Abstract
The development of novel agents has transformed the treatment paradigm for multiple myeloma, with minimal residual disease (MRD) negativity now achievable across the entire disease spectrum. Bone marrow–based technologies to assess MRD, including approaches using next-generation flow and next-generation sequencing, have provided real-time clinical tools for the sensitive detection and monitoring of MRD in patients with multiple myeloma. Complementary liquid biopsy–based assays are now quickly progressing with some, such as mass spectrometry methods, being very close to clinical use, while others utilizing nucleic acid–based technologies are still developing and will prove important to further our understanding of the biology of MRD. On the regulatory front, multiple retrospective individual patient and clinical trial level meta-analyses have already shown and will continue to assess the potential of MRD as a surrogate for patient outcome. Given all this progress, it is not surprising that a number of clinicians are now considering using MRD to inform real-world clinical care of patients across the spectrum from smoldering myeloma to relapsed refractory multiple myeloma, with each disease setting presenting key challenges and questions that will need to be addressed through clinical trials. The pace of advances in targeted and immune therapies in multiple myeloma is unprecedented, and novel MRD-driven biomarker strategies are essential to accelerate innovative clinical trials leading to regulatory approval of novel treatments and continued improvement in patient outcomes.
Translational Relevance.
The pace of advances in targeted and immune therapies in multiple myeloma is unprecedented. To keep this momentum going, a framework is proposed outlining key elements and regulatory considerations that will delineate how minimal residual disease (MRD) data could be collected to help standardize correlative analyses across clinical studies. The framework is intended for use by sponsors to incorporate into ongoing or planned trials, without compromising or interrupting their primary trial objectives. Also covered are technologies already impacting MRD assessment in myeloma and emerging approaches that sponsors should consider including in their trials. The current value of MRD to inform clinical care is presented using real-world cases of patients with smoldering multiple myeloma, newly diagnosed transplant eligible and ineligible, and relapse refractory disease, with each case summarizing what is known and questions to be addressed in clinical studies.
Introduction
The treatment paradigms in multiple myeloma have changed significantly over the past 5 years, both for initial management of newly diagnosed disease and during relapse after initial response to therapy. Increasing treatment options with novel drugs and drug combinations have led to deeper responses in multiple myeloma, associated with improved outcome for patients with newly diagnosed disease and relapsed multiple myeloma. This in turn has highlighted the inadequacy of traditional response assessment in myeloma that relied entirely on quantitation of the monoclonal protein in the serum and urine using gel electrophoresis and detection of residual protein using immunofixation techniques, along with morphologic evaluation of the marrow to define complete response (CR). CR by this conventional definition provided a false sense of disease control, because nearly all patients eventually relapsed despite achieving CR. Subsequent attempts to improve response assessment using serum free light chain assay and clonality assessment in the marrow led to designation of stringent CR (sCR), which provided only a modest degree of improvement in assessing the depth of response.
It was in this context that the International Myeloma Working Group (IMWG) updated the multiple myeloma uniform response criteria incorporating minimal residual disease (MRD) assessment as an additional level of response. The IMWG relied on available data demonstrating a prognostic value for MRD negativity in patients with newly diagnosed or relapsed multiple myeloma (1). It utilized a minimum cutoff of 10−5 cells for defining MRD negativity, based on data available at the time of the revision and the availability of technology that could reliably demonstrate residual disease only up to this level of detection. The response criteria were agnostic to the methodology utilized, as long as the method was validated for the level of sensitivity required, and specifically identified flow cytometry or a VDJ gene sequencing approach as acceptable methods. For the first time, the revised criteria also incorporated sensitive imaging techniques into the definition of MRD negativity, based on data from several randomized European trials as well as retrospective data from multiple centers. FDG-PET was the method of choice for incorporation into response criteria, given the available data and the delay in changes seen using conventional MRI compared with functional imaging using FDG-PET. Importantly, technology has continued to improve, and novel flow cytometry and next-generation sequencing (NGS) methods are able to attain sensitivity levels of 10−6 cells or lower in high-quality bone marrow (BM) samples. In addition, mass spectrometry has continued to evolve for detection of smaller amounts of monoclonal protein than immunofixation techniques. Other methods to detect single circulating tumor cells or cell-free DNA (cfDNA) in the peripheral blood are also being evaluated.
In January 2016, a collaboration of advocacy organizations, patients, research foundations, academia, government (NIH and FDA), and industry was convened by the Foundation for the National Institutes of Health (FNIH) Biomarkers Consortium to update current status of MRD in improving patient care and enhancing the development of new therapies in multiple myeloma. The resulting white paper (2) summarized state of the science and technology as well as clinical data supporting the use of MRD in multiple myeloma; and most importantly, proposed studies needed to define MRD as a surrogate endpoint for regulatory purposes and informing clinical decisions in multiple myeloma.
Significant progress has been made in the 4 years since these original discussions, both in terms of establishing MRD as a regulatory endpoint for clinical trials leading to drug approval, and in design and initiation of clinical trials that are defining the role of MRD testing in routine clinical practice. Several ongoing efforts are attempting to bring together the existing data from large phase III trials for surrogacy analysis, a requisite step to obtain acceptance from the regulatory authorities around the world for MRD to serve as a surrogate marker for longer term outcomes. Notably, a recent meta-analysis, the largest to date, reviewed data from 93 publications covering 8,098 patients and reaffirmed the importance and strong prognostic value of MRD negativity in improving long-term survival in a heterogeneous cohort of patients with multiple myeloma (3). Also, there are efforts being led by the multiple myeloma community in collaboration with industry partners which are at the stage of MRD data aggregation and analysis for submission to health authorities. We anticipate that these efforts will eventually lead to increased utilization of MRD as an endpoint for clinical trials with regulatory intent. Even more progress has been made in the design of clinical trials that incorporate MRD testing at various stages of therapy to inform subsequent treatment decisions. In our previous white paper (2), we suggested several potential clinical trial designs to evaluate the value of MRD at various stages of multiple myeloma. Many of these designs have now been implemented, and almost 50 phase III trials are currently actively enrolling using MRD-directed treatment assignment or MRD as an endpoint (Table 1). These trials are designed to ask clinically relevant questions including: defining the ideal duration of maintenance therapy after autologous stem cell transplantation (ASCT); defining whether MRD-directed treatment intensification is beneficial in patients who do not achieve MRD negativity after induction therapy with triplet combinations or those who remain MRD positive at 1 year of post-transplant maintenance. Given the considerable progress made over the 4 years since our initial white paper, we here review the progress and update recommendations regarding the current role of MRD testing in clinical trials and routine clinical practice, as well as in new drug registration.
Table 1.
Phase III clinical trials in myeloma using MRD assessment as primary or secondary endpoints.
| Trial ID | Sponsor | Eligibility | Enrollment | Interventions (Trial name) | MRD-based endpointsa | Publications |
|---|---|---|---|---|---|---|
| NCT03652064 | Janssen Research and Development, LLC | Newly diagnosed multiple myeloma (NDMM) not undergoing hematopoietic stem cell transplant (HSCT) | 395 | Daratumumab, Bortezomib, Lenalidomide, Dexamethasone (CEPHEUS) | MRD negativity [after randomization and prior to progressive disease (PD) or subsequent anti-myeloma therapyb], MRD negative rate (1 year, throughout study) | J Clin Oncol 2019;37(15 Suppl): abstr TPS8056 |
| NCT04071457 | Southwest Oncology Group | Multiple myeloma (MM) patients who have undergone systemic induction therapy and autologous stem cell transplantation (ASCT) | 1,100 | Lenalidomide, Daratumumab/rHuPH20 (DRAMMATIC) | MRD negativity (24 months from initial randomization) | |
| NCT04096066 | Fondazione Neoplasie Sangue Onlus | Patients with NDMM ≥ 65 years | 340 | Carfilzomib, Lenalidomide, Dexamethasone | MRDb, MRD negativity (5 years) | |
| NCT03941860 | NCI | Previously diagnosed patients with MM on lenalidomide maintenance post stem cell transplantation (SCT) | 510 | Ixazomib, Ixazomib Citrate, Lenalidomide (OPTIMUM) | MRD conversion rate (at 12 and 24 cycles post-randomization) | |
| NCT02406144 | PETHEMA Foundation | Patients with NDMM after ASCT | 316 | MLN9708, Lenalidomide, Dexamethasone | MRD (5 years) | |
| NCT03948035 | Wuerzburg University Hospital | Patients with NDMM eligible for ASCT | 576 | Elotuzumab, Carfilzomib, Lenalidomide, ASCT | MRD negativity rate (end of cycle 6b) | |
| NCT03710603 | European Myeloma Network | Patients with NDMM | 690 | Daratumumab, Velcade, Lenalidomide, Dexamethasone (PERSEUS) | Post-consolidation MRD negativity rate | J Clin Oncol 2019;37(15 Suppl): abstr TPS8055 |
| NCT03617731 | U. of Heidelberg Medical Center | Untreated patients with NDMM requiring systemic therapy | 662 | Lenalidomide, Bortezomib, Dexamethasone, Isatuximab (GMMG HD7) | MRD negativity (post inductionb, post therapy, during and after maintenance) | |
| NCT03562169 | U. of Leeds | Relapsed patients with MM previously treated with ASCT | 406 | Ixazomib, thalidomide, & dexamethasone (ITD), Augmented and conventional ASCT (ACCoRd) | MRD negativity rate, continuous MRD (baseline, end of re-induction, 100 days post-ASCT, After two cycles of consolidation; 8 weeks and 12 months post-randomization 2) | |
| NCT03275285 | Sanofi | Relapsed and/or refractory patients with MM previously treated with 1 to 3 lines of therapy | 302 | Isatuximab, SAR650984, carfilzomib, dexamethasone (IKEMA) | MRD negativity (36 months) | J Clin Oncol 2018;36(15 Suppl): abstr TPS8060 |
| NCT03720041 | U. of Leeds | Patients with NDMM non-eligible for SCT | 740 | Ixazomib, Lenalidomide, Dexamethasone (IRD) (FiTNEss) | MRD negativity (end of induction therapy and 12 months post randomization 2) | |
| NCT02659293 | U. of Chicago | Patients with MM after ASCT being considered for lenalidomide maintenance. | 180 | Lenalidomide, Carfilzomib, Dexamethasone | MRD negativity rate (6 and 12 months after randomization) | |
| NCT03319667 | Sanofi | Patients with NDMM not eligible for transplant | 475 | Isatuximab, Bortezomib, Lenalidomide, Dexamethasone (IMROZ) | MRD negativity rate (60 months after the FPI or scheduled assessment) | |
| NCT04091126 | GlaxoSmithKline | Transplant ineligible (TI) newly diagnosed multiple myeloma (NDMM). | 750 | Belantamab mafodotin, Bortezomib, Lenalidomide, Dexamethasone | % of patients with sustained MRD negativity (1 year) | |
| NCT02136134 | Janssen Res. & Dev., LLC | Patients with relapsed or refractory MM (RRMM) | 499 | Daratumumab, Bortezomib and Dexamethasone | MRD negativity rate (up to disease progression) | PMID: 27557302 |
| NCT03158688 | Amgen | Patients with RRMM | 466 | Dexamethasone, Daratumumab, Carfilzomib (CANDOR) | MRD[-]CR (12 and 28 months) | Blood 2019;134 (Suppl 2):LBA-6 |
| NCT02181413 | Millennium Pharmaceuticals, Inc. | Patients with NDMM responsive to induction therapy and who have received high-dose therapy and ASCT | 656 | Ixazomib Citrate | MRD conversion rate, maintenance of MRD negativity [EOT (24 months)] | PMID: 30545780 |
| NCT04181827 | Janssen Res. & Dev., LLC | Patients with relapsed and lenalidomide-refractory MM | 400 | JNJ-68284528, Pomalidomide, Bortezomib, Dexamethasone, Daratumumab (CARTITUDE-4) | MRD negativity rate overall and in participants with CR or sCR, sustained MRD negative rate (6 years) | ASH 2019; abstr 577 |
| NCT01863550 | ECOG-ACRIN Cancer Research Group | Patients with NDMM | 1,087 | Bortezomib, Carfilzomib, Dexamethasone, Lenalidomide | MRD negativity rate [post induction randomization and post-maintenance randomization (2,3 years)] | |
| NCT03748953 | Millennium Pharmaceuticals, Inc. | Patients with NDMM with CR/VGPR/PR to initial therapy and who have not undergone SCT | 105 | Ixazomib | MRD conversion rate (24 months) | |
