This review discusses the SnRK1 signaling associated with different subcellular compartments and how this compartmentalization may contribute to the multitasking role of the SnRK1 complex.
Keywords: Autophagy, plant cell, SnRK1 compartmentalization, SnRK1-interacting proteins, SnRK1/TOR signaling, stress granules
Abstract
SNF1-related protein kinase 1 (SnRK1), the plant ortholog of mammalian AMP-activated protein kinase/fungal (yeast) Sucrose Non-Fermenting 1 (AMPK/SNF1), plays a central role in metabolic responses to reduced energy levels in response to nutritional and environmental stresses. SnRK1 functions as a heterotrimeric complex composed of a catalytic α- and regulatory β- and βγ-subunits. SnRK1 is a multitasking protein involved in regulating various cellular functions, including growth, autophagy, stress response, stomatal development, pollen maturation, hormone signaling, and gene expression. However, little is known about the mechanism whereby SnRK1 ensures differential execution of downstream functions. Compartmentalization has been recently proposed as a new key mechanism for regulating SnRK1 signaling in response to stimuli. In this review, we discuss the multitasking role of SnRK1 signaling associated with different subcellular compartments.
Introduction
Plants are sessile organisms continuously exposed to a wide range of environmental cues including light, wounding, or temperature, which have a major impact on their development and productivity. Consequently, they have developed sophisticated cellular mechanisms to survive in ever-changing environments. In this regard, the evolutionarily conserved protein SNF1-related kinase 1 (SnRK1) is considered as a master regulator that integrates external signals with plant growth (Baena-Gonzalez et al., 2007; Broeckx et al., 2016). SnRK1 is activated by sugar starvation, promoting the phosphorylation of a large number of proteins (Jamsheer et al., 2021). Arabidopsis SnRK1 and its orthologs, the yeast sucrose non-fermenting-1 protein kinase (SNF1) and mammalian AMP-activated protein kinase (AMPK), operate as a heterotrimeric complex composed of a catalytic α-subunit and two regulatory subunits, β and γ. In plants, a hybrid SnRK1βγ protein (with a carbohydrate-binding domain typically found in β-subunits) functions as the γ-subunit. While the kinase α-subunit is required for activation of signaling events associated with SnRK1, β- and βγ-subunits control SnRK1α activity, localization, and substrate specificity (Jamsheer et al., 2021).
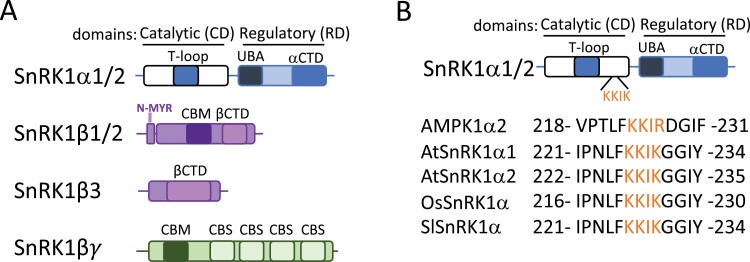
In Arabidopsis, the catalytic α-subunit of SnRK1 is encoded by three genes, SnRK1α1, SnRK1α2, and SnRK1α3 (also referred to as AKIN10/AKIN11/AKIN12 or KIN10/ KIN11/KIN12), of which α1 and α2 are partially redundant (Baena-Gonzalez et al., 2007). SnRK1α3, which is poorly expressed, is often considered to be a pseudogene (Baena-Gonzalez et al., 2007; Le et al., 2011), and SnRK1α3 cloning has not been reported yet. Notably, snrk1α1/snrk1α2 (snrk1α1/1α2) double knockout appears to be lethal, supporting the non-functionality of SnRK1α3 (Baena-Gonzalez et al., 2007). The domain architecture of the α-subunit is highly conserved and includes a Ser/Thr kinase domain (also referred to as a catalytic domain; CD) at the N-terminus followed by a regulatory domain (RD) at the C-terminus (Fig. 1A). The CD contains an activation loop (T-loop), with a conserved threonine (Arabidopsis SnRK1α1/α2T175/176), whose phosphorylation has been reported to be critical for SnRK1 activity (Baena-Gonzalez et al., 2007; Herzig and Shaw, 2018; Lin and Hardie, 2018). In mammals, the level of T-loop phosphorylation parallels AMPK kinase activity, although this correlation does not seem to be so clear in plants (Emanuelle et al., 2015; Herzig and Shaw, 2018; Lin and Hardie, 2018). Likewise, the C-terminal part of the protein includes both a ubiquitin-associated (UBA) domain and a far C-terminal (αCTD) domain. While UBA was found to be crucial for maintaining the catalytic activity of SnRK1α (Emanuelle et al., 2018), the αCTD is required for the interaction with the β- and γ-subunits (Kleinow et al., 2000).
Fig. 1.
SnRK1 subunit architecture. (A) SnRK1α subunits contain a Ser/Thr kinase domain (referred to as the catalytic domain; CD) at the N-terminus followed by a regulatory domain (referred to as the RD) at the C-terminus. The CD contains an activation loop (T-loop), while the C-terminal part includes both ubiquitin-associated (UBA) and far C-terminal (αCTD) subdomains. The regulatory β-subunits consist of an N-terminal myristoylation (N-MYR) motif, a carbohydrate-binding module (CBM), and a β-C-terminal domain (βCTD) for SnRK1β1 and SnRK1β1, and a βCTD for SnRK1β3. The regulatory βγ-subunit combines four cystathionine-β-synthase (CBS) domains at the C-terminus with an N-terminal CBM. (B) Multiple amino acid sequence alignment of the putative nuclear localization signal (NLS) of SnRK1 from Arabidopsis thaliana (AtSnRK1α), Oryza sativa (OsSnRK1α), Solanum lycopersicum (SlSnRK1α) and human (AMPK1α2). The NLS is marked in orange.
The Arabidopsis genome encodes three β-subunits, SnRK1β1, SnRK1β2, and SnRK1β3. SnRK1β1 and SnRK1β2 are constituted by an N-terminal myristoylation (N-MYR) motif, a carbohydrate-binding module (CBM), and a β-C-terminal domain (βCTD), whereas SnRK1β3 is formed exclusively by a βCTD (Fig. 1A). Although the three β-subunits are involved in SnRK1 signaling (Emanuelle et al., 2018), a lack of studies in these proteins makes it difficult to establish the specific contribution of each domain and/or isoform to the complex. To date, it has been shown that myristylation of the N-MYR motif controls AtSnRK1α activity and localization (Pierre et al., 2007; Ramon et al., 2019). In contrast to mammals or yeast, plants possess an atypical γ-subunit that combines four cystathionine-β-synthase (CBS) domains at the C-terminus with an N-terminal CBM, usually found in β-subunits (Fig. 1A), which explains why this atypical γ-subunit is referred to as the βγ-subunit in plants. The lethality of the Arabidopsis SnRK1βγ knockout mutant suggests an essential role for this gene in plants (Ramon et al., 2013). The binding of adenine nucleotides (ATP, ADP, or AMP) to AMPKγ has been reported as necessary for AMPK activity (Gowans et al., 2013). Although this regulatory mechanism is absent in plants (Emanuelle et al., 2015), several findings suggest that, similar to AMPKγ, the Arabidopsis βγ-subunit is crucial for SnRK1 signaling. For example, a reduced SnRK1βγ expression correlated well with reduced SnRK1 target gene expression (Ramon et al., 2013). On the other hand, the presence of the βγ-subunit is necessary for the heterotrimeric SnRK1α1βγβ3 complex activity in response to maltose (Ruiz-Gayosso et al., 2018).
SnRK1 has been involved in the regulation of important cellular functions, including growth, autophagy, stress response, stomatal development, pollen maturation, hormone signaling, and gene expression (Li et al., 2017; Han et al., 2020; Jamsheer et al., 2021). However, the mechanism whereby SnRK1 ensures differential execution of downstream functions remains to be determined. One possibility is that response specificity may be achieved by stimulus-specific phosphorylation of target proteins. In fact, two recent and independent phosphoproteomic studies indicated that SnRK1α regulates the phosphorylation state of ~500 proteins (Cho et al., 2016; Nukarinen et al., 2016). Another solution to achieve multitasking within the cellular space is compartmentalization. Indeed, SnRK1 has been localized in the cytoplasm, nucleus, plasma membrane, chloroplast, endoplasmic reticulum (ER), and stress granules (SGs) in response to various physiological inputs (Fragoso et al., 2009; Jamsheer et al., 2018b; Gutierrez-Beltran et al., 2021; Song et al., 2021; Sun et al., 2021). To date, all studies performed on plant SnRK1 have been focused on understanding the mechanistic implication of SnRK1 activation. However, little is known about the spatially defined SnRK1 regulation. In this review, we discuss the SnRK1 signaling associated with different subcellular compartments and how this compartmentalization may contribute to the multitasking role of the SnRK1 complex.
