Background:
Direct cardiac reprogramming of fibroblasts into cardiomyocytes has emerged as a promising strategy to remuscularize injured myocardium. However, it is insufficient to generate functional induced cardiomyocytes from human fibroblasts using conventional reprogramming cocktails, and the underlying molecular mechanisms are not well studied.
Methods:
To discover potential missing factors for human direct reprogramming, we performed transcriptomic comparison between human induced cardiomyocytes and functional cardiomyocytes.
Results:
We identified TBX20 (T-box transcription factor 20) as the top cardiac gene that is unable to be activated by the MGT133 reprogramming cocktail (MEF2C, GATA4, TBX5, and miR-133). TBX20 is required for normal heart development and cardiac function in adult cardiomyocytes, yet its role in cardiac reprogramming remains undefined. We show that the addition of TBX20 to the MGT133 cocktail (MGT+TBX20) promotes cardiac reprogramming and activates genes associated with cardiac contractility, maturation, and ventricular heart. Human induced cardiomyocytes produced with MGT+TBX20 demonstrated more frequent beating, calcium oscillation, and higher energy metabolism as evidenced by increased mitochondria numbers and mitochondrial respiration. Mechanistically, comprehensive transcriptomic, chromatin occupancy, and epigenomic studies revealed that TBX20 colocalizes with MGT reprogramming factors at cardiac gene enhancers associated with heart contraction, promotes chromatin binding and co-occupancy of MGT factors at these loci, and synergizes with MGT for more robust activation of target gene transcription.
Conclusions:
TBX20 consolidates MGT cardiac reprogramming factors to activate cardiac enhancers to promote cardiac cell fate conversion. Human induced cardiomyocytes generated with TBX20 showed enhanced cardiac function in contractility and mitochondrial respiration.
Keywords: cellular reprogramming; fibroblasts; heart; myocytes, cardiac; regeneration; transcription factors
Clinical Perspective.
What Is New?
TBX20 enhances human cardiac reprogramming and improves contractility and mitochondrial function in the reprogrammed cardiomyocytes.
TBX20 binds to cardiac enhancers, promotes enhancer activity and chromatin accessibility, and activates transcription of cardiac contractility genes in reprogrammed cardiomyocytes.
TBX20 consolidates reprogramming factors MEF2C, GATA4, and TBX5 by enhancing their genome-wide DNA binding and co-occupancy at cardiac contractility genes.
What Are the Clinical Implications?
Enhancing the efficiency and quality of direct cardiac reprogramming from human fibroblasts is a critical step in the clinical translation of this technology.
Better understanding of the synergistic regulation of key cardiac transcription factors during reprogramming will provide new insights into the genetic basis in normal and diseased hearts.
In recent years, direct lineage conversion of cardiomyocytes from fibroblasts has been achieved by cellular reprogramming. Direct cardiac reprogramming induced by overexpression of cardiac transcriptional factors holds great promise to repair the injured heart because it exhibits dual therapeutic benefits by reducing scar tissue and generating new cardiomyocytes from existing fibroblasts to improve heart function.1–4 However, the direct reprogramming technique has been restricted when applied to human cells. Current cocktails for direct cardiac reprogramming in human fibroblasts suffer from low efficiency, slow rate of conversion, and insufficient production of functional cardiomyocytes.5–8
Besides the core reprogramming factors Gata4, Mef2c, and Tbx5, a variety of cocktail combinations have been reported to produce cardiomyocytes with different efficiency and quality,9,10 highlighting the power of combinatorial transcription control of cell fate. From a developmental perspective, it has been known that a set of transcriptional factors orchestrates cardiac gene programs, including contractility components, sarcomere proteins, and ion channel units. Transcriptional factors often act in concert and form tightly controlled networks, featuring common targets among different transcriptional factors.11,12 Therefore, missing one key component during heart development could lead to heart function defects and congenital heart disease.13 Such circumstances might cause incomplete function of cardiomyocytes in the direct reprograming process. We thus reasoned that an additional factor that governs transcriptional activation of gene programs associated with sarcomere contractility and ion channel might promote acquisition of cardiomyocyte identity and functions.
We used computational analyses of transcriptomic data6,14 to discover the missing key factors and identified TBX20 as the most underexpressed transcription factor in human induced cardiomyocytes (iCMs) compared with endogenous cardiomyocytes. The addition of TBX20 to the MGT133 cocktail (human MEF2C, GATA4, TBX5, and miR-133) not only led to enhanced sarcomere and contractility gene programs along with more defined well-organized myofibrils, but also resulted in functional improvements in contractility, calcium oscillation, and action potential of human iCMs. TBX20 also promoted mitochondria metabolism switch from glycolysis to β-oxidation metabolism phenotype, which more closely resembled the adult cardiomyocyte metabolism phenotype. Mechanistically, TBX20 colocalizes with MGT factors at enhancer elements and synergizes with MGT to activate contractility genes. Therefore, our study highlights the undocumented role of TBX20 as a vital regulator of direct human cardiac reprogramming.
Methods
Detailed materials and methods are provided in the Supplemental Material. The data, analytic methods, and study materials will be made available to other researchers for purposes of reproducing the results or replicating the procedure. No animals were used in this study. The Institutional Review Board of the University of Alabama at Birmingham reviewed our use of human cells in this study and provided a determination of not human subjects research.
Data Availability
All genomic datasets have been deposited to the Gene Expression Omnibus with accession number GSE193810 and are publicly available.
Statistical Analysis
Data were analyzed using GraphPad Prism 8 or the statistical language R (www.R-project.org) using RStudio. All results are reported as mean ± SEM. Data distribution and homogeneity of variance were tested using the Shapiro-Wilk and Levene tests, respectively. Differences between groups were examined for statistical significance using the Student t test or 1-way analysis of variance followed by the Dunnett multiple comparisons test (to a single control group) or the Tukey multiple comparisons test (among groups) where appropriate. For data violating the assumption of normal distribution, the Welch t test or Welch analysis of variance was used. For multiple group comparisons with >2 variables, 2-way analysis of variance was conducted, followed by the Tukey or Holm-Šídák multiple comparisons test. Differential gene expression in the single-cell RNA sequencing data was determined using a 2-sided Wilcoxon rank-sum test. The difference in single-cell RNA sequencing trajectory was determined by a Kolmogorov-Smirnov test. Overlapping enrichment analysis was determined by the Fisher exact test. Replicates and statistical tests are described in the figure legends. P<0.05 was considered significant.
Results
TBX20 Is a Key Regulator That Enhances Cardiac Direct Reprogramming
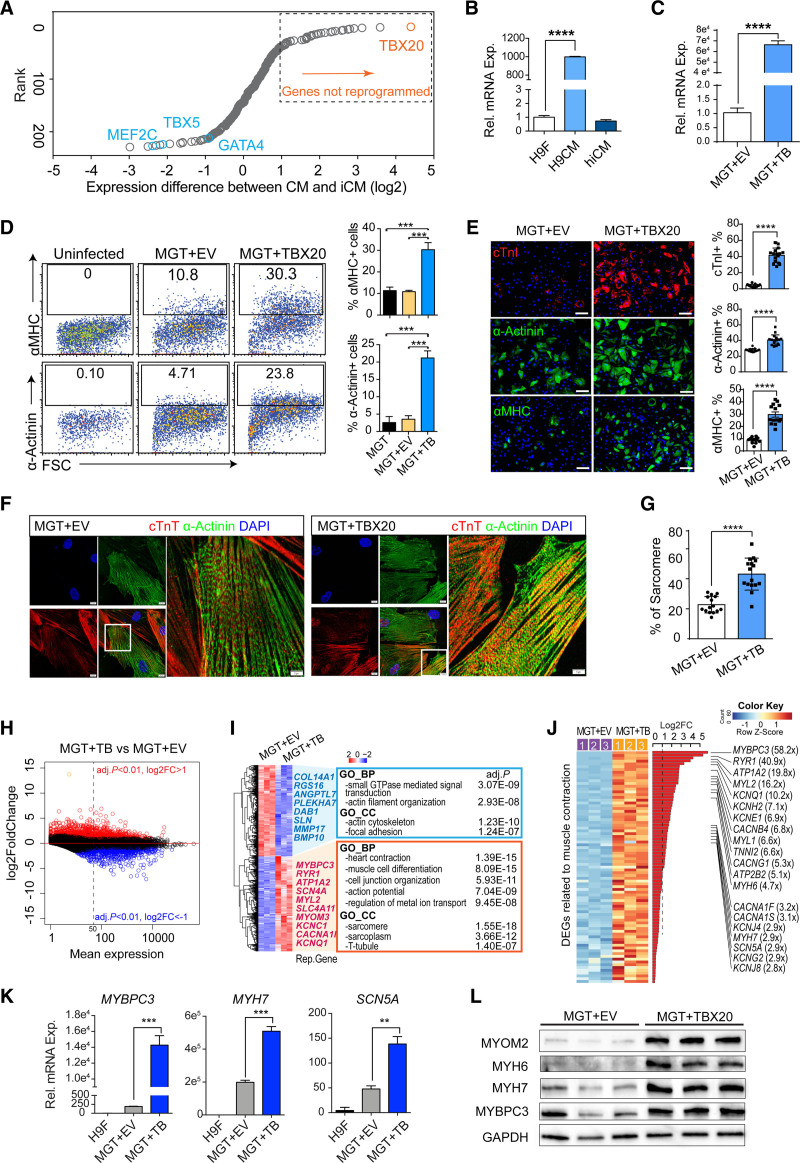
Through transcriptomic comparison, we identified that TBX20 was the most underexpressed factor during reprogramming (see Supplemental Methods, Figure S1A, and Table S1) and had roughly 100-fold lower expression in iCMs than in functional cardiomyocytes (Figure 1A and 1B). Single cell transcriptome analysis of human iCMs (hiCMs) induced by MGT+miR-133 (MGT133) also showed that TBX20 remained silenced throughout the whole process, whereas the alternative cardiogenic factor HAND2 was strongly activated (Figure S1B).
Figure 1.
