Abstract
KRAS is the most commonly mutated oncogene in NSCLC and development of direct KRAS inhibitors has renewed interest in this molecular variant. Different KRAS mutations may represent a unique biologic context with different prognostic and therapeutic impact. We sought to characterize genomic landscapes of advanced, KRAS-mutated non–small cell lung cancer (NSCLC) in a large national cohort to help guide future therapeutic development.
Molecular profiles of 17,095 NSCLC specimens were obtained using DNA next-generation sequencing of 592 genes (Caris Life Sciences) and classified on the basis of presence and subtype of KRAS mutations. Co-occurring genomic alterations, tumor mutational burden (TMB), and PD-L1 expression [22C3, tumor proportion score (TPS) score] were analyzed by KRAS mutation type.
Across the cohort, 4,706 (27.5%) samples harbored a KRAS mutation. The most common subtype was G12C (40%), followed by G12V (19%) and G12D (15%). The prevalence of KRAS mutations was 37.2% among adenocarcinomas and 4.4% in squamous cell carcinomas. Rates of high TMB (≥10 mutations/Mb) and PD-L1 expression varied across KRAS mutation subtypes. KRAS G12C was the most likely to be PD-L1 positive (65.5% TPS ≥ 1%) and PD-L1 high (41.3% TPS ≥ 50%). STK11 was mutated in 8.6% of KRAS wild-type NSCLC but more frequent in KRAS-mutant NSCLC, with the highest rate in G13 (36.2%). TP53 mutations were more frequent in KRAS wild-type NSCLC (73.6%).
KRAS mutation subtypes have different co-occurring mutations and a distinct genomic landscape. The clinical relevance of these differences in the context of specific therapeutic interventions warrants investigation.
Introduction
KRAS is the most common oncogenic driver in non–small cell lung cancer (NSCLC) identified in up to 25% of adenocarcinomas and 3% of squamous cell carcinomas (1, 2). KRAS activation results in downstream signaling to several pathways, including the RAF-MEK-ERK pathway. The prognostic value of KRAS mutations in patients with NSCLC remains unclear. Some studies have suggested worse outcomes with chemotherapy (3) while others have not (4, 5). There is similar discordance with KRAS co-mutation status and immunotherapy. A large retrospective study reported shorter progression-free survival (PFS) and overall survival (OS) with use of immunotherapy in patients with KRAS-mutant NSCLC harboring co-mutations in STK11/LKB1 genes (6). An analysis of the KEYNOTE-189 study failed to confirm these findings, with similar benefit observed from the addition of pembrolizumab to chemotherapy, independent of STK11 or KEAP1 mutation status (7).
One contributing factor to these discordant results is the heterogeneity within KRAS. There is growing recognition of vast genetic and phenotypic heterogeneity of patients with KRAS-mutated NSCLC (8–10). Most frequently, KRAS mutations (chromosome 12p12.1) involve codons 12 and 13 and less frequently codon 61. Transversion mutations, including purine to pyrimidine (i.e., G>C) or pyrimidine to purine, are more common in current or former smokers, compared with transition mutations, either purine to purine (i.e., G>A) or pyrimidine to pyrimidine (i.e., T>C) which are more common in never or light smokers (11–13). Distinct KRAS mutations can influence the specific biology and the genomic landscape of a given cancer. This in turn can have notable therapeutic implications. With the recent development of direct KRAS inhibitors, these genomic contexts are increasingly relevant. Here, we characterize a large cohort of KRAS-mutant NSCLC, describing co-mutations, tumor mutational burden (TMB), and PD-L1 expression for each KRAS mutation subtype to help frame future treatment strategies.
Materials and Methods
Patient samples
A total of 17,095 NSCLC tumors were submitted to Caris Life Sciences for next-generation sequencing (NGS) molecular profiling between February 2015 and January 2020.
NGS
NGS was performed on genomic DNA isolated from formalin-fixed paraffin-embedded (FFPE) tumor samples using the NextSeq platform (Illumina). Matched normal tissue was not sequenced. A custom-designed SureSelect XT assay was used to enrich 592 whole-gene targets (Agilent Technologies). The 592-gene list was custom designed to include cancer-related genes across all solid tumors that have been the best characterized for their functions and clinical relevance, including prognostic effects and targetability. All variants were detected with >99% confidence based on allele frequency and amplicon coverage, with an average sequencing depth of coverage of > 500× and an analytic sensitivity of 5%. Prior to molecular testing, tumor enrichment was achieved by harvesting targeted tissue using manual microdissection techniques. Variants detected were mapped to reference genome (hg19) and well-established bioinformatics tools such as BWA, SamTools, GATK, and snpFF were incorporated to perform variant calling functions; germline variants were filtered with various germline databases including 1000 genome and dbSNP genetic variants identified were interpreted by board-certified molecular geneticists and categorized as “pathogenic,” “presumed pathogenic,” “variant of unknown significance,” “presumed benign,” or “benign,” according to the American College of Medical Genetics and Genomics standards. When assessing mutation frequencies of individual genes, “pathogenic” and “presumed pathogenic” were counted as mutations.
TMB calculation
TMB was measured by counting all non-synonymous missense, nonsense, in-frame insertions/deletions, and frameshift mutations found per tumor that had not been previously described as germline alterations in dbSNP151, Genome Aggregation Database (gnomAD) or benign variants identified by Caris geneticists. A cut-off point of ≥10 mutations (mt) per MB was used (14). Caris Life Sciences is a participant in the Friends of Cancer Research TMB Harmonization Project (15).
PD-L1 expression
IHC was performed on FFPE sections of glass slides. Slides were stained using automated staining techniques, per the manufacturer's instructions, and were optimized and validated per CLIA/CAO and ISO requirements. A board-certified pathologist evaluated all IHC results independently. The primary PD-L1 antibody clone was 22c3 (Dako). Tumor proportion score (TPS) was defined as the percentage of viable tumor cells showing partial or complete membrane staining at any intensity. The tumor was considered positive if TPS ≥ 1% and high PD-L1 expression was defined as TPS ≥ 50%.
