Abstract
Many studies have shown that circular RNA (circRNA) is an important regulator mediating the malignant progression of cancer. However, the role and mechanism of circ‐EIF3I in lung cancer (LC) development are still unclear. A total 36 paired LC tumor tissues and adjacent normal tissues were enrolled. The expression of circ‐EIF3I, microRNA (miR)‐1253, and neuro‐oncological ventral antigen 2 (NOVA2) was measured by quantitative real‐time PCR. The proliferation, apoptosis, migration, and invasion of LC cells were determined by MTT assay, colony formation assay, flow cytometry, and transwell assay. Dual‐luciferase reporter assay was performed to verify the interaction between miR‐1253 and circ‐EIF3I or NOVA2. The protein levels of NOVA2 and Wnt/β‐catenin pathway‐related markers were detected by western blot analysis. Xenograft tumor was constructed to explore the function of circ‐EIF3I on LC tumor growth. Circ‐EIF3I was upregulated in LC tumor tissues and cells. Silenced circ‐EIF3I could suppress the proliferation, migration, invasion, and enhance the apoptosis of LC cells in vitro, as well as reduce LC tumor growth in vivo. Circ‐EIF3I could sponge miR‐1253, and miR‐1253 inhibitor overturned the regulation of circ‐EIF3I knockdown on LC cell progression. NOVA2 was confirmed to be a target of miR‐1253, which could reverse the inhibitory effects of miR‐1253 on LC cell progression. Further experiments showed that circ‐EIF3I regulated NOVA2 expression by sponging miR‐1253. In addition, circ‐EIF3I silencing could inhibit the activity of Wnt/β‐catenin pathway via regulating the miR‐1253/NOVA2 axis. Circ‐EIF3I might function as an oncogene in LC, which promoted LC progression by the miR‐1253/NOVA2/Wnt/β‐catenin network.
Keywords: circ‐EIF3I, lung cancer, miR‐1253, NOVA2, Wnt/β‐catenin
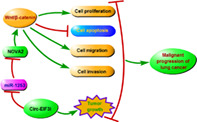
Circ‐EIF3I promoted LC cell proliferation, migration, invasion, and inhibited apoptosis, as well as enhanced LC tumor growth by regulating the miR‐1253/NOVA2 axis to mediate the activity of Wnt/β‐catenin pathway.
INTRODUCTION
Lung cancer (LC) is a malignant tumor originating in the lungs, and it has become one of the most threatening malignant tumors to the health and life of the occupational population in the world. 1 , 2 According to different pathological types, LC is often divided into two categories: non–small cell LC (NSCLC) and small cell LC (SCLC). 3 , 4 The pathogenesis of LC has not yet been fully defined, but there is evidence that it may be the result of environmental factors and genetic factors. 5 , 6 Although surgery, chemotherapy, and radiotherapy have greatly improved the cure rate of lung cancer, 7 , 8 the discovery of effective biomarkers is still of great significance for exploring the pathogenesis of LC and improving the prognosis of LC patients.
Non‐coding RNAs (ncRNAs) refer to a type of RNA that cannot encode proteins and have been proven to have important regulatory effects on the biological functions of cells. 9 , 10 Circular RNA (circRNA) is a type of ncRNA that has been intensively studied in recent years, because they have a compact circular structure and are extremely stable in organisms. 11 In terms of molecular mechanism, circRNA can also act as a sponge of microRNA (miRNA) to participate in the regulation of downstream genes. 12 In cancer‐related research, circRNA has been proven to be an important biomarker for the diagnosis, treatment, and predicted risk. 13 , 14 For example, Xu et al. 15 proposed that circTADA2A‐E6 sponged miR‐203a‐5p to repress breast cancer cell proliferation, metastasis, and clonogenicity via regulating SOCS3. Sheng et al. 16 suggested that circUBAP2 promoted ovarian cancer proliferation and migration by CHD2 through sponging miR‐144.
Hsa_circ_0011385 was derived from host gene EIF3I, so it was also called circ‐EIF3I. We used Gene Expression Omnibus (GEO) database to analyze the differentially expressed circRNA in LC tumor tissues and adjacent normal tissues and discovered that circ‐EIF3I was significantly upregulated in LC tumor tissues. In the previous study, circ‐EIF3I was found to be upregulated in thyroid cancer, which could promote cancer proliferation and metastasis. 17 , 18 However, whether the high expression of circ‐EIF3I also promotes the progression of LC has not been studied.
In this research, we aimed to reveal the role of circ‐EIF3I in LC progression and its underlying molecular mechanism. Through bioinformatics analysis, we found that circ‐EIF3I could complement with miR‐1253, and miR‐1253 could bind to the 3′UTR of neuro‐oncological ventral antigen 2 (NOVA2). Therefore, we proposed and confirmed the hypothesis that circ‐EIF3I regulated miR‐1253/NOVA2 axis to promote LC progression. Our research hopes to provide new target for LC treatment.
MATERIALS AND METHODS
Patient tissues
Thirty‐six paired LC tumor tissues and adjacent normal tissues were collected from 36 LC patients at The Fifth Affiliated Hospital of Zhengzhou University. None of the patients received any treatment before surgery. According to the tumor, nodes, and metastasis (TNM) stage (I, II and III) and whether there was lymph node metastasis (yes or no) in the patients, the LC tumor tissues were classified separately. Each patient signed written informed consent. This study was approved by the Ethics Committee of The Fifth Affiliated Hospital of Zhengzhou University and all patients were followed up for 5 years.
Cell culture and transfection
Human LC cell lines (H1650, H460, A549 and H1299) (American Type Culture Collection [ATCC]) and bronchial epithelial cell line (HBE) (Procell) were cultured in RPMI‐1640 medium (Gibco) at 37°C in 5% CO2 incubator. The medium was added with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin–streptomycin (Invitrogen) to prepare complete medium.
Lipofectamine 3000 (Invitrogen) was used for cell transfection. All plasmids and oligonucleotides were synthesized from GenePharma, including circ‐EIF3I small interfering RNA (siRNA), overexpression vector, and lentivirus short hairpin RNA (shRNA) (si‐circ‐EIF3I: 5′‐AGTATAGTGCCAAGGAAAGCAdTdT‐3′, circ‐EIF3I and sh‐circ‐EIF3I: 5′‐CCGGATAGTGCCAAGGAAAGCAGCTCTCGAGAGCTGCTTTCCTTGGCACTATTTTTTG‐3′), miR‐1253 mimic and inhibitor (miR‐1253: 5′‐AGAGAAGAAGAUCAGCCUGCA‐3′ and anti‐miR‐1253: 5′‐UGCAGGCUGAUCUUCUUCUCU‐3′), and plasmid cloning DNA (pcDNA) NOVA2 overexpression plasmid or their negative controls (si‐NC, vector, sh‐NC, miR‐NC, anti‐miR‐NC, and pcDNA).
