Highlights
-
•
F. nucleatum promoted tumour progression and induced PD-L1 expression in CRC
-
•
F. nucleatum promoted PD-L1 expression via m6A modification of IFIT1
-
•
IFIT1 was a potential oncogene in CRC
-
•
IFIT1 regulated PD-L1 protein levels through altering its ubiquitination and degradation
Keywords: F. nucleatum, Colorectal cancer, m6A, IFIT1, PD-L1
Abbreviations: CRC, colorectal cancer; F. nucleatum/ F.n, Fusobacterium nucleatum; UTR, untranslated region; MOI, multiple of infection
Abstract
Fusobacterium nucleatum (F. nucleatum) is enriched in colorectal cancer (CRC) tissues and a high amount of F. nucleatum was associated with an immunosuppressive tumor environment. PD-L1 is an important immune checkpoint expressed on tumor cells and promotes tumor immune escape. Whether PD-L1 is regulated by F. nucleatum is still unclear. We demonstrated that F. nucleatum promoted CRC progression and upregulated PD-L1 protein expression in CRC cell lines. Combined m6A-seq and RNA-seq identified m6A-modified IFIT1 mediating F. nucleatum induced PD-L1 upregulation. IFIT1 mRNA was modified with m6A modifications in 3’UTR and the m6A levels were altered by F. nucleatum treatment. Our results also indicated that IFIT1 served as a potential oncogene in CRC and regulated PD-L1 protein levels through altering PD-L1 ubiquitination. Clinical CRC data confirmed the correlation among F. nucleatum abundance, IFIT1 and PD-L1 expressions. Our work highlighted the function of F. nucleatum in stimulating PD-L1 expression through m6A-modified IFIT1 and provided new aspects for understanding F. nucleatum mediated immune escape.
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer as well as the third fatal cancer in the world [1,2]. Recent evidence has demonstrated that host-microbiota interactions play an important role in CRC progression [3,4]. F. nucleatum is an important cancer-associated bacterium contributing to cancer development especially CRC [5,6]. A high enrichment of F. nucleatum is associated with colorectal carcinogenesis, disease stages and overall survival. F. nucleatum coculture could stimulate cancer cell proliferation and F. nucleatum gavage resulted in more and larger tumors in AOM-DSS CRC model and Apcmin/+ mice [7]. Besides, F. nucleatum also promotes resistance to chemotherapy and glycolysis in CRC [8,9].
PD-L1 is an essential immune checkpoint molecule, generating an inhibitory signal and establishing an immunosuppressive environment [10]. PD-L1 is highly expressed in various cancer cells and stromal immune cells, allowing tumors to evade attacks of infiltrated T cells [11,12]. Several population-based studies have reported a higher abundance of F. nucleatum was associated with an immunosuppressive tumor environment, with lower T cell density [13,14]. The virulence factor derived from F. nucleatum, Fap2, inhibited NK cell cytotoxicity through interacting with TIGIT [15]. Whether F. nucleatum could regulate PD-L1 expression in CRC is still unclear.
N6-methyladenosine (m6A) modification is abundant and prevalent in eukaryotic cells, involving in diverse biological processes in cancer [16,17]. PD-L1 expression could be directly modified with m6A modifications and also indirectly regulated by m6A-modified target genes [18,19]. Whether RNA m6A modification participated in F. nucleatum mediated immunosuppressive tumor environment was still unclear.
In this work, we found F. nucleatum promoted CRC cell proliferation in vitro and in vivo. F. nucleatum treatment could markedly stimulate PD-L1 protein expression without mRNA levels alteration. Through combined m6A-seq and RNA-seq in CRC cells cocultured with F. nucleatum, we identified m6A-modified IFIT1 and found IFIT1 knockdown could reverse F. nucleatum induced PD-L1 expression. Mechanically, IFIT1 overexpression increased PD-L1 protein levels through inhibiting its ubiquitination. Also, we found IFIT1 was higher expressed in CRC tissues than normal colon tissues. A higher expression of IFIT1 was associated with higher cancer stage and worse survival. In addition, METTL3 and METTL14 methylated IFIT1 in 3’UTR and promoted its mRNA stability in a IGF2BP2/3-dependent manner. In clinical samples, F. nucleatum abundance, IFIT1 and PD-L1 expressions were positively correlated. Our work indicated that F. nucleatum stimulated PD-L1 protein expression via m6A modification of IFIT1 and provided new aspects for understanding F. nucleatum mediated immune escape.
Materials and methods
Cell lines
RKO cells and SW1116 cells were purchased from American Type Culture Collection (ATCC) and incubated with RPMI-1640 medium (Gibco) supplemented with 10% fetal bovine serum (FBS). HT29 cells and HCT116 cells were also purchased from ATCC and cultured with McCoy’5A (Gibco) with 10% FBS (Gibco). All cell lines were grown at 37 °C in a humidified 5% CO2 atmosphere.
Patient specimens
We studied four cohorts of CRC patients having undergone surgery in Renji Hospital affiliated with Shanghai Jiao Tong University School of Medicine. The study was approved by the ethics committee of Renji Hospital and all patients have obtained the written informed consent. Respectively, there were 36 pairs of FFPE CRC tissues and non-cancerous normal colon tissues in cohort 1, 79 FFPE cancer tissues in cohort 2, 34 pairs of FFPE cancer tissues and non-cancerous normal tissues in cohort 3 and 180 FFPE cancer tissues in cohort 4. The clinical characteristics of these cohorts were shown in Supplementary Table 1, 2, 3 and 4.
Animal studies
Four-week-old male BALB/c nude mice were housed in Shanghai Model Organism Center under specific pathogen-free conditions. To investigate the effects of F. nucleatum on tumor progression, HT29 and HCT116 cells (5×106) were injected subcutaneously into the right axilla. Mice were randomly separately into three groups after 5 days’ inoculation, receiving intratumoral injection of PBS, F. nucleatum solution and E. coli solution (10^7 CFU per mouse) separately. The injection was carried twice one week. Mice were sacrificed 20 days later and subcutaneous tumors were harvested and weighed. To investigate the effects of IFIT1, HCT116 cells were stably transfected with shIFIT1 lentivirus, pcDNA3.1_IFIT1 lentivirus and their corresponding control. HCT116 cells (5×106) were also injected subcutaneously into the right axilla. Mice were sacrificed 20 days later and subcutaneous tumors were harvested and weighed. These experiments were performed following the Institute's guidance on animal experiments.
Bacterial strains and growth conditions
Fusobacterium nucleatum ATCC 25586 and Enterotoxigenic Bacteroides fragilis were purchased from ATCC. DH5α E. coli was obtained from Tiangen Company. They were cultured as previously described [8].
Detection of bacterial abundance
QIAamp DNA FFPE Tissue Kit (QIAGEN) was used to extract the genome DNA from FFPE CRC tissue. The extracted DNA was assessed using qPCR and the F. nucleatum abundance was normalized using 16S genes. The primer for Fusobacterium nucleatum detection is “F-CAACCATTACTTTAACTCTACCATGTTCA” and “R-GT TGACTTTACAGAAGGAGATTATGTAAAAATC”. The primer for 16S detection is “F- GGTGAATACGTTCCCGG” and “R- TACGGCTACCTTGTTACGACTT”.
Cell proliferation assay
Cell proliferation was assessed with Cell Counting Kit-8 assay (Dojindo), EdU assay (RiboBio) and colony formation assay. For Cell Counting Kit-8 assay, 3000 cells were planted into 96-well plates. Cell Counting Kit-8 (Dojindo) was mixed with medium at 1:10 dilution. 100ul of the above mixture was added into the wells. Incubated for 2 hours, the absorbance at 450nm was measured. For EdU assay, cells were incubated with EdU stain for 2 hours. Cells with positive stain were counted under microscope. For the colony formation assay, 600 cells were planted into 12-well plates. After 7 to 10 days’ culture, cells were washed with PBS twice, fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The colonies were counted after washing with PBS. The above experiments were conducted independently three times.
Construction and transfection of small interfering RNAs, lentivirus shRNA and plasmids
METTL3, METTL14, IGF2BP1/2/3 and IFIT1 small interfering RNAs (siRNAs) were obtained from GenePharma (Shanghai, China). IFIT1 lentivirus shRNAs and pcDNA3.1_IFIT1 overexpressed lentivirus were also obtained from GenePharma (Shanghai, China). The control plasmid, METTL3, METTL14, IFIT1 and PD-L1 overexpressing plasmids were constructed in Generay Technologies (Shanghai, China). Cells were seeded in six-well plates overnight. For siRNA transfection, we made the transfection mixture with 5ul siRNA(50nM), 5ul Dharma FECT 1 transfection reagent and 390ul opti-MEM medium. For plasmid transfection, we added 1ug plasmid and 3ul viaFECT for one well. After six hours of incubation, the medium was replaced by normal culture media. Cells were harvested after 48h. For lentivirus shRNA transfection, cells were incubated with lentivirus at the MOI of 100. After 24 hours’ incubation, cells were treated with puromycin for two weeks. The sequences of siRNAs and shRNAs were listed in Supplementary Table 7.
