Summary
Emergence from the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has been facilitated by the rollout of effective vaccines. Successful vaccines generate high-affinity plasma blasts and long-lived protective memory B cells. Here, we show a requirement for T follicular helper (Tfh) cells and the germinal center reaction for optimal serum antibody and memory B cell formation after ChAdOx1 nCoV-19 vaccination. We found that Tfh cells play an important role in expanding antigen-specific B cells while identifying Tfh-cell-dependent and -independent memory B cell subsets. Upon secondary vaccination, germinal center B cells generated during primary immunizations can be recalled as germinal center B cells again. Likewise, primary immunization GC-Tfh cells can be recalled as either Tfh or Th1 cells, highlighting the pluripotent nature of Tfh cell memory. This study demonstrates that ChAdOx1 nCoV-19-induced germinal centers are a critical source of humoral immunity.
Keywords: germinal center, Tfh cells, CD40L, vaccine, SARS-CoV-2, ChAdOx1 nCoV-19
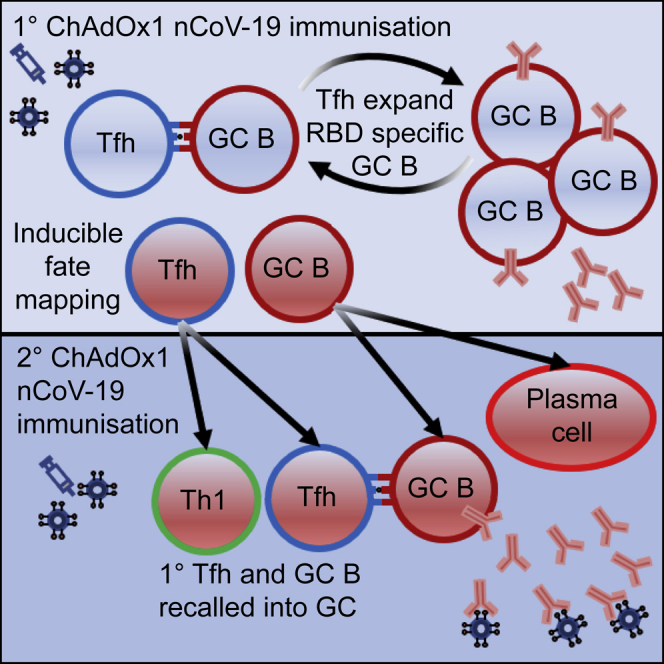
Graphical abstract
Highlights
-
•
Tfh cells are required for RBD+ B cell selection after ChAdOx1 nCoV-19 vaccination
-
•
Absence of GC B cells limits RBD+ B cell expansion and serum immunity
-
•
RBD+ B cells are recalled to the GC during secondary responses
-
•
GC Tfh cells are recalled during secondary responses as Tfh cells or as Th1 cells
Effective vaccines elicit protective humoral immunity. Foster et al. use high-parameter flow cytometry and confocal microscopy to demonstrate that vaccine-induced germinal centers are a critical source of humoral immunity following ChAdOx1 nCoV-19 vaccination.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has resulted in over 6 million confirmed deaths, 620 million confirmed infections,1 and significant global healthcare and economic disruption. The rapid development, mass production, and deployment of safe and effective SARS-CoV-2 vaccines has curtailed pandemic impact, significantly reducing coronavirus 2019 disease (COVID-19) morbidity and mortality. The ChAdOx1 nCoV-19 vaccine (AZD1222) has had two billion doses supplied to more than 170 countries, and, importantly, two thirds of these doses have been made available to low- and middle-income countries, making it a key global vaccine.2 The ChAdOx1 nCoV-19 vaccine is an efficacious vaccine that has been demonstrated to reduce the severity of infection,3 limit the transmissibility of the virus in those who become infected,4 prevent symptomatic infection,5 and, most importantly, prevent hospitalization and death caused by variants of concern.6
ChAdOx1 nCoV-19 is a replication-deficient simian adenoviral vectored vaccine that encodes the SARS-CoV-2 spike protein and stimulates both cellular and antibody-mediated immunity.7,8,9,10,11,12 In humans, neutralizing antibodies and antibody titers to both the SARS-CoV-2 spike protein and its receptor-binding domain (RBD) are associated with protection from symptomatic infection after ChAdOx1 nCoV-19 immunization, consistent with the majority of vaccines currently in use.13,14 In support of the protective role of humoral immunity to COVID-19, real-world observational studies of a SARS-CoV-2 outbreak on a fishing vessel and a longitudinal study of vaccinated healthcare workers has associated breakthrough infections with lower serum antibody titers.15,16 Moreover, animal studies have shown that transferring SARS-CoV-2-specific antibodies can provide direct protection against infection, demonstrating a causal link between antibodies and protection.17,18,19 Therefore, the successful production of humoral immunity is a key function of COVID-19 vaccines.
The humoral immune response consists of antibody-secreting cells and memory B cells, which can be quickly reactivated into plasma cells upon antigen reencounter either by natural infection or vaccination. Antibody-secreting cells and memory B cells can be generated through two pathways: either through the germinal center (GC) reaction or the extrafollicular response.20,21 GCs are microstructures that are generated inside secondary lymphoid tissues following antigen exposure and that provide a specialized niche for B cell proliferation and differentiation.20 Within the GC, antigen-specific B cell clones interact with T follicular helper (Tfh) cells, being cyclically selected and expanded based on antigen affinity. Successful delivery of Tfh-cell-derived help to GC B cells results in the formation of high-affinity, class-switched antibody-secreting cells as well as long-lived memory B cells.20,22 B cell differentiation can also occur in a GC-independent manner, and this process is an important source of humoral immunity. The extrafollicular response allows for more rapid clonal expansion of antigen-specific B cells, resulting in a burst of short-lived antibody-secreting cells as well as memory B cells,23 and can be supported by Tfh cells acting outside the B cell follicle.24 As a result of the differing kinetics of the two responses—the extrafollicular response dominating early and the GC response persisting longer term—it has largely been assumed that both the GC response and Tfh cells are essential for long-lasting humoral immunity to SARS-CoV-2. However, upon SARS-CoV-2 infection, neutralizing antibodies can be formed without Tfh cells and a GC reaction,25 similar to what has been previously reported for influenza A virus infection.26 Likewise, the formation of both immunoglobulin M (IgM)+ and class-switched memory B cells can form in animals that lack Tfh cells and GC B cells,27,28,29 clearly demonstrating redundancy in the mechanisms by which humoral immunity can be made.
Here, the aim of our study was to determine the cellular pathway(s) required for humoral immunity after ChAdOx1 nCoV-19 administration. High-dimensional flow cytometry identifies multiple subsets of RBD-binding B cells, with the majority of cells having a GC phenotype, which, at later time points, transitions into a mix of class-switched (IgD−) GC B cells and memory cells. Immunization of Tfh-cell-deficient mice showed a near absence of RBD-binding B cells and reduced titers of anti-spike and anti-RBD antibodies, with a particular defect in IgG1. The delivery of Tfh cell help to B cells depends upon CD40L, and patients with loss-of-function CD40L mutations have decreased Tfh cell numbers.30,31,32 In a patient with primary immune deficiency due to a mutated CD40L gene, RBD-binding memory B cells and SARS-CoV-2-specific antibodies are undetectable in their circulation, demonstrating that in humans, as in mice, T cell help to B cells is essential for ChAdOx1 nCoV-19-induced humoral immunity. As Tfh cells can also support B cells outside the B cell follicle, we tested the specific requirement for the GC response in facilitating humoral immunity.33,34 Genetic ablation of GC B cells results in a 10-fold loss of RBD-binding B cells, which corresponded with a log-fold reduction in IgG serum antibody levels and neutralizing antibody titers and fewer memory B cells 6 weeks after vaccination. Genetic fate mapping of GC-derived memory B cells and GC-Tfh cells showed that both were recalled after a second immunization, with GC B cells giving rise to antibody-secreting cells and reseeding new GCs. Memory Tfh cells were recalled as both Tfh and Th1 cells, highlighting the pluripotent nature of GC-derived Tfh cells. Together, this demonstrates that the GC reaction and Tfh cells are critical for high-titer antibody responses and for efficient expansion of RBD-specific B cells following ChAdOx1 nCoV-19 immunization, without which humoral immunity is severely restricted.
Results
The class-switched, antigen-specific B cell response evolves following ChAdOx1 nCoV-19 immunization
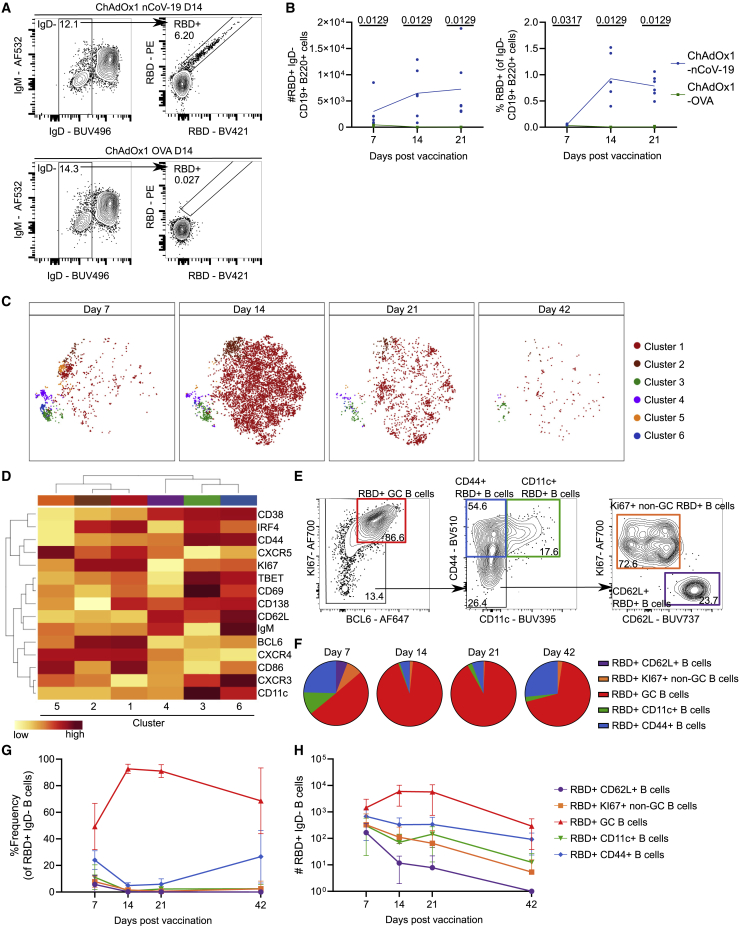
A successful humoral response to vaccination has two outputs: the formation of plasma cells that secrete pathogen-binding antibodies and the generation of memory B cells that can mount rapid recall responses upon (re)infection or booster immunization. The anti-SARS-CoV-2 antibody response to ChAdOx1 nCoV-19 has been well described previously.9,10,12,35 To characterize the antigen-specific B cell response to ChAdOx1 nCoV-19 vaccination, we immunized adult C57BL/6 mice with ChAdOx1 nCoV-19 or ChAdOx1 OVA by intramuscular injection (Figure S1A). Both vaccines induced a GC response that peaked at day 14 (Figures S1B and S1C). IgD− B cells specific for the RBD of the SARS-CoV-2 spike protein were only detected in ChAdOx1 nCoV-19-immunized mice. These increased in number and frequency from day 7 post-immunization (Figures 1A and 1B), as did the number and frequency of RBD-binding GC B cells (Figures S1D and S1E). This demonstrates that ChAdOx1 nCoV-19 vaccination induces an RBD-specific GC B cell response in the draining medial iliac lymph node (mILN).
Figure 1.