Microsatellite instability/mismatch repair determination
A combination of multiple test platforms was used to determine the microsatellite stability (MSI) or mismatch repair (MMR) status of the tumors profiled, including fragment analysis (Promega), IHC [MLH1, M1 antibody; MSH2, G2191129 antibody; MSH6, 44 antibody; and PMS2, EPR3947 antibody (16)], and NGS (for tumors tested with NextSeq platform, 7,000 target microsatellite loci were examined and compared with the reference genome hg19 from the University of California, Berkeley, CA).
Statistical plan
Molecular alterations among various KRAS-mutated groups were compared using Chi-square or Fisher exact tests [KRAS wild-type (WT) groups excluded from the comparative analyses] and a P value of <0.05 was considered a trending difference. Because of the large sample size of this study, P values were further corrected for multiple comparison using the Benjamini–Hochberg method and an adjusted P value (i.e., q value) of <0.05 was considered a significant difference. This study was conducted in accordance with guidelines of the Declaration of Helsinki, Belmont report, and U.S. Common rule. In keeping with 45 CFR 46.101(b)(4), this study was performed utilizing retrospective, deidentified clinical data and received Institutional Review Board exemption from patient consent.
Results
Clinical characteristics
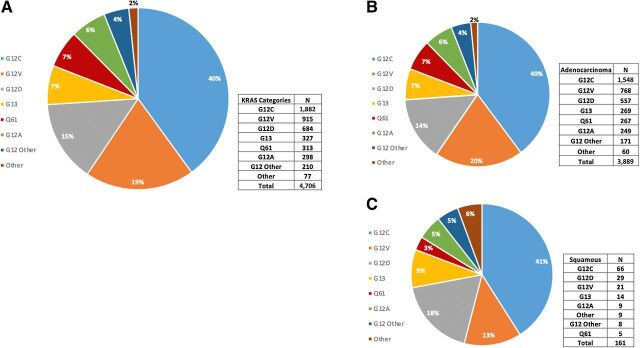
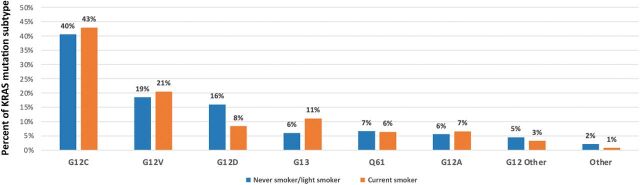
Across 17,095 NSCLC samples analyzed, 4,706 (27.5%) samples harbored a KRAS mutation (Table 1). The most common was G12C (40%), followed by G12V (19%) and G12D (15%; Fig. 1). KRAS mutations were more prevalent in female than male patients (31.35 vs. 23.7%, P < 0.0001) and there was no significant difference in age between KRAS subtypes (Table 1). The prevalence of KRAS mutations was 37.2% (3,889) among adenocarcinoma and only 4.4% (191) in squamous cell carcinoma samples; however, KRAS mutational distribution was similar in both histologies (Fig. 1). The smoking status of 1,841 patients were retrieved and categorized as never smoker, light smoker (designated as less than 15 packs per year), and current smoker (Fig. 2; Supplementary Table S1). The findings were not completely consistent with the known biology of KRAS subtypes. In our limited sample, 43% of patients with G12C mutations, 7% of patients with G12A mutations, and 8% of patients with G12D mutations were current smokers. 16% of patients with G12D mutations but only 6% of patients with G12A mutations were never or light smokers. Among KRAS subgroups, G12C mutations had the highest rate of current smokers, 43%.
Table 1.
Patient characteristics of the NSCLC cohort studied. 4,706 (27.5%) samples of the entire cohort (17,095) had a KRAS mutation. Each KRAS mutation subtype is listed along with the frequency of patients harboring each subtype, the female versus male distribution, as well as the age range and median age.
| Total N (%) | Female N (%) | Male N (%) | Median age | Age range | |
|---|---|---|---|---|---|
| KRAS WT | 12,389 (72.5) | 5,862 (47) | 6,527 (53) | 68.0 | 20–97 |
| All KRAS mutation | 4,706 (27.5) | 2,677 (57) | 2,029 (43) | 68.0 | 22–97 |
| G12C | 1,882 (40) | 1,102 (59) | 780 (41) | 68.0 | 27–95 |
| G12V | 915 (19) | 504 (55) | 411 (45) | 68.0 | 37–92 |
| G12D | 684 (15) | 386 (56) | 298 (44) | 69.0 | 22–97 |
| G13 | 327 (7) | 184 (56) | 143 (44) | 67.0 | 41–90 |
| Q61 | 313 (7) | 175 (56) | 138 (44) | 69.0 | 40–90 |
| G12A | 298 (6) | 160 (54) | 138 (46) | 70.0 | 37–90 |
| G12 Other | 210 (4) | 130 (62) | 80 (38) | 68.0 | 35–91 |
| Other | 77 (2) | 36 (47) | 41 (53) | 68.0 | 37–89 |
| Total | 17,095 | 8,539 (50) | 8,556 (50) | 68.0 | 20–97 |
Figure 1.
KRAS mutational distribution in all NSCLC (A) and adenocarcinoma (B) and squamous cell (C) NSCLC histologies. The prevalence of KRAS mutations was 37.2% among adenocarcinoma and only 4.4% in squamous cell samples, however KRAS mutational distribution was similar in both histologies.
Figure 2.
Frequency of KRAS mutation subtypes in never smokers/light smokers (<15 packs/year) and current smokers. Smoking status data was only available in 1,841 of 4,706 patients with KRAS mutations in this cohort.
