ABSTRACT
Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis are the major pathogens of the spore-forming genus Bacillus and possess an outer spore layer, the exosporium, not found in many of the nonpathogenic species. The exosporium consists of a basal layer with the ExsY, CotY, and BxpB proteins being the major structural components and an exterior nap layer containing the BclA glycoprotein. During the assembly process, the nascent exosporium basal layer is attached to the spore coat by a protein linker that includes the CotO and CotE proteins. Using transmission electron microscopy, Western blotting, immunofluorescence, and fluorescent fusion protein approaches, we examined the impact of single, double, and triple mutants of the major exosporium proteins on exosporium protein content and distribution. Plasmid-based expression of exsY and cotE resulted in increased production of exosporium lacking spores, and the former also resulted in outer spore coat disruptions. The exosporium bottlecap produced by exsY null spores was found to be more stable than previously reported, and its spore association was partially dependent on CotE. Deletion mutants of five putative spore genes (bas1131, bas1142, bas1143, bas2277, and bas3594) were created and shown not to have obvious effects on spore morphology or BclA and BxpB content. The BclC collagen-like glycoprotein was found to be present in the spore and possibly localized to the interspace region.
IMPORTANCE B. anthracis is an important zoonotic animal pathogen causing sporadic outbreaks of anthrax worldwide. Spores are the infectious form of the bacterium and can persist in soil for prolonged periods of time. The outermost B. anthracis spore layer is the exosporium, a protein shell that is the site of interactions with both the soil and with the innate immune system of infected hosts. Although much is known regarding the sporulation process among members of the genus Bacillus, significant gaps in our understanding of the exosporium assembly process exist. This study provides evidence for the properties of key exosporium basal layer structural proteins. The results of this work will guide future studies on exosporium protein-protein interactions during the assembly process.
KEYWORDS: Bacillus anthracis, CotE, assembly, electron microscopy, endospores, exosporium, mutants
INTRODUCTION
The genus Bacillus is comprised of Gram-positive endospore-forming species that can shift to an alternative developmental pathway, sporulation, when growth conditions become unfavorable (1). Spores of the Bacillus cereus family (comprising 13 species, including B. anthracis, B. cereus, B. thuringiensis, B. mycoides, and B. cytotoxicus) belong to a subset of Bacillus species that has evolved to become occasional pathogens of mammals, fish, and insects (2). Spores from these species diverge from the spore architecture of the well-characterized B. subtilis in that they contain an additional outer structure known as the exosporium (3). The exosporium is comprised of two distinct sublayers, the basal layer and the outer hair-like nap layer (4–6). The balloon-like exosporium is displaced from the spore coat by the interspace region of the spore (6). Many B. anthracis and B. cereus exosporium structural proteins have been identified (7–10). The major basal layer-associated structural proteins include ExsY, CotY, and ExsFA/BxpB (here designated BxpB) (11–19). Mutants of B. anthracis lacking the ExsY protein produce an incomplete exosporium corresponding to the mother cell-central pole of the spore (the “bottlecap” portion of the exosporium) (11, 18). ExsY thus is a major structural component of the noncap region of the exosporium basal layer (17). The cap structure in exsY mutant spores is not stably anchored to the mature spore, with loss of the cap reported to be more common in spores produced in liquid cultures, perhaps owing to shear forces in the shaken cultures (11). Mutants of B. cereus lacking ExsY produced exosporium-less spores with some small fragments of material, possibly exosporium in nature, attached to the spore surface (16). The lack of the cap structure was attributed to the spores being produced from broth cultures. CotY, a protein that is 85% identical in amino acid sequence to ExsY, is a structural component of the cap (18, 19; J. Durand-Heredia and G. C. Stewart, submitted for publication). The precursor to the bottlecap structure forms early in the sporulation process. Before the early engulfment stage is complete, a small cap-like structure appears at the mother cell side of the septum, but this structure lacks the electron density and hair-like projections of the mature cap, and the gap between it and the forespore (the interspace) is narrower than the interspace seen in the mature spore (18). Mutants unable to produce CotY do not exhibit the cap structure early in sporulation. Because the cap is thought to serve as the primer site for noncap ExsY assembly, it is expected that loss of CotY would produce spores lacking an exosporium. However, cotY mutants of B. anthracis and B. cereus were reported to produce a normal-appearing exosporium (16, 18). However, exosporium assembly was found to be delayed in B. anthracis cotY mutants and instead of synthesis initiating at the mother cell-central pole of the developing spore (the cap site), exosporium synthesis initiates principally at sites along the long axis of the spores (18).
External to the CotY and ExsY internal basal layer is an outer layer that includes BxpB and an outer “hair-like” nap layer are composed primarily of the collagen-like glycoprotein BclA (20–25; J. Durand-Heredia, H.-Y. Hsieh, K. A. Spreng, and G. C. Stewart, submitted for publication). Assembly of the BclA filaments requires both BxpB and ExsFB. Mutant spores lacking BxpB produce an exosporium with a marked reduction of BclA, mutants lacking ExsFA exhibit a modest reduction in BclA levels, and mutants lacking both BxpB and ExsFB produce an exosporium that lacks the BclA nap layer (13, 15, 24).
The components of the exosporium are produced in the mother cell and are subsequently assembled on the developing spore. Most of the exosporium genes have promoter elements specific for the σK-containing late-stage sporulation mother cell RNA polymerase (25, 26). The assembly of the intact exosporium is dependent on proteins that permit the proper attachment of the exosporium to the spore coat layers. The CotE protein is essential for attachment of the exosporium. A lack of CotE results in production of spores devoid of an exosporium and exosporium sheets that are released by lysis of the mother cells (18, 19, 27). CotE was found to anchor the developing exosporium through an interaction with CotO, and mutants lacking CotO produce spores devoid of the exosporium layer (18). Although the exosporium assembly pathway is not well defined, the basal layer proteins ExsY and CotY were shown to be able to self-assemble into two dimensional arrays when expressed in heterologous Escherichia coli host cells, and these structures are stable to heat and chemical denaturants (17).
In this study, we utilized a genetic approach including mutation analysis, complementation, and transmission electron microscopy (TEM) to examine the effects of loss of these exosporium genes on assembly of the exosporium.
RESULTS
Contributions of CotY and ExsY to exosporium assembly.
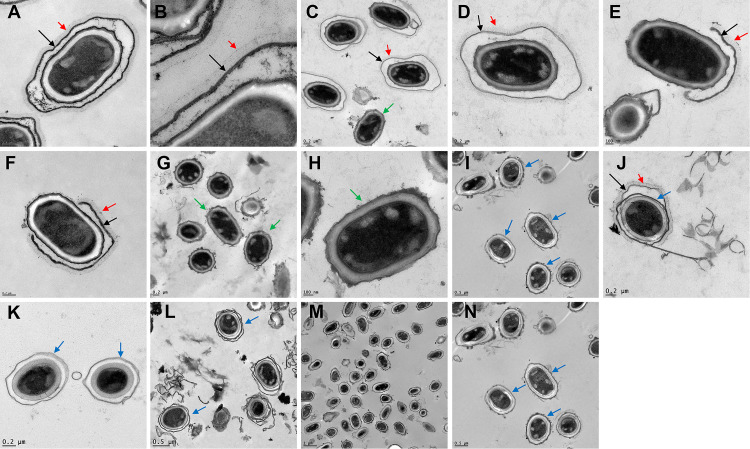
Studies with B. anthracis found that exsY mutants produced only the bottlecap portion of the exosporium, indicating that ExsY was essential for production of the noncap portion of the exosporium basal layer (11). In a study with B. cereus, exsY mutants lacked an intact exosporium but had some small fragments of material, possibly exosporium in nature, attached to the spore surface (16). Deletion of cotY resulted in spores possessing a morphologically normal-appearing exosporium with both B. cereus and B. anthracis (16, 18). Mutants of B. cereus with deletions of both exsY and cotY lacked an exosporium, but a similar mutant of B. anthracis has not been described. To further evaluate the contributions of CotY and ExsY to exosporium structure in B. anthracis, deletion mutants of cotY, exsY, and cotY and exsY in the Sterne strain were created and spore structure was evaluated by transmission electron microscopy. The results are shown in Fig. 1, and quantification of the spore phenotypes is presented in Table 1. We confirmed that CotY-negative spores possessed an intact exosporium that included the BclA nap layer (Fig. 1C and D). With the exsY deletion spores, >90% of the spores exhibited the expected cap-only exosporium (Fig. 1E), with about 5% lacking an exosporium and ~2% showing an extended cap structure (Fig. 1F). Mutant spores lacking both ExsY and CotY were devoid of an exosporium (Fig. 1G and H), similar to the findings with the B. cereus double mutant (16) but now also demonstrated with B. anthracis.
