Abstract
Background
Patients with rare diseases often undergo a diagnostic odyssey that can last many years until the diagnosis is definitively established. To improve the diagnosis and treatment of these patients, the German National Task Force for Patients With Rare Diseases (Nationales Aktionsbündnis für Menschen mit Seltenen Erkrankungen, NAMSE) has recommended the creation of Rare Disease Centers (RDCs).
Methods
As part of the joint Translate-NAMSE project, sponsored by the G-BA Innovation Fund (G-BA, German Federal Joint Committee), we investigated the performance of RDCs in establishing the diagnosis of patients suspected to have a rare disease. The results of interdisciplinary case conferences and of exome diagnostic tests were analyzed in a prospective, multicenter observational study.
Results
A total of 5652 patients (of whom 3619 were under 18 years old, and 2033 were at least 18 years old) from 10 RDCs who did not yet have a definitive diagnosis of a rare disease were included in the study. On average, those who were under 18 years old had been symptomatic for 4.5 years without receiving a diagnosis in a standard care setting; the analogous figure for adult patients was 8.2 years. Over the course of this project (2017–2021), 1682 patients (30%) received a definitive diagnosis. 193 had a common disease, 88 had a psychosomatic disease (only in patients who were at least 18 years old), and 1401 had a rare disease. 14 850 case conferences were conducted. 1599 exome analyses led to 506 definitive genetic diagnoses (32%).
Conclusion
A diagnostic evaluation with the aid of interdisciplinary case conferences and the opportunity for exome analysis can be of benefit to people with rare diseases who have not received a definitive diagnosis in a standard care setting. Further improvement of the diagnosis rate can come from whole-genome analysis and from the introduction of an international registry.
Diseases that affect no more than 1 in 2000 citizens are considered rare diseases (RD) in Europe. The group of RDs includes a large number of quite different diseases, often with a complex phenotype and not only involving one organ. Approximately 6000 different RDs are known at present (1). Overall, the prevalence of all RDs in the population is thought to be between 3.5 and 5% (1). Despite the great heterogeneity, what many patients with an RD have in common is that the diagnosis is made late—often only after a diagnostic odyssey lasting many years (2).
Over 80% of RDs can already occur in childhood. In 75% of cases, a genetic cause in the form of a monogenic disease is suspected (1). This includes more than 3000 diseases that occur less frequently than 1 in 1 million population and have often been reported only in individual families. Initial manifestation has been reported for 12% of RDs during adulthood and for 18% in adulthood or childhood. Apart from genetic RDs, patients may also present with acquired rare diseases, where the focus is on autoimmune diseases, degenerative neurological disorders, rare malignancies, and rare infectious diseases (1).
Given their diversity and complexity, RDs have only recently been classified as a separate group of diseases requiring a special interdisciplinary approach to diagnosis and clinical care. Initiated by patient advocacy groups, measures have been called for in Europe since 2009 to improve care for patients with RDs (3). In Germany, recommendations were subsequently developed by the German National Task Force for Patients With Rare Diseases (Nationales Aktionsbündnis für Menschen mit Seltenen Erkrankungen, NAMSE), primarily to establish Centers for Rare Diseases (CRDs) at university hospitals (4). Since then, more than 30 rare disease centers have been created. The NAMSE-A centers of a CRD assume coordinating tasks in this regard, focussing on interdisciplinary diagnostics for patients in whom an RD is suspected. NAMSE-B centers represent institutions with care expertise for a specific RD or group of RDs (5).
Project description and aims
The Innovation Fund Project TRANSLATE-NAMSE was designed to assess the significance and performance of these newly established CRDs and, in particular, the NAMSE-A centers. The aim was to assess the diagnosis in pediatric and adult patients with a suspected RD in whom standard care procedures had previously failed to establish a definitive diagnosis. This involved evaluating the structured diagnostic process, up to and including the implementation of exome diagnostics in daily care, where appropriate.
Methods
The Innovation Fund Project TRANSLATE-NAMSE was designed as a prospective multicenter observational study without a comparison group. It was not possible to generate adequate reference groups at the same time or in the past because of the health care structures in place, the heterogeneity of the RD diagnoses, and the rarity of some cases.
Therefore, the main endpoint of the study was defined as diagnosis made or not made.
Based on a cooperation agreement with a total of ten CRDs and participating partner health insurers, patients for whom no definitive diagnosis had previously been established in a standard care setting were recruited for TRANSLATE-NAMSE from December 2017 to March 2020. Further information on the project partners and collaborators is provided in the eMethods section. Individuals with an uncertain diagnosis (Cohort 1) and those with a definite suspected RD diagnosis (Cohort 2) from the areas of neurological movement disorders (≥18 years) and metabolic defects, congenital endocrinopathies, anemias, immunodeficiencies, and autoinflammation in pediatric patients (<18 years) were included.
In this project, the model of care involving a case conference-based diagnostic process was implemented using a patient pathway consented to by the participating CRDs. Therefore, an initial interdisciplinary case conference was conducted after reviewing and processing the available diagnostic reports. If no RD diagnosis was possible on the basis of the available findings and an RD was still suspected, supplementary special or exome diagnostics were requested by interdisciplinary consensus—with the mandatory involvement of specialists in human genetics. Here, the individual clinical picture or spectrum of the patients under discussion was documented using human phenotype ontology (HPO) (6).
In further interdisciplinary case conferences, the findings of this additional diagnostic workup were evaluated and, if possible, a definitive diagnosis was made. A diagnosis of an RD was considered established if its prevalence was less than 1 in 2000 according to the Orphanet database; RD diagnoses were documented using Orpha codes (7).
The object of the project funding was to finance staff employed for the preparation, coordination and implementation of the case conferences as well as communication with the patients (so-called pilots and coordinators of the NAMSE-A centers). The project-funded grant for a case conference (excluding the cost of expert participation) was 260 euros.
Exome diagnostics involved sequencing and bioinformatic analysis—in some cases as trio exomes, i.e., with additional sequencing of the parents. This was funded, on application, by the health insurance companies involved in the project (3000 euros) and conducted for patients of all ten sites in four partner CRDs with expertise in genomic diagnostics (Berlin, Bonn, Munich, Tübingen).
Elements characterizing the care pathway, for example, number, duration, composition of case conferences, and exome diagnostic parameters, were recorded in the project database.
Information on patient satisfaction was collected using a questionnaire on completion of the diagnostic process (ePatient Questionnaire). Data for the health economic evaluation were gathered by means of a questionnaire sent to each of the health care providers and patients, as well as through data from the health insurance funds.