Immunotherapy biomarkers
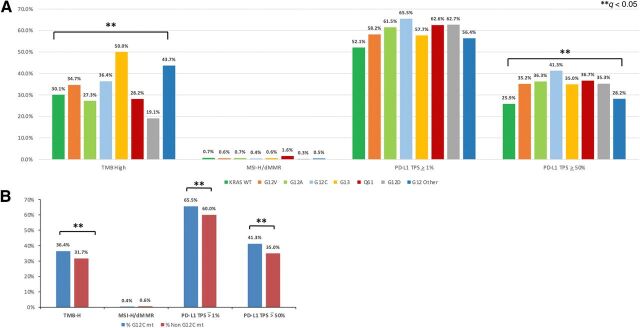
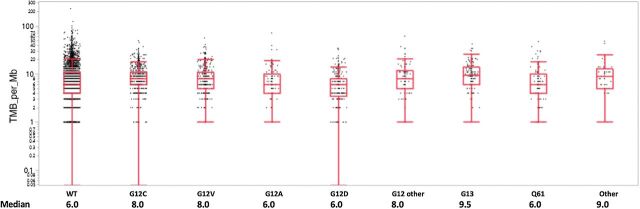
TMB varied significantly across the different KRAS mutation types (P < 0.001). High TMB was most common in G13 (50.0%) followed by G12 (43.7%) and least common in G12D mutations (19.1%; Fig. 3; Supplementary Table S2A). However, the distribution of median TMB values between different KRAS mutation types was narrow: 6.0 to 9.5 mt/Mb (Fig. 4). PD-L1 expression was also significantly different among KRAS mutations across major cutoffs, PD-L1 positive (TPS ≥ 1%), PD-L1 TPS ≥ 10%, and PD-L1 high (TPS ≥ 50%; Fig. 3A; Supplementary Table S2A). Of note, the KRAS WT population had lower PD-L1 expression than all KRAS mutations. KRAS G12C was the most likely to be PD-L1 positive, with 65.5% TPS ≥ 1%, and the most likely to be PD-L1 high, with 41.3% TPS ≥ 50% (Fig. 3A; Supplementary Table S2A). Immunotherapy biomarkers were compared in patients with KRAS G12C mutations and any other KRAS mutation subtype (Fig. 3B; Supplementary Table S2B). TMB and PD-L1 expression across major cutoffs were significantly higher in the G12C subtype compared with any other KRAS subtype. An additional analysis was performed to compare these biomarkers in patients with KRAS G12A, G12C, and G13D mutation subtypes due to different preferentially activated signaling pathways (Supplementary Table S2C; refs. 17, 18). However, additional analysis demonstrated that there were no significant differences in comparison of immune checkpoint inhibitor response markers, including PD-L1 expression, TMB, and MSI/MMR deficiency, between KRAS G12A and G12C mutations. In addition, after correction for multiple comparison, there were no significant differences for KRAS G13D compared with G12A and G12C subtypes.
Figure 3.
Immune checkpoint therapy associated markers among KRAS-mutated tumors (A) and comparison of these markers between KRAS G12C mutated (G12C mt) and all other subtypes (non G12C mt; B). Prevalence of patients with NSCLC with high TMB (defined by ≥ 10 mt/Mb), MSI-H/MMR, and PD-L1 TPS (IHC 22c3) expression across major cut offs (TPS ≥ 1%, ≥ 10% and ≥ 50%) among each KRAS mutation subtype. B, The prevalence of patient with NSCLC with G12C mutations who had tumors with a high TMB (P = 0.01; q = 0.013), PD-L1 TPS ≥1% (P < 0.001; q < 0.001), and PD-L1 TPS ≥ 50% (P < 0.001; q < 0.001) was significantly greater than any other KRAS subtype. ** represents q < 0.05 (statistically significant).
Figure 4.
TMB distribution among KRAS mutations. TMB distribution values with beeswarm plot displaying all patient data points. Median TMB displayed for KRAS WT and each KRAS mutation subtype.
Co-occurring mutations
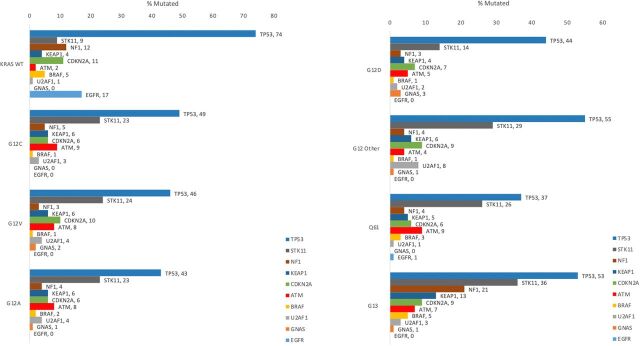
Significant differences in STK11 (LKB1), KEAP1, TP53, BRAF, U2AF1, NF1, and GNAS co-mutations were observed across KRAS mutational subtypes (P < 0.0001; Fig. 5; Supplementary Table S3). STK11 was mutated in 8.6% of KRAS WT NSCLC but more frequently noted in every KRAS subtype, with the highest rate in G13 mutations (36.2%) and the lowest in G12D (14.2%). KEAP1 was mutated most frequently with KRAS G13 (13.10%), which was more than twice the frequency in any other KRAS mutation subtype (3.70%–6.30%) or WT (4.20%) cases. TP53 mutations were more frequent in KRAS WT NSCLC (73.6%), with the highest rate among KRAS mutants at 55.4% (G12other) and the lowest at 36.8% (Q61 mutations). BRAF and U2AF1 mutations were much less common overall. BRAF mutations were most frequent in G13-mutated cases (5.20%), compared with any other KRAS mutation subtype (0.70%–0.260%), followed by WT cases (4.80%). U2AF1 was mutated most frequently in KRAS G12other cases (7.70%), which was more than twice the frequency in any other KRAS mutation subtype (1.30%–3.70%) or WT (0.60%) cases. NF1 was noted to be mutated in 21.4% of KRAS G13 cases, while all other KRAS mutations had a lower frequency of NF1 mutations (2.8%–5.1%) than KRAS WT (11.5%). GNAS mutations were observed most frequently in G12D cases (3.4%) and less frequently in other KRAS mutations and WT cases (0.3%).
