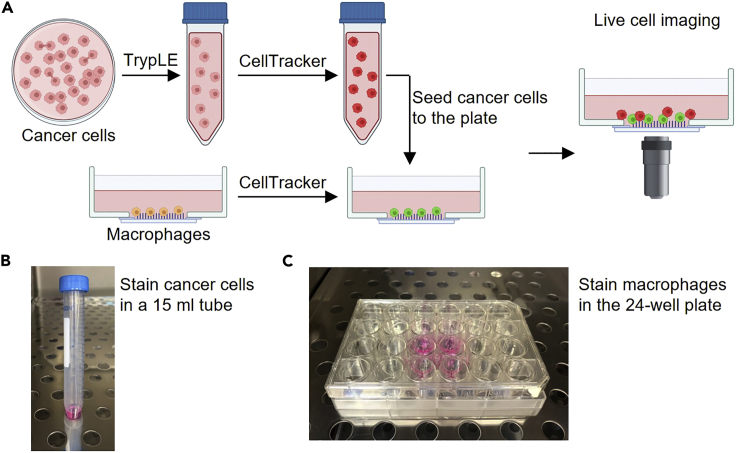
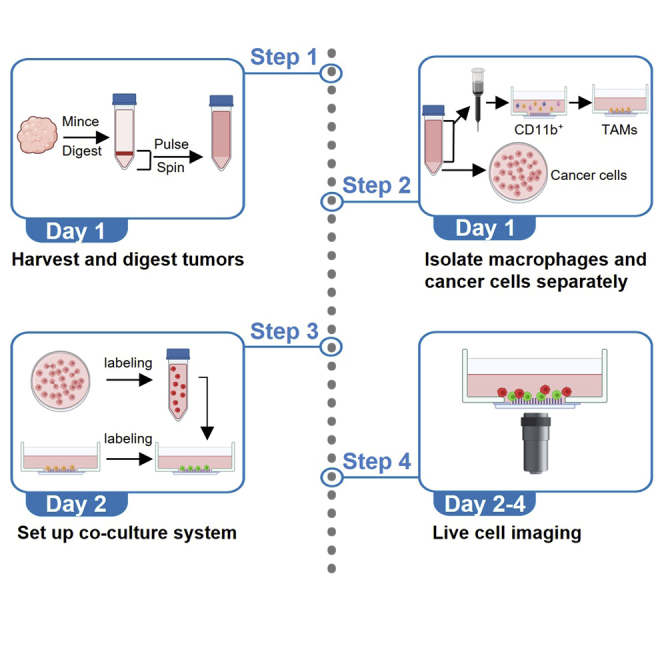
Summary
We recently established an in vitro co-culture system in which monophosphoryl lipid A + interferon-γ (MPLA+IFNγ)-treated tumor-associated macrophages (TAMs) killed cancer cells. Here, we describe a step-by-step protocol for isolating TAMs and cancer cells from mouse primary mammary carcinomas, the setup of the co-culture system, and the image acquisition approach. The technical difficulties in the co-culture assay involve isolating pure TAMs and cancer cells from the same tumor and staining them with different dyes to track the macrophages’ tumoricidal activity.
For complete details on the use and execution of this protocol, please refer to Sun et al. (2021).1
Subject areas: Cell Biology, Cell culture, Cell isolation, Cancer, Immunology, Microscopy, Molecular/Chemical Probes
Graphical abstract
Highlights
-
•
Isolate tumor-associated macrophages (TAMs) and cancer cells from the same tumor
-
•
Use CD11b MicroBeads followed by adherence method to enrich for macrophages
-
•
Establish co-cultures for tracking the TAMs’ tumoricidal activity or for other assays
-
•
May be adapted to allow isolation of and co-culturing with other immune cell types
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
We recently established an in vitro co-culture system in which monophosphoryl lipid A + interferon-γ (MPLA+IFNγ)-treated tumor-associated macrophages (TAMs) killed cancer cells. Here, we describe a step-by-step protocol for isolating TAMs and cancer cells from mouse primary mammary carcinomas, the setup of the co-culture system, and the image acquisition approach. The technical difficulties in the co-culture assay involve isolating pure TAMs and cancer cells from the same tumor and staining them with different dyes to track the macrophages’ tumoricidal activity.
Before you begin
Timing: 1 h
TAMs promote tumor growth and metastasis,2,3,4 and also contribute to the immunosuppressive microenvironment enabling tumors to evade intrinsic antitumor immune responses and therapies.5,6 However, macrophages can be polarized to kill cancer cells.7 TAM reprogramming could thus be a strategy for controlling cancer.8 To enable an in vitro test of strategies to reprogram TAMs into tumoricidal macrophages, we have developed a protocol to isolate TAMs and cancer cells from mouse mammary carcinomas and co-culture them ex vivo.
The protocol below describes the specific steps for using primary mammary carcinomas from C57BL/6-MMTV-PyMT (mouse mammary tumor virus-polyoma middle T antigen) mice (C57BL/6 background). However, we have also used this protocol in primary tumors from FVB/N-MMTV-PyMT mice (FVB/N background) and transplanted PyMT or 4T1 tumors.
We have not tested this protocol on primary tumors other than murine mammary tumors. However, we anticipate that the general protocol (Figure 1A) can be optimized for other types of solid tumors by adapting in particular the steps involving mechanical and enzymatic dissociation and the pulse centrifugation to enrich for cancer cells in the pellet and immune cells (including, but not limited to, macrophages) in the supernatant.
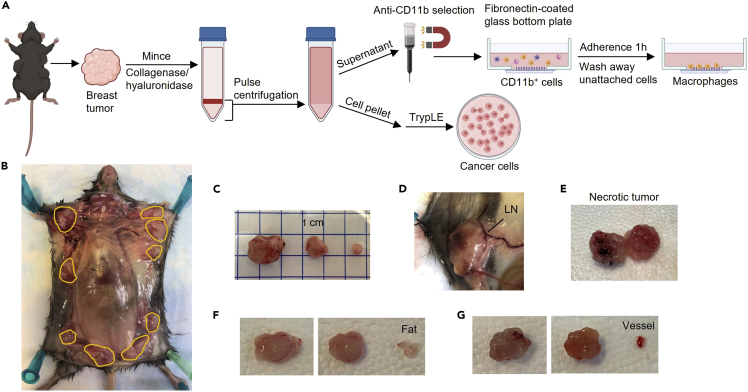
Figure 1.
Harvest the primary tumors from the MMTV-PyMT mouse
(A) Schematic diagram of the process to isolate mammary carcinoma-derived macrophages and cancer cells.
(B) Exposed mammary carcinomas. Tumors are outlined with orange lines.
(C) Mammary carcinomas of different sizes.
(D) Avoid collecting lymph nodes (LN).
(E) Necrotic tumor should not be used for the procedure.
(F) The tumor before (left) and after (right) removing fat tissue.
(G) The tumor before (left) and after (right) removing surrounding vessels.
Institutional permissions
MMTV-PyMT mice (on C57BL/6 or FVB/N background) were bred at Cold Spring Harbor Laboratory. Six- to eight-week-old female BL/6 and BALB/c host mice were purchased from Jackson Laboratory and acclimated to the animal housing facility for one week prior to performing experiments. All animal experiments were conducted in accordance with procedures approved by the Institutional Animal Care and Use Committee at CSHL and the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
-
1.Monitor the primary tumors of MMTV-PyMT mice.
-
a.Make sure the tumors are solid (not necrotic or cystic) and that there are enough tissue for cell isolation.
-
b.Determine how many tumors are needed according to the number of cells required for the specific assays to be performed and the separation efficiency (Refer to steps 3i, 4v and 5g below).
-
a.
Note: We routinely obtain 4.5 ± 0.9 × 107 cancer cells (>90% purity) and 2.2 ± 0.6 × 106 TAMs (>90% purity) from 1.0 g MMTV-PyMT tumor (two tumors, ∼10 mm in diameter, each tumor weighing ∼0.5 g).
Note: When using primary tumors from C57BL/6 background mice, we usually harvest the tumors when the mice are about 5 months old; when using the primary tumors from FVB/N background mice, we harvest the tumors when mice are about 2–3 months old as the tumors of FVB/N mice develop sooner than those of C57BL/6 mice.
Note: To obtain transplanted PyMT tumors, isolated cancer cells (follow step 1a through step 3i) can be injected at 2 × 105 cells/fat pad into the 4th mammary glands of syngeneic mice. Transplanted tumors are also harvested for isolation of macrophages and cancer cells when they reach ∼10 mm in diameter (approximately, after 1 month).