Ethics and data protection
The TRANSLATE-NAMSE project was carried out in compliance with ethical and data protection requirements, in consultation with local data protection officers and ethics committees. The initial opinion was rendered by the Ethics Committee of the Charité (EA2/140/17).
Results
During the funding period from December 2017 to March 2020, a total of 5652 patients who had previously failed to receive a definitive RD diagnosis in a standard care setting were enrolled in the project. Of these, 3619 were under 18 years of age. Prior to inclusion in the project, data from patient questionnaires showed that patients had been symptomatic for a mean of 4.5 years (pediatric patients from the age of one) or 8.2 years (adults) and had not received a diagnosis.
Overall, by the end of the project, a definitive diagnosis had been established in 1682 participants (30%)—including 1161 pediatric (32%) and 521 adult patients aged ≥18 years (26%). This involved a total of 14 850 case conferences (2.6 per involved person) with an average of 5.5 experts. The documented average duration of case conferences for pediatric and adult patients was 56 and 42 minutes, respectively, and preparation times were 43 and 96 minutes, respectively. Exome diagnostics were performed for 1599 subjects, resulting in a definitive RD diagnosis in 506 (32%) of them by the end of the project (table).
Table. Overview of results.
| Total | Patients | ||
| <18 years | ≥18 years | ||
| Patients recruited without a diagnosis | 5652 | 3619 | 2033 |
| Total number of diagnoses made | 1682 | 1161 | 521 |
| Rare diagnoses (<1:2000; orpha code) | 1401 | 1088 | 313 |
| Common diagnoses | 193 | 73 | 120 |
| Psychosomatic disorders | 88 | 0 | 88 |
| Diagnosis made using exome sequencing (a total of 1599 exome diagnostic tests) | 506 | 415 | 91 |
| Total number of case conferences | 14 850 | 8110 | 6740 |
| Case conferences per patient | 3 | 3 | 2 |
| Number of experts per case conference | 6 | 7 | 4 |
Adult patients
In Cohort 1 of adult participants with previously uncertain diagnosis, a definite diagnosis was reached in 395 of the 1768 patients (22%) by the end of the project. One hundred and eighty-three involved individuals were diagnosed with a common disorder, and 212 were diagnosed with a rare disorder. A total of 73 of the common disorders were classified under the psychosomatic spectrum of disorders, primary somatization disorder or somatoform disorder—unspecified.
In Cohort 2 of adult patients with a suspected diagnosis of a rare neurological movement disorder, a confirmed diagnosis was reached in 126 of 265 (48%) cases. One hundred and one patients were diagnosed with a rare disease and 25 with a common disease, 15 of which belonged to the group of psychosomatic disorders.
In total, 91 of the 313 RD diagnoses in adult Cohorts 1 and 2 were determined by exome examination (29%) (table).
Patients in childhood and adolescence
Of 3008 pediatric patients in Cohort 1 with a previously uncertain diagnosis, a total of 795 (24%) were found to have an SD and another 73 were found to have a common disease. No psychosomatic diagnosis was made in the pediatric participant group.
In pediatric Cohort 2, which represented 611 patients with a definite suspected diagnosis, 366 individuals received a certain RD diagnosis (60%). The indication areas showed different frequencies: anemias 83%, endocrinopathies 97%, autoinflammation 70%, immunodeficiencies 60%, metabolic defects 96%. Overall, 415 of the 1161 pediatric RD diagnoses were made using exome diagnostics (35%) (table).
Rare disease diagnosis types
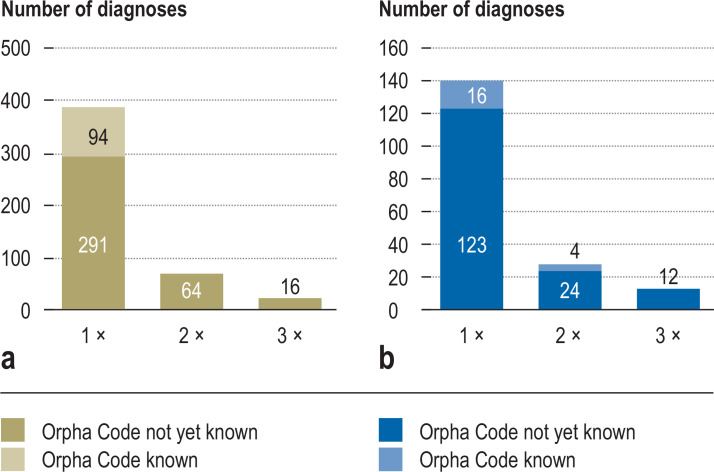
A very broad spectrum of diagnosed RDs was evident (Figure 1, Box). Overall, more than 400 RDs were diagnosed only once. These were mostly extremely rare diseases with a prevalence of <1 in 1 million. Examples are shown in Figure 1. Furthermore, exome diagnostics were used to diagnose more than 100 diseases that have only been recently reported for the first time (<3 years) and were therefore still largely unknown in standard care. No Orphanet number was available for these as yet. In both pediatric and adult patients of Cohort 1, genetically determined neurological diseases, developmental disorders, and malformation syndromes were the most common conditions.
Figure 1.
Frequency of diagnoses of rare diseases (RD), examples of diagnoses
a) RD diagnoses 0–18 years
b) RD diagnoses ≥18 years
BOX.
Examples of one-off diagnoses (0–18 years)
-
Activated PI3K delta syndrome
Orpha: 397596
-
Atypical hemolytic uremic syndrome with complement gene abnormality
Orpha: 544472
-
BICD2-related proximal spinal muscular atrophy
Orpha: 363454
-
C11ORF73-related hypomyelinating leukodystrophy
Orpha: 495844
-
DYRK1A-related intellectual disability syndrome
Orpha: 464306
-
Developmental delay and facial dysmorphism syndrome due to MED13L deficiency
Orpha: 369891
-
Kyphoscoliotic Ehlers-Danlos syndrome due to FKBP22 deficiency
Orpha: 300179
-
Loeys-Dietz syndrome
Orpha: 60030
-
Vici syndrome
Orpha: 1493
-
Walker-Warburg syndrome
Orpha: 899
Examples of one-off diagnoses (≥18 years)
-
White-Sutton syndrome
Orpha: 468687
-
Coffin-Siris syndrome 6
Orpha: 1465
-
Spinocerebellar ataxia 27
Orpha: 98764
-
Mandibulofacial dysostosis with microcephaly, Guion-Almeida type
Orpha: 79113
-
Werner syndrome
Orpha: 902
-
KBG syndrome
Orpha: 2332
-
Leukocyte adhesion deficiency (LAD) type II
Orpha: 99843
-
Speech and language disorders with orofacial dyspraxia
Orpha: 209908
-
Spastic paraplegia type 46
Orpha: 320391
-
X chromosome-related intelligence deficit hypotension movement disorder syndrome
Orpha: 457260
Avoidable diagnostic investigations
After case closure, we assessed whether previous diagnostic and care services could have been avoided for 163 patients with confirmed diagnoses, taken as examples from Cohort 1. This involved inpatient hospital stays in 31% of involved patients, specialist consults in 51%, magnetic resonance imaging in 27%, computed tomography in 10%, panel sequencing in 23%, and single gene sequencing in 13%.
