SUMMARY
Gene-based therapeutic strategies to lower ataxin-2 levels are emerging for the neurodegenerative diseases amyotrophic lateral sclerosis (ALS) and spinocerebellar ataxia type 2 (SCA2). Additional strategies to lower levels of ataxin-2 could be beneficial. Here, we perform a genome-wide arrayed small interfering RNA (siRNA) screen in human cells and identify RTN4R, the gene encoding the RTN4/NoGo-Receptor, as a potent modifier of ataxin-2 levels. RTN4R knockdown, or treatment with a peptide inhibitor, is sufficient to lower ataxin-2 protein levels in mouse and human neurons in vitro, and Rtn4r knockout mice have reduced ataxin-2 levels in vivo. We provide evidence that ataxin-2 shares a role with the RTN4/NoGo-Receptor in limiting axonal regeneration. Reduction of either protein increases axonal regrowth following axotomy. These data define the RTN4/NoGo-Receptor as a novel therapeutic target for ALS and SCA2 and implicate the targeting of ataxin-2 as a potential treatment following nerve injury.
Graphical abstract

In brief
Rodriguez et al. perform a screen for regulators of the ALS protein ataxin-2. Knockdown or peptide inhibition of RTN4/NoGo-receptor decreases levels of ataxin-2. Further study shows that ataxin-2 shares a role with RTN4/NoGo-Receptor in limiting axonal regeneration.
INTRODUCTION
Amyotrophic lateral sclerosis (ALS) is a rapidly progressive neurodegenerative disease characterized by loss of motor neurons from the brain and spinal cord. Loss of motor neurons leads to muscle weakness, paralysis, and eventual death 3–5 years following diagnosis (Taylor et al., 2016). Spinocerebellar ataxia type 2 (SCA2) is associated with loss of Purkinje neurons from the cerebellum, which affects balance and coordination and causes slow saccadic eye movements and cognitive impairment (Paulson et al., 2017). SCA2 is an autosomal dominant genetic disorder caused by long CAG repeat expansions (>34) in the ATXN2 gene, which encodes the ataxin-2 protein (Scoles and Pulst, 2018). Alternatively, intermediate length repeat expansions (27–33) in ATXN2 are a genetic risk factor for ALS (Elden et al., 2010). Therapeutic strategies to target ataxin-2 with wild-type repeat lengths in mice overexpressing human TDP-43—a protein integral in ALS pathology and disease—have shown efficacy in preclinical studies (Becker et al., 2017; Kim et al., 2020; Scoles et al., 2017), motivating the initiation of a clinical trial testing ATXN2-targeting antisense oligonucleotides (ASOs) in human patients with ALS (ClinicalTrials.gov: NCT04494256).
The role reduction of ataxin-2 plays in mitigating disease phenotypes in mice remains largely unresolved. Defective RNA processing contributes to ALS (Brown et al., 2022; Ma et al., 2022; Nussbacher et al., 2019). Ataxin-2 harbors a PAM2-domain, binds to poly(A)-binding protein, and contains an LSmdomain commonly found in splicing factors (Kozlov et al., 2001; Mangus et al., 1998; Neuwald and Koonin, 1998; Yokoshi et al., 2014). These domains likely promote ataxin-2 localization to mRNP granules and direct its role as a translational regulator (Bakthavachalu et al., 2018; Fittschen et al., 2015; Lastres-Becker et al., 2016; Lim and Allada, 2013; Nonhoff et al., 2007; Zhang et al., 2013). Ataxin-2 functions in multiple types of mRNP granules such as P-bodies and stress granules, and its association with mRNP granules in Drosophila neurons is linked to translation-dependent long-term memory (Andreev et al., 2015; Bakthavachalu et al., 2018; Paul et al., 2018). Knockdown of ataxin-2 reduces recruitment of TDP-43 to stress granules (Becker et al., 2017).
To discover targets and pathways to lower ataxin-2 levels, we developed an arrayed whole-genome small interfering RNA (siRNA) screen to specifically measure ataxin-2 levels. siRNA targets that decrease ataxin-2 were highly enriched for components of the spliceosome. Targeting one of the strongest hits, RTN4R, or peptide inhibition of RTN4R (also known as RTN4/NoGo-Receptor), a regulator of neurite development, reduced ataxin-2 levels in vitro and in vivo. Moreover, knockdown of ataxin-2 promoted axonal regeneration in primary cortical neurons following injury. These results leverage the robust effect of ataxin-2 as a disease modifier to define novel therapeutic targets.
RESULTS
Whole-genome siRNA screen identifies genes and cellular pathways that regulate ataxin-2 levels
We made a quantitative and highly sensitive reporter of endogenous ataxin-2 protein levels by inserting a HiBiT tag on the C terminus of ataxin-2 in HEK293T cells using CRISPR-Cas9 genome editing (Figure 1A). HiBiT is an 11 amino acid peptide subunit of the NanoBiT luciferase enzyme that is inert on its own but binds with high affinity to LgBiT, reconstituting NanoBiT (Dixon et al., 2016). This allows for easy and specific detection of ataxin-2 using an antibody-free blotting method (Figures 1B–1D). An in-well reaction of ataxin-2-HiBiT cell lysate with the exogenous addition of LgBiT and the furimazine substrate generates quantifiable bioluminescence as a direct measure of ataxin-2 abundance (Figure 1E). In parallel, we stably incorporated firefly luciferase (FFluc) into the ataxin-2-HiBiT line to normalize for protein abundance.
Figure 1. Generation of the ataxin-2-HiBiT cell line and overview of the whole-genome siRNA screen.

(A) We engineered an endogenous C-terminal HiBiT fusion on ataxin-2 using CRISPR-Cas9 genome editing in HEK293T cells. LgBiT compliments the HiBiT protein tag to form NanoBiT. As a control, we stably incorporated firefly luciferase (FFLuc) into this line.
(B) Antibody-based immunoblotting and HiBiT substrate-based detection on ataxin-2-HiBiT cell lysates transfected with siRNA.
(C and D) Quantification of immunoblot (A), n = 3.
(E) HiBiT signal measured via luciferase assay on cells transfected with increasing doses of siRNA, n = 5.
(F) Schematic of the whole-genome siRNA screen for regulators of ataxin-2 levels.
(G) Plot showing results of HiBiT replicates after filtering for changes in FFLuc. Datapoints in blue are primary screen hits that decrease ataxin-2. ATXN2 siRNA (red) was the strongest hit.
(H) GO-Slim Biological Process analysis of the hits from the primary screen.
(I) Summary of the major steps in pre-mRNA splicing, illustrating the involvement of the PRP19-associated complex (PRP19C) and the U1, U2, U4, U5, and U6 snRNP RNA-protein complexes.
(J) A list of the siRNAs that reduce ataxin-2 validated in the secondary screen.
(K) Ataxin-2-HiBiT cells treated with siRNAs against several splicing components, followed by a luciferase assay to measure HiBiT activity. Each splicing factor knockdown significantly reduces HiBiT signal relative to nontargeting. Two-way ANOVA with multiple comparisons: WBP11, SRSF3, and LSM5, p ≤ 0.001. ATXN2, SNRPB, SNRPC, SNRPD2, SNRPD3, SNRP70, PRPF8, ICE1, and AQR, p ≤ 0.0001.
(L) Immunoblot of unedited HEK293T cell lysates after siRNA treatment.
(C and D) Student’s t test. (E) One-way ANOVA with multiple comparisons. **p ≤ 0.01. Error bars represent ± SEM.
We used the ataxin-2-HiBiT cell line to perform a genome-wide screen using arrayed siRNA pools targeting 21,121 mRNA transcripts (Figures 1F, 1G, S1A, and S1B). We performed the screen in duplicate for both ataxin-2-HiBiT levels and FFluc levels, a control for ruling out siRNAs with nonspecific effects on gene expression. We classified an siRNA as a primary screen hit if it had an average HiBiT Z score less than −1.65, indicating a significant negative impact on ataxin-2 levels, and an average FFluc Z score greater than −1. We excluded siRNAs that significantly impacted expression levels of the FFluc control in the same direction as ataxin-2 levels—resulting in 348 primary screen hits (Table S1). We selected primary screen hits for validation based on function in shared pathways (e.g., splicing) or known roles in the nervous system. We performed a secondary validation screen using 102 siRNA pools, of which 77 validated, and we classified these as “high-confidence hits” (Figure S1C; Table S2).
Gene Ontology analysis of the primary screen hits revealed an enrichment in constituents of the pre-mRNA splicing pathway (Figures 1H and 1I), and 36 of the 77 high-confidence hits have roles in the major spliceosome complexes (Figure 1J). These proteins are enriched for the like-Sm or LSm domain (Figures S1D and S1E), a protein domain critical for complex formation and splicing activity (He and Parker, 2000). Ataxin-2 has an LSm domain that is one of the few predicted structured regions of this largely disordered protein. We independently validated several high-confidence hits using siRNAs in unedited HEK293T cells and immunoblotting for ataxin-2 (Figures 1K, 1L, and S1F). We also performed RT-qPCR and found that some of these hits affected steady-state ATXN2 mRNA levels (Figure S1G). The presence of a shared LSm domain combined with the regulation revealed by this screen suggests a potential functional connection between ataxin-2 and the spliceosome.