TBX20 enhances human direct cardiac reprogramming and promotes sarcomere gene programs. A, Dot plot ranking the genes differentially expressed in reprogramming human induced cardiomyocytes (hiCMs) when compared with functional cardiomyocytes (GSE49192; Table S1). TBX20 is the top hit, showing the greatest difference. B, Reverse transcription quantitative polymerase chain reaction validation of TBX20 expression in H9-derived fibroblasts (H9Fs), H9-derived cardiomyocytes (H9CMs), and MGT133-transduced hiCMs (n=4 per group). C, Reverse transcription quantitative polymerase chain reaction analysis of TBX20 expression after transduction of TBX20 or control empty vector (EV) together with MGT133 in H9Fs (n=4 per group). D, Representative flow plots and quantification of α-MHC (α-myosin heavy chain) or α-actinin expression in H9Fs after 14 days of transduction (n=3 per group). E, Immunofluorescence staining images and quantification of cardiac troponin I (cTnI)+, α-actinin+, and α-MHC+ cells in hiCMs generated from H9Fs. Scale bars, 100 μm (n=20 per group). F, Representative confocal images showing sarcomere structures in cardiac troponin T (cTnT)+ and α-actinin+ hiCMs derived from H9F. Scale bars, 10 μm. G, Percentage of cells with visible sarcomeres in cTnT+ hiCMs derived from H9F (n=20 per group). H, MA plot representation of the DEGs in hiCMs upon TBX20 overexpression. Red and blue dots represent the significant upregulated or downregulated differentially expressed genes (DEGs; adjusted P<0.01; log2 fold change [abs] >1). I, Transcriptomic analysis identified the top 500 DEGs between TBX20-infected and EV-infected hiCMs. Representative genes and Gene Ontology terms enriched in downregulated (blue) and upregulated genes (red) were listed. J, Heatmap expression of DEGs associated with cardiac muscle contraction in TBX20-overexpressed and control hiCMs at reprogramming day 14. K, Reverse transcription quantitative polymerase chain reaction analysis showing mRNA expression of MYBPC3, MYH7, and SCN5A in H9Fs or hiCMs generated with or without TBX20 (n=4 per group). L, Western blotting of selected cardiac markers MYOM2, MYH6, MYH7, and MYBPC3 in hiCMs transduced with or without TBX20 from H9F. GAPDH serves as a loading control. All data are expressed as mean ± SEM (C, E, and G, Student t test; B, D, and K, 1-way analysis of variance). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
We next overexpressed TBX20 in H9-derived fibroblasts (H9Fs) together with MGT133 (Figure 1C) and evaluated reprogramming efficiency. MGT+TBX20 led to >20% of cells expressing α-MHC (α-myosin heavy chain) or cardiac α-actinin, which represented 3- to 6-fold more cells when compared with the empty vector (EV)–treated group (MGT+EV; Figure 1D). We also used EGFP (enhanced green fluorescent protein)–fused TBX20 to indicate TBX20-transduced cells and found that TBX20 overexpression substantially promoted reprogramming in TBX20-expressed EGFP+ cells but not in EGFP– TBX20-null cells (Figure S1C). Immunofluorescence staining using the adult cardiomyocyte marker cardiac troponin I showed a ~10-fold increase of cardiac troponin I+ cells after adding TBX20 (~40%; Figure 1E). Reverse transcription quantitative polymerase chain reaction analysis of additional maturation markers showed consistent results (Figure S1D). Moreover, higher percentages of cells exhibited visible sarcomere structures in MGT+TBX20 hiCMs (43.2% versus 22.7%; P<0.0001; Figure 1F and 1G). TBX20 also led to more robust cardiac reprogramming in human dermal fibroblasts, as evidenced by cardiac gene expression (Figure S1E and S1F). Therefore, we demonstrated that TBX20 enhances cardiac reprogramming and promotes sarcomere formation in hiCMs.
To understand the overall effect of TBX20 on reprogramming, we performed RNA sequencing (RNA-seq) using H9F-derived hiCMs. We identified 991 TBX20 upregulated genes and 728 TBX20 downregulated genes (adjusted P<0.01, fold change >2). Many genes with relatively low expression levels in MGT+EV-transduced hiCMs (mean expression <50) were activated by TBX20 (Figure 1H), as exemplified by MYH7 and MYBPC3 (Figure S1G). Gene Ontology analysis for the top 500 most differentially expressed genes (DEGs) showed that the upregulated genes were highly associated with cardiac functions, such as heart contraction and sarcomere (Figure 1I). Furthermore, the heatmap of DEGs associated with cardiac muscle contractility (Figure 1J) and the gene set enrichment analysis demonstrated the enhanced cardiac gene programs in MGT+TBX20 hiCMs (Figure 1J and Figure S1H). The increase of sarcomere genes was validated further by reverse transcription quantitative polymerase chain reaction and Western blot analyses (Figure 1K and 1L and Figure S1I). Taken together, these data demonstrate that TBX20 promotes cardiac reprogramming through global activation of cardiac genes related to sarcomere structure, ion channels, and heart contractions.
TBX20 Is Essential for Efficient hiCM Conversion From Cardiac Fibroblasts
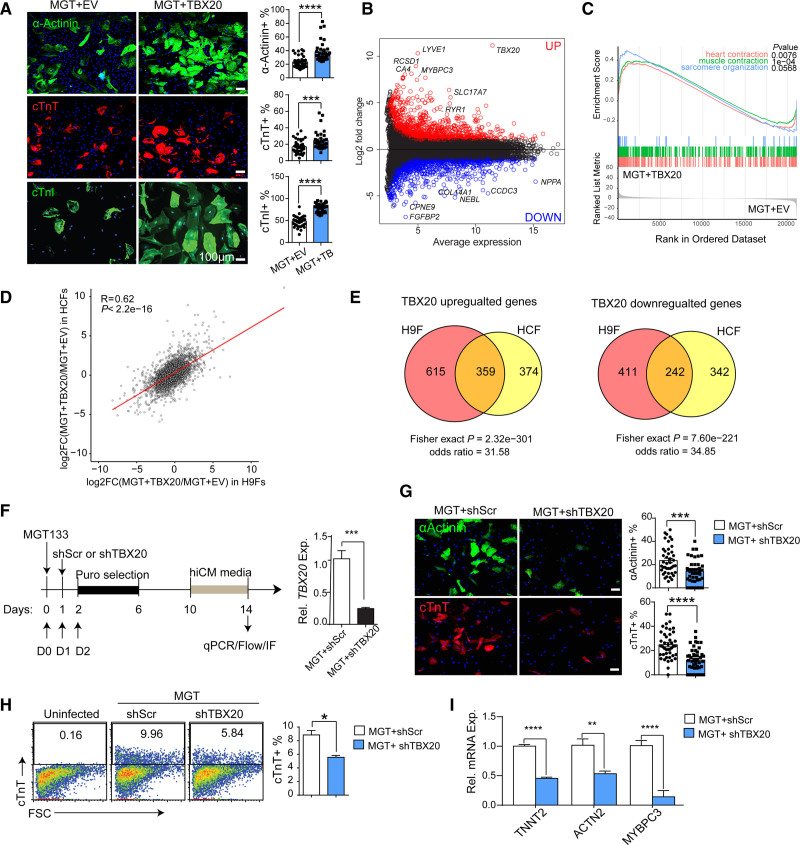
Next, we evaluated the role of TBX20 in iCM conversion of primary ventricular human cardiac fibroblasts (HCFs), the primary cell type of in vivo reprogramming. Previous reports showed that TBX20 is expressed in HCFs,15 which may also provide a unique opportunity to evaluate the requirement of TBX20 in direct reprogramming. First, we confirmed that TBX20 protein was detectable in HCFs (Figure S2A). TBX20 mRNA levels were ~30-fold higher in HCFs than in H9Fs or human dermal fibroblasts, where TBX20 was not expressed (Figure S2B and S2C), whereas H9-differentiated cardiomyocytes had ~1000-fold higher TBX20 expression than H9Fs (Figure 1B). Similar to our observation in H9Fs, MGT transduction was insufficient to upregulate TBX20 in HCFs (Figure S2D). Therefore, we performed both gain- and loss-of-function assays in HCFs. TBX20 overexpression led to further enhancement of hiCM reprogramming in HCFs with increased sarcomere proteins and global activation of cardiac gene programs (Figure 2A through 2C and Figure S2E and S2F). Furthermore, we showed a global correlation and significant overlaps of mRNA changes induced by TBX20 in H9Fs and HCFs (Figure 2D and 2E). To investigate whether TBX20 is essential in hiCM reprogramming, we performed shRNA-mediated TBX20 knockdown during HCF-derived reprogramming (Figure 2F). Knockdown of TBX20 greatly impaired reprogramming, indicated by decreased expression of cardiac genes (Figure 2G through 2I and Figure S2G). To further exclude the potential off-target effects of shRNA oligos, we applied shTBX20 in H9F cells where TBX20 was not expressed (Figure S2A through S2D). shTBX20 did not affect reprogramming efficiency in TBX20-null cells (Figure S2H through S2J). CRISPR/Cas9-mediated knockout of TBX20 also led to the reduction of sarcomere genes in HCF-derived hiCMs (Figure S2K and S2L). Therefore, TBX20 was demonstrated as an essential factor for efficient cardiac reprogramming and has the potential for therapeutic translation.
Figure 2.
TBX20 regulates hiCM generation from HCFs. A, TBX20 or empty vector (EV) was transduced along with MGT133 in human cardiac fibroblasts (HCFs). Reprogramming efficiency was measured at reprogramming day 14 by immunofluorescence (IF) staining and quantification of cardiac marker expression (n=20 per group). Scale bars, 100 μm. B, MA plot showing fold change and average gene expression between EV- or TBX20-treated human induced cardiomyocytes (hiCMs) derived from HCFs. Differentially expressed genes (DEGs) were defined by fold change >2 and adjusted P<0.01. C, Gene set enrichment analysis of DEGs using gene lists from Gene Ontology terms associated with cardiac muscle contractility. D, RNA sequencing scatterplot analysis comparing fold change of mRNA after TBX20 overexpression in H9-derived fibroblasts (H9Fs) and HCFs. Pearson correlation coefficient and P value are indicated. E, Venn diagrams showing significant overlap between upregulated and downregulated genes in H9F and HCF. P value and odds ratios were determined by Fisher exact test. F, Schematic and reverse transcription quantitative polymerase chain reaction (qPCR) validation of the shRNA-mediated TBX20 knockdown experiment. G, Representative images and quantification of immunofluorescence staining for α-actinin+ and cardiac troponin T (cTnT)+ cells after TBX20 knockdown in HCF-derived reprogramming cells (n=20 per group). H, Flow plots and quantification of cTnT expression in HCFs 14 days after MGT133 transduction with or without TBX20 knockdown (n=3 per group). I, Reverse transcription quantitative polymerase chain reaction evaluation of cardiac marker genes TNNT2, ACTN2, and MYBPC3 after TBX20 knockdown in hiCMs derived from HCFs (n=4 per group). All data are expressed as mean ± SEM (A, F through I, Student t test). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. cTnI indicates cardiac troponin I.
