ABSTRACT
In defined media supplemented with single carbon sources, Mycobacterium tuberculosis (Mtb) exhibits carbon source specific growth restriction. When supplied with glycerol as the sole carbon source at pH 5.7, Mtb establishes a metabolically active state of nonreplicating persistence known as acid growth arrest. We hypothesized that acid growth arrest on glycerol is not a metabolic restriction, but rather an adaptive response. To test this hypothesis, we selected for and identified several Mtb mutants that could grow under these restrictive conditions. All mutations were mapped to the ppe51 gene and resulted in variants with 3 different amino acid substitutions– S211R, E215K, and A228D. Expression of the ppe51 variants in Mtb promoted growth at acidic pH showing that the mutant alleles are sufficient to cause the dominant gain-of-function, Enhanced Acid Growth (EAG) phenotype. Testing growth on other single carbon sources showed the PPE51 variants specifically enhanced growth on glycerol, suggesting PPE51 plays a role in glycerol uptake. Using radiolabeled glycerol, enhanced glycerol uptake was observed in Mtb expressing the PPE51 (S211R) variant, with glycerol overaccumulation in triacylglycerol. Notably, the EAG phenotype is deleterious for growth in macrophages, where the mutants have selectively faster replication and reduced survival in activated macrophages compared to resting macrophages. Recombinant PPE51 protein exhibited differential thermostability in the wild type (WT) or S211R variants in the presence of glycerol, supporting the model that EAG substitutions alter PPE51-glycerol interactions. Together, these findings support that PPE51 variants selectively promote glycerol uptake and that slowed growth at acidic pH is an important adaptive mechanism required for macrophage pathogenesis.
IMPORTANCE It is puzzling why Mycobacterium tuberculosis (Mtb) cannot grow on glycerol at acidic pH, as it has a carbon source and oxygen, everything it needs to grow. In this study, we found that Mtb limits uptake of glycerol at acidic pH to restrict its growth and that mutations in ppe51 promote uptake of glycerol at acidic pH and enable growth. That is, Mtb can grow well at acidic pH on glycerol, but has adapted instead to stop growth. Notably, ppe51 variants exhibit enhanced replication and reduced survival in activated macrophages, supporting a role for pH-dependent slowed growth during macrophage pathogenesis.
KEYWORDS: Mycobacterium tuberculosis, genetic selection, growth regulation, nutrient acquisition, pathogenesis
INTRODUCTION
During infection, Mycobacterium tuberculosis (Mtb) senses and adapts to a variety of immune cues including hypoxia (1, 2), nutrient limitation (3, 4), pH changes (5), and nitrosative and oxidative stress (6). Exposure to these stresses can promote Mtb to establish slowed growth or a non-replicating persistent (NRP) state. NRP bacteria are tolerant to immune and antibiotic-mediated killing (7–9), therefore understanding mechanisms underlying NRP may promote new methods to shorten the course of tuberculosis (TB) therapy.
Following macrophage infection, Mtb senses the mildly acidic pH of the phagosome and broadly remodels its gene expression (10). Adaptation to acidic pH includes the induction of the PhoPR regulon, induction of ESX-1 secretion, and remodeling of central metabolism and cell envelope lipids (11). Defects in adaptation to acidic pH reduce Mtb virulence in macrophages and animals (12–15), therefore, pH-dependent adaptation is required for Mtb virulence.
Previous studies conducted by our lab sought to understand the interplay of acidic pH and Mtb central metabolism. We observed that Mtb exhibits selectivity of the carbon sources on which it can growth at pH 5.7 relative to pH 7.0. For example, Mtb incubated at acidic pH with glycerol as a sole carbon source is restricted for growth and establishes a viable, metabolically active state of NRP called acid growth arrest (11, 16, 17). Acid growth arrest is observed on a variety of other carbon sources associated with glycolysis and tricarboxylic acid (TCA) cycle. Interestingly, Mtb can resuscitate its growth at acidic pH by addition of specific carbon sources, such as pyruvate, acetate, oxaloacetate (OA) and cholesterol, which function at the intersection of glycolysis and the TCA cycle (also known as the anaplerotic node) (17). This discovery suggests that the anaplerotic node is the location of a pH-dependent metabolic switch that may promote Mtb growth on permissive carbon sources during pathogenesis at acidic pH, and that metabolic remodeling is required for pH adaptation (11).
It is puzzling that Mtb cannot grow at acidic pH on specific carbon sources, as Mtb is provided with oxygen as a terminal electron acceptor and a carbon source that is well utilized at pH 7.0. Thus, acid growth arrest is different from other NRP models, where the bacterium is missing a key factor required for growth (e.g., oxygen or nutrients in the hypoxia or starvation models of NRP, respectively). Therefore, we hypothesized that acid growth arrest is not an actual restriction on growth, but an adaptation by the bacterium to slow and arrest its growth. In a previous study, our lab sought to identify genes regulating acid growth arrest by selecting for mutants incapable of arresting their growth on minimal medium agar plates, buffered to pH 5.7 with glycerol as the carbon source. From our selection efforts, novel missense mutations were identified in ppe51 (H37Rv annotated Mtb gene, Rv3136) exhibiting what we refer to as an Enhanced Acid Growth (EAG) phenotype (16). PPE51 is part of the PE and PPE mycobacterial protein family. Named for their N-terminus motifs Pro-Glu (PE) and Pro-Pro-Glu (PPE), most of these proteins have remained largely enigmatic in their functional roles (18, 19). However, a growing body of literature in recent years has assigned diverse putative functional roles for PE and PPE proteins including immune evasion (19–23), calcium binding (24), iron utilization (25, 26), Mg2+ and PO32− transport (27), fibronectin binding (28), and lipase activity (29, 30).
Mounting evidence suggests that some PE and PPE proteins may play important roles in Mtb nutrient acquisition. Examination of pe and ppe evolution reveals an expansion of this protein family corresponding with Type VII or ESX secretion systems, where it is thought that ancestral pe and ppe genes inserted into an esx gene cluster and expanded alongside this secretion system during subsequent gene duplication events (21, 31, 32). Secretion via ESX provides a route for PE and PPE proteins to access the cell surface and nutrients in the host cell milieu, which is supported by high-throughput proteomic evidence showing direct surface localization of PE and PPE proteins (25, 27, 33–35). Mtb contains five ESX secretion systems (36), with ESX-5 contributing to the majority of PE and PPE secretion (31, 33, 37). Furthermore, ESX-5 and its cognate PE and PPE proteins have been implicated in the uptake of fatty acids and possibly the utilization of other nutrient substrates (33). ESX-3-mediated PE and PPE proteins are thought to play a role in iron acquisition, whereby they have been shown to be directly involved in mycobactin-mediated iron uptake and heme uptake (25, 38, 39). Taken together, these results provide direct examples of Mtb acquiring and utilizing host resources through secretion of PE and PPE proteins.
Based on these findings showing a role for PPE proteins in transport and that PPE51 EAG variants could grow on glycerol, we hypothesized in our 2018 study (16), “that these amino acid substitutions may increase Mtb growth by modulating mycomembrane permeability, possibly by modulating the channel size or specificity of PPE51, which may function as a porin to enhance access to glycerol or other nutrients at acidic pH”. The goal of this study was to test this hypothesis and further define the role of PPE51 in glycerol acquisition and pathogenesis. Notably, concurrent with this study, recently published studies confirmed the hypothesis that PPE51 is an exported cell surface-associated protein and linked to the nutrient acquisition of glycolytic carbon sources (27, 40, 41). Furthermore, a recent study by Babu Sait et al. showed that PPE51 is required for mediating the transport of trehalose across the mycomembrane, further corroborating its role as an important mycomembrane transporter (42). Here, we show that in a saturating genetic selection only 3 PPE51 variants, S211R, E215K and A228D were isolated as mutants with the EAG phenotype. The PPE51 variants specifically promoted growth at pH 5.7 on glycerol and no other tested carbon sources, supporting the notion that these substitutions selectively promote glycerol utilization. Radiolabeling studies show that the S211R variant has enhanced uptake of glycerol and accumulation of triacylglycerol (TAG), showing that the variants promote glycerol uptake. Differential thermal stability of WT versus S211R variant PPE51 proteins in the presence of glycerol, support the variant has direct and differential interactions with glycerol. Structural modeling supports that PPE51 forms a structure homologous with bacterial nutrient transporters, with the variants altering the predicted ligand specificity. These data are consistent with a model where PPE51 promotes uptake of glycerol across the mycomembrane by acting like a porin and that variants alter the conformation to enhance glycerol uptake. ppe51 EAG variants exhibit enhanced replication and reduced survival in activated macrophages, supporting a role for pH-dependent slowed growth during macrophage pathogenesis.
RESULTS
All isolated EAG mutants have spontaneous mutations in ppe51.
During acid growth arrest in minimal media, Mtb is provided all required nutrients including a metabolically utilized carbon source and a terminal electron acceptor. This suggests that acid growth arrest is not due to a physiological limitation presented by the growth environment but rather is a regulated process whereby Mtb adapts to its acidic environment. A previously published genetic selection tested this hypothesis using a CDC1551 transposon mutant library containing >100,000 independent mutants (16). The library was plated on MMAT defined minimal media agar with glycerol at pH 5.7 and selected for 98 transposon (Tn) mutants and 2 spontaneous mutants that could grow under these normally restrictive conditions (16). These mutants were isolated and confirmed to exhibit the EAG phenotype based on their ability to grow well compared to WT Mtb at pH 5.7 in liquid MMAT supplemented with glycerol (16). Interestingly, complementation attempts with the Tn mutants did not restore growth arrest, and whole genome sequencing identified spontaneous mutations in ppe51 in both Tn and sponteneous mutant backgrounds (16). To repeat a saturating selection, in the absence of Tn mutagenesis, a second genetic selection was performed on MMAT agar buffered to pH 5.7 with glycerol, using a larger bacterial population (4 × 109 bacteria) in the Erdman Mtb strain. From the WT Erdman selection, 98 spontaneous EAG mutants were isolated of which 52 were colony-purified and confirmed for enhanced growth under acidic conditions in liquid MMAT containing glycerol (Fig. 1A). The mutants exhibited an up to ~4-fold increase in growth compared to WT Erdman which exhibited complete growth arrest (Fig. 1A). Of these mutants, 23 were selected for whole genome sequencing. Remarkably, all isolates had single nucleotide polymorphisms (SNPs) mapping to the ppe51 gene (Table 1). All mutations were non-synonymous, were centrally located within a 50 bp region on the ppe51 gene and caused amino acid substitutions S211R, E215K, or A228D. The S211R and A228D variants were also identified in the prior Tn mutant CDC1551 selection, with E215K being a novel mutation found in the new Erdman selection.
FIG 1.