Knockdown or inhibition of the RTN4/NoGo-Receptor lowers ataxin-2 levels
Because splicing is an essential cellular process and splicing defects contribute to ALS (Lagier-Tourenne et al., 2010), we sought to identify the optimal target with therapeutic potential by first filtering high-confidence hits for essentiality (using the Dependency Map [DepMap] database, average gene effect > −1). DepMap is a resource of essential genes across a range of cell types (McFarland et al., 2018; Meyers et al., 2017), usually used to identify cancer vulnerabilities that can be exploited. However, for neurodegeneration targets, we sought to avoid essential genes and thus filtered these out of our hit list. We also filtered hits for central nervous system expression (GTEx database, cortical protein-coding transcript per million [pTPM] > 15) (GTEx Consortium, 2013) (Figure 2A). We reasoned that selecting targets with high nervous system specificity may eliminate potential negative off-target effects in nondiseased tissues. Following these filtering steps, the top hit was RTN4R, the gene that encodes RTN4/NoGo-Receptor. RTN4/NoGo-Receptor has been implicated in axon regeneration, sprouting, and plasticity (Akbik et al., 2013; Bhagat et al., 2016; Fink et al., 2015; Fournier et al., 2001; Kim et al., 2004; McGee et al., 2005; Wang et al., 2011, 2020), and targeting its ligand NoGo-A modulates mutant SOD1 in mouse models of ALS (Bros-Facer et al., 2014; Fournier et al., 2001; Jokic et al., 2006; Yang et al., 2009). RTN4/NoGo-Receptor has several glia- and neuron-derived ligands including the three gene products of the RTN4 gene—NoGo-A, -B, and -C—as well as oligodendrocyte myelin glycoprotein (OMgp) and myelin-associated glycoprotein (MAG) (Domeniconi et al., 2002; Schwab, 2010; Wang et al., 2002). Other high-affinity ligands include BAI adhesion G protein-coupled receptors (GPCRs) (Chong et al., 2018; Wang et al., 2021), LGI1 (Thomas et al., 2010), BLyS (Zhang et al., 2009), and LOTUS (Sato et al., 2011). RTN4R knockdown reduced ataxin-2 levels by HiBiT assay and immunoblot (Figures 2B–2D). We confirmed this in SH-SY5Y neuroblastoma cells (Figures S2A and S2B). Conversely, knockdown of ataxin-2 had no effect on RTN4/NoGo-Receptor levels (Figures S2C and S2D). This effect seems specific to ataxin-2 and not polyQ proteins in general because knocking down RTN4R did not affect expression levels of polyQ disease proteins huntingtin and ataxin-3 nor the ataxin-2 paralog ATXN2L (Figures S2E and S2F). As another functional readout of decreased ataxin-2 function (Becker et al., 2017), RTN4R knockdown reduced recruitment of TDP-43 to stress granules (Figures S3A and S3B). These data provide evidence that RTN4/NoGo-Receptor is required to maintain ataxin-2 levels.
Figure 2. Targeting RTN4R lowers levels of ataxin-2.

(A) High-confidence hits ranked by average HiBiT Z score, then filtered for essentiality (gene effect, DepMap) and CNS expression (GTEx).
(B) HiBiT signal measured by luciferase assay in ataxin-2-HiBiT cells transfected with designated siRNA, n = 6.
(C) Immunoblot for ataxin-2 and GAPDH on lysates derived from unedited HEK293T cells treated with designated siRNA.
(D) Ataxin-2 levels are quantified, n = 3. We performed RT-qPCR on RNA from cells treated as in (C).
(E and F) We probed for (E) RTN4R transcript (n = 4) or (F) ATXN2 transcript (n = 3) along with ACTB for normalization.
(G) We treated ataxin-2-HiBiT cells with siRNA then treated cells for 8 h with proteasome inhibitor MG-132 or DMSO and performed a luciferase assay to measure HiBiT activity (n = 6).
(H) The NEP1-40 peptide, a shared extracellular region of the NoGo proteins, binds to RTN4/NoGo-Receptor and prevents further signaling through the receptor.
(I) We treated ataxin-2-HiBiT cells for 48 h with increasing doses of NEP1-40 and performed a luciferase assay to measure HiBiT activity.
(B and I) One-way ANOVA with multiple comparisons. (G) Two-way ANOVA with multiple comparisons. (D–F) Student’s t test. *p ≤0.05, **p ≤0.01, ***p ≤0.001, ****p ≤ 0.0001. Error bars represent ± SEM.
Regulation of ataxin-2 by RTN4/NoGo-Receptor occurs at the protein level because RTN4R knockdown did not affect ATXN2 mRNA levels (Figures 2E and 2F). The HiBiT system allows for monitoring protein degradation (Riching et al., 2018). We used the ataxin-2-HiBiT line to test if RTN4R knockdown leads to ataxin-2 protein degradation by the proteasome or autophagy. Following siRNA knockdown of RTN4R, treatment of cells with a proteasome inhibitor (Figure 2G), but not an autophagy inhibitor (Figure S3C), resulted in an increase in ataxin-2 to levels comparable to the nontargeting control, indicating that ataxin-2 is degraded by the proteasome. RTN4R knockdown did not increase 20S proteasome activity (Figure S3D), suggesting a specific effect on ataxin-2 proteasomal degradation caused by RTN4R knockdown.
NEP1-40 is a peptide that acts as a competitive RTN4/NoGo-Receptor antagonist. It is a fragment of the luminal region of NoGo-A, -B, and -C that binds to RTN4/NoGo-Receptor to prevent ligand signaling (Figure 2H) (GrandPré et al., 2002). NEP1-40 treatment decreased ataxin-2 levels in the ataxin-2-Hi-BiT cells; 50 μM treatment caused a reduction in HiBiT signal to 77.52% of the 0 μM control condition (Figure 2I). Thus, targeting RTN4R by either genetic knockdown or with a peptide inhibitor leads to decreased levels of ataxin-2.
Knockdown or inhibition of the RTN4/NoGo-Receptor lowers ataxin-2 levels in mouse and human neurons
To extend our findings to neurons, we tested this interaction in mouse cortical neurons and human induced pluripotent stem cell (iPSC)-derived neurons (iNeurons) (Figures 3A and 3B) (Bieri et al., 2019). Treatment of mouse cortical neurons and human iNeurons with lentiviruses expressing short hairpin RNA (shRNA) targeting mouse or human RTN4R, respectively, resulted in about a 50% reduction of ataxin-2 (Figures 3C–3F and S4A–S4C), without affecting levels of ATXN2 mRNA (Figures S4D and S4E). Application of the NEP1-40 inhibitor peptide caused a dose-dependent reduction of ataxin-2 in both mouse cortical neurons and human iNeurons (Figures 3G–3L). Longer incubation times after the addition of shRNA-expressing lentivirus were needed to see a significant knockdown of the RTN4/NoGo-Receptor, whereas NEP1-40 addition binds and blocks the receptor effectively within hours of application (GrandPré et al., 2002), likely allowing for a more rapid effect on ataxin-2 levels. Additionally, the stability of the RTN4/NoGo-Receptor protein and how it changes between cell types (HEK293T and primary neurons) is not known; longer incubation time after RNAi treatment is likely required with a more stable protein in order to reach a sufficiently low concentration to observe the relevant outcome. To test the specificity of RTN4R knockdown, we performed RNA sequencing of iNeurons following RTN4R shRNA treatment. We found many transcript level alterations (Figure S5A; Table S3), but none of these connected to pathways highlighted by our ataxin-2 screen, suggesting a specific effect of RTN4R knockdown on ataxin-2 levels (Figure S5B). Together, these results suggest that targeting RTN4/NoGo-Receptor, either genetically or with a peptide inhibitor, is sufficient to markedly decrease levels of ataxin-2 in neurons.
Figure 3. RTN4R knockdown or inhibition of RTN4/NoGo-Receptor in mouse and human neurons and in mouse brain reduces ataxin-2 levels.

(A) Primary neurons isolated from embryonic mouse cortex. Treatment timeline for shRNA lentivirus or RTN4/NoGo-Receptor peptide inhibitor NEP1-40 in primary mouse neurons. DIV, days in vitro.
(B) Induced neuron differentiation in a human iPSC line with NGN2 stably integrated, as verified by Tuj1 (green) and NeuN (red) immunostaining. Scale bar: 20 μm. Treatment timeline for shRNA lentivirus or NEP1-40 in human induced neurons (iNeurons). DPI, days post induction.
(C) Immunoblot on lysates from mouse neurons treated with shRNA.
(D) Quantification of ataxin-2 and RTN4/NoGo-Receptor levels, n = 5–6.
(E) Immunoblot on lysates from iNeurons treated with shRNA.
(F) Quantification of ataxin-2 and RTN4/NoGo-Receptor (n = 3).
