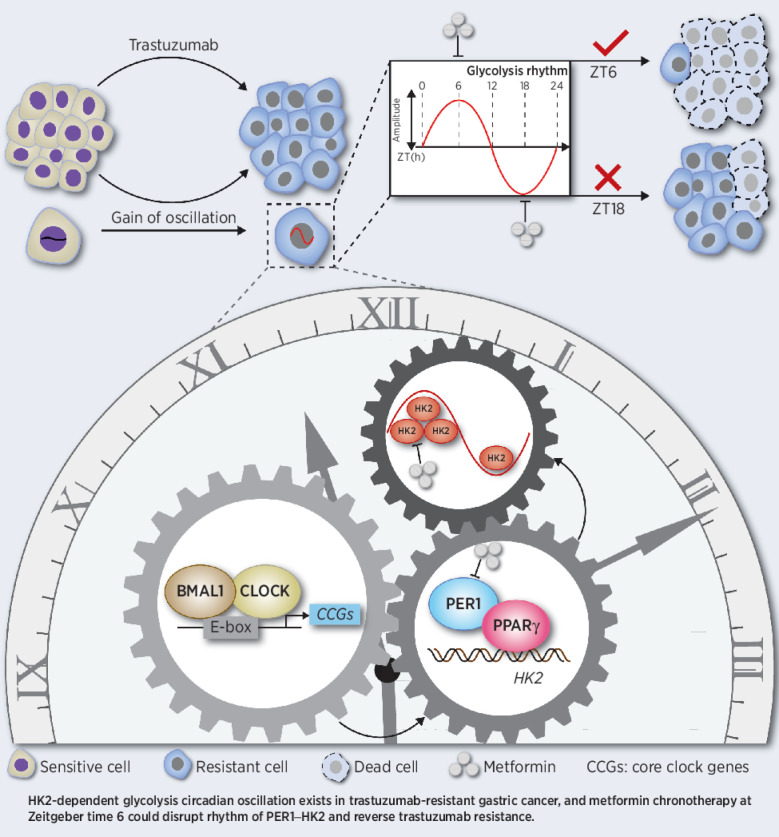
In trastuzumab-resistant HER2-positive gastric cancer, glycolysis fluctuates with a circadian oscillation regulated by the BMAL1–CLOCK–PER1–HK2 axis, which can be disrupted with a metformin-based chronotherapy to overcome trastuzumab resistance.
Abstract
Trastuzumab is the only approved targeted drug for first-line treatment of HER2-positive advanced gastric cancer, but the high rate of primary resistance and rapid emergence of secondary resistance limit its clinical benefits. We found that trastuzumab-resistant (TR) gastric cancer cells exhibited high glycolytic activity, which was controlled by hexokinase 2 (HK2)-dependent glycolysis with a circadian pattern [higher at zeitgeber time (ZT) 6, lower at ZT18]. Mechanistically, HK2 circadian oscillation was regulated by a transcriptional complex composed of PPARγ and the core clock gene PER1. In vivo and in vitro experiments demonstrated that silencing PER1 disrupted the circadian rhythm of PER1–HK2 and reversed trastuzumab resistance. Moreover, metformin, which inhibits glycolysis and PER1, combined with trastuzumab at ZT6, significantly improved trastuzumab efficacy in gastric cancer. Collectively, these data introduce the circadian clock into trastuzumab therapy and propose a potentially effective chronotherapy strategy to reverse trastuzumab resistance in gastric cancer.
Significance:
In trastuzumab-resistant HER2-positive gastric cancer, glycolysis fluctuates with a circadian oscillation regulated by the BMAL1–CLOCK–PER1–HK2 axis, which can be disrupted with a metformin-based chronotherapy to overcome trastuzumab resistance.
Graphical Abstract
Introduction
The HER2 gene is amplified in approximately 10% of advanced gastric cancers. Trastuzumab, the only HER2-targeted agent used as a standard first-line treatment for advanced HER2-positive gastric cancer, has achieved a breakthrough in this patient population (1). Unfortunately, most patients inevitably develop resistance to trastuzumab treatment, necessitating active exploration of resistance issues. Four central mechanisms are involved in the resistance process: (i) HER2 secondary mutations or structural abnormalities; (ii) co-expression of tyrosine kinase receptors, including HER3, IGFR, and c-MET; (iii) bypass signaling activation, such as the PI3K/AKT/mTOR pathway and CDK4/6 axis; (iv) other factors involved in the tumor microenvironment and metabolic processes (2, 3). Emerging agents and regimens are currently being developed to reverse trastuzumab resistance, but satisfactory outcomes are still limited (4, 5).
The circadian timing system controls most physiological processes in mammals, and an impaired circadian clock is considered an emerging hallmark of cancer that influences oncogenesis and progression (6). Importantly, most key factors determining the toxicity and efficacy of antitumor drugs are regulated by the circadian clock at the molecular level (7). Circadian rhythms are driven by transcriptional–translational feedback loops (TTFL), whereby rhythmic expressions of clock genes, including BMAL1, CLOCK, and PER1/2/3, regulate biological processes, such as cell cycle, DNA repair, metabolism, and angiogenesis, into an approximately 24 hours cycle (7–10). Studies have reported that the core clock gene BMAL1, CLOCK affects drug toxicity by regulating the transcriptional activity of drug-metabolizing enzymes, such as CYPs and ABCG2 (11). Besides, the molecular clock interacts with the cell cycle, affecting chemosensitivity and tolerance of 5-fluorouracil (5-FU) and oxaliplatin (7). This implies that the present antitumor therapeutic strategies should be considered in the circadian clock dimension. Treatment based on the circadian clock, termed as chronotherapy, showed potential in precisely targeting resistance mechanisms in a temporal dimension to overcome the treatment dilemma. During the past decades, the advantages of chemotherapeutic drug chronotherapy, which considers drug delivery timing based on the circadian rhythm, have been revealed (12, 13). Moreover, clock genes PER2 and CRY1 could induce the circadian fluctuation of VEGF expression, and antiangiogenic drug delivery based on VEGF oscillation significantly enhanced antitumor efficacy (14). Interestingly, a recent pan-cancer genomic landscape also revealed correlations between clock genes and clinically actionable drug targets, such as EGFR signaling, suggesting the possible interaction between chronotherapy strategy and targeted therapy (15).
Exploring the mechanism of trastuzumab resistance and adopting time-based medication may facilitate low toxicity and sensitive targeted therapy for gastric cancer. In HER2-amplified gastric cancer and breast cancer, glycolysis reprogramming mediated trastuzumab resistance, and combining glycolysis inhibitor 2-deoxy-D-glucose (2DG) and oxamate could partially reverse resistance (16). Furthermore, it has been reported that a glycolytic gene set, including HKDC1, exhibits a circadian expression pattern with a 24 hours cycle (17). However, the underlying association between trastuzumab resistance and glycolysis rhythm remains largely unclear. On the basis of the preliminary success of chronochemotherapy and anti-metabolism strategies, chronotherapy targeting metabolic rhythms may become a new paradigm with great potential in overcoming trastuzumab resistance. In this study, we found that glycolysis activity fluctuated with the circadian rhythm in trastuzumab-resistant (TR) HER2-positive gastric cancer. Mechanism analysis clarified that the circadian rhythm was mediated by the PER1–HK2 axis through protein–protein interactions between PER1 and PPARγ, rather than by direct binding of the heterodimeric BMAL1–CLOCK complex to the putative E-box elements. Furthermore, PER1 or HK2 expression rhythm can be considered a candidate biomarker for metformin chronotherapy, which not only inhibits glycolysis but also degrades PER1 protein to efficiently improve trastuzumab efficacy in HER2-positive gastric cancer.
Materials and Methods
Animal studies
Female BALB/c nude mice ages 4 weeks were used in this study. All animal procedures were performed under the supervision of the Institutional Animal Care and Use Committee, and all mice were housed at 23°C–25°C in the Animal Center of Nanfang Hospital and maintained under 12 hours light:12 hours dark (LD) cycles with the lights on from 8 a.m. (zeitgeber time 0, ZT0) to 8 p.m. (ZT12) and fed with food and water ad libitum. Four in vivo experiments were designed in this study. In experiment 1, to assess glycolysis circadian oscillations in trastuzumab resistance, 1×106 NCI-N87 and NCI-N87TR cells were injected into the left or right flank of nude mice, respectively. 18F-fluorodeoxyglucose (18F-FDG) PET-CT was used to measure the glycolysis levels in the subcutaneous tumors of nude mice every 4 hours within 48 hours. In experiment 2, NCI-N87TR/NC and NCI-N87TR/shPER1 (PER1 knockdown) cells were injected into the left or right flank of nude mice, respectively. 18F-FDG PET-CT was used to measure the glycolysis levels every 4 hours within 48 hours to explore the effect of PER1 on glycolysis in vivo. Similarly, in experiment 3, the mice were divided into NCI-N87TR/NC and NCI-N87TR/shPER1 groups. Two weeks after cell inoculation, trastuzumab (Roche) was administered at a dose of 10 mg/kg twice a week for a month. Tumor volumes and mice weight were measured every 3 days. Tumor volume was calculated using the formula V = 0.5×L×W2, where V is the volume, L is the length, and W is the width. After 4 weeks, the mice were euthanized to compare tumor weight and volume, and tumors were collected for immunohistochemistry staining. In experiment 4, to verify the effect of metformin combined with trastuzumab, based on the circadian clock on reversing trastuzumab resistance, 1×106 NCI-N87TR cells were injected into nude mice subcutaneously. Mice were randomly divided into 8 groups. Each group was administered one of the following: PBS, metformin (250 mg/kg, i.p., daily; Cat# S1741, Beyotime), trastuzumab (10 mg/kg, i.p., twice a week), and metformin combined with trastuzumab, at two different time points: ZT6 and ZT18. Tumor volumes and mice weights were measured and recorded, and the mice were euthanized at the indicated ZTs.
