Abstract
Background
The antiviral drug molnupiravir was licensed for treating at-risk patients with COVID-19 on the basis of data from unvaccinated adults. We aimed to evaluate the safety and virological efficacy of molnupiravir in vaccinated and unvaccinated individuals with COVID-19.
Methods
This randomised, placebo-controlled, double-blind, phase 2 trial (AGILE CST-2) was done at five National Institute for Health and Care Research sites in the UK. Eligible participants were adult (aged ≥18 years) outpatients with PCR-confirmed, mild-to-moderate SARS-CoV-2 infection who were within 5 days of symptom onset. Using permuted blocks (block size 2 or 4) and stratifying by site, participants were randomly assigned (1:1) to receive either molnupiravir (orally; 800 mg twice daily for 5 days) plus standard of care or matching placebo plus standard of care. The primary outcome was the time from randomisation to SARS-CoV-2 PCR negativity on nasopharyngeal swabs and was analysed by use of a Bayesian Cox proportional hazards model for estimating the probability of a superior virological response (hazard ratio [HR]>1) for molnupiravir versus placebo. Our primary model used a two-point prior based on equal prior probabilities (50%) that the HR was 1·0 or 1·5. We defined a priori that if the probability of a HR of more than 1 was more than 80% molnupiravir would be recommended for further testing. The primary outcome was analysed in the intention-to-treat population and safety was analysed in the safety population, comprising participants who had received at least one dose of allocated treatment. This trial is registered in ClinicalTrials.gov, NCT04746183, and the ISRCTN registry, ISRCTN27106947, and is ongoing.
Findings
Between Nov 18, 2020, and March 16, 2022, 1723 patients were assessed for eligibility, of whom 180 were randomly assigned to receive either molnupiravir (n=90) or placebo (n=90) and were included in the intention-to-treat analysis. 103 (57%) of 180 participants were female and 77 (43%) were male and 90 (50%) participants had received at least one dose of a COVID-19 vaccine. SARS-CoV-2 infections with the delta (B.1.617.2; 72 [40%] of 180), alpha (B.1.1.7; 37 [21%]), omicron (B.1.1.529; 38 [21%]), and EU1 (B.1.177; 28 [16%]) variants were represented. All 180 participants received at least one dose of treatment and four participants discontinued the study (one in the molnupiravir group and three in the placebo group). Participants in the molnupiravir group had a faster median time from randomisation to negative PCR (8 days [95% CI 8–9]) than participants in the placebo group (11 days [10–11]; HR 1·30, 95% credible interval 0·92–1·71; log-rank p=0·074). The probability of molnupiravir being superior to placebo (HR>1) was 75·4%, which was less than our threshold of 80%. 73 (81%) of 90 participants in the molnupiravir group and 68 (76%) of 90 participants in the placebo group had at least one adverse event by day 29. One participant in the molnupiravir group and three participants in the placebo group had an adverse event of a Common Terminology Criteria for Adverse Events grade 3 or higher severity. No participants died (due to any cause) during the trial.
Interpretation
We found molnupiravir to be well tolerated and, although our predefined threshold was not reached, we observed some evidence that molnupiravir has antiviral activity in vaccinated and unvaccinated individuals infected with a broad range of SARS-CoV-2 variants, although this evidence is not conclusive.
Funding
Ridgeback Biotherapeutics, the UK National Institute for Health and Care Research, the Medical Research Council, and the Wellcome Trust.
Research in context.
Evidence before this study
Molnupiravir was the first orally administered direct-acting antiviral for the treatment of SARS-CoV-2 infection, which gained conditional marketing authorisation from the UK Medicines and Healthcare products Regulatory Agency in November, 2021, and early use authorisation from the US Food and Drug Administration in December, 2021. These approvals were based on the interim analysis of the MOVe-Out study, in which 775 unvaccinated adults at high risk of developing severe COVID-19 were randomly assigned to receive 5 days of molnupiravir or placebo; molnupiravir was associated with a significant reduction in hospitalisations and deaths. We searched PubMed for articles published in English between database inception and Aug 3, 2022, using the search terms “SARS-CoV-2” AND “randomised trial” AND “molnupiravir”. A phase 2a trial (NCT04405570) with 204 participants reported faster viral clearance with molnupiravir compared with placebo, but only 53 participants received the currently approved dose of molnupiravir. The full dataset from MOVe-Out (including all 1433 participants) showed an absolute difference in hospitalisations and deaths of 3·0 percentage points (compared with 6·8 percentage points at the interim analysis) with molnupiravir versus placebo. Both these studies did not include vaccinated participants and were done before the omicron variant predominated. Preliminary data from India suggested that open-label, generic molnupiravir plus standard of care (antipyretics, ivermectin, and budesonide) reduced the incidence of hospitalisation in 1218 adults with mild COVID-19 compared with standard of care alone; no details on variants or vaccination status were provided. There is a need to confirm these previous findings in vaccinated individuals infected with contemporary SARS-CoV-2 variants.
Added value of this study
We derive data from rich, serial, nasopharyngeal sampling within a stringent, randomised, placebo-controlled trial that has enabled differences in time to PCR negativity and viral titre for molnupiravir versus placebo to be evaluated. Molnupiravir was associated with a quicker time to PCR-negativity than placebo in a population comprising both vaccinated and unvaccinated individuals infected with a broad range of SARS-CoV-2 variants.
Implications of all the available evidence
We present results that do not contradict existing evidence, showing a moderate antiviral effect, without conclusive evidence, of molnupiravir. Definitive evidence for the clinical efficacy of molnupiravir in a highly vaccinated population is anticipated from the UK's PANORAMIC trial, which has included more than 25 000 participants and is due to report its results later in 2022.
Introduction
Molnupiravir (EIDD-2801 or MK-4482) is the first orally available direct-acting antiviral licensed for the treatment of individuals with mild-to-moderate COVID-19 at high risk for severe outcomes. Within AGILE, the UK early-phase platform for experimental COVID-19 therapies, we have previously reported on an optimal dose of molnupiravir for adults with documented SARS-CoV-2 infection: 800 mg every 12 h for 5 days.1 MOVe-OUT, a double-blind, placebo-controlled trial in 1433 unvaccinated adults with at least one risk factor for severe COVID-19, reported that molnupiravir decreased clinical progression, as judged by hospitalisations and death; the risk of hospitalisation or death at day 29 was 6·8 percentage points lower with molnupiravir than with placebo at the interim analysis and 3·0 percentage points lower in the all-randomised analysis.2 Preliminary data presented at the 2022 Conference on Retroviruses and Opportunistic Infections from an open-label study from India of 1218 adults with mild COVID-19 also reported that a generic formulation of molnupiravir significantly reduced hospitalisations compared with standard of care (nine [1·5%] of 608 vs 26 [4·3%] of 610).3 Compared with standard of care, molnupiravir was also associated with a significantly higher rate of SARS-CoV-2 PCR negativity after 5 days of treatment (77·1% vs 29·3%) and at days 10 (91·3% vs 70·2%) and 14 (93·9% vs 89·0%). Details on the vaccination status of participants were not provided. These studies have shown that molnupiravir is generally well tolerated and its longer-term safety continues to be monitored via ongoing clinical studies and pharmacovigilance programmes. Molnupiravir is a prodrug and is rapidly hydrolysed to N-hydroxycytidine. Although N-hydroxycytidine did return a positive result in a bacterial reverse mutation assay (an Ames test), extensive study of molnupiravir in in-vivo whole-animal mutagenicity assays has not shown clinically significant genotoxicity.4, 5
The shifting epidemiology of SARS-CoV-2 variants globally, with the high prevalence of the omicron (B.1.1.529) BA.2 lineage and other subsequent omicron lineages, gives cause for concern, particularly with the anticipated loss of clinical effect of many monoclonal antibodies. Although direct-acting antivirals are expected to remain effective, confirmation of their continued efficacy against emerging variants is required. The AGILE platform undertook a seamless phase 1b/2a evaluation of molnupiravir in the UK by use of a Bayesian adaptive design.6 We then aimed to evaluate the safety and virological efficacy of molnupiravir in a phase 2 study among vaccinated and unvaccinated participants.
