Dietary consumption of Lactobacillus-derived exopolysaccharide induces CCR6+ CD8+ T cells in Peyer's patches, and these cells distribute into CCL20-expressing tumor tissues to augment immune checkpoint blockade therapies in mice.
Abstract
Microbes and their byproducts have been reported to regulate host health and immune functions. Here we demonstrated that microbial exopolysaccharide produced by Lactobacillus delbrueckii subsp. bulgaricus OLL1073R-1 (EPS-R1) induced CCR6+ CD8+ T cells of mice and humans. In mice, ingestion of EPS-R1 augmented antitumor effects of anti–CTLA-4 or anti–PD-1 monoclonal antibody against CCL20-expressing tumors, in which infiltrating CCR6+ CD8+ T cells were increased and produced IFNγ accompanied by a substantial immune response gene expression signature maintaining T-cell functions. Of note, the antitumor adjuvant effect of EPS-R1 was also observed in germ-free mice. Furthermore, the induction of CCR6 expression was mediated through the phosphorylated structure in EPS-R1 and a lysophosphatidic acid receptor on CD8+ T cells. Overall, we find that dietary EPS-R1 consumption induces CCR6+ CD8+ T cells in Peyer's patches, favoring a tumor microenvironment that augments the therapeutic effect of immune-checkpoint blockade depending on CCL20 production by tumors.
Significance:
Gut microbiota- and probiotic-derived metabolites are attractive agents to augment the efficacy of immunotherapies. Here we demonstrated that dietary consumption of Lactobacillus-derived exopolysaccharide induced CCR6+ CD8+ T cells in Peyer's patches and improved the tumor microenvironment to augment the therapeutic effects of immune-checkpoint blockade against CCL20-producing tumors.
See related commentary by Di Luccia and Colonna, p. 1189.
This article is highlighted in the In This Issue feature, p. 1171
INTRODUCTION
The immune-suppressive microenvironment in tumor tissue often renders CD8+ T cells, which play a pivotal role to eradicate tumors, dysfunctional (often termed exhaustion), resulting in tumor progression (1). The blockade of immune checkpoints, critical immunosuppressive mechanisms for tumors, such as CTLA-4 and PD-L1/PD-1 induces robust antitumor immune responses (2, 3). Nevertheless, the response rates to immune-checkpoint blockade (ICB) therapies still remain low (4), and some human cancers, such as most colorectal cancer (5), are resistant.
Additionally, it has been reported that tumor cells dictate immune responses in their microenvironment by producing chemokines (6), thereby controlling patient survival (7). In ICB therapy, it was reported that IFNγ-inducible chemokines (CXCL9, CXCL10, and CXCL11) and their chemokine receptor (CXCR3) were required for the enhancement of the intratumor CD8+ T-cell responses and therapeutic efficacy (8, 9). Thus, conditioning of tumor tissues to become an inflamed IFNγ-rich microenvironment (hot tumor) would be an attractive strategy to augment the antitumor effect of ICB therapies.
Host gut microbiota is now established as one of critical parameters to improve or impair the efficacy of ICB therapies (10–13). These effects were postulated to be mediated by bacterial-derived metabolites or cell wall components [such as short chain fatty acids (14), inosine (15), and peptidoglycan (16)]. It was reported that commensal bacteria modulated the tumor microenvironment (17) and further that intestinal commensal bacteria–induced IFNγ-producing CD8+ T cells augmented antitumor responses in ICB therapies (18). Moreover, it was recently reported that Bifidobacterium pseudolongum–produced inosine activated Th1 cells and antitumor effects in the presence of IFNγ or other costimulation (15). However, the immunologic role of gut-related CD8+ T cells in the tumor microenvironment and the precise mechanisms by which the antitumor effect of ICB was augmented were not clarified.
Probiotics, live microorganisms that confer a health benefit when administered at the appropriate levels (19), have been thought to affect host biological functions and reactions. Several functional molecules/mechanisms of probiotics have been previously reported (20, 21), and probiotic-derived metabolites are attractive reagents to augment the efficacy of immune therapies, particularly against cancer (22, 23). Exopolysaccharides (EPS) are high-molecular-weight polymers composed of sugar residues secreted by a microorganism into the surrounding environment. We have been examining the biological functions of EPS produced by Lactobacillus and found that these EPS induced IFNγ production by splenocytes (24). In particular, Lactobacillus delbrueckii subsp. bulgaricus OLL1073R-1 (L. bulgaricus R1) produced a substantial amount of EPS (EPS-R1)-inducing IFNγ in the fermented skim milk (24). Here, we examined the effect of oral consumption of EPS-R1 on T cells and found that CCR6+ CD8+ T cells were induced in Peyer's patches, which infiltrated into CCL20-producing tumor tissues to augment antitumor effects of ICB therapies in experimental mouse tumor models.
RESULTS
Oral Ingestion of EPS-R1 Induces CCR6+ CD8+ T Cells
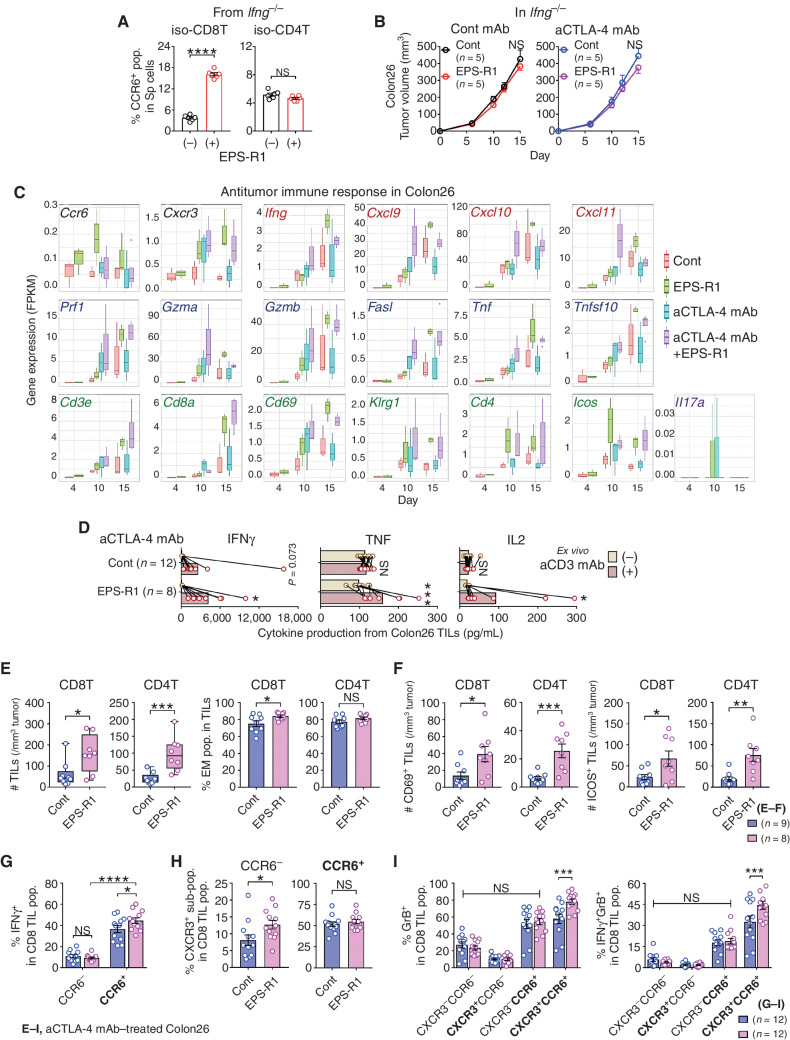
The recruitment of effector cytotoxic T lymphocytes (CTL) into the tumor is one of critical steps in the cancer–immunity cycle (25). Lymphocyte migration from the intestine into the tumor is an interesting possible mechanism by which gut microbiota, probiotics, or their metabolites might augment antitumor immune responses (26). When we examined the chemokine receptor gene expression profile of T cells in mouse Peyer's patches following oral ingestion of EPS-R1 for 6 days, we observed that Ccr6 mRNA expression was significantly augmented in CD8+ T cells, but not in CD4+ T cells (Fig. 1A). This upregulation of Ccr6 mRNA expression in CD8+, but not CD4+, T cells in Peyer's patches was confirmed by flow-cytometric analysis (increased CCR6+ population; Fig. 1B; Supplementary Fig. S1A and S1B). However, the CCR6+ population was not increased in CD8+ T cells in other intestinal compartments such as small intestinal epithelium or mesenteric lymph nodes (Fig. 1C). Flow-cytometric analysis was also consistent with the lack of change in Cxcr3 mRNA expression in CD8+ and CD4+ T cells following EPS-R1 ingestion (Fig. 1A and B). Although reduced Ccr9 mRNA expression in CD4+ T cells was demonstrated (Fig. 1A), EPS-R1 ingestion did not decrease the CCR9+ population in Peyer's patch CD4+ T cells (Fig. 1B). No inflammatory symptoms were histologically and clinically observed in the small intestine following EPS-R1 ingestion (Supplementary Fig. S1C).
Figure 1.
Oral ingestion of EPS-R1 induces CCR6+ CD8+ T cells. A and B, After BALB/c WT mice had ingested EPS-R1 for 6 days, mononuclear cells were prepared from their Peyer's patches. Following CD8+ or CD4+ T-cell isolation, mRNA gene expression of 10 chemokine receptors in CD8+ or CD4+ T cells was examined by quantitative reverse transcription PCR (RT-qPCR) methods (n = 4; A). CCR6+ or CXCR3+ populations among Peyer's patch CD8+ T cells (n = 16 in control and n = 15 in EPS-R1; all results obtained from three experiments are presented) and the CCR6+, CXCR3+, or CCR9+ populations among Peyer's patch CD4+ T cells (n = 4 for CCR6 and n = 6 for CXCR3 and CCR9) were examined by flow cytometry (B). C, After BALB/c WT mice had ingested EPS-R1 for 6 days, small intestinal intraepithelial lymphocytes (sIEL) or mesenteric lymph node cells (MLN) were prepared, and CCR6+ populations among CD8+ T cells were examined by flow cytometry (n = 6). D, Splenocytes freshly prepared from BALB/c WT mice were stimulated with EPS-R1 for 24 hours in vitro, and then CCR6+ or CXCR3+ populations among CD8+ and CD4+ T cells were examined by flow cytometry (n = 6). E and F, CD8+ or CD4+ T cells were isolated from splenocytes of BALB/c WT mice and stimulated with EPS-R1 for 24 hours in vitro. CCR6+ populations among CD8+ or CD4+ T cells were examined by flow cytometry (n = 8; E). Proliferation was examined by a CellTrace kit using flow cytometry, and the percentage of proliferating cells among CCR6+ or CCR6− populations in CD8+ T cells was presented (n = 8; F). G, Human PBMCs were obtained from three individuals and stimulated with EPS-R1 for 24 hours in vitro, and then the CCR6+ populations among CD8+ T cells were examined by flow cytometry (n = 8). H and I, EPS-R1 or water was intraduodenally administered into Wistar/ST rats, and CD8+ T cells were isolated from lymphocytes in the thoracic duct (TD) lymph 22 to 24 hours after the administration. Then, RNAs were prepared, and the gene expression of five chemokine receptors was examined by RT-qPCR methods (n = 3 in control and n = 4 in EPS-R1). Results are presented as the mean ± SEM and are depicted as scatter plots of the results of individual samples (A–G and I), and processed as the relative change of postadministration (post) mRNA expression levels compared with the preadministration (pre) controls in the same rats (I). Statistical analyses were performed by two-way ANOVA with Bonferroni correction (A and I) or Student t test (B–G). NS, not significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Similar results were obtained from two (A, B in CCR9, C and F), three (B in CXCR3 and CCR6), and five (D and E) independent experiments. CD8(4)T, CD8(4)+ T cell(s); cont, control; iso, isolated; pop., population(s); PP, Peyer's patch; Sp, spleen. Bold font in A and I is used to highlight the three most important chemokine receptors.
When splenocytes were stimulated in vitro with EPS-R1, CCR6+, but not CXCR3+, populations were significantly and selectively increased among CD8+ T cells (Fig. 1D). The CCR6+ population was also selectively increased among CD8+ T cells, when CD8+ or CD4+ T cells isolated from splenocytes were stimulated with EPS-R1 in vitro (Fig. 1E; Supplementary Fig. S1D). However, the frequency of proliferating cells was not increased among both CCR6+ and CCR6− CD8+ T cells (Fig. 1F; Supplementary Fig. S1E). In contrast, EPS prepared from the small intestinal contents of BALB/c wild-type (WT) mice did not increase the CCR6+ population among splenic CD8+ T cells in vitro (Supplementary Fig. S1F), suggesting that EPS derived from commensal microbiota was not sufficient to induce CCR6+ CD8+ T cells, even if commensal Lactobacilli were resident in the small intestine. Importantly, the CCR6+ population was increased in CD8+ T cells when human peripheral blood mononuclear cells (PBMC) were stimulated with EPS-R1 in vitro (Fig. 1G; Supplementary Fig. S1G).
Gut-preactivated CD8+ T cells were reported to migrate to the periphery (27), and that may possibly occur through the thoracic duct (28). We confirmed that rat thoracic duct CD8+ T cells expressed the five chemokine receptor genes that were dominantly expressed in mouse Peyer's patch CD8+ T cells (Supplementary Fig. S1H). The duodenal administration of EPS-R1 by catheter significantly augmented the expression of Ccr6 mRNA, but not other chemokine receptor mRNAs, in the thoracic duct CD8+ T cells compared with the control (Fig. 1H and I).
Taken together, these results suggested that EPS-R1 increased the CCR6+ population among CD8+ T cells, but not CD4+ T cells, and that these cells in Peyer's patches possibly distributed to the periphery through the thoracic duct.
CCR6 Gene Expression Correlates with Favorable Prognosis in Some Human CCL20-Producing Cancers
It was reported that tumors intrinsically shape antitumor immune responses by their chemokine production profile (6). When we examined the CCL20 gene, encoding the sole ligand for CCR6, in human cancers, a number of cancer types substantially expressed the CCL20 gene as compared with paired normal tissues (Supplementary Fig. S2A and Supplementary Table S1). Colorectal adenocarcinoma (CRAD) was demonstrated to express CCL20 at relatively higher levels compared with the other cancer types, whereas skin cutaneous melanoma expressed a low level of CCL20 transcripts (Fig. 2A).
