Abstract
Anthrax lethal toxin (LeTx), consisting of protective antigen (PA) and lethal factor (LF), rapidly kills primary mouse macrophages and macrophage-like cell lines such as RAW 264.7. LF is translocated by PA into the cytosol of target cells, where it acts as a metalloprotease to cleave mitogen-activated protein kinase kinase 1 (MEK1) and possibly other proteins. In this study, we show that proteasome inhibitors such as acetyl-Leu-Leu-norleucinal, MG132, and lactacystin efficiently block LeTx cytotoxicity, whereas other protease inhibitors do not. The inhibitor concentrations that block LF cytotoxicity are similar to those that inhibit the proteasome-dependent IκB-α degradation induced by lipopolysaccharide. The inhibitors did not interfere with the proteolytic cleavage of MEK1 in LeTx-treated cells, indicating that they do not directly block the proteolytic activity of LF. However, the proteasome inhibitors did prevent ATP depletion, an early effect of LeTx. No overall activation of the proteasome by LeTx was detected, as shown by the cleavage of fluorogenic substrates of the proteasome. All of these results suggest that the proteasome mediates a toxic process initiated by LF in the cell cytosol. This process probably involves degradation of unidentified molecules that are essential for macrophage homeostasis. Moreover, this proteasome-dependent process is an early step in LeTx intoxication, but it is downstream of the cleavage by LF of MEK1 or other putative substrates.
Bacillus anthracis, a gram-positive spore-forming bacterium, is the causative agent of anthrax. The three secreted proteins which together are called anthrax toxin are PA, LF, and EF. By interacting with a cell surface protein receptor, PA mediates the endocytosis and translocation of EF and LF into the cytosol (14, 24, 32). EF is a calmodulin-dependent adenylate cyclase (23). LF is a metalloprotease with the consensus zinc-binding site HEXXH (21). A cellular substrate for LF was recently identified as MEK1, which is inactivated after cleavage of the N-terminal seven amino acids (7, 41). The combination of LF and PA, termed LeTx, is the major contributor to virulence in infected animals, as proven by the >1,000-fold decrease in virulence when the LF gene is inactivated (33). LeTx is cytolytic for some primary mouse macrophages and macrophage-like cell lines (9, 34). It remains unknown whether cleavage of MEK1 is sufficient to cause cytolysis of macrophages or whether cleavage of other substrates accounts for the final cytolysis.
Previous studies to identify the cellular mechanism of action of LeTx have identified a series of physiological changes that precede macrophage lysis. The earliest events, beginning 45 min after toxin challenge, are an increase in permeability to Na+ and Rb+ and a conversion of ATP to ADP and AMP. Later events include alterations in membrane permeability to Ca2+, Cr2+, Cl−, SO42−, amino acids, and uridine, beginning at 60 min; inhibition of macromolecular synthesis, leakage of cellular lactate dehydrogenase, and onset of gross morphological changes, beginning at 75 min; and cell lysis, beginning at 90 min (16–18). Certain inhibitors of endopeptidases have been shown to block intoxication of macrophages by LeTx (21, 26). However, not all of these inhibit the in vitro proteolytic activity of LF (13), suggesting that they act on events downstream in the cytolytic cascade that follows the initial proteolytic cleavage event(s) catalyzed by LF. Moreover, protein synthesis has been shown to be required for expression of LeTx cytotoxicity (1). LeTx-induced cytotoxicity can be switched to apoptosis under narrow conditions when cells were preincubated with the protein phosphatase inhibitor calyculin A (25), suggesting that apoptotic and LeTx-induced death mechanisms may have similarities and may utilize some of the same cellular components.
Although the caspases are the central proteases involved in apoptosis, recent studies have shown that the proteasome also plays a role in some apoptotic pathways. The proteasome is a multicatalytic protease that accounts for the major extralysosomal degradation of cellular proteins. The 20S proteasome is a large (∼700 kDa), hollow, cylindrical complex composed of four stacked rings, each containing seven subunits. The subunits of the inner rings are associated with the proteolytic activities of the complex. The 20S proteasome forms the catalytic core of the larger 26S proteasome, which is responsible for the ATP-dependent degradation of proteins tagged for destruction by ubiquitin as well as nonubiquitinated substrates (39). The proteasome participates in a number of proteolytically mediated intracellular processes, including the rapid elimination of proteins with abnormal structures (4, 19) and the temporal reduction in levels of regulatory proteins critical for control of the cell cycle and transcription (28). The proteasome plays a role in apoptosis, as shown by the ability of proteasome inhibitors to either induce (5) or, more commonly, inhibit (12, 20, 36) apoptosis, depending on the cell type and phase of the cell cycle examined.
To identify the molecular events by which LF proteolytic action in the cytosol leads to macrophage lysis, we extended studies of protease inhibitors (21) to include recently identified agents specific for caspases and the proteasome. The specific proteasome inhibitors efficiently protected macrophages from LeTx but did not block cleavage of MEK1, the substrate of LF. This suggests that one of the downstream events following the cleavage of MEK1 or other putative substrates of LF is the degradation of certain protein molecules and that this is an essential step in the cascade leading to macrophage lysis.
