ABSTRACT
Prenatal lead (Pb) exposure is associated with adverse developmental outcomes and to epigenetic alterations such as DNA methylation and hydroxymethylation in animal models and in newborn blood. Given the importance of the placenta in foetal development, we sought to examine how prenatal Pb exposure was associated with differential placental DNA methylation and hydroxymethylation and to identify affected biological pathways linked to developmental outcomes. Maternal (n = 167) and infant (n = 172) toenail and placenta (n = 115) samples for prenatal Pb exposure were obtained from participants in a US birth cohort, and methylation and hydroxymethylation data were quantified using the Illumina Infinium MethylationEPIC BeadChip. An epigenome-wide association study was applied to identify differential methylation and hydroxymethylation associated with Pb exposure. Biological functions of the Pb-associated genes were determined by overrepresentation analysis through ConsensusPathDB. Prenatal Pb quantified from maternal toenail, infant toenail, and placenta was associated with 480, 27, and 2 differentially methylated sites (q < 0.05), respectively, with both increases and decreases associated with exposure. Alternatively, we identified 2, 1, and 14 differentially hydroxymethylated site(s) associated with maternal toenail, infant toenail, and placental Pb, respectively, with most showing increases in hydroxymethylation with exposure. Significantly overrepresented pathways amongst genes associated with differential methylation and hydroxymethylation (q < 0.10) included mechanisms pertaining to nervous system and organ development, calcium transport and regulation, and signalling activities. Our results suggest that both methylation and hydroxymethylation in the placenta can be variable based on Pb exposure and that the pathways impacted could affect placental function.
KEYWORDS: DNA methylation, hydroxymethylation, placenta, lead, epigenetics, EWAS
Introduction
Lead (Pb) is an environmental toxicant known to adversely affect human health. With regulatory guidelines in place, the once major sources of Pb, leaded gasoline and lead-based paint, are now lesser concerns as exposure sources. Yet even at low levels, exposure through consumer products, contaminated soil, or drinking water through corroding pipes and fixtures, can still pose threats to the population [1–4]. In addition, exposure to Pb in the sensitive pregnancy window has been shown to lead to numerous adverse health outcomes, including foetal growth restriction, low birth weight, neurodevelopmental deficits, behaviour issues, and cognitive and intellectual impairment [5–12]. During gestation, the placenta plays a vital role in regulating not only nutrients but also toxicant transport between the mother and infant. It also undertakes a wide variety of molecular processes in promoting normal growth and development. A potential mechanism for Pb exposure to affect development and contribute to diseases later in life may be through impacts on placental function in addition to known impacts directly to the foetus.
Epigenetic modifications are heritable changes in the control of gene expression that occur independently from changes in the original DNA sequences. DNA methylation, the addition of a methyl group at the C5 position of the cytosine (5mC), is one of the most assessed epigenetic markers [13,14]. Emerging studies have suggested altered methylation patterns upon Pb exposure [15–17]. In a zebrafish model, Pb exposure results in altered DNA methylation and impaired activity of DNA methyltransferases (DNMTs), the enzymes that catalyse the methylation of DNA[18]. Bone Pb level was negatively associated with DNA methylation within LINE-1 elements in umbilical cord blood in human studies[19]. In a human embryonic stem cell study, Pb was also associated with altered DNA methylation status of genes involved in brain development[20]. With evidence from previous studies, Pb exposure is implied to impact DNA methylation during the sensitive gestational period, and given the critical role of placenta in foetal growth and development, Pb-induced methylation changes may partially facilitate the long-term effects of gestational Pb exposure.
Hydroxymethylation, a more recently identified type of epigenetic modification, is considered as an intermediate of the active demethylation process. With enzymes from the ten-eleven translocation (TET) family, a hydrogen atom at the C5 position of the cytosine is replaced by a hydroxymethyl group, promoting oxidation of 5mC into 5hmC [21–23]. It is suggested that hydroxymethylation (5hmC) can also be linked to gene expression regulation as 5hmC is observed in enhancer and gene bodies in embryonic stem cells [24,25]. Researchers also found that 5hmC is most abundant in the brain and is likely a key factor involved in brain development [26–28]. Some studies have considered the highly tissue-specific 5hmC as a stable epigenetic mark [25]. Emerging studies have shown that early mercury exposure was linked to cord blood 5hmC changes, while Pb exposure altered peripheral blood 5hmC profile in candidate genes [29,30]. With its distinct features from 5mC, identifying epigenome-wide 5hmC pattern changes in the placental in response to Pb toxicity may provide addition mechanistic understandings on developmental effects resulted from early Pb exposure.
The placental epigenome may be subject to epigenetic modifications elicited by metal exposures, such as Pb, and result in disruption in placental functions and adverse birth and developmental outcomes. Given the research gaps in investigating human 5mC and 5hmC profiling in tissues aside from blood, and the well-described role of the placenta in foetal development and programing, our study utilized epigenome profiling in the placenta to help elucidate Pb-induced epigenetic alterations during the prenatal window. We hypothesized that prenatal Pb, quantified through maternal and infant toenails and the placenta thus reflecting prenatal exposure levels, results in differential placenta DNA methylation and hydroxymethylation, and that these Pb-associated epigenetic changes would be associated with biological mechanisms and functions in the placenta that are most relevant to foetal development.
Materials and methods
Study population
This study included mother–infant pairs enrolled in the Rhode Island Child Health Study (RICHS). Mothers at least 18 years of age, without any life-threatening medical conditions, and delivered term ( 37 weeks gestation) singletons that were free of congenital or chromosomal abnormalities were recruited from the Women & Infants Hospital in Providence, Rhode Island. RICHS oversampled for small for gestational age (SGA; lowest 10th percentile) and large for gestational age (LGA; highest 10th percentile) infants, and adequate for gestational age (AGA) infants were matched on sex, maternal age ( 2 years), and gestational age ( 3 days). Anthropometric and clinical data were obtained from structured medical record review, and exposure histories, demographic, and lifestyle information were collected through interviewer-based questionnaires. All participants provided written informed consent under appropriate protocols approved by the Institutional Review Boards at Women and Infants’ Hospital and Emory University.
Pb exposure assessment
Following hospital discharge, first toenail clippings from all toes were requested from both mother and infant, and were mailed back to the laboratory in provided envelopes. Pb levels at microgram per gram of toenail were analysed in the Dartmouth Trace Element Analysis laboratory using established inductively couple plasma mass spectrometry (ICP-MS) protocols as previously described [31].
Placenta samples were collected from all RICHS participants for molecular epigenetic studies, and for a subset of consecutive participants, samples were collected to allow for the assessment of trace metals. For molecular epigenetic studies, placental parenchyma biopsies were collected with 2 hours of delivery, excised at 1–2 cm from the umbilical cord insertion site and free of maternal decidua. Samples were then placed immediately in RNAlater (Life Technology) and stored at 4°C. After 72 hours, samples were removed from RNAlater and stored at −80°C until DNA was extracted for further examination. For trace metals analysis, placenta parenchyma samples were collected, flash-frozen in liquid nitrogen, and stored at −80°C in trace element–free tubes until analysis. The measurement of placental Pb using ICP-MS has been described previously [31]. The limit of detection (LOD) for placental Pb was 2.12 ng/g.
DNA methylation and hydroxymethylation
DNA was extracted and quantified from placental samples, and methylation and hydroxymethylation analysis were conducted using standard bisulphite (BS) and oxidative bisulphite (oxBS) modification preparations, respectively, with DNeasy Blood and Tissue Kit (Qiagen, MD, USA), Qubit Fluorometer (Thermo Fisher Scientific Life Sciences) and TrueMethyl oxBS Module (NuGen, CA, USA) following manufacturer’s protocols.
Epigenome-wide methylation and hydroxymethylation of CpG sites was assessed in paired samples using the Illumina Infinium® MethylationEPIC BeadChip (Illumina, CA, USA) at Emory University (N = 230). Normalization and background correction of the BS and oxBS-converted samples were performed with the FunNorm function in R package minfi [32–34]. To minimize batch effects, samples were randomized as pairs, across multiple batches. Ten samples were repeated as quality control, randomized across batches. Samples were filtered out if the estimated sex based on the methylation data, through the getSex function in minfi, did not match the infant sex in the phenotype data (n = 2). Samples were excluded if 2% of the probes for the sample had detection p-values > 0.01 (n = 1). Cross-reactive and poor-performing probes, based on detection p-value, were excluded resulting in 6,772 probes excluded in the standard bisulphite (BS) data, and 20,803 probes excluded in the oxidative bisulphite (ox-BS) data. Batch effects from slides were corrected using the ComBat function [35,36].