(G) Immunoblot on lysates from primary mouse neurons treated with increasing doses of NEP1-40.
(H) Quantification of (G), n = 4.
(I) Immunocytochemistry and fluorescence microscopy on mouse neurons treated with 50 μM NEP1-40. MAP2 labels neurons, DAPI labels nuclei. Scale bar: 20μm.
(J) Quantification of neuronal ataxin-2 fluorescence.
(K) Immunoblot on lysates from iNeurons treated for 48 h with increasing doses of NEP1-40.
(L) Quantification of (K), n = 2–3.
(M) Immunoblot on whole-cortex lysates from RTNR +/+, +/−, and −/− mice.
(N) Quantification of (M), n = 3–4.
(D, F, and J) Student’s t test. (H, L, and N) One-way ANOVA with multiple comparisons. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. Error bars represent ± SEM.
Since Sm-containing SNRP genes were among the strongest modifiers of ataxin-2 levels in our screen, we tested if knockdown of RTN4/NoGo-Receptor impacted SNRP gene expression. However, knocking down RTN4R did not affect SNRPB expression levels (Figures S5C and S5D). This suggests that RTN4R’s effect on ataxin-2 levels is independent of splicing pathways; however, a more systematic approach is needed to rule out every splicing factor yielded by our screen. Because ataxin-2 is a modifier of TDP-43 toxicity (Becker et al., 2017; Elden et al., 2010), we tested the effect of RTN4R knockdown on TDP-43-induced toxicity. We upregulated TDP-43 to induce degeneration and cell death in primary neurons (Maor-Nof et al., 2021), then measured caspase activation as a readout of cell death. Knockdown of RTN4R for 12 days prior to TDP-43 up-regulation prevented TDP-43-induced caspase activation (Figure S5E). These data extend our findings in HEK293T cells where reduction of RTN4R decreased TDP-43 transport into stress granules to further suggest that RTN4R knockdown can mitigate TDP-43-induced cellular toxicity.
Finally, we tested the effects of RTN4/NoGo-Receptor on ataxin-2 levels in vivo. We analyzed heterozygous and homozygous Rtn4r knockout mice. These mice are fertile and viable, with no apparent deficits in the nervous system (Kim et al., 2004). We observed a dose-dependent reduction in ataxin-2 in lysates from mouse cortex from the Rtn4r heterozygous and homozygous knockout mice compared with wild-type mice (Figures 3M and 3N), providing evidence that reduction of RTN4/NoGo-Receptor is sufficient to lower levels of ataxin-2 in the nervous system.
Knockdown of ataxin-2 improves axon regrowth after injury
RTN4/NoGo-Receptor signaling destabilizes the actin cytoskeleton leading to growth cone collapse, limiting neurite outgrowth (Chivatakarn et al., 2007; Montani et al., 2009; Schwab, 2010). Reduction or inhibition of RTN4/NoGo-Receptor or its ligands has been shown to limit axonal regeneration following injury (Fink et al., 2015; Fournier et al., 2001; Kim et al., 2004; Schwab and Strittmatter, 2014; Wang et al., 2011,2020). Importantly, our RNA sequencing results in iNeurons showed an increase in abundance of transcripts involved in axonal extension and neuronal development following RTN4R knockdown (Figure S5B). Since we have demonstrated that knocking down or inhibiting RTN4/NoGo-Receptor lowers ataxin-2, we tested if reducing ataxin-2 levels is sufficient to promote axon regeneration, perhaps functioning downstream of RTN4/NoGo-Receptor. We plated primary mouse neurons in the soma compartment of microfluidics chambers (Figure 4A), treated them with lentivirus-expressing shRNA targeting either Atxn2 or Rtn4r at day in vitro 5 (DIV5) (Figure 4B), and allowed them to mature and project axons through the microchannels and into the inner chamber of the axonal compartment. At DIV17, we performed vacuum-assisted axotomy to fully sever axons projecting into the inner chamber (Figure 4C) and permitted neurons to regenerate axons for 48 h. We analyzed Tuj1-stained axons and found that the average length of regrown axons was markedly increased in either the Rtn4r or Atxn2 knockdown conditions relative to the nontargeting control (nontargeting: 148.9 μm, Rtn4r. 188.8 μm, and Atxn2: 184.8 μm; Figures 4D and 4E). These results provide evidence of a role for ataxin-2 in limiting regeneration after nerve injury and present ataxin-2 as a potential therapeutic target following nerve injury. Additionally, this finding provides further evidence that RNA-binding proteins that drive RNA granule formation following stress can limit axon regeneration—as has been reported for TIAR-2 (Andrusiak et al., 2019; Becker et al., 2017; Boeynaems and Gitler, 2019; Liu-Yesucevitz et al., 2011).
Figure 4. Reduction of ataxin-2 increases axonal regrowth after axotomy.
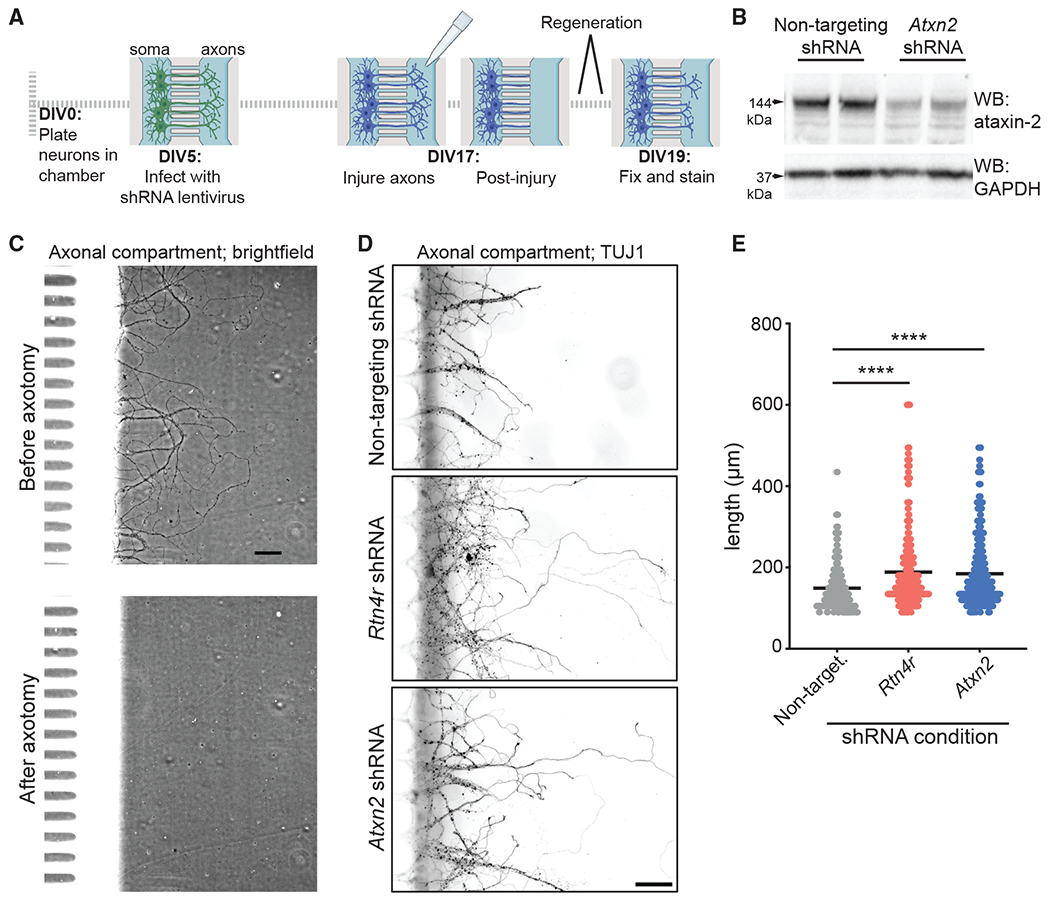
(A) Timeline for axotomy and regeneration experiment using mouse primary neurons grown in microfluidics chambers.
(B) Immunoblot on lysates from mouse neurons treated with Atxn2 shRNA.
(C) Bright-field images of an inner chamber of the axonal compartment of a microfluidics chamber before and after vacuum-assisted axotomy. Scale bar: 50 μm.
(D) Immunocytochemistry and fluorescence microscopy on the axonal compartment after 48 h of regrowth after axotomy. Tuj1 labels axons. Scale bar: 50 μm.
(E) Quantification of the length of regenerating neurites (identified by the morphological presence of a growth cone) from three separate chambers per condition, n = 234–306. One-way ANOVA with multiple comparisons. ****p ≤ 0.0001. Error bars represent ± SEM.