Note: Do not isolate cancer cells for transplantation from primary MMTV-PyMT tumors that are smaller than 6 mm in diameter, as such transplanted tumors grow much slower than those from later stage tumors, and sometimes fail to grow.
-
2.
Before the start of the experiment, prepare the dissociation buffer, wash buffer, 1% BSA (bovine serum albumin in PBS), MACS (magnetic cell sorting) buffer, and FACS (fluorescence activated cell sorting) buffer, as well as the macrophage culture medium, and cancer cell culture medium. Make sure that there is enough of all the solutions.
Note: The dissociation buffer and macrophage culture medium should be prepared fresh; other buffers can be stored at 4°C for several weeks.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BV785™ anti-mouse CD45 Antibody (1:100 dilution) | BioLegend | Cat #103149; RRID: AB_2564590 |
| APC anti-mouse/human CD11b Antibody (1:100 dilution) | BioLegend | Cat #101212; RRID: AB_312795 |
| Alexa Fluor® 488 anti-mouse F4/80 Antibody (1:100 dilution) | BioLegend | Cat #123120; RRID: AB_893479 |
| APC anti-mouse CD326 (EpCAM) Antibody (1:100 dilution) | BioLegend | Cat #118214; RRID: AB_1134102 |
| Chemicals, peptides, and recombinant proteins | ||
| L-glutamine | Gibco | Cat #25030081 |
| Sodium pyruvate | Gibco | Cat #11360070 |
| 2-mercaptoethanol | Gibco | Cat #31350010 |
| Collagenase/hyaluronidase | STEMCELL Technologies | Cat #07912 |
| DNase I | Roche | Cat #4716728001 |
| TrypLE Express Enzyme | Gibco | Cat #12605010 |
| Mouse M-CSF (macrophage colony-stimulating factor) | ProSpec | Cat #CYT-439 |
| Fibronectin | Sigma-Aldrich | Cat #F1141 |
| CellTracker™ Deep Red Dye | Invitrogen | Cat #C34565 |
| CellTracker™ Green CMFDA Dye | Invitrogen | Cat #C7025 |
| MPLA (monophosphoryl lipid A) | InvivoGen | Cat #tlrl-mpls |
| Mouse IFNγ | R&D Systems | Cat #485-MI/CF |
| Fc receptor blocker | Innovex | Cat #NB309 |
| BSA (bovine serum albumin) | Sigma-Aldrich | Cat #A3294 |
| DPBS (Dulbecco’s phosphate-buffered saline) | Gibco | Cat #14190-144 |
| FBS (fetal bovine serum) | Corning | Cat #35-010-CV |
| Penicillin-streptomycin | Gibco | Cat #15140122 |
| DMEM (Dulbecco’s Modified Eagle Medium) | Corning | Cat #10-013-CV |
| RPMI (Roswell Park Memorial Institute) 1640 | Corning | Cat #10-040-CV |
| ACK (Ammonium-Chloride-Potassium) Lysis Buffer | Gibco | Cat #2414466 |
| Experimental models: Organisms/strains | ||
| MMTV-PyMT mice (on C57BL/6 background, female) | The Jackson Laboratory | Cat #022974; RRID:IMSR_JAX:022974 |
| MMTV-PyMT mice (on FVB/N background, female) | The Jackson Laboratory | Cat #002374; RRID:IMSR_JAX:002374 |
| Software and algorithms | ||
| Micro-Manager 1.4 software | National Institutes of Health | RRID: SCR_000415 |
| Imaris software | Bitplane | RRID: SCR_007370 |
| ImageJ software | National Institutes of Health | RRID: SCR_003070 |
| Other | ||
| CD11b MicroBeads, human and mouse | Miltenyi Biotec | Cat #130-049-601 |
| 24-well glass bottom plate | MatTek | Cat #P24G-1.5-13-F |
| MidiMACS™ Separator | Miltenyi Biotec | Cat #130-042-302 |
| MACS MultiStand | Miltenyi Biotec | Cat #130-042-303 |
| Pre-Separation filters (30 μm) | Miltenyi Biotec | Cat #130-041-407 |
| LS Column | Miltenyi Biotec | Cat #130-042-401 |
| Incubator-Genie | Scientific Industries | Model #SI-1400 |
| LiveCellTM Stage Top Incubation System | Pathology Devices | N/A |
| Yokogawa spinning disk confocal microscope | Solamere Technology Group | N/A |
Materials and equipment
RPMI 1640+5% FBS
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI 1640 | N/A | 470 mL |
| FBS | 5% (vol/vol) | 25 mL |
| Penicillin-streptomycin 100× | 1× | 5 mL |
| Total | N/A | 500 mL |
Store at 4°C.
Dissociation buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI 1640+5% FBS | N/A | 9 mL |
| Collagenase/hyaluronidase 10× | 1× | 1 mL |
| DNase I (10 U/μL) | 4 U/mL | 4 μL |
| Total | N/A | 10 mL |
Prepare fresh every time.
Wash buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| DPBS | N/A | 470 mL |
| FBS | 5% (vol/vol) | 25 mL |
| Penicillin-streptomycin 100× | 1× | 5 mL |
| Total | N/A | 500 mL |
Store at 4°C. This wash buffer can be substituted with HBSS (Hanks’ Balanced Salt Solution) +5% FBS.
1% BSA solution
| Reagent | Final concentration | Amount |
|---|---|---|
| BSA | 1% (w/vol) | 0.5 g |
| PBS | N/A | 50 mL |
| Total | N/A | 50 mL |
Mix the tube on a rocker for 30 min. Filter sterilize. Store at 4°C.
MACS buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| DPBS | N/A | 490.5 mL |
| FBS | 0.5% (vol/vol) | 2.5 mL |
| EDTA (Ethylenediamine tetraacetic acid, 0.5 M stock in water) | 2 mM | 2 mL |
| Penicillin-streptomycin 100× | 1× | 5 mL |
| Total | N/A | 500 mL |
Store at 4°C.
FACS buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| DPBS | N/A | 493 mL |
| FBS | 1% (vol/vol) | 5 mL |
| NaN3 (5% stock in water) | 0.02% (w/vol) | 2 mL |
| Total | N/A | 500 mL |
Store at 4°C.
Macrophage culture medium
| Reagent | Final concentration | Amount |
|---|---|---|
| RPMI | N/A | 385 mL |
| FBS | 20% (vol/vol) | 100 mL |
| L-glutamine (200 mM) | 2 mM | 5 mL |
| Sodium pyruvate (200 mM) | 2 mM | 5 mL |
| Penicillin-streptomycin 100× | 1× | 5 mL |
| 2-mercaptoethanol | 55 μM | 1.92 μL |
| Mouse M-CSF (100 μg/mL stock in water, −20°C) | 10 ng/mL | 50 μL |
| Total | N/A | 500 mL |
Store at 4°C (except for 2-mercaptoethanol and M-CSF). Add fresh 2-mercaptoethanol and M-CSF each time.
Cancer cell culture medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM | N/A | 445 mL |
| FBS | 10% (vol/vol) | 50 mL |
| Penicillin-streptomycin 100× | 1× | 5 mL |
| Total | N/A | 500 mL |
Store at 4°C.
Fibronectin coating solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Fibronectin (1 mg/mL) | 10 μg/mL | 100 μL |
| PBS | N/A | 9.9 mL |
| Total | N/A | 10 mL |
Store at 4°C for up to one week.
Alternatives: This protocol uses an SI-1400 incubator from Scientific Industries to shake the dissociated primary tumors at 37°C. Any incubator/shaker that can hold 50 mL and 15 mL tubes, set up 37°C, and rock at a speed of 20 cycles per minute are alternatives.
Alternatives: This protocol uses a LiveCellTM Stage Top Incubation System from Pathology Devices to control the temperature, humidity, and CO2 levels of the co-culture system during time-lapse imaging. Other incubation systems that have these functions and are compatible with inverted microscopes are alternatives.
Alternatives: This protocol uses a Yokogawa Spinning Disk Scanning Unit and a Zeiss confocal microscope. Other microscopes that have the ability to rapidly acquire images with limited phototoxicity are also suitable.