| NCT03993912 | University Hospital, Lille | Elderly frail patients with NDMM ineligible for high-dose chemotherapy and ASCT | 294 | Daratumumab, Lenalidomide, Dexamethasone (IFM2017_03) | MRD negativity rate (12 months) | |
| NCT02541383 | Intergroupe Francophone du Myelome | Transplant eligible previously untreated patients with MM | 1,085 | Bortezomib, Thalidomide, Dexamethasone, Daratumumab (Cassiopeia) | MRD negativity (post-consolidation, post-ASCT) | PMID: 31171419 |
| NCT04162210 | GlaxoSmithKline | Patients with RRMM | 380 | Belantamab, Pomalidomide plus low-dose Dexamethasone | MRD negativity rate (up to 55 months) | |
| NCT02252172 | Janssen Res. & Dev., LLC | Patients with NDMM not eligible for either high-dose chemotherapy or ASCT. | 737 | Daratumumab, Lenalidomide, Dexamethasone | MRD negativity rate (any timepoint after randomization to 60 months post C1D1 with an expected average of 24 months) | PMID: 31141632 |
| NCT03539744 | AbbVie | t(11;14)-positive patients with RRMM | 244 | Pomalidomide, Dexamethasone, Venetoclax (CANOVA) | MRD negativity rate (28 months from first randomization) | |
| NCT03651128 | Celgene | Patients with RRMM | 381 | bb2121, Daratumumab, Pomalidomide, Dexamethasone, Bortezomib, Ixazomib, Lenalidomide (KarMMa-3) | MRD (5 years from randomization) | |
| NCT03180736 | European Myeloma Network; Collab.: Janssen Res & Dev, LLC | Patients with RRMM | 304 | Daratumumab, Pomalidomide, Dexamethasone | MRD negativity rate [from randomization until PD (approximately up to 3 years)] | J Clin Oncol 2018;36(15 Suppl): abstr 8059 |
| NCT03729804 | University of Chicago | Newly diagnosed untreated patients with MM requiring systemic chemotherapy | 250 | Carfilzomib, Lenalidomide, Dexamethasone, Bortezomib (COBRA) | MRD negativity at indicated timepoints of study after randomization (5 years) | |
| NCT03792620 | Grupo de Estudos Multicentricos em Onco-Hematologia | Newly diagnosed myeloma in older patients (≤65 years) without comorbidity | 20 | Cyclo Thal Dex Daratumumab (MAXDARA) | MRD negativity rate (24 months) | |
| NCT02312258 | Millennium Pharmaceuticals, Inc. | Patients with NDMM with CR, VGPR, or PR to initial therapy and who have not undergone SCT | 706 | Ixazomib (TOURMALINE-MM4) | MRD conversion rate, maintenance of MRD negativity (screening, cycle 13, and cycle 26) | TOURMALINE-MM4_Mmhub.com |
| NCT02076009 | Janssen Res. & Dev., LLC | Patients with RRMM | 569 | Daratumumab, Lenalidomide, Dexamethasone (POLLUX) | MRD negativity rate from randomization to the date of first documented evidence of PD until 3 years | J Clin Oncol 2017;35(15 Suppl): abstr 8025 |
| NCT02990338 | Sanofi | Patients with RRMM | 307 | Isatuximab, Pomalidomide, Dexamethasone (ICARIA-MM) | Number of participants with MRD (up to 76.7 weeks) | PMID: 31735560 |
| NCT02195479 | Janssen Res. & Dev., LLC | Untreated patients with MM ineligible for high-dose chemotherapy and ASCT | 706 | Velcade, Melphalan, Prednisone, Daratumumab, Dexamethasone | Percentage of participants with negative MRD [from randomization to disease progression (up to 2.4 years)] | PMID: 29231133 |
| NCT03859427 | Amgen | Patients with RRMM | 460 | Carfilzomib, Lenalidomide, Dexamethasone (ARROW2) | MRD[-]CR rate (time frame: 12 months and study completion) | Clin Lymphoma Myeloma Leuk 2019;19(Suppl 1):S332-S333 |
| NCT03901963 | Janssen Res. & Dev., LLC | Patients with NDMM who have undergone four to eight cycles of induction and/or consolidation, HDT and ASCT | 214 | Daratumumab, Lenalidomide (AURIGA) | MRD negativity rate (12b, 36 months) |
aSecondary endpoint unless otherwise noted.
bPrimary endpoint.
Scientific and Technological Considerations
A number of large professional society meetings focused on hematologic malignancies have included a point/counterpoint debate on whether MRD assessment is “ready for prime time” in clinical management of patients with lymphoid malignancies including multiple myeloma. While these sessions are useful for introducing the concepts of MRD to practitioners, they belie and obscure the fact that MRD has become a fundamental gauge for clinical management in various cancers. The concept of remission in hematologic malignancies was once based on clinical exam alone, then on the enumeration of morphologically aberrant cells in the BM or blood from a patient, then by conventional flow cytometry, which provided a sensitivity based on practical application (not on biology) of 10−4 cells, and then by analyses of immunoglobulin light or heavy chains in the blood (or urine). At each point in time when these measures were adopted for clinical care, they represented an increase in sensitivity over what had existed before. Their utility, however, was only relevant once therapeutic interventions existed that could lower the tumor burden to a level at which such increased sensitivity was necessary. Physicians treating patients with leukemia, lymphoma, or multiple myeloma have evolved their practices as each new more refined, standardized, accurate, and sensitive measure of residual disease has been introduced. Initially, the clinicians' response was often exasperation exemplified by responses to the sensitivities of current new technologies, “I should be so lucky as to have to worry whether my patient still has one malignant cell within one million normal hematopoietic cells.” Fortunately, for one after another indication, treatment has become more and more efficacious, and therapeutic options for a given patient have markedly increased. Today, it is possible to approach a patient with multiple myeloma with the prospect of long-term control. Thus, in-depth assessment of residual tumor burden and kinetics of residual tumor growth over time have become essential to patient-physician discussions of a therapeutic plan and decisions of whether to change, discontinue, or intensify a given treatment course. In fact, physicians have started incorporating MRD determination as an aid in their real time clinical management decisions (4). In this section, several advances in the determination of MRD are described. Utilization of these methods in clinical trials, in drug development, and in real-world patient management are discussed in subsequent sections.
Strengths and weaknesses of the various MRD technologies have been well described elsewhere and will not be discussed in details here. Regardless of which “next-generation” technology (next-generation flow or NGF, NGS, imaging, mass spectrometry–based paraprotein analyses, etc.) is employed, some basic principles apply to its incorporation into clinical trials and real-time patient management. Single point in time assessment of MRD is a measure of tumor burden, not tumor biology. On the other hand, achievement of such deep remission might reflect a favorable biology. Any threshold of such tumor burden (e.g., 10−4, 10−5, 10−6 cells or undetectable) is useful to provide consistency when comparing treatment arms or stratifying patient populations into different arms of therapeutic intervention and ultimately in the future in choosing treatment options in real-world practice. Such thresholds provide information at the population level on average therapeutic efficacy. However, MRD is a continuous variable and for individual patient management any MRD value must be considered in the overall context of prior treatment regimens, other diagnostic tests and risk markers, as well as all of the signs, symptoms, co-morbidities, and quality-of-life factors that are relevant to that particular person. Determination of MRD at a single point in time may have prognostic significance, but quantitative, accurate, standardized, and sensitive MRD determinations may provide greater information relevant to tumor biology and likelihood of relapse when performed in a sequential fashion over multiple timepoints during a continuum of care to establish the trend and pace of the change in tumor burden.
NGF cytometry
Multiparametric flow cytometry (MFC) is a widely available, fast, and highly applicable technique for detection and enumeration of BM plasma cells (PC). In addition, it allows clear cut and highly sensitive discrimination between normal (i.e., polyclonal) and clonal PCs based on their uniquely distinct (normal vs. aberrant) immunophenotypic characteristics, even when tumor PCs are present at very low frequencies, for example, MRD (5–7). Early studies and more recent investigations have highlighted the value of MRD monitoring by MFC for: (i) improved evaluation of response to therapy, particularly among patients that reach a CR; (ii) prognostic stratification of patients with multiple myeloma after therapy; and (iii) to modify treatment in the settings of clinical trials for both transplant eligible and transplant ineligible patients (8–22). However, none of the referenced studies have definitively demonstrated yet in a randomized fashion that modifying treatment in an MRD-positive patient converts them to MRD negative and improves outcomes, something being currently addressed through ongoing clinical trials. In parallel, these studies also showed that conventional MFC MRD approaches are associated with several important limitations when compared with molecular MRD approaches, particularly a lower sensitivity and lack of standardization (23, 24).
Highly sensitive NGF MRD approaches have been developed in the last 5 years by EuroFlow to overcome most limitations of conventional MFC MRD techniques (23, 25, 26). This novel NGF approach for MRD in multiple myeloma is based on a more efficient sample preparation protocol for acquisition of >10 million BM cells, optimized and well-tuned 8–12 antibody combinations (25), and innovative automatic data analysis strategies and software tools that have resulted in increased sensitivity (vs. conventional 4–10-color MFC approaches) of 10−6 (26). Importantly, NGF can be applied in virtually every patient with multiple myeloma, even in the absence of a diagnostic sample. Furthermore, NGF MRD also provides information about the quality of the BM sample and the potential presence of hemodilution, which is critical for early identification of false-negative MRD results (23, 26, 27). Because of these clear advantages and its standardized nature, MRD detection in multiple myeloma by the EuroFlow NGF approach has been recognized by the IMWG as the reference method to define flow MRD negativity after therapy (1). This is of utmost importance, because the clinical impact of high-sensitivity MRD detection by NGF (26) has been recently validated in the settings of both real-world patients with multiple myeloma (28) and the PETHEMA/GEM 2012MENOS65 clinical trial (24). Notably, only 7% of MRD-negative (MRD < 2 × 10−6 cells) cases in the GEM/PETHEMA trials showed disease progression (mostly related to extraosseous disease) with an 82% and 88% reduction in the risk of progression and death, respectively, and HRs of 0.18 [95% confidence interval (CI), 0.11–0.30; P < 0.001] and 0.12 (95% CI, 0.05–0.29; P < 0.001), respectively, overcoming the poor prognostic value of high-risk cytogenetics at diagnosis in cases that achieved MRD negativity by NGF (24). Although the recently extended use of anti-CD38 antibody-based therapies could interfere with CD38-based NGF detection due to bound antibodies at the surface of multiple myeloma cells, combined use of alternative (e.g., multiepitope and nanobody) CD38 conjugates and the NGF-based automatic data analysis strategies have allowed for robust detection of MRD, even in patients receiving anti-CD38 therapy (26, 29, 30). Meanwhile, occurrence of extraosseous disease in this subset of patients highlights the complementarity of BM-based MRD assessment with other approaches including MRD detection in blood by NGF (30), Qip-mass spectrometry (31) and particularly, imaging techniques (24, 32).
NGS MRD
DNA-based assessments of immune receptor clonality (e.g., Southern blot, PCR) have been employed to address clinical research questions for many years (33, 34). It has only been within the past 10 years that advances in knowledge to develop a comprehensive “immunosequencing” high-throughput method of enumerating, specifying, and quantifying each and every B and/or T cell in a sample of interest have come together to allow for a DNA sequence–based immune receptor repertoire approach to real time clinical questions. One of the first and most obvious applications is the determination of MRD in lymphoid malignancies. The technology behind NGS has been well described elsewhere (35, 36). The assay is robustly quantitative, which has been ensured by primer design and concentration adjustments, so that every possible V-J combination is amplified equivalently. The sensitivity of the assay is simply a function of the amount of genomic DNA that is analyzed. Because of the absolute specificity of most CDR3 regions, only a single event (the presence of a single sequence within the sample) is required to make a call as to the presence of a clone of interest. If one million cells are analyzed, the sensitivity of the reaction approaches 10−6 cells. At the patient level, the assay can detect MRD in >95% of extracted gDNA samples from that patient if, on average, there are approximately two or more malignant-cell equivalents per the number of total cells assessed in the patient sample. As will be discussed in the Regulatory Section below, the NGS-based assay of immune receptor repertoire profiling developed by Adaptive Biotechnologies (clonoSEQ) is currently the only assay cleared by the FDA for MRD determination in BM from patients with multiple myeloma.