Figure 2.
CCR6 gene expression correlates with favorable prognoses of some human CCL20-producing cancers. A, Fragments per kilobase of exon per million reads mapped (FPKM) of CCL20 gene expression were calculated from the RNA-sequencing results in 20 types of indicated human cancers using The Cancer Genome Atlas (TCGA) database obtained from the Human Protein Atlas. Results are presented as box plots with 10 to 90 percentiles, and outliers are shown. Lung cancer includes LUAD and LUSC, and renal cancer includes KICH, KIRC, and KIRP. B, HRs of death for high CCR6 and CCL20 gene expression in 19 types of human cancers in the TCGA database are presented as forest plots. Results are presented with 95% CIs on a logarithmic scale. Size of the marks indicates the number of samples (patients). The red bars indicate the correlation with prolonged survival, and the blue bars indicate the correlation with brief survival. A random-effects model meta-analysis was applied to pool the effect size, and the pooled effect size with the associated 95% CIs is described in a diamond. The 95% prediction interval of the pooled effect is described in the horizontal bar. Between-cancer-type heterogeneity variance (I2 value) is shown in the panel. C, OS curves for patients with a higher level of CCR6 or CCL20 gene expression in tumor than the median level (green) and patients with lower level of either gene in their tumor than the median level (purple) in CRAD, HNSC, and BRCA are presented. Survival curves are presented within day 3,000, and points censored are shown as a black bar. Results were analyzed with Kaplan–Meier survival analysis by log-rank methods, and P values are shown in the panel. NS, not significant. *, P < 0.05; **, P < 0.01. Number (n) of tumors (A) or patients (B and C) in every type of cancer is indicated. Cancer-type abbreviations are defined in Supplementary Table S1. Can., cancer. Bold font in A and B is used to highlight the most significant findings.
The expression levels of CCR6 and CCL20 appeared to variably correlate among 19 different human cancer types (Supplementary Fig. S2B). Interestingly, when we examined the correlation between prognosis and CCR6 or CCL20 gene expression in the 19 human cancer types, CCR6, but not CCL20, expression correlated with favorable prognoses in CRAD, head and neck squamous cell carcinoma (HNSC), and breast invasive carcinoma (BRCA; Fig. 2B). Moreover, patients with CRAD, HNSC, or BRCA expressing CCR6 at higher than median levels demonstrated a significantly longer overall survival (OS) when compared with patients expressing lower levels of CCR6, whereas CCL20 gene expression level did not correlate with prolongation of OS (Fig. 2C). We next performed a random-effects model meta-analysis under the assumption that the impact of CCR6 expression on prognosis may vary across cancer types. We found that the overall hazard ratio (HR) was 0.893 [95% confidence interval (CI), 0.798–0.9999; P = 0.0499], suggesting that higher expression of CCR6 was associated with better OS (Fig. 2B). Of note, we observed low to moderate between-cancer-type heterogeneity variance at an I2 value of 34.9% (95% CI, 0.0%–62.5%; P = 0.07), and the prediction interval of HR ranged from 0.67 to 1.19. This indicated that the positive impact of higher CCR6 expression on survival seen in some cancers might be due to a random effect (Fig. 2B).
Overall, these results suggested that infiltration of CCR6+ cells into tumor tissues correlated with the favorable prognosis of some human CCL20-producing cancers, although the impact of CCR6 expression on prognosis was not consistent across cancer types.
EPS-R1 Ingestion Exerts an Antitumor Adjuvant Effect for ICB Therapies in CCL20-Producing Tumors
Next, we examined experimental mouse tumors and found that CD45− cells isolated from Colon26 colon adenocarcinoma and 4T1 mammary carcinoma expressed a significantly increased CCL20 mRNA level compared with B16F10 melanoma (Fig. 3A). CCL20 protein expression in Colon26 and 4T1 tumor tissues was confirmed by IHC (Supplementary Fig. S3A). In addition, tumor-infiltrating CD45+ cells isolated from Colon26 and 4T1 tumors expressed Ccr6 mRNA at a significantly higher level compared with those isolated from B16F10 tumors (Fig. 3A). Furthermore, in the tumor-bearing mice, single administration of EPS-R1 by oral gavage began to increase the CCR6+ population among CD8+ tumor-infiltrating lymphocytes (TIL) in Colon26 tumors within 6 hours and significantly increased this population after 24 hours (Fig. 3B). These results possibly suggested that EPS-R1–induced CCR6+ CD8+ T cells distributed from Peyer's patches selectively infiltrated CCL20-expressing tumor tissues in mice.
Figure 3.
EPS-R1 augments antitumor effects of ICB therapies. A, BALB/c WT or C57BL/6 (B6) WT mice were subcutaneously inoculated with syngeneic Colon26, 4T1 (BALB/c), or B16F10 (B6) tumor cells, respectively. CD45− or CD45+ cells were isolated from single-cell suspensions prepared from growing tumors on day 15, and RNAs were obtained from these cells. Ccl20 gene expression in CD45− cells and Ccr6 gene expression in CD45+ cells were examined by RT-qPCR methods (n = 6). All results obtained from two experiments are presented. B, BALB/c WT mice were subcutaneously inoculated with syngeneic Colon26 tumor cells, and EPS-R1 was administered by oral gavage on day 7. Then, the CCR6+ population among CD8+ TILs was examined by flow cytometry 6, 24, 48, and 72 hours later [n = 5 in control (6, 24, and 48 hours) and n = 7 in control (72 hours) and in EPS-R1]. C, BALB/c WT mice were subcutaneously inoculated with syngeneic Colon26 tumor cells, and dietary ingestion of EPS-R1 was started on day 0. Some mice were intraperitoneally treated with anti–CTLA-4 mAb or anti–PD-1 mAb. Tumor volumes were periodically measured and statistically compared on day 15. Tumor growth among anti–CTLA-4 mAb– or anti–PD-1 mAb–treated mice was not significantly different with that among control mice. All results obtained from three (control Ig– and anti–CTLA-4 mAb–treated groups) or two (anti–PD-1 mAb–treated group) experiments are presented. D, BALB/c WT mice were subcutaneously inoculated with syngeneic 4T1 tumor cells, and dietary ingestion of EPS-R1 was started on day 0. Some mice were intraperitoneally treated with anti–CTLA-4 mAb. Tumor volumes were periodically measured and statistically compared on day 15. Tumor growth among anti–CTLA-4 mAb–treated mice was not significantly different with that among control mice. All results obtained from two experiments are presented. E, BALB/c WT mice were subcutaneously inoculated with syngeneic 4T1 tumor cells, and dietary ingestion of EPS-R1 was started on day 0. Some mice were intraperitoneally treated with anti–PD-1 mAb. Tumor volumes were periodically measured and statistically compared on day 15. Tumor growth among anti–PD-1 mAb–treated mice was not significantly different from that among control mice. F, BALB/c WT mice were subcutaneously inoculated with syngeneic Colon26 tumor cells, and dietary ingestion of EPS-R1 commenced on day 0. Some mice were intraperitoneally treated with anti–CTLA-4 mAb or anti–PD-1 mAb. Some EPS-R1–ingested mice were additionally treated with anti-ASGM1 Ab to deplete NK cells. Tumor volumes were periodically measured and those of EPS-R1–ingested mice treated with or without anti-ASGM1 Ab were statistically compared on day 14. Tumor growth among anti–CTLA-4 mAb– or anti–PD-1 mAb–treated mice was not significantly different from that among control mice. G, Syngeneic Colon26 tumor cells were inoculated into the cecal wall of BALB/c WT mice (orthotopic transplant model), and dietary ingestion of EPS-R1 commenced on day 0. Some mice were intraperitoneally treated with an anti–CTLA-4 mAb or anti–PD-1 mAb. Tumor volume was measured and statistically compared on day 14. Tumor growth among anti–CTLA-4 mAb– or anti–PD-1 mAb–treated mice was not significantly different from that among control mice. All results obtained from three experiments are presented. H and I, B6 WT mice were subcutaneously inoculated with syngeneic B16F10 tumor cells (H) or B16F10-CCL20 tumor cells (I), and dietary ingestion of EPS-R1 was started on day 0. All mice were intraperitoneally treated with an anti–CTLA-4 mAb (H), and some mice were intraperitoneally treated with an anti–CTLA-4 mAb or anti–PD-1 mAb (I). Tumor volumes were periodically measured and statistically compared on day 15. Tumor growth among anti–CTLA-4 mAb– or anti–PD-1 mAb–treated mice was not significantly different from that among control mice (I). J, BALB/c WT mice were subcutaneously inoculated with syngeneic Colon26 tumor cells, and some mice were dietary ingested with EPS-R1 starting on day 0. All mice were intraperitoneally treated with an anti–CTLA-4 mAb and additionally with a neutralizing anti-CCL20 mAb. Tumor volumes were periodically measured and statistically compared on day 15. K, Single-cell suspensions were prepared from some anti–CTLA-4 mAb–treated tumors on day 15 presented in C, D, H, and J. Then, CCR6-expressing populations among CD3+ CD8+ TILs were examined by flow cytometry. Results are presented as box plots with individual samples (A), mean ± SEM (C–F and H–J), and scatter plots of the results of individual samples (B, G, and K). Results are processed as the relative values of individual EPS-R1–ingested samples compared with the mean values of control samples in each time point. Number (n) of mice in every group is indicated in the panels (C–K). Statistical analyses were performed by Kruskal–Wallis test with Dunn correction (A), two-way ANOVA with Bonferroni correction (B), or Student t test (C–K). NS, not significant. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Similar results were obtained from two (A, C in anti–PD-1 mAb group, H, J, and K) or three (C in control Ig and anti–CTLA-4 mAb groups, D, E, G, and I) independent experiments. aASGM1, anti-ASGM1Ab; aCTLA-4 (PD-1, CCL20), anti–CTLA-4 (PD-1, CCL20) (mAb); CD8 TIL(s), CD8+ TIL(s); cont, control; pop., population; ortho., orthotopic.
We then examined the effect of ingestion of EPS-R1 upon tumor growth with or without ICB monoclonal antibodies (mAb). Although ingestion of EPS-R1 alone did not inhibit the growth of Colon26 or 4T1 tumors, the antitumor effect of ICB using anti-CTLA-4 mAb or anti-PD-1 mAb was significantly augmented by the ingestion of EPS-R1 (Fig. 3C–E; Supplementary Fig. S3B and S3C). Natural killer (NK) cell depletion did not diminish augmentation of the antitumor effect of ICB mAbs by EPS-R1 ingestion against Colon26 tumors (Fig. 3F). Of note, ingestion of EPS-R1 similarly augmented the antitumor effect of ICB mAbs in the orthotopic transplant model of Colon26 tumors (Fig. 3G). By contrast, ingestion of EPS-R1 did not augment the antitumor effect of the anti–CTLA-4 mAb against B16F10 tumors that did not produce CCL20 (Fig. 3H). Alternatively, ingestion of EPS-R1 augmented the antitumor effect of ICB mAbs against genetically engineered B16F10 tumors that produce CCL20 (B16F10-CCL20; Fig. 3I). Moreover, neutralizing anti-CCL20 mAb treatment abolished the increased antitumor effect of the anti–CTLA-4 mAb and EPS-R1 combination against Colon26 tumors (Fig. 3J).
Ingestion of EPS-R1 increased the CCR6-expressing population among CD8+ T cells infiltrating anti–CTLA-4 mAb–treated Colon26 and 4T1 tumors, but not B16F10 tumors (Fig. 3K; Supplementary Fig. S3D and S3E). Neutralizing anti-CCL20 mAb treatment abolished the increase of the CCR6+ population among CD8+ T cells in anti–CTLA-4 mAb–treated Colon26 tumors following EPS-R1 ingestion (Fig. 3K).
These results demonstrated that ingestion of EPS-R1 augmented the antitumor effects of ICB therapies against CCL20-producing tumors in a CCL20-dependent manner, suggesting that EPS-R1–induced CCR6+ CD8+ T cells might play some adjuvant roles in this antitumor effect.
IFNγ-Producing Cytotoxic CXCR3+ CCR6+ CD8+ T Cells Are Increased in the CCL20-Producing Tumors Treated with an Anti–CTLA-4 mAb and EPS-R1
When Colon26 or 4T1 tumor cells were cocultured with EPS-R1, EPS-R1 neither induced apoptosis or immunogenic cell death nor inhibited cell proliferation (Supplementary Fig. S4A–S4C), suggesting that EPS-R1 was not augmenting antitumor immune responses by direct effects on tumor cells (29). Although the CCR6+ population was increased when IFNγ-deficient (Ifng−/−) CD8+ T cells were stimulated with EPS-R1 in vitro (Fig. 4A), ingestion of EPS-R1 did not augment the antitumor effect of anti–CTLA-4 mAb against Colon26 tumors in Ifng−/− mice (Fig. 4B). Then, we performed RNA sequencing (RNA-seq) analysis upon Colon26 tumors (Supplementary Fig. S5A). Expression of Ccr6 transcripts was increased in the tumors of the mice that had ingested EPS-R1 (Fig. 4C). Anti–CTLA-4 mAb treatment, compared with the control on day 10, augmented the expression of Cxcr3 and the genes encoding cytotoxic molecules (Prf1, Gzma, Gzmb, Fasl, Tnf, and Tnfsf10; Fig. 4C). On day 10, the combination of anti–CTLA-4 mAb with EPS-R1 further augmented the expression of these genes and Ifng as well as the gene encoding IFNγ-inducible chemokines (Cxcl9, Cxcl10, and Cxcl11; Fig. 4C). Importantly, most of this elevated gene expression was maintained until day 15 (Fig. 4C). These results suggested that IFNγ produced in the tumor microenvironment might play important roles in the increased antitumor effect of anti–CTLA-4 mAbs with EPS-R1 ingestion.
Figure 4.
