Abstract
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in wastewater has been used to track community infections of coronavirus disease-2019 (COVID-19), providing critical information for public health interventions. Since levels in wastewater are dependent upon human inputs, we hypothesize that tracking infections can be improved by normalizing wastewater concentrations against indicators of human waste [Pepper Mild Mottle Virus (PMMoV), β-2 Microglobulin (B2M), and fecal coliform]. In this study, we analyzed SARS-CoV-2 and indicators of human waste in wastewater from two sewersheds of different scales: a University campus and a wastewater treatment plant. Wastewater data were combined with complementary COVID-19 case tracking to evaluate the efficiency of wastewater surveillance for forecasting new COVID-19 cases and, for the larger scale, hospitalizations. Results show that the normalization of SARS-CoV-2 levels by PMMoV and B2M resulted in improved correlations with COVID-19 cases for campus data using volcano second generation (V2G)-qPCR chemistry (rs = 0.69 without normalization, rs = 0.73 with normalization). Mixed results were obtained for normalization by PMMoV for samples collected at the community scale. Overall benefits from normalizing with measures of human waste depend upon qPCR chemistry and improves with smaller sewershed scale. We recommend further studies that evaluate the efficacy of additional normalization targets.
Keywords: SARS-CoV-2, COVID-19, PMMoV, B2M, fecal coliform, normalization
Short abstract
Improved correlations between wastewater SARS-CoV-2 RNA concentrations and COVID-19 cases and hospitalizations depend upon normalization target, qPCR chemistry, and watershed scale.
1.0. Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the virus responsible for the COVID-19 pandemic, continues to spread throughout the world, affecting human welfare since its initial outbreak in December 2019. Community-based clinical COVID case surveillance is the global standard for evaluating the state of the pandemic.1−3 However, clinical data has its limitations, which include restricted clinical testing capacity and access to testing.4 The efficacy of clinical tracking has been diminished due to widespread vaccinations against COVID-19 resulting in more asymptomatic or mild cases for which individuals will not seek testing.3,5,6 In contrast, wastewater-based epidemiology (WBE), also known as wastewater surveillance, has been shown to be a useful tool to predict potential outbreaks.7−15 In addition to saliva and nasal discharges, studies have shown that the RNA of the virus that causes COVID-19, SARS-CoV-2, can be found in feces and urine of both symptomatic and asymptomatic individuals.13−17 This phenomenon is like with other viruses, such as poliovirus—one of the initial focuses of WBE.18−20 Studies have shown that RNA within excreted SARS-CoV-2 viral particles can be reliably detected after sample collection and processing.21,22 A study in Brisbane indicated that wastewater SARS-CoV-2 RNA detection could predict COVID-19 outbreaks about a week before it was observed in clinical records.22 This forewarning can be used to initiate mitigation measures earlier to minimize disease transmission and to prepare communities for increases in hospitalization rates.
Wastewater is a combination of all waters that drain through a building. It includes waters from toilets, sinks, showers, dishwashing, clothes washing, and other indoor uses along with inflow and infiltration of outside water sources within the wastewater collection system. Not all sources of water to the sewer system would be expected to contain SARS-CoV-2. Human waste contributing SARS-CoV-2 to wastewater includes feces and urine, both associated with toilet water, and sputum and nasal discharge, which may be associated with water used for face washing (e.g., sink and shower water). Even these sources can have variable levels of SARS-CoV-2 depending upon the amount of water used per toilet flush or body washing. In efforts to better characterize wastewater containing human sources of SARS-CoV-2, indicators of human fecal waste or body fluids can be measured alongside SARS-CoV-2 concentrations. The contribution of SARS-CoV-2 from human sourced water can be estimated by normalizing (i.e., dividing) the measured SARS-CoV-2 concentration by the concentration of the human waste indicator. Two fecal indicators, typically examined in wastewater-based studies and used here for normalization of SARS-CoV-2 concentration, include Pepper Mild Mottle Virus (PMMoV) and fecal coliform bacteria. In addition, a human protein-coding gene, β-2 microglobulin (B2M), was assessed as it is expressed in all nucleated cells and is a marker of human origin.23 Utilizing three parameters to normalize the SARS-CoV-2 concentration in this study allowed for a more robust analysis of the wastewater data and provided this study with the capacity to compare effective normalization parameters for future wastewater-based detection of SARS-CoV-2.
PMMoV is often measured in combination with wastewater as it is known to be stable with concentrations showing little variation over time.24−29 PMMoV is an endemic plant virus with a dietary origin in humans from various pepper species,30,31 easily detectable within human feces, and also used as a human waste indicator.28,31 Moreover, PMMoV is a single-stranded RNA virus, like SARS-CoV-2, allowing for it to be processed in a similar fashion. Because of this, PMMoV has been demonstrated to be a robust indicator of fecal presence in numerous studies assessing treated and untreated wastewater as well as coastal waterways.24−27 Here, the quantified PMMoV concentrations were used to provide normalized reporting of the SARS-CoV-2 concentration to account for the proportion of human waste within the wastewater analyzed.
Fecal indicator bacteria, typically species from genus’ Enterobacter and Escherichia, are commonly found within wastewater and sewage systems and measured in laboratory settings by culturing on agar.32,33 Fecal coliforms were chosen because they have a long history of use as indicators of human waste. Their measurement also provides the potential for adding an additional normalization target not requiring molecular analyses at a reduced cost. In this study, fecal coliforms were used to normalize the SARS-CoV-2 concentration measurement to determine if the normalized parameter provides a better correlation with human COVID-19 cases.
B2M is found elevated in bodily fluids under circumstances of infection or inflammation and is typically used as a biological marker for patients with HIV or various cancers.34−36 In the case of coronavirus disease 2019 (COVID-19), researchers have shown that the virus impacts vital organs including intestinal epithelial cells, which can also release increased B2M due to inflammation. Consequently, correlations have been observed between SARS-CoV-2 and B2M shed from infected individuals.17 The B2M assay used in the current study targets the mature, spliced mRNA in human cells that have been excreted or been deposited into the drainage system by other means, such as washing.
In the current study, we evaluated relationships between COVID-19 incidence, and wastewater measurements through direct measurements of SARS-CoV-2 concentrations plus SARS-CoV-2 concentrations normalized to three targeted indicators of human fluids or wastes. Our aim was to compare normalization methods for WBE, overlay wastewater measurements with human health surveillance, and explore the efficiency of wastewater monitoring for forecasting new COVID-19 cases and hospitalizations. This study also provides comparative data between qPCR chemistries, Volcano second Generation (V2G) and Reverse Transcriptase (RT)-coupled qPCR. Since data were available from both local laboratories and an off-site commercial laboratory, results from this study also provided the opportunity to compare results between samples that were shipped versus those that were not shipped.
2.0. Materials and Methods
2.1. Sampling Sites and Wastewater Collection
Wastewater measurement data were collected through two primary sources. First was through a monitoring program that collected samples weekly from September 2020 to November 2021 from the University of Miami (UM) campus located in Coral Gables, FL USA and from January 2021 to November 2021 from Miami-Dade County’s Central District Wastewater Treatment Plant (CDWWTP). This first set of samples is referred to as the UM set. The second main source of data consisted of samples collected weekly from March 2020 to November 2021 from the Miami-Dade County (MDC) Water and Sewer Department (WASD), who contracted with Biobot to analyze wastewater from its three major wastewater treatment plants in MDC, the North District (service population of 776,150), Central District (service population of 829,725), and South District (service population of 920,528) wastewater treatment plants (WWTP). This second set of samples is referred to as the WASD set. The WASD samples were analyzed for SARS-CoV-2 and PMMoV. All WASD samples were flow-weighted composites collected using refrigerated autosamplers and were collected by MDC-WASD personnel and shipped overnight to Biobot Analytics for analysis.
The UM campus samples were collected at three locations (WG01, WG02, and WG0V) within the university representing the collective contribution from the Gables campus where the resident undergraduate students are housed and where most undergraduate courses are held. The wastewater contributions to all these sampling stations correspond to residential and administrative buildings only. The wastewater does include food processing wastes from the dormitory cafeterias and restaurants, after passing grease and solids interceptors. The undergraduate student population at the University of Miami Gables campus is 11,300 with 4200 on-campus resident students. A grab sample was collected at each of the locations at approximately the same time each Tuesday morning. The UM CDWWTP weekly samples consisted of two types, grab and composite samples. The grab sample (WC0Dg) was collected at approximately the same time each Tuesday morning. The composite sample (WC0Dc) consisted of 24, 1 h composite samples (HACH AS950 fitted with an IO9000 for flow proportional sampling) from 12 am (midnight) Monday morning to 12 pm (midnight) Monday night.
