Abstract
Tick-borne encephalitis virus (TBEV) is the most medically relevant tick-transmitted Flavivirus in Eurasia, targeting the host central nervous system and frequently causing severe encephalitis. The primary function of its capsid protein (TBEVC) is to recruit the viral RNA and form a nucleocapsid. Additional functionality of Flavivirus capsid proteins has been documented, but further investigation is needed for TBEVC. Here, we show the first capsid protein 3D structure of a member of the tick-borne flaviviruses group. The structure of monomeric Δ16-TBEVC was determined using high-resolution multidimensional NMR spectroscopy. Based on natural in vitro TBEVC homodimerization, the dimeric interfaces were identified by hydrogen deuterium exchange mass spectrometry (MS). Although the assembly of flaviviruses occurs in endoplasmic reticulum–derived vesicles, we observed that TBEVC protein also accumulated in the nuclei and nucleoli of infected cells. In addition, the predicted bipartite nuclear localization sequence in the TBEVC C-terminal part was confirmed experimentally, and we described the interface between TBEVC bipartite nuclear localization sequence and import adapter protein importin-alpha using X-ray crystallography. Furthermore, our coimmunoprecipitation coupled with MS identification revealed 214 interaction partners of TBEVC, including viral envelope and nonstructural NS5 proteins and a wide variety of host proteins involved mainly in rRNA processing and translation initiation. Metabolic labeling experiments further confirmed that TBEVC and other flaviviral capsid proteins are able to induce translational shutoff and decrease of 18S rRNA. These findings may substantially help to design a targeted therapy against TBEV.
Keywords: tick-borne flaviviruses, capsid, nucleus, nucleolus, protein structure, translational shutoff
Abbreviations: ACN, acetonitrile; AHA, l-azidohomoalanine; APS, ammonium persulfate; BMRB, Biological Magnetic Resonance Data Bank; bNLS, bipartite nuclear localization sequence; co-IP, coimmunoprecipitation; DENV, dengue virus; ER, endoplasmic reticulum; FA, formic acid; FDR, false discovery rate; gRNA, genomic RNA; GST, glutathione-S-transferase; HDX, hydrogen–deuterium exchange; HRP, horseradish peroxidase; IgG, immunoglobulin G; IMPα, importin-alpha; KFDV, Kyasanur forest disease virus; LIV, Louping ill virus; MS, mass spectrometry; PLGS, ProteinLynx Global SERVER; POWV, Powassan virus; RCSB PDB, Research Collaboratory for Structural Bioinformatics Protein Data Bank; RFU, relative fractional uptake; SEC, size-exclusion chromatography; TBEV, tick-borne encephalitis virus; TBFV, tick-borne flavivirus; TEM, transmission electron microscopy; TEMED, N, N, N', N'-tetramethylethylenediamine; TEV, tobacco etch virus; WNV, West Nile virus; XL–MS, chemical crosslinking coupled with mass spectrometric detection; ZIKV, Zika virus
Tick-borne encephalitis virus (TBEV) is an important human pathogen belonging to the tick-borne viruses from the Flaviviridae family, genus Flavivirus. Except for tick-borne flaviviruses (TBFVs), the Flavivirus genus also includes mosquito-borne flaviviruses such as dengue virus (DENV), West Nile virus (WNV), or Zika virus (ZIKV). As a neurotropic virus, TBEV targets mainly the central nervous system, and the infection in humans may result in severe encephalitis or meningoencephalitis (1).
All members of the genus Flavivirus share a similar architecture of their virions, genomic organization, and life cycle. The flavivirus genome consists of a single-stranded RNA of positive polarity, approximately 11 kilobases long. Viral genomic RNA (gRNA) contains one open reading frame encoding a single polyprotein of about 3400 amino acids. The polyprotein is cotranslationally and post-translationally processed to three structural (pre-membrane [prM/M], capsid [C], and envelope [E]) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). While the structural proteins represent the main building units of the viral particle, the nonstructural proteins participate in viral replication. In addition, the flavivirus genome includes 5′ and 3′ untranslated regions, which are characterized by the presence of many secondary structures. Flaviviruses enter the cell via receptor-mediated endocytosis, and the released viral gRNA is translated into an endoplasmic reticulum (ER)–associated transmembrane polyprotein using the host enzymes. Viral gRNA is replicated by the nonstructural proteins in ER-derived vesicular compartments. Nascent gRNA is associated with the capsid protein resulting in nucleocapsid assembly. Formed nucleocapsids are further enveloped by ER-derived membrane containing prM/M and E proteins while budding into the ER lumen. Enveloped virions are then transported along the host cell secretory pathway and released from the cell by fusion of transport vesicles with the plasma membrane (2, 3).
Despite being one of the least conserved proteins among flaviviruses (identity is lower than 40%), the 3D structure of various flaviviral C proteins is remarkably similar (4, 5). Flaviviral C proteins are highly basic proteins of approximately 100 amino acid residues forming a homodimer in solution. Based on the structural studies on DENV, ZIKV, and WNV, monomeric C protein consists of four α-helices (α1–α4) and an unstructured N-terminal domain. Dimerization of C protein is facilitated mainly by interactions of antiparallel α2–α2′ and α4–α4′ helices of the two monomeric subunits, forming thus the main dimer interface. The dimer shows asymmetric charged surface distribution: the apolar surface on one side of a molecule and the positively charged region on the other side. Hydrophobic cleft formed by α1–α1′ and α2–α2′ helices was proposed to interact with the membrane, whereas highly positively charged regions of α4–α4′ helices are supposed to interact with viral gRNA (6). The structure of TBEVC has not been experimentally described yet; however, the predictions closely follow the structures of already characterized C proteins from DENV, ZIKV, and WNV (5, 7).
The primary function of flaviviral C protein is to recruit the viral gRNA and form the nucleocapsid. Although the assembly of flaviviruses takes place in ER-derived vesicular compartments, capsid protein has also been detected to be accumulated on the surface of lipid droplets (8, 9) and in nuclei or nucleoli of infected cells (10, 11, 12, 13, 14). The nuclear/nucleolar localization of C protein suggests that, except for its structural role, it may also be an essential factor in the interactions with the host environment. Indeed, several studies described various functions of C protein linked to nuclear localization. For example, DENV and Japanese encephalitis virus C proteins were found to interfere with the ribosome biogenesis through the binding to NPM1/B23 factor (15, 16), whereas ZIKV and WNV C proteins were shown to induce apoptosis via the activation of p53 signaling cascade (14, 17). Moreover, based on the interactions with core histones, DAXX transcription factor, and UBF1 splicing regulator, DENV and ZIKV C proteins were shown to affect host gene expression and nonsense-mediated mRNA decay, respectively (12, 18, 19). Furthermore, the complex functionality of the flavivirus C protein has been further documented by the emerging evidence of indiscriminate binding to the various single- or double-stranded RNA and DNA molecules in the case of yellow fever virus and ZIKV (19, 20, 21). Recently, we have described similar findings in the case of TBEVC (22) in combination with the occurrence of TBEV-induced transcriptional and translational shutoff (23). As our observations suggest a possible link between these two phenomena, we further studied TBEVC and its biological relevance. Here, we characterize the capsid protein of a TBFV for the first time, focusing on its structure, subcellular distribution, interacting network, and biological relevance during TBEV infection in human neural cells.
Results
Structure of monomeric Δ16-TBEVC
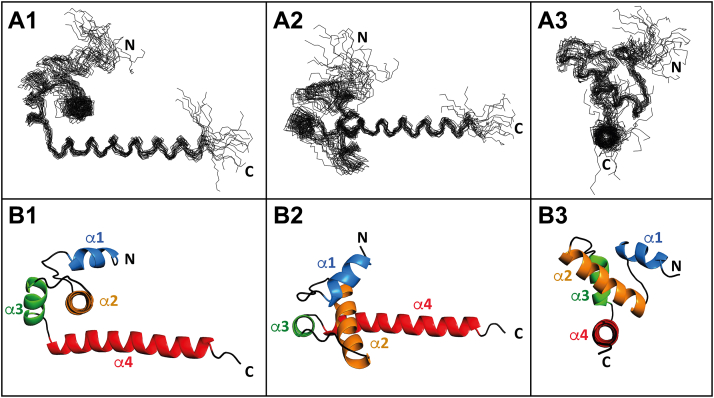
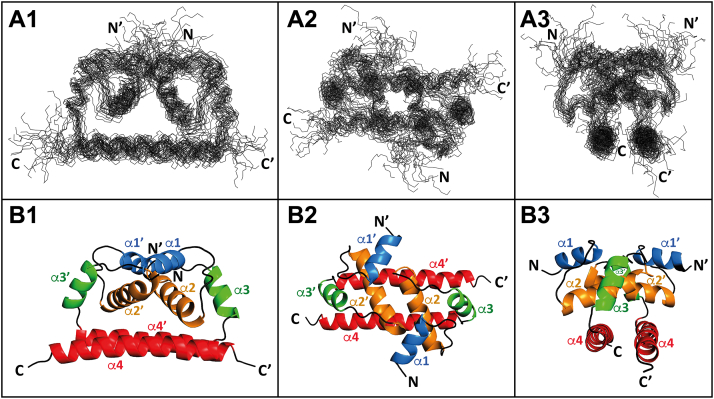
To better understand the structure–function relationship of TBEVC, we solved its 3D structure using high-resolution multidimensional NMR spectroscopy in combination with computational methods. The resonances were assigned to protein atoms and deposited into the Biological Magnetic Resonance Data Bank (BMRB) with BMRB ID: 34709. Structure calculation was based on intramolecular distance 1H–1H constraints obtained from NOE experiments and backbone dihedral angle (φ, ψ) restrictions estimated by TALOS+ (24) (Table S1). The 20 best-calculated structures of Δ16-TBEVC [23] out of 1000 were chosen and superimposed over the backbone heavy atoms of helices (Fig. 1A). The average RMSD relative to the mean heavy atom positions of the best-defined parts (all four α-helices) is 1.3 ± 0.3 Å.
Figure 1.
Structure of monomeric Δ16-TBEVC.A, superposition over the backbone heavy atoms of helices α2–α4 of the 20 monomeric Δ16-TBEVC best structures represented as lines of the main chain. B, the average structure of Δ16-TBEVC is represented as a cartoon. 1—side view, 2—top view, and 3—front view. The helices are marked α1–α4. Generated with the PyMOL program (65). TBEVC, capsid protein of tick-borne encephalitis virus.
The monomeric Δ16-TBEVC consists of four α-helices connected by loops (Fig. 1B). The individual parts of the structure were denoted as: α-helix 1 (α1), post-α1 loop, α-helix 2 (α2), post-α2 loop, α-helix 3 (α3), post-α3 loop, and α-helix 4 (α4). Such structural motives arrangement is conserved among the capsid proteins across the whole family of flaviviruses (6, 21, 25) and can be divided into three “layers”: the top one formed by short α1 and long post-α1 loop, the middle one formed by α2, and the bottom one formed by long α4 (21). The short α3 flanked by post-α2 and post-α3 loops connects the middle and bottom “layers.”
Comparison of the NMR structure of Δ16-TBEVC with previously solved ones of other flaviviral C proteins, such as Δ20-DENVC (6), Δ25-ZIKVC (21), and Δ23-WNVC (25), revealed not only structural similarities but also some minor differences. The main difference is that TBEVC N-terminal unstructured part is shorter (by 7–17 amino acids). Thus, while within Δ20-DENVC, Δ25-ZIKVC, and Δ23-WNVC, helices 2, 3, and 4 start almost at the same position, all helices in Δ16-TBEVC are sequentially shifted by more than six amino acids toward the N terminus (Table 1). Even so, the length of individual helices across structures is very similar. See a schematic comparison in the supporting information (Fig. S1A).
Table 1.
While a full-length alignment of Δ20-DENVC, Δ25-ZIKVC, and Δ23-WNVC showed 24% similarity, with 29 conserved and 19 homologous residues (Fig. S1B), the alignment to the Δ16-TBEVC sequence revealed only 2% similarity, matching at two conserved residues (W62 and A70) and 18 homologous residues (Fig. S1C).
Structure of homodimeric Δ16-TBEVC
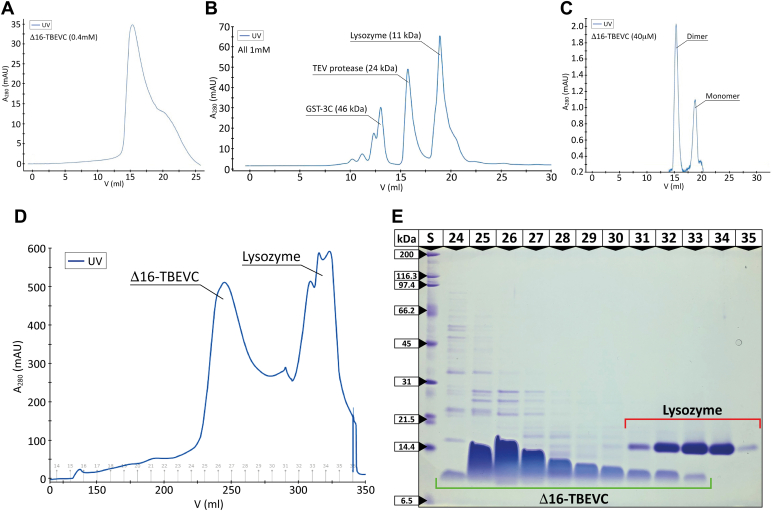
Although we determined the NMR structure of the monomeric Δ16-TBEVC protein, the structures of other flaviviral capsid proteins suggested that these proteins occur naturally as homodimers (6, 21, 25). To investigate whether Δ16-TBEVC protein forms dimerize, size-exclusion chromatography (SEC) and chemical crosslinking coupled with mass spectrometric detection (XL–MS) were employed. SEC, performed under the same conditions as the NMR experiments, demonstrated the approximate molecular weight of Δ16 TBEVC of about 24 kDa, corresponding to the dimeric form (Fig. 2. A and B). The only conditions under which we observed signal for the monomeric form was in a 10-times diluted TBEVC in the presence of 1% SDS (Fig. 2C). Thus, we concluded that under native conditions, the equilibrium between monomer and dimer was shifted toward the dimeric form. This was also consistent with the results of SEC analysis during the purification process, where Δ16-TBEVC (of molecular weight 9 kDa) eluted earlier than lysozyme, a protein of molecular weight 11 kDa (Fig. 2, D and E). This indicated that the formation of Δ16-TBEVC dimer or even its higher oligomers occurs during the protein production in bacterial cells.
Figure 2.
Analytical size-exclusion chromatography of Δ16-TBEVC.A, SEC chromatogram of 0.4 mM Δ16-TBEVC performed at pH 6.0. Equilibrium between the monomer and dimer is shifted toward the dimeric form, and no monomer is detectable. B, SEC chromatogram of calibration using GST-3C (1 mM, 46 kDa), TEV protease (1 mM, 24 kDa), and lysozyme (1 mM, 11 kDa). C, SEC chromatogram of 10-times diluted Δ16-TBEVC (final concentration of 40 μM) with 1% SDS performed at pH 6.0. Separated peaks correspond to the monomeric and dimeric forms of Δ16-TBEVC. D, SEC chromatogram of protein purification showing separation of Δ16-TBEVC and lysozyme (used to lyse bacterial cell wall). The elution volume of Δ16-TBEVC is smaller than for lysozyme, indicating that the formation of Δ16-TBEVC dimer or even its higher oligomers takes place shortly after the protein expression. E, electropherogram of Tris–tricine SDS-PAGE showing SEC purification step of Δ16-TBEVC. About 10% acrylamide gel stained by Coomassie Brilliant Blue R-250. (S) SDS-PAGE standards, broad range (Bio-Rad) (6, 21, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32); numbers of fractions from SEC (Fig. 2D). Red line marks protein zones of lysozyme, and green line marks protein zones of Δ16-TBEVC. GST, glutathione-S-transferase; SEC, size-exclusion chromatography; TEV, tobacco etch virus.
