ABSTRACT
Objective
To assess the interactions between Streptococcus mutans and Candida albicans during cariogenic biofilm formation.
Methods
The S. mutans and C. albicans duo-species biofilms were formed in 1% sucrose to mimic the high caries risk challenges. The biofilm structure was assessed using two-photon laser confocal microscopy. The transcriptome of 48h-biofilms was assessed by RNA-Seq. The expression of S. mutans and C. albicans virulence genes was examined via real-time reverse transcription-polymerase chain reaction.
Results
The morphogenesis of C. albicans-S. mutans duo-species biofilms was significantly altered when comparing to S. mutans or C. albicans single-species biofilm. Duo-species biofilms exhibited unique expression profile with a large number of differentially expressed genes (DEGs), including a higher expression of S. mutans atpD (acid-adaptive), C. albicans CHT2 (fungal cell wall chitin remodeling), and C. albicans SOD3 (cytotoxic oxygen radical destroying) (p < 0.05). KEGG pathway analyses further revealed that the majority of the up-regulated DEGs are related to microbial metabolism. Furthermore, the expressions of S. mutans and C. albicans key virulence genes (gtfB, gtfC, gtfD, ECE1, HWP1, ERG4, CHT2) were associated with sugar availability-related and time-related dynamics.
Conclusion
Cross-kingdom interactions impact S. mutans-C. albicans biofilm formations and dynamic expressions of virulence genes.
KEYWORDS: Biofilms, dental caries, transcriptome, streptococcus mutans, candida albicans, oral infection
Introduction
Microbial interactions are crucial to maintaining microbial populations, microbiome structure, and ecosystem functions [1–3]. Ranging from mutualism to antagonism, interactions between bacteria and fungi have been in the spotlight because they play an essential role in driving biochemical cycles, maintaining balance in numerous ecosystems, and contributing to health and disease [4,5]. Interaction mechanisms have been elucidated for several pathogenic bacteria–fungi relationships, such as Candida albicans and the commonly isolated bacterial species Pseudomonas aeruginosa and Staphylococcus aureus [6], the microbial secondary metabolite-mediated interaction between the plant-pathogenic bacterium Ralstonia solanacearum and two plant-pathogenic fungal organisms Fusarium fujikuroi and Botrytis cinerea [7], and the characterization of 16 different bacterial-fungal pairs, examining the impact of 8 different fungi isolated from cheese rind microbiomes and two bacterial species (Escherichia coli and a cheese-isolated Pseudomonas psychrophila) [1].
Bacteria Streptococcus mutans and fungi Candida albicans are considered pathogenic microorganisms responsible for causing and accentuating oral diseases [8]. For instance, previous studies support the clinical importance of the association between S. mutans and C. albicans in the pathogenesis of early childhood caries (ECC) [9,10]. This relationship fosters initiation and maturation of cariogenic biofilms/plaque on the tooth surface, mediating cariogenic development [11,12], and ultimately causing ECC. Furthermore, our previous study examined the oral colonization of S. mutans and C. albicans among a cohort of underserved infants and indicated the early colonization of oral C. albicans is associated with a 3.5 times higher risk of S. mutans emergence when children turn one year of age. We also revealed a positive correlation between infant salivary S. mutans abundance and the C. albicans colonization [10].
In addition to clinical evidence of the symbiotic relationship between S. mutans and C. albicans, in vitro biofilm studies and animal models further support their collective role in caries disease [12–16]. A rat model showed that S. mutans and C. albicans co-infection led to a rapid onset of severe carious lesions on the smooth surfaces of teeth. The presence of C. albicans and S. mutans dramatically enhanced the assembly of an exopolysaccharide (EPS)-rich matrix, leading to the development of thicker biofilms than those formed by S. mutans or C. albicans alone [12]. Moreover, the duo-species biofilm matrix of S. mutans and Candida spp. delayed or blocked the antimicrobial diffusion into the biofilm, making treatment much more difficult or unsuccessful [17], dual-species biofilms are more resistant to stress conditions [16].
While attention has been devoted to examining the synergistic relationship between S. mutans and C. albicans, the molecular basis of S. mutans and C. albicans interplay, especially global transcriptomic analysis, remain under-investigated. Although transcriptome analysis of S. mutans in duo-species biofilms (i) confirmed the induction of specific genes (such as comS, sigX) and (ii) the stimulation of the complete quorum sensing system of S. mutans by C. albicans [18], as well as (iii) demonstrated a dramatically altered gene expression in S. mutans in the presence of C. albicans, however, the dual transcriptome of S. mutans and C. albicans interplay in duo-species biofilms remain unclear [19]. To better understand the microbial interactions between S. mutans and C. albicans during cariogenic biofilm development, we used an in vitro biofilm model that mimics a clinical high caries risk condition. We performed transcriptomic analysis via RNA-Seq and real-time reverse transcription-polymerase chain reaction (qRT-PCR) on single- and duo-species biofilms. Our results revealed distinctive transcriptomic profiles of S. mutans and C. albicans in duo-species biofilm development compared the single-species biofilms. We also revealed the dynamic expression of several virulence genes in biofilms and culture media. This knowledge may be vital to developing better strategies to prevent and treat ECC.
Materials and methods
Bacterial strains and starter preparation
S. mutans UA159 and C. albicans SC5314 were recovered from frozen stock using Blood agar (TSA with Sheep Blood, Thermo Scientific™ R01202) and YPD agar (BD Difco™, 242,720), respectively. After a 48-h incubation, 3–5 colonies of each species were inoculated into 10 ml of broth for overnight incubation (5% CO2, 37°C). S. mutans was grown in TSBYE broth (3% Tryptic Soy, 0.5%Yeast Extract Broth, BD Bacto™ 286,220, and Gibco™ 212,750) with 1% glucose; C. albicans was grown in YPD broth (BD Difco™, 242,820). The following morning, 0.5 ml of the overnight starter was added to the fresh broth and incubated for 3–4 h until reaching the mid-exponential phase with desirable optical density (the optical density at a wavelength of 600 nm (OD600) of 1.0 for S. mutans and OD600 of 0.8 for C. albicans). The morning starters were used for biofilm formation.
