuberous and non-tuberous roots, into fragments (Figures 3H and 3I), placed the fragments in soil, and allowed them to grow and generate shoots. All of the shoots that formed on GFP-positive roots were transgene positive (Figures 3J and S5).
We next determined whether the CDB delivery system can be used to obtain gene-edited sweet potato. We first tried the CDB system with Yanshu 25, a high-quality sweet potato cultivar, using the PDS gene as the target. To achieve robust editing in the highly heterozygous hexaploid background (2n = 6X = 90) of sweet potato,16 we designed two PDS sgRNAs using GlytRNA as the connector that can target all PDS homoeoalleles. The double-editing design worked well. We performed the experiment three times. In each experiment, we used 20 cut apical stems as explants and obtained albino plants (Figures 3K and 3L). From a total of 60 explants in all three experiments, 25 gave rise to transgene-positive roots. Among them, 16 generated transgenic shoots, and 14 produced gene-edited shoots (Figure 3M). We did sequence analysis using DNA isolated from the hairy roots and albino shoots. Both revealed simultaneous editing of the intended target sites (Figure 3N and S6).
Modifying the starch composition of sweet potato for human consumption and for various industrial applications has long been a major breeding goal for sweet potato, which is one of the top starch-rich root crops grown worldwide.18 To modify the starch composition in Yanshu25, we designed CRISPR–Cas9 constructs targeting the starch synthesis gene GBSSI, SBEI, or SBEII, or targeting both SBEI and SBEII, and obtained sweet potato lines with corresponding mutations (Figure S7). A crude determination of the starch content of the sweet potato tuberous roots showed that the knockout of the GBSSI gene drastically decreased the amylose content, from 27.4% (±0.9%) in the WT to 3.5% (±0.3%) in the gene-edited line. Knocking out SBEII alone or SBEI and SBEII together increased the amylose content in tuberous roots to 31.2% (±0.9%) or 33.8% (±3.3%) in the gene-edited lines compared with 27.4% (±0.9%) in the WT tuberose roots (Figure S8A). Iodine-staining of amylose and amylopectin showed that the sweet potato starch granules in the GBSSI knockout line turned red–brown, while the sweet potato starch granules with SBEII or SBEI and SBEII knockout were slightly deep blue (Figures S8B–S8F), which were consistent with their respective amylose contents.
Efficacy of the CDB delivery system in sweet potato is not genotype dependent
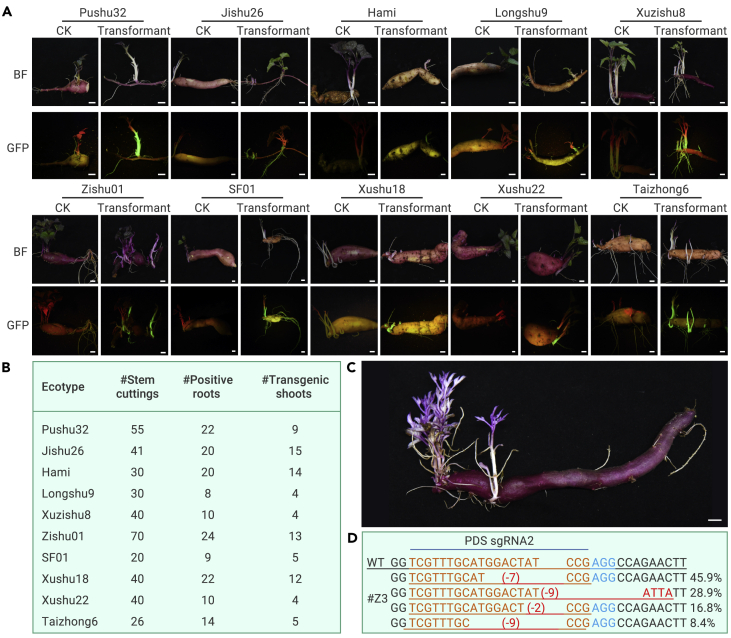
To evaluate whether the CDB delivery system works well with different cultivars of sweet potato, we tested the efficiency of the system on 10 sweet potato cultivars, including cultivar Jishu 26 and two purple sweet potato cultivars (Xuzhishu 8 and Zhishu 01), which are difficult to transform using traditional transformation methods.19 All 10 cultivars were transformed using the CDB delivery system (Figures 4A and 4B), suggesting that the CDB delivery system is not genotype dependent.
