Koestel and Batoko preview work from the Kang and Dagdas labs that reveals the existence of amphisome compartments in plant cells and identifies CFS1 as a bridging adaptor for amphisome formation.
Abstract
The fusion of autophagosomes with endocytic compartments to form amphisomes has only been described in metazoans. In this issue, Zhao et al. (2022. J. Cell Biol. https://doi.org/10.1083/jcb.202203139) demonstrate the existence of amphisomes in the plant cell and identify a plant-specific adaptor protein, CFS1, that mediates their biogenesis.
Evolutionary conserved selective and non-selective protein degradation pathways are essential for eukaryotic cell growth and survival under detrimental physiological conditions (1). For example, in response to stress, including nutrient limitation, endocytosis and macroautophagy (hereafter autophagy) allow for adaptive intracellular proteome remodeling and nutrient recycling. Endocytosed plasma membrane proteins are degraded in an endosomal sorting complex required for transport (ESCRT)-dependent manner via the multivesicular body (MVB) pathway. Thus, autophagy ensures efficient regulated degradation of a portion of the cytoplasm containing damaged or superfluous cellular components, thereby preserving cellular and organismal homeostasis. The hallmark of autophagy is a stepwise de novo formation of a double membrane vesicle called the autophagosome (2). Formation of the autophagosome involves multiple membrane remodeling processes, including initiation and nucleation of a cup-shaped precursor known as the phagophore (2, 3). The phagophore subsequently expands and closes to form the autophagosome, sequestering materials or cargo to be degraded. The outer membrane of the phagophore is decorated with autophagy-related (ATG) 8 family members, tethering factors, Rab GTPases, SNARE, ESCRT proteins, and adaptor proteins acting coordinately to seal the phagophore and mediate cytoskeleton-dependent targeting and direct fusion of the autophagosome with a lytic compartment (vacuole in yeast and plants, lysosome in metazoans). However, in metazoans, nascent autophagosome can also fuse with vesicles from the endocytic pathway to form an MVB-autophagosome intermediate hybrid compartment called amphisome (4, 5), which later fuses with the lysosome in a process known as autophagosome maturation. Whether amphisome formation and autophagosome maturation occur in other multicellular organisms, including plants, is unclear. In addition, molecular determinants of autophagosome fusion with the vacuole in plants, such as Rab GTPase, SNARE, tethering factors, or adaptor proteins, are currently unknown.
To identify adaptor proteins associated with plant autophagosomes, Zhao et al. established a differential centrifugation protocol leading to the enrichment of a microsomal fraction with intact autophagosomes from the model plant Arabidopsis thaliana. Then, they isolated autophagosomes from this microsomal fraction by affinity purification, using tagged ATG8 decorating their membranes as bait and subjected them to proteomic analysis. Surface proteome analysis of purified vesicles (proteinase K-sensitive proteins) yielded 48 polypeptides, including CFS1 (cell death-related endosomal FYVE/SYLF protein 1) which is a well-conserved plant-specific protein involved in the endocytic pathway and autophagy (6, 7). Their protease-protection assays and Western blotting experiments suggested that CFS1 associates with the inner and outer membranes of autophagosomes and is likely incorporated into these membranes at least as early as the phagophore initiation stage.
The authors subsequently used microscopy and biochemical approaches to confirm the association of CFS1 with autophagosome membranes. They generated transgenic Arabidopsis plants expressing a fluorescent and functional CFS1 chimera (fluorescent protein–tagged CFS1 variants). Using confocal microscopy, they showed that CFS1 does not colocalize with specific markers of the Golgi, the trans-Golgi network, or the endosome. However, autophagy-inducing treatments such as nitrogen starvation or salt stress induced colocalization of CFS1 with ATG8 in autophagosomes generated in response to these stresses. CFS1 also colocalized with ATG11, a core autophagy protein in plants, and with the Arabidopsis NBR1 (neighbor of BRCA1) homolog, a selective autophagy receptor. Furthermore, time-lapse confocal imaging showed that autophagic vesicles co-labeled with CFS1 and ATG8, or NBR1, move together in the root epidermal cells of Arabidopsis, confirming that, indeed, CFS1 specifically labels autophagic compartments. CFS1 contains the functional domains FYVE (Fab-1, YGL023, Vps27, and EEA1) and SYLF (SH3YL1, Ysc84p/Lsb4p, Lsb3p, and plant FYVE), through which it interacts with PI3P (phosphatidylinositol-3-phosphate) enriched in the phagophore and autophagosome membranes and with actin filaments (6). However, if CFS1 is an adaptor involved in the autophagosome fusion with a lytic compartment, then it should also interact specifically with ATG8 via a putative ATG8 interacting motif (AIM). To test this hypothesis, Zhao et al. used pull-down assays. CFS1 interacted with six of the nine Arabidopsis ATG8 in vitro, and the presence of a synthetic AIM peptide competitively inhibited this interaction, suggesting that the interaction of CFS1 with ATG8 occurs via a conserved AIM motif. The authors identified the AIM motif of CFS1 in its linear sequence WLNL between the FYVE and SYLF motifs. Indeed, mutating this motif to ALNA (WLNL-267-ALNA) and expressing this mutant CFS1AIM form in a cfs1 null mutant background substantially reduced the interaction of CFS1AIM with ATG8, as the researchers demonstrated by pull-down and co-immunoprecipitation experiments. Compared to their wild-type counterparts, CFS1AIM mutant plants were relatively more sensitive to nitrogen starvation, similar to cfs1 and atg5 mutants. At the cellular level, confocal imaging showed that CFS1AIM-expressing plants formed significantly fewer autophagic vesicles than plants expressing the wild-type protein. Residual association of CFS1AIM with autophagic compartments could be attributed to the FYVE and SYLF domain interactions with PI3P since mutating all three domains (AIM, FYVE, and SYLF) disrupted the interaction and colocalization of CFS1AIM with ATG8 and resulted in the diffuse localization of the mutant form in the cytoplasm. These results suggested that CFS1 could act as an autophagy adaptor on the autophagosome membrane and mediate autophagosome trafficking and maturation. CFS1 could also bridge PI3P-rich endomembrane compartments with autophagosomes (Fig. 1).
