Abstract
Lipid phosphoinositides are master regulators of almost all aspects of a cell’s life and death and are generated by the tightly regulated activity of phosphoinositide kinases. Although extensive efforts have focused on drugging class I phosphoinositide 3-kinases (PI3Ks), recent years have revealed opportunities for targeting almost all phosphoinositide kinases in human diseases, including cancer, immunodeficiencies, viral infection and neurodegenerative disease. This has led to widespread efforts in the clinical development of potent and selective inhibitors of phosphoinositide kinases. This Review summarizes our current understanding of the molecular basis for the involvement of phosphoinositide kinases in disease and assesses the preclinical and clinical development of phosphoinositide kinase inhibitors.
Subject terms: Mechanisms of disease, Kinases, Medicinal chemistry
The potential of therapeutically targeting phosphoinositide kinases (PIKs) beyond the class I PI3Ks is increasingly being realized. Here, Burke et al. describe the structure, function, regulation and roles in disease of all clinically relevant PIKs outside of the class I PI3Ks, assessing potent and specific small-molecule inhibitors in development.
Introduction
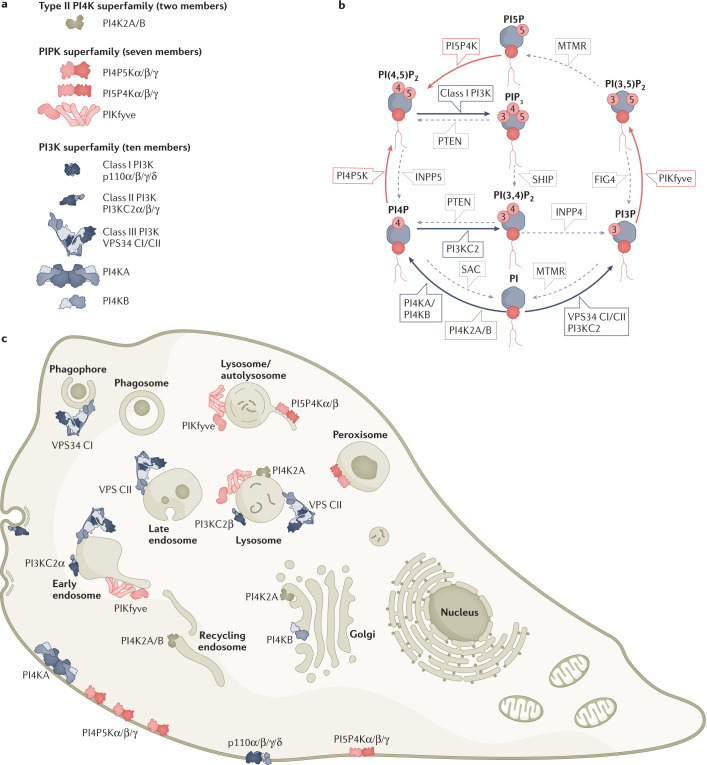
Lipid phosphoinositides in cell membranes are master regulators of myriad membrane signalling events1–3. Phosphoinositides have central roles in membrane trafficking, metabolism, growth, signalling and autophagy, with alterations in phosphoinositide metabolism being causative for many human diseases. There are seven different phosphoinositide species: three mono-phosphorylated phosphatidylinositol phosphates (PIPs), three bis-phosphorylated PIP2s and a single tris-phosphorylated PIP3 (Fig. 1). These phosphoinositides are distributed differently in unique cell types, and their levels can change dramatically upon activation of cell surface receptors or in pathogen-infected cells. Phosphoinositides are generated with spatiotemporal precision from phosphatidylinositol (PI) by the action of lipid phosphoinositide kinases (19 unique genes in mammals), and are degraded by the action of phosphoinositide phosphatases (up to 35 unique genes in mammals) (Fig. 1). This Review focuses on the phosphoinositide kinases, and readers are referred to other reviews for discussion of the roles of the various phosphoinositide species4–8. Phosphoinositide kinases can be broadly split into three general families: one family contains all classes of the phosphoinositide 3-kinases (PI3Ks; also known as phosphatidylinositol 3-kinases) and the type III PI4Ks, another family contains the PIP kinases, with the last family containing the type II PI4Ks (Fig. 1).
Fig. 1. Phosphoinositides and the phosphoinositide kinases that generate them.
a, All phosphoinositide kinases (PIKs) encoded by the human genome, grouped by evolutionary relatedness. Families were initially grouped around activity, before specific enzymes had been cloned or complete identification of the specific regio-isomer substrates and products. Hence, the phosphoinositide 3-kinase (PI3K) superfamily incorporates the type I PIKs (now known to be PI3Ks) and type III PIKs (now known to be PI4KA and PI4KB, and still referred to as PI4KIIIα and PI4KIIIβ at the protein level). Class III PI3K VPS34 exists as two distinct heterotetramers, differing in a single subunit between complexes I and II (referred to as VPS34 CI/CII). Cloning of the type II PIKs (PI4K2A, PI4K2B) revealed them to be an evolutionarily distinct family of enzymes. The phosphatidylinositol phosphate kinases (PIPKs), are now known to be three subfamilies, each catalysing a specific hydroxyl phosphorylation on different substrates. b, Substrate and catalytic activity of PIKs. The production and turnover of phosphoinositides are mediated by the coordinated action of lipid kinases, phosphatases and lipases. All phosphoinositide species are generated from phosphatidylinositol (PI). Three different hydroxyls on the inositol ring of PI can be phosphorylated, at the D3, D4 and D5 positions. This leads to the generation of seven phosphoinositides: three mono-phosphorylated PIPs, phosphatidylinositol 3-phosphate (PI3P), PI4P and PI5P; three bis-phosphorylated PIP2s, phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), PI(3,5)P2, PI(3,4)P2 and the single tris-phosphorylated PIP3 phosphatidylinositol 3,4,5-trisphosphate (PI(3,4,5)P3 or PIP3). Grey lines indicate families of phosphatases that remove indicated phosphates, usually opposing a kinase reaction. Note that, like the kinases, the phosphatases were named on the basis of activity, phenotype or homology. These often predated definitive identification of specific catalytic activity, so the names are now somewhat arbitrary. For many enzymes, alternative substrates or catalytic activities have been reported in the test tube or in cells. However, we focus on the major pathways and activities that support the biology and pathology discussed in the text. c, Subcellular distribution of PIKs. We focus on membranes where most activity is reported, which does not necessarily reflect steady-state distribution of the enzymes themselves. For example, the PI5P4K enzymes are mostly localized in the cytosol and/or nucleoplasm. FIG4, FIG4 phosphoinositide 5-OH phosphatase; INPP4, inositol polyphosphatase 4-OH phosphatase; INPP5, inositol polyphosphatase 5-OH phosphatase; MTMR, mytotubularin-related; PTEN, phosphatase and tensin homologue; SAC, SAC phosphoinositide phosphatase.
Multiple phosphoinositide kinases are therapeutic targets in various human diseases, including cancer, viral infection, neurodegenerative diseases, developmental disorders, diabetes and inflammatory diseases. Alterations in phosphoinositide metabolism are foundational in disease states, presenting multiple opportunities for therapeutic modulation of lipid kinase activity with small molecules. Mutations have been identified in several phosphoinositide kinases that either hyperactivate or inactivate lipid kinase activity, leading to disease progression. In addition, selectively targeting phosphoinositide kinases in parasites has potential for treating infection. Currently, there are many inhibitors of the class I PI3Ks that are clinically approved for the treatment of several cancers (Box 1); clinical trials are ongoing with small-molecule inhibitors that target the lipid kinase PIKfyve in cancer and viral infection, and inhibitors of the Plasmodium homologue of PI4K are in clinical trials for the treatment of malaria (Table 1).
Table 1.
Phosphoinositide kinase inhibitors outside of class I PI3Ks in the clinic
| Agent | Target | Disease indication | Current development status | Clinical trial ID/reference |
|---|---|---|---|---|
|
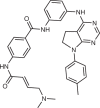
Apilimod/LAM-002A
|
PIKfyve | SARS-CoV-2, non-Hodgkin lymphoma, amyotrophic lateral sclerosis | Active phase II clinical trials | NCT04446377, NCT02594384, NCT05163886 |
|
ESK981
|
PIKfyve | Prostate cancer, renal cell carcinoma | Active phase II clinical trials | NCT03562507, NCT03456804, NCT04159896 |
|
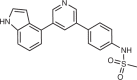
MMV390048
|
plasmodium PI4KB | Malaria infection | Phase I/II clinical trial (completed or terminated) | NCT02783833, NCT02554799, NCT02230579, NCT02783820 |
|
Enviroxime/LY122772
|
plasmodium PI4KB | Poliovirus infection, enterovirus infection | Discontinued in phase II | Phillpotts et al. (1983)168 |
PI4KB, type III phosphatidylinositol 4-kinase beta; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
This Review describes the structure–function, regulation and involvement in disease of all clinically relevant phosphoinositide kinases outside of the class I PI3Ks — including members of the PIP kinase superfamily, PI4Ks and class II and III PI3Ks — as recent reviews have described the class I PI3K path to clinical development9,10 (Box 1). The various members of the phosphoinositide kinases show a set of both commonalities and differences in their architecture and regulation (Box 2). Regarding the PI4Ks, the focus is primarily on the type III PI4Ks over type II owing to the extensive drug discovery efforts targeting PI4KA and PI4KB in viral infection, malaria and cancer, with preclinical development of type II PI4K inhibitors being at a very nascent stage11,12. The state of the art in the development of potent and specific small-molecule inhibitors for the treatment of a multitude of disease states is discussed.
Box 1 Class I PI3Ks as therapeutic targets.
The class I phosphoinositide 3-kinases (PI3Ks) generate phosphatidylinositol 3,4,5-trisphosphate (PIP3) and are master regulators of cell growth, metabolism and immune function243. The PI3K pathway is the most frequently mutated pathway in cancer, and PIK3CA is the second most frequently mutated gene in all cancers244. Somatic mutations of PI3Ks are causative in multiple human cancers, primary immunodeficiencies, developmental disorders and overgrowth syndromes.
There are four class I PI3K catalytic isoforms (class IA (p110α, p110β, p110δ, encoded by PIK3CA, PIK3CB, PIK3CD, respectively) and class IB (p110γ encoded by PIK3CG)). PI3K catalytic subunits require a regulatory subunit for biological activity, with five class IA subunits (p85α, p55α and p50α (encoded by PIK3R1), p85β (encoded by PIK3R2) and p55γ (encoded by PIK3R3)), and two class IB PI3K subunits (p84/p87 (encoded by PIK3R5) and p101 (encoded by PIK3R6)). The catalytic cores of class IA and IB PI3Ks are similar, although they differentially interact with regulatory subunits245,246. Class I PI3Ks are activated downstream of receptor tyrosine kinases (RTKs), G protein-coupled receptors (GPCRs) or Ras superfamily GTPases247. Most diseases that involve PI3K are driven by hyperactivation of PI3K catalytic activity248. However, developmental disorders249–251 and immunodeficiencies252,253 caused by inactivating PI3K mutations highlight how important it is to turn PI3K on and off at appropriate times in development and immune signalling.
All PI3Ks are therapeutic targets in human disease. The p110α isoform is activated downstream of insulin signalling166, with activating alterations in either p110α254,255 or p85α256,257 causing increased p110α signalling that leads to tumorigenesis. In cancer cells, there appears to be a crucial threshold for p110α activation, as cancer-like transcriptional remodelling in stem cells occurred only in cells homozygous for an activating mutant258. This fits with the clinical observation that cis double oncogenic mutations of PIK3CA lead to increased PI3K signalling and oncogenicity259. Activating PIK3CA mutations also occur in overgrowth syndromes called PIK3CA-related overgrowth spectrum (PROS)260,261. Although the p110β isoform is not frequently altered in cancer, it drives tumorigenesis in phosphatase and tensin homologue (PTEN)-deficient cancers262,263. The p110δ isoform is primarily expressed in immune cells, with somatic activating alterations in p110δ264–266 or p85α267–269 leading to the primary immunodeficiency activated PI3K delta syndrome (APDS). The p110γ isoform is not frequently altered in disease, but inhibition promotes antitumour immune responses270,271 and is protective in inflammatory conditions272.
Five PI3K inhibitors are FDA approved for the treatment of solid tumours and blood cancers. The only pan-PI3K inhibitor approved is copanlisib (BAY 80-6946/Aliqopa), which is approved in relapsed follicular lymphoma (FL)273. The following isoform-selective inhibitors have been approved: alpelisib (BYL719/Piqray, p110α selective)234 for hormone receptor (HR)-positive, HER2-negative, locally advanced or metastatic breast cancer with a PIK3CA mutation, with it also showing positive clinical response in patients with PROS274; idelalisib (GS-1101/CAL-101/Zydelig, p110δ selective)275–278 for B cell cancers including relapsed chronic lymphocytic leukaemia (CLL), follicular B cell non-Hodgkin lymphoma and relapsed small lymphocytic lymphoma (SLL); umbralisib (TGR-1202/RP5264, p110δ selective)279 for relapsed or refractory marginal zone lymphoma (MZL) or FL; and duvelisib (IPI-145/INK1197, p110δ/γ selective)280 for relapsed CLL, SLL and FL. All clinically approved PI3K inhibitors are associated with extensive side effects, including increased risk of infections, colitis and hyperglycaemia.
A challenge in the therapeutic exploitation of PI3K inhibitors is toxicity and pathway reactivation. Feedback can counteract PI3K inhibition by both cell-intrinsic and systematic mechanisms. Inhibition of PI3K leads to decreased activation of AKT, which relieves suppression of receptor tyrosine kinase (RTK) expression281–284, reactivating the PI3K pathway. Although this increased expression can be therapeutically exploited, for example, in the dual treatment of oestrogen receptor (ER)-positive cancers with PI3K inhibitors and anti-ER therapies, this feedback complicates effective PI3K dosage. PI3K inhibition also leads to increased blood glucose and insulin, causing reactivation of the PI3K pathway285. This suggests an opportunity to combine PI3K inhibition with dietary or pharmacological interventions to lower blood glucose levels. Other approaches to moderate toxicity could be through intermittent dosing, as PI3Kδ inhibition in solid tumours has been hampered by severe immune-related adverse events; however, these events could be minimized by a modified treatment regimen with PI3Kδ inhibitor intermittent dosing286.
A major focus in PI3K drug development is mutant-selective inhibitors, with Genentech reporting an inhibitor that leads to selective degradation of mutant p110α/p85β over wild-type p110α287, which prevented RTK-dependent pathway reactivation. In addition, LOXO Pharmaceuticals and Relay Therapeutics have reported H1047R-selective small-molecule inhibitors, which selectively target the mutant H1047R over wild-type p110α. It would be expected that these will cause decreased side effects and less inhibition-driven pathway activation. Further drug discovery efforts will be required to test whether other PI3K mutants can be selectively targeted.
The long road that PI3K inhibitors have followed to arrive at the clinic will inform the design of other phosphoinositide kinase inhibitors. Like the PI3Ks, the development of phosphoinositide kinase therapeutics will require extensive fundamental basic research to fully understand the mechanisms that underlie phosphoinositide metabolism.
Box 2 Commonalities and differences in the architecture and regulation of phosphoinositide kinases.
The phosphoinositide kinases can be split into three groups on the basis of their evolutionary history, with an evolutionary family containing all phosphatidylinositol phosphate (PIP) kinases (PIKfyve, phosphoinositide 5-phosphate 4-kinases (PI5P4Ks) and phosphoinositide 4-phosphate 5-kinases (PI4P5Ks)), an evolutionary family containing all classes of the phosphoinositide 3-kinases (PI3Ks) and the type III PI4Ks, and a final evolutionary family composed of the type II PI4Ks131,288. It is important to note that enzymes with unique evolutionary histories can generate the same lipid (that is, PI4P generated by type II PI4Ks and type III PI4Ks), which highlights the importance of PIP metabolism. Members of the PIP kinase group share a conserved evolutionarily related bi-lobal kinase domain; however, there are major differences in the structural organization of PIKfyve and the PI5P4Ks and PI4P5Ks. Dimerization has a crucial role in the regulation of the PI5P4Ks and PI4P5Ks but has no established role in regulating PIKfyve (described more below). PIKfyve is one of the largest phosphoinositide kinases at >2,000 amino acids, and it forms a large trimeric complex with regulatory subunits, whereas the PI5P4Ks/PI4P5Ks are ~400–700 amino acids and do not form stable assemblies with other regulatory subunits.
Members of the group that contains all classes of the PI3Ks and the type III PI4Ks share a conserved core composed of a helical scaffolding domain and a bi-lobal kinase domain. The N-lobe of the kinase domain for this group has a unique helical extension that, at least for the class I PI3Ks, plays a crucial part in membrane association289–292. A major difference within this group is the formation of stable assemblies with regulatory subunits, with class I and III PI3Ks, and the PI4KA isoform of type III PI4Ks forming large multi-protein complexes, whereas the class II PI3Ks and the PI4KB isoform of type III PI4Ks do not and instead are regulated by more transient protein–protein interactions.
The smallest group is the one that contains the two isoforms of the type II PI4Ks. Unique among all of the phosphoinositide kinases, the type II PI4Ks can be regulated by lipidation, with this playing a key role in their cellular localization and activity.
PIP kinase evolutionary family
PIKfyve
Structure and regulation
PIKfyve is conserved from yeast to humans and is the only protein in eukaryotes that catalyses the production of phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) from phosphatidylinositol 3-phosphate (PI3P)13. The primary pool of PI3P used by PIKfyve as a substrate is generated by the class III PI3K VPS34 (ref.14). PIKfyve also produces the main pool of PI5P in the cell, which is primarily generated indirectly through the dephosphorylation of PI(3,5)P2 by lipid phosphatases, although a small pool of PI5P may be generated by PIKfyve’s direct phosphorylation of PI to generate PI5P. The PIKfyve complex is primarily localized at endosomal membranes15,16, with PI(3,5)P2 in endosomes/lysosomes regulating the activity of ion channels, playing important parts in ion homeostasis17. PIKfyve is a crucial regulator of the endocytic pathway, with roles in endosomal trafficking14,18, cell migration19 and lysosomal function20,21.
In mammalian cells, PIKfyve is a large protein (2,098 amino acids) composed of four putative structured regions: a FYVE domain that binds to PI3P22, a CCT domain that mediates interaction with Vac14, a structurally uncharacterized CCR module, and a kinase domain with sequence homology to the PI5P4Ks and PI4P5Ks23 (Fig. 2a). In addition to lipid kinase activity, PIKfyve also has activity as a protein kinase and is regulated by inhibitory autophosphorylation24. Loss of PIKfyve protein leads to embryonic lethality in mammals25,26, and PIKfyve null fibroblasts have undetectable PI(3,5)P2 and significant depletion of PI5P13.
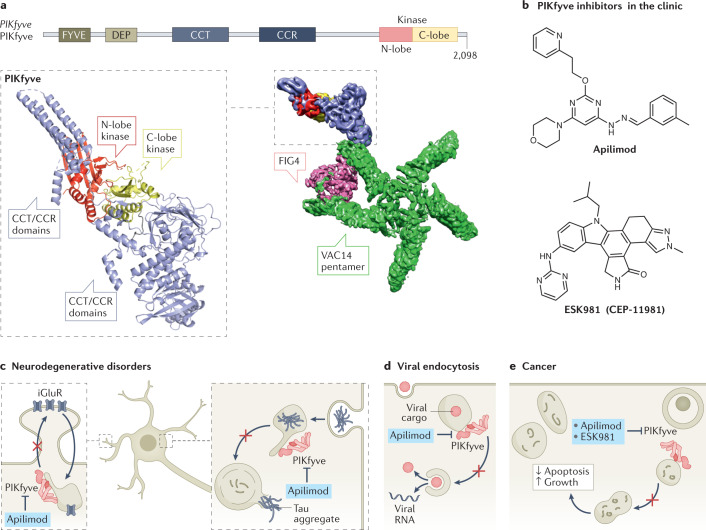
Fig. 2. Structure–function, inhibition and therapeutic targeting of PIKfyve.
a, Domain architecture of PIKfyve. The predicted structure of a fragment of the CCR/CCT and kinase domains (alphafold model of Q9Y2I7, all regions with pLDDT <50 removed)235 from the cryo-electron microscopy (cryo-EM) density is shown with the domains coloured according to the domain schematic. The cryo-EM density of the complex of PIKfyve with VAC14 and FIG4 is also shown, with the VAC14 pentamer coloured green, FIG4 coloured pink and the PIKfyve coloured as in the schematic. VAC14 in isolation forms a symmetrical pentamer, with oligomerization mediated by the C terminus236. FIG4 can form a complex with VAC14 in the absence of PIKfyve, and it binds at the end of two VAC14 arms, leading to distortion of the symmetry of the VAC14 pentamer. PIKfyve’s association with VAC14 is strongly dependent on the presence of FIG4 (ref.237). A single copy of PIKfyve binds to the opposite sides of the VAC14 arm bound to FIG4. Multiple cryo-EM maps were compiled to generate this figure (EMD: 22631, EMD: 22647, EMD: 22634). b, PIKfyve selective inhibitors (apilimod and ESK981) currently in clinical trials for cancer and viral infection. c, PIKfyve as a target for neurodegenerative disorders. Inhibition of PIKfyve prevents endocytic recycling of ionotropic glutamate receptors to the synapse, reducing excitotoxic death of glutamatergic neurons. It also prevents endocytic trafficking of tau or α-synuclein aggregates to the lysosome. d, PIKfyve as a target for viral infection. Again, disruption of PIKfyve activity prevents endocytic trafficking of endocytosed virus, preventing its escape into the cytoplasm from endolysosomes. e, PIKfyve as a target for cancer. PIKfyve inhibition prevents maturation and fusion of late autophagosomes with lysosomes, preventing the anti-apoptotic and pro-growth effects of autophagy in cancer cells.
