Summary:
Proteogenomic dentification of translated small open reading frames has revealed thousands of previously unannotated, largely uncharacterized microproteins, or polypeptides of less than 100 amino acids, and alternative proteins (alt-proteins) that are co-encoded with canonical proteins and are often larger. The subcellular localizations of microproteins and alt-proteins are generally unknown but can have significant implications for their functions. Proximity biotinylation is an attractive approach to define the protein composition of subcellular compartments in cells and animals. Here, we developed a high-throughput technology to map unannotated microproteins and alt-proteins to subcellular localizations by proximity biotinylation with TurboID (MicroID). More than 150 microproteins and alt-proteins are associated with subnuclear organelles. One alt-protein, alt-LAMA3, localizes to the nucleolus and functions in pre-rRNA transcription. We applied MicroID in a mouse model, validating expression of a conserved nuclear microprotein, and establishing MicroID for discovery of microproteins and alt-proteins in vivo.
eTOC blurb:
Na. et al. develop MicroID, a high-throughput technology to accelerate functional characterization of the trove of recently discovered, unannotated microproteins and alternative proteins. MicroID globally mapped unannotated microproteins and alternative proteins to subcellular localizations in cells and in animals, and enabled identification of an alt-protein that regulates pre-rRNA transcription.
Graphical Abstract
Introduction
Small open reading frames (smORFs) encoding short proteins (“microproteins”) of less than 100 amino acids, as well as alternative ORFs (alt-ORFs), which encode alt-proteins that overlap canonical protein coding sequences and are often longer than 100 amino acids, were historically excluded from genome annotation (Basrai et al., 1997; Brunet et al., 2018; Orr et al., 2020) due to computational constraints. It is now known that thousands of smORFs and alt-ORFs are translated in eukaryotic cells (Brunet et al., 2021b; Martinez et al., 2020; Orr et al., 2020; Slavoff et al., 2013). Microproteins and alt-proteins regulate important cellular and physiological processes in diverse organisms, including DNA repair (Arnoult et al., 2017), transcription (Koh et al., 2021), and innate immunity (Jackson et al., 2018), among others (Cao et al., 2021; Huang et al., 2021a; Lee et al., 2021; Zhang et al., 2020). Microproteins GREP1 (Prensner et al., 2021), CASIMO1 (Polycarpou-Schwarz et al., 2018) and MP31 (Huang et al., 2021b) are associated with cancer, and alt-FUS (Brunet et al., 2021a) may contribute to amyotrophic lateral sclerosis-associated protein aggregation. Hundreds of human smORFs function in cell proliferation, suggesting that they are broadly biologically significant (Chen et al., 2020; Prensner et al., 2021). Multiple recent studies suggest that alt-proteins, including longer species ranging in length from 100–300 amino acids, also play important roles in human cells (Brunet et al., 2021a; Cao et al., 2022; Cao et al., 2021; Gagnon et al., 2021). However, the vast majority of smORFs and alt-ORFs remain poorly characterized, and the question of how many of these recently identified genes play roles in biology is now paramount (Samandi et al., 2017). However, homology-based functional prediction for smORFs is challenging due to their short lengths (Keeling et al., 2019). Therefore, it is critical to develop methods to chemically fractionate the microproteome/alt-proteome to identify microproteins and alt-proteins with properties consistent with cellular functionality.
Proximity labeling was recently developed to map molecular interactions and subcellular localization in living cells (Gupta et al., 2015; Liu et al., 2018; Qin et al., 2021). Proximity biotinylation uses engineered peroxidases (APEX2 (Fazal et al., 2019; Myers et al., 2018)) and mutant biotin ligases ((Droujinine et al., 2021; Go et al., 2021)(Branon et al., 2018; Wei et al., 2021)) to label and identify endogenous proteins within a restricted radius (1–10 nm) of the enzyme inside living cells (Hung et al., 2017). Proximity labeling has been used to define the composition of protein complexes and the proteomes of organelles (Jing et al., 2015; Loh et al., 2016; Markmiller et al., 2018; Wei et al., 2021) that are difficult to purify. In addition to cultured mammalian cells, proximity labeling has been applied in vivo in yeast (Branon et al., 2018), plant protoplasts (Branon et al., 2018), parasites (Boucher et al., 2018), mouse (Droujinine et al., 2021; Skinnider et al., 2021; Takano et al., 2020), flies (Li et al., 2020) and worms (Branon et al., 2018). However, proximity biotinylation has not yet been applied to microprotein and alt-protein subcellular localization mapping.
Here we developed a method for global mapping of unannotated microproteins and alt-proteins to subcellular localizations, coupling their discovery to a dimension of functional information. We termed this technology microprotein and alt-protein TurboID, or “MicroID”. In this method, the engineered biotin ligase, TurboID, is targeted to subcellular locations of interest, enabling size selection, affinity purification and identification of peptides by liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Khitun and Slavoff, 2019; Slavoff et al., 2013), followed by use of transcriptome data to identify microproteins and alt-proteins. We applied MicroID to discovery of ~150 microproteins and alt-proteins in subnuclear organelles in cultured cells and in vivo, enabling identification of alt-LAMA3 as a ribosome biogenesis factor.
Results
Biotinylation of microproteins and alt-proteins in cell culture by MicroID
We sought to apply proximity biotinylation to reveal the spatial organization of the microproteome/alt-proteome in human cells. We focused on non-membrane-bounded subnuclear compartments with the goal of identifying microproteins and alt-proteins associated with gene expression and other essential nuclear processes. We first constructed stable cell lines expressing TurboID fusions with fibrillarin (nucleolus), H2B (chromatin), lamin B1 (nuclear envelope), a nuclear localization signal (3xNLS, whole nucleus) and untargeted TurboID as control (Figure S1A). The localization of each construct and spatially restricted protein biotinylation were confirmed by immunofluorescence microscopy and Western blotting (Figure S1B–S1C), consistent with prior reports (Go et al., 2021; Liu et al., 2018). We enriched biotinylated proteins from each sample, size-selected microproteins, alt-proteins and proteins from 5–30 kilodaltons using peptide gels, and performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, followed by searching acquired spectra against (1) the UniProt human proteome (Homo sapiens, version 2021), and (2) a 3-frame translated transcriptome database followed by exclusion of peptides that match sequences within annotated proteins as previously reported (Khitun and Slavoff, 2019). Unannotated peptide-spectral matches were subjected to manual inspection and validation (vide infra).
We first analyzed annotated proteins identified in each sample in order to validate the method. Across all samples and replicates, a total of 1611 human proteins were identified, with a length distribution that peaked between 0–300 amino acids (Figure S1D). 64.12% (1033 of 1611) of detected proteins were below 300 amino acids, consistent with the size selection. We then applied label-free quantitative (LFQ) analysis to map annotated proteins in each subnuclear compartment-targeted TurboID cell line, with untargeted (whole-cell) TurboID-expressing cells as a comparison (Figure S1E). Cutoffs for subcellular compartment assignments were enrichment of >1.5 relative to untargeted control, and p value < 0.05 (Student’s t-test). Three biological replicates were performed for each sample, and reproducibility between two selected replicates for each sample was high with a median pairwise Pearson correlation coefficient of 0.96–0.98 and excellent linearity of magnitude in protein LFQ values (Figure S1F). The gene ontology (GO) term profiles of proteins proximal to each TurboID fusion construct were unique and consistent with the respective subcellular region (Figure S1G–S1I). We then asked how many proteins within the selected molecular weight range previously reported to localize to each compartment (true positives) were identified in our dataset. We filtered the human UniProt database (Homo sapiens, version 2021), for length <300 amino acids, then for association with nucleolus, chromatin, or nuclear envelope. Intersecting the lists with our MicroID dataset revealed coverage of the <300 amino acid proteome of each compartment ranging from 21–33% (Figure 1C).
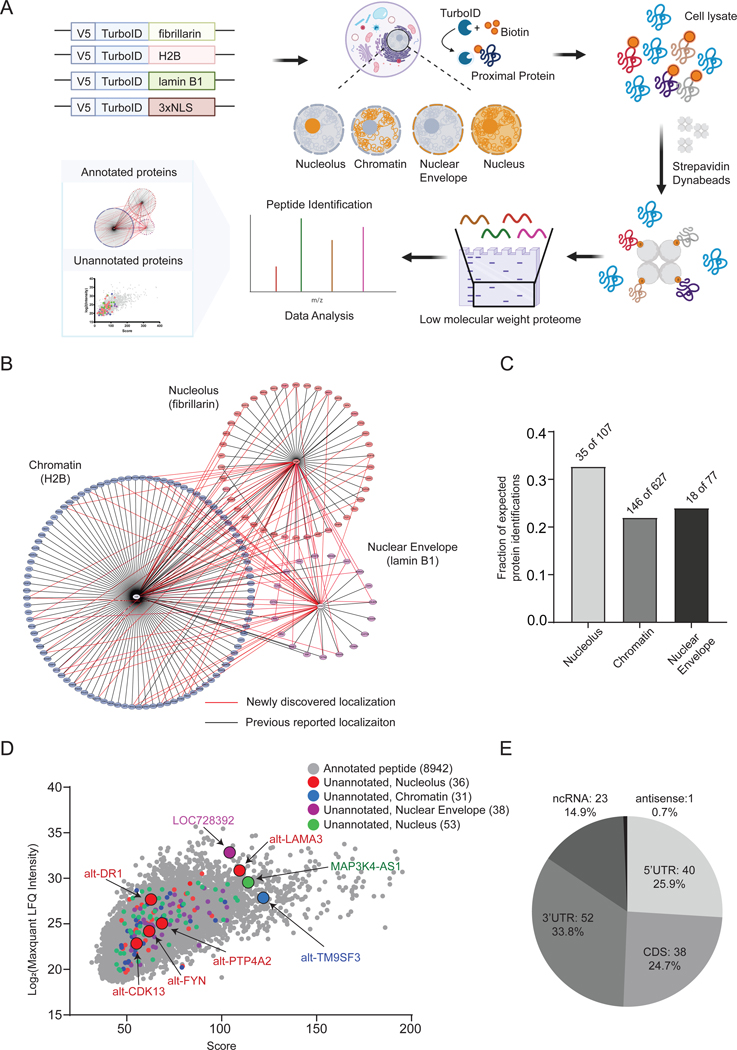
Figure 1. MicroID reveals annotated proteins and unannotated microproteins and alt-proteins in subnuclear regions in cell culture.
(A) Schematic of mapping subcellular localizations of canonical proteins (less than 300 amino acids) and unannotated microproteins and alt-proteins with MicroID. (B) Molecular context map for 3 subcellular organelles/structures profiled (nucleolus, chromatin and nuclear envelope). Subcellular compartment proteomes (below 300 amino acids) were defined by protein enrichments >1.5 relative to untargeted control, p value (T-test) < 0.05. N = 3, biological replicates. The annotated proteins identified in each region are arranged in a circle around the corresponding compartment marker. Proteins identified in more than one region are shown with connecting lines representing multiple localizations. Associations uniquely identified in this study are shown with red edges and previously known localizations with black. (C) The number of annotated Homo sapiens proteins <300 amino acids in length with current UniProt localization information for nucleolus (107), chromatin (627), and nuclear envelope (77) were compiled to generate compartment-specific true-positive proteome lists. Intersection of these true positive proteomes with annotated proteins identified in the MicroID datasets (protein enrichments >1.5 relative to untargeted control, p value (T-test) < 0.05. N = 3, biological replicates) revealed the fraction of detected vs. expected proteins <300 amino acids. (D) The label-free quantitation (LFQ) intensities of >150 unannotated microproteins and alt-proteins identified with MicroID in each compartment (red, blue, purple, and green), overlaid on LFQ intensities of annotated peptides detected in each dataset (gray). Microproteomes/alt-proteomes of each compartment were defined by at least one unique detection of an unannotated tryptic peptide in each compartment and not in control, and supported by extracted ion chromatograms (see Figure S1J–S1M). (E) Incidence of unannotated microproteins that map uniquely to open reading frames within the 5′UTR, CDS, or 3′UTR of mRNA, to noncoding RNAs (ncRNAs) and to antisense RNAs.
We then generated a <300 amino acid proteomic map of each compartment and their interactions (Figure 1B) using Cytoscape 3.8.2. We identified many known and previously unreported instances of proteins that exhibit multiple localizations. 33 proteins associated both with the nuclear envelope and chromatin; chromatin and nucleolus shared 28 proteins; and the nuclear envelope and nucleolar proteomes exhibited 12 overlaps. Some of the dual localizations have been reported previously. For example, NPM1 and NPM3 play roles in ribosome biogenesis and chromatin remodeling, and, accordingly, have previously been detected in both the nucleolus and chromatin (Okuwaki et al., 2012). We detect NPM1 and NPM3 using fibrillarin- and H2B-TurboID, as expected, as well as associated with the nuclear envelope. Pending experimental validation, these data may be useful in deriving insights about canonical proteins.
We next proceeded to identify unannotated microproteins and alt-proteins in the MicroID datasets. Searching the LC-MS/MS data against the RNA-seq database coupled with stringent score cutoffs and manual inspection of peptide-spectral matches identified 154 unannotated microprotein and alt-protein tryptic peptides (Figure 1D and Table S1). The corresponding smORFs occur in five major transcript regions or classes: 5′ untranslated region (5′ UTR); out-of-frame, same-strand overlaps within canonical protein coding sequences (CDS); 3′ untranslated region (3′ UTR); long noncoding RNAs (lncRNA); and antisense transcripts (Figure 1E). Because of their small size, detection of more than one tryptic fragment of a given microprotein or alt-proteins in more than one experimental replicate is rare (Slavoff et al., 2013). To provide additional confidence in their assignments, we chose four microproteins and alt-proteins – one from each compartment – and analyzed their MS1 extracted ion chromatograms (EICs) in the assigned compartment vs. control. In all four cases, a mass consistent with the tryptic peptide was detected within the examined retention time window for all three MicroID samples from the corresponding subnuclear compartment, and not in the untargeted control (Figure S1J–S1M). This analysis supports microprotein assignments based on detection in one MicroID experiment, though any additional microproteins listed should be confirmed experimentally.