PPE51 variants drive the EAG phenotype and exhibit phenotypic and carbon source-dependent growth differences. (A) Mutants with the EAG phenotype show distinct clustering of variant type based on relative growth. (B) Growth curve of WT Mtb (Erdman and CDC1551 strains) expressing ppe51 and EAG variants. Growth of pVV16 empty vector and pVV-ppe51-WT were compared to WT carrying the expression constructs of the mutant alleles (S211R, A228D, and E215K) in minimal media (pH 5.7 + glycerol). Expression of mutant alleles in WT Mtb results in significantly enhanced growth under acidic conditions. This experiment was repeated three times in duplicate. Error bars indicate standard deviation. (C) Statistical analysis of growth differences between spontaneous EAG variant strains containing S211R or A228D was performed using an unpaired t test (***, P < 0.001) with Welch’s correction.
TABLE 1.
Selection of ppe51 mutants with enhanced growth on glycerol at acidic pH
| Plate no. | Mutant strain | SNP location | Quality score | Nucleotide change | Amino acid change in PPE51 | Relative growth |
|---|---|---|---|---|---|---|
| 1 | eag1.7 | 3497961 | 5478 | GCC→GAC | A228D | 1.5 |
| eag1.8 | 3497911 | 6929 | AGC→AGA | S211R | 3.2 | |
| eag1.12 | 3497961 | 7373 | GCC→GAC | A228D | 1.0 | |
| eag1.14 | 3497911 | 6328 | AGC→AGA | S211R | 3.0 | |
| eag1.33 | 3497911 | 5270 | AGC→AGA | S211R | 3.3 | |
| eag1.34 | 3497911 | 5270 | AGC→AGA | S211R | 3.4 | |
| 2 | eag2.1 | 3497961 | 4417 | GCC→GAC | A228D | 2.0 |
| eag2.2 | 3497961 | 6803 | GCC→GAC | A228D | 2.5 | |
| eag2.3 | 3497911 | 5205 | AGC→AGA | S211R | 3.8 | |
| eag2.6 | 3497911 | 5976 | AGC→AGA | S211R | 2.7 | |
| eag2.8 | 3497961 | 4270 | GCC→GAC | A228D | 1.6 | |
| eag2.14 | 3497921 | 3442 | GAG→AAG | E215K | 0.9 | |
| eag2.16 | 3497911 | 4575 | AGC→AGG | S211R | 3.6 | |
| 3 | eag3.2 | 3497961 | 4140 | GCC→GAC | A228D | 1.5 |
| eag3.4 | 3497961 | 3292 | GCC→GAC | A228D | 2.1 | |
| eag3.9 | 3497911 | 3645 | AGC→AGG | S211R | 2.5 | |
| eag3.15 | 3497911 | 3784 | AGC→AGA | S211R | 3.6 | |
| eag3.23 | 3497911 | 4609 | AGC→AGG | S211R | 3.0 | |
| 4 | eag4.5 | 3497961 | 5153 | GCC→GAC | A228D | 1.7 |
| eag4.7 | 3497911 | 5107 | AGC→AGG | S211R | 2.5 | |
| eag4.12 | 3497911 | 5179 | AGC→AGG | S211R | 1.2 | |
| eag4.21 | 3497961 | 3958 | GCC→GAC | A228D | 1.9 | |
| eag4.24 | 3497911 | 5517 | AGC→AGG | S211R | 2.5 |
ppe51 mutations are sufficient to overcome growth arrest.
Given that ppe51 variants exhibit enhanced growth at acidic pH, we investigated the function of the variant alleles in the presence of the WT ppe51 allele. Expression constructs of WT or mutant ppe51 were transformed into WT CDC1551 or WT Erdman Mtb strains. Expression strains were grown in MMAT at pH 5.7 with glycerol as a carbon source. Expression of ppe51-S211R and ppe51-A228D variants in WT Mtb resulted in significantly enhanced growth under acidic conditions (Fig. 1B). In contrast, expression of WT ppe51 and the empty vector exhibited complete growth arrest at pH 5.7. Expression of the E215K allele did not overcome growth arrest under these growth conditions. Additionally, all expression strains grew equally well at pH 7.0 (Fig. S1A), showing that the observed growth phenotype is pH specific. Although the growth phenotype with A228D expression resulted in enhanced growth at acidic pH, it grew at a slower rate compared to S211R expression strains in both CDC1551 and Erdman backgrounds. We examined individual variant alleles from the selection (Fig. 1A) and observed that S211R variants significantly grouped together at a higher rate of growth compared to A228D and E215K (Fig. 1C). Together, these results demonstrate that the S221R and A228D variants confer a dominant, gain-of-function growth phenotype, and specific mutations are associated with differential strength of the phenotype.
ppe51 variants selectively promote growth on glycerol.
Based on the enhanced growth phenotype that the variants exhibit at acidic pH, we hypothesized that this phenotype may be due to ppe51 variants modulating mycomembrane permeability, resulting in enhanced nutrient uptake. To test this hypothesis, we conducted an Ethidium Bromide (EtBr) assay looking at permeability with WT Mtb expression constructs (empty vector, WT, and S211R) in both CDC1551 and Erdman backgrounds. With the EtBr assay, we did not observe differences in the rate of uptake between the WT and EAG expression strains in either CDC1551 or Erdman (Fig. S1B), suggesting that the growth phenotype is not due to a general increase in permeability. We then hypothesized that the growth phenotype may be due to nutrient-specific uptake. We explored this possibility by growing CDC1551 and Erdman expression strains (empty vector, ppe51 and ppe51-S211R) in liquid minimal media (pH 5.7) in the presence of various growth-permissive (e.g., pyruvate, acetate, cholesterol) and non-permissive (e.g., glucose, glycerol, propionate) carbon sources (17). After 20 days, we found that enhanced acid growth was only observed with ppe51-S211R in the presence of 10 mM glycerol, a normally non-permissive carbon source at pH 5.7, in both CDC1551 and Erdman (Fig. 2, Fig. S1C, and Fig. S2). All expression strains exhibited enhanced growth on permissive carbon sources in both Mtb backgrounds, which is consistent with what has been previously published (17). Notably, the ppe51 S211R variant specifically promotes growth on glycerol, but not glucose, another proposed nutrient associated with PPE51-dependent uptake (27, 40), demonstrating this variant is selective for glycerol. We also observed slightly enhanced growth for both the ppe51 and ppe51-S211R expressing strains on cholesterol, in both CDC1551 and Erdman (Fig. 2, Fig. S1C, and Fig. S2), supporting potential for ppe51 playing a role in a cholesterol-dependent physiology.
FIG 2.

Analysis of the ppe51 S211R variant growth on various carbon sources. CDC1551 expression strains were grown in MMAT medium (pH 5.7) in the presence of various growth-permissive (e.g., pyruvate, acetate, OA) and non-permissive (e.g., glucose, propionate, lactate) carbon sources. ppe51-S211R (pink bars) growth is carbon source specific and only exhibits enhanced growth on glycerol, a normally non-permissive carbon source at pH 5.7. Growth on permissive carbon sources is not impacted by ppe51-S211R. The horizontal dotted line indicates the starting density of 0.05 OD600. Similar results were observed in Erdman (Fig. S1C). Mean ± SD are shown in the bar graph.
ppe51 is not required for survival during acid growth arrest.
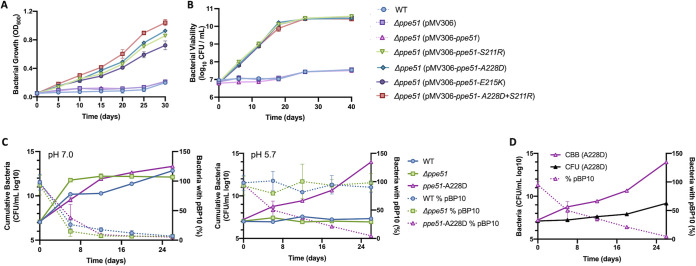
Transcriptional profiling studies previously conducted show that ppe51 is significantly induced during acid growth arrest (17). We hypothesized that ppe51 may be required for Mtb to promote survival when exposed to acid growth arresting conditions. To test this hypothesis, a Δppe51 disruption mutant strain was generated in both CDC1551 and Erdman Mtb using the mycobacteria-specific ORBIT system (43). Successful disruption of ppe51 was confirmed by sequencing the oriE and HygC junction sites of Δppe51, PCR amplification of the entire disrupted region, and qRT-PCR. Complementation constructs containing the native ppe51 promoter were introduced into Δppe51 carrying WT ppe51, variant ppe51 (S211R, A228D, E215K) and a double variant (S211R+A228D). An empty complementation vector was also introduced into Δppe51. The expression of these constructs was also confirmed via qRT-PCR. Growth curves of the complementation constructs grown in growth arresting conditions showed that WT and empty vector strains exhibit growth arrest at pH 5.7, whereas the variant complemented strains exhibited enhanced growth in both CDC1551 and Erdman (Fig. 3A and Fig. S3). Additionally, the S211R, A228D, and S211R+A228D variant complemented strains grow slightly better compared to E215K in both CDC1551 and Erdman Mtb Δppe51 mutant backgrounds, which aligns with previous relative growth data (Fig. 1). At pH 7.0, all WT and complemented strains showed similar levels of growth (Fig. S3). To examine the role of ppe51 in survival, a viability assay was performed with the previously described panel of strains. The empty vector and WT-complemented Δppe51 maintained viability over the course of 40 days at pH 5.7 (Fig. 3B, and Fig. S9A). The complemented ppe51 variants also maintain viability and replicate at pH 5.7. At pH 7.0, all WT and complemented strains maintain viability and exhibited similar increases in CFU over the course of 18 days (Fig. S3).
FIG 3.
Viability and replication dynamics of ppe51 variants. (A) Growth curve of Δppe51 complemented from its native promoter with the WT ppe51 allele or mutant alleles and performed under acid growth arrest conditions: minimal media buffered to pH 5.7 with glycerol as a carbon source. EAG variants promote Mtb growth at pH 5.7, while Δppe51 complemented with WT ppe51 maintains growth arrest. This experiment was repeated two times. n = 3. Error bars indicate standard deviation. (B) Viability of Mtb strains as measured by CFU. This experiment was repeated two times. n = 3. Error bars indicate standard deviation. (C) All CDC1551 Mtb strains, WT, Δppe51, and the spontaneous variant allele, A228D, continue to replicate at pH 7.0 in minimal media with glycerol. To estimate replication dynamics of the indicated strains, plasmid frequency data was obtained from CFU counts (right axis, dotted lines). CFU of plasmid-free and plasmid-bearing strains were then used to calculate cumulative bacterial burden (CBB) of total live, dead, or degraded Mtb (left axis, solid lines). CDC1551 Mtb containing the spontaneous variant allele, A228D, continues to replicate at pH 5.7 in minimal medium on glycerol, but WT Mtb and Δppe51 cease replication. This experiment was repeated two times. n = 3. Error bars indicate standard deviation. (D) Replication dynamics of the spontaneous A228D (CDC1551) variant, comparing CBB (cumulative bacterial burden), CFU (total CFU from nonselective plating), and % pBP10 (percentage of bacteria carrying plasmid). Note: Δppe51 mutants in this figure harbor a PDIM mutation.