FIG 1.
Transmission electron micrographs showing spores of B. anthracis. Images are of the Sterne (A and B), ΔcotY (C and D), ΔexsY (E and F), ΔcotY ΔexsY (G and H), ΔcotE (I to K), ΔcotE ΔexsY (L), and ΔcotE ΔcotY ΔexsY (M and N) strains. Black arrows indicate positions of an exosporium, red arrows indicate the BclA nap layer, green arrows indicate spores lacking an exosporium, and blue arrows indicate detached outer spore coat layers.
TABLE 1.
Spore architecture by cotE, cotY, and exsY exosporium genotype as determined by TEM
| Genotype | % of spores in indicated category |
Total no. of spores counted | ||||
|---|---|---|---|---|---|---|
| Intact exosporium | No exosporium | Cap only exosporium | Extended pole | Multilayers external to spore coat | ||
| Sterne (wild type) | 100 | 0 | 0 | 0 | 0 | 101 |
| ΔcotY | 99.4 | 0.6 | 0 | 0 | 0 | 180 |
| ΔexsY | 0 | 4.6 | 93.5 | 1.9 | 0 | 216 |
| ΔcotY ΔexsY | 0 | 100 | 0 | 0 | 0 | 100 |
| ΔcotE | 0 | 100a | 0 | 0 | 0 | 96 |
| ΔcotE ΔexsY | 0 | 100 | 0b | 0 | 0 | 130 |
| Sterne pHPS2-exsY | 19.2 | 3 | 5.1 | 2.5 | 62.1 | 198 |
| Sterne pHPS2-cotE | 36.1 | 57.7 | 0 | 6.2 | 0 | 97 |
| ΔcotE pHPS2-cotE | 24.1 | 73 | 0 | 3.0 | 0 | 237 |
| Sterne pHPS2-exsY-mcherry | 24 | 22 | 39.3 | 14.7 | 0 | 150 |
| Sterne ΔamyS::exsY | 0 | 2 | 0 | 49.5 | 48.5 | 99 |
| Sterne ΔamyS::exsY-mcherry | 100 | 0 | 0 | 0 | 0 | 46 |
| ΔexsY pHPS2-exsY | 24.5 | 1.8 | 9.1 | 10 | 54.5 | 110 |
| ΔexsY pHPS2-exsY-mcherry | 0 | 35.6 | 62.5 | 1.9 | 0 | 104 |
| ΔexsY ΔamyS::exsY | 9 | 4.2 | 7.2 | 79.6 | 0 | 167 |
| ΔexsY ΔamyS::exsY-mcherry | 0 | 93 | 3 | 4 | 0 | 100 |
| ΔcotY ΔexsY pHPS2-exsY | 47 | 0 | 15 | 34 | 4 | 100 |
| ΔcotY ΔexsY pHPS2-cotY | 0 | 97.3 | 1.8 | 0.9 | 0 | 110 |
Two spores with an adherent exosporium strip.
Free spore exosporium caps present in the spore samples (by TEM and anti-BclA fluorescence).
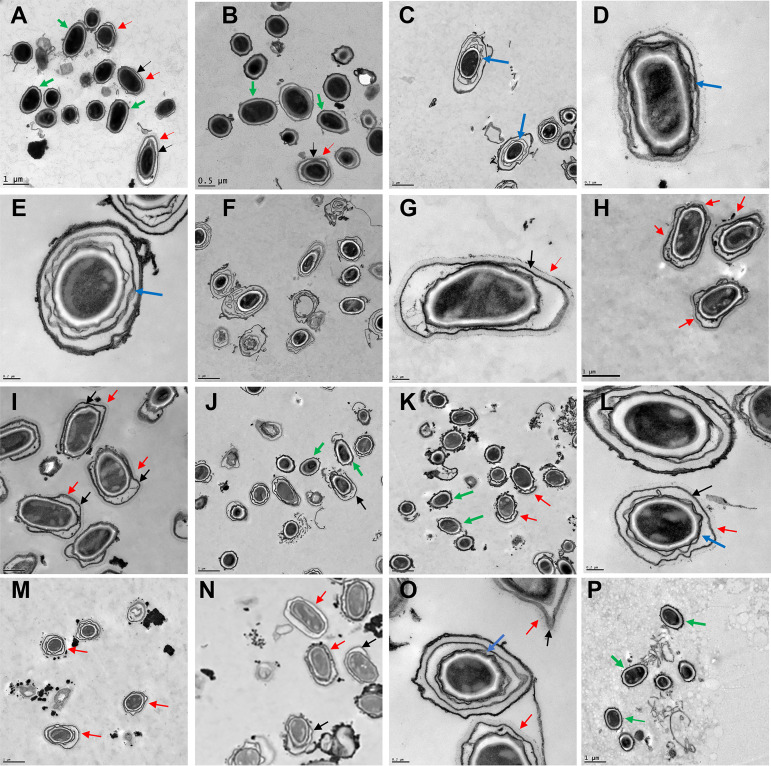
Complementation plasmids, or single-copy wild-type genes inserted at the amyS locus (Durand-Heredia and Stewart, submitted), were introduced into the Sterne and null mutant strains. The shuttle plasmid pHPS2 is a derivative of pHP13 with the chloramphenicol resistance marker replaced with a spectinomycin resistance marker (24, 28). This is a relatively low-copy-number replicon, with a copy number of ~5 in Bacillus subtilis hosts. With the Sterne strain bearing the pHPS2-exsY plasmid (pJD6265), major disruptions of the outer spore coat layer were evident and only 19% of the spores possessed an exosporium (Fig. 2C to E and Table 1). The outer spore coat was displaced, producing distinct spore coat layers separated by gaps. The increased exsY copy number negatively impacted the assembly of the outer spore coat and disrupted the anchoring of the exosporium. The effects on the spore coat morphology appeared to be dose dependent. Creation of a merodiploid strain with the insertion of exsY at the amyS locus resulted in 48.5% of the spores exhibiting displaced spore coat layers, versus 62% of the Sterne strain bearing the plasmid copy of exsY (Fig. 2J and L and Table 1). Interestingly, absence of the ExsY protein did not result in spores with visual disruptions of the outer spore coat layer (Fig. 1E and F and Table 1). Complementation of the exsY deletion differed between the plasmid and single-copy integration systems, likely reflecting dose effects on expression. The plasmid complementation construct was more effective in restoring an intact exosporium than the single-copy chromosomal insertion, with 24.5% of the spores possessing an intact exosporium plus 10% exhibiting an extended-pole exosporium versus 9% and 79.6%, respectively, with the single ectopic site copy of exsY (Fig. 2F to I and Table 1). There appeared to be positional effects on exsY expression resulting in incomplete complementation. When a plasmid-borne exsY-mcherry fusion construct was introduced into the exsY mutant, no complementation (resulting in an intact exosporium) was observed by TEM and there was a reduction in the spores exhibiting the cap-only exosporium with a corresponding increase in exosporium-less spores (Fig. 2J and K and Table 1). Production of the ExsY-mCherry fusion protein also did not lead to the spore coat disruption evident with the unfused ExsY protein in the Sterne and ΔexsY backgrounds (Fig. 2J and M). Although the ExsY-mCherry fusion protein is able to localize to the exosporium (Durand-Heredia and Stewart, submitted), it is not fully functional and cannot complement the null mutant phenotype. Production of the plasmid-borne ExsY-mCherry in Sterne cells did lead to loss of the exosporium (only 24% of the spores possessed the exosporium), reflecting a combination of the fusion protein acting as a chain terminator in ExsY assembly and being less efficient in serving as a substrate for ExsY assembly (Durand-Heredia and Stewart, submitted). Insertion of the exsY-mcherry gene into the B. anthracis Sterne chromosome did not result in visible loss of the exosporium (Fig. 2M and Table 1), probably due to lower expression levels.