Methods
Study design and participants
This randomised, placebo-controlled, double-blind, phase 2 trial (AGILE CST-2), was done at five UK National Institute for Health and Care Research (NIHR) Clinical Research Facility sites (in Liverpool, Manchester, Lancashire, Southampton, and London), coordinated by the NIHR Southampton Clinical Trials Unit, and sponsored by the University of Liverpool. Eligible participants were male and female outpatients aged at least 18 years with PCR-confirmed, mild-to-moderate (ambulant with peripheral capillary oxygen saturation >94% on room air) SARS-CoV-2 infection who were within 5 days of symptom onset, in generally good health, and free of uncontrolled chronic conditions. Individuals who had tested positive for SARS-CoV-2 infection (or who had symptoms suggestive of COVID-19) were contacted by telephone to seek consent and assess eligibility. Women of childbearing potential and men who were sexually active with women of childbearing potential were required to use two effective methods of contraception, one of which needed to be highly effective. Women were required to use contraception throughout the study and for up to 50 days after the last follow-up visit and men were required to use contraception throughout the study and for up to 100 days after the last follow-up visit. Participants were eligible irrespective of whether they were unvaccinated or had received one or multiple UK-approved vaccines. Any of the following criteria excluded participants from the study: pregnant or breastfeeding women; stage 4 or stage 5 (severe) chronic kidney disease or a requirement for dialysis; clinically significant liver dysfunction or renal impairment; a peripheral capillary oxygen saturation of less than 95% by oximetry or a lung disease that requires supplementary oxygen; an alanine aminotransferase concentration, an aspartate aminotransferase concentration, or both of more than five-times the upper limit of normal; a platelet count of less than 50 × 109 platelets per L; any grade 3 or higher (on the Common Terminology Criteria for Adverse Events [CTCAE] version 5) adverse event; previously reported hepatitis C virus infection or concurrent bacterial pneumonia; known allergy to any study medication; having received any other experimental agents within 30 days of the first dose of study drug (the use of other comedications was allowed); having taken other prohibited drugs within 30 days or five-times the half-life of enrolment (whichever was longer); participation in another trial of an investigational medicinal product; the presence of a febrile respiratory illness that included pneumonia and resulted in hospitalisation or required hospitalisation, oxygenation, mechanical ventilation, or other supportive modalities; and the presence of clinically significant end-organ disease as a result of relevant comorbidities or any condition that would put the patient at increased risk, in the opinion of the investigator.
All participants provided written, informed consent before enrolment. The study protocol was reviewed and approved by the UK Medicines and Healthcare products Regulatory Agency (EudraCT 2020-001860-27). Ethical approval was received from the Health Research Authority West Midlands—Edgbaston Research Ethics Committee (20/WM/0136). The protocol is published as appendix 1.
Randomisation and masking
Using a permuted block (block size 2 or 4) method and stratifying by site, participants were randomly assigned (1:1) to receive either molnupiravir plus standard of care or placebo plus standard of care. The randomisation sequence was generated by use of STATA (version 16) by an independent statistician (who had no further involvement in the trial) and used to prepare labelled placebo and treatment packs, which were assigned sequentially to patients on randomisation. Placebo and molnupiravir were provided in tablets of identical appearance. Participants, the staff giving and assessing the interventions, and those who analysed the data were masked to treatment allocation until the end of the study.
Procedures
Screening evaluations to determine eligibility included medical history, SARS-CoV-2 diagnostic nasopharyngeal swab, the WHO Clinical Progression Scale,7 the National Early Warning Score 2 (NEWS2), a 12-lead electrocardiogram, current use of supplementary oxygen, and blood draws for laboratory investigations.
Molnupiravir and matching placebo were provided by Ridgeback Biotherapeutics as 200 mg capsules and were administered orally at 800 mg twice daily (every 12 h; in the morning and evening with water) for 5 days on days 1–5 (or days 1–6 if the first dose was given in the evening), totalling ten 800 mg doses. Participants were required to fast for at least 2 h before dose administration and for 1 h after administration. Standard of care involved symptomatic relief, including antipyretics. Participants received the first dose of the study drug in the clinic on day 1 and underwent a 4-h period of observation before they were sent home with the remaining doses for self-administration. Participants returned to the clinic on days 3, 5, and 8, bringing their study medication with them for drug accountability. Because no specific clinical drug–drug interaction data were available at the time of this trial, the investigators were permitted to apply discretion regarding the use of concomitant medications, guided by the Liverpool COVID-19 Drug Interactions tool.
On days 1, 3, 5, 8, 11, 15, 22, and 29, data were collected from the measurement of NEWS2, clinical examinations, the WHO Clinical Progression Scale, and the assessment of oxygen use and mechanical ventilation. Baseline and follow-up laboratory assessments (eg, SARS-CoV-2 nasopharyngeal swabs) were done on days 1, 5, 11, 15, 22, and 29 from blood samples and included urea and electrolytes, full blood count, liver function tests, and estimated glomerular filtration rate. The Influenza Patient-Reported Outcome (FLU-PRO) questionnaire and viral titres by PCR from SARS-CoV-2 serial surveillance nasopharyngeal swabs were done at each visit (at screening and again [if the visits were separate] at baseline [day 1], and then days 3, 5, 8, 11, 15, 22, and 29). The swabs were sampled from the oropharynx and then the nasopharyngeal space and collected in DNA/RNA shield solution (Zymo Research; Irvine, CA, USA). Viral RNA was extracted from samples by use of the Maxwell RSC Viral Total Nucleic Acid Purification Kit (number AS1330; Promega; Madison, WI, USA) according to the manufacturer's instructions. PCR was done (blinded to treatment allocation) by use of the TaqPath COVID-19 RT-PCR Kit (ThermoFisher Scientific; Waltham, MA, USA), with readings comprising three amplicons: the spike gene, the nucleocapsid gene, and ORF1 (cycle thresholds were adjusted for each amplicon on each analysis to give a cycle threshold of 32 with a control of 25 templates per reaction).
Viral titre was quantified from the nasopharyngeal swabs. Because approved quantitative standards were not yet commercially available, we developed in-house quantitation based on estimating a viral pseudoconcentration (expressed as copies of template per reaction). Swabs dipped into a culture containing 1 × 107 plaque-forming units of a primary SARS-CoV-2 isolate from Liverpool (Pango lineage B; REMRQ0001/Human/2020/Liverpool) were serially diluted to produce a calibration curve. The limit of quantitation (published by the kit manufacturer) was 25 templates per reaction, and a control known to contain 25 copies per reaction was used to adjust the thresholds on all three templates (spike gene, nucleocapsid gene, and ORF1) to yield a cycle threshold of 32 (we did not compare the effects of variants, but instead recalibrated all samples to ensure that a readout of 25 templates per reaction was equivalent to a cycle threshold of 32). Exponential regression was then done on each calibration curve, giving three different coefficients (these coefficients were checked periodically for consistency), which were used to estimate a fold change (from the 25 copies per reaction estimate) for any threshold cycle. The mean of estimated titres across the three genes (spike gene, nucleocapsid gene, and ORF1), where available, was calculated and then transformed into log10 values. The change in SARS-CoV-2 viral load in the nasopharyngeal swabs was measured by subtracting the log10 estimated titre from the baseline titre.
For the typing of variants, viral RNA from the baseline nasopharyngeal swabs was reverse transcribed and then sequenced by use of the EasySeq RC-PCR SARS-CoV-2 whole genome sequencing kit (NimaGen; Nijmegen, the Netherlands). Sequence reads were cleaned, trimmed, and mapped to the SARS-CoV-2 Wuhan-Hu-1 reference genome (NC_045512.2).8 For each sample, genomic variants were called and filtered by quality, with high-quality variant calls being used to generate the consensus genome sequence for each sample. The consensus genome sequence was then processed by use of Pangolin,9 a widely used computational tool that assigns the most likely lineage to a given SARS-CoV-2 genome sequence according to the Pango dynamic lineage nomenclature scheme.
Concomitant medications were checked at all visits. Major protocol deviations related to the FLU-PRO questionnaire not being fully completed by 15 patients, adverse events not being identified from the FLU-PRO questionnaire, and one patient underdosing their home-administered doses.
Outcomes
The primary endpoint was the time from randomisation to a negative SARS-CoV-2 PCR test. Secondary endpoints were: the 11-point WHO Clinical Progression Scale for COVID-19 at days 15 and 29;7 the 32-item NEWS2 score (UK Royal College of Physicians, 2017), measuring acute illness, at days 15 and 29; the FLU-PRO (version 1.2) questionnaire of the presence and severity of influenza-like symptoms across six domains (nose, throat, eyes, chest and respiratory, gastrointestinal, and body and system) at days 15 and 29; overall survival (time-to-event; from randomisation to death due to any cause, with those still alive censored at the last time known to be alive); mortality at days 15 and 29; time to hospital admission; hospitalisation rates at days 15 and 29; duration of mechanical ventilation (including the incidence and duration of new mechanical ventilation use); duration of oxygen use; oxygen-free days; the incidence of a peripheral capillary oxygen saturation of less than 92% (based on at least two consecutive recordings on the same day, lasting at least 1 day) by day 29; treatment compliance; and safety. Safety was evaluated by use of CTCAE (version 5), with real-time serious adverse event reporting. Adverse events of CTCAE grade 1–2 were categorised as mild and adverse events of a CTCAE grade of 3 or more were defined as severe. Data on dose-limiting toxicities (defined as any adverse event of a CTCAE [version 5] grade ≥3 during the first 7 days) were recorded to support the findings of the previous phase 1 trial. Our prespecified exploratory virological outcome was the change in viral titre from day 1.