The stable viability of WT or Δppe51 mutant may be due to growth arrest or, alternatively, balanced growth and death. To determine if the strains are truly growth arrested, we examined replication using the pBP10 clock plasmid, transformed into WT Mtb, Δppe51, and EAG variants in CDC1551 (44). The strains were incubated in minimal media (pH 5.7 and 7.0) with glycerol for 40 days. When in vitro pBP10 growth studies were conducted at pH 7.0 with all strains, we observed similarly high rates of replication and plasmid loss across all strains (Fig. 3C). We observed that the WT and Δppe51 strains did not replicate under acid growth arrest conditions (Fig. 3C). In contrast, we observed high rates of replication in the EAG variants at pH 5.7 (Fig. 3C). We then compared EAG variants calculated cumulative bacterial burden (CBB) to total CFU counted on nonselective plates and observed that greater rates of replication in the EAG variants is associated with a high death rate, yielding a large difference between CBB and total CFU (Fig. 3D). These results show that enhanced growth at acidic pH is driven by higher replication, but this growth is offset somewhat by a higher death rate, supporting the conclusion that faster replication at acidic pH may be deleterious to Mtb survival.
Phthiocerol dimycocerosates biosynthesis is disrupted in the ppe51 deletion strains.
We found that the Δppe51 mutant generated in this study did not have the same growth defects as compared to previous studies that have generated similar disruption or knockdowns of ppe51 (27, 40, 45). We found that the Δppe51 mutant in our study grew just as well as other strains at pH 7.0 on glycerol (Fig. S3). This observation was previously made by Wang et al., who showed that mutations in phthiocerol dimycocerosates (PDIM) biosynthesis are responsible for permeabilizing the mycomembrane and compensating for the loss of functional ppe51 (27). We sequenced the genomes of Δppe51 mutants in both the CDC1551 and Erdman backgrounds and found that both Δppe51 mutants had evolved mutations in PDIM biosynthesis pathway genes (ppsC in Δppe51-CDC1551, and ppsD in Δppe51-Erdman) (Fig. S4A). We confirmed for loss of functional PDIM by radiolabeling Mtb with 14C-glycerol and 14C-acetate for 6 days and extracting total lipids for TLC analysis. As expected, we observed loss of functional PDIM accumulation in the Δppe51::pMV-EV compared to WT Mtb radiolabeled at both pH 5.7 and pH 7.0 with 14C-acetate and 14C-glycerol, respectively (Fig. S4). However, despite the occurrence of these PDIM mutations in the Δppe51 mutants, no PDIM mutations were present in the sequenced spontaneous variant mutants selected in this study or WT Mtb, highlighting that the EAG variants remain gain-of-function, dominant mutants (Fig. 1A), that selectively have enhanced growth on glycerol (Fig. 2). We also sequenced the genome of the CDC1551 EAG A228D mutant carrying the pB10 plasmid (Fig. 3), following transformation and selection for the plasmid, and did not observe any PDIM mutations.
To determine whether we could complement back PDIM synthesis, we incorporated a complementation construct containing WT ppsD and its native promoter into Δppe51 Erdman. To mitigate the possibility of selecting additional PDIM mutations via growth on glycerol as the carbon source, we selected for and grew Δppe51::pMV306+ppsD in two separate conditions: 7H9+OAC with no carbon and 7H9+OAC with pyruvate. Dextrose (i.e., glucose) was also eliminated from the media since PPE51 has been shown to act as a putative glucose transporter (27). PDIM accumulation was examined by radiolabeling with 14C-acetate at pH 5.7 for 6 days and extracting total lipids for TLC analysis. Despite incorporation of the pMV306+ppsD vector, we did not observe full complementation of PDIM in Δppe51 (Fig. S4). This indicates that expression of ppsD is not sufficient at overcoming loss-of-function mutation of ppsD in the Δppe51 background, and supports there is strong selection for PDIM mutations in a Δppe51 mutant, consistent with other similar observations of linkage between ppe51 loss-of-function mutations and PDIM mutations (27, 40, 45). Notably, there is no linkage between the EAG gain-of-function mutations and PDIM mutations, suggesting the EAG mutant phenotype is PDIM-independent.
Given the PDIM mutations in the Δppe51 mutants, we cannot draw conclusions about the null mutation of ppe51 in these studies. However, this result provided an opportunity to determine if the gain-of-function EAG phenotypes were PDIM-dependent or PDIM-independent, by comparison of the Δppe51 mutant complemented with EAG variant alleles compared to the ppe51 point mutants. We hypothesized that if phenotypes were conserved in both Δppe51 mutants expressing variant alleles and ppe51 point mutants, then the phenotypes are PDIM-independent. Thus, we have included comparison data for both Δppe51 mutants expressing EAG alleles and the spontaneous mutants isolated from the genetic selection in the following experiments.
Acidic pH limits glycerol uptake and PPE51 variants overcome this restriction.
Pyruvate rescues Mtb growth on glycerol in a concentration-dependent manner at pH 5.7 (17). However, it is unknown whether glycerol concentration affects acid growth arrest. We hypothesized that acid growth arrest may be driven by glycerol starvation and the PPE51 variants promote growth by promoting enhanced uptake of glycerol. If this is the case, we would expect to see a dependence of glycerol concentration and acidic pH on growth. To examine this, we examined dose responses combining varying pH levels (pH 6.5-5.5) and glycerol concentrations (80 mM-0.13 mM) using the panel of isogenic strains. The standard concentration of glycerol used in our acid growth arrest model is 10 mM. Growth in the wells was analyzed using optical density (OD600) and data was normalized to wells containing the highest (100%) levels of growth and wells with no carbon representing the lowest (0%) levels of growth. Growth assays were performed for 21 days, and the data shown is Day 14 which is representative for the duration of the experiment. Interestingly, we found that growth arrest appears to be both pH and glycerol concentration-dependent, with growth partially rescued at high concentrations of glycerol (~80 mM) for WT, Δppe51::pMV306 and Δppe51::pMV-ppe51 at pH 5.7 (Fig. 4 and Fig. S5A). Additionally, we observed higher levels of growth at lower glycerol concentrations (~0.82 mM) at pH 5.7 with the complemented ppe51 variants compared to the empty vector and WT ppe51 complemented strains. Growth could also be rescued with high glycerol concentration (~32 mM) for variants at pH 5.5. The presence of the double ppe51 variant (S211R+A228D) overcomes growth arrest at pH 5.5 at even lower glycerol concentrations (~5.12 mM) compared to the single variants, indicating that the presence of 2 EAG point mutations confers a slight growth advantage during acid growth arrest. Concentrations of glycerol below 0.33 mM did not rescue growth starting at pH 6.0 in any ppe51 variant expressing strains, which could be due to glycerol being fully consumed. Similarly, these observations were also made in the variants selected in the WT backgrounds of both CDC1551 and Erdman, while WT exhibited a reduced capacity for glycerol uptake (Fig. S5B). Together, these findings suggest that Mtb has reduced capacity to metabolize or uptake glycerol in a pH-dependent manner, and that PPE51 variants may function by promoting enhanced uptake of glycerol in a PDIM-independent manner.
FIG 4.
Mtb restricts growth in a pH and glycerol concentration dependent manner. Growth of WT CDC1551, Δppe51 (empty vector), and Δppe51 complemented strains in minimal media supplemented in a dose-dependent manner with glycerol and buffered to one of 5 pH levels (pH 6.5, 6.2, 6.0, 5.7, or 5.5). All strains exhibit a reduced capacity for growth starting ~2 mM glycerol compared to higher glycerol concentrations. At decreasing pH, WT, Δppe51(empty vector), and Δppe51::pMV-WT restrict their ability to uptake glycerol, whereas any variant complement is able to maintain glycerol uptake. However, restricted growth can be rescued at high concentrations of glycerol (~80 mM) at pH 5.7 for WT, Δppe51(empty vector), and Δppe51::pMV-ppe51, and pH 5.5 for variant complements. Growth analyses were performed at Day 14 following initial inoculation with data being shown as percent of the maximum well-growth. All conditions were conducted in triplicate and representative of multiple independent experiments. Note: Δppe51 mutants in this figure harbor a PDIM mutation. Similar data were observed in a spontaneous EAG mutant (Fig. S5) that do not harbor a PDIM mutation. Error bars indicate standard deviation.
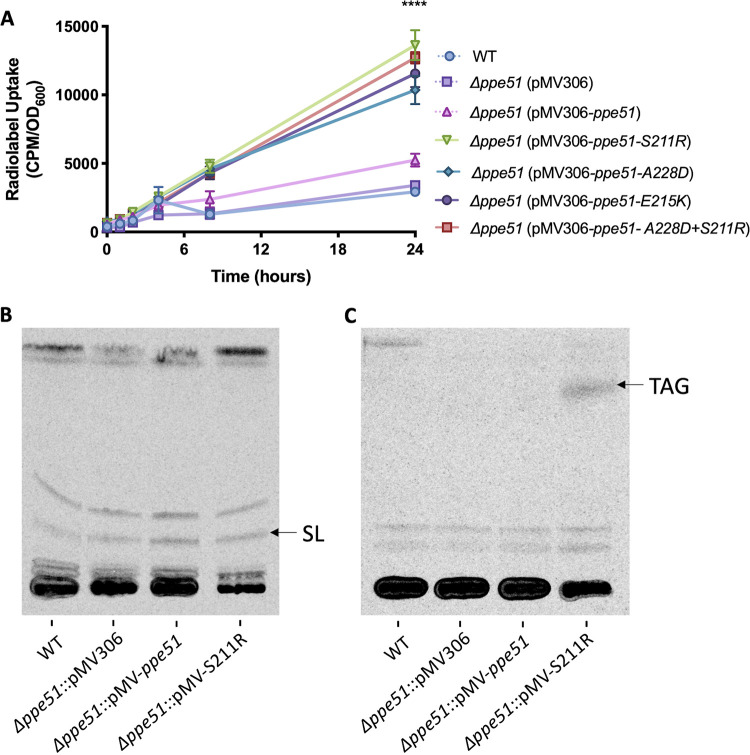
Based on these dose response findings, Mtb appears to restrict its growth on glycerol at acidic pH. Additionally, WT Mtb has been shown to completely arrest its growth at pH 5.7 on 10 mM glycerol; however, it is able to maintain viability for up to 40 days, remains metabolically active, and incorporate limited amounts of exogenous 14C-glycerol into lipids (16). To further test the hypothesis that Mtb restricts glycerol uptake at acidic pH and that ppe51 variants promote enhanced glycerol uptake, a radiolabeling experiment using 14C-glycerol was conducted with WT Erdman and the Δppe51 complemented strains previously described. Strains were pre-adapted for 3 days in MMAT (pH 5.7 or 7.0) with 10 mM glycerol and washed with PBS prior to radiolabeling with 6 μCi of 14C-glycerol. Samples were collected over the course of 24 h, washed, and analyzed for radiolabel uptake by scintillation counting. All complemented strains containing a ppe51 variant accumulated 14C-glycerol at a similarly increased rate of approximately 300% compared to the WT Mtb strain (Fig. 5A). These results are consistent with radiolabeling that was conducted with the pVV16 empty vector, S211R overexpression strain, and spontaneous ppe51-S211R variant where we observed similar enhanced glycerol uptake at ~ 60% with strains containing S211R compared to WT empty vector (Fig. S6). We also looked at glycerol uptake with WT CDC1551, empty vector, and complemented S211R at pH 7.0. We did not observe significant differences in glycerol uptake between strains, and the rate of uptake was similar to the complemented ppe51 variant strains at pH 5.7 (Fig. S6). Together, these results show that strains containing ppe51 variants have significantly enhanced glycerol uptake in a PDIM-independent manner.