DISCUSSION
Here, we performed an unbiased genome-wide screen and discovered RTN4/NoGo-Receptor as a novel regulator of ataxin-2 levels. We provide evidence that RTN4R functions up-stream of ataxin-2. Since ataxin-2 has been implicated in two neurodegenerative diseases—ALS and SCA2—efforts are underway to target it therapeutically, including the use of ASOs targeting ATXN2 (Becker et al., 2017; Scoles et al., 2017). Together with the accompanying manuscript by Kim et al. identifying v-ATPase as a target to lower ataxin-2 levels and etidronate and other bisphosphonates as potent small-molecule inhibitors of ataxin-2 levels (Kim et al., 2022), we now present an additional way to reduce ataxin-2 levels. The hits we identified from this arrayed siRNA screen differ from those presented in the accompanying manuscript using a CRISPR-based pooled screen. We discuss the potential reasons for identifying different hits in each screen in the accompanying manuscript (Kim et al., 2022), but it is notable that we provide extensive validation of the lead hits from each screen.
RTN4/NoGo-Receptor seems like an optimal target for therapeutic reduction of ataxin-2 because in addition to gene-based strategies, there are several ways in which it may be targeted including receptor inhibition—demonstrated here—as well as neutralization of its ligands (Schwab, 2010). A RTN4/NoGo-Receptor decoy (Wang et al., 2011, 2020) is being investigated in a clinical trial for chronic spinal cord injury (ClinicalTrials.gov: NCT03989440). With additional targets and strategies to lower ataxin-2 levels in hand, combination therapies can be envisioned and further developed to have maximum therapeutic benefit and to mitigate potential negative effects of relying on a single strategy.
Limitations of the study
The effectiveness of RTN4R knockdown in rescuing degeneration in a mouse model of ALS remains to be tested. However, reducing ATXN2 is a validated method for rescuing degeneration phenotypes in a mouse model of TDP-43 toxicity and motor neuron degeneration (Becker et al., 2017) and in a mouse model of SCA2 (Scoles et al., 2017). Moreover, human genetics has provided compelling validation for ATXN2 as a therapeutic target for both ALS and SCA2 (Elden et al., 2010; Scoles and Pulst, 2018). The data presented should not be considered as preclinical but rather as new directions to build upon for modulating ataxin-2 that require further validation. Future work will be needed to determine the precise method for reducing RTN4R—and subsequently ATXN2—in mouse models to test its influence on disease phenotypes. But given that clinical trials are underway to test a NoGo-Receptor inhibitor in spinal cord injury (ClinicalTrials.gov: NCT03989440), lessons learned from that trial will hopefully aid in testing this approach for ALS and SCA2.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Aaron Gitler (agitler@stanford.edu).
Materials availability
Ataxin-2-HiBiT knock-in cell line is available upon reasonable request.
Data and code availability
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Ataxin-2 antibody | Novus | Cat# NBP1-90063; RRID:AB_11028587 |
| Tuj1 antibody | Biolegend | Cat# 802001; RRID:AB_2564645 |
| Actin antibody | EMD Millipore | Cat# MAB1501, RRID:AB_2223041 |
| RTN4/NoGo-Receptor antibody | Abcam | Cat# ab184556 |
| GAPDH antibody | Sigma Aldrich | Cat# G8795; RRID:AB_1078991 |
| Goat anti-Rabbit HRP antibody | Life Technologies | Cat# 31462; RRID:AB_228338 |
| Goat anti-mouse HRP antibody | Thermo Fisher Scientific | Cat# 62–6520; RRID:AB_2533947 |
| MAP2 antibody | Synaptic Systems | Cat# 188004; RRID:AB_2138181 |
| NeuN antibody | EMD Millipore | Cat# MAB377; RRID:AB_2298772 |
| Chemicals, peptides, and recombinant proteins | ||
| NEP1-40 peptide | Tocris | Cat# 1984 |
| Critical commercial assays | ||
| Nano-Glo HiBiT Lytic Detection System | Promega | Cat# N3050 |
| ONE-GLO Luciferase Assay System | Promega | Cat# E6120 |
| TaqMan gene-specific expression assay: human ATXN2 | Thermo Fisher Scientific | Hs00268077_m1 |
| TaqMan gene-specific expression assay: human ACTB | Thermo Fisher Scientific | Hs01060665_g1 |
| TaqMan gene-specific expression assay: human RTN4R | Thermo Fisher Scientific | Hs00368533_m1 |
| TaqMan gene-specific expression assay: mouse ATXN2 | Thermo Fisher Scientific | Mm00485932_m1 |
| TaqMan gene-specific expression assay: mouse GAPDH | Thermo Fisher Scientific | Mm99999915_g1 |
| TaqMan gene-specific expression assay: mouse RTN4R | Thermo Fisher Scientific | Mm00452228_m1 |
| Human ATXN2 siRNA | Horizon Discovery | Cat# L-011772-00 |
| Non-targeting siRNA | Horizon Discovery | Cat# D-001810-10 |
| Human RTN4R siRNA | Horizon Discovery | Cat# L-008075-00 |
| Oligonucleotides | ||
| Target-specific sgRNA sequence | IDT | AGCCTTACAACTGCTGTTGG |
| Forward Primer for ATXN2 exon 25 amplification and sequencing | IDT | GCAATACTGGTGCTTGGCTAATATTTGGGG |
| Reverse Primer for ATXN2 exon 25 amplification and sequencing | IDT | CACTCTTGTTACTTCTTTTGCTAGCTGATGTG |
| Deposited data | ||
| iNeuron RNA sequencing data | GEO | GEO: GSE200530 |
| Experimental models: Cell lines | ||
| HEK293T Cells | ATCC | Cat# CRL-321; RRID:CVCL_0063 |
| SH-SY5Y | ATCC | Cat# CRL-226; RRID:CVCL_0019 |
| Experimental models: Organisms/strains | ||
| Time pregnant C57BL/6 | Charles River Labs | Cat# C57BL/6NCr; RRID:MGI:2159769 |
| Rtn4r KO mouse line | Provided by Dr. Stephen Strittmatter | N/A |
| Recombinant DNA | ||
| Mission® shRNA, mouse RTN4R | Sigma Aldrich | TRCN0000436683 |
| Mission® shRNA, human RTN4R | Sigma Aldrich | TRCN0000061558 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Mice were bred, cared for, and experimented on as approved by the Administrative Panel of Laboratory Animal Care (APLAC) of Stanford University. Mice were housed in ventilated cages with 2–5 animals per cage, with a 12-h light/dark cycle. They were fed ad libitum with standard laboratory chow. For neuron cultures, time-pregnant C57BL/6 mice were procured from Charles River Labs. Mouse embryos of both sexes were removed at embryonic day 16.5 for dissociation and culturing. For Rtn4r mouse studies, mice of both sexes were sacrificed at 20 days old for tissue harvest for all genotypes listed. Heterozygous breeders were generously provided by the laboratory of Dr. Stephen Strittmatter.
Cell lines
HEK293T and SH-SY5Y cells were obtained from ATCC. No additional authentication was performed on these lines. HEK293T cells were maintained in a 37°C incubator with 5% CO2 in DMEM with Glutamax and HEPES (Thermo Fisher Scientific, cat# 10564–029), 10% fetal bovine serum (vol/vol; Invitrogen, cat# 16000–044), and 1% Pen/Strep (vol/vol; Invitrogen, cat# 15140–122). SH-SY5Y cells were maintained in a 37°C incubator with 5% CO2 in DMEM/F12 (Thermo Fisher Scientific, cat# 11320033), 10% fetal bovine serum, and 1% Pen/Strep.
We induced neuron differentiation in human iPSC-derived neurons (iNeurons) using a previously established protocol with a Tet-On induction system that allows expression of the transcription factor NGN2 (Bieri et al., 2019). Briefly, iPS cells were maintained with mTeSR1 medium (Stemcell Technologies, cat# 85850) on Matrigel (Fisher Scientific, cat# CB-40230). On the next day of passage, NGN2 was expressed by adding doxycycline (2μg/mL) and selection with puromycin (2μg/mL) for rapid and highly efficient iNeuron induction. Three days after induction, iNeurons were dissociated and grown in Neurobasal medium containing N-2 supplement (Gibco, cat# 17502048), B-27 supplement, BDNF (R&D Systems) and GDNF (R&D Systems) on Matrigel-coated plates.
Primary cell culture
For cortical cultures, mouse embryos were removed at E16.5, and cortices were isolated and placed in ice-cold Dissociation Media (DM; PBS without Mg2+ or Ca2+, supplemented with 0.6% glucose, 10mM HEPES, and Pen/Strep). Neurons were dissociated with the Papain Dissociation System (Worthington Biochemical Corporation, cat# LK003153). Cells were seeded on Poly-L-lysine (Sigma Aldrich, cat# P4832) coated 24-well plates at a density of 350,000cells/well. They were grown in Neurobasal media supplemented with B-27 at 1:50 (Life Technologies, cat# 17504–044), Glutamax at 1:100 (Invitrogen, cat# 35050–061) and Pen/Strep. Neurons were maintained in a 37°C incubator with 5% CO2 with half media changes every 4 to 5 days.