Statistical analysis
The sample size was based on time-to-PCR-negativity (censored at 29 days), comparing molnupiravir with placebo. One formal interim analysis (with the independent data monitoring and ethics committee) was scheduled after 60 participants were enrolled to evaluate futility or efficacy; another interim analysis was added after 120 participants. We used a Bayesian adaptive approach to accelerate decision making, which was based on a hazard ratio (HR) of having PCR negativity with molnupiravir compared with placebo. Our primary model used a two-point prior based on equal prior probabilities (50%) that the HR was 1·0 (ie, no effect) or 1·5 (the threshold of effect judged to be clinically important). We defined a priori that if the probability of a HR of more than 1 was in excess of 80% molnupiravir would be recommended for further testing in a larger definitive study. If the probability was less than 0·3 at either interim analysis, the study would stop for futility. The maximum sample size of 180 was selected to ensure that the overall probability of concluding that molnupiravir was better than placebo was 0·1 when the HR was 1·0 (one-sided type I error accounting for one formal interim analysis) and that the power to recommend molnupiravir for further testing was approximately 0·77 if the HR was 1·5 (equivalent to decreasing the median time to viral clearance from 14·0 days to 9·3 days or increasing viral clearance after 28 days from 75·0% to 87·5% with molnupiravir). In addition to the two-point prior, we also ran a sensitivity analysis in which we used a continuous (uninformative) prior to estimate the probability that the HR was greater than 1 and construct 95% credible intervals (CrIs). Full details of this methodology are available.10
All trial endpoints were prespecified in the finalised and signed statistical analysis plan, including the exploratory endpoint, before final analysis and database lock. All analyses were done in the intention-to-treat population (comprising all people who were randomly assigned and had relevant data), apart from the safety analysis, which was done in the safety population comprising only participants who had received at least one dose of the allocated treatment. There was no imputation of missing data, data transformations, or adjustment for multiplicity for any of the analyses and results are presented with two-sided p values and 95% CIs or 95% CrIs (for the Bayesian analyses), unless otherwise stated. The primary and secondary analyses were done after all participants had been followed up until day 29.
The phase 2 primary analysis involved the comparison of groups on time to viral clearance by use of a Bayesian Cox proportional hazards model. Time of negativity within an amplicon was determined by the time of the first of two consecutive readings of less than the limit of detection (cycle threshold ≥32) when at least two amplicons were concordant. If all three amplicons differed in negativity status, the median time to negative PCR was used. If only two amplicons were evaluable (eg, if the third was censored), the later time of the two was used. When only one amplicon was evaluable, time to negative PCR was censored at the last PCR measurement. In the event of spike gene amplification failure, the spike gene was considered censored at day 29 and the same rules applied.
For the main analysis from day 1 to day 5 and prespecified sensitivity analyses from day 1 to day 3 or day 8, we compared mean reductions in viral loads between groups using a Student's t test. We analysed viral load reduction by vaccination status. We evaluated the pattern of viral elimination (confirmed as the mean value of at least two concordant amplicons), with patients categorised into one of four groups: (1) viral clearance (stable trajectory of viral load decline to less than the limit of quantitation); (2) transient increase in viral titre (following a viral load reduction, a subsequent increase in viral titre of at least 0·5 log10 copies per reaction and to a titre that was maintained or increased at the next consecutive sample); (3) indeterminate (following a viral load reduction, a subsequent increase in titre that was not confirmed in the next consecutive sample), and (4) non-evaluable.
Time-to-event data are presented as Kaplan–Meier curves, with secondary analyses comparing treatment groups by use of simple, unadjusted Cox regression models. Descriptive analyses of baseline characteristics and other endpoints are summarised by use of means, medians (from Kaplan–Meier curves for time-to-event data), and proportions, with corresponding SDs, IQRs, and 95% CIs, as appropriate. Statistical testing for differences between groups used two non-parametric evaluations. Initially, log-rank testing was specified but a review by the independent statistical expert in our data monitoring and ethics committee on Nov 8, 2021, led to the recommendation of including the Breslow–Gehan–Wilcoxon test as a more sensitive discriminator of differences at early timepoints, which are anticipated with antiviral therapy. Exploratory prespecified subgroup analyses of the primary outcome were done, grouping by SARS-CoV-2 variant, vaccination status, ethnicity, and sex.
All analyses are reported according to Consolidated Standards of Reporting Trials 2010 and International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use E9 guidelines on statistical principles in clinical trials. All analyses were done in SAS (version 9.4) and Stata (version 16), except the Bayesian analyses, which were done by use of packages available in R (version 4.0.2). This trial is registered in ClinicalTrials.gov, NCT04746183, and the ISRCTN registry, ISRCTN27106947.
Role of the funding source
Employees of Ridgeback Biotherapeutics, including those listed as authors, contributed to the development and implementation of this trial. The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
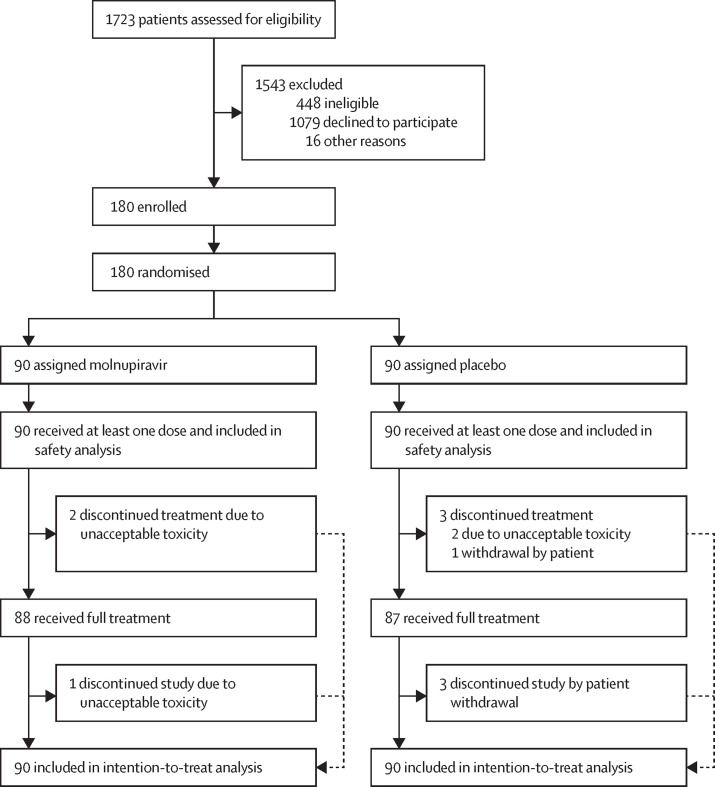
Between Nov 18, 2020, and March 16, 2022, 1723 patients were assessed for eligibility, of whom 180 were randomly assigned to receive either molnupiravir (n=90) or placebo (n=90) and were included in the intention-to-treat analysis (figure 1 ). Participants' baseline characteristics were similar across the molnupiravir and placebo groups (table 1 ; appendix 2 pp 2–5). Overall, the median age was 43 years (IQR 28–55), 103 (57%) of 180 participants were female and 77 (43%) were male, all 180 participants had a WHO Clinical Progression Scale score of 2 (ambulant and independent, with mild, symptomatic disease) on day 1, and 90 (50%) participants had received at least one dose of a COVID-19 vaccine at least 14 days before entry into the trial (table 1). The overall median time from symptom onset to randomisation was 3·0 days (IQR 3·0–4·0) and the delta (B.1.617.2), alpha (B.1.1.7), omicron, EU1 (B.1.177), and XE (a recombinant variant of BA.1 and BA.2) variants were represented among the SARS-CoV-2 infections (table 1).
Figure 1.
Trial profile
Table 1.
