The authors show that tumor-specific Cbl-b−/− CD8+ T cells are resistant to the effects of Treg cells in the tumor microenvironment via enhanced IFNγ production, supporting the potential for targeting Cbl-b in TCR-transduced or CAR T-cell therapy.
Abstract
Regulatory T cells (Treg) are an integral component of the adaptive immune system that negatively affect antitumor immunity. Here, we investigated the role of the E3 ubiquitin ligase casitas B-lineage lymphoma-b (Cbl-b) in establishing CD8+ T-cell resistance to Treg-mediated suppression to enhance antitumor immunity. Transcriptomic analyses suggested that Cbl-b regulates pathways associated with cytokine signaling and cellular proliferation. We showed that the hypersecretion of IFNγ by Cbl-b–deficient CD8+ T cells selectively attenuated CD8+ T-cell suppression by Tregs. Although IFNγ production by Cbl-b–deficient T cells contributed to phenotypic alterations in Tregs, the cytokine did not attenuate the suppressive function of Tregs. Instead, IFNγ had a profound effect on CD8+ T cells by directly upregulating interferon-stimulated genes and modulating T-cell activation. In murine models of adoptive T-cell therapy, Cbl-b–deficient T cells elicited superior antitumor immune response. Furthermore, Cbl-b–deficient CD8+ T cells were less susceptible to suppression by Tregs in the tumor through the effects of IFNγ. Collectively, this study demonstrates that the hypersecretion of IFNγ serves as a key mechanism by which Cbl-b–deficient CD8+ T cells are rendered resistant to Tregs.
See related Spotlight by Wolf and Baier, p. 370 .
Introduction
Regulatory T cells (Treg) play a pivotal role in maintaining tolerance to self-antigens and preventing autoimmunity (1, 2). Tregs can also constrain beneficial antitumor immune responses, and as a consequence tumor progression occurs (3). However, their precise role in attenuating antitumor immunity and the mechanism by which Tregs suppress T cells in a tumor is still unclear (4, 5). Acknowledging the significance of Tregs and their potential role in inhibiting antitumor immunity, strategies have been proposed to deplete and destabilize Tregs in the context of tumor immunity (3, 6–9). One major challenge with Treg depletion is the paucity of Treg-specific surface markers because many surface receptors expressed on Tregs are also present on activated lymphocytes. Furthermore, unless intratumoral Tregs are selectively targeted, depletion of Tregs at a systemic level may result in unwanted adverse events (10, 11). Thus, the concept of rendering tumor-specific T cells refractory to the suppressive effects of Tregs serves as a novel approach to eliciting superior antitumor immune response.
A variety of molecular pathways and cellular mechanisms that render T cells resistant to Treg-mediated suppression have been identified. Cytokines such as IL1β, IL2, IL4, IL6, IL7, IL15, and IL21, and intracellular molecules such as Cbl-b, Akt1, MyD88, TRAF6, and SHP-1, have been reported to render T cells resistant or sensitive to Treg-mediated suppression (9, 12). Studies involving Cbl-b–deficient T cells were among the first to demonstrate the role of intracellular molecules in rendering T cells resistant to the suppressive effects of Tregs (13, 14).
Cbl-b is an E3 ubiquitin ligase that regulates PKCθ, PLCγ1, Vav, and Nedd4 along with several other T-cell receptor (TCR) signaling molecules (15–18). Although the predominant mechanism of regulation remains context dependent, the molecule serves as a powerful negative regulator of T-cell activation (19). Cbl-b–deficient T cells produce increased levels of IL2, do not require CD28 costimulation for activation, and are less sensitive to TGFβ signaling, which culminates in these cells showing enhanced T-cell activation following TCR engagement (14, 20–23). It has been reported that Cbl-b–deficient CD4+ T cells hypersecrete IL2 to reduce their susceptibility to Treg-mediated suppression in vitro (23). Furthermore, adoptively transferred Tregs fail to suppress Cbl-b–deficient CD4+ T cells in a graft-versus-host disease model, demonstrating that Cbl-b–deficient CD4+ T cells resist the suppressive effects of Tregs in vivo (14). Similar to CD4+ T cells, Cbl-b–deficient CD8+ T cells also have robust proliferative capacity in vitro even when activated in the presence of Tregs (24). However, the precise mechanism that renders Cbl-b–deficient CD8+ T cells resistant to Tregs is unknown. Moreover, it remains unclear whether Cbl-b–deficient CD8+ T cells are also capable of withstanding the suppressive effects of Tregs in vivo, particularly in the context of tumor immunity.
In this study, we demonstrate that hypersecretion of IFNγ by Cbl-b–deficient CD8+ T cells results in resistance to the suppressive effects of Tregs in the context of antitumor immunity.
Materials and Methods
Mice and cell lines
C57BL/6 wild-type (WT), IFNγ, IFNγR1, OT-1 Thy1.1 mice were purchased from Taconic and Jackson Laboratory. Cbl-b–knockout (KO) mice on the C57BL/6 background were a kind gift from Dr. J.M. Penninger (University of British Columbia), and DEpletion of REGulatory T cells (DEREG) mice on the C57BL/6 background were a kind gift from Dr. T. Sparwasser (University Medical Center of the Johannes Gutenberg University Mainz). P14 mice, which express a transgenic TCR that recognizes H2-Db presenting the gp33 peptide of lymphocytic choriomeningitis virus (LCMV), have been previously described (25). RIP-Gp mice (26) and mice bearing spontaneous pancreatic tumors (RIP-TAg2 mice) have also been previously described (27). All mice were maintained and bred under the guidelines and policies set by UHN Animal Resource Centre and with approval of the Ontario Cancer Institute Animal Ethics Committee. Female mice ages 2 to 4 months were used for experiments.
B16F10-gp33 cells were obtained from Dr. Rolf M. Zinkernagel (University of Zurich). These cells contained a neomycin resistance gene and were cultured with G418 selection reagent (800 mg/mL, InVivogen) for 7 days prior to freezing. B16F10-gp33 cells were cultured in complete DMEM (catalog no. A4192101, Gibco) containing 10% FCS (catalog no. A15-701, PAA Laboratories), 1% L-glutamine (catalog no. 25030-81, Gibco), 1% penicillin–streptomycin (catalog no. 15140-122, Gibco), and 0.0004% 2-mercaptoethanol (catalog no. M7522, Sigma). The cells were used for tumor inoculation at 2 to 4 passages after thaw. E.G7 cells (EL4 cell line expressing Ova antigen) were acquired from Dr. D. Brooks (Princess Margaret Cancer Centre, 2018). E.G7 cells were cultured in complete RPMI (catalog no. 11875119, Invitrogen) containing 10% FCS, 1% L-glutamine, 1% penicillin–streptomycin, and 0.0004% 2-mercaptoethanol. For both cell lines, no cell line authentication occurred in the past year, and no Mycoplasma testing was performed.
Cell isolation
CD4+ or CD8+ T cells were negatively selected from spleens and lymph nodes of mice using a magnetic purification kit (catalog no. 130-104-454 and 130-104-075, Miltenyi Biotec). To separate Tregs, negatively selected CD4+ T cells were stained using anti-CD4 (catalog no. 100412, BioLegend) and anti-CD25 (catalog no. 102008, BioLegend), and a BD fluorescence-activated cell sorting (FACS) Aria was used to further separate unstimulated naïve T cells (CD4+CD25–) and Tregs (CD4+CD25high). Purity was checked through intracellular FoxP3 staining using anti-FoxP3 (catalog no. 25-5773-82, eBioscience; prepared as described in "Surface/intracellular staining and flow cytometry”). For purification of antigen-presenting cells (APC), CD5 (Ly-1) MicroBeads were used to deplete CD5+ T and B cells as per the manufacturer's instructions (catalog no. 130-049-301, Miltenyi Biotec).
In vitro T-cell stimulation
For functional assays, T-cell stimulation was performed using anti-CD3 and irradiated APCs. Prior to coculture, APCs were irradiated with a dose of 2,500 cGy using X-RAD 320 (PXi Precision X-Ray). Purified CD8+ or CD4+ T cells were stained with 10 µmol/L cell proliferation dye eFluor 450 (catalog no. 65-0842-90, eBioscience) in PBS for 20 minutes at 4°C. After three washes, 5×104 T cells were cocultured with 2×105 irradiated APCs and 1 µg/mL anti-CD3 (clone 145-2C11, catalog no. 14-0031-85, eBioscience) in complete RPMI-1640 media (catalog no. 11875119, Invitrogen) containing 10% FCS (catalog no. A15-701, PAA Laboratories), 1% L-glutamine (catalog no. 25030-81, Gibco), 1% penicillin–streptomycin (catalog no. 15140-122, Gibco), and 0.0004% 2-mercaptoethanol (catalog no. M7522, Sigma). Cells were incubated in Thermo Scientific Nunc MicroWell 96-well polystyrene microplates (catalog no. 12-565-66, Fisher Scientific) in 5% CO2 and 37°C incubation. Supernatants were harvested on day 1 or 3 after stimulation and stored at −80°C before cytokine analysis. Flow cytometry analysis was also performed on either day 1 or 3 after stimulation.
For adoptive transfer experiments, purified CD8+ T cells were stimulated through plate-bound anti-CD3/CD28 (catalog no. 14-0031-85, 16-0281-82, eBioscience) and 10 ng/mL recombinant IL2 (catalog no. 575404, BioLegend) in complete RPMI-1640 media. T cells (5 × 104) were plated per well in the 96-well polystyrene microplates for 3 days. Cells were collected and washed prior to injection.
Tumor experiments
B16F10-gp33 (3 × 105) cells or E.G7-Ova (4 × 105) cells resuspended in 100 µL PBS were subcutaneously injected into the shaved left flanks of mice between the ages of 8 to 16 weeks. Tumor growth was tracked every 3 days using a caliper. On day 10 after inoculation, mice bearing tumors of approximately 5-mm diameter were randomly distributed to each group for intravenous (i.v.) infusion of 1×106 T cells. Mice with ulceration/necrosis or tumors over 225 mm2 were euthanized and recorded as deceased. For in vivo depletion of Tregs, FoxP3+ cells were transiently ablated in DEREG mice, which are mice in which DTR-eGFP is selectively expressed under the direct control of the FoxP3 promoter. FoxP3+ cells were depleted by intraperitoneal (i.p.) administration of 10 ng per g body weight diphtheria toxin (Merck) on days 10, 11, 19, and 20 after tumor inoculation. For CD8+ T-cell depletion, monoclonal antibodies were prepared in-house using the CD8 hybridoma cells (YTS169), which were acquired from Dr. Rolf M. Zinkernagel (University of Zurich), and CellMax artificial capillary cell culture systems. For experiments involving PD-L1 blockade, 100 µg of anti–PD-L1 (catalog no. BE0101, Bio X Cell) or rat IgG2b isotype control (catalog no. BE0090, Bio X Cell) were i.p. injected on days 10, 14, and 18 after tumor inoculation.
Treg suppression assay
Upon FACS of Tregs (CD4+CD25+) and T cells (CD4+CD25– or CD8+), purified CD4+ or CD8+ T cells were stained with 10 µmol/L cell proliferation dye eFluor 450 (catalog no. 65-0842-90, eBioscience) in PBS for 20 minutes, followed by three washes in complete RPMI-1640 media (catalog no. 11875119, Invitrogen). 5 × 104 T cells (Teff) were subsequently cultured with 2 × 105 irradiated APCs (prepared as described in “In vitro T-cell stimulation”), anti-CD3 (1 µg/mL), and 5 × 104 CD4+CD25+ Tregs. For Treg suppression assays involving 1:1 to 16:1 Teff:Treg ratios, the quantity of Tregs was adjusted from 5 × 104 to 3.125 × 103 cells, respectively. For exogenous supplementation of IFNγ signaling agonist/inhibitor, recombinant IFNγ (catalog no. 14-8311-63, eBioscience; 10 ng/mL) and anti-IFNγ (catalog no. 16-7311-81, eBioscience; 10 mg/mL) were used, respectively. TNFα (catalog no. 575204) and IL17A (catalog no. 576004) were purchased from BioLegend. Cells were incubated in Thermo Scientific Nunc MicroWell 96-well polystyrene microplates (catalog no. 12-565-66, Fisher Scientific) in 5% CO2 and 37°C incubation. Unless noted otherwise, supernatants were harvested on day 1 or 3 after stimulation and stored at −80°C before cytokine analysis. Flow cytometry analysis was performed either on day 1 or 3 after stimulation.