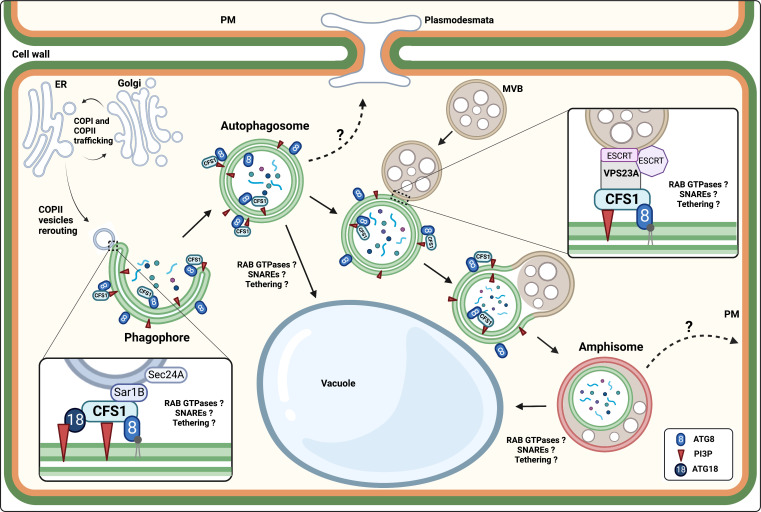
Figure 1.
The plant-specific CFS1 acts as an autophagy adaptor for autophagosome maturation and amphisome biogenesis in plants. CFS1 is associated with the phagophore and autophagosome membranes by interacting with ATG8 and PI3P. CFS1 may interact with the COPII machinery during the phagophore initiation (7). CFS1 is required for amphisome biogenesis by connecting the autophagosome to the MVB through its interaction with the ESCRT-I component Vps23A. Hypothetically (dotted arrows), the autophagosome and/or the amphisome could travel from cell to cell through plasmodesmata, and the amphisome could be involved in non-conventional secretion by fusing with the plasma membrane.
To unravel the function of CFS1 as an autophagic adaptor, Zhao et al. assessed the cellular phenotype of cfs1 mutant plants; in particular, they evaluated the autophagic flux in the presence or absence of CFS1 activity. Blocking the v-ATPase by concanamycin A (ConA) treatment causes a reduction in vacuolar acidity and inhibits the activity of vacuolar hydrolases. As a result of ConA treatment, autophagic bodies accumulate in the vacuole lumen and can be counted by fluorescence microscopy (8). Using this approach, they showed that after induction of autophagy, cfs1 mutants displayed fewer autophagic bodies than wild-type control plants. Moreover, the degradation rate of ATG8 or NBR1 in cfs1 mutant plants was lower, as measured by calculating the ratio of a free fluorescent protein that becomes hydrolyzed when it enters the vacuole (GFP) to the corresponding fusion protein (GFP-ATG8) (8). These observations suggested that, on the one hand, the absence of CFS1 activity reduces autophagic flux and, on the other hand, the lack of CFS1 does not completely inhibit the formation, transport, and fusion of autophagosomes with the vacuole. Indeed, the authors observed fewer ATG8-labeled autophagosomes in the cfs1 mutants than in the wild-type plants but not as affected as in the atg5 mutant, which in principle, is incapable of forming autophagosomes. Selective mitochondrial degradation via autophagy was also comparably affected in the cfs1 mutant. Similar observations were made with the CFS1 homolog in Marchantia polymorpha, suggesting that the function of CFS1 is conserved across plants. To understand the mechanism by which CFS1 regulates autophagy flux in plant cells, Zhao et al. performed a genome-wide yeast two-hybrid screen to analyze the CFS1 interactome. Among the CFS1 interactors, they found an essential component of the ESCRT-I complex, Vps23A, which is involved in endocytosis and the MVB pathway. Arabidopsis expresses two functionally redundant isoforms of this protein, Vps23A and Vps23B, the double mutant being lethal. Interestingly, CFS1, which colocalizes with the autophagosome markers, also partially colocalized with VPS23A and other components of the ESCRT complex, notably FREE1 and ALIX. The authors then used transmission electron microscopy to unambiguously identify the subcellular compartment where CFS1 and Vps23A partially colocalized and identified a hybrid compartment formed by the fusion of the autophagosome with the prevacuolar compartment (PVC). The PVC is the equivalent of the MVB in plant cells (9). The interactors of Vps23A contain a “PSAPP” motif, which docks into the ubiquitin E2 variant domain of Vps23 (6). Since structural models predicted the same type of interaction between