Abstract
Purpose:
The heterogeneity of high-grade serous ovarian cancer (HGSOC) is not well studied, which severely hinders clinical treatment of HGSOC. Thus, it is necessary to characterize the heterogeneity of HGSOC within its tumor microenvironment (TME).
Experimental Design:
The tumors of 7 treatment-naïve patients with HGSOC at early or late stages and five age-matched nonmalignant ovarian samples were analyzed by deep single-cell RNA sequencing (scRNA-seq).
Results:
A total of 59,324 single cells obtained from HGSOC and nonmalignant ovarian tissues were sequenced by scRNA-seq. Among those cells, tumor cells were characterized by a set of epithelial-to-mesenchymal transition (EMT)-associated gene signatures, in which a combination of NOTCH1, SNAI2, TGFBR1, and WNT11 was further selected as a genetic panel to predict the poor outcomes of patients with HGSOC. Matrix cancer-associated fibroblasts (mCAF) expressing α-SMA, vimentin, COL3A, COL10A, and MMP11 were the dominant CAFs in HGSOC tumors and could induce EMT properties of ovarian cancer cells in the coculture system. Specific immune cell subsets such as C7-APOBEC3A M1 macrophages, CD8+ TRM, and TEX cells were preferentially enriched in early-stage tumors. In addition, an immune coinhibitory receptor TIGIT was highly expressed on CD8+ TEX cells and TIGIT blockade could significantly reduce ovarian cancer tumor growth in mouse models.
Conclusions:
Our transcriptomic results analyzed by scRNA-seq delineate an ecosystemic landscape of HGSOC at early or late stages with a focus on its heterogeneity with TME. The major applications of our findings are a four–EMT gene model for prediction of HGSOC patient outcomes, mCAFs’ capability of enhancing ovarian cancer cell invasion and potential therapeutic value of anti-TIGIT treatment.
Translational Relevance.
High-grade serous ovarian cancer (HGSOC) accounts for approximately 80% of ovarian cancer total deaths. Currently, there is still a lack of effective clinical strategies for treatment of HGSOC. Our studies focus on the genetic expression features of different cell populations of HGSOC in its various tumor stages, which will contribute to develop novel treatment strategies. In the current study, deep single-cell RNA sequencing (scRNA-seq) was performed to analyze tumors from 7 treatment-naïve patients with HGSOC at different stages and five age-matched nonmalignant ovarian samples. Among the differentially expressed genes being identified by scRNA-seq, a combination of epithelial-to-mesenchymal transition (EMT) markers NOTCH1, SNAI2, TGFBR1, and WNT11 was selected and this panel could be applied to predict the poor patient outcomes. Moreover, the effects of primary cancer-associated fibroblasts were confirmed to induce tumor cell EMT. In addition, we examined the effects of immune coinhibitory receptor TIGIT on ovarian cancer and found that TIGIT blockade could alleviate tumor burden in ID8 tumor-bearing mice, suggesting a potential immunotherapy target for HGSOC treatment.
Introduction
Ovarian cancer is the most lethal gynecologic malignancy, with 184,799 female deaths annually worldwide (1, 2). Among all ovarian cancer subtypes, high-grade serous ovarian cancer (HGSOC) is the most common phenotype, which is responsible for approximately 80% of all ovarian cancer deaths (3). Because of the ambiguous symptoms, only 20% cases of this malignancy can be identified at the early stages for successful treatment. Most HGSOC progresses into the later advanced stages with poor patient outcomes, which reflects the aggressive nature of this disease (4, 5). Even after therapy, HGSOC often recurs due to the development of chemoresistance, with an overall 5-year survival probability of 31% (6, 7). Therefore, development of effective treatments for patients with HGSOC is imperative for better prognosis and overall survival (OS).
HGSOC appears several features. One of them is the epithelial-to-mesenchymal transition (EMT) process (7, 8), which is a major mechanism underlying ovarian cancer invasion and metastasis, as well as chemoresistance (9, 10). Patients with HGSOC with activated EMT transcriptional program exhibited worse outcomes (11). EMT inhibition by simvastatin suppresses cancer cell metastasis and invasion (12). Another hallmark is the tumor heterogeneity (13), characterized by its tumor microenvironment (TME; ref. 14). The single-cell RNA sequencing (scRNA-seq) is a powerful analysis tool to disclose TME information of HGSOCs. For instance, a previous study using scRNA-seq showed that the inhibition of the JAK/STAT pathway had potent antitumor activities (15). Another work provided broad characterization of the cellular composition associated with three tumor immune phenotypes (i.e., infiltrated, excluded, or desert; ref. 16). A recent report characterized certain stromal cell phenotypes in TME regulation in high-grade serous tubo-ovarian cancers, such as TGFβ-driven cancer-associated fibroblasts (CAF), lymphatic endothelial cells, and mesothelial cells (17).
One approach to regulate TME is via mediating immune cell behaviors and programmed cell death protein 1 ligand (PD-L1), which is pivotal checkpoint element of T-cell suppression (18), have been studied for immunotherapies against ovarian cancer but with limited impacts. One major reason is that PD-L1 was upregulated in only approximately 1.3% (14/1,052) of patients with ovarian epithelial carcinoma, indicating that this approach could not account for most of ovarian cancer cases (19). Thus, it is significantly beneficial to disclose cellular features at different HGSOC stages by scRNA-seq, which provides more precise information for developing appropriate immunotherapies targeting specific disease stages.
Here, we characterized in detail the comprehensive single-cell transcriptomic landscape of primary HGSOC tumors at early and advanced clinical stages. The epithelial cell developmental hierarchies were described by pseudotime analysis at different tumor stages, and properties of EMT were identified. Particularly, in the majority of CAFs in HGSOC tissues, matrix CAFs (mCAF) significantly enhanced invasive ability of ovarian cancer cells. Other immune cells, including C7-APOBEC3A M1 macrophages, CD8+ TRM cells, and CD8+ TEX cells, were mainly expressed in early-stage tumors. In addition, immune coinhibitory receptor TIGTI blockade suppressed tumor growth of ID8-derived C57BL/6 mice models. Taken together, our study reported the unique TME aspects of HGSOC and tumor cell features associated with tumor stages, which will help to develop new clinical strategies for the HGSOC treatment.
Materials and Methods
Human specimens and ethical approval
HGSOC tumor samples were collected from patients who had undergone bilateral salpingo-oophorectomy (BSO)/hysterectomy + comprehensive staging or debulking at Women's Hospital of Zhejiang University. Nonmalignant ovarian samples were collected from patients who underwent unilateral salpingo-oophorectomy or BSO, respectively and/or hysterectomy because of benign gynecologic diseases at Women's Hospital of Zhejiang University. We obtained the normal part of the nonmalignant ovaries. Fresh tissues were immediately dissected into fractions for enzymatic digestion into single cells and fixed in 4% paraformaldehyde solution followed by paraffin embedding.
This study was conducted in accordance with Declaration of Helsinki and approved by the Institutional Review Board (IRB) of Women's Hospital of Zhejiang University (IRB-20200186-R). Written informed consent was obtained from each patient.
Single-cell dissociation
scRNA-seq experiments were performed by experimental personnel in the laboratory of Novel Bio Co, Ltd. The tissues were surgically removed and kept in MACS Tissue Storage Solution (Miltenyi Biotec) until processing. Tissue samples were processed as described below. Briefly, samples were first washed with PBS, minced into small pieces (∼1 mm3) on ice, and enzymatically digested with 125 U/mL collagenase IV (Sigma), 25 U/mL collagenase I (Sigma), and 25 U/mL DNase I (Worthington) for 30 minutes at 37°C, with agitation. After digestion, samples were sieved through a 40-μm cell strainer, and then centrifuged at 300 × g for 5 minutes. The supernatant was removed and the pelleted cells were suspended in red blood cell lysis buffer (Solarbio) to lyse the red blood cells. The cells were resuspended in RPMI1640 medium containing 10% FBS and refiltered through a 35-μm cell strainer. Dissociated single cells were then stained with acridine orange/propidium iodide for viability assessment using a Countstar Fluorescence Cell Analyzer. The single-cell suspension was further enriched with a MACS dead cell removal kit (Miltenyi Biotec).