TBX20 Induces Stage-Specific Changes of Transcriptome During Reprogramming
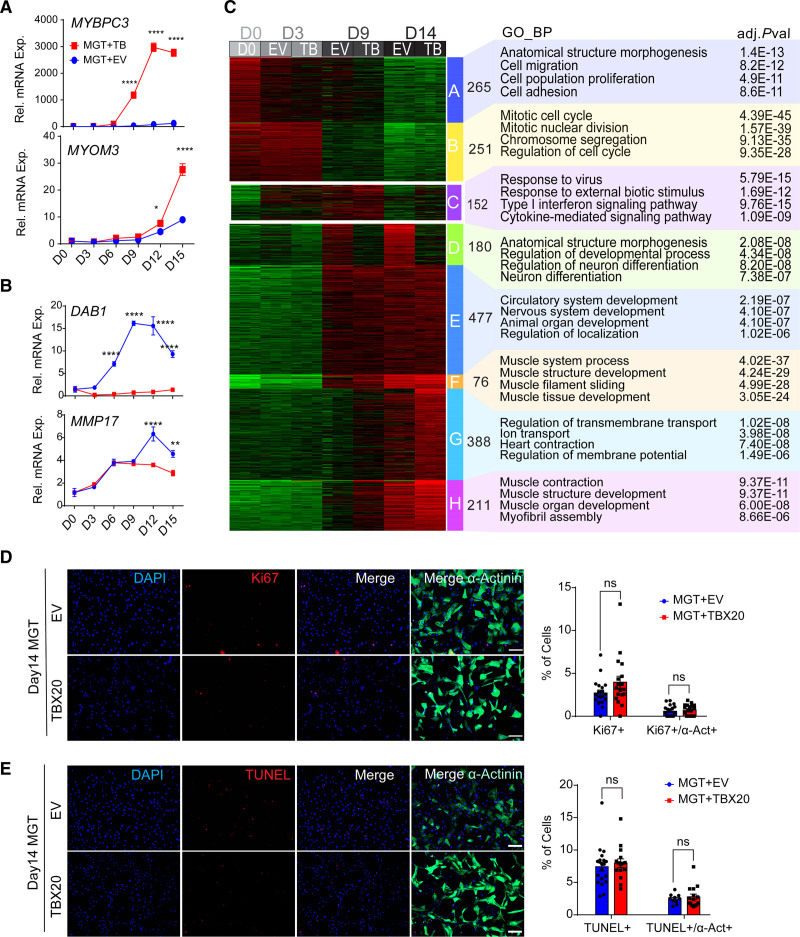
To gain a comprehensive view of the effect of TBX20 during the reprogramming process, we performed a time course gene expression analysis. We found enhanced effect of TBX20 on DEGs when reprogramming progressed (Figure 3A and 3B and Figure S3A). The number of DEGs (false discovery rate–adjusted P<0.01 and fold change >2) at reprogramming days 3, 9, and 14, both upregulated and downregulated, increased over the time of reprogramming (Figure S3B). K-means clustering analysis revealed 8 clusters of gene expression patterns, which were subsequently annotated by Gene Ontology (Figure 3C). Cluster A and B genes, which showed early repression by MGT, were enriched in cell adhesion and mitotic cell cycle. Cluster E and F genes, which were induced gradually by MGT, were associated with muscle structure development. Cluster G and H genes represented TBX20-activated genes from day 9 and were enriched in ion transport and muscle contraction. We also found that cluster C has an up-and-down expression pattern, suggesting a transient activation of genes, and these genes were involved in response to virus and type I interferon signaling pathways at the middle stage of reprogramming. Cluster D genes were suppressed continuously by TBX20 and, surprisingly, were associated with neuron differentiation. Therefore, our time course analyses suggest that TBX20 mainly functions as a transcriptional activator at the late stage of reprogramming.
Figure 3.
Time course evaluation of MGT+TBX20-induced reprogramming. A, Time course reverse transcription quantitative polymerase chain reaction to evaluate relative mRNA expression of TBX20 upregulated genes, including MYBPC3 and MYOM3, during cardiac reprogramming from H9-derived fibroblasts (H9Fs; n=4 for each point). B, Time course reverse transcription quantitative polymerase chain reaction to evaluate relative mRNA expression of TBX20 downregulated genes, including DAB1 and MMP17 (n=4 for each point). C, Time course RNA sequencing analysis of MGT+TBX20 and MGT + empty vector (EV) infected H9Fs at day 0, 3, 9, and 14 of reprogramming. Left, heatmap showing 8 clusters (A through H) of genes with distinct expression patterns during reprogramming. Right, top Gene Ontology biological processes enriched in each cluster. D, Immunofluorescence staining images and quantification of Ki67+ or Ki67+/α-actinin+ cells in human induced cardiomyocytes (hiCMs) derived from H9Fs with MGT+EV or MGT+TBX20 (n=20 per group). E, immunofluorescence staining and TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) analysis for TUNEL+ or TUNEL+/α-actinin+ cells in H9Fs at 14 days induced with MGT+EV or MGT+TBX20 (n=20 per group). All data are expressed as mean ± SEM (A and B, 2-way analysis of variance; D and E, Student t test). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. ns indicates not significant.
Although we found repressed cell cycle genes along with reprogramming in clusters A and B, it is unknown whether TBX20 increases hiCM percentage through its reported role in promoting cardiomyocyte proliferation.16–18 We thus evaluated the effect of TBX20 on cell proliferation and apoptosis at different time points of reprogramming. TBX20 overexpression did not increase cell proliferation throughout the reprogramming process in either α-actinin+ or all cultured cells, as indicated by the expression of proliferation marker Ki67 (Figure 3D and Figure S3C). Meanwhile, TBX20 overexpression did not inhibit cell apoptosis, as measured by TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling staining; Figure 3E and Figure S3D). These data suggest that TBX20 enhances cardiac reprogramming through mechanisms independent of its known role in promoting cardiomyocyte proliferation.
TBX20 Enhances Calcium Flux, Contractility, and Mitochondrial Function in hiCMs
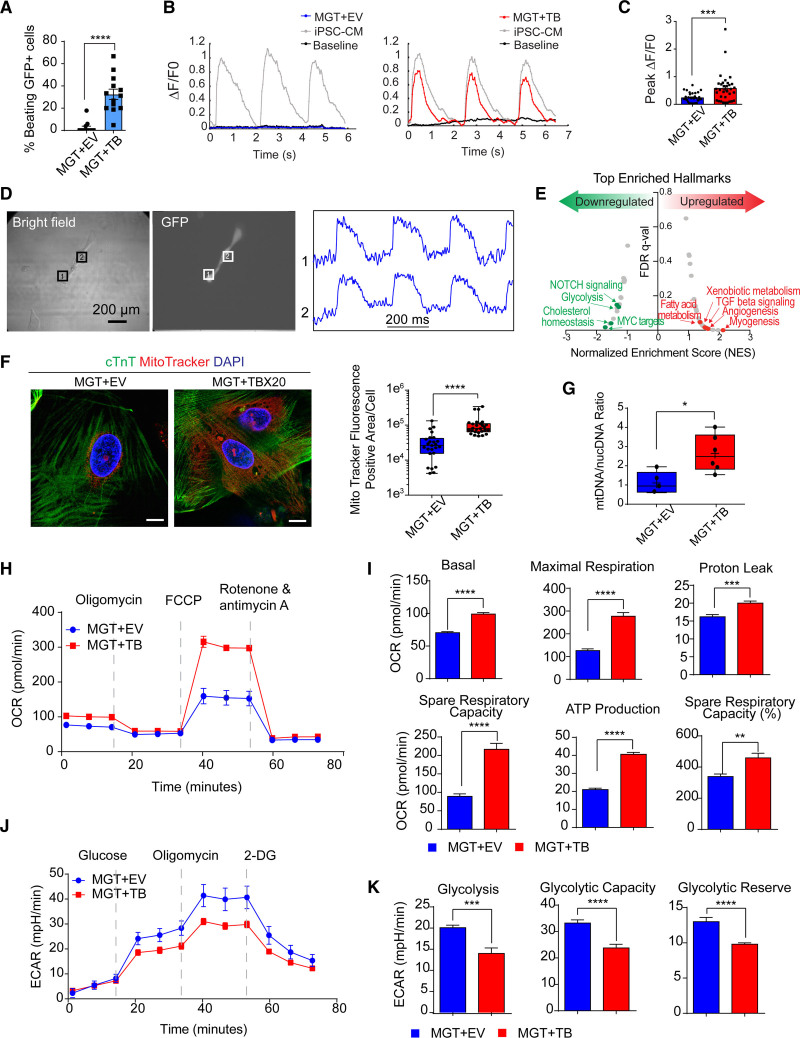
To validate the role of TBX20 on cardiac contractility functionally, we used a previously described protocol5–7 that cocultures hiCMs together with spontaneously beating cardiomyocytes derived from human induced pluripotent stem cells (hiPSC-CMs; Figure S4A). We labeled hiCMs with lentiviral GFP (green fluorescent protein) and seeded GFP+ hiCMs with beating iPSC-CMs together in a monolayer culture. After coculture for 1 month, we counted beating GFP+ cells and traced the calcium flux in hiCMs generated with or without TBX20. We observed 32.50±4.61% of GFP+ cells showing spontaneous beating in the TBX20 group versus only 2.29±1.52% of cells contracted in the control treatment (Figure 4A and Video S1). Moreover, the beating GFP+ hiCMs exhibited calcium transients in the coculture experiments (Figure 4B and Videos S2 and S3). MGT+TBX20 resulted in substantially higher peak amplitude than MGT+EV and the peak level is comparable with that in the cocultured hiPSC-CMs (Figure 4B and 4C). We also detected calcium flux in H9F- or HCF-derived hiCMs without coculture and observed increased peak amplitude after TBX20 overexpression (Figure S4B). Action potential was detected in MGT+TBX20 hiCMs isolated by puromycin selection (Figure 4D and Figure S4A). The average spontaneous conduction length from several batches was recorded as 581±436.99 ms with an average action potential duration at 50% of repolarization of 304.6±172.38 ms and an average action potential duration at 80% of repolarization of 357.8±201.17 ms (Figure S4C). In contrast, no action potential was captured in any hiCM generated with MGT+EV. These results collectively suggest that TBX20 functionally improves hiCMs by promoting calcium flux and contractility.
Figure 4.