MATERIALS AND METHODS
Abbreviations.
Abbreviations used in this paper are as follows: ALLN, acetyl-Leu-Leu-norleucinal; ALLM, acetyl-Leu-Leu-methioninal; AMC, 7-amino-4-methylcoumarin; DEVD-FMK, Asp-Glu-Val-fluoromethylketone; EF, anthrax toxin edema factor; LeTx, anthrax lethal toxin; LF, anthrax toxin lethal factor; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MEK1, mitogen-activated protein kinase kinase 1; MTT, 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl-tetrazolium bromide; PA, anthrax toxin protective antigen; SLLVY, succinyl-Leu-Leu-Val-Tyr; YVAD-CMK, Tyr-Val-Ala-Asp-chloromethylketone; Z-LLE, benzyloxycarbonyl-Leu-Leu-Glu; Z-VKM, benzyloxycarbonyl-Val-Lys-Met.
Reagents.
Lactacystin, MG132 (carbobenzyloxy-Leu-Leu-leucinal), ALLN (also called calpain inhibitor I), ALLM (also called calpain inhibitor II), and the fluorogenic proteasome substrates SLLVY-AMC, Z-LLE-AMC, and Z-VKM-AMC were purchased from Calbiochem (San Diego, Calif.). The caspase inhibitors YVAD-CMK and DEVD-FMK were from Clontech. Other protease inhibitors were from Sigma (St. Louis, Mo.). Rabbit polyclonal antibodies against the N terminus (amino acids 2 to 18) of mouse MEK1 and the C terminus of Xenopus MEK1 (amino acids 360 to 378) were purchased from Upstate Biotechnology (Lake Placid, N.Y.). Rabbit polyclonal antibodies against IκB-α and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.).
Cell culture.
Murine macrophage-like cells, RAW 264.7 (ATCC TIB-71), were obtained from the American Type Culture Collection (Manassas, Va.). The cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Cells were grown in a humidified atmosphere of 5% CO2 at 37°C.
Cytotoxicity assays.
RAW 264.7 cells were grown in 96-well plates to 70% confluence. Inhibitors were diluted serially in complete medium and applied to the cells. The PA and LF mixture in medium was immediately applied to the cells with each at a final concentration of 500 ng/ml. After incubation at 37°C for 2 h, MTT was added to the cells to a final concentration of 0.5 mg/ml. After another 2-h incubation, the medium was removed and the blue pigment was dissolved in 0.5% sodium dodecyl sulfate–40 mM HCl–90% isopropanol. The A570 was measured in a microplate reader (Molecular Devices, Menlo Park, Calif.). Positive controls were cells treated with PA and LF in absence of inhibitors, whereas negative controls were cells treated with neither toxin nor inhibitors. Cell viability was calculated as 100 × (Ax − Apc)/(Anc − Apc), where Ax, Apc, and Anc are the A570 of the sample, the positive control, and the negative control, respectively.
Preparation of cell lysates and Western blotting.
RAW 264.7 cells were grown in six-well plates to confluence and treated with inhibitors and toxins, as indicated, at 37°C. The cells were washed with cold Hanks balanced salt solution without Ca2+ and Mg2+ and lysed in 120 μl of cold lysis buffer (1% Triton X-100, 1% sodium deoxycholate, 25 mM HEPES [pH 7.5], 150 mM NaCl, 20 mM NaF, 2 mM dithiothreitol, 1 mM EDTA, 1 mM EGTA, 0.2 mM sodium vanadate, 20 mM β-glycerol phosphate, 100 μg of phenylmethylsulfonyl fluoride per ml, 2 μg of aprotinin per ml, 2 μg of leupeptin per ml, 10 μg of p-aminobenzamidine per ml). The cell lysate was centrifuged and the supernatant was mixed with sodium dodecyl sulfate sample buffer for electrophoresis. Proteins were blotted onto a nitrocellulose membrane and probed with antibodies against MEK1 N or C terminus or antibodies against IκB-α. Horseradish peroxidase conjugated secondary antibodies were used, and a blotting signal was developed with SuperSignal chemiluminescent substrate from Pierce (Rockford, Ill.).
ATP measurement.
RAW 264.7 cells were grown in 96-well plates to 70% confluence. The cells were treated with inhibitors and toxins as indicated at 37°C for 1 h. Intracellular ATP was released and determined by luciferin/luciferase with a bioluminescent somatic cell assay kit from Sigma, according to the manufacturer’s instructions. Luminescence was measure on a Monolight model 2010 luminometer (Analytical Luminescence Laboratory, San Diego, Calif.) with 2-s delay and 10-s signal integration. Concentrations of ATP were calculated by comparison to a standard curve obtained with pure ATP. Positive controls were cells treated with 10 mM 2-deoxy-d-glucose and 10 μM antimycin in Dulbecco’s phosphate-buffered saline supplemented with 2 mM Ca2+ and 1.5 mM Mg2+. Negative controls were cells with no treatment. Percentage of control was calculated as 100× (Cx − Cpc)/(Cnc − Cpc), where Cx, Cpc, and Cnc are the ATP concentrations of the sample, the positive control, and the negative control, respectively.