Patient satisfaction
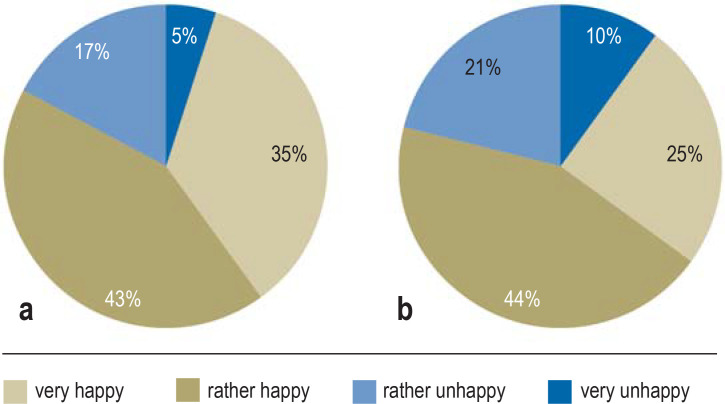
Using the patient questionnaire to evaluate the interventions from the perspective of those involved, the TRANSLATE-NAMSE process was finally assessed in a total of 1001 (21%) of the 4776 participants from Cohort 1 or their families (529 pediatric and 472 adult patients). Regardless of their age, the majority were satisfied with the care pathway; by the time a diagnosis was established, this amounted to 78% of patients. Even without a definitive diagnosis during the project period, 69% of participants were still satisfied (figure 2).
Figure 2.
Answer to the question: How happy are you with the project?
A total of 1001 assessable questionnaires
a) diagnosis established (n = 674); b) no diagnosis (n = 674)
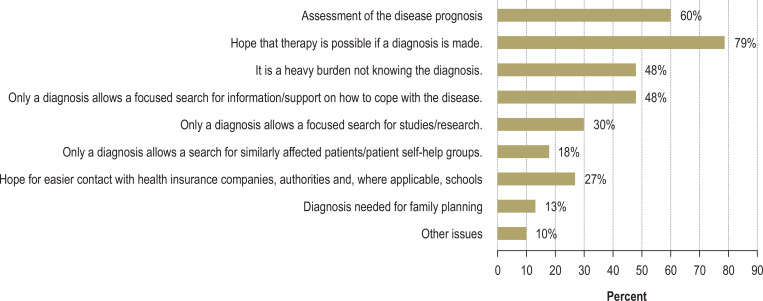
An important aspect of patient satisfaction related to the wish to know their own diagnosis. Figure 3 presents the reasons why, from the patients’ point of view, knowledge of their own diagnosis was cited in the questionnaire as being important. Apart from their hope for a treatment option (79%), 60% cited knowledge of disease prognosis as important. Forty-eight percent each cited the opportunity to gain information about their disease and the burden of not knowing the diagnosis.
Figure 3.
Answer to the question: Why is knowledge of the diagnosis important? A total of 1301 assessable questionnaires
Discussion
The TRANSLATE-NAMSE project was able to demonstrate that the measure put forward in NAMSE of an interdisciplinary approach to establishing the diagnosis at RDCs can be applied in a standard care setting and is useful for establishing the diagnosis of patients with RDs. During the study, this form of care was offered to more than 5600 participants. In total, a definitive diagnosis was established in 1682 patients during the project (32% of pediatric patients and 26% of adult patients), although all of them had not been previously diagnosed by standard care examinations over a prolonged period of time (mean 4.5 years in pediatric patients and 8.2 years in adult patients).
Interdisciplinary collaboration during case conferences was crucial for successfully establishing the diagnosis. In view of the considerable number of presenting symptoms, it was important to enlist the participation of experts from various disciplines. On average, five different specialties were involved. It also became evident that two independent teams with expertise in pediatrics and adult medicine were required to liaise with experts in different clinical areas and to coordinate their involvement in the case conferences. Given the high proportion of genetic and neurological disorders encountered, experts in human genetics and neurology or neuropediatrics were essential participants in the majority of the interdisciplinary case conferences. The proportion of nearly 20% psychosomatic diagnoses among adult patients also highlights the significant role of psychosomatic expertise in the diagnostic process involving adults.
Of the 1599 exome diagnostics performed, 506 led to a definitive diagnosis (32%) by detecting a causal mutation. This was within the expected range as seen in international study results on exome sequencing, which was between 25 and 35% for comparable, non-symptom specific cohorts (8– 10). The role of interdisciplinary case conferences for these results of exome diagnostics becomes clear on considering that variants of, initially, uncertain significance (“VUS class 3” [11]), which can only be considered causal upon further interdisciplinary discussion of the clinical picture and the specific family constellation, are frequently identified during exome sequencing.
Interestingly, this structured case conference-based diagnostic process showed a higher proportion (92 of the 212 diagnoses made, 43%) of genetically based diagnoses in the adult population than in previous comparable studies in adult-only collectives, where the proportion was only 17% (12). Particularly in the selected patient group of adults with neurological movement disorders, the presented case conference-based process was successful, with a percentage of genetically established diagnoses amounting to 46%.
The project was accepted by the involved persons and their families with high satisfaction scores. This should be viewed particularly in the light of the many years of unsuccessful diagnostic efforts that often preceded it. Patients confirmed their desire for a definitive diagnosis when completing the project questionnaires. Bogart et al. have previously shown that simply obtaining a diagnosis after years of diagnostic odyssey improved quality of life in patients with RDs (13).
Despite all efforts, a diagnosis was not established in 70% of patients in TRANSLATE-NAMSE either. A range of further measures are necessary to bring about an improvement here:
A re-evaluation of the identified sequence variants should be established after a specified time for individuals with a previously unremarkable exome diagnosis (9, 14), because the identification of new disease-associated mutations in known, or even previously unknown, disease genes can also be expected in the future (14).