Figure 5.
Mutation rates of key biomarkers in KRAS-mutated NSCLC cohorts. Frequency of the 10 most common co-occurring mutations including TP53, STK11, NF1, KEAP1, CDKN2A, ATM, BRAF, U2AF1, GNAS, and EGFR are displayed for each KRAS mutation subtype as well as KRAS WT group.
Significant differences in STK11 (P < 0.0001), KEAP1 (P < 0.0001), TP53 (P = 0.0002), and BRAF (P = 0.0002) persisted across KRAS mutational subtypes in adenocarcinoma patients alone (Supplementary Fig. S1; Supplementary Table S4A). STK11 was mutated most frequently in KRAS G13 cases (37.3%) and least frequently in G12D (15.8%). KEAP1 mutations were more frequent in G13 (12.6%) which was more than twice the frequency in any other KRAS mutation subtype (3.4%–6.2%). TP53 mutations were most frequent in G12other (51.8) followed closely by G13 (51.1%) and least frequent in Q61 mutations (36.0%). BRAF was mutated most frequently in G13 cases (4.9%) and least frequently in G12D cases (0.9%).
There were no significant differences in co-mutation frequency observed across KRAS subtypes in squamous NSCLC, only 4.4% of KRAS mutations (Supplementary Fig. S1; Supplementary Table S4B). Co-mutations that approached statistical significance included STK11, TP53, and CDKN2A. In the squamous cell carcinoma cohort, STK11 was mutated most frequently in G12other (37.5%) while no STK11 mutations were seen in G12D or Q61 cases. TP53 co-mutation was seen in all G12 other mutated cases (100%) and least frequently in Q61 (40.0%). CDKN2A was most frequently mutated in G13 (41.7%) but was not seen at all in G12 or Q61 cases. BRAF was mutated most frequently in G12A (11.1%) followed by G13 (7.7%) but was not seen at all in the other KRAS subtypes.
Further analysis of ATM and U2AF1 co-mutations was performed. Significantly different genomic alterations were found in patients with KRAS-mutated NSCLC with concomitant ATM mutations compared to ATM WT (Supplementary Fig. S2A). Mutations more frequently observed in KRAS-mutated/ATM-mutated patients include CCND1 (3.8% vs. 0.8%), FGF3 (4.2% vs. 0.8%), FGF4 (3.5% vs. 0.7%), and TP53 (18.4% vs. 49.7%) among others (P < 0.0001). There was a trend toward increased PTCH1 mutations in co-mutated ATM (2.4% vs. 0.4%; P = 0.003, Q = 0.063), conversely CDKN2A co-mutations trended higher in ATM WT compared with patients with ATM-mutant NSCLC (7.5% vs. 2.9%; P = 0.002, Q = 0.042). STK11 mutations were significantly increased in patients with KRAS mutated but U2AF1 WT NSCLC compared with KRAS- and U2AF1-mutant (23.6% vs. 9.9%, P < 0.001; Supplementary Fig. S2B). However, PD-L1–positive expression had a more frequent trend in those patients with concomitant KRAS and U2AF1 mutations compared with U2AF1 WT (72.3% vs. 60.3%; P = 0.006, Q = 0.227).
Discussion
While the role of KRAS mutations in tumorigenesis has been known for decades, no anti-cancer therapies targeting KRAS mutations have been successfully developed, until recently (19). Previous efforts to target MEK 1/2 or CDK 4/6 were ineffective (20). MEK inhibition lead to only modest efficacy with response rates of 11% and 12%, which were notably transient (21, 22). Recently, exciting phase I results have been reported with sotorasib, formerly AMG 510, a small molecule that irreversible inhibits KRAS G12C-mutant protein, demonstrating a 32.2% response rate and 88.1% disease control, in patients with KRAS G12C-mutated NSCLC (23, 24). Encouraging preclinical and clinical data has also been demonstrated using another KRAS G12C inhibitor, adagrasib, formerly MRTX849, with a recently reported response rate of 45% and disease control rate of 96.1% (25, 26). While these data are very promising for patients with KRAS-mutated NSCLC, there is significant variability in outcomes, duration of response, and mechanisms of resistance, all of which may be influenced by specific co-mutations present at diagnosis. Our study highlights the mutational heterogeneity that may explain prior inconsistent KRAS-targeted trial results and may influence future outcomes with subtype specific KRAS inhibitors and potentially with use of PD-1/PD-L1 inhibitors.
In KRAS-mutated NSCLC, there has been conflicting data on whether co-mutations influence outcomes with immunotherapy. In 2015, Skoulidis and colleagues described three major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities by analysis of gene expression profiles and co-occurring genomic alterations (8). The three major KRAS-mutant subsets were defined by co-mutations in STK11/LKB1 (KL subgroup), TP53 (KP subgroup), and CDKN2A/B inactivation as well as low expression of NKX2–1 (TTF1) transcription factor (KC subgroup). The KC subgroup had biallelic deletions of CDKN2A (encoding for the p16 tumor suppressor) and CDKN2B (encoding for the p15 tumor suppressor), both significantly enriched in this cohort. The other two subgroups had distinct immune profiles. The KP subgroup of patients with a co-mutation in TP53 had a higher tumor mutational load and characteristics of inflammatory response with increased expression of co-stimulatory (i.e., CD28) and co-inhibitory signals, including PD-L1. Therapeutic strategies using immune checkpoint inhibitors were appealing in this patient population given the reliance on PD-L/PD-L1 signaling and the increased immunogenicity with a large range of neoantigens. However, patients in the KL subgroup with a co-mutation in STK11/LKB1 were found to have more alterations in KEAP1 and ATM and had a “cold” immune microenvironment (relatively immune inert) with a lower rate of somatic mutations and anti-inflammatory signaling. Other studies have supported these immunogenic differences and response to immunotherapy in patients with KRAS-mutant NSCLC with TP53 or STK11 co-mutations (27, 28). In 2018, Skoulidis and colleagues showed that STK11/LKB1 co-mutations were associated with a shorter PFS and OS when treated with immune checkpoint inhibitors (6). Recently, their group demonstrated that STK11/LKB1 and/or KEAP1 alterations drive primary resistance to immune checkpoint inhibitors with a lack of benefit from the addition of pembrolizumab to chemotherapy and an inferior OS in patients with non-squamous NSCLC (29). Importantly, resistance persisted in PD-L1–positive patients which emphasizes the potential significance of STK11/LKB1 and/or KEAP1 co-mutations and also demonstrates the challenges with the PD-L1 biomarker. Of note, there was no consistent association between common mutant KRAS alleles (G12C, G12V, G12D) and the three expression clusters that were originally described previously (8).
