ABSTRACT
Streptococcus pyogenes, otherwise known as Group A Streptococcus (GAS), is an important and highly adaptable human pathogen with the ability to cause both superficial and severe diseases. Understanding how S. pyogenes senses and responds to its environment will likely aid in determining how it causes a breadth of diseases. One regulatory network involved in GAS’s ability to sense and respond to the changing environment is the Rgg2/3 quorum sensing (QS) system, which responds to metal and carbohydrate availability and regulates changes to the bacterial surface. To better understand the impact of Rgg2/3 QS on S. pyogenes physiology, we performed RNA-seq and tandem mass tag (TMT)-LC-MS/MS analysis on cells in which this system was induced with short hydrophobic peptide (SHP) pheromone or disrupted. Primary findings confirmed that pheromone stimulation in wild-type cultures is limited to the induction of operons whose promoters contain previously determined Rgg2/3 binding sequences. However, a deletion mutant of rgg3, a strain that endogenously produces elevated amounts of pheromone, led to extended alterations of the transcriptome and proteome, ostensibly by stress-induced pathways. Under such exaggerated pheromone conditions, a connection was identified between Rgg2/3 and the stringent response. Mutation of relA, the bifunctional guanosine tetra- and penta-phosphate nucleoside synthetase/hydrolase, and alarmone synthase genes sasA and sasB, impacted culture doubling times and disabled induction of Rgg2/3 in response to mannose, while manipulation of Rgg2/3 signaling modestly altered nucleotide levels. Our findings indicate that excessive pheromone production or exposure places stress on GAS resulting in an indirect altered proteome and transcriptome beyond primary pheromone signaling.
IMPORTANCE Streptococcus pyogenes causes several important human diseases. This study evaluates how the induction or disruption of a cell-cell communication system alters S. pyogenes’s gene expression and, in extreme conditions, its physiology. Using transcriptomic and proteomic approaches, the results define the pheromone-dependent regulon of the Rgg2/3 quorum sensing system. In addition, we find that excessive pheromone stimulation, generated by genetic disruption of the Rgg2/3 system, leads to stress responses that are associated with the stringent response. Disruption of stringent response affects the ability of the cell-cell communication system to respond under normally inducing conditions. These findings assist in the determination of how S. pyogenes is impacted by and responds to nontraditional sources of stress.
KEYWORDS: Streptococcus pyogenes, proteomics, quorum sensing, stress response, stringent response, transcriptional regulation
INTRODUCTION
Streptococcus pyogenes, otherwise known as group A Streptococcus (GAS), causes millions of infections per year worldwide (1, 2), resulting in an array of disease states, from superficial tonsilitis to severe infections such as necrotizing fasciitis (3, 4). Genetic programs that regulate virulence attributes of GAS are responsive to the microorganism’s environment, and its metabolic status, through intercellular signaling networks. Quorum sensing (QS), a form of intercellular communication, is used by GAS to regulate gene expression in response to molecular signals produced by bacteria (5–11). QS activity is often coupled to the status of the environment, where characteristics such as nutrient availability or pH can impact expression or action of QS-centric genes. A conserved network of QS pathways in GAS involves Rgg transcription factors, which are receptors for peptide pheromones. Rgg proteins are a part of a larger family of pheromone-responsive proteins known as RRNPP (Rap, Rgg, NprR, PlcR, PrgX) (10). GAS possesses four Rgg proteins, RopB (Rgg1), Rgg2 and Rgg3 (Rgg2/3), and ComR (Rgg4) (12). Several studies have documented their roles in virulence, biofilm formation, and lysozyme resistance (5, 6, 8, 9, 11, 13, 14).
Rgg2 and Rgg3 are allosteric regulators of transcription that respond indiscriminately to the short hydrophobic peptides SHP2 and SHP3 (Fig. 1). The active forms of the peptides differ by only one conserved amino acid (SHP2: DIIIIVGG; SHP3: DILIIVGG), and empirical evaluation has determined they have equivalent inducer activities; for simplicity we refer to them as SHP (5, 9, 13, 15–20). Induction of gene transcription requires Rgg2, which when bound to SHP, acts as a transcriptional activator. Deletion of rgg2 (Δrgg2) disables transcription induction, where target genes remain silent or express only at very low levels (15). In contrast, Rgg3 represses transcription until it binds to SHP, whereupon repression is abrogated (15, 16). Environmental signals also integrate into the Rgg2/3 circuit. Depleting growth media of manganese or iron, or provision of mannose as a primary carbon source, have the effect of inducing QS signaling (13). We have suggested these conditions resemble the nutrient status seen in the host where GAS communication may enhance fitness and adaptability.
FIG 1.
Schematic depicting the current understanding of the Rgg2/3 quorum sensing system and its response to SHP2/3 peptides. Upon import of the SHP2/3 via Opp, SHPs bind to Rgg2 and Rgg3. Binding of SHPs to Rgg3 and Rgg2 leads to derepression and activation of the illustrated gene clusters: the stcA locus and the spy49_0450 operon. Activation of these operons leads to production of StcA, a protein demonstrated to modulate lysozyme resistance and biofilm formation; and a yet unidentified product from the spy49_0450 operon that leads to NF-κB suppression in macrophages. Activation of the system additionally leads to increased SHP2/3 production. Additionally produced SHP2/3 is exported and processed via the Eep protease and PptAB exporter, which leads to further QS system induction in surrounding cells. An additional turn over system for SHPs also exists (not shown), in which SHPs are degraded by the endopeptidase PepO. Previous work has demonstrated that deletion of Rgg3 results in a constitutively QS-ON cell state, whereas deletion of Rgg2 leads to a constitutively QS-OFF cell state.
Two polycistronic operons are known regulatory targets of Rgg2/3, defined in part by an Rgg2/3 binding sequence that overlaps a typical −35 promoter element (Fig. 1) (16). At one locus, shp2 is followed by spy49_0414c (stcA). The other locus encodes shp3, nine putative enzymes, and a major facilitator superfamily (MFS) transporter (spy49_0450 – spy49_0460) (21). stcA is associated with enhanced lysozyme resistance and biofilm formation, whereas spy49_0450-0460 is required for QS-induced immune suppression (5, 9, 13).
Given the complex nature of identified phenotypes associated with this QS system, we wondered if other genes are under SHP and Rgg2/3 control. To assess this, we performed transcriptomic and proteomic analyses to observe responses of wild-type NZ131 and isogenic Δrgg3 and Δrgg2 mutants in the presence and absence of SHP. We find that the transcriptomic and proteomic responses of wild-type stimulated by SHP are restricted to genes previously associated with the system. However, a mutant of rgg3, which produces elevated amounts of pheromone due to its derepressed state, results in differential gene and protein expression extending beyond patterns observed for stimulated wild-type, likely the result of an unspecified stress response due to disruption of the Rgg2/3 system.
RESULTS
RNA-seq identifies the core regulon for Rgg2/3 quorum sensing.
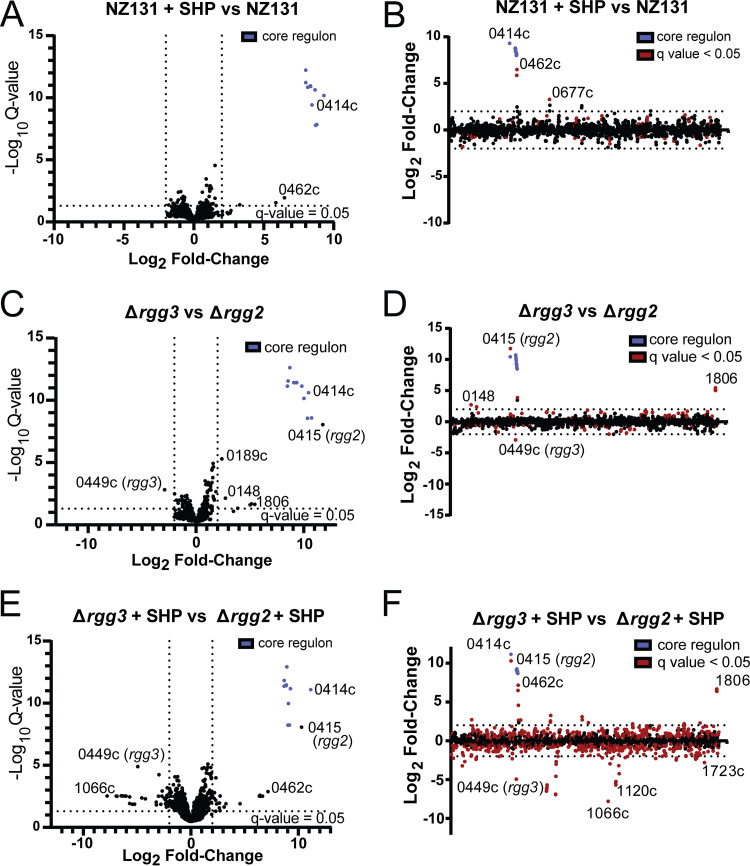
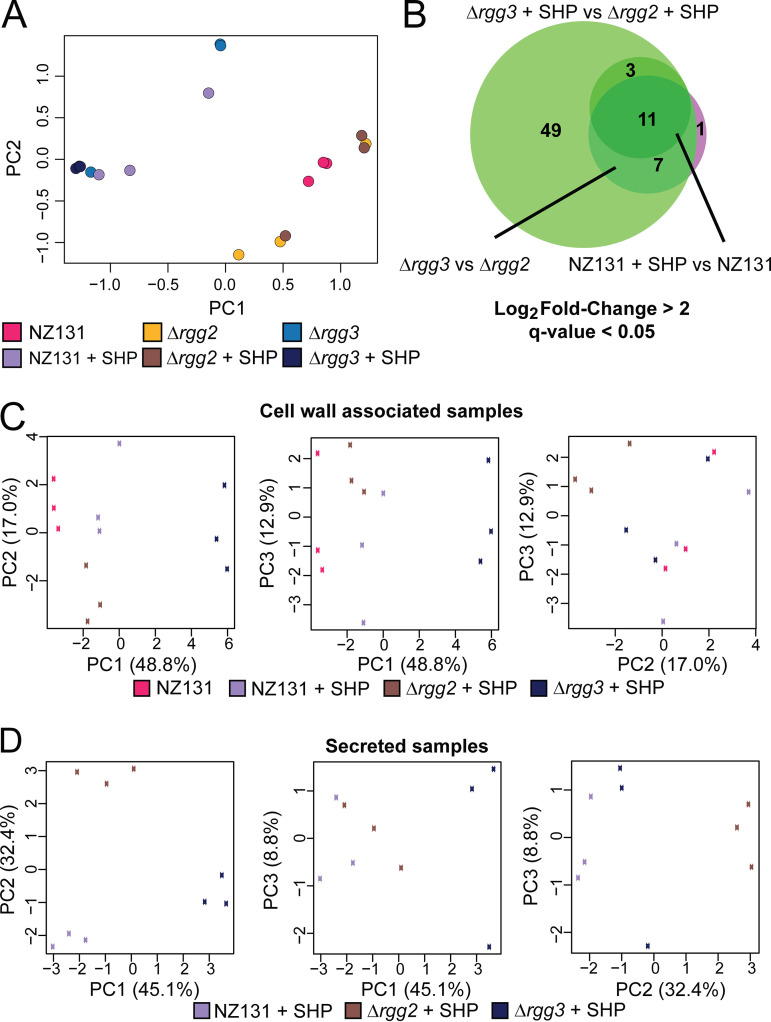
Given that multiple environmental inputs impact Rgg2/3 signaling, and because phenotypes associated with this QS system are complex (e.g., immunosuppression, lysozyme resistance) and likely to involve several genes, we evaluated gene and protein expression patterns when GAS NZ131 (serotype M49) was induced by SHP and in isogenic mutant backgrounds Δrgg2 or Δrgg3. Direct sequencing and quantitation of genomic mRNA transcripts with analysis to identify differentially expressed genes (DEGs) between unstimulated cultures and those treated with SHP led to the determination that 11 genes were induced >240-fold and constitute the core Rgg2/3 regulon (Fig. 2A and B; Table 1; Supplemental File 1). Confirming prior reports (5, 9, 15), spy49_0414c (stcA) and spy49_0450-0460 were expressed from transcription start sites directly upstream of either shp2 or shp3 and appear to be coexpressed as polycistronic operons. Three additional genes were significantly differentially regulated: spy49_0461c and 0462c are hypothetical genes encoded immediately downstream and on the opposite strand from the 0450–0460 operon, and spy49_0677c (transposase, ISS1mu1) was found to be differentially expressed ~4-fold and barely meeting statistical criteria. A similar list of genes was identified when comparing Δrgg2 to Δrgg3 strains, which are effectively noninducible and auto-inducible strains, respectively (Fig. 2C and D; Supplemental File 1). Aside from this, principal-component analysis indicates that Δrgg3 mimics wild-type induced with SHP, whereas the Δrgg2 strain replicates wild-type grown without SHP induction in CDM medium (Fig. 4A). These observations reinforce our previous suggestion that Δrgg3 exists in a QS-ON status, where Δrgg2 remains in a QS-OFF state.