CFS1 and Vps23A, and CFS1 bears a PSAPP motif in its N-terminal end, they tested the requirement of the PSAPP motif for the CFS1-Vps23A interaction and for autophagic flux. Biochemical and microscopic analyses in plant cells expressing a version of CFS1 where the PSAPP motif is mutated to alanines (CFS1PSAPP) demonstrated that the PSAPP motif of CFS1 is required for its interaction with Vps23A. Csf1 null mutants expressing CFS1PSAPP phenocopied the CFS1AIM phenotype; CFS1PSAPP-expressing plants exhibited fewer autophagic bodies in the vacuole under autophagy-inducing conditions, defective autophagic flux, and sensitivity to nitrogen starvation. These observations suggested that the CFS1-VPS23A interaction regulates the fusion of the autophagosome with the PVC/MVB and the autophagic flux in plants.
This work demonstrates for the first time the formation of amphisome during autophagosome maturation in plants and identifies CFS1 as the essential adaptor for the biogenesis of these hybrid intermediate compartments. However, whether this autophagosome maturation pathway involving amphisome formation functions independently of cell type or autophagy type remains to be clarified. CFS1 does not appear to be essential for the initiation and fusion of autophagosomes with the vacuole. The cellular phenotype of the cfs1 mutant seems to be more pronounced in some root cell types than others (7). Thus, the role of CFS1 may vary depending on the autophagosome subpopulation and cell type. Furthermore, it would be interesting to clarify in future studies whether CFS1 acts as a monomer or an oligomer and whether its role is limited to autophagy. The involvement of CFS1 in amphisome biogenesis is partly related to the PSAPP motif because the Arabidopsis CFS2 protein structurally related to CFS1 but lacking the PSAPP motif plays no role in autophagy. Leaves of the cfs1 mutant show lesions due to cell death (6), and amphisomes can be involved in virus propagation (10); thus, CFS1 may be playing other functions in plants, such as in non-conventional secretion.
Acknowledgments
This research was funded by the Belgian Funds for Scientific Research (FRS-FNRS; Research Credit grant #19516174 and Research project grant #T.0050.18 to H. Batoko). J. Koestel is supported by a PhD fellowship from the Fund for Research Training in Industry and Agriculture, and H. Batoko is a senior research associate of the FRS-FNRS. The figure was created with BioRender (BioRender.com).
The authors declare no competing financial interests.
References
- 1.Müller, M., et al. 2015. eLife. 10.7554/eLife.07736 [DOI] [Google Scholar]
- 2.Nakatogawa, H., et al. 2009. Nat. Rev. Mol. Cell Biol. 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- 3.Zhao, Y.G., and Zhang H.. 2019. J. Cell Biol. 10.1083/jcb.201810099 [DOI] [Google Scholar]
- 4.Gordon, P.B., et al. 1992. Biochem. J. 10.1042/bj2830361 [DOI] [Google Scholar]
- 5.Patel, K.K., et al. 2013. EMBO J. 10.1038/emboj.2013.233 [DOI] [Google Scholar]
- 6.Sutipatanasomboon, A., et al. 2017. Sci. Rep. 10.1038/s41598-017-08577-8 [DOI] [Google Scholar]
- 7.Kim, J.H., et al. 2022. Plant Cell. 10.1093/plcell/koab263 [DOI] [Google Scholar]
- 8.Bassham, D.C. 2015. Methods. 10.1016/j.ymeth.2014.09.003 [DOI] [Google Scholar]
- 9.Cui, Y., et al. 2016. Mol. Plant. 10.1016/j.molp.2016.01.011 [DOI] [PubMed] [Google Scholar]
- 10.Corona, A.K., et al. 2018. Cell Rep. 10.1016/j.celrep.2018.03.003 [DOI] [Google Scholar]