Summary
Background
Epidermal growth factor receptor (EGFR) is an essential target for cancer treatment. However, EGFR inhibitor erlotinib showed limited clinical benefit in pancreatic cancer therapy. Here, we showed the underlying mechanism of tumor microenvironment suppressing the sensitivity of EGFR inhibitor through the pancreatic stellate cell (PSC).
Methods
The expression of alpha-smooth muscle actin (α-SMA) and hypoxia marker in human pancreatic cancer tissues were detected by immunohistochemistry, and their correlation with overall survival was evaluated. Human immortalized PSC was constructed and used to investigate the potential effect on pancreatic cancer cell lines in hypoxia and normoxia. Luciferase reporter assay and Chromatin immunoprecipitation were performed to explore the potential mechanisms in vitro. The combined inhibition of EGFR and Met was evaluated in an orthotopic xenograft mouse model of pancreatic cancer.
Findings
We found that high expression levels of α-SMA and hypoxia markers are associated with poor prognosis of pancreatic cancer patients. Mechanistically, we demonstrated that hypoxia induced the expression and secretion of HGF in PSC via transcription factor HIF-1α. PSC-derived HGF activates Met, the HGF receptor, suppressing the sensitivity of pancreatic cancer cells to EGFR inhibitor in a KRAS-independent manner by activating the PI3K-AKT pathway. Furthermore, we found that the combination of EGFR inhibitor and Met inhibitor significantly suppressed tumor growth in an orthotopic xenograft mouse model.
Interpretation
Our study revealed a previously uncharacterized HIF1α-HGF-Met-PI3K-AKT signaling axis between PSC and cancer cells and indicated that EGFR inhibition plus Met inhibition might be a promising strategy for pancreatic cancer treatment.
Funding
This study was supported by The National Natural Science Foundation of China.
Keywords: Tumor microenvironment, Hypoxia, Hepatocyte growth factor, Met
Research in context.
Evidence before this study
Pancreatic cancer is one of the most fatal caners with few effective therapies. The refractory responses of pancreatic cancer to chemotherapy as well as targeted therapy and immunotherapy could be largely attributed to the highly desmoplastic stroma comprised of abundant activated pancreatic stellate cells (PSCs). However, the underline mechanism of PSC mediated drug resistance is not fully understood.
Added value of this study
This study revealed the underlying mechanism of PSC suppressed the sensitivity of EGFR inhibitor in pancreatic cancer under hypoxia. A previously uncharacterized HIF1α-HGF-Met-PI3K-AKT signaling axis between PSC and cancer cells mediates EGFR resistance was identified.
Implications of all the available evidence
Our study indicated high expression levels of α-SMA and hypoxia marker are associated with poor prognosis of pancreatic cancer patients. The combined inhibition of EGFR and Met signaling might be a promising strategy for pancreatic cancer treatment.
Introduction
Pancreatic cancer is currently the third leading cause of cancer death in the United States, with a five-year survival rate of 11%.1 One major characteristic of pancreatic cancer tissues is the extensive desmoplasia that leads to tissue hypoxia and restricts immune infiltration.2, 3, 4, 5, 6 Pancreatic stellate cell (PSC), the resident mesenchymal cells of the pancreas, is the primary source of cancer-associated fibroblast or myofibroblasts that contributes to the desmoplasia in pancreatic cancer.7, 8, 9 PSCs are at low numbers at quiescent statuses in normal pancreas and characteristically express alpha-smooth muscle actin (α-SMA) when activated and transform into myofibroblasts.10 Activated PSCs profoundly affect tumor cell initiation, progression, and metastasis through secreting extracellular matrix components, growth factors, amino acids, and lipids.11, 12, 13, 14, 15, 16, 17
Hypoxia is a common feature of most solid tumors, particularly pancreatic cancer.18 Hypoxia-inducible factor-1α (HIF-1α) plays a central role in hypoxic cancer by regulating its downstream targeted genes.19, 20, 21 Previous studies show increased hypoxia and activated PSC during pancreatic cancer development, but the underlying mechanisms are not fully understood.2,16,22 Hepatocyte growth factor (HGF), the ligand of the Met (mesenchymal–epithelial transition factor)-receptor, which is secreted mainly from stromal cells and plays an essential role in resistance to tyrosine kinase inhibitors (TKIs) in many cancers, including melanoma, lung cancer, and colorectal cancer.23, 24, 25, 26, 27 The expression of HGF was reported to correlate with HIF-1α, but the underlying mechanism is unclear.28,29
The epidermal growth factor receptor (EGFR) tyrosine kinase is often activated in various cancer types.30 Activated EGFR leads to the subsequent signaling cascades, such as PI3K/Akt, MAPK, and STAT signaling, which involve cell survival, apoptosis, and proliferation.31 Epidermal growth factor receptor TKIs are well-used therapeutic agents for non-small cell lung cancer and colorectal cancer in patients with EGFR mutations.32,33 However, many patients develop drug resistance after treatment due to secondary mutation or bypass signaling activation.34 The EGFR inhibitor erlotinib (ERL) has also been approved for treating pancreatic cancer but showed little effect in clinical applications. The high-frequency mutation of KRAS in pancreatic cancer has been thought to be the factor that limited the effect of erlotinib, but it could not be a predictor for therapy resistance.35, 36, 37 The limited effect of EGFR inhibitor in pancreatic cancer needs further investigation since EGFR is overexpressed in 57%–95% of pancreatic cancer and EGFR signaling is necessary for KRAS-driven tumor progression.38, 39, 40
In the present study, we found that patients with a high level of α-SMA expression and a high level of carbonic anhydrase IX (CAIX), a hypoxia marker, predict a worse prognosis in pancreatic cancer. Upon investigating the cytokine profiles of PSC conditioned medium under hypoxia, we observed that the level of HGF significantly increased. Using the luciferase reporter system and chromatin immunoprecipitation assay, we found that hypoxia-induced stabilization of HIF-1α regulates the expression and secretion of HGF in PSC, which contributes to the resistance of EGFR inhibitor treatment in pancreatic cancer in a KRAS-independent manner. EGFR inhibitor, in combination with the Met inhibitor, showed a remarkable effect in suppressing tumor growth in the pancreatic cancer orthotopic xenograft mouse model. In conclusion, this study showed a novel mechanism of stromal signaling from PSC under hypoxia that contributes to the resistance to EGFR inhibition in pancreatic cancer.
Methods
Ethics
This study was approved by the Human Ethics Committee of Tongji Hospital, Huazhong University of Science and Technology. All animal experiments were approved by the Animal Care and Use Committee of Huazhong University of Science and Technology.
Human specimens
Human pancreatic cancer specimens were obtained from the Department of Biliary-Pancreatic Surgery at Tongji Hospital (Wuhan, China). All tissue sections were stained with H&E and diagnosed by pathological examination.
Cells and cell culture
PANC-1 (#CRL-1469), BxPC-3 (#CRL-1687), SW1990 (#CRL-2172), MIA PaCa-2 (#CRM-CRL-1420), AsPC-1 (#CRL-1682), CFPAC-1 (#CRL-1918), and Panc 03.27 (#CRL-2549) cells were purchased from American Type Culture Collection. These cells were cultured following the manufacturer's instructions as described previously.41,42 Human primary pancreatic stellate cells were purchased from ScienCell (#3830; ScienCell Research Laboratories, CA, USA). The pLVSIN-CMV vector containing sequences of SV40 large T-antigen and hTERT was used for constructing immortalized human pancreatic stellate cell lines (ImPSC). The ImPSC were identified by PCR, and these cell lines did not exhibit senescence traits, even after passage 20. PSC and ImPSC were cultured in the recommended stellate cell medium (#5301; ScienCell Research Laboratories, CA, USA) with 2% fetal bovine serum (FBS, #0010, ScienCell) and 1% stellate cell growth supplement (#5352; ScienCell). All the cell lines used in the study were identified by short tandem repeat (STR) profiling.