Figure 4.
CDB delivery system is genotype-independent in sweet potato
(A) GFP was transformed into 10 sweet potato cultivars via the CDB delivery system.
(B) The number of transgenic shoots obtained from each of the 10 sweet potato cultivars.
(C) Albino shoots indicating homozygous PDS knockout in the Xuzishu8 cultivar.
(D) Sequencing results of the albino shoot of Xuzhishu8.
Bar: 1cm.
To assess the gene-editing efficiency in the difficult-to-transform Xuzhishu 8 and Jishu26, we delivered a construct targeting PDS via the CDB system. We obtained albino seedlings in both cultivars (Figures 4C, 4D, S9A, and S9B). In addition, sgRNAs were designed to target the starch-branching enzymes SBEI and SBEII in Jishu26 and were successfully used to edit both genes (Figures S9C and S9D).
Application of the CDB delivery system in woody plants and a legume
Having success with transforming TKS and different cultivars of sweet potato, we speculated that the CDB delivery system could be used to transform other plant species that has the ability to form root suckers. We decided to further test the system in some previously non-transformable or difficult-to-transform species, such as trees and leguminous pasture plants, with the ability to form root suckers.
We first tried CDB delivery system in the transformation of three woody plant species: Ailanthus altissima (Mill) Swingle from the family Simaroubaceae, Aralia elata (Miq.) Seem. from the family Araliaceae, and Clerodendrum chinense Mabb. from the family Lamiaceae. All three were selected because of their roots’ ability to generate shoots, especially from root cuttings. The wood and bulk of A. altissima can be used to produce high-value-added non-cellulose gels and for astringent traditional Chinese medicine for the treatment of diarrhea and bleeding, respectively.20,21 A. elata is rich in saponins and is widely used in Chinese traditional medicine for various pharmacological effects.22 C. chinense has been used traditionally in the treatment of inflammation owing to its analgesic, anti-inflammatory, and anti-pyretic properties.23 The genetic transformation of A. altissima or C. chinense has not been reported. Although tissue–culture-dependent Agrobacterium tumefaciens-mediated genetic transformation of A. elata was reported,22 the transformation is still very difficult. We carried out the CDB protocol with these three species using RUBY as the reporter gene for the transformation of A. altissimae and GFP as the reporter gene for the transformation of A. elata and C. chinense. Positive hairy roots were formed on the infection site of explants from all three woody plant species, and after 3–4 months, these hairy roots grew into normal-appearing roots (Figures 5A, 5B, 5E and 5H). Except for C. chinense, which can directly generate transgenic buds from the GFP-positive roots during cultivation (Figure 5I and S10A), the transgenic-positive roots of the other two woody species were cleaned and segmented to promote bud formation (Figures 5C and 5F). Transgene-positive shoots were generated from the root segments after about 2–4 weeks (Figures 5D, 5G, S10B, and S10C). These results suggest that the CDB delivery system works for different families of woody plants.
Figure 5.
CDB delivery system was effective in transforming three woody plant species and a leguminous pasture plant
(A–D) Transformation of Ailanthus altissima (Mill) Swingle (Aa) via the CDB system.
(A) Hairy root formation on Aa explants.
(B) Hairy roots of Aa developed into normal-appearing roots.
(C) Segments of transgene (RUBY)-positive Aa roots.
(D) Transgene-positive shoot formed on an Aa root segment.
(E–G) Transformation of Aralia elata Seem. (Ae) via the CDB system.
(E) Hairy root formation and development into normal-appearing roots of Ae.
(F) Segments of transgene (GFP)-positive Ae roots.
(G) Transgene-positive shoot formed on an Ae root segment.
(H and I) Transformation of Clerodendrum chinense Mabb. (Cc) via the CDB system.