FIG 2.
Transmission electron micrographs showing spores of B. anthracis bearing plasmid or chromosomal complementation genes. Images are of the Sterne pHPS2-cotE (A), ΔcotE pHPS2-cotE (B), Sterne pHPS2-exsY (C to E), ΔexsY pHPS2-exsY (F and G), ΔexsY ΔamyS::exsY (H and I), Sterne pHPS2-exsY-mcherry (J), ΔexsY pHPS-exsY-mcherry (K), Sterne ΔamyS::exsY (L), Sterne ΔamyS::exsY-mcherry (M), ΔexsY ΔcotY pHPS2-exsY (N and O), and ΔexsY ΔcotY pHPS2-cotY (P) strains. Black arrows indicate positions of an exosporium, red arrows indicate the BclA nap layer, green arrows indicate spores lacking an exosporium, and blue arrows indicate detached outer spore coat layers.
Complementation of the cotY exsY double mutant with the exsY complementing plasmid (pJD6265) resulted in 47% of the spores regaining the exosporium layer (ExsY+ and CotY− phenotype) and an additional 34% with an extended-pole exosporium (Fig. 2N and O and Table 1). However, the cotY complementation plasmid (pJD4952) poorly complemented, with approximately 97% of the spores exhibiting the double mutant exosporium-deficient phenotype and only 1.8% of the spores giving the CotY+ ExsY− phenotype of cap-only exosporium (Fig. 2P and Table 1).
CotE anchoring of the exosporium and exosporium cap.
Mutants with a deletion of the cotE gene produce strips of exosporium but the strips are not stably attached to the spore, so mature spores lack this layer. Occasionally, a fragment of the exosporium can be seen adhering to the spore surface (an example of the rare partially associated exosporium is shown in Fig. 1J). Exosporium strips, complete with the BclA nap, were observed in the spore preparations. Interpretation of the morphology of the spores lacking CotE is complicated by the fact that this protein is a spore coat morphogenic protein and loss of it results in disruption of the integrity of the outer spore coat layer of Bacillus subtilis (29). The cotE deletion strain spores exhibit an exosporium-like outer layer that is separated from the inner spore coat layer (Fig. 1J to L). The displaced outer spore coat results in a spore with what appears to be an interspace layer and an exosporium. This outermost layer lacks the BclA nap, but it cannot be conclusively determined by TEM if it is a “bald” exosporium basal layer or a detached and spatially separated outer coat layer. To distinguish between these two possibilities, we examined spores of a triple deletion mutant lacking the CotE, CotY, and ExsY proteins (Fig. 1M and N). The same outermost layer was present, and because the spores lacking CotY and ExsY fail to produce an exosporium, the outer layer must be the detached outer coat layer.
When a plasmid bearing the cotO gene was introduced into B. anthracis, the result was a loss of the exosporium layer (18). CotO’s putative binding partner in the exosporium anchoring chain is CotE. To determine if cotE on a multicopy also impacts attachment of the exosporium, we introduced pJD6177 into the Sterne and Sterne ΔcotE strains and examined their spores by TEM. The presence of pJD6177 in the cotE deletion mutant resulted in 24% of the spores possessing an exosporium, indicative of partial complementation of the cotE mutation (Fig. 2B and Table 1). However, when the plasmid was introduced into the wild-type Sterne strain, 58% of the spores were also found to be lacking the exosporium (Fig. 2A and Table 1). The inability to achieve 100% complementation with the plasmid is likely due to increased levels of the CotE protein and its impact on the exosporium attachment process.
The CotO-CotE-containing linker was shown to be important in the proper positioning of the exosporium cap at the early pre-engulfment stage of sporulation, suggesting that CotE is part of the linkage between the forespore and the cap (18). We created a cotE exsY double deletion mutant and examined the spores by TEM to assess the exosporium cap-only characteristic of the exsY null phenotype. All of the cotE exsY double mutant spores were devoid of the bottlecap exosporium structure, compared to only 4.6% of the spores lacking the cap exosporium with the exsY null mutant (Fig. 1L and Table 1). With spores prepared for immunofluorescence microscopy, which entails less rigorous washing steps, cap-possessing spores were seen with the cotE exsY double deletion mutant, but a smaller proportion of the overall spore population than with the spores from the exsY single mutant (Fig. 3). It appears that in the cotE mutant strain, production of the exosporium strips, extended beyond the cap by ExsY, cannot stably remain associated with the developing spore. However, the cap structure alone (in the cotE exsY double mutant), can remain spore associated in a subpopulation of the mature spores.
FIG 3.
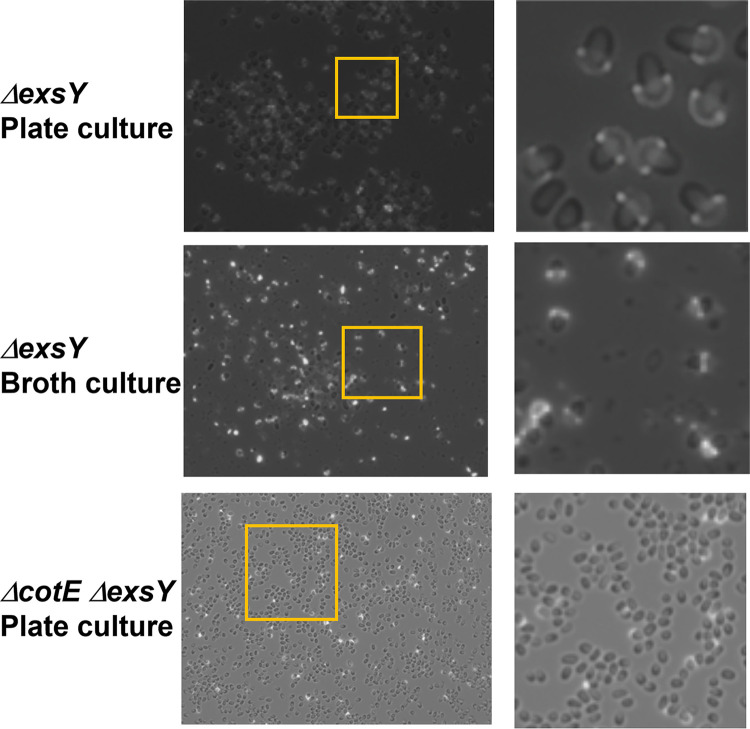
Pattern of BclA on the surface of exsY mutant spores. Spores were purified from nutrient agar plates or nutrient broth cultures. Spores were treated with rabbit anti-rBclA serum and goat anti-rabbit-Alexa Fluor 568 secondary antibodies and examined by epifluorescence microscopy. The panels on the right-hand side are enlargements of the boxed sections of the left-hand side images.
Stability of the exosporium bottlecap.
It was reported that the bottlecap exosporium of the B. anthracis exsY mutant spore is unstable, such that spores prepared in liquid media are devoid of the cap structure, likely due to shear forces present in the shake cultures (11). To evaluate the stability of the exosporium cap structure, spores were prepared from nutrient agar plate medium and from nutrient broth shake cultures. The spores were stained with anti-BclA antibodies and examined by epifluorescence microscopy. The presence of the exosporium cap structures was quantified (an example of the spore samples is presented in Fig. 3). Although the percentage of spores with exosporium caps varied somewhat from batch to batch, no significant difference in the percentage of spores with exosporium caps was obtained when liquid culture prepared spores were compared to spores harvested from plates. Given that our spore purification methods include repeated pelleting steps followed by vigorous vortex mixing to resuspend the pelleted spores, the presence of caps suggests they are more stably attached to spores than previously suggested. As determined by anti-BclA immunofluorescence microscopy, ~16% of spores from the exsY cotE double mutant possessed the cap structure (Fig. 3). This suggests that the absence of caps observed by TEM resulted from loss of the cap during the processing steps for TEM, but the gentler processing steps for immunofluorescence microscopy allowed for retention of the caps on some spores. The results indicate that CotE is involved in anchoring of the cap structure in the ΔexsY spores, in addition to the proper positioning of the cap structure earlier in the sporulation process (Durand-Heredia and Stewart, submitted).
