Abstract
Background
Lactobacillus species dominate the vaginal microflora performing a first-line defense against vaginal infections. Extracellular vesicles (EVs) released by lactobacilli are considered mediators of their beneficial effects affecting cellular communication, homeostasis, microbial balance, and host immune system pathways. Up to now, very little is known about the role played by Lactobacillus EVs in the vaginal microenvironment, and mechanisms of action remain poorly understood.
Results
Here, we hypothesized that EVs can mediate lactobacilli beneficial effects to the host by modulating the vaginal microbiota colonization. We recovered and characterized EVs produced by two vaginal strains, namely Lactobacillus crispatus BC5 and Lactobacillus gasseri BC12. EVs were isolated by ultracentrifugation and physically characterized by Nanoparticle Tracking Analysis (NTA) and Dynamic Light Scattering (DLS). EVs protein and nucleic acids (DNA and RNA) content was also evaluated. We explored the role of EVs on bacterial adhesion and colonization, using a cervical cell line (HeLa) as an in vitro model. Specifically, we evaluated the effect of EVs on the adhesion of both vaginal beneficial lactobacilli and opportunistic pathogens (i.e., Escherichia coli, Staphylococcus aureus, Streptococcus agalactiae, and Enterococcus faecalis). We demonstrated that EVs from L. crispatus BC5 and L. gasseri BC12 significantly enhanced the cellular adhesion of all tested lactobacilli, reaching the maximum stimulation effect on strains belonging to L. crispatus species (335% and 269% of average adhesion, respectively). At the same time, EVs reduced the adhesion of all tested pathogens, being EVs from L. gasseri BC12 the most efficient.
Conclusions
Our observations suggest for the first time that EVs released by symbiotic Lactobacillus strains favor healthy vaginal homeostasis by supporting the colonization of beneficial species and preventing pathogens attachment. This study reinforces the concept of EVs as valid postbiotics and opens the perspective of developing postbiotics from vaginal strains to maintain microbiota homeostasis and promote women’s health.
Keywords: Extracellular vesicles, Lactobacillus, Adhesion, Postbiotics, Vaginal homeostasis
Background
The vaginal microbiota of a healthy woman is mainly composed of Lactobacillus species which significantly affect the homeostasis of the vaginal ecosystem [1, 2]. Lactobacillus colonization of the vaginal mucosa reduces urogenital infections preventing pathogens’ adhesion, attachment, and consequent invasion to host tissues [3, 4]. Moreover, lactobacilli produce a wide range of antimicrobial metabolites such as lactic acid, hydrogen peroxide (H2O2), lectins, bacteriocins, and biosurfactants, whose activity was demonstrated towards a broad spectrum of pathogens [5–11].
Despite their thick cell wall, Lactobacillus spp. can produce extracellular vesicles (EVs), spherical lipid bilayer membrane-derived structures widespread throughout all domains of life [12]. During vesiculogenesis, EVs load different molecules, such as lipids, proteins, nucleic acids, and other compounds from various cell compartments that are exported and, subsequently, available in the environment [13]. Depending on the cargo and the surface composition, EVs are involved in various biological pathways like cell viability, nutrient uptake, antibiotic resistance, nucleic acids transfer, biofilm formation, intraspecies communication (quorum-sensing), and communication with the host (crosstalk) [14].
Regarding Lactobacillus spp., EVs play a role in mediating their beneficial effect to the host, affecting pathogen infectivity and/or modulating the host immune system. Indeed, it has been reported that EVs from lactobacilli can reduce pathogen infection by the exposure of antimicrobial molecules and/or by mediating the competitive exclusion between pathogenic and mutualistic bacteria [15–18]. In addition, Lactobacillus EVs impaired enterococci and Staphylococcus aureus infection by modulating host immune system pathways [18, 19]. As more and more studies highlight the association between EVs and the probiotic bacteria health benefits, the concept of EVs as new postbiotics is coming to consolidation [20].
Besides this evidence, only one study reported the protective role of EVs from Lactobacillus in the vaginal niche [8], pointing out the need for further investigations in this field.
In the present paper, we investigated the potential of Lactobacillus EVs in modulating the vaginal microbiota composition in favor of the host state of health. EVs were recovered from two Lactobacillus strains isolated from the healthy vagina, namely Lactobacillus crispatus BC5 and Lactobacillus gasseri BC12 [10]. First, we characterized EVs physical and chemical properties in terms of yield, size, and total protein and nucleic acids (DNA and RNA) content. We sought for the ability of EVs to modulate the adhesion of beneficial resident lactobacilli, including the producing strains themselves and other strains belonging to the species L. crispatus (BC1, BC3, and BC4) and L. gasseri (BC9, BC10, and BC11). Moreover, EVs effects were tested toward the adhesion of four vaginal opportunistic pathogens: Escherichia coli, Staphylococcus aureus, Streptococcus agalactiae, and Enterococcus faecalis.
Results
Characterization of EVs released by vaginal lactobacilli
EVs were isolated from L. crispatus BC5 and L. gasseri BC12 strains at the stationary growth phase in MRS medium. EVs concentrations and dimensions were analyzed by NTA technology, Z-potential was measured by DLS technique. EVs were also recovered from sterile MRS medium and physically characterized. Results are reported in Table 1.
Table 1.
Physical characterization of EVs isolated from L. crispatus BC5, L. gasseri BC12 and MRS medium
| Concentration (particles/mL) | Size (nm) | Ζ-potential (mV) | |
|---|---|---|---|
| L. crispatus BC5-EVs | 34.70 ± 2.31 × 109 | 89.3 ± 49.2 | − 20.3 ± 1.8 |
| L. gasseri BC12-EVs | 30.60 ± 1.05 × 109 | 129.1 ± 51.9 | − 10.4 ± 0.7 |
| MRS-EVs | 14.40 ± 0.71 × 109 | 150.9 ± 73.8 | − 26.1 ± 3.4 |
Data are reported as mean ± standard deviation (SD) (n = 2)
As reported in Table 1, L. crispatus BC5 and L. gasseri BC12 produced EVs in similar amounts, EVs average size slightly varied between the two strains (mean diameter of 89.3 nm vs 129.1 nm). EVs Z-potential resulted in negative values, L. crispatus BC5-EVs Z-potential was significantly lower (− 20.3 ± 1.8 mV) than that of L. gasseri BC12-EVs (− 10.4 ± 0.7 mV) (p < 0.05). Particles were also found in MRS medium not conditioned by microorganisms, in a concentration about 2.5-fold lower than EVs derived from bacteria. EVs from MRS medium displayed larger dimension and significantly lower Z-potential than those reported for L. crispatus BC5 and L. gasseri BC12 (p < 0.05).