Furthermore, whole genome sequencing, including non-coding sequences, should also be made available for these patients (8). While whole genome sequencing has become much easier as a result of methodological advances, interpreting the numerous variants in the non-coding sequences is a major challenge. Whole genome diagnostics in RDs is currently being prepared by the pilot project launched by the Federal Ministry of Health in accordance with Section 64e of the German Social Security Code Part V and is to be implemented in health care with funding from health insurance providers (15).
Another measure for patients without a diagnosis is the creation of a national patient registry for undiagnosed rare diseases using HPO-based phenotyping (16). Regular re-evaluation of available phenomic and genomic data, including comparisons with international patient data stored in databases, should make it possible to diagnose new disease entities (17).
Limitations
In TRANSLATE-NAMSE, only a quantitative process evaluation and a quantitative and qualitative evaluation of the satisfaction of the included patients with the outcome of the project were conducted. Nevertheless, important insights were gained into essential aspects of a structured diagnostic process in everyday care.
Conclusion
The health care project presented here demonstrates that implementation of the type of care recommended in NAMSE with the aid of interdisciplinary case conferences and the opportunity for exome sequencing is feasible in practice and improves the diagnosis of people with an RD.
The full range of healthcare services provided by case conference-based diagnostics, including exome diagnostics, should, if possible, be made available to all patients at existing NAMSE-A centers.
Supplementary Material
eMethods
Collaborators and institutions involved in TRANSLATE-NAMSE
Collaborators
At the clinical sites:
Berlin: Magdalena Danyel1–1,1–2, Sarina Schwartzmann1–1,1–2, Annemarie Bösch1–1, Denis Horn1–2,, Nadja Ehmke1–2, Felix Boschann1–2, Stefan Mundlos1–1,1–2, Angela Kaindl1–3, Christoph Bührer1–4, Tilmann Kalinich1–5, Horst v Bernuth1–5, Natalie Weinhold1–6, Phillip Buffler1–6, Dominik Müller1–6, Susanne Holzhauer1–7, Manuel Holtgrewe1–8, Charlotte Wernicke 1–1,1–9, Laura Schmidt-Pennington1–1,1–9, Joachim Spranger1–1,1–9, Annette Grüters1–10, Heiko Krude1–1,1–10
Bochum/Essen: Corinna Grasemann2–1,2–2, Nora Matar2–1,2–2, Janet Atinga2–1,2–2, Folke Brinkmann2–1,2–2, Frank Kaiser2–3,2–4, Bernhard Horsthemke2–3,2–4, Adela Della Marina2–5, Eva Manka2–3,2–6, Cordula Kiewert2–6, Martin Munteanu2–4, Alma Kuechler2–4, Raphael Hirtz2–6, Michael Schündeln2–7, Florian Stehling2–7, Sabine Hoffjan2–8, Nicole Unger2–9, Tim Hagenacker2–10, Stephan Klebe2–10, Paul Manka2–11, Freya Dröge2–12, Florian Grabellus2–13, Huu Nguyen2–8, Björn Bühring2–14
Bonn: Martin Mücke3–1, Tim Bender3–1,3–2, Nadine Weinstock3–1, Lorenz Grigull3–1, Julia Sellin3–1, Marzena Marawiec3–1, Christiane Stieber3–1,3–3, Markus M. Nöthen3–1,3–2, Axel Schmidt3–1,3–2, Martina Kreiß3–1,3–2, Elisabeth Mangold3–2, Sophia Peters3–2, Hartmut Engels3–2, Peter Krawitz3–1,3–4, Alexej Knaus3–2,3–4, Tzung-Chien Hsieh3–2,3–4, Hannah Klinkhammer3–2,3–4, Thomas Klockgether3–1,3–5, Elena Schlapakow3–5,3–6, Valentin Schäfer3–1,3–7, Pantelis Karakostas8–1,8–7, Patrick Weydt3–1,3–7, Sarah Bernsen3–1,3–8, Cornelia Kornblum3–1,3–5
Dresden: Reinhard Berner4–1,4–2, Min Ae Lee-Kirsch4–1,4–2, Andre Heinen4–1,4–2, Julia Koerholz4–1,4–2, Tanita Kretschmer4–1,4–2, Manja Unrath4–1,4–2, Victoria Tüngler 4–1,4–2
Hamburg: Christoph Schramm5–1, Franziska Rillig5–1, Kurt Ullrich5–1, Felix Braun5–1, Maximilian Groffmann5–1, Cornelia Rudolph5–1, Christina Weiler Normann5–1, Maja Hempel5–2, Theresia Herget5–2, Jasmin Lisfeld5–2, Christian Schlein5–2, Christian Kubisch5–2, Ania C. Muntau5–3
Heidelberg: Georg F. Hoffmann6–1,6–2, Daniela Choukair6–1,6–2, Pamela Okun6–1,6–2, Petra Wagenlechner6–1,6–2, Christian P. Schaaf6–1,6–3, Eva M. C. Schwalbold6–1,6–3, Peter Burgard6–1,6–2, Franziska Krause6–2
Lübeck: Tobias Bäumer7–1, 7–4, Alexander Münchau7–1, 7–4, Annekathrin Ripke7–1, Christian Himstedt7–1, Martje Pauly7–1, 7–3, Olaf Hiort7–1, 7–5, Isabel Mönig7–5, Irina Hüning 7–2, Katja Lohmann7–3, Norbert Brüggemann7–3, Christine Klein7–3, Rebecca Herzog7–1
Munich: Fabian Hauck8–1, Stella Bergemann8–1, Astrid Blaschek8–1, Ingo Borggräfe8–1, Katharina Danhauser8–1, Julia Eilenberger8–1, Matthias Griese8–1, Lisa-Maria Köhler8–1, Bärbel Lange-Sperandio8–1, Eberhard Lurz8–1, Wolfgang Müller Felber8–1, Antonia Pelshenke8–1, Karl Reiter8–1, Esther Maier8–1, Katharina Vill, Christoph Klein8–1, Thomas Meitinger8–2, Tim Strom8–2, Riccardo Berutti8–2, Matias Wagner8–2, Theresa Brunet8–2, Melanie Brugger8–2, Katharina Mayerhanser8–2, Korbinian Riedhammer8–2
Tübingen: Andrea Bevot9–1,9–2, Janine Magg9–1,9–2, Ingeborg Krägeloh-Mann9–1,9–2, Monika Glauch9–1,9–3, Martin Kehrer9–1,9–3, Holm Graessner9–1,9–3, Tobias Haack9–1,9–3, Olaf Riess9–1,9–3, Stefanie Beck-Wödl9–1,9–3, Lena Zeltner9–1,9–4, Jutta Eymann9–1, Ludger Schoels9–1,9–4, Kathrin Grundmann-Hauser9–1,9–2, Till-Karsten Hauser9–1,9–5
Evaluation Dresden: Jochen Schmitt10, Gabriele Müller10, Diana Druschke10
Evaluation Berlin: Tobias Kurth11, Kerstin Wainwright11, Sylvana Baumgarten11
AOK: Werner Wyrwich12, Katja Basso 12
Barmer: Alfred Kindshofer13, Regina Kotzbacher13, Ursula Marschall13
ACHSE e. V.: Christine Mundlos14
Participating institutions
Berlin:
1–1 Center for Rare Diseases (BCSE), Charité—Berlin University of Medicine, 13353 Berlin
1–2 Institute for Human Genetics, Charité—Berlin University of Medicine, 13353 Berlin
1–3 Department of Pediatric Neurology, Charité—Berlin University of Medicine, 13353 Berlin
1–4 Department of Neonatology, Charité—Berlin University of Medicine, 13353 Berlin
1–5 Department of Pediatric Pneumology, Immunology and Intensive Care, Charité—Berlin University of Medicine, 13353 Berlin
1–6 Department of Pediatric Gastroenterology, Nephrology and Metabolic Diseases, Charité—Berlin University of Medicine, 13353 Berlin
1–7 Department of Pediatric Oncology, and Hematology, Charité—Berlin University of Medicine, 13353 Berlin
1–8 CoreUnit Bioinformatics, Berlin Institute of Health, Charité—Berlin University of Medicine, 13353 Berlin
1–9 Department of Endocrinology and Metabolic Disorders, Charité—Berlin University of Medicine, 13353 Berlin
1–10 Institute for Experimental Pediatric Endocrinology, Charité—Berlin University of Medicine, 13353 Berlin
Bochum/Essen:
2–1 Center for Rare Diseases Ruhr, CeSER, Catholic Hospital Bochum, Ruhr University Bochum, 44791 Bochum
2–2 Department of Pediatrics, Catholic Hospital Bochum, Ruhr University Bochum, 44791 Bochum
2–3 Center for Rare Diseases Essen, EZSE, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–4 Institute for Human Genetics, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–5 Clinic for Pediatrics I, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–6 Clinic for Pediatrics II, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–7 Clinic for Pediatrics III, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–8 Institute for Human Genetics, Ruhr University Bochum, 44791 Bochum
2–9 Department for Endocrinology, Diabetes and Metabolism, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–10 Department of Neurology, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–11 Department of Internal Medicine, Knappschaftskrankenhaus Bochum, Ruhr University Bochum, 44791 Bochum
2–12 ENT Department, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–13 Institute for Pathology, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–14 “Rheumatology Center of the Ruhr District”, Ruhr University Bochum, Herne
Bonn:
3–1 Center for Rare Diseases, University Hospital Bonn, 53127 Bonn
3–2 Institute for Human Genetics, University Hospital Bonn, 53127 Bonn
3–3 Institute of General Practice and Family Medicine, University Hospital Bonn, 53127 Bonn
3–4 Institute for Genomic Statistics and Bioinformatics, University Hospital Bonn, 53127 Bonn
3–5 Department of Neurology, University Hospital Bonn, 53127 Bonn
3–6 Department of Neurology, Halle (Saale) University Hospital, 06120 Halle (Saale)
3–7 Department of Internal Medicine III, University Hospital Bonn, 53127 Bonn
3–8 Department of Neurodegenerative Diseases, University Hospital Bonn, 53127 Bonn
Dresden:
4–1 Clinic and Outpatient Clinic for Pediatric and Adolescent Medicine, University Hospital and the Carl Gustav Carus Faculty of Medicine at the Technical University Dresden, 01307 Dresden
4–2 University Center for Rare Diseases (USE), University Hospital and the Carl Gustav Carus Faculty of Medicine at the Technical University Dresden, 01307 Dresden
Hamburg:
5–1 Martin Zeitz Center for Rare Diseases, University Medical Center Hamburg-Eppendorf, 20246 Hamburg
5–2 Institute for Human Genetics, University Medical Center Hamburg-Eppendorf, 20246 Hamburg
5–3 University Pediatric Department, University Medical Center Hamburg-Eppendorf, 20246 Hamburg
Heidelberg:
6–1 Center for Pediatric and Adolescent Medicine, University Hospital Heidelberg, 69120 Heidelberg
6–2 Center for Rare Diseases, University Hospital Heidelberg, 69120 Heidelberg
6–3 Institute for Human Genetics, University Hospital Heidelberg, 69120 Heidelberg
Lübeck:
7–1 Center for Rare Diseases, University Hospital Schleswig-Holstein Lübeck, 23562 Lübeck
7–2 Institute for Human Genetics, University Hospital Schleswig-Holstein Lübeck, 23562 Lübeck
7–3 Institute for Neurogenetics, University of Lübeck, 23562 Lübeck
7–4 Institute of Systemic Motor Research, University Hospital Schleswig Holstein, Campus Lübeck, 23562 Lübeck
7–5 Department for Pediatric and Adolescent Medicine, University Hospital Schleswig Holstein, Campus Lübeck, 23562 Lübeck
Munich:
8–1 Pediatric Department, Dr. von Hauner Children’s Hospital, Ludwig Maximilian University Hospital Munich, 81377 Munich
8–2 Institute for Human Genetics, Rechts der Isar Hospital, Technical University of Munich, 80337 München
Tübingen:
9–1 Center for Rare Diseases, University Hospital Tübingen, 72076 Tübingen
9–2 Department of Pediatric Neurology and Developmental Disorders, University Hospital Tübingen, 72076 Tübingen
9–3 Institute for Medical Genetics and Applied Genomics, University Hospital Tübingen, 72076 Tübingen
9–4 Department of Neurology, University Hospital Tübingen, 72076 Tübingen
9–5 Department of Radiology, University Hospital Tübingen, 72076 Tübingen
Evaluation:
10 Center for Evidence-Based Healthcare, University Hospital and the Carl Gustav Carus Faculty of Medicine at the Technical University Dresden, 01307 Dresden
11 Institute for Public Health, Charité—Berlin University of Medicine, 10117 Berlin
Health insurance funds:
12 AOK Nordost, 13409 Berlin
13 Barmer, 81675 Munich
Patient representatives:
14 ACHSE e.V., 13359 Berlin
Acknowledgments
Translated from the original German by Dr. Grahame Larkin, MD.