Of the 154 total unannotated proteins we detected, 103 were microproteins below 100 amino acids; the rest were longer alt-proteins. We mapped 23 microproteins and 7 alt-proteins that have been previously identified with proteomics (Cao et al., 2020; Slavoff et al., 2013). For example, we detected alt-C1ORF122 (Cao et al., 2020) and alt-TM9SF3 (Slavoff et al., 2013) in chromatin for the first time. Finally, we identified and mapped 80 microproteins and 44 unannotated alt-proteins that we have not previously detected with proteomics (Table S1). MicroID can therefore map microproteins and alt-proteins that lack characterization to subcellular localizations, and MicroID may also increase sensitivity of microprotein and alt-protein detection.
Validation of unannotated microproteins and alt-proteins localizations from MicroID
We selected one microprotein or alt-protein identified by MicroID and EIC analysis in each (sub)nuclear compartment for further validation. Alt-LAMA3, located in the 5′UTR of the LAMA3 transcript, was selected from the TurboID-fibrillarin sample; alt-TM9SF3, a microprotein encoded in the 5′UTR of TM9SF3, was selected from the TurboID-H2B experiment; LOC728392, a predicted protein mapped to a long non-coding RNA (lncRNA), was selected from the TurboID-lamin B1 dataset; and MAP3K4-AS1, a microprotein encoded within a lncRNA antisense to the MAP3K4 transcript, was selected from the TurboID-3xNLS list (Figure S2A).
To validate endogenous expression, we generated Cas9-directed knock-in (KI) HEK 293T cell lines with a 3×GFP11-FLAG-HA tag appended to the 3’ end of the corresponding open reading frames (Cao et al., 2021; Na et al., 2020). Immunoreactive bands of the expected molecular weights were observed in each cell line and absent from control (Figure S2B). Immunofluorescence microscopy revealed that alt-LAMA3 co-localized with the nucleolar marker fibrillarin, alt-TM9SF3 (chromatin) and MAP3K4-AS1 (nucleus) co-localized with histone H3, and LOC728392 co-localized with lamin B1 (nuclear envelope), consistent with their respective MicroID localizations (Figure 2A). To probe the depth of our nucleolar dataset, we cloned four additional alt-proteins (alt-DR1, encoded in the 5′UTR of DR1, alt-PTP4A2, encoded in the 5′UTR of PTP4A2, alt-FYN, encoded in the 5′UTR of FYN and alt-CDK13, encoded in the 5′UTR of CDK13) (Figure S2C). Transient transfection of constructs containing the coding sequence of each target with a FLAG tag confirmed their translation (Figure S2D) and nucleolar localization (Figure S2E).
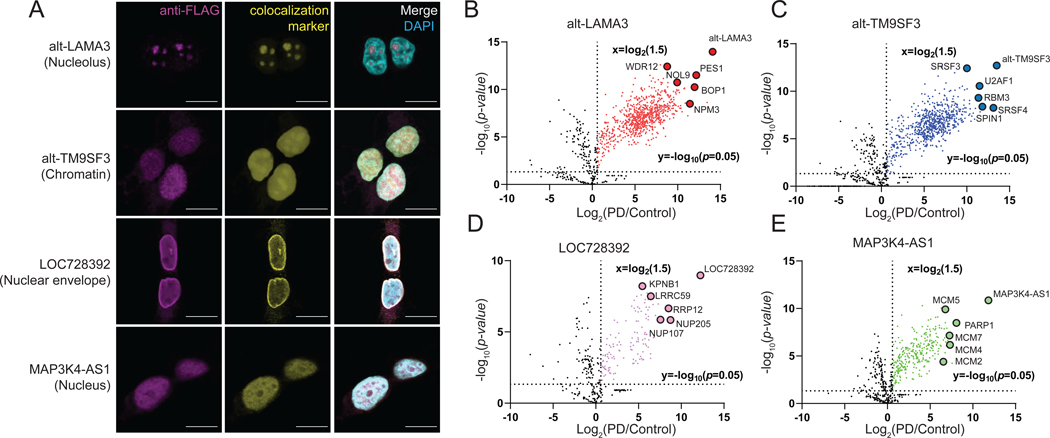
Figure 2. Unannotated microproteins and alt-proteins identified with MicroID are endogenously expressed and correctly localized to subnuclear regions of cultured human cells.
(A) HEK 293T cell lines bearing epitope tag knock-ins (KI) to a genomic copy of the indicated microprotein/alt-protein coding sequence were subjected to immunofluorescence with anti-FLAG tag (magenta), colocalization marker for each indicated (sub)nuclear region (yellow, see Methods), and DAPI (cyan). Scale bar, 10 μm. Data are representative of three biological replicates. (B-E) Volcano plot of proteins enriched by co-immunoprecipitation (co-IP) from microprotein/alt-protein FLAG tag KI cells (PD, pull-down) vs. wild-type control HEK 293T nuclear lysates with label-free quantiative (LFQ) proteomics (N = 3, biological replicates). Baits and candidate interaction partner proteins known to colocalize to the same subnuclear compartment are indicated and gene names are labeled.
We hypothesized that our selected microproteins and alt-proteins might interact with proteins that co-localize to the same region of the cell. Co-immunoprecipitation (co-IP) and quantitative proteomics from KI cell lines, with HEK 293T cells as a negative control, showed that alt-LAMA3, alt-TM9SF3, LOC728392 and MAP3K4-AS1 enriched unique protein complexes consistent with their subcellular localizations (Figure 2B–2E). For example, nucleolar alt-LAMA3 enriched the PeBoW complex (PES1, BOP1 and WDR12), which has been implicated in pre-ribosomal RNA transcription and processing (Grimm et al., 2006; Rohrmoser et al., 2007), as well as other proteins involved in ribosome biogenesis (NOL9, NPM3) (Figure 2B). Chromatin-associated alt-TM9SF3 interacted with pre-mRNA splicing factors (SRSF3, SRSF4 and U2AF1), suggesting that it may participate in co-transcriptional splicing (Anko et al., 2012; Song et al., 2019) (Figure 2C). The nuclear envelope-associated LOC728392 microprotein co-immunoprecipitated with nuclear import proteins (LRRC59 and KPNB1) (Zhen et al., 2012; Zhu et al., 2018) (Figure 2D). Nuclear MAP3K4-AS1 microprotein enriched the MCM complex (MCM2, MCM4, MCM5 and MCM7) (Meagher et al., 2019) and PARP1 (Bonfiglio et al., 2020), suggesting that it may play a role in genome replication and maintenance (Figure 2E). GO analysis of the top 50 most enriched proteins in each sample identified biological processes consistent with the bait protein’s observed subcellular localization and interaction partners (Figure S2F–S2I). Microproteins and alt-proteins mapped with MicroID are therefore associated with essential cellular protein complexes.
Alt-LAMA3 is functionally associated with pre-rRNA transcription and required for global protein synthesis
In order to establish the biological importance of an alt-protein identified with MicroID, we carried out molecular characterization of alt-LAMA3. Alt-LAMA3 is found within the 5′UTR of LAMA3 (Laminin subunit alpha-3) transcript variant 1 (NM_198129.4) (Figure 3A). The alt-LAMA3 coding sequence initiates upstream of the start codon of LAMA3, and extends partially into the LAMA3 coding sequence, but since alt-LAMA3 is translated in the +1 reading frame relative to LAMA3, the amino acid sequences of these two proteins are completely different (Figure 3A). To identify the start codon of alt-LAMA3, the cDNA sequence comprising the 5′UTR of LAMA3 transcript variant 1 (NM_198129.4) through the stop codon of alt-LAMA3 was cloned with an epitope tag appended to the 3´ end, and six candidate near-cognate start codons were individually deleted. Only deletion of A93GG abolished expression of alt-LAMA3 (Figure S3A). Mutating A93GG to A93TG increased alt-LAMA3 expression, while mutation to T93AG abolished expression, confirming that it is the alt-LAMA3 start codon (Figure S3B). Double banding may be due to proteolysis.
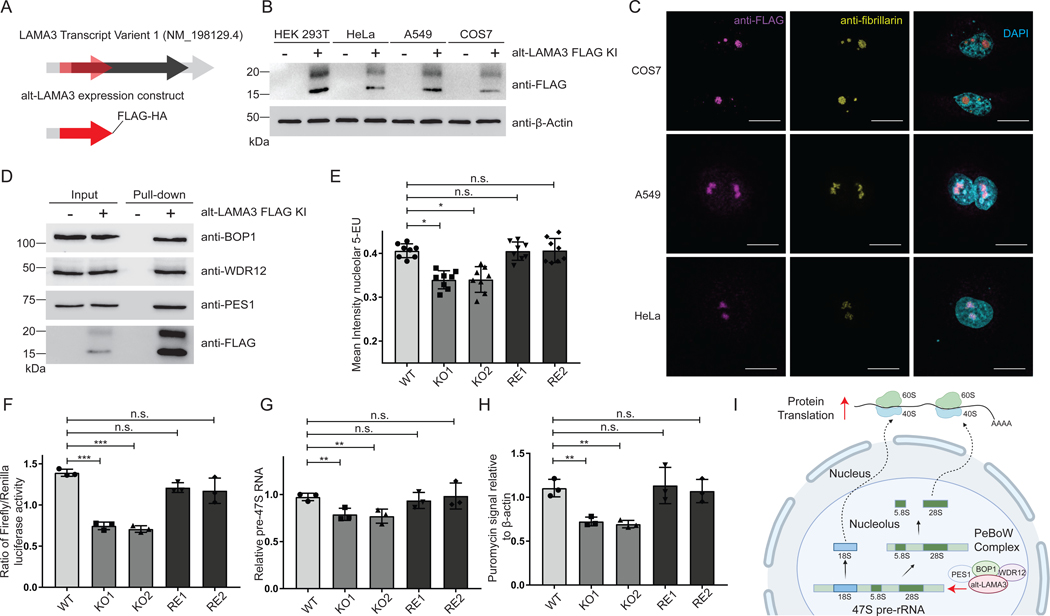
Figure 3. Alt-LAMA3 is physically and functionally associated with pre-rRNA transcription and required for global protein synthesis.
(A) Schematic representation of human LAMA3 transcript variant 1 (light gray). Black, annotated LAMA3 coding sequence; Red, alternative open reading frame (alt-ORF) encoding alt-LAMA3. (B,C) Alt-LAMA3 is endogenously expressed in KI HEK 293T, HeLa, A549 and COS7 cells. (B) Western blotting with anti-FLAG, and (C) immunofluorescence of alt-LAMA3 epitope tag KI HeLa, A549 and COS7 cell lines with anti-FLAG tag (magenta), anti-fibrillarin (yellow), and DAPI (cyan). Scale bar, 10 μm. Data are representative of three biological replicates. (D) Validation of endogenously expressed alt-LAMA3 interaction with PeBoW complex proteins (PES1, BOP1 and WDR12) by anti-FLAG immunoprecipitation from KI cells and Western blotting. (E-H) Alt-LAMA3 is required for pre-rRNA transcription by RNA polymerase I (RNAPI) and ribosome biogenesis. (E) 5-ethynyluridine (5-EU) incorporation assay for RNAPI transcription. Two independent HEK 293T alt-LAMA3 knockout (KO) cell lines and HEK 293T alt-LAMA3 rescue cell lines (RE, in which alt-LAMA3 was stably reintroduced on the KO background) were generated. HEK 293T, KO and RE cells were incubated with 1mM 5-EU for 1h, immunostained with anti-fibrillarin, and treated with a click chemistry reaction containing Alexa Fluor™ 488 Azide to visualize 5-EU-labeled nascent RNA in the nucleolus. Statistical significance for nine biological replicates was calculated using ANOVA linear regression. Error bars, standard deviation. All comparisons are relative to HEK 293T control. n.s., not significant; *P ≤ 0.05. (F) Reporter assay for RNAPI transcription. HEK 293T, KO and RE cells were transfected with plasmids containing firefly luciferase (under the control of the rDNA promoter) and Renilla luciferase (under the control of a constitutive promoter). Luminescence was quantified 24 h after transfection. Statistical significance for nine biological replicates was calculated using ANOVA linear regression. Error bars represent the mean ± the standard deviation. All comparisons are relative to HEK 293T. n.s., not significant; ***P ≤ 0.001. (G) Direct measurement of primary pre-rRNA transcript. qRT-PCR was used to quantitate the levels of the primary pre-rRNA, using the 7SL RNA as an internal control. N = 3. Error bars, standard deviation. Significance was analyzed by ANOVA linear regression. n.s., not significant; **P ≤ 0.01. (H) Measurement of global cellular protein synthesis. HEK 293T, KO and RE cells were treated with 1 μM puromycin for 1 h to label nascent peptides, followed by Western blotting with an anti-puromycin antibody. ImageJ was used to quantify puromycin incorporation for indicated samples relative to HEK 293T (N = 3). Error bars, standard deviation. Significance was analyzed by ANOVA linear regression. n.s., not significant; **P ≤ 0.01. (I) Model: alt-LAMA3 is functionally associated with pre-rRNA transcription and global protein translation.
Having ascertained the 148-amino acid alt-LAMA3 sequence, we examined its conservation. Clustal Omega alignment of conceptual +1 frame translations of mammalian LAMA3 mRNAs revealed high sequence identity in primates (Figure S3C), a degree of conservation observed for several other functional human microproteins (Denli et al., 2015)(Douka et al., 2021). We then tested expression of endogenous alt-LAMA3 in human and primate cell lines from different tissues of origin using Cas9-directed homology repair to append 3xGFP11-FLAG epitope tags to the alt-LAMA3 genomic locus. Alt-LAMA3 was expressed (Figure 3B) in the nucleolus (Figure 3C) of all three cell lines, consistent with expression in multiple human cell types and primates.