EVs samples were also investigated in terms of protein and nucleic acid content and results are reported in Table 2.
Table 2.
Chemical characterization of EVs isolated from L. crispatus BC5, L. gasseri BC12 and MRS medium
| Protein content(μg/109particles) | DNA(μg/109 particles) | RNA(μg/109 particles) | |
|---|---|---|---|
| L. crispatus BC5-EVs | 1.19 ± 0.07 | 0.22 ± 0.04 | 0.03 ± 0.01 |
| L. gasseri BC12-EVs | 1.83 ± 0.27 | 0.11 ± 0.00 | 0.08 ± 0.03 |
| MRS-EVs | 8.51 ± 0.28 | 0.28 ± 0.06 | 0.04 ± 0.01 |
Data are reported as mean ± standard deviation (SD) (n = 2)
Considering Lactobacillus-EVs cargo, protein content was similar in EVs from L. crispatus BC5 and L. gasseri BC12. Overall, EVs samples contained more DNA than RNA with slight differences in terms of quantity between Lactobacillus strains. In particular, EVs from L. crispatus BC5 showed numerically higher amount of DNA than L. gasseri BC12, while RNA amount was numerically higher in EVs from L. gasseri BC12 than L. crispatus BC5. EVs recovered from sterile culture medium also contain proteins, DNA and RNA in valuable amounts.
Effects of EVs on Lactobacillus adhesion to HeLa cells
To study the effect of EVs in supporting resident lactobacilli colonization, different vaginal Lactobacillus strains were allowed to adhere to HeLa cells in the presence of L. crispatus BC5-EVs and L. gasseri BC12-EVs and results were shown in Figs. 1 and 2, respectively. Bacterial adhesion in the absence of EVs (Phosphate-buffered saline, PBS) was used to normalize data.
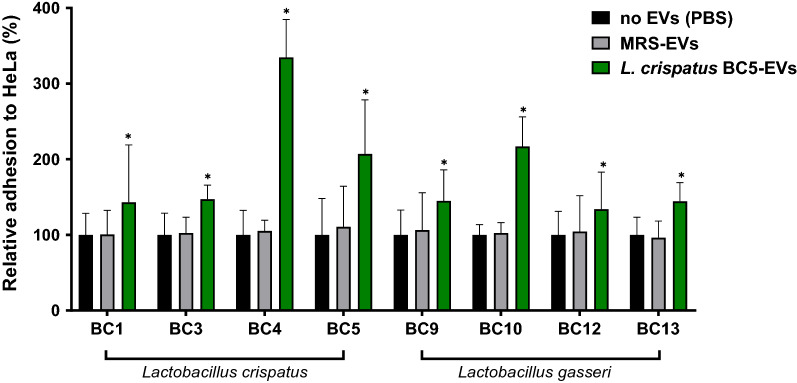
Fig. 1.
Adhesion assays of Lactobacillus strains to HeLa cells in presence of L. crispatus BC5-EVs. The adhesion rates are shown as a percentage relative to bacterial adhesion in the absence of EVs (PBS, 100%). Data are reported as mean ± SD (n = 2). *p value < 0.05
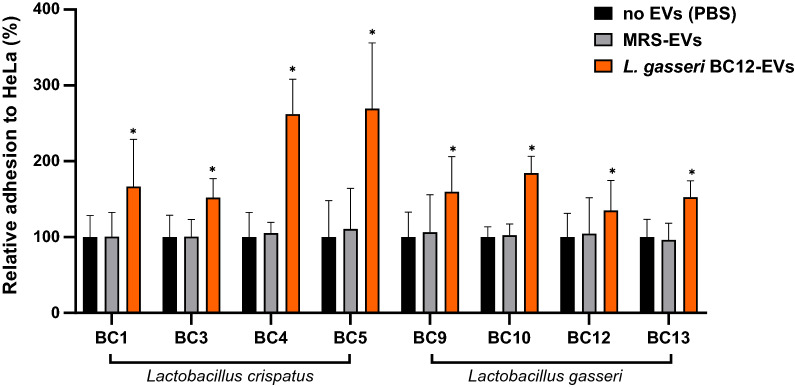
Fig. 2.
Adhesion assays of Lactobacillus strains to HeLa cells in presence of L. gasseri BC12-EVs. The adhesion rates are shown as a percentage relative to bacterial adhesion in the absence of EVs (PBS, 100%). Data are reported as mean ± SD (n = 2). *p value < 0.05
First, we excluded a possible cytotoxic effect of EVs on the cell line by the evaluation of HeLa cells morphology and integrity at the optical microscope (data not shown).
EVs effect was evaluated on the adhesion of the producer itself and of other strains belonging to the same species, namely L. crispatus BC1, BC3, BC4 and L. gasseri BC9, BC10, BC13. As shown in Figs. 1 and 2, the adhesion of Lactobacillus strains to epithelial cells significantly increased in the presence of L. crispatus BC5-EVs and L. gasseri BC12-EVs. Contrariwise, no effect on Lactobacillus adhesion was registered in the presence of EVs derived from MRS medium, suggesting that the effect exerted on Lactobacillus was specifically associated to EVs origin.
Overall, according to Fig. 1, L. crispatus BC5-EVs exerted a good stimulatory activity towards lactobacilli, with an average adhesion ranging from 134% (L. gasseri BC12) to 335% (L. crispatus BC4). Interestingly, L. crispatus BC4 was the most stimulated strain by L. crispatus BC5-EVs compared to all strains (average adhesion 335%), including L. crispatus BC5 itself (207%). In addition, the highest stimulation activity of L. crispatus BC5-EVs towards L. gasseri species was observed on strain L. gasseri BC10, reaching 217% of average adhesion (ANOVA, p < 0.05).
Regarding L. gasseri BC12-EVs, as reported in Fig. 2, a similar stimulatory activity was registered, reaching an average adhesion between 135% (L. gasseri BC12) and 269% (L. crispatus BC5). Particularly, L. gasseri BC12-EVs increased more the adhesion of L. crispatus BC4 (262%) and L. crispatus BC5 (269%) compared to the other strains, including the producer’s itself (135%) (ANOVA, p < 0.05).