M6A-seq and RNA-seq
M6A-seq was conducted according to the previous studies [20]. Briefly, polyadenylated mRNA was isolated from total RNA with GenEluteTM mRNA Miniprep Kit. The quantity and quality of mRNA were tested with agarose gel electrophoresis and NanoDropTM instruments. The isolated mRNA was fragmented into short fragments and approximately 1/10 of the RNA fragments were conserved for RNA sequencing by oebiotech(Shanghai, China). The m6A-methylated mRNAs were immunoprecipitated with anti-m6A antibodies for further meRIP-seq. The differentially m6A-RIP-enriched regions (peaks) were calculated using exomePeak.
MeRIP-qPCR
This experiment was conducted as a previous report [17]. Polyadenylated mRNA was fragmented with RNA fragmentation reagents (NEB). The anti-m6A antibody (NEB) was used to immunoprecipitate m6A-methylated mRNAs. The m6A-methylated mRNAs were extracted, purified and resolved with nuclease-free water as previously described [17]. Real-time PCR was performed to measure the m6A level in target gene transcripts. Primers in this experiment were listed in Supplementary Table 7.
Western blot assay and Immunoprecipitation
Cells were collected after lysis with RIPA added with a protease inhibitor cocktail. Cell supernatant was subjected to BCA Protein Assay Kit to measure the concentration. Same amount of protein samples was separately with 10% SDS-PAGE and then transferred to PVDF membranes (Bio-Rad). Membranes were blocked with 5% nonfat milk and then incubated with primary antibodies overnight at 4°C. Next day, membranes were incubated with secondary antibodies. The ECL Kit was used to detect the signals. Immunoprecipitation was performed as previously described. Cells were harvested with IP lysis buffer (Thermo Fisher). After centrifugation, 1/10 of the supernatant was added with 5× SDS- PAGE loading buffer, serving as an input control. The remaining was supplemented with 20ul Pierce Protein G Magnetic Beads (thermo fisher, 88847). After supplemented with 2ul primary antibodies and isotype-matched IgG, the mixture was rotated slowly at 4°C overnight. Subsequently, the precipitation was isolated using the magnetic rack (NEB), followed by washing with PBS five times. The precipitation was mixed with 50 µL 1× SDS-PAGE loading buffer and heated at 95°C for 10min. Subsequent steps are as above described. The antibodies used in the experiments are listed as follow: anti-PD-L1 (CAT#13684, CST); anti-IFIT1 (CAT#14769S, CST); anti-METTL3 (CAT#15073-1-AP, Proteintech); anti-METTL14 (CAT#51104, CST); anti-HA tag (CAT#3724, CST); anti-FLAG tag (CAT#2368, CST); anti-PI3K (CAT#4249T, CST); anti-p-AKT (CAT#9271T, CST); anti-AKT (CAT#4691T, CST); anti-p-JAK2 (CAT#ab32101, abcam); anti-JAK2 (CAT#ab108596, abcam); anti-p-STAT1 (CAT#ab109457, abcam); anti-STAT1 (CAT#ab109320, abcam); anti-p-NF-kB (CAT#3033T, CST); anti-NF-kB (CAT#8242T, CST); anti-GAPDH (KANGCHENG Technology); anti-rabbit-HRP (KANGCHENG Technology); anti-mouse-HRP (KANGCHENG Technology).
Quantitative real-time PCR
Cells were lysed and extracted with RNAiso Plus (Takara). Reverse transcription was conducted with PrimeScriptP RT Reagent Kit (Takara) and specific genes expressions were measured with SYBR Premix Ex Taq II (Takara). Primers were obtained from tsingke Company and the sequences were listed in Supplementary Table 7.
Immunofluorescence
Cells were seeded into eight-well chamber slides. After adherence and treatment, cells were fixed using 4% formaldehyde (Biosharp), permeabilized using Triton-X-100 for 10 minutes and blocked with 1% BSA for 1 hour. Following incubated with primary antibodies overnight at 4°C, cells were incubated with fluorescently-labeled conjugated secondary antibodies (Alexa Fluor 594 or 488; Invitrogen). Slides were washed with PBS, stained with DAPI and sealed with coverslips. Fluorescence was observed with Zeiss LSM710 confocal microscope. The antibodies used in the experiment are listed as follow: Rabbit anti-PD-L1 (CAT#17952-1-AP, Proteintech), 1:100 dilution; Mouse anti-IFIT1(CAT#ab118062, abcam), 1:100 dilution. The above experiments were conducted independently three times.
Immunohistochemistry
Paraffin-embedded colon tissues were deparaffinized and rehydrated. After antigen retrieval and endogenous peroxidase block, the sections were blocked with goat serum, incubated with primary antibodies overnight and incubated with secondary antibody for 30 minutes. DAB was used as chromogen and hematoxylin was used for nucleus counterstaining. Immunohistochemistry (IHC) staining score was assessed under 200X microscope. The antibodies used in the experiment are listed as follow: Rabbit anti-PD-L1 (CAT#17952-1-AP, Proteintech), 1:200 dilution; Rabbit anti- IFIT1 (CAT#ab236256, abcam), 1:50 dilution.
Flow cytometry assays
Flow cytometry assays were performed for cell surface PD-L1 detection. After harvested with trypsin, cells were stained with APC- conjugated anti-human PD-L1 antibody (1:200) (BioLegend) for 30min at 4℃ followed by washing with PBS twice. The staining process was protected from light. And then cells were analyzed with flow cytometry. The above experiments were conducted independently three times.
Detection of PD-L1 protein stability
Cells were transfected with IFIT1 siRNAs and incubated with cycloheximide (beyotime) (50 µg/mL) for specific times. The proteins were extracted and analyzed using western blot. The intensity of bands was calculated with Image J and were normalized by GAPDH intensity.
RNA stability assay
Cells were seeded in 12-well plates overnight, followed by actinomycin D (5 μg/mL) treatment for 0h, 2h, and 4h. Total RNA was extracted and qPCR was performed to assess IFIT1 mRNA levels. The half-life of IFIT1 mRNAs was estimated using linear regression analysis.
RNA immunoprecipitation (RIP) and RIP-qPCR
RIP assays were performed with Magna RIP Kit (Millipore, New Bedford, MA). Anti-IGF2BP2 (proteintech, 11601-1-AP), anti-IGF2BP3(proteintech, 14642-1-AP), and isotype-matched IgG were used for RIP. Real-time PCR was conducted to determine the interaction between IGF2BP2/3 and IFIT1 mRNA.
Statistical analysis
GraphPad Prism (8.0.2) was used to plot column bar graphs and scatter plots. Values were showed as mean±SD with three independent experiments. Two independent samples were compared with two-tailed Student's t test and more than two groups were compared using one-way ANOVA. Immunofluorescence and western blot were analyzed with ImageJ (2.1.0) software. Flow cytometry assays were analyzed with Flowjo 10.4 (Bioscience). The correlation assays were analyzed using Spearman correlation test.
Results
F. nucleatum promoted tumor progression and upregulated PD-L1 expression in CRC
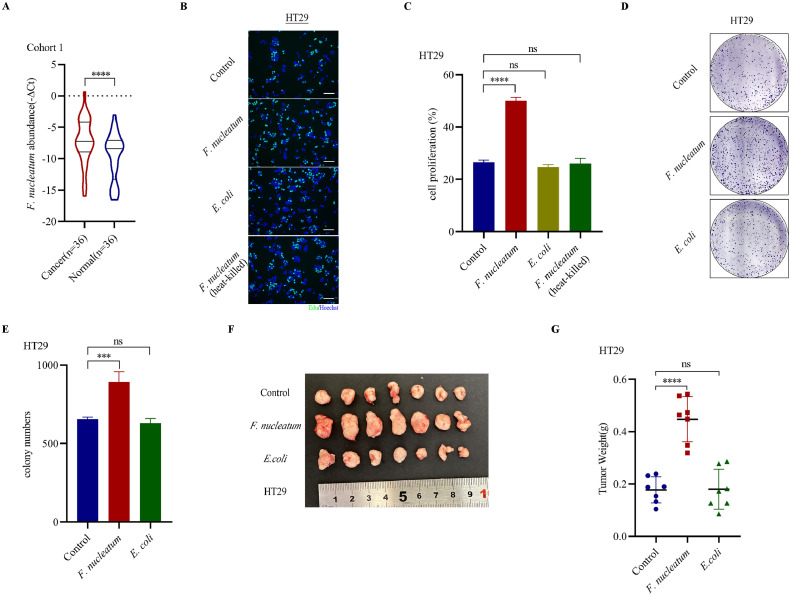
F. nucleatum is a well-known oncobacterium in cancers, especially in CRC. We measured F. nucleatum abundance in 36 pairs of formalin-fixed paraffin-embedded (FFPE) CRC tissues and normal colon tissues in cohort 1. The results showed a higher F. nucleatum abundance in cancer tissues (Fig. 1A). In vitro, F. nucleatum coculture (MOI=300, MOI, multiple of infection) remarkably enhanced HT29 and HCT116 cell proliferation compared with DH5α E. coli (MOI=300), heat-killed F. nucleatum and medium control through EdU assays (Fig. 1B-C, Supplementary Figure S1B-C). The similar results were also obtained through CCK8 assays and colony formation assays (Fig. 1D-E, Supplementary Figure S1A, D-F). In vivo, we inoculated CRC cells into nude mice and injected F. nucleatum solution intratumorally and found F. nucleatum treatment stimulated tumor growth significantly (Fig. 1F-G, Supplementary Figure S1G-H). Ki-67 is an important proliferation marker in cancer progression. In clinical CRC samples, we observed a higher F. nucleatum abundance was associated with higher Ki-67 scores (Supplementary Figure S1I). however, the difference was not significant. Various factors including histological differentiation, tumor size, lymphatic metastasis and AJCC II/III/IV stage could also influence Ki-67 score.