Footnotes
*2 Other authors and affiliations
Tobias Bäumer, Georg F. Hoffmann, Daniela Choukair, Reinhard Berner, Min Ae Lee-Kirsch, Martin Mücke, Corinna Grasemann, Annekatrin Ripke, Lena Zeltner, Gabriele Müller, Monika Glauch, Holm Graessner, Fabian Hauck, Christoph Klein, Markus M. Nöthen, Olaf Riess, Stefan Mundlos, Thomas Meitinger, Tobias Kurt, Kerstin L. Wainwright, Jochen Schmitt
Center for Rare Diseases, University Hospital Schleswig-Holstein Lübeck: Prof. Dr. med. Tobias Bäumer, Dr. med. Annekatrin Ripke
Center for Pediatric and Adolescent Medicine, University Hospital Heidelberg: Prof. Dr. med. Georg F. Hoffmann, Dr. med. Daniela Choukair
Clinic and Outpatient Clinic for Pediatric and Adolescent Medicine and University Center for Rare Diseases (USE), University Hospital and the Carl Gustav Carus Faculty of Medicine at the Technical University Dresden: Prof. Dr. med. Reinhard Berner, Prof. Dr. med. Min Ae Lee-Kirsch
Center for Rare Diseases Aachen (ZSEA), University Hospital of Aachen: Prof. Dr. med. Martin Mücke
Center for Rare Diseases Ruhr, CeSER, Clinic for Pediatrics, Catholic Hospital Bochum, Ruhr University Bochum: Prof. Dr. med. Corinna Grasemann
Center for Rare Diseases, University Hospital Tübingen: Dr. med. Lena Zeltner, Dr. rer. nat. Monika Glauch, Dr. rer. nat. Holm Graessner, Prof. Dr. med. Olaf Riess
Center for Evidence-Based Healthcare, University Hospital and the Carl Gustav Carus Faculty of Medicine at the Technical University Dresden: Dr. med. Gabriele Müller, Prof. Dr. med. Jochen Schmitt
Pediatric Department at the Dr. von Hauner Children’s Hospital and Munich Center for Rare Diseases and Ludwig Maximilian University Hospital Munich (M-ZSELMU), University Hospital of Munich: Dr. med. Fabian Hauck, Prof. Dr. med. Christoph Klein
Institute for Human Genetics and Center for Rare Diseases, University Hospital, Bonn: Prof. Dr. med. Markus M. Nöthen
Institute for Medical Genetics and Applied Genomics, University of Tübingen: Prof. Dr. med. Olaf Riess
Berlin Center for Rare Diseases (BCSE), Charité—Berlin University of Medicine: Prof. Dr. med. Stefan Mundlos, Prof. Dr. med. Heiko Krude
Institute for Human Genetics, Charité—Berlin University of Medicine: Prof. Dr. med. Stefan Mundlos
Institute of Human Genetics at the Rechts der Isar Hospital, Technical University of Munich: Prof. Dr. med. Thomas Meitinger
Institute for Public Health, Charité—Berlin University of Medicine: Prof. Dr. med. Tobias Kurt, Kerstin L. Wainwright
Martin Zeitz Center for Rare Diseases, University Medical Center Hamburg-Eppendorf: Prof. Dr. med. Christoph Schramm
The study was funded by the Innovation Fund of the Federal Joint Committee.
Conflict of interest statement Prof. Mücke received funds for consulting services from the companies TAKEDA and STADA Pharm. He has received payment for lecture events/PODCAST project from the companies TAKEDA and STADA Pharm. He has a board of management function in the working group “Centers for Rare Diseases”.
Prof. Mücke received funds from Eli Lilly & Company, Teva Pharmaceuticals, Total Energies S.E., the BMJ and Frontiers
Prof. Riess received funds for studies from Illumina.
The other authors declare that no conflicts of interest exists.
References
- 1.Nguengang Wakap S, Lambert DM, Olry A, et al. Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. Eur J Hum Genet. 2020;28:165–173. doi: 10.1038/s41431-019-0508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreira CR. The burden of rare diseases. Am J Med Genet A. 2019;179:885–892. doi: 10.1002/ajmg.a.61124. [DOI] [PubMed] [Google Scholar]
- 3.The Council of the European Union. Council recommondation on an action in the field of rare disease. eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:C:2009:151:0007:0010:EN:PDF (last accessed on 17 June 2022) 2009 [Google Scholar]
- 4.Mundlos C. [Speak up for people with rare diseases: ACHSE?: ACHSE, Rare Diseases Germany, and its network] Internist (Berl) 2018;59:1327–1334. doi: 10.1007/s00108-018-0517-z. [DOI] [PubMed] [Google Scholar]
- 5.Nationaler Aktionsplan für Menschen mit Seltenen Erkrankungen. www.namse.de (last accessed on 17 June 2022) 2013 [Google Scholar]
- 6.Köhler S, Gargano M, Matentzoglu N, et al. The human phenotype ontology in 2021. Nucleic Acids Res. 2021;49:D1207–D1217. doi: 10.1093/nar/gkaa1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orphanet. Das Portal für seltene Krankheiten und Orphan Drugs. www.orpha.net/consor/cgi-bin/Disease.php?lng=DE (last accessed on 17 June 2022) [Google Scholar]
- 8.Smedley D, Smith KR, Martin A, et al. 100,000 genomes pilot on rare-disease diagnosis in health care—preliminary report. N Engl J Med. 2021;385:1868–1880. doi: 10.1056/NEJMoa2035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shickh S, Mighton C, Uleryk E, Pechlivanoglou P, Bombard Y. The clinical utility of exome and genome sequencing across clinical indications: a systematic review. Hum Genet. 2021;140:1403–1416. doi: 10.1007/s00439-021-02331-x. [DOI] [PubMed] [Google Scholar]
- 10.Smith HS, Swint JM, Lalani SR, et al. Clinical application of genome and exome sequencing as a diagnostic tool for pediatric patients: a scoping review of the literature. Genet Med. 2019;21:3–16. doi: 10.1038/s41436-018-0024-6. [DOI] [PubMed] [Google Scholar]