Based on its PeBoW complex association, we hypothesized that alt-LAMA3 regulates ribosome biogenesis. We first confirmed the interaction of endogenous alt-LAMA3 with PeBoW complex members with Western blotting (Figure 3D). We then generated two independent alt-LAMA3 knockout (KO) HEK 293T cell lines (Ran et al., 2013). One-hundred seventy-four nucleotides surrounding the alt-LAMA3 start codon were deleted, abrogating its expression. Confirming successful KO, the region of the 5′TR of LAMA3 transcript variant 1 that encodes alt-LAMA3 was undetectable by mRNA-seq and genomic PCR in KO cells, but present in parental HEK 293T cells (Figure S3D and Figure S3E). In contrast, mRNA-seq reads covering the start codon and coding sequence of LAMA3 were detected in both KO and wild-type cells, and expression of LAMA3 was identical in KO and wild-type HEK 293T at the mRNA and protein level, confirming that deletion of alt-LAMA3 did not perturb LAMA3 expression (Figure S3F–G). To confidently assign KO phenotypes as specific to alt-LAMA3 loss, we generated rescue cells in which alt-LAMA3 was stably reintroduced in each KO (Figure S3G).
With genetic tools to dissect its cellular function in hand, we probed the role of alt-LAMA3 in ribosome biogenesis. First, we examined its expression during the cell cycle, since ribosome biogenesis increases during G1 and maximizes in S and G2 phase (Andrews et al., 2018; Hernandez-Verdun, 2011; Iyer-Bierhoff and Grummt, 2019). Alt-LAMA3 expression in synchronized HEK 293T alt-LAMA3–3xGFP11-FLAG KI cells increased at S phase, decreasing again by G2/M phase (Figure S3H), suggesting that expression of alt-LAMA3 is coordinated with ribosome biogenesis during the cell cycle.
The PeBoW complex has been implicated in pre-rRNA processing (Grimm et al., 2006; Rohrmoser et al., 2007) and rDNA transcription (Sondalle et al., 2019). We therefore examined whether alt-LAMA3 affects rDNA transcription as well. Using a miniaturized 5-ethynyl uridine (5-EU) assay for nucleolar rRNA transcription (Hayashi et al., 2018; Ogawa et al., 2021; Stamatopoulou et al., 2018)(Bryant et al., 2022), we found a significant defect in pre-rRNA synthesis in alt-LAMA3 KO cells (Figure 3E and Figure S3I). Utilizing a dual-luciferase reporter (Ghoshal et al., 2004; Sondalle et al., 2019), we observed a defect in RNA polymerase I (RNAPI) transcription in the absence of alt-LAMA3 (Figure 3F). As a third assay, we measured a decrease in 47S pre-rRNA levels using qRT-PCR (Sondalle et al., 2019) in the absence of alt-LAMA3 (Figure 3G). In all cases, alt-LAMA3 rescue cells resembled wild-type. Defects in pre-rRNA transcription inhibit ribosome biogenesis and lead to decreases in global protein translation (Farley-Barnes et al., 2018; Schmidt et al., 2009), so we hypothesized that alt-LAMA3 may be required for maximal cellular protein synthesis. Quantification of nascent proteins labeled with puromycin (Farley-Barnes et al., 2018; Schmidt et al., 2009; Sondalle et al., 2019) revealed a defect in protein synthesis in alt-LAMA3 KO cells that can be rescued by alt-LAMA3 reintroduction (Figure 3H and Figure S3J). Alt-LAMA3 therefore functions in pre-rRNA transcription and cellular protein synthesis in a human cell line, though its molecular mechanism remains to be elucidated (Figure 3I).
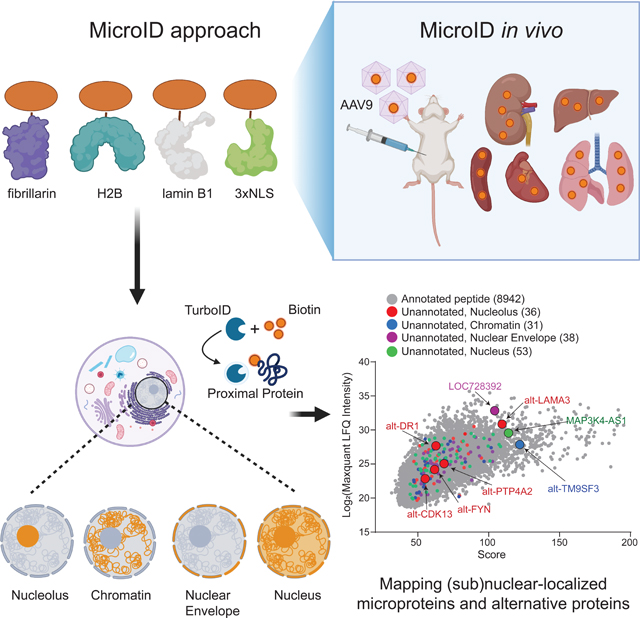
Mapping (sub)nuclear-localized microproteins in vivo with MicroID
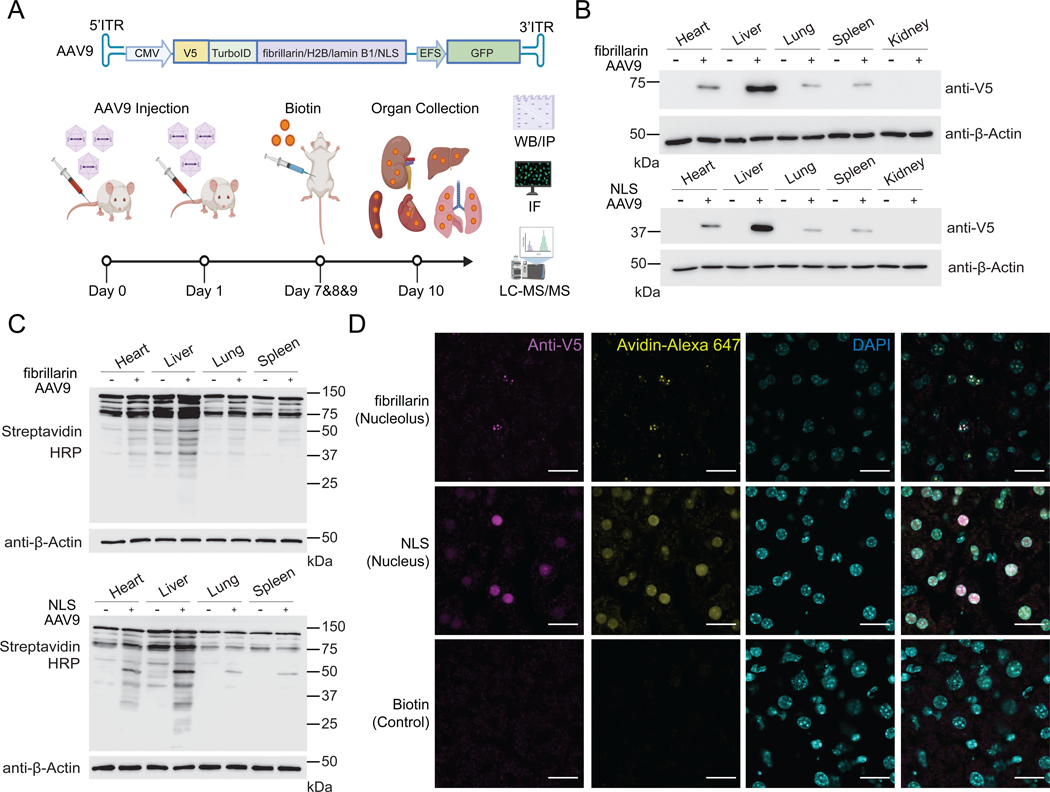
In order to enable detection of microproteins and alt-proteins in subnuclear compartments in mouse, expression, localization and activity of adeno-associated viral vector AAV9- (Chow et al., 2017; Dai et al., 2019) delivered TurboID fusion constructs were tested in Hepa1–6 cells. Western blotting (Figure S4A) and immunofluorescence (Figure S4B) confirmed expression of the epitope-tagged protein, as well as spatially restricted biotinylation in >80% of transduced cells. We then tested MicroID in C57BL/6J mice (see Methods). After transgene delivery and biotin administration, liver, spleen, lung, heart and kidney were dissected and collected for Western blotting, immunofluorescence and LC-MS/MS (Figure 4A). TurboID fusion proteins and enzyme-specific biotinylation were detected strongly in liver, while expression and biotinylation were lower in heart, lung, and spleen, and absent in kidney (Figure 4B–C). >80% of hepatocytes were transduced in virus-infected mice and low background was observed in controls (Figure 4D).
Figure 4. Application of MicroID in vivo: validation of the method.
(A) Schematic of mapping subcellular localizations of unannotated microproteins in vivo with MicroID. (B) Western blotting of TurboID-V5 expression in a panel of murine tissues following transduction with AAV9 viruses. (+) indicates AAV9 transduction, and (–) indicates no viral transduction. Top: nucleolus-targeted TurboID; Bottom: whole nucleus-targeted TurboID. (C) Streptavidin blotting of different tissue lysates from mice transduced with the indicated AAV9, which were then treated with vehicle (–) or biotin (+; 24 mg per kg per day, intraperitoneally for 3 consecutive days). Tissues were collected 24 h after the final biotin injection. Top: nucleolus-targeted biotinylation; Bottom: nucleus-targeted biotinylation. (D) Immunofluorescence of frozen liver sections from mice transduced with the indicated AAV9 virus or control mice. Scale bar, 20 μm.
We next subjected MicroID-labeled mouse tissues to lysis, streptavidin enrichment, protein size selection and LC-MS/MS proteomics. We first examined canonical proteins. In liver, we identified 1238 nucleolar proteins 1403 nuclear proteins; in heart, we identified 298 and 464, respectively, consistent with biotinylation signal intensity (Figure 5A). LFQ analysis was not performed for the in vivo samples, so comprehensive compartment microproteome/alt-proteome mapping is not possible. Regardless, GO analysis of 50 proteins with the highest ranked peptide intensity values in each sample showed that the enriched biological processes and cellular components correlate with the subcellular compartment profiled (Figure S5A–S5D), though higher background from endogenously biotinylated mitochondrial proteins was observed in vivo. Nearly 33% of annotated proteins mapped to the nucleolus in human cells were identified with TurboID-fibrillarin in mouse, and 35% of annotated proteins identified in the nucleus in human cells were identified in the nucleus in mouse (Figure S5E), validating the method in vivo.
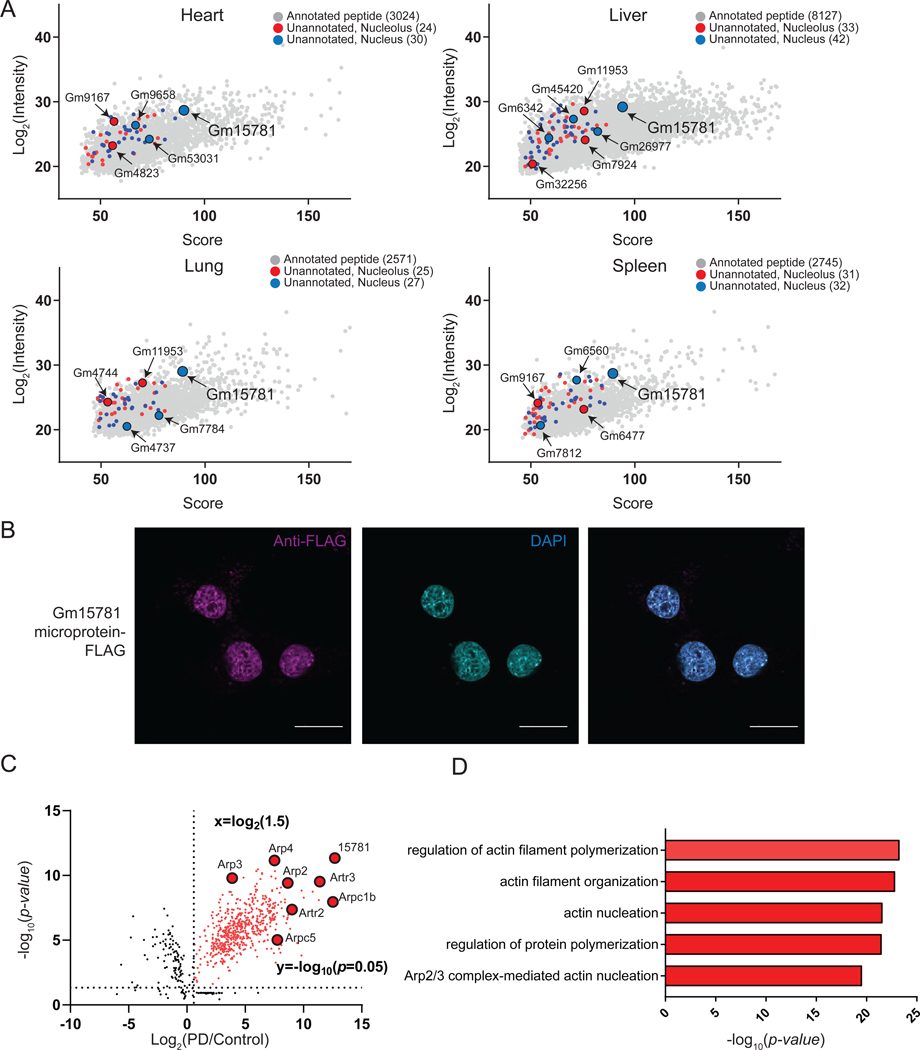
Figure 5. Application of MicroID in vivo: identification and validation of nucle(ol)ar localized microproteins and alt-proteins in various tissues.
