Abstract
As an important NAD+-dependent enzyme, SIRT6 has received significant attention since its discovery. In view of observations that SIRT6-deficient animals exhibit genomic instability and metabolic disorders and undergo early death, SIRT6 has long been considered a protein of longevity. Recently, growing evidence has demonstrated that SIRT6 functions as a deacetylase, mono-ADP-ribosyltransferase and long fatty deacylase and participates in a variety of cellular signaling pathways from DNA damage repair in the early stage to disease progression. In this review, we elaborate on the specific substrates and molecular mechanisms of SIRT6 in various physiological and pathological processes in detail, emphasizing its links to aging (genomic damage, telomere integrity, DNA repair), metabolism (glycolysis, gluconeogenesis, insulin secretion and lipid synthesis, lipolysis, thermogenesis), inflammation and cardiovascular diseases (atherosclerosis, cardiac hypertrophy, heart failure, ischemia-reperfusion injury). In addition, the most recent advances regarding SIRT6 modulators (agonists and inhibitors) as potential therapeutic agents for SIRT6-mediated diseases are reviewed.
Keywords: SIRT6, molecular network, ageing, metabolism, inflammation, cardiovascular diseases
Sirtuins, comprising a group of evolutionarily conserved nicotinamide adenine dinucleotide (NAD+)-dependent proteins, beneficially regulate lifespan and cellular senescence [1]. Interestingly, in contrast to class I, II and IV HDACs, for which Zn2+ is a catalytic cofactor, the dependence of sirtuins on NAD+ distinguishes them from other classes of HDACs; sirtuins are class III HDACs [2]. In mammalian cells, seven different sirtuin proteins have been identified (SIRT1-7) [3]. They share a conserved ~270 residue catalytic core domain composed of an NAD+-binding Rossmann subdomain and a Zn2+-binding module, with variable N- and C-terminal extensions (NTEs and CTEs, respectively) of different lengths and sequences, which account for their specific localization, unique substrates and multiple physiological functions [4]. SIRT1 and SIRT2 are present in both the nucleus and cytoplasm; SIRT3, SIRT4 and SIRT5 are exclusively found in mitochondria, and SIRT6 and SIRT7 are thought to be located in the nucleus [3]. Notably, in response to stress, SIRT6 localizes to cytoplasmic stress granules [5, 6], suggesting that SIRT6 is not exclusively a nuclear protein. Sirtuins are involved in a broad range of physiological processes, including genome stability, energy metabolism, aging, tumorigenesis, and cardiovascular biology, via their regulation of key protein activities [3, 7].
Among sirtuin family members, sirtuin 6 (SIRT6) is of particular interest and has gained more attention due to its distinctive enzymatic activities; for example, SIRT6 catalyzes deacetylation and mono-ADP-ribosylation and exhibits long-chain fatty acid (FA) deacylase activity [6]. These enzymatic activities indicate that SIRT6 is closely related to cellular biological processes, such as DNA repair, genome stability, inflammation and metabolic homeostasis [6]. Studies have revealed that dysregulation of SIRT6 activity leads to the onset and development of many diseases, including but not limited to metabolic diseases, cardiovascular diseases (CVDs), cancers and neurodegenerative diseases [8]. Therefore, a thorough and detailed understanding of the roles and regulatory mechanisms of SIRT6 may pave a new way to novel therapeutic interventions for these diseases.
In this review, we first focus on the distinctive molecular structure and biological functions of SIRT6 and then illustrate recent advances in understanding the involvement of SIRT6 in cell and molecule signaling pathways related to senescence, dysregulated metabolism, inflammation and oxidative stress. In particular, we highlight the critical roles of SIRT6 in CVDs and their related risk factors and discuss recent developments in SIRT6 modulators, their pharmacological profiles with respect to their potential use as therapeutics.
1. The structure and enzymatic activities of SIRT6
1.1 Structural features of SIRT6
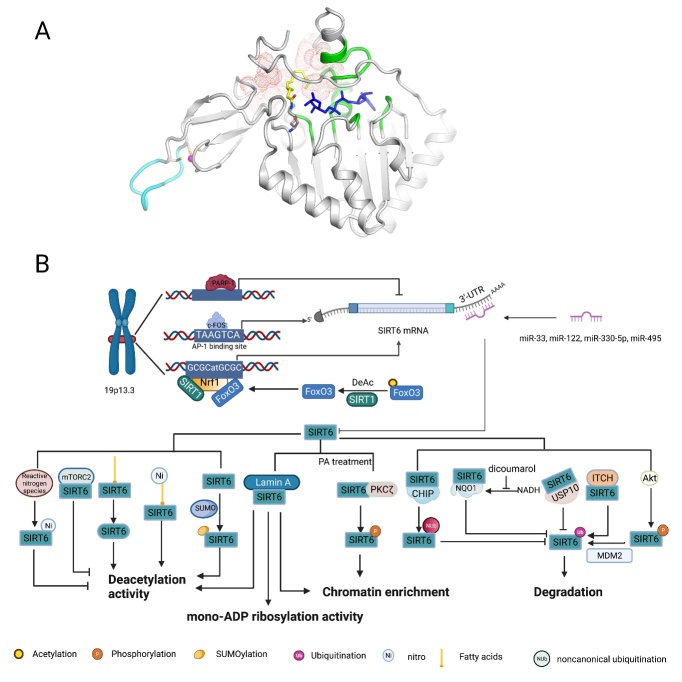
Characterizing the molecular functions of SIRT6 has been challenging, mainly because this protein can participate in three different enzymatic activities with diverse substrates [9]. Fortunately, detailed structural studies have provided in-depth insights into the characteristics of SIRT6, such as its subcellular localization, catalytic activity, and substrate preference. The N-terminus of SIRT6 is essential for chromatin association and intrinsic histone 3 lysine 9 (H3K9) and H3K56 deacetylation activity, whereas the C-terminus is required for the nuclear localization and recognition of nucleosomal DNA [10, 11]. In the catalytic core, Rossmann fold subdomain of SIRT6 contains a stable helical structure for NAD+ binding; however, most sirtuins are characterized by a highly flexible NAD+-binding loop [12]. This distinctive structure of SIRT6 favors high affinity binding with NAD+ even in the absence of acetylated substrates, which may be the reason that SIRT6 can efficiently promote mono-ADP-ribosyltransferase reactions [12]. As mentioned above, Zn2+ plays a purely structural role and is coordinated by SIRT6 Cys141, Cys144, Cys166, and Cys177 in the small and splayed zinc-binding module, in which ten residues (167-176) form a flexible loop that is unique to SIRT6 [13]. Mutation of Cys144 was experimentally shown to decrease SIRT6 deacetylase activity and significantly promote its glycolytic capacity and lead to the accumulation of extracellular lactate but did not decrease glucose transporter-1 (GLUT-1) expression, suggesting that Cys144 of SIRT6 is potentially a critical site in SIRT6 regulation of glucose metabolism [14]. Additionally, several other mutations of residues that impair SIRT6 functions have been identified: The H133Y mutation results in abrogation of SIRT6 catalytic activity and impaired SIRT6 binding with chromatin; the S56Y mutant lacks both deacetylase activity and mono-ADP-ribosyltransferase activity; R65A leads to abrogated deacetylase activity, but mono-ADP-ribosyltransferase activity is retained, while the G60A mutation has the opposite effect. In addition, a long active site is found between the Rossmann and Zn2+ subdomains of SIRT6; it is the widest hydrophobic channel pocket in sirtuins [12], which may explain the reason that SIRT6 prefers long-chain fatty acylation to acetylation, at least in vitro [15-17] (Fig. 1A).
Figure 1.
The structural features and molecular regulation of SIRT6. (A) The structural features of SIRT6: Structure of human SIRT6 in complex with H3K9-Myr (yellow) and ADP-ribose (blue) bound (PDB ID: 3ZG6). The zinc ion (pink)-binding structure is shown in the bottom left, and the unique flexible loop in the zinc-binding module is shown in azure. The stable structure of NAD+ binding with SIRT6 is colored green, and the long and wide hydrophobic-channel pocket is shown as red dots. (B) At the transcriptional level, p53 and pharmacological inhibition of PARP1 both upregulate SIRT6 expression. Similarly, c-FOS binds to the AP-1-binding site (TAAGTCA) in the SIRT6 promoter to directly promote SIRT6 expression. Under nutrient stress, SIRT1 interacts with and deacetylates FOXO3a, which is favorable for the formation of the SIRT1-FOXO3a-NRF1 (SFN) complex on the promoter of SIRT6, which upregulates SIRT6 expression. In addition, endogenous microRNAs (miRNAs), such as miR-33, miR-122, miR-330-5p and miR-495, silence translation by binding to the 3’-untranslated region (UTR) of SIRT6 mRNA. At the protein level, fatty acids (FAs) and nitrated FAs can activate SIRT6 deacetylase activity. The binding of electrophilic nitro-FAs and SIRT6 induces efficient activation (40-fold at 20 μM). In contrast, reactive nitrogen-induced nitration of tyrosine 257 (Y257) in SIRT6 causes loss of SIRT6 activity. Interaction between mTORC2 and SIRT6 suppresses SIRT6 deacetylase activity in adipose tissue. Lamin A is an endogenous activator of SIRT6, promoting SIRT6 recruitment to chromatin and activating both its deacetylase and mono-ADP ribosyltransferase activity. In addition, SUMOylation of SIRT6 specifically regulates SIRT6 deacetylation on H3K56 in vivo, and four lysine residues of SIRT6, K296, K300, K316 and K332, are thought to be SUMOylated. Under PA treatment, PKCζ binds to SIRT6 and phosphorylates SIRT6 at threonine 294 (T294) to promote SIRT6 recruitment to chromatin. Regarding stability, Akt-mediated phosphorylation of SIRT6 at serine 338 (S338) makes SIRT6 favorable for the ubiquitination via MDM2, promoting SIRT6 degradation. In contrast, SIRT6 interaction with USP10, CHIP and NQO1 blocks ubiquitin-mediated degradation of SIRT6. Among these factors, CHIP induces noncanonical ubiquitination of SIRT6 at K170, preventing canonical ubiquitination by other ubiquitin ligases. In addition, the NQO1 cofactor NADH promotes the binding of NQO1 to SIRT6, whereas DIC compromised the interaction between NQO1 and SIRT6. Ni, nitration; SUMOylation, SUMO-induced modification; Nub, noncanonical ubiquitination; Ub, ubiquitination.
1.2 Enzymatic activities of SIRT6
1.2.1 Deacetylase activity
A breakthrough in understanding the molecular functions of SIRT6 came with the discovery of its deacetylase activity against H3K9 and H3K56 [18, 19], with this discovery leading to a series of studies on the roles of SIRT6 in regulating gene expression and telomeric chromatin stability and its effects on the dynamic association of DNA repair factors with chromatin [20]. Later studies revealed that SIRT6 deacetylates H3K18, which is required for the maintenance of pericentric heterochromatin and repression of pericentric transcripts, thereby protecting against mitotic errors and cellular senescence [21]. SIRT6 also actively deacetylates H3K27, but the specific molecular mechanisms involved in this modification need to be further explored [22]. While the histone deacetylase activity of SIRT6 has been well characterized, SIRT6 has been shown to engage in deacetylase activity at a rate ~3 orders of magnitude slower than that of other sirtuins in vitro [15]. Furthermore, Feldman et al. showed that SIRT6 deacetylation activity was positively upregulated by long-chain FAs, providing an explanation for the discrepancy between its poor deacetylase activity in vitro and the clear observation that SIRT6 is a histone deacetylase in vivo [23]. Another explanation suggests that SIRT6 histone deacetylase activity depends on packaged nucleosomes, not free histone proteins; in other words, the SIRT6 deacetylase structure can be converted upon binding to nucleosomes [24]. Recently, SIRT6 has been found to directly detach acetyl groups from some nonhistone proteins, a regulatory function important to certain cellular biological processes. For example, SIRT6 directly deacetylates general control nonrepressed protein 5 (GCN5) and pyruvate kinase M2 (PKM2), thereby regulating their enzymatic activities [25, 26]. SIRT6 also increases cellular NADPH levels by directly deacetylating nicotinamide phosphoribosyltransferase (NAMPT), conferring cellular resistance to oxidative stress damage [27]. The active spliced form of X-box-binding protein 1 (XBP1) is deacetylated by SIRT6, which leads to XBP1 degradation and subsequent prevention of endoplasmic reticulum (ER) stress-induced hepatic steatosis [28]. Recently, SIRT6 has also been shown to deacetylate SMAD2 and SMAD3, attenuating the development of liver fibrosis [29, 30].
1.2.2 Mono-ADP-ribosyltransferase activity
Although the deacetylase activity of SIRT6 has been well established, its first discovered enzymatic activity is the NAD+-dependent mono-ADP-ribosyltransferase activity [31]. In an in vitro experiment, SIRT6 was shown to transfer radiologically labeled [32P] NAD+ onto itself, which indicated that SIRT6 underwent autoregulation via mono-ADP-ribosylation, however, the physiological significance of this auto-ADP ribosylation remains unknown [31]. Recent studies identified several mono-ADP-ribosylation substrates of SIRT6, including KAP1, PARP1, KDM2A and BAF170, which are mainly involved in the aging process, genome stability maintenance and oxidative stress [32-35]. Moreover, findings by Van Meter et al. suggested that the mono-ADP-ribosyltransferase activity of SIRT6 was involved in the regulation of the apoptosis of cancer cells by activating the p53 and p73 signaling cascades [36]. Notably, previous studies reported that the mono-ADP-ribosylation activity of SIRT6 was negligible in vitro, suggesting that this activity depended on specific in vivo substrates or conditions [9].
1.2.3 Long-chain FA deacylase activity
The weak deacetylation activity of SIRT6 and its preference for long-chain fatty acylation in vitro led to the speculation that SIRT6 was engaged in other enzymatic activities in addition to its established deacetylation and mono-ADP-ribosylation functions. Indeed, SIRT6 can efficiently catalyze long-chain FA deacylation, and the most notable effect of this function is associated with protein secretion [37]. One representative protein, TNF-α, is secreted and regulated by SIRT6 demyristoylation [15]. Furthermore, SIRT6 has also been shown to change the subcellular localization of R-Ras2 by regulating its depalmitoylation, inhibiting activation of the PI3K/Akt signaling pathway [38]. An in vitro experiment also showed that the FA deacylation of myristoylated, palmitoylated and octanoylated peptides in H3K9 and that of a myristoyl group on H2BK12 was an efficient function of SIRT6 [15]. Another biological study confirmed that SIRT6 effectively removed fatty acyl groups from histones H3K9, H3K18, and H3K27 and from histones H3K14, H3K36, H3K56 and H3K79 at a relatively low rate [22]. This discovered enzymatic function of SIRT6 may be involved in regulating chromatin accessibility and gene transcription, but the exact potential physiological functions remain to be further elucidated.
2. The molecular network that regulates SIRT6
The abundance of SIRT6 is subjected to intricate and precise regulation in vivo (Fig. 1B). At the transcriptional level, pharmacological inhibition of poly-ADP-ribose polymerase 1 (PARP1) significantly activates the transcription of SIRT6 [39]. Additionally, c-FOS and the SIRT1-FOXO3a-NRF1 (SFN) complex bind the SIRT6 promoter to upregulate SIRT6 expression [40, 41]; the tumor suppressor p53 has also been shown to activate the expression of SIRT6 [42]. In contrast, both high glucose levels in vitro and maternal diabetes in vivo significantly reduced the SIRT6 level in the embryo or neural stem cells [43]. Interestingly, a separate study demonstrated that high glucose exposure was associated with increased demethylation of the SIRT6 promoter and increased SIRT6 mRNA transcription [44]. Translation or posttranslational modifications might be feasible explanations for the discrepancy observed in the regulatory mechanisms of SIRT6. At the translational level, endogenous microRNAs (miRNAs), particularly miR-33a/b, miR-122, miR-330-5p and miR-495, can regulate SIRT6 translation by binding the 3’-untranslated region (UTR) of SIRT6 [45-48]. At the protein level, the stability of SIRT6 is most notably regulated through the proteasome-dependent degradation pathway. Previous studies showed that Akt-induced phosphorylation and E3 ubiquitin ligase ITCH-induced ubiquitination of SIRT6 both accelerated its degradation [49, 50]. In contrast, the ubiquitin ligase CHIP and ubiquitin-specific peptidase USP10 acted as protective regulators to maintain SIRT6 stability [51, 52]. In addition, NAD(P)H: quinone oxidoreductase 1 (NQO1) has been shown to physically interact with SIRT6 to prevent its degradation [53].
SIRT6 deacetylase activity can be activated by binding of several specific long-chain fatty FAs (myristic, oleic, and linoleic acids) in vitro, suggesting that regulatory pathways are involved in SIRT6 enzymatic activities, at least its deacetylase activity [23]. Mechanistically, FAs bind to the hydrophobic pocket of SIRT6 to induce a conformational change in SIRT6 that increases its affinity for acetylated substrates. Recent research has revealed that more efficient nitro-fatty acids (NO2-FA) binding stimulates SIRT6 deacetylase activity in a cellular environment [13]. In contrast, peroxynitrite and other nitric oxide-derived oxidants have been shown to modify tyrosine and cysteine residues in SIRT6, resulting in impaired SIRT6 deacetylase activity [54, 55]. Similarly, mTORC2 interacts with SIRT6 to suppress its deacetylation activity against FoxO1 [56]. In addition, small ubiquitin-like modifier (SUMO) has been shown to modify SIRT6 and specifically upregulate its deacetylation of H3K56 but not H3K9 [57]. Atypical protein kinase C ζ (PKCζ) phosphorylates SIRT6 to promote its enrichment on the promoters of β-oxidation-related genes [58]. Both deacetylation of SIRT6 by SIRT1 and the binding of SIRT6 to Lamin A promote the retention of SIRT6 on DNA breaks to promote DNA repair [59, 60]. In addition, Lamin A binds nuclear factor erythroid 2-related factor 2 (Nrf2) and SIRT6 and stimulates both the deacetylation and mono-ADP ribosylation activities of SIRT6, promoting the proper expression of NRF2 target genes [61].
In summary, stress conditions, cellular metabolites and other stimuli have been proven to finely tune the abundance and enzymatic activity of SIRT6 in multiple ways. A previous study showed that SIRT6 was phosphorylated at conserved residues (T294, 303, S330, and S338) within the C-terminus, suggesting that the C-terminus modification patterns may be crucial to SIRT6 functions [62]. As mentioned above, in the C-terminus, both Akt- and PKCζ-induced phosphorylation and SUMO-induced SUMOylation affected SIRT6 abundance and functions. However, the effects of other modifications at C terminal sites remain to be further studied. Considering the diverse roles of SIRT6 in numerous molecular signaling pathways, some of the aforementioned regulatory factors may be feasible intervention targets for regulating SIRT6-mediated signaling.
3. The biological functions of SIRT6
Among the diverse functions of SIRT6, its most remarkable role is an intricate regulator of cell senescence and lifespan. Owing to genome instability and dysregulated metabolism, Sirt6-deficient mice exhibit aging-associated degenerative phenotypes, including severe metabolic disorders, subcutaneous fat loss, and lordokyphosis, finally dying at about 4 weeks old; in contrast, transgenic mice overexpressing Sirt6 show a longer lifespan than their wild-type counterparts [63, 64]. Furthermore, the contributions of SIR6 to genome stability, DNA repair and telomere function highlight its important therapeutic potential for aging and aging-related diseases [9]. In addition, oxidative stress, inflammation, and metabolism, in which SIRT6 is involved, are crucial for the occurrence and development of CVDs [65]. Moreover, SIRT6 plays essential roles in protecting vessels and heart from endothelial dysfunction [66], atherosclerosis [67], pathological cardiac hypertrophy [68], myocardial fibrosis [69], heart failure [70] and ischemia/reperfusion (I/R) injury [71]. Notably, in some cases, SIRT6 appeared to be a pernicious regulator. For example, overexpression of SIRT6 significantly promoted angiogenesis and hemorrhage in carotid plaques, resulting in plaque instability [72]. In addition, in a mouse model, Sirt6 silencing reduced neutrophil infiltration, myocardial infarct size and reactive oxygen species (ROS) generation within infarcted heart tissues in the early phases of I/R [73]. Given the important and diverse enzymatic effects of SIRT6 on cellular physiological functions, it is of significance to discuss the mechanisms by which these pleiotropic SIRT6 functions extensively affect biological functions and the determination of whether SIRT6 is a feasible therapeutic target for aging-related diseases. Detailed information on SIRT6 substrates and functions is shown in Table 1.
Table 1.