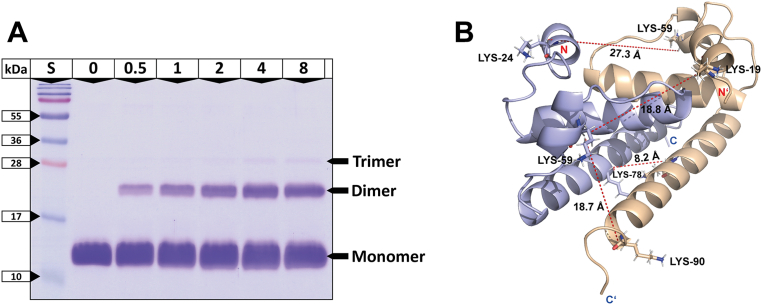
Next, we performed XL–MS using the amine-reactive homobifunctional and MS-cleavable crosslinker C2 capable to generate covalent bonds mainly between lysine residues in close proximity of up to 25 Å (26, 27). Five different protein-to-crosslinker molar ratios (1:0.5, 1:1, 1:2, 1:4, and 1:8) were applied, and crosslinking products were visualized by SDS-PAGE. Figure 3A shows the presence of both monomeric and dimeric forms, also with the indication of trimers with increasing C2 concentration.
Figure 3.
The Δ16-TBEVC naturally forms homodimers.A, electropherogram of Tris–glycine SDS-PAGE showing crosslinking products of Δ16-TBEVC using the amine-reactive homobifunctional and MS-cleavable crosslinker C2 at different protein-to-crosslinker molar ratios. About 15% gel stained by Coomassie Brilliant Blue R-250. (S) PageRuler Prestained Protein Ladder (Thermo Fisher Scientific); (0–8) Specific protein-to-crosslinker molar ratios. Black arrows point to the protein zones corresponding to the monomeric, dimeric, and trimeric states. B, predicted structure of the homodimeric Δ16-TBEVC using I-TASSER server (28, 29, 30) and ZIKVC (21) (Protein Data Bank ID: 5YGH) as a template (5). The measured distances, highlighted in red, correspond to the identified Cα−Cα distances within the dimer (Table 2). Generated with the PyMOL program (65). MS, mass spectrometry.
For further LC–MS/MS analysis, the protein-to-crosslinker molar ratios of 1:0.5, 1:1, and 1:2 were selected to target mainly the covalently bound Δ16-TBEVC dimers. The XL–MS resulted in the identification of four unique lysine–lysine linkages (Table 2) observed at all three ratios (Table S2 and Figs. S2–S5). Evaluation and visualization (Fig. 3B) of these linkages were done on a model predicted by I-TASSER server (5, 28, 29, 30). Based on the measured distances and spatial comparison, we evaluated all four linkages as intermonomeric ones. Their intramonomeric character was excluded because of the following findings (Table 2): K59–K19 and K59–K24 are not possible because of steric hindrance, K59–K90 exceeds the Cα–Cα linkable distance (26, 27) and K78–K78 cannot be observed in a Δ16-TBEVC monomer. Thus, similarly to other flaviviral capsid proteins (6, 21, 25), SEC and XL–MS experiments unambiguously confirm that Δ16-TBEVC naturally forms a homodimer.
Table 2.
Linkages identified by XL–MS and its distances within the structure
| Molar ratio reagent:protein | 1:0.5 | 1:1 | 1:2 | Cα–Cα distance within the monomer (Å) | Cα–Cα distance within the dimer (Å) |
|---|---|---|---|---|---|
| Identified linkages | K59–K19 | K59–K19 | K59–K19 | 21.1a | 18.8 |
| K78–K78 | K78–K78 | K78–K78 | — | 8.2 | |
| K59–K24 | K59–K24 | 15.9a | 27.3 | ||
| K59–K90 | 37.7 | 18.7 |
Steric hindrance.
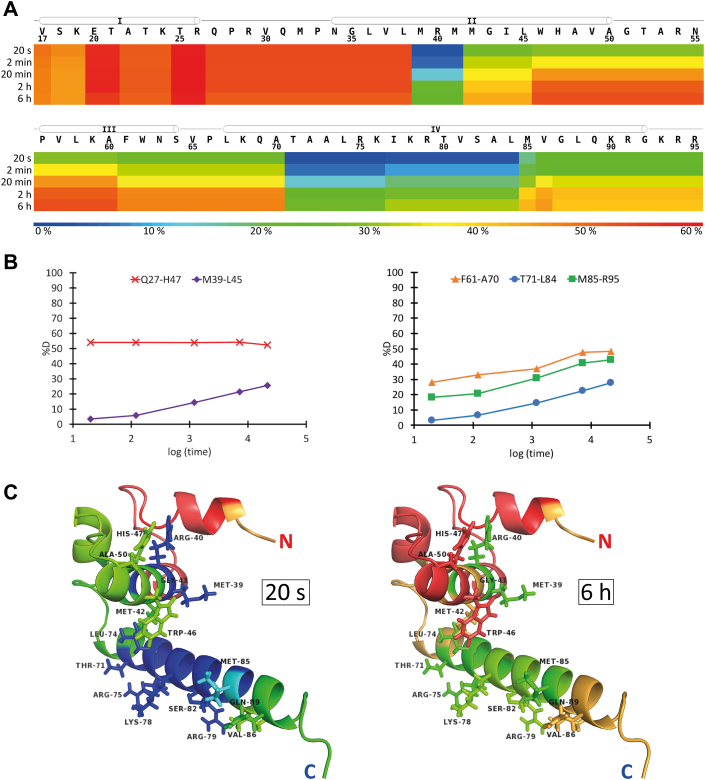
To calculate the structure of the Δ16-TBEVC dimer, we first identified the residues involved in the dimeric interface. As NMR spectroscopy provided only a few weak signals, which could not be conclusively taken as intermonomeric restraints, we performed the hydrogen–deuterium exchange experiment (HDX–MS). It was performed in three repetitions, including protein labeling at five different time points: 20 s, 2 min, 20 min, 2 h, and 6 h (Table S3 and Fig. 4A). To visualize the HDX evolution of 16 peptides, which have survived the filtering process, the web-based application MStools (31) was applied using the values of relative fractional uptake (RFU) obtained from DynamX software.
Figure 4.
Dimeric interfaces of Δ16-TBEVC homodimers.A, heat map showing the relative deuterium uptake for all Δ16-TBEVC residues using a rainbow color gradient in each labeling time point. Each line represents one time point (from top to bottom—20 s; 2 min; 20 min; 2 h; 6 h). The observed helices (NMR) are marked as cylinders above the protein sequence. Generated by MStools (31) web-based application. B, deuterium uptake plots show the time course of deuteration for selected peptides (Table S3). The plot on the left covers the post-α1 loop and 4/5 of α2. The plot on the right partially covers α3 and the entire α4. C, depicted deuteration states on the predicted monomer structure of Δ16-TBEVC. The left model depicts the deuteration state for the first time point (2 s). The right model depicts the deuteration state for the last time point (6 h). Interacting residues that have been selected for docking by Haddock (32) are represented as sticks. Generated with the PyMOL program (65).
The HDX evolution for selected peptides Q27–H47, M38–L45, F61–A70, T71–L84, and M85–R95 is shown on the deuteration uptake plots (Fig. 4B), and the deuteration profiles of full-length Δ16-TBEVC obtained at 20 s and 6 h are presented on the predicted structure of the Δ16-TBEVC monomer (Fig. 4C). The results showed that HDX was the slowest within α2 and α4, especially in their middle regions. It means that these regions were spatially poorly accessible, indicating the compactness of this part of the structural motif as an effect of the dimer formation. Thus, this experiment not only confirmed the protein dimerization but also provided information about the regions highly involved in the interaction of Δ16-TBEVC monomers within the dimer.
We selected the following interacting residues for docking by Haddock (32) based on these findings: M39, R40, M42, G43, W46, H47, A50, T71, L74, R75, K78, R79, S82, M85, V86, and Q89 (Fig. 4C). The docking was performed for all the best previously calculated 20 Δ16-TBEVC monomer structures, and the final dimeric structures were superimposed over the backbone heavy atoms of α2–α4 (Fig. 5A). The average RMSD relative to the mean heavy atom positions was 1.7 ± 0.4 Å. Finally, the resulting dimers were deposited into the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) with PDB ID: 7YWQ. The average structure of the Δ16-TBEVC homodimer (Fig. 5B) is a tightly packed and centrally symmetric molecule in which the contact surfaces of the monomers are formed predominantly by antiparallel α2/α2′ and α4/α4′ and probably also by the participation of α1/post-α1′ loop and α1’/post-α1 loop. The experimental homodimer structure of Δ16-TBEVC matches the predicted structure well, sharing the same topology with already solved structures of Δ20-DENVC (6), Δ25-ZIKVC (21), and Δ23-WNVC (25).
Figure 5.
A 3D structure of Δ16-TBEVC homodimer.A, superposition over the backbone heavy atoms of helices α2–α4 of the 20 dimeric Δ16-TBEVC dimeric structures represented as lines of the main chain. B, the average structure of the Δ16-TBEVC homodimer represented as a cartoon. 1—side view, 2—top view, and 3—front view. The helices are marked as α1–α4. Generated with the PyMOL program (65).
TBEVC protein localizes in host nucleolus
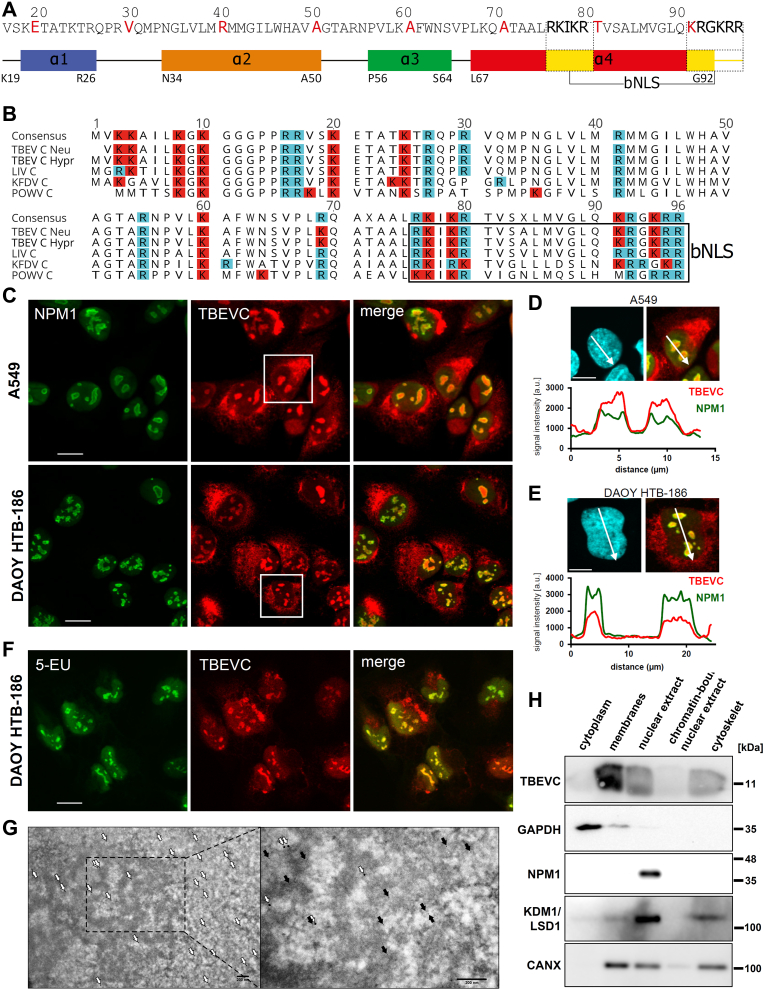
Close inspection of the helix 4 of TBEVC revealed a putative bipartite nuclear localization sequence (bNLS) 75-RKIKR-79 and 90-KRGKRR-95 (Fig. 6A). Furthermore, alignment of the amino acid sequence of capsid proteins of other TBFVs, Kyasanur forest disease virus (KFDV), Louping ill virus (LIV), and Powassan virus (POWV), to TBEVC proteins revealed similar arginine- and lysine-rich bNLS motifs at the C terminus (Fig. 6B).
Figure 6.
TBEVC contains bNLS and localizes to the nucleoli of infected cells.A, identification of putative bNLS in helix 4 of TBEVC. B, alignment (Clustal Omega) of TBFVC protein sequences with the marked C-terminal bNLSs using Geneious Prime, version 2022.0.1. Arginine (R) and lysine (K) residues are highlighted in blue and red, respectively. C, A549 and DAOY cells were infected with TBEV strain Hypr (MOI 5) and the colocalization of TBEVC protein (red) and nucleolar marker NPM1 (green) was performed at 36 h p.i. The scale bars represent 20 μm. Representative images from two independent biological replicates are shown. White squares mark areas selected for nuclear intersection measurements in D and E. D, signal quantification of nuclear intersection (white arrow) in A549 cells was performed using ImageJ. The scale bar represents 10 μm. E, signal quantification of nuclear intersection (white arrow) in DAYO HTB-186 cells was performed using ImageJ. The scale bar represents 10 μm. F, in situ metabolic labeling of nascent RNA in TBEV-infected DAOY cells (5 MOI, 36 h p.i.) using 1 mM 5-EU and its subsequent detection via Alexa Fluor 488-azide and Click reaction (green). Colocalization of TBEVC was performed using anti-TBEVC antibodies (red). The scale bars represent 20 μm. Representative images from two independent biological replicates are shown. G, transmission electron microscopy of TBEV Hypr-infected DAOY cells (5 MOI; 36 h p.i.). Immunogold colocalization of TBEVC (5 nm, black arrows) and nucleolar transcription factor MYBBP1A (15 nm, white arrows) in the nucleus of infected cell. H, subcellular fractionation of TBEV Hypr-infected DAOY cells (5 MOI) was performed at 36 h p.i. Subsequent immunodetection of TBEVC, GAPDH (cytoplasmic marker), NPM1 (nuclear marker), KDM1/LSD1 (nuclear marker), and CANX (ER/nuclear membranes marker) was performed in all five fractions. Representative results from three independent experiments are shown. bNLS, bipartite nuclear localization sequence; ER, endoplasmic reticulum; 5-EU, 5-ethynyluridine; MOI, multiplicity of infection; TBEVC, capsid protein of tick-borne encephalitis virus; TBFV, tick-borne flavivirus.
As nuclear localization was also reported for several mosquito-borne flaviviral C proteins (10, 11, 12, 13, 14), we characterized the TBEVC distribution in infected DAOY and A549 cells. Interestingly, a substantial part of TBEVC colocalized with nascent rRNA in the host nucleolus, when NPM1 (nucleolar marker) and metabolically labeled RNA were used as colocalization determinants (Fig. 6, C–F). Additional experiments further explored TBEVC colocalization with dsRNA, TBEV E, and NS3 proteins in the ER (Fig. S6A). Nuclear localization of TBEVC was present also in infected human primary neural cells, astrocytes, and neurons (Fig. S6B).
Immunogold detection of TBEVC in infected DAOY cells using transmission electron microscopy (TEM) further confirmed its nuclear localization with scarce signal also detected in mitochondria (Figs. 6G and S6C). In order to verify the phenomenon of nuclear/nucleolar localization of TBEVC, we performed subcellular fractionation of infected DAOY HTB-186 cells and subsequent detection of TBEVC in each of the fractions. As expected, the majority of TBEVC was localized to the membraneous fraction representing ER/Golgi membranes where flaviviral replication and maturation processes occur. Considerably high signal for TBEVC was, however, detected in nuclear fraction as well (Fig. 6H).