Stable cell line construction
The shRNA sequence of HGF was inserted into the pLKO.1 puro (Plasmid #8453, Addgene) using AgeI and EcoRI sites. The recombinant and control plasmid were co-transfected with lentiviral packaging plasmids into 293T cells. Lentiviral supernatants were collected and transduced to the PSC as we described previously.41 Stable cells of HGF knockdown were selected by puromycin for two weeks.
Preparation for the conditioned medium
Serum-free medium was added to the PSC at >80% confluence to form a conditioned medium under hypoxia (5% CO2, 1% O2, and 94% N2) or normoxia (5% CO2, 21% O2, and 74% N2) for 24 h. The conditioned medium (CM) was centrifuged at 3000 rpm for 10 min and filtered through the 0.45 μm PES filter. The CM was stored at −80 °C until use.
Quantitative real-time PCR
Total RNA of cells or tissues was extracted using the Trizol reagent ((#15596026, Invitrogen). The purity and concentration of RNA were detected by the NanoDrop spectrophotometer (Thermo Scientific, USA). Messenger RNA was converted to complementary DNA using the Reverse Transcriptase Kit (#RR047A, Takara, Japan) and amplification by real-time PCR reaction using an SYBR Premix Ex Taq kit (#RR420A, Takara, Japan), following the manufacturer's protocol. Comparative quantification of gene expression was calculated using the 2−ΔΔCt method. The gene-specific primer sequences used for PCR are listed in Supplementary Table S4.
Luciferase reporter assay
The pRL Renilla luciferase reporter vector with pGL3 reporter vector (RRID: Addgene_48743) containing the partial or full promoter or control pGL3 vector was co-transfected into PSC with pcDNA3-HIF-1α or a control pcDNA3 vector, then incubated for 24 h under normoxia or hypoxia. Relative luciferase activity was detected using a dual-luciferase reporter assay system per the manufacturer's protocols (#1910; Promega, USA).
Chromatin immunoprecipitation assay
Based on the manufacturers’ recommendations, the Pierce™ Agarose ChIP kit (#26156, Thermo Scientific, USA) was used for the ChIP assay. PSCs were treated with 1% formaldehyde to induce DNA-protein crosslinking. Next, incubated with glycine, lysed in lysis buffer, and sonicated to shear DNA. After centrifugation and resuspension, cell lysates were incubated with Protein A/G Agarose beads and immunoprecipitated overnight with either anti-HIF-1α or IgG antibody at 4 °C. After that, the protein-DNA complexes were pulled down by Protein A/G Agarose beads. The immunoprecipitated DNA was quantified by PCR.
Animal experiment
Five-week-old female BALB/c nude mice were purchased from HFK BioTechnology (Beijing, China). One single mouse is an experimental unit. The sample size was calculated by the “resource equation” method,43 and we used the power analysis tool PASS v16 (Utah, USA, 2018) to estimate the minimum detectable effect size of the selected sample size. In a single factor ANOVA study, the sample size of 6 mice per group and 4 groups total achieves 80% power to detect a difference of at least 3.37 standard deviation (SD) using the Dunnett (With Control) multiple comparison test at a 0.05 significance level. Since there are two different groups of cells in the animal experiment, the minimum number of mice is 48 in the animal experiment. Considering the expected attrition or death of animals, the total number of mice we used in this study is 55. PANC-1 cells (1 × 106) transduced to express luciferase with or without PSC (1 × 106) were suspended in 50 μl PBS mixed with Matrigel and were orthotopically injected into mice pancreas. Four mice were excluded from the experiment due to leakage during implantation. One week after implantation, mice injected with PANC-1 and PANC-1+PSC cells were randomly (simple randomization) divided into four groups (DMSO, ERL, Criz, ERL+Criz), six mice per group. Each cage was selected randomly from the pool of all cages. Then, we administered 50 mg/kg ERL and/or 40 mg/kg Criz or DMSO daily by gavage for seven weeks and collected tumors and tissues for detection at the endpoint of the experiment. Mouse weights were monitored twice a week. Tumor volumes were monitored by bioluminescence imaging weekly.
Statistics
Statistical analysis was performed with IBM SPSS statistics 24 or GraphPad Prism 8 software. The correlation between study variables was analyzed by the Spearman correlation test. The Survival curves were plotted using the Kaplan–Meier method, and the survival rates were compared by log-rank test. The significance of single-factor and multiple-factor on the survival time was analyzed using univariate and multivariate Cox regression analyses. The Shapiro–Wilk test and Kolmogorov–Smirnov test were used for the data normality evaluation. For the two groups' comparison, Student's t-test (parametric test) was performed for continuous variables following normal distribution, and the Mann–Whitney test (non-parametric test) was performed for the data that followed the skewed distribution. For multiple comparisons, the ANOVA test (parametric test) followed by the Bonferroni test was performed for data following normal distribution, and the Kruskal–Wallis test (non-parametric test) was performed for the data following the skewed distribution. Bonferroni correction was performed to adjust the P value in multiple comparations. A P-value <0.05 was considered statistically significant.
Role of the funding source
The funding agencies of this study had no role in the study design, data collection, analysis, interpretation of data, or manuscript writing.
Results
High levels of α-SMA and CAIX predict a worse prognosis in patients with pancreatic cancer
The pancreatic tumor microenvironment is characterized by the accumulation of α-SMA+ myofibroblasts and a large number of stromal collagen fibers in both primary tumors and metastases in the liver (Supplementary Fig. S1a). To determine the functions of cancer-associated fibroblasts in pancreatic cancer, we used immunohistochemical staining to evaluate the expression of α-SMA, CAIX, PCNA, Vimentin, TP53, and SMAD4, which play important roles in pancreatic cancer. Using a tissue microarray from a cohort of 90 patients with pancreatic cancer, we found that the expression of α-SMA was positively correlated with CAIX (Spearman r = 0.391; P < 0.0001). While PCNA, Vimentin, TP53, and SMAD4 were not correlated with either CAIX or α-SMA (P > 0.05; Supplementary Tables S1 and S2). Since CAIX is an essential enzyme of cancer cell response to the hypoxic environment, another hallmark of pancreatic cancer, we further evaluated the CAIX level in tumor tissues. To analyze the effects of α-SMA and CAIX on patients’ survival, we divided the expression of α-SMA and CAIX into high (>2.0) and low (≤2.0) groups according to immunohistochemical staining score. The staining of α-SMA was scored based on staining intensity in the stromal cells, and the CAIX was scored based on staining intensity in both stromal cells and cancer cells. We found that a high level of α-SMA was associated with a poor prognosis (median survival of 13.8 months vs. 28.0 months; P = 0.001 (log-rank); Fig. 1a). The high level of CAIX was insignificantly associated with poor prognosis (P = 0.093, Fig. 1b). When we combined α-SMA with CAIX, we found that a high level of α-SMA and a high level of CAIX predicted a worse prognosis (median survival of 9.2 months compared with 33.0 months; P = 0.003; Fig. 1c). Representative images of immunohistochemical staining of α-SMA and CAIX in pancreatic cancer tissue microarrays were shown in Fig. 1d and e.
Fig. 1.
Hypoxia promotes PSC activation and upregulates HGF level in pancreatic cancer. a–c, Overall survival of pancreatic cancer patients stratified by α-SMA expression (a), CAIX expression (b), or both α-SMA and CAIX expression (c). d and e, Representative tissue microarray images of immunohistochemistry staining for α-SMA and CAIX from a set of 90 patients with pancreatic cancer. f, Morphology in culture conditions of primary pancreatic stellate cells and immortalized pancreatic stellate cells. g and h, The immortalized human pancreatic stellate cell line was identified by RT-PCR. i, Human primary and immortalized pancreatic stellate cells were identified by immunofluorescent staining using α-SMA and GFAP (Glial fibrillary acidic protein) antibody observed under a confocal microscope. Scale bars, 100 μm j, Cytokine array analysis between HCM and NCM of PSC. Hierarchical clustering of cytokine according to their relative signal intensity. k, Cross-data analysis of TCGA, tissue data, and PCR-analyzed cell line data demonstrated that HGF was significantly up-regulated in all datasets. ∗∗∗P < 0.001.