Fig. 1.
F. nucleatum promoted tumor progression in CRC. A. PCR analysis of F. nucleatum abundance in FFPE tissues in Cohort 1(Wilcoxon matched-pairs signed-rank test). B-C. EdU assays detecting cell proliferation of HT29 cells treated with F. nucleatum (MOI=300), DH5α E. coli (MOI=300), heat-killed F. nucleatum (MOI=300) and medium control (mean± SD). D. Representative images of colony formation assays of HT29 cells treated with F. nucleatum (MOI=300), DH5α E. coli (MOI=300) and medium control. E. The summarized data of colony formation assays of HT29 cells treated with F. nucleatum (MOI=300), DH5α E. coli (MOI=300) and medium control (n=3, mean± SD, One-way ANOVA). F. Representative images of tumors (HT29 cells) in nude mice intratumorally treated with F. nucleatum, DH5α E. coli and PBS control (n=7). G. Statistical analysis of tumor weights in F (n=7, One-way ANOVA).
**p < 0.01. *** p < 0.001. **** p < 0.0001. ns, not significant. OD, optical diameter.
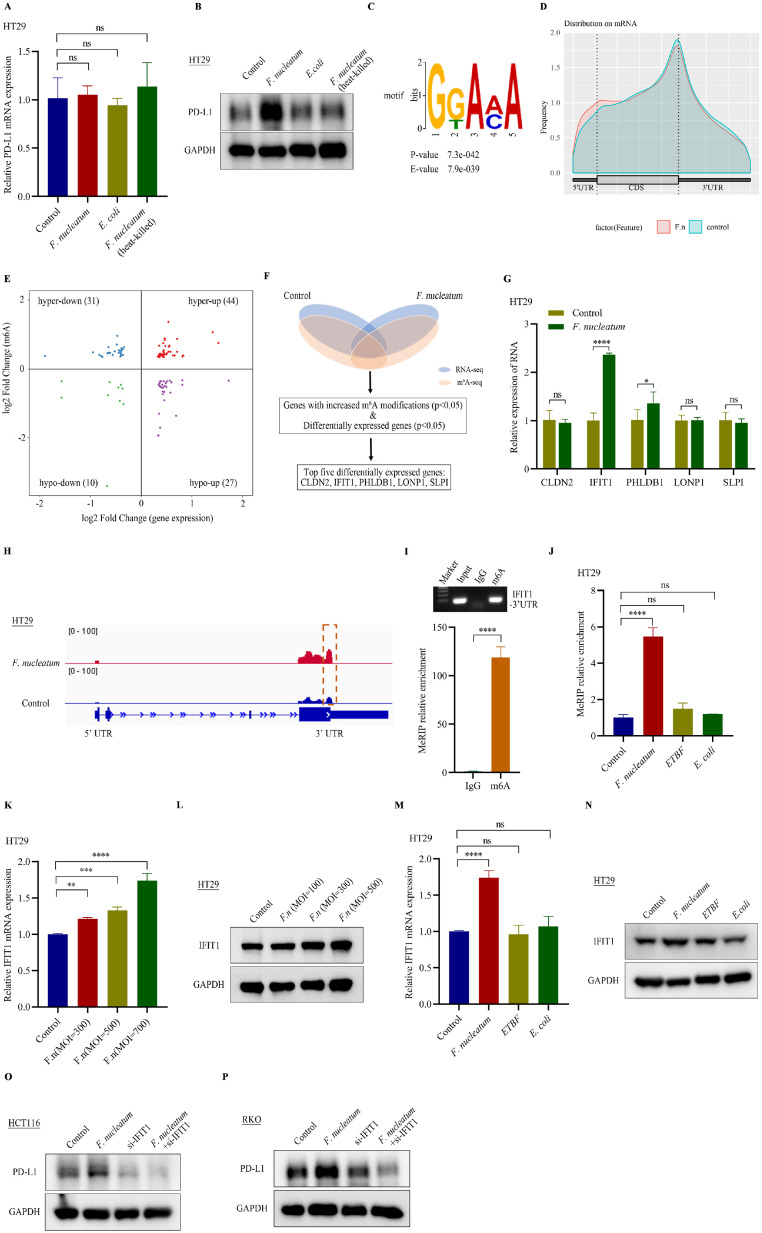
Previous researches have suggested that F. nucleatum in colorectal tissues could modulate a tumor immunosuppressive environment, accompanied with lower density of CD3+ T cells and an expansion of myeloid-derived immune cells [13,14]. PD-L1 is an important immunosuppressive molecule, dampening cancer immunity through providing suppressive signals to CD8+ T cells [10]. Whether F. nucleatum treatment could stimulate PD-L1 expression is still unclear. We treated CRC cells with F. nucleatum for 3 hours and measured PD-L1 mRNA expression using RT-qPCR. The results showed that F. nucleatum coculture didn't alter PD-L1 mRNA levels (Supplementary Figure S2A-B, D). However, western blot assays showed that F. nucleatum coculture (MOI=300) stimulated PD-L1 protein expression compared with DH5α E. coli (MOI=300), heat-killed F. nucleatum (MOI=300) and medium control in CRC cells, while mRNA levels were not altered (Fig. 2A-B). The similar results also existed in HCT116 and RKO cells (Supplementary Figure S2C, E). Altogether, our data indicated that F. nucleatum promoted CRC progression and simulated PD-L1 protein expression without significant mRNA alterations.
Fig. 2.
F. nucleatum promoted PD-L1 expression via m6A modification of IFIT1. A. PCR analysis of PD-L1 mRNA levels in HT29 cells treated with F. nucleatum (MOI=300), DH5α E. coli (MOI=300), heat-killed F. nucleatum (MOI=300) and medium control (n=3, mean± SD, One-way ANOVA). B. Western blot assays detecting PD-L1 protein expression in HT29 cells treated with F. nucleatum (MOI=300), DH5α E. coli (MOI=300), heat-killed F. nucleatum (MOI=300) and medium control. C. The top consensus m6A motif in HT29 cells. D. The distribution of m6A peaks. E. Distribution of genes with a significant change in both m6A level and gene expression level in HT29 cells treated by F. nucleatum (MOI=300) compared with medium control (p<0.05). F. The flow chart of selecting candidate target genes. G. PCR analysis of 5 candidate target genes in HT29 cells treated with F. nucleatum (MOI=300) and medium control (n=3, mean± SD, two-tailed Student's t-test). H. IGV analysis representing m6A abundances in IFIT1 transcripts in HT29 cells in response to F. nucleatum (up panel) and the medium control (bottom panel). I. Agarose electrophoresis (up panel) and meRIP-qPCR assays (down panel) of IFIT1 3’UTR m6A levels in HT29 cells (n=3, mean± SD, two-tailed Student's t-test). J. MeRIP-qPCR analysis of IFIT1 3’UTR m6A levels in HT29 cells treated with medium control, F. nucleatum (MOI=300), ETBF (MOI=300) and DH5α E coli (MOI=300) (n=3, mean± SD, One-way ANOVA). K. PCR analysis of IFIT1 mRNA levels in HT29 cells treated with different MOI of F. nucleatum (n=3, mean± SD, One-way ANOVA). L. Western blot assays detecting IFIT1 protein expression in HT29 cells treated with different MOI of F. nucleatum. M. PCR analysis of IFIT1 mRNA levels in HT29 cells treated with medium control, F. nucleatum (MOI=300), ETBF (MOI=300) and DH5α E coli (MOI=300) (n=3, mean± SD, One-way ANOVA). N. Western blot assays detecting IFIT1 protein expression in HT29 cells treated with medium control, F. nucleatum (MOI=300), ETBF (MOI=300) and DH5α E coli (MOI=300). O. Western blot assays detecting PD-L1 protein expression in IFIT1 knockdown and the control HCT116 cells treated with F. nucleatum (MOI=300). P. Western blot assays detecting PD-L1 protein expression in IFIT1 knockdown and the control RKO cells treated with F. nucleatum (MOI=300). F.n, F. nucleatum.