For analysis, division index (DI) was quantified by averaging the number of cell divisions using FlowJo software, and the percentage of suppression was calculated by normalizing T-cell proliferation in the presence of Tregs to the intrinsic proliferative capacity of the T cells (% suppression = 100 – (DI with Tregs/DI without Tregs) × 100).
RNA sequencing
RNA was extracted from 12-hour stimulated T cells (>99% CD8+ purity) using the RNeasy Mini Kit (catalog no. 74104, Qiagen) according to the manufacturer's instructions. After quantity and quality assessment using Agilent Bioanalyzer RNA Nano 6000 Kit (catalog no. 5067-1511, Agilent) and Qubit BRRNA kit (catalog no. Q10210, Thermo Fisher Scientific), RNA libraries were prepared by removing ribosomal RNA using the TruSeq Stranded Total RNA kit (catalog no. 20020596, Illumina). After cDNA preparation, the paired-end libraries were sequenced on NextSeq 500 (Illumina) using 100 bp protocol. RNA sequencing (RNA-seq) was performed by the Princess Margaret Genomics Centre (Toronto, Canada). RNA-seq FASTQ files were mapped using STAR v2.5.2 aligner (28) with mouse transcript coordinates from GENCODE release M17 (29) using mouse genome GRCm38 (mm10). Reads were summarized per gene using RSEM v1.3.0 (30). The Bioconductor (31) biomaRt (32) package was used to map the original Ensemble transcript ids to NCBI gene ids and Unigene symbols. Principal component analysis (PCA) was performed on the normalized RNA-seq count using variance-stabilizing transformation. Differential expression analysis was performed using DESeq2 comparing different experimental groups (33), and P values were adjusted for multiple testing using the FDR correction of Benjamini and Hochberg (34). DESeq2 log2 fold-change predictions were moderated using the APEGLM algorithm. All other RNA-seq visualizations were created using the R ggplot2 package.
For gene set enrichment analysis (GSEA), mouse-to-human gene homolog mapping was performed using biomaRt, and multimapping homologs resolved to produce a one-to-one mapped set of human homologs. GSEA v3.0 with MSigDB v6.2 human gene sets was used to identify enriched gene sets (FDR-adjusted P < 0.1). The Hallmark (H), Canonical Pathways (CP), Gene Ontology Biological Process (GO BP), and Molecular Function (GO MF) gene set collections were tested for enrichment. Cytoscape v3.8.0 (RRID:SCR_003032) with the EnrichmentMap v3.3.0 plugin was used to visualize and cluster GSEA results (using GLay network clustering algorithm) into network clusters of partially overlapping, upregulated gene sets. Gene set clusters of 10 or fewer gene sets were excluded from figures for clarity.
For TGFβ signaling pathway analysis, KEGG TGFβ signaling data set (source: hsa04350) and TGFβ–SMAD3 response genes identified by Delisle and colleagues (35) were utilized to create a reference gene library. All identifiable genes with NCBI IDs were included for further study (61/86 genes from the KEGG TGFβ signaling data set and 32/40 genes from the Delisle and colleagues data set). DESeq2 comparing different experimental groups was performed and selected genes in the TGFβ pathway were analyzed. Each datapoint represent log2 fold change of differentially expressed genes (P < 0.05 were annotated in red). TGFβ pathway visualization was performed using the R ggplot package.
Cytokine analysis
Coculture supernatants, stored in −80°C, were analyzed using IL2 (catalog no. 88-7024-88, eBioscience), IL3 (catalog no. RAB0295-1KT, Sigma), IFNγ (catalog no. 88-7314-86, eBioscience, catalog no. RAB0224-1KT, Sigma), Lif (catalog no. ab238261, Abcam) and TNFα ELISA kits (catalog no. 88-7324-22, eBioscience). LEGENDplex Mouse Th Cytokine Panel cytometric bead array (catalog no. 740005, BioLegend) was used for T-cell cytokine secretion profiling.
Surface/intracellular staining and flow cytometry
Individual cell suspensions were washed twice in FACS buffer (PBS supplemented with 2% FCS and 0.05% sodium azide), followed by FcR blocking (30 minutes) using anti-CD16/32 (catalog no. 14-0161-85, eBioscience) and viability staining (catalog no. 65-0865-14, Invitrogen). For surface marker analyses, cells were subsequently stained with Abs for 30 minutes on ice followed by two washes. The following antibody clones were used: CD3 (145-2C11), CD8 (53-6.7), CD4 (GK1.5), CD25 (PC61), CD69 (H1.2F3), 4-1BB (17B5), PD-1 (RMP1-30), LAG3 (C9B7W), ICOS (15F9), OX40 (OX-86), LFA-1 (H155-78), FasL (MFL3), Gal-9 (108A2), Nrp-1 (12C2), CD39 (5F2), CD73 (AD2), GITR (DTA-1), MHCII (M5/114.15.2), Thy1.1 (His51), Thy1.2 (30-H12 and 53-2.1), and Ki-67 (SolA15), all purchased from eBioscience, BioLegend, or BD Biosciences. For all surface marker staining, cells were fixed using 4% paraformaldehyde after washes.
Intracellular protein staining was performed using FoxP3 Transcription Factor Staining Buffer Set (catalog no. 00-5523-00, eBioscience) with the following antibodies: IFNγ (XMG1.2), IL2 (JES6-5H4), CTLA-4 (UC10-489), GzmB (GB12), and FoxP3 (FKJ-16s) from eBioscience or BD Bioscience. For intracellular cytokine staining, cells were stimulated using PMA/ionomycin cell stimulation cocktail (catalog no. 00-4970-93, eBioscience) and Brefeldin A (catalog no. 00-4506-51, eBioscience) for 5 hours prior to intracellular staining.
All flow cytometry data were acquired using a BD FACSCanto II Flow Cytometer (BD Biosciences) or Advanced Analyzers BD LSRFortessa 2 (BD Biosciences) and were analyzed on FlowJo software 10.5.3 (FlowJo LLC, RRID:SCR_008520).
Western blot
Single-cell suspensions were resuspended in RIPA buffer [150 mmol/L NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mmol/L Tris (pH 8), protease inhibitors (catalog no. 11-836-153-001, Roche), and phosphatase inhibitors (catalog no. 04-906-837-001, Roche)] for 20 minutes. The protein samples were quantified with a BCA protein assay kit (catalog no. 23225, Pierce). Protein samples were separated on a 4%–12% NuPAGE Bis-Tris gel (Thermo) and transferred to PVDF membranes (EMD Millipore). Membranes were incubated at 4°C overnight with primary antibodies: anti–Cbl-b (catalog no. sc-8006, Santa Cruz), anti-PKCθ (catalog no. sc-212, Santa Cruz), anti-PLCγ1 (catalog no. sc-81, Santa Cruz), and anti-actin (A2066, Sigma). Secondary antibodies included goat anti-mouse HRP (catalog no. 32430, Thermo) and goat anti-rabbit HRP (catalog no. 32460, Thermo), and blots were incubated for 1 hour at room temperature. Blot signals were detected using SuperSignal West Femto maximum sensitivity substrate (catalog no. 34094, Thermo). Western blots were scanned and imported into ImageJ (RRID:SCR_003070), where the blots were analyzed.
Statistical analysis
Statistical analysis was performed as indicated per figure legend. Statistical tests used were one-way ANOVA, two-way repeated-measure ANOVA, a log-rank test, and two-tailed paired Student t test, followed by post-analysis such as Holm–Sidak test. Results were considered statistically significant when P < 0.05. All data are presented as mean with standard error using GraphPad Prism 8 (GraphPad Software Inc., RRID:SCR_002798). All experiments are representative of two or more biological replicates.
Data availability
The data generated in this study are publicly available in Gene-Expression Omnibus at GSE189350.
Results
Cbl-b–deficient CD8+ T cells enhance antitumor immunity
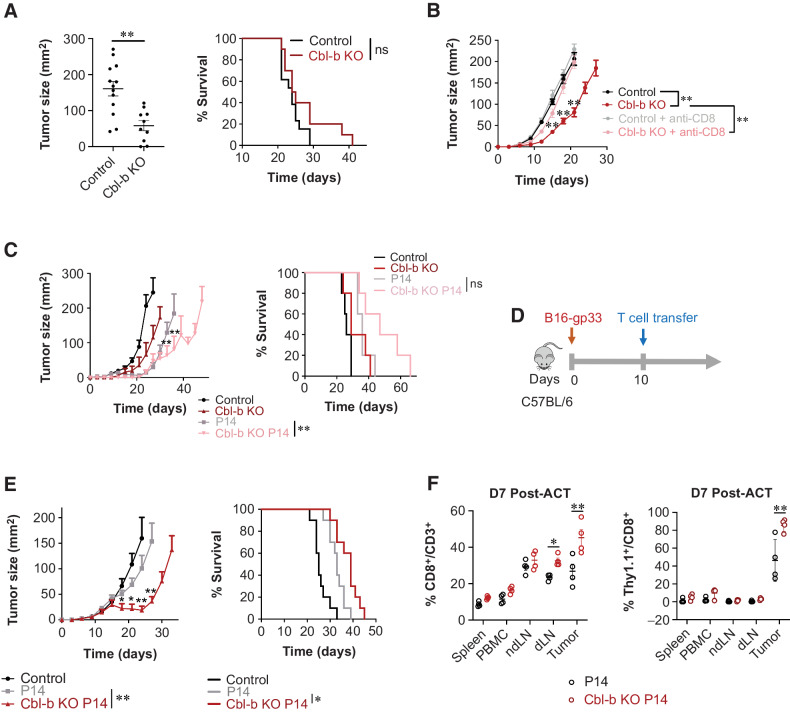
Consistent with literature suggesting that Cbl-b deficiency improves antitumor immunity in syngeneic murine models (24, 36), we found tumor growth was reduced in Cbl-b KO mice bearing B16 melanoma (Fig. 1A). Similarly, Cbl-b deficiency in spontaneous insulinoma-bearing RIP-TAg2 mice resulted in improved overall survival (Supplementary Fig. S1). To evaluate whether CD8+ T cells play a role in the improved response in the Cbl-b KO mice, we compared the tumor growth kinetics in Cbl-b KO mice with or without CD8+ T-cell depletion. CD8+ T-cell depletion abrogated the effects of Cbl-b deficiency, resulting in an increase in the overall tumor size (Fig. 1B). Furthermore, we compared the ability of Cbl-b–sufficient and –deficient P14 TCR Vα2Vβ8-transgenic mice to control the growth of B16 tumors constitutively expressing the LCMV-gp33 (KAVYNFATM) antigen recognized by P14 CD8+ T cells. P14 TCR-transgenic mice demonstrated improved antitumor immunity against B16-gp33 in comparison with littermate controls. In addition, Cbl-b KO P14 TCR-transgenic mice showed enhanced antitumor immunity, with a few mice surviving up to 60 days after tumor injection (Fig. 1C). Thus, tumor-specific CD8+ T-cell activity can be improved in the absence of Cbl-b.