Cell lines
NCI-N87 (RRID: CVCL_IL03) and SNU216 (RRID: CVCL_3946) cells were used to establish TR cells by a three-dimensional (3D) collagen model. NCI-N87 cells were obtained from the National Collection of Authenticated Cell Cultures (NCACC, Shanghai, China), and SNU216 cells were a gift from Xu RH (Sun Yat-sen University Cancer Center, Guangzhou, China). In brief, a 3D model was set up using three collagen layers (400 µL; Gibco, 2052954) in 12-well culture dishes, where middle layers contained NCI-N87 or SNU216 single cell suspension (5,000 cells). RPMI-1640 medium with 10% FBS and trastuzumab (10 µg/mL) was added on top and replaced every 3 days. After 6 months, the TR cells were established and detected by a cell ability assay. The authenticity of the four cell lines NCI-N87, NCI-N87TR, SNU216, and SNU216TR were confirmed by short tandem repeat analysis. All cells were tested regularly for Mycoplasma contamination and cultured in RPMI-1640 medium (Solarbio, 31800) with 10% FBS (Wisent) in a 5% CO2 incubator at 37°C. All experiments were performed within 3–8 passages after thawing the cells. Parental and TR cell lines were authenticated by short tandem repeat sequencing (Supplementary Table S1).
Reagents
Antibodies
Rabbit monoclonal anti-BMAL1 (Cat# 14020S, RRID:14020S), mouse monoclonal anti-DYKDDDDK (Cat# 8146, RRID:AB_10950495), rabbit polyclonal anti-cleaved caspase-3 (Cat# 9661), and rabbit monoclonal anti-PPARγ (Cat# 2443S, RRID:AB_823598) were obtained from Cell Signaling Technology (CST). Rabbit polyclonal anti-HK2 (Cat# 22029–1-AP, RRID:AB_11182717), rabbit polyclonal anti-GLUT1 (Cat# 21829–1-AP, RRID:AB_10837075), rabbit polyclonal anti-PFKFB3 (Cat# 13763–1-AP, RRID:AB_2162854), rabbit polyclonal anti-PFK1 (Cat# 55028–1-AP, RRID:AB_10858390), rabbit polyclonal anti-PKM2 (Cat# 15822–1-AP, RRID:AB_1851537), rabbit polyclonal anti-KI67 (Cat# 27309–1-AP, RRID:AB_2756525), rabbit polyclonal anti-HER2 (Cat# 18299–1-AP, RRID:AB_2099264), rabbit polyclonal anti-CD36 (Cat# 18836–1-AP, RRID:AB_10597244), mouse monoclonal anti–β-actin (Cat# 60008–1-Ig, RRID:AB_2289225), and mouse monoclonal anti-GAPDH (Cat# 60004–1-Ig, RRID:AB_2107436) were obtained from Proteintech. Rabbit polyclonal anti-LDH (Cat# WL03271), rabbit polyclonal anti-AKT (Cat# WL0003b, RRID:AB_2833233), rabbit polyclonal anti-p-AKT (Ser473; Cat# WLP001a), rabbit polyclonal anti-ERK (Cat# WL01864), rabbit polyclonal anti–p-ERK (Thr202/Tyr204; Cat# WLP1512), Rabbit polyclonal anti-AMPK (Cat# WL02254), and Rabbit polyclonal anti–p-AMPK (T182/T172; Cat# WL05103) were obtained from Wanleibio. Rabbit polyclonal anti-PER1 (Cat# bs-2350R, RRID:AB_10857437) and rabbit polyclonal anti-MCT1 (Cat# bs-10249R) were obtained from Boiss. Alexa Fluor 555–labeled Donkey Anti-Mouse IgG (Cat# A0460, RRID:AB_2890133) and Alexa Fluor 488-labeled Goat Anti-Rabbit IgG (Cat# A0423, RRID:AB_2891323) were obtained from Beyotime. Rabbit polyclonal anti-CLOCK (Cat# PB0638) was obtained from Boster, whereas mouse monoclonal anti–p-HER2 (Cat# sc-81506, RRID: AB_1126593) from Santa Cruz Biotechnology.
Chemicals
The 2DG (Cat# HY-13966), rosiglitazone (Cat# HY-17386), and pioglitazone (Cat# HY-13956) were obtained from MCE (MedChemExpress). Trastuzumab was obtained from Roche Pharma (Schweiz) Ltd., metformin (Cat# S1741) from Beyotime, and dexamethasone (Cat# D4902) from Sigma-Aldrich.
Commercial assays
Adenosine triphosphate (ATP) assay (Cat# S0026) and BCA assay (Cat# P0011) kits were obtained from Beyotime. SimpleChIP Enzymatic Chromatin IP Kit (Magnetic Beads; Cat# 9003) was obtained from CST, HK Kit (Cat# HK-1-Y) from Cominbio, Lactic Acid assay Kit (Cat# A019–2-1) from Jiancheng Bioengineering Institute, and Calcein-AM/PI Kit (Cat# BB-4126–2) from BestBio.
Circadian rhythm induction
Cells were cultured in RPMI-1640 medium with 10% FBS in a 6-well plate. Dexamethasone (100 nmol/L, Sigma-Aldrich) was added to the medium, the plate was incubated for 2 hours, and then replaced with fresh medium (this time as ZT0). Cells were harvested at the indicated time (every 4 hours, ZT2, ZT6, ZT10, etc.) for subsequent qPCR, Western blotting assay, and coimmunoprecipitation (Co-IP) analysis.
18F-FDG PET CT
NCI-N87 or NCI-N87/NC cells were injected into the left flank of nude mice, whereas NCI-N87TR or NCI-N87TR/shPER1 cells were injected into the right flank in experiments 1 and 2 (Details can be found in the Animal Studies). The mice were maintained under 12 hours light:12 hours dark (LD) cycles. When the xenograft grew to a maximum diameter of 8 ∼ 10 mm, the mice were injected with 18F-FDG (10 Ci/g) through the tail vein at the indicated time (ZT2, ZT6, etc.). After 60 minutes, the mice were anesthetized and tested by 18F-FDG PET-CT. SUV-BWmax = 6E0, SUV-BWmix = 0.
HK enzyme activity assay
Cells were harvested for HK activity detection using a HK kit (HK-1-Y, Cominbio). Briefly, 10 µL cell lysates were added with 180 µL glucose 6–phosphate and 10 µL glucose 6–phosphate dehydrogenase. The absorbance was measured at 340 nm using the SpectraMax M5 microplate reader (Molecular Devices) at 20 seconds and 5 minutes and 20 seconds, respectively. The protein level of each sample was also measured for standardization using the BCA assay (Beyotime).
Lactate level analysis
The cell supernatant was isolated for lactate level measurement using a lactic acid assay kit (Jiancheng Bioengineering Institute) according to the manufacturer's protocol, as previously described (18). The absorbance was measured at 530 nm by the SpectraMax M5 microplate reader.
Cellular ATP level analysis
The cell lysates were collected for ATP detection using an ATP assay Kit (Cat# S0026, Beyotime). Lysates (20 µL) were mixed with 10 µL ATP detection working dilution, and subsequently luminance was measured using the SpectraMax M5 microplate reader. The protein levels were measured using a BCA assay kit (Cat# P0011, Beyotime). Cellular relative ATP levels were calculated by the ratio of ATP value to protein value.
Live/dead cell vitality (calcein-AM/PI) assay
Cells were collected and stained with Calcein-AM/PI kit (Cat# BB-4126–2, BestBio). Briefly, cells were washed with PBS and incubated with 2 µmol/L Calcein-AM and 1 µmol/L PI for 15 minutes at 4°C in the dark. Photographs (Calcein-AM: emission 490/excitation 515 nm; PI: emission 488–545/excitation 617 nm) were then obtained with a fluorescence microscope (Olympus). Representative images of different fields were obtained for analysis by ImageJ (v1.8.0, RRID:SCR_003070).
RNA isolation and quantitative PCR analysis
Total RNA was extracted from the cells using the TRIzol kit (R401–01, Vazyme) according to the manufacturer's instructions. Reverse transcription was performed using the First Strand cDNA Synthesis kit (R312–01, Vazyme). qPCR was performed using the LightCycler 480 system (Roche), as described in a previous study (18). The expression levels of the target genes were normalized by subtracting the corresponding β-actin threshold cycle (Ct) value. The primer sequences are listed in Supplementary Table S2.
Western blot
The procedure of Western blot assay was described in our previous study (18). Briefly, immunoblots were visualized with a chemiluminescence (ECL) detection system or LI-COR Odyssey infrared imaging system. The following primary antibodies were used: anti-HER2 (1:1,000; 18299–1-AP, Proteintech), anti-p-HER2 (1:100; sc-81506, Santa Cruz Biotechnology), anti-AKT (1:500; WL0003b, Wanleibio), anti-p-AKT (1:500; WLP001a, Wanleibio), anti-ERK (1:500; WL01864, Wanleibio), anti–p-ERK (1:500; WLP1512, Wanleibio), anti-cleaved caspase-3 (1:1,000; Cat# 9661, CST), anti-HK2 (1:1,000; 22029–1-AP, Proteintech), anti-PFK1 (1:1,000; 55028–1-AP, Proteintech), anti-PFKFB3 (1:1,000; 13763–1-AP, Proteintech), anti-PKM2 (1:1,000; 55028–1-AP, Proteintech), anti-MCT1 (1:1,000; bs-10249R, Boiss), anti-GLUT1 (1:1,000; 21829–1-AP, Proteintech), anti-LDH (1:1,000; WL03271, Wanleibio), anti-GADPH (1:5,000; 60004–1-Ig, Proteintech), anti-CLOCK (1:1,000; PB0638, Boster), anti-BMAL1 (1:1,000; Cat# 14020S, CST), anti-PER1(1:800; bs-2350R, Boiss), anti-AMPK (1:500; WL02254, Wanleibio), anti–p-AMPK (1:500; WL05103, Wanleibio), anti-PPARγ (1:1,000; Cat# 2443S, CST), anti-CD36 (1:1,000; 18836–1-AP, Proteintech), anti-DYKDDDDK (1:1,000; Cat# 8146, CST), and anti–β-actin (1:5,000; 60008–1-Ig, Proteintech). β-Actin served as the loading control. Relative quantification was conducted by ImageJ (v1.8.0, RRID:SCR_003070).