Linking SnRK1 localization with functional output
Signal-dependent nuclear shuttling of SnRK1α as a mechanism for controlling gene expression
Numerous studies have demonstrated the presence of the catalytic α-subunit in the nucleus. In fact, a large number of proteins have been reported to interact with SnRK1α in this organelle (Fig. 2; Supplementary Table S1). Although most of these studies are based on transient expression assays in Nicotiana benthamiana and Arabidopsis protoplasts, it is well established that SnRK1α shuttles between the cytoplasm and nucleus under certain conditions. For example, low-energy stress triggers a change in localization of AtSnRK1α1 from the cytoplasm to the nucleus (Ramon et al., 2019). More recently, the nuclear interaction between Oryza sativa SnRK1α1 and the histone H3K27me3 demethylase JMJ705 was enriched under starvation stress (Wang et al., 2021). However, the mechanism that regulates the cytoplasm to nuclear translocation of SnRK1 is unknown. Studies on mammalian models have revealed that a conserved sequence localized at the N-terminus of AMPKα facilitates the signal-dependent shuttling between the cytoplasm and nucleus (Suzuki et al., 2007; Kazgan et al., 2010). In particular, the amino acid sequence KKIR located in the catalytic domain of AMPKα2 was essential for nuclear translocation in response to the hormone leptin (Suzuki et al., 2007). Considering that the minimum requirement for a monopartite nuclear localization signal (NLS) is Lys-(Lys/Arg)-X-(Lys/Arg) (Lu et al., 2021), an amino acid sequence alignment of SnRK1α proteins from several plant species showed a high conservation of the KKIK sequence (Fig. 1B), suggesting a possible conservation of the mechanism.
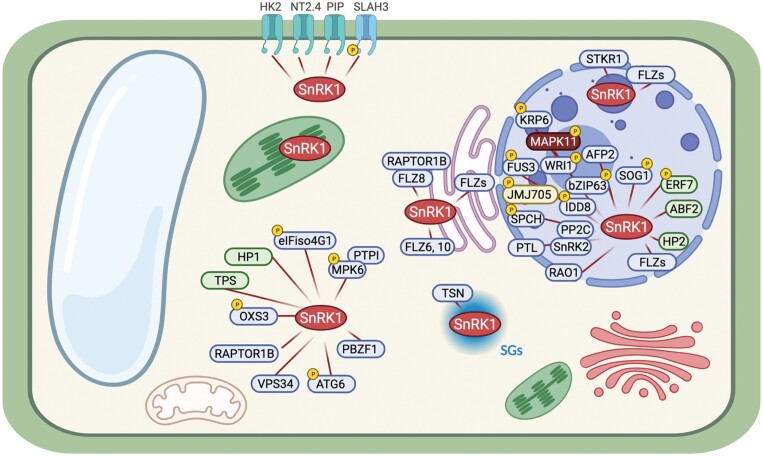
Fig. 2.
SnRK1 downstream substrates identified in plants. Subcellular localization and interaction data of SnRK1 were retrieved from the literature (see Supplementary Table S1). SnRK1 interaction proteins are marked in blue, green, yellow, or red, when the interaction is reported in Arabidopsis thaliana, Glycine max, Oryza sativa, or Solanum lycopersicum, respectively. The phosphorylation targets of SnRK1 are marked with a yellow circle with a P inside. The interactors were annotated using TAIR v10. The figure was created with BioRender.
The current model for AMPKα action/function indicates that cytoplasmically activated protein is translocated to the nucleus where it promotes phosphorylation of downstream transcriptional regulators to control gene expression (Chauhan et al., 2020). In fact, the phosphorylation of the conserved Thr172 residue is essential for the nuclear translocation of the protein (Suzuki et al., 2007). Similar to the mammalian ortholog, SnRK1α has been found to phosphorylate a large number of transcription regulators in plants, among them Arabidopsis indeterminate domain 8 (AtIDD8), WRINKLED1 (AtWR1), osJMJ705, or Glycine max AP2/ERF domain-containing protein (GsERF7) (Fig. 2; Supplementary Table S1). However, whether SnRK1α phosphorylation is required for the nuclear translocation of the protein is currently unknown. To date, a clear correlation between T-loop phosphorylation and nuclear function of SnRK1α has been established based on the following observations: (i) gene expression triggered by SnRK1α is inhibited in plants expressing the inactive mutant form AtSnRK1α1T175A (Baena-Gonzalez et al., 2007; Cho et al., 2012); (ii) SnRK1α-dependent degradation of the transcription factor AtWRI1 does not take place when AtSnRK1α1T175A is expressed (Zhai et al., 2017); (iii) similarly to the SnRK1α wild-type form, the constitutively active form AtSnRK1α1T175D is translocated to the nucleus and promotes stabilization of the transcription factor SPEECHLESS (SPCH) (Han et al., 2020); and (iv) phosphorylation of Thr175 is required for Arabidopsis gene expression (Ramon et al., 2019).
SnRK1α has been recently localized in nuclear bodies (NBs), suggesting an exciting and unexplored role for the complex in the nucleus (Blanco et al., 2019). This localization was also observed by bimolecular fluorescence complementation (BiFC) experiments, in which AtSnRK1α and interacting partners were found to interact in these structures. The AtSnRK1α-interacting proteins include several members of the FCS-like zinc fingers family (FLZs, earlier known as DUF581) and GL1 enhancer-binding protein (GeBP) (Fig. 2; Supplementary Table S1) (Nietzsche et al., 2014, 2018). NBs are biomolecular condensates whose functional role in plants remains largely unknown. However, several recent findings support the model whereby NBs have key roles in nuclear functions in response to environmental stimuli (Meyer, 2020). Abscisic acid (ABA) is a phytohormone essential for plant response to environmental stress that mediates SnRK1 signaling (Rodrigues et al., 2013; Belda-Palazon et al., 2020). Moreover, several SnRK1α-interacting proteins involved in ABA signaling have been reported to localize in NBs, such as ABA-insensitive 5 (ABI5), ABI5-binding protein (AFP), phytochrome-interacting factor 4 (PIF4), or WRKY family members (Lopez-Molina et al., 2003; Geilen and Bohmer, 2015; Hwang et al., 2019; Carianopol et al., 2020). These findings, together with the fact that SnRK1 and ABA signaling were found to regulate a common set of stress-responsive genes (Rodrigues et al., 2013), suggest a role for NB-dependent SnRK1 localization in ABA-mediated regulation of gene expression in plants. However, the biological significance of this localization is yet to be identified.
The endoplasmic reticulum as a platform for SnRK1/TOR regulation through FLZ proteins
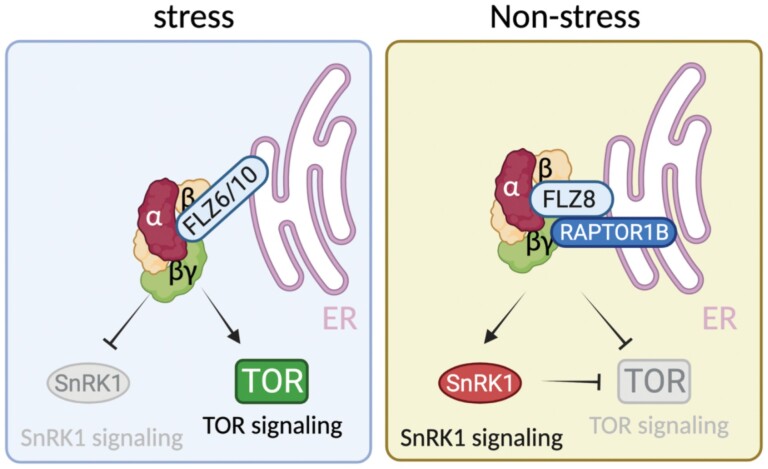
A study using both transient and stable expression in plants has shown that SnRK1α is stably associated with the ER (Blanco et al., 2019). The ER is a dynamic cellular organelle involved in protein synthesis, peptide chain folding, and trafficking (Manghwar and Li, 2022). Apart from its central role in protein synthesis, the ER is also involved in regulating the stress response in plant cells (Liu and Li, 2019). A previous study demonstrated that AtSnRK1α is able to interact with at least 10 members of the FLZ protein family in the ER (Fig. 2; Supplementary Table S1) (Jamsheer et al., 2018a, b). FLZs are small proteins with a C2–C2 FLZ domain that have been involved in the regulation of abiotic stress and ABA responses (He and Gan, 2004; Chen et al., 2013). From the FLZ family, both FLZ6 and FLZ10 were reported first to interfere with the SnRK1/target of rapamycin (TOR) signaling pathways (Jamsheer et al., 2018a). Thus, protein levels of AtSnRK1α were found to be enhanced in flz6 and flz10 single mutants, while the level of phosphorylated ribosomal protein S6 kinase (S6K), a well-established target of TOR, was found to be reduced. The authors propose a model where the interaction of SnRK1α with both FLZ6/10 proteins in the ER may mediate the antagonist signaling of the SnRK1/TOR module in plants under unfavorable conditions (Fig. 3).
Fig. 3.
Model for the SnRK1/TOR/FLZ signaling network. Under unfavorable conditions, both FLZ6 and FLZ10 repress SnRK1, allowing TOR signaling, in an ER localization manner. Under favorable conditions, FLZ8 induces TOR signaling inhibition by two different mechanisms, namely (i) promoting SnRK1 signaling through enhancing the SnRK1α1 level and (ii) stimulating RAPTOR1B–SnRK1α1 interaction in the ER. Lines with arrows indicate positive regulation and lines with bars indicate negative regulation. The figure was created with BioRender.