Preferentially infiltrating CCR6+ CD8+ T cells increase effector CD8+ T cells in anti–CTLA-4 mAb/EPS-R1–treated Colon26 tumors. A, CD8+ or CD4+ T cells were isolated from splenocytes of BALB/c Ifng−/− mice and stimulated with EPS-R1 for 24 hours in vitro. CCR6+ populations among CD8+ or CD4+ T cells were examined by flow cytometry (n = 6). B, BALB/c Ifng−/− mice were subcutaneously inoculated with Colon26 tumor cells, and dietary ingestion of EPS-R1 was started on day 0. Some mice were i.p. treated with anti–CTLA-4 mAb. Tumor volumes were periodically measured and statistically compared on day 15. Tumor growth among anti–CTLA-4 mAb–treated mice was not significantly different from that in control mice. C, RNA samples were obtained from Colon26 tumors in BALB/c WT mice treated with anti–CTLA-4 mAb and/or EPS-R1 ingestion on day 4, 10, and 15. Then, RNA-seq analysis was performed on individual samples (n = 3–4 in every group). Fragments per kilobase of exon per million reads mapped (FPKM) of 19 selected antitumor immune response–related genes are presented. D–I, CD45+ single-cell suspensions were prepared from Colon26 tumors in BALB/c WT mice treated with anti–CTLA-4 mAb with or without EPS-R1 ingestion on day 15. These cells were stimulated with anti-CD3 mAb ex vivo and cell-free culture supernatants were harvested 48 hours later, and then the amounts of IFNγ, TNF, and IL2 were examined by ELISA (n = 12 in control and n = 8 in EPS-R1; D). The number of CD8+ or CD4+ T cells per 1 mm3 tumor and the population of EM phenotype (CD44hi CD62L−) cells among CD8+ or CD4+ T cells (n = 9 in control and n = 8 in EPS-R1; E), the number of CD69- or ICOS-expressing CD8+ or CD4+ T cells per 1 mm3 tumor (n = 9 in control and n = 8 in EPS-R1; F), IFNγ+ population among CCR6-expressing CD8+ T cells (n = 12; G), CXCR3+ population among CCR6− or CCR6+ CD8+ T cells (n = 12; H), and granzyme B+ or IFNγ+ granzyme B+ (double-positive) population among CXCR3- and/or CCR6-expressing CD8+ T cells (n = 12; I) were examined by flow cytometry. Results are presented as the mean ± SEM and are depicted as scatter plots of the results of individual samples (A, E in % EM, and F–I), mean ± SEM (B), box plots (C), dot plots with mean and line connected between the results using the same samples (D), or box plots with individual samples (E in # TILs). Number (n) of mice in every group is indicated in the panels (B). Statistical analyses were performed by Student t test (A, B, E in % EM, F, and H), two-way ANOVA with Bonferroni correction (D, G, and I) or Mann–Whitney U test (E in # TILs). NS, not significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Similar results were obtained from two independent experiments (A, B, and G–I). All the results obtained from two independent experiments are presented (D). aCD3, anti-CD3ε; aCTLA-4, anti–CTLA-4; CD8(4)T, CD8(4)+ T cell(s); cont, control; GrB, granzyme B; iso, isolated; pop., population(s); Sp, spleen. Bold font in G–I is used to highlight positive expressing markers.
The higher expression levels of Cd3e, Cd8a, Cd69, and Klrg1 also suggested that EPS-R1 ingestion increased the infiltration of activated CD8+ T cells (Fig. 4C). Moreover, increased expression of Cd4 and Icos following EPS-R1 ingestion might suggest the increased infiltration of effector Th1-like cells that were previously reported to enhance the efficacy of anti–CTLA-4 mAb therapies (ref. 30; Fig. 4C). TILs obtained from Colon26 tumors treated with anti–CTLA-4 mAb/EPS-R1 produced a significantly increased amount of IFNγ, TNF, and IL2 following ex vivo anti-CD3 mAb stimulation (Fig. 4D). By contrast, TILs obtained from Colon26 tumors treated with anti–CTLA-4 mAb alone displayed only a trend toward an increased production of IFNγ (Fig. 4D). Consistently, EPS-R1 ingestion significantly increased the number of both CD8+ and CD4+ T cells and CD44high CD62L− effector memory (EM) phenotype populations among CD8+ T cells in anti–CTLA-4 mAb–treated Colon26 tumors (Fig. 4E). Moreover, CD69- or ICOS-expressing CD8+ and CD4+ T cells were increased in these tumors (Fig. 4F), supporting the results of RNA-seq analysis.
CXCR3, a chemokine receptor for IFNγ-inducible chemokines, was reported to play an important role in the efficacy of ICB therapy (9). When we examined TILs isolated from Colon26 tumors treated with an anti–CTLA-4 mAb by flow cytometry, IFNγ-producing cells were enriched in CCR6+ CD8+ TILs and that was significantly increased by EPS-R1 ingestion (Fig. 4G; Supplementary Fig. S5B). Interestingly, the CXCR3+ population was increased in CCR6− but not in CCR6+ CD8+ T cells (Fig. 4H; Supplementary Fig. S5C), suggesting the possibility that CXCR3+ CCR6− CD8+ T cells were recruited into Colon26 tumors by IFNγ-inducible chemokines. When we examined the expression of IFNγ and granzyme B along with CXCR3 and CCR6 on CD8+ T cells, ingestion of EPS-R1 significantly increased granzyme B+ cells and IFNγ+ granzyme B+ cells in CXCR3+ CCR6+ but not other CD8+ T-cell populations (Fig. 4I; Supplementary Fig. S5D and S5E).
In summary, these results suggested that CCR6+ CD8+ T cells induced by EPS-R1 ingestion infiltrated into CCL20-producing tumors and produced IFNγ, making the tumor microenvironment inflamed and maintaining T-cell functions, and that would result in enhancing the antitumor effects of anti–CTLA-4 mAb treatment.
Combination with EPS-R1 Ingestion Increases Tumor-Specific CTLs in Anti–CTLA-4 mAb–Treated Tumors
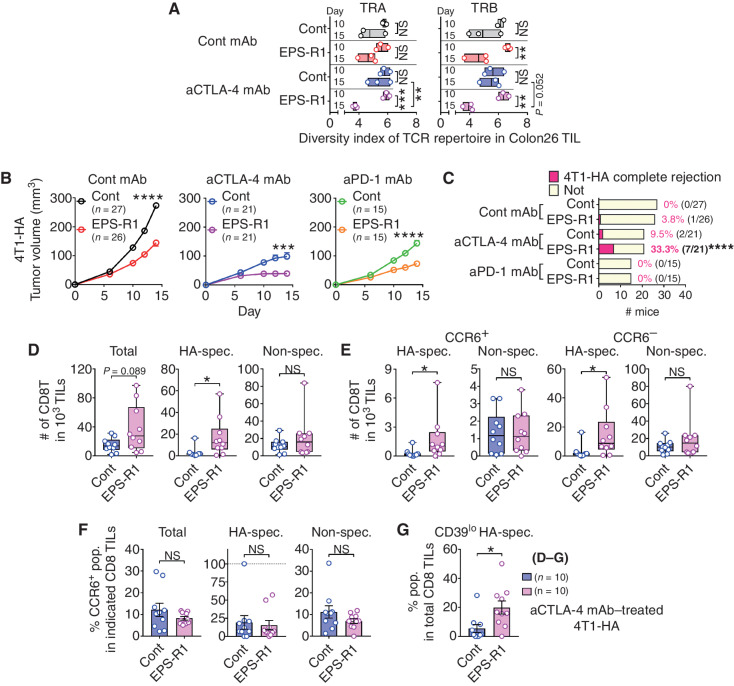
Next, we examined the T-cell receptor (TCR) repertoire of TILs in Colon26 tumors treated with anti–CTLA-4 mAb and/or EPS-R1 ingestion on days 7, 10, and 15 (Supplementary Fig. S6A and S6B). Significantly increased TCR [TCR α chain (TRA) and TCR β chain (TRB)] clonality of TILs on day 15 was demonstrated when compared with those on day 10 in anti–CTLA-4 mAb/EPS-R1–treated tumors by the Shannon–Wiener diversity index (Fig. 5A). Moreover, TRA clonality of TILs on day 15 in anti–CTLA-4 mAb/EPS-R1–treated tumors was significantly increased when compared with that in anti–CTLA-4 mAb–treated tumors (Fig. 5A). Furthermore, clonality of TRB in EPS-R1–treated tumors was also significantly increased on day 15 compared with that on day 10 (Fig. 5A). These results suggested that dietary EPS-R1 increased the clonality of intratumor T cells after day 10, and thus tumor-specific CTLs might be increased in anti–CTLA-4 mAb/EPS-R1–treated tumors by day 15 after EPS-R1 ingestion.
Figure 5.
Anti–CTLA-4 mAb/EPS-R1 treatment increases tumor-specific CTL infiltration into 4T1-HA tumors. A, RNA samples were prepared from Colon26 tumors in BALB/c WT mice treated with an anti–CTLA-4 mAb and/or EPS-R1 ingestion on day 10 (top bar) and day 15 (bottom bar), and TCR genes were analyzed by deep sequencing on individual samples. Diversity of TRA and TRB repertoire was examined by the Shannon–Wiener diversity index (n = 2–3). B and C, BALB/c WT mice were subcutaneously inoculated with 4T1-HA tumor cells, and dietary EPS-R1 was started on day 0. Some mice were intraperitoneally treated with an anti–CTLA-4 mAb or anti–PD-1 mAb. Tumor volumes were periodically measured and statistically compared on day 14 (B). Tumor growth in anti–CTLA-4 mAb– or anti–PD-1 mAb–treated mice was significantly suppressed (P < 0.0001, respectively) when compared with those in the control mice. Number (n) of mice in every group is indicated. Tumor rejection rate on day 14 is indicated and statistically compared (C). All results obtained from three (control Ig–treated group) or two (anti–CTLA-4 mAb– and anti–PD-1 mAb–treated groups) independent experiments are presented. D–F, Single-cell suspensions were prepared from 4T1-HA tumors in BALB/c WT mice treated with an anti–CTLA-4 mAb with or without EPS-R1 ingestion on day 15. Then, the number of total, HA-specific TCR+, or HA-nonspecific CD8+ T cells among 103 of TILs (D), the number of CCR6+ or CCR6− HA-specific TCR+/– CD8+ T cells among 103 of TILs (E), and the population of CCR6-expressing cells among total, HA-specific TCR+, or HA-nonspecific CD8+ T cells (F) were examined by flow cytometry (n = 10). G, CD39lo (nonexhausted) HA-specific TCR+ populations among CD8+ T cells in the single-cell suspensions prepared as described in D–F were examined by flow cytometry (n = 10). Results are presented as a floating bar graph with mean and individual samples (A), mean ± SEM (B), bar chart (C), box plots with individual samples (D and E), or dot plots with mean ± SEM (F and G). Statistical analyses were performed by two-way ANOVA with Bonferroni (between days) or Tukey (between treatments) correction (A), Student t test (B, F, and G), Chi-square residual analysis (C), or Mann–Whitney U test (D and E). NS, not significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Similar results were obtained from two independent experiments (D–G). aCTLA-4 (PD-1), anti–CTLA-4 (PD-1); CD8T, CD8+ T cell(s); cont, control; spec., specific; pop., population. Bold font in C is used to highlight statistical significance.
To examine tumor-specific CTL infiltration, we used 4T1 cells transduced with the influenza hemagglutinin (HA) antigen (4T1-HA; ref. 31). In this tumor model, either anti–CTLA-4 mAb or anti–PD-1 mAb treatment or ingestion of EPS-R1 alone exerted a significant antitumor effect and the combination of ICB mAb with EPS-R1 exerted an even greater antitumor effect resulting in the significantly increased tumor rejection rate (Fig. 5B and C; Supplementary Fig. S6C). Upon examination of TILs in anti–CTLA-4 mAb–treated 4T1-HA tumors, ingestion of EPS-R1 significantly increased HA-specific TCR-expressing CD8+ T cells but not HA-nonspecific or total CD8+ T cells (Fig. 5D; Supplementary Fig. S6D). Both CCR6+ and CCR6− populations of HA-specific, but not HA-nonspecific, CD8+ T cells were significantly increased among TILs (Fig. 5E), meaning that the frequency of the CCR6+ population was not increased in total, HA-specific, or HA-nonspecific CD8+ T cells (Fig. 5F). These results suggested that EPS-R1 ingestion increased not only CCR6+ but also CCR6− HA-specific CD8+ T cells among TILs in the highly antigenic 4T1-HA tumors. Moreover, the CD39lo nonexhausted (32) HA-specific population was significantly increased in CD8+ T cells within anti–CTLA-4 mAb/EPS-R1–treated tumors compared with those in the anti–CTLA-4 mAb alone–treated tumors (Fig. 5G; Supplementary Fig. S6E). These results implied that the tumor-specific CTLs increased in 4T1-HA tumors treated with anti–CTLA-4 mAb/EPS-R1 might not be exhausted. This assertion was consistent with the finding that TILs in anti–CTLA-4 mAb/EPS-R1–treated Colon26 tumors produced cytokines when stimulated with anti-CD3 mAb ex vivo (Fig. 4D).