Next, we wanted to assess whether nucleolar localization of the capsid protein is a common feature among TBFVs. For this purpose, we prepared expression vectors with wildtype variants of KFDV, LIV, POWV, and TBEV C proteins with a C-terminal FLAG tag (Fig. S7A). Subsequent colocalization of NPM1 and overexpressed C proteins in transfected DAOY cells documented nucleolar localization for all tested C proteins, although with different frequencies (Fig. S7B). In the case of LIVC, nucleolar/nuclear localization was documented in 84.2% of C protein–positive cells. Lower frequency was observed in the case of POWVC and TBEVC where only 54.3% and 61.8% of C protein–positive cells exhibited nucleolar/nuclear localization, respectively. Interestingly, nucleolar localization of KFDVC was observed only in 11.7% of cells expressing C protein (Fig. S7C). Intriguingly, the nuclear/nucleolar localization of TBFVC proteins was observed despite the absence of any other viral proteins or gRNA, which suggests that the host machinery only is sufficient to mediate the nuclear transport of TBFVC proteins.
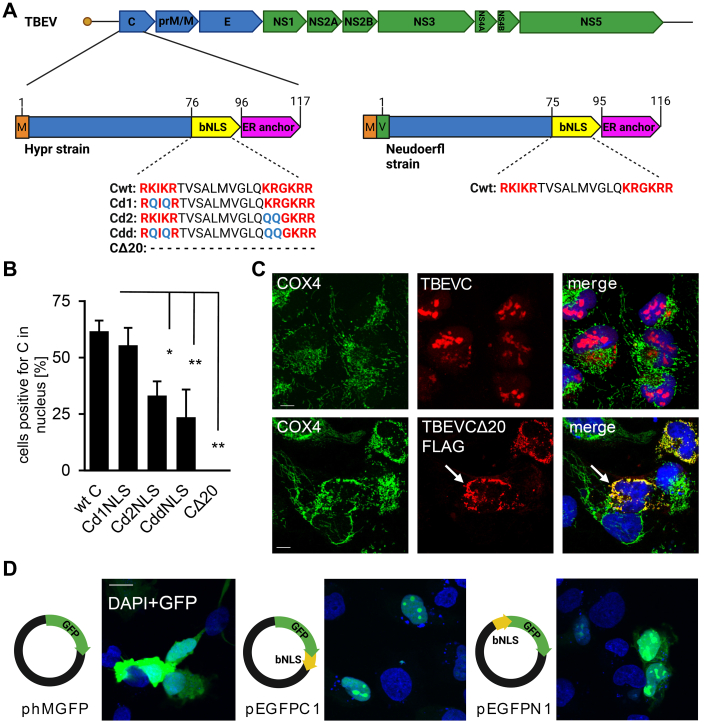
In order to verify the involvement of putative bNLS, we prepared a series of mutants carrying mutations or truncations within bNLS (d1, d2, dd, Δ20; Figs. 7A and S7A). These mutated constructs were transfected into DAOY cells, and the subcellular distribution of TBEVC proteins was assessed at 24 h post-transfection. Data in Figure 7B demonstrate that the C-terminal part of bNLS (90-KRGKRR-95) plays a crucial role in the nuclear localization of TBEVC since a significantly decreased rate of cells positive for TBEVC in the nucleus was observed for TBEVC-d2 and TBEVC-dd mutants (one-way ANOVA with post hoc Tukey's honestly significant difference test; α < 0.05). Interestingly, deletion of the last 20 amino acids (CΔ20 mutant) resulted in almost exclusive localization in mitochondria supporting the key role of identified bNLS in nuclear transport of TBEVC (Fig. 7C). To further demonstrate the role of newly identified TBEVC bNLS in nuclear transport, we constructed expression vectors containing GFP gene fused with TBEVC bNLS sequence either at the N terminus (pEGFPN1) or at the C terminus (pEGFPC1). Both variants of fused protein, bNLS-GFP and GFP-bNLS, accumulated in nuclei and nucleoli of transfected DAOY (Figs. 7D and S8A) and A549 (Fig. S8B) cells.
Figure 7.
Importance of bNLS for the TBEVC localization.A, schematic overview of TBEV gRNA with emphasis to predicted TBEVC bNLS and derived mutants TBEVC-d1NLS, TBEVC-d2NLS, TBEVC-ddNLS, and TBEVC-Δ20NLS. Differences in the C protein of TBEV strains, Hypr and Neudoerfl, used in this study, are depicted. Created with BioRender.com. B, DAOY cells were transfected with the corresponding TBEVC-FLAG plasmid, and the colocalization of TBEVC protein and nucleolar marker NPM1 was performed at 24 h p.t. The percentage of cells positive for TBEVC signal in the nucleus was determined as the ratio of nucleus-TBEVC-positive cells and the total number of TBEVC-positive cells (nwt = 83; nd1 = 162; nd2 = 145; ndd = 193; and nΔ20 = 79). Data are representative of three independent experiments, and values are expressed as mean ± SD. Significant difference from the control was calculated using one-way ANOVA with post hoc Tukey's honestly significant difference test (∗p < 0.05, ∗∗p < 0.01). C, DAOY cells were either infected (TBEV-Hypr strain; 5 MOI) or transfected with the TBEV-CΔ20-FLAG plasmid, and the colocalization of TBEVC protein (red) and mitochondrial marker COX4 (green) was performed at 36 h p.i. or 24 h p.t.; the scale bar represents 20 μm, and cell nuclei were costained by DAPI (nucleus). Representative images from two independent biological replicates are shown. D, TBEVC bNLS was cloned at the C terminus (pEGFPC1; middle) or N terminus (pEGFPN1; right) of GFP (green) and localization in DAOY HTB-186 cells transfected with 0.1 μg of an appropriate plasmid was examined 24 h post-transfection on a coverslip and compared with wt GFP expression (phMGFP, Promega; left). bNLS induced strong GFP nuclear localization (nucleus marker DAPI, blue) in comparison with the untagged GFP (phMGFP). Representative images from three independent biological replicates are shown; the scale bar represents 20 μm. Magnified image sections with merged channels are shown; for full-size images and individual channel views, see Fig. S8A. bNLS, bipartite nuclear localization sequence; DAPI, 4′,6-diamidino-2-phenylindole; gRNA, genomic RNA; MOI, multiplicity of infection; TBEVC, capsid protein of tick-borne encephalitis virus.
Structure of the TBEVC bNLS bound to the nuclear import adapter protein importin-alpha
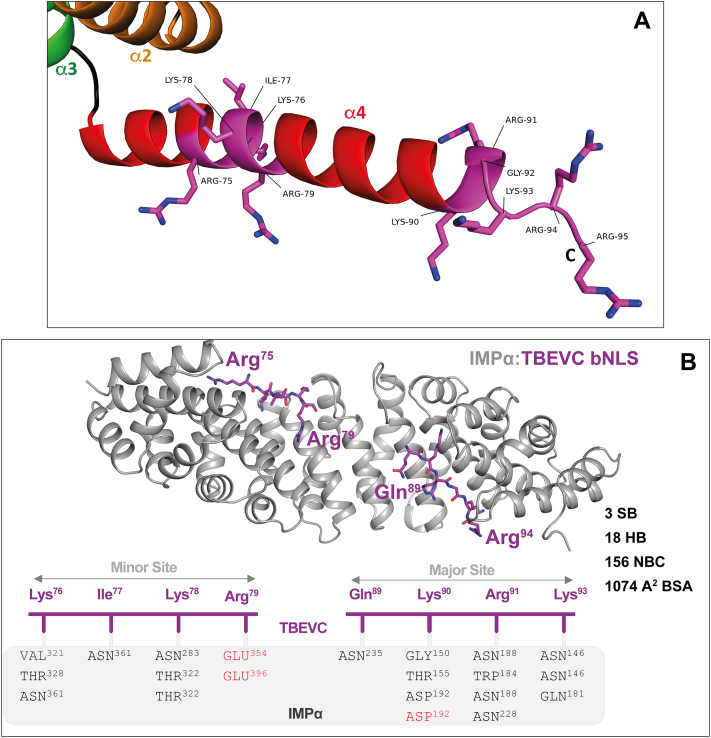
We employed X-ray crystallography of a complex between TBEVC bNLS (Fig. 8A) and the classical import adapter protein importin-alpha (IMPα) to map their putative interactions (Fig. 8B). Crystals of the TBEVC–IMPα diffracted to 2.05 Å and indexed in P212121. The final structure produced an R/Rfree value of 19.3% and 21.9%, respectively (Table S4 for data collection and refinement statistics). IMPα formed 10 repeating armadillo repeat motifs, with each motif comprised of three α-helices. We found that the bNLS of TBEVC bound at both the major and minor sites of IMPα, confirming that the NLS is bipartite (Fig. 8B). The interface was mediated by an extensive array of interactions involving both side chains and main chains of TBEVC and IMPα (Table S5). At the major site, we identified that TBEVC Gln89 bound at the P1 position of IMPα. At the P2 site, the TBEVC Lys90 side chain formed both a salt bridge with IMPα Asp192 and hydrogen bonded with Gly150 and Thr155. TBEVC Arg91 formed a hydrogen bond interaction with IMPα Trp184, Asn188, and Asn228 at the P3 site. TBEVC Lys93 formed H bond interactions with IMPα Asn146 and Gln181 at the P4 site. At the minor site, TBEVC Lys76 formed hydrogen bonds with IMPα Val321, Thr328, and Asn361. TBEVC Ile77 formed a hydrogen bond interaction with IMPα Asn361. TBEVC Lys78 formed hydrogen bond interactions with IMPα Asn283 and Thr322. Finally, TBEVC Arg79 formed a salt bridge interaction with IMPα Glu354 and Glu396. Overall, the interaction was mediated through three salt bridge interactions, 18 hydrogen bond interactions, 156 non–hydrogen-bonded contacts, and exhibited a buried surface area of 1074 Å2. The resulting TBEVC bNLS:IMPα structure was deposited to RCSB PDB with PDB ID: 8ECH.
Figure 8.
Structural analysis of bNLS interaction with IMPα.A, detail of bNLS within helix 4 of NMR Δ16-TBEVC structure. Generated with the PyMOL program (65). B, 2.05 Å resolution crystal structure of IMPα bound to the TBEVC bNLS peptide. IMPα is in light gray and ribbon mode, and the discontinuous TBEVC bNLS peptide (middle residues were not resolved) is in purple in stick mode. PyMOL software was used to generate the structure (65). Below is a simplified representation highlighting the hydrogen bonds and salt bridges (red). bNLS, bipartite nuclear localization sequence; BSA, buried surface area; HB, number of hydrogen bond; IMPα, importin-alpha; NBC, number of nonhydrogen bond; SB, number of salt bridges; TBEVC, capsid protein of tick-borne encephalitis virus.
TBEVC protein interactome
To gain a deeper insight into the biological relevance of TBEVC nuclear localization, we analyzed its host–cell interaction network using coimmunoprecipitation (co-IP) with subsequent MS.
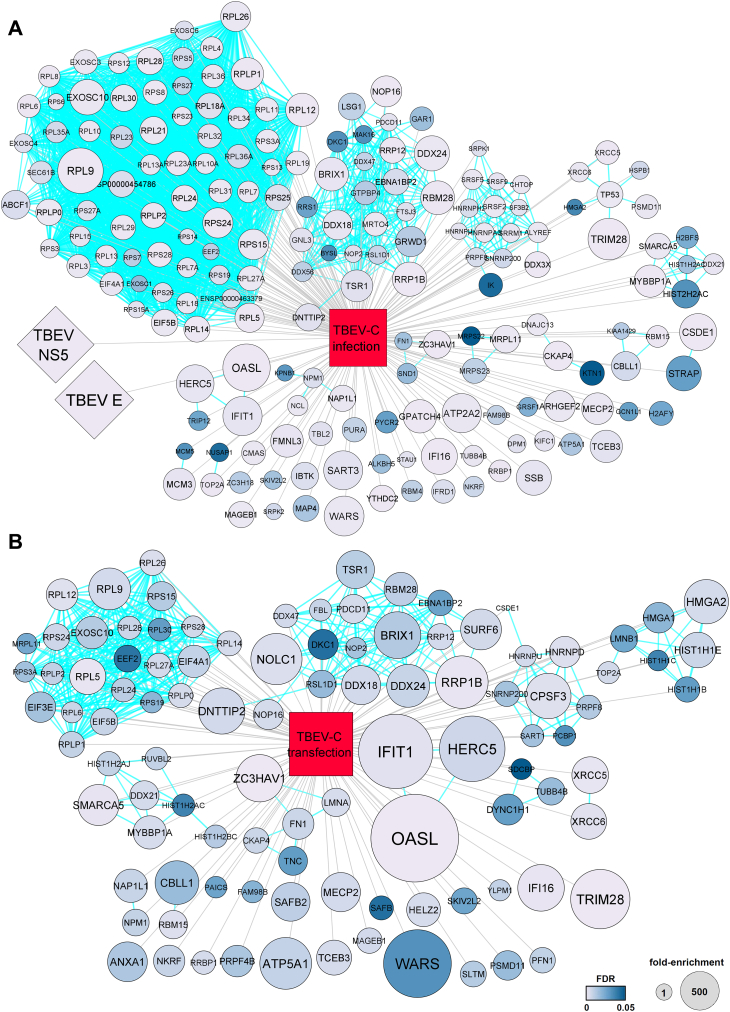
For this purpose, DAOY cells were either infected by TBEV or transfected by pCMV-C-FLAG, and total protein lysates were harvested at 36 h postinfection and 24 h post-transfection. The co-IP was performed using anti-TBEVC antibodies (Fig. S9A) with LC–MS/MS identification of prey proteins in elution fraction. In total, 186 and 103 proteins were identified as interacting with TBEVC in infected (Table S6) and transfected cells (Table S7), respectively (false discovery rate [FDR] <0.05; log2 fold enrichment >1). Of 186 identified proteins in TBEV-infected cells, 113 (60.8%) were unique to the infection process only (including viral proteins E and NS5) (Figs. 9 and S9B). For illustration, Table 3 includes the list of top-ranked proteins for both variants with detailed information.
Figure 9.
Whole-cell TBEVC interactome identified via TBEVC-specific coimmunoprecipitation. DAOY cells were either (A) infected with TBEV strain Hypr (5 MOI) or (B) transfected by TBEV-Cwt-FLAG plasmid and lysed at 36 h p.i. and 24 h p.t., respectively. Lysates were used for TBEVC-specific coimmunoprecipitation using anti-TBEVC antibodies and Dynabeads coupled with protein A. Proteins interacting with TBEVC were subsequently identified using LC–MS/MS. The interaction network (gray lines) combined with STRING database (light blue lines) and statistical evaluation was generated in CytoScape software, version 3.8.2. MOI, multiplicity of infection; TBEVC, capsid protein of tick-borne encephalitis virus.
Table 3.