Measurement of proteasome activity.
RAW 264.7 cells were dissociated from plates with cell dissociation buffer (GIBCO-BRL, Rockville, Md.). The cells were suspended and treated with or without toxin for 1 h at 37°C. After centrifugation, the cells were suspended in cytoplasmic buffer (50 mM Tris-HCl buffer [pH 8.0] containing 140 mM KCl, 10 mM glucose, 2 mM ATP, 5 mM MgCl2, 1 mM EGTA, 0.5 mM dithiothreitol, and 10% glycerol and supplemented with a protease inhibitor cocktail [100 μg of phenylmethylsulfonyl fluoride per ml, 2 μg of aprotinin per ml, 2 μg of leupeptin per ml, 1 μg of pepstatin per ml, 10 μg of p-aminobenzamidine per ml, and 50 μM E-64). The cells were sonicated in the presence or absence of 20 μM lactacystin, and 50 μl of cell extracts containing 150 μg of protein was mixed with 50 μl of an 80 μM fluorogenic substrate (SLLVY-AMC, Z-LLE-AMC, or Z-VKM-AMC). The reaction mixtures were incubated at 37°C for 15 min. The results were read on an LS-50 fluorescence spectrometer (Perkin-Elmer, Norwalk, Conn.) with excitation at 380 nm and emission at 460 nm. Specific proteasome activity was calculated as the difference between fluorescence intensities in the absence and in the presence of lactacystin.
RESULTS
Proteasome-specific inhibitors block the cytotoxicity of LeTx.
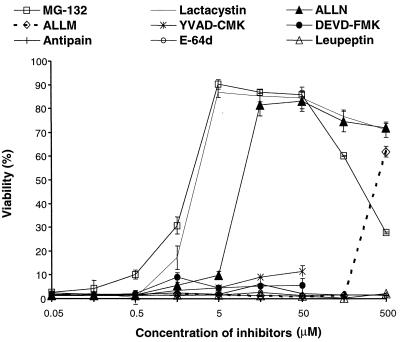
Protease inhibitors with various target specificities were serially diluted and applied to RAW 264.7 cells. PA and LF were each added at a final concentration of 500 ng/ml for 2 h, and cytotoxicity was measured by the MTT assay. As shown in Fig. 1, all of the proteasome inhibitors, i.e., MG132, lactacystin, and ALLN, were very potent in inhibiting the cytotoxicity of LeTx. MG132 alone had some toxicity to the cells at concentrations above 50 μM. Both YVAD-CMK, a caspase-1 inhibitor, and DEVD-FMK, a caspase-3 inhibitor, were ineffective at 50 μM. The cysteine and serine protease inhibitors E-64d, antipain, and leupeptin did not inhibit cytotoxicity.
FIG. 1.
Proteasome inhibitors protect RAW 264.7 cells from LeTx. Cells were exposed for 2 h to 500 ng (each) of PA and LF per ml in the presence of inhibitors at the indicated concentrations. Cytotoxicity was measured by MTT assay, and cell viability was calculated as described in Materials and Methods. Data are averages of five independent experiments.
One of the effective agents, ALLN, is also an inhibitor of calpain. Both ALLN and ALLM inhibit calpain at nanomolar concentrations, and with similar potencies (35). ALLN and ALLM also inhibit the proteasome, but at micromolar concentrations and with markedly different potencies (35). In RAW 264.7 cells, ALLM also inhibited LeTx cytotoxicity but at a concentration 100-fold higher than the effective concentration of ALLN, indicating that this inhibition is probably not due to an effect on calpain. The 50% inhibitory concentrations for MG132, lactacystin, and ALLN were approximately 3.0, 3.0, and 9.0 μM, respectively. These data indicate that proteasome activity is necessary for LeTx cytotoxicity, whereas other cellular protease activities are not.
Proteasome inhibitors inhibit the proteasome at concentrations similar to those that inhibit LeTx action.
To confirm that the inhibitors tested above block proteasome activity in RAW 264.7 cells, we utilized a cellular marker that is degraded by the proteasome. IκB-α is an inhibitor of the nuclear transcriptional factor NF-κB. Following stimulation of cells by extracellular signals such as LPS and tumor necrosis factor alpha, IκB-α undergoes phosphorylation, ubiquitination and proteasome-dependent degradation. Degradation of IκB-α releases NF-κB, allowing its translocation to the nucleus where it activates certain responsive genes (37).