We next used single-factor and multiple-factor regression analyses to assess the impact of expression of α-SMA and CAIX on survival. We noticed that both α-SMA (HR = 2.349; P = 0.003), and CAIX (HR = 1.491; P = 0.095) are correlated with worse prognosis in univariate analysis (Supplementary Table S3). A more significant effect on survival was found in combined α-SMA with CAIX expression in both univariate analysis (HR = 3.026; P = 0.004) and multivariate analysis (HR = 3.997; P = 0.001) (Supplementary Table S3). In the univariate analysis, other traditional clinical-pathological prognostic indicators were also analyzed. Among these indicators, tumor size, vascular infiltration, and tumor differentiation exhibited a significant correlation with overall survival (P < 0.05) (Supplementary Fig. S1b–h). Taken together, these results indicated that high α-SMA and hypoxia predict significantly poor prognosis in pancreatic cancer patients.
Hypoxia promotes HGF expression and secretion in pancreatic stellate cells
The pancreatic stellate cell is an important source of cancer-associated fibroblasts or α-SMA+ myofibroblasts,44 which can affect pancreatic tumor progression by secreting ECM and cytokines. Based on our findings above, we hypothesized that hypoxia-related cytokines from PSC could play an essential role during tumor progression. To this end, we used primary pancreatic stellate cells isolated from the human pancreas to construct an immortalized pancreatic stellate cell line via transfection with the SV40 large T antigen and human telomerase (hTERT) (Fig. 1f–h). The primary and immortalized PSC were identified by immunofluorescence with characterized markers of PSC (Fig. 1i).
To investigate the hypoxia-related cytokines secreted by PSC, we collected the conditioned medium under normoxia (NCM) and the conditioned medium under hypoxia (HCM). Using a cytokine microarray analysis, we found seventeen differently expressed cytokines in HCM compared with NCM, and six of these cytokines were significantly overexpressed (Fig. 1j). We further evaluated the mRNA levels of these cytokines in PSC cultured in hypoxia or normoxia. The mRNA levels of HGF, MIF, IL-6, and IL-1α were significantly higher in hypoxia compared to normoxia (Supplementary Fig. S2a). To gain insight into the clinical relevance of hypoxia-related cytokines in PSC, the RNA levels of cytokines in pancreatic cancer specimens (n = 178) were analyzed by The Cancer Genome Atlas (TCGA) database. We found that RNA levels of these six cytokines, except IL-1α, were significantly higher in pancreatic cancer tissues than adjacent tumor controls (Supplementary Fig. S2b–g). Moreover, the results from our pancreatic cancer patients’ specimens consisting of matched normal and tumor tissues (n = 30, Supplementary Fig. S2h–m) indicated a significant difference in the RNA levels of HGF (P < 0.0001) and CCL2 (P = 0.0019) in tumor tissues and controls. When considered together with the mRNA levels from the stellate cells data, the TCGA database and our tissue samples, we found that HGF level was significantly changed in pancreatic tumor tissues compared to adjacent normal control tissues (Fig. 1k). Taken together, these results indicate that α-SMA+ PSC highly expressed HGF under hypoxia and may contribute to the malignant phenotype of pancreatic cancer.
Hypoxia increases HGF expression and secretion in pancreatic stellate cells by enhancing transcriptional activity
Since HGF is mainly secreted by stromal cells and hypoxia affects the expression of a large number of genes that promote cell survival by regulating the stabilization of HIF-1α,45,46 we further investigated the association between HGF and HIF-1α under hypoxia condition. We compared the expression of HGF in PSC under normoxic and hypoxic conditions for different time points (Fig. 2a and b). We found that hypoxia exposure increased the expression of HGF in a HIF-1α dependent manner in PSC, which suggests that HGF is overexpressed in hypoxic conditions and is positively correlated with HIF-1α level. To determine the association between HGF and HIF-1α, we knocked down and overexpressed HIF-1α in PSC (Fig. 2c). Three shRNAs of HIF-1α were designed to knock down HIF-1α. Among them, shRNA1 showed the most significant inhibition efficiency in PSC (Supplementary Fig. S2n). Thus, we used shRNA1 for the subsequent experiments and named it shHIF-1α. Then, we observed that HGF was increased in PSC when the HIF-1α was upregulated, whereas HGF expression was decreased at both RNA and protein levels with HIF-1α knocked down under hypoxia (Supplementary Fig. S2o–p).
Fig. 2.
Hypoxia activates the transcription of HGF in PSC. a, HGF expression level was detected in PSC by immunofluorescence assay after treatment with hypoxia or normoxia for 24 h. b, The expression of HGF and HIF-1α in PSC was determined by Western blot after treatment with hypoxia for 0, 2, 4, 6, 12, or 24 h. c, The protein expression level of HIF-1α and HGF in PSC under hypoxia for 24 h was assayed by Western blot after transfection with pcDNA, pcDNA-HIF-1α, shControl, or shHIF-1α. d, Graphical representation of putative transcription factor binding sites in the promoter of the HGF gene. e, Relative luciferase activity assayed after co-transfection with luciferase reporter vector containing the promoter of HGF with shControl, shHIF1α, pcDNA, or pcDNA-HIF1α in PSC under hypoxia or normoxia. f, ChIP products were amplified by quantitative PCR with each designed primer. Cell lysates of PSC exposure to hypoxia for 24 h were subjected to the ChIP assay with control IgG or HIF-1α antibody. g, Luciferase activity was detected in PSC under hypoxia for 24 h after transfection with luciferase reporter vector containing HGF full promoter, a promoter with BS1, BS2, or BS3 deletion. h, ChIP-PCR assay was conducted to analyze HIF-1α binding on human HGF promoter in PSC cells. i, Schematic diagram of the luciferase reporter vector containing wild-type or mutant BS1. j, Luciferase activity was detected in PSC under hypoxia for 24 h after transfection with luciferase reporter vector containing wild-type, mutant BS1, or empty control. ns: not significant, ∗P < 0.05, ∗∗P < 0.01.
As HIF-1α could bind to the hypoxia-response elements (HRE) on the promoters of its target genes to regulate expression under hypoxia,45 here, we hypothesized that HIF-1α might activate HGF transcription. To verify this, we analyzed the promoter sequence of the HGF gene by using the JASPAR public database.47 Three putative HRE sites (BS1, BS2, and BS3) were identified in the promoter of the HGF gene on chromosome 7 (Fig. 2d). Then, we performed the luciferase reporter assay to examine whether HIF-1α could regulate the transcription activity of HGF. As expected, we found that HIF-1α significantly increased HGF promoter activity in stellate cells under hypoxic conditions compared with normoxic conditions. Accordingly, the promoter activity of HGF decreased after HIF-1α was knockdown by shRNA (Fig. 2e). Next, through the chromatin immunoprecipitation (ChIP) assay we found that HIF-1α could directly bind to the promoter of HGF, and we found that all the three putative binding sites on the promoter of HGF were detectable (Fig. 2f). To further determine which binding site is necessary for regulating the transcriptional activity of HGF, we constructed three deletion reporter vectors of these three binding sites, respectively. We observed that only BS1 deletion led to a significant change in luciferase activity (Fig. 2g). The ChIP-PCR results showed HIF-1α could directly bind to the binding site 1 of HGF promoter region (Fig. 2h). Accordingly, the mutation of BS1 showed decreased luciferase activity (Fig. 2i and j), indicating that HIF-1α increased HGF transcription by binding to its BS1 region of the promoter.