List of top 10 proteins interacting with TBEVC identified via LC–MS/MS
| Protein | UniProt code | Name | Fold change | FDR | |
|---|---|---|---|---|---|
| TBEV infection | E | Q01299 | Envelope protein (TBEV) | 475.6 | 0.0001 |
| NS5 | Q01299 | NS5 protein (TBEV) | 399.8 | 0.0001 | |
| OASL | Q15646 | 2′-5′-Oligoadenylate synthetase like | 31.3 | 0.0001 | |
| RPL9 | P32969 | 60S Ribosomal protein L9 | 26.5 | 0.0002 | |
| TRIM28 | Q13263 | Tripartite motif containing 28 | 18.9 | 0.0001 | |
| WARS | P23381 | Tryptophanyl-tRNA synthetase 1 | 16.3 | 0.0005 | |
| IFIT1 | P09914 | Interferon-induced protein with tetratricopeptide repeats 1 | 15.3 | 0.0012 | |
| ATP2A2 | P16615 | Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 | 15.3 | 0.0006 | |
| SART3 | Q15020 | Squamous cell carcinoma antigen recognized by T-cells 3 | 15.1 | 0.0020 | |
| HERC5 | Q9UII4 | HECT And RLD domain containing E3 ubiquitin protein ligase 5 | 13.5 | 0.0010 | |
| TBEVC transfection | OASL | Q15646 | 2′-5′-Oligoadenylate synthetase like | 66.2 | 0.0002 |
| IFIT1 | P09914 | Interferon-induced protein with tetratricopeptide repeats 1 | 33.0 | 0.0025 | |
| WARS | P23381 | Tryptophanyl-tRNA synthetase 1 | 23.5 | 0.0318 | |
| HERC5 | Q9UII4 | HECT And RLD domain containing E3 ubiquitin protein ligase 5 | 20.8 | 0.0064 | |
| TRIM28 | Q13263 | Tripartite motif containing 28 | 15.1 | 0.0002 | |
| RRP1B | Q14684 | Ribosomal RNA processing protein 1 homolog B | 10.6 | 0.0011 | |
| ATP5A1 | P25705 | ATP synthase subunit alpha, mitochondrial | 10.2 | 0.0085 | |
| NOLC1 | Q14978 | Nucleolar and coiled-body phosphoprotein 1 | 9.9 | 0.0039 | |
| ZC3HAV1 | Q7Z2W4 | Zinc finger CCCH-type antiviral protein 1 | 8.0 | 0.0003 | |
| BRIX1 | Q8TDN6 | Ribosome biogenesis protein BRX1 homolog | 7.8 | 0.0119 |
Based on the functional clustering of TBEVC interactome according to STRING, several clusters were identified, including ribosomal and nucleolar proteins involved in (i) ribosome biogenesis, (ii) rRNA synthesis and modification, (iii) RNA splicing, and (iv) mRNA binding. Interestingly, TBEVC-interacting proteins with the highest score included several immune response–related proteins, such as OASL, IFIT1, HERC5, IFI16, and TRIM28 (Fig. 9). Subsequent Gene Ontology enrichment analysis of TBEVC interactome using DAVID tool (33) confirmed the data from functional clustering since processes such as rRNA processing, ribosome biogenesis, and nonsense-mediated mRNA decay were identified as the most significantly affected ones (Fig. S9, C and D). Reverse co-IP using anti-OASL, anti-CSDE1, and anti-MYBBP1 antibodies confirmed the interaction with TBEVC protein (Fig. S9A).
TBEVC reduces levels of host nascent proteins and 18S rRNA
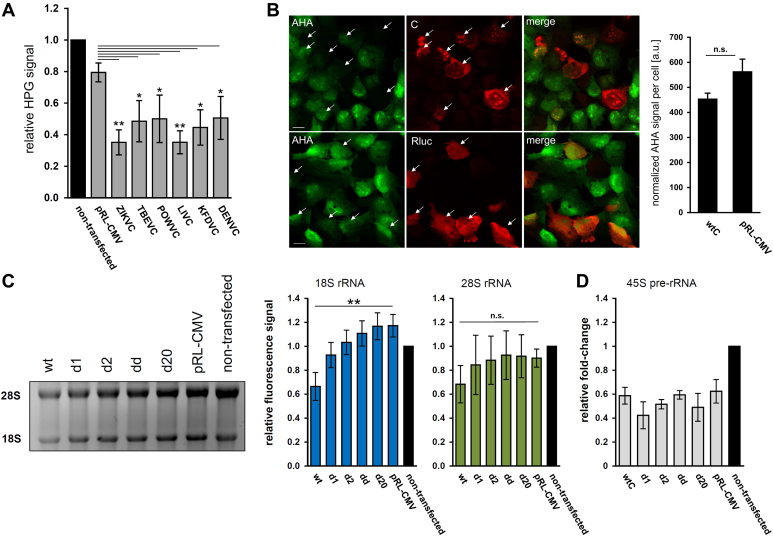
Our previous study described TBEV-induced decrease of nascent rRNA and protein production in host cells (23). Based on the nucleolar localization of TBEVC and its interacting network, we hypothesize that TBEVC may substantially contribute to the observed phenomenon of host transcriptional and translational shutoff. In order to test this, de novo protein synthesis was determined in DAOY cells transiently overexpressing DENV, KFDV, LIV, POWV, TBEV, and ZIKV C proteins using metabolic labeling with methionine analogs homopropargylglycine or l-azidohomoalanine (AHA). Surprisingly, the transfection process itself decreased the nascent protein synthesis compared with nontransfected control (Fig. 10A). Nevertheless, transient overexpression of every C protein tested resulted in a significant decrease of incorporated homopropargylglycine levels when compared with pRL-CMV control (one-way ANOVA with post hoc Holm–Sidak test; α = 0.05; Fig. 10A). Metabolic protein labeling in situ also revealed a decrease in incorporated AHA levels in TBEVC-expressing DAOY cells; however, the difference was not statistically significant (Fig. 10B). Analogous experiments were performed to reveal the TBEVC effect on rRNA production. Introduction of constructs encoding either wildtype or bNLS mutants of TBEVC (pCMV-C-wt/d1/d2//Δ20/dd-FLAG) induced a significant decrease in mature 18S rRNA levels for wildtype TBEVC only (Fig. 10C). A similar pattern was also observed in the case of mature 28S rRNA, however, without statistical significance. Based on these results, we aimed to quantify the precursor for both rRNAs, the 45S pre-rRNA, in transfected cells. However, no significant changes were documented for any of the analyzed construct pCMV-C-wt/d1/d2/dd-FLAG (Fig. 10D). These data indicate that nucleus-localized TBEVC interferes with the rRNA synthesis or processing in infected host cells but might require co-operation with other TBEV proteins.
Figure 10.
TBEVC decreases the levels of host nascent proteins and mature 18S rRNA.A, DAOY cells were transfected with pCMV-DENV/KFDV/LIV/POWV/TBEV/ZIKV-C-FLAG constructs, and metabolic labeling of nascent proteins was performed using 50 μM HPG. As a negative control, backbone plasmid pRL-CMV was used as well as nontransfected and unlabeled cells. Data in the graph summarize three independent experiments with values expressed as mean ± SD. (∗p < 0.05, ∗∗p < 0.01; one-way ANOVA with post hoc Holm–Sidak test; α = 0.05). B, in situ metabolic labeling of nascent proteins (50 μM AHA; green) in DAOY cells transfected with pCMV-wtC-FLAG (upper panel) or pRL-CMV (lower panel) (both in red). Representative images with graphical data summary from two independent experiments are shown. Data in the graph are expressed as mean ± SD. ns, not significant (unpaired two-sample Student’s t test; α = 0.05). Only cells positive for either TBEVC or Rluc signal (white arrows) were included in the quantification analyses (ImageJ). The scale bar represents 20 μm. C, total RNA from DAOY cells transfected either with one of pCMV-wtC/Cd1/Cd2/Cdd/CΔ20 constructs or pRL-CMV (backbone plasmid) was isolated at 24 h p.i. Cell viability was determined before the RNA isolation using the alamarBlue reagent. Total RNA was separated on a 1.2% Mops-buffered agarose gel and visualized using GelRed; the input amount was normalized to the cell number. The intensity of 18S and 28S rRNA bands was quantified using ImageJ. Gel image (left) is a representative of three independent experiments, and graph values (right) are expressed as mean ± SD. Significant difference from the control was calculated using one-way ANOVA with post hoc Tukey's honestly significant difference test (∗∗p < 0.01). D, total RNA was further used for the quantitative RT–PCR analysis of 45S pre-rRNA levels (relative quantification using the Δ-Ct method with normalization to the cell number). Data summarize three independent experiments performed in three technical replicates, and values in graphs are expressed as mean ± SD. AHA, l-azidohomoalanine; DENV, dengue virus; KFDV, Kyasanur forest disease virus; LIV, Louping ill virus; POWV, Powassan virus; TBEV, tick-borne encephalitis virus; TBEVC, capsid protein of tick-borne encephalitis virus; ZIKV, Zika virus.
Discussion
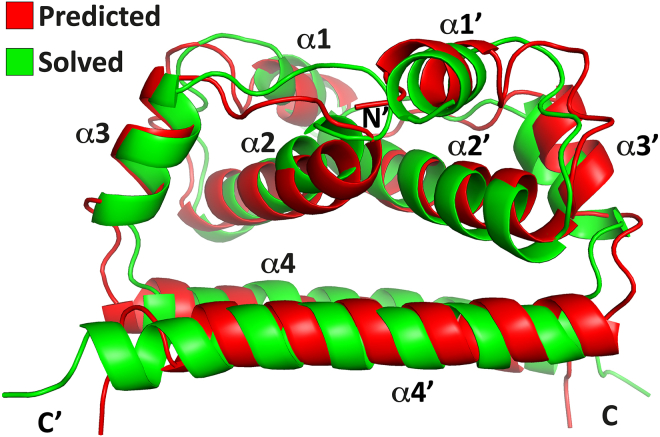
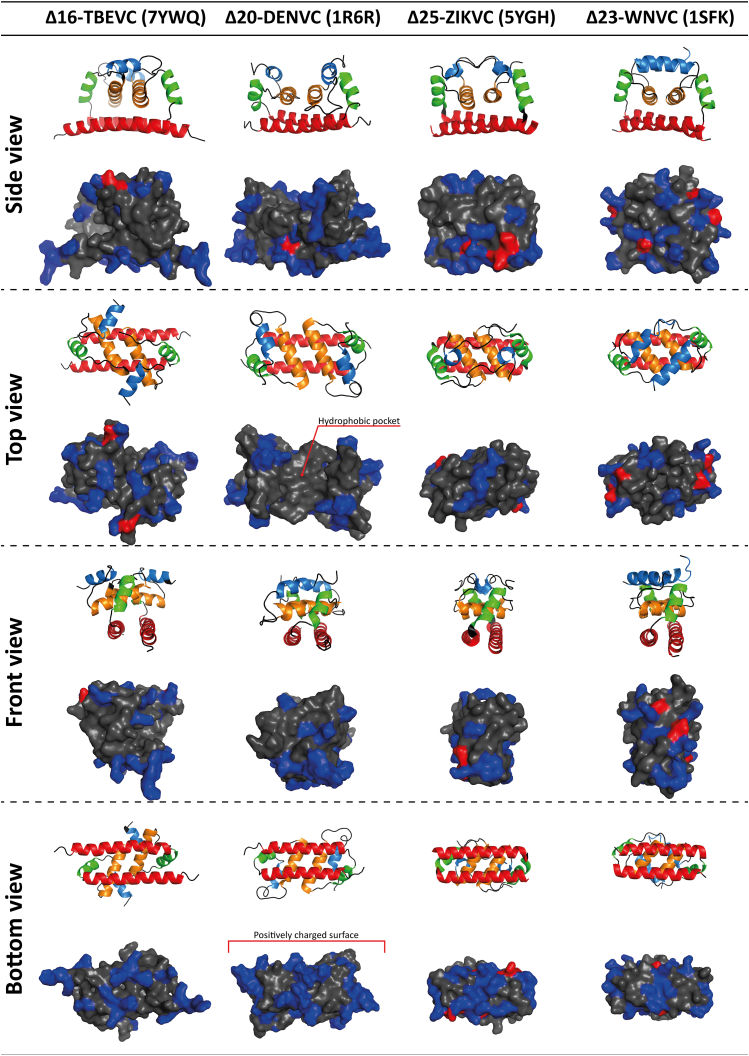
Here, we present the complete structure of Δ16-TBEVC solved by a combination of NMR spectroscopy, MS, and computational methods. The structure of monomeric Δ16-TBEVC consists of four α-helices forming the canonical structural motif, which is well preserved among the other flavivirus C proteins (6, 21, 25). The dimeric form of TBEV C and the dimer interface were identified by a combination of SEC, crosslinking, and HDX–MS analysis. Δ16-TBEVC homodimer is a tightly packed and centrally symmetric molecule with the interaction interface formed predominantly by antiparallel α2/α2′ and α4/α4′ and by the participation of α1/post-α1′ loop and α1’/post-α1 loop. The Δ16-TBEVC homodimer structure matches quite well the predicted structure (5) (Fig. 11). The resulting Δ16-TBEVC homodimer structure follows the same topology as the other solved structures of flaviviral C proteins (6, 21, 25) (Fig. 12). Flaviviral C proteins are typical for their charge distribution. This phenomenon was first described for Δ20-DENVC (6, 34) whose “bottom” side formed (Fig. S12, bottom view) by α4 is predominantly positively charged, whereas the “upper” side (Fig. S12, top view) formed by α1 and α2 forms a noticeable hydrophobic pocket, which is thought to be responsible for the interaction with the membrane (34). This charge distribution phenomenon is also observed in the remaining flavivirus C protein structures (21, 25), including Δ16-TBEVC. The situation is somewhat different on the “upper” side in the region of α1 and α2. In contrast to Δ20-DENVC, the remaining structures are more compact in this region, and this hydrophobic pocket is not present or is covered by a positive charge. Four phenylalanine residues involved in the α2/α2′ interface stabilization in DENVC, ZIKVC, and WNFVC are in the TBEVC replaced by two tryptophan and two histidine residues (Fig. S10, A and B).
Figure 11.
Comparison of predicted and solved structures of homodimeric Δ16-TBEVC.Red is the predicted structure of Δ16-TBEVC using I-TASSER server (28, 29, 30) and ZIKVC (21) (Protein Data Bank ID: 5YGH) as a template (5). Green is the solved structure using NMR spectroscopy and Haddock (32) server. Generated with the PyMOL program (65). TBEVC, capsid protein of tick-borne encephalitis virus; ZIKV, Zika virus.
Figure 12.
Comparison of flaviviral C protein structures. The first row of each view represents the protein structures in the cartoon representation. Blue—α1, orange—α2, green—α3, and red—α4. The second row of each view represents the protein structures with the shown surface. Positive residues are highlighted in blue, negative residues in red, and the remaining residues in gray. For Δ20-DENVC (Protein Data Bank ID: 1R6R), a hydrophobic pocket (top view), which is not as clearly defined in other structures, and a positively charged surface (bottom view), which is common to all structures, are indicated (34). Generated with the PyMOL program (65). DENV, dengue virus.
Protein C of flaviviruses is a promising target for antiviral drug design as was exemplified in the case of DENV (35, 36). A potent inhibitor ST148 fastens two capsid homodimers together, and the resulting C tetramers incorporated in the viral particles hinder effective uncoating of virions (36). Tailoring the inhibitor to fit the topology of the tick-borne C homodimers could yield promising drug candidates. Furthermore, the knowledge of the topology of positively charged residues on the bottom of the C homodimer might lay the cornerstone for the study of the specific interaction of TBEV C with the gRNA and the mechanism of viral particle formation, which is another target for antiviral drug development.
The primary function of flaviviral C protein is to recruit viral gRNA and form nucleocapsid. Although assembly of flaviviruses occurs in ER-derived vesicular compartments, capsid protein has also been detected on the surface of LDs (8, 9) and in nuclei or nucleoli of infected cells (10, 11, 12, 13, 14). However, all the information regarding capsid protein localization and novel functions is available only for mosquito-borne flaviviruses. Here, we report nucleolar localization of C protein of four TBFVs: POWV, LIV, KFDV, and TBEV. All analyzed TBFVC proteins were predicted to contain a C-terminal bNLS necessary for importin-mediated nuclear import. Mutational analyses of TBEVC bNLS confirmed its key role in the nucleolar localization in transfected cells. Furthermore, structural analyses (Fig. 8B) demonstrated that residues within the TBEV C bNLS form multiple specific interactions with the host IMPα nuclear import factor at the major and minor cargo-binding sites. In particular, TBEVC Lys90 forms strong interactions (three hydrogen bonds and one salt bridge) with IMPα at the canonical P2 site. Interactions between importins and NLS cargo at this P2 site have been shown to be the key determinants of binding (37), thus supporting the results of our TBEVC-d2 and TBEVC-dd mutants that confirm the importance of Lys90 for C protein nuclear import.