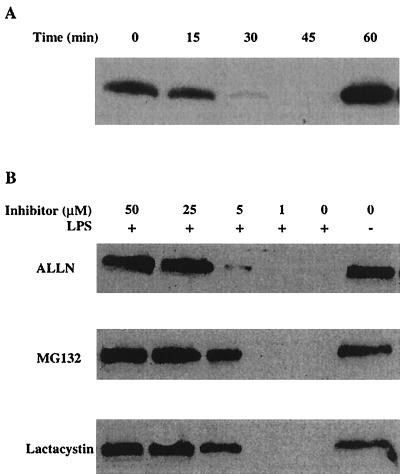
We first examined the time course of IκB-α degradation induced by LPS. As shown in Fig. 2A, IκB-α degradation was rapid and transient. Degradation peaked at 30 min after LPS treatment, and resynthesis of IκB-α occurred at 60 min. This result is consistent with previous results (2). Therefore, we selected 30 min as the time point at which to analyze IκB-α degradation. The proteasome inhibitors were incubated with RAW 264.7 cells for 30 min before addition of LPS. After 30 min at 37°C, the cells were lysed and analyzed by Western blotting with antibodies against IκB-α. As shown in Fig. 2B, ALLN, MG132, and lactacystin all inhibited the proteasome-dependent degradation of IκB-α and did so in a concentration-dependent manner. The concentrations that inhibited proteasome-dependent degradation of IκB-α were similar to those that inhibited the cytotoxicity of LeTx. These data further support the hypothesis that these inhibitors block LeTx action through inhibition of proteasome activity.
FIG. 2.
Proteasome inhibitors block degradation of IκB-α induced by LPS in RAW 264.7 cells. (A) Cells were incubated with 1 μg of LPS per ml for the indicated times and lysed for Western blotting with antibodies against IκB-α. (B) Proteasome inhibitors were incubated with cells for 30 min before LPS was added for a further 30-min incubation. Cells were then lysed for Western blotting with antibodies against IκB-α. These blots showed a single immunoreactive band of approximately 40 kDa, corresponding to IκB-α, and only this portion of the blot is shown. Results are representative of three independent experiments.
Proteasome inhibitors do not prevent proteolytic cleavage of MEK1 by LF.
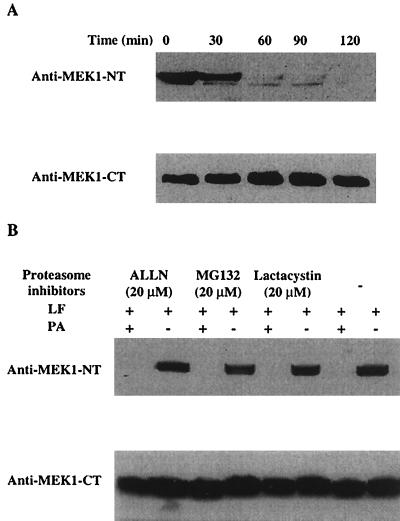
Agents which protect macrophages from LeTx might act at any step in the intoxication process. To exclude the possibility that the proteasome inhibitors block LF binding, endocytosis, translocation into the cytosol, or catalytic activity, we measured cleavage of MEK1 in LeTx-treated cells. Cleavage of MEK1 was detected by Western blotting with polyclonal antibodies raised to MEK1 residues 2 to 18. Cleavage after residue 7 by LF eliminates reactivity with this antiserum (7). The presence of native and cleaved forms of MEK1 was verified with antibodies against the C-terminal region. As shown in Fig. 3A, after 60 min of toxin treatment, no signal was detected by blotting with antibodies against the N terminus, whereas there was no change in reactivity with C terminus-specific antiserum. This indicated that MEK1 was completely cleaved after 60 min of toxin treatment. Then, RAW 264.7 cells were treated with PA and LF for 60 min in the presence or absence of the inhibitors, followed by Western blotting. As shown in Fig. 3B, MEK1 was cleaved in the presence of the proteasome inhibitors. This indicates that the proteasome inhibitors do not block any process before cleavage of cytosolic MEK1, e.g., toxin binding, internalization, or translocation. Therefore, the proteasome must be required at some stage subsequent to the initial proteolytic cleavage by LF of MEK1 or other cellular substrates.
FIG. 3.
Proteasome inhibitors do not block intracellular proteolytic cleavage of MEK1 by LF. (A) RAW 264.7 cells were treated with 500 ng (each) of PA and LF per ml for the indicated times and lysed for Western blotting with antibodies against the N terminus or C terminus of MEK1. (B) Cells were incubated with proteasome inhibitors at the indicated concentrations together with PA and LF for 1 h and lysed for Western blotting, as described for panel A. These blots showed a single immunoreactive band of 43 kDa, corresponding to MEK1, and only this part of the blot is shown. Results are representative of three independent experiments.
LeTx-induced ATP depletion is prevented by proteasome inhibitors.
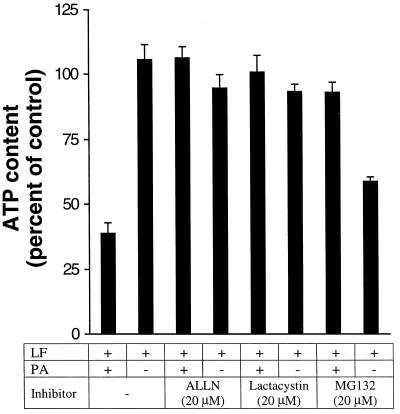
One of the early events detectable in LeTx-treated macrophages is depletion of ATP (17). To determine whether proteasome activity is necessary at an early stage, we measured the effect of proteasome inhibitors on the ATP depletion induced by LeTx. RAW 264.7 cells were treated with toxins in the presence or absence of inhibitors for 1 h at 37°C. Cells were lysed, and the released ATP was quantified by luciferin/luciferase bioluminescence. As shown in Fig. 4, LeTx reduced the intracellular ATP level while ALLN and lactacystin alone did not change the intracellular ATP level. MG132 reduced the intracellular ATP level, suggesting it may have other physiological effects. When the cells were treated with toxin in presence of ALLN or lactacystin, ATP depletion was blocked. This indicated that the proteasome is required at an early stage in LF action, before ATP depletion occurs.