F. nucleatum promoted PD-L1 expression via m6A modification of IFIT1
RNA m6A modification is a prevalent posttranscriptional mRNA modification in eukaryotic cells [20]. Previous reports have indicated that RNA m6A modification was an important regulator of PD-L1 and was also associated with tumor immune cell infiltration [21]. Whether m6A modification participated in PD-L1 expression induced by F. nucleatum was still unknown. To explore this question, we preformed combined m6A-seq and RNA-seq in HT29 cells treated with F. nucleatum (MOI=300) and attempted to find the potential genes regulating PD-L1 expression. Consistent with the previous findings [22], the m6A consensus motif GGAC was enriched in the detected peaks (Fig. 2C). The m6A peaks were mostly distributed in 5’-untranslated regions (5’UTR), coding regions and 3’-untranslated regions (3’UTR), especially abundant surrounding the stop codon (Fig. 2D, Supplementary Figure S2F). Through combined analysis, we have identified 112 genes with altered m6A modifications and expression (Fig. 2E) (Supplementary Table 5 and 6). The top five genes were listed as the candidate genes, including CLDN2, IFIT1, PHLDB1, LONP1, and SLPI (Fig. 2F). Real-time PCR was performed to confirm which candidate genes were regulated by F. nucleatum treatment. The results showed IFIT1 was the most significantly altered in HT29 and HCT116 cells after F. nucleatum infection (MOI=300) (Fig. 2G, Supplementary Figure S2G). The Integrative Genomics View (IGV) data showed a higher m6A level in 3’UTR of IFIT1 after F. nucleatum treatment (Fig. 2H). MeRIP-qPCR assays confirmed the m6A peaks located in 3’UTR of IFIT1 transcripts and the m6A levels were specifically increased after F. nucleatum infection (MOI=300) in HT29 cells, compared with treatments with ETBF (MOI=300) and DH5α E. coli (MOI=300) (Fig. 2I-J). The IFIT1 mRNA and protein expressions were induced by F. nucleatum in a MOI-dependent manner (Fig. 2K-L, Supplementary Figure S2H-I). Additionally, IFIT1 upregulation was induced specifically by F. nucleatum (MOI=300) compared with ETBF (MOI=300) and DH5α E. coli (MOI=300) (Fig. 2M-N, Supplementary Figure S2J-K). To explore whether IFIT1 mediated F. nucleatum induced PD-L1 expression, we cocultured F. nucleatum with IFIT1 knockdown CRC cells and the control cells. The results showed that IFIT1 knockdown could partially reversed F. nucleatum (MOI=300) induced PD-L1 protein upregulation (Fig. 2O-P). Previous researches have also indicated that PI3K pathway is an important signal regulating PD-L1 expression in cancer progression [23,24]. And F. nucleatum treatment could activate PI3K pathway [25,26]. To investigate whether F. nucleatum induced PD-L1 expression also depended on activating PI3K pathway, we treated CRC cells with the PI3K inhibitor, LY294002, prior to F. nucleatum coculture. The western blot results showed that F. nucleatum treatment could activate PI3K pathway in CRC cells. However, PI3K inhibition could not reverse PD-L1 upregulation induced by F. nucleatum treatment (Supplementary Figure S2L-Q). Thus, our results indicated that F. nucleatum promoted PD-L1 expression partially via m6A modification of IFIT1.
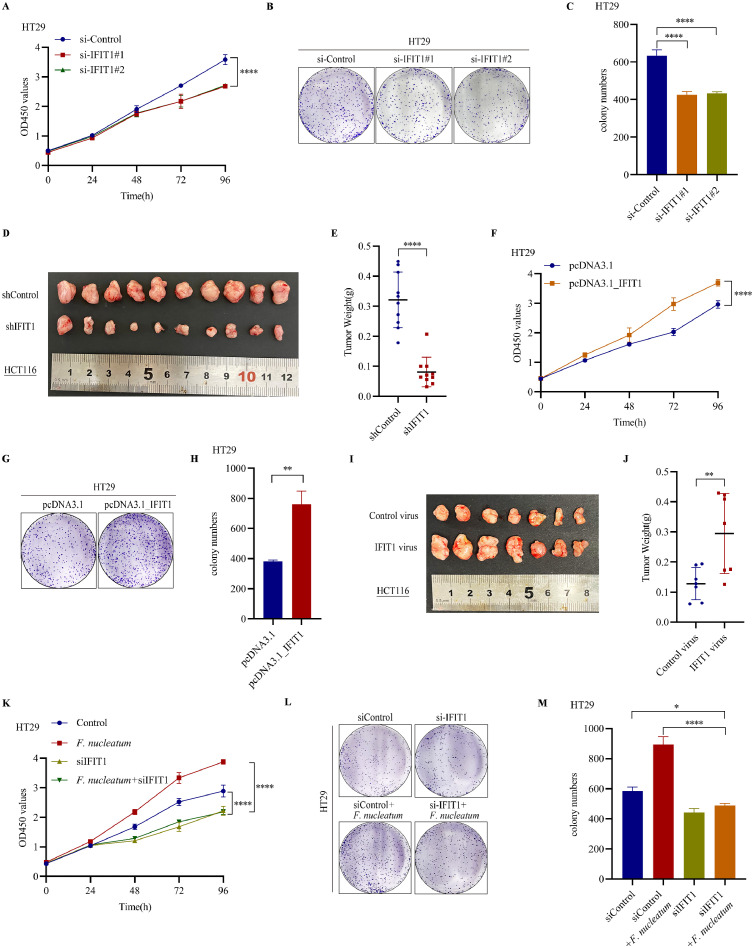
IFIT1 promoted CRC cell proliferation and participated in F. nucleatum induced CRC progression
IFIT1, interferon-induced proteins with tetratricopeptide repeats gene 1, is a member of interferon-stimulated genes (ISGs), which function as essential anti-viral proteins [27]. Emerging findings demonstrated its roles in tumor progression [28,29]. However, the roles of IFIT1 in CRC progression are still unknown. We performed CCK8 assays and colony formation assays in HT29 and HCT116 cells with IFIT1 knockdown. The results showed that IFIT1 knockdown dampened CRC proliferation and colony numbers (Fig. 3A-C, Supplementary Figure S3A-C). To confirm the results in vivo, we used lentivirus to establish HCT116 cells stably transfected with IFIT1 shRNAs. Knockdown of IFIT1 markedly reduced HCT116 tumor burden in nude mice models (Fig. 3D-E). Meanwhile, IFIT1 overexpression stimulated CRC cell proliferation and colony numbers (Fig. 3F-H, Supplementary Figure S3D-F). To confirm the results in vivo, we used lentivirus to establish HCT116 cells stably overexpressing IFIT1. Overexpression of IFIT1 dramatically increased HCT116 tumor volume and tumor weight in xenograft mice models (Fig. 3I-J). Since F. nucleatum could increase IFIT1 expression in CRC cells, whether IFIT1 participated in F. nucleatum induced CRC progression was still unknown. To explore this question, we decreased IFIT1 in HT29 and HCT116 cells using siRNAs and treated cells with F. nucleatum and performed CCK8 assay and colony formation assays. The results showed that IFIT1 knockdown could partially block F. nucleatum (MOI=300) induced cell proliferation (Fig. 3K-M, Supplementary Figure S3G-I). Altogether, IFIT1 promoted CRC proliferation in vitro and in vivo. Also, FIT1 upregulation participated in F. nucleatum induced CRC progression.
Fig. 3.
IFIT1 promoted CRC cell proliferation and participated in F. nucleatum induced CRC progression. A. CCK8 assays detecting cell growth of HT29 cells after transfection with IFIT1 siRNAs (mean± SD). B. Representative images of colony formation assays of HT29 cells after transfection with IFIT1 siRNAs. C. The summarized data of colony formation assays of HT29 cells after transfection with IFIT1 siRNAs (n=3, mean± SD, One-way ANOVA). D. Representative images of tumors in nude mice bearing HCT116 cells stably transfected with shIFIT1 lentivirus (n=10). E. Statistical analysis of tumor weights in D (n=10, two-tailed Student's t-test). F. CCK8 assays detecting cell growth of HT29 cells after transfection with pcDNA3.1_IFIT1 (mean± SD). G. Representative images of colony formation assays of HT29 cells after transfection with pcDNA3.1_IFIT1. H. The summarized data of colony formation assays of HT29 cells after transfection with pcDNA3.1_IFIT1 (n=3, mean± SD, One-way ANOVA). I. Representative images of tumors in nude mice bearing HCT116 cells stably transfected with IFIT1 plasmid (n=7). J. Statistical analysis of tumor weights in I (n=7, two-tailed Student's t-test). K. CCK8 assays detecting cell growth of HT29 cells transfected with IFIT1 siRNAs and treated with F. nucleatum (MOI=300) (mean± SD). L. Representative images of colony formation assays of HT29 cells transfected with IFIT1 siRNAs and treated with F. nucleatum (MOI=300). M. The summarized data of colony formation assays of HT29 cells transfected with IFIT1 siRNAs and treated with F. nucleatum (MOI=300) (n=3, mean± SD, One-way ANOVA).