(A) Identification and LFQ intensities of >100 microproteins and alt-proteins in indicated tissues using MicroID in nucleus (blue) and nucleolus (red). LFQ intensities of canonical, annotated proteins <300 amino acids detected in the same experiments (gray) are also plotted for reference. (B) Unannotated microprotein Gm15781 was epitope tagged in an expression vector, transfected into murine liver Hepa1–6 cells, and immunofluorescence performed with anti-FLAG (magenta) and DAPI (cyan). Scale bar, 10 μm. Data are representative of three biological replicates. (C) Volcano plot of co-IP quantitative proteomics (N = 3) from Gm15781-FLAG transfected Hepa1–6 cells (PD) or control Hepa1–6 nuclear lysates. Baits and proteins are indicated and gene names are labeled. (D) GO (biological processes) analysis of genes enriched (fold change ≥ 30) in Gm15781 pull-down over control.
We then identified unannotated tryptic peptides using a murine RNA-seq-derived database, which were assigned to 75 unannotated microproteins and alt-proteins in liver, 63 in spleen, 52 in lung and 54 in heart (Figure 5A and Table S2). 87 microproteins and alt-proteins were identified in the combined nucleolar datasets, and 96 in the nucleus (Figure 5A and Table S2). MicroID can therefore identify nucle(ol)ar microproteins and alt-proteins in mouse tissues, though comprehensive mapping of murine microproteomes/alt-proteomes will require additional controls.
The unannotated 80-amino acid microprotein Gm15781, translated from the pseudogene Gm15781 transcript, was identified in the nucleus in all four tissues, suggesting that it may be ubiquitously expressed (Figure 5A and Figure S5F). Over-expression of an expression plasmid encoding the 5′UTR and epitope-tagged microprotein coding sequence of pseudogene Gm15781 in Hepa1–6 cells revealed anti-FLAG immunoreactive bands, which were absent from control (Figure S5G), as well as immunofluorescence corresponding to microprotein Gm15781 in the nucleus, as expected (Figure 5B). Clustal Omega alignment of hypothetical microprotein Gm15781 homologs from human, mouse, and rat revealed sequence conservation (Figure S5H). Finally, we examined the interaction partners of microprotein Gm15781 using co-immunoprecipitation from transfected Hepa1–6 cells, with untransfected cells as a control. LC-MS/MS, label-free quantitation and GO analysis identified enrichment of the Arp2/3 complex and actin-related biological processes (Figure 5C–D), which have been previously implicated in homologous recombination (Hurst et al., 2019; Schrank et al., 2018). MicroID can therefore reveal microproteins associated with nuclear processes in vivo.
Discussion
Since the first report of ubiquitous translation of noncanonical yeast small open reading frames in 2009 (Ingolia et al., 2009), and the first published methods for direct proteomic detection of small open reading frame-encoded microproteins in high throughput (Slavoff et al., 2013; Vanderperre et al., 2013), thousands of human microproteins have been identified, but the vast majority remain undiscovered. While dozens of mammalian microproteins and alt-proteins are now known to have important biological functions (Couso and Patraquim, 2017), some smORFs frames may represent a process of protogene de novo evolution (Carvunis et al., 2012) and may not yet be optimized for cellular fitness effects; others, especially upstream ORFs (uORFs), likely play cis-regulatory roles in translation (Zhang et al., 2019). It is therefore difficult to predict the roles of microproteins and alt-proteins a priori, so development of experimental methods to reveal their functional properties in high throughput will be critical.
Here we introduced MicroID, a strategy for proximity biotinylation and identification of unannotated microproteins and alt-proteins in (non-)membrane-bounded organelles in cultured cells and in vivo. Our detection of 80 previously unreported microproteins and 44 longer alt-proteins suggests that biotin enrichment can enhance identification of microproteins and alt-proteins that are refractory to detection in whole-cell small proteomes. We therefore speculate that MicroID may prove powerful in identifying microproteins and alt-proteins that are intractable to detection by standard proteomic workflows, such as hydrophobic polypeptides, which may be associated with (endo)membranes (Speers and Wu, 2007). Our datasets may additionally be useful for functional hypothesis generation for the >100 microproteins and alt-proteins mapped to subnuclear locales in human cells and mouse that were not further characterized in this work, pending experimental validation.
In the future, it will be important to expand MicroID to different organelles, both membrane- and non-membrane-bounded, in order to identify microproteins and alt-proteins involved in biological processes beyond the nucleus, as well as various cell types and additional animal models. We envision that application of MicroID in comparative mode could enable identification of microproteins and alt-proteins expressed in disease. This study therefore lays the foundation to accelerate molecular and cellular characterization of the recently discovered class of microprotein and alt-protein-encoding genes more broadly.
Limitations of the study
The signal-to-noise for in vivo proximity biotinylation is lower than in vitro due to the abundance of endogenously biotinylated mitochondrial proteins in tissue; overcoming this issue with bio-orthogonal labeling strategies that circumvent biotinylation might improve microprotein and alt-protein detection sensitivity in animal models. Furthermore, this study employed trypsin digest and one-dimensional LC-MS/MS, and therefore, our coverage of the unannotated proteome was likely limited. Improved proteomic strategies including but not limited to tailored search databases derived from ribosome profiling data (Koch et al., 2014), as well as improved sensitivity from multienzyme digests (Kaulich et al., 2021), and multi-dimensional LC-MS/MS (Slavoff et al., 2013), will likely also increase the sensitivity of microprotein and alt-protein detection by MicroID. Finally, proteomic searches against expanded databases are subject to increased false positive identifications(Cargile et al., 2004), so any microproteins/alt-proteins presented in Tables S1–2 should be validated at the molecular level.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sarah A. Slavoff (sarah.slavoff@yale.edu).
Materials Availability
Reagents generated in this study are available on request with a completed Materials Transfer Agreement.
Data and Code Availability
Proteomic data are available from the PRIDE repository under accession number PXD033067. The mRNA-seq data have been deposited in the NCBI Gene Expression Omnibus under accession GSE205869. The raw imaging and original Western blotting data have been deposited at Mendeley Data. Accession numbers are listed in the key resources table. Additional data reported in this paper are available as of the date of publication.
This paper does not report original code. Previously reported code for identification of microproteins is available at 10.5281/zenodo.5921116.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key Resource Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-V5 | Cell Signaling Technology | Cat#13202S; RRID: AB_2687461 |
| Mouse monoclonal anti-FLAG | Sigma | Cat#F3165; RRID: AB_259529 |
| Rabbit monoclonal anti-FLAG | Cell Signaling Technology | Cat#14793S; RRID: AB_2572291 |
| Mouse monoclonal anti-beta actin | Invitrogen | Cat#BA3R; RRID: AB_10979409 |
| Strepavidin horseradish peroxidase conjugate | Invitrogen | Cat#S911 |
| Mouse monoclonal anti-fibrillarin | Abcam | Cat#ab4566; RRID: AB_304523 |
| Rabbit monoclonal anti-lamin B1 | Abcam | Cat#ab133741; RRID: AB_2616597 |
| Rabbit monoclonal anti-histone H3 | Cell Signaling Technology | Cat#4499S; RRID: AB_10544537 |
| Rabbit monoclonal anti-LAMA3 | Abcam | Cat#ab151715 |
| Rabbit polyclonal anti-PES1 | Bethyl Laboratories | Cat#A300-903A-2; RRID: AB_625300 |
| Rabbit polyclonal anti-BOP1 | Bethyl Laboratories | Cat#A302-148A-1; RRID: AB_1720319 |
| Rabbit polyclonal anti-WDR12 | Bethyl Laboratories | Cat#A302-650A; RRID: AB_10568823 |
| Rabbit polyclonal anti-puromycin | Kerafast | Cat#EQ0001; RRID: AB_2620162 |
| Anti-FLAG M2 affinity gel | Sigma | Cat#A2220; RRID: AB_10063035 |
| Goat anti-rabbit IgG horseradish peroxidase conjugate | Rockland | Cat#611-1302; RRID: AB_218567 |
| Goat anti-mouse IgG horseradish peroxidase conjugate | Rockland | Cat#610-1319-0500; RRID: AB_219659 |
| Goat anti-rabbit IgG Alexa FluorTM 568 | Invitrogen | Cat#A11011; RRID: AB_143157 |
| Goat anti-mouse IgG Alexa FluorTM 647 | Invitrogen | Cat#A21235; RRID: AB_2535804 |
| Strepavidin Alexa FluorTM 647 conjugate | Invitrogen | Cat#S21374; RRID: AB_2336066 |
| Bacterial and virus strains | ||
| One Shot Stbl3 Chemical Competent E. coli | Thermo Fisher Scientific | Cat#C737303 |
| Chemicals, peptides, and recombinant proteins | ||
| DPBS, no calcium, no magnesium | Gibco | Cat#14190250 |
| RPMI 1640 Medium | Gibco | Cat#1875-093 |
| DMEM, high glucose, pyruvate | Gibco | Cat#11995065 |
| Fetal Bovine Serum | Sigma Aldrich | Cat#F4135-500ML |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat#15140122 |
| 2-Mercaptoethanol | Sigma Aldrich | Cat#M6250-10ML |
| PEI MAX | Polyscience | Cat#24765-1 |
| PEG8000 | Promega | Cat#V3011 |
| RIPA buffer | Boston BioProducts | Cat#BP-115 |
| Protease inhibitor coctail | Thermo Fisher Scientific | Cat#78437 |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23227 |
| QuickExtract DNA Extraction Solution | Epicenter | Cat#QE09050 |
| QIAamp DNA Blood Mini Kit | Qiagen | Cat#51106 |
| Gibson Assembly® Master Mix | NEB | Cat#E2611 |
| Phusion Flash High-Fidelity PCR Master Mix | Thermo Fisher Scientific | Cat#F548L |
| QIAquick Gel Extraction Kit | Qiagen | Cat#28706 |
| TRIzol™ Reagent | Invitrogen | Cat#15596026 |
| RNeasy Plus mini isolation kit | Qiagen | Cat#74134 |
| 4–20% Mini-PROTEAN® TGX™ Precast Protein Gels, 10-well | BioRad | Cat#4561094 |
| Bovine Serum Albumin | Sigma Aldrich | Cat#A9418-100G |
| Pierce™ ECL Western Blotting Substrate | Thermo Fisher Scientific | Cat#32106 |
| EDTA | Sigma Aldrich | Cat#E8008-100ML |
| puromycin | Sigma Aldrich | Cat#P8833 |
| biotin | Sigma Aldrich | Cat#B4501 |
| Dynabeads™ M-280 Streptavidin | Thermo Fisher Scientific | Thermo Fisher |
| Formalin | Thermo Fisher Scientific | Cat#SF100-4 |
| DAPI | EMD Millipore | Cat#268298 |
| 3×FLAG peptide | Sigma Aldrich | Cat#F4799 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat#11668019 |
| 5-ethynyluridine | Click Chemistry Tools | Cat#1261-10 |
| BMH-21 | Cayman Chemical Company | Cat#22282-5mg |
| Alexa FluorTM 488 Azide | Click Chemistry Tools | Cat#1275-1 |
| Critical commercial assays | ||
| Dual-Luciferase® Reporter Assay System | Promega | Cat#E1910 |
| iScript gDNA Clear cDNA Synthesis Kit | Bio-Rad | Cat#172-5035 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat#172-5121 |
| Deposited data | ||
| Raw and analyzed mRNA sequencing data | This paper | GEO: GSE205869 |
| Raw proteomic data | This paper | PRIDE: PXD033067 |
| Raw imaging and Western blotting data | This paper | Mendeley: https://doi:10.17632/yh8t333wwp.1 |
| Custom code | Khitun and Slavoff, 2019 | Zenodo: https://doi.org/10.5281/zenodo.5921116 |
| Experimental models: Cell lines | ||
| Human: HEK 293T | ATCC | Cat#CRL-3216 |
| Human: HEK 293FT | Thermo Fisher Scientific | Cat# R70007 |
| Human: HeLa | ATCC | Cat#CCL-2 |
| Human: A549 | Dr. Craig Crews (Yale University) | N/A |
| Cercopithecus aethiops: COS7 | ATCC | Cat#CRL-1651 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J mice | The Jackson Laboratory | Cat#000664 |
| Oligonucleotides | ||
| Primer: alt-LAMA3 Forward: CTGGCTTTGACCTGAGCGGTGAGT | This paper | N/A |
| Primer: alt-LAMA3 Reverse: ACCAAATTCGTTTACAAGCCTCCT | This paper | N/A |
| Primer: LAMA3 Forward: CACCGGGATATTTCGGGAATC | This paper | N/A |
| Primer: LAMA3 Reverse: AGCTGTCGCAATCATCACATT | This paper | N/A |
| Primer: Primary pre-rRNA Forward: CTCCGTTATGGTAGCGCTGC | This paper | N/A |
| Primer: Primary pre-rRNA Reverse: GCGGAACCCTCGCTTCTC | This paper | N/A |
| Primer: 7SL-RNA Forward: ATCGGGTGTCCGCACTAAGTT | This paper | N/A |
| Primer: 7SL-RNA Reverse: CAGCACGGGAGTTTTGACCT | This paper | N/A |
| Recombinant DNA | ||
| pLJM1-V5-TurboID-fibrillarin | This paper | N/A |
| pLJM1-V5-TurboID-H2B | This paper | N/A |
| pLJM1-V5-TurboID-lamin B1 | This paper | N/A |
| pLJM1-V5-TurboID-3xNLS | This paper | N/A |
| pMD2.G | pMD2.G was a gift from Didier Trono | Addgene Plasmid #12259 |
| psPAX2 | psPAX2 was a gift from Didier Trono | Addgene Plasmid #12260 |
| pSpCas9(BB)-2A-GFP | pSpCas9(BB)-2A-GFP (PX458) was a gift from Feng Zhang | Addgene Plasmid # 48138 |
| pXD017-V5-TurboID-fibrillarin | This paper | N/A |
| pXD017-V5-TurboID-H2B | This paper | N/A |
| pXD017-V5-TurboID-lamin B1 | This paper | N/A |
| pXD017-V5-TurboID-3xNLS | This paper | N/A |
| pAdDeltaF6 | pAdDeltaF6 was a gift from James M. Wilson | Addgene Plasmid # 112867 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| GraphPad Prism (version 9.0) | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
| Mascot | Matrix Science | https://www.matrixscience.com/ |
| MaxQuant | MaxQuant lab | https://www.maxquant.org/ |
| Perseus | MaxQuant lab | https://maxquant.net/perseus/ |
| Python (2.7.11) | Python Software Foundation, DE | https://www.python.org/downloads/ |
| Cutadapt (1.9.1) | Marcel Martin, 2010 | https://cutadapt.readthedocs.io/en/stable/ |
| STAR (2.7.1) | Alexander Dobin et al., 2013 | http://code.google.com/p/rna-star/ |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
HEK 293T, HEK 293FT, COS7 and HeLa cells were amplified from early-stage stocks prepared from cells purchased from ATCC. HEK 293T, HEK 293FT, COS7 and HeLa cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Corning, Cat. 10–013-CV) supplemented with 10% (vol/vol) fetal bovine serum (FBS, Sigma Aldrich, Cat. F4135–500ML) and 100 U/mL penicillin-streptomycin (Sigma Aldrich, Cat. P4333–100ML). A549 cells were a gift from Craig Crews (Yale University) and were cultured in RPMI-1640 (Corning, Cat. 10–040-CV) medium containing 10% FBS and 100 U/mL penicillin-streptomycin. Hepa1–6 cells were cultured in DMEM (Corning, Cat. 10–013-CV) medium containing 10% FBS and 100 U/mL penicillin-streptomycin. Cells were incubated at 37 °C in a humidified atmosphere with 5% CO2. The ATCC Universal mycoplasma detection kit was used to confirm mycoplasma-free status of all cell lines.