Baseline characteristics in the intention-to-treat population
| Molnupiravir (n=90) | Placebo (n=90) | Total (n=180) | ||
|---|---|---|---|---|
| Demographics | ||||
| Age at consent, years | 45 (31–55) | 43 (28–54) | 43 (28–55) | |
| Sex | ||||
| Female | 52 (58%) | 51 (57%) | 103 (57%) | |
| Male | 38 (42%) | 39 (43%) | 77 (43%) | |
| Ethnicity | ||||
| White English, Welsh, Scottish, Northern Irish, or British | 73 (81%) | 78 (87%) | 151 (84%) | |
| Any other White background | 13 (14%) | 7 (8%) | 20 (11%) | |
| Asian or British Asian: Indian | 1 (1%) | 1 (1%) | 2 (1%) | |
| Mixed or part of multiple ethnic groups: White and Black African | 1 (1%) | 0 | 1 (1%) | |
| Mixed or part of multiple ethnic groups: White and Asian | 0 | 1 (1%) | 1 (1%) | |
| Black, African, Caribbean, or Black British–Caribbean | 1 (1%) | 0 | 1 (1%) | |
| Asian or British Asian: Pakistani | 0 | 1 (1%) | 1 (1%) | |
| Asian or British Asian: Chinese | 0 | 1 (1%) | 1 (1%) | |
| Any other Black, African, or Caribbean background | 1 (1%) | 0 | 1 (1%) | |
| Any other Asian background | 0 | 1 (1%) | 1 (1%) | |
| Body-mass index, kg/m2 | 28·2 (24·2–32·2) | 27·1 (23·6–31·6) | 27·4 (24·0–32·0) | |
| Disease characteristics | ||||
| Time from symptom onset to randomisation, days | 3·5 (3·0–4·0) | 3·0 (3·0–4·0) | 3·0 (3·0–4·0) | |
| National Early Warning Score 2 | 0·3 (0·6) | 0·4 (0·8) | 0·3 (0·7) | |
| SARS-CoV-2 variant | ||||
| Alpha (B.1.1.7) | 17 (19%) | 20 (22%) | 37 (21%) | |
| B.1.1.1 | 0 | 1 (1%) | 1 (1%) | |
| EU1 (B.1.177) | 15 (17%) | 13 (14%) | 28 (16%) | |
| Omicron (B.1.1.529) BA.1 | 15 (17%) | 12 (13%) | 27 (15%) | |
| Omicron (B.1.1.529) BA.2 | 5 (6%) | 6 (7%) | 11 (6%) | |
| Delta (B.1.617.2) | 37 (41%) | 35 (39%) | 72 (40%) | |
| XE | 0 | 1 (1%) | 1 (1%) | |
| Unknown | 1 (1%) | 2 (2%) | 3 (2%) | |
| Viral load | ||||
| n | 89 | 89 | 178 | |
| Mean, log10copies per reaction | 7·1 (2·7) | 7·4 (3·0) | 7·2 (2·9) | |
| Vaccinated against COVID-19* | ||||
| No | 46 (51%) | 44 (49%) | 90 (50%) | |
| Yes | 44 (49%) | 46 (51%) | 90 (50%) | |
| First vaccine received | ||||
| Pfizer–BioNTech (BNT162b2) | 21/44 (48%) | 26/46 (57%) | 47/90 (52%) | |
| AstraZeneca (ChAdOx1 nCoV-19) | 21/44 (48%) | 17/46 (37%) | 38/90 (42%) | |
| Novavax (NVX-CoV2373) | 1/44 (2%) | 0 | 1/90 (1%) | |
| Unknown | 1/44 (2%) | 3/46 (7%) | 4/90 (4%) | |
| Second vaccine received | ||||
| Pfizer–BioNTech (BNT162b2) | 15/35 (43%) | 22/34 (65%) | 37/69 (54%) | |
| AstraZeneca (ChAdOx1 nCoV-19) | 17/35 (49%) | 11/34 (32%) | 28/69 (41%) | |
| Novavax (NVX-CoV2373) | 1/35 (3%) | 0 | 1/69 (1%) | |
| Unknown | 2/35 (6%) | 1/34 (3%) | 3/69 (4%) | |
| Third vaccine received | ||||
| Pfizer–BioNTech (BNT162b2) | 12/12 (100%) | 6/7 (86%) | 18/19 (95%) | |
| Moderna (mRNA-1273) | 0 | 1/7 (14%) | 1/19 (5%) | |
Data are median (IQR), n (%), mean (SD), or n/N, unless otherwise specified.
A patient was deemed vaccinated if they had received at least one COVID-19 vaccine at least 14 days before entry into the trial.
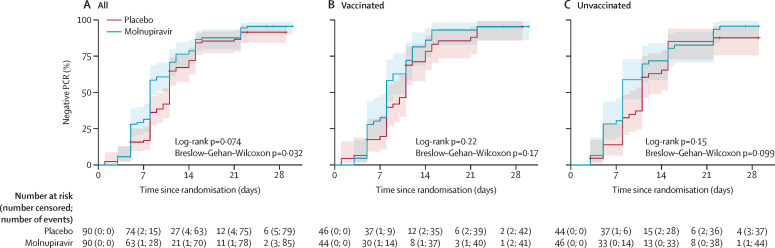
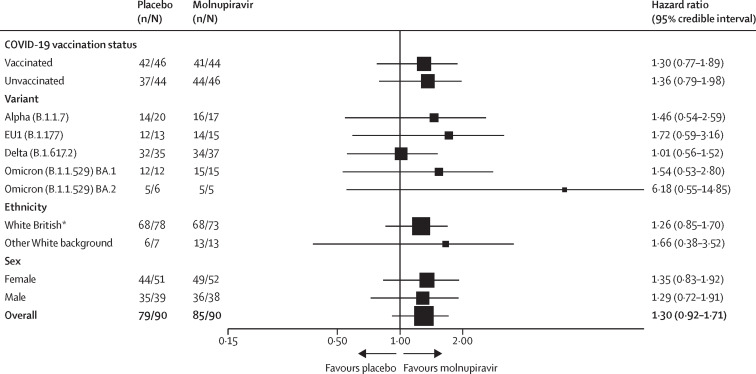
Participants in the molnupiravir group had a faster median time from randomisation to PCR negativity (8 days [95% CI 8–9]) than participants in the placebo group (11 days [10–11]; log-rank p=0·074; Breslow–Gehan–Wilcoxon p=0·032; table 2 ; figure 2 ). The Bayesian Cox proportional hazards model based on a two-point prior gave a probability of 75·4% for the HR being more than 1 (75·9% at the interim analysis for the first 60 participants and 53·5% at the interim analysis for the first 120 participants), which was less than the 80% threshold for recommending a candidate for large-scale evaluation. However, our sensitivity analysis with non-informative, continuous priors gave a corresponding probability of the HR being greater than 1 of 94·7%, with an estimated HR of 1·30 (95% CrI 0·92–1·71; appendix 2 p 9). Our exploratory analysis of time to negative PCR by vaccination status, SARS-CoV-2 variant, sex, and ethnicity is shown in figure 3 . The numbers of participants in each subgroup were too small for statistical evaluation, but HRs were similar among vaccinated versus unvaccinated participants, with a nominally (non-significant) greater effect in unvaccinated participants (figure 3). By day 29, all but nine participants had negative PCR results (seven participants did not have a day 29 PCR test result but had previously tested negative). Eight of these nine participants had discordant results across the three genes amplified (or two genes, for the four participants with spike gene amplification failure), and only one participant had all three genes detectable at this timepoint.
Table 2.
Time from randomisation to negative PCR in the intention-to-treat population
| Molnupiravir (n=90) | Placebo (n=90) | ||
|---|---|---|---|
| Median time from randomisation to negative PCR (95% CI), days | |||
| All | 8 (8–9) | 11 (10–11) | |
| Alpha (B.1.1.7) | 11 (5–22); n=17 | 11 (11–15); n=20 | |
| EU1 (B.1.177) | 8 (5–8); n=15 | 11 (8–15); n=13 | |
| Omicron (B.1.1.529) BA.1 | 8 (5–8); n=15 | 11 (5–14); n=12 | |
| Omicron (B.1.1.529) BA.2 | 8 (3–NE); n=5 | 15 (7–NE); n=6 | |
| Delta (B.1.617.2) | 11 (8–12); n=37 | 10 (8–11); n=35 | |
| Proportion with negative PCR test | |||
| Day 15 | |||
| Actual number | 77 (86%) | 73 (81%) | |
| Kaplan–Meier estimate (95% CI) | 86·5% (78·6–92·6) | 84·2% (75·6–91·0) | |
| Day 29 | |||
| Actual number | 85 (94%) | 79 (88%) | |
| Kaplan–Meier estimate (95% CI) | 95·5% (89·7–98·5) | 91·5% (84·3–96·2) | |
Data are n (%), unless otherwise specified. NE=not estimable.
Figure 2.
Kaplan–Meier plots for time from randomisation to negative PCR
(A) All patients. (B) Vaccinated patients. (C) Unvaccinated patients. Shaded areas represent 95% CIs.
Figure 3.
Forest plot for time from randomisation to negative PCR by subgroup
For our analysis by variant, we did not include patients with an unknown or unique (B.1.1.1 and XE) variant type. *People could identify as White English, Welsh, Scottish, Northern Irish, or British.
