Abstract
Potato common scab, which is mainly caused by the bacterium Streptomyces scabies, occurs in key potato growing regions worldwide. It causes necrotic or corky symptoms on potato tubers and decreases the economic value of potato. At present, there is no recommended chemical or biological control for combating potato common scab in Taiwan. It can only reduce the occurrence by cultivation control, but the efficacy is limited. Previously we found that Bacillus amyloliquefaciens Ba01 could control potato common scab in pot assay and in the field. The potential anti-S. scabies mechanism was associated with surfactin secretion, but further molecular dissection was not conducted. Thus, in this study we aimed to determine whether surfactin is the main compound active against S. scabies by knocking out the srf gene cluster in Ba01. The cloning plasmid pRY1 was transformed to Ba01 by electroporation for in-frame deletion. Two independent Δsrf mutants were obtained and confirmed by specific primers and mass spectrometry. The swarming ability and S. scabies inhibition was significantly decreased (P<0.001) in Δsrf mutants. The swarming ability of Δsrf mutants could be restored by the addition of surfactin. Furthermore, we found that Ba01 formed wrinkled biofilm in MSgg liquid medium, while Δsrf mutants formed biofilm abnormally. Furthermore, the α-amylase, protease and phosphate-solubilizing ability of Δsrf mutants was decreased, and the mutants could not inhibit the growth and sporulation of S. scabies on potato tuber slices. In conclusion, srf gene cluster of B. amyloliquefaciens Ba01 is responsible for the secretion of surfactin and inhibition of S. scabies.
Keywords: potato common scab, Streptomyces scabies, Bacillus amyloliquefaciens, Surfactin, srf gene cluster
Introduction
Potato is one of the four major food crops in the world but is easily infected with multiple diseases including common scab, late blight, bacterial wilt and soft rot. Potato common scab is mainly caused by gram positive bacteria from the Streptomyces genus including S. scabies, S. acidiscabies, S. turgidiscabies and S. ipomoeae (Li et al., 2019). The infected potato may exhibit necrotic lesions or corky symptoms on tuber surfaces, thus reducing tuber quality and marketability. Scab symptoms are mainly caused by secreted thaxtomins, such as thaxtomin A, a cellulose biosynthesis inhibitor and key pathogenicity determinant of most Streptomyces species (Lawrence et al., 1990; Loria et al., 1995). Although thaxtomin A acts as the main phytotoxin in the development of scab symptom, other phytotoxins can also play significant role in disease development (Li et al., 2019). For instance, Bignell team demonstrated that nigericin and geldanamycin secreted by Streptomyces sp. 11-1-2 were phytotoxic to potato tuber tissue (Diaz-Cruz et al., 2022). Currently, development of effective control strategies for potato common scab is challenging due to limited knowledge about the genetic diversity of S. scabies and genetic differences in multiple potato cultivars (Lerat et al., 2009). The traditional cultivation control includes crop rotation, irrigation, soil amendment to lower pH, disease-free tuber seed and disease resistant potato cultivars (Dees and Wanner, 2012). Biological control of potato common scab using Bacillus spp., Pseudomonas spp. and non-pathogenic Streptomyces spp. is an emerging area (Arseneault et al., 2016; Lin et al., 2018; Zhang et al., 2020) as their use can reduce environmental pollution produced by chemical pesticides. In our previous study, we found that B. amyloliquefaciens Ba01 attenuated the disease severity of potato common scab in pot assays (from 55.6% to 4.2%) and field trial (from 14.4% to 5.6%, achieving 61.1% control effectiveness) via secreting surfactin, iturin A and fengycin as potential antagonistic mechanisms (Lin et al., 2018). However, the study did not provide genetic evidence showing that Bacillus genes responsible for synthesis of surfactin, iturin A or fengycin are required to combat potato common scab.
Bacillus spp. can secrete many secondary metabolites including lipopeptides (surfactin, fengycin and iturin), polyketides (difficidin, bacillaene and macrolactin) and volatiles, which may directly inhibit the growth or sporulation of plant pathogens or elicit induced systemic resistance (ISR) of the plant host (Ongena and Jacques, 2008; Soni and Keharia, 2021). Surfactin or biosurfactant, a lipoheptapeptide, was first isolated in 1968 from B. subtilis (Arima et al., 1968), and its synthesis required multiple enzymes called nonribosomal peptide synthetases (Theatre et al., 2021). The surfactin family comprises various peptide moieties of surfactin and thus provides a broad spectrum of biological activity. Surfactin secretion is required for Bacillus spp. to control various plant pathogens or diseases. Luo team demonstrated that srfA of B. subtilis 916 was required for surfactin production and antifungal activity against Fusarium oxysporum (Luo et al., 2015). Nifakos team demonstrated that B. velezensis Bvel1 could control bunch rot caused by Botrytis cinerea in postharvest table grapes, and the potential mechanism was associated with secretion of surfactin and other secondary metabolites, but did not provide evidence on the gene level (Nifakos et al., 2021). Chen team showed that secretion of crude lipopeptides including surfactin, fengycin and iturin by B. velezensis FJAT-46737 exhibited antagonistic activity against Ralstonia solanacearum causing tomato bacterial wilt (Chen et al., 2020). Sarwar team revealed that surfactin A purified from Bacillus NH-100 and NH-217 possessed biocontrol activity against rice bakanae disease (Sarwar et al., 2018). Jin team proved that surfactin produced by B. velezensis HN-2 could inhibit the growth of Xanthomonas oryzae pv. oryzae causing rice disease (Jin et al., 2020). In addition to directly inhibiting the growth of plant pathogens, surfactin secreted from Bacillus also has the ability to induce plant defense responses. For example, Cawoy team showed that induced systemic resistance of the host was correlated with the amount of surfactin secreted by Bacillus (Cawoy et al., 2014). Wang team demonstrated that surfactin secreted by B. pumilus W-7 could inhibit potato late blight via induced plant defense responses by increasing the expression of biocontrol genes such as pod, pal and cat (Wang et al., 2020). However, the roles of Bacillus genes involved in surfactin biosynthesis and targeting potato common scab remain largely unknown.
In this study, we found that srf gene cluster of B. amyloliquefaciens Ba01 is required for surfactin biosynthesis, swarming ability, biofilm formation and inhibition ability against S. scabies, a bacterium that commonly causes potato common scab.
Materials and methods
Strains, chemicals, media and growth conditions
The strains and plasmids used in this study are listed in Table 1 . Streptomyces scabies PS07 was grown on yeast malt extract (YME) solid medium at 28°C. For sporulation, S. scabies PS07 was grown on solid Streptomyces sporulation medium #1 (SSM1) at 28°C for 14 days in the dark (Lin et al., 2018). Spores were collected with a sterilized cotton swab and 1% phosphate buffered saline (PBS) plus 0.1% Tween 20 solution and washed twice with ddH2O. Spore suspension was filtered through miracloth (22-25 m; Calbiochem, Billerica, MA, USA), while spore concentration was determined by serial dilution. Bacillus amyloliquefaciens Ba01 and Escherichia coli strains were cultured in Luria-Bertani (LB) broth or on LB solid plates at 37°C.
Table 1.
Strains and plasmids used in this study.
| Strain/plasmid | Description | Source |
|---|---|---|
| Strain | ||
| Bacillus amyloliquefaciens | ||
| Ba01 | Wild type | (Lin et al., 2018) |
| FRY1 | Δsrf #1 | This study |
| FRY3 | Δsrf #3 | This study |
| Streptomyces scabies | ||
| PS07 | Wild type | (Lin et al., 2017) |
| Plasmid | ||
| pMiniMad2 | E. coli−Bacillus shuttle vector, Apr, Emr | (Patrick and Kearns, 2008) |
| pRY1 | pMiniMad2 carrying 5′ and 3′ NCR of Ba01 srf gene cluster | This study |
Media and chemicals used in this study are described below. YME medium (0.4% yeast extract, 1% malt extract, 0.4% dextrose and 2% agar), SSM1 (0.1% MgSO4·7H2O, 0.05% K2HPO4, 0.1% yeast extract, 1% soluble starch, 0.1% casein and 2% agar), LB broth (0.5% yeast extract, 1% tryptone and 1% NaCl)/solid medium (1.5% agar), surfactin (dissolved in methanol; Sigma SI-S3523, St. Louis, MO, USA). When necessary, antibiotics were added at the following concentrations: 1 μg/ml erythromycin (Bio Basic, Markham, ON, Canada) plus 25 μg/ml lincomycin (Bio Basic, Markham, ON, Canada) (mls for Bacillus spp.) (Tsui et al., 2004; Patrick and Kearns, 2008), 100 μg/ml ampicillin (MDBio, New Taipei City, Taiwan) (for E. coli). Minimal salts glutamate glycerol (MSgg) medium (0.026 g KH2PO4, 0.061 g K2HPO4, 2.09 g MOPS and 0.04 g MgCl2·6H2O per 100 ml of dH2O, and adjust pH to 7.0 using KOH)/solid (1.5% agar). The MSgg base was autoclaved, cooled down to room temperature and supplemented with filtered (0.22 μm) 0.1 ml of 0.7 M CaCl2, 0.1 ml of 100 mM MnCl2, 0.1 ml of 50 mM FeCl3, 0.1 ml of 1 mM ZnCl2, 0.1 ml of 2 mM thiamine, 0.57 ml of 86% glycerol and 10 ml of 5% K-glutamate.