All components of the PIKfyve signalling complex are conserved from yeast to humans, composed of the proteins PIKfyve, the scaffolding protein VAC14 (encoded by VAC14; also referred to as ArPIKfyve)27,28 and the dual lipid–protein phosphatase FIG4 (encoded by FIG4; also referred to as SAC3)29–31. Studies of mouse mutants and knockout models, as well as clinical mutations in patients have revealed the crucial role of all members of the PIKfyve complex in the central and peripheral nervous systems. Both Vac14 (ref.32) and Fig4 (ref.33) knockout mice have extensive neurodegeneration, accompanied by decreased PI(3,5)P2 levels and enlarged vacuoles. Mice with mutations in Vac14 that prevent association with PIKfyve27, or mutations in Fig4 that prevent association with VAC14 (ref.34) show similar neurodegeneration. Mutations in FIG4 have been found in patients with the neurodegenerative diseases amyotrophic lateral sclerosis (ALS), primary lateral sclerosis (PLS)35 and Charcot–Marie–Tooth disease type 4J (CMT4J)33,36. In addition, biallelic loss-of-function mutations in VAC14 (ref.37) were found in patients with a progressive neurological disease with early childhood onset. Overall, evidence suggests that PI(3,5)P2 production, through the action of the PIKfyve–FIG4–VAC14 complex, is crucial for the proper development and maintenance of nervous system tissues.
Complicating the study of the regulation of the PIKfyve complex is that FIG4 has lipid phosphatase activity against PI(3,5)P2, yet loss of FIG4 paradoxically leads to decreased PI(3,5)P2 levels33, suggesting a key role of FIG4 in regulating PIKfyve activity. VAC14 forms a homo-pentamer, which binds to single copies of PIKfyve and FIG4 (ref.38) (Fig. 2a). Studies suggest that a conformational change occurs in the VAC14 subunit upon binding to FIG4 that alters the affinity towards PIKfyve38. A crucial role of FIG4 in increasing PIKfyve activity was identified by its ability to dephosphorylate an inhibitory autophosphorylation site in the activation loop of the kinase domain of PIKfyve (S2053)38. The current biochemical and biophysical data suggest a model in which FIG4 has two important roles in controlling PIKfyve activity: it stabilizes the association of PIKfyve with VAC14, and its protein phosphatase activity regulates PIKfyve lipid kinase activity.
Pharmacological inhibitors
Potent and selective inhibitors towards PIKfyve have been useful in defining the numerous roles of PI(3,5)P2 signalling in cells. Although structure-guided drug design has been limited by the lack of high-resolution structural data for the PIKfyve kinase domain, multiple inhibitors of PIKfyve have been developed, with the first generation entering clinical trials (Tables 1 and 2).
Table 2.
Summary of preclinical inhibitors of the PIP kinases
| Compound | Target | Target potency (Kd or IC50) | Known off-targets/selectivity | References |
|---|---|---|---|---|
|
YM201636
|
PIKfyve | IC50 33 nM | ~100-fold selectivity over class I PI3Ks | Jefferies et al. (2008)40 |
|
MF4
|
PIKfyve | IC50 23 nM | ~10- to 50-fold selectivity over class I PI3Ks | de Lartigue et al. (2009)18 |
|
APY0201
|
PIKfyve | IC50 5.2 nM | Increased selectivity over Apilimod, > 50% inhibition of ITPK1/LOK at 300 nM | Hayakawa et al. (2014)43 |
|
WX8
|
PIKfyve | Kd 0.93 nM | Kd for PI5P4Kγ of ~340 nM | Sharma et al. (2019)44 |
|
NDF
|
PIKfyve | Kd 1.6 nM | Kd for PI5P4Kγ of 24,000 nM | Sharma et al. (2019)44 |
|
MOMIPP
|
PIKfyve | Kd 5 nM | Kd for PI5P4Kγ of ~15,000 nM | Cho et al. (2018)46 |
|
Series of 4-aminopyridine derivatives (compounds 8, 20, 25)
|
Pan PI4P5K | IC50 4–90 nM | >100-fold selectivity over other lipid kinases | Andrews et al. (2022)94 |
ISA-2011B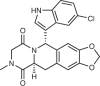
|
PI4P5Kα | ND | Weakly selective; binds class I PI3K, p110α | Semenas et al. (2014)92 |
|
UNC3230
|
PI4P5Kγ PI5P4Kγ |
Kd 51 nM Kd 4 nM |
Kd ~300 nM for MAPK10 and SGK1 | Wright et al. (2014)93 |
|
A131
|
Pan-PI5P4K | IC50 0.6 µM (PI5P4Kα) | Not reported | Kitagawa et al. (2017)103 |
|
CC260
|
Pan-PI5P4K | Kd 40 nM (PI5P4Kα), 30 nM (PI5P4Kβ) | Off-target protein and lipid kinase activity (PIKfyve, PIK3CD, PIK3CG) | Chen et al. (2021)101 |
|
THZ-P1-2 derivative compound 30
|
Pan-PI5P4K(covalent) | IC50 1340 nM (PI5P4Kα) | Highly selective | Sivakumaren et al. (2020)104, Manz et al. (2020)111 |
|
CVM-05-002 derivative compound 13
|
Pan-PI5P4K | IC50 1.96 µM (PI5P4Kα) | Highly selective | Manz et al. (2020)105 |
|
BAY-091 (Bayer)
|
PI5P4K | IC50 1.3 nM | Highly selective | Wortmann et al. (2021)107 |
SAR088/imanixil (Sanofi-Aventis) 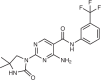
|
PI5P4Kβ | IC50 2.18 µM | 9-fold selectivity over PI5P4Kα | Voss et al. (2014)108 |
|
NCT-504
|
PI5P4Kγ (allosteric) | Kd 354 nM | Highly selective | Al-Ramahi et al. (2017)110 |
|
NIH-12848 derivative compound 40
|
PI5P4Kγ (allosteric) | Kd 68 nM | Highly selective | Boffey et al. (2022)113 |
Covalent and allosteric (non-ATP competitive) inhibitors are indicated. Half-maximal inhibitory concentration (IC50), dissociation constant (Kd) and selectivity data are from the indicated references. ND, not determined; PIP, phosphatidylinositol phosphate; PI4P5K, phosphoinositide 4-phosphate 5-kinase; PI5P4K, phosphoinositide 5-phosphate 4-kinase.
Similarly to many of the other phosphoinositide kinases, one of the first potent and selective PIKfyve inhibitors to be discovered arose from drug discovery efforts focused on the class I PI3Ks39 and led to the identification of the ATP-competitive pyridofuro-pyrimidine compound YM201636, which was highly potent towards PIKfyve, with 100-fold selectivity over the class I PI3K p110α40. This compound induced dramatically decreased PI(3,5)P2 levels and enlarged vacuoles and/or lysosomes, similar to small interfering RNA (siRNA) knock-down of PIKfyve. The chemically very similar MF4 inhibitor (lacking an amino group off the pyrimidine ring) exhibits similar potency towards PIKfyve and ~10-fold selectivity over p110α18. Although these compounds were useful for defining the functional roles of PIKfyve in multiple cell types and tissues, the weak selectivity profile over class I PI3K p110α prevented further clinical development.
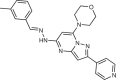
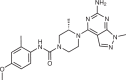
The small molecule N-[(E)-(3-methylphenyl)methylideneamino]-6-morpholin-4-yl-2-(2-pyridin-2-ylethoxy)pyrimidin-4-amine (apilimod) was originally identified as an inhibitor of IL-12 and IL-23 cytokine production through an unknown mechanism41. Apilimod underwent clinical trials for Crohn’s disease, rheumatoid arthritis and psoriasis, and was generally well tolerated, although it had limited clinical effect. The initial lack of a defined therapeutic target limited assessment of efficacy and toxicity. However, apilimod was found to be a highly potent and selective PIKfyve inhibitor42, which has shown promise as a treatment for cancer and viral infection, with ongoing clinical trials in cancer and severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) infection (Fig. 2b, described below).
Additional PIKfyve inhibitors have been discovered in high-throughput screens for anticancer and anti-inflammatory compounds. This includes the pyrazolo[1,5-a]pyrimidine inhibitor APY0201, which was identified as an inhibitor of IL-12 and IL-23 production that targets the PIKfyve complex, acting as a potent ATP-competitive inhibitor with limited effect on other kinases, G protein-coupled receptors (GPCRs) or ion channels43. The WX8 family of compounds with either a 1,3,5-triazin-2-amine or pyrimidine-4-amine core were identified to induce excess DNA replication in cancer cells compared with non-malignant cells. Although WX8 was the most potent compound in this series, it exhibited off-target inhibition on PI5P4Kγ, and NDF was the most selective compound44. The 3-(5-methoxy-2-methyl-1H-indol-3yl)-1-(4-pyridinyl)-2-propene-1-one (MOMIPP) compound was identified as a promoter of methuosis45, which is a form of nonapoptotic cell death, with PIKfyve later identified as the target46. MOMIPP is a potent ATP-competitive inhibitor of PIKfyve with weak inhibition of PI5P4Kγ and limited inhibition of any other lipid kinase.
ESK981 (11-(2-methylpropyl)-12,13-dihydro-2-methyl-8-(pyrimidin-2-ylamino)-4H-indazolo[5,4-a]pyrrolo[3,4-c]carbazol-4-one, initially named CEP-11981) was first identified as an inhibitor of the tyrosine kinases TIE2, VEGFR1, VEGFR2, VEGFR3 and FGFR1, and completed phase I clinical assessment for pharmacokinetics and safety47. However, a screen of multi-tyrosine kinase inhibitors towards a panel of prostate cancer cells identified ESK981 as a potent PIKfyve inhibitor48, with weak inhibition of PI5P4Ks. ESK981 is currently undergoing clinical trials for castration-resistant prostate cancer (Fig. 2b, described below).
PIKfyve inhibition in neurodegenerative disorders
The severe peripheral demyelinating neuropathy Charcot–Marie–Tooth disease type 4B (CMT4B) can be caused by loss-of-function mutations in a PI(3,5)P2 phosphatase/regulator, MTMR2, MTMR5 or MTMR13; recent studies have shown that apilimod can rescue the associated in vitro myelin outfoldings49,50 and improve nerve conduction velocity in mouse models of CMT4B1 (MTM2 mutants). Thus, preventing aberrant PI(3,5)P2 accumulation through PIKfyve inhibitors seems to be a promising therapy for these rare disorders.
More broadly, many sporadic neurodegenerative disorders are characterized by the accumulation of intracellular protein aggregates, such as tau in Alzheimer disease and frontotemporal dementia (FTD), and α-synuclein in Parkinson disease, that lead to neuronal death (Fig. 2c). Misfolded copies of these proteins act like prion ‘seeds’; they enter the neuronal endocytic pathway, disrupt lysosomal integrity and induce the misfolding of endogenous cellular proteins, eventually leading to cell death51. Recent studies suggest that the role of PIKfyve in endocytic trafficking of these neurodegenerative seeds makes it a promising therapeutic target.
In an in vitro model of Alzheimer disease in cultured hippocampal neurons, inhibition of PIKfyve by siRNA, YM201636 or apilimod strongly reduces the appearance of seeded tau aggregates in this system52. PIKfyve acts by preventing traffic to the acidic (lysosomal) compartments52 (Fig. 2c). PIKfyve inhibition also reduced neurodegeneration in a human organoid model of tau-induced FTD, which models the excitotoxic death observed in glutamatergic neurons in the disease53. In this system, PIKfyve did not reduce tau levels per se; apparently, it instead reduced excitotoxic death by reducing endocytic recycling of ionotropic glutamate receptors53.
PIKfyve inhibition was similarly effective in in vitro models of Parkinson disease with YM201636, apilimod and vacuolin-1 all blocking the seeded aggregation of α-synuclein in HEK293 cells54. The inhibitors did not block endocytosis of α-synuclein fibrils, but instead blocked traffic to the lysosomes; they also reduced lysosomal damage and thus escape of the seeds into the cytosol54.
PIKfyve has also been implicated as a potential target in ALS, which is caused by selective loss of motor neurons. Although mainly sporadic, around 10% of cases of ALS are caused by autosomal dominant inheritance of a heptad repeat (GGGGCC) expansion in an intron of the C9ORF72 gene. Recent evidence suggests that this causes neurodegeneration via two mechanisms55; first, neurotoxic accumulation of dipeptides occurs from non-AUG translation of the repeat, akin to the toxic protein aggregates in other neurodegenerative disorders; second, reduced C9ORF72 protein expression (that is, haploinsufficiency) reduces endocytosis of ionotropic glutamate receptors (iGluRs), leading to excitotoxicity of motor neurons55. In a patient-derived, induced motor neuron culture model of the disease, such excitotoxic death can be prevented with apilimod, YM201636 or knock-down of PIKfyve55 (Fig. 2c).
It therefore seems that despite a range of disease mechanisms, PIKfyve inhibitors prevent toxicity in neurons by disrupting endocytic traffic, be that of aggregation-inducing seeds or excess glutamate receptors. Could chronic reductions in PI(3,5)P2 levels via PIKfyve inhibition be a general treatment for neurodegenerative disease? As discussed above, loss of function of any member of the PIKfyve complex (PIKfyve–FIG4–VAC14) itself causes neurodegenerative phenotypes in mice as well as in humans, including ALS35, the peripheral neuropathy Charcot–Marie–Tooth disease32,33, and a severe neuropathy that includes hypo-myelination of central neurons56. Therefore, PIKfyve activity seems essential for central and peripheral nervous system homeostasis. The extensive vacuolation of cells after inhibition of PIKfyve has been observed in neurons from VAC14 and FIG4 mutant mice32,33. This vacuolation is reminiscent of the neuronal spongiosis observed in prion-driven spongiform encephalopathies. Recent evidence suggests that this is more than just coincidence57: mouse brains, or a human hypothalamic neuronal cell line, exhibit ablated PIKfyve expression when infected with prions. This is also observed in brains from patients who died of sporadic Creutzfeldt–Jacob disease (sCJD). Mechanistically, prion-induced endoplasmic reticulum stress appears to mis-localize two key zinc-finger acyltransferases that normally acylate and stabilize PIKfyve. Without this stabilizing acylation, PIKfyve is rapidly degraded, leading to the vacuolation and eventual death of cerebral neurons57.
Collectively, it seems that PIKfyve-regulated endocytic traffic is essential for normal neuronal homeostasis, but also contributes to pathological traffic in aggregation- and excitotoxicity-driven neurodegeneration. The therapeutic potential of selective PIKfyve compounds will therefore derive from their ability to attenuate the latter while sparing the former. As neurodegenerative diseases that are alleviated or caused by PIKfyve inhibition all develop over many years or decades, safety, tolerability and efficacy of these PIKfyve inhibitors may not be apparent in the short term, even in clinical trials. Promisingly though, whereas PIKfyve null mice are pre-implantation lethal, heterozygous mice develop normally without obvious phenotype, despite measurably reduced PI(3,5)P2 levels25. Therefore, some window of physiologically sustainable PI(3,5)P2 reduction exists. Whether this level is sufficient to exhibit meaningful attenuation of pathological traffic, while preventing its own neurodegenerative insult, will dictate the success of these approaches in the clinic.
PIKfyve inhibition in viral infection
Many viral pathogens achieve cell entry through the endolysosome pathway, where, upon encountering a host cell receptor in the endocytic pathway, this triggers membrane fusion, causing the release of the virus into the cytoplasm. Unique viruses have evolved different trafficking routes to reach the site of membrane fusion. PIKfyve has shown promise as a target for inhibiting viral infection through blocking viral entry into the host cell cytoplasm through disruption of endolysosomal trafficking (Fig. 2d).
The role of PIKfyve in viral infection was initially identified through a genome-wide haploid genetic screen for host factors in Ebola and Marburg filoviruses, which are causative agents of high-mortality viral haemorrhagic fevers. This screen uncovered multiple components involved in endolysosomal trafficking, including PIKfyve, and identified the Niemann–Pick C1 (NPC1) cholesterol transporter as the host cell receptor for filoviruses58. These findings suggested that PIKfyve inhibitors may prevent the endolysosomal trafficking required for viral entry. Apilimod blocked infection of Ebola and Marburg viruses in liver, kidney and monocyte-derived macrophage cells59, by preventing the release of the viral genome into the cytoplasm through disruption of trafficking to NPC1-positive endolysosomes, resulting in the virus being blocked from its site of fusion and preventing entry into the cell. Knock-down of one of the PIKfyve signalling complex components VAC14 or FIG4 also decreases viral entry of filoviruses, highlighting the crucial role of PI(3,5)P2 production in viral entry of these viruses60. In Zaire Ebola virus infection, apilimod caused distension of the RAB5 and RAB7 endocytic compartments into vacuoles, preventing the release of the virus from endosomal compartments61.
The coronavirus disease 2019 (COVID-19) pandemic led to extensive experiments to try to understand the mechanism of viral entry and develop inhibitors as antiviral therapeutics. Multiple lines of evidence support PIKfyve inhibitors as a potential antiviral therapeutic. SARS-CoV-2 was found to enter human cells through endocytosis, with PIKfyve playing a crucial part in viral entry in HEK293 cells stably expressing the ACE2 receptor. Treatment of these cells with either of the PIKfyve inhibitors apilimod or YM201636 dramatically decreased viral infection62. In a large-scale screen of 12,000 FDA-approved small molecules that prevent cellular infection by SARS-CoV-2, apilimod was identified as one of 13 possible molecules that inhibited viral infection at achievable therapeutic doses63. Apilimod also demonstrated antiviral efficacy in a primary human lung explant model. Analysis of SARS-CoV-2-infected cells identified altered phosphorylation in a panel of druggable protein and lipid kinases, which included PIKfyve64. Further, apilimod showed antiviral efficacy in cell studies, and clinical trials in patients with COVID-19 are ongoing (Table 1).
Although there has been a tremendous amount of enthusiasm in the use of PIKfyve inhibitors as potent host factor-specific antivirals, potential roadblocks may exist. PIKfyve is crucial in coordinating the immune response of neutrophils, being required for both the generation of reactive oxygen species and chemotaxis, which are both important in the innate immune response65. Careful analysis of the appropriate dosing of PIKfyve will be required to determine whether it is possible to decrease viral infection but not dramatically decrease immune cell function.
Inhibiting PIKfyve in cancer
PIKfyve is emerging as an attractive target for various cancers as several screens have discovered PIKfyve inhibitors to exert anticancer activity (Fig. 2e). Apilimod was first identified as an antiproliferative compound across many cancer subtypes, with B cell non-Hodgkin lymphoma (B-NHL) cells being the most sensitive to apilimod treatment compared with normal cells66. Cytotoxicity induced by apilimod in B-NHL cells is mediated through the blockade of autophagy and ultimately disruption of lysosome homeostasis. These findings provide a promising new approach for treating multiple subtypes of B-NHL as a single agent or in combination with existing therapies, which is currently under clinical evaluation.
The WX8 family has provided further supporting evidence for targeting PIKfyve as a therapy for autophagy-dependent cancers, including autophagy-addicted melanoma cells44. WX8 family inhibitors impaired several stages of lysosome homeostasis, including disrupting lysosome fission, trafficking into lysosomes and the formation of autolysosomes (Fig. 2e). Importantly, even though WX8 family drugs extensively disrupted lysosome homeostasis, they selectively killed autophagy-dependent cancer cells without affecting either the proliferation or viability of non-malignant cells. APY0201 effectively inhibited multiple myeloma (MM) cell growth in vitro as well as in ex vivo models of MM67. As previous reports have shown that MM cells are dependent on autophagy for survival68, targeting autophagy through PIKfyve inhibition may provide a feasible strategy for patients with MM. However, in contrast, inhibition of PIKfyve with YM201636 suppressed liver cancer growth by promoting autophagy, possibly through induction of EGFR expression69.
The multi-tyrosine kinase PIKfyve inhibitor ESK981 blocked tumour growth in preclinical models of castration-resistant prostate cancer48. ESK981 not only blocked autophagy but also recruited T cells to the tumours. In fact, when an immune checkpoint inhibitor was combined with ESK981, tumour growth was even further reduced. These findings reveal that targeting PIKfyve via ESK981 may be a promising approach for immunotherapy, which can convert prostate tumours from non-immunogenic ‘cold’ tumours into immune-inflamed ‘hot’ tumours. Phase II clinical trials using ESK981 alone or in combination with the immunotherapy nivolumab for metastatic castration-resistant prostate cancer are underway. Results from these studies will be imperative to better understand the therapeutic effects of inhibiting PIKfyve for cancer treatment.
PI4P5Ks and PI5P4Ks
Structure and regulation
PI(4,5)P2 is not only the most abundant bis-phosphorylated phosphoinositide in mammalian cells, but it also acts as a substrate for the fundamental cancer and metabolism kinase PI3K (Box 1). Beyond PI3K signalling, PI(4,5)P2 is an established player in numerous cellular processes, including vesicular trafficking, membrane dynamics, modulation of ion channel function, gene regulation and the generation of second messengers (that is, diacyl glycerol and inositol-1,4,5-trisphosphate downstream of phospholipase C signalling). However, there is much to be understood about the regulation of PI(4,5)P2, specifically regarding the enzymes responsible for its production. PI(4,5)P2 is produced either from the subsequent phosphorylation of mono-phosphoinositides from PI, or by dephosphorylation of PI(3,4,5)P3. The kinases responsible for generating PI(4,5)P2 are characterized into two subfamilies regarded as the type I and type II kinases. Considered the canonical pathway, the type I PI4P5Ks generate PI(4,5)P2 by phosphorylating the most abundant mono-phosphorylated species PI4P at the inositol ring D5 hydroxyl group70. The non-canonical type II PI5P4Ks phosphorylate the less abundant mono-phosphorylated PI5P at the inositol ring D4 hydroxyl group71. Although the PI4P5Ks and PI5P4Ks both produce PI(4,5)P2, the timing and localization of their activity is distinctly orchestrated. The type I pathway functions predominantly at the plasma membrane, and the type II kinases produce PI(4,5)P2 at intracellular organelle membranes72–75.