FIG. 4.
Proteasome inhibitors block ATP depletion induced by LeTx. RAW 264.7 cells were treated with 500 ng (each) of PA and LF per ml in the absence or presence of proteasome inhibitors for 1 h. ATP was released from the cells and measured by bioluminescence from luciferin/luciferase, as described in Materials and Methods. Data are averages of three independent experiments.
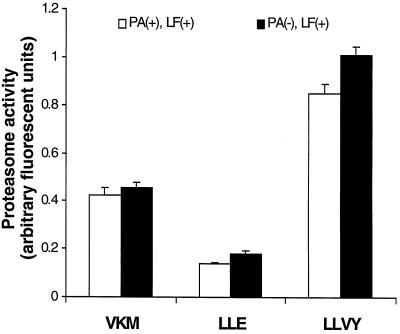
LeTx does not induce proteasome activity.
To investigate whether LeTx induces a generalized activation of the proteasome that would lead to extensive degradation of cellular proteins, we used fluorogenic substrates to measure the proteasome activity. The proteasome has at least three peptidase activities, including chymotrypsin-like activity, trypsin-like activity, and peptidylglutamyl activity (31). Fluorogenic substrates that are used to measure proteasome activities include SLLVY-AMC, Z-VKM-AMC, and Z-LLE-AMC. RAW 264.7 cells in suspension were found to be as sensitive to LeTx as those adherent to surfaces (data not shown) and were used in this experiment. RAW 264.7 cells were treated in suspension for 1 h with 500 ng (each) of PA and LF per ml. The cells were collected and disrupted by sonication. This crude cell extract was used to measure proteasome activity. Because lactacystin specifically inhibits the peptidase activities of the proteasome, specific proteasome activities were calculated as the difference in substrate hydrolysis in the absence and in the presence of 20 μM lactacystin. For all three substrates, more than 50% of the total hydrolytic activity was inhibited by lactacystin (data not shown). As shown in Fig. 5, there was no increase in proteasome activities in cells treated with PA and LF compared with untreated cells, indicating that LeTx does not increase the activity of the proteasome during its toxic process.
FIG. 5.
LeTx does not induce proteasome activity. Cells were treated with or without 500 ng (each) of PA and LF per ml for 1 h. Cell lysates were prepared and incubated with fluorogenic substrates for 15 min at 37°C. Proteasome specific activity was calculated as the difference between fluorescence in the absence and fluorescence in the presence of lactacystin. VKM, LLE, and LLVY stand for the fluorogenic substrates Z-VKM-AMC, Z-LLE-AMC, and SLLVY-AMC, respectively.
DISCUSSION
In this study, comparison of a group of protease inhibitors led to the discovery that the proteasome inhibitors ALLN, MG132, and lactacystin are very potent blockers of the cytotoxicity of LeTx (Fig. 1). Although ALLN and MG132 can strongly inhibit the cysteine proteases calpain and cathepsin B, lactacystin has no effect on any cellular protease tested other than the proteasome, including calpain I, calpain II, and cathepsin B (8). Furthermore, lactacystin does not inhibit lysosomal protein degradation (3). Lactacystin covalently binds to the catalytic subunits of the proteasome (3) so that physiological effects caused by lactacystin are highly diagnostic of proteasome involvement (8). We found that the concentrations of the proteasome inhibitors required to block LeTx-induced cytotoxicity were similar to those that blocked the intracellular activity of the proteasome, as assessed by the LPS-induced degradation of IκB-α (Fig. 2). These data indicate that functional proteasomes are indispensable to the cytotoxicity of LeTx. Because the inhibitors did not block cleavage of MEK1 by LeTx but did block the early event of ATP depletion, the proteasome must be involved in a process occurring soon after the initial cleavage by LF of its intracellular substrate(s).
Another well-characterized proteolytic process that leads to cell death is caspase-mediated programmed cell death, or apoptosis (11). Although LeTx-induced macrophage killing appears to be more necrotic than apoptotic and occurs more rapidly (90 min) than apoptotic death, we considered that LeTx might somehow recruit components of the caspase cascade in its toxic mechanism. However, the specific caspase inhibitors YVAD-CMK and DEVD-FMK did not prevent lysis when used at 50 μM, a concentration that blocks apoptosis in most cell types. Therefore, it appears that caspase activity is not essential to the action of LeTx.