ChAdOx1 nCoV-19 immunization generates a mix of RBD-specific GC and memory B cells in the draining medial iliac lymph node
(A) Median flow cytometry plots for IgD− RBD+ B cell staining, pre-gated on live, single, CD19+ B220+ cells.
(B) Total number and relative frequency of IgD− RBD+ B cells.
(C) tSNE analysis of IgD− RBD+ B cells separated by time point. FlowSOM analysis was used to identify 6 clusters of cells.
(D) Heatmap showing mean fluorescence intensity (MFI) of each marker used in (C) for clustering analysis.
(E) Flow cytometry gating of 5 IgD− RBD+ subpopulations based on a concatenated sample of all IgD− RBD+ B cells shown in (C).
(F) Pie charts showing relative frequency of subpopulations identified in (E) for each of the 4 time points.
(G and H) Line graphs showing relative frequency (G) and quantification (H) of subpopulations identified in (E).
Error bars show mean and standard deviation. For each time point and condition, n = 5 or 6 per group. For (B), multiple Mann-Whitney tests per row were used, with p values corrected for multiple comparison analysis with the Holm-Šídák method. Data are representative of two individual experiments.
To characterize the RBD-specific B cell response in detail, we used high-dimensional spectral flow cytometry coupled with t-distributed stochastic neighbor embedding (tSNE) and FlowSOM analysis of IgD− RBD+ B cells. Six tSNE clusters were identified across a 42-day ChAdOx1 nCoV-19 immunization time course (Figures 1C and 1D), five of which could be subsequently identified by manual gating (Figure 1E). As was expected during the first 42 days after vaccination, the largest cluster (tSNE cluster 1) corresponded to the GC B cells, having the highest expression of BCL6 and KI67. Cluster 2 also resembled GC B cells, with decreased expression of CD138 and CD86 compared with cluster 1 (Figure 1D). Despite this, these two clusters were not separable by conventional biaxial flow cytometry analysis using these markers, thus we grouped these two clusters together in further analysis (henceforth called RBD+ GC B cells). Cluster 3 expressed CD44 and CD11c, a phenotype corresponding to atypical memory B cells36 or age-associated B cells37 (CD11c+ RBD+ cells). Cluster 6 was closely related to cluster 3 by hierarchical clustering, with high expression of CD44, but lacked expression of CD11c; this population also increased over time (Figures 1F–1H), indicating that it is likely to be a pool of RBD-specific memory B cells (CD44+ RBD+ B cells). Clusters 4 and 5 were the rarest two subsets and were primarily found at day 7 after vaccination (Figures 1F–1H). Cluster 4 was enriched for CD62L, CD38, and IgM, indicative of a recently activated pool of B cells (CD62L+ RBD+ B cells). Cluster 5 had low expression of CD38 and expressed KI67, being hierarchically most similar to the two GC clusters (Figure 1D, clusters 1 and 2), likely representing early proliferating B cells and possible GC B cell precursors (KI67+ non-GC B cells). The spleen is also a reactive lymphoid organ following intramuscular ChAdOx1 nCoV-19 vaccination,7,8,10 and we identified similar populations of RBD-binding B cells in the spleen after immunization (Figures S2A–S2E).
For each of the four time points analyzed, we elucidated the composition, frequency, and total number of the RBD+ IgD− B cell pool’s constituent subsets using the manually gated subsets identified by tSNE analysis (Figures 1F–1H and S2F–S2H). The GC B cell population was the dominant population at all time points analyzed, indicating that the majority of RBD-binding B cells participate in the GC reaction. The CD62L+ and KI67+ non-GC RBD+ B cell subsets were present primarily at day 7 after immunization and declined to very low numbers 42 days after immunization, indicative of an early-activated B cell phenotype. The CD44+ RBD+ B cell subset was present at a higher frequency both before and after the GC peak at days 7 and 42, respectively, and its retention at day 42 indicates a memory phenotype (Figures 1G, 1H, S2G, and S2H). Likewise, the CD11c+ RBD+ B cell population is retained at day 42, consistent with an atypical memory B cell phenotype. Collectively, these data demonstrate that after immunization, the antigen-specific B cell pool evolves over time, with the GC being the dominant compartment of RBD-binding B cells with memory B cell populations observed at later time points.
Tfh cells are required for antigen-specific GC B cell formation and serum immunity
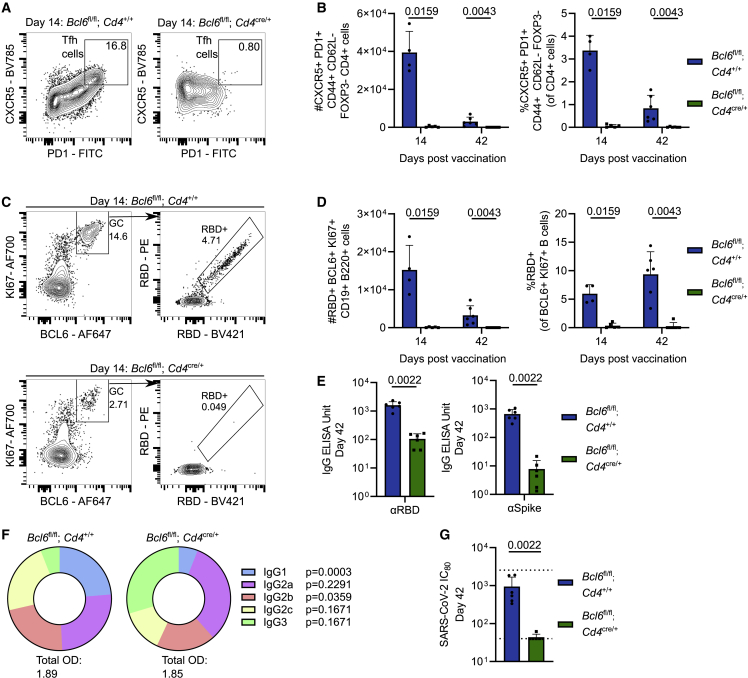
The production of vaccine-reactive memory B cells and antibodies can be dependent on help from Tfh cells. Tfh cells can support the B cell response outside the GC, for example in facilitating class-switch recombination and memory B cell formation,24,29 and within the GC, where they support the positive selection and clonal expansion of GC B cells. To test the role for Tfh cells in generating antibodies and RBD-binding B cells upon ChAdOx1 nCoV-19 immunization, Bcl6fl/fl Cd4cre/+ mice were analyzed 14 and 42 days post-vaccination. Tfh cells formed in littermate control Bcl6fl/fl mice but not in Bcl6fl/fl Cd4cre/+ animals (Figures 2A and 2B), and the absence of Tfh cells was coupled with a near absence of RBD-specific GC B cells (Figures 2C and 2D). The combined loss of Tfh cells and GC B cells resulted in an order-of-magnitude-reduced titer of anti-RBD and anti-spike IgG 42 days after immunization (Figure 2E). Analysis of the subclasses of anti-RBD IgG that did form upon immunization revealed a defect in the formation of anti-RBD IgG1 in Bcl6fl/fl Cd4cre/+ mice, indicating a particular importance for Tfh cells in supporting class switch to IgG1 upon ChAdOx1 nCoV-19 immunization (Figure 2F). Consistent with a reduced titer of RBD-binding antibodies, we observed an impaired capacity of serum from Bcl6fl/fl Cd4cre/+ mice to neutralize pseudotyped SARS-CoV-2 infection in vitro (Figure 2G).
Figure 2.
Tfh cells are required for efficient RBD-specific GC B cell generation and serum immunity
Bcl6fl/flCd4+/+ and Bcl6fl/flCd4cre/+ mice were immunized with ChAdOx1 nCoV-19 intramuscularly, with tissues taken for analysis at indicated time points.
(A) Day 14 median flow cytometry plots for mILN Tfh cell staining, pre-gated on live, single, CD4+ FOXP3−, CD44+ CD62L− cells.
(B) Total number and relative frequency of mILN Tfh cells after immunization at indicated time points.
(C) Day 14 median flow cytometry plots for mILN RBD+ GC B cell staining pre-gated on live, single, CD19+ B220+ cells.
(D) Total number and relative frequency of mILN RBD+ GC B cells after immunization at indicated time points.
(E) Serum anti-spike and anti-RBD IgG antibodies at day 42 post-immunization.
(F) Pie charts indicating the mean abundance of each IgG antibody subclass in the serum at the indicated time points after immunization.
(G) SARS-CoV-2 neutralizing antibody titers in sera were determined by micro-neutralization test, expressed as reciprocal serum dilution to inhibit pseudotyped virus entry by 80% (IC80). Dashed lines represent upper and lower detection limits.
For each time point and condition, n = 4–6, respectively, per group. For (B), (D), and (F), multiple Mann-Whitney tests per row were used, with p values corrected for multiple comparison analysis with the Holm-Šídák method. For (E) and (G), a Mann-Whitney test was used. In bar graphs, each symbol represents a biological replicate, bar height the mean, and the error bars the standard deviation. Data are representative of two individual experiments.
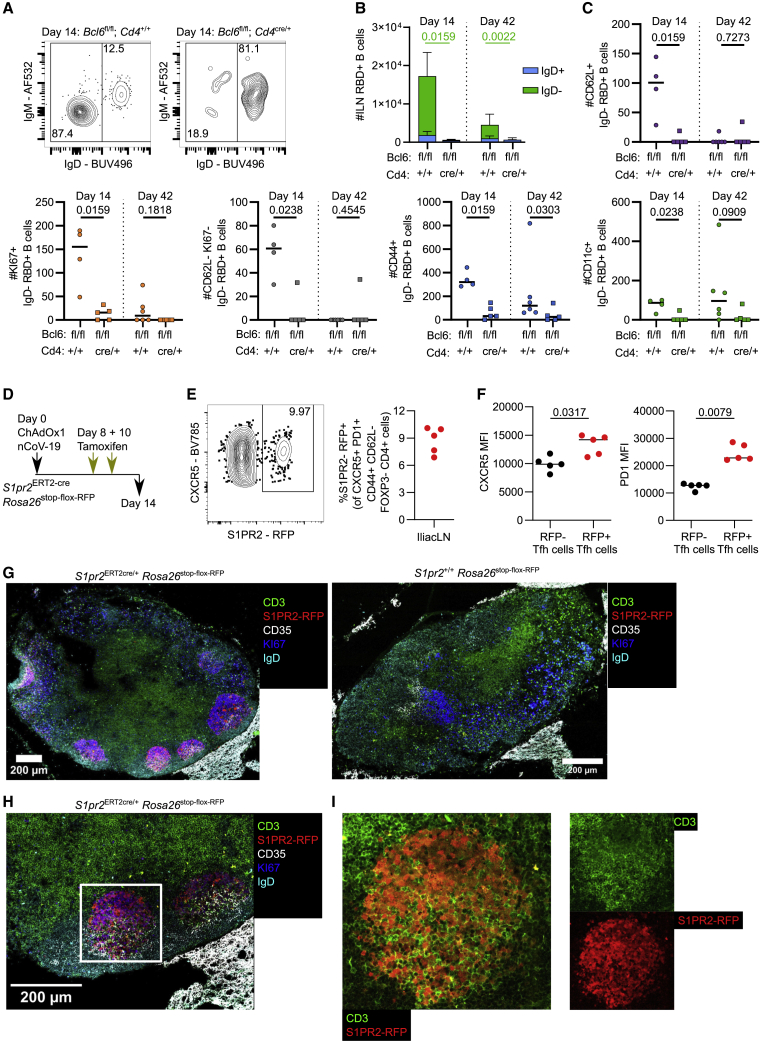
We next employed our RBD probes and multidimensional flow cytometry to understand the importance of Tfh cells in the generation of individual RBD-binding B cell subsets, specifically to understand which subsets, if any, were Tfh cell dependent. Mice lacking Tfh cells had a 90× decreased frequency of class-switched (IgD−), RBD-binding B cells at day 14 and 170-fold decreased frequency at day 42 in the mILN (Figures 3A and 3B). We then profiled the individual subsets of non-GC IgD− RBD-binding B cells using the same gating strategy as Figure 1E. Fourteen days after immunization, all non-GC RBD-binding B cell subsets in the mILN were reduced in mice lacking Tfh cells, with a deficit in CD44+ memory B cells 6 weeks after immunization (Figure 3C). In contrast to the mILN, splenic RBD-binding non-GC B cells were not significantly reduced in Tfh-cell-deficient mice, despite a clear loss of splenic GC B cells (Figures S3A–S3C). This was due to an increase in IgM+ IgD− RBD-binding B cells that were more abundant in the spleen than the mILN after immunization. These data collectively demonstrate that Tfh cells play an important role in the generation of anti-SARS-CoV-2 antibodies upon immunization. Within the mILN, Tfh cells are critical for most RBD-binding B cell subsets. In the spleen, however, a higher proportion of RBD-binding B cells are measured across various non-GC phenotypes independent of Tfh cell activity (Figure S3).