The substrates and functions of SIRT6.
| SIRT6 function | Substrates | Molecular mechanism | Physiological functions | |
|---|---|---|---|---|
| Deacetylase | Histones | H3K9 | Inhibits NF-kB target gene expression | Prevents premature aging [160] |
| Inhibits 53BP1 binding to telomeres | Prevents telomere damage and delays VSMC senescence [78] | |||
| Facilitates loading of CHD4 onto DNA damage sites | Promotes chromatin relaxation and subsequent DNA repair [108] | |||
| Inhibits Nkx3.2 expression and thereby promotes GATA5 expression | Prevents hypertension and associated cardiorenal injury [66] | |||
| Inhibits HIF1α and associated glycolytic gene expression | Improves mitochondrial respiration and inhibits glycolysis against metabolic diseases [111] | |||
| Inhibits Notch1 and Notch4 expression | Protects podocytes from injury and attenuates proteinuria [245] | |||
| Inhibits expression of the proatherogenic gene TNFSF4 | Maintains endothelial function and protects against atherosclerosis [246] | |||
| Inhibits IGF signaling-related gene expression | Restricts the development of cardiac hypertrophy [68] | |||
| Inhibits c-Jun-dependent proinflammatory gene expression | Attenuates chronic liver inflammation and fibrosis [247] | |||
| Inhibits ERK1/2 expression | Attenuates cisplatin-induced kidney injury [248] | |||
| Facilitates binding of WRN with telomeres | Protects against telomere dysfunction and premature ageing disorders [249] | |||
| Inhibits NKG2D ligand expression in ECs | Stabilizes atherosclerotic plaques and restricts atherosclerosis [179] | |||
| Inhibits Txnip expression in β cells | Maintains pancreatic β-cell function and viability [124] | |||
| H3K56 | Inhibits β-catenin-dependent pro-fibrotic gene expression | Protects against kidney fibrosis following kidney ischemia-reperfusion injury [250] | ||
| Facilitates recruitment of the chromatin remodeler SNF2H to DSBs | Promotes DSB repair and genomic stability [236] | |||
| Facilitates recruitment of DNA repair proteins | Promotes the DDR process [106] | |||
| Inhibits catalase expression | Promotes neovascular injury by increasing ROS [72] | |||
| Promotes assembly of the Nrf2-RNAPII transcription complex at the HO-1 promoter, upregulating HO-1 expression | Facilitates ROS clearance to counteract oxidative stress injury [154] | |||
| H3K18 | Reduces levels of activated RNA Pol II and H3K36me3 | Protects against mitotic errors and cellular senescence [21] | ||
| Non- histones | SMAD2 (K54) | Reduces the transcriptional activity of SMAD2 | Protects against liver fibrosis [29] | |
| SMAD3 (K333/378) | Reduces the transcriptional activity of SMAD3 | Protects against liver fibrosis [30] | ||
| P53 (K381/382) | Inhibits P53 transcriptional activity | Ameliorates aging-associated phenotypes and attenuates cellular apoptosis [182, 224] | ||
| ERRγ (K195) | Promotes ERRγ degradation, thereby reducing Cyp7a1 expression | Attenuates cholestatic liver injury and fibrosis [251] | ||
| DDB2 (K35/77) | Promotes DDB2 ubiquitination and detachment from DNA lesions | Promotes the process of NER [102] | ||
| Mtf1 | Activates Mtf1 for the induction of Mt | Reduces oxidative stress and inflammation by inducing ROS [156] | ||
| SKP2 (K73/77) | Promotes SKP2 nuclear stability, thus increasing Suv39h1 ubiquitination and exclusion from chromatin, enabling H3K9me2 and H3S10p | Promotes expression of the NF-κB inhibitor IκBα to restrict the NF-κB pathway [162] | ||
| NAMPT (K53) | Promotes NAMPT enzymatic activity | Increases cellular NADPH levels to confer cell resistance to oxidative stress damage [27] | ||
| NPM1 | Inhibits NPM1 transcriptional activity and the expression of senescent genes | Inhibits cellular senescence [252] | ||
| PKM2 (K433) | Inhibits PKM2-induced STAT3 phosphorylation and activation | Suppresses macrophage polarization towards the proinflammatory M1 phenotype, inhibiting obesity-associated tissue inflammation and metabolic disorders [26, 144] | ||
| EZH2 | Promotes EZH2 DNA binding to promote FOXC1 expression | Ameliorates ischemic stroke-induced inflammation [214] | ||
| XBP1s (K257/297) | Promotes XBP1s degradation | Suppresses ER stress [28] | ||
| Caveolin-1 | Promotes autophagic degradation of Caveolin-1 | Ameliorates hyperglycemia-induced LDL transcytosis across ECs and atherosclerotic progression [171] | ||
| NFATc4 | Promotes NFATc4 nuclear export to inhibit BNP expression | Inhibits cardiac hypertrophy [189] | ||
| FoxO1 (K423) | Promotes FoxO1 nuclear export to increase expression of the glucose-sensing genes Pdx1 and Glut2 in pancreatic β-cells; inhibits PCK1 and G6PC expression in liver cells | Maintains the GSIS ability of β-cells and deceases gluconeogenesis to maintain glucose metabolic homeostasis [42, 120] | ||
| GCN5 (K549) | Promotes GCN5 activity to acetylate PGC-1α | Reduces hepatic gluconeogenesis [117] | ||
| Mono-ADP-ribosyltransferase | Non- histones | SIRT6 | May regulate Sirt6 enzymatic activity [31] | NR |
| KAP1 | Promotes SIRT6-KAP1-HP1α complex formation and heterochromatin packaging to decrease LINE1 expression | Inhibits genomic instability and senescence[33] | ||
| PARP1 (K521) | Activates PARP1 to recruit DNA repair factors | Promotes DNA damage repair [32] | ||
| KDM2A (R2020) | Promotes the displacement of KDM2A from chromatin | Promotes DNA damage repair, ensures replication fidelity [34] | ||
| BAF170 (K312) | Removes H3K27me3 and promotes chromatin accessibility to enhance HO-1 expression | Promotes the clearance of ROS to protect against cellular oxidative stress [35] | ||
| Long-chain fatty deacylase | Histones | H3 (K9/18/27/14/36/56/79) | May inhibit gene expression | NR |
| Non-histones | TNF-α (K19/20) | Promotes TNF-α secretion | Promotes inflammation [15] | |
| R-Ras2 (K192/194/196/197) | Suppresses R-Ras2 plasma membrane translocation and activation | Inhibits cell proliferation [38] | ||
| Non- catalytic activity | Non- histones | SIRT6-TIP60-GATA4 | Recruits TIP60 to acetylate GATA4 at K328/330; in turn, GATA4 inhibits the deacetylase activity of SIRT6 to promote TIP60-induced H3K9 acetylation, increasing expression of the anti-apoptotic gene Bcl-2 | Attenuates DOX-induced cardiomyocyte apoptosis [197] |
| SIRT6-Sp1-mTOR | Decreases Sp1 transcriptional activity to represses mTOR signaling gene expression | Regulates global cellular protein synthesis to combat cardiac hypertrophy [190] | ||
53BP1: p53-binding protein 1; VSMC: vascular smooth muscle cell; CHD4: chromodomain helicase DNA-binding protein 4; Nkx3.2: NK3 homeobox 2; GATA5: GATA-binding protein 5; TNFSF4: tumour necrosis factor superfamily member 4; IGF: insulin-like growth factor; ERK1/2: extracellular signal-regulated kinase 1/2; WRN: Werner syndrome helicase-nuclease; NKG2D: natural-killer group 2, number D; Txnip: thioredoxin-interacting protein; SNF2H: SWItch/sucrose nonfermentable catalytic subunit SNF2; DSBs: double-strand breaks; DDR: DNA damage response; Nrf2: nuclear factor erythroid-2 related factor 2; HO-1: haem oxygenase-1; SMAD2/3: nuclear translocation of mothers against decapentaplegic homologue 2/3; ERRγ: oestrogen-related receptor γ; Cyp7a1: cholesterol 7 α-hydroxylase; DDB2: DNA damage-binding protein 2; Mtf: metal transcription factor; Mt: metallothionein; SKP2: S phase kinase-associated protein 2; NPM1: nucleophosmin; Suv39h1:suppressor of variegation 3-9 homologue 1; PKM: M2 isoform of pyruvate kinase; STAT3: signal transducer and activator of transcription 3; FOXC1: Forkhead box C1; XBP1s: X-box-binding protein 1 spliced transcription factor; HMGB1: high mobility group box 1; NFATc4: nuclear factor of activated T cell 4; FoxO1: forkhead box protein O1; Pdx1: pancreatic duodenal homeobox 1; Glut2: glucose transporter 2; PCK1: phosphoenolpyruvate carboxykinase; G6PC; glucose-6-phosphatase; GSIS: glucose-stimulated insulin secretion; GCN5: general control nonrepressed protein 5; PGC-1α: peroxisome proliferator-activated receptor-γ coactivator 1-α; KAP1:KRAB-interacting protein 1; PARP1:poly ADP-ribose polymerase 1; LINE1: long interspersed nuclear element 1; KDM2A:lysine-specific demethylase 2; BAF170: BRG/BRM-associated factor (BAF) chromatin remodeler subunit; NRF2: nuclear factor erythroid 2-related factor 2; NR, not reported.
3.1 SIRT6 inhibits senescence by maintaining genome stability
Although as an inevitable part of human life, cellular senescence remains a significant risk factor for age-related diseases [74]. Notably, numerous studies have shown that older mammals are more likely to suffer from DNA damage than younger mammals. However, in progeroid syndromes, defects in cellular responses to DNA damage can accelerate the aging process; that is, to some extent, DNA damage is probably a cause, not a consequence of aging, highlighting the importance of DNA damage in the aging process [75]. SIRT6 attenuates cell aging by maintaining genome stability. For example, SIRT6-mediated stabilization of repressive hetero-chromatin in subtelomeric regions results in the silencing of telomere-proximal genes, which is important to genome stability maintenance [76]. In addition, both the deacetylation and mono-ADP ribosylation activities of SIRT6 have been shown to promote DNA repair through multiple pathways [8]. Importantly, SIRT6, which maintains genome integrity, for example, by preventing DNA damage and promoting DNA repair, has been implicated in CVDs [77]. Previous works have demonstrated that SIRT6 protects endothelial cells (ECs) and vascular smooth muscle cells (VSMCs) from telomere and genomic DNA damage, thus preventing the onset of senescence [78, 79]. Therefore, we mainly elaborate on the specific mechanisms of SIRT6 in genomic damage to highlight its protective roles of SIRT6 during aging.
3.1.1 SIRT6 inhibits LINE1-induced genomic damage
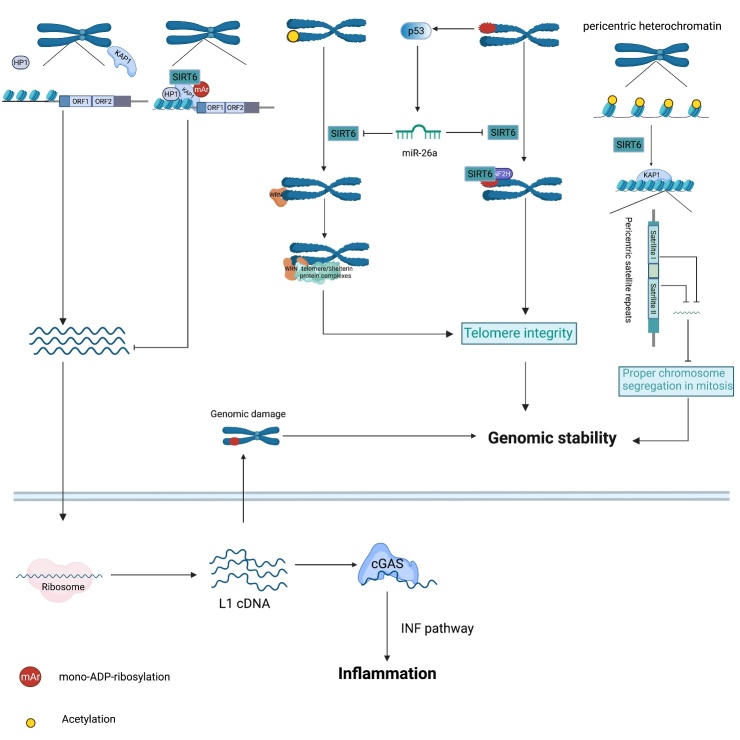
Long interspersed nuclear element 1 (LINE1), a retrotransposable element (RTE), is the only RTE capable of autonomous retrotransposition[80]. DNA breaks, insertions and site deletions are required for the replication and integration of LINE1 into host genes, which can cause genomic instability[81]. In response to these threats to genome stability, SIRT6 silences LINE1 retrotransposons through its mono-ADP-ribosyltransferase activity, stabilizing the genome [33] (Fig. 2). Notably, in response to DNA damage and/or during the course of aging, SIRT6 uncouples from LINE1 to enable DNA repair, but which largely compromises the SIRT6-mediated repression of LINE1. The relocalization of SIRT6 is thought to change LINE1 expression patterns and lead to cell dysfunction and aging-related diseases. Therefore, it was unsurprising when studies found that SIRT6-knockout (KO) cells and animals exhibited increased cytoplasmic LINE1 cDNA, which triggered DNA damage, sterile inflammation and pathological progeroid [82]. In addition, pericentric heterochromatin is thought to be associated with proper mitosis and genome stability, and SIRT6-induced H3K18 deacetylation within pericentric satellite sequences has been shown to be involved in the maintenance of pericentric heterochromatin [21, 83].
Figure 2.
SIRT6 inhibits genomic and telomeric instability. SIRT6 binds to the 5’-untranslated region (UTR) of LINE1 loci and mono-ADP-ribosylates KAP1, which promotes the interaction between KAP1 and HP1α, leading to LINE1 elements packaging into transcriptionally silent heterochromatin. In contrast, loss of SIRT6 in the 5’-UTR of LINE1 elevates LINE1 activity. Accumulation of cytoplasmic LINE1 cDNA instigates chromosomal rearrangements and provokes a strong type I interferon response and subsequent inflammation through the cytoplasmic DNA sensor cGAS. Impaired telomeres upregulate p53 expression, which activates miR-26, thereby decreasing SIRT6 content. During the S phase, SIRT6 deacetylates telomeric H3K9, enabling the efficient binding of the WRN protein to telomeres, leading to the recruitment of telomere/shelterin protein complexes to protect telomere integrity. In response to oxidative stress, SIRT6 recruits SNF2H to damaged telomeres to promote chromatin relaxation, which is associated with the maintenance of telomeres. In addition, SIRT6-mediated deacetylation of H3K18 within pericentric satellite sequences favors the retention of KAP1, repressing satellite transcription and maintaining proper chromosome segregation in mitosis.
3.1.2 SIRT6 maintains telomere integrity
Telomeres, complexes with hypoacetylated histone tails, protect linear chromosome ends from DNA degradation to maintain genome integrity. Telomeres undergo shortening, especially during the S phase, and when they reach a critical length, a cell becomes dysfunctional, entering senescence and losing the ability to proliferate [84]. Recent study has revealed that impaired telomeres can repress sirtuin expression, including SIRT6, resulting in further damage to telomere integrity and subsequent progressive deterioration, suggesting SIRT6 is closely associated with telomere integrity maintenance [85] (Fig. 2). SIRT6 deacetylates telomeric H3K9 during the S phase to stabilize the Werner helicase (WRN) in telomeric chromatin and thus prevents aberrant telomere sequence loss [18]. SIRT6 can also maintain telomere-proximal gene silencing, preventing telomere sequence loss and end-to-end chromosomal fusion [76]. In addition, SIRT6 recruits the chromatin remodeling factor SNF2H to decondense chromatin, thereby promoting directional telomere movement, which is associated with the maintenance of telomere integrity [86]. Interestingly, upon exposure to agents that cause DNA damage, SIRT6 deacetylates telomere repeat-binding factor 2 (TRF2) to promote its ubiquitylation and subsequent proteolysis, which may increase the instability of damaged telomeres and promote cell apoptosis [87].
3.1.3 SIRT6 promotes DNA repair
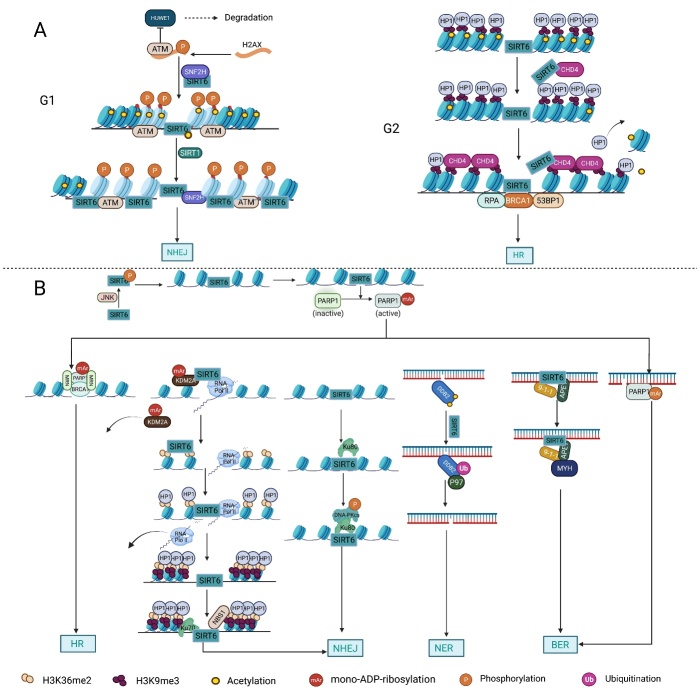
The significance of SIRT6 in DNA repair was first suggested by scientists who observed that Sirt6-deficient cells exhibited DNA damage hypersensitivity, genomic instability and progeroid-associated degenerative phenotypes, which are also linked to DNA repair and aging [88]. Additionally, in neurodegenerative studies, SIRT6 protein levels were reduced in both Alzheimer's disease patients and mouse models, and overexpression of SIRT6 in hippocampal neurons protected cells from amyloid-beta42 (Aβ42)-induced DNA damage [89]. As research has advanced in recent years, more direct and detailed evidence confirming the roles played by SIRT6 in various DNA repair pathways has been described (Fig. 3).
Figure 3.
The multitasking roles played by SIRT6 in DNA repair. (A) Chromatin accessibility: In response to DNA damage, both the deacetylation of H3K56 by SIRT6 and the interaction between SIRT6 and SNH2F are important for the recruitment of SNH2F to double-strand break (DSB) sites. The SIRT6/SNF2H complex and ATM cooperatively phosphorylate H2AX at S139 upon DSB formation to block HUWE1-induced H2AX polyubiquitination and degradation, ensuring the efficient formation of γH2AX near damaged sites. γH2AX promotes the retention of DNA repair factors, including ATM, to facilitate nonhomologous end joining (NHEJ) in the G1 phase, and in turn, ATM further phosphorylates H2AX. In addition, SIRT1-mediated deacetylation of SIRT6 at K33 enhances the interaction between SIRT6 and γH2AX. Subsequently, this interaction promotes SIRT6 retention on the chromatin flanking DSBs, enhancing the levels of deacetylation of H3K9 and H3K56 near damaged DNA sites. Similarly, SIRT6-mediated deacetylation of H3K9 and the interaction between SIRT6 and CHD4 both promote the loading of CHD4 onto H3K9me3 to competitively exclude HP1α from chromatin, leading to chromatin relaxation and the recruitment of DNA repair factors to induce efficient HR. (B) DNA repair: Under oxidative stress, SIRT6 is phosphorylated by JNK at S10, which in turn recruits PARP1 to DBSs, and SIRT6 mono-ADP-ribosylates PARP1 at K521. PARP1 mono-ADP-ribosylation is necessary for the efficient recruitment of DNA repair factors such as the MRN complex and BRCA1. SIRT6 directly binds to Ku80 and promotes the interaction between Ku80 and DNA-PKcs, which, in turn, promotes the phosphorylation of DNA-PKcs at S2056 and enhances NHEJ. In addition, SIRT6 mono-ADP-ribosylates KDM2A at R1020 within a leucine rich repeat (LRR), which facilitates displacement of KDM2A from chromatin and the subsequent increase in H3K36 dimethylation near damaged DNA sites. The accumulated H3K36me2 serves as a platform to recruit both early DNA repair components and HP1α; the former promotes NHEJ, and the latter promotes the deposition and spreading of H3K9me3 marks around DSB sites to reduce the abundance of RNA Pol II and thus ensure replication fidelity. SIRT6 deacetylates DDB2 at K35 and K77 to promote DDB2 ubiquitination, which enhances the affinity between DDB2 and ubiquitin-selective p97 segregation and subsequently releases DDB2 from damaged DNA sites. Removal of DDB2 results in relaxation of the nucleosomes around the damaged site and accumulation of downstream DNA repair factors to initiate the nucleotide excision repair (NER) cascade to repair DNA damage. The mono-ADP-ribosylation activity of SIRT6 is necessary for the functional interactions between SIRT6, Rad9-Rad1-Hus1, MYH and APE1, which promote efficient base excision repair (BER). In addition, SIRT6 promotes BER in a PARP1-dependent manner.