FIG 5.
ppe51 EAG variants exhibit enhanced 14C-glycerol uptake and incorporation into lipids. (A) Strains expressing EAG variants uptake 14C-glycerol at an enhanced rate. Mtb was pre-adapted for 3 days in MMAT (pH 5.7) with 10 mM glycerol and subsequently washed prior to the addition of radiolabeled glycerol. 14C-glycerol uptake was measured using scintillation counting at various time points over the course of 24 h. Significance was determined by two-way ANOVA (Tukey’s multiple-comparison test; ****, P < 0.0001). Error bars indicate standard deviation. (B) Incorporation of 14C-glycerol into sulfolipids at acidic pH. Sulfolipid is indicated with an arrow and accumulates at a similar rate in each strain. Strains were analyzed in duplicate with representative results being shown. (C) Incorporation of 14C-glycerol into TAG at acidic pH. TAG is indicated with an arrow and are absent from all strains except for Δppe51::pMV-S211R. Strains were analyzed in duplicate with representative results being shown. Note: Δppe51 mutants in this figure harbor a PDIM mutation. Similar results with the spontaneous ppe51 mutants isolated from the genetic selection, without PDIM mutations, are presented in Fig. S6.
While the radiolabeling strongly indicated that glycerol was being taken up by the strains, it did not answer whether glycerol was being metabolized by Mtb and incorporated into lipids or binding to the mycomembrane without uptake across the plasma membrane. To address this question, we performed lipid radiolabeling with 14C- glycerol. WT Erdman and Δppe51 complemented strains were pre-adapted for 3 days in the same culture conditions as the previously described radiolabeled uptake experiment. The operon controlling sulfolipid synthesis is induced in a phoPR-dependent and a pH-dependent manner (17, 46, 47). We examined sulfolipids by TLC, resolving them in a polar solvent system that resolves sulfolipid (13). Sulfolipid was observed to specifically accumulate at pH 5.7 (Fig. 5B) with no accumulation occurring at pH 7.0 (Fig. S6C). Triacylglycerol (TAG) has been shown to accumulate during periods of hypoxia and pH-stress (17, 48), and pathways involved in TAG synthesis play a role in reducing Mtb growth by redirecting carbon flux away from the TCA cycle (49). A nonpolar solvent system was used to separate lipids, and the bands observed migrated to a position consistent with TAG (13). We found that TAG accumulated specifically in the complemented S211R strain at pH 5.7 (Fig. 5C). In contrast, we did see similar TAG accumulation across all strains at pH 7.0 (Fig. S6D). The observation of labeled lipids, in both growth arrested and growing Mtb at acidic pH, shows that glycerol is imported and metabolized at acidic pH, with enhanced uptake in the S211R variant.
ppe51 variants have selectively reduced growth in activated macrophages.
ppe51 is induced in a pH-dependent and phoP-dependent manner within 2 h following phagocytosis by macrophages (10), suggesting that ppe51 is important for pathogenesis. We hypothesized that ppe51 or its EAG variants may be required for pathogenesis, specifically in activated macrophages, where the phagosome is acidified. To test this hypothesis, resting and activated primary murine bone marrow-derived macrophages (BMDMs) were infected with WT CDC1551 and Δppe51 mutant and complemented variant strains. In resting macrophages, we did not observe significant differences in Mtb growth between the strains (Fig. 6A), with all strains growing ~1.25-log over 9 days. In contrast, in activated macrophages, while the WT, Δppe51 empty vector, and Δppe51 WT-complemented strain still exhibit ~1.25-log increase in growth, the ppe51 complemented variants show significantly lower growth (Figure 6A and B). These results show that ppe51 variants are selectively less fit in activated macrophage environment, which is consistent with a pH-dependent phenotype that is observed in vitro. Notably, this phenotype of reduced survival is also observed in the spontaneous ppe51 EAG variant harboring the A228D substitution (Fig. 6C and Fig. S7), demonstrating the reduced survival due to the ppe51 EAG variant is not PDIM-dependent.
FIG 6.
EAG variants exhibit selectively enhanced replication and reduced survival in activated macrophages. (A) BMDMs infected with the isogenic panel of CDC1551 Δppe51 complemented strains and WT CDC1551. Growth is similar for all strains in resting macrophages, but in activated BMDMs, WT, Δppe51(empty vector), and Δppe51::pMV-ppe51 exhibit ~1.25 log increase in growth compared to variant complements which show a lower log increase in growth (~0.25-1). Data shown was conducted in triplicate and representative of three independent experiments. Error bars indicate standard deviation. (B) Statistical analysis of growth differences between Δppe51 complemented strains at Day 9 in activated BMDMs. Significance was determined by one-way ANOVA (Tukey’s multiple-comparison test; *, P < 0.05, ***, P < 0.001, ****, P < 0.0001). Mean ± SD are shown in the bar graph. (C) Activated BMDMs infected with WT CDC1551, Δppe51, and spontaneous A228D variant strains containing the pBP10 replication clock plasmid. CFU on selective plates were compared to CFU on nonselective plates and used to calculate frequency of plasmid-bearing bacteria (% pBP10), cumulative bacterial burden (CBB) of total live and dead bacteria, and total enumerated colonies on nonselective plates (CFU). Data shown was conducted in triplicate and representative of 2 independent experiments. Error bars indicate standard deviation. Note: Δppe51 mutants in this figure harbor a PDIM mutation.
Rohde et al. showed that rapid replication of intracellular Mtb is associated with greater Mtb killing by the macrophage (10). We observed in vitro that variants had enhanced death during replication at acidic pH, and we hypothesized that the EAG variants may be replicating faster than the WT in macrophages but have lower CFU due to enhanced death rates. To test this hypothesis, we infected BMDMs with CDC1551 WT, Δppe51, and spontaneous A228D variant containing the pBP10 plasmid as described previously. Infection was conducted over the course of 8 days with cells lysed and plated for viable CFU every 2 days. We observed an initial ~0.5 log decrease in viable CFU in both WT and Δppe51 around day 2 that is consistent with observations made by Rohde et al. (50), and supports their findings that Mtb exhibits delayed adaptation to survive and replicate within macrophages (Fig. 6C). Both WT and Δppe51 then replicated over the course of 8 days inside activated BMDMs as evident by their ~1 log increase in CFU starting at day 2. In contrast, the A228D variant lacks this initial adaptation period and instead show a continual ~1 log decrease in CFU over the course of 8 days. Calculating the CBB of the A228D variant shows a large difference between the CBB and CFU, demonstrating that the A228D variant is replicating at a higher rate and dying at an even greater rate. Notably, the ppe51 (A228D) variant is able to replicate and survive better in resting BMDMs compared to activated BMDMs (Fig. 6C), leading to an ~4 log, compared to ~2 log difference between CBB and CFU in activated compared to resting macrophages, respectively. We conclude that slowed growth in response to acidic pH inside activated macrophages is necessary for mycobacterial survival and that the EAG variants do not sufficiently slow their growth inside macrophages, resulting in enhanced killing. This enhanced killing is observed in strains with both WT and mutant PDIM backgrounds, showing the result is PDIM-independent. These results also support that the PPE51 variant is promoting uptake of a carbon source during macrophage infection, suggesting that Mtb may metabolize glycerol when growing in macrophages.
Differential thermal stability of PPE51 and the S211R variant proteins support direct interactions between PPE51 and glycerol.
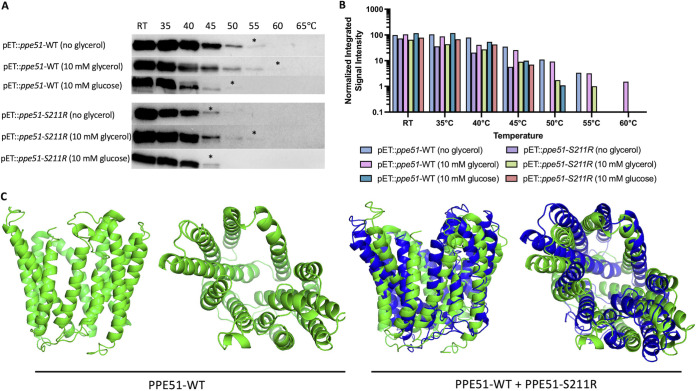
We hypothesized that the ppe51 EAG variants promote uptake of glycerol by altering PPE51 structure and its affinity for glycerol. Changes in the thermal stability of the protein would provide evidence supporting this hypothesis. C-terminal his-tagged recombinant PPE51 and PPE51 (S211R) variant proteins were produced and purified from Escherichia coli (Fig. S8). Glycerol was omitted from the reagents used in the purification process and loading dye. All reagents were buffered to pH 7.6. In the absence of glycerol, we observed differential stability between the WT and S211R PPE51 variants, with the WT and S211R proteins completely denaturing at 60°C and 50°C, respectively, supportive of a significant structural change by the amino acid substitution (Fig. 7A and B). In glycerol, the WT protein exhibited enhanced stability, completely denaturing at 65°C, a shift of 5°C, and the S211R protein completely denaturing at 60°C, a shift of 10°C. These findings support that glycerol/PPE51 interactions, with differential stability shifts dependent on the S211R substitution.
FIG 7.