Figure 3.
Tfh cells are required for all IgD− RBD-specific B cell subsets, except CD11c+ B cells in the draining medial iliac lymph node
(A) Day 14 median flow cytometry plots for IgD staining of RBD+ B cells, pre-gated on live, single, CD19+ B220+, RBD+ cells.
(B) Total number of RBD+ B cells at indicated time point, p value shown is from comparison of the number of IgD− RBD+ cells, bar height shows the mean, and the error bars the standard deviation.
(C) IgD− RBD+ B cell subsets were enumerated using the gating strategy as shown in Figure 1E.
(D) Schematic of experimental setup: S1pr2ERT2−creRosa26stop-flox-RFP mice were immunized with ChAdOx1 nCoV-19 intramuscularly, followed by tamoxifen oral gavage at 8 and 10 days post-immunization.
(E) Relative frequency of S1PR2-RFP fate-mapped Tfh cells at day 14.
(F) CXCR5 and PD1 MFI of S1PR2-RFP− and S1PR2-RFP+ fate-mapped Tfh cells, respectively.
(G) Day 14 confocal microscopy from S1pr2ERT2-creRosa26stop-flox-RFP and S1pr2+/+Rosa26stop-flox-RFP mice.
(H) 40× objective zoom of highlighted area of (G).
(I) Zoom of highlighted area of (H) showing RFP and CD3 channels together and RFP/CD3 channels individually.
For (B) and (C), multiple Mann-Whitney tests per row were used, with p values corrected for multiple comparison analysis with the Holm-Šídák method. For (F), Mann-Whitney tests were used. In dot plots, each symbol represents a biological replicate and the bar height the mean. Data are representative of two individual experiments.
Because Tfh cells can act both within and outside the GC, we next sought to understand the abundance of GC-Tfh cells after ChAdOx1 nCoV-19 immunization. We employed an S1PR2-inducible RFP fate-mapper system in which GC-Tfh cells can be labeled upon tamoxifen administration.33 Between 7% and 10% of mILN Tfh cells are RFP labeled after ChAdOx1 nCoV-19 vaccination when tamoxifen was given at days 8 and 10 post-vaccination (Figures 3D and 3E). Consistent with a GC Tfh phenotype, these RFP+ Tfh cells had higher expression of CXCR5 and PD1 than the RFP− Tfh cells from the same animal (Figure 3F). We also used confocal microscopy in order to confirm that S1PR2-RFP fate-mapped cells were found primarily within LN secondary follicles (Figures 3G and S4). We show that RFP-expressing cells are found within both the light zone of the GC (demarcated by CD35+ follicular dendritic cells) and the dark zone of the GC (Figure 3H). The GCs also contained CD3+ T cells that had internal RFP signals, although not all GC CD3+ T cells were fate mapped using this approach (Figure 3I). Tfh cells therefore support effective humoral response to ChAdOx1 nCoV-19 vaccination, with their loss leading to a reduction in all antigen-specific B cell subsets, which may occur inside or outside the GC.
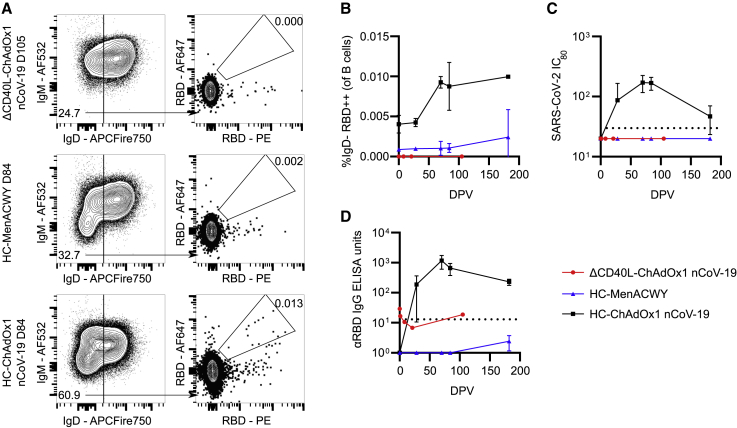
Human CD40L deficiency results in absence of RBD-specific circulating B cells and humoral immunity after ChAdOx1 nCoV-19 vaccination
To determine whether, like in the mouse, Tfh cell help is required for humoral immunity upon ChAdOx1 nCoV-19 vaccination, we examined the role of Tfh cells in a cohort of human volunteers. We longitudinally collected peripheral blood mononuclear cells (PBMCs) and serum from healthy controls (HCs), as well as a patient with primary immunodeficiency as a result of CD40L deficiency, after they received their second of two ChAdOx1 nCoV-19 vaccinations. We compared this patient and the HCs with a healthy control group who instead received the MenACWY vaccine. RBD-specific IgD− B cells were readily detectable in healthy controls vaccinated with ChAdOx1 nCoV-19 up to 182 days post-vaccination and were entirely absent in the person with CD40L deficiency (Figures 4A and 4B). Consistent with this observation, only serum from healthy controls vaccinated with ChAdOx1 nCoV-19 had SARS-CoV-2-neutralizing capacity and high RBD-specific IgG titers (Figures 4C and 4D). Thus, in both humans and mice, T cell help to B cells supports the generation of class-switched, RBD-binding B cells and antibody-mediated immunity to the ChAdOx1 nCoV-19 vaccine.
Figure 4.
Tfh-cell-mediated CD40L signaling is required for circulating RBD+ B cells and serum immunity in humans
PBMC and serum were collected from a patient with a loss-of-function CD40L mutation (ΔCD40L) or from 3 healthy controls (HCs) after their second ChAdOx1 nCoV-19 vaccination or from 3 healthy controls who received their second MenACWY (Nimenrix) vaccine at indicated timepoints.
(A) Median flow cytometry plots for circulating RBD+ IgD− B cell staining, pre-gated on live, single, CD19+ B220+ cells at indicated time points for respective groups.
(B) Frequency of IgD− RBD+ B cells in the blood.
(C) SARS-CoV-2 neutralizing antibody titers in sera were determined by micro-neutralization test, expressed as reciprocal serum dilution to inhibit pseudotyped virus entry by 80% (IC80). Dashed line represents lower detection limit.
(D) Serum anti-RBD IgG antibodies were quantified by ELISA.
In (B)–(D), symbols represent the mean, and error bars represent the standard deviation for the healthy controls that received either ChAdOx1 nCoV-19 (black) or MenACWY (blue) vaccines.
Loss of GC B cells compromises antibody and memory B cell formation after ChAdOx1 nCoV-19 immunization
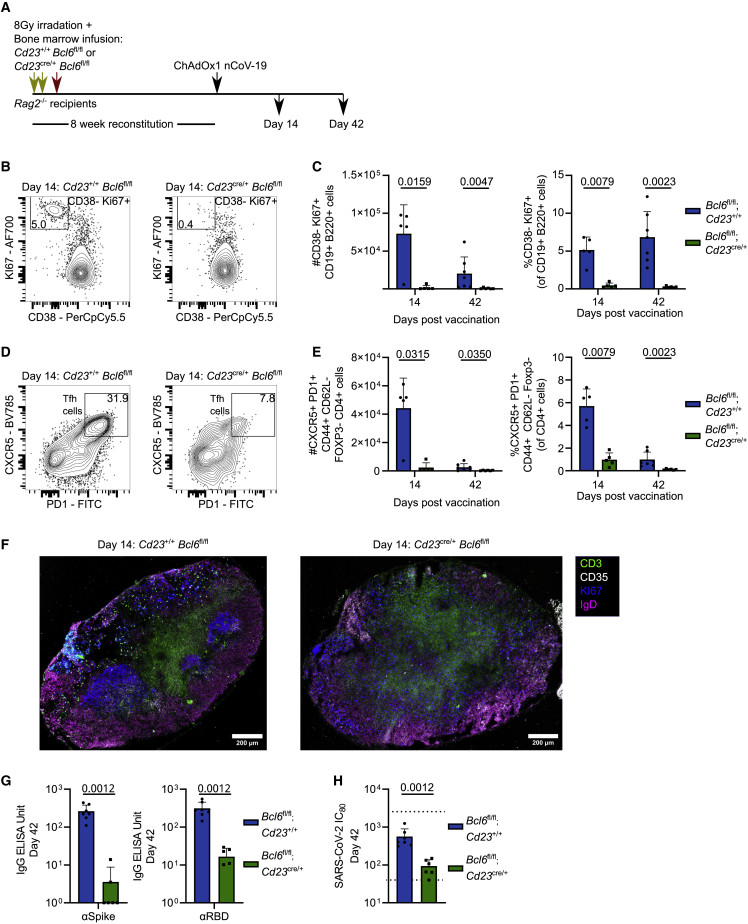
As our data indicated that GC B cells are the dominant RBD-binding subset but that only a proportion of Tfh cells are GC localized after immunization, we wanted to determine the contribution of GC B cells to the antibody- and RBD-binding B cell response to ChAdOx1 nCoV-19 immunization. In order to generate mice that cannot form GC B cells upon immunization, we reconstituted irradiated Rag2−/− recipient mice with Cd23cre/+ Bcl6fl/fl bone marrow (Figure 5A). Bcl6 is absolutely essential for GC B cell formation,38,39,40 and therefore Cd23cre/+ Bcl6fl/fl mice do not form GC B cells due to B cells’ intrinsic lack of BCL6 (Figures 5B and 5C). This is coupled with a near absence of Tfh cells, because of the interdependence of Tfh and GC B cells (Figures 5D and 5E). In Cd23cre/+ Bcl6fl/fl mice, GC-containing secondary lymphoid follicles were absent, leading to a scattering of KI67+ B cells, suggesting that any remaining Tfh cells in these animals act in an extrafollicular manner24 (Figures 5F and S5). Serum analysis showed that the anti-spike IgG, anti-RBD IgG, and neutralizing antibody response was almost completely dependent on the GC reaction (Figures 5G and 5H), highlighting the importance of this structure for generating antibody-mediated immunity upon vaccination.
Figure 5.
GC B cells are required for effective serum immunity following ChAdOx1 nCoV-19 vaccination
(A) Rag2−/− recipient bone marrow chimera mice experiment overview.
(B) Day 14 median flow cytometry plots for mILN GC B cell staining (gated as KI67+ CD38−), pre-gated on live, single, CD19+ B220+ cells.
(C) Total number and relative frequency of mILN GC B cells after immunization at indicated time points.
(D) Day 14 median flow cytometry plots for mILN TFH cell staining, pre-gated on live, single, CD4+ FOXP3−, CD44+ CD62L− cells.
(E) Total number and relative frequency of mILN TFH cells after immunization at indicated time points.