Plasmid construction and transformation
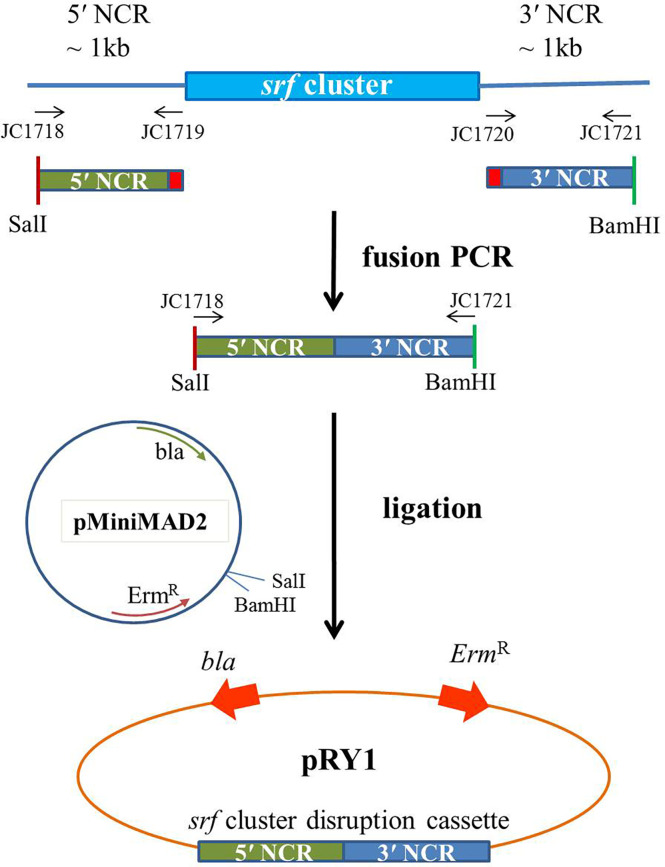
The 5′ and 3′ non-coding region (NCR) fragments (~1 kb) of srf gene cluster of Ba01 were amplified using primers JC1718/1719 and JC1720/1721, respectively. The primers JC1719/17120 have 27 bp complementary sequence for fusion PCR (Gao et al., 2017). These two fragments were fused together by fusion PCR using primers JC1718/1721, and digested with BamHI and SalI. Then the fragment was ligated into the BamHI and SalI cutted pMiniMAD2, which carried a temperature-sensitive origin of replication and an erythromycin resistance cassette to generate plasmid pRY1 ( Table 1 ; Figure 1 ). Plasmid pRY1 was transformed into Ba01 using a high osmolarity electroporation method with slight modifications (Zhang et al., 2011; Lu et al., 2012). In brief, single colony of Ba01 was grown in 3 ml LB broth overnight at 37°C with shaking at 200 rpm. Culture was 16-fold diluted with LB broth containing 0.5 M sorbitol, then incubated for about 2.5 h at 37°C with shaking in order to achieve optical density at 600 nm (OD600) 0.85 to 0.95. The culture was cooled on ice for 10 min and harvested by centrifugation for 10 min with 8,000 g at 4°C. Then, the cells were washed four times with ice-cold electroporation medium (ETM) (0.5 M sorbitol, 0.5 M mannitol and 10% glycerol), and electro-competent cells were resuspended in 1/40 (v/v) ETM (~2×1010 CFU/ml). For electroporation, 100 μl of Ba01 competent cells were mixed with 1 to 3 μg plasmid pRY1, and transferred into an ice-cold electroporation cuvette (Bio-Rad #1652086, Hercules, CA, USA). After incubation on ice for 1 min, the mixture was shocked by MicroPulser Electroporator (Bio-Rad #165-2100, Hercules, CA, USA) at 2.1 kV/cm for 4.5 to 5.0 ms. Then 1 ml of recovery medium (LB broth containing 0.5 M sorbitol and 0.38 M mannitol) was added to the mixture and incubated at 37°C for 3 h with agitation. After incubation, the culture was spread on mls agar plate (LB agar plate containing 1 μg/ml erythromycin and 25 μg/ml lincomycin) at 28°C for selecting transformants. Transformants were initially checked by colony PCR with primers JC1512/1513 by amplifying ErmR cassette on plasmid pRY1.
Figure 1.
Scheme of the plasmid construction for making Δsrf mutant in B amyloliquefaciens Ba01. Primers JC1718/1719 and JC1720/1721 were used to amplify 5′ and 3′ NCR sequences of srf gene cluster of B amyloliquefaciens Ba01, respectively, and fused together by fusion PCR using primers JC1718/1721. After enzyme digestions the ~2 kb cassette was ligated to pMiniMAD2 in order to generate disruption plasmid pRY1.
In-frame deletion of the srf gene cluster in B. amyloliquefaciens Ba01
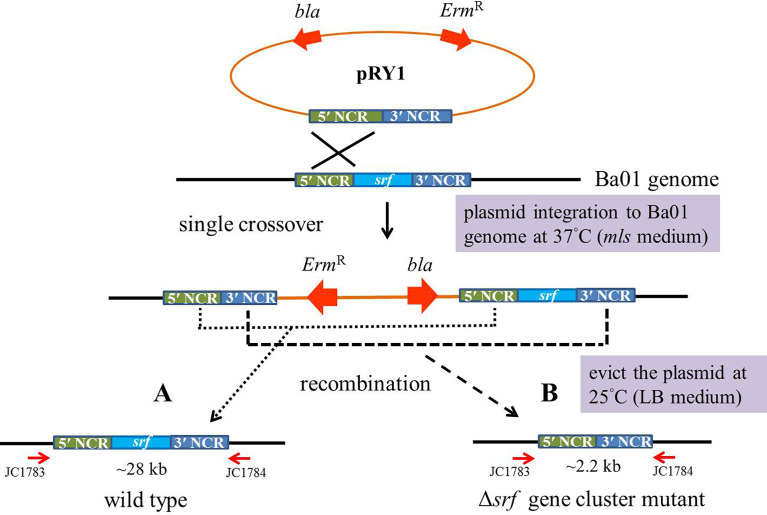
Transformants with the erythromycin resistance gene were incubated overnight in 3 ml mls media at 28°C with agitation. The cultures were diluted and plated on mls solid media for 1 day at 37°C, a restrictive temperature for plasmid integration into the Ba01 genome in order to obtain single crossover recombinants. Potential recombinants were picked and subcultured in 3 ml LB broth overnight at 25°C, then subcultured overnight in fresh LB broth at 1:100 ratio at 25°C, a permissive temperature for plasmid replication. The above procedures were repeated in order to induce second crossover in recombinants. The cultures of potential recombinants were diluted and plated on LB agar plates at 37°C for 1 day, then the colonies were patched on mls solid medium to identify mls-sensitive colonies ( Figure 2 ). Two independent srf gene cluster mutants (FRY1 and FRY3) were obtained and checked by PCR with primers JC1468/1469 (for amplification of srfAD ORF) and JC1783/1784 (for confirmation of correct integration).
Figure 2.