To accurately estimate 5mC and 5hmC in the placental epigenome, we utilized the algorithm as described in detail in Green et al [37]. In brief, maximum likelihood was used to fit the data-generating model that outputs parameters indicating the unmethylated proportion (π1), 5mC proportion (π2), and 5hmC proportion (π3). The novel approach of this model is the constraint that disallows negative proportions for the proportion parameters. This method is publicly available from the Comprehensive R Archive Network repository as the OxyBS package (http://cran.r-project.org).
Covariates
A structured, interviewer administered questionnaire was used to collect self-reported variables, while a structured medical abstraction was used to obtain clinical information on infant and mother from the hospital record. Self-reported highest obtained education status was dichotomized into more than high school or high school or less, and self-reported maternal race was coded as either white or other, given our predominantly white study population. Maternal age was treated as a continuous variable. From medical records, height and weight were used to calculate mother’s pre-pregnancy BMI, and infant sex and birth weight data were also obtained from medical chart abstraction. Additionally, infants with standardized birth weight percentiles (calculated via the Fenton growth chart) below the 10th percentile were categorized into SGA; those between the 10th and 90th percentile were classified as AGA; and those above the 90th percentile were classified as LGA [38].
Statistical analysis
Demographic characteristics of participants with available Pb exposure biomarkers (maternal toenail, infant toenail, placenta) were compared using chi-square and ANOVA tests. Characteristics for the sub-cohort of participants with available EPIC array data (N = 230) were also assessed. The mean, standard deviation, and minimum and maximum values of Pb were examined across biomarkers.
To examine the association between array-wide mean DNA methylation and hydroxymethylation and Pb biomarkers, we averaged values across all probes for each individual [39]. We performed multivariable linear regression with the three different Pb biomarkers in our study, and models were adjusted for variables including infant sex, maternal age, race, BMI, educational status, and placental cell types (only syncytiotrophoblast and hofbauer cells) [40].
Epigenome-wide association study (EWAS) methodology was performed to identify differential methylation and hydroxymethylation associated with Pb exposure biomarkers. With R package limma, we performed robust linear regressions for each CpG site, regressing the methylation beta-values on log2-transformed Pb exposure measured through the three biomarkers. Models were adjusted for cell type proportions, utilizing the planet package in R, and included are trophoblasts, stromal cells, hofbauer cells, endothelial cells, nucleated red blood cells, and syncytiotrophoblast cells, along with previously established covariates including infant sex, maternal age, and maternal race to account for potential population stratification, maternal BMI, and education status. To account for multiple testing, a false discovery rate (FDR) of 5% was considered as the statistically significant cut point for differentially methylated and hydroxymethylated site(s). Q-Q plots and genomic inflation factor (λ) were generated and assessed for each analysis and provided in Supplementary Figure S1 and Supplementary Table S2. Manhattan plots were utilized to demonstrate positions of the statistically significant CpG sites across different biomarkers from EWAS results. To identify potential differential methylated regions (DMRs), regional analysis was performed using the comb-p method in the ENmix package in R. This method analysed DMRs using chromosome coordinates and p-values derived from EWAS. Comb-p adjusted for auto-correlation among neighbouring probes using the Stouffer-Liptak-Kechris method, then to adjust for multiple comparisons, applied Sidak correction to the generated regional p-values [41]. For the comb-p argument, we used a maximum distance of 1000 base pairs to combine adjacent DMRs, and an FDR significance threshold of 0.001. DMRs containing only a single probe were excluded, and regions with a Sidak p-value < 0.05 were considered significant. Analysis of epigenomic data were performed through R v3.5.1.
To further ascertain biological functions of the differentially methylated and hydroxymethylated CpG sites, ConsensusPathDB (CPDB) was used for pathway over-representation of the top 250 sites derived from EWAS. CPDB is an online database system that utilizes 12 different source databases to understand different types of human molecular functional interactions. The web interface at http://cpdb.molgen.mpg.de demonstrates resources including protein interactions, gene regulations, and signalling and genetic interactions to provide a less biased integration of functional networks and biological pathways. The system calculates an enrichment p-value from the hypergeometric distribution of genes in the user-specified candidate list and the predefined gene set[42]. FDR was used to correct for multiple testing, and a q-value <0.10 was considered as a significantly enriched pathway or gene ontology set.
Results
The analysis strategy of this study is demonstrated in Figure 1. 230 participants had available paired EPIC array data, and the demographic and gestational characteristics of these participants that also had available Pb exposure data from the three biomarkers, placenta, maternal toenail, and infant toenail are shown in Table 1. Of the 230 participants, 54 mother–infant pairs had all three types of Pb biomarker data (placenta, maternal toenail, and infant toenail). 165 mother–infant pairs had both maternal and infant toenail data, 59 had both placental and maternal toenail data, and 55 had both placental and infant toenail data. Infant gender (female-to-male ratio is around 1:1) and maternal race (predominantly white) distributions were similar across three subsets. Participants with maternal toenail and infant toenail samples showed similar percentages of AGA infants (57.0% vs. 56.3%), post-high-school education (89%), average birth weight (3553 g vs. 3552 g), and average gestational age (39.46 weeks). Notable difference between the three biomarker subsets was observed in maternal age, with mothers with placental Pb data slightly younger (mean = 30.1 yrs), compared to those with maternal or infant toenail data (mean = 31.8 yrs; p = 0.003).
Figure 1.
Analysis strategy. RICHS study population comprised of 840 mother–infant pairs with demographic information, and 230 participants had available EPIC array data. Participants with Pb exposure data quantified from different biomarkers are included in this study: maternal toenail (N = 172), infant toenail (N = 167), and placenta (N = 115).x
Table 1.
Exposure, demographic and gestational characteristics for subsets with different Pb biomarkers in the RICHS study population.
|
With available Pb exposure data in |
|||
|---|---|---|---|
| Characteristic | Maternal toenail (N = 172) | Infant toenail (N = 167) |
Placenta (N = 115) |
| Lead (Pb) | |||
| Mean ± SD | 0.31 ± 0.58 (ug/g) | 0.97 ± 2.01 (ug/g) | 3.20 ± 3.75 (ng/g) |
| Interquartile range (IQR) | 0.23 | 0.66 | 1.80 |
| n (%) | |||
| Infant Gender | |||
| Female | 87 (50.6%) | 84 (50.3%) | 54 (47.0%) |
| Male | 85 (49.4%) | 83 (49.7%) | 61 (53.0%) |
| Birth Weight Categorya | |||
| SGA | 28 (16.3%) | 28 (16.8%) | 19 (16.5%) |
| AGA | 98 (57.0%) | 94 (56.3%) | 53 (46.1%) |
| LGA | 46 (26.7%) | 45 (26.9%) | 43 (37.4%) |
| Maternal Race | |||
| White | 134 (77.9%) | 132 (79.0%) | 83 (72.2%) |
| Other | 32 (18.6%) | 29 (17.4%) | 31 (27.0%) |
| N/A | 6 (3.5%) | 6 (3.6%) | 1 (0.9%) |
| Infant Race | |||
| White | 128 (74.4%) | 126 (75.4%) | 74 (64.3%) |
| Other | 38 (22.1%) | 35 (21.0%) | 39 (33.9%) |
| N/A | 6 (3.5%) | 6 (3.6%) | 2 (1.8%) |
| Maternal Education Status | |||
| No more than high school | 18 (10.5%) | 18 (10.8%) | 23 (20.0%) |
| Some post-high school | 154 (89.5%) | 149 (89.2%) | 92 (80.0%) |
| Mean ± SD | |||
| Birth weight (grams) | 3553 ± 643.47 | 3552 ± 648.09 | 3610 ± 703.41 |
| Gestational age (weeks) | 39.46 ± 0.93 | 39.46 ± 0.94 | 39.26 ± 0.97 |
| Maternal age (years) | 31.75 ± 4.16 | 31.78 ± 4.12 | 30.10 ± 5.46 |
| Maternal BMI (kg/m2) | 26.58 ± 6.42 | 26.61 ± 6.35 | 26.48 ± 6.48 |
aSGA: small for gestational weight; AGA: adequate for gestational weight; LGA: large for gestational weight
The mean Pb level measured in maternal toenail, infant toenail, and placental samples were 0.31 ug/g (interquartile range, IQR: 0.23), 0.97 ug/g (IQR: 0.66), and 3.20 ng/g (IQR: 1.8), respectively. Spearman correlation results between three Pb biomarkers showed the strongest correlation was observed between maternal and infant toenail Pb (r = 0.41), while correlations between placental Pb and maternal toenail and infant toenail Pb were −0.02 and 0.18, respectively. Histograms showed that Pb levels were right skewed in all three types of biomarkers, and log2-transformed values were used for the following epigenetics analysis to improve interpretation.