Biofilm models
We used a duo-species biofilm model to assess the interactions between S. mutans and C. albicans during biofilm formation; see Figure 1 for study flow. Single species biofilms of S. mutans or C. albicans alone were grown as controls. The biofilm method detailed previously [20,21] formed on the saliva-coated hydroxyapatite discs (0.50” diameter × 0.05” thickness, Clarkson Chromatography Products, Inc., South Williamsport, PA) was employed. The discs were placed vertically in a 24-well plate using a custom-made disc holder to mimic the caries-prone smooth tooth surfaces in the oral cavity [20]. The inoculation quantity of S. mutans [105 colony-forming unit (cfu)/ml] and C. albicans (103 cfu/ml) mimicked a high-risk clinical condition. Biofilms were formed in 2.8 ml of TSBYE broth with 0.1% (w/v) sucrose in a 24-well plate. The organisms were grown undisturbed to allow initial biofilm formation during the first 24 h at 5% CO2 and 37°C. At 24-h and 48-h, biofilms were transferred to a fresh cultural medium with 1% (w/v) sucrose to induce cariogenic challenge. Biofilms were collected at selected time points for biological (biofilm structure and cultural medium pH) and molecular (RNA-seq and qRT-PCR) assays. Independent assays were conducted three times with four replicated discs in each round.
Figure 1.
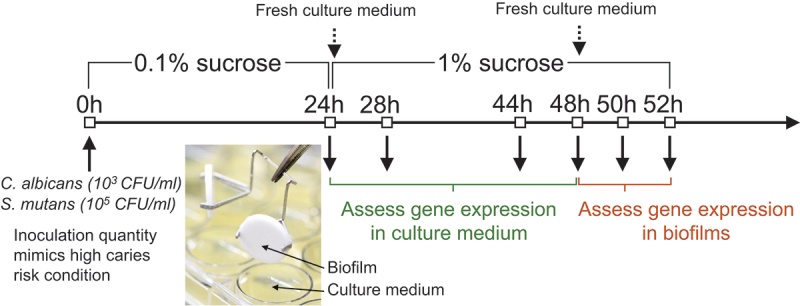
Study design The saliva-coated hydroxyapatite disc, consisting of similar components as in tooth enamel, was used as the substrate for biofilm formation. C. albicans and S. mutans were grown alone or together in 0.1% sucrose without disturbance until 24-h for initial biofilm establishment. The culture medium was changed once daily. To induce a cariogenic challenge, the culture medium was changed to 1% sucrose at 24-h and 48-h. The inoculation quantity of C. albicans (103 CFU/ml) and S. mutans (105 CFU/ml) was chosen to mimic high caries risk conditions in the clinical setting.
Laser scanning confocal fluorescence microscopy (LCSFM) imaging of biofilm matrix
We assessed two essential components of the biofilm matrix: bacteria and exopolysaccharides (EPS) using LCSFM methods detailed previously [18]. Briefly, 1 μM Alexa Fluor® 647-labeled dextran conjugate (Molecular Probes, Invitrogen Corp., Carlsbad, CA) was added to the culture medium from the beginning of and during the development of the biofilm to enable visualization of exopolysaccharides. The bacterial and fungal species were labeled by SYTO® 9 green-fluorescent nucleic acid stain (485/498 nm; Molecular Probes). The images were obtained using an Olympus FV 1000 two-photon laser scanning microscope (Olympus, Tokyo, Japan) equipped with a 10X (0.45 numerical aperture) water immersion objective lens [22]. Amira 5.0.2 (Mercury Computer Systems Inc., Chelmsford, MS) was used to create 3D renderings of EPS and bacteria of the biofilms detailed previously [22,23].
Transcriptome analysis by RNAseq
The 48 h biofilm formed by single-species (S. mutans or C. albicans) and duo-species (S. mutans+C. albicans) underwent RNA-seq analysis. The biofilms were harvested from four discs for each condition. The discs were incubated in RNALater (Applied Biosystems/Ambion, Austin, TX, USA) for 1 h before biomass collection. RNA extraction and purification were performed with MasterPure complete DNA and RNA purification kit (Epicenter, Lucigen, Wisconsin, USA). Raw RNA product was quantified by NanoDrop One Microvolume UV-Vis Spectrophotometer (Thermo Scientific™, Wilmington, DE, USA). The rRNAs were depleted with Ribozero rRNA Removal Kit (Illumina, San Diego, CA, USA). An RNA sequencing library was prepared using NEBNext Ultra RNA Library Prep Kit for Illumina by following the manufacturer’s recommended protocol (NEB, Ipswich, MA, USA). Briefly, enriched RNAs were fragmented at 94°C for 15 minutes. The cDNA fragments were end-repaired and adenylated at 3ʹends, and a universal adapter was ligated, followed by index addition and library enrichment with limited cycle PCR. Sequencing libraries were validated using the Agilent TapeStation 4200 (Agilent Technologies, Palo Alto, CA, USA) and quantified by using a Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) as well as by quantitative PCR (Applied Biosystems, Carlsbad, CA, USA).
Following the manufacturer’s instruction, the sequencing libraries were multiplexed and clustered on one flow cell lane and loaded on the Illumina HiSeq instrument. A 2 × 150 Paired-End (PE) configuration was used for sample sequencing. HiSeq Control Software (HCS) was used for image analysis and base calling. Raw sequence data generated from Illumina HiSeq was converted into FASTQ files and de-multiplexed using Illumina’s bcl2fastq 2.17 software. The sequence reads of all samples were deposited in the NCBI Sequence Read Archive (SRA) as a study under the accession number of PRJNA809829. One mismatch was allowed for index sequence identification. The sequence reads were trimmed using Trimmomatic v.0.36 and were mapped by the STAR aligner v.2.5.2b [24] to the reference genomes. Unique gene hit counts were calculated using feature Counts from the Subread package v.1.5.2. Unique reads within exon regions were counted. Gene hit counts were extracted, and the gene hit counts table was used for downstream differential expression analysis.
A comparison of gene expression between the groups of samples was performed using DESeq2. p-values and Log2 fold-changes were calculated by the Wald test. S. mutans genes with adjusted p-values (False Discovery Rate (FDR) p-values) <0.05 and absolute log2 fold changes >1 and C. albicans with adjusted p-values (False Discovery Rate (FDR) p-values) <0.05 and absolute log2 fold changes >3 were defined as differentially expressed genes (DEGs) for each comparison. The statistically significant genes underwent a gene ontology (GO) analysis by implementing the software GeneSCF v1.1 [25]. The GO list was used to cluster the set of genes based on their biological process and determine their statistical significance. The ‘plotPCA’ function within the DESeq2 R package was used for performing Principal component analysis (PCA); the first two principal components were plotted in a 2D plane for the samples. The top 500 genes, selected by highest row variance, were used to generate the plot. Volcano plots were generated by VolcaNoseR (https://huygens.science.uva.nl/VolcaNoseR) [26]. Kyoto Encyclopedia of Genes and Genomes pathways were generated by KEGG mapper (genome.jp) and Cytoscape software version 3.8.2.