(F) Day 14 confocal microscopy of mILNs from Cd23+/+Bcl6fl/fl and Cd23cre/+Bcl6fl/fl mice.
(G) Serum anti-spike and anti-RBD IgG antibodies at day 42 post-immunization.
(H) SARS-CoV-2 neutralizing antibody titers in sera were determined by micro-neutralization test, expressed as reciprocal serum dilution to inhibit pseudotyped virus entry by 80% (IC80). Dashed lines represent upper and lower detection limits.
For each time point and condition, n = 5–7, respectively, per group. For (C) and (E), multiple Mann-Whitney tests per row were used, with p values corrected for multiple comparison analysis with the Holm-Šídák method. For (G) and (H), Mann-Whitney tests were used. In bar graphs, each symbol represents a biological replicate, bar height the mean, and the error bars the standard deviation. Data are representative of two individual experiments.
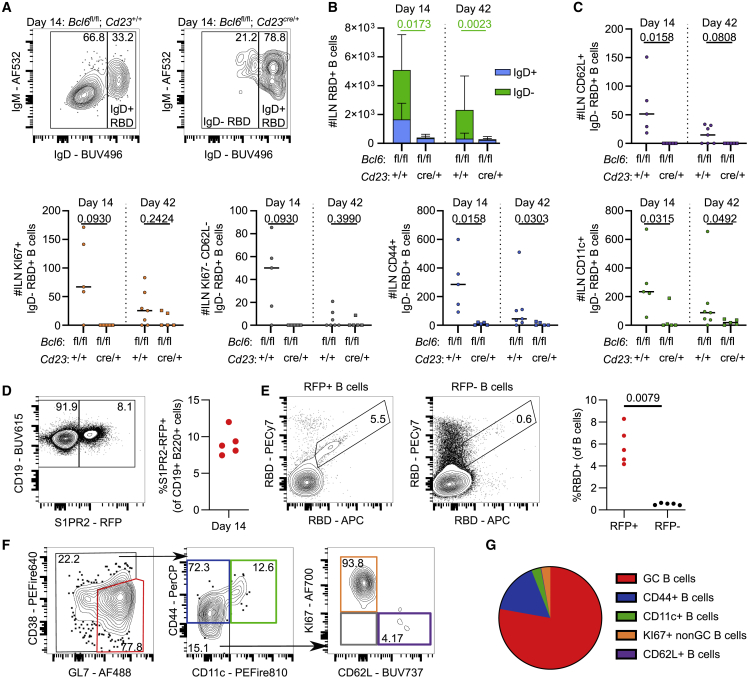
In cases where antibody titers are low, memory B cells can act as a second source of antibody-producing cells upon infection or booster immunization.41,42 To determine which RBD-specific B cell populations can exist in the absence of a GC reaction, we assessed the RBD-binding B cell subsets identified in Figure 1 in the mILN and spleen of Cd23cre/+ Bcl6fl/fl mice 14 and 42 days after ChAdOx1 nCoV-19 immunization. The inability to mount a GC B cell response resulted in a 10-fold reduction in the frequency and number of IgD− RBD-binding B cells in the mILN (Figures 6A and 6B), with all non-GC RBD+ B cell subsets being reduced in Cd23cre/+ Bcl6fl/fl mice (Figure 6C). Likewise, in the spleen, we observed a reduction in the numbers of RBD-binding B cells in the Cd23cre/+ Bcl6fl/fl mice compared with controls (Figures S6A and S6B). The CD44− subsets (KI67+ CD62L+ and CD62L− KI67−) were more abundant in the early phases of a vaccine response (Figures 1G and S1G) but were not significantly different between the Cd23cre/+ Bcl6fl/fl and Cd23+/+ Bcl6fl/fl mice, suggesting that they do not need a functioning GC B cell population to form in the spleen. Unlike in the mILN, the CD11c+ atypical B cell population was not significantly different between the two groups, indicating that splenic CD11c+ memory B cells are not absolutely dependent upon the GC for their formation. By contrast, the CD44+ memory B cell population was significantly reduced, suggesting that these cells are likely progeny of the GC reaction (Figure S6C). To further characterize the GC-derived memory B cells, we used the S1PR2 genetic fate-labeling strategy that marks GC B cells upon tamoxifen administration; after a primary immunization, around 5% of B cells were S1PR2 fate mapped (Figure 6D). The majority of RBD-binding B cells were found within the S1PR2 fate-mapped B cell pool (Figure 6E), and these S1PR2-labeled RBD+ B cells were primarily of GC (GL7+ CD38−) or CD44+ memory phenotype (Figures 6F and 6G). Thus, loss of GC B cells results in a significant reduction in antibody production and a loss of class-switched, RBD-binding B cells following vaccination. Despite this, some antigen-specific B cells are still produced; for example, CD11c+ atypical B cells within the spleen are still generated, albeit at reduced numbers, indicating that not all vaccine-reactive B cell subsets are dependent on the successful formation of the GC.
Figure 6.
GC B cells are required for IgD− RBD+ B cell amplification and can be S1PR2 fate mapped
(A) Day 14 median flow cytometry plots for IgD staining of mILN RBD+ B cells, pre-gated on live, single, CD19+ B220+, RBD+ cells.
(B) Total number of RBD+ B cells at indicated time points. p value shown is from comparison of the number of IgD− RBD+ cells, bar height shows the mean, and the error bars the standard deviation.
(C) IgD− RBD+ B cell subsets were enumerated using the gating strategy as shown in Figure 1E.
(D) Day 14 median flow cytometry plot and relative frequency of S1PR2-RFP+ fate-mapped B cells.
(E) Median flow cytometry plots of RBD staining and quantification of RBD+ B cells in S1PR2-RFP+ fate-mapped and S1PR2-RFP− fate-mapped populations, respectively.
(F) Median flow cytometry plots of RBD+ S1PR2-RFP+ fate-mapped B cell subsets.
(G) Mean frequency of RBD+ S1PR2-RFP+ fate-mapped B cell subsets.
For each time point and condition, n = 5–7, respectively, per group. For (B) and (C), multiple Mann-Whitney tests per row were used, with p values corrected for multiple comparison analysis with the Holm-Šídák method. For (E), a Mann-Whitney test was used. In dot plots, each symbol represents a biological replicate and the bar height the mean. Data are representative of two individual experiments
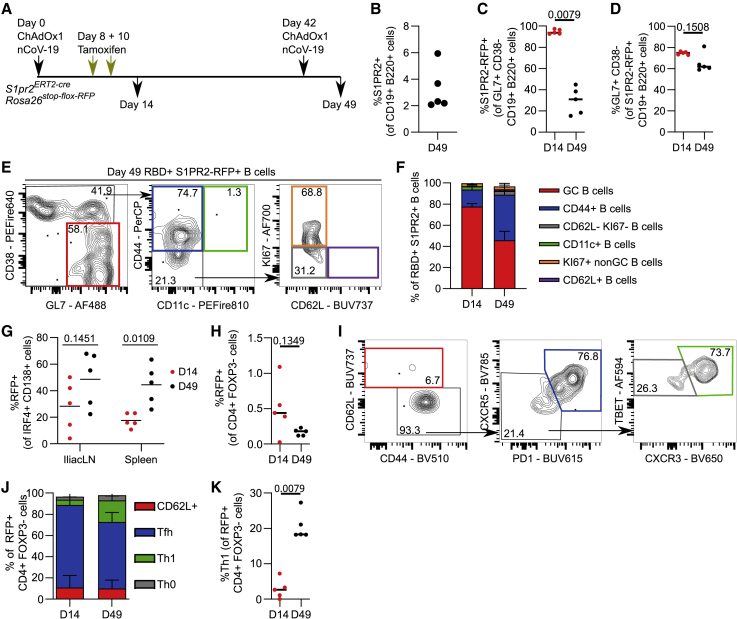
GC B cells can be recalled into secondary GCs and GC-Tfh cells can be recalled as Th1 cells following ChAdOx1 nCoV-19 prime-boost immunization
To determine how these GC-derived memory B cells are recalled upon booster immunization, we assessed the fate of S1PR2 fate-mapped cells after a prime-boost immunization strategy (Figure 7A). One week post-boost, RFP+ B cells were detectable (Figure 7B) and unlike the primary immunization, in which most of the GC B cells were RFP labeled, this was not the case—suggesting that the secondary GC is indeed primarily seeded by “new” cells (Figure 7C), consistent with previous reports.43 It should be noted, however, that the majority of RFP-labeled B cells present at day 49 did take on a GC B cell phenotype (Figure 7D), suggesting although most of the cells forming secondary GCs are not rerecruited cells, the main fate of memory B cells that do not differentiate into plasma cells is to reenter the GC. The RFP-labeled RBD+ IgD− B cell subsets consisted of GC and CD44+ CD11c− GC-derived memory B cell subsets after both prime and boost vaccinations (Figures 7E and 7F). We also explored the generation of S1PR2-RFP+ plasma cells and found that in the spleen, a significantly greater proportion of plasma cells were RFP+ after boost compared with primary immunization (Figure 7G). This suggests that memory cells that were produced during the primary response play an important role generating plasma blasts in subsequent responses, facilitating the rapid production of antibodies upon booster immunization. The S1PR2 fate-mapping system enabled us to track ex-GC-Tfh cells and determine their contribution to booster immunization; ≈0.4% of all CD4+ Foxp3− cells were S1PR2 fate mapped, with a similar proportion maintaining labeling during the recall response (Figure 7H). The majority of Foxp3− CD4 T cells labeled in the primary response were Tfh cells (Figure 7I), while in the secondary response, these were recalled as either Tfh or as Th1 cells, indicating that GC-Tfh cells form multipotent memory cells that can be recalled (Figures 7J and 7K).
Figure 7.
S1PR2 fate-mapped GC B cells and GC Tfh cells are recalled in boost immunization as GC B cells, GC-derived CD44+ memory B cells and plasma cells, or Tfh and Th1 cells, respectively
(A) S1pr2ERT2−creRosa26stop-flox-RFP mice recall experiment overview.
(B) Relative frequency of S1PR2-RFP+ fate-mapped mILN B cells at day 49.
(C) Relative frequency of S1PR2-RFP+ fate-mapped mILN GC B cells.
(D) Relative frequency of GC phenotype within S1PR2-RFP+ fate-mapped mILN B cells.
(E) Median flow cytometry plots of RBD+ S1PR2-RFP+ fate-mapped mILN B cell subsets.
(F) Relative frequency of RBD+ S1PR2-RFP+ fate-mapped mILN B cell subsets at days 14 and 49, respectively.
(G) Relative frequency of S1PR2-RFP+ fate-mapped plasma cells.
(H) Relative frequency of S1PR2-RFP+ fate-mapped CD4+ FOXP3− cells.
(I) Day 49 median flow cytometry plots of S1PR2-RFP+ fate-mapped mILN FOXP3− CD4+ cell subsets.
(J) Relative frequency of S1PR2-RFP+ fate-mapped mILN FOXP3− CD4+ cell subsets.
(K) Relative frequency of S1PR2-RFP+ fate-mapped mILN Th1 (TBET+ CXCR3+ CXCR5− PD1− CD44+ CD62L− FOXP3− CD4+) cells.
For each time point, n = 5. For (C), (D), (H), and (K), the Mann-Whitney test was used. For (G), multiple Mann-Whitney tests per row were used, with p values corrected for multiple comparison analysis with the Holm-Šídák method. In (B)–(D), (G), (H), and (K), each symbol represents a biological replicate and the bar height the mean. In (F) and (J), bar heights represent mean, and error bars represent the standard deviation. Data are representative of two individual experiments.
