SUMMARY
Murine norovirus (MNoV) is a model for human norovirus and for interrogating mechanisms of viral tropism and persistence. We previously demonstrated that the persistent strain MNoVCR6 infects tuft cells, which are dispensable for the non-persistent strain MNoVCW3. We now show that diverse MNoV strains require tuft cells for chronic enteric infection. We also demonstrate that interferon-l (IFN-l) acts directly on tuft cells to cure chronic MNoVCR6 infection and that type I and III IFNs signal together via STAT1 in tuft cells to restrict MNoVCW3 tropism. We then develop an enteroid model and find that MNoVCR6 and MNoVCW3 similarly infect tuft cells with equal IFN susceptibility, suggesting that IFN derived from non-epithelial cells signals on tuft cells in trans to restrict MNoVCW3 tropism. Thus, tuft cell tropism enables MNoV persistence and is determined by tuft cell-intrinsic factors (viral receptor expression) and -extrinsic factors (immunomodulatory signaling by non-epithelial cells).
Graphical Abstract
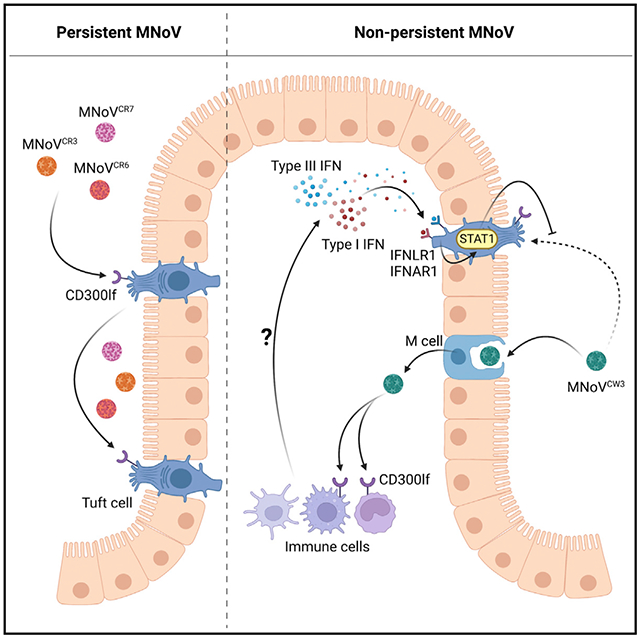
In brief
Strine et al. interrogate the determinants and consequences of tuft cell tropism on murine norovirus infection. Using enteroid and mouse models, they show that tuft cells are required for persistent enteric infection and that non-epithelial cells restrict tuft cell tropism via combinatorial type I and III interferon signaling on tuft cells.
INTRODUCTION
Human norovirus (HNoV) is a major etiological agent of viral gastroenteritis worldwide, causing up to 700 million infections and 200,000 deaths annually (Lopman et al., 2016). There are currently no approved drugs and vaccines for NoV treatment and prevention (Cortes-Penfield et al., 2017; Zhang et al., 2021). Despite recent progress in the field, the role of innate and adaptive immunity as a determinant of NoV infection represents a knowledge gap. Due to the limitations in the culture and reverse genetics systems for HNoV, murine NoV (MNoV) is used as a surrogate virus to study host-virus interactions (Graziano et al., 2020; Wobus et al., 2006). MNoV exhibits similar viral genetics, virion structure, and replication mechanisms to HNoV and recapitulates many of the key features of HNoV pathogenesis, including fecal-oral transmission, epithelial and immune cell tropism, interactions with bile acids, and the capacity to establish persistent infection in some hosts (Baldridge et al., 2016; Karst et al., 2014; Wobus et al., 2016). In contrast to HNoV, which cannot robustly replicate in small-animal models, MNoV can readily infect mice, providing a tractable in vivo model for NoV studies and facilitating the identification of host immune and viral factors regulating NoV replication and pathogenesis (Karst et al., 2003).
MNoV strains have been classified based on their distinct pathogenic properties. The non-persistent strain MNoV-1 (and its derived molecular clone MNoVCW3) causes acute systemic infection and can be cleared within 1 to 2 weeks (Thackray et al., 2007; Ward et al., 2006). MNoVCW3 infection is initiated in the distal small intestine and spreads through the large intestine, MLNs, and spleen (Karst et al., 2003). Early replication occurs in gut-associated lymphoid tissues, particularly in the macrophages, dendritic cells, B cells, and T cells (Grau et al., 2017). However, there is scant MNoVCW3 shedding in the feces (Karst et al., 2015; Nice et al., 2013; Thackray et al., 2007). Conversely, the persistent strain MNoVCR6 replicates in both the small and large intestine initially and then predominantly in the colon after several weeks post-infection. MNoVCR6 viral RNA is detected in draining MLNs, but viral replication is not observed in systemic tissues (Nice et al., 2013, 2015). Consistent with this, MNoVCR6 robustly sheds in the feces for months, if not for the lifetime of the animal (Karst et al., 2015; Nice et al., 2013, 2015; Thackray et al., 2007; Wilen et al., 2018).
Recently, we identified CD300lf as the functional receptor for diverse MNoV strains (Haga et al., 2016; Kitagawa et al., 2010; Orchard et al., 2016; Wilen et al., 2018). CD300lf, which is required for infection in mice, is expressed on intestinal tuft cells and diverse hematopoietic cells. We previously demonstrated that tuft cells are infected by MNoVCR6 but not MNoVCW3 (Graziano et al., 2021; Lee et al., 2017; Wilen et al., 2018). Tuft cells are rare epithelial cells located in the gut that have diverse and critical functions in anti-helminth and parasitic immunity, inflammation, and tumorigenesis (Strine and Wilen, 2022). The development of tuft cells requires expression of the POU domain transcription factor, POU class 2 homeobox 3 (POU2F3), which is also essential for taste receptor cells (Matsumoto et al., 2011). Both short- and long-lived tuft cells have been described (Westphalen et al., 2014). Tuft cells secrete interleukin-25 (IL-25), which acts on type 2 innate lymphoid cells to stimulate production of IL-13 that signals to intestinal epithelial stem cells to induce tuft cell hyperplasia in a positive feedback cycle (Gerbe et al., 2016; Howitt et al., 2016; Moltke et al., 2016). The cytokines IL-4, IL-13, and IL-25 also enhance MNoVCR6 replication and pathogenesis by causing an increase in tuft cell abundance (Wilen et al., 2018).
Viruses that successfully establish persistent infection must overcome both innate and adaptive immune defenses. In mice deficient in the type I interferon (IFN-α/β) receptor (IFNAR1), MNoVCW3 replicates to higher titers and causes lethal, rather than self-limiting, infection (Karst et al., 2003; Strong et al., 2012). In Ifnar1−/− mice, MNoVCR6 can spread systemically but does not cause lethal disease (Nice et al., 2016). The ability of MNoVCW3 to cause death in Ifnar1−/− mice maps to a single amino acid on the viral capsid protein (Strong et al., 2012). In contrast to type I IFN, type III IFN (IFN-λ) acts locally in the gut. Endogenous IFN-λ partially controls MNoVCR6 replication, and a single dose of exogenous IFN-λ is sufficient to cure mice persistently infected with MNoVCR6 (Nice et al., 2015). The NS1 gene of MNoVCR6 is necessary and sufficient for epithelial cell tropism and viral persistence in vivo and likely functions to counteract IFN-λ signaling (Lee et al., 2017; Nice et al., 2013, 2015). Understanding why MNoVCR6 infects tuft cells but MNoVCW3 does not has important implications for our understanding of NoV immunity and persistence. MNoVCW3 and MNoVCR6 robustly replicate in cell lines from multiple that express mouse CD300lf (Graziano et al., 2020; Nice et al., 2013; Van Winkle et al., 2018). However, while HNoV can be cultivated in human intestinal stem-cell-derived enteroid monolayer cultures (Costantini et al., 2018; Ettayebi et al., 2016; Zou et al., 2019), efforts to similarly culture MNoV in mouse enteroids have been unsuccessful. This has limited the ability to identify viral and host determinants of tuft cell tropism and persistence.
In this study, we examine the role of tuft cells in viral persistence and the role of innate immunity in dictating tuft cell tropism. Here, we show that short-lived tuft cells are the persistent reservoir for MNoV and that tuft cell tropism specifically enables chronic MNoV infection. We further demonstrate that IFN-λ acts directly on tuft cells to cure MNoVCR6 infection and that MNoVCW3 can cause persistent infection when STAT1 is ablated in tuft cells, likely by preventing the combinatorial action of type I and III IFN signaling. We then developed a mouse enteroid culture system that can support MNoV infection of tuft cells. Surprisingly, we observed that MNoVCR6 and MNoVCW3 similarly infect tuft cells in vitro. Furthermore, both strains were highly sensitive to the antiviral effects of type I, II, and III IFN signaling. These results suggest that non-epithelial cell production of type I and III IFNs in response to MNoVCW3 infection acts in trans on tuft cells via STAT1 to regulate tuft cell tropism and thus MNoV persistence.
RESULTS
Tuft cells are essential for enteric infection by diverse persistent MNoV strains
We previously demonstrated that MNoVCR6, but not MNoVCW3, infects tuft cells in the small and large intestines of immunocompetent mice (Graziano et al., 2021; Lee et al., 2017; Wilen et al., 2018). However, whether tuft cells are the sole physiologic target for oral MNoVCR6 infection, and whether tuft cells are required for MNoV persistence, was unknown. To test the role of tuft cells in MNoV tropism in vivo, we generated Pou2f3 knockout (KO) mice by CRISPR-Cas9. POU2F3 is a transcription factor essential for tuft cell development (Matsumoto et al., 2011). We validated that Pou2f3 deletion inhibits tuft cell development by immunofluorescence for the tuft cell marker DCLK1 (Figure 1A). We then perorally (PO) challenged Pou2f3 heterozygote (+/−) and KO (−/−) mice with 106 plaque-forming units (PFUs) of five MNoV strains, including three known to establish chronic infection in mice (CR6, CR3, and CR7), an acute MNoV strain (CW3), and an acute MNoV strain that can variably persist in some animals (Wu23) (Thackray et al., 2007; Walker et al., 2021). Tuft cells are required for fecal shedding of the persistent strains MNoVCR6, MNoVCR3, and MNoVCR7 at both 3 and 7 days post-infection (Figures 1B and 1C). Relative to Pou2f3+/−, viral genome copies of MNoVCR6, MNoVCR3, and MNoVCR7 are significantly reduced in the mesenteric lymph nodes (MLNs), ileum, and colon of Pou2f3−/− mice, indicating that tuft cell infection is necessary for the establishment of viral infection in the gut as well as detection in draining LNs (Figures 1D, 1F, and 1G). Consistent with previous reports, MNoVCR6, MNoVCR3, and MNoVCR7 show little to no detectable viral RNA in the spleen, even in control mice (Figure 1E) (Graziano et al., 2020). Similar to CW3, Wu23 infection is unaffected by tuft cell deletion and exhibits the highest levels of viral genome copies in the spleen and MLN (Figures 1D–1G). These results suggest that diverse persistent MNoV strains, but not acute strains, require tuft cells for enteric infection.
Figure 1. Tuft cells are essential for enteric infection by diverse persistent MNoV strains.