Scheme of in-frame markerless deletion method to construct the srf gene cluster mutants in B amyloliquefaciens Ba01. Single crossover recombinants were obtained by culturing the transformants at restrictive temperature (37°C) for plasmid pRY1 integration into Ba01 genome and using mls medium for selection. To evict the plasmid from Ba01 genome after homologous recombination, the strain was incubated overnight in 3 ml of LB broth at a permissive temperature (25°C) for plasmid replication. The cell cultures were diluted and plated on LB agar at 37°C for 24 h The colonies were patched on mls medium to select mls-sensitive colonies. The srf gene cluster mutants were checked by PCR with primers JC1783/1784.
Cell growth and endospore formation
For measuring cell growth, wild type and Δsrf mutants were incubated overnight in 5 ml LB broth at 37°C and diluted to OD600 = 1. Then 200 μl of culture was inoculated in 20 ml fresh LB broth at 37°C with agitation. For growth curve experiments, bacterial numbers were calculated by measuring OD600 and serial dilution methods after 0, 3, 6, 9, 12, 24 and 30 h. Each experiment was performed in triplicate.
For measuring endospore formation, cultures of the wild type and Δsrf mutants were serial diluted and plated on LB agar plates to calculate total bacterial numbers after incubating for 1, 3, 5 and 7 days at 37°C with agitation. Cultures were then incubated at 80°C for 10 min to kill vegetative cells, serial diluted and incubated on LB agar plates at 37°C for 24 h. The ratio of endospore and total bacterial numbers was represented as percentage of endospore production. Each experiment was performed as three replicates. Endospore staining was performed based on Reynolds team with minor modifications (Reynolds et al., 2009). In brief, 20 μl culture of wild type or Δsrf mutant were placed on a slide and heat fixed by flame, then the smear was flooded with malachite green, and steamed 5 to 10 min for staining. Then the slides were washed with ddH2O, the smear was flooded with safranin and left for 1 min before washing the slide with ddH2O in order to remove the secondary stain. The endospores were observed under optical microscope with 100× magnification.
Disk diffusion assays
One hundred microliters of 107 CFU/ml S. scabies spores were spread on YME agar plates, and 6 mm disks were placed on the plates. Then 3 μl culture of wild type or Δsrf mutant (OD600 = 1) was dropped on the disk, and the plates were incubated at 28°C for 2 days in the dark. The size of the inhibition zone was calculated as below: (diameter of the inhibition zone – diameter of the colony size)/2 (mm). Each experiment contained four replicates.
MALDI-TOF mass spectrometry analysis
The wild type and Δsrf mutants were incubated in 10 ml LB broth with agitation at 37°C for 36 h, and then centrifuged at 13,000 rpm for 5 min to collect the supernatant, which was further extracted twice with ethyl acetate, and finally dissolved in methanol for mass analysis (Han et al., 2005; Liao et al., 2016). One microliter of the sample and surfactin standard (2 mg/ml) were dropped into a sample well. The data were collected using the MALDI TOF/TOF spectrometer (Bruker Autoflex Speed, Agricultural Biotechnology Research Center, Academia Sinica) and analyzed by Bruker Compass version 1.2 software suite.
Swarming motility
The wild type and Δsrf mutants were incubated overnight in LB broth with agitation at 37°C, washed with ddH2O and diluted to OD600 = 1. Then 5 μl of each culture was spotted on the center of 0.25%, 0.5% or 0.7% solid LB agar plates, and incubated at 37°C for 48 h (Gao et al., 2017). In order to investigate whether surfactin could rescue the swarming motility of Δsrf mutants, 10 μl solution containing 1 μg, 20 μg or 100 μg of commercial surfactin (dissolved with methanol) was added on the center of 0.7% solid LB agar plate before adding 5 μl culture of Δsrf mutants (OD600 = 1) onto the top. The plates were incubated at 37°C for 48 h and the diameter of the colony was measured. Each experiment was performed with five replicates.
Biofilm formation and colony architecture
For observation of biofilm formation, 10 μl of strains (OD600 = 1) were added to 1 ml MSgg broth and incubated in 24-well polystyrene plate at 30°C for 2 days without agitation. Twenty micromolar commercial surfactin was further added into the wells in order to investigate whether surfactin could rescue biofilm formation of Δsrf mutants. For colony architecture, 3 μl of strains (OD600 = 1) were spotted on the center of MSgg solid medium, and incubated at 30°C for 2 or 4 days. To determine whether commercial surfactin could rescue the colony architecture of Δsrf mutants, 5 μl of solution containing 10 μg commercial surfactin was spotted on the center of MSgg solid medium before adding 3 μl of Δsrf mutants (OD600 = 1).
Extracellular enzyme activity and phosphate-solubilizing ability
Bacillus spp. has the ability to secrete extracellular hydrolases and possesses phosphate-solubilizing ability to defend against pathogens or help plants to absorb nutrients. To investigate whether hydrolases and phosphate-solubilizing ability of Δsrf mutants would be affected, the following assays were performed. (i) protease activity assay: 3 μl of strains (OD600 = 1) were inoculated on skimmed milk agar (1% peptone and 1.5% agar mixed with 1% skimmed milk) and incubated at 37°C for 48 h. The colorless zone represented colonies with protease-producing ability. (ii) α-amylase activity assay: 3 μl of strains (OD600 = 1) were inoculated on the medium (0.2% soluble starch, 1.5% agar, pH = 7) and incubated at 37°C for 48 h, then the plates were stained with iodine solution (0.67% potassium iodide, 0.33% iodine). The colorless zone represented the colony with α-amylase-producing ability. (iii) cellulase activity assay: 3 μl of strains (OD600 = 1) were inoculated on carboxymethyl cellulose (CMC) agar medium (1% peptone, 1% yeast extract, 1% carboxymethyl cellulose, 0.5% NaCl, 0.1% KH2PO4, 1.5% agar, pH = 7) and incubated at 37°C for 48 h, then the colonies were washed with a sterilized cotton swab and ddH2O. Plates were then flooded with 0.1% Congo red for 1 h and destained with 1 M NaCl solution for 1 h. The colorless zone represented colonies with cellulase-producing ability. (iv) phosphate-solubilizing ability assay: 3 μl of strains (OD600 = 1) were inoculated on Pikovskaya’s agar medium (Pikovskaya, 1948), and incubated at 37°C for 5 days, then the colonies were washed with a sterilized cotton swab and ddH2O. The colorless zone represented colonies with phosphate-solubilizing ability.
Tuber slice assays
Commercial G4 potato tubers of the cultivar Kennebec were used. The surfaces of potato tubers were sterilized with 400 ppm hypochlorous acid water. Cores (1.2 cm diameter) of pith tissue were removed from tubers using a puncher and sliced into pieces 0.25 cm thick. Then 25 μl (108 CFU/ml) of S. scabies PS07 spores were inoculated onto potato tuber slices, which were placed on moist filter paper in Petri dishes. After incubating the tuber slices at 28°C for 1 day in the dark, 20 μl of wild type, Δsrf mutants (OD600 = 1), commercial surfactin (0.1 mg/ml) or ddH2O were dropped onto the potato tuber slices. The samples were further incubated in a moist chamber in the dark at 28°C for 5 days before being photographed.
Results
srf gene cluster was deleted in B. amyloliquefaciens Ba01
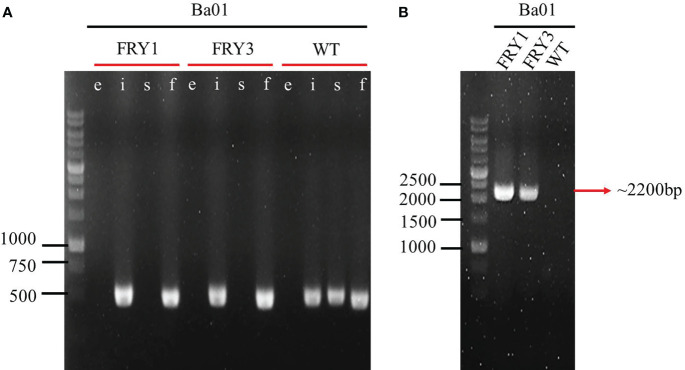
The plasmid pRY1 was constructed and transformed into B. amyloliquefaciens Ba01 by electroporation ( Figure 1 ). An in-frame markerless deletion method was used to generate Δsrf gene cluster mutants (Δsrf mutant; Figure 2 ). Two independent Δsrf mutants FRY1 and FRY3 were obtained and confirmed by PCR with primer pairs JC1512/1513, JC1466/1467, JC1468/1469 and JC1470/1471 ( Table 2 ). In Figure 3A , srfAD ORF could only be amplified with primers JC1468/1469 in the wild type, but not in the Δsrf mutants. Two Δsrf mutants were further validated by primers JC1783/1784 (SrfOUT-F/R) in order to verify the correct integration in the genome. In Figure 3B , srf gene cluster of the wild type was too long (~26 kb) to amplify, while two Δsrf mutants could amplify a band of 2.2 kb, a fragment containing only 5′ and 3′ NCR of srf gene cluster.