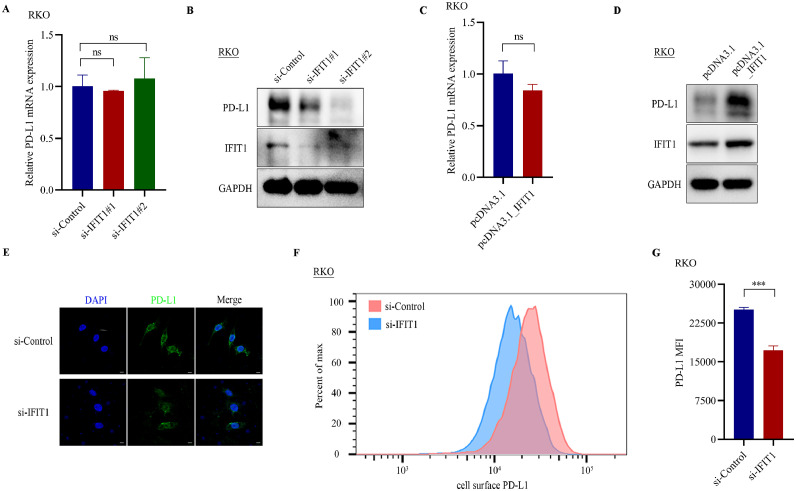
IFIT1 altered PD-L1 protein expression
Since IFIT1 knockdown could reverse F. nucleatum stimulated PD-L1 protein expression, we guessed that IFIT1 may regulate PD-L1 protein expression. First, we measured PD-L1 mRNA and protein expressions in IFIT1 siRNAs-transfected RKO cells. The results suggested that IFIT1 knockdown impaired PD-L1 protein expression but not mRNA expression (Fig. 4A-B, Supplementary Figure S4A). Ectopically overexpressed IFIT1 markedly enhanced PD-L1 protein expression without mRNA alterations in RKO cells (Fig. 4C-D, Supplementary Figure S4B). Immunofluorescence assays and flow cytometry also displayed that IFIT1 knockdown induced a reduction of PD-L1 in RKO cells (Fig. 4E-G). These results indicated that IFIT1 regulated PD-L1 protein expression without mRNA level alterations.
Fig. 4.
IFIT1 altered PD-L1 protein expression. A. PCR analysis of PD-L1 mRNA levels in RKO cells after transfection with IFIT1 siRNAs (n=3, mean± SD, One-way ANOVA). B. Western blot assays detecting PD-L1 protein expression in RKO cells after transfection with IFIT1 siRNAs. C. PCR analysis of PD-L1 mRNA levels in RKO cells after transfection with pcDNA3.1_IFIT1 (n=3, mean± SD, two-tailed Student's t-test). D. Western blot assays detecting PD-L1 protein expression in RKO cells after transfection with pcDNA3.1_IFIT1. E. Immunofluorescence assay of PD-L1 in RKO cells with IFIT1 knockdown using siRNAs. Scale bar, 5µm. F. Flow cytometry detecting cell surface PD-L1 in IFIT1 knockdown RKO cells. G Statistic analysis of mean fluorescence intensity (MFI) of cell surface PD-L1 in IFIT1 knockdown RKO cells versus control RKO cells (n=3, mean± SD, two-tailed Student's t-test).
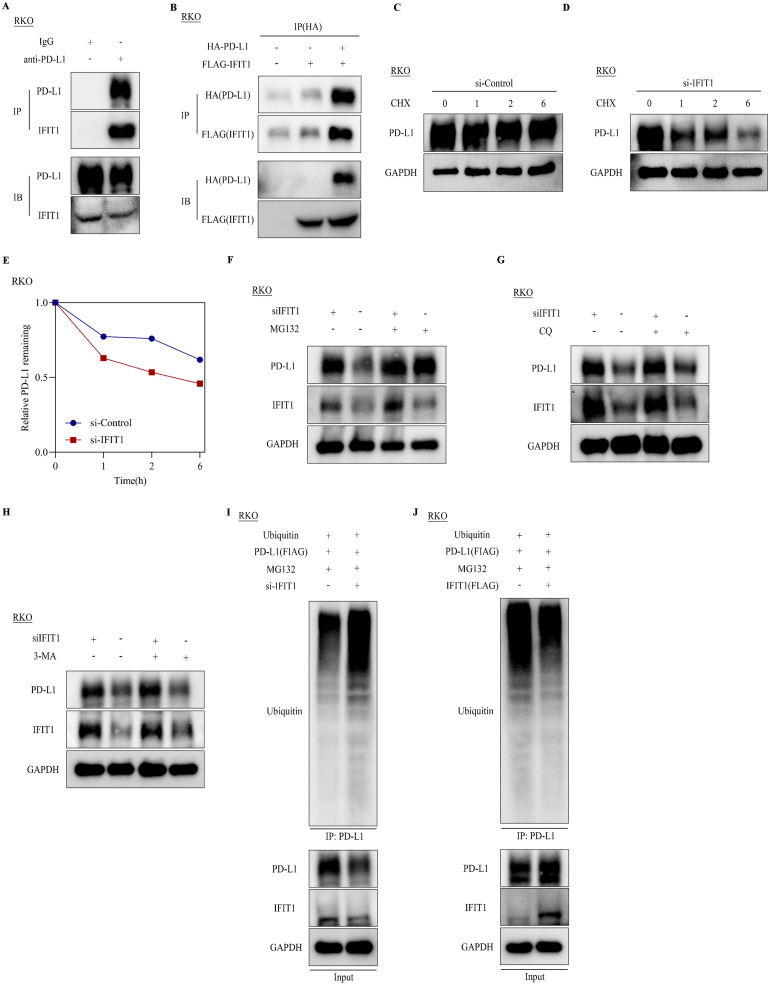
IFIT1 mediated PD-L1 stabilization via regulating PD-L1 ubiquitination
We sought to explore how IFIT1 regulated PD-L1 protein expression. NF-κB and JAK-STAT signals were two important pathways regulating PD-L1 expression [30], [31], [32]. Firstly, to investigate whether IFIT1 regulated these two pathways, we detected the expression of key components of NF-κB and JAK-STAT pathways in RKO cells with IFIT1 knockdown and overexpression using western blot assay. The results showed that IFIT1 knockdown and overexpression didn't alter these two pathways (Supplementary Figure S5A-B). Previous studies have indicated that NF-κB and JAK-STAT pathways regulate PD-L1 expression in mRNA levels. Consistent with these results and previous studies, our data showed IFIT1 altered PD-L1 protein expression not mRNA expression. In consideration of the TPR motifs of IFIT1, we hypothesized that IFIT1 may exert its effects through interacting with PD-L1. We immunoprecipitated RKO cell lysates with primary anti-PD-L1 antibodies and detected IFIT1 using western blot. The results indicated endogenous interaction of IFIT1 and PD-L1 (Fig. 5A). Next, we transfected FLAG-IFIT1 and HA-PD-L1 into RKO cells and confirmed their interaction ectopically (Fig. 5B). Since IFIT1 didn't affect PD-L1 mRNA levels, we guessed that IFIT1 affected PD-L1 protein degradation. CHX assays showed PD-L1 showed a more rapid decay after IFIT1 knockdown in RKO cells (Fig. 5C-E). Treatment with the proteasome inhibitor MG132 could reverse IFIT1 knockdown induced PD-L1 downregulation, while the lysosomal inhibitor chloroquine (CQ) and autophagy inhibitors 3-methyladenine (3-MA) displayed no such effects (Fig. 5F-H). These results indicate that IFIT1 may regulate PD-L1 expression via the ubiquitin-proteasome pathway. To investigate whether IFIT1 influenced PD-L1 ubiquitination, we treated RKO cells with PD-L1 and ubiquitin plasmids and transfected them with IFIT1 siRNA and IFIT1 plasmid separately. The results showed that knockdown of IFIT1 could increase PD-L1 ubiquitination and overexpression of IFIT1 could decrease its ubiquitination (Fig. 5I-J). These results suggested that IFIT1 upregulated PD-L1 expression via decreasing its ubiquitination and degradation.
Fig. 5.