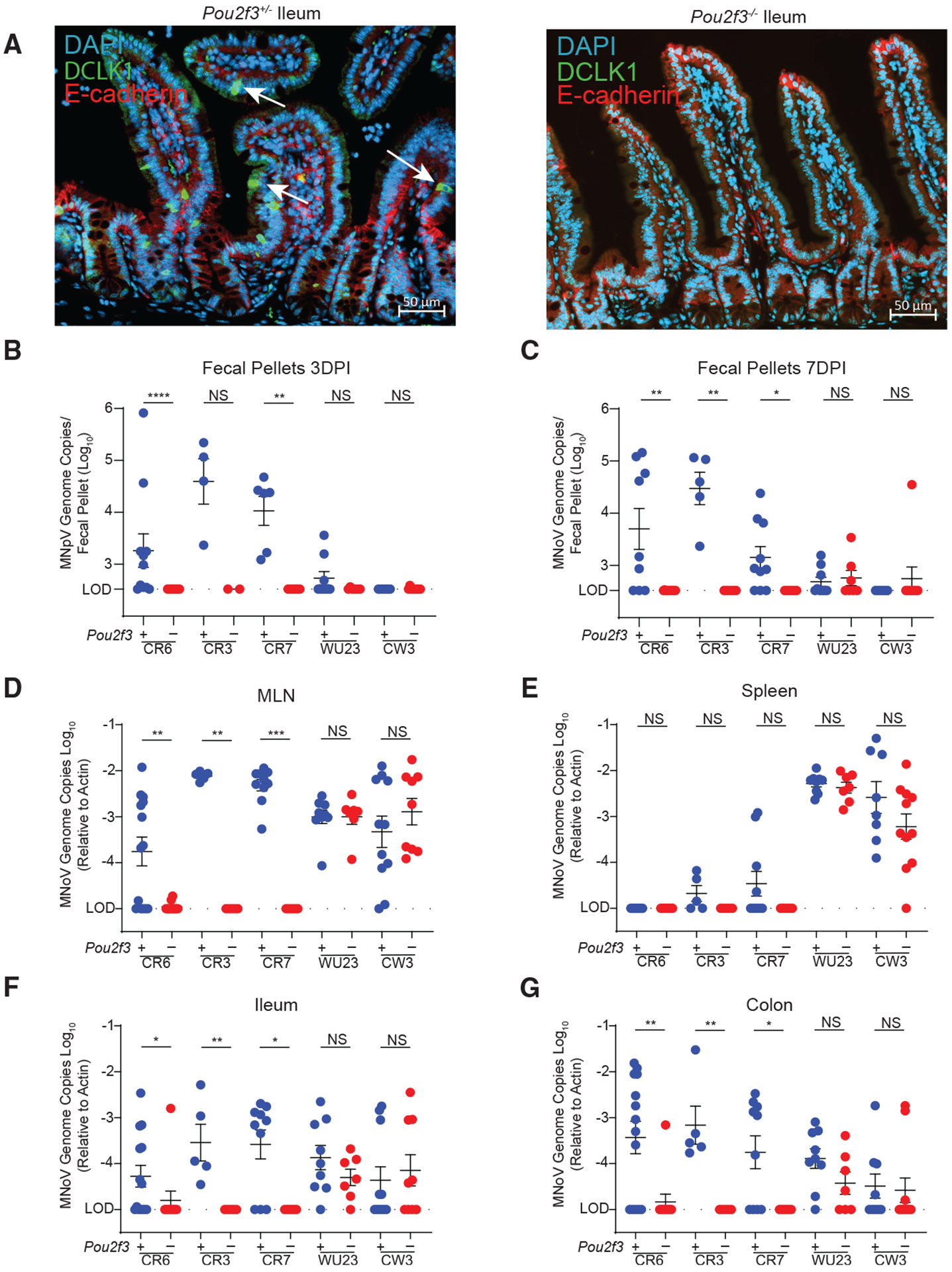
(A) Pou2f3+/− (left) and Pou2f3−/− (right) mice were generated and stained with DAPI (blue), E-cadherin (red), and DCLK1 (green) for tuft cells in the mouse ileum. Images are representative of one of at least three independent experiments.
(B–G) Pou2f3+/− and Pou2f3−/− mice were challenged perorally (PO) with 106 plaque-forming units (PFUs) of MNoV strains CR6, CR3, CR7, and Wu23. MNoV strain CW3 data are also shown and were previously published elsewhere (Grau et al., 2020). Fecal samples were collected at 3 and 7 days post-infection, and mice were sacrificed at 7 days post-infection (dpi). MNoV genome copies were quantified by qPCR in the (B and C) feces, (D) MLN, (E) spleen, (F) distal ileum, and (G) proximal colon.
Infection experiments were performed at least two independent times, and qPCR was performed in triplicate. Data in (B)–(G) are pooled from two to four independent experiments with at least three mice per group and were analyzed by Mann-Whitney test. Shown are means ± SEM. Statistical significance annotated as follows: NS, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; LOD, limit of detection.
Tuft cells are not essential for extraintestinal infection by MNoVCR6
Next, we asked whether the ability of MNoVCR6 to cause persistent infection required infection of tuft cells. Therefore, we tested whether MNoVCR6 could replicate in non-tuft cells if inoculated extra-intestinally. We challenged mice by intraperitoneal (i.p.) infection of MNoVCR6 and observed viral RNA in the MLN and spleen and, to a lesser extent, the ileum and colon at 7 days post-infection (dpi). Viral shedding was not detectable in the feces (Figures 2A–2E). Pou2f3+/− and Pou2f3−/− mice do not exhibit differences in viral RNA, suggesting that tuft cells are dispensable for extraintestinal MNoVCR6 infection (Figures 2B–2E). Next, we asked whether extraintestinal MNoVCR6 could cause chronic infection. In contrast to orally inoculated MNoVCR6, i.p. MNoVCR6 infection is undetectable in the MLN, spleen, ileum, and colon at 21 dpi (Figures 2F–2I). To investigate the role of adaptive immunity in eliminating MNoVCR6, we crossed Pou2f3−/− mice to Rag1-deficient mice that lack mature T and B lymphocytes (Mombaerts et al., 1992). In the absence of Rag1, i.p. MNoVCR6 can persist at 21 dpi (Figures 2F–2I). These data reveal that although Rag1 is nearly dispensable for PO MNoVCR6 infection, MNoVCR6 is not inherently resistant to immune-mediated clearance, and adaptive immune responses can eliminate MNoVCR6 when MNoVCR6 is not restricted to tuft cells.
Figure 2. Tuft cells are not essential for extraintestinal infection by persistent MNoV strain CR6.
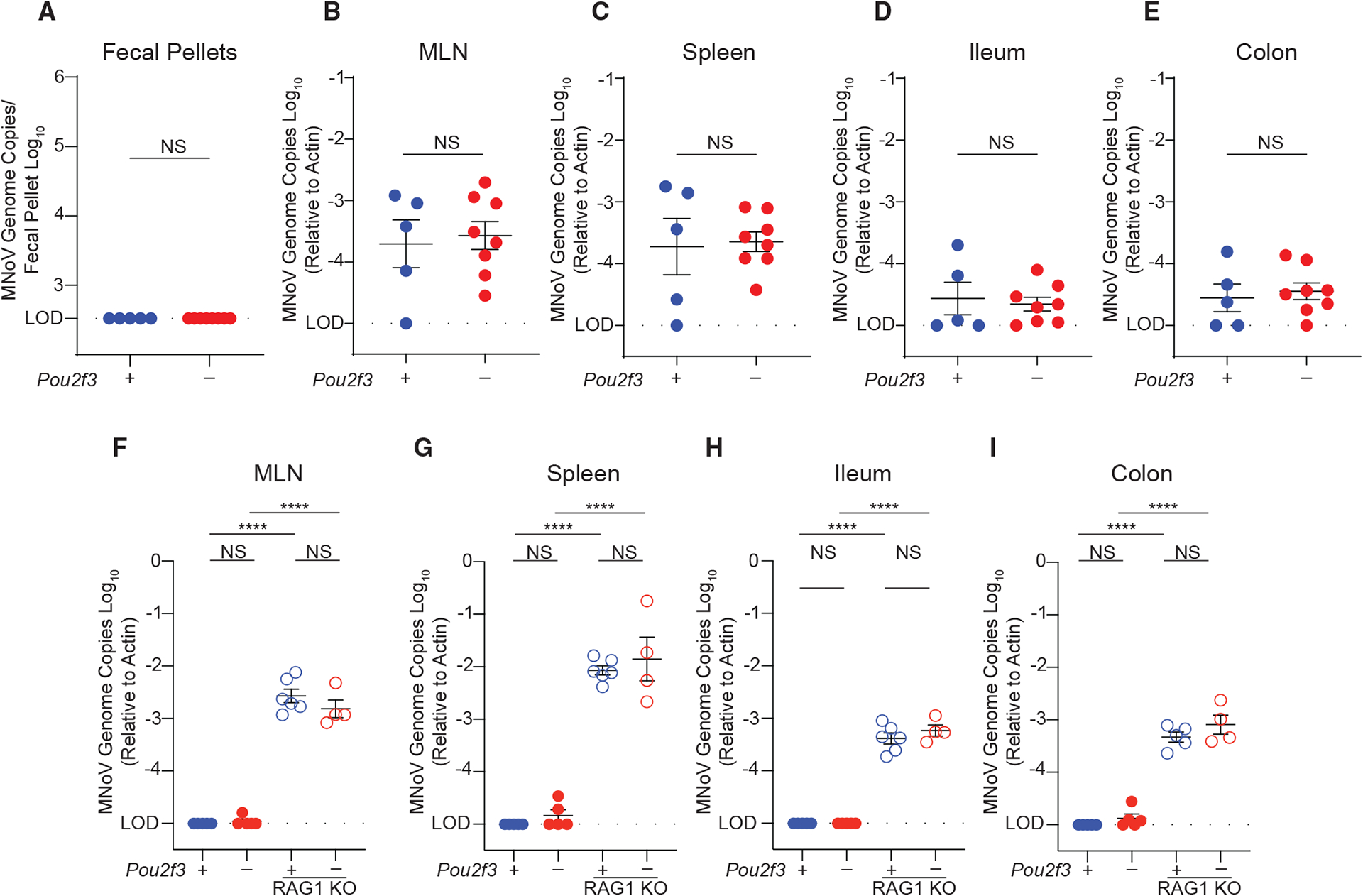
Pou2f3+/−, Pou2f3−/−, Pou2f3+/− Rag1−/−, and Pou2f3−/−Rag1−/− mice were challenged with 106 PFUs MNoVCR6 by intraperitoneal (i.p.) injection.
Mice were sacrificed at (A–E) 7 dpi or (F–I) the chronic time point of 21 dpi. MNoV genome copies were quantified by qPCR in the (A) feces, (B and F) MLN, (C and G) spleen, (D and H) distal ileum, and (E and I) proximal colon.
Experiments were performed at least two independent times with at least two mice per group, and qPCR was performed in triplicate. Data were analyzed by Mann-Whitney test (A–E) and ordinary one-way ANOVA (F–I). Shown are means ± SEM. Statistical significance annotated as follows: NS, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; LOD, limit of detection.
Short-lived tuft cells are the reservoir for persistent MNoV infection
While most tuft cells have a half-life of ~5 days, a subset of long-lived tuft cells have been described that can last for 6 months or longer (Westphalen et al., 2014). Long-lived tuft cells have quiescent stem-cell-like properties, which may render them relatively immunoprivileged (Agudo et al., 2018; Westphalen et al., 2014). Therefore, as tuft cell tropism is necessary for MNoVCR6 persistence, we asked whether chronic infection was due to serial re-infection of short-lived tuft cells or chronic infection of long-lived tuft cells. To test this hypothesis, we generated a tamoxifen-inducible CD300lf conditional KO mouse that specifically deletes CD300lf from tuft cells (Cd300lfF/F Dclk1 CreERT). We infected mice with 106 PFUs MNoVCR6 PO, and then after establishment of chronic infection (15–33 dpi), we administered tamoxifen to delete CD300lf from Dclk1-expressing tuft cells (Figure 3A). Tamoxifen administration significantly reduced MNoVCR6 in the feces within 5 days, and this effect persisted for 10 days (Figure 3B). A similar reduction in viral RNA was observed in the colon, the major site of viral persistence (Figure 3C) (Nice et al., 2013; Thackray et al., 2007). Importantly, ablation of CD300lf on tuft cells did not affect tuft cell abundance, supporting that MNoVCR6 titers were reduced as a direct consequence of restricted viral entry rather than depletion of total tuft cells (Figures 3D and 3E). These data suggest that infection of short-lived tuft cells is required for MNoVCR6 persistence and that infection is not restricted to a subset of long-lived tuft cells.
Figure 3. Short-lived tuft cells support persistent MNoV infection.