HGF derived from PSC under hypoxia suppresses the sensitivity of cancer cells to EGFR inhibitor
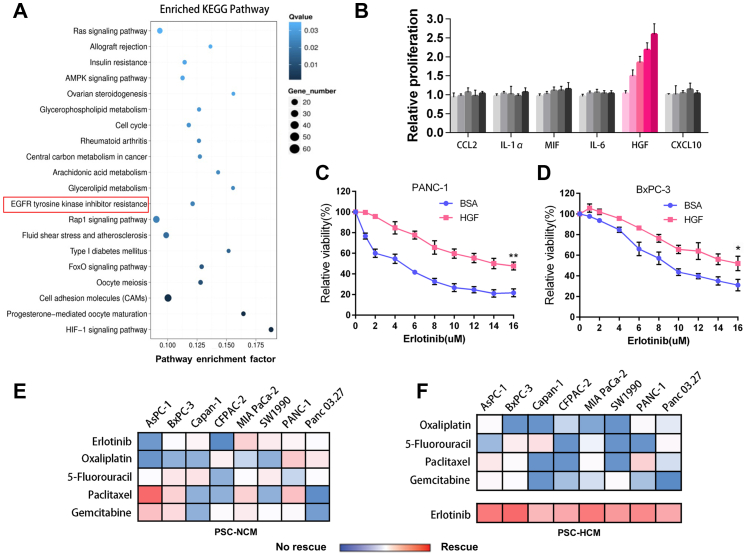
Previous studies reported that stromal-derived factors acting on pancreatic cancer cells had various effects, such as proliferation, invasion, and metastasis in a paracrine manner.48 To investigate the effects of HCM and NCM from PSC on pancreatic cancer cells, we performed a transcriptome sequencing analysis with PC cells incubated with PSC-HCM or PSC-NCM. The conditioned treatment schema was shown in Supplementary Fig. S3a. We noticed a significant change in the signaling pathway of EGFR tyrosine kinase inhibitor resistance (Fig. 3a). HGF-Met activity has been shown to affect the sensitivity of tyrosine kinase inhibitor, including the EGFR inhibitor.23,26 Thus, we postulated that PSC might affect the sensitivity of pancreatic cancer cells to EGFR inhibitors by secreting HGF. We found that only HGF could rescue pancreatic cancer cell lines from erlotinib, an EGFR inhibitor killing (Fig. 3b), while CCL2, IL-1α, MIF, IL-6, or CXCL10 could not. The effect of HGF promoting cell proliferation is insignificant in the absence of erlotinib (Supplementary Fig. S3b). The cell viability analysis showed that recombinant HGF decreased the sensitivity of pancreatic cancer cells to erlotinib (Fig. 3c and d).
Fig. 3.
HGF suppresses the sensitivity of EGFR inhibitor in pancreatic cancer cells. a, Top twenty pathways of differentially expressed genes in PANC-1 treated with PSC-NCM or PSC-HCM. b, The effect of six cytokines, each at five concentrations(0, 6.25, 12.5, 25, 50 ng/ml) on PANC-1 cells that were treated with 2 μM erlotinib. Proliferation was quantified after 5 days and was normalized to no-cytokine controls. c and d, Cell viability assay was performed to evaluate the effect of HGF (40 ng/ml) on the sensitivity (IC 50) of PANC-1 and BxPC-3 cells to erlotinib. BSA was used as a control. e and f, Hierarchical clustering of pancreatic cancer cell lines (vertical) according to cell viability rescued from anticancer drugs (horizontal) treated by PSC-NCM (e) and PSC-HCM (f) compared with control. Rescued is defined as the cell viability increased more than 1-fold compared to the control group without conditioned medium exposure. No rescue is defined as the cell viability was increased less than 1-fold compared to the control group without conditioned medium exposure. The red and blue shades represent rescue/resistance (>1-fold change) and no rescue/sensitivity (<1-fold change), respectively. ∗P < 0.05, ∗∗P < 0.01.
Based on the above results, we reasoned that HCM from PSC may serve as an intrinsic barrier to drug therapy sensitivity in pancreatic cancer. To verify this, we analyzed the effects of five widely used drugs in eight pancreatic cancer cell lines under treatment with PSC-HCM or PSC-NCM. We found that PSC-HCM significantly rescued the pancreatic cancer cell lines to erlotinib compared with PSC-NCM (Fig. 3e and f). At the same time, we found that PSC-HCM-mediated rescue was significant to erlotinib, but not oxaliplatin, 5-fluorouracil, paclitaxel, or gemcitabine. Then, we quantified the effect of exposing pancreatic cancer cell lines to the half-maximum inhibitory concentration (IC50) of the conditioned medium and cell proliferation for erlotinib. We found that PSC-HCM increased the resistance of pancreatic cancer to erlotinib, promoted proliferation, colony formation, and inhibited cell apoptosis (Fig. 4a–d; Supplementary Fig. S3c–f).
Fig. 4.
PSC-HCM induced the suppression of EGFR inhibitor sensitivity in pancreatic cancer via secreting HGF. a, Cell viability assay was tested in PANC-1 incubated with PSC-NCM or PSC-HCM in different concentrations of erlotinib for 72 h. Cell viability in the group of PSC-NCM or PSC-HCM was normalized to control (serum-free medium). IC50s were 3.5 μM of the PSC-NCM group and 4.7 μM of the PSC-HCM group. b, Cell viability assay was used to test the effects of erlotinib on the viability of PANC-1 treated with PSC-NCM or PSC-HCM for 0–4 days. Cell viability in the group of PSC-NCM or PSC-HCM was normalized to control (serum-free medium). c, Apoptosis experiment of PANC-1 treated with erlotinib or DMSO control in PSC-NCM or PSC-HCM. Cell apoptosis was assessed by flow cytometry after annexin V/PI staining. d, Colony formation experiment of PANC-1 treated with erlotinib or DMSO control in PSC-NCM or PSC-HCM. e, Colony formation experiment of PANC-1 cultured with PSC-HCM from HGF-knockdown or HGF overexpression PSC treated with erlotinib. f, Cell viability assay was used to evaluate the effect of anti-HGF neutralizing antibodies on PSC-HCM rescue. PANC-1 cultured with PSC-HCM added erlotinib for 5 days. g, Cell viability assay was used to evaluate erlotinib sensitivity when treated with HGF and or Met inhibitor alone. PANC-1 were cultured with HCM with or without HGF and treated with erlotinib with or without crizotinib. Proliferation was quantified after 5 days and was normalized to the control with no erlotinib, no HGF, and no crizotinib. h, Apoptosis experiment of PANC-1 cultured with HCM with or without HGF and treated with ERL with or without crizotinib. The control had no erlotinib, no HGF, and no crizotinib. ∗P < 0.05, ∗∗P < 0.01.
Next, we examined whether PSC promotes pancreatic cancer progress and erlotinib resistance was mediated by HGF. We found that the colony formation capacity of cancer cells in the presence of erlotinib was decreased with HGF knockdown, whereas there was an increase with HGF overexpression in PSC (Fig. 4e). However, cell colony formation, cell viability, and apoptosis in cancer cell in the absence of erlotinib did not change significantly after HGF knockdown in PSC (Supplementary Fig. S3g–i). When we used the HGF-neutralizing antibody to block HGF, and it increased pancreatic cancer cell sensitivity to erlotinib in PSC-HCM in a concentration-dependent manner (Fig. 4f; Supplementary Fig. S3j). Moreover, to validate whether HGF mediated erlotinib resistance is dependent on Met receptor, we treated cells with Crizotinib (Criz), a potent inhibitor of Met. We found the PSC-HCM-mediated drug resistance could be reversed partially by Criz (Fig. 4g and h; Supplementary Fig. S3k and l). These results indicated that PSC-HCM increased the resistance of pancreatic cancer to EGFR inhibition is mediated by HGF.