Figure 1.
Cbl-b deficiency enhances antitumor immunity. A, Mean tumor area and survival of tumor-bearing C57BL/6 (WT) and Cbl-b KO mice. B16-gp33 tumor cells were injected into the left flanks of each mouse (n = 5). Tumor size was measured on day 21 after tumor injection. B, Mean tumor area and survival of tumor-bearing C57BL/6 (WT) and Cbl-b KO mice treated with or without anti-CD8 (n = 5). Anti-CD8 was administered on days 0, 2, 14, and 16 after B16-gp33 injection. C, Mean tumor area and survival of tumor-bearing C57BL/6 (WT) P14 and Cbl-b KO P14 mice (n = 5). D, Experimental overview of adoptive T-cell transfer in B16-gp33 tumor-bearing hosts. CD8+ T cells were stimulated with anti-CD3/CD28 and IL2 (10 ng/mL) and 1.0 × 106 cells were adoptively transferred into B16-gp33 tumor-bearing mice. E, Mean tumor area and survival were measured (n = 10). F, Evaluating T-cell infiltration into the tumor by WT and Cbl-b KO P14 Thy1.1+CD8+ T cells. Tumor, ndLN), dLN, spleen, and PBMCs were isolated from tumor-bearing mice 7 days after adoptive T-cell transfer. Surface expression of CD3, CD8, and Thy1.1 was analyzed by flow cytometry (n = 5). Statistical analyses were performed using repeated measure ANOVA with Holm–Sidak test (mean tumor area), and log-rank test (survival) (P < 0. 05; *, P < 0.01; **, not significant; ns; A, B, C, and E). Two-way ANOVA with Holm–Sidak test was performed in F (*, P < 0. 05; **, P < 0.01).
To directly confirm that activated CD8+ T cells mediate antitumor immunity in the model, we stimulated purified Cbl-b–sufficient and –deficient P14 CD8+ T cells for 3 days using anti-CD3/CD28 and IL2 and adoptively transferred them into WT mice bearing palpable B16-gp33 tumors (Fig. 1D). Cbl-b–deficient P14 CD8+ T cells yielded superior tumor regression for the first 10 days of T-cell transfer and increased the overall survival of mice (Fig. 1E). These findings were recapitulated when Cbl-b–sufficient and –deficient OT-1 CD8+ T cells were adoptively transferred into WT mice bearing palpable E.G7-Ova tumors (Supplementary Fig. S2). To confirm that the Cbl-b–KO CD8+ T cells are functional and migrate into the tumor microenvironment, Cbl-b–WT and –KO P14 CD8+ T cells congenically labeled with Thy1.1 were adoptively transferred into Thy1.2 mice bearing established melanomas. On day 7 after T-cell transfer, PBMCs, spleen, nondraining lymph node (ndLN), tumor-draining lymph node (dLN), and tumor tissue were harvested. Mice treated with Cbl-b–KO P14 CD8+ T cells had a higher frequency of CD8+ T cells in both dLN and tumor, and had a higher frequency of congenically labeled T cells in the tumor (Fig. 1F). Lastly, to examine the autoimmune potential in our experimental system, we adoptively transferred prestimulated Cbl-b–WT or –deficient P14 CD8+ T cells into RIP-gp transgenic mice, which express LCMV glycoprotein under the control of rat insulin promoter. Adoptively transferred Cbl-b–deficient P14 T cells alone were insufficient to induce autoimmune hyperglycemia and weight loss in RIP-gp mice (Supplementary Fig. S3). Therefore, in our adoptive transfer model, effector CD8+ T cells play an important role in controlling tumor growth, and Cbl-b–deficient effector CD8+ T cells show improved efficacy without triggering autoimmune pathology.
Cbl-b deficiency renders CD8+ T cells resistant to the suppressive effects of Tregs
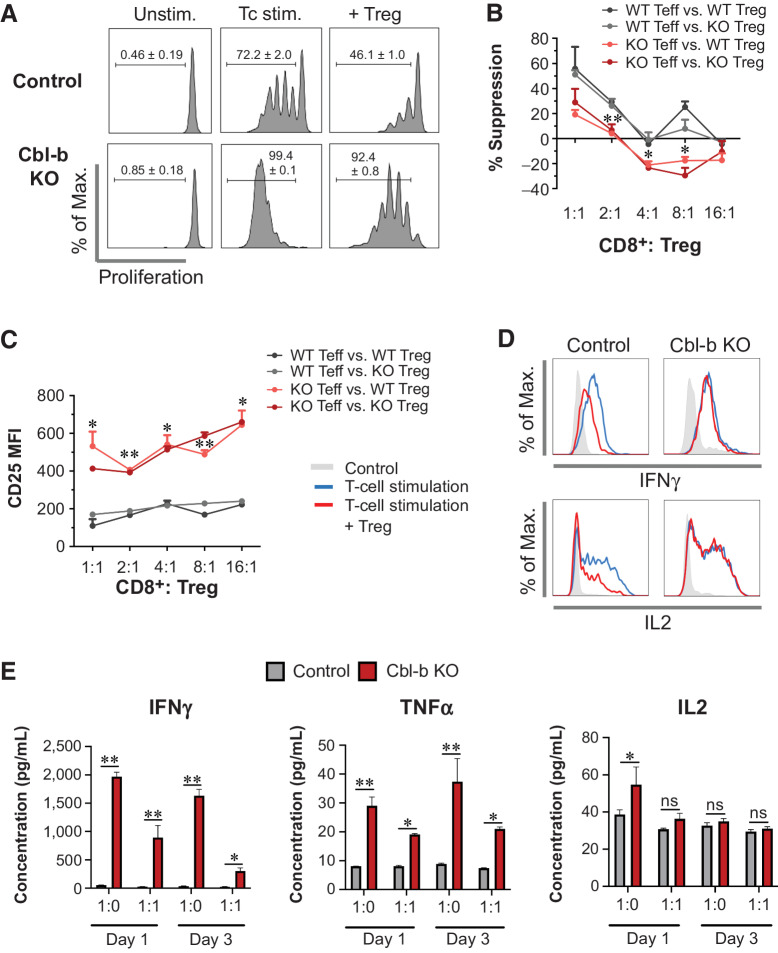
Cbl-b–deficient CD8+ T cells demonstrated enhanced antitumor immunity (Fig. 1E and F) despite the presence of Tregs in the tumor (37, 38). We hypothesized that Cbl-b deficiency renders CD8+ T cells resistant to the effects of Tregs, contributing to strengthened antitumor immunity. To explore the hypothesis, we first performed in vitro Treg suppression assays. Cbl-b–KO CD8+ effector T cells rapidly proliferated despite the presence of Tregs (Fig. 2A). As T-cell proliferation may not necessarily indicate Treg resistance, we calculated a percentage suppression score. Cbl-b–KO CD8+ T cells demonstrated significantly a lower percentage of suppression in any given ratio of Teff to Tregs in comparison with WT CD8+ T cells (Fig. 2B). Thus, CD8+ T cells from Cbl-b–KO mice were refractory to the inhibitory effects of Tregs independent of enhanced proliferation. Cbl-b–KO CD8+ T cells also expressed a higher level of the activation marker CD25, despite the presence of Tregs (Fig. 2C). Furthermore, in the Treg suppression assay, the presence of Tregs decreased intracellular expression of IFNγ and IL2 by WT CD8+ T cells (Fig. 2D). In contrast, expression of IFNγ and IL2 remained unchanged in Cbl-b–deficient CD8+ T cells despite the presence of Tregs (Fig. 2D). In addition, Cbl-b–KO CD8+ T cells secreted higher levels of IFNγ and TNFα than WT CD8+ T cells (Fig. 2E). Although the addition of Tregs reduced the absolute quantity of IFNγ and TNFα, Cbl-b KO CD8+ T cells still produced a large quantity of these cytokines despite the presence of Tregs (Fig. 2E). In summary, Cbl-b deficiency strengthens CD8+ T-cell activation and renders the cells less sensitive to the suppressive effects of Tregs.
Figure 2.
Cbl-b KO CD8+ T cells display resistance to Treg-mediated suppression. A, Proliferation of WT and Cbl-b KO CD8+ T cells in the Treg-suppression assay, analyzed on day 3 after stimulation (n = 3). B and C, The percentage of suppression (B) and CD25 expression (C) of WT and Cbl-b KO CD8+ at different Teff to Treg ratios (n = 3). D, Intracellular expression of IFNγ and IL2 by WT and Cbl-b KO CD8+ effector T cells in the presence of WT Tregs. After 24 hours of coculture, T cells were restimulated using PMA/ionomycin and Golgi block and were stained for CD8, IFNγ, and IL2 for flow cytometry. E, Cytokine secretion by WT and Cbl-b KO CD8+ T cells in the presence of Tregs. Supernatants from stimulated WT and Cbl-b KO CD8+ T cells in the presence (or absence) of Tregs were collected on days 1 and 3 after stimulation (n = 3). ELISAs were performed to assess secretion of IFNγ, IL2, and TNFα by CD8+ T cells. Statistical analyses were performed using repeated measure (B and C) or nonrepeated (E) two-way ANOVA with Holm–Sidak test (*, P < 0. 05; **, P < 0.01; ns, not significant).
Cbl-b regulates cytokine expression at the level of the T-cell transcriptome
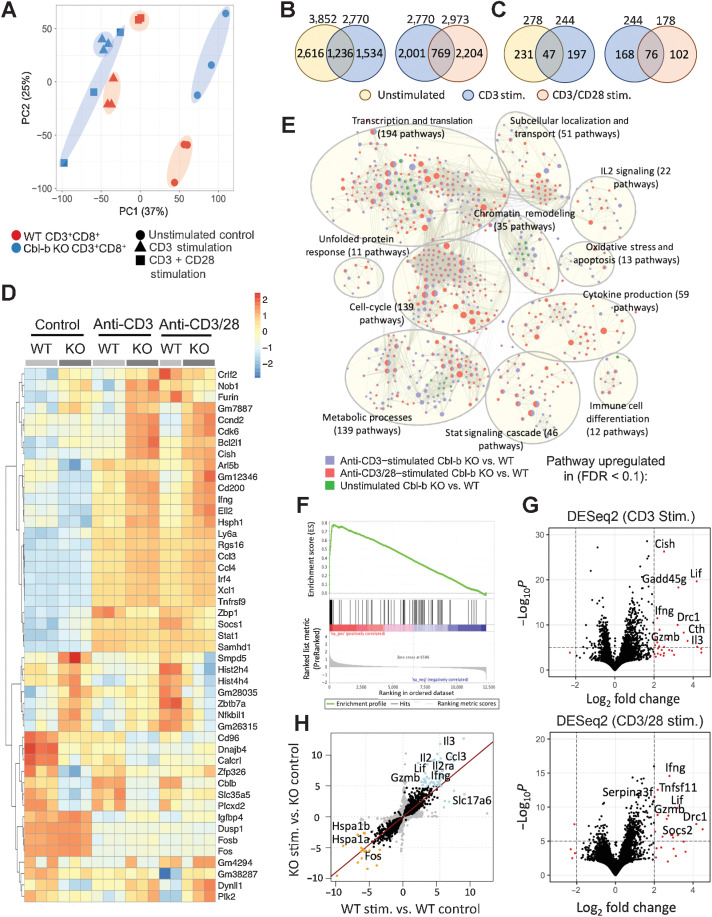
To uncover the mechanism by which Cbl-b deficiency renders CD8+ T cells refractory to Treg-mediated suppression, we performed transcriptomic analysis comparing Cbl-b–sufficient and –deficient CD8+ T cells. The cells were stimulated with anti-CD3 and/or anti-CD28 for 12 hours, followed by RNA extraction and sequencing. The timepoint of 12 hours after activation was selected because this corresponded with increased Cbl-b expression at the protein level following TCR stimulation (Supplementary Fig. S4). Unsupervised PCA showed that Cbl-b–sufficient and –deficient CD8+ T cells independently segregated for unstimulated, anti-CD3–stimulated and anti-CD3/CD28–stimulated conditions, suggesting that Cbl-b deficiency has a profound impact on the T-cell transcriptome pre- and post-TCR stimulation (Fig. 3A). A large proportion of the genes could be attributed to stimulatory condition-specific differences, but there were also similarities across the two conditions (Fig. 3B and C).