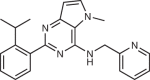
In mammals, the type I PI4P5K family consists of three isoforms (PI4P5Kα, PI4P5Kβ and PI4P5Kγ), which are encoded by the genes PIP5K1A, PIP5K1B and PIP5K1C, respectively76–78 (Fig. 3a). There are several different splice variants for each isoform; however, the roles of these variants are still poorly understood. Knockout mice for all three isoforms of PI4P5K have been generated, which have provided further evidence that the isoforms have distinct functions in vivo79. The PI4P5Ks and PI5P4Ks share conserved structural elements including a lipid kinase domain and dimerization domain, with an overall organization distinct from PI3Ks, and similar to the kinase domain of PIKfyve. Although both PI4P5Ks and PI5P4Ks are able to homo-dimerize, the dimer interface is proposed to be different between PI4P5Ks and PI5P4Ks (Fig. 3). There is still debate on the possible roles of PI4P5K–PI5P4K dimers in cell signalling.
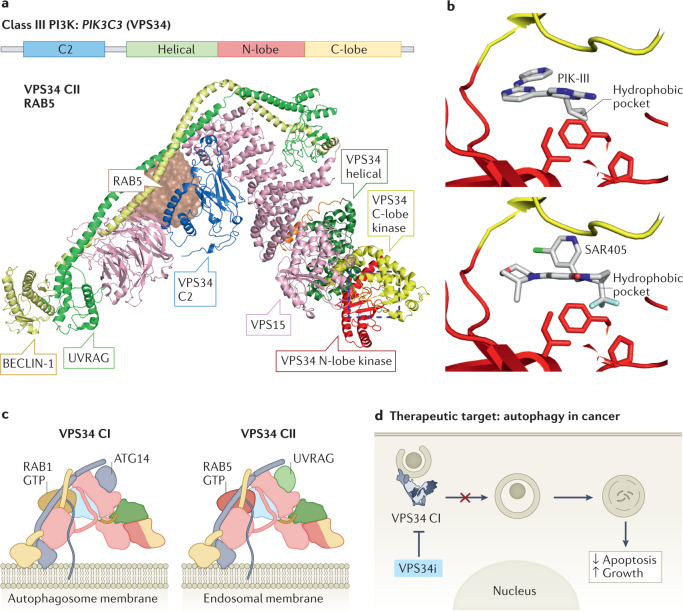
Fig. 3. Structure–function, and inhibition of the PI4P5Ks and PI5P4Ks.
a, Structure of the zebrafish homologue of phosphoinositide 4-phosphate 5-kinase-α (PI4P5Kα), with domains annotated on the figure. PI4P5Kα (PDB:4TZ7) shows a putative dimeric interface composed of the dimerization domain as well as the N-lobe of the kinase domain, which is unique compared with the phosphoinositide 5-phosphate 4-kinases (PI5P4Ks; see panel b). In contrast to the stable PI5P4K dimers, PI4P5Ks exist in a monomer–dimer equilibrium in solution, with dimerization promoted by binding to PI(4,5)P2-containing membrane surfaces, leading to enhanced catalytic efficiency238. Like all lipid kinases, the kinase domain contains an activation loop that determines substrate specificity and also has a role in membrane recruitment. Swapping the activation loops between the type I PI4P5Ks and the type II PI5P4Ks led to not only swapped substrate specificity between PI4P and PI5P but also modified subcellular localization239,240. b, Structure of PI5P4Kα dimer, in which, in contrast to the PI4P5K, dimerization is putatively mediated solely by the dimerization domain (PDB: 6YM5). For type II PI5P4Ks, catalytic activity and PI5P substrate binding is carried out by the kinase domain, while homo- and heterodimerization with other type II PI5P4Ks is driven solely by the dimerization domain80. This differs from the type I PI4P5Ks, which have a unique dimerization interface composed of both the kinase and dimerization domains, with dimerization required for PI4P5K lipid kinase activity (panel a)241. The difference in the dimerization interface between PI4P4Ks and PI5P4Ks allows for the potential formation of complexes between type I and type II phosphatidylinositol phosphate (PIP) kinases, with their roles being unknown, although preliminary evidence suggests a potential regulatory role120,242. c, Structure of PI5P4Kα bound to the selective inhibitor BAY-091, with the domains coloured according to panel b (PDB: 6YM5) and residues that make crucial interactions in determining selectivity shown as sticks.
There are three mammalian type II PI5P4K isoforms (PI5P4Kα, PI5P4Kβ and PI5P4Kγ), which are encoded by the genes PIP4K2A, PIP4K2B and PIP4K2C, respectively (Fig. 3b). At a sequence level, the PI5P4Kα and PI5P4Kβ isoforms share 83% protein homology, with the γ-isoform being less similar to either the α- or β-isoform80,81. Knockout mice for all three isoforms are viable with normal lifespans and subtle phenotypes82–84.
Kinetically, PI5P4Kα has the highest kinase activity, which is reported to be 100-fold that of PI5P4Kβ. PI5P4Kγ has the lowest kinase activity, with activity 2,000-fold lower than that of PI5P4Kβ81,85,86. This variable catalytic activity between isoforms can be explained by sequence differences in the ATP-binding G-loop80. Although both PI5P4Kα and PI5P4Kβ are catalytically active, there are still important differences in the regulation of their kinase activity. Unique among all lipid kinases, the PI5P4Kβ isoform preferentially uses GTP as a phosphate donor in contrast to ATP, with its activity proposed to act as a GTP sensor in cells87. Importantly, kinase activity may not be necessary for the cellular roles of all three isoforms, as the PI5P4Ks can function together as heterodimers. Whereas PI5P4Kα more efficiently catalyses PI(4,5)P2 from PI5P, dimerization with PI5P4Kβ or PI5P4Kγ enables spatial and temporal translocation to precise membrane locations85,88. The regulation of the PI5P4Ks is linked to the cellular demand for specific pools of PI5P and PI(4,5)P2, with phosphorylation playing a key part in this regulation. For example, it is suggested that nuclear PI5P accumulates because of catalytic inhibition of PI5P4Kβ following phosphorylation at Ser326 by MAP kinase p38 (ref.89). Inhibitory phosphorylation occurs at Ser326 and Thr322 of both PI5P4Kα and PI5P4Kβ isoforms90. Further, inhibitory phosphorylation of PI5P4Kγ at Ser324 and Ser328 occurs downstream of mTORC1 to balance basal mTORC1 homeostasis using a nutrient-dependent feedback loop91.
Pharmacological inhibitors of PI4P5Ks
A limited number of potent and specific PI4P5K inhibitors have been reported to date and are still in early stages of preclinical development (Table 2). One of the best described is the diketopiperazine fused C-1 indol-3-yl substituted tetra-hydro-isoquinoline, termed ISA-2011B92. Although ISA-2011B binds with high affinity to PI4P5Kα and inhibits its protein expression and cancer growth in multiple models as well as inhibits inflammation, it does have significant off-target effects, including potent binding to multiple other kinases, including the class IA PI3K p110α92. Further optimization of ISA-2011B, or the development of another unique chemical scaffold with increased selectivity towards PI4P5Kα, will be necessary to advance a PI4P5Kα inhibitor beyond preclinical studies.
A focused screen discovered UNC3230 (5-(cyclohexanecarboxamido)-2-(phenylamino)thiazole-4-carboxamide), which inhibited PI4P5Kγ, with no inhibition of PI4P5Kα at concentrations up to 10 μM (ref.93). However, UNC3230 showed higher potency towards PI5P4Kγ over PI4P5Kγ in in vitro lipid kinase assays, and off-target effects on multiple additional kinases. Although UNC3230 was validated as a potent PI4P5Kγ inhibitor in biochemical assays, further optimization is warranted as it does hit other lipid and/or protein kinases and also has a narrow efficacy window and low solubility in appropriate vehicles that prevented dose–response experiments both in vitro and in vivo. Nonetheless, UNC3230 was proposed to regulate nociceptive sensitization in response to diverse stimuli that cause pain93. Future studies are pertinent to validate PI4P5Kγ as an analgesic drug target in a clinical setting and assess its potential side effects.
A high-throughput screen of an AstraZeneca compound collection identified a series of high-quality 4-aminopyridine pan-isoform inhibitors of PI4P5K that showed potency against all PI4P5K isoforms and >100-fold selectivity over other lipid kinases and that can be used as in vitro tool probes to further investigate the therapeutic potential of inhibiting PI4P5Ks94. Importantly, these compounds provide a sound foundation for optimization of PI4P5K drugs with a promise of entering clinical testing in the near future.
PI4P5K inhibition in cancer and inflammatory disease
PI4P5Ks are intriguing potential targets for therapeutics in cancer and inflammation as they produce PI(4,5)P2 at the plasma membrane, which is required for activation of PI3K–AKT signalling, one of the most frequently activated pathways in human cancers and inflammation (Box 1). To date, PI4P5Kα has been shown to be expressed at higher levels in both prostate and breast cancers, which correlates with poor patient outcome92,95. The discovery of putative direct interactions between PI4P5Kα and p53 and oncogenic KRAS highlights an additional potential therapeutic benefit of targeting PI4P5Kα for the treatment of cancers with KRAS or TP53 mutations96,97. PI4P5Kα is the predominant isoform that the immune cell receptor CD28 recruits and activates at the immunological synapse in T lymphocytes98,99. Further, PI4P5Kα is a key regulator of CD28 autonomous signals that stimulate nuclear factor-κB (NF-κB) transcriptional activity and the transcription of pro-inflammatory genes99, in a PI3K-dependent manner100. Most studies examining the inhibition of PI4P5Kα and their role in cancer and inflammation have used ISA-2011B, which also potently binds to class IA PI3K p110α, complicating the analysis of its role in targeting PI4P5Kα. Although there might be a therapeutic benefit to targeting PI4P5Kα, the full validation of the therapeutic tractability of PI4P5Kα will require development of a truly potent and selective PI4P5Kα-specific inhibitor.
Pharmacological inhibitors of PI5P4Ks
The PI5P4Ks have become attractive drug targets in p53 null tumours and are implicated as key regulators of metabolism82,101,102. Early-phase small-molecule inhibitors are being explored in preclinical studies, including pan-PI5P4K inhibitors101,103–105 and isoform-specific PI5P4K inhibitors106–110 (Table 2). The optimization of drug analogues has produced new collections of tool compounds105,111; however, PI5P4K inhibitors have yet to advance to the clinical setting.
A phenotypic screen of indole acrylonitriles for agents that selectively kill cancer cells over normal cells led to the identification of the small molecule a131, which targets PI5P4Ks103. Although formulation of a131 for in vivo application is possible, a131 has poor aqueous solubility and a relatively short half-life after intravenous administration, limiting its in vivo use103,112. Biochemical screening of >5,700 small molecules identified several PI5P4Kα/β dual inhibitors with a conserved 2-amino-dihydropteridinone core, including the kinase inhibitors volasertib, palbociclib and BI-D1870. Refinement of this scaffold led to the PI5P4Kα/β dual inhibitor CC260, although further optimization is likely needed owing to several off-target activities for other protein and lipid kinases101.
THZ-P1-2 is a covalent inhibitor of PI5P4K shown to target all three isoforms irreversibly. This compound was developed from a modified backbone of an acrylamide-based JNK inhibitor, which was identified in a chemo-proteomic kinase screen104. THZ-P1-2 targets conserved cysteines outside the ATP-binding pocket of the PI5P4K kinase domain. In cancer cells, THZ-P1-2 treatment reduced proliferation and impaired autophagy, which phenocopies models that genetically target PIP4K2 (refs.74,104). Optimization of this molecule led to the derivative compound 30, which showed enhanced selectivity111. High-throughput screening identified the (Z)-5-methylene thiazolidin-4-one inhibitor CVM-05-022, which upon optimization, led to the selective PI5P4K inhibitor compound 13 (ref.105). The best and most selective PI5P4K inhibitor available is the 1,7-naphthyridine-based inhibitor BAY-091 (Fig. 3c) developed by Bayer, although its selectivity for PI5P4K isoforms is unknown107.
PI5P4K isoform-specific inhibitors are also in preclinical development. One example is the pyrimidine-2,4-diamine compound SAR088 (Imanixil; Sanofi-Aventi), which is a PI5P4Kβ-specific inhibitor108, exhibiting suitable drug properties, including no liver CYP34A inhibition, intermediate cell permeability and high metabolic stability. Importantly, pharmacodynamic evaluation of SAR088 in vivo demonstrated efficacy and bioavailability after oral administration. The quinazolin-4-amine compound NIH-12848 was identified as a PI5P4Kγ inhibitor, acting as an allosteric non-ATP-competitive inhibitor that binds to the putative PI5P substrate binding site109. Optimization of this scaffold led to compound 40, which binds an allosteric pocket composed of the activation loop, and PI5P4Kγ unique residues113. The allosteric PI5P4Kγ inhibitor NCI-504 also targets this site, increasing productive autophagic flux in fibroblasts and leading to disrupted phosphoinositide equilibrium in cells110.
Inhibiting PI5P4Ks in cancer
Therapeutic opportunities for targeting PI5P4Ks in cancer have recently been reviewed in detail114. Briefly, PIP4K2B overexpression co-occurs with Erb-B2 receptor tyrosine kinase 2 (ERBB2 or HER2)-amplified breast cancers115; however, reduced PIP4K2B expression correlated with reduced patient survival in breast cancer116, suggesting that either too high or too low levels of PIP4K2B can be involved in disease. PI5P4Kα and PI5P4Kβ are elevated in breast cancer relative to normal tissue and their genetic ablation in a Trp53 null genetic mouse model led to a dramatic reduction in tumour formation, with silencing of both the α and β isoforms inhibiting breast cancer cell proliferation in vitro and in a xenograft tumour model82. Further, inhibition of both PI5P4Kα and PI5P4Kβ impairs mitochondrial function, which consequently reduces proliferation and in vivo tumour formation101,102.
There is potential for targeting PI5P4K in haematological malignancies, as PIP4K2A and PIP4K2C transcript expression is associated with clinical outcomes of patients with acute myeloid leukaemia (AML)117,118. Early characterization of the THZ-P1-2 covalent PI5P4K inhibitor showed that AML/ALL cell lines were sensitive to pharmacological PI5P4K inhibition104. Although targeting the PI5P4Ks in preclinical cancer models is promising, this strategy may not suit all cancers. Glioblastoma (GBM) brain tumours downregulate PI5P4Kα compared with normal tissue, with PI5P4Kα playing a tumour suppressor role119. Finally, the potent and selective PI5P4K inhibitors developed by Bayer did not show antiproliferative effects on p53 null tumour cells107, suggesting either that the antitumour effect requires complete loss of both PI5P4Kα and PI5P4Kβ as found in the genetic ablation models, or that inhibitors have to potently inhibit both PI5P4Kα and PI5P4Kβ isoforms to have an antitumour effect. Continued development of well-validated isoform-selective inhibitors will be crucial in defining any therapeutic opportunities.
PI5P4K inhibition in diabetes
PI5P4Kβ is a potential target in the treatment of hyperglycaemia and type 2 diabetes mellitus. This link was evident in the first Pip4k2b genetic knockout murine model. Pip4k2b−/− animals are viable but show notably reduced body weight and adiposity and are hypersensitive to insulin84. Preclinical testing of the selective PI5P4Kβ inhibitor SAR088 confirms the genetic phenotypes, as it dramatically lowers blood glucose levels of hyperglycaemic male obese rats108. Although PI5P4Kβ inhibitors pose an opportunity for the treatment of insulin resistance, it is still unclear whether the ability of PI5P4Ks and PI4P5Ks to potentially form mixed heterodimer isoforms could impair the efficacy demonstrated by SAR088, as the PI5P4Ks demonstrate complex crosstalk at multiple nodes of insulin signalling120.
PI5P4K inhibition in immunological disease
High expression of PI5P4Ks occurs in specialized immune organs such as the lymph node and spleen121. Hyperinflammation is the primary phenotype of mice that lack PI5P4Kγ. Although transgenic mice with a germline deletion of Pip4k2c have normal viability and growth, animals have heightened T helper (TH) cell activation, decreased regulatory T (Treg) cell populations and elevated plasma pro-inflammatory cytokines83. Inhibition of PI5P4Kβ and PI5P4Kγ is efficacious in reprogramming Treg cells to attenuate immunosuppressive activity and increase immune surveillance122. Genetic depletion and treatment with the PI5P4Kγ inhibitor, NIH-12848, impaired FOXP3 expression and reduced Treg cell proliferation, while sparing CD4+ conventional T cells and TH cell differentiation122.
PI5P4K inhibition in neurodegenerative disorders
Huntington disease is an autosomal dominant neurodegenerative disorder that results from the aggregation of mutated huntingtin protein. Pharmacological inhibition of PI5P4Kγ with the selective allosteric inhibitor NCI-504 increases autophagic flux, which is thought to reduce the accumulation of mutant huntingtin protein. NCI-504 also stimulates autophagy in rat primary cortical neurons with no impact on cell viability110. Aside from Huntington disease, the accumulation of protein aggregates occurs in Alzheimer disease and Parkinson disease. Catabolism of these aggregates following induction of autophagic flux by targeting PI5P4K could potentially alleviate neuronal toxicity. In addition, genome-wide association studies associate polymorphisms of PIP4K2A with elevated risk of schizophrenia123–125. Although no causative relationship for the correlation has been found, the PI5P4Ks are enriched in neural tissues and are hypothesized to affect synaptic function126.
PI3K−type III PI4K evolutionary family
PI4KA and PI4KB
Regulation and structure
There are four mammalian phosphatidylinositol 4-kinases (PI4Ks), composed of type II (PI4KIIα and PI4KIIβ, encoded by the genes PI4K2A and PI4K2B, respectively) and type III (PI4KIIIα and PI4KIIIβ, encoded by the genes PI4KA and PI4KB127, respectively), which together generate PI4P from phosphatidylinositol128,129. PI4P has well-defined roles in multiple organelles, including the Golgi–trans-Golgi network (TGN), endosomal membranes and plasma membrane6,130. Its production at these organelles mediates membrane recruitment of proteins, modulation of integral membrane protein activity and lipid transport between organelles7. The kinase domains of type II and type III PI4Ks are structurally divergent and evolutionarily distinct. The evolution of two distinct structural folds to generate the lipid species PI4P underlies the importance of regulating PI4P metabolism in multiple aspects of membrane trafficking, signalling and lipid transport.
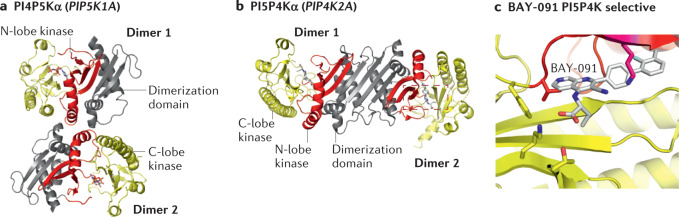
The PI4KA and PI4KB proteins have homologous helical and lipid kinase domains that are evolutionarily related to PI3Ks131 (Fig. 4a,b). However, the two proteins have divergent modes of regulation through a set of unique protein-binding partners and post-translational modifications. PI4KA generates the plasma membrane pool of PI4P132,133. PI4KA at 2,102 amino acids is the largest of the phosphoinositide kinases132,134, composed of an α-solenoid domain, a dimerization domain, and helical and lipid kinase domains homologous to those found in PI4KB and PI3Ks135. The mammalian signalling complex of PI4KA is composed of two additional regulatory proteins, the armadillo repeat protein TTC7 (either TTC7A or TTC7B)136, and the protein FAM126 (either FAM126A or FAM126B)137. The trimeric complex of PI4KA–TTC7B–FAM126A dimerizes through the dimerization domain of PI4KA, forming a ~900 kDa complex135 (Fig. 4a). The proposed primary mechanism mediating plasma membrane recruitment of the PI4KA complex is through TTC7 binding to the lipidated protein EFR3 (ref.136).
Fig. 4. Structure–function, inhibition and therapeutic targeting of PI4KA and PI4KB.
a, Domain architecture of phosphatidylinositol 4-kinase A (PI4KA). The architecture of the dimer of heterotrimers of PI4KA–TTC7–FAM126B (PDB: 6BQ1, with the solenoid region generated from the predicted α-fold model), with the domains coloured according to the domain schematic. The dimer interface between the two heterotrimers of PI4KA–TTC7–FAM126 is highlighted by a box, with regions that directly contact the other dimer unit coloured differently to highlight the dimer interface. b, Domain architecture of PI4KB. The structure of PI4KB (PDB: 4D0L), with the domains coloured according to panel domain schematic. c, Inhibition of PI4KB as an antiviral for positive-strand single-stranded RNA viruses (+ssRNA). Multiple picornaviruses require PI4KB as a host factor to generate PI4P-enriched viral replication organelles after viral infection. PI4P in these organelles recruits additional cellular machinery and restructures the lipid environment to generate a platform optimal for viral replication. Disruption of PI4KB either genetically or pharmacologically can prevent viral replication. d, Inhibition of the malarial homologue of PI4KB as an antimalarial therapeutic. The life cycle of malaria in both the vector (mosquito) and host (human) is indicated. The various life cycle stages of the Plasmodium species that cause malaria are annotated, and where malarial PI4KB inhibitors (PI4KBi; KDU691, KAI407, MMV390048) have shown efficacy are shown. PI4KBi have shown particular promise in the prevention of the multi-nucleated schizont stages in blood and liver by preventing membrane trafficking from the Golgi. e, Malarial PI4KBi currently in clinical trials. f, Inhibition of PI4KB as an anticancer therapeutic. PI4P generated by PI4KB plays a crucial part in malignant secretion of pro-tumorigenic effector proteins from cancer cells that contain a chromosome 1q region that is frequently amplified in diverse cancers. PI4P enhances secretion through activating Golgi phosphoprotein 3 (GOLPH3)-dependent vesicular release from the Golgi, with inhibition of PI4KB using highly selective PI4KIIIβ-IN-10 derivatives (PI4KBi) preventing this secretion. Panel a adapted with permission from ref.131, Elsevier.