Considering differences in activity between L. crispatus BC5-EVs and L. gasseri BC12-EVs, we observed that L. crispatus BC5-EVs were significantly more active than L. gasseri BC12-EVs regarding the adhesion of only two strains out of eight: L. crispatus BC4 (335% and 262%) and L. gasseri BC10 (217% and 184%), while L. crispatus BC5 resulted to be more stimulated by L. gasseri BC12-EVs rather than its own EVs (269% and 207%) (Student’s t-test, p < 0.05).
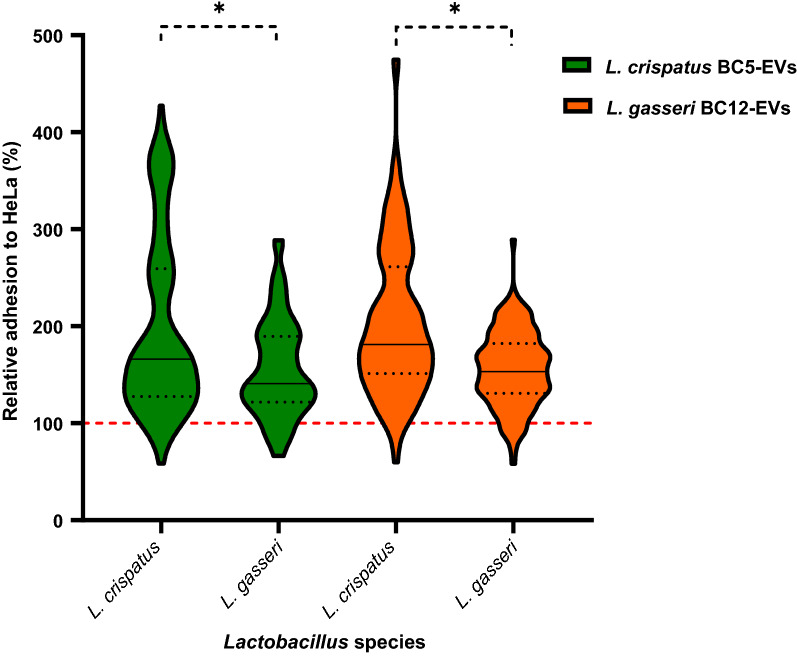
In addition, to investigate the role of EVs in species communication, Lactobacillus strains were grouped per species and the adhesion in the presence of L. crispatus BC5-EVs and L. gasseri BC12-EVs was reported in a violin plot (Fig. 3). Interestingly, EVs derived from L. crispatus BC5 and L. gasseri BC12 strains mainly affected the adhesion of L. crispatus strains than L. gasseri ones, pointing out that L. crispatus species was more sensitive to the stimulation effect of Lactobacillus-EVs. At the same time, no significant differences were found between L. crispatus BC5-EVs and L. gasseri BC12-EVs regarding the adhesion of L. crispatus or L. gasseri species, showing that EVs effect was not related to the producer species.
Fig. 3.
Violin plot of Lactobacillus adhesion grouped for species in presence of L. crispatus BC5-EVs (green) and L. gasseri BC12-EVs (orange). The adhesion rates are shown as a percentage relative to bacterial adhesion in the absence of EVs (PBS, 100%, red line). Solid and dotted black lines represent median values and quartiles, respectively. *p < 0.05
Effects of EVs on pathogens adhesion to HeLa cells
Lactobacillus crispatus BC5-EVs and L. gasseri BC12-EVs effects were also sought on the adhesion of four common vaginal pathogens, namely E. coli, S. aureus, S. agalactiae, and E. faecalis.
As reported in Figs. 4 and 5, EVs isolated from L. crispatus BC5 and L. gasseri BC12 significantly reduced the adhesion of all pathogens tested. On the other hand, no activity was observed for EVs isolated from MRS medium, indicating that the inhibitory effect of vesicles was related to EVs origin.
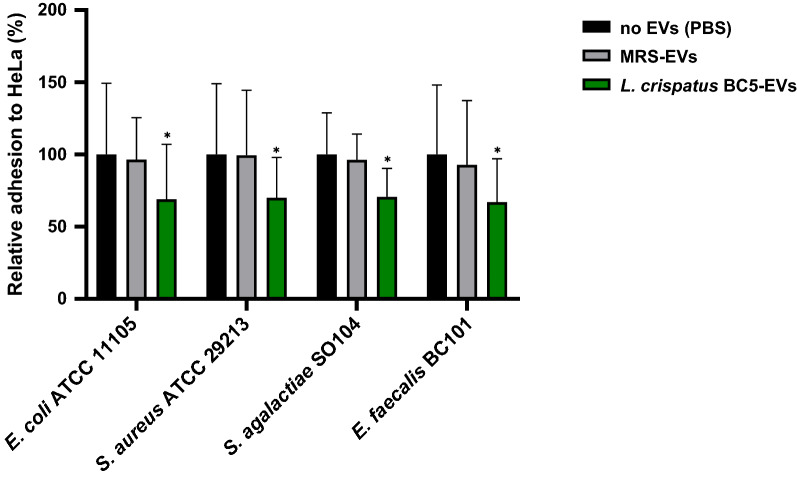
Fig. 4.
Adhesion assays of Escherichia coli ATCC 11105, Staphylococcus aureus ATCC 29213, Streptococcus agalactiae SO104, and Enterococcus faecalis BC101 to HeLa cells in presence of L. crispatus BC5-EVs. The adhesion rates are shown as a percentage relative to adhesion in the absence of EVs (PBS, 100%). Data are reported as mean ± SD (n = 2). *p value < 0.05
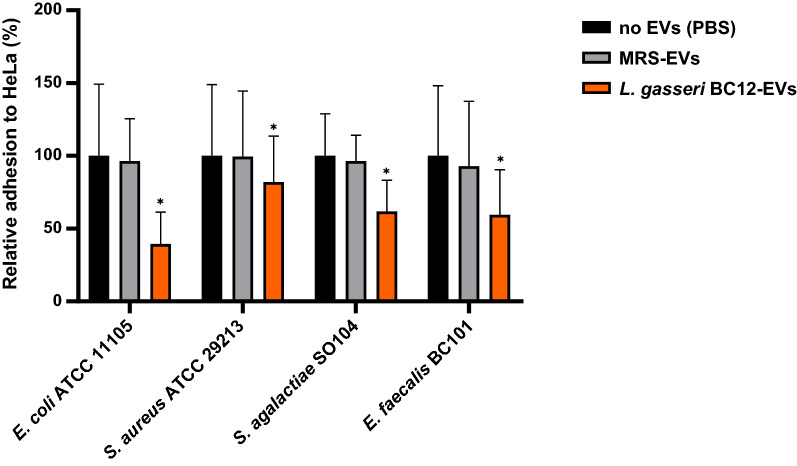
Fig. 5.