METHOD DETAILS
Cell transfection
Cells were reverse transfected on 96-well plates for luciferase assays. 25μL of Opti-MEM (Life Technologies, cat# 31985–062) with 0.12μL of Dharmafect 1 (Horizon Discovery, cat# T-2001-03) and 200nM (unless specified in figure) of ON-TARGETplus siRNA (Horizon Discovery) or non-targeting (Horizon Discovery, cat# D-001810-10) was added to an individual well and incubated at room temperature for 30 min 1.0 × 104 cells/well in 100μL of medium (without Pen/Strep) was added to the wells, and the plate was placed in the incubator for 72hr. To interrogate proteasome or autophagy regulation by HiBiT assay, 72hr after reverse transfection cells were treated for 6hr with 10μM MG-132 (Millipore Sigma, cat# M7449) or 24hr with 100μM Bafilomycin A (Sigma Aldrich, cat# B1793) or DMSO prior to lysing.
For immunoblotting, unedited HEK293T cells were maintained as above. Unedited cells were used to reduce cell line-specific effects. Cells were reverse transfected on 12-well plates. 200μL of Opti-MEM with 4μL of Dharmafect 1 and 200nM of ON-TARGETplus siRNA or non-targeting control was added to an individual well and incubated at room temperature for 30 min 2.0 × 105 cells/well were plated in 1mL of medium without Pen/Strep and placed in the incubator for 72hr prior to lysing for experimentation.
Small-scale luciferase assays
Ataxin-2-HiBiT cells were lysed in-well with 125μL of Nano-Glo® HiBiT Lytic Buffer, and lysate from one well was split and used for detection of both HiBiT- and FFLuc-generated luminescence. 25μL of lysate was placed in two wells of an opaque white, flat bottom 96-well assay plate (Sigma Aldrich, cat# CLS3990). For HiBiT detection, 25μL of HiBiT lytic reagent (1:25 substrate, 1:50 LgBiT) was added to the lysate (Promega, cat# N3050). For FFluc detection, 25μL of ONE-Glo Assay Buffer (Promega, cat# E6120) was added to the lysate. Plates were incubated in the dark with gentle rotation for 10 min, then luminescence was measured on a Tecan Spark plate reader (Tecan). For all assays, HiBiT signal was normalized to the FFluc signal for each individual well of the original transfected plate then normalized to the non-targeting/untreated control, this value is represented in bar graphs.
Genome editing
HEK293T cells (ATCC) were transfected using Lipofectamine™ CRISPRMAX™ Cas9 Transfection Reagent (Thermo Fisher Scientific, cat# cmax00003), TrueCut Cas9 Protein V2 (Thermo Fisher Technology, cat# A36497), purified sgRNA and ssDNA (IDT). Sequences are listed in the Key resources table. 72 hr later, cells were lifted and re-plated at a density of <1 cell/well in a 96-well plate. Wells were grown to confluency and split onto an additional plate. A HiBiT assay was performed on the additional plate as a preliminary screen for successful knock-in. The wells with HiBiT signal above the negative control (unedited cells) were maintained and confirmed for knock-in by Sanger sequencing (see Key resources table for primers). For firefly luciferase integration, the successful clone was transduced with lentivirus using pLenti CMV V5-LUC Blast (Addgene, cat# w567-1), followed by blasticidin selection. Transduced cells were pooled for future experimentation.
Immunoblotting
Cells were lysed in RIPA buffer (Sigma Aldrich, cat# R0278) supplemented with Complete mini protease inhibitor tablet (EMD Millipore, cat# 11836170001) and clarified by centrifugation at 21,000 x g for 15 min. Protein concentration was measured by BCA protein assay (Thermo Fisher Scientific, cat# 23225). Lysates were diluted to equal protein concentration in 1X NuPAGE® LDS Sample Buffer (Life Technologies, cat# NP0008). Lysates were boiled at 70°C for 10 min, then loaded on a NuPAGE® Novex® 4–12% Bis-Tris Protein gel (Life Technologies, cat# NP0321). Protein was transferred onto a nitrocellulose membrane (Bio-Rad, cat# 162–0115) at 4°C for 1hr and 45 min in 1X NuPAGE® Transfer Buffer (Life Technologies, cat# NP0006-1). Blocking and antibody incubation was performed in 2% BSA (Sigma Aldrich, cat# A7906) in PBS + 0.1% tween 20 (PBST). Washes were performed in PSBT. Primary antibodies were used at 1:1000, this includes: ataxin-2 antibody (Novus, cat# NBP1-90063), Tuj1 antibody (BioLegend, cat# 802001), actin antibody (EMD Millipore, cat# MAB1501), RTN4/NoGo-Receptor antibody (Abcam, ab184556), and GAPDH antibody (Sigma Aldrich, cat# G8795). Goat anti-Rabbit HRP (Life Technologies, cat# 31462) or goat anti-mouse HRP (Thermo Fisher Scientific, cat# 62–6520) secondary antibodies were used at 1:5000. Membranes were developed in ECL Prime (Sigma Aldrich, cat# GERPN2232), and imaged on a Bio-Rad ChemiDoc XRS+ imager (BioRad). HiBiT-based immunoblotting was performed according to the Nano-Glo® HiBiT Blotting System protocol (Promega, cat# n2410).
For mouse cortex, male and female mice at P20 were sacrificed for tissue harvesting. Ice-cold RIPA buffer with protease inhibitor was added and tissue was homogenized using a motorized pestle. Crude lysates were rocked at 4°C for 30 min, passed through a homogenization column (Qiagen, cat# 79656), and centrifuged at 21,000 x g for 15 min. Protein was quantified in the clarified supernatant and prepared for immunoblotting as above.
Whole-genome siRNA screen
The whole-genome siRNA screen was performed in the High-Throughput Bioscience Center (HTBC) at Stanford University. Briefly, we used the Dharmacon Human Genome siARRAY library (cat# G-005000-025) to target 21,121 genes individually in a single well of a 384-well plate with 4 pooled siRNAs. All siRNA pools were tested in duplicate for both FFluc and HiBiT on 4 separate plates. Each plate had three controls: siTOX control siRNA, nontargeting control siRNA, and ATXN2 siRNA (Horizon Discovery, cat# L-011772-00) as a toxicity, negative, and positive control, respectively. All siRNAs were reverse transfected using 10μL of Opti-MEM, 0.075μL/well of Dharmafect 1, and 10μL siRNA (final concentration of 50nM). Cells were seeded at 1000 cells/well in 30μL of media (as above without Pen/Strep) in solid white 384-well plates, then placed in a 37°C incubator with 5% CO2. After 3 days, plates were removed from the incubator. HiBiT lytic reagent was made by diluting Nano-Glo® HiBiT Lytic Substrate (1:50) and LgBiT Protein (1:100) in Nano-Glo® HiBiT Lytic Buffer. For HiBiT detection, 10 μL of HiBiT lytic reagent was dispensed into wells with a Multidrop Reagent Dispenser (Thermo Scientific), and rapidly shook for 15 s. For FFluc detection, 10μL of ONE-Glo Assay Buffer was dispensed into wells and rapidly shook for 15 s. Luminescence was read on a Tecan Infinite M1000 Pro (Tecan).
Z-scores were calculated for each siRNA replicate using the in-plate standard deviation, then averaged for both HiBiT and FFluc conditions respectively. The average Z-scores were used to filter for primary screen hits. siRNAs were only considered if they had an average FFluc Z score greater than −1, this cutoff removed any siRNA treatment that resulted in FFluc levels decreased more than one standard deviation from the mean. Since directionality of effect matters specifically when searching for a treatment that decreases ataxin-2, we implemented this cutoff for FFluc to ensure that our negative control was not similarly affected indicating a global and non-specific effect on the treated cells. siRNAs were categorized as negative regulators of ataxin-2 expression if they had an average HiBiT Z score less than −1.65. 102 of these were retested in a secondary screen by reselecting siRNAs from the original library stock. The same experimental parameters applied, except for the performance of a single replicate for FFluc detection. High confidence hits were chosen if they had a greater than 30% decrease in average HiBiT levels relative to the non-targeting siRNA control run in parallel on the same plate. High confidence hits were ranked by average HiBiT Z score, then were filtered further to determine the most optimal therapeutic target. These were filtered out if they had an average gene effect less than or equal to −1 as determined by DEMETR2 or CERES (depmap.org). Either measure shows the essentiality of a given gene across all cell lines tested for RNAi or CRISPR/Cas9 knockout respectively, where −1 is the median score for all essential genes and a score of 0 suggests the gene has no effect on survival in the cell lines tested. While such measures were developed to test genetic dependencies in cancer cell lines, we reasoned that implementing a cutoff of −1–indicating a “common essential” gene–would remove genes with the potential to cause toxicity in neural tissues or off-target toxicity elsewhere, making it an undesirable therapeutic target. The resulting hits were further filtered if they had a corresponding GTEx cortical pTPM less than 15 (proteinatlas.org).