UM samples were analyzed for SARS-CoV-2, and three normalization parameters (PMMoV, B2M, and fecal coliform) plus in field measurements of water quality including turbidity and dissolved oxygen (Xylem YSI ProDSS). Laboratory analysis of the UM samples was facilitated by a multiple laboratory collaboration called South Florida Radical (SF-RAD), with each laboratory receiving sample splits. SF-RAD employed four laboratories. These laboratories include the UM Center for Aids Research (CFAR), the UM Sylvester Comprehensive Cancer Center (SCCC) Oncogenomic Shared Resource (OGSR), the Weill Cornell Medicine (WCM) genomics laboratory (WCM), and a commercial laboratory called Source Molecular, a Luminultra Company (SM).
2.2. Wastewater Analyses
UM samples were split among four molecular laboratories (CFAR, OGSR, WCM, SM) from September 2020 through January 2021 and among three molecular laboratories (CFAR, OGSR, WCM) from February to November 2021. A comparison of SARS-CoV-2 measurements among these laboratories is described in Babler et al. included within this Special Issue.37 Of these splits, one split occurred in the field upon sample collection and used by SM and concentrated by electronegative filtration (EN) and analyzed for SARS-CoV-2 N1 and N2 targets and PMMoV. The other split was preprocessed at the University of Miami SCCC Biospecimen Share Resource (BSSR) for fecal coliform quantification and for preparing sample concentrates for subsequent molecular analysis by CFAR, OGSR, and WCM. Fecal coliform was analyzed at the BSSR using mFC agar by culturing 1 and 0.1 mL aliquots from 100:1 dilution in sterile phosphate buffered saline as per standard methods (Method 9222D, APHA 2005) with quantification by counting colonies showing characteristic growth, with units defined as colony forming units (CFUs). Concentrates for subsequent molecular analyses were prepared at the BSSR via EN and were distributed to the three molecular laboratories.38,39 In brief the EN concentrates prepared in the BSSR began by volumetrically adjusting the sample volume to 500 mL, adding an internal recovery control, human coronavirus -OC43 (OC43) to 106 genomic copies (gc) per liter, adding MgCl2 to a concentration of 50 mM, and acidifying (10% HCl) to a pH between 3.5 and 4.5. Acidified aliquots were then filtered through an electronegative membrane (0.45 μm pore size, 47 mm diameter, Millipore HAWP4700) in triplicate. The filters were folded and placed in 5 mL Eppendorf tubes containing 1.5 mL of 1X DNA/RNA Shield (Zymo) and then distributed with one filter concentrate sent to each of the three laboratories (CFAR, OGSR, WCM). Samples sent to WCM were stored.
CFAR analyzed electronegative filter samples for levels of SARS-CoV-2 using volcano second generation (V2G)-qPCR. V2G is a polymerase that is capable of reading both RNA and DNA templates. As such, RNA targets were quantified directly by qPCR without a separate cDNA synthesis step.3 It also allowed us to avoid the competitive usage of commercial kits and single-step preparation master mix kits for qPCR measurements. CFAR extracted the EN filter concentrates using a Zymo Quick-RNA Viral Kit, a silica-based spin column approach, and spiked the eluted RNA with 800 cP/μL HIV RNA for downstream quality control measurements. The purified RNA was analyzed by V2G-qPCR for five molecular targets including a predetermined target (N3) in the nucleocapsid of SARS-CoV-2, B2M, and PMMoV, as mentioned above, plus two controls: OC43 (recovery control) and HIV (inhibition control). Results are reported as genomic copies per liter of raw wastewater.
OGSR analyzed samples for levels of SARS-CoV-2 using standard methods for reverse transcriptase (RT)-qPCR. The kit employed for RNA extraction of electronegative filter concentrates was a MagMAX Viral/Pathogen II Nucleic Acid Isolation Kit IFU, and the manual method for 200 μL sample input volume was utilized for sample processing. The quantification of RNA performed at OGSR followed the Applied Biosystems TaqPath COVID-19 Combo Kit protocol (https://www.fda.gov/media/136112/download). A PerkinElmer New Coronavirus Nucleic Acid Detection Kit IFU and corresponding protocol (https://www.fda.gov/media/136410/download) at a 20 μL reaction volume was utilized for the RT-qPCR analysis; all results are reported in genomic copies per liter of raw wastewater.
The WASD samples were analyzed by Biobot Analytics for levels of SARS-CoV-2 and PMMoV; at the laboratory, samples were pasteurized, concentrated, and quantified for the two molecular targets mentioned. In brief, samples were pasteurized to inactivate the ambient SARS-CoV-2 virus (90 °C for 60 min).29 Raw wastewater samples were then concentrated using the polyethylene glycol precipitation (PEG) method,40,41 which required centrifugation of samples (5 min) before PEG8000 (Sigma-Aldrich, Burlington, MA) and sodium chloride were added. The treated wastewater samples were incubated overnight at 4 °C on a shaker.40 A single 30 min centrifugation of the sample followed the overnight incubation, and concentrates were created following elution.40 The nucleic-acid measurement approach Biobot employed was RT-qPCR coupled with a TaqMan Gene Expression Master Mix following methods developed by Torii et al. 2021; a QIAamp Viral RNA Mini Kit was first utilized to extract and purify viral RNA from the wastewater concentrate samples.40,42 PMMoV was used as the internal control for SARS-CoV-2 for the WASD data set, and all results are reported in genomic copies per liter of raw wastewater, keeping consistent with UM data.
Date ranges of when each molecular target was measured varied between the UM data set and the WASD data set. SARS-CoV-2 was measured at UM from September 30, 2020 to November 9, 2021, whereas WASD began their surveillance of SARS-CoV-2 earlier in the pandemic, from March 26, 2020 to November 4, 2021. SM results were used for validating the methodology of the UM process during the inception of the surveillance program. SM assessed both SARS-CoV-2 and PMMoV from September 30, 2020 to February 16, 2021. CFAR began measuring the normalization parameter PMMoV internally from February 23, 2021 to November 9, 2021.
2.3. Clinical Data Sources
Clinical data was available at the campus level and at the county level. The University campus level data came from the University’s COVID-19 dashboard, which reports the number of positive cases among students and faculty/staff on a day-by-day basis.43 County level data was available through the Centers for Disease Control and Prevention public database and through the COVID ActNow website.44,45 The CDC data includes COVID-19 cases and hospitalizations. CDC data were available daily from March 2020 to June 2021. County-level clinical data were available on a weekly basis from June 2021 to November 2021.
Data collected from the CDC database included the Miami-Dade County 7-day moving average of positive case data and hospitalization data. The COVID ActNow website was used to consolidate vaccination data. Statistical distributions of wastewater and human health data were analyzed using the Shapiro–Wilk test. Results indicated that the data were not normal nor log-normal distributed. Regression analysis was therefore conducting using the nonparametric Spearman’s rank correlation, rs (SPSS version 26). Correlations were considered statistically significant for p < 0.05. Correlations were deemed “weak” for rs values less than 0.2.
3.0. Results and Discussion
3.1. University of Miami Campus Data
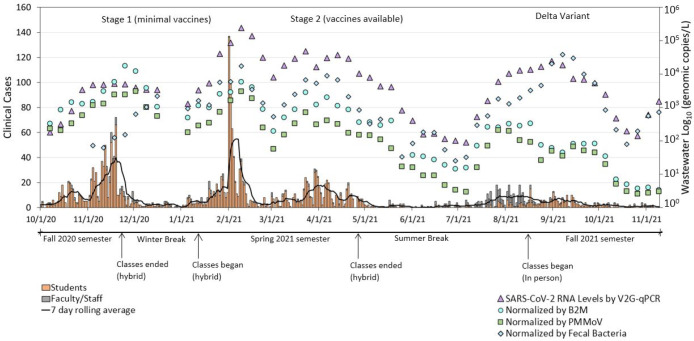
Results show that wastewater SARS-CoV-2 levels as measured by V2G-qPCR reflect the trends of COVID-19 clinical cases. These results from the current study are consistent with other studies that have demonstrated that wastewater-based surveillance is capable of estimating COVID-19 case dynamics in communities.3,4,22,46 Here, we built up the relationship between wastewater concentrations of SARS-CoV-2 and COVID-19 cases, demonstrating that viral titers in wastewater were connected showing similar trends between wastewater SARS-CoV-2 RNA levels and student and staff health data (Figure 1). During winter and summer breaks, the positive clinical cases were near zero and the wastewater SARS-CoV-2 levels were at or below detection limits of 102 gc/L. Wastewater SARS-CoV-2 trends at the UM campus also mirrored the COVID-19 clinical cases dynamic due to increases in COVID-19 cases observed at the start of the semesters. These results are consistent with a study in Ohio that found that levels of SARS-CoV-2 in wastewater coincided with the postholiday COVID-19 surge because of the increased family gathering and travel.7 It is likely that students upon returning to campus increase the probability of disease transmission due to prior travel-related exposures.