Other studies have not shown a consistent relationship between KRAS co-mutations in NSCLC and outcomes with immunotherapy; PD-L1 expression has not been well characterized in this particular patient subset. Arbour and colleagues found co-occurring KEAP1 or NFE2L2 mutations were associated with shorter OS; however, STK11 and TP53 were not associated with an OS difference (9). In addition, exploratory analysis of patients with NSCLC, not necessarily harboring a KRAS mutation, enrolled in KEYNOTE-042 and KEYNOTE-189 demonstrated better outcomes with pembrolizumab (alone or with chemotherapy, respectively) independent of STK11 or KEAP1 mutation status (7, 30). Thus, while STK11 and KEAP1 mutations alone may have prognostic value, a KRAS co-mutation may also predict immune checkpoint inhibitor resistance. In this article, we describe the molecular heterogeneity of each specific KRAS mutation subtype. Interestingly, we found patients with G13 mutations had the highest rate of STK11 and KEAP1 co-mutations (Fig. 5; Supplementary Table S3). It will be important to explore treatment outcomes with immunotherapy and chemoimmunotherapy in patients with KRAS G13-mutated NSCLC, as this subset may be a primary driver of the lack of benefit observed in some studies with PD-1 axis blockade (16, 31–36).
While several preclinical and clinical studies have described patients with KRAS-mutated NSCLC and TP53, STK11, KEAP1 or CDKN2A co-mutations (6, 8, 9, 29, 37), there has been little prior work describing the potential clinical relevance of other predominant co-mutations demonstrated in our study, including ATM and U2AF1 (38). There is evidence that ATM-deficient lung adenocarcinoma is sensitive to PARP1 and ATR inhibitors (39). Here we have shown several mutations which are more frequent in patients with KRAS-mutated NSCLC with an ATM mutation (7.5% of KRAS-mutant cohort) compared with ATM WT (Supplementary Fig. S2A). Further exploration is needed to determine any therapeutic implications of these co-mutations. The functional role of U2AF1 in NSCLC has not been completely elucidated (40). We have demonstrated an interesting trend in PD-L1, a biomarker of immunotherapy response, which was higher in patients with KRAS-mutant/U2AF1-mutant NSCLC (3% of KRAS-mutant cohort) compared with KRAS-mutant/U2AF1 WT. In contrast, STK11, a potential marker of immunotherapy resistance, was significantly higher in patients with KRAS-mutant/U2AF1 WT NSCLC (Supplementary Fig. S2B). Further studies are needed to determine the response to immune checkpoint inhibitors in these patient groups.
In terms of characterization of PD-L1 expression in KRAS mutation subtypes, we found that any KRAS mutation subtype had a greater likelihood of PD-L1 expression, across all major cutoffs, than the WT subgroup (Fig. 3A; Supplementary Table S2A). Specifically, G12C was the most likely to be PD-L1 positive, with 65.5% TPS > 1%, and the most likely to be PD-L1 high, with 41.3% TPS ≥ 50% (Fig. 3B; Supplementary Table S2B). Unfortunately, we have limited data on the patient's smoking status which is known to be associated with higher TMB and PD-L1 expression, as well as response to immunotherapy (Fig. 2; Supplementary Table S1; refs. 42, 43). In addition, “light smokers” categorized as less than 15 packs per year does not accurately capture this patient population due to the loose definition. Prior studies which have included a “light smokers” category have used up to 10 pack years (13, 41). Thus, heavy smokers may also be captured in this category. While the proportions of KRAS mutation subtypes in each smoking category resembles some prior data, there are differences with the known biology of these subtypes (11–13). Therefore, we are unable to draw any conclusions regarding the influence of smoking status, or the KRAS biology itself, based on this cohort. Our data did not show a difference in immune checkpoint response markers (PD-L1, TMB, or MSI/MMR) when comparing subtypes with varying RAF signaling pathway dependence (Supplementary Table S2C). G12C and G13D subtypes have a high intrinsic GTPase hydrolysis rate and are less RAF signaling dependent compared with G12A, which has a low GTPase hydrolysis rate and is dependent on RAF signaling (17, 18). The observed difference, or lack thereof, will need to be subsequently explored in terms of clinical treatment outcomes to better understand their significance.
KRAS mutations are relatively common events in lung adenocarcinoma. Specific KRAS mutations exist in slightly different genomic landscapes: the rates of co-mutation of various relevant genes varied by specific KRAS mutation type. PD-L1 expression was also significantly different across specific KRAS mutations. These differences likely reflect differences in the underlying biology of each KRAS subset. Future therapeutic interventions must take note of these genomic differences as we further personalize cancer care.