Table 2.
Primers used in this study.
| Primer | Use | Sequence (5′ → 3′)* |
|---|---|---|
| JC1466 | ituD ORF | aaaAGTGTATGCCGCACCCTTTT |
| JC1467 | ituD ORF | aaaGAGCGATGCGATCTCCTTGG |
| JC1468 | srfAD ORF | aaaCCGCCGTTGAGGATTTTGAA |
| JC1469 | srfAD ORF | aaaCATGTGGCCGTCCGAAAACT |
| JC1470 | fenA ORF | aaaGCGAGAGGCTGGTATTGCAT |
| JC1471 | fenA ORF | aaaGAACACCTTTCACTGGCGGA |
| JC1512 | ErmR cassette-F | TAGGCACACGAAAAACAAGTTAAGG |
| JC1513 | ErmR cassette-R | AACCGTGTGCTCTACGACC |
| JC1718 | SrfUP‐F (SalI) | GCCGTCGACATGGGAATAACTTTTTATCC |
| JC1719 | SrfUP‐R | GGCATCGATATTGCTCCAGAGATACTGTAAAC |
| JC1720 | SrfDN‐F | CAGTATCTCTGGAGCAATATCGATGCCGATCG |
| JC1721 | SrfDN‐R (BamHI) | CGCGGATCCATCTTTAACCATTAAAGGAAAAG |
| JC1783 | SrfOUT-F | GGAGGCTGTTTCTAAGGAAGAATTGAC |
| JC1784 | SrfOUT-R | GACGTTTTATTTTGCCGGTCTGTTG |
*Bold letters represent restriction sites used for the cloning; underlined letters represent overlapping PCR sequences.
Figure 3.
Confirmation of srf gene cluster mutants by PCR amplification. (A) lane e: Erm R marker (~1100 bp; JC1512/1513), lane i: ituD ORF (469 bp; JC1466/1467), lane s: srfAD ORF (485 bp; JC1468/1469), lane f: fenA ORF (455 bp; JC1470/1471). (B) Outside primer of 5′ and 3′ NCR of srf gene cluster (~2200 bp; JC1783/1784).
srf gene cluster is required for secreting surfactin in B. amyloliquefaciens Ba01
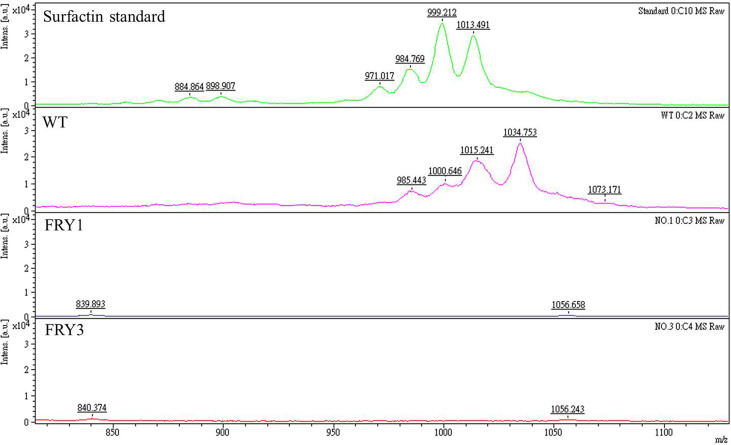
Based on mass spectrometry analysis, the peaks of surfactin were detected in the wild type and surfactin standard, but absent in Δsrf mutants ( Figure 4 ). These results indicated that srf gene cluster is essential for secreting surfactin in B. amyloliquefaciens Ba01. Besides, we found that the mass-to-charge (m/z) ratio of the surfactin from Ba01 was slightly different from the surfactin standard. We speculated that the difference might be because the surfactin standard purchased was extracted from B. subtilis, a relative of B. amyloliquefaciens.
Figure 4.
The Δsrf mutants lose the ability to secrete surfactins. The supernatants of cultures were collected and extracted twice with ethyl acetate, followed by resuspension with methanol before conducting mass spectrometry analysis. The surfactin peaks were found in the standard and wild type (WT), but deficient in Δsrf mutants (FRY1 and FRY3).
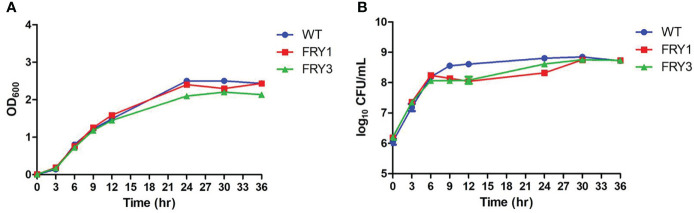
The growth kinetics of B. amyloliquefaciens Δsrf mutants were slightly decreased
B. amyloliquefaciens Ba01 wild type and Δsrf mutants were cultured in LB broth, OD600 values were measured by spectrophotometer and bacterial number (colony-forming units) was determined by serial dilution at 0, 3, 6, 9, 12, 24 and 30 h. Although there were no differences between the wild type and Δsrf mutants based on OD600 values ( Figure 5A ), the colony-forming units of two Δsrf mutants were slightly decreased at 9 and 12 h compared with the wild type (P < 0.001, t-test) ( Figure 5B ).
Figure 5.
The growth kinetics of Δsrf mutants were slightly decreased. (A) Growth curves were determined based on the OD600 values. (B) Growth curves were determined based on the viable cell counts (CFU/ml). The strains were 10-fold diluted and spread on LB agar plates. Total colony numbers were counted after 1 day growth at 37°C.
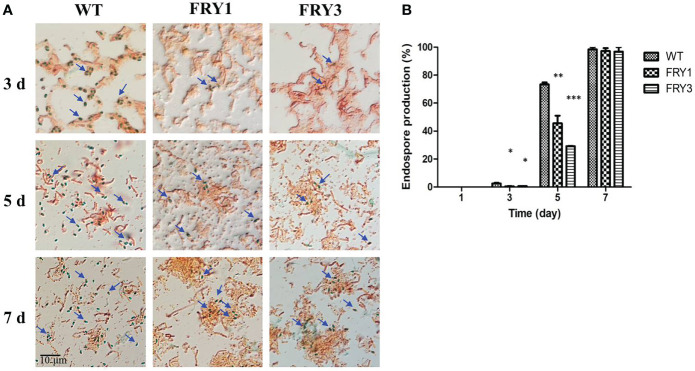
The endospore formation was decreased in Δsrf mutants of B. amyloliquefaciens Ba01
In the endospore formation test, B. amyloliquefaciens Ba01 wild type and Δsrf mutants did not form endospores after incubation for 1 day in LB broth. After 3 days, the endospore formation rate of the wild type was 2.5 ± 0.46%, while those of Δsrf mutants were significantly reduced (P<0.01) to 0.5 ± 0.09% (FRY1) and 0.7 ± 0.7% (FRY3) ( Figures 6A, B ). After 5 days, the endospore formation rate of the wild type was 73.4 ± 1.35%, while those of Δsrf mutants were also significantly reduced (P<0.01) to 45.6 ± 5.49% (FRY1) and 29.1 ± 0.13% (FRY3). Under a microscope, more endospores were observed in the wild type than in the Δsrf mutants after incubation for 3 or 5 days ( Figure 6A ). After 7 days, the endospore formation rate of the wild type was 98.3 ± 1.67%, while those of the Δsrf mutants were 97.2 ± 2.14% (FRY1) and 96.8 ± 3.2% (FRY3).
Figure 6.
The endospore formation of Δsrf mutants was decreased. (A) Endospores were stained and observed under a microscope (100×). (B) Graph of endospore formation. The strains were treated at 80°C for 10 min to kill vegetative cells and plated on LB agar to calculate the number of endospores. Endospore formation was the ratio of endospore numbers divided by total colony numbers. P values were calculated by t-test, “*”, “**” and “***” represent P < 0.05, P < 0.01 and P < 0.001 respectively as compared to the WT.