3.1.3.1 DNA double-strand break (DSB) repair
DNA double-strand breaks (DSBs) are severe DNA lesions that often impair genome integrity [90]. Recent studies have indicated that SIRT6 and DNA DSB repair, but not base excision repair (BER), coevolved with longevity systems, further highlighting the importance of SIRT6 in DSB repair [91]. In response to DSBs, SIRT6 decreases acetylated H3K9 (H3K9Ac) levels and recruits DNA-protein kinase catalytic subunit (DNA-PKcs) to chromatin near damaged DNA sites to promote DNA repair [92]. Several DSB sensors, such as PARP1, Ku 70/80 and the MRE11-RAD50-NBS1 (MRN) complex, have been identified [93, 94]. Recently, SIRT6 has been found to act as a DSB sensor to detect and as a dimer directly bind sites with damaged DNA ends prior to broad cascade activation that initiates the DNA damage response (DDR) [95]. In addition, SIRT6 is also closely associated with the three other aforementioned sensors [32, 96-98]. In response to DNA damage, c-Jun N-terminal kinase (JNK) phosphorylates SIRT6 at serine(S) 10 to promote its mobilization to DNA break sites, which is followed by PARP1 recruitment and efficient DSB repair [99]. At damaged DNA sites, SIRT6 physically interacts with PARP1 and catalyzes its mono-ADP-ribosylation at K521, stimulating PARP1 poly-ADP-ribosyltransferase activity, which promotes DSB repair [32]. In addition, overexpression of SIRT6, but not the individual or combinatory re-expression of homologous recombination (HR)-related proteins, such as NBS1, Rad51, or Rad52, efficiently attenuated the decline in HR in aging cells, and this outcome was dependent on SIRT6 mono-ADP ribosylation, emphasizing the importance of SIRT6 in coordinating these DNA repair proteins [97]. A study of induced pluripotent stem cells (iPSCs) demonstrated that SIRT6 promoted the interaction between Ku80 and DNA-PKcs, thereby facilitating DNA-PKcs auto-phosphorylation and nonhomologous end joining (NHEJ) [96]. Through its mono-ADP-ribosylation of KDM2A, SIRT6 promoted KDM2A detachment from chromatin and subsequently increased the levels of H3K36me2[34]. H3K36me2 serves as a platform for HP1α and DNA repair component accumulation, forming a transcription-silencing microenvironment and promoting NHEJ of transcribed chromatin, respectively [34]. In summary, SIRT6 dexterously averts clashes between transcription and DNA repair to promote efficient DNA repair.
3.1.3.2 Base excision repair (BER) and nucleotide excision repair (NER)
Both BER and NER are involved in DNA repair. SIRT6 improves genome stability by regulating DNA polymerase β (polβ) activity, which influences BER, but the precise mechanism by which SIRT6 affects polβ activity is still unclear [63]. A recent study found that overexpression of SIRT6 attenuated BER deficiency in aging cells in a PARP1-dependent manner [100]. In addition, under conditions of oxidative DNA damage, SIRT6, AP endonuclease 1 (APE1) and the checkpoint clamp Rad9-Rad1-Hus1 (9-1-1) cooperatively bound to the DNA glycosylase MutY homolog (MYH), forming a complex that promoted efficient BER [101]. The mono-ADP ribosylation activity of SIRT6 plays important role in its functional interactions [101]. In addition to BER, SIRT6 deacetylates damaged DNA-binding protein 2 (DDB2) to promote its ubiquitination and separation from chromatin, eventually resulting in relaxation of the nucleosomes around damaged sites to initiate NER [102] (Fig. 3).
3.1.4 SIRT6 modulates chromatin accessibility
The wrapping of eukaryotic DNA into chromatin around nucleosomes leads to an additional physical barrier that prevents access to DNA by DNA repair factors [103]; hence, chromatin relaxation is essential for DDR. It has been established that chromatin remodeling factors and posttranslational modifications of histone proteins, especially acetylation/deacetylation, can significantly affect chromatin structure and promote DNA repair [104, 105]. The interaction between SIRT6 and chromatin remodeling factors was first suggested by scientists who observed that SIRT6-induced H3K56 deacetylation promoted the recruitment of the chromatin remodeler SNF2H to DSBs, which was critical for the proper unfolding of chromatin and subsequent recruitment of DNA repair factors [106]. In addition, the SIRT6/SNF2H complex has been shown to facilitate ATM blocking of HUWE1-mediated H2AX degradation, thereby promoting H2AX stabilization and subsequent recruitment to DSBs [107]. ATM-induced formation of γH2AX foci is important for the retention of DNA repair factors including deacetylated SIRT6 to DSBs, where SIRT6 deacetylates H3K9 and H3K56 to facilitate chromatin remodeling and subsequent DNA repair [59, 107]. Specifically, SIRT6-mediated H3K9 deacetylation facilitated the CHD4 loading at a DNA damage site, where CHD4 competitively excluded HP1α from chromatin to promote chromatin relaxation and HR [108]. Additionally, SIRT6/SNF2H were also essential for NHEJ in the G1 phase, whereas SIRT6/CHD4 were indispensable for HR in the G2 phase [108]. These findings suggest that basis on cell and chromatin status, SIRT6 flexibly regulates the availability of key chromatin remodeling factors to facilitate DSB repair. In general, SIRT6 is involved in modulating chromatin accessibility by either modifying histone proteins or regulating the recruitment of chromatin remodeling factors, enabling cells to respond faster and more efficiently to DSBs (Fig. 3).
3.2 SIRT6 maintains metabolic homeostasis
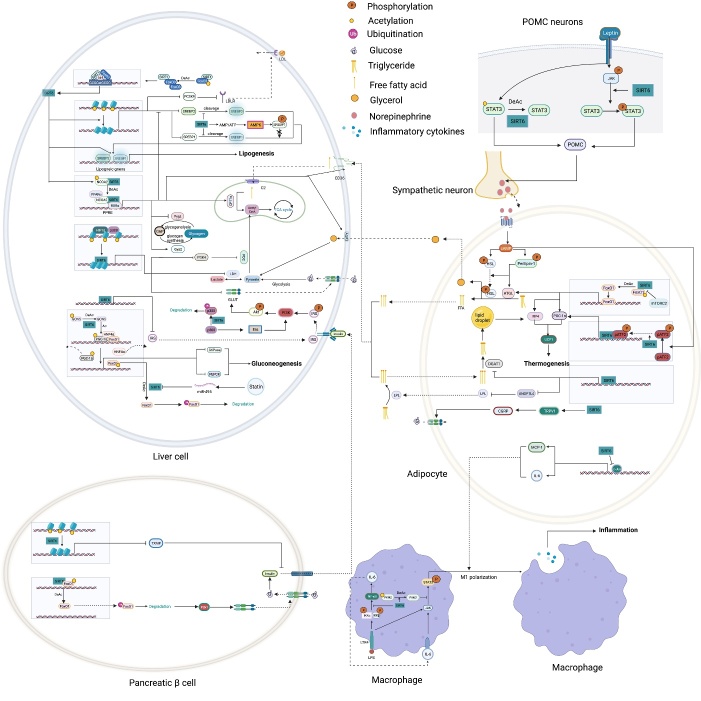
As an NAD(+)-dependent deacetylase, SIRT6 can regulate various metabolic pathways in response to cellular energy demands [109] (Fig. 4). Sirt6-deficient mouse models showed severe metabolic disorders, including elevated insulin resistance, high serum triglyceride levels and obesity, suggesting important roles of SIRT6 in glucose, lipid and energy metabolism [110].
Figure 4.
The roles played by SIRT6 in metabolic homeostasis. Glucose metabolism: By deacetylating H3K9 on HIF1α target genes, SIRT6 downregulates PDK4 expression to maintain the catalytic activity of the mitochondrial pyruvate dehydrogenase complex (PDC); LDH suppresses lactate production; and GLUT decreases glucose uptake. SIRT6 also inhibits insulin receptor and insulin receptor substrate (IRS) expression, repressing the phosphorylation of AKT at serine 473 (S473) and threonine 308 (T308) and subsequent glucose uptake. In pancreatic β-cells, SIRT6 inhibits Txnip expression to maintain cell function and survival. In addition, SIRT6-induced FoxO1 deacetylation promotes FoxO1 nuclear export and subsequent degradation, releasing the FoxO1 transcriptional repression of Pdx1 and Glut2. Increased Glut2 and Pdx1 expression promote glucose uptake by β-cells and subsequent insulin production and secretion, increasing glucose uptake and consumption by liver cells. In adipocytes, SIRT6 activates the TRPV1-CGRP-GLUT4 signaling axis to promote glucose uptake. However, in liver cells, SIRT6 downregulates p300 in a ubiquitin-proteasome system-dependent manner to reduce the expression of the estrogen receptor ERα, which restricts PI3K and AKT phosphorylation, eventually disrupting insulin signal transduction and cellular insulin sensitivity. SIRT6 deacetylates GCN5 at K549 to promote its phosphorylation and subsequent acetyltransferase activity on PGC-1α. The acetylation of PGC-1α compromises its ability to promote the expression of gluconeogenic genes, such as PCK1 and G6PC. In addition, statins increase endogenous miR-495 expression to downregulate SIRT6 expression, inhibiting PGC-1α coactivation factor FoxO1 deacetylation and subsequent ubiquitination and degradation. In a PPARα-dependent manner, SIRT6 inhibits Pygl expression to decrease glycogenolysis and promotes Gys2 expression to increase glycogen synthesis. Lipid metabolism: In liver cells, SIRT1 deacetylates FoxO3 to promote the formation of the SIRT1-FOXO3a-NRF1 (SFN) complex on the SIRT6 promoter, increasing SIRT6 protein expression levels. FoxO3 also recruits SIRT6 to the promoter of SREBP2. By deacetylating H3K9 and H3K56, SIRT6 inhibits SREBP1/2 and PCSK9 expression to suppress lipogenic gene expression and low-density lipoprotein (LDL) receptor degradation, respectively. SIRT6 inhibits the cleavage of SREBP1/2 to prevent their activation. Sirt6 also increases the AMP/ATP ratio to promote AMPK-mediated phosphorylation of SREBP1, which inhibits its cleavage. SIRT6 deacetylates NCOA2 at K780 to activate PPARα transcriptional activity. Activated PPARα binds to the retinoic acid receptor RXRα to form a heterodimer that regulates different metabolic pathways. In a PPARα-dependent manner, SIRT6 inhibits SREBP1/2 and their target gene expression to suppress cholesterol biogenesis; stimulates the expression of the FA transporter cluster of differentiation 36 (CD36), acetyl carnitine (C2) and the β-oxidation activator Cpt1α to promote fatty acid utilization; and promotes glycerol transporter Aqp3 expression, leading to increased glycerol uptake. In POMC-expressing neurons, SIRT6 maintains the leptin-induced phosphorylation of STAT3, which promotes POMC production, while SIRT6 can reduce POMC production by deacetylating STAT3. POMC promotes sympathetic activity in adipose tissues, increasing norepinephrine release to increase the cAMP level in adipocytes. An increased cAMP level promotes phosphorylation of hormone-sensitive triglyceride lipase (HSL), Perilipin-1 and ATF2. On the one hand, phosphorylated Perilipin-1 activates ATGL, and on the other hand, it transfers activated HSL from the cytoplasm to the lipid droplet surface, thereby promoting lipolysis. The breakdown of triglycerides yields free fatty acids (FFAs) and glycerol, which are then transported to liver cells through the long-chain FA transporter CD36 and the glycerol transporter aquaporin 3 (AQP3). In adipocytes, SIRT6 promotes deacetylation of FoxO1 to increase ATGL expression. In addition, SIRT6 suppresses DGAT1 expression to inhibit the synthesis of triglycerides. SIRT6 also suppresses the expression of ANGPTL4 to upregulate lipoprotein lipase (LPL) production, thereby enhancing the clearance of serum triglycerides. In brown adipocytes, SIRT6-mediated deacetylation of FoxO1 promotes interferon regulatory factor 4 (IRF4) and PGC1α expression to promote UCP1 expression, thereby enhancing thermogenesis. In addition, SIRT6 promotes p-ATF2 binding to the promoter of PGC1α, upregulating PGC1αexpression. However, SIRT6 suppresses the expression of c-JUN target genes (MCP-1 and IL-6) to inhibit inflammation in adipose tissue. In macrophages, SIRT6 not only inhibits the expression of the NF-KB target gene IL-6 but also prevents STAT3 phosphorylation by deacetylating PKM2 at K433, thereby disrupting the activation of the NF-κB-IL-6-STAT3 axis and preventing macrophage polarization and migration toward adipose tissue.
3.2.1 SIRT6 in glucose metabolism
Early studies showed that Sirt6-deficient mice presented with lethal hypoglycemia, highlighting the regulatory role played by SIRT6 in glucose metabolism [88]. In-depth studies showed that SIRT6 regulates glucose homeostasis through multiple complex processes, including glycogen synthesis and glycogenolysis, glucose uptake, insulin sensitivity, insulin secretion and gluconeogenesis at the cellular level.
3.2.1.1 Glycolysis and glucose transport
Sirt6-deficient mice presented with an intrinsic increase in glucose uptake in both brown adipose tissue (BAT) and muscle [111]. Mechanistically, SIRT6 competed with HIF1α at the promoters of several glycolytic genes, including glucose transporter 1 (GLUT1), pyruvate dehydrogenase kinase 4 (PDK4), and lactate dehydrogenase (LDH), deacetylating histone H3K9 at those promoters, which suppresses the expression of these genes [111]. In addition, SIRT6 deficiency led to the increased membrane association of GLUT1 and GLUT4 and activated the AKT signaling pathway by increasing the expression of insulin receptor substrates (IRSs), such as IRS1 and IRS2, eventually enhancing glucose uptake [112]. However, a separate study reported that overexpression of Sirt6 increased insulin-stimulated glucose uptake in skeletal muscle and liver, suggesting that SIRT6 engaged in tissue-specific regulation of glycometabolism [113]. Because SIRT6 can lower blood glucose levels by enhancing glycolysis and glucose uptake, SIRT6 inhibition has been considered a promising therapeutic treatment for hyperglycemia. Recently, the application of small-molecule SIRT6 inhibitors has been shown to be a viable strategy for regulating blood glucose levels [114, 115]. These studies with type 2 diabetes mellitus (T2DM) mice model suggested that SIRT6 inhibitors increased oral glucose tolerance by upregulating the expression of GLUT1 and GLUT4 in muscle and enhancing glycolytic activity.
3.2.1.2 Gluconeogenesis
Net hepatic glucose production reflects the net increase in glucose flux due to glycogen synthesis, glycogenolysis, gluconeogenesis, glycolysis and other metabolic activities, and dysregulated gluconeogenesis, not glycogen breakdown, is a primary cause of T2DM [116]. In vivo, hepatic SIRT6 overexpression suppressed gluconeogenesis to lower blood glucose levels in diabetic mice model [117]. Mechanistically, SIRT6 deacetylates GCN5 to upregulate its acetyltransferase activity against peroxisome proliferator-activated receptor-γ coactivator 1-α (PGC-1α) [25], thereby reducing the expression of gluconeogenic genes, such as glucose-6-phosphatase (G6PC) and phosphoenolpyruvate carboxykinase (PCK1), and blood glucose levels [116]. In addition, SIRT6-mediated FoxO1 deacetylation promotes its nuclear export, which inhibits G6PC and PCK1 expression to suppress gluconeogenesis [42]. However, some previous studies showed that liver-specific Sirt6 KO did not affect gluconeogenesis [41], and in contrast, in the context of aging, SIRT6 promoted hepatic gluconeogenic gene expression to maintain energy homeostasis [118].
3.2.1.3 Insulin sensitivity and secretion
Dysregulated insulin secretion and decreased β-cell mass contribute to the development and progression of diabetes mellitus [119]; therefore, maintaining β-cell physiological function and proper insulin secretion are important for the organismal response to changes in blood glucose. Sirt6 overexpression protected model mice from developing hyperglycemia and decreased glucose tolerance after eating a high-fat diet (HFD) [113]. In contrast, compromised glucose-stimulated insulin secretion (GSIS) has been commonly observed in Sirt6-KO pancreatic β-cells [120-122]. These data suggest that the impaired insulin sensitivity and pancreatic β-cell function may cause glucose metabolic disorder in the Sirt6-depleted mice. Specifically, SIRT6 can maintain mitochondrial function and regulate Ca2+ dynamics, affecting cell membrane depolarization and/or signaling after depolarization, regulating insulin secretion [121]. Furthermore, SIRT6-induced deacetylation of FoxO1 triggers its nuclear export and degradation, which increases pancreatic duodenal homeobox 1 (Pdx1) and glucose transporter 2 (GLUT2) expression, upregulating insulin secretion[120]. In an obese HFD-fed mice model, SIRT6 was found to activate the transient receptor potential vanilloid 1 (TRPV1)-calcitonin gene-related peptide (CGRP)-GLUT4 signaling axis to increase adipocyte glucose uptake and insulin sensitivity [123]. In addition, SIRT6 also cloud downregulate the expression of thioredoxin-interacting protein (Txnip), thereby attenuating glucose-stimulated glucotoxicity, promoting the survival of β-cells and partially increasing glucose tolerance [124]. However, a separate study found that SIRT6 disrupted insulin signal transduction and reduced insulin sensitivity in the liver through the Sirt6-p300-Erα-PI3K signaling pathway, suggesting SIRT6 regulated insulin sensitivity of liver in a gender-dependent manner [125].
3.2.1.4 Diabetic cardiomyopathy
Insulin resistance, hyperinsulinemia and diabetes mellitus are known to induce the development of diabetic cardiomyopathy (DCM) [126]. Inhibition of SIRT6 accelerates pathological processes closely related to DCM, including the inflammatory response and oxidative stress, thereby exacerbating DCM development [127]. A recent study showed that SIRT6 overexpression enabled mitochondria to maintain their functions and protect myocardial cells from DCM development by downregulating the expression of the negative Nrf2 regulator Kelch-like ECH-associated protein 1 (Keap1), which stabilized Nrf2 [128]. In addition, melatonin-mediated activation of the SIRT6-AMPK-PGC1α-AKT axis has been found to attenuate DCM development and progression and subsequently to reduce myocardial vulnerability to myocardial I/R injury [129]. Furthermore, SIRT6 has been shown to attenuate high-glucose-induced apoptosis by activating the AMPK pathway in podocytes [130]. Recent clinical studies showed that metformin treatment led to improved clinical outcomes for acute myocardial infarction patients with prediabetes by improving SIRT6 expression, which reduced inflammatory factor effects [131].
3.2.2 SIRT6 in lipid metabolism
The important roles of SIRT6 in regulating lipid metabolism have been observed in animal models. In hepatic-specific SIRT6 knockout mice, increased triglyceride accumulation, decreased β-oxidation and accelerated fatty liver formation were observed [41]. Additionally, neural-specific Sirt6-KO mice did not develop fatal hypoglycemia but presented with growth retardation and eventually became obese [132]. Transgenic mice overexpressing Sirt6, however, exhibited reduced accumulation of visceral fat, triglycerides and low-density lipoprotein cholesterol (LDL-C) after consuming a HFD [110].
3.2.2.1 Lipid synthesis
In adipocytes, SIRT6 has been shown to suppress the transcription of specific peroxisome proliferator-activated receptor γ (PPARγ)-regulated genes such as angiopoietin-like protein 4 (ANGPTL4) and diacylglycerol acyltransferase 1 (DGAT1), facilitating the clearance of serum triglycerides and reducing triglyceride synthesis, respectively [110]. SIRT6 also negatively regulates cholesterol biosynthesis in the following three ways: repressing the expression of sterol-regulatory element binding protein 1 and 2 (SREBP1 and SREBP2) and their target genes; restricting the cleavage of SREBP1/SREBP2 to prevent the formation of their active forms; and activating AMPK to inhibit the phosphorylation of SREBP1, eventually inhibiting its nuclear translocation and lipogenic program activation in the liver [133, 134]. In addition, SIRT6 activates PPARα to inhibit SREBP-dependent cholesterol synthesis [135]. Moreover, SIRT6 downregulates the proprotein convertase subtilisin/kexin type 9 (PCSK9) by deacetylating H3K9 and H3K56, preventing LDL receptor degradation and causing increased LDL uptake and decreased serum LDL-C concentrations [136]. In addition, SIRT6 activates the expression of the CDP-diacylglycerol synthase CDS1 and CDS2, promoting the de novo biosynthesis of cardiolipin (CL) and attenuating palmitic acid-induced lipid accumulation [137]. Finally, SIRT6 can remodel the chromatin structure to modulate BMAL1: CLOCK binding to DNA and thus regulate the rhythmic expression of proteins involved in FA and cholesterol metabolism [138]. Therefore, SIRT6 may be an important mediator between metabolic cues and epigenetic signaling to control circadian rhythmicity.
3.2.2.2 Lipolysis
Previous studies showed that SIRT6 increased β-oxidation by activating AMPK [45, 139]. Recently, SIRT6 has been found to deacetylate the PPARα coactivator NCOA2 to upregulate PPARα activity and the expression of its target gene carnitine palmitoyl transferase 1a (Cpt1a), eventually increasing lipid consumption [135]. A separate study found that PKCζ phosphorylated SIRT6 to promote its enrichment at the promoters of FA β-oxidation-related genes, inducing their expression [58]. In addition, SIRT6-induced deacetylation of FoxO1 increased its nuclear retention and transcriptional activity to induce the expression of the key lipolytic enzyme adipose triglyceride lipase (ATGL) to upregulate lipolysis [140]. In pro-opiomelanocortin (POMC)-expressing neurons, SIRT6 ensured activation of the leptin-induced JAK-STAT3 signaling cascade, increasing POMC production and thus promoting sympathetic nervous system (SNS) activity, which was closely related to the lipolytic activity and browning of adipose tissue [141]. Unexpectedly, SIRT6 overexpression has been shown to reduce STAT3 acetylation, which reduced POMC production, impairing SNS activity in adipose tissue [142]. These findings indicate the importance of maintaining SIRT6 activity within a physiological range.
3.2.2.3 Immunoreactions in adipose tissue
An association between continued inflammation of adipose tissue and metabolic pathway disorders has been established, making inflammatory pathways appealing targets in novel treatments of metabolic diseases and their related complications [143]. Fat-specific Sirt6 ablation increased the expression of two proinflammatory cytokines MCP-1 and IL-6 [140]. Furthermore, myeloid Sirt6 deficiency led to obesity-associated tissue inflammation and subsequent insulin resistance and macrophage infiltration[144]. Upon lipopolysaccharide (LPS) stimulation, activation of NF-κB and endogenous IL-6 production were promoted, which resulted in STAT3 activation and positive feedback signaling that stimulated NF-κB expression [144]. SIRT6 has been shown to deacetylate nuclear PKM2, leading to inhibited STAT3 phosphorylation, which disrupted NF-κB-IL-6-STAT3 axis activation, inhibiting the migration of macrophages to adipose tissue and macrophage polarization toward the proinflammatory M1 phenotype [26, 144]. These findings suggest that SIRT6 plays a protective role in controlling adipose tissue inflammation.