Glycerol differentially interacts with recombinant WT PPE51 or S211R variant proteins. (A) Recombinant WT PPE51 and S211R proteins were assessed for thermostability under no glycerol, 10 mM glycerol, and 10 mM glucose conditions. The protein was preincubated at room temperature (RT) for 20 min and subjected to eight different temperature conditions as indicated for 5 min. Following heating, samples were spun down to pellet the protein precipitate. Soluble protein was removed and analyzed by immunoblotting. (*) represents the highest temperature where soluble protein was detected. (B) Signal intensity of individual bands were measured and normalized to the pET::ppe51-WT (no glycerol) band at RT. Samples containing glycerol continue to show detectable signal intensity up to 55°C for pET::ppe51-S211R and 60°C for pET::ppe51-WT. (C) in silico protein structure modeling and function prediction for PPE51. The peptide sequence of PPE51-WT (green) and PPE51-S211R (blue) were analyzed using the Iterative Threading ASSEmbly Refinement (I-TASSER) approach (51). Both WT and variant PPE51 were modeled without constraint and appear to form a porin-like structure with an inner channel. PPE51-S211R is modeled against the WT to show the slight conformational changes that occur with the introduction of this mutation. PPE51-WT and PPE51-S211R structures received C-scores of -0.86 and -1.24, respectively, which is a measure of structure confidence on a range of -5 (low) to 2 (high) (51). A model with C-score >-1.5 usually indicates a correct fold.
We previously noted that PPE51 S211R did not promote the growth on glucose and therefore, examined the thermal stability in glucose. Compared to glycerol, we observed reduced stability of the WT protein in glucose, completely denaturing at 55°C and did not observe any differences in stability with the S211R protein in glucose, supporting the stability shifts are selectively dependent on glycerol (Fig. 7A and B). Together, these data show that glycerol selectively increases the thermal stability of PPE51, with enhanced impact on the S211R variant, lending further support for a mechanism whereby PPE51 directly binds glycerol for uptake and acquisition into the Mtb cell.
Based on the EAG phenotype and differences in thermal stability, we hypothesized that these substitutions may have a significant impact on protein structure and thus conducted in silico modeling of PPE51 using the Iterative Threading ASSEmbly Refinement (I-TASSER) server for protein structure and function prediction (51). The best fit model of PPE51-WT had a moderately high confidence score (C-score) of -0.86 on a scale of -5 (low confidence) to 2 (high confidence) (Fig. 7C). Threading of the sequence against known transporter structures produced a porin-like model with a possible channel. This model was matched to all structures in the Protein Data Bank (PDB) library. The top 10 proteins from the PDB with the closest structural similarity were all predicted to be nutrient transporter proteins (Fig. S8C) An overlay of the S211R variant (blue) with PPE51-WT model shows that the introduction of this substitution confers a noticeable conformation shift in the predicted protein structure (Fig. 7C). For PPE51-WT, the top predicted ligands were maltose and a monoacylglycerol derivative (78N), with one predicted ligand binding site for maltose being within the 18 amino acid residue range of the substitutions observed in the EAG variants, located at residue 225 (Fig. S8D). Additionally, the predicted top Gene Ontology terms for the molecular function, biological process, and cellular component are hexose:hydrogen symporter activity (GO:0009679), transmembrane transport (GO:0055085), and integral to membrane (GO:0016021), respectively (Fig. S8E). While these in silico results are predictions, they provide further support for a role with PPE51 acting as a nutrient transporter for Mtb. Furthermore, all EAG variant mutations mapped to a single alpha helix on the predicted I-TASSER model, with S211R and E215K located at the top of the predicted channel and A228D located within the center channel structure (Fig. S8F) and the substitutions, altered the modeled substrate interaction, further supporting our model for PPE51 variants acting to promote uptake of glycerol by altering the protein structure and ligand interactions.
DISCUSSION
Mtb exhibits complex regulatory and physiological adaptations when grown in acidic environments, including changes in growth rate. The underlying basis of slowed growth in mildly acidic environments is still not fully resolved, but appears to be associated with metabolic and redox stress, that may be linked to balancing cytoplasmic pH-homeostasis and respiration (11). Providing specific carbon sources, such as pyruvate or acetyl-CoA, relieve this metabolic stress and enable Mtb to grow similarly well at acidic and neutral pH (16, 17). However, it has been puzzling as to why Mtb cannot grow on glycerol at acidic pH, as it has a carbon source and oxygen, everything it needs to grow. In this study, we found that Mtb limits uptake of glycerol at acidic pH to restrict its growth and that mutations in ppe51 promote uptake of glycerol at acidic pH and enable growth. That is, Mtb can grow at acidic pH on glycerol, but has adapted instead to stop growth. It was recently shown that Mtb has reduced glyceraldehyde-3-phosphate dehydrogenase activity at acidic pH (52), and it is possible that in minimal medium with glycerol as a sole carbon source, that Mtb growth is arrested as insufficient carbon is available for glycolysis. Enhanced uptake of glycerol by the PPE51 variants may supply sufficient carbon to glycolysis to overcome this restriction.
We further show that this pH-dependent metabolic adaptation is required for pathogenesis. Selectively in activated macrophages, where the pH of the phagosome is more acidic, we observed a survival defect in strains expressing variants with an EAG phenotype. Notably, using a replication clock plasmid, we found that the variants have enhanced growth in macrophages, but even greater killing, the balance of which results in reduced fitness. Thus, slowed growth in macrophages, in an activation dependent manner is dependent on the restriction of metabolism at acidic pH, and PPE51 variants overcome this restriction to the detriment of the pathogen. This finding supports that the nutrient imported by the PPE51 variant is relevant to the macrophage environment. We showed that the variants specifically promote uptake of glycerol, therefore, it is plausible that glycerol is a key regulator of Mtb growth in the macrophage. It has been previously shown that Mtb can uptake TAG in macrophages (53), TAG is abundant in granulomas (54), and Mtb exports the TAG lipase LipY (55), therefore, it is possible glycerol is released from TAG during infection, and restriction of glycerol uptake plays an important role in slowing growth during infection. Studies examining the interactions of PPE51 EAG variants, LipY and glycerol metabolism genes during pathogenesis will be undertaken to test this hypothetical model.
It is a striking finding that all of the mutants selected were in ppe51, and that they all clustered with a highly conserved region of 18 amino acid residues (residues 211–228). Three single amino acid substitutions (S211R, A228D, and E215K) greatly altered WT PPE51 function and promoted growth under acid stress when given the non-permissible carbon source, glycerol. S211R was able to confer the greatest enhanced growth, whereas A228D conferred moderate enhanced growth and E215K exhibited the least amount of enhanced growth, comparatively (Fig. 1). The growth phenotypes of the spontaneous ppe51 mutants were further recapitulated in expression studies in a WT Mtb background as well as a Δppe51 background, where again we observed overall greater growth at acidic pH with the S211R variant compared to A228D and E215K (Fig. 2A, Fig. 4A and Fig. S8A). Given that the phenotype was conserved in PDIM containing strains (the initially isolated mutants and the expressors in the WT) and PDIM lacking strains (the ppe51 disruption mutants), demonstrates that the gain-of-function phenotype is independent of PDIM. This region of PPE51 may play a key role in protein-substrate interactions, and indeed with recombinant proteins, we observed differential stability in the variant protein and its interaction with glycerol. The structural modeling showed substitutions in this region altered the predicted ligand of the modeled transporter, supporting further study of this critical region for modulating PPE51-ligand interactions.
Another key finding of this study is that glycerol uptake is restricted at acidic pH. Data supporting this conclusion include the reduced uptake of radiolabeled glycerol at acidic pH compared to neutral pH (Fig. 5A and Fig. S6), the dependence of glycerol concentration and pH in regulating growth (Fig. 4 and Fig.S5), and the ability of PPE51 variants to enhance growth and glycerol uptake at acidic pH (Fig. 5A and Fig. S6). How Mtb restricts glycerol uptake is still not known, but it is puzzling that PPE51 is strongly induced at acidic pH and counter to a model where PPE51 promotes in glycerol uptake, but Mtb restricts glycerol uptake at acidic pH. This contradiction remains unresolved and points to a new unknown aspect of Mtb metabolism restriction at acidic pH.
We identified that the ppe51 variants selectively enabled growth on glycerol compared to WT Mtb (Fig. 2). The identification of this carbon specificity with PPE51 variants implies a putative role for PPE proteins in nutrient acquisition, a model that is strongly supported by data put forth by Ates et al. (33), Mitra et al. (25), and Wang et al. (27). These studies showed that PE and PPE proteins located at the cell envelope and cell surface play a vital role in nutrient uptake for Mtb. Ates et al. (33) provides strong evidence that the type VII secretion system, ESX-5, is essential for mycobacterial growth and nutrient uptake. In this study, essentiality of ESX-5 could be rescued by altering cell wall lipid composition or introducing the Mycobacterium smegmatis outer membrane porin, mspA, which mediates cell wall permeability and influx of hydrophilic nutrients (56–58). ESX-5 mutations in Mycobacterium marinum result in significantly reduced growth on medium with Tween 40 or Tween 80 as the sole carbon source, and the ESX-5 mutant strain exhibits significantly impaired uptake of fluorescently labeled fatty acids compared to WT and complemented strains (33). These data support ESX-5 facilitating the uptake of fatty acids to be used as a carbon source through the secretion of PE and PPE proteins. In support of Ates et al. (33) ESX-5 substrate nutrient influx hypothesis, Mitra and colleagues (25) showed direct evidence tying PE and PPE proteins to iron acquisition. Mitra identified Mtb transposon mutants that were resistant to a toxic heme analog (25). The mutants were in three previously uncharacterized genes of which 2 were PPE proteins, PPE36 and PPE62 (25). Furthermore PPE62 was shown to be surface-accessible and predicted structure indicates that it may form a β-barrel that resembles Haemophilus influenzae heme cell surface receptor (25, 59) and that heme transport is facilitated into the cell by the periplasmic lipoprotein DppA (39). Finally, the Wang et al. (27) study provided direct evidence that PPE51 is exported to the mycomembrane to promote uptake of glycerol and glucose, possibly by acting like a porin. Notably, loss-of-function ppe51 mutants have altered sensitivity to antibiotics, including pyrazinamide (45) and meropenem (60), suggesting that PPE51 mediated impacts on carbon source uptake or mycomembrane permeability play a role in drug susceptibility, supporting further studies of PPE51 as a target for potentiating antibiotics.