(J and K) Transformation of Coronilla varia L. (Cv) via the CDB system.
(L) Statistics for the three woody plant species (Aa, Ae, and Cc) and the leguminous pasture plant Cv transformed via the CDB delivery system.
Red arrow indicates transformed tissue. Bar: 1 cm.
Next, we evaluated the effectiveness of CDB delivery in transforming the leguminous pasture plant Coronilla varia L. from Fabaceae. The CDB delivery system worked well on this herbaceous plant. GFP-positive hairy roots and, subsequently, GFP-positive shoots were obtained (Figures 5J, 5K, and S10D). Taken together, these results suggest a broad applicability of the CDB delivery system in transforming a wide range of plant species.
Discussion
The delivery of genetic-improvement tools into plant cells has always been a bottleneck to gene editing and targeted crop improvement.1 Among the hundreds of thousands of higher plants in the world, only dozens of them are amenable to genetic transformation. At present, heritable gene editing can be attained for only a small number of plants and almost entirely through tissue–culture-dependent genetic transformation. The tissue–culture-based transformation efficiency is often genotype dependent and can be notoriously inconsistent, with efficiencies varying greatly among laboratories. The ideal solution is to develop a transformation system like the Arabidopsis floral dipping method that does not require tissue culture. At present, the floral dipping method can only be used to transform the model plant A. thaliana and a few close relatives without tissue culture.8
Many plant species have the ability to generate root suckers. In this study, we demonstrated that various plant species with such an ability can be stably transformed using the CDB delivery system. With the CDB system, transgenic seedlings can be obtained using an extremely simple protocol under non-sterile conditions and without tedious tissue culture procedures. In addition, the current results show that gene-editing tools can be efficiently delivered into plants via the CDB system.
It is reasonable to assume that the CDB delivery system can be used to transform and edit all plant species that can develop root suckers. If that assumption is correct, the system could facilitate the genetic improvement of numerous plants including economically important plants such as fruit trees (apple, grape, pear, peach, etc.) and berries (strawberry, blueberry, blackberry, red raspberry, etc.) that are difficult or impossible to transform using traditional transformation approaches. One limiting step in the conventional genetic improvement of fruit trees is the time required for the introgression of the corresponding alleles into elite lines after genetic crosses with lines harboring desired alleles. It takes many generations to somewhat restore the elite background, and in species with a long juvenile phase, it may require several decades for the selectable phenotype to emerge in each generation.24 Gene editing enables the direct generation of new alleles in elite varieties in a single generation, eliminating the need for repetitive backcrossing. The CDB system would also enable the creation of commercially desirable traits by generating genetic variation when no known source of variation (eg, seedless blackberries) is available.
More work is needed to apply the CDB delivery system to plants without root-suckering ability. Since root-suckering ability can be manipulated,25 it may be possible to discover regulators of root suckering. The use of such regulators may make many more plants amenable to transformation by the CBD system in the future.
Material and methods
Vector construction
All PCR clones were made using KOD-Plus-Neo DNA Polymerase (Toyobo, KOD-401s). GFP or RUBY reporters were constructed using pCAMBIA1300 as the backbone. The gene-editing Cas9 expression cassette and the AtU6 expression cassette were constructed as previously described26 between EcoR I and Hind III restriction sites in front of the reporter expression cassette. In addition, a 20-bp targeting sequence was designed and synthesized to be ligated between the Bsa I restriction sites of the gene-editing vector. For simultaneous editing of two sites within the same gene, Gly–tRNA scaffold was amplified with a specific primer containing a 20-bp targeting sequence27 and constructed into a gene-editing vector for expression driven by the promoter of the AtU6 cassette.
CDB delivery system
A. rhizogenes strain K599 was used to deliver transgene vectors or gene-editing vectors as previously described.28 The binary vector was transferred into strain K599 by electroporation. Colonies formed on the plate were selected and cultured overnight in TY liquid medium containing 10 mM CaCl2 and suitable antibiotics (28°C, 220 RPM). These clones were dispersed into 3–5 mL TY liquid medium and cultured overnight. A 300-μL aliquot of this overnight culture was spread onto TY solid medium and cultured until the medium was covered with a uniform layer of bacteria. Another 100-μL aliquot of this overnight culture was added to 100 mL TY liquid medium and was allowed to grow until the OD600 increased to one. Afterward, bacterial suspensions and bacteria harvested from solid media were used to directly infect explants without centrifugation and resuspension.