Real-time reverse transcription polymerase chain reaction assay
qRT-PCR experiments were performed to validate genes of interest and specific genes associated with the virulence factors or viability of S. mutans or C. albicans. The genes and primers used are listed in Table S1. Methods were detailed previously [20].
In each experimental condition, RNAs in biofilms were collected and extracted from four discs, and RNAs in the culture medium were collected and extracted from the liquid of two wells (4 ml) from the 24-well plate. 0.2 µg of purified RNA was used as a template for cDNA synthesis with the BioRad iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc., Hercules, CA). cDNA and negative control samples were amplified using Applied Biosystems™ PowerTrack™ SYBR Green Master Mix and a QuantStudio™ 3 Real-Time PCR System (Thermo Fisher Scientific, USA). Each 20 µl reaction contained cDNA, 10 µM of each primer, and 2× SYBR-Green mix (SYBR-Green and Taq DNA Polymerase). gyrA for S. mutans genes [27] and ACT1 for C. albicans were used as the internal reference for comparative expression calculation. Data analysis was conducted using the comparative CT method [28].
Furthermore, to determine the dynamic changes of genes of interest during duo-species biofilm formation, we performed qRT-PCR assays for biofilms and culture medium at specific stages. The time points selected for gene expression assessment were before and after culture media change associated with a change in sugar availability. For the schematic design, see Figure 1.
Statistical analysis
Dynamic expression of genes was assessed by qRT-PCR. For the genes in biofilms, 2^ (-ddCT) value of each specific time point was compared with that at 48-h. For the genes in the culture medium, 2^ (-ddCT) value of each specific time point was compared with that at 24-h. Normality tests were first performed. When comparing the duo-species biofilm to the single-species biofilm, the comparison of the 2^ (-ddCT) values was assessed using the t-test for normally distributed data and the Mann-Whitney U test for non-normal data. When comparing the gene expression of different time points among the duo-species biofilms and culture medium, repeated ANOVA with a post hoc test was used after assessing data normality. Statistical tests were two-sided, with a significant level of 5%. IBM SPSS was used for statistical analyses.
Results
Presence of both S. mutans and C. albicans enhanced biofilm formation
The 48-h duo- and single-species biofilms were visualized by a two-photon laser confocal microscope. The 3D top and cross-sectional views of microcolonies are shown in Figure 2. Compared to the single species (Figure 2A and 2B), the morphogenesis of C. albicans-S. mutans duo-species biofilms (Figure 2C) was significantly altered. Microcolonies in biofilms are considered virulent and functional structures. Larger micro-colonies and significantly more EPS were formed in duo-species biofilms when compared to S. mutans single species biofilms. Well-formed mushroom-shaped microcolonies were identified grown in the duo-species group (Figure 2C). By comparison, microcolonies formed by S. mutans alone were less structured, with less bacteria content enmeshed with EPS (Figure 2A). Not surprisingly, few microcolonies and barely no EPS were formed in the C. albicans single-species biofilms; C. albicans does not form biofilms well alone under the selected culture conditions (Figure 2B).
Figure 2.
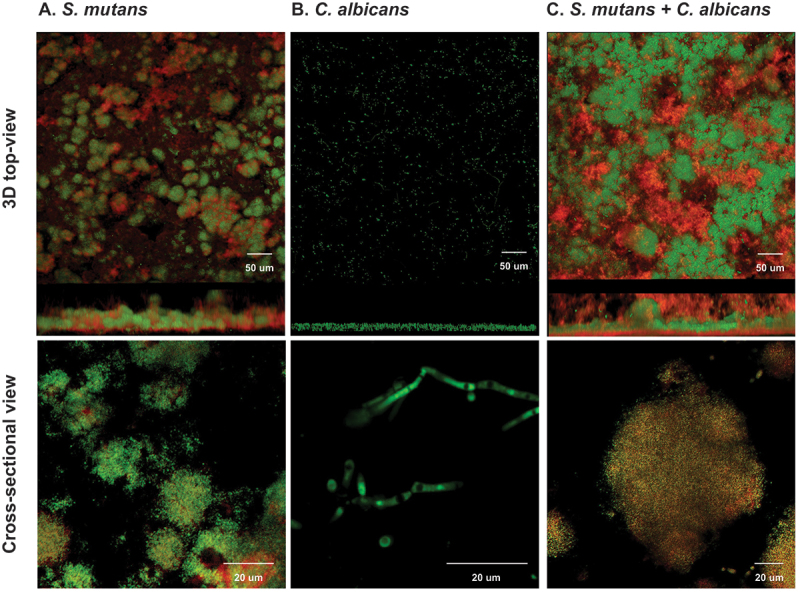
Morphogenesis of micro-colonies in 48-h biofilms (1% sucrose condition) In 1% sucrose condition, the 48-h biofilms were visualized by a two-photon laser confocal microscope. The biofilms of single species were shown in panel A (S. mutans) and panel B (C. albicans). The biofilms of duo-species (C. albicans and S. mutans) was shown in panel C. The confocal images indicate the cross-sectional and 3D-top views of biofilms; the green color indicates bacteria, and the red indicates the exopolysaccharides (EPS). Compared to the single species, the morphogenesis of C. albicans and S. mutans duo-species biofilms were significantly altered and characterized by formation of well-structured microorganism cluster enmeshed with EPS. These clusters are defined as microcolonies. In the biofilm formed by C. albicans alone, no microcolonies defined as above were identified.
Transcriptome analysis reveals interactions between S. mutans and C. albicans in biofilms
The Principal Component Analysis (PCA) (Fig S1) and the hierarchical clustering analysis (HCA) (Fig S2) indicate distinctive transcriptomic profiles of S. mutans and C. albicans in duo-species biofilm compared to those in single-species biofilm. Our data on the Q-Score distribution of RNA samples (Fig S3) support the validity of downstream analyses. The results of PCA and HCA also indicate high levels of correlation and reproducibility within the same sample types (single- and duo-species biofilms).
Volcano plots in Figure 3 indicate the transcriptomic comparison between duo- and single-species biofilms. Overall, 1915S. mutans and 6007 C. albicans genes were detected via RNA-seq. Among the detected genes, nearly half of the S. mutans and C. albicans genes were up-regulated in duo-species biofilms. We further assessed the DEGs [> (-) 1 Log2 fold change] of S. mutans genes; 49S. mutans DEGs were up-regulated and 13 were down-regulated. We also assessed the DEGs [> (-) 3 Log2 fold change] of C. albicans genes; 49 C. albicans DEGs were up-regulated and 43 were down-regulated. The qRT-PCR validation for selected genes of interest was consistent with the RNA-Seq data. The S. mutans virulence genes in the context of cariogenicity were up-regulated when S. mutans grew with C. albicans. The genes include atpD (related to acid stress tolerance response), eno (related to the degradation of carbohydrates via glycolysis), and lacC and lacG (related to galactose metabolism). Similarly, when grown with S. mutans, C. albicans genes related to fungal cell wall chitin remodeling (CHT2) and cytotoxic oxygen radicals destroying (SOD3) were significantly up-regulated [3.5 Log2 fold increase for CHT2 (p = 0.01), and 1.5 Log2 increase for SOD3 (p = 0.02)]. However, our results show that C. albicans hyphal-specific ECE1 were down-regulated in duo-species biofilms.