Discussion
In this study, we used a combination of antigen-specific B cell staining and analysis of serum following ChAdOx1 nCoV-19 vaccination to demonstrate that a functional GC is required for effective humoral immunity to ChAdOx1 nCoV-19 immunization. When either Tfh or GC B cells are removed, RBD-specific cells are greatly reduced in frequency, and the functional capacity of the resultant serum is diminished by two orders of magnitude. These observations in mice were recapitulated in humans; a patient with CD40L null mutation had no detectable RBD-specific B cells or serum neutralizing activity following vaccination; this supports the concept that Tfh cell functionality is required for humoral immunity to ChAdOx1 nCoV-19. Genetic fate-mapping experiments showed the recall of both GC B and GC-Tfh cells in subsequent booster vaccinations. The majority of S1PR2+ GC B cells are recalled as GC B cells once again or are differentiated into antibody-secreting plasma cells, while GC-Tfh cells were recalled in a pluripotent capacity as either Tfh cells again or as Th1 cells. Together, this study identifies the key role for Tfh cells and GC B cells as central mediators of humoral immunity upon vaccination.
Tfh cells are thought to be essential for GC formation and normal antibody responses following immune challenges.44 However, recent reports for SARS-CoV-2 infection, mRNA-based vaccination for SARS-CoV-2, and work using influenza A virus infection have shown the successful production of antibody in a Tfh-cell-deficient setting, calling into question their indispensability.25,26 Such antibodies are likely produced through the extrafollicular pathway, and the B cells responsible may derive their T cell help from Th1 cells, as has been shown for viral infection in the lung.45 Here, we likewise observe that splenic IgM+ RBD-binding B cells can form in the absence of Tfh cells. Our data, in both humans and mice, demonstrate that Tfh cells promote the selection and amplification of B cells specific for the immunogenic RBD domain required for SARS-CoV-2 cell entry.46 Thus, our work builds on that of Chen et al.,25 demonstrating that after immunization, although a Tfh-cell-independent antibody can be generated, it is of a lower titer compared with an antibody produced in Tfh-cell-competent mice and thus provides less neutralizing capacity in our in vitro assays. We further demonstrate that Tfh cells are required to expand IgD− RBD-binding B cells after vaccination.
Extrafollicular responses are seen in patients with chronic autoimmunity, and patients with severe SARS-CoV-2 infection also show evidence of increased extrafollicular B cell activity.47 These patients are characterized with having an expansion of CD11c+ B cells; such cells are seen to have anti-viral properties in mouse models and are similarly expanded during advanced age.36,48 CD11c+ memory B cells are likely part of the normal B cell response to vaccination, being present in both healthy individuals as well as those with malaria.49 RBD-specific CD11c+ B cells were present, in reduced numbers, in both Cd4cre/+ Bcl6fl/fl and Cd23cre/+ Bcl6fl/fl murine models described here, indicating it is possible for these cells to be of extrafollicular origin despite a previous report that these cells are Tfh cell dependent.50 Our S1PR2-fatemapping studies also show CD11c+ memory B cells can derive from GC B cell precursors, consistent with our recent human infleunza vaccination study.51 Taken together, these findings demonstrate that CD11c+ memory B cells can arise from both the extrafollicular and GC pathways. These cells were also present in the spleen, suggesting that these atypical memory B cells may be disseminating through the circulation and accumulating in the spleen, as reported in aged mice.52
In this study, we used an ipsilateral boost, and consistent with another recent report, we found that primary-vaccination-induced GC B cells can be recalled into secondary GCs.53 Previous publications have shown that while memory B cells can reseed GCs during subsequent challenges,54 primary GC B cells reentering a secondary GC is a relatively rare occurrence.43 Our data corroborate this; here, while a majority of labeled primary GC B cells were recalled as secondary GC B cells, they represented a minority of total secondary GC B cells. Thus, the majority of cells seeding the GC were new to the GC environment, and memory B cells are rapidly recalled to become antibody-secreting cells upon booster immunization.
In summary, our data suggest that the GC is indispensable for the selection and proliferation of antigen-specific B cells following ChAdOx1 nCoV-19 vaccination. Tfh cells support this GC process, and loss of a Tfh-supported GC results in a lowered serum titer and neutralization capacity in both mice and humans. Genetic fate mapping demonstrates that GC B cells from primary vaccination are recalled again as GC B cells, GC-derived memory B cells, or plasma cells, while GC-Tfh cells are recalled as either Tfh or Th1 cells. Together, this demonstrates that the production of humoral immunity to ChAdOx1 nCoV-19 requires effective GC activation in which antigen-specific cells are selected and expanded. Thus, further vaccine development should aim to optimize GC responses, for example through use of adjuvants55 and the use of vaccine vectors, which produce long-lived persistent memory responses.
Limitations of the study
A limitation of the work presented here is the lack of viral challenge following vaccination to test vaccine efficacy. We are unable to measure vaccination response metrics such as the contribution vaccine-induced effector CD8 T cells make to viral clearance following subsequent infection. Rather, here, we measure antibody titer and function as a readout of humoral immunity as this is the focus of this study.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BUV661 Rat Anti-Mouse CD19 | BD Biosciences | RRID: AB_2870243 |
| APC/Fire™ 810 anti-mouse/human CD45R/B220 Antibody | BioLegend | RRID: AB_2860603 |
| Alexa Fluor® 647 anti-mouse/human Bcl-6 Antibody | BioLegend | RRID: AB_2565299 |
| Alexa Fluor® 700 anti-mouse Ki-67 Antibody | BioLegend | RRID: AB_2564285 |
| BUV496 Rat Anti-Mouse IgD | BD Biosciences | Custom conjugation |
| IgM Monoclonal Antibody (II/41) | Thermo Fisher Scientific | RRID: AB_467582 |
| Alexa Fluor® 594 anti-mouse CD38 Antibody | BioLegend | RRID: AB_2566435 |
| Pacific Blue™ anti-IRF4 Antibody | BioLegend | RRID: AB_2814497 |
| Brilliant Violet 510™ anti-mouse/human CD44 Antibody | BioLegend | RRID: AB_2650923 |
| Brilliant Violet 785™ anti-mouse CD185 (CXCR5) Antibody | BioLegend | RRID: AB_2563981 |
| PE/Dazzle™ 594 anti-T-bet Antibody | BioLegend | RRID: AB_2565677 |
| PE/Cyanine5 anti-mouse CD69 Antibody | BioLegend | RRID: AB_313113 |
| Brilliant Violet 711™ anti-mouse CD138 (Syndecan-1) Antibody | BioLegend | RRID: AB_2562571 |
| BUV737 Rat Anti-Mouse CD62L | BD Biosciences | RRID: AB_2870155 |
| APC anti-mouse CD184 (CXCR4) Antibody | BioLegend | RRID: AB_2562785 |
| Brilliant Violet 605™ anti-mouse CD86 Antibody | BioLegend | RRID: AB_11204429 |
| PerCP/Cyanine5.5 anti-mouse CD183 (CXCR3) Antibody | BioLegend | RRID: AB_1186017 |
| BUV395 Hamster Anti-Mouse CD11c | BD Biosciences | RRID: AB_2738580 |
| CD279 (PD-1) Monoclonal Antibody (RMP1-30), FITC | Thermo Fisher Scientific | RRID: AB_465467 |
| FOXP3 Monoclonal Antibody (FJK-16s), PE-Cyanine5.5 | Thermo Fisher Scientific | RRID: AB_11218094 |
| PE/Fire(TM) 640 anti-mouse CD4 antibody | BioLegend | RRID: AB_2860585 |
| PerCP/Cyanine5.5 anti-mouse CD38 antibody | BioLegend | RRID: AB_2563333 |
| BUV615 Rat Anti-Mouse CD279 (PD-1) | BD Biosciences | RRID: AB_2875871 |
| Alexa Fluor® 594 anti-T-bet Antibody | BioLegend | RRID: AB_2728474 |
| Brilliant Violet 650™ anti-mouse CD183 (CXCR3) Antibody | BioLegend | RRID: AB_2563160 |
| BUV615 Mouse Anti-Human CD19 | BD Biosciences | RRID: AB_2875287 |
| Spark NIR™ 685 anti-human CD20 Antibody | BioLegend | RRID: AB_2860775 |
| APC/Fire™ 750 anti-human IgD Antibody | BioLegend | RRID: AB_2616988 |
| Alexa Fluor® 488 anti-mouse/human GL7 Antigen (T and B cell Activation Marker) Antibody | BioLegend | RRID: AB_2563285 |
| PE/Fire™ 640 anti-mouse CD38 Antibody | BioLegend | RRID: AB_2890673 |
| PE/Fire™ 810 anti-mouse CD11c Recombinant Antibody | BioLegend | RRID: AB_2904307 |
| PerCP anti-mouse/human CD44 | BioLegend | RRID: AB_10639933 |
| Alexa Fluor® 700 anti-mouse IgD Antibody | BioLegend | RRID: AB_2563341 |
| Pacific Blue™ anti-mouse CD3 Antibody | BioLegend | RRID: AB_493645 |
| Purified anti-mouse CD279 (PD-1) Antibody | BioLegend | RRID: AB_313418 |
| Ki-67 Monoclonal Antibody (SolA15), FITC, eBioscience™ | Thermo Fisher Scientific | RRID: AB_11151330 |
| Goat anti-Rat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 647 | Thermo Fisher Scientific | RRID: AB_141778 |
| CD21/CD35 Monoclonal Antibody (eBio8D9 (8D9)), Biotin, eBioscience™ | Thermo Fisher Scientific | RRID: AB_466390 |
| Goat Anti-Mouse IgG- Alkaline Phosphatase conjugate | Sigma-Aldrich | RRID: AB_11212223 |
| Goat anti-mouse IgG1-Alkaline Phosphatase | Southern Biotech | RRID: AB_2794425 |
| Goat anti-mouse IgG2a-Alkaline Phosphatase | Southern Biotech | RRID: AB_2794494 |
| Goat Anti-mouse IgG2b-Alkaline Phosphatase | Southern Biotech | RRID: AB_2794541 |
| Goat anti-mouse IgG2c-Alkaline Phosphatase | Southern Biotech | RRID: AB_2794461 |
| Goat anti-mouse IgG3-Alkaline Phosphatase | Abcam | RRID: AB_10674160 |
| Goat Anti-Human IgG Antibody, Alkaline Phosphatase conjugate | Sigma-Aldrich | RRID: AB_92427 |
| Bacterial and virus strains | ||
| DH5α Competent Cells | Thermo Fisher Scientific | Cat#: 18265017 |
| ChAdOx1 nCoV-19 | this manuscript | Van Doremalan Nature volume 586, pages 578–582(2020) |
| ChAdOx1 OVA | this manuscript | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| APC streptavidin | BioLegend | Cat#: 405207 |
| Alexa Fluor® 647 Streptavidin | BioLegend | Cat#: 405237 |
| Brilliant Violet 421™ Streptavidin | BioLegend | Cat#: 405225 |
| PE Streptavidin | BioLegend | Cat#: 405204 |
| PE/Cyanine7 Streptavidin | BioLegend | Cat#: 405206 |
| Streptavidin, Alexa Fluor™ 750 conjugate | Thermo Fisher Scientific | Cat#: S21384 |
| Biotin ≥99% (HPLC), lyophilized powder | Sigma-Aldrich | CAS Number: 58-85-5 |
| Ni-NTA Agarose | Qiagen | Cat#: 30210 |
| Tamoxifen | Sigma-Aldrich | CAS Number: 10540-29-1 |
| Peanut oil | Sigma-Aldrich | CAS Number: 8002-03-7 |
| Fixation Buffer | BD Biosciences | Cat#: 554655 |
| Scigen O.C.T. Compound Cryostat Embedding Medium | Thermo Fisher Scientific | Cat#: 23-730-625 |
| Normal Rat Serum | Sigma-Aldrich | Cat#: R9759 |
| Goat serum | Sigma-Aldrich | Cat#: G9023-10ML |
| Bovine serum albumin | Sigma-Aldrich | CAS Number: 9048-46-8 |
| TritonX-100 | Sigma-Aldrich | CAS Number: 9036-19-5 |
| Hydromount | National Diagnostics | Cat#: HS-106 |
| Blocker Casein in PBS | Thermo Fisher Scientific | Cat#: 37528 |
| Fetal Bovine Serum | Sigma-Aldrich | Cat#: F9665-500ML |
| Paraformaldehyde | Sigma-Aldrich | Cat#: P6148 |
| Sucrose | Sigma-Aldrich | Cat#: S0389 |
| p-Nitrophenyl Phosphate Substrate Buffer | Sigma-Aldrich | Cat#: 487664 |
| SARS-CoV-2 FL-S protein | this manuscript | Graham et al.8 |
| Critical commercial assays | ||
| EndoFree Plasmid Mega Kit | Qiagen | Cat#: 12362 |
| Zenon™ Alexa Fluor™ 532 Mouse IgG1 Labeling Kit | Thermo Fisher Scientific | RRID: AB_2736944 |
| PEI MAX® - Transfection Grade Linear Polyethylenimine Hydrochloride (MW 40,000) | Polysciences | Cat#: 24765 |
| eBioscience™ Foxp3/Transcription Factor Staining Buffer Set | Thermo Fisher Scientific | Cat#: 00-5523-00 |
| Avidin/Biotin Blocking Kit | Vector laboratories | Cat#: SP-2001 |
| HiLoad Superdex 200 pg 16/600 column | Sigma-Aldrich | GE28-9893-35 |
| Amicon® Ultra-4 Centrifugal Filter Unit | Merck | Cat#: UFC801024 |
| Deposited data | ||
| Experimental models: Cell lines | ||
| FreeStyle™ 293-F Cells | ThermoFisher Scientific | RRID: CVCL_D603 |
| 2.4G2 Hybridoma | ATCC | ATCC HB-197™ |
| Experimental models: Organisms/strains | ||
| C57BL/6J-Rag2em3Lutzy/J | The Jackson Laboratory | RRID: IMSR_JAX:033526 |
| Oligonucleotides | ||
| Recombinant DNA | ||
| SARS-CoV-2 Spike | BioBasic | Wuhan strain QHR63290.2 |
| BirA plasmid | this manuscript | N/A |
| RBD-avi-His plasmid | this manuscript | N/A |
| pcDNA3.1 | Invitrogen | Cat#: V79020 |
| p8.91 plasmid (encoding for HIV-1 gag-pol) | this manuscript | N/A |
| CSFLW (lentivirus backbone expressing a firefly luciferase reporter gene) | this manuscript | N/A |
| Human Angiotensin-converting enzyme 2 (ACE2) expression plasmid | Addgene | Plasmid#1786; RRID: Addgene_1786 |
| Software and algorithms | ||
| FlowJo v10.7 | TreeStar | RRID: SCR_008520 |
| R (version 4.0.2) | https://www.R-project.org/ | RRID: SCR_001905 |
| GraphPad Prism | GraphPad | RRID: SCR_002798 |
| Glomax-Multi detection system | Promega | RRID: SCR_015575 |
| Leica Application Suite X (LAS X) | Leica | RRID: SCR_013673 |
| SOFTmax PRO | Molecular Devices | RRID: SCR_014240 |
| ImageJ v1.53 | https://imagej.nih.gov/ij/ | RRID: SCR_003070 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Michelle Linterman (Michelle.Linterman@babraham.ac.uk).