Figure 3.
Cbl-b KO CD8+ T cells display distinct transcriptomic profiles. RNA was extracted from unstimulated, anti-CD3, and anti-CD3/CD28-stimulated Cbl-b KO and WT CD8+ T cells (n = 3) and sequenced. A, PCA of Cbl-b–sufficient and –deficient CD8+ T cells in the different stimulatory conditions. B and C, Venn diagrams representing either the total number of DEGs (B) or number of upregulated genes (C) between Cbl-b KO and WT CD8+ T cells of each condition (P < 0.05 and log2 fold change > 1). D, A heatmap representing DEGs of each group. Values were calculated using individual gene's Z-normalized log2 score (RNA-seq read count + 1). E, GSEA network clustering for the identification of highly upregulated pathways in Cbl-b–deficient CD8+ T cells of each condition. F, Enrichment plot depicting gene-expression signatures from Gene Ontology Cytokine Activities comparing anti-CD3–stimulated KO vs. WT CD8+ T cells. The barcode plot represents the position of the genes in the gene set; red and blue colors represent positive and negative Pearson correlation with Cbl-b deficiency, respectively. G, Volcano plots highlighting DEGs between Cbl-b KO and WT CD8+ T cells stimulated with anti-CD3 or anti-CD3/CD28. H, Pairwise analysis of DEGs between stimulated and unstimulated Cbl-b KO and WT CD8+ T cells. Blue and orange points indicate upregulated and downregulated genes in response to TCR stimulation, respectively.
To define key transcriptional signatures of Cbl-b–deficient CD8+ T cells, we identified differentially expressed genes (DEG) for each of the groups (Fig. 3D). Upon T-cell activation, Cbl-b–deficient CD8+ T cells upregulated genes important for cellular proliferation, including Cdk6, Ccnd2, and Plk2, while also upregulating genes important for immunologic functions, including Ifng, Ccl3, Ccl4, and Tnfrsf9 (Fig. 3D). GSEA identified cytokine activities as one of the most enriched gene sets in Cbl-b–deficient CD8+ T cells (Fig. 3E and F). More specifically, differential gene-expression analysis based on the negative binomial distribution identified Lif, Ifng, and Il3 as among the most highly expressed genes in activated Cbl-b–deficient CD8+ T cells (Fig. 3G), which was confirmed by protein expression analysis (Fig. 2E; Supplementary Fig. S5). Cbl-b deficiency also altered the transcriptomic profile of CD8+ T cells prior to TCR stimulation (Fig. 3A–D), which may serve as a confounding variable and alter the interpretation of our data. We, therefore, normalized the effect of Cbl-b prior to TCR stimulation through pairwise DESeq2 analysis. Consistent with our previous data, Cbl-b deficiency significantly upregulated the expression of cytokines including Lif, Ifng, Il2, and Il3 upon TCR stimulation (Fig. 3H). Together, these data suggest that some of the primary features of activated Cbl-b–deficient CD8+ T cells are enhanced cellular proliferation and cytokine production.
IFNγ renders Cbl-b–deficient CD8+ T cells resistant to the suppressive effects of Tregs
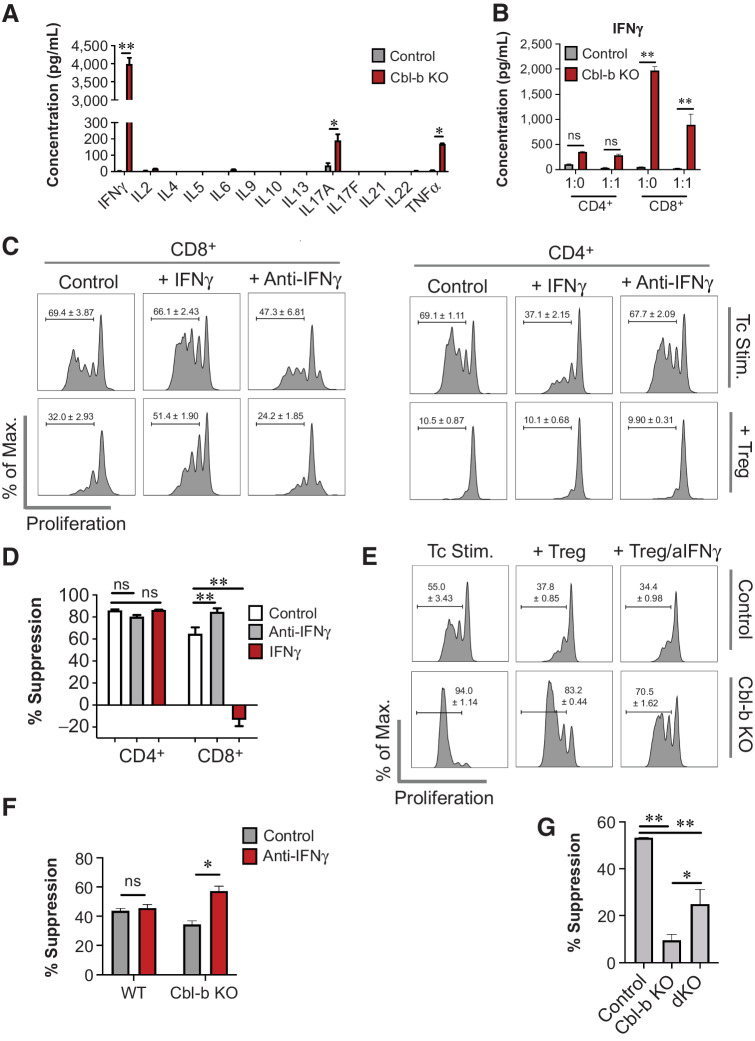
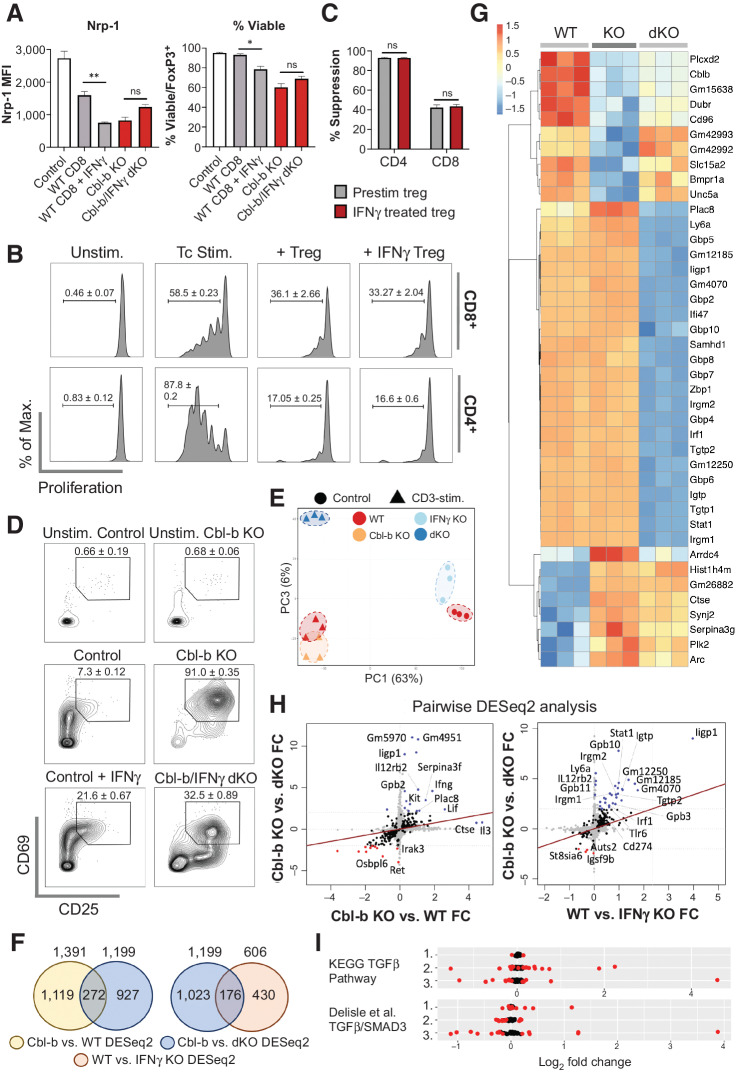
In general, cytokines play an important role in modulating T-cell responses, and this immunomodulatory capacity also occurs in the presence of Tregs (9, 12, 23). Our transcriptomic analysis demonstrated that cytokines may be highly upregulated in Cbl-b–deficient CD8+ T cells. To determine whether any of these cytokines attenuate the suppressive effects of Tregs, we analyzed cytokine levels in the supernatant of Cbl-b–sufficient and –deficient CD8+ T cells. Cbl-b deficiency increased the secretion of IFNγ, TNFα, IL17A, IL3, and Lif upon TCR stimulation (Fig. 4A; Supplementary Fig. S5). In particular, Cbl-b KO CD8+ T cells showed a 1,000-fold upregulation of IFNγ (Fig. 4A), and IFNγ hyperproduction was observed as early as 4 hours after TCR stimulation (Supplementary Fig. S6). IFNγ hypersecretion was a predominant feature in Cbl-b–KO CD8+ T cells but not in Cbl-b–KO CD4+ T cells, and Cbl-b–KO CD8+ T cells secreted high levels of IFNγ despite the presence of Tregs (Fig. 4B).
Figure 4.
IFNγ hypersecretion by Cbl-b KO CD8+ T cells induces Treg resistance. A, Cytokine secretion profile of activated WT and Cbl-b KO CD8+ T cells. Supernatants were collected on day 3 after stimulation, and LEGENDplex cytokine array was performed (n = 3). B, IFNγ secretion by WT and Cbl-b KO T cells in the presence of Tregs. Supernatants from the Treg suppression assay were collected on day 3 after stimulation, and IFNγ ELISA was performed (n = 3). C, Proliferation of WT T cells cocultured with or without Tregs in the presence of IFNγ or anti-IFNγ (n = 3). D, The percentage suppression of WT CD4+ and CD8+ T cells for each stimulatory condition (n = 3). E, Proliferation of Cbl-b KO CD8+ T cells in the presence of Tregs, with or without anti-IFNγ (n = 3). F, The percentage suppression of WT and Cbl-b KO CD8+ T cells stimulated with or without anti-IFNγ (n = 3). G, Treg suppression assay comparing percentage suppression of WT, Cbl-b KO, and Cbl-b/IFNγ double KO CD8+ T cells in response to Tregs (n = 3). Statistical analyses were performed using Student t test (A and F), and ANOVA with Holm–Sidak test (B, D, and G; *, P < 0. 05; **, P < 0.01; ns, not significant).
To evaluate whether IFNγ renders T cells resistant to the suppressive effects of Tregs, we cocultured WT T cells and Tregs in the presence of either IFNγ or anti-IFNγ. IFNγ enhanced CD8+ T-cell proliferation in the presence or absence of Tregs, whereas anti-IFNγ deceased CD8+ T-cell proliferation (Fig. 4C). In contrast, IFNγ reduced CD4+ T-cell proliferation and did not attenuate CD4+ T-cell suppression by Tregs (Fig. 4C). Because T-cell proliferation does not necessarily indicate Treg resistance, we calculated a percentage suppression score to demonstrate that IFNγ selectively decreases the percentage of suppression of CD8+ T cells (Fig. 4D). Although Cbl-b–deficient CD8+ T cells also upregulated other cytokines, including TNFα and IL17A (Fig. 4A), we found that both TNFα and IL17A failed to reduce the percentage of suppression of WT CD8+ T cells by WT Tregs (Supplementary Fig. S7). Collectively, this analysis shows that IFNγ selectively renders CD8+ T cells less sensitive to the inhibitory effects of Tregs.