3.2.2.4 Thermogenesis in adipose tissue
Nonshivering thermogenesis in BAT substantially increases energy expenditure, which makes it a promising therapeutic target for the treatment of obesity and associated diseases [145]. Recently, SIRT6 has been shown to upregulate thermogenesis in response to cold and increase β-adrenergic agonist effects in BAT [146]. SIRT6 is an adaptor protein that recruits the phosphorylated activating transcription factor 2 (p-ATF2) protein to upregulate the expression of PGC-1α and the downstream thermogenic gene UCP1 [146]. However, Sirt6-KO mice showed insulin resistance, which corresponded with decreased expression of adiponectin in white adipose tissue (WAT) and UCP1 in BAT and increased inflammatory signaling in both WAT and BAT [147]. Similarly, SIRT6 could promote UCP1 and interferon regulatory factor 4 (IRF4) expression by deacetylating FoxO1 in BAT [56]. Hence, SIRT6 has the capacity to promote nonshivering thermogenesis mediated by UCP1, which may be a promising target for treatments that maintain metabolic homeostasis.
3.3 SIRT6 regulates oxidative stress and inflammation
3.3.1 SIRT6 alleviates oxidative stress
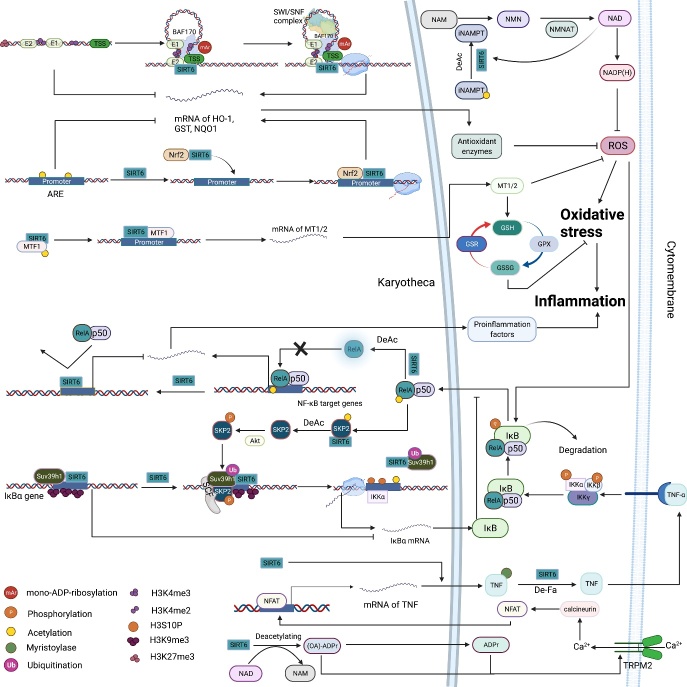
Oxidative stress, characterized by an imbalance between ROS production and clearance, is an important pathological mechanism that causes disease [148]. Previous studies have proven that oxidative stress can lead to downregulated SIRT6 activity [149, 150], and conversely, SIRT6 can alleviate oxidative stress through multiple signaling pathways, among which the SIRT6-NRF2-HO-1 pathway is the best characterized [151-153]. Specifically, SIRT6 deacetylates H3K56 at the HO-1 promoter, triggering the assembly of the NRF2-RNAP II transcription complex and upregulating HO-1 expression [154]. Interestingly, these observations seem to contradict findings indicating that SIRT6 interacts with RNA Pol II and negatively regulates the release of negative elongation factor (NELF) to stabilize RNA Pol II promoter-proximal pausing, thereby inhibiting gene expression [155]. Recently, additional research revealed that SIRT6 promotes the transcription of NRF2 target genes through its mono-ADP-ribosyltransferase activity but not deacetylase activity, and in contrast, SIRT6-induced deacetylation of H3K56 may suppress the NRF2 response [35]. Under oxidative stress conditions, SIRT6 catalyzes the mono-ADP-ribosylation of BRG/BRM-associated factor 170 (BAF170), promoting its recruitment to the E2 enhancer at the HO-1 promoter to promote chromatin remodeling. Subsequently, chromatin remodeling reduces chromain physical barrier for recruitment of RNA Pol II and NRF2. In addition, SIRT6 can deacetylate metal transcription factor (Mtf), a key transcription factor of metallothionein (Mt) 1 and 2, to counteract oxidative stress [156]. SIRT6 also directly deacetylates NAMPT to increase cellular NAD+ and NADPH levels and protect against oxidative stress [27].
3.3.2 Anti-inflammatory and proinflammatory effects of SIRT6
Inflammation is a complex pathophysiological response to damage; in general, the inflammatory cascade results in tissue damage and organ dysfunction [157]. Among proinflammatory cytokines, NF-κB and TNF-α are two important modulators of inflammation-related pathways. Recent studies found that the important roles played by SIRT6 in regulating anti-inflammatory responses were largely dependent on the regulated expression or protein functions of TNF-α and NF-κB. In macrophages, SIRT6 repressed NF-κB and endogenous IL-6 expression, thereby disrupting the crosstalk between NF-κB and STAT3 and inhibiting macrophage polarization toward the proinflammatory phenotype [144]. In myocardial I/R, SIRT6 restricted the NF-κB pathway to attenuate mitochondrial defects and cell death [158]. Under pressure overload, SIRT6 inhibited the expression of regulators downstream of NF-κB to attenuate inflammation and cardiac fibrosis[159]. Mechanistically, SIRT6 blocks the NF-κB signaling pathway at multiple levels. First, SIRT6 inhibits NF-κB-targeted gene expression by deacetylating H3K9 [160]. Second, SIRT6 restricts the transcriptional activity of the NF-κB subunit RelA via deacetylation [161]. Finally, SIRT6 accelerates negative feedback loop signaling that downregulates NF-κB activity by enhancing the expression of IκBα, an important inhibitor of NF-κB [162].
Interestingly, however, SIRT6 has also been shown to accelerate the production and secretion of the inflammatory cytokine TNF-α [15, 163, 164]. At the transcriptional level, O-acetyl-ADP ribose (OAADPr), a product of SIRT6-mediated deacetylation, or its derivative ADP ribose (ADPr) can activate transient receptor potential cation channel subfamily M, member 2 (TRPM2) to accelerate Ca2+ influx, thereby enhancing the expression of TNF-α and IL-8 [164]. Moreover, studies showed that a high intracellular NAD+ concentration can activate immune cells and promote TNF-α synthesis, but SIRT6 is the only sirtuin family member that has been shown to upregulate TNF-α mRNA translational efficiency [163]. In addition, upon LPS stimulation, SIRT6 is rapidly localized to the ER and promotes TNF-α secretion through its demyristoylase activit y[15, 165]. Overall, SIRT6 positively regulates TNF-α production and secretion at multiple levels, including at the transcriptional and translational levels and through posttranscriptional modification, demonstrating the important roles of SIRT6 in the transduction of TNF-α signaling. However, there is currently no evidence that SIRT6 exacerbates inflammatory diseases (e.g., atherosclerosis) through promoting TNF-αsecretion.
In summary, in a variety of tissue-derived cells, SIRT6 regulates oxidative stress and inflammation through multiple pathways, especially the NRF2, NF-κB and TNF-α signaling pathways (Fig. 5). Notably, discrepancies between the anti-inflammatory and proinflammatory roles played by SIRT6 may contribute to the specific effects of a cell type and stimulus; for example, SIRT6 deficiency did not influence NF-κB signaling in embryonic stem (ES) cells [111]. In summary, SIRT6 acts as a mediator in the TNF-α-NF-κB signaling axis to maintain proper reaction intensity, which is important for maintaining organismal homeostasis, to some extent. Regardless, the multifaceted roles played by SIRT6 in oxidative stress and inflammation make targeting SIRT6 a reasonable therapeutic strategy.
Figure 5.
The roles played by SIRT6 in the regulation of oxidative stress and inflammation. (A) Oxidative stress: SIRT6 deacetylates H3K56 at the promoter of Nrf2 target genes and is a bridge that recruits the NRF2-RNAP II transcription complex, thereby upregulating antioxidant enzyme expression. Furthermore, in response to oxidative stress, SIRT6 mono-ADP-ribosylates BAF170 at K312 to promote the recruitment of the SWI/SNF complex to the promoter region of HO-1, facilitating chromatin loop formation at the HO-1 locus to reduce the total physical volume between the enhancer and transcription start site. On the promoter of Mt1/2, SIRT6 physically interacts with and deacetylates Mtf1, coactivating the transcription of Mt1 and Mt2 to boost cell defenses against reactive oxygen species (ROS). Moreover, increased Mt1 and Mt2 levels promote the GSH-GSSH system to counteract oxidative stress. SIRT6 deacetylates NAMPT at K53 to promote its activity, increasing cellular NAD+ and NADPH levels, which confers cell resistance against oxidative stress damage. Increased NAD+ levels also upregulate SIRT6 deacetylase activity. (B) Inflammation: Through deacetylating H3K9 on the promoter of NF-κB target genes, SIRT6 prevents the binding of RelA to chromatin, repressing proinflammatory factor expression. In addition, SIRT6 directly interacts with and deacetylates RelA at K310, which restricts its DNA binding activity, terminating NF-κB signaling. Under TNF-α stimulation, IκBα is phosphorylated, subsequently ubiquitinated and eventually degraded, which leads to the translocation of NF-κB (RelA/p50) to the nucleus, where it binds to target gene promoters and regulates their expression. Moreover, TNF-α-induced activation of NF-κB activates SIRT6, which in turn deacetylates the E3 ubiquitin ligase SKP2 at K73 and K77, resulting in subsequent phosphorylation of SKP2. These modifications enhance the stability and nuclear content of SKP2, contributing to SKP2-mediated monoubiquitination of Suv39h1 and subsequent Suv39h1 release from the promoter of IκBα. Then, the H3K9 demethylation rate is increased, and H3S10 is phosphorylated by IKKα on the IκBα promoter, eventually promoting the transcription of IκBα. The product of SIRT6-mediated deacetylation OAADPr and its derivative ADPr activate TRPM2 to promote Ca2+ influx, thereby promoting TNF-α and IL-8 expression through the calcineurin-nuclear factor of activated T cells (NFAT) pathway. SIRT6 promotes TNF-α secretion by removing the fatty acyl groups from K19/20. SIRT6 upregulates TNF-α mRNA translation efficiency.
4. SIRT6 regulates the pathophysiology of CVDs
Among aging-related diseases, CVDs are thought to be the most important cause of human death. CVDs arise from cellular senescence, metabolic disorder, inflammation, cell death and aberrant cell growth, which can lead to the dysfunction of blood vessels and pathological cardiac remodeling in the heart. Sirtuins are extensively involved in these processes, regulating the pathophysiological conditions of the heart[166]. Here, we will focus on the roles of SIRT6 in atherosclerosis (early events, Fig. 6), cardiac hypertrophy and fibrosis (middle-stage events), heart failure (end-stage events) and I/R injury (recovery-stage events) in CVDs (Fig. 7).
Figure 6.
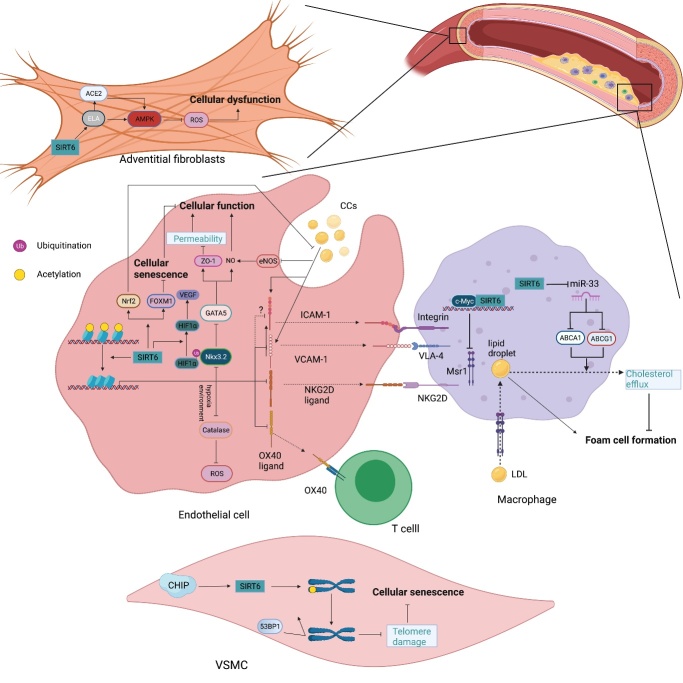
The roles played by SIRT6 in vascular atherosclerosis. In endothelial cells (ECs), SIRT6 downregulates the expression of proinflammatory cytokines, including VCAM-1, ICAM-1, NKG2D ligand and OX40 ligand, by deacetylating H3K9 on their respective promoters, reducing the number of inflammatory cells to inhibit vascular inflammation. SIRT6 also inhibits the cholesterol crystal-induced high expression of ICAM-1 and VCAM-1 by activating Nrf2. In addition, by deacetylating H3K9, SIRT6 suppresses Nkx3.2 expression to promote GATA5, which induces zonula occluden-1 (ZO-1) and NO activity, thereby decreasing cellular permeability and maintaining endothelial cell function. SIRT6 upregulates FOXM1 expression to prevent cellular senescence. Under hypoxic stress, SIRT6 deubiquitinates HIF-1α at K37 and K532 to protect it from decomposition, thus upregulating VEGF expression, which increases angiogenesis. In addition, SIRT6 inhibits catalase activity by deacetylating H3K56 on its promoter, which inhibits reactive oxygen species (ROS) clearance, further aggravating injury and hemorrhage of the neovasculature. In macrophages, SIRT6 downregulates Msr1 expression by suppressing c-Myc transcriptional activity and reducing oxidized low-density lipoprotein (ox-LDL) uptake and foam cell formation. SIRT6 also upregulates ABCA1 and ABCG1 expression by inhibiting miR-33 production and promoting cholesterol efflux. In vascular smoot muscle cells (VSMCs), SIRT6 maintains telomere stability by deacetylating H3K9 to prevent 53BP1 binding, thereby delaying cellular senescence and attenuating atherosclerosis development. In adventitial fibroblasts, SIRT6-ELA-ACE2 signaling can upregulate AMPK activity to reduce the ROS levels, thus protecting against oxidative stress and apoptosis.
Figure 7.
The roles played by SIRT6 in myocardial diseases. At c-Jun target gene promoters, SIRT6 interacts with c-Jun and deacetylates H3K9 to inhibit the expression of IGF signaling-related genes downstream of C-Jun, such as FoxO1, IGF2, IGF2R and Akt, attenuating cardiac hypertrophy and heart failure. SIRT6 also inhibits the phosphorylation of Akt to suppress its activation. Phosphorylated Akt promotes p300 phosphorylation and subsequent expression of NF-κB target genes, such as BNP and ANF, leading to the development of myocardial hypertrophy. In addition, phosphorylated Akt facilitates phosphorylation of FoxO3 and its subsequent nuclear export to inhibit autophagic gene expression, including Atg8, Atg12 and Gabarapl1, promoting pathological growth of cardiomyocytes. EGCG has been shown to upregulate SIRT6 activity by enhancing NMNAT activity and subsequently increasing NAD+ levels. In addition, the novel PARP-1 inhibitor AG-690 inhibits PARP-1 activity to maintain NAD+ intracellular levels, thereby upregulating SIRT6 activity. SIRT6 inhibits the expression and transcriptional activity STAT3 to hinder the expression of its target genes BNP and ANF. In addition, SIRT6 both inhibits NFATc4 expression and deacetylates NFATc4 to promote its nuclear export and subsequent degradation, suppressing the expression of its downstream genes BNP and ANF. On the promoters of mTOR signaling genes, SIRT6 interacts with Sp1 to repress the expression of related proteins, including mTOR, Rheb and p70S6K, thereby inhibiting both the synthesis of abnormal proteins and development of myocardial hypertrophy. In the promoter of Bcl-2, SIRT6 initially occupies this region via its property of high nucleosome-binding affinity and recruits TIP60. GATA4 recognizes the GATA sequence and subsequently interacts with SIRT6 via its C-terminal Zn-finger to form the SIRT6-TIP60-GATA4 complex. In this complex, SIRT6 deacetylase activity is repressed by GATA4, while TIP60 enhances GATA4 transcriptional activity and the acetylation level of local histones, ensuring the transcription of Blc-2. CircITCH sponges miR-330-5p to upregulate SIRT6 expression, attenuating reactive oxygen species (ROS) formation, DNA damage and cardiotoxicity. By deacetylating H3K9 and H3K56, SIRT6 inhibits the expression of TGF-β signaling-related proteins, such as SMAD3, TGF-β1 and TGF-β2. In addition, SIRT6 deacetylates SMAD3 at K333 and K378 to repress its transcriptional activity. SIRT6 deacetylates FoxO1 to promote its degradation, and SIRT6-mediated deacetylation of H3K9 within the PDK4 promoter also suppresses FoxO1 binding to this region, thereby reducing PDK4 expression and the subsequent accumulation of pyruvate and maintaining cardiac function. SIRT6 also inhibits myostatin expression to prevent the development of heart failure and its complications. After cardiac ischemia/reperfusion (I/R), SIRT6 upregulates AMPK activity and downregulates NF-κB signaling pathway activation, reducing cellular ROS levels and attenuating myocardial apoptosis. In addition, SIRT6 inhibits FoxO3 phosphorylation in an AMPK-dependent manner, and SIRT6 promotes FoxO3 nuclear translocation to induce FoxO3 target gene expression and reduce ROS production.
4.1 Atherosclerosis
Endothelial dysfunction promotes the initiation and development of atherosclerosis and its complications [167]. As a guardian that maintains genome stability, SIRT6 can protect endothelial cell (EC) from DNA and telomere damage and thus prevent premature senescence and maintain normal EC functions [79]. In addition, endothelial SIRT6 deficiency induces EC senescence associated with decreased FOXM1 expression [168]. Furthermore, SIRT6 attenuates minute cholesterol crystal (CC)-induced EC dysfunction through Nrf2 activation [169]. By deacetylating histone H3K9, SIRT6 downregulates NK3 homeobox 2 (Nkx3.2) transcription to promote GATA-binding protein 5 (GATA5) expression, thereby increasing the production of vascular endothelial nitric oxide synthase (eNOS) and preventing endothelial injury [66, 170]. SIRT6-induced deacetylation of caveolin-1 contributes to its autophagic degradation, retarding LDL transcytosis across ECs and atherosclerosis progression [171]. In VSMCs, the deacetylase activity of SIRT6 is crucial for delaying cellular senescence and inflammation because it prevents telomere damage, thereby stabilizing atherosclerotic plaque [78].
Atherosclerosis is an inflammatory disease of the arterial intima in which inflammatory cells and inflammatory responses play crucial roles [172]. Endothelial SIRT6 deficiency results in the increased expression of proinflammatory proteins, such as IL-1b and NF-κB [65]. In addition, SIRT6 inhibits the adhesion of monocytes to ECs by decreasing the expression of vascular cell adhesion molecule-1 (VCAM-1) and intracellular adhesion molecule-1(ICAM-1) to relieve atherosclerosis [173]. Additionally, the expression of tumor necrosis factor superfamily member 4 (TNFSF4, also known as OX40 L), a risk factor for atherosclerosis, was reduced by SIRT6 under normal conditions or after TNF-α stimulation [67]. The OX40L and OX40 interaction has been implicated in the activation of T cells and B cells, as well as cytokine production; therefore, SIRT6 may ameliorate atherosclerotic lesions [174]. In addition, SIRT6-induced macrophage autophagy in oxidized LDL-C (ox-LDL-C)-treated ECs increases cholesterol efflux and reduces macrophage infiltration, endothelial inflammation and foam cell formation, thereby attenuating the pathological development of atherosclerosis [175, 176].
As mentioned above, SIRT6 has been shown to accelerate the clearance of serum triglycerides and LDL-C [110, 136]; therefore, SIRT6 exerts a protective effect against atherosclerosis through the maintenance of lipid homeostasis. In contrast, ITCH ubiquitinates SIRT6, promoting its degradation, thereby disrupting FA metabolism and leading to hypercholesterolemia and hyperlipidemia and accelerating atherosclerosis progression [50]. In addition, oxidized LDL (ox-LDL) and its ligand, a scavenger receptor, contribute to the pathogenesis of atherosclerosis [177]. SIRT6 can suppress the expression of miR-33 to upregulate ABCA1 and ABCG1 expression, thereby promoting cholesterol efflux and attenuating atherosclerosis [176]. A separate study showed that SIRT6 was a corepressor of c-MYC expression, which led to the inhibition of macrophage scavenger receptor 1 (Msr1) expression, reducing ox-LDL uptake and inhibiting foam cell formation [178]. By deacetylating H3K9 and H3K56, SIRT6 also suppressed the expression of the ligand NKG2D, inhibiting immune cell recruitment and stabilizing atherosclerotic plaques [179, 180].