The PI4KA gene is essential in yeast and mammals, with either pharmacological or genetic inactivation of PI4KA leading to sudden death133. PI4KA is ubiquitously expressed in all tissues; however, it is heavily enriched in brain tissues138. Biallelic loss-of-function mutations in PI4KA lead to a spectrum of neurological conditions, including neurodevelopmental delay, developmental brain abnormalities and paraplegia139,140. Loss-of-function mutations in TTC7A (termed TTC7A deficiency) have been identified in patients with inflammatory bowel disease, intestinal atresia and combined immune defects141–143, which mimic intestinal defects that occur in mice treated with PI4KA inhibitors133. PI4KA is also a crucial host factor in hepatitis C virus (HCV)144 and encephalomyocarditis virus infection145, through viruses hijacking PI4KA, leading to the generation of PI4P-enriched viral replication organelles.
PI4KB generates PI4P at the Golgi and TGN, and it has important roles in membrane trafficking and cytokinesis. The 801 amino acid PI4KB protein is composed of a helical domain and a lipid kinase domain146, as well as four disordered regions that mediate binding to regulatory proteins (Fig. 4b). Recruitment of PI4KB to the Golgi is primarily mediated by an interaction with the Golgi-resident protein ACBD3 and the N terminus of PI4KB147,148. The activity of PI4KB can also be modulated by phosphorylation, with protein kinase D (PKD) phosphorylating the linker between the helical and kinase domains, which leads to binding of 14-3-3 proteins149,150, which putatively stabilize PI4KB. PI4KB is also able to be phosphorylated by protein kinase A (PKA) in a disordered region of the kinase domain, which increases the affinity for the armadillo repeat protein ARMH3 (ref.151), which can act as a positive regulator of PI4KB activity. In addition, the C terminus of PI4KB contains an amphipathic lipid-packing sensor motif that increases PI4KB activity at packing defects in the Golgi–TGN152.
Although knockout of the yeast homologue of PI4KB is lethal, the knockout of the PI4KB homologue in flies leads to viable progeny, with male flies being sterile owing to defects in spermatogenesis153. Characterization of a mouse knockout of PI4KB has not been published, but loss of PI4KB in Schwann cells in mice leads to aberrant myelination of peripheral nerves154. Multiple pathogenic viruses, including kobuviruses, enteroviruses and Coxsackie viruses, hijack PI4KB to generate viral replication organelles, where PI4P is required to generate a lipid environment conducive to viral replication155–157.
Pharmacological inhibitors of PI4KA
The covalent PI3K inhibitor wortmannin was essential to the original biochemical characterization and identification of both PI4KA and PI4KB158–160, with the type III PI4Ks being separated from type II PI4Ks by their sensitivity to wortmannin. The discovery of the crucial role of PI4KA in HCV infection144 led to efforts to develop potent and specific PI4KA inhibitors (Fig. 4c and Table 3). A library screen for anti-HCV inhibitors identified the 4-anilino quinazoline AL-9, which inhibited PI4KA, but with weak specificity over PI4KB and p110α161. A small-molecule compound screen for PI4KA inhibitors by Boehringer Ingelheim identified multiple PI4KA inhibitors with anti-HCV activity, with roughly 10- to 20-fold selectivity for PI4KA over PI4KB162. The most potent and selective PI4KA inhibitors currently available are molecules developed by GSK and AstraZeneca. The GSK compounds are quinazoline precursors optimized to generate the potent and selective inhibitor GSK-A1 (ref.163). The GSK-A1 compound suffered from poor pharmacokinetic properties, with the quinazoline GSK-F1 having improved pharmacokinetics and similar selectivity over PI4KB and PI3Ks, although with decreased PI4KA potency133. AstraZeneca identified a 2-aminobenzothiazole derivative that was moderately selective for PI4KA over PI4KB. Further optimization of this scaffold led to highly potent PI4KA inhibitors with increased selectivity164,165.
Table 3.
Summary of preclinical inhibitors of the type III PI4Ks and class II and III PI3Ks
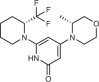
| Compound | Target | Target potency (Kd or IC50) | Known off-targets/selectivity | References |
|---|---|---|---|---|
|
GSK-A1 (GSK)
|
PI4KA | IC50 3.1 nM | >100-fold selectivity over PI4KB/class I PI3Ks | Bojjireddy et al. (2014)133 |
|
GSK-F1 (GSK)
|
PI4KA | IC50 16 nM | >100-fold selectivity over PI4KB/class I PI3Ks | Bojjireddy et al. (2014)133 |
| Aminobenzothiazole derivatives (AstraZeneca) | PI4KA | IC50 1–6 nM | >100-fold selectivity over PI4KB/class I PI3Ks | Raubo et al. (2015)164 |
|
PIK-93 derivative PI4KIIIbeta-IN-10
|
PI4KB | IC50 3.6 nM | >200-fold selectivity over PI4KA/class PI3Ks | Rutaganira et al. (2016)167 |
T-00127-HEV1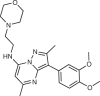
|
PI4KB | IC50 60 nM | Partial inhibition of PIK3CD at 10 µM | Arita et al. (2011)169 |
|
|
PI4KB | IC50 5.7 nM | >250-fold selectivity over PI4KA | van der Schaar (2013)170 |
|
MI356 derivative compound 10
|
PI4KB | IC50 6.1 nM | No detectable inhibition of any protein or lipid kinase at 1 µM | Mejdrova et al. (2017)172 |
|
UCB9608 (UCB Pharma)
|
PI4KB | IC50 11 nM | 15-fold selectivity over class II PI3Ks | Reuberson (2018)175 |
|
MIPS-19416
|
PI3KC2α/β | IC50 7 nM (C2α) 43 nM (C2β) | 10-fold over class I PI3K | Selvadurai et al. (2020)209 |
|
PITCOIN1
|
PI3KC2α | IC50 95 nM (C2α) | Selective for PI3KC2α | Lo et al. (2022)211 |
|
PITCOIN2
|
PI3KC2α | IC50 121 nM (C2α) | Selective for PI3KC2α | Lo et al. (2022)211 |
|
PITCOIN3
|
PI3KC2α | IC50 126 nM (C2α) | Highly selective for PI3KC2α | Lo et al. (2022)211 |
|
VPS34-IN1
|
VPS34 | IC50 25 nM | Highly selective | Bago et al. (2014)222 |
|
SAR405 (Sanofi)
|
VPS34 | IC50 1.2 nM | Highly selective | Ronan et al. (2014)224 |
|
PIK-III (Novartis)
|
VPS34 | IC50 18 nM | Highly selective | Dowdle et al. (2014)223 |
|
SB02024 (Sprint Bioscience)
|
VPS34 | Kd 1.1 nM | Highly selective | Dyczynski et al. (2018)225 |
Covalent and allosteric (non-ATP competitive) inhibitors are indicated. Half-maximal inhibitory concentration (IC50), dissociation constant (Kd) and selectivity data are from the indicated references. PI3K, phosphoinositide 3-kinase; PI4K, phosphatidylinositol 4-kinase.
Limiting the further clinical or preclinical development of PI4KA inhibitors has been the toxicity associated with PI4KA inhibition. Mice homozygous for either knockout of PI4KA or a knock-in PI4KA kinase dead mutant displayed a lethal phenotype, with extensive degeneration of the gastrointestinal tract162. Pharmacological blockade of PI4KA in mice with GSK PI4KA inhibitors led to sudden lethality at the highest doses, with animals dosed at the lowest levels remaining alive but displaying moderate to severe gastrointestinal abnormalities133. These studies suggest that any possible use of small molecules targeting PI4KA will have a very small therapeutic window owing to adverse toxicity.
Pharmacological inhibitors of PI4KB
One of the first semi-selective PI4KB inhibitors, the phenylthiazole PIK-93 was identified in structure–function studies targeting the p110α isoform of PI3K166. Although PIK-93 has been used extensively as a ‘selective’ PI4KB inhibitor, it inhibits the PI3Ks VPS34 and p110γ with almost equal IC50 values167. Structure-based drug design allowed for the design of multiple improved PIK-93 derivatives, with the inclusion of a t-butyl group on the acetamide group off the central thiazole causing similar potency, but leading to greater than 200-fold selectivity over all other phosphoinositide kinases167.
Potent and selective PI4KB inhibitors have been found to prevent viral infection (Fig. 4c). Enviroxime was discovered as a molecule that prevented poliovirus replication, with clinical trials initiated for enterovirus infection but discontinued in phase II owing to insufficient therapeutic effects168. A pyrazolo-pyrimidinamine derivative of enviroxime (T-00127-HEV1) is a potent and selective PI4KB inhibitor169. The chemically similar imidazo[1,2-a]pyrazine derivative BF738735 was identified from a kinase inhibitor library screen towards inhibitors of CVB3 infection, with PI4KB identified as the primary target170. Derivatives with an imidazo[1,2-b]pyridazine scaffold were also developed, leading to MI14 (ref.171). Using structure-based drug design from the imidazo[1,2-b]pyridazine scaffold and combining features from the most potent PIK-93 derivatives led to MI356, which is the most potent and selective PI4KB inhibitor discovered to date172.
The discovery of PI4KB as a therapeutic target in viral infection led to extensive efforts from the pharmaceutical sector to develop PI4KB inhibitors as antivirals173. However, conflicting results on toxicity and effects on the immune system have complicated its therapeutic potential. A set of highly potent and selective aminoimidazole PI4KB inhibitors developed by Novartis were able to block HCV replication, with limited cellular toxicity, but had a strong antiproliferative effect on lymphocytes174. Further supporting the role of PI4KB in immune function, it was identified as the target of a class of immunosuppressive 7-piperazin-1-ylthiazolo[5,4-d]pyrimidin-5-amine analogues, leading to the generation of the derivative UCB9608, which prevented immune rejection in mouse models of organ transplantation175. Outside of their effects on the immune system, there have been concerns about the toxicity of PI4KB inhibitors. An aminothiazole series developed by Boehringer Ingelheim and the chemically distinct PI4KB inhibitor T-00127-HEV1 were toxic in SJL mice176. However, the PI4KB inhibitor BF738735 was well tolerated in mice and had a dose-dependent protective effect in a CVB4-induced pancreatitis model170. The combination of immunosuppressive effects and conflicting reports on PI4KB inhibitor toxicity, together with the fact that enteroviruses can acquire resistance mutations that bypass the need for PI4KB in viral replication, has decreased enthusiasm for further development of PI4KB inhibitors as antiviral therapeutics.
PI4KB inhibition in parasitic infections
The most clinically advanced inhibitors of PI4KB are specific towards the parasite homologue of PI4KB, found in the causative agents of malaria and cryptosporidiosis177,178. A major challenge in the treatment of malaria is the identification of compounds that can inhibit all life cycle stages of parasite development. High-throughput screening of compounds that target the asexual blood stage of Plasmodium falciparum (causative agent of malaria) identified an imidazopyrazine-derived class of antimalarials that targeted the Plasmodium homologue of PI4KB177. This study led to the molecules KAI407 and KDU691, which both showed >1,000-fold selectivity for the Plasmodium PI4KB over any human lipid kinase, including human PI4KB177, and potent activity against all life cycle stages (Fig. 4d). Further validation of Plasmodium PI4KB as a therapeutic target was provided by a chemo-proteomics screen that identified anti-parasitic 2-aminopyridine derivatives as PI4KB inhibitors179, as well as a diversity-oriented synthetic screen that identified selective PI4KB inhibitors as antimalarials180. The 2-aminopyridine derivative inhibitor MMV390048 was efficacious against all life cycle stages, and showed efficacy in a monkey model of malarial infection181 (Fig. 4e). Owing to the roles of human PI4KB in immune function, the development of highly specific Plasmodium PI4KB inhibitors over the human variant is crucial. Initial insight into the molecular mechanism of selectivity was generated by molecular modelling of MMV390048 into the human and Plasmodium variants of PI4KB182. Clinical trials of MMV390048 are ongoing183 (Fig. 4e), with the next generation of MMV390048 derivatives being optimized for solubility and potency, leading to the more potent 2-aminopyrazine compound UCT943 (ref.184).
PI4KA and PI4KB inhibition in cancer
There are some preclinical data that highlight the potential for targeting PI4KA and PI4KB in cancer185–188. PI4KA generates the PI4P pool at the plasma membrane, which is phosphorylated into PI(4,5)P2, which can be further phosphorylated into the pro-growth signal PIP3. Some of the most frequently mutated genes in human cancer are HRAS, NRAS and KRAS, with the activation of class IA PI3K p110α to generate PIP3 being crucial in Ras-driven tumorigenesis189. An analysis of the Ras interactome identified an interaction between the PI4KA regulatory protein EFR3A and KRAS, with disruption of either PI4KA or EFR3A leading to decreased PI4P, PS and KRAS levels at the plasma membrane, with a concurrent decrease in oncogenic signalling and tumorigenesis185. Treatment of mutant KRas pancreatic cell lines with a combination of a G12C-specific Ras inhibitor (sotorasib) and the most potent 2-aminobenzothiazole AstraZeneca PI4KA inhibitor had a synergistic inhibitory effect on cancer cell growth. Another study showed that knock-down of the ORP5/8 lipid transport machinery that mediates the PI4P-driven lipid transport of PS to the plasma membrane was required for KRAS oncogenesis in a pancreatic cancer mouse model, and that PI4KA and EFR3A were upregulated in pancreatic tumours versus normal tissue186. This finding suggests that there may be a beneficial, limited therapeutic window of PI4KA inhibition in mutant KRAS-driven cancers when used in combination with either PI3K or KRAS inhibitors, although extremely careful analysis of toxicity will be essential.
PI4KB is overexpressed in various cancers, with a chromosomal region containing PI4KB being frequently amplified in many cancer types. The role of PI4KB in tumorigenesis is proposed to be mediated through its ability to recruit the oncogenic PI4P-binding protein Golgi phosphoprotein 3 (GOLPH3), leading to enhanced secretion of pro-tumorigenic effector proteins, including semaphorin 3C (SEMA3C), lysyl hydroxylase 3 (PLOD3), tissue inhibitor of metalloproteinase 1 (TIMP1), peroxiredoxin 5 (PRXD5), annexin A2 (ANXA2), clusterin (CLU) and stanniocalcin 2 (STC2), which drive increased metastasis188 (Fig. 4f). PI4KB inhibitors derived from the most potent PIK-93 scaffold with improved pharmacokinetic properties led to smaller primary tumours and fewer metastases in the contralateral lung in mice containing orthotopic chromosome 1q-amplified lung tumours188 (Fig. 4f). The clinical potential of PI4KB in chromosome 1q-amplified lung tumours may be limited, as lung cancer cells treated with PI4KB inhibitors acquire tolerance by upregulating the type II PI4K2A (PI4KIIα), providing an alternative source of Golgi-resident PI4P.
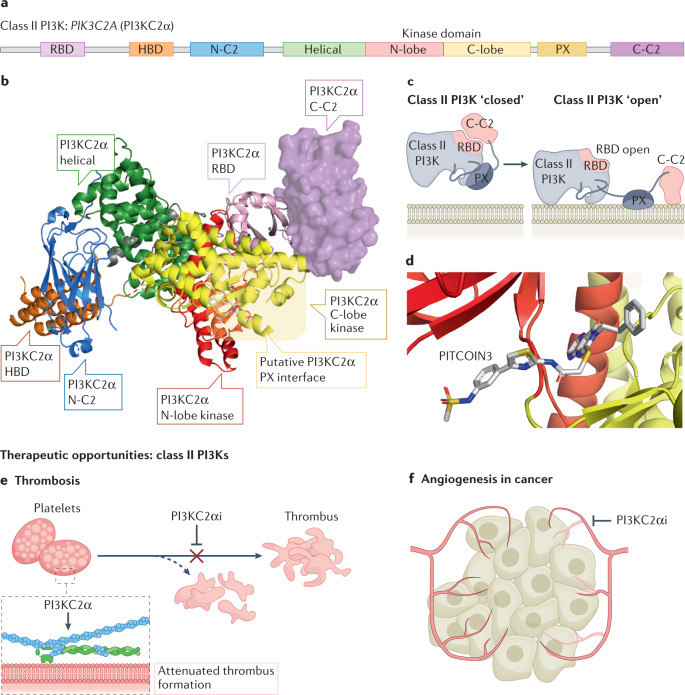
Class II PI3Ks
Structure and regulation
The class II PI3Ks, otherwise known as PI3K-C2s (herein PI3KC2s), are a family defined by a unique substrate selectivity and an extended domain architecture, which incorporates an eponymous C-terminal C2 domain (hence the ‘C2’ in PI3KC2s, which does not explicitly refer to ‘class 2’). There are three isoforms, α, β and γ, which are encoded by PIK3C2A, PIK3C2B and PIK3C2G, respectively. The extended domain architecture appears to substitute for a regulatory subunit, which has not been identified for PI3KC2s, contrasting with the constitutively associated regulatory subunits of class I and III enzymes190 (Fig. 5a). The PI3KC2s catalyse the 3-OH phosphorylation of both PI and PI4P; thus, they appear to be both an alternative to class III PI3K for PI3P synthesis and an alternative to class I PI3K and 5-OH phosphatase-mediated production of PI(3,4)P2 (Fig. 1b). Both products appear to be physiologically relevant for PI3KC2 function190, which is discussed later. A recent crystal structure of the C2α isoform reveals adjacent Arg and Lys residues in the activation loop that confer the ability to bind to PI4P, which are lacking in the PI-selective class III PI3K191.
Fig. 5. Structure–function, inhibition and therapeutic targeting of class II PI3Ks.
a, Domain architecture of class II PI3Ks. b, The combined model of PI3KC2α with the domains coloured according to the domain schematic. The model is derived from both cryogenic electron microscopy (cryo-EM) and X-ray data (compiled from PDB: 7BI2, 6BTY). The interface of the C-terminal C2 domain (C-C2) with the Ras-binding domain (RBD) was generated by docking the structure of the C-C2 (PDB: 6BTY) into the cryo-EM density of EMD-12191. The putative PX interface on the kinase domain is based on HDX-MS data. c, The model of PIK3C2α activation, in which the PX and C-C2 domains inhibit class II PI3K activity in the closed configuration, and upon lipid binding of both these domains, class II PI3K adopts an open active configuration, allowing it to bind to lipid substrate, leading to opening of both the kinase and RBD domains. There are additional protein-binding partners that have important roles in activating and switching off class II PI3K signalling, but which are omitted here for clarity of the inactive and active states. d, Structure of PI3KC2α bound to the selective inhibitor PITCOIN3 (PDB: 7Z75)211, with the domains coloured according to the domain schematic in panel a. e, Inhibition of PI3KC2α as an anti-thrombolytic: genetic or pharmacological inhibition of PI3KC2α (PI3KC2αi) is reported to disrupt attachment of the membrane skeleton in platelets, disrupting the extensive infolding of the plasma membrane known as the open canicular system. This impedes the formation of protrusions and filopodia in activated platelets, thus reducing the formation of thrombi under arterial shear stress. This has been observed in mice, as well as in humans ex vivo. f, PI3KC2α as a target for tumour angiogenesis. Genetic disruption of PI3KC2α has been shown to potently inhibit angiogenesis; in adult mice, this can severely disrupt vascularization of tumours. HBD, helical bundle domain; N-C2, N-terminal C2 domain.
The structure of the PI3KC2α isoform191 (Fig. 5b) includes the C2 domain, the Ras-binding domain (RBD), and helical and kinase domains of the catalytic core, which exhibit a similar architecture to the class I PI3Ks but are mounted atop a unique four-helix bundle in the class II enzymes. Each family member contains a long, apparently unstructured N-terminal region, containing a clathrin-binding motif in PI3KC2α192. The C-terminal PX–C2 domain module is required for optimal activity of the enzyme by facilitating binding to PI(4,5)P2-containing membranes193. Structural and biochemical analysis of the module reveals moderate binding selectivity and affinity for PI(4,5)P2 in both C2 and PX194. Crucially though, both domains are oriented in such a way that concomitant membrane binding by each domain is possible; thus, when combined with membrane interactions of the kinase core region, high avidity PI(4,5)P2-dependent membrane interaction is enabled194 (Fig. 5c).
Although the C-terminal PX–C2 module is necessary for optimal catalytic activity, there is an apparent steric inhibition of the module through binding at the RBD and kinase domain193. Mutation of an EKP motif in the kinase domain kα7–8 loop to KKT (found in class I enzymes) disrupts this interaction and leads to constitutive activation of the enzyme. Thus, PI(4,5)P2 binding to PX–C2 not only recruits PI3KC2α to the membrane, but also relieves autoinhibition, analogous to phosphotyrosine-relieved autoinhibition of class I PI3K catalytic subunits by the C-terminal SH2 domain of the regulatory subunit. The C-terminal C2 at the RBD interface potentially plays a part in preventing Ras activation191. The EKP motif does not directly bind to the C-terminal C2 domain, but instead stabilizes an unfolded conformation of the kα12 helix, permitting sufficient flexibility for the C-terminal C2 domain to dock at a second site on the RBD. Release of the PX and C2 domains allows the membrane-binding kα12 helix to position in an orientation competent for catalysis (Fig. 5c).