Adhesion assays of Escherichia coli ATCC 11105, Staphylococcus aureus ATCC 29213, Streptococcus agalactiae SO104, and Enterococcus faecalis BC101 to HeLa cells in presence of L. gasseri BC12-EVs. The adhesion rates are shown as a percentage relative to adhesion in the absence of EVs (PBS, 100%). Data are reported as mean ± SD (n = 2). *p value < 0.05
Particularly, EVs from L. crispatus BC5 reduced the adhesion of all tested pathogens with similar efficiency, resulting in an average adhesion of 67–71% (Fig. 4). A good anti-adhesive effect was also identified for L. gasseri BC12-EVs, with slight differences in pathogens’ adhesion depending on the strain tested (39–82%) (Fig. 5).
Considering differences in activity between L. crispatus BC5-EVs and L. gasseri BC12-EVs, we observed that L. gasseri BC12-EVs were significantly more active than L. crispatus BC5-EVs in reducing the adhesion of only one strain out of four: E. coli (39% and 69% of average adhesion, respectively) (Student’s t-test, p < 0.05). These results indicated that the inhibitory effect of EVs was not related to Lactobacillus producer strains, rather, it was related to pathogens’ strain sensitivity.
Discussion
The vaginal microbiota of healthy reproductive-age women is characterized by the abundance of the Lactobacillus genus, being L. crispatus, L. gasseri, L. jensenii, and L. iners the most common species [1]. Lactobacillus spp. play an important role in maintaining the woman’s state of health by regulating the microbiota homeostasis and reducing pathogens adhesion, proliferation, and consequent infections [21].
Recently, it was discovered that vaginal Lactobacillus strains release nanosized membrane particles, named extracellular vesicles (EVs), implicated in cell-to-cell and microbiota-host communications [8]. According to literature, EVs play a crucial role in a variety of physiological and pathological processes due to their capacity to carry bioactive macromolecules (i.e., proteins, DNA, and RNA) that can alter the biological properties of bacteria and/or host cells [22].
Compared to other human niches, only a few studies have been carried out on EVs released in the vaginal environment by beneficial lactobacilli, and our understanding of their biogenesis, composition, and functionality are still poor [22, 23].
Here, we evaluated for the first time the contribution of EVs released by vaginal lactobacilli in maintaining the microbiota balance by modulating the microorganism’s colonization. In particular, we investigated the ability of Lactobacillus-EVs to promote the adhesion of lactobacilli to HeLa cells and their anti-adhesive effect towards pathogens’ attachment.
EVs were recovered from two vaginal strains, belonging to L. crispatus and L. gasseri species, frequently predominant in a healthy vaginal microbiota [1]. We observed that L. crispatus BC5 and L. gasseri BC12 released nanosized vesicles, whose size and concentration were coherent with those previously reported by Palomino et al. [8]. Moreover, the dimensions of EVs from vaginal Lactobacillus were comparable to those of EVs released by other lactobacilli, isolated from different human niches (50–200 nm) [12, 16, 17, 19, 24, 25]. Regarding EVs surface charge, L. crispatus BC5 and L. gasseri BC12-EVs presented negative values of Z-potential (− 20.3 mV and − 10.4 mV, respectively), with differences between lactobacilli strains. These values are in agreement with those reported for other EVs from Gram-positive bacteria, i.e., Lactobacillus casei, Bacillus subtilis and Bacillus anthracis, whose Z-potential values are − 8.7 mV, − 18.2 mV and − 65.6 mV, respectively [17, 26]. Since EVs are structures mainly composed by negative charged phospholipids, as exosomes from eukaryotic cells, a negative value of Z-potential is expected [27]. Here, we also recovered EVs from MRS medium, but in concentration 2.5-fold lower than those reported for Lactobacillus samples. MRS-EVs average size were similar to those reported by Palomino et al. [8] (150–160 nm) and characterized by a negative Z-potential value. Since it is generally recognized that bacterial EVs effect can be associated to their cargo [22], we decided to characterize Lactobacillus-EVs composition in terms of total protein and nucleic acids content. As previously reported by Palomino et al. [8], we confirmed that L. crispatus BC5-EVs and L. gasseri BC12-EVs contain proteins and, for the first time, also DNA and RNA were recovered. Particularly, the protein content was higher than the content of DNA and RNA, in accordance with previous studies regarding vesicles from other Lactobacillus strains [17, 28]. Considering the content of nucleic acids, DNA and RNA were found in both L. crispatus BC5-EVs and L. gasseri BC12-EVs samples with slight differences in concentration between the two. The DNA concentration was higher in L. crispatus BC5-EVs sample, while RNA concentration was higher in L. gasseri BC12-EVs compared to L. crispatus BC5-EVs one. Also, EVs from MRS medium displayed to contain proteins and nucleic acids. This result is not surprising since MRS is a complex medium containing high amounts of digests, i.e. peptone, yeast extract and beef extract. From our findings, it appears that the biological macromolecules of medium ingredients are also carried by nanometric vesicles. As far as we know, there is no characterization of nucleic acids delivered by Lactobacillus-EVs and further studies are required to deeply analyze EVs composition [22].
The effect of EVs was studied on Lactobacillus adhesion considering the producer strains themselves and other strains belonging to the same species. We demonstrated that EVs released by L. crispatus BC5 and L. gasseri BC12 stimulated the adhesion of all Lactobacillus in a similar way, with some variability among strains. Notably, no effect on Lactobacillus adhesion rates was induced by MRS-EVs, pointing out that the observed activity was related to Lactobacillus EVs peculiarities rather than the mere presence of nanometric EVs, that have been also retrieved in MRS medium.
Interestingly, considering Lactobacillus species, L. crispatus adhesion was more stimulated by both L. crispatus BC5-EVs and L. gasseri BC12-EVs than L. gasseri adhesion, underlying that the stimulation effect of EVs was not related to the producer strain, on the contrary, different Lactobacillus species may have different sensitivity to EVs modulation.
The vaginal tract is naturally characterized by the coexistence of different Lactobacillus species, with the prevalence of one above the others [1]. Due to this co-inhabitance, lactobacilli have adopted a collaborative strategy rather than promoting competitive behavior to survive. Particularly, coculture experiments of L. crispatus and L. gasseri species demonstrated a cooperative behavior between the species in terms of niche colonization but L. crispatus species better colonizes the niche compared to L. gasseri, underlying the high adaptability of this species to others favoring its persistence [29].