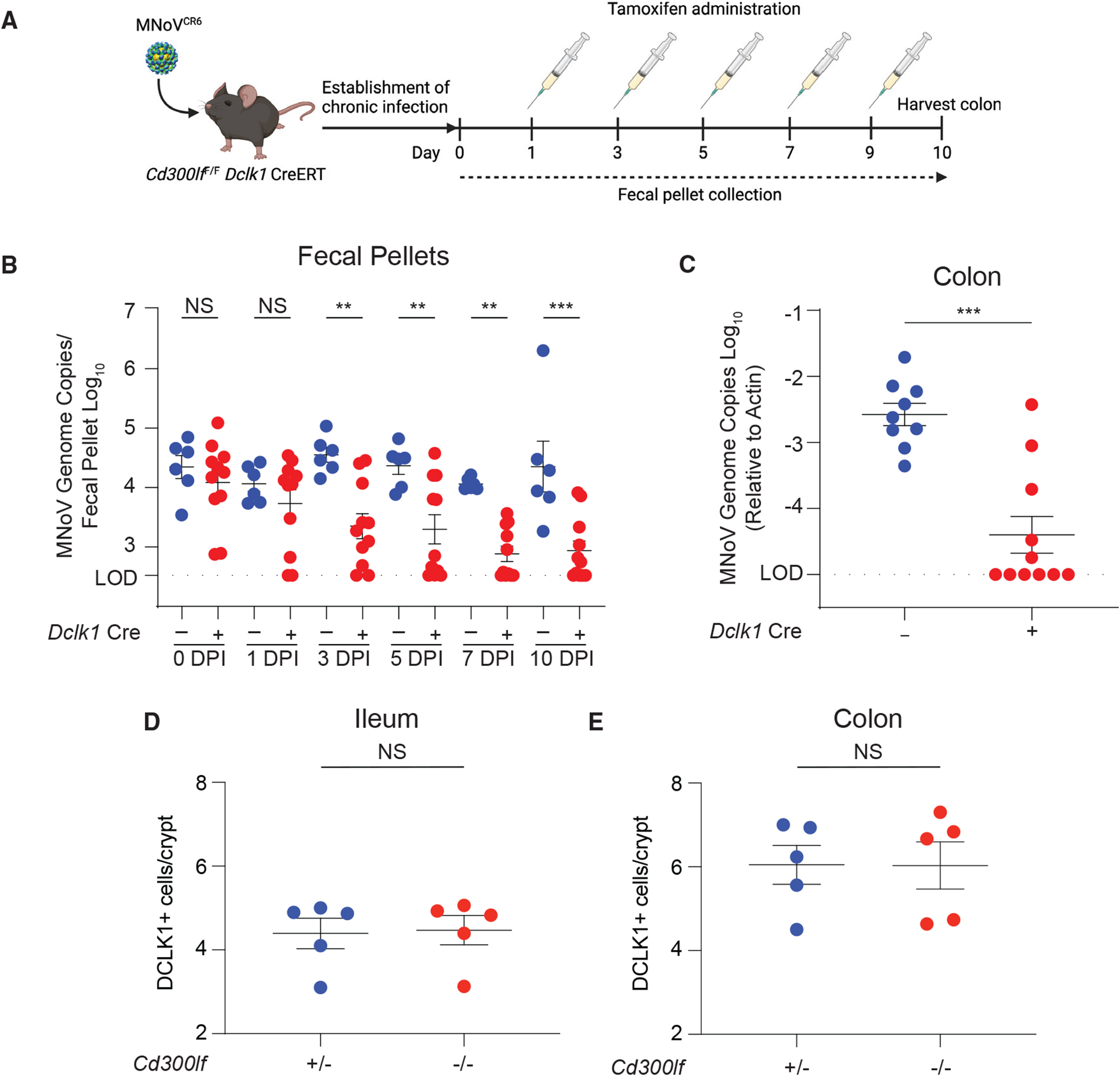
(A) Schematic depicting the infection and tamoxifen treatment used on Cd300lfF/F Dclk1 CreERT mice. Mice were infected PO with 106 PFUs MNoVCR6 for 15 to 33 days to establish chronic infection. Mice were then dosed with tamoxifen via i.p. injection every other day for 9 days to ablate CD300lf expression on newly differentiated tuft cells.
(B) Fecal samples were collected, and MNoVCR6 fecal shedding was quantified on days 1, 3, 6, 7, and 10.
(C) MNoVCR6 genome copies were assessed in the proximal colon at 10 days post-tamoxifen treatment. Genome copies were measured by qPCR and normalized to actin in tissues. Experiments were performed two independent times with at least five mice per group, and qPCR was performed in triplicate.
(D and E) Tuft cell abundance was quantified along the crypt-villus axis in the ileum (D) and colon (E) of Cd300lf+/− and Cd300lf−/− mice.
Data were analyzed by Mann-Whitney test (B, D, and E) and ordinary one-way ANOVA (C). Shown are means ± SEM. Statistical significance annotated as follows: NS, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; LOD, limit of detection.
Tuft cells are essential for enteric infection in Stat1-deficient mice
Next, we aimed to elucidate the role of innate immunity in restricting tuft cell tropism. We crossed Pou2f3+/− and Pou2f3−/− mice to mice deficient in STAT1, a transcription factor critical for IFN-stimulated gene (ISG) expression (Durbin et al., 1996). We challenged mice PO with 106 PFUs PO MNoVCW3, MNoVCR6, or MNoVCR6 with the capsid of MNoVCW3 (MNoVCR6–VP1–CW3). Consistent with prior studies, MNoVCW3, but not MNoVCR6, causes lethal infection in Stat1−/− mice, a phenotype dependent upon the capsid of MNoVCW3 (Thackray et al., 2007, 2012) (Figure 4A). While these aspects were previously demonstrated, it was unknown whether tuft cells contributed to MNoVCW3-induced lethality in the absence of Stat1. However, we now show MNoV-induced lethality was independent of tuft cell infection (Figure 4A).
Figure 4. Tuft cells are essential for enteric infection in Stat1-deficient mice.
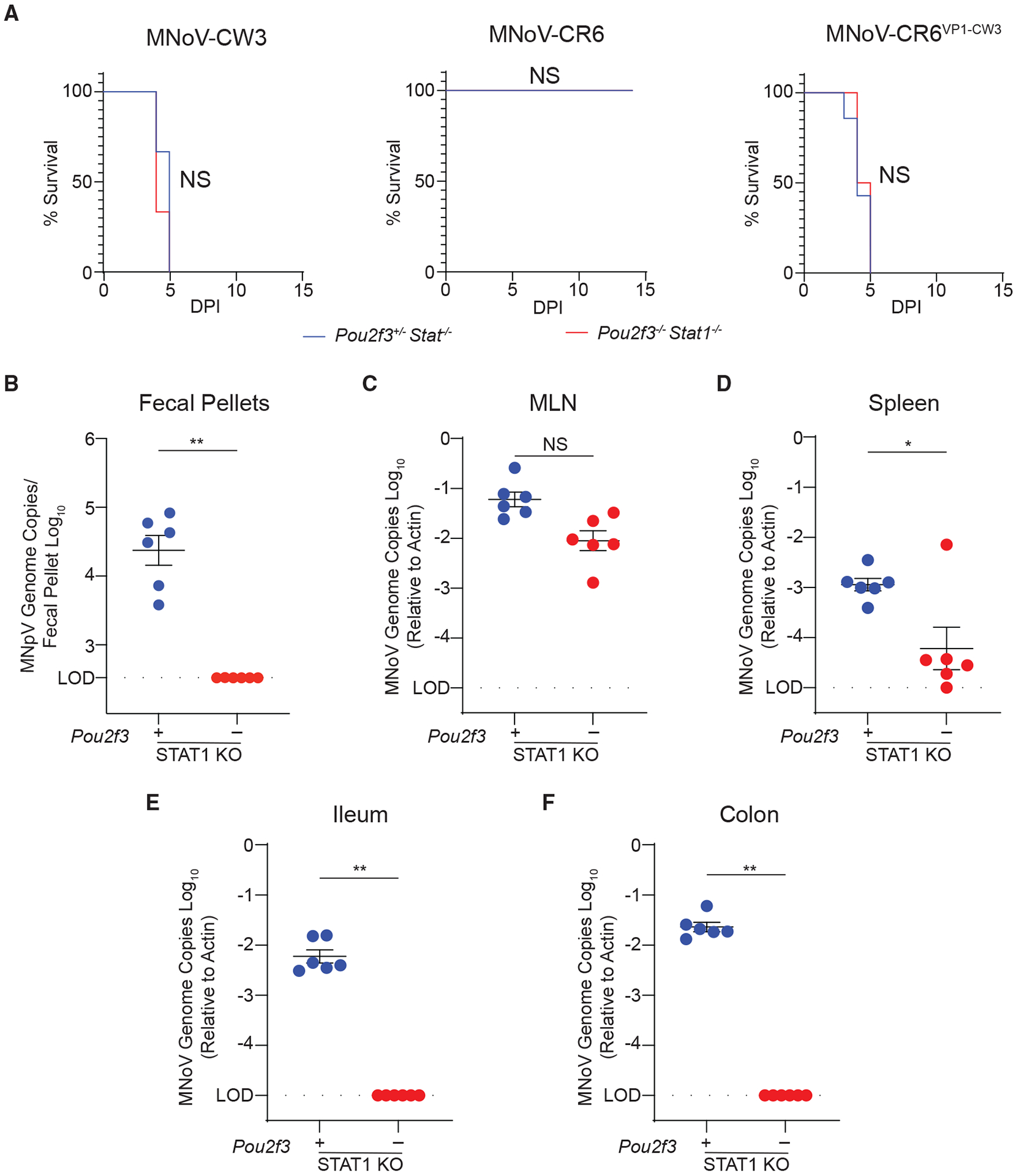
(A) Pou2f3+/− Stat1−/− or Pou2f3−/− Stat1−/− mice were infected PO with 106 PFUs MNoV strains CR6 or CW3 or the chimeric strain CR6-VP1-CW3. Mouse survival was tracked for up to 7 days.
(B–F) Mice challenged with MNoVCR6 were sacrificed and viral load was assessed by qPCR at 7 dpi for MNoV genome copies in the (B) feces, (C) MLN, (D) spleen, (E) distal ileum, and (F) proximal colon.
Experiments were performed at least two independent times with at least three mice per group and data were analyzed by Mann-Whitney. Kaplan-Meier curves were generated for survival experiments. Shown are means ± SEM. Statistical significance annotated as follows: NS, not significant; *p < 0.05; **p < 0.01; LOD, limit of detection.
We then asked whether MNoVCR6 tropism could expand in the absence of STAT1 by challenging Pou2f3−/− Stat1−/− mice or Pou2f3+/− Stat1−/− controls with 106 PFUs MNoVCR6 PO. Even in the absence of STAT1-mediated innate immunity, tuft cells were essential for viral RNA shedding in the feces, ileum, and colon and were partially necessary in the spleen (Figures 4B–4F). Interestingly, MNoVCR6 RNA levels in the MLN were similar between Pou2f3+/− and Pou2f3−/− mice regardless of Stat1 deletion (Figure 4C). These results demonstrate that tuft cells are not required for MNoV-induced lethality or systemic infection; however, tuft cells are necessary for enteric infection by MNoVCR6, even in the absence of STAT1.
Tuft-cell-intrinsic IFN signaling restricts MNoV infection
IFN-λ acts on epithelial cells to cure persistent MNoVCR6 infection (Baldridge et al., 2017; Nice et al., 2015). Whether IFN signaling is needed specifically within tuft cells to cure MNoVCR6 infection is unclear. To test this, we specifically disrupted Stat1 in tuft cells (Stat1F/F Dclk1 Cre) and infected mice with 106 PFUs MNoVCR6 PO for 21 days prior to exogenous IFN-λ treatment. In agreement with previous findings, a single dose of IFN-λ was sufficient to cure Cre mice (Figure 5A). However, in mice with tuft cells lacking Stat1, IFN-λ treatment was not curative, indicating that IFN-λ signals directly on tuft cells to clear persistent MNoVCR6 infection (Figure 5A).
Figure 5. Tuft-cell-intrinsic innate immune signaling restricts MNoV infection.