FIG 2.
Transcriptomic changes in response to pheromone stimulation. For a complete list of differentially regulated genes and RNA-seq data, see Supplemental File 1. Volcano plots (panels A, C, E) indicate transcript fold changes and q-value significance. The core Rgg2/3 regulon genes are indicted by blue points. Dotted lines indicate threshold values for log2 fold change of transcript counts (minimum 4-fold) or log10 q values (<0.05). For panels B, D, and F, log2 fold changes of transcript counts plotted as a function of the NZ131 linear genome map. Red dots indicate significant q values <0.05. Dotted line indicates threshold > 4-fold differential transcript counts. (A and B) NZ131 versus NZ131 with supplemented 100 nM SHP. (C and D) Δrgg3 versus Δrgg2. (E and F) Δrgg3 versus Δrgg2 cultures, each stimulated with 100 nM SHP.
TABLE 1.
Core regulon of Rgg2/3 QSa
| Locus tag | Predicted or known functionb | Predicted protein localizationc | Log2FCd NZ131 + SHP vs NZ131 | Log2FC Δrgg3 vs Δrgg2 | Log2FC Δrgg3 + SHP vs Δrgg2 + SHP |
|---|---|---|---|---|---|
| spy49_0414c | StcA, lysozyme resistance and biofilm formation | Extracellular | 9.31 | 10.44 | 11.15 |
| spy49_0450 | AroE.2, shikimate dehydrogenase | Unknown | 8.71 | 10.72 | 9.16 |
| spy49_0451 | Sugar isomerase | Cytoplasmic | 8.79 | 10.34 | 9.04 |
| spy49_0453 | F420-dependent γ-glutamyl ligase | Cytoplasmic | 8.45 | 10.02 | 9.07 |
| spy49_0454 | Oxidoreductase | Cytoplasmic | 8.67 | 9.81 | 9.26 |
| spy49_0455 | Archaeal metK SAM synthetase | Cytoplasmic | 8.36 | 9.34 | 8.87 |
| spy49_0456 | Hpr kinase/phosphatase domain | Unknown | 8.36 | 9.28 | 8.80 |
| spy49_0457 | Glycosyltransferase | Cytoplasmic membrane | 8.34 | 9.07 | 8.87 |
| spy49_0458 | Pseudogene; acyl carrier protein or nonribosomal peptide synthetase | Unknown | 8.01 | 8.70 | 8.92 |
| spy49_0459 | Nucleotide sugar dehydrogenase | Cytoplasmic | 8.00 | 8.54 | 8.68 |
| spy49_0460 | Putative efflux protein | Cytoplasmic membrane | 8.14 | 8.48 | 8.67 |
Genes that were observed to be differentially regulated. All genes had a greater than 2-fold log2 fold change, and a q-value of less than 0.05.
Predicted function as experimentally validated for StcA in Gogos et al. (5) as determined by protein homology in Rahbari et al. (9) or via BLASTp of protein coding sequences. For further details see Materials and Methods.
Cell localization as predicted by PSORTb, only StcA has been experimentally validated as having extracellular localization in Gogos et al. (5). For further details see Materials and Methods.
Log2FC indicates log2 fold change.
FIG 4.
PCA analysis of RNA-seq and proteomics results, and additional RNA-seq data. For a complete list of differentially expressed transcripts and proteins, see Supplemental Files 1 to 3. (A) Principle component analysis of RNA-seq results. The legend below the graph indicates the color of each sample, for which three biological replicates were harvested. (B) Venn diagram of RNA-seq results, demonstrating a core regulon of 11 genes for the Rgg2/3 system. NZ131 + SHP versus NZ131 is indicated by the darker green circle, Δrgg3 versus Δrgg2 by the pink-purple circle, and Δrgg3 + SHP versus Δrgg2 + SHP by the light green circle. Log2 fold change greater than 2 and q-value of less than 0.05 was used as a cutoff for significance. Eleven genes are found to be differentially regulated under all conditions. (C) Three-way principal-component analysis of cell wall associated samples proteomics results. The legend below the graphs indicates the corresponding sample with the box color on each graph. (D) Three-way principal-component analysis of secreted samples proteomics results. The legend below the graphs indicates the corresponding sample with the box color on each graph.
It was also of interest to learn whether stimulation of the Δrgg2 strain with SHP impacted gene expression, as the hypothesis that Rgg2 is the sole activator of the QS system has not yet been challenged. Comparing mRNA levels of the Δrgg2 strain treated with SHP to that of wild-type or unstimulated Δrgg2 cultures found minimal significant differences. We applied the same stimulation to the Δrgg3 strain. We anticipated only to find an increase in the expression level of the core regulon given that we determined that the amount of SHP increases in this strain (~200 to 300 nM for Δrgg3 versus ~100 to 150 nM for SHP-stimulated wild-type) (Fig. 3F). Instead, when we compared stimulated Δrgg3 with wild-type unstimulated cultures we found ~50 additional genes with differential expression patterns (Supplemental File 1). Some of these genes included sclB (spy49_0830), grab (protein G-related macroglobulin-binding protein, spy49_1080c), and pyrR (spy49_0650). A similar number of differentially regulated genes (~59 additional genes) was observed between SHP-treated Δrgg3 and SHP-treated Δrgg2 (Fig. 2E and F; Supplemental File 1). This included genes involved in immune regulation such as mac (spy49_0679), emm49 (spy49_1671c), and nucleotide metabolism such as xpt (spy49_0887, xanthine phosphoribosyltransferase), spy49_0888 (predicted xanthine permease), and pyrR.
FIG 3.
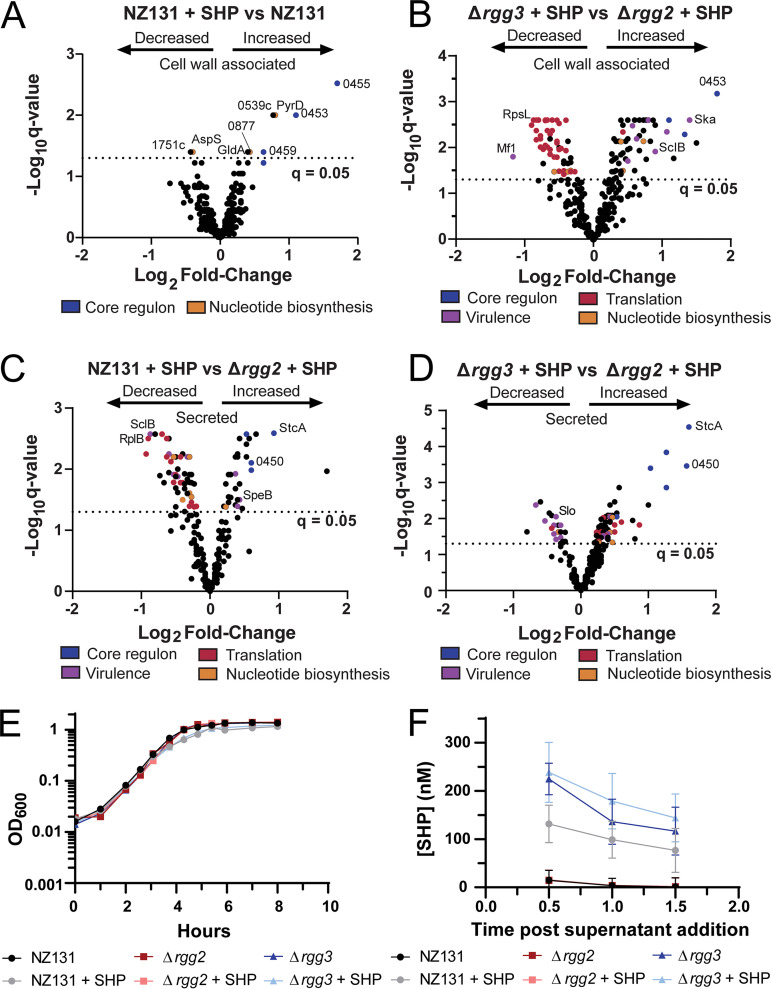
Examination of the response of S. pyogenes to the disruption or induction of the Rgg2/3 QS system. (A to D) Volcano plots of proteomics results. The legends below the graphs indicate processes or regulons in which proteins are involved. Blue, core Rgg2/3 regulon; purple, virulence; red, translation; orange, nucleotide biosynthesis. Proteins of particular interest are also indicated on each graph. For a complete list of differentially expressed proteins, TMT-LC-MS/MS results, predicted protein localization, and comparison to Wilk et al.’s (22) J. Proteome data set, see Supplemental Files 2 and 3. (A) Differentially expressed cell wall associated proteins in wild-type NZ131 + SHP versus NZ131. (B) Differentially expressed cell wall associated proteins in Δrgg3 + SHP versus Δrgg2 + SHP. (C) Differentially expressed secreted proteins in wild-type NZ131 + SHP versus Δrgg2 + SHP. (D) Differentially expressed secreted proteins in Δrgg3 + SHP versus Δrgg2 + SHP. (E) Growth curve of wild-type NZ131, Δrgg2, and Δrgg3 with or without 100 nM SHP peptide. Strains and conditions are indicated in the legend below the graph and is representative of more than three independent experiments. (F) Relative SHP concentrations from supernatants isolated from wild-type NZ131, Δrgg2, and Δrgg3 with or without 100 nM SHP peptide. Strains and conditions are indicated in the legend below the graph. Graph represents an average of three independent experiments. For further experimental details see Materials and Methods.
Effect of SHP stimulation on the S. pyogenes proteome.
As phenotypes associated with the Rgg2/3 system include enhanced lysozyme resistance, biofilm formation, and contact-dependent immunosuppression which reflect putative changes to the GAS surface (5, 9, 13), we sought to evaluate proteomic profiles of surface-associated and released proteins from cultures with differentially stimulated Rgg2/3. A published protocol for isolation of proteins from S. pyogenes for proteomic evaluation was adapted and performed with TMT labeling in conjunction with LC-MS/MS (22, 23) (Fig. S1A). Cultures of wild-type, Δrgg2, or Δrgg3 were grown with or without SHP until late exponential phase (the same point at which transcriptomic evaluations were conducted) when proteins were harvested. We note that in the preparation of both our proteomics and transcriptomics samples that SHP-stimulated wild-type and Δrgg3 cultures had a slower doubling time than unstimulated or Δrgg2 cultures (Fig. 3E; Table S2). However, total protein amounts isolated from each fraction did not differ significantly between strains (Fig. S2F). Overall, 404 unique proteins were detected between the cell-wall associate and secreted fractions, representing approximately 25% of the theoretical proteome (Fig. S1B). To further examine the results, we additionally examined the predicted protein localization of each detected protein via PSORTb and compared the proteins detected in each fraction with those previously published in Wilk et al. (22), which we based our protocol for extraction on. We find that a significant fraction of the cell wall associated proteins we detected are predicted to be cytoplasmic, indicating that the protocol for surface protein isolation utilized by Wilk et al. (22) also appears to result in isolation of cytoplasmic proteins as well. We compared the proteins detected in our cell wall associated fraction to that published in Wilk et al. (22) and found that 268 of the 318 proteins detected in our cell wall associated samples were also detected in the cell-wall fraction in Wilk et al. (22). We also note that only eight out of 318 proteins in our cell wall data set were found to not be detected in the Wilk et al. (22) proteomics cell wall fraction (Supplemental File 2). A poorer correlation between the secreted proteins detected in our analysis versus the secreted fraction from Wilk et al. (22) was observed, with only 81 of the 272 proteins detected in our sample also detected in the Wilk et al. (22) secreted fraction (Data Set S3). We have provided additional filtering of these data sets in our analysis based on this comparison, as well as unfiltered data sets. Complete lists of detected proteins and statistical analysis are provided in Supplemental Files 2 and 3.
Due to the difference in correlation between secreted proteins observed in our proteomics analysis versus Wilk et al.’s (22) M1 GAS proteome isolation, we decided to examine if any lysis was occurring between strains during growth. To examine this, we grew wild-type NZ131, Δrgg2, and Δrgg3 strains and examined if a measurable drop in CFU/mL occurred during growth in the absence or presence of 100 nM SHP. We observed no significant differences or drop in CFU/mL upon the addition of SHP peptide to strains (Fig. S3A). A small but statistically significant increase in CFU/mL for the Δrgg2 strain at 3-h post-SHP addition was observed, but this was only significant when compared with NZ131, NZ131 + SHP, and Δrgg3 + SHP conditions. Due to the observation that no overt cell lysis was observed during growth in CDM under our conditions, we conclude that detected cytoplasmic proteins in our proteomics samples were a result of the proteomics cell preparation method used. As such, we conclude that caution should be taken when interpreting results concerning proteomics preparations when cell wall fractions are prepared using enzymatic preparation such as mutanolysin digestion.