TBX20 overexpression leads to functional improvement of hiCMs. A, Quantification of observed beating human induced cardiomyocytes (hiCMs) derived from H9-derived fibroblasts (H9Fs) after 1 month of co-culture with human induced pluripotent stem cell–derived cardiomyocytes (iPSC-CMs; n=10 for each group, repeated 2 times independently). B, Tracing and measurements from microscopy imaging show calcium transients in MGT + empty vector (EV) or MGT+TBX20 co-cultured cells as indicated in A. C, Quantification of measured peak ΔF/F0 in co-cultured hiCMs (n=33, repeated 2 times independently). D, Bright field and GFP (green fluorescent protein) fluorescence images showing MGT+TBX20-induced individual hiCMs after co-culture. Right panel shows the selected optical traces of membrane potential (Vm) during spontaneous beating with a cycle length of 270 ms. E, Gene set enrichment analysis of RNA sequencing data from H9F-derived hiCMs showing the enriched hallmarks in TBX20 upregulated and downregulated genes. F, Representative images and quantification of mitochondria content labeled using MitoTracker in cardiac troponin T (cTnT)+ hiCMs derived from H9F (n=20 per group). Scale bars, 10 μm. G, Reverse transcription quantitative polymerase chain reaction results showing the relative ratio of mitochondria DNA to the total genomic DNA in H9F-derived hiCMs (n=6 per group). H, Representative oxygen consumption rate profile analyzed from mito-stress Seahorse assay in hiCMs derived from H9Fs (n=10 per group, repeated 3 times independently). I, Quantification of basal respiration, maximal respiration, proton leak, spare respiratory capacity, adenosine triphosphate (ATP) production, and spare respiratory capacity as a percentage in hiCMs treated with MGT+TBX20 or MGT+EV from H9Fs (n=10 per group). J, Representative extracellular acidification rate profile analyzed from glucose-stress Seahorse assay in TBX20-treated or EV-treated hiCMs derived from H9Fs (n=10 per group). K, Quantification of glycolysis, glycolytic capacity, and glycolytic reserve in H9F-derived hiCMs transduced with TBX20 or EV (n=10 per group). All data are expressed as mean ± SEM (A, C, F, G, I, and K, Student t test). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. 2-DG indicates 2-deoxy-glucose; ECAR‚ extracellular acidification rate; FCCP‚ fluoro-carbonyl cyanide phenylhydrazone; FDR, false discovery rate; and OCR, oxygen consumption rate.
Mitochondria function is critical for cardiomyocyte maturation and is the energetic source for contractility.19 Thus, we sought to investigate whether the mitochondrial function was improved along with enhanced contractility during MGT+TBX20-induced reprogramming. Gene set enrichment analysis of RNA-seq data identified a transcriptional alteration of metabolism genes after TBX20 overexpression. The hallmark of glycolysis was enriched in TBX20 downregulated genes, whereas TBX20 upregulated genes were highly associated with fatty acid metabolism, xenobiotic metabolism, and myogenesis (Figure 4E and Figure S4D). Next, we labeled mitochondria in cardiac troponin T+ hiCMs with MitoTracker and found greatly increased MitoTracker-positive area in MGT+TBX20 hiCMs (Figure 4F). The evaluation of mitochondrial DNA copy number by quantitative polymerase chain reaction also demonstrated increased mitochondrial number in the presence of TBX20 (Figure 4G). Furthermore, we used HCFs for reprogramming and observed increased mitochondrial volume after TBX20 overexpression (Figure S4E) but reduced MitoTracker signals upon TBX20 knockdown (Figure S4F). To examine the metabolism phenotypes associated with mitochondrial changes, we performed Seahorse metabolic flux assays of hiCMs derived from H9Fs. The oxygen consumption rate measurements showed substantially increased basal, maximal respiration rate, spare respiratory capacity, and adenosine triphosphate production in hiCMs with TBX20 (Figure 4H and 4I). Increased mitochondrial consumption could be a result of both increased mitochondrial number and increased mitochondrial activity. Moreover, we measured extracellular acidification rate and observed lower glycolysis, glycolytic capacity, and glycolytic reserve in hiCMs after TBX20 treatment (Figure 4J and 4K). Taken together, these results demonstrate reduced glycolysis and increased oxygen consumption in MGT+TBX20 hiCMs (Figure S4G and S4H), indicating that TBX20 may promote the metabolic switch into adult cardiomyocyte-like mitochondrial respiration.
Single-Cell RNA-seq Profiling of MGT+TBX20-Induced hiCMs
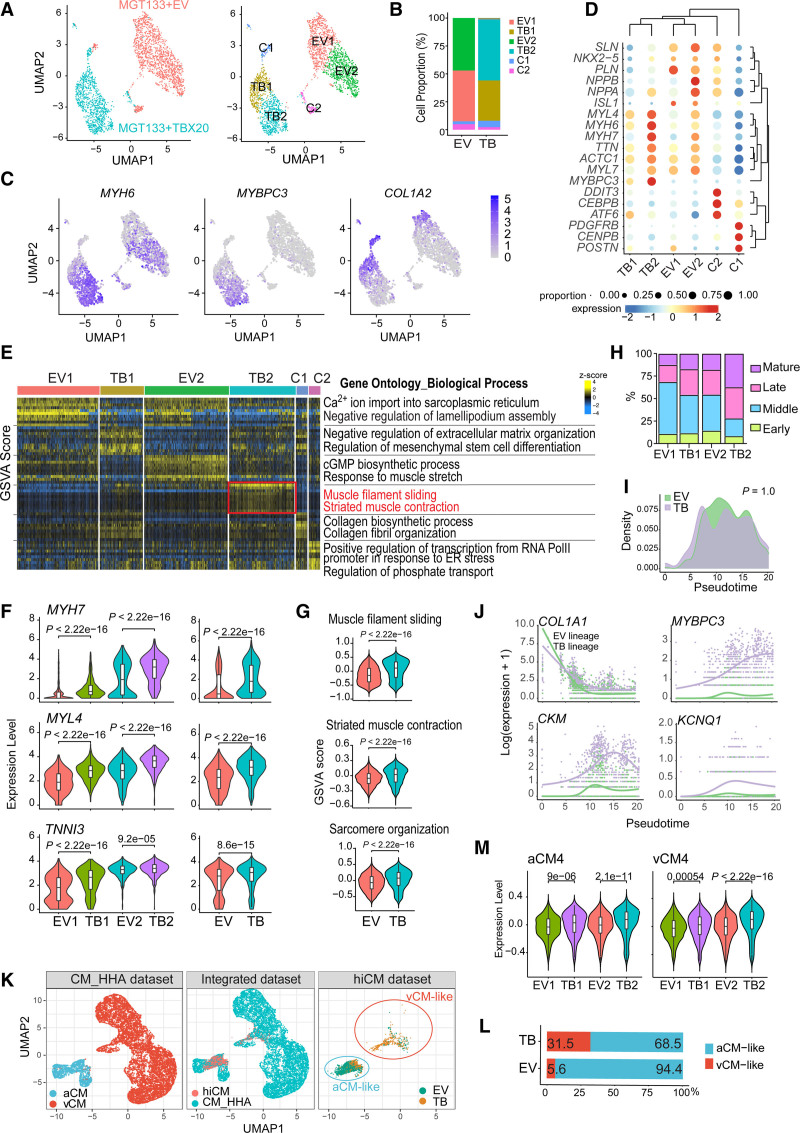
Considering the heterogeneous nature of reprogrammed cells, we performed a single-cell RNA-seq analysis for iCMs generated with MGT+EV or MGT+TBX20 to investigate the effects of TBX20 at the single cell level. The unbiased clustering of 2600 filtered cells showed 6 clusters, which were named EV1, EV2, TB1, TB2, C1, and C2 (Figure 5A and Figure S5A). Expression levels of exogenous MGT-puro and miR-133 were confirmed comparable between EV and TBX20 groups (Figure S5A through S5C). The majority of MGT+EV-induced or MGT+TBX20-induced cells were divided into 2 clusters: EV1/EV2 or TB1/TB2 (Figure 5A and 5B). EV2 and TB2 were identified as reprogrammed hiCMs with high MYH6 and low COL1A2 whereas EV1 and TB1 were partially reprogrammed cells with incomplete silence of COL1A2 (Figure 5A through 5C). The identification and annotation of the top marker genes further demonstrated the remaining fibroblast features in EV1 and TB1 and the enriched myogenesis in EV2 and TB2 (Figure 5D and 5E and Figure S5D). Gene expression violin plots consistently showed an increasing trend of MYH7, MYL4, and TNNI3 through clusters EV1, TB1, EV2, and TB2 (Figure 5F). In addition, small populations of MGT+EV (2.5% and 5%, respectively) and MGT+TBX20 (5.8% and 2.2%) formed clusters C1 and C2. Gene Ontology analysis and gene set variation analyses identified C1 as fibroblasts highly expressing genes associated with collagen fibril organization (Figure 5D and 5E and Figure S5D). The C2 cluster signatures were enriched in endoplasmic reticulum stress and lack of cardiac or fibroblast markers (Figure 5D and 5E and Figure S5D and S5E), suggesting a small number of stressed cells that were not on the reprogramming trajectory.
Figure 5.
Single-cell expression analysis of MGT+TBX20-induced hiCMs. A, Uniform manifold approximation and projection (UMAP) analysis of single-cell RNA sequencing data. Left, UMAP colored by treatment with MGT133 + empty vector (EV; red) or MGT133+TBX20 (teal). Right, UMAP visualization of the cell clusters defined as EV1, EV2, TB1, TB2, C1, and C2. B, Bar plot showing corresponding cell proportion of each cell cluster in human induced cardiomyocytes (hiCMs) generated by MGT133+EV or MGT133+TBX20. C, Feature plots showing expression levels of selected marker genes (MYH6, MYBPC3, and COL1A2) on UMAP. D, Bubble plot showing expression and cell proportion of marker genes from each cluster. E, Heatmap displaying top enriched Gene Ontology biological processes (n=10) in each cluster as determined by gene set variation analysis (GSVA). F, Violin plots showing expression of indicated cardiac genes, including MYH7, MYL4, and TNNI3, in subclusters EV1, TB1, EV2, and TB2 as well as global expression in EV and TB cell populations. G, Violin plot showing selected Gene Ontology terms and GSVA enrichment score between EV and TB cell populations. H, Stacked bar plot displaying the percentage of cells on indicated reprogramming states (early, middle, late, and mature) sets in each subcluster of hiCMs. Maturation states were defined by the state showing the highest GSVA score in each cell. I, Differential pseudotime analysis starting from partially reprogramming cells to late-stage hiCMs between EV and TB lineage. P value was calculated by a Kolmogorov-Smirnov test. J, Pseudotime analysis showing contractile genes (MYBPC3, CKM, and KCNQ1) activation by TBX20. K, UMAP plots showing reference single-cell RNA sequencing data from human heart cell atlas21 cardiomyocytes and integrated mapping using Symphony. EV and TB hiCMs were clustered into atrial cardiomyocyte (aCM)–like cells and ventricular cardiomyocyte (vCM)–like cells. L, Bar plot showing the proportion of aCM-like and vCM-like cells in EV or TB populations. M, Violin plots showing expression of human heart cell atlas cardiomyocyte subpopulation markers in hiCM clusters (EV1, EV2, TB1, and TB2). F, G, and M, P values were calculated by a 2-sided Wilcoxon rank sum test. P<2.2×10−16 represents a P value approaching 0. Box, median ± interquartile range. Whiskers, 1.5×interquartile range.