Quantitative real‐time polymerase chain reaction (qRT‐PCR)
Total RNA was extracted by RNeasy Plus Universal Kit (Qiagen). PrimeScript RT‐PCR Kit (Takara) was used to synthesize complementary DNA (cDNA). According to instructions of SYBR GreenER qPCR SuperMix Universal (Invitrogen), PCR was performed basing on the primers of circ‐EIF3I, EIF3I, miR‐1253, and NOVA2. Relative expression was normalized to GAPDH or U6 and analyzed with 2−ΔΔCT method. The primer sequences were shown: circ‐EIF3I, F 5′‐AAATCACCAGTGCTGTTTGGG‐3′, R 5′‐TTGCCCCCAAAGTCAAAACC‐3′; EIF3I, F 5′‐GGCCATGAGCGGTCCATTAC‐3′, R 5′‐ACATTGACGATAGGGTCCTTGG‐3′; miR‐1253, F 5′‐GCCGAGAGAGAAGAAGATCAGCC‐3′, R 5′‐ATCCAGTGCAGGGTCCGAGG‐3′; NOVA2, F 5′‐AAGGCGAATACTTCCTGAAGGT‐3′; R 5′‐TACTAGGCATACCCGCTCTGT‐3′; GAPDH, F 5′‐CTCTGCTCCTCCTGTTCGAC‐3′, R 5′‐CGACCAAATCCGTTGACTCC‐3′; U6, F 5′‐CTCGCTTCGGCAGCACA‐3′, R 5′‐AACGCTTCACGAATTTGCGT‐3′.
RNase R assay
After the RNA was extracted from A549 and H1299 cells, 2 μg RNA was incubated with 3 units of RNase R for 30 min at 37°C. Using RNA that had not been incubated with RNase R as a control (Mock), qRT‐PCR was used to detect the expression of circ‐EIF3I and linear EIF3I, respectively.
MTT assay
MTT Assay Kit (Beyotime) was used to measure cell proliferation. Transfected LC cells were collected and reseeded into 96‐well plates. At 24, 48, and 72 h, MTT solution was added into each well for 4 h incubation. After removing the medium of wells, formazan solvent was added into each well to dissolve formazan. The optical density (OD) value at 570 nm was measured by a microplate reader to evaluate cell viability.
Colony formation assay
Transfected LC cells were harvested and then seeded into 6‐well plates (150 cells per well). After 14 days, the surviving cells formed colonies were fixed with methanol (Zhuoli Chemical Productions) and stained with crystal violet (Beyotime). The number of colonies (>50 cells) was counted under a microscope.
Flow cytometry
Annexin V‐FITC/propidine iodide (PI) Apoptosis Detection Kit was obtained from Vazyme. In brief, A549 and H1299 cells (2 × 10 5 ) were collected and the cells were suspended with 1× binding buffer. The cell suspensions were incubated with annexin V‐FITC and PI for 10 min. The apoptosis rate of cells was assessed by a flow cytometer.
Transwell migration and invasion assays
After transfection, A549 and H1299 cells were resuspended in serum‐free medium and then seeded into the upper chamber of the 24‐well transwell plates (Corning). In addition, the bottom chamber was filled with complete medium. Twenty‐four hours later, the cells migrated to the lower layer of the upper chamber was fixed and stained. The migration cell number was counted under a microscope (100×). For cell invasion assay, an additional Matrigel (Corning) was needed to pre‐coat the upper chamber. The rest of the procedure was consistent with the cell migration assay.
Dual‐luciferase reporter assay
The binding sites between miR‐1253 and circ‐EIF3I or NOVA2 3′UTR were predicted using Circinteractome software or Targetscan software. According to this, the wild‐type (WT) and mutate‐type (MUT) sequences of circ‐EIF3I or NOVA2 3′UTR were amplified and then cloned into pGL3 reporter vector (Promega) to generate the WT and MUT reporter vector for circ‐EIF3I or NOVA2 3′UTR. A549 and H1299 cells were transfected with the above reporter vector and miR‐1253 mimic or miR‐NC. Forty‐eight hours later, the luciferase activity was tested by Dual‐Luciferase Reporter Assay System (Promega).
Western blot analysis
Radioimmunoprecipitation (RIPA) lysis buffer (Beyotime) was used to extract total protein from A549 and H1299 cells. The same amount protein was subjected to sodium dodecyl‐sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) gel and then transferred onto a polyvinylidene fluoride (PVDF) membrane (Beyotime). The membrane was blocked with 5% skimmed milk and incubated with primary antibodies including anti‐NOVA2 (1:2000, Boster), anti‐β‐catenin (1:1500, Boster), anti‐c‐Myc (1:2000, Boster), anti‐CyclinD1 (1:1000, Boster), or anti‐GAPDH (1:2000, Boster). After the membrane was hatched with secondary antibody (1:10,000, Boster), the protein blots were visualized by Ultra Sensitive ECL Chemiluminescence Substrate (Boster). The relative protein expression was analyzed using ImageJ software (National Institutes of Health).
Xenograft tumor
For lentiviral transduction, H1299 cells (reached 50% confluence) were infected by virus particles (sh‐NC or sh‐circ‐EIF3I) in media containing 8 μg/mL of polybrene. Twenty‐four hours later, the infected cells were selected by 1 μg/mL of puromycin over 72 h. H1299 cells stable infected with sh‐NC or sh‐circ‐EIF3I were inoculated subcutaneously into the back of BALB/c nude mice (n = 6, male, Vital River). After 8 days, the tumor volume was counted by measuring the tumor length and width using slide caliper every 4 days. All mice were euthanized by isoflurane inhalation followed by cervical dislocation at 23 days, and the tumors were collected for detecting the expression of circ‐EIF3I, miR‐1253, and NOVA2. All animal operations were approved by the Animal Ethics Committee of The Fifth Affiliated Hospital of Zhengzhou University.
Statistical analysis
Each experiment was performed in triplicate, and all data were expressed as means ± standard deviation. Student's t‐test or one‐way analyses of variance were used to assess the differences between groups. Statistical analyses were performed using GraphPad Prism 7 (GraphPad). The relationship between the overall survival of patients and circ‐EIF3I expression was analyzed by Kaplan–Meier analysis, and the correlation between circ‐EIF3I and miR‐1253 or miR‐1253 and NOVA2 was analyzed by Pearson correlation analysis. p < 0.05 was considered statistically significant.
