This study reveals how aberrant m6A modification of the WTAPP1 pseudogene results in increased translation of its protein-coding counterpart to promote Wnt signaling, which contributes to pancreatic cancer progression.
Abstract
Pseudogenes may play important roles in cancer. Here, we explore the mechanism and function of a pseudogene WTAPP1 in the progress of pancreatic ductal adenocarcinoma (PDAC). WTAPP1 RNA was significantly elevated in PDAC and was associated with poor prognosis in patients. Overexpression of WTAPP1 RNA promoted PDAC proliferation and invasiveness in vitro and in vivo. Mechanistically, N6-methyladenosine (m6A) modification stabilized WTAPP1 RNA via CCHC-type zinc finger nucleic-acid binding protein (CNBP), resulting in increased levels of WTAPP1 RNA in PDAC cells. Excessive WTAPP1 RNA bound its protein-coding counterpart WT1-associated protein (WTAP) mRNA and recruited more EIF3 translation initiation complex to promote WTAP translation. Increased WTAP protein enhanced the activation of Wnt signaling and provoked the malignant phenotypes of PDAC. Decreasing WTAPP1 RNA significantly suppressed the in vivo growth and metastasis of PDAC cell lines and patient-derived xenografts. These results indicate that m6A-mediated increases in WTAPP1 expression promote PDAC progression and thus may serve as a therapeutic target.
Significance:
This study reveals how aberrant m6A modification of the WTAPP1 pseudogene results in increased translation of its protein-coding counterpart to promote Wnt signaling, which contributes to pancreatic cancer progression.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) remains one of the deadliest malignancies with only approximately 10% of 5-year survival (1, 2). Most patients with PDAC eventually die of cancer progression due to lack of effective treatment modalities (2, 3), which poses a great medical challenge and requires urgent development of new target therapies. To achieve this, it is important to have better understanding of molecular mechanisms underlying the pathogenesis of PDAC.
Recent studies have shown that pseudogene-produced RNAs play an important regulatory role in many types of human cancer (4–6). RNAs produced by pseudogene are considered as a subclass of long ncRNAs (lncRNA) that develop from protein-coding genes but harbor multiple mutations that abrogate their translation into functional proteins (7). However, accumulating evidence has demonstrated that RNAs produced by pseudogene may function in regulating their protein-coding counterparts at either transcriptional or posttranscriptional level (5–8). While attempts have been made to explore the mechanism of anomalously expressed pseudogene RNAs in several types of cancer (5, 6, 8), little has been known about their roles and functions in PDAC.
It has been well known that RNAs are substantially modified during their maturation and aberrant modifications might cause their functional alterations and thus implicate in the development of diseases such as cancer (9–13). N6-methyladenosine (m6A) is the most abundant modification of RNAs that regulates RNAs at different levels such as RNA stability (9, 14–16). Recent studies have shown that some long ncRNAs are regulated by m6A modifications and are involved in tumor progression (9, 17–21). However, there are few reports on the modifications of RNAs produced by pseudogene and their resultant roles in PDAC.
In this study, we have demonstrated a m6A-modified RNA produced by the pseudogene WTAPP1 as an oncogenic regulator in pancreatic cancer. We have found that CCHC-type zinc finger nucleic acid binding protein (CNBP) can recognize m6A-modified WTAPP1 RNA and increase its stability. Excessive WTAPP1 RNA enhances WTAP translation through recruiting EIF3B to WTAP RNA. Increased WTAP level may evoke oncogenic Wnt signaling and promote PDAC progression. Furthermore, we have treated mouse models with WTAPP1 inhibitor and the results suggest that WTAPP1 may be a potential therapeutic target for PDAC.
Materials and Methods
Tissue specimens
Surgically removed PDAC samples and their corresponding adjacent normal tissues were collected from patients at Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China (N = 158) and Chinese Academy of Medical Sciences Cancer Hospital, Beijing, China (N = 73) between 2010 and 2016 (Supplementary Table S1). PDAC were confirmed histopathologically by three independent pathologists. None of the patients included in this study received any preoperative radiotherapy or chemotherapy. The patient survival time was the interval between the date of diagnosis and the date of last contact (death or last follow-up). Death information was obtained from inpatient and outpatient records or follow-up telephone calls. Written informed consent was obtained from each patient, and this study was approved by the Institutional Review Board of the Sun Yat-sen Memorial Hospital and Chinese Academy of Medical Sciences Cancer Hospital.
Pseudogene detection and differential expression analysis
We performed RNA-sequencing (RNA-seq) using 65 PDAC tumors and 33 normal tissue samples. Adaptors and low-quality bases of reads were trimmed by Cutadapt (v1.16; ref. 22), and reads shorter than 20 nucleotides were discarded. RNA-seq by expectation maximization (RSEM; ref. 23) was used to quantify gene expression. To perform a comprehensive survey of pseudogenes, we obtained the genomic information of human pseudogenes through GTF file from the GENCODE database (version 25; ref. 24) and quantified pseudogene expression as reads per kilobase per million mapped reads (RPKM). The pseudogenes with detectable expression were defined as those with an RPKM ≥ 0.1 across at least 2 samples. Finally, we identified 1,958 pseudogenes in our PDAC samples. We then used DESeq2 (25) for further differential expression analysis.
Cell lines and cell culture
Human PDAC (SW1990, Capan-2 and PANC-1) and embryonic kidney (293T) cell lines were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences, Shanghai Institute of Biochemistry and Cell Biology. All cell lines were authenticated by the DNA finger printing analysis and tested to be free of mycoplasma infection. Cells were routinely cultured in DMEM medium supplemented with 10% FBS in an atmosphere of 5% CO2 and 99% relative humidity at 37°C.
Small interfering and guide RNAs
siRNA targeting the transcriptional products of the 10 top-candidate pseudogenes and WTAP, METTL3, CNBP, EIF3B genes were purchased from GenePharma (Supplementary Table S2). Transfection of siRNA (75 nmol/L) was performed with lipofectamine 2000 (Life Technologies). Two single-guide RNAs (sgRNA) adjacent to the WTAPP1 m6A site were designed (Supplementary Table S2). All guide RNAs (gRNA) were subjected to NCBI BLAST (see URLs) to avoid mismatch to unexpected RNAs in the human genome.
RNA extraction and qRT-PCR analysis
Details are provided in Supplementary Materials and Methods and the primer sequences used for qRT-PCR and other assays are shown in Supplementary Table S3.
Northern blot analysis
Approximately 50 μg of total RNA isolated from PDAC cell lines was subjected to formaldehyde gel electrophoresis and transferred to a Biodyne Nylon membrane (Pall). The RNA probes labeled with digoxigenin (Supplementary Table S4) were synthesized by Bersinbio. After prehybridization for 30 minutes, the membrane was hybridized for 12 hours at 68 °C in buffer containing the denatured probes. After washing, signal on the membrane was detected using an Odyssey infrared scanner (Li-Cor, Lincoln).
RNA immunoprecipitation assays
To determine the m6A levels in WTAPP1 RNA, total RNA from tissues or cells was extracted and subjected to fragmentation using the RNA fragmentation reagents (Ambion). Precipitation was performed using an anti-m6A antibody (Synaptic Systems, 202003) previously bound to magnetic beads in RIP immunoprecipitation buffer (Magna RIP Kit, Millipore) and incubated with fragmented RNAs. After treating with proteinase K (10 mg/mL), RNAs was extracted with phenol/chloroform/isoamyl alcohol and subjected to qRT-PCR using the primers for WTAPP1 RNA around m6A site, which was normalized to input.
RNA immunoprecipitation (RIP) assays were performed using the Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore). Antibody against CNBP, HuR, or EIF3B were used. Total RNA for each antibody were assayed simultaneously and coprecipitated RNA was detected by qRT-PCR also normalized to input.
For MS2-based RIP assays, pcDNA3.1-MS2–6X, pcDNA3.1-MS2-WTAPP1-WT, or pcDNA3.1-MS2-WTAPP1-Δ (Umine Biotechnology) was cotransfected with pMS2-GFP (Addgene) into SW1990 or Capan-2 cells with Lipofectamine 2000. RIP assays were performed using GFP antibody.
Immunoprecipitation and immunoblotting assays
PDAC cells were lysed with 1 × RIPA buffer supplemented with Protease/Phosphatase Inhibitor Cocktail (Pierce). Lysate was briefly centrifugated and the supernatant was treated with RNase A (20 μg/mL) or RNase inhibitor (200 U/mL, New England Biolab) prior to immunoprecipitation or immunoblotting with CNBP antibody. The immunoblot signal was detected using Clarity Western ECL Substrate (Thermo).
RNA pulldown and mass spectrometry analysis
RNA pulldown was performed with the Pierce Magnetic RNA-Protein Pull-Down Kit (20164, Thermo). Biotin-labeled fragment containing 50-bp WTAPP1 RNA sequences with or without m6A modification were commercially synthesized and incubated with cellular protein extracts from SW1990 and Capan-2 cells. Streptavidin beads were then added and total proteins associated was subjected to mass spectrometry (MS) or Western blot analysis. Similarly, biotinylated full-length WTAPP1 RNA or its antisense was pulled down with the same approach and analyzed by MS or Western blotting.
RNA electrophoretic mobility shift assays
Assays were performed using the LightShift Chemiluminescent RNA EMSA Kit (Life Technologies). Biotin-labeled RNA oligonucleotides (4 nmol/L final concentration) were incubated in a 20-μL system containing binding buffer (10 mmol/L HEPES pH 7.3, 20 mmol/L KCl, 1 mmol/L MgCl2, 1 mmol/L DTT, 5% glycerol, and 40 U/mL RNasin) and various concentrations of recombinant CNBP proteins (0–10 μmol/L) at room temperature for 20 minutes. The RNA-protein mixture was separated in 8% native polyacrylamide gels (in 0.5 × Tris-borate-EDTA buffer) at 4°C for 60 minutes. Complexes were then transferred to a nylon membrane, cross-linked to the membrane using the UVP cross-linker (120 mJ/cm2 of 254 nm UV) and detected by chemiluminescence.