The relevance of the ER for SnRK1/TOR signaling is also strengthened by the fact that three members of the TOR complex (TORC), namely TOR, regulatory-associated protein of TOR (RAPTOR), and lethal with SEC13 protein 8 (LST8) have been localized in the ER in distant lineages such as animals or algae (Liu and Zheng, 2007; Diaz-Troya et al., 2008; Yadav et al., 2013). In fact, the ER localization of mammalian TOR (mTOR) has been reported to be crucial for its activity (Liu and Zheng, 2007). Jamsheer et al. (2022) have recently suggested that FLZ8, another member of the FLZ family, may act as a scaffold protein regulating SnRK1/TOR activity in plants. They found that FLZ8 negatively regulates TOR signaling by two different mechanisms: (i) stimulating antagonistic SnRK1α1 signaling and (ii) promoting SnRK1α1/RAPTOR1B association (Fig. 3). Notably, the FLZ8–SnRK1α–RAPTOR1B association was found to take place in the ER (Fig. 2; Supplementary Table S1). Interestingly, the potential role of FLZ family proteins as scaffolds has been recently highlighted in a new study (Bortlik et al., 2022, Preprint). In this work, the authors found that FLZ3 inhibits SnRK1 activity by interfering with the upstream activating kinase GRIK2. Moreover, FLZ3 was found to localize in the ER (Jamsheer et al., 2018b). Collectively, these studies suggest a possible role for the ER as a hub for SnRK1/TOR regulation mediated by FLZ proteins in plants, although further studies are needed to demonstrate the specific contribution of individual FLZ proteins to the regulation of SnRK1/TOR signaling.
Chloroplasts, a hub for SnRK1-mediated starch metabolism regulation?
Both Arabidopsis SnRK1α1 and SnRK1α2 isoforms were found to be localized inside and around the chloroplast (Fragoso et al., 2009; Ruiz-Gayosso et al., 2018; Blanco et al., 2019). Although the functionality of this localization is still an open question, several studies indicate the existence of a convincing link between SnRK1α signaling and the organelle. A quantitative phosphoproteomic study indicated that the phosphorylation status of several proteins with a known role in chloroplast light reactions was down-regulated in the snrk1α1/1α2 double mutant compared with wild-type plants (Nukarinen et al., 2016). Further works using protein–protein interaction approximations found a clear link between SnRK1 and chloroplast function and development (Rohila et al., 2009; Carianopol et al., 2020). A recent study showed that treatment with DCMU [3-(3,4-dichlorophenyl)-1,1-dimethylurea], a known inhibitor of chloroplast electron transport, causes a profound effect on SnRK1α localization, showing a re-localization from the non-nuclear to the nuclear fraction (Blanco et al., 2019). Accordingly, activation of AtSnRK1 kinase activity has been reported under energy deprivation triggered by both DCMU treatment and prolonged darkness (Baena-Gonzalez et al., 2007; Kim et al., 2017). The latter scenario is known to promote degradation of chloroplast proteins and chlorophyll, leading to a misregulation of the chloroplast function and an imbalance in the cellular redox state (Dietz et al., 2016; Kim et al., 2017). In close agreement, SnRK1α activity has been recently reported to be strongly dependent on the redox state (Wurzinger et al., 2017). All these results point to a functional connection of SnRK1 activity with the chloroplast, but whether it is direct or indirect is unknown.
Besides SnRK1α isoforms, the regulatory β- and βγ-subunits have been localized in the chloroplast (Fragoso et al., 2009; Avila-Castaneda et al., 2014; Ruiz-Gayosso et al., 2018). Among them, SnRK1β1, SnRK1β2, and SnRK1βγ share a CBM domain (Fig. 1A), a domain known to inhibit AMPK activity when bound to glycogen (Koay et al., 2010). Starch, the plant analog of glycogen, is stored inside the chloroplast as a transitory polysaccharide granule. Initially, the CBMs from SnRK1β2 and SnRK1βγ were described to bind starch in vitro (Avila-Castaneda et al., 2014). However, a later study using AMPKβ subunits as positive controls reported that SnRK1 CBMs cannot bind to this polysaccharide (Emanuelle et al., 2015). A subsequent study reported that maltose, the main product of starch degradation at night, binds to SnRK1β1 and SnRK1β2 subunits, and to the SnRK1βγ/β3 complex in vitro (Ruiz-Gayosso et al., 2018). Given that the SnRK1β3 subunit lacks a CBM domain, its capacity to bind maltose might be facilitated by forming a complex with the βγ-subunit (Ruiz-Gayosso et al., 2018). Curiously, when the impact of maltose binding on SnRK1 activity was analyzed, only the complex formed by α1/β3/βγ was stimulated, indicating a possible level of control depending on which subunit is assembled. Based on these results, Ruiz-Gayoss et al. proposed a model in which the accumulation of maltose at night promotes the increase of SnRK1 activity, inducing maltose metabolism via an as yet undefined mechanism. This finding, together with other studies, suggests that the SnRK1 complex might promote the carbon flux from starch to degradation products (Thelander et al., 2004; Baena-Gonzalez et al., 2007). However, although these findings suggest a possible role for the SnRK1 complex in starch metabolism, both maltose binding and chloroplast localization should be further confirmed.
The cytoplasm, a meeting place for SnRK1/TOR signaling, stress granules, and autophagy
The cytoplasm is the major intracellular fluid where a plethora of important biological reactions take place. As part of the cytosolic pool, SnRK1 occupies a key position involved in numerous reactions that include involvement in protein synthesis and degradation or stress response signaling. As an example of the latter, SnRK1α has been reported to interact in the cytosol with proteins involved in both biotic and abiotic stress responses (Cho et al., 2016; Chen et al., 2021; Gutierrez-Beltran et al., 2021). For instance, SnRK1α was found to interact with MPK6 and its regulator [protein tyrosine phosphatase 1 (PTP1)] in the cytoplasm under hypoxia caused by submergence (Fig. 2; Supplementary Table S1) (Bartels et al., 2009). This observation suggested the existence of an SnRK1α–PTI1–MPK6 cascade during submergence, which was later confirmed by Cho et al. (2016). This study found that SnRK1-induced phosphorylation of PTP1 disrupted the PTP1–MPK6 association, promoting the activation of nuclear target genes dependent on MPK6. On the other hand, IFiso4G1 and eIFiso4G2, two translation initiation factors, were also reported to be cytosolic partners of SnRK1α1 during submergence (Cho et al., 2019). The phosphorylation of both translation initiation factors via SnRK1α1 promoted the cytosolic translation of core hypoxia and stress response genes during submergence.
The assembly of SGs takes place in the cytosol. SGs are cytoplasmic biomolecular condensates that assemble transiently in response to both environmental and internal signals as an adaptive survival mechanism (Alberti and Carra, 2018; Hofmann et al., 2021). SGs typically contain translationally arrested mRNAs, small ribosomal subunits, various translation initiation factors (eIFs), poly(A)-binding proteins (PABs), and a variety of RNA-binding proteins and non-RNA-binding proteins (Protter and Parker, 2016). SnRK1α has been recently reported to be among the multiple proteins associated with SGs, and both SnRK1α1 and SnRK1α2 isoforms from Arabidopsis have been shown to interact with Tudor staphylococcal nuclease (TSN) in these membraneless organelles (Gutierrez-Beltran et al., 2021). TSN is a scaffold protein required for the proper assembly of plant SGs (Gutierrez-Beltran et al., 2016). The formation of SGs and the presence of TSN are required for the activation of SnRK1 signaling in response to heat stress (Gutierrez-Beltran et al., 2021). Although the link between SnRK1 and plant SG assembly is still poorly understood, the role of their yeast and animal homologs in SG biogenesis is well known. Thereby, the presence of AMPKα or SNF1 is required for the proper assembly of SGs in largely divergent organisms such as Caenorhabditis elegans, Saccharomyces cerevisiae, or mammals (Hofmann et al., 2012; Mahboubi et al., 2015a; Kuo et al., 2020). Furthermore, the pharmacological activation of AMPKα affects key aspects of SG biology, including assembly and fusion (Mahboubi et al., 2015a, 2016). In this respect, both β and γ regulatory subunits have been also localized in SGs in mammalian cells (Mahboubi et al., 2015b). Given the pro-survival role of SGs, the effect of SnRK1/AMPK/SNF1 on SG biogenesis may be considered as a new avenue for modulating cell survival in response to stress.