There are numerous studies that demonstrate the utility of this assay for the determination of response to therapy, prognosis, and monitoring of disease status. For example, in recent study using this assay for the assessment of patients with multiple myeloma pre or post maintenance demonstrated that the level of MRD correlates with outcome, and that the deeper the level of MRD either pre or post maintenance (down to a level of less than 10−6 cells), the better the prognosis (37). In this study, more patients in the ASCT treatment arm achieved NGS-based MRD negativity than in the control arm. However, the best prognostic marker was the achievement of NGS-MRD negativity, regardless of the treatment arm to which a patient was assigned. Similar findings have emerged from other clinical trials. For example, in a comparative study of patients stratified to receive either VMP alone or daratumumab-VMP, more patients in the arm including daratumumab achieved NGS-MRD negativity; however, the outcome for all patients achieving NGS-MRD negativity was the same (38). This again supports the concept that achieving MRD negativity may be more important than how one reached that state.
It should also be mentioned that the same chemistry and algorithmic principles described here for NGS-based MRD analyses can be applied to any situation in which elaborating, specifying, and quantifying the immune response is important. This broadens the application of NGS-based immune receptor repertoire analysis to indications in immunotherapy, infectious disease, autoimmunity, vaccine response, aging, and general considerations of individual or population health status (39–41).
Toward blood-based MRD approaches
Advancements in technologies to assess MRD, including NGF and NGS, have provided real-time clinical tools for the sensitive detection and monitoring of MRD in patients with multiple myeloma (28, 42). However, the routinely used MRD assays rely on information obtained from repeated BM aspirates, which are invasive and may contribute to false negatives due to the multifocal “patchy” nature of the disease in the BM. Liquid biopsy–based assays could be very useful to implement into MRD assessment as they can: (i) provide accessible repeatable measurements for routine monitoring, (ii) detect total tumor burden reflective of disseminated disease and undetected lesions to stratify patients by recurrence risk, and (iii) provide comprehensive information on heterogeneous genetic alterations that can help guide therapy for earlier intervention (43). While some blood-based approaches, such as mass spectrometry methods, are now very close to prime time, others utilizing nucleic acid-based technologies are still developing and will prove to be important complementary tools to assess the biology of minimal residual disease.
Mass spectrometry methods
Mass spectrometry methods to detect the M-protein in blood offer a promising, less-invasive alternative to BM-based MRD testing. These methods can detect the M-protein at lower concentrations than serum immunofixation, better distinguish therapeutic antibodies from M-proteins, and detect post-translational modifications (i.e., glycosylation), which may be relevant for identifying amyloidosis (44, 45). Importantly, mass spectrometry methods have the ability to distinguish between myeloma-derived monoclonal proteins and therapeutic mAbs.
Mass spectrometry methods are based on the fact that each immunoglobulin has a unique amino acid sequence and therefore a unique mass. Although a variety of methods have been developed, they all involve the same general steps: enriching immunoglobulins from serum, processing the enriched proteins into smaller components, and measuring the mass of the components. Data are analyzed for the presence of the patient's M protein–specific mass, which is used as a marker of disease and is constant over time.
Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-TOF MS) is a rapid, high-throughput technique that has the potential to replace current electrophoretic methods for M-protein detection. The Mayo Clinic currently offers the MASS-FIX assay as a laboratory-developed test. This method is more analytically sensitive and specific than serum protein electrophoresis and immunofixation (46, 47), and provides similar clinical sensitivity for the detection of monoclonal gammopathies (48). Another similar method is being developed by The Binding Site with the goal of producing an FDA-approved, commercially available assay. Both methods follow the signal from intact light chains to detect M-proteins. However, they differ in technical details, particularly the immunoglobulin purification step, and a head-to-head comparison of these techniques has not yet been reported.
More sensitive LC/MS methods have also been developed to detect M-protein but remain in the research realm. A method termed monoclonal immunoglobulin rapid accurate mass measurement follows intact light chains, and was reported to be about 100 times more sensitive than serum immunofixation (49). Other LC/MS methods track clonotypic peptides as a marker of disease. In these methods, enriched immunoglobulins are digested with an endoproteinase, most commonly trypsin, and patient-specific peptides from the immunoglobulin heavy and light chain variable regions are monitored by LC/MS. Reported detection limits of these clonotypic peptide methods range from 0.01 to 4 mg/dL (45, 50–52).
Currently, the field is still developing these techniques and evaluating how to best use them. Some studies comparing mass spectrometry results to BM-based MRD assays have found that the M-protein can be detected when BM-based tests are negative (53–56). And, a recent study found LC/MS to be a better predictor of progression-free survival (PFS; ref. 56). Prior to the incorporation of these methods into clinical laboratories, work is needed to better understand the clinical utility of low-level M-protein measurements. Questions remain regarding the half-life of immunoglobulins and the level of sensitivity needed for detecting the M-protein in the setting of MRD.
Circulating cfDNA for MRD assessment
Liquid biopsies that track tumor mutations present in circulating cfDNA isolated from blood plasma have become a promising minimally invasive tool to characterize and monitor many cancers (57–60). Multiple groups have executed studies that revealed cfDNA dynamics and high concordance of clonal somatic mutations and copy-number alterations between BM and cfDNA of patients with multiple myeloma (61–64). However, extending these findings to implement cfDNA profiling for MRD assessment has remained challenging due to limitations in detection of the low abundance of circulating tumor DNA (ctDNA) from normal cfDNA derived from peripheral blood cells (65). Ultimately, many current methods lack the capacity to detect MRD when the fraction of ctDNA in the bloodstream is lower than twice the inverse of the number of copies of each gene in a given sample, referred to as the genomic equivalent (GE) limit (66).
Ultradeep targeted sequencing has significantly improved the detection of ctDNA present at low concentrations, although sensitivity is driven by the number of tumor mutations available to track. To date, the validity of cfDNA profiling in the MRD setting in multiple myeloma has been limited to a small subset of clinical studies (42, 67, 68). Most recently, the first comparative prospective study tracked all clonal immunoglobulin gene rearrangements using NGS in paired cfDNA and BM samples for MRD assessment in 42 patients with multiple myeloma (42). The study demonstrated there was no correlation for MRD between cfDNA and BM, with only 49% consistency between the paired samples. The most frequent discrepancy observed was undetectable MRD in plasma, which was positive in the BM. This suggests different NGS strategies are needed to increase the sensitivity of detection of low-frequency variants without compromising specificity (69, 70). An alternative strategy that remains to be tested in multiple myeloma is tracking a larger number of patient-specific tumor mutations based on a tumor-derived mutation fingerprint. This strategy has successfully been shown to increase the likelihood that the targeted mutations will be captured when the tumor fractions in the blood are below the GE limit (66). Alternatively, large, targeted sequencing panels that incorporate frequently mutated genes, frequent copy-number alterations, V(D)J rearrangements and translocations, as well as minimize template DNA losses during library preparation and suppress background errors, may also be applicable and have yet to be described (70–72).
To validate the utility of ctDNA in the MRD setting, comparative studies are needed that incorporate both BM and cfDNA and assess MRD with different methodologies. Additional efforts toward the design of targeted sequencing assays and bioinformatics techniques are likely required before they can be proposed as standard approaches. Sufficiently powered prospective investigations will also be needed to prove that ctDNA detects a window of intervention before current BM-based methods or is complementary in some way, and that clearance of ctDNA levels is reflective of disease control (73).
Single-cell RNA sequencing
Single-cell RNA sequencing (scRNA-seq) offers high-throughput and high-resolution analysis of gene transcription on a cell by cell basis. The single-cell approach provides benefit by allowing resolution of malignant PCs from stromal and immune cells of the microenvironment compared with bulk sequencing, which yields an aggregate of transcriptional activity from unknown cells from a specimen (74–78). Therefore, this sensitivity facilitates the detection of rare malignant cells that constitute MRD. The power of the technology lies in the combination of single-cell resolution with transcriptional activity. Molecular signatures of the evolution and progression of disease have been found in PCs, as well as the nontumor cells of the BM microenvironment (79–82). Therefore, scRNA-seq may be used to select appropriate targeted therapies according to the identification of resistant or sensitive populations of cells, based on specific gene mutations, splice variants, and common rearrangements. For example, t(11;14) and high BCL2 mRNA appear to be predictive markers for response to venetoclax (83), and certain gene expression signatures have been found to predict for response to proteasome inhibition (84).
However, several challenges need to be overcome before scRNA-seq can enter routine clinical practice (85). At the technical level, sample preparation is time consuming and loss of polyadenylated RNA at low levels of gene expression may lead to amplification bias (86). Therefore, there needs to be a standardization and simplification of the workflow both in sample preparation and bioinformatic analysis to ensure accuracy and reproducibility among patients and centers. Furthermore, evidence to support the use of targeted agents in response to specific transcriptional signatures is in its infancy.
Despite these considerations, scRNA-seq has the potential not only to be a highly sensitive technique for MRD determination, but also to provide detailed information to guide subsequent therapeutic intervention. In essence, it can identify the disease at the earliest opportunity and provide a solution in the same test. The use of scRNA-seq to monitor intraclonal heterogeneity in individual patients has the potential to take the care of patients with myeloma one step closer to true precision medicine.
Imaging
For the longest time, imaging has been limited to assessment of myeloma bone disease. However, because a multitude of studies have shown that modern cross-sectional whole body imaging techniques like PET with CT (PET/CT) and MRI provide a comprehensive overview of the tumor burden beyond osteolytic lesions including extramedullary disease (EMD), these techniques have been investigated as methods to assess residual disease after therapy (87). This is particularly important because other techniques, which rely on BM specimens are by nature limited to sampling a very small area of the body. Besides the patchy infiltration of BM PCs (BMPC) and the presence of EMD (88), recent prospective studies serially monitoring patients with functional imaging and focal lesions (FL) biopsies demonstrated that multiple myeloma entails spatial heterogeneity, with possible coexistence of different disease clones with different genomic profiles in the BM versus FLs (89). The larger the FL size, the greater the heterogeneity (90). Therefore, whole body imaging provides important complementary information about residual disease after therapy, and also about early relapse. EM sites of clonal proliferating PCs in a context of BM MRD negativity are more frequent in patients with EMD at diagnosis (5%–10%) or with paramedullary plasmacytomas (91).
18Fluorodeoxyglucose (18F-FDG)-PET is an excellent imaging tool to assess tumor metabolic activity and monitor response to treatment, due to its ability to distinguish between active and inactive (e.g., fibrotic) disease. In addition, low-dose chemotherapy, which is typically done for localization along with FDG-PET, constitutes a precise screen for bone and extramedullary findings (92).
Several studies have demonstrated an unfavorable prognostic role for PET-positive lesions after completion of therapy (93–96). Conversely, in patients achieving complete remission (CR), FDG-PET/CT negativity after ASCT predicted a lower risk of progression or death in patients with conventionally defined CR than in patients with metabolically active sites of disease (97, 98). Moreover, in patients achieving MRD negativity by flow cytometry with a sensitivity of 10−5 cells, imaging either by PET/CT or whole-body diffusion-weighted MRI (WB-DWI-MRI) was positive in 12% of the cases and associated with a shorter PFS (99). In contrast, patients achieving an MRD-negative CR during salvage therapy frequently had FLs (50%). Also, it has recently been shown that patients obtaining PET FL normalization upon therapy have comparable prognosis to patients without baseline increased metabolism, suggesting the value of treating until suppression of glucose metabolism (100). The complementarity between imaging (either FDG-PET/CT or WB-DWI-MRI) and BM techniques in defining the prognosis of patients was demonstrated by two prospective studies, using flow cytometry with a sensitivity of 10−4 (10) and 10−5 cells (99).
On the basis of the above-reported results, 18F-FDG PET/CT is currently considered the preferred imaging technique for evaluating and monitoring metabolic response to therapy (87). However, it has been reported that in 10% to 15% of cases, PCs may not be 18F-FDG-avid due to lack of hexokinase enzyme, which is responsible for FDG trapping in the cells (101). In these patients, FDG PET/CT is not an appropriate tool to evaluate metabolic response to therapy. In addition to 18F-FDG, new PET/CT tracers targeting different metabolic pathways or receptors expressed by PCs may represent potentially more sensitive and specific molecular imaging biomarkers, and have been preliminarily investigated in limited series of patients with multiple myeloma or in mouse models (102). PET imaging targeted to CXCR4 (103, 104), CD38 (105, 106), and VLA-4 (NCT03804424) has advanced into translational clinical trials, bringing us closer to powerful imaging options for multiple myeloma. Radiolabeled antibody imaging may be advantageous, as it is imaging tumor cells with antigenic expression regardless of metabolic processes, resulting in earlier and more specific assessment of response. For example, all multiple myeloma cells express CD38, making it an excellent focus for targeted imaging and therapy. Daratumumab is an already FDA-approved monoclonal antibody therapy for multiple myeloma that targets CD38. Conjugating daratumumab with the positron emitting radioisotopes Copper-64 (64Cu) and Zirconium-89 (89Zr) has allowed for the creation of immunoPET tracers for multiple myeloma imaging. 89Zr-Daratumumab has demonstrated the ability to detect multiple myeloma in early clinical trials (107) when it was not detected by FDG-PET/CT and other clinically standard imaging methods. More advanced clinical trials for these immunoPET agents are planned.