Aminomethyltrioxsalen crosslink assays
PDAC cells were suspended in PBS with or without 0.5 mg/mL of 4′-Aminomethyltrioxsalen (AMT) hydrochloride (Sigma) at a concentration of 2 × 107 cells/mL and incubated on ice for 15 minutes and crosslinked twice with 150 mJ/cm2 of UV light (254 nm). Cells were pelleted for RNA isolation after cross-linking. To pull-down the WTAP mRNAs, 25 pmol in vitro transcribed and biotin-labeled WTAPP1 RNA probes were denatured to 90°C for 2 minutes and transferred immediately on ice. Then probes and 10 μg RNA were mixed in binding buffer (50 mmol/L Tris-HCl pH 7.5, 150 mmol/L NaCl, 0.5% NP40, and 1 mmol/L ribonucleoside vanadyl complexes) and transferred to a 37°C thermomixer, shaking at 1,200 rpm for 2 hours, followed by the addition of 30 μL washed Streptavidin beads and incubated for 2 hours at room temperature. After washing, precipitated RNA was extracted by TRIzol and detected by qRT-PCR and normalized to input.
Chromatin isolation by RNA purification assays
Antisense biotin-labeled WTAP ssDNA probes were designed online (see URLs) and synthesized (Supplementary Table S4). The 13 probes were separated into 2 pools named “odd” and “even”. PDAC cells (4 × 107) with WTAPP1 overexpression or knockdown were used for ChIRP experiment. Cell harvesting, lysis, disruption, and ChIRP were performed as manuals of Magna ChIRP RNA Interactome Kits (Millipore). LacZ probes were used as negative probes and β-ACTIN as negative retrieved RNA and protein. WTAP mRNA was detected by qRT-PCR while EIF3B was detected by Western blotting.
RNA FISH and immunofluorescence
Fixed and permeabilized PDAC cells or dewaxing tumor sections were hybridized with WTAPP1 RNA or WTAP RNA probes (Genepharma), or primary antibody against EIF3B or CNBP, overnight in a humidified chamber at 37°C in the dark. Cell nuclei were counterstained with DAPI. Images were taken with Olympus FV1000 confocal microscope (Olympus) for cells and with Polaris Automatic Digital Slide Scanner (Akoya Biosciences) for tissue samples.
IHC staining and scoring
Tissue microarrays purchased from Outdo Biotech Co. were IHC stained using antibody against WTAP. The IHC scoring criteria is described in Supplementary Materials and Methods.
Establishment of mouse xenograft models
Female BALB/c nude mice aged 5 weeks (Beijing Vital River Laboratory Animal Technology) were randomly grouped and injected subcutaneously with 0.1 mL of cell suspension containing 2 × 106 PDAC cells in the back flank. When palpable, the tumor was measured every 5 days and tumor volume was calculated by length × width2 × 0.5. We also implanted luciferase-labeled PDAC cells (2 × 106) to the pancreas of mouse by surgical injection (26). Tumor volume was monitored twice a week by bioluminescence imaging with a Living Image system (Perkin Elmer) and the quantitative data were expressed as photon flux. The pancreas, lung, liver, and intestine of each mouse were removed when animal was sacrificed for further pathologic examination.
For mouse patient-derived xenograft (PDX) models, fresh PDAC samples obtained from 3 patients who underwent surgery were propagated as subcutaneous tumors in 4-week-old NOD/SCID gamma (NSG) mice (F1). Xenografts from F1 mice were cut into small pieces and then implanted into other mice (F2). When tumors grew up to about 1,500 mm3, they were excised and cut again into small pieces and transplanted to other mice (F3; ref. 27).
Treatment of xenografts in mice
Seven days after orthotopic implantation or PDX grew up to about 200 mm3, mice were divided into 2 groups (n = 5 per group) and intravenously injected with in vivo optimized siControl or siWTAPP1 (75 mg/kg; RiboBio; Supplementary Table S2) dissolved in saline. Treatment was performed once a day in the first 3 days and after that, once 3 days for 3 weeks (28). Tumor burden in orthotopically implantated mice was monitored twice a week by bioluminescence imaging and the PDX volume (length × width2 × 0.5) was monitored every 3 days. The animal survival time was recorded from the day of tumor implantation to the date of death.
Animal experiments were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University Cancer Center (reference no. L102042019080W) and the animals were handled in accordance with institutional guidelines.
m6A individual-nucleotide resolution cross-linking and immunoprecipitation sequencing
Total RNA from PANC-1 cells was digested with DNase I and then subjected to 2 rounds of RiboMinus (Illumina) treatment to eliminate ribosomal RNAs. The resultant RNA (200 μg) was then fragmented to about 100 nucleotides in length, incubated with 10 μg of anti-m6A antibody (Synaptic Systems, 202003) before cross-linking with 150 mJ/cm2 254-nm UV light. After incubating with Dynabeads protein A/G (Millipore) at 4°C overnight, m6A-modified RNA was treated with T4 PNK (NEB) on beads, followed by proteinase K treatment, acidic phenol/chloroform extraction, and ethanol precipitation. RNA was subsequently used for library construction with NEBNext small RNA library prep kit (E7330S, NEB) and sequenced on Illumina Hiseq4000.
Identification of m6A modification by iCLIP-sequencing
Read preprocessing was performed essentially as previously reported (29). Adaptors and low-quality bases were trimmed by cutadapt (v1.16; see URLs), and reads shorter than 20 nt were discarded. Reads demultiplexing based on their experimental barcode and sequence-based removal of PCR duplicates were then performed using the pyCRAC tool suite (30). The reverse reads were reversely complemented and processed in the same way as the forward counterparts. Reads were then mapped to human genome (hg38) with BWA (v0.7.15; ref. 31), with parameters “bwa aln -n 0.06 -q 20”, as recommended by the online CTK Documentation (see URLs). We detected cross-linking–induced mutation sites (CIMS) and cross-linking–induced truncation sites (CITS) using CLIP Tool Kit (CTK; ref. 32). To identify the m6A locus, the mode of mutation calling was performed as previously reported (33). For each mutation position, the coverages of unique tag (k) and mutations (m) were determined by CIMS.pl script of CLIP Tool Kit. We next removed the known SNPs (dbSNP 147) from all the mutation positions. Sequentially, the C > T mutation positions within m/k ≤ 50% and only mutation positions at the +1 position of adenosines were identified as CIMS-based m6A residues, as previously reported. Truncation sites with a significance value of P ≤ 0.05 that occurred neighbor adenosines were retained to yield a list of CITS-based m6A residues, as online CTK Documentation recommended.
CNBP CLIP-sequencing
In brief, PANC-1 cells were washed with ice-cold PBS, cross-linked with 150 mJ/cm2 of 254 nm UV light and harvested on ice. Cell extraction was isolated and sonicated, followed by treating with DNase I and low-dilution RNase A. Prewashed Dynabeads protein A/G conjugated with anti-CNBP antibody was incubated with the extraction at 4°C overnight with rotating. RNA was then treated with proteinase K, followed by 3′ linker ligation, acidic phenol/chloroform extraction, and ethanol precipitation. Purified RNA was reverse transcribed with Superscript III reverse transcriptase (Life Technologies) and size-selected on an 8% 1 ris-borate EDTA (TBE)–Urea gel (Life Technologies).
Identification of CNBP binding site
For the identification of CNBP-binding site, we utilized the peak calling mode of CTK with default parameters. To finally visualize each CLIP-seq dataset, we counted positional coverage across the genome by bam2wig and viewing in IGV (34).
Pancreatic cancer-cell RNA-seq and GO pathway analysis
Raw-reads processing and quantification of gene-expression level were performed as described for tissue samples. We normalized RNA-seq data as transcripts per million (TPM) for further analysis. |Foldchange| ≥ 2 (WTAPP1 knockdown versus control) were set as the cutoffs for differentially expression genes (DEG). The clusterProfiler package (35) in R was used to perform functional enrichment analysis of DEGs.
Statistical analysis
For functional analysis, results were presented as mean ± SEM of 3 or more repeated experiments. We used Student t test to examine the difference of mean between 2 groups. Data in abnormal distribution were analyzed by nonparametric test. Kaplan–Meier method was used to compare the survival time by different levels of WTAPP1 RNA. Pearson correlations were calculated between WTAPP1 RNA and their m6A levels or WTAP protein levels. Correlations were considered significant and positive when P < 0.05 and r > 0.30. All statistical analyses were performed using the SPSS software package (version 20.0; IBM SPSS) and GraphPad Prism (version 8.0.0). P < 0.05 was considered significant for all statistical analyses.
Data availability
The RNA-seq data containing 65 tumor and 33 normal tissue samples in this paper have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers PRCJA005425 that are publicly accessible at https://bigd.big.ac.cn/gsa. RNA-seq, m6A individual-nucleotide resolution cross-linking and immunoprecipitation sequencing (miCLIP), and CNBP CLIP data have been deposited in the NCBI Short Read Archive with the BioProject ID- PRJNA693621 that are publicly accessible at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA693621.
For additional experimental procedures and URLs, please refer to Supplementary Materials and Methods.
Results
Identification of the pseudogene WTAPP1 involved in PDAC
We performed RNA-seq in 65 PDAC tumor and 33 normal samples and found 104 differentially expressed pseudogenes (log2 fold change < –0.58 or log2 fold change > 0.58, FDR < 0.05; see methods) and selected 10 top pseudogenes according to the rank of their FDR values for further investigation (Fig. 1A). We treated 2 frequently used PDAC cell lines, SW1990 and Capan-2, with siRNAs targeting these 10 pseudogenes and the resultant cell proliferation and migration changes were monitored with xCELLigence technology. The results suggested that, only siWTAPP1 was able to suppress cell proliferation and migration in both cell lines (Fig. 1B). We then examined the WTAPP1 RNA levels in Cohort 1 (N = 158) and Cohort 2 (N = 73) patients with PDAC (Supplementary Table S1) and the results showed that the levels of WTAPP1 RNA were significantly higher in tumors than in adjacent normal tissues (Fig. 1C) and in stages III/IV PDACs than in stages I/II PDACs (Fig. 1D). Kaplan–Meier analysis showed that PDAC patients with high WTAPP1 RNA level (≥ median) had worse prognosis and shorter overall survival time than those with low WTAPP1 level (< median; Fig. 1E). We also analyzed The Cancer Genome Atlas (TCGA) data and the Genotype-Tissue Expression (GTEx) data from the GEPIA online database (see URLs) and also found that the WTAPP1 RNA levels were significantly upregulated in PDAC (Supplementary Fig. S1A) and high levels were correlated with short overall survival time (Supplementary Fig. S1B).