MTT assay
Cells were plated in 96-well plates, transfected with a plasmid, and exposed to different concentrations of trastuzumab, 2DG, or metformin for 48 hours. Thiazolyl blue (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT) was added to the cells that were incubated for 4–6 hours, and after washing, dimethyl sulfoxide 150 µL per well were added, as previously described (18). Absorbance was measured at 570 nm using the SpectraMax M5 microplate reader.
Colony formation
Cells were seeded into 12-well plates (1,000 cells per well) and cultured for 2 weeks. The medium was replaced every 3 days. The colonies were washed using PBS, fixed with 4% paraformaldehyde for 30 minutes, stained with 0.1% crystal violet for 30 minutes, and finally photographed for quantitative analysis by ImageJ (v1.8.0, RRID:SCR_003070).
Immunofluorescence staining
Immunofluorescence was performed as previously described (18). Briefly, the cells or tissues were directly fixed with 4% paraformaldehyde, permeabilized with 0.3% Triton X-100, and incubated with the primary antibodies anti-FLAG (1:200; Cat# 8146, CST), anti-PPARγ (1:200; Cat# 2443S CST), and anti-HK2 (1:200; 22029–1-AP, Proteintech) overnight at 4°C. Next, the cells or tissues were incubated with the secondary antibodies Alexa Fluor 555-labeled Donkey Anti-Mouse IgG (1:200; Beyotime) and Alexa Fluor 488–labeled Goat anti-Rabbit IgG (1:200; Beyotime) for 1 hour at room temperature. Finally, the cells or tissues were incubated for 10 minutes with 4′,6-diamidino-2-phenylindole diluted with methanol to stain the nucleus. Images were obtained with fluorescence and laser confocal microscopes (A1-DUVB-2, Nikon).
IHC staining
Tissues were fixed in 4% formaldehyde overnight and embedded in paraffin. Serial sections (4 µm) were incubated with the horseradish peroxidase–labeled Polymer anti-Rabbit or anti-Mouse antibody and 3,3′-diaminobenzidine from DAKO. IHC staining was used to detect the expression levels of HK2 (1:500; 22029–1-AP, Proteintech), PER1 (1:500; bs-2350R, Boiss), Ki67 (1:5,000;27309–1-AP, Proteintech), and cleaved caspase-3 (1:400; Cat# 9661, CST). Images were obtained using a microscope (BX51, Olympus).
Co-IP analysis
A total of 1 × 107 cells were plated in 10-cm dishes, without treatment or transfected with PER1-FLAG plasmid and then harvested after 48 hours. Cells in another experiment were synchronized by dexamethasone (100 nmol/L) for 2 hours, which was replaced by fresh medium (this time as ZT0), and then harvested from 2 to 46 hours with 4 hours intervals. Next, cell lysates were incubated with a specific primary antibody, anti-PPARγ (1:200; Cat# 2443S CST), anti-FLAG (1:200; Cat# 8146, CST), or anti-PER1 (1:100; sc-398890, Santa Cruz Biotechnology), and mixed with protein A/G-Sepharose beads (7 Sea Biotech) overnight at 4°C. After extensive washing, the beads were boiled in 4×SDS-PAGE loading buffer for 5 minutes and analyzed by immunoblotting with specific antibodies to detect the protein complex.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed using a SimpleChIP Enzymatic Chromatin IP kit (Cat#9003, CST). Briefly, 1×107 cells were plated in 10-cm dishes, washed in PBS, and then incubated with 1% formaldehyde for 10 minutes at room temperature. The fragmented chromatin was treated with nuclease, sonicated, and incubated with rabbit monoclonal anti-BMAL1 (1:50; Cat#14020S, CST) or anti-PPARγ (1:80; Cat# 2443S, CST), or rabbit anti-histone 3, and rabbit anti-IgG (for the negative control) antibodies overnight at 4°C. Target-bound DNA fragments were reverse crosslinked and purified, then amplified by qPCR and measured using gel electrophoresis. Quantitative data were obtained as the ratio of DNA-antibody complex to input DNA. Primer pairs for the ChIP assay are listed in Supplementary Table S3.
DNA constructs and siRNA transfection
For gene knockdown of PER1, cells were stably transfected with GV248-hU6-MCS-Ubiquitin-EGFP-IRES-Puro-PER1 constructs (Genechem). Transfected cell lines were selected with 1 µg/mL puromycin medium (Invitrogen). Overexpressing PER1 or HK2 and empty vector (GV141-CMV-MCS-3FLAG-SV40-Neomycin) plasmids were purchased from Genechem Company. Ablation of clock genes and other genes was performed by transfection with siRNA duplex oligos, which were synthesized by Ribobio Company. Cell transfection was performed with Lipofectamine 2000 (Invitrogen), as described previously in the manufacturer's protocol. The sequences used in this study are listed in Supplementary Table S4. qPCR and Western blotting were used to verify the efficiency of the sequence.
Statistical analysis
Each experiment was repeated at least three times. Differences between experimental groups were assessed using Student t test or one-way ANOVA. All data were analyzed using GraphPad Prism 7.0 software (RRID:SCR_002798) or SPSS 20.0 software (RRID:SCR_002865). All values are presented as mean ± SD, and statistical significance was noted as a P value of < 0.05.
Data availability statement
The data generated in this study are available within the article and its Supplementary Data Files.
Results
Glycolysis activity is upregulated and undergoes circadian oscillations in TR HER2-positive gastric cancer
To investigate the mechanism of trastuzumab resistance, HER2-positive gastric cancer cell lines NCI-N87 and SNU216 (wild-type, WT) were used to develop TR cells using a 3D collagen model (Supplementary Fig. S1A; ref. 19). After being cultured with trastuzumab for more than 6 months, gastric cancer cells that maintained cell viability were generated as TR cells. WT cells were cultured in normal medium and remained sensitive to trastuzumab (Supplementary Fig. S1B). Consistent with previous studies (2), phosphorylation of resistance-related proteins, such as HER2, AKT, and ERK, was also activated in TR cells (Supplementary Fig. S1C). Moreover, TR cells with continuous trastuzumab treatment had a stronger clone formation ability than that of WT cells (Supplementary Fig. S1D), confirming the successful construction of drug-resistant cell lines. The circadian rhythm of the key receptor tyrosine kinase (RTK) pathway and its application value in the chronological dosing strategy have elicited research attention (20, 21). We evaluated the circadian rhythms of RTKs, such as p-HER2, p-ERK, and p-AKT, in two consecutive circadian cycles in TR cells (Supplementary Fig. S2A). Western blot results showed that the protein expression of p-HER2, p-ERK, and p-AKT exhibited rhythmic oscillations in SNU216TR, but this was not obvious in NCI-N87TR.
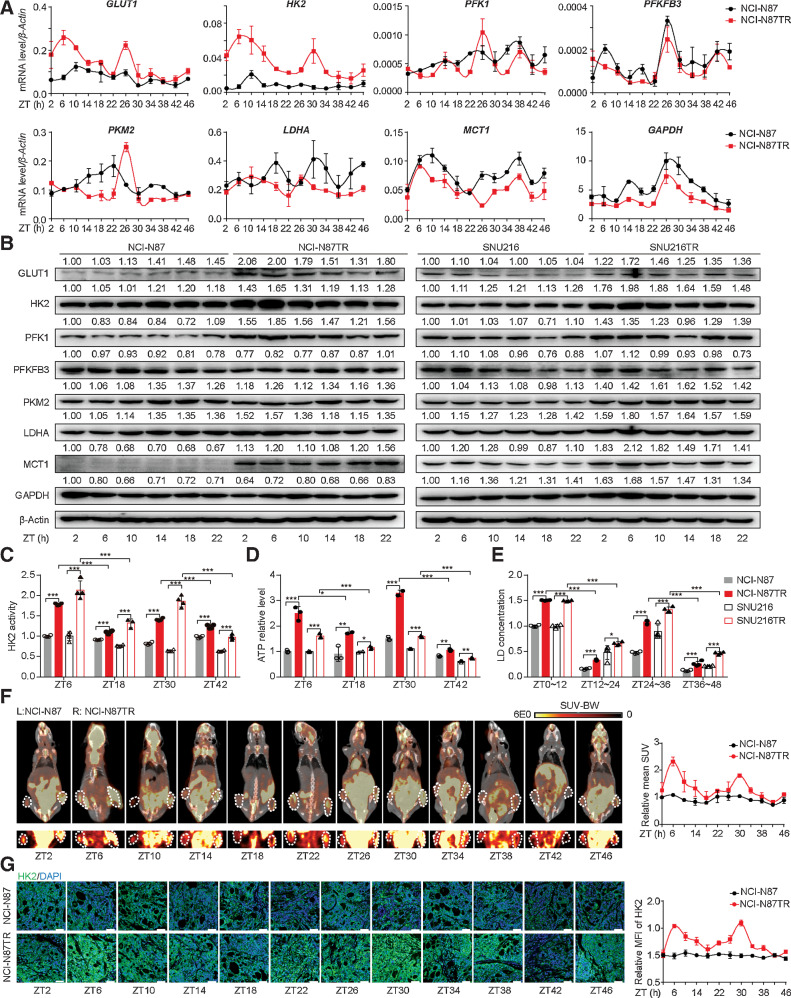
The high glycolytic activity in gastric cancer cells is a critical factor determining the occurrence of trastuzumab resistance (22). To further address the potential interaction between glycolysis and the circadian clock, we identified the expression of glycolysis-related genes at different ZTs by qPCR and Western blotting. We first confirmed that trastuzumab resistance led to an increase in the expression of glycolysis-related genes with circadian rhythm compared with that in trastuzumab-sensitive cells (Fig. 1A and B; Supplementary Fig. S2B and S2C). The expression levels of these genes, particularly HK2, underwent a 24 hours phase shift in circadian oscillations and were higher at earlier ZTs (e.g., ZT6 or ZT30) and lower at ZT18 or ZT42. HK2 is a crucial enzyme in glucose metabolism, which catalyzes the irreversible rate-limiting step in glycolysis in an ATP-dependent manner (23). Besides the expression level, the enzymatic activity of HK2 also increased and exhibited circadian oscillations with peak values at ZT6 and ZT30 (Fig. 1C). As expected, the products of glycolysis, including ATP and lactic acid, increased during the first half of the 24 hours circadian phase, indicating that glycolysis was activated in a circadian oscillation in TR cells (Fig. 1D and E). Moreover, we performed 18F-FDG PET-CT to measure the glycolysis levels in subcutaneous tumors of nude mice and observed a higher signal with injection of TR cells, particularly at ZT6 or ZT30 (Fig. 1F). The expression of HK2 in subcutaneous tumors presented a similar circadian oscillation, as shown by immunofluorescence (Fig. 1G). These results revealed that trastuzumab resistance triggered the circadian oscillations of glycolysis in HER2-positive gastric cancer cells.