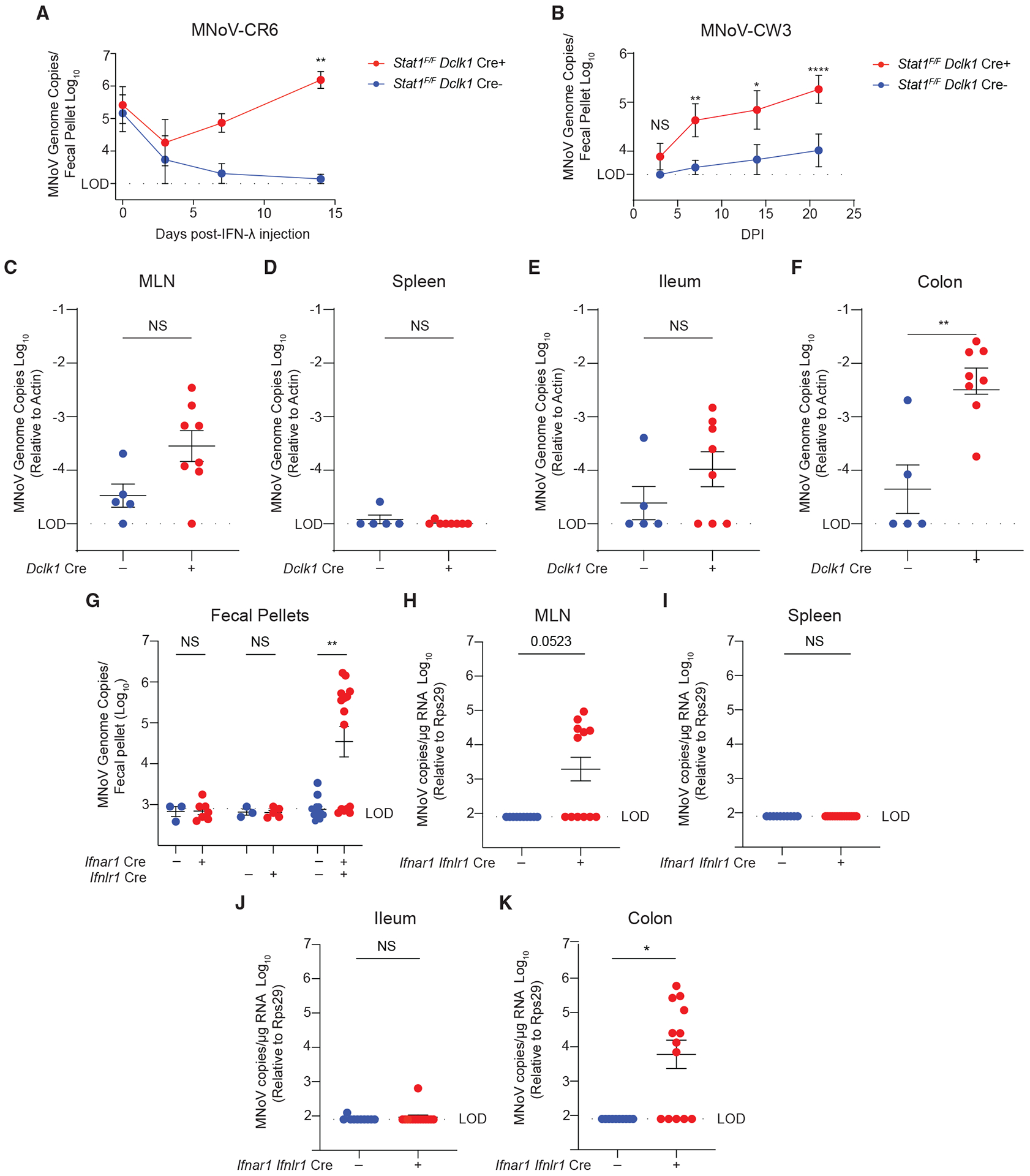
(A) Stat1F/F Dclk1 Cre mice were infected with 106 PFUs PO MNoVCR6 to establish chronic infection. Mice were treated with IFN-λ for 2 weeks. Fecal pellets were collected longitudinally at days 0, 3, 7, and 14 post-IFN-λ.
(B–F) Stat1F/F Dclk1 Cre mice were infected with 106 PFUs PO MNoVCW3. Fecal pellets were collected at (B) 4, 7, 14, and 21 dpi and (C) MLN, (D) spleen, (E) ileum, and (F) colon at 21 dpi, mice were sacrificed, and tissues were harvested.
(G–K) Ifnar1F/F Villin Cre, Ifnlr1F/F Villin Cre , and Ifnar F/F Ifnlr1F/F Villin Cre mice were infected with 106 PFUs PO MNoVCW3, and (G) fecal pellets, (H) MLN, (I) spleen, (J) ileum, and (K) colon were harvested at 14 dpi. Viral loads were assessed by qPCR for MNoV genome copies.
Experiments were performed two independent times, and data were analyzed by Mann Whitney or two-way ANOVA. Shown are means ± SEM. Statistical significance annotated as follows: NS, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; LOD, limit of detection.
We next assessed the role of tuft-cell-intrinsic Stat1 signaling in MNoVCW3 pathogenesis. We challenged Stat1F/F Dclk1 Cre mice with 106 PFUs PO MNoVCW3 and longitudinally assessed MNoVCW3 RNA in the feces, MLN, spleen, proximal ileum, and colon at 21 dpi (Figures 5B–5F). Surprisingly, we observed MNoVCW3 fecal shedding, chronic infection, and viral RNA in the ileum, colon, and MLN of Stat1F/F Dclk1 Cre+ mice relative to Cre controls (Figures 5B–5F). This finding indicates that MNoVCW3 can phenocopy MNoVCR6 intestinal pathogenesis and persistence in the absence of tuft-cell-intrinsic IFN signaling. To clarify the cytokines acting upstream of STAT1 to restrict MNoVCW3 persistence, we ablated type I and III IFN signaling by conditionally deleting Ifnar1, Ifnlr1, or both Ifnar1 and Ifnlr1 in epithelial cells using Cre recombinase operating under the Villin promoter. In single Ifnar1F/F Villin Cre+ and Ifnlr1F/F Villin Cre+ mice, MNoVCW3 caused acute infection with no fecal shedding (Figure 5G). However, MNoVCW3 was shed in the feces and persisted in the colon in double IfnarF/F Ifnlr1F/F Villin Cre+ mice with modest viral loads in the MLN but not the spleen or ileum, suggesting that together type I and III IFNs restrict MNoVCW3 tropism for tuft cells in vivo (Figures 5G–5K).
Development of an enteroid culture system reveals MNoVCW3 can infect tuft cells in vitro
Small-animal models have helped elucidate the contributions of host and viral restriction factors that determine cellular tropism. However, a complete understanding of MNoV-tuft cell interactions is limited by the lack of an in vitro tuft cell culture system. Here, we overcome these technical limitations by generating an in vitro enteroid culture system that supports tuft cell development and MNoV replication. Murine enteroids recapitulate the mouse intestinal epithelium but lack hematopoietic and stromal compartments (Sato et al., 2009). We isolated intestinal stem cells from the small intestinal epithelium of adult wild-type C57BL/6J mice, expanded and differentiated enteroids, and stimulated tuft cell expansion with recombinant IL-4 (Figure 6A) (Gerbe et al., 2016; Howitt et al., 2016; Moltke et al., 2016). In mice, tuft cells express CD300lf on their apical surface, which faces the intestinal lumen and enables MNoVCR6 infection (Wilen et al., 2018). In the three-dimensional (3D) enteroid structure, the lumen and apical surface of tuft cells are situated facing inward (Figure 6A). As a result, CD300lf on tuft cells in 3D is inaccessible for MNoVCR6 binding and entry, which would render enteroids resistant to MNoVCR6 infection (Figure 6A). We resolve this constraining aspect of enteroid structure by polarizing the cells in a monolayer across a Transwell membrane, exposing the apical CD300lf-expressing surface of tuft cells to MNoV (Figure 6B). We added MNoVCR6 to the apical side of the monolayer cultures and observed productive infection measured by plaque assay. Viral production was higher in the apical, relative to basolateral, compartment, consistent with polarized release of progeny virus (Figure 6B). Infection was also IL-4-dependent, consistent with the ability of IL-4 to induce tuft cells both in enteroids and in mice (Figure 6B) (Wilen et al., 2018). By polarizing tuft cells in a monolayer on a Transwell, MNoVCR6 can productively infect enteroid cultures with growth kinetics peaking at 24 hours post-infection (hpi) and clearance between 48 and 72 hpi independent of viral inoculum (Figure 6C). Next, we leveraged this enteroid culture system to assess the role of IFNs in tuft cell infection independent of non-epithelial cells such as immune cells, which may regulate tuft cell tropism in vivo (Figures 6D and 6E). We pre-treated enteroids with type I, II, or III IFNs, and, as expected, we observed significant upregulation of canonical ISGs CXCL10 and ISG15 (Figure 6D). To determine the role of innate immune signaling on MNoV infection of intestinal tuft cells, we administered type I, II, and III IFNs to MNoV-infected enteroid cultures (Figure 6E). We observed that both MNoVCR6 and MNoVCW3 were highly susceptible to type I, II, and III IFNs (Figure 6E). More surprisingly, we found that MNoVCW3—which infects hematopoietic cells rather than tuft cells in immunocompetent mice—could also replicate in enteroids (Figure 6E). Furthermore, inoculation of Pou2f3−/− enteroids failed to support MNoV infection, demonstrating that tuft cells are required for both MNoVCR6 and MNoVCW3 to replicate in enteroid cultures (Figure 6F). This demonstrates that both persistent and non-persistent MNoV strains similarly replicate in tuft cells in vitro and exhibit comparable susceptibility to IFN signaling. Taken together, our data suggest that epithelial cell-extrinsic factors, likely in the form of type I and III IFNs acting via STAT1 signaling on tuft cells, restrict MNoVCW3 tropism for tuft cells in vivo.
Figure 6. Development of an enteroid culture system reveals MNoVCR6 and MNoVCW3 can infect tuft cells in vitro.
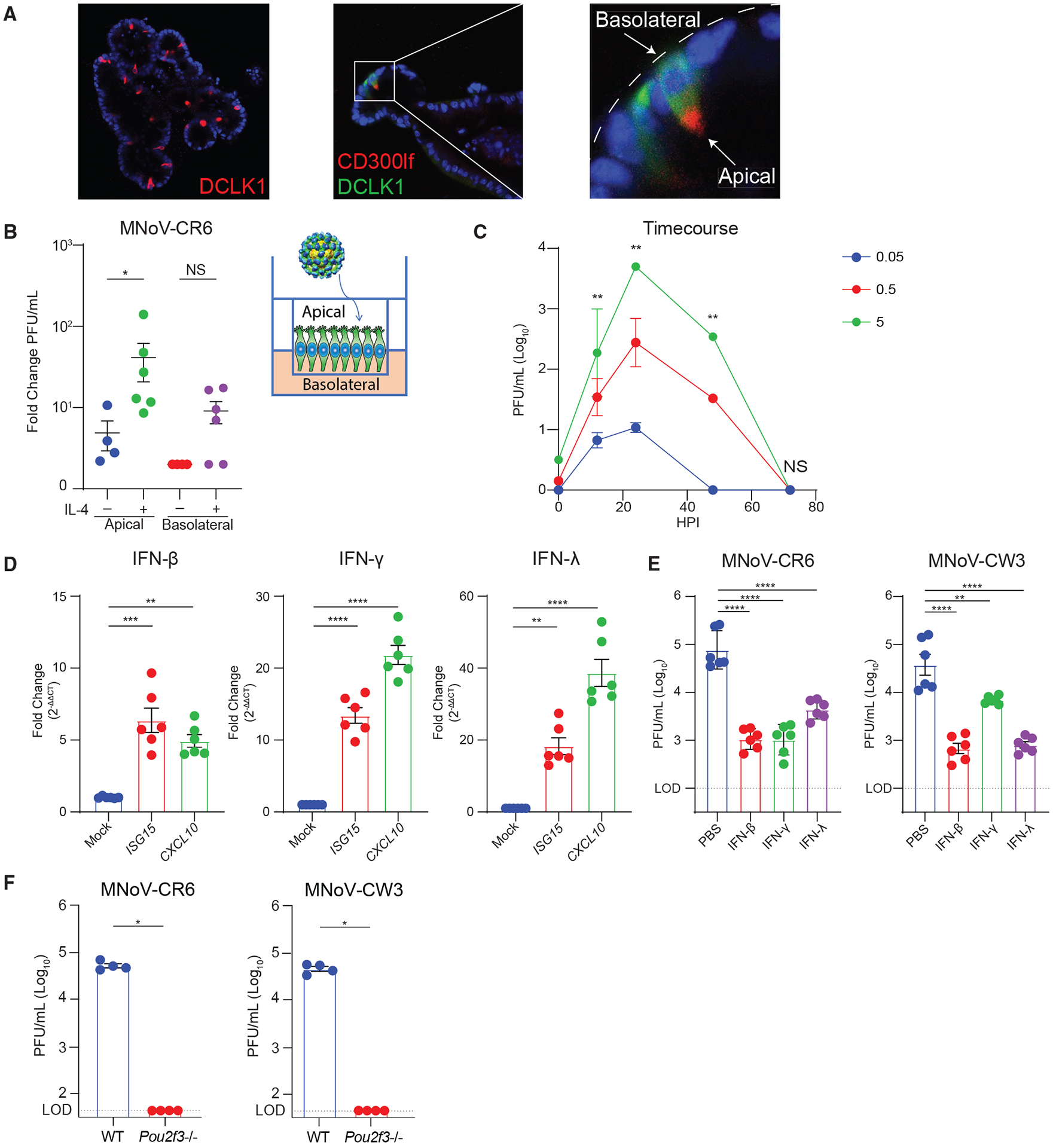
Intestinal epithelial stem cells (IECs) were isolated from crypts of wild-type mice. IECs were differentiated and treated with recombinant IL-4 (rIL-4) to induce tuft cell differentiation.
(A) Immunofluorescence staining for the tuft cell markers DCLK1 and CD300lf, showing apical distribution of CD300lf facing the interior lumen of the 2D enteroid. Images are representative of one of at least three independent experiments.