Single-cell sequencing
The scRNA-seq libraries were generated using the 10× Genomics Chromium Controller Instrument and Chromium Single Cell 3′ V3 Reagent Kits (10× Genomics). Briefly, cells were concentrated to 1,000 cells/μL and approximately 8,000–10,000 cells were loaded into each channel to generate single-cell gel bead-in-emulsions (GEM), which resulted in the expected mRNA barcoding of 3,000–8,000 single cells for each sample. After the reverse transcription step, GEMs were broken and barcoded cDNA was purified and amplified. The amplified barcoded cDNA was fragmented, A-tailed, ligated with adaptors and index PCR amplified. The final libraries were quantified using a Qubit High Sensitivity DNA assay (Thermo Fisher Scientific) and the size distribution of these libraries was determined by a High Sensitivity DNA chip on a Bioanalyzer 2200 (Agilent). All libraries were then sequenced by an Illumina sequencer (Illumina) on a 150 bp paired-end run.
Single-cell RNA statistical analysis
scRNA-seq data analysis was performed by NovelBio Co. Ltd. with the NovelBrain Cloud Analysis Platform (www.novelbrain.com). We applied fastp with default parameter filtering of the adaptor sequence and removed the low-quality reads and short reads to obtain clean data (20). Then, feature-barcode matrices were obtained by aligning reads to the human genome (GRCh38 Ensemble: version 91) using CellRanger v3.1.0 and determining the real cell parameters expect for those with cell counts equal to 10,000 by considering the UMI and Cell-UMI Slope. Furthermore, to solve the bias caused by unbalanced sequencing, we applied the downsample analysis by the CellRanger Aggr function among samples sequenced according to the mapped barcoded reads per cell of each sample and finally achieved the aggregated matrix. Cells containing over 200 expressed genes and a tissue specific mitochondrial UMI rate (below 40%) passed cell quality filtering, and mitochondrial genes were removed from the expression table. The Seurat package (version: 3.1.4, https://satijalab.org/seurat/) was used for cell normalization and regression based on the expression table according to the UMI counts of each sample and the percentage of mitochondria to obtain the scaled data. Principal component analysis was constructed on the basis of the scaled data with the top 2,000 highly variable genes, and the top 10 principals were used for uniform manifold approximation and projection (UMAP) construction. Canonical correlation analysis (CCA) in the Seurat package was applied for batch effect removal and primary clustering. Utilizing the graph-based cluster method, we acquired the unsupervised cell cluster result based on the CCA top 10 principal and we calculated the marker genes by the FindAllMarkers function with Wilcoxon rank-sum test algorithm under the following criteria: (i) logFC > 0.25; (ii) P < 0.05; and (iii) min.PCT > 0.1. Cell type was defined by knowledge-based biomarkers. For the subclustering of each cell type, considering that the CCA algorithm performed worse among the batch effect algorithms caused by overcorrection, we applied the MNN algorithm from the scran package (http://www.bioconductor.org/packages/release/bioc/html/scran.html) for subclustering batch effect analysis with a k-value equal to 5. On the basis of the MNN results, graph-based clustering with optimal parameters and marker analysis as above was utilized for cell clustering and identification. We applied GSVA (1.32.0; ref. 21) for immune-related scoring analysis, and utilized the Wilcox rank-sum test to calculate the significance between early-stage and later-stage tumors. Padj values <0.01 were selected as statistically significant.
CAFs and ovarian cancer cells transwell coculture analysis
The human ovarian cancer cell line OVCAR3 was obtained from the ATCC, and A2780 was obtained from Sigma. These cell lines were verified by short tandem repeat and without Mycoplasma contamination. OVCAR3 cells were grown in DMEM containing 10% FBS and 1% penicillin/streptomycin. A2780 cells were grown in RPMI1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin.
To determine the effect of CAFs on ovarian cancer cell EMT via a Transwell coculture system (Corning, 3460), 5 × 104 CAFs were placed in the upper insert with 0.4-μm pore size polycarbonate membranes, and the same amount of ovarian cancer cells was seeded beneath the transwell insert of the 12-well plates. Cells were cocultured for 48 hours. The ovarian cancer cells were further examined by Western blot analysis for the expression of ZEB1 (Cell Signaling Technology, 3396, 1:1,000), vimentin (Cell Signaling Technology, 5741, 1:5,000), snail (Cell Signaling Technology, 3879, 1:1,000), and GAPDH (Santa Cruz Biotechnology, sc47727, 1:4,000). The protein bands on the blots were quantified by Quantity One software (Bio-Rad Laboratories, Inc). The data represent three independent experiments (mean ± SD).
For the transwell assay, 5 × 104 CAFs were first cultured for 24 hours. The same amount of ovarian cancer cells was seeded into the upper well of 8-μm pore size PET track-etched membranes (FALCON, 353097) coated with Matrigel (Corning, 356234). After coincubation for 24 hours, the cells that invaded to the opposite side of the filter were stained with 0.5% crystal violet, and counted under the macroscope (Leica DMI 4000B). The data represent three independent experiments (mean ± SD).
Survival analysis in The Cancer Genome Atlas and Gene Expression Omnibus datasets
To assess the prognostic effect of individual genes or each set of signature genes derived from specific clusters, TCGA-OV data, GSE26712 (22), GSE13876 (23), and GSE9891 (24) were employed. The TCGA-OV gene expression data and survival data were downloaded from UCSC Xena (https://xenabrowser.net/datapages/). While evaluating the effect of each set of signature genes on survival, the mean expression of signature genes was scaled by zscores to represent the relative expression level of signature genes. For all survival analyses, patients were grouped into high and low expression groups by the optimal cutoff. Kaplan–Meier survival curves were drawn in R-4.0.3 with the R package survminer (25).
Animal study
All experimental mice were housed in specific pathogen-free conditions and all animal procedures were approved by the Institutional Animal Care and Use Committee of Zhejiang Chinese Medical University (20211129-21). Six to eight weeks old female C57BL/6 mice were inoculated subcutaneously with 5 × 106 ID8 cells. The ID8 cells were gifted from the laboratory of Prof. Chen Dong (Qinghua University, Beijing, P.R. China). Tumor growth was measured with calipers regularly and the volume was calculated as 0.5 × length × width2. On day 56 (average tumors reached 100 mm3), mice were randomized into treatment group and treated with anti-TIGIT (200 μg, clone:1B4, Absolute Antibody) or isotype-matched control antibody (200 μg) by intraperitoneal injection five times (once every 3 days). On day 76, mice were sacrificed for downstream analyses. Tumor tissues were minced, and digested with collagenase IV (1 mg/mL, Sigma) and DNase I (1 mg/mL, Roche) in RPMI1640 (Gibco) for 1 hour at 37°C. The resulting cell suspensions were filtered through a 70-μm cell strainer prior to centrifugation on a discontinuous Percoll gradient (GE Healthcare Life Sciences). Isolated cells were then used in flow cytometry.
Data availability
scRNA-seq data that support the findings of this study have been deposited on Gene Expression Omnibus (GEO) platform under the accession code "GSE184880."
Statistical analyses
For cellular experiments and animal study, statistical analysis was performed using GraphPad Prism 9.2. Comparisons were assessed using Student t test or one-way ANOVA. Two-way ANOVA was used to compare the tumor growth curves. Data are presented as mean ± SD. P <0.05 was considered as statistically significant.