Step-by-step method details
Isolate macrophages and cancer cells from mouse primary mammary carcinomas – Day 1
Timing: 6–7 h
MMTV-PyMT is well established breast cancer mouse model9,10 and the immune profile of these tumors has been identified and compared with other breast cancer models.11 The primary tumors of MMTV-PyMT mice (on C57BL/6 background) are mechanically dissociated and digested with the collagenase/hyaluronidase solution containing DNase I at 37°C for 45 min. After pulse centrifugation (450 × g), the supernatant (immune cells and single cancer cells) and cell pellet (cancer cell aggregates) are separated and collected. CD11b MicroBeads are used to enrich CD11b+ cells from the supernatant, and macrophages are obtained by allowing the CD11b+ cells to adhere to fibronectin-coated glass bottom plate for 1 h (non-adherent cells are removed by washing with PBS after 1 h). Cancer cells are isolated from the tumor cell pellets by dissociating the cell aggregates into single cancer cells with TrypLE Express Enzyme containing DNase I at 37°C for 10 min (Figure 1A).
Note: The tumor can be any size (Figures 1B and 1C). Choose the size of the tumor according to your purpose, such as an early-stage tumor (tumor size <5 mm) or an advanced-stage tumor (tumor size >12 mm). This protocol uses 1.0 g tumor (two tumors, ∼10 mm in diameter, each tumor weighing ∼0.5 g) per 10 mL dissociation buffer in a 50 mL tube and per 5 mL TrypLE Express Enzyme in a 15 mL tube. If more tumors are needed, scale up all reagent volumes and tube numbers (e.g., for 2.0 g tumors, use two 50 mL tubes with 10 mL of dissociation buffer in each tube, and two 15 mL tubes with 5 mL of TrypLE Express Enzyme in each tube).
Note: Pulse centrifugation is used to separate single cells (including immune cells, cancer cells and other cell types) from cell aggregates by difference in masses/sedimentation rates. Centrifugation (with acceleration/deceleration set at 9/9) is set to 450 × g and stopped immediately (in 5 s) when the speed reaches 450 × g. The longer the centrifugation time at 450 × g, the more single cells will move from the supernatant to the pellet with cell aggregates, and the yield of macrophages isolated from the supernatant may therefore decrease. The yield of macrophages from the supernatant after pulse centrifugation and from the whole tumor (centrifugation at 450 × g for 10 min to collect all the cells) are approximately the same. We can obtain more than enough cancer cells from the cell aggregates in the pellet. With pulse centrifugation, we can simultaneously isolate immune and cancer cells from the same tumors, isolating a large amount of immune cells from the supernatant and more than sufficient cancer cells from pellet.
-
1.
Harvest the primary tumors.
Before harvesting the tumors, thaw 10× collagenase/hyaluronidase (stored at −20°C) and warm up 1% BSA solution to 20°C–25°C in a tissue culture room.-
a.Put one 10 cm Petri dish with cold PBS on ice. Place surgery tools (tweezers and scissors) on the surgery table and turn on the dry glass bead sterilizer.
-
b.Disinfect the open bench for the tumor harvest.12
-
i.Wipe down the dissection station with 70% ethanol.
-
ii.Clean all surgical supplies, a dissection board (a Styrofoam board covered with a sterile, new blue pad), and pins with 70% ethanol.
-
iii.Disinfect the surgery tools by heat in the glass bead sterilizer at 280°C for 1 min. Cool the tools on the blue pad.
-
i.
-
c.Euthanize the mouse in a CO2-saturated chamber, followed by cervical dislocation.
-
d.With the carcass on its back, pin it to the dissection board. Then spray with 70% ethanol until the fur is soaking wet.
-
e.Pinch a small piece of the skin with a tweezer, and make a small incision on the abdominal skin with sharp scissors, then extend the cut along the ventral midline up to the neck. Be careful to cut only the skin and not the peritoneum underneath.
-
f.Extend the cuts laterally towards the forelimbs and hind limbs. Use the tweezer and a cotton swab to release the skin from the peritoneum. Pull the skin away and stretch it tight before pinning it down (Figure 1B).
-
g.Collect and pool the primary tumors in the Petri dish.
 CRITICAL: To obtain an optimal tumor-associated macrophage purity and yield, avoid collecting the lymph nodes (Figure 1D) and necrotic tumors (Figure 1E) and remove the fat and vessels around the tumor (Figures 1F and 1G). This is done because macrophages from lymph nodes may contaminate TAMs; necrotic tumors contain a high percentage of neutrophils; vessels with excess amount of erythrocytes demand increased ACK lysis time in step 4c, which in turn cause damage to the cells.
CRITICAL: To obtain an optimal tumor-associated macrophage purity and yield, avoid collecting the lymph nodes (Figure 1D) and necrotic tumors (Figure 1E) and remove the fat and vessels around the tumor (Figures 1F and 1G). This is done because macrophages from lymph nodes may contaminate TAMs; necrotic tumors contain a high percentage of neutrophils; vessels with excess amount of erythrocytes demand increased ACK lysis time in step 4c, which in turn cause damage to the cells. -
h.Bring the dissected tumors to a tissue culture hood.
-
a.
-
2.Digest the tumors with dissociation buffer.Note: For this step, the pipettes should be pre-coated with 1% BSA to increase cellular yield, as tumor tissues easily stick to the un-coated pipette and are hard to be removed. To coat the inner surface of the pipettes, pipette BSA solution up and down once before use. A short coating with BSA is sufficient.
-
a.Prepare the dissociation buffer (refer to “materials and equipment”) and turn on the 37°C shaker. You will need 10 mL of buffer in a 50 mL tube for 1.0 g tumor.
 CRITICAL: Collagenase/hyaluronidase 10× solution should be freshly diluted with RPMI 1640+5% FBS to a 1× solution, and DNase I (final concentration 4 U/mL) is added to the 1× solution. DNase I will digest the viscous genomic DNA leaking out from the dead cells during digestion and will thus make the solution clearer and less sticky and increase the cellular yield.
CRITICAL: Collagenase/hyaluronidase 10× solution should be freshly diluted with RPMI 1640+5% FBS to a 1× solution, and DNase I (final concentration 4 U/mL) is added to the 1× solution. DNase I will digest the viscous genomic DNA leaking out from the dead cells during digestion and will thus make the solution clearer and less sticky and increase the cellular yield. -
b.Rinse the tumors with PBS and transfer them to a new 10 cm Petri dish. Add 1 mL of dissociation buffer to the tumors (Figure 2A).
 CRITICAL: Do not mince the tumors without the buffer.
CRITICAL: Do not mince the tumors without the buffer. -
c.Cut the tumors into small pieces with a razor blade, and mince them until they are paste-like (Figure 2B).Note: Cut one tumor into 2–4 pieces, and let the cross section touch the dish to prevent the tumor from sliding around.
 CRITICAL: Do not use scissors which may damage the cells by squeezing the tumor.
CRITICAL: Do not use scissors which may damage the cells by squeezing the tumor. -
d.Transfer the minced tissue into the 50 mL tube with a BSA-coated 10 mL pipette (Figures 2C and 2D). Place the tube horizontally in a 37°C shaker to ensure even mixing (Figure 2E). Shake for 45 min at a speed of 20 cycles/min.Note: A longer incubation time will not increase the yield of macrophages and will digest the cell surface antigens.Note: While waiting for digestion, warm up the wash buffer (refer to “materials and equipment”) and the TrypLE Express Enzyme to 20°C–25°C.
-
e.After digestion (Figure 2F), pipette the cell suspension up and down about 30 times using a BSA-coated 5 mL pipette. Add 10 mL of wash buffer to each 50 mL tube.Note: It is very important to resuspend the cells completely, as erythrocytes tend to attach to myeloid cells.
-
f.Spin down cells at 450 × g for 10 min at 20°C–25°C.
-
g.Discard the supernatant (Figure 2G). Resuspend the pellet with 10 mL of wash buffer. Transfer the suspension to a 15 mL tube.
-
h.Pulse to 450 × g. Stop immediately when the speed reaches 450 × g. Collect the supernatant for macrophage isolation (Figure 2H).
-
i.Resuspend the pellet in 10 mL of wash buffer.
-
j.Repeat wash/pulse spins (steps 2h and 2i) 3–5 times until the supernatant is clear (not red/pink, Figure 2H). Pool all the non-clear supernatants in one 50 mL tube for macrophage isolation (step 4 below). When the supernatant is clear, use the pellet for tumor cell isolation.Note: The supernatant contains immune cells and single cancer cells. The pellet contains cancer cell aggregates.Note: Place the 50 mL tube with the pooled, non-clear supernatants on ice.