Phosphate-Containing Moiety in EPS-R1 Is the Functional Component that Induces CCR6+ CD8+ T Cells through Lysophosphatidic Acid Receptor 2
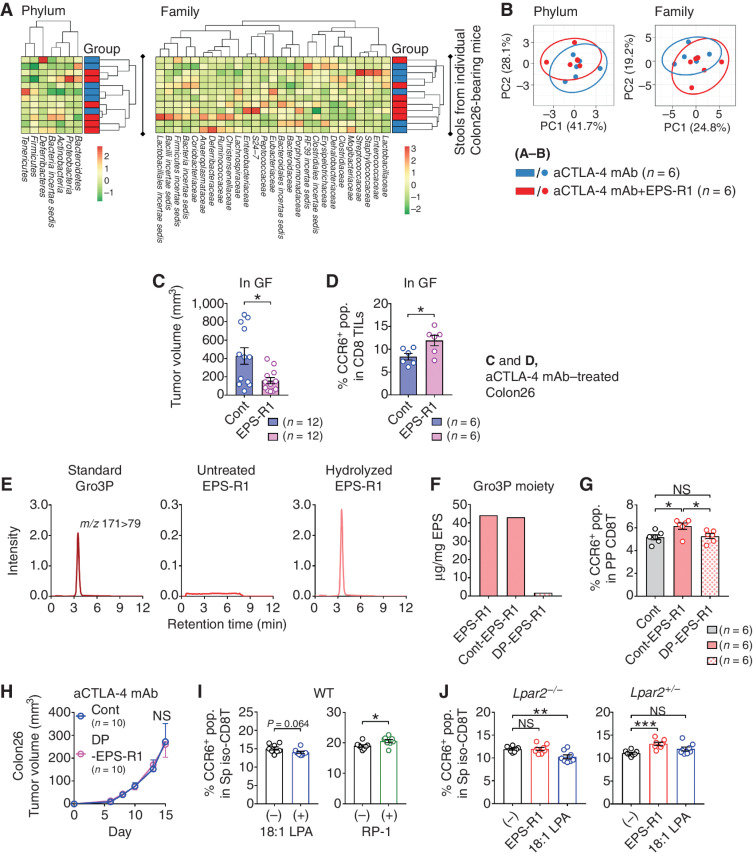
We further examined gut microbiota by 16S ribosomal RNA-seq (rRNA-seq) analysis of stools. Ingestion of EPS-R1 did not alter the gut microbiota of Colon26 tumor–bearing mice treated with an anti–CTLA-4 mAb (Fig. 6A and B). Furthermore, the ingestion of EPS-R1 by germ-free (GF) mice still augmented the antitumor effect of anti–CTLA-4 mAb (Fig. 6C) and increased the CCR6+ population among CD8+ T cells infiltrating Colon26 tumors treated with an anti–CTLA-4 mAb (Fig. 6D). These results indicated that dietary intake of EPS-R1 augmented the antitumor effect of anti–CTLA-4 mAbs via the induction of CCR6+ CD8+ T cells independently of host gut microbiota.
Figure 6.
The Gro3P moiety of EPS-R1 and lysophosphatidic acid (LPA) receptor 2 (LPA2) is involved in EPS-R1–induced CCR6 expression on CD8+ T cells. A, BALB/c WT mice were subcutaneously inoculated with Colon26 tumor cells and intraperitoneally treated with anti–CTLA-4 mAb with or without ingestion of EPS-R1. Stools were collected from the rectum on day 15, and microbiota of each mouse were examined by 16S rRNA-seq analysis (n = 6). Relative abundance of phyla and families was clustered for all mice in a heat map. B, Relative abundance of phyla and families demonstrated in A was assessed with principal component analysis (PCA) with prediction ellipses (95%) and contribution rate of PC1 and PC2. C, GF BALB/c WT mice were subcutaneously inoculated with Colon26 tumor cells and intraperitoneally treated with an anti–CTLA-4 mAb (n = 12). Dietary EPS-R1 was started on day 0. Tumor volumes were statistically compared on day 15. D, Single-cell suspensions were prepared from Colon26 tumors in GF BALB/c WT mice treated with an anti–CTLA-4 mAb with or without EPS-R1 ingestion on day 15 (n = 6). Then, the CCR6+ population among CD8+ T cells in TILs was analyzed by flow cytometry. E, Gro3P (standard), untreated EPS-R1, and hydrolyzed EPS-R1 were analyzed by LC-MS/MS to detect Gro3P. F, EPS-R1 was treated with HF for 10 minutes to prepare non-dephosphorylated EPS-R1 as the control (Cont-EPS-R1) or for 3 days to obtain dephosphorylated EPS-R1 (DP-EPS-R1). Then Gro3P content in these different EPS-R1 was examined by LC-MS/MS. Contents of Gro3P are presented as μg/mg in EPS. G, BALB/c WT mice were orally administered Cont-EPS-R1 or DP-EPS-R1 for 6 days, and then CCR6+ populations among Peyer's patch CD8+ T cells were examined by flow cytometry (n = 6). H, BALB/c WT mice were subcutaneously inoculated with syngeneic Colon26 tumor cells, and dietary ingestion of DP-EPS-R1 commenced on day 0. All mice were intraperitoneally treated with an anti–CTLA-4 mAb. Tumor volumes were periodically measured and statistically compared on day 15. I, CD8+ T cells were freshly isolated from splenic mononuclear cells (MNC) of BALB/c WT mice and stimulated with 18:1 LPA or RP-1 in vitro. After 24-hour incubation, the CCR6+ populations were examined by flow cytometry (n = 8). J, CD8+ T cells were freshly isolated from splenic MNCs of Lpar2−/− or Lpar2+/− ICR mice and stimulated with EPS-R1 or 18:1 LPA in vitro. After 24-hour incubation, the CCR6+ populations were examined by flow cytometry (n = 8). Results are presented as dot plots with mean ± SEM (C, D, G, I, and J), chromatograms (E), or mean ± SEM (H). Statistical analyses were performed by Student t test (C, D, H, and I) or one-way ANOVA with Tukey correction (G) or Dunnett correction (J). NS, not significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001. All the results obtained from two independent experiments are demonstrated (C), and similar results were obtained from two (A, B, D–H, and J) or three (I) independent experiments. aCTLA-4, anti–CTLA-4; CD8 TILs, CD8+ TILs; CD8T, CD8+ T cell(s); cont, control; iso., isolated; m/z, mass-to-charge ratio; pop., population; PP, Peyer's patch; Sp, spleen.
It was shown that EPS-R1 could be divided into neutral and acidic EPS subtypes, and acidic EPS-R1 induced IFNγ production from splenocytes (33). It was also reported that acidic EPS-R1 showed mitogenic activity on splenocytes, which was abolished by dephosphorylation of EPS-R1 (34). Thus, the phosphate group in EPS-R1 appeared to be important for its immunomodulatory activities. We examined the presence of glycerol-3-phosphate (Gro3P) moiety in EPS-R1, because the previous structural analyses indicated the presence of Gro3P moiety in EPS derived from some Lactobacillus strains (35). We now showed the release of Gro3P, as detected by liquid chromatography tandem mass spectrometry (LC-MS/MS), when EPS-R1 was subjected to acid hydrolysis (Fig. 6E). When mice were orally administered EPS-R1 dephosphorylated by hydrofluoric acid (HF) treatment (DP-EPS-R1; Fig. 6F), the CCR6+ population was not increased among Peyer's patch CD8+ T cells (Fig. 6G), and consistently that did not augment the antitumor effect when combined with anti–CTLA-4 mAb treatment (Fig. 6H). Furthermore, EPS from skim milk fermented with other L. bulgaricus strains contained Gro3P moiety (Supplementary Fig. S7A), and these EPS, such as EPS-R1, also increased the CCR6+ population among in vitro–cultured splenic CD8+ T cells (Supplementary Fig. S7B). These results suggested that Gro3P moiety is the functional component of L. bulgaricus–produced EPS that increased CCR6+ CD8+ T cells.
Signaling through G protein–coupled receptors (GPCR) has been reported as an underlying mechanism for some metabolites of commensal microbiota to modulate biological functions of the host (36). The structure of Gro3P is present in all glycerophospholipids, including lysophosphatidic acid (LPA, 1- or 2-acyl-sn-glycerol 3-phosphate), like EPS-R1 (Supplementary Fig. S7C). LPA is a typical lysophospholipid mediator and induces various immunologic responses through its specific GPCRs (37). Interestingly, the Gro3P moiety is a unique structure of LPA but not other lysophospholipid mediators such as lysophosphatidylcholine (LPC) or sphingosine 1-phosphate (S1P; ref. 38). Moreover, it was reported that dephosphorylation diminished biological activities of LPA (39), similarly to its impact upon the ability of dietary Gro3P-containing EPS-R1 to induce CCR6 expression on Peyer's patch CD8+ T cells and to augment the efficacy of anti–CTLA-4 mAb treatment. Thus, it is a reasonable hypothesis that LPA receptors contribute to the immunologic effects of EPS-R1.
Interestingly, the CCR6+ population was not statistically changed when splenic CD8+ T cells were stimulated with 18:1 LPA in vitro, but by contrast these cells were increased when stimulated with radioprotectin-1 (RP-1), a nonlipid LPA receptor 2 (LPA2)–specific agonist (Fig. 6I). Moreover, when splenic CD8+ T cells of Lpar2-deficient (Lpar2−/−) mice were stimulated with EPS-R1 or 18:1 LPA in vitro, the CCR6+ population was not increased by EPS-R1 stimulation but was decreased by 18:1 LPA stimulation (Fig. 6J; Supplementary Fig. S7D). Furthermore, stimulation with EPS-R1, but not 18:1 LPA, increased the CCR6+ population in control Lpar2+/− mouse–derived splenic CD8+ T cells (Fig. 6J; Supplementary Fig. S7D). These results would suggest that LPA2 mediates the EPS-R1–induced CCR6 expression on CD8+ T cells.
DISCUSSION
The gut microbiota and microbial metabolites are attractive substances to augment the efficacy of ICB therapies in cancer (10–13). In this report, we demonstrated that dietary consumption of EPS-R1 (one of metabolites of L. bulgaricus R1) increased CCR6+ CD8+ T cells in Peyer's patches. These EPS-R1–induced T cells were capable of infiltrating CCL20-producing tumors and producing IFNγ, supporting an immune response gene expression signature that maintains tumor-specific CTL function in the tumor. Hence, dietary consumption of EPS-R1 augments the effect of ICB therapies in CCL20-producing tumors in mice (Supplementary Fig. S8). Moreover, our results suggested that the Gro3P structure of EPS contributes to EPS-induced CCR6 expression on CD8+ T cells possibly through LPA2. These results provide one effector mechanism as to how Lactobacillus modulates the host immune system as a probiotic.
CXCR3+ CD8+ T cells and IFNγ-inducible chemokines were reported as critical for the therapeutic efficacy of ICB (8, 9). Among CD8+ TILs in the anti–CTLA-4 mAb–treated Colon26 tumors, EPS-R1 ingestion increased IFNγ+ granzyme B+ populations in CXCR3+ CCR6+, but not CXCR3+ CCR6−, CD8+ T cells. Thus, the majority of effector CXCR3+ CD8+ T cells in anti–CTLA-4 mAb/EPS-R1–treated Colon26 tumors likely originated from CCR6+ CD8+ T cells that were reinvigorated by tumor antigen recognition in the IFNγ-rich tumor microenvironment following CCL20-dependent infiltration. Decreased diversity of the TCR repertoire of TILs on day 15 after EPS-R1 ingestion, compared with that on day 10, supports this possibility, because Ccr6 expression was already increased in the tumors by day 10. On the other hand, EPS-R1 ingestion increased both CCR6+ and CCR6− HA-specific CD8+ T cells in anti–CTLA-4 mAb–treated highly antigenic 4T1-HA tumors on day 15. Thus, it is likely that the increase of HA-specific CD8+ T cells in 4T1-HA tumors was due to an increase of both the CCR6+ population, reinvigorated by HA antigen recognition in the tumor, and CXCR3-expressing CCR6− CD8+ T cells recruited in an IFNγ-dependent manner. It was recently reported that an IFNγ-dependent recruitment of CXCR3-expressing CD8+ T cells by CXCL9-producing CD103+ dendritic cells in the tumor was a critical event for anti–PD-1 mAb efficacy (8). In this context, EPS-R1–induced IFNγ-producing CCR6+ CD8+ T cells might infiltrate into CCL20-expressing tumors and stimulate CD103+ dendritic cells to recruit CXCR3+ CCR6− CD8+ T cells. Therefore, EPS-R1–induced CCR6+ CD8+ T cells may not only directly exert an antitumor effect by themselves, but they may recruit ICB-induced CXCR3+ CCR6− antigen-specific CD8+ T cells in the tumors. Overall, these events would result in an increase of total antigen-specific CD8+ T cells, particularly in the highly antigenic tumors. The contribution of CCR6+ CD8+ T cells to antitumor immune responses may vary among different tumor types, possibly depending on tumor antigenicity, other infiltrating immune cells, the amount and variety of chemokines produced by the tumors, and the time after ICB treatment. Further studies are required to reveal the mechanisms that increase tumor-specific effector CTLs and maintain their function in the tumor, although modification of the tumor microenvironment by both ICB and CCR6+ CD8+ T cells appears to play critical roles.
LPA is a typical lysophospholipid mediator and its specific GPCRs, LPA2, 5, and 6, were reported to be expressed on lymphocytes (37). The Gro3P moiety is a basic component of both LPA and EPS-R1, and thus it is possible that the LPA receptors are involved in the induction of CCR6 expression on CD8+ T cells by EPS-R1. It was reported that LPA inhibited CD8+ T-cell activation through LPA5 but not LPA2 (40). Furthermore, LPA enhanced the invasion of T-cell lymphomas into tissue substrates depending on LPA2 (41). We have now shown that an LPA2-specific agonist, RP-1, like EPS-R1, increased the CCR6+ population among CD8+ T cells, and EPS-R1 did not increase the CCR6+ population among Lpar2−/− CD8+ T cells in vitro. Interestingly, 18:1 LPA, an agonist of LPA receptors, decreased the CCR6-expressing population in CD8+ T cells of Lpar2−/− mice in vitro. Thus, EPS-R1 may stimulate LPA2 to induce CCR6 expression or to block the reduction in CCR6 expression caused by endogenous ligands binding inhibitory receptors such as LPA5. It was reported that affinity to each LPA receptor is variable among LPAs and LPA-similar compounds (42). Thus, the different affinity of LPA receptors to EPS-R1 might result in the different immunologic effects of LPAs; however, we did not exclude the possibility that other receptors may also mediate the signals by EPS-R1. The contribution of other cells to induce CCR6 expression on CD8+ T cells was not completely excluded, particularly when EPS-R1 was orally ingested. Notably, RP-1 was reported to enhance the survival of intestinal stem cells and protect from gastrointestinal acute radiation syndrome (43). Thus, oral EPS-R1 ingestion might modulate the condition of the intestinal epithelium to augment the effects of EPS-R1 on CD8+ T cells in Peyer's patches. Further detailed studies, including structural analysis of EPS-R1, are required to determine the pathways by which EPS-R1 and other EPS stimulate CD8+ T cells. These studies will provide an understanding of the mechanisms by which dietary ingestion of some Gro3P-containing EPS produced by L. bulgaricus strains exerts their immune-health benefits.