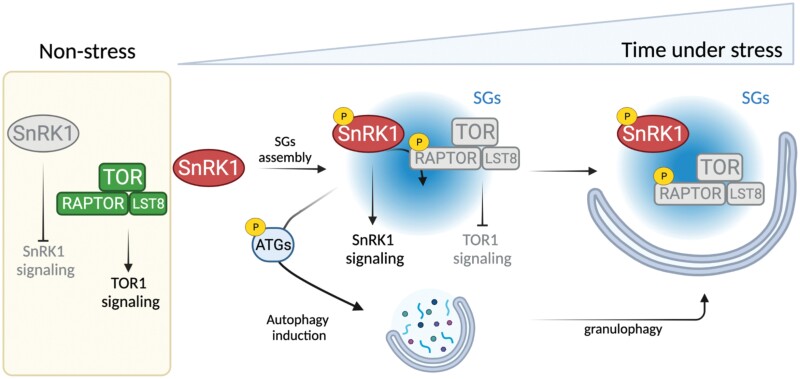
A previous study reported that Arabidopsis SnRK1α1 phosphorylates (in vitro) and interacts with RAPTOR1B in the cytoplasm (Nukarinen et al., 2016). RAPTOR1B is part of the TORC in plants, which also includes LST8. The Arabidopsis genome contains two copies for RAPTOR (RAPTOR1A and RAPTOR1B) and LST8 (LST8-1 and LST8-2) genes, although LST8-2 shows undetectable transcript levels (Anderson et al., 2005; Moreau et al., 2012). In contrast to plants, mammalian cells contain two different TOR complexes, mTORC1 (homolog to plant TORC) and mTORC2. The latter is formed by the association of rapamycin-insensitive companion of TOR (RICTOR) and mammalian stress-activated protein kinase-interacting protein 1 (mSIN1). In yeast and mammalian models, AMPK/SNF1 are well-established upstream negative regulators of TORC1. While phosphorylation of mammalian RAPTOR (mRAPTOR) via AMPKα promotes the inhibition of mTOR kinase activity (Gwinn et al., 2008), this link is not so obvious in yeast (Hughes Hallett et al., 2015). In Arabidopsis, a recent study revealed that the cytoplasmic interaction between SnRK1α1 and TOR is required for TOR inhibition in response to stress (Belda-Palazon et al., 2020, 2022). However, whether this control is mediated by RAPTOR is still an open question. Together with a previous study showing TOR inhibition by stress-induced phosphorylation of RAPTOR1B in Arabidopsis by SnRK2 (Wang et al., 2018), these findings strongly suggest the existence of a SnRK1–RAPTOR–TOR regulatory network in plants. In this respect, SGs might operate as a platform for this signaling module in plants. Both RAPTOR and mTOR are bona fide SG components in the mammalian system (Rehbein et al., 2021). Growing evidence indicates that SGs constitute a cytoplasmic compartment in which mTORC1 is inhibited under stress through several mechanisms, which include sequestration of both RAPTOR and mTOR proteins (Thedieck et al., 2013; Wippich et al., 2013; Mediani et al., 2021; Prentzell et al., 2021). Given that stress-induced localization of SnRK1α in SGs promotes its activation (Gutierrez-Beltran et al., 2021), one possibility is that SnRK1α regulates TOR signaling inhibition by phosphorylation of SG-localized RAPTOR1B (Fig. 4). It is very well established that TOR acts as a central metabolic regulator playing largely antagonistic roles to SnRK1 (Margalha et al., 2019). Therefore, the association of SnRK1α–TOR with SGs may imply a checkpoint for the activation/inhibition of these signaling pathways which fully depends on the cellular homeostasis (see Fig. 4 for a hypothetical model).
Fig. 4.
Hypothetical model for SG-dependent regulation of the SnRK1/TOR signaling network. Under favorable conditions, SnRK1 activity is repressed while TOR signaling is activated, promoting processes associated with cell proliferation and growth. Following stress perception, sequestration and activation of SnRK1 in SGs might contribute to TOR signaling inhibition by SnRK1-dependent phosphorylation of RAPTOR1B. At the same time, SG-dependent SnRK1 activation induces stress responses, which might include activation of autophagy (via ATG phosphorylation). Finally, autophagy might mediate SG degradation via granulophagy. Lines with arrows indicate positive regulation and lines with bars indicate negative regulation. The figure was created with BioRender.
It is well known that AMPK and TOR antagonistically regulate autophagy in mammalian cells (Gonzalez et al., 2020). While TOR has been postulated to act as a negative regulator, AMPK plays a positive role in autophagy dynamics. In contrast to the mammalian model, the molecular mechanism of SnRK1/TOR-mediated control of autophagy in plants is still under study. Several recent findings point to the presence of a common nexus between the SnRK1/TOR module and autophagy via autophagy-related (ATG) proteins (Liao and Bassham, 2020). Hence, the phosphorylation of Arabidopsis ATG13 is considered as a key modification whereby TOR mediates the inhibition of autophagy (Son et al., 2018). Regarding SnRK1, a recent study has shown that the phosphorylation and interaction with ATG6 in cytoplasmic foci promotes autophagy during prolonged carbon starvation in Arabidopsis (Fig. 2; Supplementary Table S1) (Huang et al., 2019). Previously, it was demonstrated that overexpression of AtSnRK1α1 enhanced both autophagosome formation and ATG1a phosphorylation in vivo (Chen et al., 2017). Among ATG proteins, ATG1, ATG13, or ATG6 are required for earlier events of autophagy induction, suggesting that the SnRK1/TOR module might act early in autophagy regulation (Huang et al., 2019). Based on the fact that mammalian AMPK controls both autophagy induction and SG dynamics, we hypothesize that SG-localized SnRK1 may be involved in autophagy activation via phosphorylation of key ATG proteins (Fig. 4). In mammals, autophagy controls SG disassembly through a process known as granulophagy (Seguin et al., 2014; Hofmann et al., 2021). Indeed, ULK1 and ULK2 proteins, the mammalian orthologs of ATG1, have been shown to promote SG disassembly (Wang et al., 2019). In plants, granulophagy has been described to control SG disassembly during extended hypoxia (Field et al., 2021, Preprint). However, whether activation of autophagy under stress-induced SG assembly conditions as well as granulophagy is controlled by the SnRK1/TOR module is totally unknown.
The plasma membrane links SnRK1 and channel regulation
Arabidopsis SnRK1α1 interacts with the cytosolic C-terminal region of the plasma membrane (PM) protein SLAC1 homolog 3 (SLAH3) (Fig. 2; Supplementary Table S1) (Sun et al., 2021). SLAH3 is an anion channel involved in the efflux of NO3– under high-NH4+/low- NO3– conditions as a mechanism of ammonium detoxicity in plants (Zheng et al., 2015). It has been proposed that under physiological growth conditions, the cytosol-localized AtSnRK1α1 interacts with and phosphorylates SLAH3 to inhibit its activity, preventing nitrate loss (Zheng et al., 2015). When the concentration of NH4+ is high, active AtSnRK1α1 migrates to the nucleus, which releases the inhibition of SLAH3 and leads to nitrate efflux. This is the first evidence showing the regulation of channel activity via SnRK1 in plants. However, previous studies identified other channels or transporters as SnRK1 interactor partners. For example, a yeast two-hybrid (Y2H) assay showed that both aquaporin PIP1 and nitrate transporter 2.4 (NT2.4) interact with soybean SnRK1α (Song et al., 2019). More recently, protein–protein interaction analysis using a Y2H assay revealed the interaction of AtSnRK1α with several cyclic nucleotide-gated channels (CNGCs), including CNGC12, CNGC13, and CNGC18, as well as channels involved in phosphate transport such as phosphate transporter 1;4 (PHT1; 4) and PHO1 homolog 7 (PHO1; H7) (Carianopol et al., 2020; Jamsheer et al., 2021).
Similar to plants, several studies in mammals have shown that AMPK directly or indirectly alters the activities of various channels (Lang and Foller, 2014). For example, AMPKα phosphorylates and inhibits BKCa, a voltage-gated potassium channel (Wyatt et al., 2007). Furthermore, AMPKα also controls the channel activity via intermediates. For example, AMPKα stimulates Nedd4.2, a ubiquitin ligase that mediates the down-regulation of the epithelial Na+ channel ENaC (Bhalla et al., 2006). However, it remains unknown whether this undirect mechanism of regulation exists in plants. In both plants and mammalian models, the interaction between AMPK/SnRK1 and channels was detected at the PM (Fig. 2; Supplementary Table S1) (Lang and Foller, 2014; Sun et al., 2021). Membrane localization of the AMPK/SnRK1 complex has been reported to be controlled by N-terminal myristoylation of the regulatory β-subunits (Lin et al., 2003; Oakhill et al., 2010; Ramon et al., 2019). One possibility is that the phosphorylation/interaction of SnRK1 with channels is mediated via myristoylation, although this has not been explored. Taken together, these studies suggest that control of channel activity via SnRK1 may be a common feature in plants. However, a thorough analysis should be performed to demonstrate the biological significance of these interactions.
Is the plant vacuole a key hub for SnRK1/TOR signaling?
The lysosome (or vacuole in yeast and plants) is a membrane-bound organelle that facilitates the digestion of macromolecules. However, lysosomes have been also proposed to have a key role in other cellular processes including cellular differentiation, metabolism, or signaling regulation (Lim and Zoncu, 2016; Young et al., 2016; Abu-Remaileh et al., 2017). Moreover, several studies have linked lysosomes as a hub for the mechanistic regulation of AMPK/mTOR via a v-ATPase-Ragulator complex (Carroll and Dunlop, 2017). Thus, under glucose starvation, the v-ATPase promotes assembly of an AXIN–liver kinase B1 (LKB1) complex at the lysosome surface to activate AMPK (Zhang et al., 2014). At the same time, v-ATPase facilitates the release of mTORC1 from the lysosome surface, leading to the inhibition of mTORC1 activity (Zhang et al., 2014). A growing body of evidence indicates now that SGs might also be involved in the lysosomal regulation of mTORC1 activity via the core SG marker G3BP1 (Rehbein et al., 2021). Indeed, G3BP1 has been reported to anchor the tuberous sclerosis complex (TSC) to lysosomes and suppress mTORC1 signaling (Prentzell et al., 2021). In the budding yeast, glucose starvation has been reported to increase the threshold for TORC1 activation when Kog1/RAPTOR is re-localized from the vacuolar membrane to a single body near the edge of the organelle, in an event dependent on SNF1 (Hughes Hallett et al., 2015). These findings reveal a key hub role for the lysosome/vacuole organelle in AMPK/TOR regulation. Whether such mechanisms of regulation via the vacuole exist in plants requires further investigation.