The best-studied class II isoform is PI3KC2α, which has crucial roles in development, with PI(3,4)P2 generated by PI3KC2α acting as a key driver of endocytosis195. Loss of C2α expression196,197 or catalytic activity198 is embryonic lethal in mice, associated at least in part with defective primary cilia signalling, with a loss of a PI3KC2α-generated pool of PI3P at the pericentriolar recycling compartment, causing disruption of the membrane traffic required for ciliogenesis197. The enzyme has also been found to be essential for primary cilia transduction of shear stress, which induces autophagy, which is required to regulate cell size in kidney tubule epithelia199. Such cilia-derived phenotypes have been described in human fibroblasts from patients with homozygous loss-of-function PI3KC2A mutations200. Clinically, these patients present with an array of neurological manifestations, short stature, facial dysmorphia and congenital cataracts with secondary glaucoma200. Although most of these phenotypes may be ascribed to primary cilia dysfunction, recent evidence has revealed a novel role for PI3KC2α in cell division: the high incidence of cataracts is due to aberrant abscission in lens epithelial cells, leading to elevated levels of senescence201. Mechanistically, PI3KC2α is localized to the midbody of the cytokinetic bridge by γ-tubulin, where it generates a pool of PI(3,4)P2 necessary for the recruitment of VPS36 — a core component of the ESCRT complex that drives the final membrane scission between daughter cells. Despite this essential role for PI3KC2α in primary cilia function and human development, mice heterozygous for an inactivated version of the enzyme are healthy and fertile, even though loss of 50% of the PI3K activity is apparent in vivo198. Therefore, there appears to be promise of tolerability of PI3KC2α-directed therapy.
Preclinical data for the other PI3KC2 isoforms are less extensive. Mice homozygous for kinase dead mutations in PI3KC2β show enhanced glucose tolerance. This occurs as a result of enhanced insulin sensitivity in metabolic tissues202. In hepatocytes, PI3KC2β inactivation decreased endosomal PI3P levels and increases APPL1 endosome number, causing a failure of insulin receptor endocytosis, thus providing a mechanistic basis for increased insulin sensitivity. These early indications suggest that this enzyme could be a target in type 2 diabetes. Notably, these insulin-sensitizing effects seem to be independent of the canonical class I PI3K signalling pathway downstream of insulin receptors202. PI3KC2β inactivation also decreases ischaemic reperfusion injury by maintaining endothelial integrity; the mechanism echoes the effects in hepatocytes, where decreased endosomal maturation seems to enhance endocytic recycling of the crucial endothelial adhesion molecule, VE-cadherin203. PI3KC2β also stimulates focal adhesion turnover by recruiting a RhoA GAP, facilitating cell migration and potentially playing a part in metastasis204. Finally, muscle-specific knockout of PI3KC2β has also been shown to alleviate the elevated PI3P levels association with myotubular myopathy, caused by loss-of-function mutations in the PI3P phosphatase gene, MTM1 (ref.205). Therefore, there seems to be scope for pharmacological targeting of PI3KC2β in various diseases. Development of novel isoform-selective inhibitors will be crucial to test these roles. However, a concerning finding regarding the potential utility of PI3KC2β-specific inhibitors relates to the role of the enzyme in inhibiting TORC1 signalling206: loss-of-function mutations in PI3KC2B were recently identified, and through elevated mTOR signalling, were found to drive focal epilepsy in humans and mice207. Therefore, even acute inhibition of the enzyme may have severe neurological adverse effects in patients.
Pharmacology of class II PI3Ks
The first class II PI3K to be cloned was PI3KC2α208, characterized by a remarkable insensitivity to the generic PI3K inhibitors wortmannin and LY294002. There was little attention paid to selectively drugging these enzymes until very recently, as the importance of the enzymes in human health is becoming apparent190. Initial inhibitors for PI3KC2s were developed from derivatives of the class I PI3K inhibitor PIK-90, leading to the 4-aminonicotinamide derivative MIPS-19416, which display potency against PI3KC2α and PI3KC2β, but with only modest selectivity over class I enzymes209 (Table 3). A combination of high-throughput screening and iterative medicinal chemistry optimization led to the development of the highly selective PI3KC2α inhibitors (PITCOINs)201,210,211. The most selective of these molecules (PITCOIN3) is exquisitely sensitive for PI3KC2α, with no inhibition of >100 other tested human lipid and protein kinases, and high selectivity over other class II PI3Ks. The structure of PITCOIN3 bound to PI3KC2α showed non-conserved interactions of the N-phenylmethanesulfonamide of PITCOIN3 in driving selectivity over other class II PI3Ks (Fig. 5d). Treatment of platelets with PITCOIN3 recapitulated the anti-thrombotic effect observed in PI3KC2α knockout cells, suggesting that it may be an intriguing starting point for anti-thrombotics without the side effect of increased bleeding.
Therapeutic indications for class II PI3Ks
Although there have only been very recent advances in the development of selective compounds that target PI3KC2s, there is considerable genetic evidence for therapeutic potential. Mouse experiments that induced reduction of PI3KC2α expression212 or catalytic inactivation of a single allele213 showed the intriguing potential of PI3KC2α as an anti-thrombosis target, with comparatively mild effects on haemostasis. In either case, basal PI3P levels are reduced in platelets212,213, correlating with a swollen and expanded infolding of the plasma membrane, known as the open canalicular system (OCS). Mechanistically, this seems to stem from a loss of attachment of components of the membrane skeleton, including spectrin, myosin and moesin, leading to stiffer platelets unable to form stable thrombi under arterial sheer213 (Fig. 5e). Intriguingly, the development of MIPS-21335 showed that acute pharmacological inhibition could rapidly and reversibly recapitulate these effects on platelet OCS and thrombus formation, and was even more potent in human and mouse assays of thrombosis than gold standard aspirin and P2Y12 antagonists209.
Roles for PI3KC2α have also been identified in cancer. The helical bundle domain was reported to target PI3KC2α to the mitotic spindle193, where it assembles with clathrin and TACC3 to bridge the kinetochore fibres. Loss of PIK3C2A expression in cancer cells thus increases aneuploidy and sensitizes cancer cells to microtubule-based taxane therapies214. However, it should be noted that the scaffold function of PI3KC2α in the clathrin–TACC3 complex has not been observed in other cell types215. A catalytic function of PI3KC2α has also been reported, wherein the helical bundle domain targets the kinase to focal adhesions in breast cancer cells, where the resulting PI(3,4)P2 stimulates turnover, promoting metastasis210. Promisingly, this enhanced metastatic potential can be blocked with a PI3KC2α inhibitor (PITCOIN1). Finally, there are indications for a role of PI3KC2α in tumour angiogenesis. Endothelial cell-specific knockout of PIK3C2A recapitulates the embryonic lethality seen in the global knockout owing to a failure in angiogenesis196. This appears to occur as a result of deficits in an endosomal PI3P pool required for VE-cadherin and VEGF receptor trafficking. However, endothelial-selective PIK3C2A knockout in adult mice inhibited angiogenesis and tumour growth of both lung carcinoma and melanoma models, revealing a potential therapeutic use of PI3KC2α inhibitors196 (Fig. 5f).
Class III PI3K: VPS34
Structure and regulation
The class III PI3K, VPS34 (encoded by PIK3C3 in humans) is the primordial PI3K enzyme in eukaryotes. It exclusively catalyses PI3P production from PI in vitro and in vivo (Fig. 1) and is found as two distinct tetrameric complexes, complex I and complex II. Both complexes are composed of VPS34, the pseudokinase VPS15 and the BARA domain containing BECLIN-1. Complex I is defined by a unique fourth component, ATG14; additionally, it is activated by a fifth subunit, NRBF2. In complex II, the fourth subunit is UVRAG (Fig. 6a). The two complexes are functionally distinct, with complex I initiating macroautophagy, and complex II controlling flux through the endocytic pathway216.
Fig. 6. Structure–function, inhibition and therapeutic targeting of class III PI3Ks.
a, Domain architecture of VPS34. The architecture of the tetramer of complex II (CII) of VPS34 is shown (PDB: 7BL1), with VPS34 domains coloured according to the domain schematic. The additional UVRAG, BECLIN-1 and VPS15 subunits are indicated, with the binding site of the VPS34 CII activator RAB5 shown (RAB5 shown as a transparent surface). b, Structure of VPS34 bound to the class III PI3K selective inhibitors PIK-III and SAR405 (PDB: 4PH4, 4OYS). The hydrophobic pocket surrounding the -CF3 (SAR405) or cyclo-propyl (PIK-III) groups that is crucial in VPS34 selectivity is annotated. c, Complex-selective activation of the VPS34 CI and VPS34 CII by either RAB1 or RAB5 GTPase, respectively. RAB1, present at the autophagosome, recruits and activates VPS34 CI, whereas RAB5, present on endosomal membranes, recruits and activates VPS34 CII. d, VPS34 inhibitors (VPS34i) target autophagy in cancer. Pharmacological block of VPS34 disrupts VPS34 CI, disrupting autophagosome biogenesis. This precludes a necessary adaptation and mechanism of resistance for the cancer cell in the stressful environment of the tumour bed.
The crystal structure of complex II revealed a V-shaped assembly with the VPS34 C2 domain at its nexus. The remainder of VPS34 is intertwined with the VPS15 α-solenoid and kinase domains along one arm, with both the VPS15 and VPS34 kinase domains at the tip217. The other arm of the V consists of the VPS15 WD40 domain, flanked by coiled-coil domains from BECLIN-1 and UVRAG (Fig. 6a), which present their membrane-binding domains at the tip of the arm. It was thus implicit that the active enzyme would orient on the membrane by the tips of the V, with the nexus protruding from the membrane surface217. Complex I exhibits a largely similar architecture, with ATG14 replacing UVRAG, again presenting its membrane-binding BATS domain at the tip of the arm218. In the complex II structure, key elements in the VPS34 kinase domain, including the activation loop, interact with VPS15 in a manner incompatible with substrate access217. Intriguingly, the complex I structure showed several conformational classes, some of which showed poor density for the VPS34 kinase domain, indicating mobility. Such mobility was essential for catalysis: a ‘leashed’ domain with a shortened C2–HELCAT linker only able to adopt the closed V structure was inactive218. Thus, release of the VPS34 kinase domain from VPS15 appears to be essential for catalysis.
A further structure of complex I with the fifth subunit NRBF2 showed how the release of the inhibited VPS34 kinase can occur. The binding of an NRBF2 MIT domain occurs at the nexus of the complex, causing the VPS15 α-solenoid to bend, displacing the VPS34 kinase domain from its inhibited interaction with VPS15; the VPS34 kinase domain then swings out by 25˚. Binding of a second NRBF2 MIT domain is then required for full activation of the enzyme by a mechanism not yet structurally defined219. Since then, the interaction of complex II with the master regulator of early endosome function, the small GTPase RAB5, has been defined220 (Fig. 6a). RAB5 binds with the UVRAG/BECLIN-1 arm of the complex, in a pocket formed by the VPS15 SGD and WD40 domains, along with the VPS34 C2 domain helical hairpin. Similarly to NRBF2 binding to complex I, RAB5 binding releases the VPS34 kinase domain from its inhibitory interaction with VPS15. Intriguingly, a similar binding and activation mechanism occurs with complex I upon binding of RAB1, revealing a novel mechanism of regulation in macroautophagy220 (Fig. 6c).
The RAB5–complex II interaction with liposomes can adopt a range of orientations, suggesting a striking mechanism for catalysis of both complex I and complex II bound to Rab proteins220: the membrane-interacting domains of the BECLlN-1 arm, together with the tethered GTPase hypervariable region, anchor this arm of the complex to the membrane surface. This allows the complex to ‘scoot’ across the membrane, facilitating processivity. Meanwhile, the VPS34 arm swings up and down, ‘pecking’ at the surface of the membrane and phosphorylating PI lipids.
A key lesson from this elegant structural characterization is that both complexes share essentially the same activation mechanisms: releasing the VPS34 domain from an inhibited conformation by a series of interactions unique to each complex. However, the mode of catalysis and orientation of the VPS34 domain appears highly conserved in each. It is not possible for an ATP-competitive kinase inhibitor to distinguish complexes I and II. Instead, future efforts to selectively inhibit complex I or complex II will likely need to target the different allosteric activation sites located in the more distal regions of the complexes.
Pharmacology of class III PI3K
The significance of inhibiting VPS34 was recognized more than two decades ago when an inhibitor of autophagy, 3-methyladenine (3-MA), was discovered to block this enzyme221. However, 3-MA is an extremely non-selective compound, inhibiting the other classes of PI3K221. More recently, several potent and highly selective VPS34 inhibitors have been developed (Fig. 6b and Table 3). These include the bipyrimidinamine derivative VPS34-IN1 from the University of Dundee222, the chemically similar 4-aminopyridine PIK-III from Novartis223 and the tetrahydropyrimido-pyrimidinone derivative SAR405 from Sanofi224. More recently, SB02024 (Sprint)225 and dihydropyrazolopyrazinone derivatives from Genentech have been reported226. All have nanomolar potency and are selective for VPS34 over other classes of lipid kinase by several orders of magnitude. A common feature of their selectivity is the exploitation of a somewhat larger hydrophobic pocket in the ATP-binding region of the VPS34 kinase domain, adjacent to the P-loop. This pocket is occupied by a cyclo-pentyl group in VPS34-IN1 and PIK-III, a trifluoromethyl group in SAR405 and an isopropyl group in the Genentech series, which would not be accommodated in class I PI3K. In short, excellent, high potency and exquisitely selective compounds are available and in use for preclinical studies — with the caveat that they inhibit both complex I and complex II.
Therapeutic indications for class III PI3K
The central involvement of VPS34 complex I in autophagy has attracted the most attention for therapeutically targeting this enzyme. Macroautophagy maintains cellular homeostasis and is generally thought of as tumour suppressing in healthy tissue. However, once tumours form, the capacity of macroautophagy to support cell survival in the stressful and nutrient-poor tumour bed causes the process to become increasingly tumour promoting227 (Fig. 6d). PIK-III, SAR405, SB02024 and VPS34-IN1 all inhibit autophagic flux222–225. Recent preclinical studies have also shown indications for these compounds to be effective in cancer treatment.
Combined inhibition of VPS34 and the class I PI3K, PI3Kδ, enhanced the cytotoxicity of various cellular models of B cell malignancies, including CLL and AML228. Enhanced efficacy was achieved by combining VPS34-IN1 with a clinically approved compound, CAL-101 (idelalisib), or by using a novel dual-selectivity compound, PI3K/V-IN-01 (ref.228). VPS34-IN1 and PIK-III also increased apoptosis in AML cell lines, an effect attributed at least in part to the inhibition of autophagy229.
SB02024 reduced growth of breast cancer cell xenografts in mice225. SB02024 and SAR405 also decrease tumour growth and improve survival in murine models of melanoma and colorectal cancer230. Mechanistically, VPS34 inhibition enhanced tumour cell STAT1–IRF7 signalling, increasing CCL5 and CXCL10 expression, and causing enhanced infiltration of CD8+ T cells and natural killer cells into the tumour bed. Interestingly, VPS34 inhibition enhanced the expression of programmed cell death 1 (PD1) and programmed death 1 ligand 1 (PDL1), a key target of immunotherapy. Indeed, VPS34 inhibitors sensitized these tumour models to anti-PD1/PDL1 therapy230.
Macroautophagy can be a mechanism of resistance in cancer, helping to prevent the induction of apoptosis227. In fact, SAR405 restored the sensitivity of several cisplatin-resistant bladder cancer cell lines to cisplatin231. The effect could be recapitulated with 3-MA or autophagy-blocking chloroquine. Furthermore, SB02024 increased the sensitivity of breast cancer cell lines to the approved tyrosine kinase inhibitors erlotinib and sunitinib in vitro225. Therefore, there seems to be some promise for the use of VPS34 inhibitors as combinatorial therapies (Fig. 6d).
However, the promise of VPS34 inhibitors in the clinic must be tempered by the fact that both complex I and II core functions, namely macroautophagy and endocytosis, are essential cellular housekeeping functions; severe on-target adverse effects are to be anticipated in vivo. In fact, recent work testing high-potency VPS34 inhibitors from both GSK232 and Genentech226 revealed severe toxicity in rats. On the other hand, no such toxicities were reported over several weeks in mice treated with SB02024 or SAR405 (refs.225,230). Clues to this discrepancy come from mice carrying targeted inactivation of the VPS34 kinase domain233. Mice with two inactive kinase domains die in early embryogenesis, whereas mice with a single active allele display mildly attenuated autophagy and beneficial metabolic enhancements. It therefore seems that there may be a beneficial therapeutic window of attenuated VPS34 activity that reduces unwanted autophagy but protects essential cellular housekeeping. The promise of VPS34 inhibitors in the clinic will likely rest on their pharmacodynamic and pharmacokinetic capacity to exploit this relatively narrow window.
Outlook
Numerous phosphoinositide kinases are implicated in various haematological, neurological and infectious diseases, along with an assortment of cancers. They have proved amenable to the development of small-molecule inhibitors, portending great potential as bona fide therapeutic targets. Highly potent and selective inhibitors have been developed for most of the PI3K superfamily, along with PIKfyve; these are at the stage of advanced preclinical research or even entering clinical trials (Tables 1–3). Following behind, initial promising tool compounds have been developed against the PI4P5Ks and PI5P4Ks, as well as against the type II PI4Ks (Table 1). Therefore, the scene is set for accelerating preclinical and clinical development, following the paradigm established by the class I PI3Ks (Box 1).
That said, some key challenges remain. Whereas class I PI3Ks are exclusively activated by extracellular stimuli, the rest of the phosphoinositide kinases appear to be constitutively active, facilitating essential cellular ‘housekeeping’ functions. It follows that on-target toxicities are to be anticipated with small-molecule inhibitors; we have discussed specific examples for the PI4Ks133 and class III PI3Ks226,232. Likewise, loss-of-function mutations of several phosphoinositide kinases have been shown to cause human monogenic inherited diseases33,200, heralding potential long-term consequences of enzyme inhibition. Therefore, it seems that the most immediately promising targets are diseases in which acute enzyme inhibition with a course of perhaps a few days can have a therapeutic effect and the best tolerability, for example, the potential of PIKfyve inhibitors in viral infection. Notably, targeting phosphoinositide kinases as pathogen host factors is a promising avenue. Selective targeting of phosphoinositide kinases of the parasitic eukaryotic pathogen also seems potentially promising, as host activity can be completely spared in this context, with the development of antimalarial and anti-cryptosporidiosis PI4K inhibitors as key examples177,178.
Inhibiting many phosphoinositide kinases likely requires exploitation of a therapeutically beneficial window, where pathological function can be disrupted, but sufficient ‘housekeeping’ activity remains to maintain cell functions throughout the body. Although this will undoubtedly prove to be a big challenge, it is worth noting that such challenges were also present when developing inhibitors of the class I PI3Ks (Box 1). For example, the PI3Kα inhibitor alpelisib presents a range of on-target adverse effects but nonetheless has proven clinical benefit to patients with breast cancer and PIK3CA-related overgrowth syndromes234. Therefore, it would be folly to treat such challenges as insurmountable in developing therapeutic applications for phosphoinositide kinase inhibitors. Another lesson from the class I PI3Ks has been the crucial role of isoform-selective inhibitors to minimize on-target adverse effects. This is likely to apply to most of the families discussed here.
Collectively, it is clear that many phosphoinositide kinases are feasible and potentially very useful targets for therapeutics. The next few years of preclinical and clinical development will likely see many successes — and inevitable failures. However, in surveying the state of the field, the potential gains clearly justify ongoing, substantial investment in basic and preclinical research to maximize the eventual success in the clinic.
Acknowledgements
J.E.B. is supported by the Natural Science and Engineering Research Council (Discovery grant NSERC-2020-04241), the Michael Smith Foundation for Health Research (Scholar Award 17686), the Canadian Institute of Health Research (CIHR Project grant 168998) and the Cancer Research Society (operating grant CRS-843232). J.T. is supported by the Swiss National Science Foundation (#310030_207635). B.M.E. receives research funding from NCI (R01 CA237536, CBC under contract no. 75N91019D00024, task order no. 75N91020F00003) and ACS (RSG-20-064-01-TBE, TLC-21-156-01). G.R.V.H. receives funding from NIGMS (R35 GM119412) and NCATS (R03TR003624). The authors are grateful to R. Wills, A. Shaw and C. Weckerly for valuable discussions and suggestions for the manuscript. They apologize to all authors whose work could not be mentioned because of the limitations on the number of references that could be cited.
Glossary
- Acute myeloid leukaemia
(AML). A fast-growing cancer of the blood and bone marrow.
- Alzheimer disease
A progressive brain disorder that affects memory and other cognitive abilities.
- Amyotrophic lateral sclerosis
(ALS). A rare neurological disease that affects neurons responsible for controlling voluntary muscle movement.
- B cell non-Hodgkin lymphoma
(B-NHL). Cancer that arises when B cells, a type of lymphocyte that normally fights infections by producing antibodies to neutralize foreign invaders, start to grow uncontrollably, eventually overwhelming healthy cells.
- Charcot–Marie–Tooth disease
A group of inherited disorders that cause nerve damage. Also referred to as hereditary motor and sensory neuropathy.
- Creutzfeldt–Jacob disease
(CJD). A rapidly progressive degenerative brain disorder that leads to dementia and death.
- Cryptosporidiosis
A diarrhoeal disease caused by a microscopic parasite called Cryptosporidium, with particular prevalence in children and immunocompromised individuals.