Nevertheless, we examined the data obtained from our proteomics samples. We observe that in our data members of the core Rgg2/3 regulon (stcA and those encoded by genes 0450 to 0460) consistently displayed increased amounts in all samples in which quorum sensing was stimulated compared with unstimulated. A larger number of proteins (Fig. S1C and D; Supplemental Files 2 and 3) were altered between SHP-stimulated cultures of Δrgg3 and Δrgg2 (Fig. 3B); these changes were not seen between stimulated and unstimulated cultures of wild-type for the cell-wall-associated fraction (Fig. 3A). Examination of data sets with additional filtering based on Wilk et al. (22) did not alter these trends (Supplemental Files 2 and 3). The expanded number of differentially produced proteins in the mutant-strain comparisons are primarily due to the SHP-stimulated Δrgg3 condition, as these samples formed a cluster separate from others in the primary dimension when evaluated by principal-component analysis (Fig. 4C and D; unstimulated Δrgg3 and rgg2 cultures were not processed due to limited resources). In addition to core-regulon targets being differentially expressed, virulence factors, ribosomal and translation-associated proteins, and nucleotide biosynthesis protein levels were altered in the cell wall associated fraction (Fig. 3B; Supplemental File 2). A similar trend in ribosomal and translation-related proteins was observed when comparing the stimulated Δrgg3 condition to wild-type with or without SHP (Fig. S2A and B), which was not observed in stimulated Δrgg2 versus wild-type (Fig. S2C and D).
We note that compared with unstimulated wild-type, the stimulated Δrgg2 strain produced additional changes in the proteome that were not observed in the stimulated wild-type strain (Fig. S2C; Fig. 3A). These changes also included virulence factors and proteins involved in nucleotide biosynthesis (SclB, Gmk, PyrD). This was initially unexpected due to the absence of small number of transcriptional changes in the Δrgg2 + SHP versus wild-type condition (Supplemental File 1). It is feasible that imbalance of the Rgg2/3 system (i.e., deletion of one of the regulatory proteins) and subsequent stimulation with SHP could have off-target effects on the proteome. However, we do caution that the preparation method used for isolating the proteomics samples could have inadvertently affected these results and conclude that further experimentation should verify these changes. Overall, however, we find that genetic disruption of this system and subsequent stimulation with SHP peptides produces otherwise unforeseen alterations in the S. pyogenes proteome.
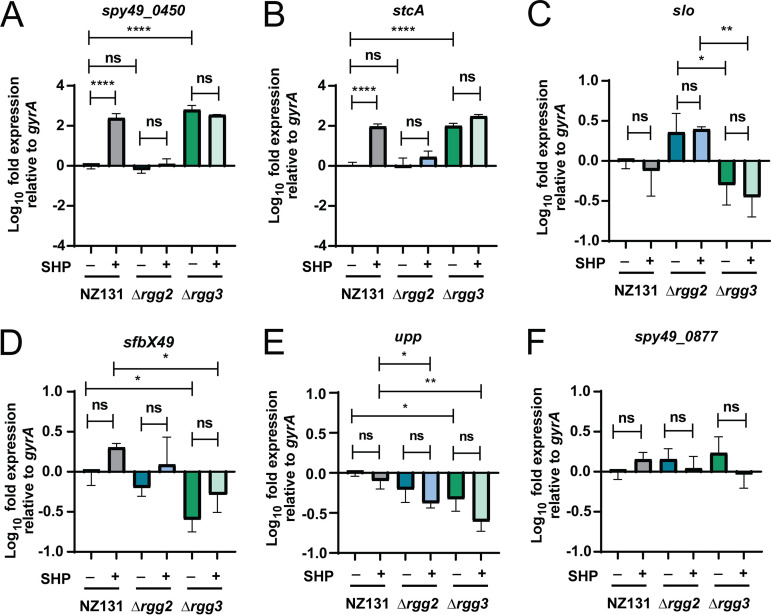
Given that previous work has found that up to 60% of proteins may display poor correlation between cellular concentrations and their corresponding RNA transcripts (24, 25), we attempted to validate RNA-seq and proteomic results from additional biological replicates using targeted qRT-PCR and Western blotting. Transcripts of the primary Rgg2/3 targets spy49_0450 and stcA were confirmed as substantially upregulated (Fig. 5A and B). Several additional subjects were selected based on their roles as virulence factors or differential expression pattern in either the RNA-seq and/or the proteomic data sets. These included: slo, upp, spy49_0877, plr, prtF, ska, sfbX49, and sclB (Fig. 5; Fig. S3C to F). Half of these targets (slo, upp, prtF, sfbX49) displayed transcriptional changes between conditions, but the others could not be statistically validated (spy49_0877, plr, ska, sclB) (Fig. 5; Fig. S3C to F). We observed that these targets (slo, upp, prtF, sfbX49) were also altered transcriptionally in our RNA-seq data set, although they did not necessarily meet the criteria for statistical significance for all conditions in this analysis (Supplemental File 1).
FIG 5.
qRT-PCR results verifying RNA-seq and examining if any proteomics targets have altered transcript levels. Wild-type NZ131, Δrgg2, and Δrgg3 strains were grown in biological triplicate with or without 100 nM SHP peptide (indicated by + or – sign below each graph) and RNA was harvested late exponential phase. RNA was processed for qRT-PCR and transcript levels were determine relative to the gyrA reference gene. Significance of transcript level changes were determined using a one-way ANOVA with Multiple Comparisons Posttest. *, P < 0.05; **, P < 0.005; ****, P < 0.0005; ns, nonsignificant. For further experimental details see Materials and Methods. (A) Relative transcript levels of spy49_0450 (aroE.2). (B) Relative transcript levels of stcA (spy49_0414c). (C) Relative transcript levels of slo (spy49_0146). (D) Relative transcript levels of sfbX49 (spy49_1683c). (E) Relative transcript levels of upp (spy49_0322). (F) Relative transcript levels of spy49_0877.
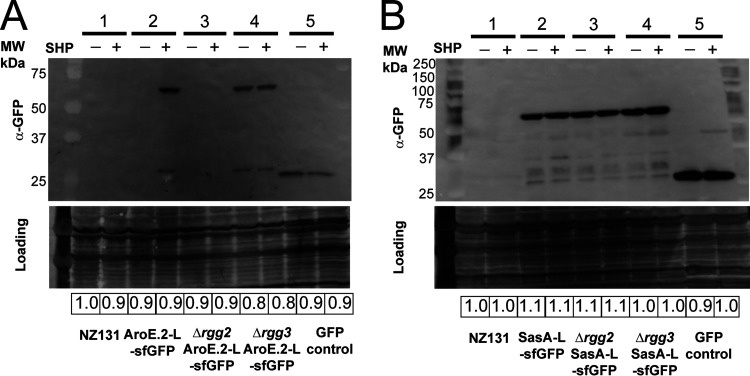
To evaluate changes in protein levels, we constructed sfGFP fusions to two proteins of interest: AroE.2 (Spy49_0450) and Spy49_0877. AroE.2 was only detectable in the stimulated wild-type or in Δrgg3 but not in unstimulated wild-type or either Δrgg2 condition (Fig. 6A). For Spy49_0877, the SHP-induced change observed in the proteomics data set was slight but significant (~1.3 ratio increase in expression; Supplemental File 2); however, the fusion to sfGFP showed no significant differences in levels between strains or conditions (Fig. 6B). Taken together, validation was confirmed for the core targets of Rgg2/3 regulation, whereas small changes observed in transcriptomic and proteomic data sets are likely stochastic fluctuations resulting from apparent stress induced responses.
FIG 6.
Western blots of selected proteins validating proteomics data. Blots were probed with α-GFP to detect GFP-linked proteins or a positive vector control expressing GFP (GFP control). India ink stain was used to control for Western blot loading, and relative loading is shown below for each blot. Shown beside the blot is the corresponding molecular weight to the ladder. The expected size of AroE.2-L-sfGFP is 63.5 kDa, SasA-L-sfGFP is 54.3 kDa, sfGFP is 27.0 kDA, and GFP is 26.8 kDA. All experiments were performed three times. (A) Western blot examining expression of AroE.2-L-sfGFP in NZ131 wild-type background, Δrgg2, or Δrgg3 background, with (+) or without (–) 100 nM SHP peptide. 1: NZ131 wild type; 2: NZ131 pBER40 (pLZ12-spec-aroE.2-L-sfGFP shp3[ATG→GGG]); 3: NZ131 Δrgg2 pBER40; 4: NZ131 Δrgg3 pBER40; 5: GFP control (HSC5 pFED630). (B) Western blot examining expression of SasA-L-sfGFP in NZ131 wild-type background, Δrgg2, or Δrgg3 background, with (+) or without (–) 100 nM SHP peptide. 1: NZ131 wild type; 2: NZ131 pBER41 (pLZ12-spec-sasA-L-sfGFP); 3: NZ131 Δrgg2 pBER41; 4: NZ131 Δrgg3 pBER41; 5: GFP control (HSC5 pFED630).
Rgg2/3 stimulation impacts growth rate and has a link to stringent response.
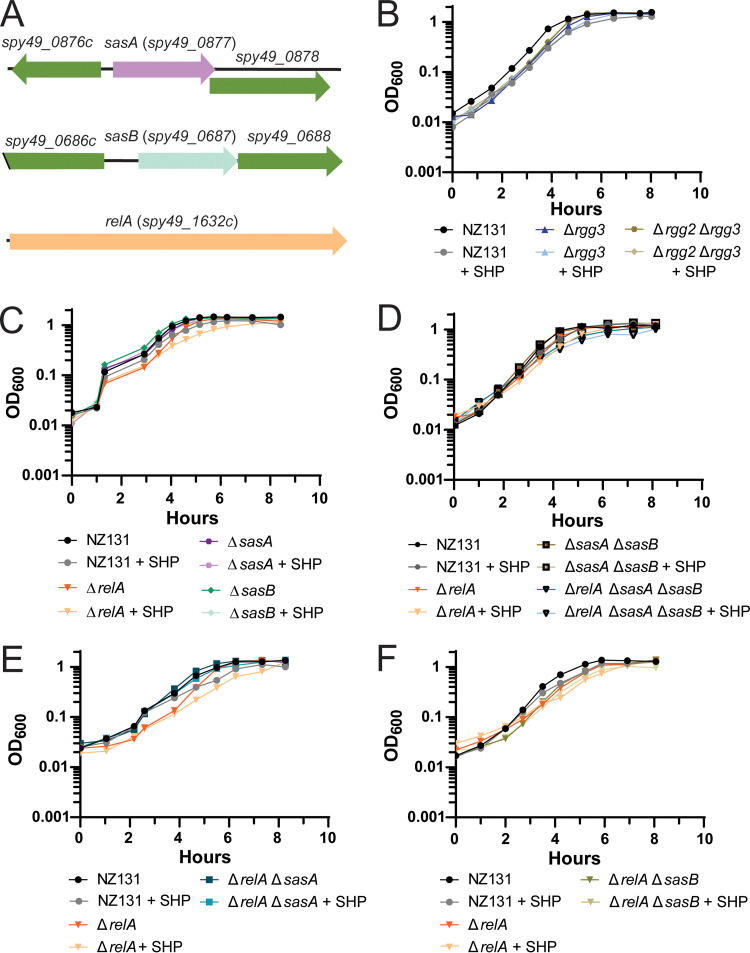
Several proteins displaying altered levels in the proteomics data set are involved in nucleotide biosynthesis, most notably those involved in stringent response or purine biosynthesis (PurA, PurH, GuaB, Spy49_0877) (Supplemental File 2). Additionally, differential regulation of transcripts related to purine or pyrimidine biosynthesis were also observed under extreme conditions (Δrgg3 + SHP versus Δrgg2 + SHP): such as spy49_0887 (xpt, xanthine phosphoribosyltransferase), spy49_0888 (xanthine permease), pyrR (master regulator of de novo pyrimidine nucleotide biosynthesis), and its according regulon. As regulatory links have been documented between some quorum sensing systems and the stringent response (26, 27), we sought to test a possible connection between the stress-response system and Rgg2/3. In prior studies, Steiner and Malke found that RelA (spy49_1632c) was responsible for the majority of (p)ppGpp accumulation under amino acid starvation in S. pyogenes, but they also identified two additional putative alarmone synthases (spy49_0877 and spy49_0687 in NZ131, spy_1125 and spy_0873 in SF370, respectively) (28, 29). RelA is predicted to have two enzymatic activities, serving as a bifunctional synthetase and hydrolase, whereas spy49_0877 and spy49_0687 are predicted to encode small alarmone synthases (SASs) that we have named sasA and sasB, respectively, based on orthologous genes in B. subtilis (30, 31). As sasA and sasB have remained untested in S. pyogenes, we took the opportunity to construct deletions of each synthase gene, as well as double and triple knockout combinations with relA (Fig. 7). Growth-rate phenotypes were evaluated for each mutant and were compared with wild-type, Δrgg2, and Δrgg3 strains upon stimulation with SHP. Doubling times of SHP-stimulated NZ131 were longer than unstimulated (36 ± 1 min unstimulated versus 47 ± 1 min stimulated) and addition of a reversed-sequence SHP peptide did not delay growth (Fig. 3E; Table S2). The Δrgg2 strain was unresponsive to SHP, growing at the same rate as unstimulated wild-type, whereas SHP-supplemented Δrgg3 grew slower when treated with SHP (38 ± 2 min unstimulated versus 52 ± 2 min stimulated), consistent with the idea that Rgg2/3 induction poses a stress to cells (Fig. 3E, Table S2). We additionally examined a Δrgg2 Δrgg3 knockout strain and found that the deletion of both abrogated the SHP-induced increase in doubling time (40 ± 3 min unstimulated versus 36 ± 3 min stimulated) (Fig. 7B). This indicates that the presence of Rgg2, or the ability to induce the Rgg2/3 QS circuit, is required for this increase in doubling time.