Our finding suggests that IFNγ selectively renders CD8+ T cells resistant to Treg-mediated suppression, but it does not demonstrate that Cbl-b–deficient CD8+ T cells rely on this particular mechanism. Therefore, we performed Treg suppression assays with Cbl-b–sufficient and –deficient CD8+ T cells with or without anti-IFNγ. IFNγ blockade decreased proliferation of Cbl-b–deficient CD8+ T cells activated in the presence of Tregs (Fig. 4E). The use of anti-IFNγ also increased the susceptibility of Cbl-b KO CD8+ T cells to suppression by Tregs, as demonstrated through increased percentage of suppression, whereas the effect of anti-IFNγ was negligible with WT CD8+ T cells (Fig. 4F).
To further confirm the role of IFNγ on Cbl-b–deficient CD8+ cells, Cbl-b/IFNγ double KO mice were generated. Consistent with the previous finding, using Cbl-b/IFNγ double KO CD8+ T cells resulted in higher percentage of suppression in comparison with using Cbl-b–KO CD8+ T cells, but these cells were less sensitive to suppression in comparison with WT CD8+ T cells (Fig. 4G). Together, these data suggest that hypersecretion of IFNγ by Cbl-b–KO CD8+ T cells plays an important role in attenuating the suppressive effects of Tregs.
IFNγ directly modulates Cbl-b–deficient CD8+ T-cell response
Next, we examined whether IFNγ may disrupt the stability of Tregs and thereby reduce suppression, as suggested in a previous study (7). We found that Tregs cocultured with Cbl-b KO CD8+ T cells had a similar pattern of surface marker expression as Tregs stimulated with IFNγ (Supplementary Fig. S8). In both conditions, Tregs downregulated markers including Nrp-1, a surrogate for Treg stability (Supplementary Fig. S8). To specifically examine whether IFNγ secretion by Cbl-b KO CD8+ T cells influenced the Treg phenotype, Tregs were cocultured with WT (with or without additional IFNγ), Cbl-b KO, and Cbl-b/IFNγ double KO CD8+ T cells. Consistent with Supplementary Fig. S8, supplementing WT CD8+ T cells with exogenous IFNγ significantly reduced Nrp-1 expression and reduced the percentage of viability (Fig. 5A). However, although Nrp-1 expression and the percentage of viability were higher for Tregs cocultured with Cbl-b/IFNγ dKO CD8+ T cells in comparison with Cbl-b–KO CD8+ T cells, the differences were not statistically significant (Fig. 5A; Supplementary Fig. S9). Lastly, we examined the functional consequences of IFNγ signaling on Tregs by prestimulating Tregs with or without IFNγ for 48 hours, prior to the Treg suppression assay. Both Tregs prestimulated with or without IFNγ potently inhibited T-cell proliferation and resulted in comparable percentage suppression (Fig. 5B and C). In summary, although IFNγ contributed to phenotypic changes in the Treg compartment, the cytokine did not attenuate the suppressive function of Tregs.
Figure 5.
IFNγ directly modulates Cbl-b KO CD8+ T-cell responses. A, The effect of IFNγ on Tregs in suppression assay. Purified CD25+FoxP3+ Tregs were cocultured with WT CD8+ T cells (with or without IFNγ), Cbl-b KO, or Cbl-b/IFNγ double KO CD8+ T cells for 3 days; Nrp-1 expression on viable FoxP3+ cells and the percentage of viable cells within FoxP3+ cells were measured (n = 3). B, Proliferation of WT CD8+ and WT CD4+ T cells in the presence of preactivated Tregs treated with or without IFNγ (n = 3). C, The percentage suppression of WT CD4+ and CD8+ T cells cocultured with Tregs treated with or without IFNγ. D, CD69 and CD25 expression on activated WT, Cbl-b KO, and Cbl-b/IFNγ double KO CD8+ T cells (without Tregs). T cells were stimulated for 24 hours. E–I, Transcriptomic analyses of WT, Cbl-b KO, and Cbl-b/IFNγ double KO CD8+ T cells (n = 3). E, PCA of CD8+ T cells from different stimulatory conditions. F, Venn diagrams representing the total number of unique and overlapping differentially expressed genes for each DESeq2. G, A heatmap representing differentially expressed genes of each stimulated group. Values were calculated using individual gene's Z-normalized log2 score (RNA-seq read count + 1). H, Pairwise analyses comparing differentially expressed genes between Cbl-b KO vs. Cbl-b/IFNγ double KO DESeq2, Cbl-b KO vs. WT DESeq2 and WT vs. IFNγ KO DESeq2. Blue and red points indicate upregulated and downregulated genes in each condition, respectively. I, TGFβ signaling pathway analysis using KEGG and Delisle et al. data set (35). Genes represented in each TGFβ signaling dataset were analyzed in WT vs. IFNγ KO (1), Cbl-b KO vs. WT (2), and Cbl-b KO vs. Cbl-b/IFNγ double KO (3) DESeq2 datasets. DEGs with P < 0.05 were annotated in red.
To examine the role of IFNγ on CD8+ T cells, WT (with or without additional IFNγ), Cbl-b–KO, and Cbl-b/IFNγ double KO CD8+ T cells were stimulated, and CD69 and CD25 expression was measured. Supplementing WT CD8+ T cells with IFNγ increased the proportion of CD69+CD25+ cells from 7.3% ± 0.12% to 21.6% ± 0.67%, whereas deletion of IFNγ in Cbl-b–deficient T cells decreased the proportion of CD69+CD25+ cells from 91.0% ± 0.35% to 32.5% ± 0.89% (Fig. 5D). Our findings suggest that IFNγ secretion by Cbl-b–deficient T cells plays an important role in T-cell activation.
To further understand the role of IFNγ on CD8+ T cells, we stimulated WT, Cbl-b KO, and Cbl-b/IFNγ dKO CD8+ T cells with anti-CD3 for 12 hours and performed transcriptomic analyses. We also examined naïve WT and IFNγ KO CD8+ T cells to understand the baseline alteration in T-cell transcriptome prior to TCR engagement. First, we performed unsupervised PCA and found that IFNγ deficiency has a profound impact on Cbl-b KO CD8+ T cells post-TCR stimulation (Fig. 5E; Supplementary Fig. S10). Naïve WT and IFNγ KO T cells also had different transcriptomes (Fig. 5E; Supplementary Fig. S10). To quantify the differences among each group, we examined the number of DEGs between Cbl-b KO versus WT, Cbl-b KO versus dKO and unstimulated WT versus IFNγ KO CD8+ T cells. We found 272 DEGs that were modulated in Cbl-b deficiency but reversed upon IFNγ deletion (Fig. 5F). The key transcription signatures distinguishing Cbl-b KO and Cbl-b/IFNγ double KO CD8+ T cells were interferon-stimulated genes (ISG) such as Irgm1, Stat1, Tgtp1, Igtp, Irf1, and Iigp1 (Fig. 5G). Pairwise DESeq2 analysis comparing log2-fold change of Cbl-b KO versus WT CD8+ T cells with log2-fold change of Cbl-b KO versus Cbl-b-IFNγ dKO CD8+ T cells showed that the top genes upregulated in both conditions included ISGs such as Iigp1 and Gbp2, and Il12rb2, which play an important role in IL12 signaling (Fig. 5H). When we compared log2-fold change of Cbl-b KO versus Cbl-b/IFNγ double KO CD8+ T cells with log2-fold change of WT versus IFNγ KO CD8+ T cells, many ISGs upregulated in Cbl-b–deficient T cells were also mildly upregulated in the unstimulated control experiment (Fig. 5H). In summary, IFNγ production by Cbl-b KO CD8+ T cells primarily upregulates ISGs in the T cells; however, some of these genes are also modulated to a limited degree in naïve T cells prior to TCR stimulation.
As previous studies have suggested that Cbl-b regulates TGFβ sensitivity through posttranscriptional regulation of SMAD7 in primary T cells (39) and that IFNγ antagonizes the effects of TGFβ through regulation of SMAD family proteins (40, 41), we investigated whether Cbl-b deficiency and IFNγ production regulates a specific set of genes associated with TGFβ signaling. Gene sets were extracted from either KEGG TGFβ signaling data set (source: hsa04350) or TGFβ–SMAD3 response genes identified by Delisle and colleagues (35) and were specifically examined in DEG analysis of Cbl-b KO versus WT CD8+ T cells and Cbl-b KO versus Cbl-b/IFNγ dKO CD8+ T cells (Fig. 5I). In Cbl-b KO versus Cbl-b/IFNγ dKO CD8+ T-cell DESeq2 analysis, 23% of genes from the KEGG TGFβ pathway were differentially expressed (P < 0.05), whereas 47% of TGFβ–SMAD3 pathway genes from Delisle and colleagues (35) were differentially expressed, suggesting that Cbl-b deficiency and IFNγ production contribute to differential expression of genes involved in TGFβ signaling. In summary, our data suggest a link between the interferon response and TGFβ signaling in Cbl-b–deficient T cells.
Tregs inhibit adoptively transferred CD8+ T cells in vivo
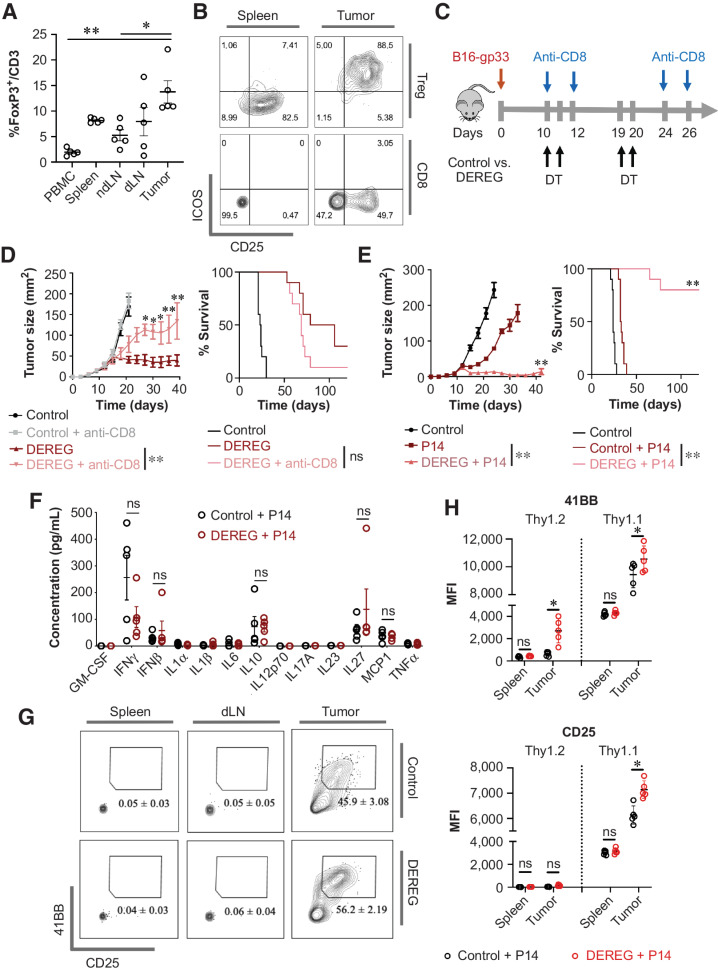
To evaluate whether Cbl-b–deficient CD8+ T cells are less susceptible to negative regulation by Tregs in vivo, we established a system to capture the interaction between tumor-specific CD8+ T cells and Tregs in the tumor. We first confirmed that immunosuppressive Tregs are abundant in B16 melanomas by allowing the tumors to grow in WT mice for 10 days and then analyzing various secondary lymphoid organs and the tumor. Approximately 13.8% of CD3+ cells in the tumor expressed FoxP3, whereas secondary lymphoid organs including spleen and dLN had a lower frequency of CD3+FoxP3+ cells (Fig. 6A). Tregs highly expressed costimulatory molecules such as ICOS in the tumor but not in the spleen, suggesting that intratumoral Tregs had increased immunosuppressive properties, which is consistent with previous reports (Fig. 6B; refs. 42–44). These findings were recapitulated in the E.G7-Ova tumor model, where approximately 21% of CD3+ cells in the tumor expressed FoxP3, the majority of which also highly expressed ICOS (Supplementary Fig. S11A and S11B). To directly confirm the role of Tregs in inhibiting antitumor CD8+ T cells and consequently in promoting tumor progression, we utilized DEREG mice, which allow for selective and potent depletion of FoxP3+ cells upon systemic administration of diphtheria toxin (Supplementary Fig. S11C; ref. 45). B16-gp33 tumor-bearing control or DEREG mice were injected with diphtheria toxin with or without anti-CD8 (Fig. 6C). Depletion of FoxP3+ cells resulted in controlled tumor growth, and over 50% of mice survived up to 100 days after tumor injection (Fig. 6D). Administration of anti-CD8 partially attenuated the effect of Treg depletion, as demonstrated through increased tumor growth and reduced overall survival (Fig. 6D). Similarly, depletion of FoxP3+ cells resulted in the disappearance of E.G7-Ova tumors and survival of 100% of the mice, and depletion of CD8+ cells reduced the efficacy of Treg depletion (Supplementary Fig. S11D and S11E). Taken together, these data show that Tregs are highly abundant in the tumor microenvironment in this model and play an active role in regulating the antitumor CD8+ T-cell response.