IFIT1 mediated PD-L1 stabilization via regulating PD-L1 ubiquitination. A. Co-immunoprecipitation (Co-IP) assay detecting IFIT1 in RKO cell lysates immunoprecipitated with anti-PD-L1. B. RKO cells were overexpressed with FLAG_IFIT1 and HA_PD-L1. Co-IP assay detecting exogenous IFIT1 (FLAG) in RKO cell lysates immunoprecipitated with anti-HA. C-D. RKO cells transfected with control siRNAs and IFIT1 siRNAs were treated with cycloheximide (CHX) (50 µg/mL) at the indicated time points. Immunoblot of PD-L1 at indicated time points. E. Quantification of relative remaining PD-L1 in C-D. F. Immunoblot of PD-L1 in RKO cells transfected with IFIT1 siRNAs and treated with MG132 (10uM, 6 hours). G. Immunoblot of PD-L1 in RKO cells transfected with IFIT1 siRNAs and treated with CQ (25uM, 6 hours). H. Immunoblot of PD-L1 in RKO cells transfected with IFIT1 siRNAs and treated with 3-MA (6 hours). I-J. Immunoblot of cell lysates immunoprecipitated with anti-PD-L1 in RKO cells transfected with the indicated constructs.
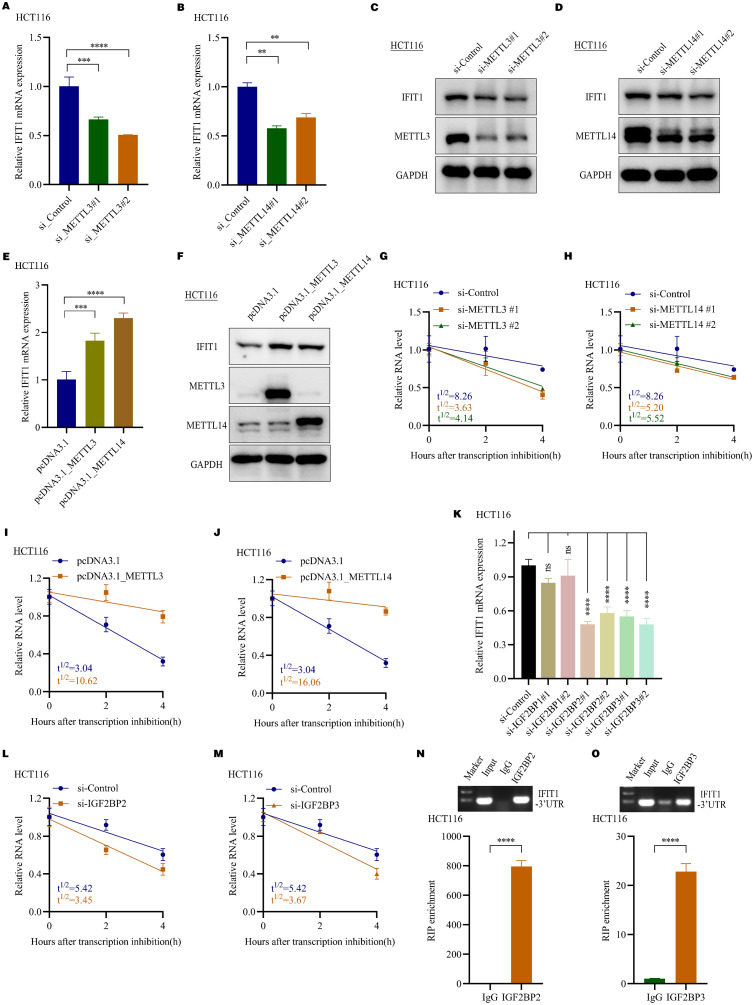
IFIT1 was methylated by METTL3 and METTL14 and recognized by IGF2BP2/3
The above results suggested obvious m6A peaks in 3’UTR of IFIT1 mRNAs. We attempted to investigate which m6A regulators mediated m6A modifications in IFIT1 mRNAs and what functions m6A modifications have. In biological validation assays, IFIT1 mRNA and protein levels were markedly dampened in METTL3 and METTL14 knockdown CRC cells (Fig. 6A-D, Supplementary Figure S6A-B). Overexpression of METTL3 and METTL14 increased IFIT1 expression (Fig. 6E-F, Supplementary Figure S6C-D). Since METTL3 and METTL14 positively regulated IFIT1 expression, we guessed that METTL3 and METTL14 may increase IFIT1 mRNAs stability. RNA stability assays showed that IFIT1 transcripts were less stable when METTL3 and METTL14 knockdown and more stable when METTL3 and METTL14 overexpressed in HCT116 cells (Fig. 6G-J).
Fig. 6.
IFIT1 was methylated by METTL3 and METTL14 and recognized by IGF2BP2/3. A. PCR analysis of IFIT1 mRNA levels in HCT116 cells after transfection with METTL3 siRNAs (n=3, mean± SD, One-way ANOVA). B. PCR analysis of IFIT1 mRNA levels in HCT116 cells after transfection with METTL14 siRNAs (n=3, mean± SD, One-way ANOVA). C. Western blot assays detecting IFIT1 protein expression in HCT116 cells after transfection with METTL3 siRNAs. D. Western blot assays detecting IFIT1 protein expression in HCT116 cells after transfection with METTL14 siRNAs. E. PCR analysis of IFIT1 mRNA levels in HCT116 cells after transfection with pcDNA3.1_METTL3 and pcDNA3.1_METTL14 (n=3, mean± SD, two-tailed Student's t-test). F. Western blot assays detecting IFIT1 protein expression in HCT116 cells after transfection with pcDNA3.1_METTL3 and pcDNA3.1_METTL14. G. The detection of half-life(t1/2) of IFIT1 mRNAs in HCT116 cells transfected with control siRNAs and METTL3 siRNAs using real-time PCR, linear regression analysis. H. The detection of half-life(t1/2) of IFIT1 mRNAs in HCT116 cells transfected with control siRNAs and METTL14 siRNAs using real-time PCR, linear regression analysis. I. The detection of half-life(t1/2) of IFIT1 mRNAs in HCT116 cells with and without METTL3 overexpression using real-time PCR, linear regression analysis. J. The detection of half-life(t1/2) of IFIT1 mRNAs in HCT116 cells with and without METTL14 overexpression using real-time PCR, linear regression analysis. K. PCR analysis of IFIT1 mRNA level in HCT116 cells with IGF2BP1, IGF2BP2 and IGF2BP3 siRNAs transfection (n=3, mean± SEM, One-way ANOVA). L. The detection of half-life(t1/2) of IFIT1 mRNAs in HCT116 cells transfected with control siRNAs and IGF2BP2 siRNAs using real-time PCR, linear regression analysis. M. The detection of half-life(t1/2) of IFIT1 mRNAs in HCT116 cells transfected with control siRNAs and IGF2BP3 siRNAs using real-time PCR, linear regression analysis. N. The direct binding of IGF2BP2 with IFIT1 3’UTR using Agarose electrophoresis and RIP-qPCR assays in HCT116 cells (n=3, mean± SD, two-tailed Student's t-test). O. The direct binding of IGF2BP3 with IFIT1 3’UTR using Agarose electrophoresis and RIP-qPCR assays in HCT116 cells (n=3, mean± SD, two-tailed Student's t-test).
M6A readers recognize and bind methylated mRNAs and mediate corresponding functions. IGF2BP family, including IGF2BP1, IGF2BP2 and IGF2BP3, are responsible for mRNA stability while other readers including YTHDF2 and YTHDF3 are responsible for mRNA decay [33,34]. Considering that METTL3 and METTL14 positively regulated IFIT1 expression and increased IFIT1 mRNA stability, we guessed that IGF2BP family may recognize IFIT1 mRNA and increase mRNAs stability. To investigate this question, we designed separately two siRNAs targeting IGF2BP1, IGF2BP2, and IGF2BP3 to knockdown their expression and detected IFIT1 mRNA expression alterations. The results showed that IFIT1 mRNA expression was remarkably suppressed after IGF2BP2/3 knockdown, but not IGF2BP1 knockdown in CRC cells (Fig. 6K, Supplementary Figure S6E). Furthermore, RNA stability assays showed that the half-life of IFIT1 transcripts was shortened after IGF2BP2 and IGF2BP3 knockdown HCT116 cells (Fig. 6L-M). RIP assays validated that IGF2BP2 and IGF2BP3 could directly bind the 3’UTR of IFIT1 mRNA (Fig. 6N-O). These data demonstrate that METTL3 and METTL14 methylated IFIT1 mRNAs and increased its expression through IGF2BP2/3-dependent mRNA stability regulation.