Some tumor-infiltrating CCR6+ cells have previously been suggested as tumor-promoting cells (44–46). It was reported that diet-induced obesity increased IL6-induced macrophages that produced CCL20 to recruit CCR6+ tumor-promoting cells (44). The CCL20–CCR6 axis was also reported to recruit protumor IL22-producing CD4+ T cells into colon tumors and increased cancer stemness (45). Moreover, the IL6/CCL20/CCR6 axis was also reported to contribute to the mammary tumor progression through IL17A (46). Recently, IL17A-producing CCR6+ TCF+ CD8+ T cells (Tc17) were reported to contribute to poor ICB response (47). When we analyzed correlation between CCR6 expression and response to ICB therapies in publicly available study cohorts, CCR6-high expression did not correlate with good response to ICB therapies in some human cancers (Supplementary Fig. S9). Rather CCR6-low expression appeared to correlate with better response to ICB therapies in urothelial cancers (Supplementary Fig. S9). These results suggested that a majority of CCR6+ cells recruited into CCL20-producing tumors might be tumor-promoting cells and inhibit the efficacy of ICB in human cancers in the absence of EPS-R1 ingestion. Of note, EPS-R1 ingestion augmented Ifng, but not Il17a, expression in anti–CTLA-4 mAb–treated Colon26 tumors (Fig. 4C). Thus, EPS-R1 ingestion might bias CCR6+ T cells toward an IFNγ-producing CD8+ T cell–rich population that inflamed tumor tissue and augmented the antitumor effect of ICB against CCL20-producing tumors.
It might be interesting to examine the effect of dietary ingestion of EPS-R1 on ICB therapies against human CCL20-producing cancers in clinical trials, as EPS-R1 increased the CCR6+ population in CD8+ T cells of human PBMCs in vitro. Herein, the CCR6 gene expression level correlated with prolonged OS in patients with CRAD, HNSC, and BRCA, but the impact of CCR6 expression on prognosis in these cancer types might be a random effect due to the heterogeneity across human cancers. Perhaps PD-L1/PD-1 therapy–resistant colorectal cancer would be an interesting target indication (5). Exposure to fermented dairy foods has been previously reported to be associated with a reduced risk of colorectal cancer (48, 49). It will also be interesting to analyze whether long-term dietary ingestion of fermented skim milk containing EPS-R1 might also reduce the risk of colorectal cancer. No doubt the cross-talk between microbiota and the host is much more complicated in humans, who are genetically diverse and more diverse microbe–exposed compared with experimental rodents. Thus, before clinical trials of dietary ingestion of EPS-R1 can be tested against human cancer, further detailed assessments into the nature of any human IFNγ-producing CCR6+ CD8+ T cells induced by EPS-R1 are required.
METHODS
Study Design
Animals were randomly assigned to experimental groups, but experimenters were not blinded. Sample size was not determined with a power calculation. More than five animals per group were typically used for antitumor efficacy studies and flow cytometry analyses. Figure legends contain sample sizes, replicate information, and the statistical tests used.
EPS Preparation and Dietary Ingestion
EPS was respectively prepared from six L. bulgaricus strains kept in bacterial libraries of Meiji Co., Ltd., including Lactobacillus delbrueckii subsp. bulgaricus OLL1073R-1 or small intestinal contents from three BALB/c WT mice following the previously described methods (24). Briefly, fermented skim milk or intestinal contents were mixed with trichloroacetic acid to remove protein, and ethanol precipitation was performed. Crude EPS was dialyzed (molecular weight cutoff: 6,000–8,000 Da) and incubated with RNase, DNase, and proteinase K. After ethanol precipitation and dialysis again, EPS was obtained by freeze-drying (Supplementary Fig. S10A and S10B). In the mouse experiments, EPS-R1 was dissolved in double-distilled water (DDW; 25 μg/mL) and freely ingested by the mice from the water bottle. Mice generally ingested EPS-R1 at 75 to 125 μg/body/day. DDW was used as the control. In some experiments, EPS-R1 was dissolved in the DDW and administered into the Colon26 tumor–bearing mice 7 days after tumor inoculation by a single oral gavage (400 μg/400 μL/body).
Antibodies and Reagents
Antagonistic anti-mouse CTLA-4 (CD152) mAb (UC10-4F10; kindly provided by Dr. Jeffrey A. Bluestone; ref. 50) and antagonistic anti-mouse PD-1 (CD279) mAb (RMP1-14; ref. 51) were prepared and purified in our laboratory as previously described (52). Neutralizing anti-mouse CCL20 mAb (2F5-5) was kindly provided by KAN Research Institute, Inc. (Kobe; ref. 53). 18:1 LPA was purchased from Avanti Polar Lipids (54), RP-1 was synthesized by Abama Chemicals (43), and autotaxin inhibitor (ATXi), 2-(4-chlorophenyl)-8-hexyl-7-methylimidazo[1,2-a]pyrimidin-5(8H)-one (Compound 30), was synthesized as previously described (55). RP-1 and ATXi were prediluted with dimethyl sulfoxide (DMSO). Next, these three reagents were dissolved in RPMI culture medium (RPMI 1640 medium supplemented with 10% FBS and 1% penicillin–streptomycin; Sigma-Aldrich). ATXi (1 μmol/L) was used in RP-1–stimulating experiments to avoid generation of LPAs from endogenous LPCs.
Mice and Rats
Four- to seven-week-old BALB/c WT and B6 WT mice were purchased from Charles River Japan Inc. BALB/c Ifng−/− mice were derived as described previously (52). To obtain ICR Lpar2−/− and Lpar2+/− mice, B6 Lpar2−/− mice were obtained (56) and backcrossed to ICR mice for four generations. Mice were maintained under specific pathogen–free (SPF) conditions at Juntendo University, Meiji Innovation Center (Hachioji, Japan) and Tohoku University (Sendai, Japan). GF BALB/c WT mice were purchased from CLEA Japan and maintained in sterile conditions in the Meiji Innovation Center facility. Frozen whole small intestines obtained from 6-week-old BALB/c WT mice were purchased from CLEA Japan. All mice were used in accordance with the institutional guidelines based on the approval of Juntendo University, Meiji Innovation Center, Tohoku University, and CLEA Japan Animal Experimental Ethics Committee. Eight-week-old SPF Wistar/ST rats were purchased from Japan SLC and were maintained for 2 weeks at Iwate University and used in accordance with the institutional guidelines based on the approval of Iwate University.
Tumor Cells
Colon26 colon adenocarcinoma cells were kindly provided by the Japanese Foundation for Cancer Research (Tokyo, Japan), and 4T1 mammary carcinoma cells and B16F10 melanoma cells were purchased from ATCC. 4T1 mammary carcinoma cells expressing the influenza HA (4T1-HA) were generated as previously reported (31). Mouse Ccl20 (mCcl20)-transduced B16F10 (B16F10-CCL20) cells were generated according to the manufacturer's instructions with some modifications. Briefly, mCcl20 cDNA (NM_016960) was amplified from Colon26 cells by RT-PCR using the following primers with the XhoI or NotI cloning site: sense, 5′-ATACTCGAGCACCATGGCCTGCGGTGGCAAGCG-3′; anti-sense, 5′-ATAGCGGCCGCTTACATCTTCTTGACTCTTAGGCTGAG-3′. The cDNA was ligated into the pMX-IRES-GFP (pMX-IG) retroviral vector (Cell Biolabs) to generate mCCL20/pMX-IG. Platinum-E (Plat-E, Cell Biolabs) cells were transfected with control pMX-IG or mCCL20/pMX-IG using Lipofectamin 3000 (Thermo Fisher), and the supernatant at day 2 was mixed with the B16F10 cell suspension in the presence of 8 mg/mL polybrene (Sigma-Aldrich). After transduction, cells that expressed GFP were sorted on a FACSMelody cell sorter (BD Biosciences). We confirmed CCL20 production by B16F10-CCL20, but not B16F10, tumor cells in culture supernatants by a mouse CCL20/MIP-3α–specific ELISA (R&D Systems). All cells were authenticated by short tandem repeat analysis and cultured in RPMI 1640 culture medium. All cells were used at low passage numbers and were regularly tested for Mycoplasma.
Coincubation of Tumor Cells with EPS-R1
Colon26 or 4T1 tumor cells were coincubated with EPS-R1 (5 or 25 μg/mL) or doxorubicin hydrochloride (1 or 10 μmol/L; Wako) in RPMI culture medium (37°C, 5% CO2) in a 96-well flat-bottom plate (Corning). Cell apoptosis was examined by staining with 7-AAD/annexin V (FITC Annexin V Apoptosis Detection Kit; BioLegend) and analyzed using FACSVerse flow cytometer (BD Biosciences) 24 hours after coincubation. Immunogenic cell death was examined by detecting secretory damage-associated molecular patterns (DAMP) ATP and HMGB1 (29). ATP was examined by the RealTime-Glo Extracellular ATP Assay (Promega) and analyzed using Cytation 1 (BioTek) during 24-hour coincubation. HMGB1 was examined by the HMGB1 Detection Kit (Chondrex) or HMGB1 Measuring Kit FUSO (Fuso Pharmaceutical Industries, Ltd.) and analyzed using SYNERGY H1 (BioTek) 24 hours after coincubation. Cell proliferation was examined by staining with CellTrace Far Red (Thermo Fisher) and analyzed using FACSVerse flow cytometer 4 days after coincubation; 2 × 105 cells/well (cell apoptosis) or 1 × 104 cells/well (immunogenic cell death and cell proliferation) were assessed, respectively. All of these tests were performed according to the manufacturer's instructions. Doxorubicin hydrochloride was prediluted with DMSO.
Therapy of Transplanted Tumors
Tumor cells (3–6 × 105/body) were s.c. inoculated in the left flank of mice, and the tumor size was measured periodically with a caliper. Tumor volumes were calculated as long diameter × (short diameter)2 × 1/2 (mm3). In the orthotopic transplant model for colorectal cancer, a Colon26 tumor cell suspension (5 × 105/20 μL) was injected into the cecal wall, and the tumor volumes were measured on day 14. Groups of mice were administered i.p. with 150 μg of anti–CTLA-4 mAb, anti–PD-1 mAb, control hamster IgG, or control rat IgG (Sigma-Aldrich) in phosphate-buffered saline (PBS) (−) on days 4, 7, and 10. Some mice were orally administered EPS-R1 (25 μg/mL) dissolved in DDW from day 0. In some experiments, mice were additionally treated with 500 μg of neutralizing anti-mouse CCL20 mAb in PBS (−) on days −3, 0, 3, 7, 10, and 14. In some experiments, mice were additionally treated with 50 μg of polyclonal rabbit anti-asialo GM1 (ASGM1) Ab (Wako) in PBS (−) to deplete NK cells on days −3, 0, 4, 8, and 12 (57). We have confirmed that in these treatment models, tumors rejected by day 14 or 15 never later regrew and tumors detected on day 14 or 15 were never later rejected.
Mononuclear Cell Preparation and In Vitro Stimulation
Peyer's patches, mesenteric lymph nodes, and spleens were removed from mice. Mononuclear cells (MNC) were prepared aseptically following the described procedure (58). Briefly, these were ground with MAS-coated slide glasses (Matsunami Glass), filtrated through nylon mesh (70 μm; Kyosin Rikoh), and washed 3 times with RPMI 1640 culture medium. Red blood cells (RBC) were lysed with ammonium–chloride–potassium (ACK) buffer. Intraepithelial lymphocytes were prepared from the whole small intestine of mice as described previously (59). In some experiments, CD8+ and CD4+ T cells were prepared from MNCs using a mouse CD8α+ T Cell Isolation Kit or a CD4+ T Cell Isolation Kit (Miltenyi Biotec) on MACS Manual Separators (Miltenyi Biotec) according to the manufacturer's instructions. Splenic MNCs, PBMCs (5.0 × 106 cells/mL), or magnetically isolated T cells (0.2–1.0 × 106 cells/mL) were stimulated with EPS (150 μg/mL), 18:1 LPA (100 μmol/L), or RP-1 (25 μmol/L) for 24 hours in RPMI culture medium (37°C, 5% CO2).
Collection of Lymphocytes from the Thoracic Duct
Lymph was collected from the thoracic duct of rats as described previously (60). Briefly, rats were implanted with a catheter into the thoracic duct and duodenum, and placed individually in Bollman-type restraining cages. Then, a single dose of EPS-R1–dissolved DDW (1.5 mg/body) or DDW was administered intraduodenally during collection of the lymph. CD8+ T cells were collected using rat CD8α MicroBeads (Miltenyi Biotec) on MACS Manual Separators according to the manufacturer's instructions.
Preparation of Single-Cell Suspensions from Tumor Masses
Single-cell suspensions from solid tumors were prepared by mincing them with a scalpel and agitating them in the RPMI culture medium containing 2 mg/mL collagenase D (from Clostridium histolyticum; Roche) and 200 μg/mL DNase I (grade II from bovine pancreas; Roche) for 60 minutes at 37°C. Then, following RBC lysis, the cells were strained (100 μm) and washed 2 times with RPMI culture medium. In some experiments, CD45+ cells and CD45− cells were isolated using mouse CD45 (TIL) MicroBeads (Miltenyi Biotec) on a MACS Manual Separator according to the manufacturer's instructions.