Figure 1.
The glycolysis level is upregulated and undergoes circadian oscillations in trastuzumab-resistant HER2-positive gastric cancer. A, Time-series (every 4 hours) expression of glycolysis-related genes determined by qPCR in synchronized NCI-N87 and NCI-N87TR gastric cancer cells. Cells were synchronized by dexamethasone (100 nmol/L) for 2 hours, which was replaced with fresh medium (this time as ZT0) and then harvested from 2 to 46 hours with 4 hours intervals. B, Western blot analysis of glycolysis-related genes in WT and TR cells at the indicated ZT2 to ZT22. β-Actin served as the loading control. C–E, The HK enzyme activity (C), ATP level (D), and lactic acid concentration (E) in the synchronized WT and TR cells were detected at ZT6, ZT18, ZT30, and ZT42. F, NCI-N87 WT (left) and TR (right) cells were subcutaneously injected into both flanks of nude mice for 1 month. Glycolysis level in tumors was measured by 18F-FDG PET-CT at ZT2 to ZT46 (left), and mean standard uptake value (SUV) was quantified (right; n = 2). MicroPET-CT images of whole nude mice are shown above, whereas PET images of 18F-FDG uptake by the tumor are presented below. White circles, tumor location. G, IF analysis for HK2 expression in WT and TR tumor tissues. Representative IF images (left) and the quantification (right) of mean fluorescence intensity (MFI) are shown (n = 4). Scale bar, 50 µm. Statistics were calculated with 3 (A and D) or 4 (C and E) samples. Student t test was performed in C–E. Data are presented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant, P > 0.05.
The BMAL1–CLOCK–PER1 axis triggers trastuzumab resistance via regulating HK2 circadian oscillation in TR cells
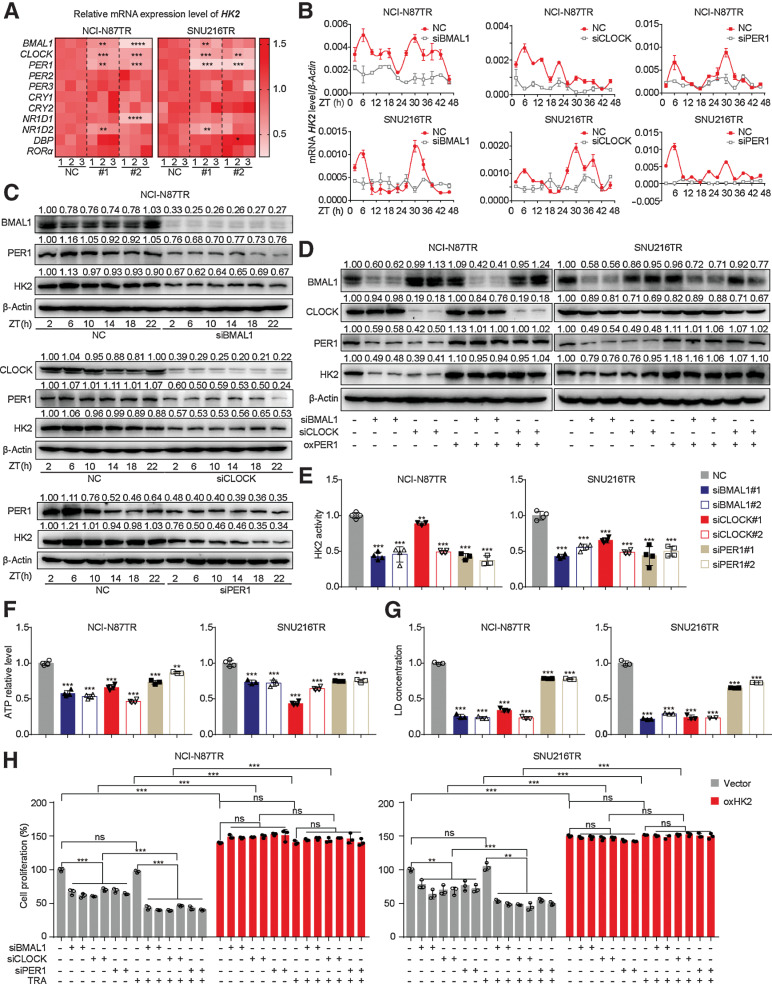
TTFLs of circadian rhythms comprise the transcription factor BMAL1–CLOCK heterodimer, which induces the transcription of downstream genes PERs/CRYs/NR1D1/2 (24). In turn, PERs/CRYs proteins are transferred into the nucleus to repress the formation of the BMAL1–CLOCK complex, creating a negative feedback loop (25, 26). We noted that HK2 was expressed in a circadian oscillating manner in TR cells, indicating the underlying relationship between HK2 and the circadian clock. Furthermore, we sought to determine the core clock genes that regulate HK2 transcription. First, the cells were synchronized by dexamethasone for 2 hours and harvested at different ZTs to detect the circadian rhythm of the expression of 11 core clock genes (Supplementary Fig. S3A). The circadian oscillation in PER1 expression was similar to that in HK2. In vivo experiments showed similar results (Fig. 1G; Supplementary Fig. S3B). Then, a silence library of core clock genes was established to assess the changes in HK2 expression in transfected TR cells (Supplementary Fig. S4A). Silencing BMAL1, CLOCK, or PER1 significantly downregulated the mRNA expression and disrupted the circadian oscillation of HK2 (Fig. 2A–C; Supplementary Fig. S4B and S4C). Collectively, these results demonstrated that the clock genes BMAL1, CLOCK, and PER1 could induce the expression and circadian rhythm of HK2.
Figure 2.
The BMAL1–CLOCK–PER1 axis triggers trastuzumab resistance via regulating HK2 circadian oscillation in TR cells. A, Heatmap of mRNA expression level of HK2 in TR cells with siRNA transfection of 11 core clock genes. NC, normal contrast as the control group of siRNA, #1 or #2. The silencing group with one siRNA sequence of genes. B, qPCR analysis of mRNA levels of HK2 in synchronized TR cells transfected with siRNA of BMAL1, CLOCK, and PER1. C, Western blot analysis of HK2 and PER1 proteins in synchronized NCI-N87 TR cells infected with siRNA of BMAL1, CLOCK, and PER1. D, Western blot analysis of PER1 and HK2 proteins in PER1-overexpressing TR cells infected with siRNA of BMAL1 or CLOCK. E–G, The HK2 enzyme activity (E), ATP level (F), and lactic acid concentration (G) of TR cells infected with siRNA of BMAL1, CLOCK, and PER1. H, CCK8 assay was used to assess cell proliferation of TR cells treated with trastuzumab (TRA; 10 µg/mL) after transfection with siRNA of BMAL1, CLOCK, and PER1 with or without overexpressing HK2. Statistical analysis was performed using 3 (A, B, and F) or 4 (E, G, and H) samples. Student t test was performed in (A and E–H). Data are presented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant, P > 0.05.
It has been reported that the BMAL1–CLOCK heterodimer binds to E-boxes in the promoters of PER1 genes and activates its transcription, which further regulates the transcription of other clock-controlled genes to maintain rhythmicity at the cellular and organismal levels (27, 28). Therefore, we speculated that the BMAL1–CLOCK complex could regulate the circadian oscillation of HK2 through PER1. To verify this possibility, the siRNA of BMAL1 and CLOCK and the plasmid of PER1 were cotransfected into TR cells. Notably, overexpression of PER1 significantly reversed the inhibitory effect of siBMAL1 or siCLOCK on HK2 expression (Fig. 2D). The silencing of BMAL1, CLOCK, and PER1 impaired HK2 activity, ATP, and lactic acid production (Fig. 2E–G). These results indicated that inhibiting the BMAL1–CLOCK–PER1 axis reduces HK2 expression and suppresses glycolysis in TR cells.
As mentioned, a high glycolysis activity can result in trastuzumab resistance in gastric cancer. However, the effect of the circadian clock on the resistance process is still unknown. The cell vitality of TR cells was markedly reduced after trastuzumab administration at different concentrations, suggesting that the silencing of BMAL1, CLOCK, or PER1 could enhance the trastuzumab response of TR cells (Supplementary Fig. S4D). We further investigated whether the BMAL1–CLOCK–PER1 axis induced trastuzumab resistance by regulating HK2 in TR cells. MTT assays showed that overexpression of HK2 completely reversed the decreased cell viability mediated by the downregulation of the BMAL1–CLOCK–PER1 axis in TR cells (Supplementary Fig. S4E). The same results were obtained from the detection of the proliferation rate by the CCK8 assay (Fig. 2H). Collectively, the BMAL1–CLOCK–PER1 axis triggered trastuzumab resistance in TR cells through upregulation of HK2-dependent glycolysis.