Results
Single-cell profiling of nonmalignant ovarian and primary HGSOC tumor ecosystems
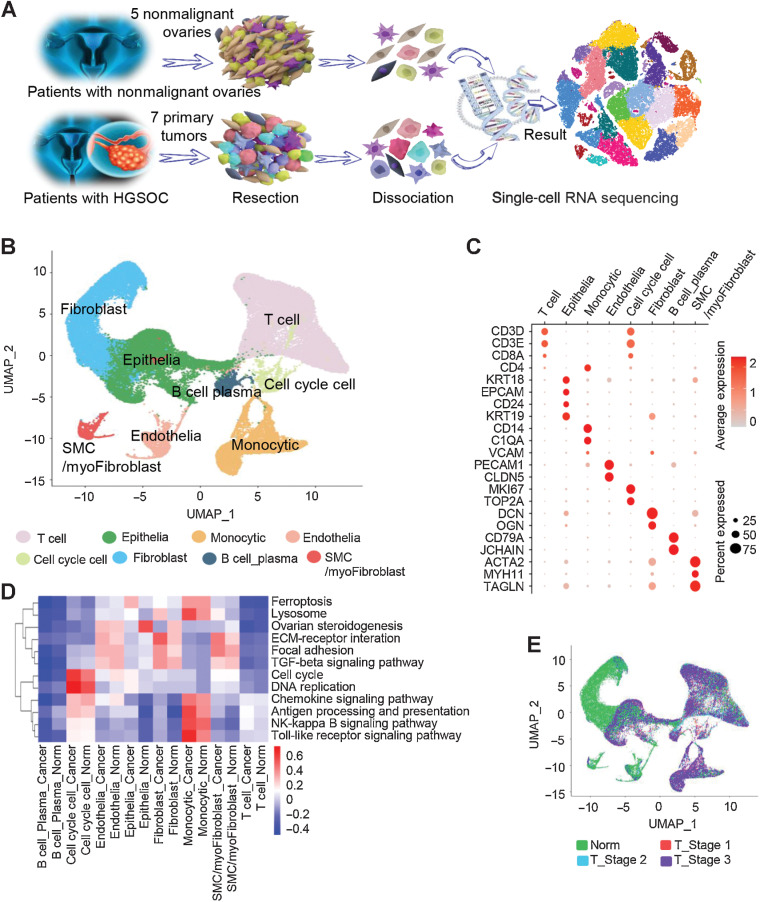
To systematically interrogate the intratumoral heterogeneity of HGSOC, we performed deep scRNA-seq on individual ovarian cells from 12 treatment-naïve patients, including 7 patients with HGSOC and 5 age-matched patients with nonmalignant ovaries (Supplementary Table S1). The nonmalignant ovaries analyzed here were from perimenopausal or postmenopausal women as suitable age-matched HGSOC controls. A total of 59,324 cells were acquired from these samples following standard procedures (Materials and Methods). Of these, 33,264 cells (56%) were from HGSOC tumors and 26,060 (44%) were from nonmalignant ovaries (Fig. 1A;Supplementary Fig. S1A and S1B; Supplementary Table S2). These cells were then clustered (Materials and Methods) and annotated according to the established gene marker list. Visualization of the cells was performed using UMAP approaches, as shown in Fig. 1B. Furthermore, Fig. 1C demonstrates the identification of ecosystematic cell types, including T-cell lineages (marked by CD3D, CD3E, and CD8A), epithelial cells (marked by KRT18, EPCAM, CD24, and KRT19), monocytes (CD14 and C1QA), endothelial cell types (PECAM1 and CLDN5), cell-cycle cells (MKI67 and TOP2A), fibroblast cells (DCN and OGN), B cell and plasma cell types (CD79A and JCHAIN), and smooth muscle cells/myofibroblasts (featuring as ACTA2, MYH11, and TAGLN).
Figure 1.
Diverse cell types in HGSOC and nonmalignant ovarian tissues delineated by single-cell transcriptomic analysis. A, Workflow depicting the collection and processing of specimens of HGSOC tumors and nonmalignant ovarian tissues for scRNA-seq. B, The UMAP plot demonstrates the main cell types in HGSOC and control ovarian tissues. C, Dot plots showing the expression levels of specific marker genes in each cell type. The size of dots indicates the proportion of cells expressing the particular marker gene. The spectrum of color represents the mean expression levels of the marker genes. D, Heatmap showing differentially activated pathways of each cell type in the HGSOC and nonmalignant groups. E, Cell composition distribution for each group with different clinical stages.
The cell subset types are similar in HGSOC tumors and nonmalignant ovaries (Supplementary Fig. S1C), while the cellular distribution was quite different. For instance, the nonmalignant ovaries had a predominance of fibroblasts, consistent with the increased ovarian fibrosis in aged ovaries (26). In contrast, the tumors contained more T cells or epithelial cells. The Kyoto Encyclopedia of Genes and Genomes pathway analysis was then applied to reveal the features for each cell subset (Fig. 1D). For the epithelial cells of HGSOC tumors, the analysis showed an enrichment of genes involved in ferroptosis, which is consistent with previous findings (27, 28). For those in nonmalignant ovarian tissues, our work showed gene enrichment in ovarian steroidogenesis.
Next, we correlated cell clusters with tumor features such as clinical stages. The proportion of T-cell cluster was decreased as the tumor stage went along (Fig. 1E; Supplementary Fig. S1D). To verify this, we examined the expression levels of T-cell marker CD3 in 40 cases of tissues from nonmalignant ovary and HGSOC tumor stage 1, stage 2, and stage 3 (n = 10 in each group) using IHC. Our results confirmed that the proportions of CD3-positive immune cells were decreased from early to late tumor stages (Supplementary Fig. S1E).
Ecosystematic epithelial cell features in different tumor stages
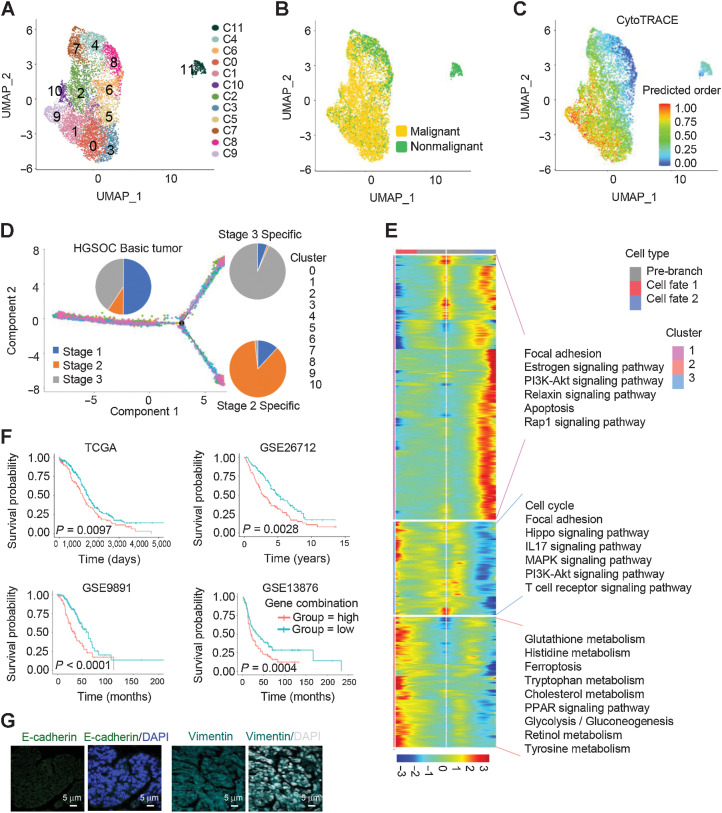
We next characterized the features of tumor epithelial cells. Somatic copy-number variations (CNV) for each cell type were analyzed using the R package infercnv (v0.8.2), and malignant epithelial cells were determined with CNV signals above 0.05 and CNV correlations above 0.5 (Supplementary Fig. S2). A total of 14,636 ovarian epithelial cells were collected across all tissues and divided into a diverse set of 12 clusters, including 8,192 cells from HGSOC tumors and 6,444 cells from nonmalignant ovarian tissues (Fig. 2A and B). CytoTRACE (29) was then applied to predict the differentiation states of these epithelial cells and to identify quiescent stem cells in HGSOC (Fig. 2C; Supplementary Table S3). To further explore the protumor immune signaling network for these cells, we performed immune-related scoring in early and late tumor stages (Supplementary Table S4; Supplementary Fig. S3A). Strikingly, most of ligand genes responsible for immune checkpoint inhibition were strongly enriched at early tumor stage 1 but significantly less at the late tumor stage. The levels of the PD-1 and CTAL-4 ligand genes CD274 (PD-L1) and CD80/86 (B7-1/2) were fairly low in all stages of HGSOC, which is consistent with previous clinical reports of low therapeutic response when blockade against PD-1 or CTLA-4 was applied in ovarian cancer (30, 31). Differently, VTCN1 expression was significantly increased in the late tumor stage 3 (Supplementary Fig. S3A).