Flow-Cytometric Analysis
Flow-cytometric analyses were performed on FACSVerse following immunofluorescence staining. After single-cell preparation, cells were preincubated with anti-mouse CD16/32 (2.4G2) mAb (Mouse BD Fc Block, BD Biosciences) to avoid nonspecific binding of mAbs to Fcγ receptors. Then, cells were stained with the following as indicated: fluorescein isothiocyanate (FITC)–conjugated anti-mouse CD3 molecular complex mAb (17A2; BD Biosciences), phycoerythrin (PE)–cyanin (Cy) 7–conjugated anti-mouse CD4 mAb (RM4-5; Invitrogen), allophycocyanin (APC)–Cy7–conjugated anti-mouse CD8α mAb (53-6.7; BD Biosciences), BD Horizon brilliant violet (BV) 421–conjugated anti-mouse CCR6 mAb (140706; BD Biosciences), APC-conjugated anti-mouse CXCR3 mAb (CXCR3-173; BD Biosciences), PE-conjugated anti-mouse CD199 (CCR9) mAb (9B1; BioLegend), peridinin chlorophyll protein (PerCP)–Cy5.5–conjugated anti-mouse CD3 molecular complex mAb (17A2; BD Biosciences), APC-Cy7 anti-mouse CD3ε mAb (145-2C11; BD Biosciences), PE-conjugated anti-mouse CD8α mAb (53-6.7; BD Biosciences), APC-conjugated anti-mouse CD69 mAb (H1.2F3; Invitrogen), BV421-conjugated anti-mouse CD279 (ICOS) mAb (7E.17G9; BD Biosciences), PerCP–Cy5.5–conjugated anti-mouse CD44 mAb (IM7; BioLegend), APC-conjugated anti-mouse CD62L mAb (MEL-14; BioLegend), PE–Cy7–conjugated anti-mouse CD39 mAb (Duha59; BioLegend), T-Select H-2Kd Influenza HA Tetramer-IYSTVASSL-PE (Medical and Biological Laboratories), or isotype-matched control mAbs (BD Biosciences, Invitrogen, and BioLegend) and control PE-conjugated tetramer (T-Select H-2Kd EGFP Tetramer-HYLSTQSAL-PE, Medical and Biological Laboratories). In some experiments, CellTrace Far Red was used to trace the division of cells. After incubation for 4 to 6 hours (37°C, 5% CO2), a Fixation/Permeabilization Solution Kit with BD GolgiPlug (BD Biosciences) was used to perform intracellular staining for IFNγ by PE–Cy7–conjugated anti-mouse IFNγ mAb (XMG1.2; eBioscience) and granzyme B by FITC-conjugated anti-mouse granzyme B mAb (NGZB; Invitrogen) according to the manufacturer's instructions. Cultured PBMCs (normal human PBMC, purified, characterized; Precision for Medicine) were preincubated with Human BD Fc Block (BD Biosciences) to avoid nonspecific binding of mAbs to Fcγ receptors. Cells were stained with the following mAbs as indicated: violet (V) 500–conjugated anti-human CD3ε mAb (UCHT1; BD Biosciences), APC–H7–conjugated anti-human CD8α mAb (SK1; BD Biosciences), BV421-conjugated anti-human CCR6 mAb (11A9; BD Biosciences), or isotype-matched control mAbs (BD Biosciences). Dead cells were excluded by 7-AAD (BD Biosciences) or Fixable Viability Dye eFluor 506 (FVD506; eBioscience). Data were acquired by BD FACSuite (BD Biosciences) and analyzed in FlowJo (BD Biosciences).
Histologic Examination
The jejunum and ileum were obtained from the BALB/c WT mice ingested with or without EPS-R1 (25 μg/mL) for 6 days (n = 4) and analyzed histopathologically blinded by two pathologists using hematoxylin and eosin staining. Colon26 or 4T1 tumor cells were inoculated in the left flank of mice. Tumor masses were harvested on day 15, fixed in 10% formaldehyde neutral buffer solution (Wako), and then embedded in paraffin. Paraffin sections were incubated with rabbit anti-CCL20 polyclonal Ab (5.0 μg/mL; ab9829, Abcam) or rabbit immunoglobulin fraction (Dako) as the control. CCL20 was detected by Histostar (Rb) for Mouse tissue (Medical and Biological Laboratories) for manual immunostaining by using a Super Sensitive DAB (BioGenex). Finally, sections were counterstained with hematoxylin. All samples were scanned in an All-in-One Fluorescence Microscope (BZ-X810, Keyence). The histologic examination was supported by Advantec Co., Ltd. and New Histo Science Laboratory.
ELISA
CD45+ single-cell suspensions were prepared from Colon26 tumors in anti–CTLA-4 mAb–treated mice on day 15, and the live-cell number was measured using NucleoCounter NC-100 (ChemoMetec) according to the manufacturer's instructions. For anti-CD3 mAb stimulation, the cells were incubated (1 × 106 live cells/mL) with or without functional grade anti-CD3ε mAb (145-2C11; eBioscience) for 48 hours, and then cell-free supernatants were analyzed for ELISA to measure IFNγ, TNF, and IL2 by the appropriate BD OptEIA Mouse ELISA Set (BD Biosciences) using SYNERGY H1 with Gen5 software (BioTek).
RT-qPCR
Total RNA was isolated from samples prepared from Peyer's patch MNCs, thoracic duct lymphocytes, or single-cell suspensions from tumors using NucleoSpin RNA (MACHEREY-NAGEL GmbH & Co. KG). First-stranded cDNA was prepared using PrimeScript RT Master Mix (Perfect Real Time; Takara Bio) following the manufacturer's instructions. Gene expression of mouse 10 chemokine receptors in Peyer's patch CD8+ and CD4+ T cells, and Ccl20 in CD45− and Ccr6 in CD45+ cells in tumor suspensions, was examined by RT-qPCR methods performed using TB Green Premix Ex Taq (Tli RNaseH Plus; Takara Bio) following the manufacturer's instructions: Mouse target genes were Ccr1 (MA102191), Ccr2 (MA104375), Ccr3 (MA097020), Ccr4 (MA112706), Ccr5 (MA173758), Ccr6 (MA167481), Ccr9 (MA124740), Ccr10 (MA122912), Cxcr3 (MA125723), Cxcr6 (MA109145), Ccl20 (MA032304), or endogenous control Gapdh (MA050371). Gene expression of five rat chemokine receptors in thoracic duct CD8+ T cells was also examined by RT-qPCR methods. Rat target genes were Cxcr6 (RA071759), Ccr6 (RA065511), Cxcr3 (RA054984), Ccr5 (RA035868), Ccr9 (RA051266), or endogenous control Gapdh (RA015380). qPCR was performed by a 7300 Real-Time PCR System (Applied Biosystems) or QuantStudio 3 and Real-Time PCR Systems (Applied Biosystems), and the expression levels of respective genes were measured as a ratio compared with Gapdh in the same sample by calculation of cycle threshold (Ct) value in amplification plots using 7300 SDS System Software v1.4.0 (Applied Biosystems) or QuantStudio Design and Analysis Software v1.4 (Applied Biosystems), respectively. All primer sets were obtained from a Perfect Real-Time Support System (Takara Bio) according to the indicated Primer Set ID.
RNA-seq
RNA-seq analysis was performed in Repertoire Genesis Inc. Total RNA was prepared from tumors, and the quality of total RNA was assessed using the Agilent Bioanalyzer 2100 system (Agilent Technologies). Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs) following the manufacturer's recommendations. The libraries were sequenced on Illumina NovaSeq 6000 (Illumina) using a 2 × 150 paired-end configuration.
RNA-seq Data Processing
RNA-seq processing was performed in Repertoire Genesis Inc. Sequenced reads were filtered to exclude adaptor and low-quality (N > 10% and Q score ≤5) portions. The resulting reads were aligned to the UCSC mouse reference genome (mm10; http://genome.ucsc.edu; ref. 61) with TopHat (62) and assembled with Cufflinks by referring gene structures described in NCBI Reference Sequence (RefSeq) genes (63). For quantification, fragments per kilobase of exon per million reads mapped (FPKM) were calculated using Cuffquant function in the Cufflinks (63). Immune-related genes were selected as described previously (64) and using the Molecular Signatures Database (MSigDB; ref. 65) on the keywords (immune, lymphocyte, and macrophage).
TCR Sequencing and Data Analyses
TCR sequencing and data analysis were performed by Repertoire Genesis Inc. as described previously (66). The copy number and percentage occurrence frequencies of identical unique sequence reads in each sample were calculated. TRA and TRB diversity among anti–CTLA-4 mAb–and/or EPS-R1–treated tumors on days 10 and 15 was statistically examined by two-way analysis of variance (ANOVA) with Tukey correction, respectively, and TRA and TRB diversity of tumors in each group on day 15 compared with that on day 10 was statistically examined by two-way ANOVA with Bonferroni correction by a Shannon–Wiener diversity index using GraphPad Prism 7 (GraphPad software). Abundance of each sequence read of TRA and TRB in anti–CTLA-4 mAb– and/or EPS-R1–treated tumors on days 10 and 15 was transformed as log10 (X + 1), and unit variance scaling was applied to each sequence read. The compensated abundances are shown as heat map clustered using correlation distance and average linkage method using ClustVis, a web tool for visualizing clustering of multivariate data (67).
Structural Analysis of EPS
The Gro3P structure in EPS was analyzed and quantified by an LC-MS/MS system. At first, EPS-dissolved (1 mg/mL) water (50 μL) was mixed with 5 M hydrochloric acid (HCl; 5 μL) and heated at 90°C for 30 minutes to cleave the glycosidic bond. Then, 1 M sodium hydrogen carbonate (NaHCO3; 50 μL) was added to the hydrolyzed EPS sample. Chromatographic separation of Gro3P was achieved on a Mastro C18 column (Shimadzu GLC). Detection was conducted on a tandem mass spectrometer with negative electrospray ionization, and multiple reaction monitoring was used at m/z 171 to 79 for Gro3P. Quantitation plots of Gro3P peak area obtained from EPS samples were examined in comparison with standard [n-glycerol 3-phosphate bis (cyclohexylammonium) salt (Sigma-Aldrich)] solutions. The method was supported by Shimadzu Techno-Research, Inc.
Dephosphorylation of EPS-R1 by HF Treatment
EPS-R1 was dephosphorylated by HF treatment following the previously described methods (34). Briefly, EPS-R1 was agitated in cold HF (Wako) for 3 days, and then HF was excluded by chloroform (DP-EPS-R1). For a control, EPS-R1 was agitated in cold HF for 10 minutes (Cont-EPS-R1). By LC-MS/MS analysis, more than 95% Gro3P was dephosphorylated in DP-EPS-R1 compared with Cont-EPS-R1. DP-EPS-R1 was also ingested in some BALB/c WT mice following the same protocol for EPS-R1 ingestion.
16S rRNA-seq Analysis
Stool samples were collected from the rectum of anti–CTLA-4 mAb-treated Colon26-bearing mice on day 15. DNA extraction from stools was performed by QIAamp DNA Stool Mini Kit (QIAGEN) according to the manufacturer's instructions with the following modifications: 20 mg fecal sample was mixed with 1.4 mL ASL buffer in a tube containing 0.7 g (diameter, 0.1 mm) and 0.2 g (diameter, 0.3 mm) sterile zirconia/silica beads (Carl Roth), and thoroughly vortexed. Samples were then processed by a TissueLyser (Qiagen Retsch) for 15 minutes at 25 Hz, and lysis was completed (70°C, 15 minutes; ref. 68). A 16S rRNA-seq library was constructed according to the 16S Metagenomic Sequencing Library Preparation protocol (Illumina) targeting the V3 and V4 hypervariable regions of the 16S rRNA gene. Samples were sequenced on the MiSeq sequencing platform using a 2 × 300 cycle v3 kit (Illumina), and analyzed using the open-source software package QIIME to obtain abundances of fecal bacteria (69). After unit variance scaling was applied to each phyla and families, the data were assessed by heat map (rows are centered, rows and columns are clustered by correlation distance and average linkage method) and principal component analysis (singular value decomposition with imputation was used to calculate principal components, prediction ellipses with 95%), respectively, using ClustVis.
Human Cancer Analysis
For comparison between human cancer tissues and normal tissues, CCL20 gene expression (transcripts per million) in 31 human cancer tissues and paired normal tissues was obtained from the Gene Expression Profiling Interactive Analysis (GEPIA; ref. 70) utilizing data from The Cancer Genome Atlas (TCGA; cancer tissues; ref. 71) and the Genotype-Tissue Expression (GTEx; normal tissues; ref. 72) project, respectively. For the differential analysis of types of paired tissues, higher than preset log2 (fold change) value (= 1) and lower than preset Q-value (= 0.01) were considered statistically different in the four-way ANOVA method (using sex, age, ethnicity, and disease state). For survival analyses, CCR6 and CCL20 gene expression data (FPKM) of TCGA cancer samples were obtained from the Human Protein Atlas (http://www.proteinatlas.org; ref. 73), and clinical data were obtained from the Broad Institute TCGA Genome Data Analysis Center (GDAC; https://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/). HRs were obtained by univariate Cox regression analysis and presented as forest plots using JMP v.15 (SAS Inc.). Testicular germ cell tumor was omitted in Fig. 2B, because 95% CI did not take a finite value. OS curves were presented and examined with Kaplan–Meier survival analysis by log-rank methods using GraphPad Prism 7.
A random-effects model was used to pool effect sizes in meta-analysis of the impact of CCR6 and CCL20 expression on survival. The Paule–Mandel estimator was used to calculate the heterogeneity variance, and Knapp–Hartung adjustments was used to calculate the 95% CI around the pooled effect (74). Between-cancer-type heterogeneity variance was quantified by calculating I2 statistic (75), and the range of true effects was estimated by calculating prediction interval (76). For the analysis of ICB response and CCR6 expression, publicly available study cohorts providing both gene expression data and ICB treatment response data were included (77–82). Of note, gene expression data provided by the HWANG_SCIREP_2020 study cohort were based on the panel sequencing of 395 immune-related genes [Oncomine Immune Response Research Assay (Thermo Fisher)]. Other study cohorts were all based on whole-transcriptome sequence data. Normalized expression data provided by the authors were used when available. Count data were normalized by variance-stabilizing transformation using DESeq2 R package (1.26.0). Within each study cohort, patients were grouped into two by CCR6 expression higher than median or not. Odds ratios of ICB treatment response were calculated (CCR6 high vs. low) for each study cohort. Partial response and complete response were included in responders, and stable disease and progressive disease were included in nonresponders. Effect sizes of each study cohort were calculated using esc R package (0.5.1). A random-effects model was used to pool effect sizes in meta-analysis. Meta-analyses were performed using meta R package (4.19.0) in R 3.6.1 (https://www.R-project.org/).