FIG 7.
Examination of the potential connection between the Rgg2/3 system and stringent response. All experiments were performed a minimum of three times. (A) Genetic loci of sasA (spy49_0877), sasB (spy49_0687), and relA (spy49_1632c), shown to relative scale. (B) Growth curve of wild-type NZ131, Δrgg3, and Δrgg2 Δrgg3 with or without 100 nM SHP peptide. Strains and conditions are indicated in the legend below the graph. (C) Growth curve of wild-type NZ131, ΔrelA, ΔsasA, and ΔsasB with or without 100 nM SHP peptide. Strains and conditions are indicated in the legend below the graph. (D) Growth curve of single deletion of relA, compared to double and triple deletions. Wild-type NZ131, ΔrelA, ΔsasA ΔsasB, and ΔrelA ΔsasA ΔsasB were grown with or without 100 nM SHP peptide. Strains and conditions are indicated in the legend below the graph. (E) Growth curve of single deletion of relA compared to double deletion strain. NZ131, ΔrelA, and ΔrelA ΔsasA were grown with or without 100 nM SHP peptide. Strains and conditions are indicated in the legend below the graph. (F) Growth curve of single deletion of relA compared to double relA sasB deletion. NZ131, ΔrelA, and ΔrelA ΔsasB were grown with or without 100 nM SHP peptide. Strains and conditions are indicated in the legend below the graph.
In observing growth rates of the guanosine-phosphate synthases mutants, SHP stimulation had little effect on the sasA and sasB deletions but significantly slowed the growth of the relA deletion (Fig. 7C; Table S2). Complementing relA restored its growth to a wild-type rate (Fig. S4A). We also observed that presence of a second copy of sasA or relA but not sasB abrogated the SHP-dependent increase in doubling time compared to wild-type, although a slight increase in doubling time was still observed for these strains but was nonsignificant (Table S2; Fig. S4A to C). We then examined growth of double and triple knockout strains. Although growth rates were unaffected for double-knockout strains (ΔrelA ΔsasA, ΔrelA ΔsasB, and ΔsasA ΔsasB) (Table S2; Fig. 7D to F), the triple-knockout (ΔrelA ΔsasA ΔsasB) displayed a longer doubling time for both stimulated and unstimulated conditions (62 ± 8 min unstimulated and 72 ± 5 min stimulated) (Table S2; Fig. 7D).
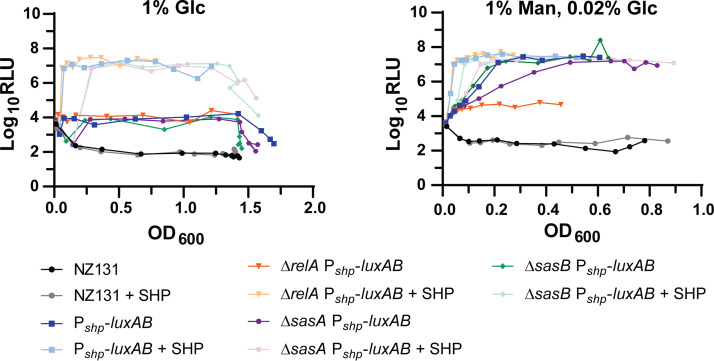
Given the compounding effects on growth rate seen in mutants stimulated with SHP, we investigated whether the two regulatory systems (QS and stringent response) had reciprocal regulatory impacts on one another. While transcriptional expression of sasA and sasB were unaffected by Rgg2/3 activity, as determined by luciferase reporters (Fig. S4E and F, unfortunately we were unable to generate a PrelA-luxAB reporter), the impact of alarmone synthase mutants on QS activity was apparent under certain conditions. Previous studies (13, 32) have demonstrated that induction of Rgg2/3 is autonomous in the presence of mannose, as can be observed using luciferase fusions to the shp promoter (Fig. 8). Mannose-dependent induction was eliminated in the relA mutant and partially incapacitated in the sasA mutant, but not affected by sasB (Fig. 8).
FIG 8.
Effect of deletion of Rel homologs on Pshp activity in different carbon sources. Luciferase assays of Pshp reporter strains in the presence of 1% glucose (1% Glc) or 1% mannose supplemented with 0.02% glucose (1% Man, 0.02% Glc). The assay in 1% Glc was performed twice with similar results, whereas the assay in 1% Man, 0.02% Glc was performed three times. Wild-type, Pshp-luxAB, ΔrelA Pshp-luxAB, ΔsasA Pshp-luxAB, and ΔsasB Pshp-luxAB were grown in the above conditions with or without 100 nM SHP peptide. Strains and conditions are indicated in the legend below the graph.
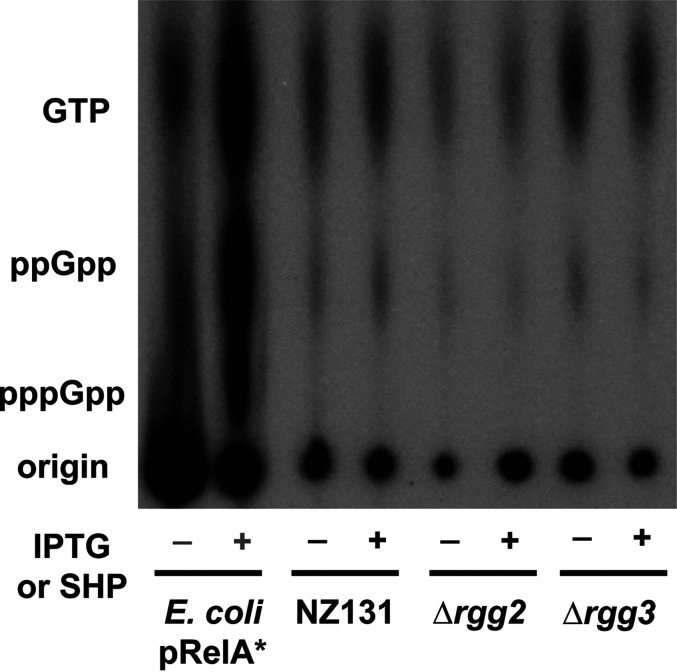
Additionally, to observe if relative GTP and (p)ppGpp levels were altered upon induction or disruption of the Rgg2/3 system, thin-layer-chromatography (TLC) was conducted following incorporation of radiolabeled 32P-orthophosphate during culture growth. We observed minimal differences in the levels of ppGpp and GTP, with overall nucleotide levels appearing to drop slightly in the Δrgg2 strain (Fig. 9). Further analysis would be required to verify any changes or this drop in the total nucleotide pool in the Δrgg2 strain. No overt induction of (p)ppGpp was observed upon SHP induction or disruption of the Rgg2/3 system (Fig. 9).
FIG 9.
Examination of the impact of SHP induction and disruption of the Rgg2/3 system on nucleotide production. Thin layer chromatography (TLC) examining nucleotide levels in cells grown in MOPS-CDM with 150 μCi/mL 32P-orthophosphate. As a control, an IPTG-inducible constitutive form of RelA from E. coli was run to provide standards for GTP, ppGpp, and pppGpp production; – indicates no IPTG added, + indicates 100 mM IPTG added. Wild-type NZ131 and derivative strains were grown with (+) or without (–) 100 nM SHP peptide. Labels beside the TLC indicate the location of GTP, ppGpp, and pppGpp as based on the IPTG induced E. coli pRelA* standard. This experiment was performed three times. Image shown was evenly adjusted for contrast and brightness.
Finally, we examined the possibility that induction of SHP pheromones could lead to depletion of branched-chain amino acids. Curiously, SHP pheromones are rich in branched-chain amino acids (SHP2: MKKVNKALLFTLIMDILIIVGG; SHP3: MKKISKFLPILILAMDIIIIVGG) and we suspected that excessive induction of the system could lead to depletion of chain amino acid (BCAA) pools via exaggerated expression and translation of SHPs. This could in turn affect the activity of the global transcriptional regulator CodY, thus modulating the activity of RelA homologs or directly inhibit (p)ppGpp hydrolysis by Rel via BCAA depletion (33–38). We reasoned that supplementation of BCAAs in SHP-stimulated cultures might be able to restore the SHP-induced growth defect in wild-type and Δrgg3 strains. However, we observed little to no effect of additional BCAA supplementation on SHP-induced cultures (Fig. S3G), indicating that depletion of BCAAs is an unlikely reason for SHP-induced growth defect.
Given these data and the finding ΔrelA was unable to respond to mannose conditions, we conclude that an indirect connection between the Rgg2/3 system and stringent response exists, possibly as an adaptation to exposure of high concentrations of the hydrophobic SHP peptide, the products produced by the Rgg2/3 operon, or to dysregulation of nutritional status monitoring.
DISCUSSION
The objective of this study was to determine the genome-wide impact on gene and protein expression as a function of signaling by the Rgg2/3 quorum sensing system in S. pyogenes. Our ongoing mechanistic understanding of gene regulation governed by two transcriptional regulators, Rgg2 and Rgg3, was further validated through analysis of responses to SHP pheromone and in use of null mutants. Stimulation of wild-type with SHP confirmed that two operons, whose transcription start sites include the Rgg2/3 bindings sequence, are the primary and direct targets of this regulatory network. This is not surprising, as previous work identified sequences required for Rgg2/3 binding are proximal to the shp2 and shp3 promoters and needed for robust regulation by the two transcriptional regulators in response to pheromone (16). The sequences occur only twice in the S. pyogenes genome, at the primary targets of the system, and are found conserved in other streptococcal species, including S. agalactiae, S. dysgalactiae, and S. pneumoniae where orthologs of Rgg2 or Rgg3 are present (39). As deletion of rgg3 removes the repressive impact on target promoters, and therefore increases production of SHP pheromones, Δrgg3 mutants express Rgg2/3 target genes in a manner like that of wild-type cultures of NZ131 responding to exogenous SHP. Conversely, mutants of rgg2 no longer have the transcriptional activator necessary for promoter induction; therefore, Δrgg2 mutants are incapable of turning the circuit on, even with exogenously supplied SHP, as the results presented herein substantiate. The equivocation of Δrgg3 as being QS-ON, and Δrgg2 as QS-OFF is upheld by results presented here.
Recently we employed Δrgg2 and Δrgg3 mutants generated in three different GAS serotypes and studied their ability to colonize the murine upper respiratory tract. Although the three serotypes colonized to different extents and for different lengths of time, in each case the Δrgg3 mutant (QS-ON) colonized as well or to higher numbers than wild-type. The Δrgg2 mutant (QS-OFF) were all poor colonizers, indicating the Rgg2/3 system contributes to colonization ability (20). We also observed that QS-ON and QS-OFF mutants held opposite abilities at inhibiting immune activation of macrophages. The QS-ON cells (Δrgg3, or wild-type stimulated with SHP) suppressed macrophage activation as reported by NF-κB activity (9). Thus, the genes encoded in the core regulon should be the focus of future studies aimed at determining the mechanisms providing for tissue colonization and immunosuppression.