PER1 interacts with PPARγ to regulate the expression and rhythm of HK2
Considering the high synchronization of the expression rhythm of HK2 at the mRNA and protein levels in TR cells, we intended to delineate the transcriptional regulation mechanism by which the BMAL1–CLOCK–PER1 axis disrupts HK2 expression and oscillation. Researchers have found that the BMAL1–CLOCK heterodimer drives the transcription of circadian clock genes by binding to the E-boxes located in the promoter of the target genes; thus, inducing their expression (27). We first tested the possibility of BMAL1–CLOCK heterodimer as a transcription factor directly binding to the HK2 promoter region. We first studied the structure of the HK2 gene and identified six noncanonical E-boxes on the HK2 promoter site (a region of 2,000 bp upstream from the HK2 coding sequences; Supplementary Fig. S5A). Next, we analyzed their potential binding to sites in the HK2 promoter by ChIP and found that none of these sites could obviously be bound to HK2 (Supplementary Fig. S5B and S5C). These results suggest that BMAL1–CLOCK heterodimers may not directly regulate HK2 by binding E-boxes. We then focused on the regulatory mechanism of PER1.
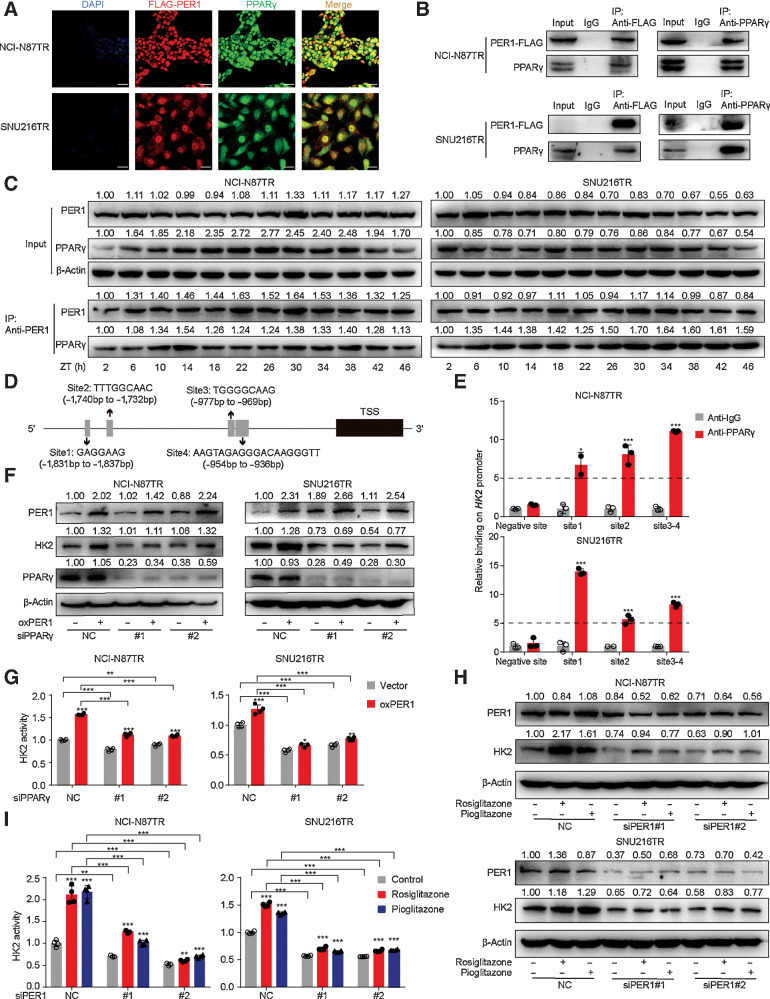
Although PER1 is related to transcription pathways in gene ontology annotations, PER1 as a transcription factor for the regulation of downstream target genes has not been elucidated. However, previous studies have shown that PER1 interacts with PPARγ and affects its transcriptional activity (29). To further investigate the association between PER1 and PPARγ, we performed immunofluorescence assays and found that PER1 and PPARγ were colocalized in the nucleus (Fig. 3A). Co-IP assays of cells transiently expressing PER1–FLAG protein and PPARγ confirmed the presence of the PER1–PPARγ complex (Fig. 3B). Further experiments were conducted with the dexamethasone-synchronized TR cells using PER1 (IP antibody) to detect the coprecipitation of PPARγ and PER1 (Fig. 3C). We found that in the Input group, TR cells showed conservative PER1 rhythmic oscillation. However, the expression of PPARγ protein in the Input group did not have an obvious circadian rhythm, whereas the PPARγ protein in the IP group showed consistent expression variation due to PER1. The above results suggested that the transcriptional activity of PER1–PPARγ might follow a circadian rhythm, which could be driven by the expression of PER1 protein rather than PPARγ. Furthermore, CD36, widely known as a downstream molecule of PPARγ, was significantly downregulated in the PPARγ knockdown cell lines (Supplementary Fig. S5D and S5E). Interestingly, neither the knockdown nor the overexpression of PER1 affected PPARγ at the mRNA and protein levels, whereas CD36 was significantly decreased (Supplementary Fig. S5F and S5G). This suggests that PER1 binds to PPARγ to affect its activity, not its expression.
Figure 3.
PER1 interacts with PPARγ to regulate the expression and rhythm of HK2. A, Representative IF images of PPARγ (green), FLAG-tagged PER1 (red), and DAPI (blue) in TR cells transfected with FLAG-tagged PER1 plasmid. Scale bar, 75 µm. B, Co-IP assay showing the presence of a complex containing PER1 and PPARγ. Top, PER1–FLAG antibody coprecipitating PPARγ. Bottom, PPARγ antibody coprecipitating PER1-FLAG. Input, protein expression in cell lysates detected by Western blot. IgG, negative control. IP, expression of compound coprecipitated by PER1-FLAG or PPARγ antibody. C, Cells were synchronized by dexamethasone (100 nmol/L) for 2 hours, which was replaced by fresh medium (this time as ZT0), and then cells were harvested from 2 to 46 hours with 4 hours intervals. Co-IP assay shows the level of PER1 and PPARγ protein expression of cell lysates (Input). β-Actin served as the loading control (top). The expressions of PER1 and PPARγ in compounds coprecipitated by PER1 antibody (IP) are shown (bottom). D, A schematic plot of transcription factor-binding sites of PPARγ on HK2 upstream promoter (2 Kb). E, TR cells were harvested for ChIP assay to detect the enrichment of PPARγ around the HK2 promoter. Immunoprecipitated DNA was analyzed by qPCR with specific primers. Anti-IgG antibody was used as the negative control (n = 3). F and G, Western blot (F) and HK2 enzyme activity (G) in TR cells after overexpression of PER1 and interference of PPARγ. H and I, Western blot (H) and HK2 enzyme activity (I) in TR cells after interfering PER1 with or without rosiglitazone (20 µmol/L), pioglitazone (20 µmol/L). Statistical analysis was performed using 4 samples in G and I. Student t test was performed in E, G, and I. Data are graphed as the mean ± SD; *, P < 0.05, **, P < 0.01; ***, P < 0.001; ns, nonsignificant, P > 0.05.
It has been reported that PPARγ plays an important role in HK2 and PKM2 glycolytic isozyme gene transcription (30). Our results revealed that silencing PPARγ in TR gastric cancer cells could significantly inhibit HK2 expression at the mRNA and protein levels (Supplementary Fig. S5D and S5E). Therefore, we hypothesized that PPARγ might participate in the transcriptional activity of HK2. First, we used the hTFtarget database (http://bioinfo.life.hust.edu.cn/hTFtarget#!/) to predict the binding sites of PPARγ on the HK2 promoter. There were four predicted binding sites located in the upstream of HK2 coding sequences (Fig. 3D). Because site 3 and site 4 are spatially close, we designed three pairs of primers covering site 1, site 2, and site 3–4, respectively (Supplementary Table S3). The ChIP analysis showed that PPARγ was enriched in sites 1, 2, and 3–4 of the HK2 promoter, demonstrating that PPARγ was directly bound to the promoter regions of HK2 (Fig. 3E). To further determine the role of PPARγ, siRNA was used to knockdown PPARγ in PER1-overexpressed TR cells. In Western blot and qPCR assays, the knockdown of PPARγ significantly reduced HK2 expression level and activity (Fig. 3F and G). We also used a selective PPAR agonist (rosiglitazone or pioglitazone) in PER1-silenced cells to verify the alteration in HK2 expression and enzymatic activity. The addition of rosiglitazone or pioglitazone partially restored the HK2 translation level and enzyme activity after knocking down PER1 (Fig. 3H and I). Collectively, these results revealed that PER1 binds to PPARγ, acts as a transcriptional activator, and regulates the expression and rhythm of HK2.