Authors’ Disclosures
J. Judd reports non-financial support from Caris Life Sciences during the conduct of the study. N. Abdel Karim reports grants from BMS, Exelixis, and Pfizer outside the submitted work. J. Xiu reports other support from Caris Life Sciences during the conduct of the study. A.M. VanderWalde reports personal fees from Bristol Myers Squibb, Mirati Therapeutics, Caris Life Sciences, Elsevier, and George Clinical; non-financial support from Roche/Genentech and AstraZeneca; and grants from Amgen outside the submitted work. H. Mamdani reports personal fees from AstraZeneca and Zentalis outside the submitted work. L.E. Raez reports grants from BMS, Loxo, Lilly, Pfizer, Merck, Amgem, Genentech, Syndax, and Nanth Health during the conduct of the study. M. Nagasaka reports personal fees from AstraZeneca, Daiichi Sankyo, Takeda, Novartis, EMD Serono, Janssen, Pfizer, Caris Life Sciences, Blueprint Medicines, and Lilly, and non-financial support from An Heart outside the submitted work. S.G. Pai reports grants from University of South Alabama during the conduct of the study; personal fees from AstraZeneca outside the submitted work. M.A. Socinski reports grants and personal fees from Genentech, AstraZeneca, Merck, and Novartis; personal fees from Jazz, Lilly, Mirati, Regeneron, Guardant, AbbVie, Blueprint, and Janssen; grants from Spectrum, BeiGene, and Daiichi Sankyo during the conduct of the study; grants and personal fees from Genentech, AstraZeneca, Merck, and Novartis outside the submitted work. J.J. Nieva reports grants from Merck, and Genentech; personal fees from AstraZeneca, Fujirebio, Western Oncolytics, and Naveris outside the submitted work. C. Kim reports grants from AstraZeneca, BMS, Regeneron, Tesaro, Karyopharm, Debiopharm, Mirati, Genentech, Spectrum, and Merck; grants and personal fees from Novartis and Janssen; and personal fees from PierianDx outside the submitted work. A.J. Wozniak reports personal fees from Premier, Beigene, Incyte, Novocure, Janssen, GlaxoSmithKline, and Regeneron; other support from BeyondSpring and Odronate outside the submitted work. G. de Lima Lopes reports CARIS has provided research payments to my institution. A.I. Spira reports grants and personal fees from Amgen and Mirati; personal fees from Novartis, Pfizer, Merck, and AstraZeneca outside the submitted work. W.M. Korn reports personal fees from Merck outside the submitted work; and reports employment with Caris Life Sciences (stock ownership). E.S. Kim reports personal fees from AstraZeneca, Boehringer Ingelheim, and Genentech during the conduct of the study. S.V. Liu reports personal fees from AstraZeneca, Beigene, Daiichi Sankyo, G1 Therapeutics, Guardant Health, Inivata, Janssen, Jazz Pharmaceuticals, PharmaMar, Regeneron, Takeda, Novartis, and Amgen; grants and personal fees from Blueprint, Bristol Myers Squibb, Genentech, Lilly, Merck, Pfizer, Elevation Oncology, and Turning Point Therapeutics; grants from Alkermes, Bayer, Merus, Rain Therapeutics, and RAPT outside the submitted work. H. Borghaei reports grants and personal fees from BMS, Lilly, and Amgen; personal fees from Genentech, Pfizer, EMD-Serono, AstraZeneca, Novartis, Genmab, Regeneron, Pharmamar, and Daiichi; other support from Merck, Boehringer-Ingelheim, Huya Bio, Miratti, Incyte, Sonnetbio, Rgenix, and Nucleai; personal fees and other support from Takeda outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Supplemental Table 1. Frequency of KRAS mutation subtypes in never smokers/ light smokers (<15 packs/year) and current smokers
Supplemental Table 2. Immune checkpoint therapy associated markers among KRAS-mutated tumors (A), comparison of these markers between KRAS G12C and all other subtypes (B) and further comparison between G12C, G12A and G13D subtypes (C). * represents p or q values <0.05.
Supplemental Table 3. Mutation rates of key biomarkers in KRAS mutated NSCLC cohorts
Supplemental Figure 1. Key Biomarkers for KRAS mutation subtypes in adenocarcinoma and squamous cell NSCLC histologies. Frequency of the 10 most common co-occurring mutations including TP53, STK11, ATM, KEAP1, CDKN2A, U2AF1, BRAF, NFE3L2, RAF1 and EGFR are displayed for each KRAS mutation subtype in adenocarcinoma and squamous cell NSCLC cohorts.
Supplemental Table 4. Key Biomarkers for KRAS mutation subtypes in adenocarcinoma (A) and squamous cell (B) NSCLC histologies.
Supplemental Figure 2 . Differences in co-mutations and immune markers between patients with KRAS mutated (KRAS MT) NSCLC with or without an ATM mutation (ATM MT; A) and between patients with KRAS mutated (KRAS MT) NSCLC with or without a U2AF1 co-mutation (U2AF1 MT; B). (A) Frequency of the 10 most common co-occurring mutations as well as TMB high in patients with tumors harboring KRAS mutations with or without an ATM mutation. (B) Fequency of the 10 most common co-occurring mutation in patients with tumors harboring both KRAS mutations with or without U2AF1 mutations.
Data set used for analysis.
Acknowledgments
The authors would like to acknowledge the significant help and support received from the entire Caris team (Caris Life Sciences) in compilation of these data and in preparation of this article.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article is featured in Highlights of This Issue, p. 2315
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Contributions
J. Judd: Conceptualization, investigation, visualization, writing–original draft, writing–review and editing. N. Abdel Karim: Conceptualization, investigation, writing–review and editing. H. Khan: Conceptualization, investigation, writing–review and editing. A.R. Naqash: Investigation, writing–review and editing. Y. Baca: Data curation, software, formal analysis, visualization. J. Xiu: Data curation, software, formal analysis, validation, visualization. A.M. VanderWalde: Investigation, writing–review and editing. H. Mamdani: Investigation, writing–review and editing. L.E. Raez: Investigation, writing–review and editing. M. Nagasaka: Investigation, writing–review and editing. S.G. Pai: Investigation, writing–review and editing. M.A. Socinski: Investigation, writing–review and editing. J.J. Nieva: Investigation, writing–review and editing. C. Kim: Investigation, writing–review and editing. A.J. Wozniak: Investigation, writing–review and editing. C. Ikpeazu: Investigation, writing–review and editing. G. de Lima Lopes Jr: Investigation, writing–review and editing. A.I. Spira: Investigation, writing–review and editing. W.M. Korn: Investigation, writing–review and editing. E.S. Kim: Investigation, writing–review and editing. S.V. Liu: Conceptualization, investigation, visualization, writing–review and editing. H. Borghaei: Conceptualization, investigation, writing–review and editing.