EWAS analysis results at q < 0.05 are summarized in Table 2 (DNA methylation) and Table 3 (DNA hydroxymethylation), and scatterplots of these results are depicted in Supplementary Figures S2 and S3. After adjusting for cell-type proportions and covariates, we observed 480 differentially methylated sites (range of estimates: −0.02 ~ 0.04) associated with maternal toenail Pb, with 313 (65.2%) sites revealing decreased methylation levels with increasing Pb concentrations. For infant toenail Pb, 27 sites were differentially methylated (range of estimates: −0.02 ~ 0.02) and 24 (88.9%) showed decreased methylation levels with increasing Pb. Only the top 10 most robust findings (smallest FDR) from the toenail Pb EWAS are included in Table 2 (complete lists of differentially methylated CpG sites associated with toenail Pb are shown in Supplementary Table S3 and Table S4). Pb quantified from placenta was associated with decreased placental methylation at two sites. Hydroxymethylation results showed that two (one increased and one decreased) and one (increased hydroxymethylation) differentially hydroxymethylated site(s) were associated with maternal and infant toenail Pb, respectively. Placental Pb results showed 14 differentially hydroxymethylated sites (range of estimates: −0.01 ~ 0.01) at the statistically significant level, and among them two sites showed decreased hydroxymethylation (Table 3). Pairwise correlations between effect estimates of the methylation and hydroxymethylation results by biomarker showed weak correlations for maternal toenail Pb (r = 0.21), infant toenail Pb (r = 0.18), and placental Pb (r = 0.16). We did not observe any significant (p < 0.05) associations for array-wide mean DNA methylation or hydroxymethylation values and Pb measured in placenta, maternal toenail or infant toenail.
Table 2.
Epigenome-wide association study results for differential methylation (q < 0.05) at CpG sites associated with Pb quantified in maternal toenail (ng/g), infant toenail (ng/g), and placenta (ug/g) samples.
| Probe | Position | Gene | Estimate | SE | p-value | q-value |
|---|---|---|---|---|---|---|
| Maternal toenail+ | ||||||
| cg16285217 | chr2:20,095,489 | −0.0022 | 0.0004 | 7.57E-09 | 0.0053 | |
| cg11374425 | chr3:62,359,677 | FEZF2 | 0.0033 | 0.0006 | 7.20E-08 | 0.0101 |
| cg10012394 | chr5:63,461,371 | RNF180 | 0.0114 | 0.0020 | 1.95E-08 | 0.0063 |
| cg06821993 | chr8:114,449,011 | CSMD3 | 0.0322 | 0.0056 | 4.70E-08 | 0.0082 |
| cg19902005 | chr11:57,545,678 | TMX2-CTNND1; CTNND1 | 0.0089 | 0.0016 | 1.81E-07 | 0.0114 |
| cg06090833 | chr14:32,288,033 | NUBPL | −0.0047 | 0.0009 | 1.89E-07 | 0.0114 |
| cg00049033 | chr16:49,317,080 | CBLN1 | 0.0251 | 0.0045 | 1.33E-07 | 0.0114 |
| cg10018294 | chr17:911,609 | ABR | −0.0080 | 0.0014 | 2.69E-08 | 0.0063 |
| cg04295815 | chr17:27,045,188 | RAB34 | −0.0054 | 0.0010 | 1.04E-07 | 0.0114 |
| cg21667047 | chr20:9,495,726 | LAMP5-AS1; LAMP5 | 0.0207 | 0.0038 | 1.99E-07 | 0.0114 |
| Infant toenail+ | ||||||
| cg10814131 | chr1:203,009,651 | PPFIA4 | −0.0087 | 0.0016 | 1.23E-07 | 0.0108 |
| cg11365072 | chr5:154,392,274 | KIF4B | −0.0031 | 0.0005 | 3.53E-08 | 0.0058 |
| cg10299585 | chr12:54,321,717 | 0.0147 | 0.0026 | 4.95E-08 | 0.0058 | |
| cg07136023 | chr16:86,537,316 | 0.0137 | 0.0024 | 4.70E-08 | 0.0058 | |
| cg09422806 | chr19:3,364,015 | NFIC | −0.0149 | 0.0027 | 2.12E-07 | 0.0148 |
| cg04640975 | chr19:3,464,991 | −0.0129 | 0.0021 | 9.39E-09 | 0.0033 | |
| cg02859421 | chr19:3,465,071 | −0.0091 | 0.0016 | 7.02E-08 | 0.0070 | |
| cg13822446 | chr19:46,235,676 | BHMG1 | −0.0042 | 0.0008 | 1.74E-07 | 0.0135 |
| cg03359161 | chr20:39,191,619 | −0.0020 | 0.0003 | 4.46E-08 | 0.0058 | |
| cg15445952 | chr22:43,653,574 | SCUBE1 | −0.0203 | 0.0030 | 2.29E-10 | 0.0002 |
| Placenta | ||||||
| cg04773990 | chr6:32,820,410 | TAP1 | −0.0039 | 0.0005 | 2.78E-11 | <0.0001 |
| cg09465791 | chr20:19,633,301 | SLC24A3 | −0.0106 | 0.0018 | 4.55E-08 | 0.0159 |
+ Top 10 sites are shown.
Note: Estimates reflect the change in DNA methylation per doubling (log2) of Pb concentration.
Table 3.
Epigenome-wide association study results for differential hydroxymethylation (q < 0.05) at CpG sites associated with Pb quantified in maternal toenail (ng/g), infant toenail (ng/g), and placenta (ug/g) samples.
| Probe | Position | Gene | Estimate | SE | p-value | q-value |
|---|---|---|---|---|---|---|
| Maternal toenail | ||||||
| cg15320238 | chr16:51,091,402 | −0.0027 | 0.0004 | 1.14E-08 | 0.0079 | |
| cg15591645 | chr19:46,147,397 | EML2 | 0.0052 | 0.0009 | 1.21E-07 | 0.0417 |
| Infant toenail | ||||||
| cg20800997 | chr21:47,018,175 | 0.0036 | 0.0006 | 7.16E-09 | 0.0049 | |
| Placenta | ||||||
| cg09869950 | chr1:59,783,505 | FGGY | 0.0060 | 0.0011 | 8.40E-07 | 0.0441 |
| cg11036421 | chr1:61,510,287 | 0.0100 | 0.0019 | 4.94E-07 | 0.0401 | |
| cg25805115 | chr1:81,964,207 | −0.0128 | 0.0023 | 1.29E-07 | 0.0295 | |
| cg19519828 | chr1:206,664,973 | IKBKE | 0.0027 | 0.0004 | 1.38E-08 | 0.0062 |
| cg25195477 | chr3:148,847,134 | HPS3 | 0.0005 | 0.0001 | 6.79E-07 | 0.0401 |
| cg25687585 | chr5:42,756,950 | CCDC152 | 0.0075 | 0.0014 | 2.48E-07 | 0.0401 |
| cg13694662 | chr9:140,214,551 | EXD3 | −0.0107 | 0.0020 | 3.76E-07 | 0.0401 |
| cg26252498 | chr10:34,865,917 | PARD3 | 0.0063 | 0.0012 | 5.57E-07 | 0.0401 |
| cg16119628 | chr10:88,282,035 | WAPAL | 0.0005 | 0.0001 | 8.98E-07 | 0.0441 |
| cg09847717 | chr11:126,318,677 | KIRREL3 | 0.0049 | 0.0009 | 7.01E-07 | 0.0401 |
| cg15574100 | chr12:27,168,121 | TM7SF3 | 0.0048 | 0.0009 | 4.69E-07 | 0.0401 |
| cg19704558 | chr17:35,296,732 | LHX1 | 0.0069 | 0.0011 | 1.81E-08 | 0.0062 |
| cg05279901 | chr19:42,498,662 | ATP1A3 | 0.0024 | 0.0005 | 6.34E-07 | 0.0401 |
| cg24118151 | chr20:42,354,839 | GTSF1L | 0.0048 | 0.0009 | 3.36E-07 | 0.0401 |
Note: Estimates reflect the change in DNA methylation per doubling (log2) of Pb concentration.