Animals
C57BL/6J mice 6–8 weeks old were purchased from the Jackson Laboratory. For animal studies, male littermates were randomly assigned to experimental or control groups. We do not expect sexual dimorphism to affect the results of this proof-of-principle study, but future biological applications should be performed in both sexes. All animal work, maintenance and care were approved by Yale University’s Institutional Animal Care and Use Committee (IACUC) and performed with approved protocols (2018–20068).
METHOD DETAILS
Lentivirus production and infection
Lentivirus was produced as previously described (Tiscornia et al., 2006). Briefly, HEK 293T cells were co-transfected with expression construct pLJM1-V5-TurboID-fibrillarin/H2B/laminB1/3xNLS and pLJM1-V5-TurboID (untargeted control), along with pMD2.G and psPAX2, and growth media were replaced after 6 h. After 48 h post-transfection, media containing viruses was harvested, filtered through a 0.45 μm filter, concentrated, aliquoted and flash frozen. Stable cell lines were generated by transducing wild type HEK 293T with 50 μl of lentiviral particle suspension at 60% confluency in T25 flask followed by selection with 2 ng/ml puromycin (Sigma-Aldrich, cat. No. P8833) for 3 days. Cells were harvested and stable transgene expression validated by Western blotting.
Biotin labeling with MicroID in live mammalian cells, streptavidin pull-down and proteomics
For labeling of HEK 293T V5-TurboID-fibrillarin/H2B/laminB1/3xNLS and V5-TurboID (untargeted control) stable cell lines (grown in 15 cm dishes), we used a final concentration of 500 μM biotin for 30 min at 37 °C following a reported protocol (Branon et al., 2018). Labeling was stopped after the desired time period by transferring the cells to ice and washing five times with cold PBS buffer. For negative controls, we omitted exogenous biotin. Cells were detached from the flask by gently pipetting of PBS directly onto the cells, then pellets were collected by centrifuging the resulting cell suspension at 1,600 r.p.m. for 5 min. Cells were suspended in 1 mL nuclear isolation buffer (10 mM Hepes pH 7.4, 100 mM KCl, 5 mM MgCl2 with 0.5% NP40 and Roche Complete protease inhibitor cocktail tablets (Roche, Cat. No.11873580001), and incubated on ice for 10 min, followed by centrifugation 3 min at 4°C by 3,000 rpm. The nuclear pellets were suspended in 1 mL RIPA lysis buffer by gentle pipetting and sonication at 4 °C. Lysates were clarified by centrifugation at 15,000 r.p.m. for 30 min at 4 °C. To enrich biotinylated proteins, 300 μL Dynabeads™ M-280 Streptavidin (Thermo Fisher, Cat. #11205D) were washed twice with RIPA buffer, incubated with cell lysates containing ~10 mg protein for each sample and rotated at 4 °C overnight. The beads were subsequently washed twice with 1 mL of RIPA lysis buffer, once with 1 mL of 1 M KCl, once with 1 mL of 0.1 M Na2CO3, once with 1 mL of 2 M urea in 10 mM Tris-HCl (pH 8.0), and twice with 1 mL RIPA lysis buffer. Bound proteins were eluted by boiling in 2×SDS loading buffer containing 20 mM DTT and 2 mM biotin for 15 min. After elution, proteins were electrophoresed on a Tricine-SDS-PAGE gel, and gel bands corresponding in the 2–25 kDa size range were excised for proteomic analysis according to a published protocol (Cao et al., 2021). Briefly, LC-MS/MS analysis was performed on a Thermo Scientific Q Exactive Plus equipped with a Waters nanoAcquity UPLC system utilizing a binary solvent system (Solvent A: 100% water, 0.1% formic acid; Solvent B: 100% acetonitrile, 0.1% formic acid). Peptides were separated using an analytical PicoFrit column packed with 1.9 μm ReproSil-Pur 120Å C18-AQ resin (Dr. Maisch) using ACQUITY UPLC M-Class connected to a Q Exactive Plus and eluted at 250 nl/min with the following gradient: 99% Solvent A for 40 min, 2 min linear gradient to 94% Solvent A, 58 min to 76% Solvent A, 5 min to 52% Solvent A, 5 min to 26% Solvent A, 5 min to 10% Solvent A, 5 min back to 99% Solvent A, and final 10 min hold at 99% Solvent A. Set Q Exactive with a nanospray source at an electrospray potential of 1.6 kV. MS: 30,000 resolution, 3 × 106 AGC target, 298–1,750 m/z scan range. MS/MS data was collected using a top 10 high-collisional energy dissociation method in data-dependent mode with a normalized collision energy of 33.0 eV and a 2.0 m/z isolation window. The first mass was 100 m/z in fixed mode. MS/MS resolution was 7500 and dynamic exclusion was 60 s.
For identification of annotated and unannotated microproteins, ProteoWizard MS Convert was used for peak picking and files were analyzed using Mascot algorithm (version 2.8) (Matrix Science). Oxidation of methionine and N-terminal acetylation were set as variable modifications, and a previously reported three-frame translation of assembled transcripts from HEK 293T mRNA-seq was used as the database (Slavoff et al., 2013). The search parameters included tryptic digestion with up to 2 missed cleavages, peptide charge set to 2+, 3+ and 4+, 20 ppm precursor mass tolerance and 0.02 Da fragment mass tolerance. Normal and decoy database searches were run, with the confidence level set to 95% (p<0.05). The false discovery rate (FDR) was set to 1% on both peptide and protein levels. The minimum required peptide length was eight amino acids.
For quantification, raw data were analyzed using MaxQuant (version 1.6.8.0), oxidation of methionine and N-terminal acetylation were set as variable modifications, and human UniProt (https://www.uniprot.org/, version 2021) was used as the database for searching annotated proteins. For identification of unannotated microproteins and alt-proteins, a modified database, which contained both human UniProt (https://www.uniprot.org/, version 2021) and microproteins/alt-proteins protein sequences from Table S1, was used. For all analyses, a mass deviation of 20 p.p.m. was set for MS1 peaks, and 0.02 Da was set as maximum allowed MS/MS peaks with a maximum of two missed cleavages. Maximum false discovery rates (FDR) were set to 1% both on peptide and protein levels. Protein interaction networks were constructed from MaxQuant LFQ data that were imported into Cytoscape 3.8.2 (Liu et al., 2018). Cutoffs for subcellular compartment assignments of annotated proteins were enrichment of >1.5 and significance (p value, T-test) of p value = 0.05) for each subcellular compartment relative to the untargeted control.
The extracted ion chromatograms (EICs) quantitation of microprotein peptide was accomplished via spectral counting (For alt-LAMA3, peptide: PGRGGEDLGHR, observed mass: 384.2028, mass window for EIC: 384.20–384.21, from TurboID-fibrillarin rep1/rep2/rep3 versus untargeted TurboID only rep1/rep2/rep3 (control); For alt-TM9SF3, peptide: AVAAAAAAAPDPGGR, observed mass: 633.3333, mass window for EIC: 633.33–633.34, from TurboID-H2B rep1/rep2/rep3 versus untargeted TurboID only rep1/rep2/rep3 (control); For LOC728392, peptide: GLEQIRPDPESEGLFDKPPPEDPPAAR, observed mass: 740.1235, mass window for EIC: 740.12–740.13, from TurboID-lamin B1 rep1/rep2/rep3 versus untargeted TurboID only rep1/rep2/rep3 (control); For MAP3K4-AS1, peptide: PSGPTEFGPGPAPLSASDR, observed mass: 920.4468, mass window for EIC: 920.44–920.45, from TurboID-3xNLS rep1/rep2/rep3 versus untargeted TurboID only rep1/rep2/rep3 (control)), followed by comparing the MS1 extracted ion chromatograph (EIC) peak intensity in TurboID-fibrillarin/H2B/laminB1/3xNLS cell line versus untargeted TurboID only (control) cell line using Xcalibur 4.3 (Thermo Scientific).
Immunofluorescence
HEK 293T V5-TurboID-fibrillarin/H2B/laminB1/3xNLS cells (2×104 cells per well) and mouse Hepa 1–6 V5-TurboID-fibrillarin/H2B/laminB1/3xNLS cells (2×104 cells per well) were grown on fibronectin-coated glass coverslips (AmScope, CS-R18–100, 18 mm diameter round microscope glass cover slides) in a 12-well plate to 70% confluency. For transgene overexpression, HEK 293T cells in 12-well plate to 40% confluency transfected using Lipofectamine 2000 with plasmids encoding the full-length coding sequencing of alt-CDK13, alt-FYN, alt-DR1 and alt-PTP4A2 with a FLAG tag appended to the C-terminus of the smORF/alt-ORF. Medium was replaced and immunofluorescence performed 24h later. Cells were fixed with 10% neutral buffered formalin (Fisher Scientific), permeabilized with methanol at −20 °C, and blocked with blocking buffer (3% BSA in PBS) for 1 h at room temperature. Cells were stained with rabbit monoclonal anti-V5 (Cell Signaling Technology, 13202S) at a 1:1000 dilution (volume: volume) in blocking buffer overnight at 4 °C, followed by 3 washes with PBS. Goat anti-rabbit Alexa Fluor™ 568 (Life Technologies, A-11011) was subsequently applied at a 1:1000 dilution in blocking buffer for 1 to 4 hours at room temperature in the dark, followed by 5 with PBS washes. Cells were post-fixed with 10% buffered formalin, nuclei were stained with DAPI (EMD Millipore, Cat. 268298, 1:20000 dilution in 1x PBS), and imaging was performed on a laser scanning confocal microscope (Leica TCS SP8) with PL (field planarity) APO (apochromatic) 63x/1.40 oil, CS2 and PL APO 100x/1.44 oil, CORR (correction collar) CS (confocal scanning).
Generation of knock-in cell lines.
Alt-LAMA3-, alt-TM9SF3-, LOC728392- and MAP3K4-AS1–3xGFP11-FLAG-HA knock-in (KI) HEK 293T cells were generated using CRISPR-Cas9-directed homologous recombination. Guide RNAs (gRNAs) were designed with the guide design tool from the Zhang lab (crispr.mit.edu) to target the desired genomic region (Ran et al., 2013). Double-stranded DNA oligonucleotides corresponding to the gRNAs were inserted into pSpCas9(BB)-2A-GFP vector (Addgene, as a gift from F. Zhang, MIT, Cambridge, MA). A donor plasmid containing 300 bp homology left-arm and 300 bp homology right-arm sequences around the stop codon of alt-LAMA3, alt-TM9SF3, LOC728392 or MAP3K4-AS1, separated by an encoded 3xGFP11-FLAG-HA tag and BamHI/KpnI restriction sites, was synthesized by GenScript, and a DNA sequence containing hPGK promoter and puromycin resistance genes were subcloned into the donor plasmid using the BamH1 and KpnI restriction sites. An equal mixture of the gRNA and donor plasmids were transfected into HEK 293T cells using polyethyleneimine (Polysciences, Cat. No, 23966), and 2 ng/ml puromycin (Sigma-Aldrich, cat. No. P8833) selection was performed for 48h. Cells were harvested and KI were confirmed by genomic DNA PCR and Sanger sequencing, as well as Western blotting.
Immunoprecipitation.
HEK 293T cells or KI cells were grown to 80–90% confluency in 15 cm dishes. Cells were harvested and suspended in 1 mL nuclear isolation buffer (10 mM Hepes pH 7.4, 100 mM KCl, 5 mM MgCl2 with 0.5% NP40 and Roche Complete protease inhibitor cocktail tablets (Roche, Cat. No.11873580001)), and incubated on ice for 10 min, followed by centrifugation 3min at 4°C by 3,000 rpm. The nuclear pellets were suspended in 1 mL RIPA lysis buffer, followed by sonication and immunoprecipitation as previously described (Cao et al., 2021). After the final wash with PBS buffer, elution was in 50 μL of 3× FLAG peptide (Sigma-Aldrich, Cat. No. F4799) in RIPA lysis buffer at 4°C for 1 h. The eluted proteins were subjected to SDS-PAGE gel electrophoresis and the whole lane was excised and digested for proteomic analysis. Data were analyzed by using MaxQuant (version 1.6.8.0), oxidation of methionine and N-terminal acetylation were set as variable modifications, and human UniProt (https://www.uniprot.org/, version 2021) plus protein sequences of alt-LAMA3, alt-TM9SF3, LOC728392 and MAP3K4-AS1, was used as the database for searching.