References
- 1. Kris MG, Johnson BE, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Aronson SL, et al. Identification of driver mutations in tumor specimens from 1,000 patients with lung adenocarcinoma: The NCI's Lung Cancer Mutation Consortium (LCMC). J Clin Oncol 2011;29:CRA7506. [Google Scholar]
- 2. Hammerman PS, Lawrence MS, Voet D, Jing R, Cibulskis K, Sivachenko A, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shepherd FA, Domerg C, Hainaut P, Janne PA, Pignon JP, Graziano S, et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol 2013;31:2173–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rodenhuis S, Boerrigter L, Top B, Slebos RJ, Mooi WJ, van't Veer L, et al. Mutational activation of the K-ras oncogene and the effect of chemotherapy in advanced adenocarcinoma of the lung: a prospective study. J Clin Oncol 1997;15:285–91. [DOI] [PubMed] [Google Scholar]
- 5. Kalikaki A, Koutsopoulos A, Hatzidaki D, Trypaki M, Kontopodis E, Stathopoulos E, et al. Clinical outcome of patients with non-small cell lung cancer receiving front-line chemotherapy according to EGFR and K-RAS mutation status. Lung Cancer 2010;69:110–5. [DOI] [PubMed] [Google Scholar]
- 6. Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov 2018;8:822–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gadgeel SM, Rodriguez-Abreu D, Felip E, Esteban E, Speranza G, Reck M, et al. Pembrolizumab plus pemetrexed and platinum vs placebo plus pemetrexed and platinum as first-line therapy for metastatic nonsquamous NSCLC: analysis of KEYNOTE-189 by STK11 and KEAP1 status [abstract]. In: Proceedings of the Annual Meeting of the American Association for Cancer Research 2020; 2020 Apr 27–28 and Jun 22–24. Philadelphia (PA): AACR; Cancer Res 2020;80(16 Suppl):Abstract nr LB-397. [Google Scholar]
- 8. Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5:860–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arbour KC, Jordan E, Kim HR, Dienstag J, Yu HA, Sanchez-Vega F, et al. Effects of co-occurring genomic alterations on outcomes in patients with KRAS-mutant non-small cell lung cancer. Clin Cancer Res 2018;24:334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scheffler M, Ihle MA, Hein R, Merkelbach-Bruse S, Scheel AH, Siemanowski J, et al. K-ras mutation subtypes in NSCLC and associated co-occuring mutations in other oncogenic pathways. J Thorac Oncol 2019;14:606–16. [DOI] [PubMed] [Google Scholar]
- 11. Le Calvez F, Mukeria A, Hunt JD, Kelm O, Hung RJ, Taniere P, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res 2005;65:5076–83. [DOI] [PubMed] [Google Scholar]
- 12. Riely GJ, Kris MG, Rosenbaum D, Marks J, Li A, Chitale DA, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res 2008;14:5731–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dogan S, Shen R, Ang DC, Johnson ML, D'Angelo SP, Paik PK, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res 2012;18:6169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol 2020;21:1353–65. [DOI] [PubMed] [Google Scholar]
- 15. Merino DM, McShane LM, Fabrizio D, Funari V, Chen SJ, White JR, et al. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the Friends of Cancer Research TMB Harmonization Project. J Immunother Cancer 2020;8:e000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, et al. Durvalumab after chemoradiotherapy in stage III non-small-cell lung cancer. N Engl J Med 2017;377:1919–29. [DOI] [PubMed] [Google Scholar]
- 17. Tomasini P, Walia P, Labbe C, Jao K, Leighl NB.: Targeting the KRAS pathway in non-small cell lung cancer. Oncologist 2016;21:1450–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD.: Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol Cancer Res 2015;13:1325–35. [DOI] [PubMed] [Google Scholar]
- 19. McCormick F. K-Ras protein as a drug target. J Mol Med 2016;94:253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roman M, Baraibar I, Lopez I, Nadal E, Rolfo C, Vicent S, et al. KRAS oncogene in non-small cell lung cancer: clinical perspectives on the treatment of an old target. Mol Cancer 2018;17:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lopez-Chavez A, Thomas A, Rajan A, Raffeld M, Morrow B, Kelly R, et al. Molecular profiling and targeted therapy for advanced thoracic malignancies: a biomarker-derived, multiarm, multihistology phase II basket trial. J Clin Oncol 2015;33:1000–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blumenschein GR Jr, Smit EF, Planchard D, Kim DW, Cadranel J, De Pas T, et al. A randomized phase II study of the MEK1/MEK2 inhibitor trametinib (GSK1120212) compared with docetaxel in KRAS-mutant advanced non-small-cell lung cancer (NSCLC)dagger. Ann Oncol 2015;26:894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fakih M, O'Neil B, Price TJ, Falchook GS, Desai J, Kuo J, et al. Phase 1 study evaluating the safety, tolerability, pharmacokinetics (PK), and efficacy of AMG 510, a novel small molecule KRASG12C inhibitor, in advanced solid tumors. J Clin Oncol 37:15s, 2019. (suppl; abstr 3003). [Google Scholar]
- 24. Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, et al. KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N Engl J Med 2020;383:1207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRAS(G12C) Inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov 2020;10:54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jänne PA, Rybkin II, Spira AI, Riely GJ, Papadopoulos KP, Sabari JK, et al. KRYSTAL-1: Activity and Safety of Adagrasib (MRTX849) in Advanced/ Metastatic Non-Small-Cell Lung Cancer (NSCLC) Harboring KRAS G12C Mutation. European Journal of Cancer 138:S1-S2, 2020. [Google Scholar]