F. nucleatum abundance, IFIT1 and PD-L1 expression were positively correlated in CRC patients
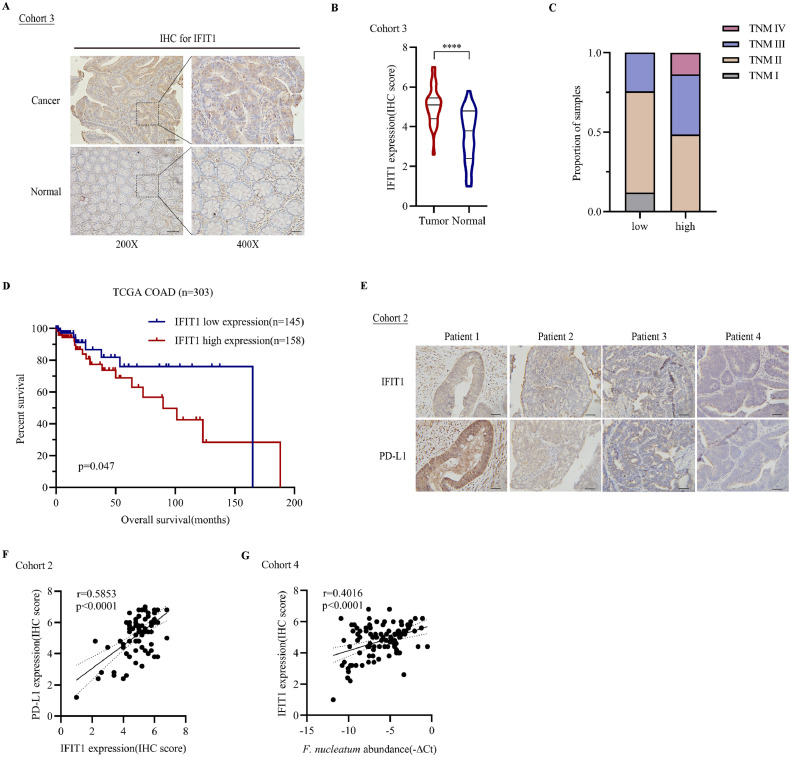
Our results have suggested that F. nucleatum stimulated PD-L1 protein expression via m6A-modified IFIT1. The function of PD-L1 has been extensively studied in clinical samples. The functions of IFIT1 have not been studied in CRC samples. we measured IFIT1 expression in 34 pairs of CRC tissues and non-cancerous normal colon tissues in cohort 3. The results showed that IFIT1 was significantly upregulated in CRC tissues (Fig. 7A-B). Also, we measured IFIT1 expression in clinical CRC tissues using immunohistochemistry and found that IFIT1 expression was more highly expressed in TNM stage III/IV CRC than in stage I/II (Fig. 7C). Kaplan–Meier analysis showed higher IFIT1 expression correlated with worse overall survival based on TCGA COAD database (Fig. 7D). In cohort 2, there was a strong correlation between IFIT1 and PD-L1 expression (r=0.5853, p<0.0001) (Fig. 7E-F). F. nucleatum abundance was also positively correlated with IFIT1 expression (Fig. 7G). Our data showed that IFIT1 was higher expressed in tumor tissues and F. nucleatum abundance, IFIT1 and PD-L1 expression were positively correlated in CRC patients.
Fig. 7.
F. nucleatum abundance, IFIT1 and PD-L1 expression were positively correlated in CRC patients. A. Immunohistochemical staining for IFIT1 in colorectal cancer tissues and para-cancerous normal tissues in cohort 3, scale bar, 100µm in 200X, 50µm in 400X. B. Statistical analysis of IFIT1 IHC score in cohort 3 (paired two-tailed Student's t-test). C. TNM stages of samples in cohort 2 grouped by IFIT1 expression. D. Overall survival analysis of TCGA_COAD samples grouped by IFIT1 expression. E. Representative images of IFIT1 and PD-L1 staining of samples in cohort 2. F. Statistical analysis of the correlation of IFIT1 and PD-L1 expression (r=0.5853, p<0.0001, two-sided Pearson's correlation). G. Statistical analysis of the correlation among F. nucleatum abundance and the expression of IFIT1 (r=0.4016, p<0.0001, two-sided Pearson's correlation).
Discussion
Increasing evidence suggests that microorganisms are strongly associated with colorectal carcinogenesis [3,35]. F. nucleatum is reported as a pathogenic bacterium in CRC progression [36]. Emerging evidence has indicated that higher F. nucleatum amount was associated with an immunosuppressive tumor environment [14]. In Apcmin/+ mice, F. nucleatum gavage resulted in an expansion of myeloid cells in colon tumors, which represented tumor-promoting functions including myeloid-derived suppressor cells (MDSC) and tumor-associated macrophages (TAM). A population-based study suggested that higher F. nucleatum amount in tumors was associated with a lower CD3+ T cell density [13,37]. Also, F. nucleatum could inhibited NK cell activity through its virulence factor Fap2, which directly interacted with the inhibitory receptor TIGIT [15]. PD-L1 is an important immune checkpoint expressed on tumor cells and promotes tumor immune escape [38]. Here, we found F. nucleatum treatment promoted CRC progression and stimulated PD-L1 protein expression. The PD-L1 upregulation didn't depend on F. nucleatum treatment activated PI3K pathway. Combined m6A-seq and RNA-seq identified m6A modified IFIT1 mediating F. nucleatum induced PD-L1 upregulation. IFIT1 regulated PD-L1 expression without altering mRNA level through interacting with PD-L1 protein and influencing its ubiquitination.
The regulatory mechanisms of PD-L1 expression are complex, including genetic alterations, transcriptional regulations, post-transcriptional regulations and post-translational modifications [10,39]. Emerging researches have indicated RNA m6A modification could regulated PD-L1 expression directly and indirectly [18,19]. PD-L1 can be directly modified with m6A modifications and indirectly regulated by m6A modified target genes. Here, we performed combined m6A-seq and RNA-seq in F. nucleatum-treated HT29 cells and identified m6A-modified IFIT1 mediated F. nucleatum stimulated PD-L1 upregulation. IFIT1 altered PD-L1 protein expression without affecting its mRNA levels. IFIT1 could interact with PD-L1 protein and its overexpression inhibited PD-L1 ubiquitination and decreased its degradation.
IFIT1 is a member of interferon-stimulated genes (ISGs) which function as essential anti-viral proteins [27]. Emerging evidence has indicated its roles in carcinogenesis. IFIT1 and IFIT3 were reported as oncogenes in oral squamous cell carcinoma with the function to promote epithelial-mesenchymal transition and activate EGFR signaling [28]. IFIT1 and IFIT3 were also overexpressed in hepatocellular carcinoma tissues and mediated cell migration [29]. Here, we found IFIT1 was higher expressed in CRC tissues and its higher expression indicated worse survival. Inhibition of IFIT1 dampened CRC progression in vitro and in vivo. IFIT1 was found to interact with PD-L1 protein and regulate its ubiquitination. In addition, IFIT1 was modified with m6A peaks located in 3’UTR and the m6A levels were altered by F. nucleatum treatment. IFIT1 was methylated by METTL3 and METTL14 and m6A-modified IFIT1 was more stable dependent on IGF2BP2/3 recognition. Our data suggested that IFIT1 may be a potential oncogene in CRC and IFIT1 may participate in tumor immune escape.
In conclusion, F. nucleatum promoted CRC progression in vitro and in vivo and upregulated PD-L1 expression. Combined m6A-seq and RNA-seq identified m6A-modified IFIT1 mediated F. nucleatum induced PD-L1 upregulation. IFIT1 was modified with m6A in 3’UTR and the m6A levels were altered by F. nucleatum treatment. IFIT1 served as a potential oncogene in CRC and regulated PD-L1 expression through regulating PD-L1 ubiquitination. Our work suggested that F. nucleatum promoted PD-L1 protein expression via m6A modification of IFIT1 and provided new aspects for understanding F. nucleatum mediated immune escape.
Author contributions
Y.Q.G., T.H.Z., P.P.X., J.H., H.Y.C., and J.-Y.F. designed the experiments. Y.Q.G., T.H.Z., P.P.X., Y.C.W., and Y.J. performed the experiments and analyzed the data. Y.Q.G. wrote the manuscript. Y.-X.C., H.J., H.Y.C. and J.-Y.F. reviewed and revised the manuscript.
Data availability
The raw sequencing data are available in the Gene Expression Omnibus database under the accession number GSE191257. All the other data are available in the article and supplemental information.
Declaration of Competing Interest
The authors declare no potential conflicts of interest.
Acknowledgements
This project was supported by grants from the National Natural Science Foundation of China (31970718, 82002622).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100850.
Contributor Information
Haoyan Chen, Email: haoyanchen@sjtu.edu.cn.
Jie Hong, Email: jiehong97@sjtu.edu.cn.
Jing-Yuan Fang, Email: jingyuanfang@sjtu.edu.cn.