Materials availability
This study did not generate new unique reagents.
Method details
Human sample collection
Recruitment of the patient with CD40L deficiency was approved by the National Research Ethics Committee and Health Research Authority (East of England – Cambridge Research Ethics Committee (‘‘NIHR BioResource’’ 17/EE/0025)).
Healthy controls were recruited as part of the COV001 and COV002 clinical trials, with all samples coming from the standard dose/standard dose arm of the trials.5 Briefly, healthy adult participants were enrolled after screening to exclude those with pre-existing health conditions. Participants were randomly assigned 1:1 to receive ChAdOx1 nCoV-19 at a dose of 5 × 1010 viral particles (standard dose), measured using spectrophotometry, or meningococcal group A, C, W, and Y conjugate vaccine (MenACWY) as control.
Mouse housing and husbandry
C57BL/6 mice and genetically modified mice on the C57BL/6 background used in this study were derivatives of the following: Rag2−/− (The Jackson Laboratory, Stock No. 033526), Cd4cre,56 Bcl6fl/fl,57 Tg(Fcer2acre)58 and S1PR2ERT2-Cre59 strains. Mice were bred and maintained in the Babraham Institute Biological Support Unit. No primary pathogens or additional agents listed in the FELASA recommendations were detected during health monitoring surveys of the stock holding rooms. Ambient temperature was 19–21°C and relative humidity 52%. Lighting was provided on a 12 h light: 12 h dark cycle including 15 min ‘dawn’ and ‘dusk’ periods of subdued lighting. After weaning, mice were transferred to individually ventilated cages with 1–5 mice per cage. Mice were fed CRM (P) VP diet (Special Diet Services) ad libitum and received seeds (e.g. sunflower, millet) at the time of cage-cleaning as part of their environmental enrichment. All mouse experimentation was approved by the Babraham Institute Animal Welfare and Ethical Review Body. Animal husbandry and experimentation complied with existing European Union and United Kingdom Home Office legislation and local standards (PPL: P4D4AF812). All mice were immunised at between 8 and 12 weeks old, except for bone marrow chimera experiments, for which mice were reconstituted between 8 and 12 weeks old, and immunised at 16-20 weeks old respectively.
Immunisation, tamoxifen gavage and tissue sampling
Mice were immunised in the right quadriceps femoris muscle with 50μL of either 108 infectious units of ChAdOx1 nCoV-19 or ChAdOx1 OVA in phosphate buffered saline (PBS). Where indicated, mice were given an oral gavage of Tamoxifen dissolved in ethanol, emulsified in peanut oil for a final concentration of 50 mg/mL, administered as 4 μL of tamoxifen per 1g of bodyweight. At the indicated timepoints post vaccination, blood, the right mILN and spleen were taken for analysis.
Generation of fluorescent RBD-specific B cell probes
Biotinylated SARS-CoV-2 RBD monomers were generated as follows; BirA and RBD-avi-His plasmids were subcloned in DH5α Competent Cells (Invitrogen™, Cat# 18265017) and purified using EndoFree Plasmid Mega Kit (Qiagen, Cat# 12381) following manufacturer’s protocols. Then, biotinylated SARS-CoV-2 RBD with C-terminal Avi and hexahistidine tags was expressed by transient co-transfection of RBD-avi-His and BirA expression plasmids in FreeStyle™ 293-F Cells (ThermoFisher Scientific Cat# R79007) using PEI MAX (Polysciences Cat# 24765). 1.2L of culture at a density of 1.0 × 106 cells/mL was supplemented with 175 mM biotin (Sigma-Aldrich Cat# B4501) and transfected with 600 μg total DNA at a 4:1 ratio (RBD:BirA) using 12 mL PEI MAX (1 mg/mL). Culture medium was harvested 6 days post-transfection and protein purified using Ni-NTA agarose beads (Qiagen Cat# 30210). Eluted protein was further purified by size-exclusion chromatography on a HiLoad Superdex 200 pg 16/600 column (Cytiva Cat# 28989335) equilibrated in 1X PBS. Peak fractions were pooled, concentrated in a 10 kDa MWCO centrifugal filter (Merck Cat# UFC801024) and snap frozen in liquid nitrogen.
RBD monomers were combined with fluorescently labelled streptavidin (BioLegend) at a 3.95:1 molecular ratio, to ensure complete tetramerisation of streptavidin molecules. RBD was added to streptavidin in 10% increments, with a 10-min interval between each addition, and was gently mixed throughout the tetramerisation process. Tetramerisation was carried out at room temperature, before storage at 4°C.
Cell preparation and flow cytometry
A single cell suspension was prepared from cryopreserved human peripheral blood mononuclear cells (PBMC) samples as follows: 1 mL PBMC samples were defrosted in a 37°C water bath, and then immediately diluted into 9 mL of pre-warmed RPMI+10% Fetal Bovine serum (FBS). Cells were washed twice with 10 mL of FACS buffer (PBS containing 2% FBS and 1 mM EDTA). Cells were then resuspended in 500 μL of FACS buffer and cell numbers and viability were determined using a Countess™ automated cell counter (Invitrogen). 5×106 viable cells were transferred to 96-well plates for antibody staining.
For murine flow cytometric staining a single cell suspension was prepared from either the mILN or half the spleen by pressing the tissues through a 70 μm mesh and washing through with FACS buffer. Cell numbers and viability were determined using a CASY TT Cell Counter (Roche). 2×106 cells were transferred to 96-well plates for antibody staining.
Cells were then washed once with FACS buffer, and stained with 100 μL of surface antibody mix (including RBD probes) for 2 h at 4°C. Cells were then washed twice with FACS buffer, and fixed with the eBiosciences Foxp3/Transcription Factor Staining Buffer (#00-5323-00) for 30 min at 4°C. For experiments with S1PR2-RFP reporter mice, cells were instead fixed with 1% Paraformaldehyde for 20 min at room temperature. Cells were then washed with 1× Permeabilisation buffer (eBioscience #00-8333-56) twice and stained with intracellular antibody mix in permeabilisation buffer at 4°C overnight. For mouse cell staining, the permeabilisaton buffer was supplemented with 20% 2.4G2 hybridoma (ATCC hb-197) tissue culture supernatant. Following overnight staining, samples were washed twice with 1× permeabilisation buffer and once with FACS buffer and acquired on a Cytek™ Aurora. Cells for single colour controls were prepared in the same manner as the fully stained samples. The antibodies used for surface and overnight staining are listed in the Key Resources Table.
Manual gating of flow cytometry data was done using FlowJo v10.7 software (Tree Star). tSNE, FlowSOM and heatmap analysis were performed using R (version 4.0.2) using code that has previously been described.60
Confocal microscopy staining and acquisition
Medial iliac lymph nodes were harvested, before being fixed in BD Cytofix™ Fixation Buffer (BD Biosciences) diluted 1:3 in PBS for 5 h at 4°C. Samples were then washed 3 times, by being submerged in 2mls of PBS. Following this, samples were gently dried with a paper towel, and dehydrated by being submerged in a 30% sucrose solution (30 g sucrose in 100 mL PBS) for 18 h. Samples were then gently dried with a paper towel, and embedded in Scigen O.C.T. Compound Cryostat Embedding Medium (ThermoFisher) as previously published.61 Samples were cut to 14 μM using a Leica CM3050 cryostat (Leica Biosystems) onto Superfrost Plus microscope slides (VWR). A hydrophobic barrier was drawn using an ImmEdge® Hydrophobic Barrier PAP Pen H-4000 (Vector laboratories). All proceeding staining was performed in the dark. Samples were stained at room temperature for 2 h with 2% normal goat serum (Merck), 1% Bovine serum albumin and 10% Rat serum (Merck) in PBS to block non-specific interactions. Samples were then washed for 5 min in PBS +0.5% Tween 20 (PBS-T), then permeabilised with 2% TritonX-100 (Sigma Aldrich) in PBS for 30 min. Samples were then washed for 5 min in PBS-T, and endogenous biotin/streptavidin interactions were blocked using the Avidin/Biotin Blocking Kit (Vector laboratories) before being washed in PBS-T for 5 min. PD-1 was stained for using a two-step reaction at room temperature (1.5hrs staining with Purified anti-mouse CD279 (PD-1) Antibody (BioLegend Cat # 109101), then washed in PBS-T, followed by Goat anti-Rat IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 647 (ThermoFisher Scientific) for 1 h. Samples were then washed with PBS-T for 5 min. The remaining antibodies (CD3, CD35, B220, IgD and KI67) were then added, and stained for 16 h at 4°C. Samples were then washed with PBS-T for 5 min. Finally, samples were stained with Streptavidin, Alexa Fluor™ 750 conjugate (ThermoFisher Scientific) for 1 h at room temperature. Samples were then washed twice quickly with PBS-T followed by PBS, before being mounted with microscope cover slips (Thermo Fisher Scientific) using Hydromount mounting medium (National diagnostics). Slides were imaged with a Leica STELLARIS 8 microscope and Leica Application Suite X (LAS X) software (Leica microsystems) and processed using ImageJ v1.53.