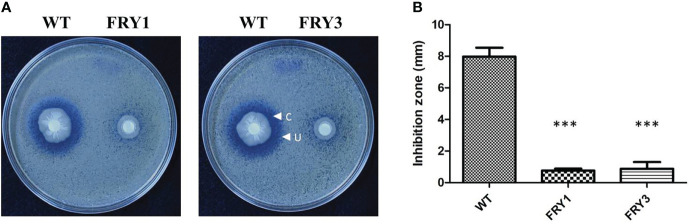
The Δsrf mutants showed reduced inhibition activity against S. scabies
In disk diffusion assays, B. amyloliquefaciens Ba01 wild type could effectively inhibit the growth of S. scabies PS07 and formed clear inhibition/undifferentiating zones after incubation for 2 days, while Δsrf mutants could only form an undifferentiating but not a clear inhibition zone ( Figure 7A ). The clear and undifferentiating inhibition zones produced by the wild type were 4.63 ± 0.49 mm and 7.97 ± 0.57 mm, respectively, while undifferentiating zones produced by Δsrf mutants were 0.77 ± 0.12 mm (FRY1) and 0.88 ± 0.42 mm (FRY3), showing significantly reduced (P<0.001) as compared to the wild type ( Figure 7B ).
Figure 7.
The Δsrf mutants reduced the inhibition activity against S. scabies. (A) Disk diffusion assay. The 106 spores of S. scabies PS07 were spread on YME agar plates, and a disk containing 3 μl WT or Δsrf mutant culture (OD600 = 1) was put on the surface of plates. The plates were incubated at 28°C for 2 days. “C” represents clear inhibition zone, and “U” represents undifferentiated zone. (B) Graph of (A). P values were calculated by t-test, “***” represents P < 0.001 as compared to the WT.
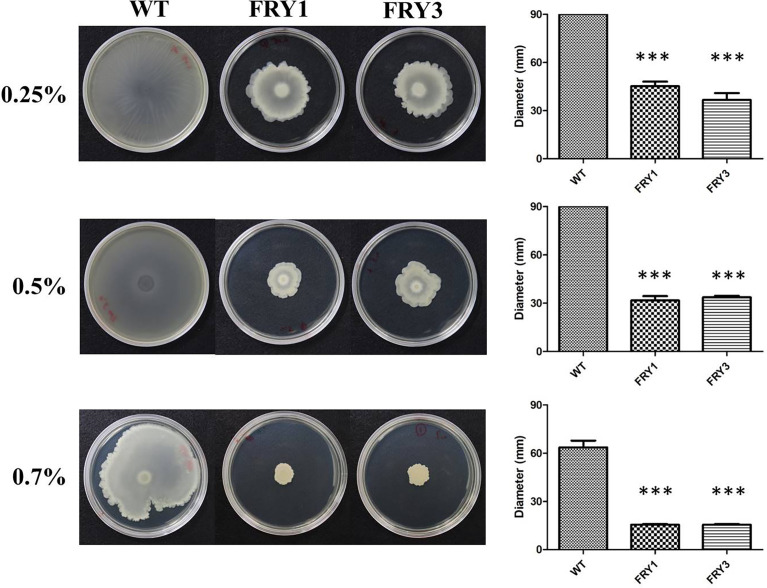
Swarming motility was attenuated in Δsrf mutants of B. amyloliquefaciens Ba01
The wild type could swarm over the 0.25% and 0.5% LB agar plates, while the swarming motility of Δsrf mutants was reduced to 45.2 ± 2.8 mm (FRY1) and 36.6 ± 4.31 mm (FRY3) on 0.25% LB agar plates, and 31.7 ± 2.68 mm (FRY1) and 33.7 ± 0.89 mm (FRY3) on 0.5% LB agar plates ( Figure 8 ). In addition, the wild type and Δsrf mutants swarmed 63.6 ± 4.19 mm, 15.5 ± 0.46 mm (FRY1) and 15.5 ± 0.48 mm (FRY3), respectively, on 0.7% LB agar plates. Overall, the swarming motility of Δsrf mutants on 0.25%, 0.5% and 0.7% LB agar plates were significantly decreased compared with the wild type (P < 0.001, Figure 8 ).
Figure 8.
The Δsrf mutants showed attenuated swarming motility. Five microliters of strains (OD600 = 1) were inoculated on 0.25%, 0.5% or 0.7% solid LB agar at 37°C for 48 h. P values were calculated by t-test, “***” represents P < 0.001 as compared to the WT.
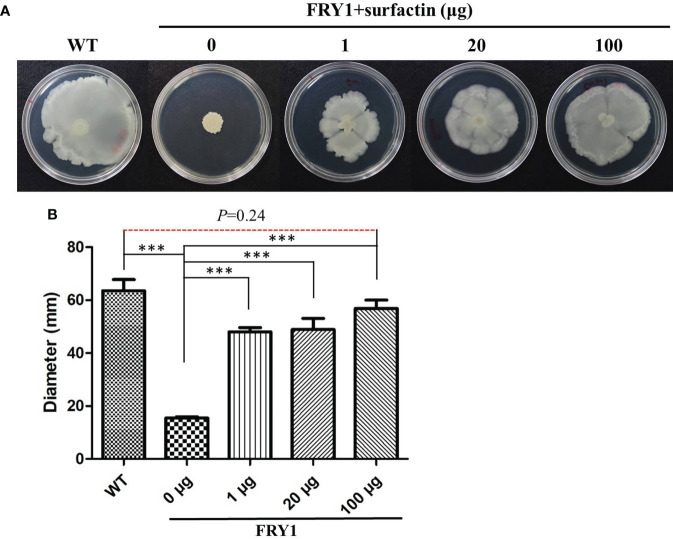
Surfactin could rescue the swarming motility of Δsrf mutant in B. amyloliquefaciens Ba01
In order to confirm that the defect of swarming motility in Δsrf mutants was associated with surfactin acquirement, a surfactin supplementing experiment was conducted. After adding 1, 20 or 100 μg of commercial surfactin on 0.7% LB agar plates, the swarming motility of Δsrf mutant (FRY1) was significantly increased compared to Δsrf mutant without adding surfactin ( Figures 9A, B , P < 0.001). In addition, there was no difference in the swarming motility between the wild type and Δsrf mutant supplemented with 100 μg surfactin ( Figures 9A, B ), indicating that 100 μg of surfactin was sufficient to restore the swarming motility of Δsrf mutant to similar to the wild type.
Figure 9.
Surfactin could rescue the swarming motility of Δsrf mutant. (A) Solution of 10 μl containing 1 μg, 20 μg or 100 μg of surfactin were spotted on the center of 0.7% solid LB medium before dropping 5 μl of strains (OD600 = 1) onto the top, and then incubated at 37°C for 48 h (B) Graph of (A). P values were calculated by t-test, “***” represents P < 0.001 as compared to the Δsrf mutant (FRY1) in the absence of surfactin.
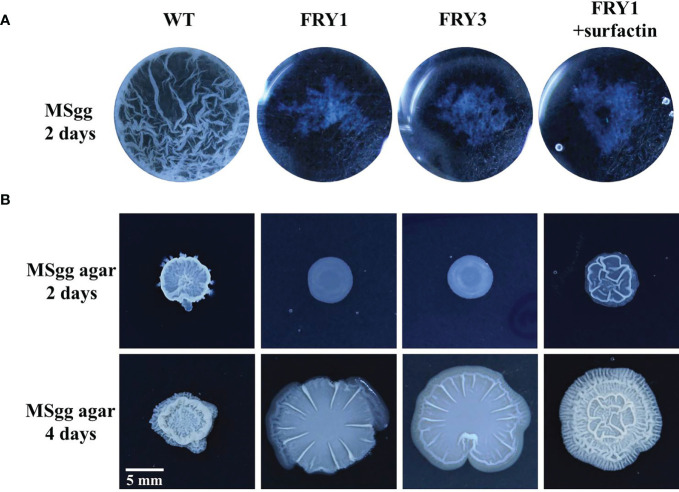
Biofilm formation was impaired in Δsrf mutants of B. amyloliquefaciens Ba01
The wild type could form thick and wrinkled biofilm in liquid MSgg medium after incubation for 2 days, while Δsrf mutants exhibited impaired biofilm formation ( Figure 10A ). Interestingly, the biofilm formation of Δsrf mutant (FRY1) could not be restored by supplementing surfactin ( Figure 10A ). On solid MSgg medium, Δsrf mutants formed less structured and flattered colonies than the wild type after incubation for 2 or 4 days, while supplementing surfactin could partially rescue the colony architecture of Δsrf mutant (FRY1) ( Figure 10B ).