Although our structure of TBEV C demonstrates that bipartite NLS residues are located within a structured motif (alpha-helix 4; Fig. 8A), the complex of TBEV C NLS peptide with IMPα demonstrates this region is unstructured during this interaction with its nuclear import receptor (Fig. 8B). It is well established that helical structures may unwind when interacting with IMPα cargo-binding sites; a phenomenon exemplified by the importin-beta binding domain at the N terminus of IMPα. Crystal structures of the importin-beta binding domain bound to importin-beta (i.e., PDB ID: 1QGK) demonstrate that this region exists as an alpha helix (38). However, when this region is bound to the cargo-binding site of IMPα in its autoinhibited apo state (PDB ID: 1IAL) (39) or during export from the nucleus (PDB ID: 1WA5) (40), it is resolved as a linear structure. Thus, our observed differences in folding of the TBEV C terminus are not unexpected.
However, the frequency of cells with nucleolar distribution varied among the investigated TBFVC proteins; LIVC, POWVC, and TBEVC reached 55 to 85% positivity whereas KFDVC only 11.7%. Thus, another factor(s), except for the already confirmed bNLS-importin system (41), seems to regulate the nuclear import, especially for KFDVC. These may include additional viral proteins or post-translational modifications, such as Ser/Thr phosphorylation, as was shown in the case of WNVC (42).
Regarding the medical relevance, we further focused on the detailed characterization of TBEVC biological and structural features. In order to describe the interaction network of TBEVC in host cells, we performed a whole-cell TBEVC-specific pull down from TBEV-infected or TBEVC-transfected cells. The obtained data revealed 213 interacting proteins, including viral proteins E and NS5. Strikingly, already described interaction partners of DENV, WNV, and ZIKVC proteins were not identified in the case of TBEVC. These include UPF1 and PYM1 (factors functionally involved in nonsense-mediated mRNA decay) (19, 43), APOE and PLIN3 (surface proteins of lipid droplets) (44, 45), and SEC3p (negative regulator of flavivirus transcription and translation) (46). The only exception is the NPM1/B23 protein, a nucleolar protein involved in ribosome biogenesis and p53 signaling cascade, which was already shown to interact with DENV and JEVC proteins (15, 16).
The co-IP further identified OASL and IFIT1 proteins among the top-ranked TBEVC-interacting partners. Both proteins are well-described antiviral effectors in the case of flaviviruses (47, 48, 49). Therefore, the binding of TBEVC to these proteins may negatively affect their antiviral functions and thus interfere with antiviral response in infected cells. Nevertheless, OASL and IFIT1 were also shown to bind MECP2 and SIN3A, respectively (50, 51). MECP2 and SIN3A are transcriptional corepressors regulating histone acetylation; thus, except for interference with host antiviral response, TBEVC may dysregulate the expression of host genes via interactions with OASL and IFIT1. MECP2 itself was identified as a TBEVC-binding protein as well.
Our previous study documented a TBEV-induced decrease of de novo synthesized proteins as well as decrease of mature (18S and 28S) and nascent 45S pre-rRNA levels in infected cells (23). Based on the colocalization of nascent rRNA and TBEVC protein, nature of TBEVC protein interactome, and high nonspecific binding affinity of TBEVC to various DNA/RNA molecules (22), we hypothesized that TBEVC might be associated with the phenomena of reduced host protein or rRNA levels. Indeed, the presence of TBEVC, as well as other flaviviral C proteins, resulted in reduced host translation rate (Fig. 10, A and B). In addition, only the wildtype TBEVC was observed to decrease the levels of mature 18S and 28S rRNAs, although only 18S rRNA was reduced significantly (Fig. 10, C and D). In addition, no changes were observed in the case of 45S pre-rRNA transcripts. Hence, TBEVC seems to interfere with post-transcriptional processing of 45S pre-rRNA rather than downregulating the expression of rDNA gene clusters. This hypothesis is further supported by the identification of several RNA splicing factors (CPSF3, SART1, SNRNP20, and RBM28) as binding partners of TBEVC. Except for the splicing of 45S pre-rRNA, chemical modifications of rRNA, including its methylation, represent another important step in post-transcriptional rRNA processing. The rRNA methylation process could be another target of TBEVC-induced deregulation of the host environment since NOP2 (m5C-methyltransferase) and FBL (2′-O-methyltransferase) were identified as TBEVC-interacting proteins. In addition to rRNA methylation, TBEVC was further described to bind proteins involved in ribosome biogenesis, such as BRIX1, TSR1, and RRP1B. The depletion of these crucial factors by binding to TBEVC in combination with impaired post-transcriptional processing of rRNA may eventually result in decreased formation of functional ribosomes.
Altogether, we have characterized the structure of TBEVC and the formation of a stable homodimer, revealed TBEVC trafficking to the host nucleus via interaction with the IMPα, and described its localization within the nucleolus, site of rDNA transcription and ribosome biogenesis. TBEVC alone was sufficient to reduce the amount of de novo synthesized 18S rRNA making it partially responsible for the transcriptional shutoff described earlier. That was also supported by the array of identified interacting partners involved in rRNA processing, ribosome biogenesis, and nonsense-mediated mRNA decay. This identifies TBEVC as an important factor in TBEV pathogenesis and a promising target for future research.
Experimental procedures
Cell lines
The human medulloblastoma (DAOY HTB-186; American Type Culture Collection) and human lung adenocarcinoma (A549; a gift from R. Randall, University of St Andrews) cell lines were grown in low-glucose Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% antibiotics–antimycotics (amphotericin B 0.25 μg/ml, penicillin G 100 units/ml, and streptomycin 100 μg/ml), and 1% l-alanyl-l-glutamine. DAOY cell line is derived from desmoplastic cerebellar medulloblastoma of a 4-year-old Caucasian male (52). A549s are derived from a lung cancerous tissue (alveolar basal epithelial cells) of a 58-year-old Caucasian male (53).
Viruses and infection
A representative of the European TBEV subtype strain Hypr was used for all infection experiments. In more detail, low passage TBEV Hypr (fourth passage in suckling mice brains; GenBank accession no.: U39292) (54) was used and is available at the Faculty of Science, University of South Bohemia, České Budějovice, Czech Republic. The virus was handled under biosafety level 3 conditions.
One day postseeding, cells were incubated with TBEV for 2 h, washed with PBS, and a fresh prewarmed medium was added. Brain suspension from uninfected suckling mice was used as a negative control.
Plasmid construction and transfection
For C protein cloning, the sequences coding for the ER anchor were not included. The complementary DNA sequences of KFDV (55), LIV, and POWV were provided from the Arbovirus collection of the Biology Centre Collections of Organisms by Prof Ruzek. DENV2C was recloned from pET22b (22). For ZIKVC cloning, complementary DNA from ZIKV PE243-infected A594 cells was used (56). Briefly, PCR-amplified fragment of the respective C protein with 5′-NheI restriction site and 3′-FLAG tag-NotI restriction site was used for the ligation into NheI/NotI-linearized pRL-CMV vector (Promega), where renilla luciferase (Rluc) gene was exchanged for flaviC-FLAG tag fragment. TBEVC-Δ20 mutant was prepared analogously using specific primers for C-linked truncation (Table S8). TBEVC mutants d1, d2, and dd were prepared using PCR site-directed mutagenesis, and subsequent fragment fusion was performed using the Gibson assembly kit (NEB; catalog no.: E2611). 5′-NheI and 3′-FLAG tag-NotI sequences were added to the resulting fragments via PCR and ligated into the linearized pRL-CMV vector as in the case of TBEV Hypr C-wt. To fuse TBEVC bNLS to GFP, pEGFP (Clontech) vectors were used. A set of complementary oligonucleotides encoding TBEVC bNLS with cohesive ends compatible for ligation into NheI–HindIII pEGFPN1 and HindIII–BamHI pEGFPC1, respectively, were mixed at the final concentration of 2 μM, denatured by heating at 95 °C, and cooled slowly to laboratory temperature. The double-stranded oligonucleotides were then used for ligation into pEGFPC1 or pEGFPN1 vectors and cleaved with the appropriate restriction enzymes. Primers used are listed in Table S8. All the resulting constructs were verified via Sanger sequencing. The resulting capsid gene sequences confirmed 100% identity with KFDV strain W-377 (GenBank no.: JF416960.1), LB strain of POWV (GenBank no.: NC_003687.1), and 99.65% identity (279G → 279A) with LIV isolate SCO_31_1931 (GenBank no.: MK007532.1).
PolyJet In Vitro Transfection Reagent (SignaGen; catalog no.: SL100688) was used for all the transfection experiments. The procedure was carried out according to the manufacturer’s protocol.
Antibodies and reagents
The following primary antibodies were used: anti-TBEVC polyclonal antibody (23), monoclonal FLAG M2 antibody (Sigma–Aldrich; catalog no.: F1804-200UG), anti-NPM1 polyclonal antibody (Life Technologies; catalog no.: PA5-12446), anti-COX4 monoclonal antibody (Life Technologies; catalog no.: MA5-15078), anti-calnexin monoclonal antibody (Abcam; catalog no.: ab22595), Anti-E Flavivirus Group Antigen Antibody (Merck Millipore; catalog no.: MAB10216), anti-dsRNA mAb SCICONS J2 (Biocompare; catalog no.: 10010200), anti-TBEV NS3 polyclonal antibody (Dr M. Bloom, NIAID; (23)), anti-RPL9 antibody (EP13752) (Abcam; catalog no.: ab182556), anti-KAP1/TRIM28 antibody (Abcam; catalog no.: ab190178), anti-OASL Polyclonal Antibody (Life Technologies; catalog no.: PA5-31317), anti-CSDE1/NRU antibody (EPR17413) (Abcam; catalog no.: ab200663), and anti-MYBBP1A antibody (Abcam; catalog no.: ab99361).
The following secondary antibodies were used: goat anti–guinea pig horseradish peroxidase (HRP) (Life Technologies; catalog no.: A18769), anti-rabbit DyLight 488 (Vector Laboratories; catalog no.: DI-1488), antichicken DyLight 488 (Abcam; catalog no.: ab96947), anti–guinea pig Alexa Fluor 594 (Abcam; catalog no.: ab150188), antimouse DyLight 488 (Vector Laboratories; catalog no.: DI-2488), and antimouse DyLight 594 (Abcam; catalog no.: ab96881).
Subcellular fractionation
Subcellular fractionation was performed using Subcellular Protein Fractionation Kit for Cultured Cells (Life Technologies; catalog no.: 78840) according to the manufacturer’s protocol. Briefly, infected or transfected DAOY cells (5 × 105 cells per well) were first washed with ice-cold PBS and harvested using trypsin–EDTA solution and then centrifuged at 500g for 5 min. Subsequently, the cell pellet was washed with ice-cold PBS and further processed using CEB, MEB, NEB, and PEB buffers supplemented with Halt Protease Inhibitor Cocktail. The resulting fractions were directly used for Western blot analysis.
Co-IP
Infected or transfected DAOY cells were first washed with ice-cold PBS and subsequently lysed in IP lysis buffer (25 mM Tris–HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 1 mM EDTA, and 5% glycerol) for 15 min on ice, scraped, and resulting lysates were directly used for the co-IP assay.
Prepared lysates (430 μg per sample) were incubated with the respective antibody for 60 min at room temperature. Magnetic beads conjugated with protein A (Invitrogen; catalog no.: 10002D) were first washed with co-IP buffer and subsequently added to the lysate/antibody mixture (1.5 mg of beads per sample). Co-IP was performed overnight at 4 °C. Beads with bound proteins were washed twice with complete co-IP buffer and twice with co-IP buffer without NP-40 detergent. The resulting bead pellets were resuspended in 50 μl of co-IP buffer without NP-40 detergent and used directly for either MS or Western blot analyses.
MS analysis
On-bead digestion
Following IP washes, bead-bound protein complexes were digested directly on beads by addition of 0.75 μg (1 μg/μl) of trypsin (sequencing grade; Promega) in 50 mM NaHCO3 buffer. Beads were gently tapped to ensure even suspension of trypsin solution and incubated at 37 °C with mild agitation for 2 h. Beads were then vortexed, and the partially digested sample was transferred to clean tubes to separate it from the beads and incubated at 37 °C for 16 h without agitation. The resulting peptides were extracted into LC–MS vials by 2.5% formic acid (FA) in 50% acetonitrile (ACN) and 100% ACN with the addition of polyethylene glycol (20,000 dilution; final concentration of 0.001%) and concentrated in a SpeedVac concentrator (Thermo Fisher Scientific).
LC–MS analysis of peptides
LC–MS/MS analyses of peptide mixtures were done using an Ultimate 3000 RSLCnano system connected to Orbitrap Q-Exactive HF-X mass spectrometer (Thermo Fisher Scientific). Prior to LC separation, tryptic digests were online concentrated and desalted using a trapping column (300 μm × 5 mm, μ-precolumn, 5 μm particles, Acclaim PepMap100 C18; Thermo Fisher Scientific) at 40 ºC. After washing of trapping column with 0.1% FA, the peptides were eluted (flow rate of 300 nl/min) from the trapping column onto an analytical column (Acclaim Pepmap100 C18, 3 μm particles, 75 μm × 500 mm; Thermo Fisher Scientific) at 40 ºC by a 120 min linear gradient program (2–35% of mobile phase B; mobile phase A: 0.1% FA in water; mobile phase B: 0.1% FA in 80% ACN). Equilibration of the trapping column and the analytical column was done prior to sample injection to the sample loop. The analytical column outlet was directly connected to the Digital PicoView 550 (New Objective) ion source with sheath gas option and SilicaTip emitter (New Objective; FS360-20-15-N-20-C12) utilization. Active background ion reduction device (ESI Source Solutions) was installed.
MS data were acquired in a data-dependent strategy. The survey scan range was set to m/z 350 to 2000 with the resolution of 120,000 (at m/z 200) with a target value of 3 × 106 ions and maximum injection time of 100 ms. Higher energy collisional dissociation MS/MS (27% relative fragmentation energy) spectra were acquired with a target value of 1.0 × 105 and resolution of 15,000 (at m/z 200) and a maximum injection time of 250 ms. Dynamic exclusion was enabled for 40 s. The isolation window for MS/MS fragmentation was set to m/z 1.2.
The mass spectrometric RAW data files were analyzed using the MaxQuant software (version 1.6.10.43) (57) using default settings unless otherwise noted. MS/MS ion searches were done against a modified cRAP database (based on http://www.thegpm.org/crap) containing protein contaminants like keratin, trypsin, and others, and UniProtKB protein database for Homo sapiens (ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/reference_proteomes/Eukaryota/UP000005640_9606.fasta.gz; downloaded September 2018, version 2018/08, number of protein sequences: 21,053). Oxidation of methionine and proline, deamidation (N, Q), and acetylation (protein N terminus) as optional modification, and trypsin/P enzyme with two allowed miss cleavages were set. Peptides and proteins with FDR threshold <0.01 and proteins having at least one unique or razor peptide were considered only. The match between runs option was set among all analyzed samples. Protein abundance was assessed using protein intensities calculated by MaxQuant.