HGF suppresses the sensitivity of EGFR inhibition in pancreatic cancer by re-activating the PI3K-AKT pathway
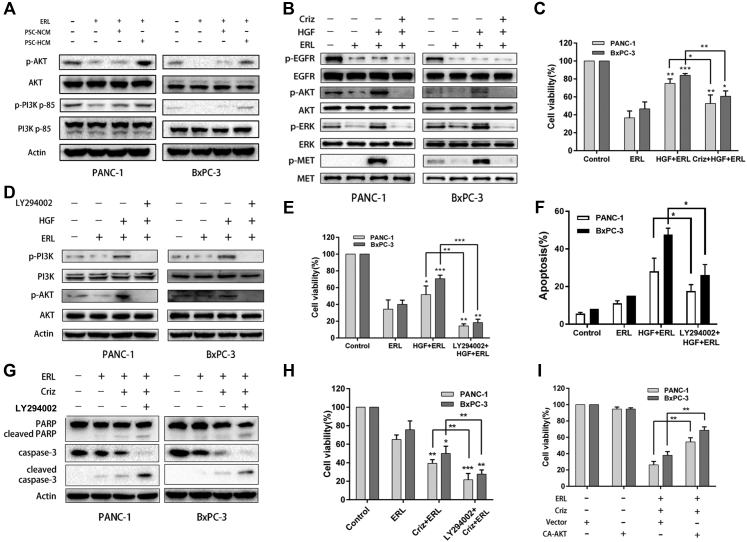
To investigate the mechanism of PSC-HCM derived HGF mediated EGFR inhibitor resistance, we assessed the status of three downstream survival signaling pathways activated by EGF that have been reported in previous studies: the MAPK, PI3K-AKT, and STAT3 pathways.49 We first used PSC-HCM or PSC-NCM to treat pancreatic cancer cell lines with or without erlotinib and then detected the activity of ERK, AKT, and STAT3 pathways. We found that PSC-HCM led to the significant activation of the PI3K/AKT pathway upon treatment with erlotinib compared with PSC-NCM (Fig. 5a). Similar activations of the MAPK and STAT3 pathways were also observed (Supplementary Fig. S4a and b).
Fig. 5.
Pancreatic stellate cells derived HGF suppressed the sensitivity of cancer cells to EGFR inhibitor through the re-activating PI3K-AKT pathway. a, The effect of CM on pancreatic cell lines treated with erlotinib or DMSO control. PI3K-AKT pathway activation was assessed by Western blot analysis after 24 h of treatment. b, Immunoblots showing the effect of Met kinase inhibition ± HGF on EGFR, Met, AKT, and ERK phosphorylation. Cells were co-treated with erlotinib. c, CCK-8 assay showing the relative cell proliferation of pancreatic cancer cells. Cells were co-treated with erlotinib ± HGF ± crizotinib. d, The effect of LY294002 on pancreatic cell lines treated with erlotinib or DMSO control, with or without HGF. PI3K-AKT pathway activation was assessed by Western blot analysis after 24 h of treatment. e, The cell viability of pancreatic cancer cell lines treated with the PI3K inhibitor. Cells were co-treated with erlotinib ± HGF. f, The apoptosis rates of PANC-1 and BxPC-3 cells, which were incubated with erlotinib ± HGF ± PI3K inhibitor and stained with Hoechst 33258. g, Cell apoptosis was measured by Western blot assay of PANC-1 and BxPC-3 cells treated with erlotinib ± Met inhibitor ± PI3K inhibitor. Cells were co-treated with HGF. h, Cell viability was measured by CCK-8 assay of PANC-1 and BxPC-3 cells treated with erlotinib ± Met inhibitor ± PI3K inhibitor. Cells were co-treated with HGF. i, Cell viability was determined after cells were transfected with the vehicle plasmid or the constitutively active AKT plasmid (CA-AKT) in the presence or absence of erlotinib and crizotinib. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Interestingly, we found that HGF activated downstream survival signaling of EGFR in the presence of erlotinib, and this was largely abolished by Met inhibitor (Fig. 5b, Supplementary Fig. S4c). Here, these results indicated that MAPK, PI3K-AKT, and STAT3 pathways re-activation was not induced by EGF, as EGFR tyrosine kinase phosphorylation was suppressed by erlotinib, indicating that they were re-activated by the presence of HGF. As expected, when the Met activity was inhibited by crizotinib, HGF-mediated pathway activation was abolished, followed by increased sensitivity of cancer cells to erlotinib (Fig. 5c). To determine whether these three signaling pathway activations are necessary for HGF-induced intrinsic resistance, we used signal pathway inhibitors for further study. We found that LY294002, a PI3K inhibitor, potently reversed the resistance to erlotinib induced by HGF treatment (Supplementary Fig S4d, Fig. 5d–h). We also found that pretreatment with LY294002 suppressed apoptosis and increased viability of pancreatic cancer cells upon combined co-treatment with EGFR inhibitor and Met inhibitor (Supplementary Fig. S4e). These findings indicated the critical role of the PI3K-AKT pathway in HGF-mediated suppression of EGFR inhibitor sensitivity for cancer cells. We next used constitutively active AKT to assess the role of the PI3K-AKT pathway in the HGF-mediated suppression of EGFR inhibitor sensitivity. As expected, overexpression of constitutively active AKT significantly reversed cell death induced by co-treatment of EGFR inhibitor and Met inhibitor in pancreatic cancer cells (Fig. 5i). When we pretreated cells with PD184352 (MEK1/2 inhibitor) and BP-1-102 (STAT3 inhibitor), it did not significantly affect the cell death induced by combination treatment (Supplementary Fig S4f and g). These results indicated that the MAPK and STAT3 signaling pathways may be involved but are not sufficient for the regulation of HGF-mediated suppression of EGFR inhibitor sensitivity. The above findings suggest that HGF-mediated suppression of EGFR inhibitor sensitivity in pancreatic cancer cells occurs via re-activation of the PI3K/AKT signaling pathway.
The combination of EGFR inhibitor and Met inhibitor significantly suppresses pancreatic cancer growth
To further confirm the role of HGF-Met signaling in pancreatic cancer resistance to erlotinib, we established an orthotopic model by implanting PANC-1 Luc cells or PANC-1 Luc cells plus PSC into the pancreases of immunodeficiency mice. The schema of the animal model is shown in Fig. 6a. After the first bioluminescence imaging on the seventh day of implantation, EGFR inhibitor, Met inhibitor, EGFR inhibitor plus Met inhibitor, or vehicle control was administered to mice in each group daily for seven weeks. We observed that ERL monotherapy inhibited tumor growth in both the mono-transplantation co-transplantation models (Supplementary Fig. S5a and b, Fig. 6b–e). ERL in combination with Criz significantly inhibited tumor growth in the co-transplantation model compared with the Criz alone or ERL alone groups (Fig. 6b–e). Consistent with the previous reports, the co-transplantation group's tumors grew faster than those in the mono-transplantation group. There was no significant toxicity observed (Supplementary Fig. S5c). These results showed that the combination of EGFR inhibitor and Met inhibitor significantly suppresses pancreatic cancer growth in orthotopic mouse model.
Fig. 6.
Met inhibitor in combination with the EGFR inhibitor showed a remarkable effect in suppressing pancreatic cancer growth in vivo. a, The tumor pancreatic orthotopic formation assay in BALB/c mouse model. PANC-1 Luciferase (Luc) cells (control group, n = 6) or PANC-1 Luc mixed with PSC (co-injection group, n = 6) were implanted into the pancreata of each immunodeficient mouse. b, The tumor volume curves of indicated group were monitored by bioluminescence image analysis. c, Tumor volumes were assessed in orthotopically implanted mice on the eighth week after all mice were euthanized. d, Representative bioluminescence images of indicated groups. e, Tumor weight were assessed in orthotopically implanted mice on the eight week after all mice were euthanized. f, Schematic diagram of how PSC-derived HGF suppresses the sensitivity of EGFR inhibitor in pancreatic cancer. Left: Hypoxia promotes the transcriptional activation of HGF gene expression by HIF-1α in the pancreatic stellate cell, then the overexpressed HGF contributed to the resistance of pancreatic cancer cells to EGFR inhibitor through re-activating the PI3K-AKT pathway. Right: Met inhibitor inhibited HGF mediated downstream signaling and increased the sensitivity of pancreatic cancer cells to EGFR inhibitor. ns: not significant, ∗P < 0.05, ∗∗P < 0.01.