The interference of PER1 reverses trastuzumab resistance in gastric cancer
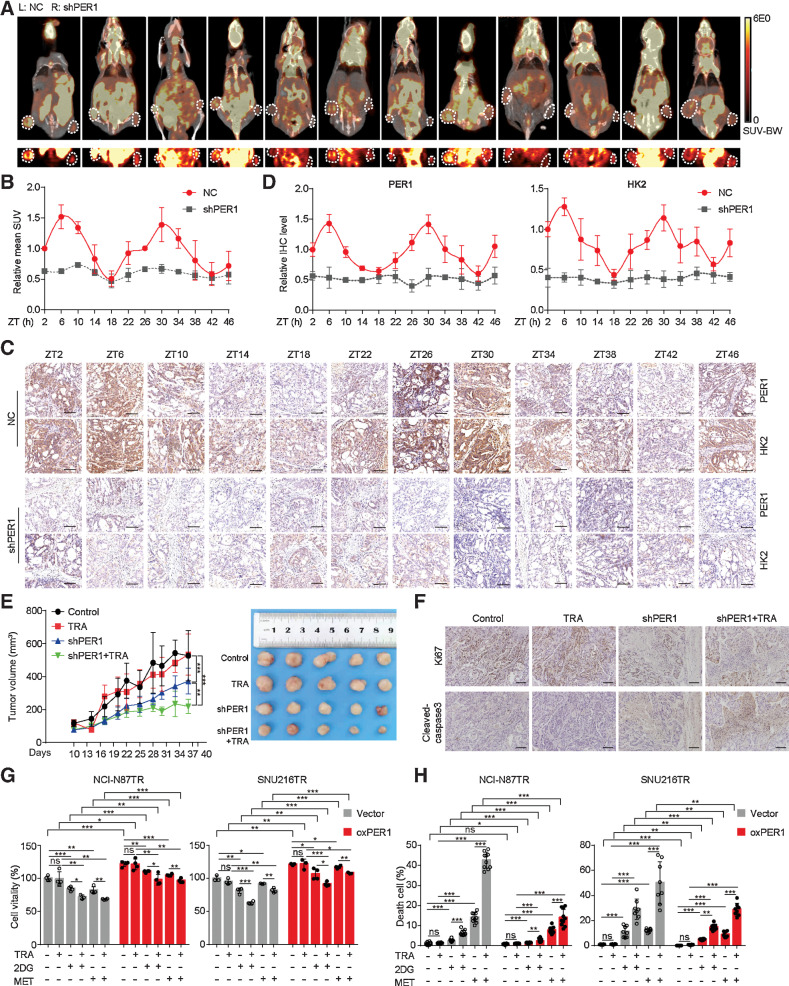
So far, we have demonstrated that the clock gene PER1, but not the BMAL1–CLOCK heterodimer, was capable of directly regulating the expression and oscillation of HK2 in HER2-positive gastric cancer in vitro. Next, we conducted in vivo experiments to explore the function of PER1 and glycolysis fluctuations in subcutaneous tumor models. PER1-knockdown TR cells were injected subcutaneously into the nude mice. Consistent with our data above, PET-CT showed that glycolysis activity in the control group displayed circadian oscillations, which were higher at ZT6 or ZT30 and lower at ZT18 or ZT42 (Fig. 4A and B). In addition, in tumors with PER1 knockdown, the glycolysis activity was generally reduced and lost rhythmic oscillation, suggesting that the glycolysis rhythm was disrupted through PER1 interference. IHC staining of HK2 and PER1 on serial sections also showed the same trend as that of glucose metabolism (Fig. 4C and D). Next, we treated tumor-bearing mice with trastuzumab. The tumor volume in the PER1-knockdown group was reduced, verifying that PER1 was indeed associated with trastuzumab resistance (Fig. 4E). Remarkably, the knockdown of PER1 in TR cells significantly increased the sensitivity to trastuzumab administration in vivo. IHC staining of proliferation marker Ki67 and apoptosis marker cleaved caspase-3 showed that trastuzumab significantly inhibited tumor cell proliferation and promoted apoptosis in the PER1-knockdown group (Fig. 4F). In general, we confirmed that blocking PER1 expression reversed trastuzumab resistance in vivo.
Figure 4.
The interference of PER1 reverses trastuzumab resistance in gastric cancer. A, Stably transfected NCI-N87TR cells with control (left) or PER1 knockdown (right) were subcutaneously injected into nude mice. Glycolysis level was measured by 18F-FDG PET-CT at ZT2 to ZT46. MicroPET-CT images of whole nude mice are shown above, whereas PET images of 18F-FDG uptake by tumors is shown below. White circles, tumor location. B, Quantification of mean standard uptake value (SUV; n = 3). C, Representative IHC images of PER1 and HK2 expression on serial sections were presented at indicated ZTs. Scale bar, 100 µm. D, Quantitative analysis of relative IHC level of PER1 and HK2 (n = 4). E, Tumor growth (left) and representative images (right) of transplanted subcutaneous tumors with PER1 knockdown and trastuzumab treatment (TRA; 10 mg/kg, twice a week). F, Representative IHC images of Ki67 and cleaved-caspase-3 on serial sections of subcutaneous tumors of xenograft nude mice. Scale bar, 140 µm. G and H, Growth inhibition (G) was detected by MTT assay and cell death (H) was detected by Calcein-AM/PI assay in TR cells treated with trastuzumab (TRA; 10 µg/mL), 2DG (4 mmol/L), and metformin (MET; 10 mmol/L) after overexpressing PER1. Statistical analysis was performed using 3 (A), 4 (B and G), 5 (E), or 8–10 (H) samples. The Student t test was performed in G and H. One-way ANOVA was performed in E. Data are graphed as the mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, nonsignificant, P > 0.05.
As a key enzyme in glucose metabolism, HK2 can partly characterize the glycolysis activity (23). Therefore, we suggested that the BMAL1–CLOCK–PER1 axis triggers trastuzumab resistance via HK2-mediated glycolysis circadian oscillations in TR cells. In the growth inhibition and cell death assays, TR cells were treated with the glycolysis inhibitor 2DG or metformin. We observed that 2DG and metformin treatment combined with trastuzumab inhibited TR cell viability and enhanced cell death, thus ameliorating trastuzumab resistance. Notably, PER1 overexpression reversed these effects, indicating that PER1 induced trastuzumab resistance by regulating glycolysis (Fig. 4G and H). Interference with both PER1 and glycolysis has the potential to reverse the drug resistance of TR cells.
Administering the glycolysis inhibitor metformin based on HK2 circadian rhythm improves trastuzumab efficacy
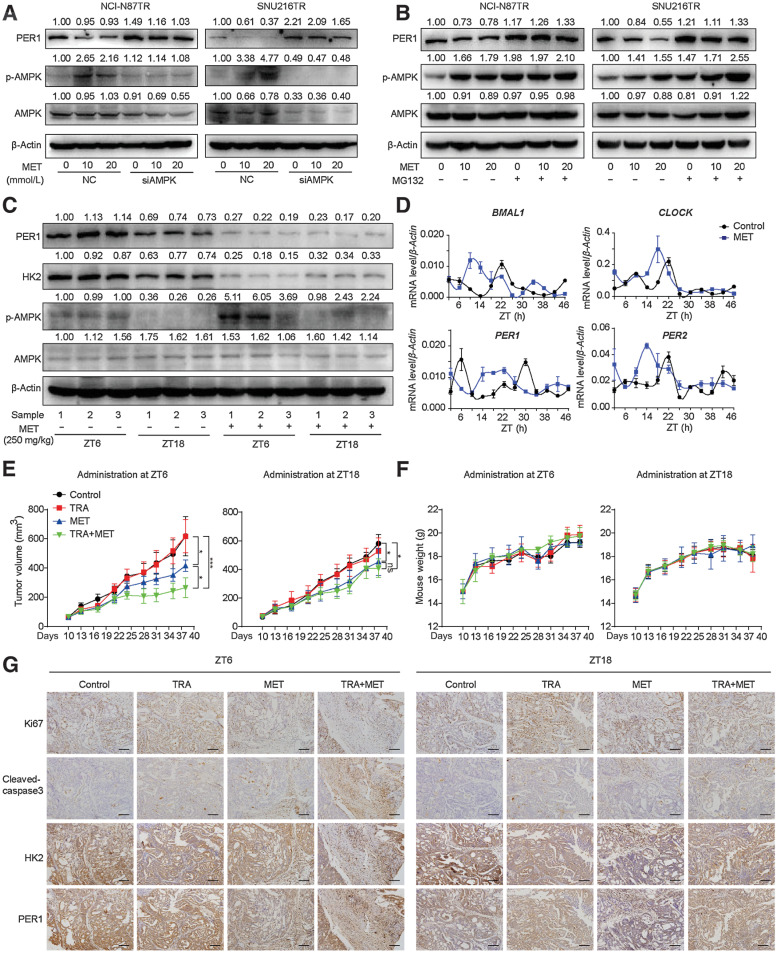
Metformin is a widely used oral antidiabetic drug with relative safety and pleiotropy (31). Previous studies demonstrated that metformin activates adenosine monophosphate-activated protein kinase (AMPK) and inhibits glucose phosphorylation and glycolysis through an AMPK-independent pathway (32, 33). In addition, metformin has been shown to serve as a clock regulator and rebuilds the circadian clock via the activation of AMPK (34–36). In this study, we treated TR cells with metformin to detect the expression of AMPK and PER1 (Fig. 5A). Using the Western blot assay, we found that metformin increased the phosphorylation of AMPK and inhibited the expression of PER1 protein. This phenomenon was reversed after AMPK was silenced, and PER1 expression was significantly restored, suggesting that metformin inhibited the expression of PER1 by activating AMPK. Furthermore, we used MG132 to block the proteasome pathway and found that the inhibitory effect of metformin on PER1 was reversed (Fig. 5B). Moreover, in vivo, we verified that metformin significantly upregulated the expression of phosphorylated AMPK and inhibited HK2 and PER1 in ZT6; however, this effect was not observed in metformin administration at ZT18 (Fig. 5C). Next, we investigated the effect of metformin on the circadian clock. The qPCR analysis was performed to detect the mRNA levels of core clock genes in tumor tissues in the control and metformin monotherapy groups (Fig. 5D). Metformin rearranged the expression rhythm of clock genes, including BMAL1, CLOCK, and PER1/2, in vivo. Accordingly, we proposed the dual function of metformin, particularly in the inhibition of glycolysis and suppression of PER1 expression through the proteasome pathway.
Figure 5.
Administering the glycolysis inhibitor metformin based on HK2 circadian rhythm improves trastuzumab efficacy. A and B, Western blot analysis of PER1, AMPK, and p-AMPK proteins in TR cells treated with MET with or without AMPK knockdown (A) or MG132 treatment (B). C, NCI-N87TR cells were subcutaneously injected in nude mice under a 12:12 h light/dark cycle with the lights on from ZT0 to ZT12. Mice were treated with PBS, trastuzumab (TRA; 10 mg/kg), metformin (MET; 250 mg/kg), or TRA combined with MET at ZT6 or ZT18. Western blot analysis was performed to detect PER1, HK2, AMPK, and p-AMPK in PBS and MET groups at ZT6 or ZT18. D, Nude mice were killed at 4 hours intervals across the 24 hours light/dark cycle for two consecutive days. qPCR analysis of relative mRNA levels of clock genes BMAL1, CLOCK, PER1, and PER2 in PBS and MET (250 mg/kg) groups are shown (n = 3). E and F, Tumor volumes (E) and weight (F) of nude mice were calculated (n = 5). One-way ANOVA was used to calculate P values. G, Representative IHC images of Ki67, cleaved-caspase-3, PER1, and HK2 performed on serial sections of subcutaneous tumors of xenograft nude mice. Scale bar, 140 µm. Data are graphed as the mean ± SD; *, P < 0.05; ***, P < 0.001; ns, nonsignificant, P > 0.05.