Factors affecting the subcellular localization of SnRK1
N-Terminal myristoylation is a key process controlling SnRK1 localization
The Arabidopsis SnRK1β1 and SnRK1β2 subunits, but not SnRK1β3, have an N-MYR motif and are myristoylated in vivo on a conserved glycine residue at position 2 (Gly2) (Fig. 1A). N-myristoylation is catalyzed by N-myristoyltransferase (NMT) and consists of the addition of the 14-carbon fatty acid, myristate, to the N-terminus via a covalent amide bond. This post-translational modification facilitates the association of proteins with cellular membranes. In the case of SnRK1, N-myristoylation of β-subunits has been reported to control both SnRK1 localization and activity (Lin et al., 2003; Pierre et al., 2007; Oakhill et al., 2010; Broeckx et al., 2016; Ramon et al., 2019). Thus, the N-myristoylation of both regulatory β-subunits has been reported to negatively regulate the nuclear translocation of SnRK1α1, whose localization is required for SnRK1-induced target gene activation during metabolic stress (Ramon et al., 2019). A previous work found that loss of NMT activity leads to an enhancement of SnRK1-associated kinase activity, providing evidence of N-myristoylation-dependent activation of SnRK1 (Pierre et al., 2007). However, whether this phenotype is caused by a defect in SnRK1β-dependent recruitment of the α-subunit to membranes is still an open question. In mammals, N-terminal myristoylation of the β-subunits has been shown to suppress AMPKα activity, keeping AMPKα in an inactive state at the membrane (Warden et al., 2001; Oakhill et al., 2010). Nevertheless, the mechanism whereby N-terminal myristoylation mediates SnRK1 activity is still under study.
The subcellular localization of SnRK1 changes in a stimulus-dependent manner
As discussed above, the SnRK1α subunit is localized at the cytoplasm, nucleus, chloroplast, ER, or SGs, and this localization seems to be stimulus dependent in some cases. For example, under non-stress conditions, the α1 isoform exhibits a nuclear localization that is particularly prominent in Arabidopsis root meristem cells, and it delocalizes to the cytoplasm in response to ABA (Belda-Palazón et al., 2022). This phenomenon appears to be required for the cytoplasmic control of TOR activity in response to ABA (Belda-Palazon et al., 2020, 2022). The cytoplasm to nucleus migration of SnRK1α1 has been described as a mechanism to induce, but not repress, target gene expression under metabolic, hypoxia, DCMU, dark, or high-ammonium stresses (Ramon et al., 2019; Sun et al., 2021; Wang et al., 2021). In another work, the localization in SGs of both α1 and α2 isoforms was described to be heat stress dependent (Gutierrez-Beltran et al., 2021). Notably, the heat-induced SG localization was linked with both T-loop activation and gene expression. Apart from the stress type, the degree of the stress has been also found to generate a response in the compartmentalized pools of AMPK. Thus, a recent study in mouse embryonic fibroblasts (MEFs) reported that compartmentalized AMPKs undergo a hierarchical activation, which fully depend on the intensity of the stress (Zong et al., 2019). Whether this level of regulation exists in plants is completely unknown. In contrast to plants, the activity of compartmentalized AMPK pools has been extensively studied. For instance, the design of biosensors has largely contributed to monitor the spatiotemporal activation of AMPK across multiple organelles in response to stress (Tsou et al., 2011; Miyamoto et al., 2015).
Differential SnRK1 heterotrimeric complex assembly and expression as a level of specificity
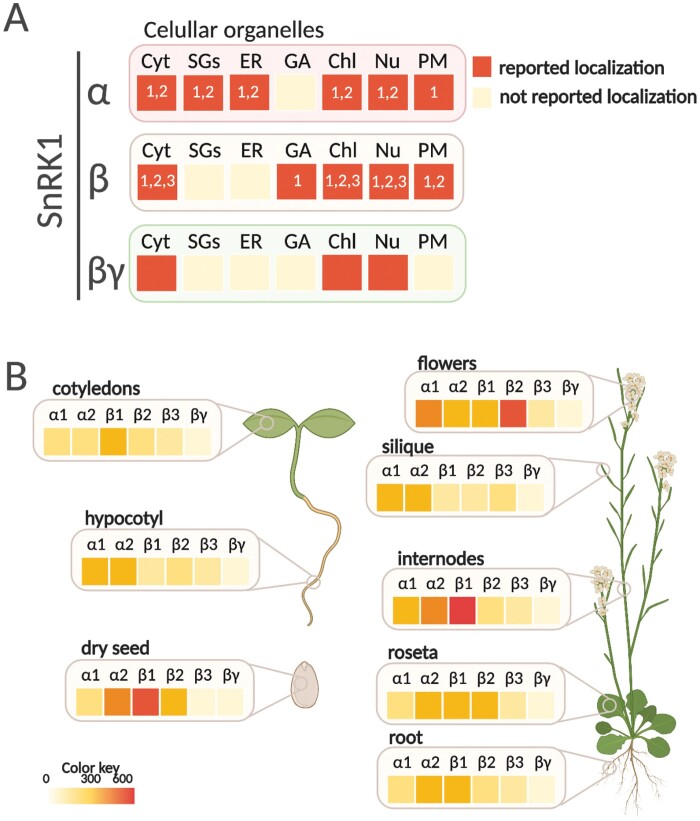
A protein complex can play multiple roles by changing the members of its modular constituents. In the case of Arabidopsis SnRK1, the heterotrimeric complex is composed of α-, β- and βγ-subunits, with α and β having different isoforms (Broeckx et al., 2016). According to this protein composition, six different heterotrimeric complexes are possible. In fact, a previous study showed that all six combinations are assembled in vitro (Emanuelle et al., 2015), although it is not clear whether all combinations exist in vivo. For example, the differential localization described for each of the subunits make some combinations impossible (Fig. 5A; Supplementary Table S2). Thus, under physiological conditions, SnRK1α isoforms, SnRK1β3 and SnRK1βγ, are predominantly localized to the cytoplasm and nucleus, while both SnRK1β1 and SnRK1β2 are limited to the cytoplasm (Gissot et al., 2006; Bitrian et al., 2011; Gao et al., 2016; Ramon et al., 2019). A more recent study, indeed, shows that SnRK1β1 is localized in the Golgi under transient expression in N. benthamiana epidermal cells (Fig. 5A) (Wang et al., 2020). A higher level of complexity is observed based on the tissue- or cell type-specific and subcellular localization. For example, under physiological conditions, α1- and βγ-subunits show preferential nuclear localization in Arabidopsis meristematic cells (Bitrian et al., 2011; Belda-Palazón et al., 2022). Similarly, SnRK1α1 is mainly localized in the nucleus in both guard and stomatal Arabidopsis cells (Han et al., 2020). In stigmata and pistils of young flowers, SnRK1α1 accumulated in the cytoplasm, whereas SnRK1βγ is detected predominantly in nuclei (Bitrian et al., 2011; Gao et al., 2016). Notably, it should not be ruled out that the stress signal may trigger a localization change, thus allowing a differential heterotrimeric assembly. In this respect, yeast α- and all three β-subunits change from cytoplasm to specific cellular compartments when glucose becomes limiting (Hedbacker and Carlson, 2008).
Fig. 5.
Subcellular localization and expression pattern of SnRK1 subunits. (A) SnRK1 subunits have been visualized in the cytoplasm (Cyt), stress granules (SGs), endoplasmic reticulum (ER), Golgi (AG), chloroplast (Chl), nucleus (Nu), and plasma membrane (PM) in plants. The numbering inside the square indicates the type of subunit. Although subcellular localization of SnRK1 subunits has been retrieved from the literature (see Supplementary Table S2), some of them are controversial and should be corroborated by additional studies (e.g. chloroplast localization of SnRK1α). (B) In silico analysis of SnRK1 subunit gene expression using Tissue Specific Root eFP tool (http://bar.utoronto.ca/eplant/). The figure was created with BioRender.
The limited studies in plants make it difficult to establish which complexes are assembled in vivo. As an exception, a previous study showed the interaction between AtSnRK1βγ and both AtSnRK1β2 and AtSnRK1β3 subunits in the cytoplasm and nucleus when these proteins are overexpressed (Gissot et al., 2006). In mammals, structures for several functional AMPK heterotrimers have been resolved, including α1β1γ1, α1β2γ1, α2β1γ1, and α2β2γ1 (Xiao et al., 2013; Calabrese et al., 2014; Li et al., 2015; Ngoei et al., 2018). In plants, few types of complexes have been proposed to be functional. One of them was SnRK1α1βγβ3, whose kinase activity was enhanced in vitro in the presence of maltose compared with α1βγβ1 and α1βγβ1 (Maya-Bernal et al., 2017; Ruiz-Gayosso et al., 2018). Further studies are required to identify downstream targets specific to one particular heterotrimeric combination and isoform-specific effects on SnRK1 function in plants.
Several studies performed in mammals indicated that the gene expression pattern of AMPK subunits vary across tissues and cells, introducing a new level of heterotrimeric assembly complexity (Trefts and Shaw, 2021). Although a more exhaustive analysis of SnRK1 is required, a similar pattern has been observed in plants. For example, a β-glucuronidase (GUS) promoter analysis found that SnRK1β3 is preferably expressed in developing pollen, ovules, and seeds, while β1- and β2-subunits are ubiquitously expressed (Polge et al., 2008). A more recent study using the same approach showed that the expression pattern of SnRK1α2 was more restricted than that of SnRK1α1, whose expression was detected almost ubiquitously in the full plant (Williams et al., 2014). An in silico analysis of SnRK1 subunit expression using Tissue Specific Root eFP (http://bar.utoronto.ca/eplant/) revealed that both catalytic α-subunits and the regulatory SnRK1β1 are expressed throughout development and in different tissues (Fig. 5B). Regarding the regulatory subunits, SnRK1β2 showed a high level of expression in flowers, while the expression level of SnRK1β3 was moderate compared with the rest of the subunits (Fig. 5B). This scenario is presumably different when the expression pattern of SnRK1 subunits is analyzed under different stimuli (Baena-Gonzalez et al., 2007; Polge et al., 2008; Williams et al., 2014).