-
a.
-
3.Isolate cancer cells from the pellet.
-
a.Wash the pellet with 10 mL of PBS to remove residual FBS from the wash buffer (steps 2i and 2j). Spin down cells at 450 × g for 5 min at 20°C–25°C and discard the supernatant.Note: Residual FBS inhibits TrypLE Express Enzyme activity.
-
b.Resuspend the pellet in 5 mL of TrypLE Express Enzyme, and add 2 μL DNase I (final concentration 4 U/mL).
-
c.Place the 15 mL tube horizontally in the 37°C shaker (Figure 3A). Incubate the tube for 10 min with constant low agitation (20 cycles/min).Note: While waiting for the digestion, warm up the cancer cell culture medium to 20°C–25°C.
-
d.After digestion (Figure 3B), add 9–10 mL of wash buffer to each 15 mL tube. Spin down the cells at 450 × g for 5 min at 20°C–25°C.
-
e.Discard the supernatant (Figure 3C). Wash the pellet with 10 mL of wash buffer. Spin down cells at 450 × g for 5 min at 20°C–25°C.Note: Do not touch the pellet when removing the supernatant, as the loose pellet is easily removed together with the supernatant.
-
f.Resuspend the cell pellet in wash buffer and transfer to a 50 mL falcon tube. Adjust the volume to 20–30 mL with additional wash buffer.
-
g.Filter the cell suspension through a 70 μm cell strainer to a new 50 mL tube.
-
h.Filter the cell suspension through a 40 μm cell strainer to a new 50 mL tube (Figure 3D).
-
i.Count the cells. Usually, we can obtain 4.5 ± 0.9 × 107 viable cancer cells (∼90% viability, based on n=12 isolations) from 1.0 g tumor, and the purity of EpCAM+ cells is >90% (Figure 3E).Optional: Take 1–2 × 106 cells and stain with APC anti-mouse CD326 (EpCAM) antibody for purity analysis by flow cytometry.Note: Tumors of different sizes have different numbers of cancer cells. This predicted cancer cell number is from two ∼10 mm MMTV-PyMT tumors (C57BL/6 background).
-
j.Spin down 5 × 106 cancer cells at 450 × g for 5 min and plate them onto one 10 cm tissue culture dish.
- k.
-
a.
-
4.Isolate CD11b+ cells from the supernatant.Note: Some macrophages have low F4/80 expression, therefore we do not use F4/80 magnetic separation, but rather, the CD11b separation and adherence method. In the mouse, the CD11b antigen is expressed on monocytes/macrophages, and to a lower extent, on granulocytes, NK cells and a subset of dendritic cells.Sample preparation:
-
a.Place ACK lysis buffer and MACS buffer (refer to “materials and equipment”) on ice.
-
b.Spin down the collected, pooled supernatant at 300 × g for 10 min at 4°C.
 CRITICAL: Do not centrifuge higher than 300 × g when working with immune cells, or the viability of immune cells will be decreased. Keep the cells cold.
CRITICAL: Do not centrifuge higher than 300 × g when working with immune cells, or the viability of immune cells will be decreased. Keep the cells cold. -
c.Discard the supernatant (Figure 4A). Resuspend the cell pellet in 5–10 mL of cold ACK lysis buffer to lyse erythrocytes. Keep on ice for 5–10 min.Note: The ACK lysis buffer can also be used at 20°C–25°C with shorter lysis time (3–5 min).Note: The cell pellet should be resuspended immediately after adding ACK lysis buffer to reduce the risk of damaging the macrophages.
-
d.Add 10 mL of cold MACS buffer.
-
e.Spin down the cells at 300 × g for 10 min at 4°C (Figure 4B). If lysis is incomplete, as evidenced by pink or red color of the pellet, repeat steps 4c and 4d.Note: Usually we do not need to repeat ACK lysis when we follow the step 1g carefully.
-
f.Resuspend the cell pellet in 20–30 mL of MACS buffer.
-
g.Filter the cell suspension through a 40 μm cell strainer to a new 50 mL tube.
-
h.Filter the cell suspension through a 30 μm pre-separation filter to a new 50 mL tube (Figure 4C).
 CRITICAL: For optimal performance, it is important to obtain a single cell suspension before magnetic separation. Pass cells through a 30 μm pre-separation filter to remove cell clumps that may clog the column.Note: To increase the speed of pre-separation filtering, use a 40 μm cell strainer first to remove larger clumps.
CRITICAL: For optimal performance, it is important to obtain a single cell suspension before magnetic separation. Pass cells through a 30 μm pre-separation filter to remove cell clumps that may clog the column.Note: To increase the speed of pre-separation filtering, use a 40 μm cell strainer first to remove larger clumps. -
i.Determine the cell number. Usually, we can obtain 4.8 ± 1.0 × 107 viable pre-CD11b cells (∼88% viability, based on n=15 isolations) from 1.0 g tumor.Optional: Take 1–2 × 106 cells and stain with BV785™ anti-mouse CD45 antibody and APC anti-mouse/human CD11b antibody for flow cytometry (Figure 4D).Note: Tumors of different sizes have different percentages of immune cells and different numbers of pre-CD11b cells. This predicted pre-CD11b cell number is from two ∼10 mm tumors.
Magnetic labeling (positive selection of CD11b+ cells, following the manufacturer’s instructions https://www.miltenyibiotec.com/upload/assets/IM0001265.PDF): CRITICAL: For the following magnetic labeling steps, work fast, keep the cells cold, and use pre-cooled solutions. This approach will reduce the unspecific binding of antibodies on the cell surface.
CRITICAL: For the following magnetic labeling steps, work fast, keep the cells cold, and use pre-cooled solutions. This approach will reduce the unspecific binding of antibodies on the cell surface.-
j.Centrifuge the cell suspension at 300 × g for 10 min at 4°C. Pipette off the supernatant completely.
-
k.Resuspend the cell pellet in 90 μL of MACS buffer per 107 total cells.Note: When working with higher cell numbers, scale up all reagent volumes and total volumes accordingly (e.g., for 2 × 107 total cells, use twice the volume of all indicated reagent volumes and total volumes).
-
l.Add 10 μL of CD11b MicroBeads per 107 total cells.
 CRITICAL: Vortex the MicroBeads thoroughly before use (e.g., pulse vortex multiple times).
CRITICAL: Vortex the MicroBeads thoroughly before use (e.g., pulse vortex multiple times). -
m.Mix well (Figure 4E) and incubate for 15 min at 4–8°C (in a refrigerator).Note: Working on ice may require increased incubation times. Higher temperatures and/or longer incubation times lead to non-specific cell labeling.
-
n.Wash cells by adding 1–2 mL of buffer per 107 cells and centrifuge at 300 × g for 10 min at 4°C. Pipette off the supernatant completely.
-
o.Resuspend up to 108 cells in 500 μL of MACS buffer.Note: For higher cell numbers, scale up the buffer volume accordingly.
Magnetic separation: CRITICAL: The maximum number of loaded cells for each LS column is 108. If you load more than 108 cells, use more LS columns, or the column will be clogged and unlabeled cells may not be washed away.
CRITICAL: The maximum number of loaded cells for each LS column is 108. If you load more than 108 cells, use more LS columns, or the column will be clogged and unlabeled cells may not be washed away.-
p.Place the LS column in the magnetic field of the MACS separator (see Figure 4F).
-
q.Rinse the LS column with 3 mL of MACS buffer.Note: Steps 4p and 4q can be done during centrifugation in step 4n.
-
r.Load the cell suspension onto the column.
-
s.Wash the column three times with MACS buffer (3 × 3 mL, Figure 4F).
 CRITICAL: Add buffer each time the column reservoir is empty.Note: Unlabeled cells will pass through to the 15 mL tube.
CRITICAL: Add buffer each time the column reservoir is empty.Note: Unlabeled cells will pass through to the 15 mL tube. -
t.Remove the column from the separator and place it partially in a new 15 mL collection tube.
-
u.Pipette 5 mL of MACS buffer onto the column. Immediately flush out the fraction with the magnetically labeled cells by firmly applying the plunger supplied with the column (Figure 4G).
-
v.Count the cells. Usually, we can obtain 6.7 ± 1.7 × 106 viable CD11b+ cells (>90% viability, based on n=12 isolations) from 1.0 g tumor (two ∼10 mm tumors), and the purity of CD11b+ cells is >90% (Figure 4H).Optional: Take 1–2 × 106 cells and stain with BV785™ anti-mouse CD45 antibody and APC anti-mouse/human CD11b antibody for flow cytometry to determine purity.