Figure 1.
Time series of human case data (students and faculty/staff) shown by stacked bar plots referencing the left arithmetic scale against SARS-CoV-2 wastewater data (raw and normalized by PMMoV, B2M, and fecal coliform) shown by symbols and referencing the right logarithmic scale. Seven-day moving average of clinical data shown by a gray line. Three-week moving average shown for wastewater SARS-CoV-2 data.
For the current study, four waves of elevated COVID-19 cases were observed on campus, one corresponding to the Fall 2020 semester, two corresponding to the Spring 2021 semester, and one corresponding to the Fall 2021 semester, which were reflected more strongly in the wastewater concentration than in the clinical case incidence. Wastewater SARS-CoV-2 levels reflected these four waves by showing increases and decreases that coincided with the human cases. In mid-April 2021, the University of Miami started to provide vaccines to all students and faculty. Wastewater levels, normalized RNA concentration, and positive cases dropped quickly after that time point and also coincided with the summer break during which fewer students resided on campus. In August 2021, the SARS-CoV-2 levels increased most likely due to the spread of the Delta variant in Miami. The wave observed during the end of July 2021 through September was more predominant among the faculty/staff than among the students, perhaps reflecting differences in susceptibilities to the Delta variant among these different populations and employees returning to work immediately prior to the start of the Fall 2021 semester. The wastewater SARS-CoV-2 concentration had a significant increase at the end of July 2021 as compared to positive clinical cases. However, the wastewater SARS-CoV-2 concentration continued to increase despite stabilization of new case counts. We believe that the relationship between SARS-CoV-2 in wastewater versus COVID-19 may have changed in July possibly due to the combined effects of vaccination and/or due to changes in shedding rates caused by the predominance of different variants. Regardless of the changing dynamics of SARS-CoV-2 concentrations and case study data, overall, the correlations between wastewater and COVID-19 cases were significant (rs = 0.69, p < 0.001) when wastewater was measured using V2G-qPCR. Measurements of wastewater by RT-qPCR also resulted in significant correlations but weaker than for V2G-qPCR (rs = 0.39 for N1 and 0.40 for ORF1ab) (Table 1).
Table 1. Spearman Correlation Coefficients (rs) and Significance p Values of Wastewater Measures (3-week moving average) against Clinical Case Data (7-day moving average) for UM Campus Dataa.
| rs | p | |
|---|---|---|
| V2G-qPCR | ||
| SARS-CoV-2 (N3) | 0.69 | <0.001 |
| PMMoV | –0.08 | 0.598 |
| B2M | –0.14 | 0.316 |
| fecal coliform by culture | 0.16 | 0.267 |
| SARS-CoV-2/PMMoV | 0.73 | <0.001 |
| SARS-CoV-2/B2M | 0.71 | <0.001 |
| SARS-CoV-2/fecal coliform | 0.35 | 0.012 |
| RT-qPCR | ||
| SARS-CoV-2 (N1) | 0.39 | 0.024 |
| SARS-CoV-2 (N1)/PMMoV | 0.36 | 0.010 |
| SARS-CoV-2 (N1)/B2M | 0.30 | 0.088 |
| SARS-CoV-2 (N1)/fecal coliform | 0.31 | 0.077 |
| SARS-CoV-2 (ORF1ab) | 0.40 | 0.015 |
| SARS-CoV-2 (ORF1ab)/PMMoV | 0.38 | 0.024 |
| SARS-CoV-2 (ORF1ab)/B2M | 0.31 | 0.064 |
| SARS-CoV-2 (ORF1ab)/fecal coliform | 0.41 | 0.014 |
B2M and PMMoV analyzed by V2G-qPCR.
Studies conducted at other Universities have shown that building level wastewater surveillance could lead to the identification of COVID-19 cases with sensitivities as low as one asymptomatic case per 200 resident population.47,48 Wastewater samples obtained daily from manholes serving dormitories at the University of Arizona (UA) were determined to be positive for SARS-CoV-2 at an average concentration of 1.1 × 106 gc/L at a roughly 1% positivity rate.48 The collective concentration for the entire UM university campus reached 2.5 × 105 gc/L on February 9, 2021. Given the relationships observed at the UA, less than 42 cases would have been observed at UM during the peak. A model developed for the UM campus (percent positivity = 0.9 × ln(C), where C = gc/L for SARS-CoV-2 in wastewater)3 suggests that, on the basis of the Fall 2020 data, about 470 cases would have been observed among the UM student resident population. The results from UA and UM are just outside an order of magnitude of one another. At its peak, the maximum one-day positive detection was at 137 on February 1 with a maximum 7-day moving average of 55, which is low compared to the case counts predicted from the Fall 2020 UM model and high compared to the UA data. All results were generally consistent. For University campuses showing SARS-CoV-2 levels in wastewater between 105 and 106 gc/L, between a 1% to 12% positivity rate would be expected among residents contributing to the wastewater. This is a wide range of positivity rates. Since levels of SARS-CoV-2 in wastewater are dependent upon wastewater flows, future studies should include measures of wastewater flow rates in order to compare fluxes of SARS-CoV-2 (gc per unit time) as opposed to concentrations (gc per volume of wastewater). The inclusion of flow data may help to reduce the uncertainty in estimating positivity rates from wastewater measurements.
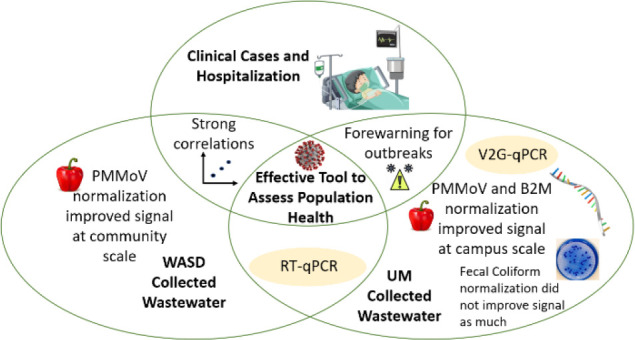
We also compared the raw PMMoV, B2M, and fecal coliform trends with COVID-19 cases at the UM campus, but the wastewater concentration waves appeared after the peaks in the clinical cases; correlations were low (rs ≤ 0.16) between the fecal/human indicators and clinical cases, and these correlations were not significant (p > 0.05) (Table 1). This is not surprising as only individuals infected with COVID-19 shed SARS-CoV-2 whereas all individuals shed their waste. As such, improvements were observed when the fecal/human indicators were used for normalizing the SARS-CoV-2 measures. When normalizing the SARS-CoV-2 concentrations by the fecal/human waste indicators, results show as expected that the trend of normalized SARS-CoV-2 measured from wastewater almost mirrors the dynamics of the clinical cases in the community (Figure 1). At first, the trends were similar between non-normalized and normalized SARS-CoV-2, especially for the normalization by PMMoV and B2M. Normalization by fecal coliform tends to result in deviations from the clinical case study data during the October to November 2020 time frame. The SARS-CoV-2 levels normalized by fecal coliform provided low correlations (rs = 0.35); this observation illustrated that the normalization method utilizing fecal coliform was not as useful for estimating clinical cases using wastewater SARS-CoV-2 concentrations. However, when wastewater SARS-CoV-2 concentrations were normalized by PMMoV and B2M, its correlations with COVID-19 cases at the campus scale increased (rs = 0.73 for SARS-CoV-2 normalized by PMMoV, and rs = 0.71 for SARS-CoV-2 normalized by B2M) as compared to non-normalized SARS-CoV-2 (rs = 0.69). Of particular interest is the usefulness of the PMMoV and B2M normalization toward the end of the period of record, between July and November 2021, when the clinical case counts dropped. This drop was not reflected by the non-normalized SARS-CoV-2 concentration and thus drove the higher correlations between clinical case data and SARS-CoV-2 normalized by PMMoV and B2M. Given the improvement in correlations, the use of these two-normalization targets for future campus level wastewater-based surveillance research is recommended.
Among the three fecal/human indicator targets evaluated in this study, only one (PMMoV) has been used to normalize pathogenic viral levels concentrated from wastewater. We are unaware of studies that use either B2M or fecal coliform to normalize SARS-CoV-2 wastewater concentrations. For PMMoV, one study found a strong positive correlation between the recovery loads of murine hepatitis virus (MHV) and PMMoV (R2 = 0.70, p < 0.01), indicating that PMMoV has potential as an indicator for the recovery efficiency of MHV. The advantages of monitoring the PMMoV are its simplicity and practicality.41 Like SARS-CoV-2, PMMoV is a positive-sense, single-stranded RNA virus so extracted RNA from wastewater samples can be used for both targets using very similar qPCR techniques.