Figure 6.
Tregs inhibit antitumor effector CD8+ T cells in vivo. A and B, Evaluating Treg infiltration in B16-gp33 tumors. Tumor, ndLN, dLN, spleen, and PBMCs were isolated from tumor-bearing mice (day 10 post-tumor injection; n = 5). T-cell lineage and surface markers were analyzed to acquire the proportion of FoxP3+/CD3+ cells (A), and expression of ICOS and CD25 on Treg and CD8+ T cells (B). C, Experimental overview of Treg and CD8+ T-cell depletion in tumor-bearing hosts. D, Mean tumor area and survival of tumor-bearing hosts were measured (n = 10). E–H, Evaluating the effect of Tregs in regulating tumor-specific effector CD8+ T cells. Prestimulated P14 CD8+ T cells were adoptively transferred into control or DEREG mice administered with DT (n = 5; E). Three days after adoptive T-cell transfer, serum was collected from each mouse for LEGENDplex cytokine array (n = 5; F). Prestimulated P14 Thy1.1+CD8+ T cells were adoptively transferred with or without Treg depletion; spleen, dLN, and tumor were isolated on day 7 post–T cell transfer, and adoptively transferred cells were gated for analysis (n = 5; G). 4-1BB and CD25 expression on Thy1.1+ and Thy1.2+ CD8+ T cells were represented in H. Statistical analyses were performed using ANOVA with Holm–Sidak test (A and H), repeated-measure ANOVA with Holm–Sidak test (mean tumor area; D and E), log-rank test (survival; D and E), and multiple Student t test (F; *, P < 0. 05; **, P < 0.01; ns, not significant).
To establish whether Tregs specifically regulate effector CD8+ T cells in the context of tumor immunity, we investigated the ability of Tregs to suppress adoptively transferred prestimulated tumor-specific CD8+ T cells. First, the adoptive transfer of activated P14 CD8+ T cells resulted in delayed tumor growth as anticipated; however, T-cell transfer alone was insufficient to maintain antitumor immunity and only increased survival by approximately 10 days. In contrast, effector CD8+ T-cell transfer in conjunction with Treg depletion eliminated palpable tumors by day 40 and 80% of the mice survived for more than 100 days after tumor inoculation (Fig. 6E), suggesting that the potency of the T-cell therapy may be limited by the presence of Tregs. In our T-cell transfer model, depletion of Tregs did not increase inflammatory cytokines in the serum (Fig. 6F), suggesting that the antitumor immune response likely resulted from local attenuation of immune suppression, rather than bolstering of T-cell responses triggered by systemic inflammation. To conclusively demonstrate that Tregs suppress effector CD8+ T cells in the tumor, activated Thy1.1 P14 CD8+ T cells were adoptively transferred into B16-gp33 tumor-bearing WT or DEREG mice treated with diphtheria toxin, and various organs were harvested on day 7 after T-cell transfer for surface marker analyses. Depletion of Tregs did not increase the expression of 4-1BB or CD25 on adoptively transferred T cells found in the spleen (Fig. 6G and H). In contrast, Treg depletion selectively increased the expression of 4-1BB and CD25 on adoptively transferred P14 CD8+ T cells found in the tumor (Fig. 6G and H), demonstrating the increased activation of tumor-specific T cells directly in the tumor microenvironment. In summary, we demonstrate that effector CD8+ T cells in the tumor microenvironment are susceptible to suppression by Tregs.
Cbl-b deficiency renders effector CD8+ T cells resistant to Tregs in vivo
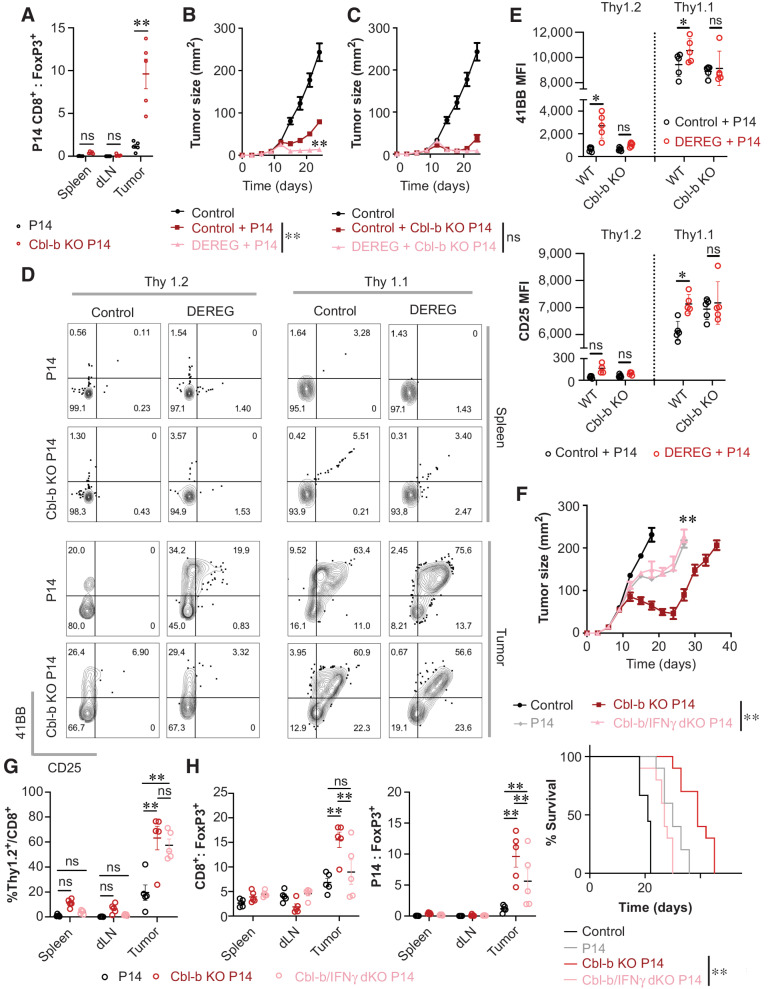
Adoptive transfer of Cbl-b KO P14 CD8+ T cells increased cell infiltration in the tumor and prolonged survival of tumor-bearing mice, in comparison with WT P14 CD8+ T cells (Fig. 1E and F). Furthermore, the average ratio of P14 CD8+ T cells to Tregs increased from 1.16 to 9.62 when using Cbl-b–deficient P14 CD8+ T cells (Fig. 7A). To demonstrate that Cbl-b–deficient CD8+ T cells are less sensitive to the suppressive effects of Tregs in the tumor, Cbl-b–deficient or –sufficient P14 CD8+ T cells were adoptively transferred into B16-gp33 tumor-bearing control or DEREG mice treated with diphtheria toxin and tumor size was monitored. Ablation of FoxP3+ cells further limited the progression of tumor growth when mice were given WT P14 CD8+ T cells (Figs. 6E and 7B). In contrast, the effect of FoxP3+-cell depletion was negligible when tumor-bearing mice were treated with Cbl-b KO P14 CD8+ T cells (Fig. 7C). However, this experiment cannot discriminate the difference between a hyperactive T-cell response and attenuation of immune suppression by Tregs. Thus, we adoptively transferred congenically labeled Cbl-b–deficient or –sufficient P14 CD8+ T cells into tumor-bearing WT or DEREG mice to profile immune infiltrates. Unlike T cells present in the spleen, 61.22% ± 3.49% of adoptively transferred WT P14 CD8+ T cells in the tumor acquired CD25 and 4-1BB coexpression, compared with 58.3% ± 2.63% of Cbl-b KO P14 CD8+ T cells (Fig. 7D). Transient depletion of FoxP3+ cells increased the percentage of CD25+4–1BB+ coexpressing WT P14 Thy1.1+CD8+ cells in the tumor from 61.22% ± 3.49% to 71.78% ± 2.85% (Fig. 7D), whereas the effect of FoxP3+-cell depletion was negligible with Cbl-b KO P14 Thy1.1+CD8+ cells (58.3% ± 2.63% with Tregs; 60.7% ± 6.23% without Tregs; Fig. 7D). Similarly, the median fluorescence intensity (MFI) of CD25 and 4-1BB on WT P14 Thy1.1+CD8+ T cells in the tumor increased in response to depletion of FoxP3+ cells, whereas Treg depletion did not increase the MFI of CD25 and 4-1BB on Cbl-b KO P14 Thy1.1+CD8+ T cells (Fig. 7E). These data suggest that unlike WT T cells, which are susceptible to regulation by Tregs in the tumor, Cbl-b–deficient CD8+ T cells function independently of Tregs.
Figure 7.
Cbl-b KO CD8+ T cells induce robust antitumor immunity in the presence of Tregs through IFNγ. A, The ratio between adoptively transferred tumor-specific CD8+ T cells and Tregs in the tumor. Congenically labeled WT and Cbl-b KO P14 CD8+ T cells were injected into B16-gp33 tumor-bearing WT mice, and spleen, dLN, and tumor were harvested on day 7 after T-cell transfer (n = 5). B and C, Evaluating the role of Tregs in regulating adoptively transferred Cbl-b KO P14 CD8+ T cells. B16-gp33 tumor-bearing C57BL/6 and DEREG mice were treated with WT (B) or Cbl-b KO P14 (C) CD8+ T cells on day 10 after tumor inoculation. Mean tumor area of tumor-bearing hosts was measured (n = 10). D and E, Evaluating the role of Tregs in regulating surface marker expression on adoptively transferred Cbl-b KO P14 CD8+ T cells in tumor. Congenically labeled WT and Cbl-b KO P14 CD8+ T cells were adoptively transferred with or without the depletion of Tregs; spleen and tumor were harvested on day 7 after T-cell transfer to evaluate the proportion of 4-1BB+CD25+ cells among transferred and endogenous CD8+ T cells (n = 5; D). MFI of 4-1BB and CD25 expression on endogenous and transferred CD8+ T cells in the tumor (E). F, The effect of adoptively transferred WT, Cbl-b KO, and Cbl-b/IFNγ double KO P14 CD8+ T cells in B16-gp33 tumor-bearing mice. Mean tumor area and survival of tumor-bearing hosts were measured (n = 10). G and H, Evaluating the infiltration status and T cell-to-Tregs ratio of adoptively transferred P14 Thy1.2+CD8+ T cells. % Thy1.2+/CD8+ (G), total CD8+-to-Treg ratio, and Thy1.2+CD8+-to-Treg ratio (H) were measured 7 days after T-cell transfer. Statistical analyses were performed using Student t test (A) repeated-measure ANOVA with Holm–Sidak test to compare the mean tumor area (B, C, and F), log-rank test (survival; F), and two-way ANOVA with Holm–Sidak test (E, G, and H; *, P < 0. 05; **, P < 0.01; ns, not significant).