Generation of CRISPR knock out (KO) cells
Alt-LAMA3 KO HEK 293T cells were generated using guide RNAs (gRNAs) designed with the guide design tool from the Zhang lab (crispr.mit.edu) to target the alt-LAMA3 genomic region (gRNAs: 5’-TAGTCCTGGCGCTGCAGGTC-3’ and 5’-GCGGGAGAGACGCCGTCTGC-3’). Double-stranded DNA oligonucleotides corresponding to the gRNAs were inserted into pSpCas9(BB)-2A-GFP vector (Addgene, as a gift from F. Zhang, MIT, Cambridge, MA) (Ran et al., 2013). In each case, an equal mixture of the two gRNA plasmids were transfected into HEK 293T cells using Lipofectamine 2000 (Thermo Fisher, Cat. 11668019) according to the manufacturer’s instructions, and GFP-positive cells were sorted with flow cytometry. Loss of alt-LAMA3 expression was confirmed by genomic DNA PCR, Sanger sequencing and mRNA-seq. In the alt-LAMA3 KO cell lines used in this study, the two alleles were disrupted by a 116-nt homozygous deletion. Rescue (RE) cells reintroducing alt-LAMA3-FLAG on the KO background were generated with lentiviral transduction and selection as described above.
Ribosomal RNA synthesis by 5-Ethynyluridine labelling
Ribosomal RNA synthesis was assayed with 5-ethynyluridine labelling as previously described (Bryant, 2021; Jao and Salic, 2008). Briefly, HEK 293T, HEK 293T alt-LAMA3 KO and HEK 293T alt-LAMA3 rescue (RE, alt-LAMA3 stable reintroduction in KO) cells were grown to 70–80% confluency in 6 well plates, then cells were treated with 1 mM 5-ethynyluridine (Click Chemistry Tools, Cat. 1261–10) or 1 uM BMH-21 (Cayman Chemical Company, Cat. 22282–5mg) as positive control. Cells were incubated at 37 °C for 1 h to label nascent RNA. Cells were fixed with 10% neutral buffered formalin (Fisher Scientific), permeabilized with 0.5% Triton in TBS-T for 5 min, and blocked with blocking buffer (3% BSA in PBS) for 1 h at room temperature. Cells were stained with mouse monoclonal anti-fibrillarin (Abcam, ab4566) at a 1:1000 dilution (volume:volume) in blocking buffer overnight at 4 °C, followed by 3 consecutive washes with PBS. Goat anti-mouse IgG Alexa Fluor™ 647 (Invitrogen, A21235) was subsequently applied at a 1:1000 dilution in blocking buffer for 1 hour at room temperature in the dark, followed by 5 PBS washes. A solution of 5 μM Alexa Fluor™ 488 Azide (Click Chemistry Tools, Cat. #1275–1), 0.5 mg/mL CuSO4 (Sigma-Aldrich, Cat. No. C8027) and 20 mg/mL sodium L-ascorbate (Sigma-Aldrich, Cat. No. A4034) was then added to the cells and incubated for 1 hour at room temperature in the dark, followed by 3 PBS washes. Nuclei were stained with DAPI (EMD Millipore, Cat. 268298, 1:20000 dilution in 1x PBS), and imaging was performed on a laser scanning confocal microscope (Leica TCS SP8) with PL (field planarity) APO (apochromatic) 63x/1.40 oil, CS2 and PL APO 100x/1.44 oil, CORR (correction collar) CS (confocal scanning). Image analysis was conducted with a custom pipeline for CellProfiler 3.1.9 (Bryant, 2021; Carpenter et al., 2006; McQuin et al., 2018).
Dual Luciferase Assay
The dual luciferase assay for RNAPI activity followed a published protocol (Freed et al., 2012; Ghoshal et al., 2004). Briefly, HEK 293T, HEK 293T alt-LAMA3 KO and HEK 293T alt-LAMA3 RE cells were grown to 70–80% confluency in 6 well plates, then transfected with 400 ng of pHrD-internal ribosome entry site (IRES)-luciferase plasmid and 1 ng of Renilla luciferase plasmid using Lipofectamine 2000 (Cat. No. L3000015; Life Technologies). After 24h, cells were harvested and luciferase activities quantified following the manufacturer’s protocol (Dual-Luciferase® Reporter Assay System, Promega Cat. E1910). The ratio of pHrD-IRES-luciferase/Renilla activity was calculated to control for transfection efficiency.
qRT-PCR Analysis
Total RNA was extracted from HEK 293T, HEK 293T alt-LAMA3 KO and HEK 293T alt-LAMA3 RE and purified with TRIzol (Cat. No. 15596026; Life Technologies) per the manufacturer’s instructions. cDNA was synthesized using the iScript gDNA Clear cDNA Synthesis Kit (Bio-Rad, Cat. 172–5035) following the manufacturer’s instructions. Reactions for qPCR were set up on ice according to the manufacturer’s instructions using the iTaq Universal SYBR Green Supermix (Bio-Rad, Cat. 172–5121). Amplification of the 7SL RNA was used as an internal control, and relative expression between samples was calculated with the comparative CT (2−ΔΔCt) method as previously reported (Sondalle et al., 2019).
Global protein synthesis assay
Global protein synthesis was quantified as previously reported (Farley-Barnes et al., 2018; Schmidt et al., 2009). HEK 293T, HEK 293T alt-LAMA3 KO and HEK 293T alt-LAMA3 RE cells were grown to 70–80% confluency in 6 well plates, then growth media was replaced with media containing 1 μM puromycin and cells were incubated at 37 °C for 1 h to label nascent peptides. The cells were washed 3 times with PBS, harvested and analyzed with Western blotting. Image J was used for lane densitometry.
AAV production and purification
For the generation of AAV TurboID-fibrillarin/H2B/laminB1/3xNLS-GFP expression vector (pZK1–4), CMV-TuboID-fibrillarin/H2B/laminB1/NLS expression cassette was synthesized and subcloned into an AAV-GFP backbone (pXD017) containing inverted terminal repeats by using Gibson Assembly® (NEB) as previously reported (Dai et al., 2019).
We produced AAV9 by transfecting HEK 293 FT cells (Thermo Fisher) in 15-cm tissue culture dishes (Corning). For transfection, AAV transgene vectors, packaging (pDF6) plasmid, and AAV9 serotype plasmid were pooled together at 1:2:1.7 ratio with polyethyleneimine. Transfected cells were collected with PBS 72 h after transfection. For the AAV purification, transfected cells were mixed with pure chloroform (1:10 volume) and incubated at 37 °C with vigorous shaking for 1 h. NaCl was added to a final concentration of 1 M, and then the samples were centrifuged at 20,000g at 4 °C for 15 min. The chloroform layer was discarded while the aqueous layer was transferred to another tube. PEG8000 was added to 10% (w/v) and shaken until dissolved. The mixture was incubated at 4 °C for 1 h and then centrifuged at 20,000g at 4 °C for 15 min. The supernatant was discarded and the pellet was suspended using PBS with MgCl2, treated with universal nuclease (Thermo Fisher), and incubated at 37 °C for 30 min. Chloroform (1:1 volume) was then added, shaken, and centrifuged at 12,000 rpm at 4 °C for 15 min. The aqueous layer was isolated and concentrated through a 100-kDa molecular-weight cutoff filter (Millipore). Virus was titered by quantitative PCR using custom Taqman assays (Thermo Fisher) targeted to promoter EFS (Dai et al., 2019).
Viral transduction in vivo
C57BL/6J mice between 6–8 weeks old were used and injected with TurboID-fibrillarin/NLS AAV9 viruses via tail vein at a dose of 1011 genome copies (GC) per mouse for 2 days. One week after transduction with AAV9 viruses, biotin was administered by intraperitoneal injection (24 mg/ml, in a solution of 18:1:1 saline:Kolliphor EL:DMSO, final volume of 200 μl per mouse per day) for 3 consecutive days by following previously protocol (Wei et al., 2021). Twenty-four hours after the final biotin dose, liver, spleen, lung, heart and kidney were dissected and collected into Eppendorf tubes and immediately frozen on dry ice or stored at −80 °C.
Mouse tissue preparation for molecular biology
Mouse tissues were homogenized in 0.5 ml of cold RIPA buffer using 24-well polyethylene vials with metal beads in a GenoGrinder machine (OPS Diagnostics) (Wang et al., 2019). Homogenized tissue was centrifuged at 13,000 rpm, 10 min, 4 °C. For validation of TurboID-fibrillarin/3xNLS expression, supernatant was transferred to nitrocellulose membranes followed by a standard Western blotting protocol. Briefly, immunoblots were blocked with 3% BSA in TBS-T for 1 hour at RT and probed with primary antibodies against V5 (Cell Signaling Technology, 13202S) in 3% BSA in TBS-T overnight at 4 °C. The membrane was washed three times with TBS-T, probed with secondary antibodies in 3% BSA in TBS-T for 1 hour at RT, and washed three times with TBS-T before development with Clarity ECL Western Blotting Substrate (Bio-Rad, Cat. 1705060) and imaging.
To enrich remaining biotinylated tissue protein samples for proteomic study, 200 μl Dynabeads MyOne Streptavidin T1 magnetic beads (ThermoFisher, Cat. 65602) were washed twice with washing buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% SDS, 0.5% sodium), incubated with clarified lysates containing ~ 10 mg protein for each sample with rotation for 1 h at room temperature, then moved to 4 °C and incubated overnight with rotation. The beads were subsequently washed twice with 1 mL of RIPA lysis buffer, once with 1 mL of 1 M KCl, once with 1 mL of 0.1 M Na2CO3, once with 1 mL of 2 M urea in 10 mM Tris-HCl (pH 8.0), and twice with 1 mL RIPA lysis buffer. The beads were removed and biotinylated proteins were eluted by boiling the beads in 75 μL of 2× protein loading buffer supplemented with 20 mM DTT and 2 mM biotin. The eluted proteins were separated on a Tricine-SDS-PAGE gel and proteomic analyses were performed as described above. Searches were against translated mRNA-seq assembled transcripts from Ensembl GRCm39 (Mus musculus, https://useast.ensembl.org/index.html) as databases.
Immunofluorescence of liver sections
Freshly dissected livers were embedded in OCT and sectioned at a thickness of 10 μm on a freezing-sliding microtome (Chow et al., 2017). Sections were collected using Superfrost Plus Stain slides (Fisher Scientific). Slides were rehydrated with PBS for 10 min. Sections were fixed with 10% neutral buffered formalin (Fisher Scientific), permeabilized with 0.5% Triton in TBS-T for 5 min, and blocked with blocking buffer (3% BSA in PBS) for 1 h at room temperature. Cells were stained with rabbit monoclonal anti-V5 (Cell Signaling Technology, Cat. 13202S) at a 1:1000 dilution (volume:volume) in blocking buffer overnight at 4 °C, followed by 3 consecutive washes with PBS. Goat anti-rabbit Alexa Fluor™ 568 (Life Technologies, A-11011) was subsequently applied at a 1:1000 dilution in blocking buffer for 1 hour at room temperature in the dark, followed by 5 PBS washes. Strepavidin-Alexa Fluor™ 647 (Invitrogen, S21374) was then added at 1:1000 dilution and incubated for 1 hour, followed by 3 PBS washes. Sections were post-fixed with 10% buffered formalin, nuclei were stained with DAPI (EMD Millipore, Cat. 268298, 1:20000 dilution in 1x PBS), and imaging was performed on a laser scanning confocal microscope (Leica TCS SP8) with PL (field planarity) APO (apochromatic) 63x/1.40 oil, CS2 and PL APO 100x/1.44 oil, CORR (correction collar) CS (confocal scanning).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis of results was performed using two-tailed Student’s t-test, or analysis of variance (ANOVA), followed by Dunnett’s test, as stated in the figure legends. All analyses were done using GraphPad Prism, version 9.3.0. For experiments in cell culture, n refers to one well or dish in which cells are grown; for in vivo experiments, n refers to one animal. Exact values for n are presented in each figure legend. In all figures, the center represents the mean and error bars represent standard deviation. Statistical significance is represented in all figures, as follows: p-value of <0.0001: ****, p-value of 0.0001 to 0.001: ***, p-value of 0.001 to 0.01: **, p-value of 0.01 to 0.05: * and p-value of ≥0.05: not significant.
Supplementary Material
Table S1: Full list of unannoated microproteins and alt-proteins identified in cell culture by MicroID (related to Figure 1). Alt-proteins with lengths greater than 100 amino acids are highlighted in yellow.
Table S2: Full list of unannoated microproteins and alt-proteins mapped in vivo by MicroID (related to Figure 5). Alt-proteins with lengths greater than 100 amino acids are highlighted in yellow.
Highlights:
1. MicroID reveals microproteins and alternative proteins in subnuclear compartments
2. Alt-LAMA3 is physically and functionally associated with pre-rRNA transcription
3. MicroID detects unannotated microproteins and alternative proteins in vivo
Acknowledgments
We thank all members of the Slavoff lab, Baserga lab and Chen lab for helpful discussions. This work was supported by the Searle Scholars Program, the Richard and Susan Smith Family Foundation, the NIH (R01GM122984), and Yale University West Campus (to S. A. S.). X.D. is supported by a Revson Fellowship. X.C. was supported in part by a Rudolph J. Anderson postdoctoral fellowship from Yale University. S.J.B, C.M.H. and A. B. were supported by R35 GM131687.