Interestingly, however, a separate innovative study showed a pernicious roles of SIRT6 in the development of hemorrhage in unstable carotid plaques [72]. SIRT6 deubiquitinated HIF-1α, protecting it from decomposition, and through its stabilization, HIF-1α upregulated the expression of vascular endothelial growth factor (VEGF) and promoted subsequent angiogenesis under hypoxic stress [72]. Moreover, SIRT6 deacetylated H3K56, downregulating the expression of the ROS scavenger Catalase, which aggravated the accumulation of ROS and subsequent neovascular injury and hemorrhage [72]. SIRT6 induces continuous angiogenesis, and hemorrhage of the neovasculature contributes to the instability of atherosclerotic plaques. In general, SIRT6 exerts an antiatherosclerotic effect, but the adverse effects of SIRT6 overexpression on atherosclerotic plaques are notable. In addition, these findings provide guidance for the development and use of SIRT6 modulators based on the different stages of diseases.
4.2 Cardiac hypertrophy and fibrosis
The heart undergoes hypertrophy in response to pressure-overload stimuli, but this hypertrophy generally progresses to heart failure through pathological cardiac remodeling [181]. As an anti-senescence molecule, SIRT6 was shown to attenuate cardiac aging and aging-associated cardiac hypertrophy [182]. In addition, cardiac-specific deletion of Sirt6 significantly increased left ventricular wall thickness, interstitial fibrosis and cardiomyocyte cross-sectional area, whereas overexpression of Sirt6 alleviates pressure-overload-induced cardiac hypertrophy and cardiac abnormalities [68, 183]. Indeed, SIRT6 serves as a negative regulator of IGF-Akt signaling, whose constitutive activation ultimately contributes to cardiac hypertrophy [68]. Additionally, SIRT6 reduces Akt protein levels to promote FoxO3 dephosphorylation and nuclear retention, inducing autophagic gene expression and attenuating isoproterenol (ISO)-induced hypertrophy [184]. Inhibition of NF-κB activation could inhibit or even reverse pressure-overload-induced cardiac remodeling and hypertrophy [181]. At the pharmaceutical level, epigallocatechin-3-gallate (EGCG) blocked NF-κB DNA-binding activity to protect against cardiac hypertrophy mediated through the PSMB5-Nmnat2-SIRT6 axis [185, 186]. Furthermore, AG-690/11026014 (6014), a novel PARP-1 inhibitor, has been shown to block Ang II-induced cardiac hypertrophy, which was partially attributed to activation of SIRT6 deacetylase activity through upregulating the intracellular NAD+ levels [187]. Furthermore, SIRT6 inhibits phenylephrine (PE)-induced induction of the fetal gene programme through inhibiting STAT3 expression and phosphorylation [188]. Similarly, SIRT6 downregulated the expression of NFATc4 and deacetylated NFATc4 to promote its nuclear export, thereby reducing BNP expression and protecting against cardiac hypertrophy [189]. Cardiac hypertrophy requires an increased rate of protein synthesis, and mTOR signaling controls protein synthesis and degradation [181]. At promoters of mTOR signaling genes, SIRT6 interacted with Sp1 to repress mTOR-related gene expression and subsequent protein synthesis, protecting against abnormal enlargement of cardiomyocyte [190].
Cardiac fibrosis, characterized by the transformation of fibroblasts into myofibroblasts, aggravates the development of pathologic cardiac remodeling and heart failure, and TGF-β/AMSD3 signaling is the key pathway in fibrogenesis [191, 192]. By deacetylating H3K9 and H3K56, SIRT6 suppresses the expression of TGF-β/SMAD3 signaling pathway genes [193]. In addition, SIRT6 can deacetylate SMAD3 to decrease its DNA-binding affinity [193]. Additionally, SIRT6 was reported to inhibit the phenotypic conversion of cardiac fibroblasts to myofibroblasts through blocking NF-κB signaling, thereby inhibiting cardiac fibrosis [69].
4.3 Heart failure
The metabolic network of cardiomyocytes contains a complex set of interacting pathways. Disordered energy metabolism in cardiomyocytes, such as a decrease in mitochondrial oxidative capacity, abnormal accumulation of substrate and dysfunctional intermediate utilization, are thought to be important pathogenic mechanisms in heart failure [194]. In Sirt6-null cells, HIF1-α expression was upregulated, which increased the expression of glycolytic genes and diminished mitochondrial respiration to shift glucose metabolism toward glycolysis [111]. Intriguingly, however, later research found no noticeable upregulation of HIF-1α-targeted glycolytic gene expression or glucose uptake in Sirt6-KO hearts, suggesting that SIRT6 deficiency-induced alterations of systemic glucose homeostasis in cardiomyocytes was not dependent on HIF-1α [68]. Recent findings proved that impaired mitochondrial oxidation in Sirt6-KO hearts was mediated by FoxO1 but not HIF-1α [70]. In cardiomyocytes, SIRT6 inhibited PDK4 expression by repressing the FoxO1 occupancy at its promoter, which contributed to enhanced PDH1 activity and subsequent metabolic shifting toward increased glucose oxidation to meet cellular energy requirements [70]. Hence, SIRT6 is indispensable for producing glycolytic substrates in the mitochondria for cardiac oxidation/utilization, and is, at least partially, critical for heart failure in cardiac-specific Sirt6-KO mice. In addition, SIRT6 attenuated myocardial lipid accumulation and the development of cardiac dysfunctions by reducing fatty acid uptake [195]. Mechanistically, SIRT6 inhibited the transcriptional activity of PPARγ to negatively regulate the expression of several critical fatty acid transporters, such as caveolin-1 and CD36 [195]. In summary, these studies showed that SIRT6 maintains the cellular energy supply by regulating glucose oxidation and lipid metabolism, thereby enhancing cardiac functions.
In addition to regulating metabolism, SIRT6 also maintains cardiac functions in other ways. For example, SIRT6 inhibits myostatin expression by mitigating NF-κB binding to the myostatin promoter, which is beneficial for maintaining muscle function and preventing chronic heart failure-induced cachexia [196]. In addition, the circular RNA ITCH (CircITCH) has been shown to prevent miR-330-5p-mediated degradation of SIRT6 to ameliorate doxorubicin-induced cardiotoxicity (DOXIC) [47]. Similarly, the SIRT6-TIP60-GATA4 axis couples gene transcription and epigenetic activation, protecting against DOXIC by regulating anti-apoptotic pathway signaling [197]. The nonenzymatic functions of SIRT6 in maintaining cardiac function provide a new perspective for better understanding the significance of SIRT6 in CVDs.
4.4 Cardiac ischemia/reperfusion (I/R) injury
The protective roles of SIRT6 in I/R injury have been established. For example, SIRT6 has been found to attenuate cerebral I/R by activating NRF2 signaling to reduce oxidative stress; in contrast, miR-370-induced inhibition of SIRT6 expression aggravates cerebral I/R by affecting the NRF2/ARE signaling pathway [198, 199]. A recent study found that EC-specific depletion of Sirt6 increased cell death after I/R injury and impaired blood-brain barrier integrity by activating caspase-3 [200]. In addition, cardiomyocytes overexpressing SIRT6 exhibited higher tolerance to hypoxia via reducing ROS production [201]. Subsequent findings showed that SIRT6 could attenuate cellular oxidative stress, myocardial damage, and ventricular remodeling in mice with myocardial I/R injury [71]. Specifically, I/R stress significantly increased the SIRT6 level in the cytoplasm to enhance AMPK activity through upregulating AMP/ATP and then promoting the transcriptional activity of FoxO3α. Subsequently, FoxO3α initiates the expression of antioxidant-encoding genes, such as catalase and superoxide dismutase (SOD2) to accelerate the clearance of ROS and impede cellular apoptosis, improving function and survival of cardiomyocytes in the ischemic heart. In a rabbit model, massive cardiomyocyte apoptosis was observed after myocardial I/R, and even a small increase in the apoptosis rate was considered sufficient to cause heart failure [202]. Hence, SIRT6-inhibited myocardial cell death is crucial for the maintenance of cardiac function after I/R.
Interestingly, silencing SIRT6 expression and inhibiting NAMPT activity reduced the biosynthesis of the neutrophil chemoattractant CXCL8 in mononuclear cells, thereby attenuating neutrophil infiltration and limiting myocardial infarct size during reperfusion [73]. In view of the protective effects of SIRT6 on myocardial I/R described above, it is surprising that SIRT6 promotes neutrophil infiltration during myocardial I/R. Notably, the authors of the previous study observed infarct size and neutrophil accumulation within 24 h of reperfusion, but long-term cardiac function was not evaluated in this study. New evidence has also shown that neutrophil infiltration is critical for tissue repair because it promotes debris clearance, cytokine scavenging, endothelial recovery, and angiogenesis [203]. Therefore, the potential effects of silencing SIRT6 expression on cardiac function preservation over long periods deserve further observation.
5. SIRT6 modulators
Given the important roles of SIRT6 in CVDs and multiple SIRT6-related molecular pathways, including pathways involved in maintaining genome stability and in inflammation and metabolism, regulation of SIRT6 expression and enzymatic activities may provide therapeutic benefits. In recent years, on the basis of details on the SIRT6 structural features, an increasing number of SIRT6 regulatory compounds with finely tuned regulatory effects on SIRT6 and related signaling pathways have been identified [204].
5.1 SIRT6 activators
First, some natural plant extracts have been proven to modulate SIRT6 expression or activity. For example, icariin and quercetin, both polyphenols, are known SIRT1 activators [205, 206], and recent studies have shown that they both stimulate SIRT6 deacetylation activity in a dose-dependent manner [198, 207]. In addition, quercetin inhibits SIRT6-dependent demyristoylation, suggesting that it has pleiotropic nonspecific effects [207]. The quercetin derivative cyanidin has been reported to be an effective activator of SIRT6 that upregulates its deacetylation activity and expression [208]. Ergothioneine (Egt) stimulated SIRT6 enzymatic activity and restricted NF-κB signaling, reducing ROS levels and inhibiting inflammation and senescence [209]. Chrysophanol has been shown to upregulate SIRT6 expression, alleviating metabolic syndromes mediated through the SIRT6/AMPK signaling pathway [210].
Second, many synthesized chemical compounds have been developed to activate SIRT6. For example, UBCS039 increased the deacetylation activity of SIRT6 to enhance autophagy-dependent cell death under oxidative stress [211]. 2-(1-Benzofuran-2-yl)-N-(diphenylmethyl) quinoline-4-carboxamide is another promising activator of SIRT6 that has shown excellent selectivity and biological anticancer functions [212]. Recently, several allosteric SIRT6 deacetylase activators have been identified, such as MDL-800, MDL-801and MDL-811 [213, 214]. Among these compounds, MDL-800 is notable because its permeability coefficient shows that it can be appropriately internalized and accumulated in cells. By activating SIRT6, MDL-800 has been shown to improve genome stability by promoting NHEJ and BER and attenuating hepatocyte injury and fibrosis [29, 215]. MDL-811 activates SIRT6 to attenuate inflammation and ischemic injury via the EZH2-FOXC1 axis [214]. Finally, an increasing number of studies have found that the pansirtuin activator nicotinamide riboside (NR), which upregulates NAD+ levels, may be a potential drug for the treatment of SIRT6-related diseases [216].
Third, clinical drugs have been shown to modulate SIRT6 to exert protective effects. A GLP-1 receptor agonist (GLP-1RA) and an SGLT2 inhibitor (SGLT2i) were shown to stabilize plaques in diabetic patients by upregulating SIRT6 [217, 218]. In addition, the hypolipidemic agent fluvastatin regulates cholesterol homeostasis by upregulating the deacetylase activity of SIRT6 [219, 220]. Rosiglitazone and rolipram have been shown to upregulate SIRT6 expression, ameliorating metabolic disorder [221, 222]. Detailed information on SIRT6 activators and their effects is presented in Table 2.
Table 2.
The activators of SIRT6.
| Categories and names | Mechanisms | Functions | Ref. | ||
|---|---|---|---|---|---|
| Natural products | Icariin
|
Promotes SIRT6 expression and enzymatic activity; inhibits NF-κB signaling | Attenuate senescence and cardiac inflammation | [198] | |
Egt
|
Promotes SIRT6 expression; inhibits NF-κB signaling | Attenuates high glucose-induced endothelial senescence | [209] | ||
| Flavonoids | Quercetin

|
Binds at a distal region of the acyl channel to promote SIRT6 deacetylase activity (EC50=990±250 μM) | Inhibits frizzled 4 (FZD4) expression to repress proliferation and invasion of hepatoblastoma cells | [207, 253] | |
Cyanidin
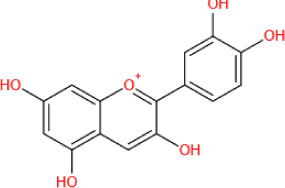
|
Binds at a distal region of the acyl channel to promote SIRT6 expression and deacetylase activity (EC50=460±20 μM); promotes FoxO3α, reduces GLUT1 and NF-κB signaling | Reduces inflammatory reactions | [208, 254] | ||
Chrysophanol
|
Promotes SIRT6 expression; promotes SIRT6/AMPK signaling | Alleviates metabolic disorders | [210] | ||
| Chemical synthesized compounds | UBCS039
|
Binds at a distal region of the acyl channel and “C site” of SIRT6 to promote SIRT6 deacetylase activity (EC50=38±13 μM) | Promotes autophagy under oxidative stress | [207, 211, 255] | |
2-(1-benzofuran-2-yl)-N-(diphenylmethyl) quinoline-4-carboxamide

|
Binds the surface allosteric site of SIRT6 to promote deacetylase (EC50=5.35±0.69 μM) and demyristoylase (EC50= 0.72 ± 0.25 μM) activities | Inhibits tumor growth | [212] | ||
MDL-800
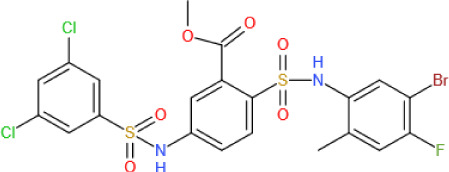
|
Binds the surface allosteric site of SIRT6 to promote SIRT6 deacetylase activity (EC50=10.3 μM); activates NHEJ and BER | Attenuates hepatocyte injury and fibrosis | [29, 213, 215] | ||
MDL-811
|
Binds the surface allosteric site of SIRT6 to promote SIRT6 deacetylation activity (EC50=5.7±0.8 μM); promotes EZH2 deacetylation and further FOXC1 expression; inhibits CYP24A1 expression | Attenuates ischemic injury; inhibits the proliferation of colorectal cancer cells | [214, 256] | ||
Nicotinamide riboside
|
Promotes sirtuin deacetylase activity by increasing NAD+ levels | Attenuates SIRT6 (or sirtuin)-related diseases | [216] | ||
| Clinical drugs | GLP-1RA and SGLT2i | Promotes SIRT6 expression | Inhibits inflammation and oxidative in ECs and stabilizes atherosclerotic plaque | [217, 218] | |
Fluvastatin
|
Binds the exit of the SIRT6 acyl channel; increases the phosphorylation of SREBP1 and AMPKα | Maintains cholesterol homeostasis | [219, 220] | ||
Rosiglitazone
|
Promotes SIRT6 expression by activating PPARγ | Ameliorates hepatic lipid accumulation | [222] | ||
Rolipram
|
Promotes SIRT6 expression | Improves adipose distribution and metabolic disorder | [221] | ||
5.2 SIRT6 inhibitors
Loss and gain of SIRT6 function result in paradoxical phenotypes in many diseases, such as altered metabolism and inflammation, making it difficult to ascertain whether inhibiting or activating SIRT6 actions is preferable for maintaining homeostasis [15, 73, 111, 160]. Therefore, several SIRT6 inhibitors playing protective roles through their intervention of SIRT6 activity under certain conditions have been developed. Compounds with a quinazoline dione-based structure and OSS-128167 impaired the binding of competing ligands, reducing SIRT6 deacetylase activity [223]. Among these compounds, SYN17739303 lowered blood glucose and inhibited inflammatory responses [114], while OSS-128167 upregulated glucose uptake and inhibited TNF-α activity in a time-dependent manner, sensitizing cancer cells to DNA-damaging agents [223].
Recently, trichostatin A (TSA), a known inhibitor of class I/II HDACs, has been shown to inhibit SIRT6 deacetylase activity with higher specificity and affinity than other known inhibitors [224, 225]. TSA increased p53K382Ac levels, accelerating the p53-mediated apoptosis [224]. Similar to quercetin, the quercetin derivative catechin gallate (CG) occupies the acyl channel of SIRT6, but powerfully inhibits SIRT6 activity [207]. By screening DNA-encoded chemical libraries, scientists found that the compound A127-(CONHPr)-B178 effectively inhibited SIRT6 activity, as shown by increased DNA damage, accelerated apoptosis and decreased TNF-α secretion [226]. Synthesized peptides harboring a different Nε-modified lysine residue mimicked endogenous substrates to competitively occupy substrate-binding regions, thereby inhibiting SIRT6 deacetylase and/or demyristoylase activity [227-230]. In a mouse model of T2DM, the compound 5-(4-methylpiperazin-1-yl)-2-nitroaniline upregulated GLUT1 expression to improve blood glucose levels by inhibiting SIRT6 deacetylase activity [115]. SIRT6 inhibitors and their effects are presented in Table 3.
Table 3.
The inhibitors of SIRT6.
| Categories and names | Mechanisms | Functions | Ref. | |
|---|---|---|---|---|
| Quinazolinediones | SYN17739303
|
All quinazolinediones bind the NAM-binding pocket, inhibiting the deacetylase activity of SIRT6; increase GLUT1 expression and reduce TNF-α secretion | Improves oral glucose tolerance, lowers blood glucose in T2DM; inhibits inflammatory responses | [114, 223, 257, 258] |
| Salicylates | OSS-128167
|
The salicylic moiety interacts with the NAM-binding pocket of SIRT6, inhibiting its deacetylase activity; reduces TNF-α secretion | Increases glucose uptake; sensitizes cancer cells to DNA damage agents; promotes cellular apoptosis in diffuse large B-cell lymphoma cells through inhibiting SIRT6-induced activation of PI3K/Akt/mTOR signaling pathway | [223, 259] |
| Trichostatin A |
|
Binds the NAM pocket and acyl channel to inhibit SIRT6 deacetylase activity; increases p53K382 acetylation level | Increasing cellular apoptosis under stress resistance | [224, 225] |
| Catechin gallate |
|
Binds the acyl channel to inhibit SIRT6 deacetylase activity | Not reported | [207] |
| A127-(CONHPr)-B178 |
|
Binds the NAD+ pocket to inhibit demyristoylase activity (IC50=6.7 μmol) | Promotes DNA damage, senescence and cell apoptosis | [226] |
| Lysine-based peptides | BH-TM4
|
A modified lysine residue binds the Nε-acetyl lysine-binding site, inhibiting both the deacetylase (IC50=8.2 μmol) and demyristoylase (IC50=1.7 μmol) activities of SIRT6 | Not reported | [229] |
Cyclic peptide
|
Inhibits the demyristoylase activity (IC50=0.319 μmol) of SIRT6 | Not reported | [230] | |
| 5-(4-Methylpiperazin-1-yl)-2-nitroaniline |
|
Inhibits SIRT6 deacetylase activity (IC50= 4.93 μmol); promotes GLUT1 levels | Reduces blood glucose | [115] |
Overall, the development of modulators targeting SIRT6 provides novel and promising strategies for the treatment of human diseases. Despite the limited number of clinical trials on SIRT6 modulators, several modulators, such as MDL-800, DML-811 and quinazoline dione analogs, have been shown to exert therapeutic effects in animal experiments. In addition, animal experiments and clinical studies have shown that moderate exercise and calorie restriction (CR) stimulated SIRT6 expression, improving age-related pathological processes such as inflammation, metabolic disorders, and susceptibility to CVDs [231-235]. Therefore, a healthy lifestyle, such as moderate physical exercise and CR, may be potential tools for preventing cellular senescence and aging by regulating SIRT6. In summary, these available interventions can pave the way for scientists to discover additional potent SIRT6 modulators with excellent drug-like properties.
6. Future perspectives
A decade of studies on SIRT6 has revealed its expansive functions and simultaneously shown the limitations of our current understanding of SIRT6. SIRT6 regulates chromatin status during transcription and DNA repair [155, 236], as well as transcriptional silencing during DNA repair[34], suggesting that it balances transcription and repair pathways in unexplored ways. A close relationship has been identified between SIRT1 and SIRT6, such as their synergistic effect on DNA repair and antagonistic effect on cancer [59, 237]. In addition, SIRT3 and SIRT6 have been found to maintain each other’s levels, attenuating the development of DCM [128]. Nevertheless, understanding the crosstalk between SIRT6 and other sirtuins, including possible links and cellular mechanisms in various diseases, remains a challenge in this field. In DNA damage repair and CVDs, SIRT6 is closely related to p53: p53 activates the expression of SIRT6 to protect against DNA damage, whereas SIRT6 attenuates doxorubicin (DOX)-induced cardiotoxicity by inhibiting p53 transcriptional activity [238, 239]. Studies have also shown that SIRT6 inhibits p53 transcriptional activity by deacetylating p53 at K381 and K382 [182, 224]. However, further studies are needed to determine whether SIRT6-induced deacetylation of p53 and crosstalk between SIRT6 and p53 play roles in other CVDs. Notably, both glucose deprivation and I/R significantly reduce SIRT6 chromatin enrichment and nuclear content [71, 155], suggesting that the roles of SIRT6 extend outside the nucleus, which is consistent with the roles played by SIRT6 in TNF-α secretion and cytoplasmic stress granule formation [5, 15]. According to these findings, results from further studies on the roles of SIRT6 in regulating the interaction between intranuclear and cytoplasmic factors will be significant.