We next performed further comparisons among EV1, EV2, TB1, and TB2 reprogramming cells to determine the effect of TBX20 on relevant biological processes. Unbiased Gene Ontology analysis demonstrated the enrichment of gene sets including striated muscle contraction and sarcomere organization in the TBX20-overexpressed cell population (Figure 5E and 5G), further supporting the phenotype we discovered. In addition, TBX20 led to altered maturation status of hiCMs with increased late and mature stage genes and reduced early and middle stage genes20 (Figure 5H and Figure S6A and S6B). We also found the involvement of inflammatory pathways in TBX20-mediated reprogramming at the single cell level by gene set enrichment analysis results (Figure S6C and S6D). The increase of immune genes was confirmed by reverse transcription quantitative polymerase chain reaction analysis when comparing MGT+TBX20 with MGT+dsRed (Figure S6E). Next, using the samples from the end point reprogramming day 15, we reconstructed the reprogramming trajectory (Figure S6F and S6H). Although TBX20 did not affect the overall reprogramming progression (Figure 5I), the contractility genes, such as MYBPC3, CKM, and KCNQ1, were activated progressively along pseudotime only after TBX20 overexpression (Figure 5J and Figure S6I). These results highlight the role of TBX20 in promoting hiCM contractility instead of modulating the route of direct reprogramming.
To gain insight into how MGT+EV/TBX20-induced hiCMs resemble endogenous cardiomyocytes, we compared our data with the publicly available human heart single-cell RNA-seq data in the human heart cell atlas. 21 First, TBX20-regulated genes were enriched in cardiomyocytes rather than other cardiac cell types (Figure S7A and S7B), suggesting a cardiac lineage–specific role of TBX20. Next, we mapped our single-cell RNA sequencing data into cardiomyocytes from human heart cell atlas using Symphony.22 We found that the majority of EV cells (94.4%) were projected to the atrial cardiomyocytes (aCMs) cluster, whereas only 5.6% of the cells were more like ventricular cardiomyocytes, which is consistent with the previous report that MGT-generated iCMs were more atrial-like.2 However, when TBX20 was added, an increasing number of cells (31.5%) were clustered to ventricular cardiomyocytes (Figure 5K and 5L). Because the human heart cell atlas classified 5 different cardiomyocyte subtypes (CM1 through CM5), we also evaluated gene expression of CM1 through CM5 markers in each hiCM cluster and found that TBX20 substantially activated genes associated with CM4 population (Figure 5M and Figure S7C and S7D). CM4 cells were identified in both aCM and ventricular cardiomyocyte populations, showing enriched expression of nuclear-encoded mitochondrial genes and denoting high energetic state and metabolic activity,21 which further suggested the involvement of TBX20 in regulating cardiomyocyte energy metabolism.
In addition, we employed weighted correlation network analysis 23 to explore the coexpression network and key modules regulated by TBX20 (Figure S7E). Among the 5 identified coexpression modules, the TBX20-activated module (blue) was highly associated with muscle contraction, ATPase activity, and calcium channel complex, whereas the TBX20-repressed module (yellow) was associated with skeletal development and neuron projection (Figure S7F). The hub-gene-network analysis showed that the TBX20-activated module contains TBX20 itself and several muscle function hub genes, such as MYBPC3, CKM, and ENO3, whereas the TBX20-repressed module included a few atrial chamber–specific genes such as NTM and SLN (Figure S7G and S7H). To predict the upstream regulators of the TBX20-activated module network, which was closely associated with TBX20 function in promoting heart contraction, we performed motif analysis for the blue module gene-associated cis-regulatory regions identified from ATAC-seq (assay for transposase-accessible chromatin with high-throughput sequencing) in MGT+TBX20 hiCMs from H9Fs (Figure S7I). The motifs of TBX20 and MGT factors were greatly enriched in the TBX20-activated module network (Figure S7I), suggesting a potential cooperation between TBX20 and MGT factors in promoting activation of these genes.
TBX20 Binds to and Activates Cardiac Gene Enhancers
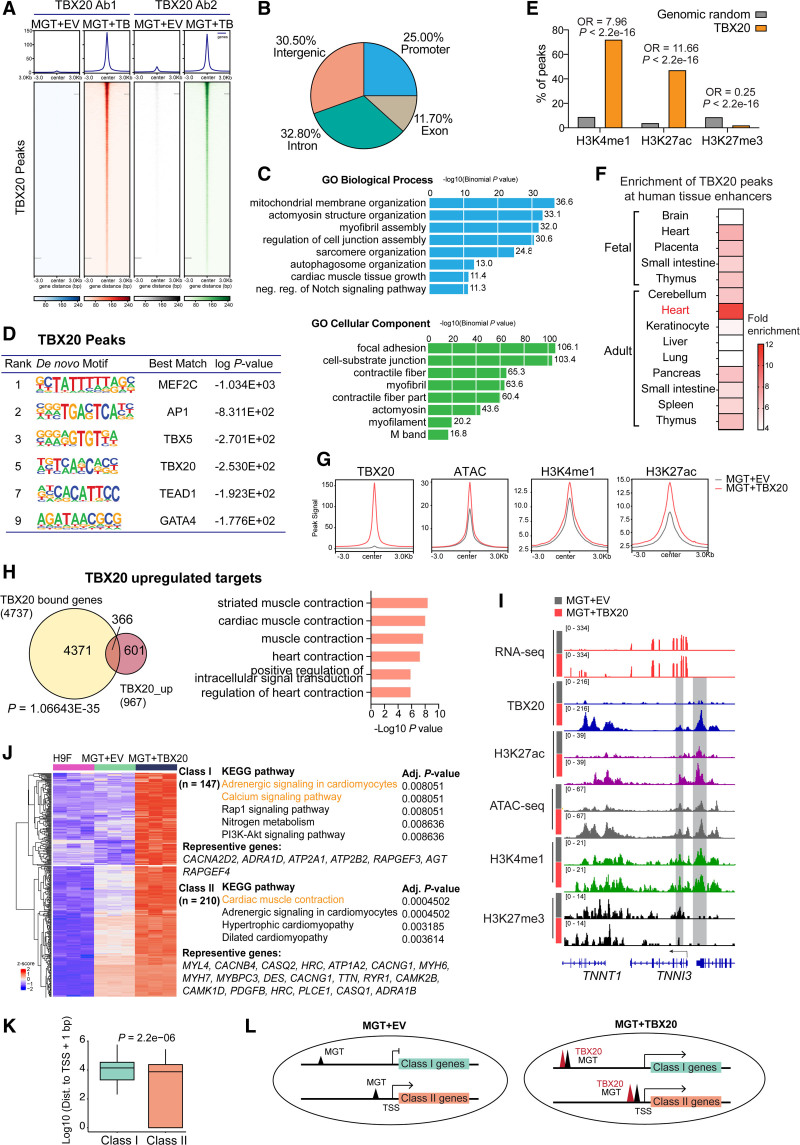
To identify the downstream targets of TBX20 and define the mechanism by which TBX20 enhances reprogramming, we performed the chromatin occupancy profiling of TBX20 using CUT&Tag (cleavage under targets and tagmentation) followed by next-generation sequencing in hiCMs 2 weeks after transduction of MGT133 with or without TBX20 from H9Fs. Consistent TBX20 chromatin-binding profiles were identified by using 2 independent antibodies (Figure 6A) and selected TBX20 binding loci were validated by quantitative polymerase chain reaction (Figure S8A). Overall, the majority of TBX20 bound sites are promoters (25%), introns (32.8%), intergenic regions (30.5%), or exons (11.7%; Figure 6B). The TBX20 peak–associated genes showed significant enrichment in mitochondrial membrane organization, actomyosin structure organization, and cellular components including contractile fiber and actomyosin (Figure 6C). The de novo motif analysis of TBX20 peaks showed enriched consensus motifs of TBX20 and other reprogramming factors MEF2C, TBX5, and GATA4 (Figure 6D), suggesting a potential multifactor core formed on TBX20 targets. We also found TBX20 peaks showed enriched motifs of other transcription factors, such as AP-1 and TEAD1, which are both known as regulators in heart regeneration and mitochondrial function.24,25
Figure 6.
TBX20 directly binds to enhancers of cardiac function genes. A, CUT&Tag (cleavage under targets and tagmentation) peak heatmap showing specific TBX20 binding loci by using 2 independent TBX20 antibodies. B, Pie chart showing the distribution of TBX20 peaks in different genomic regions as indicated. C, GREAT (Genomic Regions Enrichment of Annotations Tool) analysis of TBX20 peaks. D, De novo motif analysis identified from TBX20 peaks. E, Statistical analysis using Fisher exact test showed that significantly more enhancer regions were enriched in TBX20 peak sequences than randomly generated sequences. P<2.2×10−16 represents a P value approaching 0. F, Heatmap showing enrichment of TBX20 peaks in human tissue enhancers collected in EnhancerAtlas 2.0.35 G, Density plots showing signals of ATAC-seq (assay for transposase-accessible chromatin with high-throughput sequencing), H3K4me1, and H3K27ac at TBX20 binding sites. H, Venn diagram showing overlapping of genes bound and upregulated by TBX20. P values were calculated by Fisher exact test. Right panel, Gene Ontology terms enriched on TBX20 upregulated gene targets. I, IGV tracks showing RNA sequencing, TBX20 CUT&Tag, ATAC-seq, and CUT&RUN (cleavage under targets and release using nuclease) signals for H3K27ac, H3K4me1, and H3K27me3 at TNNI3 gene locus between MGT + empty vector (EV) and MGT+TBX20 human induced cardiomyocytes (hiCMs). J, Heatmap displaying class I and class II TBX20 upregulated targets and corresponding representative genes and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment terms. Class I genes were defined as de novo upregulated genes by MGT+TBX20 and class II genes were defined as further activated by the addition of TBX20. K, Boxplot showing TBX20 binding peak distance to transcript start site for class I and class II genes. P value was calculated by a 2-sided Wilcoxon rank sum test. Box, median ± interquartile range. Whiskers, 1.5 × interquartile range. L, Schematic diagram showing the possible mechanism for the regulation of gene expression by TBX20. OR indicates odds ratio.