Figure 1.
WTAPP1 is overexpressed in PDAC and is associated with PDAC-cell phenotypes and clinical outcomes. A, Heatmap of the 10 top differentially expressed pseudogenes in 65 PDAC-tumor and 33 normal tissue samples (All FDR <0.05). Red (higher expression) or blue (lower expression) represents the normalized expression value of indicated pseudogenes. B, Real-time analysis of proliferation and migration of SW1990 and Capan-2 cells at time point of 96 hours (proliferation) and 18 hours (migration), which were treated with negative-control RNA or siRNA of selected 10 top differentially expressed pseudogenes. Data are mean ± SEM from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 of Student t test. C and D, Expression levels of WTAPP1 RNA in surgically removed PDAC samples compared with paired nontumor tissue samples (C) and in advanced III/IV stages compared with early I/II stages (D) in two cohorts and combined sample. *, P < 0.05; **, P < 0.01; ***, P < 0.001 of Wilcoxon rank-sum tests in C and Mann–Whitney tests in D. E, Kaplan–Meier estimates of survival time in two PDAC patient cohorts and combined sample by different WTAPP1 RNA levels in tumors with HR = 1.84 [95% confidence interval (CI) = 1.29–2.64] for Cohort 1; HR = 1.89 (95% CI = 1.11–3.21) for Cohort 2; and HR = 1.88 (95% CI = 1.40–2.53) for combined sample.
We then performed experiments to characterize WTAPP1 RNA in PDAC cell lines and found that it presents at about 100 copies per cell and dominantly located in the cytoplasm (Supplementary Fig. S1C–S1E). Coding potential calculator based on sequence intrinsic features (CPC, see URLs) identified WTAPP1 as a noncoding transcript (Supplementary Table S5). UniProt human-protein profile of 20 PDAC cell lines in Cancer Cell Line Encyclopedia (CCLE) identified no peptides matching WTAPP1 ORFs (Supplementary Fig. S1F, Supplementary Table S5). Experimentally, FLAG-fused tagging of putative protein assays confirmed that the WTAPP1–FLAG fusion proteins did not exhibit the predicted relative molecular weight determined by Western blotting. The assays included MYC (FLAG-tagged) as a positive control and lncRNA without coding potential, WSPAR (FLAG-tagged), as a negative control (Supplementary Fig. S1G; ref. 36). Furthermore, ribosome sedimentation analysis revealed that similar to classic noncoding RNA XIST, WTAPP1 RNA has low affinity for polysome (Supplementary Fig. S1H), suggesting that it may not have protein-producing ability and is a bona fide pseudogene involved in PDAC prognosis.
WTAPP1 RNA promotes PDAC cells proliferation and invasiveness
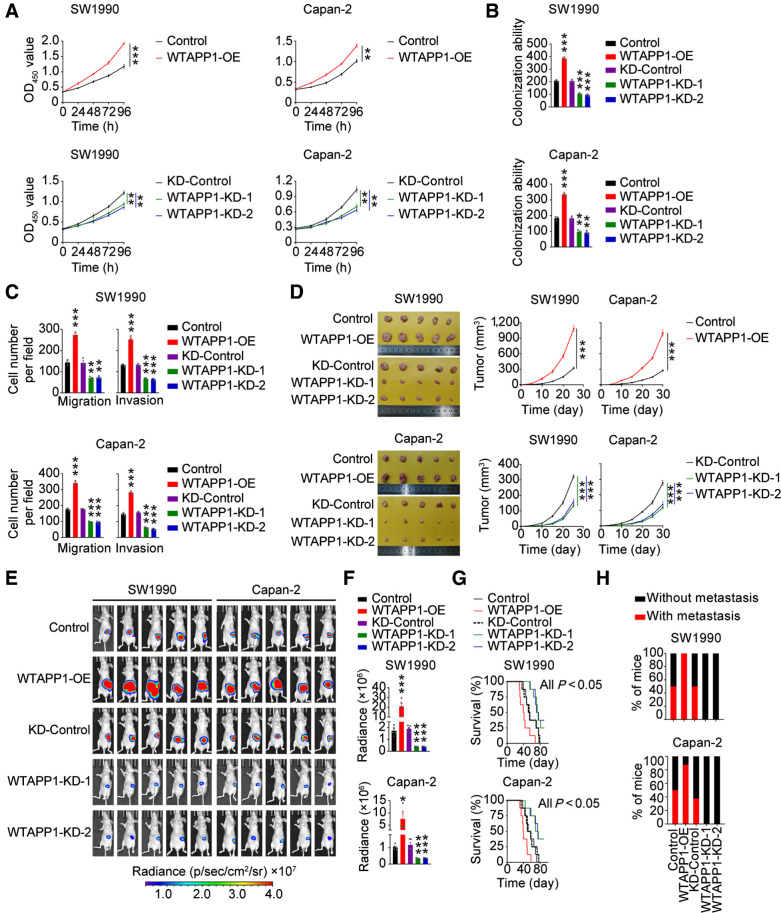
We then investigated the role of WTAPP1 RNA by changing its expression in PDAC cell lines (Supplementary Fig. S2A). Overexpressing WTAPP1 substantially enhanced the abilities of PDAC-cell proliferation, colony formation, migration, and invasion in vitro, while silencing WTAPP1 had opposite effects (Fig. 2A–C and Supplementary Fig. S2B and S2C). These effects were further verified in vivo in mouse xenograft models. Subcutaneous xenografts derived from PDAC cells overexpressing WTAPP1 had significantly increased growth rates while tumors derived from WTAPP1 silencing PDAC cells had significantly reduced growth rates compared with each control (Fig. 2D). We also implanted PDAC cells in mice pancreas and tested the effect of WTAPP1 on tumor metastasis. The results showed that WTAPP1 overexpression significantly increased distant metastasis of PDAC cells and reduced animal survival time, while WTAPP1 silence had opposite effects (Fig. 2E–H; and Supplementary Fig. S2D, Supplementary Table S6). Taken together, these results indicate that WTAPP1 is an oncogenic pseudogene capable of promoting proliferation and metastasis of PDAC cells.
Figure 2.
WTAPP1 promotes malignant phenotypes of PDAC in vitro and in vivo. A–C, Effects of WTAPP1 overexpression or knockdown on PDAC-cell proliferation (A), colony formation (B), and migration or invasion (C). Data are mean ± SEM from at least three independent experiments. D, Effects of WTAPP1 expression change on subcutaneous PDAC xenograft growth in mice. Images of xenograft tumors (left) and tumor growth curves over time (right). Data represent mean ± SEM (N = 5). E and F, Effects of WTAPP1 expression change on the growth of PDAC xenograft transplanted in mouse pancreas. Luminescence images (E) and quantification (F) of radiance intensity. Data represent mean ± SEM. G, Effect of WTAPP1 expression change on survival time of mice with PDAC xenograft in the pancreas (N = 8). H, Percentage of mice with or without metastasis in each group (N = 8). All statistical examinations in this figure are Student t tests unless specified. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
m6A modification of WTAPP1 RNA and the consequences
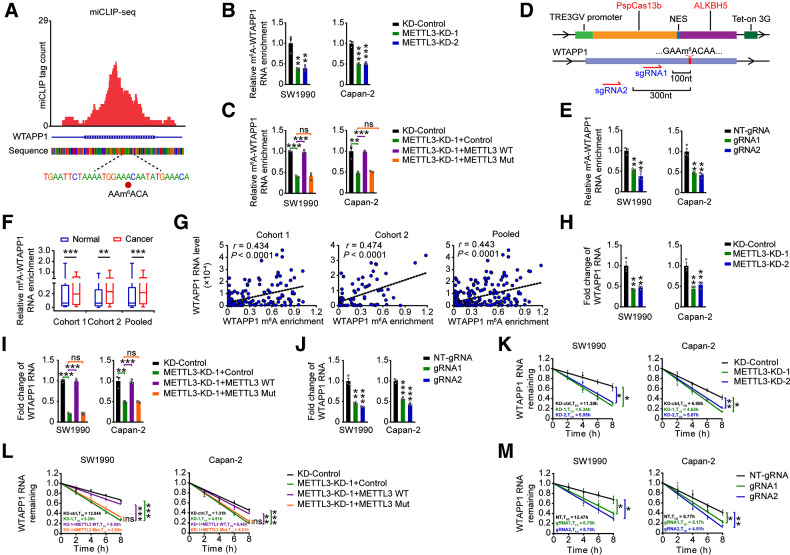
To explore why WTAPP1 is overexpressed in PDAC, we analyzed genomic alterations including mutations and CNAs of WTAPP1 in PDAC tissues derived from different datasets and the results were negative (Supplementary Fig. S3A). We did not find any CpG islands in the promoter region of WTAPP1 and the nearest DNA methylation probes is about 10,000 base pairs away from the transcription start site. It has been shown that posttranscriptional RNA m6A modification may affect RNA stability and then its level (16, 37). By using miCLIP-seq, we identified a m6A site at exon 6 (1076A) of WTAPP1 RNA (Fig. 3A). Subsequent m6A-specific RNA immunoprecipitation coupled qRT-PCR analysis showed that m6A levels of WTAPP1 RNA were decreased in cells with downregulation of METTL3 (Fig. 3B and Supplementary Fig. S3B), a well-known m6A writer. In PDAC cells with METTL3 silenced, overexpression of wild-typed METTL3 but not its catalytic mutant (38, 39) could restore m6A-WTAPP1 RNA level (Fig. 3C; and Supplementary Fig. S3C). Since recent studies showed that dCas13b-ALKBH5 combined with sgRNAs successfully demethylated specific m6A-RNAs (40, 41), we constructed a similar dm6ACRISPR system (dCas13b-ALKBH5 and gRNAs adjacent to the m6A site of WTAPP1 RNA) and the results further verified the presence of m6A in WTAPP1 RNA (Fig. 3D–E; Supplementary Fig. S3D).