It has been reported that Lactobacillus-EVs metabolites, nucleic acids, and protein content can be associated to a peculiar biological role of EVs [18, 22]. Regarding L. crispatus BC5-EVs and L. gasseri BC12-EVs protein content, Palomino et al. identified some adhesins (i.e., enolases, elongation factor-TU, 30S ribosomal protein, pyruvate kinase, chaperon proteins) involved in Lactobacillus attachment to human cell receptors [8, 30–32]. Moreover, EVs from L. casei delivered an exclusive adhesin protein absent in cell extracts, suggesting that EVs may affect Lactobacillus-host cell interfaces [17].
Beneficial effect exerted by lactobacilli is, at least in part, related to the ability of reducing pathogens adhesion to host cell surfaces [5, 33]. In this regard, supernatants from Lactobacillus were found to reduce pathogens adhesion, suggesting that Lactobacillus derivatives can exert an anti-adhesive activity [34].
Here, we hypothesized that EVs could mediate Lactobacillus anti-adhesive properties in the vaginal ecosystem. Lactobacillus-EVs were evaluated towards the adhesion of four vaginal opportunistic pathogens, i.e., E coli, S. aureus, E. faecalis, and S. agalactiae. Surprisingly, L. crispatus BC5-EVs and L. gasseri BC12-EVs were able to reduce the adhesion of all pathogens tested with similar effects.
As reported above, the EVs inhibitory mechanisms can be associated to some Lactobacillus adhesins found in L. crispatus BC5-EVs and L. gasseri BC12-EVs by Palomino et al. [8]. These adhesins are involved not only in Lactobacillus adhesion but also in preventing pathogens’ interactions to host receptors by competitive inhibition [31, 35]. In particular, EVs from L. crispatus BC5 and L. gasseri BC12 delivered enolase-1 that resulted to reduce Neisseria ghonorreae attachment to host cells [8, 36, 37]. Moreover, L. gasseri BC12-EVs transported two more adhesins (i.e., enolase-2 and elongation factor-TU) that inhibited E. coli adhesion to host mucosa [8, 35, 38]. Our data, supported by literature, suggested that Lactobacillus-EVs could prevent pathogens adhesion by the saturation of host adhesins receptors.
Besides these suggestions, many more aspects of Lactobacillus-EVs activity might be elucidated. In this regard, a possible effect of EVs on cervical cells can also be considered. Until now, it is known that EVs mediate the Lactobacillus crosstalk communication with the human host cells and trigger some cell signaling cascades, specially related to the host immune system [12, 13, 18, 19, 23, 24, 28]. Previous studies reported that vaginal lactobacilli were able to modify HeLa cell plasma membrane in terms of lipid composition, fluidity, and protein exposure, making the host less permissive to Candida albicans and Chlamydia trachomatis attachment and infection [39, 40]. Moreover, as well as EVs from pathogenic bacteria, EVs from beneficial Lactobacillus can deliver DNA and RNA to host cells, possibly affecting gene expression [41–43]. Even if the molecular mechanism is not fully understood, since EVs cargo is composed of molecules from different cell compartments, we can’t exclude possible EVs-induced modifications on host pathways [44]. In this respect, a modification in membrane fluidity and/or gene expression in host cells could alter the rotational and lateral motion and/or expression of receptors affecting their availability for bacterial recognition [45].
Whether the effect of Lactobacillus-EVs on bacteria adhesion is the result of one or a combination of the mechanisms proposed remains an open and, in some ways, tough question to answer. A deeper characterization of Lactobacillus-EVs structure and the purification of EVs components could allow a wider understanding of EVs mode of action. In this perspective, we are planning to further characterize EVs physical and chemical properties by super-resolution and electron microscopy, as well as the nucleic acids quality and integrity by high-resolution sequencing techniques.
As a matter of fact, our discovery provides new insights into the role of Lactobacillus-EVs in modulating the vaginal ecosystem and gives further information on their functionality within this ecological niche.
In this perspective, our results reinforce the association between Lactobacillus EVs and health benefits [8, 15, 18, 20], opening to the idea of using EVs derived from vaginal strains as potential postbiotics to support the vaginal balance in favor of the host well-being.
Methods
Bacterial cultures and growth conditions
Lactobacillus strains used in this study were previously isolated from vaginal swabs of healthy premenopausal women, according to the protocol of the Ethics Committee of the University of Bologna (52/2014/U/Tess) [10]. Here, we selected strains belonging to two species highly represented in the vaginal niche: L. crispatus (BC1, BC3, BC4, and BC5) and L. gasseri (BC9, BC10, BC12, and BC13). Lactobacillus strains were cultured anaerobically at 37 °C in de Man, Rogosa, and Sharpe (MRS) (Beckton, Dickinson, and Co., MI, Italy) broth with the supplement of 0.05% L-cysteine (Sigma-Aldrich, MI, Italy). The anaerobic conditions were reached through jars containing GasPak EZ (Beckton, Dickinson, and Co.).
Pathogenic bacteria used for the anti-adhesive study were Escherichia coli ATCC 11105, Staphylococcus aureus ATCC 29213, Streptococcus agalactiae SO104, and Enterococcus faecalis BC101. E. coli and S. aureus were cultured aerobically in Nutrient Broth (NB) (Beckton, Dickinson, and Co.) at 37 °C. S. agalactiae SO104 was isolated from vaginal swabs in the Microbiology Laboratory of Sant’ Orsola-Malpighi University Hospital of Bologna (Italy) during routine diagnostic procedures. E. faecalis BC101 belongs to the Department of Pharmacy and Biotechnology, University of Bologna (Italy) [11]. S. agalactiae and E. faecalis were cultured in Brain Heart Infusion (BHI) (Beckton, Dickinson, and Co.) broth, in 5% CO2 at 37 °C.
For each microorganism, two sequential 24 h-cultures were carried out, then 1 × 109 CFU/mL bacterial suspensions were prepared in sterile saline and used in adhesion assays.