Conclusions
SnRK1 signaling is an extremely complex pathway that remains poorly understood in plants. Extensive studies have provided resolution of SnRK1 signal transduction under a set of cellular and environmental cues. These studies include the identification of a plethora of phosphorylated and interacting targets. However, it remains largely unknown how SnRK1 can release a stimulus-type-specific response. This review provides a survey on how localized protein interaction can invoke targeted signaling programs. SnRK1 has been found to interact with targets in such different organelles as the cytoplasm, nucleus, chloroplast, ER, and SGs. Moreover, differential heterotrimeric assembly and the subunit expression pattern can add an extra level of specificity in the downstream SnRK1 response. Although several recent works have provided new insights into cellular compartmentalization of SnRK1, further studies are required to unravel the interplay between the spatiotemporal SnRK1 localization and its downstream signaling mechanisms.
Supplementary data
The following supplementary data are available at JXB online.
Table S1. SnRK1-interacting proteins shown in Fig. 1.
Table S2. Subcellular localization of SnRK1 subunits shown in Fig. 5A.
Acknowledgements
We are grateful to the editor and reviewers for their comments and suggestions.
Contributor Information
Emilio Gutierrez-Beltran, Instituto de Bioquimica Vegetal y Fotosintesis, Consejo Superior de Investigaciones Cientificas (CSIC)-Universidad de Sevilla, Sevilla, Spain; Departamento de Bioquimica Vegetal y Biologia Molecular, Facultad de Biologia, Universidad de Sevilla, Sevilla, Spain.
Jose L Crespo, Instituto de Bioquimica Vegetal y Fotosintesis, Consejo Superior de Investigaciones Cientificas (CSIC)-Universidad de Sevilla, Sevilla, Spain.
Elena Baena-González, Instituto Gulbenkian de Ciência, Portugal.
Author contributions
EG-B: conceptualization, writing, visualization (figure preparation), and funding acquisition; JLC: review.
Conflict of interest
We have no conflicts of interest associated with this publication.
Funding
This work was supported by grant PID2020-119737GA-I00 funded by the Ministerio de Ciencia e Innovacion (MCIN/AEI/10.13039/501100011033) to EG-B, and grant PGC2018-099048-B-100 funded by the Ministerio de Economia y Competitividad to JLC.
References
- Abu-Remaileh M, Wyant GA, Kim C, Laqtom NN, Abbasi M, Chan SH, Freinkman E, Sabatini DM.. 2017. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Carra S.. 2018. Quality control of membraneless organelles. Journal of Molecular Biology 430, 4711–4729. [DOI] [PubMed] [Google Scholar]
- Anderson GH, Veit B, Hanson MR.. 2005. The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biology 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Castaneda A, Gutierrez-Granados N, Ruiz-Gayosso A, Sosa-Peinado A, Martinez-Barajas E, Coello P.. 2014. Structural and functional basis for starch binding in the SnRK1 subunits AKINβ2 and AKINβγ. Frontiers in Plant Science 5, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J.. 2007. A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. [DOI] [PubMed] [Google Scholar]
- Bartels S, Anderson JC, Gonzalez Besteiro MA, Carreri A, Hirt H, Buchala A, Metraux JP, Peck SC, Ulm R.. 2009. MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. The Plant Cell 21, 2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazon B, Adamo M, Valerio C, Ferreira LJ, Confraria A, Reis-Barata D, Rodrigues A, Meyer C, Rodriguez PL, Baena-Gonzalez E.. 2020. A dual function of SnRK2 kinases in the regulation of SnRK1 and plant growth. Nature Plants 6, 1345–1353. [DOI] [PubMed] [Google Scholar]
- Belda-Palazón B, Costa M, Beeckman T, Rolland F, Baena-González E.. 2022. ABA represses TOR and root meristem activity through nuclear exit of the SnRK1 kinase. Proceedings of the National Academy of Sciences, USA 119, e2204862119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla V, Oyster NM, Fitch AC, Wijngaarden MA, Neumann D, Schlattner U, Pearce D, Hallows KR.. 2006. AMP-activated kinase inhibits the epithelial Na+ channel through functional regulation of the ubiquitin ligase Nedd4-2. Journal of Biological Chemistry 281, 26159–26169. [DOI] [PubMed] [Google Scholar]
- Bitrian M, Roodbarkelari F, Horvath M, Koncz C.. 2011. BAC-recombineering for studying plant gene regulation: developmental control and cellular localization of SnRK1 kinase subunits. The Plant Journal 65, 829–842. [DOI] [PubMed] [Google Scholar]
- Blanco NE, Liebsch D, Guinea Diaz M, Strand A, Whelan J.. 2019. Dual and dynamic intracellular localization of Arabidopsis thaliana SnRK1.1. Journal of Experimental Botany 70, 2325–2338. [DOI] [PubMed] [Google Scholar]
- Bortlik J, Alseekh S, Weiste C, Fernie AR, Droge-Laser W, Bornke F.. 2022. DUF581-9 (At2g44670; FLZ3) negatively regulates SnRK1 activity by interference with T-loop phosphorylation. bioRxiv doi: 10.1101/2022.03.17.484690 [Preprint]. [DOI] [Google Scholar]
- Broeckx T, Hulsmans S, Rolland F.. 2016. The plant energy sensor: evolutionary conservation and divergence of SnRK1 structure, regulation, and function. Journal of Experimental Botany 67, 6215–6252. [DOI] [PubMed] [Google Scholar]
- Calabrese MF, Rajamohan F, Harris MS, et al. 2014. Structural basis for AMPK activation: natural and synthetic ligands regulate kinase activity from opposite poles by different molecular mechanisms. Structure 22, 1161–1172. [DOI] [PubMed] [Google Scholar]
- Carianopol CS, Chan AL, Dong S, Provart NJ, Lumba S, Gazzarrini S.. 2020. An abscisic acid-responsive protein interaction network for sucrose non-fermenting related kinase1 in abiotic stress response. Communications Biology 3, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll B, Dunlop EA.. 2017. The lysosome: a crucial hub for AMPK and mTORC1 signalling. Biochemical Journal 474, 1453–1466. [DOI] [PubMed] [Google Scholar]
- Chauhan AS, Zhuang L, Gan B.. 2020. Spatial control of AMPK signaling at subcellular compartments. Critical Reviews in Biochemistry and Molecular Biology 55, 17–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Li Y, Yan R, et al. 2021. SnRK1.1-mediated resistance of Arabidopsis thaliana to clubroot disease is inhibited by the novel Plasmodiophora brassicae effector PBZF1. Molecular Plant Pathology 22, 1057–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Su ZZ, Huang L, Xia FN, Qi H, Xie LJ, Xiao S, Chen QF.. 2017. The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Frontiers in Plant Science 8, 1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang Z, Visser RG, Broekgaarden C, Vosman B.. 2013. Overexpression of IRM1 enhances resistance to aphids in Arabidopsis thaliana. PLoS One 8, e70914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Hong JW, Kim EC, Yoo SD.. 2012. Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiology 158, 1955–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HY, Lu MJ, Shih MC.. 2019. The SnRK1–eIFiso4G1 signaling relay regulates the translation of specific mRNAs in Arabidopsis under submergence. New Phytologist 222, 366–381. [DOI] [PubMed] [Google Scholar]
- Cho HY, Wen TN, Wang YT, Shih MC.. 2016. Quantitative phosphoproteomics of protein kinase SnRK1 regulated protein phosphorylation in Arabidopsis under submergence. Journal of Experimental Botany 67, 2745–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Troya S, Florencio FJ, Crespo JL.. 2008. Target of rapamycin and LST8 proteins associate with membranes from the endoplasmic reticulum in the unicellular green alga Chlamydomonas reinhardtii. Eukaryotic Cell 7, 212–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ, Turkan I, Krieger-Liszkay A.. 2016. Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiology 171, 1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelle S, Doblin MS, Gooley PR, Gentry MS.. 2018. The UBA domain of SnRK1 promotes activation and maintains catalytic activity. Biochemical and Biophysical Research Communications 497, 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelle S, Hossain MI, Moller IE, et al. 2015. SnRK1 from Arabidopsis thaliana is an atypical AMPK. The Plant Journal 82, 183–192. [DOI] [PubMed] [Google Scholar]
- Field S, Gulledge W, Roberts DM.. 2021. The Arabidopsis calcium sensor calmodulin-like 38 regulates stress granule autophagy and dynamics during low oxygen stress and re-aeration recovery. bioRxiv doi: 10.1101/2021.01.10.426134 [Preprint]. [DOI] [Google Scholar]
- Fragoso S, Espindola L, Paez-Valencia J, Gamboa A, Camacho Y, Martinez-Barajas E, Coello P.. 2009. SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiology 149, 1906–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao XQ, Liu CZ, Li DD, Zhao TT, Li F, Jia XN, Zhao XY, Zhang XS.. 2016. The Arabidopsis KINβγ subunit of the snrk1 complex regulates pollen hydration on the stigma by mediating the level of reactive oxygen species in pollen. PLoS Genetics 12, e1006228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geilen K, Bohmer M.. 2015. Dynamic subnuclear relocalisation of WRKY40 in response to abscisic acid in Arabidopsis thaliana. Science Reports 5, 13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissot L, Polge C, Jossier M, Girin T, Bouly JP, Kreis M, Thomas M.. 2006. AKINβγ contributes to SnRK1 heterotrimeric complexes and interacts with two proteins implicated in plant pathogen resistance through its KIS/GBD sequence. Plant Physiology 142, 931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez A, Hall MN, Lin SC, Hardie DG.. 2020. AMPK and TOR: the yin and yang of cellular nutrient sensing and growth control. Cell Metabolism 31, 472–492. [DOI] [PubMed] [Google Scholar]