Our analyses also revealed that further stimulation of Δrgg3 with exogenously supplied 100 nM SHP had a more severe impact on gene and protein expression than simply inducing the system in wild-type or in unstimulated Δrgg3. To a lesser extent, protein expression changes were also observed in stimulated Δrgg2. These changes were in addition to those observed in the core regulon. However, we caution overinterpretation of the proteomics results, as we find in our comparison of our proteomics cell surface associated and secreted fractions that cytoplasmic proteins were also observed alongside expected cell surface and secreted proteins. When we compared our proteomics results to the Wilk et al. (22) protocol which we based our proteomic samples isolation on, we found overall good agreement between their results and ours, especially for the cell surface associated protein fraction. Due to this and the observation that no overt cell lysis was occurring during growth of our samples, we conclude results obtained from mutanolysin digestion and preparation of cell surface proteomics samples should be compared with that of other publications and interpretated cautiously.
Nevertheless, we observed that the increased differential in expression and in numbers of genes and proteins, when compared with wild-type, wild-type + SHP or Δrgg2 + SHP, was primarily due to changes seen in the Δrgg3 + SHP samples, best visualized by principle component analysis (Fig. 2E and F; Fig. 3B to D; Fig. 4; Fig. S1). This extreme condition affected the expression of important virulence factors (prtF, sfbX49, slo) and proteins involved in translation and nucleotide biosynthesis, and we suspected this impact might be due to excessive levels of the hydrophobic SHP peptide (Fig. 3F) or an imbalance in Rgg2/3 signaling. The observed shift in expression of genes related to nucleotide biosynthesis/acquisition (xpt, spy49_0888, pyrR, and its regulon), increase in SasA protein amounts, and a general downshift in translation-related proteins in SHP stimulated Δrgg3 cultures led us to hypothesize an interplay between the Rgg2/3 system and stringent response. To test this hypothesis we constructed individual and combinatorial gene deletions of the three enzymes predicted or demonstrated as involved in stringent response (28). While minimal effects on Rgg2/3 signaling were seen in knockouts of sasA and sasB, the ΔrelA strain enhanced the SHP-induced growth phenotype, and more remarkably, blocked the ability of mannose to induce Rgg2/3 signaling. Our lab recently demonstrated that Rgg2/3 QS induction in mannose is connected to the phosphorylation status of HPr, specifically via its impact on Mga phosphorylation (32). In E. coli, the activity of the primary ppGpp hydrolase SpoT is impacted by HPr phosphorylation status and carbon source via its interaction with the protein Rsd (40). Although there is no homolog of rsd in the S. pyogenes genome, a similar connection between HPr phosphorylation status and RelA hydrolytic activity could be envisioned and would provide an indirect connection between mannose induction of Rgg2/3 QS and stringent response.
We also examined if any change in GTP or (p)ppGpp levels occurred upon stimulation or disruption of the Rgg2/3 system. No effect on (p)ppGpp induction was observed, and disruption of the Rgg2/3 system had minimal impact on overall nucleotide levels. These data support an indirect connection between the two systems and indicates that SHP stimulation has a minimal impact on GTP and (p)ppGpp levels under the conditions used. Finally, we also investigated if the increase in doubling time observed in SHP-stimulated cultures was a result of an increased need for BCAAs due to increased SHP biosynthesis. We reasoned this could potentially deplete the BCAA pool and result in CodY repression of various targets. However, upon supplementing with BCAAs during SHP-induction, no restoration of growth was observed (Fig. S3G), indicating this likely is not the mechanism by which SHPs alter cellular growth. A secondary hypothesis arises from the observation that the Rgg2/3 system induces a large potential biosynthetic operon. This operon has been connected to suppression of NF-κB response in macrophages (9), and we are currently investigating what product(s) or cellular modifications it might produce. It is feasible that the observed slow-down in growth could be due to increased metabolic stress or need because of induction of this operon.
Although the mechanism behind this remains unclear, these emerging signs of stress and delayed growth are consistent with the idea that excessive induction during imbalance of the Rgg2/3 system has a negative impact on cellular physiology (Fig. 10). We conclude that whatever feedback is occurring between the Rgg2/3 system and stringent response is likely indirect and seen under artificial conditions. Nevertheless, the observed impact we document should be considered in future laboratory studies in which signaling systems are subject to exogenous and genetic manipulations. In total, this work provides a clearer picture of the impact of the Rgg2/3 system on the proteome and transcriptome of S. pyogenes and provides a framework for further understanding how this QS system affects cell physiology.
FIG 10.
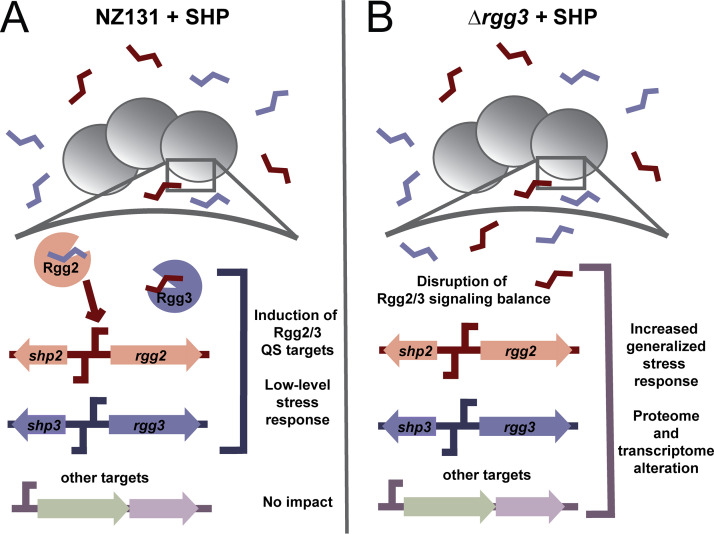
Model illustrating the impact of disruption of the Rgg2/3 QS system. Panel A illustrates what occurs when SHPs are added in a wild-type NZ131 background. Rgg3 binds SHPs and is released from target promoters, while Rgg2 binds SHPs and activates target promoters. This includes the SHP2/Rgg2 and SHP3/Rgg3 loci, as well as others such as the spy49_0450 operon and stcA. Normal Rgg2/3 QS occurs and a small number of target transcripts are affected. Panel B illustrates what occurs in the absence of Rgg3 with the addition of SHP peptide. Disruption of the system leads to an imbalance in QS activation, and this response is dependent on the presence of Rgg2. This results in a generalized stress response that alters the transcriptome and proteome of NZ131 cells.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table S1. S. pyogenes strains were derived from NZ131 or HSC5 (15, 41) and were grown on Todd-Hewitt plates (TH; BD Biosciences) with 1.4 Bacto-Agar (BD Biosciences) and 0.2% yeast (BD Biosciences), in Todd-Hewitt broth with 0.2% yeast (THY), in chemically defined medium (CDM) plus 1% glucose or 1% mannose with 0.02% glucose, or in MOPS-CDM (see next section) at 37°C. The components and recipe for CDM used were as described previously (13, 15). E. coli strains were grown on Luria-Bertani (BD Biosciences) plates with 1.4% Bacto-Agar at 37°C or in Luria-Bertani (LB) broth with shaking at 175 rpm. When required, ampicillin (100 μg/mL, E. coli only), spectinomycin (100 to 200 μg/mL), erythromycin (0.5 μg/mL for S. pyogenes, 500 μg/mL for E. coli), kanamycin (50 to 100 μg/mL), and chloramphenicol (3 to 6 μg/mL) were added to S. pyogenes or E. coli culture media.
Preparation of MOPS-CDM.
MOPS-CDM recipe was based on Kazmierczak et al. (42). In brief, MOPS-CDM replaces potassium phosphate and sodium phosphate in the original CDM recipe with 40 mM MOPS, 4 mM Tricine, 0.28 mM K2SO4, and 50 mM NaCl. All other components were kept the same as in the original CDM recipe with 1% glucose added (15).
Construction of plasmids in E. coli.
To construct plasmids one of the four following methods were used: restriction digest and ligation, Gibson assembly, a combination of restriction digest and Gibson assembly, or quick-change mutagenesis. Method used for construction of each plasmid, primers and additional information is listed in Table S1. All plasmids were propagated in DH5α cells and extracted using a GeneJET Plasmid MiniPrep Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. All PCR amplifications were carried out with Phusion High-Fidelity DNA polymerase (New England Biolabs [NEB]), and resulting fragments were purified with DNA Clean & Concentrator kit (Zymo Research). Restriction digests (NEB) were carried out according to the manufacturer’s instructions. All final plasmid constructs were electroporated into E. coli DH5α using a Bio-Rad Gene Pulser II Electroporation system (Bio-Rad) with the following parameters: 2.0 V, 200 Ω, 250 μF. Constructed plasmids were confirmed by restriction digest, PCR and/or Sanger sequencing.
To construct plasmids by restriction digest and ligation, PCR obtained linear fragments or plasmid vectors were digested with restriction enzymes as indicated in Table S1. Resulting insert and vectors were purified and ligated together using T4 DNA ligase (NEB) according to the manufacturer’s instructions. To construct plasmids by Gibson assembly, plasmid backbone and inserts were obtained from templates via PCR. Resulting fragments were purified and assembled via Gibson assembly using NEBuilder HiFi DNA Assembly MasterMix (NEB). To construct plasmids by combination of restriction digest and Gibson assembly, inserts for plasmids were obtained via PCR and assembled via Gibson assembly. Resulting inserts and plasmid vectors were digested with restriction enzymes as outlined in Table S1 and ligated together with T4 DNA ligase. For pBER18, an intermediate plasmid was required for construction. The 3′ flanking region near spy49_0877 was amplified, digested, and ligated into pFED760. The resulting intermediate plasmid and the 5′ flanking region near spy49_0877 was then PCR amplified and assembled via Gibson assembly to obtain the final pBER18 plasmid. For quick-change mutagenesis, required quick change primers as indicated in Table S1 were used, following the protocol for Phusion site-directed mutagenesis from Thermo Fisher Scientific. The resulting linearized quick-change plasmid was then digested with DpnI (NEB) to eliminate the parent plasmid and ligated using T4 DNA ligase.
Construction of S. pyogenes strains with markerless deletions.
To construct markerless deletions of relA, sasA, sasB, and combinations thereof, the following procedure based on the protocol for Gene Replacement by Allelic Exchange published by Le Breton and McIver was used (43). All strains and their derivatives are listed in Table S1. Required knockout plasmids (pBER16, pBER17, or pBER18) were propagated in E. coli DH5α and extracted using a GeneJET Plasmid MiniPrep Kit. Then, 500 ng of purified plasmids were electroporated into NZ131 using the following parameters: 1.75 V, 400 Ω, 250 μF. After electroporation, cells were allowed to recover in THY at 30°C for 2 h and were then plated on THY + Erm plates and incubated for 2 days at 30°C. Resulting colonies were patched on THY + Erm plates to confirm resistance and growth at 30°C and to confirm inability to grow at 37°C. Colonies were then grown overnight at 30°C with Erm, and additionally stored as “unintegrated plasmid” stocks with 20% glycerol at −70°C. Resulting stocks were diluted 1:100 into fresh THY + Erm and grown at 30°C for 2 h. After 2 h, cultures were shifted to 37°C and grown for 3 h to allow the plasmid to integrate into the chromosome. Cultures were consecutively diluted 1:10 into THY, plated on THY + Erm, and incubated at 37°C overnight. Individual colonies were streaked out THY + Erm plates and incubated at 37°C again. Resulting colonies were PCR confirmed for plasmid integration, and overnight cultures from these colonies were diluted 1:100 into fresh THY (without Erm). Cultures were passaged 1:100 into fresh THY 5 times, consecutively diluted 1:10 and plated on THY plates. Individual colonies were patched on THY and THY + Erm plates and confirmed for loss of ErmR at 37°C. Resulting ErmS colonies were then grown overnight at 37°C in THY and stored as final markerless knockout stocks at −70°C with 20% glycerol. Genomic DNA from resulting strains was then isolated and strains were confirmed by PCR and/or sequencing.
Construction of other S. pyogenes strains.
To construct other S. pyogenes strains in NZ131 or HSC5, 500 ng of required plasmid (Table S1) was electroporated into the parent strain as previously mentioned. Resulting strains were single-colony isolated, stored as 20% glycerol stocks at −70°C and confirmed by PCR and/or sequencing. All strains and their derivatives are listed in Table S1.
Synthesis of SHP3-C8 (SHP) and reverse SHP3-C8 (revSHP) peptides.
Synthetic peptides were purchased from ABClonal (Woburn, MA) and have been described previously (15, 16). Purities and preparations used in assays were greater than 70%. All peptides were reconstituted as 10 mM stocks in DMSO and stored in aliquots at −20°C. Subsequent dilutions for working stocks (100 μM) were made in DMSO and stored at −20°C.
Isolation of RNA from bacterial cultures for qRT-PCR and RNA-seq.