TKS explants were obtained from seedlings. TKS seeds were planted in soil. After about 3 weeks, seedlings (including the roots) were pulled from the soil. After soil and other impurities were removed from the roots with water, the lateral roots and most leaves were removed; only the main root and two to four young leaves were retained. The seedlings were cut with a blade near where the soil surface contacted the main root, and dipped with A. rhizogenes from a solid medium. The upper part of the cut seedling was inserted into a culture box containing moist vermiculite. Each stem was inoculated with 3–5 mL of the liquid Agrobacterium suspension, which flowed down the stem to the base. The inoculated TKS explant was incubated at 24°C and with a 16/18-h light/dark cycle. After about 2 weeks, transgenic-positive hairy roots had formed and were transferred to larger pots. After about 2 months, the root systems were rinsed with tap water to remove soil and other impurities. The roots were then cut into segments of about 2 cm with a scalpel, and the segments were placed on the surface of moist vermiculite and were kept at 24°C with an 18-h light/dark cycle until shoots emerged. The young shoots were transplanted into soil for normal vegetative and reproductive growth.
Sweet potato explants used in this research were apical buds that emerged and were cut from tuberous roots at 15 days after planting. Most leaves on the explants were removed to reduce water loss. The resulting explants had two nodes and two young leaves. After the cut site was coated with A. rhizogenes collected from a solid medium, the explant was planted in a culture box containing moist vermiculite. Immediately after planting, 3–5 mL of a liquid Agrobacterium suspension was added dropwise and was allowed to flow into the soil along the stem to the cut site. The inoculated explants were cultured at 26°C and with a 12/12-h light/dark cycle. After about 3 weeks, transgenic hairy roots had formed on the explants, which were transferred to a large pot and watered as needed. After about 2–3 months, the roots were washed with water and cut into multiple root segments, which were placed on moist vermiculite. Transgenic shoots emerged after several weeks.
For the woody plants A. altissima, A. elata, and C. chinense, young shoots were used as explants. The inoculation and cultivation methods were basically the same as with TKS and sweet potato. For A. altissima and A. elata, when the hairy roots grew into normal-appearing roots, they were cut into root segments of about 2–10 cm. These root segments were then placed on moist vermiculite for bud formation and shoot growth. For C. chinense, buds formed on GFP-positive roots without first segmenting the roots. The plants were cultivated according to the growth habits of the species.
C. varia explants were from 2-week-old seedlings. GFP-positive hairy roots formed 1–2 weeks after inoculation of the explant. After 2–3 months, buds formed on GFP-positive roots without first segmenting the roots.
DNA extraction and amplicon sequencing
Genomic DNA was extracted from plant tissues with the CTAB method. To determine the editing efficiency, the hairy roots were collected. The flanking sequence of the target site was amplified by PCR and subjected to Sanger sequencing directly or after TA cloning. Leaf tissues that grew from the root segments were also used to extract genomic DNA for determining gene-editing type and efficiency. The leaf genomic DNA was also used to verify the presence of the binary vector and root-inducing plasmid in the genome. The transgene insertion site was identified by three rounds of TAIL-PCR as previously described.29
Determination of amylose content
Starch and amylose were extracted and quantified from sweet potato tissue as previously described.18 Tuberous roots were kept at room temperature for 1 week before they were mashed and suspended in aqueous ethanol (80% v/v). After vigorous shaking, the suspension was passed through a 200-mesh sieve. After the filtrate had dried, 25 mg was dissolved in water, heated in boiling water for 10 min, and adjusted to a volume of 50 mL by the addition of distilled water. Amylose content was measured using a spectrophotometer at a wavelength of 710 nm. Pure potato amylose and amylopectin (Sigma, Shanghai, China) were used for constructing standard curves. For iodine staining of starch, 5 g iodine and 10 g potassium iodide were dissolved in 85 mL distilled water and diluted 80 times.30
Statistical analysis
The statistical tests, sample sizes, and replication are reported in the figure legends and tables.