Proteomic analysis of the exosporium from a bxpB exsFB null mutant.
To investigate if other proteins could be identified in the exosporium layer, we took advantage of the report indicating that the exosporium of the bxpB exsFB double mutant is more fragile than an intact exosporium (15). Spores were prepared from the Sterne wild type and the bxpB exsFB double deletion mutant strains. Purified spores were then subjected to extensive vortexing, exosporium fragments were isolated, and proteins were boiled in SDS plus fresh β-mercaptoethanol and electrophoresed on a 4 to 20% SDS polyacrylamide gel. No proteins were detected in the Sterne samples (data not shown), but five proteins were present in exosporium samples from the bxpB exsFB mutant spores (Fig. 4). N-terminal sequence analysis of the proteins unambiguously identified only the smallest of the proteins. The N-terminal sequence (AKHELPNLP) corresponds to amino acids 2 to 10 of the SodA1 superoxide dismutase (bas4177; molecular weight [MW] = 22,664), a known spore-associated protein (12, 30). To identify the other protein bands, a mass spectrometry-based proteomic approach was utilized. The 25-kDa species was confirmed to be SodA1. The largest species was identified as the EA1 S-layer protein (bas0842; MW = 91,362). The other bands were found to contain peptides corresponding to the ExsY and CotY proteins. Both of these exosporium basal layer proteins have been shown to be capable of self-assembly into sheet-like structures which are resistant to dissociation by heat and reducing agents (17). These protein bands, shown in Fig. 4, likely represent oligomeric forms of the proteins that are stable to the extraction conditions used. Because the molecular masses of the two proteins are similar (16.8 kDa for CotY versus 16.2 kDa for ExsY), it is not possible to conclude that these bands represent homopolymers of each protein which comigrate or if the band is a polymer containing both exosporium basal layer proteins. Nonetheless, the results do indicate that the ExsY and CotY proteins represent the major components of the residual exosporium basal layer in spores of the bxpB exsFB double deletion mutant.
FIG 4.
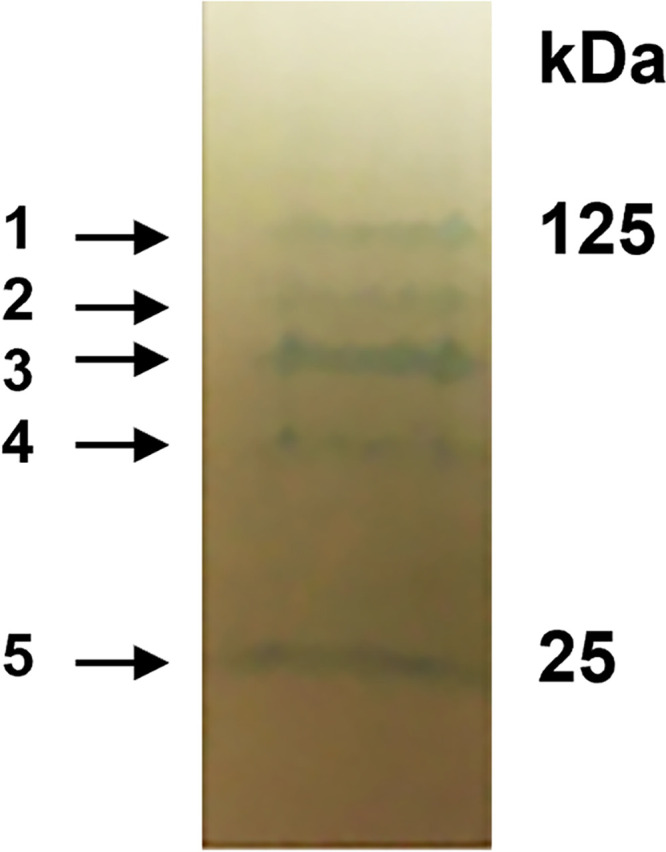
Proteins extracted from spores of the bxpB exsFB double mutant. Spores were extracted by boiling in buffer containing SDS, 8 M urea, and β-mercaptoethanol and electrophoresed in SDS-PAGE gradient gels. Proteins were electrophoretically transferred to a PVDF membrane and stained with Coomassie brilliant blue. The five proteins indicated by the arrows were subjected to identification by liquid chromatography-mass spectroscopy.
Investigation of roles of exosporium proteins of unknown function in exosporium assembly.
In our proteomic analysis of wild-type exosporium composition (data not shown), certain proteins were identified that have not been well characterized. The genes are all transcribed at time points consistent with sporulation genes (31) (Fig. 5A). To assess whether these genes play a role in exosporium assembly, deletion mutants of these genes were created and impacts on exosporium production and BclA and BxpB assembly were assessed. The results are presented in Fig. 5 and 6 and Table 2. bas1131 is one of four putative glycosyl transferase genes flanking the bclA gene (bas1130) in the B. anthracis genome. It was the only one, however, which reproducibly was identified in the exosporium preparations. The bas3957 gene encodes a small protein of unknown function present in the exosporium preparations. The bas1142 and bas1143 genes contain small open reading frames (ORFs), and although we did not detect them in our proteomic studies, the genes reside in the B. anthracis genome between the exsY and the bxpB exosporium-associated genes (Fig. 6A). Deletion mutants representing each of these genes produced spores with a normal-appearing exosporium (visualized by TEM) and possessed normal contents of BclA and BxpB (Fig. 5 and 6).
FIG 5.
Phenotypes of bas3957, bas1131, and bclC null mutants of B. anthracis. (A) Transcription of the B. anthracis putative and known exosporium genes (microarray data from reference 31). Expression of these sporulation-associated genes occurred at 240 min and later in this in vitro study. (B) Western blot of spore extracts probed with antiserum against rBclA (left) and rBxpB (right). Blots were first probed with anti-rBclA and then stripped and reacted with the anti-rBxpB antiserum. Lane M, protein size markers; lane 1, Sterne; 2, Sterne Δbas3957; lane 3, Sterne Δbas1131. (C) Transmission electron micrograph of spores of the Sterne ΔbclC (bas3557) strain; the red arrows indicate the presence of the exosporium nap layer attached to the basal layer. (D to G) Fluorescence micrographs of sporulating Sterne cells harboring the pBT1812 plasmid bearing bclC-egfp (D) and the mature spores from this culture (E). Panels F and G show sporulating cells and spores from Sterne expressing BclA-eGFP, which localizes to the exosporium nap layer. (H) Bright-field image of a wet mount of sporulating cells of the Sterne Δbas2277 mutant. Cells were plated on nutrient agar plates and incubated for 1 week at 30°C followed by 1 week at room temperature. Sporulation was complete, but most of the spores remained inside mother cell filament “ghosts” (indicated by the arrows).
FIG 6.
(A) Organization of the cotO (bas1140) to cotY (bas1145) exosporium gene cluster of the B. anthracis chromosome. The arrows indicate the direction of transcription of each of the genes. The stem-loop structure between cotY and bxpB represents a strong rho-independent terminator 15 bases distal to the cotY open reading frame stop codon (shown in bold font). The bxpB gene has its own promoter. (B) Western blot of spore extracts probed with anti-BclA serum (left) and anti-BxpB serum (right). Lane 1, Sterne; lane 2, Δbas1142 strain (clone 1); lane 3, Δbas1142 strain (clone 2); lane 4, Δbas1143 strain; lane 5, Δbas1142 Δbas1143 strain (clone 1); lane 6, Δbas1142 Δbas1143 strain (clone 2). (C) Immunofluorescence of spores from Sterne, Sterne Δbas1142, Sterne Δbas1143, and Sterne Δbas1142 Δbas1143 treated with antiserum against rBclA. Left-hand images are bright-field images and right-hand images are the fluorescent images of the same field.
TABLE 2.