Glycerol stocks of bacterial strains at −70°C were inoculated into THY broth and incubated overnight at 37°C. The next morning, strains were diluted 1:100 into 6 mL of prewarmed CDM and grown at 37°C with OD600 observed every 45 min to 1 h with a GENESYS 30 Vis spectrophotometer (Thermo Fisher Scientific). When strains reached OD600, ~0.05 to 0.1, 100 nM SHP peptide was added to required tubes. Cultures were grown until OD600 reached ~0.8 to 1.0. At this time, 1.5 mL of cultures were harvested and centrifuged at 2,700 × g for 10 min. Supernatant was discarded and cells were resuspended in 750 μL RNA-later solution (Thermo Fisher Scientific, AM7020), and allowed to sit for 10 min at room temperature. Cells were then centrifuged at 21,130 × g for 1 min, supernatant was discarded, and cells were frozen at −70°C. RNA was extracted from frozen cell pellets using the RiboPure-Bacteria kit (Thermo Fisher Scientific, AM1925) according to manufacturer’s instructions. Samples were DNase treated with DNase I from the RiboPure-Bacteria kit according to the manufacturer’s instructions, and RNA concentration and quality was assessed via spectrophotometer (Nanodrop 1000 Spectrophotometer). Samples were additionally run on a 0.9% agarose gel via electrophoresis to verify the presence of 23S and 16S rRNA bands, to confirm samples had not degraded.
RNA quality determination, preparation of cDNA libraries for DNA sequencing, and total RNA-seq.
RNA-seq was conducted in the Northwestern University NUSeq Core Facility. RNA quantity was determined via Qubit fluorometer. Total RNA examples were also checked for fragment sizing using a Bioanalyzer 2100 (Agilent). The Illumina Stranded Total RNA Library Preparation Kit (Illumina) was used to prepare sequencing libraries from 500 ng of total RNA samples, according to manufacturer’s instructions. This procedure includes rRNA depletion with RiboZero Gold, cDNA synthesis, 3′ end adenylation, adapter ligation, and library PCR amplification and validation. lllumina HiSeq 4000 Sequencer (Illumina) was used to sequence the libraries with the production of single-end, 50-bp reads.
RNA-seq analysis.
The raw RNA sequencing data were processed using the RStudio platform (RStudio 1.4, Boston, MA), with the “Bioconductor” (44, 45) packages “Rsubread” (46) and “Rsamtools” (47) to align RNA-sequencing reads to the S. pyogenes strain NZ131 reference genome (GenBank assembly accession: GCA_000018125.1). Annotated feature counts were then produced from the aligned read data, which was subsequently filtered and normalized to generate the final count data for differential expression (DE) analysis using the “edgeR” (48, 49) and “qvalue” (50) packages from “Bioconductor.” Venn diagrams illustrating sample analysis were created using the BioVenn online platform (51).
Prediction of gene function and predicted protein localization for the core regulon of Rgg2/3 QS and significantly differentially expressed genes in RNA-seq.
The predicted gene functions of the spy49_0450 operon have been previously reported (9), aside from spy49_0458. To identify the predicted function of this gene, the protein coding sequence for spy49_0458 was input into the NCBI blastp online suite (52) with the following search parameters: database, nonredundant protein sequences (nr), excluding Streptococcus pyogenes (taxid:1314); all other parameters set to default.
To predict the protein localization, NCBI protein reference sequences for Spy49_0450 to Spy49_0460 were input into the PSORTb ver. 3.0.3 online server (https://www.psort.org/psortb/index.html) (53) with the following parameters: organism, bacteria; Gram stain, positive; output format, normal. The function and protein localization of StcA (spy49_0414c) have been previously reported (5). To predict or identify the function of genes significantly differentially regulated in RNA-seq, the protein coding sequences for each gene were input into the NCBI blastp online suite as detailed above.
Preparation of cDNA for qRT-PCR and qRT-PCR experiments.
Total RNA from S. pyogenes strains was used to prepare cDNA using the Superscript III first-strand synthesis system (Thermo Fisher Scientific) according to the manufacturer’s instructions, including treatment with RNase H. Random hexamer primers were used for cDNA synthesis. All qRT-PCR primers used are listed in Table S1. cDNA was diluted 1:10 and qRT-PCR was performed using 1× Fast SYBR green Master Mix (Thermo Fisher Scientific) with gene-specific primers according to the manufacturer's instructions and performed on a Life Technologies Viia7 real-time PCR system (Thermo Fisher Scientific). As a reference gene, spy49_0905 (gyrA) was used, as this gene has been previously reported to not have differential expression during growth (54). Nontemplate controls were included to confirm the absence of primer-dimer formation, and all RNA samples were run a minimum of one time as a control to verify absence of high levels of contaminating genomic DNA. All samples were run in triplicate biological and technical replicates on a single plate. Relative gene expression was determined using the 2-ΔΔCT method (55). Data were plotted and statistical significance between expression levels was determined via one-way ANOVA with Tukey’s Multiple Comparisons Posttest with Graph Pad Prism 9.2.0 (GraphPad Software).
Sample isolation for TMT-LC-MS/MS.
To isolate samples for TMT-LC-MS/MS, the following procedure was used and adapted from Wilk et al. (22). Overnight cultures were diluted 1:100 into 50 mL CDM and grown at 37°C until OD600 ~0.05 to 0.10, at which time 100 nM SHP peptide was added to required cultures and cells were placed back at 37°C. When cells reached late exponential/early stationary phase (OD600 ~0.8 to 1.0), cultures were transferred to 15 mL conical tubes and centrifuged at 2,700 × g for 15 min at 4°C. After centrifugation, supernatants were separated from cell pellets into new 15-mL conical tubes.
Supernatants were then filtered through a 0.2-μM pore size filter via syringe (VWR) and Pierce Protease Inhibitor Tablets, EDTA-free (Thermo Fisher Scientific, A32965) were added to 30-mL aliquots of filtered supernatants. Supernatants were placed on ice and subsequently concentrated using Amicon Ultra-4 Centrifugal Filter Units with a 10 kDA MW cutoff (Millipore-Sigma) by centrifugation continually at 2,700 × g for 15 min at 4°C until samples reached 1-mL volume. After samples were concentrated to 1 mL, samples were diluted three times with 5 mL of ice-cold PBS and concentrated back down to 1 mL each time. After final concentration to 1 mL with ice-cold PBS, samples were collected, and further concentrated to 250 μL with Amicon Ultra-0.5 Centrifugal Filter Units with a 10 kDA MW cutoff (Millipore-Sigma). Final concentrated supernatants were stored at −70°C until proceeding to further sample prep for TMT-LC-MS/MS.
Pellets from isolated samples were washed with 5 mL of ice-cold PBS and centrifuged at 2,700 × g for 15 min at 4°C. Supernatants were discarded and samples were washed twice in 5 mL of chilled TE buffer (50 mM Tris-HCl, pH 8.0; 1 mM EDTA). After the second wash, pellets were resuspended in 1 mL of ice-cold Enzyme Buffer (50 mM Tris-HCl, pH 8.0; 1 mM EDTA; 20% [wt/vol] sucrose; Pierce Protease Inhibitor Tablets [EDTA-free]) with 100 units of mutanolysin (Sigma-Aldrich, M9901). Resuspended pellets in enzyme mix were transferred to sterile microcentrifuge tubes and incubated at 37°C for 18 h with shaking (200 rpm). After digestion with mutanolysin, tubes were centrifuged at 21,130 × g for 5 min at room temperature. Supernatants (solubilized cell wall proteins) were collected and stored at −70°C until proceeding to further sample prep.
After obtaining secreted and solubilized cell wall proteins, total protein concentrations of each sample were measured using the Bio-Rad DC Protein Assay (Bio-Rad, 5000112) according to the manufacturer’s instructions. Then, 100 μg of each protein sample was submitted to Mass Spectrometry Core in Research Resources Center of University of Illinois at Chicago.
Sample preparation and digestion for TMT-LC-MS/MS.
Filter-aided sample preparation (FASP) was performed with samples submitted to the Mass Spectrometry Core. Protein samples were dissolved by 8 M urea in 0.1 M Tris-HCl, pH 8.5, and then filtered with a 0.22-μM membrane (Millipore). The flow-through was collected and transferred into a 1.5-mL MicroconYM-30 centrifugal unit (Millipore). Protein reduction, alkylation, and tryptic digestion were performed step-by-step in the centrifugal unit. After overnight digestion at 37°C, the peptides were eluted twice with 50 μL 0.1% formic acid (FA). The concentration of proteins and peptides collected in each step was measured using a Nanodrop ONE (Thermo Fisher Scientific). The digested peptides were then desalted, dried, and stored at −80°C until further use.
Peptide labeling by TMT, sample fractionation, and TMT-LC-MS/MS data processing and analysis.
For isobaric labeling, peptides per sample for 6-plex TMT were labeled using TMTsixplex Isobaric Mass Tagging Kit (Thermo Fisher Scientific, #90064) according to the manufacturers' instructions. After labeling excess reagents and detergents were removed by reversed phase solid-phase extraction (Oasis HLB C18 SPE, Waters). The samples were dried and resulting pellets stored in −80°C.
Samples were then thawed and further fractionated by high pH reversed-phase chromatography using an Agilent 1260 HPLC (Agilent) and a Waters XBridge BEH C18 column (130 Å, 3.5 μm, 4.6 × 150 mm). A total of 90 fractions were collected and combined into 10 fractions, followed by desalting using Nestgroup C18 tips (Southborough, MA). Fractionated peptides were dried and redissolved in 0.1% FA for LC-MS/MS analysis.
For LC-MS/MS, fractions were run on Thermo Fisher Orbitrap Velos Pro coupled with Agilent NanoLC system (Agilent) over a 60 min gradient on an Aglient Zorbax 300SB-C18 LC column (15 cm × 75 μm ID, Aglient). Samples were run via a 60-min linear gradient (0% to 35% acetonitrile with 0.1% FA) and data were acquired in a data dependent manner, in which MS/MS fragmentation was performed on the top 12 intense peaks of every full MS scan. RAW files were converted into .mgf files using MSConvert (56). A database search was carried out using Mascot Server (v2.6, Matrix Science) and Mascot Daemon Toolbox (Matrix Science) for peptide matches and protein searches against the Streptococcus pyogenes NZ131 genomic database UniProtKB (Proteome ID: UP000001039). For TMT reporter ion detection, the fragment mass tolerance was set to 20 ppm. Fixed modifications were defined as carbamidomethylation (57.02146) on cysteine and TMT modification (229.1629) on N-terminus and lysine. Miscleavage was set to 2. Search results from 18 runs were imported into Scaffold (Proteome Software) for quantitative analysis.
Peptide identifications were accepted if they could be established at 1% FDR probability as specified by the Peptide Prophet algorithm (57) and as protein identification probability with 99% or greater confidence as determined by Scaffold. Scaffold performed quantitative analysis, including extract of the TMT reporter ion intensities from the MS/MS, correction of isotope contamination, and normalization. As equal amounts of total protein were loaded across all TMT channels, reporter ion intensities in each of the channels were normalized by the sum of all reported ion intensities of the corresponding channel. Normalized reporter ion intensities were used for relative protein abundance calculations. Comparisons between groups were made by use of multiple-sample ANOVA test in MaxQuant 1.61 software (58). Statistical significance was set at adjusted q value < 0.05.
The final metadata from the Mass Spectrometry Core in Research Resources Center of University of Illinois at Chicago was submitted to UIC Research Informatics Core. Differential protein expression analysis was then performed using limma package (59). A one-way ANOVA was used to perform multigroup analysis, as well as pairwise comparisons. P values were adjusted for multiple testing using the false discovery rate (FDR) correction of Benjamini and Hochberg (60). Protein level differences were considered statistically significant if q values < 0.05. Venn diagrams illustrating sample analysis were created using the BioVenn online platform (51). Resulting detected proteins were also examined for their predicted cell localization using the PSORTb ver. 3.0.3 (https://www.psort.org/psortb/), with settings as the following: organism, bacteria; Gram stain, positive; output format, long format (tab delineated) (53). Additional analysis of detected proteins in cell-associated and secreted fractions was performed to compare with proteins detected in Wilk et al.’s proteomics study (22). Proteins were classified as “detected (detected, homolog detected, or detected but low signal to noise), not detected, no homolog in M1 GAS, or not annotated in Wilk et al.” Homologs were defined as proteins that could be found in the same cellular fraction in Wilk et al. (22) that had at least >50% identity and >50% coverage by examining the UniProt entry for each protein or via input of the amino acid sequence into BLASTp (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) with organism selected as “Streptococcus pyogenes M1 GAS (taxid:160490)” (52, 61). No homolog was defined as no protein found in Wilk et al. (22). with more than >50% identity and >50% coverage in M1 GAS. Not annotated in Wilk et al. was classified as a protein that was not included in the original proteome analysis by Wilk et al. These criteria were used to provide a “filtered” data set analysis based on those proteins also detected in Wilk et al. (22). This is included in Supplemental Files 2 and 3.