B2M has the same advantages as PMMoV, as it is also an RNA molecule that copurifies with SARS-CoV-2 RNA. Thus, the extracted wastewater RNA prepared for SARS-CoV-2 analysis can also be analyzed by qPCR for the B2M target. An important aspect of B2M is that it is found in most cells (e.g., skin, hair, blood) and in body fluids such as saliva where SARS-CoV-2 is also known to accumulate.36,49 In addition to indicating fecal and urine contributions, B2M would indicate discharges from saliva (mouth washing in sinks), plus surface skin and hair washing.50 B2M differs from PMMoV in that PMMoV is a purely fecal indicator and linked to the dietary traditions (i.e., consumption of peppers) of the community contributing wastewater.26,28 B2M comes from a larger range of body fluids and is elevated in humans when they are experiencing inflammation, immune response, and illness.34−36,50 Given these characteristics and the promising results observed in the current study, B2M should be further considered as a possible human indicator in wastewater for normalizing SARS-CoV-2 RNA concentrations. It is possible that, for WBE, B2M may be a more direct indicator of human influence than the more traditional fecal indicators.
Fecal coliform, on the contrary, did not provide a benefit for the normalization of SARS-CoV-2 concentrations. Fecal coliform in this study was quantified by counting colonies grown on agar. Additional drawbacks from culture-based fecal coliform measurements are that fecal bacteria can multiply in the environment,51,52 including within sewer systems. The wastewater collected at the campus level did contain settled food waste from cafeterias, and it is possible that, in addition to fecal material, this food waste could serve as a substrate for bacterial growth. It is also well-known that culture-based detection of fecal coliform is susceptible to chlorination.53−55 The water supply of Miami-Dade County comes from centralized plants that provide a combined chlorine residual of at least 0.2 mg/L at the point of use. It is well-known that these levels of chlorine can inactivate fecal coliform especially when fecal inputs are low.53 It was common for wastewater harvested from the campus-level sanitary sewer system to be free of fecal coliform colonies, suggesting levels less than 100 CFU/L. Water quality within the sewers was highly variable with turbidities ranging from 4 to 3400 nephelometric turbidity units and dissolved oxygen from 0.8 to 8.3 mg/L (see Sharkey et al. 2021 and Babler et al. in this special issue for additional water quality data),3,37 suggesting that some samples have low organic inputs especially for water samples with low turbidity and high dissolved oxygen levels. It is likely that the chlorine in the tap water servicing MD-WASD (or other disinfectants used for cleaning) inactivated the fecal coliform, interfering with its ability to serve as an indicator at low levels of feces, especially for small sewersheds that are subjected to a large range of water quality variations with highly variable levels of fecal inputs. This is contrasted by the RNA of SARS-CoV-2. Although the SARS-CoV-2 virus is inactivated by chlorine,56 the measurement technique employed in this study does not require that the virus remain viable for detection. The RNA can be detected from both nonviable and viable SARS-CoV-2. Thus, it is likely, especially at low levels of fecal inputs, that the fecal coliform were inactivated due to chlorination whereas SARS-CoV-2 persisted.
Therefore, among the fecal/human waste indicators evaluated in this study, PMMoV and B2M are recommended as normalization parameters at the campus watershed scale. Their quantification should be included when evaluating campus level sized sewersheds, as they have the potential for providing more accurate estimates of human case counts when combined with measures of SARS-CoV-2.
3.2. WASD Data from Miami-Dade County
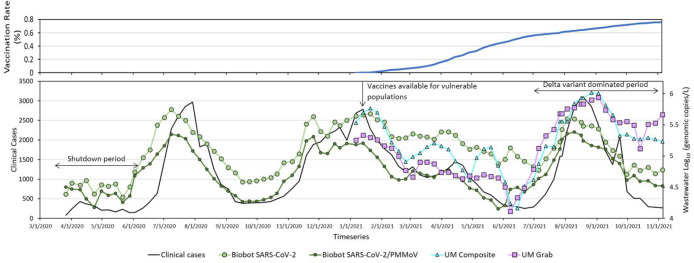
The exponential rise in clinical cases in Miami-Dade County at the first stage of the pandemic started in June 2020, followed by a rapid decline through August, with cases rising again in the winter toward November 2020, remaining elevated through April 2021, and dipping during the April through July time frame until the next peak was observed during August 2021. The comparison of WASD wastewater data showed similar waves of SARS-CoV-2 levels. For the first and second waves of SARS-CoV-2, increases were about 2 weeks before the peak of COVID 19 cases. After the second wave, the wastewater data generally mirrored trends of clinical data likely due to increased COVID-19 testing in the county (Figure 2). Quantitatively, human case numbers were significantly correlated (p < 0.001) with all combinations of the WASD wastewater measures, whether they were normalized by PMMoV or evaluated for the full or partial period of record corresponding to the UM data set (Table 2). The correlation coefficients were similar for the shorter period of WASD data set (rs = 0.86) compared to the longer period data set (rs = 0.88). For the WASD data set, normalization with PMMoV does not appear to have improved the correlation with human case data with rs values of 0.76 and 0.79 for the shorter versus the longer period of record (Table 2).
Figure 2.
Miami-Dade County full (two shots) vaccination rate (top smaller panel) and case data (7-day moving average shown by heavy black line). Three-day moving average wastewater data shown on bottom panel, including WASD raw SARS-CoV-2 RNA concentrations, WASD normalized SARS-CoV-2 RNA concentrations, and UM raw SARS-CoV-2 RNA concentrations for composite and grab samples. WASD data corresponds to weighted average SARS-CoV-2 for measurements taken at all three wastewater treatment plants, whereas the UM SARS-CoV-2 corresponds to measurements taken at the CDWWTP only.
Table 2. Spearman Correlation Coefficients (rs) and Significance p Values of Wastewater Measures (3-week moving average) against Clinical Case Data (7-day moving average) for WASD and UM County-Level Dataa.
| UM composite (Jan 2021-Nov 2021) |
UM grab (Jan 2021-Nov 2021) |
|||
|---|---|---|---|---|
| wastewater measure | rs | p value | rs | p value |
| V2G-qPCR | ||||
| SARS-CoV-2 (N3) | 0.69 | <0.001 | 0.49 | <0.001 |
| PMMoV | 0.07 | 0.642 | 0.16 | 0.308 |
| B2M | 0.01 | 0.925 | 0.06 | 0.710 |
| fecal coliform | 0.00 | 1.00 | 0.13 | 0.396 |
| SARS-CoV-2/PMMoV | 0.76 | <0.001 | 0.54 | <0.001 |
| SARS-CoV-2/B2M | 0.86 | <0.001 | 0.71 | <0.001 |
| SARS-CoV-2/fecal coliform | 0.59 | <0.001 | 0.17 | 0.259 |
| RT-qPCR | ||||
| SARS-CoV-2 (N1) | 0.38 | 0.012 | 0.56 | <0.001 |
| SARS-CoV-2 (N1)/PMMoV | 0.24 | 0.129 | 0.52 | <0.001 |
| SARS-CoV-2 (N1)/B2M | 0.17 | 0.298 | 0.50 | <0.001 |
| SARS-CoV-2 (N1)/fecal coliform | 0.35 | 0.022 | 0.34 | 0.026 |
| SARS-CoV-2 (ORF1ab) | 0.35 | 0.022 | 0.54 | <0.001 |
| SARS-CoV-2 (ORF1ab)/PMMoV | 0.27 | 0.090 | 0.48 | 0.001 |
| SARS-CoV-2 (ORF1ab)/B2M | 0.22 | 0.168 | 0.48 | 0.001 |
| SARS-CoV-2 (ORF1ab)/fecal coliform | 0.36 | 0.021 | 0.28 | 0.071 |
| WASD Composite (March 2020- Nov 2021) | WASD Composite (Jan 2021-Nov 2021) | |||
|---|---|---|---|---|
| SARS-CoV-2 | 0.88 | <0.001 | 0.86 | <0.001 |
| SARS-CoV-2/PMMoV | 0.79 | <0.001 | 0.76 | <0.001 |
Comparisons provided for composite versus grab samples for UM samples and between consistent periods of record for both UM and WASD data (January 2021 to November 2021). The statistics are also provided for the complete period of record (March 2020 to November 2021) for the WASD data. B2M and PMMoV analyzed by V2G-qPCR. WASD SARS-CoV-2 levels correspond to weighted average (by population served) for all three major wastewater treatment plants in the county, whereas the UM data corresponds to SARS-CoV-2 levels measured at the CDWWTP only.