Lentivirus production
HEK293T cells were grown on 10cm culture dishes to 80–90% confluency. Cells were transfected using Lipofectamine 3000. Briefly, 10μg of shRNA vector (Mission® shRNA, Sigma Aldrich), 2.9μg pRSV-REV, 5.8μg pMDLg/pRRE, 3.5μg pMD2.G, and 40μL of P3000 was added to 1mL of Opti-MEM. This was combined with another 1mL of Opti-MEM with 40μL of Lipofectamine 3000. This mixture was incubated at room temperature for 10 min then added to cells in media without antibiotics. 48hr after transfection, media was removed from cells, passed through a 0.45μm PES filter, and combined with Lenti-X concentrator (Takara, cat# 631232). The mixture was incubated over night at 4°C. This was centrifuged at 1,500 x g for 45 min at 4°C. The pellet was resuspended in 1mL Neurobasal Medium (Life Technologies, cat # 21103–049), 50μL was added directly to cells for transduction.
Primary neuron treatment
At DIV5, neurons were transduced with virus as above (Mission® shRNA, mouse RTN4R: TRCN0000436683). 24hr later, a half media change was performed. Neurons were maintained for 12 days after transduction prior to harvesting for experimentation. For caspase activity assays, this was followed by re-treatment with lentivirus expressing GFP or TDP-43 (Maor-Nof et al., 2021). Three or five days later, cells were lysed and the Caspase-Glo 3/7 assay (Promega, cat# G8090) was performed according to manufacturer protocol. For NEP1-40 (Tocris, cat# 1984) treatment, 1mg of NEP1-40 was diluted in 21.62μL of DMSO and 843.2μL of nuclease-free water (Thermo Fisher Scientific, cat# AM9937) to a working concentration of 250μM. The peptide was added to the final concentration specified along with vehicle (0μM). Cells were maintained for 48hr prior to experimentation.
For microfluidics chamber experiments, primary neurons were isolated as above and plated on one side of a Poly-D-lysine coated microfluidics chamber (Xona Microfluidics, cat# XC450) with a microchannel length of 450μm at a density of 200,000cells/chamber. Media was changed the next day, then half-changed every 3 days following. Lentivirus was added to the somatic compartment as above, then maintained for 12 days prior to axotomy. Vacuum-assisted axotomy was achieved by complete aspiration of media from the axonal compartment/inner chamber, allowing for an air bubble to dislodge and shear axons(Tong et al., 2015). Media was replaced and completely aspirated a second time. Media was replaced carefully to prevent the inclusion of any air bubbles in the inner chamber. A half media change was performed the next day. Neurons were allowed to regrow axons for a total of 48hr prior to fixation.
iNeuron treatment
At 7 days post neural induction, iNeurons were transduced with virus as above (Mission® shRNA, human RTN4R: TRCN0000061558). 24hr later, a half media change was performed. iNeurons were maintained for 3 days before being re-transduced with a half dose of virus, then maintained for another 5 days prior to harvesting for experimentation. For NEP1-40 treatment, 1mg of NEP1-40 was diluted as above. The peptide was added at 7 days post induction to the final concentration specified along with vehicle (0μM). Cells were maintained for 48hr prior to lysis.
Immunocytochemistry and fluorescence microscopy
Cells were fixed for 10 min in a solution of 4% paraformaldehyde/4% sucrose in PBS-MC (PBS with 1mM MgCl2 and 0.1 mM CaCl2). Then cells were washed in PBS-MC, and permeabilized for 10 min in 0.1% Triton X- in PBS-MC. Cells were blocked in 2% BSA for an hour, then incubated in primary antibody for at least 2 h at room temperature [ataxin-2 (Novus), MAP2 (Synaptic Systems, cat# 188004), Tuj1 (BioLegend, cat# 802001), NeuN (EMD Millipore, cat# MAB377)]. Cells were washed 3 times with PBS-MC, followed by incubation in species-specific Alexa Fluor®-labeled secondary antibody (Thermo Fisher Scientific) for 1 h at 1:1000. Cells were subsequently washed and placed in ProLong Gold Antifade with DAPI (Thermo Fisher Scientific, cat# P36931), and imaged using a Leica DMI6000B fluorescent microscope.
Images were processed in ImageJ. For neuronal expression, individual MAP2 labeled soma were selected as regions of interest (ROIs), in which ataxin-2 fluorescence was measured and averaged within each frame. This was performed for 4 frames per biological replicate (4 replicates for the NEP1-40 treatment and 3 for the vehicle), totaling 16 frames for the NEP1-40 condition and 12 for the vehicle. For stress granule analysis, stress granules were automatically determined based on shape and size using the Analyze Particles function in ImageJ in the G3BP channel. Each stress granule was made into an ROI in which TDP-43 fluorescence was measured and averaged for every stress granule in the frame. This was performed for 6 frames per biological duplicate, totaling 12 frames per condition. For axonal regrowth after axotomy, entire axonal compartments were imaged for three microfluidics chambers per shRNA condition (9 total). A grid image was superimposed onto each image with demarcations every 15μm after the end of the microchannels. Regenerating neurites were surveyed beginning 75μm from the end of the microchannel before which individual neurites are difficult to parse and there is a build-up of sheared neurites without the presence of growth cones. The length of every Tuj1-labeled neurite with a growth cone was determined by their crossing point on the grid and binned into lengths. Neurites with punctate Tuj1 staining (similar to beads on a string) were excluded from analysis, as these neurites were likely undergoing degeneration. The average of all neurites from each condition was taken.
RNA quantification and sequencing
Cells were reverse transfected with siRNAin 12-well plates as described above. RNA was isolated using the PureLink® RNA Mini Kit according to the kit protocol with DNase digestion (Thermo Fisher Scientific, cat# 12183025). 500ng of RNA was used to make cDNA using the Applied Biosystems High-Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, cat# 4368813). For RNA sequencing, polyA-selected libraries were prepared in biological duplicate for each shRNA condition using the TruSeq Stranded mRNA kit from Illumina and sequenced on a HiSeq 4000 (Illumina) with paired end, 75 nucleotide-long reads. GEO accession number GSE200530. qPCR was performed in a 20μL reaction using TaqMan™ Universal Master Mix II (Thermo Fisher Scientific, cat# 4440040) and 25ng of RNA. 1μL of 20X TaqMan gene-specific expression assay was added to the reaction (Thermo Fisher Scientific; human ATXN2: Hs00268077_m1, human ACTB: Hs01060665_g1, human RTN4R: Hs00368533_m1, mouse ATXN2: Mm00485932_m1, mouse GAPDH: Mm99999915_g1, mouse RTN4R: Mm00452228_m1). All conditions were run in technical triplicates that were averaged to account for each of the biological triplicates. Thermocycler was programmed according to the suggested protocol. Relative expression was calculated using the ΔΔCt method.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed in GraphPad Prism v.9, except the screen data calculations, which was performed in MATLAB (MathWorks). An unpaired Student’s t-test (two tailed) with a 95% confidence interval (CI) was performed for all assays comparing two experimental groups. A two-way analysis of variance (ANOVA) with a 95% CI was performed for the HiBiT assays with multiple siRNA conditions compared to respective non-targeting controls run in parallel and siRNA/drug treatment. A one-way ANOVA with multiple comparisons was performed for all other experiments with more than two experimental conditions. A Fisher’s LSD test was performed for the NEP1-40 treated HiBiT assay. A Dunnett’s multiple comparisons test was performed for all others. All bar graphs show the mean ± SEM. All samples were randomly assigned to experimental groups. No statistical methods were used to predetermine sample sizes.
Supplementary Material
Highlights.
Whole-genome siRNA screen in human cells identifies regulators of ataxin-2 levels
siRNA targets that decrease ataxin-2 are enriched for proteins with an LSm domain
RTN4/NoGo-Receptor knockdown or peptide inhibition in neurons reduces ataxin-2
Knockdown of ataxin-2 in primary neurons increases regeneration after axotomy
ACKNOWLEDGMENTS
This work was supported by NIH grants R35NS097263 (A.D.G.), R01AG064690 (A.D.G.), R35NS097283 (S.M.S.), F32NS116208 (C.M.R.), T32NS007280 (G.L.J.), T32AG047126 (T.A.), and F31NS125681 (G.K.); the Robert Packard Center for ALS Research at Johns Hopkins (A.D.G.); Target ALS (A.D.G.); and the Brain Rejuvenation Project of the Wu Tsai Neurosciences Institute (A.D.G.). G.K. is supported by a fellowship from the Stanford Knight-Hennessy Scholars Program. T.A. is supported by a fellowship from the Takeda Science Foundation. The whole-genome siRNA screen was performed with the expertise and resources in the High-Throughput Bioscience Center at Stanford University. Some of the figures were created with BioRender.com. RNA sequencing library preparation and sequencing was performed by the Stanford University Genome Sequencing Service Center.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111505.
DECLARATION OF INTERESTS
A.D.G. is a scientific founder of Maze Therapeutics. S.M.S. is a founder and equity holder in ReNetX Bio, Inc., which seeks clinical development of the decoy receptor, NgR1-Fc (AXER-204), for chronic spinal cord injury treatment. Stanford University has filed a provisional patent (63/388,086) on methods described in this manuscript for treatment of neurodegenerative diseases through the inhibition of ataxin-2.