Protein intensities reported in the proteinGroups.txt file (output of MaxQuant) were further processed using the software container environment (https://github.com/OmicsWorkflows). Processing workflow is available upon request. Briefly, it covered (a) removal of decoy hits and contaminant protein groups, (b) protein group intensities log2 transformation, (c) LoessF normalization, (d) interacting partners were filtered out (with at least two peptides in at least one replicate of a single sample type), and (e) differential expression using LIMMA statistical test. Proteins with adjusted p value <0.05 and log2 fold change >1 were considered as differentially expressed proteins for a given comparison.
Western blotting
Samples from co-IP or subcellular fractionation experiments were separated on a 12% denaturing polyacrylamide stain-free gels and blotted onto polyvinylidene difluoride membranes. Membranes were blocked (5% skimmed milk in PBS with Tween-20) and incubated with primary and secondary antibodies; membranes were washed three times in PBS with Tween-20 between each staining step. Chemiluminescent signal was developed using the WesternBright Quantum kit for HRP (Advansta; catalog no.: K-12042-D20). The signal was subsequently quantified using the Fiji software (58).
Immunofluorescence
DAOY and A549 cells were seeded in chamber slides (0.3 cm2/well; 1 × 104 cells/well) and at the indicated time intervals p.i. processed as previously described (59). As the primary antibodies, guinea pig anti-TBEVC (1:1500 dilution), mouse anti-FLAG tag (1:200 dilution), rabbit anti-NPM1 (1:200 dilution), mouse anti-flavivirus E (1:200 dilution), chicken anti-NS3 (1:5000 dilution), mouse anti-dsRNA J2 (1:250 dilution), rabbit anticalnexin (1:200 dilution), and rabbit anti-COX4 (1:200 dilution) were used. As the secondary antibodies, anti-rabbit DyLight 488 (1:500 diluiton), antichicken DyLight 488 (Abcam; 1:500 dilution), antiguinea pig Alexa Fluor 594 (1:500 dilution), antimouse DyLight 594 (1:500 dilution), and antimouse DyLight 488 (1:500 dilution) were used. Slides were eventually mounted in Vectashield mounting medium (Vector Laboratories). The Olympus Fluoview FV10i confocal microscope was used for imaging, and images were exported in FV10-ASW software (Olympus Life Science; version 1.7).
Metabolic labeling
Metabolic labeling experiments were performed as previously described utilizing the Click chemistry (copper(I)-catalyzed azide–alkyne cycloaddition) with AHA and 5-ethynyluridine as analogs for methionine and uridine, respectively (23, 60). For in situ metabolic labeling, Alexa Fluor 488-azide was used to visualize the incorporated 5-ethynyluridine, and Olympus Fluoview FV10i confocal microscope was used for imaging, and subsequent export of images was done in FV10-ASW software (version 1.7). In the case of Click-on-membrane labeling experiments, the signal was developed using WesternBright Quantum kit for HRP (Advansta; catalog no.: K-12042-D20). AHA signal from both types of experiments was quantified using Fiji software (58).
Ribosomal RNA quantification
Total cellular RNA was isolated using Trizol-based RNA Blue reagent (Top-Bio; catalog no.: R013) according to the manufacturer’s instructions. RNA pellets were dissolved in diethyl pyrocarbonate–treated water and directly used for either real-time quantitative RT–PCR or analysis on an RNA gel.
Quantification of mature 18S and 28S rRNA
Total RNA samples were separated on a 1.2% agarose Mops-buffered gel. The input amount of each sample was normalized to cell viability measured by alamarBlue (Thermo Fisher Scientific) as described previously (23). RNA was visualized using the GelRed dye (Biotium). The 18S and 28S rRNA signal was quantified using the Fiji software (58) and normalized to the nontransfected cells.
Quantification of 45S pre-rRNA
KAPA SYBR FAST One-Step Kit (Sigma–Aldrich) was used for quantitative RT–PCR analyses according to the manufacturer’s protocol. The obtained data were processed via relative quantification using the delta Ct (Δ-Ct) method; the amount of RNA was adjusted to the cell number instead of the Ct values of the housekeeping reference gene as described (23). All samples were treated with dsDNase (Life Technologies; catalog no.: EN0771) and subsequently 5× diluted in RNAse-free water before the real-time PCR analysis. All samples were analyzed in technical triplicates. Primers used for 45S pre-rRNA quantification were designed according to Yan et al. (61) and described in Table S8.
TEM
Cells were fixed in 4% formaldehyde and 0.1% glutaraldehyde in 0.1 M Hepes for 1 h at room temperature. After washing in 0.01 M glycine in Hepes, cell pellets were embedded into 10% gelatine at 37 °C and left for 4 days on a rotating wheel in 2.3 M sucrose at 4 °C. Samples were frozen by immersion in liquid nitrogen. Ultrathin cryosections were cut using an ultramicrotome Leica EM FCS equipped with Leica UCT cryochamber (Leica Microsystems). Sections were picked up on a drop of sucrose/methylcellulose (one part of 2.3 M sucrose and one part of 2% methylcellulose, 25 cP) and transferred onto a Formvar–carbon-coated TEM grid. Sections were blocked in a solution containing 1% fish skin gelatine in Hepes and incubated with guinea pig immunoglobulin G (IgG) directed against the TBEVC and anti-MYBBP1 rabbit IgG molecules (both 1:50 dilution) for 45 min at room temperature. After washing in the blocking solution, sections were incubated for 1 h in goat anti-rabbit IgG conjugated to 15 nm gold particles and anti–guinea pig IgG 5 nm (both from British Biocell International) with 1:50 dilution in 0.5% fish skin gelatine. Sections were washed in Hepes, distilled water, contrasted, and dried using 2% methylcellulose with 3% aqueous uranyl acetate solution diluted at 9:1. Sections were examined with 80 kV JEOL 1010 TEM. Background labeling was tested by a negative control (in the absence of primary antibody) under the conditions described previously and labeling of wt-TBEVC-transfected cells.
Δ16-TBEVC production and purification
An expression vector encoding Δ16-TBEVC (TBEV Neudoerfl strain; prepared as previously described (22)) was transformed into Escherichia coli BL21(DE3) CodonPlus-RIL (Invitrogen). The transformed cells were used to inoculate 2.4 l of LB medium (Sigma–Aldrich) or 2.4 l of M9 minimal medium (supplemented with [U-13C]d-glucose as a sole source of carbon and [U-15N](NH4)2SO4 as a sole source of nitrogen). Cells were grown at 37 °C and 270 rpm. Protein expression was induced by the addition of IPTG (final concentration of 0.4 mM), followed by postinduction incubation for another 4 h at 37 °C and 270 rpm or overnight at 16 °C and 270 rpm in the case of M9 minimal medium. The biomass was harvested by centrifugation at 10,000g for 10 min.
The cell pellet obtained from 2.4 l of medium (LB or M9) was resuspended in 144 ml of wash buffer (50 mM sodium phosphate, 150 mM NaCl, 1 mM EDTA, pH 6.0) with the addition of protein inhibitor mix and β-mercaptoethanol to the final concentration of 0.1%. Cells were disrupted by 30 min incubation with 10 mg of lysozyme (Sigma–Aldrich) on ice, followed by sonication (probe KE76, 4 × 45 s, total energy 6 kJ) and another 30 min incubation on ice with sodium deoxycholate added at a final concentration of 0.1%. The cell lysate was then centrifuged at 35,000g for 20 min at 4 °C. Supernatant S1 was discarded. Then, three subsequent centrifugation steps (35,000g for 20 min) were performed: each pellet was resuspended in 72 ml of wash buffer containing 0.5% Triton X-100, 0.5% β-mercaptoethanol, and gradually increasing concentration of NaCl 0.5 M, 1 M, and 2 M. The obtained supernatants S2–S4 were precipitated with ammonium sulfate (80% saturation) and centrifuged at 35,000g for 20 min.
The final pellets were resuspended in a total of 5 ml of SEC buffer (50 mM sodium phosphate, 1 M NaCl, pH 6.0), dialyzed against the same buffer, and centrifuged at 70,000g for 20 min. The supernatant was loaded onto a size-exclusion column HiLoad 26/600 Superdex 200 pg (GE Healthcare Life Science) equilibrated with the SEC buffer at a flow rate of 0.2 ml/min.
The fractions containing Δ16-TBEVC were combined, diluted 10× with SEC buffer, and dialyzed against the CATEX-B buffer (50 mM sodium phosphate, 200 mM NaCl, 20% glycerol, pH 6.0). The sample was then centrifuged at 30,000g for 20 min and loaded onto a HiTrap SP HP cation-exchange column (GE Healthcare Life Science) equilibrated with the CATEX-B buffer at a flow rate of 5 ml/min. The bound Δ16-TBEVC was eluted with 300 ml of CATEX-E buffer (50 mM sodium phosphate, 2 M NaCl, 20% glycerol, pH 6.0) at a flow rate of 5 ml/min.
To ensure that the protein is purified as much as possible from nucleic acids, we decided to include also anion-exchange chromatography. The fractions from cation-exchange chromatography, containing Δ16-TBEVC, were combined and dialyzed against the ANEX-B buffer (50 mM sodium phosphate, 100 mM NaCl, 5% glycerol, pH 6.0). The sample was centrifuged at 30,000g for 20 min and loaded onto a HiTrap Q HP anion-exchange column (GE Healthcare Life Science) equilibrated with the ANEX-B buffer at a flow rate of 5 ml/min. Δ16-TBEVC from the flow-through was dialyzed against the S buffer (50 mM sodium phosphate, 50 mM NaCl, 5% glycerol, pH 6.0) and concentrated by Amicon Ultra-4 Ultra-3K (Merck Millipore). The concentration of the final Δ16-TBEVC was determined spectrophotometrically by measurement of the absorbance at 280 nm and calculated by using the molar extinction coefficient of 11,000 M−1 cm−1. The Δ16-TBEVC was aliquoted and stored at −80 °C. Throughout the process, samples of the supernatants, pellets, and fractions were analyzed by Tris–tricine SDS-PAGE.
NMR spectroscopy
All NMR experiments were measured at 298 K on a Bruker AvanceIII 600 MHz NMR spectrometer equipped with a cryoprobe. Backbone assignments were made by using standard triple-resonance experiments: 3D HNCACB, CBCA(CO)NH, HNCA, HN(CO)CA, HNCO, and HN(CA)CO; in combination with 2D experiment 1H–15N heteronuclear single quantum coherence. Side-chain assignments were obtained from 3D hCCH-TOCSY and HcCH-TOCSY experiments in combination with 2D 1H–13C heteronuclear single quantum coherence. Proton–proton distance restraints used for structure calculation of Δ16-TBEVC were obtained from 13C- and 15N-edited NOESY experiments. NMR data were processed in TopSpin 3.5 (Bruker BioSpin, GmbH) and further analyzed in CcpNmr Analysis 2.5.2 program (62).
Structure determination
The structure calculation of monomeric form of Δ16-TBEVC was based on pair-wise proton–proton distance restraints obtained from NOESY spectra and on backbone dihedral angles φ and ψ, which were estimated by TALOS+ (24) from chemical shifts of 1HN, 13CO, 13Cα, 13Cβ, and 15NH. Distance restraints were calculated from NOE contacts using r--6 distance summation with CcpNMR Analysis 2.5.2 program (62). The regularity of α-helices was later reinforced by the addition of hydrogen bond restraints. The structures were calculated in the Xplor-NIH, version 3.2 program (63, 64) using a protocol for simulated annealing with torsion angle dynamics. Finally, 20 best calculated structures out of 1000 were chosen for further analysis. All structures were visualized in PyMOL, version 1.7.4.5 program (65) and validated using the PROSESS server (66). RMSD of the structures was calculated by Visual Molecular Dynamics program (67). Finally, homodimeric forms of the Δ16-TBEVC were created by docking monomers using the Haddock server (32, 68).
SEC
SEC was performed on a Superdex 75 10/300 GL column (GE Healthcare Life Science) using an Äkta pure chromatography system. Samples were run in a mobile phase (50 mM sodium phosphate, 50 mM NaCl, 5% glycerol, pH 6.0 or pH 7.4, alternatively with the addition of SDS to the final concentration of 1%) at a flow rate of 0.5 ml/min. The column system was calibrated using 500 μl of human rhinovirus 3C protease in fusion with glutathione-S-transferase (GST) tag (GST-3C, 1 mM, 46 kDa), tobacco etch virus (TEV) protease (1 mM, 24 kDa), and lysozyme (1 mM, 11 kDa). About 500 μl of 16-TBEVC were applied to the column at 0.4 mM (3.6 mg/ml) protein concentration or 10 times diluted. Detection was carried out using UV absorbance at 280 nm. All SEC experiments were performed at room temperature.
Protein crosslinking and digestion
The 0.5 mM Δ16-TBEVC protein solution was treated by 0.5-, 1-; 2-, 4-, and 8-fold molar excess of bis(2,5-dioxopyrrolidin-1-yl)2,2'-(carbonyl bis(azanediyl))diacetate (C2; Urea crosslinker-C2-arm, NHS ester; CF Plus Chemicals), respectively, for 30 min to find the molar excess that generates the sufficient amount of covalently linked dimer and thus also crosslinked peptides. The reaction was quenched by the addition of 50-fold molar excess of 1 M Tris–HCl of the concentration of C2. Subsequently, the crosslinking products were visualized by Tris–glycine SDS-PAGE. Because of the absence of cysteine residues in the Δ16-TBEVC sequence, the crosslinked samples were not treated by any reduction and alkylation agent, and they were immediately digested in solution at 37 °C for 16 h by Pierce MS-grade trypsin (Thermo Fisher Scientific), using the trypsin-to-substrate ratio of 1:100 (w/w). The digestion was terminated by the addition of TFA to the final concentration of 0.5% (v/v). The obtained peptide mixture was then desalted by Pierce C18 Spin Columns (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Nano-UHPLC/nano-ESI-Orbitrap MS
The obtained crosslinked peptide mixtures were analyzed on an Ultimate 3000 RSLC nano-HPLC system (Dionex) coupled to the EASY-Spray NG nano-ESI source of an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher Scientific). The peptides were dissolved in 0.1% TFA and separated using reversed-phase C18 columns (μ-precolumn: Acclaim PepMap 100 C18, 300 μm × 5 mm, 5 μm; separation column: Acclaim PepMap RSLC C18, 75 μm × 250 mm, 2 μm; Thermo Fisher Scientific). Peptides were eluted and separated using gradients from 5% to 12% B (varying between 0 and 3 min), 12% to 35% (varying between 3 and 52 min), 35% to 90% B (1 min), 90% B (53–57 min), 90% to 5% B (1 min), and 5% B (58–65 min) with the constant flow rate of 300 nl/min, where solvent A was 0.1% FA and solvent B was 100% ACN containing 0.1% FA.
MS data were acquired in the data-dependent MS/MS mode with the higher energy collisional dissociation fragmentation technique. Each high-resolution full scan (m/z 350–2000 Da, R = 60,000) in the Orbitrap was followed by product-ion scans (R = 30,000) also in the Orbitrap, starting with the most intense signal in the full-scan mass spectrum (an isolation window of m/z 1.6). The automatic gain control value was set to 400,000 (MS) and 50,000 (MS/MS), whereas the maximum accumulation time was set to 50 ms (MS and MS/MS). Precursor ions with charge states from 2+ to 6+ were selected for the fragmentation. Dynamic exclusion (exclusion duration of 10 s and exclusion window of 10 ppm) was enabled.