In terms of the timing of the clinical versus wastewater peaks, results are consistent with a study conducted in the Boston area that showed different relationships between the first and second waves. In the first pandemic phase in Boston, the clinical cases peaked shortly after wastewater peaked, while the approximately 6-day time lag between them did not present in the second wave.4 In Miami-Dade County, WASD wastewater concentration also peaked before clinical cases and showed no obvious early peak in the second wave. However, in the third wave, wastewater presented an early wave shortly before the clinical cases, which might be due to the dynamics of the Delta variant (Figure 2).
We also used the wastewater and human health data to compare the local policy changes and COVID-19 variant spread. The public health measures like the mandatory indoor masking, online work or class, temporarily closing restaurants and stores, etc., efficiently prevented the spread of infection (shutdown period-initiated March 17 in MDC).57 However, with reopening of the economy (i.e., restaurants and gyms, reopening-initiated June 4),57 cases increased and peaked in July 2020. It is not clear why the levels declined during August 2020. Beginning in January 2021, vaccination was available; gradually rising vaccination rates in the county lead to the decrease of clinical cases and wastewater SARS-CoV-2 RNA concentrations (gc/L) through May 2021. Because of the dominance of the new variant, Delta, case numbers rose quickly after June in 2021.
3.3. UM Central District Wastewater Data
The comparison of non-normalized and normalized composite and grab data for UM CDWWTP samples shows a similar trend with MDC human COVID-19 cases (Figure 2, Table 2). In general, strong rs values were observed for the raw SARS-CoV-2 concentration (rs = 0.69 for UM composite samples and rs = 0.49 for UM grab samples) as measured by V2G-qPCR. Correlations based upon RT-qPCR were not as strong (e.g., rs = 0.38 and 0.56 for the N1 gene target composite and grab samples, respectively, and rs = 0.35 and 0.54 for the ORF1ab composite and grab samples, respectively). Overall, V2G-qPCR composite samples provide a better correlation with human case data than did the grab samples. However, the opposite was observed for the RT-qPCR results, which in general provided lower rs values than for the V2G-qPCR. Results show that normalization of SARS-CoV-2 concentrations resulted in an increase in the predictive ability of the SARS-CoV-2 to indicate clinical case data. For example, for the composite sample, the rs increased to 0.76 for normalization with PMMoV, was 0.86 for normalization with B2M, and decreased to 0.59 for normalization with fecal coliform. In this case, the use of the fecal/human waste indicators PMMoV and B2M improved the correlations with human health, whereas normalization with fecal coliform reduced correlations. As mentioned above, fecal coliform has added disadvantages due to its susceptibility to chlorination in the tap water, and consistent with the above, provided the poorest correlations. Overall results show that the highest rs values for UM samples collected at the UM Central District Wastewater Treatment Plant correspond to V2G-qPCR results for composite samples normalized by B2M.
A comparison of UM against WASD data suggests that both provided similar results. Using the sample period of record (January to November 2021) and comparing composite samples (WASD only analyzed composite samples), the correlation coefficients were the same for SARS-CoV-2 normalized by PMMoV (rs were 0.76 for both). When comparing the non-normalized values, the rs for the UM sample set was 0.69, whereas that for the WASD set was 0.88. In the case of UM, unlike WASD, normalization of the SARS-CoV-2 concentration by PMMoV did result in a benefit, increasing the correlation between the PMMoV normalized SARS-CoV-2 concentration to a value of 0.76. Such results suggest that, for the large community sewershed level, normalization by PMMoV or B2M may be beneficial for increasing the correlation between wastewater measures and human clinical cases. In the case of UM, the analysis process benefited from the normalization. The WASD samples did not benefit from the normalization. Overall correlations between the UM set and the WASD set were comparable, indicating that locally analyzed samples and shipped samples provided similar results.
3.4. Hospitalization
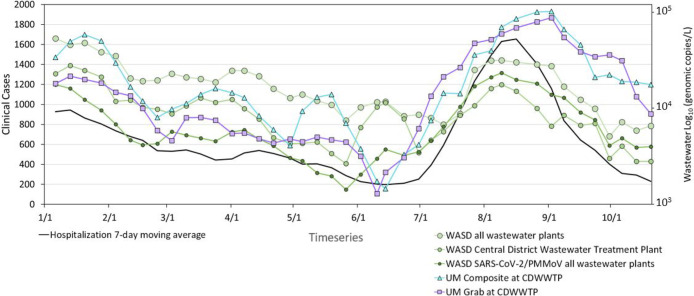
The hospitalization data shows decreasing COVID bed use in hospitals due to the rising rate of vaccination, while in the summer 2021 (July), the hospitalization incidence went up almost exponentially (Figure 3) due to the spread of the Delta variant, which resulted in hospitalizations of the primarily unvaccinated subpopulation.
Figure 3.
Comparison of moving average values between hospitalization (7-day moving average) and wastewater (3-week moving average) in Miami-Dade County detected by WASD and UM laboratories. WASD MDC wastewater includes three wastewater treatment plans in Miami-Dade County and the WASD Central wastewater data only sampled from the Central District Wastewater Treatment plan.
These hospitalization trends match with the raw wastewater concentration data of WASD and UM V2G all with statistically significant correlations (p < 0.001) and a Spearman coefficient as high as 0.76 for the non-normalized UM V2G-qPCR data set. The non-normalized UM RT-qPCR data set provided weaker correlations (rs from 0.21 to 0.23). The WASD raw SARS-CoV-2 data had strong correlations with the weighted average of the PMMoV normalized SARS-CoV-2 data for all three wastewater plants providing a correlation of 0.75. If the data came from only the CDWWTP, the correlation was lower at 0.61.
When the wastewater data were normalized by indicators of human waste, the correlations with hospitalizations increased for the WASD data to rs values as high as 0.90. However, PMMoV and fecal coliform normalization did not help for the UM data set, where correlations with hospitalization rates decreased when SARS-CoV-2 was divided by the PMMoV or fecal coliform measurements (Table 3). However, correlations did increase when the UM V2G-qPCR was normalized by B2M, increasing the rs to 0.83. These results are consistent with those described in the prior section showing that correlations between hospitalizations and the UM wastewater improved by normalization of the wastewater SARS-CoV-2 concentration with B2M.
Table 3. Spearman Correlation Coefficients (rs) and Significance p Values of Wastewater Measures (composite and grab samples) against Hospitalization in Miami-Dade County (7-day average)a.
| composite |
grab |
|||
|---|---|---|---|---|
| wastewater measure | rs | p value | rs | p value |
| WASD SARS-CoV-2 for three Miami-Dade County WWTP | ||||
| SARS-CoV-2 | 0.75 | <0.001 | NA | NA |
| SARS-CoV-2/PMMoV | 0.90 | <0.001 | NA | NA |
| WASD SARS-CoV-2 for CDWWTP | ||||
| SARS-CoV-2 | 0.61 | <0.001 | NA | NA |
| SARS-CoV-2/PMMoV | 0.76 | <0.001 | NA | NA |
| UM SARS-CoV-2 for CDWWTP | ||||
| SARS-CoV-2 (N3, V2G) | 0.76 | <0.001 | 0.77 | <0.001 |
| SARS-CoV-2 (N3, V2G)/PMMoV | 0.66 | <0.001 | 0.72 | <0.001 |
| SARS-CoV-2 (N3, V2G)/B2M | 0.83 | <0.001 | 0.87 | <0.001 |
| SARS-CoV-2 (N3, V2G)/fecal coliform | 0.26 | 0.102 | 0.34 | 0.030 |
| SARS-CoV-2 (N1, RT) | 0.21 | 0.175 | 0.30 | 0.056 |
| SARS-CoV-2 (N1, RT)/PMMoV | 0.00 | 0.998 | 0.36 | 0.024 |
| SARS-CoV-2 (N1, RT)/B2M | –0.04 | 0.788 | 0.32 | 0.044 |
| SARS-CoV-2 (N1, RT)/fecal coliform | 0.33 | 0.041 | 0.21 | 0.200 |
| SARS-CoV-2 (ORF1ab, RT) | 0.23 | 0.145 | 0.30 | 0.052 |
| SARS-CoV-2 (ORF1ab, RT)/PMMoV | 0.06 | 0.72 | 0.33 | 0.040 |
| SARS-CoV-2 (ORF1ab, RT)/B2M | 0.02 | 0.908 | 0.29 | 0.072 |
| SARS-CoV-2 (ORF1ab, RT)/fecal coliform | 0.26 | 0.102 | 0.14 | 0.383 |
Wastewater data correspond to three-week moving average. Period of record for all correlation analyses corresponds to January to November 2021. B2M and PMMoV analyzed by V2G-qPCR.