On the basis of our data, both the interference of PER1 and inhibition of glycolysis can efficiently improve the efficacy or reverse trastuzumab resistance in HER2-positive gastric cancer. Nevertheless, as a core clock gene, PER1 is involved in numerous physiological and pathological processes, and simple interference might disrupt other normal pathways besides glycolysis (37, 38), indicating that the direct inhibition of glycolysis may be more suitable for obtaining better therapeutic effects. Therefore, metformin was used in this study to treat trastuzumab resistance in consideration of its clinical applicability and dual suppression of PER1 and glycolysis. As the glycolysis level characterized by HK2 reached its peak at ZT6 and the bottom at ZT18, we further investigated whether glycolysis inhibition by metformin based on the circadian clock can reverse trastuzumab resistance. The nude mice were subcutaneously injected with TR cells and treated with metformin combined with trastuzumab at ZT6 and ZT18. Trastuzumab monotherapy could not reduce tumor volume in the TR cell-constructed model, confirming in vivo resistance. As expected, because trastuzumab has a relatively long half-life (28∼38 days), its administration at different timepoints did not affect the outcome (Fig. 5E). However, the intraperitoneal injection of metformin at ZT6 and ZT18 partially reduced tumor volumes. Notably, the combination of metformin and trastuzumab administrated at ZT6 achieved a better antitumor effect, compared with administration at ZT18, which preliminarily confirmed the effectiveness of timed therapeutic strategy. In addition, metformin and trastuzumab did not influence the weight of the mice (Fig. 5F). Subsequently, adjacent serial sections of tumor tissues were used to perform IHC staining, and there was no obvious difference in Ki67, cleaved caspase-3, HK2, and PER1 after trastuzumab monotherapy (Fig. 5G). After additional metformin treatment at ZT6, the expression of HK2, PER1, and Ki-67 was decreased, whereas that of cleaved caspase-3 was increased. In contrast, the antitumor effects of the metformin and trastuzumab combination were relatively weaker at ZT18. These results suggest that metformin treatment at high glycolysis levels (ZT6) achieved optimal efficacy in TR mice. In summary, trastuzumab combined with metformin reverses trastuzumab resistance, and its administration based on the glycolysis circadian rhythm emerges as a promising strategy for targeted therapy.
Discussion
The mechanism of trastuzumab resistance in gastric cancer is not well understood. Limited studies have revealed the role of metabolic reprogramming, such as the Warburg effect, in driving trastuzumab resistance (22). Through establishing TR gastric cancer cells by continuous chronic exposure to trastuzumab in a 3D culture model, we revealed that TR gastric cancer cells not only exhibited higher levels of glycolysis but also, more importantly, were accompanied by regular HK2-dependent glycolysis rhythm. Our findings preliminarily clarify that metabolic rhythm reprogramming may be an important mechanism for trastuzumab resistance. In the past, glycolysis inhibitors lacked wide clinical application due to obvious side effects (39), but circadian rhythm–based chronotherapy provides the possibility for the development of less toxic and highly efficient antitumor treatment strategies (25). Accordingly, our data suggest that targeting glycolysis rhythm may be a novel strategy to benefit patients with trastuzumab resistance.
As mentioned earlier, TTFLs are the main performers of circadian rhythms in mammals (10). In humans and mice, 50%–80% of protein-coding genes are controlled by TTFLs, highlighting their role as key mediators of the glycolysis rhythm in TR gastric cancer cells (25). Using a silence library of core clock genes, we discovered that the silencing of BMAL1, CLOCK, and PER1 downregulated the expression levels of HK2 and disrupted its circadian rhythm, rather than CRY or NR1D1/2. At the same time, silencing BMAL1, CLOCK, and PER1 can inhibit glycolysis and restore the sensitivity to trastuzumab in HK2-overexpressed TR cells. These results supported the assumption that the BMAL1–CLOCK–PER1 axis triggered trastuzumab resistance via regulation of HK2 circadian oscillations.
At present, the mechanism by which clock genes regulate drug resistance in tumor therapy remains unclear. For instance, studies have reported that overexpression of BMAL1 increased the sensitivity of paclitaxel in tongue squamous cell carcinoma and the sensitivity of oxaliplatin and bevacizumab in colorectal cancer (40–42). However, silencing BMAL1 can also increase the sensitivity of breast cancer cells to cisplatin and adriamycin (43). These contradictory results may be attributed to the extensive and complex interactions of the downstream targets of core clock genes. Thus, it is better to identify and target molecular rhythms directly downstream. Notably, the BMAL1–CLOCK complex exerts its effects mainly by driving the PER1 rhythm. In our study, comparing HK2 and clock gene rhythms in WT and TR gastric cancer cells, we noticed that the amplitude and phase of PER1 and HK2 rhythms displayed the closest correlation. Overexpression of PER1 reversed the negative regulation of BMAL1 and CLOCK in HK2. Meanwhile, in vivo experiments showed that the loss of PER1 could directly increase the sensitivity to trastuzumab. Therefore, PER1 was related to HK2-dependent trastuzumab resistance and may be a predictive factor for poor trastuzumab response in patients with gastric cancer.
The direct transcription regulation mechanism of the clock gene BMAL1–CLOCK is to bind the E-box sequence in the promoter region of the target gene and activate its transcription (44, 45). However, through ChIP–qPCR experiments, we found that BMAL1–CLOCK is not a direct transcription factor driving the rhythm of HK2. Previous studies have shown that clock genes can also regulate target genes at the transcriptional level by binding to other transcription factors. PPARγ is a member of the nuclear hormone receptor superfamily, which plays an important role in fat formation, glycolipid metabolism, and the immune system. It has been confirmed that PPARγ is associated with the occurrence and development of various diseases, such as diabetes, obesity, and cancer (46, 47). An increasing number of studies have shown that nuclear receptors are regarded as intermediaries between the circadian clock and cellular metabolism. Researchers have found that PPAR family members exhibit circadian oscillation expression in the liver, skeletal muscle, and adipose tissue (48). As a core clock gene, PER1 interacts with PPARγ and enhances its transcriptional activity (29). In addition, PPARγ directly regulates HK2 expression as a transcription factor (30). In this study, we demonstrate that PPARγ serves as a medium between PER1 and HK2, which can physically bind PER1 to form protein–protein complexes and subsequently enhance glycolysis via HK2.
Nowadays, second-line clinical studies of metformin combined with trastuzumab have been conducted for HER2-positive breast cancer (49). We also showed that combining metformin with trastuzumab significantly restored trastuzumab sensitivity at ZT6 (characterized by high HK2 expression and glycolysis level) in the TR gastric cancer model. This provides evidence that adjusting the administration time of metformin can improve the efficacy of antitumor drugs and reduce side effects. On the basis of the intra-individual and inter-individual variability of the molecular clock, treatment at the best tolerance or response time point can achieve optimal efficacy. Studies have shown that chemotherapy also follows a circadian rhythm. Patients with metastatic adenocarcinoma were unable to tolerate 5-FU between 3:00 and 4:00 o'clock in the morning, but could benefit from the same drug from 9:00 to 10:00 o'clock in the evening (50). In addition, different drugs present diverse circadian rhythms, making it difficult to select a uniform time for combined treatment. Thus, we proposed a novel approach to examine drug resistance, focused on the circadian rhythm of tumor metabolism. Applying a glycolysis inhibitor at a specific time based on glycolysis rhythm resulted in the best efficacy and reversed trastuzumab resistance.
In our study, metformin not only inhibited HK2 but also induced the degradation of PER1 protein in cells and mice. Previous reports have demonstrated that metformin acts as a clock regulator and rebuilds the clock, depending on the activation of AMPK (34–36). For instance, metformin treatment caused AMPK activation and mPer1/2 degradation in mouse fibroblasts and shortened the phase of the circadian cycle. Injection of metformin into the peripheral tissues of WT mice promoted mPer2 degradation and phase pattern alteration of the clock gene rhythm, but this was not observed in Ampkα knockout mice (36). Our results show that in dexamethasone-synchronized TR cells, metformin can shorten the oscillation cycle of PER1 by activating AMPK. Meanwhile, the expression peaks of BMAL1 and CLOCK were advanced, confirming that metformin can regulate the molecular circadian clock.
In summary, we revealed that the clock gene PER1 induces trastuzumab resistance by driving the HK2-dependent glycolysis circadian rhythm. In addition, combining metformin with trastuzumab at a high glycolysis level is a potential chronological strategy to reverse trastuzumab resistance. Further research on the biological clock and RTKs, as well as immunity and metabolism, will ensure the development of more promising chronotherapies for tumor treatment with low toxicity and high efficiency. Finally, the development of noninvasive methods to evaluate the circadian rhythm of cancer will enable rhythm-based chronotherapy to achieve true clinical transformation.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (82073325), Wu Jieping Medical Foundation (3206750202011018), and the Chinese Society of Clinical Oncology Foundation (Y-HR2018-192; to M. Shi).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
J. Wang: Data curation, methodology, writing–review and editing. Q. Huang: Data curation, writing–original draft, writing–review and editing. X. Hu: Data curation, methodology, writing–original draft. S. Zhang: Conceptualization, methodology. Y. Jiang: Writing–original draft, writing–review and editing. G. Yao: Resources, investigation. K. Hu: Resources, validation, methodology. X. Xu: Data curation, methodology. B. Liang: Data curation, methodology. Q. Wu: Investigation, writing–original draft. Z. Ma: Software, methodology. Y. Wang: Validation, methodology. C. Wang: Formal analysis, methodology. Z. Wu: Software, investigation. X. Rong: Supervision, investigation. W. Liao: Resources, data curation, supervision. M. Shi: Conceptualization, resources, supervision, writing–review and editing.