Manhattan plots for all three Pb biomarkers showed the position of the genes annotated to the differentially methylated (Figures 2a-2c) and hydroxymethylated (Figures 2d-2f) sites (q < 0.05). The differentially methylated probe with the lowest p-value (p = 2.29E-10) was cg15445952, where a doubling of infant toenail Pb was associated with 0.0203 percentage lower DNA methylation. This probe was annotated to the SCUBE1 gene, which was also identified among the differentially methylated sites associated with maternal toenail Pb.
Figure 2.
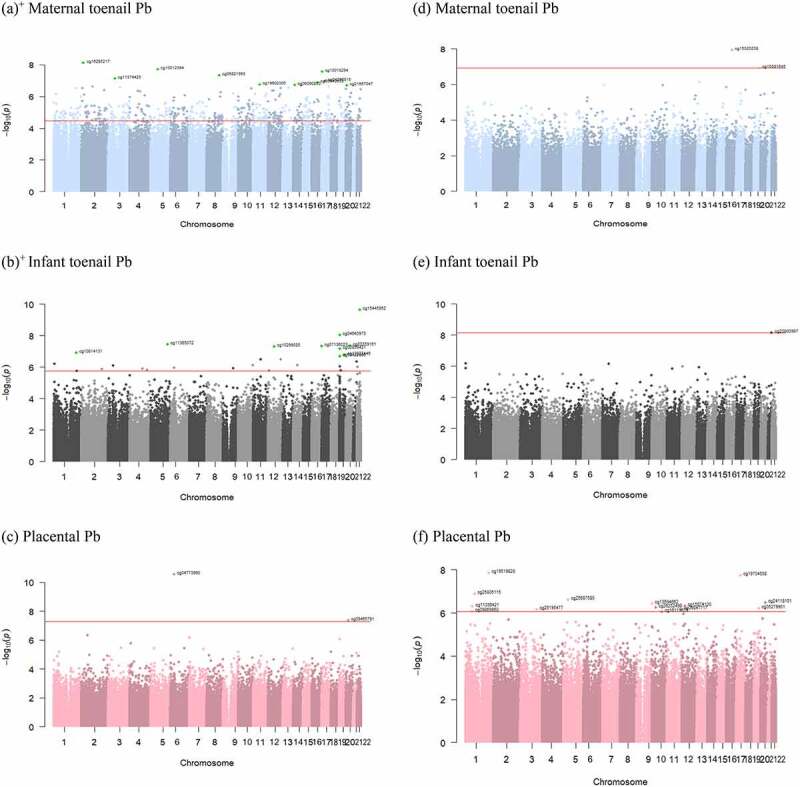
Manhattan plots showing the positions of the differentially methylated (a)-(c) and hydroxymethylated (d)-(f) CpG sites (q < 0.05). X-axis represents the genomic location of the probes and the y-axis represents -log10(p-value) for the CpG sites associated with Pb biomarkers: maternal toenail [(a)+ and (d)], infant toenail [(b)+ and (e)], and placenta [(c) and (f)]. FDR cut-off is indicated as the horizontal red line. + Top 10 sites are labelled.
Based on the Manhattan plots for the Pb-associated differentially methylated and hydroxymethylated probes across the genome, we postulated that there may be differentially methylated region(s) (DMRs) associated with maternal toenail Pb, and so focused on this particular model to perform a regional analysis. According to our results, there are 60 DMRs associated with maternal toenail Pb (Sidak p < 0.05), with the most significant DMR identified at chromosome 20 and annotated to the GNAS gene (Supplementary Table S5).
Using genes annotated to the top 250 methylated and hydroxymethylated sites from EWAS in the candidate gene list for ConsensusPathDB, we observed significantly enriched pathways (q < 0.1) across Pb quantified in maternal toenail, infant toenail and placenta samples. Among the three Pb biomarkers, more pathways and gene ontology (GO) terms were overrepresented among genes annotated from differential methylated and hydroxymethylated sites by infant toenail Pb. Summarized in Table 4 for methylation and Table 5 for hydroxymethylation, we found overrepresented pathways across the three Pb biomarkers that shared similar biological and functional characteristics. Significant GO terms associated with both maternal and infant toenail Pb include were related to nervous system development (GO:0007399) and calcium ion binding (GO:0005509). Calcium transport and regulation pathways were also identified among placental Pb-associated genes. Multiple pathways involved in development of the brain (cerebellum cortex, hindbrain) and neuron were also linked to infant toenail Pb-associated genes. Signalling pathways and GO terms including Hedgehog and TGF- superfamily-related (activin binding, SMAD binding) were also overrepresented among genes differentially methylated or hydroxymethylated upon prenatal Pb exposure. The complete list of significantly enriched pathways and GO terms is shown in Supplementary Tables S6–S11.
Table 4.
Summary of significantly enriched pathways and GO terms (q < 0.10) involved in neuronal, developmental, and cellular signalling processes. Candidate gene set comprised of genes annotated to the top 250 differentially methylated CpGs by Pb quantified in maternal toenail, infant toenail and placenta.
| Pathway ID/GO Term | Pathway description/Term name | q-value |
|---|---|---|
| Maternal toenail | ||
| GO:0005509 | Calcium ion binding | <0.0000 |
| GO:0043169 | Cation binding | <0.0001 |
| GO:0046872 | Metal ion binding | 0.0001 |
| GO:0007399 | Nervous system development | 0.0092 |
| Infant toenail | ||
| R-HSA-1266738a | Developmental Biology | 0.0600 |
| R-HSA-5610787a | Hedgehog, off, state | 0.0600 |
| R-HSA-5358351a | Signalling by Hedgehog | 0.0600 |
| R-HSA-5635851a | GLI proteins bind promoters of Hh responsive genes to promote transcription | 0.0916 |
| path:hsa04340b | Hedgehog signalling pathway – Homo sapiens (human) | 0.0916 |
| GO:0005509 | Calcium ion binding | 0.0000 |
| GO:0048856 | Anatomical structure development | 0.0005 |
| GO:0007399 | Nervous system development | 0.0014 |
| GO:0048731 | System development | 0.0027 |
| GO:0021695 | Cerebellar cortex development | 0.0029 |
| GO:0021549 | Cerebellum development | 0.0042 |
| GO:0030902 | Hindbrain development | 0.0046 |
| GO:0007507 | Heart development | 0.0130 |
| GO:0030182 | Neuron differentiation | 0.0130 |
| GO:0048666 | Neuron development | 0.0214 |
| GO:0048568 | Embryonic organ development | 0.0294 |
| GO:0022008 | Neurogenesis | 0.0294 |
| Placenta | ||
| GO:0051279 | Regulation of release of sequestered calcium ion into cytosol | 0.0225 |
| GO:0010522 | Regulation of calcium ion transport into cytosol | 0.0402 |
| GO:0008528 | G protein-coupled peptide receptor activity | 0.0417 |
| GO:0051282 | Regulation of sequestering of calcium ion | 0.0574 |
| GO:0070410 | Co-SMAD binding | 0.0629 |
| GO:0051208 | Sequestering of calcium ion | 0.0800 |
a Pathway source: Reactome; b Pathway source: KEGG
Table 5.
Summary of significantly enriched pathways and GO terms (q < 0.10) involved in neuronal, developmental, and cellular signalling processes. Candidate gene set comprised of genes annotated to the top 250 differentially hydroxymethylated CpGs by Pb quantified in maternal toenail, infant toenail, and placenta.
| Pathway ID/GO Term | Pathway description/Term name | q-value |
|---|---|---|
| Maternal toenail | ||
| GO:0098794 | Postsynapse | 0.0799 |
| GO:0050807 | Regulation of synapse organization | 0.0835 |
| GO:0050803 | Regulation of synapse structure or activity | 0.0885 |
| Infant toenail | ||
| WP4816a | TGF-beta receptor signalling in skeletal dysplasias | 0.0680 |
| path:hsa05220b | Chronic myeloid leukaemia – Homo sapiens (human) | 0.0680 |
| WP560a | TGF-beta Receptor Signalling | 0.0680 |
| GO:0034673 | Inhibin-betaglycan-ActRII complex | 0.0193 |
| GO:0048185 | Activin binding | 0.0218 |
| GO:0046332 | SMAD binding | 0.0254 |
| GO:0004674 | Protein serine/threonine kinase activity | 0.0264 |
| GO:0004675 | Transmembrane receptor protein serine/threonine kinase activity | 0.0279 |
| Placenta | ||
| GO:0035904 | Aorta development | 0.0218 |
aPathway source: Wikipathways; b Pathway source: KEGG
Discussion
Our study explored epigenetic modifications associated with prenatal Pb exposure that was measured through maternal and infant toenails and placental tissue in the RICHS study population. We found that Pb measured in all three biomarkers was linked to differential methylation and hydroxymethylation in several CpG sites. Pb exposure quantified through toenail samples resulted in more differentially methylated sites, while placental Pb was linked to higher numbers of differentially hydroxymethylated sites. We also demonstrated various common themes of biological functions and pathways, including 1) nervous system and organ development, 2) calcium regulation and neuronal activity, and 3) signalling pathways such as Hedgehog and TGF-β receptor signalling, that were overrepresented by genes annotated to the differentially methylated and hydroxymethylated sites associated with Pb exposure.