A comparison of the wastewater SARS-CoV-2 RNA concentration (gc/L) and hospitalization shows a surprisingly closer relationship than it did with clinical cases. This higher correlation is driven by the large peak in hospitalizations associated with the Delta variant during the July to August 2021 time frame. During this period, the wastewater SARS-CoV-2 levels, as measured by WASD and UM, also corresponded to very high levels, mirroring the impacts to the hospitals. As observed for the campus data (Figure 1), the last wave of COVID-19 associated with the Delta variant had much higher levels of SARS-CoV-2 in the wastewater than what would have been anticipated from the clinical case data. But, this last wave of SARS-CoV-2 associated with the Delta variant was associated with unprecedented levels of hospitalizations, especially among the unvaccinated due to the presumed higher virulence of disease.58 For this reason, the stronger increase in SARS-CoV-2 in the wastewater facilitated the higher correlations with hospitalizations.
Overall results show that wastewater surveillance can help hospitals prepare for the peak hospitalization capacity, by prioritizing hospital beds and distribution of medical supplies on the basis of the levels observed within wastewater. Normalization was observed to improve the correlations with hospitalizations. Normalization with PMMoV resulted in improvements for the WASD data set, and normalization with B2M resulted in improvements for the UM data set.
4.0. Conclusion
Wastewater monitoring is an important tool for COVID-19 tracking, and the relationships between wastewater SARS-CoV-2 levels and clinical cases change over time on the basis of public health response and possibly due to changes in variant dynamics. In this study, the wave of the wastewater SARS-CoV-2 levels during the early stages of the pandemic was about a week earlier than the clinical cases wave, and in the following pandemic period, the timing was more closely tied to the timing of the human health data. Wastewater SARS-CoV-2 RNA concentration at the campus levels benefited from PMMoV or B2M normalization methods to provide a more accurate tracking of COVID-19 cases. At the community level, normalization of the SARS-CoV-2 concentration had mixed results in its efficacy to track COVID-19 cases. For the samples collected at a major wastewater treatment plant, those analyzed locally and immediately upon collection (UM set) benefited from normalization whereas samples that required shipping and analysis (WASD set) did not benefit from normalization. Overall results from both sets of samples provided similar correlations with human health suggesting that samples analyzed locally provided similar results to those that were shipped.
Hospitalization data was more strongly correlated with the wastewater SARS-CoV-2 RNA concentration data. Normalization with PMMoV improved the correlations observed from the WASD data set. Normalization with B2M improved the V2G correlations observed from the local UM laboratory analyses. Overall, wastewater monitoring can be used to estimate COVID-19 clinical cases and can forecast hospitalization peaks, providing critical preparation time for communities to respond to an outbreak. This forecasting ability can be improved through the normalization of the raw SARS-CoV-2 measurements.
Limitations of this study include possible incomplete human testing results (not all individuals were tested or reported positive tests), thereby introducing bias toward under-estimating the degree to which the community is infected. This study would be strengthened from the inclusion of wastewater flow measurements at the campus level when developing the model relating case positivity to wastewater concentration measures. In addition, the three-week average for wastewater data limits the predictive capability of the wastewater measure. Future studies should include daily measurements of the SARS-CoV-2 signal to allow for averaging over shorter time scales. To further develop normalization procedures for WBE purposes, the evaluation of additional indicators of human fecal waste is recommended. Additional indicators can include the DNA virus crAssphage or human Bacteroidales marker HF183,27,59 among other fecal indicators that have been included in other studies. A more comprehensive study of potential normalization parameters is recommended to develop standards for normalizing microbial shedding of pathogens into wastewater as a means to estimate population infection rates.
Acknowledgments
The research reported in this publication was supported by the National Institute on Drug Abuse of the National Institutes of Health (NIH) under Award Number U01DA053941. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Additional support for this study was received from the UM administration, with in-kind contributions from University Facilities, University Environmental Health and Safety, and UM Health Safety Division. Laboratory facilities and support were made available in-kind through the Sylvester Comprehensive Cancer Center, the Miami Center for AIDS Research, the Miami Clinical and Translational Science Institute, and the UM College of Engineering Laboratories. The authors are thankful to Felix Nguyen and Olutosin Lawal for assistance with literature reviews. The authors are also thankful to Gabriella Cosculluela, Dannie Mackler, Erik Lamm, Chris Moree, Helen Kattoura, Ashley Lewis, Shashana Fiedler, and Thomas Stone for assistance in the field and laboratory.
The authors declare no competing financial interest.
References
- Black J.; Aung P.; Nolan M.; Roney E.; Poon R.; Hennessy D.; Crosbie N. D.; Deere D.; Jex A. R.; John N.; Baker L.; Scales P. J.; Usher S. P.; McCarthy D. T.; Schang C.; Schmidt J.; Myers S.; Begue N.; Kaucner C.; Thorley B.; Druce J.; Monis P.; Lau M.; Sarkis S. Epidemiological evaluation of sewage surveillance as a tool to detect the presence of COVID-19 cases in a low case load setting. Sci. Total Environ. 2021, 786, 147469. 10.1016/j.scitotenv.2021.147469. [DOI] [Google Scholar]
- Juel M. A. I.; Stark N.; Nicolosi B.; Lontai J.; Lambirth K.; Schlueter J.; Gibas C.; Munir M. Performance evaluation of virus concentration methods for implementing SARS-CoV-2 wastewater based epidemiology emphasizing quick data turnaround. Sci. Total Environ. 2021, 801, 149656. 10.1016/j.scitotenv.2021.149656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey M. E.; Kumar N.; Mantero A. M. A.; Babler K. M.; Boone M. M.; Cardentey Y.; Cortizas E. M.; Grills G. S.; Herrin J.; Kemper J. M.; Kenney R.; Kobetz E.; Laine J.; Lamar W. E.; Mader C. C.; Mason C. E.; Quintero A. Z.; Reding B. D.; Roca M. A.; Ryon K.; Solle N. S.; Schurer S. C.; Shukla B.; Stevenson M.; Stone T.; Tallon J. J. Jr; Venkatapuram S. S.; Vidovic D.; Williams S. L.; Young B.; Solo-Gabriele H. M. Lessons learned from SARS-CoV-2 measurements in wastewater. Sci. Total Environ. 2021, 798, 149177. 10.1016/j.scitotenv.2021.149177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A.; Wu F.; Bushman M.; Zhang J.; Imakaev M.; Chai P. R.; Duvallet C.; Endo N.; Erickson T. B.; Armas F.; Arnold B.; Chen H.; Chandra F.; Ghaeli N.; Gu X.; Hanage W. P.; Lee W. L.; Matus M.; McElroy K. A.; Moniz K.; Rhode S. F.; Thompson J.; Alm E. J. Metrics to relate COVID-19 wastewater data to clinical testing dynamics. medRxiv 2022, 212, 118070. 10.1016/j.watres.2022.118070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaTurner Z. W.; Zong D. M.; Kalvapalle P.; Gamas K. R.; Terwilliger A.; Crosby T.; Ali P.; Avadhanula V.; Santos H. H.; Weesner K.; Hopkins L.; Piedra P. A.; Maresso A. W.; Stadler L. B. Evaluating recovery, cost, and throughput of different concentration methods for SARS-CoV-2 wastewater-based epidemiology. Water Res. 2021, 197, 117043. 10.1016/j.watres.2021.117043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo D.; Quintela-Baluja M.; Corbishley A.; Jones D. L.; Singer A. C.; Graham D. W.; Romalde J. L. Making waves: Wastewater-based epidemiology for COVID-19 - approaches and challenges for surveillance and prediction. Water Res. 2020, 186, 116404. 