(B) Enteroids were polarized and differentiated in monolayers on Transwell inserts to expose the apical surface of tuft cells. Monolayers were infected with MNoVCR6 (MOI = 5) at the apical surface ± rIL-4 administered to the apical or basolateral surface. Data are shown as fold change (increase) in viral titer from 1 to 24 hours post-infection (hpi).
(C) Growth kinetics of MNoVCR6 (MOI = 0.05, 0.5, or 5) demonstrating efficient viral replication in the polarized tuft cell enteroid culture system.
(D) Enteroids are susceptible to ISG induction, shown by upregulation of ISG15 and CXCL10 after treatment with 1,000 ng/mL IFN-β1, IFN-γ, or IFN-λ for 24 h.
(E) Enteroids were pre-treated for 24 h with 1,000 ng/mL IFN-β1, IFN-γ, or IFN-λ and infected with MNoVCR6 or MNoVCW3.
(F) Tuft cells are required for infection by MNoVCR6 and MNoVCW3 in enteroids.
(D–F) Viral replication was quantified by plaque assay on BV2 cells at (D) 12, 24, 48, and 72 or (E and F) 24 hpi of enteroid culture.
Data are pooled from two to four independent experiments and analyzed by Mann-Whitney (C and F) and one-way ANOVA (D and E). Shown are means ± SEM. Statistical significance annotated as follows: NS, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; LOD, limit of detection.
DISCUSSION
Elucidating the mechanisms governing cellular tropism of NoV has important implications for viral pathogenesis and immunity. Cell tropism requires a cell to be both susceptible to viral entry (i.e., express the viral receptor) and permissive to intracellular viral replication, which necessitates overcoming host barriers to infection. We previously showed that the persistent strain MNoVCR6 infects CD300lf-expressing tuft cells in the intestinal epithelium and that tuft cell tropism determines augmented MNoVCR6 pathogenesis by type II cytokines (Orchard et al., 2016; Wilen et al., 2018). However, it was unknown whether tuft cells are essential for the pathogenesis of MNoVCR6 and other diverse NoV strains, what factors can regulate tuft cell tropism beyond expression of the CD300lf receptor, and what interactions exists between tuft cells and MNoVCW3. Here, we leverage a tuft cell-deficient mouse model to interrogate the role of tuft cells in MNoV pathogenesis. We demonstrate that tuft cells are required for oral infection by diverse persistent MNoV strains, including MNoVCR6, MNoVCR3, and MNoVCR7. In contrast, tuft cells are dispensable for oral MNoVCW3 and MNoVWu23 challenges. Tuft cells are also not essential for MNoVCR6 when the enteric transmission route is bypassed by i.p. inoculation. Extra-intestinal inoculation of MNoVCR6 results in pathogenesis similar to oral MNoVCW3 infection, characterized by non-persistent systemic infection rather than persistent enteric infection. The clearance of extra-intestinal MNoVCR6 is RAG1 dependent and thus mediated by adaptive immunity. We further demonstrate that inducing Cd300lf deletion on tuft cells cures infection with a half-life of approximately 5 days, high-lighting short-lived tuft cells as the primary reservoir for persistent MNoVCR6 infection and suggesting continuous re-infection of permissive tuft cells as a mechanism for persistent infection. Notably, these results indicate that virus shed from tuft cells must be reinfecting other tuft cells.
The mechanisms by which MNoVCR6 evades adaptive immunity are also unclear. MNoVCR6 can elicit antiviral CD8+ T cells and antibody responses, although neither are sufficient for viral clearance (Lee et al., 2019; Nice et al., 2013; Tomov et al., 2013, 2017). It was unclear why MNoVCR6 is inherently resistant to adaptive immune clearance or whether tuft cells are immunoprivileged, or some combination thereof. Here, we establish that after i.p. challenge, MNoVCR6 can be cleared by adaptive immunity. This finding demonstrates that MNoVCR6 is not inherently able to escape adaptive immunity and instead supports that tuft cells may serve as an immunoprivileged niche that mediates MNoVCR6 immune evasion (Lee et al., 2019; Nice et al., 2013; Tomov et al., 2013, 2017). The majority of tuft cells are post-mitotic with a half-life of approximately 4 to 5 days (Gerbe et al., 2011; Nakanishi et al., 2013; Westphalen et al., 2014). Long-lived DCLK1+ intestinal tuft cells marked with a half-life of >6 months have been described (Chandrakesan et al., 2015; Westphalen et al., 2014). Tuft cells with such longevity may offer a reservoir for persistent MNoVCR6 infection. However, we show that conditional deletion of Cd300lf from tuft cells after establishment of persistent infection is sufficient to cure mice in approximately 1 week. This suggests that serial infection of short-lived tuft cells rather than persistent infection of long-lived tuft cells is the major mechanism of MNoVCR6 persistence.
We then investigated the contribution of innate immunity to tuft-cell-mediated persistence using global or conditional Stat1 deletion. The transcription factor STAT1 controls systemic spread of MNoVCR6, and a single dose of recombinant IFN-λ can cure persistent MNoVCR6 infection (Nice et al., 2015). However, the cellular target(s) of IFN-λ, and whether innate immune control of tuft cell infection was tuft cell intrinsic or extrinsic, are unclear. To this end, we demonstrate that loss of Stat1 expands MNoVCR6 tissue tropism to extra-intestinal sites, whereas enteric infection and fecal shedding still depend on tuft cells. We also show that tuft cells are the direct target of exogenous IFN-λ in curing chronic MNoVCR6 infection. This supports a model in which tuft-cell-intrinsic innate immunity in combination with CD300lf expression determines tropism and host susceptibility to infection (Graziano et al., 2021). Persistent MNoV strains require tuft cells for enteric infection, and chronic infection of tuft cells is ineffectively controlled by innate and adaptive immunity, which invites the question of how tuft cell tropism enables immune evasion. In the case of innate immunity, caspase-mediated cleavage and secretion of MNoVCR6 NS1 is required for tuft cell infection of immunocompetent mice (Lee et al., 2019; Robinson et al., 2019). It is thought that secreted NS1 counteracts IFN-λ-mediated restriction of MNoVCR6 by globally suppressing intestinal IFN responses by a yet-to-be-determined mechanism (Lee et al., 2019). However, NS1/2 cleavage has also been shown to promote persistence independently of IFN antagonism (Robinson et al., 2019). Interestingly, our observation that MNoVCR6 is susceptible to IFN-λ in an enteroid culture system suggests that secreted NS1 is not simply counteracting IFN-λ activity on a tuft cell. Rather, the data are consistent with secreted NS1 preventing IFN induction from non-tuft cells.
We next addressed the role of tuft cells in MNoVCW3 pathogenesis. We previously demonstrated that tuft cells are not essential for MNoVCW3 in immunocompetent mice and here show that tuft cells do not contribute to MNoVCW3-induced lethality in the absence of Stat1 (Grau et al., 2020). Using a chimeric virus (MNoVCR6–VP1–CW3), we confirm that the capsid protein VP1 of MNoVCW3 drives lethality and tropism for non-tuft cells during oral infection, suggesting both host and viral determinants of cell and tissue tropism for MNoV (Robinson et al., 2019; Strong et al., 2012). Unexpectedly, while only persistent MNoV strains can detectably infect tuft cells in wild-type mice in vivo, our in vitro enteroid culture system supports similar productive infection by both acute (MNoVCW3) and persistent (MNoVCR6) strains, with both exhibiting sensitivity to type I, II, and III IFNs. These data raise the question as to why MNoVCW3 can infect tuft cells in vitro but not in vivo. Similar to MNoVCR6, MNoVCW3 uses CD300lf for entry (Graziano et al., 2020, 2021; Orchard et al., 2016). In multiple cell lines from different species, CD300lf expression is sufficient for infection, suggesting a post-entry restriction of MNoVCW3 in tuft cells in mice (Haga et al., 2016; Hosmillo et al., 2019; Orchard et al., 2016). Furthermore, we show that Stat1 deletion in tuft cells supports MNoVCW3 persistence in mice. MNoVCW3 induces a more potent IFN response in the MLNs and Peyer’s matches of mice compared with MNoVCR6, which is mediated, at least in part, by NS1/2 (Bouziat et al., 2018; Nice et al., 2015; Robinson et al., 2019). Similar to previous data, we show that ablating Ifnar1 in epithelial cells is not sufficient to support MNoVCW3 persistence (Nice et al., 2016). Instead, we demonstrate that combinatorial signaling through IFNAR1 and IFNLR1 on epithelial cells, rather than type I or III IFN alone, restricts MNoVCW3 persistence in vivo. Importantly, MNoVCW3 replication in enteroids, which lack hematopoietic cells, suggests that type I and III IFNs must derive from a non-epithelial source. Therefore, together our in vitro and in vivo data support a model in which MNoVCW3 induces a robust IFN response governed by NS1/2 and VP1 during infection of hematopoietic cells that acts in trans on tuft cells to restrict MNoVCW3 infection in vivo. However, the cellular source(s) of IFNs that restrict tuft cell tropism in vivo and whether these IFNs result from direct infection or bystander effects are important unanswered questions. It is also possible that additional contributors to MNoVCW3 persistence exist, given that MNoVCW3 failed to establish persistence in some IfnarF/F Ifnlr1F/F Villin Cre mice. Such factors include IFN-independent STAT1 signaling and variations in the microbiota (Baldridge et al., 2015). Nonetheless, robust MNoV replication in our reductionist enteroid model will enable future studies to further characterize host-pathogen interactions of infected tuft cells and between tuft cells and select immune cells via co-culture. Our work highlights that cellular tropism is determined by both viral receptor expression and immune determinants, namely IFNs, that restrict productive viral infection in a cell-type-specific manner.
These findings have important implications for the pathogenesis of other enteric viruses beyond MNoV. Specifically, rotavirus was recently demonstrated to infect tuft cells, yet rotavirus does not cause persistent infection in mice; this suggests that tuft cells are not a universal reservoir for viral persistence (Bomidi et al., 2021). Also, while the target cells for diverse HNoV strains are incompletely understood, HNoV can replicate in vitro in intestinal epithelial cells and, to a limited extent, in B cell-like lines (Ettayebi et al., 2016; Jones et al., 2015). Viral antigen-positive enterocytes and enteroendocrine cells have also been described from human biopsies (Green et al., 2020; Karandikar et al., 2016). Also, similar to MNoVCR6, HnoV can persistently and asymptomatically be shed in the stool (Atmar et al., 2008; Barrabeig et al., 2010; Schmid et al., 2011; Sukhrie et al., 2010). Whether HNoV can also infect tuft cells and whether mechanisms of persistence are shared between HNoV and MNoV represent important future directions.
Limitations of the study
Our study was performed primarily in mice and enteroids derived from the C57BL/6 background and has not been validated in other in-bred laboratory mouse strains. While we defined that type I and III IFNs act directly on tuft cells via STAT1 to restrict MNoVCW3 tropism, we did not identify the tissue or cellular source of IFNs or the causative ISGs. We also did not explore IFN-independent determinants of MNoV persistence, given that MNoVCW3 did not cause persistent infection in all IfnarF/F IfnlrF/F Villin Cre mice. Whether similar tropism patterns and mechanisms underlying viral persistence apply to HNoV remains to be seen.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources or reagents should be directed to and will be fulfilled by the lead contact, Craig B. Wilen (craig.wilen@yale.edu).
Materials availability
Pou2f3−/− (C57BL/6J-Pou2f3em1Cbwi/J, JAX Stock No. 037040) and Cd300lfF/F (C57BL/6?-Cd300lfem2Cbwi/J, JAX Stock No. 037039) mice generated in this study have been deposited at The Jackson Laboratory.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse strains and animal ethics
All mouse strains were derived from a C57BL/6J background (Jackson Laboratory) and bred in-house. Pou2f3−/− , Cd300lfF/F, Ifnar1F/F Villin Cre, and Ifnlr1F/F Villin Cre mice were generated as previously described (Baldridge et al., 2017; Grau et al., 2020; Graziano et al., 2021; Nice et al., 2016; Walker et al., 2021). Pou2f3−/− (C57BL/6J-Pou2f3em1Cbwi/J, JAX Stock No. 037040) and Cd300lfF/F (C57BL/6?-Cd300lfem2Cbwi/J, JAX Stock No. 037039) mice have been deposited at Jackson Laboratory. Pou2f3−/− mice were crossed to Stat1−/− (B6.129S(Cg)-Stat1tm1Div/J, Jackson Laboratory) and Cd300lfF/F mice were crossed to an existing tamoxifen inducible Dclk1 CreERT background (Graziano et al., 2021; Westphalen et al., 2014). Double Ifnar F/F Inflr1F/F Villin Cre mice were generated by crossing Ifnar1F/F to Ifnlr1F/F Villin Cre mice (Baldridge et al., 2017; Nice et al., 2016). All experiments employed littermate controls, were sex-balanced, and performed at least two independent times. Mice were infected at 6–10 weeks of age and co-housed unless otherwise indicated. Genotyping was performed by real-time PCR as previously described or by Transnetyx, Inc. (Orchard et al., 2016). The care and use of all animals were approved in accordance with the Yale Animal Resource Center and Institution Animal Care and Use Committee (#2021–20198) in agreement with the standards set by the Animal Welfare Act.