Figure 3.
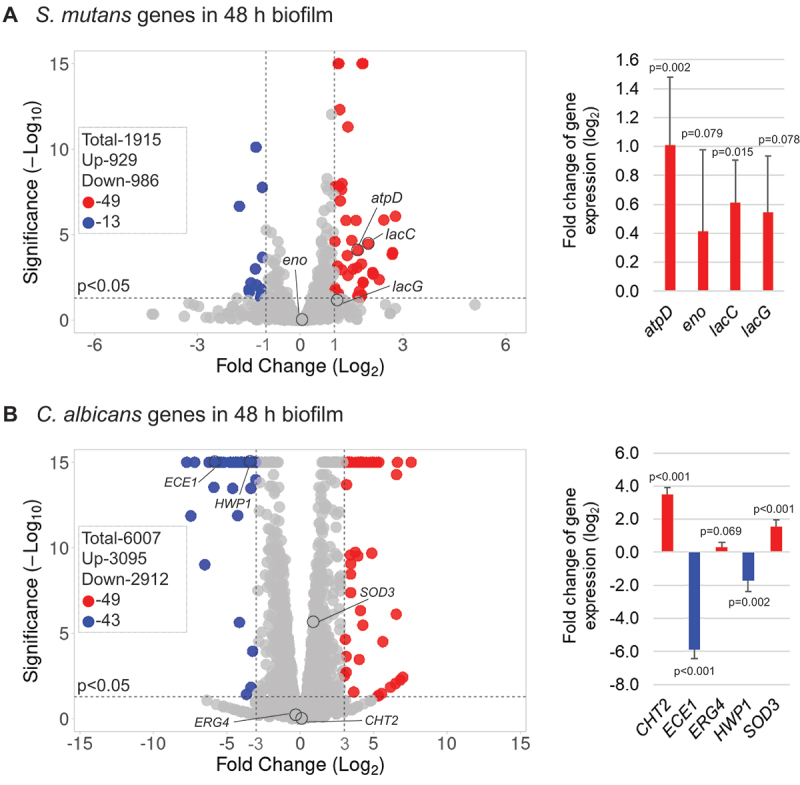
Comparison of transcriptomic profiling between duo-species biofilms with single-species biofilms (A) Volcano plots from transcriptome analysis of S. mutans in duo- (S. mutans + C. albicans) biofilm (48-h, 1% sucrose) compared to S. mutans in single-species biofilm. (B) C. albicans in duo-species biofilm (48-h, 1% sucrose) compared to C. albicans in single-species biofilm. Data represent three independent biological replicates of each condition. qRT-PCR validation results of selected genes are shown on the right side of each volcano plots.
We further performed KEGG pathway analyses to understand the functional association with gene expression. KEGG PATHWAY is a collection of pathway maps representing the knowledge of the molecular interaction, reaction and relation networks for metabolism, genetic information processing, environmental information processing, cellular processes, organismal systems, human diseases and drug development. mutans DEGs [> (-) 1 Log2 fold change] and 92 C. albicans DEGs [> (-) 3 Log2 fold change] were input into KEGG mapper for analysis. 17 pathways for S. mutans and 36 pathways for C. albicans were mapped in Figure 4 and Figure 5. KEGG pathway analyses showed that nearly all the S. mutans and C. albicans DEGs that related to metabolism pathways were up-regulated.
Figure 4.
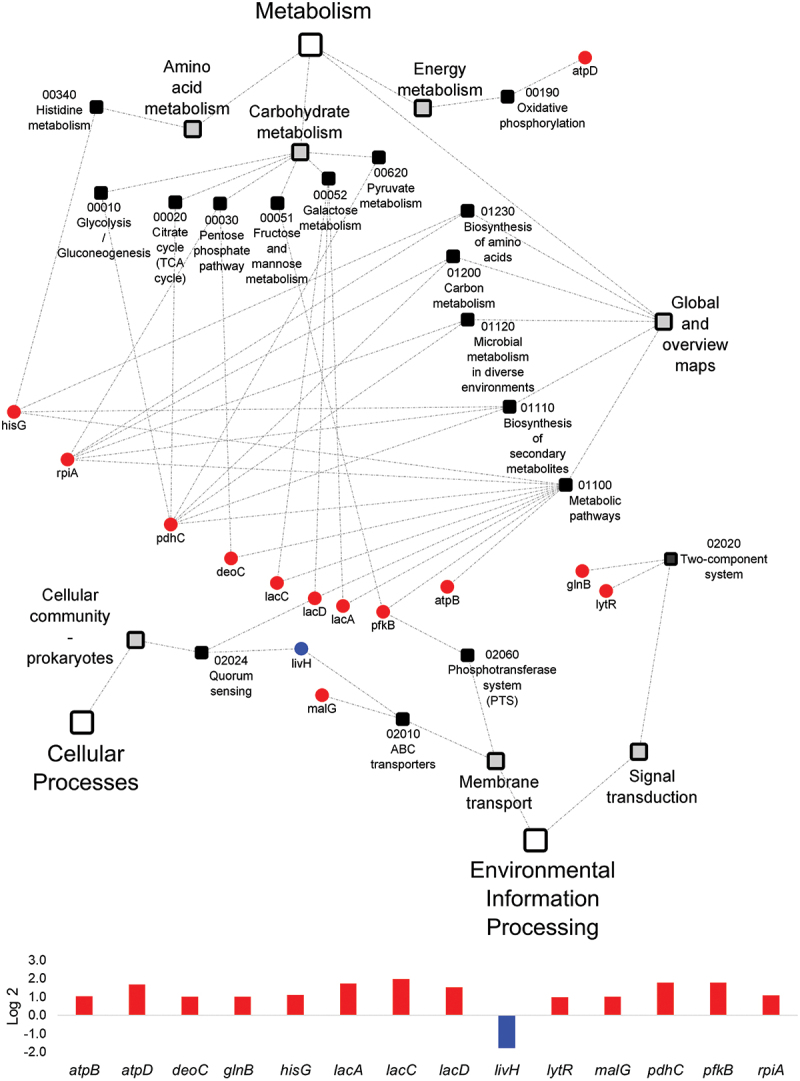
KEGG pathway network for S. mutans differentially expressed genes between the duo-species and single-species biofilms The genes of S. mutans that are differentially expressed between the comparison groups with FDR p-values < 0.05 and log2 fold changes > 1 were defined as S. mutans DEGs and listed in Supplementary Table 4. 17 impacted pathways were found for S. mutans DEGs. The fold change of the DEGs involved in the identified pathways are shown in the lower panel.