Pb is a well-documented environmental toxicant for its’ adverse health effects, including impacts on foetal growth and development, neurodevelopment, and neurologic functions [7–9,11], Particularly toxic to the vulnerable children’s population due to environmental Pb exposure sources (i.e., surface dust, soil, deteriorating paint chips) and hand-to-mouth activity, this heavy metal has been regulated by government agencies, and CDC has established a reference range upper value of 5 ug/dL for children’s blood Pb level (BLLs) [1]. However, the general population is still exposed to Pb, albeit at lower concentrations, through sources such as consumer products, drinking water and diet, as well as remaining contamination of painted surfaces and soil. Compared with other U.S. study populations, the average Pb level measured in infant toenail in this study (0.97 ug/g) was slightly higher than that measured in children’s toenails in the Cincinnati Childhood Allergy and Air Pollution Study (CCAAPS) (0.66 ug/g), and the New Hampshire Birth Cohort Study (NHBCS) (0.31 ug/g) [43,44]. The mean placental Pb level observed in RICHS was similar to several epidemiologic studies in non-occupationally exposed settings in the USA and around the world, and generally on the lower end among the studies included in a review of placental toxic metals by Esteban-Vasallo et al [45].
A growing body of research has investigated the association between prenatal Pb exposure and differential methylation. Findings from animal studies established a negative association between Pb exposure and nervous tissue DNA methylation levels, and demonstrated Pb interfered with DNA methyltransferase activities, albeit at exposure levels beyond what would be observed in our study [15,46,47]. EWAS results from a Project Viva study reported Pb exposure measured in maternal first trimester red blood cells was associated with lower cord blood methylation level at one CpG site, and in the ELEMENT study group, maternal bone Pb was negatively associated with Alu and LINE-1 methylation in cord blood [19,48]. To the best of our knowledge, this is the first human population study investigating the association between toenail and placental Pb concentration and placental differential DNA methylation and hydroxymethylation. In our methylation EWAS models, we observed greater numbers of hypomethylated sites associated with all three types of Pb biomarkers. As we compare across different Pb biomarkers, we note a potential limitation in the smaller sample size of placental Pb data from RICHS participants as this may contribute to the smaller number of differentially methylated sites. Nevertheless, placental samples provide the concentration of Pb within the placenta at the time of collection and may provide evidence of prenatal Pb exposure levels, given the ability of Pb to transport across the placenta. Maternal toenails, on the other hand, likely reflect a longer-term measure of Pb exposure, while infant toenails may similarly provide a longer term integrated measure of Pb exposure that reached the foetus during the later course of gestation.
From our hydroxymethylation (5hmC) results, we found in total, a lower number of differentially hydroxymethylated sites associated with Pb exposure, but in contrast to methylation results, placenta Pb was associated with the most differentially hydroxymethylated sites across the three biomarkers. 5hmC levels are highly tissue-specific, with the highest percentage found in brain tissue [26,49,50]. In RICHS, patterns of 5hmC were measured in the less well-explored placenta, compared to the comprehensive 5hmC profiling first conducted in brain tissue or embryonic stem cells [50–52]. Our finding of a greater number of differentially hydroxymethylated sites associated with placental Pb, compared to differentially methylated sites, may suggest that while Pb readily passes through the placenta and is not as well sequestered in the tissue compared to other toxic metals (i.e., cadmium) [53], Pb toxicity could still result through effects on placental 5hmC dysregulation before the metal is transported from the placenta.
Epigenome-wide association studies on prenatal Pb exposure and placental 5hmC are scarce. In the ELEMENT cohort, Rygiel et al. found that trimester-specific blood Pb level was associated with gene-specific 5mC and 5hmC measured in peripheral blood in 11- to 18-year-old Mexican adolescents [30]. Among the four candidate genes, pre-selected based on their developmental- and neurological-related features, gestational Pb was significantly associated with 5mC in HCN2 and 5hmC in NINJ2 across all three trimesters. Similar to the direction of their finding, our study also found a positive association between maternal toenail Pb and placental 5mC in HCN2 (estimate = 0.0035; FDR = 0.182). From our placental 5hmC profiling, cg19692784, annotated to gene NINJ2, was not significantly associated with any of the Pb biomarkers. Rygiel et al. also assayed 5mC and 5hmC at RAB5A from the RAS oncogene family, but no significant associations with blood Pb were observed in the ELEMENT cohort. We observed a CpG site within the body of a different gene from this oncogene family, RAB34, having a negative association between maternal toenail Pb and 5mC (estimate = −0.005; FDR = 0.012).
Several studies have explored the associations between Pb exposure and DNA methylation (DNAm) in different tissues. Rygiel et al. showed that cord blood DNAm at cg02901723 and cg18515027 was associated with second trimester maternal blood Pb levels in an ELEMENT cohort study; however, we did not observe associations (FDR > 0.05) in our study results for these two probes and any of the Pb biomarkers [54]. In a Korean exposome study, Park et al. found that among male infants, cord blood DNAm at cg11110036 was associated with late pregnancy maternal Pb, and 11 other CpG sites were associated with early pregnancy maternal Pb [55]. 4 of the 11 sites were not in our dataset, and we did not observe an FDR-significant association for cg11110036 or the remaining 7 differentially methylated sites (cg01167615, cg22410544, cg04862633, cg01995958, cg23900741, cg17984508, cg05374263) with maternal toenail, infant toenail or placental Pb in our results. Engström et al. found that maternal urinary Pb was inversely associated with cord blood DNAm at several sites, for instance, cg23796967 (annotated to gene GP6) [56]. While we also observed decreased methylation levels at this probe in association to maternal and infant toenail Pb, our results did not reach our pre-defined FDR cut-off. Montrose et al. identified 33 differentially methylated sites (q < 0.2) associated with Pb measured from neonatal dried bloodspots and stated 82% showed decreased methylation levels [57]. Our present study findings on differentially methylated sites did not overlap, though we also observed higher numbers of hypomethylated CpG sites associated with maternal and infant toenail Pb. On the other hand, Heiss et al. did not report significant associations between cord blood Pb, maternal blood Pb or maternal bone Pb and cord blood DNAm in the PROGRESS cohort [58]. In an EARLI study, Aung et al. observed 11 hypermethylated CpG sites that were associated with first or second trimester maternal blood Pb [59]. Our results did not overlap with theirs, with the exception of two sites near two of the identified genes, ASCL2 and FAT1. We observed maternal toenail Pb was associated with differential DNA methylation at cg24057514 (annotated to gene ASCL2; FDR = 0.02) and cg05723219 (annotated to gene FAT1; FDR = 0.04). In summary, due to the wide variety of tissues utilized in measuring prenatal or early postnatal Pb exposure and quantifying DNA methylation, along with varying sample collection techniques and time periods, there is limited overlap across study results in regards to the identified probes or genes associated with Pb exposure.
Development of the nervous system and major organs (i.e., brain, cerebellar cortex, and hindbrain) was one of the common themes of overrepresented pathways associated with toenail Pb. As a well-established neurotoxicant that mainly targets the CNS, Pb is known to adversely impact neurodevelopment, behaviour, motor activities and cognitive functions in newborns and children, with longitudinal studies establishing persistent effects into early adolescent [4,7,60–63]. Given the critical role of the placenta in foetal development and programming, and the activity of a number of pathways also present in the brain, studies have suggested that differentially methylated genes in the placenta act as biomarkers for adverse foetal developmental outcomes [64,65]. In this study, the identified developmental pathways significantly enriched for Pb-associated genes provided additional understanding on biological functions affected by placental DNA methylation and may suggest an additional way in which Pb can lead to developmental and behavioural outcomes and phenotypes later in life. This expands and adds to the evidence that Pb-elicited differential epigenetic changes during the sensitive gestational period may perturb placental pathways and functions, in some cases paralleling effects seen in other tissues, and that those impacts on placental function could consequently alter foetal development.