RESULTS
Circ‐EIF3I was highly expressed in LC and was associated with poor prognosis of LC patients
According to the cut‐off criteria (|log2 fold change| >1 and p < 0.05), 10 differentially expressed circRNA was screened in 5 paired LC tumor tissues and adjacent normal tissues in GEO database (GSE101586) (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE101586) (Figure 1(a)). Among them, hsa_circRNA_100 146 showed a significant difference and was selected for this study. Basing on the probe sequence, the CircBase ID of hsa_circRNA_100 146 aligned in the CircBase database was hsa_circ_0011385, also termed as circ‐EIF3I. In our study, we measured circ‐EIF3I expression in 36 paired LC tumor tissues and adjacent normal tissues, and found that circ‐EIF3I was markedly increased in LC tumor tissues (Figure 1(b)). Additionally, circ‐EIF3I was significantly upregulated in four LC cells (H1650, H460, A549, and H1299) compared to HBE cells (Figure 1(c)). In addition, we found that circ‐EIF3I expression in the tumor tissues of LC patients in stage III was remarkably higher than that in stages I + II (Figure 1(d)), and it was also obviously higher in the LC patients with lymph node metastasis than in LC patients without metastasis (Figure 1(e)). According to the median expression of circ‐EIF3I in LC tumor tissues, we divided LC patients into high circ‐EIF3I expression group (n = 18) and low circ‐EIF3I expression group (n = 18). The results of Kaplan–Meier analysis showed that the overall survival rate of LC patients in the high circ‐EIF3I expression group was lower than that of the low circ‐EIF3I expression group (Figure 1(f)). These data showed that high circ‐EIF3I was related to the poor prognosis of LC patients. To assess the stability of circ‐EIF3I, RNase R assay was carried out and the results showed that circ‐EIF3I expression was not affected by RNase R, whereas its linear EIF3I could be degraded by RNase R in A549 and H1299 cells (Figure 1(g),(h)). These results confirmed that circ‐EIF3I was a circRNA with highly expression in LC, which was related to the poor prognosis of patients.
FIGURE 1.
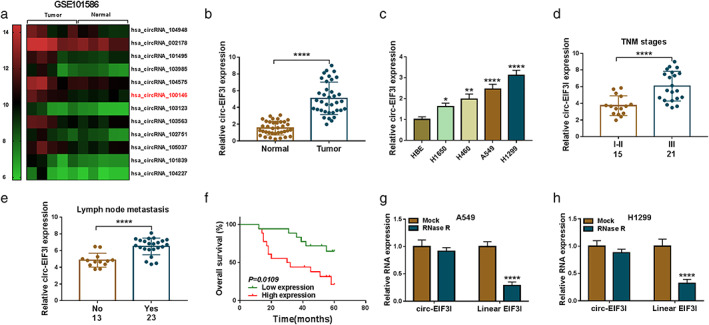
Circ‐EIF3I was highly expressed in LC and was associated with poor prognosis of LC patients. (a) The 10 differentially expressed circRNA screened in five paired LC tumor tissues and adjacent normal tissues in GEO database (GSE101586) were shown. (b) Circ‐EIF3I expression in 36 paired LC tumor tissues and adjacent normal tissues was detected using qRT‐PCR. (c) qRT‐PCR was used to measure the expression of circ‐EIF3I in HBE cells and four LC cells (H1650, H460, A549 and H1299). (d) Circ‐EIF3I expression in the tumor tissues of LC patients in stage III and stages I + II was determined using qRT‐PCR. (e) qRT‐PCR was used to assess circ‐EIF3I expression in LC patients with or without lymph node metastasis. (f) Kaplan–Meier analysis was performed to analyze the relationship between the overall survival rate of LC patients and circ‐EIF3I expression. (g), (h) RNase R assay was used to assess the stability of circ‐EIF3I in A549 and H1299 cells. *p < 0.05, ***p < 0.001, ****p < 0.0001.LC, lung cancer; circRNA, circular RNA; GEO, Gene Expression Omnibus; qRT‐PCR, quantitative real‐time polymerase chain reaction; HBE, human bronchial epithelial cell line
Knockdown of circ‐EIF3I inhibited the proliferation, metastasis, and promoted the apoptosis of LC cells
To validate the hypothesis that circ‐EIF3I had a pro‐cancer role in LC, A549 and H1299 cells were transfected with siRNA of circ‐EIF3I. After transfection, the expression of circ‐EIF3I was notably reduced in A549 and H1299 cells, indicating that the transfection of si‐circ‐EIF3I was successful (Figure 2(a)). Through MTT assay and colony formation assay, we found that circ‐EIF3I knockdown could decrease the viabilities and colony numbers of A549 and H1299 cells (Figure 2(b)–(d)), which suggested that circ‐EIF3I could promote LC cell proliferation. The results of flow cytometry showed that the apoptosis rate of A549 and H1299 cells was significantly enhanced in the presence of si‐circ‐EIF3I (Figure 2(e)). In addition, silenced circ‐EIF3I also repressed the numbers of migratory and invasive A549 and H1299 cells (Figure 2(f),(g)). These data suggested that circ‐EIF3I might contribute to LC progression.
FIGURE 2.
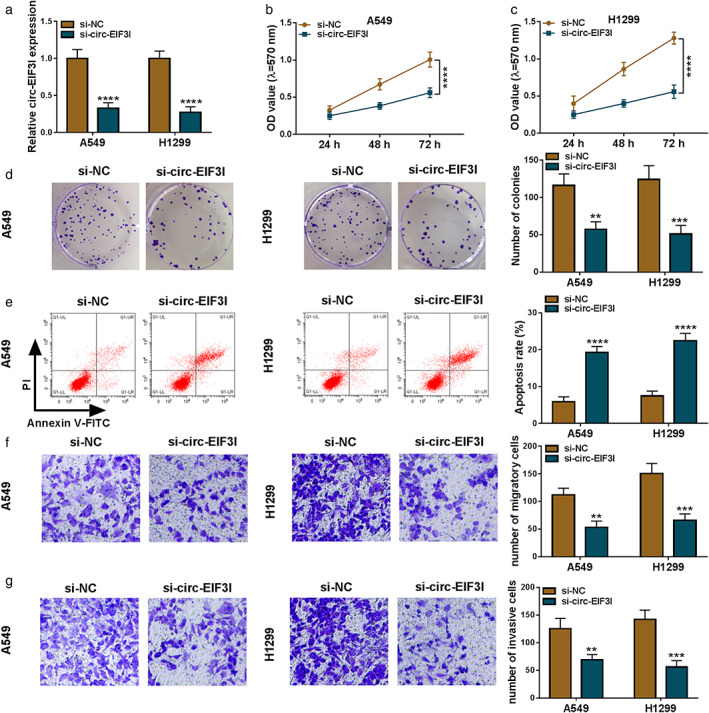
Knockdown of circ‐EIF3I inhibited LC cell progression. A549 and H1299 cells were transfected with si‐NC or si‐circ‐EIF3I. (a) The expression of circ‐EIF3I was detected by qRT‐PCR. MTT assay (b), (c), colony formation assay (d), flow cytometry (e), and transwell assay (f), (g) were used to measure the viability, colony number, apoptosis, migration, and invasion of cells. **p < 0.01, ***p < 0.001, ****p < 0.0001. LC, lung cancer; qRT‐PCR, quantitative real‐time polymerase chain reaction
Circ‐EIF3I interacted with miR‐1253
To explore the mechanism of circ‐EIF3I in LC, we performed the bioinformatics analysis. The Circinteractome software predicted that miR‐1253 had complementary sites with circ‐EIF3I (Figure 3(a)). The results of dual‐luciferase reporter assay further revealed that the luciferase activity of WT‐circ‐EIF3I vector could be decreased by miR‐1253 overexpression, whereas that of the MUT‐circ‐EIF3I vector was not changed by miR‐1253 or miR‐NC (Figure 3(b),(c)). In LC tumor tissues, miR‐1253 was lowly expressed compared with that in adjacent normal tissues (Figure 3(d)), and its expression was negatively correlated with circ‐EIF3I expression (Figure 3(e)). Moreover, the expression of miR‐1253 also was found to be downregulated in A549 and H1299 cells compared to HBE cells (Figure 3(f)). In addition, circ‐EIF3I overexpression vector was built and the promoting effect of it on circ‐EIF3I expression was confirmed (Figure 3(g)). In A549 and H1299 cells transfected with si‐circ‐EIF3I or circ‐EIF3I overexpression vector, we discovered that the expression of miR‐1253 could be increased by circ‐EIF3I knockdown, whereas decreased by circ‐EIF3I overexpression (Figure 3(h)). The above data indicated that circ‐EIF3I could sponge miR‐1253 to negatively regulate miR‐1253 expression.