Viral Stocks.
Molecular clones of MNoVCW3 (GenBank accession EF014462.1) MNoVCR6 (GenBank accession JQ237823), and MNoVCR6–VP1–CW3 were generated as previously described and used to produce working stocks of infectious virus (Strong et al., 2012). Plasmids encoding infectious molecular clones were transfected into 293T cells (ATCC) to generate P0 stocks as described elsewhere (Arias et al., 2012; Orchard et al., 2016; Strong et al., 2012). Briefly, P1 stocks were produced by passaging the P0 stock in mouse microglial-like BV2 cells (gift of H.W. Virgin). The P1 stocks were inoculated onto BV2 cells at a multiplicity of infection (MOI) of 0.05 for 48 h and the cells were mechanically lysed by freeze/thaw at −80°C. Cellular debris was clarified by centrifugation at 1200 × g for 5 min (min). Supernatant was filtered through a 0.22 μm filter, and virus was concentrated through a 100,000 MWCO by tangential flow filtration or Amicon Ultra filtration. MNoVCR3 (GenBank accession EU004676.1), MNoVCR7 (GenBank accession EU004677), and MNoVWU23 (GenBank accession EU004668) were not derived from molecular clones. A seed stock previously generated in RAW cells was expanded once in BV2 cells, clarified, and concentrated as described above. Each stock titer was quantified by plaque assay as previously described in at least three independent replicates.
Crypt isolation and enteroid maintenance
The mouse ileum was harvested and cleared of fat and connective tissue from male or female mice. The tissue was flushed with cold 1X PBS, cut longitudinally, and vortexed in 1X PBS. The ileum was diced and crypts were freed from the small intestine by incubation at 37°C for 30 min in crypt culture medium (DMEM/F12 with HEPES (Sigma), 1X L-Glutamine (Gibco), 1X Penicillin/Streptomycin, 10% heat-inactivated FBS (VWR) supplemented with 2 mg/mL collagenase type I (Invitrogen) and 50 μM gentamicin (Sigma). Isolated crypts were filtered through a 40 μM cell strainer and washed with unsupplemented crypt culture medium. Crypts were pelleted at 500 × g for 5 min and washed once more to reduce contamination with mesenchymal cells. Purified crypts were pelleted at 500 × g for 5 min and suspended in 30 μL of Matrigel (Corning) per well of a 24-well tissue plate. Plates containing Matrigel containing enteroids were inverted and polymerized for ~10 min at 37°C. Then, 600 μL 50% L-WRN CM was added per well of a 24-well plate for culture. Media was changed every 2–3 days and enteroids were split 1:8 every 7 days. For 3D enteroid passage, culture media were aspirated and each well was added with 500 μL of 1X PBS with 5 mM EDTA. Matrigel/enteroid bubble was gently scratched off on the plate using a pipette tip and was transferred to a 15 mL conical tube. Harvested samples were centrifuged at 500 × g for 5 min and PBS-EDTA was aspirated as much as possible. 0.05% Trypsin-EDTA (Gibco) was added and tubes were placed in a 37°C water bath for ~1–2 min. Enteroids were pipetted and neutralized with DMEM with 10% FBS. Collected cells were spanned for 5 min at 500 × g and medium was aspirated gently. Cell pellets were resuspended in 30 μL Matrigel per well of a 24-well plate and grown in 50% L-WRN CM, maintained and processed as indicated below.
METHOD DETAILS
Immunofluorescence
Segments from the ileum of Pou2f3+/−, Pou2f3−/−, Cd300lf+/−, and Cd300lf−/− mice were harvested and fixed in 10% neutral buffered formalin. Tissue samples were processed and paraffin embedded. Afterward, slides were deparaffinized and antigen retrieval was performed using a citrate buffer (DAKO). Tissue samples were blocked for 1 h with 2% normal goat serum in 1X tris-buffered saline with Tween 20 (TBST). Primary antibodies were incubated overnight at 4°C. Tuft cells were detected using a DCLK1 antibody (1:200, Abcam), and epithelial cellular adhesion was shown by using the e-cadherin antibody (1:500, BD Bioscience). For differentiated 3D mouse ileal enteroids were harvested and fixed in 4% paraformaldehyde for ~30 min at RT. Cells were submitted for histology and processed as mentioned above. In addition to DCLK staining, CD300lf antibody (1:200, R&D Systems) was added to check expression and location of the MNoV receptor. The immunofluorescence was detected with Zeiss Axio Imager 2 microscope and photos were processed using the ZenPro program.
Viral quantification by plaque assay
BV2 cells were seeded into 6-well plates at a concentration of 2 × 106 cells/well in DMEM (Gibco), 10% Fetal Bovine Serum (FBS) (BioWest), 1% Penicillin/Streptomycin (Gibco) and incubated overnight at 37°C and 5% CO2. After 24 h, medium was removed and BV2 cells and each well was inoculated with individual serial dilutions of virus. Plates were gently rocked at room temperature for 1 h to allow virus binding. Inocula were then removed and 2 mL overlay medium (MEM) (Gibco) containing 10% FBS, 1% methylcellulose (Sigma), 1% HEPES (Gibco), 1% Glutamax (Gibco), and 1% Penicillin/Streptomycin) was added to each well. Plates were incubated for 48 h with overlay. To visualize plaques, the overlay medium was aspirated after 48 h and wells were stained with 1 mL crystal violet solution (20% ethanol and 0.2% (w/v) crystal violet (Sigma)) for 30 min at room temperature with gentle rocking.
In vivo MNoV infections, tamoxifen treatment, and IFN-λ administration
Mice were perorally (PO) inoculated with 106 plaque forming units (PFU) in 25 μL total volume. Intraperitoneal (IP) infections were administered via insulin syringe with 106 PFU MNoV in 200 μL in 1X phosphate buffered saline (PBS) (Gibco) and were injected into the left lower quadrant of the intraperitoneal cavity. To drive conditional Cd300lf deletion on tuft cells, mice were treated at chronic stages of infection (15–35 dpi) every other day for ten days via IP injection with 1 mg tamoxifen (Sigma) dissolved in 100 μL corn oil (Sigma). For mice treated with cytokines, mice were treated with 25 μg recombinant murine IFN-λ (R&D Systems) diluted 1X PBS via IP injection in a total volume of 200 μL.
Tissue processing and quantitative PCR
MNoV genome copies in fecal pellets and tissues were quantified as previously described (Orchard et al., 2016; Wilen et al., 2018). Briefly, intestinal tissues were harvested as 1 cm tissue from the distal small intestine or proximal colon. Viral RNA from fecal pellets was extracted according to the manufacturer’s protocol using the Quick-RNA Viral 96 kit (Zymo Research). Tissues, approximately 15–25mg each, were homogenized in TRIzol (Life Technologies) and RNA extraction was performed with Direct-zol RNA MiniPrep Plus according to manufacturer’s instructions (Zymo Research). A cDNA template was synthesized in two steps with 5 μL RNA, random hexamers, and Improm-II Reverse Transcriptase (Promega). TaqMan quantitative PCR (qPCR) was performed in technical duplicate or triplicate for each sample. Standard curves were generated using MNoV-specific oligonucleotides (Thermo Fisher Scientific): Probe: 5′ 6FAM-CGCTTTGGAACAATG-MGBNFQ 3’; Forward primer: 5′ CACGCCACCGATCTGTTCTG-3′; Reverse primer: 5′ GCGCTGCGCCATCACTC-3′. MNoV genome copies in each tissue were normalized to the housekeeping gene b-actin using murine b-actin-specific primer and probes as follows (IDT): Probe: 6-FAM-CACCAGTTC/ZEN/GCCATGGATGACGA-IABkFQ 3’; Forward primer: 5′ GCTCCTTCGTTGCCGGTCCA-3′; Reverse primer: 5′ TTGCACATGCCGGAGCCGTT-3′. The limit of detection for MNoV and b-actin were 10 and 100 copies/μl, respectively. In tissue samples with undetectable MNoV genome copies, values were plotted at 0.0001 copies relative to actin. In Ifnar1F/F, Ifnlr1F/F, Ifnar1F/F Ifnlr1F/F Villin Cre mice, MNoV genome copies were normalized to ribosomal protein S29 (Rps29) instead of b-actin using SYBR green qPCR with the following primers: Forward primer: 5′-AGC AGC TCT ACT GGA GTC ACC-3′; Reverse primer: 5′ AGG TCG CTT AGT CCA ACT TAA TG -3′ at a concentration of 0.2 μM in 1X Power SYBR green master mix (Applied Biosystems). Limit of detection was set to 1.9log10 based on Rps29. Cycling parameters were identical to those for TaqMan qPCR with the inclusion of an additional melting curve analysis.
Preparation of conditioned media for enteroid culture
Conditioned media for enteroids was prepared as described previously (Miyoshi and Stappenbeck, 2013; VanDussen et al., 2019). Briefly, L-WRN cells (gift from T. Stappenbeck) were cultured in high glucose DMEM containing 1% Penicillin/Streptomycin and 10% FBS (L-cell media). When confluent, cells were passaged and were transferred to 150 cm2 flasks containing 500 μg/mL G418 (Gibco) and 2mM hygromycin (InvivoGen). Cells were grown to be overly confluent. Then cells were washed twice with 20 mL of 1X PBS and once with 10 mL of primary culture media (Advanced DMEMF/12, 100X L-glutamine, 1.25%Penicillin/Streptomycin, 4% FBS). Fresh primary culture medium was added to each flask and incubated. Conditioned media (CM) was collected every 24 h. The CM fractions were stored at 4°C, pooled, clarified by centrifugation (500 × g for 5 min), and filtered (0.22 μM). To make the 50% L-WRN CM, the collected CM was mixed with an equal volume of fresh primary culture medium. CM were aliquoted and stored at −80°C until further use.
Transwell culture of mouse enteroids and MNoV enteroid infections
For the rIL-4 experiment, 3D enteroids were processed as described above. Trypsinized single cells were plated directly into the upper chamber TranswellⓇ inserts (CostarⓇ, 6.5 mm insert, 0.4 μm polyester membrane, 3470) and cultured with growth 50% L-WRN CM + 10 mM Y27632 in the upper and lower compartments for 7 days. Media were then removed from the upper chamber and cells were cultured at the air-liquid interface (ALI) for another 7 days, changing the media in the lower chamber every other day. After 7 days, fresh 50% L-WRN CM supplemented with or without 50 ng/mL rIL-4 was added to both the apical or basolateral chambers. ALI cultures were treated with or without rIL-4 for 4 days, changing media every other day. Cultures were infected apically with 5.625 × 106 PFU MNoVCR6 diluted in 50% L-WRN CM + Y27632 inhibitor (Tocris) and supplemented with 0.025% bovine bile (Sigma) in a total volume of 200 μL. Virus was adhered at 37°C for 1 h and then carefully removed by washing the upper and lower chambers 4x with 400 μL media. Then 200 μL and 500 μL fresh 50% L-WRN CM + Y27632 were added to the upper and lower compartments, respectively. From upper and lower compartments, 100 μL supernatant was harvested at 1 hpi and 24 hpi. Supernatants were serially diluted and virus replication was quantified by plaque assay.