No participants had a peripheral capillary oxygen saturation of less than 92%. No participants in the molnupiravir group were hospitalised. Four participants in the placebo group were hospitalised, one of whom received 2 days of oxygen (the only patient in the trial who received any oxygen). Three (3%) of 90 people in the placebo group were hospitalised at day 15 and four (4%) people in the placebo group were hospitalised at day 29. For time to hospitalisation, group medians and a HR could not be calculated due to the low number of events. The median number of oxygen-free days for each group was 29 days (IQR 29–29). No patient required mechanical ventilation of any kind. The WHO Clinical Progression Scale score, NEWS2, and the FLU-PRO overall score were similar in each group at day 15 and day 29, with 73 (42%) of 172 participants having a WHO score of 0 or 1 at day 15 and 122 (71%) of 173 participants having a WHO score of 0 or 1 at day 29 (appendix 2 pp 6–7). NEWS2 (mean 0·3 [SD 0·7] at day 15 and 0·3 [0·6] at day 29) and FLU-PRO scores (mean 0·2 [SD 0·3] at day 15 and 0·1 [0·2] at day 29) were very low at days 15 and 29 (appendix 2 pp 6–7). All 180 participants received at least one dose of treatment, with 88 (98%) of 90 fully completing molnupiravir treatment and 87 (97%) of 90 fully completing placebo treatment (figure 1). Both groups received a median of ten doses (IQR 10–10) during a median of 5 days (5–6) on treatment. Five (3%) of 180 participants ended treatment early; two participants in each group ended treatment early due to adverse events (nausea [n=1] and vomiting [n=1] in the placebo group; hypertension [n=1] and no specified side-effect [n=1] in the molnupiravir group) and one participant in the placebo group withdrew from treatment. No participants died (due to any cause) during the trial.
73 (81%) of 90 participants in the molnupiravir group and 68 (76%) of 90 participants in the placebo group had at least one adverse event (at ≥grade 1) by day 29 (table 3 ). One person had a dose-limiting toxicity (hypertension) in the molnupiravir group and three people had dose-limiting toxicities in the placebo group (hypomagnesaemia and hypocalcaemia [n=1]; nausea and vomiting [n=1]; and gallstone pancreatitis, increased alanine aminotransferase, increased blood bilirubin, and increased γ-glutamyl transferase [n=1]) in the placebo group. There were 200 adverse events (199 grade 1–2 and one grade ≥3) in the molnupiravir group and 219 adverse events (211 grade 1–2 and eight grade ≥3) in the placebo group. One participant in the molnupiravir group (grade 3 hypertension) and three participants in the placebo group (eight events; grade 3 vomiting [n=1], grade 3 nausea [n=1], grade 3 gallstone pancreatitis [n=1], a grade 3 increase in blood bilirubin concentration [n=1], a grade 3 increase in alanine aminotransferase concentration [n=1], grade 3 hypocalcaemia [n=1], a grade 3 increase in γ-glutamyl transferase [n=1], and grade 4 hypomagnesaemia [n=1]) had an adverse event of a grade 3 or higher severity. Two of these four participants were vaccinated and two were unvaccinated. Four (4%) of 90 participants (three unvaccinated and one who had received two doses of the Pfizer–BioNTech BNT162b2 vaccine) in the placebo group had serious adverse events (gallstone pancreatitis [n=1], vomiting [n=1], hypocalcaemia and hypomagnesaemia [n=1], and breathlessness requiring oxygen therapy [n=1]), all of which led to hospitalisation. No patients in the molnupiravir group had a serious adverse event.
Table 3.
Adverse events by system organ class in the safety population
| Molnupiravir (n=90) | Placebo (n=90) | Total (n=180) | ||
|---|---|---|---|---|
| At least one adverse event | 73 (81%) | 68 (76%) | 141 (78%) | |
| Blood and lymphatic system disorders | 1 (1%) | 2 (2%) | 3 (2%) | |
| Anaemia | 0 | 1 (1%) | 1 (1%) | |
| Left axillary painful lymph node with no swelling noted | 0 | 1 (1%) | 1 (1%) | |
| Lymphopenia | 1 (1%) | 0 | 1 (1%) | |
| Right axillary lymph node painless swelling | 0 | 1 (1%) | 1 (1%) | |
| Cardiac disorders | 5 (6%) | 1 (1%) | 6 (3%) | |
| Cardiac chest pain | 2 (2%) | 0 | 2 (1%) | |
| Chest tightness | 2 (2%) | 0 | 2 (1%) | |
| Palpitations | 1 (1%) | 1 (1%) | 2 (1%) | |
| Sinus bradycardia | 1 (1%) | 0 | 1 (1%) | |
| Ear and labyrinth disorders | 3 (3%) | 1 (1%) | 4 (2%) | |
| Ear pain | 2 (2%) | 0 | 2 (1%) | |
| Ear and labyrinth disorder (itching left ear) | 1 (1%) | 0 | 1 (1%) | |
| Vertigo | 0 | 1 (1%) | 1 (1%) | |
| Eye disorders | 4 (4%) | 6 (7%) | 10 (6%) | |
| Blurred vision | 2 (2%) | 2 (2%) | 4 (2%) | |
| Eye pain | 0 | 1 (1%) | 1 (1%) | |
| Erythema on right eyelid | 0 | 1 (1%) | 1 (1%) | |
| Eyes sensitive to light | 1 (1%) | 0 | 1 (1%) | |
| Itchy eyes | 1 (1%) | 0 | 1 (1%) | |
| Photophobia | 0 | 1 (1%) | 1 (1%) | |
| Watering eyes | 0 | 2 (2%) | 2 (1%) | |
| Gastrointestinal disorders | 27 (30%) | 37 (41%) | 64 (36%) | |
| Abdominal pain | 6 (7%) | 7 (8%) | 13 (7%) | |
| Bloating | 0 | 1 (1%) | 1 (1%) | |
| Constipation | 1 (1%) | 0 | 1 (1%) | |
| Diarrhoea | 9 (10%) | 16 (18%) | 25 (14%) | |
| Dry mouth | 1 (1%) | 1 (1%) | 2 (1%) | |
| Dyspepsia | 0 | 1 (1%) | 1 (1%) | |
| Dysphagia | 0 | 3 (3%) | 3 (2%) | |
| Flatulence | 1 (1%) | 0 | 1 (1%) | |
| Gastro-oesophageal reflux disease | 0 | 1 (1%) | 1 (1%) | |
| Gingival pain | 1 (1%) | 0 | 1 (1%) | |
| Nausea | 6 (7%) | 14 (16%) | 20 (11%) | |
| Abdominal cramping and loose stools | 0 | 1 (1%) | 1 (1%) | |
| Burning in throat | 0 | 1 (1%) | 1 (1%) | |
| Gallstone pancreatitis | 0 | 1 (1%) | 1 (1%) | |
| Indigestion | 0 | 1 (1%) | 1 (1%) | |
| Loose stools | 1 (1%) | 1 (1%) | 2 (1%) | |
| Tongue pain | 1 (1%) | 0 | 1 (1%) | |
| Stomach pain | 3 (3%) | 3 (3%) | 6 (3%) | |
| Vomiting | 2 (2%) | 9 (10%) | 11 (6%) | |
| General disorders and administration site conditions | 10 (11%) | 13 (14%) | 23 (13%) | |
| Chills | 2 (2%) | 3 (3%) | 5 (3%) | |
| Face oedema | 0 | 1 (1%) | 1 (1%) | |
| Fatigue | 2 (2%) | 2 (2%) | 4 (2%) | |
| Fever | 1 (1%) | 1 (1%) | 2 (1%) | |
| Influenza-like symptoms | 0 | 1 (1%) | 1 (1%) | |
| Non-cardiac chest pain | 5 (6%) | 5 (6%) | 10 (6%) | |
| Immune system disorders | 2 (2%) | 1 (1%) | 3 (2%) | |
| Allergic reaction | 0 | 1 (1%) | 1 (1%) | |