FIGURE 3.
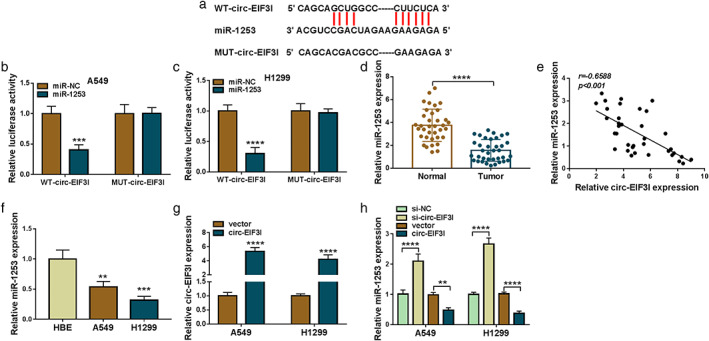
Circ‐EIF3I interacted with miR‐1253. (a) The binding sites between circ‐EIF3I and miR‐1253 were shown. (b), (c) Dual‐luciferase reporter assay was performed to assess the interaction between circ‐EIF3I and miR‐1253. (d) The expression of miR‐1253 in 36 paired LC tumor tissues and adjacent normal tissues was measured by qRT‐PCR. (e) Pearson correlation analysis was used to evaluate the correlation between circ‐EIF3I and miR‐1253 in LC tumor tissues. (f) qRT‐PCR was used to test miR‐1253 expression in HBE cells and LC cells (A549 and H1299). (g) Circ‐EIF3I expression was determined by qRT‐PCR in A549 and H1299 cells transfected with vector or circ‐EIF3I overexpression vector. (h) A549 and H1299 cells were transfected with si‐NC, si‐circ‐EIF3I, vector or circ‐EIF3I. The expression of miR‐1253 was examined by qRT‐PCR. **p < 0.01, ***p < 0.001, ****p < 0.0001. LC, lung cancer; qRT‐PCR, quantitative real‐time polymerase chain reaction; HBE, human bronchial epithelial cell line
Circ‐EIF3I regulated the proliferation, apoptosis, and metastasis of LC cells by sponging miR‐1253
The rescue experiments were performed to investigate whether circ‐EIF3I sponged miR‐1253 to regulate LC cell progression. A549 and H1299 cells were co‐transfected with si‐circ‐EIF3I and anti‐miR‐1253. The increasing effect of si‐circ‐EIF3I on miR‐1253 expression could be reversed by anti‐miR‐1253 (Figure 4(a)). Subsequently, the biological functions of A549 and H1299 cells were measured. Our data showed that the inhibitory effects of circ‐EIF3I silencing on the viabilities and colony numbers of A549 and H1299 cells could be abolished by miR‐1253 inhibitor (Figure 4(b)–(d)). Additionally, miR‐1253 inhibitor also overturned the promotion effect of circ‐EIF3I knockdown on the apoptosis (Figure 4(e)), and the suppressive effect on the migration and invasion of A549 and H1299 cells (Figure 4(f),(g)). All data revealed that circ‐EIF3I might sponge miR‐1253 to promote LC progression.
FIGURE 4.
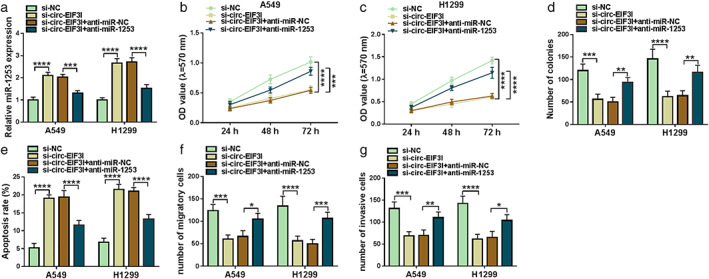
Circ‐EIF3I regulated LC cell progression by sponging miR‐1253. A549 and H1299 cells were transfected with si‐NC, si‐circ‐EIF3I, si‐circ‐EIF3I + anti‐miR‐NC or si‐circ‐EIF3I + anti‐miR‐1253. (a) MiR‐1253 expression was determined using qRT‐PCR. The viability, colony number, apoptosis, migration and invasion of cells were detected using MTT assay (b), (c), colony formation assay (d), flow cytometry (e), and transwell assay (f), (g). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. LC, lung cancer; qRT‐PCR, quantitative real‐time polymerase chain reaction
MiR‐1253 could directly target NOVA2
To search for the downstream targets of miR‐1253, the Targetscan software was used and the results suggested that the 3′UTR of NOVA2 could directly bind to miR‐1253 (Figure 5(a)). Moreover, dual‐luciferase reporter assay results showed that miR‐1253 overexpression specially inhibited the luciferase activity of NOVA2 3′UTR‐WT vector without affecting that of the NOVA2 3′UTR‐MUT vector (Figure 5(b),(c)). Furthermore, we discovered that NOVA2 messenger RNA (mRNA) expression in LC tumor tissues was higher than that in adjacent normal tissues and was negatively correlated with miR‐1253 expression (Figure 5(d),(e)). In addition, we also found significantly high expression of NOVA2 in LC tumor tissues and cells at the protein levels (Figure 5(f),(g)). To further determine the regulation of miR‐1253 on NOVA2 expression, miR‐1253 mimic or inhibitor were transfected into LC cells. The markedly high and low expression of miR‐1253 confirmed the transfection efficiency of miR‐1253 mimic and inhibitor, respectively (Figure 5(h)). The detection results of NOVA2 protein expression revealed that miR‐1253 overexpression could inhibit NOVA2 expression, whereas its inhibitor had an opposite effect (Figure 5(i)). These results indicated that NOVA2 was a target of miR‐1253 and its expression was negatively regulated by miR‐1253.