Multiple pathways in relation to calcium homoeostasis and transport (calcium ion binding, regulation and release of sequestered calcium ion) and neuronal activities (neuron differentiation and neurogenesis) were also significantly enriched for Pb-associated genes. Pb is known to mimic and compete with calcium, a critical and essential metal required for maintaining regular neuron signalling and activity [66–68]. A study by Lafond et al. showed that maternal blood Pb concentration was significantly linked to a decrease in calcium uptake by placental syncytiotrophoblast cells. Their results suggest that, in the placenta, Pb exposure has the potential to decrease foetal calcium supply [69]. During gestation, high demand of calcium during foetal development can lead to maternal skeletal calcium mobilization, which could also permit the release of Pb sequestered in maternal bone into the circulating system and thus elevate exposures to the placenta and foetus [70–72]. Pb-elicited disruptions to foetal calcium homoeostasis can lead to alterations in synapse structure and regulation and disruption in neurotransmitter release, which then may result in lasting adverse outcomes later in life such as memory and learning impairments [68,73–75]. Interfering with calcium status in placental cells can also lead to generation of reactive oxygen species (ROS), subsequently reflected by the overrepresentation of programmed cell death and apoptosis pathways related to prenatal Pb exposure in our findings [76,77].
Our study also identified overrepresented Hedgehog signalling pathways associated with prenatal Pb exposure. Hedgehog signalling is involved in cell signalling, proliferation and differentiation, and the pathway is also in charge of regulating organ development and patterning during embryogenesis [78–80]. There is limited research on how Pb exposure impacts Hedgehog signalling, however, previous research has linked hedgehog regulators from the GLI family zinc-finger genes, GLI2 and GLI3, to arsenic exposure [81–83]. GLI3 gene expression in human placenta was also found to be negatively associated with arsenic exposure in NHBCS [84]. From our EWAS results, we found prenatal Pb exposure was associated with differential methylation of several Hedgehog pathway associated genes, such as GLI2, GLI3, and HHAT (hedgehog acyltransferase), which given the consistency with findings from arsenic exposures could indicate that this pathway is impacted by a number of toxic metals.
We observed the TGF-β signalling pathway was also enriched for Pb-associated gene methylation variation. Transforming growth factor-β (TGF-β) is a major player in central regulator of growth, proliferation, differentiation, and apoptosis [85–87]. TGF-β superfamily members are also critical regulators in placental development and functions for supporting healthy pregnancies and foetal development [88]. Dysregulated TGF-β members have been implicated in aberrant placental angiogenesis related to foetal growth restriction and preeclamptic pregnancies [89–91]. A potential mechanism of aberrant TGF-β signalling may be attributed to oxidative stress induced by Pb exposure [92–95]. From our EWAS results, genes identified in these pathways (i.e., SMAD3, SMAD9, TGFBR2, TGFBR3) were not significantly associated with Pb exposure after FDR adjustment. This may be due to our generally low-to-moderate levels of Pb-biomarkers measured in the study population. Further investigation is needed to better understand the plausible mechanism of dysregulated TGF-β signalling as a consequence of Pb-induced epigenetic modifications on target genes of the pathway.
A potential limitation with our EWAS analysis is the higher genomic inflation observed in the 5mC and maternal toenail Pb model compared to the other models, which could lead in inflation in test statistics. Although we observed some differential hydroxymethylated sites associated with prenatal Pb exposure, the associations were not as robust as those findings of 5mC. This may be partially explained by the generally low toenail Pb levels, along with low abundance of placental 5hmC and individual variability in 5hmC across our study participants. Alternatively, we took note of the challenges of understanding the stability and role of 5hmC within different tissues and cell types. As pointed out by a preliminary study looking at 5hmC landscape in RICHS placenta, we are also aware that while 5hmC acts as an intermediate in the demethylation process, it is possible that the measurable levels are stable 5hmC modifications, whereas some degree of 5hmC during the rapid differentiation or reprogramming periods may not be properly detected[37]. Regulation of TET enzymes that catalyse the demethylation process may be indirectly affected by metal exposures such as Pb. It is suggested that Pb-associated oxidative stress increased TET activity through the accumulation of TET enzyme cofactor alpha ketoglutarate (α-KG), at least partially contributes to increased 5hmC upon Pb exposure [21,96,97]. This aligned with our finding that among the differentially hydroxymethylated sites associated with placental Pb, 12 out of 14 sites showed increased 5hmC levels. Future studies are required to investigate more about epigenome-wide 5hmC pattern changes in response to Pb and other toxicant exposures, and to elucidate how such changes may affect biological mechanisms, mediate regulatory responses in genes, and contribute to disease progression.
A strength of this study is utilizing data prepared with the algorithm that calculated and prevented any of the 5mC, 5hmC or unmethylated proportions to be below 0, refining our 5hmC profiling in the RICHS placental epigenome, as previous approaches to 5hmC estimation may result in negative numbers due to the subtraction method [37]. Another advantage of this study is leveraging 5mC and 5hmC data quantified through placental tissue in a human population, which provided a more analogous interpretation on prenatal Pb toxicity and epigenetic dysregulation in the sensitive developmental window. Existing literature on how toxicant exposure alters 5hmC levels mainly focused on animal models, with few studies exploring 5hmC levels in human tissue such as stem cells or blood [29,98]. Although brain tissue is of particular interest when researchers assess Pb toxicity, utilizing non-invasive and surrogate tissues like the placenta captures information unique to the pregnancy period. Given the placenta serves as an interface for toxicant regulation between the mother and infant during the gestation period, we suggest that perturbation to the placental epigenome by prenatal Pb exposure likely led to impaired cellular functions and resulted in adverse developmental outcomes.
Conclusion
In summary, our study shows that prenatal exposure to Pb, a known developmental toxicant, is associated with differential placental DNA methylation and hydroxymethylation, and demonstrates Pb-induced epigenetic changes are related to multiple biological pathways and functions, including developmental, calcium transport and regulation, and signalling activities. With our placental epigenetic profiling, we provide evidence that not only DNA methylation but also hydroxymethylation, may serve as potential response markers to environmental toxicant exposures during the gestation period.
Supplementary Material
Acknowledgments
We acknowledge the cooperation of the participants enrolled in RICHS, and the contributions of the research staff who also worked on RICHS. This work was supported by the National Institutes of Health [NIH-NIEHS R21ES028226, NIH-NIEHS R24 ES028507, NIH-NIEHS P30 ES019776, NIH-NIGMS P20 GM104416 and NIH-NIEHS P01 ES022832, NIH-NICHD K99HD104991] and the U.S. Environmental Protection Agency [US EPA grant RD83544201]. Its contents are solely the responsibility of the grantee and do not necessarily represent the official views of the U.S. EPA. Further, the U.S. EPA does not endorse the purchase of any commercial products or services mentioned in the presentation.
Funding Statement
This work was supported by the National Institutes of Health [K99HD104991]; National Institutes of Health [P20 GM104416, P01 ES022832]; National Institutes of Health [R21ES028226, R24ES028507, P30ES019776]; US Environmental Protection Agency [RD83544201].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2022.2126087.
Data availability statement
RICHS EPIC array methylation/hydroxymethylation data and accompanying exposure/covariate data are available through NCBI GEO: GSE144129. Code for the analyses performed is available through Emory Dataverse https://doi.org/10.15139/S3/UYJYSI.