Isolation of extracellular vesicles (EVs) from Lactobacillus
EVs were recovered from L. crispatus BC5 and L. gasseri BC12 growth cultures. EVs were isolated according to Ñahui Palomino et al. 2019, with some modifications [8, 46]. MRS medium and phosphate-buffered saline pH 7.4 (PBS) used for EVs isolation were previously autoclaved (120 °C for 30 min) and filtered with 0.22 μm polyethersulfonate PES vacuum filters (Membrane Solutions, LLC, Auburn, WA, USA) to remove large particles and possible contaminants. Lactobacillus were cultured anaerobically for 24 h in filtered MRS and then subcultured in fresh medium for additional 24 h. 200 mL of bacterial suspensions (1 × 109 CFU/mL) were centrifuged at 3600×g for 15 min at 4 °C (Sartorius Centrisart® D-16C, Sartorius, Goettingen, Germany). Supernatants were collected and subsequently filtered with 0.22 μm cellulose acetate filters to eliminate any remaining bacteria. Afterward, the filtered supernatants were centrifugated at 10000×g for 30 min to eliminate any cell debris. EVs were precipitated from obtained supernatants by ultracentrifugation at 100,000×g for 70 min at 4 °C (Beckman Optima L-90 K, Rotor: SW 28 Ti Swinging-Bucket, capacity 8 × 38.5 mL, Beckman Coulter, Inc., Brea, USA) and washed with PBS using the same centrifugation setting. EVs pellet was resuspended in PBS (final volume of 1.5 mL) and stored at − 80 °C until use. The same protocol was applied to isolate particles from filtered MRS medium.
Physical and chemical characterization of EVs
EVs samples were characterized for their physical properties in terms of yield and size by Nanoparticle Tracking Analysis (NTA) technology (NanoSight 49 NS300, Malvern Panalytical, Grovewood Road, Malvern, UK). Samples were diluted at 1:100 in PBS and videos were recorded at 30 frames per second using a 20× objective. Measurements of EVs Zeta-potential were performed with the instrument Malvern Zetasizer 3000 HS (Malvern Panalytical Ltd., Malvern, UK) set at 25 °C by diluting EVs samples 1:2 in a MilliQ water. EVs protein content was measured by Bradford assay (BioRad Laboratories, Inc., CA, USA) after EVs lysis. EVs were lysed with RIPA buffer according to Prabal Subedi et al., with some modifications [47]. RIPA buffer 5× was prepared as follows: 50 mM Tris–HCl (pH 8.0), 5 mM EDTA, 2.5 mM EGTA, 5% Triton X-100, 0.5% sodium deoxycholate, 0.5% SDS, 140 mM NaCl, aliquoted and stored at − 20 °C until use. Briefly, 10 μL of EVs suspension were incubated with 2.5 μL of RIPA 5× at 4 °C for 30 min, afterwards, samples were placed in an ice-cold sonication bath for 30 s. This step was followed by a gentle agitation on ice for 15 min. Protein concentration was considered to normalize EVs treatments in adhesion assays.
DNA and RNA were isolated from EVs samples through TRIzol™ Reagent (Invitrogen, Thermo Fisher Scientific Inc, Massachusetts, USA) according to the manufacturer’s instructions. Before the isolation procedure, approximately 1.5 mL of EVs were pelleted as previously described and concentrated in a volume of 500 μL. EVs were treated with 1.5 mL of TRIzol™ Reagent and the manufacturer’s protocol was followed. Briefly, after the addition of TRIzol™ Reagent, 300 μL of chloroform were added and samples centrifuged for 15 min at 12,000×g at 4 °C. The mixture separated into a lower red phenol–chloroform, a gel interphase and a colorless aqueous upper phase. The DNA was extracted from the interphase and the lower phase while the RNA was recovered from the aqueous phase. The DNA was precipitated in ethanol 100% (v/v), washed once in 0.1 M sodium citrate and resuspended in ethanol 75% (v/v). Afterwards, the DNA was pelleted and resuspended in 200 μL of 8 mM NaOH, 1 mM EDTA and then buffered to pH 7.0 with 0.1 M HEPES. Starting from the aqueous phase, RNA was precipitated in isopropanol 99% (v/v) and resuspended in ethanol 75% (v/v). Afterwards, the RNA was pelleted, resuspended in 20 μL of 0.1 mM EDTA and incubated for 15 min in a 55 °C water bath, to allow RNA solubilization. Samples of DNA were stored at − 20 °C while RNA was stored at − 80 °C. DNA and RNA quantification was assessed by NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The contents of protein, DNA and RNA were normalized on 1 × 109 particles.
Cell cultures
HeLa cell line was routinely grown in 25 cm2 tissue culture flasks, at 37 °C in 5% CO2 in Dulbecco’s Modified Eagle medium (DMEM, Lonza Group Ltd, Basel, Switzerland) supplemented with 10% Fetal Serum Bovine (FBS) and 1% l-glutamine. For adhesion experiments, cells were seeded at a density of 2 × 104 cells/cm2 on sterile round coverslips in 24-wells cell culture plates (Sarstedt AG & Co., Nümbrecht, Germany) and allowed to grow to 80% confluence (approx. 3 days). Before adhesion assay, exhausted medium was replaced with fresh complete medium (0.2 mL per well).
Adhesion assays
A certain volume of Lactobacillus EVs corresponding to 20 μg of proteins was used to pretreat 1 × 105 HeLa cells for 1 h, in 5% CO2 at 37 °C. Afterwards, bacterial suspensions (1 × 109 CFU/mL in sterile saline) were added to HeLa monolayer, applying a ratio of 100:1 (bacteria: HeLa cells), and plates were incubated in 5% CO2 at 37 °C for an additional hour. Hela cells pretreated with PBS were used to evaluate basal bacterial adhesion (100%). For each sample, at least two independent experiments were carried out. Bacteria adherent to HeLa cell monolayers were stained by May-Grunwald/Giemsa protocol as previously reported [10]. Adherent bacteria were counted at optical microscope Nikon Eclipse 21 (Objective 100×, Nikon, Tokyo, Japan) considering at least 30 microscopic fields per sample and adhesion was expressed as the percentage of adherent bacteria compared to the control.
Statistical analysis
Statistical analysis was performed using Student’s t-test for two means comparison and ordinary one-way ANOVA for multiple comparisons (GraphPad Prism version 8.0.1, GraphPad Prism Software Inc, San Diego, CA, USA). Results were expressed as mean ± standard deviation (SD) and differences were deemed significant for p < 0.05.
Acknowledgements
Fondazione CARISBO (Bologna, Italy) is gratefully acknowledged.
Author contributions
VC, CP and BV designed the study; CP and SF supervised the experiments; VC and BG performed the experiments; CF provided microbial strains; VC, CP, BG and BV analyzed the data; BV supervised the study; VC wrote the first draft of the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded under the Grant (#19038) by Fondazione CARISBO (Bologna, Italy), project “Strategia bioterapeutica per contrastare le infezioni ginecologiche basata su vescicole extracellulari di lattobacilli probiotici” coordinated by B. Vitali.