FIGURE 5.
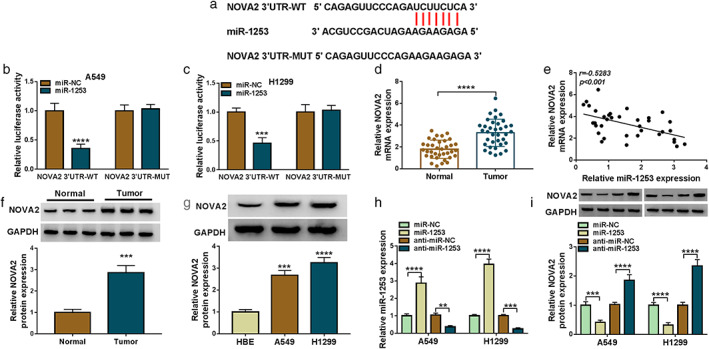
MiR‐1253 could directly target NOVA2. (a) The binding sites between miR‐1253 and NOVA2 3′UTR were presented. (b), (c) The interaction between miR‐1253 and NOVA2 was confirmed using dual‐luciferase reporter assay. (d) The mRNA expression of NOVA2 in 36 paired LC tumor tissues and adjacent normal tissues was detected by qRT‐PCR. (e) The correlation between miR‐1253 and NOVA2 in LC tumor tissues was analyzed by Pearson correlation analysis. (f) WB analysis was used to assess NOVA2 protein expression in three paired LC tumor tissues and adjacent normal tissues. (g) The protein expression of NOVA2 in HBE cells and LC cells (A549 and H1299) was determined by WB analysis. (h) The transfection efficiencies of miR‐1253 mimic and inhibitor were evaluated by detecting miR‐1253 expression using qRT‐PCR. (i) A549 and H1299 cells were transfected with miR‐NC, miR‐1253, anti‐miR‐NC or anti‐miR‐1253. The protein expression of NOVA2 was tested by WB analysis. **p < 0.01, ***p < 0.001, ****p < 0.0001. NOVA2, neuro‐oncological ventral antigen 2; mRNA, messenger RNA; LC, lung cancer; qRT‐PCR, quantitative real‐time polymerase chain reaction; WB, western blot
NOVA2 reversed the suppressive effect of miR‐1253 on LC cell progression
Furthermore, miR‐1253 mimic and pcDNA NOVA2 overexpression plasmid were co‐transfected into A549 and H1299 cells to confirm whether miR‐1253 targeted NOVA2 to regulate LC cell progression. In LC cells, the decreasing effect of miR‐1253 on NOVA2 protein expression could be abolished by NOVA2 overexpression (Figure 6(a)). The results of MTT assay and colony formation assay indicated that miR‐1253 could reduce the viabilities and colony number of A549 and H1299 cells, whereas these effects could be reversed by NOVA2 overexpression (Figure 6(b)–(d)). Overexpressed NOVA2 could recover the enhancing effect of miR‐1253 on the apoptosis of LC cells (Figure 6(e)), and reverse the inhibitory effect on the migration and invasion of LC cells (Figure 6(f),(g)). These data revealed that miR‐1253 suppressed LC progression by targeting NOVA2.
FIGURE 6.
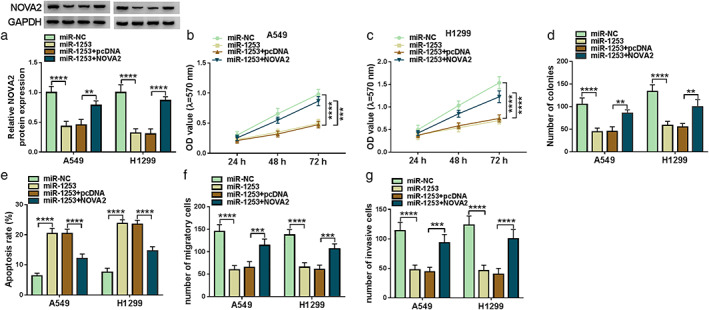
NOVA2 reversed the suppressive effect of miR‐1253 on LC cell progression. A549 and H1299 cells were transfected with miR‐NC, miR‐1253, miR‐1253 + pcDNA, or miR‐1253 + NOVA2. (A) The protein expression of NOVA2 was detected by WB analysis. MTT assay (b), (c), colony formation assay (d), flow cytometry (e), and transwell assay (f), (g) were performed to determine the viability, colony number, apoptosis, migration and invasion of cells. **p < 0.01, ***p < 0.001, ****p < 0.0001. NOVA2, neuro‐oncological ventral antigen 2; LC, lung cancer; WB, western blot
Circ‐EIF3I regulated NOVA2 expression by sponging miR‐1253
The above results demonstrated that circ‐EIF3I could sponge miR‐1253 and miR‐1253 could target NOVA2. To explore whether circ‐EIF3I sponged miR‐1253 to regulate NOVA2, we detected NOVA2 expression in A549 and H1299 cells transfected with si‐circ‐EIF3I and anti‐miR‐1253. As presented in Figure 7(a),(b), we discovered that circ‐EIF3I knockdown significantly inhibited the mRNA and protein expression of NOVA2 in A549 and H1299 cells. However, miR‐1253 inhibitor could reverse the suppressive effect of circ‐EIF3I silencing on NOVA2 expression. Therefore, our data revealed that circ‐EIF3I sponged miR‐1253 to positively regulate NOVA2 expression.
FIGURE 7.