Deletion mutants in putative exosporium genes
| Gene | Properties |
|---|---|
| bas1131 | Encodes a 370-aaa protein; gene adjacent to bclA; annotated as glycosyl transferase, group 2 family protein; identified in exosporium proteome; deletion mutant unaffected in exosporium production and spore content of BxpB and BclA |
| bas1142 | Encodes a 119-aa protein; deletion mutant unaffected in spore content of BxpB and BclA |
| bas1143 | Encodes a 61-aa protein; deletion mutant unaffected in spore content of BxpB and BclA |
| bas2277 | Annotated as an N-acetylmuramoyl-l-alanine amidase family 3 protein of 245 aa; expressed late in sporulation (31); deletion results in a defect in mother cell lysis and spore release |
| bas3557 (bclC) | Encodes a 481-aa collagen-like protein that lacks the N-terminal exosporium targeting domain. Gene transcribed with similar kinetics to that of exsY and bxpB. Deletion mutant displays no obvious defect in spore architecture in TEM analysis |
| bas3957 | Encodes a 120-aa protein; expression pattern parallels that of bxpB (31); deletion mutant unaffected in spore content of BxpB and BclA |
aa, amino acids.
The bclC gene encodes one of the collagen-like proteins of B. anthracis (32). It is expressed at times consistent with it being a sporulation gene. However, it lacks the N-terminal exosporium localization motif found on the BclA, BclB, BetA, and BclE (bas4623) proteins (25, 33, 34). Using a bclC-egfp fusion construct, it was found that this protein is expressed in sporulating cells, is positioned around the periphery of the spore, and is present in mature spores (Fig. 5C). The “ground glass” appearance of the fluorescence is characteristic of fluorescent fusions of proteins found in the interspace region of B. anthracis spores (6). The appearance of BclA-enhanced green fluorescent protein (eGFP), which localizes to the exosporium nap layer, is shown in comparison to the BclC-eGFP image (Fig. 5). With the collagen-like proteins potentially playing a structural role in the spores, a bclC deletion mutant was created and TEM of its spores was carried out. The loss of BclC had no apparent impact on the interspace or presence of the exosporium basal layer and attached nap (Fig. 5C).
bas2277 is a protein encoded by a gene which maps close to the bclB spore gene (bas2281). Deletion of the bas2277 gene resulted in a strain that sporulated but exhibited a defect in mother cell lysis and spore release (Fig. 5D). The gene is annotated as a putative N-acetylmuramoyl-l-alanine amidase family 3 protein, which may be involved in the breakdown of the mother cell peptidoglycan layer at the end of sporulation. The gene is expressed relatively late in the sporulation process (31) (Fig. 5A).
DISCUSSION
Assembly of the exosporium during sporulation by B. anthracis is a complex process. It initiates through the formation of a cap structure that occurs early in the sporulation process, shortly after the engulfment phase (18). Cap production occurs at the mother cell-central pole of the spore and requires CotY, which is its primary structural protein. Positioning of the cap requires the CotE and CotO proteins (18, 27). In the absence of the anchoring proteins, the exosporium appears in sheets located near the mother cell-central pole of the spore and does not properly associate with the spore. Remarkably, we observed that the cap structure can still form and be pole associated in the absence of the CotE protein, as shown with the cotE exsY double mutant spores. The basal layer of the exosporium is assembled later than the first appearance of the cap, at a time coincident with spore coat assembly. Exosporium assembly initiates at the cap, which serves as the primer for basal layer extension to form the noncap portion of the exosporium. The major structural protein for this noncap assembly is ExsY (11, 17). Exosporium assembly progresses from the mother cell-central (cap) pole of the spore toward the mother cell-distal spore pole. Lagging slightly behind the noncap basal layer assembly is the assembly of the outer basal layer of the exosporium, which includes the BxpB (ExsFA) and BclA proteins. These proteins assemble into high-MW complexes in the cytoplasm of the mother cell, and the complexes are then positioned on the developing exosporium (3, 24, 33). The BclA glycoprotein is cleaved, and presumably covalently attached to the basal layer, when the outer exosporium basal layer has largely encircled the spore and with the BclA cleavage progressing from the cap pole to the noncap pole (33).
Mutants of B. anthracis that do not produce the ExsY protein form only the exosporium cap structure (11). The B. cereus (strain NRRL B-569; ATCC 10876) exsY mutant produced spores lacking an exosporium but with adherent blebs of presumed exosporium, but no caps were observed (16). The explanation given was that the spores were prepared in liquid culture, and based on the findings of Boydston et al. (11), it was speculated that the lack of caps was due to the spores being prepared from liquid cultures (16). We found the B. anthracis caps to be more stably attached to the spores than initially reported, raising the possibility that the B. cereus exsY mutant failed to produce an organized cap structure. However, a recent study found that exsY mutants of the same B. cereus strain possessed the bottlecap exosporium structures on a subpopulation of the spores (19). Mutants of both B. anthracis and B. cereus lacking CotY are both capable of producing spores with an intact exosporium bearing BclA on their surface (16, 18, 19). This result is consistent with the findings of Boone et al. (18), who discovered that in the cotY mutant, which does not produce the cap structure which primes noncap ExsY assembly, production of the ExsY-containing basal layer is delayed and initiates more frequently along the lateral spore surfaces. In cotY mutant sporangia, forespores with a complete coat but no exosporium, or only partially complete exosporium, were readily observed (18). Despite the presence of an intact exosporium in the mature spores of the cotY mutant, the organization of the basal layer is altered, as evidenced by reduced reactivity with the anti-BxpB antiserum relative to that of the parent Sterne strain spores (Durand-Heredia et al., submitted).
Complementation of exosporium gene mutants was found to be incomplete, both with plasmid complementation systems and with single-copy chromosomal integration at an ectopic site. Plasmid-based complementation results in presumed overexpression of the proteins, which affects the ratio of the structural proteins and defects in exosporium assembly. Overexpression of ExsY or CotE in the Sterne wild-type strain resulted in production of a subpopulation of spores lacking the exosporium, and expression in the homologous gene deletion mutation only partially restored the wild-type phenotype. This was true even when the complementing gene was inserted into the bacterial chromosome in single copy but at an ectopic site. Altered expression levels of ExsY resulted in loss of the exosporium in a subpopulation of spores and disruptions of the outer spore coat layer. The spore coat defects may contribute to the exosporium assembly defects. ExsY fluorescent fusion proteins have been used to study ExsY assembly into spores. However, the complementation studies in this investigation that suggest the fusion protein is not fully functional in basal layer synthesis. The ExsY-eGFP fusion protein in single copy did not result in loss of the exosporium layer in the Sterne spores, and the multicopy expression resulted in only 75% of the spores lacking the exosporium layer. The plasmid-borne unfused ExsY gene resulted in all spores lacking the exosporium. The ExsY overexpression-associated spore coat defects were not observed with the ExsY-eGFP fusion protein expression, and the coat defects were observed only when ExsY was overexpressed (even with just two chromosomal copies) and not with exsY null mutant spores.
Attachment of the exosporium to the spore coat involves the CotO and CotE proteins (18, 19). When cotO was introduced on a plasmid in B. anthracis Sterne cells, loss of an attached exosporium was observed. CotO is thought to partner with CotE as part of this attachment structure. We found that when Sterne harbored a plasmid bearing cotE, a subpopulation of exosporium-negative spores was obtained. Each of the linkage proteins bridging the outer spore coat and exosporium has two binding partners. Overproduction of one of these proteins could result in proteins bound to the coat side of the chain, but not the exosporium, or vice versa. This breakage of the linkage chain would result in a failure of the exosporium to be properly oriented in the developing spore to be properly assembled around the spore. Elongation of the cap by ExsY results in the appearance of exosporium strips in the cotE null mother cell cytoplasm (27). The lack of CotE results in a failure of the exosporium to properly align with the forespore surface to generate the intact exosporium. However, in the absence of ExsY, elongation of the cap does not occur and under these conditions, a subpopulation of spores possessing the cap structure is obtained. These caps, however, appear to be less stably attached to the spore than those from the strain lacking ExsY but possessing CotE. CotO and CotE are also spore coat structural proteins, and thus, their absence also affects the composition of the spore coat outer layer, in addition to the effects on exosporium assembly.