For time course and interferon experiments, TranswellⓇ inserts were pre-coated with 100 μL of 10% Matrigel solution in sterile 1X PBS and incubated at 37°C for at ~20 min. Enteroids in Matrigel bubbles were dissociated to enable plating on TranswellⓇ. 2–3 wells from a 24-well plate 3D enteroid culture were used per TranswellⓇ insert. Briefly, 500 μL of PBS-EDTA was added per well of a 24-well plate, Matrigel/enteroid bubbles were scratched with a pipette tip, manually pipetted up and down several times, prior to transferring to a 15 mL conical tube. Cells were centrifuged at 500 × g for 5 min. The PBS-EDTA solution was aspirated and 200 ul of 0.05% pre-warmed Trypsin-EDTA was added per bubble. Cells were incubated at 37°C water bath for ~1–2 min. Spheroids were further disrupted by pipetting the cells up and down to fully dissociate cells. Trypsin was neutralized by adding DMEM containing 10% FBS. Cells were pelleted at 500 × g for 5 min and supernatant was aspirated. Single cells were suspended in 50% L-WRN CM with 10 mM Y27632. The Matrigel/PBS-EDTA solution was removed from the pre-incubated TranswellⓇ inserts, and 100 ul of suspended cells was added in the upper TranswellⓇ compartment and 600 ul of 50% L-WRN CM with 10 mM Y27632 in the lower compartment. The next day, fresh mouse IntestiCult Organoid Growth Medium (STEMCELL) was added into the upper and lower compartments of the TranswellⓇ. Switching from L-WRN CM to IntestiCult media was needed to enable tuft cell differentiation. Media were replaced every 1–2 days. Cells were maintained for ~14 days before use for experiments. For the time course experiment, cells were infected apically with 100 μL of MNoVCR6 at different MOIs (0.05, 0.5, 5), then unbound virus were washed after 1 h, harvested at 0, 12, 24, 48, 72 hpi and plaque assay was performed for virus titration. MOI was estimated based on 106 cells/ml from previous experiments. For the interferon experiment, cells were pre-treated with 1000 ng/mL of IFN-β1, IFN-γ and IFN-λ for 24 h, and infected with MOI = 5 of MNoVCR6 and MNoVCW3. Samples were collected at 24 hpi and plaque assay was performed.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed in Prism version 9 (San Diego, CA). Error bars indicate the standard error of the mean unless otherwise indicated. Normally distributed data was analyzed using unpaired Student’s t-tests while Mann-Whitney tests were performed for non-normally distributed data. Kaplan-Meier curves were utilized to analyze survival data. p values of <0.05 were considered statistically significant (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-DCLK1 (Rabbit polyclonal) | Abcam | Cat# ab37994 RRID:AB_873538 |
| Anti-CD300lf (Clone 305835) | R&D | Cat# MAB27881 RRID:AB_2074115 |
| Anti-E-cadherin (clone 36) | BD Bioscience | Cat# 610181 RRID:AB_397580 |
| Goat anti-rabbit AF488 | Jackson ImmunoResearch | Cat# 111-545-144 RRID:AB_2338052 |
| Goat anti-mouse AF594 | Jackson ImmunoResearch | Cat# 115-585-003 RRID:AB_2338871 |
| Goat anti-rat AF594 | Jackson ImmunoResearch | Cat# 112-585-167 RRID: AB_2338383 |
| Bacterial and virus strains | ||
| MNoVCW3 | Strong et al. (2012) | GenBank accession EF014462.1 |
| MNoVCR6 | Strong et al. (2012) | GenBank accession JQ237823 |
| MNoVCR6–VP1–CW3 | Strong et al. (2012) | N/A |
| MNoVCR3 | Thackray et al. (2007) | GenBank accession EU004676.1 |
| MNoVCR7 | Thackray et al. (2007) | GenBank accession EU004677 |
| MNoVWU23 | Thackray et al. (2007) | GenBank accession EU004668 |
| Chemicals, peptides, and recombinant proteins | ||
| 10X Target Antigen Retrieval Solution, DAKO | Agilent Pathology Solutions | Cat# S236784-2 |
| 10X Tris-Buffered Saline | Bio-Rad | Cat# P1706435 |
| Tween 20 | Sigma | Cat# P7949 |
| Normal Goat Serum | Jackson ImmunoReserach | Cat# 005-000-121 |
| 10% Neutral Buffered Formalin | Sigma | Cat# 252549 |
| 1X PBS | Gibco | Cat# 14287080 |
| MEM | Gibco | Cat# 11095072 |
| Methylcellulose | Sigma | Cat# M0511 |
| HEPES | Gibco | Cat# 15630080 |
| GlutaMax | Gibco | Cat# 35050061 |
| Penicillin/Streptomycin | Gibco | Cat# 15140122 |
| Gentamicin | Gibco | Cat# 10131035 |
| Hygromycin | InvivoGen | Cat# ant-hg-1 |
| Geneticin (G418 Sulfate) | Gibco | Cat# 10131035 |
| Fetal Bovine Serum | VWR | Cat# S1620 |
| DMEM | Gibco | Cat# 11966025 |
| DMEM, high glucose | Gibco | Cat# 11965092 |
| DMEMF/12 | Sigma | Cat# 51445C |
| L-Glutamine | Gibco | Cat# 25030081 |
| EDTA, 0.5M solution, pH 8.0 | AmericanBio | Cat# AB00502 |
| Trypsin-EDTA (0.05%) | Gibco | Cat# 25300062 |
| Trypsin (0.05%) | Gibco | Cat# 25300054 |
| Collagenase type I | ThermoFisher Scientific | Cat# 17018029 |
| Matrigel Matrix | Corning | Cat# 356234 |
| IntestiCult Organoid Growth Medium | STEMCELL Technologies | Cat# 06005 |
| Bovine bile | Sigma | Cat# B3883 |
| Crystal violet | Sigma | |
| Tamoxifen | Sigma-Aldrich | Cat# T5648 |
| Corn oil | Sigma | Cat# C8267 |
| Y27632 ROCK Inhibitor | Tocris | Cat# 1254 |
| Mouse IFN-λ2 | Peprotech | Cat# 250-33 |
| Mouse IFN-β1 | BioLegend | Cat# 581304 |
| Mouse IFN-γ | BioLegend | Cat# 575302 |
| Critical commercial assays | ||
| Quick RNA Viral 96 Kit | Zymo Research | Cat# R1041 |
| Direct-zol RNA MiniPrep Plus | Zymo Research | Cat# R2056 |
| TRIzol Reagent | Life Technologies | Cat# 15596018 |
| AmpliTaq Gold | Applied Biosystems | Cat# N8080241 |
| ImpromII Reverse Transcriptase | Promega | Cat# PRA3803 |
| SYBR Green Master Mix | Applied Biosystems | Cat# 4309155 |
| Experimental models: Cell lines | ||
| BV2 cells | Gift of Herbert Virgin | N/A |
| HEK293T cells | ATCC | Cat# CRL-11268 |
| L-WRN cells | Gift of T. Stappenbeck | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL6/J | Jackson Laboratory | Stock# 000664 RRID:IMSR_JAX:000664 |
| Mouse: Pou2f3−/− (C57BL/6J-Pou2f3em1Cbwi/J) | This study | Stock# 037040 |
| Mouse: CD300lfF/F (C57BL/6?-Cd300lfem2Cbwi/J) | This study | Stock# 037039 |
| Mouse: Dclk1 CreERT |
Graziano et al. (2021); Westphalen et al. (2014) |
N/A |
| Mouse: Villin Cre | Jackson Laboratory | Stock# 004586 RRID:IMSR_JAX:004586 |
| Mouse: Ifnar1F/F (B6(Cg)-Ifnar1tm1.1Ees/J) | Jackson Laboratory | Stock# 028256 RRID:IMSR_JAX:028256 |
| Mouse: Ifnlr1F/F (Ifnlr1tm1a(EUCOMM)Wtsi) | Baldridge et al. (2017) | N/A |
| Mouse: Rag1−/− (B6.129S7-Rag1tm1Mom/J) | Jackson Laboratory | Stock# 002216 RRID:IMSR_JAX:002216 |
| Mouse: Stat1−/−(B6.129S(Cg)-Stat1tm1Div/J) | Jackson Laboratory | Stock# 012606 RRID:IMSR_JAX:012606 |
| Mouse: CD300lf−/− (B6.CD300lfem1Cbwi/J) | Orchard et al. (2016) | N/A |
| Mouse: Dclk1 Cre |
Graziano et al. (2021); Westphalen et al. (2014) |
N/A |
| Mouse: Stat1F/F(B6;129S-Stat1tm1Mam/Mmjax) | Jackson Laboratory | Stock# 032054-JAX RRID:MMRRC_032054-JAX |
| Oligonucleotides | ||
| MNoV qPCR probe: 5′ 6FAM-CGCTTTGGAACAATG-MGBNFQ 3′ | ThermoFisher Scientific | N/A |
| MNoV qPCR forward primer: 5′ CACGCCACCGATCTGTTCTG-3′ | IDT | N/A |
| MNoV qPCR reverse primer: 5′ GCGCTGCGCCATCACTC −3′ | IDT | N/A |
| Actin qPCR probe: 6-FAM-CACCAGTTC/ZEN/GCCATGGATGACGA-IABkFQ 3 | IDT | N/A |
| Actin qPCR forward primer: 5′ GCTCCTTCGTTGCCGGTCCA-3′ | IDT | N/A |
| Actin qPCR reverse primer: 5′ TTGCACATGCCGGAGCCGTT-3′ | IDT | N/A |
| Rps29 qPCR forward primer: 5′- AGC AGC TCT ACT GGA GTC ACC-3′ | IDT | N/A |
| Rps29 qPCR reverse primer: 5′ AGGTCGCTTAG TCCAACTTAATG-3′ | IDT | N/A |
| Software and algorithms | ||
| Prism 9.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Zeiss Axio Imager 2 | Zeiss | N/A |
| ZenPro | Zeiss | https://www.zeiss.com/microscopy/en/products/software/zeiss-zen.html |
| Other | ||
| Transwell® 6.5 mm insert, 0.4 μm polyester membrane | Corning/Fisher Scientific | Cat# 3470 |
Highlights.