Discussion
In the present study, we found that high levels of α-SMA+ myofibroblasts and hypoxia predict poor prognosis in pancreatic cancer. Further studies indicated that pancreatic stellate cell-derived HGF contributes to the limited sensitivity of EGFR inhibitor in pancreatic cancer cells by re-activating the Met-PI3K-AKT pathway (Fig. 6f). Although some studies have shown the tumor-suppressing role of stroma on tumor growth and metastasis, there is continuously growing evidence supporting the tumor-promoting role of PSC.50, 51, 52, 53 On one hand, α-SMA+ myofibroblasts promote tumor progression by secreting cytokines; on the other hand, α-SMA+ myofibroblasts suppress tumor proliferation and metastasis via secreting ECM, creating hypoxia and nutrient-deficient microenvironments.54 Recently, Öhlund et al. revealed the heterogeneity of CAF in pancreatic cancer, as they found that two types of CAF coexist with significant differences in space and function in pancreatic cancer, which provides new information on the controversial CAF function in pancreatic cancer.55 Although the role of PSC in pancreatic cancer remains controversial, our findings provide a potential mechanism of EGFR inhibitor resistance mediated by PSC under hypoxia.
HGF, also called scatter factor, is a mesenchymal cell-derived cytokine that activates a tyrosine kinase signalling cascade via binding to the Met receptor.56 The Met proto-oncogene plays a pivotal role in tumor invasion and metastasis, but the abnormal HGF/Met signaling in the human tumor microenvironment is not fully understood.57,58 Pennacchietti et al. reported that hypoxia increases the expression of Met, which can partly account for the overexpressed Met in human neoplastic lesions.59 McDonald et al. found pancreatic cancer cells with activated KRAS upregulate CAIX via stabilizing HIF-1α in response to hypoxia.60 Xu et al. demonstrated HGF/c-Met pathway is a crucial element of pancreatic cancer chemotherapy that limits primary tumor growth and eliminates metastasis.61 Our present results showed that hypoxia could also promote the transcriptional activation of HGF gene expression in the stromal cells of pancreatic cancer. Considering the critical role of HGF-Met signaling in human cancer, our study further explored the underlying mechanism between hypoxia and HGF-Met signaling in pancreatic cancer.
Drug resistance is a big challenge for patients with pancreatic cancer.62, 63, 64, 65 Wilson et al. found that growth factors can drive the resistance of cancer cells to kinase inhibitors.66 Straussman et al. demonstrated that the tumor microenvironment promoted innate resistance to RAF inhibitors in melanoma by secreting HGF.25 Joosten et al. reported that EGFR inhibition could be overcome by HGF/Met signaling in colorectal cancer.23 The EGFR-targeted inhibitor erlotinib has also been approved for treating pancreatic cancer but has had little effect in clinical applications.39 The overall results were not sufficiently significant for the FDA to recommend the combination of gemcitabine with erlotinib as a standard of care. Overexpression of EGFR in pancreatic cancer provides a basis for using EGFR-targeted inhibitors for erlotinib in this disease. Many studies have shown that the main reason for the poor efficacy of EGFR-targeted inhibitors in pancreatic cancer is that KRAS is abundantly mutated in pancreatic cancer cells, and EGFR is located at upstream of KRAS.67,68 The inhibition of EGFR cannot prevent the downstream oncogene signaling pathway driven by KRAS mutation. However, EGFR signaling is necessary for KRAS-driven pancreatic tumorigenesis and progression, with an unknown mechanism. Recently, Yang et al. found that Rhein, a medicinal herb, could potentiate human pancreatic cancer cells to EGFR inhibitors by suppressing the STAT3 signaling pathway.69 Our results indicate that the Met-PI3K-AKT pathway is specifically against EGFR-targeted inhibitor-induced apoptosis in a KRAS-independent manner, regardless of KRAS mutation status. A similar erlotinib resistance effect was demonstrated in the KRAS mutant PANC-1 cell line and the KRAS wild-type BxPC-3 cell line. We suspect that the intrinsic resistance of pancreatic cancer to erlotinib is at least partially caused by HGF secreted from the pancreatic tumor microenvironment. Whether HGF also plays a role in erlotinib-acquired drug-resistant pancreatic cancer remains to be determined.
Several small molecule inhibitors or antibodies of HGF or Met are in clinical development or have been approved by the FDA for use in other diseases.70 Given the resistance of these drugs to EGFR inhibitors, joint clinical trials of pancreatic cancer and other tumor types should be considered. Our findings indicate that the microenvironment is an important source of anticancer drug resistance. We recognize that this study has limitations: 1) The cohort of 90 patients is from a single center which may somewhat limit the generalizability of the study findings. 2) The effect of ERL combined with Criz against tumor growth was evaluated only in mice with orthotopic xenograft tumors. However, the results of the present study indicated that the mechanism of resistance could be revealed by systematically dissecting the interaction between tumor cells and the microenvironment.
In conclusion, we showed that hypoxia increased the secretion of HGF from stromal cells in pancreatic cancer, contributing to the limited effect of EGFR inhibitor in a KRAS-independent manner. We found that hypoxia promoted HGF expression and secretion in pancreatic stellate cells via transcriptional activation. HGF acts on Met, an HGF receptor, suppressing the sensitivity of EGFR inhibitor in pancreatic cancer cells by re-activating the PI3K-AKT pathway. The combination of EGFR inhibitor and Met inhibitor significantly suppressed pancreatic cancer growth in an orthotopic xenograft mouse model. These results indicate that the combined inhibition of EGFR and Met might be a promising strategy for pancreatic cancer therapy.
Contributors
Conceptualisation: XS, MW, YZ, ML, and RQ; Data curation, formal analysis, and methodology: XS, MW, YZ, XG, ML, ZZ, YZ, RH, YG, YL, SP, MZ, CZ, TY, XL, HW, JY, FZ, ML, and RQ; Software and formal analysis: XS, MW, YZ, XG, ML, ZZ, YZ, RH, YG, YL, SP, MZ, CZ, TY, XL, HW, JY, FZ, ML, and RQ; Validation, visualisation, writing – original draft: XS, MW, and YZ. Funding acquisition: RQ and MW; Project administration and supervision: ML and RQ; Writing – review & editing: XS, MW, YZ, ML, and RQ. All authors read and approved the final manuscript.
Data sharing statement
All data are available upon request to the corresponding authors.
Declaration of interests
The authors declare that they have no competing interests.
Acknowledgments
This work was supported by The National Natural Science Foundation of China grants 81772950 (to RQ) and 81773160 (to MW).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2022.104352.
Contributor Information
Min Li, Email: Min-Li@ouhsc.edu.
Renyi Qin, Email: ryqin@tjh.tjmu.edu.cn.