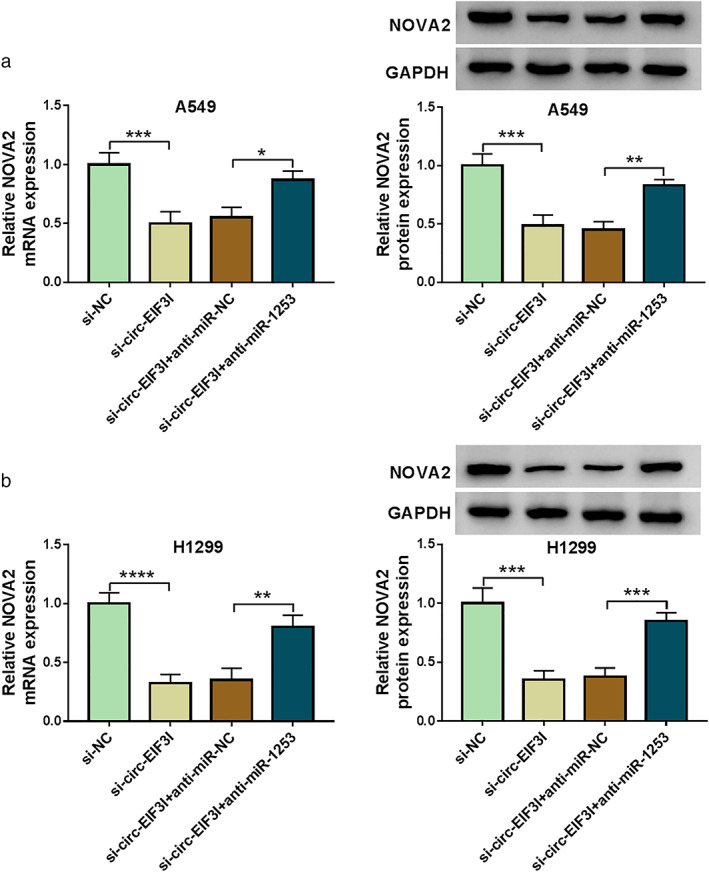
Circ‐EIF3I regulated NOVA2 expression by sponging miR‐1253. A549 and H1299 cells were transfected with si‐NC, si‐circ‐EIF3I, si‐circ‐EIF3I + anti‐miR‐NC or si‐circ‐EIF3I + anti‐miR‐1253. (a), (b) The mRNA and protein expression levels of NOVA2 were detected by qRT‐PCR and WB analysis. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. NOVA2, neuro‐oncological ventral antigen 2; mRNA, messenger RNA; qRT‐PCR, quantitative real‐time polymerase chain reaction; WB, western blot
Silenced circ‐EIF3I repressed LC tumor growth in vivo
Animal experiments were carried out to further confirm the role of circ‐EIF3I in LC. In H1299 cells transfected with sh‐circ‐EIF3I, we found that the expression of circ‐EIF3I was remarkably decreased (Figure 8(a)). H1299 cells transfected with sh‐circ‐EIF3I or sh‐NC were injected into the back of nude mice. By monitoring the volume of subcutaneous xenograft tumors, we discovered that the tumor volume of mice in sh‐circ‐EIF3I group was significantly lower than that of the control group (Figure 8(b)). The tumor weight in the sh‐circ‐EIF3I group was smaller than that in the control group (Figure 8(c)). In addition, circ‐EIF3I expression was indeed inhibited, miR‐1253 expression was enhanced, and NOVA2 protein expression was markedly repressed in the tumor tissues of the sh‐circ‐EIF3I group (Figure 8(d),(e)). These results showed that circ‐EIF3I silencing reduced LC tumor growth by regulating the miR‐1253/NOVA2 axis.
FIGURE 8.
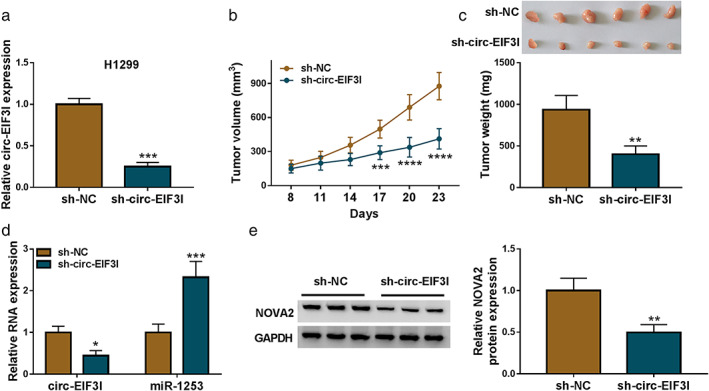
Silenced circ‐EIF3I repressed LC tumor growth in vivo. (a) Circ‐EIF3I expression in H1299 cells transfected with si‐NC or sh‐circ‐EIF3I was detected by qRT‐PCR. Tumor volume (b) and tumor weight (c) were determined in the mice of the sh‐NC group and sh‐circ‐EIF3I group. (d) The expression of circ‐EIF3I and miR‐1253 was detected by qRT‐PCR in the tumor tissues of the sh‐NC group and sh‐circ‐EIF3I group. (e) WB analysis was used to measure NOVA2 protein expression in the tumor tissues of the sh‐NC group and sh‐circ‐EIF3I group. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. LC, lung cancer; qRT‐PCR, quantitative real‐time polymerase chain reaction; WB, western blot
Circ‐EIF3I/miR‐1253/NOVA2 axis regulated the activity of Wnt/β‐catenin pathway
Wnt/β‐catenin pathway plays an important role in embryonic development, organogenesis, and tumor genesis. 19 , 20 In many cancer‐related studies, Wnt/β‐catenin pathway has been shown to be closely related to the malignant progression of cancers. 21 , 22 Here, we examined the protein expression of Wnt/β‐catenin pathway markers. The results showed that circ‐EIF3I knockdown could repress the protein levels of β‐catenin, c‐Myc, and CyclinD1 in A549 and H1299 cells, and these effects could be reversed by miR‐1253 inhibitor or NOVA2 overexpression (Figure 9(a),(b)). Our results suggested that circ‐EIF3I regulated the activity of Wnt/β‐catenin pathway via the miR‐1253/NOVA2 axis.
FIGURE 9.
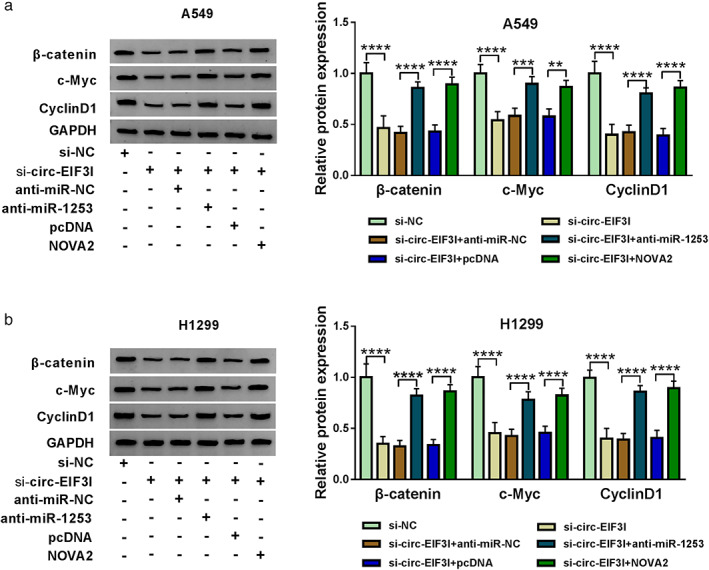
Circ‐EIF3I/miR‐1253/NOVA2 axis regulated the activity of Wnt/β‐catenin pathway. A549 and H1299 cells were transfected with si‐NC, si‐circ‐EIF3I, si‐circ‐EIF3I + anti‐miR‐NC, si‐circ‐EIF3I + anti‐miR‐1253, si‐circ‐EIF3I + pcDNA or si‐circ‐EIF3I + NOVA2. (a), (b) The protein expression levels of β‐catenin, c‐Myc and cyclin D1 were detected by WB analysis. **p < 0.01, ***p < 0.001, ****p < 0.0001. NOVA2, neuro‐oncological ventral antigen 2; WB, western blot
DISCUSSION
The important role of circRNA in LC development has been clarified by many studies. For instance, hsa_circ_100395 was lowly expressed in LC, and it could sponge miR‐1228 to promote TCF21 expression, thereby inhibiting LC proliferation and metastasis. 23 Han et al. 24 showed that circ‐BANP had an upregulated expression in LC, which could promote LC progression by increasing cell proliferation and metastasis via the miR‐503/LARP axis. In addition, inhibition of circ_100 146 also was confirmed to inhibit NSCLC proliferation and invasion through sponging miR‐361‐3p and miR‐615‐5p. 25 Our study confirmed that circ‐EIF3I was highly expressed in LC tissues, especially in LC patients with advanced TNM and lymph node metastasis, suggesting that high circ‐EIF3I expression was associated with poor prognosis in LC patients. Function experiments showed that circ‐EIF3I silencing repressed LC cell proliferation, migration, invasion, and enhanced apoptosis in vitro, and inhibited LC tumor growth in vivo. These suggested that circ‐EIF3I might play an oncogene role in LC, which was consistent with its role in other cancers. 17 , 18 Further, many studies have confirmed that abnormal activation of the Wnt/β‐catenin pathway can promote LC malignant progression. 26 , 27 Here, we discovered that circ‐EIF3I knockdown also decreased the activity of Wnt/β‐catenin pathway, indicating that circ‐EIF3I contributed to LC progression by activating the Wnt/β‐catenin pathway.