- 11.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shickh S, Gutierrez Salazar M, Zakoor KR, et al. Exome and genome sequencing in adults with undiagnosed disease: a prospective cohort study. J Med Genet. 2021;58:275–283. doi: 10.1136/jmedgenet-2020-106936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bogart KR, Irvin VL. Health-related quality of life among adults with diverse rare disorders. Orphanet J Rare Dis. 2017;12 doi: 10.1186/s13023-017-0730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan NB, Stapleton R, Stark Z, et al. Evaluating systematic reanalysis of clinical genomic data in rare disease from single center experience and literature review. Mol Genet Genomic Med. 2020;8 doi: 10.1002/mgg3.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.§ 64e Modellvorhaben zur umfassenden Diagnostik und Therapiefindung mittels Genomsequenzierung bei seltenen und bei onkologischen Erkrankungen, Verordnungsermächtigung. www.gesetze-im-internet.de/sgb_5/__64e.html (last accessed on 17 June 2022) [Google Scholar]
- 16.Berger A, Rustemeier AK, Göbel J, et al. How to design a registry for undiagnosed patients in the framework of rare disease diagnosis: suggestions on software, data set and coding system. Orphanet J Rare Dis. 2021;16 doi: 10.1186/s13023-021-01831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taruscio D, Groft SC, Cederroth H, et al. Undiagnosed Diseases Network International (UDNI): white paper for global actions to meet patient needs. Mol Genet Metab. 2015;116:223–225. doi: 10.1016/j.ymgme.2015.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods
Collaborators and institutions involved in TRANSLATE-NAMSE
Collaborators
At the clinical sites:
Berlin: Magdalena Danyel1–1,1–2, Sarina Schwartzmann1–1,1–2, Annemarie Bösch1–1, Denis Horn1–2,, Nadja Ehmke1–2, Felix Boschann1–2, Stefan Mundlos1–1,1–2, Angela Kaindl1–3, Christoph Bührer1–4, Tilmann Kalinich1–5, Horst v Bernuth1–5, Natalie Weinhold1–6, Phillip Buffler1–6, Dominik Müller1–6, Susanne Holzhauer1–7, Manuel Holtgrewe1–8, Charlotte Wernicke 1–1,1–9, Laura Schmidt-Pennington1–1,1–9, Joachim Spranger1–1,1–9, Annette Grüters1–10, Heiko Krude1–1,1–10
Bochum/Essen: Corinna Grasemann2–1,2–2, Nora Matar2–1,2–2, Janet Atinga2–1,2–2, Folke Brinkmann2–1,2–2, Frank Kaiser2–3,2–4, Bernhard Horsthemke2–3,2–4, Adela Della Marina2–5, Eva Manka2–3,2–6, Cordula Kiewert2–6, Martin Munteanu2–4, Alma Kuechler2–4, Raphael Hirtz2–6, Michael Schündeln2–7, Florian Stehling2–7, Sabine Hoffjan2–8, Nicole Unger2–9, Tim Hagenacker2–10, Stephan Klebe2–10, Paul Manka2–11, Freya Dröge2–12, Florian Grabellus2–13, Huu Nguyen2–8, Björn Bühring2–14
Bonn: Martin Mücke3–1, Tim Bender3–1,3–2, Nadine Weinstock3–1, Lorenz Grigull3–1, Julia Sellin3–1, Marzena Marawiec3–1, Christiane Stieber3–1,3–3, Markus M. Nöthen3–1,3–2, Axel Schmidt3–1,3–2, Martina Kreiß3–1,3–2, Elisabeth Mangold3–2, Sophia Peters3–2, Hartmut Engels3–2, Peter Krawitz3–1,3–4, Alexej Knaus3–2,3–4, Tzung-Chien Hsieh3–2,3–4, Hannah Klinkhammer3–2,3–4, Thomas Klockgether3–1,3–5, Elena Schlapakow3–5,3–6, Valentin Schäfer3–1,3–7, Pantelis Karakostas8–1,8–7, Patrick Weydt3–1,3–7, Sarah Bernsen3–1,3–8, Cornelia Kornblum3–1,3–5
Dresden: Reinhard Berner4–1,4–2, Min Ae Lee-Kirsch4–1,4–2, Andre Heinen4–1,4–2, Julia Koerholz4–1,4–2, Tanita Kretschmer4–1,4–2, Manja Unrath4–1,4–2, Victoria Tüngler 4–1,4–2
Hamburg: Christoph Schramm5–1, Franziska Rillig5–1, Kurt Ullrich5–1, Felix Braun5–1, Maximilian Groffmann5–1, Cornelia Rudolph5–1, Christina Weiler Normann5–1, Maja Hempel5–2, Theresia Herget5–2, Jasmin Lisfeld5–2, Christian Schlein5–2, Christian Kubisch5–2, Ania C. Muntau5–3
Heidelberg: Georg F. Hoffmann6–1,6–2, Daniela Choukair6–1,6–2, Pamela Okun6–1,6–2, Petra Wagenlechner6–1,6–2, Christian P. Schaaf6–1,6–3, Eva M. C. Schwalbold6–1,6–3, Peter Burgard6–1,6–2, Franziska Krause6–2
Lübeck: Tobias Bäumer7–1, 7–4, Alexander Münchau7–1, 7–4, Annekathrin Ripke7–1, Christian Himstedt7–1, Martje Pauly7–1, 7–3, Olaf Hiort7–1, 7–5, Isabel Mönig7–5, Irina Hüning 7–2, Katja Lohmann7–3, Norbert Brüggemann7–3, Christine Klein7–3, Rebecca Herzog7–1
Munich: Fabian Hauck8–1, Stella Bergemann8–1, Astrid Blaschek8–1, Ingo Borggräfe8–1, Katharina Danhauser8–1, Julia Eilenberger8–1, Matthias Griese8–1, Lisa-Maria Köhler8–1, Bärbel Lange-Sperandio8–1, Eberhard Lurz8–1, Wolfgang Müller Felber8–1, Antonia Pelshenke8–1, Karl Reiter8–1, Esther Maier8–1, Katharina Vill, Christoph Klein8–1, Thomas Meitinger8–2, Tim Strom8–2, Riccardo Berutti8–2, Matias Wagner8–2, Theresa Brunet8–2, Melanie Brugger8–2, Katharina Mayerhanser8–2, Korbinian Riedhammer8–2
Tübingen: Andrea Bevot9–1,9–2, Janine Magg9–1,9–2, Ingeborg Krägeloh-Mann9–1,9–2, Monika Glauch9–1,9–3, Martin Kehrer9–1,9–3, Holm Graessner9–1,9–3, Tobias Haack9–1,9–3, Olaf Riess9–1,9–3, Stefanie Beck-Wödl9–1,9–3, Lena Zeltner9–1,9–4, Jutta Eymann9–1, Ludger Schoels9–1,9–4, Kathrin Grundmann-Hauser9–1,9–2, Till-Karsten Hauser9–1,9–5
Evaluation Dresden: Jochen Schmitt10, Gabriele Müller10, Diana Druschke10
Evaluation Berlin: Tobias Kurth11, Kerstin Wainwright11, Sylvana Baumgarten11
AOK: Werner Wyrwich12, Katja Basso 12
Barmer: Alfred Kindshofer13, Regina Kotzbacher13, Ursula Marschall13
ACHSE e. V.: Christine Mundlos14
Participating institutions
Berlin:
1–1 Center for Rare Diseases (BCSE), Charité—Berlin University of Medicine, 13353 Berlin