Statistical Analysis
Statistical analyses, except TCR sequencing analysis, 16S rRNA-seq analysis, and human cancer analysis, are explained in this section. Statistical analyses except Chi-square residual analysis were performed using GraphPad Prism 7. Two-tailed unpaired Student t test or two-tailed Mann–Whitney U test (T-cell number comparison in Colon26 or 4T1-HA tumors) were used to compare two groups. In multiple comparisons (e.g., multiple chemokine receptors) between two groups, two-way ANOVA with unpaired Bonferroni correction was performed. Concerning cytokine production, comparison between matched CD45+ cell suspensions was analyzed by two-way ANOVA with paired Bonferroni correction, and concerning ATP secretion, comparison between stimulants was analyzed by two-way ANOVA with paired Dunnett correction. For analysis of more than three groups, gene expression comparison of three types of mouse tumors was examined by a Kruskal–Wallis test with a Dunn test. CCR6 expression comparison of three mouse groups of Peyer's patch CD8+ T cells was examined by one-way ANOVA with Tukey correction. Comparison of CCR6 expression stimulated with EPS obtained from six L. bulgaricus strains or small intestinal contents of three mice, comparison of CCR6 expression in stimulated Lpar2−/− or Lpar2+/− CD8+ T cells, and comparison of the effect of coculture of tumor cells with EPS-R1 was examined by one-way ANOVA with Dunnett correction. Comparison of complete rejection rate of 4T1-HA tumor was examined by Chi-square residual analysis using BellCurve for Excel (Social Survey Research Information Co., Ltd.). P values less than 0.05 were considered significant.
Data and Materials Availability
All data associated with this study are presented in the article or the Supplementary Materials. All sequencing data (RNA-seq, TCR sequencing, and 16S rRNA-seq) presented in the current study have been deposited at DNA Data Bank of Japan Sequenced Read Archive under the accession numbers DRA013461, DRA013454, and DRA013453. Correspondence and request for materials generated in our laboratories should be addressed to K. Takeda.
Supplementary Material
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant numbers 15K14410, 18K19483, and 18H02695; to K. Takeda). J. Aoki was supported by JSPS KAKENHI (grant number 19H03366) and by AMED-LEAP (grant number 20gm0010004h9904). M.J. Smyth was supported by a National Health and Medical Research Council of Australia Research Investigator Grant (1173958) and Program Grant (1132519). We are grateful to Repertoire Genesis (Osaka, Japan) for their technical support, Junko Nishimura-Uemura (Fukushima University) for HF treatment, and Shin Fujiwara (Meiji Co., Ltd.) for genomic analysis support. We are also grateful to the staff in the animal facility of Juntendo University, Meiji Innovation Center, and the animal physiology laboratory of Iwate University. We thank Yun-Gi Kim (Keio University) for discussion, KAN Research Institute for providing anti-mouse CCL20 mAb, and Meiji Co., Ltd. (Tokyo, Japan) for providing all Lactobacillus strains used in the study. The results reported here are in part based upon data generated by the TCGA Research Network (https://www.cancer.gov/tcga).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
H. Kawanabe-Matsuda reports a patent for WO/2019/240218 pending. K. Takeda reports grants from the Japan Society for the Promotion of Science KAKENHI and Meiji Holdings Co., Ltd. during the conduct of the study. M. Nakamura reports a patent for WO/2019/240218 pending. K. Kakimi reports that the Department of Immunotherapeutics of The University of Tokyo Hospital is endowed by Takara Bio. M. Nishimukai reports other support from Meiji Holdings Co., Ltd. during the conduct of the study. I. Gomperts Boneca reports grants from Meiji Holdings Co., Ltd. during the conduct of the study. G. Eberl reports grants from Meiji Holdings Co., Ltd during the conduct of the study. M.J. Smyth reports grants from the National Health and Medical Research Council of Australia during the conduct of the study, as well as personal fees from Bristol Myers Squibb and Tizona Therapeutics outside the submitted work. K. Okumura reports grants from Meiji Holdings Co., Ltd. during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
H. Kawanabe-Matsuda: Conceptualization, resources, data curation, software, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. K. Takeda: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, writing–review and editing. M. Nakamura: Resources, data curation, formal analysis, investigation, methodology. S. Makino: Resources, data curation, investigation, methodology. T. Karasaki: Data curation, software, formal analysis, investigation, writing–review and editing. K. Kakimi: Data curation, software, formal analysis, investigation, writing–review and editing. M. Nishimukai: Resources, data curation, formal analysis, methodology. T. Ohno: Resources, investigation, methodology. J. Omi: Resources, data curation, formal analysis, investigation. K. Kano: Resources, data curation, formal analysis, investigation. A. Uwamizu: Resources, data curation, formal analysis, investigation. H. Yagita: Resources. I. Gomperts Boneca: Conceptualization, writing–review and editing. G. Eberl: Conceptualization, writing–review and editing. J. Aoki: Conceptualization, writing–review and editing. M.J. Smyth: Conceptualization, funding acquisition, writing–review and editing. K. Okumura: Conceptualization, writing–review and editing.
References
- 1. Speiser DE, Ho PC, Verdeil G. Regulatory circuits of T cell function in cancer. Nat Rev Immunol 2016;16:599–611. [DOI] [PubMed] [Google Scholar]
- 2. Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med 2012;366:2455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gide TN, Wilmott JS, Scolyer RA, Long GV. Primary and acquired resistance to immune checkpoint inhibitors in metastatic melanoma. Clin Cancer Res 2018;24:1260–70. [DOI] [PubMed] [Google Scholar]
- 5. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 2015;372:2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol 2017;17:559–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 2004;10:942–9. [DOI] [PubMed] [Google Scholar]
- 8. Chow MT, Ozga AJ, Servis RL, Frederick DT, Lo JA, Fisher DE, et al. Intratumoral activity of the CXCR3 chemokine system is required for the efficacy of anti–PD-1 therapy. Immunity 2019;50:1498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. House IG, Savas P, Lai J, Chen AXY, Oliver AJ, Teo ZL, et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res 2020;26:487–504. [DOI] [PubMed] [Google Scholar]
- 10. Routy B, Gopalakrishnan V, Daillère R, Zitvogel L, Wargo JA, Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol 2018;15:382–96. [DOI] [PubMed] [Google Scholar]
- 11. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91–7. [DOI] [PubMed] [Google Scholar]
- 13. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coutzac C, Jouniaux JM, Paci A, Schmidt J, Mallardo D, Seck A, et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat Commun 2020;11:2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mager LF, Burkhard R, Pett N, Cooke NCA, Brown K, Ramay H, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020;369:1481–9. [DOI] [PubMed] [Google Scholar]
- 16. Griffin ME, Espinosa J, Becker JL, Luo JD, Carroll TS, Jha JK, et al. Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science 2021;373:1040–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013;342:967–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 2019;565:600–5. [DOI] [PubMed] [Google Scholar]
- 19. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 2014;11:506–14. [DOI] [PubMed] [Google Scholar]
- 20. Rangan KJ, Pedicord VA, Wang YC, Kim B, Lu Y, Shaham S, et al. A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science 2016;353:1434–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, et al. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host Microbe 2013;14:26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med 2019;25:377–88. [DOI] [PubMed] [Google Scholar]
- 23. Klaenhammer TR, Kleerebezem M, Kopp MV, Rescigno M. The impact of probiotics and prebiotics on the immune system. Nat Rev Immunol 2012;12:728–34. [DOI] [PubMed] [Google Scholar]
- 24. Makino S, Sato A, Goto A, Nakamura M, Ogawa M, Chiba Y, et al. Enhanced natural killer cell activation by exopolysaccharides derived from yogurt fermented with Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1. J Dairy Sci 2016;99:915–23. [DOI] [PubMed] [Google Scholar]
- 25. Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1–10. [DOI] [PubMed] [Google Scholar]
- 26. Ansaldo E, Belkaid Y. How microbiota improve immunotherapy. Science 2021;373:966–7. [DOI] [PubMed] [Google Scholar]
- 27. Akrami M, Menzies R, Chamoto K, Miyajima M, Suzuki R, Sato H, et al. Circulation of gut-preactivated naive CD8+ T cells enhances antitumor immunity in B cell-defective mice. Proc Natl Acad Sci U S A 2020;117:23674–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hunter MC, Teijeira A, Halin CT. Cell trafficking through lymphatic vessels. Front Immunol 2016;7:613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kepp O, Senovilla L, Vitale I, Vacchelli E, Adjemian S, Agostinis P, et al. Consensus guidelines for the detection of immunogenic cell death. Oncoimmunology 2014;3:e955691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fu T, He Q, Sharma P. The ICOS/ICOSL pathway is required for optimal antitumor responses mediated by anti-CTLA-4 therapy. Cancer Res 2011;71:5445–54. [DOI] [PubMed] [Google Scholar]
- 31. Takeda K, Nakayama M, Hayakawa Y, Kojima Y, Ikeda H, Imai N, et al. IFN-γ is required for cytotoxic T cell-dependent cancer genome immunoediting. Nat Commun 2017;8:14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Canale FP, Ramello MC, Núñez N, Araujo Furlan CL, Bossio SN, Gorosito Serran M, et al. CD39 expression defines cell exhaustion in tumor-infiltrating CD8+ T cells. Cancer Res 2018;78:115–28. [DOI] [PubMed] [Google Scholar]
- 33. Makino S, Ikegami S, Kano H, Sashihara T, Sugano H, Horiuchi H, et al. Immunomodulatory effects of polysaccharides produced by Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1. J Dairy Sci 2006;89:2873–81. [DOI] [PubMed] [Google Scholar]
- 34. Kitazawa H, Harata T, Uemura J, Saito T, Kaneko T, Itoh T. Phosphate group requirement for mitogenic activation of lymphocytes by an extracellular phosphopolysaccharide from Lactobacillus delbrueckii ssp. bulgaricus. Int J Food Microbiol 1998;40:169–75. [DOI] [PubMed] [Google Scholar]
- 35. De Vuyst L, Degeest B. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol Rev 1999;23:153–77. [DOI] [PubMed] [Google Scholar]
- 36. Cohen LJ, Esterhazy D, Kim SH, Lemetre C, Aguilar RR, Gordon EA, et al. Commensal bacteria make GPCR ligands that mimic human signalling molecules. Nature 2017;549:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Knowlden S, Georas SN. The autotaxin-LPA axis emerges as a novel regulator of lymphocyte homing and inflammation. J Immunol 2014;192:851–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Makide K, Uwamizu A, Shinjo Y, Ishiguro J, Okutani M, Inoue A, et al. Novel lysophosphoplipid receptors: their structure and function. J Lipid Res 2014;55:1986–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang X, Benesch MG, Brindley DN. Lipid phosphate phosphatases and their roles in mammalian physiology and pathology. J Lipid Res 2015;56:2048–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oda SK, Strauch P, Fujiwara Y, Al-Shami A, Oravecz T, Tigyi G, et al. Lysophosphatidic acid inhibits CD8 T cell activation and control of tumor progression. Cancer Immunol Res 2013;1:245–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zheng Y, Kong Y, Goetzl EJ. Lysophosphatidic acid receptor-selective effects on Jurkat T cell migration through a Matrigel model basement membrane. J Immunol 2001;166:2317–22. [DOI] [PubMed] [Google Scholar]