10.1016/j.watres.2020.116404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai Y.; Davis A.; Jones D.; Lemeshow S.; Tu H.; He F.; Ru P.; Pan X.; Bohrerova Z.; Lee J. Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Sci. Total Environ. 2021, 801, 149757. 10.1016/j.scitotenv.2021.149757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldovin T.; Amoruso I.; Fonzo M.; Buja A.; Baldo V.; Cocchio S.; Bertoncello C. SARS-CoV-2 RNA detection and persistence in wastewater samples: An experimental network for COVID-19 environmental surveillance in Padua, Veneto Region (NE Italy). Sci. Total Environ. 2021, 760, 143329. 10.1016/j.scitotenv.2020.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcelo D. Wastewater-Based epidemiology to monitor COVID-19 outbreak: Present and future diagnostic methods to be in your radar. Case Studies in Chem. and Env Eng. 2020, 2, 100042. 10.1016/j.cscee.2020.100042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W. Q.; Schmitz B. W.; Innes G. K.; Prasek S. M.; Pogreba Brown K. M.; Stark E. R.; Foster A. R.; Sprissler R. S.; Harris D. T.; Sherchan S. P.; Gerba C. P.; Pepper I. L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021, 779, 146408. 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R.; Curtis K.; Bivins A.; Bibby K.; Weir M. H.; Yetka K.; Thompson H.; Keeling D.; Mitchell J.; Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020, 186, 116296. 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay L.; Amoah I. D.; Deepnarain N.; Pillay K.; Awolusi O. O.; Kumari S.; Bux F. Monitoring changes in COVID-19 infection using wastewater-based epidemiology: A South African perspective. Sci. Total Environ. 2021, 786, 147273. 10.1016/j.scitotenv.2021.147273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W.; Cuevas-Ferrando E.; Sanjuan R.; Domingo-Calap P.; Sanchez G. Metropolitan wastewater analysis for COVID-19 epidemiological surveillance. Int. J. Hyg Environ. Health 2020, 230, 113621. 10.1016/j.ijheh.2020.113621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan S. P.; Shahin S.; Ward L. M.; Tandukar S.; Aw T. G.; Schmitz B.; Ahmed W.; Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Sci. Total Environ. 2020, 743, 140621. 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z.; Reinke R.; Hoxsey M.; Jackson J.; Krikorian E.; Melitas N.; Rosso D.; Jiang S. Detection of SARS-CoV-2 in wastewater: Community variability, temporal dynamics, and genotype diversity. Environ. Sci. Technol. 2021, 1 (8), 1816–1825. 10.1021/acsestwater.1c00119. [DOI] [Google Scholar]
- Holshue M. L.; DeBolt C.; Lindquist S.; Lofy K. H.; Wiesman J.; Bruce H.; Spitters C.; Ericson K.; Wilkerson S.; Tural A.; Diaz G.; Cohn A.; Fox L.; Patel A.; Gerber S. I.; Kim L.; Tong S.; Lu X.; Lindstrom S.; Pallansch M. A.; Weldon W. C.; Biggs H. M.; Uyeki T. M.; Pillai S. K. First Case of 2019 Novel Coronavirus in the United States. N Engl J. Med. 2020, 382 (10), 929–936. 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z.; Xiaoyun S.; Luoying W.; Xingcheng Y.; Yue Q.; Xirui Y.; Xuan Z.; et al. The molecular mechanism of multiple organ dysfunction and targeted intervention of COVID-19 based on time-order transcriptomic analysis. Front. Immunol. 2021, 12, 729776. 10.3389/fimmu.2021.729776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer A. F.; Eisenberg J. N. S.; Pomeroy C. D.; Shulman L. M.; Hindiyeh M.; Manor Y.; Grotto I.; Koopman J. S.; Eisenberg M. C. Epidemiology of the silent polio outbreak in Rahat, Israel, based on modeling of environmental surveillance data. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (45), E10625–E10633. 10.1073/pnas.1808798115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman L. M.; Manor Y.; Handsher R.; Delpeyroux F.; McDonough M. J.; Halmut T.; Silberstein I.; Alfandari J.; Quay J.; Fisher T.; Robinov J.; Kew O. M.; Crainic R.; Mendelson E. Molecular and antigenic characterization of a highly evolved derivative of the type 2 oral poliovaccine strain isolated from sewage in Israel. J. Clin Microbiol 2000, 38 (10), 3729–34. 10.1128/JCM.38.10.3729-3734.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman L. M.; Manor Y.; Sofer D.; Handsher R.; Swartz T.; Delpeyroux F.; Mendelson E. Neurovirulent vaccine-derived polioviruses in sewage from highly immune populations. PLoS One 2006, 1, e69 10.1371/journal.pone.0000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W.; Bertsch P. M.; Bibby K.; Haramoto E.; Hewitt J.; Huygens F.; Gyawali P.; Korajkic A.; Riddell S.; Sherchan S. P.; Simpson S. L.; Sirikanchana K.; Symonds E. M.; Verhagen R.; Vasan S. S.; Kitajima M.; Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020, 191, 110092. 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W.; Bertsch P. M.; Bivins A.; Bibby K.; Farkas K.; Gathercole A.; Haramoto E.; Gyawali P.; Korajkic A.; McMinn B. R.; Mueller J. F.; Simpson S. L.; Smith W. J. M.; Symonds E. M.; Thomas K. V.; Verhagen R.; Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020, 739, 139960. 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussow D.; Rein R.; Ginjaar I.; Hochstenbach F.; Seemann G.; Kottman A.; Ploegh H. L. The human beta 2-microglobulin gene. Primary structure and definition of the transcriptional unit. J. Immunol 1987, 139 (9), 3132–3138. [PubMed] [Google Scholar]
- D’Aoust P. M.; Mercier E.; Montpetit D.; Jia J. J.; Alexandrov I.; Neault N.; Baig A. T.; Mayne J.; Zhang X.; Alain T.; Langlois M. A.; Servos M. R.; MacKenzie M.; Figeys D.; MacKenzie A. E.; Graber T. E.; Delatolla R. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021, 188, 116560. 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrity D.; Papp K.; Stoker M.; Sims A.; Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in Southern Nevada: Methodology, occurrence, and incidence/prevalence considerations. Water Res. X 2021, 10, 100086. 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyawali P.; Croucher D.; Ahmed W.; Devane M.; Hewitt J. Evaluation of pepper mild mottle virus as an indicator of human faecal pollution in shellfish and growing waters. Water Res. 2019, 154, 370–376. 10.1016/j.watres.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Kitamura K.; Sadamasu K.; Muramatsu M.; Yoshida H. Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci. Total Environ. 2021, 763, 144587. 10.1016/j.scitotenv.2020.144587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E. M.; Sinigalliano C.; Gidley M.; Ahmed W.; McQuaig-Ulrich S. M.; Breitbart M. Faecal pollution along the southeastern coast of Florida and insight into the use of pepper mild mottle virus as an indicator. J. Appl. Microbiol. 2016, 121 (5), 1469–1481. 10.1111/jam.13252. [DOI] [PubMed] [Google Scholar]
- Wu F.; Zhang J.; Xiao A.; Gu X.; Lee W. L.; Armas F.; Kauffman K.; Hanage W.; Matus M.; Ghaeli N.; Endo N.; Duvallet C.; Poyet M.; Moniz K.; Washburne A. D.; Erickson T. B.; Chai P. R.; Thompson J.; Alm E. J. SARS-CoV-2 Titers in Wastewater Are Higher than Expected from Clinically Confirmed Cases. mSystems 2020, 5 (4), e00614. 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K.; Symonds E. M.; Sinigalliano C.; Stewart J.; Breitbart M. Pepper mild mottle virus as an indicator of fecal pollution. Appl. Environ. Microbiol. 2009, 75, 7261–7267. 10.1128/AEM.00410-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.; Breitbart M.; Lee W. H.; Run J. Q.; Wei C. L.; Soh S. W.; Hibberd M. L.; Liu E. T.; Rohwer F.; Ruan Y. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2005, 4 (1), e3 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoodi R.; Pirsaheb M.; Karimyan K.; Gupta V. K.; Takhtshahi A. R.; Sharafi H.; Moradi M. Data for distribution of various species of fecal coliforms in urban, rural and private drinking water sources in ten years period - A case study: Kermanshah, Iran. Data Brief 2018, 18, 1544–1550. 10.1016/j.dib.2018.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.; Chen L.; Shen Z. Impacts of rapid urbanization on characteristics, sources and variation of fecal coliform at watershed scale. J. Environ. Manage 2021, 286, 112195. 10.1016/j.jenvman.2021.112195. [DOI] [PubMed] [Google Scholar]