Figure 10.
The Δsrf mutants lost the ability to form biofilm. (A) WT formed a thick and wrinkled biofilm, while Δsrf mutants did not. Surfactin did not rescue the biofilm formation of Δsrf mutant in liquid MSgg medium. For biofilm formation, 10 μl of strains (OD600 = 1) were added to 1 ml MSgg broth and incubated in a 24-well plate at 30°C for 2 days. The addition of 20 μM of surfactin was to determine if surfactin could rescue biofilm formation. (B) For colony morphology, 3 μl of strains (OD600 = 1) were spotted on the center of MSgg solid medium, and then incubated at 30°C for 2 or 4 days. To determine if surfactin could rescue colony morphology, solution of 5 μl containing 10 μg of surfactin was spotted on the center of MSgg solid medium before adding 3 μl of FRY1 (OD600 = 1).
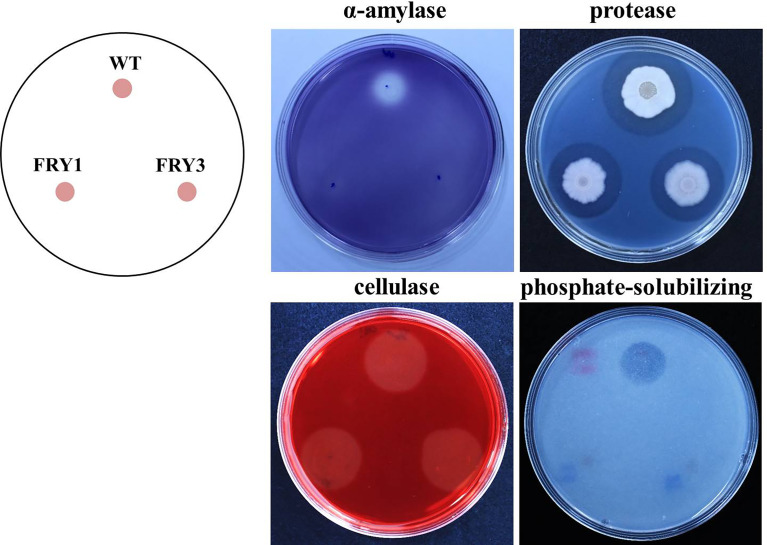
Secretion of extracellular enzymes and solubilizing phosphates was decreased in Δsrf mutants of B. amyloliquefaciens Ba01
In extracellular α-amylase activity and phosphate-solubilizing ability tests, we found that two Δsrf mutants could not grow normally and form clear zone in the same way as the wild type, indicating that α-amylase secretion and solubilizing phosphate activity was impaired in Δsrf mutants ( Figure 11 ). In the protease test, although two Δsrf mutants could form clear zones [6.0 ± 0.16 mm (FRY1) and 5.5 ± 0.39 mm (FRY3)], they were significantly smaller than that of the wild type (7.4 ± 0.13 mm) based on t-test statistical analysis. However, in the cellulase test, wild type and Δsrf mutants grew normally and exhibited no difference in forming a clear zone ( Figure 11 ).
Figure 11.
The enzyme activity of α-amylase and protease, and phosphate-solubilizing ability of Δsrf mutants were decreased. Three microliters of strains (OD600 = 1) were inoculated on media testing α-amylase, protease, cellulase and phosphate-solubilizing ability and incubated at 37°C for 2 days (α-amylase, protease, cellulase) or 5 days (phosphate-solubilizing).
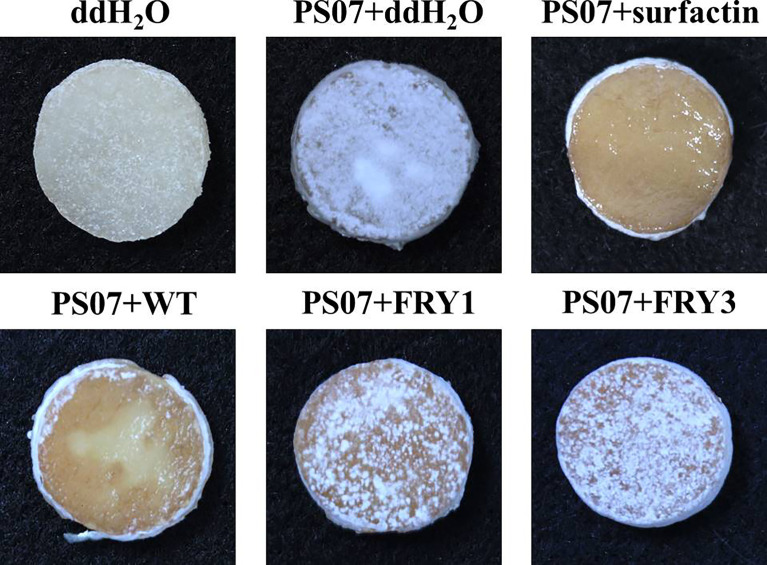
The Δsrf mutants could not inhibit growth and sporulation of S. scabies on potato tuber slices
In potato tuber slice assays, tuber slices treated with S. scabies PS07 were covered with S. scabies colonies and white spores; however, supplementing surfactin or adding B. amyloliquefaciens Ba01 effectively inhibited the growth and sporulation of S. scabies PS07 on potato tuber slices after incubation for 5 days at 28°C ( Figure 12 ). In contrast, the two Δsrf mutants (FRY1 and FRY3) could not effectively inhibit the growth and sporulation of S. scabies PS07 on potato tuber slices ( Figure 12 ).
Figure 12.
Two Δsrf mutants did not effectively inhibit the growth and sporulation of S. scabies PS07 on potato tuber slices. The 25 μl 108 spores/ml of S. scabies PS07 spores were inoculated on potato tuber slices, incubated at 28°C for 1 day in the dark, and then 20 μl of Ba01 WT, FRY1, FRY3 (OD600 = 1), ddH2O or surfactin (0.1 mg/ml) were added to potato tuber slices. The samples were further incubated in the moist chamber at 28°C for 5 days before photographing.
Discussion
Currently, many strategies have been developed to control potato common scab. Our previous study showed that B. amyloliquefaciens Ba01 can control common scab in the pot assay and field trial, but detailed mechanism remains unclear (Lin et al., 2018). In this study, we further demonstrated that the mechanism of Ba01 used to inhibit the growth of S. scabies is at least in part via secretion of surfactin since the deletion of Ba01 srf gene cluster resulted in reduced inhibition activity against S. scabies. The practical use of biocontrol agents to combat common scab is still present, although it is limited by various environmental or plant host factors. The environmental factors include temperature, humidity and soil pH/microbiota, etc., while host factors contain plant resistance to common scab causing pathogens. Besides biocontrol strategy, foliar application of auxin (2,4-dichlorophenoxyacetic acid) has been used to induce potato defense response against common scab disease and its mechanism is associated with enhanced tolerance to thaxtomin A (Tegg et al., 2008). This result demonstrates that auxin can ameliorate thaxtomin A toxicity. However, the role of surfactin secreted by Bacillus species on biosynthesis of thaxtomins is unclear, indicating a potential research topic in the future.
Bacillus species are important biocontrol agents in agriculture, and gene manipulation can be used as a tool to understand the mechanisms through which they combat plant diseases. However, there are relatively few studies using mutants of B. amyloliquefaciens, mainly due to its low transformation efficiency (Zhang et al., 2013; Zhang et al., 2015). B. amyloliquefaciens, which is a gram positive bacterium, has a thick cell wall that acts as a natural barrier when introducing plasmids into the cell. Moreover, B. amyloliquefaciens possesses a BamHI restriction-modification system, which prevents incorporation of exogenous DNA (Zhang et al., 2013; Zhang et al., 2015). In addition, whether plasmids can maintain and stably replicate also significantly influences the result of transformation. In comparison to domesticated strains, the strain used in this study, B. amyloliquefaciens Ba01, is a wild strain isolated from potato tuber, which makes it particularly difficult to transform. In this study, we tried three methods to carry out transformation, the first method was making competent cells according to Konkol team (Konkol et al., 2013); the second method was conjugation, using E. coli S17-1 as donor (Rachinger et al., 2013); and the third method was electroporation. Although we finally succeeded in transforming Ba01 and obtaining Δsrf mutants via electroporation, all three methods mentioned above had low transformation efficiency, with false positive transformants that did not result in plasmids growing on media containing antibiotics, or easy loss of plasmids during subculturing. Therefore, for future studies, an improved transformation method will be required to establish a stable method by which to study gene functions of Ba01.