Availability of data and materials
All data generated or analyzed during this study are included in this published article [and its additional files].
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amabebe E, Anumba DOC. The vaginal microenvironment: the physiologic role of lactobacilli. Front Med. 2018;5:181. doi: 10.3389/fmed.2018.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boris S, Suárez JE, Vázquez F, Barbés C. Adherence of human vaginal lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect Immun. 1998;66(5):1985–1989. doi: 10.1128/iai.66.5.1985-1989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yadav AK, Tyagi A, Kumar A, Panwar S, Grover S, Saklani AC, et al. Adhesion of lactobacilli and their anti-infectivity potential. Crit Rev Food Sci Nutr. 2017;57(10):2042–2056. doi: 10.1080/10408398.2014.918533. [DOI] [PubMed] [Google Scholar]
- 5.De Gregorio PR, Parolin C, Abruzzo A, Luppi B, Protti M, Mercolini L, et al. Biosurfactant from vaginal Lactobacillus crispatus BC1 as a promising agent to interfere with Candida adhesion. Microb Cell Factories. 2020;19(1):133. doi: 10.1186/s12934-020-01390-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foschi C, Salvo M, Cevenini R, Parolin C, Vitali B, Marangoni A. Vaginal lactobacilli reduce Neisseria gonorrhoeae viability through multiple strategies: an in vitro study. Front Cell Infect Microbiol. 2017;7:502. doi: 10.3389/fcimb.2017.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marziali G, Foschi C, Parolin C, Vitali B, Marangoni A. In-vitro effect of vaginal lactobacilli against group B Streptococcus. Microb Pathog. 2019;136:103692. doi: 10.1016/j.micpath.2019.103692. [DOI] [PubMed] [Google Scholar]
- 8.Ñahui Palomino RA, Vanpouille C, Laghi L, Parolin C, Melikov K, Backlund P, et al. Extracellular vesicles from symbiotic vaginal lactobacilli inhibit HIV-1 infection of human tissues. Nat Commun. 2019;11:10. doi: 10.1038/s41467-019-13468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nardini P, Ñahui Palomino RA, Parolin C, Laghi L, Foschi C, Cevenini R, et al. Lactobacillus crispatus inhibits the infectivity of Chlamydia trachomatis elementary bodies, in vitro study. Sci Rep. 2016;29:6. doi: 10.1038/srep29024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parolin C, Marangoni A, Laghi L, Foschi C, Ñahui Palomino RA, Calonghi N, et al. Isolation of vaginal lactobacilli and characterization of anti-candida activity. PLoS ONE. 2015;10(6):e0131220. doi: 10.1371/journal.pone.0131220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siroli L, Patrignani F, Serrazanetti DI, Parolin C, Ñahui Palomino RA, Vitali B, et al. Determination of antibacterial and technological properties of vaginal lactobacilli for their potential application in dairy products. Front Microbiol. 2017;8:166. doi: 10.3389/fmicb.2017.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Nedawi K, Mian MF, Hossain N, Karimi K, Mao YK, Forsythe P, et al. Gut commensal microvesicles reproduce parent bacterial signals to host immune and enteric nervous systems. FASEB J. 2015;29(2):684–695. doi: 10.1096/fj.14-259721. [DOI] [PubMed] [Google Scholar]
- 13.Briaud P, Carroll RK. Extracellular vesicle biogenesis and functions in gram-positive bacteria. Infect Immun. 2020 doi: 10.1128/IAI.00433-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orench-Rivera N, Kuehn MJ. Environmentally controlled bacterial vesicle-mediated export. Cell Microbiol. 2016;18(11):1525–1536. doi: 10.1111/cmi.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean SN, Rimmer MA, Turner KB, Phillips DA, Caruana JC, Hervey WJ, et al. Lactobacillus acidophilus membrane vesicles as a vehicle of bacteriocin delivery. Front Microbiol. 2020;11:710. doi: 10.3389/fmicb.2020.00710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean SN, Leary DH, Sullivan CJ, Oh E, Walper SA. Isolation and characterization of Lactobacillus-derived membrane vesicles. Sci Rep. 2019;9:877. doi: 10.1038/s41598-018-37120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domínguez Rubio AP, Martínez JH, Martínez Casillas DC, Coluccio Leskow F, Piuri M, Pérez OE. Lactobacillus casei BL23 produces microvesicles carrying proteins that have been associated with its probiotic effect. Front Microbiol. 2017;8:1783. doi: 10.3389/fmicb.2017.01783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim MH, Choi SJ, Choi HI, Choi JP, Park HK, Kim EK, et al. Lactobacillus plantarum-derived extracellular vesicles protect atopic dermatitis induced by Staphylococcus aureus-derived extracellular vesicles. Allergy Asthma Immunol Res. 2018;10(5):516–532. doi: 10.4168/aair.2018.10.5.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li M, Lee K, Hsu M, Nau G, Mylonakis E, Ramratnam B. Lactobacillus-derived extracellular vesicles enhance host immune responses against vancomycin-resistant enterococci. BMC Microbiol. 2017;17(1):66. doi: 10.1186/s12866-017-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wegh CAM, Geerlings SY, Knol J, Roeselers G, Belzer C. Postbiotics and their potential applications in early life nutrition and beyond. Int J Mol Sci. 2019;20(19):4673. doi: 10.3390/ijms20194673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta S, Kakkar V, Bhushan I. Crosstalk between vaginal microbiome and female health: a review. Microb Pathog. 2019;136:103696. doi: 10.1016/j.micpath.2019.103696. [DOI] [PubMed] [Google Scholar]