In many cancers, miR‐1253 has been shown to regulate cancer progression as a tumor suppressor, including prostate cancer 28 and pancreatic ductal adenocarcinoma. 29 Wang et al. 30 suggested that miR‐1253 could inhibit proliferation and promote apoptosis in vascular smooth muscle cells to accelerate the progression of atherosclerosis. In the previous study, miR‐1253 was confirmed to suppress the proliferation and invasion of NSCLC. 31 , 32 In this, we uncovered that miR‐1253 was targeted by circ‐EIF3I and its expression was negatively regulated by circ‐EIF3I in vitro and in vivo. The downregulated miR‐1253 expression in LC tissues and cells also was verified in our results. In the rescue experiments, we found that the inhibitory effect of silenced circ‐EIF3I on LC cell progression and the Wnt/β‐catenin pathway could be reversed by miR‐1253 inhibitor, showing that circ‐EIF3I sponged miR‐1253 to mediate the activity of Wnt/β‐catenin pathway therefore regulating LC progression.
In addition, NOVA2 was found to be a target of miR‐1253. NOVA2 is a member of the NOVA superfamily. In past studies, NOVA2 has been associated with neurodevelopment and vascularization. 33 , 34 The researchers also found abnormal overexpression of NOVA2 in many tumors, including glioma 35 and colorectal cancer. 36 Importantly, in many LC‐related studies, NOVA2 has been found to be significantly highly expressed and promote the malignant progression of LC. 37 , 38 , 39 Consistent with this, our date determined that NOVA2 was upregulated in LC tissues and cells. Furthermore, we proposed that miR‐1253 inhibited LC cell progression via targeting NOVA2. The positive regulation of circ‐EIF3I on NOVA2 expression revealed that circ‐EIF3I could regulate NOVA2 expression through acting as a sponge for miR‐1253. We also showed that circ‐EIF3I regulated the activity of Wnt/β‐catenin pathway by regulating NOVA2. This confirmed the existence of the circ‐EIF3I/miR‐1253/NOVA2 axis in LC.
CONCLUSION
In conclusion, our data proposed that circ‐EIF3I played an active role in LC, which could promote LC progression by regulating the miR‐1253/NOVA2 axis to mediate the activity of Wnt/β‐catenin pathway. These findings supported the hypothesis that circ‐EIF3I might be a potential therapeutic target for LC and had breakthrough significance for LC targeted treatment.
DISCLOSURE OF INTEREST
The authors declare that they have no financial conflicts of interest.
Chen T, Feng G, Xing Z, Gao X. Circ‐EIF3I facilitates proliferation, migration, and invasion of lung cancer via regulating the activity of Wnt/β‐catenin pathway through the miR‐1253/NOVA2 axis. Thorac Cancer. 2022;13(22):3133–3144. 10.1111/1759-7714.14665
REFERENCES
- 1. Mao Y, Yang D, He J, Krasna MJ. Epidemiology of lung cancer. Surg Oncol Clin N Am. 2016;25(3):439–45. 10.1016/j.soc.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 2. Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. 2019;103(3):463–73. 10.1016/j.mcna.2018.12.006 [DOI] [PubMed] [Google Scholar]
- 3. Hensing T, Chawla A, Batra R, et al. A personalized treatment for lung cancer: molecular pathways, targeted therapies, and genomic characterization. Adv Exp Med Biol. 2014;799:85–117. 10.1007/978-1-4614-8778-4_5 [DOI] [PubMed] [Google Scholar]
- 4. E L, Lu L, Li L, et al. Radiomics for classification of lung cancer histological subtypes based on nonenhanced computed tomography. Acad Radiol. 2019;26(9):1245–52. 10.1016/j.acra.2018.10.013 [DOI] [PubMed] [Google Scholar]
- 5. Akhtar N, Bansal JG. Risk factors of lung cancer in nonsmoker. Curr Probl Cancer. 2017;41(5):328–39. 10.1016/j.currproblcancer.2017.07.002 [DOI] [PubMed] [Google Scholar]
- 6. Romaszko AM, Doboszyńska A. Multiple primary lung cancer: A literature review. Adv Clin Exp Med. 2018;27(5):725–30. 10.17219/acem/68631 [DOI] [PubMed] [Google Scholar]
- 7. Lemjabbar‐Alaoui H, Hassan OU, Yang YW, Buchanan P. Lung cancer: biology and treatment options. Biochim Biophys Acta. 2015;1856(2):189–210. 10.1016/j.bbcan.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crawford J, Wheatley‐Price P, Feliciano JL. Treatment of lung cancer in medically compromised patients. Am Soc Clin Oncol Educ Book. 2016;35:e484–91. 10.1200/edbk_158713 [DOI] [PubMed] [Google Scholar]
- 9. Esteller M. Non‐coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–74. 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- 10. Mattick JS, Makunin IV. Non‐coding RNA. Hum Mol Genet. 2006;15:R17–29. 10.1093/hmg/ddl046 [DOI] [PubMed] [Google Scholar]
- 11. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–91. 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 12. Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79. 10.1007/978-981-13-1426-1_6 [DOI] [PubMed] [Google Scholar]
- 13. Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. 10.1186/s12943-017-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ojha R, Nandani R, Chatterjee N, et al. Emerging role of circular RNAs as potential biomarkers for the diagnosis of human diseases. Adv Exp Med Biol. 2018;1087:141–57. 10.1007/978-981-13-1426-1_12 [DOI] [PubMed] [Google Scholar]
- 15. Xu JZ, Shao CC, Wang XJ, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR‐203a‐3p/SOCS3 axis. Cell Death Dis. 2019;10(3):175. 10.1038/s41419-019-1382-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sheng M, Wei N, Yang HY, Yan M, Zhao QX, Jing LJ. CircRNA UBAP2 promotes the progression of ovarian cancer by sponging microRNA‐144. Eur Rev Med Pharmacol Sci. 2019;23(17):7283–94. 10.26355/eurrev_201909_18833 [DOI] [PubMed] [Google Scholar]