Here, we present a model that integrates the current understanding of PE and PPE nutrient acquisition with our findings (Fig. 8), wherein PPE51 embeds itself into the outermost layer of the cell envelope and is surface-accessible to glycerol (27, 41, 61). Gene expression profiling data supports induction of ppe51 by phoP and acidic pH (15, 17). Phylogenetic evidence shows that PPE51 is duplicated alongside ESX-5 (31), which has been shown to mediate the secretion of most PE/PPE proteins in M. marinum, including PPE51 (33, 37). We propose that an unknown periplasmic nutrient transporter helps mediate the import of glycerol across the plasma membrane and into the cell from initial import by PPE51. pe/ppe families have high variation rates between M. tuberculosis complex (MTBC) genomes with ppe51 being an exception in showing almost no variation (62). However, under the specific pressure of our genetic selection, we have shown that we can select for mutations that enhance PPE51’s proposed uptake of glycerol (Fig. 8). Furthermore, our initial in silico modeling of PPE51 suggests that it can form a porin-like structure consistent with a role in transport and ligand binding sites for carbon nutrient sources (Fig. 7C). Based on these data, we further propose a model, whereby the EAG phenotype amino acid substitutions introduce conformational changes that allow for a possible PPE51-porin structure to widen or enhance the binding the glycerol, allowing enhanced transport through the mycomembrane.
FIG 8.
A proposed model for the role of ppe51 and EAG variants in glycerol acquisition. Presented is a hypothetical model, in which ppe51 expression is induced by PhoP under acidic conditions. PPE51 is thought to be secreted through ESX-5 and embeds itself into the mycomembrane, making itself surface-accessible. At this interface, it could interact with glycerol and promote transport across the mycomembrane (WT Pathway). PPE51 variants may function by having an altered channel opening or ligand binding surface, allowing for enhanced glycerol transport across the mycomembrane and leading to the enhanced growth phenotype observed during acid growth arrest (Mutant Pathway).
This study has focused on the role of the PPE51 variants, and not the Δppe51 mutant, due to confounding mutations in PDIM in the deletion strains. It is interesting that both deletion mutants (in Erdman and CDC1551) evolved these mutations during the construction of the mutants and suggests there may have been a selective advantage for the mutations. Indeed, Wang et al. (27), showed that loss-of-function ppe51 mutants only had a glycerol uptake phenotype when the PDIM was restored in the mutant. Furthermore, Babu Sait et al. (42) found that all their ppe51 clones except for one harbored mutation in different genes involved in PDIM biosynthesis; however, resistance was attributed to the ppe51 mutations themselves irrespective of the PDIM mutations (42). These findings are consistent with our observation that the Δppe51 mutants in this study did not have a growth defect in glycerol, presumably due to the lack of PDIM, whereas the ppe51 mutants in the Wang et al. (27) study were defective for growth. Given the conservation of the EAG phenotype in strains expressing variants with or without PDIM, we conclude that PDIM levels do not appreciably impact the enhanced uptake of glycerol in EAG variants. However, it is also possible that differences for the PPE51 mutants between this study and the others may be driven by genomic differences. Wang et al. (27), Korycka-Machała et al. (40), and Babu Sait et al. (42) used the H37Rv Mtb strain for their mutant or CRISPRi knockdown studies. However, sequence analysis of the region directly upstream of ppe51 in both CDC1551 and Erdman compared to H37Rv shows an almost total deletion of the ppe50 gene preceding ppe51. The ppe50 sequence is also not present anywhere else in the CDC1551 or Erdman genome except for a matching 66 bp sequence that precedes ppe51 in both genomes. The large sequence difference in the ppe51 promoter region between strains could imply an additional reason why we see strong phenotypic growth differences between our respective Δppe51 mutants.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All experiments were performed with Mtb strains Erdman and CDC1551. Mtb was grown at 37°C and 5% CO2 in vented T-25 culture flasks containing Middlebrook 7H9 media with 10% oleic acid-albumin-dextrose-catalase (OADC), 0.05% Tween 80, and 0.2% glycerol. For acid stress and single carbon source experiments, MMAT defined minimal media was used as described by Lee et al. (63): 1 g/L KH2PO4, 2.5 g/L Na2PO4, 0.5 g/L (NH4)2SO4, 0.17 g/L L-Asparagine monohydrate, 10 mg/L MgSO4, 50 mg/L ferric ammonium citrate, 0.1 mg/L ZnSO4, 0.5 mg/L CaCl2, and 0.05% Tyloxapol. MMAT media was buffered with 100 mM 3-(N-morpholino)propanesulfonic acid (MOPS) for experiments requiring pH 6.6–7.0 and 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) for experiments requiring pH 5.5–6.5 (64). For growth curve experiments, Mtb was grown to mid-late log phase (OD600 0.6–1.0) and seeded in MMAT at a starting OD600 of 0.05. Optical density measurements were conducted by removing 500 μL of samples at each time point. Viability assays were performed in a similar manner with samples being diluted 10-fold in PBS + 0.05% Tween 80 and plated for viable CFU on 7H10 + 10% OADC agar plates.
Genetic selection and sequencing.
A wild type Erdman Mtb population of 4x109 bacteria was plated on MMAT agar plates (1 g/L KH2PO4, 2.5 g/L Na2PO4, 0.5 g/L [NH4]2SO4, 0.17 g/L L-Asparagine monohydrate, 10 mg/L MgSO4, 50 mg/L ferric ammonium citrate, 0.1 mg/L ZnSO4, 0.5 mg/L CaCl2, and 0.05% Tyloxapol) supplemented with 10 mM glycerol as the sole carbon source and buffered to pH 5.7 with 100 mM MES (64). Plates were incubated at 37°C with spontaneous mutants appearing around week 8 and isolated for growth. Single-colony isolates were confirmed as EAG mutants under acidic conditions in liquid MMAT (pH 5.7) media amended with 10 mM glycerol. Whole genome sequencing (WGS) was performed on genomic DNA isolated from mutants representing various levels of enhanced growth as well as a wild type Erdman control. Samples were sequenced using the Illumina MiSeq in a 2 × 250-bp paired end format. Base calling was done by Illumina Real Time Analysis v1.18.54, demultiplexed, and converted to FastQ using Illumina Bcl2fastq v2.19.1. Low-quality bases were trimmed and adapter sequences were removed using Trimmomatic (v0.36) (65) and aligned sequence reads to the Erdman reference genome using BWA-MEM (66). SNPs and indels were identified using Genome Analysis ToolKit (GATK) (67). All identified mutations in the EAG mutants are presented in Table S2.
Generation and analysis of ppe51 disrupted strains.
The ppe51 gene was disrupted with a hygromycin resistance (HygR) cassette in both Erdman and CDC1551 Mtb strain backgrounds using a new chromosomal engineering system called ORBIT (for “oligonucleotide-mediated recombineering followed by Bxb1 integrase targeting”) that combines site-specific recombination with homologous recombination (43). An ORBIT recombineering plasmid (pKM444) expressing RecT annealase and Bxb1 integrase from the anhydrotetracycline (ATc)-inducible Ptet promoter and containing a kanamycin resistance (KanR) cassette was first transformed into Mtb, selected for KanR, induced with ATc, and generated into electrocompetent cells. Electrocompetent cells were then transformed with an integration plasmid (pKM464) harboring HygR and targeting oligonucleotide. Hygromycin-resistant colonies were isolated and cured of the kanamycin-containing recombineering plasmid. Genomic DNA was extracted from transformants and the 5′ and 3′ junction sites of the disruption were confirmed by PCR and sequencing using ORBIT target-specific and ppe51-specific primers (Fig. S7B to D and Table S1). Gene replacement was further verified via quantitative real-time PCR (qRT-PCR) (Fig. S7E). Δppe51 was complemented with WT and variant ppe51 from their native promoter and confirmed by qRT-PCR (Fig. S7F). Later in this study, we discovered PDIM mutations (Fig. S11), ppsC and ppsD, present in both the CDC1551 and Erdman backgrounds of the Δppe51 mutants, respectively.
pH and glycerol dose response combination growth assays.
Mtb cultures were incubated in a range of pH buffered MMAT media (pH 5.0-pH 7.0) at a starting OD600 of 0.2 in 96-well plates. Cultures were treated with 2.5-fold dose response (0.13-80 mM) of glycerol and incubated over the course of 21 days, with growth assessed by optical density. Bacterial viability was assessed by diluting wells 10-fold and plating for viable CFU on 7H10 + 10% OADC agar plates. Optical density data was converted to percent of maximum well-growth and normalized based on no carbon control at pH 5.5 (0%) and maximum Mtb growth on glycerol at pH 6.5 (100%). Each condition and time point experiment was conducted in triplicate and representative of multiple individual experiments.
Radiolabeled glycerol uptake assay.
Mtb Erdman cultures were pre-adapted for 3 days in MMAT media (pH 5.7 or pH 7.0) containing 10 mM glycerol. Following adaptation, Mtb was washed twice with PBS + 0.05% Tween 80 and resuspended in the same buffered MMAT media amended with 10 mM glycerol and 6 μCi of [U-14C] Glycerol. Samples were removed over the course of 24 h, fixed with 4% paraformaldehyde, and assessed for total radioactivity using scintillation counting. All strains used for radiolabel uptake experiments were repeated in 2 biological replicates.
Analysis of metabolism of radiolabeled lipids into Mtb lipids.
Mtb Erdman cultures were pre-adapted as described above for the uptake experiments. Following pre-adaptation, cultures were seeded at a starting OD600 of 0.2 in MMAT media (pH 5.7 or pH 7.0) + 10 mM glycerol and set up in 2 biological replicates. Lipids were labeled with 6 μCi of [U-14C] Glycerol or 6 μCi of [U-14C] Acetate for 6 days, and samples were pelleted and washed with PBS before lipid extraction. Total lipids were extracted and Folch washed as previously described (17) and 14C-incorporation was measured using scintillation counting. For thin-layer chromatography (TLC), 5,000 cpm (CPM) of each pH 5.7 sample and 10,000 CPM of each pH 7.0 sample was loaded on a 100-cm2 high-performance TLC silica gel 60 aluminum sheet (EMD Millipore) and analyzed with a chloroform:methanol:water (90:10:1 vol/vol/vol) solvent system (46). Sulfolipids, TAG, and PDIM were separated as previously described (13, 17, 46) and quantified using a phosphor screen and Typhoon imager and ImageJ software (68).
Replication during acid growth arrest.
For measurement of replication during acid growth arrest, WT Mtb, Δppe51, and spontaneous ppe51 variants in both CDC1551 and Erdman backgrounds carrying the pBP10 plasmid (69) were inoculated into MMAT media (pH 5.7 and pH 7.0) + 10 mM glycerol and in the absence of kanamycin. Plasmid loss and percentage of bacteria still containing the pBP10 was determined by plating for CFU on 7H10 + 10% OADC agar plates ±25 μg/μL kanamycin. Rates of growth, death, and cumulative bacterial burden were quantified using equations as previously described (44). Specifically, equations 10, 11, and 13, as detailed in the supplemental materials of Gill et al. (44) were used to calculate rate of replication, rate of death, and numbers of dead bacteria, respectively. The Mtb segregation constant (s = 0.18 ± 0.023) – the frequency of Mtb daughter cells losing plasmid per generation as previously determined by Gill et al. (44) – was used for calculations in this study.