Surfactin, which is a cyclic lipopeptide, has been found to be commonly secreted by several Bacillus species, and it plays an important role in Bacillus biology. According to previous studies, colonies of surfactin-deficient Bacillus mutants may become flat and unwrinkled, indicating that this molecule is required for normal colony morphology in some strains (Zeriouh et al., 2014; Luo et al., 2015; Zhang et al., 2022). Also, surfactin has been found to act as a signal molecule and triggers quorum sensing, which further facilitates multicellular behaviors such as swarming and biofilm formation (Shank and Kolter, 2011; Zeriouh et al., 2014; Luo et al., 2015). In this study, although addition of extracellular surfactin could restore the ability of Δsrf mutant to swarm and form normal colonies, biofilm formation was still impaired. Failure of additional surfactin to rescue biofilm formation was also found in Bacillus subtilis ΔsrfAA mutant obtained by Luo team (Luo et al., 2015), while Zeriouh team discovered that the ability of B. subtilis ΔsrfAB mutant to form biofilm could be restored by adding commercial surfactin (Zeriouh et al., 2014). This difference suggests the possibility that there might be some other proteins which regulate biofilm formation cooperatively with srfAA gene or other genes such as srfAC and srfAD in the srf gene cluster, and surfactin may play a role as a signal triggering the biofilm formation pathway.
Besides acting as a signal molecule, surfactin is also a strong biosurfactant, with the function of promoting moisture and reducing surface tension. In addition, according to our experimental results, surfactin is related to the sporulation ability of B. amyloliquefaciens Ba01, with Δsrf mutants having poor sporulation rate. Together with stimulation of biofilm formation, these functions facilitate swarming to nutrient rich niches and stable colonization, and enhance survival rate in the field, which are determinant factors that lead to efficient biocontrol capability of Bacillus species (Zeriouh et al., 2014; Aleti et al., 2016). In addition, there have been previous reports that indicated that surfactin is an ISR elicitor, which stimulates plant defense response in a wide range of crops (Cawoy et al., 2014; Chowdhury et al., 2015). This study also showed that surfactin may play an important role in plant-growth promotion, in which this compound seems to contribute to the enzyme activity of α-amylase and protease, and the phosphate-solubilizing ability of its producer. Consequently, we postulated that by producing surfactin, Ba01 could promote plant-growth and indirectly increase plant immunity to enhance the resistance of host plants against pathogens. All of the findings above suggest that surfactin production is essential for improving biocontrol ability of biocontrol microorganisms.
In conclusion, this study revealed that surfactin is a versatile compound. Besides its direct antagonistic activity against S. scabies, it is also related to swarming, biofilm formation, sporulation, phosphate solubilization and enzyme activity of α-amylase and protease. With these functions, B. amyloliquefaciens Ba01 is a potential candidate for controlling potato common scab. Combined with the findings of previous reports, there is also a chance that Ba01 could induce a systemic response in potato, providing another mechanism for combating potato common scab, which is worth further investigation. If surfactin does trigger induced systemic resistance in potato plants, besides using Ba01 as a biocontrol agent, direct application of surfactin may serve as an option as well, since the compound possesses pathogen growth inhibition and plant immunity enhancement ability simultaneously.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
R-YF designed and conducted the experiments. Y-HC wrote the manuscript. CL, C-HT and Y-LY assisted in some experimental sections. Y-LC supervised the experiments and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work is financially supported by grant NTU110L7811 from the Higher Education Sprout Project, Ministry of Education, and grants MOST 110-2923-B-002-002-MY3 and 110-2320-B-002-050-MY3 from the Ministry of Science and Technology, Taiwan.
Acknowledgments
We are grateful to Miranda Loney for language editing, and Drs. Shih-Tung Liu and Wan-Ju Ke at Chang Gung University for providing Bacillus transformation protocol. This work is financially supported by grant NTU110L7811 from the Higher Education Sprout Project, Ministry of Education, and grants MOST 110-2923-B-002-002-MY3 and 110-2320-B-002-050-MY3 from the Ministry of Science and Technology, Taiwan.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aleti G., Lehner S., Bacher M., Compant S., Nikolic B., Plesko M., et al. (2016). Surfactin variants mediate species-specific biofilm formation and root colonization in Bacillus . Environ. Microbiol. 18 (8), 2634–2645. doi: 10.1111/1462-2920.13405 [DOI] [PubMed] [Google Scholar]
- Arima K., Kakinuma A., Tamura G. (1968). Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: isolation, characterization and its inhibition of fibrin clot formation. Biochem. Biophys. Res. Commun. 31 (3), 488–494. doi: 10.1016/0006-291X(68)90503-2 [DOI] [PubMed] [Google Scholar]
- Arseneault T., Goyer C., Filion M. (2016). Biocontrol of potato common scab is associated with high Pseudomonas fluorescens LBUM223 populations and phenazine-1-Carboxylic acid biosynthetic transcript accumulation in the potato geocaulosphere. Phytopathology 106 (9), 963–970. doi: 10.1094/PHYTO-01-16-0019-R [DOI] [PubMed] [Google Scholar]
- Cawoy H., Mariutto M., Henry G., Fisher C., Vasilyeva N., Thonart P., et al. (2014). Plant defense stimulation by natural isolates of Bacillus depends on efficient surfactin production. Mol. Plant Microbe Interact. 27 (2), 87–100. doi: 10.1094/MPMI-09-13-0262-R [DOI] [PubMed] [Google Scholar]
- Chen M., Wang J., Liu B., Zhu Y., Xiao R., Yang W., et al. (2020). Biocontrol of tomato bacterial wilt by the new strain Bacillus velezensis FJAT-46737 and its lipopeptides. BMC Microbiol. 20 (1), 160. doi: 10.1186/s12866-020-01851-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S. P., Hartmann A., Gao X., Borriss R. (2015). Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42 - a review. Front. Microbiol. 6, 780. doi: 10.3389/fmicb.2015.00780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dees M. W., Wanner L. A. (2012). In search of better management of potato common scab. Potato. Res. 55 (3), 249–268. doi: 10.1007/s11540-012-9206-9 [DOI] [Google Scholar]
- Diaz-Cruz G. A., Liu J., Tahlan K., Bignell D. R. D. (2022). Nigericin and geldanamycin are phytotoxic specialized metabolites produced by the plant pathogen streptomyces sp. 11-1-2. Microbiol. Spectr. 10 (2), e0231421. doi: 10.1128/spectrum.02314-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Liu F., Zhang W., Quan Y., Dang Y., Feng J., et al. (2017). Mutations in genes encoding antibiotic substances increase the synthesis of poly-gamma-glutamic acid in Bacillus amyloliquefaciens LL3. Microbiologyopen 6 (1), e00398. doi: 10.1002/mbo3.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. S., Cheng J. H., Yoon T. M., Song J., Rajkarnikar A., Kim W. G., et al. (2005). Biological control agent of common scab disease by antagonistic strain bacillus sp. sunhua. J. Appl. Microbiol. 99 (1), 213–221. doi: 10.1111/j.1365-2672.2005.02614.x [DOI] [PubMed] [Google Scholar]
- Jin P., Wang Y., Tan Z., Liu W., Miao W. (2020). Antibacterial activity and rice-induced resistance, mediated by C15surfactin a, in controlling rice disease caused by Xanthomonas oryzae pv. oryzae . Pesticide. Biochem. Physiol. 169, 104669. doi: 10.1016/j.pestbp.2020.104669 [DOI] [PubMed] [Google Scholar]