Figure 2.
Differential gene expression signatures and impact of EMT-associated genes on HGSOC tumors. A, UMAP plot with clusters demarcated by colors demonstrating 12 distinct clusters based on gene expression differences for 14,636 epithelial cells passing quality control. B, The UMAP plot demarcated by colors showing the two groups of HGSOC tumors (malignant) and nonmalignant ovarian tissues. C, CytoTRACE analysis of epithelial cells. D, Pseudotime analysis of malignant epithelial cells inferred by Monocle2. Each point corresponds to a single cell. Cluster and stage information is shown. E, The differentially expressed genes (rows) along the pseudotime (columns) were hierarchically clustered into three subclusters. The representative annotated pathways of each subcluster are provided. F, The combination of NOTCH1, SNAI2, WNT11, and TGFBR1 expression was associated with worse patient OS in TCGA HGSOC cohort, GSE26712 HGSOC cohort, GES9891 serous ovarian cancer cohort, and GSE13876 serous ovarian cancer cohort, respectively. P values were calculated by a log-rank test. G, IF staining with anti–E-cadherin and vimentin antibodies in HGSOC tissue sections.
To better understand the roles of epithelial cells in tumor development, the Monocle algorithm was applied in pseudotime analysis for malignant epithelial cells to project their developmental trajectories. Eleven clusters of the cells were aggregated on the basis of gene expression similarities, and were projected into a pseudotime process defined as HGSOC basic, stage 2, and stage 3 tumors (Fig. 2D). Along the trajectory, the gene signature of cluster 1 cells at stage 3 was characterized as certain functional pathways, including focal adhesion, the estrogen signaling pathway, the PI3K-AKT signaling pathway, the relaxin signaling pathway, apoptosis, and the rap1 signaling pathway (Fig. 2E). This cluster also expressed a set of EMT-associated genes, such as IGF2 (32), WNT7A (33), and HBEGF (ref. 34; Supplementary Fig. S3B), suggesting an induction of EMT. Interestingly, the gene signature of cluster 3 with HGSOC basic tumor stage demonstrated a strong association of metabolic pathways as shown in Fig. 2E. The beam genes in this cluster included SOX9, CXCL10, and WNT6 (Supplementary Fig. S3C).
We further examined the expression of genes related to EMT in HGSOC tumor cells at different stages. Thirty-eight EMT markers were differentially expressed in HGSOC cells compared with nonmalignant control cells (Supplementary Fig. S4A). To check the association of these EMT markers with patient survival, TCGA HGSOC dataset, GEO HGSOC dataset (GSE no. 26712), and two serous ovarian cancer datasets (GSE no. 13876 and no. 9891) were assessed using TCGA and GEO online analyses with available OS results, in which 1,202 tumor samples were evaluated. Our analysis showed that four genes, including NOTCH1, SNAI2, WNT11, and TGFBR1, were significantly associated with poor outcome in at least three cohorts of these four bulk expression datasets (Supplementary Fig. S4B–S4S4E). Of note, the combination of these four genes was associated with worse OS in all these four datasets (Fig. 2F). Moreover, other EMT marker genes CDH1 and VIM were further validated in HGSOC tissues by immunofluorescence (IF) analysis, and the results suggested that a robust group of cells carry potential EMT functions (Fig. 2G).
Diversity of stromal mesenchymal stem cells and features of CAFs
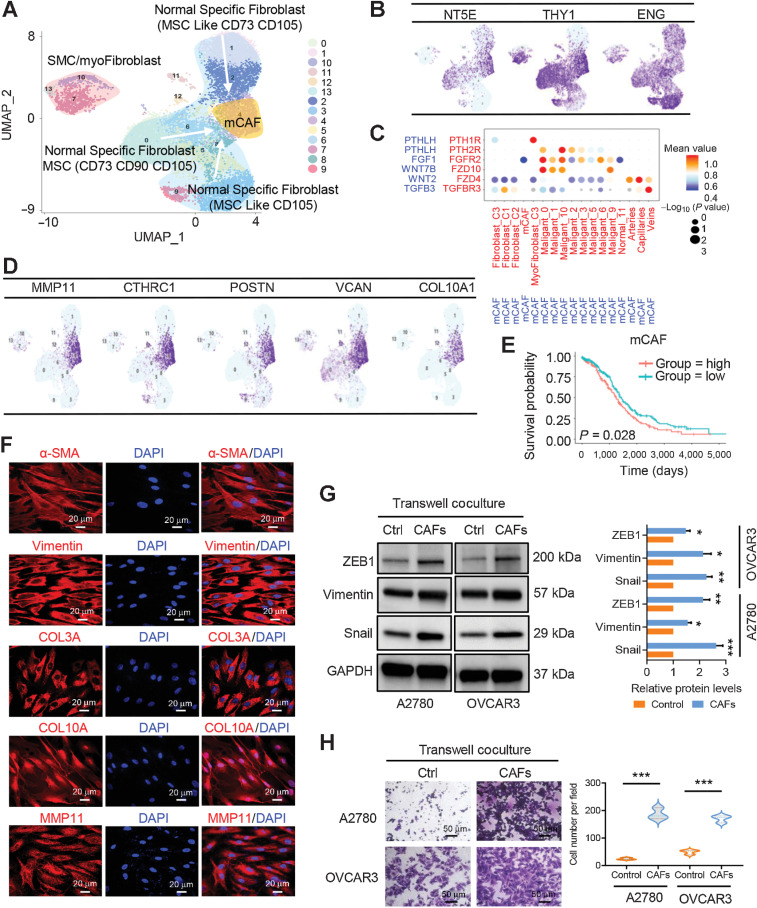
We continued to study the features of stromal fibroblasts. Of the 13,201 fibroblasts, 14 cellular clusters emerged (Fig. 3A). The properties of mesenchymal stem cells (MSC) were observed in many nonmalignant ovary-specific fibroblasts (Fig. 3A). Three subclusters of nonmalignant fibroblasts were grouped on the basis of each gene expression features. MSC subclusters 1, 2, and 3 were referred to as NT5E/THY1/ENG+ MSCs, NT5E/ENG+ MSC-like cells, and ENG+ MSC-like cells, respectively (Fig. 3A and B).
Figure 3.
Fibroblast clusters in nonmalignant ovarian tissues and HGSOC tumors. A, UMAP plot with clusters demarcated by colors demonstrating 14 distinct clusters based on gene expression differences for 13,201 fibroblasts. B, UMAP plot color coded for the expression (blue to purple) of marker genes for the clusters of nonmalignant fibroblasts as indicated. C, Dot plot of the cross-compartment chemokine ligand and corresponding chemokine receptor expression by the cell type of the mCAF and TME. The color intensity of each dot represents the mean scran-normalized expression across all patients. The size of dots indicates the proportion of cells that express a gene relative to the total number of cells in that cell type. D, UMAP plot color coded for the expression (blue to purple) of marker genes for the mCAFs. E, Kaplan–Meier OS curves of patients with TCGA HGSOC grouped by the top 10–gene signature of mCAF markers. P values were calculated by a log-rank test. F, IF staining of α-SMA, vimentin, COL3A, COL10A, and MMP11 in primary CAFs derived from HGSOC ascites. G, Protein expression levels of mesenchymal biomarkers including ZEB1, vimentin, and snail were analyzed in ovarian cancer A2780 and OVCAR3 cells alone or transwell cocultured with primary CAFs by Western blot analysis. The protein expression levels were normalized with GAPDH. The normalized value of the control group was set to 1, and the relative protein levels of the sample group are shown as mean ± SD. The results were averaged from three independent experiments. H, Representative images for the invasion analysis of A2780 and OVCAR3 cells alone or transwell cocultured with primary CAFs. Data are mean ± SD from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Among malignant fibroblasts, mCAFs with a strong extracellular matrix signature, such as PTHLH, FGF1, WNT7B, WNT2, and TGFB3, were the dominant CAFs in HGSOC tumors (Fig. 3C). In addition to those, mCAFs, largely from Cancer 6, exhibited remarkably high levels of MMP11, THRC1, POSTN, VCAN, and COL10A1 (Fig. 3D). Consistently, Fig. 3E showed a correlation between high expression of these top marker genes and worse patient prognosis, evaluated by survival analysis using TCGA HGSOC data. The presence of mCAFs was also confirmed in ascites of 5 patients with late-stage HGSOC. IF staining demonstrated that mCAFs at late-stage HGSOC were positive for the canonical CAF markers α-SMA, vimentin, and COL3A, as well as the mCAF markers COL10A and MMP11 (Fig. 3F). These mCAFs possessed certain pro-EMT properties, evidenced by upregulation of mesenchymal biomarkers such as ZEB1, vimentin, and snail at protein levels and an increase of tumor cell invasion in a CAF/ovarian cancer cell (A2780 or OVCAR3) transwell coculture system (Fig. 3G and H).