- Endocytosis
A cellular process by which cells internalize substances from their external environment.
- Frontotemporal dementia
(FTD). An uncommon type of dementia that is a result of damage to neurons in the frontal and temporal lobes of the brain, which causes problems in behaviour and language.
- Glioblastoma
(GBM). A fast-growing type of central nervous system tumour that forms from the glial tissue of the brain and spinal cord.
- Macroautophagy
A fundamental degradative pathway for cytosolic components, such as proteins, RNA, DNA, lipids and organelles. It involves the sequestering of such cytosolic material in double-membraned structures called autophagosomes, followed by membrane trafficking to the lysosome for degradation and recycling.
- Malaria
A disease transmitted by the bite of infected female mosquitoes caused by Plasmodium parasites (five species infect humans, with two species, Plasmodium vivax and Plasmodium falciparum, posing the largest threat).
- Parkinson disease
A central nervous system disorder which is associated with a deficiency of the neurotransmitter dopamine and affects movement often resulting in tremors.
- Primary lateral sclerosis
(PLS). A rare neuromuscular disease that affects the central motor neurons and is characterized by the gradual weakness and stiffness of muscles.
- p53 null tumours
Tumours that are deficient in either the tumour suppressor TP53 gene or the functional p53 protein product, which induces apoptosis, cell cycle arrest or senescence in response to distinct stimuli.
- TTC7A deficiency
A rare genetic disorder caused by dysfunction of a gene (TTC7A), which causes diarrhoea, inflammation of the intestines, bowel obstructions, immune dysfunction and an inability to absorb nutrients.
- Viral pathogens
Viruses that can infect and replicate within human cells and cause disease.
Competing interests
J.E.B. reports personal fees from Scorpion Therapeutics and Olema Oncology; and research grants from Novartis. J.T., B.M.E. and G.R.V.H. declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
John E. Burke, Email: jeburke@uvic.ca
Brooke M. Emerling, Email: bemerling@sbpdiscovery.org
Gerald R. V. Hammond, Email: ghammond@pitt.edu
References
- 1.Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dickson EJ, Hille B. Understanding phosphoinositides: rare, dynamic, and essential membrane phospholipids. Biochem. J. 2019;476:1–23. doi: 10.1042/BCJ20180022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schink KO, Tan K-W, Stenmark H. Phosphoinositides in control of membrane dynamics. Annu. Rev. Cell Dev. Biol. 2016;32:143–171. doi: 10.1146/annurev-cellbio-111315-125349. [DOI] [PubMed] [Google Scholar]
- 4.Hasegawa J, Strunk BS, Weisman LS. PI5P and PI(3,5)P2: minor, but essential phosphoinositides. Cell Struct. Funct. 2017;42:49–60. doi: 10.1247/csf.17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gozzelino L, De Santis MC, Gulluni F, Hirsch E, Martini M. PI(3,4)P2 signaling in cancer and metabolism. Front. Oncol. 2020;10:360. doi: 10.3389/fonc.2020.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batrouni AG, Baskin JM. The chemistry and biology of phosphatidylinositol 4-phosphate at the plasma membrane. Bioorg. Med. Chem. 2021;40:116190. doi: 10.1016/j.bmc.2021.116190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammond GRV, Burke JE. Novel roles of phosphoinositides in signaling, lipid transport, and disease. Curr. Opin. Cell Biol. 2020;63:57–67. doi: 10.1016/j.ceb.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baba T, Balla T. Emerging roles of phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate as regulators of multiple steps in autophagy. J. Biochem. 2020;168:329–336. doi: 10.1093/jb/mvaa089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanhaesebroeck B, Perry MWD, Brown JR, André F, Okkenhaug K. PI3K inhibitors are finally coming of age. Nat. Rev. Drug Discov. 2021;20:741–769. doi: 10.1038/s41573-021-00209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasan N, Cantley LC. At a crossroads: how to translate the roles of PI3K in oncogenic and metabolic signalling into improvements in cancer therapy. Nat. Rev. Clin. Oncol. 2022;19:471–485. doi: 10.1038/s41571-022-00633-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, et al. PI-273, a substrate-competitive, specific small molecule inhibitor of PI4KIIα, inhibits the growth of breast cancer cells. Cancer Res. 2017;77:6253–6266. doi: 10.1158/0008-5472.CAN-17-0484. [DOI] [PubMed] [Google Scholar]
- 12.Sengupta N, et al. A large scale high-throughput screen identifies chemical inhibitors of phosphatidylinositol 4-kinase type II alpha. J. Lipid Res. 2019;60:683–693. doi: 10.1194/jlr.D090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zolov SN, et al. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. Natl Acad. Sci. USA. 2012;109:17472–17477. doi: 10.1073/pnas.1203106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giridharan SSP, et al. Lipid kinases VPS34 and PIKfyve coordinate a phosphoinositide cascade to regulate retriever-mediated recycling on endosomes. eLife. 2022;11:e69709. doi: 10.7554/eLife.69709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rutherford AC, et al. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J. Cell Sci. 2006;119:3944–3957. doi: 10.1242/jcs.03153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabezas A, Pattni K, Stenmark H. Cloning and subcellular localization of a human phosphatidylinositol 3-phosphate 5-kinase, PIKfyve/Fab1. Gene. 2006;371:34–41. doi: 10.1016/j.gene.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Dong X, et al. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat. Commun. 2010;1:38. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lartigue J, et al. PIKfyve regulation of endosome-linked pathways. Traffic. 2009;10:883–893. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oppelt A, et al. Production of phosphatidylinositol 5-phosphate via PIKfyve and MTMR3 regulates cell migration. EMBO Rep. 2013;14:57–64. doi: 10.1038/embor.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bissig C, Hurbain I, Raposo G, van Niel G. PIKfyve activity regulates reformation of terminal storage lysosomes from endolysosomes. Traffic. 2017;18:747–757. doi: 10.1111/tra.12525. [DOI] [PubMed] [Google Scholar]
- 21.Choy CH, et al. Lysosome enlargement during inhibition of the lipid kinase PIKfyve proceeds through lysosome coalescence. J. Cell Sci. 2018;131:jcs213587. doi: 10.1242/jcs.213587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sbrissa D, Ikonomov OC, Shisheva A. Phosphatidylinositol 3-phosphate-interacting domains in PIKfyve. Binding specificity and role in PIKfyve. Endomenbrane localization. J. Biol. Chem. 2002;277:6073–6079. doi: 10.1074/jbc.M110194200. [DOI] [PubMed] [Google Scholar]
- 23.Shisheva A, Sbrissa D, Ikonomov O. Cloning, characterization, and expression of a novel Zn2+-binding FYVE finger-containing phosphoinositide kinase in insulin-sensitive cells. Mol. Cell Biol. 1999;19:623–634. doi: 10.1128/MCB.19.1.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sbrissa D, Ikonomov OC, Shisheva A. PIKfyve lipid kinase is a protein kinase: downregulation of 5′-phosphoinositide product formation by autophosphorylation. Biochemistry. 2000;39:15980–15989. doi: 10.1021/bi001897f. [DOI] [PubMed] [Google Scholar]
- 25.Ikonomov OC, et al. The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve−/− embryos but normality of PIKfyve+/− mice. J. Biol. Chem. 2011;286:13404–13413. doi: 10.1074/jbc.M111.222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takasuga S, et al. Critical roles of type III phosphatidylinositol phosphate kinase in murine embryonic visceral endoderm and adult intestine. Proc. Natl Acad. Sci. USA. 2013;110:1726–1731. doi: 10.1073/pnas.1213212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin N, et al. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse. EMBO J. 2008;27:3221–3234. doi: 10.1038/emboj.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sbrissa D, et al. A mammalian ortholog of Saccharomyces cerevisiae Vac14 that associates with and up-regulates PIKfyve phosphoinositide 5-kinase activity. Mol. Cell Biol. 2004;24:10437–10447. doi: 10.1128/MCB.24.23.10437-10447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sbrissa D, et al. Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J. Biol. Chem. 2007;282:23878–23891. doi: 10.1074/jbc.M611678200. [DOI] [PubMed] [Google Scholar]
- 30.Duex JE, Tang F, Weisman LS. The Vac14p-Fig4p complex acts independently of Vac7p and couples PI3,5P2 synthesis and turnover. J. Cell Biol. 2006;172:693–704. doi: 10.1083/jcb.200512105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rudge SA, Anderson DM, Emr SD. Vacuole size control: regulation of PtdIns(3,5)P2 levels by the vacuole-associated Vac14-Fig4 complex, a PtdIns(3,5)P2-specific phosphatase. Mol. Biol. Cell. 2004;15:24–36. doi: 10.1091/mbc.e03-05-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc. Natl Acad. Sci. USA. 2007;104:17518–17523. doi: 10.1073/pnas.0702275104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chow CY, et al. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature. 2007;448:68–72. doi: 10.1038/nature05876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenk GM, et al. Pathogenic mechanism of the FIG4 mutation responsible for Charcot-Marie-Tooth disease CMT4J. PLoS Genet. 2011;7:e1002104. doi: 10.1371/journal.pgen.1002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chow CY, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am. J. Hum. Genet. 2009;84:85–88. doi: 10.1016/j.ajhg.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, et al. Mutation of FIG4 causes a rapidly progressive, asymmetric neuronal degeneration. Brain. 2008;131:1990–2001. doi: 10.1093/brain/awn114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lenk GM, et al. Biallelic mutations of VAC14 in pediatric-onset neurological disease. Am. J. Hum. Genet. 2016;99:188–194. doi: 10.1016/j.ajhg.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lees JA, Li P, Kumar N, Weisman LS, Reinisch KM. Insights into lysosomal PI(3,5)P2 homeostasis from a structural-biochemical analysis of the PIKfyve lipid kinase complex. Mol. Cell. 2020;80:736–743.e4. doi: 10.1016/j.molcel.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayakawa M, et al. Synthesis and biological evaluation of 4-morpholino-2-phenylquinazolines and related derivatives as novel PI3 kinase p110alpha inhibitors. Bioorg. Med. Chem. 2006;14:6847–6858. doi: 10.1016/j.bmc.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 40.Jefferies HBJ, et al. A selective PIKfyve inhibitor blocks PtdIns(3,5)P2 production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 2008;9:164–170. doi: 10.1038/sj.embor.7401155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wada Y, et al. Selective abrogation of Th1 response by STA-5326, a potent IL-12/IL-23 inhibitor. Blood. 2007;109:1156–1164. doi: 10.1182/blood-2006-04-019398. [DOI] [PubMed] [Google Scholar]
- 42.Cai X, et al. PIKfyve, a class III PI kinase, is the target of the small molecular IL-12/IL-23 inhibitor apilimod and a player in Toll-like receptor signaling. Chem. Biol. 2013;20:912–921. doi: 10.1016/j.chembiol.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayakawa N, et al. Structure-activity relationship study, target identification, and pharmacological characterization of a small molecular IL-12/23 inhibitor, APY0201. Bioorg. Med. Chem. 2014;22:3021–3029. doi: 10.1016/j.bmc.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 44.Sharma G, et al. A family of PIKFYVE inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupt multiple events in lysosome homeostasis. Autophagy. 2019;15:1694–1718. doi: 10.1080/15548627.2019.1586257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trabbic CJ, et al. Synthesis and biological evaluation of indolyl-pyridinyl-propenones having either methuosis or microtubule disruption activity. J. Med. Chem. 2015;58:2489–2512. doi: 10.1021/jm501997q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho H, et al. Indolyl-pyridinyl-propenone-induced methuosis through the inhibition of PIKFYVE. ACS Omega. 2018;3:6097–6103. doi: 10.1021/acsomega.8b00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hudkins RL, et al. Synthesis and biological profile of the pan-vascular endothelial growth factor receptor/tyrosine kinase with immunoglobulin and epidermal growth factor-like homology domains 2 (VEGF-R/TIE-2) inhibitor 11-(2-methylpropyl)-12,13-dihydro-2-methyl-8-(pyrimidin-2-ylamino)-4H-indazolo[5,4-a]pyrrolo[3,4-c]carbazol-4-one (CEP-11981): a novel oncology therapeutic agent. J. Med. Chem. 2012;55:903–913. doi: 10.1021/jm201449n. [DOI] [PubMed] [Google Scholar]
- 48.Qiao Y, et al. Autophagy inhibition by targeting PIKfyve potentiates response to immune checkpoint blockade in prostate cancer. Nat. Cancer. 2021;2:978–993. doi: 10.1038/s43018-021-00237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guerrero-Valero M, et al. Dysregulation of myelin synthesis and actomyosin function underlies aberrant myelin in CMT4B1 neuropathy. Proc. Natl Acad. Sci. USA. 2021;118:e2009469118. doi: 10.1073/pnas.2009469118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sawade L, et al. Rab35-regulated lipid turnover by myotubularins represses mTORC1 activity and controls myelin growth. Nat. Commun. 2020;11:2835. doi: 10.1038/s41467-020-16696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soto C, Pritzkow S. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci. 2018;21:1332–1340. doi: 10.1038/s41593-018-0235-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soares AC, et al. PIKfyve activity is required for lysosomal trafficking of tau aggregates and tau seeding. J. Biol. Chem. 2021;296:100636. doi: 10.1016/j.jbc.2021.100636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bowles KR, et al. ELAVL4, splicing, and glutamatergic dysfunction precede neuron loss in MAPT mutation cerebral organoids. Cell. 2021;184:4547–4563.e17. doi: 10.1016/j.cell.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.See, S. K. et al. PIKfyve inhibition blocks endolysosomal escape of α-synuclein fibrils and spread of α-synuclein aggregation. Preprint available at bioRxiv10.1101/2021.01.21.427704 (2021).
- 55.Shi Y, et al. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat. Med. 2018;24:313–325. doi: 10.1038/nm.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lenk GM, et al. Cerebral hypomyelination associated with biallelic variants of FIG4. Hum. Mutat. 2019;40:619–630. doi: 10.1002/humu.23720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lakkaraju AKK, et al. Loss of PIKfyve drives the spongiform degeneration in prion diseases. EMBO Mol. Med. 2021;13:e14714. doi: 10.15252/emmm.202114714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carette JE, et al. Ebola virus entry requires the cholesterol transporter Niemann–Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nelson EA, et al. The phosphatidylinositol-3-phosphate 5-kinase inhibitor apilimod blocks filoviral entry and infection. PLoS Negl. Trop. Dis. 2017;11:e0005540. doi: 10.1371/journal.pntd.0005540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiu S, et al. Ebola virus requires phosphatidylinositol (3,5) bisphosphate production for efficient viral entry. Virology. 2018;513:17–28. doi: 10.1016/j.virol.2017.09.028. [DOI] [PubMed] [Google Scholar]
- 61.Kang Y-L, et al. Inhibition of PIKfyve kinase prevents infection by Zaire Ebolavirus and SARS-CoV-2. Proc. Natl Acad. Sci. USA. 2020;117:20803–20813. doi: 10.1073/pnas.2007837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ou X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Riva L, et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bouhaddou M, et al. The global phosphorylation landscape of SARS-CoV-2 Infection. Cell. 2020;182:685–712.e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dayam RM, et al. The lipid kinase PIKfyve coordinates the neutrophil immune response through the activation of the Rac GTPase. J. Immunol. 2017;199:2096–2105. doi: 10.4049/jimmunol.1601466. [DOI] [PubMed] [Google Scholar]
- 66.Gayle S, et al. Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma. Blood. 2017;129:1768–1778. doi: 10.1182/blood-2016-09-736892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Campos CB, et al. Identification of PIKfyve kinase as a target in multiple myeloma. Haematologica. 2020;105:1641–1649. doi: 10.3324/haematol.2019.222729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Milan E, Fabbri M, Cenci S. Autophagy in plasma cell ontogeny and malignancy. J. Clin. Immunol. 2016;36(Suppl. 1):18–24. doi: 10.1007/s10875-016-0254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hou J-Z, et al. Inhibition of PIKfyve using YM201636 suppresses the growth of liver cancer via the induction of autophagy. Oncol. Rep. 2019;41:1971–1979. doi: 10.3892/or.2018.6928. [DOI] [PubMed] [Google Scholar]
- 70.Loijens JC, Boronenkov IV, Parker GJ, Anderson RA. The phosphatidylinositol 4-phosphate 5-kinase family. Adv. Enzym. Regul. 1996;36:115–140. doi: 10.1016/0065-2571(95)00005-4. [DOI] [PubMed] [Google Scholar]
- 71.Rameh LE, Tolias KF, Duckworth BC, Cantley LC. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- 72.Bazenet CE, Ruano AR, Brockman JL, Anderson RA. The human erythrocyte contains two forms of phosphatidylinositol-4-phosphate 5-kinase which are differentially active toward membranes. J. Biol. Chem. 1990;265:18012–18022. doi: 10.1016/S0021-9258(18)38264-4. [DOI] [PubMed] [Google Scholar]
- 73.Ling LE, Schulz JT, Cantley LC. Characterization and purification of membrane-associated phosphatidylinositol-4-phosphate kinase from human red blood cells. J. Biol. Chem. 1989;264:5080–5088. doi: 10.1016/S0021-9258(18)83702-4. [DOI] [PubMed] [Google Scholar]
- 74.Lundquist MR, et al. Phosphatidylinositol-5-phosphate 4-kinases regulate cellular lipid metabolism by facilitating autophagy. Mol. Cell. 2018;70:531–544.e9. doi: 10.1016/j.molcel.2018.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu A, et al. PIP4K2A regulates intracellular cholesterol transport through modulating PI(4,5)P2 homeostasis. J. Lipid Res. 2018;59:507–514. doi: 10.1194/jlr.M082149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishihara H, et al. Cloning of cDNAs encoding two isoforms of 68-kDa type I phosphatidylinositol-4-phosphate 5-kinase. J. Biol. Chem. 1996;271:23611–23614. doi: 10.1074/jbc.271.39.23611. [DOI] [PubMed] [Google Scholar]
- 77.Loijens JC, Anderson RA. Type I phosphatidylinositol-4-phosphate 5-kinases are distinct members of this novel lipid kinase family. J. Biol. Chem. 1996;271:32937–32943. doi: 10.1074/jbc.271.51.32937. [DOI] [PubMed] [Google Scholar]
- 78.Oude Weernink PA, Schmidt M, Jakobs KH. Regulation and cellular roles of phosphoinositide 5-kinases. Eur. J. Pharmacol. 2004;500:87–99. doi: 10.1016/j.ejphar.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 79.van den Bout I, Divecha N. PIP5K-driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J. Cell. Sci. 2009;122:3837–3850. doi: 10.1242/jcs.056127. [DOI] [PubMed] [Google Scholar]
- 80.Rao VD, Misra S, Boronenkov IV, Anderson RA, Hurley JH. Structure of type II beta phosphatidylinositol phosphate kinase: a protein kinase fold flattened for interfacial phosphorylation. Cell. 1998;94:829–839. doi: 10.1016/S0092-8674(00)81741-9. [DOI] [PubMed] [Google Scholar]
- 81.Clarke JH, Irvine RF. The activity, evolution and association of phosphatidylinositol 5-phosphate 4-kinases. Adv. Biol. Regul. 2012;52:40–45. doi: 10.1016/j.advenzreg.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 82.Emerling BM, et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell. 2013;155:844–857. doi: 10.1016/j.cell.2013.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shim H, et al. Deletion of the gene Pip4k2c, a novel phosphatidylinositol kinase, results in hyperactivation of the immune system. Proc. Natl Acad. Sci. USA. 2016;113:7596–7601. doi: 10.1073/pnas.1600934113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lamia KA, et al. Increased insulin sensitivity and reduced adiposity in phosphatidylinositol 5-phosphate 4-kinase beta-/- mice. Mol. Cell Biol. 2004;24:5080–5087. doi: 10.1128/MCB.24.11.5080-5087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Clarke JH, Irvine RF. Evolutionarily conserved structural changes in phosphatidylinositol 5-phosphate 4-kinase (PI5P4K) isoforms are responsible for differences in enzyme activity and localization. Biochem. J. 2013;454:49–57. doi: 10.1042/BJ20130488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bulley SJ, et al. In B cells, phosphatidylinositol 5-phosphate 4-kinase-α synthesizes PI(4,5)P2 to impact mTORC2 and Akt signaling. Proc. Natl Acad. Sci. USA. 2016;113:10571–10576. doi: 10.1073/pnas.1522478113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sumita K, et al. The lipid kinase PI5P4Kβ is an intracellular GTP sensor for metabolism and tumorigenesis. Mol. Cell. 2016;61:187–198. doi: 10.1016/j.molcel.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bultsma Y, Keune W-J, Divecha N. PIP4Kbeta interacts with and modulates nuclear localization of the high-activity PtdIns5P-4-kinase isoform PIP4Kalpha. Biochem. J. 2010;430:223–235. doi: 10.1042/BJ20100341. [DOI] [PubMed] [Google Scholar]
- 89.Jones DR, et al. Nuclear PtdIns5P as a transducer of stress signaling: an in vivo role for PIP4Kbeta. Mol. Cell. 2006;23:685–695. doi: 10.1016/j.molcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 90.Keune W-J, et al. Regulation of phosphatidylinositol-5-phosphate signaling by Pin1 determines sensitivity to oxidative stress. Sci. Signal. 2012;5:ra86. doi: 10.1126/scisignal.2003223. [DOI] [PubMed] [Google Scholar]