Figure 5.
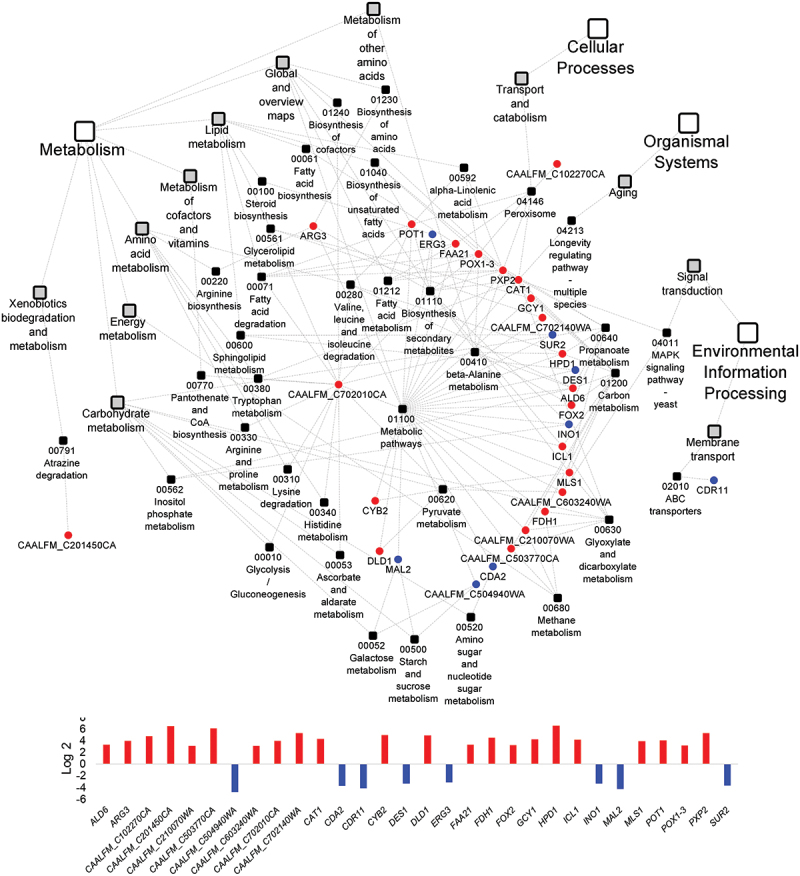
KEGG pathway network for C. albicans differentially expressed genes between the duo-species and single-species biofilms The genes of C. albicans that are differentially expressed between the comparison groups with FDR p-values < 0.05 and log2 fold changes > 3 were defined as C. albicans DEGs and listed in Supplementary Table 4. 36 impacted pathways were found for 92 C. albicans DEGs. The fold change of the DEGs involved in the identified pathways are shown in the lower panel.
Overall, S. mutans genes involved in metabolism were up-regulated, including carbohydrate metabolism, galactose metabolism, glycolysis/gluconeogenesis, and global and overall metabolism (Figure 4). For example, atpD, associated with acid stress tolerance response, was 1.7-fold up-regulated. The majority of C. albicans DEGs were up-regulated in the following pathways (Figure 5): metabolism, cellular processes, and organismal systems (aging process that was included in the organismal systems was involved). For instance, a 4-fold up-regulation of ARG3 in the pathway of arginine biosynthesis and a 5.4-fold up-regulation of PXP2 of fatty acid degradation was seen in duo-species biofilms. A limited number of C. albicans DEGs that are associated with environmental process were down-regulated; for instance, CDR11 was 4.1-fold down-regulated, which is an ATP-binding cassette transporter and drug efflux gene, essential for phenotypic expression. MAL2, a C. albicans promoter, was 4.2-fold down-regulated in the pathways of galactose metabolism and starch and sucrose metabolism. MAL2 is inducible by maltose and repressible by glucose [29].
Dynamic expression of genes of interest in duo-species biofilms and culture media
Compared to the 48-h biofilms (Figure 6), S. mutans genes related to EPS formation (gtfB, gtfC, and gtfD) were up-regulated after culture medium change; for example, gtfB was 26-fold up-regulated at 50-h and 6.5-fold up-regulated at 52-h, when comparing to the value of 48-h. S. mutans atpD (related to acid stress tolerance response) and eno (related to the degradation of carbohydrates via glycolysis) were also up-regulated at 50-h and 52-h. In contrast, genes related to lactose metabolism (lacC and lacG) were slightly down-regulated at 50-h and 52-h. Meanwhile, C. albicans genes related to C. albicans hyphal formation (ECE1 and HWP1) were significantly up-regulated following culture medium changes: ECE1 with a 50-fold change at 50-h and an 11.5-fold change at 52-h, and HWP1 with a 15.8-fold change at 50-h and a 7.2-fold change at 52-h. On the contrary, C. albicans ERG4 (related to antifungal medication resistance), SOD3 (related to cytotoxic oxygen radicals destroying), and CHT2 (related to fungal cell wall chitin remodeling) were slightly down-regulated at 50-h and 52-h.
Figure 6.
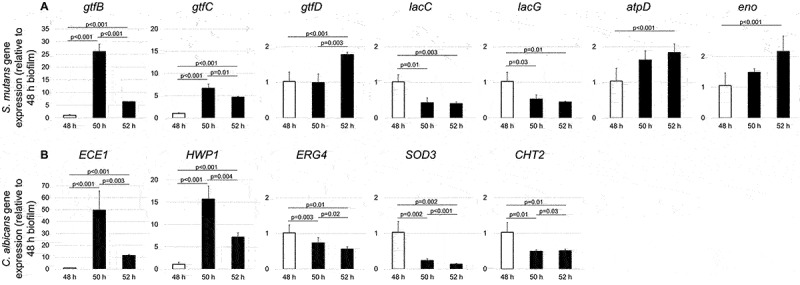
Dynamic expression of S. mutans and C. albicans genes in duo-species biofilms To determine the dynamic transcriptional changes in genes of interest during specific stages of biofilms formation, qRT-PCR was performed for biofilms at 50-h and 52-h. We compared S. mutans and C. albicans gene expression in the duo-species biofilm (S. mutans + C. albicans). Panel A indicated dynamic expression of S. mutans genes, and panel B indicated dynamic expression of C. albicans genes.