Figure 3.
m6A modification increases WTAPP1 RNA level in PDAC. A, Identification of m6A site by miCLIP-seq in PDAC cells. The m6A residue was detected by cross-linking–induced mutation site in WTAPP1 RNA. Red tracks of miCLIP-seq are unique tag coverage and filled red circle denotes miCLIP-called m6A site. B, Effects of METTL3 knockdown on the levels of m6A-WTAPP1 RNA in PDAC cells. m6A-WTAPP1 RNA was detected by immunoprecipitation, followed by qRT-PCR analysis. C, m6A-WTAPP1 RNA levels in cells with METTL3-knockdown but transfected again with plasmids encoding WT METTL3 or its catalytic mutant (aa395–398, DPPW→APPA). D, Schematic representation of the domain organization of dCas13b-ALKBH5 expression cassette (top) and the positions of m6A site within WTAPP1 RNA and regions targeted by two gRNAs (bottom). E, m6A-WTAPP1 RNA levels in PDAC cells cotransfected with doxycycline-inducible dCas13b-ALKBH5 plasmid and NT-gRNA (control) or gRNAs with doxycycline pretreatment. F, m6A-WTAPP1 RNA levels in PDAC compared with paired nontumor tissues in the two cohorts and combined sample. Data displayed are minimum to maximum boxplots and difference was tested by Wilcoxon rank-sum test. G, Correlations between WTAPP1 RNA levels and their m6A levels in PDAC tumors of patients. The correlations were analyzed by Pearson test. H, Effects of METTL3 knockdown on WTAPP1 RNA levels in PDAC cells. I,WTAPP1 RNA levels in cells with METTL3 knockdown but transfected with plasmids encoding wild-type METTL3 or its catalytic mutant. J,WTAPP1 RNA levels in PDAC cells cotransfected with doxycycline-inducible dCas13b-ALKBH5 plasmid and NT-gRNA (control) or gRNAs with doxycycline pretreatment. K, Half-life of WTAPP1 RNA in cells with METTL3 knockdown. L, Half-life of WTAPP1 RNA in cells with METTL3 knockdown but transfected with plasmids encoding WT METTL3 or its catalytic mutant. M,WTAPP1 RNA stability in PDAC cells cotransfected with doxycycline-inducible dCas13b-ALKBH5 plasmid and NT-gRNA (control) or gRNAs with doxycycline pretreatment. Student t tests were used to examine the difference between two means unless specifically indicated. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
We next investigated the m6A-WTAPP1 RNA levels in PDAC of our patient cohorts and found that tumors had significantly higher m6A-WTAPP1 RNA levels than their adjacent normal tissues (Fig. 3F) and advanced PDACs had significantly higher m6A-WTAPP1 RNA levels than early PDACs (Supplementary Fig. S3E). Furthermore, patients having high m6A-WTAPP1 RNA level had significantly shorter survival time than those having low m6A-WTAPP1 RNA level (Supplementary Fig. S3F). In addition, we grouped PDAC patients into 4 based on the expression patterns of WTAPP1 m6A and RNA level in PDAC tissues and performed survival analysis and revealed that patients with low WTAPP1 RNA and m6A levels had better survival compared with those with high WTAPP1 RNA and/or high m6A levels (Supplementary Fig. S3G). Similar results were obtained when patients were stratified by METTL3 and WTAPP1 expression levels (Supplementary Fig. S3H). Furthermore, we also found that WTAPP1 RNA level was positively correlated with its m6A modification level (Fig. 3G).
We then examined the effect of METTL3-mediated m6A formation on the WTAPP1 levels in cells and found that knockdown of METTL3 significantly reduced the WTAPP1 RNA levels (Fig. 3H); however, restoration of wild-type (WT) METTL3 but not its inactive mutant in METTL3-knockdown cells could recover the WTAPP1 RNA levels (Fig. 3I). Furthermore, using the dm6ACRISPR system also significantly diminished the WTAPP1 RNA level in cells (Fig. 3J). These results indicate that METTL3-mediated m6A modification may have significant impact on WTAPP1 RNA stability. Indeed, we explored the stability of WTAPP1 and found that downregulation of METTL3 significantly reduced the stability of WTAPP1, which could be rescued when WT but not mutant METTL3 expression was recovered in cells (Fig. 3K and L). In addition, the dm6ACRISPR system also decreased the WTAPP1 stability (Fig. 3M). Together, these results suggest that higher WTAPP1 RNA level in PDAC might be attributed to the enhanced WTAPP1 RNA stability due to m6A modification.
CNBP recognizes m6A modification of WTAPP1 and accelerates WTAPP1 stability
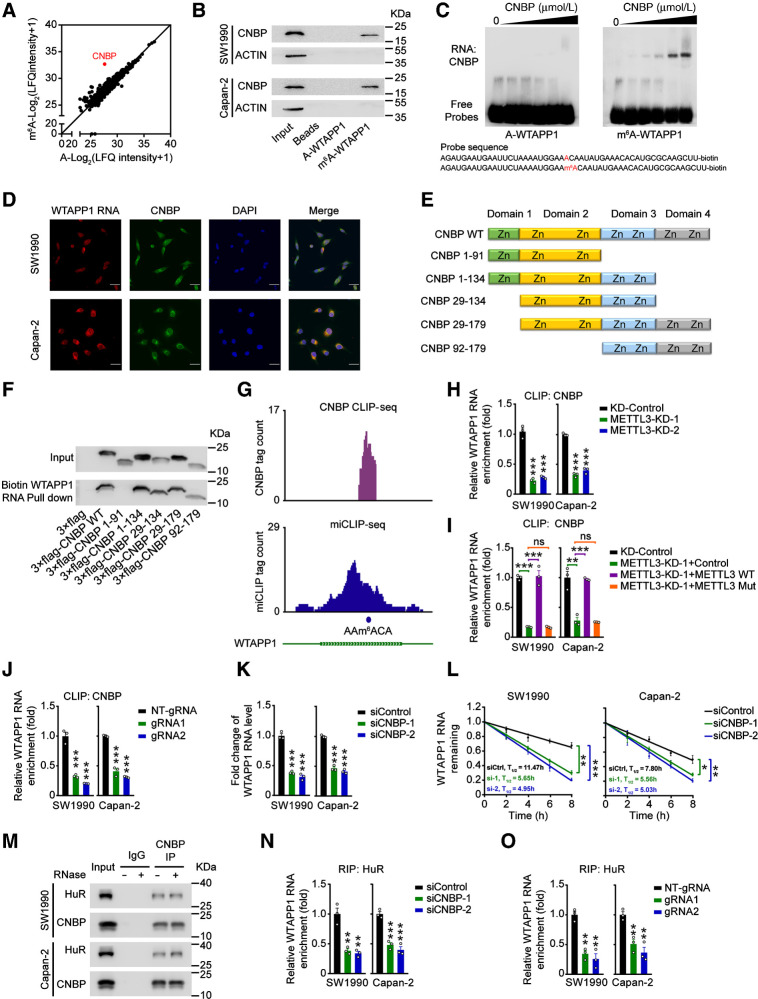
It is well known that deposit of m6A requires a reader to recognize and process the modified RNAs and several proteins such as IGF2BPs and YTHDFs have impact on m6A-modified RNA stability (15, 37). To explore the proteins that may recognize m6A in WTAPP1 RNA, we performed MS analysis of proteins obtained by RNA pulldown using biotin labeled 50-bp WTAPP1 or m6A-WTAPP1 RNA probes and the results showed that in 2 PDAC cell lines, CNBP was the most preferential protein bound to the m6A-WTAPP1 RNA probe but not WTAPP1 RNA probe without m6A modification (Fig. 4A and Supplementary Data 1), which was verified by Western blot assays (Fig. 4B), RNA electrophoretic mobility shift assays (Fig. 4C), and immunofluorescence (IF) assays in PDAC cells (Fig. 4D).
Figure 4.
CNBP is a m6A mediator that stabilizes WTAPP1 RNA. A, Scatter plot of proteins in SW1990 and Capan-2 cells that bound to m6A-modified or nonmodified WTAPP1 probe, both of which were 50-bp oligonucleotides centering on WTAPP1 RNA m6A site. Unique peptide of ≥ 5 was selected as filter criterium. The diagonal line marks the middle values of x and y axes. The red point represents CNBP protein. B, Western blot analysis showed specific association of m6A-WTAPP1 RNA with CNBP identified by proteomic screening. C, Electrophoretic mobility shift assays of recombinant CNBP with m6A-modified or unmodified WTAPP1 RNA probes. The reactions were carried out with a constant level of probes and various amount of recombinant CNBP (0–10 μmol/L). D, Colocalization of WTAPP1 RNA and CNBP protein in cells shown by IF. Scale bars, 30 μm. E and F, Pull-down assays with in vitro–transcribed biotin-labeled WTAPP1 RNA showed binding of WTAPP1 RNA to whole CNBP or truncated CNBP. Schematic of the domain structures of CNBP protein is shown in E and Western blot analysis of FLAG-tagged full-length CNBP or truncated CNBP in 293T cells is shown in F. G, IGV snapshots showed the CNBP CLIP-seq and miCLIP-seq reads distribution on WTAPP1 RNA. Purple tracks are unique tag coverage of CNBP CLIP-seq and blue tracks are unique tag coverage of miCLIP-seq, respectively. Filled blue circle denotes miCLIP-called m6A site. H, The levels of CNBP bound to WTAPP1 RNA in PDAC cells with or without METTL3 knockdown. I, The levels of CNBP bound to WTAPP1 RNA in PDAC cells with METTL3 knockdown but restored by transfection with WT METTL3 or its mutant. J, Binding levels of CNBP with WTAPP1 RNA in PDAC cells cotransfected with dCas13b-ALKBH5 and NT-gRNA or sgRNAs. K and L, Comparison of WTAPP1 RNA level and its stability in cells with or without CNBP silence. M, Immunoprecipitation (IP) assays show CNBP and HuR interaction in PDAC cells. N, The levels of HuR bound to WTAPP1 RNA in cells with or without CNBP silence. O, The levels of HuR bound to WTAPP1 RNA in cells with dm6ACRISPR system. All data are mean ± SEM from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant of Student t test.