Appendix A. Supplementary data
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Erkan M., Reiser-Erkan C., Michalski C.W., et al. Cancer-stellate cell interactions perpetuate the hypoxia-fibrosis cycle in pancreatic ductal adenocarcinoma. Neoplasia. 2009;11(5):497–508. doi: 10.1593/neo.81618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vonderheide R.H., Bayne L.J. Inflammatory networks and immune surveillance of pancreatic carcinoma. Curr Opin Immunol. 2013;25(2):200–205. doi: 10.1016/j.coi.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherman M.H., Yu R.T., Tseng T.W., et al. Stromal cues regulate the pancreatic cancer epigenome and metabolome. Proc Natl Acad Sci U S A. 2017;114(5):1129–1134. doi: 10.1073/pnas.1620164114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carstens J.L., Correa de Sampaio P., Yang D., et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun. 2017;8 doi: 10.1038/ncomms15095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fan Y.F., Shang W.T., Lu G.H., et al. Decreasing hyaluronic acid combined with drug-loaded nanoprobes improve the delivery and efficacy of chemotherapeutic drugs for pancreatic cancer. Cancer Lett. 2021;523:1–9. doi: 10.1016/j.canlet.2021.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Endo S., Nakata K., Ohuchida K., et al. Autophagy is required for activation of pancreatic stellate cells, associated with pancreatic cancer progression and promotes growth of pancreatic tumors in mice. Gastroenterology. 2017;152(6):1492. doi: 10.1053/j.gastro.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 8.Neesse A., Bauer C.A., Ohlund D., et al. Stromal biology and therapy in pancreatic cancer: ready for clinical translation? Gut. 2019;68(1):159–171. doi: 10.1136/gutjnl-2018-316451. [DOI] [PubMed] [Google Scholar]
- 9.Cai W., Sun X., Jin F., et al. PERK-eIF2alpha-ERK1/2 axis drives mesenchymal-endothelial transition of cancer-associated fibroblasts in pancreatic cancer. Cancer Lett. 2021;515:86–95. doi: 10.1016/j.canlet.2021.05.021. [DOI] [PubMed] [Google Scholar]
- 10.Kuninty P.R., Bansal R., De Geus S.W.L., et al. ITGA5 inhibition in pancreatic stellate cells attenuates desmoplasia and potentiates efficacy of chemotherapy in pancreatic cancer. Sci Adv. 2019;5(9) doi: 10.1126/sciadv.aax2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee J.W., Stone M.L., Porrett P.M., et al. Hepatocytes direct the formation of a pro-metastatic niche in the liver. Nature. 2019;567(7747):249–252. doi: 10.1038/s41586-019-1004-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tape C.J., Ling S., Dimitriadi M., et al. Oncogenic KRAS regulates tumor cell signaling via stromal reciprocation. Cell. 2016;165(4):910–920. doi: 10.1016/j.cell.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sousa C.M., Biancur D.E., Wang X., et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536(7617):479–483. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auciello F.R., Bulusu V., Oon C., et al. A stromal lysolipid-autotaxin signaling axis promotes pancreatic tumor progression. Cancer Discov. 2019;9(5):617–627. doi: 10.1158/2159-8290.CD-18-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y., Gao W., Lytle N.K., et al. Targeting LIF-mediated paracrine interaction for pancreatic cancer therapy and monitoring. Nature. 2019;569(7754):131–135. doi: 10.1038/s41586-019-1130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ide T., Kitajima Y., Miyoshi A., et al. Tumor-stromal cell interaction under hypoxia increases the invasiveness of pancreatic cancer cells through the hepatocyte growth factor/c-Met pathway. Int J Cancer. 2006;119(12):2750–2759. doi: 10.1002/ijc.22178. [DOI] [PubMed] [Google Scholar]
- 17.Tang H.W., Weng J.H., Lee W.X., et al. mTORC1-chaperonin CCT signaling regulates m(6)A RNA methylation to suppress autophagy. Proc Natl Acad Sci U S A. 2021;118(10) doi: 10.1073/pnas.2021945118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAleese C.E., Choudhury C., Butcher N.J., Minchin R.F. Hypoxia-mediated drug resistance in breast cancers. Cancer Lett. 2021;502:189–199. doi: 10.1016/j.canlet.2020.11.045. [DOI] [PubMed] [Google Scholar]
- 19.Choudhry H., Harris A.L. Advances in hypoxia-inducible factor biology. Cell Metab. 2018;27(2):281–298. doi: 10.1016/j.cmet.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Ye L., Jin K., Liao Z., et al. Hypoxia-reprogrammed regulatory group 2 innate lymphoid cells promote immunosuppression in pancreatic cancer. EBioMedicine. 2022;79 doi: 10.1016/j.ebiom.2022.104016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., Bao Y., Song Y., et al. Hypoxia-alleviated nanoplatform to enhance chemosensitivity and sonodynamic effect in pancreatic cancer. Cancer Lett. 2021;520:100–108. doi: 10.1016/j.canlet.2021.07.008. [DOI] [PubMed] [Google Scholar]
- 22.Li X., Lee Y., Kang Y., et al. Hypoxia-induced autophagy of stellate cells inhibits expression and secretion of lumican into microenvironment of pancreatic ductal adenocarcinoma. Cell Death Differ. 2019;26(2):382–393. doi: 10.1038/s41418-018-0207-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Joosten S.P.J., Mizutani T., Spaargaren M., Clevers H., Pals S.T. MET signaling overcomes epidermal growth factor receptor inhibition in normal and colorectal cancer stem cells causing drug resistance. Gastroenterology. 2019;157(4):1153–1155.e1. doi: 10.1053/j.gastro.2019.06.029. [DOI] [PubMed] [Google Scholar]
- 24.Cascone T., Xu L., Lin H.Y., et al. The HGF/c-MET pathway is a driver and biomarker of VEGFR-inhibitor resistance and vascular remodeling in non-small cell lung cancer. Clin Cancer Res. 2017;23(18):5489–5501. doi: 10.1158/1078-0432.CCR-16-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Straussman R., Morikawa T., Shee K., et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487(7408):500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Apicella M., Giannoni E., Fiore S., et al. Increased lactate secretion by cancer cells sustains non-cell-autonomous adaptive resistance to MET and EGFR targeted therapies. Cell Metab. 2018;28(6):848–865.e6. doi: 10.1016/j.cmet.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Mori S., Akita H., Kobayashi S., et al. Inhibition of c-MET reverses radiation-induced malignant potential in pancreatic cancer. Cancer Lett. 2021;512:51–59. doi: 10.1016/j.canlet.2021.04.029. [DOI] [PubMed] [Google Scholar]
- 28.Yu F., Lin Y., Zhan T., Chen L., Guo S. HGF expression induced by HIF-1alpha promote the proliferation and tube formation of endothelial progenitor cells. Cell Biol Int. 2015;39(3):310–317. doi: 10.1002/cbin.10397. [DOI] [PubMed] [Google Scholar]
- 29.Gluck A.A., Orlando E., Leiser D., et al. Identification of a MET-eIF4G1 translational regulation axis that controls HIF-1alpha levels under hypoxia. Oncogene. 2018;37(30):4181–4196. doi: 10.1038/s41388-018-0256-6. [DOI] [PubMed] [Google Scholar]
- 30.Yu S., Li A., Liu Q., et al. Recent advances of bispecific antibodies in solid tumors. J Hematol Oncol. 2017;10(1):155. doi: 10.1186/s13045-017-0522-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsao M.S., Sakurada A., Cutz J.C., et al. Erlotinib in lung cancer - molecular and clinical predictors of outcome. N Engl J Med. 2005;353(2):133–144. doi: 10.1056/NEJMoa050736. [DOI] [PubMed] [Google Scholar]
- 33.Karapetis C.S., Khambata-Ford S., Jonker D.J., et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359(17):1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 34.Liu Q., Yu S., Zhao W., Qin S., Chu Q., Wu K. EGFR-TKIs resistance via EGFR-independent signaling pathways. Mol Cancer. 2018;17(1):53. doi: 10.1186/s12943-018-0793-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.da Cunha Santos G., Dhani N., Tu D., et al. Molecular predictors of outcome in a phase 3 study of gemcitabine and erlotinib therapy in patients with advanced pancreatic cancer: National Cancer Institute of Canada Clinical Trials Group Study PA.3. Cancer. 2010;116(24):5599–5607. doi: 10.1002/cncr.25393. [DOI] [PubMed] [Google Scholar]
- 36.Yan H.H., Jung K.H., Lee J.E., et al. ANGPTL4 accelerates KRAS(G12D)-induced acinar to ductal metaplasia and pancreatic carcinogenesis. Cancer Lett. 2021;519:185–198. doi: 10.1016/j.canlet.2021.07.036. [DOI] [PubMed] [Google Scholar]