In addition to assessing gene expression in S. mutans and C. albicans in biofilm conditions, we further assessed the dynamic changes of these microorganism genes in the culture medium at the early stage of biofilm formation (scheme shown in Figure 1). When the culture medium at the later time points (28-h, 44-h, and 48-h) was compared to the culture medium at 24-h, S. mutans gtfB, lacC, and lacG were down-regulated, however, gtfD, atpD, and eno were up-regulated (Figure 7). gtfC was up-regulated a short time (28-h) after the culture medium change, followed by a down-regulation at 44-h and 48-h. S. mutans atpD (related to acid stress tolerance response) was 5.6-fold up-regulated in the 44-h culture medium. On the contrary, lacC and lacG (related to galactose metabolism) were significantly down-regulated at all time points after 24-h. For C. albicans, the expression of ECE1, HWP1, ERG4, and CHT2 in the culture medium was down-regulated at 28-h, 44-h, and 48-h (Figure 7). Interestingly, C. albicans SOD3 (related to cytotoxic oxygen radicals destroy) was significantly up-regulated with an 11.8-fold change at 44-h and a 9.5-fold change at 48-h.
Figure 7.
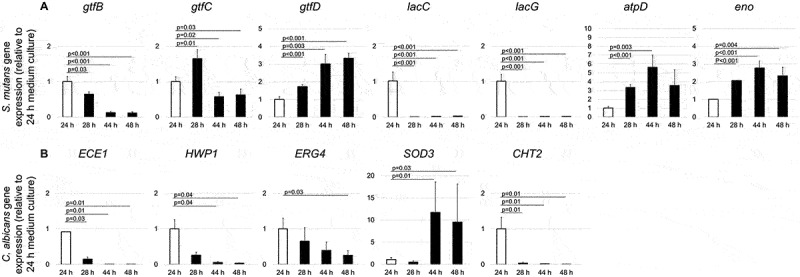
Dynamic expression of S. mutans and C. albicans genes in culture medium To determine the dynamic transcriptional changes in genes of interest during the earlier stages of biofilms formation, qRT-PCR was performed for culture medium at 24-h, 28-h, 44-h, and 48-h. We compared S. mutans and C. albicans gene expression of the culture medium of duo-species biofilm (S. mutans + C. albicans). Panel A indicated dynamic expression of S. mutans genes, and panel B indicated dynamic expression of C. albicans genes.
Discussion
The impact of the duo-species cross-kingdom interplay on S. mutans
Understanding the molecular basis for microbial interactions is critical to developing a new therapeutic strategy for oral polymicrobial infectious diseases, such as dental caries related to S. mutans and C. albicans colonization. In recent years, various studies focused on the role/contributions of S. mutans-derived exoenzyme glucosyltransferase B (gtfB), synergistic carbohydrate metabolism, and enhanced sugar metabolism in S. mutans and C. albicans mixed-species biofilms. This mixed-species biofilm contains an extensive matrix of extracellular α-glucans, which is produced by S. mutans gtfB. gtfB gene readily binds to C. albicans cells strongly and stably in an active form [30]. Mannans locates on the outer surface of C. albicans cell-wall mediates gtfB gene binding, enhancing glucan-matrix production and modulating bacterial-fungal association within the biofilm. S. mutans-secreted gtfB gene binds to the mannan layer of C. albicans to promote extracellular matrix formation and their co-existence within biofilms [31].
In the present study, the dynamic expression of the gtfB supports the key role of gtfB in developing duo-species biofilms over time. The gtfB in biofilms was significantly up-regulated at 50-h and 52-h (compared to 48-h). The highest biofilm gtfB expression occurred at 50-h, a short time after the culture medium was changed. Thus, gtfB expression might be induced by fresh culture medium, which is nutrient-rich and contains the sucrose necessary for production of α-glucan by gtfB. However, gtfB was down-regulated in the culture medium at 28-h, 44-h, and 48-h (compared to 24-h), suggesting gtfB is more highly expressed in biofilms than in the culture medium at these later time points. In agreement with our observations, gtfB expression is typically enhanced in biofilm versus planktonic modes of growth, consistent with its role in biofilm matrix formation [31].
Previous studies demonstrated that C. albicans stimulates S. mutans micro-colony development via mixed-species biofilm-derived metabolites. Consistently, in our study, all S. mutans DEGs (> (-) 1 Log2 fold change) related to metabolism pathways were up-regulated during the formation of duo-species biofilms. For instance, we observed the upregulation of S. mutans lacC and lacG (galactose metabolism) and atpD (related to acid stress tolerance response).
Compared to a previous study that showed the presence of C. albicans dramatically altered gene expression in mixed-species biofilms, our results for lacC and lacG expression was consistent [19]. He et al. revealed elevated pyruvate and galactose metabolism when S. mutans and C. albicans were grown together, suggesting co-cultivation influenced the carbohydrate utilization by S. mutans [19]. Among S. mutans DEGs, we found that the amount of up-regulated S. mutans DEGs in the duo-species biofilms was 3.8-fold higher than that of the down-regulated DEGs. This observation may indicate enhancement in the metabolism of S. mutans when grown with C. albicans.
We also found that S. mutans lytR and glnB, which are involved in a two-component system pathway related to environmental information processing, were up-regulated in duo-species biofilm. The LytR protein, encoded by ORF0317, belongs to the LytR/CpsA/Psr protein family. This family plays a putative role in maintaining cell wall structure. The lytR mutant S. mutans strain has a defect in cell division, and it shows increased autolysin activity [32]. The transcriptomic up-regulation of lytR expression during duo-species biofilms formation in this study indicated a lower autolytic activity and cell-wall re-modeling when co-culturing with C. albicans. Therefore, S. mutans likely has a more stable cell-wall structure in duo-species biofilms. GlnB belongs to the PII type signal transduction protein family, which regulates enzyme activity and gene expression and is involved in nitrogen regulation/metabolism, as well as glutamine synthesis activities in bacterial species. The PII protein encoded by glnB also regulates the expression of the putative ammonium transporter [33]. Our results confirm that when grown together, C. albicans augments expression of S. mutans genes involved in biofilm formation, virulence expression, and metabolism, which offers a mutual benefit to duo-species biofilm.