We constructed various truncated CNBP (Fig. 4E) to examine the CNBP domains that bind WTAPP1 RNA, and found that the amino acid residues 92–134 in CNBP are required for the binding (Fig. 4F). CLIP-sequencing of CNBP revealed that CNBP binds to the WTAPP1 RNA m6A site (Fig. 4G). Furthermore, we observed a high binding intensity for CNBP centering at m6A residues and vice versa for the m6A sites. The m6A sites obtained by miCLIP substantially overlapped with CNBP-binding sites obtained by CNBP CLIP-seq (Supplementary Fig. S4A), indicating that CNBP may also recognize m6A sites in RNAs other than m6A-WTAPP1 RNA.
Next, we examined whether the m6A modification is necessary for the association between CNBP and WTAPP1 RNA. We found that METTL3 depletion substantially reduced the interaction of CNBP with WTAPP1 RNA in cells where CNBP protein-expression level was not altered (Fig. 4H; Supplementary Fig. S4B) and restoring WT METTL3 but not its mutant enhanced the level of CNBP and WTAPP1 RNA association (Fig. 4I). In addition, cells transferred with the dm6ACRISPR system also showed decreased level of CNBP and WTAPP1 RNA association (Fig. 4J). We also found that CNBP was aberrantly overexpressed in PDAC tumors compared with normal tissues (Supplementary Fig. S4C), which showed the same expression tendency with WTAPP1 in PDAC tissues. All these results suggest that the association between CNBP and WTAPP1 RNA is dependent on the m6A modification. Similarly, CNBP knockdown substantially reduced the level and stability of WTAPP1 RNA in cells (Fig. 4K, L and Supplementary Fig. S4D) and consequently diminished PDAC cell proliferation, migration, and invasion abilities (Supplementary Fig. S4E–S4G), indicating that CNBP is a m6A mediator that plays a role in enhancing the stability and eventually the high level of WTAPP1 RNA and aggressive phenotype in PDAC cells.
Because m6A readers may enhance the stability of their host RNAs by recruiting cofactors (16), we speculated that the stabilization of WTAPP1 RNA by CNBP might have the same mechanism. Interestingly, we found that HuR, an RNA stabilizer, was identified by pull-down coupled MS analysis (Supplementary Data S1). Further immunoprecipitation assays confirmed a persisting CNBP and HuR interaction independent on RNA (Fig. 4M); RIP assays with HuR antibody revealed that CNBP silence substantially decreased HuR bound to WTAPP1 RNA (Fig. 4N). We also performed RIP assays with HuR antibody in PDAC cells with dm6ACRISPR system and the results showed that decreased WTAPP1 RNA m6A modification significantly diminished binding of HuR to WTAPP1 (Fig. 4O). These results suggest that CNBP probably recruits RNA stabilizers HuR to promote WTAPP1 RNA stability.
WTAPP1 promotes WTAP translation by recruiting translation initiation factor EIF3 complex
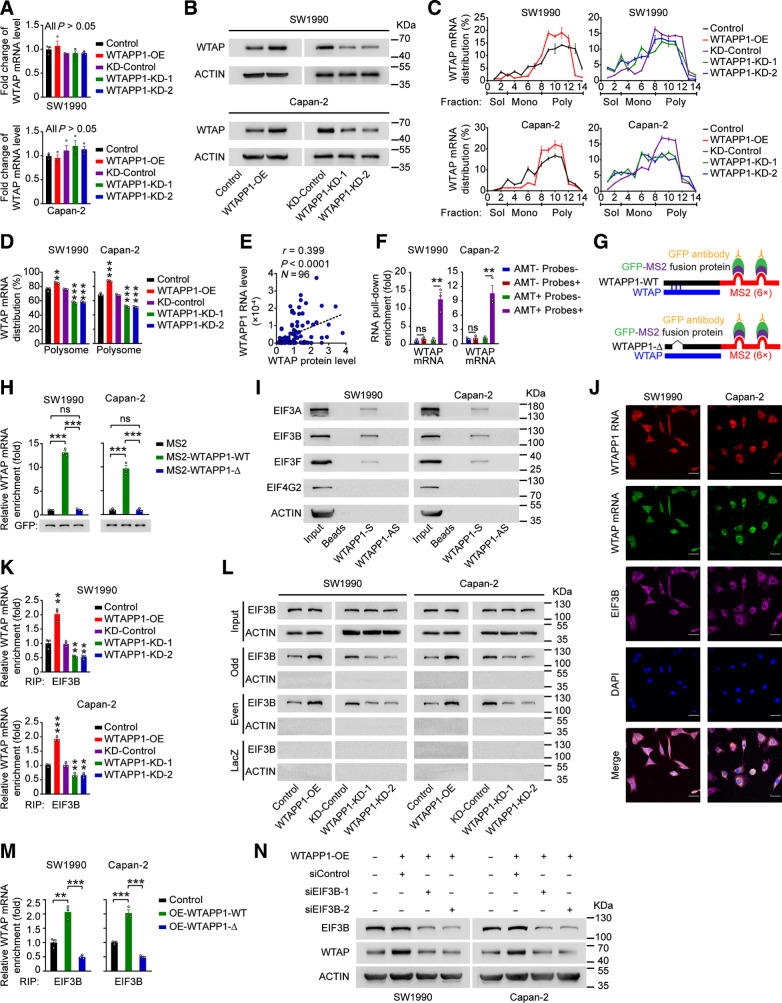
We next wanted to seek the functional mechanism underlying the oncogenic role of overexpressed WTAPP1 RNA in PDAC. Since pseudogenes may be involved in regulating their protein-coding counterparts and WTAPP1 RNA is mainly distributed in cytoplasm of PDAC cells, we speculated that WTAPP1 RNA might disturb the post-transcriptional regulation or protein translation of its protein-coding counterpart WTAP RNA. We found that changing the level of WTAPP1 RNA did not significantly altered the levels of WTAP mRNA; however, it significantly increased WTAP protein level (Fig. 5A and B), implying that overexpression of WTAPP1 RNA may promote protein translation of WTAP. We then performed a series of polysome profile analyses and the results showed that WTAPP1 overexpression increased but knockdown decreased the proportion of WTAP mRNA distribution in polysome fraction, without altering the distribution of mRNAs produced by other genes (Fig. 5C and D; and Supplementary Fig. S5A–S5C), indicating that WTAPP1 RNA specifically increases the translational efficiency of WTAP. A significant positive correlation between the WTAPP1 RNA and WTAP protein levels was detected in our 96 clinical PDAC tissue samples (Fig. 5E; and Supplementary Fig. S5D).
Figure 5.
WTAPP1 RNA promotes WTAP translation through interacting with WTAP mRNA to recruit EIF3 complex. A,WTAP mRNA levels in PDAC cells with WTAPP1 expression change. B, Western blot analysis of the change of WTAP protein levels in cells with WTAPP1 overexpression or knockdown. C and D, Polysome fraction analysis in cells with WTAPP1 expression change. The level of WTAP mRNA in each gradient fraction was measured by qRT-PCR and plotted as a percentage of total WTAP mRNA level in that sample (C). The translational activity associated with each fraction is indicated as untranslated (Sol, soluble), moderately translated (Mono, monosome), and actively translated (Poly, polysome). Polysome fractions are shown as bar graphs in D. E, Pearson correlation of the WTAPP1 RNA and WTAP protein levels in PDAC tissues (N = 96). F, Enrichment of WTAP mRNA in RNA pull-down with WTAPP1 RNA from PDAC cells. Biotinylated WTAPP1 RNA probes were incubated with AMT-crosslinked or untreated RNA, the interaction with WTAPP1 was quantified by qRT-PCR. G, Schematic diagram of MS2-RIP assay. Full length (WTAPP1-WT; top) or binding sites–deleted WTAPP1 (WTAPP1-Δ, bottom) linked to the MS2 fragments. H, MS2-RIP assay shows the levels of WTAP mRNA bound to WTAPP1 RNA in cells with ectopic expression of WT WTAPP1 (MS2-WTAPP1-WT) or WTAPP1 with binding sites deletion (MS2-WTAPP1-Δ). I, Western blot showed proteins identified by MS in RNA pull-down assay with biotinylated WTAPP1 sense or antisense probes. J, Colocalization of WTAPP1 RNA, WTAP mRNA, and EIF3B protein revealed by IF assays. Scale bars, 30 μm. K, The levels of EIF3B bound to WTAP mRNA in cells with WTAPP1 expression change. L, ChIRP assays showed proteins retrieved by WTAP mRNA probes labeled with biotin and divided into the odd or even group. After incubation with streptavidin beads, proteins were analyzed by Western blot. LacZ served as the probe negative control while β-ACTIN served as retrieved-protein negative control. M, The levels of EIF3B bound to WTAP mRNA in cells with ectopic expression of WT WTAPP1 or binding sites–deleted WTAPP1. N, Western blot analysis of the WTAP protein level in PDAC cells with WTAPP1 overexpression and EIF3B silence. All measurement data are from three independent experiments and are represented as mean ± SEM. Student t test was used except for the correlation analysis (E), which was Pearson test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
We then investigated how WTAPP1 RNA may affect WTAP translation. Based on the public ENCORI data (see URLs) suggesting that WTAPP1 RNA may interact with WTAP mRNA, we assumed that WTAPP1 RNA might recruit certain proteins to promote WTAP translation via interacting with WTAP mRNA. To test this hypothesis, we firstly performed the 4′-AMT assays to verify the interaction of these 2 RNAs. As shown in Fig. 5F, WTAP mRNA could be retrieved from WTAPP1 RNA probes in AMT-treated PDAC cells, suggesting that WTAP mRNA interacts with WTAPP1 RNA. Next, we analyzed the binding region of WTAPP1 using the bibiserv website (see URLs) and the results suggested that a region at exon 1–2 of WTAPP1 may interact with WTAP mRNA (Supplementary Fig. S5E). We then performed MS2-based RIP assays in cells transfected with plasmid containing full length or binding sites delated WTAPP1 and GFP-MS2 fusion protein (Fig. 5G; Supplementary Fig. S5F) and qRT-PCR analysis showed that the WTAP mRNA level was greatly elevated in cells with MS2-WTAPP1-WT compared with that in cells with empty MS2 or MS2-WTAPP1-Δ (Fig. 5H), confirming the binding sites of WTAPP1 is required for its interaction with WTAP mRNA.