In our in vitro studies, Cbl-b–deficient CD8+ T cells hypersecreted IFNγ, which played an important role in attenuating the suppressive effects of Tregs (Fig. 4F and G). To examine the effect of IFNγ hypersecretion in vivo, we adoptively transferred WT, Cbl-b KO, or Cbl-b/IFNγ double KO P14 CD8+ T cells into B16-gp33 tumor-bearing WT mice. The effect of Cbl-b deficiency was abrogated with the deletion of IFNγ, as exemplified through accelerated tumor growth and a decreased survival rate (Fig. 7F). Next, we examined the immune infiltrates by adoptively transferring Thy1.2+ WT, Cbl-b KO, or Cbl-b/IFNγ double KO P14 CD8+ T cells into tumor-bearing Thy1.1+ mice. The proportion of transferred Cbl-b KO and Cbl-b/IFNγ dKO P14 T cells within intratumoral CD8+ T cells was comparable between the two groups and were significantly higher than WT P14 T-cell transfer (Fig. 7G). Thus, the absence of IFNγ in Cbl-b/IFNγ dKO CD8+ T cells does not impair the ability of T cells to infiltrate into the tumor. Instead, IFNγ deletion significantly reduced the ratio between CD8+ T cells and Tregs (Fig. 7H), which demonstrates that one of the main biological consequences of IFNγ overproduction is to improve the balance between T cells and Tregs.
To examine how IFNγ production by Cbl-b–deficient P14 T cells affects the balance between T cells and Tregs in the tumor, we first measured Ki-67 as a surrogate for cellular proliferation. The proportion of Ki-67+Thy1.2+ cells in the tumor were comparable between Cbl-b KO T cells and Cbl-b/IFNγ dKO T cells (Supplementary Fig. S12A and S12B), which suggests that the propensity of the T cells to proliferate in the tumor does not change with IFNγ deficiency. Other studies have suggested that IFNγ may directly modulate antitumor T-cell responses by abrogating the function and/or stability of Tregs (7, 46, 47). Thus, we investigated the effect of adoptively transferred T cells on FoxP3+ Tregs. We found that the percentage of FoxP3+ cells within CD4+ T cells and the percentage of Ki67+ Tregs remained consistent across all experimental groups (Supplementary Fig. S12C–S12E). Furthermore, we examined the expression of Nrp-1 on T cells and Tregs and found that although the production of IFNγ by Cbl-b–deficient T cells downregulated Nrp-1 on the transferred T cells, Nrp-1 expression did not change in FoxP3+ cells (Supplementary Fig. S13A). These data suggest that Cbl-b KO CD8+ T cells hypersecrete IFNγ and counteract the suppressive effects of Tregs without impairing Tregs in the tumor. In summary, we demonstrate that Cbl-b–deficient CD8+ T cells are refractory to the suppressive effects of Tregs in tumor, and the effect of Cbl-b deficiency is dependent on hypersecretion of IFNγ.
Beyond the role of IFNγ in modulating the CD8+ T-cell and Treg balance and promoting potent antitumor immunity, the pleiotropic cytokine is also known to regulate the PD-1/PD-L1 axis on tumor cells as well as other immune infiltrates. Thus, we investigated whether adoptive T-cell therapy using Cbl-b–deficient T cells would benefit from PD-L1 blockade in the B16 tumor model. We examined PD-1 expression on adoptively transferred T cells in the tumor and found that Cbl-b–deficient T cells within the tumor expressed a significantly lower level of PD-1 in comparison with their WT counterparts and that IFNγ deletion partially reversed the effects of Cbl-b deficiency (Supplementary Fig. S14A). Next, we performed adoptive T-cell transfer of WT and Cbl-b KO T cells with or without anti–PD-L1. Cbl-b–deficient T cells enhanced antitumor immunity in comparison with their WT counterparts (Supplementary Fig. S14B); however, anti–PD-L1 failed to improve the therapeutic response (Supplementary Fig. S14C). In summary, we demonstrate that the PD-1/PD-L1 axis plays a limited role in constraining the antitumor effects of Cbl-b–deficient T cells in B16 melanoma.
Discussion
Despite our limited understanding of how Tregs directly affect antitumor CD8+ T cells in vivo, numerous strategies have been proposed to disrupt the stability or function of intratumoral Tregs, or to attenuate the inhibitory signals processed by CD8+ T cells (9, 12). In our study, we explored the underlying mechanism of how Cbl-b–deficient CD8+ T cells are resistant to the suppressive effects of Tregs both in vitro and in vivo. We showed that ablation of Cbl-b results in modification in the transcriptome of CD8+ T cells, favoring cellular division and cytokine secretion. In conjunction with their hyperproliferative capacity, hypersecretion of IFNγ was shown to be a key factor that attenuated the suppressive signals by Tregs. IFNγ produced by Cbl-b–deficient T cells had marked effects on the T-cell transcriptome and activation, whereas the cytokine had marginal effects on the suppressor function of Tregs. To explore these findings in vivo, we first confirmed that immunosuppressive intratumoral Tregs suppress effector CD8+ T cells, illuminating what was previously an assumption. To abrogate the suppressive effects of these Tregs, we deleted Cbl-b in effector CD8+ T cells in an adoptive transfer model. These cells performed well independent of Tregs and induced robust antitumor immunity through the secretion of IFNγ.
The biology of rendering T cells refractory to Treg-mediated suppression is poorly understood, because enhanced T-cell activation cannot be easily distinguished from attenuation of immune suppression. Our data suggest that ablation of Cbl-b simultaneously strengthens CD8+ T-cell activation, while rendering them less sensitive to the suppressive effects of Tregs. Among the studies that have specifically examined CD8+ T-cell resistance to Tregs, Loeser and colleagues also found that Cbl-b KO CD8+ T cells are less sensitive to the suppressive effects of Tregs in an in vitro Treg suppression assay (24). Similarly, ablation of SHP-1, a protein tyrosine phosphatase that dephosphorylates Cbl-b upon TCR stimulation to prevent ubiquitination and degradation of Cbl-b, renders CD8+ T cells less sensitive to Treg-mediated suppression (48). In these studies, CD8+ T cells have also increased T-cell proliferation while decreasing the percentage of suppression score (24, 48). Such observations are consistent with the literature, which suggests that Cbl-b amplifies the TCR signaling pathway while abrogating specific inhibitory signaling pathways (9). For instance, Cbl-b ubiquitinates and subsequently downregulates SMAD7, an attenuator of TGFβ receptor signaling (39). Cbl-b–deficient T cells display reduced sensitivity to TGFβ-mediated inhibition (13, 22), and deletion of SMAD7 in Cbl-b–deficient T cells restored sensitivity to TGFβ (39). Thus, the ability of CD8+ T cells to attenuate inhibitory signals likely synergizes with enhanced T-cell activation to overcome Treg-mediated suppression. Our study has taken further steps to demonstrate that this particular phenotype is dependent on IFNγ.
Evidence from studies including ours collectively demonstrates that Cbl-b serves as a powerful regulator of T-cell activation and controls the susceptibility to suppression by Tregs (9, 13, 14, 22, 23). However, the mechanism of how Cbl-b regulates T-cell activation is often generalized for both CD4+ and CD8+ T cells. A recent study published by our group specifically focuses on the role of Cbl-b in CD4+ T-cell resistance to Tregs (23). In the study, Cbl-b–deficient CD4+ T cells display amplified TCR signaling and reduced sensitivity to Treg-mediated suppression (23). Although a few other groups also suggest that Cbl-b–deficient CD4+ T cells upregulate IL2 (14, 20, 21), our group has specifically demonstrated that increased IL2 is the key mechanism mediating Treg resistance in Cbl-b KO CD4+ T cells (23). Cbl-b–deficient CD8+ T cells share some characteristics with Cbl-b–deficient CD4+ T cells including increased proliferation, decreased sensitivity to Treg-mediated suppression and hypersecretion of cytokines. However, the mechanism by which Cbl-b deficiency renders CD8+ T cells resistant to Tregs is exclusive to CD8+ T cells. Previous studies have shown that CD8+ T cells deficient in Cbl-b upregulate IFNγ (36, 49, 50). Here, we demonstrate that IFNγ selectively renders CD8+ T cells resistant to Treg-mediated suppression. This is in contrast to CD4+ T cells where increased IFNγ does not alter proliferation in the presence of Tregs. In summary, although Cbl-b serves as a promising target to strengthen T-cell activation and attenuate inhibitory signals, the biology of Cbl-b may not always be generalizable across both CD4+ and CD8+ T-cell compartments.
Adoptive T-cell therapy (ACT) is being used to treat multiple malignancies. However, it remains unclear whether adoptively transferred tumor-specific T cells, previously stimulated ex vivo, are susceptible to regulation by Tregs in vivo. In a clinical setting in which nonmyeloablative chemotherapy was followed by tumor-infiltrating lymphocyte (TIL) transfer and IL2 administration, the proportion of reconstituted CD4+FoxP3+ Tregs in blood after treatment was found to negatively correlate with responsiveness to the therapy (51). In mouse studies, Tregs regulate antitumor CD8+ T-cell responses in syngeneic tumor models (37, 38, 52). Although the differentiation of effector CD8+ T cells can be attenuated through the modulation of IL2 homeostasis by Tregs (53, 54), the question remains whether ex vivo–primed effector CD8+ T cells are also susceptible to regulation by Tregs in the context of tumor immunity. This scenario is relevant for ACT, including ACT using TCR-transduced T cells and CAR-T cells. Here, we demonstrate that adoptively transferred tumor-specific effector CD8+ T cells acquire the effector phenotype of CD25 and 41BB expression specifically in the tumor, and their regulation by Tregs also take place in the tumor. By establishing an in vivo system to capture the interaction between effector CD8+ T cells and Tregs in the tumor microenvironment, we demonstrate that ablation of Cbl-b renders effector CD8+ T cells impartial to the suppressive effects of Tregs through the effects of IFNγ.
In summary, we have defined a mechanism that renders CD8+ T cells resistant to the inhibitory effects of Tregs by targeting Cbl-b in CD8+ T cells. Such a strategy could be potentially incorporated in cancer treatment modalities including CAR-T cell therapy or TCR-engineering–based immunotherapy to generate robust effector T-cell responses and antitumor immunity.