Enzyme-linked immunosorbent assay (ELISA)
Standardised ELISA was performed to detect SARS-CoV-2 FL-S, or SARS-CoV-2 RBD – specific antibodies in sera. MaxiSorp plates (Nunc) were coated with 100 ng/well protein overnight at 4°C for detection of IgG, prior to washing in PBS/Tween (0.05% v/v) and blocking with Blocker Casein in PBS (Thermo Fisher Scientific) for 1 h at room temperature (RT). Standard positive serum (pool of serum with high endpoint titre against FL-S protein), individual serum samples, negative and an internal control (diluted in casein) were incubated for 2 h at RT for detection of specific IgG. Following washing, bound antibodies were detected by addition of AP-conjugated goat anti-mouse IgG or AP-conjugated goat anti-human IgG (Sigma-Aldrich) for 1 h at RT and addition of pNPP substrate (Sigma-Aldrich). An arbitrary number of ELISA units were assigned to the reference pool and OD values of each dilution were fitted to a 4-parameter logistic curve using SOFTmax PRO software. ELISA units were calculated for each sample using the OD values of the sample and the parameters of the standard curve.
The IgG subclass ELISA were performed according to the protocol described above. In addition, all serum samples were diluted to 1 total IgG ELISA unit and then detected with anti-mouse IgG subclass-specific secondary antibodies (Southern Biotech or Abcam). The results of the IgG subclass ELISA are presented as pie charts using OD values instead of the ELISA units used for the total IgG ELISA.
Micro-neutralisation test using lentiviral-based pseudotypes bearing the SARS-CoV-2 spike
Lentiviral-based SARS-CoV-2 pseudotyped viruses were generated in HEK293T cells incubated at 37°C, 5% CO2 as previously described.62 Briefly, cells were seeded at a density of 7.5 × 105 in 6 well dishes, before being transfected with plasmids as follows: 500 ng of SARS-CoV-2 spike, 600 ng p8.91 (encoding for HIV-1 gag-pol), 600 ng CSFLW (lentivirus backbone expressing a firefly luciferase reporter gene), in Opti-MEM (Gibco) along with 10 μL PEI (1 μg/mL) transfection reagent. A ‘no glycoprotein’ control was also set up using the pcDNA3.1 vector instead of the SARS-CoV-2 S expressing plasmid. The following day, the transfection mix was replaced with 3 mL DMEM with 10% FBS (DMEM-10%) and incubated for 48 and 72 h, after which supernatants containing pseudotyped SARS-CoV-2 (SARS-CoV-2 pps) were harvested, pooled and centrifuged at 1,300 × g for 10 min at 4°C to remove cellular debris. Target HEK293T cells, previously transfected with 500 ng of a human ACE2 expression plasmid (Addgene, Cambridge, MA, USA) were seeded at a density of 2 × 104 in 100 μL DMEM-10% in a white flat-bottomed 96-well plate one day prior to harvesting SARS-CoV-2 pps. The following day, SARS-CoV-2 pseudotyped viruses were titrated 10-fold on target cells, and the remainder stored at −80°C. For micro neutralisation tests, sera were diluted 1:20 in serum-free media and 50 μL was added to a 96-well plate in triplicate and titrated 2-fold. A fixed titred volume of SARS-CoV-2 pps was added at a dilution equivalent to 105 signal luciferase units in 50 μL DMEM-10% and incubated with sera for 1 h at 37°C, 5% CO2 (giving a final sera dilution of 1:40). Target cells expressing human ACE2 were then added at a density of 2 × 104 in 100 μL and incubated at 37°C, 5% CO2 for 72 h. Firefly luciferase activity was then measured with BrightGlo luciferase reagent and a Glomax-Multi+ Detection System (Promega, Southampton, UK). Pseudovirus neutralisation titres were expressed as the reciprocal of the serum dilution that inhibited luciferase expression by 80% (IC80).
Quantification and statistical analysis
All mouse experiments were performed either twice or three times with 3–8 mice per group. Data was first tested for gaussian distribution using a Shapiro-Wilk test. Then data that was consistent with a normal distribution was analyzed with either a Student’s t test for comparing two data set, or one-way ANOVA test for data with multiple groups. If the data did not follow a normal distribution, then a Mann-Whitney test was used for comparing two datasets and a Kruskal Wallis test for multiple comparisons. All p values shown are adjusted for multiple comparisons where multiple tests were performed on the same data. Analyses were performed within the Prism v9 software (GraphPad).
Acknowledgments
We thank M. Busslinger (Research Institute of Molecular Pathology) for the Tg(Fcer2acre) mice and A. Dent for the Bcl6flox/flox mice and staff within the Biological Support Unit and Flow Cytometry Facility at the Babraham Institute for research support. We also thank Helen Sanders, Reece Mabbett, and Federica Cappucini of the Jenner Institute. This study was supported by funding from the Biotechnology and Biological Sciences Research Council (BBSRC; BBS/E/B/000C0427, BBS/E/B/000C0428, and the Campus Capability Core Grant to the Babraham Institute); the Lister institute of Preventative Medicine; the EPSRC VaxHub (EP/RO13756/1); and Innovate UK (biEBOV: 971615). H.J.S. is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (109407) and a BBSRC institutional program grant (BBS/E/B/000C0433). T.L. and A.J.S. are Jenner Investigators. M.A.L. and H.J.S. are EMBO Young Investigators and Lister Institute Prize Fellows. J.L.L. is supported by a National Science Scholarship (PhD) by the Agency for Science, Technology and Research, Singapore. J.E.T., J.C.Y.-P., and E.C.H. are supported by the MRC (MC_UU_0025/12) and the Medical Research Foundation (MRF-057-0002-RG-THAV-C0798). N.T., J.N., and D.B. were supported by the MRC (MR/W005611/1) and BBSRC (BBS/E/I/COV07001, BBS/E/I/00007031, and BB/T008784/1).
Author contributions
Conceptualization, W.S.F., M.A.L., and T.L.; methodology, W.S.F., J.L.L., A.J.S., S.D., D.W., L.G., N.T., J.N., I.M.H., S.I., J.C.Y.-P., E.C.H., H.J.S., J.E.T., D.B., T.L., and M.A.L.; investigation, W.S.F., J.L.L., A.J.S., S.D., D.W., L.G., N.T., J.N., I.M.H., and S.I.; resources, J.E.T., J.C.Y.-P., and T.L.; writing – original draft, M.A.L. and W.S.F.; writing – review & editing, all authors; project administration, W.S.F., M.A.L., and T.L.; funding acquisition, M.A.L., J.E.T., D.B., and T.L.
Declaration of interests
T.L. is named on a patent application covering ChAdOx1 nCoV-19. The funders played no role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research.
Published: November 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100845.
Contributor Information
Teresa Lambe, Email: teresa.lambe@paediatrics.ox.ac.uk.
Michelle A. Linterman, Email: michelle.linterman@babraham.ac.uk.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report the original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.
References
- 1.WHO Coronavirus (COVID-19) Dashboard. (2021). https://covid19.who.int/.
- 2.AstraZeneca . 2021. Two billion doses of AstraZeneca’s COVID-19 vaccine supplied to countries across the world less than 12 months after first approval.https://www.astrazeneca.com/media-centre/press-releases/2021/two-billion-doses-of-astrazenecas-covid-19-vaccine-supplied-to-countries-across-the-world-less-than-12-months-after-first-approval.html [Google Scholar]
- 3.Lopez Bernal J., Andrews N., Gower C., Robertson C., Stowe J., Tessier E., Simmons R., Cottrell S., Roberts R., O'Doherty M., et al. Effectiveness of the Pfizer-BioNTech and Oxford-AstraZeneca vaccines on covid-19 related symptoms, hospital admissions, and mortality in older adults in England: test negative case-control study. BMJ. 2021;373:n1088. doi: 10.1136/bmj.n1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris R.J., Hall J.A., Zaidi A., Andrews N.J., Dunbar J.K., Dabrera G. Effect of vaccination on household transmission of SARS-CoV-2 in England. N. Engl. J. Med. 2021;385:759–760. doi: 10.1056/NEJMc2107717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nasreen S., Chung H., He S., Brown K.A., Gubbay J.B., Buchan S.A., Fell D.B., Austin P.C., Schwartz K.L., Sundaram M.E., et al. Effectiveness of COVID-19 vaccines against symptomatic SARS-CoV-2 infection and severe outcomes with variants of concern in Ontario. Nat. Microbiol. 2022;7:379–385. doi: 10.1038/s41564-021-01053-0. [DOI] [PubMed] [Google Scholar]
- 7.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graham S.P., McLean R.K., Spencer A.J., Belij-Rammerstorfer S., Wright D., Ulaszewska M., Edwards J.C., Hayes J.W.P., Martini V., Thakur N., et al. Evaluation of the immunogenicity of prime-boost vaccination with the replication-deficient viral vectored COVID-19 vaccine candidate ChAdOx1 nCoV-19. NPJ Vaccines. 2020;5:69. doi: 10.1038/s41541-020-00221-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva-Cayetano A., Foster W.S., Innocentin S., Belij-Rammerstorfer S., Spencer A.J., Burton O.T., Fra-Bidó S., Le Lee J., Thakur N., Conceicao C., et al. A booster dose enhances immunogenicity of the COVID-19 vaccine candidate ChAdOx1 nCoV-19 in aged mice. Med (N Y) 2021;2:243–262.e8. doi: 10.1016/j.medj.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewer K.J., Barrett J.R., Belij-Rammerstorfer S., Sharpe H., Makinson R., Morter R., Flaxman A., Wright D., Bellamy D., Bittaye M., et al. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat. Med. 2021;27:270–278. doi: 10.1038/s41591-020-01194-5. [DOI] [PubMed] [Google Scholar]
- 12.Barrett J.R., Belij-Rammerstorfer S., Dold C., Ewer K.J., Folegatti P.M., Gilbride C., Halkerston R., Hill J., Jenkin D., Stockdale L., et al. Phase 1/2 trial of SARS-CoV-2 vaccine ChAdOx1 nCoV-19 with a booster dose induces multifunctional antibody responses. Nat. Med. 2021;27:279–288. doi: 10.1038/s41591-020-01179-4. [DOI] [PubMed] [Google Scholar]
- 13.Plotkin S.A. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng S., Phillips D.J., White T., Sayal H., Aley P.K., Bibi S., Dold C., Fuskova M., Gilbert S.C., Hirsch I., et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:2032–2040. doi: 10.1038/s41591-021-01540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergwerk M., Gonen T., Lustig Y., Amit S., Lipsitch M., Cohen C., Mandelboim M., Levin E.G., Rubin C., Indenbaum V., et al. Covid-19 breakthrough infections in vaccinated health care workers. N. Engl. J. Med. 2021;385:1474–1484. doi: 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Addetia A., Crawford K.H.D., Dingens A., Zhu H., Roychoudhury P., Huang M.L., Jerome K.R., Bloom J.D., Greninger A.L. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.02107-20. e02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- 18.Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Victora G.D., Nussenzweig M.C. Germinal centers. Annu. Rev. Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 21.MacLennan I.C.M., Toellner K.M., Cunningham A.F., Serre K., Sze D.M.Y., Zúñiga E., Cook M.C., Vinuesa C.G. Extrafollicular antibody responses. Immunol. Rev. 2003;194:8–18. doi: 10.1034/j.1600-065x.2003.00058.x. [DOI] [PubMed] [Google Scholar]