Two of the major structural proteins of the basal layer, ExsY and to a lesser extent CotY, have been shown in B. cereus to be capable of self-assembly (17). Because the amino acid sequence of these proteins is highly conserved among the B. cereus sensu lato bacteria, it is reasonable to assume that self-assembly is a common feature of Bacillus exosporium basal layer assembly. The outer sublayer components of the basal layer, consisting of BxpB, ExsFB, BclA, and possibly other proteins, assemble into high-MW complexes in the mother cell cytoplasm and then are positioned for assembly onto the CotY/ExsY layer, initiating at the mother cell-central (cap) pole (3, 24, 25, 33). We determined that the BxpB/BclA outer basal layer can assemble into a spore-shaped sacculus in the absence of the CotY/ExsY inner basal layer and that in a triple deletion mutant lacking CotE, CotY, and ExsY, these ghost structures are not spore associated (Durand-Heredia et al., submitted). Thus, BxpB/ExsFB assembly into a cylindrical shell is, at least in part, independent of formation of the underlying basal layer.
The exosporium fragments that are released from the spores lacking BxpB and ExsFB contain oligomeric forms of ExsY and CotY and two likely nonstructural proteins, the SodA1 superoxide dismutase and the EA1 surface layer glycoprotein. The former is one of four superoxide dismutases that contribute to protections against oxidative stress in B. anthracis spores (35). SodA1 was shown to be a major protectant from intracellular superoxide stress (36). Because the exosporium is not degraded during germination and outgrowth, the presence of the SodA1 protein might provide a protective microenvironment against oxidative stress. During the initial stages of infection, the germination step within macrophages or dendritic cells constitutes the time when the bacterium is most sensitive to host innate defense mechanisms, until the cell can begin to synthesize and secrete toxins.
The EA1 glycoprotein is commonly reported for spore and exosporium preparations of B. anthracis. Williams and Turnbough (37) provided evidence that EA1 is a contaminant that can be removed with rigorous washing steps. Whether EA1 is a contaminant or a true, but relatively unstable, spore component, the eag gene is actively expressed during sporulation (Fig. 5A). Its transcription pattern is that of increased mRNA in late logarithmic phase (during which time EA1 replaces SAP as the cell’s S-layer component) (38, 39). A second wave of expression occurs during the sporulation phase at a time coincident with expression of spore coat and exosporium genes. The eag gene is expressed from a σH promoter (38) and also contains a putative σK promoter (AC-16 bp-CATAGGCTT) 647 bp upstream of the EA1 coding sequence. The sporulation phase expression would explain its presence in our exosporium preparation, whether as a contaminant from the mother cell expression or possibly as an actual component of the mature spore.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains used in this study are listed in Table 3. The B. anthracis strains used in this study were derived from the Sterne (pXO1+, pXO2−) vaccine strain. All strains lacked the capsule-encoding pXO2 plasmid and are therefore attenuated for virulence and exempt from tier 1 select agent guidelines. The studies were conducted under biosafety level 2 biocontainment conditions. B. anthracis strains were cultured in brain heart infusion (BHI) broth or brain heart infusion agar plates (Difco) at 37°C unless indicated otherwise. E. coli strains were cultured as described previously (33).
TABLE 3.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) or descriptiona | Reference or source |
|---|---|---|
| Bacillus anthracis strains | ||
| MUS8135 | Sterne bxpB::kan ΔexsFB | Durand-Heredia et al., submitted |
| MUS8228 | Sterne ΔexsY | Durand-Heredia and Stewart, submitted |
| MUS8229 | Sterne ΔcotY | Durand-Heredia and Stewart, submitted |
| MUS8230 | Sterne ΔcotY ΔexsY | |
| MUS8231 | Sterne ΔcotE::kan ΔexsY | This study |
| MUS8232 | Sterne ΔcotE::kan ΔcotY ΔexsY | Durand-Heredia and Stewart, submitted |
| MUS8233 | Sterne ΔamyS::exsY | This study |
| MUS8234 | Sterne ΔamyS::exsY-mcherry | This study |
| MUS8236 | Sterne Δbas1141 | This study |
| MUS8237 | Sterne Δbas1142 | This study |
| MUS8238 | Sterne Δbas1141 Δ1142 | This study |
| MUS8239 | Sterne Δbas1131 | This study |
| MUS8240 | Sterne Δbas3957::spc | This study |
| MUS8241 | Sterne ΔbclC::spc | This study |
| RG56 | Sterne ΔcotE::kan | 27 |
| Sterne | pXO1+ pXO2− | Lab stock |
| E. coli | ||
| DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK−) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Lab stock |
| GM48 | F– thr leu thi lacY galK galT ara fhuA tsx dam dcm glnV44 | Lab stock |
| Plasmids | ||
| pBT1812 | pMK4-bclC-egfp | This study |
| pDG4099 | pMK4 egfp fusion vector | 18 |
| pDG4100 | pMK4 mcherry fusion vector | 41 |
| pGS4294 | Allele replacement ts shuttle vector; Ampr Eryr | Lab stock |
| pHP13 | Shuttle vector; Cmr Eryr | 28 |
| pHPS2 | Shuttle vector; Spcr Eryr | 24 |
| pJD6139 | pHPS2-PbxpB-bxpB-mcherry | This study |
| pJD6173 | pHPS2-cotY | This study |
| pJD6177 | pHPS2-cotE | This study |
| pJD6265 | pHPS2-exsY | This study |
| pJD6270 | pHPS2-exsY-mcherry | This study |
| pMK4 | Shuttle plasmid; Ampr Cmr | 42 |
Ampr, Cmr, Eryr, Kanr, and Spcr, resistance to 100 μg of ampicillin mL−1, 10 μg of chloramphenicol mL−1, 5 μg of erythromycin mL−1, 50 μg of kanamycin mL−1, and 100 μg of spectinomycin mL−1, respectively.
DNA purification.
The Wizard SV miniprep kit (Promega) was used to isolate plasmid DNA. Genomic DNA was isolated using the Wizard genomic DNA purification kit (Promega). With B. anthracis, the cell pellets were frozen at −80°C overnight and thawed at 37°C prior to plasmid or genomic DNA extraction.
Generation of B. anthracis deletion mutants, electroporation of B. anthracis, and construction of the fluorescent reporter gene fusions.
Procedures were performed as described previously (Durand-Heredia et al., submitted). Plasmids to be introduced into B. anthracis were first passaged through E. coli strain GM48 to produce DNA lacking Dam methylation.
Construction of complementation plasmids.
Complementation plasmids were generated for the cotE, cotY, and exsY open reading frames by amplifying the ORF with its native promoter using primers that added EcoRI or SalI sites to the ends. The resulting amplicons were then cloned into the E. coli-Bacillus shuttle plasmid pHPS2 and sequence verified.
Production of spores.
B. anthracis cells from a BHI broth culture were swab inoculated onto the surface of 150- by 15-mm Oxoid nutrient agar plates with the appropriate antibiotics. The cultures were incubated at 30°C for 5 days. The surface layer of bacterial growth was harvested with a sterile cotton swab and the spores were dispersed into phosphate-buffered saline (PBS). The spores were pelleted by centrifugation at 15,000 rpm, and the upper pellet layer containing lysed cell debris was removed by aspiration and discarded. The was process was repeated until there was no evidence of vegetative cells or cell debris present. Dried spore pellets were weighed to determine weight of spores per volume, and the purified spore pellets were adjusted to a concentration of 10 μg/μL using PBS and stored at room temperature. For spores produced in broth culture, the cells were inoculated into 50 mL of nutrient broth and incubated at 30°C for 5 days with shaking at 240 rpm. Sporulation was greater than 90% as assessed by phase-contrast microscopy. Spores were harvested and purified as described above.
SDS-PAGE and electrotransfer to Immobilon membranes.
Protein samples were loaded onto a 15-well mini-Protean TGX gel (Bio-Rad) and electrophoresed at 190 V in SDS buffer (25 mM Tris, 192 mM glycine, 0.1% SDS [pH 8.3]). The proteins were electrotransferred to Immobilon membranes (Millipore Sigma) for 1 h on ice at 250 mA in 25 mM Tris, 192 mM glycine, and 10% methanol.