However, the lower availability of these newer tracers, interpatient tumor heterogeneity regarding specific targets, as well as the lack of prognostic data and standard reporting, prevent any definite conclusion from being drawn at this time. As with BM techniques, standardization of imaging criteria and definition of cutoffs for positivity/negativity, highly important to allow for data reproducibility and harmonization, is currently ongoing. A standardized definition of PET complete metabolic response has been proposed considering the uptake of the liver as threshold, and is currently under confirmation in independent prospective series of patients (108).
Conventional MRI without contrast agents has been used for response assessment in several clinical trials, in addition to serologic and BM-derived parameters. Two studies using MRI of the spine and pelvis and whole body MRI in a total of 711 patients treated with high-dose chemotherapy protocols showed that residual lesions after completion of the most aggressive treatment had a significant adverse prognostic significance (109, 110). A later study again comparing axial MRI (including spine and pelvis) with PET/CT in 134 patients with multiple myeloma treated in a multi-center trial showed that PET/CT was superior to MRI with regards to prognostic significance after therapy. The most likely explanation has been examined in the first study of this kind showing that treatment response in MRI appears delayed, with FLs of multiple myeloma disappearing more slowly because MRI is not able to differentiate between vital and necrotic tissue within preexisting osteolytic lesions (109). Interestingly, a change of lesions into a more liquid or cystic appearance was associated with a higher rate of CRs, but also with a higher proliferation index in gene expression profiling (111). A further development of MRI is DWI, which measures the movement of water molecules in the investigated tissue. This Brownian movement is in part limited by lipophilic cell membranes and promoted by tissue perfusion, which in turn gives DWI the ability to assess cellularity and microcirculation in the BM (112). Recent studies suggest that whole body DWI might be equivalent or even slightly superior to FDG PET/CT in the assessment of MRD (113).
In summary, at this time PET/CT is the most widely available and most reliable imaging technique for the assessment of residual disease in areas outside of the BM, and should be added to assess remission, especially within clinical trials.
Regulatory Considerations
FDA perspective on regulatory considerations for MRD assessment and incorporation into clinical trials
There is an interest in the multiple myeloma community to evaluate new sensitive markers of response, like MRD, to provide an improved and early estimate of activity of drugs to expedite drug development. While available evidence suggests that MRD as a general measure of tumor burden in patients with multiple myeloma has multiple potential regulatory and clinical uses as a biomarker, the clinical and regulatory uses are very different. For MRD to function as an effective regulatory tool, several aspects within each context of use need to be addressed, such as underlying disease, patient heterogeneity, therapeutic context, target of therapy, or a combination of disease parameters.
Regardless of the registrational intent for a trial, it is important that the trial is adequate to meet its stated objectives [21 CFR 312.42 (b) (2)(ii)]. If MRD assessment is a key component of the trial objectives, the MRD assessment should provide interpretable data. A review of applications submitted to the FDA's Division of Hematology Products between 2014 and 2016 that included MRD data highlighted several issues with MRD assessments in regulatory submissions. In more than half of the submissions, data were considered inadequate (31%), or were not proposed for inclusion (23%) because of missing or disparate data points, high amounts of test failure rate, incomplete test characteristics, and incomplete planned statistical analysis (114).
The recent FDA guidance “Hematologic Malignancies: Regulatory Considerations for Use of Minimal Residual Disease in Development of Drug and Biological Products for Treatment Guidance for Industry” was published to help sponsors planning to use MRD as a biomarker in clinical trials conducted under an investigational new drug application or to support marketing approval of drugs and biological products for treating specific hematologic malignancies (115). This guidance addresses several aspects including assay considerations, the strength of evidence and data that are required to support MRD as a surrogate endpoint, and considerations for using MRD as a patient selection factor.
Specifically, with respect to assay selection, FDA is agnostic as to which technology platform is used in clinical trials assessing MRD as long as the assay to be used is analytically validated for its context of use in the trial, that is, the assay demonstrates acceptable sensitivity, specificity, accuracy, precision, and other relevant performance characteristics as defined under a specified technical protocol. The assay validation considerations may differ based on the specific technology platform, for example, cellular versus molecular platform. If the assay is not approved or cleared for the intended use and the trial is considered a significant risk device trial, an investigational device exemption (IDE) may be required to use the assay in the clinical trial. For a trial considered a nonsignificant risk device study, the sponsor should submit abbreviated information about the assay, as stated in the MRD guidance (116). It is important to note that FDA cleared or approved assays have been analytically validated for the specific context of use. As described above, the adaptive clonoSEQ assay is an in vitro diagnostic that uses multiplex PCR and NGS to identify and quantify certain gene sequences in DNA extracted from BM of patients with acute lymphoblastic leukemia or multiple myeloma, and blood or BM from patients with chronic lymphocytic leukemia. The clonoSEQ assay's intended use is to measure MRD to monitor changes in burden of disease during and after treatment. The use of an approved assay outside of its intended use, for example, use of the approved clonoSEQ assay to monitor MRD in peripheral blood samples of patients with multiple myeloma, may dictate additional assay and informed consent considerations. It is important for Sponsors to interact with the regulatory agency when considering incorporating MRD assessments into a clinical trial (117).
In multiple myeloma clinical trials, MRD can be used in a variety of ways that may expedite drug development. MRD can be used for patient selection, stratification or enrichment of the trial population, to guide treatment decisions, or as an endpoint. There are specific considerations and potential risks associated with each use of MRD in the clinical trial setting. With patient selection or enrichment, MRD may be used to identify a patient population that is at high risk of relapse or poor outcome. In this setting, it would be important to identify the MRD threshold which best portends poor outcome and merits further intervention. When using MRD to guide treatment decisions, the threshold for intervention should be adequately supported, but there are additional considerations for these trial designs. Randomized intervention based on MRD status (e.g., randomization of MRD-positive patients to additional therapy vs. observation) permits a more robust analysis compared with single-arm intervention studies (e.g., all MRD-positive patients receive additional therapy). Again, in certain circumstances, the MRD assay may represent a significant risk device, which requires further discussion with the FDA and may require an IDE. As an endpoint, MRD can provide useful information regarding the activity of the investigational therapeutic. However, there are several statistical and clinical considerations when using MRD as a clinical trial endpoint. As such, the Agency recommends that sponsors discuss with the Agency the use of MRD when considering it as a key secondary or primary endpoint.
The FDA guidance on MRD states, “the strength of evidence to support surrogacy depends on (i) the biological plausibility of the relationship, (ii) demonstration in epidemiologic studies of the prognostic value of the surrogate endpoint for the clinical outcome, and (iii) evidence from clinical trials that the treatment effects on the surrogate endpoint correspond to effects on the clinical outcome” (118). The biological plausibility and prognostic value of MRD are well understood and have been supported by multiple studies. What remains to be elucidated is the evidence from clinical trials regarding the relationship between the treatment effect on MRD and the treatment effect on clinical outcomes of interest. There are current efforts to gather evidence from clinical trials to allow for further evaluation of the treatment effect relationship between MRD and clinical outcomes such as PFS and overall survival (OS) on both a trial level and at an individual patient level. These data will supplement our understanding of MRD and how to most effectively use MRD to expedite drug development.
With development of more sensitive assays, there may be opportunity for better disease quantification and discrimination of patients that are in CR and a potential for use of MRD as a drug development tool. However, data collection and assay performance characteristics should be of sufficient rigor and completeness to allow for comprehensive assessments. Including MRD assessments in prospective multiple myeloma clinical trials can help strengthen the available evidence for the use of this biomarker in multiple myeloma.
Industry perspectives on the regulatory importance of MRD as a surrogate endpoint
There have been many advancements in the development of novel and more effective therapies for the treatment of multiple myeloma in recent years, providing meaningful clinical benefit and prolonging patient survival. This has resulted in an increasingly challenging situation for bringing new drugs to patients with multiple myeloma in a timely manner. This challenge is especially true in the case of frontline trials, in which the time to reach the traditional endpoints of PFS or OS is becoming prohibitively long as patients are experiencing progressively longer PFS and OS. Despite these advances, there continues to be a significant unmet medical need in patients with multiple myeloma. Hence, novel therapies and regimens continue to be required to further enhance PFS and OS and to ultimately lead to a cure. Given the dramatic improvements in PFS with current therapies, clinical trials will require formidable sample sizes and unrealistic follow-up time to detect clinically meaningful treatment effects with sufficient statistical power. This in turn is making it increasingly difficult for companies to conduct clinical trials using traditional clinically relevant endpoints to achieve regulatory approval. Conversely, if an intervention is not likely to be beneficial, patients may stay on therapy for a long period before these studies reveal the lack of benefit. In short, using PFS or OS as a primary endpoint is becoming unsustainable. Moreover, because the multiple myeloma landscape is continuously evolving, the longer these clinical trials take to reach their endpoints, the more likely it is that the comparator arms become obsolete by the time the studies are analyzed. To maintain the speed and progress of new drug development in multiple myeloma, there is a need for a surrogate endpoint that (i) provides an earlier assessment of efficacy and (ii) correlates strongly with clinically relevant endpoints. Improved methods to evaluate response in support of early registration and access to therapies are needed. Evaluation of MRD has come to the forefront of these efforts in multiple myeloma, where MRD negativity status is associated with significantly improved survival outcomes.
While the need for a surrogate endpoint is most urgent in frontline trials, MRD endpoints will similarly aid patients and drug development efforts in the relapsed/refractory multiple myeloma (RRMM) setting.
A considerable amount of MRD data has been prospectively generated and is available from numerous multiple myeloma clinical trials in both the frontline and RRMM settings, with many more trials currently underway collecting MRD data. MRD negativity is the primary or co-primary endpoint for the early assessment of response in many currently ongoing clinical trials. As such, MRD assessment of response as a primary endpoint in multiple myeloma offers the potential to promptly deliver new therapies to patients with an unmet need.
It would be beneficial for regulatory agencies to use existing data from completed multiple myeloma trials to provide industry with guidance on specific timepoints that can be used in prospective multiple myeloma trials for evaluating MRD status. Such guidance will of course evolve, but if we wait for future clinical trials to provide definitive validation of MRD as a surrogate endpoint, valuable years will be lost as we wait for these studies to progress and these data to mature. Often, novel drugs are most likely to benefit patients with multiple myeloma in early lines more than in the late lines of treatment, where they are first evaluated, and the availability of MRD as a surrogate endpoint will expedite these clinical trials and their application. It is vitally important for the continuation and advancement of drug development in multiple myeloma that regulatory agencies and industry collaborate in the near term to establish a faster, accepted, surrogate endpoint for the future treatment of patients with multiple myeloma.
Considerations for Clinical Use of MRD in Different Patient Populations
The accumulated data over the past decade, especially in the recent years with introduction of highly effective therapeutic combinations, have clearly shown the value of MRD testing in multiple myeloma. As described earlier in the article, MRD testing in multiple myeloma has several important roles to play. At any stage in the disease evolution, achievement of MRD negativity will predict for a better outcome compared with patients at a similar stage who have not achieved the same depth of response with any given therapy. As stated earlier, clinicians are starting to incorporate MRD determination in routine clinical management decisions. In this section, we have used patient scenarios to outline the potential clinical utility of MRD, and remaining gaps in our knowledge to be addressed in ongoing and future clinical trials. The aim here is more to highlight the potential of MRD in various disease settings rather than making specific recommendations, something that is starting to being addressed through international harmonization efforts (119).