Enrichment of M1 macrophages indicates a favorable prognosis in the early stage of HGSOC
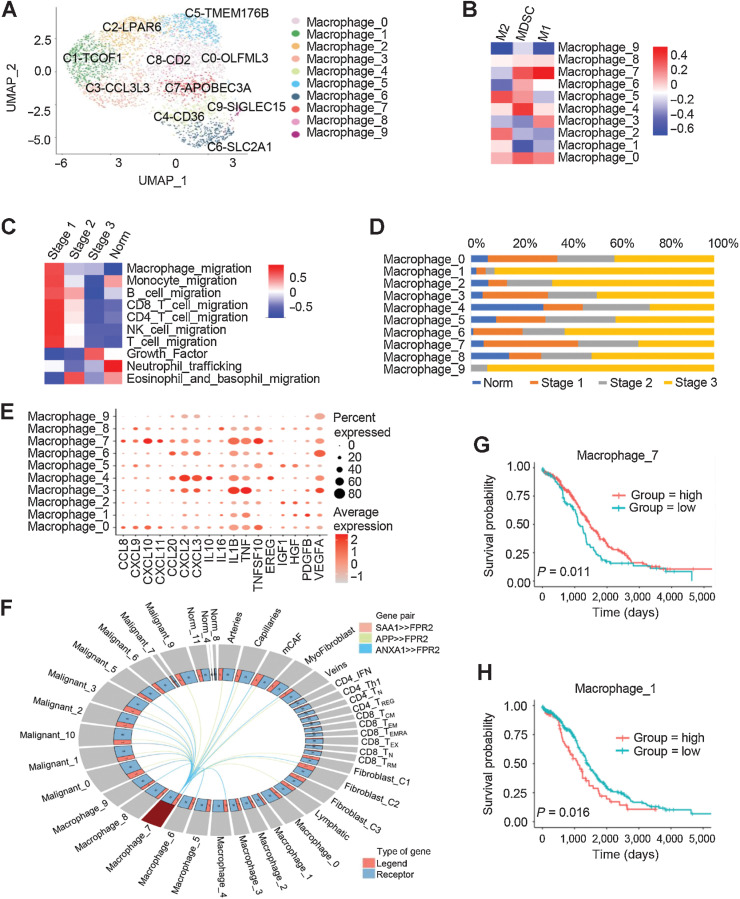
We rearranged the macrophage clusters into 10 clusters by the MNN clustering method to deeply analyze the cell features (Fig. 4A). The gene expression pattern of these clusters were further compared with the classical ones for M1, myeloid-derived suppressor cell (MDSC), and M2 macrophages (Fig. 4B). Our results showed that the cluster C7-APOBEC3A cells highly expressed M1 macrophage-related genes, including IFI6, ISG20, LY6E, IFIT3, CXCL10, and IL1RN (Fig. 4B; Supplementary S5A). The cluster C0-OLFML3 cells have all three macrophage subtypes characteristics, suggesting the dynamic transformation among M1, MDSC, and M2 macrophages in the TME of HGSOC tumors (Fig. 4B).
Figure 4.
Characteristics of macrophages in different tumor stages. A, UMAPs of macrophages from all patients, colored by the identified cell subpopulations. B, Heatmap depicting the gene enrichment for classical cell types M1, M2, and MDSCs in comparison with the macrophage subclusters. C, Heatmap showing differentially activated pathways of each clinical stage in the HGSOC and nonmalignant groups. D, Macrophage cell-type fractions relative to the total macrophage cell count in each clinical stage group. Each stacked bar represents a cluster for which the total macrophage cell count was scaled to 1. E, Dot plots showing the expression levels of specific chemokine genes in each macrophage cluster. F, Model of the cross-compartment chemokine ligand-receptor interactions between macrophage_7 and the TME. OS for patients further stratified according to macrophage_7 (G) and macrophage_1 (H) signature expression. Log-rank P values are shown.
The capabilities of macrophages interact with other immune cell types were then examined in the HGSOC tissues. The extent of the migration of monocytes, B cells, T cells, and natural killer cells were weakened at late tumor stages, suggesting that macrophages lose their attraction to other immune cells (Fig. 4C). Instead, many genes representing growth factor secretion in macrophages were significantly induced at the late tumor stages (Fig. 4C). These results indicated that macrophages undergo malignant transformation during this process, although macrophages appear antitumor behaviors at stage 1.
In early stage 1, macrophages had a strong ability to recruit immune cells, suggesting that stage 1 is more suitable for the regulation of the tumor immune response (Fig. 4C). The cluster C7-APOBEC3A cells have been shown M1 macrophage possessing antitumor activities (Fig. 4D). Figure 4E shows that cluster C7-APOBEC3A displayed enhanced secretion of chemokines including CCL8, CXCL10, CXCL11, and TNFSF10. This cluster also specifically expressed high levels of IDO1 (Supplementary Fig. S5B). Consistently, IDO1 expression levels were much higher in early tumor stage 1 than stages 2 or 3 (Supplementary Fig. S5C). The induction of SAA1, APP, and ANXA1, as well as FPR2 was observed in C7-APOBEC3A macrophages, which represented macrophage activation (Fig. 4F). This activation could be strengthened by CCL2, CCL7, and CCL8, which are produced by other cell types in the TME, such as myofibroblasts, fibroblast-C2 cells, and macrophages themselves (Supplementary Fig. S5D). The gene signature of cluster C7-APOBEC3A was associated with a better prognosis (Fig. 4G), while the one for cluster C1-TCOF1 was associated with a poor prognosis (Fig. 4H).