-
a.
-
5.Separate the macrophages from CD11b+ cells using the adherence method.Note: Pre-coat a 24-well glass bottom plate with fibronectin during centrifugation in step 4j. Dilute the fibronectin with PBS to 10 μg/mL. Add 300 μL of diluted fibronectin to each well. Incubate for 45 min–1 h at 37°C. The plate can also be pre-coated for 12–16 h at 4°C. Fibronectin coated plates can be stored for 2–4 weeks at 4°C in a closed sterile container or sterile sealable bags.Note: Vortexing or excessive agitation of fibronectin solutions should be avoided.Note: Warm up the macrophage culture medium to 20°C–25°C.
 CRITICAL: Add fresh 2-mercaptoethanol (final concentration: 55 μM) and M-CSF (final concentration: 10 ng/mL) to the macrophage culture medium (refer to “materials and equipment”). 2-mercaptoethanol is a reducing agent to prevent toxic levels of oxygen radicals. M-CSF plays an important role in enhancing the effector functions of macrophages.
CRITICAL: Add fresh 2-mercaptoethanol (final concentration: 55 μM) and M-CSF (final concentration: 10 ng/mL) to the macrophage culture medium (refer to “materials and equipment”). 2-mercaptoethanol is a reducing agent to prevent toxic levels of oxygen radicals. M-CSF plays an important role in enhancing the effector functions of macrophages.-
a.Centrifuge the CD11b+ cell suspension at 300 × g for 5 min at 4°C. Pipette off the supernatant.
-
b.Resuspend the cell pellet in 500 μL of macrophage culture medium per 6 × 105 cells.
-
c.Carefully aspirate the remaining fibronectin solution from the plate.
 CRITICAL: Work fast and do not let the wells completely dry before moving to next step, as over-drying affects the attachment of cells.
CRITICAL: Work fast and do not let the wells completely dry before moving to next step, as over-drying affects the attachment of cells. -
d.Rinse the plate once with PBS.
 CRITICAL: Work fast and do not let the wells completely dry before moving to next step.
CRITICAL: Work fast and do not let the wells completely dry before moving to next step. - e.
-
f.After 1 h of attachment,13 remove the unattached cells (granulocytes, NK cells and dendritic cells) by pipetting out the supernatant (medium) of the wells.
-
g.Wash the attached cells 3 times with PBS to remove any remaining leukocytes.Note: Depending on the percentage of macrophages out of the CD11b+ cells (∼40% for MMTV-PyMT tumors) and the loss of cells in these steps, we can obtain ∼1.5 × 105 macrophages from 6 × 105 CD11b+ cells per well. The purity of macrophages is >90% (Figure 4J).Optional: Detach the adherent cells with Trysin+0.5 M EDTA at 37°C until the cells become round (∼10 min) and scrape the round cells (macrophages) gently with a cell lifter. Take 1–2 × 106 cells and stain with BV785™ anti-mouse CD45 antibody and Alexa Fluor® 488 anti-mouse F4/80 antibody for flow cytometry.
-
h.Culture the macrophages for 12–16 h in the incubator at 37°C with 5% CO2. The morphology of the macrophages is shown in Figure 4K.
-
a.
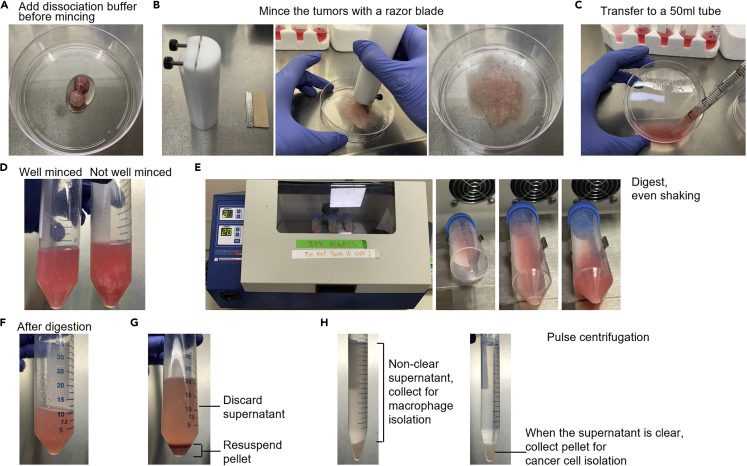
Figure 2.
Digest the tumors with collagenase/hyaluronidase dissociation buffer
(A) Add 1 mL of dissociation buffer to the tumors before mincing.
(B) Mince the tumors with a sterilized razor blade until it is paste-like.
(C) Transfer the minced tissue into a 50 mL tube with a 10 mL pipette. Pipettes are pre-coated with 0.1% BSA.
(D) Comparison of a well-minced tumor (left) and a not well-minced tumor (right).
(E) Digest the minced tumors with dissociation buffer in a 37°C shaker, horizontally. Shake for 45 min at a speed of 20 cycles/min.
(F) The tumors after digestion.
(G) After centrifugation, discard the supernatant and resuspend the pellet with wash buffer.
(H) Use pulse centrifugation to harvest the unclear supernatant (left) for macrophage isolation. Repeat pulse centrifugation until the supernatant is clear (right), and then collect the pellet for cancer cell isolation.
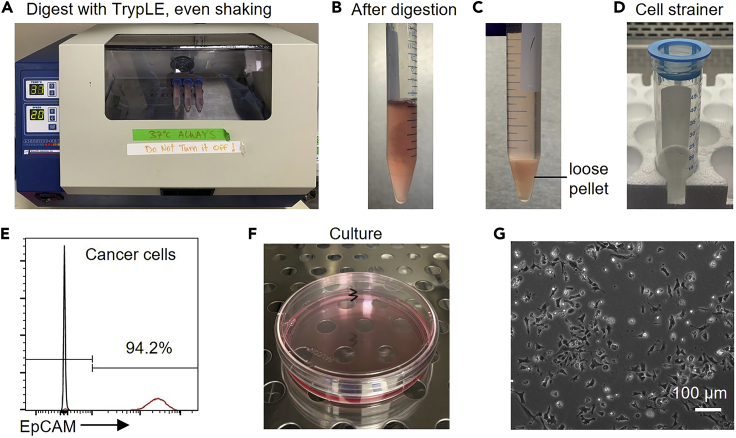
Figure 3.
Isolate cancer cells from the pellet
(A) Digest the pellet with TrypLE Express Enzyme in a 37°C shaker horizontally. Shake for 10 min at a speed of 20 cycles/min.
(B) The tumors after TrypLE digestion.
(C) Loose pellet after centrifugation. Discard the supernatant carefully and resuspend the pellet.
(D) Filter cell suspension using cell strainers.
(E) Purity of cancer cells (EpCAM+, CD326) determined by flow cytometry.
(F) Culture cancer cells for 12–16 h in the incubator.
(G) Morphology of cancer cells (MMTV-PyMT).
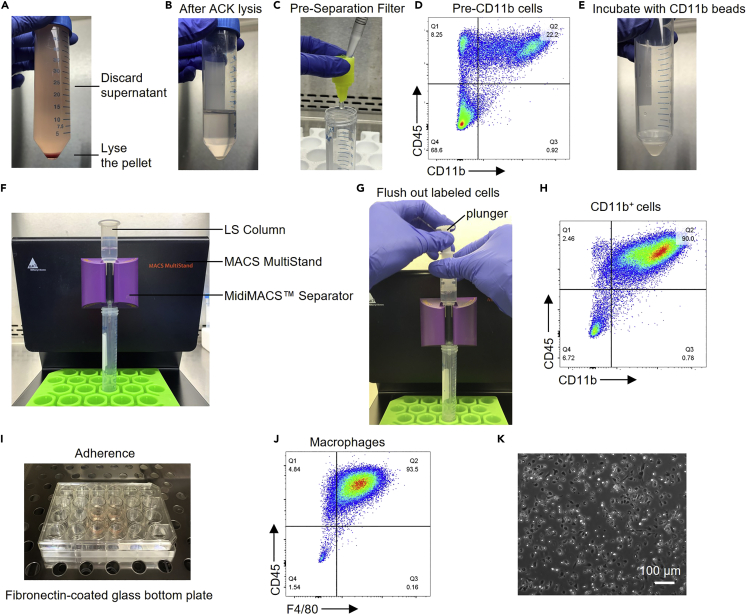
Figure 4.