Smoldering multiple myeloma
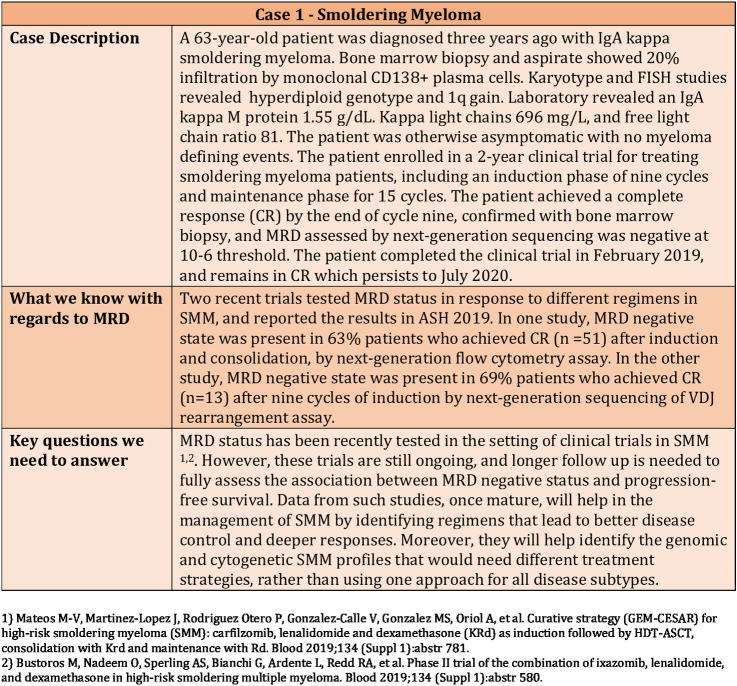
The standard of care and the general approach is not to treat patients with smoldering multiple myeloma (SMM). Translational and clinical research is ongoing to identify those patients needing intervention, and what regimens would be the best to prevent disease progression and end-organ damage, along with deeper responses and minimal toxicities. In SMM, MRD testing is currently only performed in a clinical trial setting (Fig. 1).
Figure 1.
Clinical Case 1—Use of MRD in a patient with smoldering myeloma.
Newly diagnosed multiple myeloma
The current standard of care for patients with newly diagnosed multiple myeloma who are eligible to undergo an ASCT is to have four to six cycles of induction therapy with a triplet containing a proteasome inhibitor and an immunomodulatory imide drug (PI-IMiD), followed by a single ASCT and lenalidomide maintenance following transplant. Recent trials have focused on development of quadruplets with addition of mAbs to the PI-IMiD triplets.
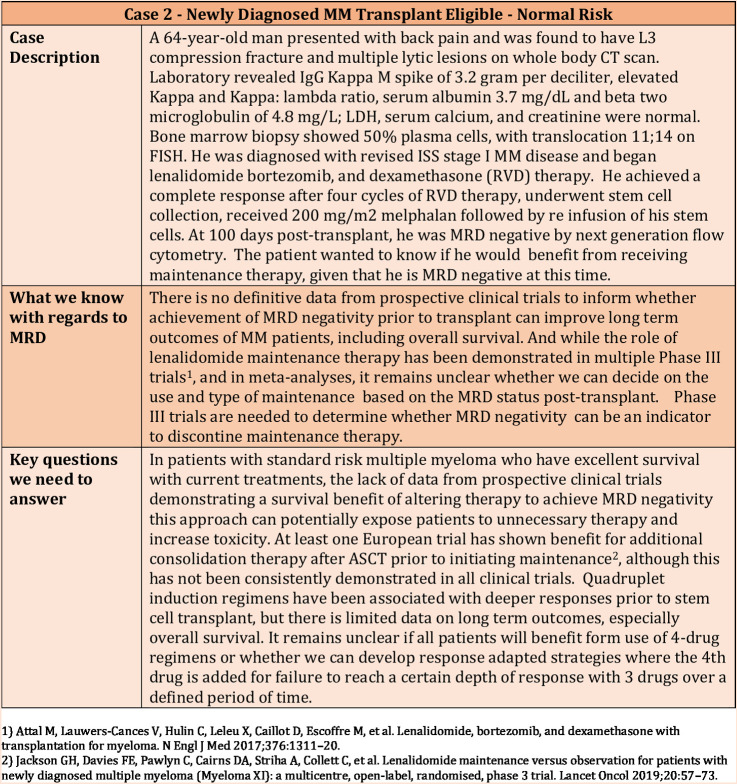
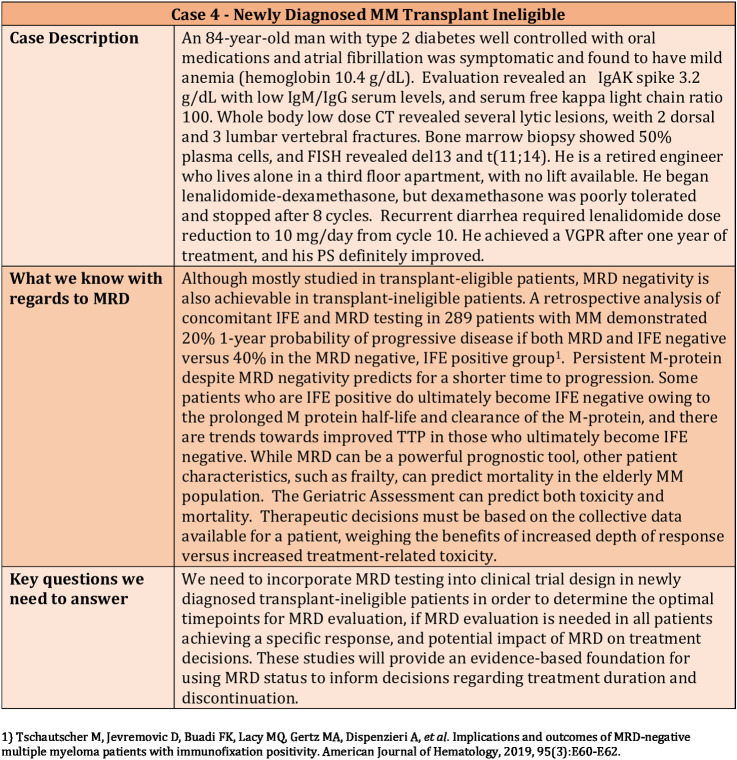
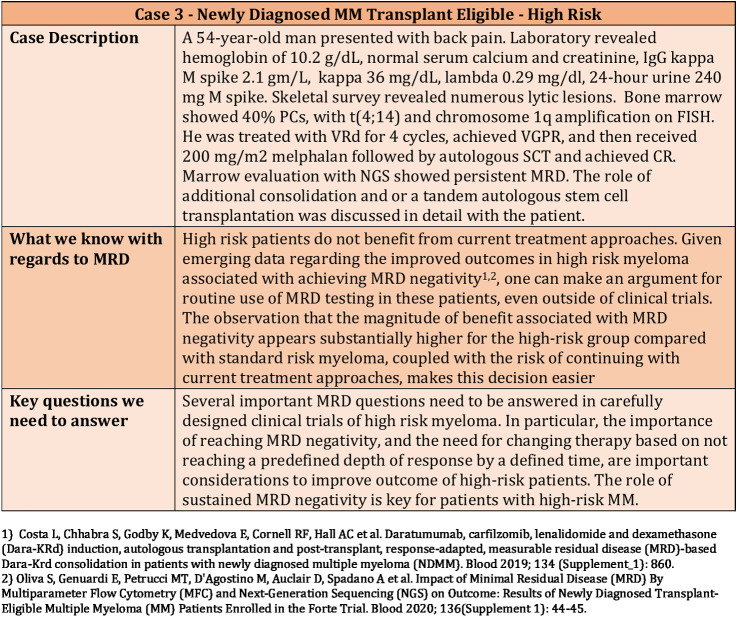
The role of stem cell transplantation and lenalidomide maintenance for multiple myeloma has been based on large randomized trials that were performed both in the context of older therapies as well as newer induction regimens. There is a possibility that MRD status at the end of induction therapy can help guide this decision-making, and similarly, that we can decide on the use of maintenance or the specific regimen to be used for maintenance based on the MRD status. However, at this time, data from prospective clinical trials are not available to make truly informed decisions based on MRD testing. Regardless, this is an issue that clinicians encounter routinely in daily practice as exemplified in Figs. 2–4.
Figure 2.
Clinical Case 2—Use of MRD in a newly diagnosed transplant-eligible normal-risk patient.
Figure 4.
Clinical Case 4—Use of MRD in a newly diagnosed transplant-ineligible patient.
Figure 3.
Clinical Case 3—Use of MRD in a newly diagnosed transplant-eligible high-risk patient.
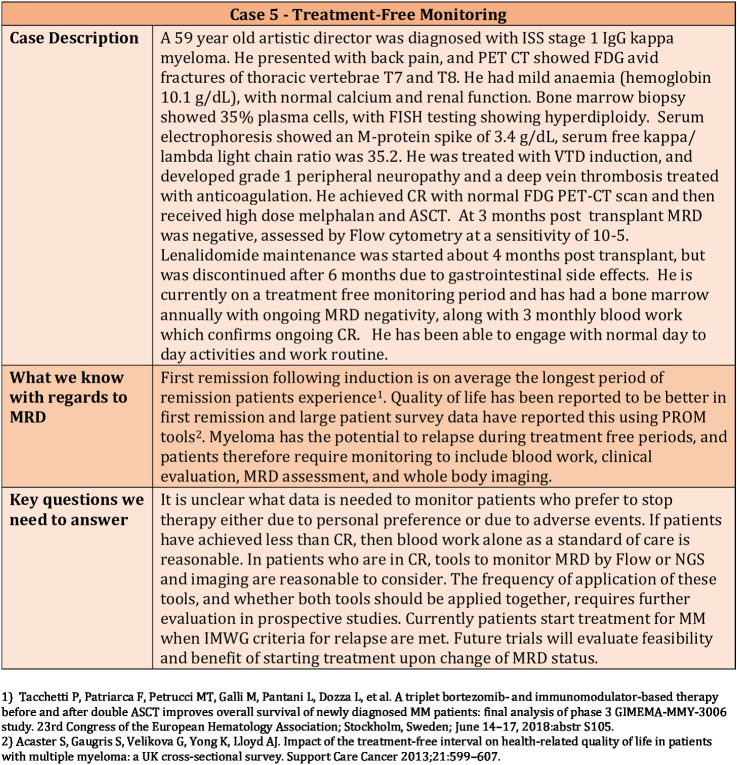
First remission following induction is on average the longest period of remission that patients will experience. Quality of life has been reported to be better in first remission, and large patient survey data have reported this using PROM tools. Lenalidomide is now an established standard of care as post-transplant maintenance, but 36% of patients stopped lenalidomide maintenance treatment in the Myeloma XI trial due to adverse events or personal preference. It is therefore important to have sensitive reliable tools like MRD to assess disease during treatment-free monitoring (Fig. 5).
Figure 5.
Clinical Case 5—Use of MRD for treatment-free monitoring.
Relapsed refractory myeloma
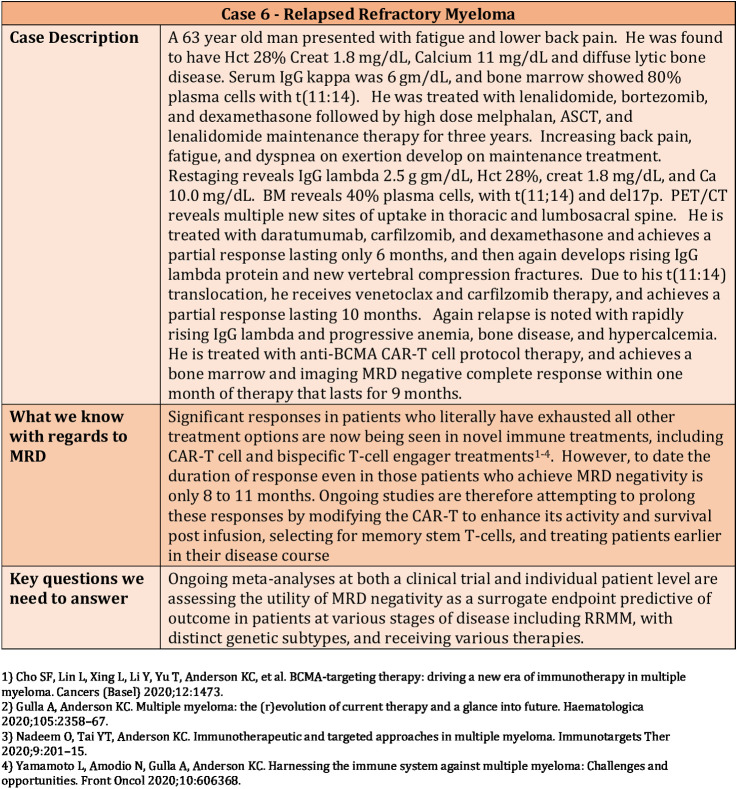
Novel immune therapies have a remarkable ability to achieve rapid MRD-negative CRs even in the context of multiply relapsed, genetically high-risk disease. The value of MRD negativity as a surrogate marker in RRMM may be context dependent and vary with stage of disease and treatment modality (Fig. 6).
Figure 6.
Clinical Case 6—Use of MRD in a patient with relapsed refractory myeloma.