- 27. Schabath MB, Welsh EA, Fulp WJ, Chen L, Teer JK, Thompson ZJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene 2016;35:3209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Biton J, Mansuet-Lupo A, Pecuchet N, Alifano M, Ouakrim H, Arrondeau J, et al. TP53, STK11, and EGFR mutations predict tumor immune profile and the response to anti-PD-1 in lung adenocarcinoma. Clin Cancer Res 2018;24:5710–23. [DOI] [PubMed] [Google Scholar]
- 29. Skoulidis F, Arbour KC, Hellmann MD, Patil PD, Marmarelis ME, Awad MM, et al. Association of STK11/LKB1 genomic alterations with lack of benefit from the addition of pembrolizumab to platinum doublet chemotherapy in non-squamous non-small cell lung cancer. J Clin Oncol 37:15s, 2019. (suupl; abstr 102). [Google Scholar]
- 30. Cho BC. Genetic Mutations and Immunotherapy vs Chemotherapy in KEYNOTE-042. AACR Virtual Annual Meeting 2020; 2020. [Google Scholar]
- 31. Gandhi L, Rodriguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med 2018;378:2078–92. [DOI] [PubMed] [Google Scholar]
- 32. Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gumus M, Mazieres J, et al. Pembrolizumab plus chemotherapy for squamous non-small-cell lung cancer. N Engl J Med 2018;379:2040–51. [DOI] [PubMed] [Google Scholar]
- 33. Socinski MA, Jotte RM, Cappuzzo F, Orlandi F, Stroyakovskiy D, Nogami N, et al. Atezolizumab for first-line treatment of metastatic nonsquamous NSCLC. N Engl J Med 2018;378:2288–301. [DOI] [PubMed] [Google Scholar]
- 34. Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016;375:1823–33. [DOI] [PubMed] [Google Scholar]
- 35. Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A, et al. Updated analysis of KEYNOTE-024: pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer With PD-L1 tumor proportion score of 50% or greater. J Clin Oncol 2019;37:537–46. [DOI] [PubMed] [Google Scholar]
- 36. Antonia SJ, Villegas A, Daniel D, Vicente D, Murakami S, Hui R, et al. Overall survival with durvalumab after chemoradiotherapy in stage III NSCLC. N Engl J Med 2018;379:2342–50. [DOI] [PubMed] [Google Scholar]
- 37. Karim NFA, Onuoha C, Ahmad I, Eldessouki I, Ma PC, Feldman R, et al. Correlation between K-RAS mutant subsets, TP53 mutation, and PD-L1 status in non-small cell lung cancer (NSCLC). J Clin Oncol 37:15s, 2019. (suppl; abstr e14272). [Google Scholar]
- 38. Smida M, de la Cruz FF, Kerzendorfer C, Uras IZ, Mair B, Mazouzi A, et al. MEK inhibitors block growth of lung tumours with mutations in ataxia-telangiectasia mutated. Nat Commun 2016;7:13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmitt A, Knittel G, Welcker D, Yang T-P, George J, Nowak M, et al. ATM deficiency is associated with sensitivity to PARP1- and ATR inhibitors in lung adenocarcinoma. Cancer Res 2017;77:3040–56. [DOI] [PubMed] [Google Scholar]
- 40. Esfahani MS, Lee LJ, Jeon YJ, Flynn RA, Stehr H, Hui AB, et al. Functional significance of U2AF1 S34F mutations in lung adenocarcinomas. Nat Commun 2019;10:5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gainor JF, Rizvi H, Jimenez Aguilar E, Skoulidis F, Yeap BY, Naidoo J, et al. Clinical activity of programmed cell death 1 (PD-1) blockade in never, light, and heavy smokers with non-small-cell lung cancer and PD-L1 expression ≥50. Ann Oncol 2020;31:404–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ng TL, Liu Y, Dimou A, Patil T, Aisner DL, Dong Z, et al. Predictive value of oncogenic driver subtype, programmed death-1 ligand (PD-L1) score, and smoking status on the efficacy of PD-1/PD-L1 inhibitors in patients with oncogene-driven non-small cell lung cancer. Cancer 2019;125:1038–49. [DOI] [PubMed] [Google Scholar]
- 43. Willis C, Fiander M, Tran D, Korytowsky B, Thomas JM, Calderon F, et al. Tumor mutational burden in lung cancer: a systematic literature review. Oncotarget 2019;10:6604–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Frequency of KRAS mutation subtypes in never smokers/ light smokers (<15 packs/year) and current smokers
Supplemental Table 2. Immune checkpoint therapy associated markers among KRAS-mutated tumors (A), comparison of these markers between KRAS G12C and all other subtypes (B) and further comparison between G12C, G12A and G13D subtypes (C). * represents p or q values <0.05.
Supplemental Table 3. Mutation rates of key biomarkers in KRAS mutated NSCLC cohorts
Supplemental Figure 1. Key Biomarkers for KRAS mutation subtypes in adenocarcinoma and squamous cell NSCLC histologies. Frequency of the 10 most common co-occurring mutations including TP53, STK11, ATM, KEAP1, CDKN2A, U2AF1, BRAF, NFE3L2, RAF1 and EGFR are displayed for each KRAS mutation subtype in adenocarcinoma and squamous cell NSCLC cohorts.
Supplemental Table 4. Key Biomarkers for KRAS mutation subtypes in adenocarcinoma (A) and squamous cell (B) NSCLC histologies.
Supplemental Figure 2 . Differences in co-mutations and immune markers between patients with KRAS mutated (KRAS MT) NSCLC with or without an ATM mutation (ATM MT; A) and between patients with KRAS mutated (KRAS MT) NSCLC with or without a U2AF1 co-mutation (U2AF1 MT; B). (A) Frequency of the 10 most common co-occurring mutations as well as TMB high in patients with tumors harboring KRAS mutations with or without an ATM mutation. (B) Fequency of the 10 most common co-occurring mutation in patients with tumors harboring both KRAS mutations with or without U2AF1 mutations.
Data set used for analysis.