Western blot analysis.
For Western blot analysis, 10 mg of spores was pelleted in an Eppendorf 5425 microcentrifuge at 15,000 rpm for 2 min and the resulting pellets were then resuspended in 100 μL of urea SDS-PAGE buffer (8 M urea, 50 mM Tris-HCl [pH 10], 2% SDS, 0.002% bromophenol blue, 0.71 M β-mercaptoethanol, 10% glycerol) and boiled for 10 min. Spores were pelleted by centrifugation for 10 min, and 15 μL of the extract was subjected to SDS-PAGE and transfer to Immobilon membranes. Membranes were blocked overnight at 4°C using SuperBlock (Thermo Fisher) and stained with a 1:20,000 dilution of anti-rBclA or anti-rBxpB rabbit polyclonal antibody at room temperature for 1 h. The membranes were then washed six times with 0.1% Tween 20 in PBS, 5 min each with agitation. Secondary staining was done with 1:20,000 anti-rabbit IgG-horseradish peroxidase (HRP) conjugate (Invitrogen) at room temperature for 1 h, and the membrane was washed eight times with 0.1% Tween 20 in PBS for 5 min with agitation. The membrane was processed using Pierce ECL Western blotting substrate and then exposed to autoradiography film (Midwest Scientific).
Immunofluorescence microscopy.
Ten milligrams of spores was resuspended in 750 μL of SuperBlock blocking buffer (Thermo Scientific) and incubated for at least 20 min at room temperature. The spores were then harvested by centrifugation, and the spore pellet was resuspended in 250 μL of SuperBlock blocking buffer with 1 μL of primary antibody and incubated at room temperature for 20 min (with mixing every 5 min). Rabbit polyclonal anti-rBclA antibodies (24) were used. Following incubation with the primary antibody, the spores were harvested by centrifugation and washed with 750 μL of SuperBlock blocking buffer. The pellet was then resuspended in 250 μL of SuperBlock blocking buffer with secondary antibody conjugate (1:250 goat anti-rabbit IgG-Alexa Fluor 568; Invitrogen). The spores were incubated at room temperature for 20 min, pelleted, and washed with 750 μL of SuperBlock blocking buffer, followed by three washes with 750 μL of PBS and, finally, resuspension in 250 μL of PBS. The spores examined by epifluorescence microscopy using a Nikon E600 epifluorescence microscope and the mCherry filter set.
TEM.
Spores were fixed in 1 mL of 2% glutaraldehyde-100 mM sodium cacodylate solution containing 0.1% ruthenium red (Sigma-Aldrich) for 1 h at 37°C (40). Each spore pellet was then washed in 100 mM sodium cacodylate buffer, embedded in Histogel, and washed with 100 mM sodium cacodylate buffer containing 0.01 M 2-mercaptoethanol and 0.13 M sucrose (2-ME buffer). Samples underwent secondary fixation in 1% osmium tetroxide (Electron Microscopy Sciences). Spores were washed in 2-ME buffer and dehydrated with sequential treatments of 20, 50, 75, 90, and 100% acetone. Polymerization occurred at 60°C in Epon/Spurrs resin after extended resin infiltration. Samples were cut into 85-nm-thick sections, and the sections were mounted on 200-mesh nickel grids and stained with 2% uranyl acetate (Electron Microscopy Sciences) for 20 min at room temperature. The samples were treated with lead for 5 min at room temperature. Grids were washed in ultrapure water and observed by transmission electron microscopy (TEM) using a JEOL 1400 electron microscope at the University of Missouri Electron Microscopy Core Facility.
Spore image quantification.
To determine the percentage of spores that fall into different categories of exosporium expression, fluorescent images or TEM micrographs were examined. To be counted, the spore had to be positioned such that its long axis was parallel to the surface of the slide (fluorescence microscopy) or the plane of sectioning was through the long axis near the middle of the spore (TEM). This was done to ensure that the exosporium was properly classified (intact, cap-only, extended cap, or absent) or that the fluorescence pattern was properly characterized.
Isolation of exosporium fragments.
Spores from the ΔbxpB ΔexsFB mutant were prepared as described above and suspended in PBS in 1.5-mL microcentrifuge tubes. The ΔbxpB ΔexsFB spores were subjected to shear forces by additional vortex mixing. The samples were filtered through a 0.45-μ syringe filter and the spore-free filtrate was collected. The exosporium fragment-containing samples were then analyzed for protein composition by SDS-PAGE.
Protein identification.
For N-terminal sequence identification, proteins were resolved by SDS-PAGE and electroblotted onto a polyvinylidene difluoride (PVDF) membrane (Immobilon, EMD-Millipore). The filter containing the individual protein bands were submitted to the Iowa State University Protein Facility for N-terminal sequence analysis by the Edman method using a Shimadzu PPSQ-53A gradient protein sequencer (https://www.protein.iastate.edu/nsequencePPSQ.html). For protein identification by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis, the protein bands from the SDS-PAGE gel were excised and submitted to the University of Missouri Gehrke Proteomics Facility for trypsin digestion and protein identification (http://proteomics.missouri.edu/).
ACKNOWLEDGMENTS
This work was supported by NIAID grants R21AI101093 and R21AI112725 and the University of Missouri McKee Microbial Pathogenesis endowment fund to G.C.S.
Contributor Information
George C. Stewart, Email: stewartgc@missouri.edu.
Tina M. Henkin, Ohio State University
REFERENCES
- 1.Henriques AO, Moran CP, Jr.. 2007. Structure, assembly, and function of the spore surface layers. Annu Rev Microbiol 61:555–588. 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- 2.Fayad N, Kallassy Awad M, Mahillon J. 2019. Diversity of Bacillus cereus sensu lato mobilome. BMC Genomics 20:436. 10.1186/s12864-019-5764-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart GC. 2015. The exosporium layer of bacterial spores: a connection to the environment and the infected host. Microbiol Mol Biol Rev 79:437–457. 10.1128/MMBR.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driks A. 2002. Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol 10:251–254. 10.1016/s0966-842x(02)02373-9. [DOI] [PubMed] [Google Scholar]
- 5.Hachisuka Y, Kojima K, Sato T. 1966. Fine filaments on the outside of the exosporium of Bacillus anthracis spores. J Bacteriol 91:2382–2384. 10.1128/jb.91.6.2382-2384.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giorno R, Mallozzi M, Bozue J, Moody K-S, Slack A, Qiu D, Wang R, Friedlander A, Welkos S, Driks A. 2009. Localization and assembly of proteins comprising the outer structures of the Bacillus anthracis spore. Microbiology (Reading) 155:1133–1145. 10.1099/mic.0.023333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlton S, Moir AJ, Baillie L, Moir A. 1999. Characterization of the exosporium of Bacillus cereus. J Appl Microbiol 87:241–245. 10.1046/j.1365-2672.1999.00878.x. [DOI] [PubMed] [Google Scholar]
- 8.Lai E, Phadke ND, Kachman MT, Giorno R, Vazquez S, Vazquez JA, Maddock JR, Driks A. 2003. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J Bacteriol 185:1443–1454. 10.1128/JB.185.4.1443-1454.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Redmond C, Baillie LWJ, Hibbs S, Moir AJG, Moir A. 2004. Identification of proteins in the exosporium of Bacillus anthracis. Microbiology (Reading) 150:355–363. 10.1099/mic.0.26681-0. [DOI] [PubMed] [Google Scholar]