TMT-LC-MS/MS data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (62) with the data set identifier PXD033703.
Growth curves of strains.
Overnight cultures were diluted 1:100 into 6 mL of prewarmed CDM and grown at 37°C, with OD600 observed every 45 min to 1 h with a GENESYS 30 Vis spectrophotometer. Alternatively, glycerol starter cultures frozen at OD600 ~0.6 were washed twice with CDM and resuspended to OD600 ~0.01 in prewarmed CDM and grown/monitored as described above. When strains reached OD600 ~0.05 to 0.1, 100 nM SHP peptide was added to required tubes. For experiments to examine addition of increasing amounts of SHP peptide, 100 nM, 200 nM, or 500 nM SHP peptide were added to required tubes. For experiments to examine the effect of additional supplementation of branched-chain amino acids, ~1× CDM concentration (381 μM) of L-Ile, L-Val, and L-Leu (approximate amount normally present in CDM: 381 μM for L-Ile and L-Leu, 427 μM for L-Val) were added to required cultures at the same time. OD600 was continued to be observed every 45 min to 1 h with a GENESYS 30 Vis spectrophotometer. Resulting data from growth curves was plotted and doubling times were calculated by taking OD600s ~0.015 to 0.9 and performing a nonlinear regression analysis with exponential growth equation fitting with Graph Pad Prism 9.2.0 (GraphPad Software). Statistical significance between doubling times were determined by one-way ANOVA with Šidák’s or Dunnett’s Multiple Comparisons Posttest with Graph Pad Prism 9.2.0.
Luciferase assays.
Overnight cultures were diluted 1:100 into prewarmed CDM with either 1% glucose or 1% mannose. When cells reached OD600 ~0.05 to 0.1, 100 nM SHP peptide was added to required tubes. At each time point OD600 measurements were taken with a GENESYS 30 Vis spectrophotometer and luciferase measurements were conducted by removing 100 μL aliquots from each strain and condition and transferred to an opaque 96-well plate. Samples were exposed to decyl aldehyde (Sigma-Aldrich) fumes for 1 min and counts per second (CPS) were measured using a Veritas microplate luminometer (Turner Biosystems). Relative light units (RLU) were calculated by normalizing CPS to OD600. Data from experiments was plotted and analyzed using Graph Pad Prism 9.2.0.
Determination of relative SHP concentrations from cell supernatants.
To determine relative SHP concentrations from cell supernatants, glycerol starter cultures frozen at OD600 ~0.6 were washed twice with CDM and resuspended to OD600 ~0.01 in prewarmed CDM. When cells reached OD600 ~0.05 to 0.1, 100 nM SHP peptide was added if required. Growth was monitored every 45 min by a GENESYS 30 Vis spectrophotometer as previously mentioned, and when cells reached an OD600 ~0.8 to 1.0, cultures were harvested. Cultures were centrifuged at 2700 × g for 10 min, and supernatants were harvested and filtered via a 0.2 μM PES syringe filter (Thermo Fisher Scientific). At the same time as harvest of supernatants, a reporter strain lacking the ability to synthesize native SHP peptides (BNL206; Table S1) was thawed from a glycerol starter culture and resuspended into 3 mL CDM to OD600 ~0.1. To according tubes, 3 mL of harvested supernatants from each strain or CDM with increasing concentrations of SHP (10 nM, 50 nM, 100 nM, and 500 nM) were added to the reporter strain (final OD600 ~0.05). Cultures were then monitored for luciferase activity and growth as outlined in Luciferase assays. Data from the reporter strain with increasing concentrations of SHP was plotted as a standard curve (RLU versus SHP concentration) for each time point. Linear regression analysis in Graph Pad Prism 9.2.0 was used to determine the best-fit line to obtain the equation for the standard curve. This equation was used to determine the concentration of SHP in each supernatant, with the value being multiplied by 2 to account for one-half dilution of the supernatant with resuspended cells. The resulting concentrations for each supernatant were plotted and analyzed with GraphPad Prism 9.2.0 as a function of time. Replicate concentrations were analyzed via a one-way ANOVA with Tukey’s Multiple Comparisons Posttest to determine if concentrations were significantly different between supernatants.
CFU/mL experiments.
To determine if SHP peptide addition resulted in cell lysis, glycerol starter cultures frozen at OD600 ~0.6 were washed twice with CDM and resuspended to OD600 ~0.01 in prewarmed CDM. When cells reached OD600 ~0.05 to 0.1, 100 nM SHP peptide was added to required tubes. At this time (time point 0), cells were diluted to 10−7 in THY, and plated in triplicate for CFU/mL on THY plates. Cells were additionally plated in at 0.5-h, 1-h, and 3-h post-SHP induction. The next day, plates were enumerated for CFU/mL, and data were plotted and analyzed using GraphPad Prism 9.2.0.
Western blotting.
Samples were grown as detailed in the “Isolation of RNA from bacterial cultures for qRT-PCR and RNA-seq” section, with the following exceptions. To overnight growth of cultures, antibiotics were added if required to select for reporter plasmid propagation. The next day cells were grown in 6- to 10-mL CDM and when cells reached OD600 ~0.05 to 0.1, 100 to 120 nM SHP peptide was added if required. Cultures were then grown until OD600 reached ~0.7 to 1.0. At this time, cultures were harvested and centrifuged at 2,700 × g for 10 min. Supernatants were discarded and cells were resuspended in 250–500 μL Western blot lysis buffer (20 mM Tris-HCl, pH 7.5; 200 mM NaCl), transferred to screwcap tubes with ~150 to 250 mg 0.1-mM diameter zirconia/silica beads (BioSpec Products), and subsequently lysed for 10 min in a Mini-Beadbeater-16 (BioSpec Products). After lysis, cells were spun down at 21,130 × g for 1 min. Supernatant was harvested and transferred to 1.5-mL Eppendorf tubes on ice. Samples were subsequently measured for total protein concentration using the Bio-Rad DC Protein Assay (Bio-Rad).
After determining protein concentration, samples were diluted 1:1 with 2× Laemmli sample buffer (120 mM Tris-HCl, pH 6.8; 4% SDS, 20% Glycerol, trace bromophenol blue, 1.43 M β-mercaptoethanol) and boiled for 10 min at 100°C. Then, 15 μg of each sample was loaded on a 12% SDS-PAGE gel, along with 5 μL of Precision Plus Protein Dual Color Protein Standard (Bio-Rad, #1610374). Loaded SDS-PAGE gels were run with 1 × SDS-PAGE Running Buffer (25 mM Tris-HCl, pH 8.3; 192 mM glycine; 0.1% SDS) at 150 V for 1.5 h on a Mini-PROTEAN Tetra Vertical Electrophoresis Cell (Bio-Rad) with a PowerPac Basic Power Supply (Bio-Rad). Proteins separated by SDS-PAGE were transferred to nitrocellulose membrane (0.45 μm, Thermo Fisher Scientific, #88018) at 350 mA for 1.5 h using a Mini Trans-Blot Module (Bio-Rad) hooked up to the PowerPac Basic Power Supply. Membranes were subsequently washed twice with TBST (50 mM Tris-HCl, pH 7.5; 150 mM NaCl, 0.05% Tween 20) and blocked with TBST + 5% bovine serum albumin (BSA, A2153) for 30 min. Blots were then rinsed briefly twice with TBST and incubated with 1:4,000 rabbit anti-GFP-Tag pAb (ABClonal, #AE011) in TBST + 5% BSA overnight at 4°C. The next morning, blots were washed three times for 10 min with 10 mL TBST, and then incubated with 1:100,000 goat anti-rabbit IgG (H+L) secondary antibody conjugated to HRP (Thermo Fisher Scientific, #31460) for 1 h. Blots were washed again with 10 mL TBST three times for 10 min, and subsequently incubated with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, #34094) according to the manufacturer's instructions. Blots were then imaged using a ProteinSimple FluorChem R Imaging System (ProteinSimple, Biotechne) on the chemiluminescence setting for 1 to 5 min. After imaging, blots were briefly washed twice with 10 mL TBST and stained for protein loading with India ink (Thermo Fisher Scientific, R21518) in TBST. India ink stained blots were imaged using a G:BOX Chemi XRQ gel doc system (Syngene) and GeneSys (ver. 1.5.6.0) software on the white light setting for 175 ms. Relative loading between samples was then estimated using ImageJ (63).
Preparation of radiolabeled (p)ppGpp standards from E. coli.
E. coli (p)ppGpp standards were prepared as follows, and the procedure was adapted from Kaczmierczak et al. (42). Strain TX2737 (Table S1) was streaked to single colony isolation from frozen glycerol stocks at −70°C on LB + Amp plates, and grown overnight at 37°C. The next day, five to six colonies were suspended in 200 μL of MOPS-CDM 150 μCi/mL [32P] orthophosphate (Perkin Elmer, NEX053H001MC) and with or without 100 mM IPTG. Cells were incubated at 37°C for 30 min, and at this time, 25-μL culture aliquots were harvested and placed into tubes containing 25 μL ice-cold 13 M formic acid. Following this, three consecutive freeze thaw cycles alternating cells on dry ice versus incubation at 37°C were performed. After the final freeze-thaw, cells were spun down at 8,000 × g for 5 min. After centrifugation, supernatant was carefully removed without resuspending cellular debris and transferred to new tubes. Standards were diluted 1:1 with 13 M formic acid and stored at −20°C for up to 2 weeks.
Nonuniform labeling of (p)ppGpp from S. pyogenes and TLC.
S. pyogenes was grown and radiolabeled nucleotides were extracted as follows, adapting the procedure from Kaczmierczak et al. (42). Prestored glycerol stocks of bacterial strains at −70°C were inoculated into THY broth and incubated overnight at 37°C. The next morning, overnight cultures were centrifuged at 2,700 × g for 10 min and washed twice with 1 mL of prewarmed CDM. Strains were diluted 1:200 into 6 mL of fresh prewarmed CDM and grown at 37°C, until they reached OD600 ~0.05 to 1.0, and then 100 nM SHP was added to required cultures. Cells were grown for another 1.5 h to 2 h (when wild-type reached OD600 ~0.3 to 0.5) and cells were centrifuged again at 2,700 × g for 10 min. After centrifugation supernatants were discarded, and cells were resuspended in 1 mL of MOPS-CDM. Cultures were centrifuged again at 2,700 × g for 10 min, supernatants were discarded, and cultures were resuspended in 6 mL MOPS-CDM or MOPS-CDM containing 100 nM SHP peptide. OD600 was measured using a spectrophotometer and cells were adjusted to the same density (around OD600 ~0.05) in 1 mL volumes. Then, 92.5 μL of each culture were transferred to new Eppendorf tubes, and 7.5 μL of 2 μCi/μL (final concentration: 150 μCi/mL) [32P] orthophosphate was added to each culture. Cultures were incubated at 37°C for 30 min (according to the maximal induction of (p)ppGpp seen by Steiner and Malke [28]), and then cultures were harvested and transferred to Eppendorf tubes containing 100-μL ice-cold 13 M formic acid. Following this, three consecutive freeze thaw cycles alternating cells on dry ice versus incubation at 37°C were performed. After the final freeze-thaw, cells were spun down at 8,000 × g for 5 min. After centrifugation, supernatant was carefully removed without resuspending cellular debris and transferred to new tubes that were kept on ice or at −20°C until running on TLC.
For TLC separation, a PEI-cellulose TLC plate (Millipore-Sigma, #Z122882) prerun in distilled H2O and dried was obtained. To the TLC plate, 2 to 3 μL of each sample and E. coli standards were added. TLC was developed with 1.5 M KH2PO4, pH 3.4, removed from the chromatography chamber, and allowed to dry overnight. The next morning, the plate was exposed to a storage phosphor screen (Molecular Dynamics) for at least 4 h. The storage phosphor screen was subsequently imaged using an Amersham Typhoon scanner (GE) set to Phosphor Imaging scanning mode with the following settings: 4000 sensitivity, 200 μM pxl size. Resulting images were contrast and brightness adjusted using ImageJ (63).
ACKNOWLEDGMENTS
We acknowledge the following research cores for their help in data acquisition: RNA-seq library preparation and data collection was supported by the Northwestern University NUSeq Core Facility; TMT-LC-MS/MS data acquisition and partial analysis was performed by the Mass Spectrometry Core in the Research Resources Center of University of Illinois at Chicago; and additional bioinformatics analysis of TMT-LC-MS/MS was performed by the UIC Research Informatics Core, supported in part by NCATS through Grant UL1TR002003. We also thank M.E. Winkler and H.T. Tsui for the kind gift of the E. coli strain TX2737, and S. Mankin and N. Vázquez-Laslop for the gift of pY71-sfGFP. Additionally, we thank the members of the Federle lab for helpful discussions and proofreading of the manuscript.