Footnotes
Declaration of interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Andrews JO, Conway W, Cho WK, Narayanan A, Spille JH, Jayanth N, Inoue T, Mullen S, Thaler J, and Cisse II (2018). qSR: a quantitative super-resolution analysis tool reveals the cell-cycle dependent organization of RNA Polymerase I in live human cells. Sci Rep 8, 7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, and Neugebauer KM (2012). The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol 13, R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult N, Correia A, Ma J, Merlo A, Garcia-Gomez S, Maric M, Tognetti M, Benner CW, Boulton SJ, Saghatelian A, et al. (2017). Regulation of DNA repair pathway choice in S and G2 phases by the NHEJ inhibitor CYREN. Nature 549, 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basrai MA, Hieter P, and Boeke JD (1997). Small open reading frames: beautiful needles in the haystack. Genome Res 7, 768–771. [DOI] [PubMed] [Google Scholar]
- Bonfiglio JJ, Leidecker O, Dauben H, Longarini EJ, Colby T, San Segundo-Acosta P, Perez KA, and Matic I. (2020). An HPF1/PARP1-Based Chemical Biology Strategy for Exploring ADP-Ribosylation. Cell 183, 1086–1102 e1023. [DOI] [PubMed] [Google Scholar]
- Boucher MJ, Ghosh S, Zhang L, Lal A, Jang SW, Ju A, Zhang S, Wang X, Ralph SA, Zou J, et al. (2018). Integrative proteomics and bioinformatic prediction enable a high-confidence apicoplast proteome in malaria parasites. PLoS Biol 16, e2005895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branon TC., Bosch JA., Sanchez AD., Udeshi ND., Svinkina T., Carr SA., Feldman JL., Perrimon N., and Ting AY. (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36, 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet MA, Jacques JF, Nassari S, Tyzack GE, McGoldrick P, Zinman L, Jean S, Robertson J, Patani R, and Roucou X. (2021a). The FUS gene is dual-coding with both proteins contributing to FUS-mediated toxicity. EMBO Rep 22, e50640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet MA, Levesque SA, Hunting DJ, Cohen AA, and Roucou X. (2018). Recognition of the polycistronic nature of human genes is critical to understanding the genotype-phenotype relationship. Genome Res 28, 609–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet MA, Lucier JF, Levesque M, Leblanc S, Jacques JF, Al-Saedi HRH, Guilloy N, Grenier F, Avino M, Fournier I, et al. (2021b). OpenProt 2021: deeper functional annotation of the coding potential of eukaryotic genomes. Nucleic Acids Res 49, D380–D388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant CJ, McCool MA, Abriola L, Surovtseva YV, and Baserga SJ (2022). A high-throughput assay for directly monitoring nucleolar rRNA biogenesis. Open Biol 12, 210305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant CJ, McCool MA, Abriola L, Surovtseva YV & Baserga SJ (2021). A high-throughput assay for directly monitoring nucleolar rRNA biogenesis. bioRxiv, 2021.2007.2019.452935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Khitun A, Harold CM, Bryant CJ, Zheng SJ, Baserga SJ, and Slavoff SA (2022). Nascent alt-protein chemoproteomics reveals a pre-60S assembly checkpoint inhibitor. Nat Chem Biol 18, 643–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Khitun A, Luo Y, Na Z, Phoodokmai T, Sappakhaw K, Olatunji E, Uttamapinant C, and Slavoff SA (2021). Alt-RPL36 downregulates the PI3K-AKT-mTOR signaling pathway by interacting with TMEM24. Nat Commun 12, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Khitun A, Na Z, Dumitrescu DG, Kubica M, Olatunji E, and Slavoff SA (2020). Comparative Proteomic Profiling of Unannotated Microproteins and Alternative Proteins in Human Cell Lines. J Proteome Res 19, 3418–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargile BJ, Bundy JL, and Stephenson JL Jr. (2004). Potential for false positive identifications from large databases through tandem mass spectrometry. J Proteome Res 3, 1082–1085. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, et al. (2012). Proto-genes and de novo gene birth. Nature 487, 370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Brunner AD, Cogan JZ, Nunez JK, Fields AP, Adamson B, Itzhak DN, Li JY, Mann M, Leonetti MD, et al. (2020). Pervasive functional translation of noncanonical human open reading frames. Science 367, 1140–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow RD, Guzman CD, Wang G, Schmidt F, Youngblood MW, Ye L, Errami Y, Dong MB, Martinez MA, Zhang S, et al. (2017). AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat Neurosci 20, 1329–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couso JP, and Patraquim P. (2017). Classification and function of small open reading frames. Nat Rev Mol Cell Biol 18, 575–589. [DOI] [PubMed] [Google Scholar]
- Dai X, Park JJ, Du Y, Kim HR, Wang G, Errami Y, and Chen S. (2019). One-step generation of modular CAR-T cells with AAV-Cpf1. Nat Methods 16, 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Narvaiza I, Kerman BE, Pena M, Benner C, Marchetto MC, Diedrich JK, Aslanian A, Ma J, Moresco JJ, et al. (2015). Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell 163, 583–593. [DOI] [PubMed] [Google Scholar]
- Douka K, Birds I, Wang D, Kosteletos A, Clayton S, Byford A, Vasconcelos EJR, O’Connell MJ, Deuchars J, Whitehouse A, et al. (2021). Cytoplasmic long noncoding RNAs are differentially regulated and translated during human neuronal differentiation. RNA 27, 1082–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droujinine IA., Meyer AS., Wang D., Udeshi ND., Hu Y., Rocco D., McMahon JA., Yang R., Guo J., Mu L., et al. (2021). Proteomics of protein trafficking by in vivo tissue-specific labeling. Nat Commun 12, 2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley-Barnes KI, McCann KL, Ogawa LM, Merkel J, Surovtseva YV, and Baserga SJ (2018). Diverse Regulators of Human Ribosome Biogenesis Discovered by Changes in Nucleolar Number. Cell Rep 22, 1923–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazal FM, Han S, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, Chang HY, and Ting AY (2019). Atlas of Subcellular RNA Localization Revealed by APEX-Seq. Cell 178, 473–490 e426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EF, Prieto JL, McCann KL, McStay B, and Baserga SJ (2012). NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing. PLoS Genet 8, e1002892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon M, Savard M, Jacques JF, Bkaily G, Geha S, Roucou X, and Gobeil F. (2021). Potentiation of B2 receptor signaling by AltB2R, a newly identified alternative protein encoded in the human bradykinin B2 receptor gene. J Biol Chem 296, 100329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal K, Majumder S, Datta J, Motiwala T, Bai S, Sharma SM, Frankel W, and Jacob ST (2004). Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem 279, 6783–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go CD, Knight JDR, Rajasekharan A, Rathod B, Hesketh GG, Abe KT, Youn JY, Samavarchi-Tehrani P, Zhang H, Zhu LY, et al. (2021). A proximity-dependent biotinylation map of a human cell. Nature 595, 120–124. [DOI] [PubMed] [Google Scholar]
- Grimm T, Holzel M, Rohrmoser M, Harasim T, Malamoussi A, Gruber-Eber A, Kremmer E, and Eick D. (2006). Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res 34, 3030–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GD, Coyaud E, Goncalves J, Mojarad BA, Liu Y, Wu Q, Gheiratmand L, Comartin D, Tkach JM, Cheung SW, et al. (2015). A Dynamic Protein Interaction Landscape of the Human Centrosome-Cilium Interface. Cell 163, 1484–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y, Fujimura A, Kato K, Udagawa R, Hirota T, and Kimura K. (2018). Nucleolar integrity during interphase supports faithful Cdk1 activation and mitotic entry. Sci Adv 4, eaap7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Verdun D. (2011). Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2, 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang N, Li F, Zhang M, Zhou H, Chen Z, Ma X, Yang L, Wu X, Zhong J, Xiao F, et al. (2021a). An Upstream Open Reading Frame in Phosphatase and Tensin Homolog Encodes a Circuit Breaker of Lactate Metabolism. Cell Metab 33, 454. [DOI] [PubMed] [Google Scholar]
- Huang N, Li F, Zhang M, Zhou H, Chen Z, Ma X, Yang L, Wu X, Zhong J, Xiao F, et al. (2021b). An Upstream Open Reading Frame in Phosphatase and Tensin Homolog Encodes a Circuit Breaker of Lactate Metabolism. Cell Metab 33, 128–144 e129. [DOI] [PubMed] [Google Scholar]
- Hung V, Lam SS, Udeshi ND, Svinkina T, Guzman G, Mootha VK, Carr SA, and Ting AY (2017). Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst V, Shimada K, and Gasser SM (2019). Nuclear Actin and Actin-Binding Proteins in DNA Repair. Trends Cell Biol 29, 462–476. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, and Weissman JS (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer-Bierhoff A, and Grummt I. (2019). Stop-and-Go: Dynamics of Nucleolar Transcription During the Cell Cycle. Epigenet Insights 12, 2516865719849090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R, Kroehling L, Khitun A, Bailis W, Jarret A, York AG, Khan OM, Brewer JR, Skadow MH, Duizer C, et al. (2018). The translation of non-canonical open reading frames controls mucosal immunity. Nature 564, 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao CY., and Salic A. (2008). Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci U S A 105, 15779–-1578.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing J, He L, Sun A, Quintana A, Ding Y, Ma G, Tan P, Liang X, Zheng X, Chen L, et al. (2015). Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca(2)(+) influx. Nat Cell Biol 17, 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaulich PT, Cassidy L, Bartel J, Schmitz RA, and Tholey A. (2021). Multi-protease Approach for the Improved Identification and Molecular Characterization of Small Proteins and Short Open Reading Frame-Encoded Peptides. J Proteome Res 20, 2895–2903. [DOI] [PubMed] [Google Scholar]
- Keeling DM, Garza P, Nartey CM, and Carvunis AR (2019). The meanings of ‘function’ in biology and the problematic case of de novo gene emergence. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khitun A, and Slavoff SA (2019). Proteomic Detection and Validation of Translated Small Open Reading Frames. Curr Protoc Chem Biol 11, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A, Gawron D, Steyaert S, Ndah E, Crappe J, De Keulenaer S, De Meester E, Ma M, Shen B, Gevaert K, et al. (2014). A proteogenomics approach integrating proteomics and ribosome profiling increases the efficiency of protein identification and enables the discovery of alternative translation start sites. Proteomics 14, 2688–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh M, Ahmad I, Ko Y, Zhang Y, Martinez TF, Diedrich JK, Chu Q, Moresco JJ, Erb MA, Saghatelian A, et al. (2021). A short ORF-encoded transcriptional regulator. Proc Natl Acad Sci U S A 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CQE, Kerouanton B, Chothani S, Zhang S, Chen Y, Mantri CK, Hock DH, Lim R, Nadkarni R, Huynh VT, et al. (2021). Coding and non-coding roles of MOCCI (C15ORF48) coordinate to regulate host inflammation and immunity. Nat Commun 12, 2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Han S, Li H, Udeshi ND, Svinkina T, Mani DR, Xu C, Guajardo R, Xie Q, Li T, et al. (2020). Cell-Surface Proteomic Profiling in the Fly Brain Uncovers Wiring Regulators. Cell 180, 373–386 e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Salokas K, Tamene F, Jiu Y, Weldatsadik RG, Ohman T, and Varjosalo M. (2018). An AP-MS-and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat Commun 9, 1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KH, Stawski PS, Draycott AS, Udeshi ND, Lehrman EK, Wilton DK, Svinkina T, Deerinck TJ, Ellisman MH, Stevens B, et al. (2016). Proteomic Analysis of Unbounded Cellular Compartments: Synaptic Clefts. Cell 166, 1295–1307 e1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, et al. (2018). Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 172, 590–604 e513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez TF, Chu Q, Donaldson C, Tan D, Shokhirev MN, and Saghatelian A. (2020). Accurate annotation of human protein-coding small open reading frames. Nat Chem Biol 16, 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuin C, Goodman A, Chernyshev V, Kamentsky L, Cimini BA, Karhohs KW, Doan M, Ding L, Rafelski SM, Thirstrup D, et al. (2018). CellProfiler 3.0: Next-generation image processing for biology. PLoS Biol 16, e2005970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meagher M, Epling LB, and Enemark EJ (2019). DNA translocation mechanism of the MCM complex and implications for replication initiation. Nat Commun 10, 3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SA, Wright J, Peckner R, Kalish BT, Zhang F, and Carr SA (2018). Discovery of proteins associated with a predefined genomic locus via dCas9-APEX-mediated proximity labeling. Nat Methods 15, 437–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na Z, Luo Y, Schofield JA, Smelyansky S, Khitun A, Muthukumar S, Valkov E, Simon MD, and Slavoff SA (2020). The NBDY Microprotein Regulates Cellular RNA Decapping. Biochemistry 59, 4131–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa LM., Buhagiar AF., Abriola L., Leland BA., Surovtseva YV., and Baserga SJ. (2021). Increased numbers of nucleoli in a genome-wide RNAi screen reveal proteins that link the cell cycle to RNA polymerase I transcription. Mol Biol Cell 32, 956–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuwaki M, Sumi A, Hisaoka M, Saotome-Nakamura A, Akashi S, Nishimura Y, and Nagata K. (2012). Function of homo- and hetero-oligomers of human nucleoplasmin/nucleophosmin family proteins NPM1, NPM2 and NPM3 during sperm chromatin remodeling. Nucleic Acids Res 40, 4861–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MW, Mao Y, Storz G, and Qian SB (2020). Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res 48, 1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polycarpou-Schwarz M, Gross M, Mestdagh P, Schott J, Grund SE, Hildenbrand C, Rom J, Aulmann S, Sinn HP, Vandesompele J, et al. (2018). The cancer-associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene 37, 4750–4768. [DOI] [PubMed] [Google Scholar]