- 10.Todd SJ, Moir AJG, Johnson MJ, Moir A. 2003. Genes of Bacillus cereus and Bacillus anthracis encoding proteins of the exosporium. J Bacteriol 185:3373–3378. 10.1128/JB.185.11.3373-3378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boydston JA, Yue L, Kearney JF, Turnbough CL, Jr.. 2006. The ExsY protein is required for complete formation of the exosporium of Bacillus anthracis. J Bacteriol 188:7440–7448. 10.1128/JB.00639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steichen C, Chen P, Kearney JF, Turnbough CL, Jr.. 2003. Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J Bacteriol 185:1903–1910. 10.1128/JB.185.6.1903-1910.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steichen CT, Kearney JF, Turnbough CL, Jr.. 2005. Characterization of the exosporium basal layer protein BxpB of Bacillus anthracis. J Bacteriol 187:5868–5876. 10.1128/JB.187.17.5868-5876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swiecki MK, Lisanby MW, Shu F, Turnbough CL, Jr, Kearney JF. 2006. Monoclonal antibodies for Bacillus anthracis spore detection and functional analyses of spore germination and outgrowth. J Immunol 176:6076–6084. 10.4049/jimmunol.176.10.6076. [DOI] [PubMed] [Google Scholar]
- 15.Sylvestre P, Couture-Tosi E, Mock M. 2005. Contribution of ExsFA and ExsFB proteins to the localization of BclA on the spore surface and to the stability of the Bacillus anthracis exosporium. J Bacteriol 187:5122–5128. 10.1128/JB.187.15.5122-5128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson MJ, Todd SJ, Ball DA, Shepherd AM, Sylvestre P, Moir A. 2006. ExsY and CotY are required for the correct assembly of the exosporium and spore coat of Bacillus cereus. J Bacteriol 188:7905–7913. 10.1128/JB.00997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terry C, Jiang S, Radford DS, Wan Q, Tzokov S, Moir A, Bullough PA. 2017. Molecular tiling on the surface of a bacterial spore—the exosporium of the Bacillus anthracis/cereus/thuringiensis group. Mol Microbiol 104:539–552. 10.1111/mmi.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boone TJ, Mallozzi M, Nelson A, Thompson B, Khemmani M, Lehmann D, Dunkle A, Hoeprich P, Rasley A, Stewart G, Driks A. 2018. Coordinated assembly of the Bacillus anthracis coat and exosporium during bacterial spore outer layer formation. mBio 9:e01166-18. 10.1128/mBio.01166-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lablaine A, Serrano M, Bressuire-Isoard C, Chamot S, Bornard I, Carlin F, Henriques AO, Broussolle V. 2021. The morphogenetic protein CotE positions exosporium proteins CotY and ExsY during sporulation of Bacillus cereus. mSphere 6:e00007-21. 10.1128/mSphere.00007-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodenburg CM, McPherson SA, Turnbough CL, Jr, Dokland T. 2014. Cryo-EM analysis of the organization of BclA and BxpB in the Bacillus anthracis exosporium. J Struct Biol 186:181–187. 10.1016/j.jsb.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sylvestre P, Couture-Tosi E, Mock M. 2002. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol Microbiol 45:169–178. 10.1046/j.1365-2958.2000.03000.x. [DOI] [PubMed] [Google Scholar]
- 22.Sylvestre P, Couture-Tosi E, Mock M. 2003. Polymorphism in the collagen-like region of the Bacillus anthracis BclA protein leads to variation in exosporium filament length. J Bacteriol 185:1555–1563. 10.1128/JB.185.5.1555-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daubenspeck JM, Zeng H, Chen P, Dong S, Steichen CT, Krishna NR, Pritchard DG, Turnbough CL, Jr.. 2004. Novel oligosaccharide side chains of the collagen-like region of BclA, the major glycoprotein of the Bacillus anthracis exosporium. J Biol Chem 279:30945–30953. 10.1074/jbc.M401613200. [DOI] [PubMed] [Google Scholar]
- 24.Thompson BM, Hsieh HY, Spreng KA, Stewart GC. 2011. The co-dependence of BxpB/ExsFA and BclA for proper incorporation into the exosporium of Bacillus anthracis. Mol Microbiol 79:799–813. 10.1111/j.1365-2958.2010.07488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thompson BM, Hoelscher BC, Driks A, Stewart GC. 2012. Assembly of the BclB glycoprotein into the exosporium and evidence for its role in the formation of the exosporium ‘cap’ structure in Bacillus anthracis. Mol Microbiol 86:1073–1084. 10.1111/mmi.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng Q, Kao G, Qu N, Zhang J, Li J, Song F. 2016. The regulation of exosporium-related genes in Bacillus thuringiensis. Sci Rep 6:19005. 10.1038/srep19005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giorno R, Bozue J, Cote C, Wenzel T, Moody KS, Mallozzi M, Ryan M, Wang R, Zielke R, Maddock JR, Friedlander A, Welkos S, Driks A. 2007. Morphogenesis of the Bacillus anthracis spore. J Bacteriol 189:691–705. 10.1128/JB.00921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haima P, Bron S, Venema G. 1987. The effect of restriction on shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol Gen Genet 209:335–342. 10.1007/BF00329663. [DOI] [PubMed] [Google Scholar]
- 29.Zheng LB, Donovan WP, Fitz-James PC, Losick R. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev 2:1047–1054. 10.1101/gad.2.8.1047. [DOI] [PubMed] [Google Scholar]
- 30.Liu H, Bergman NH, Thomason B, Shallom S, Hazen A, Crossno J, Rasko DA, Ravel J, Read TD, Peterson SN, Yates J, III, Hanna PC. 2004. Formation and composition of the Bacillus anthracis endospore. J Bacteriol 186:164–178. 10.1128/JB.186.1.164-178.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergman NH, Anderson EC, Swenson EE, Niemeyer MM, Miyoshi AD, Hanna PC. 2006. Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J Bacteriol 188:6092–6100. 10.1128/JB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leski TA, Caswell CC, Pawlowski M, Klinke DJ, Bujnicki JM, Hart SJ, Lukomski S. 2009. Identification and classification of bcl genes and proteins of Bacillus cereus group organisms and their application in Bacillus anthracis detection and fingerprinting. Appl Environ Microbiol 75:7163–7172. 10.1128/AEM.01069-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson BM, Stewart GC. 2008. Targeting of the BclA and BclB proteins to the Bacillus anthracis spore surface. Mol Microbiol 70:421–434. 10.1111/j.1365-2958.2008.06420.x. [DOI] [PubMed] [Google Scholar]
- 34.Thompson BM, Hoelscher BC, Driks A, Stewart GC. 2011. Localization and assembly of the novel exosporium protein BetA of Bacillus anthracis. J Bacteriol 193:5098–5104. 10.1128/JB.05658-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cybulski RJ, Jr, Sanz P, Alem F, Stibitz S, Bull RL, O’Brien AD. 2009. Four superoxide dismutases contribute to Bacillus anthracis virulence and provide spores with redundant protection from oxidative stress. Infect Immun 77:274–285. 10.1128/IAI.00515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Passalacqua KD, Bergman NH, Herring-Palmer A, Hanna P. 2006. The superoxide dismutases of Bacillus anthracis do not cooperatively protect against endogenous superoxide stress. J Bacteriol 188:3837–3848. 10.1128/JB.00239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams DD, Turnbough CL, Jr.. 2004. Surface layer protein EA1 is not a component of Bacillus anthracis spores but is a persistent contaminant in spore preparations. J Bacteriol 186:566–569. 10.1128/JB.186.2.566-569.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mignot T, Mesnage S, Couture-Tosi E, Mock M, Fouet A. 2002. Developmental switch of S-layer protein synthesis in Bacillus anthracis. Mol Microbiol 43:1615–1627. 10.1046/j.1365-2958.2002.02852.x. [DOI] [PubMed] [Google Scholar]
- 39.Couture-Tosi E, Delacroix H, Mignot T, Mesnage S, Chami M, Fouet A, Mosser G. 2002. Structural analysis and evidence for dynamic emergence of Bacillus anthracis S-layer networks. J Bacteriol 184:6448–6456. 10.1128/JB.184.23.6448-6456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waller LN, Fox N, Fox KF, Fox A, Price RL. 2004. Ruthenium red staining for ultrastructural visualization of a glycoprotein layer surrounding the spore of Bacillus anthracis and Bacillus subtilis. J Microbiol Methods 58:23–30. 10.1016/j.mimet.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Hermanas TM, Subramanian S, Dann CE, III, Stewart GC. 2021. Spore-associated proteins involved in c-di-GMP synthesis and degradation of Bacillus anthracis. J Bacteriol 203:e00135-21. 10.1128/JB.00135-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan MA, Yasbin RE, Young FE. 1984. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene 29:21–26. 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]