Macrophage infection studies.
Bone Marrow-derived macrophages (BMDMs) were extracted and infected with the panel of complemented strains built into the CDC1551 Δppe51 mutant background at a multiplicity of infection (MOI) of 1:1 using previously described methods (70). BMDMs were activated by treating with 100 units/mL IFN-γ overnight, followed by treatment with 10 ng/mL lipopolysaccharide overnight. Infected BMDMs were lysed at days 0, 3, 6, and 9 and intracellular bacterial lysates were serially diluted and enumerated on 7H10 + 10% OADC agar plates. Each strain for each time point was performed in triplicate. BMDMs were also infected with CDC1551 strains containing the pBP10 replication clock plasmid as described in the pBP10 in vitro experiments using the same macrophage infection methods described above with minor modifications. BMDMs infected with pBP10-containing strains were lysed at days 0, 2, 4, 6, and 8 and enumerated on 7H10 + 10% OADC agar plates ±25 μg/μL kanamycin selection. Calculations for pBP10 plasmid loss and replication dynamics were performed as described in the in vitro pBP10 experiments.
Recombinant PPE51 protein production and purification.
The ORF of PPE51 was amplified using pET23::ppe51_FWD and pET23::ppe51_REV primers (Table S1) and cloned into the pET23a+ vector containing a C-terminal polyhistidine (His)-tag (Clontech). The cloned gene has a deletion of the final four C-terminal amino acids. Transformants propagated in E. coli BL21(DE3) were selected on LB agar plates containing ampicillin. The S211R mutation was introduced into the pET::ppe51-WT construct using the site-directed mutagenesis primers PPE51-S211R_FWD and PPE51-S211R_REV (Table S1) and the QuikChange site-directed mutagenesis kit (Stratagene). Overnight cultures were expanded into 250 mL of fresh LB media with ampicillin at an initial inoculum OD600 of 0.05 and grown to an OD600 of 0.6 at 37°C with shaking at 200 rpm. Proteins were then induced with 1 mM isopropyl-ß-d-thiogalactopyranoside (IPTG) at 18°C for 20 h. Culture was then harvested via centrifugation at 4000 rpm for 25 min at 4°C. Pellets were then lysed for 30 min on ice with occasional vortexing in ice-cold lysis buffer (50 mM phosphate buffer [pH 7.6], 200 mM NaCl, 0.1% Triton X-100, 0.1 mg/mL PMSF, 0.5 mg/mL lysozyme). Because PPE51 possibly interacts with glycerol, glycerol was completely removed from all buffers used during the purification process. Cells were further lysed by sonication and cell lysate was clarified via centrifugation at 14,000 rpm for 30 min at 4°C. Supernatant was loaded onto a nickel ion-containing affinity resin column and bound overnight with shaking at 4°C. Protein was washed first with wash buffer containing no imidazole and a second time with wash buffer containing 50 mM imidazole. PPE51 protein was then eluted into (4) 1 mL fractions with elution buffer containing 200 mM imidazole. Recombinant PPE51 was quantified using the Qubit assay.
PPE51 protein thermostability assay.
The thermostability assay was performed as previously described (71) with 13.5 μL of 0.635 mg/mL of batch-purified PPE51 samples aliquoted into PCR tube containing 1.5 μL of 100 mM glycerol, yielding a final glycerol concentration of 10 mM. Samples were incubated for 20 min at room temperature and transferred to PCR thermocyclers where they were incubated for an additional 5 min at the following temperatures: 35, 40, 45, 50, 55, 60, and 65°C. Samples were then centrifuged at 4000 rpm for 10 min to pellet precipitated protein. After centrifugation, soluble protein was removed from the tubes and detected in immunoblots using mouse anti-His tag monoclonal antibody followed by HRP-conjugated anti-mouse IgG secondary antibody. Enhanced chemiluminescence (ECL) immunoblotting substrate (Pierce) was used for immunoblot detection. The AI600 Chemiluminescent Imager was used to visualize and analyze immunoblot results.
ACKNOWLEDGMENTS
We thank members of the Abramovitch lab for critical reading of the manuscript and David Sherman for sharing the pBP10 clock plasmid. This research was supported by a grant from the NIH-NIAID (R01AI116605) and AgBioResearch.
S.J.D., J.J.B., and R.B.A. conceived the project. S.J.D. performed all of the experimental studies. M.M. conducted thermal stability assay studies. S.J.D. and R.B.A. wrote the manuscript. All authors reviewed the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Robert B. Abramovitch, Email: abramov5@msu.edu.
Patricia A. Champion, University of Notre Dame
REFERENCES
- 1.Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, Taylor K, Klein E, Manjunatha U, Gonzales J, Lee EG, Park SK, Raleigh JA, Cho SN, McMurray DN, Flynn JL, Barry CE. 2008. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun 76:2333–2340. 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Converse PJ, Karakousis PC, Klinkenberg LG, Kesavan AK, Ly LH, Allen SS, Grosset JH, Jain SK, Lamichhane G, Manabe YC, McMurray DN, Nuermberger EL, Bishai WR. 2009. Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infect Immun 77:1230–1237. 10.1128/IAI.01117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munoz-Elias EJ, McKinney JD. 2006. Carbon metabolism of intracellular bacteria. Cell Microbiol 8:10–22. 10.1111/j.1462-5822.2005.00648.x. [DOI] [PubMed] [Google Scholar]
- 4.Loebel RO, Shorr E, Richardson HB. 1933. The influence of adverse conditions upon the respiratory metabolism and growth of human tubercle bacilli. J Bacteriol 26:167–200. 10.1128/jb.26.2.167-200.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vandal OH, Nathan CF, Ehrt S. 2009. Acid resistance in Mycobacterium tuberculosis. J Bacteriol 191:4714–4721. 10.1128/JB.00305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voskuil MI, Bartek IL, Visconti K, Schoolnik GK. 2011. The response of mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front Microbiol 2:105. 10.3389/fmicb.2011.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wayne LG, Sohaskey CD. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu Rev Microbiol 55:139–163. 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y. 2004. Persistent and dormant tubercle bacilli and latent tuberculosis. Front Biosci 9:1136–1156. 10.2741/1291. [DOI] [PubMed] [Google Scholar]
- 9.Deb C, Lee C-M, Dubey VS, Daniel J, Abomoelak B, Sirakova TD, Pawar S, Rogers L, Kolattukudy PE. 2009. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One 4:e6077. 10.1371/journal.pone.0006077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rohde KH, Abramovitch RB, Russell DG. 2007. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2:352–364. 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Baker JJ, Dechow SJ, Abramovitch RB. 2019. Acid fasting: modulation of Mycobacterium tuberculosis metabolism at acidic pH. Trends Microbiol 27:942–953. 10.1016/j.tim.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vandal OH, Pierini LM, Schnappinger D, Nathan CF, Ehrt S. 2008. A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat Med 14:849–854. 10.1038/nm.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abramovitch RB, Rohde KH, Hsu FF, Russell DG. 2011. aprABC: a Mycobacterium tuberculosis complex-specific locus that modulates pH-driven adaptation to the macrophage phagosome. Mol Microbiol 80:678–694. 10.1111/j.1365-2958.2011.07601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez E, Samper S, Bordas Y, Guilhot C, Gicquel B, Martín C. 2001. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol Microbiol 41:179–187. 10.1046/j.1365-2958.2001.02500.x. [DOI] [PubMed] [Google Scholar]
- 15.Johnson BK, Colvin CJ, Needle DB, Mba Medie F, Champion PAD, Abramovitch RB. 2015. The carbonic anhydrase inhibitor ethoxzolamide inhibits the Mycobacterium tuberculosis phoPR regulon and esx-1 secretion and attenuates virulence. Antimicrob Agents Chemother 59:4436–4445. 10.1128/AAC.00719-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker JJ, Abramovitch RB. 2018. Genetic and metabolic regulation of Mycobacterium tuberculosis acid growth arrest. Sci Rep 8:4168. 10.1038/s41598-018-22343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker JJ, Johnson BK, Abramovitch RB. 2014. Slow growth of Mycobacterium tuberculosis at acidic pH is regulated by phoPR and host-associated carbon sources. Mol Microbiol 94:56–69. 10.1111/mmi.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camus JC, Pryor MJ, Medigue C, Cole ST. 2002. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology (Reading) 148:2967–2973. 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 19.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537–544. 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 20.Karboul A, Mazza A, Gey van Pittius NC, Ho JL, Brousseau R, Mardassi H. 2008. Frequent homologous recombination events in Mycobacterium tuberculosis PE/PPE multigene families: potential role in antigenic variability. J Bacteriol 190:7838–7846. 10.1128/JB.00827-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akhter Y, Ehebauer MT, Mukhopadhyay S, Hasnain SE. 2012. The PE/PPE multigene family codes for virulence factors and is a possible source of mycobacterial antigenic variation: perhaps more? Biochimie 94:110–116. 10.1016/j.biochi.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 22.Mukhopadhyay S, Nair S, Ghosh S. 2012. Pathogenesis in tuberculosis: transcriptomic approaches to unraveling virulence mechanisms and finding new drug targets. FEMS Microbiol Rev 36:463–485. 10.1111/j.1574-6976.2011.00302.x. [DOI] [PubMed] [Google Scholar]
- 23.Bhat KH, Ahmed A, Kumar S, Sharma P, Mukhopadhyay S. 2012. Role of PPE18 protein in intracellular survival and pathogenicity of Mycobacterium tuberculosis in mice. PLoS One 7:e52601. 10.1371/journal.pone.0052601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. 2006. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA 103:8060–8065. 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitra A, Speer A, Lin K, Ehrt S, Niederweis M. 2017. PPE surface proteins are required for heme utilization by Mycobacterium tuberculosis. mBio 8:e01720-16. 10.1128/mBio.01720-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Hendrickson RC, Meikle V, Lefkowitz EJ, Ioerger TR, Niederweis M. 2020. Comprehensive analysis of iron utilization by Mycobacterium tuberculosis. PLoS Pathog 16:e1008337. 10.1371/journal.ppat.1008337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q, Boshoff HIM, Harrison JR, Ray PC, Green SR, Wyatt PG, Barry CE. 2020. PE/PPE proteins mediate nutrient transport across the outer membrane of Mycobacterium tuberculosis. Science 367:1147–1151. 10.1126/science.aav5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espitia C, Laclette JP, Mondragón- Palomino M, Amador A, Campuzano J, Martens A, Singh M, Cicero R, Zhang Y, Moreno C. 1999. The PE-PGRS glycine-rich proteins of Mycobacterium tuberculosis: a new family of fibronectin-binding proteins? Microbiology (Reading) 145:3487–3495. 10.1099/00221287-145-12-3487. [DOI] [PubMed] [Google Scholar]