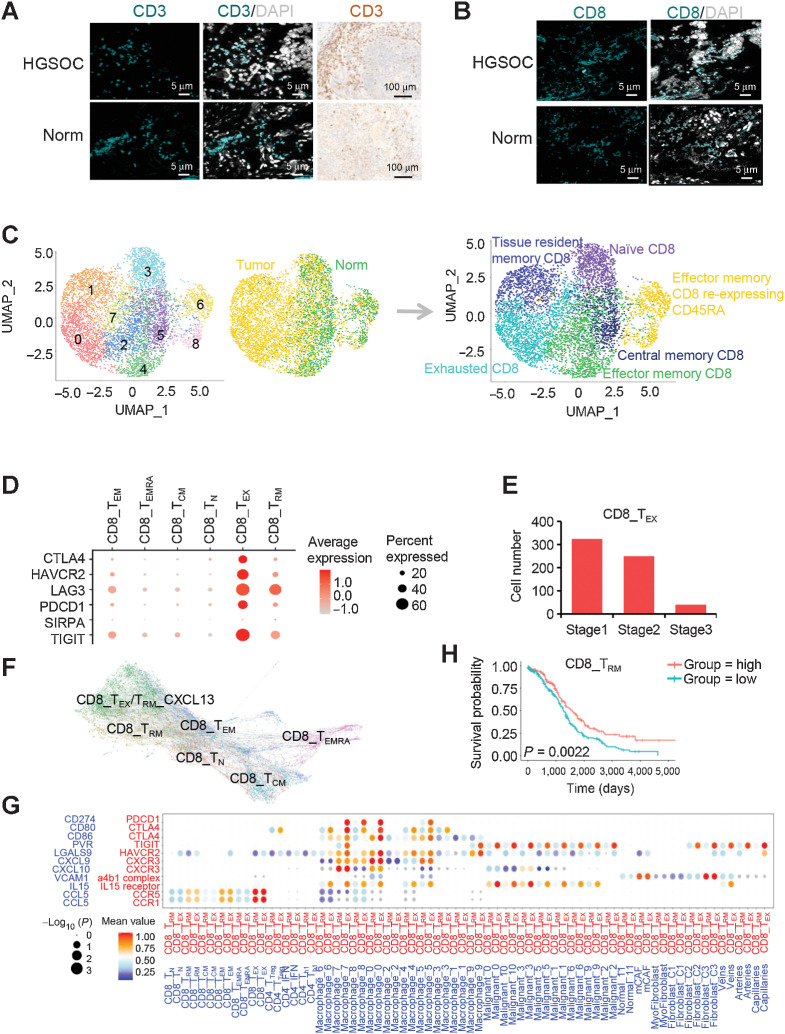
Infiltrated CD8+ T-cell states are shaped by cross-compartment interactions in the HGSOC TME
In addition to the ones of macrophages, the features and distribution of lymphocytes were characterized in HGSOC tumors compared with nonmalignant tissues. The presence of infiltrating lymphocytes in HGSOC and nonmalignant ovarian tissues was confirmed by IHC and IF with CD3 antibody (Fig. 5A). Because the T-cell lineage carried CD8A gene enrichment (Fig. 1C), we confirmed its presence in HGSOC tumors by IF analysis with CD8A antibody (Fig. 5B). The CD8A+ T cells were regrouped by MNN clustering method into nine clusters (Fig. 5C; Supplementary S6A), which appeared to be distributed in different tissues (Fig. 5C; Supplementary S6B). For example, tissue resident memory CD8+ T cells (CD8+ TRM cells), represented by the CD8-C1-IFIT3 signature, were more localized in tumor tissues. Central memory CD8+ T cells (CD8+ TCM cells), characterized by a CD8-C5-DNAJB1 gene pattern, were mainly from nonmalignant ovarian cells (Fig. 5C; Supplementary S6B). Notably, exhausted T cells (CD8+ TEX cells), marked by CD8-C0-CXCL13 or CD8-C7-TNFRSF4, populated with cells from tumors.
Figure 5.
Cell clustering and functional annotation of CD8+ T cells in HGSOC and nonmalignant ovarian tissues. A, Detection of CD3 (green) in T cells by IF staining in HGSOC and nonmalignant ovarian tissues. Nuclei were stained with DAPI (gray). The validation of CD3 was also confirmed by IHC. B, Detection of CD8 (green) in T cells by IF staining in HGSOC and nonmalignant ovarian tissues. Nuclei were stained with DAPI (gray). C, UMAPs of CD8+ T cells from all patients, colored by the identified cell subpopulations. D, Dot plot of the average expression of CTLA4, HAVCR2, LAG3, PDCD1, SIRPA, and TIGIT in CD8+ T-cell subpopulations. E, Bar plots showing the fraction of CD8+ TEX cells relative to the total CD8+ T count grouped by tumor stage. F, PAGA pseudospatial trajectory analysis of six colored CD8+ T-cell subclusters. G, Dot plot of the cross-compartment chemokine ligand and corresponding receptor expression by cell type of the CD8+ TRM /TEX and TME. H, Kaplan–Meier survival curves for OS from n = 373 primary HGSOCs showing significant prognostic separation according to the CD8+ TRM marker gene signature derived from single-cell data. Log-rank P values are shown.
We next focused on the two cell types enriched in HGSOC tissues, including CD8+ TRM and CD8+ TEX cells. To better understand the roles of CD8+ TEX cells in HGSOC, the gene expression pattern of CD8+ TEX cells was deeply explored (Supplementary Table S5). Among these exhaustion markers, CTLA4, HAVCR2, LAG3, PDCD1, SIRPA, and TIGIT were among the top-ranked genes (Fig. 5D). Of note, the coinhibitory receptor TIGIT was expressed at higher levels in HGSOC tissues. Figure 5E showed that the numbers of CD8+ TEX cells were high at early stage 1 but decreased at late stages. Interestingly, the PAGA pseudospatial trajectory analysis showed that the preferential enrichment of exhausted T cells (CD8+ TEX cells), also named CD8+ TRM-CXCL13 cells, was located at the end of CD8+ TRM and TEM cell differentiation (Fig. 5F).
CD8+ TRM cells contributed to local immune protection in early-stage HGSOC tumors. To explain how CD8+ TRM cells form in early-stage HGSOC tumors, we checked the expression of survival factors, such as IL15, IL17, and NOTCH ligands, which are known to determine TRM cell formation and persistence (35–37). One of them, IL15 was expressed in HGSOC-derived malignant epithelial cells and induced the formation of CD8+ TRM cells and CD8+ TEX cells (Fig. 5G). In addition to epithelial cells, M1 macrophages in HGSOC tumors predominantly expressed CXCL9 and CXCL10 to recruit CD8+ TRM cells via the CXCL9/CXCL10–CXCR3 interaction (Fig. 5G). We also checked the association of the gene signature of CD8+ TRM cells with patient survival, and demonstrated that the signature of CD8+ TRM cells was significantly associated with improved patient survival (Fig. 5H; Supplementary S6C). Overall, our findings strongly suggested a potential interactive mechanism among CD8+ TRM/TEX cells and epithelial cells as well as macrophages in early-stage HGSOC tumors.
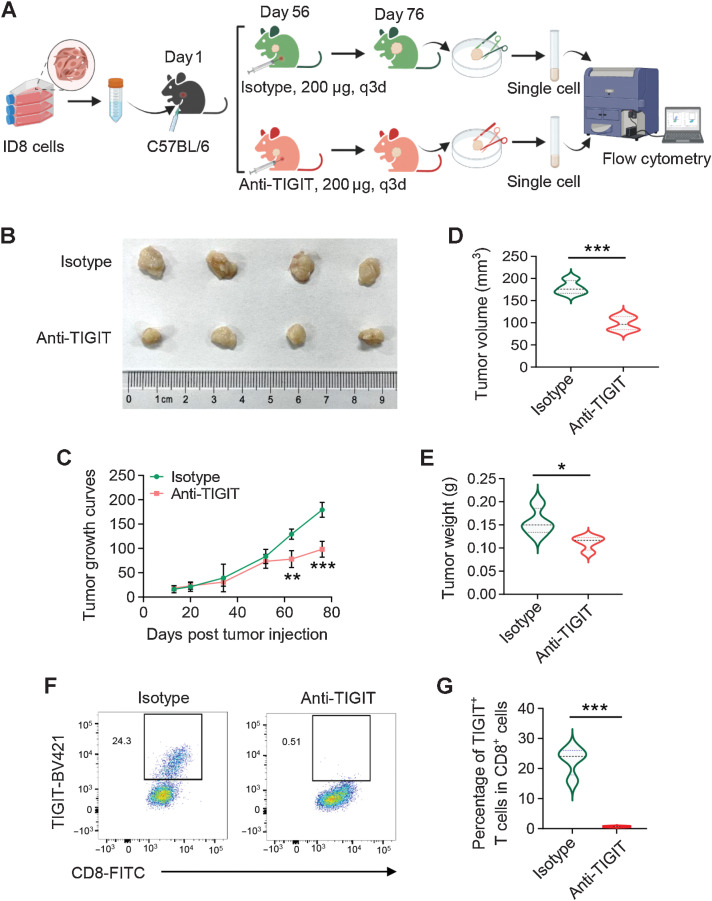
The roles of TIGIT blockade to ovarian cancer tumorigenesis
The immune coinhibitory receptor TIGIT is known to regulate antitumor CD8+ T cells responses (38, 39), which was also highly expressed on CD8+ TEX cells in our study. Next, we tested whether TIGIT blockade could contribute to ovarian cancer growth, ID8 cells were subcutaneously injected into 8 female C57BL/6 mice. The ID8 tumor-bearing mice were treated with anti-TIGIT or isotype-matched control antibody after 8 weeks (Fig. 6A). As expected, anti–TIGIT-treated mice showed reduced tumor burden (Fig. 6B), as shown by retarded tumor growth (Fig. 6C), lower tumor volume and tumor weight at the endpoint (Fig. 6D and E). TIGIT blockade significantly suppressed the frequency of TIGIT+-CD8+ T cells in tumors (Fig. 6F and G). These results demonstrated that the TIGIT neutralization by anti-TIGIT antibody on CD8+ T cells alleviated the tumor burden in ID8 tumor-bearing mice.