Previously, protease and aminopeptidase inhibitors including bestatin and l-phenylalaninamide were shown to inhibit the cytotoxicity of LeTx (21). These agents do not inhibit the cleavage of MEK1 in toxin-treated cells (data not shown) and therefore do not directly interfere with the proteolytic activity of LF. We considered whether they might act, like lactacystin and ALLN, to inhibit proteasome activity. However, two lines of evidence argue against this mechanism. First, they did not block the proteolytic cleavage of fluorogenic proteasome substrates by cellular extracts. Second, they did not cause accumulation of polyubiquitinated proteins as did other proteasome inhibitors (data not shown). Therefore, bestatin and phenylalaninamide do not inhibit the proteasome and must act on some other cellular component required for LeTx action.
This study was undertaken to begin to define the molecular steps leading to LF-mediated macrophage lysis. The recent demonstration that LF cleaves MEK1 within cells (7, 41) constitutes an important step toward defining the process of LeTx action. The demonstration that the proteasome is essential to this process helps to limit the number of possible mechanisms for LeTx action but leaves many possibilities to consider. We and others (6, 15, 38) have discussed whether MEK1 cleavage alone could lead to the rapid lysis of mouse macrophages (90 min) and the even more rapid death of Fischer 344 rats (38 min). It has been difficult to identify mechanisms through which MEK1 cleavage could lead to rapid cell lysis, and therefore it is possible that other important proteins in macrophages are targets of LF cleavage.
In seeking to explain how LF and the proteasome might interact to cause cell lysis, it must be considered that inhibition of proteasome activity has secondary effects on cells that might indirectly make them resistant to LeTx. For example, MG132 is reported to induce a shock response and the JNK pathway (27). However, these effects occur slowly in comparison to the rapid action of LeTx and therefore are unlikely to account for the protective effects of proteasome inhibitors.
We also considered the possibility that LeTx action might cause a general and nonspecific increase in proteasome activity that leads to destruction of many proteins essential to homeostasis. However, LeTx-treated cells did not have increased ability to cleave three fluorogenic proteasome substrates (Fig. 5). Therefore, it is probable that the proteasome contributes to LeTx toxicity by cleavage of a small number of proteins that are not normal proteasome substrates. Such proteins must be so essential for homeostasis of macrophage-like cells that degradation of them will result in cell lysis. These could be either structural proteins required for membrane integrity or “monitoring” proteins that prevent activation or release from specialized compartments of the toxic molecules macrophages use to kill phagocytosed bacteria.
In light of the established findings that LF cleaves and inactivates MEK1, it can be expected that a downstream substrate(s) of MEK1 will be dephosphorylated. The well-characterized substrates downstream of MEK1 include the Erk1/2 MAPKs involved in transcriptional regulation. However, there may be additional proteins downstream of MEK1 that are highly expressed or uniquely essential in macrophages. The dephosphorylation of such a molecule(s) could decrease its stability and cause its rapid degradation by the proteasome. Both phosphorylation and dephosphorylation have been reported to make certain proteins susceptible to ubiquitination and degradation by the proteasome. An example that comes from within the same MAPK cascade as MEK1 is the proteasome-mediated degradation of c-Mos that occurs following dephosphorylation of a serine close to the N terminus (30). Another kinase that is made more susceptible to proteasome action by dephosphorylation is protein kinase C (22). In contrast, a protein at the end of the MAPK cascade, c-Jun, is stabilized by phosphorylation (10, 29). Although events of this type may contribute in some way to the rapid cell lysis caused by LeTx, it is difficult to cite examples in which blocking of a protein kinase cascade leads directly to such rapid and cytolytic events. It seems more likely that alterations to the kinase cascades could contribute to a broader disruption in homeostasis in which other events are dominant.
One direct way in which LF might interact with the proteasome is by exposing a previously cryptic signal, resulting in degradation of a particular cytosolic protein by the proteasome. For certain cytosolic proteins, the half-life is determined by the identity of the N-terminal amino acid. According to the N-end rule formulated by Varshavsky (40), a stable cytosolic protein can be converted to a highly unstable protein by removing a few amino acids so that the new N terminus is a destabilizing residue which targets the protein for ubiquitination and proteasome degradation. Thus, one could imagine that LF cleaves an essential protein near the N terminus (as it does with MEK1), thereby making it a preferred substrate for proteasome action.
To our knowledge, this is the first report that a functional proteasome in the target cell is essential for a bacterial protein toxin to accomplish its toxic process. Because LF is itself a protease, the series of events leading to macrophage lysis depends on the activity of both a bacterial (pathogen) protease and a cellular one. Presumably, other cellular components are needed to initiate and amplify the cascade that leads to cell lysis.
During preparation of this paper, a gene designated Ltx1, which determines the susceptibility of mouse macrophages to LeTx, was mapped to chromosome 11 by analysis of crosses between resistant and susceptible mouse strains (34). Susceptibility was dominant to resistance, suggesting that resistance is caused by an absence of, or polymorphism in, a molecule that acts downstream of the activity of LF. This finding supports our view that LeTx action involves cascades in which cellular components are altered to lead to cell lysis. Identification of these components will clarify the pathogenic mechanisms used by B. anthracis and may identify targets for therapy.