Acknowledgments
This work was supported by Shandong Shunfeng Biotechnology Co., Ltd., Jinan, China. We are grateful to the Xuzhou Sweet Potato Research Center for providing sweet potato varieties XuZishu8, Xushu18, and Xushu22, to Prof. Zhe Sun of Tai’an Academy of Agricultural Sciences for providing the sweet potato variety Taizhong No. 6, and to Prof. Xuchu Wang of Hainan Normal University for providing the TKS seeds.
Author contributions
X.C., H.X., M.S., Z.L., G.L. and J.-K.Z. designed the research, X.C., H.X., M.S., J.L., P.M., B.H., M.W., and Y.T. carried out the experiments. X.C., H.X., M.S., F.C., J.P., Z.L., G.L., and J.-K.Z. analyzed the data. X.C., G.L., and J.-K.Z. wrote the paper.
Declaration of interests
The authors declare no competing interests.
Published Online: October 25, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xinn.2022.100345.
Contributor Information
Guofu Li, Email: liguofu@bellagen.cn.
Jian-Kang Zhu, Email: zhujk@sustech.edu.cn.
Lead contact website
Jian-Kang Zhu: https://iab.sustech.edu.cn/Research-details-pid-6-typeid-2.html
Supplemental information
References
- 1.Mao Y., Botella J.R., Liu Y., Zhu J.K. Gene editing in plants: progress and challenges. Natl. Sci. Rev. 2019;6:421–437. doi: 10.1093/nsr/nwz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ji X., Yang B., Wang D. Achieving plant genome editing while bypassing tissue culture. Trends Plant Sci. 2020;25:427–429. doi: 10.1016/j.tplants.2020.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Lowe K., La Rota M., Hoerster G., et al. Rapid genotype "independent" Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell Dev. Biol. Plant. 2018;54:240–252. doi: 10.1007/s11627-018-9905-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mookkan M., Nelson-Vasilchik K., Hague J., et al. Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators BABY BOOM and WUSCHEL2. Plant Cell Rep. 2017;36:1477–1491. doi: 10.1007/s00299-017-2169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang K., Shi L., Liang X., et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants. 2022;8:110–117. doi: 10.1038/s41477-021-01085-8. [DOI] [PubMed] [Google Scholar]
- 6.Debernardi J.M., Tricoli D.M., Ercoli M.F., et al. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 2020;38:1274–1279. doi: 10.1038/s41587-020-0703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong J., Martin-Ortigosa S., Finer J., et al. Overexpression of the transcription factor GROWTH-REGULATING FACTOR5 improves transformation of dicot and monocot species. Front. Plant Sci. 2020;11 doi: 10.3389/fpls.2020.572319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 9.Maher M.F., Nasti R.A., Vollbrecht M., et al. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 2020;38:84–89. doi: 10.1038/s41587-019-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T., Hu J., Sun Y., et al. Highly efficient heritable genome editing in wheat using an RNA virus and bypassing tissue culture. Mol. Plant. 2021;14:1787–1798. doi: 10.1016/j.molp.2021.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Ma X., Zhang X., Liu H., Li Z. Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9. Nat. Plants. 2020;6:773–779. doi: 10.1038/s41477-020-0704-5. [DOI] [PubMed] [Google Scholar]
- 12.Klimes L., Klimesova J., Hendriks R., Van Groenendael J. Clonal plant architecture: a comparative analysis of form and function. Ecology and Evolution of Clonal Plants. 1997:1–29. [Google Scholar]
- 13.Lin T., Xu X., Ruan J., et al. Genome analysis of Taraxacum kok-saghyz Rodin provides new insights into rubber biosynthesis. Natl. Sci. Rev. 2017;5:78–87. [Google Scholar]