Conclusion, Summary of Recommendations, and Call to Action
The development of novel agents has transformed the treatment paradigm for multiple myeloma, with MRD negative response now achievable both in newly diagnosed and relapsed disease. NGS and NGF are now most commonly utilized to measure BM MRD, along with PET/CT imaging to assess extramedullary disease. However, additional technologies including mass spectrometry-based paraprotein analyses, cfDNA, scRNA-seq, as well as whole body DWI MRI and novel tracers for imaging are rapidly evolving and may be used to measure MRD in the future. Section 3 of this article outlines regulatory considerations for both MRD assessment and incorporation into clinical trials. Although multiple retrospective individual patient and clinical trial level meta-analyses have already shown and will continue to assess MRD as a surrogate for patient outcome, designing current and future trials incorporating these regulatory considerations is necessary to assure continued rapid new drug development in multiple myeloma. The current value of MRD to inform clinical patient care is also illustrated using real-world cases of patients with SMM, newly diagnosed transplant eligible multiple myeloma, newly diagnosed transplant ineligible multiple myeloma, and RRMM, in each case summarizing what is known and key questions to be addressed in clinical trials. Most importantly, broad input from international academic clinical researchers and caregivers, FNIH, FDA, biotechnology and pharmaceutical industries formulated this white paper and call to action and represent the team necessary to implement these recommendations and assure progress utilizing MRD both as a regulatory endpoint and to guide clinical care. The pace of advances in targeted and immune therapies in multiple myeloma is unprecedented, and this collaborative effort, inspired by a shared commitment to patients, will assure that these advances translate to clinical trials leading to regulatory approval of novel treatments and continued improvement in patient outcomes.
Authors' Disclosures
K.C. Anderson reports personal fees from Pfizer, AstraZeneca, Janssen, Precision Biosciences, Amgen, Windmill, Mana, Starton, Raqia, Oncopep, and C4Therapeutics outside the submitted work. A. Agarwal reports other support from BMS outside the submitted work, and A. Agarwal is an employee of BMS. M. Anderson reports personal fees from Takeda Pharmaceuticals outside the submitted work. M. Bustoros reports personal fees from Takeda, Bristol Myers Squibb, Janssen, and Axiom Healthcare outside the submitted work. D.E. Connors reports stock ownership with Adaptive Biotechnologies. A. Dash reports other support from Takeda Pharmaceuticals outside the submitted work. A. Di Bacco reports other support from Takeda Pharmaceutical outside the submitted work. L. Du reports other support from GlaxoSmithKline outside the submitted work. T. Facon reports personal fees from BMS, Takeda, Janssen, Karyopharm, Roche, Amgen, Oncopeptides, and AbbVie outside the submitted work. J. Flores-Montero reports grants from International Myeloma Foundation during the conduct of the study; in addition, J. Flores-Montero has a patent for Methods reagents, and kit for detecting minimal residual disease issued, licensed, and with royalties paid from Cytognos SL. F. Gay reports personal fees from Amgen, Celgene, Janssen, Takeda, Bristol-Myers Squibb, AbbVie, GSK, Roche, Adaptive Biotechnologies, Oncopeptides, and Bluebird Bio outside the submitted work. I.M. Ghobrial reports grants and personal fees from BMS, Janssen, Takeda, and Sanofi and personal fees from GSK during the conduct of the study. I. Gupta reports other support from GlaxoSmithKline during the conduct of the study, as well as other support from GlaxoSmithKline outside the submitted work. H. Higley reports other support from CCS Associates outside the submitted work. J. Hillengass reports personal fees from Amgen, Adaptive, BMS, Celgene, GSK, Janssen, Oncopeptides, Oncotracker, Sanofi, Skyline, and Takeda outside the submitted work. D. Kazandjian reports personal fees from Bristol Myers Squibb outside the submitted work. I.R. Kirsch reports personal fees from Adaptive Biotechnologies during the conduct of the study, as well as personal fees from Adaptive Biotechnologies outside the submitted work; in addition, I.R. Kirsch is full-time employee of Adaptive Biotechnologies. B. Kremer reports other support from GlaxoSmithKline during the conduct of the study, as well as other support from GlaxoSmithKline outside the submitted work. O. Landgren reports grants and personal fees from Amgen, Celgene, and Janssen; other support from Takeda, Janssen, and Merck; personal fees from Glenmark, Seattle Genetics, Karyopharm, Adaptive Biotech, Bristol Myers Squibb, Cellectis, and Oncopeptides; and grants from Multiple Myeloma Research Foundation, Perelman Family Foundation, NCI, and FDA outside the submitted work. S. Lonial reports personal fees from Takeda, Celgene, BMS, Amgen, Janssen, Genentech, and GSK during the conduct of the study, as well as other support from TG Therapeutics outside the submitted work. M.-V. Mateos reports personal fees from Janssen, BMS-Celgene, Amgen, Takeda, GSK, AbbVie, Oncopeptides, Sanofi, Pfizer, Regeneron, Roche, Sea-Gen, Bluebird bio, and Adaptive outside the submitted work. R. Montes de Oca reports other support from GlaxoSmithKline outside the submitted work. N.C. Munshi reports personal fees from Takeda, BMS, Oncopep, Janssen, Amgen, AbbVie, Adaptive Biotechnology, Karyopharm, and Legend Biotech during the conduct of the study; in addition, N.C. Munshi has a patent for Oncopep licensed. A. Orfao reports other support from Cytognos SL, Becton/Dickinson Biosciences, and Blu Print and personal fees and other support from Amgen, Kite-Gilead, Alexion, Janssen, and Novartis outside the submitted work; in addition, A. Orfao has a patent for PCT WO 2013/187765A2 (Methods, reagents and kits for detecting minimal residual disease) issued and licensed to Cytognos SL. B. Paiva reports other support from Amgen, Adaptive, Becton Dickinson, Creative Biolabs, Janssen, Takeda, and Sanofi; grants from Roche; and grants and other support from GSK and BMS-Celgene during the conduct of the study. T.J. Pugh reports a patent for Hybrid-capture sequencing for determining immune cell clonality pending to University Health Network. K. Ramasamy reports grants and personal fees from Adaptive Biotech outside the submitted work and receives other support from Janssen, BMS (Celgene), Amgen, Takeda, and GSK (grants, advisory board and speaker honoraria) and other support from AbbVie, Karyopharm, Oncopeptides, and Sanofi (advisory board and speaker honoraria). J. Ray reports personal fees from Roche outside the submitted work. M. Roshal reports other support from Auron and grants from NGM, Genentech, Agios, Bayer, and Cellularity outside the submitted work. J.A. Ross reports employment at AbbVie and holds stock or other options. K.L. Thoren reports grants and personal fees from Sebia and non-financial support from The Binding Site outside the submitted work. N. Valente is an employee of Genentech. B.M. Weiss reports other support from Janssen Research & Development outside the submitted work. E. Zamagni reports personal fees from BMS, Sanofi, Janssen, Amgen, Oncopeptide, and GSK outside the submitted work. S.K. Kumar reports grants and other support from AbbVie, Takeda, BMS, Amgen, Janssen, AstraZeneca, Roche-Genentech, and Novartis; grants from Carsgen and TeneBio; and other support from Oncopeptides during the conduct of the study. No disclosures were reported by the other authors.
References
- 1. Kumar S, Paiva B, Anderson KC, Durie B, Landgren O, Moreau P, et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol 2016;17:e328–e46. [DOI] [PubMed] [Google Scholar]
- 2. Anderson KC, Auclair D, Kelloff GJ, Sigman CC, Avet-Loiseau H, Farrell AT, et al. The role of minimal residual disease testing in myeloma treatment selection and drug development: current value and future applications. Clin Cancer Res 2017;23:3980–93. [DOI] [PubMed] [Google Scholar]
- 3. Munshi NC, Avet-Loiseau H, Anderson KC, Neri P, Paiva B, Samur M, et al. A large meta-analysis establishes the role of MRD negativity in long-term survival outcomes in patients with multiple myeloma. Blood Adv 2020;4:5988–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martinez-Lopez J, Wong SW, Shah N, Bahri N, Zhou K, Sheng Y, et al. Clinical value of measurable residual disease testing for assessing depth, duration, and direction of response in multiple myeloma. Blood Adv 2020;4:3295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Flores-Montero J, Sanoja L, Pérez JJ, Pojero F, Puig N, Vidriales MB, et al. Plasma cell disorders. In: Detrick B, Schmitz JL, Hamilton RG, editors. Manual of molecular and clinical laboratory immunology. Washington, DC: ASM Press; 2016. p. 235–50. [Google Scholar]
- 6. Rawstron AC, Orfao A, Beksac M, Bezdickova L, Brooimans RA, Bumbea H, et al. Report of the European Myeloma Network on multiparametric flow cytometry in multiple myeloma and related disorders. Haematologica 2008;93:431–8. [DOI] [PubMed] [Google Scholar]
- 7. Paiva B, Almeida J, Pérez-Andrés M, Mateo G, López A, Rasillo A, et al. Utility of flow cytometry immunophenotyping in multiple myeloma and other clonal plasma cell-related disorders. Cytometry B Clin Cytom 2010;78:239–52. [DOI] [PubMed] [Google Scholar]
- 8. Rawstron AC, Gregory WM, de Tute RM, Davies FE, Bell SE, Drayson MT, et al. Minimal residual disease in myeloma by flow cytometry: independent prediction of survival benefit per log reduction. Blood 2015;125:1932–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rawstron AC, Child JA, de Tute RM, Davies FE, Gregory WM, Bell SE, et al. Minimal residual disease assessed by multiparameter flow cytometry in multiple myeloma: impact on outcome in the Medical Research Council Myeloma IX Study. J Clin Oncol 2013;31:2540–7. [DOI] [PubMed] [Google Scholar]
- 10. Rawstron AC, Davies FE, DasGupta R, Ashcroft AJ, Patmore R, Drayson MT, et al. Flow cytometric disease monitoring in multiple myeloma: the relationship between normal and neoplastic plasma cells predicts outcome after transplantation. Blood 2002;100:3095–100. [DOI] [PubMed] [Google Scholar]
- 11. Roussel M, Lauwers-Cances V, Robillard N, Hulin C, Leleu X, Benboubker L, et al. Front-line transplantation program with lenalidomide, bortezomib, and dexamethasone combination as induction and consolidation followed by lenalidomide maintenance in patients with multiple myeloma: a phase II study by the Intergroupe Francophone du Myélome. J Clin Oncol 2014;32:2712–7. [DOI] [PubMed] [Google Scholar]
- 12. Paiva B, Gutiérrez NC, Rosiñol L, Vídriales MB, Montalbán M, Martínez-López J, et al. High-risk cytogenetics and persistent minimal residual disease by multiparameter flow cytometry predict unsustained complete response after autologous stem cell transplantation in multiple myeloma. Blood 2012;119:687–91. [DOI] [PubMed] [Google Scholar]
- 13. Paiva B, Vidriales MB, Cerveró J, Mateo G, Pérez JJ, Montalbán MA, et al. Multiparameter flow cytometric remission is the most relevant prognostic factor for multiple myeloma patients who undergo autologous stem cell transplantation. Blood 2008;112:4017–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. San Miguel JF, Almeida J, Mateo G, Bladé J, López-Berges C, Caballero D, et al. Immunophenotypic evaluation of the plasma cell compartment in multiple myeloma: a tool for comparing the efficacy of different treatment strategies and predicting outcome. Blood 2002;99:1853–6. [DOI] [PubMed] [Google Scholar]
- 15. Mateos MV, Oriol A, Martínez-López J, Teruel AI, López de la Guía A, López J, et al. GEM2005 trial update comparing VMP/VTP as induction in elderly multiple myeloma patients: do we still need alkylators? Blood 2014;124:1887–93. [DOI] [PubMed] [Google Scholar]
- 16. Paiva B, Martinez-Lopez J, Vidriales MB, Mateos MV, Montalban MA, Fernandez-Redondo E, et al. Comparison of immunofixation, serum free light chain, and immunophenotyping for response evaluation and prognostication in multiple myeloma. J Clin Oncol 2011;29:1627–33. [DOI] [PubMed] [Google Scholar]
- 17. Paiva B, Cedena MT, Puig N, Arana P, Vidriales MB, Cordon L, et al. Minimal residual disease monitoring and immune profiling in multiple myeloma in elderly patients. Blood 2016;127:3165–74. [DOI] [PubMed] [Google Scholar]
- 18. Korde N, Roschewski M, Zingone A, Kwok M, Manasanch EE, Bhutani M, et al. Treatment with carfilzomib-lenalidomide-dexamethasone with lenalidomide extension in patients with smoldering or newly diagnosed multiple myeloma. JAMA Oncol 2015;1:746–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paiva B, Merino J, San Miguel JF. Utility of flow cytometry studies in the management of patients with multiple myeloma. Curr Opin Oncol 2016;28:511–17. [DOI] [PubMed] [Google Scholar]
- 20. Munshi NC, Avet-Loiseau H, Rawstron AC, Owen RG, Child JA, Thakurta A, et al. Association of minimal residual disease with superior survival outcomes in patients with multiple myeloma: a meta-analysis. JAMA Oncol 2017;3:28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Landgren O, Devlin S, Boulad M, Mailankody S. Role of MRD status in relation to clinical outcomes in newly diagnosed multiple myeloma patients: a meta-analysis. Bone Marrow Transplant 2016;51:1565–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Paiva B, Puig N, García-Sanz R, San Miguel JF. Is this the time to introduce minimal residual disease in multiple myeloma clinical practice? Clin Cancer Res 2015;21:2001–8. [DOI] [PubMed] [Google Scholar]
- 23. Paiva B, van Dongen JJ, Orfao A. New criteria for response assessment: role of minimal residual disease in multiple myeloma. Blood 2015;125:3059–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paiva B, Puig N, Cedena MT, Rosiñol L, Cordón L, Vidriales MB, et al. Measurable residual disease by next-generation flow cytometry in multiple myeloma. J Clin Oncol 2020;38:784–92. [DOI] [PubMed] [Google Scholar]
- 25. Berger N, Kim-Schulze S, Parekh S. Minimal residual disease in multiple myeloma: impact on response assessment, prognosis and tumor heterogeneity. Adv Exp Med Biol 2018;1100:141–59. [DOI] [PubMed] [Google Scholar]
- 26. Flores-Montero J, Sanoja-Flores L, Paiva B, Puig N, García-Sánchez O, Böttcher S, et al. Next generation flow for highly sensitive and standardized detection of minimal residual disease in multiple myeloma. Leukemia 2017;31:2094–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Van Dongen JJM, Orfao de Matos Correia e Vale JA, Flores Montero JA, Almeida Parra JM, Van der Velden VHJ, Boettcher S, et al. Methods, reagents and kits for detecting minimal residual disease. Patent number: WO2013187765. Publication date: 2013-12-19. Application number: WO2013-NL50420. Application date: 2013-06-14. Kind code: A2. Assignee: Erasmus University Medical Center Rotterdam, Netherlands.