- 17. Wang YF, Li MY, Tang YF, Jia M, Liu Z, Li HQ. Circular RNA circEIF3I promotes papillary thyroid carcinoma progression through competitively binding to miR‐149 and upregulating KIF2A expression. Am J Cancer Res. 2020;10(4):1130–9. [PMC free article] [PubMed] [Google Scholar]
- 18. Xia F, Chen Y, Jiang B, Bai N, Li X. Hsa_circ_0011385 accelerates the progression of thyroid cancer by targeting miR‐361‐3p. Cancer Cell Int. 2020;20:49. 10.1186/s12935-020-1120-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nusse R, Clevers H. Wnt/β‐catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169(6):985–99. 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 20. Clevers H. Wnt/beta‐catenin signaling in development and disease. Cell. 2006;127(3):469–80. 10.1016/j.cell.2006.10.018 [DOI] [PubMed] [Google Scholar]
- 21. Cheng X, Xu X, Chen D, et al. Therapeutic potential of targeting the Wnt/β‐catenin signaling pathway in colorectal cancer. Biomed Pharmacother. 2019;110:473–81. 10.1016/j.biopha.2018.11.082 [DOI] [PubMed] [Google Scholar]
- 22. Shang S, Hua F, Hu ZW. The regulation of β‐catenin activity and function in cancer: therapeutic opportunities. Oncotarget. 2017;8(20):33972–89. 10.18632/oncotarget.15687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen D, Ma W, Ke Z, Xie F. CircRNA hsa_circ_100395 regulates miR‐1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle. 2018;17(16):2080–90. 10.1080/15384101.2018.1515553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han J, Zhao G, Ma X, Dong Q, Zhang H, Wang Y, et al. CircRNA circ‐BANP‐mediated miR‐503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun. 2018;503(4):2429–35. 10.1016/j.bbrc.2018.06.172 [DOI] [PubMed] [Google Scholar]
- 25. Chen L, Nan A, Zhang N, Jia Y, Li X, Ling Y, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR‐361‐3p and miR‐615‐5p in non‐small cell lung cancer. Mol Cancer. 2019;18(1):13. 10.1186/s12943-019-0943-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wan L, Zhang L, Fan K, et al. Circular RNA‐ITCH suppresses lung cancer proliferation via inhibiting the Wnt/β‐catenin pathway. 2016;2016:1579490. 10.1155/2016/1579490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li XY, Liu YR, Zhou JH, Li W, Guo HH, Ma HP. Enhanced expression of circular RNA hsa_circ_000984 promotes cells proliferation and metastasis in non‐small cell lung cancer by modulating Wnt/β‐catenin pathway. Eur Rev Med Pharmacol Sci. 2019;23(8):3366–74. 10.26355/eurrev_201904_17700 [DOI] [PubMed] [Google Scholar]
- 28. Chen Y, Gu M, Liu C, et al. Long noncoding RNA FOXC2‐AS1 facilitates the proliferation and progression of prostate cancer via targeting miR‐1253/EZH2. Gene. 2019;686:37–42. 10.1016/j.gene.2018.10.085 [DOI] [PubMed] [Google Scholar]
- 29. Xu Y, Yao Y, Gao P, Cui Y. Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR‐1253 and miR‐1294. Biochem Biophys Res Commun. 2019;509(1):138–42. 10.1016/j.bbrc.2018.12.088 [DOI] [PubMed] [Google Scholar]
- 30. Wang YQ, Xu ZM, Wang XL, Zheng JK, du Q, Yang JX, et al. LncRNA FOXC2‐AS1 regulated proliferation and apoptosis of vascular smooth muscle cell through targeting miR‐1253/FOXF1 axis in atherosclerosis. Eur Rev Med Pharmacol Sci. 2020;24(6):3302–14. 10.26355/eurrev_202003_20698 [DOI] [PubMed] [Google Scholar]
- 31. Liu M, Zhang Y, Zhang J, et al. MicroRNA‐1253 suppresses cell proliferation and invasion of non‐small‐cell lung carcinoma by targeting WNT5A. 2018;9(2):189. 10.1038/s41419-017-0218-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li L, Wan K, Xiong L, et al. CircRNA hsa_circ_0087862 acts as an oncogene in non‐small cell lung cancer by targeting miR‐1253/RAB3D Axis. Onco Targets Ther. 2020;13:2873–86. 10.2147/ott.s243533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mattioli F, Hayot G, Drouot N, Isidor B, Courraud J, Hinckelmann MV, et al. De novo frameshift variants in the neuronal splicing factor NOVA2 result in a common C‐terminal extension and cause a severe form of neurodevelopmental disorder. Am J Hum Genet. 2020;106(4):438–52. 10.1016/j.ajhg.2020.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Giampietro C, Deflorian G, Gallo S, et al. The alternative splicing factor Nova2 regulates vascular development and lumen formation. 2015;6:8479. 10.1038/ncomms9479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gallo S, Arcidiacono MV, Tisato V, et al. Upregulation of the alternative splicing factor NOVA2 in colorectal cancer vasculature. Onco Targets Ther. 2018;11:6049–56. 10.2147/ott.s171678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li G, Huang M, Cai Y, et al. Circ‐U2AF1 promotes human glioma via derepressing neuro‐oncological ventral antigen 2 by sponging hsa‐miR‐7‐5p. J Cell Physiol. 2019;234(6):9144–55. 10.1002/jcp.27591 [DOI] [PubMed] [Google Scholar]
- 37. Tan Z, Cao F, Jia B, et al. Circ_0072088 promotes the development of non‐small cell lung cancer via the miR‐377‐5p/NOVA2 axis. Thorac Cancer. 2020;11(8):2224–36. 10.1111/1759-7714.13529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xiao H. MiR‐7‐5p suppresses tumor metastasis of non‐small cell lung cancer by targeting NOVA2. Cell Mol Biol Lett. 2019;24:60. 10.1186/s11658-019-0188-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li C, Liu H, Niu Q, et al. Circ_0000376, a novel circRNA, promotes the progression of Non‐small cell lung cancer through regulating the miR‐1182/NOVA2 network. Cancer Manage Res. 2020;12:7635–47. 10.2147/cmar.s258340 [DOI] [PMC free article] [PubMed] [Google Scholar]