- 91.Mackey AM, Sarkes DA, Bettencourt I, Asara JM, Rameh LE. PIP4kγ is a substrate for mTORC1 that maintains basal mTORC1 signaling during starvation. Sci. Signal. 2014;7:ra104–ra104. doi: 10.1126/scisignal.2005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Semenas J, et al. The role of PI3K/AKT-related PIP5K1α and the discovery of its selective inhibitor for treatment of advanced prostate cancer. Proc. Natl Acad. Sci. USA. 2014;111:E3689–E3698. doi: 10.1073/pnas.1405801111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wright BD, et al. The lipid kinase PIP5K1C regulates pain signaling and sensitization. Neuron. 2014;82:836–847. doi: 10.1016/j.neuron.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Andrews DM, et al. Identification and optimization of a novel series of selective PIP5K inhibitors. Bioorg. Med. Chem. 2022;54:116557. doi: 10.1016/j.bmc.2021.116557. [DOI] [PubMed] [Google Scholar]
- 95.Sarwar M, et al. The role of PIP5K1α/pAKT and targeted inhibition of growth of subtypes of breast cancer using PIP5K1α inhibitor. Oncogene. 2019;38:375–389. doi: 10.1038/s41388-018-0438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Adhikari H, Counter CM. Interrogating the protein interactomes of RAS isoforms identifies PIP5K1A as a KRAS-specific vulnerability. Nat. Commun. 2018;9:3646. doi: 10.1038/s41467-018-05692-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Choi S, Chen M, Cryns VL, Anderson RA. A nuclear phosphoinositide kinase complex regulates p53. Nat. Cell Biol. 2019;21:462–475. doi: 10.1038/s41556-019-0297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Muscolini M, et al. Phosphatidylinositol 4-phosphate 5-kinase α activation critically contributes to CD28-dependent signaling responses. J. Immunol. 2013;190:5279–5286. doi: 10.4049/jimmunol.1203157. [DOI] [PubMed] [Google Scholar]
- 99.Muscolini M, et al. Phosphatidylinositol 4-phosphate 5-kinase α and Vav1 mutual cooperation in CD28-mediated actin remodeling and signaling functions. J. Immunol. 2015;194:1323–1333. doi: 10.4049/jimmunol.1401643. [DOI] [PubMed] [Google Scholar]
- 100.Camperio C, et al. CD28 ligation in the absence of TCR stimulation up-regulates IL-17A and pro-inflammatory cytokines in relapsing-remitting multiple sclerosis T lymphocytes. Immunol. Lett. 2014;158:134–142. doi: 10.1016/j.imlet.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 101.Chen S, et al. Pharmacological inhibition of PI5P4Kα/β disrupts cell energy metabolism and selectively kills p53-null tumor cells. Proc. Natl Acad. Sci. USA. 2021;118:e2002486118. doi: 10.1073/pnas.2002486118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ravi A, et al. PI5P4Ks drive metabolic homeostasis through peroxisome-mitochondria interplay. Dev. Cell. 2021;56:1661–1676.e10. doi: 10.1016/j.devcel.2021.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kitagawa M, et al. Dual blockade of the lipid kinase PIP4Ks and mitotic pathways leads to cancer-selective lethality. Nat. Commun. 2017;8:2200. doi: 10.1038/s41467-017-02287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sivakumaren SC, et al. Targeting the PI5P4K lipid kinase family in cancer using covalent inhibitors. Cell Chem. Biol. 2020;27:525–537.e6. doi: 10.1016/j.chembiol.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Manz TD, et al. Discovery and structure-activity relationship study of (Z)-5-methylenethiazolidin-4-one derivatives as potent and selective Pan-phosphatidylinositol 5-phosphate 4-kinase inhibitors. J. Med. Chem. 2020;63:4880–4895. doi: 10.1021/acs.jmedchem.0c00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Davis MI, et al. A homogeneous, high-throughput assay for phosphatidylinositol 5-phosphate 4-kinase with a novel, rapid substrate preparation. PLoS ONE. 2013;8:e54127. doi: 10.1371/journal.pone.0054127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wortmann L, et al. Discovery and characterization of the potent and highly selective 1,7-naphthyridine-based inhibitors BAY-091 and BAY-297 of the kinase PIP4K2A. J. Med. Chem. 2021;64:15883–15911. doi: 10.1021/acs.jmedchem.1c01245. [DOI] [PubMed] [Google Scholar]
- 108.Voss MD, et al. Discovery and pharmacological characterization of a novel small molecule inhibitor of phosphatidylinositol-5-phosphate 4-kinase, type II, beta. Biochem. Biophys. Res. Commun. 2014;449:327–331. doi: 10.1016/j.bbrc.2014.05.024. [DOI] [PubMed] [Google Scholar]
- 109.Clarke JH, et al. The function of phosphatidylinositol 5-phosphate 4-kinase γ (PI5P4Kγ) explored using a specific inhibitor that targets the PI5P-binding site. Biochem. J. 2015;466:359–367. doi: 10.1042/BJ20141333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Al-Ramahi I, et al. Inhibition of PIP4Kγ ameliorates the pathological effects of mutant huntingtin protein. eLife. 2017;6:e29123. doi: 10.7554/eLife.29123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Manz TD, et al. Structure-activity relationship study of covalent Pan-phosphatidylinositol 5-phosphate 4-kinase inhibitors. ACS Med. Chem. Lett. 2020;11:346–352. doi: 10.1021/acsmedchemlett.9b00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.See CS, et al. Discovery of the cancer cell selective dual acting anti-cancer agent (Z)-2-(1H-indol-3-yl)-3-(isoquinolin-5-yl)acrylonitrile (A131) Eur. J. Med. Chem. 2018;156:344–367. doi: 10.1016/j.ejmech.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 113.Boffey HK, et al. Development of selective phosphatidylinositol 5-phosphate 4-kinase γ inhibitors with a non-ATP-competitive, allosteric binding mode. J. Med. Chem. 2022;65:3359–3370. doi: 10.1021/acs.jmedchem.1c01819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Arora GK, Palamiuc L, Emerling BM. Expanding role of PI5P4Ks in cancer: a promising druggable target. FEBS Lett. 2022;596:3–16. doi: 10.1002/1873-3468.14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Luoh S-W, Venkatesan N, Tripathi R. Overexpression of the amplified Pip4k2beta gene from 17q11-12 in breast cancer cells confers proliferation advantage. Oncogene. 2004;23:1354–1363. doi: 10.1038/sj.onc.1207251. [DOI] [PubMed] [Google Scholar]
- 116.Keune W-J, et al. Low PIP4K2B expression in human breast tumors correlates with reduced patient survival: a role for PIP4K2B in the regulation of E-cadherin expression. Cancer Res. 2013;73:6913–6925. doi: 10.1158/0008-5472.CAN-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lima K, et al. PIP4K2A and PIP4K2C transcript levels are associated with cytogenetic risk and survival outcomes in acute myeloid leukemia. Cancer Genet. 2019;233–234:56–66. doi: 10.1016/j.cancergen.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 118.Jude JG, et al. A targeted knockdown screen of genes coding for phosphoinositide modulators identifies PIP4K2A as required for acute myeloid leukemia cell proliferation and survival. Oncogene. 2015;34:1253–1262. doi: 10.1038/onc.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shin YJ, et al. PIP4K2A as a negative regulator of PI3K in PTEN-deficient glioblastoma. J. Exp. Med. 2019;216:1120–1134. doi: 10.1084/jem.20172170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang DG, et al. PIP4Ks suppress insulin signaling through a catalytic-independent mechanism. Cell Rep. 2019;27:1991–2001.e5. doi: 10.1016/j.celrep.2019.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fagerberg L, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteom. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Poli A, et al. PIP4Ks impact on PI3K, FOXP3, and UHRF1 signaling and modulate human regulatory T cell proliferation and immunosuppressive activity. Proc. Natl Acad. Sci. USA. 2021;118:e2010053118. doi: 10.1073/pnas.2010053118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.He Z, et al. The PIP5K2A gene and schizophrenia in the Chinese population-a case-control study. Schizophr. Res. 2007;94:359–365. doi: 10.1016/j.schres.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 124.Thiselton DL, et al. Association analysis of the PIP4K2A gene on chromosome 10p12 and schizophrenia in the Irish study of high density schizophrenia families (ISHDSF) and the Irish case-control study of schizophrenia (ICCSS) Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153B:323–331. doi: 10.1002/ajmg.b.30982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schwab SG, et al. Evidence for association of DNA sequence variants in the phosphatidylinositol-4-phosphate 5-kinase IIalpha gene (PIP5K2A) with schizophrenia. Mol. Psychiatry. 2006;11:837–846. doi: 10.1038/sj.mp.4001864. [DOI] [PubMed] [Google Scholar]
- 126.Noch EK, Yim I, Milner TA, Cantley LC. Distribution and localization of phosphatidylinositol 5-phosphate, 4-kinase alpha and beta in the brain. J. Comp. Neurol. 2021;529:434–449. doi: 10.1002/cne.24956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Balla T, et al. Isolation and molecular cloning of wortmannin-sensitive bovine type III phosphatidylinositol 4-kinases. J. Biol. Chem. 1997;272:18358–18366. doi: 10.1074/jbc.272.29.18358. [DOI] [PubMed] [Google Scholar]
- 128.Boura E, Nencka R. Phosphatidylinositol 4-kinases: function, structure, and inhibition. Exp. Cell Res. 2015;337:136–145. doi: 10.1016/j.yexcr.2015.03.028. [DOI] [PubMed] [Google Scholar]
- 129.Dornan GL, McPhail JA, Burke JE. Type III phosphatidylinositol 4 kinases: structure, function, regulation, signalling and involvement in disease. Biochemical Soc. Trans. 2016;44:260–266. doi: 10.1042/BST20150219. [DOI] [PubMed] [Google Scholar]
- 130.Tan J, Brill JA. Cinderella story: PI4P goes from precursor to key signaling molecule. Crit. Rev. Biochem. Mol. Biol. 2014;49:33–58. doi: 10.3109/10409238.2013.853024. [DOI] [PubMed] [Google Scholar]
- 131.Burke JE. Structural basis for regulation of phosphoinositide kinases and their involvement in human disease. Mol. Cell. 2018;71:653–673. doi: 10.1016/j.molcel.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 132.Nakatsu F, et al. PtdIns4P synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol. 2012;199:1003–1016. doi: 10.1083/jcb.201206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bojjireddy N, et al. Pharmacological and genetic targeting of the PI4KA enzyme reveals its important role in maintaining plasma membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate levels. J. Biol. Chem. 2014;289:6120–6132. doi: 10.1074/jbc.M113.531426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nakagawa T, Goto K, Kondo H. Cloning, expression, and localization of 230-kDa phosphatidylinositol 4-kinase. J. Biol. Chem. 1996;271:12088–12094. doi: 10.1074/jbc.271.20.12088. [DOI] [PubMed] [Google Scholar]
- 135.Lees JA, et al. Architecture of the human PI4KIIIα lipid kinase complex. Proc. Natl Acad. Sci. USA. 2017;114:13720–13725. doi: 10.1073/pnas.1718471115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wu X, et al. Structural insights into assembly and regulation of the plasma membrane phosphatidylinositol 4-kinase complex. Dev. Cell. 2014;28:19–29. doi: 10.1016/j.devcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Baskin JM, et al. The leukodystrophy protein FAM126A (hyccin) regulates PtdIns(4)P synthesis at the plasma membrane. Nat. Cell Biol. 2016;18:132–138. doi: 10.1038/ncb3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Raghu P, Joseph A, Krishnan H, Singh P, Saha S. Phosphoinositides: regulators of nervous system function in health and disease. Front. Mol. Neurosci. 2019;12:208. doi: 10.3389/fnmol.2019.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pagnamenta AT, et al. Germline recessive mutations in PI4KA are associated with perisylvian polymicrogyria, cerebellar hypoplasia and arthrogryposis. Hum. Mol. Genet. 2015;24:3732–3741. doi: 10.1093/hmg/ddv117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Salter CG, et al. Biallelic PI4KA variants cause neurological, intestinal and immunological disease. Brain. 2021;144:3597–3610. doi: 10.1093/brain/awab313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bigorgne AE, et al. TTC7A mutations disrupt intestinal epithelial apicobasal polarity. J. Clin. Invest. 2014;124:328–337. doi: 10.1172/JCI71471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Avitzur Y, et al. Mutations in tetratricopeptide repeat domain 7A result in a severe form of very early onset inflammatory bowel disease. Gastroenterology. 2014;146:1028–1039. doi: 10.1053/j.gastro.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen R, et al. Whole-exome sequencing identifies tetratricopeptide repeat domain 7A (TTC7A) mutations for combined immunodeficiency with intestinal atresias. J. Allergy Clin. Immunol. 2013;132:656–664.e17. doi: 10.1016/j.jaci.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Berger KL, et al. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc. Natl Acad. Sci. USA. 2009;106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dorobantu CM, et al. Modulation of the host lipid landscape to promote RNA virus replication: the picornavirus encephalomyocarditis virus converges on the pathway used by hepatitis C virus. PLoS Pathog. 2015;11:e1005185. doi: 10.1371/journal.ppat.1005185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Burke JE, et al. Structures of PI4KIIIβ complexes show simultaneous recruitment of Rab11 and its effectors. Science. 2014;344:1035–1038. doi: 10.1126/science.1253397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McPhail JA, Ottosen EH, Jenkins ML, Burke JE. The molecular basis of Aichi virus 3A protein activation of phosphatidylinositol 4 kinase IIIβ, PI4KB, through ACBD3. Structure. 2017;25:121–131. doi: 10.1016/j.str.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 148.Klima M, et al. Structural insights and in vitro reconstitution of membrane targeting and activation of human PI4KB by the ACBD3 protein. Sci. Rep. 2016;6:23641. doi: 10.1038/srep23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hausser A, et al. Protein kinase D regulates vesicular transport by phosphorylating and activating phosphatidylinositol-4 kinase IIIbeta at the Golgi complex. Nat. Cell Biol. 2005;7:880–886. doi: 10.1038/ncb1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chalupská D, et al. Phosphatidylinositol 4-kinase IIIβ (PI4KB) forms highly flexible heterocomplexes that include ACBD3, 14-3-3, and Rab11 proteins. Sci. Rep. 2019;9:567. doi: 10.1038/s41598-018-37158-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.McPhail JA, et al. Characterization of the c10orf76-PI4KB complex and its necessity for Golgi PI4P levels and enterovirus replication. EMBO Rep. 2020;21:e48441. doi: 10.15252/embr.201948441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mesmin B, et al. Sterol transfer, PI4P consumption, and control of membrane lipid order by endogenous OSBP. EMBO J. 2017;36:3156–3174. doi: 10.15252/embj.201796687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Brill JA, Hime GR, Scharer-Schuksz M, Fuller MT. A phospholipid kinase regulates actin organization and intercellular bridge formation during germline cytokinesis. Development. 2000;127:3855–3864. doi: 10.1242/dev.127.17.3855. [DOI] [PubMed] [Google Scholar]
- 154.Baba T, et al. Myelination of peripheral nerves is controlled by PI4KB through regulation of Schwann cell Golgi function. Proc. Natl Acad. Sci. USA. 2020;117:28102–28113. doi: 10.1073/pnas.2007432117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.van der Schaar HM, Dorobantu CM, Albulescu L, Strating JRPM, van Kuppeveld FJM. Fat(al) attraction: picornaviruses usurp lipid transfer at membrane contact sites to create replication organelles. Trends Microbiol. 2016;24:535–546. doi: 10.1016/j.tim.2016.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Altan-Bonnet N, Balla T. Phosphatidylinositol 4-kinases: hostages harnessed to build panviral replication platforms. Trends Biochem. Sci. 2012;37:293–302. doi: 10.1016/j.tibs.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hsu N-Y, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nakanishi S, Catt KJ, Balla T. A wortmannin-sensitive phosphatidylinositol 4-kinase that regulates hormone-sensitive pools of inositolphospholipids. Proc. Natl Acad. Sci. USA. 1995;92:5317–5321. doi: 10.1073/pnas.92.12.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nakanishi S, Catt KJ, Balla T. A wortmannin-sensitive phosphatidylinositol 4-kinase that regulates hormone-sensitive pools of inositolphospholipids. Proc. Natl Acad. Sci. USA. 1995;92:5317–5321. doi: 10.1073/pnas.92.12.5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Meyers R, Cantley LC. Cloning and characterization of a wortmannin-sensitive human phosphatidylinositol 4-kinase. J. Biol. Chem. 1997;272:4384–4390. doi: 10.1074/jbc.272.7.4384. [DOI] [PubMed] [Google Scholar]
- 161.Bianco A, et al. Metabolism of phosphatidylinositol 4-kinase IIIα-dependent PI4P Is subverted by HCV and is targeted by a 4-anilino quinazoline with antiviral activity. PLoS Pathog. 2012;8:e1002576. doi: 10.1371/journal.ppat.1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Vaillancourt FH, et al. Evaluation of phosphatidylinositol-4-kinase IIIα as a hepatitis C virus drug target. J. Virol. 2012;86:11595–11607. doi: 10.1128/JVI.01320-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Leivers AL, et al. Discovery of selective small molecule type III phosphatidylinositol 4-kinase alpha (PI4KIIIα) inhibitors as anti hepatitis C (HCV) agents. J. Med. Chem. 2014;57:2091–2106. doi: 10.1021/jm400781h. [DOI] [PubMed] [Google Scholar]
- 164.Raubo P, et al. Discovery of potent, selective small molecule inhibitors of α-subtype of type III phosphatidylinositol-4-kinase (PI4KIIIα) Bioorg. Med. Chem. Lett. 2015;25:3189–3193. doi: 10.1016/j.bmcl.2015.05.093. [DOI] [PubMed] [Google Scholar]
- 165.Waring MJ, et al. Potent, selective small molecule inhibitors of type III phosphatidylinositol-4-kinase α- but not β-inhibit the phosphatidylinositol signaling cascade and cancer cell proliferation. Chem. Commun. 2014;50:5388–5390. doi: 10.1039/C3CC48391F. [DOI] [PubMed] [Google Scholar]
- 166.Knight Z, et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rutaganira FU, et al. Design and structural characterization of potent and selective inhibitors of phosphatidylinositol 4 kinase IIIβ. J. Med. Chem. 2016;59:1830–1839. doi: 10.1021/acs.jmedchem.5b01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Phillpotts RJ, Wallace J, Tyrrell DA, Tagart VB. Therapeutic activity of enviroxime against rhinovirus infection in volunteers. Antimicrob. Agents Chemother. 1983;23:671–675. doi: 10.1128/AAC.23.5.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Arita M, et al. Phosphatidylinositol 4-kinase III beta is a target of enviroxime-like compounds for antipoliovirus activity. J. Virol. 2011;85:2364–2372. doi: 10.1128/JVI.02249-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.van der Schaar HM, et al. A novel, broad-spectrum inhibitor of enterovirus replication that targets host cell factor phosphatidylinositol 4-kinase IIIβ. Antimicrob. Agents Chemother. 2013;57:4971–4981. doi: 10.1128/AAC.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mejdrová I, et al. Highly selective phosphatidylinositol 4-Kinase IIIβ inhibitors and structural insight into their mode of action. J. Med. Chem. 2015;58:3767–3793. doi: 10.1021/acs.jmedchem.5b00499. [DOI] [PubMed] [Google Scholar]
- 172.Mejdrová I, et al. Rational design of novel highly potent and selective phosphatidylinositol 4-Kinase IIIβ (PI4KB) inhibitors as broad-spectrum antiviral agents and tools for chemical biology. J. Med. Chem. 2017;60:100–118. doi: 10.1021/acs.jmedchem.6b01465. [DOI] [PubMed] [Google Scholar]
- 173.McPhail JA, Burke JE. Molecular mechanisms of PI4K regulation and their involvement in viral replication. Traffic. 2022 doi: 10.1111/tra.12841. [DOI] [PubMed] [Google Scholar]
- 174.Lamarche MJ, et al. Anti-hepatitis C virus activity and toxicity of type III phosphatidylinositol-4-kinase beta inhibitors. Antimicrob. Agents Chemother. 2012;56:5149–5156. doi: 10.1128/AAC.00946-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Reuberson J, et al. Discovery of a potent, orally bioavailable PI4KIIIβ inhibitor (UCB9608) able to significantly prolong allogeneic organ engraftment in vivo. J. Med. Chem. 2018;61:6705–6723. doi: 10.1021/acs.jmedchem.8b00521. [DOI] [PubMed] [Google Scholar]
- 176.Spickler C, et al. Phosphatidylinositol 4-kinase III beta is essential for replication of human rhinovirus and its inhibition causes a lethal phenotype in vivo. Antimicrob. Agents Chemother. 2013;57:3358–3368. doi: 10.1128/AAC.00303-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.McNamara CW, et al. Targeting Plasmodium PI(4)K to eliminate malaria. Nature. 2013;504:248–253. doi: 10.1038/nature12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Manjunatha UH, et al. A Cryptosporidium PI(4)K inhibitor is a drug candidate for cryptosporidiosis. Nature. 2017;546:376–380. doi: 10.1038/nature22337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ghidelli-Disse S, et al. Identification of Plasmodium PI4 kinase as target of MMV390048 by chemoproteomics. Malar. J. 2014;13:P38. doi: 10.1186/1475-2875-13-S1-P38. [DOI] [Google Scholar]
- 180.Kato N, et al. Diversity-oriented synthesis yields novel multistage antimalarial inhibitors. Nature. 2016;538:344–349. doi: 10.1038/nature19804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Paquet T, et al. Antimalarial efficacy of MMV390048, an inhibitor of Plasmodium phosphatidylinositol 4-kinase. Sci. Transl. Med. 2017;9:eaad9735. doi: 10.1126/scitranslmed.aad9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fienberg S, et al. Structural basis for inhibitor potency and selectivity of Plasmodium falciparum phosphatidylinositol 4-kinase inhibitors. ACS Infect. Dis. 2020;6:3048–3063. doi: 10.1021/acsinfecdis.0c00566. [DOI] [PubMed] [Google Scholar]