Appendix. Supplementary materials
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 3.Janney A, Powrie F, Mann EH. Host-microbiota maladaptation in colorectal cancer. Nature. 2020;585:509–517. doi: 10.1038/s41586-020-2729-3. [DOI] [PubMed] [Google Scholar]
- 4.Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16:690–704. doi: 10.1038/s41575-019-0209-8. [DOI] [PubMed] [Google Scholar]
- 5.Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25:968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 6.Mehta RS, Nishihara R, Cao Y, Song M, Mima K, Qian ZR, Nowak JA, Kosumi K, Hamada T, Masugi Y, et al. Association of Dietary Patterns With Risk of Colorectal Cancer Subtypes Classified by Fusobacterium nucleatum in Tumor Tissue. JAMA Oncol. 2017;3:921–927. doi: 10.1001/jamaoncol.2016.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21. Gastroenterology. 2017;152 doi: 10.1053/j.gastro.2016.11.018. 851-866 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170 doi: 10.1016/j.cell.2017.07.008. 548-563 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong J, Guo F, Lu SY, Shen C, Ma D, Zhang X, Xie Y, Yan T, Yu T, Sun T, et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut. 2021;70:2123–2137. doi: 10.1136/gutjnl-2020-322780. [DOI] [PubMed] [Google Scholar]
- 10.Sun C, Mezzadra R, Schumacher TN. Regulation and Function of the PD-L1 Checkpoint. Immunity. 2018;48:434–452. doi: 10.1016/j.immuni.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren Y, Qian Y, Ai L, Xie Y, Gao Y, Zhuang Z, Chen J, Chen YX, Fang JY. TRAPPC4 regulates the intracellular trafficking of PD-L1 and antitumor immunity. Nat Commun. 2021;12:5405. doi: 10.1038/s41467-021-25662-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C, Chi H, Deng S, Wang H, Yao H, Wang Y, Chen D, Guo X, Fang JY, He F, et al. THADA drives Golgi residency and upregulation of PD-L1 in cancer cells and provides promising target for immunotherapy. J Immunother Cancer. 2021;9 doi: 10.1136/jitc-2021-002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K, et al. Fusobacterium nucleatumand T Cells in Colorectal Carcinoma. JAMA Oncology. 2015;1 doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X, Zhang X, Cao Y, Ma D, Zhu X, et al. m(6)A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19:72. doi: 10.1186/s12943-020-01190-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu X, Yang S, Wang S, Wu J, Zheng B, Wang K, Shen S, Jeong S, Li Z, Zhu Y, et al. M(6)A Demethylase ALKBH5 Regulates PD-L1 Expression and Tumor Immunoenvironment in Intrahepatic Cholangiocarcinoma. Cancer Res. 2021;81:4778–4793. doi: 10.1158/0008-5472.CAN-21-0468. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, Wang T, She Y, Wu K, Gu S, Li L, Dong C, Chen C, Zhou Y. N(6)-methyladenosine-modified circIGF2BP3 inhibits CD8(+) T-cell responses to facilitate tumor immune evasion by promoting the deubiquitination of PD-L1 in non-small cell lung cancer. Mol Cancer. 2021;20:105. doi: 10.1186/s12943-021-01398-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Gao S, Liu W, Wong CC, Wu J, Wu J, Liu D, Gou H, Kang W, Zhai J, et al. RNA N(6)-Methyladenosine Methyltransferase METTL3 Facilitates Colorectal Cancer by Activating the m(6)A-GLUT1-mTORC1 Axis and Is a Therapeutic Target. Gastroenterology. 2021;160 doi: 10.1053/j.gastro.2020.11.013. 1284-1300 e16. [DOI] [PubMed] [Google Scholar]
- 21.Guo W, Tan F, Huai Q, Wang Z, Shao F, Zhang G, Yang Z, Li R, Xue Q, Gao S, et al. Comprehensive Analysis of PD-L1 Expression, Immune Infiltrates, and m6A RNA Methylation Regulators in Esophageal Squamous Cell Carcinoma. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.669750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ugai T, Zhao M, Shimizu T, Akimoto N, Shi S, Takashima Y, Zhong R, Lau MC, Haruki K, Arima K, et al. Association of PIK3CA mutation and PTEN loss with expression of CD274 (PD-L1) in colorectal carcinoma. Oncoimmunology. 2021;10 doi: 10.1080/2162402X.2021.1956173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y, Yang J, Cai Y, Fu S, Zhang N, Fu X, Li L. IFN-gamma-mediated inhibition of lung cancer correlates with PD-L1 expression and is regulated by PI3K-AKT signaling. Int J Cancer. 2018;143:931–943. doi: 10.1002/ijc.31357. [DOI] [PubMed] [Google Scholar]
- 25.Kang W, Jia Z, Tang D, Zhang Z, Gao H, He K, Feng Q. Fusobacterium nucleatum Facilitates Apoptosis, ROS Generation, and Inflammatory Cytokine Production by Activating AKT/MAPK and NF-kappaB Signaling Pathways in Human Gingival Fibroblasts. Oxid Med Cell Longev. 2019;2019 doi: 10.1155/2019/1681972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsueh CY, Huang Q, Gong H, Shen Y, Sun J, Lau HC, Zhang D, Tang D, Wu C, Guo Y, et al. A positive feed-forward loop between Fusobacterium nucleatum and ethanol metabolism reprogramming drives laryngeal cancer progression and metastasis. iScience. 2022;25 doi: 10.1016/j.isci.2022.103829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson B, VanBlargan LA, Xu W, White JP, Shan C, Shi PY, Zhang R, Adhikari J, Gross ML, Leung DW, et al. Human IFIT3 Modulates IFIT1 RNA Binding Specificity and Protein Stability. Immunity. 2018;48 doi: 10.1016/j.immuni.2018.01.014. 487-499 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pidugu VK, Wu MM, Yen AH, Pidugu HB, Chang KW, Liu CJ, Lee TC. IFIT1 and IFIT3 promote oral squamous cell carcinoma metastasis and contribute to the anti-tumor effect of gefitinib via enhancing p-EGFR recycling. Oncogene. 2019;38:3232–3247. doi: 10.1038/s41388-018-0662-9. [DOI] [PubMed] [Google Scholar]
- 29.Liu G, Sun J, Yang ZF, Zhou C, Zhou PY, Guan RY, Sun BY, Wang ZT, Zhou J, Fan J, et al. Cancer-associated fibroblast-derived CXCL11 modulates hepatocellular carcinoma cell migration and tumor metastasis through the circUBAP2/miR-4756/IFIT1/3 axis. Cell Death Dis. 2021;12:260. doi: 10.1038/s41419-021-03545-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mimura K, Teh JL, Okayama H, Shiraishi K, Kua LF, Koh V, Smoot DT, Ashktorab H, Oike T, Suzuki Y, et al. PD-L1 expression is mainly regulated by interferon gamma associated with JAK-STAT pathway in gastric cancer. Cancer Sci. 2018;109:43–53. doi: 10.1111/cas.13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asgarova A, Asgarov K, Godet Y, Peixoto P, Nadaradjane A, Boyer-Guittaut M, Galaine J, Guenat D, Mougey V, Perrard J, et al. PD-L1 expression is regulated by both DNA methylation and NF-kB during EMT signaling in non-small cell lung carcinoma. Oncoimmunology. 2018;7 doi: 10.1080/2162402X.2017.1423170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge J, Wang J, Xiong F, Jiang X, Zhu K, Wang Y, Mo Y, Gong Z, Zhang S, He Y, et al. Epstein-Barr Virus-Encoded Circular RNA CircBART2.2 Promotes Immune Escape of Nasopharyngeal Carcinoma by Regulating PD-L1. Cancer Res. 2021;81:5074–5088. doi: 10.1158/0008-5472.CAN-20-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, Jiang Z, Zhang Y, Xu G, Zhang J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193–1205. doi: 10.1136/gutjnl-2019-319639. [DOI] [PubMed] [Google Scholar]
- 35.Tilg H, Adolph TE, Gerner RR, Moschen AR. The Intestinal Microbiota in Colorectal Cancer. Cancer Cell. 2018;33:954–964. doi: 10.1016/j.ccell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65:1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu C, Fan L, Lin Y, Shen W, Qi Y, Zhang Y, Chen Z, Wang L, Long Y, Hou T, et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes. 2021;13 doi: 10.1080/19490976.2021.1980347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Yao H, Li C, Shi H, Lan J, Li Z, Zhang Y, Liang L, Fang JY, Xu J. HIP1R targets PD-L1 to lysosomal degradation to alter T cell-mediated cytotoxicity. Nat Chem Biol. 2019;15:42–50. doi: 10.1038/s41589-018-0161-x. [DOI] [PubMed] [Google Scholar]
- 39.Zhang H, Zhu C, He Z, Chen S, Li L, Sun C. LncRNA PSMB8-AS1 contributes to pancreatic cancer progression via modulating miR-382-3p/STAT1/PD-L1 axis. J Exp Clin Cancer Res. 2020;39:179. doi: 10.1186/s13046-020-01687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data are available in the Gene Expression Omnibus database under the accession number GSE191257. All the other data are available in the article and supplemental information.