Figure 6.
Blockade of TIGIT inhibits tumor growth in syngeneic mice. A, Workflow showing the experimental process of the animal study. B, C57BL/6 mice were subcutaneously injected with ID8 cells (5 × 106 per mouse) and treated with 200 μg anti-TIGIT or isotype-matched control antibody via intraperitoneal injection as indicated. Tumor growth curves (C), tumor volume (D), and tumor weight (E) at the endpoint were measured. *, P < 0.05; **, P < 0.01; ***, P < 0.001. F and G, Frequency of TIGIT+-CD8+ T cells from tumors in anti-TIGIT or isotype-matched control antibody-treated mice by flow cytometry. Data are mean ± SD (n = 4 mice/group). ***, P < 0.001.
Discussion
Our scRNA-seq study presented a high-resolution depiction of the cellular interaction network in early- or late-stage HGSOC tumors and nonmalignant ovarian tissues. The clinical stage-specific features of the HGSOC cellular ecosystem summarized in Supplementary Fig. S7. Within the early-stage of HGSOC ecosystem, C7-APOBEC3A tumor-associated macrophages (TAM) were activated to produce chemokines to activate CD8_TEX cells and to express high levels of immune checkpoint ligand genes to suppress effector T cells. In contrast, the numbers of C7-APOBEC3A TAMs as well as CXCL13-TRM cells were reduced in late-stage tumors. mCAFs derived from late-stage HGSOC ascites induced the EMT properties of ovarian cancer cells. In addition, we confirmed the suppressive effects of TIGIT blockade on ovarian cancer tumor growth in mouse models.
The transcriptome-wide ecosystematic zonation of HGSOC tumors suggests that different TME cell types cooperate to carry out essential functions. One of our key observations is the identification of an EMT programme with 38 genes differentially expressed in HGSOC tumor cells compared with nonmalignant ovarian cells. Consistent with our studies, previous studies reported that EMT promoted ovarian cancer cell invasion and metastasis (8, 9, 11). Moreover, we determined the association of these EMT markers with patient survival using four TCGA and GEO datasets. The expression levels of NOTCH1, SNAI2, TGFBR1, and WNT11 were associated with poor survival in at least three cohorts. This four-gene combination was significantly associated with worse patient OS in all TCGA and GEO serous ovarian cancer cohorts, which may reflect a novel therapeutic opportunity for the treatment of ovarian cancer.
Immunosuppressive nature of TMEs is likely due to induction of inhibitory immune checkpoints and T-cell exhaustion. Inhibition of this induction by blockade of key coinhibitory receptors such as PD1 or CTLA4, reactivated exhausted antitumor immune responses (40, 41). However, these treatments did not work for HGSOC. In our study, CD8+ T cells were the major effector cells. However, we observed a highly immunosuppressive microenvironment of HGSOC in which most infiltrating tumor-specific CD8+ T cells became exhausted and the effector function was severely impaired. We identified CTLA4, HAVCR2, LAG3, PDCD1, SIRPA, and TIGIT as the main T-cell coinhibitory receptors. Among them, TIGIT was the most highly expressed coinhibitory receptor on CD8+ TEX cells. TIGIT has been described previously as an inhibitor of CD4+ regulatory T cells (Treg) (42). It was reported that antibody targeting TIGIT reduced the proportion of CD4+ Tregs and improve the survival rate of ovarian cancer mice (43). In recent years, TIGIT has emerged as a critical regulator of antitumor CD8+ T-cell responses in a set of tumors, such as hepatocellular carcinoma, head and neck squamous cell carcinoma, and gastric cancer (38, 39, 44). It has been shown that antibody coblockade of TIGIT and PD-L1 could synergistically enhance CD8+ T-cell effector function (45). Consistent with these recent findings, we showed that TIGIT blockade could inhibit ovarian cancer tumor growth in mouse models and significantly suppressed the frequency of TIGIT+-CD8+ T cells in tumors. Our results will provide valuable insights for developing novel immunotherapies in HGSOC.
Our macrophage data showed that the macrophage capabilities of immune cell attraction were gradually weakened, whereas the effects of growth factor secretion were significantly increased when the stages go along, indicating that the malignant transformation of macrophages occurred in this process. Notably, C7-APOBEC3A M1 macrophages were mainly in early stage 1 and displayed enhanced chemokine secretion activities. Consistently, C7-APOBEC3A M1 macrophages were associated with better survival outcomes. These findings uncovered an enrichment of C7-APOBEC3A M1 macrophages in the early stage of HGSOC with a favorable prognosis.
Finally, we investigated the interaction among the cellular components of the tumor, immune, and stromal cells to shape the tumor continuum. One important finding was that malignant cells potentially mediate TRM cell recruitment via the IL15-IL15R axis, which is required for the formation and persistence of TRM cells (35, 46). Another way to recruit CD8+TRM cells was via the CXCL9/CXCL10–CXCR3 interaction, while the predominant source of CXCL9 and CXCL10 was M1 macrophages in HGSOC tumors. Thus, our findings showed that both malignant and M1 macrophages contributed to the presence of CD8+ TRM cells in the HGSOC TME. In addition to immune cells, we confirmed that primary mCAFs could also interact with tumor cells to promote EMT. Our observations further support the hypothesis that specific CAFs contribute to the malignant progression of ovarian cancers (15, 47, 48).
In conclusion, our work provides important insights into HGSOC biology and an atlas of tumor, immune, and stromal cells that discloses the complexity of the HGSOC TME in different tumor stages. Such complexity can be reflected by our findings of an EMT programme, mCAF-induced tumor EMT, early stage–related M1 macrophages, and CD8+ TRM and TEX infiltration, and the cross compartment interaction, which can help to guide future treatment therapies.