The impact of the duo-species cross-kingdom interplay on C. albicans
Most studies have focused on the effect of C. albicans on S. mutans virulence and biofilm formation. Therefore, we also focused on the molecular changes in C. albicans during duo-species interactions. We found that the transcriptomic profile of C. albicans was significantly altered when co-cultured with S. mutans in duo-species biofilms. Compared to C. albicans single species-biofilms, we identified 92 C. albicans DEGs (with over 3-fold Log2 fold change) in the duo-species biofilms. The majority of C. albicans DEGs that related to metabolism pathways were up-regulated, such as ARG3, PXP2, and CAT1. ARG3 is related to arginine biosynthesis. The critical roles of the C. albicans arginine biosynthesis pathway in its cross-kingdom interactions with Actinomyces viscosus in root caries were identified recently, and the study results indicated that targeting this pathway was a practical way to treat root caries caused by multiple species [34]. PXP2 relates to fatty acid degradation and the peroxisome pathway. The peroxisome plays an essential role in eukaryotic cellular metabolism, including beta-oxidation of fatty acids and detoxification of hydrogen peroxide [35]. CAT1 is a catalase-specific inhibitor that can suppress the hyphal growth of wild-type cells, and it is also involved in C. albicans peroxisome, MAPK signaling, and longevity regulating pathways.
We also pinpointed several down-regulated C. albicans genes in duo-species biofilms. For instance, CDR11 was down-regulated in the pathways of the environmental process. The expression of genes for major ABC transporters (CDRs) has been reported as being positively correlated with increased azole resistance in C. albicans isolates and is up-regulated during biofilm growth [36]. However, in this study, the transcriptional level of efflux gene CDR11 was decreased by 4.1-fold in duo-species biofilm, which might suggest that the presence of S. mutans represses the drug efflux activity of C. albicans, although further investigation is needed. It is plausible that the added benefit of growing with S. mutans may circumnavigate the need for high efflux pump expression. Namely, it has been demonstrated that co-culture with S. mutans can greatly increase biofilm size and matrix deposition, which acts a diffusion barrier to antimicrobials [37], which may reduce the need to efflux pumps.
In addition, the expression of C. albicans hyphal formation ECE1 and HWP1 in the culture medium was significantly repressed at 28-h, 44-h, and 48-h (compared to 24-h culture medium), while these genes were significantly induced in biofilms at 50-h and 52-h (compared to 48-h biofilm). This finding indicates that a biofilm condition is favorable for the expression of ECE1 and HWP1. This is consistent with previous findings showing a large portion of the S.mutans-C. albicans duo-species biofilm is comprised of C. albicans hyphae [12].
SOD3, a C. albicans gene associated with cytotoxic oxygen radical defense, was significantly induced in the culture medium at 44-h and 48-h, at a relative long period after culture medium change. This may reflect a response to increasing levels of oxidative stress in the culture media as these two organisms are growing together. Microbial respiration is a significant source of oxidative radicals [38]. SOD3 overexpression from a conditional promoter could substantially restore the cells’ oxidative stress resistance [39]. Co-culturing with S. mutans at a relatively long-time stage might enhance C. albicans’ tolerance for oxidative stress.
Overall, our results show the effects of duo-species biofilm represent a ‘two-way street’ where the presence of S. mutans influences C. albicans metabolism and behavior and vice versa. The following limitations are recognized with the intriguing finding: although our study results indicated the interactions between S. mutans and C. albicans during the formation of the duo-species biofilms at the transcriptomic level, further confirmation of these interactions needs to be conducted with mutant strains with the deletion of specific genes. Second, our study has not assessed the interactions at the protein level expression, which needs to be confirmed in the future. Third, cariogenic assessment of the duo-species interactions in the animal models was not conducted in our study; however, it deserves further investigation. This study is among the first to characterize the molecular mechanisms of duo-species biofilm formation and represents a launching point for future investigation.
Conclusions
The cross-kingdom interplay characterized here impacts S. mutans and impacts C. albicans metabolism, cellular processes, and organismal systems pathways. Furthermore, expression is not static and changes over time with the relative level of available carbohydrate (e.g. sucrose) and development stage. There are distinct differences between adherent biofilm cells and the planktonic milieu. It is clear from this study and others that the relationship between these two organisms is unique and enhances overall biofilm formation and virulence of these organisms, which likely contributes to ECC. Further targeting these interactions could be an ideal strategy for combating ECC.
Supplementary Material
Acknowledgments
Dr. Xiao’s research was supported by NIDCR (K23DE027412 and R01DE031025). The funding agencies had no role in the study design, data collection, analyses, decision to publish, or preparation of the manuscript.
Funding Statement
This work was supported by the National Institute of Dental and Craniofacial
Author contributions
YZ and JX contributed to the conception, design, data acquisition, analysis, and interpretation, drafting and critically revising the manuscript; ER and TTW contributed to data acquisition, analysis, and interpretation, drafting and critically revising the manuscript; XH contributed to data acquisition and data interpretation; MF was involved in critical revision of the manuscript. All authors have read and approved the final version of the manuscript and agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/20002297.2022.2144047
References
- [1].Pierce EC, Morin M, Little JC, et al. Bacterial-fungal interactions revealed by genome-wide analysis of bacterial mutant fitness. Nat Microbiol. 2021;6(1):87–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Deveau A, Bonito G, Uehling J, et al. Bacterial-fungal interactions: ecology, mechanisms and challenges. Fems Microbiol Rev. 2018;42(3):335–352 [DOI] [PubMed] [Google Scholar]
- [3].Scherlach K, Hertweck C.. Chemical mediators at the bacterial-fungal interface. In: Gottesman S, editor. Annual Review of Microbiology. Vol. 74 (Palo Alto: Annual Reviews; ). 2020. pp. 267–290. [DOI] [PubMed] [Google Scholar]
- [4].Kruger W, Vielreicher S, Kapitan M, et al. Fungal-bacterial interactions in health and disease. Pathogens. 2019;8(2):41. PubMed PMID: WOS:000475287800029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Santus W, Devlin JR, Behnsen J. Crossing kingdoms: how the mycobiota and fungal-bacterial interactions impact host health and disease. Infect Immun. 2021;89(4):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lindsay AK, Hogan DA. Candida albicans: molecular interactions with Pseudomonas aeruginosa and Staphylococcus aureus. Fungal Biol Rev. 2014;28(4):85–96. [Google Scholar]