To gain a comprehensive view of epigenomic alterations in MGT+TBX20 hiCMs, we performed ATAC-seq and CUT&RUN (cleavage under targets and release using nuclease) sequencing for H3K4me1, H3K27ac, and H3K27me3. TBX20 peaks were significantly enriched at H3K4me1-demarcated and H3K27ac-demarcated regions (Fisher exact test; odds ratio, 7.96 and 11.66, respectively; both P<2.2e-16; Figure 6E), suggesting that enhancer is one of the primary targets of TBX20. In addition, enrichment analysis showed that TBX20 peaks were highly enriched at enhancers identified in the adult heart rather than other tissues or fetal heart (Figure 6F). To determine the effect of TBX20 on enhancer status and activity, we compared the signals of chromatin accessibility (ATAC), enhancer status (H3K4me1), and enhancer activity (H3K27ac) at TBX20 binding sites in hiCMs generated with or without TBX20. The overall peak density plots demonstrated that the binding of TBX20 led to a significant gain of chromatin accessibility and enhancer activity (Figure 6G). Furthermore, we found that TBX20 peak–associated genes overlapped substantially with TBX20-upregulated genes and were consistently enriched in genes related to cardiac muscle contraction (Figure 6H), such as TNNI3, MYBPC3, and MYH7, which were also co-occupied with active enhancer hallmarks and featured with open chromatin accessibility (Figure 6I and Figure S8B).
By comparing gene expression in the starting H9F fibroblasts, we identified 2 categories of TBX20-upregulated target genes: targets that were not activated by MGT cocktail but could be activated when TBX20 was included (class I) and targets that were activated by MGT and boosted further by TBX20 (class II; see Supplemental Methods). Class I genes were associated with adrenergic signaling in cardiomyocyte and calcium signaling pathway (Figure 6J). Class II category included genes that were enriched in cardiac muscle contraction (Figure 6J). We next investigated the genomic and epigenomic features of class I and II genes that may explain their differential response to MGT. Although MGT factors colocalized with TBX20 at these genes and their binding was enhanced by TBX20, there were no significant differences in the binding of TBX20, MEF2C, GATA4, or TBX5 between class I and class II genes (Figure S9A and S9B). Further analyses of chromatin accessibility regions and occupancy of H3K4me1, H3K27ac, and H3K27me3 on class I and II genes also showed no significant differences (Figure S9C through S9F). However, we found that TBX20 binding sites of class I genes were substantially farther from the transcript start site than those of class II genes (Figure 6K). Our data suggest that MGT binding at proximal enhancers or promoters may be sufficient to activate target transcription and can be enhanced further by TBX20 (class II genes), whereas MGT binding at long-distance distal enhancers did not exert gene activation effects and would require TBX20 to initiate gene activation (class I genes), indicating the role of TBX20 in facilitating long-distance enhancer regulation with MGT factors (Figure 6L).
TBX20 Requires MGT Factors to Activate Cardiac Genes
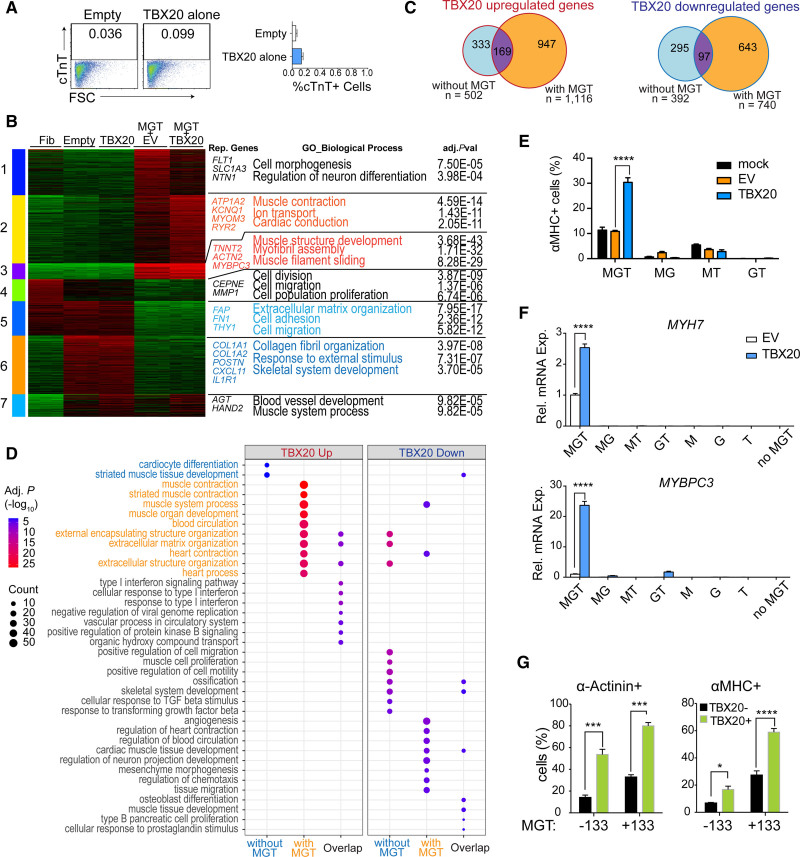
To better understand the role of TBX20 and its regulatory mechanisms for cell fate determination, we generated RNA-seq in H9Fs treated with TBX20 alone and performed comparative analyses among empty control, TBX20 alone, MGT+EV, and MGT+TBX20 treatment groups. We found that TBX20 alone was not sufficient to induce cardiac cell fate conversion (Figure 7A and 7B), with cardiac markers not activated (Figure S10A and S10B). Instead, TBX20 alone mainly regulated genes associated with cell migration and proliferation (Figure 7B). To determine the context-dependent gene regulation of TBX20, we overlapped TBX20-induced DEGs in the conditions with or without MGT, followed by Gene Ontology analysis (Figure 7C and 7D). Whereas TBX20 alone was able to upregulate genes associated with cardiocyte differentiation and muscle development and downregulate genes associated with cell migration and proliferation, the role of TBX20 in activating heart contraction genes could only be observed in the presence of MGT (Figure 7D). To determine whether all 3 MGT factors were required for TBX20 function, we performed reprogramming experiments using part of the MGT factors. TBX20 plus the cocktails missing a single reprogramming factor (MG, MT, and GT) substantially reduced reprogramming efficacy compared with TBX20+MGT (Figure 7E and 7F). However, TBX20 and MGT without miR-133 could also lead to a similar fold increase in reprogramming efficiency, suggesting a weak contribution of miR-133 to TBX20-mediated regulation (Figure 7G and Figure S10C). Together, our findings demonstrate that TBX20 requires intact MGT factors to promote direct reprogramming and activate cardiac genes and support the notion that direct cardiac reprogramming is a synergistically regulated process orchestrated by multiple factors.26
Figure 7.
TBX20 coordinates with MGT to bind on cardiac loci. A, Representative flow cytometry plot and quantification of cardiac troponin T (cTnT)+ cells after 14 days of transduction with TBX20 alone or empty control vector in H9-derived fibroblasts (H9Fs; n=3 per group). B, Heatmap showing 7 k-means clusters of genes as well as representative genes and Gene Ontology biological processes enrichment terms in H9F fibroblast cells (Fib) and H9F cells infected with empty vector (Empty), TBX20 alone (TBX20), MGT + empty vector (EV), and MGT+TBX20. C, Venn diagrams showing the numbers of TBX20 upregulated genes and TBX20 downregulated genes with or without MGT and their overlapping results. D, Bubble plot showing enriched Gene Ontology terms in TBX20 upregulated and downregulated genes in different contexts. E, Percentages of α-MHC (α-myosin heavy chain)+ cells were determined by flow cytometry after transduction of indicated cocktails with or without TBX20 in H9Fs (n=3 per group). F, Reverse transcription quantitative polymerase chain reaction evaluation of MYH7 and MYBPC3 expression in H9Fs 14 days after transduction of indicated triple, dual, or single TFs together with empty vector or TBX20 (n=4 per group). G, Flow cytometry analyses for expression of α-actinin and α-MHC in TBX20-GFP– and TBX20-GFP+ human induced cardiomyocytes (hiCMs) 14 days after transduction of MGT+TBX20-EGFP with or without miR-133. All data are expressed as mean ± SEM (A, F, and G, Student t test; E, 1-way analysis of variance). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
TBX20 Augments the Synergy of MGT+TBX20 on Promoting Cardiac Enhancer Activity
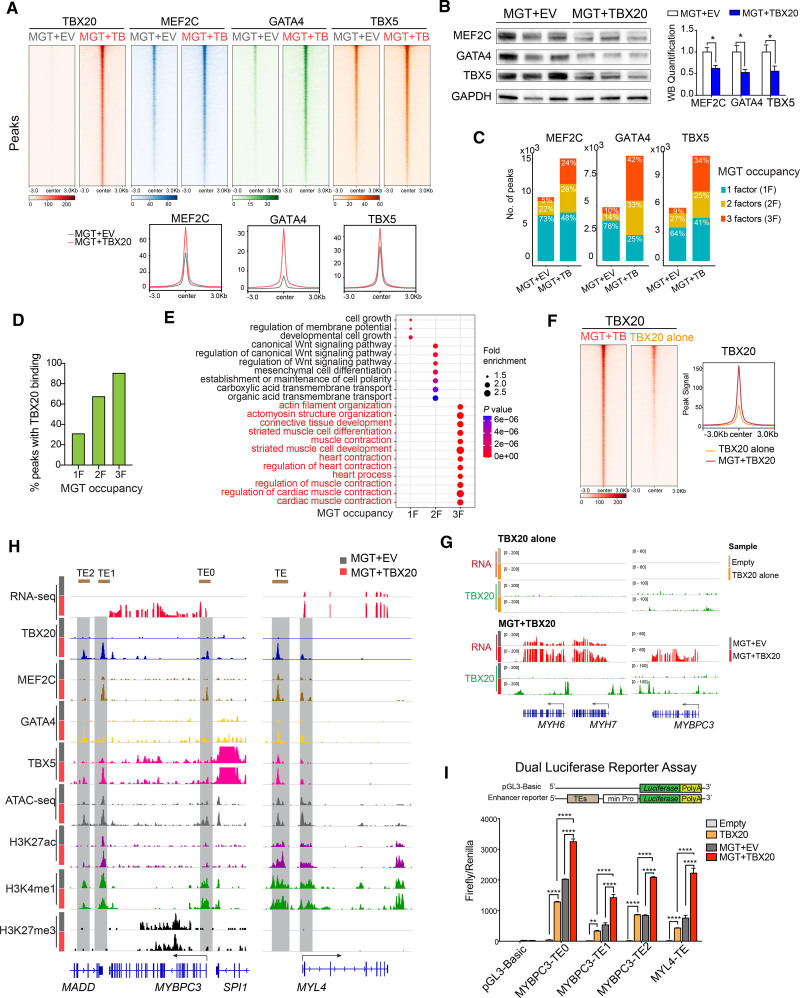
Our motif analysis of the TBX20-activated gene module suggests potential coordination between TBX20 and MGT factors (Figure S7I). To understand the molecular mechanism underlying the synergistic effects in activating gene transcription, we profiled genomic bindings of MEF2C, GATA4, and TBX5 in H9F-derived hiCMs. TBX20 peaks showed significant overlapping with peaks of MEF2C (55.7%), GATA4 (61.8%), and TBX5 (66.3%; Figure S10D). The addition of TBX20 into the MGT cocktail strongly enhanced the binding of each of the reprogramming factors to their targets (Figure 8A). Furthermore, we identified peaks with differential MGT binding after TBX20 overexpression, 93.3% to 99.6% of which showed increased occupancy (Figure S10E), suggesting that TBX20 led to a global increase rather than a redistribution of MGT binding in iCM cells. We also confirmed that the enrichment of MGT on chromatin was not simply attributable to increased MGT expression or protein stability (Figure 8B and Figure S10F). In line with the increased MGT binding to the genome, TBX20 overexpression led to an increased total number of binding peaks of MEF2C, GATA4, and TBX5 (Figure 8C). A substantially higher frequency of MGT co-binding events was observed in MEF2C (24% versus 5%), GATA4 (42% versus 10%), and TBX5 (34% versus 9%) peaks in the presence of TBX20, supporting the role of TBX20 in promoting MGT co-occupancy (Figure 8C). TBX20 binding was mostly enriched in the peaks co-occupied by all 3 MGT factors compared with the peaks bound by only 1 or 2 factors (Figure 8D). Genes with MGT factor co-occupied peaks were substantially enriched in muscle contraction, highlighting the role of TBX20 in promoting MGT co-occupancy at cardiac contraction genes (Figure 8E). On the other hand, the presence of MGT also strongly promoted the genomic occupancy of TBX20 (Figure 8F). In the absence of MGT, TBX20 could not bind to the cardiac loci, such as MYH6, MYH7, and MYBPC3, which is consistent with the inability to activate these genes by TBX20 alone (Figure 8G).