- 28. Sanoja-Flores L, Flores-Montero J, Puig N, Contreras-Sanfeliciano T, Pontes R, Corral-Mateos A, et al. Blood monitoring of circulating tumor plasma cells by next generation flow in multiple myeloma after therapy. Blood 2019;134:2218–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moreau P, Attal M, Hulin C, Arnulf B, Belhadj K, Benboubker L, et al. Bortezomib, thalidomide, and dexamethasone with or without daratumumab before and after autologous stem-cell transplantation for newly diagnosed multiple myeloma (CASSIOPEIA): a randomised, open-label, phase 3 study. Lancet 2019;394:29–38. [DOI] [PubMed] [Google Scholar]
- 30. Oberle A, Brandt A, Alawi M, Langebrake C, Janjetovic S, Wolschke C, et al. Long-term CD38 saturation by daratumumab interferes with diagnostic myeloma cell detection. Haematologica 2017;102:e368–e70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Puig N, Mateos M-V, Contreras T, Paiva B, Cedena MT, Pérez JJ, et al. Qip-mass spectrometry in high risk smoldering multiple myeloma patients included in the GEM-CESAR trial: comparison with conventional and minimal residual disease IMWG response assessment. Blood 2019;134:581. [Google Scholar]
- 32. Moreau P, Zamagni E. MRD in multiple myeloma: more questions than answers? Blood Cancer J 2017;7:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Korsmeyer SJ, Arnold A, Bakhshi A, Ravetch JV, Siebenlist U, Hieter PA, et al. Immunoglobulin gene rearrangement and cell surface antigen expression in acute lymphocytic leukemias of T cell and B cell precursor origins. J Clin Invest 1983;71:301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van Dongen JJ, Langerak AW, Brüggemann M, Evans PA, Hummel M, Lavender FL, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98–3936. Leukemia 2003;17:2257–317. [DOI] [PubMed] [Google Scholar]
- 35. Carlson CS, Emerson RO, Sherwood AM, Desmarais C, Chung MW, Parsons JM, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun 2013;4:2680. [DOI] [PubMed] [Google Scholar]
- 36. Ching T, Duncan ME, Newman-Eerkes T, McWhorter MME, Tracy JM, Steen MS, et al. Analytical evaluation of the clonoSEQ Assay for establishing measurable (minimal) residual disease in acute lymphoblastic leukemia, chronic lymphocytic leukemia, and multiple myeloma. BMC Cancer 2020;20:612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perrot A, Lauwers-Cances V, Corre J, Robillard N, Hulin C, Chretien ML, et al. Minimal residual disease negativity using deep sequencing is a major prognostic factor in multiple myeloma. Blood 2018;132:2456–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mateos MV, Cavo M, Blade J, Dimopoulos MA, Suzuki K, Jakubowiak A, et al. Overall survival with daratumumab, bortezomib, melphalan, and prednisone in newly diagnosed multiple myeloma (ALCYONE): a randomised, open-label, phase 3 trial. Lancet 2020;395:132–41. [DOI] [PubMed] [Google Scholar]
- 39. Kirsch I, Vignali M, Robins H. T-cell receptor profiling in cancer. Mol Oncol 2015;9:2063–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Emerson RO, DeWitt WS, Vignali M, Gravley J, Hu JK, Osborne EJ, et al. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet 2017;49:659–65. [DOI] [PubMed] [Google Scholar]
- 41. Gittelman RM, Lavezzo E, Snyder TM, Zahid HJ, Elyanow R, Dalai S, et al. Diagnosis and tracking of past SARS-CoV-2 infection in a large study of Vo', Italy through T-cell receptor sequencing. Preprint. medRxiv 2020.11.09.20228023; doi: 10.1101/2020.11.09.20228023 [DOI]
- 42. Mazzotti C, Buisson L, Maheo S, Perrot A, Chretien ML, Leleu X, et al. Myeloma MRD by deep sequencing from circulating tumor DNA does not correlate with results obtained in the bone marrow. Blood Adv 2018;2:2811–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cescon DW, Bratman SV, Chan SM, Siu LL. Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer 2020;1:276–90. [DOI] [PubMed] [Google Scholar]
- 44. Thoren KL. Mass spectrometry methods for detecting monoclonal immunoglobulins in multiple myeloma minimal residual disease. Semin Hematol 2018;55:41–43. [DOI] [PubMed] [Google Scholar]
- 45. Chapman JR, Thoren KL. Tracking of low disease burden in multiple myeloma: using mass spectrometry assays in peripheral blood. Best Pract Res Clin Haematol 2020;33:101142. [DOI] [PubMed] [Google Scholar]
- 46. Mills JR, Kohlhagen MC, Dasari S, Vanderboom PM, Kyle RA, Katzmann JA, et al. Comprehensive assessment of M-proteins using nanobody enrichment coupled to MALDI-TOF mass spectrometry. Clin Chem 2016;62:1334–44. [DOI] [PubMed] [Google Scholar]
- 47. Moore LM, Cho S, Thoren KL. MALDI-TOF mass spectrometry distinguishes daratumumab from M-proteins. Clin Chim Acta 2019;492:91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Milani P, Murray DL, Barnidge DR, Kohlhagen MC, Mills JR, Merlini G, et al. The utility of MASS-FIX to detect and monitor monoclonal proteins in the clinic. Am J Hematol 2017;92:772–79. [DOI] [PubMed] [Google Scholar]
- 49. Barnidge DR, Dasari S, Botz CM, Murray DH, Snyder MR, Katzmann JA, et al. Using mass spectrometry to monitor monoclonal immunoglobulins in patients with a monoclonal gammopathy. J Proteome Res 2014;13:1419–27. [DOI] [PubMed] [Google Scholar]
- 50. Bergen HR 3rd, Dasari S, Dispenzieri A, Mills JR, Ramirez-Alvarado M, Tschumper RC, et al. Clonotypic light chain peptides identified for monitoring minimal residual disease in multiple myeloma without bone marrow aspiration. Clin Chem 2016;62:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barnidge DR, Tschumper RC, Theis JD, Snyder MR, Jelinek DF, Katzmann JA, et al. Monitoring M-proteins in patients with multiple myeloma using heavy-chain variable region clonotypic peptides and LC-MS/MS. J Proteome Res 2014;13:1905–10. [DOI] [PubMed] [Google Scholar]
- 52. Zajec M, Jacobs JFM, Groenen P, de K Angelino CM, Stingl C., et al. Development of a targeted mass-spectrometry serum assay to quantify M-protein in the presence of therapeutic monoclonal antibodies. J Proteome Res 2018;17:1326–33. [DOI] [PubMed] [Google Scholar]
- 53. Martins CO, Huet S, Yi SS, Ritorto MS, Landgren O, Dogan A, et al. Mass spectrometry-based method targeting Ig variable regions for assessment of minimal residual disease in multiple myeloma. J Mol Diagn 2020;22:901–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eveillard M, Rustad E, Roshal M, Zhang Y, Ciardiello A, Korde N, et al. Comparison of MALDI-TOF mass spectrometry analysis of peripheral blood and bone marrow-based flow cytometry for tracking measurable residual disease in patients with multiple myeloma. Br J Haematol 2020;189:904–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mills JR, Barnidge DR, Dispenzieri A, Murray DL.. High sensitivity blood-based M-protein detection in sCR patients with multiple myeloma. Blood Cancer J 2017;7:e590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Derman BA, Stefka AT, Jiang K, McIver A, Kubicki T, Jasielec JK, et al. Measurable residual disease assessed by mass spectrometry in peripheral blood in multiple myeloma in a phase II trial of carfilzomib, lenalidomide, dexamethasone and autologous stem cell transplantation. Blood Cancer J 2021;11:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Burgener JM, Rostami A, De Carvalho DD, Bratman SV. Cell-free DNA as a post-treatment surveillance strategy: current status. Semin Oncol 2017;44:330–46. [DOI] [PubMed] [Google Scholar]
- 58. Scherer F, Kurtz DM, Diehn M, Alizadeh AA. High-throughput sequencing for noninvasive disease detection in hematologic malignancies. Blood 2017;130:440–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Heitzer E, Haque IS, Roberts CES, Speicher MR. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet 2019;20:71–88. [DOI] [PubMed] [Google Scholar]
- 60. Pantel K, Alix-Panabières C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol 2019;16:409–24. [DOI] [PubMed] [Google Scholar]
- 61. Kis O, Kaedbey R, Chow S, Danesh A, Dowar M, Li T, et al. Circulating tumour DNA sequence analysis as an alternative to multiple myeloma bone marrow aspirates. Nat Commun 2017;8:15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Manier S, Park J, Capelletti M, Bustoros M, Freeman SS, Ha G, et al. Whole-exome sequencing of cell-free DNA and circulating tumor cells in multiple myeloma. Nat Commun 2018;9:1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vij R, Mazumder A, Klinger M, O'Dea D, Paasch J, Martin T, et al. Deep sequencing reveals myeloma cells in peripheral blood in majority of multiple myeloma patients. Clin Lymphoma Myeloma Leuk 2014;14:131–39.e1. [DOI] [PubMed] [Google Scholar]
- 64. Guo G, Raje NS, Seifer C, Kloeber J, Isenhart R, Ha G, et al. Genomic discovery and clonal tracking in multiple myeloma by cell-free DNA sequencing. Leukemia 2018;32:1838–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC - challenges to implementing ctDNA-based screening and MRD detection. Nat Rev Clin Oncol 2018;15:577–86. [DOI] [PubMed] [Google Scholar]
- 66. Parsons HA, Rhoades J, Reed SC, Gydush G, Ram P, Exman P, et al. Sensitive detection of minimal residual disease in patients treated for early-stage breast cancer. Clin Cancer Res 2020;26:2556–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Oberle A, Brandt A, Voigtlaender M, Thiele B, Radloff J, Schulenkorf A, et al. Monitoring multiple myeloma by next-generation sequencing of V(D)J rearrangements from circulating myeloma cells and cell-free myeloma DNA. Haematologica 2017;102:1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Biancon G, Gimondi S, Vendramin A, Carniti C, Corradini P. Noninvasive molecular monitoring in multiple myeloma patients using cell-free tumor DNA: a pilot study. J Mol Diagn 2018;20:859–70. [DOI] [PubMed] [Google Scholar]
- 69. Leal A, van Grieken NCT, Palsgrove DN, Phallen J, Medina JE, Hruban C, et al. White blood cell and cell-free DNA analyses for detection of residual disease in gastric cancer. Nat Commun 2020;11:525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. McDonald BR, Contente-Cuomo T, Sammut SJ, Odenheimer-Bergman A, Ernst B, Perdigones N, et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med 2019;11:eaax7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang TT, Abelson S, Zou J, Li T, Zhao Z, Dick JE, et al. High efficiency error suppression for accurate detection of low-frequency variants. Nucleic Acids Res 2019;47:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pugh TJ. Circulating tumour DNA for detecting minimal residual disease in multiple myeloma. Semin Hematol 2018;55:38–40. [DOI] [PubMed] [Google Scholar]
- 73. Martello M, Poletti A, Borsi E, Solli V, Armuzzi S, Vigliotta I, et al. Usefulness of circulating cell-free DNA to define and tracking how multiple myeloma spread and disseminate outside the bone marrow Blood 2020;136:3178. [Google Scholar]
- 74. Paíno T, Paiva B, Sayagués JM, Mota I, Carvalheiro T, Corchete LA, et al. Phenotypic identification of subclones in multiple myeloma with different chemoresistant, cytogenetic and clonogenic potential. Leukemia 2015;29:1186–94. [DOI] [PubMed] [Google Scholar]
- 75. McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017;168:613–28. [DOI] [PubMed] [Google Scholar]