- 14.Cherian S., Ryu S.B., Cornish K. Natural rubber biosynthesis in plants, the rubber transferase complex, and metabolic engineering progress and prospects. Plant Biotechnol. J. 2019;17:2041–2061. doi: 10.1111/pbi.13181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Y., Zhang T., Sun H., et al. A reporter for noninvasively monitoring gene expression and plant transformation. Hortic. Res. 2020;7:152. doi: 10.1038/s41438-020-00390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J., Moeinzadeh M.H., Kuhl H., et al. Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants. 2017;3:696–703. doi: 10.1038/s41477-017-0002-z. [DOI] [PubMed] [Google Scholar]
- 17.Zierer W., Rüscher D., Sonnewald U., Sonnewald S. Tuber and tuberous root development. Annu. Rev. Plant Biol. 2021;72:551–580. doi: 10.1146/annurev-arplant-080720-084456. [DOI] [PubMed] [Google Scholar]
- 18.Wang H., Wu Y., Zhang Y., et al. CRISPR/Cas9-Based mutagenesis of starch biosynthetic genes in sweet potato (Ipomoea batatas) for the improvement of starch quality. Int. J. Mol. Sci. 2019;20:4702. doi: 10.3390/ijms20194702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan L.P., Yu S.L., Chen C.J., et al. Cloning a peanut resveratrol synthase gene and its expression in purple sweet potato. Plant Cell Rep. 2012;31:121–131. doi: 10.1007/s00299-011-1145-4. [DOI] [PubMed] [Google Scholar]
- 20.Hong Z.L., Xiong J., Wu S.B., et al. Tetracyclic triterpenoids and terpenylated coumarins from the bark of Ailanthus altissima ("Tree of Heaven") Phytochemistry. 2013;86:159–167. doi: 10.1016/j.phytochem.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Almeida R.O., Ramos A., Alves L., et al. Production of nanocellulose gels and films from invasive tree species. Int. J. Biol. Macromol. 2021;188:1003–1011. doi: 10.1016/j.ijbiomac.2021.08.015. [DOI] [PubMed] [Google Scholar]
- 22.Liu W., Guo W., Chen S., et al. A high-quality reference genome sequence and genetic transformation system of Aralia elata. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.822942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wahba H.M., AbouZid S.F., Sleem A.A., et al. Chemical and biological investigation of some Clerodendrum species cultivated in Egypt. Pharm. Biol. 2011;49:66–72. doi: 10.3109/13880209.2010.494674. [DOI] [PubMed] [Google Scholar]
- 24.Alvarez D., Cerda-Bennasser P., Stowe E., et al. Fruit crops in the era of genome editing: closing the regulatory gap. Plant Cell Rep. 2021;40:915–930. doi: 10.1007/s00299-021-02664-x. [DOI] [PubMed] [Google Scholar]
- 25.Wan X., Landhäusser S.M., Lieffers V.J., Zwiazek J.J. Signals controlling root suckering and adventitious shoot formation in aspen (Populus tremuloides) Tree Physiol. 2006;26:681–687. doi: 10.1093/treephys/26.5.681. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z., Mao Y., Ha S., et al. A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 2016;35:1519–1533. doi: 10.1007/s00299-015-1900-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M., Mao Y., Lu Y., et al. Multiplex gene editing in rice with simplified CRISPR-Cpf1 and CRISPR-Cas9 systems. J. Integr. Plant Biol. 2018;60:626–631. doi: 10.1111/jipb.12667. [DOI] [PubMed] [Google Scholar]
- 28.Fan Y.L., Zhang X.H., Zhong L.J., et al. One-step generation of composite soybean plants with transgenic roots by Agrobacterium rhizogenes-mediated transformation. BMC Plant Biol. 2020;20:208. doi: 10.1186/s12870-020-02421-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y.G., Chen Y. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques. 2007;43 doi: 10.2144/000112601. 649-650, 652, 654 passim. [DOI] [PubMed] [Google Scholar]
- 30.Huang X., Su F., Huang S., et al. Novel Wx alleles generated by base editing for improvement of rice grain quality. J. Integr. Plant Biol. 2021;63:1632–1638. doi: 10.1111/jipb.13098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.