- Prensner JR, Enache OM, Luria V, Krug K, Clauser KR, Dempster JM, Karger A, Wang L, Stumbraite K, Wang VM, et al. (2021). Noncanonical open reading frames encode functional proteins essential for cancer cell survival. Nat Biotechnol 39, 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Cho KF, Cavanagh PE, and Ting AY (2021). Deciphering molecular interactions by proximity labeling. Nat Methods 18, 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrmoser M, Holzel M, Grimm T, Malamoussi A, Harasim T, Orban M, Pfisterer I, Gruber-Eber A, Kremmer E, and Eick D. (2007). Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol Cell Biol 27, 3682–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samandi S, Roy AV, Delcourt V, Lucier JF, Gagnon J, Beaudoin MC, Vanderperre B, Breton MA, Motard J, Jacques JF, et al. (2017). Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EK, Clavarino G, Ceppi M, and Pierre P. (2009). SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6, 275–277. [DOI] [PubMed] [Google Scholar]
- Schrank BR, Aparicio T, Li Y, Chang W, Chait BT, Gundersen GG, Gottesman ME, and Gautier J. (2018). Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 559, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinnider MA, Scott NE, Prudova A, Kerr CH, Stoynov N, Stacey RG, Chan QWT, Rattray D, Gsponer J, and Foster LJ (2021). An atlas of protein-protein interactions across mouse tissues. Cell 184, 4073–4089 e4017. [DOI] [PubMed] [Google Scholar]
- Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, and Saghatelian A. (2013). Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol 9, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondalle SB, Longerich S, Ogawa LM, Sung P, and Baserga SJ (2019). Fanconi anemia protein FANCI functions in ribosome biogenesis. Proc Natl Acad Sci U S A 116, 2561–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Wan X, Huang T, Zeng C, Sastry N, Wu B, James CD, Horbinski C, Nakano I, Zhang W, et al. (2019). SRSF3-Regulated RNA Alternative Splicing Promotes Glioblastoma Tumorigenicity by Affecting Multiple Cellular Processes. Cancer Res 79, 5288–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speers AE, and Wu CC (2007). Proteomics of integral membrane proteins--theory and application. Chem Rev 107, 3687–3714. [DOI] [PubMed] [Google Scholar]
- Stamatopoulou V, Parisot P, De Vleeschouwer C, and Lafontaine DLJ (2018). Use of the iNo score to discriminate normal from altered nucleolar morphology, with applications in basic cell biology and potential in human disease diagnostics. Nat Protoc 13, 2387–2406. [DOI] [PubMed] [Google Scholar]
- Takano T., Wallace JT., Baldwin KT., Purkey AM., Uezu A., Courtland JL., Soderblom EJ., Shimogori T., Maness PF., Eroglu C., et al. (202. 0). Chemico-genetic discovery of astrocytic control of inhibition in vivo. Nature 588, 296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G, Singer O, and Verma IM (2006). Production and purification of lentiviral vectors. Nat Protoc 1, 241–245. [DOI] [PubMed] [Google Scholar]
- Vanderperre B, Lucier JF, Bissonnette C, Motard J, Tremblay G, Vanderperre S, Wisztorski M, Salzet M, Boisvert FM, and Roucou X. (2013). Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLoS One 8, e70698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Chow RD, Bai Z, Zhu L, Errami Y, Dai X, Dong MB, Ye L, Zhang X, Renauer PA, et al. (2019). Multiplexed activation of endogenous genes by CRISPRa elicits potent antitumor immunity. Nat Immunol 20, 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Riley NM, Yang AC, Kim JT, Terrell SM, Li VL, Garcia-Contreras M, Bertozzi CR, and Long JZ (2021). Cell type-selective secretome profiling in vivo. Nat Chem Biol 17, 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Wang Y, and Lu J. (2019). Function and Evolution of Upstream ORFs in Eukaryotes. Trends Biochem Sci 44, 782–794. [DOI] [PubMed] [Google Scholar]
- Zhang S, Reljic B, Liang C, Kerouanton B, Francisco JC, Peh JH, Mary C, Jagannathan NS, Olexiouk V, Tang C, et al. (2020). Mitochondrial peptide BRAWNIN is essential for vertebrate respiratory complex III assembly. Nat Commun 11, 1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y, Sorensen V, Skjerpen CS, Haugsten EM, Jin Y, Walchli S, Olsnes S, and Wiedlocha A. (2012). Nuclear import of exogenous FGF1 requires the ER-protein LRRC59 and the importins Kpnalpha1 and Kpnbeta1. Traffic 13, 650–664. [DOI] [PubMed] [Google Scholar]
- Zhu ZC, Liu JW, Li K, Zheng J, and Xiong ZQ (2018). KPNB1 inhibition disrupts proteostasis and triggers unfolded protein response-mediated apoptosis in glioblastoma cells. Oncogene 37, 2936–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Full list of unannoated microproteins and alt-proteins identified in cell culture by MicroID (related to Figure 1). Alt-proteins with lengths greater than 100 amino acids are highlighted in yellow.
Table S2: Full list of unannoated microproteins and alt-proteins mapped in vivo by MicroID (related to Figure 5). Alt-proteins with lengths greater than 100 amino acids are highlighted in yellow.
Data Availability Statement
Proteomic data are available from the PRIDE repository under accession number PXD033067. The mRNA-seq data have been deposited in the NCBI Gene Expression Omnibus under accession GSE205869. The raw imaging and original Western blotting data have been deposited at Mendeley Data. Accession numbers are listed in the key resources table. Additional data reported in this paper are available as of the date of publication.
This paper does not report original code. Previously reported code for identification of microproteins is available at 10.5281/zenodo.5921116.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Key Resource Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-V5 | Cell Signaling Technology | Cat#13202S; RRID: AB_2687461 |
| Mouse monoclonal anti-FLAG | Sigma | Cat#F3165; RRID: AB_259529 |
| Rabbit monoclonal anti-FLAG | Cell Signaling Technology | Cat#14793S; RRID: AB_2572291 |
| Mouse monoclonal anti-beta actin | Invitrogen | Cat#BA3R; RRID: AB_10979409 |
| Strepavidin horseradish peroxidase conjugate | Invitrogen | Cat#S911 |
| Mouse monoclonal anti-fibrillarin | Abcam | Cat#ab4566; RRID: AB_304523 |
| Rabbit monoclonal anti-lamin B1 | Abcam | Cat#ab133741; RRID: AB_2616597 |
| Rabbit monoclonal anti-histone H3 | Cell Signaling Technology | Cat#4499S; RRID: AB_10544537 |
| Rabbit monoclonal anti-LAMA3 | Abcam | Cat#ab151715 |
| Rabbit polyclonal anti-PES1 | Bethyl Laboratories | Cat#A300-903A-2; RRID: AB_625300 |
| Rabbit polyclonal anti-BOP1 | Bethyl Laboratories | Cat#A302-148A-1; RRID: AB_1720319 |
| Rabbit polyclonal anti-WDR12 | Bethyl Laboratories | Cat#A302-650A; RRID: AB_10568823 |
| Rabbit polyclonal anti-puromycin | Kerafast | Cat#EQ0001; RRID: AB_2620162 |
| Anti-FLAG M2 affinity gel | Sigma | Cat#A2220; RRID: AB_10063035 |
| Goat anti-rabbit IgG horseradish peroxidase conjugate | Rockland | Cat#611-1302; RRID: AB_218567 |
| Goat anti-mouse IgG horseradish peroxidase conjugate | Rockland | Cat#610-1319-0500; RRID: AB_219659 |
| Goat anti-rabbit IgG Alexa FluorTM 568 | Invitrogen | Cat#A11011; RRID: AB_143157 |
| Goat anti-mouse IgG Alexa FluorTM 647 | Invitrogen | Cat#A21235; RRID: AB_2535804 |
| Strepavidin Alexa FluorTM 647 conjugate | Invitrogen | Cat#S21374; RRID: AB_2336066 |
| Bacterial and virus strains | ||
| One Shot Stbl3 Chemical Competent E. coli | Thermo Fisher Scientific | Cat#C737303 |
| Chemicals, peptides, and recombinant proteins | ||
| DPBS, no calcium, no magnesium | Gibco | Cat#14190250 |
| RPMI 1640 Medium | Gibco | Cat#1875-093 |
| DMEM, high glucose, pyruvate | Gibco | Cat#11995065 |
| Fetal Bovine Serum | Sigma Aldrich | Cat#F4135-500ML |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat#15140122 |
| 2-Mercaptoethanol | Sigma Aldrich | Cat#M6250-10ML |
| PEI MAX | Polyscience | Cat#24765-1 |
| PEG8000 | Promega | Cat#V3011 |
| RIPA buffer | Boston BioProducts | Cat#BP-115 |
| Protease inhibitor coctail | Thermo Fisher Scientific | Cat#78437 |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23227 |
| QuickExtract DNA Extraction Solution | Epicenter | Cat#QE09050 |
| QIAamp DNA Blood Mini Kit | Qiagen | Cat#51106 |
| Gibson Assembly® Master Mix | NEB | Cat#E2611 |
| Phusion Flash High-Fidelity PCR Master Mix | Thermo Fisher Scientific | Cat#F548L |
| QIAquick Gel Extraction Kit | Qiagen | Cat#28706 |
| TRIzol™ Reagent | Invitrogen | Cat#15596026 |
| RNeasy Plus mini isolation kit | Qiagen | Cat#74134 |
| 4–20% Mini-PROTEAN® TGX™ Precast Protein Gels, 10-well | BioRad | Cat#4561094 |
| Bovine Serum Albumin | Sigma Aldrich | Cat#A9418-100G |
| Pierce™ ECL Western Blotting Substrate | Thermo Fisher Scientific | Cat#32106 |
| EDTA | Sigma Aldrich | Cat#E8008-100ML |
| puromycin | Sigma Aldrich | Cat#P8833 |
| biotin | Sigma Aldrich | Cat#B4501 |
| Dynabeads™ M-280 Streptavidin | Thermo Fisher Scientific | Thermo Fisher |
| Formalin | Thermo Fisher Scientific | Cat#SF100-4 |
| DAPI | EMD Millipore | Cat#268298 |
| 3×FLAG peptide | Sigma Aldrich | Cat#F4799 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat#11668019 |
| 5-ethynyluridine | Click Chemistry Tools | Cat#1261-10 |
| BMH-21 | Cayman Chemical Company | Cat#22282-5mg |
| Alexa FluorTM 488 Azide | Click Chemistry Tools | Cat#1275-1 |
| Critical commercial assays | ||
| Dual-Luciferase® Reporter Assay System | Promega | Cat#E1910 |
| iScript gDNA Clear cDNA Synthesis Kit | Bio-Rad | Cat#172-5035 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat#172-5121 |
| Deposited data | ||
| Raw and analyzed mRNA sequencing data | This paper | GEO: GSE205869 |
| Raw proteomic data | This paper | PRIDE: PXD033067 |
| Raw imaging and Western blotting data | This paper | Mendeley: https://doi:10.17632/yh8t333wwp.1 |
| Custom code | Khitun and Slavoff, 2019 | Zenodo: https://doi.org/10.5281/zenodo.5921116 |
| Experimental models: Cell lines | ||
| Human: HEK 293T | ATCC | Cat#CRL-3216 |
| Human: HEK 293FT | Thermo Fisher Scientific | Cat# R70007 |
| Human: HeLa | ATCC | Cat#CCL-2 |
| Human: A549 | Dr. Craig Crews (Yale University) | N/A |
| Cercopithecus aethiops: COS7 | ATCC | Cat#CRL-1651 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J mice | The Jackson Laboratory | Cat#000664 |
| Oligonucleotides | ||
| Primer: alt-LAMA3 Forward: CTGGCTTTGACCTGAGCGGTGAGT | This paper | N/A |
| Primer: alt-LAMA3 Reverse: ACCAAATTCGTTTACAAGCCTCCT | This paper | N/A |
| Primer: LAMA3 Forward: CACCGGGATATTTCGGGAATC | This paper | N/A |
| Primer: LAMA3 Reverse: AGCTGTCGCAATCATCACATT | This paper | N/A |
| Primer: Primary pre-rRNA Forward: CTCCGTTATGGTAGCGCTGC | This paper | N/A |
| Primer: Primary pre-rRNA Reverse: GCGGAACCCTCGCTTCTC | This paper | N/A |
| Primer: 7SL-RNA Forward: ATCGGGTGTCCGCACTAAGTT | This paper | N/A |
| Primer: 7SL-RNA Reverse: CAGCACGGGAGTTTTGACCT | This paper | N/A |
| Recombinant DNA | ||
| pLJM1-V5-TurboID-fibrillarin | This paper | N/A |
| pLJM1-V5-TurboID-H2B | This paper | N/A |
| pLJM1-V5-TurboID-lamin B1 | This paper | N/A |
| pLJM1-V5-TurboID-3xNLS | This paper | N/A |
| pMD2.G | pMD2.G was a gift from Didier Trono | Addgene Plasmid #12259 |
| psPAX2 | psPAX2 was a gift from Didier Trono | Addgene Plasmid #12260 |
| pSpCas9(BB)-2A-GFP | pSpCas9(BB)-2A-GFP (PX458) was a gift from Feng Zhang | Addgene Plasmid # 48138 |
| pXD017-V5-TurboID-fibrillarin | This paper | N/A |
| pXD017-V5-TurboID-H2B | This paper | N/A |
| pXD017-V5-TurboID-lamin B1 | This paper | N/A |
| pXD017-V5-TurboID-3xNLS | This paper | N/A |
| pAdDeltaF6 | pAdDeltaF6 was a gift from James M. Wilson | Addgene Plasmid # 112867 |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools | Li et al., 2009 | http://samtools.sourceforge.net/ |
| GraphPad Prism (version 9.0) | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
| Mascot | Matrix Science | https://www.matrixscience.com/ |
| MaxQuant | MaxQuant lab | https://www.maxquant.org/ |
| Perseus | MaxQuant lab | https://maxquant.net/perseus/ |
| Python (2.7.11) | Python Software Foundation, DE | https://www.python.org/downloads/ |
| Cutadapt (1.9.1) | Marcel Martin, 2010 | https://cutadapt.readthedocs.io/en/stable/ |
| STAR (2.7.1) | Alexander Dobin et al., 2013 | http://code.google.com/p/rna-star/ |