- Gowans GJ, Hawley SA, Ross FA, Hardie DG.. 2013. AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metabolism 18, 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Beltran E, Denisenko TV, Zhivotovsky B, Bozhkov PV.. 2016. Tudor staphylococcal nuclease: biochemistry and functions. Cell Death & Differentiation 23, 1739–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Beltran E, Elander PH, Dalman K, Dayhoff GW 2nd, Moschou PN, Uversky VN, Crespo JL, Bozhkov PV.. 2021. Tudor staphylococcal nuclease is a docking platform for stress granule components and is essential for SnRK1 activation in Arabidopsis. The EMBO Journal 40, e105043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ.. 2008. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Molecular Cell 30, 214–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Liu Y, Shi W, et al. 2020. KIN10 promotes stomatal development through stabilization of the SPEECHLESS transcription factor. Nature Communications 11, 4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Gan S.. 2004. A novel zinc-finger protein with a proline-rich domain mediates ABA-regulated seed dormancy in Arabidopsis. Plant Molecular Biology 54, 1–9. [DOI] [PubMed] [Google Scholar]
- Hedbacker K, Carlson M.. 2008. SNF1/AMPK pathways in yeast. Frontiers in Bioscience 13, 2408–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzig S, Shaw RJ.. 2018. AMPK: guardian of metabolism and mitochondrial homeostasis. Nature Reviews. Molecular Cell Biology 19, 121–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Cherkasova V, Bankhead P, Bukau B, Stoecklin G.. 2012. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Molecular Biology of the Cell 23, 3786–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S, Kedersha N, Anderson P, Ivanov P.. 2021. Molecular mechanisms of stress granule assembly and disassembly. Biochimica et Biophysica Acta 1868, 118876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zheng C, Liu F, et al. 2019. Genetic analyses of the Arabidopsis ATG1 kinase complex reveal both kinase-dependent and independent autophagic routes during fixed-carbon starvation. The Plant Cell 31, 2973–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes Hallett JE, Luo X, Capaldi AP.. 2015. Snf1/AMPK promotes the formation of Kog1/Raptor-bodies to increase the activation threshold of TORC1 in budding yeast. eLife 4, e09181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang G, Kim S, Cho JY, Paik I, Kim JI, Oh E.. 2019. Trehalose-6-phosphate signaling regulates thermoresponsive hypocotyl growth in Arabidopsis thaliana. EMBO Reports 20, e47828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsheer KM, Jindal S, Sharma M, Awasthi P, S S, Sharma M, Mannully CT, Laxmi A.. 2022. A negative feedback loop of TOR signaling balances growth and stress–response trade-offs in plants. Cell Reports 39, 110631. [DOI] [PubMed] [Google Scholar]
- Jamsheer KM, Kumar M, Srivastava V.. 2021. SNF1-related protein kinase 1: the many-faced signaling hub regulating developmental plasticity in plants. Journal of Experimental Botany 72, 6042–6065. [DOI] [PubMed] [Google Scholar]
- Jamsheer KM, Sharma M, Singh D, Mannully CT, Jindal S, Shukla BN, Laxmi A.. 2018a. FCS-like zinc finger 6 and 10 repress SnRK1 signalling in Arabidopsis. The Plant Journal 94, 232–245. [DOI] [PubMed] [Google Scholar]
- Jamsheer KM, Shukla BN, Jindal S, Gopan N, Mannully CT, Laxmi A.. 2018b. The FCS-like zinc finger scaffold of the kinase SnRK1 is formed by the coordinated actions of the FLZ domain and intrinsically disordered regions. Journal of Biological Chemistry 293, 13134–13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazgan N, Williams T, Forsberg LJ, Brenman JE.. 2010. Identification of a nuclear export signal in the catalytic subunit of AMP-activated protein kinase. Molecular Biology of the Cell 21, 3433–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GD, Cho YH, Yoo SD.. 2017. Regulatory functions of cellular energy sensor SNF1-related kinase1 for leaf senescence delay through ETHYLENE- INSENSITIVE3 repression. Science Reports 7, 3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinow T, Bhalerao R, Breuer F, Umeda M, Salchert K, Koncz C.. 2000. Functional identification of an Arabidopsis snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. The Plant Journal 23, 115–122. [DOI] [PubMed] [Google Scholar]
- Koay A, Woodcroft B, Petrie EJ, et al. 2010. AMPK beta subunits display isoform specific affinities for carbohydrates. FEBS Letters 584, 3499–3503. [DOI] [PubMed] [Google Scholar]
- Kuo CT, You GT, Jian YJ, Chen TS, Siao YC, Hsu AL, Ching TT.. 2020. AMPK-mediated formation of stress granules is required for dietary restriction-induced longevity in Caenorhabditis elegans. Aging Cell 19, e13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang F, Foller M.. 2014. Regulation of ion channels and transporters by AMP-activated kinase (AMPK). Channels (Austin) 8, 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS.. 2011. Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Research 18, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DD, Guan H, Li F, Liu CZ, Dong YX, Zhang XS, Gao XQ.. 2017. Arabidopsis shaker pollen inward K+ channel SPIK functions in SnRK1 complex-regulated pollen hydration on the stigma. Journal of Integrative Plant Biology 59, 604–611. [DOI] [PubMed] [Google Scholar]
- Li X, Wang L, Zhou XE, et al. 2015. Structural basis of AMPK regulation by adenine nucleotides and glycogen. Cell Research 25, 50–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CY, Bassham DC.. 2020. Combating stress: the interplay between hormone signaling and autophagy in plants. Journal of Experimental Botany 71, 1723–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CY, Zoncu R.. 2016. The lysosome as a command-and-control center for cellular metabolism. Journal of Cell Biology 214, 653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Hardie DG.. 2018. AMPK: sensing glucose as well as cellular energy status. Cell Metabolism 27, 299–313. [DOI] [PubMed] [Google Scholar]
- Lin SS, Manchester JK, Gordon JI.. 2003. Sip2, an N-myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. Journal of Biological Chemistry 278, 13390–13397. [DOI] [PubMed] [Google Scholar]
- Liu L, Li J.. 2019. Communications between the endoplasmic reticulum and other organelles during abiotic stress response in plants. Frontiers in Plant Science 10, 749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zheng XF.. 2007. Endoplasmic reticulum and Golgi localization sequences for mammalian target of rapamycin. Molecular Biology of the Cell 18, 1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH.. 2003. AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes & Development 17, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Wu T, Zhang B, Liu S, Song W, Qiao J, Ruan H.. 2021. Types of nuclear localization signals and mechanisms of protein import into the nucleus. Cell Communication and Signaling 19, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahboubi H, Barise R, Stochaj U.. 2015a. 5ʹ-AMP-activated protein kinase alpha regulates stress granule biogenesis. Biochimica et Biophysica Acta 1853, 1725–1737. [DOI] [PubMed] [Google Scholar]
- Mahboubi H, Barise R, Stochaj U.. 2015b. Data in support of 5ʹAMP-activated protein kinase alpha regulates stress granule biogenesis. Data Brief 4, 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahboubi H, Koromilas AE, Stochaj U.. 2016. AMP kinase activation alters oxidant-induced stress granule assembly by modulating cell signaling and microtubule organization. Molecular Pharmacology 90, 460–468. [DOI] [PubMed] [Google Scholar]
- Manghwar H, Li J.. 2022. Endoplasmic reticulum stress and unfolded protein response signaling in plants. International Journal of Molecular Sciences 23, 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalha L, Confraria A, Baena-Gonzalez E.. 2019. SnRK1 and TOR: modulating growth–defense trade-offs in plant stress responses. Journal of Experimental Botany 70, 2261–2274. [DOI] [PubMed] [Google Scholar]
- Maya-Bernal JL, Avila A, Ruiz-Gayosso A, Trejo-Fregoso R, Pulido N, Sosa-Peinado A, Zuniga-Sanchez E, Martinez-Barajas E, Rodriguez-Sotres R, Coello P.. 2017. Expression of recombinant SnRK1 in E. coli. Characterization of adenine nucleotide binding to the SnRK1.1/AKINβγ-β3 complex. Plant Science 263, 116–125. [DOI] [PubMed] [Google Scholar]
- Mediani L, Antoniani F, Galli V, et al. 2021. Hsp90-mediated regulation of DYRK3 couples stress granule disassembly and growth via mTORC1 signaling. EMBO Reports 22, e51740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HM. 2020. In search of function: nuclear bodies and their possible roles as plant environmental sensors. Current Opinion in Plant Biology 58, 33–40. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Rho E, Sample V, et al. 2015. Compartmentalized AMPK signaling illuminated by genetically encoded molecular sensors and actuators. Cell Reports 11, 657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau M, Azzopardi M, Clement G, et al. 2012. Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. The Plant Cell 24, 463–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngoei KRW, Langendorf CG, Ling NXY, et al. 2018. Structural determinants for small-molecule activation of skeletal muscle AMPK α2β2γ1 by the glucose importagog SC4. Cell Chemical Biology 25, 728–737 e729. [DOI] [PubMed] [Google Scholar]