Supplementary Material
Acknowledgments
This research was supported by a Foundation Grant from Canadian Institute for Health Research (CIHR-FDN 143220) to P.S. Ohashi. We thank the Animal Resource Centre as well as Princess Margaret Cancer Centre flow facility for technical support. Special acknowledgments are made for Dr. Tak Mak and Dr. Naoto Hirano for their suggestions and insights for this project. The authors declare no competing financial interests.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
S. Han: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. Z. Liu: Data curation, formal analysis, validation, investigation, visualization. D.C. Chung: Data curation, formal analysis, visualization. M. St. Paul: Resources, formal analysis, investigation, writing–review and editing. C.R. Garcia-Batres: Data curation, formal analysis, validation, investigation, visualization, methodology. A. Sayad: Data curation, software, formal analysis, validation, visualization, methodology. A.R. Elford: Resources, methodology. M.J. Gold: Resources, methodology. N. Grimshaw: Resources, methodology. P.S. Ohashi: Conceptualization, resources, supervision, funding acquisition, project administration, writing–review and editing.
References
- 1. Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell 2008;133:775–87. [DOI] [PubMed] [Google Scholar]
- 2. Sakaguchi S, Mikami N, Wing JB, Tanaka A, Ichiyama K, Ohkura N. Regulatory T cells and human disease. Annu Rev Immunol 2020;38:541–66. [DOI] [PubMed] [Google Scholar]
- 3. Togashi Y, Shitara K, Nishikawa H. Regulatory T cells in cancer immunosuppression—implications for anticancer therapy. Nat Rev Clin Oncol 2019;16:356–71. [DOI] [PubMed] [Google Scholar]
- 4. Nishikawa H, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Curr Opin Immunol 2014;27:1–7. [DOI] [PubMed] [Google Scholar]
- 5. Shevach EM. Foxp3+ T regulatory cells: still many unanswered questions—a perspective after 20 years of study. Front Immunol 2018;9:1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arce Vargas F, Furness AJS, Solomon I, Joshi K, Mekkaoui L, Lesko MH, et al. Fc-optimized anti-CD25 depletes tumor-infiltrating regulatory T cells and synergizes with PD-1 blockade to eradicate established tumors. Immunity 2017;46:577–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Overacre-Delgoffe AE, Chikina M, Dadey RE, Yano H, Brunazzi EA, Shayan G, et al. Interferon-γ drives Treg fragility to promote anti-tumor immunity. Cell 2017;169:1130–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang D, Quiros J, Mahuron K, Pai CC, Ranzani V, Young A, et al. Targeting EZH2 reprograms intratumoral regulatory T cells to enhance cancer immunity. Cell Rep 2018;23:3262–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Han S, Toker A, Liu ZQ, Ohashi PS. Turning the tide against regulatory T cells. Front Oncol 2019;9:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stephens LA, Gray D, Anderton SM. CD4+CD25+ regulatory T cells limit the risk of autoimmune disease arising from T cell receptor crossreactivity. Proc Natl Acad Sci U S A 2005;102:17418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim J, Lahl K, Hori S, Loddenkemper C, Chaudhry A, DeRoos P, et al. Cutting edge: depletion of Foxp3+ cells leads to induction of autoimmunity by specific ablation of regulatory T cells in genetically targeted mice. J Immunol 2009;183:7631–4. [DOI] [PubMed] [Google Scholar]
- 12. Mercadante ER, Lorenz UM. Breaking free of control: How conventional T cells overcome regulatory T cell suppression. Front Immunol 2016;7:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wohlfert EA, Callahan MK, Clark RB. Resistance to CD4+CD25+ regulatory T cells and TGF-beta in Cbl-b-/- mice. J Immunol 2004;173:1059–65. [DOI] [PubMed] [Google Scholar]
- 14. Adams CO, Housley WJ, Bhowmick S, Cone RE, Rajan TV, Forouhar F, et al. Cbl-b(-/-) T cells demonstrate in vivo resistance to regulatory T cells but a context-dependent resistance to TGF-beta. J Immunol 2010;185:2051–8. [DOI] [PubMed] [Google Scholar]
- 15. Fang D, Liu Y-C. Proteolysis-independent regulation of PI3K by Cbl-b–mediated ubiquitination in T cells. Nat Immunol 2001;2:870–5. [DOI] [PubMed] [Google Scholar]
- 16. Liu Y-C. Ubiquitin ligases and the immune response. Annu Rev Immunol 2004;22:81–127. [DOI] [PubMed] [Google Scholar]
- 17. Qiao G, Li Z, Molinero L, Alegre M-L, Ying H, Sun Z, et al. T-cell receptor-induced NF-B activation is negatively regulated by E3 ubiquitin ligase Cbl-b. Mol Cell Biol 2008;28:2470–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo H, Qiao G, Ying H, Li Z, Zhao Y, Liang Y, et al. E3 Ubiquitin ligase Cbl-b regulates Pten via Nedd4 in T cells independently of its ubiquitin ligase activity. Cell Rep 2012;1:472–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lutz-Nicoladoni C, Wolf D, Sopper S. Modulation of immune cell functions by the E3 ligase Cbl-b. Front Oncol 2015;5:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bachmaier K, Krawczyk C, Kozieradzki I, Kong Y-Y, Sasaki T, Oliveira-dos-Santos A, et al. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature 2000;403:211–6. [DOI] [PubMed] [Google Scholar]
- 21. Chiang YJ, Kole HK, Brown K, Naramura M, Fukuhara S, Hu R-J, et al. Cbl-b regulates the CD28 dependence of T-cell activation. Nature 2000;403:216–20. [DOI] [PubMed] [Google Scholar]
- 22. Wohlfert EA, Gorelik L, Mittler R, Flavell RA, Clark RB. Cutting edge: deficiency in the E3 Ubiquitin ligase Cbl-b results in a multifunctional defect in T cell TGF-beta sensitivity in vitro and in vivo. J Immunol 2006;176:1316–20. [DOI] [PubMed] [Google Scholar]
- 23. Han S, Chung DC, St. Paul M, Liu ZQ, Garcia-Batres C, Tran CW, et al. Overproduction of IL-2 by Cbl-b deficient CD4+ T cells provides resistance against regulatory T cells. Oncoimmunology 2020;9:e1737368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loeser S, Loser K, Bijker MS, Rangachari M, van der Burg SH, Wada T, et al. Spontaneous tumor rejection by cbl-b–deficient CD8+ T cells. J Exp Med 2007;204:879–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pircher H, Bürki K, Lang R, Hengartner H, Zinkernagel RM. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature 1989;342:559–61. [DOI] [PubMed] [Google Scholar]
- 26. Ohashi P, Oehen S, Buerki K, Pircher H, Ohashi CT, Odermatt B, et al. Ablation of “tolerance” and induction of diabetes by virus infection in viral antigen transgenic mice. Cell 1991;65:305–17. [DOI] [PubMed] [Google Scholar]
- 27. Hanahan D. Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature 1985;315:115–22. [DOI] [PubMed] [Google Scholar]
- 28. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019;47:D766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 2015;12:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 2009;4:1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 1995;57:289–300. [Google Scholar]
- 35. Delisle J-S, Giroux M, Boucher G, Landry J-R, Hardy M-P, Lemieux S, et al. The TGF-β-Smad3 pathway inhibits CD28-dependent cell growth and proliferation of CD4 T cells. Genes Immun 2013;14:115–26. [DOI] [PubMed] [Google Scholar]
- 36. Chiang JY, Jang IK, Hodes R, Gu H. Ablation of Cbl-b provides protection against transplanted and spontaneous tumors. J Clin Invest 2007;117:1029–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klages K, Mayer CT, Lahl K, Loddenkemper C, Teng MWL, Ngiow SF, et al. Selective depletion of Foxp3+ regulatory T cells improves effective therapeutic vaccination against established melanoma. Cancer Res 2010;70:7788–99. [DOI] [PubMed] [Google Scholar]
- 38. Teng MWL, Ngiow SF, Von Scheidt B, Mclaughlin N, Sparwasser T, Smyth MJ. Conditional regulatory T-cell depletion releases adaptive immunity preventing carcinogenesis and suppressing established tumor growth. Cancer Res 2010;70:7800–9. [DOI] [PubMed] [Google Scholar]
- 39. Gruber T, Hinterleitner R, Hermann-Kleiter N, Meisel M, Kleiter I, Wang CM, et al. Cbl-b mediates TGFβ sensitivity by downregulating inhibitory SMAD7 in primary T cells. J Mol Cell Biol 2013;5:358–68. [DOI] [PubMed] [Google Scholar]
- 40. Ulloa L, Doody J, Massagué J. Inhibition of transforming growth factor-β/SMAD signalling by the interferon-γ/STAT pathway. Nature 1999;397:710–3. [DOI] [PubMed] [Google Scholar]
- 41. Higashi K, Inagaki Y, Fujimori K, Nakao A, Kaneko H, Nakatsuka I. Interferon-gamma interferes with transforming growth factor-beta signaling through direct interaction of YB-1 with Smad3. J Biol Chem 2003;278:43470–9. [DOI] [PubMed] [Google Scholar]
- 42. De Simone M, Arrigoni A, Rossetti G, Gruarin P, Ranzani V, Politano C, et al. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity 2016;45:1135–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Plitas G, Konopacki C, Wu K, Bos PD, Morrow M, Putintseva EVV, et al. Regulatory T cells exhibit distinct features in human breast cancer. Immunity. 2016;45:1122–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Toker A, Nguyen LT, Stone SC, Cindy Yang S, Rachel Katz S, Shaw PA, et al. Regulatory T cells in ovarian cancer are characterized by a highly activated phenotype distinct from that in melanoma. Clin Cancer Res 2018;24:5685–96. [DOI] [PubMed] [Google Scholar]
- 45. Lahl K, Sparwasser T. In vivo depletion of FoxP3+ Tregs using the DEREG mouse model. Methods Mol Biol. 2011;707:157–72. [DOI] [PubMed] [Google Scholar]
- 46. Caretto D, Katzman SD, Villarino AV, Gallo E, Abbas AK. Cutting edge: the Th1 response inhibits the generation of peripheral regulatory T cells. J Immunol 2010;184:30–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Castro F, Cardoso AP, Gonçalves RM, Serre K, Oliveira MJ. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front Immunol 2018;9:847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mercadante ER, Lorenz UM. T cells deficient in the tyrosine phosphatase SHP-1 resist suppression by regulatory T cells. J Immunol 2017;199:129–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shamim M, Nanjappa SG, Singh A, Plisch EH, LeBlanc SE, Walent J, et al. Cbl-b regulates antigen-induced TCR down-regulation and IFN-gamma production by effector CD8 T cells without affecting functional avidity. J Immunol 2007;179:7233–43. [DOI] [PubMed] [Google Scholar]
- 50. Stromnes IM, Blattman JN, Tan X, Jeevanjee S, Gu H, Greenberg PD. Abrogating Cbl-b in effector CD8(+) T cells improves the efficacy of adoptive therapy of leukemia in mice. J Clin Invest 2010;120:3722–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yao X, Ahmadzadeh M, Lu Y-C, Liewehr DJ, Dudley ME, Liu F, et al. Levels of peripheral CD4(+)FoxP3(+) regulatory T cells are negatively associated with clinical response to adoptive immunotherapy of human cancer. Blood 2012;119:5688–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Onizuka S, Tawara I, Shimizu J, Sakaguchi S, Fujita T, Nakayama E. Tumor rejection by in vivo administration of anti-CD25 (interleukin-2 receptor alpha) monoclonal antibody. Cancer Res 1999;59:3128–33. [PubMed] [Google Scholar]
- 53. McNally A, Hill GR, Sparwasser T, Thomas R, Steptoe RJ. CD4+CD25+ regulatory T cells control CD8+ T-cell effector differentiation by modulating IL-2 homeostasis. Proc Natl Acad Sci U S A 2011;108:7529–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chinen T, Kannan AK, Levine AG, Fan X, Klein U, Zheng Y, et al. An essential role for the IL-2 receptor in Treg cell function. Nat Immunol 2016;17:1322–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are publicly available in Gene-Expression Omnibus at GSE189350.