| Exacerbation of allergic rhinitis | 1 (1%) | 0 | 1 (1%) | |
| Tonsillitis | 1 (1%) | 0 | 1 (1%) | |
| Infections and infestations | 4 (4%) | 6 (7%) | 10 (6%) | |
| Conjunctivitis | 0 | 1 (1%) | 1 (1%) | |
| Lung infection | 0 | 2 (2%) | 2 (1%) | |
| Cold sore | 0 | 1 (1%) | 1 (1%) | |
| Upper respiratory tract infection | 1 (1%) | 0 | 1 (1%) | |
| Otitis externa | 1 (1%) | 1 (1%) | 2 (1%) | |
| Infective rhinitis | 0 | 1 (1%) | 1 (1%) | |
| Skin infection | 1 (1%) | 0 | 1 (1%) | |
| Tooth infection | 1 (1%) | 0 | 1 (1%) | |
| Urinary tract infection | 0 | 1 (1%) | 1 (1%) | |
| Injury, poisoning, and procedural complications | 2 (2%) | 0 | 2 (1%) | |
| Fall | 1 (1%) | 0 | 1 (1%) | |
| Sunburn | 1 (1%) | 0 | 1 (1%) | |
| Investigations | 4 (4%) | 4 (4%) | 8 (4%) | |
| Increased alanine aminotransferase | 2 (2%) | 1 (1%) | 3 (2%) | |
| Increased alkaline phosphatase | 0 | 1 (1%) | 1 (1%) | |
| Increased blood bilirubin | 1 (1%) | 1 (1%) | 2 (1%) | |
| Increased creatinine | 1 (1%) | 0 | 1 (1%) | |
| Prolonged heart-rate corrected QT interval | 0 | 1 (1%) | 1 (1%) | |
| Increased γ-glutamyl transferase | 1 (1%) | 2 (2%) | 3 (2%) | |
| Decreased neutrophil count | 1 (1%) | 1 (1%) | 2 (1%) | |
| Metabolism and nutrition disorders | 3 (3%) | 1 (1%) | 4 (2%) | |
| Anorexia | 1 (1%) | 0 | 1 (1%) | |
| Hypocalcaemia | 0 | 1 (1%) | 1 (1%) | |
| Hypokalaemia | 0 | 1 (1%) | 1 (1%) | |
| Hypomagnesaemia | 0 | 1 (1%) | 1 (1%) | |
| Hypophosphataemia | 1 (1%) | 0 | 1 (1%) | |
| Loss of appetite | 1 (1%) | 0 | 1 (1%) | |
| Musculoskeletal and connective tissue disorders | 11 (12%) | 12 (13%) | 23 (13%) | |
| Arthralgia | 1 (1%) | 0 | 1 (1%) | |
| Arthritis | 0 | 1 (1%) | 1 (1%) | |
| Back pain | 1 (1%) | 1 (1%) | 2 (1%) | |
| Chest wall pain | 1 (1%) | 1 (1%) | 2 (1%) | |
| Flank pain | 1 (1%) | 0 | 1 (1%) | |
| Myalgia | 2 (2%) | 5 (6%) | 7 (4%) | |
| Neck pain | 1 (1%) | 0 | 1 (1%) | |
| Body aches and pains | 1 (1%) | 0 | 1 (1%) | |
| Knee pain | 1 (1%) | 0 | 1 (1%) | |
| Left shoulder pain | 0 | 1 (1%) | 1 (1%) | |
| Lower back pain | 1 (1%) | 0 | 1 (1%) | |
| Reduced power to right thumb | 0 | 1 (1%) | 1 (1%) | |
| Right foot pain | 1 (1%) | 0 | 1 (1%) | |
| Pain in arms or legs | 2 (2%) | 2 (2%) | 4 (2%) | |
| Nervous system disorders | 37 (41%) | 34 (38%) | 71 (39%) | |
| Anosmia | 5 (6%) | 5 (6%) | 10 (6%) | |
| Dizziness | 12 (13%) | 2 (2%) | 14 (8%) | |
| Dysgeusia | 5 (6%) | 2 (2%) | 7 (4%) | |
| Headache | 16 (18%) | 19 (21%) | 35 (19%) | |
| Hypersomnia | 3 (3%) | 0 | 3 (2%) | |
| Lethargy | 0 | 1 (1%) | 1 (1%) | |
| Ageusia | 0 | 1 (1%) | 1 (1%) | |
| Feeling light-headed and dizzy | 0 | 1 (1%) | 1 (1%) | |
| Feeling weakness in lower limbs | 0 | 1 (1%) | 1 (1%) | |
| Feeling hot | 0 | 1 (1%) | 1 (1%) | |
| Head congestion | 0 | 2 (2%) | 2 (1%) | |
| Light-headedness | 0 | 1 (1%) | 1 (1%) | |
| Loss of smell and loss of taste | 1 (1%) | 0 | 1 (1%) | |
| Numbness and dryness of third and fourth fingers on right hand | 1 (1%) | 0 | 1 (1%) | |
| Paresthesia | 2 (2%) | 2 (2%) | 4 (2%) | |
| Presyncope | 3 (3%) | 0 | 3 (2%) | |
| Syncope | 0 | 2 (2%) | 2 (1%) | |
| Tremor | 1 (1%) | 1 (1%) | 2 (1%) | |
| Renal and urinary disorders | 2 (2%) | 0 | 2 (1%) | |
| Haematuria | 1 (1%) | 0 | 1 (1%) | |
| Cystitis | 1 (1%) | 0 | 1 (1%) | |
| Increased urinary frequency | 1 (1%) | 0 | 1 (1%) | |
| Reproductive system and breast disorders | 4 (4%) | 3 (3%) | 7 (4%) | |
| Menorrhoea | 0 | 1 (1%) | 1 (1%) | |
| Vaginal bleeding (menses) | 0 | 1 (1%) | 1 (1%) | |
| Other, specify | 1 (1%) | 0 | 1 (1%) | |
| Vaginal discharge | 1 (1%) | 1 (1%) | 2 (1%) | |
| Vaginal haemorrhage | 2 (2%) | 0 | 2 (1%) | |
| Respiratory, thoracic, and mediastinal disorders | 29 (32%) | 28 (31%) | 57 (32%) | |
| Allergic rhinitis | 1 (1%) | 0 | 1 (1%) | |
| Cough | 3 (3%) | 7 (8%) | 10 (6%) | |
| Dyspnoea | 5 (6%) | 4 (4%) | 9 (5%) | |
| Hoarseness | 0 | 1 (1%) | 1 (1%) | |
| Nasal congestion | 3 (3%) | 3 (3%) | 6 (3%) | |
| Chest congestion | 2 (2%) | 3 (3%) | 5 (3%) | |
| Chest congestion or chest tightness | 0 | 1 (1%) | 1 (1%) | |
| Chest tightness | 4 (4%) | 4 (4%) | 8 (4%) | |
| Chest tightness and congestion | 1 (1%) | 0 | 1 (1%) | |
| Dry cough | 1 (1%) | 0 | 1 (1%) | |
| Increased breathlessness and chest tightness | 0 | 1 (1%) | 1 (1%) | |
| Pneumonia | 0 | 1 (1%) | 1 (1%) | |
| Scratchy or itchy throat | 0 | 1 (1%) | 1 (1%) | |
| Scratchy, itchy throat | 2 (2%) | 0 | 2 (1%) | |
| Trouble breathing | 0 | 1 (1%) | 1 (1%) | |
| Wheeze on chest examination | 1 (1%) | 0 | 1 (1%) | |
| Pleuritic pain | 1 (1%) | 0 | 1 (1%) | |
| Productive cough | 6 (7%) | 5 (6%) | 11 (6%) | |
| Rhinorrhoea | 3 (3%) | 0 | 3 (2%) | |
| Sinus pain | 1 (1%) | 2 (2%) | 3 (2%) | |
| Sneezing | 1 (1%) | 4 (4%) | 5 (3%) | |
| Sore throat | 6 (7%) | 6 (7%) | 12 (7%) | |
| Skin and subcutaneous tissue disorders | 7 (8%) | 8 (9%) | 15 (8%) | |
| Hyperhidrosis | 2 (2%) | 3 (3%) | 5 (3%) | |
| Itchy skin rash | 1 (1%) | 0 | 1 (1%) | |
| Leukonychia on fifth finger of right hand | 1 (1%) | 0 | 1 (1%) | |
| Nipple sensitivity | 0 | 1 (1%) | 1 (1%) | |
| Rosacea acne around chin (flare up) | 0 | 1 (1%) | 1 (1%) | |
| Skin blisters | 1 (1%) | 0 | 1 (1%) | |
| Skin rash | 1 (1%) | 1 (1%) | 2 (1%) | |
| Erythematous rash on both cheeks and nose | 0 | 1 (1%) | 1 (1%) | |
| Photosensitivity | 1 (1%) | 0 | 1 (1%) | |
| Maculopapular rash | 0 | 1 (1%) | 1 (1%) | |
| Vascular disorders | 3 (3%) | 0 | 3 (2%) | |
| Hot flushes | 1 (1%) | 0 | 1 (1%) | |
| Hypertension | 2 (2%) | 0 | 2 (1%) | |
| Unclassified | 4 (4%) | 3 (3%) | 7 (4%) | |
| Abdominal cramps | 1 (1%) | 0 | 1 (1%) | |
| Abdominal discomfort | 0 | 1 (1%) | 1 (1%) | |
| Blocked nose | 0 | 1 (1%) | 1 (1%) | |
| Chest tightness | 2 (2%) | 0 | 2 (1%) | |
| Coryzal symptoms | 1 (1%) | 0 | 1 (1%) | |
| Loss of sense of taste | 1 (1%) | 0 | 1 (1%) | |
| Loss of smell | 0 | 1 (1%) | 1 (1%) | |
| Loss of taste | 0 | 1 (1%) | 1 (1%) | |
| Muscular soreness over sternum | 0 | 1 (1%) | 1 (1%) | |
| Reduction in appetite | 1 (1%) | 0 | 1 (1%) | |
| Reduction in taste | 1 (1%) | 0 | 1 (1%) | |
| Vaginal thrush | 0 | 1 (1%) | 1 (1%) | |
| Viral gastroenteritis | 0 | 1 (1%) | 1 (1%) | |
Data are n (%). Within each system organ class, a participant can have more than one adverse event.