References
- 1. Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687–97. [DOI] [PubMed] [Google Scholar]
- 2. Vernieri C, Milano M, Brambilla M, Mennitto A, Maggi C, Cona MS, et al. Resistance mechanisms to anti-HER2 therapies in HER2-positive breast cancer: current knowledge, new research directions and therapeutic perspectives. Crit Rev Oncol Hematol 2019;139:53–66. [DOI] [PubMed] [Google Scholar]
- 3. Mitani S, Kawakami H. Emerging targeted therapies for HER2 positive gastric cancer that can overcome trastuzumab resistance. Cancers 2020;12:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Oh DY, Bang YJ. HER2-targeted therapies—a role beyond breast cancer. Nat Rev Clin Oncol 2020;17:33–48. [DOI] [PubMed] [Google Scholar]
- 5. Makiyama A, Sukawa Y, Kashiwada T, Kawada J, Hosokawa A, Horie Y, et al. Randomized, phase II study of trastuzumab beyond progression in patients with HER2-positive advanced gastric or gastroesophageal junction cancer: WJOG7112G (T-ACT study). J Clin Oncol 2020;38:1919–27. [DOI] [PubMed] [Google Scholar]
- 6. El-Athman R, Relógio A. Escaping circadian regulation: an emerging hallmark of cancer? Cell Syst 2018;6:266–7. [DOI] [PubMed] [Google Scholar]
- 7. Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol 2007;47:593–628. [DOI] [PubMed] [Google Scholar]
- 8. Shostak A. Circadian clock, cell division, and cancer: from molecules to organism. Int J Mol Sci 2017;18:873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kinouchi K, Sassone-Corsi P. Metabolic rivalry: circadian homeostasis and tumorigenesis. Nat Rev Cancer 2020;20:645–61. [DOI] [PubMed] [Google Scholar]
- 10. Hurley JM, Loros JJ, Dunlap JC. Circadian oscillators: around the transcription–translation feedback loop and on to output. Trends Biochem Sci 2016;41:834–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu D, Zhao M, Chen M, Wu B. Circadian clock-controlled drug metabolism: implications for chronotherapeutics. Drug Metab Dispos 2020;48:395–406. [DOI] [PubMed] [Google Scholar]
- 12. Dallmann R, Okyar A, Lévi F. Dosing-time makes the poison: circadian regulation and pharmacotherapy. Trends Mol Med 2016;22:430–45. [DOI] [PubMed] [Google Scholar]
- 13. Innominato PF, Karaboué A, Focan C, Chollet P, Giacchetti S, Bouchahda M, et al. Efficacy and safety of chronomodulated irinotecan, oxaliplatin, 5-fluorouracil and leucovorin combination as first- or second-line treatment against metastatic colorectal cancer: results from the international EORTC 05011 trial. Int J Cancer 2020;148:2512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koyanagi S, Kuramoto Y, Nakagawa H, Aramaki H, Ohdo S, Soeda S, et al. A molecular mechanism regulating circadian expression of vascular endothelial growth factor in tumor cells. Cancer Res 2003;63:7277–83. [PubMed] [Google Scholar]
- 15. Ye Y, Xiang Y, Ozguc FM, Kim Y, Liu CJ, Park PK, et al. The genomic landscape and pharmacogenomic interactions of clock genes in cancer chronotherapy. Cell Syst 2018;6:314–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao Y, Liu H, Liu Z, Ding Y, Ledoux SP, Wilson GL, et al. Overcoming trastuzumab resistance in breast cancer by targeting dysregulated glucose metabolism. Cancer Res 2011;71:4585–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuhr L, El-Athman R, Scrima R, Cela O, Carbone A, Knoop H, et al. The circadian clock regulates metabolic phenotype rewiring via HKDC1 and modulates tumor progression and drug response in colorectal cancer. EBioMedicine 2018;33:105–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang Q, Li S, Hu X, Sun M, Wu Q, Dai H, et al. Shear stress activates ATOH8 via autocrine VEGF promoting glycolysis dependent-survival of colorectal cancer cells in the circulation. J Exp Clin Cancer Res 2020;39:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat Med 2017;23:1331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lauriola M, Enuka Y, Zeisel A, D'Uva G, Roth L, Sharon-Sevilla M, et al. Diurnal suppression of EGFR signalling by glucocorticoids and implications for tumour progression and treatment. Nat Commun 2014;5:5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Robles MS, Humphrey SJ, Mann M. Phosphorylation is a central mechanism for circadian control of metabolism and physiology. Cell Metab 2017;25:118–27. [DOI] [PubMed] [Google Scholar]
- 22. Liu J, Pan C, Guo L, Wu M, Guo J, Peng S, et al. A new mechanism of trastuzumab resistance in gastric cancer: MACC1 promotes the Warburg effect via activation of the PI3K/AKT signaling pathway. J Hematol Oncol 2016;9:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: cancer's double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene 2006;25:4777–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang N, Chelliah Y, Shan Y, Taylor CA, Yoo SH, Partch C, et al. Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex. Science 2012;337:189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sancar A, Van Gelder RN. Clocks, cancer, and chronochemotherapy. Science 2021;371:eabb0738. [DOI] [PubMed] [Google Scholar]
- 26. Patke A, Young MW, Axelrod S. Molecular mechanisms and physiological importance of circadian rhythms. Nat Rev Mol Cell Biol 2020;21:67–84. [DOI] [PubMed] [Google Scholar]
- 27. Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science 1998;280:1564–9. [DOI] [PubMed] [Google Scholar]
- 28. Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012;338:349–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang T, Wang Z, Yang P, Xia L, Zhou M, Wang S, et al. PER1 prevents excessive innate immune response during endotoxin-induced liver injury through regulation of macrophage recruitment in mice. Cell Death Dis 2016;7:e2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Panasyuk G, Espeillac C, Chauvin C, Pradelli LA, Horie Y, Suzuki A, et al. PPARγ contributes to PKM2 and HK2 expression in fatty liver. Nat Commun 2012;3:672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mallik R, Chowdhury TA. Metformin in cancer. Diabetes Res Clin Pract 2018:409–19. doi: 10.1016/j.diabres.2018.05.023. Epub 2018 May 26. [DOI] [PubMed]
- 32. Thakur S, Daley B, Klubo-Gwiezdzinska J. The role of an anti-diabetic drug metformin in the treatment of endocrine tumors. J Mol Endocrinol 2019;63:R17–r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest 2001;108:1167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barnea M, Cohen-Yogev T, Chapnik N, Madar Z, Froy O. Effect of metformin and lipid emulsion on the circadian gene expression in muscle cells. Int J Biochem Cell Biol 2014;53:151–61. [DOI] [PubMed] [Google Scholar]
- 35. Viollet B, Guigas B, Sanz Garcia N, Leclerc J, Foretz M, Andreelli F. Cellular and molecular mechanisms of metformin: an overview. Clin Sci 2012;122:253–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, et al. Activation of 5'-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem 2007;282:20794–8. [DOI] [PubMed] [Google Scholar]
- 37. Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell 2006;22:375–82. [DOI] [PubMed] [Google Scholar]
- 38. Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature 2002;418:935–41. [DOI] [PubMed] [Google Scholar]
- 39. Fan T, Sun G. Tumor energy metabolism and potential of 3-bromopyruvate as an inhibitor of aerobic glycolysis: implications in tumor treatment. Cancers 2019;11:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tang Q, Cheng B, Xie M, Chen Y, Zhao J, Zhou X, et al. Circadian clock gene bmal1 inhibits tumorigenesis and increases paclitaxel sensitivity in tongue squamous cell carcinoma. Cancer Res 2017;77:532–44. [DOI] [PubMed] [Google Scholar]
- 41. Zeng ZL, Wu MW, Sun J, Sun YL, Cai YC, Huang YJ, et al. Effects of the biological clock gene Bmal1 on tumour growth and anti-cancer drug activity. J Biochem 2010;148:319–26. [DOI] [PubMed] [Google Scholar]
- 42. Burgermeister E, Battaglin F, Eladly F, Wu W, Herweck F, Schulte N, et al. Aryl hydrocarbon receptor nuclear translocator-like (ARNTL/BMAL1) is associated with bevacizumab resistance in colorectal cancer via regulation of vascular endothelial growth factor A. EBioMedicine 2019;45:139–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Korkmaz T, Aygenli F, Emisoglu H, Ozcelik G, Canturk A, Yilmaz S, et al. Opposite carcinogenic effects of circadian clock gene BMAL1. Sci Rep 2018;8:16023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell 2008;134:728–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sulli G, Manoogian ENC, Taub PR, Panda S. Training the circadian clock, clocking the drugs, and drugging the clock to prevent, manage, and treat chronic diseases. Trends Pharmacol Sci 2018;39:812–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, et al. PPARγ signaling and metabolism: the good, the bad and the future. Nat Med 2013;19:557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ha X, Wang J, Chen K, Deng Y, Zhang X, Feng J, et al. Free fatty acids promote the development of prostate cancer by upregulating peroxisome proliferator-activated receptor gamma. Cancer Manag Res 2020;12:1355–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell 2006;126:801–10. [DOI] [PubMed] [Google Scholar]
- 49. Rocca A, Cortesi P, Cortesi L, Gianni L, Matteucci F, Fantini L, et al. Phase II study of liposomal doxorubicin, docetaxel and trastuzumab in combination with metformin as neoadjuvant therapy for HER2-positive breast cancer. Ther Adv Med Oncol 2021. doi: 10.1177/1758835920985632. [DOI] [PMC free article] [PubMed]
- 50. Bjarnason GA, Kerr IG, Doyle N, Macdonald M, Sone M. Phase I study of 5-fluorouracil and leucovorin by a 14-day circadian infusion in metastatic adenocarcinoma patients. Cancer Chemother Pharmacol 1993;33:221–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its Supplementary Data Files.