- [7].Spraker JE, Wiemann P, Baccile JA, et al. Conserved responses in a war of small molecules between a plant-pathogenic bacterium and fungi. mBio. 2018;9(3):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Koo H, Andes DR, Krysan DJ. Candida-streptococcal interactions in biofilm-associated oral diseases. PLoS Pathog. 2018;14(12):e1007342. PubMed PMID: WOS:000454721500002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Koo H, Bowen WH. Candida albicans and Streptococcus mutans: a potential synergistic alliance to cause virulent tooth decay in children. Future Microbiol. 2014;9(12):1295–1297. PubMed PMID: WOS:000346478500001. [DOI] [PubMed] [Google Scholar]
- [10].Alkhars N, Zeng Y, Alomeir N, et al. Oral Candida Predicts Streptococcus mutans Emergence in Underserved US Infants. J Dent Res. 2022;101(1):54–62. PubMed PMID: WOS:000681130400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Metwalli KH, Khan SA, Krom BP, et al. Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS Pathog. 2013;9(10):5. PubMed PMID: WOS:000330383800009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Falsetta ML, Klein MI, Colonne PM, et al. Symbiotic relationship between streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 2014;82(5):1968–1981. PubMed PMID: WOS:000334666200024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kim D, Sengupta A, Niepa THR, et al. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci Rep. 2017;7:14. PubMed PMID: WOS:000392853400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ellepola K, Truong T, Liu Y, et al. Multi-omics analyses reveal synergistic carbohydrate metabolism in streptococcus mutans-candida albicans mixed-species biofilms. Infect Immun. 2019;87(10):20. PubMed PMID: WOS:000486620000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kim HE, Liu Y, Dhall A, et al. Synergism of Streptococcus mutans and Candida albicans reinforces biofilm maturation and acidogenicity in saliva: an in vitro study. Front Cell Infect Microbiol. 2021;10:11. PubMed PMID: WOS:000625515500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lobo CIV, Rinaldi TB, Christiano CMS, et al. Dual-species biofilms of Streptococcus mutans and Candida albicans exhibit more biomass and are mutually beneficial compared with single-species biofilms. J Oral Microbiology. 2019;11(1):15. PubMed PMID: WOS:000461908700001.;():. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Salehi B, Kregiel D, Mahady G, et al. Management of Streptococcus mutans-candida spp. oral biofilms’ infections: paving the way for effective clinical interventions. J Clin Med. 2020;9(2):16. PubMed PMID: WOS:000518823000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sztajer H, Szafranski SP, Tomasch J, et al. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. Isme J. 2014;8(11):2256–2271. PubMed PMID: WOS:000344454000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].He JZ, Kim D, Zhou XD, et al. RNA-Seq Reveals Enhanced Sugar Metabolism in Streptococcus mutans Co-cultured with Candida albicans within Mixed-Species Biofilms. Front Microbiol. 2017;8:15. PubMed PMID: WOS:000402856600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Xiao J, Klein MI, Falsetta ML, et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8(4):e1002623. PubMed PMID: 22496649; PubMed Central PMCID: PMCPMC3320608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zeng Y, Fadaak A, Alomeir N, et al. Lactobacillus plantarum Disrupts S. mutans-C. albicans Cross-Kingdom Biofilms. Front Cell Infect Microbiol. 2022;12:872012. PubMed PMID: WOS:000779553700001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Xiao J, Koo H. Structural organization and dynamics of exopolysaccharide matrix and microcolonies formation by Streptococcus mutans in biofilms. J Appl Microbiol. 2010;108(6):2103–2113. PubMed PMID: 19941630. [DOI] [PubMed] [Google Scholar]
- [23].Klein MI, Xiao J, Heydorn A, et al. An analytical tool-box for comprehensive biochemical, structural and transcriptome evaluation of oral biofilms mediated by mutans streptococci. J Vis Exp. 2011. 47. PubMed PMID: 21304472; PubMed Central PMCID: PMCPMC3182667. DOI: 10.3791/2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. PubMed PMID: WOS:000312654600003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Subhash S, Kanduri C. GeneSCF: a real-time based functional enrichment tool with support for multiple organisms. Bmc Bioinformatics. 2016;17(1):PubMed PMID: WOS:000383403300004. DOI: 10.1186/s12859-016-1250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Goedhart J, Luijsterburg MS. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci Rep. 2020;10(1):PubMed PMID: WOS:000596297800002. DOI: 10.1038/s41598-020-76603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zeng L, Burne RA. Comprehensive mutational analysis of sucrose-metabolizing pathways in streptococcus mutans reveals novel roles for the sucrose phosphotransferase system permease. J Bacteriol. 2013;195(4):833–843. PubMed PMID: WOS:000316961200021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C-T method. Nat Protoc. 2008;3(6):1101–1108. PubMed PMID: WOS:000258423700018. [DOI] [PubMed] [Google Scholar]
- [29].Brown DH, Slobodkin K IV. Stable transformation and regulated expression of an inducible reporter construct in Candida albicans using restriction enzyme mediated integration. Mol Gen Genet. 1996;251(1):75–80. [DOI] [PubMed] [Google Scholar]
- [30].Hwang G, Marsh G, Gao L, et al. Binding Force Dynamics of Streptococcus mutans-glucosyltransferase B to Candida albicans. J Dent Res. 2015;94(9):1310–1317. PubMed PMID: WOS:000360188900017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hwang G, Liu Y, Kim D, et al. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 2017;13(6):25. PubMed PMID: WOS:000404511700019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chatfield CH, Koo H, Quivey RG. The putative autolysin regulator LytR in Streptococcus mutans plays a role in cell division and is growth-phase regulated. Microbiology-(UK). 2005;151(2):625–631. PubMed PMID: WOS:000227031000028. [DOI] [PubMed] [Google Scholar]
- [33].Ardin AC, Fujita K, Nagayama K, et al. Identification and Functional Analysis of an Ammonium Transporter in Streptococcus mutans. PLoS One. 2014;9(9):e107569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Xiong KX, Zhu HL, Li YY, et al. The Arginine Biosynthesis Pathway of Candida albicans Regulates Its Cross-Kingdom Interaction with Actinomyces viscosus to Promote Root Caries Microbiology Spectrum 2022. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen YL, Yu QL, Wang HG, et al. The malfunction of peroxisome has an impact on the oxidative stress sensitivity in Candida albicans. Fungal Genet Biol. 2016;95:1–12. PubMed PMID: WOS:000384871800001. [DOI] [PubMed] [Google Scholar]
- [36].Ramage G, Bachmann S, Patterson TF, et al. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J Antimicrob Chemother. 2002;49(6):973–980. PubMed PMID: WOS:000175911600012. [DOI] [PubMed] [Google Scholar]
- [37].Cloete TE. Resistance mechanisms of bacteria to antimicrobial compounds. Int Biodeterior Biodegrad. 2003;51(4):277–282. PubMed PMID: WOS:000183480800009. [Google Scholar]
- [38].Fasnacht M, Polacek N. Oxidative stress in bacteria and the central dogma of molecular biology. Front Mol Biosci. 2021;8. DOI: 10.3389/fmolb.2021.671037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Liu NN, Uppuluri P, Broggi A, et al. Intersection of phosphate transport, oxidative stress and TOR signalling in Candida albicans virulence. PLoS Pathog. 2018;14(7):23. PubMed PMID: WOS:000440351300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