Isolate macrophages from the non-clear supernatant
(A) After centrifugation of the pooled, non-clear supernatants, discard the supernatant and lyse the red pellet with ACK lysis buffer.
(B) Cells after red blood cell lysis.
(C) Filter the cell suspension with a pre-separation filter.
(D) The percentage of CD11b+ cells before magnetic separation determined by flow cytometry.
(E) Incubate the pre-CD11b cells with CD11b MicroBeads.
(F) After incubation with microbeads, load the cells onto the LS column and wash away magnetically antibody unlabeled cells.
(G) Flush out the magnetically antibody labeled cells by firmly applying the plunger.
(H) The percentage of CD11b+ cells (CD45+CD11b+) after magnetic separation determined by flow cytometry.
(I) Culture CD11b+ cells in the incubator for 1 h, allowing macrophages to attach.
(J) Purity of macrophages (CD45+F4/80+) determined by flow cytometry.
(K) Morphology of macrophages.
Set up a co-culture system for the killing assay – Day 2
Timing: 3–4 h
Detach the cancer cells from the tissue culture dish with TrypLE Express Enzyme and label them with CellTracker™ CMFDA (5-chloromethylfluorescein diacetate) Dye (1 μM). Label the macrophages with CellTracker™ Deep Red Dye (1 μM). After labeling, seed the cancer cells to the macrophage-cultured 24-well glass bottom plate and allow them to attach for 2 h. Then, start applying different treatments to the co-culture system (vehicle/MPLA+IFNγ/MPLA/IFNγ) (Figure 5A). The final concentration of MPLA is 100 ng/mL, and the final concentration of IFNγ is 33 ng/mL. MPLA stock solution is at 100 μg/mL in 0.2% triethylamine, and IFNγ stock solution is at 100 μg/mL in PBS. Vehicle contains 0.2%∗10−3 triethylamine.
Note: Cancer cells and macrophages can be stained with other CellTracker™ probes if the probes have different excitation/emission spectra so we can distinguish them.
-
6.Stain cancer cells with CellTracker™ CMFDA Dye.Note: Warm up the TrypLE Express Enzyme, serum-free DMEM, and cancer cell culture medium to 20°C–25°C.
-
a.Aspirate the medium from the cancer cell culture dish. Wash with PBS to remove residual FBS.
-
b.Add 3 mL of TrypLE Express Enzyme to a 10 cm dish. Incubate 5–10 min at 37°C until the cells detach from the dish when rocked.
-
c.Add FBS to stop TrypLE Express Enzyme digestion.Note: Avoid using Trypsin since the primary cancer cells are more fragile than cancer cell lines.
-
d.Spin down the cancer cells at 300 × g for 5 min at 20°C–25°C. Aspirate the supernatant.
-
e.Resuspend the cell pellet in serum-free DMEM. Count the cells.Note: The medium can be DMEM, RPMI 1640, or any other serum-free medium.
 CRITICAL: Do not use serum-containing medium. Serum reduces staining efficacy.
CRITICAL: Do not use serum-containing medium. Serum reduces staining efficacy. -
f.Dilute the cancer cells to 1.5 × 105 per 500 μL of serum-free medium.
-
g.Add CMFDA Dye to the cells in a 15 mL tube (final concentration 1 μM) and mix by pipetting gently up and down 3–5 times. Stain for 30 min in the incubator at 37°C with 5% CO2 (Figure 5B).
-
h.Add at least 3 volumes of FBS to the stained cancer cells and centrifuge the cells at 300 × g for 5 min at 20°C–25°C.Note: FBS is critical for maintaining the high viability of the stained cells.
-
i.Resuspend the cell pellet in the cancer cell culture medium.
-
a.
-
7.Stain the macrophages with CellTracker™ Deep Red Dye.
-
a.While waiting for the cancer cell staining in step 6g, add Deep Red Dye to serum-free medium to prepare the Deep Red Dye working solution (final concentration 1 μM).
-
b.Aspirate the macrophage culture medium from the 24-well plate (from step 5h).
-
c.Wash with PBS once.
-
d.Add the Deep Red Dye working solution to the macrophages (500 μL per well). Stain the macrophages for 30 min in the incubator at 37°C with 5% CO2 (Figure 5C).
-
e.Add at least 3 volumes of FBS to the stained macrophages and aspirate the solution.
-
f.Add fresh macrophage culture medium to the plate (500 μL per well).
-
a.
-
8.Co-culture the cancer cells and macrophages.
-
a.Seed the stained cancer cells (from step 6i) to the 24-well plate. Seed 5 × 104 cancer cells in 500 μL of cancer cell culture medium per well.
-
b.After 2 h of attachment, add 1 mL of macrophage culture medium to each well (for 2 mL medium in total for each well).
-
a.
Note: To test the effects of macrophage activating drugs, add e.g., MPLA or IFNγ alone or in combination to the 1 mL of macrophage medium so that the final concentration (in 2 mL) is 100 ng/mL of MPLA and/or 33 ng/mL of IFNγ. Mixing at this step is not necessary.
Figure 5.
Stain cancer cells and macrophages with CellTracker™ dyes and co-culture them
(A) Schematic diagram of staining and co-culturing TAMs and cancer cells.
(B) Stain cancer cells with CMFDA Dye in a 15 mL tube.
(C) Stain macrophages with Deep Red Dye in the 24-well glass bottom plate.
Live cell imaging – Day 2
Timing: 2–3 days
The tumoricidal activity of macrophages is monitored by time-lapse imaging. The 24-well glass bottom plate, with the macrophage and cancer cell co-cultures, is placed on the stage of the spinning disk confocal microscope and incubated within the LiveCellTM Stage Top Incubation System. Turn on the 488 nm laser (for CMFDA Dye) and the 647 nm laser (for Deep Red Dye), and pick 5 fields of view for each condition. When the incubation system is ready (37°C, 5% CO2, 75%–85% relative humidity), start imaging: take images every 20 min for 60 h in total. Images can be analyzed using Imaris and ImageJ software.
-
9.
Set up the LiveCellTM Stage Top Incubation System.
Note: Place the co-cultured 24-well plate in the stage and turn on the LiveCellTM Incubation System. Wait until the system is stabilized (∼2 h, 37°C, 5% CO2, 75%–85% relative humidity).
CRITICAL: Use other detectors, such as a thermometer to measure the temperature and make sure that the temperature is not higher than 38°C or the cells will die.
CRITICAL: Check CO2 tank and make sure that there is sufficient CO2 for 3 days imaging.
-
10.
Operate the Yokogawa spinning disk confocal microscope.
Note: While waiting for the incubation system to stabilize in step 9, pick 5 fields of view (top, middle, bottom, left, and right) for each well.
-
11.Perform time-lapse imaging.
-
a.When the incubation system is ready, go through all the fields and refocus them.
-
b.Start imaging: take images every 20 min for 60 h, yielding 180 images in total.Note: We recommend to refocus every 12 h.
-
c.Analyze the images using Imaris and ImageJ software.Note: Export videos using Imaris software.Note: Analyze images at specific time points with ImageJ software.
-
a.
Expected outcomes
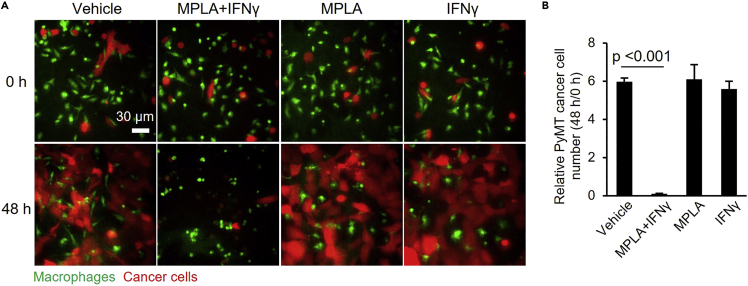
Using the above described protocol, we have shown that TAMs isolated from mouse mammary carcinomas and treated with IFNγ and MPLA can be repolarized ex vivo,1 resulting in the killing of ∼90% of cancer cells in 48 h in the co-cultures (Figure 6, Table 1, Methods videos S1 and S2). MPLA or IFNγ alone did not induce the tumoricidal activity of the macrophages (Figure 6).
Figure 6.