Diverse persistent MNoV strains require tuft cells for enteric infection
Serial infection of short-lived tuft cells enables MNoV immune escape
Tuft-cell-intrinsic STAT1 restricts tropism of non-persistent MNoVCW3
Type I and III interferons signal on tuft cells in trans to restrict MNoVCW3 tropism
ACKNOWLEDGMENTS
This work was supported by the Burroughs Wellcome Fund (C.B.W.); the Robert E. Leet and Clara Guthrie Patterson Trust (C.B.W.); the Smith Family Awards Program for Excellence in Biomedical Research (S.L.); NSF DGE1752134 (M.S.S.); and NIH grants K08 A1128043(C.B.W.), R01 AI148467 (C.B.W.), R00DK116666 (R.C.O.), R00 AI141683 (S.L.), R01 AI127552 (M.T.B.), and R01 AI139314 (M.T.B.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Agudo J, Park E, Rose S, Alibo E, Sweeney R, Dhainaut M, Kobayashi K, Sachidanandam R, Baccarini A, Merad M, and Brown B (2018). Quiescent tissue stem cells evade immune surveillance. Immunity 48, 271–285.e5. 10.1016/j.immuni.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias A, Bailey D, Chaudhry Y, and Goodfellow I (2012). Development of a reverse-genetics system for murine norovirus 3: long-term persistence occurs in the caecum and colon. J. Gen. Virol 93, 1432–1441. 10.1099/vir.0.042176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmar R, Opekun A, Gilger M, Estes M, Crawford S, Neill F, and Graham D (2008). Norwalk virus shedding after experimental human infection. Emerg. Infect. Dis 14, 1553–1557. 10.3201/eid1410.080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge M, Lee S, Brown J, McAllister N, Urbanek K, Dermody T, Nice N, and Virgin H (2017). Expression of Ifnlr1 on intestinal epithelial cells is critical to the antiviral effects of interferon lambda against norovirus and reovirus. J. Virol 91, e02079. 10.1128/JVI.02079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge M, Nice T, McCune B, Yokoyama C, Kambal A, Wheadon M, Diamond M, Ivanova Y, Artyomov M, and Virgin H (2015). Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science 347, 266–269. 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge M, Turula H, and Wobus C (2016). Norovirus regulation by host and microbe. Trends Mol. Med 22, 1047–1059. 10.1016/j.molmed.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrabeig I, Rovira A, Buesa J, Bartolomé R, Pintó R, Prellezo H, and Domínguez A (2010). Foodborne norovirus outbreak: the role of an asymptomatic food handler. BMC Infect. Dis 10, 269. 10.1186/1471-2334-10-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomidi C, Robertson M, Coarfa C, Estes M, and Blutt S (2021). Single-cell sequencing of rotavirus-infected intestinal epithelium reveals cell-type specific epithelial repair and tuft cell infection. Prod. of the Natl. Acad. Sci. USA 118, e2112814118. 10.1073/pnas.2112814118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouziat R, Biering S, Kouame E, Sangani K, Kang S, Ernest J, Varma M, Brown J, Urbanek K, Dermody T, et al. (2018). Murine norovirus infection induces T H 1 inflammatory responses to dietary antigens. Cell Host Microbe 24, 677–688.e5. 10.1016/j.chom.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrakesan P, May R, Qu D, Weygant N, Taylor V, Li J, Ali N, Sureban S, Qante M, Wang T, et al. (2015). Dclk1+ small intestinal epithelial tuft cells display the hallmarks of quiescence and self-renewal. Oncotarget 6, 30876–30886. 10.18632/oncotarget.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Penfield N, Ramani S, Estes M, and Atmar R (2017). Prospects and challenges in the development of a norovirus vaccine. Clin. Therapeut 39, 1537–1549. 10.1016/j.clinthera.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini V, Morantz E, Browne H, Ettayebi K, Zeng X, Atmar R, Estes M, and Vinjé J (2018). Human norovirus replication in human intestinal enteroids as model to evaluate virus inactivation. Emerg. Infect. Dis 24, 1453–1464. 10.3201/eid2408.180126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin J, Hackenmiller R, Simon M, and Levy D (1996). Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84, 443–450. 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- Ettayebi K, Crawford S, Murakami K, Broughman J, Karandikar U, Tenge T, Neill F, Blutt S, Zeng X, Qu L, et al. (2016). Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393. 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F, Es J.v., Makrini L, Brulin B, Mellitzer G, Robine S, Romagnolo B, Shroyer N, Bourgaux J, Pignodel C, et al. (2011). DistinctATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J. Cell Biol 192, 767–780. 10.1083/jcb.201010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F, Sidot E, Smyth D, Ohmoto M, Matsumoto I, Dardalhon V, Cesses P, Garnier L, Pouzolles M, Brulin B, et al. (2016). Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 529, 226–230. 10.1038/nature16527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau K, Roth A, Zhu S, Hernandez A, Colliou N, DiVita B, Philip D, Riffe C, Giasson B, Wallet S, et al. (2017). The major targets of acute norovirus infection are immune cells in the gut-associated lymphoid tissue. Nat. Microbiol 2, 1586–1591. 10.1038/s41564-017-0057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau K, Zhu S, Peterson S, Helm E, Philip D, Phillips M, Hernandez A, Turula H, Frasse P, Graziano V, et al. (2020). The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon. Nat. Microbiol 5, 84–92. 10.1038/s41564-019-0602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano V, Alfajaro M, Schmitz C, Filler R, Strine M, Wei J, Hsieh L, Baldridge M, Nice T, Lee S, et al. (2021). CD300lf conditional knockout mouse reveals strain-specific cellular tropism of murine norovirus. J. Virol 95, e01652–20. 10.1128/JVI.01652-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano V, Walker F, Kennedy E, Wei J, EttayebI K, Strine M, Filler R, Hassan E, Hsieh L, Kim A, et al. (2020). CD300lf is the primary physiologic receptor of murine norovirus but not human norovirus. PLoS Pathog. 16, e1008242. 10.1371/journal.ppat.1008242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K, Kaufman S, Nagata B, Chaimongkol N, Kim D, Levenson E, Tin C, Yardley A, Johnson J, Barletta A, et al. (2020). Human norovirus targets enteroendocrine epithelial cells in the small intestine. Nat. Commun 11, 2759. 10.1038/s41467-020-16491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K, Fujimoto A, Takai-Todaka R, Miki M, Doan Y, Murakami K, Yokoyama M, Murata K, Nakanishi A, and Katayama K (2016). Functional receptor molecules CD300lf and CD300ld within the CD300 family enable murine noroviruses to infect cells. Prod. of the Natl. Acad. Sci. USA 113, E6248–E6255. 10.1073/pnas.1605575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmillo M, Lu J, McAllaster M, Eaglesham J, Wang X, Emmott E, Domingues P, Chaudhry Y, Fitzmaurice T, Tung M, et al. (2019). Noroviruses subvert the core stress granule component G3BP1 to promote viral VPg-dependent translation. Elife 8, e46681. 10.7554/eLife.46681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt M, Lavoie S, Michaud M, Blum A, Tran S, Weinstock J, Gallini C, Redding K, Margolskee R, Osborne L, et al. (2016). Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science 351, 1329–1333. 10.1126/science.aaf1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MK, Grau KR, Costantini V, Kolawole AO, de Graaf M, Freiden P, Graves CL, Koopmans M, Wallet SM, Tibbetts SA, et al. (2015). Human norovirus culture in B cells. Nat. Protoc 10, 1939–1947. 10.1038/nprot.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karandikar U, Crawford S, Ajami N, Murakami K, Kou B, Ettayebi K, Papanicolaou G, Jongwutiwes U, Perales M, Shia J, et al. (2016). Detection of human norovirus in intestinal biopsies from immunocompromised transplant patients. J. Gen. Virol 97, 2291–2300. 10.1099/jgv.0.000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst S, Wobus C, Goodfellow I, Green K, and Virgin H (2014). Advances in Norovirus Biology. Cell Host Microbe 15, 668–680. 10.1016/j.chom.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst S, Wobus C, Lay M, Davidson J, and Virgin H (2003). STAT1-dependent innate immunity to a norwalk-like virus. Science 299, 1575–1578. 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- Karst S, Zhu S, and Goodfellow I (2015). The molecular pathology of noroviruses. J. Pathol 235, 206–216. 10.1002/path.4463. [DOI] [PubMed] [Google Scholar]
- Kitagawa Y, Tohya Y, Ike F, Kajita A, Park S, Ishii Y, Kyuwa S, and Yoshikawa Y (2010). Indirect ELISA and indirect immunofluorescent antibody assay for detecting the antibody against murine norovirus S7 in mice. Exp. Anim 59, 47–55. 10.1538/expanim.59.47. [DOI] [PubMed] [Google Scholar]
- Lee S, Liu H, Wilen C, Sychev Z, Desai C, Hykes B, Orchard R, McCune B, Kim K, Nice T, et al. (2019). A secreted viral nonstructural protein determines intestinal norovirus pathogenesis. Cell Host Microbe 25, 845–857.e5. 10.1016/j.chom.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Wilen C, Orvedahl A, McCune B, Kim K, Orchard R, Peterson S, Nice T, Baldridge M, and Virgin H (2017). Norovirus cell tropism is determined by combinatorial action of a viral non-structural protein and host cytokine. Cell Host Microbe 22, 449–459.e4. 10.1016/j.chom.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopman B, Steele D, Kirkwood C, and Parashar U (2016). The vast and varied global burden of norovirus: prospects for prevention and control. PLoS Med. 13, e1001999. 10.1371/journal.pmed.1001999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto I, Ohmoto M, Narukawa M, Yoshihara Y, and Abe K (2011). Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nat. Neurosci 14, 685–687. 10.1038/nn.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, and Stappenbeck T (2013). In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc 8, 2471–2482. 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moltke J.v., Ji M, Liang H, and Locksley R (2016). Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–225. 10.1038/nature16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Iacomini J, Johnson R, Herrup K, Tonegawa S, and Papaioannou V (1992). RAG-1-deficient mice have no mature B and T lymphocytes. Cell 68, 869–877. 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, Nakanishi N, Kanda K, Komekado H, Kawada M, et al. (2013). Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat. Genet 45, 98–103. 10.1038/ng.2481. [DOI] [PubMed] [Google Scholar]
- Nice T, Baldridge M, McCune B, Norman J, Lazear H, Artyomov M, Diamond M, and Virgin H (2015). Interferon-λ cures persistent murine norovirus infection in the absence of adaptive immunity. Science 347, 269–273. 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice T, Osborne L, Tomov V, Artis D, Wherry E, and Virgin H (2016). Type I interferon receptor deficiency in dendritic cells facilitates systemic murine norovirus persistence despite enhanced adaptive immunity. PLoS Pathog. 12, e1005684. 10.1371/journal.ppat.1005684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice T, Strong D, McCune B, Pohl C, and Virgin H (2013). A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. J. Virol 87, 327–334. 10.1128/JVI.01864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard R, Wilen C, Doench J, Baldridge M, McCune B, Lee Y, Lee S, Pruett-Miller S, Nelson C, Fremont D, and Virgin H (2016). Discovery of a proteinaceous cellular receptor for a norovirus. Science 353, 933–936. 10.1126/science.aaf1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson B, Winkle JV, McCune B, Peters A, and Nice T (2019). Caspase-mediated cleavage of murine norovirus NS1/2 potentiates apoptosis and is required for persistent infection of intestinal epithelial cells. PLoS Pathog. 15, e1007940. 10.1371/journal.ppat.1007940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries R, Snippert H, Wetering M.v.d., Barker N, Stange D, Es J.v., Abo A, Kujala P, Peters P, and Clevers H (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Schmid D, Kuo H, Hell M, Kasper S, Lederer I, Mikula C, Springer B, and Allerberger F (2011). Foodborne gastroenteritis outbreak in an Austrian healthcare facility caused by asymptomatic, norovirus-excreting kitchen staff. J. Hosp. Infect 77, 237–241. 10.1016/j.jhin.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Strine M, and Wilen C (2022). Tuft cells are key mediators of interkingdom interactions at mucosal barrier surfaces. PLoS Pathog. 18, e1010318. 10.1371/journal.ppat.1010318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong D, Thackray L, Smith T, and Virgin H (2012). Protruding domain of capsid protein is necessary and sufficient to determine murine norovirus replication and pathogenesis in vivo. J. Virol 86, 2950–2958. 10.1128/JVI.07038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhrie F, Siebenga J, Beersma M, and Koopmans M (2010). Chronic shedders as reservoir for nosocomial transmission of norovirus. J. Clin. Microbiol 48, 4303–4305. 10.1128/JCM.01308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray L, Duan E, Lazear H, Kambal A, Schreiber R, Diamond M, and Virgin H (2012). Critical role for interferon regulatory factor 3 (IRF-3) and IRF-7 in type I interferon-mediated control of murine norovirus replication. J. Virol 86, 13515–13523. 10.1128/JVI.01824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray L, Wobus C, Chachu K, Liu B, Alegre E, Henderson K, Kelley S, and Virgin H (2007). Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol 81, 10460–10473. 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomov V, Osborne L, Dolfi D, Sonnenberg G, Monticelli L, Mansfield K, Virgin H, Artis D, and Wherry E (2013). Persistent enteric murine norovirus infection is associated with functionally suboptimal virus-specific CD8 T cell responses. J. Virol 87, 7015–7031. 10.1128/JVI.03389-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomov V, Palko O, Lau C, Pattekar A, Sun Y, Tacheva R, Bengsch B, Manne S, Cosma G, Eisenlohr L, et al. (2017). Differentiation and protective capacity of virus-specific CD8 + T cells suggest murine norovirus persistence in an immune-privileged enteric niche. Immunity 47, 723–739.e5. 10.1016/j.immuni.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Winkle J, Robinson B, Peters A, Li L, Nouboussi R, Mack M, and Nice T (2018). Persistence of systemic murine norovirus is maintained by inflammatory recruitment of susceptible myeloid cells. Cell Host Microbe 24, 665–676.e4. 10.1016/j.chom.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDussen K, Sonnek N, and Stappenbeck T (2019). L-WRN conditioned medium for gastrointestinal epithelial stem cell culture shows replicable batch-to-batch activity levels across multiple research teams. Stem Cell Res. 37, 101430. 10.1016/j.scr.2019.101430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker F, Hassan E, Peterson S, Rodgers R, Schriefer L, Thompson C, Li Y, Kalugotla G, Blum-Johnston C, Lawrence D, et al. (2021). Norovirus evolution in immunodeficient mice reveals potentiated pathogenicity via a single nucleotide change in the viral capsid. PLoS Pathog. 17, e1009402. 10.1371/journal.ppat.1009402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J, Wobus C, Thackray L, Erexson C, Faucette L, Belliot G, Barron E, Sosnovtsev S, and Green K (2006). Pathology of immunodeficient mice with naturally occurring murine norovirus infection. Toxicol. Pathol 34, 708–715. 10.1080/01926230600918876. [DOI] [PubMed] [Google Scholar]
- Westphalen C, Asfaha S, Hayakawa Y, Takemoto Y, Lukin D, Nuber A, Brandtner A, Setlik W, Remotti H, Muley A, et al. (2014). Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J. Clin. Investig 124, 1283–1295. 10.1172/JCI73434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilen C, Lee S, Hsieh L, Orchard R, Desai C, Hykes B, McAllaster M, Balce D, Feehley T, Brestoff J, et al. (2018). Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science 360, 204–208. 10.1126/science.aar3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus C, Cunha J, Elftman M, and Kolawole A (2016). Viral Gastroenteritis: Chapter 3.4 Animal Models of Norovirus Infection (Academic Press; ). 10.1016/b978-0-12-802241-2.00019-5. [DOI] [Google Scholar]
- Wobus C, Thackray L, and Virgin H (2006). Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol 80, 5104–5112. 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Fu M, and Hu Q (2021). Advances in Human Norovirus Vaccine Research, Vaccines 9. 10.3390/vaccines9070732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Blutt S, Crawford S, Ettayebi K, Zeng X, Saxena K, Ramani S, Karandikar U, Zachos N, and Estes M (2019). Human intestinal enteroids: new models to study gastrointestinal virus infections. Methods Mol. Biol 1576, 229–247. 10.1007/7651_2017_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