- 22.MacLennan I.C. Germinal centers. Annu. Rev. Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 23.Weisel F.J., Zuccarino-Catania G.V., Chikina M., Shlomchik M.J. A temporal switch in the germinal center determines differential output of memory B and plasma cells. Immunity. 2016;44:116–130. doi: 10.1016/j.immuni.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S.K., Rigby R.J., Zotos D., Tsai L.M., Kawamoto S., Marshall J.L., Ramiscal R.R., Chan T.D., Gatto D., Brink R., et al. B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. J. Exp. Med. 2011;208:1377–1388. doi: 10.1084/jem.20102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J.S., Chow R.D., Song E., Mao T., Israelow B., Kamath K., Bozekowski J., Haynes W.A., Filler R.B., Menasche B.L., et al. High-affinity, neutralizing antibodies to SARS-CoV-2 can be made without T follicular helper cells. Sci. Immunol. 2022;7:eabl5652. doi: 10.1126/sciimmunol.abl5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyauchi K., Sugimoto-Ishige A., Harada Y., Adachi Y., Usami Y., Kaji T., Inoue K., Hasegawa H., Watanabe T., Hijikata A., et al. Protective neutralizing influenza antibody response in the absence of T follicular helper cells. Nat. Immunol. 2016;17:1447–1458. doi: 10.1038/ni.3563. [DOI] [PubMed] [Google Scholar]
- 27.Kaji T., Ishige A., Hikida M., Taka J., Hijikata A., Kubo M., Nagashima T., Takahashi Y., Kurosaki T., Okada M., et al. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J. Exp. Med. 2012;209:2079–2097. doi: 10.1084/jem.20120127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toyama H., Okada S., Hatano M., Takahashi Y., Takeda N., Ichii H., Takemori T., Kuroda Y., Tokuhisa T. Memory B cells without somatic hypermutation are generated from Bcl6-deficient B cells. Immunity. 2002;17:329–339. doi: 10.1016/s1074-7613(02)00387-4. [DOI] [PubMed] [Google Scholar]
- 29.Roco J.A., Mesin L., Binder S.C., Nefzger C., Gonzalez-Figueroa P., Canete P.F., Ellyard J., Shen Q., Robert P.A., Cappello J., et al. Class-switch recombination occurs infrequently in germinal centers. Immunity. 2019;51:337–350.e7. doi: 10.1016/j.immuni.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McAdam A.J., Greenwald R.J., Levin M.A., Chernova T., Malenkovich N., Ling V., Freeman G.J., Sharpe A.H. ICOS is critical for CD40-mediated antibody class switching. Nature. 2001;409:102–105. doi: 10.1038/35051107. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe M., Fujihara C., Radtke A.J., Chiang Y.J., Bhatia S., Germain R.N., Hodes R.J. Co-stimulatory function in primary germinal center responses: CD40 and B7 are required on distinct antigen-presenting cells. J. Exp. Med. 2017;214:2795–2810. doi: 10.1084/jem.20161955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cicalese M.P., Gerosa J., Baronio M., Montin D., Licciardi F., Soresina A., Dellepiane R.M., Miano M., Baselli L.A., Volpi S., et al. Circulating follicular helper and follicular regulatory T cells are severely compromised in human CD40 deficiency: a case report. Front. Immunol. 2018;9:1761. doi: 10.3389/fimmu.2018.01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh C.H., Finney J., Okada T., Kurosaki T., Kelsoe G. Primary germinal center-resident T follicular helper cells are a physiologically distinct subset of CXCR5(hi)PD-1(hi) T follicular helper cells. Immunity. 2022;55:272–289.e7. doi: 10.1016/j.immuni.2021.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vinuesa C.G., Linterman M.A., Yu D., MacLennan I.C.M. Follicular helper T cells. Annu. Rev. Immunol. 2016;34:335–368. doi: 10.1146/annurev-immunol-041015-055605. [DOI] [PubMed] [Google Scholar]
- 35.Barros-Martins J., Hammerschmidt S.I., Cossmann A., Odak I., Stankov M.V., Morillas Ramos G., Dopfer-Jablonka A., Heidemann A., Ritter C., Friedrichsen M., et al. Immune responses against SARS-CoV-2 variants after heterologous and homologous ChAdOx1 nCoV-19/BNT162b2 vaccination. Nat. Med. 2021;27:1525–1529. doi: 10.1038/s41591-021-01449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubtsov A.V., Rubtsova K., Fischer A., Meehan R.T., Gillis J.Z., Kappler J.W., Marrack P. Toll-like receptor 7 (TLR7)-driven accumulation of a novel CD11c(+) B-cell population is important for the development of autoimmunity. Blood. 2011;118:1305–1315. doi: 10.1182/blood-2011-01-331462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du S.W., Arkatkar T., Al Qureshah F., Jacobs H.M., Thouvenel C.D., Chiang K., Largent A.D., Li Q.Z., Hou B., Rawlings D.J., Jackson S.W. Functional characterization of CD11c(+) age-associated B cells as memory B cells. J. Immunol. 2019;203:2817–2826. doi: 10.4049/jimmunol.1900404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye B.H., Cattoretti G., Shen Q., Zhang J., Hawe N., de Waard R., Leung C., Nouri-Shirazi M., Orazi A., Chaganti R.S., et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 39.Fukuda T., Yoshida T., Okada S., Hatano M., Miki T., Ishibashi K., Okabe S., Koseki H., Hirosawa S., Taniguchi M., et al. Disruption of the Bcl6 gene results in an impaired germinal center formation. J. Exp. Med. 1997;186:439–448. doi: 10.1084/jem.186.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dent A.L., Shaffer A.L., Yu X., Allman D., Staudt L.M. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 41.Taylor J.J., Pape K.A., Jenkins M.K. A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J. Exp. Med. 2012;209:597–606. doi: 10.1084/jem.20111696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berkowska M.A., Driessen G.J.A., Bikos V., Grosserichter-Wagener C., Stamatopoulos K., Cerutti A., He B., Biermann K., Lange J.F., van der Burg M., et al. Human memory B cells originate from three distinct germinal center-dependent and -independent maturation pathways. Blood. 2011;118:2150–2158. doi: 10.1182/blood-2011-04-345579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mesin L., Schiepers A., Ersching J., Barbulescu A., Cavazzoni C.B., Angelini A., Okada T., Kurosaki T., Victora G.D. Restricted clonality and limited germinal center reentry characterize memory B cell reactivation by boosting. Cell. 2020;180:92–106.e11. doi: 10.1016/j.cell.2019.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendoza A., Yewdell W.T., Hoyos B., Schizas M., Bou-Puerto R., Michaels A.J., Brown C.C., Chaudhuri J., Rudensky A.Y. Assembly of a spatial circuit of T-bet-expressing T and B lymphocytes is required for antiviral humoral immunity. Sci. Immunol. 2021;6:eabi4710. doi: 10.1126/sciimmunol.abi4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S., Ley A.M., Kyu S., Howell J.C., Ozturk T., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubtsova K., Rubtsov A.V., Cancro M.P., Marrack P. Age-associated B cells: a T-bet-dependent effector with roles in protective and pathogenic immunity. J. Immunol. 2015;195:1933–1937. doi: 10.4049/jimmunol.1501209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutton H.J., Aye R., Idris A.H., Vistein R., Nduati E., Kai O., Mwacharo J., Li X., Gao X., Andrews T.D., et al. Atypical B cells are part of an alternative lineage of B cells that participates in responses to vaccination and infection in humans. Cell Rep. 2021;34:108684. doi: 10.1016/j.celrep.2020.108684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song W., Antao O.Q., Condiff E., Sanchez G.M., Chernova I., Zembrzuski K., Steach H., Rubtsova K., Angeletti D., Lemenze A., et al. Development of Tbet- and CD11c-expressing B cells in a viral infection requires T follicular helper cells outside of germinal centers. Immunity. 2022;55:290–307.e5. doi: 10.1016/j.immuni.2022.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burton A.R., Guillaume S.M., Foster W.S., Wheatley A.K., Hill D.L., Carr E.J., et al. The memory B cell response to influenza vaccination is impaired in older persons. Cell Reports. 2022;41:111613. doi: 10.1016/j.celrep.2022.111613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cancro M.P. Age-associated B cells. Annu. Rev. Immunol. 2020;38:315–340. doi: 10.1146/annurev-immunol-092419-031130. [DOI] [PubMed] [Google Scholar]
- 53.Kuraoka M., Yeh C.H., Bajic G., Kotaki R., Song S., Windsor I., Harrison S.C., Kelsoe G. Recall of B cell memory depends on relative locations of prime and boost immunization. Sci. Immunol. 2022;7:eabn5311. doi: 10.1126/sciimmunol.abn5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuccarino-Catania G.V., Sadanand S., Weisel F.J., Tomayko M.M., Meng H., Kleinstein S.H., Good-Jacobson K.L., Shlomchik M.J. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat. Immunol. 2014;15:631–637. doi: 10.1038/ni.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Denton A.E., Dooley J., Cinti I., Silva-Cayetano A., Fra-Bido S., Innocentin S., Hill D.L., Carr E.J., McKenzie A.N.J., Liston A., Linterman M.A. Targeting TLR4 during vaccination boosts MAdCAM-1(+) lymphoid stromal cell activation and promotes the aged germinal center response. Sci. Immunol. 2022;7:eabk0018. doi: 10.1126/sciimmunol.abk0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee P.P., Fitzpatrick D.R., Beard C., Jessup H.K., Lehar S., Makar K.W., Pérez-Melgosa M., Sweetser M.T., Schlissel M.S., Nguyen S., et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 57.Hollister K., Kusam S., Wu H., Clegg N., Mondal A., Sawant D.V., Dent A.L. Insights into the role of Bcl6 in follicular Th cells using a new conditional mutant mouse model. J. Immunol. 2013;191:3705–3711. doi: 10.4049/jimmunol.1300378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwon K., Hutter C., Sun Q., Bilic I., Cobaleda C., Malin S., Busslinger M. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 59.Shinnakasu R., Inoue T., Kometani K., Moriyama S., Adachi Y., Nakayama M., Takahashi Y., Fukuyama H., Okada T., Kurosaki T. Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol. 2016;17:861–869. doi: 10.1038/ni.3460. [DOI] [PubMed] [Google Scholar]
- 60.Pasciuto E., Burton O.T., Roca C.P., Lagou V., Rajan W.D., Theys T., Mancuso R., Tito R.Y., Kouser L., Callaerts-Vegh Z., et al. Microglia require CD4 T cells to complete the fetal-to-adult transition. Cell. 2020;182:625–640.e24. doi: 10.1016/j.cell.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fra-Bido S., Walker S.A., Innocentin S., Linterman M.A. Optimized immunofluorescence staining protocol for imaging germinal centers in secondary lymphoid tissues of vaccinated mice. STAR Protoc. 2021;2:100499. doi: 10.1016/j.xpro.2021.100499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Newman J., Thakur N., Peacock T.P., Bialy D., Elrefaey A.M.E., Bogaardt C., Horton D.L., Ho S., Kankeyan T., Carr C., et al. Neutralizing antibody activity against 21 SARS-CoV-2 variants in older adults vaccinated with BNT162b2. Nat. Microbiol. 2022;7:1180–1188. doi: 10.1038/s41564-022-01163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report the original code.
-
•
Any additional information required to reanalyse the data reported in this paper is available from the lead contact upon request.