- 183.Arendse LB, Wyllie S, Chibale K, Gilbert IH. Plasmodium kinases as potential drug targets for malaria: challenges and opportunities. ACS Infect. Dis. 2021;7:518–534. doi: 10.1021/acsinfecdis.0c00724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Brunschwig C, et al. UCT943, a next-generation Plasmodium falciparum PI4K inhibitor preclinical candidate for the treatment of malaria. Antimicrob. Agents Chemother. 2018;62:62. doi: 10.1128/AAC.00012-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Adhikari H, et al. Oncogenic KRAS is dependent upon an EFR3A-PI4KA signaling axis for potent tumorigenic activity. Nat. Commun. 2021;12:5248. doi: 10.1038/s41467-021-25523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kattan WE, et al. Components of the phosphatidylserine endoplasmic reticulum to plasma membrane transport mechanism as targets for KRAS inhibition in pancreatic cancer. Proc. Natl Acad. Sci. USA. 2021;118:e2114126118. doi: 10.1073/pnas.2114126118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shi L, et al. Addiction to Golgi-resident PI4P synthesis in chromosome 1q21.3-amplified lung adenocarcinoma cells. Proc. Natl Acad. Sci. USA. 2021;118:e2023537118. doi: 10.1073/pnas.2023537118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tan X, et al. PI4KIIIβ is a therapeutic target in chromosome 1q-amplified lung adenocarcinoma. Sci. Transl. Med. 2020;12:eaax3772. doi: 10.1126/scitranslmed.aax3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gupta S, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 190.Gulluni F, De Santis MC, Margaria JP, Martini M, Hirsch E. Class II PI3K functions in cell biology and disease. Trends Cell Biol. 2019;29:339–359. doi: 10.1016/j.tcb.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 191.Lo W-T, et al. Structural basis of phosphatidylinositol 3-kinase C2α function. Nat. Struct. Mol. Biol. 2022;29:218–228. doi: 10.1038/s41594-022-00730-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Gaidarov I, Smith ME, Domin J, Keen JH. The class II phosphoinositide 3-kinase C2alpha is activated by clathrin and regulates clathrin-mediated membrane trafficking. Mol. Cell. 2001;7:443–449. doi: 10.1016/S1097-2765(01)00191-5. [DOI] [PubMed] [Google Scholar]
- 193.Wang H, et al. Autoregulation of class II Alpha PI3K activity by its lipid-binding PX-C2 domain module. Mol. Cell. 2018;71:343–351.e4. doi: 10.1016/j.molcel.2018.06.042. [DOI] [PubMed] [Google Scholar]
- 194.Chen K-E, Tillu VA, Chandra M, Collins BM. Molecular basis for membrane recruitment by the PX and C2 domains of class II phosphoinositide 3-kinase-C2α. Structure. 2018;26:1612–1625.e4. doi: 10.1016/j.str.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 195.Posor Y, et al. Spatiotemporal control of endocytosis by phosphatidylinositol-3, 4-bisphosphate. Nature. 2013;499:233–237. doi: 10.1038/nature12360. [DOI] [PubMed] [Google Scholar]
- 196.Yoshioka K, et al. Endothelial PI3K-C2α, a class II PI3K, has an essential role in angiogenesis and vascular barrier function. Nat. Med. 2012;18:1560–1569. doi: 10.1038/nm.2928. [DOI] [PubMed] [Google Scholar]
- 197.Franco I, et al. PI3K class II α controls spatially restricted endosomal PtdIns3P and Rab11 activation to promote primary cilium function. Dev. Cell. 2014;28:647–658. doi: 10.1016/j.devcel.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Alliouachene S, et al. Inactivation of class II PI3K-C2α induces leptin resistance, age-dependent insulin resistance and obesity in male mice. Diabetologia. 2016;59:1503–1512. doi: 10.1007/s00125-016-3963-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Boukhalfa A, et al. PI3KC2α-dependent and VPS34-independent generation of PI3P controls primary cilium-mediated autophagy in response to shear stress. Nat. Commun. 2020;11:294. doi: 10.1038/s41467-019-14086-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Tiosano D, et al. Mutations in PIK3C2A cause syndromic short stature, skeletal abnormalities, and cataracts associated with ciliary dysfunction. PLoS Genet. 2019;15:e1008088. doi: 10.1371/journal.pgen.1008088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gulluni F, et al. PI(3,4)P2-mediated cytokinetic abscission prevents early senescence and cataract formation. Science. 2021;374:eabk0410. doi: 10.1126/science.abk0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Alliouachene S, et al. Inactivation of the class II PI3K-C2β potentiates insulin signaling and sensitivity. Cell Rep. 2015;13:1881–1894. doi: 10.1016/j.celrep.2015.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Anquetil T, et al. PI3KC2β inactivation stabilizes VE-cadherin junctions and preserves vascular integrity. EMBO Rep. 2021;22:e51299. doi: 10.15252/embr.202051299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Posor Y, et al. Local synthesis of the phosphatidylinositol-3,4-bisphosphate lipid drives focal adhesion turnover. Dev. Cell. 2022;57:1694–1711.e7. doi: 10.1016/j.devcel.2022.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sabha N, et al. PIK3C2B inhibition improves function and prolongs survival in myotubular myopathy animal models. J. Clin. Invest. 2016;126:3613–3625. doi: 10.1172/JCI86841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Marat AL, et al. mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science. 2017;356:968–972. doi: 10.1126/science.aaf8310. [DOI] [PubMed] [Google Scholar]
- 207.Gozzelino L, et al. Defective lipid signalling caused by mutations in PIK3C2B underlies focal epilepsy. Brain. 2022;145:2313–2331. doi: 10.1093/brain/awac082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Domin J, et al. Cloning of a human phosphoinositide 3-kinase with a C2 domain that displays reduced sensitivity to the inhibitor wortmannin. Biochem. J. 1997;326:139–147. doi: 10.1042/bj3260139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Selvadurai MV, et al. Disrupting the platelet internal membrane via PI3KC2α inhibition impairs thrombosis independently of canonical platelet activation. Sci. Transl. Med. 2020;12:eaar8430. doi: 10.1126/scitranslmed.aar8430. [DOI] [PubMed] [Google Scholar]
- 210.Li H, et al. Phosphoinositide conversion inactivates R-RAS and drives metastases in breast cancer. Adv. Sci. 2022;9:e2103249. doi: 10.1002/advs.202103249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lo W-T, et al. Development of selective inhibitors of phosphatidylinositol 3-kinase C2α. Nat. Chem. Biol. 2022 doi: 10.1038/s41589-022-01118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mountford JK, et al. The class II PI 3-kinase, PI3KC2α, links platelet internal membrane structure to shear-dependent adhesive function. Nat. Commun. 2015;6:6535. doi: 10.1038/ncomms7535. [DOI] [PubMed] [Google Scholar]
- 213.Valet C, et al. Essential role of class II PI3K-C2α in platelet membrane morphology. Blood. 2015;126:1128–1137. doi: 10.1182/blood-2015-03-636670. [DOI] [PubMed] [Google Scholar]
- 214.Gulluni F, et al. Mitotic spindle assembly and genomic stability in breast cancer require PI3K-C2α scaffolding function. Cancer Cell. 2017;32:444–459.e7. doi: 10.1016/j.ccell.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 215.Ryan EL, Shelford J, Massam-Wu T, Bayliss R, Royle SJ. Defining endogenous TACC3-chTOG-clathrin-GTSE1 interactions at the mitotic spindle using induced relocalization. J. Cell Sci. 2021;134:jcs255794. doi: 10.1242/jcs.255794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ohashi Y, Tremel S, Williams RL. VPS34 complexes from a structural perspective. J. Lipid Res. 2019;60:229–241. doi: 10.1194/jlr.R089490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Rostislavleva K, et al. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 2015;350:aac7365. doi: 10.1126/science.aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Stjepanovic G, Baskaran S, Lin MG, Hurley JH. Vps34 kinase domain dynamics regulate the autophagic PI 3-kinase complex. Mol. Cell. 2017;67:528–534.e3. doi: 10.1016/j.molcel.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Young LN, Goerdeler F, Hurley JH. Structural pathway for allosteric activation of the autophagic PI 3-kinase complex I. Proc. Natl Acad. Sci. USA. 2019;116:21508–21513. doi: 10.1073/pnas.1911612116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Tremel S, et al. Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes. Nat. Commun. 2021;12:1564. doi: 10.1038/s41467-021-21695-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3’-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 222.Bago R, et al. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem. J. 2014;463:413–427. doi: 10.1042/BJ20140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Dowdle WE, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 2014;16:1069–1079. doi: 10.1038/ncb3053. [DOI] [PubMed] [Google Scholar]
- 224.Ronan B, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 2014;10:1013–1019. doi: 10.1038/nchembio.1681. [DOI] [PubMed] [Google Scholar]
- 225.Dyczynski M, et al. Targeting autophagy by small molecule inhibitors of vacuolar protein sorting 34 (Vps34) improves the sensitivity of breast cancer cells to Sunitinib. Cancer Lett. 2018;435:32–43. doi: 10.1016/j.canlet.2018.07.028. [DOI] [PubMed] [Google Scholar]
- 226.Hu DX, et al. Structure-based design of potent, selective, and orally bioavailable VPS34 kinase inhibitors. J. Med. Chem. 2022;65:11500–11512. doi: 10.1021/acs.jmedchem.1c01180. [DOI] [PubMed] [Google Scholar]
- 227.Klionsky DJ, et al. Autophagy in major human diseases. EMBO J. 2021;40:e108863. doi: 10.15252/embj.2021108863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Liu X, et al. Simultaneous inhibition of Vps34 kinase would enhance PI3Kδ inhibitor cytotoxicity in the B-cell malignancies. Oncotarget. 2016;7:53515–53525. doi: 10.18632/oncotarget.10650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Meunier G, et al. Antileukemic activity of the VPS34-IN1 inhibitor in acute myeloid leukemia. Oncogenesis. 2020;9:94. doi: 10.1038/s41389-020-00278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Noman MZ, et al. Inhibition of Vps34 reprograms cold into hot inflamed tumors and improves anti–PD-1/PD-L1 immunotherapy. Sci. Adv. 2020;6:eaax7881. doi: 10.1126/sciadv.aax7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Schlütermann D, et al. Targeting urothelial carcinoma cells by combining cisplatin with a specific inhibitor of the autophagy-inducing class III PtdIns3K complex. Urol. Oncol. 2018;36:160.e1–160.e13. doi: 10.1016/j.urolonc.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 232.Henley ZA, et al. Optimization of orally bioavailable PI3Kδ inhibitors and identification of Vps34 as a key selectivity target. J. Med. Chem. 2020;63:638–655. doi: 10.1021/acs.jmedchem.9b01585. [DOI] [PubMed] [Google Scholar]
- 233.Bilanges B, et al. Vps34 PI 3-kinase inactivation enhances insulin sensitivity through reprogramming of mitochondrial metabolism. Nat. Commun. 2017;8:1804. doi: 10.1038/s41467-017-01969-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.André F, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N. Engl. J. Med. 2019;380:1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 235.Varadi M, et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ikonomov OC, Sbrissa D, Fenner H, Shisheva A. PIKfyve-ArPIKfyve-Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis. J. Biol. Chem. 2009;284:35794–35806. doi: 10.1074/jbc.M109.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Botelho RJ, Efe JA, Teis D, Emr SD. Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase. Mol. Biol. Cell. 2008;19:4273–4286. doi: 10.1091/mbc.e08-04-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Hansen SD, Lee AA, Duewell BR, Groves JT. Membrane-mediated dimerization potentiates PIP5K lipid kinase activity. eLife. 2022;11:e73747. doi: 10.7554/eLife.73747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kunz J, et al. The activation loop of phosphatidylinositol phosphate kinases determines signaling specificity. Mol. Cell. 2000;5:1–11. doi: 10.1016/S1097-2765(00)80398-6. [DOI] [PubMed] [Google Scholar]
- 240.Kunz J, Fuelling A, Kolbe L, Anderson RA. Stereo-specific substrate recognition by phosphatidylinositol phosphate kinases is swapped by changing a single amino acid residue. J. Biol. Chem. 2002;277:5611–5619. doi: 10.1074/jbc.M110775200. [DOI] [PubMed] [Google Scholar]
- 241.Hu J, et al. Resolution of structure of PIP5K1A reveals molecular mechanism for its regulation by dimerization and dishevelled. Nat. Commun. 2015;6:8205. doi: 10.1038/ncomms9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Wills RC, et al. A novel homeostatic mechanism tunes PI(4,5)P 2-dependent signaling at the plasma membrane. bioRxiv. 2022 doi: 10.1101/2022.06.30.498262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Fruman DA, et al. The PI3K pathway in human disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Rathinaswamy MK, et al. Structure of the phosphoinositide 3-kinase (PI3K) p110γ-p101 complex reveals molecular mechanism of GPCR activation. Sci. Adv. 2021;7:eabj4282. doi: 10.1126/sciadv.abj4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Vadas O, Burke JE, Zhang X, Berndt A, Williams RL. Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci. Signal. 2011;4:1–13. doi: 10.1126/scisignal.2002165. [DOI] [PubMed] [Google Scholar]
- 247.Burke JE, Williams RL. Synergy in activating class I PI3Ks. Trends Biochem. Sci. 2015;40:88–100. doi: 10.1016/j.tibs.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 248.Dornan GL, Burke JE. Molecular mechanisms of human disease mediated by oncogenic and primary immunodeficiency mutations in class IA phosphoinositide 3-kinases. Front. Immunol. 2018;9:575. doi: 10.3389/fimmu.2018.00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Dyment DA, et al. Mutations in PIK3R1 cause SHORT syndrome. Am. J. Hum. Genet. 2013;93:158–166. doi: 10.1016/j.ajhg.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Thauvin-Robinet C, et al. PIK3R1 mutations cause syndromic insulin resistance with lipoatrophy. Am. J. Hum. Genet. 2013;93:141–149. doi: 10.1016/j.ajhg.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Chudasama KK, et al. SHORT syndrome with partial lipodystrophy due to impaired phosphatidylinositol 3 kinase signaling. Am. J. Hum. Genet. 2013;93:150–157. doi: 10.1016/j.ajhg.2013.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Thian M, et al. Germline biallelic PIK3CG mutations in a multifaceted immunodeficiency with immune dysregulation. Haematologica. 2020;105:e488. doi: 10.3324/haematol.2019.231399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Takeda AJ, et al. Human PI3Kγ deficiency and its microbiota-dependent mouse model reveal immunodeficiency and tissue immunopathology. Nat. Commun. 2019;10:4364. doi: 10.1038/s41467-019-12311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Samuels Y, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 255.Samuels Y, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 256.Urick ME, et al. PIK3R1 (p85α) is somatically mutated at high frequency in primary endometrial cancer. Cancer Res. 2011;71:4061–4067. doi: 10.1158/0008-5472.CAN-11-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Jaiswal BS, et al. Somatic mutations in p85α promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16:463–474. doi: 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Madsen RR, et al. Oncogenic PIK3CA promotes cellular stemness in an allele dose-dependent manner. Proc. Natl Acad. Sci. USA. 2019;116:8380–8389. doi: 10.1073/pnas.1821093116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Vasan N, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors. Science. 2019;366:714–723. doi: 10.1126/science.aaw9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Castillo SD, et al. Somatic activating mutations in Pik3ca cause sporadic venous malformations in mice and humans. Sci. Transl. Med. 2016;8:332ra43. doi: 10.1126/scitranslmed.aad9982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lindhurst MJ, et al. Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nat. Genet. 2012;44:928–933. doi: 10.1038/ng.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Jia S, et al. Essential roles of PI(3)K-p110beta in cell growth, metabolism and tumorigenesis. Nature. 2008;454:776–779. doi: 10.1038/nature07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wee S, et al. PTEN-deficient cancers depend on PIK3CB. Proc. Natl Acad. Sci. USA. 2008;105:13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Angulo I, et al. Phosphoinositide 3-kinase δ gene mutation predisposes to respiratory infection and airway damage. Science. 2013;342:866–871. doi: 10.1126/science.1243292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Lucas CL, et al. Dominant-activating germline mutations in the gene encoding the PI(3)K catalytic subunit p110δ result in T cell senescence and human immunodeficiency. Nat. Immunol. 2014;15:88–97. doi: 10.1038/ni.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Takeda AJ, et al. Novel PIK3CD mutations affecting N-terminal residues of p110δ cause activated PI3Kδ syndrome (APDS) in humans. J. Allergy Clin. Immunol. 2017;140:1152–1156.e10. doi: 10.1016/j.jaci.2017.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lucas CL, et al. Heterozygous splice mutation in PIK3R1 causes human immunodeficiency with lymphoproliferation due to dominant activation of PI3K. J. Exp. Med. 2014;211:2537–2547. doi: 10.1084/jem.20141759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Deau M-C, et al. A human immunodeficiency caused by mutations in the PIK3R1 gene. J. Clin. Invest. 2014;124:3923–3928. doi: 10.1172/JCI75746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Dornan GL, et al. Conformational disruption of PI3Kδ regulation by immunodeficiency mutations in PIK3CD and PIK3R1. Proc. Natl Acad. Sci. Usa. 2017;114:1982–1987. doi: 10.1073/pnas.1617244114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Kaneda MM, et al. PI3Kγ is a molecular switch that controls immune suppression. Nature. 2016;539:437–442. doi: 10.1038/nature19834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.De Henau O, et al. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kγ in myeloid cells. Nature. 2016;539:443–447. doi: 10.1038/nature20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Campa CC, et al. Inhalation of the prodrug PI3K inhibitor CL27c improves lung function in asthma and fibrosis. Nat. Commun. 2018;9:5232. doi: 10.1038/s41467-018-07698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Liu N, et al. BAY 80-6946 is a highly selective intravenous PI3K inhibitor with potent p110α and p110δ activities in tumor cell lines and xenograft models. Mol. Cancer Ther. 2013;12:2319–2330. doi: 10.1158/1535-7163.MCT-12-0993-T. [DOI] [PubMed] [Google Scholar]
- 274.Venot Q, et al. Targeted therapy in patients with PIK3CA-related overgrowth syndrome. Nature. 2018;558:540–546. doi: 10.1038/s41586-018-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Gopal AK, et al. PI3Kδ inhibition by idelalisib in patients with relapsed indolent lymphoma. N. Engl. J. Med. 2014;370:1008–1018. doi: 10.1056/NEJMoa1314583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Brown JR, et al. Idelalisib, an inhibitor of phosphatidylinositol 3 kinase p110δ, for relapsed/refractory chronic lymphocytic leukemia. Blood. 2014;123:3390–3397. doi: 10.1182/blood-2013-11-535047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Flinn IW, et al. Idelalisib, a selective inhibitor of phosphatidylinositol 3-kinase-δ, as therapy for previously treated indolent non-Hodgkin lymphoma. Blood. 2014;123:3406–3413. doi: 10.1182/blood-2013-11-538546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kahl BS, et al. Results of a phase I study of idelalisib, a PI3Kδ inhibitor, in patients with relapsed or refractory mantle cell lymphoma (MCL) Blood. 2014;123:3398–3405. doi: 10.1182/blood-2013-11-537555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Fowler NH, et al. Umbralisib, a dual PI3Kδ/CK1ε inhibitor in patients with relapsed or refractory indolent lymphoma. J. Clin. Oncol. 2021;39:1609–1618. doi: 10.1200/JCO.20.03433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Flinn IW, et al. Duvelisib, a novel oral dual inhibitor of PI3K-δ,γ, is clinically active in advanced hematologic malignancies. Blood. 2018;131:877–887. doi: 10.1182/blood-2017-05-786566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Chandarlapaty S, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Chakrabarty A, Sánchez V, Kuba MG, Rinehart C, Arteaga CL. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc. Natl Acad. Sci. USA. 2012;109:2718–2723. doi: 10.1073/pnas.1018001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Bosch A, et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor–positive breast cancer. Sci. Transl. Med. 2015;7:283ra51. doi: 10.1126/scitranslmed.aaa4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Toska E, et al. PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science. 2017;355:1324–1330. doi: 10.1126/science.aah6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Hopkins BD, et al. Suppression of insulin feedback enhances the efficacy of PI3K inhibitors. Nature. 2018;560:499–503. doi: 10.1038/s41586-018-0343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Eschweiler S, et al. Intermittent PI3Kδ inhibition sustains anti-tumour immunity and curbs irAEs. Nature. 2022;605:741–746. doi: 10.1038/s41586-022-04685-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Song KW, et al. RTK-dependent inducible degradation of mutant PI3Ka drives GDC-0077 (Inavolisib) efficacy. Cancer Discov. 2022;12:204–219. doi: 10.1158/2159-8290.CD-21-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Brown JR, Auger KR. Phylogenomics of phosphoinositide lipid kinases: perspectives on the evolution of second messenger signaling and drug discovery. BMC Evolut. Biol. 2011;11:4. doi: 10.1186/1471-2148-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Burke JE, et al. Dynamics of the phosphoinositide 3-kinase p110δ interaction with p85α and membranes reveals aspects of regulation distinct from p110α. Structure. 2011;19:1127–1137. doi: 10.1016/j.str.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110α (PIK3CA) Proc. Natl Acad. Sci. USA. 2012;109:15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Dbouk HA, et al. G protein-coupled receptor-mediated activation of p110β by Gβγ is required for cellular transformation and invasiveness. Sci. Signal. 2012;5:ra89. doi: 10.1126/scisignal.2003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Vadas O, et al. Molecular determinants of PI3Kγ-mediated activation downstream of G-protein-coupled receptors (GPCRs) Proc. Natl Acad. Sci. USA. 2013;110:18862–18867. doi: 10.1073/pnas.1304801110. [DOI] [PMC free article] [PubMed] [Google Scholar]