References
- [1].ATSDR . Agency for Toxic Substances and Disease Registry (ATSDR) - Toxicological profile for lead. 2020. Accessed 5 February 2022. https://www.atsdr.cdc.gov/ToxProfiles/tp13.pdf
- [2].Dignam T, Kaufmann RB, LeStourgeon L, et al. Control of lead sources in the United States, 1970-2017: public health progress and current challenges to eliminating lead exposure. J Public Health Manag Pract. 2019;25(Suppl 1 LEAD POISONING PREVENTION):S13–S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nriagu JO. The rise and fall of leaded gasoline. Sci Total Environ. 1990;92:13–28. [Google Scholar]
- [4].Lanphear BP, Hornung R, Khoury J, et al. Low-level environmental lead exposure and children’s intellectual function: an international pooled analysis. Environ Health Perspect. 2005;113(7):894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Llanos MN, Ronco AM.. Fetal growth restriction is related to placental levels of cadmium, lead and arsenic but not with antioxidant activities. Reprod Toxicol. 2009;27(1):88–92. [DOI] [PubMed] [Google Scholar]
- [6].Jelliffe-Pawlowski LL, Miles SQ, Courtney JG, et al. Effect of magnitude and timing of maternal pregnancy blood lead (Pb) levels on birth outcomes. J Perinatol. 2006;26(3):154–162. [DOI] [PubMed] [Google Scholar]
- [7].Bellinger DC. Very low lead exposures and children’s neurodevelopment. Curr Opin Pediatr. 2008;20(2):172–177. [DOI] [PubMed] [Google Scholar]
- [8].Claus Henn B, Schnaas L, Ettinger AS, et al. Associations of early childhood manganese and lead coexposure with neurodevelopment. Environ Health Perspect. 2012;120(1):126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kordas K, Ardoino G, Coffman DL, et al. Patterns of exposure to multiple metals and associations with neurodevelopment of preschool children from Montevideo, Uruguay. J Environ Public Health. 2015;2015:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tung PW, Burt A, Karagas M, et al. Association between placental toxic metal exposure and NICU Network Neurobehavioral Scales (NNNS) profiles in the Rhode Island Child Health Study (RICHS). Environ Res. 2022;204:111939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Canfield RL, Henderson CR, Cory-Slechta DA, et al. Intellectual Impairment in children with blood lead concentrations below 10 µg per Deciliter. N Engl J Med. 2003;348(16):1517–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wasserman GA, Liu X, Popovac D, et al. The Yugoslavia Prospective Lead Study: contributions of prenatal and postnatal lead exposure to early intelligence. Neurotoxicol Teratol. 2000;22(6):811–818. [DOI] [PubMed] [Google Scholar]
- [13].Weinhold B. Epigenetics: the Science of Change. Environ Health Perspect. 2006;114(3):A160–A167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Relton CL, Hartwig FP, Davey Smith G. From stem cells to the law courts: DNA methylation, the forensic epigenome and the possibility of a biosocial archive. Int J Epidemiol. 2015;44(4):1083–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dou JF, Farooqui Z, Faulk CD, et al. Perinatal lead (Pb) exposure and cortical neuron-specific DNA methylation in male mice. Genes (Basel). 2019;10(4):274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wu S, Hivert MF, Cardenas A, et al. Exposure to low levels of lead in utero and umbilical cord blood DNA methylation in project viva: an epigenome-wide association study. Environ Health Perspect. 2017;125(8):087019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zeng Z, Huo X, Zhang Y, et al. Differential DNA methylation in newborns with maternal exposure to heavy metals from an e-waste recycling area. Environ Res. 2019;171:536–545. [DOI] [PubMed] [Google Scholar]
- [18].Sanchez OF, Lee J, Yu King Hing N, et al. Lead (Pb) exposure reduces global DNA methylation level by non-competitive inhibition and alteration of dnmt expression†. Metallomics. 2017;9(2):149–160. [DOI] [PubMed] [Google Scholar]
- [19].Pilsner JR, Hu H, Ettinger A, et al. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect. 2009;117(9):1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Senut MC, Sen A, Cingolani P, et al. Lead exposure disrupts global DNA methylation in human embryonic stem cells and alters their neuronal differentiation. Toxicol Sci. 2014;139(1):142–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dao T, Cheng RYS, Revelo MP, et al. Hydroxymethylation as a novel environmental biosensor. Curr Environ Health Rep. 2014;1(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Szulwach KE, Li X, Li Y, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7(6):e1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bachman M, Uribe-Lewis S, Yang X, et al. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat Chem. 2014;6(12):1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kinney SM, Chin HG, Vaisvila R, et al. Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes*. J Biol Chem. 2011;286(28):24685–24693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids. 2011;2011:870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Santiago M, Antunes C, Guedes M, et al. TET enzymes and DNA hydroxymethylation in neural development and function — How critical are they? Genomics. 2014;104(5):334–340. [DOI] [PubMed] [Google Scholar]
- [29].Cardenas A, Rifas-Shiman SL, Godderis L, et al. Prenatal Exposure to mercury: associations with global DNA methylation and hydroxymethylation in cord blood and in childhood. Environ Health Perspect. 2017;125(8):087022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rygiel CA, Goodrich JM, Solano -González M, et al. Prenatal lead (Pb) exposure and peripheral blood DNA methylation (5mC) and hydroxymethylation (5hmC) in Mexican adolescents from the ELEMENT birth cohort. Environ Health Perspect. 2021;129(6):067002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Punshon T, Li Z, Marsit CJ, et al. Placental metal concentrations in relation to maternal and infant toenails in a US cohort. Environ Sci Technol. 2016;50(3):1587–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fortin JP, Triche TJ, Hansen KD. Preprocessing, normalization and integration of the illumina HumanMethylationEPIC array with minfi. Bioinformatics. 2017;33(4):558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Triche TJ, Weisenberger DJ, Van Den Berg D, et al. Low-level processing of illumina infinium DNA methylation beadArrays. Nucleic Acids Res. 2013;41(7):e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. [DOI] [PubMed] [Google Scholar]
- [36].Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Green BB, Houseman EA, Johnson KC, et al. Hydroxymethylation is uniquely distributed within term placenta, and is associated with gene expression. FASEB J. 2016;30(8):2874–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fenton TR. A new growth chart for preterm babies: babson and Benda’s chart updated with recent data and a new format. BMC Pediatr. 2003;3(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bakulski KM, Dou JF, Feinberg JI, et al. Prenatal multivitamin use and MTHFR genotype are associated with newborn cord blood DNA methylation. Int J Environ Res Public Health. 2020;17(24):9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Dou JF, Middleton LYM, Zhu Y, et al. Prenatal vitamin intake in first month of pregnancy and DNA methylation in cord blood and placenta in two prospective cohorts. Epigenetics Chromatin. 2022;15(1):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pedersen BS, Schwartz DA, Yang IV, et al. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics. 2012;28(22):2986–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kamburov A, Wierling C, Lehrach H, et al. ConsensusPathDB—a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37(suppl_1):D623–D628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dantzer J, Ryan P, Yolton K, et al. A comparison of blood and toenails as biomarkers of children’s exposure to lead and their correlation with cognitive function. Sci Total Environ. 2020;700:134519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Farzan SF, Howe CG, Chen Y, et al. Prenatal lead exposure and elevated blood pressure in children. Environ Int. 2018;121(Pt 2):1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Esteban-Vasallo MD, Aragonés N, Pollan M, et al. Mercury, cadmium, and lead levels in human placenta: a systematic review. Environ Health Perspect. 2012;120(10):1369–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Eid A, Bihaqi SW, Renehan WE, et al. Developmental lead exposure and lifespan alterations in epigenetic regulators and their correspondence to biomarkers of Alzheimer’s disease. Alzheimers Dement (Amst). 2016;2(1):123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schneider JS, Kidd S, Anderson DW. Influence of developmental lead exposure on expression of DNA methyltransferases and methyl cytosine-binding proteins in hippocampus. Toxicol Lett. 2013;217(1):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Bozack AK, Rifas-Shiman SL, Coull BA, et al. Prenatal metal exposure, cord blood DNA methylation and persistence in childhood: an epigenome-wide association study of 12 metals. Clin Epigenet. 2021;13(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Globisch D, Münzel M, Müller M, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLOS ONE. 2010;5(12):e15367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wen L, Tang F. Genomic distribution and possible functions of DNA hydroxymethylation in the brain. Genomics. 2014;104(5):341–346. [DOI] [PubMed] [Google Scholar]