- Konkol M. A., Blair K. M., Kearns D. B. (2013). Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis . J. Bacteriol. 195 (18), 4085–4093. doi: 10.1128/JB.00696-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. H., Clark M. C., King R. R. (1990). Induction of common scab symptoms in aseptically cultured potato tubers by the vivotoxin, thaxtomin. Phytopathology 80 (7), 606–608. doi: 10.1094/Phyto-80-606 [DOI] [Google Scholar]
- Lerat S., Simao-Beaunoir A. M., Beaulieu C. (2009). Genetic and physiological determinants of Streptomyces scabies pathogenicity. Mol. Plant Pathol. 10 (5), 579–585. doi: 10.1111/j.1364-3703.2009.00561.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J. H., Chen P. Y., Yang Y. L., Kan S. C., Hsieh F. C., Liu Y. C. (2016). Clarification of the antagonistic effect of the lipopeptides produced by Bacillus amyloliquefaciens BPD1 against Pyricularia oryzae via in situ MALDI-TOF IMS analysis. Molecules 21 (12), 1670. doi: 10.3390/molecules21121670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu J., Diaz-Cruz G., Cheng Z., Bignell D. R. D. (2019). Virulence mechanisms of plant-pathogenic Streptomyces species: an updated review. Microbiol. (Reading). 165 (10), 1025–1040. doi: 10.1099/mic.0.000818 [DOI] [PubMed] [Google Scholar]
- Lin C., Fong R. Y., Tsai C. H., Chen Y. L. (2017). The fungicide tebuconazole inhibits potato common scab caused by Streptomyces scabies . J. Plant Med. 59 (3), 31–37. doi: 10.6716/JPM.201709_59(3).0004 [DOI] [Google Scholar]
- Lin C., Tsai C. H., Chen P. Y., Wu C. Y., Chang Y. L., Yang Y. L., et al. (2018). Biological control of potato common scab by Bacillus amyloliquefaciens Ba01. PloS One 13 (4), e0196520. doi: 10.1371/journal.pone.0196520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria R., Bukhalid R., Creath R., Leiner R., Olivier M., Steffens J. (1995). Differential production of thaxtomins by pathogenic Streptomyces species in vitro . Phytopathology 85, 537–541. doi: 10.1094/Phyto-85-537 [DOI] [Google Scholar]
- Luo C., Liu X., Zhou H., Wang X., Chen Z. (2015). Nonribosomal peptide synthase gene clusters for lipopeptide biosynthesis in Bacillus subtilis 916 and their phenotypic functions. Appl. Environ. Microbiol. 81 (1), 422–431. doi: 10.1128/AEM.02921-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. P., Zhang C., Lv F. X., Bie X. M., Lu Z. X. (2012). Study on the electro-transformation conditions of improving transformation efficiency for Bacillus subtilis . Lett. Appl. Microbiol. 55 (1), 9–14. doi: 10.1111/j.1472-765X.2012.03249.x [DOI] [PubMed] [Google Scholar]
- Nifakos K., Tsalgatidou P. C., Thomloudi E. E., Skagia A., Kotopoulis D., Baira E., et al. (2021). Genomic analysis and secondary metabolites production of the endophytic Bacillus velezensis Bvel1: A biocontrol agent against Botrytis cinerea causing bunch rot in post-harvest table grapes. Plants (Basel). 10 (8), 1716. doi: 10.3390/plants10081716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongena M., Jacques P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16 (3), 115–125. doi: 10.1016/j.tim.2007.12.009 [DOI] [PubMed] [Google Scholar]
- Patrick J. E., Kearns D. B. (2008). MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis . Mol. Microbiol. 70 (5), 1166–1179. doi: 10.1111/j.1365-2958.2008.06469.x [DOI] [PubMed] [Google Scholar]
- Pikovskaya R. I. (1948). Mobilization of phosphorus in soil connection with the vital activity of some microbial species. Microbiology 17, 362–370. [Google Scholar]
- Rachinger M., Bauch M., Strittmatter A., Bongaerts J., Evers S., Maurer K. H., et al. (2013). Size unlimited markerless deletions by a transconjugative plasmid-system in Bacillus licheniformis . J. Biotechnol. 167 (4), 365–369. doi: 10.1016/j.jbiotec.2013.07.026 [DOI] [PubMed] [Google Scholar]
- Reynolds J., Moyes R., Breakwell D. P. (2009). Differential staining of bacteria: endospore stain. Curr. Protoc. Microbiol. Appendix 3: 15, A.13J.11–A.13J.15. doi: 10.1002/9780471729259.mca03js15 [DOI] [PubMed] [Google Scholar]
- Sarwar A., Hassan M. N., Imran M., Iqbal M., Majeed S., Brader G., et al. (2018). Biocontrol activity of surfactin a purified from Bacillus NH-100 and NH-217 against rice bakanae disease. Microbiol. Res. 209, 1–13. doi: 10.1016/j.micres.2018.01.006 [DOI] [PubMed] [Google Scholar]
- Shank E. A., Kolter R. (2011). Extracellular signaling and multicellularity in Bacillus subtilis . Curr. Opin. Microbiol. 14 (6), 741–747. doi: 10.1016/j.mib.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni R., Keharia H. (2021). Phytostimulation and biocontrol potential of gram-positive endospore-forming bacilli. Planta 254 (3), 49. doi: 10.1007/s00425-021-03695-0 [DOI] [PubMed] [Google Scholar]
- Tegg R. S., Gill W. M., Thompson H. K., Davies N. W., Ross J. J., Wilson C. R. (2008). Auxin-induced resistance to common scab disease of potato linked to inhibition of thaxtomin a toxicity. Plant Dis. 92 (9), 1321–1328. doi: 10.1094/PDIS-92-9-1321 [DOI] [PubMed] [Google Scholar]
- Theatre A., Cano-Prieto C., Bartolini M., Laurin Y., Deleu M., Niehren J., et al. (2021). The surfactin-like lipopeptides from Bacillus spp.: Natural biodiversity and synthetic biology for a broader application range. Front. Bioeng. Biotechnol. 9, 623701. doi: 10.3389/fbioe.2021.623701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui W. H., Yim G., Wang H. H., Mcclure J. E., Surette M. G., Davies J. (2004). Dual effects of MLS antibiotics: transcriptional modulation and interactions on the ribosome. Chem. Biol. 11 (9), 1307–1316. doi: 10.1016/j.chembiol.2004.07.010 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang C., Liang J., Wang L., Gao W., Jiang J., et al. (2020). Surfactin and fengycin b extracted from Bacillus pumilus W-7 provide protection against potato late blight via distinct and synergistic mechanisms. Appl. Microbiol. Biotechnol. 104 (17), 7467–7481. doi: 10.1007/s00253-020-10773-y [DOI] [PubMed] [Google Scholar]
- Zeriouh H., De Vicente A., Perez-Garcia A., Romero D. (2014). Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ. Microbiol. 16 (7), 2196–2211. doi: 10.1111/1462-2920.12271 [DOI] [PubMed] [Google Scholar]
- Zhang G. Q., Bao P., Zhang Y., Deng A. H., Chen N., Wen T. Y. (2011). Enhancing electro-transformation competency of recalcitrant Bacillus amyloliquefaciens by combining cell-wall weakening and cell-membrane fluidity disturbing. Anal. Biochem. 409 (1), 130–137. doi: 10.1016/j.ab.2010.10.013 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Ding Z. T., Shu D., Luo D., Tan H. (2015). Development of an efficient electroporation method for iturin a-producing Bacillus subtilis ZK. Int. J. Mol. Sci. 16 (4), 7334–7351. doi: 10.3390/ijms16047334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Li C., Hao J. J., Li Y. C., Li D. Z., Zhang D. M., et al. (2020). A novel Streptomyces sp. strain PBSH9 for controlling potato common scab caused by Streptomyces galilaeus . Plant Dis. 104 (7), 1986–1993. doi: 10.1094/PDIS-07-19-1469-RE [DOI] [PubMed] [Google Scholar]
- Zhang Y., Qi J., Wang Y., Wen J., Zhao X., Qi G. (2022). Comparative study of the role of surfactin-triggered signalling in biofilm formation among different Bacillus species. Microbiol. Res. 254, 126920. doi: 10.1016/j.micres.2021.126920 [DOI] [PubMed] [Google Scholar]
- Zhang W., Xie H., He Y., Feng J., Gao W., Gu Y., et al. (2013). Chromosome integration of the vitreoscilla hemoglobin gene (vgb) mediated by temperature-sensitive plasmid enhances gamma-PGA production in Bacillus amyloliquefaciens . FEMS Microbiol. Lett. 343 (2), 127–134. doi: 10.1111/1574-6968.12139 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.