Identification of crosslinked products
The measured spectra were analyzed by the MeroX (version 2.0.1.4) search engine to identify the crosslinked peptides. The input data format MGF (Mascot generic format) obtained from raw data by MSConvert (ProteoWizard) was used, and the search parameters were set as follows: three potential missed cleavage sites on the C terminus of K and R; the cleavage site on the C terminus of K blocked when it forms a crosslink; oxidation of methionines as variable modification (maximum of two); the minimum peptide length—two; C2 as crosslinker with the K and the N-terminal amine group as the first linkable site; the N-terminal amine group, K, S, T, or Y as the second linkable site; 5 ppm as MS mass tolerance; 10 ppm as MS/MS mass tolerance; the lower mass limit of 200 Da; the upper mass limit of 8000 Da; the S/N ratio of 2.0; the possible ion types a, b, and y with the maximum of three neutral losses per crosslink; the application of the Prescore with the value above 10% intensity as the minimum fraction of signal intensity identified, compared with the total-ion current. The results were filtered with the FDR cutoff of 5%, only taking into account the candidate scans providing a fragment ion of the linkage as well as the duplet of C2 characteristic fragments (mass deviation of 26 Da) and the minimum crosslink sequence coverage of 50%.
HDX experiments and LC–MS analysis (HDX–MS)
For HDX labeling reaction, samples of Δ16-TEBVC were mixed with D2O in ratio of protein:D2O 1:9 and incubated at room temperature for 0, 20, 120, 1200, 7200, and 21,600 s. The unwanted back-exchange was minimized, and protein was denatured by the addition of quench solution (8 M urea, 1 M glycine, 200 mM Tris(2-carboxyethyl)phosphine, pH 2.51) to a final concentration of urea 4 mol/l and immediately freezed in liquid nitrogen before LC–MS/MS analysis. Samples were thawed and passed through an immobilized nepenthesin-II column (Affipro) at 100 μl/min (0.1% v/v FA) for 3 min. The generated peptides were trapped and desalted on a 2.1 mm × 5 mm Waters Acquity UPLC BEH C18 1.7 μm VanGuard precolumn and gradient eluted (10–35% ACN v/v with 0.1% v/v FA; varying between 2 and 13 min) across a 1 mm × 100 mm Waters Acquity UPLC 1.7 μm BEH C18 analytical column with column washing and equilibrating procedure (35% to 85% B [13–14 min], 85% B [14–18 min], and 85% to 10% B [18–25 min]). Protein digestion and peptide desalting and separation were conducted at 4 °C. The eluted peptides were then subjected to Synapt G2 mass spectrometer (ESI-Q/TOF; Waters). Data were acquired continually in sensitivity mode—positive polarity, mass range 50 to 2000 Da, and scan time 0.4 s. To initiate fragmentation, the high trap collision energy 23 to 50 V was applied. During all MS analyses, the Leu-enkephalin was used as a lock mass with mass of 556.2771 Da/e for charge state of 1, mass window of ±0.5 Da, scan time of 0.4 s, interval of 30 s, and number of scans to average 3. The HDX experiments were performed in triplicates for each labeling time point.
Peptide identification
To identify Δ16-TBEVC peptides obtained from nondeuterated samples and assign their mass in labeled samples, the ProteinLynx Global SERVER (PLGS; Waters; version 3.0.2)/DynamX (Waters; version 3.0) workflow was performed. PLGS processing parameters for MS/MS data were used as follows: chromatographic peak width—automatic; MS TOF resolution—automatic; lock mass for charge 1 to 556.2771 Da/e; lock mass window—0.25 Da; low energy threshold—135.0 counts; elevated energy threshold–30.0 counts; intensity threshold—750.0 counts, and PLGS workflow parameters were used as follows: searching against fasta file containing normal and reverse sequences of Δ16-TBEVC, Nepenthesine-II (UniProt code: Q766C2), and standard contaminants; peptide tolerance and fragment tolerance—automatic; minimum fragment ion matches per peptide—three; minimum fragment ion matches per protein—seven; minimum peptide matches per protein—one; primary digest reagent—nonspecific; number of missed cleavages—one; fixed modifier reagent—12C d0 C; oxidation of methionines as a variable modifier reagent; FDR of four; monoisotopic mass of peptides with charge 1+; and instrument type ESI-QUAD-TOF.
Peptide mass assignment and visualization of results
The list of all Δ16-TBEVC peptides identified by PLGS searching process in at least three from four nondeuterated samples was then imported to the DynamX search engine for peptide mass assignment and peptide filtering. The filtering was performed using peptide thresholds as follows: minimum intensity of 3000; maximum sequence length of 25; and minimum products per amino acid of 0.1. For processing of MS files, the parameters were settled as follows: 556.2771 Da as a lock mass for charge 1; lock mass window of 0.3; low energy threshold of 10 as its lowest limit, and elution time range to search in for the data is 9 to 21 min. No DynamX advanced processing parameters were applied.
The HDX data from all overlapping peptides were consolidated to individual amino acid values using a residue averaging approach. The data processed in DynamX resulted in the value of RFU for each residue in Δ16-TBEVC sequence, where the RFU considers the peptide length and its maximum deuteration and scales the absolute uptake as a percentage of this value. The values of absolute uptake of each amino acid in individual labeling time points were visualized as a heat map by using Draw Map tool available at MS tools (31) website.
X-ray crystallography of TBEVC–IMPα complex
Construct design
A gene fragment encoding TBEVC residues (75-RKIKRTVSALMVGLQKRGKRR-95) was codon optimized for E. coli expression and cloned into pGEX4T-1 using the BamHI site. An N-terminal TEV protease site was incorporated, allowing cleavage of the GST affinity tag. The gene encoding IMPα has been described previously (69).
Recombinant expression and purification
IMPα expression in E. coli pLysS cells was undertaken using the autoinduction method (70). The protein was purified as described previously (71). Briefly, the cells were harvested by centrifugation at 6000g for 30 min and resuspended in His buffer A (50 mM phosphate buffer, 300 mM NaCl, 20 mM imidazole, pH 8). Lysis was achieved through two freeze–thaw cycles. The TBEVC-GST fusion protein was expressed identically to the IMPα protein, and resuspended in GST buffer A (50 mM Tris, 125 mM NaCl, pH 8). Lysis was achieved through two freeze–thaw cycles, the extracts of IMPα and TBEVC were mixed, and lysis further performed through the addition of 1 ml of 20 mg/ml lysozyme (Sigma–Aldrich). To reduce viscosity of the sample, the extract was treated with 10 μl of 50 mg/ml DNAse (Sigma–Aldrich) and incubated for 1 h at room temperature. The extract was centrifuged at 12,000g for 30 min, filtered through a 0.45 μm filter, and applied to a 5 ml HisTrap HP column (GE Healthcare Life Science) pre-equilibrated in His buffer A. The column was washed extensively with His buffer A to remove unbound proteins (approximately 20 column volumes) and eluted by applying a concentration gradient of His buffer B (50 mM phosphate buffer, 300 mM NaCl, 500 mM imidazole, pH 8). Samples were collected, pooled, and treated with TEV protease to cleave the affinity tag. The sample was purified further via SEC using a HiLoad 26/60 Superdex 200 column (GE Healthcare Life Science) and pre-equilibrated in GST buffer A. Protein fractions were pooled and passed over a 5 ml GSTtrap HP column (GE Healthcare Life Science) to further remove the cleaved GST tag from the complex. Sample was concentrated to 28.23 mg/ml using an Amicon ultracentrifugal filter unit molecular weight cutoff 10 kDa (Merck Millipore), aliquoted, and stored at −80 °C.
Crystallization of TBEVC and IMPα complex
The TBEVC–IMPα protein complex was crystallized by mixing 1.5 μl of the protein with 1.5 μl of a reservoir solution containing 0.7 M sodium citrate, 0.1 M Hepes, pH 6.5. The solution was placed over the reservoir solution and crystallized using hanging drop vapor diffusion at 22 °C. Crystals appeared after approximately 2 days and cryoprotected in a solution comprised of the reservoir solution incorporating 20% glycerol, prior to being flash cooled in liquid nitrogen.
Data collection and structure determination
A single crystal was used to collect X-ray diffraction data at the Australian Synchrotron on the MX2 beamline (72). Diffraction data were indexed and merged in iMOSFLM (73) and then scaled and merged in Aimless (74, 75). Phasing was undertaken by molecular replacement using Phaser (Phaser Scientific Software) (76) and the PDB structure 6BW0 (77) as a search model. Model rebuilding and subsequent refinement were undertaken using Coot (78, 79) and Phenix (80), respectively.
SDS-PAGE
Gel preparation
Solutions for making gels were prepared as follows.
Tris–tricine SDS-PAGE
Separating gel mixture
33.5% Tris buffer (3 M Tris–HCl, 0.3% SDS [w/v], pH 8.45) (v/v), acrylamide mix (10% T, 3% C), 10% glycerol (w/v), 1% ammonium persulfate (APS), and 0.05% N, N, N', N'-tetramethylethylenediamine (TEMED).
Stacking gel mixture
25% Tris buffer (3 M Tris–HCl, 0.3% SDS [w/v], pH 8.45) (v/v), acrylamide mix (4% T, 3% C), 1% APS, and 0.1% TEMED.
Tris–glycine SDS-PAGE
Separating gel mixture
25% Tris buffer (1.5 M Tris–HCl, pH 8.8) (v/v), acrylamide mix (15% T, 3% C), 0.1% SDS (w/v), 1% APS, and 0.1% TEMED.
Stacking gel mixture
12.5% Tris buffer (0.5 M Tris–HCl, pH 6.8) (v/v), acrylamide mix (5% T, 3% C), 0.1% SDS (w/v), 1% APS, and 0.2% TEMED. The gel mixtures were poured into the casting modules. The separating gel mixture (approximately 5 cm high) was carefully overlaid with protective layer of water (Tris–tricine SDS-PAGE) or butanol (Tris–glycine SDS-PAGE). After polymerization, the protective layer was replaced by stacking gel mixture (approximately 2 cm high) with comb.
Sample preparation
Protein solutions were mixed with 2× loading buffer (100 mM Tris–HCl, pH 6.8, 4% SDS [w/v], 24% glycerol [w/v], 0.02% Coomassie Brilliant Blue R-250 [w/v], and 0.72% β-mercaptoethanol [v/v]) in volume ratio 1:1 and heated at 100 °C for 10 min. Samples (10–15 μl) and standards (5 μl) were applied into the wells.
Run conditions
Electrophoresis was performed at room temperature using a voltage stepped procedure.
Tris–tricine SDS-PAGE
Cathode buffer (0.1 M Tris–HCl, 0.1 M tricine, 0.1% SDS [w/v]), and anode buffer (0.2 M Tris–HCl, pH 8.9), 40 V for 30 min followed by 120 V until the tracking dye reached the bottom of the gel.
Tris–glycine SDS-PAGE
Tris–glycine buffer (25 mM Tris–HCl, 0.25 M glycine, 0.1% SDS [w/v]), 90 V for 15 min followed by 180 V until the tracking dye reached the bottom of the gel.
Staining and destaining
Immediately after electrophoresis, gels were stained in 45% methanol (v/v) and 10% acetic acid (v/v) with 0.25% Coomassie Brilliant Blue R-250 (w/v) for 45 min at room temperature while gently shaking. Destaining of gels was accomplished in 25% methanol (v/v) with 10% acetic acid (v/v), shaking for 30 min at room temperature, repeated at least three times.
Data availability
All data are contained within the article or supporting information. Both the NMR and X-ray structural data were deposited into the Worldwide Protein Data Bank (wwPDB) via the OneDep system. The resonance assignment is listed in the BMRB with accession number BMRB ID: 34709. Coordinates and structure factors are located in the RCSB PDB with accession numbers PDB ID: 7YWQ (NMR-resolved 3D structure of d16TBEVC) and 8ECH (X-ray crystallography of TBEVC–IMPα complex).
Ethics
Experiments including mice were performed in compliance with the European Union (EU) directive 2010/63/EU and in agreement with Czech national law guidelines (Act no.: 246/1992 Coll.). The project was approved by the Committee on the Ethics of Animal Experimentation of the Institute of Parasitology and of the Departmental Expert Committee for the Approval of Projects of Experiments on Animals of the Czech Academy of Sciences (permit no. 113/2013).
Supporting information
This article contains supporting information (6, 21, 25, 65, 81, 82).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Z. Vavrušková (Institute of Parasitology, Biology Centre CAS, Czech Republic) for the assistance in derivation of anti-TBEVC antibodies and I. Křížová (Department of Biotechnology, University of Chemistry and Technology Prague, Czech Republic) for preparation of GFP vectors. We thank Prof D. Růžek and Dr V. Hönig (Institute of Parasitology, Biology Centre CAS, Czech Republic) for providing us the complementary DNA of KFDV, LIV, and POWV from the Arbovirus collection of the Biology Centre Collections of Organisms (https://arboviruscollection.bcco.cz/). We acknowledge CEITEC Proteomics Core Facility of CIISB, Instruct-CZ Centre, supported by the Ministry of Education, Youth and Sports of the Czech Republic (grant no.: LM201812).
Author contributions
M. S., R. N., M. E. B., Z. Z., L. G., J. K. F., R. H., M. R., and J. Š. conceptualization; M. S., R. N., J. S., J. A. R., H. T., G. S. R., J. K. F., R. H., M. R., and J. Š. methodology; M. S., R. N., M. E. B., L. G., J. K. F., R. H., M. R., and J. Š. formal analysis; M. S., R. N., J. S., J. A. R., H. T., G. S. R., M. V., K. J., and F. K. investigation; Z. Z., L. G., J. K. F., R. H., M. R., and J. Š. resources; M. S., R. N., J. A. R., H. T., J. K. F., R. H., M. R., and J. Š. writing–original draft; M. S., R. N., J. S., J. A. R., H. T., G. S. R., M. V., K. J., F. K., L. G., J. K. F., R. H., M. R., and J. Š. writing–review and editing; M. E. B., Z. Z., L. G., J. K. F., R. H., M. R., and J. Š. supervision; M. R. and J. Š. funding acquisition.
Funding and additional information
Computational resources were supplied by the project “e-Infrastruktura CZ” (e-INFRA CZ LM2018140; to R. N.) supported by the Ministry of Education, Youth and Sports of the Czech Republic, which also supported electron microscopy core facility of Biology Centre CAS (grant no.: LM2018129; to M. V.). This work was supported by the project National Institute of virology and bacteriology (Programme EXCELES, ID project no.: LX22NPO5103, to M. R.), funded by the European Union—Next Generation European Union. The work was supported from European Regional Development Fund; OP RDE; Project: “ChemBioDrug” (no. CZ.02.1.01/0.0/0.0/16_019/0000729; to J. S.), the Czech Grant Agency (grant no.: 18-27204S and 22-25042S; to M. S., H. T., K. J., L. G., and M. R.), and Ministry of Education, Youth and Sports of the Czech Republic (grant no.: LTAUSA 18040; to M. S., H. T., K. J., L. G., and J. Š.). M. E. B. was supported by the Intramural Research Program of National Institute of Allergy and Infectious Diseases/National Institutes of Health. We acknowledge CEITEC Proteomics Core Facility of CIISB, Instruct-CZ Centre, supported by the Ministry of Education, Youth and Sports of the Czech Republic (LM2018127; to Z.Z.). Z. Z. was supported from the European Regional Development Fund—project no.: CZ.02.1.01/0.0/0.0/16_026/0008446.