MPLA+IFNγ induces tumoricidal activities of mouse mammary carcinoma-derived macrophages
(A) Macrophages and PyMT cancer cells at 0 h and 48 h after the start of imaging. Macrophages were stained with CellTracker™ Deep Red Dye, while cancer cells were stained with CellTracker™ CMFDA Dye. Deep Red Dye is falsely colored green for clarity.
(B) Statistical analysis of relative PyMT cancer cell numbers for panel A. One-way ANOVA was performed (n=3, equal variances). Data are represented as mean ± SD. See also Methods videos S1 and S2.
Table 1.
Analysis of raw data from the MPLA+IFNγ group
| Experiment | Field of view | Raw data- cancer cell number (0 h) | Raw data- cancer cell number (48 h) | Relative cancer cell number (48 h/0 h) | Average | Average of all experiments | Standard deviation (SD) |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 13 | 2 | 0.154 | 0.090 | 0.105 | 0.027 |
| 2 | 14 | 1 | 0.071 | ||||
| 3 | 12 | 1 | 0.083 | ||||
| 4 | 14 | 2 | 0.143 | ||||
| 5 | 13 | 0 | 0 | ||||
| 2 | 1 | 13 | 1 | 0.077 | 0.135 | ||
| 2 | 12 | 2 | 0.167 | ||||
| 3 | 11 | 1 | 0.091 | ||||
| 4 | 10 | 2 | 0.2 | ||||
| 5 | 7 | 1 | 0.143 | ||||
| 3 | 1 | 9 | 1 | 0.111 | 0.088 | ||
| 2 | 8 | 1 | 0.125 | ||||
| 3 | 15 | 2 | 0.133 | ||||
| 4 | 12 | 0 | 0 | ||||
| 5 | 14 | 1 | 0.071 |
Macrophages were stained with CellTracker™ Deep Red Dye, while tumor cells were stained with CellTracker™ CMFDA Dye. Deep Red Dye is falsely colored green for clarity. Images were recorded using time-lapse microscopy over 48 h. Images were taken every 20 min. The time indicated is the time after imaging was initiated.
Macrophages were stained with CellTracker™ Deep Red Dye, while tumor cells were stained with CellTracker™ CMFDA Dye. Deep Red Dye is falsely colored green for clarity. MPLA with mouse IFNγ was added 2 h before imaging was initiated. Images were recorded using time-lapse microscopy over 48 h. Images were taken every 20 min. The time indicated is the time after imaging was initiated.
iNOS is required for macrophages’ tumoricidal activity14 and only the combination treatment MPLA+IFNγ significantly induced iNOS expression, whereas treatment with MPLA or IFNγ alone did not.1 Nos2 (the gene coding for iNOS) expression is regulated by four transcription factors (NF-kB, AP-1, STAT1, IRF1).15,16,17,18 Signaling from Toll like receptor 4, the MPLA receptor, can activate NF-kB and AP-1 via MyD88,19 and IFNγ activates the JAK/STAT1 pathway, leading to phosphorylated STAT1 dimer formation and IRF1 activation.20 Our previous results show that MPLA and IFNγ induce separate, complementary signaling pathways that are required for maximal iNOS expression and thus tumoricidal activity.1
iNOS metabolizes arginine to generate NO (nitric oxide), which diffuse into adjacent cells and cause cell death.21,22 However, NO has a half-life of just 4–6 s,21 and therefore, cell-cell contact between TAMs and cancer cells is required for macrophages’ tumoricidal killing. The MPLA+IFNγ-treated TAMs kill cancer cells first and then engulf cell debris (Methods video S2). We observed that the macrophages die after eliminating the cancer cells, whereas treatment with vehicle or MPLA or IFNγ alone did not affect the viability of the macrophages during the duration of the co-culture assays.
We propose that our protocol can be used to test other modulators than MPLA+IFNγ for ability to reprogram TAMs, including for abilities to induce tumoricidal activities. We anticipate that modulators that can induce tumoricidal activities will also be inducing iNOS expression. In addition, the protocol could be adapted for studies of drugs or antibodies that regulate the ability of macrophages to phagocytose cancer cells.23
Although our protocol has been specifically optimized for mouse mammary carcinomas, the spirit and general outline of the protocol should be readily adaptable for other tumor types, including mechanical and enzymatic dissociation of the tumor, pulse centrifugal separation of populations enriched for immune cells (supernatant) and cancer cells (pellet), and finally co-culture of the isolated cells. We expect that the key steps to optimize for other tumor types are the tumor dissociation steps, where different tumor types are likely to require different buffers, enzymes and incubation times. Other immune cells than macrophages can readily be isolated from the supernatant collected after pulse centrifugation by additional purification steps, for example cell separation beads, and co-cultures consisting of more than two cell types can be established by e.g., seeding additional cell types in cell culture inserts. For example, we envision that the protocol could be adapted with ease, to study interactions between macrophages, cancer cells and T cells isolated from the same tumor. Therefore, we have documented a specific outcome using the co-cultures – the ability to reprogram TAMs to achieve tumoricidal activities – and we envision that the protocol readily can be adapted for other studies of immune cell-cancer cell interactions.
Quantification and statistical analysis
For all conditions (vehicle/MPLA+IFNγ/MPLA/IFNγ), cancer cell numbers at 0 h and 48 h were determined by counting the cells in 5 fields of view, and the cell numbers at 48 h relative to those at 0 h were then calculated and reported. Here, we show, as an example, how to analyze the data from only the MPLA+IFNγ group.
Limitations
One limitation of the current protocol is that it has only been optimized for mouse mammary carcinomas and not for other primary tumors. Conditions, e.g., enzymes used, will need to be changed/optimized depending on the tumors/tissues of interest. Likewise, another limitation is that the breast cancer cell culture medium might not be suitable for other cancer cells. Users will need to find the best medium to culture the cancer cells of interest. This protocol could not separate monocytes (Ly6C+Ly6G-) from TAMs (F4/80+) because both of them can attach to the plate in step 5e. However, the percentage of monocytes after adherence is <5% of total cells (data not shown). The low ratio of monocytes to TAMs (>90% of total cells, Figure 4J) suggests that the presence of monocytes have little influence on the activity of TAMs.
Troubleshooting
Problem 1
The purity of cancer cells is low at step 3i.
Potential solution
First, when harvesting tumors, remove the fat and vessels around the tumors (Figures 1F and 1G). Second, mince the tumors well, to around 1 mm3 pieces (Figures 2B and 2D). Third, wash and pulse-centrifuge the dissociated tumors until the supernatant is clear (Figure 2H). Endothelial cells and immune cells will be removed through these steps. To purify cancer cells further, select cancer cells with CD326 (EpCAM) MicroBeads (mouse, cat #130-105-958) after step 3h. Other cells, including fibroblasts, will be washed away.
Problem 2
The purity of the TAMs is low at step 5g.
Potential solution
When harvesting tumors, do not collect necrotic tumors, which have many neutrophils, or lymph nodes (Figures 1D and 1E). The main source of contamination of the macrophages is usually cancer cells, which also attach to the plate at step 5e. To reduce cancer cell contamination, it is critical to use a 30 μm pre-separation filter to discard cancer cell aggregates (Figure 4C) before CD11b+ cell isolation. Work fast, keep the cells cold, use pre-cooled solutions during magnetic labeling, and use an appropriate number of LS columns during magnetic separation. Furthermore, the cells from step 4u can be passed over a new, freshly prepared column to increase the purity of CD11b+ cells and subsequently macrophages.
Problem 3
Cancer cells die during time-lapse imaging at step 11.
Potential solution
As primary cells, many cancer cells die or become quiescent if cultured for longer than day 3 after harvest. Therefore, stain and co-culture the cancer cells no later than on day 2. Furthermore, add at least 3 volumes of FBS to the stained cancer cells to stop staining and to maintain high viability of cancer cells at step 6h. Use a thermometer to ensure that the temperature of the LiveCellTM Stage Top Incubation System is 37°C. Finally, check the CO2 tank during live cell imaging.
Problem 4
MPLA+IFNγ-treated mammary carcinoma-derived macrophages do not kill cancer cells at step 11.
Potential solution
This protocol involves many steps, and small differences at any step can lead to changes in macrophage activity. It is therefore imperative to be consistent and accurate at all steps. Use bone-marrow-derived macrophages (BMDMs, unpolarized macrophages), which can be easily activated by MPLA+IFNγ, as a positive control. If the BMDMs cannot kill cancer cells, MPLA and IFNγ may have lost their activity, due to e.g., excessive freeze-thaws (more than 5 times) of the reagents. To avoid such problems, aliquot the stocks and use a new aliquot for each experiment.