We evaluated changes in viral titre as an exploratory efficacy endpoint (appendix 2 p 10). Mean baseline titres were 7·1 log10 copies per reaction (SD 2·7) for the molnupiravir group and 7·4 log10 copies per reaction (3·0) for the placebo group. Compared with baseline, viral load decreased by a mean of 4·8 log10 copies per reaction (SD 2·6) in the molnupiravir group and 3·9 log10 copies per reaction (3·2) in the placebo group at day 5 (p=0·042; appendix 2 p 8). Among vaccinated individuals, the mean reduction in viral titre between baseline and day 5 was 5·4 log10 copies per reaction (SD 2·1) in the molnupiravir group and 4·1 log10 copies per reaction (3·1) in the placebo group (p=0·027). Among unvaccinated individuals, the mean reduction in viral titre between baseline and day 5 was 4·2 log10 copies per reaction (2·9) in the molnupiravir group and 3·6 log10 copies per reaction (3·4) in the placebo group (p=0·38). In our sensitivity analyses, mean reductions in viral titres between baseline and day 3 or day 8 were similar in both the molnupiravir group and placebo group, with day 3 decreases being smaller than day 5 decreases (appendix 2 p 8). Different patterns of viral elimination were seen during the 29 days' follow-up. We observed a transient increase in viral titre in seven (4%) of 180 participants (three on molnupiravir and four on placebo), viral clearance in 134 (74%; 68 on molnupiravir and 66 on placebo), an indeterminate pattern in 34 (19%; 18 on molnupiravir and 16 on placebo), and non-evaluable titres in five (3%; one on molnupiravir and four on placebo). Transient increases were observed at some point during the period of virological sampling between day 1 and day 28, were not associated with any return or worsening of clinical symptoms, and probably reflect the natural history of viral infection.
Discussion
Molnupiravir received conditional marketing authorisation from the UK Medicines and Healthcare products Regulatory Agency and early use authorisation from the US Food and Drug Administration on the basis of data from the MOVe-OUT study2 in unvaccinated individuals at high risk of severe disease who were infected with the SARS-CoV-2 variants in circulation between May and October, 2021.2 MOVe-OUT found that molnupiravir had good tolerability and reduced the number of hospitalisations and deaths by about 50% at the interim evaluation, falling to around 30% after all 1433 patients had been analysed. An evaluation of the effect of molnupiravir on virological response by SARS-CoV-2 variant or in vaccinated patients within a randomised controlled trial has not been previously published.
In our phase 2 study, patients in the molnupiravir group had a faster median time from randomisation to PCR negativity than did patients in the placebo group. We used a Bayesian framework to facilitate decision making. Using a two-point prior approach, the probability of the HR for PCR negativity being more than 1 (ie, in favour of molnupiravir vs placebo) was 75·4%, which was less than the 80% threshold we had set a priori for a clear decision to progress clinical evaluation. However, when a continuous, non-informative prior was used, molnupiravir (vs placebo) was predicted to have a high (94·7%) likelihood of having a HR for PCR negativity of more than 1. Our subgroup analyses did not have sufficient power for statistical comparison, but there was no obvious loss of effect by vaccination status or with the omicron variant. The persistence of PCR positivity at day 29 in a small minority of participants is in keeping with other published reports11 and might represent low-level detection of viral nucleic acid fragments rather than viable and infectious virus.
Participants receiving molnupiravir also had a significantly greater mean reduction in viral load from baseline to the end of treatment (5 days) compared with participants receiving placebo. This significant difference was retained when evaluating vaccinated participants only but was not maintained when evaluating unvaccinated patients only. The transient increase (following an initial decline) in viral titres observed in seven participants (three in the molnupiravir group and four in the placebo group) occurred during follow-up, was not associated with any return or worsening of clinical symptoms, and probably reflects the natural history of viral infection.
Our study has several limitations and strengths. The clinical tools used (the WHO Clinical Progression Scale and FLU-PRO) were not sensitive enough to detect small changes in our ambulatory cohort of patients. Caution should be exercised when interpreting outcomes for subgroups for which the sample size was small. We did not culture virus, and time to PCR negativity might be an insensitive marker for tracking any effect of molnupiravir, given its known mechanism of action. However, we were able to conduct serial sampling of nasopharyngeal swabs to characterise viral elimination rates and trajectories in participants who were within 5 days of symptom onset. Our study also included data in vaccinated participants and in participants with the omicron SARS-CoV-2 variant, which have been key gaps in clinical evidence for molnupiravir thus far. The efficacy of molnupiravir against these newer SARS-CoV-2 variants will be evaluated in the PANORAMIC trial (ISRCTN30448031), which will be the largest randomised evaluation of molnupiravir to date.
The use of virological responses as endpoints is a topic of much debate, but has been recommended for early-phase trials in guidance for industry from the US Food and Drug Administration.12 Viral responses are an imperfect proxy for clinical disease as viral burden in the upper airways might not fully reflect bronchial epithelial infection and host factors (eg, immune status, pre-existing comorbidities, age, and obesity) strongly influence clinical progression. What viral clearance reveals is the killing power of an antiviral; for the purposes of an early-phase trial, this measure is the most appropriate proxy for efficacy. We observed antiviral effects early in follow-up, suggesting that comparisons that give more weight to early versus late events (eg, the Breslow–Gehan–Wilcoxon test) might be more appropriate when non-proportional hazards are anticipated and viral surveillance is extended to 29 days.
We found molnupiravir to be well tolerated during 29 days of assessment. Most adverse events were mild (grade 1–2) and probably related to COVID-19, with only one patient having a severe (grade ≥3) adverse event and no patients having a serious adverse event in the molnupiravir group. No participants in the molnupiravir group were hospitalised, but four participants were hospitalised in the placebo group.
In conclusion, we have presented results showing that molnupiravir has a moderate antiviral effect, but our evidence is not conclusive. Although numbers were small and exploratory, viral load reductions were observed in both vaccinated and unvaccinated individuals. Participants infected with a broad range of SARS-CoV-2 variants were enrolled, including participants infected by omicron. Our data are consistent with preliminary observational data from Hong Kong13 and add to the growing evidence of generalisability of trial findings to newer variants.
AGILE CST-2 Study Group
This online publication has been corrected. The corrected version first appeared at thelancet.com/infection on November 14, 2022
Data sharing
The AGILE Trial Steering Committee will consider all reasonable requests by health-care providers, investigators, and researchers to provide anonymised data to address specific scientific or clinical objectives. Requests can be made at khoo@liverpool.ac.uk. The AGILE investigators are committed to reviewing requests from researchers for access to clinical trial protocols, deidentified patient-level clinical trial data, and study-level clinical trial data. Data will be assigned a digital object identifier through deposition in the University of Liverpool Research Data Catalogue (rdm@liverpool.ac.uk) and shared under a data transfer agreement (or equivalent—eg, as part of a research collaboration agreement or a confidentiality disclosure agreement). No date restrictions apply to data sharing.
Declaration of interests
SHK has received research funding from ViiV Healthcare and Merck for the Liverpool HIV Drug Interactions programme and for unrelated clinical studies. GG has received funding from Janssen-Cilag, AstraZeneca, Novartis, Astex, Roche, Heartflow, Celldex, BMS, BionTech, Cancer Research UK, the NIHR, the British Lung Foundation, Unitaid, and GSK for unrelated academic clinical trials and programme funding. WG has received funding from the Wellcome Trust. WH is a cofounder and owner of, and adviser for, Ridgeback Biotherapeutics. WP is employed by Ridgeback Biotherapeutics. JP has received honoraria for lectures from Gilead and London International Patient Services UK. JAH has received research funding from the US Food and Drug Administration and payment for expert testimony from DAC Beachcroft and Clyde and Co. TE has received unrelated research funding from the Medical Research Council, the Wellcome Trust, and Bloomsbury SET and travel grants from the European Society of Clinical Microbiology and Infectious Diseases. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
The AGILE trial is an academic, non-commercial trial sponsored by the University of Liverpool. Ridgeback Biotherapeutics provided funding for AGILE candidate-specific trial 2 and supplied molnupiravir and matched placebo. The AGILE platform infrastructure is supported by the Medical Research Council (grant number MR/V028391/1) and the Wellcome Trust (grant number 221590/Z/20/Z). Core funding from the NIHR supported the Southampton Clinical Trials Unit and clinical research facilities in Liverpool, Southampton, Manchester, Lancashire, and King's College Hospital. JAH is funded by the US Food and Drug Administration Medical Countermeasures Initiative contract (75F40120C00085). This Article reflects the views of the authors and does not represent the views or policies of the US Food and Drug Administration. Additional support was given by the NIHR Health Protection Research Unit in Emerging and Zoonotic Infections (award 200907) and the UK Medical Research Council (MR/W005611/1). TJ also received funding from the UK Medical Research Council (MC_UU_00002/14). The Bayesian adaptive analysis method used in this study was co-designed by PM, who is supported by the NIHR through the NIHR Advanced Fellowship (NIHR300576). We are deeply grateful to all participants for taking part in this trial. We also thank Sara Yates, Kiera Fines, Megan Lawrence, and Andrea Corkhill from the Southampton Clinical Trials Unit, Southampton, UK; Helen Berrington from the University of Lancaster, Lancaster, UK; Rosalind Szurko and Mathew Anuj from the NIHR Lancashire Clinical Research Facility, Preston, UK; Ling-Pei Ho from the University of Oxford, Oxford, UK; Vicky Toomey from Southampton City Council, Southampton, UK; and Sophie Kelly, Callum Rutherford, and Richard Jones from Liverpool City Council, Liverpool, UK.