Edited by Craig Cameron
Supporting information
References
- 1.Gould E.A., Solomon T. Pathogenic flaviviruses. Lancet (London, England) 2008;371:500–509. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]
- 2.Barrows N.J., Campos R.K., Liao K.C., Prasanth K.R., Soto-Acosta R., Yeh S.C., et al. Biochemistry and molecular biology of flaviviruses. Chem. Rev. 2018;118:4448–4482. doi: 10.1021/acs.chemrev.7b00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neufeldt C.J., Cortese M., Acosta E.G., Bartenschlager R. Rewiring cellular networks by members of the Flaviviridae family. Nat. Rev. Microbiol. 2018;16:125–142. doi: 10.1038/nrmicro.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byk L.A., Gamarnik A.V. Properties and functions of the dengue virus capsid protein. Annu. Rev. Virol. 2016;3:263–281. doi: 10.1146/annurev-virology-110615-042334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pulkkinen L.I.A., Butcher S.J., Anastasina M. Tick-borne encephalitis virus: a structural view. Viruses. 2018;10:350. doi: 10.3390/v10070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma L., Jones C.T., Groesch T.D., Kuhn R.J., Post C.B. Solution structure of dengue virus capsid protein reveals another fold. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3414–3419. doi: 10.1073/pnas.0305892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sotcheff S., Routh A. Understanding flavivirus capsid protein functions: the tip of the iceberg. Pathogens (Basel, Switzerland) 2020;9:42. doi: 10.3390/pathogens9010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samsa M.M., Mondotte J.A., Iglesias N.G., Assuncao-Miranda I., Barbosa-Lima G., Da Poian A.T., et al. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coyaud E., Ranadheera C., Cheng D., Gonçalves J., Dyakov B.J.A., Laurent E.M.N., et al. Global interactomics uncovers extensive organellar targeting by Zika virus. Mol. Cell. Proteomics. 2018;17:2242–2255. doi: 10.1074/mcp.TIR118.000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulich R., Aaskov J.G. Nuclear localization of dengue 2 virus core protein detected with monoclonal antibodies. J. Gen. Virol. 1992;73:2999–3003. doi: 10.1099/0022-1317-73-11-2999. [DOI] [PubMed] [Google Scholar]
- 11.Westaway E.G., Khromykh A.A., Kenney M.T., Mackenzie J.M., Jones M.K. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology. 1997;234:31–41. doi: 10.1006/viro.1997.8629. [DOI] [PubMed] [Google Scholar]
- 12.Colpitts T.M., Barthel S., Wang P., Fikrig E. Dengue virus capsid protein binds core histones and inhibits nucleosome formation in human liver cells. PLoS One. 2011;6:e24365. doi: 10.1371/journal.pone.0024365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori Y., Okabayashi T., Yamashita T., Zhao Z., Wakita T., Yasui K., et al. Nuclear localization of Japanese encephalitis virus core protein enhances viral replication. J. Virol. 2005;79:3448–3458. doi: 10.1128/JVI.79.6.3448-3458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slomnicki L.P., Chung D.H., Parker A., Hermann T., Boyd N.L., Hetman M. Ribosomal stress and Tp53-mediated neuronal apoptosis in response to capsid protein of the Zika virus. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-16952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balinsky C.A., Schmeisser H., Ganesan S., Singh K., Pierson T.C., Zoon K.C. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J. Virol. 2013;87:13094–13106. doi: 10.1128/JVI.00704-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuda Y., Mori Y., Abe T., Yamashita T., Okamoto T., Ichimura T., et al. Nucleolar protein B23 interacts with Japanese encephalitis virus core protein and participates in viral replication. Microbiol. Immunol. 2006;50:225–234. doi: 10.1111/j.1348-0421.2006.tb03789.x. [DOI] [PubMed] [Google Scholar]
- 17.Yang M.R., Lee S.R., Oh W., Lee E.W., Yeh J.Y., Nah J.J., et al. West Nile virus capsid protein induces p53-mediated apoptosis via the sequestration of HDM2 to the nucleolus. Cell Microbiol. 2008;10:165–176. doi: 10.1111/j.1462-5822.2007.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Netsawang J., Noisakran S., Puttikhunt C., Kasinrerk W., Wongwiwat W., Malasit P., et al. Nuclear localization of dengue virus capsid protein is required for DAXX interaction and apoptosis. Virus Res. 2010;147:275–283. doi: 10.1016/j.virusres.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 19.Fontaine K.A., Leon K.E., Khalid M.M., Tomar S., Jimenez-Morales D., Dunlap M., et al. The cellular NMD pathway restricts Zika virus infection and is targeted by the viral capsid protein. mBio. 2018;9 doi: 10.1128/mBio.02126-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samuel G.H., Wiley M.R., Badawi A., Adelman Z.N., Myles K.M. Yellow fever virus capsid protein is a potent suppressor of RNA silencing that binds double-stranded RNA. Proc. Natl. Acad. Sci. U. S. A. 2016;113:13863–13868. doi: 10.1073/pnas.1600544113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shang Z., Song H., Shi Y., Qi J., Gao G.F. Crystal structure of the capsid protein from Zika virus. J. Mol. Biol. 2018;430:948–962. doi: 10.1016/j.jmb.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Kaufman F., Dostálková A., Pekárek L., Thanh T.D., Kapisheva M., Hadravová R., et al. Characterization and in vitro assembly of tick-borne encephalitis virus C protein. FEBS Lett. 2020;594:1989–2004. doi: 10.1002/1873-3468.13857. [DOI] [PubMed] [Google Scholar]
- 23.Selinger M., Tykalova H., Sterba J., Vechtova P., Vavruskova Z., Lieskovska J., et al. Tick-borne encephalitis virus inhibits rRNA synthesis and host protein production in human cells of neural origin. PLoS Negl. Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen Y., Delaglio F., Cornilescu G., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dokland T., Walsh M., Mackenzie J.M., Khromykh A.A., Ee K.-H., Wang S. West Nile virus core protein: tetramer structure and ribbon formation. Structure. 2004;12:1157–1163. doi: 10.1016/j.str.2004.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ihling C.H., Piersimoni L., Kipping M., Sinz A. Cross-linking/Mass spectrometry combined with ion mobility on a timsTOF Pro instrument for structural Proteomics. bioRxiv. 2021 doi: 10.1101/2021.03.26.437136. [preprint] [DOI] [PubMed] [Google Scholar]
- 27.Merkley E.D., Rysavy S., Kahraman A., Hafen R.P., Daggett V., Adkins J.N. Distance restraints from crosslinking mass spectrometry: Mining a molecular dynamics simulation database to evaluate lysine–lysine distances. Protein Sci. 2014;23:747–759. doi: 10.1002/pro.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat. Met. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J., Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucl. Acids Res. 2015;43:W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kavan D., Man P. MSTools—web based application for visualization and presentation of HXMS data. Int. J. Mass Spectrom. 2011;302:53–58. [Google Scholar]
- 32.Van Zundert G., Rodrigues J., Trellet M., Schmitz C., Kastritis P., Karaca E., et al. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Oliveira E.R., Mohana-Borges R., de Alencastro R.B., Horta B.A. The flavivirus capsid protein: structure, function and perspectives towards drug design. Virus Res. 2017;227:115–123. doi: 10.1016/j.virusres.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Byrd C.M., Dai D., Grosenbach D.W., Berhanu A., Jones K.F., Cardwell K.B., et al. A novel inhibitor of dengue virus replication that targets the capsid protein. Antimicrob. Agents Chemother. 2013;57:15–25. doi: 10.1128/AAC.01429-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia H., Xie X., Zou J., Noble C.G., Russell W.K., Holthauzen L.M.F., et al. A cocrystal structure of dengue capsid protein in complex of inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2020;117:17992–18001. doi: 10.1073/pnas.2003056117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodel M.R., Corbett A.H., Hodel A.E. Dissection of a nuclear localization signal. J. Biol. Chem. 2001;276:1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- 38.Cingolani G., Petosa C., Weis K., Müller C.W. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- 39.Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 1999;6:388–397. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- 40.Matsuura Y., Stewart M. Structural basis for the assembly of a nuclear export complex. Nature. 2004;432:872–877. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- 41.Bhuvanakantham R., Chong M.K., Ng M.L. Specific interaction of capsid protein and importin-alpha/beta influences West Nile virus production. Biochem. Biophys. Res. Commun. 2009;389:63–69. doi: 10.1016/j.bbrc.2009.08.108. [DOI] [PubMed] [Google Scholar]
- 42.Bhuvanakantham R., Cheong Y.K., Ng M.L. West Nile virus capsid protein interaction with importin and HDM2 protein is regulated by protein kinase C-mediated phosphorylation. Microbes Infect. 2010;12:615–625. doi: 10.1016/j.micinf.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Li M., Johnson J.R., Truong B., Kim G., Weinbren N., Dittmar M., et al. Identification of antiviral roles for the exon-junction complex and nonsense-mediated decay in flaviviral infection. Nat. Microbiol. 2019;4:985–995. doi: 10.1038/s41564-019-0375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carvalho F.A., Carneiro F.A., Martins I.C., Assunção-Miranda I., Faustino A.F., Pereira R.M., et al. Dengue virus capsid protein binding to hepatic lipid droplets (LD) is potassium ion dependent and is mediated by LD surface proteins. J. Virol. 2012;86:2096–2108. doi: 10.1128/JVI.06796-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martins A.S., Carvalho F.A., Faustino A.F., Martins I.C., Santos N.C. West Nile virus capsid protein interacts with biologically relevant host lipid systems. Front. Cell Infect. Microbiol. 2019;9:8. doi: 10.3389/fcimb.2019.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhuvanakantham R., Ng M.L. West Nile virus and dengue virus capsid protein negates the antiviral activity of human Sec3 protein through the proteasome pathway. Cell Microbiol. 2013;15:1688–1706. doi: 10.1111/cmi.12143. [DOI] [PubMed] [Google Scholar]
- 47.Yakub I., Lillibridge K.M., Moran A., Gonzalez O.Y., Belmont J., Gibbs R.A., et al. Single nucleotide polymorphisms in genes for 2'-5'-oligoadenylate synthetase and RNase L inpatients hospitalized with West Nile virus infection. J. Infect. Dis. 2005;192:1741–1748. doi: 10.1086/497340. [DOI] [PubMed] [Google Scholar]
- 48.Szretter K.J., Daniels B.P., Cho H., Gainey M.D., Yokoyama W.M., Gale M., Jr., et al. 2'-O methylation of the viral mRNA cap by West Nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura T., Katoh H., Kayama H., Saiga H., Okuyama M., Okamoto T., et al. Ifit1 inhibits Japanese encephalitis virus replication through binding to 5' capped 2'-O unmethylated RNA. J. Virol. 2013;87:9997–10003. doi: 10.1128/JVI.00883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersen J.B., Strandbygård D.J., Hartmann R., Justesen J. Interaction between the 2'-5' oligoadenylate synthetase-like protein p59 OASL and the transcriptional repressor methyl CpG-binding protein 1. Eur. J. Biochem. 2004;271:628–636. doi: 10.1046/j.1432-1033.2003.03966.x. [DOI] [PubMed] [Google Scholar]
- 51.John S.P., Sun J., Carlson R.J., Cao B., Bradfield C.J., Song J., et al. IFIT1 exerts opposing regulatory effects on the inflammatory and interferon gene programs in LPS-activated human macrophages. Cell Rep. 2018;25:95–106.e106. doi: 10.1016/j.celrep.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacobsen P.F., Jenkyn D.J., Papadimitriou J.M. Establishment of a human medulloblastoma cell line and its heterotransplantation into nude mice. J. Neuropathol. Exp. Neurol. 1985;44:472–485. doi: 10.1097/00005072-198509000-00003. [DOI] [PubMed] [Google Scholar]
- 53.Giard D.J., Aaronson S.A., Todaro G.J., Arnstein P., Kersey J.H., Dosik H., et al. In vitro cultivation of human tumors: Establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 54.Pospisil L., Jandasek L., Pesek J. Isolation of new strains of tick-borne encephalitis virus, Brno region, summer 1953. Lek List. 1954;9:3–5. [PubMed] [Google Scholar]
- 55.Sirmarova J., Salat J., Palus M., Hönig V., Langhansova H., Holbrook M.R., et al. Kyasanur Forest disease virus infection activates human vascular endothelial cells and monocyte-derived dendritic cells. Emerg. Microbes Infect. 2018;7:175. doi: 10.1038/s41426-018-0177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Donald C.L., Brennan B., Cumberworth S.L., Rezelj V.V., Clark J.J., Cordeiro M.T., et al. Full genome sequence and sfRNA interferon antagonist activity of Zika virus from recife, Brazil. PLoS Negl. Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0005048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protocols. 2016;11(12):2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 58.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selinger M., Wilkie G.S., Tong L., Gu Q., Schnettler E., Grubhoffer L., et al. Analysis of tick-borne encephalitis virus-induced host responses in human cells of neuronal origin and interferon-mediated protection. J. Gen. Virol. 2017;98:2043–2060. doi: 10.1099/jgv.0.000853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sterbova J., Kocova P., Pekarek L., Selinger M., Ondrus J., Grubhoffer L., et al. Click-on-Membrane for detection of metabolically labelled proteins and RNA. Chem. Listy. 2021;115:662–668. [Google Scholar]
- 61.Yan Y., Du Y., Wang G., Li K. Non-structural protein 1 of H3N2 influenza A virus induces nucleolar stress via interaction with nucleolin. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-18087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M., et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins: Struct. Funct. Bioinform. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 63.Schwieters C.D., Kuszewski J.J., Clore G.M. Using Xplor–NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 2006;48:47–62. [Google Scholar]
- 64.Schwieters C.D., Kuszewski J.J., Tjandra N., Clore G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 65.Schrödinger L., DeLano W. PyMOL; 2020. http://www.pymol.org/pymol Available at: [Google Scholar]
- 66.Berjanskii M., Liang Y., Zhou J., Tang P., Stothard P., Zhou Y., et al. PROSESS: a protein structure evaluation suite and server. Nucl. Acids Res. 2010;38:W633–W640. doi: 10.1093/nar/gkq375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 68.Honorato R.V., Koukos P.I., Jiménez-García B., Tsaregorodtsev A., Verlato M., Giachetti A., et al. Structural biology in the clouds: the WeNMR-EOSC ecosystem. Front. Mol. Biosci. 2021;8:729513. doi: 10.3389/fmolb.2021.729513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Teh T., Tiganis T., Kobe B. Crystallization of importin alpha, the nuclear-import receptor. Acta Crystallogr. Sect. D, Biol. Crystallogr. 1999;55:561–563. doi: 10.1107/s0907444998012943. [DOI] [PubMed] [Google Scholar]
- 70.Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 71.Patterson E.I., Dombrovski A.K., Swarbrick C.M., Raidal S.R., Forwood J.K. Structural determination of importin alpha in complex with beak and feather disease virus capsid nuclear localization signal. Biochem. Biophys. Res. Commun. 2013;438:680–685. doi: 10.1016/j.bbrc.2013.07.122. [DOI] [PubMed] [Google Scholar]
- 72.McPhillips T.M., McPhillips S.E., Chiu H.J., Cohen A.E., Deacon A.M., Ellis P.J., et al. Blu-ice and the distributed control system: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 73.Battye T.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. Sect D, Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Evans P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D, Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 75.Evans P.R. An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr. Sect. D, Biol. Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith K.M., Tsimbalyuk S., Edwards M.R., Cross E.M., Batra J., Soares da Costa T.P., et al. Structural basis for importin alpha 3 specificity of W proteins in Hendra and Nipah viruses. Nat. Commun. 2018;9:3703. doi: 10.1038/s41467-018-05928-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D, Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 79.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D, Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., et al. Phenix: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D, Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucl. Acids Res. 2019;47:W636–w641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucl. Acids Res. 2014;42:W320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the article or supporting information. Both the NMR and X-ray structural data were deposited into the Worldwide Protein Data Bank (wwPDB) via the OneDep system. The resonance assignment is listed in the BMRB with accession number BMRB ID: 34709. Coordinates and structure factors are located in the RCSB PDB with accession numbers PDB ID: 7YWQ (NMR-resolved 3D structure of d16TBEVC) and 8ECH (X-ray crystallography of TBEVC–IMPα complex).