REFERENCES
- 1.Bhatnagar R, Friedlander A M. Protein synthesis is required for expression of anthrax lethal toxin cytotoxicity. Infect Immun. 1994;62:2958–2962. doi: 10.1128/iai.62.7.2958-2962.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 3.Craiu A, Gaczynska M, Akopian T, Gramm C F, Fenteany G, Goldberg A L, Rock K L. Lactacystin and clasto-lactacystin beta-lactone modify multiple proteasome beta-subunits and inhibit intracellular protein degradation and major histocompatibility complex class I antigen presentation. J Biol Chem. 1997;272:13437–13445. doi: 10.1074/jbc.272.20.13437. [DOI] [PubMed] [Google Scholar]
- 4.DeMartino G N, McCullough M L, Reckelhoff J F, Croall D E, Ciechanover A, McGuire M J. ATP-stimulated degradation of endogenous proteins in cell-free extracts of BHK 21/C13 fibroblasts. A key role for the proteinase, macropain, in the ubiquitin-dependent degradation of short-lived proteins. Biochim Biophys Acta. 1991;1073:299–308. doi: 10.1016/0304-4165(91)90135-4. [DOI] [PubMed] [Google Scholar]
- 5.Drexler H C. Activation of the cell death program by inhibition of proteasome function. Proc Natl Acad Sci USA. 1997;94:855–860. doi: 10.1073/pnas.94.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duesbery N, Vande Woude G, Leppla S. How anthrax kills: response. Science. 1998;280:1673–1674. [Google Scholar]
- 7.Duesbery N S, Webb C P, Leppla S H, Gordon V M, Klimpel K R, Copeland T D, Ahn N G, Oskarsson M K, Fukasawa K, Paull K D, Vande Woude G F. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- 8.Fenteany G, Schreiber S L. Lactacystin, proteasome function, and cell fate. J Biol Chem. 1998;273:8545–8548. doi: 10.1074/jbc.273.15.8545. [DOI] [PubMed] [Google Scholar]
- 9.Friedlander A M. Macrophages are sensitive to anthrax lethal toxin through an acid-dependent process. J Biol Chem. 1986;261:7123–7126. [PubMed] [Google Scholar]
- 10.Fuchs S Y, Dolan L, Davis R J, Ronai Z. Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene. 1996;13:1531–1535. [PubMed] [Google Scholar]
- 11.Green D R. Apoptotic pathways: the roads to ruin. Cell. 1998;94:695–698. doi: 10.1016/s0092-8674(00)81728-6. [DOI] [PubMed] [Google Scholar]
- 12.Grimm L M, Goldberg A L, Poirier G G, Schwartz L M, Osborne B A. Proteasomes play an essential role in thymocyte apoptosis. EMBO J. 1996;15:3835–3844. [PMC free article] [PubMed] [Google Scholar]
- 13.Hammond S E, Hanna P C. Lethal factor active-site mutations affect catalytic activity in vitro. Infect Immun. 1998;66:2374–2378. doi: 10.1128/iai.66.5.2374-2378.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanna P. Anthrax pathogenesis and host response. Curr Top Microbiol Immunol. 1998;225:13–35. doi: 10.1007/978-3-642-80451-9_2. [DOI] [PubMed] [Google Scholar]
- 15.Hanna P. How anthrax kills. Science. 1998;280:1671–1672. doi: 10.1126/science.280.5370.1671c. [DOI] [PubMed] [Google Scholar]
- 16.Hanna P C, Acosta D, Collier R J. On the role of macrophages in anthrax. Proc Natl Acad Sci USA. 1993;90:10198–10201. doi: 10.1073/pnas.90.21.10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanna P C, Kochi S, Collier R J. Biochemical and physiological changes induced by anthrax lethal toxin in J774 macrophage-like cells. Mol Biol Cell. 1992;3:1269–1277. doi: 10.1091/mbc.3.11.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanna P C, Kruskal B A, Ezekowitz R A, Bloom B R, Collier R J. Role of macrophage oxidative burst in the action of anthrax lethal toxin. Mol Med. 1994;1:7–18. [PMC free article] [PubMed] [Google Scholar]
- 19.Heinemeyer W, Kleinschmidt J A, Saidowsky J, Escher C, Wolf D H. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. EMBO J. 1991;10:555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch T, Dallaporta B, Zamzami N, Susin S A, Ravagnan L, Marzo I, Brenner C, Kroemer G. Proteasome activation occurs at an early, premitochondrial step of thymocyte apoptosis. J Immunol. 1998;161:35–40. [PubMed] [Google Scholar]
- 21.Klimpel K R, Arora N, Leppla S H. Anthrax toxin lethal factor contains a zinc metalloprotease consensus sequence which is required for lethal toxin activity. Mol Microbiol. 1994;13:1093–1100. doi: 10.1111/j.1365-2958.1994.tb00500.x. [DOI] [PubMed] [Google Scholar]
- 22.Lee H W, Smith L, Pettit G R, Smith J B. Bryostatin 1 and phorbol ester down-modulate protein kinase C-alpha and -epsilon via the ubiquitin/proteasome pathway in human fibroblasts. Mol Pharmacol. 1997;51:439–447. [PubMed] [Google Scholar]