- 29.Sultana R, Tanneeru K, Guruprasad L. 2011. The PE-PPE domain in Mycobacterium reveals a serine alpha/beta hydrolase fold and function: an in-silico analysis. PLoS One 6:e16745. 10.1371/journal.pone.0016745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrett CK, Broadwell LJ, Hayne CK, Neher SB. 2015. Modulation of the activity of Mycobacterium tuberculosis LipY by its PE domain. PLoS One 10:e0135447. 10.1371/journal.pone.0135447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gey van Pittius NC, Sampson SL, Lee H, Kim Y, van Helden PD, Warren RM. 2006. Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions. BMC Evol Biol 6:95. 10.1186/1471-2148-6-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simeone R, Bottai D, Frigui W, Majlessi L, Brosch R. 2015. ESX/type VII secretion systems of mycobacteria: Insights into evolution, pathogenicity and protection. Tuberculosis (Edinb) 95 Suppl 1:S150–S154. 10.1016/j.tube.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 33.Ates LS, Ummels R, Commandeur S, van de Weerd R, van der Weerd R, Sparrius M, Weerdenburg E, Alber M, Kalscheuer R, Piersma SR, Abdallah AM, Abd El Ghany M, Abdel-Haleem AM, Pain A, Jiménez CR, Bitter W, Houben ENG. 2015. Essential role of the ESX-5 secretion system in outer membrane permeability of pathogenic mycobacteria. PLoS Genet 11:e1005190. 10.1371/journal.pgen.1005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sani M, Houben ENG, Geurtsen J, Pierson J, de Punder K, van Zon M, Wever B, Piersma SR, Jiménez CR, Daffé M, Appelmelk BJ, Bitter W, van der Wel N, Peters PJ. 2010. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog 6:e1000794. 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sampson SL. 2011. Mycobacterial PE/PPE proteins at the host-pathogen interface. Clin Dev Immunol 2011:497203. 10.1155/2011/497203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gey Van Pittius NC, Gamieldien J, Hide W, Brown GD, Siezen RJ, Beyers AD. 2001. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol 2:Research0044. 10.1186/gb-2001-2-10-research0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abdallah AM, Verboom T, Weerdenburg EM, Gey van Pittius NC, Mahasha PW, Jiménez C, Parra M, Cadieux N, Brennan MJ, Appelmelk BJ, Bitter W. 2009. PPE and PE_PGRS proteins of Mycobacterium marinum are transported via the type VII secretion system ESX-5. Mol Microbiol 73:329–340. 10.1111/j.1365-2958.2009.06783.x. [DOI] [PubMed] [Google Scholar]
- 38.Tufariello JM, Chapman JR, Kerantzas CA, Wong K-W, Vilchèze C, Jones CM, Cole LE, Tinaztepe E, Thompson V, Fenyö D, Niederweis M, Ueberheide B, Philips JA, Jacobs WR. 2016. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc Natl Acad Sci USA 113:E348–E357. 10.1073/pnas.1523321113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitra A, Ko YH, Cingolani G, Niederweis M. 2019. Heme and hemoglobin utilization by Mycobacterium tuberculosis. Nat Commun 10:4260. 10.1038/s41467-019-12109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korycka-Machała M, Pawełczyk J, Borówka P, Dziadek B, Brzostek A, Kawka M, Bekier A, Rykowski S, Olejniczak AB, Strapagiel D, Witczak Z, Dziadek J. 2020. PPE51 is involved in the uptake of disaccharides by Mycobacterium tuberculosis. Cells 9:603. 10.3390/cells9030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malen H, De Souza GA, Pathak S, Softeland T, Wiker HG. 2011. Comparison of membrane proteins of Mycobacterium tuberculosis H37Rv and H37Ra strains. BMC Microbiol 11:18. 10.1186/1471-2180-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Babu Sait MR, Koliwer-Brandl H, Stewart JA, Swarts BM, Jacobsen M, Ioerger TR, Kalscheuer R. 2022. PPE51 mediates uptake of trehalose across the mycomembrane of Mycobacterium tuberculosis. Sci Rep 12 10.1038/s41598-022-06109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy KC, Nelson SJ, Nambi S, Papavinasasundaram K, Baer CE, Sassetti CM. 2018. ORBIT: a new paradigm for genetic engineering of mycobacterial chromosomes. mBio 9:e01467-18. 10.1128/mBio.01467-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gill WP, Harik NS, Whiddon MR, Liao RP, Mittler JE, Sherman DR. 2009. A replication clock for Mycobacterium tuberculosis. Nat Med 15:211–214. 10.1038/nm.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bellerose MM, Baek S-H, Huang C-C, Moss CE, Koh E-I, Proulx MK, Smith CM, Baker RE, Lee JS, Eum S, Shin SJ, Cho S-N, Murray M, Sassetti CM. 2019. Common variants in the glycerol kinase gene reduce tuberculosis drug efficacy. mBio 10:e00663-19. 10.1128/mBio.00663-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Asensio JG, Maia C, Ferrer NL, Barilone N, Laval F, Soto CY, Winter N, Daffé M, Gicquel B, Martín C, Jackson M. 2006. The virulence-associated two-component phoP-phoR system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. J Biological Chemistry 281:1313–1316. 10.1074/jbc.C500388200. [DOI] [PubMed] [Google Scholar]
- 47.Walters SB, Dubnau E, Kolesnikova I, Laval F, Daffe M, Smith I. 2006. The Mycobacterium tuberculosis phoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol Microbiol 60:312–330. 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 48.Sirakova TD, Dubey VS, Deb C, Daniel J, Korotkova TA, Abomoelak B, Kolattukudy PE. 2006. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology (Reading, Engl) 152:2717–2725. 10.1099/mic.0.28993-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baek S-H, Li AH, Sassetti CM. 2011. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol 9:e1001065 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rohde KH, Veiga DFT, Caldwell S, Balázsi G, Russell DG. 2012. Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog 8:e1002769. 10.1371/journal.ppat.1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gouzy A, Healy C, Black KA, Rhee KY, Ehrt S. 2021. Growth of Mycobacterium tuberculosis at acidic pH depends on lipid assimilation and is accompanied by reduced GAPDH activity. Proc Natl Acad Sci USA 118:e2024571118. 10.1073/pnas.2024571118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. 2011. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog 7:e1002093. 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guerrini V, Prideaux B, Blanc L, Bruiners N, Arrigucci R, Singh S, Ho-Liang HP, Salamon H, Chen P-Y, Lakehal K, Subbian S, O'Brien P, Via LE, Barry CE, Dartois V, Gennaro ML. 2018. Storage lipid studies in tuberculosis reveal that foam cell biogenesis is disease-specific. PLoS Pathog 14:e1007223. 10.1371/journal.ppat.1007223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mishra KC, de Chastellier C, Narayana Y, Bifani P, Brown AK, Besra GS, Katoch VM, Joshi B, Balaji KN, Kremer L. 2008. Functional role of the PE domain and immunogenicity of the Mycobacterium tuberculosis triacylglycerol hydrolase LipY. Infect Immun 76:127–140. 10.1128/IAI.00410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stahl C, Kubetzko S, Kaps I, Seeber S, Engelhardt H, Niederweis M. 2001. MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis. Mol Microbiol 40:451–464. 10.1046/j.1365-2958.2001.02394.x. [DOI] [PubMed] [Google Scholar]
- 57.Stephan J, Bender J, Wolschendorf F, Hoffmann C, Roth E, Mailänder C, Engelhardt H, Niederweis M. 2005. The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol Microbiol 58:714–730. 10.1111/j.1365-2958.2005.04878.x. [DOI] [PubMed] [Google Scholar]
- 58.Danilchanka O, Pavlenok M, Niederweis M. 2008. Role of porins for uptake of antibiotics by Mycobacterium smegmatis. Antimicrob Agents Chemother 52:3127–3134. 10.1128/AAC.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zambolin S, Clantin B, Chami M, Hoos S, Haouz A, Villeret V, Delepelaire P. 2016. Structural basis for haem piracy from host haemopexin by Haemophilus influenzae. Nat Commun 7:11590. 10.1038/ncomms11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu W, DeJesus MA, Rücker N, Engelhart CA, Wright MG, Healy C, Lin K, Wang R, Park SW, Ioerger TR, Schnappinger D, Ehrt S. 2017. Chemical genetic interaction profiling reveals determinants of intrinsic antibiotic resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 61:e01334-17. 10.1128/AAC.01334-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolfe LM, Mahaffey SB, Kruh NA, Dobos KM. 2010. Proteomic definition of the cell wall of Mycobacterium tuberculosis. J Proteome Res 9:5816–5826. 10.1021/pr1005873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McEvoy CRE, Cloete R, Müller B, Schürch AC, van Helden PD, Gagneux S, Warren RM, Gey van Pittius NC. 2012. Comparative analysis of Mycobacterium tuberculosis pe and ppe genes reveals high Sequence variation and an apparent absence of selective constraints. PLoS One 7:e30593. 10.1371/journal.pone.0030593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee W, VanderVen BC, Fahey RJ, Russell DG. 2013. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J Biol Chem 288:6788–6800. 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Piddington DL, Kashkouli A, Buchmeier NA. 2000. Growth of Mycobacterium tuberculosis in a defined medium is very restricted by acid pH and Mg(2+) levels. Infect Immun 68:4518–4522. 10.1128/IAI.68.8.4518-4522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. 2012. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res 40:W622–W627. 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 10.48550/arXiv.1303.3997. [DOI]
- 67.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bachrach G, Colston MJ, Bercovier H, Bar-Nir D, Anderson C, Papavinasasundaram KG. 2000. A new single-copy mycobacterial plasmid, pMF1, from Mycobacterium fortuitum which is compatible with the pAL5000 replicon. Microbiology (Reading 146:297–303. 10.1099/00221287-146-2-297. [DOI] [PubMed] [Google Scholar]
- 70.Johnson BK, Abramovitch RB. 2015. Macrophage infection models for Mycobacterium tuberculosis. Methods Mol Biol 1285:329–341. 10.1007/978-1-4939-2450-9_20. [DOI] [PubMed] [Google Scholar]
- 71.Park Y, Ahn Y-M, Jonnala S, Oh S, Fisher JM, Goodwin MB, Ioerger TR, Via LE, Bayliss T, Green SR, Ray PC, Wyatt PG, Barry CE, Boshoff HI. 2019. Inhibition of CorA-dependent magnesium homeostasis is cidal in Mycobacterium tuberculosis. Antimicrob Agents Chemother 63:e01006-19. 10.1128/AAC.01006-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S8 and Table S1. Download jb.00212-22-s0001.pdf, PDF file, 3.7 MB (3.7MB, pdf)
Table S2. Download jb.00212-22-s0002.xlsx, XLSX file, 0.05 MB (56.3KB, xlsx)