Problem 5
Cells are damaged after ACK lysis at step 4c.
Potential solution
The volume of ACK lysis buffer and the lysis time are determined by the number of erythrocyte. To decrease the amount of erythrocyte, follow the step 1g carefully: remove the vessels around the tumors and do not collect necrotic tumors. Furthermore, resuspend the cell pellet immediately after adding ACK lysis buffer to decrease the potential cell damage.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mikala Egeblad (egeblad@cshl.edu).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This work was supported by grants from the National Cancer Institute (1R01CA237413), New York State Department of Health to M.E. (DOH01-C31850GG-3450000; the opinions, results, findings, and/or interpretations of data contained therein do not necessarily represent the opinions, interpretations, or policy of the State); and Swim Across America funds to M.E.; METAvivor Research and Support, Inc. to L.S.; and by funds from the CSHL Cancer Center (P30-CA045508). The schematic diagrams and graphical abstract were created with BioRender (agreement number NB24JGYFB5).
Author contributions
Conceptualization, L.S. and M.E.; methodology, L.S.; investigation, L.S. and X.H.; writing – original draft, L.S.; writing – review & editing, L.S., X.H., and M.E.; funding acquisition, L.S. and M.E.; supervision, M.E.
Declaration of interests
M.E. is a member of the research advisory board for brensocatib for Insmed; a member of the scientific advisory board for Vividion Therapeutics; a consultant for Protalix; and holds shares in Agios.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101833.
Contributor Information
Lijuan Sun, Email: sun@cshl.edu.
Mikala Egeblad, Email: egeblad@cshl.edu.
Data and code availability
This study did not generate/analyze datasets or code.
References
- 1.Sun L., Kees T., Almeida A.S., Liu B., He X.-Y., Ng D., Han X., Spector D.L., McNeish I.A., Gimotty P., et al. Activating a collaborative innate-adaptive immune response to control metastasis. Cancer Cell. 2021;39:1361–1374.e9. doi: 10.1016/j.ccell.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arwert E.N., Harney A.S., Entenberg D., Wang Y., Sahai E., Pollard J.W., Condeelis J.S. A unidirectional transition from migratory to perivascular macrophage is required for tumor cell intravasation. Cell Rep. 2018;23:1239–1248. doi: 10.1016/j.celrep.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitamura T., Doughty-Shenton D., Cassetta L., Fragkogianni S., Brownlie D., Kato Y., Carragher N., Pollard J.W. Monocytes differentiate to immune suppressive precursors of metastasis-associated macrophages in mouse models of metastatic breast cancer. Front. Immunol. 2018;8:2004. doi: 10.3389/fimmu.2017.02004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin M., Li X., Tan S., Zhou H.J., Ji W., Bellone S., Xu X., Zhang H., Santin A.D., Lou G., Min W. Tumor-associated macrophages drive spheroid formation during early transcoelomic metastasis of ovarian cancer. J. Clin. Invest. 2016;126:4157–4173. doi: 10.1172/JCI87252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peranzoni E., Lemoine J., Vimeux L., Feuillet V., Barrin S., Kantari-Mimoun C., Bercovici N., Guérin M., Biton J., Ouakrim H., et al. Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti–PD-1 treatment. Proc. Natl. Acad. Sci. USA. 2018;115:E4041–E4050. doi: 10.1073/pnas.1720948115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruffell B., Chang-Strachan D., Chan V., Rosenbusch A., Ho C.M.T., Pryer N., Daniel D., Hwang E.S., Rugo H.S., Coussens L.M. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 2014;26:623–637. doi: 10.1016/j.ccell.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wynn T.A., Chawla A., Pollard J.W. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kowal J., Kornete M., Joyce J.A. Re-education of macrophages as a therapeutic strategy in cancer. Immunotherapy. 2019;11:677–689. doi: 10.2217/imt-2018-0156. [DOI] [PubMed] [Google Scholar]
- 9.Guy C.T., Cardiff R.D., Muller W.J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954-961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin E.Y., Jones J.G., Li P., Zhu L., Whitney K.D., Muller W.J., Pollard J.W. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim I.S., Gao Y., Welte T., Wang H., Liu J., Janghorban M., Sheng K., Niu Y., Goldstein A., Zhao N., et al. Immuno-subtyping of breast cancer reveals distinct myeloid cell profiles and immunotherapy resistance mechanisms. Nat. Cell Biol. 2019;21:1113–1126. doi: 10.1038/s41556-019-0373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Y., Deng R., You H., Lu P. 3D in vitro culture system to study collective migration in mammary organoid epithelium. STAR Protoc. 2021;2:100778. doi: 10.1016/j.xpro.2021.100778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaczmarek M., Nowicka A., Kozłowska M., Żurawski J., Batura-Gabryel H., Sikora J. Evaluation of the phenotype pattern of macrophages isolated from malignant and non-malignant pleural effusions. Tumour Biol. 2011;32:1123–1132. doi: 10.1007/s13277-011-0214-1. [DOI] [PubMed] [Google Scholar]
- 14.Dinapoli M.R., Calderon C.L., Lopez D.M. The altered tumoricidal capacity of macrophages isolated from tumor-bearing mice is related to reduce expression of the inducible nitric oxide synthase gene. J. Exp. Med. 1996;183:1323–1329. doi: 10.1084/jem.183.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J., Morrison D.C., Parmely T.J., Russell S.W., Murphy W.J. An interferon-γ-activated site (GAS) is necessary for full expression of the mouse iNOS gene in response to interferon-γ and lipopolysaccharide. J. Biol. Chem. 1997;272:1226–1230. doi: 10.1074/jbc.272.2.1226. [DOI] [PubMed] [Google Scholar]
- 16.Martin E., Nathan C., Xie Q.W. Role of interferon regulatory factor 1 in induction of nitric oxide synthase. J. Exp. Med. 1994;180:977–984. doi: 10.1084/jem.180.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salim T., Sershen C.L., May E.E. Investigating the role of TNF-alpha and IFN-gamma activation on the dynamics of iNOS gene expression in LPS stimulated macrophages. PLoS One. 2016;11:e0153289. doi: 10.1371/journal.pone.0153289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Q.W., Whisnant R., Nathan C. Promoter of the mouse gene encoding calcium-independent nitric oxide synthase confers inducibility by interferon gamma and bacterial lipopolysaccharide. J. Exp. Med. 1993;177:1779–1784. doi: 10.1084/jem.177.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Y.-C., Yeh W.-C., Ohashi P.S. LPS/TLR4 signal transduction pathway. Cytokine. 2008;42:145–151. doi: 10.1016/j.cyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Castro F., Cardoso A.P., Gonçalves R.M., Serre K., Oliveira M.J. Interferon-Gamma at the crossroads of tumor immune surveillance or evasion. Front. Immunol. 2018;9:847. doi: 10.3389/fimmu.2018.00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelm M. Nitric oxide metabolism and breakdown. Biochim. Biophys. Acta. 1999;1411:273–289. doi: 10.1016/S0005-2728(99)00020-1. [DOI] [PubMed] [Google Scholar]
- 22.Stuehr D.J., Nathan C.F. Nitric oxide. A macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J. Exp. Med. 1989;169:1543–1555. doi: 10.1084/jem.169.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamber R.A., Nishiga Y., Morton B., Banuelos A.M., Barkal A.A., Vences-Catalán F., Gu M., Fernandez D., Seoane J.A., Yao D., et al. Inter-cellular CRISPR screens reveal regulators of cancer cell phagocytosis. Nature. 2021;597:549–554. doi: 10.1038/s41586-021-03879-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Macrophages were stained with CellTracker™ Deep Red Dye, while tumor cells were stained with CellTracker™ CMFDA Dye. Deep Red Dye is falsely colored green for clarity. Images were recorded using time-lapse microscopy over 48 h. Images were taken every 20 min. The time indicated is the time after imaging was initiated.
Macrophages were stained with CellTracker™ Deep Red Dye, while tumor cells were stained with CellTracker™ CMFDA Dye. Deep Red Dye is falsely colored green for clarity. MPLA with mouse IFNγ was added 2 h before imaging was initiated. Images were recorded using time-lapse microscopy over 48 h. Images were taken every 20 min. The time indicated is the time after imaging was initiated.
Data Availability Statement
This study did not generate/analyze datasets or code.