We then performed RNA pulldown using WTAPP1 RNA or its antisense probe followed by MS analysis and revealed several translation initiation factors including the components of the EIF3 complex, EIF3B, EIF3A, and EIF3F (Supplementary Data S2), which were confirmed by Western blotting assays (Fig. 5I). EIF3 complex consisting of 13 subunits (EIF3A to EIF3M) is essential for initiation of protein synthesis and EIF3B serves as a core scaffolding protein (42–44). Through IF assays, we detected an association of EIF3B with WTAPP1 and WTAP RNAs in the cytoplasm of PDAC cells (Fig. 5J). Notably, RIP assays using EIF3B antibody revealed that WTAPP1 overexpression increased but silence decreased the interaction between EIF3B and WTAP mRNA (Fig. 5K). Consistent with this result, ChIRP assays also showed that forced change of WTAPP1 expression specifically increased the interaction of EIF3B with WTAP mRNA (Fig. 5L; and Supplementary Fig. S5G). Furthermore, we found that overexpression of full-length but not binding site-deleted WTAPP1 increased EIF3B and WTAP mRNA interaction (Fig. 5M), suggesting that the interaction of these two RNAs facilitated EIF3B bound to WTAP mRNA. Rescue assays verified that the enhanced WTAP translation by WTAPP1 RNA was EIF3B-dependent since Western blot assays showed that EIF3B knockdown significantly decreased the WTAP protein levels in PDAC cells with WTAPP1 overexpressed (Fig. 5N). Furthermore, we analyzed WTAP protein levels by IHC staining and WTAPP1 and WTAP RNAs by FISH in clinical tissue samples and found that PDAC with high WTAP protein level also had high WTAPP1 RNA level, and the 2 types of RNAs were colocalized (Supplementary Fig. S5H).
Collectively, these results demonstrate that overexpressed WTAPP1 RNA may enhance the recruitment of EIF3B to form more EIF3 complex that promotes WTAP translation.
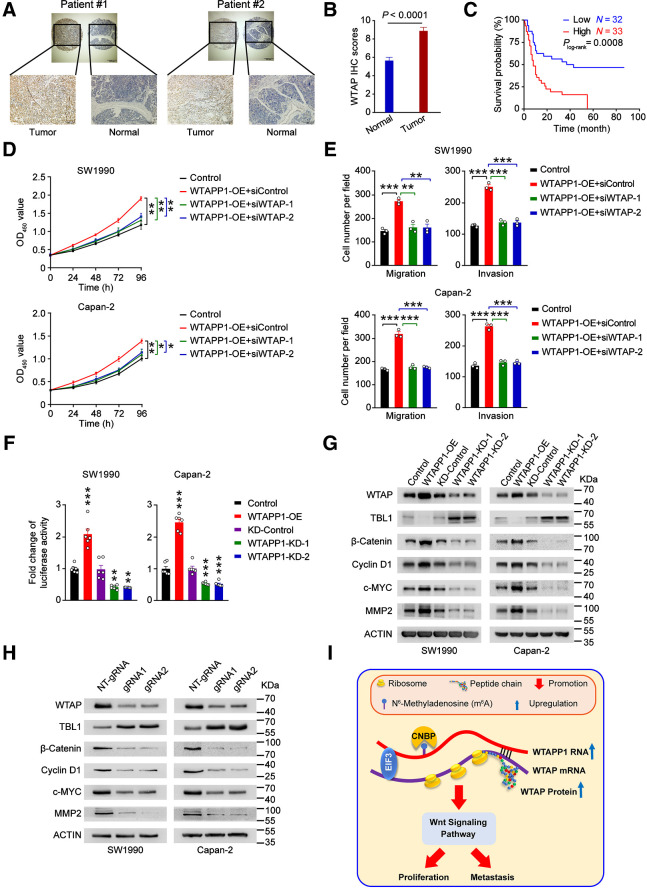
WTAPP1 is required for WTAP-initiated Wnt signaling activation
WTAP has recently been recognized as an oncoprotein in various types of cancer including PDAC (45–48). By IHC staining of 65 samples (Fig. 6A; Supplementary Data 3), we found that WTAP levels were significantly higher in tumors than in paired normal tissues (Fig. 6B). Patients with high WTAP level in tumor had significantly shorter survival time than patients with low WTAP level in tumor (Fig. 6C). We also grouped PDAC patients into 4 based on the WTAPP1 RNA and WTAP protein levels and found that patients with low WTAPP1 RNA and low WTAP protein levels had the best prognosis compared with other groups (Supplementary Fig. S6A). We also performed in vitro assays to examine the oncogenic effects of WTAP in PDAC cell lines and revealed that knockdown of WTAP in cells with WTAPP1 overexpression significantly inhibited cell proliferation, migration, and invasion (Fig. 6D and E; Supplementary Fig. S6B), suggesting that the effect of WTAPP1 RNA on malignant phenotypes of PDAC cells is dependent on WTAP.
Figure 6.
Wnt signaling is excessively activated in PDAC with WTAPP1 overexpression. A, Representive IHC staining of WTAP proteins in PDAC and paired nontumor tissue samples. Scale bars, 500 μm (top) and 200 μm (bottom). B, Quantitative statistics of IHC staining of WTAP in 65 paired PDAC tumor and nontumor tissue samples. P, Wilcoxon rank-sum test. C, Kaplan–Meier estimates of survival time in PDAC patients by WTAP protein IHC scores in tumor. The adjusted HR for death of high WTAP IHC score was 2.95 (95% CI = 1.57–5.57). D and E, Effects of WTAP expression knockdown on PDAC cell proliferation (D) or migration and invasion (E) resulted from WTAPP1 overexpression. F, Reporter gene assays with TCF/LEF1 luciferase and Renilla reporter plasmid showed the Wnt signaling activity was significantly altered in PDAC cells when WTAPP1 expression was changed. G and H, Western blot analysis demonstrated the expression alteration of signaling molecules downstream of WTAP in PDAC cells when WTAPP1 expression was changed (G) or transfected with dm6ACRISPR system (H). Data in D–F are mean ± SEM from three independent experiments. **, P < 0.01; ***, P < 0.001 of Student t test. I, A proposed action model for excessive WTAPP1 RNA level caused by the RNA m6A modification in the progression of PDAC.
We next wanted to elucidate the action mechanism in which WTAPP1 provokes PDAC cell growth and invasiveness by looking at the transcriptomic alterations in PDAC cells with WTAPP1 knockdown and pathway enrichment of DEGs. Gene ontology analysis suggested that the Wnt signaling pathway was strongly enriched in cells with WTAPP1 decrease (Supplementary Fig. S6C). Because WTAP has been shown linking to the WT1–TBL1–Wnt signaling axis (48), WTAPP1 may evoke Wnt signaling. We first tested this notion by analyzing the TCGA data and the results showed that WTAPP1 is positively correlated with positive regulators (JAG1, MYC) and negatively correlated with negative regulators (HDAC2 and NKD1) in the Wnt signaling pathway (Supplementary Fig. S6D). Luciferase reporter assays revealed that increasing WTAPP1 expression substantially activated the Wnt signaling while suppressing WTAPP1 expression substantially inhibited this signaling (Fig. 6F). Consistent with previous findings that Wnt signaling can target numerous oncogenes and contribute to tumor progression (49–52), we also observed that WTAPP1 overexpression significantly upregulated the expression of genes downstream the Wnt signaling pathway except for TBL-1, which was downregulated as reported before (48). Opposite results were observed in cells with WTAPP1 knockdown (Fig. 6G). Similarly, WTAP knockdown significantly inhibited the expression of genes downstream the Wnt signaling pathway in cells with WTAPP1 overexpression (Supplementary Fig. S6E). Furthermore, we found that PDAC cells treated with the dm6ACRISPR system, which depressed WTAPP1 expression, had substantially lower expression of genes downstream the Wnt signaling pathway compared with control cells (Fig. 6H). Overall, these results combined with the results presented above suggest that high level of WTAPP1 RNA resulted from the excessive m6A modification may activate the Wnt signaling in PDAC pathogenic processes (Fig. 6I).
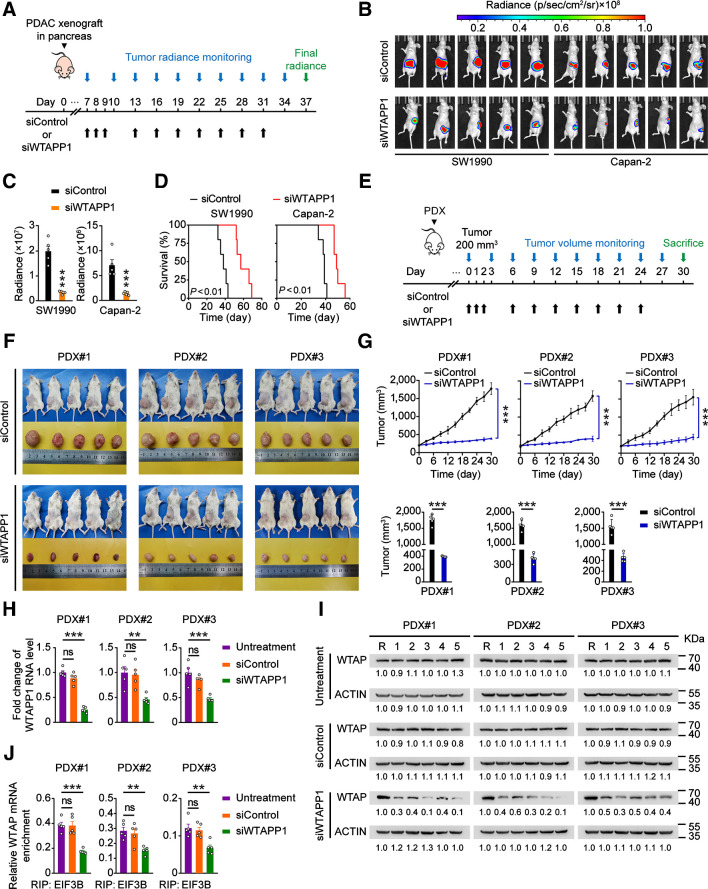
WTAPP1 is a therapeutic target in mouse xenograft tumors
Because WTAPP1 shows a strong oncogenic effect in PDAC cells, we established xenografts derived from WTAPP1-upregulated PDAC luciferase-expressing cells in mice and treated the animals with in vivo–optimized siWTAPP1 (Fig. 7A). We found that treatment of siWTAPP1 (75 mg/kg, i.v.) significantly reduced tumor burden and prolonged survival time of mice compared with siControl (Fig. 7B–D). We also established PDXs in mice and performed similar siWTAPP1 treatment (Fig. 7E). The results showed that administration of siWTAPP1 significantly reduced PDX growth rates compared with administration of siControl. The siWTAPP1 reagent showed no apparent toxicity to mice since the animals had no significant bodyweight reduction (Fig. 7F and G; Supplementary Fig. S6F). The qRT-PCR and Western blot analyses confirmed that treatment of siWTAPP1 effectively inhibited WTAPP1 RNA and WTAP protein expressions and reduced the binding ability of EIF3B with WTAP mRNA in PDXs (Fig. 7H–J). These results strong suggest that WTAPP1 may be a therapeutic target for PDAC treatment.