- Nietzsche M, Guerra T, Alseekh S, Wiermer M, Sonnewald S, Fernie AR, Bornke F.. 2018. STOREKEEPER RELATED1/G-element binding protein (STKR1) interacts with protein kinase SnRK1. Plant Physiology 176, 1773–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nietzsche M, Schiessl I, Bornke F.. 2014. The complex becomes more complex: protein–protein interactions of SnRK1 with DUF581 family proteins provide a framework for cell- and stimulus type-specific SnRK1 signaling in plants. Frontiers in Plant Science 5, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukarinen E, Nagele T, Pedrotti L, et al. 2016. Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Science Reports 6, 31697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakhill JS, Chen ZP, Scott JW, Steel R, Castelli LA, Ling N, Macaulay SL, Kemp BE.. 2010. beta-Subunit myristoylation is the gatekeeper for initiating metabolic stress sensing by AMP-activated protein kinase (AMPK). Proceedings of the National Academy of Sciences, USA 107, 19237–19241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre M, Traverso JA, Boisson B, Domenichini S, Bouchez D, Giglione C, Meinnel T.. 2007. N-myristoylation regulates the SnRK1 pathway in Arabidopsis. The Plant Cell 19, 2804–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C, Jossier M, Crozet P, Gissot L, Thomas M.. 2008. Beta-subunits of the SnRK1 complexes share a common ancestral function together with expression and function specificities; physical interaction with nitrate reductase specifically occurs via AKINbeta1-subunit. Plant Physiology 148, 1570–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentzell MT, Rehbein U, Cadena Sandoval M, et al. 2021. G3BPs tether the TSC complex to lysosomes and suppress mTORC1 signaling. Cell 184, 655–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DS, Parker R.. 2016. Principles and properties of stress granules. Trends in Cell Biology 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M, Dang TVT, Broeckx T, Hulsmans S, Crepin N, Sheen J, Rolland F.. 2019. Default activation and nuclear translocation of the plant cellular energy sensor SnRK1 regulate metabolic stress responses and development. The Plant Cell 31, 1614–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M, Ruelens P, Li Y, Sheen J, Geuten K, Rolland F.. 2013. The hybrid four-CBS-domain KINβγ subunit functions as the canonical γ subunit of the plant energy sensor SnRK1. The Plant Journal 75, 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehbein U, Prentzell MT, Cadena Sandoval M, Heberle AM, Henske EP, Opitz CA, Thedieck K.. 2021. The TSC complex–mTORC1 axis: from lysosomes to stress granules and back. Frontiers in Cell and Developmental Biology 9, 751892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A, Adamo M, Crozet P, et al. 2013. ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. The Plant Cell 25, 3871–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohila JS, Chen M, Chen S, et al. 2009. Protein–protein interactions of tandem affinity purified protein kinases from rice. PLoS One 4, e6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Gayosso A, Rodriguez-Sotres R, Martinez-Barajas E, Coello P.. 2018. A role for the carbohydrate-binding module (CBM) in regulatory SnRK1 subunits: the effect of maltose on SnRK1 activity. The Plant Journal 96, 163–175. [DOI] [PubMed] [Google Scholar]
- Seguin SJ, Morelli FF, Vinet J, Amore D, De Biasi S, Poletti A, Rubinsztein DC, Carra S.. 2014. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death & Differentiation 21, 1838–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son O, Kim S, Kim D, Hur YS, Kim J, Cheon CI.. 2018. Involvement of TOR signaling motif in the regulation of plant autophagy. Biochemical and Biophysical Research Communications 501, 643–647. [DOI] [PubMed] [Google Scholar]
- Song J, Shang L, Wang X, Xing Y, Xu W, Zhang Y, Wang T, Li H, Zhang J, Ye Z.. 2021. MAPK11 regulates seed germination and ABA signaling in tomato by phosphorylating SnRKs. Journal of Experimental Botany 72, 1677–1690. [DOI] [PubMed] [Google Scholar]
- Song Y, Zhang H, You H, et al. 2019. Identification of novel interactors and potential phosphorylation substrates of GsSnRK1 from wild soybean (Glycine soja). Plant, Cell & Environment 42, 145–157. [DOI] [PubMed] [Google Scholar]
- Sun D, Fang X, Xiao C, et al. 2021. Kinase SnRK1.1 regulates nitrate channel SLAH3 engaged in nitrate-dependent alleviation of ammonium toxicity. Plant Physiology 186, 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Okamoto S, Lee S, Saito K, Shiuchi T, Minokoshi Y.. 2007. Leptin stimulates fatty acid oxidation and peroxisome proliferator-activated receptor α gene expression in mouse C2C12 myoblasts by changing the subcellular localization of the α2 form of AMP-activated protein kinase. Molecular and Cellular Biology 27, 4317–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thedieck K, Holzwarth B, Prentzell MT, et al. 2013. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell 154, 859–874. [DOI] [PubMed] [Google Scholar]
- Thelander M, Olsson T, Ronne H.. 2004. Snf1-related protein kinase 1 is needed for growth in a normal day–night light cycle. EMBO Journal 23, 1900–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trefts E, Shaw RJ.. 2021. AMPK: restoring metabolic homeostasis over space and time. Molecular Cell 81, 3677–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou P, Zheng B, Hsu CH, Sasaki AT, Cantley LC.. 2011. A fluorescent reporter of AMPK activity and cellular energy stress. Cell Metabolism 13, 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Maxwell BA, Joo JH, et al. 2019. ULK1 and ULK2 regulate stress granule disassembly through phosphorylation and activation of VCP/p97. Molecular Cell 74, 742–757 e748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhao Y, Li Z, et al. 2018. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Molecular Cell 69, 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lu Y, Li J, Zhang X, Hu F, Zhao Y, Zhou DX.. 2021. SnRK1 stimulates the histone H3K27me3 demethylase JMJ705 to regulate a transcriptional switch to control energy homeostasis. The Plant Cell 33, 3721–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang L, Micallef BJ, Tetlow IJ, Mullen RT, Feil R, Lunn JE, Emes MJ.. 2020. AKINβ1, a subunit of SnRK1, regulates organic acid metabolism and acts as a global modulator of genes involved in carbon, lipid, and nitrogen metabolism. Journal of Experimental Botany 71, 1010–1028. [DOI] [PubMed] [Google Scholar]
- Warden SM, Richardson C, O’Donnell J Jr., Stapleton D, Kemp BE, Witters LA.. 2001. Post-translational modifications of the beta-1 subunit of AMP-activated protein kinase affect enzyme activity and cellular localization. Biochemical Journal 354, 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SP, Rangarajan P, Donahue JL, Hess JE, Gillaspy GE.. 2014. Regulation of sucrose non-fermenting related kinase 1 genes in Arabidopsis thaliana. Frontiers in Plant Science 5, 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, Pelkmans L.. 2013. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152, 791–805. [DOI] [PubMed] [Google Scholar]
- Wurzinger B, Mair A, Fischer-Schrader K, Nukarinen E, Roustan V, Weckwerth W, Teige M.. 2017. Redox state-dependent modulation of plant SnRK1 kinase activity differs from AMPK regulation in animals. FEBS Letters 591, 3625–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt CN, Mustard KJ, Pearson SA, Dallas ML, Atkinson L, Kumar P, Peers C, Hardie DG, Evans AM.. 2007. AMP-activated protein kinase mediates carotid body excitation by hypoxia. Journal of Biological Chemistry 282, 8092–8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Sanders MJ, Carmena D, et al. 2013. Structural basis of AMPK regulation by small molecule activators. Nature Communications 4, 3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav RB, Burgos P, Parker AW, Iadevaia V, Proud CG, Allen RA, O’Connell JP, Jeshtadi A, Stubbs CD, Botchway SW.. 2013. mTOR direct interactions with Rheb-GTPase and raptor: sub-cellular localization using fluorescence lifetime imaging. BMC Cell Biology 14, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NP, Kamireddy A, Van Nostrand JL, Eichner LJ, Shokhirev MN, Dayn Y, Shaw RJ.. 2016. AMPK governs lineage specification through Tfeb-dependent regulation of lysosomes. Genes & Development 30, 535–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Liu H, Shanklin J.. 2017. Phosphorylation of WRINKLED1 by KIN10 results in its proteasomal degradation, providing a link between energy homeostasis and lipid biosynthesis. The Plant Cell 29, 871–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CS, Jiang B, Li M, et al. 2014. The lysosomal v-ATPase–Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metabolism 20, 526–540. [DOI] [PubMed] [Google Scholar]
- Zheng X, He K, Kleist T, Chen F, Luan S.. 2015. Anion channel SLAH3 functions in nitrate-dependent alleviation of ammonium toxicity in Arabidopsis. Plant, Cell & Environment 38, 474–486. [DOI] [PubMed] [Google Scholar]
- Zong Y, Zhang CS, Li M, et al. 2019. Hierarchical activation of compartmentalized pools of AMPK depends on severity of nutrient or energy stress. Cell Research 29, 460–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.