REFERENCES
- Akbik FV, Bhagat SM, Patel PR, Cafferty WB, and Strittmatter SM (2013). Anatomical plasticity of adult brain is titrated by Nogo Receptor 1. Neuron 77, 859–866. 10.1016/j.neuron.2012.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, O’Connor PB, Fahey C, Kenny EM, Terenin IM, Dmitriev SE, Cormican P, Morris DW, Shatsky IN, and Baranov PV (2015). Translation of 5’ leaders is pervasive in genes resistant to eIF2 repression. Elife 4, e03971. 10.7554/eLife.03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrusiak MG, Sharifnia P, Lyu X, Wang Z, Dickey AM, Wu Z, Chisholm AD, and Jin Y (2019). Inhibition of axon regeneration by liquid-like TIAR-2 granules. Neuron 104, 290–304.e298. 10.1016/j.neuron.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavachalu B, Huelsmeier J, Sudhakaran IP, Hillebrand J, Singh A, Petrauskas A, Thiagarajan D, Sankaranarayanan M, Mizoue L, Anderson EN, et al. (2018). RNP-granule assembly via ataxin-2 disordered domains is required for long-term memory and neurodegeneration. Neuron 98, 754–766.e754. 10.1016/j.neuron.2018.04.032. [DOI] [PubMed] [Google Scholar]
- Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, Messing J, Kim HJ, Soriano A, Auburger G, et al. (2017). Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544, 367–371. 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat SM, Butler SS, Taylor JR, McEwen BS, and Strittmatter SM (2016). Erasure of fear memories is prevented by Nogo Receptor 1 in adulthood. Mol. Psychiatry 21, 1281–1289. 10.1038/mp.2015.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieri G, Brahic M, Bousset L, Couthouis J, Kramer NJ, Ma R, Nakayama L, Monbureau M, Defensor E, Schüle B, et al. (2019). LRRK2 modifies α-syn pathology and spread in mouse models and human neurons. Acta Neuropathol. 137, 961–980. 10.1007/s00401-019-01995-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, and Gitler AD (2019). Axons gonna ride ’til they can’t no more. Neuron 104, 179–181. 10.1016/j.neuron.2019.09.029. [DOI] [PubMed] [Google Scholar]
- Bros-Facer V, Krull D, Taylor A, Dick JR, Bates SA, Cleveland MS, Prinjha RK, and Greensmith L (2014). Treatment with an antibody directed against Nogo-A delays disease progression in the SOD1G93A mouse model of Amyotrophic lateral sclerosis. Hum. Mol. Genet 23, 4187–4200. 10.1093/hmg/ddu136. [DOI] [PubMed] [Google Scholar]
- Brown AL, Wilkins OG, Keuss MJ, Hill SE, Zanovello M, Lee WC, Bampton A, Lee FCY, Masino L, Qi YA, et al. (2022). TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature 603, 131–137. 10.1038/s41586-022-04436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivatakarn O, Kaneko S, He Z, Tessier-Lavigne M, and Giger RJ (2007). The Nogo-66 receptor NgR1 is required only for the acute growth cone-collapsing but not the chronic growth-inhibitory actions of myelin inhibitors. J. Neurosci 27, 7117–7124. 10.1523/jneurosci.1541-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong ZS, Ohnishi S,Yusa K, and Wright GJ (2018). Pooled extracellular receptor-ligand interaction screening using CRISPR activation. Genome Biol. 19, 205. 10.1186/s13059-018-1581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA, et al. (2016). Nano-Luc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol 11, 400–408. 10.1021/ac-schembio.5b00753. [DOI] [PubMed] [Google Scholar]
- Domeniconi M, Cao Z, Spencer T, Sivasankaran R, Wang K, Nikulina E, Kimura N, Cai H, Deng K, Gao Y, et al. (2002). Myelin-associated glycoprotein interacts with the Nogo66 receptorto inhibit neurite outgrowth. Neuron 35, 283–290. 10.1016/s0896-6273(02)00770-5. [DOI] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. (2010). Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075. 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink KL, Strittmatter SM, and Cafferty WB (2015). Comprehensive corticospinal labeling with mu-crystallin transgene reveals axon regeneration after spinal cord trauma in ngr1−/− mice. J. Neurosci 35, 15403–15418. 10.1523/jneurosci.3165-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fittschen M, Lastres-Becker I, Halbach MV, Damrath E, Gispert S, Azizov M, Walter M, Muller S, and Auburger G (2015). Genetic ablation of ataxin-2 increases several global translation factors in their transcript abundance but decreases translation rate. Neurogenetics 16, 181–192. 10.1007/s10048-015-0441-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier AE, GrandPre T, and Strittmatter SM (2001). Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature 409, 341–346. 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium (2013). The genotype-tissue expression (GTEx) project. Nat. Genet 45, 580–585. 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GrandPré T, Li S, and Strittmatter SM (2002). Nogo-66 receptor antagonist peptide promotes axonal regeneration. Nature 417, 547–551. 10.1038/417547a. [DOI] [PubMed] [Google Scholar]
- He W, and Parker R (2000). Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol 12, 346–350. 10.1016/s0955-0674(00)00098-3. [DOI] [PubMed] [Google Scholar]
- Jokic N, Gonzalez de Aguilar JL, Dimou L, Lin S, Fergani A, Ruegg MA, Schwab ME, Dupuis L, and Loeffler JP (2006). The neurite outgrowth inhibitor Nogo-A promotes denervation in an amyotrophic lateral sclerosis model. EMBO Rep. 7, 1162–1167. 10.1038/sj.embor.7400826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G, Gautier O, Tassoni-Tsuchida E, Ma XR, and Gitler AD (2020). ALS genetics: gains, losses, and implications for future therapies. Neuron 108, 822–842. 10.1016/j.neuron.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Liu BP, Park JH, and Strittmatter SM (2004). Nogo-66 receptor prevents raphespinal and rubrospinal axon regeneration and limits functional recovery from spinal cord injury. Neuron 44, 439–451. 10.1016/j.neuron.2004.10.015. [DOI] [PubMed] [Google Scholar]
- Kim G, Nakayama L, Blum JA, Akiyama T, Boeynaems S, Chakraborty M, Couthouis J, Tassoni-Tsuchida E, Rodriguez CM, Bassik MC, and Gitler AD (2022). Genome-wide CRISPR screen reveals v-ATPase as a drug target to lower levels of ALS protein ataxin-2. Cell Rep. 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Trempe JF, Khaleghpour K, Kahvejian A, Ekiel I, and Gehring K (2001). Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. USA 98, 4409–4413. 10.1073/pnas.071024998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, and Cleveland DW (2010). TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet 19, R46–R64. 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastres-Becker I, Nonis D, Eich F, Klinkenberg M, Gorospe M, Kotter P, Klein FA, Kedersha N, and Auburger G (2016). Mammalian ataxin-2 modulates translation control at the pre-initiation complex via PI3K/mTOR and is induced by starvation. Biochim. Biophys. Acta 1862, 1558–1569. 10.1016/j.bbadis.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, and Allada R (2013). ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science (New York, N.Y.) 340, 875–879. 10.1126/science.1234785. [DOI] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, and Wolozin B (2011). Local RNA translation at the synapse and in disease. J. Neurosci 31, 16086–16093. 10.1523/jneurosci.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XR, Prudencio M, Koike Y, Vatsavayai SC, Kim G, Harbinski F, Briner A, Rodriguez CM, Guo C, Akiyama T, et al. (2022). TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A. Nature 603, 124–130. 10.1038/s41586-022-04424-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Amrani N, and Jacobson A (1998). Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol 18, 7383–7396. 10.1128/mcb.18.12.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maor-Nof M, Shipony Z, Lopez-Gonzalez R, Nakayama L, Zhang YJ, Couthouis J, Blum JA, Castruita PA, Linares GR, Ruan K, et al. (2021). p53 is a central regulator driving neurodegeneration caused by C9orf72 poly(PR). Cell 184, 689–708.e620. 10.1016/j.cell.2020.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland JM, Ho ZV, Kugener G, Dempster JM, Montgomery PG, Bryan JG, Krill-Burger JM, Green TM, Vazquez F, Boehm JS, et al. (2018). Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat. Commun 9, 4610. 10.1038/s41467-018-06916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee AW, Yang Y, Fischer QS, Daw NW, and Strittmatter SM (2005). Experience-driven plasticity of visual cortex limited by myelin and Nogo receptor. Science 309, 2222–2226. 