Editorial note: The Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
AGILE Trial Steering Committee: Nicholas Paton (chair), Fred Hayden, Janet Darbyshire, Amy Lucas, and Ulrika Lorch. AGILE Independent Data Monitoring and Ethics Committee: Andrew Freedman (chair), Richard Knight, and Steven Julious. Liverpool School of Tropical Medicine, Liverpool, UK: Rachel Byrne, Ana Cubas Atienzar, Jayne Jones, and Chris Williams. Southampton Clinical Trials Unit, Southampton, UK: Anna Song, Jan Dixon, Anja Alexandersson, Parys Hatchard, and Emma Tilt. University of Lancaster, Lancaster, UK: Andrew Titman. University of Liverpool, Liverpool, UK: Ale Doce Carracedo, Vatsi Chandran Gorner, Andrea Davies, Louis Woodhouse, and Nicola Carlucci. NIHR Liverpool Clinical Research Facility, Liverpool, UK: Emmanuel Okenyi, Marcin Bula, Kate Dodd, Jennifer Gibney, Lesley Dry, Zalina Rashid Gardner, Amin Sammour, Christine Cole, Tim Rowland, Maria Tsakiroglu, Vincent Yip, Rostam Osanlou, and Anna Stewart. NIHR Manchester Clinical Research Facility, Manchester, UK: Ben Parker, Tolga Turgut, Afshan Ahmed, Kay Starkey, Sujamole Subin, Jennifer Stockdale, and Lisa Herring. NIHR Southampton Clinical Research Facility, Southampton, UK: Jonathan Baker, Abigail Oliver, Mihaela Pacurar, Dan Owens, Alistair Munro, Gavin Babbage, Saul Faust, Matthew Harvey, and Danny Pratt. NIHR King's Clinical Research Facility, London, UK: Deepak Nagra. NIHR Lancashire Clinical Research Facility, Preston, UK: Aashish Vyas.
Contributors
SHK, GG, TJ, RF, GS, SE, PM, and MJ contributed to study design. SHK, GG, TJ, GS, SE, PM, and JN contributed to data analysis and interpretation. RF, SA, CJE, DH, and JP led clinical conduct as principal investigators of the clinical sites. TF, LW, AB, DC, and RL participated in clinical assessment and data collection. WG, LL-L, KB, VS, TE, CH, JAH, and ID-B contributed to study bioanalysis. MT, DGL, MJ, JC, EK, ND, HR, CW, and CM contributed to study management and execution. WH and WP contributed preclinical and safety data on molnupiravir. The manuscript was written by the authors, with SHK and GG as the overall lead authors. No one who is not an author contributed to writing the manuscript. GS, SE, and JN directly accessed and verified the underlying data reported in this manuscript. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication. The authors assume responsibility for the accuracy and completeness of the data and for the fidelity of the trial to the protocol. All authors read and approved the final version of the manuscript.
Contributor Information
AGILE CST-2 Study Group:
Nicholas Paton, Fred Hayden, Janet Darbyshire, Amy Lucas, Ulrika Lorch, Andrew Freedman, Richard Knight, Stevan Julious, Rachel Byrne, Ana Cubas Atienzar, Jayne Jones, Chris Williams, Anna Song, Jan Dixon, Anja Alexandersson, Parys Hatchard, Emma Tilt, Andrew Titman, Ale Doce Carracedo, Vatsi Chandran Gorner, Andrea Davies, Louis Woodhouse, Nicola Carlucci, Emmanuel Okenyi, Marcin Bula, Kate Dodd, Jennifer Gibney, Lesley Dry, Zalina Rashid Gardner, Amin Sammour, Christine Cole, Tim Rowland, Maria Tsakiroglu, Vincent Yip, Rostam Osanlou, Anna Stewart, Ben Parker, Tolga Turgut, Afshan Ahmed, Kay Starkey, Sujamole Subin, Jennifer Stockdale, Lisa Herring, Jonathon Baker, Abigail Oliver, Mihaela Pacurar, Dan Owens, Alistair Munro, Gavin Babbage, Saul Faust, Matthew Harvey, Danny Pratt, Deepak Nagra, and Aashish Vyas
Supplementary Materials
References
- 1.Khoo SH, Fitzgerald R, Fletcher T, et al. Optimal dose and safety of molnupiravir in patients with early SARS-CoV-2: a phase I, open-label, dose-escalating, randomized controlled study. J Antimicrob Chemother. 2021;76:3286–3295. doi: 10.1093/jac/dkab318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jayk Bernal A, Gomes da Silva MM, Musungaie DB, et al. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N Engl J Med. 2022;386:509–520. doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumarasamy N, Saha B, Jindal A, et al. Phase 3 trial of molnupiravir in adults with mild SARS-CoV-2 infection in India. Conference on Retroviruses and Opportunistic Infections; Feb 15, 2022 (abstract O-9).
- 4.Troth S, Butterton J, DeAnda CS, et al. Letter to the editor in response to Zhou et al. J Infect Dis. 2021;224:1442–1443. doi: 10.1093/infdis/jiab362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.US Food and Drug Administration Center for Drug Evaluation and Research. FDA briefing document. Antimicrobial drugs advisory committee meeting. Nov 30, 2021. https://www.fda.gov/media/154418/download
- 6.Griffiths GO, FitzGerald R, Jaki T, et al. AGILE: a seamless phase I/IIa platform for the rapid evaluation of candidates for COVID-19 treatment: an update to the structured summary of a study protocol for a randomised platform trial letter. Trials. 2021;22:487. doi: 10.1186/s13063-021-05458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect Dis. 2020;20:e192–e197. doi: 10.1016/S1473-3099(20)30483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coolen JPM, Wolters F, Tostmann A, et al. SARS-CoV-2 whole-genome sequencing using reverse complement PCR: for easy, fast and accurate outbreak and variant analysis. J Clin Virol. 2021;144:104993. doi: 10.1016/j.jcv.2021.104993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Toole Á, Scher E, Underwood A, et al. Assignment of epidemiological lineages in an emerging pandemic using the Pangolin tool. Virus Evol. 2021;7:veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ewings S, Saunders G, Jaki T, Mozgunov P. Practical recommendations for implementing a Bayesian adaptive phase I design during a pandemic. BMC Med Res Methodol. 2022;22:25. doi: 10.1186/s12874-022-01512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–675. doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 12.US Food and Drug Administration Guidance document. COVID-19: developing drugs and biological products for treatment or prevention. May, 2020. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/covid-19-developing-drugs-and-biological-products-treatment-or-prevention
- 13.Wong CKH, Au ICH, Lau KTK, Lau EHY, Cowling BJ, Leung GM. Real-world effectiveness of early molnupiravir or nirmatrelvir–ritonavir in hospitalised patients with COVID-19 without supplemental oxygen requirement on admission during Hong Kong's omicron BA.2 wave: a retrospective cohort study. Lancet Infect Dis. 2022 doi: 10.1016/S1473-3099(22)00507-2. published online Aug 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The AGILE Trial Steering Committee will consider all reasonable requests by health-care providers, investigators, and researchers to provide anonymised data to address specific scientific or clinical objectives. Requests can be made at khoo@liverpool.ac.uk. The AGILE investigators are committed to reviewing requests from researchers for access to clinical trial protocols, deidentified patient-level clinical trial data, and study-level clinical trial data. Data will be assigned a digital object identifier through deposition in the University of Liverpool Research Data Catalogue (rdm@liverpool.ac.uk) and shared under a data transfer agreement (or equivalent—eg, as part of a research collaboration agreement or a confidentiality disclosure agreement). No date restrictions apply to data sharing.