- 42. Williams JR, Khandoga AL, Goyal P, Fells JI, Perygin DH, Siess W, et al. Unique ligand selectivity of the GPR92/LPA5 lysophosphatidate receptor indicates role in human platelet activation. J Biol Chem 2009;284:17304–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuo B, Szabo E, Lee SC, Balogh A, Norman D, Inoue A, et al. The LPA2 receptor agonist radioprotectin-1 spares Lgr5-positive intestinal stem cells from radiation injury in murine enteroids. Cell Signal 2018;51:23–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wunderlich CM, Ackermann PJ, Ostermann AL, Adams-Quack P, Vogt MC, Tran ML, et al. Obesity exacerbates colitis-associated cancer via IL-6-regulated macrophage polarisation and CCL-20/CCR-6-mediated lymphocyte recruitment. Nat Commun 2018;9:1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kryczek I, Lin Y, Nagarsheth N, Peng D, Zhao L, Zhao E, et al. IL-22+ CD4+ T cells promote colorectal cancer stemness via STAT3 transcription factor activation and induction of the methyltransferase DOT1L. Immunity 2014;40:772–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Benevides L, da Fonseca DM, Donate PB, Tiezzi DG, De Carvalho DD, de Andrade JM, et al. IL17 promotes mammary tumor progression by changing the behavior of tumor cells and eliciting tumorigenic neutrophils recruitment. Cancer Res 2015;75:3788–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Burger ML, Cruz AM, Crossland GE, Gaglia G, Ritch CC, Blatt SE, et al. Antigen dominance hierarchies shape TCF1+ progenitor CD8 T cell phenotypes in tumors. Cell 2021;184:4996–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang K, Dai H, Liang W, Zhang L, Deng Z. Fermented dairy foods intake and risk of cancer. Int J Cancer 2019;144:2099–108. [DOI] [PubMed] [Google Scholar]
- 49. Pala V, Sieri S, Berrino F, Vineis P, Sacerdote C, Palli D, et al. Yogurt consumption and risk of colorectal cancer in the Italian European prospective investigation into cancer and nutrition cohort. Int J Cancer 2011;129:2712–9. [DOI] [PubMed] [Google Scholar]
- 50. Walunas TL, Lenschow DJ, Bakker CY, Linsley PS, Freeman GJ, Green JM, et al. CTLA-4 can function as a negative regulator of T cell activation. Immunity 1994;1:405–13. [DOI] [PubMed] [Google Scholar]
- 51. Matsumoto K, Inoue H, Nakano T, Tsuda M, Yoshiura Y, Fukuyama S, et al. B7-DC regulates asthmatic response by an IFN-γ-dependent mechanism. J Immunol 2004;172:2530–41. [DOI] [PubMed] [Google Scholar]
- 52. Takeda K, Yamaguchi N, Akiba H, Kojima Y, Hayakawa Y, Tanner JE, et al. Induction of tumor-specific T cell immunity by anti-DR5 antibody therapy. J Exp Med 2004;199:437–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kitagawa Y, Kikuchi S, Arita Y, Nishimura M, Mizuno K, Ogasawara H, et al. Inhibition of CCL20 increases mortality in models of mouse sepsis with intestinal apoptosis. Surgery 2013;154:78–88. [DOI] [PubMed] [Google Scholar]
- 54. Umezu-Goto M, Kishi Y, Taira A, Hama K, Dohmae N, Takio K, et al. Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. J Cell Biol 2002;158:227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kawaguchi M, Okabe T, Okudaira S, Hama K, Kano K, Nishimasu H, et al. Identification of potent in vivo autotaxin inhibitors that bind to both hydrophobic pockets and channels in the catalytic domain. J Med Chem 2020;63:3188–204. [DOI] [PubMed] [Google Scholar]
- 56. Contos JJ, Ishii I, Fukushima N, Kingsbury MA, Ye X, Kawamura S, et al. Characterization of lpa2 (Edg4) and lpa1/lpa2 (Edg2/Edg4) lysophosphatidic acid receptor knockout mice: signaling deficits without obvious phenotypic abnormality attributable to lpa2. Mol Cell Biol 2002;22:6921–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ogasawara K, Takeda K, Hashimoto W, Satoh M, Okuyama R, Yanai N, et al. Involvement of NK1+ T cells and their IFN-γ production in the generalized Shwartzman reaction. J Immunol 1998;160:3522–7. [PubMed] [Google Scholar]
- 58. Kramer DR, Cebra JJ. Early appearance of "natural" mucosal IgA responses and germinal centers in suckling mice developing in the absence of maternal antibodies. J Immunol 1995;154:2051–62. [PubMed] [Google Scholar]
- 59. Hoytema van Konijnenburg DP, Reis BS, Pedicord VA, Farache J, Victora GD, Mucida D. Intestinal epithelial and intraepithelial T cell crosstalk mediates a dynamic response to infection. Cell 2017;171:783–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nishimukai M, Yamashita M, Watanabe Y, Yamazaki Y, Nezu T, Maeba R, et al. Lymphatic absorption of choline plasmalogen is much higher than that of ethanolamine plasmalogen in rats. Eur J Nutr 2011;50:427–36. [DOI] [PubMed] [Google Scholar]
- 61. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 2011;12:R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013;31:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, et al. The immune landscape of cancer. Immunity 2018;48:812–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kitaura K, Shini T, Matsutani T, Suzuki R. A new high-throughput sequencing method for determining diversity and similarity of T cell receptor (TCR) alpha and beta repertoires and identifying potential new invariant TCR alpha chains. BMC Immunol 2016;17:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res 2015;43:W566–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smith B, Li N, Andersen AS, Slotved HC, Krogfelt KA. Optimising bacterial DNA extraction from faecal samples: comparison of three methods. Open Microbiol J 2011;5:14–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cancer Genome Atlas Research Network; Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. GTEx Consortium; Laboratory, Data Analysis & Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; et al. Genetic effects on gene expression across human tissues. Nature 2017;550:204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 74. Hartung J, Knapp G. A refined method for the meta-analysis of controlled clinical trials with binary outcome. Stat Med 2001;20:3875–89. [DOI] [PubMed] [Google Scholar]
- 75. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539–58. [DOI] [PubMed] [Google Scholar]
- 76. IntHout J, Ioannidis JP, Rovers MM, Goeman JJ. Plea for routinely presenting prediction intervals in meta-analysis. BMJ Open 2016;6:e010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti–PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Snyder A, Nathanson T, Funt SA, Ahuja A, Buros Novik J, Hellmann MD, et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: an exploratory multi-omic analysis. PLoS Med 2017;14:e1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018;554:544–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hwang S, Kwon AY, Jeong JY, Kim S, Kang H, Park J, et al. Immune gene signatures for predicting durable clinical benefit of anti–PD-1 immunotherapy in patients with non-small cell lung cancer. Sci Rep 2020;10:643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



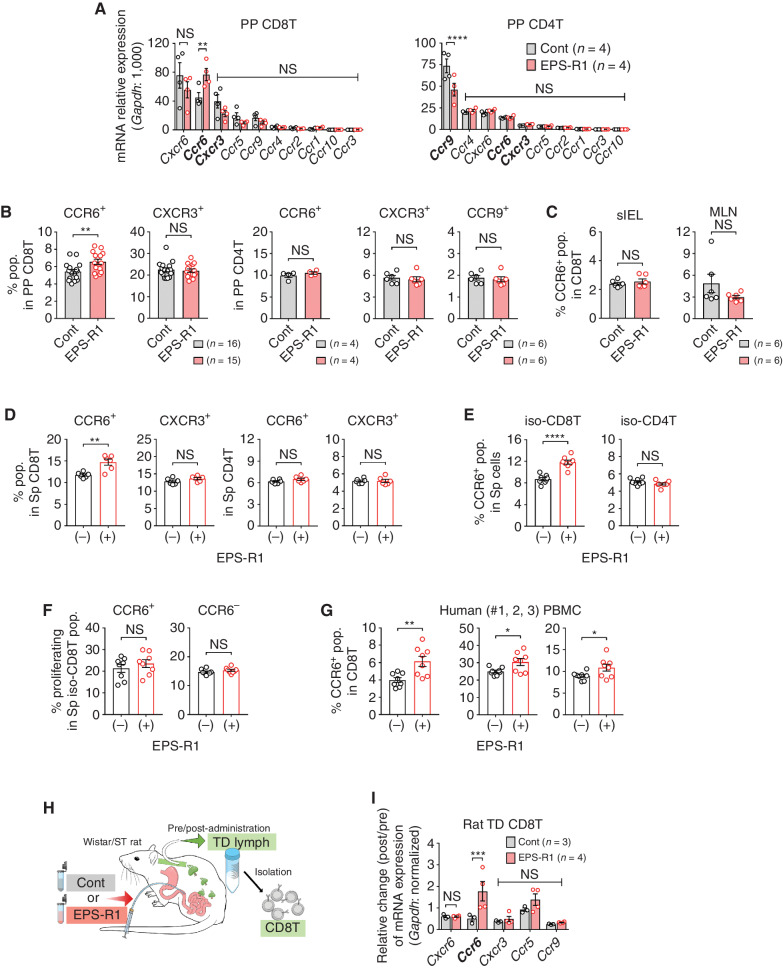
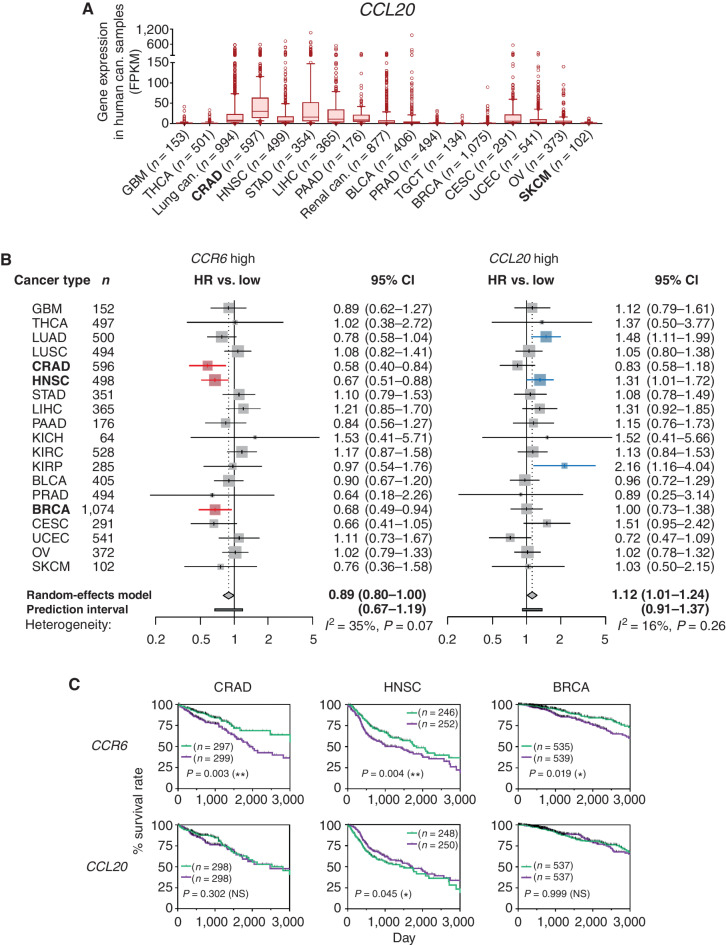
![Figure 3. EPS-R1 augments antitumor effects of ICB therapies. A, BALB/c WT or C57BL/6 (B6) WT mice were s.c. inoculated with syngeneic Colon26, 4T1 (BALB/c), or B16F10 (B6) tumor cells, respectively. CD45− or CD45+ cells were isolated from single-cell suspensions prepared from growing tumors on day 15, and RNAs were obtained from these cells. Ccl20 gene expression in CD45− cells and Ccr6 gene expression in CD45+ cells were examined by RT-qPCR methods (n = 6). All results obtained from two experiments are presented. B, BALB/c WT mice were s.c. inoculated with syngeneic Colon26 tumor cells, and EPS-R1 was administered by oral gavage on day 7. Then, the CCR6+ population among CD8+ TILs was examined by flow cytometry 6, 24, 48, and 72 hours later [n = 5 in control (6, 24, and 48 hours) and n = 7 in control (72 hours) and in EPS-R1]. C, BALB/c WT mice were s.c. inoculated with syngeneic Colon26 tumor cells, and dietary ingestion of EPS-R1 was started on day 0. Some mice were i.p. treated with anti–CTLA-4 mAb or anti–PD-1 mAb. Tumor volumes were periodically measured and statistically compared on day 15. Tumor growth among anti–CTLA-4 mAb- or anti–PD-1 mAb-treated mice was not significantly different with that among control mice. All results obtained from three (control Ig- and anti–CTLA-4 mAb-treated groups) or two (anti–PD-1 mAb-treated group) experiments are presented. D, BALB/c WT mice were s.c. inoculated with syngeneic 4T1 tumor cells, and dietary ingestion of EPS-R1 was started on day 0. Some mice were i.p. treated with anti–CTLA-4 mAb. Tumor volumes were periodically measured and statistically compared on day 15. Tumor growth among anti–CTLA-4 mAb-treated mice was not significantly different with that among control mice. All results obtained from two experiments are presented. E, BALB/c WT mice were s.c. inoculated with syngeneic 4T1 tumor cells, and dietary ingestion of EPS-R1 was started on day 0. Some mice were i.p. treated with anti–PD-1 mAb. Tumor volumes were periodically measured and statistically compared on day 15. Tumor growth among anti–PD-1 mAb-treated mice was not significantly different from that among control mice. F, BALB/c WT mice were s.c. inoculated with syngeneic Colon26 tumor cells, and dietary ingestion of EPS-R1 commenced on day 0. Some mice were i.p. treated with anti–CTLA-4 mAb or anti–PD-1 mAb. Some EPS-R1–ingested mice were additionally treated with anti-ASGM1 Ab to deplete NK cells. Tumor volumes were periodically measured and those of EPS-R1–ingested mice treated with or without anti-ASGM1 Ab were statistically compared on day 14. Tumor growth among anti–CTLA-4 mAb- or anti–PD-1 mAb-treated mice was not significantly different from that among control mice. G, Syngeneic Colon26 tumor cells were inoculated into the cecal wall of BALB/c WT mice (orthotopic transplant model), and dietary ingestion of EPS-R1 commenced on day 0. Some mice were i.p. treated with anti–CTLA-4 mAb or anti–PD-1 mAb. Tumor volume was measured and statistically compared on day 14. Tumor growth among anti–CTLA-4 mAb- or anti–PD-1 mAb-treated mice was not significantly different from that among control mice. All results obtained from three experiments are presented. H and I, B6 WT mice were s.c. inoculated with syngeneic B16F10 tumor cells (H) or B16F10-CCL20 tumor cells (I), and dietary ingestion of EPS-R1 was started on day 0. All mice were i.p. treated with anti–CTLA-4 mAb (H) and some mice were i.p. treated with anti–CTLA-4 mAb or anti–PD-1 mAb (I). Tumor volumes were periodically measured and statistically compared on day 15. Tumor growth among anti–CTLA-4 mAb- or anti–PD-1 mAb-treated mice was not significantly different from that among control mice (I). J, BALB/c WT mice were s.c. inoculated with syngeneic Colon26 tumor cells, and some mice were dietary ingested with EPS-R1 starting on day 0. All mice were i.p. treated with anti–CTLA-4 mAb and additionally neutralizing anti-CCL20 mAb. Tumor volumes were periodically measured and statistically compared on day 15. K, Single-cell suspensions were prepared from some anti–CTLA-4 mAb-treated tumors on day 15 presented in C, D, H, and J. Then, CCR6-expressing populations among CD3+ CD8+ TILs were examined by flow cytometry. Results are presented as box plots with individual samples (A), mean ± SEM (C–F and H–J) and depicted at scatter plots of the results of individual samples (B, G, and K). Results are processed as the relative values of individual EPS-R1–ingested samples compared with the mean values of control samples in each time point. Number (n) of mice in every group is indicated in the panels (C–K). Statistical analyses were performed by Kruskal–Wallis test with Dunn correction (A), two-way ANOVA with Bonferroni correction (B), or Student t test (C–K). NS, not significant. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Similar results were obtained from two (A, C in anti–PD-1 mAb group, H, J, and K) or three (C in control Ig and anti–CTLA-4 mAb groups, D, E, G, and I) independent experiments. cont, control; aCTLA-4 (PD-1, CCL20), anti–CTLA-4 (PD-1, CCL20); pop., population; CD8 TIL(s), CD8+ TIL(s); aCTLA-4(PD-1, CCL20, ASGM1), anti–CTLA-4(PD-1, CCL20, ASGM1) (mAb); ortho., orthotopic.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/10af/9662940/559c9b758cc9/1336fig3.jpg)