- Bethea M.; Forman D. T. Beta 2-microglobulin: its significance and clinical usefulness. Ann. Clin Lab Sci. 1990, 20 (3), 163–168. [PubMed] [Google Scholar]
- Palumbo A.; Avet-Loiseau H.; Oliva S.; Lokhorst H. M.; Goldschmidt H.; Rosinol L.; Richardson P.; Caltagirone S.; Lahuerta J. J.; Facon T.; Bringhen S.; Gay F.; Attal M.; Passera R.; Spencer A.; Offidani M.; Kumar S.; Musto P.; Lonial S.; Petrucci M. T.; Orlowski R. Z.; Zamagni E.; Morgan G.; Dimopoulos M. A.; Durie B. G.; Anderson K. C.; Sonneveld P.; San Miguel J.; Cavo M.; Rajkumar S. V.; Moreau P. Revised International Staging System for Multiple Myeloma: A Report From International Myeloma Working Group. J. Clin Oncol 2015, 33 (26), 2863–9. 10.1200/JCO.2015.61.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugan Y.; Korkmaz H.; Dogru A.; Koca Y. S.; Balkarli A.; Aylak F.; Tunc S. E. The significance of urinary beta-2 microglobulin level for differential diagnosis of familial Mediterranean fever and acute appendicitis. Clin Rheumatol 2016, 35 (7), 1669–72. 10.1007/s10067-016-3211-3. [DOI] [PubMed] [Google Scholar]
- Babler K. M.; Amirali A.; Sharkey M. E.; Williams S. L.; Boone M. M.; Cosculluela G. A.; Currall B. B.; Grills G. S.; Laine J.; Mason C. E.; Reding B. D.; Schurer S. C.; Stevenson M.; Vidovic D.; Solo-Gabriele H. M.. Comparison of electronegative filtration to magnetic-bead based concentration and V2G-qPCR to RT-qPCR for quantifying viral SARS-CoV-2 from wastewater. ACS ES&T Water, 2022, Article ASAP. 10.1021/acsestwater.2c00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelzaher A. M.; Solo-Gabriele H. M.; Palmer C. J.; Scott T. M. Simultaneous concentration of Enterococci and coliphage from marine waters using a dual layer filtration system. J. Environ. Qual 2009, 38 (6), 2468–73. 10.2134/jeq2008.0488. [DOI] [PubMed] [Google Scholar]
- Ahmed W.; Tscharke B.; Bertsch P. M.; Bibby K.; Bivins A.; Choi P.; Clarke L.; Dwyer J.; Edson J.; Nguyen T. M. H.; O’Brien J. W.; Simpson S. L.; Sherman P.; Thomas K. V.; Verhagen R.; Zaugg J.; Mueller J. F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: A temporal case study. Sci. Total Environ. 2021, 761, 144216. 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii S.; Furumai H.; Katayama H. Applicability of polyethylene glycol precipitation followed by acid guanidinium thiocyanate-phenol-chloroform extraction for the detection of SARS-CoV-2 RNA from municipal wastewater. Sci. Total Environ. 2021, 756, 143067. 10.1016/j.scitotenv.2020.143067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii S.; Oishi W.; Zhu Y.; Thakali O.; Malla B.; Yu Z.; Zhao B.; Arakawa C.; Kitajima M.; Hata A.; Ihara M.; Kyuwa S.; Sano D.; Haramoto E.; Katayama H. Comparison of five polyethylene glycol precipitation procedures for the RT-qPCR based recovery of murine hepatitis virus, bacteriophage phi6, and pepper mild mottle virus as a surrogate for SARS-CoV-2 from wastewater. Sci. Total Environ. 2022, 807 (Pt 2), 150722. 10.1016/j.scitotenv.2021.150722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. L.; Imakaev M.; Armas F.; McElroy K. A.; Gu X.; Duvallet C.; Chandra F.; Chen H.; Leifels M.; Mendola S.; Floyd-O’Sullivan R.; Powell M. M.; Wilson S. T.; Berge K. L.; Lim C. Y. J.; Wu F.; Xiao A.; Moniz K.; Ghaeli N.; Matus M.; Thompson J.; Alm E. Quantitative SARS-CoV-2 alpha variant B.1.1.7 tracking in wastewater by allele-specific RT-qPCR. Environ. Sci. Technol. 2021, 8, 675–682. 10.1021/acs.estlett.1c00375. [DOI] [Google Scholar]
- University of Miami Coronavirus (COVID-19) Information. Community Dashboard. https://coronavirus.miami.edu/dashboard/ (accessed on 2022-04-25).
- Centers for Disease Control and Prevention . COVID Data Tracker. https://covid.cdc.gov/covid-data-tracker (accessed on 2022-04-25).
- Covid Act Now . U.S. COVID Risk and Vaccine Tracker. https://www.covidactnow.org/us/florida-fl/?s=32822103 (accessed on 2022-04-25).
- Schmitz B. W.; Innes G. K.; Prasek S. M.; Betancourt W. Q.; Stark E. R.; Foster A. R.; Abraham A. G.; Gerba C. P.; Pepper I. L. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology. Sci. Total Environ. 2021, 801, 149794. 10.1016/j.scitotenv.2021.149794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C.; Lambirth K.; Mittal N.; Juel M. A. I.; Barua V. B.; Roppolo Brazell L.; Hinton K.; Lontai J.; Stark N.; Young I.; Quach C.; Russ M.; Kauer J.; Nicolosi B.; Chen D.; Akella S.; Tang W.; Schlueter J.; Munir M. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021, 782, 146749. 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo V.; Tillett R. L.; Chang C. L.; Gerrity D.; Betancourt W. Q.; Oh E. C. SARS-CoV-2 variant detection at a university dormitory using wastewater genomic tools. Sci. Total Environ. 2022, 805, 149930. 10.1016/j.scitotenv.2021.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butowt R.; Bilinska K. SARS-CoV-2: Olfaction, Brain Infection, and the Urgent Need for Clinical Samples Allowing Earlier Virus Detection. ACS Chem. Neurosci. 2020, 11 (9), 1200–1203. 10.1021/acschemneuro.0c00172. [DOI] [PubMed] [Google Scholar]
- Labcorp . Beta-2 Microglobulin Tumor Marker. https://www.labcorp.com/help/patient-test-info/beta-2-microglobulin-tumor-marker (accessed on 2022-04-25).
- Desmarais T. R.; Solo-Gabriele H. M.; Palmer C. J. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 2002, 68 (3), 1165–72. 10.1128/AEM.68.3.1165-1172.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solo-Gabriele H. M.; Wolfert M. A.; Desmarais T. R.; Palmer C. J. Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 2000, 66 (1), 230–7. 10.1128/AEM.66.1.230-237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmund G. K.; Allen M. J.; Rice E. W. Comparison of Escherichia coli, Total Coliform, and Fecal Coliform Populations as Indicators of Wastewater Treatment Efficiency. Water Environ. Res. 1999, 71 (3), 332–339. 10.2175/106143098X121752. [DOI] [Google Scholar]
- Huang J. J.; Hu H. Y.; Tang F.; Li Y.; Lu S. Q.; Lu Y. Inactivation and reactivation of antibiotic-resistant bacteria by chlorination in secondary effluents of a municipal wastewater treatment plant. Water Res. 2011, 45 (9), 2775–81. 10.1016/j.watres.2011.02.026. [DOI] [PubMed] [Google Scholar]
- Liu S. S.; Qu H. M.; Yang D.; Hu H.; Liu W. L.; Qiu Z. G.; Hou A. M.; Guo J.; Li J. W.; Shen Z. Q.; Jin M. Chlorine disinfection increases both intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res. 2018, 136, 131–136. 10.1016/j.watres.2018.02.036. [DOI] [PubMed] [Google Scholar]
- Xiling G.; Yin C.; Ling W.; Xiaosong W.; Jingjing F.; Fang L.; Xiaoyan Z.; Yiyue G.; Ying C.; Lunbiao C.; Liubo Z.; Hong S.; Yan X. In vitro inactivation of SARS-CoV-2 by commonly used disinfection products and methods. Sci. Rep 2021, 11 (1), 2418. 10.1038/s41598-021-82148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miami-Dade County . Emergency Orders. https://www.miamidade.gov/global/initiatives/coronavirus/emergency-orders.page (accessed on 2022-04-25).
- Saito A.; Irie T.; Suzuki R.; Maemura T.; Nasser H.; Uriu K.; Kosugi Y.; Shirakawa K.; Sadamasu K.; Kimura I.; Ito J.; Wu J.; Iwatsuki-Horimoto K.; Ito M.; Yamayoshi S.; Loeber S.; Tsuda M.; Wang L.; Ozono S.; Butlertanaka E. P.; Tanaka Y. L.; Shimizu R.; Shimizu K.; Yoshimatsu K.; Kawabata R.; Sakaguchi T.; Tokunaga K.; Yoshida I.; Asakura H.; Nagashima M.; Kazuma Y.; Nomura R.; Horisawa Y.; Yoshimura K.; Takaori-Kondo A.; Imai M.; Genotype to Phenotype Japan C.; Tanaka S.; Nakagawa S.; Ikeda T.; Fukuhara T.; Kawaoka Y.; Sato K.; et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 602 (7896), 300–306. 10.1038/s41586-021-04266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A. B.; Van De Werfhorst L. C.; Griffith J. F.; Holden P. A.; Jay J. A.; Shanks O. C.; Wang D.; Weisberg S. B. Performance of forty-one microbial source tracking methods: a twenty-seven lab evaluation study. Water Res. 2013, 47 (18), 6812–28. 10.1016/j.watres.2012.12.046. [DOI] [PubMed] [Google Scholar]