1–2 Institute for Human Genetics, Charité—Berlin University of Medicine, 13353 Berlin
1–3 Department of Pediatric Neurology, Charité—Berlin University of Medicine, 13353 Berlin
1–4 Department of Neonatology, Charité—Berlin University of Medicine, 13353 Berlin
1–5 Department of Pediatric Pneumology, Immunology and Intensive Care, Charité—Berlin University of Medicine, 13353 Berlin
1–6 Department of Pediatric Gastroenterology, Nephrology and Metabolic Diseases, Charité—Berlin University of Medicine, 13353 Berlin
1–7 Department of Pediatric Oncology, and Hematology, Charité—Berlin University of Medicine, 13353 Berlin
1–8 CoreUnit Bioinformatics, Berlin Institute of Health, Charité—Berlin University of Medicine, 13353 Berlin
1–9 Department of Endocrinology and Metabolic Disorders, Charité—Berlin University of Medicine, 13353 Berlin
1–10 Institute for Experimental Pediatric Endocrinology, Charité—Berlin University of Medicine, 13353 Berlin
Bochum/Essen:
2–1 Center for Rare Diseases Ruhr, CeSER, Catholic Hospital Bochum, Ruhr University Bochum, 44791 Bochum
2–2 Department of Pediatrics, Catholic Hospital Bochum, Ruhr University Bochum, 44791 Bochum
2–3 Center for Rare Diseases Essen, EZSE, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–4 Institute for Human Genetics, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–5 Clinic for Pediatrics I, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–6 Clinic for Pediatrics II, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–7 Clinic for Pediatrics III, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–8 Institute for Human Genetics, Ruhr University Bochum, 44791 Bochum
2–9 Department for Endocrinology, Diabetes and Metabolism, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–10 Department of Neurology, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–11 Department of Internal Medicine, Knappschaftskrankenhaus Bochum, Ruhr University Bochum, 44791 Bochum
2–12 ENT Department, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–13 Institute for Pathology, University Hospital Essen, University of Duisburg-Essen, 45147 Essen
2–14 “Rheumatology Center of the Ruhr District”, Ruhr University Bochum, Herne
Bonn:
3–1 Center for Rare Diseases, University Hospital Bonn, 53127 Bonn
3–2 Institute for Human Genetics, University Hospital Bonn, 53127 Bonn
3–3 Institute of General Practice and Family Medicine, University Hospital Bonn, 53127 Bonn
3–4 Institute for Genomic Statistics and Bioinformatics, University Hospital Bonn, 53127 Bonn
3–5 Department of Neurology, University Hospital Bonn, 53127 Bonn
3–6 Department of Neurology, Halle (Saale) University Hospital, 06120 Halle (Saale)
3–7 Department of Internal Medicine III, University Hospital Bonn, 53127 Bonn
3–8 Department of Neurodegenerative Diseases, University Hospital Bonn, 53127 Bonn
Dresden:
4–1 Clinic and Outpatient Clinic for Pediatric and Adolescent Medicine, University Hospital and the Carl Gustav Carus Faculty of Medicine at the Technical University Dresden, 01307 Dresden
4–2 University Center for Rare Diseases (USE), University Hospital and the Carl Gustav Carus Faculty of Medicine at the Technical University Dresden, 01307 Dresden
Hamburg:
5–1 Martin Zeitz Center for Rare Diseases, University Medical Center Hamburg-Eppendorf, 20246 Hamburg
5–2 Institute for Human Genetics, University Medical Center Hamburg-Eppendorf, 20246 Hamburg
5–3 University Pediatric Department, University Medical Center Hamburg-Eppendorf, 20246 Hamburg
Heidelberg:
6–1 Center for Pediatric and Adolescent Medicine, University Hospital Heidelberg, 69120 Heidelberg
6–2 Center for Rare Diseases, University Hospital Heidelberg, 69120 Heidelberg
6–3 Institute for Human Genetics, University Hospital Heidelberg, 69120 Heidelberg
Lübeck:
7–1 Center for Rare Diseases, University Hospital Schleswig-Holstein Lübeck, 23562 Lübeck
7–2 Institute for Human Genetics, University Hospital Schleswig-Holstein Lübeck, 23562 Lübeck
7–3 Institute for Neurogenetics, University of Lübeck, 23562 Lübeck
7–4 Institute of Systemic Motor Research, University Hospital Schleswig Holstein, Campus Lübeck, 23562 Lübeck
7–5 Department for Pediatric and Adolescent Medicine, University Hospital Schleswig Holstein, Campus Lübeck, 23562 Lübeck
Munich:
8–1 Pediatric Department, Dr. von Hauner Children’s Hospital, Ludwig Maximilian University Hospital Munich, 81377 Munich
8–2 Institute for Human Genetics, Rechts der Isar Hospital, Technical University of Munich, 80337 München
Tübingen:
9–1 Center for Rare Diseases, University Hospital Tübingen, 72076 Tübingen
9–2 Department of Pediatric Neurology and Developmental Disorders, University Hospital Tübingen, 72076 Tübingen
9–3 Institute for Medical Genetics and Applied Genomics, University Hospital Tübingen, 72076 Tübingen
9–4 Department of Neurology, University Hospital Tübingen, 72076 Tübingen
9–5 Department of Radiology, University Hospital Tübingen, 72076 Tübingen
Evaluation:
10 Center for Evidence-Based Healthcare, University Hospital and the Carl Gustav Carus Faculty of Medicine at the Technical University Dresden, 01307 Dresden
11 Institute for Public Health, Charité—Berlin University of Medicine, 10117 Berlin
Health insurance funds:
12 AOK Nordost, 13409 Berlin
13 Barmer, 81675 Munich
Patient representatives:
14 ACHSE e.V., 13359 Berlin