Figure 8.
TBX20 promotes M/G/T genomic binding and activates cardiac enhancers. A, Top panels: heatmaps showing MEF2C, GATA4, and TBX5 CUT&Tag (cleavage under targets and tagmentation) signals on TBX20 binding sites in MGT + empty vector (EV) and MGT+TBX20 human induced cardiomyocytes (hiCMs). Bottom panels: density plots showing enhanced binding of MGT factors on TBX20 peaks. B, Western blot images and quantification (n=3 per group) showing expression of MEF2C, GATA4, and TBX5 in MGT+EV-induced or MGT+TBX20-induced hiCMs. C, Stacked bar plots showing the number of MEF2C, GATA4, and TBX5 binding sites and the proportion of co-binding sites by MEF2C, GATA4, or TBX5 in MGT+EV and MGT+TBX20 infected hiCMs. D, Percentage of TBX20 binding on the peaks occupied with 1, 2, or 3 factors of MGT. E, Bubble plot showing the Gene Ontology annotation for genes associated with peaks occupied with 1, 2, or 3 factors of MGT. F, Heatmaps and density plot showing TBX20 peaks in H9-derived fibroblasts (H9Fs) transduced with MGT+TBX20 or TBX20 alone. G, IGV browser tracks showing RNA sequencing signals and TBX20 peaks in H9Fs transduced with TBX20 alone or MGT+TBX20. H, IGV tracks showing read signals of RNA sequencing, CUT&Tag of MEF2C, GATA4, TBX5 and TBX20, ATAC-seq (assay for transposase-accessible chromatin with high-throughput sequencing), and CUT&RUN (cleavage under targets and release using nuclease) of H3K27ac, H3K4me1, and H3K27me3 at selective cardiac marker gene loci in MGT+EV or MGT+TBX20 infected hiCMs. The brown bars on the top of the graph indicate transcription enhancers (TE) used for reporter assay. I, Construct schematics for reporter assay. MYBPC3 and MYL4 reporter activation in 293T cells after cotransfection of reporter constructs together with empty vector, TBX20 alone, MGT+EV, or MGT+TBX20 (n = at least 3). pGL3-Basic served as negative controls. All data are expressed as mean ± SEM (B, Student t test; I, 2-way analysis of variance). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
As exemplified by MYBPC3 and MYL4 gene loci, TBX20 promoted MGT binding, chromatin accessibility, and enhancer activity at cardiac enhancers (Figure 8H). To validate functionally the effect of TBX20 and MGT on these enhancers, we performed reporter assays using 4 putative enhancers cloned from these loci (Figure 8H) and found that TBX20 and MGT synergistically activated all examined enhancers (Figure 8I). Our genomic and reporter assays revealed a strong synergy between TBX20 and MGT, which leads to enhanced chromatin targeting and colocalization of these cardiac transcription factors for more robust enhancer activation of cardiac contractility genes.
Discussion
We identified TBX20 as an important factor in human cardiac reprogramming. TBX20 improves hiCM quality in sarcomere structure, contractility potential, and mitochondrial function. We also demonstrated that TBX20 consistently promotes hiCM reprogramming regardless of the reprogramming factors used (Figure S10G). Through the genome-wide CUT&Tag data analyses, we identified a pool of cardiac enhancers enriched in TBX20 targeting sites and experimentally validated their enhancer activity. In the presence of MGT, TBX20 not only enhanced chromatin occupancy of MGT but also its co-occupancy at cardiac genes. More recently, a histone reader PHF7 has been identified to promote direct reprogramming by targeting cardiac superenhancers.27 TBX20 was among the top 50 upregulated genes after PHF7 treatment, implying a potential TBX20-dependent mechanism underlying PHF7-mediated reprogramming. Further investigations on PHF7-mediated TBX20 activation may help clarify the epigenetic mechanisms by which TBX20 cannot be activated by canonical reprogramming factors.
TBX20 has been studied extensively in both developing hearts and adult cardiomyocytes, showing a context-sensitive manner in its role and regulation.16,28–31 Our results showed decreased cell proliferation after TBX20 overexpression (Figure 3D), which is distinct from its role in fetal cardiomyocytes.32 We revealed direct regulation of TBX20 on cardiac contractility genes, indicating that TBX20 function in the direct reprogramming setting is more similar to that in adult cardiomyocytes.31 Additional mechanisms may also contribute to a TBX20-induced phenotype. For example, our gene expression analysis demonstrated that activation of inflammation genes was associated with TBX20 overexpression. Because inflammation is associated with reprogramming processes,33,34 its role in mediating the effects of TBX20 warrants further studies.
We observed spontaneous beating and action potential generation in a small portion of hiCMs generated with MGT+TBX20 but not in those without TBX20 after coculture, suggesting the improved contractility potential in hiCMs. However, it is challenging to obtain beating cells efficiently in hiCM culture alone. Further examinations of excitability of hiCMs would need more comprehensive electrophysiologic analyses of action potentials and ion current measured by whole-cell patch clamp in hiCMs at the single cell level. Another limitation of this study is the lack of in vivo evaluation of the effect of TBX20 in direct reprogramming, which represents an important area of future investigation.
Our results not only demonstrate TBX20 as a critical factor that promotes hiCM function in cardiac contractility, maturation, and energy metabolism, but also reveals an important mechanism by which TBX20 synergizes with canonical reprogramming factors to activate cardiac enhancers to reconstruct cardiomyocyte identity and contractility.
Article Information
Acknowledgments
The authors thank Shanrun Liu at the University of Alabama at Birmingham (UAB) Comprehensive Flow Cytometry Core for helping with 10x Genomics sample collection and library preparation. The Seahorse analysis was performed with help from Kelley E. Smith-Johnston and Melissa J. Sammy at the UAB Bioanalytical Redox Biology Core, Diabetes Research Center (National Institute of Diabetes and Digestive and Kidney Diseases grant DK P30DK079626), Nutrition Obesity Research Center (National Institute of Diabetes and Digestive and Kidney Diseases grant DK056336), UAB Center for Exercise Medicine, UAB Comprehensive Diabetes Center, Center for Free Radical Biology, and Comprehensive Neuroscience Center. Y.T. designed experiments, conducted the experiments, analyzed the data, and wrote the manuscript. S.A. analyzed the sequencing data, prepared the figures, and wrote the manuscript. X.G.‚ X.Z.‚ and V.G.F. conducted experiments. J.Z. revised the manuscript and provided technical and financial support. R.L. designed the experiment, analyzed the data, and wrote the manuscript. Y.Z. supervised the project, designed the experiments, analyzed the data, and wrote the manuscript.
Sources of Funding
This work was supported in part by National Institutes of Health grants R01 HL153220 (to Y.Z.), R01 CA259480 (to R.L.), and R01 HL149137 and HL131017 (to J.Z.) and American Heart Association Transformational Project Award 969529 (to Y.Z.). Dr Lu is an American Society of Hematology Scholar in Basic Science and is supported by the Concern Foundation and Mark Foundation for Cancer Research.
Disclosures
None.
Supplemental Material
Supplemental Methods
Figures S1–S10
Tables S1–S3
Videos S1–S3
Supplementary Material
Nonstandard Abbreviations and Acronyms
- α-MHC
- α-myosin heavy chain
- aCM
- atrial cardiomyocyte
- ATAC-seq
- assay for transposase-accessible chromatin with high-throughput sequencing
- CUT&RUN
- cleavage under targets and release using nuclease
- CUT&Tag
- cleavage under targets and tagmentation
- DEG
- differentially expressed gene
- EGFP
- enhanced green fluorescent protein
- EV
- empty vector
- GFP
- green fluorescent protein
- H9F
- H9-derived fibroblast
- HCF
- human cardiac fibroblast
- hiCM
- human induced cardiomyocyte
- hiPSC-CM
- human induced pluripotent stem cell–derived cardiomyocyte
- iCM
- induced cardiomyocyte
- MGT133
- human MEF2C, GATA4, TBX5, and miR-133
- RNA-seq
- RNA sequencing
Y. Tang and S. Aryal contributed equally.
R. Lu and Y. Zhou contributed equally.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCULATIONAHA.122.059713.
For Sources of Funding and Disclosures, see page 1534.