- 23.Leppla S H. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc Natl Acad Sci USA. 1982;79:3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leppla S H. Anthrax toxins. In: Moss J, Iglewski B, Vaughan M, Tu A, editors. Bacterial toxins and virulence factors in disease. Handbook of natural toxins. Vol. 8. New York, N.Y: Marcel Dekker, Inc.; 1995. pp. 543–572. [Google Scholar]
- 25.Lin C G, Kao Y T, Liu W T, Huang H H, Chen K C, Wang T M, Lin H C. Cytotoxic effects of anthrax lethal toxin on macrophage-like cell line J774A.1. Curr Microbiol. 1996;33:224–227. doi: 10.1007/s002849900104. [DOI] [PubMed] [Google Scholar]
- 26.Menard A, Papini E, Mock M, Montecucco C. The cytotoxic activity of Bacillus anthracis lethal factor is inhibited by leukotriene A4 hydrolase and metallopeptidase inhibitors. Biochem J. 1996;320:687–691. doi: 10.1042/bj3200687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meriin A B, Gabai V L, Yaglom J, Shifrin V I, Sherman M Y. Proteasome inhibitors activate stress kinases and induce Hsp72. Diverse effects on apoptosis. J Biol Chem. 1998;273:6373–6379. doi: 10.1074/jbc.273.11.6373. [DOI] [PubMed] [Google Scholar]
- 28.Murray A. Cyclin ubiquitination: the destructive end of mitosis. Cell. 1995;81:149–152. doi: 10.1016/0092-8674(95)90322-4. [DOI] [PubMed] [Google Scholar]
- 29.Musti A M, Treier M, Bohmann D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science. 1997;275:400–402. doi: 10.1126/science.275.5298.400. [DOI] [PubMed] [Google Scholar]
- 30.Nishizawa M, Okazaki K, Furuno N, Watanabe N, Sagata N. The “second-codon rule” and autophosphorylation govern the stability and activity of Mos during the meiotic cell cycle in Xenopus oocytes. EMBO J. 1992;11:2433–2446. doi: 10.1002/j.1460-2075.1992.tb05308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orlowski M, Cardozo C, Michaud C. Evidence for the presence of five distinct proteolytic components in the pituitary multicatalytic proteinase complex. Properties of two components cleaving bonds on the carboxyl side of branched chain and small neutral amino acids. Biochemistry. 1993;32:1563–1572. doi: 10.1021/bi00057a022. [DOI] [PubMed] [Google Scholar]
- 32.Petosa C, Collier R J, Klimpel K R, Leppla S H, Liddington R C. Crystal structure of the anthrax toxin protective antigen. Nature. 1997;385:833–838. doi: 10.1038/385833a0. [DOI] [PubMed] [Google Scholar]
- 33.Pezard C, Berche P, Mock M. Contribution of individual toxin components to virulence of Bacillus anthracis. Infect Immun. 1991;59:3472–3477. doi: 10.1128/iai.59.10.3472-3477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts J E, Watters J W, Ballard J D, Dietrich W F. Ltx1, a mouse locus that influences the susceptibility of macrophages to cytolysis caused by intoxication with Bacillus anthracis lethal factor, maps to chromosome 11. Mol Microbiol. 1998;29:581–591. doi: 10.1046/j.1365-2958.1998.00953.x. [DOI] [PubMed] [Google Scholar]
- 35.Rock K L, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg A L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 36.Sadoul R, Fernandez P A, Quiquerez A L, Martinou I, Maki M, Schroter M, Becherer J D, Irmler M, Tschopp J, Martinou J C. Involvement of the proteasome in the programmed cell death of NGF-deprived sympathetic neurons. EMBO J. 1996;15:3845–3852. [PMC free article] [PubMed] [Google Scholar]
- 37.Stancovski I, Baltimore D. NF-kappaB activation: the I kappaB kinase revealed? Cell. 1997;91:299–302. doi: 10.1016/s0092-8674(00)80413-4. [DOI] [PubMed] [Google Scholar]
- 38.Strauss E. Microbiology: new clue to how anthrax kills. Science. 1998;280:676. doi: 10.1126/science.280.5364.676. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka K, Tsurumi C. The 26S proteasome: subunits and functions. Mol Biol Rep. 1997;24:3–11. doi: 10.1023/a:1006876904158. [DOI] [PubMed] [Google Scholar]
- 40.Varshavsky A. The N-end rule pathway of protein degradation. Genes Cells. 1997;2:13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- 41.Vitale G, Pellizzari R, Recchi C, Napolitani G, Mock M, Montecucco C. Anthrax lethal factor cleaves the N-terminus of MAPKKs and induces tyrosine/threonine phosphorylation of MAPKs in cultured macrophages. Biochem Biophys Res Commun. 1998;248:706–711. doi: 10.1006/bbrc.1998.9040. [DOI] [PubMed] [Google Scholar]