To date, three distinctive enzymatic activities of SIRT6 have been elucidated: deacetylation, mono-ADP-ribosylation and deacylation of long-chain fatty acyl groups, such as myristoyl. The histone deacetylase activity of SIRT6 has been well characterized even though it is weaker in vitro than that of other sirtuins, which may be attributed to improved methods of detecting acetylated substrates [9]. Although studies have found that SIRT6 preferentially engages in long chain fatty acylation and that SIRT6 exhibits more powerful fatty acid deacylase activity than deacetylation activity in vitro, only a few substrates of SIRT6 fatty deacylation have been detected in vivo [15, 23, 38]. This dearth in substrate identification is partially due to the lack of efficient enrichment methods for lysine fatty-acylated peptides, which has limited the detection of these substrates to some extent. Therefore, developing pan antibodies against fatty acyl groups or identifying efficient fatty acyl-binding proteins may promote the development of new enrichment methods to detect substrates of SIRT6 fatty deacylation. In addition, activating the mono-ADP-ribosylation activity of SIRT6 may require specific conditions in vivo[9], as indicated by studies reporting that SIRT6 seems more likely to exhibit this activity against nuclear proteins under oxidative stress [32-35]. This requirement raises the following two questions: is the mono-ADP-ribosylation activity of SIRT6 related to its subcellular localization or stress conditions, and how does SIRT6 balance its three enzymatic activities and the crosstalk among the related substrates under physiological or stress conditions? Finally, recent reports have shown that the nonenzymatic activities of SIRT6 are also important [190, 195, 197]. Therefore, further studies are needed to reveal other significant deacetylase-independent roles played by SIRT6.
Although the significance of SIRT6 in human diseases is known to a certain extent, the number of SIRT6 modulators is still tiny. Indeed, only limited regulatory activity and efficacy in cells and animal models have been successfully observed because these modulators do not have optimal drug properties. Among known activators, pansirtuin activators may not be suitable for human use, as SIRT4 may exert detrimental effects in people with CVDs, such as aggravation of pathological cardiac hypertrophy [240]. Additionally, other phenomena need to be further studied: depletion and supraphysiological expression of SIRT6 in POMC neurons that disrupt both POMC production and POMC-related metabolism [141, 142], and overexpression of SIRT6 that aggravates the instability of atherosclerotic plaques [9, 72]. In addition, activators of SIRT6 have been described as potential tools for treating certain cancers; however, SIRT6 acts as a tumor promoter in some cancer types [241]. Therefore, SIRT6 may engage in cell- and/or tissue-specific regulation of transcription factors to regulate cellular biological processes. The findings have suggested that cell and/or tissue specificity, SIRT6 expression level and disease and disease stage specificity need to be taken into account when SIRT6 modulators are used. Therefore, future studies should explore how drug delivery systems can be utilized to specifically deliver SIRT6 modulators to targeted cells or tissues as needed. Certain SIRT6 inhibitors have been proven effective in lowering blood glucose in a T2DM animal model [114, 223]; however, their effects on other physiological functions within the body, such as aging, inflammation and susceptibility to CVDs, have not been evaluated. More importantly, reduced SIRT6-induced increases in glycolysis leads to tumorigenesis [242] (Supplementary Fig. 1; the detailed information on SIRT6 in cancers and neurodegenerative diseases in the Supplementary Materials). Therefore, the application of SIRT6 inhibitors in the CVD field should be carefully and further investigated. In addition, it is known that inhibition of SIRT6 activity increases the sensitivity of tumor cells to chemotherapeutic agents [243, 244], suggesting that the combination of SIRT6 inhibitors and other drugs is a feasible strategy for treating cancers.
7. Conclusions
In summary, the essential roles of SIRT6 in regulating chromatin and nuclear-cytoplasmic signaling pathways important for cellular homeostasis have been well characterized. In terms of genome stability, SIRT6 enhances DNA repair and maintains telomere integrity by regulating DNA repair and chromatin-associated factors, such as PARP1, DDB2, SNF2H and WRN. With respect to cellular metabolism, SIRT6 regulates multiple metabolic processes, including glycolysis, gluconeogenesis, insulin secretion, lipid synthesis, lipolysis and thermogenesis, mainly by regulating the multiple transcriptional activities of HIF1α, FOXO proteins and the PPAR family of transcription factors. In addition, SIRT6 maintains an appropriate inflammatory response by regulating the TNF-α and NF-κB signaling pathways. These functions can influence cellular senescence and aging-related diseases, including CVDs, cancer, and neurodegenerative diseases. Therefore, studying the biological functions of SIRT6 in different diseases is valuable and helpful for the identification of highly specific SIRT6 cellular targets. With a deeper understanding of SIRT6, certain SIRT6 regulatory compounds have been identified, offering novel and promising therapeutic options for aging-related diseases. Notably, recent studies have suggested that SIRT6 exerts adverse effects in some diseases, which are intriguing findings and worthy of further study. Therefore, when SIRT6-targeted therapeutic strategies or modulators are designed, the multifaceted features and cell specificity of SIRT6 need to be considered.
Supplementary Materials
The Supplementary data can be found online at: www.aginganddisease.org/EN/10.14336/AD.2022.0413.
Acknowledgements
We thank Dr. Lulan Chen, Na Wu, Zheyan Fang, Yiqing Hu, Runyang Feng and Xueting Yu for comments on the manuscript. This work was supported by the National Natural Science Foundation of China (81870182, 82000273, 82170334), China Postdoctoral Science Foundation (BX20200092) and the National Key Basic Research Programme (2020YFC1316700).
Footnotes
Competing interests
There is no conflict of interest involved in this review.
References
- [1].Imai S, Armstrong CM, Kaeberlein M, Guarente L (2000). Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature, 403:795-800. [DOI] [PubMed] [Google Scholar]
- [2].Seto E, Yoshida M (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol, 6:a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Finkel T, Deng CX, Mostoslavsky R (2009). Recent progress in the biology and physiology of sirtuins. Nature, 460:587-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Schutkowski M, Fischer F, Roessler C, Steegborn C (2014). New assays and approaches for discovery and design of Sirtuin modulators. Expert Opin Drug Discov, 9:183-199. [DOI] [PubMed] [Google Scholar]
- [5].Simeoni F, Tasselli L, Tanaka S, Villanova L, Hayashi M, Kubota K, et al. (2013). Proteomic analysis of the SIRT6 interactome: novel links to genome maintenance and cellular stress signaling. 3:3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kugel S, Mostoslavsky RJTibs (2014). Chromatin and beyond: the multitasking roles for SIRT6. 39:72-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Navas-Enamorado I, Bernier M, Brea-Calvo G, de Cabo RJArr (2017). Influence of anaerobic and aerobic exercise on age-related pathways in skeletal muscle. 37:39-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Vitiello M, Zullo A, Servillo L, Mancini FP, Borriello A, Giovane A, et al. (2017). Multiple pathways of SIRT6 at the crossroads in the control of longevity, cancer, and cardiovascular diseases. Ageing Res Rev, 35:301-311. [DOI] [PubMed] [Google Scholar]
- [9].Tasselli L, Zheng W, Chua KF (2017). SIRT6: Novel Mechanisms and Links to Aging and Disease. Trends Endocrinol Metab, 28:168-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tennen RI, Chua KF (2011). Chromatin regulation and genome maintenance by mammalian SIRT6. Trends Biochem Sci, 36:39-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu WH, Zheng J, Feldman JL, Klein MA, Kuznetsov VI, Peterson CL, et al. (2020). Multivalent interactions drive nucleosome binding and efficient chromatin deacetylation by SIRT6. Nat Commun, 11:5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM (2011). Structure and biochemical functions of SIRT6. J Biol Chem, 286:14575-14587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Carreño M, Bresque M, Machado MR, Santos L, Durán R, Vitturi DA, et al. (2020). Nitro-fatty acids as activators of hSIRT6 deacetylase activity. J Biol Chem, 295:18355-18366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Long D, Wu H, Tsang AW, Poole LB, Yoza BK, Wang X, et al. (2017). The Oxidative State of Cysteine Thiol 144 Regulates the SIRT6 Glucose Homeostat. Sci Rep, 7:11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, et al. (2013). SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature, 496:110-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, et al. (2014). Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab, 19:605-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dai H, Sinclair DA, Ellis JL, Steegborn C (2018). Sirtuin activators and inhibitors: Promises, achievements, and challenges. Pharmacol Ther, 188:140-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, et al. (2008). SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature, 452:492-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, et al. (2009). Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle, 8:2664-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kugel S, Mostoslavsky R (2014). Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem Sci, 39:72-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tasselli L, Xi Y, Zheng W, Tennen RI, Odrowaz Z, Simeoni F, et al. (2016). SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol, 23:434-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang WW, Zeng Y, Wu B, Deiters A, Liu WR (2016). A Chemical Biology Approach to Reveal Sirt6-targeted Histone H3 Sites in Nucleosomes. ACS Chem Biol, 11:1973-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Feldman JL, Baeza J, Denu JM (2013). Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem, 288:31350-31356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gil R, Barth S, Kanfi Y, Cohen HY (2013). SIRT6 exhibits nucleosome-dependent deacetylase activity. Nucleic Acids Res, 41:8537-8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dominy JE Jr., Lee Y, Jedrychowski MP, Chim H, Jurczak MJ, Camporez JP, et al. (2012). The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell, 48:900-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bhardwaj A, Das S (2016). SIRT6 deacetylates PKM2 to suppress its nuclear localization and oncogenic functions. Proc Natl Acad Sci U S A, 113:E538-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sociali G, Grozio A, Caffa I, Schuster S, Becherini P, Damonte P, et al. (2019). SIRT6 deacetylase activity regulates NAMPT activity and NAD(P)(H) pools in cancer cells. Faseb j, 33:3704-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bang IH, Kwon OK, Hao L, Park D, Chung MJ, Oh BC, et al. (2019). Deacetylation of XBP1s by sirtuin 6 confers resistance to ER stress-induced hepatic steatosis. Exp Mol Med, 51:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhang J, Li Y, Liu Q, Huang Y, Li R, Wu T, et al. (2021). Sirt6 Alleviated Liver Fibrosis by Deacetylating Conserved Lysine 54 on Smad2 in Hepatic Stellate Cells. Hepatology, 73:1140-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhong X, Huang M, Kim HG, Zhang Y, Chowdhury K, Cai W, et al. (2020). SIRT6 Protects Against Liver Fibrosis by Deacetylation and Suppression of SMAD3 in Hepatic Stellate Cells. Cell Mol Gastroenterol Hepatol, 10:341-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liszt G, Ford E, Kurtev M, Guarente L (2005). Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem, 280:21313-21320. [DOI] [PubMed] [Google Scholar]
- [32].Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, et al. (2011). SIRT6 promotes DNA repair under stress by activating PARP1. Science, 332:1443-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, et al. (2014). SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun, 5:5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rezazadeh S, Yang D, Biashad SA, Firsanov D, Takasugi M, Gilbert M, et al. (2020). SIRT6 mono-ADP ribosylates KDM2A to locally increase H3K36me2 at DNA damage sites to inhibit transcription and promote repair. Aging (Albany NY), 12:11165-11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rezazadeh S, Yang D, Tombline G, Simon M, Regan SP, Seluanov A, et al. (2019). SIRT6 promotes transcription of a subset of NRF2 targets by mono-ADP-ribosylating BAF170. Nucleic Acids Res, 47:7914-7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Van Meter M, Mao Z, Gorbunova V, Seluanov A (2011). SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle, 10:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang X, Khan S, Jiang H, Antonyak MA, Chen X, Spiegelman NA, et al. (2016). Identifying the functional contribution of the defatty-acylase activity of SIRT6. Nat Chem Biol, 12:614-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang X, Spiegelman NA, Nelson OD, Jing H, Lin H (2017). SIRT6 regulates Ras-related protein R-Ras2 by lysine defatty-acylation. Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wencel PL, Lukiw WJ, Strosznajder JB, Strosznajder RP (2018). Inhibition of Poly(ADP-ribose) Polymerase-1 Enhances Gene Expression of Selected Sirtuins and APP Cleaving Enzymes in Amyloid Beta Cytotoxicity. Mol Neurobiol, 55:4612-4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Min L, Ji Y, Bakiri L, Qiu Z, Cen J, Chen X, et al. (2012). Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat Cell Biol, 14:1203-1211. [DOI] [PubMed] [Google Scholar]
- [41].Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, et al. (2010). Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab, 12:224-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang P, Tu B, Wang H, Cao Z, Tang M, Zhang C, et al. (2014). Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proc Natl Acad Sci U S A, 111:10684-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yu J, Wu Y, Yang P (2016). High glucose-induced oxidative stress represses sirtuin deacetylase expression and increases histone acetylation leading to neural tube defects. J Neurochem, 137:371-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Scisciola L, Rizzo MR, Marfella R, Cataldo V, Fontanella RA, Boccalone E, et al. (2021). New insight in molecular mechanisms regulating SIRT6 expression in diabetes: Hyperglycaemia effects on SIRT6 DNA methylation. J Cell Physiol, 236:4604-4613. [DOI] [PubMed] [Google Scholar]
- [45].Elhanati S, Ben-Hamo R, Kanfi Y, Varvak A, Glazz R, Lerrer B, et al. (2016). Reciprocal Regulation between SIRT6 and miR-122 Controls Liver Metabolism and Predicts Hepatocarcinoma Prognosis. Cell Rep, 14:234-242. [DOI] [PubMed] [Google Scholar]
- [46].Dávalos A, Goedeke L, Smibert P, Ramírez CM, Warrier NP, Andreo U, et al. (2011). miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A, 108:9232-9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Han D, Wang Y, Wang Y, Dai X, Zhou T, Chen J, et al. (2020). The Tumor-Suppressive Human Circular RNA CircITCH Sponges miR-330-5p to Ameliorate Doxorubicin-Induced Cardiotoxicity Through Upregulating SIRT6, Survivin, and SERCA2a. Circ Res, 127:e108-e125. [DOI] [PubMed] [Google Scholar]
- [48].Shi MY, Bang IH, Han CY, Lee DH, Park BH, Bae EJ (2020). Statin suppresses sirtuin 6 through miR-495, increasing FoxO1-dependent hepatic gluconeogenesis. Theranostics, 10:11416-11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Thirumurthi U, Shen J, Xia W, LaBaff AM, Wei Y, Li CW, et al. (2014). MDM2-mediated degradation of SIRT6 phosphorylated by AKT1 promotes tumorigenesis and trastuzumab resistance in breast cancer. Sci Signal, 7:ra71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Stöhr R, Mavilio M, Marino A, Casagrande V, Kappel B, Möllmann J, et al. (2015). ITCH modulates SIRT6 and SREBP2 to influence lipid metabolism and atherosclerosis in ApoE null mice. Sci Rep, 5:9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ronnebaum SM, Wu Y, McDonough H, Patterson C (2013). The ubiquitin ligase CHIP prevents SirT6 degradation through noncanonical ubiquitination. Mol Cell Biol, 33:4461-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lin Z, Yang H, Tan C, Li J, Liu Z, Quan Q, et al. (2013). USP10 antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell Rep, 5:1639-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Zhou HZ, Zeng HQ, Yuan D, Ren JH, Cheng ST, Yu HB, et al. (2019). NQO1 potentiates apoptosis evasion and upregulates XIAP via inhibiting proteasome-mediated degradation SIRT6 in hepatocellular carcinoma. Cell Commun Signal, 17:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hu S, Liu H, Ha Y, Luo X, Motamedi M, Gupta MP, et al. (2015). Posttranslational modification of Sirt6 activity by peroxynitrite. Free Radic Biol Med, 79:176-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kalous KS, Wynia-Smith SL, Summers SB, Smith BC (2020). Human sirtuins are differentially sensitive to inhibition by nitrosating agents and other cysteine oxidants. J Biol Chem, 295:8524-8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Jung SM, Hung CM, Hildebrand SR, Sanchez-Gurmaches J, Martinez-Pastor B, Gengatharan JM, et al. (2019). Non-canonical mTORC2 Signaling Regulates Brown Adipocyte Lipid Catabolism through SIRT6-FoxO1. Mol Cell, 75:807-822.e808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Cai J, Zuo Y, Wang T, Cao Y, Cai R, Chen FL, et al. (2016). A crucial role of SUMOylation in modulating Sirt6 deacetylation of H3 at lysine 56 and its tumor suppressive activity. Oncogene, 35:4949-4956. [DOI] [PubMed] [Google Scholar]
- [58].Gao T, Li M, Mu G, Hou T, Zhu WG, Yang Y (2019). PKCζ Phosphorylates SIRT6 to Mediate Fatty Acid β-Oxidation in Colon Cancer Cells. Neoplasia, 21:61-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Meng F, Qian M, Peng B, Peng L, Wang X, Zheng K, et al. (2020). Synergy between SIRT1 and SIRT6 helps recognize DNA breaks and potentiates the DNA damage response and repair in humans and mice. Elife, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ghosh S, Liu B, Wang Y, Hao Q, Zhou ZJCr (2015). Lamin A Is an Endogenous SIRT6 Activator and Promotes SIRT6-Mediated DNA Repair. 13:1396-1406. [DOI] [PubMed] [Google Scholar]
- [61].Gorbunova V, Rezazadeh S, Seluanov A (2016). Dangerous Entrapment for NRF2. Cell, 165:1312-1313. [DOI] [PubMed] [Google Scholar]
- [62].Miteva YV, Cristea IM (2014). A proteomic perspective of Sirtuin 6 (SIRT6) phosphorylation and interactions and their dependence on its catalytic activity. Mol Cell Proteomics, 13:168-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. (2006). Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell, 124:315-329. [DOI] [PubMed] [Google Scholar]
- [64].Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, et al. (2012). The sirtuin SIRT6 regulates lifespan in male mice. Nature, 483:218-221. [DOI] [PubMed] [Google Scholar]
- [65].Winnik S, Auwerx J, Sinclair DA, Matter CM (2015). Protective effects of sirtuins in cardiovascular diseases: from bench to bedside. Eur Heart J, 36:3404-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Guo J, Wang Z, Wu J, Liu M, Li M, Sun Y, et al. (2019). Endothelial SIRT6 Is Vital to Prevent Hypertension and Associated Cardiorenal Injury Through Targeting Nkx3.2-GATA5 Signaling. Circ Res, 124:1448-1461. [DOI] [PubMed] [Google Scholar]
- [67].Xu S, Yin M, Koroleva M, Mastrangelo MA, Zhang W, Bai P, et al. (2016). SIRT6 protects against endothelial dysfunction and atherosclerosis in mice. Aging (Albany NY), 8:1064-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sundaresan NR, Vasudevan P, Zhong L, Kim G, Samant S, Parekh V, et al. (2012). The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat Med, 18:1643-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Tian K, Liu Z, Wang J, Xu S, You T, Liu P (2015). Sirtuin-6 inhibits cardiac fibroblasts differentiation into myofibroblasts via inactivation of nuclear factor κB signaling. Transl Res, 165:374-386. [DOI] [PubMed] [Google Scholar]
- [70].Khan D, Sarikhani M, Dasgupta S, Maniyadath B, Pandit AS, Mishra S, et al. (2018). SIRT6 deacetylase transcriptionally regulates glucose metabolism in heart. J Cell Physiol, 233:5478-5489. [DOI] [PubMed] [Google Scholar]
- [71].Wang XX, Wang XL, Tong MM, Gan L, Chen H, Wu SS, et al. (2016). SIRT6 protects cardiomyocytes against ischemia/reperfusion injury by augmenting FoxO3α-dependent antioxidant defense mechanisms. Basic Res Cardiol, 111:13. [DOI] [PubMed] [Google Scholar]