- 22.Domínguez Rubio AP, D’Antoni CL, Piuri M, Pérez OE. Probiotics, their extracellular vesicles and infectious diseases. Front Microbiol. 2022;13:864720. doi: 10.3389/fmicb.2022.864720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ñahui Palomino RA, Vanpouille C, Costantini PE, Margolis L. Microbiota–host communications: bacterial extracellular vesicles as a common language. PLoS Pathog. 2021;17(5):e1009508. doi: 10.1371/journal.ppat.1009508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behzadi E, Mahmoodzadeh Hosseini H, Imani Fooladi AA. The inhibitory impacts of Lactobacillus rhamnosus GG-derived extracellular vesicles on the growth of hepatic cancer cells. Microb Pathog. 2017;110:1–6. doi: 10.1016/j.micpath.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 25.Grande R, Celia C, Mincione G, Stringaro A, Di Marzio L, Colone M, et al. Detection and physicochemical characterization of membrane vesicles (MVs) of Lactobacillus reuteri DSM 17938. Front Microbiol. 2017;13:8. doi: 10.3389/fmicb.2017.01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivera J, Cordero RJB, Nakouzi AS, Frases S, Nicola A, Casadevall A. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci USA. 2010;107(44):19002–19007. doi: 10.1073/pnas.1008843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maas SLN, de Vrij J, van der Vlist EJ, Geragousian B, van Bloois L, Mastrobattista E, et al. Possibilities and limitations of current technologies for quantification of biological extracellular vesicles and synthetic mimics. J Controlled Release. 2015;200:87–96. doi: 10.1016/j.jconrel.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu R, Lin H, Wang M, Zhao Y, Liu H, Min Y, et al. Lactobacillus reuteri-derived extracellular vesicles maintain intestinal immune homeostasis against lipopolysaccharide-induced inflammatory responses in broilers. J Anim Sci Biotechnol. 2021;12:25. doi: 10.1186/s40104-020-00532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Argentini C, Fontana F, Alessandri G, Lugli GA, Mancabelli L, Ossiprandi MC, et al. Evaluation of modulatory activities of Lactobacillus crispatus strains in the context of the vaginal microbiota. Microbiol Spectr. 2022;10(2):e02733–e2821. doi: 10.1128/spectrum.02733-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castaldo C, Vastano V, Siciliano RA, Candela M, Vici M, Muscariello L, et al. Surface displaced alfa-enolase of Lactobacillus plantarum is a fibronectin binding protein. Microb Cell Factories. 2009;8:14. doi: 10.1186/1475-2859-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waśko A, Polak-Berecka M, Paduch R, Jóźwiak K. The effect of moonlighting proteins on the adhesion and aggregation ability of Lactobacillus helveticus. Anaerobe. 2014;30:161–168. doi: 10.1016/j.anaerobe.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Celebioglu HU, Svensson B. Dietary nutrients, proteomes, and adhesion of probiotic Lactobacilli to mucin and host epithelial cells. Microorganisms. 2018;6(3):E90. doi: 10.3390/microorganisms6030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fracchia L, Cavallo M, Allegrone G, Martinotti MG. A Lactobacillus-derived biosurfactant inhibits biofilm formation of human pathogenic Candida albicans biofilm producers. Appl Microbiol Biotechnol. 2010;12.
- 34.Matsuda Y, Cho O, Sugita T, Ogishima D, Takeda S. Culture supernatants of Lactobacillus gasseri and L. crispatus inhibit Candida albicans biofilm formation and adhesion to HeLa cells. Mycopathologia. 2018;183(4):691–700. doi: 10.1007/s11046-018-0259-4. [DOI] [PubMed] [Google Scholar]
- 35.Dhanani AS, Bagchi T. The expression of adhesin EF-Tu in response to mucin and its role in Lactobacillus adhesion and competitive inhibition of enteropathogens to mucin. J Appl Microbiol. 2013;115(2):546–54. doi: 10.1111/jam.12249. [DOI] [PubMed] [Google Scholar]
- 36.Spurbeck RR, Harris PT, Raghunathan K, Arvidson DN, Arvidson CG. A moonlighting enolase from Lactobacillus gasseri does not require enzymatic activity to inhibit Neisseria gonorrhoeae adherence to epithelial cells. Probiotics Antimicrob Proteins. 2015;7(3):193–202. doi: 10.1007/s12602-015-9192-8. [DOI] [PubMed] [Google Scholar]
- 37.Płaczkiewicz J, Chmiel P, Malinowska E, Bącal P, Kwiatek A. Lactobacillus crispatus and its enolase and glutamine synthetase influence interactions between Neisseria gonorrhoeae and human epithelial cells. J Microbiol Seoul Korea. 2020;58(5):405–414. doi: 10.1007/s12275-020-9505-9. [DOI] [PubMed] [Google Scholar]
- 38.Dhanani AS, Gaudana SB, Bagchi T. The ability of Lactobacillus adhesin EF-Tu to interfere with pathogen adhesion. Eur Food Res Technol. 2011;232(5):777–785. [Google Scholar]
- 39.Calonghi N, Parolin C, Sartor G, Verardi L, Giordani B, Frisco G, et al. Interaction of vaginal Lactobacillus strains with HeLa cells plasma membrane. Benef Microbes. 2017;8(4):625–633. doi: 10.3920/BM2016.0212. [DOI] [PubMed] [Google Scholar]
- 40.Parolin C, Frisco G, Foschi C, Giordani B, Salvo M, Vitali B, et al. Lactobacillus crispatus BC5 interferes with chlamydia trachomatis infectivity through integrin modulation in cervical cells. Front Microbiol. 2018;9:2630. doi: 10.3389/fmicb.2018.02630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koeppen K, Hampton TH, Jarek M, Scharfe M, Gerber SA, Mielcarz DW, et al. A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathog. 2016;12(6):e1005672. doi: 10.1371/journal.ppat.1005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.da Rocha IFM, Amatuzzi RF, Lucena ACR, Faoro H, Alves LR. Cross-kingdom extracellular vesicles EV-RNA communication as a mechanism for host-pathogen interaction. Front Cell Infect Microbiol. 2020;10:593160. doi: 10.3389/fcimb.2020.593160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee HJ. Microbe-host communication by small RNAs in extracellular vesicles: vehicles for transkingdom RNA transportation. Int J Mol Sci. 2019;20(6):1487. doi: 10.3390/ijms20061487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martín C, Fernández-Vega I, Suárez JE, Quirós LM. Adherence of Lactobacillus salivarius to HeLa Cells promotes changes in the expression of the genes involved in biosynthesis of their ligands. Front Immunol. 2020;9(10):3019. doi: 10.3389/fimmu.2019.03019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarashi S, Zamani MS, Omrani MD, Fateh A, Moshiri A, Saedisomeolia A, et al. Commensal and pathogenic bacterial-derived extracellular vesicles in host-bacterial and interbacterial dialogues: two sides of the same coin. J Immunol Res. 2022 doi: 10.1155/2022/8092170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;30(1):3.22.1–3.22.29. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 47.Subedi P, Schneider M, Philipp J, Azimzadeh O, Metzger F, Moertl S, et al. Comparison of methods to isolate proteins from extracellular vesicles for mass spectrometry-based proteomic analyses. Anal Biochem. 2019;1(584):113390. doi: 10.1016/j.ab.2019.113390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article [and its additional files].