This study was supported by the National Institutes of Health (R01-AI091779 to M.J.F. and 5K12 GM139186/NIGMS/NIH to B.E.R.) and the DoD’s Science, Mathematics and Research for Transformation (SMART) Scholarship for Service Program to C.M.A.
Footnotes
Supplemental material is available online only.
Contributor Information
Michael J. Federle, Email: mfederle@uic.edu.
Tina M. Henkin, Ohio State University
REFERENCES
- 1.Luo R, Sickler J, Vahidnia F, Lee YC, Frogner B, Thompson M. 2019. Diagnosis and management of group A Streptococcal pharyngitis in the United States, 2011–2015. BMC Infect Dis 19. 10.1186/s12879-019-3835-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sims Sanyahumbi A, Colquhoun S, Wyber R, Carapetis JF. 2016. Global disease burden of group A Streptococcus. In Ferretti JJ, Stevens DL, Fischetti VA. (ed.), Streptococcus pyogenes: basic biology to clinical manifestations [Internet]. University of Oklahoma Health Sciences Center, Oklahoma City, OK. [PubMed] [Google Scholar]
- 3.Brouwer S, Barnett TC, Rivera-Hernandez T, Rohde M, Walker MJ. 2016. Streptococcus pyogenes adhesion and colonization. FEBS Lett 590:3739–3757. 10.1002/1873-3468.12254. [DOI] [PubMed] [Google Scholar]
- 4.Wilkening Rv, Federle MJ. 2017. Evolutionary constraints shaping Streptococcus pyogenes–host interactions. Trends Microbiol 25:562–572. 10.1016/j.tim.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gogos A, Jimenez JC, Chang JC, Wilkening Rv, Federle MJ. 2018. A quorum sensing-regulated protein binds cell wall components and enhances lysozyme resistance in Streptococcus pyogenes. J Bacteriol 200. 10.1128/JB.00701-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makthal N, Do H, VanderWal AR, Olsen RJ, Musser JM, Kumaraswami M. 2018. Signaling by a conserved quorum sensing pathway contributes to growth ex vivo and oropharyngeal colonization of human pathogen group A streptococcus. Infect Immun 86:e00169-18. 10.1128/IAI.00169-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hertzog BB, Kaufman Y, Biswas D, Ravins M, Ambalavanan P, Wiener R, Angeli V, Chen SL, Hanski E. 2018. A sub-population of group A Streptococcus elicits a population-wide production of bacteriocins to establish dominance in the host. Cell Host Microbe 23:P312–P323. 10.1016/j.chom.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Do H, Makthal N, VanderWal AR, Saavedra MO, Olsen RJ, Musser JM, Kumaraswami M. 2019. Environmental pH and peptide signaling control virulence of Streptococcus pyogenes via a quorum-sensing pathway. Nat Commun 10:2586. 10.1038/s41467-019-10556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahbari KM, Chang JC, Federle MJ. 2021. A Streptococcus quorum sensing system enables suppression of innate immunity. mBio 12:e03400-20. 10.1128/mBio.03400-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neiditch MB, Capodagli GC, Prehna G, Federle MJ. 2017. Genetic and structural analyses of RRNPP intercellular peptide signaling of Gram-positive bacteria. Annu Rev Genet 51:311–333. 10.1146/annurev-genet-120116-023507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dmitriev Av, McDowell EJ, Kappeler Kv, Chaussee MA, Rieck LD, Chaussee MS. 2006. The Rgg regulator of Streptococcus pyogenes influences utilization of nonglucose carbohydrates, prophage induction, and expression of the NAD-glycohydrolase virulence operon. J Bacteriol 188:7230–7241. 10.1128/JB.00877-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jimenez JC, Federle MJ. 2014. Quorum sensing in group A Streptococcus. Front Cell Infect Microbiol 4:127. 10.3389/fcimb.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang JC, Jimenez JC, Federle MJ. 2015. Induction of a quorum sensing pathway by environmental signals enhances group A streptococcal resistance to lysozyme. Mol Microbiol 97:1097–1113. 10.1111/mmi.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do H, Makthal N, VanderWal AR, Rettel M, Savitski MM, Peschek N, Papenfort K, Olsen RJ, Musser JM, Kumaraswami M. 2017. Leaderless secreted peptide signaling molecule alters global gene expression and increases virulence of a human bacterial pathogen. Proc Natl Acad Sci USA 114:E8498–E8507. 10.1073/pnas.1705972114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. 2011. Two group a streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog 7:e1002190. 10.1371/journal.ppat.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lasarre B, Aggarwal C, Federle MJ. 2012. Antagonistic Rgg regulators mediate quorum sensing via competitive DNA binding in Streptococcus pyogenes. mBio 3:e00333-12. 10.1128/mBio.00333-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkening Rv, Chang JC, Federle MJ. 2016. PepO, a CovRS-controlled endopeptidase, disrupts Streptococcus pyogenes quorum sensing. Mol Microbiol 99:71–87. 10.1111/mmi.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang JC, Federle MJ. 2016. PptAB exports Rgg quorum-sensing peptides in Streptococcus. PLoS One 11:e0168461. 10.1371/journal.pone.0168461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Capodagli GC, Tylor KM, Kaelber JT, Petrou VI, Federle MJ. 2020. Structure–function studies of Rgg binding to pheromones and target promoters reveal a model of transcription factor interplay. Proc Natl Acad Sci USA 9:202008427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gogos A, Federle MJ. 2020. Colonization of the murine oropharynx by Streptococcus pyogenes is governed by the Rgg2/3 quorum sensing system. Infect Immun 88:e00464-20. 10.1128/IAI.00464-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang JC, Wilkening Rv, Rahbari KM, Federle MJ. 2022. Quorum Sensing Regulation of a Major Facilitator Superfamily Transporter Affects Multiple Streptococcal Virulence Factors. J Bacteriol 204. 10.1128/jb.00176-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilk L, Happonen L, Malmström J, Herwald H. 2018. Comprehensive mass spectrometric survey of Streptococcus pyogenes subcellular proteomes. J Proteome Res 17:600–617. 10.1021/acs.jproteome.7b00701. [DOI] [PubMed] [Google Scholar]
- 23.Wilk L. 2020. Protocol for proteome analysis of group A Streptococcus. Methods Mol Biol 2136:135–144. 10.1007/978-1-0716-0467-0_9. [DOI] [PubMed] [Google Scholar]
- 24.Vogel C, Marcotte EM. 2012. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13:227–232. 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buccitelli C, Selbach M. 2020. mRNAs, proteins and the emerging principles of gene expression control. Nat Rev Genet 21:630–644. 10.1038/s41576-020-0258-4. [DOI] [PubMed] [Google Scholar]
- 26.van Delden C, Comte R, Bally AM. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J Bacteriol 183:5376–5384. 10.1128/JB.183.18.5376-5384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaspar J, Kim JN, Ahn SJ, Burne RA. 2016. An essential role for (p)ppGpp in the integration of stress tolerance, peptide signaling, and competence development in Streptococcus mutans. Front Microbiol 10.3389/fmicb.2016.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steiner K, Malke H. 2000. Life in protein-rich environments: the relA-independent response of Streptococcus pyogenes to amino acid starvation. Mol Microbiol 38:1004–1016. 10.1046/j.1365-2958.2000.02203.x. [DOI] [PubMed] [Google Scholar]
- 29.Steiner K, Malke H. 2001. relA-independent amino acid starvation response network of Streptococcus pyogenes. J Bacteriol 183:7354–7364. 10.1128/JB.183.24.7354-7364.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson BW, Fung DK, Wang JD. 2021. Regulatory themes and variations by the stress-signaling nucleotide alarmones (p)ppGpp in bacteria. Annu Rev Genet 55:115–133. 10.1146/annurev-genet-021821-025827. [DOI] [PubMed] [Google Scholar]
- 31.Fung DK, Yang J, Stevenson DM, Amador-Noguez D, Wang JD. 2020. Small Alarmone synthetase SasA expression leads to concomitant accumulation of pGpp, ppApp, and AppppA in Bacillus subtilis. Front Microbiol 11:2083. 10.3389/fmicb.2020.02083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woo JKK, McIver KS, Federle MJ. 2022. Carbon catabolite repression on the Rgg2/3 quorum sensing system in Streptococcus pyogenes is mediated by PTS Man and Mga. Mol Microbiol 117:525–538. 10.1111/mmi.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geiger T, Wolz C. 2014. Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. Int J Med Microbiol 304:150–155. 10.1016/j.ijmm.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 34.Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol 8:203–207. 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Kaiser JC, Heinrichs DE. 2018. Branching out: alterations in bacterial physiology and virulence due to branched-chain amino acid deprivation. mBio 9:e01188-18. 10.1128/mBio.01188-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hendriksen WT, Bootsma HJ, Estevão S, Hoogenboezem T, de Jong A, de Groot R, Kuipers OP, Hermans PWM. 2008. CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J Bacteriol 190:590–601. 10.1128/JB.00917-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemos JA, Nascimento MM, Lin VK, Abranches J, Burne RA. 2008. Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J Bacteriol 190:5291–5299. 10.1128/JB.00288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang M, Bauer CE. 2018. Regulation of stringent factor by branched-chain amino acids. Proc Natl Acad Sci USA 115:6446–6451. 10.1073/pnas.1803220115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cook LC, LaSarre B, Federle MJ. 2013. Interspecies communication among commensal and pathogenic streptococci. mBio 4:e00382-13. 10.1128/mBio.00382-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J-W, Park Y-H, Seok Y-J. 2018. Rsd balances (p)ppGpp level by stimulating the hydrolase activity of SpoT during carbon source downshift in Escherichia coli. Proc Natl Acad Sci USA 115:E6845–E6854. 10.1073/pnas.1722514115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Port GC, Paluscio E, Caparon MG. 2013. Complete genome sequence of emm type 14 Streptococcus pyogenes strain HSC5. Genome Announc 1:e00612-13. 10.1128/genomeA.00612-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kazmierczak KM, Wayne KJ, Rechtsteiner A, Winkler ME. 2009. Roles of relSpn in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol Microbiol 82:590–611. 10.1111/j.1365-2958.2009.06669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.le Breton Y, McIver KS. 2013. Genetic manipulation of Streptococcus pyogenes (the group A Streptococcus, GAS. Curr Protoc Microbiol 30:9D.3.1–9D.3.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oleś AK, Pagès H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. 2015. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12:115–121. 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao Y, Smyth GK, Shi W. 2019. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res 47:e47–e47. 10.1093/nar/gkz114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morgan M, Pages H, Obenchain V, Hayden N. 2021. Rsamtools: binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. 2.8.0. R package.
- 48.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40:4288–4297. 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storey J, Bass A, Dabney A, Robinson D. 2021. qvalue: Q-value estimation for false discovery rate control. 2.24.0. R package.
- 51.Hulsen T, de Vlieg J, Alkema W. 2008. BioVenn – a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9:488. 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 53.Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, Dao P, Sahinalp SC, Ester M, Foster LJ, Brinkman FSL. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615. 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaussee MS, Watson RO, Smoot JC, Musser JM. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect Immun 69:822–831. 10.1128/IAI.69.2.822-831.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 56.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak M-Y, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. 2012. A cross-platform toolkit for mass spectrometry and proteomics. Nat Biotechnol 30:918–920. 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74:5383–5392. 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 58.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 59.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc: Series B (Methodological) 57:289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 61.Bateman A, Martin M-J, Orchard S, Magrane M, Agivetova R, Ahmad S, Alpi E, Bowler-Barnett EH, Britto R, Bursteinas B, Bye-A-Jee H, Coetzee R, Cukura A, da Silva A, Denny P, Dogan T, Ebenezer T, Fan J, Castro LG, Garmiri P, Georghiou G, Gonzales L, Hatton-Ellis E, Hussein A, Ignatchenko A, Insana G, Ishtiaq R, Jokinen P, Joshi V, Jyothi D, Lock A, Lopez R, Luciani A, Luo J, Lussi Y, MacDougall A, Madeira F, Mahmoudy M, Menchi M, Mishra A, Moulang K, Nightingale A, Oliveira CS, Pundir S, Qi G, Raj S, Rice D, Lopez MR, Saidi R, Sampson J, The UniProt Consortium , et al. 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49:D480–D489. 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yılmaz Ş, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47:D442–D450. 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4 and Tables S1 and S2. Download jb.00175-22-s0001.pdf, PDF file, 1.0 MB (1MB, pdf)
Data Set S3. Download jb.00175-22-s0002.xlsx, XLSX file, 10.8 MB (10.8MB, xlsx)
Data Set S1. Download jb.00175-22-s0003.xlsx, XLSX file, 1.8 MB (1.8MB, xlsx)
Data Set S2. Download jb.00175-22-s0004.xlsx, XLSX file, 0.2 MB (256.4KB, xlsx)