- [72].Yang Z, Huang Y, Zhu L, Yang K, Liang K, Tan J, et al. (2021). SIRT6 promotes angiogenesis and hemorrhage of carotid plaque via regulating HIF-1α and reactive oxygen species. Cell Death Dis, 12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Montecucco F, Bauer I, Braunersreuther V, Bruzzone S, Akhmedov A, Lüscher TF, et al. (2013). Inhibition of nicotinamide phosphoribosyltransferase reduces neutrophil-mediated injury in myocardial infarction. Antioxid Redox Signal, 18:630-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Childs BG, Durik M, Baker DJ, van Deursen JM (2015). Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med, 21:1424-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Niedernhofer LJ, Gurkar AU, Wang Y, Vijg J, Hoeijmakers JHJ, Robbins PD (2018). Nuclear Genomic Instability and Aging. Annu Rev Biochem, 87:295-322. [DOI] [PubMed] [Google Scholar]
- [76].Tennen RI, Bua DJ, Wright WE, Chua KF (2011). SIRT6 is required for maintenance of telomere position effect in human cells. Nat Commun, 2:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Yepuri G, Ramasamy R (2019). Significance and Mechanistic Relevance of SIRT6-Mediated Endothelial Dysfunction in Cardiovascular Disease Progression. Circ Res, 124:1408-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Grootaert MOJ, Finigan A, Figg NL, Uryga AK, Bennett MR (2021). SIRT6 Protects Smooth Muscle Cells From Senescence and Reduces Atherosclerosis. Circ Res, 128:474-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Cardus A, Uryga AK, Walters G, Erusalimsky JD (2013). SIRT6 protects human endothelial cells from DNA damage, telomere dysfunction, and senescence. Cardiovasc Res, 97:571-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].De Cecco M, Ito T, Petrashen AP, Elias AE, Skvir NJ, Criscione SW, et al. (2019). L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature, 566:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Gilbert N, Lutz-Prigge S, Moran JV (2002). Genomic deletions created upon LINE-1 retrotransposition. Cell, 110:315-325. [DOI] [PubMed] [Google Scholar]
- [82].Simon M, Van Meter M, Ablaeva J, Ke Z, Gonzalez RS, Taguchi T, et al. (2019). LINE1 Derepression in Aged Wild-Type and SIRT6-Deficient Mice Drives Inflammation. Cell Metab, 29:871-885.e875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Taddei A, Maison C, Roche D, Almouzni G (2001). Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol, 3:114-120. [DOI] [PubMed] [Google Scholar]
- [84].Campisi J, d'Adda di Fagagna F (2007). Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol, 8:729-740. [DOI] [PubMed] [Google Scholar]
- [85].Amano H, Chaudhury A, Rodriguez-Aguayo C, Lu L, Akhanov V, Catic A, et al. (2019). Telomere Dysfunction Induces Sirtuin Repression that Drives Telomere-Dependent Disease. Cell Metab, 29:1274-1290.e1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Gao Y, Tan J, Jin J, Ma H, Chen X, Leger B, et al. (2018). SIRT6 facilitates directional telomere movement upon oxidative damage. Sci Rep, 8:5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Rizzo A, Iachettini S, Salvati E, Zizza P, Maresca C, D'Angelo C, et al. (2017). SIRT6 interacts with TRF2 and promotes its degradation in response to DNA damage. Nucleic Acids Res, 45:1820-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Mostoslavsky R, Chua K, Lombard D, Pang W, Fischer M, Gellon L, et al. (2006). Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. 124:315-329. [DOI] [PubMed] [Google Scholar]
- [89].Jung ES, Choi H, Song H, Hwang YJ, Kim A, Ryu H, et al. (2016). p53-dependent SIRT6 expression protects Aβ42-induced DNA damage. Sci Rep, 6:25628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Britton S, Dernoncourt E, Delteil C, Froment C, Schiltz O, Salles B, et al. (2014). DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res, 42:9047-9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Tian X, Firsanov D, Zhang Z, Cheng Y, Luo L, Tombline G, et al. (2019). SIRT6 Is Responsible for More Efficient DNA Double-Strand Break Repair in Long-Lived Species. Cell, 177:622-638.e622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, et al. (2009). SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany NY), 1:109-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Yang G, Liu C, Chen SH, Kassab MA, Hoff JD, Walter NG, et al. (2018). Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res, 46:3446-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Lavin MF (2007). ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene, 26:7749-7758. [DOI] [PubMed] [Google Scholar]
- [95].Onn L, Portillo M, Ilic S, Cleitman G, Stein D, Kaluski S, et al. (2020). SIRT6 is a DNA double-strand break sensor. Elife, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Chen W, Liu N, Zhang H, Zhang H, Qiao J, Jia W, et al. (2017). Sirt6 Promotes DNA End Joining in iPSCs Derived from Old Mice. Cell Rep, 18:2880-2892. [DOI] [PubMed] [Google Scholar]
- [97].Mao Z, Tian X, Van Meter M, Ke Z, Gorbunova V, Seluanov A (2012). Sirtuin 6 (SIRT6) rescues the decline of homologous recombination repair during replicative senescence. Proc Natl Acad Sci U S A, 109:11800-11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zhang L, Bai L, Ren Q, Sun G, Si Y (2018). Protective effects of SIRT6 against lipopolysaccharide (LPS) are mediated by deacetylation of Ku70. Mol Immunol, 101:312-318. [DOI] [PubMed] [Google Scholar]
- [99].Van Meter M, Simon M, Tombline G, May A, Morello TD, Hubbard BP, et al. (2016). JNK Phosphorylates SIRT6 to Stimulate DNA Double-Strand Break Repair in Response to Oxidative Stress by Recruiting PARP1 to DNA Breaks. Cell Rep, 16:2641-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Xu Z, Zhang L, Zhang W, Meng D, Zhang H, Jiang Y, et al. (2015). SIRT6 rescues the age related decline in base excision repair in a PARP1-dependent manner. Cell Cycle, 14:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Hwang BJ, Jin J, Gao Y, Shi G, Madabushi A, Yan A, et al. (2015). SIRT6 protein deacetylase interacts with MYH DNA glycosylase, APE1 endonuclease, and Rad9-Rad1-Hus1 checkpoint clamp. BMC Mol Biol, 16:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Geng A, Tang H, Huang J, Qian Z, Qin N, Yao Y, et al. (2020). The deacetylase SIRT6 promotes the repair of UV-induced DNA damage by targeting DDB2. Nucleic Acids Res, 48:9181-9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Price BD, D'Andrea AD (2013). Chromatin remodeling at DNA double-strand breaks. Cell, 152:1344-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Tessarz P, Kouzarides T (2014). Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol, 15:703-708. [DOI] [PubMed] [Google Scholar]
- [105].Cohn MA, D'Andrea AD (2008). Chromatin recruitment of DNA repair proteins: lessons from the fanconi anemia and double-strand break repair pathways. Mol Cell, 32:306-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, et al. (2013). SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell, 51:454-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Atsumi Y, Minakawa Y, Ono M, Dobashi S, Shinohe K, Shinohara A, et al. (2015). ATM and SIRT6/SNF2H Mediate Transient H2AX Stabilization When DSBs Form by Blocking HUWE1 to Allow Efficient γH2AX Foci Formation. Cell Rep, 13:2728-2740. [DOI] [PubMed] [Google Scholar]
- [108].Hou T, Cao Z, Zhang J, Tang M, Tian Y, Li Y, et al. (2020). SIRT6 coordinates with CHD4 to promote chromatin relaxation and DNA repair. Nucleic Acids Res, 48:2982-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chalkiadaki A, Guarente L (2012). Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat Rev Endocrinol, 8:287-296. [DOI] [PubMed] [Google Scholar]
- [110].Kanfi Y, Peshti V, Gil R, Naiman S, Nahum L, Levin E, et al. (2010). SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell, 9:162-173. [DOI] [PubMed] [Google Scholar]
- [111].Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, et al. (2010). The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell, 140:280-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Xiao C, Kim HS, Lahusen T, Wang RH, Xu X, Gavrilova O, et al. (2010). SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J Biol Chem, 285:36776-36784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Anderson JG, Ramadori G, Ioris RM, Galiè M, Berglund ED, Coate KC, et al. (2015). Enhanced insulin sensitivity in skeletal muscle and liver by physiological overexpression of SIRT6. Mol Metab, 4:846-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Sociali G, Magnone M, Ravera S, Damonte P, Vigliarolo T, Von Holtey M, et al. (2017). Pharmacological Sirt6 inhibition improves glucose tolerance in a type 2 diabetes mouse model. Faseb j, 31:3138-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Sun W, Chen X, Huang S, Li W, Tian C, Yang S, et al. (2020). Discovery of 5-(4-methylpiperazin-1-yl)-2-nitroaniline derivatives as a new class of SIRT6 inhibitors. Bioorg Med Chem Lett, 30:127215. [DOI] [PubMed] [Google Scholar]
- [116].Sharabi K, Lin H, Tavares CDJ, Dominy JE, Camporez JP, Perry RJ, et al. (2017). Selective Chemical Inhibition of PGC-1α Gluconeogenic Activity Ameliorates Type 2 Diabetes. Cell, 169:148-160.e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Dominy J, Lee Y, Jedrychowski M, Chim H, Jurczak M, Camporez J, et al. (2012). The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. 48:900-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Roichman A, Elhanati S, Aon MA, Abramovich I, Di Francesco A, Shahar Y, et al. (2021). Restoration of energy homeostasis by SIRT6 extends healthy lifespan. Nat Commun, 12:3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Bell GI, Polonsky KS (2001). Diabetes mellitus and genetically programmed defects in beta-cell function. Nature, 414:788-791. [DOI] [PubMed] [Google Scholar]
- [120].Song MY, Wang J, Ka SO, Bae EJ, Park BH (2016). Insulin secretion impairment in Sirt6 knockout pancreatic β cells is mediated by suppression of the FoxO1-Pdx1-Glut2 pathway. Sci Rep, 6:30321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Xiong X, Wang G, Tao R, Wu P, Kono T, Li K, et al. (2016). Sirtuin 6 regulates glucose-stimulated insulin secretion in mouse pancreatic beta cells. Diabetologia, 59:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Xiong X, Sun X, Wang Q, Qian X, Zhang Y, Pan X, et al. (2016). SIRT6 protects against palmitate-induced pancreatic β-cell dysfunction and apoptosis. J Endocrinol, 231:159-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Tang W, Fan Y (2019). SIRT6 as a potential target for treating insulin resistance. Life Sci, 231:116558. [DOI] [PubMed] [Google Scholar]
- [124].Qin K, Zhang N, Zhang Z, Nipper M, Zhu Z, Leighton J, et al. (2018). SIRT6-mediated transcriptional suppression of Txnip is critical for pancreatic beta cell function and survival in mice. Diabetologia, 61:906-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Tang C, Liu P, Zhou Y, Jiang B, Song Y, Sheng L (2019). Sirt6 deletion in hepatocytes increases insulin sensitivity of female mice by enhancing ERα expression. J Cell Physiol, 234:18615-18625. [DOI] [PubMed] [Google Scholar]
- [126].Tan Y, Zhang Z, Zheng C, Wintergerst KA, Keller BB, Cai L (2020). Mechanisms of diabetic cardiomyopathy and potential therapeutic strategies: preclinical and clinical evidence. Nat Rev Cardiol, 17:585-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Huang Y, Zhang J, Xu D, Peng Y, Jin Y, Zhang L (2021). SIRT6-specific inhibitor OSS-128167 exacerbates diabetic cardiomyopathy by aggravating inflammation and oxidative stress. Mol Med Rep, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Kanwal A, Pillai VB, Samant S, Gupta M, Gupta MP (2019). The nuclear and mitochondrial sirtuins, Sirt6 and Sirt3, regulate each other's activity and protect the heart from developing obesity-mediated diabetic cardiomyopathy. Faseb j, 33:10872-10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Yu LM, Dong X, Xue XD, Xu S, Zhang X, Xu YL, et al. (2021). Melatonin attenuates diabetic cardiomyopathy and reduces myocardial vulnerability to ischemia-reperfusion injury by improving mitochondrial quality control: Role of SIRT6. J Pineal Res, 70:e12698. [DOI] [PubMed] [Google Scholar]
- [130].Fan Y, Yang Q, Yang Y, Gao Z, Ma Y, Zhang L, et al. (2019). Sirt6 Suppresses High Glucose-Induced Mitochondrial Dysfunction and Apoptosis in Podocytes through AMPK Activation. Int J Biol Sci, 15:701-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Sardu C, D'Onofrio N, Torella M, Portoghese M, Mureddu S, Loreni F, et al. (2021). Metformin Therapy Effects on the Expression of Sodium-Glucose Cotransporter 2, Leptin, and SIRT6 Levels in Pericoronary Fat Excised from Pre-Diabetic Patients with Acute Myocardial Infarction. Biomedicines, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Schwer B, Schumacher B, Lombard DB, Xiao C, Kurtev MV, Gao J, et al. (2010). Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc Natl Acad Sci U S A, 107:21790-21794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Elhanati S, Kanfi Y, Varvak A, Roichman A, Carmel-Gross I, Barth S, et al. (2013). Multiple regulatory layers of SREBP1/2 by SIRT6. Cell Rep, 4:905-912. [DOI] [PubMed] [Google Scholar]
- [134].Tao R, Xiong X, DePinho RA, Deng CX, Dong XC (2013). Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J Lipid Res, 54:2745-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Naiman S, Huynh FK, Gil R, Glick Y, Shahar Y, Touitou N, et al. (2019). SIRT6 Promotes Hepatic Beta-Oxidation via Activation of PPARα. Cell Rep, 29:4127-4143.e4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Tao R, Xiong X, DePinho RA, Deng CX, Dong XC (2013). FoxO3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. J Biol Chem, 288:29252-29259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Li M, Hou T, Gao T, Lu X, Yang Q, Zhu Q, et al. (2018). p53 cooperates with SIRT6 to regulate cardiolipin de novo biosynthesis. Cell Death Dis, 9:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, et al. (2014). Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell, 158:659-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Cui X, Yao L, Yang X, Gao Y, Fang F, Zhang J, et al. (2017). SIRT6 regulates metabolic homeostasis in skeletal muscle through activation of AMPK. Am J Physiol Endocrinol Metab, 313:E493-e505. [DOI] [PubMed] [Google Scholar]
- [140].Kuang J, Zhang Y, Liu Q, Shen J, Pu S, Cheng S, et al. (2017). Fat-Specific Sirt6 Ablation Sensitizes Mice to High-Fat Diet-Induced Obesity and Insulin Resistance by Inhibiting Lipolysis. Diabetes, 66:1159-1171. [DOI] [PubMed] [Google Scholar]
- [141].Tang Q, Gao Y, Liu Q, Yang X, Wu T, Huang C, et al. (2020). Sirt6 in pro-opiomelanocortin neurons controls energy metabolism by modulating leptin signaling. Mol Metab, 37:100994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Tang Q, Liu Q, Yang X, Wu T, Huang C, Zhang J, et al. (2021). Sirtuin 6 supra-physiological overexpression in hypothalamic pro-opiomelanocortin neurons promotes obesity via the hypothalamus-adipose axis. Faseb j, 35:e21408. [DOI] [PubMed] [Google Scholar]
- [143].Reilly SM, Saltiel AR (2017). Adapting to obesity with adipose tissue inflammation. Nat Rev Endocrinol, 13:633-643. [DOI] [PubMed] [Google Scholar]
- [144].Lee Y, Ka SO, Cha HN, Chae YN, Kim MK, Park SY, et al. (2017). Myeloid Sirtuin 6 Deficiency Causes Insulin Resistance in High-Fat Diet-Fed Mice by Eliciting Macrophage Polarization Toward an M1 Phenotype. Diabetes, 66:2659-2668. [DOI] [PubMed] [Google Scholar]
- [145].Betz MJ, Enerbäck S (2018). Targeting thermogenesis in brown fat and muscle to treat obesity and metabolic disease. Nat Rev Endocrinol, 14:77-87. [DOI] [PubMed] [Google Scholar]
- [146].Yao L, Cui X, Chen Q, Yang X, Fang F, Zhang J, et al. (2017). Cold-Inducible SIRT6 Regulates Thermogenesis of Brown and Beige Fat. Cell Rep, 20:641-654. [DOI] [PubMed] [Google Scholar]
- [147].Xiong X, Zhang C, Zhang Y, Fan R, Qian X, Dong XC (2017). Fabp4-Cre-mediated Sirt6 deletion impairs adipose tissue function and metabolic homeostasis in mice. J Endocrinol, 233:307-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Münzel T, Camici GG, Maack C, Bonetti NR, Fuster V, Kovacic JC (2017). Impact of Oxidative Stress on the Heart and Vasculature: Part 2 of a 3-Part Series. J Am Coll Cardiol, 70:212-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Carreira MC, Izquierdo AG, Amil M, Casanueva FF, Crujeiras AB (2018). Oxidative Stress Induced by Excess of Adiposity Is Related to a Downregulation of Hepatic SIRT6 Expression in Obese Individuals. Oxid Med Cell Longev, 2018:6256052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Liu R, Liu H, Ha Y, Tilton RG, Zhang W (2014). Oxidative stress induces endothelial cell senescence via downregulation of Sirt6. Biomed Res Int, 2014:902842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Ka SO, Bang IH, Bae EJ, Park BH (2017). Hepatocyte-specific sirtuin 6 deletion predisposes to nonalcoholic steatohepatitis by up-regulation of Bach1, an Nrf2 repressor. Faseb j, 31:3999-4010. [DOI] [PubMed] [Google Scholar]
- [152].Zhang W, Wei R, Zhang L, Tan Y, Qian C (2017). Sirtuin 6 protects the brain from cerebral ischemia/reperfusion injury through NRF2 activation. Neuroscience, 366:95-104. [DOI] [PubMed] [Google Scholar]
- [153].Yang Y, Tian T, Wang Y, Li Z, Xing K, Tian G (2019). SIRT6 protects vascular endothelial cells from angiotensin II-induced apoptosis and oxidative stress by promoting the activation of Nrf2/ARE signaling. Eur J Pharmacol, 859:172516. [DOI] [PubMed] [Google Scholar]
- [154].Pan H, Guan D, Liu X, Li J, Wang L, Wu J, et al. (2016). SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res, 26:190-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Etchegaray JP, Zhong L, Li C, Henriques T, Ablondi E, Nakadai T, et al. (2019). The Histone Deacetylase SIRT6 Restrains Transcription Elongation via Promoter-Proximal Pausing. Mol Cell, 75:683-699.e687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Kim HG, Huang M, Xin Y, Zhang Y, Zhang X, Wang G, et al. (2019). The epigenetic regulator SIRT6 protects the liver from alcohol-induced tissue injury by reducing oxidative stress in mice. J Hepatol, 71:960-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Ruparelia N, Chai JT, Fisher EA, Choudhury RP (2017). Inflammatory processes in cardiovascular disease: a route to targeted therapies. Nat Rev Cardiol, 14:133-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Cheng MY, Cheng YW, Yan J, Hu XQ, Zhang H, Wang ZR, et al. (2016). SIRT6 suppresses mitochondrial defects and cell death via the NF-κB pathway in myocardial hypoxia/reoxygenation induced injury. Am J Transl Res, 8:5005-5015. [PMC free article] [PubMed] [Google Scholar]
- [159].Zhang Q, Tu W, Tian K, Han L, Wang Q, Chen P, et al. (2019). Sirtuin 6 inhibits myofibroblast differentiation via inactivating transforming growth factor-β1/Smad2 and nuclear factor-κB signaling pathways in human fetal lung fibroblasts. J Cell Biochem, 120:93-104. [DOI] [PubMed] [Google Scholar]
- [160].Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, et al. (2009). SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell, 136:62-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Sun H, Wu Y, Fu D, Liu Y, Huang C (2014). SIRT6 regulates osteogenic differentiation of rat bone marrow mesenchymal stem cells partially via suppressing the nuclear factor-κB signaling pathway. Stem Cells, 32:1943-1955. [DOI] [PubMed] [Google Scholar]
- [162].Santos-Barriopedro I, Bosch-Presegué L, Marazuela-Duque A, de la Torre C, Colomer C, Vazquez BN, et al. (2018). SIRT6-dependent cysteine monoubiquitination in the PRE-SET domain of Suv39h1 regulates the NF-κB pathway. Nat Commun, 9:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Van Gool F, Gallí M, Gueydan C, Kruys V, Prevot PP, Bedalov A, et al. (2009). Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nat Med, 15:206-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Bauer I, Grozio A, Lasigliè D, Basile G, Sturla L, Magnone M, et al. (2012). The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. 287:40924-40937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Bresque M, Cal K, Pérez-Torrado V, Colman L, Rodríguez-Duarte J, Vilaseca C, et al. (2022). SIRT6 stabilization and cytoplasmic localization in macrophages regulates acute and chronic inflammation in mice. J Biol Chem: 101711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Li P, Ge J, Li H (2020). Lysine acetyltransferases and lysine deacetylases as targets for cardiovascular disease. Nat Rev Cardiol, 17:96-115. [DOI] [PubMed] [Google Scholar]
- [167].Gimbrone MA Jr., García-Cardeña G (2016). Endothelial Cell Dysfunction and the Pathobiology of Atherosclerosis. Circ Res, 118:620-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Lee OH, Woo YM, Moon S, Lee J, Park H, Jang H, et al. (2020). Sirtuin 6 deficiency induces endothelial cell senescence via downregulation of forkhead box M1 expression. Aging (Albany NY), 12:20946-20967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Jin Z, Xiao Y, Yao F, Wang B, Zheng Z, Gao H, et al. (2020). SIRT6 inhibits cholesterol crystal-induced vascular endothelial dysfunction via Nrf2 activation. Exp Cell Res, 387:111744. [DOI] [PubMed] [Google Scholar]