Figure 7.
WTAPP1 RNA is a therapeutic target in mouse PDAC xenografts. A, Timeline schematic for treatment of mice carrying PDAC xenograft in the pancreas with siWTAPP1. Colored arrows indicate the times when different events occurred. B and C, Administration of siWTAPP1 via tail vein significantly suppressed tumor growth and metastasis. Shown are the radiant images of xenograft in mice (N = 5; B) and tumor growth rate expressed as radiance intensity (C). D, Survival times of mice treated with or without siWTAPP1. E, Timeline schematic for treatment of mice with PDX. Colored arrows indicate the times when different events occurred. F, Shown are the images of PDXs from three patients in 5 mice. G, Tumor growth curves (top) and tumor volumes in the end of experiment (bottom), with administration of siWTAPP1 via tail vein. H–J, Effects of siWTAPP1 treatment on WTAPP1 RNA level determined by qRT-PCR (H), WTAP protein level analyzed by Western blotting (I), and the ability of EIF3B bound to WTAP mRNA detected by EIF3B RIP-qPCR (J). R in I stands for the same positive reference sample for loading adjustment on each gel. Each protein band was semiquantified by gray density and the value for each band is relative to density of corresponding band of R. Data in G represent mean ± SD and other measurements are mean ± SEM. **, P < 0.01; ***, P < 0.001; ns, not significant of Student t test.
Discussion
In this study, we have identified a panel of aberrantly expressed pseudogenes in PDAC and among them, WTAPP1 has the strongest effect on PDAC-malignant phenotypes as demonstrated by our function screening. We found that WTAPP1 RNA level is significantly higher in PDAC and the increased level is associated with shorter survival time in patients. WTAPP1 RNA is stabilized by m6A modification and the excessive WTAPP1 RNA level may promote the EIF3 complex formation to enhance oncoprotein WTAP translation, which excessively activates Wnt signaling that plays a role in PDAC. Several novel findings have been achieved in this study. First, we have found for the first time that pseudogene WTAPP1 RNA level is elevated by aberrant m6A modification and plays an oncogenic role in human PDAC. Second, we have identified CNBP as the potential m6A reader that stabilizes WTAPP1 RNA. Third, we have revealed that WTAPP1 RNA can interact with EIF3B to promote the formation of translation initiation complex that enhance WTAP translation, which links to the activation of Wnt signaling in PDAC progression.
Accumulating evidence has suggested that pseudogene produced RNAs may play diverse roles in physiological and pathological processes, especially in cancer (5, 6). Up to date, there have been two studies reporting WTAPP1 produced RNA that plays an oncogenic role in laryngeal and lung cancer (53, 54). Here, we report that WTAPP1 RNA is significantly upregulated in PDAC, which is associated with poor prognosis of PDAC patients. Therefore, our findings in the current study provide additional evidence supporting WTAPP1 as an oncogenic pseudogene in cancer. More importantly, we have performed comprehensive functional analyses to discover the underlying molecular mechanism for the action of this oncogenic pseudogene in PDAC. The results have extended our knowledge about the role of this class of genomic makeup. Furthermore, we have demonstrated that the oncogenic role of WTAPP1 RNA in PDAC can be targeted and suppressed by siWTAPP1. This information might also be interesting for the drug and treatment option development of PDAC.
We have revealed that the elevated level of WTAPP1 RNA in PDAC is resulted from aberrant m6A modification, which significantly increases the RNA stability. Several previous studies have also suggested this mechanism for some oncogenic RNA (10, 15, 16, 37); however, little has been known about the m6A modification in PDAC (12, 13). In view of our findings that WTAPP1 RNA is aberrantly m6A-modified, it would be interesting and important to fully characterize the m6A modification at whole-transcriptome level in PDAC and to further discover the underlying mechanism for the formation and the consequences of this epigenetic modification. In addition to this, there might be other mechanisms such as microRNA interaction attributable to the aberrant WTAPP1 level in PDAC, which would be interesting to investigate.
The biological importance of m6A modification relies on m6A-binding proteins (readers) and previous studies have reported several RNA-binding proteins as m6A readers, such as YTH domain-containing proteins and IGF2BP proteins (16, 37). In the present study, we have shown that CNBP preferentially interacts with and stabilizes m6A-modified WTAPP1 RNA. CNBP is an RNA-binding protein and transcription factor that contains seven highly conserved zinc finger domains involving in RNA transcription, stabilization, and translation (55, 56). It has been shown that under the guidance of lncRNA LAST, CNBP binds to and stabilizes CCND1 mRNA (55). In this study, we have found by several experimental setting that CNBP is likely an m6A reader that recognizes the m6A modification in WTAPP1 RNA. By analyzing the published data (57), we have also revealed that CNBP preferentially binds the GGm6ACU site in the probes in both HeLa cells and mouse 3T3 cells. The consistency of our results with others indicates that CNBP may be a common reader recognizing the m6A sites in various RNAs, which is warranted to further investigation.
ncRNAs transcribed by pseudogenes play diverse roles in posttranscriptional regulation of many protein-coding genes as antisense RNAs, endogenous siRNAs, and competing endogenous RNAs (5, 6, 8). For example, it has been shown that neural nitric oxide synthase (nNOS) mRNA can directly bind RNA produced by its pseudogene to form double-strand RNA–RNA duplex and consequently diminish nNOS mRNA translation (58). In this study, we have demonstrated that WTAPP1 RNA has ability to promote WTAP translation, providing a new acting model for pseudogenes.
WTAP has increasingly attracted the research attention because of its oncogenic role in various types of malignancies including PDAC (45–48). In addition to its function as a RNA methyltransferase, WTAP also plays a carcinogenic role through activating signaling pathways. For instance, it has been shown that WTAP may activate the Wnt signaling pathway by inhibiting TBL1 transcription and reducing β-catenin degradation in colorectal cancer (48). Because excessive WTAPP1 RNA promotes WTAP protein translation, one may expect that Wnt signaling would be aberrantly enhanced in PDAC, which has previously been observed in PDAC (49, 50). We have really demonstrated that increased WTAPP1 RNA significantly evokes the Wnt signaling via increasing WTAP and suppressing TBL-1, which is probably a molecular mechanism for PDAC development and progression. Because the Wnt signaling pathway plays an important role in many other cancers, it would be interesting to explore whether WTAPP1 has the same function in other types of cancer.
Because WTAPP1 RNA shows oncogenic effect and its levels increase in a disease stage–dependent manner, we assumed that it may be a valuable therapeutic target for PDAC. Indeed, in xenograft models, we are able to show that administration of siWTAPP1 significantly reduced the growth rate and distant metastasis of PDAC in mice. The efficacy of siWTAPP1 administration in xenografts may suggest a potential option for the development of PDAC treatment although further investigations are needed such as systematical evaluation of the potential toxicity caused by WTAPP1 inhibition.
In conclusion, we have identified WTAPP1 as an oncogenic pseudogene in PDAC acting via promoting the translation of WTAP protein, a molecule that activates Wnt signaling. WTAPP1 RNA can be stabilized by aberrant m6A modification mediated by CNBP. PDAC cell growth and invasiveness in vitro and in vivo can be suppressed by siWTAPP1, highlighting that WTAPP1 may serve as a potential novel therapeutic target for PDAC.