Circulation is available at www.ahajournals.org/journal/circ
References
- 1.Srivastava D, DeWitt N. In-vivo cellular reprogramming: the next generation. Cell. 2016;166:1386–1396. doi: 10.1016/j.cell.2016.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu J-d, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song K, Nam Y-J, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wada R, Muraoka N, Inagawa K, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Kaneda R, Suzuki T, Kamiya K, et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc Natl Acad Sci USA. 2013;110:12667–12672. doi: 10.1073/pnas.1304053110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu JD, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, Delgado-Olguin P, Ding S, Bruneau BG, Srivastava D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 2013;1:235–247. doi: 10.1016/j.stemcr.2013.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone NR, Gifford CA, Thomas R, Pratt KJB, Samse-Knapp K, Mohamed TMA, Radzinsky EM, Schricker A, Ye L, Yu P, et al. Context-specific transcription factor functions regulate epigenomic and transcriptional dynamics during cardiac reprogramming. Cell Stem Cell. 2019;25:87–102.e9. doi: 10.1016/j.stem.2019.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Addis RC, Ifkovits JL, Pinto F, Kellam LD, Esteso P, Rentschler S, Christoforou N, Epstein JA, Gearhart JD. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol. 2013;60:97–106. doi: 10.1016/j.yjmcc.2013.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nam YJ, Lubczyk C, Bhakta M, Zang T, Fernandez-Perez A, McAnally J, Bassel-Duby R, Olson EN, Munshi NV. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development. 2014;141:4267–4278. doi: 10.1242/dev.114025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biggin MD. Animal transcription networks as highly connected, quantitative continua. Dev Cell. 2011;21:611–626. doi: 10.1016/j.devcel.2011.09.008 [DOI] [PubMed] [Google Scholar]
- 12.He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci USA. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furtado MB, Costa MW, Pranoto EA, Salimova E, Pinto AR, Lam NT, Park A, Snider P, Chandran A, Harvey RP, et al. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ Res. 2014;114:1422–1434. doi: 10.1161/CIRCRESAHA.114.302530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakraborty S, Sengupta A, Yutzey KE. Tbx20 promotes cardiomyocyte proliferation and persistence of fetal characteristics in adult mouse hearts. J Mol Cell Cardiol. 2013;62:203–213. doi: 10.1016/j.yjmcc.2013.05.018 [DOI] [PubMed] [Google Scholar]
- 17.Chakraborty S, Yutzey KE. Tbx20 regulation of cardiac cell proliferation and lineage specialization during embryonic and fetal development in vivo. Dev Biol. 2012;363:234–246. doi: 10.1016/j.ydbio.2011.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang FL, Guo M, Yutzey KE. Overexpression of Tbx20 in adult cardiomyocytes promotes proliferation and improves cardiac function after myocardial infarction. Circulation. 2016;133:1081–1092. doi: 10.1161/CIRCULATIONAHA.115.019357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopaschuk GD, Jaswal JS. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. J Cardiovasc Pharmacol. 2010;56:130–140. doi: 10.1097/FJC.0b013e3181e74a14 [DOI] [PubMed] [Google Scholar]
- 20.Uosaki H, Cahan P, Lee DI, Wang S, Miyamoto M, Fernandez L, Kass DA, Kwon C. Transcriptional landscape of cardiomyocyte maturation. Cell Rep. 2015;13:1705–1716. doi: 10.1016/j.celrep.2015.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litvinukova M, Talavera-Lopez C, Maatz H, Reichart D, Worth CL, Lindberg EL, Kanda M, Polanski K, Heinig M, Lee M, et al. Cells of the adult human heart. Nature. 2020;588:466–472. doi: 10.1038/s41586-020-2797-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang JB, Nathan A, Weinand K, Zhang F, Millard N, Rumker L, Moody DB, Korsunsky I, Raychaudhuri S. Efficient and precise single-cell reference atlas mapping with Symphony. Nat Commun. 2021;12:5890. doi: 10.1038/s41467-021-25957-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beisaw A, Kuenne C, Guenther S, Dallmann J, Wu CC, Bentsen M, Looso M, Stainier DYR. AP-1 contributes to chromatin accessibility to promote sarcomere disassembly and cardiomyocyte protrusion during zebrafish heart regeneration. Circ Res. 2020;126:1760–1778. doi: 10.1161/CIRCRESAHA.119.316167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu RY, Jagannathan R, Sun LF, Li F, Yang P, Lee J, Negi V, Perez-Garcia EM, Shiva S, Yechoor VK, et al. Tead1 is essential for mitochondrial function in cardiomyocytes. Am J Physiol Heart Circ Physiol. 2020;319:H89–H99. doi: 10.1152/ajpheart.00732.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashimoto H, Wang Z, Garry GA, Malladi VS, Botten GA, Ye W, Zhou H, Osterwalder M, Dickel DE, Visel A, et al. Cardiac reprogramming factors synergistically activate genome-wide cardiogenic stage-specific enhancers. Cell Stem Cell. 2019;25:69–86.e5. doi: 10.1016/j.stem.2019.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garry GA, Bezprozvannaya S, Chen K, Zhou H, Hashimoto H, Morales MG, Liu N, Bassel-Duby R, Olson EN. The histone reader PHF7 cooperates with the SWI/SNF complex at cardiac super enhancers to promote direct reprogramming. Nat Cell Biol. 2021;23:467–475. doi: 10.1038/s41556-021-00668-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsenstein M, Georges R, Davidson L, Mo R, et al. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827 [DOI] [PubMed] [Google Scholar]
- 29.Singh MK, Christoffels VM, Dias JM, Trowe MO, Petry M, Schuster-Gossler K, Burger A, Ericson J, Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005;132:2697–2707. doi: 10.1242/dev.01854 [DOI] [PubMed] [Google Scholar]
- 30.Cai CL. T-box genes coordinate regional rates of proliferation and regional specification during cardiogenesis. Development. 2005;132:2475–2487. doi: 10.1242/dev.01832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen T, Aneas I, Sakabe N, Dirschinger RJ, Wang G, Smemo S, Westlund JM, Cheng H, Dalton N, Gu Y, et al. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J Clin Invest. 2011;121:4640–4654. doi: 10.1172/JCI59472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boogerd CJ, Zhu X, Aneas I, Sakabe N, Zhang L, Sobreira DR, Montefiori L, Bogomolovas J, Joslin AC, Zhou B, et al. Tbx20 is required in mid-gestation cardiomyocytes and plays a central role in atrial development. Circ Res. 2018;123:428–442. doi: 10.1161/CIRCRESAHA.118.311339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J, Sayed N, Hunter A, Au KF, Wong WH, Mocarski ES, Pera RR, Yakubov E, Cooke JP. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, Liu Z, Welch JD, Gao X, Wang L, Garbutt T, Keepers B, Ma H, Prins JF, Shen W, et al. Single-cell transcriptomic analyses of cell fate transitions during human cardiac reprogramming. Cell Stem Cell. 2019;25:149–164.e9. doi: 10.1016/j.stem.2019.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao T, Qian J. EnhancerAtlas 2.0: an updated resource with enhancer annotation in 586 tissue/cell types across nine species. Nucleic Acids Res. 2020;48:D58–D64. doi: 10.1093/nar/gkz980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao M, Tang Y, Zhou Y, Zhang J. Deciphering role of Wnt signalling in cardiac mesoderm and cardiomyocyte differentiation from human iPSCs: four-dimensional control of Wnt pathway for hiPSC-CMs differentiation. Sci Rep. 2019;9:19389. doi: 10.1038/s41598-019-55620-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sowell B, Fast VG. Ionic mechanism of shock-induced arrhythmias: role of intracellular calcium. Heart Rhythm. 2012;9:96–104. doi: 10.1016/j.hrthm.2011.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hao Y Hao S Andersen-Nissen E Mauck WM 3rd, Zheng S Butler A Lee MJ Wilk AJ Darby C Zager M et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Bézieux HR, Van den Berge K, Street K, Dudoit S. Trajectory inference across multiple conditions with condiments: differential topology, progression, differentiation, and expression. bioRxiv. Preprint posted online March 10, 2021. doi: 10.1101/2021.03.09.433671 [Google Scholar]
- 42.Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van den Berge K, Roux de Bezieux H, Street K, Saelens W, Cannoodt R, Saeys Y, Dudoit S, Clement L. Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun. 2020;11:1201. doi: 10.1038/s41467-020-14766-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun. 2019;10:1930. doi: 10.1038/s41467-019-09982-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Zhang Y, Zhou X, Wright S, Hyle J, Zhao L, An J, Zhao X, Shao Y, Xu B, et al. Functional interrogation of HOXA9 regulome in MLLr leukemia via reporter-based CRISPR/Cas9 screen. Elife. 2020;9: doi: 10.7554/eLife.57858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang Y, Zhao L, Yu X, Zhang J, Qian L, Jin J, Lu R, Zhou Y. Inhibition of EZH2 primes the cardiac gene activation via removal of epigenetic repression during human direct cardiac reprogramming. Stem Cell Res. 2021;53:102365. doi: 10.1016/j.scr.2021.102365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu R, Wang P, Parton T, Zhou Y, Chrysovergis K, Rockowitz S, Chen WY, Abdel-Wahab O, Wade PA, Zheng D, et al. Epigenetic perturbations by Arg882-mutated DNMT3A potentiate aberrant stem cell gene-expression program and acute leukemia development. Cancer Cell. 2016;30:92–107. doi: 10.1016/j.ccell.2016.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meers MP, Tenenbaum D, Henikoff S. Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin. 2019;12:42. doi: 10.1186/s13072-019-0287-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao L, Zhang P, Galbo PM, Zhou X, Aryal S, Qiu S, Zhang H, Zhou Y, Li C, Zheng D, et al. Transcription factor MEF2D is required for the maintenance of MLL-rearranged acute myeloid leukemia. Blood Adv. 2021;5:4727–4740. doi: 10.1182/bloodadvances.2021004469 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic datasets have been deposited to the Gene Expression Omnibus with accession number GSE193810 and are publicly available.