Supplementary Material
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Universities (no. 2019QNA7035 and 2021FZZX001-43, to J. Xu), the Beijing Kanghua Foundation for the Development of Traditional Chinese and Western Medicine (KH-2021-LLZX-016, to J. Xu) and the National Natural Science Foundation of China (no. 82103505, W. Fei). Figure 1A was created with 3DS Max. Figure 6A and Supplementary Figure S7 were created with BioRender.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
J. Xu: Conceptualization, resources, supervision, funding acquisition, validation, investigation, writing–original draft, writing–review and editing. Y. Fang: Conceptualization, resources, validation, investigation. K. Chen: Validation. S. Li: Validation, investigation. S. Tang: Validation, investigation. Y. Ren: Validation. Y. Cen: Validation, investigation. W. Fei: Investigation. B. Zhang: Investigation, methodology. Y. Shen: Conceptualization, supervision, writing–review and editing. W. Lu: Conceptualization, supervision, writing–review and editing.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Maenhoudt N, Defraye C, Boretto M, Jan Z, Heremans R, Boeckx B, et al. Developing organoids from ovarian cancer as experimental and preclinical models. Stem Cell Reports 2020;14:717–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seidman JD, Horkayne-Szakaly I, Haiba M, Boice CR, Kurman RJ, Ronnett BM. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int J Gynecol Pathol 2004;23:41–4. [DOI] [PubMed] [Google Scholar]
- 4. Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin 2018;68:284–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baldwin LA, Huang B, Miller RW, Tucker T, Goodrich ST, Podzielinski I, et al. Ten-year relative survival for epithelial ovarian cancer. Obstet Gynecol 2012;120:612–8. [DOI] [PubMed] [Google Scholar]
- 6. Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin 2009;59:225–49. [DOI] [PubMed] [Google Scholar]
- 7. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loret N, Denys H, Tummers P, Berx G. The role of epithelial-to-mesenchymal plasticity in ovarian cancer progression and therapy resistance. Cancers 2019;11:838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vergara D, Merlot B, Lucot JP, Collinet P, Vinatier D, Fournier I, et al. Epithelial-mesenchymal transition in ovarian cancer. Cancer Lett 2010;291:59–66. [DOI] [PubMed] [Google Scholar]
- 10. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 2019;20:69–84. [DOI] [PubMed] [Google Scholar]
- 11. Sohn MH, Kim SI, Shin JY, Kim HS, Chung HH, Kim JW, et al. Classification of high-grade serous ovarian carcinoma by epithelial-to-mesenchymal transition signature and homologous recombination repair genes. Genes 2021;12:1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Babaei G, Aziz SG, Jaghi NZZ. EMT, cancer stem cells and autophagy; The three main axes of metastasis. Biomed Pharmacother 2021;133:110909. [DOI] [PubMed] [Google Scholar]
- 13. Winterhoff B, Talukdar S, Chang Z, Wang J, Starr TK. Single-cell sequencing in ovarian cancer: a new frontier in precision medicine. Curr Opin Obstet Gynecol 2019;31:49–55. [DOI] [PubMed] [Google Scholar]
- 14. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 15. Izar B, Tirosh I, Stover EH, Wakiro I, Cuoco MS, Alter I, et al. A single-cell landscape of high-grade serous ovarian cancer. Nat Med 2020;26:1271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hornburg M, Desbois M, Lu S, Guan Y, Lo AA, Kaufman S, et al. Single-cell dissection of cellular components and interactions shaping the tumor immune phenotypes in ovarian cancer. Cancer Cell 2021;39:928–44. [DOI] [PubMed] [Google Scholar]
- 17. Olbrecht S, Busschaert P, Qian J, Vanderstichele A, Loverix L, Van Gorp T, et al. High-grade serous tubo-ovarian cancer refined with single-cell RNA sequencing: specific cell subtypes influence survival and determine molecular subtype classification. Genome Med 2021;13:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov 2015;14:561–84. [DOI] [PubMed] [Google Scholar]
- 19. Goodman AM, Piccioni D, Kato S, Boichard A, Wang HY, Frampton G, et al. Prevalence of PDL1 amplification and preliminary response to immune checkpoint blockade in solid tumors. JAMA Oncol 2018;4:1237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018;34:i884–i90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res 2008;14:5198–208. [DOI] [PubMed] [Google Scholar]
- 23. Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res 2008;68:5478–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Crijns AP, Fehrmann RS, de Jong S, Gerbens F, Meersma GJ, Klip HG, et al. Survival-related profile, pathways, and transcription factors in ovarian cancer. PLoS Med 2009;6:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ito K, Murphy D. Application of ggplot2 to pharmacometric graphics. CPT Pharmacometrics Syst Pharmacol 2013;2:e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang S, Zheng Y, Li J, Yu Y, Zhang W, Song M, et al. Single-cell transcriptomic atlas of primate ovarian aging. Cell 2020;180:585–600. [DOI] [PubMed] [Google Scholar]
- 27. Basuli D, Tesfay L, Deng Z, Paul B, Yamamoto Y, Ning G, et al. Iron addiction: a novel therapeutic target in ovarian cancer. Oncogene 2017;36:4089–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zou Y, Henry WS, Ricq EL, Graham ET, Phadnis VV, Maretich P, et al. Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature 2020;585:603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gulati GS, Sikandar SS, Wesche DJ, Manjunath A, Bharadwaj A, Berger MJ, et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 2020;367:405–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Disis ML, Taylor MH, Kelly K, Beck JT, Gordon M, Moore KM, et al. Efficacy and safety of avelumab for patients with recurrent or refractory ovarian cancer: phase 1b results from the JAVELIN solid tumor trial. JAMA Oncol 2019;5:393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hamanishi J, Mandai M, Ikeda T, Minami M, Kawaguchi A, Murayama T, et al. Safety and antitumor activity of anti-PD-1 antibody, nivolumab, in patients with platinum-resistant ovarian cancer. J Clin Oncol 2015;33:4015–22. [DOI] [PubMed] [Google Scholar]
- 32. Cevenini A, Orru S, Mancini A, Alfieri A, Buono P, Imperlini E. Molecular signatures of the insulin-like growth factor 1-mediated epithelial-mesenchymal transition in breast, lung and gastric cancers. Int J Mol Sci 2018;19:2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang X, Zhu H, Gao Z, Li J, Zhuang J, Dong Y, et al. Wnt7a activates canonical Wnt signaling, promotes bladder cancer cell invasion, and is suppressed by miR-370-3p. J Biol Chem 2018;293:6693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eapen MS, Sharma P, Thompson IE, Lu W, Myers S, Hansbro PM, et al. Heparin-binding epidermal growth factor (HB-EGF) drives EMT in patients with COPD: implications for disease pathogenesis and novel therapies. Lab Invest 2019;99:150–7. [DOI] [PubMed] [Google Scholar]
- 35. Mazzoni A, Maggi L, Montaini G, Ramazzotti M, Capone M, Vanni A, et al. Human T cells interacting with HNSCC-derived mesenchymal stromal cells acquire tissue-resident memory like properties. Eur J Immunol 2020;50:1571–9. [DOI] [PubMed] [Google Scholar]
- 36. Holz LE, Prier JE, Freestone D, Steiner TM, English K, Johnson DN, et al. CD8(+) T cell activation leads to constitutive formation of liver tissue-resident memory T cells that seed a large and flexible niche in the liver. Cell Rep 2018;25:68–79. [DOI] [PubMed] [Google Scholar]
- 37. Mackay LK, Wynne-Jones E, Freestone D, Pellicci DG, Mielke LA, Newman DM, et al. T-box transcription factors combine with the cytokines TGF-beta and IL-15 to control tissue-resident memory T cell fate. Immunity 2015;43:1101–11. [DOI] [PubMed] [Google Scholar]
- 38. He W, Zhang H, Han F, Chen X, Lin R, Wang W, et al. CD155T/TIGIT signaling regulates CD8(+) T-cell metabolism and promotes tumor progression in human gastric cancer. Cancer Res 2017;77:6375–88. [DOI] [PubMed] [Google Scholar]
- 39. Zhang C, Wang Y, Xun X, Wang S, Xiang X, Hu S, et al. TIGIT can exert immunosuppressive effects on CD8+ T cells by the CD155/TIGIT signaling pathway for hepatocellular carcinoma in vitro. J Immunother 2020;43:236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med 2010;207:2187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1–10. [DOI] [PubMed] [Google Scholar]
- 42. Lucca LE, Dominguez-Villar M. Modulation of regulatory T cell function and stability by co-inhibitory receptors. Nat Rev Immunol 2020;20:680–93. [DOI] [PubMed] [Google Scholar]
- 43. Chen F, Xu Y, Chen Y, Shan S. TIGIT enhances CD4(+) regulatory T-cell response and mediates immune suppression in a murine ovarian cancer model. Cancer Med 2020;9:3584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu L, Mao L, Liu JF, Chen L, Yu GT, Yang LL, et al. Blockade of TIGIT/CD155 signaling reverses T-cell exhaustion and enhances antitumor capability in head and neck squamous cell carcinoma. Cancer Immunol Res 2019;7:1700–13. [DOI] [PubMed] [Google Scholar]
- 45. Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell 2014;26:923–37. [DOI] [PubMed] [Google Scholar]
- 46. Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol 2013;14:1294–301. [DOI] [PubMed] [Google Scholar]
- 47. Eckert MA, Coscia F, Chryplewicz A, Chang JW, Hernandez KM, Pan S, et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature 2019;569:723–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hussain A, Voisin V, Poon S, Karamboulas C, Bui NHB, Meens J, et al. Distinct fibroblast functional states drive clinical outcomes in ovarian cancer and are regulated by TCF21. J Exp Med 2020;217:e20191094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq data that support the findings of this study have been deposited on Gene Expression Omnibus (GEO) platform under the accession code "GSE184880."