- [51].Pastor WA, Pape UJ, Huang Y, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473(7347):394–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Richa R, Sinha RP. Hydroxymethylation of DNA: an epigenetic marker. EXCLI J. 2014;13:592–610. [PMC free article] [PubMed] [Google Scholar]
- [53].Iyengar GV, Rapp A. Human placenta as a “dual” biomarker for monitoring fetal and maternal environment with special reference to potentially toxic trace elements. Part 3: toxic trace elements in placenta and placenta as a biomarker for these elements. Sci Total Environ. 2001;280(1–3):221–238. [DOI] [PubMed] [Google Scholar]
- [54].Rygiel CA, Dolinoy DC, Bakulski KM, et al. DNA methylation at birth potentially mediates the association between prenatal lead (Pb) exposure and infant neurodevelopmental outcomes. Environ Epigenet. 2021;7(1):dvab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Park J, Kim J, Kim E, et al. Prenatal lead exposure and cord blood DNA methylation in the Korean exposome study. Environ Res. 2021;195:110767. [DOI] [PubMed] [Google Scholar]
- [56].Engström K, Rydbeck F, Kippler M, et al. Prenatal lead exposure is associated with decreased cord blood DNA methylation of the glycoprotein VI gene involved in platelet activation and thrombus formation. Environ Epigenet. 2015;1(1):dvv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Montrose L, Goodrich JM, Morishita M, et al. Neonatal lead (Pb) exposure and DNA methylation profiles in dried bloodspots. Int J Environ Res Public Health. 2020;17(18):6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Heiss JA, Téllez-Rojo MM, Estrada-Gutiérrez G, et al. Prenatal lead exposure and cord blood DNA methylation in PROGRESS: an epigenome-wide association study. Environ Epigenet. 2020;6(1):dvaa014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Aung MT, Kelly M, Bakulski K, et al. Maternal blood metal concentrations and whole blood DNA methylation during pregnancy in the Early Autism Risk Longitudinal Investigation (EARLI). Epigenetics. 2022;17(3):253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hong SB, Im MH, Kim JW, et al. Environmental lead exposure and attention deficit/hyperactivity disorder symptom domains in a community sample of South Korean school-age children. Environ Health Perspect. 2015;123(3):271–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Liu J, Liu X, Wang W, et al. Blood lead levels and children’s behavioral and emotional problems: a cohort study. JAMA Pediatr. 2014;168(8):737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ris MD, Dietrich KN, Succop PA, et al. Early exposure to lead and neuropsychological outcome in adolescence. J Int Neuropsychol Soc. 2004;10(2):261–270. [DOI] [PubMed] [Google Scholar]
- [63].Mason LH, Harp JP, Han DY. Pb neurotoxicity: neuropsychological effects of lead toxicity. Biomed Res Int. 2014;2014:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Koukoura O, Sifakis S, Spandidos DA. DNA methylation in the human placenta and fetal growth (Review). Mol Med Rep. 2012;5(4):883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Vlahos A, Mansell T, Saffery R, et al. Human placental methylome in the interplay of adverse placental health, environmental exposure, and pregnancy outcome. PLoS Genet. 2019;15(8):e1008236. Bartolomei MS, ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bridges CC, Zalups RK. Molecular and ionic mimicry and the transport of toxic metals. Toxicol Appl Pharmacol. 2005;204(3):274–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Brini M, Calì T, Ottolini D, et al. Neuronal calcium signaling: function and dysfunction. Cell Mol Life Sci. 2014;71(15):2787–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kawamoto EM, Vivar C, Camandola S. Physiology and pathology of calcium signaling in the Brain. Front Pharmacol. 2012;3:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Lafond J, Hamel A, Takser L, et al. Low environmental contamination by lead in pregnant women: effect on calcium transfer in human placental syncytiotrophoblasts. J Toxicol Environ Health A. 2004;67(14):1069–1079. [DOI] [PubMed] [Google Scholar]
- [70].Goyer RA. Transplacental transport of lead. Environ Health Perspect. 1990;89:101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kovacs CS, Kronenberg HM. Maternal-fetal calcium and bone metabolism during pregnancy, puerperium, and lactation*. Endocr Rev. 1997;18(6):832–872. [DOI] [PubMed] [Google Scholar]
- [72].Téllez-Rojo MM, Hernández-Avila M, Lamadrid-Figueroa H, et al. Impact of bone lead and bone resorption on plasma and whole blood lead levels during pregnancy. Am J Epidemiol. 2004;160(7):668–678. [DOI] [PubMed] [Google Scholar]
- [73].Neal AP, Guilarte TR. Molecular neurobiology of lead (Pb2+): effects on synaptic function. Mol Neurobiol. 2010;42(3):151–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Cory-Slechta DA. Relationships between lead-induced learning impairments and changes in dopaminergic, cholinergic, and glutamatergic neurotransmitter system functions. Annu Rev Pharmacol Toxicol. 1995;35(1):391–415. [DOI] [PubMed] [Google Scholar]
- [75].Alkondon M, Costa ACS, Radhakrishnan V, et al. Selective blockade of NMDA-activated channel currents may be implicated in learning deficits caused by lead. FEBS Lett. 1990;261(1):124–130. [DOI] [PubMed] [Google Scholar]
- [76].Ercal N, Gurer-Orhan H, Aykin-Burns N. Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage. Curr Top Med Chem. 2001;1(6):529–539. [DOI] [PubMed] [Google Scholar]
- [77].Görlach A, Bertram K, Hudecova S, et al. Calcium and ROS: a mutual interplay. Redox Biol. 2015;6:260–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Sasaki H, Nishizaki Y, Hui C, et al. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126(17):3915–3924. [DOI] [PubMed] [Google Scholar]
- [79].Briscoe J, Thérond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14(7):416–429. [DOI] [PubMed] [Google Scholar]
- [80].Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12(6):393–406. [DOI] [PubMed] [Google Scholar]
- [81].Ruiz I Altaba A. Catching a Gli-mpse of Hedgehog. Cell. 1997;90(2):193–196. [DOI] [PubMed] [Google Scholar]
- [82].Fei DL, Li H, Kozul CD, et al. Activation of Hedgehog Signaling by the Environmental Toxicant Arsenic May Contribute to the Etiology of Arsenic-Induced Tumors. Cancer Res. 2010;70(5):1981–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kim J, Lee JJ, Kim J, et al. Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Pnas. 2010;107(30):13432–13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Winterbottom EF, Fei DL, Koestler DC, et al. GLI3 links environmental arsenic exposure and human fetal growth. EBioMedicine. 2015;2(6):536–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Morikawa M, Derynck R, Miyazono K. TGF-β and the TGF-β family: context-dependent roles in cell and tissue physiology. Cold Spring Harb Perspect Biol. 2016;8(5):a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Massagué J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103(2):295–309. [DOI] [PubMed] [Google Scholar]
- [87].David CJ, Massagué J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat Rev Mol Cell Biol. 2018;19(7):419–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Jones RL, Stoikos C, Findlay JK, et al. TGF-β superfamily expression and actions in the endometrium and placenta. Reproduction. 2006;132(2):217–232. [DOI] [PubMed] [Google Scholar]
- [89].Gunatillake T, Yong HEJ, Dunk CE, et al. Homeobox gene TGIF-1 is increased in placental endothelial cells of human fetal growth restriction. Reproduction. 2016;152(5):457–465. [DOI] [PubMed] [Google Scholar]
- [90].Caniggia I, Grisaru-Gravnosky S, Kuliszewsky M, et al. Inhibition of TGF-β3 restores the invasive capability of extravillous trophoblasts in preeclamptic pregnancies. J Clin Invest. 1999;103(12):1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Goumans MJ, Liu Z, ten Dijke P. TGF-β signaling in vascular biology and dysfunction. Cell Res. 2009;19(1):116–127. [DOI] [PubMed] [Google Scholar]
- [92].Zuscik MJ, Ma L, Buckley T, et al. Lead induces chondrogenesis and alters transforming growth factor-β and bone morphogenetic protein signaling in mesenchymal cell populations. Environ Health Perspect. 2007;115(9):1276–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Beier EE, Jen ST, Dang D, et al. Heavy metal ion regulation of gene expression. J Biol Chem. 2015;290(29):18216–18226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Liu RM, Desai LP. Reciprocal regulation of TGF-β and reactive oxygen species: a perverse cycle for fibrosis. Redox Biol. 2015;6:565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Krstić J, Trivanović D, Mojsilović S, et al. Transforming growth factor-beta and oxidative stress interplay: implications in tumorigenesis and cancer progression. Oxid Med Cell Longev. 2015;2015:654594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Chia N, Wang L, Lu X, et al. Hypothesis: environmental regulation of 5-hydroxymethylcytosine by oxidative stress. Epigenetics. 2011;6(7):853–856. [DOI] [PubMed] [Google Scholar]
- [97].Tretter L, Adam-Vizi V. Inhibition of α-ketoglutarate dehydrogenase due to H2O2-induced oxidative stress in nerve terminals. Ann N Y Acad Sci. 1999;893(1):412–416. [DOI] [PubMed] [Google Scholar]
- [98].Sen A, Cingolani P, Senut MC, et al. Lead exposure induces changes in 5-hydroxymethylcytosine clusters in CpG islands in human embryonic stem cells and umbilical cord blood. Epigenetics. 2015;10(7):607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RICHS EPIC array methylation/hydroxymethylation data and accompanying exposure/covariate data are available through NCBI GEO: GSE144129. Code for the analyses performed is available through Emory Dataverse https://doi.org/10.15139/S3/UYJYSI.