10.1126/science.1114362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, Dharia NV, Montgomery PG, Cowley GS, Pantel S, et al. (2017). Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet 49, 1779–1784. 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montani L, Gerrits B, Gehrig P, Kempf A, Dimou L, Wollscheid B, and Schwab ME (2009). Neuronal Nogo-A modulates growth cone motility via Rho-GTP/LIMK1/cofilin in the unlesioned adult nervous system. J. Biol. Chem 284, 10793–10807. 10.1074/jbc.M808297200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, and Koonin EV (1998). Ataxin-2, global regulators of bacterial gene expression, and spliceosomal snRNP proteins share a conserved domain. J. Mol. Med. (Berl.) 76, 3–5. [DOI] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, and Krobitsch S (2007). Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell 18, 1385–1396. 10.1091/mbc.e06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbacher JK, Tabet R, Yeo GW, and Lagier-Tourenne C (2019). Disruption of RNA metabolism in neurological diseases and emerging therapeutic interventions. Neuron 102, 294–320. 10.1016/j.neuron.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Dansithong W, Figueroa KP, Scoles DR, and Pulst SM (2018). Staufen1 links RNA stress granules and autophagy in a model of neurodegeneration. Nat. Commun 9, 3648. 10.1038/s41467-018-06041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson HL, Shakkottai VG, Clark HB, and Orr HT (2017). Polyglutamine spinocerebellar ataxias - from genes to potential treatments. Nat. Rev. Neurosci 18, 613–626. 10.1038/nrn.2017.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riching KM, Mahan S, Corona CR, McDougall M, Vasta JD, Robers MB, Urh M, and Daniels DL (2018). Quantitative live-cell kinetic degradation and mechanistic profiling of PROTAC mode of action. ACS Chem. Biol 13, 2758–2770. 10.1021/acschembio.8b00692. [DOI] [PubMed] [Google Scholar]
- Sato Y, Iketani M, Kurihara Y, Yamaguchi M, Yamashita N, Nakamura F, Arie Y, Kawasaki T, Hirata T, Abe T, et al. (2011). Cartilage acidic protein-1B (LOTUS), an endogenous Nogo receptor antagonist for axon tract formation. Science 333, 769–773. 10.1126/science.1204144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab ME (2010). Functions of Nogo proteins and their receptors in the nervous system. Nat. Rev. Neurosci 11, 799–811. 10.1038/nrn2936. [DOI] [PubMed] [Google Scholar]
- Schwab ME, and Strittmatter SM (2014). Nogo limits neural plasticity and recovery from injury. Curr. Opin. Neurobiol 27, 53–60. 10.1016/j.conb.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoles DR, Meera P, Schneider MD, Paul S, Dansithong W, Figueroa KP, Hung G, Rigo F, Bennett CF, Otis TS, and Pulst SM (2017). Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 544, 362–366. 10.1038/nature22044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoles DR, and Pulst SM (2018). Spinocerebellar ataxia type 2. Adv. Exp. Med. Biol 1049, 175–195. 10.1007/978-3-319-71779-1_8. [DOI] [PubMed] [Google Scholar]
- Taylor JP, Brown RH Jr., and Cleveland DW (2016). Decoding ALS: from genes to mechanism. Nature 539, 197–206. 10.1038/na-ture20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Favell K, Morante-Redolat J, Pool M, Kent C, Wright M, Daignault K, Ferraro GB, Montcalm S, Durocher Y, et al. (2010). LGI1 is a Nogo receptor 1 ligand that antagonizes myelin-based growth inhibition. J. Neurosci 30, 6607–6612. 10.1523/jneurosci.5147-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Z, Segura-Feliu M, Seira O, Homs Corbera A, Antonio J, Rio J, Ade J, and Samitier J (2015). A microfluidic neuronal platform for neuron axotomy and controlled regenerative studies. RSC Adv. 5, 0–1. 10.1039/C5RA11522A. [DOI] [Google Scholar]
- Wang KC, Kim JA, Sivasankaran R, Segal R, and He Z (2002). P75 interacts with the Nogo receptor as a co-receptor for Nogo, MAG and OMgp. Nature 420, 74–78. 10.1038/nature01176. [DOI] [PubMed] [Google Scholar]
- Wang X, Duffy P, McGee AW, Hasan O, Gould G, Tu N, Harel NY, Huang Y, Carson RE, Weinzimmer D, et al. (2011). Recovery from chronic spinal cord contusion after Nogo receptor intervention. Ann. Neurol 70, 805–821. 10.1002/ana.22527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhou T, Maynard GD, Terse PS, Cafferty WB, Kocsis JD, and Strittmatter SM (2020). Nogo receptor decoy promotes recovery and corticospinal growth in non-human primate spinal cord injury. Brain 143, 1697–1713. 10.1093/brain/awaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Miao Y, Wicklein R, Sun Z, Wang J, Jude KM, Fernandes RA, Merrill SA, Wernig M, Garcia KC, and Südhof TC (2021). RTN4/NoGo-receptor binding to Bai adhesion-GPCRs regulates neuronal development. Cell 184. 10.1016/j.cell.2021.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YS, Harel NY, and Strittmatter SM (2009). Reticulon-4A (Nogo-A) redistributes protein disulfide isomerase to protect mice from SOD1-dependent amyotrophic lateral sclerosis. J. Neurosci 29, 13850–13859. 10.1523/jneurosci.2312-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoshi M, Li Q, Yamamoto M, Okada H, Suzuki Y, and Kawahara Y (2014). Direct binding of Ataxin-2 to distinct elements in 3’ UTRs promotes mRNA stability and protein expression. Mol. Cell 55, 186–198. 10.1016/j.molcel.2014.05.022. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zheng S, Wu H, Wu Y, Liu S, Fan M, and Zhang J (2009). Identification of BLyS (B lymphocyte stimulator), a non-myelin-associated protein, as a functional ligand for Nogo-66 receptor. J. Neurosci 29, 6348–6352. 10.1523/jneurosci.5040-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ling J, Yuan C, Dubruille R, and Emery P (2013). A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science (New York, N.Y.) 340, 879–882. 10.1126/science.1234746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Ataxin-2 antibody | Novus | Cat# NBP1-90063; RRID:AB_11028587 |
| Tuj1 antibody | Biolegend | Cat# 802001; RRID:AB_2564645 |
| Actin antibody | EMD Millipore | Cat# MAB1501, RRID:AB_2223041 |
| RTN4/NoGo-Receptor antibody | Abcam | Cat# ab184556 |
| GAPDH antibody | Sigma Aldrich | Cat# G8795; RRID:AB_1078991 |
| Goat anti-Rabbit HRP antibody | Life Technologies | Cat# 31462; RRID:AB_228338 |
| Goat anti-mouse HRP antibody | Thermo Fisher Scientific | Cat# 62–6520; RRID:AB_2533947 |
| MAP2 antibody | Synaptic Systems | Cat# 188004; RRID:AB_2138181 |
| NeuN antibody | EMD Millipore | Cat# MAB377; RRID:AB_2298772 |
| Chemicals, peptides, and recombinant proteins | ||
| NEP1-40 peptide | Tocris | Cat# 1984 |
| Critical commercial assays | ||
| Nano-Glo HiBiT Lytic Detection System | Promega | Cat# N3050 |
| ONE-GLO Luciferase Assay System | Promega | Cat# E6120 |
| TaqMan gene-specific expression assay: human ATXN2 | Thermo Fisher Scientific | Hs00268077_m1 |
| TaqMan gene-specific expression assay: human ACTB | Thermo Fisher Scientific | Hs01060665_g1 |
| TaqMan gene-specific expression assay: human RTN4R | Thermo Fisher Scientific | Hs00368533_m1 |
| TaqMan gene-specific expression assay: mouse ATXN2 | Thermo Fisher Scientific | Mm00485932_m1 |
| TaqMan gene-specific expression assay: mouse GAPDH | Thermo Fisher Scientific | Mm99999915_g1 |
| TaqMan gene-specific expression assay: mouse RTN4R | Thermo Fisher Scientific | Mm00452228_m1 |
| Human ATXN2 siRNA | Horizon Discovery | Cat# L-011772-00 |
| Non-targeting siRNA | Horizon Discovery | Cat# D-001810-10 |
| Human RTN4R siRNA | Horizon Discovery | Cat# L-008075-00 |
| Oligonucleotides | ||
| Target-specific sgRNA sequence | IDT | AGCCTTACAACTGCTGTTGG |
| Forward Primer for ATXN2 exon 25 amplification and sequencing | IDT | GCAATACTGGTGCTTGGCTAATATTTGGGG |
| Reverse Primer for ATXN2 exon 25 amplification and sequencing | IDT | CACTCTTGTTACTTCTTTTGCTAGCTGATGTG |
| Deposited data | ||
| iNeuron RNA sequencing data | GEO | GEO: GSE200530 |
| Experimental models: Cell lines | ||
| HEK293T Cells | ATCC | Cat# CRL-321; RRID:CVCL_0063 |
| SH-SY5Y | ATCC | Cat# CRL-226; RRID:CVCL_0019 |
| Experimental models: Organisms/strains | ||
| Time pregnant C57BL/6 | Charles River Labs | Cat# C57BL/6NCr; RRID:MGI:2159769 |
| Rtn4r KO mouse line | Provided by Dr. Stephen Strittmatter | N/A |
| Recombinant DNA | ||
| Mission® shRNA, mouse RTN4R | Sigma Aldrich | TRCN0000436683 |
| Mission® shRNA, human RTN4R | Sigma Aldrich | TRCN0000061558 |