- [170].Messaoudi S, He Y, Gutsol A, Wight A, Hébert RL, Vilmundarson RO, et al. (2015). Endothelial Gata5 transcription factor regulates blood pressure. Nat Commun, 6:8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Zhao Y, Jia X, Yang X, Bai X, Lu Y, Zhu L, et al. (2022). Deacetylation of Caveolin-1 by Sirt6 induces autophagy and retards high glucose-stimulated LDL transcytosis and atherosclerosis formation. Metabolism: 155162. [DOI] [PubMed] [Google Scholar]
- [172].Bäck M, Yurdagul A Jr., Tabas I, Öörni K, Kovanen PT (2019). Inflammation and its resolution in atherosclerosis: mediators and therapeutic opportunities. Nat Rev Cardiol, 16:389-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Liu Z, Wang J, Huang X, Li Z, Liu P (2016). Deletion of sirtuin 6 accelerates endothelial dysfunction and atherosclerosis in apolipoprotein E-deficient mice. Transl Res, 172:18-29.e12. [DOI] [PubMed] [Google Scholar]
- [174].Wang X, Ria M, Kelmenson PM, Eriksson P, Higgins DC, Samnegård A, et al. (2005). Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat Genet, 37:365-372. [DOI] [PubMed] [Google Scholar]
- [175].Zi Y, Yi-An Y, Bing J, Yan L, Jing T, Chun-Yu G, et al. (2019). Sirt6-induced autophagy restricted TREM-1-mediated pyroptosis in ox-LDL-treated endothelial cells: relevance to prognostication of patients with acute myocardial infarction. Cell Death Discov, 5:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].He J, Zhang G, Pang Q, Yu C, Xiong J, Zhu J, et al. (2017). SIRT6 reduces macrophage foam cell formation by inducing autophagy and cholesterol efflux under ox-LDL condition. Febs j, 284:1324-1337. [DOI] [PubMed] [Google Scholar]
- [177].Mineo C (2020). Lipoprotein receptor signalling in atherosclerosis. Cardiovasc Res, 116:1254-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Arsiwala T, Pahla J, van Tits LJ, Bisceglie L, Gaul DS, Costantino S, et al. (2020). Sirt6 deletion in bone marrow-derived cells increases atherosclerosis - Central role of macrophage scavenger receptor 1. J Mol Cell Cardiol, 139:24-32. [DOI] [PubMed] [Google Scholar]
- [179].Zhang ZQ, Ren SC, Tan Y, Li ZZ, Tang X, Wang TT, et al. (2016). Epigenetic regulation of NKG2D ligands is involved in exacerbated atherosclerosis development in Sirt6 heterozygous mice. Sci Rep, 6:23912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Xia M, Guerra N, Sukhova GK, Yang K, Miller CK, Shi GP, et al. (2011). Immune activation resulting from NKG2D/ligand interaction promotes atherosclerosis. Circulation, 124:2933-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Nakamura M, Sadoshima J (2018). Mechanisms of physiological and pathological cardiac hypertrophy. Nat Rev Cardiol, 15:387-407. [DOI] [PubMed] [Google Scholar]
- [182].Ghosh S, Wong SK, Jiang Z, Liu B, Wang Y, Hao Q, et al. (2018). Haploinsufficiency of Trp53 dramatically extends the lifespan of Sirt6-deficient mice. Elife, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Li Y, Meng X, Wang W, Liu F, Hao Z, Yang Y, et al. (2017). Cardioprotective Effects of SIRT6 in a Mouse Model of Transverse Aortic Constriction-Induced Heart Failure. Front Physiol, 8:394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Lu J, Sun D, Liu Z, Li M, Hong H, Liu C, et al. (2016). SIRT6 suppresses isoproterenol-induced cardiac hypertrophy through activation of autophagy. Transl Res, 172:96-112.e116. [DOI] [PubMed] [Google Scholar]
- [185].Cai Y, Yu SS, Chen SR, Pi RB, Gao S, Li H, et al. (2012). Nmnat2 protects cardiomyocytes from hypertrophy via activation of SIRT6. FEBS Lett, 586:866-874. [DOI] [PubMed] [Google Scholar]
- [186].Cai Y, Yu SS, He Y, Bi XY, Gao S, Yan TD, et al. (2020). EGCG inhibits pressure overload-induced cardiac hypertrophy via the PSMB5/Nmnat2/SIRT6-dependent signalling pathways. Acta Physiol (Oxf):e13602. [DOI] [PubMed] [Google Scholar]
- [187].Liu M, Li Z, Chen GW, Li ZM, Wang LP, Ye JT, et al. (2014). AG-690/11026014, a novel PARP-1 inhibitor, protects cardiomyocytes from AngII-induced hypertrophy. Mol Cell Endocrinol, 392:14-22. [DOI] [PubMed] [Google Scholar]
- [188].Zhang X, Li W, Shen P, Feng X, Yue Z, Lu J, et al. (2016). STAT3 Suppression Is Involved in the Protective Effect of SIRT6 Against Cardiomyocyte Hypertrophy. J Cardiovasc Pharmacol, 68:204-214. [DOI] [PubMed] [Google Scholar]
- [189].Li Z, Zhang X, Guo Z, Zhong Y, Wang P, Li J, et al. (2018). SIRT6 Suppresses NFATc4 Expression and Activation in Cardiomyocyte Hypertrophy. Front Pharmacol, 9:1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Ravi V, Jain A, Khan D, Ahamed F, Mishra S, Giri M, et al. (2019). SIRT6 transcriptionally regulates global protein synthesis through transcription factor Sp1 independent of its deacetylase activity. Nucleic Acids Res, 47:9115-9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC (2016). Cardiac Fibrosis: The Fibroblast Awakens. Circ Res, 118:1021-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Su SA, Yang D, Wu Y, Xie Y, Zhu W, Cai Z, et al. (2017). EphrinB2 Regulates Cardiac Fibrosis Through Modulating the Interaction of Stat3 and TGF-β/Smad3 Signaling. Circ Res, 121:617-627. [DOI] [PubMed] [Google Scholar]
- [193].Maity S, Muhamed J, Sarikhani M, Kumar S, Ahamed F, Spurthi KM, et al. (2020). Sirtuin 6 deficiency transcriptionally up-regulates TGF-β signaling and induces fibrosis in mice. J Biol Chem, 295:415-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Lopaschuk GD, Karwi QG, Tian R, Wende AR, Abel ED (2021). Cardiac Energy Metabolism in Heart Failure. Circ Res, 128:1487-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Khan D, Ara T, Ravi V, Rajagopal R, Tandon H, Parvathy J, et al. (2021). SIRT6 transcriptionally regulates fatty acid transport by suppressing PPARγ. Cell Rep, 35:109190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Samant SA, Kanwal A, Pillai VB, Bao R, Gupta MP (2017). The histone deacetylase SIRT6 blocks myostatin expression and development of muscle atrophy. Sci Rep, 7:11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Peng L, Qian M, Liu Z, Tang X, Sun J, Jiang Y, et al. (2020). Deacetylase-independent function of SIRT6 couples GATA4 transcription factor and epigenetic activation against cardiomyocyte apoptosis. Nucleic Acids Res, 48:4992-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Chen Y, Sun T, Wu J, Kalionis B, Zhang C, Yuan D, et al. (2015). Icariin intervenes in cardiac inflammaging through upregulation of SIRT6 enzyme activity and inhibition of the NF-kappa B pathway. Biomed Res Int, 2015:895976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Ruan ZF, Xie M, Gui SJ, Lan F, Wan J, Li Y (2020). MiR-370 accelerated cerebral ischemia reperfusion injury via targeting SIRT6 and regulating Nrf2/ARE signal pathway. Kaohsiung J Med Sci, 36:741-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Liberale L, Gaul DS, Akhmedov A, Bonetti NR, Nageswaran V, Costantino S, et al. (2020). Endothelial SIRT6 blunts stroke size and neurological deficit by preserving blood-brain barrier integrity: a translational study. Eur Heart J, 41:1575-1587. [DOI] [PubMed] [Google Scholar]
- [201].Maksin-Matveev A, Kanfi Y, Hochhauser E, Isak A, Cohen HY, Shainberg A (2015). Sirtuin 6 protects the heart from hypoxic damage. Exp Cell Res, 330:81-90. [DOI] [PubMed] [Google Scholar]
- [202].Wencker D, Chandra M, Nguyen K, Miao W, Garantziotis S, Factor SM, et al. (2003). A mechanistic role for cardiac myocyte apoptosis in heart failure. J Clin Invest, 111:1497-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Silvestre-Roig C, Braster Q, Ortega-Gomez A, Soehnlein O (2020). Neutrophils as regulators of cardiovascular inflammation. Nat Rev Cardiol, 17:327-340. [DOI] [PubMed] [Google Scholar]
- [204].Fiorentino F, Mai A, Rotili D (2021). Emerging Therapeutic Potential of SIRT6 Modulators. J Med Chem, 64:9732-9758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Cătană CS, Atanasov AG, Berindan-Neagoe I (2018). Natural products with anti-aging potential: Affected targets and molecular mechanisms. Biotechnol Adv, 36:1649-1656. [DOI] [PubMed] [Google Scholar]
- [206].Wu B, Feng JY, Yu LM, Wang YC, Chen YQ, Wei Y, et al. (2018). Icariin protects cardiomyocytes against ischaemia/reperfusion injury by attenuating sirtuin 1-dependent mitochondrial oxidative damage. Br J Pharmacol, 175:4137-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].You W, Zheng W, Weiss S, Chua KF, Steegborn C (2019). Structural basis for the activation and inhibition of Sirtuin 6 by quercetin and its derivatives. Sci Rep, 9:19176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Rahnasto-Rilla M, Tyni J, Huovinen M, Jarho E, Kulikowicz T, Ravichandran S, et al. (2018). Natural polyphenols as sirtuin 6 modulators. Sci Rep, 8:4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].D'Onofrio N, Servillo L, Giovane A, Casale R, Vitiello M, Marfella R, et al. (2016). Ergothioneine oxidation in the protection against high-glucose induced endothelial senescence: Involvement of SIRT1 and SIRT6. Free Radic Biol Med, 96:211-222. [DOI] [PubMed] [Google Scholar]
- [210].Liu X, Yang Z, Li H, Luo W, Duan W, Zhang J, et al. (2020). Chrysophanol Alleviates Metabolic Syndrome by Activating the SIRT6/AMPK Signaling Pathway in Brown Adipocytes. Oxid Med Cell Longev, 2020:7374086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Iachettini S, Trisciuoglio D, Rotili D, Lucidi A, Salvati E, Zizza P, et al. (2018). Pharmacological activation of SIRT6 triggers lethal autophagy in human cancer cells. Cell Death Dis, 9:996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Chen X, Sun W, Huang S, Zhang H, Lin G, Li H, et al. (2020). Discovery of Potent Small-Molecule SIRT6 Activators: Structure-Activity Relationship and Anti-Pancreatic Ductal Adenocarcinoma Activity. J Med Chem, 63:10474-10495. [DOI] [PubMed] [Google Scholar]
- [213].Huang Z, Zhao J, Deng W, Chen Y, Shang J, Song K, et al. (2018). Identification of a cellularly active SIRT6 allosteric activator. Nat Chem Biol, 14:1118-1126. [DOI] [PubMed] [Google Scholar]
- [214].He T, Shang J, Gao C, Guan X, Chen Y, Zhu L, et al. (2021). A novel SIRT6 activator ameliorates neuroinflammation and ischemic brain injury via EZH2/FOXC1 axis. Acta Pharm Sin B, 11:708-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Chen Y, Chen J, Sun X, Yu J, Qian Z, Wu L, et al. (2020). The SIRT6 activator MDL-800 improves genomic stability and pluripotency of old murine-derived iPS cells. Aging Cell, 19:e13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Bonkowski MS, Sinclair DA (2016). Slowing ageing by design: the rise of NAD(+) and sirtuin-activating compounds. Nat Rev Mol Cell Biol, 17:679-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Balestrieri ML, Rizzo MR, Barbieri M, Paolisso P, D'Onofrio N, Giovane A, et al. (2015). Sirtuin 6 expression and inflammatory activity in diabetic atherosclerotic plaques: effects of incretin treatment. Diabetes, 64:1395-1406. [DOI] [PubMed] [Google Scholar]
- [218].D'Onofrio N, Sardu C, Trotta MC, Scisciola L, Turriziani F, Ferraraccio F, et al. (2021). Sodium-glucose co-transporter2 expression and inflammatory activity in diabetic atherosclerotic plaques: Effects of sodium-glucose co-transporter2 inhibitor treatment. Mol Metab, 54:101337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].You W, Steegborn C (2020). Structural Basis for Activation of Human Sirtuin 6 by Fluvastatin. ACS Med Chem Lett, 11:2285-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Kim JH, Lee JM, Kim JH, Kim KR (2018). Fluvastatin activates sirtuin 6 to regulate sterol regulatory element-binding proteins and AMP-activated protein kinase in HepG2 cells. Biochem Biophys Res Commun, 503:1415-1421. [DOI] [PubMed] [Google Scholar]
- [221].Wang Z, Liang Y, Zhang L, Zhang N, Liu Q, Wang Z (2018). Phosphodiesterase 4 inhibitor activates AMPK-SIRT6 pathway to prevent aging-related adipose deposition induced by metabolic disorder. Aging (Albany NY), 10:2394-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Yang SJ, Choi JM, Chae SW, Kim WJ, Park SE, Rhee EJ, et al. (2011). Activation of peroxisome proliferator-activated receptor gamma by rosiglitazone increases sirt6 expression and ameliorates hepatic steatosis in rats. PLoS One, 6:e17057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Parenti MD, Grozio A, Bauer I, Galeno L, Damonte P, Millo E, et al. (2014). Discovery of novel and selective SIRT6 inhibitors. J Med Chem, 57:4796-4804. [DOI] [PubMed] [Google Scholar]
- [224].Wood M, Rymarchyk S, Zheng S, Cen Y (2018). Trichostatin A inhibits deacetylation of histone H3 and p53 by SIRT6. Arch Biochem Biophys, 638:8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].You W, Steegborn C (2018). Structural Basis of Sirtuin 6 Inhibition by the Hydroxamate Trichostatin A: Implications for Protein Deacylase Drug Development. J Med Chem, 61:10922-10928. [DOI] [PubMed] [Google Scholar]
- [226].Yuen LH, Dana S, Liu Y, Bloom SI, Thorsell AG, Neri D, et al. (2019). A Focused DNA-Encoded Chemical Library for the Discovery of Inhibitors of NAD(+)-Dependent Enzymes. J Am Chem Soc, 141:5169-5181. [DOI] [PubMed] [Google Scholar]
- [227].Kokkonen P, Rahnasto-Rilla M, Kiviranta PH, Huhtiniemi T, Laitinen T, Poso A, et al. (2012). Peptides and Pseudopeptides as SIRT6 Deacetylation Inhibitors. ACS Med Chem Lett, 3:969-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].He Y, Yan L, Zang W, Zheng W (2015). Novel sirtuin inhibitory warheads derived from the N(ε)-acetyl-lysine analog L-2-amino-7-carboxamidoheptanoic acid. Org Biomol Chem, 13:10442-10450. [DOI] [PubMed] [Google Scholar]
- [229].He B, Hu J, Zhang X, Lin H (2014). Thiomyristoyl peptides as cell-permeable Sirt6 inhibitors. Org Biomol Chem, 12:7498-7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Liu J, Zheng W (2016). Cyclic peptide-based potent human SIRT6 inhibitors. Org Biomol Chem, 14:5928-5935. [DOI] [PubMed] [Google Scholar]
- [231].Zhang N, Li Z, Mu W, Li L, Liang Y, Lu M, et al. (2016). Calorie restriction-induced SIRT6 activation delays aging by suppressing NF-κB signaling. Cell Cycle, 15:1009-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Koltai E, Szabo Z, Atalay M, Boldogh I, Naito H, Goto S, et al. (2010). Exercise alters SIRT1, SIRT6, NAD and NAMPT levels in skeletal muscle of aged rats. Mech Ageing Dev, 131:21-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Mohammadkhani R, Khaledi N, Rajabi H, Salehi I, Komaki A (2020). Influence of the maternal high-intensity-interval-training on the cardiac Sirt6 and lipid profile of the adult male offspring in rats. PLoS One, 15:e0237148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Hooshmand-Moghadam B, Eskandari M, Golestani F, Rezae S, Mahmoudi N, Gaeini AA (2020). The effect of 12-week resistance exercise training on serum levels of cellular aging process parameters in elderly men. Exp Gerontol, 141:111090. [DOI] [PubMed] [Google Scholar]
- [235].Bétry C, Meugnier E, Pflieger M, Grenet G, Hercberg S, Galan P, et al. (2019). High expression of CPT1b in skeletal muscle in metabolically healthy older subjects. Diabetes Metab, 45:152-159. [DOI] [PubMed] [Google Scholar]
- [236].Toiber D, Erdel F, Bouazoune K, Silberman D, Zhong L, Mulligan P, et al. (2013). SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. 51:454-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Park JJ, Hah YS, Ryu S, Cheon SY, Won SJ, Lee JS, et al. (2021). MDM2-dependent Sirt1 degradation is a prerequisite for Sirt6-mediated cell death in head and neck cancers. Exp Mol Med, 53:422-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Wu S, Lan J, Li L, Wang X, Tong M, Fu L, et al. (2021). Sirt6 protects cardiomyocytes against doxorubicin-induced cardiotoxicity by inhibiting P53/Fas-dependent cell death and augmenting endogenous antioxidant defense mechanisms. Cell Biol Toxicol. [DOI] [PubMed] [Google Scholar]
- [239].(2000). Technology status evaluation report. High resolution and high-magnification endoscopy. Gastrointest Endosc, 52:864-866. [PubMed] [Google Scholar]
- [240].Luo YX, Tang X, An XZ, Xie XM, Chen XF, Zhao X, et al. (2017). SIRT4 accelerates Ang II-induced pathological cardiac hypertrophy by inhibiting manganese superoxide dismutase activity. Eur Heart J, 38:1389-1398. [DOI] [PubMed] [Google Scholar]
- [241].Fiorentino F, Carafa V, Favale G, Altucci L, Mai A, Rotili D (2021). The Two-Faced Role of SIRT6 in Cancer. Cancers(Basel), 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Sebastián C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, et al. (2012). The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell, 151:1185-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Cagnetta A, Soncini D, Orecchioni S, Talarico G, Minetto P, Guolo F, et al. (2018). Depletion of SIRT6 enzymatic activity increases acute myeloid leukemia cells' vulnerability to DNA-damaging agents. Haematologica, 103:80-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Cea M, Cagnetta A, Adamia S, Acharya C, Tai YT, Fulciniti M, et al. (2016). Evidence for a role of the histone deacetylase SIRT6 in DNA damage response of multiple myeloma cells. Blood, 127:1138-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Liu M, Liang K, Zhen J, Zhou M, Wang X, Wang Z, et al. (2017). Sirt6 deficiency exacerbates podocyte injury and proteinuria through targeting Notch signaling. Nat Commun, 8:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Xu S, Yin M, Koroleva M, Mastrangelo M, Zhang W, Bai P, et al. (2016). SIRT6 protects against endothelial dysfunction and atherosclerosis in mice. 8:1064-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Xiao C, Wang RH, Lahusen TJ, Park O, Bertola A, Maruyama T, et al. (2012). Progression of chronic liver inflammation and fibrosis driven by activation of c-JUN signaling in Sirt6 mutant mice. J Biol Chem, 287:41903-41913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Li Z, Xu K, Zhang N, Amador G, Wang Y, Zhao S, et al. (2018). Overexpressed SIRT6 attenuates cisplatin-induced acute kidney injury by inhibiting ERK1/2 signaling. Kidney Int, 93:881-892. [DOI] [PubMed] [Google Scholar]
- [249].Michishita E, McCord R, Berber E, Kioi M, Padilla-Nash H, Damian M, et al. (2008). SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. 452:492-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Cai J, Liu Z, Huang X, Shu S, Hu X, Zheng M, et al. (2020). The deacetylase sirtuin 6 protects against kidney fibrosis by epigenetically blocking β-catenin target gene expression. Kidney Int, 97:106-118. [DOI] [PubMed] [Google Scholar]
- [251].Hao L, Bang IH, Wang J, Mao Y, Yang JD, Na SY, et al. (2020). ERRγ suppression by Sirt6 alleviates cholestatic liver injury and fibrosis. JCI Insight, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Lee N, Kim DK, Kim ES, Park SJ, Kwon JH, Shin J, et al. (2014). Comparative interactomes of SIRT6 and SIRT7: Implication of functional links to aging. Proteomics, 14:1610-1622. [DOI] [PubMed] [Google Scholar]
- [253].Liu T, Li Z, Tian F (2021). Quercetin inhibited the proliferation and invasion of hepatoblastoma cells through facilitating SIRT6-medicated FZD4 silence. Hum Exp Toxicol, 40:S96-s107. [DOI] [PubMed] [Google Scholar]
- [254].Jiang C, Sun ZM, Hu JN, Jin Y, Guo Q, Xu JJ, et al. (2019). Cyanidin ameliorates the progression of osteoarthritis via the Sirt6/NF-κB axis in vitro and in vivo. Food Funct, 10:5873-5885. [DOI] [PubMed] [Google Scholar]
- [255].You W, Rotili D, Li TM, Kambach C, Meleshin M, Schutkowski M, et al. (2017). Structural Basis of Sirtuin 6 Activation by Synthetic Small Molecules. Angew Chem Int Ed Engl, 56:1007-1011. [DOI] [PubMed] [Google Scholar]
- [256].Shang J, Zhu Z, Chen Y, Song J, Huang Y, Song K, et al. (2020). Small-molecule activating SIRT6 elicits therapeutic effects and synergistically promotes anti-tumor activity of vitamin D(3) in colorectal cancer. Theranostics, 10:5845-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Sociali G, Galeno L, Parenti MD, Grozio A, Bauer I, Passalacqua M, et al. (2015). Quinazolinedione SIRT6 inhibitors sensitize cancer cells to chemotherapeutics. Eur J Med Chem, 102:530-539. [DOI] [PubMed] [Google Scholar]
- [258].Ferrara G, Benzi A, Sturla L, Marubbi D, Frumento D, Spinelli S, et al. (2020). Sirt6 inhibition delays the onset of experimental autoimmune encephalomyelitis by reducing dendritic cell migration. J Neuroinflammation, 17:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Yang J, Li Y, Zhang Y, Fang X, Chen N, Zhou X, et al. (2020). Sirt6 promotes tumorigenesis and drug resistance of diffuse large B-cell lymphoma by mediating PI3K/Akt signaling. J Exp Clin Cancer Res, 39:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplementary data can be found online at: www.aginganddisease.org/EN/10.14336/AD.2022.0413.