- 76. Turajlic S, Sottoriva A, Graham T, Swanton C. Resolving genetic heterogeneity in cancer. Nat Rev Genet 2019;20:404–16. [DOI] [PubMed] [Google Scholar]
- 77. De Smedt E, Lui H, Maes K, De Veirman K, Menu E, Vanderkerken K, et al. The epigenome in multiple myeloma: impact on tumor cell plasticity and drug response. Front Oncol 2018;8:566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lomas OC, Tahri S, Ghobrial IM. The microenvironment in myeloma. Curr Opin Oncol 2020;32:170–75. [DOI] [PubMed] [Google Scholar]
- 79. Ryu D, Kim SJ, Hong Y, Jo A, Kim N, Kim HJ, et al. Alterations in the transcriptional programs of myeloma cells and the microenvironment during extramedullary progression affect proliferation and immune evasion. Clin Cancer Res 2020;26:935–44. [DOI] [PubMed] [Google Scholar]
- 80. Jang JS, Li Y, Mitra AK, Bi L, Abyzov A, van Wijnen AJ, et al. Molecular signatures of multiple myeloma progression through single cell RNA-Seq. Blood Cancer J 2019;9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ledergor G, Weiner A, Zada M, Wang SY, Cohen YC, Gatt ME, et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat Med 2018;24:1867–76. [DOI] [PubMed] [Google Scholar]
- 82. Zavidij O, Haradhvala NJ, Mouhieddine TH, Sklavenitis-Pistofidis R, Cai S, Reidy M, et al. Single-cell RNA sequencing reveals compromised immune microenvironment in precursor stages of multiple myeloma. Nat Cancer 2020;1:493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kumar SK, Harrison SJ, Cavo M, de la Rubia J, Popat R, Gasparetto C, et al. Venetoclax or placebo in combination with bortezomib and dexamethasone in patients with relapsed or refractory multiple myeloma (BELLINI): a randomised, double-blind, multicentre, phase 3 trial. Lancet Oncol 2020;21:1630–42. [DOI] [PubMed] [Google Scholar]
- 84. Mitra AK, Harding T, Mukherjee UK, Jang JS, Li Y, HongZheng R, et al. A gene expression signature distinguishes innate response and resistance to proteasome inhibitors in multiple myeloma. Blood Cancer J 2017;7:e581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Shalek AK, Benson M. Single-cell analyses to tailor treatments. Sci Transl Med 2017;9:eaan4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kim JK, Kolodziejczyk AA, Ilicic T, Teichmann SA, Marioni JC. Characterizing noise structure in single-cell RNA-seq distinguishes genuine from technical stochastic allelic expression. Nat Commun 2015;6:8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zamagni E, Tacchetti P, Cavo M. Imaging in multiple myeloma: how? when? Blood 2019;133:644–51. [DOI] [PubMed] [Google Scholar]
- 88. Lu YY, Chen JH, Lin WY, Liang JA, Wang HY, Tsai SC, et al. FDG PET or PET/CT for detecting intramedullary and extramedullary lesions in multiple myeloma: a systematic review and meta-analysis. Clin Nucl Med 2012;37:833–7. [DOI] [PubMed] [Google Scholar]
- 89. Rasche L, Chavan SS, Stephens OW, Patel PH, Tytarenko R, Ashby C, et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat Commun 2017;8:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Rasche L, Angtuaco EJ, Alpe TL, Gershner GH, McDonald JE, Samant RS, et al. The presence of large focal lesions is a strong independent prognostic factor in multiple myeloma. Blood 2018;132:59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Paiva B, Puig N, Cedena MT, Cordon L, Vidriales M-B, Burgos L, et al. Impact of next-generation flow (NGF) minimal residual disease (MRD) monitoring in multiple myeloma (MM): results from the PETHEMA/GEM2012 trial. Blood 2017;130:905. [Google Scholar]
- 92. Cavo M, Terpos E, Nanni C, Moreau P, Lentzsch S, Zweegman S, et al. Role of (18)F-FDG PET/CT in the diagnosis and management of multiple myeloma and other plasma cell disorders: a consensus statement by the International Myeloma Working Group. Lancet Oncol 2017;18:e206–e17. [DOI] [PubMed] [Google Scholar]
- 93. Bartel TB, Haessler J, Brown TL, Shaughnessy JD Jr, van Rhee F, Anaissie E, et al. F18-fluorodeoxyglucose positron emission tomography in the context of other imaging techniques and prognostic factors in multiple myeloma. Blood 2009;114:2068–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zamagni E, Patriarca F, Nanni C, Zannetti B, Englaro E, Pezzi A, et al. Prognostic relevance of 18-F FDG PET/CT in newly diagnosed multiple myeloma patients treated with up-front autologous transplantation. Blood 2011;118:5989–95. [DOI] [PubMed] [Google Scholar]
- 95. Usmani SZ, Mitchell A, Waheed S, Crowley J, Hoering A, Petty N, et al. Prognostic implications of serial 18-fluoro-deoxyglucose emission tomography in multiple myeloma treated with total therapy 3. Blood 2013;121:1819–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Moreau P, Attal M, Caillot D, Macro M, Karlin L, Garderet L, et al. Prospective evaluation of magnetic resonance imaging and [(18)F]fluorodeoxyglucose positron emission tomography-computed tomography at diagnosis and before maintenance therapy in symptomatic patients with multiple myeloma included in the IFM/DFCI 2009 trial: results of the IMAJEM study. J Clin Oncol 2017;35:2911–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zamagni E, Nanni C, Mancuso K, Tacchetti P, Pezzi A, Pantani L, et al. PET/CT improves the definition of complete response and allows to detect otherwise unidentifiable skeletal progression in multiple myeloma. Clin Cancer Res 2015;21:4384–90. [DOI] [PubMed] [Google Scholar]
- 98. Rasche L, Alapat D, Kumar M, Gershner G, McDonald J, Wardell CP, et al. Combination of flow cytometry and functional imaging for monitoring of residual disease in myeloma. Leukemia 2019;33:1713–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Davies FE, Rosenthal A, Rasche L, Petty NM, McDonald JE, Ntambi JA, et al. Treatment to suppression of focal lesions on positron emission tomography-computed tomography is a therapeutic goal in newly diagnosed multiple myeloma. Haematologica 2018;103:1047–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hillengass J, Usmani S, Rajkumar SV, Durie BGM, Mateos MV, Lonial S, et al. International myeloma working group consensus recommendations on imaging in monoclonal plasma cell disorders. Lancet Oncol 2019;20:e302–e12. [DOI] [PubMed] [Google Scholar]
- 101. Rasche L, Angtuaco E, McDonald JE, Buros A, Stein C, Pawlyn C, et al. Low expression of hexokinase-2 is associated with false-negative FDG-positron emission tomography in multiple myeloma. Blood 2017;130:30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Pandit-Taskar N. Functional imaging methods for assessment of minimal residual disease in multiple myeloma: Current status and novel immunoPET based methods. Semin Hematol 2018;55:22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Herrmann K, Schottelius M, Lapa C, Osl T, Poschenrieder A, Hänscheid H, et al. First-in-human experience of CXCR4-directed endoradiotherapy with 177Lu- and 90Y-labeled pentixather in advanced-stage multiple myeloma with extensive intra- and extramedullary disease. J Nucl Med 2016;57:248–51. [DOI] [PubMed] [Google Scholar]
- 104. Lapa C, Herrmann K, Schirbel A, Hänscheid H, Lückerath K, Schottelius M, et al. CXCR4-directed endoradiotherapy induces high response rates in extramedullary relapsed multiple myeloma. Theranostics 2017;7:1589–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Caserta E, Chea J, Minnix M, Poku EK, Viola D, Vonderfecht S, et al. Copper 64-labeled daratumumab as a PET/CT imaging tracer for multiple myeloma. Blood 2018;131:741–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ghai A, Maji D, Cho N, Chanswangphuwana C, Rettig M, Shen D, et al. Preclinical development of CD38-targeted [(89)Zr]Zr-DFO-daratumumab for imaging multiple myeloma. J Nucl Med 2018;59:216–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Ulaner GA, Sobol NB, O'Donoghue JA, Kirov AS, Riedl CC, Min R, et al. CD38-targeted immuno-PET of multiple myeloma: From xenograft models to first-in-human imaging. Radiology 2020;295:606–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Zamagni E, Nanni C, Dozza L, Carlier T, Bailly C, Tacchetti P, et al. Standardization of (18)F-FDG-PET/CT according to Deauville criteria for metabolic complete response definition in newly diagnosed multiple myeloma. J Clin Oncol 2021;39:116–25. [DOI] [PubMed] [Google Scholar]
- 109. Walker R, Barlogie B, Haessler J, Tricot G, Anaissie E, Shaughnessy JD Jr, et al. Magnetic resonance imaging in multiple myeloma: diagnostic and clinical implications. J Clin Oncol 2007;25:1121–8. [DOI] [PubMed] [Google Scholar]
- 110. Hillengass J, Ayyaz S, Kilk K, Weber MA, Hielscher T, Shah R, et al. Changes in magnetic resonance imaging before and after autologous stem cell transplantation correlate with response and survival in multiple myeloma. Haematologica 2012;97:1757–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Merz M, Hielscher T, Mai EK, Seckinger A, Hose D, Jauch A, et al. Cystic transformation of focal lesions after therapy is associated with remission but adverse outcome in myeloma. Blood Cancer J 2019;9:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Le Bihan D, Breton E, Lallemand D, Aubin ML, Vignaud J, Laval-Jeantet M. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology 1988;168:497–505. [DOI] [PubMed] [Google Scholar]
- 113. Pawlyn C, Fowkes L, Otero S, Jones JR, Boyd KD, Davies FE, et al. Whole-body diffusion-weighted MRI: a new gold standard for assessing disease burden in patients with multiple myeloma? Leukemia 2016;30:1446–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Gormley N, Bhatnagar V, Ehrlich LA, Kanapuru B, Lee H-Z, McKee AE, et al. FDA analysis of MRD data in hematologic malignancy applications. J Clin Oncol 35: 15s, 2017. (suppl; abst 2541). [Google Scholar]
- 115. U.S Food and Drug Administration. Hematologic malignancies: regulatory considerations for use of minimal residual disease in development of drug and biological products for treatment—guidance for industry; 2020. Available from: https://www.fda.gov/media/134605/download.
- 116. U.S Food and Drug Administration. FDA authorizes first next generation sequencing-based test to detect very low levels of remaining cancer cells in patients with acute lymphoblastic leukemia or multiple myeloma. FDA news release; 2018. Available from: https://www.fda.gov/news-events/press-announcements/fda-authorizes-first-next-generation-sequencing-based-test-detect-very-low-levels-remaining-cancer.
- 117. U.S Food and Drug Administration. Requests for feedback and meetings for medical device submissions: the Q-Submission Program. Guidance for Industry and Food and Drug Administration Staff; 2019. Available from: https://www.fda.gov/media/114034/download.
- 118. U.S Food and Drug Administration. Formal meetings between the FDA and sponsors or applicants of PDUFA products guidance for industry. Guidance for industry. Draft guidance; 2017. Available from: https://www.fda.gov/media/109951/download.
- 119. Costa LJ, Derman BA, Bal S, Sidana S, Chhabra S, Silbermann S, et al. International harmonization in performing and reporting minimal residual disease assessment in multiple myeloma trials. Leukemia 2021;35:18–30. [DOI] [PubMed] [Google Scholar]