- 37.Chang W.H., Nguyen T.T., Hsu C.H., et al. KRAS-dependent cancer cells promote survival by producing exosomes enriched in Survivin. Cancer Lett. 2021;517:66–77. doi: 10.1016/j.canlet.2021.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ardito C.M., Gruner B.M., Takeuchi K.K., et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell. 2012;22(3):304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore M.J., Goldstein D., Hamm J., et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol. 2007;25(15):1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- 40.Xiong H.Q., Rosenberg A., LoBuglio A., et al. Cetuximab, a monoclonal antibody targeting the epidermal growth factor receptor, in combination with gemcitabine for advanced pancreatic cancer: a multicenter phase II trial. J Clin Oncol. 2004;22(13):2610–2616. doi: 10.1200/JCO.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 41.Shi X., Yang J., Liu M., et al. Circular RNA ANAPC7 inhibits tumor growth and muscle wasting via PHLPP2-AKT-TGF-beta signaling axis in pancreatic cancer. Gastroenterology. 2022;162(7):2004–20017.e2. doi: 10.1053/j.gastro.2022.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J., Zhang Z., Zhang Y., et al. ZIP4 promotes muscle wasting and cachexia in mice with orthotopic pancreatic tumors by stimulating RAB27B-regulated release of extracellular vesicles from cancer cells. Gastroenterology. 2019;156(3):722–734.e6. doi: 10.1053/j.gastro.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Charan J., Kantharia N.D. How to calculate sample size in animal studies? J Pharmacol Pharmacother. 2013;4(4):303–306. doi: 10.4103/0976-500X.119726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Q., Zhang B., Hu Q., et al. The impact of cancer-associated fibroblasts on major hallmarks of pancreatic cancer. Theranostics. 2018;8(18):5072–5087. doi: 10.7150/thno.26546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Intlekofer Andrew M., Dematteo Raymond G., Venneti S., et al. Hypoxia induces production of L-2-hydroxyglutarate. Cell Metab. 2015;22(2):304–311. doi: 10.1016/j.cmet.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel M.B., Pothula S.P., Xu Z., et al. The role of the hepatocyte growth factor/c-MET pathway in pancreatic stellate cell-endothelial cell interactions: antiangiogenic implications in pancreatic cancer. Carcinogenesis. 2014;35(8):1891–1900. doi: 10.1093/carcin/bgu122. [DOI] [PubMed] [Google Scholar]
- 47.Khan A., Fornes O., Stigliani A., et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46(D1):D260–d6. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X., Wang Z., Ma Q., et al. Sonic hedgehog paracrine signaling activates stromal cells to promote perineural invasion in pancreatic cancer. Clin Cancer Res. 2014;20(16):4326–4338. doi: 10.1158/1078-0432.CCR-13-3426. [DOI] [PubMed] [Google Scholar]
- 49.Scaltriti M., Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res. 2006;12(18):5268–5272. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 50.Ozdemir B.C., Pentcheva-Hoang T., Carstens J.L., et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25(6):719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhim A.D., Oberstein P.E., Thomas D.H., et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell. 2014;25(6):735–747. doi: 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schnittert J., Bansal R., Prakash J. Targeting pancreatic stellate cells in cancer. Trends Cancer. 2019;5(2):128–142. doi: 10.1016/j.trecan.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 53.Iwamoto C., Ohuchida K., Shinkawa T., et al. Bone marrow-derived macrophages converted into cancer-associated fibroblast-like cells promote pancreatic cancer progression. Cancer Lett. 2021;512:15–27. doi: 10.1016/j.canlet.2021.04.013. [DOI] [PubMed] [Google Scholar]
- 54.Neesse A., Algul H., Tuveson D.A., Gress T.M. Stromal biology and therapy in pancreatic cancer: a changing paradigm. Gut. 2015;64(9):1476–1484. doi: 10.1136/gutjnl-2015-309304. [DOI] [PubMed] [Google Scholar]
- 55.Ohlund D., Handly-Santana A., Biffi G., et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214(3):579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Y., Zhang H., Han X., et al. STAT3 mediated upregulation of C-MET signaling acts as a compensatory survival mechanism upon EGFR family inhibition in chemoresistant breast cancer cells. Cancer Lett. 2021;519:328–342. doi: 10.1016/j.canlet.2021.07.048. [DOI] [PubMed] [Google Scholar]
- 57.Rajadurai C.V., Havrylov S., Zaoui K., et al. Met receptor tyrosine kinase signals through a cortactin-Gab1 scaffold complex, to mediate invadopodia. J Cell Sci. 2012;125(12):2940–2953. doi: 10.1242/jcs.100834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gu Z., Du Y., Zhao X., Wang C. Tumor microenvironment and metabolic remodeling in gemcitabine-based chemoresistance of pancreatic cancer. Cancer Lett. 2021;521:98–108. doi: 10.1016/j.canlet.2021.08.029. [DOI] [PubMed] [Google Scholar]
- 59.Pennacchietti S., Michieli P., Galluzzo M., Mazzone M., Giordano S., Comoglio P.M. Hypoxia promotes invasive growth by transcriptional activation of the met protooncogene. Cancer Cell. 2003;3(4):347–361. doi: 10.1016/s1535-6108(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 60.McDonald P.C., Chafe S.C., Brown W.S., et al. Regulation of pH by carbonic anhydrase 9 mediates survival of pancreatic cancer cells with activated KRAS in response to hypoxia. Gastroenterology. 2019;157(3):823–837. doi: 10.1053/j.gastro.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 61.Xu Z., Pang T.C.Y., Liu A.C., et al. Targeting the HGF/c-MET pathway in advanced pancreatic cancer: a key element of treatment that limits primary tumour growth and eliminates metastasis. Br J Cancer. 2020;122(10):1486–1495. doi: 10.1038/s41416-020-0782-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu M., Zhang Y., Yang J., et al. ZIP4 increases expression of transcription factor ZEB1 to PROMOTE INTegrin alpha3beta1 signaling and inhibit expression of the gemcitabine transporter ENT1 in pancreatic cancer cells. Gastroenterology. 2020;158(3):679–692e1. doi: 10.1053/j.gastro.2019.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shukla S.K., Purohit V., Mehla K., et al. MUC1 and HIF-1alpha signaling crosstalk induces anabolic glucose metabolism to impart gemcitabine resistance to pancreatic cancer. Cancer Cell. 2017;32(3):392. doi: 10.1016/j.ccell.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 64.Yu X., Liu W., Wang Z., et al. CD73 induces gemcitabine resistance in pancreatic ductal adenocarcinoma: a promising target with non-canonical mechanisms. Cancer Lett. 2021;519:289–303. doi: 10.1016/j.canlet.2021.07.024. [DOI] [PubMed] [Google Scholar]
- 65.Lee J.E., Kang Y.W., Jung K.H., et al. Intracellular KRAS-specific antibody enhances the anti-tumor efficacy of gemcitabine in pancreatic cancer by inducing endosomal escape. Cancer Lett. 2021;507:97–111. doi: 10.1016/j.canlet.2021.03.015. [DOI] [PubMed] [Google Scholar]
- 66.Wilson T.R., Fridlyand J., Yan Y., et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487(7408):505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yarden Y., Sliwkowski M.X. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2(2):127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 68.Navas C., Hernandez-Porras I., Schuhmacher A.J., Sibilia M., Guerra C., Barbacid M. EGF receptor signaling is essential for k-ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22(3):318–330. doi: 10.1016/j.ccr.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang L., Lin S., Kang Y., et al. Rhein sensitizes human pancreatic cancer cells to EGFR inhibitors by inhibiting STAT3 pathway. J Exp Clin Cancer Res. 2019;38(1):31. doi: 10.1186/s13046-018-1015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu J., Su X., Li Z., et al. HGF/c-MET pathway in cancer: from molecular characterization to clinical evidence. Oncogene. 2021;40(28):4625–4651. doi: 10.1038/s41388-021-01863-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.