Authors' Disclosures
X. Huang reports grants from Natural Science Foundation of China during the conduct of the study. D. Lin reports grants from Natural Science Foundation of China during the conduct of the study. J. Zheng reports grants from Natural Science Foundation of China during the conduct of the study. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Materials and Methods
Supplementary Figure S1-S6 and corresponding Supplementary Figure legends
Supplementary Table S1-S6
WTAPP1-m6A/WTAPP1 50bp RNA probe based Pull-down mass spectrum data
WTAPP1 full length RNA pull down with mass spectrum
Clinical information for patients with IHC
Acknowledgments
This study was supported by the Natural Science Foundation of China (81772586, 82072617 to J. Zheng; 81802407 to X. Huang; and 91753142 to D. Lin), Program for Guangdong Introducing Innovative and Entrepreneurial Teams (2017ZT07S096 to D. Lin), National Young Top-notch Talent Support Program (to J. Zheng), Natural Science Foundation of Guangdong Province (2018A030313327 to X. Huang), and Sun Yat-sen University Intramural Funds (to D. Lin and to J. Zheng).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
J. Deng: Data curation, formal analysis, validation, visualization, methodology, writing–original draft, writing–review and editing. J. Zhang: Formal analysis, validation, visualization, methodology. Y. Ye: Data curation, software, formal analysis, visualization, methodology. K. Liu: Methodology. L. Zeng: Methodology. J. Huang: Data curation, formal analysis. L. Pan: Methodology. M. Li: Methodology. R. Bai: Methodology. L. Zhuang: Methodology. X. Huang: Funding acquisition, methodology. G. Wu: Methodology. L. Wei: Methodology. Y. Zheng: Methodology. J. Su: Methodology. S. Zhang: Methodology. R. Chen: Conceptualization, supervision, project administration. D. Lin: Conceptualization, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing. J. Zheng: Conceptualization, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Grossberg AJ, Chu LC, Deig CR, Fishman EK, Hwang WL, Maitra A, et al. Multidisciplinary standards of care and recent progress in pancreatic ductal adenocarcinoma. CA Cancer J Clin 2020;70:375–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet 2020;395:2008–20. [DOI] [PubMed] [Google Scholar]
- 4. Kalyana-Sundaram S, Kumar-Sinha C, Shankar S, Robinson DR, Wu YM, Cao X, et al. Expressed pseudogenes in the transcriptional landscape of human cancers. Cell 2012;149:1622–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu X, Gao A, Ji L, Xu J. Pseudogene in cancer: real functions and promising signature. J Med Genet 2015;52:17–24. [DOI] [PubMed] [Google Scholar]
- 6. Karreth FA, Reschke M, Ruocco A, Ng C, Chapuy B, Leopold V, et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell 2015;161:319–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheetham SW, Faulkner GJ, Dinger ME. Overcoming challenges and dogmas to understand the functions of pseudogenes. Nat Rev Genet 2020;21:191–201. [DOI] [PubMed] [Google Scholar]
- 8. Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010;465:1033–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang H, Weng H, Chen J. m(6)A Modification in coding and non-coding RNAs: Roles and therapeutic implications in cancer. Cancer Cell 2020;37:270–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N (6)-methyladenosine modification in cancers: current status and perspectives. Cell Res 2018;28:507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, et al. m(6)A Demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 2017;31:591–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. He Y, Hu H, Wang Y, Yuan H, Lu Z, Wu P, et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem 2018;48:838–46. [DOI] [PubMed] [Google Scholar]
- 13. Zhang J, Bai R, Li M, Ye H, Wu C, Wang C, et al. Excessive miR-25–3p maturation via N (6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun 2019;10:1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 2019;20:608–24. [DOI] [PubMed] [Google Scholar]
- 15. Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 2018;28:616–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of RNA N (6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol 2018;20:285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pan T. N6-methyl-adenosine modification in messenger and long non-coding RNA. Trends Biochem Sci 2013;38:204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang D, Qiao J, Wang G, Lan Y, Li G, Guo X, et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res 2018;46:3906–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016;537:369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res 2017;45:6051–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou KI, Parisien M, Dai Q, Liu N, Diatchenko L, Sachleben JR, et al. N(6)-methyladenosine modification in a long noncoding RNA hairpin predisposes its conformation to protein binding. J Mol Biol 2016;428:822–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 2011;17:10–2. [Google Scholar]
- 23. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019;47:D766–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mueller MT, Hermann PC, Witthauer J, Rubio-Viqueira B, Leicht SF, Huber S, et al. Combined targeted treatment to eliminate tumorigenic cancer stem cells in human pancreatic cancer. Gastroenterology 2009;137:1102–13. [DOI] [PubMed] [Google Scholar]
- 27. Rubio-Viqueira B, Jimeno A, Cusatis G, Zhang X, Iacobuzio-Donahue C, Karikari C, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin Cancer Res 2006;12:4652–61. [DOI] [PubMed] [Google Scholar]
- 28. Tadros S, Shukla SK, King RJ, Gunda V, Vernucci E, Abrego J, et al. De novo lipid synthesis facilitates gemcitabine resistance through endoplasmic reticulum stress in pancreatic cancer. Cancer Res 2017;77:5503–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. Mapping argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc 2014;9:263–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Webb S, Hector RD, Kudla G, Granneman S. PAR-CLIP data indicate that Nrd1-Nab3-dependent transcription termination regulates expression of hundreds of protein coding genes in yeast. Genome Biol 2014;15:R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shah A, Qian Y, Weyn-Vanhentenryck SM, Zhang C. CLIP Tool Kit (CTK): a flexible and robust pipeline to analyze CLIP sequencing data. Bioinformatics 2017;33:566–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 2015;12:767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol 2011;29:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 2015;16:413–25. [DOI] [PubMed] [Google Scholar]
- 37. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014;505:117–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell 2016;62:335–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2020;69:1193–205. [DOI] [PubMed] [Google Scholar]
- 40. Li Z, Peng Y, Li J, Chen Z, Chen F, Tu J, et al. N (6)-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat Commun 2020;11:2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li J, Chen Z, Chen F, Xie G, Ling Y, Peng Y, et al. Targeted mRNA demethylation using an engineered dCas13b-ALKBH5 fusion protein. Nucleic Acids Res 2020;48:5684–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys 2004;37:197–284. [DOI] [PubMed] [Google Scholar]
- 43. Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 2006;31:553–62. [DOI] [PubMed] [Google Scholar]
- 44. ElAntak L, Tzakos AG, Locker N, Lukavsky PJ. Structure of eIF3b RNA recognition motif and its interaction with eIF3j: structural insights into the recruitment of eIF3b to the 40 S ribosomal subunit. J Biol Chem 2007;282:8165–74. [DOI] [PubMed] [Google Scholar]
- 45. Li BQ, Huang S, Shao QQ, Sun J, Zhou L, You L, et al. WT1-associated protein is a novel prognostic factor in pancreatic ductal adenocarcinoma. Oncol Lett 2017;13:2531–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li BQ, Liang ZY, Seery S, Liu QF, You L, Zhang TP, et al. WT1 associated protein promotes metastasis and chemo-resistance to gemcitabine by stabilizing Fak mRNA in pancreatic cancer. Cancer Lett 2019;451:48–57. [DOI] [PubMed] [Google Scholar]
- 47. Chen Y, Peng C, Chen J, Chen D, Yang B, He B, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer 2019;18:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang J, Tsoi H, Li X, Wang H, Gao J, Wang K, et al. Carbonic anhydrase IV inhibits colon cancer development by inhibiting the Wnt signalling pathway through targeting the WTAP-WT1-TBL1 axis. Gut 2016;65:1482–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li L, Li Z, Kong X, Xie D, Jia Z, Jiang W, et al. Down-regulation of microRNA-494 via loss of SMAD4 increases FOXM1 and beta-catenin signaling in pancreatic ductal adenocarcinoma cells. Gastroenterology 2014;147:485–97. [DOI] [PubMed] [Google Scholar]
- 50. Morris JP 4th, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer 2010;10:683–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell 2012;149:1192–205. [DOI] [PubMed] [Google Scholar]
- 52. Peng K, Kou L, Yu L, Bai C, Li M, Mo P, et al. Histone demethylase JMJD2D Interacts With beta-catenin to induce transcription and activate colorectal cancer cell proliferation and tumor growth in mice. Gastroenterology 2019;156:1112–26. [DOI] [PubMed] [Google Scholar]
- 53. Shi XY, Lin JJ, Ge XJ, Shi Y. LncRNA WTAPP1 promotes proliferation of laryngeal carcinoma cells through regulating microRNA-592. Eur Rev Med Pharmacol Sci 2020;24:9532–40. [DOI] [PubMed] [Google Scholar]
- 54. Zhang L, Jin C, Yang G, Wang B, Hua P, Zhang Y. LncRNA WTAPP1 promotes cancer cell invasion and migration in NSCLC by downregulating lncRNA HAND2-AS1. BMC Pulm Med 2020;20:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cao L, Zhang P, Li J, Wu M. LAST, a c-Myc-inducible long noncoding RNA, cooperates with CNBP to promote CCND1 mRNA stability in human cells. Elife 2017;6:e30433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee E, Lee TA, Kim JH, Park A, Ra EA, Kang S, et al. CNBP acts as a key transcriptional regulator of sustained expression of interleukin-6. Nucleic Acids Res 2017;45:3280–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, et al. N (6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol 2017;24:870–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Korneev SA, Park JH, O'Shea M. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J Neurosci 1999;19:7711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Supplementary Figure S1-S6 and corresponding Supplementary Figure legends
Supplementary Table S1-S6
WTAPP1-m6A/WTAPP1 50bp RNA probe based Pull-down mass spectrum data
WTAPP1 full length RNA pull down with mass spectrum
Clinical information for patients with IHC
Data Availability Statement
The RNA-seq data containing 65 tumor and 33 normal tissue samples in this paper have been deposited in the Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers PRCJA005425 that are publicly accessible at https://bigd.big.ac.cn/gsa. RNA-seq, m6A individual-nucleotide resolution cross-linking and immunoprecipitation sequencing (miCLIP), and CNBP CLIP data have been deposited in the NCBI Short Read Archive with the BioProject ID- PRJNA693621 that are publicly accessible at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA693621.
For additional experimental procedures and URLs, please refer to Supplementary Materials and Methods.



![Figure 1. WTAPP1 is overexpressed in PDAC and is associated with PDAC-cell phenotypes and clinical outcomes. A, Heatmap of the 10 top differentially expressed pseudogenes in 65 PDAC-tumor and 33 normal tissue samples (All FDR <0.05). Red (higher expression) or blue (lower expression) represents the normalized expression value of indicated pseudogenes. B, Real-time analysis of proliferation and migration of SW1990 and Capan-2 cells at time point of 96 hours (proliferation) and 18 hours (migration), which were treated with negative-control RNA or siRNA of selected 10 top differentially expressed pseudogenes. Data are mean ± SEM from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 of Student t test. C and D, Expression levels of WTAPP1 RNA in surgically removed PDAC samples compared with paired nontumor tissue samples (C) and in advanced III/IV stages compared with early I/II stages (D) in two cohorts and combined sample. *, P < 0.05; **, P < 0.01; ***, P < 0.001 of Wilcoxon rank-sum tests in C and Mann–Whitney tests in D. E, Kaplan–Meier estimates of survival time in two PDAC patient cohorts and combined sample by different WTAPP1 RNA levels in tumors with HR = 1.84 [95% confidence interval (CI) = 1.29–2.64] for Cohort 1; HR = 1.89 (95% CI = 1.11–3.21) for Cohort 2; and HR = 1.88 (95% CI = 1.40–2.53) for combined sample.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/fb8b/9662857/4d8329735b55/5268fig1.jpg)